Abstract
Neutrophils play essential anti-microbial and inflammatory roles in host defense, however, their activities require tight regulation as dysfunction often leads to detrimental inflammatory and autoimmune diseases. Here we show that the adhesion molecule GPR97 allosterically activates CD177-associated membrane proteinase 3 (mPR3), and in conjugation with several protein interaction partners leads to neutrophil activation in humans. Crystallographic and deletion analysis of the GPR97 extracellular region identified two independent mPR3-binding domains. Mechanistically, the efficient binding and activation of mPR3 by GPR97 requires the macromolecular CD177/GPR97/PAR2/CD16b complex and induces the activation of PAR2, a G protein-coupled receptor known for its function in inflammation. Triggering PAR2 by the upstream complex leads to strong inflammatory activation, prompting anti-microbial activities and endothelial dysfunction. The role of the complex in pathologic inflammation is underscored by the finding that both GPR97 and mPR3 are upregulated on the surface of disease-associated neutrophils. In summary, we identify a PAR2 activation mechanism that directs neutrophil activation, and thus inflammation. The PR3/CD177/GPR97/PAR2/CD16b protein complex, therefore, represents a potential therapeutic target for neutrophil-mediated inflammatory diseases.
Subject terms: Neutrophils, Inflammation, Cell signalling, G protein-coupled receptors
Activation of neutrophil leukocytes is tightly regulated, and it is important to understand the molecular mechanisms of their response to physiological and pathological stimuli. Here authors show that the adhesion molecule G protein-coupled receptor 97 and its interaction partners play pivotal roles in neutrophil leukocyte activation both in anti-microbial response and in inflammatory diseases.
Introduction
Neutrophils express a plethora of immune effectors including proteases, cytokines, and receptors which contribute significantly to both the innate and adaptive immune responses1. However, these neutrophil-derived molecules require stringent regulation as their uncontrolled activities are often associated with detrimental inflammatory and autoimmune diseases2. Neutrophil serine proteases (NSPs) are azurophilic granule proteins which play critical roles in anti-microbial and inflammatory responses3,4. Proteinase 3 (PR3) is unique among NSPs as it also represents the designated auto-antigen of granulomatosis with polyangiitis (GPA), an autoimmune disorder characterised by the inflammatory necrosis of small/medium-sized blood vessels and the common presence of anti-neutrophil cytoplasmic antibodies (ANCA) against PR35. Interestingly, a fraction of PR3 is exocytosed and tethered to neutrophil membrane (termed mPR3) mainly by specific binding to the glycosylphosphatidylinositol-anchored CD1776. Although increased plasma PR3 and mPR3 levels are positively associated with GPA pathogenesis, the normal cellular function of mPR3 remains enigmatic7,8.
G protein-coupled receptors (GPCRs) are critically involved in the activation and chemotaxis of neutrophils, and deregulated GPCR activities in neutrophils usually result in immune dysfunction and clinical disease9. Protease-activated receptors (PARs) represent a group of unique GPCRs that are activated by a tethered peptide ligand exposed by the specific proteolytic processing of their N-terminal region10. Activation of PAR2, the predominant PAR expressed by neutrophils, is involved in tissue inflammation and prenatal death associated with the autoimmune antiphospholipid syndrome11,12. Although trypsin is the prototypic activator of PAR2, many other proteinases are similarly able to activate or disarm PAR213. Intriguingly, PAR2 can also be transactivated by PAR1 by the formation of PAR1-PAR2 heterodimers14. To date, it is not fully understood how PAR2 activation is triggered in human neutrophils and whether PAR2 transactivation occurs in neutrophils.
Adhesion GPCRs (aGPCRs) are atypical GPCRs, having a large extracellular region (ECR) before the seven-transmembrane domain15. Most aGPCRs are expressed as a non-covalent bipartite complex due to the auto-proteolytic cleavage of ECR at the GPCR proteolysis site (GPS) within the GPCR autoproteolysis-inducing (GAIN) domain15. Consequently, many aGPCRs are activated by a tethered agonism mechanism remarkably similar to that of PARs which involves the unmasking of an internal agonistic peptide following the dissociation/dislocation of ECR upon binding to its cellular ligand(s)16. Human neutrophils express several aGPCRs including EMR2/ADGRE2, EMR3/ADGRE3, CD97/ADGRE5, and GPR97/ADGRG317,18. Although an anti-microbial role has been demonstrated for GPR97 previously, the lack of specific ligands has hindered the mechanistic understanding of its function.
Here we show that GPR97 is the binding partner and allosteric activator of mPR3 through the macromolecular CD177/GPR97/PAR2/CD16b receptor complex. Intriguingly, GPR97-augmented mPR3 enzymatic activity cleaves and activates PAR2 leading to robust neutrophil activation. In conclusion, our results uncover an aGPCR-GPCR activation mechanism in human neutrophils that contributes to inflammatory responses.
Results
Activated neutrophils up-regulate GPR97 expression which correlates with the inflammation and/or disease status of various inflammatory disorders
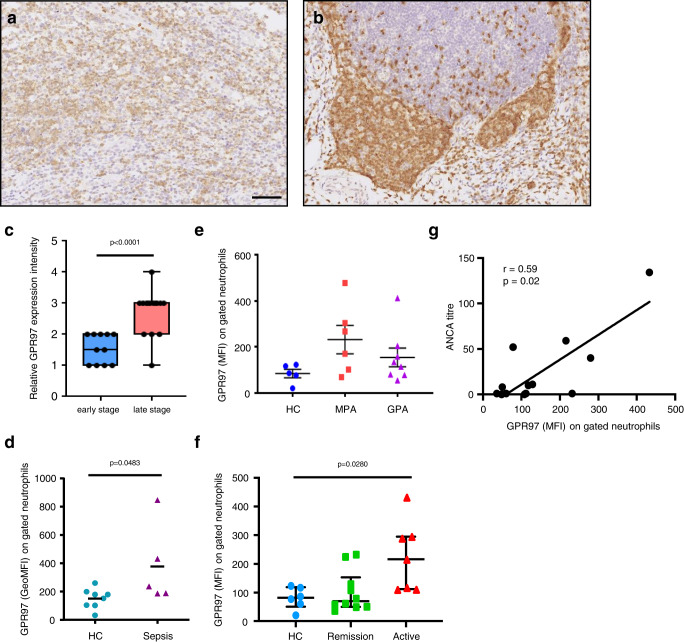
Up-regulated GPR97 transcript and protein have been identified in blood neutrophils and tissue-infiltrating neutrophils of diverse inflammatory disorders18. To further confirm the relationship of neutrophil GPR97 expression and tissue inflammatory status, we compared GPR97 expression in early- and late-stage appendicitis. Specific GPR97 expression was consistently detected in tissue-infiltrating neutrophils which were more numerous in late-stage appendicitis (Fig. 1a, b). Moreover, much higher GPR97 staining intensities were identified in the late-stage than the early-stage appendicitis (Fig. 1c).
Fig. 1. Up-regulated GPR97 expression in neutrophils correlates with the disease status of inflammatory disorders.
a, b The immunohistochemical analyses of GPR97 expression in tissue sections of the early- a and late-stage b appendicitis. Reactivity of the anti-GPR97 mAb to GPR97 was shown as brownish staining. Scale bar, 100 μm. c The relative GPR97 staining intensities in tissue-infiltrating neutrophils of the early- (n = 11) and late-stage (n = 16) appendicitis tissues. Data are means ± SEM and p value was determined by one-sided unpaired student’s t-test. d Flow cytometry analyses of surface GPR97 levels of gated neutrophils of HC (n = 8) and bacterial sepsis patients (n = 5). Data are means ± SEM and p value was determined by two-sided unpaired student’s t-test. e, f Flow cytometry analyses of surface GPR97 levels of gated neutrophils of HC (n = 5 in e, 6 in f) and patients based on the disease category (GPA n = 8, MPA n = 6) e and status (remission n = 10, active n = 7) f. g The positive correlation of GPR97 levels with the ANCA titers of GPA (n = 8) and MPA (n = 6) patients. Data are means ± SEM and p value was determined by one-way ANOVA. HC healthy control, GPA granulomatosis with polyangiitis, MPA microscopic polyangiitis. Source data are provided in the Source Data file.
Similarly, higher surface GPR97 levels were detected in blood neutrophils of bacterial sepsis patients than those of healthy controls by flow cytometry analyses (Fig. 1d). Thereafter, we enrolled patients of GPA and microscopic polyangiitis (MPA), two distinct autoimmune diseases characterized by ANCA mostly directed toward PR3 and myeloperoxidase (MPO), respectively5. No significant differences in GPR97 expression were observed among healthy controls and patients (Fig. 1e). However, when patients were divided into remission and active disease groups, much higher GPR97 levels were detected in neutrophils of patients with signs of active disease (Fig. 1f). Moreover, GPR97 expression levels also correlated positively to the ANCA titers of patients (Fig. 1g). Altogether, up-regulated GPR97 expression in activated neutrophils is positively associated with disease activity of various inflammatory disorders.
Neutrophils are activated by GPR97 binding to a putative ligand expressed exclusively in a human neutrophil subpopulation
Given that many aGPCRs have cell-associated ligands, a panel of cell lines and primary cells were screened via flow cytometry for potential GPR97 ligands19. A chimeric GPR97E-mFc protein containing the human GPR97-ECR fused with a mouse fragment crystallizable (Fc) was used as the probe (Fig. 2a, and Supplementary Fig. 1a). From over 30 different cell samples screened, a putative GPR97-ligand was successfully identified which was expressed in a unique bimodal pattern exclusively on human neutrophil surfaces (Fig. 2b, c, and Supplementary Fig. 1b). Intriguingly, the GPR97-ligand+ neutrophil subpopulation ranged from 40–90% among different donors but remained rather constant in the same individuals over time (Fig. 2d). Ligand-binding was not detected in human neutrophils probed by mouse GPR97E-mFc, nor in mouse neutrophils probed by human or mouse GPR97E-mFc probes (Supplementary Fig. 1c). Hence, the interaction of GPR97 and its putative ligand is a human neutrophil-specific feature and the expression of GPR97-ligand is strictly regulated in neutrophils.
Fig. 2. Identification of a putative GPR97-ligand in a distinct human neutrophil subpopulation that is activated upon interaction with GPR97E-mFc.
a Schematics of GPR97 receptor and GPR97E-mFc probe. b List of cell samples screened for the expression of GPR97 and the putative GPR97-ligand. * indicates a subpopulation of primary human neutrophils. c A representative bimodal surface expression profile of the GPR97-ligand in human neutrophils detected by the GPR97E-mFc probe. d Highly variable GPR97-ligand− (blue) and GPR97-ligand+ (pink) neutrophil subsets in bloods of different donors (samples A to N). e, f Morphological changes of neutrophils induced by GPR97E-mFc as observed by inverted light microscope e and Wright-Giemsa stain f. Scale bar, 20 μm. Experiments were repeated 3 times with similar results. g–j Phenotypic analyses of activated neutrophils induced by GPR97E-mFc included up-regulated ROS production g, MPO activity (h, n = 6), expressional changes of specific CD markers (i, n = 7 for CD54 and n = 4 for others) and IL-8 production (j, n = 3) of independent samples. The mFc protein and fMLF were used as a negative and a positive control, respectively. Data are presented as means ± SEM and p value was determined by two-way ANOVA in h, j and one-way ANOVA in i. Source data are provided in the Source Data file.
Importantly, incubation of resting neutrophils with GPR97E-mFc triggered robust activation, resulting in several distinct phenotypic manifestations. These include cell shape changes, enhanced production of reactive oxygen species (ROS) and interleukin-8 (IL-8), increased MPO activity, as well as expressional changes of multiple neutrophil activation markers such as CD62L, Mac-1, CD54, and CXCR1 (Fig. 2e–j, and Supplementary Fig. 1d–g). Notably, IL-8 production was up-regulated comparably by neutrophils treated with soluble or immobilized GPR97E-mFc in a dose- and time-dependent manner (Fig. 2j, and Supplementary Fig. 1h). In summary, a putative GPR97-specific ligand is identified exclusively in a distinct human neutrophil subset whose interaction with GPR97E-mFc induces inflammatory activation.
GPR97 is a specific binding partner and allosteric activator of neutrophil mPR3
The known ligands of aGPCRs include lipids, glycosaminoglycans, and proteins15. To delineate the putative GPR97-ligand, far-western blot analysis of neutrophil proteins was performed using GPR97E-mFc as a probe. Three distinct signals were detected specifically in the membrane fraction, including a prominent ~29 kDa protein and two weaker signals of ~52 and ~140 kDa proteins (Fig. 3a), suggesting that the GPR97-ligand is likely a membrane protein. To date, CD177 (~50–60 kDa), PR3 (~29 kDa), and olfactomedin 4 (OLFM4)(~57 kDa) are all known to show a bimodal profile in human neutrophils hence are strong GPR97-ligand candidates20. The glycosylphosphatidylinositol-linked CD177 is a neutrophil-restricted Ly6 receptor family member expressed in a characteristic 0–100% pattern in different individuals due to unique genetic mechanisms21,22. The intracellular elastase-like PR3 is somewhat exocytosed and forms mPR3 on resting neutrophils mainly by binding to CD177, thus displaying a CD177-like expression pattern6. By contrast, OLFM4 is a specific granule resident protein which is secreted upon neutrophil activation and is not known to tether on the neutrophil membrane23.
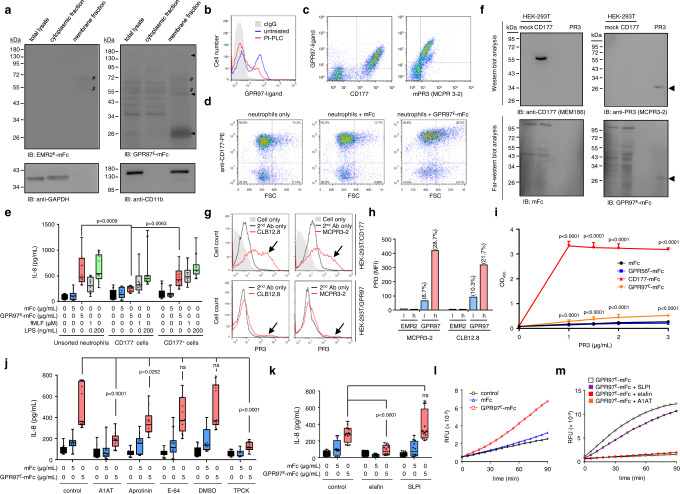
Fig. 3. GPR97 is a binding partner and allosteric activator of mPR3.
a Far-western blot analyses of the putative GPR97-ligand(s)(black arrowheads). The EMR2E-mFc protein was used as a negative control. Anti-CD11b and anti-GADPH Abs detected the corresponding protein markers of the membranous and cytoplasmic fractions, respectively. #, non-specific signals. Experiments were repeated 3 times with similar results. b FACS-based ligand-binding analysis of neutrophils pre-treated without or with phosphoinositide phospholipase C (PI-PLC). c Dot-plots of neutrophils double-stained with GPR97E-mFc and anti-CD177 or anti-PR3 mAb. d Dot-plots (FSC vs. surface CD177 levels) of neutrophils incubated with GPR97E-mFc. e IL-8 produced by unsorted and sorted neutrophils incubated with various reagents as indicated. n = 5 independent experiments. Cells treated with fMLF and LPS were included as positive controls. f Far-western blot analysis of specific GPR97-PR3 binding. Samples included total lysates of mock- and CD177-transfected HEK-293T cells as well as purified PR3. The arrowheads indicated the specific signals detected by the anti-PR3 mAb and the GPR97E-mFc probe. Experiments were repeated 3 times with similar results. g, h The FACS-based PR3-binding assay of unsorted g and FACS-sorted h HEK-293T cells expressing CD177 or GPR97. The black arrows in g indicated the specific PR3-binding. FACS-sorted GPR97high h and GPR97low l HEK-293T cells were examined. Numbers on top of the bars indicated the percentage of PR3-binding cell populations. i ELISA-like PR3-binding analyses. CD177-mFc and GPR56-mFc proteins were included as the positive and negative controls, respectively. n = 6 biologically independent samples. j, k ELISA analysis of IL-8 secreted by neutrophils treated without or with indicated protease inhibitors. n = 5 j, 4 k independent experiments. Data are presented as means ± SEM and p value was determined by two-way ANOVA in e, i–k. ns non-significant. l, m The ex vivo mPR3 enzymatic activity of neutrophils incubated in the absence or presence of mFc or GPR97-mFc protein probe without or with protease inhibitors as indicated. Source data are provided in the Source Data file.
Interestingly, the putative GPR97-ligand was sensitive to digestion by phosphoinositide phospholipase C that cleaves the glycosylphosphatidylinositol-linker (Fig. 3b, and Supplementary Fig. 2a). Moreover, the GPR97-ligand was detected exclusively in the CD177+ and mPR3+ neutrophil subsets, showing closely matched expression profiles and strong co-localization with CD177 and mPR3 (Fig. 3c, and Supplementary Fig. 2b). By contrast, only 20–40% of neutrophils expressing OLFM4 or glycosylphosphatidylinositol-linked CD55 were GPR97-ligand+ (Supplementary Fig. 2c). Finally, morphological changes and IL-8 up-regulation induced by GPR97E-mFc were detected solely in the CD177+ but not CD177− neutrophil subset (Fig. 3d, e, and Supplementary Fig. 2d). These results suggested strongly a direct interaction of GPR97E-mFc with either CD177 or mPR3. Thereafter, far-western blot analyses revealed specific binding of GPR97E-mFc to PR3 but not CD177 (Fig. 3f). We subsequently verified the specific GPR97-PR3 interaction using two different PR3-binding assays, namely the FACS-based ligand-binding assay performed in CD177- and GPR97-expressing HEK-293T cells and the enzyme-linked immunosorbent assay (ELISA)-like protein-protein binding assay (Fig. 3g–i, and Supplementary Fig. 2e, f). Both assays showed that GPR97, like CD177, binds to PR3 specifically, albeit much less efficiently. Altogether, we conclude that GPR97-ECR interacts specifically with the PR3 moiety of the mPR3 (PR3-CD177) complex on human neutrophil surfaces.
The enzymatic activity of mPR3 is much reduced compared to soluble PR3 due to the PR3-CD177 interaction24. This prompted us to investigate the potential role of mPR3 activity in GPR97-induced neutrophil activation and the possible effect of GPR97-mPR3 binding to its proteolytic activity. Surprisingly, GPR97-induced neutrophil activation was significantly attenuated in the presence of broad-spectrum serine protease inhibitors including α1-antitrypsin (A1AT), aprotinin, and TPCK, but not the cysteine protease inhibitor E-64 (Fig. 3j, and Supplementary Fig. 3a). Accordingly, two endogenous NSP inhibitors, elafin and secretory leukocyte peptidase inhibitor (SLPI), were employed next. SLPI only inhibits neutrophil elastase (NE) and cathepsin G (CG), while elafin inhibits NE, CG, and PR325. Interestingly, IL-8 up-regulation was dramatically inhibited in elafin- but not SLPI-treated neutrophils (Fig. 3k). These findings established that mPR3 protease activity is essential for GPR97-induced neutrophil activation and implied that GPR97 binding to mPR3 likely promotes its proteolytic activity. We therefore determined the ex vivo enzymatic activity of mPR3 using a PR3-specific FRET-based substrate26. As expected, incubation of neutrophils with GPR97E-mFc resulted in augmented mPR3 activities which were inhibited by A1AT and elafin, but not SLPI without apparent degranulation of azurophilic granules (Fig. 3l, m, and Supplementary Fig. 3b–d). Thus, we conclude that GPR97 is a binding partner and allosteric activator of mPR3.
Atomic resolution of the GPR97 extracellular region and mapping of the mPR3-binding domains
The ECR of most aGPCRs contains an extended N-terminal segment followed by a GAIN domain both of which are able to interact with distinct ligands15. Previous structural studies have shown that the canonical GAIN domain is evolutionarily conserved and includes A and B subdomains27. Subdomain A typically consists of 6 α-helices, while subdomain B comprises a twisted β-sandwich made of 13 β-strands and 2 small α-helices27. Nevertheless, structural analyses of GPR56/ADGRG1 and GPR126/ADGRG6 have revealed a smaller subdomain A with fewer α-helices28,29. Interestingly, our bioinformatic analyses predicted that GPR97 contains an even smaller GAIN domain than GPR56 and GPR126.
To gain a better insight into the structural organization and mPR3-binding characteristics of GPR97-ECR, we determined its atomic structure (residues 28–260) at 3.37 Å resolution using X-ray crystallography (Fig. 4a–c, and Supplementary Fig. 4a–e). The structural analyses showed the GPR97-GAIN domain retains a complete subdomain B but contains only one α-helix in subdomain A, approximately where α6 of the canonical GAIN-subdomain A would be located (Supplementary Fig. 4f). The electron density map also confirmed that the GPR97 GAIN domain is mostly auto-proteolysed at its GPS motif, as expected and observed for other GAIN domains (Supplementary Fig. 1a)18,27–29. Interestingly, we identified a small N-terminal domain (NTD) adjacent to the GAIN domain, which contains four cysteine residues (C32, C50, C54, and C62) that are highly conserved among GPR97 orthologs (Supplementary Fig. 5a). Sulphur-SAD phasing was hence used to precisely locate the cysteine residues and the resulting model shows that the single helix of the subdomain A (helix α6 in canonical GAIN domains) packs against subdomain B and acts as an anchor for the upstream helix of NTD. A cysteine bridge stabilises the packing of the NTD helix against the subdomain A helix (C54-C62) (Fig. 4a, b). The NTD helix is further stabilised by extensive hydrophobic interactions with the GAIN domain, involving the residues Y39, L44, F51 from the NTD and L67, Y70, W71, Y74, H77 and F91 from the GAIN domain, and a cysteine bridge between the helix and the N-terminal loop of the NTD (C32-C50) (Fig. 4c). These hydrophobic residues are highly conserved specifically among GPR97 orthologs (Supplementary Fig. 5a,b). Structural comparison with a model calculated by alpha-fold30 shows that the GAIN domain is similar, but the NTD-helix is off-set compared to our experimental model. Alpha-fold predicts residues Q21-G27 to be flexible (Supplementary Fig. 4i). Taken together, the GPR97-ECR consists of a small cysteine-stabilised helical NTD followed by an unusually small GAIN domain that consists of a subdomain A with one α-helix and a typical subdomain B with 13 β-strands (Fig. 4a).
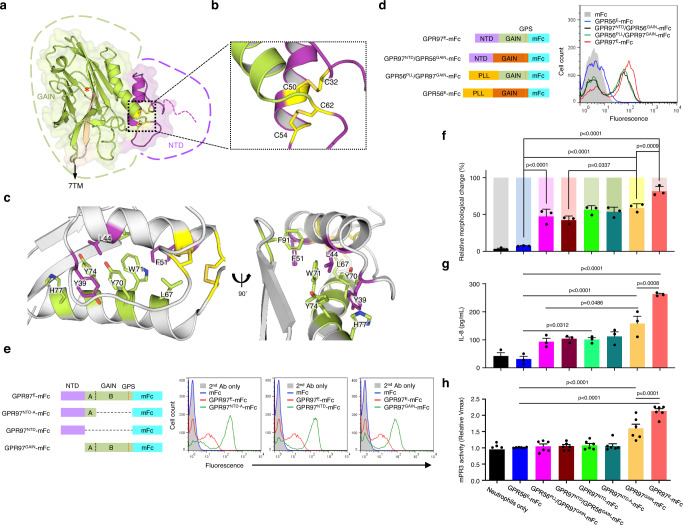
Fig. 4. Structural analysis of GPR97-ECR and mapping of the mPR3-binding regions.
a Cartoon representation of the GPR97-ECR structure obtained from X-ray crystallography experiments. The NTD is depicted in purple and the GAIN domain in green and beige (the 13th β-strand). Cysteine residues involved in linking and stabilising the NTD are shown as yellow sticks. The red asterisk indicates the GPS cleavage site. b Close-up of the two disulphide bridges, C32-C50 and C54-C62, that are involved in the stabilisation of the NTD and GAIN domains. c Close-up on the extensive hydrophobic interactions stabilising the packing of the NTD against the GAIN domain. Residues from the NTD are depicted in purple and those from the GAIN domain are depicted in green. d, e Mapping of the mPR3-binding region(s) using the domain-swapped d and domain-truncated e GPR97-mFc probes as indicated. n = 5 d and 6 e independent experiments. f–h. Activated neutrophil phenotypes, namely morphological changes f, IL-8 production g, and the ex vivo mPR3 enzymatic activity h treated with various GPR97-mFc probes as indicated. n = 3 f, 4 g, and 6 h independent experiments. The bright and dim colors in f represent the percentage of irregular-shaped and round-shaped neutrophils, respectively. The results shown in h are relative Vmax of the ex vivo mPR3 enzymatic activity. Data are presented as means ± SEM and p value was determined by two-way ANOVA. Source data are provided in the Source Data file.
To map the mPR3-binding region(s), we generated domain-swapped mFc-fusion probes by interchanging the GAIN domains of GPR97 and GPR56, two closely related ADGRG subfamily members (Fig. 4d, and Supplementary Fig. 6a). A similar ligand-binding signal was detected by the GPR97NTD/GPR56GAIN-mFc and GPR56PLL/GPR97GAIN-mFc probes as did GPR97E-mFc, while GPR56E-mFc showed nil binding (Fig. 4d). These results suggested that both the GPR97-NTD and -GAIN domains contain one independent mPR3-binding region, respectively. Domain-truncated GPR97-mFc probes were used subsequently to confirm the two different domains can indeed bind to mPR3 individually (Fig. 4e). Unexpectedly however, these domain-swapped and -truncated mFc-fusion proteins induced much weaker neutrophil activation phenotypes compared to the full-length GPR97E-mFc (Fig. 4f, g, and Supplementary Fig. 6b). Similarly, only GPR97GAIN-mFc among the various GPR97-mFc probes stimulated a moderately-increased mPR3 activity (Fig. 4h). We conclude that the maximal GPR97-induced neutrophil activation is achieved when mPR3 is bound simultaneously by the two ligand-binding regions in the GPR97-NTD and -GAIN domains.
GPR97-mPR3 interaction requires a macromolecular CD177/GPR97/PAR2/CD16b receptor complex and triggers PAR2 activation in neutrophils
As CD177 is incapable of direct signaling due to the lack of a transmembrane moiety, the results above implicated strongly the involvement of an undefined signaling molecule triggered by the GPR97-augmented mPR3 in inducing neutrophil activation. In order to identify this signal transducer, a heterologous HEK-293T cell transient expression system was established. Unexpectedly, while exogenous PR3 bound readily to CD177-expressing HEK-293T cells hence forming mPR3, no GPR97E-mFc binding was ever detected in these cells (Fig. 5a). We thus speculated that additional co-receptor(s) expressed in neutrophils but not HEK-293T cells is needed for efficient GPR97-mPR3 binding and likely represents the signaling molecule of interest.
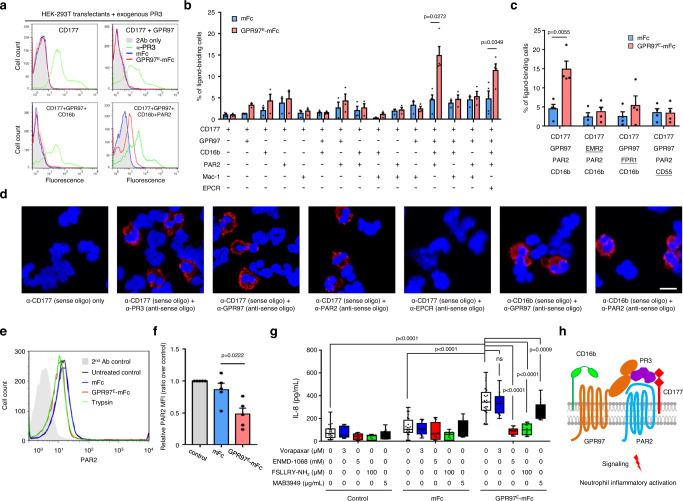
Fig. 5. The CD177/GPR97/PAR2/CD16b receptor complex is required for efficient GPR97-mPR3 interaction that induces PAR2 activation.
a–c Flow cytometry analyses of GPR97-mPR3 interaction in HEK-293T cells expressing various receptors as indicated. a Surface levels of membrane-bound PR3 (green), GPR97E-mFc (red), and mFc (blue) were detected using appropriate Abs. b, c The percentage of transfectants displaying the positive GPR97E-mFc binding signal in the ligand-binding assay. n = 3 b, 4 c independent experiments. The underlined receptor in c indicates the substituted component of the core receptor complex. Data are means ± SEM and p value was determined by two-sided unpaired student’s t-test. d The PLA analyses showed the interaction signals of specific receptor pairs of the CD177-associated complex in resting neutrophils. Scale bar, 10 μm. Experiments were repeated 3 times with similar results. e, f Flow cytometry analyses of PAR2 proteolysis in neutrophils e and HEK-293T cells expressing the CD177-associated receptor complex f. Trypsin-treated neutrophils and mFc were included as a positive and a negative control, respectively. n = 5 independent experiments. Data are means ± SEM and p value was determined by one-way ANOVA. g ELISA analyses of IL-8 secreted by neutrophils incubated with the indicated reagents. n = 4 independent experiments. Data are means ± SEM and p value was determined by two-way ANOVA. ns, non-significant. h Schematic diagram of the PR3/CD177/GPR97/PAR2/CD16b complex in neutrophils. CD16, a green-colored receptor with two connected ovals; GPR97, a brown-colored 7TM receptor with two extracellular ovals; PAR2, a blue-colored 7TM receptor; PR3, a purple-colored soluble molecule; CD177, a red-colored receptor with two diamonds. Source data are provided in the Source Data file.
To date, several interacting proteins/substrates of CD177 and PR3, including the endothelial protein C receptor (EPCR), CD16b (FcγRIIIb), Mac-1 (CD11b/CD18), and PAR2 have been identified in neutrophils31–34. Hence, we co-expressed CD177 and these proteins in different combinations in HEK-293T cells. Surprisingly, a positive GPR97-mPR3 binding signal was only detected in cells co-expressing the specific combination of CD177, GPR97, PAR2 and CD16b (Fig. 5a, b). Moreover, the mPR3-binding signal was lost when any member of the four-receptor complex was replaced with a different protein of the same receptor type, indicating that every co-receptor is indispensable for efficient GPR97-mPR3 binding (Fig. 5c). Next, the proximity-ligation assay (PLA) was carried out to verify the close association of various receptor-pairs of the CD177/GPR97/PAR2/CD16b complex on the neutrophil membrane (Fig. 5d, and Supplementary Fig. 3e). These results indicate that a clustered CD177/GPR97/PAR2/CD16b receptor complex is minimally required for efficient GPR97-mPR3 binding.
PAR2 is the predominant PAR expressed by human neutrophils and its activation is known to induce activation phenotypes similar to those triggered by GPR97E-mFc12. PAR2 hence seemed the most likely signaling transducer activated by GPR97-augmented mPR3. Indeed, the surface PAR2 levels of both neutrophils and PR3-bound HEK-293T transfectants were significantly reduced when incubated with GPR97E-mFc (Fig. 5e, f). Most critically, enhanced IL-8 production by GPR97E-mFc-treated neutrophils was significantly and specifically attenuated in the presence of a functional-blocking anti-PAR2 mAb and PAR2-specific antagonists, while the PAR1 antagonist had no inhibitory effect (Fig. 5g). These results clearly demonstrated that PAR2 is not only the essential component of the CD177-associated complex, but also is the signaling receptor targeted by the GPR97-augmented mPR3. Thus, we propose a GPR97-PAR2 activation mechanism that involves PR3 and the CD177/GPR97/PAR2/CD16b complex. More precisely, GPR97 binds specifically to mPR3 presented by the CD177-associated receptor complex and promotes its enzymatic activity which in turn cleaves and activates PAR2 (Fig. 5h).
Up-regulated GPR97 and PAR2 expression induces GPR97-PAR2 activation in activated neutrophils
Resting human neutrophils are generally inert suggesting no competent induction of GPR97-PAR2 activation which is almost certainly regulated by the expression levels of individual members of the CD177-associated receptor complex. Indeed, our results showed that while CD16b, mPR3, and CD177 were highly expressed in all or some (for mPR3 and CD177) resting neutrophils, GPR97 and PAR2 were weakly, sometimes barely, expressed (Fig. 6a, and Supplementary Fig. 7a). Interestingly, we detected increased PAR2, but not GPR97 expression only in neutrophils treated with certain inflammatory stimulants such as IFN-γ, indicating a stringent expressional regulation of GPR97 and PAR2 expression (Supplementary Fig. 7b, c). It is likely that low expression levels of GPR97 and PAR2 limit the initiation of GPR97-PAR2 activation and the spontaneous self-activation of resting neutrophils.
Fig. 6. Up-regulated GPR97 and PAR2 expression in activated neutrophils induces GPR97-PAR2 activation.
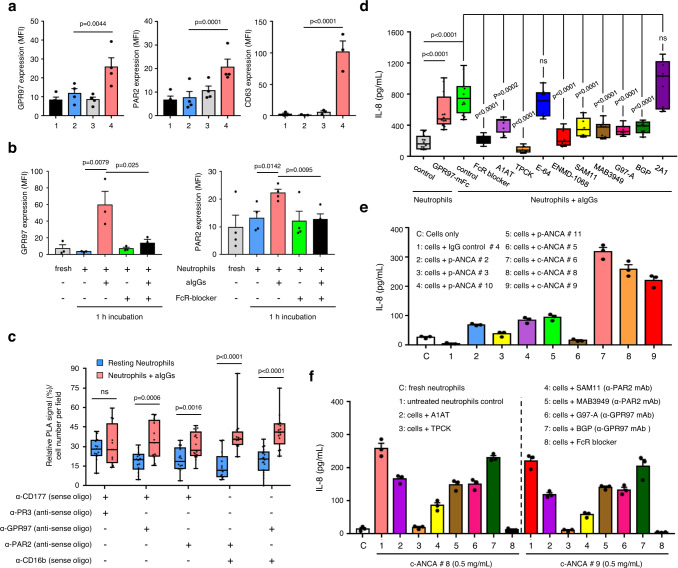
a, b Flow cytometry analyses of GPR97 and PAR2 expression in resting neutrophils treated without or with the azurophil granule degranulation stimulants (n = 4, except CD63 n = 3) a and aIgGs (GPR97 n = 3, PAR2 n = 4) b as indicated. a Samples include: 1, fresh neutrophils; 2, untreated neutrophils; 3, neutrophils treated with fMLF (1 μM) for 15 min; 4, neutrophils treated with cytochalasin B (cytoB, 5 μM) for 5 min, followed by fMLF (1 μM) for 10 min. CD63 expression was used as a degranulation marker of azurophil granule. c Relative PLA signals of indicated receptor pairs in resting (n = 3) and aIgG-treated (n = 3) neutrophils. d ELISA analyses of IL-8 secreted by neutrophils incubated for 3 h at 37 °C as indicated (n = 3). Data in a–d are presented as means ± SEM. P value was determined by two-way ANOVA in a, b, d and by one-sided unpaired student’s t-test in c. ns non-significant. e, f ELISA analyses of IL-8 secreted by neutrophils that were incubated with purified IgGs (0.5 mg/mL) from different PR3-ANCA and MPO-ANCA patients for 3 h at 37 °C as indicated. Data are presented as means ± SEM of one representative experiment done in triplicate. ENMD-1068: PAR2 antagonist; SAM11 and MAB3949: anti-PAR2 mAbs; G97-A and BGP: anti-GPR97 mAbs; 2A1: anti-EMR2 mAb. Source data are provided in the Source Data file.
In line with previous results, significant up-regulation of both GPR97 and PAR2 was identified when resting neutrophils were stimulated by degranulation stimulants of azurophilic granules or by heat-aggregated IgGs (aIgGs) via an Fc receptor (FcR)-dependent mechanism (Fig. 6a, b). Consequently, PLA analyses revealed significantly increased interactions between receptor pairs of the CD177-associated complex in aIgG-activated neutrophils, compared to resting controls (Fig. 6c). Importantly, less IL-8 was produced in aIgG-stimulated neutrophils in the presence of FcR blocker, serine protease inhibitors, PAR2 antagonists, or functional blocking anti-PAR2 and anti-GPR97 mAbs, suggesting a requirement for GPR97-PAR2 activation (Fig. 6d).
To evaluate the clinical relevance of FcR-induced GPR97-PAR2 activation, neutrophils were incubated with purified PR3-ANCA IgGs which activate neutrophils via FcγRs35. Indeed, increased GPR97 and PAR2 expression was detected in resting neutrophils incubated with PR3-ANCA, but not MPO-ANCA IgGs, in a FcR-dependent manner (Supplementary Fig. 8a, b). Critically, PR3-ANCA IgGs caused extensive neutrophil aggregates and led to increased production of IL-8. This effect was also attenuated in the presence of FcR blocker, serine protease inhibitors, or functional blocking anti-PAR2 and anti-GPR97 mAbs (Fig. 6e, f, and Supplementary Fig. 8c,d). Altogether, we conclude that most PAR2 and GPR97 are stored in azurophilic granules and translocated to cell surfaces when neutrophils received specific activation signals. The increased presence of PAR2 and GPR97 at the cell surface promotes clustering of CD177-associated receptors and induces GPR97-PAR2 activation, leading to further neutrophil activation.
GPR97-PAR2 activation promotes neutrophil-mediated anti-microbial activity and endothelial cell activation and dysfunction
To investigate the potential innate immune function of GPR97-PAR2 activation, the effects of GPR97E-mFc on the bacterial uptake and killing abilities of neutrophils were examined. As shown, the engulfment of several bacteria species including E. coli, S. typhimurium, and S. aureus was enhanced in neutrophils incubated with GPR97E-mFc, but not the mFc control (Fig. 7a). Similarly, the bacterial killing assay showed that GPR97E-mFc treatment significantly facilitates the elimination of S. typhimurium and S. aureus by neutrophils (Fig. 7b). We compared neutrophil activation phenotypes after application of PAR2-specific agonists and found that GPR97-mediated PAR2 activation is more effective overall, suggesting that it is a predominant PAR2 activation mechanism in neutrophils (Supplementary Fig. 9).
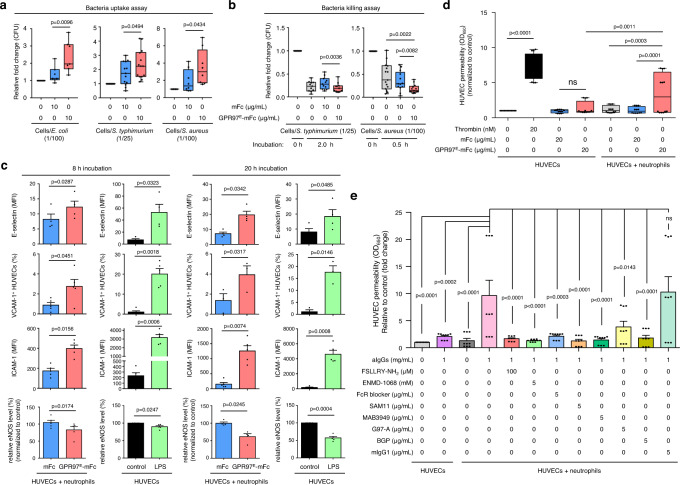
Fig. 7. GPR97-mediated PAR2 activation enhances neutrophil-mediated bacterial phagocytosis and killing as well as endothelial cell activation and dysfunction.
a, b Phagocytosis a and killing b of live bacteria (E. coli n = 10, S. typhimurium n = 13 for phagocytosis assay n = 11 for killing assay, and S. aureus n = 8 for phagocytosis assay n = 11 for killing assay) by neutrophils incubated without or with GPR97E-mFc. mFc was included as a negative control. Data are means ± SEM. P value was determined by one-way ANOVA. c The expressional analyses of cell activation markers (E-selectin, ICAM-1, VCAM-1 and eNOS) of HUVECs co-cultured with neutrophils in the absence or presence of GPR97E-mFc. mFc was included as a negative control. HUVECs treated without or with LPS were used as a negative and a positive control, respectively. n = 3 independent experiments. Data are means ± SEM and p value was determined by two-sided unpaired student’s t-test. d, e Endothelial cell permeability assays of HUVECs co-cultured with neutrophils. d Assays were done in HUVEC-neutrophil co-culture in the absence or presence of GPR97E-mFc for 20 h at 37 °C. mFc was included as a negative control. HUVECs alone treated without or with mFc were negative control groups, while those treated with thrombin were the positive control. n = 4 independent experiments. e The HUVEC-neutrophil co-culture was treated without or with aIgGs in the absence or presence of protease inhibitors/PAR2 antagonists/blocking Abs as indicated. HUVECs alone treated without or with aIgGs were included as controls. n = 3 independent experiments. Data are means ± SEM and p value was determined by two-way ANOVA. ns, non-significant. Source data are provided in the Source Data file.
Uncontrolled and excess activated neutrophils are associated with adverse tissue damage seen in various inflammatory disorders36. To delineate the effect of GPR97-PAR2 activation on the vasculature, the activation and dysfunction of endothelial cells were investigated using the HUVEC-neutrophil co-culture system. HUVECs were significantly activated when co-cultured with neutrophils in the presence of GPR97E-mFc (Fig. 7c). Moreover, GPR97E-mFc treatment promoted endothelial permeability and dysfunction (Fig. 7d). Similar increased endothelial permeability was identified in HUVEC-neutrophil co-culture in the presence of aIgGs. FcR blocker, PAR2 antagonists, or functional blocking anti-PAR2 and anti-GPR97 mAbs attenuated this effect (Fig. 7e). We conclude that mPR3-mediated GPR97-PAR2 activation in neutrophils plays a role in anti-microbial responses and endothelial cell activation and dysfunction.
Discussion
In this study, GPR97 was identified as the binding partner and allosteric activator of mPR3 which in turn activated PAR2 on human neutrophils (Figs. 2 and 3). The mPR3-mediated GPR97-PAR2 activation adds a significant component to the repertoire of neutrophil activation mechanisms (Figs. 5–7).
PAR2 activation is normally induced by proteolytic enzymes including soluble proteases such as trypsin and microbial proteases as well as single-transmembrane or glycosylphosphatidylinositol-linked membrane sheddases such as matriptase and testisin37,38. The active coagulation protein complex containing tissue factor (TF)/factor VIIa (FVIIa)/factor Xa (FXa) could also act as a potent PAR2 activator39. In this case, PAR2 is transactivated indirectly by the TF/FVIIa/FXa complex via its proteolytic activation of the matriptase zymogen40. By contrast, PAR1-PAR2 transactivation was achieved via interaction of PAR2 with the thrombin-exposed tethered ligand of PAR114. Interestingly, the GPR97-PAR2 activation process also involves a unique protein complex, here containing a serine protease, two glycosylphosphatidylinositol-linked receptors, and an aGPCR (Fig. 5h).
PAR2 is expressed ubiquitously and its activation has been shown to play multiple immune-regulatory roles including neutrophil activation and inflammatory cytokine production41–43. Critically, neutrophil activation via the TF/FVIIa/PAR2 axis resulted in trophoblast injury and fetal death in an animal model of autoimmune antiphospholipid syndrome11. Our results herein show that neutrophil PAR2 activation, hereby GPR97/mPR3, plays a role in endothelial cell activation/dysfunction as well as bacterial uptake and less effectively in bacterial killing (Fig. 7). Our findings of the up-regulation of GPR97 and PAR2 expression in activated neutrophils suggested that the GPR97-PAR2 activation mode is likely a common neutrophil activation mechanism associated with inflammatory diseases (Figs. 1 and 6).
The absolute requirement of the clustered PR3/CD177/GPR97/PAR2/CD16b complex for efficient GPR97-PAR2 activation is surprising (Fig. 5), but it is not without precedent. In fact, it is highly analogous to the selective involvement of the glycosylphosphatidylinositol-anchored RECK and GPR124/ADGRA2 receptor complex in mediating the Frizzled 4 (FZD4)/Low-density lipoprotein receptor-related protein 5 (LRP5)-dependent Wnt7a-specific bioactivity for CNS angiogenesis and blood-brain barrier integrity44,45. Coincidentally, the two aGPCR-GPCR protein complexes all consist of one soluble protein (PR3/Wnt7) and 4 receptor molecules including one glycosylphosphatidylinositol-linked receptor (CD177/RECK), one aGPCR (GPR97/GPR124), and one GPCR (PAR2/FZD4).
GPCR transactivation is an efficient means to expand the signaling outputs and usually involves the activation of another receptor type such as receptor tyrosine kinases (RTKs) by GPCRs46. Nevertheless, recent studies have shown that transactivation of GPCRs can also be induced by RTKs and other GPCRs47,48. GPCR transactivation of RTKs typically involves the activation of extracellular metalloproteases, which cleave and release membrane-bound protein ligands of RTKs49. In line with this, it is intriguing to note the involvement of GPR97-augmented mPR3 enzymatic activity in the PAR2 activation process (Fig. 3). Interestingly, approximate analogies were also noted in the Wnt7a-RECK-GPR124 complex in which RECK acted as a competitive inhibitor of Wnt7a signaling by specifically ligating and segregating Wnt7a from FZD4. GPR124 engagement efficiently realigned the RECK-Wnt7a complex to be available for FZD4 recognition hence permitting signaling45. With these in mind, we suggest that the aGPCR-GPCR activation mechanism symbolizes a previously unappreciated GPCR activation paradigm beyond the classical ligand-induced receptor activation.
GPR97 belongs to the ADGRG subgroup of the aGPCR family which contains some of the smaller-sized members including GPR56 and GPR11450. Although the GAIN domain has been identified as an evolutionarily conserved structural fold sufficient for GPS auto-proteolysis, the minimal domain structure needed for the unusual auto-proteolytic reaction remained to be fully elucidated27. Our analyses of GPR97-ECR showed that a well-conserved subdomain B plus a small one α-helix-containing subdomain A are competent for efficient GPS auto-proteolysis (Fig. 4). Recently, the cryo-electron microscopy structures of glucocorticoid-bound GPR97-Go complexes have revealed the first 3D structure of the seven-transmembrane region of an aGPCR51. With the structural resolution of GPR97-ECR reported here, it is possible now to build a complete structural feature of a full-length GPR97.
PR3 is the most abundant protease of azurophilic granules with versatile intracellular and extracellular functions4. Specific binding of mPR3 by PR3-ANCA which ligated and activated FcγRs was considered as the major disease mechanism of neutrophil activation in GPA35. Despite these, little is known of the physiological function of mPR3. Our findings of the tunable enzymatic activity of mPR3 by GPR97 and the subsequent PAR2 activation hence uncover a role for mPR3 in immune regulation (Figs. 3–7). Paradoxically, the outcomes of PR3-mediated PAR2 cleavage seemed to depend critically on the cell types studied. Indeed, PR3 digestion was shown to disarm PAR2 for subsequent trypsin-mediated activation in kidney epithelial cells52. On the other hand, PAR2 was efficiently activated by PR3 in immature dendritic cells41 and by the IL-32γ/PR3 complex in THP-1 cells53. In the future, it could be a valuable advance to reveal the mechanistic detail of GPR97-PAR2 activation and to survey other potential protein substrates of the GPR97-augmented mPR3 activity.
In view of the unique human neutrophil-specific feature of GPR97-mPR3 interaction (Fig. 2, and Supplementary Fig. 1c), it was surprising to find interspecies differences in several members of the PR3/CD177/GPR97/PAR2/CD16b complex. Human PR3 differed from gibbon and murine PR3s by containing a distinctive hydrophobic patch which mediated its interaction with CD17754. Unlike the neutrophil-restricted expression of GPR97 in human, Adgrg3/Gpr97 was involved in B-lymphocyte fate decision55, obesity-associated macrophage inflammation56, and lymphatic endothelial cell migration in mice57. Finally, as CD16b is the neutrophil-specific glycosylphosphatidylinositol-anchored FcγRIII expressed only in human and not in mice58, it suggests that GPR97-PAR2 activation is a human neutrophil-restricted phenomenon thereby highlighting the requirement for caution when extrapolating mouse data. Importantly, the strict regulation of GPR97 and PAR2 expression in neutrophils indicates that the GPR97-PAR2 activation reaction is induced only by unique inflammatory triggers, such as FcR-mediated signalling, that up-regulate surface GPR97 and PAR2 expression to a significant level. We believe the reason for having the PR3/CD177/GPR97/PAR2/CD16b complex, consequently a GPR97-PAR2 activation mechanism, is to provide an extra layer of control that acts on primed/activated neutrophils. Once induced, it allows for full neutrophil activation. Our results suggest that this is important for an effective anti-microbial response and immune effector function. Moreover, as proteolytic enzymes and GPCRs belong to two major families of molecular drug targets, the PR3/CD177/GPR97/PAR2/CD16b receptor complex represents a potential multi-target complex for the development of therapeutics to modulate human neutrophil-mediated inflammatory diseases59,60.
Methods
Reagents and antibodies
All chemicals and reagents were purchased from Sigma (St. Louis, MO, USA) and Invitrogen (CA, USA) unless specified otherwise. E-64 (#78434) was from Thermo Fisher Scientific. Human proteinase 3 (PR3) (#16-14-161820) was purchased from Athens Research & Technology, Inc. (Athens, Georgia, USA). Human Fc receptor blocker (Human BD Fc Block™, #564220) was from BD Biosciences (New Jersey, USA). Antibodies used in this study are listed in Supplementary Table 1.
Cell culture
All cell culture media and supplements, including fetal calf serum (FCS), L-glutamine, penicillin and streptomycin were purchased from Invitrogen. Cell lines used in this study (listed in Supplementary Table 2) were purchased from the American Type Culture Collection (Manassas, VA, USA) or Bioresource Collection and Research Center (Hsinchu, Taiwan) and cultured in conditions as suggested.
Construction of expression vectors and cell transfection
Mammalian expression constructs encoding the mFc only as well as various GPR97-, GPR56-, and EMR2-mFc fusion proteins were generated in the pcDNA3.1-mFc vector using standard molecular biology technologies as described previously19. Gene-specific oligonucleotide primers used to construct the mFc-fusion protein expression vectors are listed in Supplementary Table 3. For the construction of expression vectors encoding GPR97-TM7-EGFP and GPR97-myc, the full-length human GPR97 cDNA was cloned into the pEGFP-N1 (Clontech) and pcDNA3.1/myc-His A (Invitrogen) vectors, respectively. Expression vectors encoding the PAR2, EPCR, Mac-1 (CD11b/CD18), CD16b, and CD55 receptors were purchased from Sino Biological Inc. (Beijing, China). The CD177 and FPR1 expression constructs were gifts from Dr. Ralph Kettritz of Charité of Universitätsmedizin Berlin, Germany and Dr. TL Hwang of Chang Gung University, Taiwan, respectively. The EMR2 expression construct was described previously61. For transient cell transfection, HEK-293T cells were transfected with purified plasmid DNAs using LipofectamineTM or LipofectamineTM 2000 (Invitrogen) as described elsewhere.
Generation and purification of the mFc-fusion proteins
All recombinant mFc-fusion proteins were purified from conditioned media of transiently transfected HEK-293T cells using the Protein A-Sepharose affinity chromatography as described previously19. Briefly, HEK-293T cells were transfected with the expression constructs of interest using the calcium phosphate-based transfection method. Following transfection, cells were cultured in serum-free OPTI-MEM medium for 4–5 days and conditioned media was collected, centrifuged, and passed through the Protein A-Sepharose affinity column (nProtein A SepharoseTM 4 Fast Flow, GE Healthcare), followed by extensive washes with washing buffer (50 mM Tris-HCl pH 7.4, 10 mM CaCl2, 150 mM NaCl). The mFc-fusion protein was eluted and subjected to dialysis in the Slide-A-Lyzer® Dialysis Cassettes (Thermo Scientific) in washing buffer for 24 h at 4 °C, filtered, and stored at −80 °C until use.
Purification of blood neutrophils
Human neutrophils were isolated from fresh venous blood donated by healthy volunteers and diseased patients who have all given the signed informed consent and receive no participant compensation. All experimental procedures were approved by the Chang Gung Memorial Hospital Ethics Committee (CGMH IRB No: 201701852B0, 201901358B0, 201901293B0, and 202002255B0) and performed according to the guidelines set by the Committee. In brief, blood samples were drawn into the collection tubes coated with sodium heparin and neutrophil isolation was performed using PolymorphprepTM density gradient centrifugation (Axis-Shield, Oslo, Norway) as described previously18. For the isolation of murine neutrophils, blood was collected by cardiac puncture and separated by Ficoll (GE Healthcare) gradient separation. Following the lysis of the remnant erythrocytes, isolated neutrophils were checked for purity by flow cytometry with specific surface markers and resuspended in RPMI medium containing 10% FCS for all following experiments unless otherwise denoted. The isolation procedure routinely produced a > 95% pure and viable neutrophil population. When indicated, isolated human neutrophils were further subjected to the magnetic cell sorting (MACS) separation of CD177+ and CD177− sub-populations using PE-conjugated anti-CD177 Ab coupled to the MACS MicroBeads and Separator (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s recommendations.
Immunofluorescence assay and immunohistochemistry
Freshly isolated human neutrophils (1 × 106 cells/mL) were treated as described and then fixed with 4% paraformaldehyde/PBS for 20 min at 4 °C. Cells were then incubated with the blocking buffer (1% BSA, 5% normal goat serum (NGS) in PBS) for 1 h at 4 °C, then with indicated primary antibody (5 μg/mL) diluted in blocking buffer for 1 h at 4 °C. Following extensive washes in cold PBS, cells were incubated for 1 h at 4 °C with fluorescence-labeled secondary antibody diluted in blocking buffer at a pre-determined optimal concentration. Cells were washed thoroughly before being spun onto slides. Fluorescence images were taken by Fluoview FV10i (Olympus) or the LSM780 confocal microscope system (ZEISS). Immunohistochemistry was carried out on formalin-fixed, paraffin-embedded tissue sections (4 μm) using G97-A anti-GPR97 mAb (5 μg/mL) as described previously18. All tissue staining procedures were performed in an automated immunostainer (BOND-MAXTM, Leica Biosystem). Immunoreactivity was assessed independently by two expert pathologists. Neutrophils in capillaries were found to display strong GPR97 expression constantly. Therefore, the GPR97 expression level of tissue infiltrating neutrophils was scored as follows: 1 + , if GPR97 expression was far weaker than that of intravascular neutrophils; 2 + , if GPR97 expression was weaker than that of intravascular neutrophils; 3 + , if the GPR97 expression intensity was equal to that of intravascular neutrophils; 4 + , if GRP97 expression was stronger than that of intravascular neutrophils.
Flow cytometry analysis and the FACS-based ligand-binding assay
All procedures were performed at 4 °C. Cells were blocked in ice-cold blocking buffer (1% BSA/5% NGS in PBS) for 1 h. For the standard flow cytometry analysis, cells were incubated sequentially with the primary Ab of interest and appropriate fluorescence-labeled secondary Ab diluted in blocking buffer at pre-determined optimal concentrations for 1 h. For the analysis of GPR97 expression in blood neutrophils of normal controls and diseased patients, blood leukocytes were obtained following RBC lysis with ACK buffer and washed twice with ice-cold PBS. Cells (1 × 106 cells/reaction) were stained with a mixture of aqua fluorescent reactive dye (MAN0006891, Life Technologies), anti-CD16b-FITC (130-126-529, Miltenyi Biotec), anti-CD66b-PerCP-Cy5.5 (305108, Biolegend), and G97-A conjugated with APC (Ab201807, Abcam) for 20 min. Fluorescent minus one controls include GPR97 and aqua fluorescent reactive dye only control were included for background determination. The fluorescence-labeled cells were analyzed by BD FACSCanto II flow cytometry system. For the cellular ligand-binding assay, cells were incubated with the mFc-fusion protein probe (10 μg/mL) in blocking buffer for 2 h. Where indicated, cells were pretreated or treated simultaneously with various reagents in the presence of the probes. Fluorescence-conjugated goat anti-mouse IgG (2 μg/mL) was used as the 2nd Ab. Following extensive washes between each incubation step, cells were washed lastly in cold PBS and subjected to analysis by FACSCalibur flow cytometer (BD Biosciences). Positive ligand-binding was determined as the shift of fluorescence signals. The mFc protein was always included as a negative control.
Phenotypic analyses of neutrophils
Freshly isolated human neutrophils (1 × 106 cells/mL) were suspended in RPMI complete medium and incubated with soluble GPR97E-mFc, control mFc protein (10 μg/mL), or f-MLF (10−6 M) for 3 h at 37 °C. For the expressional analysis of CD markers, cells were fixed by 4% paraformaldehyde/PBS for 20 min at 4 °C and stained with fluorescence-conjugated CD markers as indicated using the standard flow cytometry protocol. For the analysis of neutrophil morphological changes, live cells were either collected at 1 h after incubation and photographed under the inverted microscopy, or analyzed directly by FACSCalibur flow cytometer and shown as FSC/SSC. Alternatively, cells were fixed and subjected to the Wright-Giemsa stain, or permeabilized and blocked in the blocking buffer (1% BSA, 5% NGS, 0.1% saponin in PBS) for 30 min, then incubated with Phalloidin-TRITC (10 μg/mL) and analyzed by flow cytometer as well as Fluoview FV10i (Olympus). For IL-8 production, culture supernatant was collected at 3 h incubation and subjected to the ELISA analysis.
Analyses of reactive oxygen species production and myeloperoxidase activity
Freshly isolated human neutrophils (2 × 106 cells/mL) were resuspended in PBS supplemented with 0.2% BSA and 5 mM glucose, and incubated with 2 μM dihydrorhodamine-123 (DHR123; Molecular Probes #D23806) for 30 min at room temperature (RT). Cells were then incubated respectively with GPR97E-mFc or control mFc-fusion protein (10 μg/mL) for 30 min at 37 °C. Cells were then stimulated with or without fMLF (10−6 M) for 15 min before being placed on ice to stop the reaction. The accumulation of ROS represented by fluorescent oxidized DHR123 was immediately measured by flow cytometry as described previously18.
The MPO enzyme activity was analyzed using the colorimetric MPO activity assay kit (BioVision) performed exactly as suggested by the manufacturer. Briefly, freshly isolated human neutrophils (2.5 × 106 cells/mL) were resuspended in PBS supplemented with 0.2% BSA and 5 mM glucose. Cells were then incubated with GPR97E-mFc or control mFc fusion protein (10 μg/mL) for 30 min at 37 °C. When necessary, cells were first incubated with GPR97E-mFc, followed by stimulation with or without fMLF (10−6 M) for 15 min. Cells were lysed with the MPO assay buffer and then centrifuged at 10,000 g for 10 min at 4 °C. Lysate supernatant (10 μL/sample) was added to 40 μL of MPO assay buffer in a 96-well plate. The reaction was initiated by the addition of 10 μL MPO substrate solution to each well and incubated for 60 min at RT. Finally, 2 μL of stop solution was added, followed immediately with 50 μL of Ellman’s reagent (5,5′-dithiobis-(2-nitrobenzoic acid), DTNB) for the colorimetric analysis of the MPO activity at 412 nm.
Measurement of IL-8 by sandwich ELISA
Freshly isolated human neutrophils (1 × 106 cells/mL) were cultured in RPMI/10% FBS in the presence of fMLF (10−6 M), mFc or GPR97E-mFc (10 μg/mL) for 3 h at 37 °C. When indicated, culture plates were coated with the mFc fusion proteins at 4 °C overnight before incubation with neutrophils. When necessary, neutrophils were pretreated with various reagents/inhibitors such as α1-antitrypsin (5 mg/mL), aprotinin (50 μg/mL), E-64 (50 μg/mL) and N-p-Tosyl-L-phenylalanine chloromethyl ketone (TPCK, 10 μM) for 1 h at 37 °C or incubated with elafin (4 μM) and SLPI (4 μM) simultaneously. Cell-free supernatants were collected to determine the concentration of IL-8 by using the ELISA kit (DuoSet® ELISA, R&D Systems, DY208) according to the protocols suggested by the manufacturer.
Structural analysis of GPR97-ECR
For crystallisation experiments, human GPR97-ECR (UniProt Q86Y34, residues 1 to 264) was cloned into the EcoRI-KpnI cloning sites of vectors from the pHLsec family with its native secretion signal sequence and a C-terminal His6 tag. The cDNA encoding GPR97 possesses a SNP at residue 447 (M→V). GPR97-ECR was expressed in a secreted form in GlnTI-deficient HEK-293S cells using previously described protocols62. Briefly, plasmid DNAs were transfected with PEI in a 1:2 ratio into 3 L of 90–100% confluent HEK-293S cells. After 10 days, the cell culture medium containing the secreted GPR97-ECR was clarified by centrifugation and filtration prior to diafiltration into PBS, 20 mM Tris pH 7.5 and 150 mM NaCl. GPR97-ECR was then purified by immobilized metal affinity chromatography (HiTrapTM HP, GE Healthcare) and size-exclusion chromatography (Superdex 200 16/600, GE Healthcare) in 20 mM Tris pH 7.5 and 300 mM NaCl. The protein purity was assessed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and concentrated to 40 mg/mL using 10,000 MWCO concentrators (Amicon Ultra Centrifugal Filters).
GPR97-ECR crystals grew in 100 nL + 100 nL sitting nanodrops by the vapour diffusion method at 18 °C in 1.8 M tri-ammonium citrate pH 7. The crystals were harvested and cryo-protected in 3 M tri-ammonium citrate pH 7. A native dataset was collected at a wavelength of 0.9763 Å at the European Synchrotron Radiation Facility beamline ID30B. 180° of data were collected with an exposure of 0.02 s per 0.1° rotation on a PILATUS3 6 M (Dectris). The data was processed using DIALS63 and AIMLESS64. A S-SAD dataset was also collected at a wavelength of 2.75 Å on the long-wavelength beamline I2365 at Diamond Light Source, UK. 360° of data were collected with an exposure of 0.1 s per 0.1° rotation on a PILATUS 12 M detector (Dectris). The data showed sign of anisotropy and was therefore processed using AUTOPROC66 and STARANISO (Tickle, I.J. et al. (2018) STARANISO. Cambridge, United Kingdom: Global Phasing Ltd.) (Supplementary Table 4).
The flexible loops of the GAIN domains of Lphn1, BAI3 and GPR56 (PDB accession numbers 4DLQ, 4DLO, 5KVM respectively) were removed and an ensemble of those trimmed GAIN domains was used as a search model for molecular replacement using the native dataset and PHASER67. The solution consisted of two molecules in the asymmetric unit and displayed additional density on top of the GAIN domain, accounting for the small N-terminal domain. To help build this N-terminal domain, molecular replacement using the S-SAD dataset and PHASER67 was carried out with the partial model consisting of the GPR97 GAIN domain. The resulting solution was subjected to one round of refinement in autoBUSTER (Bricogne G., et al (2017) BUSTER version 2.10.3. Cambridge, United Kingdom: Global Phasing Ltd.)68. Electron density maps were improved in PARROT69 revealing the missing N-terminal domain. Anomalous difference maps calculated with ANODE70 were used to locate sulphur atoms and help manual building in COOT71. All-atom refinement with autoBUSTER (Bricogne G., et al (2017) BUSTER version 2.10.3. Cambridge, United Kingdom: Global Phasing Ltd.)68 resulted in the final model containing residues 28–260.
Western and far-western immunoblotting analysis
Cells were lysed in RIPA lysis buffer (20 mM Tris-HCl pH7.4, 5 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P-40, and 1× Complete Protease Inhibitors) supplemented with 1 mM sodium orthovanadate, 1 mM AEBSF, and 5 mM Levamisole. Lysate proteins were quantified using the Bicinchoninic acid (BCA) protein assay kit (Pierce). Protein samples were separated in 10% SDS or native PAGE gels and transferred to PVDF membranes. Membranes were then soaked in blocking buffer (PBS containing 5% nonfat skimmed milk and 0.1% Tween 20) for 1 h at RT. Next, the membranes were incubated with indicated primary Ab or GPR97E-mFc (5 μg/mL) diluted in blocking buffer overnight at 4 °C. EMR2E-mFc or mFc (5 μg/mL) protein was used as negative controls. Following extensive washes in washing buffer (0.1% Tween 20 in PBS), membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse Fc Ab (1:2000) in blocking buffer. Finally, the specific binding of Ab and mFc-fusion proteins on membrane blots was revealed with chemiluminescent HRP Substrate (Millipore WBKLS0500).
Protein extraction of the cytoplasmic and membrane fractions
All protein isolation procedures are performed at 4 °C. Freshly isolated neutrophils (2 × 107 cells) were washed three times with cold PBS, re-suspended in RIPA buffer and passed through a 30-gauge needle 20 times. Cell lysates were then centrifuged at 700 g for 3 min at 4 °C, and the supernatant was collected and subjected to ultracentrifugation at 10,000 g for 40 min at 4 °C. The resulting supernatant was collected as the cytoplasmic fraction and the pellet was the membrane fraction. All protein samples were stored at −80 °C until use.
FACS-based PR3-binding assay
HEK-293T cells were transfected with various expression constructs as indicated. HEK-293T transfectants were collected, washed, and incubated in blocking buffer (1% BSA, 5% NGS/PBS) for 1 h at 4 °C before incubating with commercially available purified PR3 (5 μg/mL) diluted in blocking buffer for 1 h at 4 °C. Cells were subjected to extensive washes in cold PBS, and then incubated with the anti-PR3 mAb (5 μg/mL)(clone MCPR3-2, Thermo Fisher Scientific MA5-11945) or PeliCluster ANCA (5 μg/mL)(clone CLB-12.8, Sanquin M1574) for 1 h at 4 °C. Cells were extensively washed and then incubated with fluorescence-labeled secondary antibody for 1 h at 4 °C. Finally, cells were washed three times in cold PBS and subjected to analysis by FACSCalibur flow cytometer (BD Biosciences). When indicated, HEK-293T cells were first transfected with the GPR97-TM7-EGFP construct. Transfected cells were then subjected to FACS sorting by FACSAriaTM II (BD Biosciences) to select the top 30% highest GPR97-expressing (GPR97high) and low GPR97-expressing (GPR97low) cells based on the GFP expression levels. The sorted GPR97high and GPR97low HEK-293T cells were used to examine the PR3-binding ability as described above. Similar EMR2high and EMR2low HEK-293T transfectants were included as negative controls.
For the analysis of GPR97-mPR3 binding in transfected HEK-293T cells, cells were transfected as described. Half of the transfected cells were checked by the flow cytometry analysis using receptor-specific Abs for the expression of the specific receptors. The other half of the transfected cells were incubated first with purified PR3 (5 μg/mL) diluted in blocking buffer for 1 h at 4 °C, washed, and followed by incubation with mFc (10 μg/mL) or GPR97E-mFc (10 μg/mL) diluted in blocking buffer for 2 h at 4 °C. Cells were washed and reacted with fluorescence-labeled goat anti-mouse IgG for 1 h at 4 °C. Finally, cells were washed extensively in cold PBS and subjected to analysis by FACSCalibur flow cytometer (BD Biosciences).
ELISA-like PR3-binding assay
Saturated mFc-fusion proteins (10 μg/mL), including CD177-mFc, GPR56E-mFc, GPR97E-mFc, and mFc were separately coated onto 96-well plates for 16 h at 4 °C. Coated plates were washed with the wash buffer (0.05% Tween 20 in PBS) three times, and blocked with the blocking buffer (5% BSA in PBS) for 1 h at 4 °C. Purified PR3 protein (1, 1.5, 2, and 3 μg/mL in blocking buffer) was added for 2 h at 4 °C. Wells were then washed extensively and incubated with the MCPR3-2 anti-PR3 mAb (0.4 μg/mL in blocking buffer) for 2 h at 4 °C. Wells were washed extensively and incubated with goat anti-mouse κ chain HRP (1 μg/mL) for 1 h at 4 °C. Following extensive washes, tetramethylbenzidine (TMB) substrate (100 μL/well) was added for 20 min at 4 °C before adding the stop solution (2 N H2SO4)(50 μL/well). Reaction signals were detected by the ELISA reader at OD450.
The ex vivo membrane PR3 activity assay
The ex vivo neutrophil membrane PR3 activity was measured using the PR3-specific FRET substrate, Abz-VADnV-RDRQ-EDDNP (Cayman, MI, USA) exactly as described previously26. Briefly, freshly isolated neutrophils (1 × 106 cells/well) were washed twice with PBS. Cells were then suspended in 150 μL PBS and incubated with recombinant mFc-fusion proteins of interest in appropriate concentration for 10 min at 37 °C before adding the PR3-specific FRET substrate (20 μM). Fluorescence intensity was detected by the SpectraMax M2e ELISA reader (Molecular Devices, CA, USA) for at least 90 min during the reaction. When necessary, cells were treated with various protease inhibitors as indicated. The corresponding Vmax was analyzed by the Softmax Pro 5.3 software.
The proximity-ligation assay
To detect the close interaction of CD177, PR3, GPR97, PAR2, and CD16b in human neutrophils, the Duolink in situ PLA (Sigma-Aldrich®, DUO92013) was employed according to the manufacturer’s protocol. Purified human neutrophils were cytospun on Poly-D-Lysine-coated coverslips and blocked in blocking buffer (1% BSA/5% NGS in PBS) for 60 min at 4 °C. Cells were incubated with the receptor-specific Abs (15 μg/mL) pre-conjugated with the PLA-PLUS or -MINUS oligonucleotide probe (Sigma-Aldrich®, DUO92009 and DUO92010) for 60 min at RT. The enzymatic ligation of the PLA-PLUS and –MINUS probes were performed for 30 min at 37 °C, followed by circle amplification for 90 min at 37 °C and finally incubated with Hoechst 33342 (Invitrogen) for nuclear staining. Cells were observed by the LSM780 confocal microscope system (ZEISS) at 40× magnification.
Bacteria uptake and killing assays
Freshly isolated neutrophils (1.25 × 106 cells/mL) were suspended in RPMI and incubated without or with GPR97E-mFc or control mFc fusion protein (10 μg/mL) respectively for 30 min at 37 °C. Neutrophils were then incubated with live E. coli (DH10B)(MOI 1:100), S. typhimurium (MOI 1:25), and S. aureus (MOI 1:100) in logarithmic-phase for 1 h at 37 °C. For the bacteria uptake assay, extracellular bacteria were removed by extensive washing in PBS, followed by incubation for 30 min in RPMI-1640 medium containing 50 μg/mL gentamicin. Afterwards, neutrophils were washed twice with PBS and lysed with 0.05% Triton X-100 in PBS. For the bacteria killing assay, neutrophils were collected and lysed with 0.05% Triton X-100 in PBS at desired time points after the bacteria uptake and the removal of extracellular bacteria by gentamicin. Serial dilutions of the neutrophil lysate were plated on LB plates and incubated overnight at 37 °C to determine the bacterial colony-forming units (CFU) the next day.
Neutrophil-HUVEC co-culture
Human umbilical vein endothelial cells (HUVECs) were maintained in Medium 199 supplemented with 10% FBS, L-glutamine, penicillin, streptomycin, 25 U/mL heparin and 30 μg/mL endothelial cell growth supplement. HUVECs (6 × 105 cells/well) were plated on 1% gelatin-coated culture dishes (6 mm) until confluence. HUVECs were incubated with freshly isolated human neutrophils (6 × 106 cells/well) without or with indicated mFc-fusion proteins (10 μg/mL) for 8 or 20 h at 37 °C. HUVECs incubated with LPS (1 μg/mL) were used as a positive control. Cells were washed with ice-cold PBS twice and collected for analysis using flow cytometry by staining with CD62E-APC (E-selectin, BD Pharmingen), CD54-APC (ICAM-1, BD Pharmingen), CD106-PE (VCAM-1, BD Pharmingen) and eNOS-PE (BD Pharmingen). For the endothelial cell permeability assay, HUVECs (5 × 104 cells/well) were plated on 1% gelatin-coated Transwell plates (12 mm)(Costar, Corning) containing polycarbonate membranes (3 μm pore size) until confluence. Cells were incubated with freshly isolated human neutrophils (1 × 106 cells/well) without or with indicated mFc-fusion proteins (20 μg/mL) for 24 h. HUVECs incubated with thrombin (20 nM) for 30 min were used as a positive control. Alternatively, HUVECs were incubated with human aIgGs (1 mg/mL) without or with PAR2 antagonists (FSLLRY-NH2 and ENMD-1068)(100 μM), PAR2 antibodies (SAM11 and MAB3949)(5 μg/mL), GPR97 antibodies (G97-A and BGP)(5 μg/mL) and mouse IgG1 as indicated for 8 h. The lower chambers were replenished with fresh M199 growth medium while the upper chambers were filled with fresh medium containing 4% BSA and Evans blue (0.67 mg/mL). Cells were incubated for 30 min at 37 °C and the optical density of the medium from lower chambers was measured at 650 nm in a spectrophotometer.
Statistics and reproducibility
No statistical method was used to predetermine sample size and no data were excluded from the analyses. All results were analyzed using GraphPad Prism (version 6.0 and 7.0; GraphPad Software, San Diego, CA, USA) and expressed as means ± standard error of the mean (SEM) with the number of experimental replicates (n) provided. Differences between groups were determined by student’s t-test, one-way, and two-way ANOVA as indicated. In all cases, a probability (p) value of <0.05 was accepted to reject the null hypothesis and considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Dr. Jörg Hamann and Dr. Gin-Wen Chang for their critical reading of the manuscript. The authors acknowledge the technical assistance from the Microscopy and Instrumentation Centers, Chang Gung University, Taoyuan, Taiwan. C.Z.G. was funded by the Wellcome Trust DPhil in Structural Biology programme. E.S. was funded by the Wellcome Trust (202827/Z/16/Z), the M.R.C. (MR/L018039/1), and the EMBO Young Investigator programme. H.H.L. was funded by Chang Gung Memorial Hospital (CMRPD1M0031, CMRPD1M0511, CMRPD1M0521, and CMRPD1M0321) and the Ministry of Science and Technology (MOST), Taiwan (MOST-107-2320-B-182-006 and MOST-110-2320-B-182-024).
Source data
Author contributions
T.Y.C., C.Z.-G., K.Y.H., Y.C.C., Y.W.C., K.Y.I., Y.L.L., N.Y.C., H.Y.C., W.Y.T., C.Y.S., Y.M.W., Y.S.P., C.H.H., T.C.C., K.E.O., M.A., and S.R.H. performed experiments, M.S., S.G., C.Y.L., and A.S. provided advice and critical reagents, T.Y.C., C.Z.-G., E.S., and H.H.L. designed experiments and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Dimitrios Fotiadis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
All data needed to evaluate the conclusions in the paper are present in the main manuscript, the Supplementary Information and the Source data. Source data are provided with this paper and comprise all relevant raw data from each figure in the main manuscript and in the Supplementary Information. The X-ray structure data of GPR97 extracellular domain generated in this study have been deposited in the database of Protein Data Bank in Europe (PDBe) under accession code PDB ID 7QU8 (Deposition ID: D_1292120102). Source data are provided in this paper.
Code availability
No custom codes/softwares were generated in this study. The following publicly available softwares were used for structural analysis. DIALS 1.9.3 (https://dials.github.io/installation.html), Coot 0.9.6, Aimless 0.7.3, Phaser 2.8.3, Parrot 1.0.4, and CCP4 suite 7.1 (https://www.ccp4.ac.uk/download/#os=mac). ANODE 2013/1 is available through the SHELX software suite at https://www.ccp4.ac.uk/download/#os=mac. autoPROC 1.0.5 and autoBUSTER 2.10.4 are from https://www.globalphasing.com/. STARANISO V3.347 is accessed at https://staraniso.globalphasing.org/. Structural figures and structural alignments were generated using PyMOL v.2.3.4. (https://pymol.org/2/). The AlphaFold model of GPR97 is available at https://alphafold.ebi.ac.uk/entry/Q86Y34. FlowJo v7.6.1 (https://www.flowjo.com/solutions/flowjo/downloads), ZEISS ZEN 2011 (https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html#downloads), Olympus FluoView FV10i olympus fluoview fv10i (https://www.olympus-lifescience.com/en/support/downloads/), and Graphpad prism v6 and v7 (https://www.graphpad.com/scientific-software/prism/) were used for the analysis of cellular phenotypes.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tai-Ying Chu, Céline Zheng-Gérard.
These authors jointly supervised this work: Elena Seiradake, Hsi-Hsien Lin.
Contributor Information
Elena Seiradake, Email: elena.seiradake@bioch.ox.ac.uk.
Hsi-Hsien Lin, Email: hhlin@mail.cgu.edu.tw.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-34083-1.
References
- 1.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 2.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 3.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 2010;62:726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 5.Kitching AR, et al. ANCA-associated vasculitis. Nat. Rev. Dis. Prim. 2020;6:71. doi: 10.1038/s41572-020-0204-y. [DOI] [PubMed] [Google Scholar]
- 6.von Vietinghoff S, et al. NB1 mediates surface expression of the ANCA antigen proteinase 3 on human neutrophils. Blood. 2007;109:4487–4493. doi: 10.1182/blood-2006-10-055327. [DOI] [PubMed] [Google Scholar]
- 7.Hu N, Westra J, Kallenberg CG. Membrane-bound proteinase 3 and its receptors: relevance for the pathogenesis of Wegener’s Granulomatosis. Autoimmun. Rev. 2009;8:510–514. doi: 10.1016/j.autrev.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Rarok AA, Stegeman CA, Limburg PC, Kallenberg CG. Neutrophil membrane expression of proteinase 3 (PR3) is related to relapse in PR3-ANCA-associated vasculitis. J. Am. Soc. Nephrol. 2002;13:2232–2238. doi: 10.1097/01.asn.0000028642.26222.00. [DOI] [PubMed] [Google Scholar]
- 9.Futosi K, Fodor S, Mocsai A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 2013;17:638–650. doi: 10.1016/j.intimp.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramachandran R, Noorbakhsh F, Defea K, Hollenberg MD. Targeting proteinase-activated receptors: therapeutic potential and challenges. Nat. Rev. Drug Discov. 2012;11:69–86. doi: 10.1038/nrd3615. [DOI] [PubMed] [Google Scholar]
- 11.Redecha P, Franzke CW, Ruf W, Mackman N, Girardi G. Neutrophil activation by the tissue factor/Factor VIIa/PAR2 axis mediates fetal death in a mouse model of antiphospholipid syndrome. J. Clin. Investig. 2008;118:3453–3461. doi: 10.1172/JCI36089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shpacovitch V, Feld M, Hollenberg MD, Luger TA, Steinhoff M. Role of protease-activated receptors in inflammatory responses, innate and adaptive immunity. J. Leukoc. Biol. 2008;83:1309–1322. doi: 10.1189/jlb.0108001. [DOI] [PubMed] [Google Scholar]
- 13.Shpacovitch V, Feld M, Bunnett NW, Steinhoff M. Protease-activated receptors: novel PARtners in innate immunity. Trends Immunol. 2007;28:541–550. doi: 10.1016/j.it.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Lin H, Trejo J. Transactivation of the PAR1-PAR2 heterodimer by thrombin elicits beta-arrestin-mediated endosomal signaling. J. Biol. Chem. 2013;288:11203–11215. doi: 10.1074/jbc.M112.439950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamann J, et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoneberg T, Liebscher I, Luo R, Monk KR, Piao X. Tethered agonists: a new mechanism underlying adhesion G protein-coupled receptor activation. J. receptor signal Transduct. Res. 2015;35:220–223. doi: 10.3109/10799893.2015.1072978. [DOI] [PubMed] [Google Scholar]
- 17.Hamann J, Hsiao CC, Lee CS, Ravichandran KS, Lin HH. Adhesion GPCRs as modulators of immune cell function. Handb. Exp. Pharmacol. 2016;234:329–350. doi: 10.1007/978-3-319-41523-9_15. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao CC, et al. The adhesion G protein-coupled receptor GPR97/ADGRG3 is expressed in human granulocytes and triggers antimicrobial effector functions. Front. Immunol. 2018;9:2830. doi: 10.3389/fimmu.2018.02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang NY, et al. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J. Biol. Chem. 2011;286:14215–14225. doi: 10.1074/jbc.M110.183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beyrau M, Bodkin JV, Nourshargh S. Neutrophil heterogeneity in health and disease: a revitalized avenue in inflammation and immunity. Open Biol. 2012;2:120134. doi: 10.1098/rsob.120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulenberg-Gustavus C, Bahring S, Maass PG, Luft FC, Kettritz R. Gene silencing and a novel monoallelic expression pattern in distinct CD177 neutrophil subsets. J. Exp. Med. 2017;214:2089–2101. doi: 10.1084/jem.20161093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flesch BK, et al. Multicenter study on differential human neutrophil antigen 2 expression and underlying molecular mechanisms. Transfus. Med. hemotherapy: offizielles Organ der Dtsch. Ges. fur Transfusionsmedizin und Immunhamatologie. 2020;47:385–395. doi: 10.1159/000505523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemmensen SN, et al. Olfactomedin 4 defines a subset of human neutrophils. J. Leukoc. Biol. 2012;91:495–500. doi: 10.1189/jlb.0811417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerke U, Marino SF, Daumke O, Kettritz R. Characterization of the CD177 interaction with the ANCA antigen proteinase 3. Sci. Rep. 2017;7:43328. doi: 10.1038/srep43328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korkmaz B, et al. Inhibitors and antibody fragments as potential anti-inflammatory therapeutics targeting neutrophil proteinase 3 in human disease. Pharmacol. Rev. 2016;68:603–630. doi: 10.1124/pr.115.012104. [DOI] [PubMed] [Google Scholar]
- 26.Korkmaz B, et al. Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nat. Protoc. 2008;3:991–1000. doi: 10.1038/nprot.2008.63. [DOI] [PubMed] [Google Scholar]
- 27.Arac D, et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzman GS, et al. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 2016;91:1292–1304. doi: 10.1016/j.neuron.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leon K, et al. Structural basis for adhesion G protein-coupled receptor Gpr126 function. Nat. Commun. 2020;11:194. doi: 10.1038/s41467-019-14040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David A, Fridlich R, Aviram I. The presence of membrane Proteinase 3 in neutrophil lipid rafts and its colocalization with FcgammaRIIIb and cytochrome b558. Exp. cell Res. 2005;308:156–165. doi: 10.1016/j.yexcr.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 32.Jerke U, et al. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J. Biol. Chem. 2011;286:7070–7081. doi: 10.1074/jbc.M110.171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YC, et al. Membrane-bound proteinase 3 and PAR2 mediate phagocytosis of non-opsonized bacteria in human neutrophils. Mol. Immunol. 2011;48:1966–1974. doi: 10.1016/j.molimm.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 34.Kurosawa S, Esmon CT, Stearns-Kurosawa DJ. The soluble endothelial protein C receptor binds to activated neutrophils: involvement of proteinase-3 and CD11b/CD18. J. Immunol. 2000;165:4697–4703. doi: 10.4049/jimmunol.165.8.4697. [DOI] [PubMed] [Google Scholar]
- 35.Granel J, et al. Pathogenicity of Proteinase 3-Anti-Neutrophil Cytoplasmic Antibody in Granulomatosis With Polyangiitis: Implications as Biomarker and Future Therapies. Front. Immunol. 2021;12:571933. doi: 10.3389/fimmu.2021.571933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruger P, et al. Neutrophils: between host defence, immune modulation, and tissue injury. PLoS Pathog. 2015;11:e1004651. doi: 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawar NR, Buzza MS, Antalis TM. Membrane-anchored serine proteases and protease-activated receptor-2-mediated signaling: co-conspirators in cancer progression. Cancer Res. 2019;79:301–310. doi: 10.1158/0008-5472.CAN-18-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothmeier AS, Ruf W. Protease-activated receptor 2 signaling in inflammation. Semin. Immunopathol. 2012;34:133–149. doi: 10.1007/s00281-011-0289-1. [DOI] [PubMed] [Google Scholar]
- 39.Camerer E, Huang W, Coughlin SR. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc. Natl Acad. Sci. USA. 2000;97:5255–5260. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Gall SM, et al. Matriptase activation connects tissue factor-dependent coagulation initiation to epithelial proteolysis and signaling. Blood. 2016;127:3260–3269. doi: 10.1182/blood-2015-11-683110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Csernok E, et al. Wegener autoantigen induces maturation of dendritic cells and licenses them for Th1 priming via the protease-activated receptor-2 pathway. Blood. 2006;107:4440–4448. doi: 10.1182/blood-2005-05-1875. [DOI] [PubMed] [Google Scholar]
- 42.Heuberger DM, Schuepbach RA. Protease-activated receptors (PARs): mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thrombosis J. 2019;17:4. doi: 10.1186/s12959-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saffarzadeh M, et al. Macrophage protease-activated receptor 2 regulates fetal liver erythropoiesis in mice. Blood Adv. 2020;4:5810–5824. doi: 10.1182/bloodadvances.2020003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho C, Smallwood PM, Nathans J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and Blood-Brain Barrier Regulation. Neuron. 2017;95:1221–1225. doi: 10.1016/j.neuron.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 45.Eubelen, M. et al. A molecular mechanism for Wnt ligand-specific signaling. Science361, (2018). [DOI] [PubMed]
- 46.Wang W, Qiao Y, Li Z. New Insights into Modes of GPCR Activation. Trends Pharmacol. Sci. 2018;39:367–386. doi: 10.1016/j.tips.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Cattaneo F, et al. Cell-surface receptors transactivation mediated by g protein-coupled receptors. Int. J. Mol. Sci. 2014;15:19700–19728. doi: 10.3390/ijms151119700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin H, Liu AP, Smith TH, Trejo J. Cofactoring and dimerization of proteinase-activated receptors. Pharmacol. Rev. 2013;65:1198–1213. doi: 10.1124/pr.111.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schafer AE, Blaxall BC. G protein coupled receptor-mediated transactivation of extracellular proteases. J. Cardiovascular Pharmacol. 2017;70:10–15. doi: 10.1097/FJC.0000000000000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin HH, et al. Adhesion GPCRs in regulating immune responses and inflammation. Adv. Immunol. 2017;136:163–201. doi: 10.1016/bs.ai.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Ping YQ, et al. Structures of the glucocorticoid-bound adhesion receptor GPR97-Go complex. Nature. 2021;589:620–626. doi: 10.1038/s41586-020-03083-w. [DOI] [PubMed] [Google Scholar]
- 52.Ramachandran R, et al. Neutrophil elastase acts as a biased agonist for proteinase-activated receptor-2 (PAR2) J. Biol. Chem. 2011;286:24638–24648. doi: 10.1074/jbc.M110.201988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakayama M, et al. IL-32-PAR2 axis is an innate immunity sensor providing alternative signaling for LPS-TRIF axis. Sci. Rep. 2013;3:2960. doi: 10.1038/srep02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korkmaz B, Kuhl A, Bayat B, Santoso S, Jenne DE. A hydrophobic patch on proteinase 3, the target of autoantibodies in Wegener granulomatosis, mediates membrane binding via NB1 receptors. J. Biol. Chem. 2008;283:35976–35982. doi: 10.1074/jbc.M806754200. [DOI] [PubMed] [Google Scholar]
- 55.Wang JJ, et al. Gpr97 is essential for the follicular versus marginal zone B-lymphocyte fate decision. Cell Death Dis. 2013;4:e853. doi: 10.1038/cddis.2013.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi J, et al. Gpr97 is dispensable for metabolic syndrome but is involved in macrophage inflammation in high-fat diet-induced obesity in mice. Sci. Rep. 2016;6:24649. doi: 10.1038/srep24649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valtcheva N, Primorac A, Jurisic G, Hollmen M, Detmar M. The orphan adhesion G protein-coupled receptor GPR97 regulates migration of lymphatic endothelial cells via the small GTPases RhoA and Cdc42. J. Biol. Chem. 2013;288:35736–35748. doi: 10.1074/jbc.M113.512954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unkeless JC, Shen Z, Lin CW, DeBeus E. Function of human Fc gamma RIIA and Fc gamma RIIIB. Semin. Immunol. 1995;7:37–44. doi: 10.1016/1044-5323(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 59.Robertson JG. Enzymes as a special class of therapeutic target: clinical drugs and modes of action. Curr. Opin. Struct. Biol. 2007;17:674–679. doi: 10.1016/j.sbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 60.Santos R, et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin HH, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 62.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 63.Winter G, et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D., Struct. Biol. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. Sect. D. Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner A, Duman R, Henderson K, Mykhaylyk V. In-vacuum long-wavelength macromolecular crystallography. Acta Crystallogr. Sect. D., Struct. Biol. 2016;72:430–439. doi: 10.1107/S2059798316001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D., Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. Sect. D., Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cowtan K. Recent developments in classical density modification. Acta Crystallogr. Sect. D., Biol. Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thorn A, Sheldrick GM. ANODE: anomalous and heavy-atom density calculation. J. Appl. Crystallogr. 2011;44:1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the main manuscript, the Supplementary Information and the Source data. Source data are provided with this paper and comprise all relevant raw data from each figure in the main manuscript and in the Supplementary Information. The X-ray structure data of GPR97 extracellular domain generated in this study have been deposited in the database of Protein Data Bank in Europe (PDBe) under accession code PDB ID 7QU8 (Deposition ID: D_1292120102). Source data are provided in this paper.
No custom codes/softwares were generated in this study. The following publicly available softwares were used for structural analysis. DIALS 1.9.3 (https://dials.github.io/installation.html), Coot 0.9.6, Aimless 0.7.3, Phaser 2.8.3, Parrot 1.0.4, and CCP4 suite 7.1 (https://www.ccp4.ac.uk/download/#os=mac). ANODE 2013/1 is available through the SHELX software suite at https://www.ccp4.ac.uk/download/#os=mac. autoPROC 1.0.5 and autoBUSTER 2.10.4 are from https://www.globalphasing.com/. STARANISO V3.347 is accessed at https://staraniso.globalphasing.org/. Structural figures and structural alignments were generated using PyMOL v.2.3.4. (https://pymol.org/2/). The AlphaFold model of GPR97 is available at https://alphafold.ebi.ac.uk/entry/Q86Y34. FlowJo v7.6.1 (https://www.flowjo.com/solutions/flowjo/downloads), ZEISS ZEN 2011 (https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html#downloads), Olympus FluoView FV10i olympus fluoview fv10i (https://www.olympus-lifescience.com/en/support/downloads/), and Graphpad prism v6 and v7 (https://www.graphpad.com/scientific-software/prism/) were used for the analysis of cellular phenotypes.