Abstract
Screening a large Escherichia coli collection for P-fimbrial adhesin classes identified 20 unclassifiable strains. Cloning and sequencing of papG from an unclassifiable strain identified another G allele. The novel adhesin gene has 65% identity to the class I adhesin gene, 44% identity to the class II adhesin gene, and 43% identity to the class III adhesin gene.
P-fimbrial adhesins in Escherichia coli enable the colonization of host tissues. By mediating attachment to P-blood group antigens on uroepithelial cells (6, 12, 16), P fimbriae play a critical role in the development of urinary tract infections (UTIs). P fimbria production is regulated by a chromosomal pap operon, containing 11 genes (4). The P-fimbrial-tip adhesin, which is encoded by papG, attaches directly to host cells (7, 9). The three adhesin classes (papGJ96 [class I], papGAD/IA2 [class II], and prsGJ96 [class III]) were characterized based on their capacities for binding to specific Gal(α1-4)Gal-containing glycolipids (14). prf (pap-related fimbriae) is generalized nomenclature that includes all gene clusters encoding P fimbriae. The prf probe contains the most conserved genes and thus hybridizes to all of these gene clusters (1). Class I adhesins have 45% identity at the amino acid level to class II and 46% identity to class III, while class II adhesins have 56% identity to class III. Minor papG variants have been reported (GenBank accession numbers AAF61952 [J. R. Johnson, N. Kaster, T. T. O'Bryan, and A. L. Stell, unpublished data], AAF61956 [5], AAD13607 [3], and AAA59216 [10]) with homology to the three papG alleles, ranging in identity from 89 to 96%. We present the discovery of a new G allele of P fimbriae with less than 65% identity to known adhesin classes.
Three E. coli collections were studied: 313 isolates from college women aged 18 through 39 at the University of Michigan or University of Texas at Austin between 1992 and 1995 with a first-time UTI (first UTI), 51 isolates from a subset of these same women reporting a second UTI within 6 months of the first (second UTI), and 377 fecal and 74 periurethral isolates from healthy women presenting to the University of Michigan Student Health Service for gynecological exams between February and March 1996. All E. coli isolates were cultured and processed as previously described (1, 17).
A total of 815 E. coli strains were screened by dot blot hybridization for the presence of prf, a cluster of gene sequences specific to P-related fimbriae, as described previously (1). Sequence homology to prf was detected in 332 (41%) of the strains tested. The P-fimbrial adhesin class was also determined by screening those strains positive for prf with the three class-specific DNA probes using dot blot hybridization (Table 1). The adhesin class-specific probes (papGJ96 [class I], papGAD [class II], and prsGJ96 [class III]) were derived from published sequences (8, 13, 14) and isolated from control strain J96 or C1212 by PCR (1, 2). Strain J96 contains papGJ96 and prsGJ96, while control strain C1212 contains papGAD/IA2. We confirmed dot blot results with PCR using unique primers for each adhesin class. Identical PCR conditions, except the annealing temperatures, were used for each adhesin class (30 cycles of 94°C for 60 s and 73°C for 40 s, with an annealing time of 35 s). Table 2 lists the annealing temperatures and PCR primers. Based on these results, 20 strains positive for prf and negative for all three adhesin classes were identified (Table 1), suggesting the presence of papG variants.
TABLE 1.
P-fimbrial adhesin classes by strain source
| Population | No. (%) of prf-positive strains | No. (%) of isolates carrying adhesin
genea
|
|||
|---|---|---|---|---|---|
| papGJ96 (class I) | papGAD/IA2 (class II) | prsGJ96 (class III) | Unknown class | ||
| First UTI (n = 313) | 153 (48.9) | 5 (1.6) | 85 (27.2) | 62 (19.8) | 8 (3.3) |
| Second UTI (n = 51) | 24 (47.1) | 0 (0) | 10 (19.6) | 10 (19.6) | 1 (2.4) |
| Fecal (n = 377) | 128 (34.0) | 0 (0) | 88 (23.3) | 31 (8.2) | 11 (4.1) |
| Periurethral (n = 73b) | 27 (37.0) | 0 (0) | 13 (17.8) | 13 (17.8) | 0 (0) |
The subclass totals exceed the prf-positive totals for the first-UTI and fecal populations because some of these strains have more than one adhesin class.
Data missing for one periurethral strain.
TABLE 2.
Oligonucleotides used in PCR typing
| PCR primer | Oligonucleotide | Strand | Size (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| Conserved | 5′-ACCACGGCCAGTATGAGCATG-3′ | + | ||
| Class I (papGJ96) | 5′-CAGGATAGAAACATATACGGGCA-3′ | − | 449 | 58 |
| Class II (papGAD) | 5′-AATCTGGCGTTCAGGGTAACAC-3′ | − | 551 | 62 |
| Class III (prsGJ96) | 5′-CCAAGTAACTCGGGAAATGAC-3′ | − | 637 | 61 |
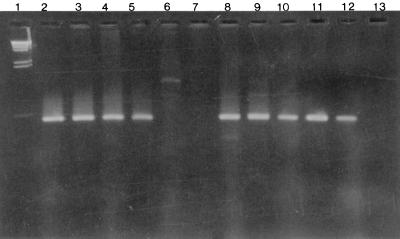
Pulsed-field gel electrophoresis analysis showed that all 20 strains differ by three or more bands (15) and therefore do not represent a clonal grouping (data not shown). Because papF is conserved among the three known adhesin classes (13), we assessed whether it could be amplified using PCR in the 20 strains (Fig. 1). A 502-bp fragment was amplified in control strains J96 and C1212 and in 11 of 20 (55%) strains with an unknown papG adhesin class.
FIG. 1.
PCR amplification of papF. Lane 1, DNA lambda digested with HindIII; lane 2, J96; lanes 3 to 12, unknown-adhesin strains BF31, BF56, BF1163, BF141, BF192, BF115, BF1160, BF166, and BF268; lane 13, negative control. PCR amplification of papF was detected using primers 5′-ATCGTTGCTTCTGACATCGG-3′ and 5′-GTCAATAAGTAATCCCATA CTG-3′ (30 cycles of 94°C for 60 s, 56°C for 30 s, and 74°C for 30 s).
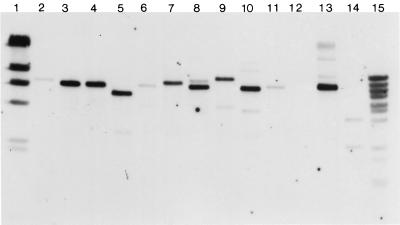
In order to determine whether any of these strains contained novel P-fimbrial G alleles, we cloned and sequenced papG from two randomly selected strains (BF1163 and BF31). Southern blot hybridization using a 502-bp papF PCR probe labeled with digoxigenin (Genius System kit; Boehringer Mannheim, Indianapolis, Ind.) detected DNA fragments of 6.5 kb for fecal strain BF31 (Fig. 2), and of 4 and 2.3 kb for UTI strain BF1163 digested with BsaB1 and Psp14061 (data not shown), respectively. Both the 6.5- and the 4-kb DNA fragments were purified following gel electrophoresis and cloned using the pZErO-1 vector (Invitrogen, San Diego, Calif.) with TOP10F′ (Invitrogen) as the recipient strain by methods described previously (17). Plasmid DNA was isolated using a plasmid preparation kit (Qiagen, Chatsworth, Calif.). Restriction enzyme digestion, nuclease treatment, and ligation were performed according to standard protocols (11).
FIG. 2.
Restriction fragment length polymorphisms of E. coli strains digested with BsaB1 and probed with papF (502 bp). Lanes 1 and 15; γ DNA markers digested with HindIII and BstEII; lanes 2, 3, 4, 5, 7, 8, 9, l0, and 12, unknown-adhesin fecal strains BF6, BF31, BF54, BF56, BF164, BF166, BF191, BF224, and BF370; lanes 6, 11, and 13, unknown-adhesin UTI strains BF115, BF268, and BF1009; lane 14, control strain J96.
The double-stranded DNA sequences of both clones were determined at the University of Michigan Biology Core Facility with an Applied Biosystems model 373A automated sequencer using primers T7 and SP6. Fecal strain BF31 contained a novel papG allele (papGBF31), whereas UTI strain BF1163 contained a variant of papG with a deletion. papGBF31 had amino acid sequence identities of 65% to papGJ96 (class I), 46% to papGAD/IA2 (class II), and 45% to prsGJ96 (class III) (Fig. 3). We refer to papGBF31 as the P-fimbrial class IV adhesin gene. BF1163 was most similar (70%) to papGJ96; however, the open reading frame was truncated at bp 290.
FIG. 3.
Amino acid comparison of the new class IV adhesin (encoded by papGBF31) with the three known adhesin classes. DNAStar (Madison, Wis.) and Genetics Computer Group (Madison, Wis.) software was used for DNA and amino acid analyses.
In order to estimate the prevalence of papGBF31 in other E. coli strains, we screened a sample of strains (n = 308) positive for prf by dot blot hybridization using a 371-bp probe specific to papGBF31. papGBF31 occurred with similar frequency in each collection. The numbers of strains positive for papGBF31 were as follows: 21 (15%) among the first UTI collection (n = 144), 36 (15%) among the second UTI collection (n = 20), 20 (17%) among the fecal collection (n = 120), and 3 (13%) among the periurethral collection (n = 24). papGBF31 is positively associated with aer and drb and is negatively associated with prsGJ96, hly, cnfl, ompT, and sfa (Table 3). Among isolates positive for papGBF31, papGAD was present in 55%.
TABLE 3.
Prevalence of virulence factor genes among the entire collection and among the subset positive for papGBF31
| Sample | No. (%) positive for selected virulence
factor gene
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| papGAD | papGJ96 | prsGJ96b | aerb | kpsMT | hlyb | cnf1b | ompTb | drbb | sfab | fim | |
| Total collection (n = 308)a | 183 (59) | 5 (2) | 105 (34) | 142 (46) | 275 (89) | 152 (49) | 105 (34) | 282 (92) | 19 (6) | 112 (36) | 308 (100) |
| papGBF31(n = 47) | 26 (55) | 2 (4) | 1 (2) | 35 (74) | 40 (85) | 10 (21) | 2 (4) | 30 (64) | 11 (22) | 4 (9) | 47 (100) |
The virulence factor genes encode the following: aerobactin (aer), group II capsules (kpsMT), α-hemolysin (hly), cytotoxic necrotizing factor 1 (cnf1), outer membrane protease T (ompT), afimbrial adhesins I to IV and F1845 pili (drb), S fimbriae (sfa), and type 1 fimbriae (fim). The data are the numbers (percentages) of isolates containing each gene (e.g., among those strains positive for papGBF31, 55% were also positive for papGAD).
The proportion positive for a selected gene among isolates with papGBF31 is significantly (P < 0.0001) different from the proportion in the total collection.
PCR was performed on the remaining 19 prf-positive, class I-, II-, and III-negative strains using primers 5′-GACTATTCTGGTTATGATTC-3′ and 5′-CAATGAATTAAGGTTTAG-3′ (30 cycles of 95°C for 60 s, 46°C for 40 s and 73°C for 23 s), taken from a unique coding region of papGBF31. A 371-bp fragment was amplified in 8 of the 19 (42%) strains, suggesting that other novel G allele variants may exist.
The novel class IV adhesin gene shows 45 to 65% similarity at the amino acid level to the three adhesin classes, thereby representing a unique adhesin class that is found equally among UTI and fecal E. coli strains. Thus, class IV adhesins are not exclusively associated with UTIs, although they could be associated with the pathogenesis of other important diseases.
The large prf probe used in this study hybridized to strains containing novel papG alleles as well as inactive papG variants or variants with deletions. Because a PCR fragment specific to the class IV gene was not amplified in 11 of the strains without class I, II, or III adhesins, it is possible that other novel molecular variants of papG exist. Future work should include hemagglutination assays to determine whether papGBF31 is functional and to identify other novel papG variants and assess their role in UTIs or other diseases.
Nucleotide sequence accession number.
The GenBank accession number for the papGBF31 nucleotide sequence is AF304159.
Acknowledgments
We thank Charlotte Williams, Ronald Mulder, and the laboratory and gynecologic clinic staffs at the University of Michigan Student Health Service as well as Scott Spear, Shirley Arldt, Nell Curtis, Barbara Locke, and the staff at the University of Texas at Austin Student Health Service for collecting bacterial specimens, and we thank Karin Palin for her intellectual contributions.
This work was supported by Public Health Service grants DK-35368 and DK-55496 from the National Institutes of Health.
REFERENCES
- 1.Foxman B, Zhang L, Palin K, Tallman P, Marrs C F. Bacterial virulence characteristics of Escherichia coliisolates from first-time urinary tract infection. J Infect Dis. 1995;171:1514–1521. doi: 10.1093/infdis/171.6.1514. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B, Zhang L, Tallman P, Andree B C, Geiger A M, Koopman J S, Gillespie B W, Palin K A, Sobel J D, Rode C K, Bloch C A, Marrs C F. Transmission of uropathogens between sex partners. J Infect Dis. 1997;175:989–992. doi: 10.1086/514007. [DOI] [PubMed] [Google Scholar]
- 3.Hull R A, Rudy D C, Donovan W H, Wieser I E, Stewart C, Darouiche R O. Virulence properties of Escherichia coli83972, a prototype strain associated with asymptomatic bacteriuria. Infect Immun. 1999;67:429–432. doi: 10.1128/iai.67.1.429-432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hull R A, Gill R E, Hsu P, Minshew B H, Falkow S. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coliisolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson J R, O'Bryan T T, Low D A, Ling G, Delavari P, Fasching C, Russo T A, Carlino U, Stell A L. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papGallele III. Infect Immun. 2000;68:3327–3336. doi: 10.1128/iai.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Källenius G, Mollby R, Svensson S B, Winberg J, Lundblad A, Svensson S, Cedergren B. The Pk antigen as receptor for the haemagglutinin of pyelonephritic Escherichia coli. FEMS Microbiol Lett. 1980;7:297–302. [Google Scholar]
- 7.Lindberg A A, Brown J E, Stromberg N, Westling-Ryd M, Schultz J E, Karlsson K A. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriaetype 1. J Biol Chem. 1987;262:1779–1785. [PubMed] [Google Scholar]
- 8.Lund B, Lindberg F, Normark S. Structure and antigenic properties of the tip-located P pilus proteins of uropathogenic Escherichia coli. J Bacterial. 1988;170:1887–1894. doi: 10.1128/jb.170.4.1887-1894.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund B, Lindberg F, Marklund B I, Normark S. The PapG protein is the alpha-d-galactopyranosyl-(1---4)-beta-d-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1987;84:5898–5902. doi: 10.1073/pnas.84.16.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiti S N, DesGroseillers L, Fairbrother J M, Harel J. Analysis of genes coding for the major and minor fimbrial subunits of the Prs-like fimbriae F165(1) of porcine septicemic Escherichia colistrain 4787. Microb Pathog. 1994;16:15–25. doi: 10.1006/mpat.1994.1002. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 12.Marcus D M, Kundu S, Suzuki A. The P blood group system: recent progress in immunochemistry and genetics. Semin Hematol. 1981;18:65–78. [PubMed] [Google Scholar]
- 13.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prspili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 14.Strömberg N, Marklund B I, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson K A, Normark S. Host-specificity of uropathogenic Escherichia colidepends on differences in binding specificity to Galα1-4Gal-containing isoreceptors. EMBO J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaisanen V, Elo J, Tallgren L G, Siitonen A, Makela P H, Svanborg-Eden C, Kallenius G, Svenson S B, Hultberg H, Korhonen T. Mannose-resistant haemagglutination and P antigen recognition are characteristics of Escherichia colicausing primary pyelonephritis. Lancet. 1981;ii:1366–1369. doi: 10.1016/s0140-6736(81)92796-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Foxman B, Tallman P, Cladera E, Le Bouguenec C, Marrs C F. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coliisolates and identification of new subtypes. Infect Immun. 1997;65:2011–2018. doi: 10.1128/iai.65.6.2011-2018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]