Abstract
Wastewater-Based Epidemiological Monitoring (WBEM) is an efficient surveillance tool during the COVID-19 pandemic as it meets all requirements of a complete monitoring system including early warning, tracking the current trend, prevalence of the disease, detection of genetic diversity as well asthe up-surging SARS-CoV-2 new variants with mutations from the wastewater samples. Subsequently, Clinical Diagnostic Test is widely acknowledged as the global gold standard method for disease monitoring, despite several drawbacks such as high diagnosis cost, reporting bias, and the difficulty of tracking asymptomatic patients (silent spreaders of the COVID-19 infection who manifest nosymptoms of the disease). In this current reviewand opinion-based study, we first propose a combined approach) for detecting COVID-19 infection in communities using wastewater and clinical sample testing, which may be feasible and effective as an emerging public health tool for the long-term nationwide surveillance system. The viral concentrations in wastewater samples can be used as indicatorsto monitor ongoing SARS-CoV-2 trends, predict asymptomatic carriers, and detect COVID-19 hotspot areas, while clinical sampleshelp in detecting mostlysymptomaticindividuals for isolating positive cases in communities and validate WBEM protocol for mass vaccination including booster doses for COVID-19.
Keywords: SARS-CoV-2, Wastewater-Based Epidemiological Monitoring, Clinical Diagnostic Test, Genetic diversity, New variants and mutations, Mass vaccination, Booster doses
Graphical abstract
1. Introduction
The current ongoing coronavirus disease-2019 (COVID-19) having flu-like symptoms caused by a causative agent,severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has wreaked havoc, on global economics, businesses,communities, and public health due to widespread infection, with 43.7 million confirmed cases and1.17 million deaths in 218 countries as of March 31, 2022 (WH0 2022; JHU 2022) (1). Two major obstaclesto managingSARS-CoV-2 rapid infections aredifficulties identifyinginfected people based on their signs and symptoms as well as preventingthe spread of viral infections in the communities(2). Over the past 18 months, the SARS-CoV-2 virus has diversified through multiple new mutationsand various genetic variants such as Alpha (B.1.1.7) with seven, Beta (B.1.351) with nine, Gamma (P.1) with 12, Delta (B.1.6, B.1.6.2) with 17 new mutations in spike protein gene, the latestOmicron (B.1.1.529) and Neocov variants, have been discovered and propagated into different parts of the world as variants of interest to concern(3, 4, 5). These altered genetic factors increased transmissibility, virulence, disease severity, and mortalities while they also decreased the effectiveness of current therapeutics and vaccines (6, 7, 8).
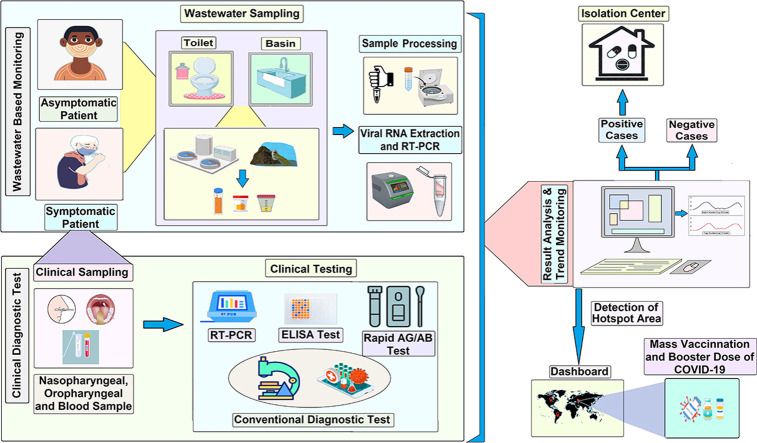
Since the beginning of the COVID-19 pandemic, the standard clinicaldiagnostic testing (CDT) is a recognized, valid system for monitoring infectious diseaseslike COVID-19 with some negative sides (9), which relies on the patient's signs and symptoms. As an alternative to individual tracking,wastewater-based epidemiology (WBE) has been used on a wide scale across theworld (10∗, 11∗, 12) to monitor the prevalence of COVID-19 patients. Despite the simplicity of wastewater sampling and transportation on time, viral RNA concentration and extraction are very difficult for low RNA quantities (13,14). Hence, it is verycrucial to establish a unified system incorporating CDT and WBEM to identify infectedindividuals with COVID-19 hotspots while discovering new variants and mutations, monitoringthe current pandemic scenario, followed by anticipating future waves (15). CDT and WBEM can be used synergistically to track localCOVID-19 epidemics where clinical samples will be used to identify the SARS-CoV-2 symptomatic patients and WBEM will be optimizedas a validated method for further analyses of wastewater released into communal drains from individual household drains and public places (e.g. bus and rail stands, airports, rivers, and market) (16∗, 17∗, 18∗, 19∗, 20). As a result, adapting the collective approach combiningWBEM and CDT could relieve burdens on the public health system, while it assists in making informed decisions for better and proper timely treatments, receiving vaccines, or booster dosages of vaccines for COVID-19.
.Based on our experience, it is worth mentioning that continuing WBEM without proper sanitation systems is strenuous, especially for low-middle income countries or non-WASH (Water, Sanitation and Hygiene) countries (1,21,22). However, adapting a combined approach can be the best model for both the developing and developed world (25).
In developing countries, WBEM of COVID-19 is more challenging without CDT, as the majority of households are not connected to sewerage systems. The CDT for SARS-CoV-2 can detect the viral genetic markers of the viral RNA in-between 7–14 days following the exposure and are unable to detect asymptomatic individuals (silent spreaders of COVID-19) within the communities (26,27). WBEM is an approach for tracking the pandemic through the identification of severely infected areas (COVID-19 hotspot zone) and monitoring of the infection trends (24,27). However, the recovery of the genetic biomarkers of the SARS-CoV-2 viral RNA in wastewater is very challenging due to differential stability in sewage streams, various environmental factors such as rainfall and temperature, as well as the presence of inhibitory substances (Ribonuclease Enzyme-RNase) (29,30). As a result performing a well-structured surveillance system combining both CDT and WBEM for symptomatic, asymptomatic, and paucisymptomatic carriers with unusual mutations as well asidentifying early warning for new variants are also helpful in vaccine development. From our recent completed 30 days follow-up on the quantitative analyses of SARS-CoV-2 genetic materials in wastewater from the residence of a positive patient family (Islam et al., 2022 under revision STOTEN-D-22-08275R1), SARS-CoV-2 positive patient number was identified as the lowest when the CT value was observed the highest (lowest copy number) in wastewater samples. On the other hand, the number of positive patients was found the highest with the corresponding lowest CT value (the highest copy number) in sewage samples as detected in the same study. In addition, increased signals of the SARS-CoV-2 genetic materials were noticed earlier in WS compared to positive patient viral load in clinical samples.
There are limited studies that link the concentation of SARS-CoV-2 viral biomarkers in wastewater with the identification of clinical cases in a specific residential area lacking wastewater treatment plants in developing countries (31, 32, 33). The combined CDT and WBEM follow-up study was performed in our laboratory to determine the relationship between the positive cases of SARS-CoV-2 infections and their discharged wastewater viral loads from one single house enrolling the entire family members’ clinical sample in a developing country without having a proper sewage system. The research findings demonstrated that a wastewater sample monitoring system tailored to a specific location could be established as a tool to identify SARS-CoV-2 infection and complement the clinically testing. This review emphasizes the combined monitoring of SARS-CoV-2 using CDT and WBE systems can guide the way forward for effective surveillance of the prevalence of the infectious disease such as COVID-19.
2. Clinical Diagnostic Test (CDT) and Wastewater-Based Epidemiologic Monitoring (WBEM)
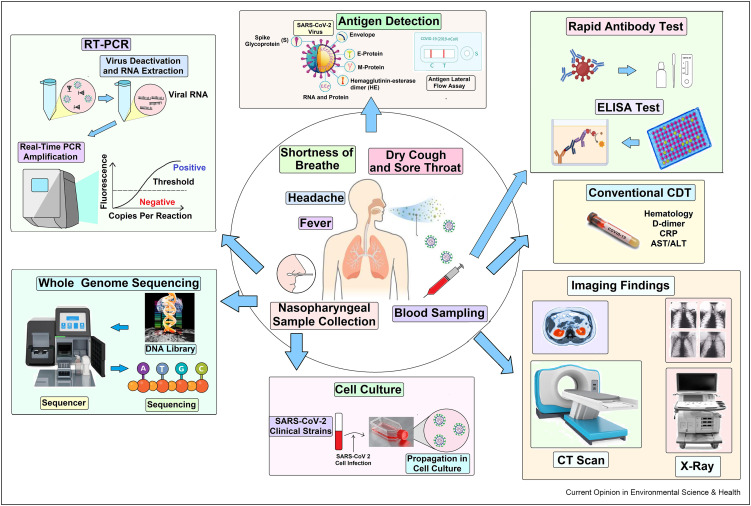
The accurate and rapid clinicaldiagnostic tests are essential for identifying the SARS-CoV-2 positive cases, contact tracing, and making public health decisions (9). Clinical testing is a conventional method for monitoring the status of COVID-19, notably, the clinical signs and symptoms of a patient such as fever, dry cough, headache, and shortness of breath usually develop 2-14 days after the exposureto SARS-CoV-2 (5,25) (Supplementary Table ST 1). The CDT is recommended for the diagnosis of any diseases based on the patient's specific signs and symptoms (35) (Figure 1 , Supplementary Figure SF1). However, the maximum COVID-19 positive individuals are asymptomatic (36,37), and clinical data may be limited due to testing capacity, reagent costs, laboratory facilities with proper instruments, expert hands, and availability issues.
Figure 1.
COVID-19 patients’ clinical diagnostic tests include general clinical signs and symptoms, imaging findings, and laboratory markers (38). In vitro, diagnostic, and clinical laboratory tests include molecular techniques e.g., viral antigen detection, antibody tests; nucleic acid amplification tests (NAAT); real-time polymerase chain reaction test (RT-PCR); next-generation sequencing (NGS); cell culture; enzyme-linked immunosorbent assay (ELISA); and traditional clinical tests: neutrophil-lymphocyte ratio (NLR); c-reactive protein test (CRP); erythrocyte sedimentation rate test (ESR,); IL-6/interleukin-6; lactate dehydrogenase test (LDH); aspartate aminotransferase test (AST); alanine aminotransferase test (ALT);imaging method: computed tomography (CT); ultrasound sonography (USG), X-ray.
The specific catchment area or community wastewater can be used to identify and observe COVID-19 infection scenarios in the same area, in the same manner, that was previously used for eradication of poliovirus, and this is recognized as wastewater-based epidemiology (WBE) (39). The SARS-CoV-2 genetic materials havebeen identified in feces from pre-symptomatic persons even1–5 days before the positive clinical test(29, 30) and in people with mild signs and symptoms (41). Recent investigationsof a few WBE studies have detected COVID-19 patients before the onset of clinical symptoms from feces samples, and 48-67% of diseased people had SARS-CoV-2 viral RNA in their stool, which survives in wastewater and can last up to >33 days (41, 42, 43, 44). Previous findings showed anassociation between wastewater viral concentration and COVID-19 confirmed cases where SARS-CoV-2 viral RNA limit between 2.0 and 6.0 log10 gc/L (genomic copies per liter), which is similar to our recent research findings (36, 38, 41). In addition, the accuracy of WBE was found in many studies (35, 36, 37, 38) where Betancourt et al. (2021c), confirmed the WBEM has a sensitivity of 76.0%, specificity of 90.7%, positive and negative predictive value of 79.8%, and 88.6%, respectively when findings of wastewater samples were compared with clinical samples (46). Furthermore, according to previous studies, SARS-CoV-2 RNA prevalence in stool was higher (48.1%) than in patients detected with gastrointestinal symptoms (17%) (42). Although WBEM is capable to detect SARS-CoV-2 RNA genetic biomarkers for monitoring the pandemic, there is an ongoing debate over how wastewater data should be used and to what extent the approaches are useful to public health decisions.
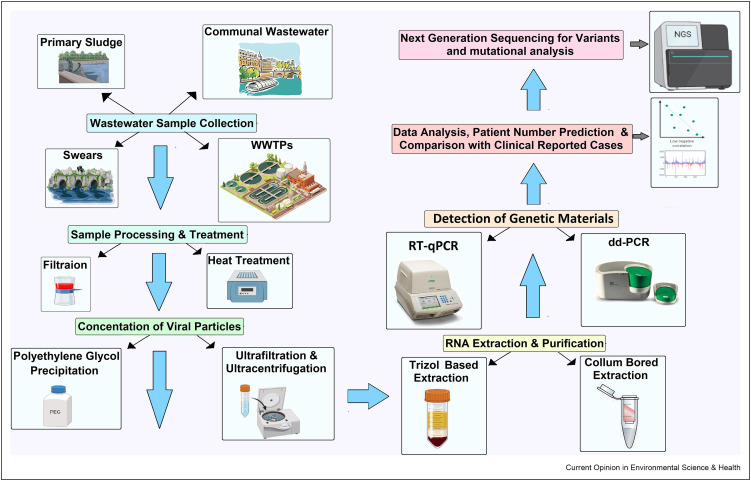
As the COVID-19 pandemic continues, individual clinical diagnostic testing (CDT) did not represent itself as a holistic approach to community health status determination. One major concern with COVID-19 pandemic is that most cases in the United States, patients were generally asymptomatic and pre-symptomatic, allowing infected people to spread the virus as healthy carriers (47). Moreover, a significant percentage of COVID-19 survivors might still be carrying and shedding the virus (48). Hence, in addition to the clinical test, wastewater surveillance should be used together with clinical data to infer the average virus shedding patterns at a population level (49,50). Figure 2 depicts a high-level overview of the WBEM system from sample collection to result in interpretations. The selection of WS collection sources playsan important role as a proper catchment area (wastewater treatment plants, sewer drains, primary networking system or communal watershed, river courses, bus stand, airports) (22). Heat treatment (600C, 30 minutes), filtration (to remove large particles), or chemical treatment with NaOCl can be used for sample processing and disintegrating the viruses (19). Several methods are already used in various studies for concentrating viral biomarkers like polyethylene glycol (PEG), ultrafiltration, ultracentrifugation, centrifugation, or skim milk procedure (51). Viral nucleic acid can be extracted using manually (Trizol Based) or kit-based (Qiagen, Thermo Fisher) (52). For calculating viral copy number maximum studies have used equations 1 and 2.
Figure 2.
Overview of WBEM procedure. Wastewater sample collection and processing, viral RNA concentration, nucleic acid/RNA extraction from the concentrated samples, interpretation of RT-PCR results, and monitoring the trend of disease outbreak compared with CDT and Whole Genome Sequencing (WGS) for new variants and mutations.
Number of infected individuals = Equation 1 (51).
Number of infected individuals = Equation 2 (53).
Positive, negative, no-template, and extraction control controls should be used with standard curve calculation as well as PCR inhibitors should check accordingly to MIQE rules(54). Major roles and drawbacks of both WBEM and CDT are given in Table 1 . Considering the previous research outcomes on WBEM and CDT activities with various validated methods and comparisons of the two approaches, the following future recommendations should be done for (1) disease burden correlation (2) trend analysis compared with clinical tests (3) observing the effectiveness of the intervention with the declining number of patients, and (4) detection of hotspot areas with COVID-19 cases for vaccination and booster doses (55).
Table 1.
A comparison of the major advantages and drawbacks of CDT and WBEM.
| Parameters | Advantages | Disadvantages | Reference |
|---|---|---|---|
| CDT |
|
|
(59), (60),(61),(62), (63) |
| WBEM |
|
|
(64), (65) (23), (66), (67) |
One previous study in Massachusetts between March and May 2020 found similar trends of the SARS-CoV-2 virus in wastewater with the number of affected patients (56). Another study in Utah used 9-week wastewater sampling and found a link between a community outbreak and an increase in SARS-CoV-2 RNA (57). The SARS-CoV-2 virus concentrations in wastewater samples in Ottawa, Canada surged by more than 400% just 48 hours after a 300% or greater rise in recognized cases (58), and in Utah showed a strong link between community outbreaks and an increase in SARS-CoV-2 RNA in wastewater (57). Environmental parameters are also linked to SARS-CoV-2 genetic materials, as evidenced by an increase in wastewater temperature resulting from a decrease in viral gene copy numbers (2).
3. Examples of combined wastewater-based monitoring with clinical diagnostic tests
The previous WBS studies have found a direct correlation between CDT-confirmed COVID-19 cases and wastewater SARS-CoV-2 viral concentration (68,69). Various findings reflected how SARS-CoV-2 WBEM provided early warnings in the population analyzed, and detected viral RNA in WS before CDT (65,66,67). Viral RNA was found in wastewater samples in Milan, Italy few days after the first confirmed COVID-19 patient by clinical test (70), in Australia (Brisbane), when there were hundreds of clinical cases (71); in Japan (Yamanashi Prefecture), when clinical test results were at their peak (72); and in Spain (Murcia), when the COVID-19 cases were the least in the Iberian Peninsula (73) (Table 2 ). The detection of SARS-CoV-2 RNA in the wastewater treatment plant was also reported initially in Louisiana, USA (61), Gujarat, India (74), Dubai (75), Gothenburg, Sweden (76), and in the Southeast England of the United Kingdom (77). Medema et al. (2021) (78) successfully detected SARS-CoV-2 viral RNA from the city wastewater in the Netherlands six days before the first confirmed clinical case (79), and another group of researchers from northeastern of the United States reported that viral titers in WS indicated COVID-19 infections were higher than clinical reports (80).
Table 2.
Salient examples of integration of wastewater monitoring with clinical testing data.
| Country | Area/Population/Time | Sampling Site, type | Concentration method | RNA/Nucleic Acid extraction Kit Name | RT-PCR Kit and Covered gene | Viral load range (gc/L)/CT value | Clinical cases/range | Main Findings (Correlation of Waste water result with clinical data) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Qatar | Doha 2.8 million January 2021 |
WWTP Influent Raw wastewater composite |
PEG | Quick-RNA Viral Kit | Bio-Radi Taq Universal Probes One-Step Kit | 7.8×103 to 5.4×105 | 31,181 to 542,313 | A similar trend as both WS and CS were decreasing at the same time | (90)) |
| Germany | North
Rhine-Westphalia 3,725,633 August 2020 |
Filtration | RNA Blue Kit | N gene, S gene, and ORF1ab gene) | NF 6.16 × 1014 |
1-10000 | WS could be used as an early warning tool. | (91) | |
| USA | Utah 1597460 July 2021 |
Influent, activated and anaerobic ally digested sludge, composite | Filtration Centrifugation | AllPrep Power Viral DNA/RNA kit | TaqPathTM 1-Step
RT-qPCR Master Mix N1, N2 |
1.0 × 105 to 1.0 × 106 GC/L. | NF | Primary sludge can be used to predict disease prevalence | (92) |
| USA | New Orleans
290,321 June 2020 |
3.1×103 and 4.3 ×103 copies/L | Ultrafiltration and adsorption-eluting using electronegative membrane | ZR Viral RNA Kit | 0-7000 | 3.1 × 103–7.5 × 103/L | PerfecTa qPCR ToughMix (Quantabio, Beverly, MA) | Detection of viral RNA in WS after first caseby clinical diagnostic test. | (61) |
| Mexico | 3.8 million | WWTP Grab |
NF | RNeasy Mini Kit | QuantiNova®
SYBR® Green PCR Kits (Qiagen, USA) N1, N2 |
1.9 ×103 to 3.5×106 | 74 to 82,690 | WS detected SARS-CoV-2 2–7 days earlierthan clinical reports. | (93) |
| India | Ahmedabad NF September 2020 |
WWTP Grab Influent wastewater |
PEG | NucleoSpin® RNA Virus isolation kit | TaqPathTM
Covid-19 RT-PCR Kit ORF1ab, N, and S |
(∼10,729
copies/L) > September (∼3047 copies/L) > October (∼454 copies/L) |
50-650 | Identification of COVID-19 hotspots | (94) |
| USA | Boston(Massachusetts) NR June, 2021 |
WWTP raw sewage composite | Pasteurization, Filtration | RNeasey PowerSoil Total RNA Kit, | Amicon Ultra-15 centrifugal ultrafiltration units (Millipore UFC903096) | Log(10+1)(4, 5, 6)/l 200000-1200000 | NF N1, N2 |
WS detected SARS-CoV-2 RNA before CS in the first wave. | (95) |
| France | Nancy 250,000 April2020 |
raw WWTP | NucliSENS® lysis buffer | One-Step RT-ddPCR™ Kit for Probes | Two
concentration procedures, based on ultrafiltration and on
PEG 6000 precipitation, r |
2.1 × 107 ± 1.1 × 107 gc/L and 1.6 × 107 ± 1.4 × 107 gc/L uncon and conc | 100-1000 | Decrease viral load during lockdown with decreasing patient's number of COVID-19. | (96) |
| UK | Gwynedd,
Cardiff,Liverpool,Manchester, Wirral, Wrexham ∼3 million July 2020 |
Untreated WWTP Influent; Composite and Grab samples | Centrifugation,Ultrafiltration | NucliSENSeasyMag | Ultrasense Reaction Mix,Enzyme Mix; N1 and E | <1.2×103-1.5×104/100mL | 5000-20,000 | Both positive and negative correlation | (83) |
| UK | South East
England 4 million April 2020 |
Raw WWTP influent; Composite | Filtration | High Pure Viral RNA kit | Invitrogen SuperScript III One-Step RT-PCR System | 3.5-5.27 log
10gc/l 3.1×103 -6.0× 105 |
0-10,000 | Able to detect prevalence variant by WS sequencing | (97) |
| Netherlands | Amsterdam,
DenHaag, Utrect, Apeldoorn, Amersfoot, Tilburg and Schiphol
2,802,800 July 2020 |
WWTP Composite |
Ultrafiltration | Biomerieux Nuclisens kit | Neasy PowerMicrobiome Kit (Qiagen, Hilden, Germany) |
2.6−30 gene copies per mL | 20-140 | WS detected RNA prior to CS. | (98) |
| South Africa | NF Durban April 2021 |
WWTP composite and Grab Raw samples | Ultrafiltration, Adsorption |
RNeaseyPowerSoil Total RNA Kit, | Primer, Probe | 1.55×105-7.32×106 | 95,000 to 2.3 million | WS viral load similar with CS | (99) |
| Australia | Brisbane 934,000 November,2020 |
WWTP Composite |
Filtration | RNeasy Mini Kit | iTaq™
UniversalProbesOne-Step Reaction Mix N1, N2, E |
135 to 11,992 gene copies (GC)/100 mL | 0-40 | No correlation | (100) |
| India | Chennai 9.6 million January2021 |
WWTP, Composite wastewater | UV,
Pasteurization, Filtration Corning Spin X-ultrafiltration |
Manually (TRIzol for RNA extraction) | 2019-nCoV CDC EUA Kit | 1.41×104-1.99×104 | 3983- 5523 | Higher viral load than CS | (101) |
| USA | Virginia 1,700,000 March 2020 |
Raw
wastewater WWTP Grab samples NI |
Innova Prep Concentrating method | Nucli SEN Seasy Mag | RT-ddPCR, One-Step RT-ddPCR Advanced Kit, N1, N2, N3 |
102-105 | Trend correlated with clinical data | (101) | |
| USA | Indiana NF November2020 |
University Sewer Manhole,Raw sewage, Grab | Filtration,Centrifugation | QIAmp Viral Mini Kit | RT-PCR master
mix N1, N2 |
1.0× 104-1.0× 106 | 0-120 | Early detected than clinical | (102) |
| USA | Las
Vegas 1,060,000 March 2020 |
WWTP Grab samples |
Raw influent
wastewater and primary effluent composite and grab sample |
RNeasy Mini Kit | i Script™ Select Kit, E,N1,N2,ORF1a | 104-106 | 20 and 200 | Correlation with clinical data | (103) |
| USA | Charlotte >2550 November,2020 |
University
plumbing cleanouts & manhole, composite Raw |
Electronegative filtration | QIAamp viral mini kit | iTaq™ UniversalProbesOne-Step Reaction Mix N1, | 394-2,990,271 | 400-13,00 | Asymptomatic patients detection | (104) |
| Brazil | Belo
Horizonte ∼2,000,000 August 2020 |
WWTP Influent Composite |
Adsorption | RT-qPCR PrepPowerViral DNA/RNA, |
iTaq™ Universal
probes One Step reaction mix N1 |
5.6× 101 -2.1×105 | 0-1200 | A similar trend with hospital cases affected by COVID-19 | (105) |
| Sweden | Gothenburg 755,940 July 2020 |
WWTP Influent, Composite & grab |
Adsorption, Filtration | DNeasy Blood and Tissue kit | qPCR Reaction Mix (Invitrogen) | 6.7×103 -1.8× 106 | 0-90) | WS trend peaked when hospitalized COVID-19 increased | (106) |
| Qatar | Doha 2,503,457 August 2020 |
Influent WWTP Composite |
PEG | RT-qPCRSARS-CoV-2(2019-nCoV)kit N1,N2 |
Quick-RNA Viral
Kits (Zymo Research, Irvine CA, USA Cat. No. R1041) |
7.8×103 -5.4× 105 | 500-2500 | Higher than CS | (107) |
| Brazil | Florianopolis
∼5000 March 2020 |
Raw sewage | Adsorption | QIAamp Viral RNA Mini kit | One-Step qPCR
Quantinovakit, Seegene Allplex™2019-nCoV N1,S,RdRp |
Avg 3.1× 105-4.8× 106 |
NR | WS detected prior to CDT | (108) |
| Italy | Milan, Turin Bologna 4,998,600 Feb,2020 | WWTP Composite raw sewage |
Ultracentrifugation | Nucli SEN Smini MAG | Super Fi Green
PCR Master Mix RdRp, ORF1ab |
2.9× 102 -5.6× 104 | 3095 in Latium Region and 2186 in the province of Rome | WS dtected before first case by CDT | (109) |
| Spain | Murcia 716,388 April 2020 |
Influent, WWTPsComposite | Aluminum
hydroxide adsorption– precipitation method with 3% beef extract |
Nucleo Spin RNA virus kit | TaqMan one-step
master mix (N1, N2, and E) |
1×105 to 3.4 × 105 | 12-622 | Detected RNA in low prevalence area | (110) |
| USA | Virginia, 143,000 September 2020 |
WWTP, Hospitals,Dormitoriecomposite, and Grab Influent wastewater |
Filtration, PEG |
QiaAmp viral RNA
mini kit and NucleoSpin RNA Plus kit |
NF N1, N2, RP |
Ct value(30.6-41.9) | NR | Consistency with clinical data | (111) |
| Japan | Ishikawa and
Toyama 465,243 April 2020 |
Influent wastewater WWTP grab | PEG and NaCl | QIAamp Viral RNA Mini Kit | Prime
Script™ N2,N3 |
1×100 - 3.5 × 104 | 5-30 | Higher than clinical data | (112) |
| Japan | Yamanashi
Prefecture NF May 2020 |
WWTP Grab | Adsorption-elution, Electronegative filtration | RNeasy Power Water Kit | Probe qPCR Mix
with UNG N1, N2 |
qPCR Mix with UNG (Takara Bio, Kusatsu, Japan) | (1.4 × 102–2.5 × 103 copies/L) | Similar to clinical data | (113) |
| UAE | June,2020 | Raw sewage WWT Composite | Ultrafiltration
columns, PEG/TRIzol |
ABIOpure Viral DNA/RNA Extraction kits | GENESIG COVID-19 kits RdRP | 7.5×102 - 3.4 × 104 | 0-800 | Decrease of WS load related to CS decline | (114) |
| France | Montpellier≈
470,000 July 2020 |
WWTP raw composite | . Filtration, Centrifugation |
NucleoSpin RNA Virus kit (Macherey-Nagel) | Primer, Probe
based detection N1, N3 |
100-10,000/100ml | 0-75 | WS detected SARS-CoV-2 genes before CS | (115) |
| Spain | Barcelona 2.7 million July 2020 |
WWTP Composite and grab raw samples |
Polyethylene glycol-6000 | NucliSENSminiMAG extraction system | NF RdRp,IP2, IP4,E,N1 |
103-105 | 2000-8000 | WS Detected prior to CDT | (116) |
| USA | Boston(Massachusetts) NF July 2020 |
WTPcomposite and Grab samples | PEG+NaCL | TRIzol-chloroform | PCR is used by New England Biolab Master Mix | 0Log10-3Log10 | 0Log10-1Log10 | SARS-CoV-2 WS titer higher than CS | (117) |
Padilla-Reyes et al. (2022a) found that the concentration of genomic copies of SARS-CoV-2 viral biomarkers (103 and 106 gc/L) were compatible with the reported clinical cases of COVID-19 in three out of four wastewater treatment plants (WTP). The study also revealed that WBEM was capable of giving a signal 2–7 days in advance as early warning, which might be helpful in low-income countries. Another study performed in Mexico found an increasing number of SARS-CoV-2 viral genes in WS two weeks before the clinical cases were raised (82). According to Hillary et al. (2021a), Giraud-Billoud et al. (2021) and Peccia et al. (2020), wastewater viral RNA detection precedes clinical reports by two to five days, three to six days, and six to eight days respectively (65,84,85,88). In another study, Zhang et al. (2020) (86) claimed that the SARS-CoV-2 viral concentration in wastewater was well correlated with COVID-19 clinical cases when samples were collected on day-to-day basis for monitoring the pandemic. According to Nemudryi et al. (2020), the SARS-CoV-2 viral RNA concentration in wastewater samples correlated with dates from sample collection to RT-PCR detection, whereviral genesare detectable in the wastewater samples 5–8 days after collection (68). Zhang et al. (2020) claimed that SARS-CoV-2 in the stool specimen was found significantly elevated than in the serum/blood specimenor nasal swab samples (86).
A recent WBE study conducted over 40 US cities found that a weekly incidence might not be sufficient to support the interpretation of viral concentration in the wastewater (88). Wu et al. (2021) recommended that at least two wastewater samples in a week are necessary to keep the accuracy in COVID-19 trend analysis (88). In another study, Petala et al. (2022) suggested that a weekly-based sampling method for viral quantification with fixed sampling times could be scheduled to reduce the day-to-day deviation (89). In addition, they strongly proposed that wastewater sample test results should be validated with clinical data.
4. Conclusion
WBEM has the potential to detect hotspots, identify the prevalence, and predict early warning. On the other hand, CDT can be used to isolate positive patients, mass vaccination, and quarantine measures to limit direct, indirect, or close contact. In the context of making the CDT method more cost-effective and efficient, it is important to improve in terms of rapidness, sensitivity, and portability of the analyses to demonstrate a functional diagnostic tool for detecting cases of positivity. It is also noteworthy that, the presence of SARS-CoV-2 in the community will be detected earlier by the WBEM than by the CDT. Hence, the dual monitoring of COVID-19 by using WBEM and CDT will immensely help control the spread and threat of the COVID-19 global pandemic.
5. Ethical Statement
No ethical statement is required for this review article.
6. Funding sources
This study was supported by the Water Aid Bangladesh, North South University, International Training Network of Bangladesh University of Engineering and Technology (ITN-BUET) - Centre, Noakhali Science and Technology University. PB and MTI would gratefully acknowledge the Life Science Technology Platform, Science for Life Laboratory for the seed funding to initiate the wastewater-based epidemiological studies for SARS-CoV-2 in Bangladesh.
Uncited reference
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Water Aid Bangladesh, North South University, International Training Network of Bangladesh University of Engineering and Technology (ITN-BUET) - Centre, Noakhali Science and Technology University. PB and MTI acknowledge the Life Science Technology Platform, Science for Life Laboratory for the seed funding to initiate the wastewater-based epidemiological studies for SARS-CoV-2 in Bangladesh. We would like to acknowledge the two anonymous reviewers for their critical comments as well as their thoughtful insights which has significantly improved the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.coesh.2022.100396.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Islam M.A., Al Marzan A., Islam M.S., Sultana S., Parvej M.I., Hossain M.S., Amin M.T., Hossain F.E., Barek M.A., Hossen M.S., et al. Sex-specific epidemiological and clinical characteristics of COVID-19 patients in the southeast region of Bangladesh. medRxiv. 2021 [Google Scholar]
- 2.Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci Total Environ. 2020:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakib M.M.H., Nishat A.A., Islam M.T., Uddin M.A.R., Iqbal M.S., Hossen F.F.B., Ahmed M.I., Bashir M.S., Hossain T., Tohura U.S., et al. Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2. Comput Biol Med. 2021:104759. doi: 10.1016/j.compbiomed.2021.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hossain M, Huq TS, Rahman A, Islam MA,Tabassum SN,Hasan KN,Khaleque A, Sadique A, Hossain MS, Bahadur NM, et al.:Novel mutations identified from whole-genome sequencing of SARS-CoV-2 isolated from Noakhali, Bangladesh. Research Square 2021.
- 5.Islam M.A., Haque M.A., Rahman M.A., Hossen F., Reza M., Barua A., Marzan A.A., Das T., Baral S.K., He C., et al. A Review on Measures to Rejuvenate Immune System: Natural Mode of Protection Against Coronavirus Infection. Front Immunol. 2022:837290. doi: 10.3389/fimmu.2022.837290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J Infect. 2021;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y‐mutated SARS‐CoV‐2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.037. 2348-2361.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., DittrichS, Yansouni C.P. Diagnostic Testing for Severe Acute Respiratory Syndrome–Related Coronavirus 2: A Narrative Review. Ann Intern Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi M., Patel A.K., Joshi C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: A temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ Res. 2021:110946. doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article discussed about early warning of COVID-19 pandemic situation using wastewater and clinical data.
- Kumblathan T., Liu Y., Uppal G.K., Hrudey S.E., Li X.-F. Wastewater-based epidemiology for community monitoring of SARS-CoV-2: Progress and challenges. ACS Environ Au. 2021;1:18–31. doi: 10.1021/acsenvironau.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper discusses about early warning system using wastewater in developing countries with help of clinical data analysis.
- 12.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mailepessov D., Arivalan S., Kong M., Griffiths J., Low S.L., Chen H., Hapuarachchi H.C., Gu X., Lee W.L., Alm E.J., et al. Development of an efficient wastewater testing protocol for high-throughput country-wide SARS-CoV-2 monitoring. Sci Total Environ. 2022:154024. doi: 10.1016/j.scitotenv.2022.154024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – Suitability for COVID-19 surveillance and potential transmission risks. Sci Total Environ. 2021:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakib SH, Masum S., Patwari MRI, Fahima RA, Farhana A, Islam MA: Design and development of a low cost ultraviolet disinfection system to reduce the cross infection of SARS-CoV-2 in ambulances. International Conference on Electronics, Communications and Information Technology (ICECIT), 14-16 September 2021, Khulna, Bangladesh. https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=9641131
- Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 Omicron variant at Frankfurt airport but limited spread in the city of Frankfurt, Germany. Microbiol ResourAnnounc. November 2021;2022 doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper successfully confirmed Omicron variant from airport watewater sample that was similar to clinical sample variants in specific area.
- Thongpradit S., Prasongtanakij S., Srisala S., Kumsang Y., Chanprasertyothin S., Boonkongchuen P., Pitidhammabhorn D., Manomaipiboon P., Somchaiyanon P., Chandanachulaka S., et al. A simple method to detect SARS-CoV-2 in wastewater at low virus concentration. J Environ Public Health. 2022:4867626. doi: 10.1155/2022/4867626. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article discusses the wastewater surveillance from market places with simple wastewater method.
- Acosta N., Bautista M.A., Hollman J., McCalder J., Beaudet A.B., Man L., Waddell B.J., Chen J., Li C., Kuzma D., et al. A multicenter study investigating SARS-CoV-2 in tertiary-care hospital wastewater. viral burden correlates with increasing hospitalized cases as well as hospital-associated transmissions and outbreaks. Water Res. 2021:117369. doi: 10.1016/j.watres.2021.117369. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper discussesthe use of hospital wastewater for tracing patient numbers correlated with hospitalized patient numbers.
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci Total Environ. 2020:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reported the first detection of SARS-CoV-2 viral RNA in wastewater and their trends with the reported was same as clinical cases.
- 20.Yao Y., Pan J., Liu Z., Meng X., Wang W., Kan H., Wang W. No association of COVID-19 transmission with temperature or UV radiation in Chinese cities. Eur Respir J. 2020:2000517. doi: 10.1183/13993003.00517-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., et al. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci Total Environ. 2021:145724. doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reports the first detection of SARS-CoV-2 genetic materials from Bangladesh and the viral gene copies were similar with isolation center patient number. In this study clinical samples were used for extraction control.
- 22.Jakariya M., Ahmed F., Islam M.A., Ahmed T., Marzan A Al, Hossain M., Reza H.M., Bhattacharya P., Hossain A., Nahla T., et al. Wastewater based surveillance system to detect SARS-CoV-2 genetic material for countries with on-site sanitation facilities: An experience from Bangladesh. medRxiv. 2021 [Google Scholar]
- Haque R., Moe C.L., Raj S.J., Ong L., Cherles K., Ross A.G., Shirin T., Raqib R., Sarker P., Rahman M., et al. Wastewater surveillance of SARS-CoV-2 in Bangladesh: Opportunities and challenges. Curr Opin Environ Sci Health. 2022:100334. doi: 10.1016/j.coesh.2022.100334. [DOI] [PMC free article] [PubMed] [Google Scholar]; This published review discussed major challenges and opportunities for wastewatersurveillance in Bangladesh, where they indicated to use clinical data with wastewater based surveillance.
- 24.Bhattacharya P, Kumar M, Islam MT, Haque R, Chakraborty S, Ahmad A, Niazi NK, Cetecioglu Z, Nilsson D, Ijumulana J, van der Voorn T, Jakariya M, Hossain M, Ahmed F, Rahman M, Akter N, Johnston D, Ahmed KM: Prevalence of SARS-CoV-2 in communities through wastewater surveillance – A potential approach for estimation of disease burden. Current Poll Rep 7: 160-166. 10.1007/s40726-021-00178-4 [DOI] [PMC free article] [PubMed]
- 25.Jakariya M., Ahmed F., Islam M.A., Al Marzan A., Hasan M.N., Hossain M., Ahmed T., Hossain A., Reza H.M., Hossen F.…Bhattacharya P. Wastewater-based epidemiological surveillance to monitor the prevalence of SARS-CoV-2 in developing countries with onsite sanitation facilities. Environ Pollut. 2022:119679. doi: 10.1016/j.envpol.2022.119679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg S., Kim L., Whitaker M., O’Halloran A., Cummings C., Holstein R., Prill M., Chai S.J., Kirley P.D., Alden N.B., et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 — COVID-NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biggerstaff M., Jhung M.A., Reed C., Fry A.M., Balluz L., Finelli L. Influenza-like illness, the time to seek healthcare, and influenza antiviral receipt during the 2010-2011 influenza season - United States. J Infect Dis. 2014;210:535–544. doi: 10.1093/infdis/jiu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mainardi PH, Bidoia ED:Challenges and emerging perspectives of an international SARS-CoV-2 epidemiological surveillance in wastewater. An Acad Bras Cienc 2021: e20210163. [DOI] [PubMed]
- 29.Farkas A., Pap B., Kondorosi É., Maróti G. Antimicrobial activity of NCR plant peptides strongly depends on the test assays. Front Microbiol. 2018:2600. doi: 10.3389/fmicb.2018.02600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: Wastewater-based epidemiology for COVID-19 – approaches and challenges for surveillance and prediction. Water Res. 2020:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci Total Environ. 2021:144587. doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., MarescaM LongobardiC., ManconA RomeriF., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci Total Environ. 2020:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakib SH. Design of a cost-effective Ultraviolet Disinfection unit to minimize the cross-contamination of COVID-19 in transport. 2022; 2–7.
- 34.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardian Y., Kosasih H., Karyana M., Neal A., Lau C.-Y. Review of current COVID-19 diagnostics and opportunities for further development. Front Med. 2021:615099. doi: 10.3389/fmed.2021.615099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection:Anarrative review. Ann Intern Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H., Waugh D.W., Orbe C., Chen G. Dependence of atmospheric transport into the arctic on the meridional extent of the Hadley cell. Geophys Res Lett. 2020 [Google Scholar]
- 38.Rezaei M., RazaviBazaz S., Zhand S., Sayyadi N., Jin D., Stewart M.P., Warkiani M.E. Point of care diagnostics in the age of COVID-19. Diagnostics. 2020;11:9. doi: 10.3390/diagnostics11010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kassebaum N.J., Bertozzi-Villa A., Coggeshall M.S., Shackelford K.A., Steiner C., Heuton K.R., Gonzalez-Medina D., Barber R., Huynh C., Dicker D., et al. Global, regional, and national levels and causes of maternal mortality during 1990-2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:980–1004. doi: 10.1016/S0140-6736(14)60696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., Duvallet C., Endo N., Erickson T.B., Armas F., et al. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022:118070. doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper stated that wastewater viral concentration related with patients’ clinical data.
- 41.Wong J, Goh QY, Tan Z, Lie SA, Tay YC, Ng SY, Soh CR:Preparing for a COVID-19 pandemic: A review of operating room outbreak response measures in a large tertiary hospital in Singapore. Can J Anaesth2020, 67:732–745. [DOI] [PMC free article] [PubMed]
- 42.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam AR. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: Systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., SpadacciniM, Colombo M., Gabbiadini R., Artifon E.L.A., et al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients withcoronavirus disease 2019. JAMA Netw Open. 2020 doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J., Pang J. Wastewater surveillance to infer COVID-19 transmission: A systematic review. Sci Total Environ. 2022:150060. doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., SprisslerRS, Harris D.T., Sherchan S.P., et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci Total Environ. 2021:146408. doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moghadas S.M., Shoukat A., Fitzpatrick M.C., Wells C.R., Sah P., Pandey A., Sachs J.D., Wang Z., Meyers L.A., Singer B.H., et al. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc Natl Acad Sci USA. 2020;117:9122–9126. doi: 10.1073/pnas.2004064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landi S.M., Viswanathan P., Serene S., Freiwald W.A. A fast link between face perception and memory in the temporal pole. Science. 2021;373:581–585. doi: 10.1126/science.abi6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article highlights the use of wastewater samples for SARS-CoV-2 surveillence and showed how the generic materials in untreated wastewater can be used as a indicator for prediction of the number of patients.
- 52.Monteiro S., Rente D., Cunha M.V., Gomes M.C., Marques T.A., Lourenço A.B., Cardoso E., Álvaro P., Silva M., Coelho N., et al. A wastewater-based epidemiology tool for COVID-19 surveillance in Portugal. Sci Total Environ. 2022:150264. doi: 10.1016/j.scitotenv.2021.150264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng Y., Xu X., Zheng X., Ding J., Li S., Chui H., Wong T.K., Poon L.L.M., Zhang T., et al. Use of sewage surveillance for COVID-19 to guide public health response: A case study in Hong Kong. Sci Total Environ. 2022:153250. doi: 10.1016/j.scitotenv.2022.153250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R., Hellemans J., Kubista M., Mueller R.D., Nolan T., et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 55.Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ Int. 2020:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bi P., Wang J., Hiller J.E. Weather: driving force behind the transmission of severe acute respiratory syndrome in China? Intern Med J. 2007;37:550–554. doi: 10.1111/j.1445-5994.2007.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci Total Environ. 2021:145790. doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., BaigAT, Mayne J., Zhang X., Alain T., et al. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci Total Environ. 2021:145319. doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaferani AH, Dehghan MG, Atashi HA, Rezaei Nejad A, Ghorani SM, Abolghasemi S, Bahrani M, Khaledian H, Savodji PB, Hoseinian M, et al.: Understanding the clinical and demographic characteristics of second coronavirus spike in 192 patients in Tehran, Iran: A retrospective study. PLoS One 2021:e0246314. [DOI] [PMC free article] [PubMed]
- 60.Zheng X., Deng Y., Xu X., Li S., Zhang Y., Ding J., On H.Y., Lai J.C.C., YauCI, Chin A.W.H., et al. Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci Total Environ. 2022:153687. doi: 10.1016/j.scitotenv.2022.153687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci Total Environ. 2020:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed F., Aminul Islam M., Kumar M., Hossain M., Bhattacharya P., Tahmidul Islam M., et al. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre through wastewater surveillance in Bangladesh. medRxiv. 2020 doi: 10.1101/2020.09.14.20194696. [Internet] 2020.09.14.20194696. Available from: [DOI] [Google Scholar]
- 63.Haque M.A., Wang F., Chen Y., Hossen F., Islam M.A., Hossain M.A., Siddique N., He C., Ahmed F. Bacillus spp. contamination: A novel risk originated from animal feed to human food chains in South-Eastern Bangladesh. Front Microbiol. 2022:783103. doi: 10.3389/fmicb.2021.783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sangkham S. A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral fragments. J Environ Manage. 2021:113563. doi: 10.1016/j.jenvman.2021.113563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., et al. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021:117214. doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper discusses that wastewater can be used to predict COVID-19 lockdown comparing with clinical cases.
- 66.Kierkegaard P., McLister A., Buckle P. Rapid point-of-care testing for COVID-19: Quality of supportive information for lateral flow serology assays. BMJ Open. 2021 doi: 10.1136/bmjopen-2020-047163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anand U., Li X., Sunita K., Lokhandwala S., Gautam P., Suresh S., Sarma H., Vellingiri B., Dey A., Bontempi E., et al. SARS-CoV-2 and other pathogens in municipal wastewater, landfill leachate, and solid waste: A review about virus surveillance, infectivity, and inactivation. Environ Res. 2022:111839. doi: 10.1016/j.envres.2021.111839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., VanderwoodKK, Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep Med. 2020:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bar-Or I., Weil M., Indenbaum V., Bucris E., Bar-Ilan D., Elul M., Levi N., AguvaevI, Cohen Z., Shirazi R., et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci Total Environ. 2021:148002. doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowen A.C., Mubareka S., Steel J., Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoSPathog. 2007;3:1470–1476. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed W, Bivins A, Bertsch PM, Bibby K, Choi PM, Farkas K,GyawaliP, Hamilton KA, HaramotoE, Kitajima M,et al.:Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. CurrOpin Environ Sci Health2020, 17:82–93. [DOI] [PMC free article] [PubMed]
- 72.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci Total Environ. 2020:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonçalves J., Koritnik T., Mioč V., Trkov M., Bolješič M., Berginc N., ProsencK KotarT., Paragi M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci Total Environ. 2021:143226. doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar M., Mohapatra S., Mazumder P., Singh A., Honda R., Lin C., Kumari R., Goswami R., Jha P.K., Vithanage M., et al. Making Waves Perspectives of Modelling and Monitoring of SARS-CoV-2 in Aquatic Environment for COVID-19 Pandemic. CurrPollut Rep. 2020;6:468–479. doi: 10.1007/s40726-020-00161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Albastaki A., Naji M., Lootah R., Almeheiri R., Almulla H., Almarri I., AlreyamiA, Aden A., Alghafri R. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: The use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci Total Environ. 2021:143350. doi: 10.1016/j.scitotenv.2020.143350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amereh F., Negahban-Azar M., Isazadeh S., Dabiri H., Masihi N., Jahangiri-Rad M., Rafiee M. Sewage systems surveillance for sars-cov-2: Identification of knowledge gaps, emerging threats, and future research needs. Pathogens. 2021:946. doi: 10.3390/pathogens10080946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin L., White M.P., Hunt A., Richardson M., Pahl S., Burt J. Nature contact, nature connectedness and associations with health, wellbeing and pro-environmental behaviours. J Environ Psychol. 2020:101389. [Google Scholar]
- 78.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. (Supporting information) [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]; This paper discussed the strength of wastewater to predict patients’ number as early warning tool.
- 80.Wu F.Q., Xiao A., Zhang J.B., Gu X.Q., Lee W.L., Kauffman K., Hanage W.P., Matus M., Ghaeli N., Endo N., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Padilla-Reyes D.A., Álvarez M.M., Mora A., Cervantes-Avilés P.A., Kumar M., Loge F.J., Mahlknecht J. Acquired insights from the long-term surveillance of SARS-CoV-2 RNA for COVID-19 monitoring: The case of Monterrey Metropolitan Area (Mexico) Environ Res. 2022:112967. doi: 10.1016/j.envres.2022.112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sosa-Hernández J.E., Oyervides-Muñoz M.A., Melchor-Martínez E.M., Driver E.M., Bowes D.A., Kraberger S., Lucero-Saucedo S.L., Fontenele R.S., Parra-Arroyo L., Holland L.A., et al. Extensive wastewater-based epidemiology as a resourceful tool for SARS-CoV-2 surveillance in a low-to-middle-income country through a successful collaborative quest: WBE, mobility, and clinical tests. Water. 2022:1842. [Google Scholar]
- 83.Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., et al. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021:117214. doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giraud-Billoud M., Cuervo P., Altamirano J.C., Pizarro M., Aranibar J.N., Catapano A., Cuello H., Masachessi G., Vega I.A. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci Total Environ. 2021:148887. doi: 10.1016/j.scitotenv.2021.148887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. NatBiotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript discussed the use of wastewater to monitoring community infection dynamics using clinical data.
- 86.Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., et al. SARS-CoV-2 RNA in Wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ Sci Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., et al. Wastewater surveillance of SARS-CoV-2 across 40 U.S. states from February to June 2020. Water. 2021:117400. doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study discussed used both wastewater samples to track patients’ number with the use of clinical data.
- 89.Petala M., Kostoglou M., Karapantsios T., Dovas C.I., Lytras T., Paraskevis D., Koutsolioutsou-Benaki A., Panagiotakopoulos G., Sypsa V., MetallidisS, et al. Relating SARS-CoV-2 shedding rate in wastewater to daily positive tests data: A consistent model based approach. Sci Total Environ. 2022:150838. doi: 10.1016/j.scitotenv.2021.150838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saththasivam J., El-Malah S.S., Gomez T.A., Jabbar K.A., Remanan R., Krishnankutty A.K., OgunbiyiO, Rasool K., AshhabS Rashkeev S., et al. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci Total Environ. 2021:145608. doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci Rep. 2021:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhattarai B., Sahulka S.Q., Podder A., Hong S., Li H., Gilcrease E., Beams A., Steed R., Goel R. Prevalence of SARS-CoV-2 genes in water reclamation facilities: From influent to anaerobic digester. Sci Total Environ. 2021:148905. doi: 10.1016/j.scitotenv.2021.148905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Padilla-Reyes D.A., Álvarez M.M., Mora A., Cervantes-Avilés P.A., Kumar M., Loge F.J., et al. Acquired insights from the long-term surveillance of SARS-CoV-2 RNA for COVID-19 monitoring: The case of Monterrey Metropolitan Area (Mexico) Environ Res. 2022:112967. doi: 10.1016/j.envres.2022.112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar M., Joshi M., Shah A.V., Srivastava V., Dave S. Wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: A perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. Sci Total Environ. 2021;792:148367. doi: 10.1016/j.scitotenv.2021.148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., et al. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article highlighted that the time lag and transfer function analysis of the wastewater data preceeded the clinically reported cases in the first wave of the pandemic, but did not serve as a leading indicator in the second wave due to increased testing capacity for case detection and reporting.
- 96.Bertrand I., Challant J., Jeulin H., Hartard C., Mathieu L., Lopez S. Scientific Interest Group Obépine, Schvoerer E, Courtois S, Gantzer C: Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: Comparison of ultrafiltration- and protein precipitation-based methods. Int J Hyg Environ Health. 2021:113692. doi: 10.1016/j.ijheh.2021.113692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin J., Klapsa D., Wilton T., Zambon M., Bentley E., BujakiE, Fritzsche M., Mate R., Majumdar M. Tracking SARS-CoV-2 in sewage: Evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses. 2020:1144. doi: 10.3390/v12101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ Sci Technol Lett. 2020 Jul 14;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. https://pubs.acs.org/doi/10.1021/acs.estlett.0c00357 [Internet] [DOI] [PubMed] [Google Scholar]; This paper discussed the strength of wastewater for SARS-CoV-2 surveillance as early warning tool and to predict the number of patients in sewersheds.
- 99.Pillay L., Amoah I.D., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. Monitoring changes in COVID-19 infection using wastewater-based epidemiology: A South African perspective. Sci Total Environ. 2021:147273. doi: 10.1016/j.scitotenv.2021.147273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Leah Clarke L., Dwyer J., Edson J., Nguyen T.M.H., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci Total Environ. 2021:144216. doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study also confirmed that clinical patient’s numbers can be tracked with the help of wastewater and clinical data.
- 102.Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., SprisslerRS, Harris D.T., SherchanSP, et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci Total Environ. 2021:146408. doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: Methodology, occurrence, and incidence/prevalence considerations. Water Res X. 2021:100086. doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., RoppoloBrazell L., Hinton K., LontaiJ, Stark N., Young I., et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci Total Environ. 2021:146749. doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mota C.R., Bressani-Ribeiro T., Araújo J.C., Leal C.D., Leroy-Freitas D., Machado E.C., Espinosa M.F., Fernandes L., LeãoTL, Chamhum-Silva L., et al. Assessing spatial distribution of COVID-19 prevalence in Brazil using decentralised sewage monitoring. Water Res. 2021:117388. doi: 10.1016/j.watres.2021.117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., DavidssonF DotevallL., MattssonA, Trybala E., et al. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021:116620. doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saththasivam J., El-Malah S.S., Gomez T.A., Jabbar K.A., Remanan R., Krishnankutty A.K., et al. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci Total Environ. 2021:145608. doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fongaro G, Rogovski P, Savi BP, Cadamuro RD, Pereira JVF, Anna IHS,Rodrigues IH, Souza DSM, Saravia EGT, Rodríguez-Lázaro D,et al.:SARS-CoV-2 in human sewage and river water from a remote and vulnerable area as a surveillance tool in Brazil. Food Environ Virol 2021,1-4. [DOI] [PMC free article] [PubMed]
- 109.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int J Hyg Environ Health. 2020:113621. doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article narrates the use of wastewater as an early warning tool in conjunction with the clinical cases.
- 111.Colosi L.M., Barry K.E., Kotay S.M., Porter M.D., Poulter M.D., Ratliff C., Simmons W., Steinberg L.I., Wilson D.D., Morse R., et al. Development of wastewater pooled surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from congregate living settings. Appl Environ Microbiol. 2021 doi: 10.1128/AEM.00433-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci Total Environ. 2021:143578. doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci Total Environ. 2020:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., Alsafar H., Yousef A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Sci Total Environ. 2021:142929. doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trottier J., Darques R., AitMouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020:100157. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., ParairaGalofré B., Sánchez G., Pintó R.M., Bosch A. Time Evolution of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Wastewater during the First Pandemic Wave of COVID-19 in the Metropolitan Area of Barcelona, Spain. Appl Environ Microbiol. 2021 doi: 10.1128/AEM.02750-20. 027500-e2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.