Abstract
Infection with Trichinella nematodes elicits non-specific and specific immune responses; these depend on the dose of infection, the nematode, and the host species. Few studies have examined the presence of specific antibodies against Trichinella spp. in the meat juice of wild animals. The aims of the study were to determine the prevalence of antibodies against Trichinella spp. in meat juice and to identify the specific proteins reacting with the meat juice from free-living carnivores naturally infected with the parasite. Meat juice samples were taken from foxes, badgers, raccoon dogs, and martens and tested with indirect ELISA. Antibodies against Trichinella spp. were detected in 10% of foxes and 46% of raccoon dogs. The ELISA results were confirmed by immunoblot, which revealed different protein patterns in meat juice from red foxes, raccoon dogs, and badgers. The most frequently observed bands were sent for further analysis by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) for the detection of Trichinella britovi immunogenic proteins. The results confirm the presence of proteins such as serine protease and heat shock proteins associated with Trichinella infection. These findings provide that meat juice is a useful matrix for proteomic analysis.
Keywords: Trichinella britovi, wildlife, antibodies, meat juice immunology
1. Introduction
The parasitic nematodes of the genus Trichinella have a global reach, where they circulate in both domestic and sylvatic cycles [1]. In Poland, and Europe in general, the most common Trichinella species observed in the sylvatic cycle is Trichinella britovi, which is found in animals such as red foxes, wolves, lynxes, martens, badgers, golden jackals, raccoons, and raccoon dogs [2,3,4,5,6,7,8,9,10,11,12,13]. Their life cycle comprises three major stages, viz. muscle larvae, adult worms, and new-born larvae, which are observed in a single host.
All developmental stages of Trichinella elicit an immune response, including IgG antibodies, which can be used for the serological detection of Trichinella spp. infection [14]. Infection with nematodes of the genus Trichinella causes both non-specific and specific immune responses, which can vary depending on the dose of infection, and the species of Trichinella and host. The presence of anti-Trichinella antibodies and specific proteins or antigens may be observed in serum or plasma and meat juice. Both serum and plasma are easy to collect and are widely used as sources of proteins in a highly soluble form [15]; indeed, there are thousands of proteins that are released by cells and tissues [16]. Serum is a complex biological matrix, and the general health status of the specimen and the presence of parasitic, bacterial, or viral infections are reflected in changes in its proteomic pattern [16]. However, in many cases, it is not possible to collect serum samples from wild animals. In such cases, meat juice, consisting of a mixture of intra- and extracellular fluid, blood, and lymph, might provide an alternative matrix that can be easily obtained for post-mortem analysis from carcasses of free-living animals [17]. It has been demonstrated that meat juice samples may be successfully used as a matrix in immunological surveillance of parasitic (e.g., Toxoplasma gondii, Neospora caninum) [18,19,20], bacterial (e.g., Salmonella, Brucella) [21,22], and viral (e.g., Japanese encephalitis virus, hepatitis E virus) [23] infections in animals. Moreover, meat juice samples have been also used as a matrix for immunological monitoring of the occurrence of anti-Trichinella IgG antibodies in domestic and wild animals [24,25,26,27,28,29]. When performing ELISA, the dilution factor has to be 10 times higher for the serum than for the meat juice sample; however, both sample types can use the same cut-off value to categorize immunopositive from immunonegative animals [21].
Although T. spiralis has typically been regarded as the most significant pathogen related to public health, there is a need for further studies on the antigenic profile associated with T. britovi infection [30], which occurs more frequently among wild carnivores [1,12,31,32]. In natural environments, carnivorous animals play an important role in the transmission of Trichinella nematodes, including T. britovi. Carcasses of infected animals may become a source of invasion for other animals, especially for wild boars, for which unexamined meat is known to be a source of Trichinella infection in humans [33,34,35].
Although in the domestic cycle the immune response during Trichinella infection has been widely studied in pigs, under controlled conditions with a known infection dose and species of Trichinella [14,30,36,37,38,39], the immunoprevalence of anti-Trichinella antibodies has been also examined in many animals species connected to the sylvatic cycle [13,24,26,40,41,42,43,44,45,46] as the main reservoir of Trichinella nematodes in the environment. Infections in wildlife occur in a natural way—the dose of infection, the timing, and whether they occur once or repeatedly remain unknown. Therefore, it is important to gain a deeper understanding of immune response during Trichinella infection in free-living animals.
As the present literature contains little or no data concerning the presence of specific proteins/antigens related to Trichinella infection in the meat juice of infected free-living animals, the aims of the present study were twofold: (1) to determine the prevalence of antibodies against Trichinella spp. in meat juice; (2) to examine the protein profile using meat juice collected from free-living animals.
2. Results
2.1. ELISA and Immunoblot Results
The ELISA testing found 14 of the 88 examined meat juice samples to be positive for the presence of Trichinella antibodies. Antibodies against Trichinella spp. were detected in two meat juice samples from foxes (10%) and 12 meat juice samples from raccoon dogs (46%). Additionally, one fox sample had a borderline result in ELISA. The positive and borderline results from ELISA were confirmed by immunoblot, which revealed a Trichinella-specific protein pattern recognized by meat juice obtained from red foxes (n = 2), raccoon dogs (n = 12), and a badger (n = 1). The characteristic bands typically migrated with a molecular weight (MW) between 50 and 250 kDa (Figure 1).
Figure 1.
The immunoblot analysis of T. britovi ML C antigens incubated with meat juice samples taken from different animals. Lanes 1–3—meat juice samples taken from foxes; lanes 4–15—meat juice samples taken from raccoon dogs; lane 16—meat juice sample taken from a badger; lanes 17–20—meat juice samples taken from badger, marten, fox, and raccoon dog (respectively), which were used as a negative control for immunoblot. Lines marked with + were recognized as positive in ELISA and immunoblot.
The presence of T. britovi larvae was confirmed in all animals recognized as immunopositive by both methods: ELISA and immunoblot (Figure 1, lanes 2–15), with different intensity of infection: animal 2: 1.61 larvae per gram (LPG); animal 3: 0.29 LPG, animal 4: 37.05 LPG, animal 5: 165.67 LPG, animal 6: 17.93 LPG, animal 7: 31.13 LPG, animal 8: 2.27 LPG, animal 9: 7.03 LPG, animal 10: 14.21 LPG, animal 11: 331.26 LPG, animal 12: 19.04 LPG, animal 13: 180,39 LPG, animal 14: 3.08 LPG, and animal 15: 4.85 LPG (the number of animals corresponds with the number of lanes in Figure 1). The results concerning the intensity of infection in raccoon dogs were partially published by Cybulska et al. (2019) [12]. Moreover, the result was borderline by ELISA, but negative by immunoblot in the case of one fox (Figure 1, lane 1), which was also T. britovi-positive in digestion. The intensity of infection was around one LPG. Immunoblot confirmed also the presence of anti-Trichinella antibodies in a meat juice sample from one badger that was not positive in ELISA (Figure 1, lane 16). Interestingly, this badger was infected with T. britovi, with a low intensity of infection (0.13 LPG).
Lanes 17–20 (Figure 1) represent meat juice samples used as a negative control for immunoblot. These samples were taken from badger, marten, fox, and raccoon dog (respectively), which were negative in digestion and in ELISA.
2.2. Immunoblot Analysis
Two meat juice samples from red foxes (F1, F2), and three from raccoon dogs (R1, R2, R3) were classified for further analysis to detect the specific proteins in the characteristic bands (Figure 2). The immunoreactive bands matched the corresponding protein bands on silver-stained gels and were selected for further LC-MS/MS analysis.
Figure 2.
(A) The immunoblot analysis of T. britovi ML C antigens incubated with selected meat juice samples. (B) Signal intensity and relative migration values for T. britovi ML C antigens incubated with examined meat juice samples. Lanes 1–2—meat juice samples from foxes (F1, F2, respectively); Lanes 3–5—meat juice samples from raccoon dogs (R1, R2, R3, respectively). No. of bands—A, B, C, D, E, F, G.
2.3. LC-MS/MS Analysis
Three gel pieces were obtained for both examined meat juice samples from red foxes (bands: A, B, C), four gel pieces for the two meat juice samples from raccoon dogs (bands: A, B, C, D), and seven gel pieces for the single meat juice sample from another raccoon dog (bands: A, B, C, D, E, F, G) (Figure 2). LC-MS/MS analysis revealed the presence of 279 proteins in the bands recognized by meat juice from fox F1; 209 proteins in the bands recognized by meat juice from fox F2; 357 proteins in the bands recognized by meat juice from raccoon dog R1; 339 proteins in the bands recognized by meat juice from raccoon dog R2; and 393 proteins in the bands recognized by meat juice from raccoon dog R3. More detailed information is presented in Supplementary Material Table S1 (foxes) and Table S2 (raccoon dogs). A comparative analysis of the proteins identified from the five examined meat juice samples indicates that all samples share 117 proteins, including heat shock proteins or paramyosin, which are known immunomodulators (Table 1). In addition, samples from foxes share 136 proteins, and those from raccoon dogs share 238 proteins. Several proteins were identified from multiple bands; these may correspond to protein isoforms or post-translational modifications (Tables S1 and S2).
Table 1.
Alphabetical list of 117 proteins common for five examined meat juice samples.
| Protein | Access to Genbank | Protein | Access to Genbank |
|---|---|---|---|
| 1,5-anhydro-D-fructose reductase | KRY43899.1 | Lysosomal aspartic protease, partial | KRY49432.1 |
| 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | KRY48325.1 | Malate dehydrogenase, cytoplasmic | KRY57329.1 |
| 26S protease regulatory subunit 4, partial | KRY52921.1 | Mannose-6-phosphate isomerase, partial | KRY55073.1 |
| 26S proteasome non-ATPase regulatory subunit 6, partial | KRY50659.1 | Mediator of RNA polymerase II transcription subunit 23 | KRY54660.1 |
| 32 kDa beta-galactoside-binding lectin | KRY59236.1 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial, partial | KRY52707.1 |
| 3-ketoacyl-CoA thiolase, mitochondrial | KRY51064.1 | Mitochondrial succinyl-CoA ligase subunit beta-like protein | KRY60682.1 |
| 4-hydroxybutyrate coenzyme A transferase, partial | KRY56869.1 | Mitochondrial-processing peptidase subunit alpha | KRY50357.1 |
| 60 kDa heat shock protein, mitochondrial | KRY51676.1 | Mitochondrial-processing peptidase subunit beta | KRY53080.1 |
| 60S acidic ribosomal protein P0 | KRY54528.1 | Mitochondrial isocitrate dehydrogenase, partial | KRY60163.1 |
| 60S ribosomal protein L4 | KRY61057.1 | Myophilin | KRY53866.1 |
| Actin-related protein 2/3 complex subunit 2 | KRY56438.1 | NADP-dependent malic enzyme, mitochondrial, partial | KRY59664.1 |
| Adducin-related protein 1 | KRY47021.1 | Obg-like ATPase 1 | KRY54762.1 |
| Adenylosuccinate lyase | KRY47589.1 | Paramyosin, partial | KRY49321.1 |
| Adenylosuccinate synthetase, partial | KRY50054.1 | Peroxiredoxin-2, partial | KRY49990.1 |
| ADP-ribose pyrophosphatase, mitochondrial | KRY48334.1 | Phosphate carrier protein, mitochondrial, partial | KRY57035.1 |
| Alpha-L-fucosidase | KRY54685.1 | Phosphoglucomutase-1 | KRY58984.1 |
| Aspartate aminotransferase, mitochondrial, partial | KRY49869.1 | Phosphoinositide 3-kinase regulatory subunit 4, partial | KRY53638.1 |
| Aspartate--tRNA ligase, cytoplasmic | KRY49144.1 | Polyubiquitin | KRY46475.1 |
| ATP synthase subunit alpha, mitochondrial, partial | KRY49394.1 | Polyubiquitin-like protein, partial | KRY24237.1 |
| Calpain clp-1 | KRY60775.1 | Polyubiquitin-B, partial | KRY44570.1 |
| Calreticulin, partial | KRY53845.1 | Polyubiquitin-C | KRY55776.1 |
| cAMP-dependent protein kinase regulatory subunit | KRY58719.1 | Proliferation-associated protein 2G4, partial | KRY53054.1 |
| Chymotrypsin-like elastase family member 2A, partial | KRY59723.1 | Propionyl-CoA carboxylase alpha chain, mitochondrial | KRY51819.1 |
| Cleavage and polyadenylation specificity factor subunit 2 | KRY53654.1 | Propionyl-CoA carboxylase beta chain, mitochondrial, partial | KRY52668.1 |
| Cytochrome b-c1 complex subunit 2, mitochondrial, partial | KRY49815.1 | Protein arginine N-methyltransferase 1, partial | KRY51927.1 |
| Cytosol aminopeptidase | KRY54953.1 | Protein ERGIC-53, partial | KRY52865.1 |
| Deoxyribonuclease-2-alpha | KRY53116.1 | putative [pyruvate dehydrogenase (acetyl-transferring)] kinase, mitochondrial, partial | KRY57184.1 |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | KRY46167.1 | putative aconitate hydratase, mitochondrial | KRY56796.1 |
| Dihydropyrimidinase-related protein 3 | KRY48683.1 | putative aminopeptidase W07G4.4 | KRY54333.1 |
| Diphthine--ammonia ligase, partial | KRY47980.1 | putative arginine kinase F46H5.3 | KRY58392.1 |
| DnaJ-like protein dnj-20 | KRY45252.1 | putative oxidoreductase-like protein | KRY46863.1 |
| Dynein heavy chain, cytoplasmic | KRY59544.1 | putative phosphoglycerate kinase | KRY46311.1 |
| Elongation factor G, mitochondrial | KRY55228.1 | putative pyruvate dehydrogenase E1 component subunit alpha, mitochondrial | KRY58181.1 |
| Endoplasmic reticulum resident protein 44 | KRY50742.1 | Rab GDP dissociation inhibitor alpha | KRY54650.1 |
| Endoplasmin, partial | KRY54651.1 | secretion antigen precursor | CAD86782.1 |
| Eukaryotic initiation factor 4A, partial | KRY53637.1 | Serine hydroxymethyltransferase | KRY56265.1 |
| Eukaryotic initiation factor 4A-III | KRY49326.1 | Serine protease 30 | KRY58838.1 |
| Far upstream element-binding protein 2 | KRY57083.1 | Snake venom 5’-nucleotidase | KRY49427.1 |
| Fructose-bisphosphate aldolase 1, partial | KRY52953.1 | Solute carrier family 2, facilitated glucose transporter member 3 | KRY47169.1 |
| Glucose-6-phosphate isomerase | KRY50483.1 | Splicing factor 3A subunit 3 | KRY56136.1 |
| Glyceraldehyde-3-phosphate dehydrogenase 1 | KRY59635.1 | Stress-induced-phosphoprotein 1 | KRY50814.1 |
| Glycogen phosphorylase, partial | KRY47089.1 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | KRY54543.1 |
| Glycogenin-1 | KRY61196.1 | Succinate-semialdehyde dehydrogenase, mitochondrial | KRY54971.1 |
| Heat shock cognate 71 kDa protein, partial | KRY58599.1 | Sulfide:quinone oxidoreductase, mitochondrial | KRY46292.1 |
| Heat shock protein 83, partial | KRY48983.1 | T-complex protein 1 subunit alpha | KRY49588.1 |
| Histone-lysine N-methyltransferase EHMT2, partial | KRY52481.1 | T-complex protein 1 subunit beta | KRY52365.1 |
| hypothetical protein T03_13827 | KRY56487.1 | T-complex protein 1 subunit epsilon | KRY50385.1 |
| hypothetical protein T03_17187 | KRY50177.1 | T-complex protein 1 subunit eta | KRY48081.1 |
| hypothetical protein T03_539 | KRY55455.1 | T-complex protein 1 subunit gamma | KRY47292.1 |
| hypothetical protein T03_7459 | KRY60639.1 | T-complex protein 1 subunit theta | KRY50910.1 |
| hypothetical protein T03_8666, partial | KRY58917.1 | Transketolase | KRY48205.1 |
| hypothetical protein T03_8694 | KRY46541.1 | Transmembrane protease serine 5 | KRY50806.1 |
| hypothetical protein T03_9489 | KRY46491.1 | Transmembrane protease serine 9 | KRY58843.1 |
| Ig-like and fibronectin type-III domain-containing protein C25G4.10 | KRY58616.1 | Trifunctional enzyme subunit beta, mitochondrial | KRY48050.1 |
| Intermediate filament protein B, partial | KRY59373.1 | Tubulin alpha-3 chain | KRY50534.1 |
| Intermediate filament protein ifa-1 | KRY45949.1 | Type I inositol 1,4,5-trisphosphate 5-phosphatase, partial | KRY50866.1 |
| Kynurenine--oxoglutarate transaminase 3 | KRY54139.1 | Ubiquitin carboxyl-terminal hydrolase 14, partial | KRY58645.1 |
| L-2-hydroxyglutarate dehydrogenase, mitochondrial | KRY56323.1 | Uncharacterized protein T03_9851 | KRY58607.1 |
| Leukocyte elastase inhibitor C, partial | KRY55578.1 |
2.4. Gene Ontology (GO) Analysis
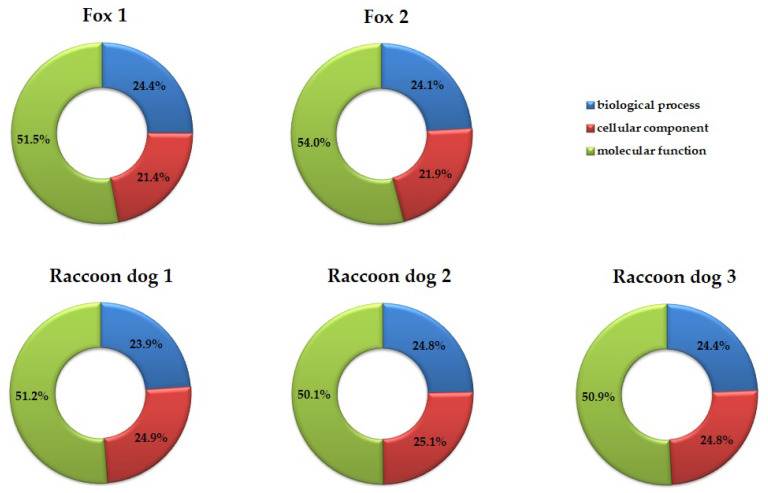
Gene Ontology (GO) analysis was used to identify proteins reacting with antibodies present in examined meat juice samples, and these proteins were categorized according to their molecular function, cellular component, and biological process. The percentage share of the number of identified proteins by categories is shown in Figure 3. The data indicates that 222 proteins are associated with biological processes for F1, 159 for F2, 284 for R1, 282 for R2, and 312 for R3. Regarding biological processes, the highest number of proteins are involved in translation and protein folding processes. Briefly, 220 proteins were associated with cellular components for F1, 145 for F2, 295 for R1, 186 for R2, and 317 for R3. The identified proteins are mainly associated with organelles (nucleus, ribosome) and cell parts (cytoplasm, components of membrane). Molecular functions were also found: 469 proteins for F1, 357 for F2, 608 for R1, 571 for R2, and 651 for R3. The predominant types are related to binding (e.g., ion binding, ATP binding, RNA binding) and catalytic activity (e.g., ATP hydrolysis activity, GTPase activity).
Figure 3.
The percentage share of the number of proteins derived from three categories: biological process, cellular component, and molecular function.
3. Discussion
Trichinellosis is a disease with a global range. Unsurprisingly, there is a great interest in identifying peptide or protein biomarkers that could be used as markers to identify early Trichinella infection in humans and domestic animals (diagnostic properties) or as vaccines against Trichinella infection, e.g., in pigs (immunoprotective properties). Although recent studies have focused on the immunoreactive proteins in host serum and plasma during Trichinella infection, they have mainly been based on 2DE-electrophoresis examination coupled with LC-MS/MS, or 2DE electrophoresis followed by MALDI-TOF mass spectrometry of human, mouse, or pig samples [30,36,37,38,39,47,48,49,50,51,52,53]. In contrast, the present paper employs standard 1DE electrophoresis coupled with LC-MS/MS analysis to obtain preliminary findings; it is the first proteomic analysis of proteins recognized by meat juice samples collected from wildlife. These findings provide new insight into the immune response to Trichinella infection in wild animals. Free-living animals constitute a natural reservoir for various parasites, including Trichinella nematodes, and unlike laboratory-based studies, the infection dose and the number of potential re-infections are unknown. Therefore, a thorough understanding of the protein profiles present in free-living animals is needed to provide an accurate picture of the course of a parasite infection.
The entire life cycle of the Trichinella nematodes takes place in a single host, following the ingestion of meat containing infectious Trichinella larvae. Trichinella displays three main antigenic stages: muscle larvae (ML), adult worms (Ad), and newborn larvae (NBL) [14]. The muscle larvae are released in the stomach, from where they migrate to the epithelial cells of the small intestine, where they molt and transform into adult worms. After coupling, the females start to deliver NBLs, which move mainly through the blood circulation to reach the striated muscle, where they finally develop into MLs [1]. All developmental stages of Trichinella stimulate an immune response and produce antigens which can be used for the serological detection of Trichinella spp. infection. Additionally, some research suggests that the Trichinella antigens produced by the three developmental stages are stage-specific [14] and they stimulate or suppress the host immune response to varying degrees.
Most studies have used serum/plasma samples to determine the protein profile characteristic of Trichinella infection [14,30,36,37,38,39,47,48,49,51,52,53]. However, while serum or plasma are not always available for analysis, meat juice samples provide an adequate, alternative matrix that can be easily obtained for post-mortem analysis, especially from free-living animals. The use of meat juice also offers the added benefit of studying protein profiles at the exact location where Trichinella larvae localize, thus providing potential new insight into parasite infection, especially when investigating stage-specific antigens for muscle larvae.
In the present studies, different proteins recognized by meat juice samples were observed not only between two of the examined animal species but also between those of the same species (Figure 2A, Table 1). The obtained results may suggest that the examined animals were infected with different doses of the parasite with a different duration of infection. However, in the present paper, examined meat juice samples were taken from animals infected with T. britovi in the late stage of infection, with larvae present in the muscle. It needs to be underlined that the intensity of infection in examined animals differs between specimens. Additionally, in immunoblot, the characteristic bands typically migrated between 50 and 250 kDa, with some differences between animals (Figure 1). In another study, performed with sera of raccoon dogs infected with T. spiralis or T. nativa the bands were observed at around 100 kDa, with a group of bands between 76 and 40 kDa [40]. Contrary to this, research conducted with the use of the serum of foxes infected with Trichinella nativa, revealed that most bands migrate between 40 and 60 kDa [41].
Although most trichinellosis in humans is caused by T. spiralis, some cases of T. britovi infection have recently been observed [54,55,56,57]. Furthermore, several papers have described the analysis of crude and excretory-secretory proteins characteristic of T. britovi [14,30,36,38,39,48]. These studies, conducted using human or T. britovi-infected pig serum, identified some proteins characteristic of T. britovi infection, including those that play a role in the migration of NBL in host tissue or those with immunomodulatory potential.
The aim of the present study was to determine the pattern of proteins reacting with IgG antibodies in meat juice samples collected from free-living animals infected with T. britovi. It was found that antibodies present in meat juice samples react with proteins that are associated with T. britovi infection; in addition, the majority of them appear to be involved in metabolic processes and host–parasite interaction (e.g., the mechanisms of the parasite invasion of host tissue, larval migration, larval molting, immunomodulation), such as heat shock proteins, a serine protease, paramyosin, a poly-cysteine tailed protein, and deoxyribonuclease-2-alpha [14,30,36,38,39]. The identified proteins are compared with the literature data on proteins specific to T. britovi infection in Table 2.
Table 2.
A comparison of the proteins obtained in the present study with the literature data on proteins specific to T. britovi infection. The literature data given in the table were obtained from studies relating to antigens extracted from the ML stage.
| Protein | Animal | GenBank Accession Number | Reference |
|---|---|---|---|
| Hypothetical protein T03_17187 | fox | KRY50178.1 | this study |
| raccoon dog | this study | ||
| human | [48] | ||
| pig | [14] | ||
| Deoxyribonuclease-2-alpha | fox | KRY53318.1 | this study |
| raccoon dog | this study | ||
| human | KRX47308.1 | [48] | |
| pig | [14] | ||
| Transmembrane protease serine 9 | fox | KRY59262.1 | this study |
| raccoon dog | this study | ||
| human | KRY20911.1 | [48] | |
| human | KRY21930.1 | [48] | |
| pig | KRY58843.1 | [38] | |
| Intermediate filament protein B, partial | fox | KRY59373.1 | this study |
| raccoon dog | this study | ||
| pig | KRY11608.1 | [38] | |
| pig | [14] | ||
| Intermediate filament protein ifa-1 | fox | KRY45949.1 | this study |
| raccoon dog | this study | ||
| pig | KRY09282.1 | [38] | |
| pig | [14] | ||
| Serine protease 30 | fox | KRY58841.1 | this study |
| raccoon dog | this study | ||
| pig | KRX47705.1 | [38] | |
| pig | [14] | ||
| Hypothetical protein T03_8694 | fox | KRY46541.1 | this study |
| raccoon dog | this study | ||
| pig | [38] | ||
| Cuticlin-1 | fox | KRY58751.1 | this study |
| raccoon dog | this study | ||
| Cuticlin-1, partial | pig | KRY01407.1 | [14] |
| pig | KRY57444.1 | [38] | |
| Paramyosin, partial | fox | KRY49321.1 | this study |
| raccoon dog | this study | ||
| pig | KRY49322.1 | [38] | |
| Myosin-4, partial | fox | KRY50587.1 | this study |
| raccoon dog | this study | ||
| pig | [38] | ||
| 26S protease regulatory subunit 10B | fox | KRY59566.1 | this study |
| raccoon dog | this study | ||
| pig | [38] | ||
| Adenylosuccinate lyase | fox | KRY47589.1 | this study |
| raccoon dog | this study | ||
| pig | [38] | ||
| 32 kDa beta-galactoside-binding lectin | fox | KRY59236.1 | this study |
| raccoon dog | this study | ||
| pig | [38] | ||
| Peroxiredoxin-2, partial | fox | KRY49990.1 | this study |
| raccoon dog | this study | ||
| pig | KRX46812.1 | [14] | |
| Poly-cysteine and histidine-tailed protein, partial | raccoon dog | KRY58369.1 | this study |
| pig | KRY11984.1 | [14] | |
| Calponin-like protein OV9M, partial | fox | KRY56610.1 | this study |
| raccoon dog | this study | ||
| pig | KRX28313.1 | [14] | |
| Propionyl-CoA carboxylase alpha chain, mitochondrial | fox | KRY51819.1 | this study |
| raccoon dog | this study | ||
| pig | KRZ50222.1 | [14] | |
| 1,5-anhydro-D-fructose reductase | fox | KRY43899.1 | this study |
| raccoon dog | this study | ||
| pig | KRY07641.1 | [14] | |
| Mitochondrial-processing peptidase subunit beta | fox | KRY53080.1 | this study |
| raccoon dog | this study | ||
| Mitochondrial-processing peptidase subunit beta, partial | pig | KRZ09733.1 | [14] |
| putative histone-binding protein Caf1 | fox | KRY53239.1 | this study |
| raccoon dog | this study | ||
| pig | KRZ17128.1 | [14] | |
| Rab GDP dissociation inhibitor alpha | fox | KRY54650.1 | this study |
| raccoon dog | this study | ||
| pig | KRY13378.1 | [14] |
LC-MS/MS analysis allowed to reveal the presence of a wide range of proteins known to take part in tissue invasion, larval migration, larval molting, immune modulation, and metabolic processes. For example, the subunits of 26S protease have been found to be involved in cell cycle and transcription regulation, apoptosis, and the oxidative stress response [38]. Deoxyribonuclease-2-alpha plays a role in the invasion of host tissue and in evading host defense [58,59]. In addition, intermediate filament proteins were observed in all examined samples. These proteins are known as structural elements of the cellular cytoskeleton and are responsible for worm growth [60]. As well as the filament proteins, myosin-4 and paramyosin also play a role in parasite growth. The first is responsible for cellular component organization and actin filament depolymerization, thus facilitating parasite growth and development [14], while the second influences muscle length and stability. It has been observed that paramyosin from helminths serves not only as a structural protein but also as an immunomodulatory agent [61].
In addition, peroxiredoxin-2 protects nematodes from hazardous reactive oxygen species (ROS) during invasion [62], and serine protease 30 plays a crucial role in the host tissues during cell invasions and larval molting [14]. Interestingly, all examined samples were found to contain hypothetical protein T03_17187; this protein has been found to share more than 90% identity with the multi-cystatin-like domain protein (CLP), which is a promising immunoreactive protein [48,63,64].
These proteins are known to be involved in the biological processes of the parasite, such as intracellular protein transport, stress response, glycolytic processes, tricarboxylic acid cycle, protein folding, and translation; they also form parts of important cellular components, such as the cytosol, mitochondrion, proteasome complex, small ribosomal subunit, ribosome, nucleus, cytoplasm, and integral membrane component. They are also involved in significant molecular functions, mainly as catalytic activity (ATP hydrolysis activity, GTPase activity, transferase activity, hydrolase activity, kinase activity) and binding activity to ions, ATP, and RNA. However, it has to be emphasized that differences in the protein profile were observed not only between two examined animal species but also between specimens of the same species (Table S3). This is not surprising, as it has been proposed that the host immune response may vary depending on the dose and phase of infection, parasite, and host species [65,66,67]. Interestingly, the percentage share of the number of proteins derived by three categories, viz. biological process, cellular component, and molecular function, remained comparable between the five examined meat juice samples (Figure 3).
The present study addresses the previous lack of data regarding the presence of T. britovi antibodies in meat juice from free-living animals. T. britovi is known to be widespread among wild animal species and can be transmitted to humans. Hence, there is a pressing need to identify T. britovi-specific antigens which may be used to diagnose infection in humans or support vaccine development [64]. Indeed, searching for markers or proteins with vaccine potential among free-living animals infected with the parasite is in line with the One Health Concept. Nevertheless, there is a need to extend the study using more advanced proteomics techniques such as 2DE electrophoresis coupled with LC-MS/MS analysis.
4. Materials and Methods
4.1. Collection of Material and Research Scheme
The study was conducted on 20 red foxes (Vulpes vulpes), 26 raccoon dogs (Nyctereutes procyonoides), 24 martens (Martes spp.), and 18 badgers (Meles meles). The animals were acquired between 2013 and 2016 through hunting activities within Project Life+ (NAT/PL/428). Meat juice samples were collected on the occasion of the digestion method for the presence of Trichinella spp. larvae according to EC Regulation No. 2075/2005 [68], and the Trichinella species was determined using multiplex PCR described by Zarlenga et al. 1999 [69], with some modification by Cybulska et al. 2019 [12]. Carcasses were transported to the Witold Stefański Institute of Parasitology, Polish Academy of Sciences (Warsaw, Poland). Muscle samples were taken during dissection and then stored at −20 °C for further analysis. The day before digestion, muscle samples (diaphragms and limb muscles) were thawed at room temperature (RT) and meat juice samples were collected individually into sterile tubes. Next, the obtained samples were stored at −20 °C for further tests: ELISA and immunoblotting.
4.2. ELISA
The presence of antibodies against Trichinella from the meat juice samples was detected using a commercial ELISA kit (ID Screen Trichinella Indirect Multi-species, IDvet, Grabels, France), according to the manufacturer’s instructions. The meat juice sample dilution was 1:2, according to the protocol. The optical density (O.D.) was measured at a wavelength of 450 nm using an EL*800 ELISA automated plate reader (BioTek, Winooski, VT, USA). The cutoff was calculated based on the sample-to-positive (S/P) percentage according to the formula S/P = [optical density (OD) sample − OD negative control (NC)/OD positive control (PC) − OD NC] × 100. Samples with S/P ≥ 30% were considered as positive; with 25% ≤ S/P% ≤ 30% were considered as borderline; and with S/P% ≤ 25% were considered as negative.
4.3. Preparation of Crude Antigens from Muscle Larvae of T. britovi
The Balb/C mice were orally infected with 500 T. britovi (ISS002) muscle larvae (ML), then maintained for at least three months, and then euthanized. Next, to prepare muscle larvae crude antigens (ML C antigens), the MLs were isolated using digestion. The recovered MLs were subsequently purified several times with water through succeeding steps of sedimentation in cylinders. After the final sedimentation, the MLs were collected into 1.5 mL tubes. The larval pellet was extensively washed three times in phosphate-buffered saline (PBS). After that, the larval pellet was washed three times and mixed with lysis buffer as described previously by Bień et al. 2013 [50]. The mixture was homogenized in a Potter-Elvehjem Tissue Homogenizer and disintegrated by sonication (OMNI International Tissue Homogenizer). Next, the solution was centrifuged at 14,000 RPM at 4 °C for 10 min.
4.4. SDS–Polyacrylamide Gel Electrophoresis (SDS–PAGE) and Immunoblot
The ML C antigens (approximately 10 μg per well) were run on a 12% SDS-PAGE gel with 4% stacking gel at 180 V constant voltage for 50 min. After electrophoresis, the proteins were transferred from the gel to a nitrocellulose sheet (Bio-Rad, Hercules, CA, USA) at 95V for 55 min using Mini-Protean Tetra Cell (Bio-Rad, Hercules, CA, USA). Nitrocellulose strips (NCS) were blocked with Pierce Protein-Free T20 Blocking Buffer (Thermo Fisher Scientific, Waltham, MA, USA) for one hour at RT, and then washed three times in PBS–Tween buffer (pH = 7.2). Following this, the NCSs were incubated with shaking for one hour at RT with meat juice samples diluted 1:20 in Pierce Protein-Free T20 Blocking Buffer (Thermo Fisher Scientific, Waltham, MA, USA). Afterward, the NCSs were washed and then incubated with shaking for one hour at RT with secondary antibody, i.e., horse radish-peroxidase-conjugated Anti-dog IgG (Sigma-Aldrich, Saint Louis, MO, USA), diluted by 1:25,000. Finally, after washing, the NCSs were developed using SIGMAFAST™ 3,3′-diaminobenzidine (Sigma-Aldrich, Saint Louis, MO, USA) and visualized with the Chemi Doc MP Imaging System (Bio-Rad, Hercules, CA, USA).
4.5. Proteomic Analyses—Mass Spectrometry (MS) and Protein Identification
Bands of interest were manually excised from the gel and analyzed by liquid chromatography coupled to a mass spectrometer (LC-MS/MS) in the Laboratory of Mass Spectrometry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland). The obtained peptide masses and fragmentation spectra were matched to the NCBIProt database, with a Nematoda filter using the Mascot search engine. All proteins identified in the Mascot search were compared withthe UniProtKB database (https://www.uniprot.org/; accessed on 29 August 2022) and QuickGO (http://www.ebi.ac.uk/QuickGO/; accessed on 29 August 2022) and classified in gene ontology (GO) in accordance with their molecular functions, biological processes, and cellular component information.
5. Conclusions
The findings show that meat juice is a useful material for studying antibodies from animals naturally infected with T. britovi. The use of proteomic analysis reveals the presence of proteins known to take part in tissue invasion, larval migration, larval molting, immune modulation, and metabolic processes. Additionally, the mentioned proteins are immunogenic, and they are recognized by host antibodies. The results indicate also that the IgG-response may differ between host species in wildlife. A comprehensive understanding of immune response in free-living animals may provide an accurate picture of parasite infection.
Acknowledgments
The author would like to thank Sylwia Grzelak, Justyna Bień-Kalinowska, and Bożena Moskwa from the Witold Stefański Institute of Parasitology, Polish Academy of Sciences for their priceless comments and valuable help during the creation of this paper.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11101155/s1, Table S1: Results of the LC-MS/MS analysis of selected gel fragments for two examined foxes: F1 and F2; Table S2: Results of the LC-MS/MS analysis of selected gel fragments for three examined raccoon dogs: R1, R2, and R3; Table S3: Gene Ontology (GO) database analysis outcomes for identified proteins from two foxes and three raccoon dogs. The proteins were categorized according to molecular function, cellular component, and biological process.
Institutional Review Board Statement
The protocol was approved by the First Local Ethical Committee for Scientific Experiments on Animals in Warsaw, Poland (resolution No.: 020/2016, 23 March 2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflict of interest.
Funding Statement
This research was partially funded by the Witold Stefański Institute of Parasitology, Polish Academy of Sciences, Warsaw, Poland, as part of the Internal Project for Young Scientists.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gottstein B., Pozio E., Nöckler K. Epidemiology, diagnosis, treatment, and control of trichinellosis. Clin. Microbiol. Rev. 2009;22:127–145. doi: 10.1128/CMR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaga R., Gherman C., Cozma V., Zocevic A., Pozio E., Boireau P. Trichinella species circulating among wild and domestic animals in Romania. Vet. Parasitol. 2009;159:218–221. doi: 10.1016/j.vetpar.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 3.Pozio E., Rinaldi L., Marucci G., Musella V., Galati F., Cringoli G., Boireau P., La Rosa G. Hosts and habitats of Trichinella spiralis and Trichinella britovi in Europe. Int. J. Parasitol. 2009;39:71–79. doi: 10.1016/j.ijpara.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Pozio E., Hoberg E., La Rosa G., Zarlenga D.S. Molecular taxonomy, phylogeny and biogeography of nematodes belonging to the Trichinella genus. Infect. Genet. Evol. 2009;9:606–616. doi: 10.1016/j.meegid.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Chmurzyńska E., Różycki M., Bilska-Zając E., Nöckler K., Mayer-Scholl A., Pozio E., Cencek T., Karamon J. Trichinella nativa in red foxes (Vulpes vulpes) of Germany and Poland: Possible different origins. Vet. Parasitol. 2013;198:254–257. doi: 10.1016/j.vetpar.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 6.Bień J., Moskwa B., Goździk K., Cybulska A., Kornacka A., Welc M., Popiołek M., Cabaj W. The occurrence of nematodes of the genus Trichinella in wolves (Canis lupus) from the Bieszczady Mountains and Augustowska Forest in Poland. Vet. Parasitol. 2016;231:115–117. doi: 10.1016/j.vetpar.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Deksne G., Segliņa Z., Jahundoviča I., Esīte Z., Bakasejevs E., Bagrade G., Keidāne D., Interisano M., Marucci G., Tonanzi D., et al. High prevalence of Trichinella spp. in sylvatic carnivore mammals of Latvia. Vet. Parasitol. 2016;231:118–123. doi: 10.1016/j.vetpar.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Kirjušina M., Bakasejevs E., Pezzotti P., Pozio E. Trichinella britovi biomass in naturally infected pine martens (Martes martes) of Latvia. Vet. Parasitol. 2016;231:110–114. doi: 10.1016/j.vetpar.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Dmitric M., Vidanovic D., Vaskovic N., Matovic K., Sekler M., Debeljak Z., Karabasil N. Trichinella infections in red foxes (Vulpes vulpes) and golden jackals (Canis aureus) in six districts of Serbia. J. Zoo Wildl. Med. 2017;48:703–707. doi: 10.1638/2016-0169.1. [DOI] [PubMed] [Google Scholar]
- 10.Kärssin A., Häkkinen L., Niin E., Peik K., Vilem A., Jokelainen P., Lassen B. Trichinella spp. biomass has increased in raccoon dogs (Nyctereutes procyonoides) and red foxes (Vulpes vulpes) in Estonia. Parasite Vectors. 2017;10:609. doi: 10.1186/s13071-017-2571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cybulska A., Skopek R., Kornacka A., Popiołek M., Piróg A., Laskowski Z., Moskwa B. First detection of Trichinella pseudospiralis infection in raccoon (Procyon lotor) in Central Europe. Vet. Parasitol. 2018;254:114–119. doi: 10.1016/j.vetpar.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Cybulska A., Kornacka A., Moskwa B. The occurrence and muscle distribution of Trichinella britovi in raccoon dogs (Nyctereutes procyonoides) in wildlife in the Głęboki Bród Forest District, Poland. Int. J. Parasitol. Parasites Wildl. 2019;9:149–153. doi: 10.1016/j.ijppaw.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherman C.M., Boros Z., Băieș M.H., Cozma-Petruț A., Cozma V. A review of Trichinella species infection in wild animals in Romania. Food Waterborne Parasitol. 2022;28:e00178. doi: 10.1016/j.fawpar.2022.e00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grzelak S., Moskwa B., Bień J. Trichinella britovi muscle larvae and adult worms: Stage-specific and common antigens detected by two-dimensional gel electrophoresis-based immunoblotting. Parasites Vectors. 2018;11:584. doi: 10.1186/s13071-018-3177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller I., Wait R., Sipos W., Gemeiner M. A proteomic reference map for pig serum proteins as a prerequisite for diagnostic applications. Res. Vet. Sci. 2009;86:362–367. doi: 10.1016/j.rvsc.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Gondek M., Herosimczyk A., Knysz P., Ożgo M., Lepczyński A., Szkucik K. Comparative Proteomic Analysis of Serum from Pigs Experimentally Infected with Trichinella spiralis, Trichinella britovi, and Trichinella pseudospiralis. Pathogens. 2020;9:55. doi: 10.3390/pathogens9010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallander C., Frössling J., Vågsholm I., Burrells A., Lundén A. “Meat juice” is not a homogeneous serological matrix. Foodborne Pathog. Dis. 2015;12:280–288. doi: 10.1089/fpd.2014.1863. [DOI] [PubMed] [Google Scholar]
- 18.Berger-Schoch A.E., Bernet D., Doherr M.G., Gottstein B., Frey C.F. Toxoplasma gondii in Switzerland: A serosurvey based on meat juice analysis of slaughtered pigs, wild boar, sheep and cattle. Zoonoses Public Health. 2011;58:472–478. doi: 10.1111/j.1863-2378.2011.01395.x. [DOI] [PubMed] [Google Scholar]
- 19.Kornacka A., Cybulska A., Bień J., Goździk K., Moskwa B. The usefulness of direct agglutination test, enzyme-linked immunosorbent assay and polymerase chain reaction for the detection of Toxoplasma gondii in wild animals. Vet. Parasitol. 2016;228:85–89. doi: 10.1016/j.vetpar.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Kornacka A., Cybulska A., Popiołek M., Kuśmierek N., Moskwa B. Survey of Toxoplasma gondii and Neospora caninum in raccoons (Procyon lotor) from the Czech Republic, Germany and Poland. Vet. Parasitol. 2018;262:47–50. doi: 10.1016/j.vetpar.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Vico J.P., Mainar-Jaime R.C. The use of meat juice or blood serum for the diagnosis of Salmonella infection in pigs and its possible implications on Salmonella control programs. J. Vet. Diagn. Investig. 2011;23:528–531. doi: 10.1177/1040638711403432. [DOI] [PubMed] [Google Scholar]
- 22.Szulowski K., Pilaszek J., Iwaniak W. Application of meat juice in diagnosis of brucellosis in hares and wild boars by ELISA. Bull. Vet. Inst. Pulawy. 2000;44:45–52. [Google Scholar]
- 23.Yonemitsu K., Minami S., Noguchi K., Kuwata R., Shimoda H., Maeda K. Detection of anti-viral antibodies from meat juice of wild boars. J. Vet. Med. Sci. 2019;81:155–159. doi: 10.1292/jvms.18-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cybulska A., Kornacka A., Popiołek M., Bień-Kalinowska J., Moskwa B. Use of meat juice from racoons (Procyon lotor) collected from Central Europe for immunological detection of Trichinella spp. Vet. Parasitol. 2021;297:109066. doi: 10.1016/j.vetpar.2020.109066. [DOI] [PubMed] [Google Scholar]
- 25.Winter M., Abate S.D., Pasqualetti M.I., Fariña F.A., Ercole M.E., Pardini L., Moré G., Venturini M.C., Perera N., Corominas M.J., et al. Toxoplasma gondii and Trichinella infections in wild boars (Sus scrofa) from Northeastern Patagonia, Argentina. Prev. Vet. Med. 2019;168:75–80. doi: 10.1016/j.prevetmed.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Kärssin A., Velström K., Gómez-Morales M.A., Saar T., Jokelainen P., Lassen B. Cross-Sectional Study of Anti-Trichinella Antibody Prevalence in Domestic Pigs and Hunted Wild Boars in Estonia. Vector Borne Zoonotic Dis. 2016;16:604–610. doi: 10.1089/vbz.2016.1943. [DOI] [PubMed] [Google Scholar]
- 27.Gómez-Morales M.A., Ludovisi A., Amati M., Bandino E., Capelli G., Corrias F., Gelmini L., Nardi A., Sacchi C., Cherchi S., et al. Indirect versus direct detection methods of Trichinella spp. infection in wild boar (Sus scrofa) Parasite Vectors. 2014;7:171. doi: 10.1186/1756-3305-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nöckler K., Hamidi A., Fries R., Heidrich J., Beck R., Marinculic A. Influence of methods for Trichinella detection in pigs from endemic and non-endemic European region. J. Vet. Med. B Infect Dis. Vet. Public Health. 2004;51:297–301. doi: 10.1111/j.1439-0450.2004.00770.x. [DOI] [PubMed] [Google Scholar]
- 29.Bessi C., Ercole M.E., Fariña F.A., Ribicich M.M., Montalvo F., Acerbo M., Krivokapich S.J., Pasqualetti M.I. Study of Trichinella patagoniensis in wild boars. Vet. Parasitol. 2021;297:109166. doi: 10.1016/j.vetpar.2020.109166. [DOI] [PubMed] [Google Scholar]
- 30.Grzelak S., Stachyra A., Bień-Kalinowska J. The first analysis of Trichinella spiralis and Trichinella britovi adult worm excretory-secretory proteins by two-dimensional electrophoresis coupled with LC-MS/MS. Vet. Parasitol. 2021;297:109096. doi: 10.1016/j.vetpar.2020.109096. [DOI] [PubMed] [Google Scholar]
- 31.Cybulska A., Kornacka A., Skopek R., Moskwa B. Trichinella britovi infection and muscle distribution in free-living martens (Martes spp.) from the Głęboki Bród Forest District, Poland. Int. J. Parasitol. Parasites Wildl. 2020;12:176–180. doi: 10.1016/j.ijppaw.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antolová D., Fecková M., Valentová D., Hurníková Z., Miklisová D., Avdičová M., Halánová M. Trichinellosis in Slovakia–epidemiological situation in humans and animals (2009-2018) Ann. Agric. Environ. Med. 2020;27:361–367. doi: 10.26444/aaem/125194. [DOI] [PubMed] [Google Scholar]
- 33.Rostami A., Gamble H.R., Dupouy-Camet J., Khazan H., Bruschi F. Meat sources of infection for outbreaks of human trichinellosis. Food Microbiol. 2017;64:65–71. doi: 10.1016/j.fm.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Turiac I.A., Cappelli M.G., Olivieri R., Angelillis R., Martinelli D., Prato R., Fortunato F. Trichinellosis outbreak due to wild boar meat consumption in southern Italy. Parasite Vectors. 2017;10:107. doi: 10.1186/s13071-017-2052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavic S., Andric A., Sofronic-Milosavljevic L.J., Gnjatovic M., Mitić I., Vasilev S., Sparic R., Pavic A. Trichinella britovi outbreak: Epidemiological, clinical, and biological features. Med. Mal. Infect. 2020;50:520–524. doi: 10.1016/j.medmal.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Bień J., Näreaho A., Varmanen P., Goździk K., Moskwa B., Cabaj W., Nyman T.A., Savijoki K. Comparative analysis of excretory-secretory antigens of Trichinella spiralis and Trichinella britovi muscle larvae by two-dimensional difference gel electrophoresis and immunoblotting. Proteome Sci. 2012;10:10. doi: 10.1186/1477-5956-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gondek M., Knysz P., Pomorska-Mól M., Ziomek M., Bień-Kalinowska J. Acute phase protein pattern and antibody response in pigs experimentally infected with a moderate dose of Trichinella spiralis, T. britovi, and T. pseudospiralis. Vet. Parasitol. 2020;288:109277. doi: 10.1016/j.vetpar.2020.109277. [DOI] [PubMed] [Google Scholar]
- 38.Grzelak S., Stachyra A., Moskwa B., Bień-Kalinowska J. Exploiting the potential of 2D DIGE and 2DE immunoblotting for comparative analysis of crude extract of Trichinella britovi and Trichinella spiralis muscle larvae proteomes. Vet. Parasitol. 2021;289:109323. doi: 10.1016/j.vetpar.2020.109323. [DOI] [PubMed] [Google Scholar]
- 39.Gondek M., Grzelak S., Pyz-Łukasik R., Knysz P., Ziomek M., Bień-Kalinowska J. Insight into Trichinella britovi Infection in Pigs: Effect of Various Infectious Doses on Larvae Density and Spatial Larvae Distribution in Carcasses and Comparison of the Detection of Anti-T. britovi IgG of Three Different Commercial ELISA Tests and Immunoblot Assay. Pathogens. 2022;11:735. doi: 10.3390/pathogens11070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukura A., Näreaho A., Mikkonen T., Niemi M., Oivanen L. Trichinella nativa and T. spiralis induce distinguishable histopathologic and humoral responses in the raccoon dog (Nyctereutes procyonoides) Vet. Pathol. 2002;39:257–265. doi: 10.1354/vp.39-2-257. [DOI] [PubMed] [Google Scholar]
- 41.Davidson R.K., Ørpetveit I., Møller L., Kapel C.M. Serological detection of anti-Trichinella antibodies in wild foxes and experimentally infected farmed foxes in Norway. Vet. Parasitol. 2009;163:93–100. doi: 10.1016/j.vetpar.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 42.Gómez-Morales M.A., Selmi M., Ludovisi A., Amati M., Fiorentino E., Breviglieri L., Poglayen G., Pozio E. Hunting dogs as sentinel animals for monitoring infections with Trichinella spp. in wildlife. Parasite Vectors. 2016;9:154. doi: 10.1186/s13071-016-1437-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miterpáková M., Antolová D., Hurníková Z., Březinová N., Čabanová V., Reiterová K. Seroprevalence of Trichinella infections in domestic dogs from Slovakia. J. Helminthol. 2017;91:549–554. doi: 10.1017/S0022149X16000602. [DOI] [PubMed] [Google Scholar]
- 44.Grzybek M., Cybulska A., Tołkacz K., Alsarraf M., Behnke-Borowczyk J., Szczepaniak K., Strachecka A., Paleolog J., Moskwa B., Behnke J.M., et al. Seroprevalence of Trichinella spp. infection in bank voles (Myodes glareolus)–A long term study. Int. J. Parasitol. Parasites Wildl. 2019;9:144–148. doi: 10.1016/j.ijppaw.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boros Z., Vallée I., Panait L.C., Gherman C.M., Chevillot A., Boireau P., Cozma V. Seroprevalance of Trichinella Spp. in Wild Boars (Sus Scrofa) from Bihor County, Western Romania. Helminthologia. 2020;57:235–240. doi: 10.2478/helm-2020-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilfold N.W., Richardson E.S., Ellis J., Jenkins E., Scandrett W.B., Hernández-Ortiz A., Buhler K., McGeachy D., Al-Adhami B., Konecsni K., et al. Long-term increases in pathogen seroprevalence in polar bears (Ursus maritimus) influenced by climate change. Glob. Chang. Biol. 2021;27:4481–4497. doi: 10.1111/gcb.15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gondek M., Bień J., Nowakowski Z. Use of ELISA and Western blot for serological detection of antibodies to E-S antigens of Trichinella spiralis muscle larvae in sera of swine experimentally infected with Trichinella spiralis. Vet. Immunol. Immunopathol. 2018;203:13–20. doi: 10.1016/j.vetimm.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Grzelak S., Stachyra A., Stefaniak J., Mrówka K., Moskwa B., Bień-Kalinowska J. Immunoproteomic analysis of Trichinella spiralis and Trichinella britovi excretory-secretory muscle larvae proteins recognized by sera from humans infected with Trichinella. PLoS ONE. 2020;15:e0241918. doi: 10.1371/journal.pone.0241918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z.Q., Liu R.D., Sun G.G., Song Y.Y., Jiang P., Zhang X., Cui J. Proteomic Analysis of Trichinella spiralis Adult Worm Excretory-Secretory Proteins Recognized by Sera of Patients with Early Trichinellosis. Front. Microbiol. 2017;8:986. doi: 10.3389/fmicb.2017.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bień J., Cabaj W., Moskwa B. Recognition of antigens of three different stages of the Trichinella spiralis by antibodies from pigs infected with T. spiralis. Exp. Parasitol. 2013;134:129–137. doi: 10.1016/j.exppara.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Cui J., Hu D.D., Liu R.D., Wang Z.Q. Identification of early diagnostic antigens from major excretory-secretory proteins of Trichinella spiralis muscle larvae using immunoproteomics. Parasite Vectors. 2014;7:40. doi: 10.1186/1756-3305-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui J., Liu R.D., Wang L., Zhang X., Jiang P., Liu M.Y., Wang Z.Q. Proteomic analysis of surface proteins of Trichinella spiralis muscle larvae by two-dimensional gel electrophoresis and mass spectrometry. Parasite Vectors. 2013;6:355. doi: 10.1186/1756-3305-6-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thawornkuno C., Nogrado K., Adisakwattana P., Thiangtrongjit T., Reamtong O. Identification and profiling of Trichinella spiralis circulating antigens and proteins in sera of mice with trichinellosis. PLoS ONE. 2022;17:e0265013. doi: 10.1371/journal.pone.0265013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Troiano G., Nante N. Human Trichinellosis in Italy: An epidemiological review since 1989. J. Prev. Med. Hyg. 2019;60:E71–E75. doi: 10.15167/2421-4248/jpmh2019.60.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krivokapich S.J., Gatti G.M., Gonzalez Prous C.L., Degese M.F., Arbusti P.A., Ayesa G.E., Bello G.V., Salomón M.C. Detection of Trichinella britovi in pork sausage suspected to be implicated in a human outbreak in Mendoza, Argentina. Parasitol. Int. 2019;71:53–55. doi: 10.1016/j.parint.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Dimzas D., Diakou A., Koutras C., Gómez Morales M.A., Psalla D., Keryttopoulos P., Deligianni D., Kontotasios K., Pozio E. Human trichinellosis caused by Trichinella britovi in Greece, and literature review. J. Helminthol. 2019;94:e33. doi: 10.1017/S0022149X19000075. [DOI] [PubMed] [Google Scholar]
- 57.Stroffolini G., Rossi L., Lupia T., Faraoni S., Paltrinieri G., Lipani F., Calcagno A., Bonora S., Di Perri G., Calleri G. Trichinella britovi outbreak in Piedmont, North-West Italy, 2019-2020: Clinical and epidemiological insights in the one health perspective. Travel Med. Infect. Dis. 2022;47:102308. doi: 10.1016/j.tmaid.2022.102308. [DOI] [PubMed] [Google Scholar]
- 58.Liao C., Liu M., Bai X., Liu P., Wang X., Li T., Tang B., Gao H., Sun Q., Liu X., et al. Characterisation of a plancitoxin-1-like DNase II gene in Trichinella spiralis. PLoS Negl. Trop. Dis. 2014;8:e3097. doi: 10.1371/journal.pntd.0003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi X., Yue X., Han Y., Jiang P., Yang F., Lei J.J., Liu R.D., Zhang X., Wang Z.Q., Cui J. Characterization of Two Trichinella spiralis Adult-Specific DNase II and Their Capacity to Induce Protective Immunity. Front. Microbiol. 2018;9:2504. doi: 10.3389/fmicb.2018.02504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang N., Stamenović D. Contribution of intermediate filaments to cell stiffness, stiffening, and growth. Am. J. Physiol. Cell Physiol. 2000;279:188–194. doi: 10.1152/ajpcell.2000.279.1.C188. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z., Yang J., Wei J., Yang Y., Chen X., Zhao X., Gu Y., Cui S., Zhu X. Trichinella spiralis paramyosin binds to C8 and C9 and protects the tissue-dwelling nematode from being attacked by host complement. PLoS Negl. Trop. Dis. 2011;5:e1225. doi: 10.1371/journal.pntd.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gretes M.C., Poole L.B., Karplus P.A. Peroxiredoxins in parasites. Antioxid. Redox Signal. 2012;17:608–633. doi: 10.1089/ars.2011.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang B., Liu M., Wang L., Yu S., Shi H., Boireau P., Cozma V., Wu X., Liu X. Characterisation of a high-frequency gene encoding a strongly antigenic cystatin-like protein from Trichinella spiralis at its early invasion stage. Parasite Vectors. 2015;8:78. doi: 10.1186/s13071-015-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stachyra A., Zawistowska-Deniziak A., Basałaj K., Grzelak S., Gondek M., Bień –Kalinowska J. The Immunological Properties of Recombinant Multi-Cystatin-Like Domain Protein from Trichinella Britovi Produced in Yeast. Front. Immunol. 2019;10:2420. doi: 10.3389/fimmu.2019.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wakelinn D. Immunity to Parasites: How Animals Control Parasitic Infections, London, UK. 1st ed. Edoward Arnold; Baltimore, MD, USA: 1984. [Google Scholar]
- 66.Gómez-Morales M.A., Ludovisi A., Amati M., Cherchi S., Tonanzi D., Pozio E. Differentiation of Trichinella species (Trichinella spiralis/Trichinella britovi versus Trichinella pseudospiralis) using western blot. Parasite Vectors. 2018;11:631. doi: 10.1186/s13071-018-3244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pozio E., Varese P., Morales M.A., Croppo G.P., Pelliccia D., Bruschi F. Comparison of human trichinellosis caused by Trichinella spiralis and by Trichinella britovi. Am. J. Trop. Med. Hyg. 1993;48:568–575. doi: 10.4269/ajtmh.1993.48.568. [DOI] [PubMed] [Google Scholar]
- 68.European Commission Regulation (EC) No. 2075/2005 of the European Parliament and of the Council of 5 December 2005 laying down specific rules on official controls for Trichinella in meat. Off. J. Eur. Union. 2005;L338:60–82. [Google Scholar]
- 69.Zarlenga D.S., Chute M.B., Martin A., Kapel C.M. A multiplex PCR for unequivocal differentiation of all encapsulated and non-encapsulated genotypes of Trichinella. Int. J. Parasitol. 1999;29:1859–1867. doi: 10.1016/S0020-7519(99)00107-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.