Abstract
An Escherichia coli O157:H7 strain isolated from a patient with hemorrhagic colitis was found to exhibit two slightly different colony morphology types on differential medium. Each morphological type, designated TT12A and TT12B, was isolated, and serological testing using various assays confirmed that both strains carried the O157 and the H7 antigens. Biochemical testing showed that the strains had identical profiles on AP120E analysis and, like typical O157:H7 strains, did not ferment sorbitol or exhibit β-glucuronidase activity. Analysis with a multiplex PCR assay showed that TT12B did not carry the gene for either Shiga toxin 1 (Stx1) or Stx2, whereas these genes were present in TT12A and the toxins were produced. Apart from that, both strains carried the +93 gusA mutation, the cluster I ehxA gene for enterohemolysin, and the eae gene for γ-intimin, which are all characteristics of the O157:H7 serotype. Phenotypic assays confirmed that both strains exhibited enterohemolysin activity and the attachment and effacing lesion on HeLa cells. Multilocus enzyme electrophoresis analysis showed that the strains are closely related genetically and belong in the same clonal group. Pulsed-field gel electrophoresis (PFGE) typing of XbaI-digested genomic DNA revealed that the two strains differed by two bands but shared 90% similarity and clustered in the same clade. All other non-Stx-producing O157:H7 strains examined clustered in a major clade that was distinct from that of Stx-producing O157:H7 strains. The findings that TT12B was identical to TT12A, except for Stx production, and its PFGE profile is also more closely related to that of Stx-producing O157:H7 strains suggest that TT12B was derived from TT12A by the loss of both stx genes.
Serotype O157:H7 is the prototypic enterohemorrhagic Escherichia coli (EHEC) strain and the predominant serotype implicated in food-borne infections worldwide. The EHEC group is characterized by its unique virulence factors. These include the eae gene, which encodes intimin, a protein essential for the cellular attachment and effacing (A/E) lesion, and a 60-mDa plasmid that carries the ehxA gene, which encodes enterohemolysin. However, the production of Shiga toxins (Stx), particularly Stx1 and Stx2, encoded by the phage-borne stx1 and stx2 genes, is probably the most defining trait of EHEC, as these two toxins are most often implicated in disease in humans.
Isogenic, non-Stx-producing variants of EHEC provide useful controls in studying the role of Stx in pathogenesis in animal models. The production of Stx can be eliminated from strains by inducing the Stx-encoding phages with sublethal dosages of UV or antibiotics, although prophage release often results in cell lysis (16). Alternatively, Gunzer et al. (12) deleted the Stx2-encoding sequences from a wild-type O157:H7 strain with a suicide vector and thus constructed a toxin deletion isogen that did not produce Stx. In addition, Karch et al. (14) reported spontaneous loss of Stx production by EHEC strains other than O157:H7 during routine subculture. This process, which did not appear to be medium dependent, suggested that the loss of stx genes occurs frequently in Stx-producing E. coli (14). In contrast, O157:H7 strains seem to be an exception, since the stx genes in O157:H7 appear to be more stably maintained, so that non-Stx-producing O157:H7 strains are rarely encountered (18).
In our laboratory and during routine culturing of an O157:H7 isolate (strain TT12) on Rainbow agar (Biolog, Inc., Hayward, Calif.), a selective and differential plating medium for Stx-producing E. coli, two slightly different colony morphology types were observed. The strain designated TT12A had dark gray colonies, whereas colonies of the strain designated TT12B had a lighter, cream-colored appearance. In this study, we characterized these two strains and obtained data which suggest that TT12B was derived from TT12A by the loss of the stx1 and stx2 genes.
MATERIALS AND METHODS
Bacterial strains.
The following three strains of serotype O157:H7 are all patient isolates from sporadic cases of hemorrhagic colitis or hemolytic-uremic syndrome in Japan between 1992 and 1994: strain TT12, which produces both Stx1 and Stx2; strain TT7, which produces only Stx1; and strain TT8, which produces only Stx2. Strains ATCC 35150 and ATCC 43895 are both O157:H7 strains that produce Stx1 and Stx2 and were obtained from the American Type Culture Collection (ATCC), Manassas, Va. Strains MA9, MA11, and MA36 are non-Stx-producing O157:H7 strains isolated from meats in Asia and were obtained from M. Nishibuchi, Kyoto, Japan. Strains CV261 and CV267 are also non-Stx-producing O157:H7 strains isolated from cattle in France and were obtained from C. Vernozy-Roland, Lyon, France (20). Strain CDCG5244 is the O157:H7 reference strain used by the Centers for Disease Control and Prevention (CDC), Atlanta, Ga., for pulsed-field gel electrophoresis (PFGE) typing.
Biochemical characterization.
Strains TT12A and TT12B were analyzed for biochemical phenotypes using the AP120E test kit (bioMerieux, Marcy l'Etoile, France) and were also examined for several trait O157:H7 phenotypes. The inability to ferment sorbitol was determined using cefixime tellurite sorbitol MacConkey (CT-SMAC) agar (22). The absence of β-glucuronidase (GUD) activity was also tested on CT-SMAC plates using a modification of the methylumbelliferone-β-d-glucuronide (MUG) assay (10). Briefly, after streak plating of the isolates, a ColiComplete disk (BioControl, Bellevue, Wash.) saturated with the MUG substrate was aseptically placed on the streak zone in which the heaviest growth was expected. After overnight incubation at 37°C, the plates were examined under long-wave UV light for bluish fluorescence caused by GUD activity on MUG in the medium around the disk.
Serological characterization.
Strains were tested serologically for the O157 antigen using various assays (Table 1). The presence of the H7 antigen was determined with a latex agglutination test (RIM E. coli O157:H7; Remel, Lenexa, Kans.), and the production of Stx1 and Stx2 toxins was tested with a reverse passive latex agglutination test (Verotox-F; Denka Seiken, Japan). Most of these tests, listed in Table 1, are commercially available and were performed according to the manufacturers' instructions.
TABLE 1.
Biochemical, serological, and genetic analyses of strains TT12A and TT12B
| Test | Manufacturer | Reaction
|
|
|---|---|---|---|
| TT12A | TT12B | ||
| Biochemical | |||
| AP120E | bioMerieux | 7144162b | 7144162 |
| Sorbitol fermentation | NAa | − | − |
| GUD | BioControl | − | − |
| growth on CT-SMAC | NA | + | + |
| Serological | |||
| RIM anti-O157 | Remel | + | + |
| RIM anti-H7 | Remel | + | + |
| VIDAS E. coli O157 | bioMerieux | + | + |
| Reveal O157 | Neogen | + | + |
| VIP EHEC O157 | BioControl | + | + |
| Verotox-F Stx1 | Denka Seiken | + | − |
| Verotox-F Stx2 | Denka Seiken | + | − |
| Genetic | |||
| Probe | |||
| DIG-stx1 | NA | + | − |
| DIG-stx2 | NA | + | − |
| PCR | |||
| stx1 | NA | + | − |
| stx2 | NA | + | − |
| gusA | NA | + | + |
| γ-eae | NA | + | + |
| ehxA | NA | + | + |
NA, not applicable.
Analytical profile index number.
Genetic characterization.
Strains TT12A and TT12B were examined by multiplex PCR for the presence of five trait EHEC genes and virulence factors (9). The gene targets, with the expected sizes of the amplification products in parentheses, are stx1 (348 bp); stx2 (584 bp); eae, which encodes a γ-intimin derivative (397 bp); cluster 1 ehxA (166 bp), which encodes enterohemolysin; and gusA (252 bp), which encodes GUD. The last marker is a single-base mutation at +93 in gusA that is unique to O157:H7 (5) and detected by mismatch PCR. The multiplex PCR for these five gene products was set up as described previously (9), and amplification was done for 25 cycles using AmpliTaq Gold polymerase enzyme (PE Applied Biosystems, Foster City, Calif.), with an annealing temperature of 56°C for 1 min. The PCR products were examined by agarose (1%) gel electrophoresis in 1× Tris-borate-EDTA buffer, pH 8.2.
Both strains were also examined by multilocus enzyme electrophoresis (MLEE) for genetic relatedness. MLEE looks at the electrophoretic mobilities of 20 E. coli housekeeping enzymes for variations that are indicative of charge-altering genetic changes. The MLEE analysis was performed by T. S. Whittam (Michigan State University) using previously described procedures (19).
Phenotypic assays for virulence factors.
The enterohemolytic activities of strains TT12A and TT12B were tested on washed sheep blood agar plates containing calcium. The medium was prepared by supplementing tryptic soy agar with 10 mM CaCl2 (pH 7.3) and 5% defibrinated sheep blood that had been washed three times in phosphate-buffered saline (PBS). Both strains were streak plated onto the medium, incubated overnight at 37°C, and checked for hemolysis surrounding the colonies.
The expression of the A/E lesion by TT12A and TT12B was examined with the in vitro fluorescence actin staining assay using HeLa cells. Briefly, about 105 HeLa cells, maintained in Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum, were seeded on 12-mm-round glass coverslips and grown at 37°C in a 5% CO2 incubator. The monolayers were infected with strain TT12A or TT12B for 3.5 h, washed four times with PBS to remove nonadherent bacteria, and incubated for another 3.5 h in fresh Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum. Postinfection, the HeLa monolayers were washed three times with PBS, fixed with 3.0% paraformaldehyde in PBS (pH 7.2) for 30 min, and then washed three more times with PBS. To stain filamentous actin, the fixed cells were permeabilized with 20 μl of 0.1% Triton X-100 in PBS in the presence of phalloidin-Texas red. Stained samples were visualized by phase-contrast and immunofluorescence microscopy and photographed as described elsewhere (11).
PFGE and Southern blotting.
To look for similarities in genetic fingerprint patterns, genomic DNA from strains TT12A and TT12B was digested with XbaI or ApaI and analyzed by PFGE. The PFGE profiles of strains TT12A and TT12B, obtained with XbaI-digested genomic DNA, were also compared to the profiles of other Stx-producing and non-Stx-producing O157:H7 strains. Agarose plugs of bacterial strains were prepared and digested as described previously (4), and PFGE was performed with the CHEF MAPPER system (Bio-Rad Laboratories, Hercules, Calif.), in a 1% agarose gel and 0.5× Tris-borate-EDTA buffer at 14°C. The running time was 18 h at 6 V/cm, with initial and final switch times of 2.16 and 54.17 s, respectively. Genomic DNA of strain CDCG5244, also digested with XbaI, was used as the reference marker along with the λ ladder CHEF DNA size standard. The gel was stained with ethidium bromide (1 μg/ml), and the gel image, captured with the Gel Doc 1000 system (Bio-Rad), was analyzed with Molecular Analyst fingerprinting software (Bio-Rad).
To verify the presence or absence of stx genes in the various O157:H7 strains and to size the XbaI DNA fragments carrying the stx genes, the PFGE gel was transferred by Southern blotting onto a nylon membrane and sequentially probed with stx1- and stx2-specific DNA probes that were generated by PCR and labeled with digoxigenin (DIG)-11-dUTP during amplification (21). The stx1 and stx2 probes are 173 and 364 bp, respectively, in size, and both probes are specific for stx gene regions that do not overlap with that amplified by the stx primers used in the multiplex PCR described above. Procedures for hybridization and chemiluminescent detection of specific reactions were performed as described previously (21).
RESULTS
Biochemical characterization.
Strains TT12A and TT12B showed identical biochemical profiles on AP120E analysis, and both had characteristic traits of O157:H7 strains in that they grew on CT-SMAC medium and did not ferment sorbitol or exhibit GUD activity (Table 1). These two strains therefore were indistinguishable by standard biochemical assays used to identify O157:H7 strains.
Serological characterization.
Serological typing by various kits confirmed that TT12A and TT12B expressed O157 and H7 antigens and hence are isolates of the O157:H7 serotype. However, TT12B was found not to produce either Stx1 or Stx2, while both toxins were produced by TT12A (Table 1).
Genetic characterization.
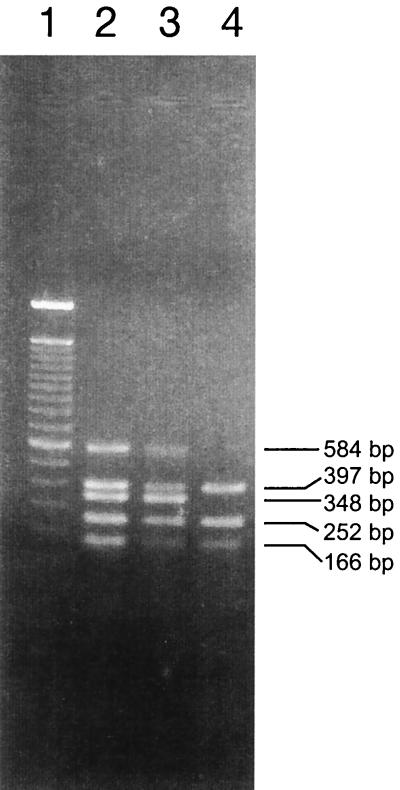
Similarly, PCR analysis showed that TT12A and TT12B carried characteristic O157:H7 genes and virulence factors, with the exception that TT12B did not have either of the stx genes (Fig. 1 and Table 1). Like an O157:H7 control strain (Fig. 1, lane 2), TT12A and TT12B carried the +93 T-to-G-base change in the gusA gene (uidA) that is unique to O157:H7. Both had the eae gene for the γ-intimin derivative that is found in the EHEC I clonal type (17), which consists mostly of the two closely related O157:H7 and O55:H7 serotypes (7). Furthermore, both strains also carried the ehxA gene sequence present in cluster I Stx-producing strains, which includes the O157:H7 serotype (3). The only distinction between the two strains was the absence of stx1 and stx2 gene sequences in TT12B (Fig. 1, lane 4). This is consistent with the serological data showing that TT12B did not produce either toxin. Multiplex PCR analysis of other non-Stx-producing O157:H7 strains showed that they also carried all the O157:H7 markers tested, except for the stx1 and stx2 genes (data not shown).
FIG. 1.
Agarose gel electrophoresis of DNA fragments amplified by multiplex PCR for EHEC virulence genes and other markers of the O157:H7 serotype. Lanes: 1, 100-bp DNA ladder (Life Technologies, Rockville, Md.); 2, ATCC 35150 (O157:H7 serotype), positive control; 3, TT12A; 4, TT12B. The sizes of amplified products are shown at the right of the gel. The products are (from top to bottom) stx2, eae, stx1, gusA, and ehxA.
Analysis of TT12A and TT12B by MLEE also showed the strains to be closely related genetically, as both exhibited electrophoretic type 1 profiles typical of O157:H7 strains (7).
Phenotypic assays.
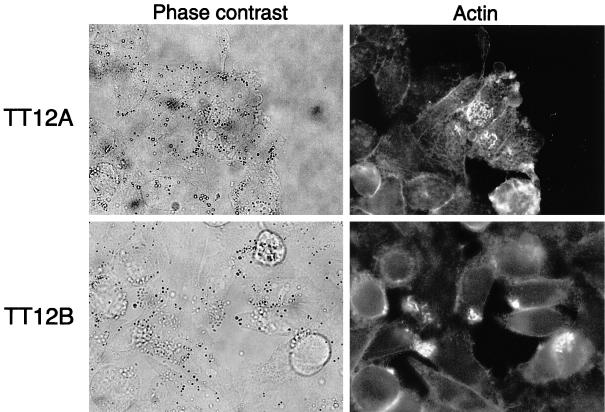
The TT12A and TT12B strains exhibited enterohemolysin activity on blood agar plates, and therefore express the ehxA gene. Infection studies on HeLa cell monolayers showed that both strains adhered to HeLa cells and induced the accumulation of cytoskeletal actin beneath the adherent bacteria (Fig. 2). These results indicate that the TT12A and TT12B strains express eae to produce the intimin protein and have the ability to adhere to HeLa cells and cause the A/E lesion.
FIG. 2.
Photographs showing the infection by and the A/E lesion on HeLa cell monolayers caused by TT12A and TT12B. Shown are bacterial adherence (phase-contrast) and the resulting accumulation of cytoskeletal actin beneath the adherent bacteria as viewed under immunofluorescent microscopy.
PFGE and Southern blotting.
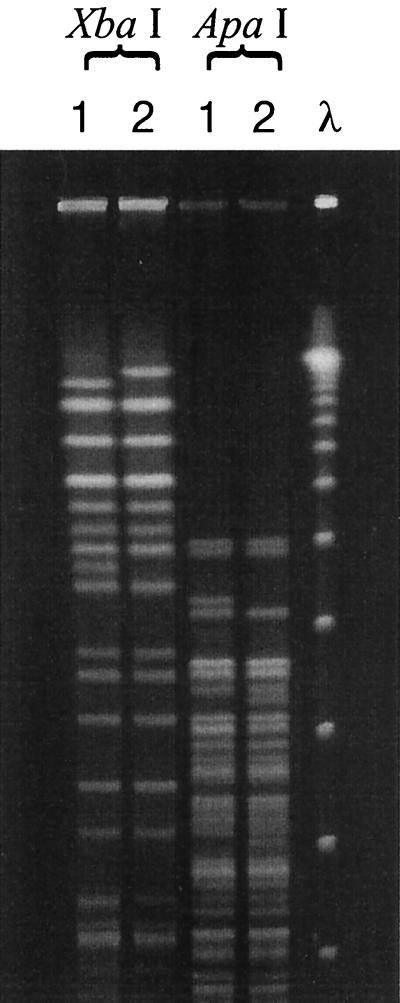
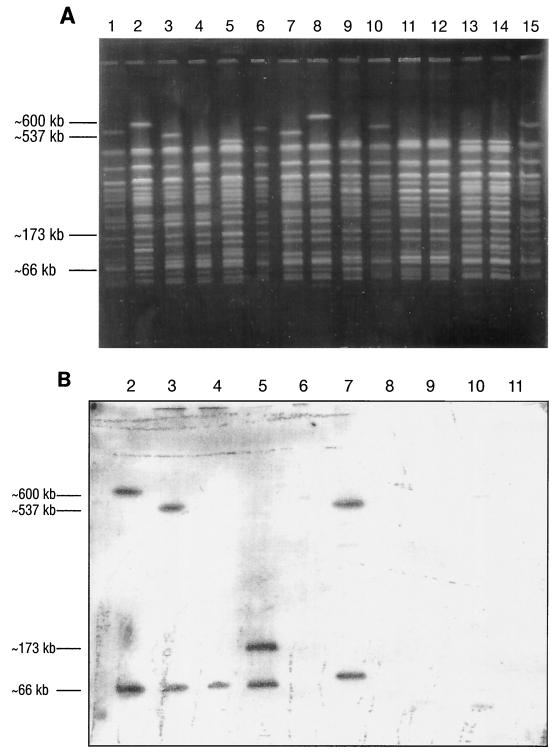
Since TT12A and TT12B are indistinguishable by biochemical assays or by phenotypic, serologic, or PCR analysis, they were examined by PFGE and their profiles were compared to those of other Stx-producing and non-Stx-producing O157:H7 strains. Analysis of XbaI-digested DNA revealed that the genomic profiles of TT12A and TT12B differed by only two fragments (Fig. 3). Analysis of ApaI-digested DNA also showed close similarities between these two strains, with only few band differences (Fig. 3). Comparison of the XbaI profiles of these two strains with those of other Stx-producing and non-Stx-producing O157:H7 strains showed that the profiles of TT12A (Fig. 4A, lane 7) and TT12B (Fig. 4A, lane 8) resembled those of strains ATCC 35150 (Fig. 4A, lane 3) and ATCC 43895 (Fig. 4A, lane 2), respectively, but the profiles of all these strains differed from those of the non-Stx-producing O157:H7 strains examined (Fig. 4A, lanes 9 and 11 to 14).
FIG. 3.
Agarose gel of PFGE of XbaI- and ApaI-digested genomic DNA of the Stx-producing TT12A (lanes 1) and the non-Stx producing TT12B (lanes 2) strains. Lane λ shows the λ ladder CHEF DNA size standard (Bio-Rad).
FIG. 4.
(A) Agarose gel of PFGE of XbaI digestion patterns of Stx-producing and non-Stx-producing E. coli O157:H7 strains. The samples, with the Stx phenotype shown in parentheses, are as follows: lanes 1, 6, 10, and 15, CDCG5244, PFGE reference strain (Stx1, Stx2); lane 2, ATCC 43895 (Stx1, Stx2); lane 3, ATCC 35150 (Stx1, Stx2); lane 4, TT7 (Stx1); lane 5, TT8 (Stx2); lane 7, TT12A (Stx1, Stx2); lane 8, TT12B (no Stx); lane 9, MA9 (no Stx); lane 11, MA11 (no Stx); lane 12, MA36 (no Stx); lane 13, CV261 (no Stx); and lane 14, CV267 (no Stx). The fragment sizes shown at the left are transposed from panel B and are the estimated sizes of the fragments that hybridized with stx1 and stx2 probes on a Southern blot (panel B). (B) Southern blot of panel A, sequentially hybridized with DIG-labeled stx1 and stx2 DNA probes. For most E. coli O157:H7 strains, the stx2 and stx1 genes resided on the larger (∼500- to 600-kb) and smaller (∼66-kb) XbaI bands, respectively. The lane numbers correspond to numbers in panel A. The fragment sizes, shown at the left, were estimated using the λ ladder CHEF DNA size standard (Bio-Rad).
A Southern blot of the PFGE gel showed that in most Stx1-producing O157:H7 strains, an XbaI fragment of about 66 kb hybridized with the stx1 probe (Fig. 4B, lanes 2 to 4). In contrast, the stx1 gene of strain TT12A was on a fragment of about 75 kb (Fig. 4B, lane 7). Subsequent hybridization with the stx2 probe showed that the stx2 gene resided on a 600-kb XbaI fragment in strain ATCC 43895 (Fig. 4B, lane 2), but in strains ATCC 35150 (Fig. 4B, lane 3) and TT12A (Fig. 4B, lane 7), the gene was on a fragment of about 537 kb. Strain TT12B (Fig. 4B, lane 8) and the other non-Stx-producing O157:H7 strains (Fig. 4B, lanes 9 and 11) did not hybridize with either probe, hence confirming that these strains did not carry stx1 or stx2 gene sequences. The CDC reference strain CDCG5244 produces both Stx1 and Stx2, and PCR analysis verified the presence of both toxin genes in this strain (data not shown). In the Southern blot of CDCG5244 (Fig. 4B, lanes 6 and 10), the stx2 gene resided on an XbaI fragment that was slightly larger than the 537-kb fragment observed for TT12A (Fig. 4B, lane 7), but the stx1 gene was on a much smaller band, one of less than 60 kb. Both of these hybridization products were only weakly visible on the original blot; as a result, they did not reproduce well in Fig. 4. The DNA fragments in the PFGE profiles of CDCG5244 (Fig. 4A, lanes 6 and 10) were not as intense as those of other strains, suggesting that these samples had smaller amounts of DNA. Hence, it is possible that an insufficient amount of CDCG5244 DNA was transferred onto the blot to yield clearly visible hybridization products. Strain TT8 is an O157:H7 strain that produces only Stx2, and initial probing with DIG-stx1 confirmed the absence of the stx1 gene in this strain (data not shown). However, in the subsequent probing with DIG-stx2, two smaller XbaI fragments, of 173 and 66 kb, were found to react with the stx2 probe (Fig. 4B, lane 5).
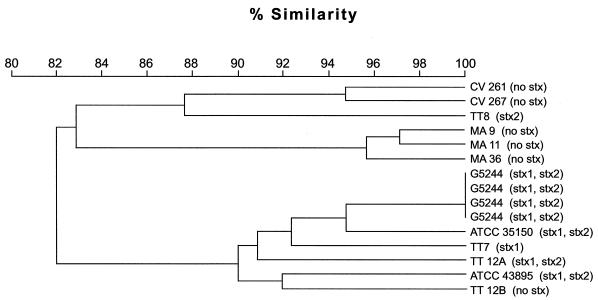
Cluster analysis of PFGE profiles supported the differences noted in the visual observations that there are distinct similarities among strains, as the non-Stx-producing O157:H7 strains were found to cluster into a major clade, while the Stx-producing O157:H7 strains clustered into another clade. Exceptions were the Stx2-producing TT8 strain, which clustered with the non-Stx-producing strains, and the non-Stx-producing TT12B strain, which clustered with the Stx-producing O157:H7 strains (Fig. 5).
FIG. 5.
Dendrogram comparing strain relationships of various Stx-producing and non-Stx-producing E. coli O157:H7 strains. The cluster tree was constructed using Molecular Analyst fingerprinting software (Bio-Rad), from comparisons of XbaI-digested PFGE patterns. The Shiga toxin phenotype is shown in parentheses at the right of each strain.
DISCUSSION
Strains TT12A and TT12B were indistinguishable, as they exhibited identical biochemical profiles and had typical O157:H7 characteristics, including growth on CT-SMAC agar. The tellurite used in CT-SMAC inhibits the growth of many bacteria, including the pathogenic, sorbitol-fermenting O157:H− variants from Germany (2, 18). Hence, growth on this medium appears to be fairly selective for O157:H7 and may also serve as an indicative trait for isolates of this serotype.
Serological tests showed that TT12A and TT12B are isolates of the O157:H7 serotype. Genetic and phenotypic assays further confirmed that except for the absence of the stx1 and stx2 genes in TT12B, both strains carried and expressed O157:H7 markers and virulence factors, including the cluster I ehxA sequences, the eae gene for γ-intimin, and the +93 gusA mutation that is unique to O157:H7 and its phenotypic variants (5, 6, 8). Interestingly, although both strains exhibited the A/E phenotype on HeLa monolayers, the extent of cell damage caused by TT12B was minimal compared to that caused by TT12A. This difference may be attributed to the cytotoxic effects on HeLa cells of Stx1 and Stx2, which are produced by TT12A, and serves as further evidence that neither toxin is produced by TT12B. Results of these various assays show that these two strains are identical except for the Stx phenotype.
Consistent with all the above findings, MLEE analysis showed that TT12A and TT12B are closely related genetically and are in the same clonal group as the other O157:H7 strains. Since these two strains are indistinguishable except for their toxin profiles, they were examined by PFGE. Comparison of XbaI- and ApaI-digested DNA showed that TT12A and TT12B differed by few bands, which may be due to the loss of the stx genes by TT12B. Loss of the Stx genotype has been reported to cause shifts in PFGE profiles of related strains (1, 15). Overall comparison, however, showed that TT12A and TT12B shared 90% similarity in profiles (Fig. 5); and moreover, both strains clustered into the same major clade with other Stx-producing O157:H7 strains. These results support our other findings that these two strains are closely related.
Strain TT12B is indistinguishable phenotypically and serologically from the other non-Stx-producing O157:H7 strains examined. These strains were also identical by PCR, as all had the unique +93 gusA mutation, the eae gene for γ-intimin, and the cluster I ehxA gene sequence, which are typical markers for the O157:H7 serotype (data not shown). However, comparison of PFGE profiles showed distinct differences, as the non-Stx-producing O157:H7 strains clustered into a major clade, while TT12B clustered with the Stx-producing O157:H7 strains. Previously, Itoh et al. (13) characterized four O157:H7 strains, isolated from cattle carcasses, that did not produce Stx but did carry other trait EHEC virulence factors. These strains were found to be closely related genotypically, and PFGE analysis of XbaI-digested DNA showed that they also shared similar profiles, hence suggesting that these non-Stx-producing O157:H7 strains had the same clonal descent (13). This is consistent with the results of our PFGE analysis, which showed similar clustering of non-Stx-producing O157:H7 strains. The two strains from France (CV261 and CV267) were closely related, as were the three strains from Japan (MA9, MA11, and MA36), and all of these strains clustered into a major clade that was distinct from the Stx-producing O157:H7 strains (Fig. 5). One exception was TT8, an Stx2-producing O157:H7 strain that clustered with the non-Stx-producing strains. Southern blot analysis showed that in the TT8 strain, the stx2 gene was present on two much smaller fragments than in most O157:H7 strains, where the stx2 gene resided on a 500- to 600-kb XbaI fragment. This suggests that TT8 is genetically distinct from the other Stx-producing O157:H7 strains, as it appears to carry deletions, additional XbaI sites in the stx2 gene region, or even multiple copies of stx2, which may have shifted its PFGE profiles and caused TT8 to cluster with the non-Stx-producing O157:H7 strains.
Since non-Stx-producing O157:H7 strains tend to cluster together, the fact that TT12B, a non-Stx-producing strain, clustered with the Stx-producing O157:H7 strains suggests that it is closely related to the latter group. This observation, coupled with the evidence that TT12B is identical to TT12A except for Stx production, tends to suggest that it is derived from TT12A by the loss of the stx1 and stx2 genes. Stx-producing E. coli can frequently lose stx genes during routine culturing in media, but this process does not appear to be common with strains of the O157:H7 serotype (18). The mechanism by which these stx genes are lost has not been determined (14).
In conclusion, we characterized a pair of Stx-producing (TT12A) and non-Stx-producing (TT12B) strains of O157:H7 which were identical and exhibited typical traits for O157:H7, except for the absence of Stx in TT12B. The PFGE profiles of these strains were also similar, and both strains clustered in the same clade with other Stx-producing O157:H7 strains; these findings suggest that TT12B is a progeny strain that arose from the parental strain (TT12A) by losing the stx1 and stx2 genes. These isogenic, toxin deletion variants may be useful in studying the pathogenesis of Stx in O157:H7 infections.
ACKNOWLEDGMENTS
We thank T. S. Whittam for performing MLEE analysis on the isogens, S. Weagant for providing the DIG-labeled stx probes, M. Nishibuchi and C. Vernozy-Roland for providing strains, S. Monday and T. S. Whittam for critical reading of the manuscript, and M. Meltzer for graphics.
REFERENCES
- 1.Akiba M, Sameshima T, Nakazawa M. The shift of genetic subtypes of Escherichia coli O157:H7 isolates from cattle. Epidemiol Infect. 1999;122:343–346. doi: 10.1017/s0950268899002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammon A, Petersen L R, Karch H. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H−. J Infect Dis. 1999;179:1274–1277. doi: 10.1086/314715. [DOI] [PubMed] [Google Scholar]
- 3.Boerlin P, Chen S, Colbourne J K, Johnson R, De Grandis S, Gyles C. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect Immun. 1998;66:2553–2561. doi: 10.1128/iai.66.6.2553-2561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: CDC training manual. Atlanta, Ga: Foodborne and Diarrheal Diseases Branch, Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 5.Feng P. Identification of Escherichia coli serotype O157:H7 by DNA probe specific for an allele of uidA gene. Mol Cell Probes. 1993;7:151–154. doi: 10.1006/mcpr.1993.1021. [DOI] [PubMed] [Google Scholar]
- 6.Feng P, Fields P I, Swaminathan B, Whittam T S. Characterization of nonmotile variants of Escherichia coli O157 and other serotypes by using an antiflagellin monoclonal antibody. J Clin Microbiol. 1996;34:2856–2859. doi: 10.1128/jcm.34.11.2856-2859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng P, Lampel K A, Karch H, Whittam T S. Sequential genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 8.Feng P, Sandlin R C, Park C H, Wilson R A, Nishibuchi M. Identification of a rough strain of Escherichia coli O157:H7 that produces no detectable O157 antigen. J Clin Microbiol. 1998;36:2339–2341. doi: 10.1128/jcm.36.8.2339-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng P, Monday S R. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol Cell Probes. 2000;14:333–337. doi: 10.1006/mcpr.2000.0323. [DOI] [PubMed] [Google Scholar]
- 10.Feng P C S, Hartman P A. Fluorogenic assays for immediate confirmation of Escherichia coli. Appl Environ Microbiol. 1982;43:1320–1329. doi: 10.1128/aem.43.6.1320-1329.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay B B, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J Cell Sci. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 12.Gunzer F, Bohn U, Fuchs S, Mühldorfer I, Hacker J, Tzipori S, Donohue-Rolfe A. Construction and characterization of an isogenic slt-ii deletion mutant of enterohemorrhagic Escherichia coli. Infect Immun. 1998;66:2337–2341. doi: 10.1128/iai.66.5.2337-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh Y, Hayashi N, Katoh M, Yamamoto A, Hayashi S, Maeda S, Ezaki T. The characterization of Shiga toxin-non-producing Escherichia coli serotype O157:H7 isolated from carcasses of cattle at a slaughter house. Microbiol Immunol. 1999;43:699–703. doi: 10.1111/j.1348-0421.1999.tb02458.x. [DOI] [PubMed] [Google Scholar]
- 14.Karch H, Meyer T, Rüssmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karch H, Rüssmann H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–1605. doi: 10.1128/jcm.33.6.1602-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushiro A, Sato K, Miyamoto H, Yamamura T, Honda T. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J Bacteriol. 1999;181:2257–2260. doi: 10.1128/jb.181.7.2257-2260.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid S D, Betting D J, Whittam T S. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J Clin Microbiol. 1999;37:2719–2722. doi: 10.1128/jcm.37.8.2719-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt H, Scheeff J, Huppertz H I, Frosch M, Karch H. Escherichia coli O157:H7 and O157:H− strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3491–3496. doi: 10.1128/jcm.37.11.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vernozy-Roland C, Feng P, Montet M-P, Ray-Gueniot S, Villard L, Boutrand-Loei S, Bavai C, Mazuy C, Atrache V. Detection of Escherichia coli O157:H7 in heifers' faecal samples using an automated immuno-concentration system. Lett Appl Microbiol. 2000;30:217–222. doi: 10.1046/j.1472-765x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- 21.Weagant S D, Jagow J, Jinneman K C, Omiecinski C J, Kaysner C A, Hill W E. Development of digoxigenin-labeled PCR amplicon probes for use in the detection and identification of enteropathogenic Yersinia and Shiga toxin-producing Escherichia coli from foods. J Food Prot. 1999;62:438–443. doi: 10.4315/0362-028x-62.5.438. [DOI] [PubMed] [Google Scholar]
- 22.Zadik P M, Chapman P A, Siddons C A. Use of tellurite for the selection of verotoxigenic Escherichia coli O157. J Med Microbiol. 1993;39:155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]