Abstract
Protein S-nitrosylation is known to regulate enzymatic function. Here, we report that nitric oxide (NO)-related species can contribute to Alzheimer’s disease (AD) by S-nitrosylating the lysosomal protease cathepsin B (forming SNO-CTSB), thereby inhibiting CTSB activity. This posttranslational modification inhibited autophagic flux, increased autolysosomal vesicles, and led to accumulation of protein aggregates. CA-074Me, a CTSB chemical inhibitor, also inhibited autophagic flux and resulted in accumulation of protein aggregates similar to the effect of SNO-CTSB. Inhibition of CTSB activity also induced caspase-dependent neuronal apoptosis in mouse cerebrocortical cultures. To examine which cysteine residue(s) in CTSB are S-nitrosylated, we mutated candidate cysteines and found that three cysteines were susceptible to S-nitrosylation. Finally, we observed an increase in SNO-CTSB in both 5XFAD transgenic mouse and flash-frozen postmortem human AD brains. These results suggest that S-nitrosylation of CTSB inhibits enzymatic activity, blocks autophagic flux, and thus contributes to AD pathogenesis.
Subject terms: Neural ageing, Neuroscience, Macroautophagy
Introduction
It is well known that inhibition of autophagic flux and the ubiquitin-proteasome system (UPS) occurs in brains manifesting neurodegenerative diseases, contributing to the accumulation of abnormal protein aggregates, including amyloid-β peptide (Aβ), phospho-tau, α-synuclein, and mutant huntingtin, as found in Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD), respectively [1–7]. The UPS degrades normal proteins targeted for disposal, while autophagy removes defective organelles, e.g., during starvation [8, 9]. The UPS and autophagy are essential for protein quality control. UPS activity often becomes inadequate in neurodegenerative diseases, leading to autophagy activation in an attempt to remove abnormal proteins, especially aggregated forms [10, 11]. As macroautophagy (hereafter termed autophagy) is initiated, an isolated double-membrane structure called a phagophore forms, and then elongates and engulfs a portion of the cytoplasm to form a vesicle autophagosome (AP). The AP fuses with a lysosome to produce an autolysosome (AL) in which acidic lysosomal proteases digest the engulfed contents [9]. Autophagy is particularly important to neuronal maintenance. Mice deficient in autophagy machinery exhibit neuronal accumulation of aggregate-prone proteins and neurodegeneration, demonstrating the critical role of autophagy in neurons [12, 13]. Moreover, the lysosome has emerged as an important organelle in neurodegenerative diseases. For example, most lysosomal storage disorders (LSD) cause progressive neurodegeneration. Although all cell types are affected by genetic defects in LSD, neurons are particularly susceptible [14–16]. One reason neurons appear to be so vulnerable to lysosomal dysfunction may be explained by the fact that neurons cannot divide and thus dilute out or eliminate accumulated waste [17].
Reactive nitrogen species (RNS) such as nitric oxide (NO) are known to contribute to neurodegenerative diseases, including AD and PD [18–22]. NO is a ubiquitous cellular messenger and synthesized from l-arginine by a family of NO synthases (NOS). The function of many proteins can be regulated by NO-related species via S-nitrosylation of critical cysteine residues; one reaction mechanism for this likely involves thiolate anion (RS-) of a target cysteine residue, as a nucleophile, performing a reversible nucleophilic attack on a nitroso nitrogen to form an S-nitroso (SNO)-protein adduct [23]. Under physiological conditions, S-nitrosylation represents an important posttranslational modification, in some ways akin to phosphorylation. In contrast, aberrant S-nitrosylation reactions, engendered by excessive RNS, can induce protein misfolding, mitochondrial fragmentation, synaptic dysfunction, apoptosis, and autophagy inhibition [24–30]. In autophagic processes, RNS have previously been shown to act on the c-Jun N-terminal kinase 1 (JNK1)/Bcl-2/Beclin 1 and the IκB kinase (IΚΚ)/AMPK/mTORC cascades [31]. Inhibition of JNK1 by RNS reduces Bcl-2 phosphorylation and subsequently increases Bcl-2/Beclin 1 complex formation, resulting in attenuation in the initiation of autophagy. Additionally, RNS inhibit IKK, reduce AMPK phosphorylation, and activate mTORC1 in sequential order, which inhibits the initiation of autophagy. In fact, RNS have been reported to impair the initiation of autophagy via inhibition of JNK1 and IKK in models of neurodegenerative disorders [31]. Further linking autophagy to neurodegenerative diseases, several pieces of evidence are accumulating that autophagic flux or lysosomal function that are downstream from initiation are also critical points of potential intervention to affect the autophagic process [32, 33]. Therefore, in the present study, we investigated the mechanistic role of RNS on autophagic flux and lysosomal function.
Materials and methods
Cultures of mouse cerebrocortical neurons, H4 and HEK cells
Mouse cortical cultures were prepared from embryonic day 13–14 mice. Dissociated cortical cells were plated onto poly-l-lysine (Sigma-Aldrich, St. Louis, MO, USA) coated plates (SPL life sciences, Pocheon, South Korea), ten hemispheres per 6-well plate, in plating medium [Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Waltham, MA, USA) with 20 mM glucose, 40 mM sodium bicarbonate, 2 mM glutamine, 5% fetal bovine serum (FBS), and 5% horse serum (HS)] [34]. All experiments were performed at days in vitro (DIV) 10-11. H4 glioma cells, H4 cells stably expressing GFP-LC3 (GL-H4), H4 cells stably expressing RFP-GFP-LC3 (RGL-H4), and HEK cells stably expressing neuronal NOS (nNOS-HEK) were maintained in growth medium [DMEM (Gibco) with 4 mM glutamine, 10% FBS (Welgene, Gyeongsan, South Korea), 100 unit/ml penicillin, and 100 μg/ml streptomycin (Welgene)] at 37 °C in a 5% CO2 atmosphere. In the case of nNOS-HEK cells, 250 μg/ml geneticin was treated to select nNOS-overexpressing cells.
DNA constructs and transient transfection
HEK293 cells were transfected at 70–80% confluence with mutant huntingtin (mHtt)-GFP, α-synuclein (A53T)-GFP, tau(P301L)-GFP, Aβ1-42-GFP, wild-type (WT)-cathepsin B (CTSB), or mutant CTSB (C29A, C240A, Cp42A, C29A/C240A, Cp42A/C29A, Cp42A/C240A, Cp42A/C29A/C240A) DNA using a lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The expression plasmid for pEGFP-C1-mHtt-GFP was a generous gift from Dr. David C Rubinsztein (University of Cambridge, Cambridge, UK), and tau(P301L)-GFP plasmid was obtained from Dr. Jung-Jin Hwang (University of Ulsan College of Medicine, Seoul, South Korea). The pEGFP-C1-Aβ1-42 plasmid was purchased from Addgene (Cambridge, MA, USA). CTSB point mutation was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit, following the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA).
Synthesis of S-nitrosocysteine (SNOC)
For the fresh 100 mM SNOC solution, 12.1 mg of L-cysteine (Sigma-Aldrich) and 6.9 mg of sodium nitrite (Sigma-Aldrich) were introduced into 950 μl of distilled water and incubated in the ice for 5 min. And then, 50 μl HCl was added into the SNOC solution, used within 1 min. For the ‘old’-SNOC (o-SNOC) solution, we used the same solution, but it was made at least 24 h before the treatment [35].
Exposure to drugs
Before drug treatment, cortical neuronal cultures at DIV10-11 were washed with serum-free medium [minimum essential medium (MEM, Gibco)] and H4, GL-H4, or nNOS-HEK cells were washed with 1% FBS in MEM (low-serum medium). We used 200 μM SNOC, 100 nM Rapamycin (Calbiochem, San Diego, CA, USA), 100 nM Bafilomycin-A1 (Baf-A1, Enzo, Farmingdalem NY, USA), 10 μM 4-bromo A-23187 (A23187; Invitrogen), 20 μM CA074 methyl ester (CA-074Me, Calbiochem), 10 μM 7-nitroindazole (7-NI, Sigma-Aldrich), 100 μM 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox, Merck Millipore, Burlington, MA, USA), 1 mg/ml cycloheximide (CHX, Sigma-Aldrich), or 100 μM z-VAD-FMK (zVAD, MPbio, Solon, OH, USA).
Cell death assays
Cell death was detected by propidium iodide (PI, Sigma-Aldrich) or Hoechst-33258 staining. For PI staining, 2.5 μg/ml PI dye was added directly to the medium at 18 h after CA-074Me treatment. After 5 min later, cultures were washed with fresh MEM to remove the excessive PI dye.
To evaluate apoptotic cell death, cultured neurons were stained with 2 μg/ml Hoechst-33258 for 10 min, and cells exhibiting condensed chromatin nuclei were considered apoptotic cell death. Stained cells were analyzed using a digital inverted fluorescence microscope (EVOS, Waltham, MA, USA). We quantified lactate dehydrogenase (LDH) release into bathing medium to monitor both necrosis and apoptosis (with subsequent secondary necrosis and consequent LDH release), as we have previously described after exposure to NO [36, 37].
Western blot assays
Cell lysates were prepared in RIPA lysis buffer (50 mM Tris; pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Na-Doc, 0.1% SDS, 5 mM EDTA) with freshly prepared protein protease and phosphatase inhibitor (2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mM phenyl-methylsulfonyl fluoride, 1 mM Na3VO4, 5 mM NaF and 10 mM Na4P2O7). Thirty micrograms of total protein were separated by SDS-PAGE (10-15%) under reducing conditions and immunoblotted with the appropriate antibody [anti-GFP (Santa Cruz Biotechnologies, Dallas, TX, USA, sc-8334), anti-LC3 (Novus, NB100-2220), anti-SQSTM1 (p62, Santa Cruz Biotechnologies, sc-28359), anti-caspase-3 (Cell Signaling Technology, Danvers, MA, USA, #9662), anti-cleaved caspase-3 (Cell Signaling Technology, #9661), anti-tau (Cusabio, Houston, TX, USA, CSB-PA004235), anti-p-tau S404 (Cusabio, CSB-PA963344), anti-p-tau S214 (Signalway Antibody, Baltimore, MD, USA, #11109), or anti-actin (Abbkine, Wuhan, China, A01011)].
For western blot of protein aggregates, cell lysates were prepared using lysis buffer (50 mM Tris-HCl; pH7.5, 100 mM NaCl, 3 mM EDTA, 0.5% Triton X-100) with freshly prepared protein protease and phosphatase inhibitor. A total of thirty microgram protein was separated by SDS-PAGE (8–10%) under reducing condition and immunoblotted with the appropriate antibody [anti-GFP, anti-polyglutamine (Merck Millipore, MAB1574), anti-α-synuclein (BD Bioscience, Franklin Lakes, NJ, USA, 610787), anti-Aβ (6E10, BioLegend, San Diego, CA, USA, 803001), or anti-actin (Abbkine, Wuhan, China, A01011)]. Actin was used as a loading control. Normalized band intensity was quantified using Image J software. For quantitative analysis of SNO-CTSB, bands on the biotin-switch immunoblots were normalized against corresponding input control bands, as previously described [38–40]. Full and uncropped western blots are shown in Supplementary Fig. 3.
Biotin-switch assays
The detection of S-nitrosylation modification in proteins was performed with the biotin-switch assay [41]. In general, 500 μg of protein were used in biotin-switch assays and separated by SDS-PAGE (12%). To detect protein bands, we used their cognate antibodies [anti-CTSB (Cell Signaling Technology, 31718 S), anti-cathepsin L (CTSL; Abcam, Cambridge, UK, ab6314), anti-cathepsin D (CTSD; Santa Cruz Biotechnologies, sc-10725), or anti-V5 (Invitrogen, 46-1157)].
Fluorescence microscopy and confocal imaging
mHtt-GFP or α-synuclein (A53T)-GFP was transiently transfected into HEK cells. After SNOC exposure or drug treatment, cells were fixed in 4% paraformaldehyde at 4 °C for 15 min. Microscopic images were observed under a fluorescence microscope (EVOS cell imaging system). For quantification of mHtt-GFP puncta, the intensity of GFP fluorescence in the microscopic field was measured using Image J software.
For the confocal images, GL-H4 or RGL-H4 cells were cultured on cover glass (circle, 12 mm), and the fluorescence signal was observed under a laser scanning microscope (Carl Zeiss, Baden-Würrttemberg, Germany). For quantification of autophagic vesicles, GFP-LC3, or RFP-GFP-LC3 dots were counted from more than 30 cells in every experiment and repeated at least three independent experiments.
Cathepsin enzymatic activity assays
Cell lysates were prepared in lysis buffer (400 mM sodium phosphate; pH 6.0, 75 mM NaCl, 4 mM EDTA, and 0.25% Triton X-100). Forty micrograms of protein were used for the CTSB activity assay in reaction buffer (352 mM potassium phosphate buffer; pH 6.0, 48 mM sodium phosphate, and 4 mM EDTA). To detect the enzymatic activity of CTSB, the fluorescence signal was measured using a fluorometer (Gemini EM, Molecular Devices, San Jose, CA, USA) after cleavage of the specific substrate for CTSB, Z-RR-AMC (Calbiochem) in 0.1% Brij 35 (Sigma-Aldrich) solution. We used the Sensolyte®520 cathepsin D assay kit (Fluorimetric, Anaspec, Fremont, CA, USA) to assess CTSD activity and the InnoZyme™ Cathepsin L activity kit (Fluorogenic, Calbiochem) for detection of CTSL activity.
Mouse and human AD patient brain samples
5XFAD female mice (B6SJL-Tg[APPSwFlLon, PSEN1*M146L*L286V]6799Vas/Mmjax) were obtained from Dr. Inhee Mook-Jung (Seoul National University, Seoul, South Korea) and used for experiments at six-months of age. Age and sex matched samples were randomly chosen and assigned to different experimental groups. All animal experimental procedures were approved by the Animal Care and Use Committee of Sejong University and were conducted following the guidelines of the Care and Use of Laboratory Animals. Human brain samples were analyzed with institutional permission under the state of California and NIH guidelines. Informed consent was obtained according to procedures approved by Institutional Review Boards at the University of California, San Diego, School of Medicine, and The Scripps Research Institute.
Statistical analysis
All statistical comparisons were performed by two-tailed Student’s t-test for single comparisons or ANOVA with an appropriate post-hoc test for multiple comparisons as indicated in the figure legend. A p value of less than 0.05 was considered statistically significant. A Power Analysis of our prior data was used to determine the number of replicates needed for statistical purposes. Data analyses were performed in a blinded fashion, and no data were excluded.
Results
RNS inhibit autophagic flux, resulting in accumulation of protein aggregates
We used SNOC as a physiological NO donor/transnitrosylation agent versus o-SNOC, from which NO had been dissipated, as a negative control. To activate autophagy, we substituted culture medium containing 10% FBS for low-serum medium with 1% FBS (Supplementary Fig. 1). Initially, we observed that SNOC affected the number and size of autophagic vesicles (AV) in H4 glioma cells stably expressing GFP-LC3 (GL-H4 cells). After 2 h, the number and size of GFP-LC3 puncta, reflecting AV, were diminished by SNOC compared to control (Fig. 1A, B). This result is consistent with prior studies that showed NO inhibits AP formation [31]. However, at later time points, e.g., 8 h after exposure to SNOC, both the number and size of GFP-LC3 puncta were markedly increased (Fig. 1A, B). Next, to verify whether the increase in GFP-LC3 puncta is a result of inhibition rather than induction of autophagy, we added SNOC to cells after a 2-h incubation in low-serum medium. In this set of experiments, we also consistently found increased levels of LC3-II on immunoblots at 8 h after exposure to SNOC (Fig. 1C). Since we observed that RNS attenuated the initiation of autophagy, it is unlikely that AV accumulation results from promoting autophagy. Rather, we considered that RNS might block a critical step during autophagic flux. In support of this, we observed that p62 was maintained in immunoblots after SNOC exposure (Fig. 1C). Here, since we activated autophagy using low-serum medium prior to exposure to SNOC, the maintenance of the p62 level implied that SNOC inhibited p62 degradation. Therefore, we hypothesized that RNS play a critical role not only in the initial stage of autophagy but also in the final decomposition process, thereby inhibiting autophagic flux.
Fig. 1. RNS suppresses autophagic flux.
A Fluorescence confocal microscope image of GL-H4 (H4 glioma stably expressing GFP-LC3) cells 2 or 8 h after sham wash with low-serum medium (control, CTRL), or after exposure to S-nitrosocysteine (SNOC, 200 μM) or old SNOC (o-SNOC, 200 μM). Scale bar, 100 μm. B Quantitative analysis of GFP-LC3 puncta observed at 2 h or 8 h after exposure to CTRL, SNOC, or o-SNOC. Quantification of mean number (left) and size (right) of GFP-LC3 puncta per cell (mean ± SEM, n = 10 different fields taken from ≥4 independent biological replicate experiments). ***p < 0.001 by ANOVA with Bonferroni correction for multiple comparisons. C Western blot analysis for endogenous LC3 and p62 in GL-H4 cells. Protein samples were prepared at the indicated time points after exposure to sham wash (CTRL), SNOC, or o-SNOC. To activate autophagy, cultures were incubated in low-serum medium (1% FBS in MEM) for 2 h prior to SNOC exposure. For lanes 2, 3, 5, and 6, protein samples were extracted immediately after exposure to SNOC or o-SNOC. Actin was used for loading control. Quantification of conversion to LC3-II (left) and expression level of p62 (right) at the indicated time points was made in ≥3 independent biological replicate experiments. *p < 0.05, **p < 0.01, or ***p < 0.001 vs. sham wash control at baseline, and #p < 0.05, or ###p < 0.001 vs. corresponding SNOC group by ANOVA with post-hoc Dunnett’s test. D Fluorescence confocal microscope images of H4 cells stably expressing RFP-GFP-LC3 (RGL-H4 cells) 12 h after sham wash, or exposure to rapamycin (Rapa, 100 nM) or SNOC in the presence or absence of Bafilomycin A1 (Baf-A1, 100 nM). Scale bar, 20 μm. E Quantitative analysis of RFP-GFP-LC3 puncta observed 12 h after exposure to Rapa or SNOC in the presence or absence of Baf-A1. Quantification of the number of red-only or yellow puncta per cell (mean ± SEM, n = 9 different fields taken from ≥3 independent biological replicate experiments). ***p < 0.001 vs. red-only puncta of sham control, #p < 0.05, or ###p < 0.001 vs. yellow puncta of sham control, †††p < 0.001 vs. red-only puncta of Rapa or SNOC alone, and §§§p < 0.001 vs. yellow puncta of rapamycin or SNOC alone by ANOVA with post-hoc Dunnett’s test. F Western blot analysis for endogenous LC3 and p62 in RGL-H4 cells. Protein samples were prepared 12 h after the induction of autophagy triggered by incubation in low-serum medium. Quantification of conversion to LC3-II (left) and expression levels of p62 (right) from ≥3 independent biological replicate experiments. *p < 0.05, **p < 0.01 or ***p < 0.001 vs. sham wash control, and #p < 0.05, or ##p < 0.01 vs. Rapa or SNOC alone. NS: not significant by ANOVA with post-hoc Dunnett’s test.
To verify the inhibitory effect of RNS on autophagic flux, we used H4 glioma cells stably expressing RFP-GFP-LC3 (RGL-H4 cells) [42]. Whereas GFP (green) fluorescence is rapidly quenched in acidic compartments as found in lysosomes, RFP (red) fluorescence is more stable under these conditions. Therefore, the presence of yellow dots (merged green and red fluorescence) signifies AP, while ‘red-only’ dots imply AL fusion between AP and lysosomes. We found that exposure to RNS significantly enhanced the number of AV, including both yellow dots and red-only dots (Fig. 1D and third bar of Fig. 1E). To confirm that RNS blocked autophagic flux, we next used Baf-A1, a chemical inhibitor of the v-ATPase. Since Baf-A1 blocks autophagic flux by inhibiting fusion between AP and lysosomes, AP, fluorescing yellow, accumulate and AL, exhibiting red, disappear after Baf-A1 exposure (Fig. 1D and fourth bar of Fig. 1E). Rapamycin, an inducer of autophagy, significantly increased AV, and co-treatment with Baf-A1 resulted in further accumulation of yellow dots (Fig. 1D and second and fifth bars of Fig. 1E). However, co-exposure to SNOC and Baf-A1 did not result in accumulation of AP (yellow) or AL (red) (Fig. 1D and fourth vs. sixth bars of Fig. 1E). This result suggests that the RNS-induced increase in AV was due to inhibition of the autophagic process at or downstream of AL formation rather than activation of the initial autophagic process. Supporting this premise, we found that SNOC failed to further increase expression of LC3-II and p62 when autophagy was blocked by Baf-A1 (Fig. 1F, lanes 2 and 6). Taken together with prior evidence, RNS may inhibit both the initiation of autophagy and later phases of autophagy, the latter leading to accumulation of AL with the blockade of autophagic flux.
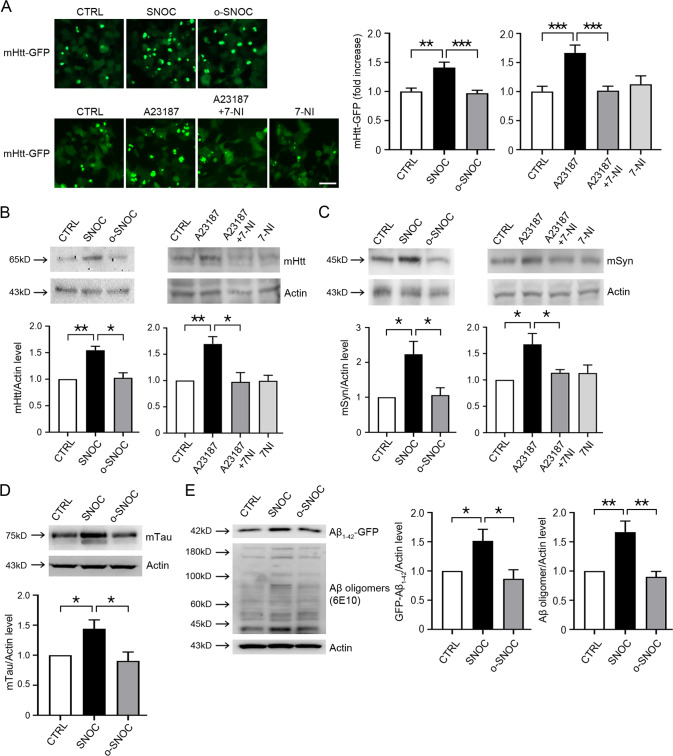
Next, we examined whether RNS led to accumulation of protein aggregates. For this purpose, we transiently transfected mHtt-GFP plasmid into nNOS-HEK cells. In this assay, we used HEK cells here because of their relatively higher transfection efficiency compared to H4 glioma cells. Exposure to SNOC but not o-SNOC markedly increased mHtt-GFP protein aggregates (Fig. 2A). Moreover, addition of A23187, a Ca2+ ionophore that activates nNOS and thus increases NO/RNS, resulted in accumulation of protein aggregates, which was reversed by 7-NI, a NOS inhibitor (Fig. 2A). To confirm whether RNS result in the accumulation of mutant protein, we also performed western blot analysis after transient transfection with mHtt-GFP, mutant A53T α-synuclein (mSyn), or mutant P301L Tau (mTau). The A53T mutation of SNCA (the gene encoding mSyn) and the P301L mutation of Tau are known to be associated with PD and AD, respectively. We observed that exposure to SNOC increased the protein levels of mHtt, mSyn, and mTau (Fig. 2B–D). We also transiently transfected amyloid-β (GFP-Aβ1-42) plasmids into HEK cells, and observed that exposure to SNOC increased the protein levels of GFP-Aβ1-42 (Fig. 2E). In GFP-Aβ1-42 overexpressing cells, we also detected an increase in Aβ oligomers (Fig. 2E, 6E10). These findings are consistent with the notion that RNS inhibit autophagic flux during the later stages of autophagy.
Fig. 2. RNS augment protein aggregates.
A Fluorescence microscope image of GFP-mHtt aggregates in nNOS-HEK cells. GFP-mHtt plasmid was transiently transfected into nNOS-HEK cells two days before drug treatment. Cells were evaluated 2 h after exposure to 200 μM SNOC or o-SNOC, or 7 h after addition of 10 μM A23187 alone or A23187 plus 10 μM 7-Nitroindazole (7-NI). Scale bar, 50 μm. Bar graphs represent quantitative analysis of GFP-mHtt fluorescence intensity. The mean intensity of GFP-mHtt fluorescence was quantified in a given microscopic field using Image J software (mean ± SEM, n = 8 different fields taken from ≥3 independent biological replicate experiments). **p < 0.01 or ***p < 0.001 by ANOVA with Dunnett’s test for post-hoc analysis. B Western blot analysis for accumulation of mHtt. GFP-mHtt plasmid was transiently transfected into nNOS-HEK cells two days before drug treatment. Protein samples were prepared 2 h after sham wash (CTRL) or exposure to SNOC or o-SNOC, or 7 h after exposure to A23187 alone or A23187 plus 7-NI. Quantification of the GFP-mHtt level from ≥3 independent biological replicate experiments. *p < 0.05 or **p < 0.01 by ANOVA with Dunnett’s test for post-hoc analysis. C Western blot analysis for accumulation of mutant α-synuclein (mSyn). A plasmid encoding mt-α-synuclein was transiently transfected into nNOS-HEK cells two days before drug treatment. Protein samples were prepared 2 h after sham wash (CTRL) or exposure to SNOC or o-SNOC, or 7 h after exposure to A23187 alone or A23187 plus 7-NI. Quantification of α-Syn level from ≥3 independent biological replicate experiments. *p < 0.05 by ANOVA with Dunnett’s test for post-hoc analysis. D Western blot analysis of accumulation of mutant tau(P301L). Tau P301L-GFP plasmid was transiently transfected into HEK cells one day prior to exposure to SNOC. Protein samples were prepared 2 h after sham wash (CTRL), or exposure to 200 μM SNOC or o-SNOC. Quantification of tau P301L-GFP from ≥3 biological replicate experiments, *p < 0.05 by ANOVA with Dunnett’s test for post-hoc analysis. E Western blot analysis for an accumulation of Aβ1-42. Aβ1-42-GFP plasmid was transiently transfected into HEK cells two days prior to SNOC exposure. Protein samples were prepared 2 h after sham wash (CTRL), or exposure to SNOC or o-SNOC. Quantification of Aβ1-42-GFP or Aβ1-42 oligomers (6E10) from ≥ 3 biological replicate experiments, *p < 0.05 or **p < 0.01 by ANOVA with Dunnett’s test for post-hoc analysis. Oligomers were quantified by densitometry of multiple bands.
S-Nitrosylation of CTSB inhibits its protease activity
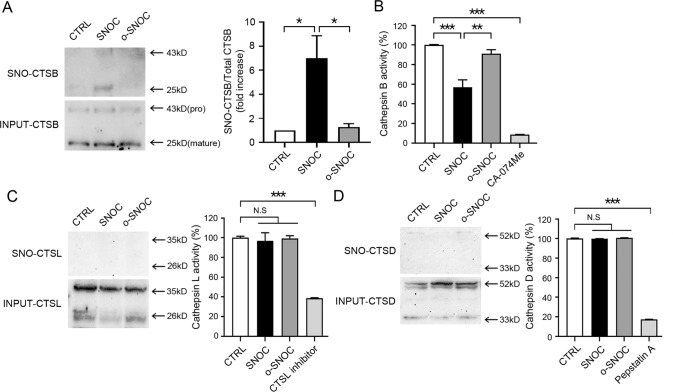
Given that RNS induced accumulation of aggregated proteins, we next examined the possibility that lysosomal function may be regulated by RNS. Previous studies had suggested that RNS could result in inhibition of cathepsin K (CTSK), a lysosomal protease, thus attenuating CTSK activity, although formation of a glutathione or sulfenic acid adduct was thought to be responsible for this action [43]. More relevant to our study, S-nitrosylation was reported to inhibit CTSB activity in vitro [44, 45], and a non-catalytic cysteine residue had been identified as a target of S-nitrosylation in CTSD in a proteomic screen [46]. To show that RNS regulate lysosomal protease activity in our paradigm, we studied the effect of SNOC on representative lysosomal proteases, including CTSD, CTSB, and CTSL because these cathepsins are thought to be primarily responsible lysosomal protease activity in neuronal cells [47, 48]. We found that under our conditions among this group of enzymes, only CTSB could be S-nitrosylated (Fig. 3A and Supplementary Fig. 2). Furthermore, CTSB enzymatic activity, but not that of CTSL or CTSD, was significantly inhibited by SNOC (Fig. 3B–D). These results are consistent with the notion that RNS can regulate CTSB protease activity via S-nitrosylation.
Fig. 3. SNOC regulates cathepsin B activity by S-nitrosylation.
A Biotin-switch assay for detection of S-nitrosylated cathepsin B (SNO-CTSB) in HEK293 cells. Protein samples were prepared 45 min after sham wash control (CTRL) or exposure to 200 μM SNOC or o-SNOC. The upper bands (43 kD) represent the pro-form of CTSB, and the lower bands (25 kD), the mature form of CTSB. The ratio of SNO-CTSB (mature form) to total input CTSB was quantified as a fold increase in ≥3 independent biological replicate experiments *p < 0.05 by ANOVA with Dunnett’s test for post-hoc analysis. B Enzyme activity assay of CTSB in HEK cells. Protein samples were prepared 45 min after sham wash control or exposure to SNOC or o-SNOC (n = 3, *p < 0.05, **p < 0.01 or ***p < 0.001 by ANOVA with Dunnett’s test for post-hoc analysis. The specific inhibitor of CTSB, CA-074Me, was used as a control. C Biotin-switch assay for detection of S-nitrosylated cathepsin L (SNO-CTSL) (left) and enzyme activity assay of CTSL (right) in HEK cells. Protein samples were prepared 45 min after sham wash control (CTRL) or exposure to SNOC or o-SNOC (n = 3, ***p < 0.001 compared to sham wash control by ANOVA with Dunnett’s test for post-hoc analysis). Specific inhibitors for CTSL were used as controls. D Biotin-switch assay for detection of S-nitrosylated cathepsin D (SNO-CTSD) (left) and enzyme activity assay of CTSD (right) in HEK cells. Protein samples were prepared 45 min after sham wash control (CTRL) or exposure to SNOC or o-SNOC (n = 3, ***p < 0.001 by ANOVA with Dunnett’s test for post-hoc analysis). Specific inhibitor for CTSD (pepstatin A) was used as control.
Chemical inhibition of CTSB mimics RNS-induced blockade of autophagic flux and resultant accumulation of protein aggregates
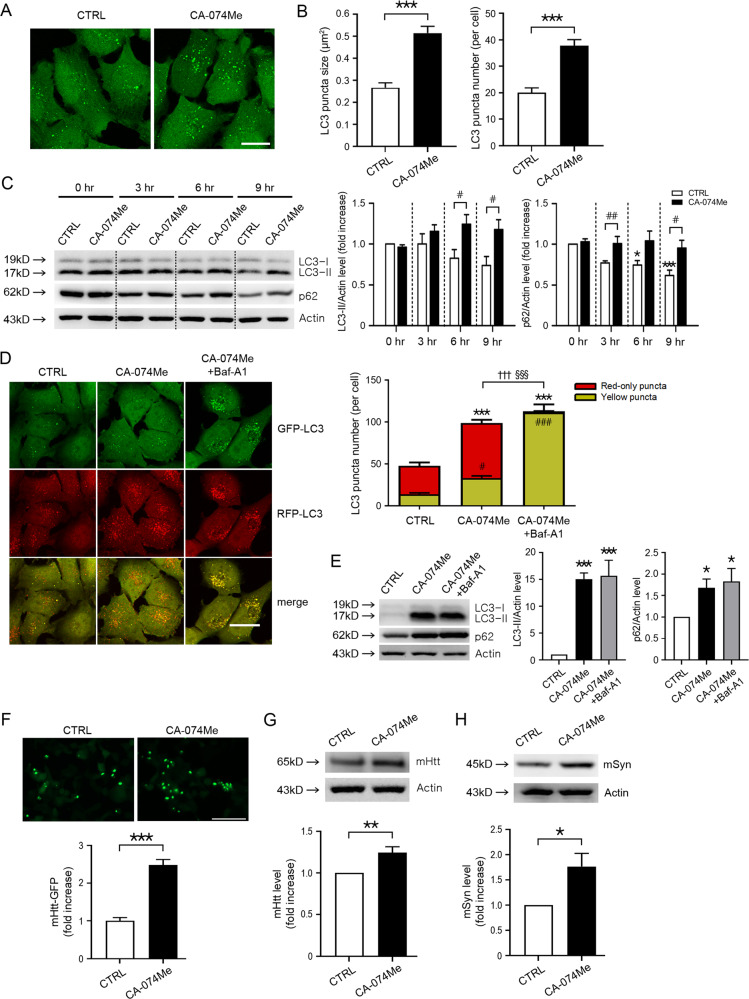
Since we found that SNOC suppressed autophagic flux and decreased CTSB enzymatic activity, we tested whether chemical inhibition of CTSB could mimic the effect of RNS-induced inhibition of autophagic flux. We observed that CA-074 methyl ester (CA-074Me), a specific CTSB inhibitor, significantly increased the size and number of GFP-LC3 puncta compared to control in GL-H4 cells (Fig. 4A, B). Similar to the effect of SNOC, CA-074Me also increased LC3-II conversion, p62 levels (Fig. 4C), and LC3 puncta in RFP-GFP-LC3 overexpressing H4 cells (Fig. 4D). RFP-GFP-LC3 analyses further revealed that CA-074Me increased both yellow and red dots due to inhibition of lysosomal function, which occurs after fusion of AP and lysosomes. In contrast, inhibition of autophagic flux at the fusion stage using Baf-A1 significantly increased yellow APs in cells exposed to CA-074Me (Fig. 4D). Moreover, Baf-A1 did not further increase LC3-II and p62 levels in cells exposed to CA-074A (Fig. 4E). These results are consistent with the notion that Baf-A1 blocks autophagic flux upstream of CA-074Me-mediated lysosomal inhibition. Additionally, CA074-Me markedly increased mHtt-GFP protein aggregates (Fig. 4F). Exposure to CA-074Me also increased the level of mHtt and mSyn on immunoblots (Fig. 4G, H). Therefore, this series of experiments show that chemical inhibition of CTSB mimics the effect of RNS on autophagic flux and protein levels.
Fig. 4. The CTSB inhibitor, CA-074Me, inhibits autophagic flux mimicking the effect of RNS.
A Fluorescence confocal microscope image of GL-H4 cells 12 h after sham wash (CTRL) or exposure to 20 μM CA-074Me. Scale bar, 20 μm. B Quantitative analysis of GFP-LC3 puncta observed 12 h after CA-074Me treatment. Mean size (left) and number (right) of GFP-LC3 puncta per cell in a given microscopic field were quantified (mean ± SEM, n = 7 different fields taken from ≥4 independent biological replicate experiments). ***p < 0.001 by two-tailed Student’s t-test. C Western blot analysis of LC3, and p62 in GL-H4 cells. Protein samples were prepared at the indicated time points after sham wash (CTRL) or exposure to 20 μM CA-074Me. Quantification of LC3-II (left) and p62 (right) at the indicated time points in ≥ 3 independent biological replicate experiments. *p < 0.05 or ***p < 0.001 vs. 0 h sham wash control, and #p < 0.05 or ##p < 0.01 vs. sham wash control of the same time point by ANOVA with post-hoc Dunnett’s test. D Fluorescence confocal microscope image of RGL-H4 cells. CA-074Me exposure for 12 h. scale bar, 20 μm. Quantification of mean number of red-only or yellow puncta per cell in a given microscopic field (mean ± SEM, n = 9 different fields taken from ≥3 independent biological replicate experiments). ***p < 0.001 vs. red-only puncta of control, #p < 0.05 or ###p < 0.001 vs. yellow puncta of control, †††p < 0.001 vs. red-only puncta of CA-074Me, and §§§p < 0.001 vs. yellow puncta of CA-074Me by ANOVA with post-hoc Dunnett’s test. E Western blot analysis for endogenous LC3 and p62 in RGL-H4 cells. Protein samples were prepared 12 h after exposure to sham wash or CA-074Me in the presence or absence of Bafilomycin A1 (Baf-A1). Quantification of conversion to LC3-II (left) and expression level of p62 (right) at 12 h was made in ≥3 independent biological replicate experiments. *p < 0.05, or ***p < 0.001 vs. sham wash control by ANOVA with Dunnett’s test for post-hoc analysis. F Fluorescence microscope image of GFP-mHtt aggregates in nNOS-HEK cells. nNOS-HEK cells transiently transfected with GFP-mHtt plasmid were exposed to 20 μM CA-074Me for 12 h. Scale bar, 200 μm. Mean intensity of GFP-mHtt in a given microscopic field was quantified using Image J software (mean ± SEM, n = 8 different photographic images in at least three independent experiments). ***p < 0.001 by two-tailed Student’s t-test. G, H Western blot analysis of accumulation of mHtt (G) and mt-α-synuclein (H). GFP-mHtt or mt-α-synuclein plasmid was transiently transfected into nNOS-HEK cells two days prior to drug treatment. Protein samples were prepared 12 h after sham wash (CTRL) or exposure to CA-074Me. Quantification of GFP-mHtt and mt-α-synuclein levels in ≥3 independent biological replicate experiments. *p < 0.05 or **P < 0.01 by two-tailed Student’s t-test.
Chemical inhibition of CTSB induces apoptotic neuronal cell death in mouse cerebrocortical cultures
Prior publications showed that cytoplasmic CTSB can activate caspase-3, inducing caspase-dependent apoptosis [49–51]. CTSB has also been reported to act as a candidate β-secretase, leading to an increase in amyloid-β peptide (Aβ1-40 or Aβ1-42) [52–54]. Since we found that SNOC inhibited CTSB and autophagic flux, we next examined whether inhibition of CTSB affects cell survival in mouse cortical neuronal cultures. Inhibition of CTSB with CA-074Me increased neuronal cell death (Fig. 5). zVAD, a pan-inhibitor of caspases, significantly attenuated CA-074Me-induced cell death (Fig. 5A and B). Additionally, prior studies have demonstrated that apoptotic pathways in post-mitotic cells such as neurons require de novo synthesis of pro-apoptotic proteins [55–57]. Accordingly, we found that CHX, a chemical inhibitor of protein synthesis, also markedly reduced cell death, whereas Trolox, an antioxidant, had no effect in this system (Fig. 5A, B). Moreover, activation of caspase-3 by CA-074Me was almost entirely blocked by CHX or zVAD but not by Trolox (Fig. 5C). Notably, the same concentration of SNOC that induced S-nitrosylation of CTSB and blocked autophagic flux (Figs. 1 and 3) also triggered neuronal cell death (Fig. 5D) [58–62]. Thus, these results are consistent with the notion that chemical or RNS-mediated inhibition of CTSB contributes to neuronal cell death.
Fig. 5. CTSB inhibition induces neuronal cell apoptosis in mouse cerebrocortical cultures.
A Microscopic images of propidium iodide (PI)-positive dead cells (upper) and Hoechst 33258-stained total nuclei (lower) in mouse cerebrocortical cultures 18 h after the addition of 20 µM CA-074Me in the presence or absence of 100 µM Trolox, 1 µg/ml cycloheximide (CHX), or 100 µM zVAD. CA-074Me markedly increased PI-positive dead cells and Hoechst stain revealed their condensed nuclei. Arrowheads indicate typical morphology of small, bright nuclei of apoptotic cells. The scale bar of PI images is 100 μm, and Hoechst 33258 is 25 μm. B Quantification of PI-positive dead cells (left), Hoechst 33258-stained condensed apoptotic nuclei (middle), and LDH release (right) 18 h after the addition of CA-074Me in the presence or absence of Trolox, cycloheximide, or zVAD to mouse cerebrocortical cultures (mean ± SEM, n = 3 cultures), *p < 0.05, **p < 0.01, or ***p < 0.001 by ANOVA with post-hoc Dunnett’s test. CHX and zVAD decreased nuclear pyknosis induced by CA-074Me, whereas Trolox did not affect CA-074Me-induced apoptotic neuronal cell death. C Western blot analysis for cleaved/active caspase-3 in mouse cerebrocortical cultures. Protein samples were prepared 18 h after sham wash (CTRL) or exposure to 20 μM CA-074Me in the presence or absence of Trolox, CHX, or zVAD. CA-074Me induced caspase-3 activation, which was markedly attenuated by CHX or zVAD, but not by Trolox. D Quantification of LDH release resulting from neuronal cell death 9 h after exposure to SNOC (mean ± SEM, n = 3 cultures), ***p < 0.001 by two-tailed Student’s t-test.
Three free cysteine residues in CTSB are candidate sites for S-nitrosylation
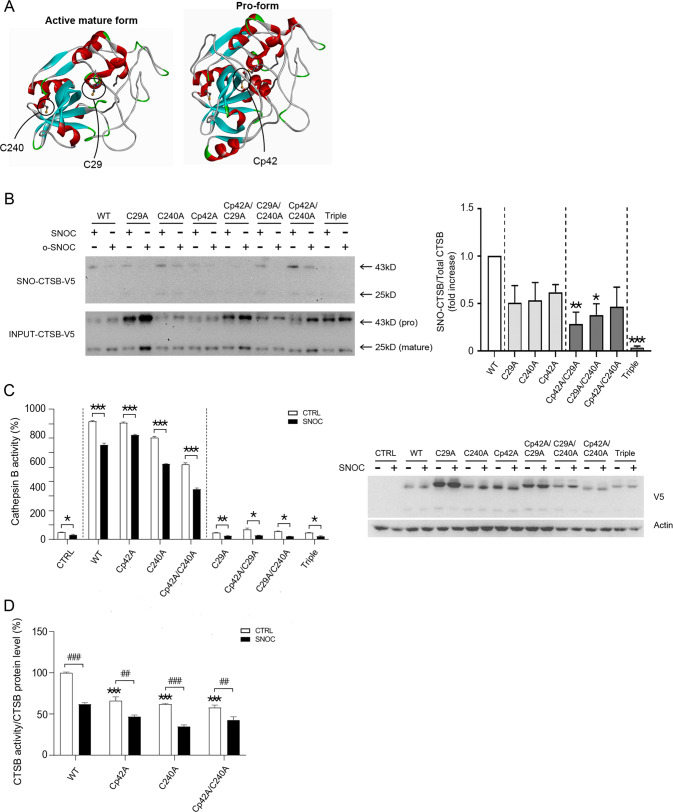
Next, we investigated the cysteine residues of CTSB that may be candidate sites for S-nitrosylation. CTSB is synthesized as a pre-proenzyme, consisting of 339 amino-acid residues [63–65]. Among them, 17 amino acids represent a signal peptide, and 62 amino acids are located in a pro-domain that is removed in the mature form of CTSB [65]. Three cysteines residues not participating in disulfide bond formation were of particular interest to us as candidate sites for S-nitrosylation (Fig. 6A)––one of these cysteine residues is located in the pro-domain (p) of CTSB at residue Cp42, another, at C29, is critical for enzymatic action [65, 66], while the third, C240, is located at a regulatory site for CTSB dimer interaction [65]. We substituted these cysteines for alanines and found that the ratio of SNO-CTSB/total input CTSB was affected by all three of these cysteine residues, as assessed with biotin-switch assay (Fig. 6B).
Fig. 6. S-Nitrosylated cysteine residues of CTSB.
A Crystal structures of the active-mature form (top; PDBID: 1HUC) and pro-form (bottom; PDBID: 1PBH) of CTSB. The side chains of the three free cysteine residues (C29, C240, and Cp42) that do not form structural disulfide bonds are circled. B Biotin-switch assay of CTSB mutants in HEK cells. Seven different CTSB cysteine mutants were transiently transfected into HEK cells two days prior to sham wash or exposure to SNOC; 45 min later, cells were lysed for analysis via biotin-switch assay. The ratio of SNO-CTSB to total input CTSB, including both pro- and mature forms, was quantified as fold increase in ≥3 independent biological replicate experiments. *p < 0.05, **p < 0.01, or ***p < 0.001 by ANOVA with post-hoc Dunnett’s test. C CTSB enzymatic activity (left graphs) and expression levels (right western blot) of CTSB mutants in HEK cells. Seven different cysteine mutants were transiently transfected into HEK cells two days prior to exposure to 200 µM SNOC; 45 min later protein samples were prepared for CTSB enzymatic assay (n = 3 independent biological replicate experiments, *p < 0.05, **p < 0.01, or ***p < 0.001 compared to sham wash control). V5 antibody was used to monitor exogenous CTSB expression levels. D Normalization of enzymatic activity of non-catalytic CTSB Cys mutants to their expression levels. ***p < 0.001 compared to sham wash control of WT, and ##p < 0.01 or ###p < 0.001 vs. sham wash control of the corresponding mutant by ANOVA with post-hoc Dunnett’s test.
Active CTSB induces cleavage of its pro-form. Therefore, catalytically inactive mutant CTSB (C29A), which abrogates protease function, would be expected to maintain high levels of the pro-form [67]. Also, since the pro-form of CTSB displays a greater degree of stability than the active form, expression of mutant CTSB (C29A) would be expected to foster high expression of the pro-form (Figs. 6B, C). Given that none of our individual mutants totally blocked S-nitrosylation, we next made double mutations (Cp42A/C29A, C29A/C240A, and Cp42A/C240A). In the double amino-acid mutants, S-nitrosylation was still present (Fig. 6B). However, the triple amino-acid mutation (Cp42A/C29A/C240A) rendered S-nitrosylation undetectable (Fig. 6B). Taken together, these results are consistent with the notion that all three of these cysteine residues can be S-nitrosylated at least to some degree.
Next, we examined the effects of these mutants on CTSB activity. Overexpression of Cp42A, C240A, or WT CTSB showed ~800% increase in CTSB activity over endogenous activity (Fig. 6C). After exposure to SNOC, CTSB activity for each of these three overexpressed constructs (or the combination of Cp42A/C240A) decreased 10–30% (Fig. 6C, D). In contrast, overexpression of any combination of mutants containing the active site mutation C29A inhibited CTSB activity back down to the endogenous level (as expected, since these cells also contained endogenous, non-mutated CTSB) (Fig. 6C). Interestingly, Cp42A, C240A, and Cp42A/C240A mutant CTSB manifested decreased activity compared to WT CTSB, and SNOC attenuated this activity by an additional ~30% (Fig. 6D). Taken together, these findings suggest that C240 and Cp42 are important in allosterically regulating CTSB enzyme activity and S-nitrosylation of these sites can modulate this activity. Moreover, these results are consistent with the notion that S-nitrosylation of the active site cysteine (C29) plays a role in RNS-dependent inhibition of CTSB activity.
Brains of 5X FAD mice exhibit an increase in SNO-CTSB and decrease in CTSB activity
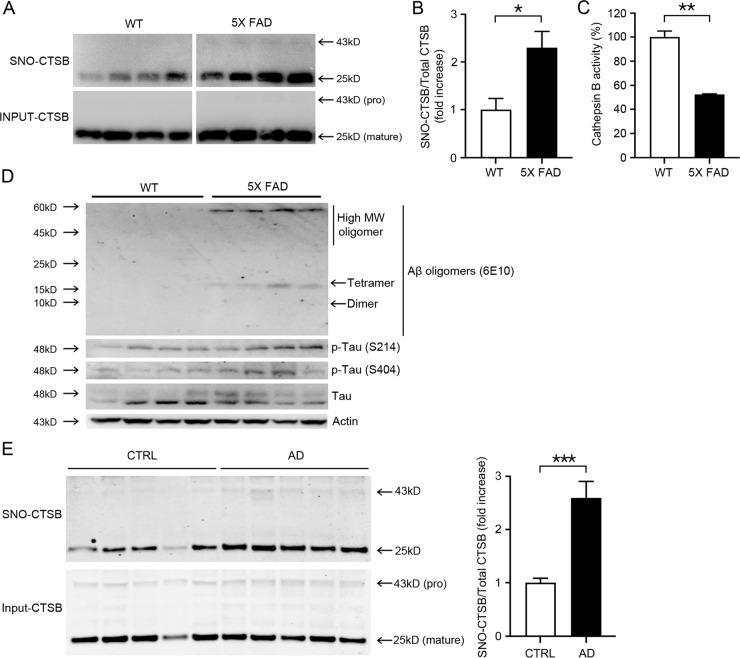
Prior work has shown that oxidative and nitrosative stress are increased and autophagic flux inhibited in several neurodegenerative diseases, including AD, PD, and HD. Therefore, we investigated whether S-nitrosylation of CTSB and resultant attenuation of enzyme activity would occur in the brains of transgenic AD mice. Biotin-switch assays of 6-month-old 5XFAD mouse brain revealed a significant increase in S-nitrosylation, particularly of the active form of CTSB (Fig. 7A, B). CTSB activity was also reduced by 47 ± 0.4% in 5XFAD mouse brain (Fig. 7C). We also confirmed the accumulation of Aβ oligomers, phospho-tau (S214 and S404), and Tau in 5XFAD mice brains (Fig. 7D). Collectively, our results are consistent with the notion that S-nitrosylation of CTSB increases the level of misfolded proteins at least in part due to inhibition of CTSB and autophagic flux.
Fig. 7. S-Nitrosylation and enzymatic inhibition of CTSB in 5X FAD mouse and human AD brains.
A Biotin-switch assay for CTSB in mouse brains. Protein samples were prepared from the cerebrocortices of wild-type (WT) and 5XFAD littermate mice. B Bars depict fold increase in relative S-nitrosylated CTSB in 5XFAD mice brains vs. WT (≥3 biological replicates, *p < 0.05 by Student’s t-test). Both the pro- and mature forms of CTSB were used for quantification. C CTSB enzymatic activity in WT and 5XFAD mouse brains. Protein samples were prepared from the cerebrocortices of WT and 5XFAD mice (n = 4 biological replicates, **p < 0.01 compared to WT mice (CTRL) by Student’s t-test). D Western blot analysis of Aβ oligomers, p-tau (S214, or S404), and tau in WT and 5XFAD mouse brains. Protein samples were prepared from cerebrocortices of WT and 5XFAD littermate mice. E Biotin-switch assay for CTSB was performed on human and control postmortem brains. Graph depicts fold increase in relative SNO-CTSB in human AD brain compared to Controls (n = 5 brains each for AD and Controls tested in ≥3 biological replicate experiments, ***p < 0.001 by two-tailed Student’s t-test). Both the pro- and mature forms of CTSB were used for quantification.
Increase in SNO-CTSB in human AD patient brains
Next, we examined whether S-nitrosylation of CTSB was also present in human AD patient brains. Compared to control human brains, AD patient brains manifested a significant increase in S-nitrosylation of CTSB by biotin-switch assay (Fig. 7E; Supplementary Table 1). Importantly, the relative level of S-nitrosylated CTSB in human AD brain, expressed as the ratio of SNO-CTSB/total input CTSB, was similar or greater than that found in our AD mouse brain models (viz. Figures 7E–B), confirming the pathophysiological relevance of our findings using these model systems.
Discussion
In this study, we present evidence that RNS contribute to aberrant protein accumulation in neurodegenerative diseases such as AD, PD, and HD via S-nitrosylation of CTSB and consequent inhibition of autophagic flux. Initially, we found that the addition of the exogenous NO donor and transnitrosylating agent SNOC decreased autophagy both at an initial step (as previously shown) and also later in the process after the autophagosome/lysosome fusion step, resulting in inhibition of autophagic flux (as demonstrated for the first time here). As evidence of a decrease in the initiation of autophagy, we observed a decrease of GFP-LC3 puncta and LC3-II conversion within 2 h after exposure to SNOC and low-serum medium conditions. However, at later time points, up to 8 h post SNOC exposure, we observed an increase in GFP-LC3 puncta, LC3-II, and p62 protein levels. Moreover, we saw a dramatic increase in RFP red fluorescence after exposure to SNOC in RFP-GFP-LC3 overexpressing cells, suggesting that RNS inhibited lysosomal function. Further along these lines, while co-exposure to rapamycin and Baf-A1 significantly increased LC3-II levels compared to Baf-Al alone (Fig. 1F, lanes 2 and 4), incubation in both SNOC and Baf-A1 did not increase LC3-II levels on immunoblots (Fig. 1F, lanes 2 and 6). These findings are consistent with the notion that excessive RNS, rather than inducing autophagy, caused inhibition of autophagic flux at a step distal to fusion of the autophagosome and lysosome.
Additionally, exposure to SNOC and Baf-A1 resulted in a significant decrease in the number of LC3 puncta compared to exposure to SNOC alone (Fig. 1D, E, third and sixth histogram bars). Nonetheless, LC3-II levels were increased under both of these conditions (Fig. 1F, lanes 5 and 6). Note in Fig. 1D, E, there were far more red puncta (signifying AL/AP fusion) after SNOC exposure, while the addition of Baf-A1 resulted in more yellow puncta (indicating inhibition of autophagic flux at the AP/AL fusion step). An increase in LC3-II levels by itself, as others have shown [68] and as monitored in Fig. 1F, cannot distinguish between inhibition of AL/AP fusion and inhibition of lysosomal function. Importantly, p62 protein levels, which measure autophagic flux more reliably than LC3-II expression, remained unaltered after simultaneous exposure to SNOC and Baf-A1 compared to the group treated with Baf-A1 alone (Fig. 1F, lanes 2 and 6), consistent with the notion that SNOC had already inhibited autophagic flux. As further evidence for this, we also observed that SNOC exposure led to an increase in aggregated protein complexes, including mHtt, mSyn, mTau, and Aβ1-42.
As a potential mechanism for inhibition of autophagic flux by RNS, we focused on lysosomal dysfunction. We found that S-nitrosylation of CTSB resulted in inhibition of its enzymatic activity. Moreover, the specific CTSB chemical inhibitor, CA-074Me, also blocked autophagic flux, mimicking exposure to SNOC. Finally, we observed an increase in S-nitrosylated CTSB and a decrease in CTSB activity in vivo in the 5XFAD transgenic mouse model of AD and in human AD brains. Collectively, these findings suggest that S-nitrosylation of CTSB inhibits autophagic flux and lysosomal degradative function, thus potentially contributing to the pathogenesis of neurodegenerative disorders such as AD. Additionally, we have recently shown that S-nitrosylation of p62 may also contribute to inhibition of autophagic flux under these conditions [69]. While previous studies had focused on the inhibitory mechanism by RNS at the initiation of autophagy [31], we now show that RNS can also inhibit autophagic flux at later stages of the process through regulation of lysosomal activity.
A prior study using quantitative proteomic methods suggested that a specific cysteine residue in CTSD could undergo protein S-nitrosylation after exposure to the transnitrosylating agent S-nitrosoglutathione [46]. In contrast, under our conditions, we did not observe S-nitrosylation of CTSD or inhibition of its enzymatic activity after exposure to SNOC (Fig. 3D). The discrepancy in these results may have arisen for several reasons. First, both different RNS and cell types were used. Second, mass spectrometry detection of S-nitrosylated peptides may have detected very low levels of SNO-CTSD [46], lower than that observed by biotin-switch assay. While evidence for SNO-CTSD was also presented by biotin-switch assay, this only occurred in HEK293T cells overexpressing CTSD rather than for endogenous CTSD, as studied in our experiments. Taken together, the disparate findings are consistent with the difference in detection being due to the level of sensitivity of the various assays.
In the present study, we report that three cysteine residues in CTSB can undergo S-nitrosylation to affect enzyme activity. In toto, CTSB has 15 cysteines, but 12 of these participate in disulfide bond formation, and only 3 have free thiols/thiolate anions with the potential of being regulated by S-nitrosylation [65, 66]. Among these, C29 is an essential cysteine for catalytic activity and is located at the active site. In fact, when C29 was substituted with alanine and transfected into cells, we could detect only endogenous CTSB activity, as expected.
In the biotin-switch assay, the double cysteine mutant CTSB(Cp42A/C240A) still manifested an increase in S-nitrosylation after exposure to SNOC, although this level was lower than that of WT CTSB. To totally eliminate S-nitrosylation of CTSB, three cysteine residues had to be mutated (Cp42A/C29A/C240A). Coupled with our CTSB activity assays, our data indicate that S-nitrosylation of active site C29 inhibited CTSB enzymatic activity, while S-nitrosylation of the other cysteine sites modulated this activity.
Prior studies have reported that activation of CTSB can contribute to apoptosis by inducing caspase-3 activity but other pathways were also invoked [70]. In the present study, we examined whether inhibition of CTSB alone could induce neuronal cell death. In mouse cerebrocortical neurons, after treatment with CA-074Me, a specific chemical inhibitor of CTSB, we observed typical apoptotic morphology and caspase-3 activation. The addition of the pan-caspase inhibitor, zVAD, or a protein synthesis inhibitor, CHX, nearly totally abrogated neuronal cell death. These results are consistent with the notion that inhibition of CTSB in neurons can induce caspase-mediated apoptosis and suggest that this pathway may occur in neurodegenerative disorders such as AD in which CTSB is inhibited by protein S-nitrosylation.
In conclusion, we observed an increase in S-nitrosylation and a consequent decrease in activity of CTSB in cell-based as well as in the 5XFAD transgenic mouse model of AD. Moreover, the relative level of SNO-CTSB was found to be similar in human AD patient brains as in these model systems, implying that our findings could contribute to inhibition of autophagic flux, accumulation of aberrant/misfolded proteins, and thus progression of neurodegenerative disorders such as AD in the human brain. These findings may also have therapeutic implications, as prevention of S-nitrosylation of CTSB and its resulting enzymatic inhibition would be expected to enhance autophagic function and therefore clearance of misfolded proteins.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Eliezer Masliah (UC San Diego/NIA) for providing human brain tissues. This work was supported in part by NIH grants R35 AG071734, R01 NS086890, R01 DA048882, DP1 DA041722, RF1 AG057409, and R01 AG056259 (to SAL), R01 AG061845, RF1 NS123298, and R61 NS122098 (to TN), and National Research Foundation of Korea (NRF) grants NRF-2017M3C7A1028945, NRF-2018R1D1A1B07049746, and NRF-2021R1A2C2008234 (to YHK).
Author contributions
YHK and SAL conceived the project and designed the experiments. KRK, EJC, JWE, SSO, TN, and CKO performed experiments. KRK, TN, CKO, SAL and YHK analyzed and interpreted results. KRK, TN, CKO, YHK, and SAL wrote the manuscript.
Data availability
Correspondence and requests for materials should be addressed to YHK or SAL.
Competing interests
The authors declare that YHK is a shareholder of Zincure Corp., and that KRK is currently employed by Zincure Corp. SAL is a scientific founder of Adamas Pharmaceuticals, Inc., EuMentis Therapeutics, Inc., and InflaMED Therapeutics, LLC.
Ethical statement and consent to participate
Our studies did not include human participants or human data. Archived human brain samples were analyzed with institutional permission under the state of California and NIH guidelines. Informed consent was obtained according to procedures approved by Institutional Review Boards at the University of California, San Diego, School of Medicine, and The Scripps Research Institute. All animal experimental procedures were approved by the Animal Care and Use Committee of Sejong University and were conducted following the guidelines of the Care and Use of Laboratory Animals.
Footnotes
Edited by G Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stuart A. Lipton, Email: slipton@scripps.edu
Yang-Hee Kim, Email: yhkim@sejong.ac.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01004-0.
References
- 1.de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–70.. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 3.Upadhya SC, Hegde AN. Ubiquitin-proteasome pathway components as therapeutic targets for CNS maladies. Curr Pharm Des. 2005;11:3807–28.. doi: 10.2174/138161205774580651. [DOI] [PubMed] [Google Scholar]
- 4.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17.. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 7.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 8.Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. J Biosci. 2006;31:137–55.. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 10.Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–22.. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 11.Juenemann K, Schipper-Krom S, Wiemhoefer A, Kloss A, Sanz Sanz A, Reits EA. Expanded polyglutamine-containing N-terminal huntingtin fragments are entirely degraded by mammalian proteasomes. J Biol Chem. 2013;288:27068–84.. doi: 10.1074/jbc.M113.486076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 14.Walkley SU, Suzuki K. Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim Biophys Acta. 2004;1685:48–62. doi: 10.1016/j.bbalip.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Tessitore A, del PMM, Sano R, Ma Y, Mann L, Ingrassia A, et al. GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gangliosidosis. Mol Cell. 2004;15:753–66.. doi: 10.1016/j.molcel.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Sleat DE, Wiseman JA, El-Banna M, Kim KH, Mao Q, Price S, et al. A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J Neurosci. 2004;24:9117–26.. doi: 10.1523/JNEUROSCI.2729-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anda FC, Madabhushi R, Rei D, Meng J, Graff J, Durak O, et al. Cortical neurons gradually attain a post-mitotic state. Cell Res. 2016;26:1033–47.. doi: 10.1038/cr.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabrier PE, Demerle-Pallardy C, Auguet M. Nitric oxide synthases: targets for therapeutic strategies in neurological diseases. Cell Mol Life Sci. 1999;55:1029–35. doi: 10.1007/s000180050353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torreilles F, Salman-Tabcheh S, Guerin M, Torreilles J. Neurodegenerative disorders: the role of peroxynitrite. Brain Res Brain Res Rev. 1999;30:153–63.. doi: 10.1016/S0165-0173(99)00014-4. [DOI] [PubMed] [Google Scholar]
- 20.Miranda S, Opazo C, Larrondo LF, Munoz FJ, Ruiz F, Leighton F, et al. The role of oxidative stress in the toxicity induced by amyloid β-peptide in Alzheimer’s disease. Prog Neurobiol. 2000;62:633–48. doi: 10.1016/S0301-0082(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 21.Simic G, Lucassen PJ, Krsnik Z, Kruslin B, Kostovic I, Winblad B, et al. nNOS expression in reactive astrocytes correlates with increased cell death related DNA damage in the hippocampus and entorhinal cortex in Alzheimer’s disease. Exp Neurol. 2000;165:12–26. doi: 10.1006/exnr.2000.7448. [DOI] [PubMed] [Google Scholar]
- 22.LaVoie MJ, Hastings TG. Peroxynitrite- and nitrite-induced oxidation of dopamine: implications for nitric oxide in dopaminergic cell loss. J Neurochem. 1999;73:2546–54.. doi: 10.1046/j.1471-4159.1999.0732546.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkubo Y, Nakato R, Uehara T. Regulation of unfolded protein response via protein S-nitrosylation. Yakugaku Zasshi. 2016;136:801–4. doi: 10.1248/yakushi.15-00292-1. [DOI] [PubMed] [Google Scholar]
- 25.Nakato R, Ohkubo Y, Konishi A, Shibata M, Kaneko Y, Iwawaki T, et al. Regulation of the unfolded protein response via S-nitrosylation of sensors of endoplasmic reticulum stress. Sci Rep. 2015;5:14812. doi: 10.1038/srep14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Barthwal MK, Dikshit M. Cdk2 nitrosylation and loss of mitochondrial potential mediate NO-dependent biphasic effect on HL-60 cell cycle. Free Radic Biol Med. 2010;48:851–61. doi: 10.1016/j.freeradbiomed.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, et al. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 28.Umanah GKE, Ghasemi M, Yin X, Chang M, Kim JW, Zhang J, et al. AMPA receptor surface expression is regulated by S-nitrosylation of thorase and transnitrosylation of NSF. Cell Rep. 2020;33:108329. doi: 10.1016/j.celrep.2020.108329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melino G, Bernassola F, Knight RA, Corasaniti MT, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–3. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 30.Zhu L, Zhang C, Liu Q. PTEN S-nitrosylation by NOS1 inhibits autophagy in NPC cells. Cell Death Dis. 2019;10:306. doi: 10.1038/s41419-019-1542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, et al. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darios F, Stevanin G. Impairment of lysosome function and autophagy in rare neurodegenerative diseases. J Mol Biol. 2020;432:2714–34.. doi: 10.1016/j.jmb.2020.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–36. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- 34.Kim YH, Kim EY, Gwag BJ, Sohn S, Koh JY. Zinc-induced cortical neuronal death with features of apoptosis and necrosis: mediation by free radicals. Neuroscience. 1999;89:175–82. doi: 10.1016/S0306-4522(98)00313-3. [DOI] [PubMed] [Google Scholar]
- 35.Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, et al. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8:1087–99.. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 36.Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-d-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc Natl Acad Sci USA. 1995;92:7162–6. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh JY, Choi DW. Quantitative determination of glutamate mediated cortical neuronal injury in cell culture by lactate dehydrogenase efflux assay. J Neurosci Methods. 1987;20:83–90. doi: 10.1016/0165-0270(87)90041-0. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, Oh CK, Liao L, Zhang X, Lopez KM, Gibbs D, et al. Noncanonical transnitrosylation network contributes to synapse loss in Alzheimer’s disease. Science. 2021;371:eaaw0843. doi: 10.1126/science.aaw0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh CK, Sultan A, Platzer J, Dolatabadi N, Soldner F, McClatchy DB, et al. S-Nitrosylation of PINK1 attenuates PINK1/parkin-dependent mitophagy in hiPSC-based Parkinson’s disease models. Cell Rep. 2017;21:2171–82.. doi: 10.1016/j.celrep.2017.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–7. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 41.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–7. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26.. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Percival MD, Ouellet M, Campagnolo C, Claveau D, Li C. Inhibition of cathepsin K by nitric oxide donors: evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574–83. doi: 10.1021/bi991028u. [DOI] [PubMed] [Google Scholar]
- 44.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, et al. S-Nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci USA. 1992;89:444–8. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, et al. Exposure of sulphydryl-containing proteins to nitric oxide and endothelium-derived relaxing factor confers novel bioactivity and modulates their intrinsic functional properties. In: Moncada S, Marletta MA, J. B. Hibbs J, Higgs EA, editors. The Biology of Nitric Oxide. London: Portland Press; 1992. p. 20–23.
- 46.Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, et al. Chemoproteomic strategy to quantitatively monitor transnitrosation uncovers functionally relevant S-nitrosations sites on cathepsin D and HADH2. Cell Chem Biol. 2016;23:727–37. doi: 10.1016/j.chembiol.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotzl JK, Colombo AV, Fellerer K, Reifschneider A, Werner G, Tahirovic S, et al. Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol Neurodegener. 2018;13:48. doi: 10.1186/s13024-018-0281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pislar A, Kos J. Cysteine cathepsins in neurological disorders. Mol Neurobiol. 2014;49:1017–30. doi: 10.1007/s12035-013-8576-6. [DOI] [PubMed] [Google Scholar]
- 49.Bhoopathi P, Chetty C, Gujrati M, Dinh DH, Rao JS, Lakka S. Cathepsin B facilitates autophagy-mediated apoptosis in SPARC overexpressed primitive neuroectodermal tumor cells. Cell Death Differ. 2010;17:1529–39. doi: 10.1038/cdd.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vancompernolle K, Van Herreweghe F, Pynaert G, Van de Craen M, De Vos K, Totty N, et al. Atractyloside-induced release of cathepsin B, a protease with caspase-processing activity. FEBS Lett. 1998;438:150–8. doi: 10.1016/S0014-5793(98)01275-7. [DOI] [PubMed] [Google Scholar]
- 51.Balboula AZ, Yamanaka K, Sakatani M, Kawahara M, Hegab AO, Zaabel SM, et al. Cathepsin B activity has a crucial role in the developmental competence of bovine cumulus-oocyte complexes exposed to heat shock during in vitro maturation. Reproduction. 2013;146:407–17. doi: 10.1530/REP-13-0179. [DOI] [PubMed] [Google Scholar]
- 52.Hook V, Toneff T, Bogyo M, Greenbaum D, Medzihradszky KF, Neveu J, et al. Inhibition of cathepsin B reduces β-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate β-secretase of Alzheimer’s disease. Biol Chem. 2005;386:931–40. doi: 10.1515/BC.2005.151. [DOI] [PubMed] [Google Scholar]
- 53.Kindy MS, Yu J, Zhu H, El-Amouri SS, Hook V, Hook GR. Deletion of the cathepsin B gene improves memory deficits in a transgenic ALZHeimer’s disease mouse model expressing AβPP containing the wild-type β-secretase site sequence. J Alzheimers Dis. 2012;29:827–40. doi: 10.3233/JAD-2012-111604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hook G, Yu J, Toneff T, Kindy M, Hook V. Brain pyroglutamate amyloid-β is produced by cathepsin B and is reduced by the cysteine protease inhibitor E64d, representing a potential Alzheimer’s disease therapeutic. J Alzheimers Dis. 2014;41:129–49. doi: 10.3233/JAD-131370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annis RP, Swahari V, Nakamura A, Xie AX, Hammond SM, Deshmukh M. Mature neurons dynamically restrict apoptosis via redundant premitochondrial brakes. FEBS J. 2016;283:4569–82.. doi: 10.1111/febs.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, et al. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29:615–28.. doi: 10.1016/S0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 57.Lee JM, Kim YJ, Ra H, Kang SJ, Han S, Koh JY, et al. The involvement of caspase-11 in TPEN-induced apoptosis. FEBS Lett. 2008;582:1871–6. doi: 10.1016/j.febslet.2008.04.056. [DOI] [PubMed] [Google Scholar]
- 58.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, et al. S-Nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–90.. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 59.D’Emilia DM, Lipton SA. Ratio of S-nitrosohomocyst(e)ine to homocyst(e)ine or other thiols determines neurotoxicity in rat cerebrocortical cultures. Neurosci Lett. 1999;265:103–6. doi: 10.1016/S0304-3940(99)00210-4. [DOI] [PubMed] [Google Scholar]
- 60.Kashii S, Mandai M, Kikuchi M, Honda Y, Tamura Y, Kaneda K, et al. Dual actions of nitric oxide in N-methyl-d-aspartate receptor-mediated neurotoxicity in cultured retinal neurons. Brain Res. 1996;711:93–101. doi: 10.1016/0006-8993(95)01330-X. [DOI] [PubMed] [Google Scholar]
- 61.Shimohama S, Akaike A, Kimura J. Nicotine-induced protection against glutamate cytotoxicity. Nicotinic cholinergic receptor-mediated inhibition of nitric oxide formation. Ann NY Acad Sci. 1996;777:356–61.. doi: 10.1111/j.1749-6632.1996.tb34445.x. [DOI] [PubMed] [Google Scholar]
- 62.Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, et al. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–32. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 63.Chan SJ, San Segundo B, McCormick MB, Steiner DF. Nucleotide and predicted amino acid sequences of cloned human and mouse preprocathepsin B cDNAs. Proc Natl Acad Sci USA. 1986;83:7721–5. doi: 10.1073/pnas.83.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fong D, Calhoun DH, Hsieh WT, Lee B, Wells RD. Isolation of a cDNA clone for the human lysosomal proteinase cathepsin B. Proc Natl Acad Sci USA. 1986;83:2909–13. doi: 10.1073/pnas.83.9.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Musil D, Zucic D, Turk D, Engh RA, Mayr I, Huber R, et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 1991;10:2321–30. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chagas JR, Ferrer-Di Martino M, Gauthier F, Lalmanach G. Inhibition of cathepsin B by its propeptide: use of overlapping peptides to identify a critical segment. FEBS Lett. 1996;392:233–6. doi: 10.1016/0014-5793(96)00822-8. [DOI] [PubMed] [Google Scholar]
- 67.Song J, Xu P, Xiang H, Su Z, Storer AC, Ni F. The active-site residue Cys-29 is responsible for the neutral-pH inactivation and the refolding barrier of human cathepsin B. FEBS Lett. 2000;475:157–62.. doi: 10.1016/S0014-5793(00)01644-6. [DOI] [PubMed] [Google Scholar]
- 68.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 69.Oh CK, Dolatabadi N, Cieplak P, Diaz-Meco MT, Moscat J, Nolan JP, et al. S-Nitrosylation of p62 inhibits autophagic flux to promote α-synuclein secretion and spread in Parkinson’s disease and Lewy body dementia. J Neurosci. 14:JN-RM-1508-21. 10.1523/JNEUROSCI.1508-21.2022. [DOI] [PMC free article] [PubMed]
- 70.Oberle C, Huai J, Reinheckel T, Tacke M, Rassner M, Ekert PG, et al. Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 2010;17:1167–78. doi: 10.1038/cdd.2009.214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Correspondence and requests for materials should be addressed to YHK or SAL.