Abstract
Small cell lung cancer (SCLC) is a particularly lethal subtype of lung cancer. Metastatic lung tumours lead to most deaths from lung cancer. Predicting and preventing tumour metastasis is crucially essential for patient survivability. Hence, in the current study, we focused on a comprehensive analysis of lung cancer patients' differentially expressed genes (DEGs) on brain metastasis cell lines. DEGs are analysed through KEGG and GO databases for the most critical biological processes and pathways for enriched DEGs. Additionally, we performed protein–protein interaction (PPI), GeneMANIA, and Kaplan–Meier survival analyses on our DEGs. This article focused on mRNA and lncRNA DEGs for LC patients with brain metastasis and underlying molecular mechanisms. The expression data was gathered from the Gene Expression Omnibus database (GSE161968). We demonstrate that 30 distinct genes are up-expressed in brain metastatic SCLC patients, and 31 genes are down-expressed. All our analyses show that these genes are involved in metastatic SCLC. PPI analysis revealed two hub genes (CAT and APP). The results of this article present three lncRNAs, Including XLOC_l2_000941, LOC100507481, and XLOC_l2_007062, also notable mRNAs, have a close relation with brain metastasis in lung cancer and may have a role in the epithelial-mesenchymal transition (EMT) in tumour cells.
Subject terms: Cancer, Computational biology and bioinformatics, Genetics
Introduction
Lung cancer (LC) accounts for the most cancer-related death globally. In 2020, nearly 2.21 million patients were diagnosed with lung cancer, and 1.80 million deaths were estimated to be related to lung cancer (https://gco.iarc.fr)1. The Brain is the most distant metastasis in small-cell lung cancer (SCLC) patients, and brain metastasis (BM) increases to 50% for patients who survive after two years of initial diagnosis2; therefore, the overall survival of 5 years is 1–5%3. An early lung cancer diagnosis is crucial since surgery is ineffective in the late stages, especially in patients with metastasis. SCLC is an aggressive type of LC with the early development of widespread metastases; this leads to SCLC with a tremendously low prognosis for patients with the disease4. SCLC is highly relevant to smoking, with only 2% of patients diagnosed with this subtype never smoking in their lifetime5. Rb1 and Trp53 are the most genetic inactivation forces for SCLC; however, the most efficient genomic alterations remain unclear at6,7. Brain metastases occur in approximately 20% of all cancer patients, with most cases in people with lung, breast, and colorectal malignancies, melanoma, or renal cell carcinoma8–11. Brain metastases (BMs) were found in more than 25% of SCLC patients, with an average survival of 9 months following total therapy.
However, as imaging methods and systemic medicines advance, the increased incidence of BMs increases year after year as patients live longer. In individuals with advanced cancer, the presence of BMs continues to significantly contribute to overall cancer mortality12. Although more initial disease stage shift is a great result, changing the early stages of the disease is a significant result; the leading indicator of the effectiveness of screening is the reduction of mortality13. It is now clear that most parts of the human genome are non-coding. Long non-coding RNA (lncRNA) is the common term for transcriptomes with 200 or more nucleotides. About a decade ago, investigators figured out that lncRNAs play an essential role in cancer biology14–17. Dysregulation of lncRNA is the most critical evidence in different cancer types.
For instance, lncRNA HOXB-AS3 can exacerbate lung cancer migration, proliferation, and invasion. The mechanisms underlying these changes are by activating the PI3K‐AKT pathway. The PI3K/Akt/mTOR pathway impacts tumorigenesis and cancer progression18–20. This breakthrough helps us to propose a potential biomarker for cancer predictivity. This study aimed to detect genes (mRNAs and lncRNAs) differentially expressed between SCLC patients with brain metastasis and SCLC patients without brain metastasis. We also performed an enrichment analysis to find correlations between specific mRNAs and lncRNAs of our two cancer patient groups. However, considering a gene as a biomarker requires deep, long, precise, and comprehensive studies.
Methods
Data predation and analysis
Data for lncRNA and mRNA microchip array extracted from peripheral blood mononuclear cells of SCLC patients were downloaded through Gene Expression Omnibus (GEO)21. The data was designed as brain metastasis (BM) patients compared to non-BM patients for lncRNAs in relation to BM in LC samples2 (Accession: GSE161968, Platform: GPL20115).
All six replicates of the samples were used for multiple analyses. Statistical analyses were performed in R (v3.5.1). All the scripts and algorithms were uploaded to the GitHub webserver.
After constructing a tidy data set of gene expression matrix with samples and probes IDs, we analysed the mean expression and probable correlations through boxplot and heatmap to ensure denormalisation and prevent any false correlation. Afterwards, by scaling the expression matrix on the mean expression of each gene, differentially expressed genes (DEGs) among samples arise in a MA plot. Moreover, principle component analysis was performed on our data sets' samples. Clusters were trimmed by ignoring six samples from both BM and non-BM groups. Subsequently, we manipulate the K-mean algorithm for re-clustering our data set for a strong validation. Limma package (v3.52.2) was used to calculate the log2foldchange (LogFC) parameter threshold to benefit the highest number of expression data well22. Hence LogFC ≥ 1.75 and LogFC ≤ -1.75 were applied to our DEG results as up and down-regulated genes, respectively. Genes with a P-value of ≤ 0.05 were considered significant. We consider 1.75 for log2foldchange as our cutoff value because a number outside this range would give us an inappropriate number of candidates. Alternatively, genes with higher expression in BM than non-BM samples were categorised as up-regulated and down-regulated. A workflow of our approaches in this study is provided (Fig. 1).
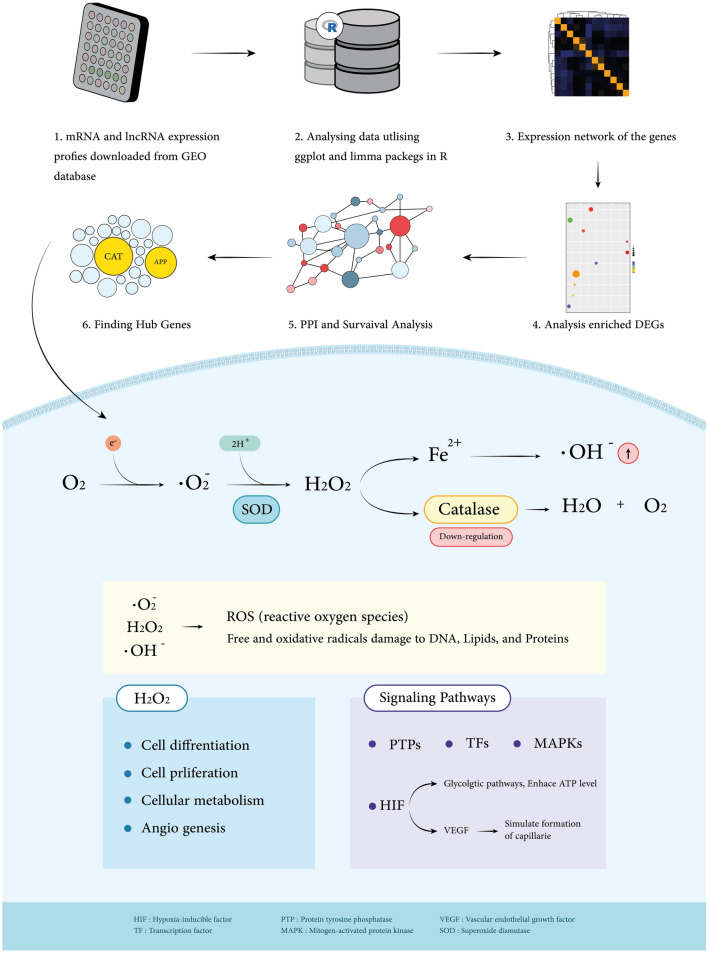
Figure 1.
Article section workflow. Workflow represents a comprehensive review summarising this study's data gathering, trimming, and analysing approaches. CAT (catalase), for instance, is one down-regulated hub gene that shows significant roles in critical pathways.
Enrichment analysis
Understanding the underlying biological roles of various genes or proteins is best accomplished through Gene-Set Enrichment. Through enrichment analysis, the interpretability of biological insights is enhanced, and the complexity of molecular data is reduced23. The basic idea of Gene Set Enrichment Analysis (GESA) is to explore groups of genes considerably over-represented in a given list of genes compared to a control set of genes. These gene sets are often, but not always, made up of genes that collaborate in a recognised biological pathway24. Enrichr (https://maayanlab.cloud/Enrichr/), a comprehensive resource for curated gene sets and a search engine provided by Maayan Megalab25, was applied, with results extracted from various databases for enrichment analysis. After exporting the results of our gene dataset, the P-value was set to ≤ 0.05.
CBioPortal (https://www.cbioportal.org/) is a database that provides visualisation, analysis, and large-scale cancer genomics data sets26. It is a tool for the interactive examination of multidimensional cancer genomics data sets, which was first created at Memorial Sloan Kettering Cancer Center27. This database was used to find mutations in the candidate genes; then, all small cell lung cancer datasets were selected, including Small Cell Lung Cancer CLCGP (Nat Genet 2012), Johns Hopkins (Nat Genet 2012), U Cologne (Nature 2015), and Multi-Institute (Cancer Cell 2017). Abundance and types of mutations for these genes were found.
Gene and protein network analysis
GeneMANIA (http://genemania.org/) plugin in Cytoscape (https://cytoscape.org/) version 3.9.1 was applied to show the relations between candidate up and down-regulated genes. GeneMANIA is a user-friendly online service that allows the generation of gene function hypotheses, analysis of gene lists, and prioritise genes for functional experiments. GeneMANIA will automatically weight data sources based on their predictive value for rebuilding the query list, provided that the list is big enough (currently five or more genes). For instance, GeneMANIA will heavily weigh protein domain similarity networks and recommend more genes with a similar domain structure when provided with query genes with comparable protein domain structures. GeneMANIA frequently discovers new members of a protein complex based on query genes that are a part of that complex and gives high weight to actual or anticipated physical interactions28. Cytoscape is a free, open-source software project that combines biomolecular interaction networks, high-throughput expression data, and other molecular states into a cohesive conceptual framework.
Protein–protein interactions (PPI) also were performed from the String protein database plugin (https://string-db.org/) using Cytoscape to indicate networks of proteins of these genes. The STRING database attempts to combine all known and expected protein–protein interactions, including physical and functional ones29. For the past 19 years, the STRING database has been in steady expansion, and the current edition includes protein interaction data for approximately 2000 species; nonetheless, all interactions are strictly intra-species. For the first time, we incorporate cross-species interactions in the STRING database in this study. STRING reported that PPIs are functional connections between proteins. These connections are not restricted to physical interactions; they may involve interactions like transcription factor binding or represent that the linked proteins are found in the same biological pathway30.
Panther (Protein Analysis Through Evolutionary Relationships) (http://www.pantherdb.org/) classification systems were applied to classify our candidate protein-coding genes. PANTHER is a comprehensive knowledge base of evolutionary and functional relationships between protein-coding genes (often referred to as proteins) and tools for utilising the classifications to analyse large-scale genomics data. Evolutionary groupings (Protein Class, Protein Family, Subfamily) and functional groupings are the main categories used to classify proteins (Gene Ontology and pathways)31. The type of protein classes and their percentage were obtained.
Survival analysis
We employed the Kaplan–Meier plotter database (https://kmplot.com/analysis/) to perform a survival analysis of the hub genes. It is a web-based tool that allows for pooled survival analysis by combining different datasets from the cancer Biomedical Informatics Grid (caBIG), GEO, and TCGA repositories32. Up to April 24th, 2022, this database presented the overall survival analysis data of 1925 lung cancer patients. We input our hub genes separately and consider a p-value threshold of 0.05 for a meaningful result. Although KM-Plotter has many filters for focusing the study on specific criteria like stage, gender, age, surgery success, histology, chemotherapy, and radiotherapy, we performed the analysis on default restrictions, making our results more comprehensive and general.
Results
After DEGs evaluation, we need to understand molecular mechanisms that would be affected by these genomic dysregulations and how the disturbances impact them. DEGs in SCLC BM and non-BM patients are related to transcription factors, non-coding RNAs, mutations, and signaling pathways. To understand the expression patterns in our gene dataset, we candidate 30 up-regulated and 31 down-regulated genes, including mRNAs and lncRNAs, in BM and non-BM patients (Table 1). The relations and overlap of our candidate genes with transcription factors, miRNAs, histone modifications, signalling pathways, cell types and tissues, and Top 10 correlated diseases acquired from specific databases for each group. (See Tables S2, S3, S4, S7, S8, and S9). In addition, BM shows different protein classes for up-regulated (16 protein-coding genes) and down-regulated (14 protein-coding genes) in BM, as shown in Fig. 6. Each gene with its class protein is demonstrated In Tables S5 and S6 for up and down genes.
Table 1.
Candidate genes with different expressions in brain metastasis patients with SCLC. p-value ≤ 0.05.
| Up-regulate genes | logFC | P-value | Down-regulate genes | logFC | P-value |
|---|---|---|---|---|---|
| PRSS33 | 3.383007303 | 0.004560695 | OCLN | − 3.566468452 | 0.022160353 |
| WHAMMP2 | 2.99733975 | 0.005815325 | XLOC_l2_007062 | − 3.329664096 | 0.013558958 |
| XLOC_l2_000941 | 2.687330175 | 4.1957237415278E-05 | ZIM2 | − 2.854825242 | 0.028619212 |
| MYOM2 | 2.601569126 | 0.021263756 | PNMA2 | − 2.660709775 | 0.01010599 |
| C1orf87 | 2.59361413 | 1.62961553882741E-05 | MPO | − 2.603630721 | 0.034713463 |
| PIGC | 2.568682566 | 0.04592453 | ANKRD34B | − 2.435841569 | 0.049785848 |
| C1orf105 | 2.537416642 | 0.004661527 | ABCA13 | − 2.425532635 | 0.010254371 |
| ALOX15 | 2.372469038 | 0.006367303 | LOC646627 | − 2.409993098 | 0.037554779 |
| IGIP | 2.276075691 | 0.000645279 | SLC14A1 | − 2.380643274 | 0.001828896 |
| SPAG6 | 2.242164253 | 0.025026256 | ITLN1 | − 2.341855873 | 0.003294184 |
| LPAR3 | 2.233936235 | 0.019032771 | CRB1 | − 2.310587746 | 0.014682181 |
| ZNF416 | 2.102556319 | 0.000160004 | PGK2.00 | − 2.111562308 | 0.019712849 |
| EMR4P | 2.088110399 | 0.006628096 | KRT1 | − 2.043848253 | 0.023066534 |
| TRIP13 | 2.06727697 | 0.000330399 | SLC2A5 | − 1.999212172 | 0.031409788 |
| IL5RA | 2.061453481 | 0.006381159 | PGLYRP1 | − 1.943771474 | 0.032295719 |
| TM4SF19 | 2.012294101 | 0.005361822 | FAM83A | − 1.926935845 | 0.002982599 |
| PCDHGA9 | 2.009416266 | 0.00140228 | XK | − 1.915279305 | 0.008257932 |
| PIGW | 1.995038208 | 0.038464331 | DPCD | − 1.83374777 | 0.040212628 |
| ZNF599 | 1.985801014 | 0.001438597 | AMFR | − 1.828192394 | 0.000293755 |
| LGSN | 1.972203834 | 0.006590149 | TMEM158 | − 1.802895011 | 0.001143757 |
| TMEM176B | 1.953156305 | 0.023746468 | DACH2 | − 1.800987665 | 0.00095326 |
| TMEM176A | 1.935063753 | 0.030140607 | OSBP2 | − 1.790583828 | 0.044198947 |
| TTC39B | 1.907722674 | 0.014295311 | CRAT | − 1.788273793 | 0.012034785 |
| SIGLEC8 | 1.861465376 | 0.004428622 | ZNF608 | − 1.78743558 | 0.002590702 |
| PRAMEF20 | 1.856405961 | 0.02697871 | CYBRD1 | − 1.776635568 | 0.001412057 |
| CLC | 1.853046847 | 0.010293129 | CTSB | − 1.776261192 | 0.012873076 |
| EGR3 | 1.843233413 | 0.003359614 | ANXA3 | − 1.767371637 | 0.010699241 |
| LOC100507481 | 1.77545701 | 0.001358476 | RIOK3 | − 1.762783691 | 0.006681225 |
| ZNF827 | 1.764674234 | 0.01536046 | MAOB | − 1.758125128 | 0.00024905 |
| HSD17B3 | 1.757453949 | 0.0062133 | LOC729870 | − 1.757276671 | 0.035413402 |
| IFT52 | − 1.752051545 | 0.014110306 |
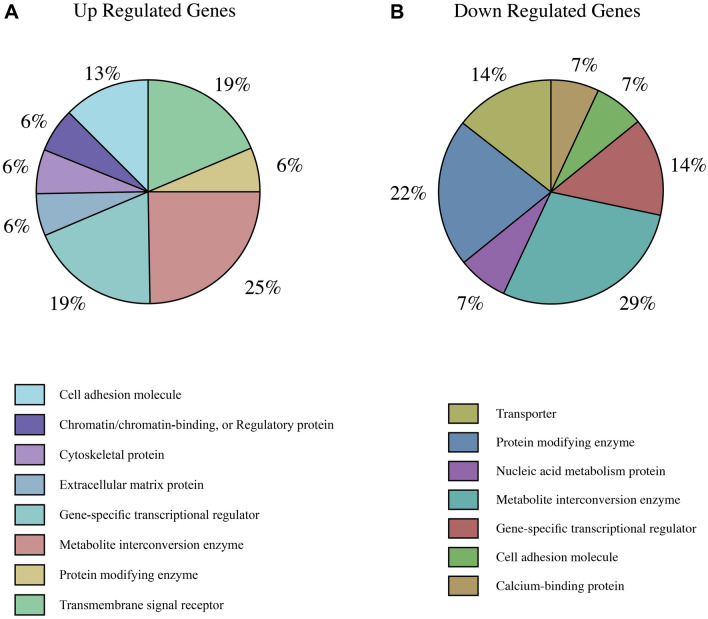
Figure 6.
Protein classes for candidate genes in BM SCLC patients. Different categories of up and down-regulated genes are shown in (A and B), respectively.
Non-coding RNAs might be directly related to brain metastasis in SCLC
Enrichment analysis displayed that our candidate genes are involved in essential cell signalling pathways. Some of these genes are important enough that we bring them up in the discussion section, such as IL5RA, EGR3, and ALOX15 as up-expressed genes, and OCLN, MAOB, CTSB, and MPO as down-expressed genes in BM SCLC. In the category of up-regulated genes, we find LOC100507481 and XLOC_l2_000941 as lncRNAs; WHAMMP2, as a pseudogene encodes 5246 nucleotide transcript length; ADGRE4P, also a pseudogene encodes 2732 nucleotide transcript length; TTC39B is a protein-coding but encodes non-coding RNA with 7023 nucleotide transcript length; ZNF827 another protein-coding that encodes 3675 nucleotide transcript and HSD17B3 protein-coding genes that encode a 3559 nucleotide transcript length. For down-regulated genes, we find XLOC_l2_007062 and LOC729870 as a lncRNA; ZIM2 is a protein-coding gene that encodes four non-coding RNA with approximately 2300 nucleotide transcripts length; Similar to this, ABCA13 also is a protein-coding gene that encodes five non-coding RNA with approximately 15,000 nucleotide transcripts length; ZNF608 encodes non-coding RNA about 6000 nucleotides, ANXA3 encodes 1056 nucleotide transcripts and RIOK3 about 3500 nucleotides, also are protein-coding genes. Our ultimate goal is to demonstrate the lncRNAs related to BM in SCLC patients. To assess our targeted lncRNAs, we utilised lncHUB, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and trimmed our dataset based on P-Value (p < 0.05)33–35. Since our microarray data was performed on lncRNAs and mRNAs, we utilised multiple databases to discover the correlated lncRNA and mRNAs to conduct a comprehensive study on pathways and biosystems with relatively close lung cancer brain metastasis.
Investigation of gene ontology and pathways
In order to investigate candidate genes' functions in the cell, it is essential to show their signalling pathways to find their role in cancer; The KEGG database is required to achieve this aim. KEGG (2021) pathway enrichment analysis showed three significant genes; ALOX15, PIGC, and PIGW, which correlated in Ferroptosis and Linoleic acid metabolism pathways and Glycosylphosphatidylinositol (GPI)-anchor biosynthesis. (Table S1 shows the result of the KEGG analysis). Gene ontology (GO) enrichment analysis is commonly used to assign functions to gene sets and obtain insights into the biological processes in which they are engaged31. GO is a collection of gene function-controlled vocabularies arranged into three ontologies: biological process, cellular component, and molecular function. Due to community initiatives, many genomes have been annotated with GO keywords, allowing GO enrichment analysis on multiple gene sets. An enrichment study, for example, identifies GO keywords that are over-represented for a specific collection of human genes, allowing for biological interpretation.
On the other hand, KEGG pathway mapping provides more information about how genes or gene products interact in pathways, although the coverage of genes is less extensive than GO. GO (2021) enrich analysis was performed on our candidate genes from DEGs results. The top five GO terms with desirable P-Value and Combined Scores are listed in Table S1. Moreover, among all GO databases results on our DEGs, the top 10 molecular function and biological process terms of both up and down-regulated genes are due to the rich factor of each term shown in Fig. 2. We utilise ggplot2 (v3.3.6) to visualise critical biological processes36.
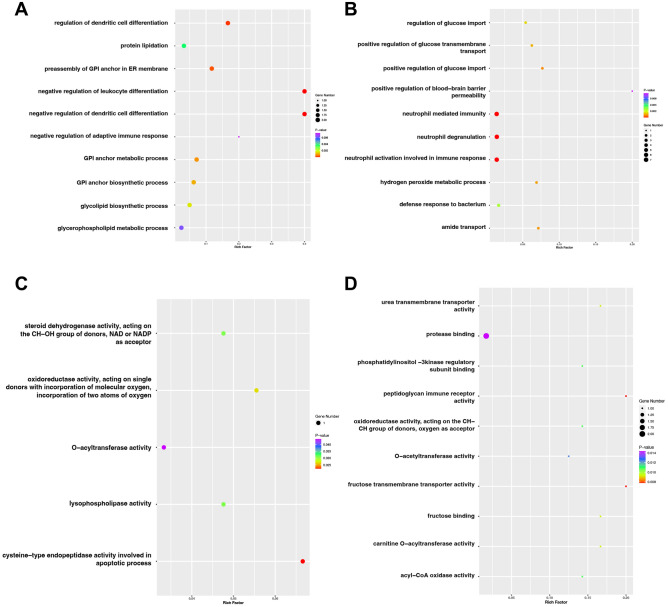
Figure 2.
Gene ontology (GO) enrichment analysis. (A) Top 10 most significant changes in the GO biological process for up-regulated genes. (B) Top 10 most significant changes in the GO biological process for down-regulated genes. (C) Top 5 most significant changes in the GO molecular functions for up-regulated genes. (D) Top 10 most significant changes in the GO molecular functions for down-regulated genes.
Pathway and cell type and tissue analysis
Pathways influenced by these candidate genes are briefly explained in Tables S2 and S3; SLC14A1, SLC2A5, MAOB, CRAT, ANXA3 down genes; PIGC, PIGW, LPAR3, IL5RA, HSD17B3, EGR3 up genes, involved in different essential pathways. Our study focuses on brain metastasis; hence in table S4, Key cell types and tissues related to up and down-regulated genes in brain metastasis patients demonstrated, as TMEM176B, ALOX15, TMEM176A up and SLC14A1, OSBP2 XK, KRT1 down genes are significant. At last, for comprehensive analysis, some other diseases related to brain metastasis SCLC genes and SCLC without brain metastasis genes are presented in Table S9 that LPAR3, TTC39B, TMEM176B, IL5RA up expressed genes and ZNF608, DACH2, ZIM2 down expressed, are related.
PIGW, a critical lncRNA
After analysing our candidate's genes, we monitor IGIP; PCDHGA9, TMEM176B; TMEM176A are up expressed and OCLN; CRB1; PNMA2, ITLN1, and ANXA3 are down-expressed genes (shown in Table 1) has correlations with noticeable lncRNAs. For example, PIGW observed concerning multiple pathways in previous analysis has a co-expression relation with LINC01386 and might have a critical molecular mechanism in brain metastasis in lung cancer. To survey key lncRNAs related to candidate genes, we utilised an enrichment analysis tool, which has presumed a constant p-value affiliated with input genes and its datasets. lncRNA co-expression analysis performed on the lncHUB database for both up and down-regulated genes, and the p-values equal 0.00983 for up-regulated genes and 0.010472 for down-regulated genes, respectively (Table 2)37.
Table 2.
Key LncRNAs related to up and down-regulated genes in brain metastasis patients (lncHUB lncRNA Co-Expression database).
| LncRNA | Up-regulated genes | LncRNA | Up-regulated genes | LncRNA | Down-regulated genes |
|---|---|---|---|---|---|
| BDNF-AS | IGIP; PCDHGA9 | LINC02712 | SIGLEC8; PCDHGA9 | FGF10-AS1 | AMFR; CYBRD1 |
| RASSF8-AS1 | IGIP; PCDHGA9 | LINC00501 | TMEM176B; TMEM176A | LINC02115 | AMFR; CYBRD1 |
| TMEM161B-AS1 | IGIP; PCDHGA9 | LACTB2-AS1 | TMEM176B; TMEM176A | LINC01716 | AMFR; ZIM2 |
| NNT-AS1 | IGIP; PCDHGA9 | C1RL-AS1 | TMEM176B; TMEM176A | MYO16-AS1 | ANXA3; FAM83A |
| SEPTIN7-AS1 | IGIP; PCDHGA9 | LINC02068 | TMEM176B; TMEM176A | LINC00424 | ANXA3; ITLN1 |
| PSMG3-AS1 | IGIP; PCDHGA9 | LINC01853 | TMEM176B; TMEM176A | LINC00560 | ANXA3; ITLN1 |
| BACE1-AS | IGIP; PCDHGA9 | LINC01504 | TMEM176B; TMEM176A | CTNNA2-AS1 | CRB1; PNMA2 |
| GRID1-AS1 | IGIP; PCDHGA9 | FBXO3-DT | TMEM176B; TMEM176A | LINC01114 | CRB1; PNMA2 |
| LINC00909 | IGIP; PCDHGA9 | LINC01843 | TMEM176B; TMEM176A | LINC02520 | CRB1; ZIM2 |
| LINC01511 | LGSN; MYOM2 | LINC02384 | TMEM176B; TMEM176A | LINC01701 | FAM83A; CTSB |
| LINC02864 | LGSN; TRIP13 | OVOL1-AS1 | TMEM176B; TMEM176A | LINC01288 | MAOB; ZIM2 |
| LPP-AS2 | LPAR3; ZNF599 | SMIM2-AS1 | TMEM176B; TMEM176A | ARHGEF2-AS1 | OCLN; ANXA3 |
| TMCO1-AS1 | PIGC; ZNF599 | LBX2-AS1 | TMEM176B; TMEM176A | NKX2-1-AS1 | OCLN; CTSB |
| LINC00448 | PRAMEF20; MYOM2 | CPNE8-AS1 | TMEM176B; TMEM176A | LINC01806 | OCLN; CTSB |
| LINC01736 | SIGLEC8; PCDHGA9 | ERVK9-11 | TMEM176B; TMEM176A | SCN1A-AS1 | PNMA2; ABCA13 |
| LINC02015 | TMEM176B; TMEM176A | LINC02712 | SIGLEC8; PCDHGA9 | SAP30-DT | PNMA2; SLC2A5 |
| LINC01880 | TMEM176B; TMEM176A | LINC01999 | TMEM176B; TMEM176A | HHLA3-AS1 | XK; ITLN1 |
| LINC02244 | TMEM176B; TMEM176A | LINC02538 | TMEM176B; TMEM176A | LINC02703 | ZNF608; SLC2A5 |
| LINC02011 | TMEM176B; TMEM176A | HNF1A-AS1 | TMEM176B; TMEM176A | ||
| SMIM2-IT1 | TMEM176B; TMEM176A | LINC01386 | TRIP13; PIGW | ||
| LINC01781 | TMEM176B; TMEM176A | SMAD9-IT1 | ZNF827; IGIP | ||
| LINC01506 | TMEM176B; TMEM176A | EDNRB-AS1 | ZNF827; PCDHGA9 | ||
| LINC02532 | TMEM176B; TMEM176A |
Gene network and PPI analysis
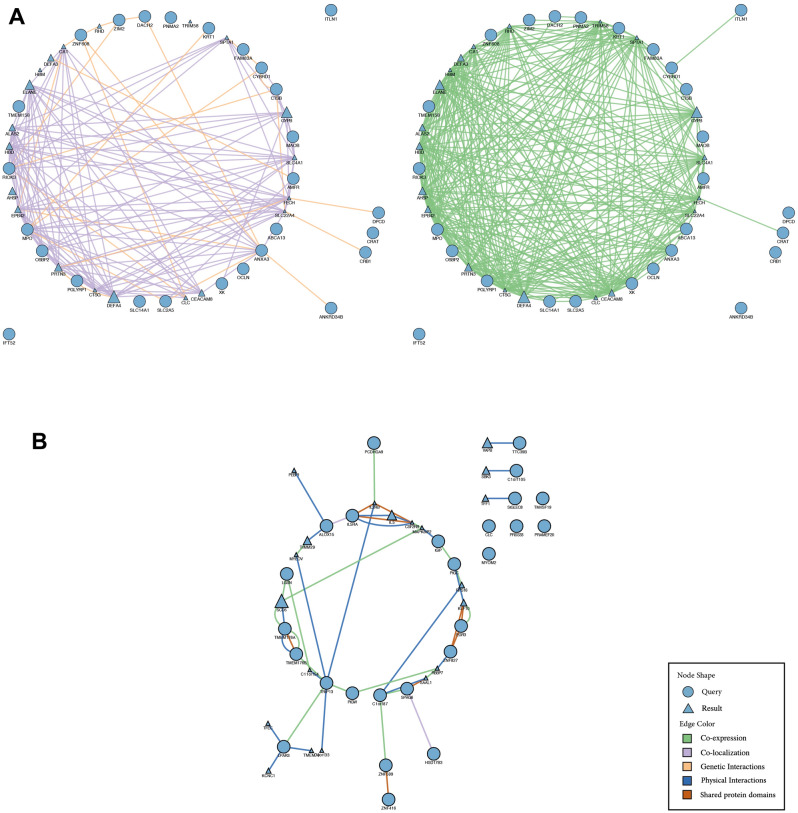
Gene network analysis co-expression and co-localisation between up and down-regulated genes were performed and provided in Fig. 3.
Figure 3.
Candidate genes in BM SCLC patients—gene networks from GeneMANIA. (A) Up-regulated genes network. Physical interactions are shown in blue lines, Co-expression genes are shown in green lines, Co-localization is shown in purple lines and shared protein domains are shown in the orange lines. The higher scores are shown as a larger circle. Intermediate genes involved were also demonstrated. (B) Down-regulated genes network. Co-expression genes are shown in green lines, Co-localization is shown in purple lines, and genetic interactions are shown in orange lines. The higher scores are shown in black as a larger circle. Intermediate genes involved were also demonstrated.
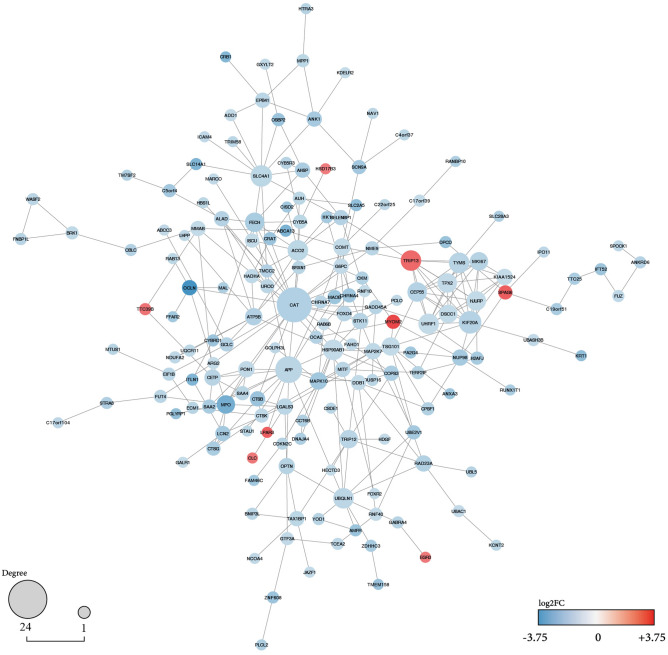
We used the STRING plugin in Cytoscape (version:3.9.1) for protein–protein interactions to construct a network between up and down-regulated genes according to our DEGs results38,39. For complete results, we filter our DEGs to |log2FC|> 1. Hence, the resulting graph showed us 169 nodes and 300 edges. Additionally, the node's colour is illustrated by log2fold change. Utilising the CentiScaPe plugin, every node's degree greater than 15 is considered hub genes, two genes (CAT:24 and APP:16)40 (Fig. 4). We managed to study our hub geans survival analysis (Fig. 5).
Figure 4.
Protein–protein interactions network with DEGs for brain metastasis in SCLC. Up and down-regulated candidate genes are coloured on the scale of logFC. Log2FC is an indicator of the expression level of these genes since logFC = 3.75 for the most up-regulated gene in BM SCLC patients and logFC = − 3.75 for the most down-regulated gene. The nodes' size characterises the connectivity degree for identifying the critical hub genes (CAT and APP). Unconnected nodes are trimmed from the figure.
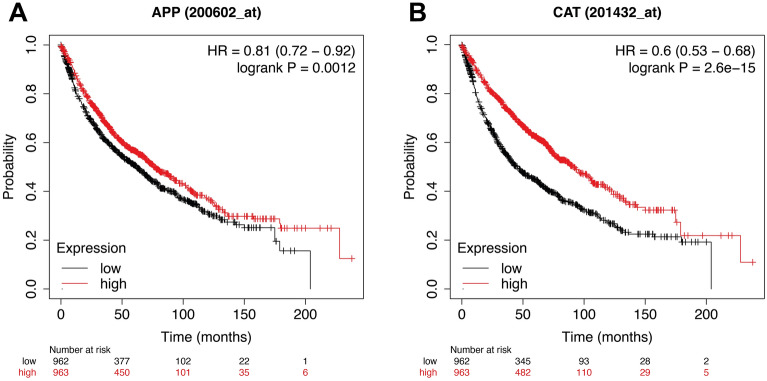
Figure 5.
Prognostic values of two hub genes in the brain metastases from SCLC patients. Kaplan–Meier survival analysis of 2 hub genes in patients with small-cell lung cancer. The prognostic value of (A) APP and (B) CAT for determining the prognostic value of APP and CAT mRNA expression were evaluated using the Kaplan–Meier plotter. The Affymetrix IDs of the genes are as follows: 200602_at (APP) and 201432_at (CAT). Logrank test for Kaplan–Meier analysis reveals the p-value for both APP and CAT survival analysis. APP and CAT were specifically represented among lung metastasis samples. The patients survive probability with a high expression of hub genes indicated as red-line, and patients survive probability with a low expression of hub genes indicated as black-line. The high and low-expression cohorts were divided by the median survival time.
We discovered that 20% of BM patients have mutations in 28 up-regulated genes in 47 individuals in 249 samples. Figure S1 depicts this. MYOM2 contains the most modifications (6%). This gene has mutations in I-set: Immunoglobulin I-set domains and Ig_2: Immunoglobulin domain.
In 77 cases, 32% of those queried had mutations in 27 down-regulated genes. Further analysis showed MPO had the most significant mutations (2.5%) in SCLC patient samples indicated in Figure S2.
CAT and ACC are significant genes in lung cancer patients' survival
As we mentioned before, CAT and ACC are considered hub genes. Initial KP-Plotter analysis showed that the results were meaningful due to top values. The overall survival plots are provided in Fig. 5. CAT and ACC's hazard ratio (HR) were 0.6 and 0.81, respectively. HR < 1 means that the up-regulation of genes drives high survival probability in LC patients. Our hub genes have a negative log2fold change in brain metastasis SCLC patients making these two potential biomarker genes. Fortunately, there is a meaningful relationship between DEGs and KP-Plots, validating our prediction. The hub genes CAT (HR = 0.6, P-value = 2.6e − 15, log2FC = − 1.3802) and APP (HR = 0.81, P-value = 0.0012, log2FC = − 1.1716) were significantly associated with an unfavorable overall survival in lung cancer patients.
Our candidate proteins' coding genes are involved in constructing metabolic interconversion enzymes, transmembrane signal receptors, cell-adhesion molecules, protein-modifying enzymes and gene-specific transcriptional regulators, as shown in Fig. 6. Knowing the class of proteins can help us to explore the differences between BM and non-BM lung cancer patients.
Discussion
Many lncRNAs showed in epigenetic changes like gene imprinting, modifications, and regulations. Beyond lncRNA, there is yet another type of non-coding RNA with much less information associated with cancer biology. Small-non-coding RNA (snoRNA) has 60 to 300 nucleotides. Recent studies have shown the migration and progression of lung cancer tumours mainly through EMT and MMPs regulated by lncRNAs. Additionally, the immune system has cytotoxic CD8 + T cells to restrict tumour cell growth in cancers. Furthermore, overexpression of tumour-promoting lncRNAs can manage immune evasion of LC cells41. However, this paper will focus on cancer-related lncRNAs and mRNAs.
XLOC_l2_000941 third up-regulated lncRNA gene in DEGs results, with log FC = 2.68 (Table 1), is related to MYC transcription factor (TF) based on Peng-ran Sun et al. studies42. MYC, also known as c-myc, is a multifunctional TF that plays a vital role in cell cycle progression, apoptosis, and cellular transformation43. c-myc expression is also up-regulated in ovarian endometriotic cyst stromal cells, and its expression can be suppressed by miR-196b44. The expression of c-MYC–regulated genes are associated with a higher risk for brain metastasis in breast cancer patients45. This paper demonstrates the different expressions of XLOC_l2_000941 in BM SCLC individuals.
MYOM2, a fourth high expression gene with Log FC = 2.60 in BM patients with the highest rate of missense mutations, is a protein-coding gene that usually plays a role in synthesising titin-associated protein fibronectin type III and immunoglobulin C2 domains46. Fibronectin is a cell adhesion protein that could affect metastasis47. MYOM2 may associate with the pharmacology of Tumor necrosis factor (TNF) blockers48, a multifunctional cytokine that plays critical roles in diverse cellular events such as cell survival, proliferation, differentiation, and death alter cancer metastasis49. As we show in Table 2, MYOM2 is associated with LINC01511 and LINC00448 lncRNAs. LINC01511 is up-regulated in triple-negative breast cancer and might play a fundamental role in the mechanism of EGFR exon 19 deletions in lung adenocarcinoma50.
WHAMMP2 (WAS protein homolog associated with actin, Golgi membranes, and microtubules pseudogene 2) is a pseudogene highly expressed in BM SCLC similar to Thymocytes in Thymus in brain metastasis patients mentioned in table S4.
TMEM176B and TMEM176A have an extensive correlation with different lncRNAs, as shown in Table 2. Some of these lncRNAs get involved in cancers; for instance, LINC00501 prohibits lung cancer development and metastasis by mediating miR-129-5p/HMGB1 and up-regulated in non-small cell lung cancer (NSCLC) patients51. Contrariwise, LBX2-AS1 promotes cell proliferation and metastasis through Notch signalling in NSCLC52. Also, we find that these two genes have physical interactions, co-expression relations, and shared protein domains, demonstrated in Fig. 3B. Transmembrane proteins (TMEMs) are membrane proteins with at least one transmembrane structure; They are also known as integral membrane proteins. TMEM176 plays diverse functions in distinct malignancies. A range of membrane protein functions is intimately associated with the proliferation, invasion, and metastasis of malignant tumours53,54. In human hepatocellular carcinoma, epigenetic silencing of TMEM176A promotes ERK signalling55.
Similarly, high expression of TMEM16A plays a role in gastrointestinal stromal tumours56. TMEM176A in glioblastoma can inhibit Bcl2 expression and cause apoptosis57,58. Increased TMEM176B protein levels have been found in various tumours, and inhibition of TMEM176B has been shown to increase CD8 + T cell-mediated tumour growth control, improving cancer therapy effectiveness59. TMEM176B represses adenosine triphosphate (ATP) and nigericin-induced NLRP3 inflammasome activation in dendritic cells and macrophages through ionic mechanisms. Targeting TMEM176B can emphasise anti-tumoral CD8+ T cell responses in an inflammasome-dependent manner while directly killing malignant cells60,61.
Phosphatidylinositol glycan anchor biosynthesis class C and W, respectively (PIGC and PIGW), are critical genes in the glycosylphosphatidylinositol (GPI)-anchor biosynthesis pathway, encode an endoplasmic reticulum (ER)-associated protein that is significant for the biosynthesis of glycosylphosphatidylinositol (GPI)62. This process happens in the endoplasmic reticulum and is a step in the glycosylphosphatidylinositol (GPI) biosynthesis, anchoring many cell surface proteins to the membrane. Defects in the PIGW gene cause the age-dependent epileptic encephalopathy West syndrome and a syndrome exhibiting hypophosphatasia and cognitive disability (HPMRS5)63. PIGC was up-regulated and associated with the transferase activity of cancer cells such as Hepatocellular carcinoma (HCC)62. Since cell adhesion can be altered in metastasis, it can indicate that in SCLC brain metastasis, GPI binding changes because of the high expression of PIGC and PIGW.
We indicate that ABCA13, an ATP-binding cassette (ABC) transporter, has the most mutations (9%) in lung cancer patients, as shown in Figure S2, Suggesting that alteration of this gene can affect the lung cancer state.
AMFR is a gene that encodes ubiquitin-protein ligase. It also correlates with lncRNAs, including FGF10-AS1, LINC02115, and LINC01716 (Table 2). Overexpression of gp78/AMFR in human cancers is related to low survival and the high stage of the tumours64,65. Thus, It can attribute narrow expression of AMFR in non-BM SCLC despite BM SCLC to a biomarker of this type of cancer metastasis. CRAT encodes an acetyltransferase and is involved in specific pathways among Beta-oxidation of very long-chain fatty acids, Beta-oxidation of pristanoyl-CoA, and Peroxisomal lipid metabolism (Table S3). CRAT is required to transport acetyl-CoA from peroxisomes or mitochondria into the cytoplasm to activate calcium/calmodulin-dependent kinase II (CaMKII). A decrease in CRAT protein levels reduces cell proliferation and low cell migration; it also is necessary for CaMKII activation66. OCLN is the most downregulated gene in DEGs results, with Log FC = − 3.56 (Table 1); naturally encoded transmembrane proteins play an essential role in regulating cell permeability barriers formed by protein tight junctions and Are an Integral Component of the blood–brain barrier (BBB)67.
We demonstrate that low expression of this gene may affect brain metastasis since prior studies reveal that CeRNA regulates OCLN mRNA, which plays a vital function in the BBB. PIK3CG is a component of the PI3K-Akt signalling pathway, controlling essential cell functions such as cell proliferation, differentiation, and metabolism68,69. OCLN also relates to three lncRNAs such as ARHGEF2-AS1, NKX2-1-AS1, and LINC01806 (Table 2). Two hub genes, including CAT and APP, are involved in different cancers; for Instance, CAT, positioned specially in peroxisomes, decomposes H2O2, a spinoff of fatty acid oxidation, to oxygen and water. Thus, CAT confers safety in opposition to the deadly consequences of H2O2 without producing intermediate unfastened radicals, and the ensuing oxygen is applied to different metabolic processes70. Catalase is likewise often downregulated in tumour tissues like non-small-cell lung cancer than in normal tissues of the exact origin71. H2O2 is a mediator of several physiological processes, including cell differentiation and proliferation, cellular metabolism, survival, and immune response, through ROS (reactive oxygen species) intracellular signalling (Fig. 1)72.
Amyloid precursor protein (APP) is a protein that has a role in the progression of Alzheimer's disease. Its cytoplasmic region contains numerous phosphorylation sites. APP has also been suggested as a molecule involved in cell proliferation and invasion in various human cancers, including non-small-cell lung cancer73–75. Our study reveals that these genes are downregulated In brain metastasis SCLC and suggests that they can influence the cancer stage and survival rate, as shown in Fig. 5. We suggest these genes mentioned above can be associated with brain metastasis in SCLC and might be employed as new therapeutic targets and potential prognosis biomarkers. However, considering these genes as a Biomarker require deep and comprehensive studies yet clinical validations. Since we performed only bioinformatical analysis, deficiencies still demand clinical experiments and data validation. Furthermore, these candidates' metastasis genes' functions and mechanisms are still to be investigated.
In conclusion, brain metastasis SCLC shows a different gene expression than non-BM SCLC. We demonstrated up and down-regulation of specific lncRNAs and mRNAs and their networks to evaluate their predictive values. Preventing cancer metastasis with reliable biomarkers can be vital; however, it is yet unknown to find the exact role of these distinct lncRNAs and how they influence cancer metastasis.
Supplementary Information
Author contributions
A.M.N. and M.J. collected the data and performed the analysis. M.J. checked the associated database and raw data. M.J. and A.M.N. wrote the original manuscript. A.M.N. and M.J. designed figures and Scientific illustrations. M.M., A.A.K., H.H.M., and M.S. edited the manuscript. A.M.N, M.J, Z.M., and M.M., revised the manuscript. A.M. supervised the work.
Funding
This work was funded by Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences (32840).
Data availability
The expression data was gathered from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) (ACCESSION NUMBER: GSE161968) and enrich analysis performed utilising KEGG (https://www.genome.jp/kegg/), GO (http://geneontology.org/), and Kaplan–Meier survival analysis (https://kmplot.com/analysis/) databases. The analysed data and R codes are available to the corresponding author with no restriction upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arsham Mikaeili Namini and Motahareh Jahangir.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-22252-7.
References
- 1.Fois SS, et al. Molecular epidemiology of the main druggable genetic alterations in non-small cell lung cancer. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms22020612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, et al. Downregulation of lncRNA XR_429159.1 linked to brain metastasis in patients with limited-stage small-cell lung cancer. Front. Oncol. 2021;11:603271. doi: 10.3389/fonc.2021.603271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson AG, et al. The international association for the study of lung cancer lung cancer staging project: Proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016;11:300–311. doi: 10.1016/j.jtho.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: Implications for therapy. Nat. Rev. Clin. Oncol. 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varghese AM, et al. Small-cell lung cancers in patients who never smoked cigarettes. J. Thorac. Oncol. 2014;9:892–896. doi: 10.1097/JTO.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foggetti G, et al. Genetic determinants of EGFR-driven lung cancer growth and therapeutic response in vivo. Cancer Discov. 2021;11:1736–1753. doi: 10.1158/2159-8290.CD-20-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultheis AM, et al. Genomic characterization of small cell carcinomas of the uterine cervix. Mol. Oncol. 2022;16:833–845. doi: 10.1002/1878-0261.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achrol AS, et al. Brain metastases. Nat. Rev. Dis. Primers. 2019;5:5. doi: 10.1038/s41572-018-0055-y. [DOI] [PubMed] [Google Scholar]
- 9.Boire A, Brastianos PK, Garzia L, Valiente M. Brain metastasis. Nat. Rev. Cancer. 2020;20:4–11. doi: 10.1038/s41568-019-0220-y. [DOI] [PubMed] [Google Scholar]
- 10.Valiente M, et al. The evolving landscape of brain metastasis. Trends Cancer. 2018;4:176–196. doi: 10.1016/j.trecan.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousefi M, et al. Lung cancer-associated brain metastasis: Molecular mechanisms and therapeutic options. Cell Oncol. (Dordr) 2017;40:419–441. doi: 10.1007/s13402-017-0345-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Exploration of spatial distribution of brain metastasis from small cell lung cancer and identification of metastatic risk level of brain regions: A multicenter, retrospective study. Cancer Imaging. 2021;21:41. doi: 10.1186/s40644-021-00410-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blandin Knight S, et al. Progress and prospects of early detection in lung cancer. Open Biol. 2017 doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Wu J. Expression profiling of long noncoding RNAs in neonatal and adult mouse testis. Data Brief. 2015;4:322–327. doi: 10.1016/j.dib.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact. Mater. 2022;13:23–36. doi: 10.1016/j.bioactmat.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan AC. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC) Thorac. Cancer. 2020;11:511–518. doi: 10.1111/1759-7714.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2022;80:1–17. doi: 10.1016/j.semcancer.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Sanaei MJ, et al. The PI3K/Akt/mTOR axis in colorectal cancer: Oncogenic alterations, non-coding RNAs, therapeutic opportunities, and the emerging role of nanoparticles. J. Cell. Physiol. 2022;237:1720–1752. doi: 10.1002/jcp.30655. [DOI] [PubMed] [Google Scholar]
- 21.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bu D, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simillion C, Liechti R, Lischer HE, Ioannidis V, Bruggmann R. Avoiding the pitfalls of gene set enrichment analysis with SetRank. BMC Bioinform. 2017;18:151. doi: 10.1186/s12859-017-1571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuleshov MV, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou H, et al. Distinctive targetable genotypes of younger patients with lung adenocarcinoma: A cBioPortal for cancer genomics data base analysis. Cancer Biol. Ther. 2020;21:26–33. doi: 10.1080/15384047.2019.1665392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franz M, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook HV, Doncheva NT, Szklarczyk D, von Mering C, Jensen LJ. Viruses.STRING: A virus-host protein-protein interaction database. Viruses. 2018 doi: 10.3390/v10100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, et al. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49:D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanczky A, Gyorffy B. Web-Based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham, H. in Use R!, 1 online resource (XVI, 260 pages 232 illustrations, 140 illustrations in color (Springer International Publishing : Imprint: Springer,, Cham, 2016).
- 37.Lachmann A, et al. Geneshot: Search engine for ranking genes from arbitrary text queries. Nucleic Acids Res. 2019;47:W571–W577. doi: 10.1093/nar/gkz393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Mering C, et al. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 40.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Entezari M, et al. Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed. Pharmacother. 2022;150:112963. doi: 10.1016/j.biopha.2022.112963. [DOI] [PubMed] [Google Scholar]
- 42.Sun PR, Jia SZ, Lin H, Leng JH, Lang JH. Genome-wide profiling of long noncoding ribonucleic acid expression patterns in ovarian endometriosis by microarray. Fertil. Steril. 2014;101:1038–1046. doi: 10.1016/j.fertnstert.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Dang CV, et al. The c-Myc target gene network. Semin. Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 44.Meola J, et al. Differentially expressed genes in eutopic and ectopic endometrium of women with endometriosis. Fertil. Steril. 2010;93:1750–1773. doi: 10.1016/j.fertnstert.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 45.Lee HY, et al. c-MYC drives breast cancer metastasis to the brain, but promotes synthetic lethality with TRAIL. Mol. Cancer Res. 2019;17:544–554. doi: 10.1158/1541-7786.MCR-18-0630. [DOI] [PubMed] [Google Scholar]
- 46.Auxerre-Plantie E, et al. Identification of MYOM2 as a candidate gene in hypertrophic cardiomyopathy and Tetralogy of Fallot, and its functional evaluation in the Drosophila heart. Dis. Model Mech. 2020 doi: 10.1242/dmm.045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin TC, et al. Fibronectin in cancer: Friend or foe. Cells. 2019 doi: 10.3390/cells9010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, et al. IgG Galactosylation status combined with MYOM2-rs2294066 precisely predicts anti-TNF response in ankylosing spondylitis. Mol. Med. 2019;25:25. doi: 10.1186/s10020-019-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol. Sin. 2008;29:1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Chen W, Chen J, Pan Q, Pan J. LncRNA expression profiles of EGFR exon 19 deletions in lung adenocarcinoma ascertained by using microarray analysis. Med. Oncol. 2014;31:137. doi: 10.1007/s12032-014-0137-y. [DOI] [PubMed] [Google Scholar]
- 51.Wang RT, Zhang Y, Yao SY, Tan XG. LINC00501 inhibits the growth and metastasis of lung cancer by mediating miR-129-5p/HMGB1. Onco. Targets Ther. 2020;13:7137–7149. doi: 10.2147/OTT.S254735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang LX, et al. Novel long non-coding RNA LBX2-AS1 indicates poor prognosis and promotes cell proliferation and metastasis through Notch signaling in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:7419–7429. doi: 10.26355/eurrev_201909_18851. [DOI] [PubMed] [Google Scholar]
- 53.Miao Qiuju XX. Role of transmembrane proteins in malignant tumors. J. Int. Oncol. 2020;47:355–359. doi: 10.3760/cma.j.cn371439-20191227-00035. [DOI] [Google Scholar]
- 54.Marx S, et al. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin. Cancer Biol. 2020;60:96–106. doi: 10.1016/j.semcancer.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 55.Li H, et al. Epigenetic silencing of TMEM176A activates ERK signaling in human hepatocellular carcinoma. Clin Epigenet. 2018;10:137. doi: 10.1186/s13148-018-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J, et al. TMEM206 promotes the malignancy of colorectal cancer cells by interacting with AKT and extracellular signal-regulated kinase signaling pathways. J. Cell Physiol. 2019;234:10888–10898. doi: 10.1002/jcp.27751. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, et al. Potential targets of TMEM176A in the growth of glioblastoma cells. Onco Targets Ther. 2018;11:7763–7775. doi: 10.2147/OTT.S179725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cacabelos R. Pharmacoepigenetics. Academic Press; 2019. [Google Scholar]
- 59.Ustinova M, et al. Whole-blood transcriptome profiling reveals signatures of metformin and its therapeutic response. PLoS ONE. 2020;15:e0237400. doi: 10.1371/journal.pone.0237400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segovia M, Russo S, Girotti MR, Rabinovich GA, Hill M. Role of inflammasome activation in tumor immunity triggered by immune checkpoint blockers. Clin. Exp. Immunol. 2020;200:155–162. doi: 10.1111/cei.13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai P, et al. gimap5 inhibits lung cancer growth by interacting with M6PR. Front. Oncol. 2021;11:699847. doi: 10.3389/fonc.2021.699847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng X, et al. Upregulation of phosphatidylinositol glycan anchor biosynthesis class C is associated with unfavorable survival prognosis in patients with hepatocellular carcinoma. Oncol. Lett. 2021;21:237. doi: 10.3892/ol.2021.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, et al. Identification of the transcriptional networks and the involvement in Angiotensin II-induced injury after CRISPR/Cas9-mediated knockdown of Cyr61 in HEK293T cells. Mediat. Inflamm. 2019;2019:8697257. doi: 10.1155/2019/8697257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghanavat M, et al. Digging deeper through glucose metabolism and its regulators in cancer and metastasis. Life Sci. 2021;264:118603. doi: 10.1016/j.lfs.2020.118603. [DOI] [PubMed] [Google Scholar]
- 65.Domenichini A, Adamska A, Falasca M. ABC transporters as cancer drivers: Potential functions in cancer development. Biochim. Biophys. Acta Gen. Subj. 1863;52–60:2019. doi: 10.1016/j.bbagen.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 66.Yu G, et al. Organelle-derived acetyl-CoA promotes prostate cancer cell survival, migration, and metastasis via activation of calmodulin kinase II. Cancer Res. 2018;78:2490–2502. doi: 10.1158/0008-5472.CAN-17-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q, et al. Exosomes containing miR-122-5p secreted by LPS-induced neutrophils regulate the apoptosis and permeability of brain microvascular endothelial cells by targeting OCLN. Am. J. Transl. Res. 2021;13:4167–4181. [PMC free article] [PubMed] [Google Scholar]
- 68.Ma SC, et al. CLDN5 affects lncRNAs acting as ceRNA dynamics contributing to regulating bloodbrain barrier permeability in tumor brain metastasis. Oncol. Rep. 2018;39:1441–1453. doi: 10.3892/or.2018.6208. [DOI] [PubMed] [Google Scholar]
- 69.Shin SB, Jang HR, Xu R, Won JY, Yim H. Active PLK1-driven metastasis is amplified by TGF-beta signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2020;39:767–785. doi: 10.1038/s41388-019-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zalewska-Ziob M, et al. Activity of antioxidant enzymes in the tumor and adjacent noncancerous tissues of non-small-cell lung cancer. Oxid. Med. Cell Longev. 2019;2019:2901840. doi: 10.1155/2019/2901840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glorieux C, et al. Evaluation of potential mechanisms controlling the catalase expression in breast cancer cells. Oxid. Med. Cell Longev. 2018;2018:5351967. doi: 10.1155/2018/5351967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Marzo N, Chisci E, Giovannoni R. the role of hydrogen peroxide in redox-dependent signaling: Homeostatic and pathological responses in mammalian cells. Cells. 2018 doi: 10.3390/cells7100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ito S, et al. Amyloid precursor protein and its phosphorylated form in non-small cell lung carcinoma. Pathol. Res. Pract. 2019;215:152463. doi: 10.1016/j.prp.2019.152463. [DOI] [PubMed] [Google Scholar]
- 74.Asiedu MK, et al. Pathways impacted by genomic alterations in pulmonary carcinoid tumors. Clin. Cancer Res. 2018;24:1691–1704. doi: 10.1158/1078-0432.CCR-17-0252. [DOI] [PubMed] [Google Scholar]
- 75.Guo Y, Wang Q, Chen S, Xu C. Functions of amyloid precursor protein in metabolic diseases. Metabolism. 2021;115:154454. doi: 10.1016/j.metabol.2020.154454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression data was gathered from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) (ACCESSION NUMBER: GSE161968) and enrich analysis performed utilising KEGG (https://www.genome.jp/kegg/), GO (http://geneontology.org/), and Kaplan–Meier survival analysis (https://kmplot.com/analysis/) databases. The analysed data and R codes are available to the corresponding author with no restriction upon request.