Abstract
Naked mole rats (NMRs, Heterocephalus glaber) are long-lived mammals that possess a natural resistance to cancer and other age-related pathologies, maintaining a healthy life span for >30 years. Here, using immunohistochemical and RNAseq analyses, we compare skin morphology, cellular composition and global transcriptome signatures between young and aged (3–4 versus 19–23-year-old) NMRs. We demonstrate that similar to human skin, aging in NMRs is accompanied by a decrease of epidermal thickness, keratinocyte proliferation, and a decline in the number of Merkel cells, T-cells, antigen-presenting cells and melanocytes. Similar to human skin aging, expression levels of dermal collagens are decreased, while Mmp-9 and Mmp-11 levels increased in aged versus young NMR skin. RNAseq analyses reveal that in contrast to human or mouse skin aging, the transcript levels of several longevity-associated (Igfbp3, Igf2bp3, Ing2) and tumor-suppressor genes (Btg2, Cdkn1a, Cdkn2c, Dnmt3a, Hic1, Socs3, Sfrp1, Sfrp5, Thbs1, Tsc1, Zfp36) are increased in aged NMR skin. Overall, these data suggest that specific features in the NMR skin aging transcriptome might contribute to the resistance of NMRs to spontaneous skin carcinogenesis and provide a platform for further investigations of NMRs as a model organism for studying the biology and disease resistance of human skin.
Introduction
The skin forms an interface between the external environment and internal milieu that protects mammalian organisms against uncontrolled water loss and numerous environmental stressors, including mechanical injury, UV irradiation, thermal/chemical insults, harmful microorganisms and viruses (Chuong et al., 2002, Larsen et al., 2020, Menon and Kligman, 2009, Slominski et al., 2012). To fulfil such complex functions, different cell lineages of epithelial, mesenchymal and neuro-ectodermal origin interact with each other and form a robust and plastic biological system capable of maintaining homeostasis and responding efficiently to environmental challenges (Rognoni and Watt, 2018).
Similar to other organs, the skin undergoes both intrinsic (chronological) and extrinsic (environmental) aging associated with changes in visual appearance and loss of functional capacity and regenerative potentials (Rittie and Fisher, 2015, Yaar et al., 2002). Major age-related changes in the skin include wrinkling, dryness, pigmentation abnormalities and the development of a variety of benign neoplasms (Yaar et al., 2002). Microscopically, skin aging is accompanied by the decrease of epidermal thickness and keratinocyte proliferation, decline in the number of melanocytes and Langerhans cells, flattening of the dermal-epidermal junction, decline of dermal volume and cellularity, fragmentation of collagen and elastic fibers, decrease of dermal blood/lymphatic vessels, cutaneous innervation and sensory perception (Rittie and Fisher, 2015, Yaar et al., 2002).
At the molecular level, aging of the epidermis is accompanied by alterations of gene expression in the keratinocyte-specific gene loci, loss of terminal differentiation-associated calcium gradient, decreased lipid synthesis and hyaluronic acid content, cytokine imbalance, and results in compromised epidermal barrier function (He et al., 2020). Thinning and flattening of the dermal-epidermal junction in aged skin is accompanied by a significant reduction in the levels and distribution of collagens IV/VII/XVII, integrin β4, and laminin-332 (Langton et al., 2016). Dermal changes in aged skin also include fragmentation of both collagen and elastic fibers, a decrease in the levels of major extracellular matrix components (decorin, versican), and upregulation of the matrix metallopeptidases (MMP1/3/9) that cleave collagen and elastic fibrils (Cole et al., 2018).
Age-associated alterations in the skin immune system include a decrease in the antigen-presenting and migratory capacities of Langerhans and dendritic cells, resulting in alterations of the barrier immunity and development of chronic low-grade inflammation (inflammaging) (Pilkington et al., 2021). Accumulation of senescent cells in the aged epidermis and dermis (Gunin et al., 2014, Victorelli et al., 2019, Waaijer et al., 2016) contributes to the “inflammaging” phenotype due to their production of pro-inflammatory cytokines as a part of Senescent-Associated Secretory Phenotype (SASP) (Toutfaire et al., 2017).
There are quite legitimate concerns among many researchers raising the question of whether mechanisms of aging identified in short-lived mammals, such as mice or rats, adequately reflect the complexity of longevity pathways that regulate aging in long-lived species like humans (Dammann, 2017). Naked mole rats (NMRs, Heterocephalus glaber) are long-lived rodents possessing remarkable resistance to spontaneous carcinogenesis, certain noxious stimuli and hypoxia and maintain sustained healthy life-span span for over 30 years (Buffenstein, 2005, Gorbunova et al., 2014, Seluanov et al., 2018, Smith et al., 2020). Comparative genome analyses have revealed that the NMR genome shows higher similarity (93% synteny) to the human genome compared to that of mice (83%) or rats (80%) (Gladyshev et al., 2011, Kim et al., 2011). Many aspects of the NMR biology have recently been extensively discussed (Braude et al., 2021, Buffenstein et al., 2021) leading to the conclusion that exceptional resistance of NMRs to aging-associated pathologies is mediated by several mechanisms, including more robust DNA repair and genome stability compared to mice, a unique organization of the tumor suppressor Ink4a/b locus, production of large amounts of higher molecular weight hyaluronic acid with unusual properties, altered IGF receptor signaling and increased proteasome activity and protein stability (Brohus et al., 2015, Del Marmol et al., 2021, Evdokimov et al., 2018, Gorbunova et al., 2014, Keyes et al., 2013, Kulaberoglu et al., 2019, MacRae et al., 2015, Rodriguez et al., 2016, Seluanov et al., 2018, Takasugi et al., 2020).
The skin of the NMR displays distinctive morphological features associated with adaptation of these animals to the subterranean environment: relatively thick epidermis with unusually thick corneal layer, presence of pigment-containing cells in the dermis, lack of hair follicles on most of the skin (Daly and Buffenstein, 1998, Menon et al., 2019, Thigpen, 1940). However, it is unclear how the NMR skin changes during aging and whether there are any particular features in the NMR skin transcriptome that underlie the remarkable natural resistance of NMRs to spontaneous skin carcinogenesis.
In this manuscript, we show that skin aging in the NMR resembles many features of human skin aging, including histological and biochemical changes in both the epidermis and dermis. However, in contrast to human skin, transcripts and proteins of several longevity-associated and tumor suppressor genes are increased in the NMR skin during aging. These data suggest that these features of the NMR skin aging transcriptome might contribute to the resistance of NMRs to spontaneous skin carcinogenesis, thus reaffirming the potential of the NMR as a model organism for studying both the biology and disease of human skin.
Results
Epidermis of aged NMRs shows decreased thickness, cell proliferation and reduced expression of the keratinocyte differentiation markers, Col17a1 and Merkel cells
To assess the impact of aging on NMR skin, we first compared visual appearance, morphological parameters and expression of established keratinocyte differentiation markers between the dorsal skin epidermis of young (2–4.5 year-old) and aged (19–23-year-old) animals (Fig. 1 A). Consistent with data published previously (Daly and Buffenstein, 1998, Tucker, 1981), the dorsal skin of NMRs has a wrinkled and saggy macroscopic appearance, and a lack of any skin appendages (Fig. 1 B). Single unpigmented hairs are only visible at the lateral part of the body, while vibrissa hairs are numerous at the facial skin (Fig. 1 A). In contrast to the skin of young NMRs (YNMRs), the skin of old NMRs (ONMRs) appeared thinner, more translucent and less pigmented (Fig. 1 B). Histologically, the dorsal skin of YNMRs presented a relatively thick multilayered epidermis with a thick stratum corneum (Menon et al., 2019), while in ONMRs, the epidermal thickness was significantly reduced (Fig. 1 C). The majority of Ki67+ cells were located in the basal epidermal layer, while their number was markedly decreased in ONMRs versus YNMRs (Fig. 1 D).
Figure 1. Visual appearance of the skin and age-related changes in the epidermis of NMRs.
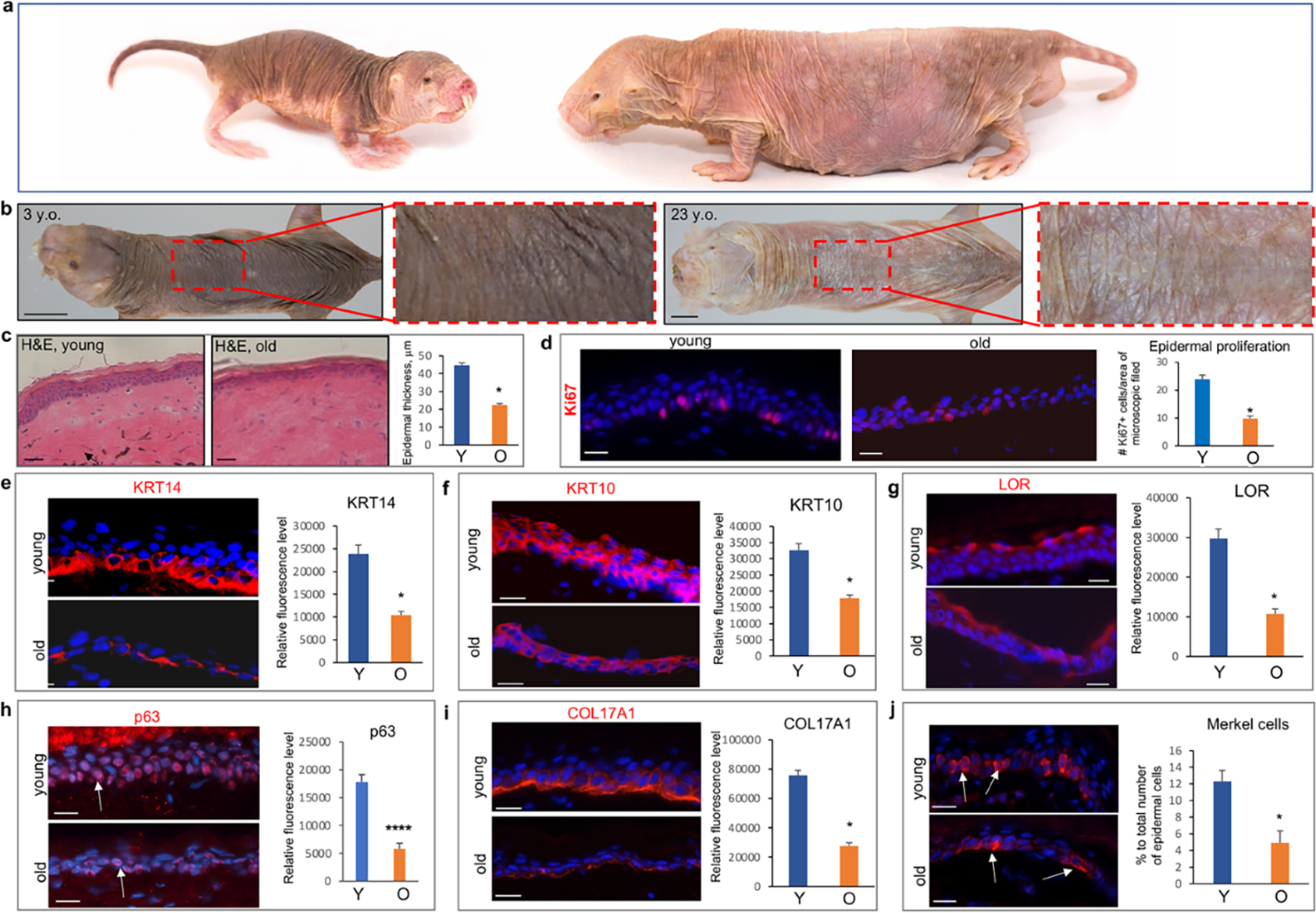
a – Images of the skin in 3- and 23-year-old animals, note the more translucent and less pigmented dorsal skin in aged NMR. b – Hematoxylin/eosin staining of the young and old skin: presence of the dermal pigment (arrow) in young skin and significantly reduced epidermal thickness in old animals. c – Significant decrease in Ki67+ cells in aged versus young NMRs. d-f - Significantly decreased immunofluorescence intensity of K14 in the basal layer (d), K10 in the spinous layer (e), and Loricrin in the granular layer (f) in the epidermis of old NMRs. g - Significant decrease in the immunofluorescence intensity of p63 in epidermal keratinocytes of old versus young NMRs. h – Significant decline in the expression of COL17A1 in basal epidermal keratinocytes and dermal-epidermal basement membrane of old animals. i – Significant decrease in the number of KRT20+ Merkel cells in the epidermis of old NMRs (arrows). (mean ± SD, *p<0.05, Student’s t-test). Scale bars: 1b – 1 cm; 1c – 50 um; 1d-j – 25 um. Y – young animal, O – old animal.
In the NMR epidermis, Krt14 expression was mainly confined to the basal layer, while Krt10 and Loricrin (Lor) were seen in the spinous and granular layers, respectively (Fig. 1 E–G). However, the immunofluorescence intensity for all three markers, as well as for p63 transcription factor, a master regulator of epidermal development and differentiation (Koster et al., 2007), was significantly decreased in the epidermis of ONMRs compared to YNMRs (Fig. 1 E–H).
The expression of Col17a1, an established marker of epidermal aging (Liu et al., 2019), was also decreased in basal epidermal keratinocytes and dermal-epidermal basement membrane of ONMRs, compared to YNMRs (Fig. 1 I). In addition, the number of Krt20+ Merkel cells, specialized sensory cells involved in mechanotransduction (Jenkins et al., 2019), was significantly lower in the epidermis of ONMRs versus YNMRs (Fig. 1 J). However, epidermal Caspase 3+ apoptotic cells did not reveal any differences in their number between YNMRs and ONMRs (Supplementary Figure S1 A).
Age-associated changes in the NMR dermis include decreased Col1a1 expression, an increase in Mmp9 and Mmp11, and loss of melanocytes
The NMR epidermis forms invaginations or buds elongating into the dermis (Tucker, 1981). Their numbers was significantly decreased in ONMRs contributing to age-associated flattening of the epidermal-dermal interface (Fig. 2 A). Similar to human skin (Langton et al., 2010), Fibrillin-2-enriched elastic fibers in the papillary dermis of NMRs formed candelabra-like cascades ascending towards the dermal-epidermal junction and epidermal buds, while their number were not changed between YNMRs and ONMRs (Fig. 2 B). Moreover, the distribution of dermal elastic fibers and Elastin expression levels were quite similar in the dermis of YNMRs and ONMRs (Supplementary Figure S1 B).
Figure 2. Age-associated changes in the NMR dermis.
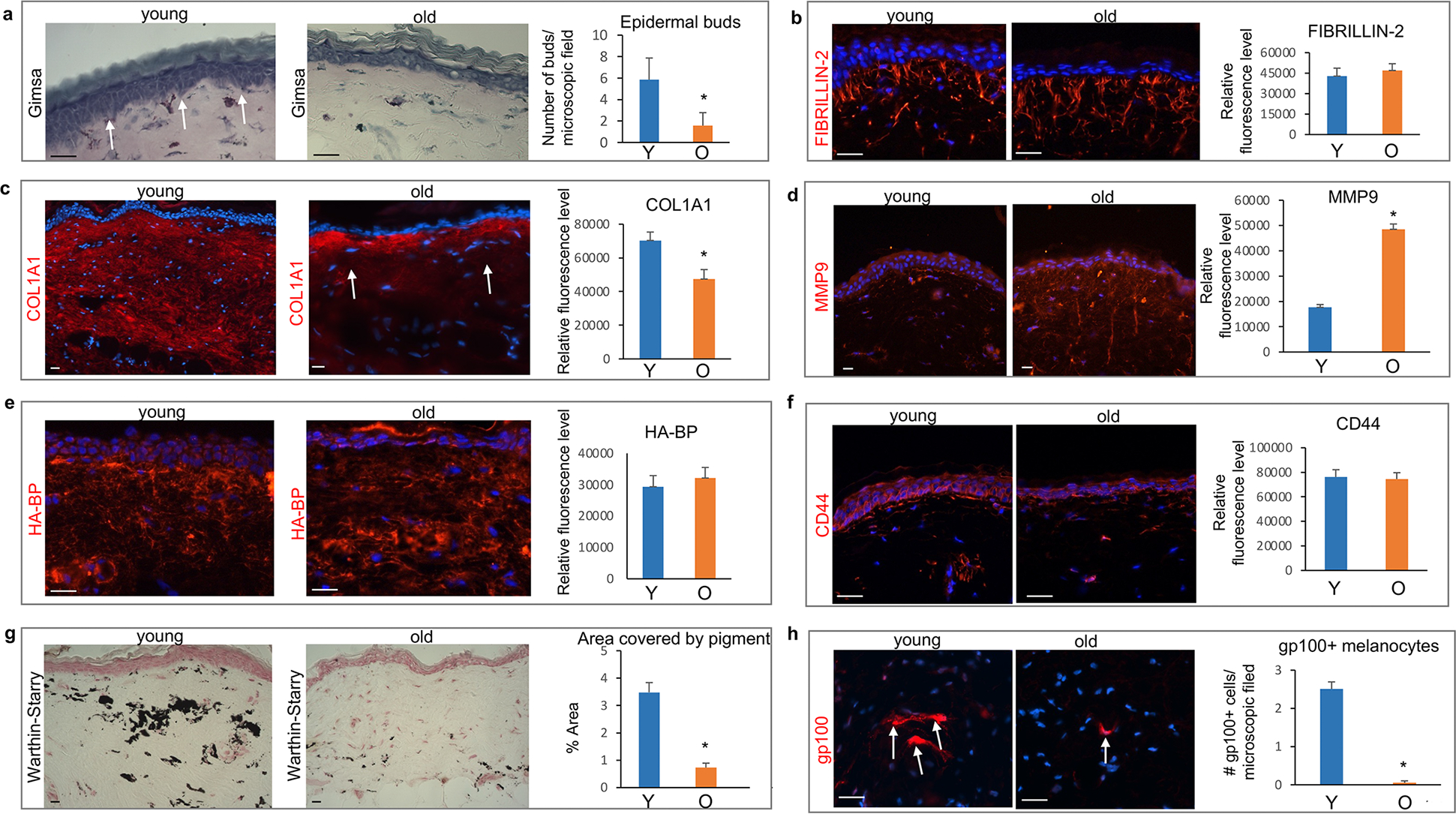
(a) Significantly reduced number of epidermal buds elongating into the dermis (arrows) in the aged NMR skin. (b) Similar distribution of Fibrillin-2+ fibers in young and old NMRs. (c) COL1A1 is broadly expressed in the papillary and reticular dermis of young NMRs, while COL1A1 expression is significantly decreased in the reticular dermis in aged skin. (d) Significant increase in the MMP9 immunofluorescence intensity in the dermis of old NMRs compared to young animals. (e) No differences in the hyaluronan-binding protein (HA-BP) binding pattern or degree of binding between the dermis of young and old NMRs. (f) Similar expression of the HA receptor CD44 in the skin of young and old NMRs. (g) Warthin-Starry stain shows a dramatic decrease in the melanin containing areas in the dermis of old compared to young NMRs, which is accompanied by a significant decrease in the number of gp100+ pigment-producing dermal melanocytes in the dermis of aged animals (h, arrows). Mean ± SD, *p<0.05, Student’s t-test. Scale bars: 25 μm. Y – young animal, O – old animal.
Col1a1, an important component of the mature collagen fibers providing tensile strength to the skin (Cole et al., 2018), was broadly expressed in the papillary and reticular dermis of YNMRs (Fig. 2 C). In ONMRs, dermal Col1a1 expression declined compared to YNMRs (Fig. 2 C), while expression levels of Col3a1, a marker of immature collagen fibers, did not show significant differences between YNMRs and ONMRs (Supplementary Figure S1 C).
Matrix metalloproteinases (Mmps) promote age-associated extracellular matrix remodeling (Cole et al., 2018). Immunohistochemical revealed a marked increase of Mmp9 and Mmp11 immunofluorescence in the dermis of ONMRs compared to YNMRs (Fig. 2 D, Supplementary Figure S1 E). However, Mmp1 levels did not show significant changes in ONMRs versus YNMRs (Supplementary Figure S1 D).
The NMR extracellular matrix contains hyaluronic acid (HA), which unlike mouse HA forms highly folded structures that may contribute to the elasticity of NMR skin (Kulaberoglu et al., 2019). Moreover, HA appears to be present in larger amounts and has a higher molecular weight in NMR than mouse or guinea pig (Del Marmol et al., 2021, Keyes et al., 2013). However, Hyaluronan-Binding Protein (HABP) immunostaining used previously for analyses of the HA content in the skin (Kulaberoglu et al., 2019) and the HA receptor Cd44 levels did not show any differences between the dermis of YNMRs and ONMRs (Fig. 2 E, F).
One of the characteristic features of the NMR dorsal skin is the presence of a large number of pigmented melanocytes in the dermis (Daly and Buffenstein, 1998). Consistently with the visual loss of skin pigmentation in old NMRs (Fig. 1 A), Warthin-Starry staining of melanin (Joly-Tonetti et al., 2016) revealed a dramatic decrease of the areas covered by pigment in the dermis of ONMRs compared to YNMRs (Fig. 2 G). Also, the number of melanocytes expressing gp100 that is expressed in melanogenically active melanocytes (Watt et al., 2013), was markedly reduced in the dermis of ONMRs (Fig. 2 H).
Aged NMR skin exhibits decreased Cd3ε T-cells, MHC class II antigen-presenting cells, mast cells and increase of senescent cells
To characterize changes in the skin immune system occurring during aging in NMRs, we used a panel of primary antibodies established for analysis of immune cell markers in NMRs (Shebzukhov et al., 2019). Comparative studies of the immune cell markers revealed that ONMRs show a significant decrease in the number of Cd3ε-positive T-cells and MHC II-positive cells in the epidermis and dermis (Fig. 3 A, B; Supplementary Figure S1 F). A quantitative analysis of Cd8+cells and Cd11b+ (macrophages) cells in the dermis did not reveal any differences between ONMRs and YNMRs (Fig. 3 C, Fig. 3 D). In contrast, the number of mast cells was significantly lower in the dermis/subcutis of old NMRs compared to young animals (Supplementary Figure S1 G).
Figure 3. Aging-associated changes in the number of immune and senescent cells in the NMR skin.
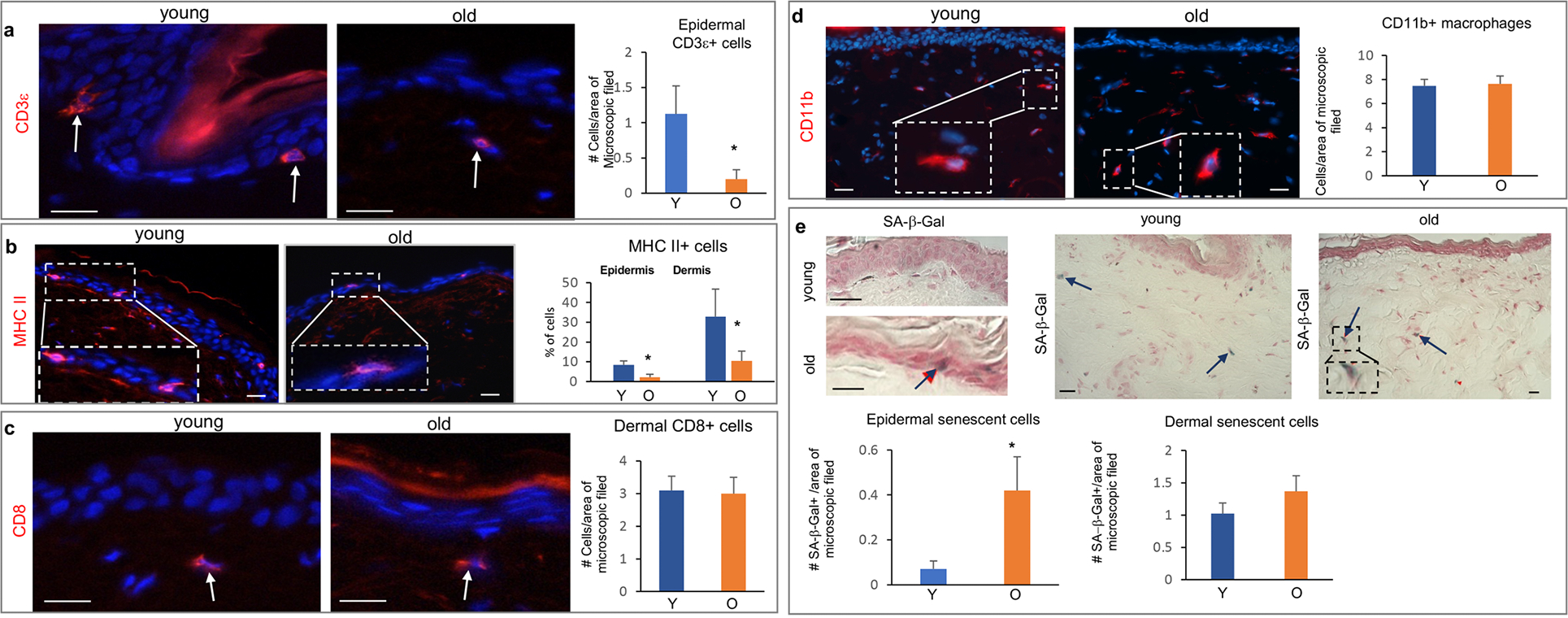
(a) Significant decrease in the number of CD3ε+ T-cells in the epidermis of old NMRs. (b) Significant decrease in the number of MHC II+ cells detected by an anti-27E7 antibody in the epidermis and dermis of old NMRs. (c) No changes in the number of CD8+ cells in the dermis of aged versus young NMRs (arrows). (d) The number of dermal CD11b+ macrophages is similar in the young and old NMRs. (e) Significant increase in the number of senescent SA-β-gal+ cells (arrow) in the epidermis of old NMRs. A tendentious, but not significant, increase in the number of SA-β-gal+ cells (arrows) in the aged dermis (p=0.082). Mean ± SD, *p<0.05, Student’s t-test. Scale bars: 25 μm. Y – young animal, O – old animal.
The presence of senescent cells in the skin of NMRs was previously reported (Zhao et al., 2018). Senescence-associated β-galactosidase (SA-β-gal) staining revealed a significant increase in the number of SA-β-gal-positive cells in the aged epidermis, and only a tendentious increase in the dermis of ONMRs, which did not reach statistical significance (p=0.082, Fig. 3 E). The increase in the number of senescent cells in the ONMR skin was accompanied by the elevated expression of p16INK4a transcript as an important marker of senescent cells (Ressler et al., 2006) that was determined by qRT-PCR in full-thickness skin samples (Supplementary Figure S1 H). However, the expression of both p15INK4b and pALTINK4a/b transcripts remained unaltered in old versus young skin (Supplementary Figure S1 H).
Transcriptome analysis of NMR aging skin reveals changes in the expression of extracellular matrix-associated genes, components of the IGF signaling pathway, regulators of glucose metabolism and cell proliferation
To further characterize changes occurring in the NMR skin during aging on the molecular level, RNAseq analyses of the full-thickness skin of 4-year-old and 19-year-old animals were preformed and revealed significant (two-fold and higher) changes in the expression of 768 genes between the skin of YNMRs and ONMRs (Fig. 4 A, B). Overall, RNAseq data were concordant with our histomorphology/immunohistochemistry results (Fig. 1, Fig. 2, Supplementary Table S3) and identified the decline in expression of dermal collagen genes (Col1a1, Col1a2), genes encoding T-cell and Langerhans cell markers (Thy1, Cd3d, Cd207), as well as of melanocyte/melanogenesis-associated markers (Dct, Tyrp1, Slc24a5) in ONMR skin compared to YNMRs (Fig. 4 C, Supplementary Tables S2, S3). Consistent with the immunohistochemistry results (Fig. 2 D, Supplementary Figure S1 D), RNAseq analyses also detected an increase in Mmp9 and Mmp11 gene expression in ONMR versus YNMR skin (Fig. 4 C, Supplementary Table S1).
Figure 4. RNAseq analyses of the age-associated changes in the cutaneous NMR transcriptome.
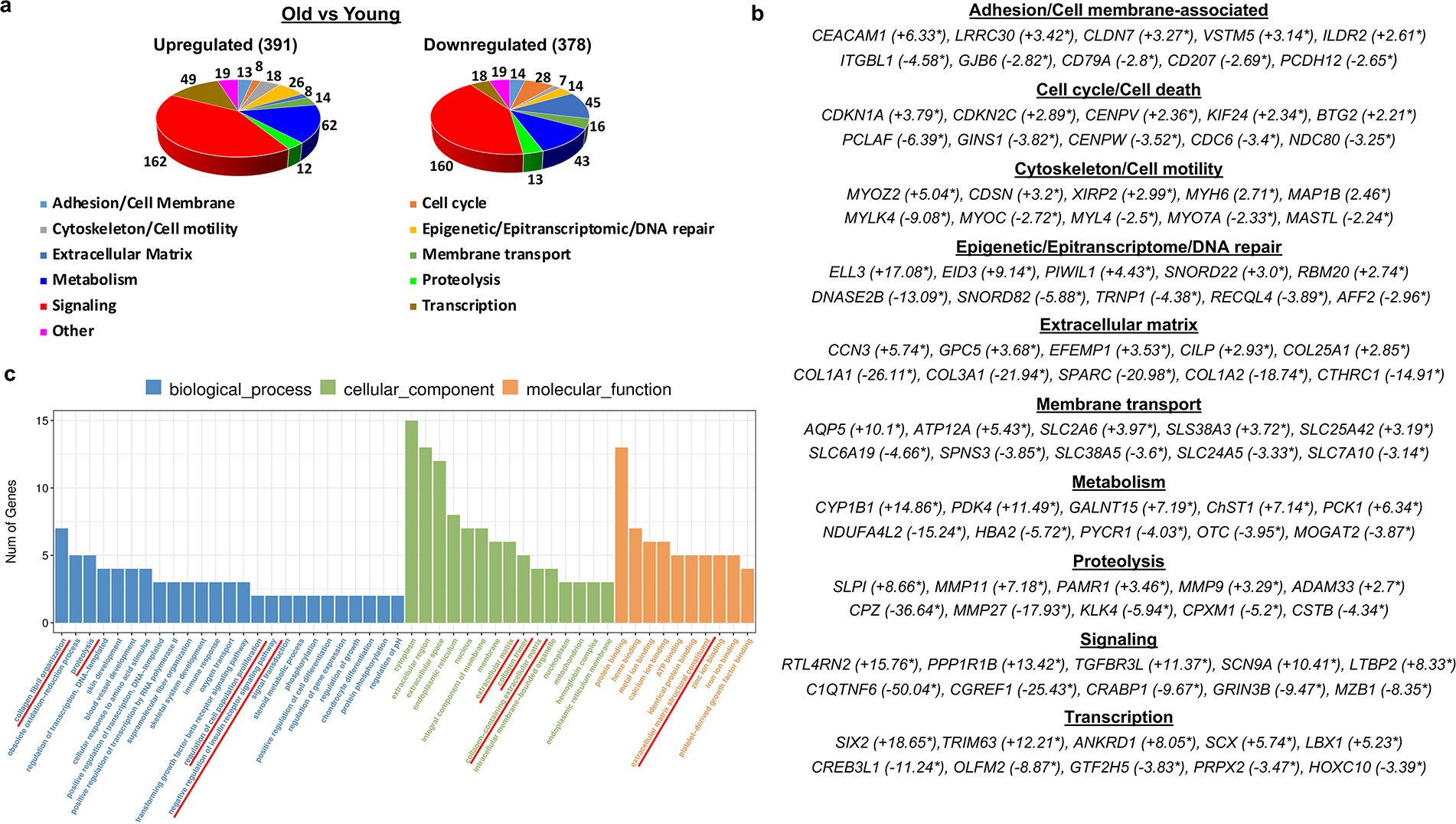
(a) Functional annotation of the differentially expressed genes based on QIAGEN Ingenuity Pathway Analysis database and manually curated functional gene sub-categories. (b) A list of five top differentially expressed genes in each functional group (fold-change expression values are indicated by asterisks). (c) GO enrichment analysis showed significant over-representation of the extracellular matrix-associated genes, components of the insulin growth factor (IGF) signaling pathway, regulators of the glucose metabolism and cell proliferation.
Interestingly, among the group of extracellular matrix-associated genes differentially expressed in the ONMR skin, we found a marked downregulation of numerous collagen genes, including Col1a1, Col1a3, Col3a1, genes encoding the enzymes involved in collagen synthesis (Pcolce, Pcolce2), and fibrillogenesis (Cthrc1, Sparc) (Fig. 4 A–C, Supplementary Table S2). Also, several genes encoding extracellular matrix-degrading enzymes (Mmp11, Mmp9, Pm20d2, Adamts5) were upregulated in the skin of ONMRs, while Mmp27 and Mmp19 were downregulated compared to YNMRs (Fig. 4 A–C, Supplementary Tables S1, S2). Although it is known that MMP9 and MMP11 can be processed by other MMPs, such as MMP2 (Bonnans et al., 2014), we observed no difference in expression levels of Mmp2, as well as of transcript of other enzyme involved in the MMP processing/cleavage Furin between ONMR and YNMR skin (data not shown).
A group of genes involved in the regulation of glucose metabolism included Pdk4, which encodes pyruvate dehydrogenase kinase 4 promoting a switch from oxidative energy metabolism to glycolysis (Stacpoole, 2012). Pdk4 was strongly upregulated in the skin of ONMRs, as well as the Pck1 gene, a critical regulator of gluconeogenesis, Acacb encoding a key enzyme in fatty acid synthesis, and Slc2a6 encoding the glucose transporter Glut6 (Barron et al., 2016).
IGF signaling is implicated in the control of longevity (Brohus et al., 2015) including skin aging (Lewis et al., 2010). RNAseq analysis revealed that expression of Igf1 was decreased in ONMR skin compared to YNMRs, while expression levels of transcripts for the IGF-1/2 binding proteins Igfbp3 and Igf2bp3 were increased (Supplementary Tables S1, S2).
The group of genes involved in cell cycle regulation that show differences in expression between ONMR and YNMR skin included cyclin-dependent kinase inhibitors (Cdkn1a, Cdkn2c) and member of the anti-proliferative BTG/TOB family (Btg2), whose expression patterns in aged skin were upregulated (Fig. 4 C, Supplementary Table S1). These changes were accompanied by decreased expression of genes positively regulating cell proliferation including Pcna, Ccne1 gene encoding cyclin E1, centromere-associated protein genes (Cenph, Cenpm, Cenpn, Cenpw, Cenpx), genes controlling mitotic cell division (Ncapg2, Pole1, Tk1) and cell cycle-associated transcription machinery (E2f1) (Fig. 4 C, Supplementary Table S2). The significant increase of Cdkn1a, Btg2 and Tob1 at the protein level in the NMR aged epidermis were further confirmed by quantitative immunofluorescence analysis (Supplementary Figure S3). These data were consistent with the results demonstrating the decrease of epidermal proliferation in ONMRs compared to YNMRs (Fig. 1 D).
Species-specific differences in aging transcriptomes hint to a potential contribution to longevity and cancer resistance in NMRs
To correlate the age-associated changes in the NMR skin transcriptome to human or mouse skin aging, we compared our data with four publicly available RNAseq skin aging datasets obtained from either full-thickness skin or epidermis of human and mouse skin (young versus aged) (Aramillo Irizar et al., 2018, Barth et al., 2019, Ge et al., 2020, Raddatz et al., 2013). This analysis revealed that only a relatively small number of genes whose expression was changed in the ONMR skin (11.7% of upregulated and 16.9% of downregulated genes), showed a similar trajectory in the aged human and mouse skin. Surprisingly, most genes differentially expressed during NMR skin aging were not changed in either human or mouse skin aging, while small number of differentially expressed genes (11.0% and 14.5% of up- and downregulated genes, respectively) in the NMR aging transcriptome showed opposite dynamics to aged human or mouse skin (Supplementary Figure S2, Supplementary Tables S4, S5).
To further correlate the relevance of age-associated changes in the NMR cutaneous transcriptome to other age-related datasets, we used the Human Ageing Genome Resource (HAGR) database platform (Tacutu et al., 2018). We found that 113 genes differentially expressed in ONMR skin (16.3% of the upregulated and 12.9% of downregulated genes, defined as NMR aging signature genes) were present in one or more HAGR datasets (Fig. 5 A; Supplementary Table S6). According to the HAGR databases, substantially more NMR aging signature genes were relevant to human aging than to the animal aging (33 versus 12, respectively), while 8 NMR genes belonged to both datasets (GeneAgeHuman, GeneAgeModel; Supplementary Table S6).
Figure 5. Comparison of age-associated changes in the transcriptome of NMR, human, and mouse skin.
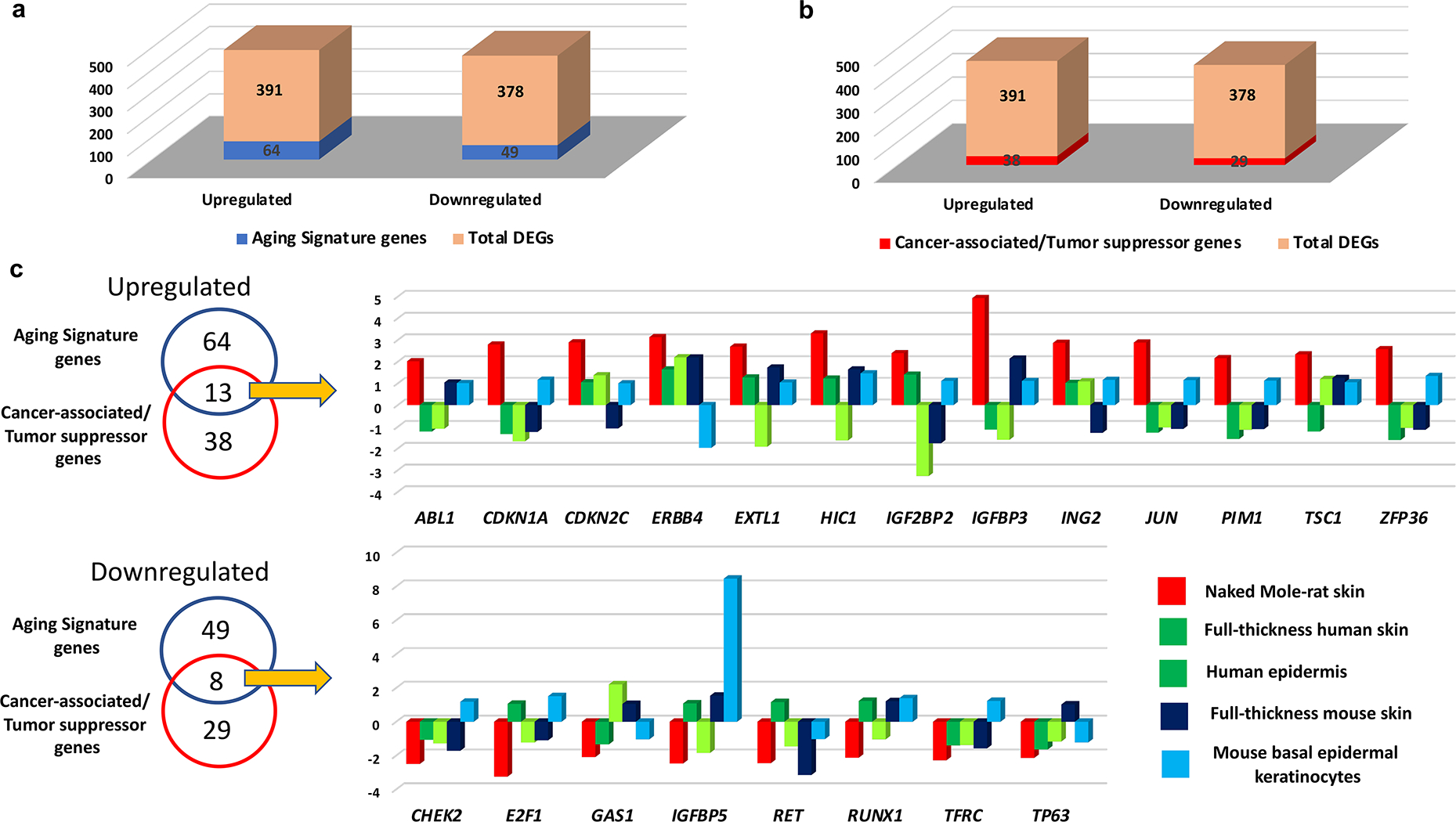
(a) The number of differentially expressed genes common between the NMR skin aging transcriptome and the Human Ageing Genome Resource (HAGR) genes (here denoted as aging signature genes) relative to the total number of differentially expressed genes; (b) The number of differentially expressed genes common between the NMR skin aging transcriptome and the Cancer Gene Census (CGC)/Tumor Suppressor Genes (TSG) datasets (here denoted as cancer signature genes) relative to the total number of differentially expressed genes; (c) A Venn diagram shows the differentially expressed genes in the NMR skin aging transcriptome that are shared with the human HAGR and CGC/TSG databases. Comparative analyses of gene expression (old versus young) in the skin of three species: NMR (total skin), human (total skin, epidermis) and mouse (total skin, FACS-sorted basal epidermal keratinocytes). The differences in gene expression are shown as the FPKM fold-change.
In ONMR skin, RNAseq detected elevated expression of Fos and Jun (Supplementary Table S1), encoding essential components of the AP-1 transcription factor, stimulating the expression of MMPs during UV-induced photoaging in human skin (Rittie and Fisher, 2002, 2015). Also, among the genes upregulated in ONMR skin was Igfbp3 implicated in the control of longevity and counteracting the life-shortening effects of IGF signaling (Martins et al., 2016), as well as anti-proliferative genes Cdkn1a and Cdkn2c (Supplementary Table S6). Protein-protein interaction network analysis performed using the STRING software tool (http://string-db.org, vision 11.0) (Crosara et al., 2018) revealed links between the Fos/Jun genes with tumor suppressor genes Cdkn1a/Cdkn2c/Zfp36 upregulated in ONMR skin (Supplementary Figure S4). On the other hand, the longevity-related genes downregulated in ONMR skin formed network controlling extracellular matrix remodeling, IGF signaling and DNA replication/cell division (Supplementary Figure S5).
Because NMRs show remarkable resistance to spontaneous carcinogenesis, we intersected the age-associated NMR transcriptome with the human Cancer Gene Census (CGC) database (Welcome Sanger/EBI, Hinxton, UK) (Sondka et al., 2018) and Tumor Suppressor Genes (TSG) database (University of Texas, Houston, TX). We found that 67 differentially expressed genes in the ONMR skin were present in the CGC/TSG databases, representing the NMR cancer-related/TSG signature genes (Fig. 5 B). Interestingly that this group of genes contained not only inhibitors of IGF pathway, but also inhibitors of the JAK/STAT (Sosc3) and Wnt pathways (Sfrp1 and Sfrp5) (Supplementary Table S7).
After merge of the longevity-associated and cancer-related/TSGs genes, we found 21 common genes seen in the HAGR and CGC/TSG databases (Fig. 5 C), thus demonstrating their relevance to both longevity and cancer development/resistance. The group of the longevity/cancer-related/TSG signature genes upregulated in ONMR skin (Fig. 5 C) included cell cycle inhibitors Cdkn1a/Cdkn2c (Winkler, 2010), tumor suppressor Hic1 gene encoding transcriptional repressor downregulated in many cancers (Rood and Leprince, 2013), as well as Tsc1 gene mutated in Tuberous Sclerosis Complex disease characterized by formation of tumor-like lesions (hamartomas) in the skin (Rosset et al., 2017). Also, expression of Zfp36 gene encoding RNA-binding protein tristetraprolin (TTP) inhibiting formation of squamous cell carcinomas (Assabban et al., 2021) was upregulated in ONMR skin (Fig. 5 C). In addition, expression of Dnmt3a regulating de-novo DNA methylation (Parry et al., 2021) increased in ONMR skin compared to YNMRs (Supplementary Table S7).
Among the longevity-associated/cancer-related genes downregulated in ONMR skin (Fig. 5 C) were proto-oncogene Ret, transcription factor E2f1 promoting cell proliferation and upregulated in chemically-induced squamous cell carcinomas (Balasubramanian et al., 1999), and Tp63 downregulated in human skin during aging (Fig. 5 C). Protein-protein interaction network of the cancer-related/TSG genes revealed additional links between a group of tumor suppressors (Btg2, Sfrp1, Socs3, Thbs1) and oncogenes (Abl1, Flt3, Kras, Sparc), as well as between DNA methyltransferase Dnmt3a, components of the FGF signaling pathway Flt3/Fgfr4 and longevity-related Tp63 (Supplementary Figure S6).
The comparative analyses of the longevity-associated/cancer-related/TSG signature genes in the NMR aging transcriptome with human or mouse skin aging datasets (Aramillo Irizar et al., 2018, Barth et al., 2019, Ge et al., 2020, Raddatz et al., 2013) revealed that none of the 13 genes of this category upregulated in ONMR skin were significantly elevated in aged human or mouse skin, while expression of only 1 out of 8 genes downregulated in ONMR skin showed similar changes in human or mouse skin (Fig. 5 C, Supplementary Tables S6, S7). These data suggest that age-associated changes in the cancer-related NMR transcriptome show unique features that may underlie the remarkable resistance of NMRs to spontaneous skin carcinogenesis.
Discussion
In this report, we provide evidence that skin aging in NMRs shows many similar features to the aging process in human skin, including: 1) Decrease of epidermal thickness, keratinocyte proliferation and expression of epidermal keratins; 2) Decline in the number of Merkel cells and increase of senescent cells in the epidermis; 3) Flattening of the epidermal/dermal border, decrease in expression levels of dermal collagens and upregulation of Mmp-9 and Mmp-11; 4) Loss of skin pigmentation and decline in the number of dermal melanocytes; 5) Decline in the number of epidermal/dermal T-cells and antigen-presenting cells.
Our data showing morphological and biochemical changes in the skin of 19–23 year-old NMRs appears to be quite different from the data obtained from 11 year-old animals (Savina et al., 2020) and demonstrate that skin aging in NMRs occurs during last trimester of life. Age-associated changes in the NMR epidermis are consistent with decline in the expression of the p63 transcription factor controlling keratinocyte proliferation, differentiation and preventing premature skin aging (Botchkarev and Flores, 2014, Koster et al., 2007, Su et al., 2009). Signaling/transcription factor-regulated and epigenetic mechanisms operate in concert in controlling epidermal differentiation (Ahmed et al., 2014, Botchkarev, 2017, Botchkarev et al., 2012, Fessing et al., 2011, Kouwenhoven et al., 2015, Mardaryev et al., 2016, Qu et al., 2019, Rapisarda et al., 2017). Skin aging in NMRs is accompanied by accumulation of DNA methylation at specific aging-associated differentially methylated CpGs (Horvath et al., 2022, Lowe et al., 2020). In this context, upregulation of genes encoding DNA methyltransferase Dnmt3a and methyl-CpG binding protein Mecp2 (Table S1) might contribute to the changes in aging-associated methylome and gene transcription in the skin of ONMRs.
Similarly to human skin (Waaijer et al., 2012, Waaijer et al., 2018), aged NMRs also show an increase in the number of senescent cells associated with the upregulation of p16INK4a in the skin. Senescent cells are present in the NMR skin, and NMR dermal fibroblasts are more resistant to induction of stress-induced premature senescence than mouse fibroblasts (Zhao et al., 2018). In aging human skin, senescent cells produce pro-inflammatory cytokines and contribute to cutaneous inflammaging (Pilkington et al., 2021, Toutfaire et al., 2017). However, detailed transcriptome/proteome analyses of the distinct cell lineages (epithelial, mesenchymal, immune/vascular, pigment, etc.) are required to define the contribution of the senescent cells and their secretory products to the development of age-associated changes in the NMR skin, including cutaneous immune surveillance and inflammatory response.
Despite their remarkable longevity, NMRs also exhibit a potent resistance to spontaneously developing cancers (Seluanov et al., 2018), with a lack of any reported skin cancer incidence, including basal/squamous cell carcinomas and malignant melanomas (Delaney et al., 2013). Our immunohistochemical data showing a lack of decline in the levels of HA and Cd44 receptor in aged NMR skin suggest that cytoprotective and anti-cancer properties of HA (Keyes et al., 2013, Takasugi et al., 2020) are likely not compromised during aging. Comparative transcriptome analyses reveal that in contrast to human or mouse skin, skin aging in NMRs is accompanied by upregulation of several classes of tumor suppressor genes: secreted inhibitors of the IGF, JAK/STAT and Wnt signaling pathways (Igfbp3, Igf2bp2, Socs3, Sfrp1, Sfrp5), cell cycle inhibitors (Btg2, Cdkn1a, Cdkn2c), transcriptional repressor Hic1, RNA-binding protein Zfp36 and epigenetic regulators (Dnmt3a, Mecp2). We speculate that together with other longevity-associated genes, these genes form a multi-level regulatory network mediating cancer resistance in the NMR skin.
Undoubtedly, further analyses are required to fully understand the roles of signaling/transcription factor-mediated and epigenetic regulatory mechanisms controlling the age-associated increase in the expression of the longevity-associated and tumor suppressor genes, as well as downregulation of cancer-related genes in the skin of NMRs. Furthermore, age-associated changes in the NMR cutaneous transcriptome need to be correlated with the proteome and protein activity due to post-transcriptional and post-translational modifications (Buccitelli and Selbach, 2020). These analyses will help define the unique features of the NMR skin aging transcriptome that might contribute to the resistance NMRs exhibit to spontaneous skin carcinogenesis.
Identification of these mechanisms and their relevance to human skin aging might be beneficial for developing novel approaches for the management of age-associated skin conditions, including skin cancer in humans. Taken together, these data provide a platform for further exploration of NMRs as an innovative model organism for studying the biology of human skin and serve as the starting point in the identification of novel mechanisms mediating skin resistance to age-associated pathologies in these unique mammals.
Materials and Methods
Animals and tissue collection
All experiments were conducted in accordance with the United Kingdom Animal (Scientific Procedures) Act 1986 Amendment Regulations 2012 under a Project Licenses granted to A.M. (University of Bradford) and to E. St. J. S. (University of Cambridge) by the Home Office. Animals were maintained in a custom-made caging system with conventional mouse/rat cages connected by different lengths of tunnel. The room was warmed to 28 °C, 50–60% humidity levels was maintained, and bedding, nesting material and water-enriched food were provided. Four young adult NMRs (3–4.5 year-old) and three aged NMRs (19–23 year-old) of both sexes were used in this study. Samples were collected from the dorsal skin, covered in Tissue-Tek medium (VWR, UK), snap-frozen in liquid nitrogen, and stored in −80 °C freezer (Botchkarev et al., 1998, Botchkarev et al., 1999).
Histology, histomorphometry and immunohistochemistry
For histology, 10 um cryosections were fixed in 4% PFA followed by haematoxylin/eosin staining, or by Giemsa staining for mast cell visualization, or by the modified Warthin-Starry staining for melanin detection (Joly-Tonetti et al., 2016). To detect senescent cells (Dimri et al., 1995), Senescence Associated β-galactosidase staining kit (Cell Signaling, Danvers, MA) was used. The number of mast cells and senescent cells, as well as semi-automated skin pigmentation analysis were performed per 200X microscopic field using ImageJ software (NIH, Bethesda, MD).
For immunofluorescence, 10 um cryosections were fixed and incubated with primary antibodies (Supplementary Table S8), followed by application of the secondary antibodies, as described previously (Botchkareva et al., 1999, Muller-Rover et al., 1998, Sharov et al., 2003). Quantification of immunofluorescence intensity or corrected total cell fluorescence (CTCF) was determined using ImageJ software, as described previously (Rapisarda et al., 2017). Statistical analysis was performed using unpaired Student’s t-test; differences were deemed significant if p<0.05.
RNA-seq, qRT-PCR and bioinformatics analyses
Total RNA was extracted from full-thickness dorsal skin samples obtained from young and old animals (4 year-old skin, n=2; 19 year-old skin, n=2) and purified using Direct-zol RNA purification kit (Zymo Research; Irvine, CA) following by DNAse treatment. Integrity of RNA was assessed on 2100 Bioanalyzer Instrument (Agilent Technologies, Inc.), only samples with RIN factor above 7.5 were used for NGS library preparation. For RNA sequencing, approximately 10 ug of total RNA was used to remove ribosomal RNA according to the manuscript of the Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, USA). The pair-end 2×150bp sequencing was performed on an Illumina Hiseq 4000 platform in the LC Sciences (Houston, TX).
qRT-PCR was performed with iQ SYBR Green Supermix (Bio-Rad Laboratories), as described previously (Mardaryev et al., 2014, Mardaryev et al., 2016). The primer sequences (Supplementary Table S9) were obtained from previously published datasets (Tian et al., 2015). Differences between samples were calculated based on the Ct (ΔΔCt) method and normalized to β-actin. Statistical analysis was performed using an unpaired Student’s t test.
The detailed description of the bioinformatics and protein-protein interaction network analyses is provided in Supplementary Text.
Data Availability Statement
Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200736, hosted at GEO (accession number GSE200736).
Supplementary Material
Acknowledgments
Authors thank L. Faulkes for help in preparation of the NMR images for this manuscript. This study was supported in part by the grants from NIAMS to VAB, AS and VG (R61AR078093), NIA to VG and AS, Dunhill Medical Trust (RPGF2002\188) to EStJS. HAGR is supported by a Biotechnology and Biological Sciences Research Council UK (BB/R014949/1) grant to JPM. AG was supported by the Leibniz Science Campus Chronic Inflammation (A.G., www.chronische-entzuendung.org).
Footnotes
Conflict of Interest The authors state no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed MI, Alam M, Emelianov VU, Poterlowicz K, Patel A, Sharov AA, et al. MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J Cell Biol 2014;207(4):549–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramillo Irizar P, Schauble S, Esser D, Groth M, Frahm C, Priebe S, et al. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat Commun 2018;9(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assabban A, Dubois-Vedrenne I, Van Maele L, Salcedo R, Snyder BL, Zhou L, et al. Tristetraprolin expression by keratinocytes protects against skin carcinogenesis. JCI Insight 2021;6(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Ahmad N, Mukhtar H. Upregulation of E2F transcription factors in chemically induced mouse skin tumors. Int J Oncol 1999;15(2):387–90. [DOI] [PubMed] [Google Scholar]
- Barron CC, Bilan PJ, Tsakiridis T, Tsiani E. Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism 2016;65(2):124–39. [DOI] [PubMed] [Google Scholar]
- Barth E, Srivastava A, Stojiljkovic M, Frahm C, Axer H, Witte OW, et al. Conserved aging-related signatures of senescence and inflammation in different tissues and species. Aging (Albany NY) 2019;11(19):8556–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 2014;15(12):786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA. The Molecular Revolution in Cutaneous Biology: Chromosomal Territories, Higher-Order Chromatin Remodeling, and the Control of Gene Expression in Keratinocytes. J Invest Dermatol 2017;137(5):e93–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkarev NV, Albers KM, van der Veen C, Lewin GR, Paus R. Neurotrophin-3 involvement in the regulation of hair follicle morphogenesis. J Invest Dermatol 1998;111(2):279–85. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol 1999;1(3):158–64. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Flores ER. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb Perspect Med 2014;4(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol 2012;132(11):2505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkareva NV, Botchkarev VA, Chen LH, Lindner G, Paus R. A role for p75 neurotrophin receptor in the control of hair follicle morphogenesis. Dev Biol 1999;216(1):135–53. [DOI] [PubMed] [Google Scholar]
- Braude S, Holtze S, Begall S, Brenmoehl J, Burda H, Dammann P, et al. Surprisingly long survival of premature conclusions about naked mole-rat biology. Biol Rev Camb Philos Soc 2021;96(2):376–93. [DOI] [PubMed] [Google Scholar]
- Brohus M, Gorbunova V, Faulkes CG, Overgaard MT, Conover CA. The Insulin-Like Growth Factor System in the Long-Lived Naked Mole-Rat. PLoS One 2015;10(12):e0145587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccitelli C, Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 2020;21(10):630–44. [DOI] [PubMed] [Google Scholar]
- Buffenstein R The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci 2005;60(11):1369–77. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Amoroso V, Andziak B, Avdieiev S, Azpurua J, Barker AJ, et al. The naked truth: a comprehensive clarification and classification of current ‘myths’ in naked mole-rat biology. Biol Rev Camb Philos Soc 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong CM, Nickoloff BJ, Elias PM, Goldsmith LA, Macher E, Maderson PA, et al. What is the ‘true’ function of skin? Exp Dermatol 2002;11(2):159–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MA, Quan T, Voorhees JJ, Fisher GJ. Extracellular matrix regulation of fibroblast function: redefining our perspective on skin aging. J Cell Commun Signal 2018;12(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosara KTB, Moffa EB, Xiao Y, Siqueira WL. Merging in-silico and in vitro salivary protein complex partners using the STRING database: A tutorial. J Proteomics 2018;171:87–94. [DOI] [PubMed] [Google Scholar]
- Daly TJ, Buffenstein R. Skin morphology and its role in thermoregulation in mole-rats, Heterocephalus glaber and Cryptomys hottentotus. J Anat 1998;193 ( Pt 4):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Marmol D, Holtze S, Kichler N, Sahm A, Bihin B, Bourguignon V, et al. Abundance and size of hyaluronan in naked mole-rat tissues and plasma. Sci Rep 2021;11(1):7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): a retrospective survey of lesions in a zoo population. Vet Pathol 2013;50(4):607–21. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 1995;92(20):9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov A, Kutuzov M, Petruseva I, Lukjanchikova N, Kashina E, Kolova E, et al. Naked mole rat cells display more efficient excision repair than mouse cells. Aging (Albany NY) 2018;10(6):1454–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol 2011;194(6):825–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Miao Y, Gur-Cohen S, Gomez N, Yang H, Nikolova M, et al. The aging skin microenvironment dictates stem cell behavior. Proc Natl Acad Sci U S A 2020;117(10):5339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN, Zhang G, Wang J. The naked mole rat genome: understanding aging through genome analysis. Aging (Albany NY) 2011;3(12):1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat Rev Genet 2014;15(8):531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunin AG, Petrov VV, Golubtzova NN, Vasilieva OV, Kornilova NK. Age-related changes in angiogenesis in human dermis. Exp Gerontol 2014;55:143–51. [DOI] [PubMed] [Google Scholar]
- He B, Zhu R, Yang H, Lu Q, Wang W, Song L, et al. Assessing the Impact of Data Preprocessing on Analyzing Next Generation Sequencing Data. Front Bioeng Biotechnol 2020;8:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Haghani A, Macoretta N, Ablaeva J, Zoller JA, Li CZ, et al. DNA methylation clocks tick in naked mole rats but queens age more slowly than nonbreeders. Nature Aging 2022;2:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BA, Fontecilla NM, Lu CP, Fuchs E, Lumpkin EA. The cellular basis of mechanosensory Merkel-cell innervation during development. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly-Tonetti N, Wibawa JI, Bell M, Tobin D. Melanin fate in the human epidermis: a reassessment of how best to detect and analyse histologically. Exp Dermatol 2016;25(7):501–4. [DOI] [PubMed] [Google Scholar]
- Keyes BE, Segal JP, Heller E, Lien WH, Chang CY, Guo X, et al. Nfatc1 orchestrates aging in hair follicle stem cells. Proc Natl Acad Sci U S A 2013;110(51):E4950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 2011;479(7372):223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Roop DR. Conflicting roles for p63 in skin development and carcinogenesis. Cell Cycle 2007;6(3):269–73. [DOI] [PubMed] [Google Scholar]
- Kouwenhoven EN, Oti M, Niehues H, van Heeringen SJ, Schalkwijk J, Stunnenberg HG, et al. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep 2015;16(7):863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaberoglu Y, Bhushan B, Hadi F, Chakrabarti S, Khaled WT, Rankin KS, et al. The material properties of naked mole-rat hyaluronan. Sci Rep 2019;9(1):6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton AK, Graham HK, Watson RE. Diverse methodologies for assessing photoaged skin. Br J Dermatol 2016;174(3):487–8. [DOI] [PubMed] [Google Scholar]
- Langton AK, Sherratt MJ, Griffiths CE, Watson RE. A new wrinkle on old skin: the role of elastic fibres in skin ageing. Int J Cosmet Sci 2010;32(5):330–9. [DOI] [PubMed] [Google Scholar]
- Larsen SB, Cowley CJ, Fuchs E. Epithelial cells: liaisons of immunity. Curr Opin Immunol 2020;62:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Travers JB, Somani AK, Spandau DF. The IGF-1/IGF-1R signaling axis in the skin: a new role for the dermis in aging-associated skin cancer. Oncogene 2010;29(10):1475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Matsumura H, Kato T, Ichinose S, Takada A, Namiki T, et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature 2019;568(7752):344–50. [DOI] [PubMed] [Google Scholar]
- Lowe R, Danson AF, Rakyan VK, Yildizoglu S, Saldmann F, Viltard M, et al. DNA methylation clocks as a predictor for ageing and age estimation in naked mole-rats, Heterocephalus glaber. Aging (Albany NY) 2020;12(5):4394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae SL, Zhang Q, Lemetre C, Seim I, Calder RB, Hoeijmakers J, et al. Comparative analysis of genome maintenance genes in naked mole rat, mouse, and human. Aging Cell 2015;14(2):288–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev AN, Gdula MR, Yarker JL, Emelianov VU, Poterlowicz K, Sharov AA, et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 2014;141(1):101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev AN, Liu B, Rapisarda V, Poterlowicz K, Malashchuk I, Rudolf J, et al. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol 2016;212(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016;15(2):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon GK, Catania KC, Crumrine D, Bradley C, Mauldin EA. Unique features of the skin barrier in naked mole rats reflect adaptations to their fossorial habitat. J Morphol 2019;280(12):1871–80. [DOI] [PubMed] [Google Scholar]
- Menon GK, Kligman AM. Barrier functions of human skin: a holistic view. Skin Pharmacol Physiol 2009;22(4):178–89. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, Peters EJ, Botchkarev VA, Panteleyev A, Paus R. Distinct patterns of NCAM expression are associated with defined stages of murine hair follicle morphogenesis and regression. J Histochem Cytochem 1998;46(12):1401–10. [DOI] [PubMed] [Google Scholar]
- Parry A, Rulands S, Reik W. Active turnover of DNA methylation during cell fate decisions. Nat Rev Genet 2021;22(1):59–66. [DOI] [PubMed] [Google Scholar]
- Pilkington SM, Bulfone-Paus S, Griffiths CEM, Watson REB. Inflammaging and the Skin. J Invest Dermatol 2021;141(4S):1087–95. [DOI] [PubMed] [Google Scholar]
- Qu J, Yi G, Zhou H. p63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes. Epigenetics Chromatin 2019;12(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Hagemann S, Aran D, Sohle J, Kulkarni PP, Kaderali L, et al. Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics Chromatin 2013;6(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapisarda V, Malashchuk I, Asamaowei IE, Poterlowicz K, Fessing MY, Sharov AA, et al. p63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope-Associated Genes in Epidermal Keratinocytes. J Invest Dermatol 2017;137(10):2157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P, et al. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell 2006;5(5):379–89. [DOI] [PubMed] [Google Scholar]
- Rittie L, Fisher GJ. UV-light-induced signal cascades and skin aging. Ageing Res Rev 2002;1(4):705–20. [DOI] [PubMed] [Google Scholar]
- Rittie L, Fisher GJ. Natural and sun-induced aging of human skin. Cold Spring Harb Perspect Med 2015;5(1):a015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KA, Valentine JM, Kramer DA, Gelfond JA, Kristan DM, Nevo E, et al. Determinants of rodent longevity in the chaperone-protein degradation network. Cell Stress Chaperones 2016;21(3):453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni E, Watt FM. Skin Cell Heterogeneity in Development, Wound Healing, and Cancer. Trends Cell Biol 2018;28(9):709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood BR, Leprince D. Deciphering HIC1 control pathways to reveal new avenues in cancer therapeutics. Expert Opin Ther Targets 2013;17(7):811–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset C, Netto CBO, Ashton-Prolla P. TSC1 and TSC2 gene mutations and their implications for treatment in Tuberous Sclerosis Complex: a review. Genet Mol Biol 2017;40(1):69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Jaffredo T, Saldmann F, Faulkes CG, Moguelet P, Leroy C, et al. Epidermal stem cell compartment remains unaffected through aging in naked mole-rats. BioRxiv 2020:doi: 10.1101/2020.11.13.381061. [DOI] [Google Scholar]
- Seluanov A, Gladyshev VN, Vijg J, Gorbunova V. Mechanisms of cancer resistance in long-lived mammals. Nat Rev Cancer 2018;18(7):433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Li GZ, Palkina TN, Sharova TY, Gilchrest BA, Botchkarev VA. Fas and c-kit are involved in the control of hair follicle melanocyte apoptosis and migration in chemotherapy-induced hair loss. J Invest Dermatol 2003;120(1):27–35. [DOI] [PubMed] [Google Scholar]
- Shebzukhov Y, Holtze S, Hirseland H, Schafer H, Radbruch A, Hildebrandt T, et al. Identification of cross-reactive antibodies for the detection of lymphocytes, myeloid cells and haematopoietic precursors in the naked mole rat. Eur J Immunol 2019;49(11):2103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski AT, Zmijewski MA, Skobowiat C, Zbytek B, Slominski RM, Steketee JD. Sensing the environment: regulation of local and global homeostasis by the skin’s neuroendocrine system. Adv Anat Embryol Cell Biol 2012;212:v, vii, 1–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ESJ, Park TJ, Lewin GR. Independent evolution of pain insensitivity in African mole-rats: origins and mechanisms. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 2020;206(3):313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18(11):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacpoole PW. The pyruvate dehydrogenase complex as a therapeutic target for age-related diseases. Aging Cell 2012;11(3):371–7. [DOI] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 2009;5(1):64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, Craig T, et al. Human Ageing Genomic Resources: new and updated databases. Nucleic Acids Res 2018;46(D1):D1083–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasugi M, Firsanov D, Tombline G, Ning H, Ablaeva J, Seluanov A, et al. Naked mole-rat very-high-molecular-mass hyaluronan exhibits superior cytoprotective properties. Nat Commun 2020;11(1):2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thigpen LW. Histology of the Skin of a Normally Hairless Rodent. J Mammalogy 1940;21(4):449–56. [Google Scholar]
- Tian X, Azpurua J, Ke Z, Augereau A, Zhang ZD, Vijg J, et al. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc Natl Acad Sci U S A 2015;112(4):1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutfaire M, Bauwens E, Debacq-Chainiaux F. The impact of cellular senescence in skin ageing: A notion of mosaic and therapeutic strategies. Biochem Pharmacol 2017;142:1–12. [DOI] [PubMed] [Google Scholar]
- Tucker R The digging behavior and skin differentiations in Heterocephalus glaber. J Morphol 1981;168(1):51–71. [DOI] [PubMed] [Google Scholar]
- Victorelli S, Lagnado A, Halim J, Moore W, Talbot D, Barrett K, et al. Senescent human melanocytes drive skin ageing via paracrine telomere dysfunction. EMBO J 2019;38(23):e101982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijer ME, Gunn DA, Adams PD, Pawlikowski JS, Griffiths CE, van Heemst D, et al. P16INK4a Positive Cells in Human Skin Are Indicative of Local Elastic Fiber Morphology, Facial Wrinkling, and Perceived Age. J Gerontol A Biol Sci Med Sci 2016;71(8):1022–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijer ME, Parish WE, Strongitharm BH, van Heemst D, Slagboom PE, de Craen AJ, et al. The number of p16INK4a positive cells in human skin reflects biological age. Aging Cell 2012;11(4):722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijer MEC, Gunn DA, van Heemst D, Slagboom PE, Sedivy JM, Dirks RW, et al. Do senescence markers correlate in vitro and in situ within individual human donors? Aging (Albany NY) 2018;10(2):278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt B, van Niel G, Raposo G, Marks MS. PMEL: a pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res 2013;26(3):300–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol 2010;222(1):66–72. [DOI] [PubMed] [Google Scholar]
- Yaar M, Eller MS, Gilchrest BA. Fifty years of skin aging. J Investig Dermatol Symp Proc 2002;7(1):51–8. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tyshkovskiy A, Munoz-Espin D, Tian X, Serrano M, de Magalhaes JP, et al. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proc Natl Acad Sci U S A 2018;115(8):1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200736, hosted at GEO (accession number GSE200736).


