Abstract
Background
Hospital-onset (HO) methicillin-resistant Staphylococcus aureus (MRSA) infections have declined over the past decade due to infection control strategies; community-onset (CO) and healthcare-associated community-onset (HACO) MRSA, particularly USA300, has declined less. We examined the role of community strains to explain the difference.
Methods
We performed whole-genome sequencing (WGS) on MRSA clinical isolates from Cook County Health patients during 2011–2014. We defined infections as CO, HO, or HACO epidemiologically. We integrated genomic, community exposure, and statewide hospital discharge data to infer MRSA origin.
Results
Among 1020 individuals with available WGS, most were USA300 wound infections (580 CO, 143 HO, 297 HACO). USA300 HO, CO, and HACO infections were intermixed on the USA300 phylogeny, consistent with common strains circulating across community and healthcare settings. Community exposures (eg, substance abuse, incarceration, homelessness) were associated with HACO and HO infections, and genetically linked individuals from both groups had little overlap in healthcare facilities, supporting community origins. Most repeat infections—over months to years—occurred in individuals persistently carrying their own strains. These individuals were more likely to have genetic linkages, suggesting a role of persistent colonization in transmission.
Conclusions
Efforts to reduce presumed nosocomial USA300 spread may require understanding and controlling community sources and transmission networks, particularly for repeat infections.
Keywords: genomic epidemiology, MRSA, healthcare-associated community-onset, community transmission, repeat infection
We used genomic epidemiology and statewide data to infer origins of methicillin-resistant Staphylococcus aureus(MRSA). Among individuals with hospital-onset and healthcare-associated community-onset MRSA, we identified community exposures associated with infection, but little evidence of shared exposure in healthcare settings.
Methicillin-resistant Staphylococcus aureus (MRSA) was once limited to infections among hospitalized patients with known risk factors [1]. In the United States, healthcare-associated infections were caused predominantly by USA100 MRSA, as defined by pulsed-field gel electrophoresis, and occasionally USA200, USA500, USA600, and USA800 [2]. However, around 1999, a new lineage of MRSA, USA300, emerged as a cause of infections among otherwise healthy individuals with no healthcare exposures, mostly in congregate settings, including jails and prisons [3, 4], sports teams [5, 6], military barracks [7], and in children [8, 9]. Since emerging, USA300 is the most common cause of skin and soft tissue infections presenting to the emergency department (ED) [10] but has also infiltrated the hospital as a common cause of hospital-onset bacteremia [11].
To predict where MRSA was acquired—where intervention is needed—MRSA is defined as community-onset (CO) if infection occurs within 72 hours of hospitalization and hospital-onset (HO) if infection occurs later. However, as USA300 is now a common cause of community and healthcare-associated infections, there is a “graying” of what defines hospital or community MRSA [12]. Adding nuance to epidemiological definitions to capture the potential role of healthcare exposures in CO-MRSA, the Centers for Disease Control and Prevention Emerging Infections Program coined the term healthcare-associated community-onset (HACO)—that is, a community-onset infection but healthcare exposures in the prior year or previous MRSA [13]. However, genomic and other epidemiological studies suggest that definitions based on timing of infection might inaccurately categorize where MRSA was acquired because of prolonged asymptomatic colonization before infection [14–17].
A further complexity is that epidemiological definitions may be strain dependent. For example, from 2005 to 2013, USA100 MRSA significantly decreased for all 3 onset types—HO, CO, and HACO—while among USA300, only HO-MRSA cases declined [18, 19]. It was hypothesized that decline of MRSA cases was due to increased infection prevention for catheter-related infections and antibiotic-resistant pathogens [19]. While hospital-centric interventions had widespread effects on reducing USA100 incidence rate, USA300 declined only in hospitals, and to a lesser degree than USA100 HO incidence rates. This suggests that the remaining burden of USA300 across all onset types may be driven by community transmission, and thus interventions in the healthcare setting have less of an effect on USA300 MRSA cases [18]. To make further strides in reducing MRSA rates, more detailed understanding is required of where MRSA transmission is actually occurring.
Here, we sought to understand the interplay of community and healthcare exposures in USA300 MRSA spread. We analyzed a comprehensive collection of MRSA clinical isolates among patients presenting to Cook County Health (CCH) during 2011–2014. Using genomic epidemiology approaches and a detailed database of hospital discharge data throughout Illinois, we directly evaluated the role of healthcare and community exposures in MRSA spread.
MATERIALS AND METHODS
Study Design
We examined existing clinical MRSA isolates from the period 2011–2014 from patients at CCH, the major public healthcare network in Chicago, Illinois. Comprehensive sample collection was conducted during these years. We performed electronic and manual chart review for community (eg, homelessness, illicit drug use, incarceration) and healthcare exposures, demographic information, and comorbidities. Outpatient (ED visits and ambulatory surgery) and inpatient visits from discharge data in Illinois from 2013 to 2017 were queried by the Illinois Department of Public Health (IDPH).
Isolates were defined as HO if the collection date was 72 hours after hospitalization; HACO if onset was within 72 hours of hospitalization and the individual had prior healthcare exposure (hospitalization, surgery, dialysis, long-term care) in the past year or MRSA infection or nares colonization in the prior 6 months; and CO if none of these exposures.
Repeat infections were defined as a repeat culture >30 days after the first culture; otherwise, repeat cultures were excluded. We defined pediatric as <13 years to include the teenage population in our epidemiologic analysis.
Statistical Tests
We compared epidemiological factors for HACO-infected individuals to HO- and CO- infected individuals using 2 independent Fisher exact tests (epitools package in R). We report odds ratio (OR) and 95% Wald confidence interval (epitools). Null data were removed; denominators are specified. Infection type, ethnicity, and race were binarized. We compared time in facilities with a Wilcoxon rank-sum test using the base R function wilcox.test.
Whole-Genome Sequencing
Genomic DNA extracted from MRSA isolates was prepared for sequencing using a Nextera DNA Flex library preparation kit (Illumina, San Diego, California) or NEBNext Ultra II FS DNA Library Prep Kit (Illumina) Library Preparation kit according to manufacturer instructions. Sequencing was performed on an Illumina MiSeq or Illumina NovaSeq instrument using a high-output kit with paired-end 2 × 250 or 2 × 150 base reads, respectively. Library preparation and sequencing were performed at the Microbiome Core and Advanced Sequencing Core, respectively, at the University of Michigan. Variant calling pipeline can be found on Github (https://github.com/Snitkin-Lab-Umich/variant_calling_pipeline). We conducted multilocus sequence typing using ARIBA [20] and further classified CC8 isolates using in silico sequencing probes provided by Bowers et al [21]. The size of the USA300 core genome was 2.58 Mb. Raw sequence data were deposited under Bioproject PRJNA734638. Details on sequences are shown in Supplementary Table 1.
Identifying Putative Transmission Links
We classified individuals plausibly involved in recent transmission based on a single-nucleotide variant (SNV) threshold of ≤20 to prioritize minimizing false positives [22, 23].
Phylogenetic Analysis
After generating the whole-genome alignment, we masked sites identified as recombinant by Gubbins [24] and used this masked whole-genome alignment to build a maximum likelihood phylogeny with IQ-TREE [25]. Non-USA300 tips were dropped using the drop.tip function in ape [26]. We overlayed metadata on the phylogenetic tree using ggtree, gheatmap, and ggnewscale [27].
Defining Healthcare Exposure in the IDPH Data
The IDPH data contained monthly level inpatient and outpatient exposures. For visualization, we added 1 and log10-transformed the data (eg, log10 value of 0 indicates no healthcare exposures). With monthly level resolution, an exposure occurring the month of MRSA could have happened before or after the MRSA diagnosis or might represent the visit where MRSA was diagnosed.
The study was approved with waiver of consent by the CCH Institutional Review Board.
RESULTS
Study Population and Clinical Cultures
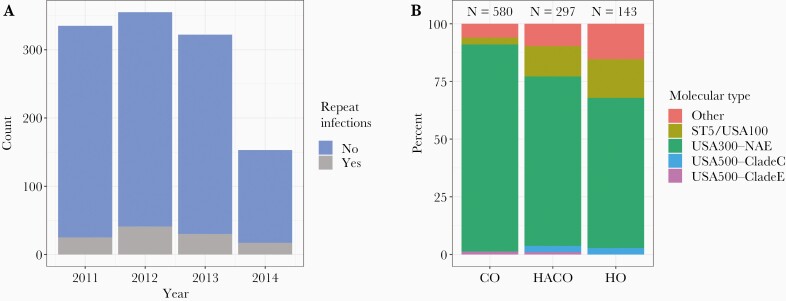
Archived clinical isolates comprising a comprehensive sampling of MRSA infections presenting to CCH were collected from 2011 to 2014 (N = 1203 total samples). The number of clinical MRSA isolates was stable over 2011 to 2013, but declined in 2014 (Figure 1A). Age ranged from 0 to >89 years. Including the pediatric population, there were 113 repeat infections from 49 individuals; 38 samples from 37 individuals were repeated cultures collected <30 days apart. Thus, there was a total of 1165 clinical isolates from 1101 individuals over the 4 years. Limiting to samples for which genomes pass quality control and restricting to the first isolate from each patient resulted in a final data set including genomes from 1020 clinical isolates. Overall, wound infections were the most common (81.2%), followed by blood (7.7%) and respiratory (6.2%).
Figure 1.
Summary of data. A, Number of clinical cultures over time. Repeated cultures from the same episode of infection (cultures <30 days apart) are not shown. Repeat infections (infections that occur >30 days apart) are colored in gray. Whole-genome sequences were available for all but 1 culture in 2011, 2013, and 2014 and all but 91 in 2012. B, Per onset type, percentage of clinical cultures of each molecular type. Abbreviations: CO, community-onset; HACO, healthcare-associated community-onset; HO, hospital-onset; NAE, North American epidemic.
Epidemiological Factors Associated With Onset Type
We assessed epidemiological factors among those with HO-MRSA and CO-MRSA compared to those with HACO-MRSA. Compared to HO infections, HACO infections were more likely to have wound cultures (OR, 4.68; P < .001; Table 1). Individuals with HACO- and HO-defined infections had exposures to the healthcare system, but individuals with HACO infections had fewer intensive care unit encounters before MRSA infection (OR, 0.13; P < .001; Table 1) compared to those with HO infections. Individuals with HACO infections had more inpatient, outpatient, and ED visits before MRSA infection compared to those with HO infection (Table 1). Individuals with HACO and HO infections had similar levels of drug use, history of incarceration, and homelessness (Table 1).
Table 1.
Comparison of Epidemiological Factors Among USA300 Healthcare-Associated Community-Onset, Community-Onset, and Hospital-Onset Infections
| Characteristic | Individuals With a Particular Epidemiological Factor, No. (%) | HACO vs HO | HACO vs CO | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HO (n = 94) | HACO (n = 221) | CO (n = 523) | OR | (95% CI) | P Value | OR | (95% CI) | P Value | |
| Demographics | |||||||||
| Wound (vs other) | 50 (53) | 186 (84) | 476 (91) | 4.68 | (2.72–8.05) | <.001 | 0.52 | (.33–.84) | .01 |
| Black/African American race (vs other) | 57 (61) | 143 (65) | 305 (58) | 1.19 | (.72–1.96) | .52 | 1.31 | (.95–1.82) | .12 |
| Hispanic ethnicity | 16a (18) | 41 (19) | 129a (25) | 1.07 | (.56–2.02) | 1 | 0.69 | (.47–1.03) | .07 |
| Pediatric individual (< 13 years old) | 4 (4.3) | 10 (4.5) | 32 (6.1) | 1.07 | (.33–3.49) | 1 | 0.73 | (.35–1.51) | .49 |
| Community factors | |||||||||
| Currently homeless | 10a (11) | 30a (14) | 26a (5) | 1.29 | (.6–2.77) | .58 | 2.97 | (1.71–5.15) | <.001 |
| History of illicit drug use | 53a (58) | 133a (61) | 215a (44) | 1.11 | (.67–1.82) | .7 | 1.95 | (1.41–2.7) | <.001 |
| Illicit drug use in past 3 mo | 21a (23) | 59a (27) | 63a (13) | 1.21 | (.68–2.15) | .57 | 2.47 | (1.66–3.68) | <.001 |
| History of injection drug use | 14a (15) | 29a (13) | 36a (7.4) | 0.84 | (.42–1.67) | .59 | 1.91 | (1.14–3.2) | .02 |
| Current injection drug user | 8a (8.9) | 19a (8.7) | 25a (5.2) | 0.97 | (.41–2.31) | 1 | 1.74 | (.94–3.24) | .09 |
| Incarceration in past year | 18 (19) | 55 (25) | 0 | 1.4 | (.77–2.54) | .31 | … | … | <.001 |
| History of incarceration | 29 (31) | 70 (32) | 58 (11) | 1.04 | (.62–1.75) | 1 | 3.72 | (2.51–5.51) | <.001 |
| Healthcare exposures and comorbidities | |||||||||
| Hospitalization in prior year | 46 (49) | 171 (77) | 0 | 3.57 | (2.14–5.96) | <.001 | … | … | <.001 |
| Surgery in prior year | 53 (56) | 107 (48) | 0 | 0.73 | (.45–1.18) | .22 | … | … | <.001 |
| Outpatient in prior year | 41 (44) | 162 (73) | 198 (38) | 3.55 | (2.14–5.88) | <.001 | 4.51 | (3.19–6.37) | <.001 |
| ED visit in prior year | 34 (36) | 127 (57) | 188 (36) | 2.38 | (1.45–3.92) | <.001 | 2.41 | (1.75–3.32) | <.001 |
| ICU 2 wk prior to MRSA | 17 (18) | 6 (2.7) | 2 (0.38) | 0.13 | (.05–.33) | <.001 | 7.27 | (1.46–36.3) | .01 |
| ICU 2 wk after MRSA | 6 (6.4) | 7 (3.2) | 0 | 0.48 | (.16–1.47) | .22 | … | … | <.001 |
| Diabetes | 34 (36) | 82 (37) | 96 (18) | 1.04 | (.63–1.72) | .9 | 2.62 | (1.85–3.73) | <.001 |
| HIV | 4 (4.3) | 31 (14) | 34 (6.5) | 3.67 | (1.26–10.71) | .01 | 2.35 | (1.4–3.93) | .002 |
ORs and 95% Wald CIs and P values from Fisher exact test calculated using epitools package in R.
Abbreviations: CI, confidence interval; CO, community-onset; ED, emergency department; HACO, healthcare-associated community-onset; HIV, human immunodeficiency virus; HO, hospital-onset; ICU, intensive care unit; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio.
Denominator differs from total number presented in the column header due to missing epidemiological data.
By definition, individuals with HACO infections were more likely to have healthcare exposures compared to CO infections. Surprisingly, individuals with HACO infections were more likely to have community exposures such as illicit drug use (OR, 2.47; P < .001), history of incarceration (OR, 3.72; P < .001), and homelessness (OR, 2.97; P < .001) compared to those with CO infections. No individuals with CO infections had been incarcerated in the past year of MRSA infection and only 11% had been incarcerated ever compared to 31% and 32% in individuals with HO- and HACO-infected individuals.
Genomic Epidemiology Across Onset Type
Extracting molecular type of genomic sequences revealed that most isolates were USA300 (n = 832) followed by those closely related to USA100/ST5 (n = 80), USA500 including clades C and E/early branching USA300 (n = 22), and other (n = 86). Onset-type classifications were 580 CO, 297 HACO, and 143 HO isolates. USA300 was the most common clinical culture across all onset types. However, a higher percentage of isolates per onset type were USA100 in HO and HACO than in CO, consistent with USA100 as a healthcare-associated pathogen (Figure 1B).
Evidence of Potential Transmission Among Infections
We next identified pairs of isolates plausibly involved in direct or indirect transmission by applying a SNV threshold of 20 (Supplementary Figure 1) [22, 23]. This yielded a total of 329 pairs of USA300 isolates, making up clusters of size 2–23. Of these 329 pairs, 35 involved a pediatric patient (≤13 years of age), with all but 1 pair involving a pediatric patient with an adult patient. Of note, 22 of the 35 pairs involved a pediatric patient under the age of 1, and only 2 of the 35 pairs shared a zip code.
For the remainder of the analyses, we removed individuals <13 years of age because of differing epidemiology among adults and children. Removing pediatric patients left 294 pairs of USA300 isolates making up clusters of size 2–21 (Supplementary Figure 1). Of note, despite the lack of sampling of asymptomatically colonized individuals, we still identified many individuals potentially related by recent direct or indirect transmission using only clinical cultures. The proportion of CO, HO, and HACO isolates genetically linked to another isolate is similar to the proportion not genetically linked, with HACOs being slightly more frequent in transmission clusters (P = .05) (Supplementary Figure 1C).
Intermixing of Onset Type Among Genetically Linked Pairs
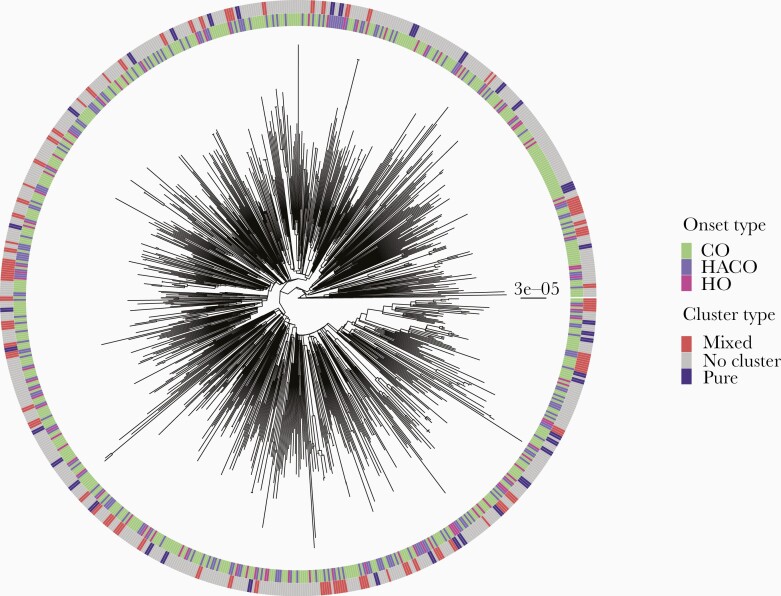
Within USA300, the dominant molecular type, we assessed intermixing among CO, HO, and HACO isolates on the USA300 phylogeny, or if different sublineages of USA300 preferentially spread in community or hospital settings. Previous work by our group focusing on bloodstream infections from 2009 to 2013 indicated no separate sublineages for CO or HO infections [16]. Expanding our analysis here to a more diverse and comprehensive sampling of clinical isolates supported this previous finding, with onset types showing no evidence of clustering on the USA300 whole-genome phylogeny (Figure 2). Moreover, even when focusing on clusters of isolates that were most closely related, and thereby most recently linked by direct or indirect transmission, most were of mixed onset type (Supplementary Figure 2). This suggests that transmission networks of USA300 MRSA in the healthcare and the community setting are overlapping.
Figure 2.
USA300 phylogeny with onset type and clustering. Maximum likelihood tree of USA300 isolates. One sample per individual shown. Scale bar indicates substitutions per site. Inner ring indicates onset type. Outer ring indicates if the isolate is in a cluster or not defined by a 20 single-nucleotide variant threshold, and if the cluster is of mixed or pure (eg, all community-onset methicillin-resistant Staphylococcus aureus) onset type. Abbreviations: CO, community-onset; HACO, healthcare-associated community-onset; HO, hospital-onset.
Epidemiological Factors Associated With Harboring MRSA Genetically Linked to Another Isolate
To identify putative community and healthcare exposures underlying detected genetic linkages, we tested if there were epidemiological factors associated with being genetically linked to another isolate. We found that MRSA from Black/African American individuals was negatively associated with being linked to another isolate whereas Hispanic ethnicity was positively associated (Table 2). Community exposures that were more likely among those with genetic linkages included history of incarceration (OR, 1.61; P = .02), illicit drug use (OR, 1.7; P = .009), and homelessness (OR, 1.8; P = .04) (Table 2). Healthcare exposures that were significantly more likely among individuals with genetic linkages included hospitalization, inpatient, and ED admissions in the past year (Table 2).
Table 2.
Epidemiological Factors Associated With Genetic Linkages of USA300 Methicillin-Resistant Staphylococcus aureus Among Adults
| Characteristic | Percentage of Individuals With a Particular Epidemiologic Factor Among Those in a Cluster (n = 210) |
Percentage of Individuals With a Particular Epidemiologic Factor Among Those Not in a Cluster (n = 576) |
OR | (95% CI) | P Value |
|---|---|---|---|---|---|
| Demographics | |||||
| Wound (vs other) | 178 (85) | 491 (85) | 0.96 | (.62–1.5) | .91 |
| Black/ African American race (vs other) | 106 (50) | 361 (63) | 0.61 | (.44–.84) | .002 |
| Hispanic ethnicity | 60a (29) | 115a (20) | 1.62 | (1.13–2.33) | .01 |
| Community factors | |||||
| Currently homeless | 25a (12) | 40a (7.1) | 1.8 | (1.06–3.05) | .04 |
| History of illicit drug use | 122a (61) | 275a (50) | 1.54 | (1.1–2.14) | .01 |
| Illicit drug use in past 3 mo | 51a (26) | 91a (17) | 1.7 | (1.15–2.51) | .009 |
| History of injection drug use | 31a (16) | 48a (8.8) | 1.9 | (1.17–3.08) | .01 |
| Current injection drug user | 22a (11) | 30a (5.5) | 2.13 | (1.19–3.78) | .01 |
| Incarceration in past year | 27 (13) | 46 (8) | 1.7 | (1.03–2.81) | .05 |
| History of incarceration | 54 (26) | 102 (18) | 1.61 | (1.1–2.34) | .02 |
| Healthcare exposures and comorbidities | |||||
| Hospitalization in prior year | 70 (33) | 135 (23) | 1.63 | (1.16–2.31) | .006 |
| Surgery in prior year | 45 (21) | 105 (18) | 1.22 | (.83–1.81) | .31 |
| Dialysis in prior year | 2 (0.95) | 2 (0.35) | 2.76 | (.39–19.72) | .29 |
| Outpatient in prior year | 109 (52) | 269 (47) | 1.23 | (.9–1.69) | .2 |
| ED visit in prior year | 103 (49) | 227 (39) | 1.48 | (1.08–2.03) | .02 |
| ICU 2 wk prior to MRSA | 8 (3.8) | 15 (2.6) | 1.48 | (.62–3.55) | .35 |
| ICU 2 wk after MRSA | 3 (1.4) | 10 (1.7) | 0.82 | (.22–3.01) | 1 |
| Diabetes | 50 (24) | 160 (28) | 0.81 | (.56–1.17) | .28 |
| HIV | 20 (9.5) | 46 (8) | 1.21 | (.7–2.1) | .47 |
Data are presented as No. (%) unless otherwise indicated. ORs and 95% Wald CIs and P values from Fisher exact test calculated using epitools package in R.
Abbreviations: CI, confidence interval; ED, emergency department; HIV, human immunodeficiency virus; ICU, intensive care unit; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio.
Denominator differs from total number presented in the column header due to missing epidemiological data.
Overlap in Healthcare Setting Does Not Explain USA300 MRSA Transmission
We next sought to identify evidence for specific locations where transmission events underlying the observed genomic linkages could be occurring. We focused on common healthcare exposures as we had access to high-resolution healthcare exposure data from IDPH with 90 inpatient and 96 outpatient (EDs and licensed ambulatory surgical centers) facilities across the state of Illinois from 2013 to 2017. The number of genetically linked USA300 pairs where both individuals were diagnosed with MRSA in 2013 or 2014 was 68 of 294 total pairs. We therefore subsetted our analysis to these genetically linked pairs to reflect the data available from IDPH.
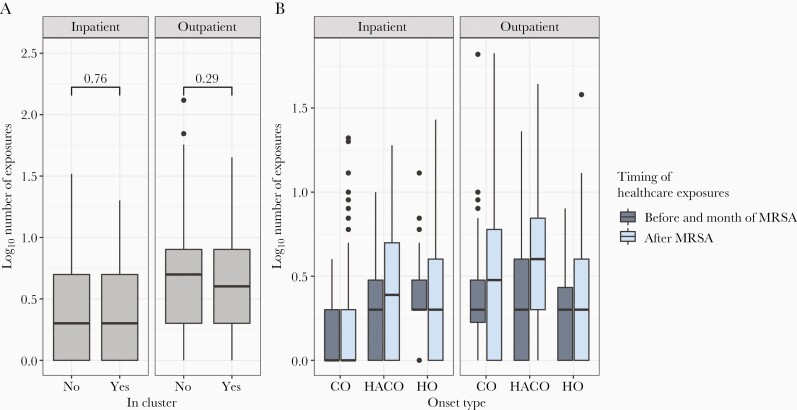
Individuals with isolates related to another isolate by 20 SNVs did not have more inpatient or outpatient exposures than those lacking genomic linkages (Figure 3A). We next sought to assess if there was indirect or direct overlap in the healthcare setting among genetically linked pairs of individuals. Despite high-resolution hospital exposure data, only 5 of 68 (7.3%) pairs overlapped in an inpatient facility in the same month and only 8 of 68 (12.7%) pairs overlapped in an ED or ambulatory surgical center in the same month. All but 1 of these overlaps occurred in the facility of MRSA diagnosis, and though infrequent, occurred more than random chance (Supplementary Figure 3). Indirect overlap was defined by attending the same facility before the date of the latest MRSA diagnosis in the pair. Seventeen of 68 pairs and 12 of 68 pairs indirectly overlapped in outpatient or inpatient facilities, respectively. Again, all but 2 indirect overlaps occurred at the facility of MRSA diagnosis and occurred more than random chance (Supplementary Figure 3), despite accounting for a small proportion of overall genetic linkages.
Figure 3.
Exposure to the healthcare system before and after methicillin-resistant Staphylococcus aureus (MRSA) diagnosis. A, Includes only individuals who were diagnosed with MRSA infection in 2013 or 2014 (n = 344). Individuals in transmission clusters (n = 86) do not have more total inpatient or outpatient healthcare exposures than those not in a transmission cluster (n = 258) as indicated by Wilcoxon rank-sum test. Numbers above brackets indicate P values from Wilcoxon-rank sum test. B, Inpatient and outpatient exposures before and after MRSA by onset type (n = 208 community-onset, 94 healthcare-associated community-onset, 42 hospital-onset) reveal that individuals often have exposure to the healthcare system before and after MRSA diagnosis and that some individuals with so-called community-onset MRSA have inpatient exposures outside of Cook County Health before MRSA diagnosis. Abbreviations: CO, community-onset; HACO, healthcare-associated community-onset; HO, hospital-onset; MRSA, methicillin-resistant Staphylococcus aureus.
Individuals Use the Healthcare System Before and After MRSA
Though we found no evidence of extensive transmission in the healthcare setting, individuals do have numerous exposures to the healthcare setting both before and after MRSA diagnosis across all onset types (Figure 3B). While the median healthcare exposures among CO-MRSA before diagnosis was 0, there were 19 of 208 individuals with CO infections who had at least 1 inpatient exposure before MRSA diagnosis, indicating that when considering the more comprehensive healthcare exposures in the hospital discharge dataset, some of these CO-MRSA isolates are actually HACO-MRSA. This number may be higher because we could not differentiate ambulatory surgery from ED visits in the outpatient IDPH data. Of note, individuals within high-risk community social networks including those with a history of illicit drug use, incarceration, and homelessness often had exposure to the healthcare setting, including the ED, before developing infection (Table 1).
Individuals With Repeat Infections Tend to Be Involved in More Transmission
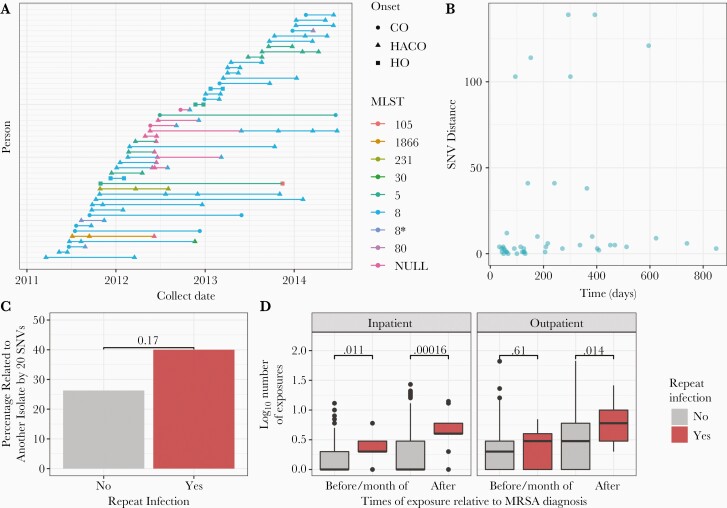
There were many repeat infections among individuals and we sought to infer if these infections were due to the same strain or acquisition of new strains. Excluding pediatric cases, there were 108 repeat infections among 47 individuals. Individuals had 2–5 repeat infections during the study period.
A total of 40 of the 47 individuals with repeat infections have multiple genomes available. Of those 40 individuals, 34 (85%) have an infection of the same multilocus sequence type over time, with 29 having repeat USA300 infections over time (Figure 4A). Most individuals with repeat USA300 infections have an infection of the same strain; only 5 of 29 individuals had at least 1 USA300 repeat infection that was greater than 20 SNVs from a previous infection, indicating infection from differing strains and 25 of 29 had at least 1 USA300 repeat infection of the same strain (within 20 SNVs) (Figure 4B). Individuals had a repeat infection with the same strain of USA300 MRSA (within 20 SNVs) up to 849 days apart (Figure 4B). Individuals with repeat infections have 1.9 times higher odds of being related to another isolate by 20 SNVs than those without repeat infections (P = .17) (Figure 4C), although the association did not attain significance. Furthermore, individuals with repeat infections tend to have more inpatient and outpatient exposures before and after MRSA (Figure 4D), suggesting that these individuals are frequent users of the healthcare system.
Figure 4.
Analysis of repeat infections. A, Individuals with repeat infections, colored by multilocus sequence type. Shape indicates onset type. B, Single-nucleotide variant (SNV) distance between USA300 repeat infections over time. C, Percentage of individuals with repeat USA300 infections that are related to another isolate by 20 SNVs (n = 25) compared to those without repeat infection of the same strain (n = 761). P value from χ2 test indicates significance. D, Number of healthcare exposures before and month of methicillin-resistant Staphylococcus aureus (MRSA) diagnosis and after MRSA diagnosis, colored by repeat adult USA300 infection compared to those with no repeat USA300 infection. Data were subsetted to those infected in 2013 or 2014 (n = 13 repeat USA300 infections of the same strain, n = 331 with no repeat USA300 infection of the same strain). P values from Wilcoxon rank-sum tests indicate significantly higher number of exposures among those with repeat infections across all inpatient, outpatient, before and after MRSA. Abbreviations: CO, community-onset; HACO, healthcare-associated community-onset; HO, hospital-onset; MLST, multilocus sequence type; MRSA, methicillin-resistant Staphylococcus aureus; SNV, single-nucleotide variant.
DISCUSSION
Infection prevention efforts and antimicrobial stewardship in healthcare settings have led to significant MRSA infection rates reductions [19], but progress has mainly occurred in USA100 MRSA and less for USA300 MRSA [18]. We sought to understand where USA300 MRSA acquisition is occurring, and whether commonly used epidemiologic-onset definitions are useful in guiding infection prevention efforts. Our observation of a lack of healthcare overlap among individuals with genetically linked isolates, combined with the observed capacity for individuals to be colonized with the same strain for years, argue against location and timing of onset or recent healthcare exposure as sufficient to attribute the hospital as a source of infection among USA300 MRSA and call into question the utility of these epidemiological onset type definitions in predicting sites of MRSA acquisition.
Consistent with previous work, we showed that there are not separate lineages of USA300 MRSA circulating in the healthcare and community settings [16]. Moreover, we observed community exposures known to increase MRSA colonization and infection risk (ie, drug use, homelessness, and prior incarceration) among healthcare-associated infections and found an association of these factors with involvement in any MRSA transmission cluster [28, 29]. Furthermore, we failed to identify direct or indirect healthcare overlap among most putative transmission pairs involving HO- or HACO-MRSA. Taken together, these observations support that both USA300 HACO- and HO-MRSA infections often are due to colonization acquired in the community. Moreover, these findings help account for the larger impact of hospital-based interventions on USA100 infection rates compared to rates of USA300 infections [18].
While we observed little evidence of USA300 healthcare transmission, individuals with MRSA infection often have exposures to the inpatient and outpatient setting before and after MRSA. Importantly, these individuals with healthcare exposures also had community exposures previously found to be associated with MRSA risk including incarceration history, illicit drug use, and homelessness [28, 29]. Thus, enhanced infection control strategies could be implemented during healthcare visits or at hospital discharge to attempt to prevent subsequent MRSA infections in the community [30]. In addition to interventions targeting community spread, continuance of healthcare interventions to reduce transmission and guidelines to prevent procedure- or device-related infections are crucial as decline of healthcare-associated MRSA has slowed in recent years [31].
We also noted that individuals with repeat infections often were infected with a persistent strain and more likely to be genetically linked to a MRSA isolate from another individual, suggesting that persistent colonization and/or repeated MRSA infection is associated with MRSA transmission risk. Importantly, these individuals have more inpatient and outpatient exposures before and after diagnosis, indicating potential points of intervention, and as such, suggests potential value of an intervention such as decolonization at hospital discharge as examined by Huang et al [30].
Our study was limited in that we only collected clinical isolates. While the lack of surveillance isolates could hinder our capacity to detect healthcare transmission due to missing intermediates in the network, we were able to detect potential transmission links with genomics. A second limitation is that the healthcare exposure data from IDPH were collected at monthly and facility resolution and therefore we cannot discern the healthcare exposure more specifically in relation to the incident infection. However, exposures were largely at different institutions than CCH and thus, likely represent healthcare encounters distinct from when the culture was collected. Also, we a priori wanted to investigate healthcare exposure burden—both before and after an encounter as a means of future targeting of infection prevention interventions—and therefore included this variable. In addition, even if higher-resolution data were available, this would only decrease the number of putative overlapping exposures, as the criteria for overlap would be more stringent. We sampled data from 2011 to 2014 because there was a comprehensive collection of MRSA clinical isolates; that being said, there was an unexplained decline in MRSA cultures in 2014. We speculate that this is more likely to be a consequence of sampling and possibly not a significant decline in MRSA. In addition, there is a chance that factors influencing community transmission could have changed since 2014; however, more recent work has consistently noted these community factors as highly relevant for MRSA spread. Finally, varying definitions for HACO exist, some with [13, 32] and some without [19] the inclusion of prior MRSA colonization and infection in the definition. In our data, 4% (n = 12) of USA300 isolates classified as HACO were classified as such solely because of a prior MRSA infection. Regardless of how HACO is defined, our data suggest that neither a history of MRSA nor healthcare exposure equates to healthcare acquisition. However, a prior MRSA designation typically results in a healthcare encounter and hence an opportunity for potential intervention.
In conclusion, our results suggest that most USA300 infections are acquired outside of the healthcare setting. Further work is warranted to understand how community transmission is occurring and optimal intervention points. Our prior work has demonstrated how integration of genomics with epidemiological data can significantly enhance knowledge of pathogen spread [29]. We have extended prior work by now including statewide healthcare exposures, enhancing our ability to delineate transmission networks for MRSA in a large urban area. More studies using epidemiological data and whole-genome sequencing are crucial to understand community reservoirs of MRSA and routes of transmission and to move the needle on risk of USA300 infections.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We wish to thank Ali Pirani at the University of Michigan for bioinformatics support, Lisa Diep at Cook County Health for informatics support, and Michael Schoeny, PhD, at Rush University for his assistance with biostatistical analysis.
Financial support. This project was supported by the Centers for Disease Control and Prevention Broad Agency Announcement: Genomic Epidemiology of Community-Onset Invasive USA300 MRSA Infections (contract ID: 75D30118C02923). S. N. T. was supported by the National Institutes of Health Molecular Mechanisms in Microbial Pathogenesis training grant (T32 AI007528) and the University of Michigan and Rackham Predoctoral Fellowship.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Stephanie N Thiede, University of Michigan, Ann Arbor, Michigan, USA.
Evan S Snitkin, University of Michigan, Ann Arbor, Michigan, USA.
William Trick, Cook County Health, Rush University Medical Center, Chicago, Illinois, USA.
Darjai Payne, Rush University Medical Center, Chicago, Illinois, USA.
Alla Aroutcheva, Rush University Medical Center/Cook County Health, Chicago, Illinois, USA.
Robert A Weinstein, Rush University Medical Center/Cook County Health, Chicago, Illinois, USA.
Kyle J Popovich, Rush University Medical Center/Cook County Health, Chicago, Illinois, USA.
References
- 1. Fridkin SK, Hageman JC, Morrison M, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005; 352:1436–44. [DOI] [PubMed] [Google Scholar]
- 2. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC.. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 2003; 41:5113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus infections in correctional facilities—Georgia, California, and Texas, 2001-2003. MMWR Morb Mortal Wkly Rep 2003; 52:992–6. [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison—Mississippi, 2000. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5042a2.htm. Accessed 19 August 2018. [PubMed]
- 5. Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants—Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5233a4.htm. Accessed 19 August 2018.
- 6. Kazakova SV, Hageman JC, Matava M, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 2005; 352:468–75. [DOI] [PubMed] [Google Scholar]
- 7. Zinderman CE, Conner B, Malakooti MA, LaMar JE, Armstrong A, Bohnker BK.. Community-acquired methicillin-resistant Staphylococcus aureus among military recruits. Emerg Infect Dis 2004; 10:941–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herold BC. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 1998; 279:593–8. [DOI] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm4832a2.htm. Accessed 16 September 2017. [PubMed]
- 10. Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. [DOI] [PubMed] [Google Scholar]
- 11. Popovich KJ, Weinstein RA, Hota B.. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis 2008; 46:787–94. [DOI] [PubMed] [Google Scholar]
- 12. Popovich KJ, Weinstein RA.. The graying of methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 2009; 30:9–12. [DOI] [PubMed] [Google Scholar]
- 13. Klevens RM, Morrison MA, Fridkin SK, et al. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg Infect Dis 2006; 12:1991–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Golubchik T, Batty EM, Miller RR, et al. Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS One 2013; 8:e61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Eiff C, Becker K, Machka K, Stammer H, Peters G.. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study group. N Engl J Med 2001; 344:11–6. [DOI] [PubMed] [Google Scholar]
- 16. Popovich KJ, Snitkin ES, Hota B, et al. Genomic and epidemiological evidence for community origins of hospital-onset methicillin-resistant Staphylococcus aureus bloodstream infections. J Infect Dis 2017; 215:1640–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Long SW, Beres SB, Olsen RJ, Musser JM.. Absence of patient-to-patient intrahospital transmission of Staphylococcus aureus as determined by whole-genome sequencing. mBio 2014; 5:e01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. See I, Mu Y, Albrecht V, et al. Trends in incidence of methicillin-resistant Staphylococcus aureus bloodstream infections differ by strain type and healthcare exposure, United States, 2005–2013. Clin Infect Dis 2020; 70:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dantes R, Mu Y, Belflower R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013; 173:1970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunt M, Mather AE, Sánchez-Busó L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics 2017; 3:e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bowers JR, Driebe EM, Albrecht V, et al. Improved subtyping of Staphylococcus aureus clonal complex 8 strains based on whole-genome phylogenetic analysis. mSphere 2018; 3:e00464–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall MD, Holden MT, Srisomang P, et al. Improved characterisation of MRSA transmission using within-host bacterial sequence diversity. eLife 2019; 8:e46402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Järvenpää M, Sater MRA, Lagoudas GK, et al. A Bayesian model of acquisition and clearance of bacterial colonization incorporating within-host variation. PLoS Comput Biol 2019; 15:e1006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paradis E, Schliep K.. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019; 35:526–8. [DOI] [PubMed] [Google Scholar]
- 27. Yu G. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinforma 2020; 69:e96. [DOI] [PubMed] [Google Scholar]
- 28. Frazee BW, Lynn J, Charlebois ED, Lambert L, Lowery D, Perdreau-Remington F.. High prevalence of methicillin-resistant Staphylococcus aureus in emergency department skin and soft tissue infections. Ann Emerg Med 2005; 45:311–20. [DOI] [PubMed] [Google Scholar]
- 29. Popovich KJ, Snitkin E, Green SJ, et al. Genomic epidemiology of USA300 methicillin-resistant Staphylococcus aureus in an urban community. Clin Infect Dis 2016; 62:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang SS, Singh R, McKinnell JA, et al. Decolonization to reduce postdischarge infection risk among MRSA carriers. N Engl J Med 2019; 380:638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kourtis AP. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections—United States. MMWR Morb Mortal Wkly Rep 2019; 68:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2003; 290:2976–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.