Abstract
The processes leading from disturbed B-cell development to adult B-cell progenitor acute lymphoblastic leukemia (BCP-ALL) remain poorly understood. Here, we describe Irf4−/− mice as prone to developing BCP-ALL with age. Irf4−/− preB-I cells exhibited impaired differentiation but enhanced proliferation in response to IL-7, along with reduced retention in the IL-7 providing bone marrow niche due to decreased CXCL12 responsiveness. Thus selected, preB-I cells acquired Jak3 mutations, probably following irregular AID activity, resulting in malignant transformation. We demonstrate heightened IL-7 sensitivity due to Jak3 mutants, devise a model to explain it, and describe structural and functional similarities to Jak2 mutations often occurring in human Ph-like ALL. Finally, targeting JAK signaling with Ruxolitinib in vivo prolonged survival of mice bearing established Irf4−/− leukemia. Intriguingly, organ infiltration including leukemic meningeosis was selectively reduced without affecting blood blast counts. In this work, we present spontaneous leukemogenesis following IRF4 deficiency with potential implications for high-risk BCP-ALL in adult humans.
Subject terms: B cells, Cancer models
Introduction
Two signaling pathways via the Interleukin-7 receptor (IL-7R) and the preB cell receptor (preBCR) ensure an orderly progression of B lymphopoiesis [1–3]. ProB cells adhere to bone marrow (BM) stromal cells (SCs) expressing CXCL12 and VCAM-1 through CXCR4 and VLA-4 respectively, while SC-derived IL-7 induces their proliferation [4]. The formation of the preBCR composed of Igμ protein and the surrogate light chain (ψL), consisting of λ5 and VPREB, marks the entrance to the preB cell stage. Signaling via the preBCR in turn induces the transcription factor (TF) interferon regulatory factor 4 (IRF4) which is also critical during T-cell differentiation [5, 6]. In preB cells, IRF4 halts cycling and facilitates recombination of the light chain locus by RAG1/2 [1]. Despite its importance, Irf4−/− mice still develop, albeit less, surface (s)Igμ+ mature B cells [7], likely due to a partially redundant function of IRF8. Accordingly, Irf4,8−/− B progenitors are completely arrested at the preB cell stage [8].
Disruption of this developmental track can provoke B-cell progenitor acute lymphoblastic leukemia (BCP-ALL). In humans, this disease preferentially affects children (age 0–19), while most deaths however occur in the adult population [9]. Cases affecting adolescents and young adults (AYA) display a different set of driver mutations compared to childhood BCP-ALL [10–13].
Herein, we report that adult Irf4−/− mice spontaneously develop BCP-ALL with age and delineate the steps from disturbed Irf4−/− B lymphopoiesis to overt leukemia.
Results
Irf4−/− mice spontaneously develop preB cell leukemia
Following the serendipitous finding, that some aged Irf4−/− mice developed tumors and died, we systematically observed 80 Irf4–/– mice over time. We detected 14 tumors (incidence 17.5%), that spontaneously appeared in lymph node (LN) areas (mean age: 268d, median: 238d, Fig. 1a). Tumors were neither detected in mice younger than 150d nor in C57BL/6 wild-type (wt) mice housed in the same room.
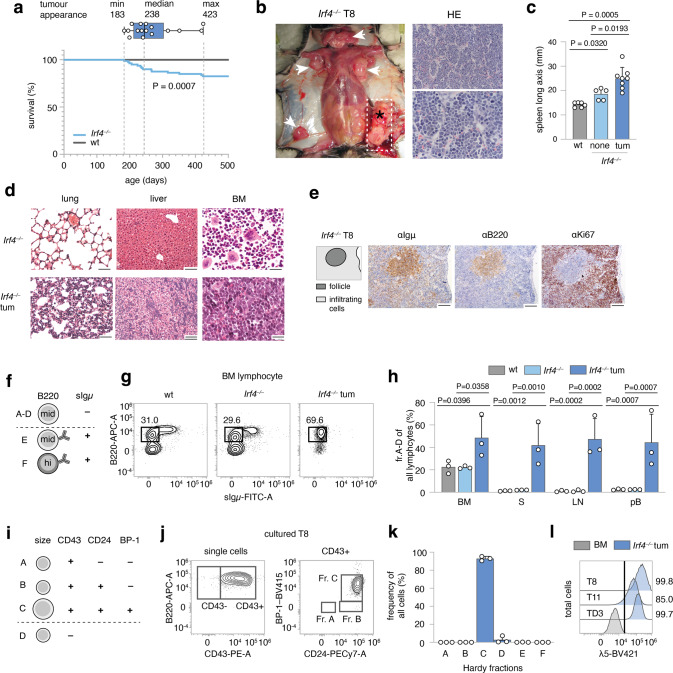
Fig. 1. Spontaneous emergence of preB-I cell BCP-ALL in adult Irf4−/− mice.
a A cohort of 80 Irf4−/− and wt mice was observed over 500 days for tumor development. Kaplan–Meier plot of survival. Box and whisker plot indicates minimum, maximum and median age at tumor appearance of the 14 affected mice. b Macroscopic appearance of an exemplary tumor (asterisk) and LNs (arrowheads) in an Irf4−/− mouse. Right: Hematoxylin-Eosin (HE) staining from the tumor. Scale bars: top: 50 µm, bottom: 20 µm. c Longitudinal spleen (S) axis (mm) of Irf4−/− mice with (n = 8) or without (n = 5) tumor and control wt mice (n = 6). tum = tumor. d HE stainings of lung, liver, and BM of tumor mouse T8 and a healthy Irf4−/− mouse. Scale Bars: 50 µm (lung and liver), 20 µm (BM). e IHC-stainings of T8 mouse spleen for Igμ, B220, and Ki67. Scale bars: 100 µm. f Schematic representation of gross Hardy fractioning by surface B220 and Igμ expression. g Whole BM cells from wt, Irf4−/−, and tumor mice were stained for B220 and sIgμ expression and analyzed by flow cytometry. h Quantification of cell frequencies gated as in (g) for BM, S, LN, and pB (peripheral blood) of n = 3 mice per group. i Tabular representation of Hardy fr. A-D by size, CD43-, CD24-, and BP-1-surface expression. j Surface expression of markers as in (i) of in vitro cultured T8 tumor cells k quantification of cell frequencies gated as in (j) for three tumors (T8, T11, TD3) l surface λ5 expression on T8, T11, and TD3 by flow cytometry in comparison to whole BM cells from Irf4−/− mice (BM) as a negative control. Statistical significance testing was performed with (c) one-way Welch-ANOVA followed by Dunnet’s T3 multiple comparison test and (h) with two-way ANOVA followed by pair-wise Tukey corrected comparisons within each organ. Bars depict mean ± SD, dots indicate mice (h) or distinct Irf4−/− tumors (k).
All tumors (Fig. 1b shows a representative tumor in situ) were accompanied by lymphadenopathy (arrowheads) and increased spleen size (Fig. 1c). The suspected lymphomatous origin was corroborated microscopically (Fig. 1b, right panels), with infiltration of mononucleated cells into the BM, lung, and liver (Fig. 1d). Due to the known impaired maturation of Irf4−/− preB cells [7], spontaneous eruption of preB-leukemia seemed plausible: In spleen sections (Fig. 1e), infiltrating cells stained positive for both B220 and Igμ (although less than untransformed “follicle” B cells) and Ki67. By flow cytometry, BM samples from tumor mice harbored an expanded pro/preB cell compartment (Hardy fraction (fr.)A-D [14], B220midsIgμ–) (Fig. 1f, g). Fr.A-D cells were detected also in peripheral lymphoid organs and blood of tumor mice (Fig. 1h). Following the Hardy classification (Fig. 1i), we determined tumor cells to be fr.C preB cells (B220midsIgμ–CD43+CD24+BP-1+) (Fig. 1j, k, sFig. 1a). In addition, Igμ, but not Igκ/λ was detected intracellularly (sFig. 1b). Lastly, tumors stained positive for surface λ5, part of the ψL (Fig. 1l). These attributes characterize the disease as preB-I cell BCP-ALL.
To prove clonality, we sequenced the VDJ junctions of the IgH region in three tumors (Supplementary Table 1). Almost all sequences per tumor were identical, demonstrating clonality. The tumors (three examples) further displayed copy number variations (CNV) (sFig. 1h), targeting differing genomic regions. Finally, transfer of tumor cells robustly elicited leukemia in wt acceptor mice (sFig. 1d–g) with as little as 500 transferred cells (sFig. 1f, g), indicating bona fide malignancy.
B lymphopoiesis in Irf4−/− mice harbors a hyperproliferative preB-I cell compartment
The uniform appearance of BCP-ALL in Irf4−/− mice suggested a defined preleukemic pro/preB cell state vulnerable to immortalization. Dimensional reduction of BM samples stained for B-cell differentiation markers, identified an enlarged fr.C preB cell compartment already in healthy Irf4−/− mice (Fig. 2a–c, sFig. 2a). This disturbed, but productive B-cell maturation confirms and extends previous reports [7]. Expression analysis of IL-7Rα and of CD2 (sFig. 2b, c), which accompanies cytosolic Igμ expression [15] further showed an increased frequency of CD2-/dimIL-7Rα+B220+sIgμ− preB cells in Irf4−/− mice.
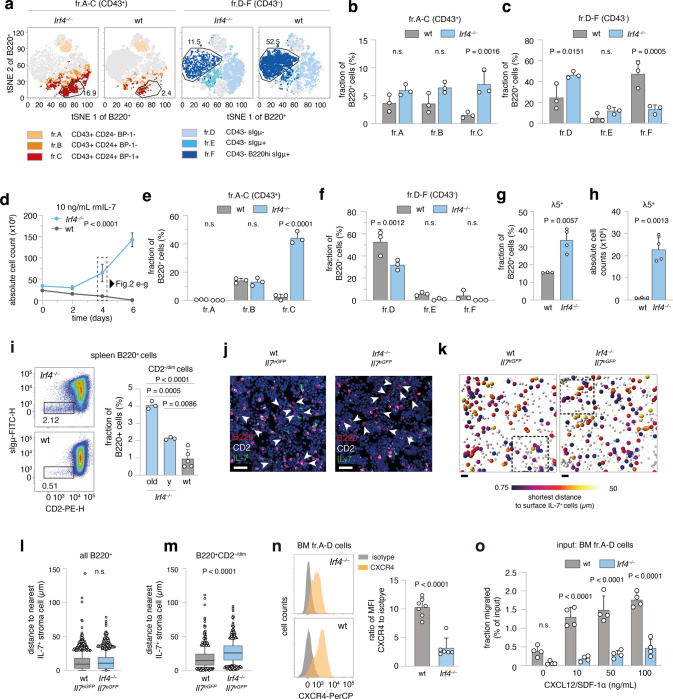
Fig. 2. Irf4−/− B lymphopoiesis is preleukemically altered.
a–c flow cytometric analysis of BM cells for Hardy markers as in Fig. 1j. a tSNE of BM cells gated on B220+ cells. Colors correspond to Hardy fractions identified by the markers detailed in the legend. b and c quantification of Hardy fraction frequencies for n = 3 mice per genotype. d–h BM cells from Irf4−/− and wt mice were cultured in the presence of 10 ng/mL rmIL-7 for 6 days and d counted every two days. e, f After 4 days, cells were stained as in a–c and Hardy fractions quantified. g frequency and h absolute cell counts of λ5+ cells on day 4. i spleen cells from Irf4−/− (“y” = young: 6–10 weeks and old: >6 months) and wt mice were analyzed for the presence of CD2–/dimsIgμ– cells within the B220+ gate. one-way ANOVA, Tukey post hoc. j–m 7 µm cryosections from Irf4−/−Il-7eGFP and wt Il-7eGFP mice were stained for B220, CD2, GFP, and DAPI. j exemplary regions of BM cryosections. Arrowheads indicate B220+CD2–/dim cells. Scale bars = 15 µm. k automated B220+ cell detection: gray spheres indicate B220+ cells, larger spheres B220+CD2–/dim cells, color-coded for their distance to GFP+ cells. Rectangles indicate magnified areas in j. Scale bars = 40 µm. l, m quantification of distances to IL-7+ cells for l all B220+ and m B220+CD2–/dim cells. (n = 4 mice per genotype, one cryosection from femur metaphysis per mouse analyzed). Box and whiskers indicate mean and 95-IQR, dots indicate cells outside 95-IQR. n BM cells from Irf4−/− and wt mice were gated on B220+sIgμ– fr.A-D cells and analyzed for CXCR4 expression (left panels as representative staining). Data is presented for n = 7 (wt) and n = 6 (Irf4−/−) mice as the ratio of geometric mean for CXCR4 to isotype staining. o MACS-purified fr.A-D cells from BM were placed in the top insert of a Boyden chamber and left to migrate towards differing concentrations of CXCL12 for 16 h. Dots represent n = 4 biologically independent experiments, presented as migrated percentage of input cells. Two-Way ANOVA, Sidak post hoc for (b, c, e, f, o), Two-tailed unpaired t test for (g, h, l–n).
Purified BM B220+ cells from Irf4−/− and wt mice were cultured with IL-7 (Fig. 2d–g) to compare proliferative capacities. After 6d, Irf4−/− cells had expanded roughly three-fold, whereas wt cell numbers decreased. Phenotypically, Irf4−/− cells accumulated at the fr.C stage (Fig. 2e, f) and expressed surface λ5 (Fig. 2g, h); exactly like Irf4−/− leukemia. In contrast, wt cells differentiated further, losing surface CD43 (making them fr.D) (Fig. 2e, f) with some cells expressing sIgμ (fr.E). Thus, IL-7 unmasked the leukemic potential of the fr.C compartment in Irf4−/− mice with both unchecked proliferation and a reinforced differentiation block. Notably, IL-7 dependent Irf4−/− preB-I cell proliferation was blocked by NIBR3049 and Ruxolitinib, inhibitors of the IL-7R downstream actors JAK3 and JAK1 respectively (sFig. 2d).
Irf4−/− B-cell progenitors exhibit reduced retention to the BM niche
As overt leukemia is characterized by systemic presence, we tested whether already preleukemic Irf4−/− B-cell progenitors would leak from the BM. To reduce the complex Hardy classification, we identified early B-cell progenitors, approximately until the preB-I stage, by B220+CD2–/dim expression (sFig. 2b). We detected higher frequencies of splenic B220+CD2–/dim cells in Irf4−/− than in wt mice (Fig. 2h), which accumulated with age. Thus, premature BM evasion adds to the impaired differentiation and hyperproliferation that characterize Irf4−/− preleukemia.
Potentially, this finding represented a systemic consequence of reduced vicinity to BM niche cells. We, therefore, analyzed the proximity of Irf4−/− and wt B220+CD2–/dim cells to IL-7+ BMSCs in situ using Il-7eGFP reporter mice (Fig. 2j–m, sFig. 2e, f, Supplementary Movie 1) [16]. In femur cryosections, the B220+CD2–/dim subset (Fig. 2j, arrowheads) but not the whole Irf4−/− B220+ cell compartment was on average located further away from IL-7+ BMSCs, compared to wt control (Fig. 2l, m). We excluded differences in IL-7+ BMSC abundance between genotypes (sFig. 2g).
B progenitor retention to BM is secured via the interaction of CXCR4 on pro/preB cells with the IL-7+ BMSC-derived chemokine CXCL12 [17, 18]. Notably, Irf4−/− pro/preB cells expressed markedly lower levels of CXCR4 compared to wt cells (Fig. 2n). Chemokine migration assays with Hardy fr.A-D cells showed that Irf4−/− cells indeed migrated significantly less towards CXCL12 (Fig. 2o). Thus, reduced CXCR4-CXCL12 interaction likely induces the systemic seeding of Irf4−/− B progeny. Inversely, direct cell interactions are likely not responsible, because Irf4−/− and wt fr.A-D cells adhered equally to monolayers of OP-9 cells in vitro (sFig. 2h).
The IL-7-JAK-STAT-axis is recurrently altered in Irf4−/− leukemia
Most likely, a second, acquired genetic alteration was necessary for bona fide leukemia development and arose with low frequency per time, explaining the affected age and relatively low penetrance. Importantly, IL-7 deprivation of BM-evaded pro/preB cells should create strong survival stress and potential selection pressure for bona fide leukemogenesis. To identify somatically acquired mutations, we performed whole-exome sequencing (WES) of three independent tumors (T8, T10, T11) compared to sorted B220+sIgµ− cells from Irf4−/− BM. Comparisons of the single nucleotide variants (SNVs) between the three samples identified nine genes affected in all three tumor samples (Fig. 3a). Out of these, SNVs in four genes (Rrs1, Jak3, AW82073, and Duxf3) showed alternate base frequencies close to 0.5 or 1 (Fig. 3b), suggesting that they could be present on one or both alleles of all leukemic cells. Although we did not exclude the oncogenic potential of the other three genes, we focused on Jak3, because it is associated with IL-7R signaling. We detected Jak3 mutations also in other tumors TD1, TD2, TD3, and T14 by Sanger- and RNA-sequencing (Fig. 3c, Supplementary Table 2). Thus, seven out of seven tested tumors carried Jak3 mutations (“JAK3mut”). All mutations targeted either the active kinase domain or the pseudokinase domain regulating JAK3 activity. Some of these SNVs have been described before [19]. Further, using two different classifiers, no gene fusions could be detected (see methods). Analysis of typical BCP-ALL genes [20] identified some respective mutations at low frequencies, indicative of subclonal events (Fig. 3d). Among these, mutations in Jak1, the partner to JAK3 in IL-7R signaling, were detected in both T8 and T11.
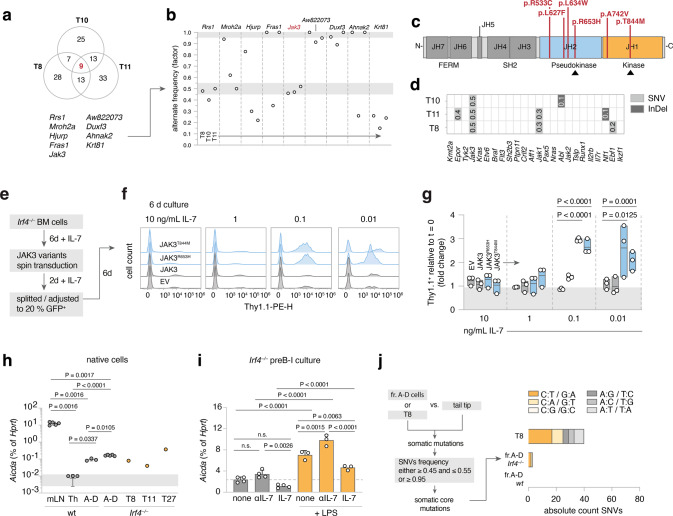
Fig. 3. leukemia-derived Jak3 mutations heighten IL-7 sensitivity of Irf4−/− preB-I cells.
a Venn diagram of shared mutated genes among WES from three Irf4−/− leukemia samples (T8, T10, T11) b the nine shared genes were filtered for SNV frequency. gray areas: >0.95 and 0.45–0.55 margins as core mutation filters. c the five detected distinct Jak3 SNVs were mapped onto JAK3 primary structure (JH = Jak homology domain). d Irf4−/− leukemia WES were analyzed for mutations (SNV or InDel = insertions/deletions) in genes commonly altered in human BCP-ALL. Numbers indicate rounded frequencies of alteration. e–g Irf4−/− BM cells were cultured for 6d with 10 ng/mL rmIL-7, transduced with control or JAK3mut coding RVs, rested for 2 days, and then split into decreasing IL-7 concentrations. f Histograms for the Thy1.1 RV infection marker 6 days after splitting. g Quantification of Thy1.1+ cells after 6 days relative to start of culture (t = 0). Dots indicate n = 3 independent experiments, plotted as floating bars. EV = empty vector. h qRT-PCR from wt and Irf4−/− cells for Aicda mRNA expression, relative to Hprt expression for n = 3 (Th (= T helper) and sorted fr.A-D wt BM cells), n = 5 (mLN (= mesenteric lymph node) and sorted fr.A-D Irf4−/− BM cells), n = 1 per tumor T8, T11, T27. i Irf4−/− preB-I cell cultures from whole BM cells were cultured in combinations of IL-7, αIL-7, and LPS for 24 h, as indicated, and analyzed for Aicda levels by qRT-PCR for n = 4 (no LPS) and n = 3 (LPS) samples. j WES from fr.A-D cells and T8 were compared to tail-tip samples to identify SNVs. Filtering on SNV frequency “0.45–0.55 or ≥0.95” yielded putative “core mutations”. Absolute numbers of nucleotide exchanges are presented as stacked bars, colors give the type of nucleotide exchange. Two-way ANOVA, Sidak post hoc for g–i.
To analyze the role of the JAK3mut, we transduced Irf4−/− preB-I cell cultures with retroviruses (RVs) encoding no or wt JAK3 or the JAK3mut R653H and T844M. Culturing transduced cells in the presence of αIL-7 to test for IL-7 independency unexpectedly resulted in cell death after a few days with no benefit for cells expressing JAK3mut (sFig. 3a, b). To test if JAK3mut would confer advantages with limited IL-7, RV-infected Irf4−/− preB-I cell cultures were exposed to decreasing IL-7 concentrations (Fig. 3e–g). At 0.1 and 0.01 ng/ml IL-7, both JAK3mut-, but not JAK3wt-RV led to the outgrowth of transduced over untransduced cells after 6d of culture (Fig. 3f, g). Thus, JAK3mut confer IL-7-hypersensitivity, but not -independency. Accordingly, ex vivo cultured Jak3-mutated T8 and T11 cells also still depended on IL-7 (sFig. 3c, d). However, the tumor cells exhibited increased proliferation (sFig. 3e) and λ5 surface retention (sFig. f, g) in decreased IL-7 concentrations when compared to wt or Irf4−/− preB cell culture.
Aicda is upregulated in Irf4−/− preB cells by LPS and deprivation of IL-7
Because six out of seven Jak3 mutations were C to T base exchanges (Table 2), we suspected a specific mutagenic agent. DNA-editing enzymes including the APOBEC family member AID can deamidate cytosines, e.g., during somatic hypermutation [21, 22]. Repair mechanisms most often ultimately cause C to T conversions [23, 24]. Notably, AID is induced in wt preB cells by IL-7 withdrawal and LPS stimulation and acts as a facilitator of human BCP-ALL [25]. Therefore, we compared Aicda expression in sorted Irf4−/− and wt fr.A-D cells to that of wt mesenteric (m)LN- and CD4+ TH1-cells as controls and to individual leukemia samples. While mLN cells highly expressed Aicda, fr.A-D preB- and leukemia cells, but not TH1-cells, also expressed readily detectable amounts (Fig. 3h).
Furthermore, like their wt counterpart [25], in vitro expanded Irf4–/– preB-I cells upregulated Aicda further under LPS treatment and during IL-7 withdrawal (Fig. 3i). This finding can explain how BM evasion and exposure to pathogens might cooperatively initiate mutagenic processes via AID in vulnerable Irf4−/− preB-I cells.
To test if T8 exhibited signs of previous AID activity on a global level, we analyzed C:T/G:A-transition frequencies in WES of T8, as well as BM-sorted Irf4−/− and wt fr.A-D cells compared with matched tail-tip samples. Indeed, we found a marked preponderance of C:T/G:A-transitions in T8, when filtering on putative somatic core SNVs (Fig. 3j).
Jak3 mutations in mice mirror Jak2 mutations in human Ph-like ALL
Next, we compared Irf4−/− leukemia to the complex landscape of human BCP-ALL subtypes (reviewed in refs. [11, 26, 27]), using a published human BCP-ALL cohort for which a random forest classifier had been established (Methods for details) [28]. Only mildly (potentially due to the interspecies comparison) elevated prediction scores were generated for Ph+, Ph-like, KMT2a- and DUX4-rearranged human BCP-ALL (sFig. 4a, b). Since all of these except Ph-like are defined by specific gene rearrangements, that we had not detected in Irf4−/− mouse leukemia, we excluded them as comparable candidates.
Ph-like ALL harbors recurrent genetic alterations in signaling molecules, especially in CRLF2 and JAK2 [20]. While BCP-ALL overall preferentially affects children, the incidence of the Ph-like subtype increases from 10% in children to above 25% in AYA and adults [20, 29], reminiscent of the older age of Irf4−/− leukemic mice. Furthermore, a published dataset of 154 Ph-like BCP-ALL cases exhibited 10-fold reduced IRF4 transcripts, when compared to other BCP-ALL subtypes [20].
While in human Ph-like ALL, Jak2 is commonly mutated, we report recurrent Jak3 mutations in Irf4−/− mice. As both proteins are part of distinct but similar signaling complexes in B-cell progenitors (Fig. 4a), we investigated structural and functional similarities between the specific Jak3 and Jak2 mutations. Comparisons of amino-acid sequences revealed high protein-wide interspecies and intermolecular similarities for both proteins (Fig. 4b). Mapping the two amino acids R653 and T844 (mutated in Irf4−/− mice) onto JAK3 structure predictions, generated by the alpha-fold algorithm [30], revealed that the two amino acids are in direct contact at an interface of JH1-JH2 domains (Fig. 4c). This interface specifically is highly conserved in JAK2 compared to JAK3 (Fig. 4d, f, sFig. 4c, d). Intriguingly, R683 (corresponding to R653 in JAK3) is by far the most commonly mutated amino acid in JAK2 in Ph-like ALL, while mutations targeting T875 (corresponding to T844) also have been described [31]. These findings suggest that mutations in human JAK2 and mouse JAK3 affect a highly similar functional hotspot.
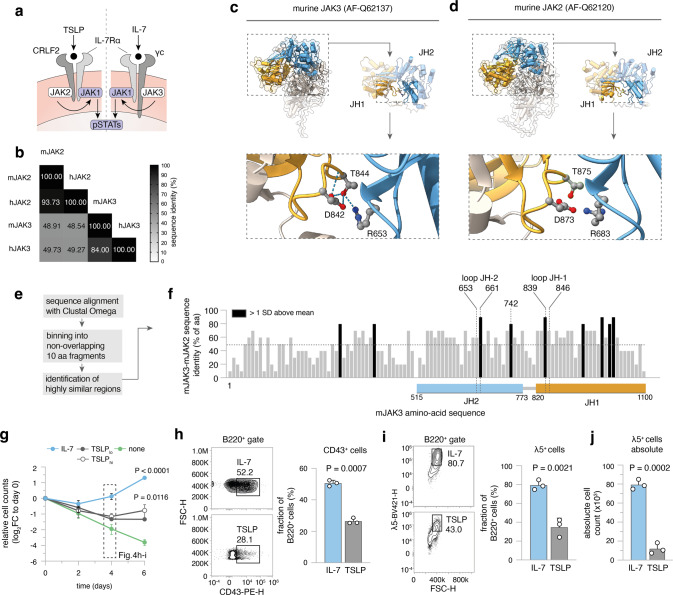
Fig. 4. Ph-like ALL in humans harbors Jak2 mutation corresponding to Jak3 mutations in Irf4−/− BCP-ALL.
a Cartoon depicting IL-7 and TSLP receptor components. b Multiple sequence alignment results (using Clustal Omega) of mouse (m) and human (h) JAK3 and JAK2 amino-acid sequences are presented as a matrix. Numbers and shade indicate sequence identity as percentage of amino acids. c–d Alpha-fold structure predictions of murine c JAK3 and d JAK2 are presented. Colors indicate domains: orange = JH1, blue = JH2. JH1-JH2 interface is magnified and T844/T875, R653/R683, D842/D873 amino acids are highlighted as ball-and-sticks representations. Dotted lines = hydrogen bonds. e Overview of analysis for f: Sequences of mJAK2 and mJAK3 were aligned using Clustal Omega. The sequence of mJAK3 was binned into 10 non-overlapping amino-acid fragments and the sequence identity to mJAK2 plotted along the mJAK3 sequence. Dotted line = mean protein-wide sequence identity, black bars = areas with sequence identity greater than 1 SD above mean. JH1 and JH2 loop regions are mapped onto the sequence, JH1 and JH2 domain regions are indicated by colored rectangles below. g–j Irf4−/− BM cells were cultured for 6 days in the presence of 10 ng/mL rmIL-7, 10, or 100 ng/mL rmTSLP(lo/hi) or no cytokine (none). g log2 of cell counts relative to day 0 for n = 3 independent experiments plotted as means ± SD. One-way ANOVA, Sidak post hoc comparing cytokine effect. h–i On day 4, h CD43+ and i λ5+ cells within B220+ cells were recorded for IL-7 and TSLP treated cultures. Numbers indicate percentages within the depicted gates of B220+ cells. j Absolute counts of λ5+ cells at day 4. Dots in h–j indicate n = 3 independent experiments, presented as bars (mean ± SD). Unpaired two-tailed t test for h–j.
As mentioned above, JAK2 and JAK3 are part of distinct, but similar receptors: JAK3 binds the common γ-chain involved in IL-7 signaling, while JAK2 associates with CRLF2 involved in TSLP signaling. Both signals involve the IL-7Rα chain and the same downstream pathways (STATs, PI3K) [32]. Therefore, the alternative presence of JAK3/JAK2 mutations between mouse and human BCP-ALL might reflect different cytokine preferences. Human proB/preB cells proliferate in response to both TSLP and IL-7 [33]. However, in Irf4−/− BM cells IL-7, but not TSLP induced robust proliferation (Fig. 4g) as well as high frequencies and absolute counts of CD43+ (Fig. 4h) and λ5+ preB cells (Fig. 4i, j).
IRF4 re-expression leads to cell death and differentiation
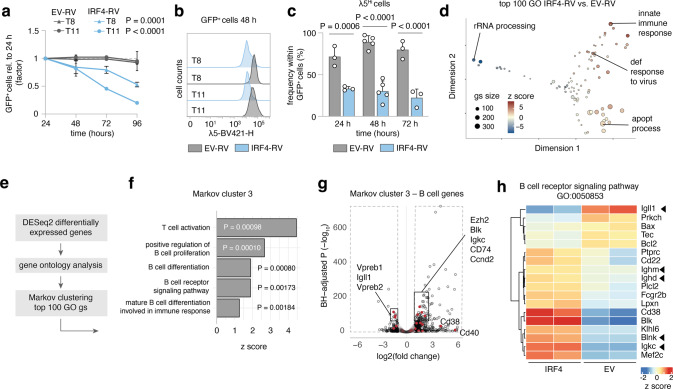
As Irf4 deletion was a prerequisite for leukemia in our model, we examined the effect of forced IRF4 re-expression, using RVs coding for GFP alone (EV-RV) or plus IRF4 (IRF4-RV). When re-introducing IRF4 into T8 or T11, GFP+ IRF4-expressing-, but not GFP+ control cells gradually disappeared over time (Fig. 5a). AnnexinV/PI stainings confirmed apoptosis (not shown). Further, we noted the loss of surface λ5-expression induced by IRF4-RV (Fig. 5b, c). Comparing the transcriptomes of still viable cells 24 h after transduction revealed strong induction of “apoptotic process” and “innate immune response” gene ontology (GO) gene-sets (gs) (Fig. 5d). Markov clustering of GO gs affected by IRF4 re-expression (Fig. 5e, f) further identified several coregulated B-cell differentiation gs (Fig. 5f), with downregulated ψL components Igll1, Vpreb1, and Vpreb2, but upregulated differentiation genes including Igμ, Igκ, and Blnk (Fig. 5g, h, sFig. 5a, b). Similar results were obtained for T11 (sFig. 5c, d). Therefore, fully transformed leukemia remained targetable by IRF4 re-expression.
Fig. 5. IRF4 re-expression results in apoptosis and differentiation of leukemia cells.
a–c T8 and T11 cells were transduced with IRF4-RV or control empty vector (EV)-RV. a GFP+ cell frequency normalized to 24 h after transduction was recorded. One-Way ANOVA, Sidak post hoc for RV effect per tumor. Mean ± SD of n = 3 independent experiments. b Representative histogram of λ5 surface expression of GFP+ cells at 48 h. c Pooled Quantification of λ5Hi cells for three (24, 72 h) to five (48 h) independent experiments for T8 and T11. d–h T8 cells were collected in duplicates at 24 h after EV-RV and IRF4-RV transduction and subjected to bulk RNAseq. d MDS plot of top 100 gene ontology (GO) gene-sets varying between EV and IRF4 transduced T8. Representative gene sets annotated. Size of circles = number of genes, color = z score. e Analysis strategy for GO gene-set clustering using Markov clustering. f Gene-sets from Markov cluster 3 and corresponding P values and z scores. g Volcano plot of B-cell genes from Markov cluster 3 (red) highlighted within all differentially regulated genes (black). h Heat map of B-cell receptor signaling GO gene-set. Immunoglobulin genes and the tumor suppressor Blnk are marked. Color = z score.
Small compound agents affecting Irf4−/− leukemia cells in vitro
Next, we screened a collection of kinase inhibitors for their capacity to kill Irf4−/− leukemia cells in vitro. We included NIBR3049 targeting JAK3, Ruxolitinib, an inhibitor of JAK1/2 (downstream of JAK3), and Dexamethasone, a cornerstone for treating lymphomatous malignancies. Furthermore, we included inhibitors of NFκB (IKK, TAK1), JNK, MEK, ERK, PP2A, GFI1, FAK, and the Bruton tyrosine kinase (BTK) acting downstream of the BCR.
A variety of these substances potently killed tumor cells (sFig. 6a), implying the involvement of multiple pathways in leukemia cell survival. The efficacy of Ruxolitinib and NIBR3049 corroborated our results concerning Jak3 driver mutations. Furthermore, inhibitors of GFI1 and PP2A, as well as NFκB and JNK, were potent. In contrast, inhibiting BTK, MEK and ERK had no impact.
In vivo therapy of established Irf4−/− B-ALL
Next, we implemented JAK inhibition as in vivo treatment for Irf4−/− leukemia. We began induction therapy with Dexamethasone around day 12 after adoptively transferring 3 × 105 T8.1 cells i.p. into wt mice (Fig. 6a), when overt leukemia was noted in peripheral blood (pB) (Fig. 6b “pre”). After 7d of treatment, leukemic cell numbers in pB were robustly reduced (Fig. 6b “post”), although few cells reproducibly remained detectable (Fig. 6c). Maintenance therapy was continued with Ruxolitinib or vehicle control by oral gavage twice daily for the following 12 days (Fig. 6a, c). Importantly, the half-life of Ruxolitinib in mice is only 0.8 h (“Australian Public Assessment Report for Ruxolitinib”, Australian Government), implying that any observed in vivo effectiveness might be underestimated.
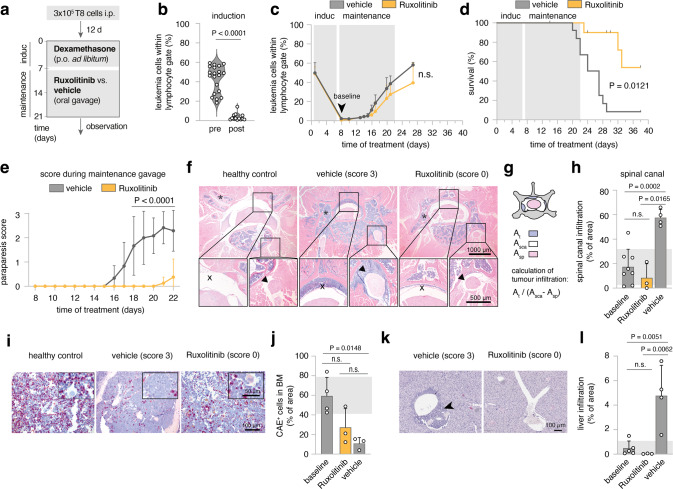
Fig. 6. Ruxolitinib reduces leukemic meningeosis and organ infiltration in vivo.
a Schematic overview of experimental design. Day 0: injection of mice with 2 × 105 T8 cells. After 12 days initiation of Dexamethasone induction therapy supplied in drinking water for seven days. Maintenance therapy comprised either Ruxolitinib-phosphate (11 mice) or vehicle control gavage (13 mice) twice daily for 14 days. Mice were scored daily and blood sampling was performed regularly. b Leukemia cell frequency (B220+sIgμ-) within lymphocyte gate before and after induction with Dexamethasone. Two-tailed unpaired t test c time-course of leukemia cell frequencies in peripheral blood for Ruxolitinib and vehicle-treated mice. d Survival as Kaplan–Meier plot analyzed with Log-rank test. In the Ruxolitinib group, four mice were excluded and censored due to intervention-related adverse reactions or due to their use in the analysis described in f–l. e Disease scores, determined as described in methods. Mean ± SD of the scores per indicated treatment group analyzed by two-way ANOVA, Sidak post hoc. n = 2 replicate experiments for b–e with similar outcome. f Exemplary histopathology (HE) of healthy or leukemia bearing mice (score 3, vehicle-treated or score 0, Ruxolitinib-treated). One representative mouse per condition. Bar size in the bottom right corners. Top panels: an overview of cross-sectioned lumbar vertebrum, bottom inserts from spinal canal (left) and spinal nerve root (right). g Schematic representation of the calculation of tumor infiltration into the spinal canal. (At: area of tumor infiltration, Asca: area of total spinal canal, Asp: area of the spinal cord). h Quantification of spinal canal infiltration according to g for n = 8 after induction (baseline), n = 3 score 0 (Ruxolitinib) and n = 4 score 3 (vehicle) mice. i Representative CAE stainings from vertebral BM for score 0 and score 3 mice. j Quantification of area occupied by CAE+ cells relative to total BM area for n = 4 (baseline), n = 3 (score 0, Ruxolitinib) and n = 4 (score 3, vehicle) mice. k Representative HE stainings from liver tissue for score 0 and score 3 mice, Scale bar bottom right. l Quantification of tumor infiltrated area relative to whole liver area for n = 5 (baseline), n = 3 (score 0, Ruxolitinib) and n = 4 (score 3, vehicle) mice. Each dot represents measurements of three complete liver cross-sections per mouse.
Despite maintenance therapy, leukemic cells in pB reappeared, with no significant difference between treatment groups (Fig. 6c). However, treatment with Ruxolitinib resulted in a clear survival benefit (Fig. 6d) and marked improvement of a prominent neurological symptom: in sham-treated animals, temporary limpness of the tail and hind legs occurred seconds after gavage, which we quantified using a newly established scoring system (ranging from 0 to 3, see Methods).
Mechanistically, ultrasound imaging revealed an echogenic paravertebral mass (sFig. 7a, b) in score 3, but not score 0 mice. By histology, score 3 correlated with severe infiltration of blasts into the spinal canal (X in Fig. 6f), extending into spinal nerve roots (arrowhead in Fig. 6f). Therefore, paraparesis likely represented a manifestation of mouse leukemic meningeosis, exacerbated by gavage-induced increases in intraabdominal pressure.
Paraparesis was reproducibly relieved during Ruxolitinib treatment (Fig. 6e), correlating with the suppression of perimyelon infiltration that ensued in vehicle-treated mice after the end of induction therapy (Fig. 6f, h). In contrast, the severely impaired hematopoiesis in sham-treated mice, indicated by low CAE+ cell frequencies, was not significantly ameliorated by Ruxolitinib (Fig. 6i, j).
These findings raised the possibility that Ruxolitinib preferentially targets infiltration of solid organs rather than BM or pB. Accordingly, Ruxolitinib fully blocked the liver infiltration as observed in sham-treated mice (Fig. 6k, l). As tissue infiltration is regulated by homing receptors, we treated T8.1 and T8.2 cells with Ruxolitinib in vitro and recorded the expression of CD29 (integrin β1), which pairs with various integrin alpha chains involved in cell- and tissue adhesion [34, 35]. Notably, on T8.1 and T8.2, Ruxolitinib reduced CD29 expression dose-dependently (sFig. 6c–e) while it even slightly increased expression of MHC I molecules (H2Db, H2Kb), stained as a specificity control.
Discussion
The herein described spontaneous leukemogenesis in Irf4−/− mouse stresses the particular vulnerability of preB-I cells. Our data provide insights for (a) conditions promoting leukemogenesis, (b) functional consequences of Jak mutations, (c) parallels of mouse and human BCP-ALL and (d) potential in vivo treatment:
(a) We provide evidence for a two-hit leukemogenesis model: The first hit (Irf4 loss) resulted in reduced differentiation, IL-7-dependent hyperproliferation, and impaired retention to the BM niche (Fig. 7). A second hit (targeting Jak3 in our model) created a dominant survival signal, probably founding overt preB-I leukemia.
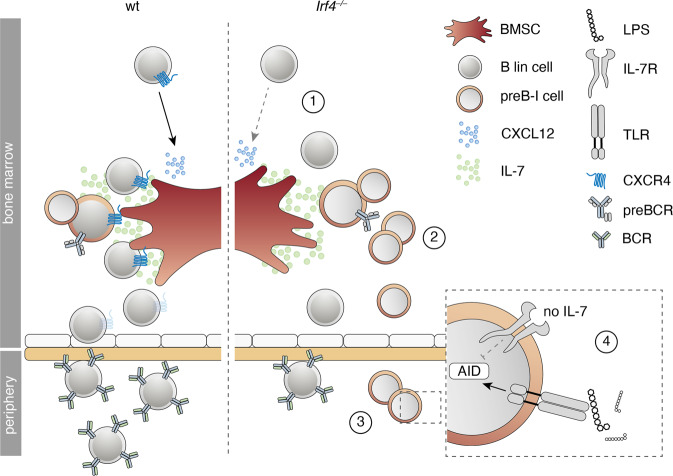
Fig. 7. Summary of the preB-I preleukemic state induced by IRF4 deficiency.
Cartoon summarizing the findings for IRF4 deficient compared to wt B lymphopoiesis. B lineage (lin) cells are less responsive to BMSC-derived CXCL12 due to reduced surface CXCR4 expression (1). Irf4−/− preB-I cells exhibit impaired differentiation and IL-7 dependent hyperproliferation (2). Irf4−/− preB cells escape into the periphery (3), where a combination of IL-7 deprivation and danger-associated molecular patterns (such as LPS) might induce AID expression (4), fueling mutagenesis.
The induction of BCP-ALL in Irf4−/− mice are similar to Ikzf1 and Pax5 mutated mouse models [36–38], implying similarities between these TF-alterations. Probably, one shared mechanism is the differentiative impairment. Importantly, for Irf4−/− fr.A-D cells we even detect slightly higher levels of Pax5 compared to wt fr.A-D cells (sFig. 8), ruling out that the findings in Irf4−/− mice merely mirror those of Pax5 deficiency. The reverse remains conceivable; that Ikzf1 and Pax5 mutations converge in lowering IRF4 expression.
In addition to mice mutated in Pax5 or Ikzf1, Irf4/Irf8−/−, and Irf4/Spi1−/− mice have been shown to develop leukemia early in life at a high incidence [39, 40]. Contrasting these studies, we report that a single deficiency for IRF4 fully suffices for leukemogenesis. We excluded secondary alterations in Irf8, Spi1 in our model: we found unchanged expression and gene sequence of IRF8 (not shown) and normal amounts of Spi1 transcripts (sFig. 8a) in Irf4−/− fr.A-D cells. The single IRF4 deficiency models potential clonal initiating events better than Irf4/Irf8−/− or Irf4/Spi1−/− mice, because Irf4−/− mice harbor productive B-cell development.
We newly describe that a preleukemic alteration can lead to reduced BM retention, presenting a tentative explanation for the induction of mutagenic signals, as deprivation from IL-7 and exposure to bacterial compounds can cooperatively induce the mutagenic agent AID [25].
(b) Why do Jak3 mutations only lead to enhanced sensitivity to, but not complete independence of IL-7? Analysis of JAK3 and JAK2 structure implied that mutations of R683/R653 and T875/T844 might decrease JH1-JH2 interaction strength. This would imply reduced auto-inhibition as the GOF mechanism—in line with findings for the JAK family member TYK2 [41]. This alone cannot explain cytokine independency, owing to the receptor biology: The two preassembled receptor chains keep JAKs intracellularly separated [42]. Ligand binding is needed for a conformational change that brings JAKs into the proximity needed for cross-phosphorylation.
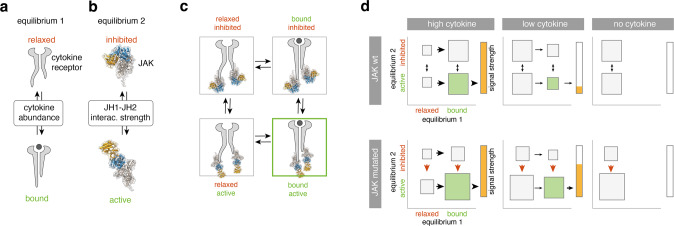
To explain our observations, we propose an oncogene model with two equilibria (Fig. 8a, b): the first is determined by cytokine concentration and dictates the probability of receptor conformation change (Fig. 8a). The second, independent equilibrium (Fig. 8b), is determined by the interaction strength at the JH1-JH2 interface and dictates the probability of JH1 and JH2 dissociation. Only the combination of the “bound” and “active” state (Fig. 8c) would result in the elicitation of a signal (Fig. 8c, green frame).
Fig. 8. A “two-equilibrium model” explains JAK mutant effects in primary preB-I cells.
a Equilibrium 1 is determined by cytokine abundance and dictates the cytokine receptor state (bound vs. relaxed). b Equilibrium 2 is determined by the interaction strength between JH1 and JH2 domains in JAKs and dictates the JAK state (inhibited vs. active). c Equilibria 1 and 2 interact to create four possible states. Green lettering indicates signaling favoring state. Green frame indicates the actively signaling state. d For high (left), low (middle), and no (right) cytokine in the presence or absence of JAK3 mutations, hypothetical probabilities of equilibria states as in c are presented. Size of rectangles signifies likelihood of state relative to others. Arrows indicate shifts of equilibria, red arrows indicate the effect of JAK mutations. Green rectangle = active signaling state (bound and active). Bars to the right of each panel indicate signal strength as a direct result of the two equilibria adjacent to it.
In this model, JAK mutations would only affect the second equilibrium (Fig. 8d, red arrows). Sporadic ligand binding would still be needed for elicitation of signaling. The model stringently predicts the better exploitation of low cytokine concentrations for JAKmut that we observed in vitro. Figure 8d depicts theoretical probabilities of receptor states in the presence (top row) or absence (bottom row) of JAK mutations.
Our findings that JAK3mut confer heightened cytokine sensitivity, but not -independence, is in contrast to what has been found for JAK2-R683G mutants expressed in the commonly used BaF3 cell line [43]. However, BaF3 cells depend on IL-3 and not IL-7Rα cytokines (i.e., IL-7 or TSLP). Therefore, it remains conceivable, that the IL-3 receptor provides a different physiology, which may deviate from the IL-7R physiology in primary B progenitors.
(c) The finding that Irf4−/− preB-I cells respond preferentially to IL-7 over TSLP presents a possible explanation, why mouse models of BCP-ALL acquire Jak3 mutations, and human Ph-like ALL typically harbors Jak2 mutations. Our comparison of JAK structure predictions yielded corresponding mutations likely to elicit similar downstream effects.
(d) Lastly, our in vivo experiments reinforce Ruxolitinib as a potential treatment for JAK-driven BCP-ALL. The compound represents an important therapeutic agent in myeloproliferative disease and is already studied for the treatment of Ph-like-ALL [44, 45]. We describe a preferential effect of Ruxolitinib on CNS- and organ infiltration, potentially due to reductions in integrin expression on leukemia cells. These effects are of potential translational importance because current CNS-targeted therapies for ALL remain toxic.
Methods
Mice
C57Bl/6 mice were purchased from Charles River, Sulzfeld, Germany. Irf4−/− mice [7] and Il-7eGFP mice [46] (provided by Koji Tokoyoda, DRFZ Berlin) were bred on the C57Bl/6 background and housed in the animal facility of the Biomedical Research Center at the University of Marburg, Germany. If not stated otherwise, all mice used in the presented experiments were 8–12 weeks old and sex-matched.
Tumor cell lines and cell culture
Stable tumor cell lines T8.1, T8.2, and T11 were established from primary Irf4–/– leukemia cells (derived from primary tumor 8, i.e., T8, or tumor 11 (T11)) by culturing them on a monolayer of irradiated (30 Gy) ST2 stromal cells [47] grown to confluency in Opti-MEM medium (31985070, ThermoFisher Scientific) supplied with 1% cell culture supernatant from JIL-7.6 J558 cells [48] (a gift from Fritz Melchers, Berlin) as a source of IL-7. After several passages, T8 and T11 cells grew independently of ST2 cells. For in vitro inhibitor experiments, 2.5 × 105 T8.1 or T8.2 cells (or T11 cells) were cultured in 500 µL RPMI medium in 48 well plates in the presence of the indicated concentrations of inhibitors. To determine the percentage of viable cells, samples were stained using Annexin V and propidium iodide (PI) (see below) after 48 h. Substances used include Defactinib (S7654, Selleckchem), Oxocaenol (O9890, Sigma), GANT61 (Sigma, G9048), SP203580 (EI-286-0001, Enzo), SP600125 (EI-305-0010, Enzo), PD98059, Promega), Ibrutinib (S2680, Selleckchem), BAY11-7082 (ALX-270-219, Alexis), Dexamethasone (PZN 08704491, mibe GmbH) and Ocadaic acid (O4511, Sigma).
Murine pro/preB cell cultures
Femur and tibia bones from 8 to 12 weeks old mice were explanted and cleaned from adherent tissues. Cells were extracted via centrifugation at 11 × 103 RPM for 10 s. Total BM cells were enriched for B220+ (sIgµ−) B lineage cells using an in-house magnetic-activated cell sorting protocol. Briefly, whole bone marrow cells were stained with a mix of FITC-conjugated antibodies to (Igµ), CD11b, B220, Ter119, CD49b, CD4, and CD8 (all from eBioscience), followed by incubation with an anti FITC/streptavidin/biotin/magnetic bead complex (Miltenyi Biotec) and magnetic sorting using a microcentrifugation tube stand (Miltenyi Biotec) [49]. Sorting efficiency, as confirmed by flow cytometry, routinely exceeded 90%. Cells were seeded at a density of 1 × 105 cells per well in 200 µL RPMI complete (96-well plates, Greiner). Pro/preB cell cultures were propagated with 10 ng/mL rmIL-7 (217-17, Peprotech) in RPMI-1640 medium complete (R8758, Sigma-Aldrich, supplemented with: 10% FCS (Sigma-Aldrich), 2 mM l-glutamine (Biochrom), 50 µM β-mercaptoethanole (Sigma-Aldrich), 0.03/0.05 g per 500 mL Penicillin G/Streptomycin Sulfate, 1% non-essential amino acids (PAA Laboratories)). In some experiments, pro/preB cells (1.25 × 106/mL medium) were treated for 24 h with LPS (Sigma, 1 µg/ml), anti-IL-7 (BioXCell, 10 µg/ml), rmIL-7, or respective combinations, before generating mRNA for qRT-PCR.
Transwell migration assay and OP-9 adhesion assay
For the transwell migration assays, Hardy fr.A-D cells were magnetically sorted from BM of wt and Irf4−/− mice as described above (with addition of FITC-conjugated anti-Igµ antibody), and 2 × 105 cells in RPMI (without additives, FCS-free) containing 10 ng/mL rmIL-7 seeded in 50 µL in the top chamber of 96-well 5 µm pore uncoated 96-well transwell plates (HTS transwell® Corning). The bottom chamber was flooded with 200 µL RPMI containing indicated concentrations of rmCXCL12 (Peprotech). After 16 h, inserts were removed, cells in the bottom chamber were collected, counted, and analyzed for B220 surface expression using flow cytometry. The fraction of migrated cells was calculated as n(migrated) × freqB220(migrated)/n(input) × freqB220(input). Normalization to B220+ cells reduced interexperimental differences due to differences in cell purity after magnetic selection. For OP-9 adhesion assays, 5 × 103 OP-9 cells (a gift from Hyun-Dong Chang, DRZF Berlin) were seeded in 96-well microtiter plates 24 h before the assay. On the day of the assay, fr.A-D cells were purified as above and 2 × 105 fr.A-D cells were seeded on top of OP-9 monolayers in RPMI complete + 10 ng/mL rmIL-7. Plates were centrifuged briefly to accelerate cell descension. After 1 h, suspended cells were collected in the supernatant and by washing OP-9 monolayers two times with PBS.
Flow cytometry and cell sorting
For surface staining of B lineage markers, cells were harvested, resuspended in PBS/1% FCS and stained with anti-B220 (RA3-6B2, Biolegend), anti-Igµ (II/41, BD Bioscience), anti-CD43 (RM2-5, Biolegend), anti-CD24 (M1/69, invitrogen), anti-BP-1 (BP-1, BD Bioscience), anti-CD2 (RM2-5, Biolegend), anti-CXCR4 (L276F12, Biolegend), anti-CD127 (=IL-7Rα) (A7R34, BD Bioscience), anti-CD179b (=λ5) (LM34, BD Bioscience) as indicated (20 min at room temperature in the dark). All antibodies were employed at a dilution of 1:500. Fluorescence was recorded using either a FACS Aria III (BD) or an Attune NxT (Thermo-Fisher) analyzer. Data analysis was performed using the FlowJo V10 software (BD). For dimensional reduction, we used the t-Distributed Stochastic Neighbor Embedding (tSNE) [50] algorithm built into FlowJo V10. Epitopes on BM cells from Irf4−/− and wt control mice used for dimensional reduction analysis comprised B220, sIgµ, CD43, CD24, BP-1. For RNA and WES analyses, BM cells were surface labeled for B220 and sIgµ expression, and B220+sIgµ− cells were sorted using a FACS Aria III (BD Bioscience). Sorting efficiency was routinely above 95%. To determine cell viability, AnnexinV/PI staining was performed using 5 µL AnnexinV (640905, Biolegend) per 500 µL HBSS. After 20 min of incubation at room temperature in the dark, 1 µL PI (421301, Biolegend) was added, and cells were immediately measured.
CNVs analysis
CNVs were analyzed in tumor samples 8, 10, and 14 and compared to Irf4−/− normal tail tissue. Whole DNA was extracted from 5 × 106 cells per sample using the Macherey-Nagel NucleoSpin Tissue kit (REF 740952.50) according to the manufacturer’s protocol. Library preparation was performed using the Illumina Nextera DNA kit according to the manufacturer’s instructions. Sequencing was performed on an Illumina-HiSeq-1500 platform in rapid-run mode at the Genomics Core Facility of Philipps-University Marburg. Fastq quality control was performed using custom scripts. Raw sequenced reads were aligned to the Ensembl Mus musculus reference (revision 79) using Bowtie2 (version 2.0.0) [51] with standard parameterization. Analysis of CNVs was performed using the cn.mops (Copy Number estimation by a Mixture Of PoissonS) package (version 1.18.1) [52] with the following parametrization: prior impact = 1, lower threshold −0.9, upper threshold = 0.5 minimum width = 4. Window length was set to 10000 and the algorithm was run in unpaired mode.
BM cryosections and analysis of B progenitor vicinity to IL-7+ BMSCs
Mouse femora from Irf4−/− or wt il-7eGFP reporter mice were explanted, cleaned from soft tissues, and fixated overnight in 4% PFA PBS (Alfa Aesar). Samples were then dehydrated by incubation in 30% sucrose in PBS for 24 h. Dried and dehydrated femora were snap-frozen in cryomolds® (Tissue-Tek) using O.C.T freezing medium (Tissue-Tek) by being placed in a beaker of Hexan, surrounded by a beaker of Acetone and dry ice. Samples were stored at −20 °C until processing. Cryosections of 7 µm were generated with a Leica cryostat (DB80 LX microtome blades, Leica) using Kawamoto tape [53] (Section-lab) as described before [54]. Cryosections were stained with antibodies against B220 (RA3-6B2, Biolegend), CD2 (14-0021-85, eBioscience, conjugated to AF555 using lightning-Link kit, Abcam), GFP (Rockland goat polyclonal anti-GFP, 600-101-215) with secondary rabbit anti-goat F(ab’)2 AF488 (thermo-scientific A21222). Samples were then mounted in DAPI ProLong Gold Antifade (ThermoFisher Scientific). Images were recorded using a Leica confocal (SP8i) microscope. Image analysis was performed in IMARIS (version 9.7.2).
Histological analyses
Tissue samples were immediately fixed in 4% PFA PBS solution. Histological analysis was performed on 3 µm thick sections from paraffin-embedded tissue as described previously [55]. Briefly, rehydrated paraffin sections were first blocked with 0.3% H2O2 and goat normal serum. For immunohistochemical (IHC) stainings, rat antibodies against CD45R/B220 (clone RA3-6B2, BD) and KI67 (clone TEC-3, Dako) were then incubated on the tissue slices and the bound antibody was detected with biotinylated goat anti-rat IgG (Southern Biotechnology). Bound antibody was visualized with the Vectastain-kit (Vector Laboratories) according to the manufacturer’s protocol. Hematoxylin-Eosin (HE) stainings were performed according to standard procedures. Cells of the granulocytic lineage were stained on paraffin-embedded tissues with the Naphthol AS-D Chloracetate (Specific Esterase, CAE) Kit (Ref: 91C-1KT, Sigma-Aldrich) according to the manufacturer's protocol.
In the in vivo therapeutic experiments, we calculated the narrowing of the spinal cord using the equation At/(Asca–Asp), where Asca is the area of the spinal canal, At that of the tumor, and Asp that of the spinal cord area. Two different cross-sections per animal were examined. The infiltration of the liver was calculated by dividing the tumor area in the liver by the whole area of the liver section. Three whole liver sections were analyzed per animal. All measurements were performed using Fijii [56].
Whole-exome sequencing and biostatistical analysis
To determine SNV within leukemia samples, genomic (g)DNA was extracted both from primary Irf4−/− tumors as well as FACS-sorted control B220+sIgµ− BM fr.A-D cells using the High Pure PCR Template Preparation kit from Roche (11796828001). The integrity of the resultant gDNA was confirmed in a 2% Agarose gel. Macrogen in Seoul performed SureSelect All Exon V6 library preparation and sequenced exons on a NovaSeq platform producing 2 × 150 bp reads at a coverage of 100× (50× on-target coverage). Fastq quality control was performed using FASTQC (version 0.11.9). Raw sequenced reads were aligned to the Ensembl Mus musculus reference (revision 96) using STAR (version 2.6.1d) using default parametrization. Soft-clipped aligned reads were then subjected to variant calling analysis. Position-wise pile-up files were generated using samtools (version 1.9) with the mpileup option and a pile-up quality threshold of 15, both for single sample and matched variant calling. Subsequently, variant calling was performed for SNP and InDel detection using VarScan2 (version 2.3.9) on single samples with the following parametrization: sampling depth = 100,000, minimum variant frequency = 0.05, minimum coverage = 8, minimum variant reads = 2, minimum average read quality = 15 and a p value threshold was set to 0.05. Only primary alignments were considered, the strand filter was enabled, and duplicates were removed. As a comparison, matched tumor-normal variant calling was performed with VarScan as well using an identical parameter setting with the somatic p value threshold set to 0.05.
For Fig. 3n raw sequenced reads were aligned to the Ensembl Mus musculus reference (revision 96) using Burrows-Wheeler Aligner (BWA version 0.7.17) using default parametrization [57]. Prior to variant calling, aligned reads were filtered using a custom filter that excludes reads with more than three mismatches, more than two indels, or a mapping quality below 20 using pysam (version 0.16.0.1). Duplicates were marked and removed using Picard (GATK version 4.1.6.0) [58]. Filtered aligned reads were then subjected to variant calling analysis. Position-wise pile-up files were generated using samtools (version 1.9) with the mpileup option and a minimal base quality threshold of 20. Subsequently, variant calling performed for SNP detection using VarScan2 (version 2.4.4) using matched tumor-normal (somatic) mode with the following parametrization: sampling depth = 100,000, minimum variant frequency = 0.2, minimum coverage = 8, minimum variant supporting reads = 5, minimum average read quality = 20 and a somatic p value threshold was set to 0.05. Only primary alignments were considered, and the strand filter was enabled. SNP calls were filtered to high confidence somatic mutations using VarScan’s somaticFilter method, SNPs with a variant allele frequency above 0 in the matched reference sample were excluded.
Sanger sequencing and polymerase chain reaction
SNVs in the JAK3 gene were confirmed by Sanger sequencing of PCR fragments spanning the Jak3 pseudokinase and kinase region (primers used for PCR amplification and Sanger Sequencing: mJAK3 for, mJAK3 rev s. Supplemental Data). Sequencing services were provided by Microsynth Seqlab. To determine the clonality of tumor cells, the VµH region was amplified by PCR. Amplicons were run on an agarose gel and extracted using the QIAquick Gel Extraction Kit (Qiagen). DNA fragments were then cloned into the vector pJet1.2 (Thermo Scientific) and transformed into DH10B E. coli. The indicated numbers of clones (Fig. 1g) for each PCR amplicon were sequenced and aligned with software from IMGT/V-quest [59].
Retroviral transduction of Jak3-mutants and IL-7 independency assay
The coding sequence of murine Jak3 was amplified from pCineo-Jak3 (a gift from Olli Silvennoinen from Tampere-university in Finland) and cloned into the pMSCV-Thy1.1 expression plasmid using BamHI and SalI restriction digestion. Site-directed mutagenesis was performed following the manufacturer’s protocol using the Quick-Change II site-directed mutagenesis kit (Agilent Technologies; primers employed are listed in the Supplemental Materials). Viral supernatant from mutated pMSCV-Thy1.1-Jak3 constructs was produced as described previously [49]. For viral transduction, 5 × 105 IL-7 dependent primary Irf4−/− preB-I cell cultures were resuspended in 400 µL RPMI medium (D5030, Sigma-Aldrich) with 600 µL viral supernatant and 1.5 µL polybrene and spun in culture plates at 2700 rpm for 90 min at 37 °C. Cells were then replenished with a conditioned medium and rested for 24 h. Transduction efficiency was measured by flow cytometry using surface staining for Thy1.1 (OX-70, Biolegend). For the IL-7 independency assay (Fig. 3b), transduced cells were split and cultured with either recombinant murine (rm)IL-7 or 10 µg/ml neutralizing anti-IL-7 antibody (BE0048, Bio X Cell).
RNA-sequencing and biostatistical analysis
RNA extraction from primary tumor samples and FACS-sorted B220+ sIgµ− pro/preB cells was performed using Trizol extraction. Quality control was performed using the Bioanalyzer RNA 6000 NanoChip (Agilent Technologies). Library preparation was performed at the Institute for Immunology, University Medical Center of the Johannes Gutenberg-University Mainz using the NEBNext Ultra Library Prep kit (New England Biolabs). For deep sequencing, the Illumina-HiSeq- 4000 platform was used (Beijing Genomic Institute). Quality control on the sequencing data were performed with the FastQC tool (version 0.11.2, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA-sequencing reads were aligned to the ENSEMBL Mus_musculus.GRCm38 reference genome. The corresponding annotation (ENSEMBL v76) was also retrieved from ENSEMBL FTP website. The STAR aligner (version 2.4.0j) was used to perform mapping to the reference genome. Alignments were processed with the featureCounts function [60] of the Rsubread package, using the annotation file also used for supporting the alignment. Exploratory Data Analysis was performed with the pcaExplorer package [61]. Differential expression analysis was performed with DESeq2 package [62], setting the false discovery rate (FDR) cutoff to 0.1. DESeq2 datasets were analyzed using the GeneTonic [63] and pcaExplorer packages. To assess the possible occurrence of gene fusions, we applied two different methods, Star-Fusion (version 1.10.1) and Arriba (version 2.1.0). For STAR-Fusion, required meta reference files were created from the Ensembl Mus musculus reverence (revision 100) as recommended in the STAR-Fusion manual. In case of Arriba, we used the mm10 + GENCODEM25 assembly. In each case, we used the dockerized versions of the tools. Raw fastq files were used as an input for both tools. Subsequently, raw reads were mapped using the recommended alternative STAR settings recommended in the tools manual to leverage chimeric reads from the alignments. Default filters as recommended by the STAR-Fusion and Arriba manuals were applied to limit the false-positive rate. For the same reason, known blacklisted regions as provided by the Arriba release were excluded from the analysis.
BCP-ALL subtype predictions using random forest classifier
Human genes (GRCh38.p13, v104) with annotated orthologous genes in mice were extracted from ensembl database using the BiomaRt online tool. Gene counts from RNA-sequencing of a previously published human BCP-ALL cohort [28] and of murine tumor samples were subsetted to include only human-mouse orthologous genes. The resulting gene counts were normalized by variant stabilization transformation using the R package DESeq2 version 1.32.0. Allocation of the murine tumor samples to human BCP-ALL molecular subtypes was performed based on gene expression using a random forest machine learning algorithm (R package caret version 6.0-88) trained on the human cohort. Predictions were plotted using R package pheatmap version 1.0.12. Differential gene expression was analyzed in R package DESeq2 and resulting gene lists ranked by log2-fold-change were analyzed in GSEA version 4.1.0.
JAK structure and sequence analysis
Mouse JAK2 (AF-Q62120) and JAK3 (AF-Q62137) structure predictions were acquired from the AlphaFold protein structure database [64] and visualized in UCSF ChimeraX (version 1.2.5) [65]. Multiple sequence alignments were performed using the EMBL-EBI Clustal Omega tool.
Quantitative real-time (qRT-)PCR
Total RNA was extracted both from primary Irf4−/− tumors as well as FACS-sorted control B220+sIgµ− BM fr.A-D cells of either Irf4−/− or wt animals using the Gdansk extractme kit (EM09.1) according to the manufacturer’s protocol. cDNA was prepared from whole RNA samples using the RevertAid cDNA kit from Thermo Fisher (K1621). qRT-PCR for Aicda, Spi1, and Pax5 was performed using the SybrGreen MasterMix reagent (4385612, AppliedBiosystems) in a StepOnePlus cycler (AppliedBiosystems). Data presented as percentage of HPRT using the formula x = 1/2(cyclesAicda − cyclesHPRT) × 100.
In vivo therapeutic studies and ultrasound imaging
Mice were injected with 3 × 105 T8.1 cells intraperitoneally and monitored daily for clinical symptoms. When mice began showing signs of general morbidity, leukemia was confirmed by FACS analysis of tail vein blood for B220+ sIgµ− blast cells. When blast cells in pB reached 25 (mean 50)%, therapy was initiated with oral Dexamethasone (Jenapharm) at 6 mg/L supplied ad libitum in the drinking water for seven days. Maintenance therapy comprised either Ruxolitinib-phosphate (S5243, Sellekchem) 1 mg (in 2% DMSO, 30% PEG300 in H2O, as proposed by the manufacturer), Defactinib (S7654, Sellekchem) 1.2 mg (in 5% DMSO, 50% PEG300, 5% Tween 80 in H2O, as proposed by the manufacturer) or vehicle control (5% DMSO, 50% PEG300, 5% Tween 80 in H2O) administered twice daily via oral gavage. During the course of the disease, this treatment led to paraparesis of the hind legs and tail. A clinical scoring system was established according to the extent of paraparesis and mice were scored daily accordingly: Scores 0–3: (0) no paraparesis, (1) paraparesis induced by treatment intervention, resolves within 30 s, (2) paraparesis induced by treatment intervention, does not resolve within 30 s, (3) persistent paraparesis, independent of treatment intervention. Score 3 prompted sacrification of affected mice. High-resolution ultrasound imaging was performed using a Visual Sonics Vevo 2100 System (FUJIFILM VisualSonics, Toronto, Canada) with microscan transducer MS-550-D, 22–55 MHz (FUJIFILM VisualSonics, Toronto, Canada) as described previously [66].
Statistical analysis
Statistical analysis was performed using the GraphPad 9.0 software. Data are commonly presented as mean ± SD. Prior to significance testing, normal distribution and homogeneity of variances were confirmed by Shapiro–Wilk test and Brown–Forsythe testing. Statistical significance when comparing two normally distributed groups was evaluated using two-tailed unpaired t tests. In case of significant differences in variances between groups, Welch’s correction was applied to account for non-norminal distribution of data. When comparing multiple groups, a one-way or two-way analysis of variance was performed, depending on the number of variables that differed between compared groups. This was followed by a Tukey’s Sidak, or Dunnett’s post hoc test, as indicated in figure legends. An alpha level of P < 0.05 was employed for significance testing. In the in vivo experiment, all animals were included in the analyses.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors want to thank Koji Tokoyoda (DRFZ, Berlin) for supplying Il-7eGFP mice for breeding, Olli Silvennoinen (Tampere-university, Finland) for supplying us with the JAK3 construct, and Fritz Melchers (DRFZ Berlin) for the JIL-7.6 J558 and ST2 cells. Further, Hyun-Dong Chang and Anja Hauser (both DRFZ, Berlin) for supplying OP-9 cells and Kawamoto materials, respectively.
Author contributions
DDG, CP, NS, MB, FH, ML designed experiments; DDG, CP, NS, MB, DS, LM, BC, FH, LH performed experiments; DDG, CP, MB, HR, ER, PD, MW, MM, AN, UMB, FM, FH, MB, HMJ, AN, AB, MK, TB, TS, AP, LB, AH, CB, ML analyzed data; DDG and ML prepared the manuscript.
Funding
D.D.G. received personal funding through the German Cancer Aid, Mildred-Scheel doctoral scholarship (70112922). M.L. was funded by the Deutsche Forschungsgemeinschaft (DFG) (LO 396/8-1) and the Else Kröner-Fresenius-Stiftung. Open Access funding enabled and organized by Projekt DEAL.
Data availability
The RNAseq datasets generated during the current study have been deposited in the Gene Expression Omnibus (GEO) archive and are available under the accession number GSE192424. The WES datasets can be accessed under PRJNA706650 in the sequencing read archive (SRA).
Competing interests
The authors declare no competing interests.
Study approval
All animal experiments were approved by the local government (Regierungspräsidium Gießen, G49/2018, G34/2021) and conducted according to the German animal protection law.
Footnotes
Edited by T Mak
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01005-z.
References
- 1.Johnson K, Hashimshony T, Sawai CM, Pongubala JMR, Skok JA, Aifantis I, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:335–45. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 2.Geier JK, Schlissel MS. Pre-BCR signals and the control of Ig gene rearrangements. Semin Immunol. 2006;18:31–9. doi: 10.1016/j.smim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 4.Fistonich C, Zehentmeier S, Bednarski JJ, Miao R, Schjerven H, Sleckman BP, et al. Cell circuits between B cell progenitors and IL-7+ mesenchymal progenitor cells control B cell development cell circuits control B cell development. J Exp Med. 2018;215:2586–99. doi: 10.1084/jem.20180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber M, Lohoff M. IRF4 at the crossroads of effector T-cell fate decision. Eur J Immunol. 2014;44:1886–95.. doi: 10.1002/eji.201344279. [DOI] [PubMed] [Google Scholar]
- 6.Lohoff M, Mak TW. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–35. doi: 10.1038/nri1552. [DOI] [PubMed] [Google Scholar]
- 7.Mittrücker H-W, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–3. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 8.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Gene Dev. 2003;17:1703–8. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz AJ, Chia VM, Schoonen WM, Kelsh MA. Acute lymphoblastic leukemia: an assessment of international incidence, survival, and disease burden. Cancer Cause Control. 2015;26:1627–42.. doi: 10.1007/s10552-015-0657-6. [DOI] [PubMed] [Google Scholar]
- 10.Tasian SK, Hurtz C, Wertheim GB, Bailey NG, Lim MS, Harvey RC, et al. High incidence of Philadelphia chromosome-like acute lymphoblastic leukemia in older adults with B-ALL. Leukemia. 2016;31:981–4. doi: 10.1038/leu.2016.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts KG. Genetics and prognosis of ALL in children vs adults. Hematol Am Soc Hematol Educ Program. 2018;2018:137–45.. doi: 10.1182/asheducation-2018.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:314–24.. doi: 10.1053/j.seminhematol.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-F, Wang B-Y, Zhang W-N, Huang J-Y, Li B-S, Zhang M, et al. Genomic profiling of adult and pediatric b-cell acute lymphoblastic leukemia. Ebiomedicine. 2016;8:173–83.. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–25. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen J, Arceci RJ, Jones W, Burakoff SJ. Expression and ontogeny of murine CD2. Eur J Immunol. 1989;19:1297–302. doi: 10.1002/eji.1830190722. [DOI] [PubMed] [Google Scholar]
- 16.Zehentmeier S, Pereira JP. Cell circuits and niches controlling B cell development. Immunol Rev. 2019;289:142–57.. doi: 10.1111/imr.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokoyoda K, Egawa T, Sugiyama T, Choi B-I, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–18. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Ma Q, Jones D, Springer TA. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999;10:463–71. doi: 10.1016/S1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- 19.Batista CR, Lim M, Laramée A-S, Abu-Sardanah F, Xu LS, Hossain R, et al. Driver mutations in Janus kinases in a mouse model of B-cell leukemia induced by deletion of PU.1 and Spi-B. Blood Adv. 2018;2:2798–810.. doi: 10.1182/bloodadvances.2018019950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang Y-L, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl J Med. 2014;371:1005–15. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 23.Noia JD, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–8. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 24.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, et al. MSH2–MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–45. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan S, Klemm L, Park E, Papaemmanuil E, Ford A, Kweon S-M, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–74. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malard F, Mohty M. Acute lymphoblastic leukaemia. Lancet. 2020;395:1146–62.. doi: 10.1016/S0140-6736(19)33018-1. [DOI] [PubMed] [Google Scholar]
- 27.Mullighan CG. How advanced are we in targeting novel subtypes of ALL? Best Pract Res Clin Haematol. 2019;32:101095. doi: 10.1016/j.beha.2019.101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bastian L, Schroeder MP, Eckert C, Schlee C, Tanchez JO, Kämpf S, et al. PAX5 biallelic genomic alterations define a novel subgroup of B-cell precursor acute lymphoblastic leukemia. Leukemia. 2019;33:1895–909.. doi: 10.1038/s41375-019-0430-z. [DOI] [PubMed] [Google Scholar]
- 29.Jain N, Roberts KG, Jabbour E, Patel K, Eterovic AK, Chen K, et al. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017;129:572–81.. doi: 10.1182/blood-2016-07-726588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold T, Schneider S, Metzeler K, Neumann M, Hartmann L, Roberts KG, et al. Philadelphia chromosome-like acute lymphoblastic leukemia in adults have frequent IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2016;102:130–8. [DOI] [PMC free article] [PubMed]
- 32.Corfe SA, Paige CJ. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin Immunol. 2012;24:198–208. doi: 10.1016/j.smim.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Milford TM, Su RJ, Francis OL, Baez I, Martinez SR, Coats JS, et al. TSLP or IL-7 provide an IL-7Rα signal that is critical for human B lymphopoiesis. Eur J Immunol. 2016;46:2155–61.. doi: 10.1002/eji.201646307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Härzschel A, Zucchetto A, Gattei V, Hartmann TN. VLA-4 expression and activation in B cell malignancies: functional and clinical aspects. Int J Mol Sci. 2020;21:2206. doi: 10.3390/ijms21062206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Springer TA. Adhesion receptors of the immune system. Nature. 1990;346:425–34.. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 36.Tijchon E, Havinga J, Leeuwen FNvan, Scheijen B. B-lineage transcription factors and cooperating gene lesions required for leukemia development. Leukemia. 2013;27:541–52. doi: 10.1038/leu.2012.293. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Lorenzo A, Hauer J, Vicente-Duenas C, Auer F, Gonzalez-Herrero I, Garcia-Ramirez I, et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discov. 2015;5:1328–43.. doi: 10.1158/2159-8290.CD-15-0892. [DOI] [PubMed] [Google Scholar]
- 38.Schjerven H, Ayongaba EF, Aghajanirefah A, McLaughlin J, Cheng D, Geng H, et al. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre–B ALLIkaros tumor suppressor function in Ph+ pre–B ALL. J Exp Med. 2017;214:793–814. doi: 10.1084/jem.20160049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang SHM, Minnich M, Gangatirkar P, Zheng Z, Ebert A, Song G, et al. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia. 2016;30:1375–87.. doi: 10.1038/leu.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S-H, Schatz JH, Acquaviva J, Singh H, Ren R. Cooperation between deficiencies of IRF-4 and IRF-8 promotes both myeloid and lymphoid tumorigenesis. Blood. 2010;116:2759–67. doi: 10.1182/blood-2009-07-234559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupardus PJ, Ultsch M, Wallweber H, Kohli PB, Johnson AR, Eigenbrot C. Structure of the pseudokinase–kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci. 2014;111:8025–30.. doi: 10.1073/pnas.1401180111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McElroy CA, Holland PJ, Zhao P, Lim J-M, Wells L, Eisenstein E, et al. Structural reorganization of the interleukin-7 signaling complex. Proc Natl Acad Sci. 2012;109:2503–8. doi: 10.1073/pnas.1116582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–17. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 44.Tasian SK, Assad A, Hunter DS, Du Y, Loh ML. A Phase 2 study of ruxolitinib with chemotherapy in children with philadelphia chromosome-like acute lymphoblastic leukemia (INCB18424-269/AALL1521): dose-finding results from the part 1 safety phase. Blood. 2018;132:555–555. doi: 10.1182/blood-2018-99-110221. [DOI] [Google Scholar]
- 45.Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hara T, Shitara S, Imai K, Miyachi H, Kitano S, Yao H, et al. Identification of IL-7–producing cells in primary and secondary lymphoid organs using IL-7–GFP knock-in mice. J Immunol. 2012;189:1577–84.. doi: 10.4049/jimmunol.1200586. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa M, Nishikawa S, Ikuta K, Yamamura F, Naito M, Takahashi K, et al. B cell ontogeny in murine embryo studied by a culture system with the monolayer of a stromal cell clone, ST2: B cell progenitor develops first in the embryonal body rather than in the yolk sac. Embo J. 1988;7:1337–43. doi: 10.1002/j.1460-2075.1988.tb02949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceredig R, Boekel Eten, Rolink A, Melchers F, Andersson J. Fetal liver organ cultures allow the proliferative expansion of pre-B receptor-expressing pre-B-II cells and the differentiation of immature and mature B cells in vitro. Int Immunol. 1998;10:49–59. doi: 10.1093/intimm/10.1.49. [DOI] [PubMed] [Google Scholar]
- 49.Kang CH, Hartmann E, Menke L, Staudenraus D, Abass E-F, Raifer H, et al. A hyperactive mutant of interferon-regulatory factor 4. Eur J Immunol. 2018;49:812–5. doi: 10.1002/eji.201847530. [DOI] [PubMed] [Google Scholar]
- 50.Maaten LVD, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–625. [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klambauer G, Schwarzbauer K, Mayr A, Clevert D-A, Mitterecker A, Bodenhofer U, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 2003;66:123–43.. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- 54.Zehentmeier S, Roth K, Cseresnyes Z, Sercan Ö, Horn K, Niesner RA, et al. Static and dynamic components synergize to form a stable survival niche for bone marrow plasma cells. Eur J Immunol. 2014;44:2306–17. doi: 10.1002/eji.201344313. [DOI] [PubMed] [Google Scholar]
- 55.Canene-Adams K. Preparation of formalin-fixed paraffin-embedded tissue for immunohistochemistry. Methods Enzymol. 2013;533:225–33. doi: 10.1016/B978-0-12-420067-8.00015-5. [DOI] [PubMed] [Google Scholar]
- 56.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 61.Marini F, Binder H. pcaExplorer: an R/Bioconductor package for interacting with RNA-seq principal components. BMC Bioinformatics. 2019;20:331. doi: 10.1186/s12859-019-2879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marini F, Ludt A, Linke J, Strauch K. GeneTonic: an R/Bioconductor package for streamlining the interpretation of RNA-seq data. Biorxiv. 2021; https://www.biorxiv.org/content/10.1101/2021.05.19.444862v1. [DOI] [PMC free article] [PubMed]
- 64.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021;50:D439–D444. [DOI] [PMC free article] [PubMed]
- 65.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchholz SM, Goetze RG, Singh SK, Ammer-Herrmenau C, Richards FM, Jodrell DI, et al. Depletion of macrophages improves therapeutic response to gemcitabine in murine pancreas cancer. Cancers. 2020;12:1978. doi: 10.3390/cancers12071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq datasets generated during the current study have been deposited in the Gene Expression Omnibus (GEO) archive and are available under the accession number GSE192424. The WES datasets can be accessed under PRJNA706650 in the sequencing read archive (SRA).