Abstract
Recent genome-wide association studies have reported multiple schizophrenia risk loci, yet the functional variants and their roles in schizophrenia remain to be characterized. Here we identify a functional single nucleotide polymorphism (rs2270363: G>A) at the schizophrenia risk locus 16p13.3. rs2270363 lies in the E-box element of the promoter of NMRAL1 and disrupts binding of the basic helix–loop–helix leucine zipper family proteins, including USF1, MAX and MXI1.
We validated the regulatory effects of rs2270363 using reporter gene assays and electrophoretic mobility shift assay. Besides, expression quantitative trait loci analysis showed that the risk allele (A) of rs2270363 was significantly associated with elevated NMRAL1 expression in the human brain. Transcription factors knockdown and CRISPR-Cas9-mediated editing further confirmed the regulatory effects of the genomic region containing rs2270363 on NMRAL1. Intriguingly, NMRAL1 was significantly downregulated in the brain of schizophrenia patients compared with healthy subjects, and knockdown of Nmral1 expression affected proliferation and differentiation of mouse neural stem cells, as well as genes and pathways associated with brain development and synaptic transmission. Of note, Nmral1 knockdown resulted in significant decrease of dendritic spine density, revealing the potential pathophysiological mechanisms of NMRAL1 in schizophrenia. Finally, we independently confirmed the association between rs2270363 and schizophrenia in the Chinese population and found that the risk allele of rs2270363 was the same in European and Chinese populations.
These lines of evidence suggest that rs2270363 may confer schizophrenia risk by regulating NMRAL1, a gene whose expression dysregulation might be involved in the pathogenesis of schizophrenia by affecting neurodevelopment and synaptic plasticity.
Keywords: schizophrenia, genome-wide association studies, rs2270363, NMRAL1, dendritic spine density
Luo et al. show that the functional variant rs2270363 confers schizophrenia risk by modulating the expression of the schizophrenia risk gene NMRAL1. Experiments in neural stem cells of mice and neurons of rats show that knockdown of Nmral1 affects brain development and synaptic transmission, and reduces dendritic spine density.
Introduction
Schizophrenia affects around 0.5–1% of the general population worldwide and the clinical symptoms of most cases manifest in late adolescence or early adulthood.1,2 Epidemiological studies show a 2–3-fold increase in mortality rate of patients with schizophrenia,3 which imposes huge economic and mental burden on the patient’s family and society.4,5 The heritability of schizophrenia was estimated at about 81%,6 indicating that genetic factors play a dominant role in schizophrenia pathogenesis.6–8 Although multiple schizophrenia risk loci have been identified by genome-wide association studies (GWASs) in recent years,9–13 identifying potential pathogenic genetic variants in the reported risk loci and elucidating their roles in schizophrenia remain major challenges in the post-GWAS era.
Most of the disease-associated risk variants are located in non-coding regions,13 suggesting that they may be involved in the pathogenesis of schizophrenia by influencing transcriptional regulation. Functional genomics studies help to identify risk SNPs with regulatory effects or functional consequences in the reported risk loci.14,15 In our previous study, we identified 132 schizophrenia risk single nucleotide polymorphisms (SNPs) that disrupt binding of transcription factors.14 To prioritize and distil the most promising functional variants for further mechanistic dissection and functional characterization, we interrogated these 132 transcription factor binding-disrupting SNPs in detail with a series of analyses, including expression quantitative trait loci (eQTL) analysis, genomic location mapping, RegulomeDB annotation16 and differential gene expression analysis ('Material and methods' section, Supplementary Fig. 1). Our analyses suggested that rs2270363 [located in the E-box element of the NMRAL1 promoter and disrupted binding of basic helix–loop–helix leucine zipper (bHLHZ) proteins, including USF1, MAX and MXI1]14,17 may represent a potential causal variant at the schizophrenia risk locus 16p13.3. Elucidating the role and regulatory mechanism of rs2270363 will not only help to demonstrate genetic mechanisms of complex diseases, but also facilitate understanding of the pathophysiology of diseases, bringing opportunities for the discovery of new therapeutic targets and treatments.
To elucidate the role of rs2270363 in schizophrenia pathogenesis, in this study, we systematically characterized the regulatory mechanisms of rs2270363 with a series of analyses and experiments. We firstly confirmed the association between rs2270363 and schizophrenia in the Chinese population. We then validated the regulatory effect of rs2270363 with reporter gene assays and our electrophoretic mobility shift assay (EMSA) demonstrated that rs2270363 affects transcription factors (including USF1) binding. We further showed that the A allele (risk allele) of rs2270363 was significantly associated with elevated NMRAL1 expression in the human brain. Moreover, we found that rs2270363 and its binding transcription factors (i.e. USF1 and MAX) could regulate NMRAL1, a gene whose expression level was significantly downregulated in brains of schizophrenia cases compared with controls. To explore the potential role of NMRAL1 in schizophrenia pathogenesis, we investigated the role of NMRAL1 in neurodevelopment (proliferation and differentiation of neural stem cells). Finally, we examined the effect of NMRAL1 on density of dendritic spines. Our study demonstrated that the functional risk variant rs2270363 (located in the NMRAL1 promoter) might contribute to schizophrenia susceptibility by regulating expression of NMRAL1, a gene whose expression downregulation affected neurodevelopment and density of dendritic spines (Supplementary Fig. 2).
Materials and methods
Expression quantitative trait loci analysis
We explored whether the 132 identified transcription factor binding-disrupting SNPs were associated with gene expression in the human brain using eQTL data from the CommonMind Consortium (CMC) and Lieber Institute for Brain Development (LIBD).14,18,19 Briefly, brain tissues (the dorsolateral prefrontal cortex) of 467 subjects (CMC) and 412 subjects (LIBD) were used for eQTL analysis and detailed information can be found in the original publication.18,19
Mapping of SNP locations
We then inspected the genomic locations of these 97 SNPs, as previous studies have demonstrated the pivotal roles of functional variants in the promoter region (a core regulatory region for gene expression) in disease susceptibility. For example, a functional variant in the promoter of MDM2 attenuates the p53 tumour suppressor pathway and accelerates tumour formation,20 and a functional variant in the microRNA-146a promoter confers risk of systemic lupus erythematosus by modulating microRNA-146a expression.21 In addition, a functional variant in the CHRNA1 promoter has also been reported to confer disease risk by regulating CHRNA1 expression in thymus.22 SNPs are considered to be located in the promoter region in this study if they meet following two criteria: (i) located 2 kb upstream and 200 bp downstream of the transcription start site; and (ii) marked with H3K4me3 (according to data from UCSC genome browser). Promoter regions were determined as upstream 2 kb and 200 bp downstream of the transcription start site and marked with H3K4me3 (a marker for active promoters) in UCSC genome browser (http://genome.ucsc.edu/).
RegulomeDB annotation
We used RegulomeDB to further annotate and prioritize the most possible functional SNPs located in promoter regions.16 RegulomeDB is a well-curated database that annotates SNPs according to multiple high-throughput data sets, including eQTL, chromatin immunoprecipitation sequencing, matched transcription factor motif, matched DNase footprint and so on. RegulomeDB assigns a numerical rank score to each SNP and the rank score reflects the possibility of functionality. The RegulomeDB rank scores range from 1 to 7 (1a–f, 2a–c, 3a, 3b, 4, 5, 6, 7) and SNPs with lower RegulomeDB scores are more likely to be functional.16 Detailed information about the RegulomeDB scores and supporting data are provided in Supplementary Table 1.
Differential gene expression analysis in schizophrenia cases and controls
We performed expression analysis to explore if the eQTL genes of the five high-confidence functional SNPs (located in promoter regions and having a RegulomeDB score < 2) showed differential expression in schizophrenia cases compared with controls using the expression data from CMC (n = 258 cases and n = 279 controls).18
Schizophrenia cases and controls used for genetic analysis in this study
A total of 3718 schizophrenia cases and 7829 healthy controls were included in this study. Detailed information about the included subjects has been described in our previous studies.23,24 Briefly, schizophrenia cases were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria, with the use of Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I). The diagnoses were completed independently by at least two experienced psychiatrists. All available information, including age of onset, symptoms, medication and hospitalization history, medical records, interviews with the patient, communications with family members and all other related information were evaluated to reach a consensus diagnosis. Controls were recruited from local volunteers and screened for potential psychiatric disorders. Individuals with a family history of mental disorders, drug and alcohol abuse, head injuries and epilepsy were excluded in this study. The ages of cases and controls were 42 ± 15.75 and 36 ± 9.59 years, respectively; 49.1% of cases and 65.2% of controls were males, respectively. Informed consent was obtained from all participants and this study was approved by the internal review board of Kunming Institute of Zoology, Chinese Academy of Sciences.
Genotyping
Whole venous blood was collected with an anticoagulant vacuum tube. DNA was extracted using the phenol/chloroform method or QIAamp DNA Blood Mini Kit (Qiagen, Cat. No. 51104). Genotyping was performed using the SNaPshot method as previously described.23,25 Detailed information about genotyping assays is provided in the Supplementary material.
Genetic association analysis and meta-analysis
The association between rs2270363 and schizophrenia was assessed with PLINK software (version 1.07).26 We performed a meta-analysis (using PLINK) by combining our results with a previously study.27 In total, 60 136 cases and 86 647 controls were included in the meta-analysis (detailed information about the studies is provided in Supplementary material). A fixed-effect model was used for meta-analysis and a heterogeneity test (I2 statistic) was performed using the PLINK software.
Cell culture
HEK-293T, SH-SY5Y and SK-N-SH cell lines were purchased from the Kunming Cell Bank (Kunming institute of Zoology, CAS), and cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% foetal bovine serum (FBS), 1% penicillin/streptavidin (Gibco, Cat. No. 15140-122). HEK-293T cells are derived from kidney and are mainly used for virus packaging. SH-SY5Y and SK-N-SH cells are neuroblastoma cell lines that are mainly used in the research of brain diseases and cancer.28–31 The mouse neural stem cells (mNSCs) and rat primary cortical neurons were isolated and cultured as previously described.23,32–34 No mycoplasma contamination was detected in this study.
Dual-luciferase reporter gene assays
The 861-bp DNA fragments containing rs2270363 (A allele) were amplified with specific primers and were inserted into the pGL3-Enhancer vector or pGL4.11 vector (Supplementary Fig. 3) using the TreliefTM SoSoo Cloning Kit (TSINGKE, Cat. No. TSV-S1). Polymerase chain reaction (PCR)-mediated point mutation was used to obtain the vector containing the G allele of rs2270363. HEK-293T, SH-SY5Y and SK-N-SH cells were used in luciferase reporter gene assays. Luciferase reporter gene assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Cat. No. E1980). More detailed information is provided in the Supplementary material.
EMSA
Nuclear extracts of SH-SY5Y cells were extracted with the nuclear and cytoplasmic protein extraction kit (Beyotime, Cat. No. P0028) and quantified by bicinchoninic acid (BCA) protein assay kit (Pierce, Cat. No. 23227). Single-strand oligonucleotides (35 bp) containing the G and A alleles of rs2270363 were synthesized and the sequences are provided in the Supplementary Table 2. More detailed information about the EMSA is provided in the Supplementary material.
Knockout of the genomic region containing rs2270363
The sgRNA (designed using https://zlab.bio/guide-design-resources; Supplementary Table 3) were cloned into the pSpCas9(BB)-2A-Puro (PX459) V2.0 vector. HEK293T cells (cultured in six-well plates) were transfected with 2.0 μg recombinant plasmid using Lipofectamine™ 3000 (Invitrogen, Cat. No. L3000-015). After transfecting for 48 h, 2 μg/ml puromycin (Sigma, Cat. No. 540222) was added to kill the untransfected cells for 2 days. The selected cells were then re-plated at a low density and allowed to grow for about 7 days. A single colony was picked and plated into 24-well plates for scale-up culture, and the culture medium was replaced every 2 days.
Knockdown experiments
The sequences of short hairpin RNAs (shRNAs) are provided in Supplementary Table 3. The regions targeted by the designed shRNAs are provided in Supplementary Fig. 4. Detailed information about knockdown assays is provided in the Supplementary material.
Overexpression experiments
The coding sequences of human NMRAL1 was cloned into plenti-CAG-IRES-GFP (Addgene, Cat. No. 69047). 1× FLAG was fused at the 3′-end of NMRAL1 gene. The primers used for cloning are provided in Supplementary Table 4. Detailed information about knockdown assays is provided in the Supplementary material.
Western blotting
The harvested cells were lysed with Radio-Immunoprecipitation Assay (RIPA) buffer (Thermo Scientific, Cat. No. 89901) containing phenylmethylsulfonyl fluoride (PMSF) buffer (100×; Beyotime, Cat. No. ST506-2), and supernatants were boiled at 95°C for 5 min. Equal amounts of total proteins were subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE; SurePAGE™, Bis–Tris, 4–20%, Cat. No. M00657) and transferred onto polyvinylidene difluoride (PVDF) membranes. These membranes were blocked in Protein Free Rapid Blocking Buffer (5×; EpiZyme, Cat. No. PS108) for 30 min, probed with primary antibodies and incubated overnight at 4°C, then incubated with goat anti-rabbit secondary antibodies (Proteintech, Cat. No. SA00001-2, 1:5000) after washing with phosphate-buffered saline with Tween (PBST). The dilutions of the primary antibodies were as follows: FLAG (Sigma, Cat. No. F7425-2mg, 1:1000), β-actin (CST, Cat. No. 13E5, 1:1000).
Quantitative real-time PCR
TRIzol reagents (Life Technologies, Cat. No. 15596018) were used to extract total RNA, and cDNA were synthesized using the PrimeScript RT reagent kit with gDNA Eraser (Takala, Cat. No. RR047B). Quantitative real-time polymerase chain reaction (qPCR) were performed using SYBR-green fluorescence quantification system (TB Green® Premix Ex Taq™ II, Takala, Cat. No. RR820B). Relative mRNA expression levels were normalized to ACTB (human) or Actb (mouse and rat), and fold change were determined using the 2−ΔΔCt method.35 All qPCR primer sequences are listed in Supplementary Table 5.
RNA sequencing
Detailed information is provided in the Supplementary material. Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were then performed using Metascape (https://metascape.org).36
5-Ethynyl-2′-deoxyuridine proliferation and Cell Counting Kit-8 assays
EdU (5-ethynyl-2′-deoxyuridine) incorporation and Cell Counting Kit-8 (CCK-8) assays were conducted as previously described.23,37,38 Detailed procedures of EdU and CCK-8 assays are provided in Supplementary material.
Spontaneous differentiation of mouse neural stem cells
The mNSCs were plated into 24-well plates [pre-coated with Laminin (Sigma, 20 µg/ml, Cat. No. L2020-1 mg)] at a density of 2 × 105 cells/well. The next day, proliferation medium was replaced by differentiation medium.39,40 After differentiating for 72 h, cells were fixed for immunofluorescence staining.
Analysis of density of dendritic spines
Morphologies of dendritic spines were analysed as previously described.34,41,42 Briefly, rat primary cortical neurons were isolated from brains of embryonic rat (E17.5–E18.5) and cultured for 14–15 days and then were co-transfected with 3.0 μg recombinant pSicoR-Ef1a-mCh-Puro plasmid and 1.0 μg Venus control plasmid using Lipofectamine™ 3000 (Invitrogen, Cat. No. L3000-015). Three days post-transfection, the neurons were fixed with 4% paraformaldehyde (with 4% sucrose) for immunofluorescence staining. NeuronStudio was used for dendritic spine analyses.34,43 Three subtypes of spines (stubby, thin and mushroom) of the secondary or tertiary dendrites of each neuron were used for analysis (length of 50–125 μm per dendrite).
Immunofluorescence staining
Detailed information about immunofluorescence staining assays and corresponding antibodies is provided in the Supplementary material.
Data availability
The data that support the findings of this study are available in CMC, LIBD, ENCODE, PsychENCODE, UCSC genome browser, SZDB, RegulomeDB, GTEx Portal and the Human Protein Atlas. These data were derived from the following resources available in the public domain: CMC, https://www.nimhgenetics.org/resources/commonmind; LIBD, http://eqtl.brainseq.org/; ENCODE, https://www.encodeproject.org/; PsychENCODE, http://resource.psychencode.org/; UCSC genome browser, http://genome.ucsc.edu/; SZDB, http://www.szdb.org/; RegulomeDB, https://www.regulomedb.org/; GTEx Portal, https://www.gtexportal.org/home/; the Human Protein Atlas, https://www.proteinatlas.org/. Other data can be available from the corresponding author upon reasonable request.
Results
Prioritization of rs2270363 as a potential causal variant
Our functional genomics identified 132 SNPs that disrupted binding of transcription factors,14 implying that these SNPs are functional or potential causal variants. To further prioritize the most promising and possible functional SNPs from the 132 transcription factor binding-disrupting SNPs, we utilized eQTL data and performed additional analyses (Supplementary Fig. 1).18 In brief, eQTL analysis distilled 97 SNPs (among the 132 transcription factor binding-disrupting SNPs).14 SNP location mapping showed that 30 of 97 SNPs are located in the promoter region and marked with H3K4me3 (a marker for active promoters). RegulomeDB functional annotation further prioritized five high-confidence (RegulomeDB rank score <2) functional SNPs in the promoter region, indicating that these SNPs may represent authentic functional variants (Supplementary Table 6). Finally, differential expression of the eQTL genes (Supplementary Table 7) of the five SNPs (located in the promoter region) were examined in schizophrenia cases and controls.18 The expression of the eQTL genes of rs223387 did not show significant change in schizophrenia cases and controls (Supplementary Table 8). We noticed that rs2269524 is in high linkage disequilibrium (r2 > 0.95) with rs1801311 (Supplementary Table 9), a functional SNP whose regulatory mechanisms in schizophrenia have been recently reported.24 Thus, we did not investigate rs1801311 in this study. Two eQTL genes (FAM109B and LINC00634) of rs2269524, two eQTL genes (NDUFA2 and SRA1) of rs3822346 and two eQTL genes (NMRAL1 and CORO7) of rs2270363 showed differential expression in schizophrenia cases compared with controls (Supplementary Table 8). However, we noted that three SNPs (rs2269524, rs1801311 and rs3822346) are far away from their differentially expressed eQTL genes and are not located in the promoter regions of their differentially expressed eQTL genes (Supplementary Fig. 5), suggesting that they are unlikely to regulate the expression of these genes by affecting the promoter activity. For example, our previous study showed that rs1801311, which is located in the first exon of NDUFA6, may confer schizophrenia risk by regulating the expression of the distal gene NAGA (an eQTL gene of rs1801311) rather than NDUFA6.24 Interestingly, rs2270363, which is located in the NMRAL1 promoter region, was associated with NMRAL1 expression in the human brain, and NMRAL1 expression was downregulated in schizophrenia cases compared with controls. This suggests that rs2270363 may regulate the expression of NMRAL1 by modulating promoter activity. These lines of evidence suggested rs2270363 as a plausible causal variant for schizophrenia at the 16p13.3 risk locus.
Confirmation of association between rs2270363 and schizophrenia in the Chinese population
To further confirm the association between rs2270363 and schizophrenia, we performed a genetic association study in a large Chinese sample (3718 cases and 7829 controls). We found that rs2270363 was also significantly associated with schizophrenia in the Chinese population (P = 3.17 × 10−4, ORA = 1.11; Table 1), with the same risk allele (A) as in Europeans. Of note, the P-value of rs2270363 in PGC2 (34 241 cases, 45 604 controls and 1235 parent affected-offspring trios) was 3.33 × 10−5 (ORA = 1.05),13 and in EAS + EUR (56 418 cases and 78 818 controls), the P was 4.59 × 10−6 (ORA = 1.05).27 Further trans-ancestry meta-analysis (by combing our association result with previously reported associations in Europeans and East Asians) showed that rs2270363 reached genome-wide significance level (P = 3.98 × 10−8, ORA = 1.05, 60 136 cases and 86 647 controls; Table 1).27 These data provide robust evidence supporting the association between rs2270363 and schizophrenia.
Table 1.
rs2270363 is associated with schizophrenia
| SNP ID | Sample | Sample size (cases/controls) | A12 | ORa | P |
|---|---|---|---|---|---|
| rs2270363 | This study | 3718/7829 | A/G | 1.11 | 3.17 × 10−4 |
| European | 33 640/43 456 | A/G | 1.05 | 1.35 × 10−4 | |
| East Asian | 22 778/35 362 | A/G | 1.05 | 1.07 × 10−2 | |
| Combined | 60 136/86 847 | A/G | 1.05 | 3.98 × 10−8 |
Odds ratio (OR) is based on A1.
Validation of the regulatory effect of rs2270363 with reporter gene assays and EMSA
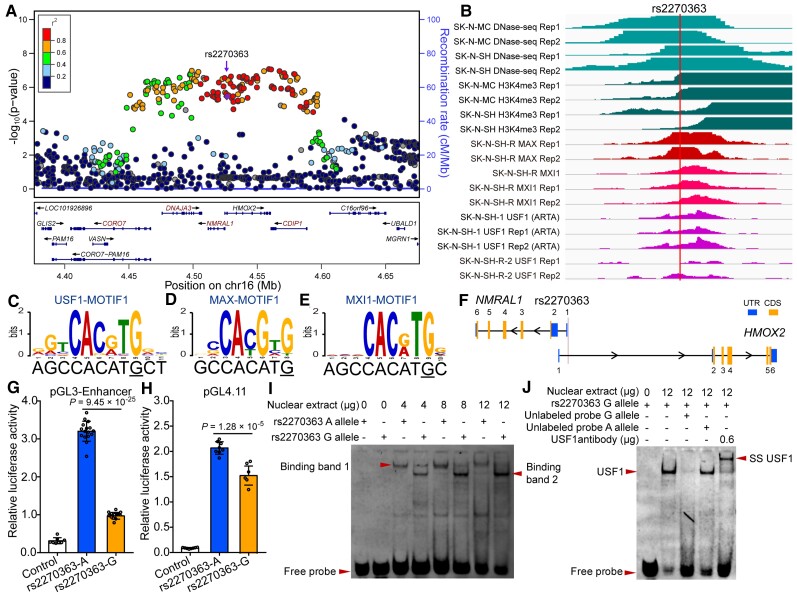
Our previous functional genomics study identified rs2270363 as a functional SNP at the 16p13.3 schizophrenia risk locus (Fig. 1A and B).14 DNase-Seq data showed that rs2270363 is located in open chromatin regions, with histone H3K4me3 modification (a marker for active promoters; Fig. 1B). Consistently, the genomic region containing rs2270363 was also marked with H3K4me3 modification in primary cultured neuronal cells derived from olfactory neuroepithelium (CNON).17 In addition, the genomic region containing rs2270363 was also marked with H3K4me3 (a marker for promoter) and H3K27ac (a marker for enhancer) signals in human microglia, neurons and oligodendrocytes (Supplementary Fig. 6).44 Data from ENCODE showed that rs2270363 is located in the promoter region (marked with H3K4me3) in multiple human brain tissues and neural cells (including neuroblastoma cell lines; Supplementary Figs 7 and 8).45 These results indicated that rs2270363 is located in an active regulatory region (marked with an active promoter H3K4me3 signal), implying the potential functionality of rs2270363. Further analysis showed that rs2270363 lies in the E-box motif (CANNTG).46 USF1, MAX and MXI1 are members of the bHLHZ protein family, which bind to the E-box sequences.46–49 Of note, ChIP-Seq data clearly showed that transcription factors MAX, USF1 and MXI1 could bind to the genomic region containing rs2270363 in neural cells (Fig. 1B), and motif analysis revealed that rs2270363 disrupts binding of USF1, MAX and MXI1 (Fig. 1C–E). These data suggested that rs2270363 is a functional SNP with potential regulatory effects (through affecting binding of TFs).
Figure 1.
Validation of regulatory effects of rs2270363 at the 16p13.3 risk locus. (A) The Locus zoom plot showed the associations between variants in genomic region (300 kb) containing rs2270363 and schizophrenia in the PGC2 + CLOZUK study. (B) ChIP-Seq data showed that transcription factors USF1, MAX and MXI1 could bind to genomic sequence containing rs2270363 (data from ENCODE). (C–E) rs2270363 disrupts binding motif (i.e. position weight matrix, PWM) of USF1, MAX and MXI1. (F) SNP rs2270363 is located in the promoter of NMRAL1 gene. (G and H) Dual-luciferase reporter gene assays showed that rs2270363 affected promoter activity in SH-SY5Y cells regardless of its orientation. The vector containing A allele of rs2270363 exhibited significant higher luciferase activity compared with G allele. (I) EMSA showed that rs2270363 affected binding of transcription factors. One binding band (band 1, marked with arrowhead) was detected for the A allele, while two binding bands (band 1 and 2) were detected for the G allele. (J) Super-shift and competitive experiments for EMSA. Super-shift assay showed that rs2270363 binds to USF1 (the super-shift band was detected in the lane with added USF1 antibody) and competitive experiments revealed that the G allele had stronger binding affinity compared with the A allele. n = 8 for control group in G, n = 16 for experimental group in G, n = 8 for H. Unpaired two-tailed Student’s t-test was used to test if the difference reached significance level (P = 0.05). Data are presented as mean ± SD.
As rs2270363 is located in the E-box element of NMRAL1 promoter (Fig. 1F), to further validate whether rs2270363 plays a role in transcriptional regulation, we performed dual-luciferase reporter assays. We found that the genomic sequence (861 bp) containing rs2270363 exhibited a strong promoter activity in the SH-SY5Y (Fig. 1G and H), HEK-293T and SK-N-SH cell lines (Supplementary Fig. 9), and the A (risk) allele of rs2270363 conferred significant higher activity compared with the G allele (Fig. 1G and H). These results confirmed that rs2270363 is a functional SNP with regulatory effect.
We next performed EMSA to test if rs2270363 affects transcription factor binding. The A allele has one binding band while the G allele has two binding bands (Fig. 1I), and we also found that the G allele prefers to bind the transcription factor of band 2. These data indicated that different alleles of rs2270363 could bind to different transcription factors (arrowhead in the last lane in Fig. 1I). The result of the competitive experiment also indicated significant differences in binding affinities between the two alleles, i.e. the transcription factor in the binding band 2 prefers to bind the G allele (Fig. 1J). Finally, the super-shift assay showed that the transcription factor in the binding band 2 is USF1 (Fig. 1J), as a super-shift band was detected in the electrophoresis lane with anti-USF1 antibody (SS USF1, arrowhead on the right side of Fig. 1J). Interestingly, sequence conservation analysis showed that the regulatory element containing rs2270363 is not highly conserved in vertebrates (Supplementary Fig. 10), suggesting this regulatory event (in terms of regulation of NMRAL1) is primate-specific [as no orthologous sequences (corresponding to the genomic sequence containing rs2270363 in human) were identified in many vertebrate species (Supplementary Fig. 10)]. Taken together, these results validated that rs2270363 (which resides in the E-box binding motif of NMRAL1 promoter) is a functional SNP and different alleles of rs2270363 could affect USF1 binding.
rs2270363 is associated with NMRAL1 expression in the human brain
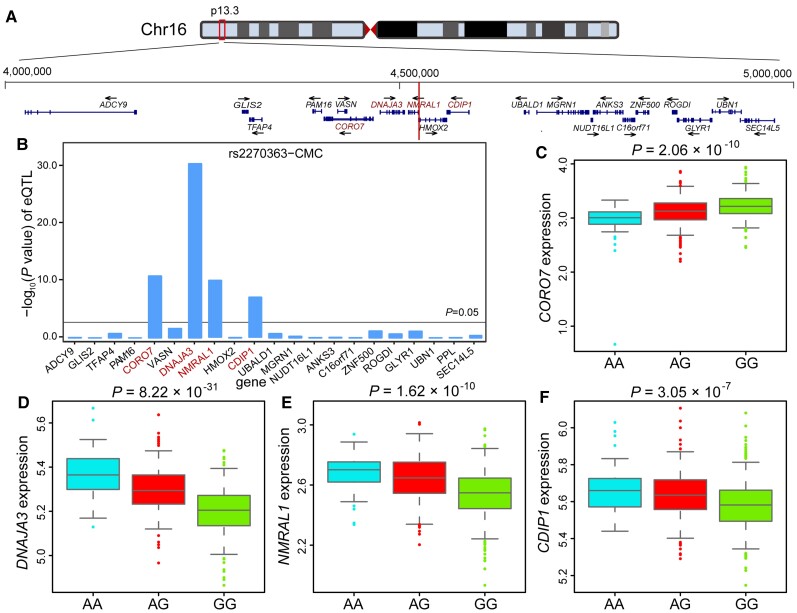
We have validated the regulatory effect of rs2270363 and demonstrated the regulatory mechanisms of this functional SNP. To further identify the potential target gene of rs2270363, we conducted eQTL analysis and found that rs2270363 was significantly associated with the expression of CORO7, DNAJA3, NMRAL1 and CDIP1 (Fig. 2A and B) in the human brain (CMC eQTL dataset).18 The A allele of rs2270363 is associated with higher expression of DNAJA3, NMRAL1 and CDIP1 and with lower expression of CORO7 in CMC data set (Fig. 2C–F). These eQTL results were successfully replicated in the LIBD brain eQTL data set (Supplementary Fig. 11A–E).19 These results suggest that rs2270363 might confer schizophrenia risk by regulating CORO7, DNAJA3, NMRAL1 and CDIP1.
Figure 2.
rs2270363 was associated with NMRAL1 expression in the human brain. (A) Genomic location of rs2270363 (1 Mb). rs2270363 lies in the promoter region of NMRAL1. (B) eQTL analysis showed the associations between rs2270363 and expression of its nearby genes (1 Mb window) in the human brain (data from CMC, n = 478).18 (C–F) The box-plot of eQTL results. The A allele is associated with lower CORO7 expression and higher expression of NMRAL1, DNAJA3, CDIP1 (data from CMC, n = 467).18
rs2270363 regulates NMRAL1 expression
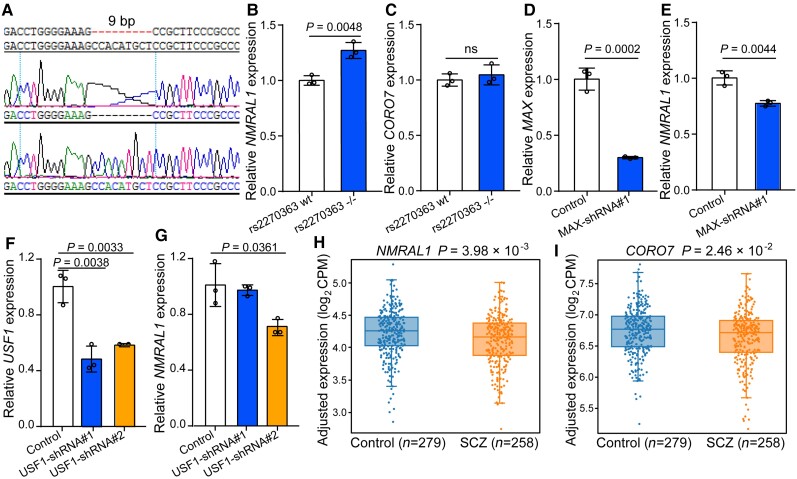
To further investigate if rs2270363 could regulate expression of its eQTL genes (i.e. NMRAL1, CORO7, DNAJA3 and CDIP1), we deleted a 9-bp genomic sequence containing rs2270363 using CRISPR/Cas9-mediated genome editing. Due to the low transfection efficiency of SH-SY5Y, we failed to obtain edited (i.e. rs2270363 deletion) SH-SY5Y monoclonal cells. However, we successfully knocked out a 9-bp fragment (containing rs2270363) in HEK-293T cells (Fig. 3A). We also examined whether the expression of four eQTL genes of rs2270363 varied in tissues or cell types, and found that NMRAL1, CORO7, DNAJA3 and CDIP1 are widely and abundantly expressed in different tissues and cell types (Supplementary Figs 12A–D and 13A–D). Importantly, all four genes are expressed in SH-SY5Y and HEK 293 cells (HEK 293T cells are derived from HEK 293 cells, which are human renal epithelial cell lines; Supplementary Fig. 13A–D), suggesting that the regulatory effect of rs2270363 might be similar in HEK293T and SH-SY5Y cells. We found that rs2270363 deletion resulted in a significant increase of NMRAL1 expression (Fig. 3B). However, expression of CORO7, DNAJA3 and CDIP did not show significant change (Fig. 3C, Supplementary Fig. 11F and G). Besides, rs2270363 resides in the first intron of HMOX2 (Fig. 1F; the genomic sequence surrounding NMRAL1 promoter overlaps with the first intron of HMOX2), which is widely expressed in different tissues and cell types (Supplementary Figs 12E and 13E). Therefore, we also investigated the effect of rs2270363 deletion on HMOX2 expression and found that deletion of the genomic sequence containing rs2270363 (9 bp) did not affect HMOX2 expression level (Supplementary Fig. 11H). These results indicated that rs2270363 can regulate the expression of NMRAL1 and prioritized NMRAL1 as the potential target gene of rs2270363.
Figure 3.
rs2270363 regulates NMRAL1 expression. (A) CRISPR-Cas9-mediated deletion of the 9-bp sequence containing rs2270363. (B) Deletion of rs2270363 (9 bp) led to upregulation of NMRAL1. However, CORO7 expression was not altered (C). (D and E) MAX knockdown altered NMRAL1 expression in neuronal cells (SH-SY5Y). (F and G) USF1 knockdown affected NMRAL1 expression in neuronal cells. Unpaired two-tailed Student’s t-test was used to determine if the difference reached significance level. n = 3. Data are presented as mean ± SD. ns = not significant. (H and I) Expression of NMRAL1 and CORO7 showed significant downregulation in the brains of schizophrenia cases compared with controls (http://www.szdb.org/).50 Schizophrenia cases, n = 258; controls, n = 279.
USF1 and MAX regulate NMRAL1 expression
To further explore if USF1 and MAX (whose binding were disrupted by rs2270363) regulate NMRAL1 expression, we knocked down USF1 and MAX in SH-SY5Y cells. Knockdown of USF1 or MAX led to significant downregulation of NMRAL1 (Fig. 3D–G), indicating that rs2270363 regulates NMRAL1 expression by disrupting binding of USF1 and MAX (Fig. 1B, I and J).
Dysregulation of NMRAL1 expression in schizophrenia cases
We have shown that the functional SNP rs2270363 might confer risk of schizophrenia by modulating NMRAL1 expression. To further explore if NMRAL1 expression in brains of schizophrenia cases was dysregulated, we examined NMRAL1 expression and the result showed that NMRAL1 was significantly downregulated in brains of schizophrenia cases compared with controls (P = 3.98 × 10−3, 258 cases and 279 controls, CMC expression data from SZDB2.0; Fig. 3H).50 In addition, we also examined other eQTL genes of rs2270363 (CORO7, DNAJA3 and CDIPI). Among these three genes, only CORO7 showed a significant downregulation in schizophrenia cases compared with controls (Fig. 3I). DNAJA3 and CDIP1 did not show significant change (Supplementary Fig. 14). Nevertheless, CORO7 showed a less-significant level of differential expression (P = 2.46 × 10−2) compared with NMRAL1 (Fig. 3I), and rs2270363 is located in the NMRAL1 promoter (but not in CORO7 promoter). These results collectively prioritized NMRAL1 as the most plausible candidate risk gene for schizophrenia at the 16p13.3 risk locus.
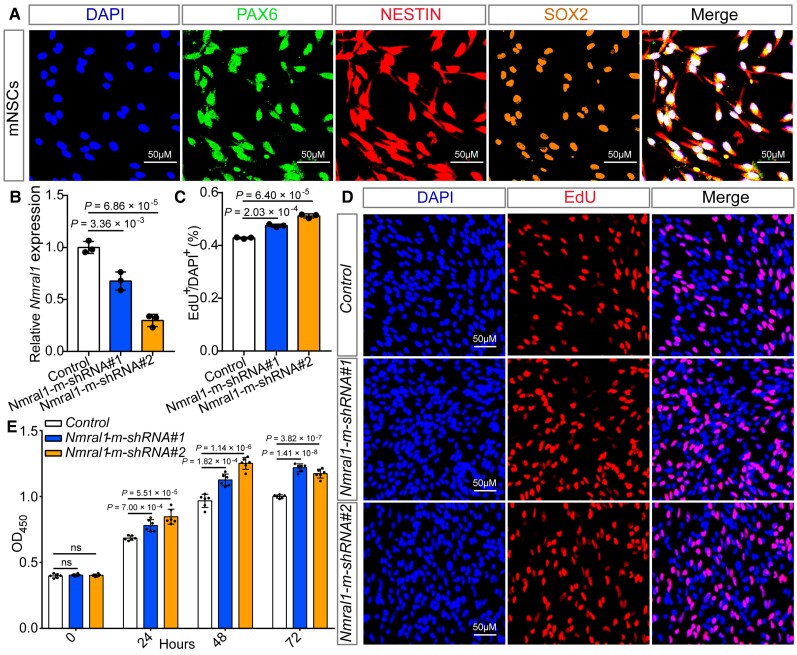
Figure 4.
Nmral1 knockdown promotes proliferation of mNSCs. (A) Immunofluorescence staining using three well-characterized markers (SOX2, PAX6 and NESTIN) for NSCs validated the identity of the isolated mNSCs. (B) Nmral1 expression was efficiently knocked-down by the shRNAs for mouse. (C and D) The results of EdU incorporation assay. (C) The quantification results of the EdU incorporation assay. (D) Immunofluorescence staining for EdU incorporation assay. Red indicates EdU-positive cells and DAPI was used to stain the nucleus (blue). (E) The results of CCK-8 assay. Data were measured at four different time points: 0, 24, 48 and 72 h. Unpaired two-tailed Student’s t-test was used to compare if the difference was significant. n = 3 for B–D; n = 6 for E. Data are presented as mean ± SD.
Integrative analysis supports NMRAL1 as a schizophrenia risk gene
To further explore if NMRAL1 is a schizophrenia risk gene, we conducted Sherlock integrative analysis by integrating GWAS summary statistics and brain eQTL data.18,27,51 Sherlock integrative analysis supported that NMRAL1 is a schizophrenia risk gene whose expression perturbation might have a role in schizophrenia (P = 1.58 × 10−6). In line with this results, SMR analysis also showed that NMRAL1 is a schizophrenia risk gene.52,53 Collectively, these convergent lines of evidence prioritized NMRAL1 as the most plausible candidate at the 16p13.3 risk locus.
NMRAL1 affects proliferation and differentiation of mouse neural stem cells
We provided lines of evidence that support NMRAL1 may be a novel schizophrenia risk gene whose expression is regulated by rs2270363. To further investigate the role of NMRAL1 in schizophrenia, we utilized the neural stem cell model (a model that is widely used to investigate the role of schizophrenia risk gene in the brain).54–56 We first validated the identity of the isolated mNSCs using three well-characterized makers for NSCs (PAX6, NESTIN and SOX2; Fig. 4A). We then knocked down Nmral1 expression in mNSCs using shRNAs (Fig. 4B). EdU incorporation assays showed that the number of EdU-positive cells was significantly increased in Nmral1 knocked-down mNSCs (Fig. 4C and D). Consistently, CCK-8 assays also revealed that Nmral1 knockdown promoted the proliferation rate of the mNSCs (Fig. 4E). As the risk allele (A) of rs2270363 was associated with elevated NMRAL1 expression (Fig. 2E), we also explored the effect of NMRAL1 overexpression on proliferation of mNSCs (Supplementary Fig. 15A and B). Both EdU and CCK-8 assays showed that NMRAL1 overexpression inhibited the proliferation rate of the mNSCs (Supplementary Fig. 15C–E). These results demonstrated that Nmral1 could regulate the proliferation of mNSCs.
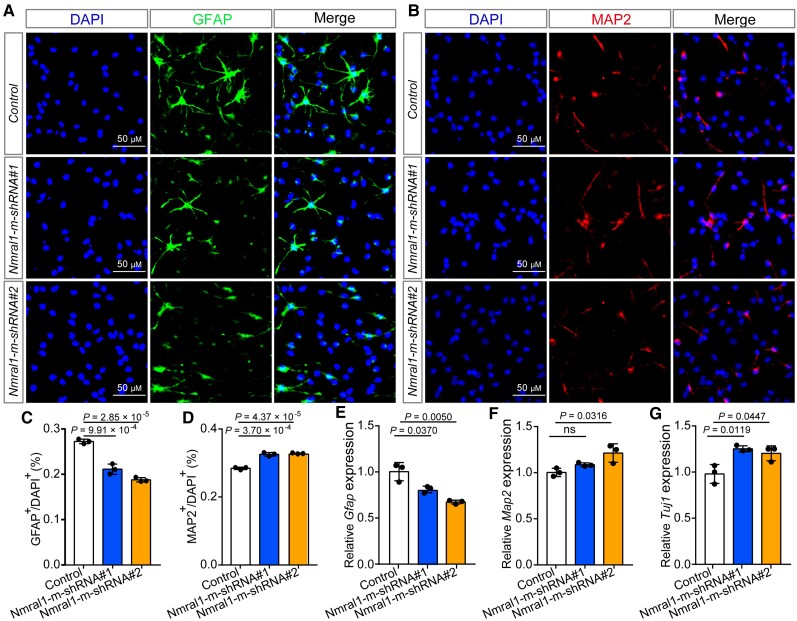
In addition, we examined the effect of Nmral1 on differentiation of mNSCs. Immunofluorescence results showed that Nmral1 knockdown affects the differentiation of mNSCs (Fig. 5A–D). The proportion of glial fibrillary acidic protein (GFAP)-positive (a marker for glia cells) cells was significantly decreased in Nmral1 knocked-down mNSCs compared with controls (Fig. 5A and C). By contrast, the proportion of MAP2-positive (a marker for mature neurons) cells was significantly increased in Nmral1 knocked-down mNSCs compared with controls (Fig. 5B and D). Using qPCR, we again found that the expression level of Gfap was significantly downregulated in Nmral1 knocked-down mNSCs compared with controls, while the expression of Map2 and Tuj1 (a marker for newly generated post-mitotic neurons) were significantly increased (Fig. 5E–G). Finally, we examined the effect of human NMRAL1 overexpression on differentiation of mNSCs. We found that NMRAL1 overexpression did not affect differentiation of mNSCs (Supplementary Fig. 16).
Figure 5.
Nmral1 knockdown affects differentiation of mNSCs. (A and B) Representative immunofluorescence staining images for glia cells and neurons differentiated from mNSCs. GFAP-positive cells were marked with green, MAP2-positive cells with red and the nuclei were stained with DAPI. (C) Quantification for the proportion of GFAP-positive glia cells. (D) Quantification for the proportion of MAP2-positive neurons. (E–G) qPCR showed the relative expression level of Gfap, Map2 and Tuj1 in control and Nmral1 knocked-down mNSCs. Unpaired two-tailed Student’s t-test was used to compare if the difference was significant. n = 3, data are presented as mean ± SD. ns = not significant.
Figure 6.
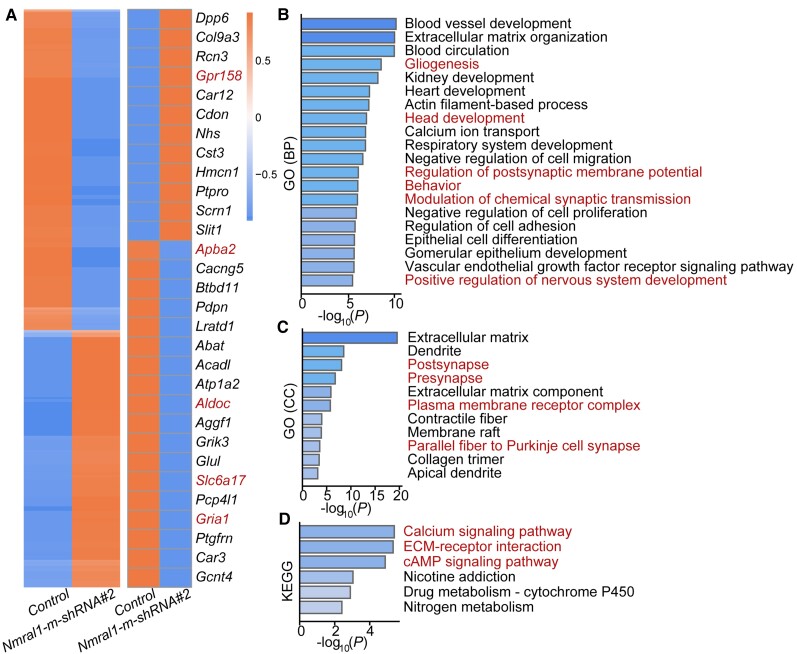
Nmral1 regulates schizophrenia-associated biological processes and signalling pathways. (A) Expression heat map of all the DEGs (n = 991) identified in Nmral1 knocked-down mNSCs compared with controls (left). Heat map plot of the top 30 DEGs (right). Five genes (marked by red colour) were selected for qPCR verification. (B) GO (biological processes) analysis of all the DEGs. (C) GO (cellular components) analysis of all the DEGs. (D) KEGG analysis of all the DEGs. Red indicates schizophrenia-associated biological processes, cellular components and signalling pathways for B–D.
Collectively, these results demonstrate the important roles of Nmral1 in neurodevelopment, a key process that was frequently reported to be dysregulated in schizophrenia.57–60 Our findings also support the neurodevelopmental hypothesis of schizophrenia.58,60
NMRAL1 regulates schizophrenia-related biological processes and signalling pathways
To identify genes and pathways regulated by Nmral1 (in mNSCs), we performed a transcriptome analysis to determine the differentially expressed genes (DEGs). We identified 991 DEGs (|fold change| > 1.5, Padj < 0.01). Among them, ∼45% (441 genes) were upregulated and ∼55% (550 genes) were downregulated in Nmral1 knocked-down mNSCs (Fig. 6A). We validated the mRNA alternations of several genes in Nmral1 knocked-down mNSCs using qPCR (Supplementary Fig. 17A–E).
GO (biological processes) analysis showed that these DEGs were significantly enriched in neurodevelopment processes, including gliogenesis, head development and nervous system development (Fig. 6B). In addition, the DEGs were also showed enrichments in cell adhesion, proliferation, migration and differentiation pathways (Fig. 6B). Of note, modulation of chemical synaptic transmission process was also enriched among the DEGs (Fig. 6B). GO analysis (cell components) slowed that the DEGs were enriched in synapse component, further supporting the potential role of NMRAL1 in synaptic transmission (Fig. 6C).
KEGG enrichment analysis indicated that the DEGs were significantly enriched in schizophrenia-associated signalling pathways (Fig. 6D), including calcium signalling pathway,61–63 extracellular matrix (ECM)–receptor interaction64,65 and cyclic adenosine monophosphate (cAMP) signalling pathway.66,67 The downregulated genes in Nmral1 knocked-down mNSCs were mainly enriched in the cAMP and calcium signalling pathways (Supplementary Fig. 17F) and the upregulated genes were mainly enriched in the ECM–receptor interaction signalling pathway (Supplementary Fig. 17G). Collectively, our transcriptome analysis provides further evidence that supports the important role of Nmral1 in neurodevelopment, and also suggests that Nmral1 may confer risk of schizophrenia through regulating schizophrenia-associated biological processes and signalling pathways.
Nmral1 regulates density of dendritic spines
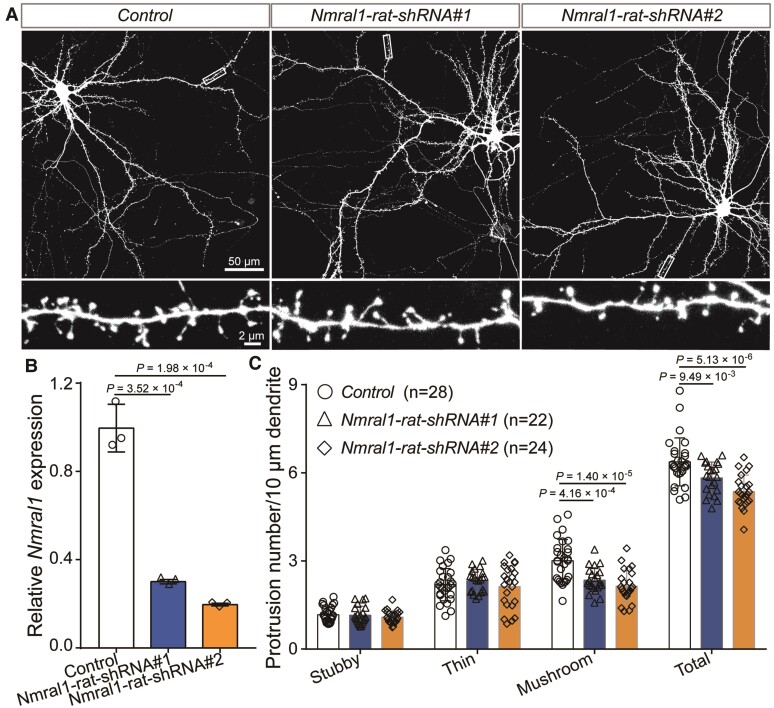
The pathophysiology of schizophrenia remains elusive. However, synaptic dysfunction has been repeatedly reported in subjects with schizophrenia.68–70 In addition, the density of dendritic spines (layer 3 pyramidal neurons in the dorsolateral prefrontal cortex) was significantly decreased in schizophrenia cases compared with controls.68,71,72 Of note, GO analysis showed that Nmral1 regulates pathways associated with synaptic transmission (Fig. 6B) and the DEGs were enriched in synapse-associated components, including post-synapse, pre-synapse, plasma membrane receptor complex and parallel fibre to Purkinje cell synapse (Fig. 6C). These results suggest the potential role of NMRAL1 in morphogenesis of dendritic spines. Therefore, we knocked-down Nmral1 in rat primary cortical neurons. Dendritic spines analysis showed no significant changes for the density of thin and stubby spines (immature spines). However, Nmral1 knockdown resulted in significant decrease of density of mushroom spines (Fig. 7A–C), which can form stable and mature synaptic connection.73–75 These results revealed the important role of NMRAL1 in dendritic spine development. Besides, these findings also suggested that NMRAL1 might contribute to schizophrenia susceptibility by affecting the density and function of dendritic spines.
Figure 7.
Nmral1 knockdown decreased dendritic spine density. (A) Representative immunofluorescence staining images for neuronal morphologies and dendrites. (B) Nmral1 expression in rat primary neurons was efficiently knocked-down by the shRNAs. (C) Dendritic spine density analysis for stubby, thin and mushroom spines. P-values were calculated using two-tailed Student’s t-test. n = 28 for control; n = 22 for Nmral1-rat-shRNA#1; n = 24 Nmral1-rat-shRNA#2.
In summary, these lines of evidence suggest that rs2270363 may confer schizophrenia risk by modulating expression of NMRAL1, a gene which has a pivotal role in neurodevelopment and dendritic spine morphogenesis (two featured characteristics of schizophrenia pathophysiology).
Discussion
In this study, we demonstrated the complex regulatory mechanisms of rs2270363, a functional schizophrenia risk SNP that disrupts binding of three bHLHZ transcription factors (USF1, MAX and MXI1)76–83 and was significantly associated with schizophrenia. Previous study has shown changes of USF activity during the conversion from proliferation to differentiation.48 Moreover, the binding strength of USF1 to the sequence containing rs2270363 was enhanced in all-trans retinoic acid-induced differentiation of SK-N-SH cells (Fig. 1B; data from ENCODE). GO (cellular components) analysis also showed that the DEGs were enriched in synaptic transmission-associated components, including post-synapse, pre-synapse, plasma membrane receptor complex and parallel fibre to Purkinje cell synapse (Fig. 6C), indicating the important role of NMRAL1 in the central nervous system. MAX can form different dimers (MAX/MXI1, MAX/MAX, MAX/MAD and MAX/MYC) and play different roles in proliferation and differentiation.47,77,83 The MAX/MYC heterocomplex was gradually replaced by the MAX/MAD heterocomplex during phorbol 12-myristate 13-acetate-induced differentiation.47 In addition, there is a competitive combination between MAX homodimers or heterodimers and USF.84,85 These results indicated that the binding of bHLHZ proteins to rs2270363 is a dynamic process during neurodevelopment. Here, we found that MAX or USF1 knockdown resulted in downregulation of NMRAL1 in SH-SY5Y cells, suggesting that MAX and USF1 are involved in the regulation of NMRAL1 expression (and this regulation is likely mediated by rs2270363, as rs2270363 disrupts binding of MAX and USF1). In addition, our reporter gene assays showed that the transcriptional activity of the A allele of rs2270363 was significantly stronger than the G allele, which was consistent with the observation that the A allele corresponds to higher expression of NMRAL in eQTL analysis. We also validated the regulatory effect of rs2270363 with EMSA, which revealed that different alleles of rs2270363 affect USF1 binding affinity.
In addition to elucidating the regulatory mechanisms of rs2270363, our study also showed that rs2270363 may confer schizophrenia risk through regulating expression of NMRAL1, a gene associated with risk of schizophrenia14,52,53,86 and with unknown function in the human brain. NMRAL1 (also known as HSCARG) encodes NmrA like redox sensor 1, an NmrA-like transcriptional regulator whose subcellular localization and protein conformation is affected by changes in cellular metabolism status (e.g. cellular redox status, ammonia concentration and immune status).87–90 NMRAL1 is an nicotinamide adenine dinucleotide phosphate (NADPH) sensor protein that preferentially binds to NADPH.87,91 Previous studies have revealed that NMRAL1 is also involved in many biological processes, including cellular antiviral response (by negatively regulating the interferon response factor 3-mediated expression of interferon beta),89,92–94 redox homeostasis,88,95 DNA damage response,96 innate immunity and cancer91 (e.g. NMRAL1 suppresses polyubiquitination of NF-κB essential modulator by interacting with the deubiquitinase USP7 to inhibit tumour necrosis factor and interleukin 1-induced NF-κB activation).93
For the first time, we showed that NMRAL1 plays pivotal roles in neurodevelopment and dendritic spine morphogenesis. Our findings support the neurodevelopmental hypothesis of schizophrenia and suggest that rs2270363 may exert functional impacts through regulating expression of NMRAL1, a gene whose expression perturbation affects neurodevelopment and dendritic spine density. Although the pathogenesis of schizophrenia remains to be elucidated, lines of evidence suggested the pivotal role of dendritic spine deficit (dendritic spine pathology) in schizophrenia. Our study not only identified NMRAL1 as a new schizophrenia risk gene, but also demonstrated the potential role of NMRAL1 in schizophrenia pathogenesis (i.e. NMRAL1 may confer risk of schizophrenia by affecting neurodevelopment and dendritic spine morphogenesis). Considering that the risk allele of rs2270363 was associated with higher NMRAL1 expression and NMRAL1 overexpression inhibited proliferation rate of the mNSCs, it is possible that NMRAL1 overexpression is a potential pathogenic mechanism for schizophrenia. However, we mainly investigated the role of NMRAL1 knockdown on neurodevelopment in the present study. More work is needed to explore if NMRAL1 overexpression has a potential pathogenic role for schizophrenia.
We also explored the transcript expression of NMRAL1 at differential transcript expression and differential transcript usage levels using PsychENCODE data.97 However, no differential transcript expression or differential transcript usage of NMRAL1 was detected (false discovery rate < 0.1) between schizophrenia and healthy controls.
Considering the important role of NMRAL1 in neurodevelopment, we also explored if this gene was associated with other neurodevelopmental disorders, including autism spectrum disorder (18 382 cases and 27 969 controls),98 attention deficit and hyperactivity disorder (20 183 cases and 35 191 controls) and bipolar disorder (20 352 cases and 31 358 controls).99,100 Genetic variants near NMRAL1 (including rs2270363) did not show suggestive associations with autism spectrum disorder, attention deficit and hyperactivity disorder and bipolar disorder (Supplementary Fig. 18). Further differential expression data (using expression data from PsychENCODE) showed that NMRAL1 did not show differential expression in brains of autism spectrum disorder cases (n = 51) compared with controls (n = 936; P = 0.14).97 However, NMRAL1 was significantly upregulated in brains of bipolar disorder cases (n = 222) compared with controls (n = 936; P = 0.00017).97 These results suggest that NMRAL1 may also have a role in bipolar disorder.
In addition to the above evidence, the biological function of NMRAL1 also supports the potential role of this gene in schizophrenia. NMRAL1 has pivotal roles in regulating immunity. For example, NMRAL1 negatively regulates innate immunity by downregulating the activities of RIG-I like receptor (RLR) and NF-κB pathways.91 Dysregulation of immune system has been frequently observed in schizophrenia.101,102 In fact, the genetic variants in the major histocompatibility complex region showed the strongest associations with schizophrenia,103 suggesting the important role of immunity-associated genes in schizophrenia. Besides, NMRAL1 regulates redox homeostasis, an important biological process that has been frequently reported to be dysregulated in schizophrenia.104 Elevated oxidative stress was repeatedly reported in schizophrenia.105,106 A recent study by Steullet et al.107 also revealed that oxidative stress-driven parvalbumin interneuron impairment is a common mechanism in models of schizophrenia. Based on the above observations, the oxidative stress hypothesis of schizophrenia and the immune hypothesis of schizophrenia have been proposed.102,106 It is possible that NMRAL1 participates in neurodevelopment by regulating redox homeostasis and NF-κB pathways. Nevertheless, more work is needed to demonstrated the role of NMRAL1 in schizophrenia.
Pinpointing the causal (or functional) variants from the reported schizophrenia risk loci remains a major challenge in the post-GWAS era. As a core regulatory region, the promoter plays a pivotal role in regulating gene expression. Multiple previous studies have shown that genetic variants in the promoter region contribute to disease susceptibility by affecting the expression level of risk genes. For example, rs11200638, a major genetic risk factor for wet age-related macular degeneration which lies in the HTRA1 promoter, modulates promoter activity and HTRA1 expression through affecting transcription factors AP2α and serum response factor (SRF) binding.108 A lung cancer risk-associated SNP rs4142441, located in the promoter of OSER1-AS1, regulates tumour suppressor gene OSER1-AS1 expression.109 rs213237 affected the ZNF323 promoter activity and confers schizophrenia risk by regulating ZNF323 expression.110 A SNP in the MDM2 promoter was reported to attenuate the p53 tumour suppressor pathway and accelerate tumour formation in humans.20 These findings highlighted the crucial role of functional variants in promoter region in disease susceptibility. In this study, we identified a functional variant (rs2270363) in the NMRAL1 promoter. We demonstrated the regulatory mechanism of this variant in regulating expression of NMRAL1. Our study not only provides a feasible framework to locate and identify the functional variants from the reported risk loci, but also demonstrates the regulatory mechanisms of rs2270363 and the potential pathophysiology role of NMRAL1 in schizophrenia.
Based on the reporter gene and eQTL results [i.e. the risk allele (A) of rs2270363 was associated with elevated expression of NMRAL1; Figs 1 and 2], NMRAL1 was predicted to be upregulated in schizophrenia cases compared with controls. However, expression analysis showed that NMRAL1 was downregulated in schizophrenia cases compared with controls. This inconsistency might be due to the following reasons. First, considering that the individuals used for gene expression and eQTL analysis were different, the number of subjects with the rs2270363 AA genotype in schizophrenia cases might be less than in controls in gene expression analysis, which will result in the observation of downregulation of NMRAL1 in schizophrenia cases. In fact, we examined the genotypic frequency of rs2270363 in tissue samples (258 cases, 279 controls) used for differential expression analysis.18 The genotypic frequency of AA (risk allele of rs2270363 is A) was 5.86% in schizophrenia cases and 8.60% in controls, the genotypic frequency of AG was 40.23% in schizophrenia cases and 32.62% in controls and the genotypic frequency of GG was 53.91% in schizophrenia cases and 58.78% in controls (Supplementary Table 10). The genotypic frequency of AA (risk allele of rs2270363 is A, which was associated with elevated NMRAL1 expression) is less in schizophrenia cases (5.86%) compared with controls (8.60%). As the A allele of rs2270363 is associated with elevated NMRAL1 expression, larger genotypic frequency of AA in controls might result in the observation of NMRAL1 downregulation in schizophrenia cases (compared with controls). Second, gene expression regulation is a complex process involving many factors (including multiple genetic variants and transcription factors). It is possible that other functional variants and rs2270363 act synergistically to regulate NMRAL1 expression. Finally, antipsychotics medication might also affect gene expression. Thus, the NMRAL1 expression level observed in schizophrenia cases might not reflect the genetic effects of rs2270363 accurately. More work is needed to investigate if NMRAL1 was dysregulated in schizophrenia cases.
Of note, rs2270363 was not among the top hits in the schizophrenia GWASs. The P of rs2270363 in recent two large-scale GWASs [i.e. PGC2 (34 241 cases, 45 604 controls and 1235 parent affected-offspring trios) and EAS + EUR (56 418 cases and 78 818 controls)] was 3.33 × 10−5 (ORA = 1.05) and 4.59 × 10−6 (ORA = 1.05), respectively.13,27 However, we noticed that the P-value of rs2270363 decreases with increasing sample size, suggesting that rs2270363 is a true risk variant. In addition, we found that the P-value of the index SNP (i.e. rs6500602) of the risk locus 16p13.3 (where rs2270363 is located) reached the genome-wide significance level in EAS + EUR (P = 2.39 × 10−9). Consistent with this, rs2270363 also showed genome-wide significant association with schizophrenia in our meta-analysis (P = 3.98 × 10−8, 60 136 cases and 86 647 controls; Table 1). These results indicate that rs2270363 is a true risk variant for schizophrenia and demonstrate that integrating genetic and functional findings will facilitate the identification of bona fide risk variants for schizophrenia. Besides, we noticed that the effect size of rs2270363 was small (ORA in European samples was 1.05), indicating the complexity of the genomic landscape of schizophrenia. Further studies considering both single and multiple variants are needed to elucidate the complex genetic aetiology of schizophrenia. In summary, our study demonstrates that rs2270363 may confer schizophrenia risk by modulating the expression of NMRAL1 (by dynamic binding to the bHLHZ proteins), a schizophrenia risk gene whose expression dysregulation might have pivotal roles in schizophrenia by affecting neurodevelopment and synaptic plasticity (Supplementary Fig. 19).
Supplementary Material
Acknowledgements
One of the brain eQTL datasets used in this study was generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: Pamela Sklar, Joseph Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Keisuke Hirai, Hiroyoshi Toyoshiba (Takeda Pharmaceuticals Company Limited), Enrico Domenici, Laurent Essioux (F. Hoffman-La Roche Ltd), Lara Mangravite, Mette Peters (Sage Bionetworks), Thomas Lehner, and Barbara Lipska (NIMH). The Genotype–Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. We thank Miss Qian Li for her technical assistance.
Abbreviations
- DEG
differentially expressed gene
- EMSA
electrophoretic mobility shift assay
- eQTL
expression quantitative trait loci
- GWAS
genome-wide association study
- mNSCs
mouse neural stem cells
- qPCR
quantitative real-time polymerase chain reaction
- SNP
single nucleotide polymorphism
Contributor Information
Junyang Wang, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Shiwu Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Xiaoyan Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Jiewei Liu, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Jinfeng Yang, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Yifan Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Wenqiang Li, Henan Mental Hospital, The Second Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan 453002, China; Henan Key Lab of Biological Psychiatry, International Joint Research Laboratory for Psychiatry and Neuroscience of Henan, Xinxiang Medical University, Xinxiang, Henan 453002, China.
Yongfeng Yang, Henan Mental Hospital, The Second Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan 453002, China; Henan Key Lab of Biological Psychiatry, International Joint Research Laboratory for Psychiatry and Neuroscience of Henan, Xinxiang Medical University, Xinxiang, Henan 453002, China.
Jiao Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Rui Chen, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Kaiqin Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Di Huang, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Yixing Liu, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China.
Luxian Lv, Henan Mental Hospital, The Second Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan 453002, China; Henan Key Lab of Biological Psychiatry, International Joint Research Laboratory for Psychiatry and Neuroscience of Henan, Xinxiang Medical University, Xinxiang, Henan 453002, China.
Ming Li, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Xiao Xiao, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Xiong Jian Luo, Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China; Zhongda Hospital, School of Life Sciences and Technology, Advanced Institute for Life and Health, Southeast University, Nanjing, Jiangsu 210096, China; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Funding
This study was supported by the National Nature Science Foundation of China (U2102205 and 31970561 to X.J.L., U1904130 to W.Q.L.), the Distinguished Young Scientists grant of the Yunnan Province (202001AV070006), the Key Research Project of the Yunnan Province (202101AS070055), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB17) and the Western Light Innovative Research Team of Chinese Academy of Sciences.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Thaker GK, Carpenter WT. Advances in schizophrenia. Nat Med. 2001;7(6):667–671. [DOI] [PubMed] [Google Scholar]
- 2. Kessler RC, Amminger GP, Aguilar-Gaxiola S, Alonso J, Lee S, Ustun TB. Age of onset of mental disorders: A review of recent literature. Curr Opin Psychiatry. 2007;20(4):359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saha S, Chant D, McGrath J. A systematic review of mortality in schizophrenia—Is the differential mortality gap worsening over time? Arch Gen Psychiatry. 2007;64(10):1123–1131. [DOI] [PubMed] [Google Scholar]
- 4. Crown WH, Neslusan C, Russo PA, Holzer S, Ozminkowski R, Croghan T. Hospitalization and total medical costs for privately insured persons with schizophrenia. Adm Policy Ment Health. 2001;28(5):335–351. [DOI] [PubMed] [Google Scholar]
- 5. Jungbauer J, Wittmund B, Dietrich S, Angermeyer MC. Subjective burden over 12 months in parents of patients with schizophrenia. Arch Psychiatr Nurs. 2003;17(3):126–134. [DOI] [PubMed] [Google Scholar]
- 6. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait—Evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. [DOI] [PubMed] [Google Scholar]
- 7. McGuffin P, Asherson P, Owen M, Farmer A. The strength of the genetic effect. Is there room for an environmental influence in the aetiology of schizophrenia? Br J Psychiatry. 1994;164(5):593–599. [DOI] [PubMed] [Google Scholar]
- 8. Kahn RS, Sommer IE, Murray RM, et al. Schizophrenia. Nat Rev Dis Primers. 2015;1:15067. [DOI] [PubMed] [Google Scholar]
- 9. O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. [DOI] [PubMed] [Google Scholar]
- 10. Pardinas AF, Holmans P, Pocklington AJ, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu H, Yan H, Li J, et al. Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol Psychiatry. 2017;22(7):954–960. [DOI] [PubMed] [Google Scholar]
- 12. Li ZQ, Chen JH, Yu H, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576–1583. [DOI] [PubMed] [Google Scholar]
- 13. Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014:511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huo YX, Li SW, Liu JW, Li XY, Luo XJ. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat Commun. 2019;10:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang H, Cai X, Li H-J, et al. Functional genomics identify a regulatory risk variation rs4420550 in the 16p11.2 schizophrenia-associated locus. Biol Psychiatry. 2020;89(3):246–255. [DOI] [PubMed] [Google Scholar]
- 16. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhie SK, Schreiner S, Witt H, et al. Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci Adv. 2018;4(12):eaav8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaffe AE, Straub RE, Shin JH, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21(8):1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bond GL, Hu WW, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602. [DOI] [PubMed] [Google Scholar]
- 21. Luo XB, Yang WL, Ye DQ, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7(6):e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giraud M, Taubert R, Vandiedonck C, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448(7156):934–937. [DOI] [PubMed] [Google Scholar]
- 23. Li K, Li Y, Wang J, et al. A functional missense variant in ITIH3 affects protein expression and neurodevelopment and confers schizophrenia risk in the Han Chinese population. J Genet Genomics. 2020;47(5):233–248. [DOI] [PubMed] [Google Scholar]
- 24. Li YF, Ma CG, Li WQ, et al. A missense variant in NDUFA6 confers schizophrenia risk by affecting YY1 binding and NAGA expression. Mol Psychiatry. 2021;6(11):6896–6911. [DOI] [PubMed] [Google Scholar]
- 25. Luo XJ, Diao HB, Wang JK, Zhang H, Zhao ZM, Su B. Association of haplotypes spanning PDZ-GEF2, LOC728637 and ACSL6 with schizophrenia in Han Chinese. J Med Genet. 2008;45(12):818–826. [DOI] [PubMed] [Google Scholar]
- 26. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lam M, Chen CY, Li ZQ, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sepulveda-Diaz JE, Naini SMA, Huynh MB, et al. HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer's disease-related tau pathology. Brain. 2015;138:1339–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung E, Choi Y, Park J, et al. Intracellular delivery of Parkin rescues neurons from accumulation of damaged mitochondria and pathological alpha-synuclein. Sci Adv. 2020;6(18):eaba1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaw KTY, Utsuki T, Rogers J, et al. Phenserine regulates translation of beta-amyloid precursor protein mRNA by a putative interleukin-1 responsive element, a target for drug development. Proc Natl Acad Sci USA. 2001;98(13):7605–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagy Z, Seneviratne JA, Kanikevich M, et al. An ALYREF–MYCN coactivator complex drives neuroblastoma tumorigenesis through effects on USP3 and MYCN stability. Nat Commun. 2021;12(1):1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sailer MHM, Sarvepalli D, Bregere C, et al. An enzyme- and serum-free neural stem cell culture model for EMT investigation suited for drug discovery. J Vis Exp. 2016;114:54018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azari H, Sharififar S, Rahman M, Ansari S, Reynolds BA. Establishing embryonic mouse neural stem cell culture using the neurosphere assay. J Vis Exp. 2011;47:e2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang ZH, Zhou DY, Li HJ, et al. The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function, and mushroom dendritic spine. Mol Psychiatry. 2020;25(1):48–66. [DOI] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 36. Zhou YY, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y, Li S, Li X, et al. Convergent lines of evidence support NOTCH4 as a schizophrenia risk gene. J Med Genet. 2021;58:666–678. [DOI] [PubMed] [Google Scholar]
- 38. Wang JY, Li XY, Li HJ, et al. Integrative analyses followed by functional characterization reveal TMEM180 as a schizophrenia risk gene. Schizophr Bull. 2021;47:1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kong H, Fan Y, Xie J, et al. AQP4 knockout impairs proliferation, migration and neuronal differentiation of adult neural stem cells. J Cell Sci. 2008;121(24):4029–4036. [DOI] [PubMed] [Google Scholar]
- 40. Yang CP, Li XY, Wu Y, et al. Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun. 2018;9:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cai X, Yang ZH, Li HJ, Xiao X, Li M, Chang H. A human-specific schizophrenia risk tandem repeat affects alternative splicing of a human-unique isoform AS3MTd2d3 and mushroom dendritic spine density. Schizophr Bull. 2021;47(1):219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Srivastava DP, Woolfrey KM, Penzes P. Analysis of dendritic spine morphology in cultured CNS neurons. J Vis Exp. 2011;53:e2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 2008;3(4):e1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nott A, Holtman IR, Coufal NG, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366(6469):1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis CA, Hitz BC, Sloan CA, et al. The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corre S, Galibert MD. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18(5):337–348. [DOI] [PubMed] [Google Scholar]
- 47. Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7(11):2110–2119. [DOI] [PubMed] [Google Scholar]
- 48. Wood MA, Walker WH. USF1/2 transcription factor DNA-binding activity is induced during rat Sertoli cell differentiation. Biol Reprod. 2009;80(1):24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc–Max recognition sites. Cell. 1993;72(2):223–232. [DOI] [PubMed] [Google Scholar]
- 50. Wu Y, Li XY, Liu JW, Luo XJ, Yao YG. Schizophrenia DB2.0: An updated comprehensive resource for schizophrenia research. Hum Genet. 2020;139(10):1285–1297. [DOI] [PubMed] [Google Scholar]
- 51. He X, Fuller CK, Song Y, et al. Sherlock: Detecting gene–disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92(5):667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu ZH, Zhang FT, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 53. Ma CG, Gu CJ, Huo YX, Li XY, Luo XJ. The integrated landscape of causal genes and pathways in schizophrenia. Transl Psychiatry. 2018;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim JY, Liu CY, Zhang FY, et al. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell. 2012;148(5):1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kang EC, Burdick KE, Kim JY, et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72(4):559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ishizuka K, Kamiya A, Oh EC, et al. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473(7345):92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35(3):528–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. [DOI] [PubMed] [Google Scholar]
- 59. Owen MJ, O’Donovan MC, Thapar A, Craddock N. Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry. 2011;198(3):173–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry. 2012;17(12):1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berridge MJ. Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion. 2013;7(1):2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hertzberg L, Katsel P, Roussos P, Haroutunian V, Domany E. Integration of gene expression and GWAS results supports involvement of calcium signaling in schizophrenia. Schizophr Res. 2015;164(1–3):92–99. [DOI] [PubMed] [Google Scholar]
- 63. Martins-de-Souza D, Gattaz WF, Schmitt A, et al. Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J Neural Transm. 2009;116(3):275–289. [DOI] [PubMed] [Google Scholar]
- 64. McCarthy NS, Melton PE, Ward SV, et al. Exome array analysis suggests an increased variant burden in families with schizophrenia. Schizophr Res. 2017;185:9–16. [DOI] [PubMed] [Google Scholar]
- 65. Berezin V, Walmod PS, Filippov M, Dityatev A. Targeting of ECM molecules and their metabolizing enzymes and receptors for the treatment of CNS diseases. Prog Brain Res. 2014;214:353–388. [DOI] [PubMed] [Google Scholar]
- 66. Pawelczyk T, Grancow-Grabka M, Trafalska E, Szemraj J, Zurner N, Pawelczyk A. An increase in plasma brain derived neurotrophic factor levels is related to n-3 polyunsaturated fatty acid efficacy in first episode schizophrenia: secondary outcome analysis of the OFFER randomized clinical trial. Psychopharmacology. 2019;236(9):2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Funk AJ, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacol. 2012;37(4):896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57(1):65–73. [DOI] [PubMed] [Google Scholar]
- 69. Osimo EF, Beck K, Marques TR, Howes OD, Reis Marques T. Synaptic loss in schizophrenia: A meta-analysis and systematic review of synaptic protein and mRNA measures. Mol Psychiatry. 2019;24(4):549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van Berlekom AB, Muflihah CH, Snijders GJLJ, et al. Synapse pathology in schizophrenia: A meta-analysis of postsynaptic elements in postmortem brain studies. Schizophr Bull. 2020;46(2):374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71(12):1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12(7):2685–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Holtmaat AJGD, Trachtenberg JT, Wilbrecht L, et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45(2):279–291. [DOI] [PubMed] [Google Scholar]
- 75. Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9(9):1117–1124. [DOI] [PubMed] [Google Scholar]
- 76. Ayer DE, Kretzner L, Eisenman RN. Mad—A heterodimeric partner for max that antagonizes myc transcriptional activity. Cell. 1993;72(2):211–222. [DOI] [PubMed] [Google Scholar]
- 77. Blackwood EM, Eisenman RN. Max—A helix–loop–helix zipper protein that forms a sequence-specific DNA-binding complex with myc. Science. 1991:251(4998):1211–1217. [DOI] [PubMed] [Google Scholar]
- 78. Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22(39):8042–8047. [DOI] [PubMed] [Google Scholar]
- 79. Howcroft TK, Murphy C, Weissman JD, Huber SJ, Sawadogo M, Singer DS. Upstream stimulatory factor regulates major histocompatibility complex class I gene expression: The U2 Delta E4 splice variant abrogates E-box activity. Mol Cell Biol. 1999;19(7):4788–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pognonec P, Boulukos KE, Aperlo C, et al. Cross-family interaction between the bHLHZip USF and bZip Fra1 proteins results in down-regulation of AP1 activity. Oncogene. 1997;14(17):2091–2098. [DOI] [PubMed] [Google Scholar]
- 81. Rodriguez CI, Girones N, Fresno M. Cha, a basic helix–loop–helix transcription factor involved in the regulation of upstream stimulatory factor activity. J Biol Chem. 2003;278(44):43135–43145. [DOI] [PubMed] [Google Scholar]
- 82. Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an Inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16(23):7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu SJ, Pena A, Korcz A, Soprano DR, Soprano KJ. Overexpression of Mxi1 inhibits the induction of the human ornithine decarboxylase gene by the Myc/Max protein complex. Oncogene. 1996;12(3):621–629. [PubMed] [Google Scholar]
- 84. Fisher F, Crouch DH, Jayaraman PS, Clark W, Gillespie DAF, Goding CR. Transcription activation by Myc and Max—Flanking sequences target activation to a subset of CACGTG motifs in-vivo. EMBO J. 1993;12(13):5075–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gregor PD, Sawadogo M, Roeder RG. The adenovirus major late transcription factor USF is a member of the helix loop helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4(10):1730–1740. [DOI] [PubMed] [Google Scholar]
- 86. Gusev A, Mancuso N, Won H, et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50(4):538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zheng XF, Dai XY, Zhao YM, et al. Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein. Proc Natl Acad Sci USA. 2007;104(21):8809–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhao YM, Zhang JF, Li HY, et al. An NADPH sensor protein (HSCARG) down-regulates nitric oxide synthesis by association with argininosuccinate synthetase and is essential for epithelial cell viability. J Biol Chem. 2008;283(16):11004–11013. [DOI] [PubMed] [Google Scholar]
- 89. Peng YY, Xu RD, Zheng XF. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 2014;10(4):e1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang M, Hu B, Li TT, et al. A CRM1-dependent nuclear export signal controls nucleocytoplasmic translocation of HSCARG, which regulates NF-κB activity. Traffic. 2012;13(6):790–799. [DOI] [PubMed] [Google Scholar]
- 91. Zang W, Zheng X. Structure and functions of cellular redox sensor HSCARG/NMRAL1, a linkage among redox status, innate immunity, DNA damage response, and cancer. Free Radic Biol Med. 2020;160:768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fitzgerald KA, McWhirter SM, Faia KL, et al. IKK epsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–496. [DOI] [PubMed] [Google Scholar]
- 93. Gan QN, Li TT, Hu B, Lian M, Zheng XF. HSCARG inhibits activation of NF-kappa B by interacting with I kappa B kinase-beta. J Cell Sci. 2009;122(22):4081–4088. [DOI] [PubMed] [Google Scholar]
- 94. Saha SK, Pietras EM, He JQ, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25(14):3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xiao WC, Peng YY, Liu Y, Li Z, Li SL, Zheng XF. HSCARG inhibits NADPH oxidase activity through regulation of the expression of p47phox. PLoS One. 2013;8(3):e59301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Huang TT, Nijman SMB, Mirchandani KD, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8(4):339–347. [DOI] [PubMed] [Google Scholar]
- 97. Gandal MJ, Zhang P, Hadjimichael E, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018:362(6420):eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Stahl EA, Breen G, Forstner AJ, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Noto C, Maes M, Ota VK, et al. High predictive value of immune-inflammatory biomarkers for schizophrenia diagnosis and association with treatment resistance. World J Biol Psychiatry. 2015;16(6):422–429. [DOI] [PubMed] [Google Scholar]
- 102. Smyth AM, Lawrie SM. The neuroimmunology of schizophrenia. Clin Psychopharmacol Neurosci. 2013;11(3):107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bergen SE, O’Dushlaine CT, Ripke S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17(9):880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Erjavec GN, Konjevod M, Perkovic MN, et al. Short overview on metabolomic approach and redox changes in psychiatric disorders. Redox Biol. 2018;14:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. O’Donnell P. Cortical interneurons, immune factors and oxidative stress as early targets for schizophrenia. Eur J Neurosci. 2012;35(12):1866–1870. [DOI] [PubMed] [Google Scholar]
- 106. Smythies JR. Oxidative reactions and schizophrenia: A review–discussion. Schizophr Res. 1997;24(3):357–364. [DOI] [PubMed] [Google Scholar]
- 107. Steullet P, Cabungcal H, Coyle J, et al. Oxidative stress-driven parvalbumin interneuron impairment as a common mechanism in models of schizophrenia. Mol Psychiatry. 2017;22(7):936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. [DOI] [PubMed] [Google Scholar]
- 109. Xie WJ, Wang YH, Zhang Y, et al. Single-nucleotide polymorphism rs4142441 and MYC co-modulated long non-coding RNA OSER1-AS1 suppresses non-small cell lung cancer by sequestering ELAVL1. Cancer Sci. 2021;112:2272–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li SW, Li XY, Liu JW, et al. Functional variants fine-mapping and gene function characterization provide insights into the role of ZNF323 in schizophrenia pathogenesis. Am J Med Genet B. 2021;186(1):28–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in CMC, LIBD, ENCODE, PsychENCODE, UCSC genome browser, SZDB, RegulomeDB, GTEx Portal and the Human Protein Atlas. These data were derived from the following resources available in the public domain: CMC, https://www.nimhgenetics.org/resources/commonmind; LIBD, http://eqtl.brainseq.org/; ENCODE, https://www.encodeproject.org/; PsychENCODE, http://resource.psychencode.org/; UCSC genome browser, http://genome.ucsc.edu/; SZDB, http://www.szdb.org/; RegulomeDB, https://www.regulomedb.org/; GTEx Portal, https://www.gtexportal.org/home/; the Human Protein Atlas, https://www.proteinatlas.org/. Other data can be available from the corresponding author upon reasonable request.