Abstract
Background & Aims:
Pancreatic ductal adenocarcinoma (PDAC) incidence is rising worldwide, and majority of patients present with an unresectable disease at initial diagnosis. Measurement of carbohydrate antigen 19-9 (CA19-9) levels lack adequate sensitivity and specificity for early detection; hence, there is an unmet need to develop alternate molecular diagnostic biomarkers for PDAC. Emerging evidence suggests that tumor-derived exosomal cargo, particularly miRNAs, offer an attractive platform for the development of cancer-specific biomarkers. Herein, genomewide profiling in blood specimens was performed to develop an exosome-based transcriptomic signature for noninvasive and early detection of PDAC.
Methods:
Small RNA-sequencing was undertaken in a cohort of 44 patients with an early-stage PDAC and 57 non-disease controls. Using machine-learning algorithms, a panel of cell-free (cf) and exosomal (exo) miRNAs was prioritized that discriminated PDAC patients from control subjects. Subsequently, the performance of the biomarkers was trained and validated in independent cohorts (n=191) using quantitative real time PCR (qRT-PCR) assays.
Results:
The sequencing analysis initially identified a panel of 30 overexpressed miRNAs in PDAC. Subsequently using qRT-PCR assays, the panel was reduced to 13 markers (5 cf- and 8 exo-miRNAs), which successfully identified patients with all stages of PDAC (AUC=0.98 training cohort; AUC=0.93 validation cohort); but more importantly, was equally robust for the identification of early-stage PDAC (stages 1&II; AUC=0.93). Furthermore, this transcriptomic signature successfully identified CA19-9 negative cases (<37 U/ml; AUC=0.96), when analyzed in combination with CA19-9 levels, significantly improved the overall diagnostic accuracy (AUC=0.99 vs. AUC=0.86 for CA19-9 alone).
Conclusions:
In this study, an exosome-based liquid-biopsy signature for the noninvasive and robust detection of patients with PDAC was developed.
Keywords: miRNA signature, Exosome, Pancreatic cancer, Diagnostic biomarker, Liquid biopsy
Graphical Abstract
LAY SUMMARY:
Our exosome-based transcriptomic signature that combines cell-free and exosomal microRNAs has the potential to identify patients with pancreatic ductal adenocarcinoma with high diagnostic accuracy, and offers an important noninvasive assay for early detection of this fatal malignancy.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the most common malignancy of the pancreas1–3. PDAC presents a substantial health problem with rising incidence and is predicted to become the second leading cause of cancer-associated mortality within the next decade in United States4, 5. As per the standard treatment strategies, surgical removal of the localized tumor offers the only potential curative option for this disease1, 6. Clinical findings in recent years have unequivocally established that surgery followed with modern adjuvant chemotherapy significantly improves patient outcomes, with a median overall survival ranging from 46 to 54 months in PDAC patients7, 8. Unfortunately, however, the patients who present with localized, resectable, and potentially curable tumors at initial diagnosis are only less than 15-20% of all cases, whereas the remainder have a more advanced unresectable or metastatic disease1, 3. This is reflected in the data that despite recent advances in treatment modalities, the 5-year survival rates in PDAC patients have essentially not improved significantly in the recent decades9–11.
One of the themes that has emerged in the recent years is that earlier diagnosis of disease offers a promising opportunity for a timely intervention and subsequent improvement in survival of patients suffering from this fatal malignancy. In this context, to date, several blood-based biomarkers have been evaluated for their clinical usefulness for early diagnosis of PDAC. Serum carbohydrate antigen 19-9 (CA19-9) remains the most well-documented and widely used serum biomarker in patients with PDAC2, 3, 12. Although CA19-9 is commonly used to monitor disease progression and therapeutic response, it lacks satisfactory sensitivity or specificity for screening and early detection of patients with PDAC13, 14. Importantly, 15-25% of patients with PDAC, including those at early-stages, often exhibit CA19-9 levels less than 37 U/ml which is considered as normal15, 16. Furthermore, 5-10% of the general population is Lewis antigen-negative with no or low secretion of CA19-917. These clinical challenges highlight the urgent need to develop robust alternate, preferably non-invasive, biomarkers for the early diagnosis of PDAC.
With recent advances in high-throughput molecular profiling technologies, the use of blood-based biomarkers for cancer diagnosis has gained significant momentum in the form of circulating proteins, DNA and various RNA molecules18–21. Within the transcriptomic landscape, microRNAs (miRNAs) represent single-stranded RNAs that are 18–25 nucleotides long, that are involved in gene regulation, oncogenesis and are frequently dysregulated in different cancers including PDAC22, 23. Since miRNAs are resistant to nuclease-mediated degradation and its abundance in tissues, blood, and other body fluids due to their small size, they have emerged as promising candidates for liquid-biopsy based molecular biomarkers in human cancers23, 24. However, it is well-recognized that not only tumor cells, but multiple other sources, including apoptotic and immune cells release cell-free miRNAs (cf-miRNAs) in circulation25. Thus, there is some debate in the field with regards to the potential heterogeneity associated with the origin of cf-miRNAs.
The recent discovery of exosomes – small membranous microvesicles, 40-140 nm in size, which inherit molecular signatures from their cells-of-origin, has brought a great degree of enthusiasm to the cancer biomarker arena26–28. Initially, exosomes were thought to be involved in the disposal of cellular garbage, but recent data offers compelling evidence that they play an important role in cell-to-cell communication, through the transfer of their molecular cargo (e.g., proteins and nucleic acids) within the tumor microenvironment. Cancer cells, analogous to healthy cells, secrete exosomes and carry distinct pathogenic miliue27. Exosomes excreted by cancer cells appear to possess specific exosomal cargos including miRNAs and offer abundant representation of tumor-derived miRNAs in systemic circulation; and hence provide an attractive paradigm for more specific detection of miRNA biomarkers in blood26, 27. Given that cf-miRNAs offer excellent sensitivity and exo-miRNAs are highly tissue-specific, a combination of the two could offer an optimal mix of sensitivity and specificity – an approach that is unique, merits attention and has not been previously explored for the early detection of pancreatic cancer29, 30.
In this present study, we performed a systematic and comprehensive genomewide transcriptomic profiling of a large number of clinical specimens from patients with early-stage PDAC (stages I and II) and appropriate non-disease control subjects to discover a novel cell-free and exosomal miRNA signature that facilitates early detection of patients with earliest stages of PDAC. Following biomarker discovery, we rigorously evaluated and validated the performance of this noninvasive circulating signature in multiple independent clinical cohorts to assess its diagnostic performance for the early detection of patients with PDAC.
MATERIALS AND METHODS
Patient cohorts
In this study, a total of 292 subjects (168 PDAC patients and 124 non-disease controls) which were segregated into a biomarker discovery cohort that was subjected to small RNA sequencing, as well as clinical training and validation cohorts were enrolled. The detailed clinicopathological characteristics of all PDAC cases and non-disease controls are presented in Supplementary Table 1. For the biomarker discovery phase, small RNA sequencing was performed in a total of 101 plasma and serum specimens which included 44 patients with early-stage PDAC (stages I and II) and 57 non-disease controls, who were enrolled at the Samsung Medical Center and Asan Medical Center, Seoul, Korea, between 2009 and 2017. In the clinical training and validation phases, qRT-PCR assays was performed to examine the expression levels of candidate cell-free and exosomal-miRNAs in 191 specimens from 124 patients with PDAC and 67 non-disease controls, who were enrolled at the Ochsner Clinic Foundation (New Orleans, LA, USA) and Nagoya University Hospital (Nagoya, Japan) between 2016 and 2020. To minimize any potential bias between these two patient populations, the two cohorts were combined and randomly divided into two cohorts (training cohort, n=96; validation cohort, n=95) for qRT-PCR based performance evaluation. This study was conducted in accordance with the Declaration of Helsinki. A written informed consent was obtained from all the subjects, and the study was approved by respective institutional review boards.
Exosomal and cell-free RNA extraction
To prepare libraries for small RNA sequencing, total exosomal RNA and cell-free RNA were isolated from 400 μL of plasma, using exoRNeasy Midi Kit and miRNeasy kit (Qiagen, Valencia, CA, USA), respectively. For qRT-PCR analysis, exosomes were first isolated from 200 μL plasma was performed using Total Exosome Isolation Kit (Invitrogen, Waltham, MA, USA), followed by RNA extraction using miRNeasy Kit (Qiagen). Cell-free total RNA was isolated from 200 μL plasma using miRNeasy Kit (Qiagen).
Small RNA sequencing and discovery analysis for identification of miRNA candidates Exosomal and cell-free RNA was prepared for library preparation by using the NEXTflex™ Small RNA-Seq Kit v3 (PerkinElmer, Waltham, MA, USA). Following size exclusion and quality assessment, the sequencing libraries were pooled, and paired-end sequencing was performed using an Illumina NovaSeq platform. The Cutadapt (v2.2) pipeline was used to trim the adapters and reads with low quality were removed. Next, the miRDeep2 module was used to align miRNA sequences (against miRBase release 22) and quantify miRNA expression. The miRNA abundance was calculated based on counts per million (CPM). The R package, limma, was used to perform differential expression analysis to identify miRNA candidates between early-stage PDAC patients and controls.
Real-time quantitative reverse transcription polymerase chain reaction
The cell-free and exosomal RNAs were first subjected to cDNA synthesis followed by LNA miRNA PCR assays using miRCURY LNA RT Kit (Qiagen). The expression of miRNAs was quantified by a SensiFAST™ SYBR Lo-ROX Kit (Bioline, London, UK) using QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The expression of miR-16-5p and miR103-3p were used as endogenous controls for data normalization. The expression of each miRNA was calculated using 2-ΔCt method. Normalized expression values were further log10 transformed.
Measurement of CA19-9 levels
The serum levels of CA19-9 were measured for all specimens analyzed for qRT-PCR based clinical training and validation by using the enzyme-linked immunosorbent assay (ELISA) kits from Alpha Diagnostic International (San Antonio, TX, USA), as per manufacturer’s instructions.
miRNA-mRNA regulatory network analysis
miRNA-mRNA regulatory network analysis was performed based on miRNA-target interactions predicted by at least three programs using starBase31 (https://starbase.sysu.edu.cn/). For higher specificity, the network was further filtered to retain target genes (log2-fold change > 0.5 and adjusted P value < 0.05) that were differentially expressed between PDAC and normal tissue samples using the GSE62452 dataset which included 69 pancreatic tumors and 61 adjacent nontumor tissues. Furthermore, functional annotation was performed on the miRNA target genes in the network based on KEGG pathways and cancer hallmark gene sets in the MSigDB32 database using the “clusterProfiler” package33.
Statistical analysis
Statistical analyses were performed using R (version 4.0.3, https://cran.r-project.org/), MedCalc Statistical Software version 20.009 (MedCalc Software Ltd, Ostend, Belgium) and GraphPad Prism 8 software (La Jolla, CA). Area under the curve values (AUCs) derived from the receiver operating characteristic (ROC) curves were calculated with confidence intervals (CIs) using the pROC package in R. All ROC curves presented in the results were represented along with 95% CI. The CI values were calculated by 2,000 bootstrap replicates. The optimal cutoff thresholds for the ROC curves were determined using Youden’s index. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), precision and accuracy of the miRNA signatures were calculated across all the cohorts using pROC package. P value of <0.05 was considered as statistically significant. The Wilcoxon test was used to compare risk scores from qRT-PCR experiments between different stages and healthy controls. Multivariate logistic regression was employed to derive a formula to predict risk for development of PDAC.
RESULTS
Genomewide profiling identifies a panel of cell-free and exosomal-miRNA biomarkers for the identification of patients with early-stage PDAC
The primary objective of this study was to identify clinically relevant cell-free miRNAs (cf-miRNAs) and exosomal miRNAs (exo-miRNAs) as biomarkers for the identification of patients with PDAC, but more importantly, for those with an early-stage disease. Towards these efforts, a systematic comprehensive, unbiased, genomewide small RNA-sequencing based biomarker discovery effort in total plasma and exosomal RNA specimens from patients with early-stage PDAC (stages I and II) and non-disease controls was performed – an approach, which has not been undertaken previously. Following sequencing, rigorous bioinformatic and statistical analyses was performed to identify candidate miRNAs that were significantly and differentially expressed between PDAC patients (n=44) vs. non-disease controls (n=57). The non-disease controls subjects were defined as asymptomatic individuals with normal abdominal CTs or negative screening endoscopies. The following criteria was used for prioritizing candidate miRNAs: for cf-miRNAs an AUC value of >0.7 and a log fold change of >1; and for exosomal miRNAs an AUC value of >0.8 and log fold change of >1. The robustness of identified candidate biomarkers was ensured by performing, 1000 time cross-validation and the markers that were most stable in such iterative analysis were selected for further study.
Bioinformatic and statistical analyses resulted in the identification of a panel of 13 cf-miRNAs (let-7e-5p, let-7f-5p, miR-30c-5p, miR-369-3p, miR-125a-5p, miR-26a-5p, miR-495-3p, miR-223-3p, miR-142-3p, miR-340-5p, miR-340-3p miR-335-5p, and miR-23b-3p) and 17 exo-miRNAs (miR-1260a, miR-1260b, miR-141-3p, miR-143-3p, miR-145-5p, miR-148a-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-216a-5p, miR-216b-5p, miR-217-5p, miR-34a-5p, miR-375-3p, miR-429, miR-199a-5p, and miR-145-3p) – all of which were significantly upregulated in patients with stage I/II PDAC vs. non-disease controls (Figure 1A and Figure 1B). While these individual markers were quite robust, in order to evaluate the performance of these candidates as combinatorial panels, a logistic regression model using the coefficients derived from each of the 13 cf- and 17 exo-miRNAs in this discovery cohort was developed. It was interesting to observe that the predictive probabilities deduced from the logistic regression model demonstrated an excellent diagnostic potential for these two types of miRNA panels; where the cf-miRNA panel exhibited an AUC value of 0.96 (95% CI, 0.93-1.00; sensitivity of 92%, specificity of 93%, PPV of 94%, and NPV of 91%) and the exo-miRNA panel yielded an AUC of 1.00 (CI, 0.99-1.00; sensitivity of 98%, specificity of 100%, PPV of 100%, and NPV of 98%). However, more importantly, the performance of these individual panels were significantly enhanced as a combination signature (AUC, 1.00; 95% CI, 1.00-1.00; sensitivity of 100%, specificity of 100%, PPV of 100%, and NPV of 100%). In summary, these biomarker discovery findings supported the original hypothesis that while both cf-and exo-miRNA markers were quite robust, their cumulative analysis offers a superior combination of sensitivity and specificity for the robust identification of patients with early-stage PDAC.
Figure 1.
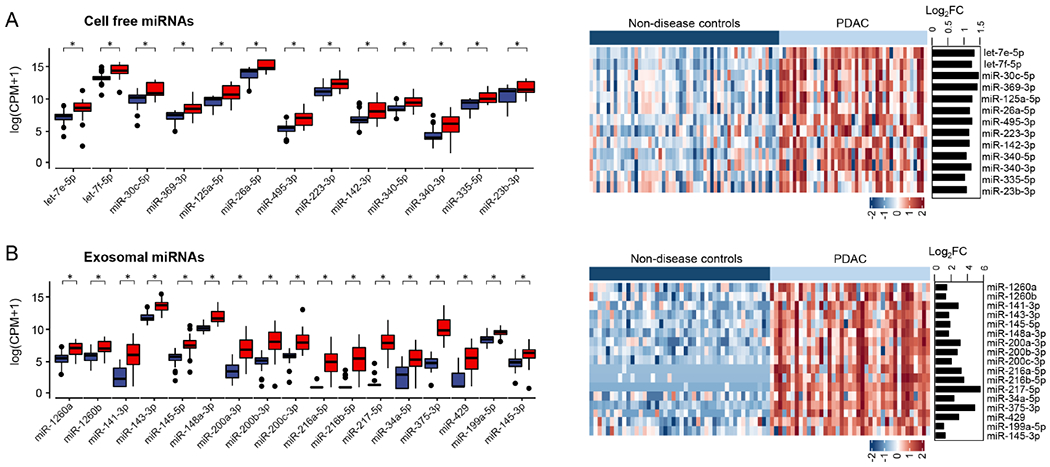
Expression level of identified cell-free and exosomal miRNA candidates for the diagnosis of patients with early-stages of PDAC obtained from genome-wide small RNA sequencing. (A) Expression level of candidate cf-miRNAs and representative heatmap in patients with early-stage of PDAC (Stage I-II) versus non-disease control samples. (B) Expression level of candidate exo-miRNAs and representative heatmap in patients with PDAC (Stage I-II) versus non-disease control samples. The miRNA expression profile was z-normalized. [miRNA: micro RNA; CPM: counts per million; FC: fold change; PDAC: pancreatic ductal adenocarcinoma (light blue) and Non-disease controls (dark blue)].
Training of a miRNA-based risk prediction model for the early detection of patients with PDAC To establish a diagnostic assay for routine clinical implementation based upon discovered miRNA biomarkers, qRT-PCR based assays in blood specimens obtained from patients in independent clinical cohorts were performed. It was quite reassuring to notice that even in qRT-PCR analysis, 9 of 13 cf-miRNAs (let-7e-5p, miR-30c-5p, miR-26a-5p, miR-223-3p, miR-142-3p, miR-340-5p, miR-340-3p, miR-335-5p, and miR-23b-3p) and 15 of 17 exo-miRNAs (miR-1260a, miR-1260b, miR-141-3p, miR-143-3p, miR-145-5p, miR-148a-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-216a-5p, miR-216b-5p, miR-217-5p, miR-34a-5p, miR-429, and miR-145-3p) were readily detectable; underscoring the robustness of the biomarker discovery efforts. The significantly upregulated (log fold change >1 and p< 0.05) cf- and exo-miRNA candidates between patients with PDAC and non-disease controls were represented as heat maps in Figure 2A and Figure 2B, respectively. The diagnostic performance of each miRNA candidate in terms of their AUC values, diagnostic accuracy, NPV, PPV, sensitivity and specificity were summarized in Table 1.
Figure 2.
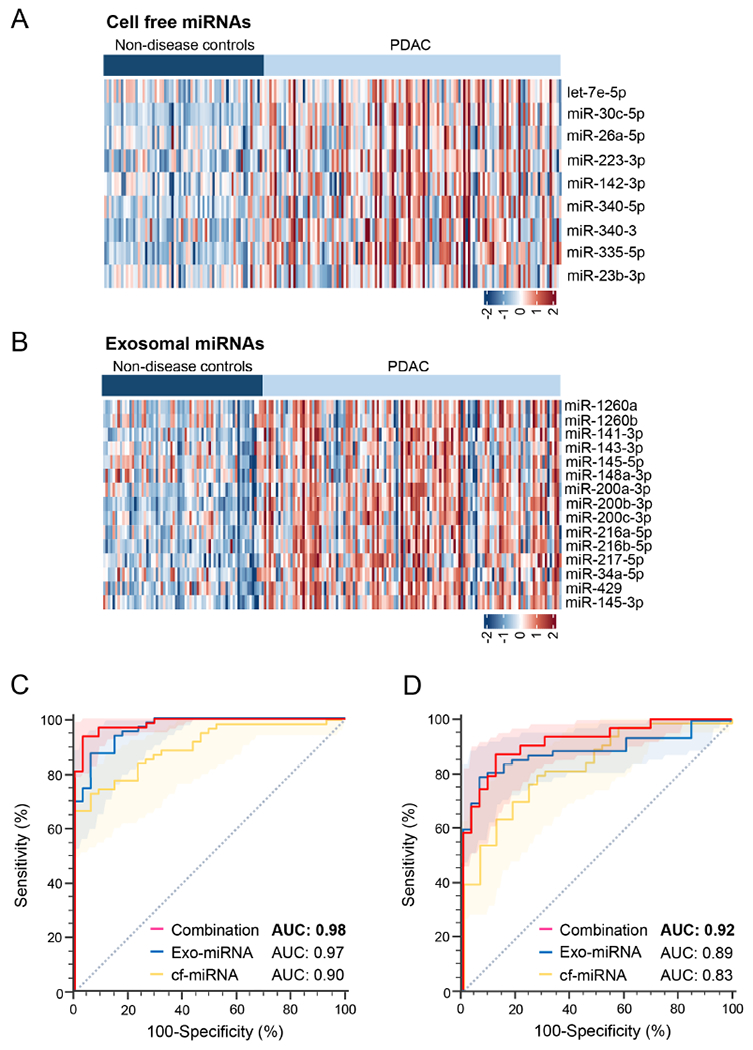
Performance evaluation of cell-free and exosomal miRNA biomarker panel in clinical cohorts by qRT-PCR. Representative heatmap of statistically significant and upregulated candidate (A) cf-miRNAs and (B) exo-miRNAs in patients with PDAC versus non-disease controls. (C) ROC curves analysis for the cf-miRNA, exo-miRNA or cf- and exocombination panel in the training cohort. (D) ROC curves analysis for the cf-miRNA, exo-miRNA and cf and exosomal combination panels in the validation cohort. [Exo: exosomal; miRNA: micro RNA; qRT-PCR: Quantitative Reverse transcription polymerase chain reaction; cf: cell-free; AUC: Area under the curve; ROC: receiver operating characteristic]
Table 1.
Summary of diagnostic performance of cell free and exosomal miRNA-based biomarkers.
| miRNA | AUC (95% CI) | Accuracy (%) (95% CI) | PPV (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | NPV (%) (95% CI) |
|---|---|---|---|---|---|---|
| cf-miR30c-5p | 0.79 (0.72-0.85) | 75 (69-81) | 91 (86-96) | 68 (59-77) | 88 (79-96) | 60 (53-67) |
| cf-let7e-5p | 0.67 (0.59-0.75) | 65 (59-72) | 79 (72-86) | 64 (56-72) | 69 (57-79) | 51 (44-58) |
| cf-miR340-5p | 0.67 (0.60-0.75) | 66 (60-73) | 87 (80-93) | 57 (48-66) | 84 (75-91) | 51 (46-58) |
| cf-miR223-3p | 0.73 (0.66-0.81) | 70 (64-76) | 77 (72-82) | 77 (69-84) | 58 (46-69) | 57 (49-67) |
| cf-miR26a-5p | 0.60 (0.52-0.68) | 57 (51-63) | 92 (84-98) | 37 (29-45) | 94 (88-99) | 45 (41-48) |
| cf-miR340-3p | 0.71 (0.64-0.79) | 69 (62-75) | 80 (75-86) | 69 (60-77) | 69 (58-79) | 54 (47-62) |
| cf-miR335-5p | 0.77 (0.71-0.84) | 73 (67-79) | 90 (85-95) | 66 (58-74) | 87 (78-94) | 58 (52-65) |
| cf-miR23b-3p | 0.57 (0.49-0.65) | 52 (47-57) | 92 (84-100) | 28 (21-36) | 96 (91-100) | 42 (39-45) |
| cf-miR142-3p | 0.65 (0.58-0.73) | 62 (57-69) | 92 (85-98) | 46 (37-55) | 93 (87-99) | 48 (44-53) |
| exo-miR200c-3p | 0.69 (0.62-0.77) | 61 (55-68) | 91 (84-97) | 45 (37-54) | 91 (85-97) | 47 (43-52) |
| exo-miR148a-3p | 0.59 (0.50-0.67) | 68 (61-73) | 71 (67-76) | 84 (77-90) | 37 (27-49) | 56 (43-69) |
| exo-miR216a-5p | 0.71 (0.64-0.78) | 65 (59-72) | 82 (75-88) | 60 (51-68) | 75 (64-85) | 50 (44-57) |
| exo-miR145-5p | 0.59 (0.51-0.67) | 62 (55-69) | 74 (68-79) | 65 (56-73) | 57 (45-69) | 47 (39-55) |
| exo-miR200b-3p | 0.87 (0.81-0.92) | 80 (74-85) | 94 (89-98) | 73 (65-81) | 91 (84-97) | 65 (58-72) |
| exo-miR143-3p | 0.63 (0.55-0.71) | 58 (51-65) | 79 (71-87) | 48 (40-57) | 76 (66-85) | 44 (39-50) |
| exo-miR34a-5p | 0.66 (0.58-0.74) | 66 (59-73) | 79 (73-86) | 65 (56-73) | 69 (57-79) | 51 (44-59) |
| exo-miR429 | 0.76 (0.68-0.83) | 70 (63-76) | 83 (77-89) | 68 (60-76) | 75 (64-85) | 56 (48-63) |
| exo-miR141-3p | 0.78 (0.72-0.85) | 72 (66-79) | 93 (87-98) | 62 (54-71) | 91 (84-97) | 57 (51-63) |
| exo-miR1260b | 0.60 (0.52-0.69) | 60 (53-67) | 74 (67-80) | 60 (52-69) | 60 (48-72) | 45 (38-52) |
| exo-miR145-3p | 0.80 (0.74-0.86) | 75 (69-81) | 91 (86-97) | 68 (60-76) | 88 (81-96) | 60 (54-67) |
| exo-miR216b-5p | 0.79 (0.72-0.85) | 74 (68-80) | 91 (85-96) | 66 (57-74) | 88 (79-96) | 59 (52-65) |
| exo-miR200a-3p | 0.76 (0.69-0.82) | 67 (61-73) | 89 (82-95) | 56 (48-65) | 87 (79-94) | 52 (47-58) |
| exo-miR1260a | 0.69 (0.61-0.76) | 67 (60-73) | 79 (72-85) | 68 (60-76) | 66 (54-76) | 53 (45-60) |
| exo-miR217-5p | 0.79 (0.73-0.85) | 72 (66-78) | 92 (86-97) | 63 (54-71) | 90 (82-97) | 57 (51-63) |
[miRNA: micro RNA; cf: cell-free miRNA; Exo: exosomal; AUC: area under the curve; PPV: positive predictive value; NPV: negative predictive value]
Based upon these findings, the final lists of 24 candidate miRNA biomarkers were subsequently included in a logistic regression analysis for training a risk-prediction model for the identification of patients with PDAC in a cohort of 62 PDAC patients and 34 non-disease controls. During this model development, the coefficients and constants derived from the logistic regression equation was applied to calculate risk scores for each of the markers within the cf- and exo-miRNA panels for their ability to diagnose any patient with PDAC, as follows: cf-miRNA panel-[(3.907893 X cf-miR30c-5p) + (−0.13495 X cf-let7e) + (0.979111 X cf-miR340-5p) + (−0.01936 X cf-miR223-3p) + (−0.38275 X cf-miR26a-5p) + (0.195893 X cf-miR340-3p) + (0.718554 X cf-miR335-5p) + (−1.75009 X cf-miR23b-3p) + (−1.3107 X cf-miR142-3p) + 1.501128]; and exo-miRNA panel-[(−0.03638 X exo-miR200c-3p) + (1.087279 X exo-miR148a-3p) + (1.04184 X exo-miR216a-5p) + (−3.1777 X exo-miR145-5p) + (4.112727 X exo-miR200b-3p) + (0.057046 X exo-miR143-3p) + (0.118696 X exo-miR34a-5p) + (−0.92937 X exo-miR429) + (−0.57374 X exo-miR141-3p) + (−4.10024 X exo-miR1260b) + (2.42878 X exo-miR145-3p) + (0.962229 X exo-miR216b-5p) + (1.189049 X exo-miR200a-3p) + (1.691518 X exo-miR1260a) + (1.622576 X exo-miR217-5p) + 2.981741]. While the performance of most individual markers was quite remarkable, it was noted that the combined analysis of these markers within each panel was significantly superior in terms of their overall diagnostic accuracy (9 cf-miRNA panel AUC, 0.90; sensitivity, 73% and specificity, 94%; 15 exo-miRNA panel AUC, 0.97; sensitivity, 87% and specificity, 94%; Figure 2C and Table 2). Subsequently, risk scores derived from cf- and exo-miRNA panels were combined and the diagnostic potential of this combined transcriptomic signature was evaluated. In accordance with previous findings from the discovery cohort, once again it was noted that the diagnostic performance of the trained model for this combined signature was significantly superior vis-a-vis individual panels, as this yielded an AUC value of 0.98 with a corresponding sensitivity of 94% and a specificity of 97% (Figure 2C and Table 2). Importantly, the overall diagnostic accuracy (95%), PPV (98%) and NPV (89%) of this combined signature was significantly superior to that of cf- and exo-miRNA panels individually, which was consistent with the findings from the biomarker discovery cohort. Taken together, this genomewide transcriptomic profiling efforts yielded clinically relevant miRNA biomarkers that further allowed for successful establishment and training of a risk-prediction model for cf- and exo-miRNAs individually, as well as their combination, for the robust identification of patients with PDAC.
Table 2.
Summary of diagnostic performance of cell-free and exosomal miRNA-based biomarker panel in the training and validation cohorts.
| Cohort | Signature | Control | Cancer | AUC (95% CI) | Accuracy (%) (95% CI) | PPV (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | NPV (%) (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| All miRNA candidates | Training cohort (n=96) | cf-miRNA panel | 34 | 62 | 0.90 (0.84-0.96) | 80 (73-88) | 96 (90-100) | 73 (61-82) | 94 (85-100) | 65 (57-75) |
| exo-miRNA panel | 34 | 62 | 0.97 (0.94-1.00) | 90 (83-96) | 96 (92-100) | 87 (77-95) | 94 (85-100) | 80 (70-91) | ||
| combined signature | 34 | 62 | 0.98 (0.97-1.00) | 95 (90-99) | 98 (95-100) | 94 (87-98) | 97 (91-100) | 89 (80-97) | ||
|
| ||||||||||
| Validation cohort (n=95) | cf-miRNA panel | 33 | 62 | 0.83 (0.75-0.91) | 77 (68-85) | 85 (77-93) | 79 (68-89) | 73 (58-88) | 65 (53-78) | |
| exo-miRNA panel | 33 | 62 | 0.89 (0.82-0.95) | 84 (77-92) | 96 (91-100) | 79 (68-89) | 94 (85-100) | 71 (61-82) | ||
| combined signature | 33 | 62 | 0.92 (0.87-0.97) | 87 (80-94) | 93 (87-98) | 87 (77-95) | 88 (76-97) | 79 (68-90) | ||
|
| ||||||||||
| Reduced miRNA candidates | Training cohort (n=96) | cf-miRNA panel | 34 | 62 | 0.90 (0.85-0.96) | 84 (77-91) | 91 (84-98) | 84 (74-92) | 85 (73-97) | 74 (64-86) |
| exo-miRNA panel | 34 | 62 | 0.96 (0.92-0.99) | 90 (83-96) | 95 (90-100) | 89 (81-95) | 91 (82-100) | 82 (71-92) | ||
| combined signature | 34 | 62 | 0.98 (0.95-1.00) | 94 (89-98) | 98 (95-100) | 92 (85-98) | 97 (91-100) | 87 (78-97) | ||
|
| ||||||||||
| Validation cohort (n=95) | cf-miRNA panel | 33 | 62 | 0.84 (0.76-0.92) | 80 (72-87) | 92 (85-98) | 76 (65-85) | 88 (76-97) | 66 (56-78) | |
| exo-miRNA panel | 33 | 62 | 0.89 (0.82-0.96) | 85 (78-92) | 96 (91-100) | 81 (69-90) | 94 (85-100) | 72 (62-83) | ||
| combined signature | 33 | 62 | 0.93 (0.88-0.98) | 83 (76-91) | 94 (88-100) | 79 (69-89) | 91 (79-100) | 70 (60-81) | ||
[miRNA: micro RNA; cf: cell-free miRNA; Exo: exosomal; AUC: area under the curve; PPV: positive predictive value; NPV: negative predictive value]
Successful validation of the circulating miRNA diagnostic signature in an independent cohort of patients with PDAC
Next, the diagnostic potential of miRNA assay was interrogated in an independent validation cohort of 62 PDAC patients and 33 non-disease controls by performing qRT-PCR based assays in plasma specimens. In this validation effort, the same logistic regression equation and the coefficients of each individual miRNAs and constants obtained from the training cohort model was used and the risk scores were calculated. In accordance with the data obtained from the training cohort, it was encouraging to note that the diagnostic potential for the cf-miRNA panel (AUC=0.83, Sensitivity=79% and Specificity=73%) and exo-miRNA panel (AUC=0.89, Sensitivity=79% and Specificity=94%; Figure 2D and Table 2), was quite comparable even in this independent cohorts of PDAC patients and controls. Likewise, as was the case in the training cohort, the combined cf- and exo miRNA signature exhibited a superior diagnostic performance with an AUC value of 0.92, Sensitivity of 87%, Specificity of 88%, PPV of 93%, and NPV of 79%; Figure 2D and Table 2) in this validation cohort. Collectively, the successful validation of the diagnostic performance of the biomarker panels and training of risk-prediction model in an independent cohort of patients with PDAC was performed. These results highlighted that while individual cf and exo-miRNA panels were quite robust, the combined transcriptomic signature demonstrated a superior diagnostic performance for the identification of patients with PDAC.
Establishment of a clinically feasible signature using a reduced number of biomarkers for the noninvasive identification of patients with PDAC
In order to develop a clinically feasible and cost-effective assay that includes only the minimal number of markers required for maintaining the overall diagnostic performance of the cf- and exo-miRNAs, the miRNA biomarker candidates were prioritized using a systematic backward elimination approach. This statistical strategy resulted in a reduced panel of 13 markers, which included 5 cf-miRNAs (miR30c-5p, miR340-5p, miR335-5p, miR23b-3p and miR142-3p) and 8 exo-miRNA candidates (miR145-5p, miR200b-3p, miR429, miR1260b, miR145-3p, miR216b-5p, miR200a-3p and miR217-5p). The performance of these reduced marker panels individually, and their combination, to discriminate PDAC patients from non-disease controls in both the clinical training and validation cohorts were summarized in Table 2.
Next, this reduced number of 13 candidate miRNA biomarkers were used for developing logistic regression equation to re-calibrate the final risk-prediction model in the training cohort of patients, as follows: cf-miRNA panel-[(3.8758 X cf-miR30c-5p) + (0.9970 X cf-miR340-5p) + (0.8286 X cf-miR335-5p) + (−1.9845 X cf-miR23b-3p) + (−1.4499 X cf-miR142-3p) + 1.5448] and exo-miRNA panel-[(−2.9317 X exo-miR145-5p) + (3.2009 X exo-miR200b-3p) + (−1.2140 X exo-miR429) + (−1.3622 X exo-miR1260b) + (2.9393 X exo-miR145-3p) + (0.9949 X exo-miR216b) + (1.5168 X exo-miR200a-3p) + (1.4536 X exo-miR217-5p) + 2.3454]. Using this recalibrated model, it was observed that in the training cohort, the diagnostic AUC values for the cf- and exo-miRNA signatures were 0.90 and 0.96 respectively (Figure 3A and Table 2), which were consistent with the diagnostic performance of the larger pool of 24 markers trained and validated earlier (9 cf-miRNAs and 15 exo-miRNAs). Furthermore, the performance of the combined transcriptomic signature using this reduced panel of biomarkers exhibited an improved overall diagnostic performance with an AUC of 0.98, accuracy of 94%, PPV of 98%, sensitivity of 92% and a specificity of 97% (Figure 3A and Table 2).
Figure 3.
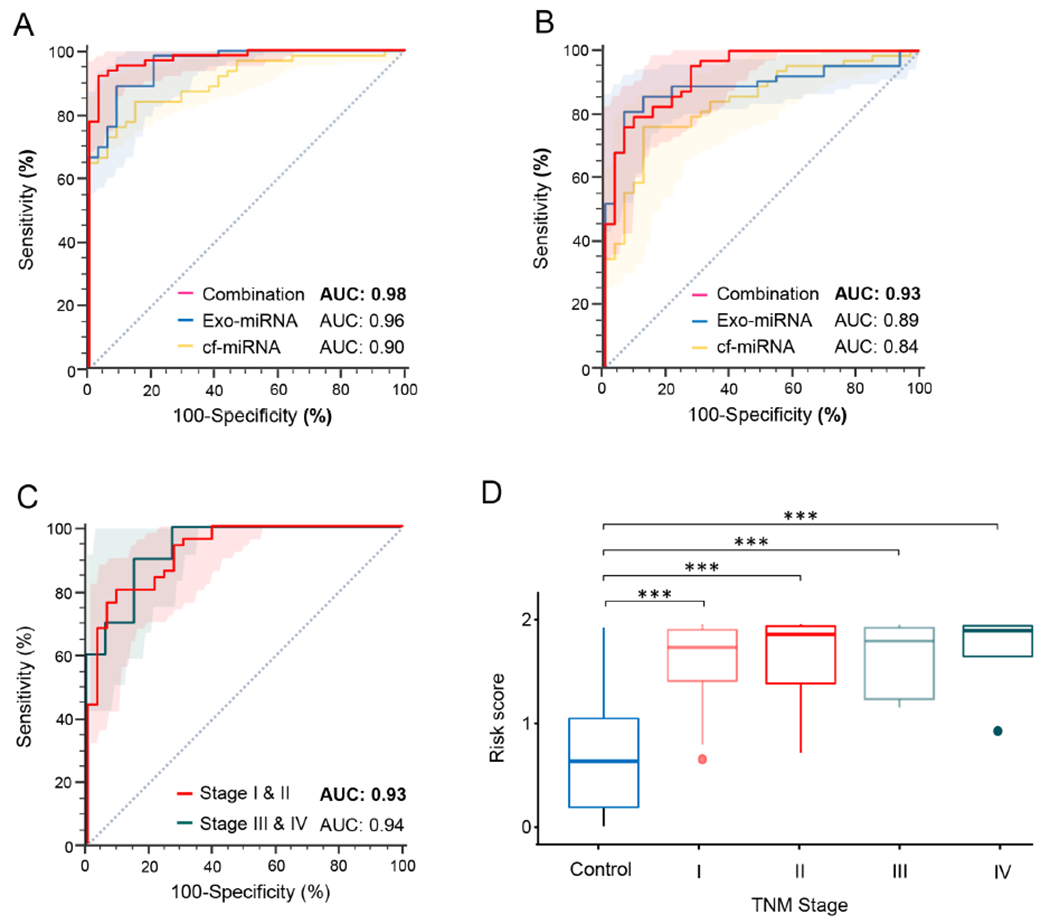
Prioritization and performance evaluation of cell-free and exosomal miRNA biomarker panel in clinical cohorts. (A) ROC curve analysis for the cf-miRNA, exo-miRNA or cf and exosomal combination panel in the training cohort. (B) ROC curve analysis for the cf-miRNA, exo-miRNA or cf and exosomal combination panels in the validation cohort (C) ROC curve analysis to identify early-stages (stage I and II) and late stages (stage III and IV) PDAC patients from non-disease controls in validation cohort. (D) Risk score analysis in all stages PDAC patients and non-disease controls in the validation cohort. ROC curves are shown with 95% CIs. [*p< 0.001, Exo: exosomal; miRNA: micro RNA; cf: cell-free; AUC: Area under the curve; TNM: tumor-node-metastasis; PDAC: pancreatic ductal adenocarcinoma; ROC: receiver operating characteristic]
Likewise, this risk-prediction model derived from training cohort was applied to the independent clinical validation cohort and it was observed that the combined transcriptomic signature offered superior diagnostic performance with an AUC value of 0.93 vs. the cf-miRNA panel (AUC, 0.84) and exo-miRNA panel (AUC, 0.89; Figure 3B). Taken together, these results were quite encouraging, and highlighted the reduced transcriptomic panel was quite robust and offered a clinically attractive and inexpensive assay for the early detection of patients with PDAC.
The optimized transcriptomic signature performs robustly even for the identification of patients with the early-stage PDAC
While previous studies have attempted to develop early detection biomarkers in PDAC patients, a majority of these studies have not focused in evaluating the performance of these assays in earliest disease stages, which is as essential criterion for improving the prognosis of patients suffering from this fatal malignancy. Therefore, in the present study, the performance of optimized 13 miRNA-based transcriptomic assay for the diagnosis of patients with early-stage PDAC (stages I and II) was evaluated. It was observed that within the clinical validation cohort, not only the patient cohort with stage III & IV cancers performed remarkably well (AUC 0.94; Sensitivity, 90%; Specificity, 85%; PPV, 64%; NPV, 97% and Accuracy 86%) for the identification of patients, but also this optimized combination transcriptomic assay performed remarkably well even for the identification of patients with stage I & II cancers (AUC, 0.93; Sensitivity, 80%; Specificity, 91%; PPV, 93%; NPV, 76% and Accuracy 84%; Figure 3C).
Furthermore, the performance of this assay was compared in individual disease stages of PDAC patients vs. non-disease control subjects. It was observed that the transcriptomic signature exhibited high risk score in all stages of patients with PDAC even in the early-stage lesions in the validation cohort (Mean risk score: non-disease controls, 0.68; stage I, 1.62; stage II, 1.67; stage III, 1.64; stage IV, 1.73; p<0.001; Figure 3D). These results once again highlighted the clinical significance of exosome-based transcriptomic assay, which performed equally robust in earliest stages of PDAC, presenting an ideal potential option for clinical translation for non-invasive identification of patients with this malignancy.
Exosome-based transcriptomic signature and CA19-9 levels in blood significantly improve diagnostic accuracy for PDAC
In routine clinical practice, CA19-9 is the only available blood-based biomarker for the management of patients with PDAC; however, it lacks sensitivity and specificity required for the early detection of patients in general population. Therefore, combination of transcriptomic signature together with this glycoprotein was explored to further improve the diagnostic performance in clinical settings. Accordingly, serum CA19-9 levels in all clinical specimens were measured, the diagnostic performance of CA19-9 by itself, and in conjunction with our transcriptomic signature was also evaluated. It was quite interesting to observe that while the CA19-9 by itself yielded an AUC value of 0.88, in all stages of PDAC patients, while combining it with optimized signature, resulted in a significant improvement in the overall diagnostic performance as evidenced by a superior AUC value of 0.99 (Figure 4A). Even more noteworthy was the finding that this diagnostic performance was equally remarkable even in early-stage PDAC patients with stage I & II lesions (AUC for CA19-9, 0.86 vs. AUC, 0.99 in combination with transcriptomic signature; Figure 4B).
Figure 4.
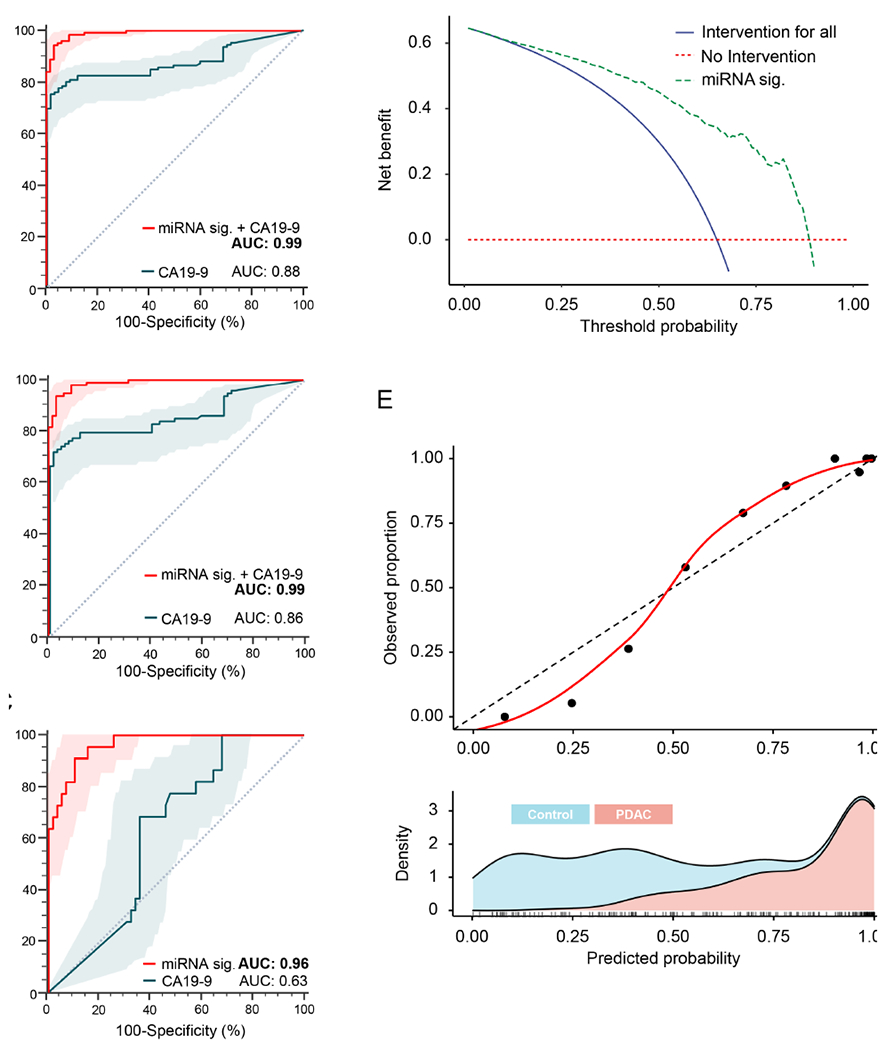
Performance evaluation of the miRNA signature in combination with CA19-9, and diagnostic potential evaluation by decision curve analysis and calibration curve analysis. (A) ROC analysis to compare diagnostic performances between cf and exosomal combination miRNA signature and CA19-9 in all stages of PDAC patients. (B) ROC analysis to compare diagnostic performance between cf and exosomal combination miRNA signature and CA19-9 in early-stages (Stage I and II) of PDAC patients. (C) Performance of cf and exosomal combination miRNA signature in the cohort of 81 participant (22 PDAC and 59 non-disease controls) who presented with al CA19-9 level less than 37 U/mL. (D) Decision curve analysis and (E) Calibration curve analysis to evaluate the performance of the combined miRNA biomarker panel. ROC curves are shown with 95% CIs. [miRNA: micro RNA; CA19-9: carbohydrate antigen 19-9; AUC: Area under the curve; Exo: exosomal; cf: cell-free; ROC: receiver operating characteristic; PDAC: pancreatic ductal adenocarcinoma]
Notably, in this study it was observed that 22 PDAC patients had CA19-9 levels lower than 37 U/ml, a cut-off threshold below which individuals are generally deemed negative for PDAC in clinical settings16, 34. However, it is well known that 15-25% of pancreatic cancer patients present with a normal CA19-915, 16, and about 5-10% of the general population is Lewis antigen-negative with no or low secretion of CA19-917. These clinical challenges further prompted to perform a subgroup analysis for evaluating the performance of the transcriptomic signature in a sub group of 81 subjects (22 PDAC and 59 non-disease controls) who presented with CA19-9 levels lower than 37 U/ml. Consistent with the previous findings, it was observed that, the transcriptomic signature exhibited an excellent diagnostic performance with AUC value of 0.96, sensitivity of 91% and a specificity of 90%, whereas CA19-9 exhibited poor performance with an AUC value of 0.63, sensitivity of 68% and a specificity of 64% in this sub group (Figure 4C). Interestingly, it was observed that miRNA transcriptomic signature could identify 20 out of 22 patients with PDAC (91%) who were considered CA19-9 negative with the CA19-9 levels lower than 37 U/ml. Collectively, these results were encouraging, and highlighted that this transcriptomic signature can offer a diagnostic strategy with improved diagnostic potential for pancreatic cancer screening that can be complemented with CA19-9.
Next, the diagnostic performance of CA19-9 and transcriptomic signature in combination with CA19-9 levels was explored after locking down the specificity of the assay at 95% and 99%, respectively. In the case of CA19-9 levels alone at a fixed specificity of 95% and 99%, a significantly lower sensitivity of 77% and 72% in all stages of PDAC patients and 74% and 68% in early-stage of patients with PDAC, respectively was observed. However, combining the final transcriptomic signature together with CA19-9 levels yielded a remarkably high sensitivity of 95% and 86% respectively, for all stages of PDAC patients, and a sensitivity of 93% and 84% in patients even in the early-stage PDAC patients (Table 3). These findings once again highlight that while this exosome-based transcriptomic assay was quite robust on its own, when combined together with CA19-9 levels, it results in a significant improvement in the overall diagnostic accuracy highlighting its potential translation into the clinic for early detection of patients with PDAC.
Table 3.
The diagnostic performance of CA19-9 alone and miRNA signature in combination with CA19-9 after fixing their specificity at 95% and 99%.
| Controls | Cancer | AUC (95% CI) | Sensitivity (%) (95% CI) | Sens@95%Spec (%) (95% CI) | Sens@99%Spec (%) (95% CI) | |
|---|---|---|---|---|---|---|
| CA19-9 (All stages) | 67 | 124 | 0.88 (0.83-0.92) | 75 (68-82) | 77 (69-85) | 72 (62-81) |
| miRNA sig + CA19-9 (All stages) | 67 | 124 | 0.99 (0.98-1.00) | 94 (90-98) | 95 (85-99) | 86 (77-97) |
| CA19-9 (Early-stages) | 67 | 91 | 0.86 (0.80-0.92) | 71 (63-80) | 74 (63-84) | 68 (56-79) |
| miRNA sig + CA19-9 (Early-stages) | 67 | 91 | 0.99 (0.98-1.00) | 93 (88-0.98) | 93 (81-0.99) | 84 (74-0.96) |
[miRNA: micro RNA; CA19-9: carbohydrate antigen 19-9; AUC: Area under the curve; Exo: exosomal; cf: cell-free; Sens@95%Spec: sensitivity after fixing the specificity at 95%; Sens@99%Spec: sensitivity after fixing the specificity at 99%; ROC: receiver operating characteristic]
An exosome-based liquid biopsy assay offers a significant benefit vs. current treatment approaches used in the clinic for the early detection of patients with PDAC
In current clinical practice, diagnosis of PDAC patients were achieved by computed tomography or an invasive biopsy followed by surgery if it is resectable. Accordingly, false positive or false negative cases based on current clinical practice would be detrimental to subjects undergoing this screening. Thus, the clinical usefulness of screening strategies should be estimated by the trade-off between the harm and diagnosis. To further examine the clinical significance of our transcriptomic assay, decision curve analysis (DCA) and calibration curve analysis were performed. As shown in Figure 4D, the X-axis represents the threshold probability for diagnosis of PDAC and the Y-axis represents the net benefit achieved. The DCA curve revealed that the exosome-based transcriptomic signature achieved a higher net benefit across most ranges of threshold probability in comparison to diagnosing all PDAC patients or none of the patients. (Figure 4D). For instance, at threshold probability of 0.50, this transcriptomic signature exhibited a significantly higher net benefit of 0.45 for the diagnosis of PDAC, vs. diagnosing all PDAC patients based on the strategy of intervention for all cases with a significantly lower net benefit of approximately 0.30. These findings suggested that transcriptomic signature offered markedly higher clinical benefit compared to intervention for all cases or none of the cases, in terms of the viewpoint of the avoidance of harm and misdiagnosis. In addition, the calibration plots showed a good agreement between the observed vs. predicted probability across all ranges (Figure 4E). A slight underestimation for diagnosis of PDAC was observed when the predicted probabilities were between 0.50-0.75 range. As the risk score increases from 0 to 1, the proportion of cancer patients were also increased, highlighting that exosome-based liquid biopsy signature exhibited robust diagnostic potential for identification of patients with PDAC.
DISCUSSION
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy, with a rising incidence, but lacks adequate approaches for its early detection. Accumulating evidence indicates that miRNA-based liquid biopsy assays offer a promising strategy for the early detection of multiple human cancers including pancreatic cancer – primarily, due to their small size, resistance to nuclease-mediated degradation and their abundant and tissue-specific presence in various bodily fluids22, 23, 35, 36. However, there is some debate whether cell-free (cf)-miRNAs, which are often released in systemic circulation from multiple cellular sources, represent adequate diagnostic specificity for cancer detection. Recently, there has been burgeoning interest in studying tumor-derived exosomes and their cargo which contains various proteins and nucleic acids, particularly specific miRNAs (exo-miRNAs), which might provide an additional measure of specificity required for improving their overall diagnostic accuracy in cancer patients26, 27. Herein, in this study it has been hypothesized that a combination of cf- and exo-miRNAs might offer an attractive approach that maximizes the sensitivity and specificity of miRNA signatures in a liquid biopsy assay for diagnosis of PDAC. Accordingly, in the present study, a systematic and comprehensive biomarker discovery approach by small RNA sequencing in patients with early-stage PDAC (stages I and II) and non-disease control subjects was performed to discover and subsequently validate a transcriptomic signature for noninvasive identification of patients with PDAC.
A previous study has explored the diagnostic performance of a cf-miRNA signature for detection of PDAC and observed that miRNA panel could diagnose patients with PDAC with an AUC value of 0.93 - 0.97 in the training cohort and 0.81 - 0.83 in the validation cohort35. In line with this previous study, in this study the cf-miRNA panel also demonstrated robust performance for the detection of PDAC with an AUC value of 0.90 in the training cohort and 0.83 in the validation cohort. However, there were several potential challenges associated with this previous study including the lack of a genomewide biomarker discovery effort, inclusion of PDAC specimens mostly from advanced stage patients, and the inclusion of the patients with similar race and ethnicity. To mitigate these potential challenges in the present study small RNA sequencing was performed by specifically analyzing only early-stage of PDAC specimens (stages I and II) for the discovery of candidate miRNAs, as well as trained and validated the performance of the candidate biomarkers in multiple, independent clinical cohorts of patients from diverse populations of PDAC patients – with the discovery cohort specimens from Korea and training and validation specimens from Japan and USA, respectively.
In this study, while the cf- and exo-miRNA biomarker panels performed remarkable well on their own, it was observed that a combined miRNA signature demonstrated a superior diagnostic performance for its ability to identify PDAC patients with an AUC value of 0.98 in the training cohort and 0.92 in the validation cohort. Thus, it has been speculated that, this novel strategy could overcome the limitations associated with the analysis of conventional analysis of cf-miRNAs35. Furthermore, in order to develop a clinically feasible and cost-effective assay that includes only the minimal number of markers required for maintaining the overall diagnostic performance of the cf- and exo-miRNAs, the candidates were prioritized by using a systematic backward elimination approach which led to identification of a 13 miRNA signature (5 cf- and 8 exo-miRNAs), which performed equally robustly with an AUC of 0.97 and 0.92 in the clinical training and validation cohorts, respectively. Even more importantly, this reduced and optimized exosome-based transcriptomic signature exhibited an impressive diagnostic accuracy (AUC, 0.96) for the identification of patients with early-stage (stages I and II) PDAC patients.
To better understand the functional relevance of these candidate miRNAs, a miRNA-mRNA regulatory network was constructed (Supplementary Figure 1A) based on miRNA-target interactions predicted by StarBase. The network was further filtered to retain 165 target genes based on their log2-fold change >0.5 and adjusted P < 0.05 that were differentially expressed between PDAC and normal tissue samples using the GSE62452 dataset. More than half of the miRNA targets (54.5%) were cancer-related genes annotated in the cancer hallmark gene sets in the MSigDB database (version 7.0). Furthermore, functional annotation based on KEGG pathways and cancer hallmark gene sets in the MSigDB database showed that the miRNA target genes were significantly enriched in cancer-related signaling pathways such as epithelial mesenchymal transition, pathways in cancer, mTORC1 signaling etc. highlighting their strong functional relevance in pathogenesis of PDAC (Supplementary Figure 1B).
In this study, the performance of combined miRNA signature with respect to conventional serological tumor marker CA19-9, which is the most widely used biomarker for the diagnosis of pancreatic cancer was also evaluated2, 3. Several previous studies have demonstrated that CA19-9 has a diagnostic potential for the detection of PDAC12, 35. However, for the early diagnosis of patients with pancreatic cancer, CA19-9 lacks sufficient diagnostic performance to be used as definitive molecular biomarker13. In ideal scenario, sensitivity is particularly important for cancer screening because the screening strategy should provide maximum sensitivity to minimize the failure of identifying the disease. In this present study, it was observed that CA19-9 alone has only 71% of sensitivity for the diagnosis of early-stage PDAC with false-negative results observed in PDAC specimens. On the other hand, when it was combined with transcriptomic signature, the final diagnostic model showed a superior diagnostic performance with a sensitivity of 95% which was significantly higher as compared to CA19-9 alone. Moreover, at fixed specificity of 95% or 99%, the miRNA signature combined with CA19-9 could successfully maintain high sensitivity. These findings highlighted that exosome based transcriptomic signature has the potential to improve the diagnostic performance for pancreatic cancer screening that can be complemented with CA19-9.
Although CA19-9 is currently the most important biomarker for pancreatic cancer37, 15-25% of pancreatic cancer patients present with a normal CA19-915, 16. Moreover, since approximately 5-10% of the population are Lewis antigen-negative with no or low secretion of CA19-917. These findings have been considered as one of the major weaknesses for CA19-9 to be used as a diagnostic biomarker for PDAC patients. Considering the limitation of CA19-9 in pancreatic cancer detection, the development of biomarkers that can complement CA19-9 in the management of Lewis negative pancreatic cancer is urgently needed17. In the present study, it was observed that 22 PDAC patients had CA19-9 levels lower than normal limit (37 U/ml), these individuals are generally considered negative for PDAC in clinic. While performing the subset analysis in patients with normal CA19-9 levels, it was observed that miRNA signature could distinguish 91% of patients with PDAC from this cohort. This result highlighted that the miRNA signature has a potential to be a novel diagnostic strategy for PDAC patients with a normal CA19-9 or Lewis antigen-negative. Together, these results highlighted that the transcriptomic signature was able to correctly classify them as PDAC, once again underscoring its importance for potential application for screening of PDAC.
Since most of the PDAC patients are diagnosed at an advanced stage, early diagnosis of PDAC is essential for improving their prognosis. However, routine screening for PDAC is not recommended for general population due to its lower disease incidence, and because currently available serological markers including CA19-9 lacks sufficient diagnostic performance for early detection of PDAC38. In this study, the exosome-based transcriptomic signature showed remarkable diagnostic accuracy for its potential to improve disease diagnosis and potential use for the early detection of PDAC in specific risk-groups. In the current clinical practice, high-risk individuals with family history of PDAC or specific hereditary background including hereditary pancreatitis or Peutz-Jeghers syndrome, and symptomatic individuals are recommended as candidates for routine screening, whereas such screening is not recommended for asymptomatic individuals38–41. Based upon our findings, we propose and highlight the potential intended use of exosome-based miRNA signature for screening of PDAC is illustrated in Supplementary Figure 2. The individuals who harbor multiple risk factors for developing PDAC can be eligible for such a non-invasive and inexpensive exosome based transcriptomic assay alongwith conventional serological tumor marker, CA19-9. In such a scenario, if a specific individual is diagnosed as “high-risk” by this screening strategy further imaging-based screening test can then be recommended for further follow-up clinical management and intervention.
Previously, several studies have reported various exosomal miRNAs42–44 (miR-21, miR-4525, miR-196a, miR-451a etc), mutant KRAS gene45 and proteins46 (CD44v6, Tspan8, EpCAM etc.) as candidate biomarkers for diagnosis, recurrence prediction and prognosis in PDAC. Majority of these studies primarily performed in relatively smaller sample size and lack of validation in independent clinical cohorts. On the other hand, this study was conducted at multiple institutions and included multiple specimens from multiple ethic background. Moreover, to the best of knowledge, this is the first study that reported the performance of combined cf- and exo-miRNA panel in PDAC diagnosis. However, it is important to acknowledge a few potential limitations of this study. Firstly, the sample size of the patient cohorts used in this study was relatively modest, although the cohorts included multiple races and ethnicities. Therefore, further prospective studies using larger patient cohorts are needed to successfully translate these findings into the routine clinical settings. Secondly, the non-disease cohorts consisted of somewhat younger population as compared to patients with PDAC. To avoid this potential confounder, age-matched controls and PDAC specimens would have been ideal for developing diagnostic strategy for PDAC.
In conclusion, using a systematic and comprehensive biomarker discovery followed by successful clinical validation, this study provides a promising evidence for the clinical significance of an exosome-based transcriptomic signature for a noninvasive, liquid biopsy assay for the early detection of patients with pancreatic ductal adenocarcinoma.
Supplementary Material
WHAT YOU NEED TO KNOW:
Background and context:
Majority of the pancreatic ductal adenocarcinoma patients are diagnosed at an advanced disease when the disease is incurable; hence, there is a clear unmet clinical need to develop biomarkers for its early detection.
New findings:
We have established an exosome-based transcriptomic signature that combines cell-free and exosomal microRNAs that can robustly identify patients with ppancreatic ductal adenocarcinoma and offer a liquid biopsy test that is superior to CA19-9 measurement.
Limitations:
Although our cohort included multiple patient populations, the sample size of our patient cohorts was modest and it was a retrospective study; hence, future prospective studies using larger patient cohorts will be needed.
Impact:
Our exosomal transcriptomic signature has the potential to transform the clinical practice by allowing non-invasive and early detection of patients with pancreatic ductal adenocarcinoma.
Funding:
This work was supported by a grant (U01 CA214254) from National Cancer Institute, and National Institutes of Health, a start-up fund (4937084) by the Chinese University of Hong Kong, Guangdong Basic and Applied Basic Research Foundation (2019B030302012), and the Young Scientists Fund of the National Natural Science Foundation of China (81802384) awarded to Xin Wang.
Abbreviations:
- AUC
Area under the curve
- CA19-9
Carbohydrate antigen 19-9
- cf
cell-free
- Exo
Exosomal
- miRNA
Micro RNA
- NPV
Negative predictive value
- OR
Odds ratio
- PDAC
Pancreatic ductal adenocarcinoma
- ROC
Receiver operating characteristic
- PPV
Positive predictive value
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- SD
Standard deviation
- TNM
Tumor-node-metastasis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose.
Ethical approval and consent to participate: The study was conducted in accordance with the Declaration of Helsinki. All participants provided informed written consent, and the study protocol was approved by the Institutional Review Board of Samsung Medical Center, Nagoya University Graduate School of Medicine and Ochsner Clinic Foundation.
Consent for publication: All subjects have written informed consent.
Competing interests: The authors declare that they have no competing interests.
REFERNECES
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039–49. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 3.Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. The Lancet 2020;395:2008–2020. [DOI] [PubMed] [Google Scholar]
- 4.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 5.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 6.Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. The Lancet 2011;378:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uesaka K, Boku N, Fukutomi A, et al. Adjuvant chemotherapy of S-1 versus gemcitabine for resected pancreatic cancer: a phase 3, open-label, randomised, non-inferiority trial (JASPAC 01). The Lancet 2016;388:248–257. [DOI] [PubMed] [Google Scholar]
- 8.Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N Engl J Med 2018;379:2395–2406. [DOI] [PubMed] [Google Scholar]
- 9.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817–25. [DOI] [PubMed] [Google Scholar]
- 10.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golan T, Hammel P, Reni M, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med 2019;381:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahrmann JF, Schmidt CM, Mao X, et al. Lead-Time Trajectory of CA19-9 as an Anchor Marker for Pancreatic Cancer Early Detection. Gastroenterology 2021;160:1373–1383 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder S, Taylor WR, Foote PH, et al. High Detection Rates of Pancreatic Cancer Across Stages by Plasma Assay of Novel Methylated DNA Markers and CA19-9. Clin Cancer Res 2021;27:2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gui JC, Yan WL, Liu XD. CA19-9 and CA242 as tumor markers for the diagnosis of pancreatic cancer: a meta-analysis. Clin Exp Med 2014;14:225–33. [DOI] [PubMed] [Google Scholar]
- 15.Tempero MA, Uchida E, Takasaki H, et al. Relationship of carbohydrate antigen 19-9 and Lewis antigens in pancreatic cancer. Cancer Res 1987;47:5501–3. [PubMed] [Google Scholar]
- 16.Martin LK, Wei L, Trolli E, et al. Elevated baseline CA19-9 levels correlate with adverse prognosis in patients with early- or advanced-stage pancreas cancer. Med Oncol 2012;29:3101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo G, Liu C, Guo M, et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann Surg 2017;265:800–805. [DOI] [PubMed] [Google Scholar]
- 18.Cescon DW, Bratman SV, Chan SM, et al. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 2020;1:276–290. [DOI] [PubMed] [Google Scholar]
- 19.Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409–424. [DOI] [PubMed] [Google Scholar]
- 20.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472–84. [DOI] [PubMed] [Google Scholar]
- 21.Kilgour E, Rothwell DG, Brady G, et al. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020;37:485–495. [DOI] [PubMed] [Google Scholar]
- 22.Toiyama Y, Okugawa Y, Fleshman J, et al. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta Rev Cancer 2018;1870:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigeyasu K, Toden S, Zumwalt TJ, et al. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res 2017;23:2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. Jama 2007;297:1901–8. [DOI] [PubMed] [Google Scholar]
- 25.Nishiwada S, Cui Y, Sho M, et al. Transcriptomic Profiling Identifies an Exosomal microRNA Signature for Predicting Recurrence Following Surgery in Patients with Pancreatic Ductal Adenocarcinoma. Ann Surg 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin X, Chen Y, Chen H, et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res 2017;23:5311–5319. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, Hurley J, Roberts D, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol 2021;32:466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Annals of Oncology 2018;29:700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.San Lucas FA, Allenson K, Bernard V, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol 2016;27:635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingenito F, Roscigno G, Affinito A, et al. The Role of Exo-miRNAs in Cancer: A Focus on Therapeutic and Diagnostic Applications. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J-H, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research 2013;42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphris JL, Chang DK, Johns AL, et al. The prognostic and predictive value of serum CA19.9 in pancreatic cancer. Ann Oncol 2012;23:1713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. Jama 2014;311:392–404. [DOI] [PubMed] [Google Scholar]
- 36.So JBY, Kapoor R, Zhu F, et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021;70:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien DP, Sandanayake NS, Jenkinson C, et al. Serum CA19-9 is significantly upregulated up to 2 years before diagnosis with pancreatic cancer: implications for early disease detection. Clin Cancer Res 2015;21:622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Xu R, Wang C, et al. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. 2021;41:1257–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poruk KE, Firpo MA, Adler DG, et al. Screening for Pancreatic Cancer: Why, How, and Who? 2013;257:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhoda A, Lu L, Clerkin BM, et al. Current Approaches to Pancreatic Cancer Screening. The American Journal of Pathology 2019;189:22–35. [DOI] [PubMed] [Google Scholar]
- 41.Force UPST. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA 2019;322:438–444. [DOI] [PubMed] [Google Scholar]
- 42.Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191, - 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018;18:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahasi K, Iinuma H, Wada K, et al. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci 2018;25:155–161. [DOI] [PubMed] [Google Scholar]
- 44.Kawamura S, Iinuma H, Wada K, et al. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J Hepatobiliary Pancreat Sci 2019;26:63–72. [DOI] [PubMed] [Google Scholar]
- 45.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015;136:2616–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


