Abstract
No biomarkers are available to predict toxicities induced by VEGFR TKIs. This study aimed to identify markers of toxicities induced by these drugs using a discovery-validation approach. The discovery set included 140 sorafenib-treated cancer patients (TARGET study) genotyped for SNPs in 56 genes. The most significant SNPs associated with grade ≥2 hypertension, diarrhea, dermatologic toxicities, and composite toxicity (any one of the toxicities) were tested for association with grade ≥2 toxicity in a validation set of 201 sorafenib-treated patients (Alliance/CALGB 80802). The validated SNP was tested for association with grade ≥2 toxicity in 107 (LCCC 1029) and 82 (Italian cohort) regorafenib-treated patients. SNP-toxicity associations were evaluated using logistic regression, and a meta-analysis between the studies was performed by inverse variance. Variant rs4864950 in KDR increased the risk of grade ≥2 composite toxicity in TARGET, Alliance/CALGB 80802, and the Italian cohort (meta-analysis p=6.79×10−4, OR=2.01, 95% CI 1.34–3.01). We identified a predictor of toxicities induced by VEGFR TKIs.
Keywords: Sorafenib, regorafenib, KDR, hypertension, diarrhea, dermatologic toxicities
Introduction
Vascular endothelial growth factor receptor (VEGFR) tyrosine kinase inhibitors (TKIs) are active drugs approved for the treatment of several types of cancer, including renal cell carcinoma (RCC), hepatocellular carcinoma (HCC), and colorectal cancer (CRC) 1. VEGFR TKIs (sorafenib, regorafenib, sunitinib, axitinib, pazopanib, cabozantinib, lenvatinib, and vandetanib) have a similar mechanism of action related to the inhibition of the VEGF pathway involved in endothelial survival, vascular permeability, and tumor angiogenesis.
VEGFR TKIs also share a similar spectrum of common side effects, including hypertension, diarrhea, and dermatologic toxicities, such as hand-foot syndrome (HFS) and rash. For example, a meta-analysis of metastatic (m)RCC patients treated with sorafenib reported a prevalence of any grade and grade ≥3 hypertension in 26.4% and 4.3%, HFS in 57.2% and 11.8%, and diarrhea in 55.7% and 4.1% patients, respectively 2. For patients with different advanced solid tumors treated with regorafenib, the prevalence of any grade and grade ≥3 hypertension was 31% and 13%, HFS 54% and 16%, and diarrhea 33% and 4%, respectively 3. The primary proposed mechanism of these toxicities involves the blockage of the nitric oxide (NO) pathway. The VEGF signaling pathway augments the transcription of endothelial NO synthase. The blockage of NO production leads to vasoconstriction, blood flow reduction, increased vascular tone, and hence hypertension 4. The vasoconstriction and blood flow reduction also disrupt endothelial and vascular repair mechanisms and lead to epithelial ischemia and hypoxia, which induces dermatologic toxicities 5 and diarrhea 4.
Currently, no biomarkers predictive of toxicity have been validated for any of the VEGFR TKIs. As angiogenesis is primarily a host-mediated process 6,7, germline variants are likely to be associated with VEGFR TKI toxicity. The investigation of germline variants that could predict toxicities induced by different VEGF-pathway inhibitors has clinical implications. Although the efficacy of VEGFR TKIs has been established as monotherapy and more recently in combination with immune checkpoint inhibitors for different types of cancers 8, dose modifications, interruptions and discontinuation of the VEGFR TKI due to toxicities are very frequent with the potential to negatively impact efficacy 9–11.
The objective of this study was to identify and validate germline variants that can predict which patients are at risk of developing toxicities when treated with two different VEGFR TKIs, sorafenib and regorafenib. Using data from four different clinical studies of patients treated with sorafenib and regorafenib, we have applied a discovery-validation approach. Sorafenib-treated mRCC patients from the phase 3 Treatment Approaches in Renal Cancer Global Evaluation Trial (TARGET) were used as the discovery set, while a phase III clinical trial of sorafenib-treated HCC patients from the Cancer and Leukemia Group B (CALGB, now Alliance for Clinical Trials in Oncology, Alliance), and two clinical studies including regorafenib-treated CRC patients were used as validation sets. Statistical analyses were conducted by the Alliance Statistics and Data Management Center.
Methods
Candidate gene analysis for SNP discovery in the TARGET study
TARGET was a double-blind, randomized, placebo-controlled phase III trial of patients with mRCC treated with 400 mg sorafenib orally twice daily or placebo 12.
Toxicities were recorded according to Common Toxicity Criteria for Adverse Events (CTCAE) version 3.0, by protocol. In this analysis, we focused on the common toxicities of hypertension, diarrhea, and dermatologic (defined as the occurrence of either HFS, rash or desquamation, pruritus, or alopecia). Composite toxicity is defined as the occurrence of any one of these three toxicities.
TARGET genotyping and statistical analysis
DNA was obtained from the peripheral blood of 140 patients treated with sorafenib. A total of 1,536 single-nucleotide polymorphisms (SNPs) in 56 candidate genes (mainly from VEGF-pathway signaling genes) were genotyped using the Illumina GoldenGate assay as previously described 6. SNPs were excluded if the genotype call rate was <97.5% and were selected based upon a minor allele frequency (MAF) >0.05 in Europeans from the 1,000 Genomes Project, and other criteria 6. After quality control (Figure 1), a total of 973 SNPs were used in this study. Tests for association between SNPs and grade 2–3 or grade 3 toxicity (no grade 4 toxicity was reported) were performed in the sorafenib-treated patients by calculating the odds ratio (OR) from a logistic regression analysis under an additive genetic model 13. Age (continuous) and gender were used as covariates. Race was not included as a covariate as it was not available for the majority of patients. Codes used for analyses will be provided upon request.
Figure 1. CONSORT and quality control flowchart for the TARGET study, CALGB 80802, LCCC 1029, and the Italian cohort.
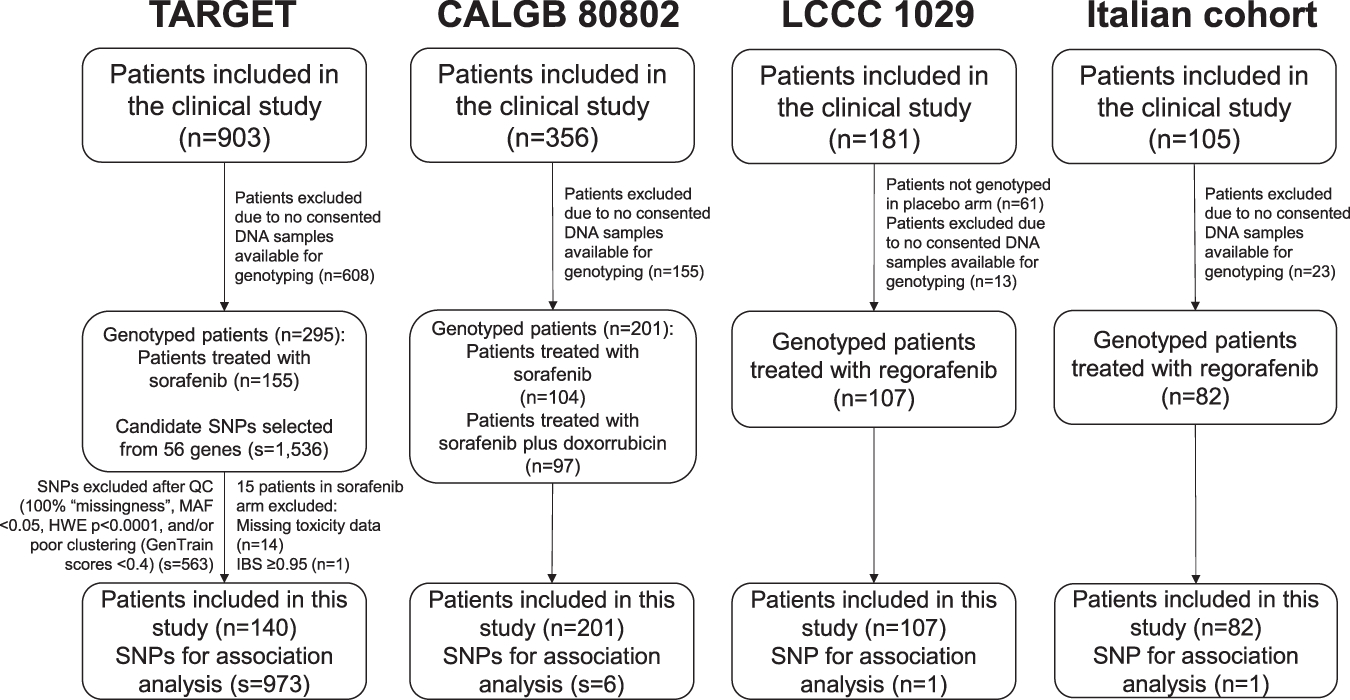
IBS: identical by state, MAF: minor allele frequency, HWE: Hardy-Weinberg Equilibrium, QC: quality control, SNP: single nucleotide polymorphism.
Selection of SNPs for testing in additional cohorts
After excluding SNPs in linkage disequilibrium (LD) with r2≥0.5, the most statistically significant SNP associated with grade 2–3 hypertension, the most statistically significant SNP associated with grade 2–3 diarrhea, the most statistically significant SNP associated with grade 2–3 dermatologic toxicity, and the three most statistically significant SNPs associated with grade 2–3 composite toxicity were selected for testing in the validation set.
Validation of SNPs in CALGB 80802 (Alliance)
CALGB 80802 (Alliance) was a randomized phase III trial of patients with advanced or mHCC treated with either 400 mg of sorafenib orally twice daily alone or 400 mg of sorafenib orally twice daily plus with 60 mg/m2 of doxorubicin intravenously every 21 days 14. Toxicities were recorded according to CTCAE version 4.0, by protocol. In this analysis, we focused on the common toxicities of hypertension, diarrhea, and HFS. Composite toxicity was defined as the occurrence of any one of these three toxicities.
CALGB 80802: genotyping and statistical analysis
DNA was obtained from the peripheral blood of 201 patients treated with sorafenib. Genotyping was performed using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA). Tests for association between SNPs and grade 2–3 or grade 3 toxicity (no grade 4 toxicity was reported) were performed across both arms by calculating the OR from a logistic regression analysis, under a dominant model based upon the distribution of the three genotypes relative to the effect observed in TARGET for each SNP. Age (continuous), gender, race/ethnicity (self-reported), and treatment arm (sorafenib alone versus sorafenib plus doxorubicin) were included as covariates. Race/ethnicity groups used here were non-Hispanic White, non-Hispanic Black, non-Hispanic other (including Asian, multi-racial, other race groups). The validation criteria were p-value <0.05 and a concordant direction of effect (such as increased or reduced risk of the minor frequency allele) between the discovery and validation cohorts for either grade 2–3 or grade 3 composite toxicity.
Validation of a SNP in clinical studies of regorafenib: LCCC 1029 and Italian cohort
Because sorafenib and regorafenib share a similar mechanism of action and have a similar spectrum of toxicities 2,3, we aimed to test if the SNP associated with sorafenib toxicity identified in the TARGET study and validated in CALGB 80802 could predict toxicities induced by regorafenib. For this purpose, two clinical studies were used. LCCC 1029 is a randomized, double-blinded, placebo-controlled, phase II trial of patients with mCRC treated with FOLFIRI given as per standard of care on days 1–3 and 15–17 of every 4-week cycle in combination with 160 mg daily regorafenib or placebo on days 4–10 and 18–24 15. The “Italian cohort” study was conducted in patients with refractory mCRC treated with 160 mg regorafenib daily following a 3-week-on, 1-week-off-cycle at a center in Italy 16. Toxicities were recorded according to CTCAE v4.0 in both studies. In this analysis, we focused on the common toxicities of hypertension, diarrhea, and HFS for LCCC 1029, and of hypertension, diarrhea, and skin rash (as it was the only dermatologic toxicity recorded) for the Italian cohort. In both studies, composite toxicity is defined as the occurrence of any one of these three toxicities.
Regorafenib clinical studies: genotyping and statistical analysis
DNA was obtained from peripheral blood of 107 patients treated with regorafenib in the LCCC 1029 trial and 82 patients treated with regorafenib in the Italian cohort. Genotyping was performed using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA). Tests for association between SNP and grade 2–3 or grade 3 toxicity were performed by calculating the OR from a logistic regression analysis under a dominant model. Age (continuous), gender, and race (white or non-white) were included as covariates for LCCC 1029, and age (continuous) and gender were included as covariates for the Italian cohort. The validation criteria were p-value <0.05 and a concordant direction of effect (such as increased or reduced risk of the minor frequency allele) between the sorafenib and the regorafenib studies for either grade 2–3 or grade 3 composite toxicity.
Downstream analyses of the validated SNP
Meta-analysis
For the SNP validated in CALGB 80802 and tested for association in LCCC 1029 and in the Italian cohort, the inverse variance formula under a fixed-effect model was used to combine the SNP effect to obtain the meta-analysis OR of the SNP-toxicity association between the different studies. The heterogeneity across studies was examined by Cochran’s Q test, and a Cochran’s Q p-value <0.20 indicates statistically significant heterogeneity.
Functional analysis
SNPs were analyzed by LDlink for analyses of LD 17. The UCSC Genome Browser (http://genome.ucsc.edu/) 18, RegulomeDB 19 and Haploreg v4 20 were used for functional inference. The Genotype-Tissue Expression project (GTEx v7) 21 was used for analyses of expression quantitative trait loci (eQTL).
Results
Patient’s characteristics and toxicities in TARGET
A total of 140 mRCC patients treated with sorafenib in TARGET were included in the discovery analysis (Figure 1). Patient demographics and the prevalence of grade 2–3 and grade 3 toxicities are shown in Table 1. No grade 4 toxicities were reported.
Table 1.
Patients treated with sorafenib in TARGET and CALGB 80802 and patients treated with regorafenib in LCCC 1029 and Italian cohort.
|
|
||||
|---|---|---|---|---|
| TARGET (n=140) | CALGB 80802 (n=201) | LCCC 1029 (n=107) | Italian cohort (n=82) | |
|
| ||||
| Study details | ||||
|
| ||||
| Cancer type | mRCC | Advanced or mHCC | mCRC | mCRC |
|
| ||||
| Treatment | 400 mg sorafenib orally twice daily | 400 mg of sorafenib orally twice daily alone or combined with 60 mg/m2 of doxorubicin | FOLFIRI on days 1–3 and 15–17 of every 4-week cycle with 160 mg regorafenib daily | 160 mg regorafenib daily following a 3-week-on, 1-weekoff-cycle |
|
| ||||
| Demographics | ||||
|
| ||||
| Age (years)–Mean (SD) | 59.6 (9.8) | 62.3 (10.4) | 61.7 (12.2) | 61.2 (9.5) |
| Male – n (%) | 105 (75.0%) | 168 (83.6%) | 19 (17.8%) | 46 (56.1%) |
| Female – n (%) | 35 (25.0%) | 33 (16.4%) | 88 (82.2%) | 36 (43.9%) |
|
| ||||
| Toxicities* (n, %) | ||||
|
| ||||
| Hypertension | ||||
| Grade ≥2 | 12 (8.6) | 61 (29.9%) | 16 (15.0%) | 23 (28%) |
| Grade 3 | 5 (3.6) | 18 (8.8%) | 8 (7.5%) | 6 (7.3%) |
|
| ||||
| Diarrhea | ||||
| Grade ≥2 | 26 (18.6%) | 42 (20.6%) | 26 (24.3%) | 15 (18.3%) |
| Grade 3 | 5 (3.6%) | 15 (7.4%) | 13 (12.2%) | 7 (8.5%) |
|
| ||||
| Dermatologic toxicity** | ||||
| Grade ≥2 | 42 (30.0%) | 55 (27.0%) | 16 (15.0%) | 25 (29.1%) |
| Grade 3 | 15 (10.7%) | 25 (12.3%) | 6 (5.6%) | 18 (20.9%) |
|
| ||||
| Composite toxicity | ||||
| Grade ≥2 | 63 (45%) | 113 (55.4%) | 43 (40.2%) | 49 (59.8%) |
| Grade 3 | 24 (17.1%) | 55 (27.0%) | 24 (22.4%) | 29 (35.4%) |
CALGB: Cancer and Leukemia Group B, mRCC: metastatic renal cell carcinoma, mHCC: metastatic hepatocellular carcinoma, mCRC: metastatic colorectal cancer, SD: standard deviation, HFS: hand-foot syndrome.
CTCAE v3.0 for TARGET and CTCAE v4.0 for CALGB 80802, LCCC 1029 and Italian cohort
dermatologic toxicity in TARGET (HFS + rash or desquamation + pruritus + alopecia), CALGB 80802 (HFS), LCCC 1029 (HFS), and Italian cohort (rash).
SNPs associated with toxicities in TARGET study (discovery set)
The most statistically significant SNP associated with grade 2–3 hypertension was rs444904 (G>A, MAF 0.14) in PIK3R5, with the A allele associated with a higher risk (p=0.0056, OR=3.88, 95% CI: 1.49–10.12) (Table 2). For grade 3 hypertension, the A allele of rs444904 was also associated with an increased risk (Table 2).
Table 2.
Top 5 SNPs ranked by adjusted (for age and gender) p-value for association with grade 2–3 hypertension, diarrhea, dermatologic toxicity, and composite toxicity in the TARGET study (discovery set) under an additive genetic model.
| SNP | Chr | Gene | Feature | Base change | MAF | grade 2–3 OR (95% CI) | grade 2–3 p-value | grade 3 OR (95% CI) | grade 3 p-value |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Hypertension | |||||||||
|
| |||||||||
| rs444904 | 17 | PIK3R5 | Intron 5 | G>A | 0.14 | 3.88 (1.49–10.12) | 0.0056 | 27.30 (3.16–236.20) | 0.0027 |
| rs1346563 | 16 | ADAMTS18 | Intron 3 | C>T | 0.30 | 3.50 (1.43–8.59) | 0.0062 | 5.25 (1.30–21.15) | 0.0197 |
| rs11125039 | 2 | PRKCE | Intron 2 | A>G | 0.24 | 3.49 (1.31–9.25) | 0.0121 | 1.29 (0.31–5.45) | 0.7292 |
| rs2330951 | 7 | EGFR | Intron 1 | A>C | 0.23 | 4.16 (1.37–12.68) | 0.0121 | 5.72 (1.08–30.35) | 0.0404 |
| rs56367980 | 7 | EGFR | Intron 1 | G>A | 0.12 | 3.34 (1.22–9.13) | 0.0185 | 1.66 (0.635–7.99) | 0.5253 |
|
| |||||||||
| Diarrhea | |||||||||
|
| |||||||||
| rs917881 | 7 | EGFR | Intron 1 | G>A | 0.18 | 3.61 (1.68–7.73) | 0.0010 | 3.94 (0.80–19.46) | 0.0921 |
| rs17682789 | 16 | CDH13 | Intron 2 | T>C | 0.31 | 0.27 (0.11–0.64) | 0.0030 | 0.38 (0.08–1.87) | 0.2339 |
| rs11651488 | 17 | MAP2K6 | Intron 1 | T>C | 0.27 | 2.54 (1.30–4.95) | 0.0064 | 2.75 (0.77–9.82) | 0.1186 |
| rs9927200 | 16 | WWOX | Intron 5 | C>A | 0.44 | 2.48 (1.24–4.95) | 0.0099 | 2.64 (0.55–12.61) | 0.2223 |
| rs2716212 | 17 | MAP2K6 | Intron 2 | T>C | 0.42 | 2.25 (1.21–4.18) | 0.0105 | 6.95 (1.32–36.70) | 0.0223 |
|
| |||||||||
| Dermatologic toxicity (HFS + rash or desquamation + pruritus + alopecia) | |||||||||
|
| |||||||||
| rs12366035 | 11 | VEGFB | Exon 5 (syn) | C>T | 0.34 | 2.91 (1.55–5.46) | 0.0009 | 1.94 (0.84–4.50) | 0.1233 |
| rs1868089 | 2 | EPAS1 | Intron 1 | T>C | 0.44 | 2.65 (1.43–4.89) | 0.0020 | 1.80 (0.77–4.21) | 0.1764 |
| rs7557402 | 2 | EPAS1 | Intron 8 | C>G | 0.48 | 2.49 (1.37–4.55) | 0.0029 | 4.35 (1.67–11.37) | 0.0027 |
| rs10958704 | 8 | NA | 2kb up FGFR1 | A>G | 0.36 | 2.06 (1.20–3.56) | 0.0091 | 2.23 (1.03–4.87) | 0.0429 |
| rs12948059 | 17 | MAP2K6 | Intron 1 | A>G | 0.18 | 2.32 (1.22–4.46) | 0.0107 | 2.15 (0.91–5.09) | 0.0816 |
|
| |||||||||
| Composite toxicity | |||||||||
|
| |||||||||
| rs12366035 | 11 | VEGFB | Exon 5 (syn) | C>T | 0.34 | 2.78 (1.54–5.02) | 0.0007 | 1.65 (0.82–3.33) | 0.1637 |
| rs4035887 | 2 | EPAS1 | Intron 1 | G>A | 0.49 | 2.33 (1.36–4.00) | 0.0021 | 1.47 (0.73–2.94) | 0.2779 |
| rs4864950 | 4 | KDR | Intron 29 | T>A | 0.23 | 2.41 (1.29–4.51) | 0.0058 | 1.44 (0.65–3.19) | 0.3626 |
| rs315498 | 17 | NA | RNU6–1134P | T>C | 0.33 | 0.46 (0.26–0.80) | 0.0061 | 0.31 (0.13–0.73) | 0.0076 |
| rs9973653 | 4 | EPAS1 | Intron 1 | G>T | 0.34 | 0.47 (0.27–0.82) | 0.0080 | 0.80 (0.39–1.63) | 0.5326 |
The SNPs in bold are the ones selected for testing in the validation set. Chr: chromosome, CI: confidence interval NA: intergenic SNP, HFS: hand-foot syndrome, MAF: minor allele frequency, OR: odds ratio, SNP: single nucleotide polymorphism, syn: synonymous; up: upstream.
The most statistically significant SNP associated with grade 2–3 diarrhea was rs917881 (G>A, MAF 0.18) in EGFR, with the A allele associated with a higher risk (p=0.0010, OR=3.61, 95% CI: 1.68–7.73) (Table 2). For grade 3 diarrhea, the A allele of rs917881 was associated with an increased risk but without reaching statistical significance (Table 2).
The most statistically significant SNP associated with grade 2–3 dermatologic toxicity was rs12366035 (C>T, MAF 0.34) in VEGFB, with the T allele associated with a higher risk (p=0.0009, OR=2.91, 95% CI: 1.55–5.46) (Table 2). For grade 3 dermatologic toxicity, the T allele of rs12366035 was associated with an increased risk but without reaching statistical significance (Table 2).
The three most statistically significant SNPs associated with increased risk of grade 2–3 composite toxicity were rs12366035 (C>T, MAF 0.34) in VEGFB (p=0.0007, OR=2.78, 95% CI: 1.54–5.02), rs4035887 (G>A, MAF 0.49) in EPAS1 (p=0.0021, OR=2.33, 95% CI: 1.36–4.00), and rs4864950 (T>A, MAF 0.23) in KDR (p=0.0058, OR=2.41, 95% CI: 1.29–4.51, Figure 2A) (Table 2). For grade 3 composite toxicity, the variant alleles of each gene was associated with an increased risk but without reaching statistical significance (Table 2).
Figure 2. Frequency of rs4864950 AA, TA, and TT genotypes associated with (A) sorafenib-induced grade 2–3 composite toxicity in TARGET study, (B) sorafenib-induced grade 3 composite toxicity in CALGB 80802, and (C) regorafenib-induced grade 2–3 composite toxicity in the Italian cohort.
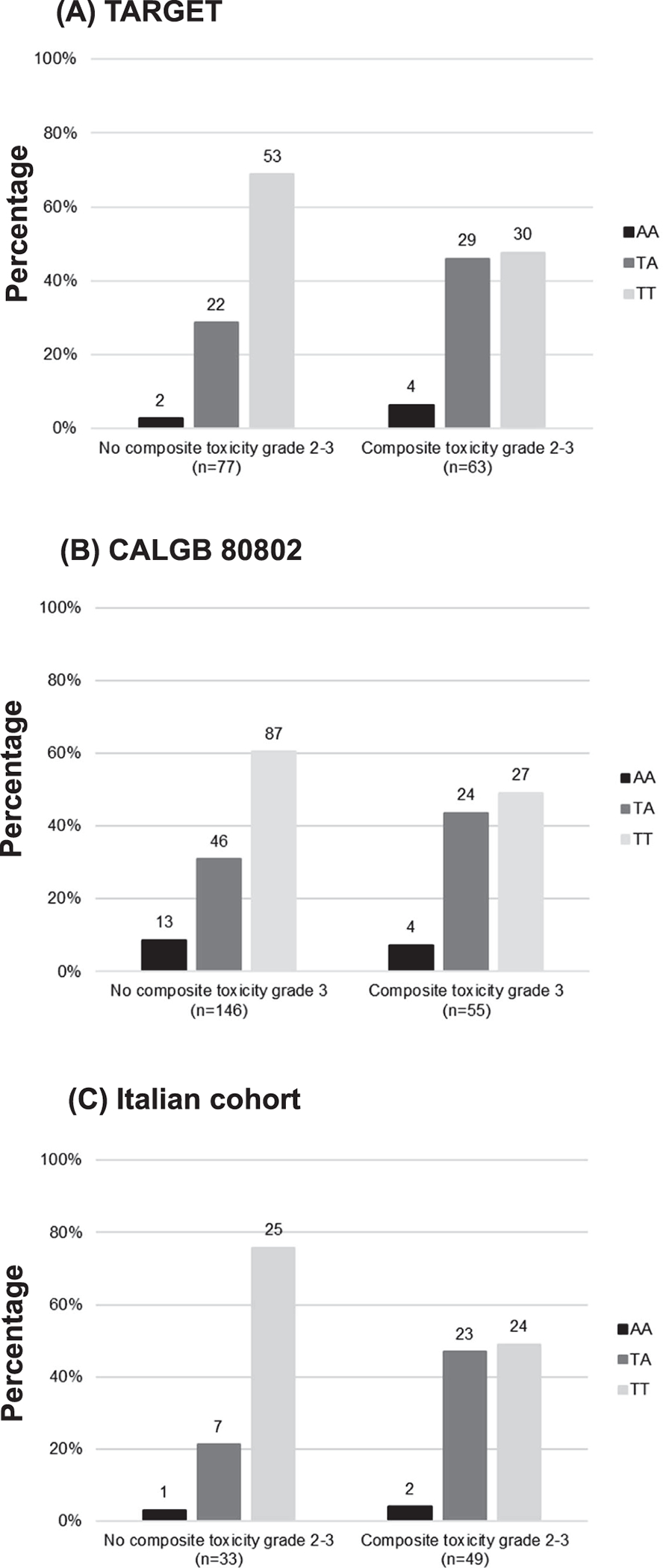
The number above each bar represents the number of patients in each genotype.
Patient characteristics and toxicities in CALGB 80802
A total of 201 HCC patients treated with sorafenib in CALGB 80802 were included in the validation set (Figure 1). Out of the 201 patients, 104 (52%) were treated with sorafenib alone and 97 (48%) were treated with sorafenib plus doxorubicin. Patient demographics and the prevalence of grade 2–3 and grade 3 toxicities are shown in Table 1. No grade 4 toxicities were reported.
SNPs associated with toxicities in CALGB 80802
A total of 6 SNPs identified in TARGET were tested in CALGB 80802 (Table 3). The only SNP that was validated in CALGB 80802 was rs4864950 (T>A) in KDR, as the A allele was associated with a higher risk of grade 3 composite toxicity (p=0.044, OR=2.03, 95% CI: 1.02–4.06, Figure 2B), similar to the increased risk of grade 2–3 composite toxicity in TARGET (p=0.0058, OR=2.41, 95% CI: 1.29–4.51, Table 2).
Table 3.
Association between SNPs and grade 2–3 and grade 3 hypertension, diarrhea, hand- foot syndrome (HFS), and composite toxicity adjusted for age (continuous), gender, race/ethnicity, and treatment arm in CALGB 80802 (validation set) under a dominant genetic model.
| SNP | MAF | grade 2–3 OR (95% CI) | grade 2–3 p-value | grade 3 OR (95% CI) | grade 3 p-value |
|---|---|---|---|---|---|
|
| |||||
| Hypertension | |||||
| rs444904 (G>A) in PIK3R5 | 0.15 | 0.42 (0.19–0.92) | 0.029 | 0.39 (0.10–1.49) | 0.17 |
|
| |||||
| Diarrhea | |||||
| rs917881 (G>A) in EGFR | 0.15 | 0.32 (0.13–0.80) | 0.015 | 0.39 (0.10–1.48) | 0.17 |
|
| |||||
| Dermatologic toxicity (HFS) | |||||
| rs12366035 (C>T) in VEGFB | 0.25 | 1.31 (0.66–2.59) | 0.44 | 1.75 (0.70–4.38) | 0.23 |
|
| |||||
| Composite toxicity | |||||
| rs12366035 (C>T) in VEGFB | 0.25 | 0.80 (0.43–1.49) | 0.47 | 1.18 (0.60–2.35) | 0.63 |
| rs4035887 (G>A) in EPAS1 | 0.44 | 1.42 (0.74–2.74) | 0.29 | 1.32 (0.62–2.79) | 0.47 |
| rs4864950 (T>A) in KDR | 0.25 | 1.39 (0.75–2.58) | 0.30 | 2.03 (1.02–4.06) | 0.044 |
The SNP in bold is the one replicated with the same direction of effect, CI: confidence interval, HFS: hand-foot syndrome, MAF: minor allele frequency, OR: odds ratio, SNP: single nucleotide polymorphism.
Patient’s characteristics and toxicities in regorafenib clinical studies: LCCC 1029 and Italian cohort
A total of 107 CRC patients treated with regorafenib in LCCC 1029 and 82 patients treated with regorafenib in the Italian cohort were included in the analysis (Figure 1). Demographics and the prevalence of grade 2–3 and grade 3 toxicities are shown in Table 1. No grade 4 toxicities were reported.
rs4864950 (T>A) in KDR associated with regorafenib-induced composite toxicity
Variant rs4864950 (T>A) in KDR was also tested in patients treated with regorafenib in LCCC 1029 and the Italian cohort. The A allele of rs4864950 (T>A, MAF 0.22) was associated with a higher risk of grade 2–3 composite toxicity in the Italian cohort (p=0.019, OR=3.35, 95% CI: 1.26–9.61, Figure 2C). For grade 3 composite toxicity, the A allele of rs4864950 was associated with an increased risk of composite toxicity but without reaching statistical significance in the Italian cohort (p=0.381, OR=1.53, 95% CI: 0.59–3.97). The A allele of rs4864950 (T>A, MAF 0.20) was not associated with the risk of grade 2–3 (p=0.421, OR=0.68, 95% CI: 0.26–1.73) or grade 3 (p=0.136, OR=0.40, 95% CI: 0.11–1.26) composite toxicity in LCCC 1029.
Meta-analysis of rs4864950 (T>A) in KDR
In the meta-analysis of all four clinical studies, the A allele of rs4864950 was associated with a higher risk of grade 2–3 composite toxicity (p=0.0050, OR=1.70, 95% CI: 1.17–2.46) and grade 3 composite toxicity without reaching statistical significance (p=0.1100, OR=1.42, 95% CI: 0.92–2.17) (Table 4). In the meta-analysis of only studies where rs4864950 was associated (p<0.05) with composite toxicity (TARGET, CALGB 80802, and Italian cohort), the A allele of rs4864950 associated with a higher risk of grade 2–3 (p=6.79×10−4, OR=2.01, 95% CI: 1.34–3.01) and grade 3 (p=0.0230, OR=1.70, 95% CI: 1.08–2.68) composite toxicity (Table 4).
Table 4.
Meta-analysis of rs4864950 (T>A) in KDR and grade 2–3 and grade 3 composite toxicity.
| TARGET and CALGB 80802 (sorafenib studies) | LCCC 1029 and Italian cohort (regorafenib studies) | TARGET, CALGB 80802, LCCC 1029, and Italian cohort | TARGET, CALGB 80802, and Italian cohort | ||
|---|---|---|---|---|---|
| Grade 2–3 | OR (95% CI) | 1.82 (1.17–2.83) | 1.43 (0.72–2.85) | 1.70(1.17–2.46) | 2.01 (1.34–3.01) |
| p-value | 0.0074 | 0.306 | 0.0050 | 6.79×10−4 | |
| Cochran’s Q p-value | 0.2190 | 0.0235 | 0.0726 | 0.2615 | |
| Grade 3 | OR (95% CI) | 1.75 (1.04–2.95) | 0.91 (0.43–1.93) | 1.42 (0.92–2.17) | 1.70 (1.08–2.68) |
| p-value | 0.0346 | 0.816 | 0.1100 | 0.0230 | |
| Cochran’s Q p-value | 0.5258 | 0.0868 | 0.1512 | 0.7928 |
CI: confidence interval, OR: odds
Regulatory evidence of rs4864950 (T>A) in KDR
Variant rs4864950 (T>A) in KDR is located in intron 29, approximately 55 bp from exon 30 in a DNaseI sensitivity region for endothelial cells, including dermis blood vessel endothelial cell, H3K4me1 and H3K27ac enhancers and H3K9ac promoter in vascular cells. There is no evidence for eQTLs for rs4864950 (T>A) in KDR or SNPs in high LD with it (r2>0.8).
Discussion
We have identified a common intronic SNP (rs4864950) located in KDR that increased the risk of composite toxicity in patients treated with the VEGFR TKIs sorafenib and regorafenib. To the best of our knowledge, this is the first validated demonstration of a genetic marker associated with increased risk of common toxicities induced by two different VEGFR TKIs, which could also suggest a potential class effect.
Sorafenib and regorafenib are both oral VEGFR TKIs with a similar chemical structure. The chemical structure of regorafenib differs from sorafenib by the addition of a fluorine atom in the proximal phenyl ring 22. Both drugs are potent inhibitors of VEGFR2, which is the main VEGFR involved in angiogenesis and it is highly expressed in endothelial cells 23. VEGFR2 is coded by KDR, and we have found an association between rs4864950 (T>A) in KDR and a higher risk of composite toxicity in patients treated with sorafenib and regorafenib. Bioinformatic analyses of rs4864950 (T>A) in KDR and SNPs in high LD (r2>0.8) with it show that they are located in putative regulatory regions in endothelial cells enriched for peaks of histone modifications linked to gene activation (H3K4me1, H3K27ac, and H3K9ac). In addition, rs4864950 (T>A) in KDR and SNPs in high LD (r2>0.8) with it are located in regions of DNAseI hypersensitivity peaks representing regions of open chromatin that are accessible to transcription factors, specifically in endothelial cells. These data suggest that rs4864950 (T>A) might regulate KDR expression through post-transcriptional histone modifications, facilitate the access of cis-regulatory elements, such as transcription factors, and affect the function of VEGFR2 in endothelial cells, which, during VEGFR2 inhibition, increases the risk of composite toxicity. The mechanism underlying the regulation of KDR expression by rs4864950 (T>A) needs to be further explored. Variant rs4864950 has a frequency of about 20% in Europeans and a global frequency of 24% 24, and it might impact a significant proportion of patients treated with these VEGFR TKIs.
Previous genetic studies of sorafenib and regorafenib-induced toxicities have focused on a few SNPs in genes of the VEGF pathway and genes encoding transporters and metabolic enzymes such as ABCB1, UGT1A9, BCRP, and OATP1B1 (for a review 25). Genetic variants in KDR other than rs4864950 (T>A) have also previously been tested for association with sorafenib-induced hypertension, diarrhea, and HFS, but no association was found 26. A review of the literature showed that rs4864950 (T>A) in KDR and SNPs in high LD (r2>0.8) with it had not been investigated for association with toxicities in patients treated with any VEGFR TKIs before. No study has reported a single marker of composite toxicity of hypertension, diarrhea, and dermatologic toxicities for either sorafenib or regorafenib, highlighting the novelty of this study.
Variant rs4864950 (T>A) in KDR was associated with composite toxicity in three out of the four studies. The three studies (TARGET, CALGB 80802, and the Italian cohort) included patients treated with VEGFR TKIs monotherapy. In LCCC 1029, the only study where rs4864950 (T>A) in KDR was not associated with composite toxicity, patients have received regorafenib in combination with FOLFIRI. Diarrhea is one of the most common toxicities induced by both 5-FU and irinotecan, with an incidence of grade ≥3 in 11–14% of patients treated with FOLFIRI 27. HFS is also frequently associated with 5-FU, with an incidence of any grade HFS varying between 2.6% and 18% 28. The mechanism of diarrhea induced by 5-FU and irinotecan involves damage in the intestine mucosa by apoptosis and inflammation of the bowel wall 27, while HFS appears to be related to the accumulation of 5-FU metabolites in the skin 29, differing from than mechanisms of diarrhea and HFS induced by VEGFR TKIs. This similar spectrum of toxicities with different mechanisms might have added confounding effects in our analysis, and we could not observe an association between rs4864950 (T>A) in KDR and composite toxicity in LCCC 1029. In fact, the Cochran’s Q p-values <0.20 when LCCC 1029 was included in the meta-analyses (Table 4) show that LCCC 1029 is the clinical study that differs the most among the four studies.
The limitations of this study include the differences between the cohorts included in the analysis, such as tumor types, treatment regimens, different covariates, and toxicity data available. Although the dermatologic toxicities data available were not concordant for the four studies, the current known mechanisms underlying these toxicities among VEGFR TKIs are the same, which allowed us to include them in the same group of toxicity. We have identified associations between rs4864950 (T>A) in KDR and grade 2–3 composite toxicity in TARGET and the Italian cohort, but grade 3 in CALGB 80802. TARGET and the Italian cohort included only patients treated with VEGFR TKIs as monotherapy, while CALGB 80802 also included patients treated with the VEGFR TKI plus doxorubicin. This difference in treatment regimens might have contributed to the difference in the severity of composite toxicity associated with the SNP. However, the same direction of effect was observed for both grade 2–3 and grade 3 in the three studies, and the meta-analysis indicates that rs4864950 (T>A) in KDR is associated with both grade 2–3 and grade 3 composite toxicity, with no statistically significant heterogeneity (Cochran’s Q p-value >0.20) among the three studies. Lastly, we do not have access to data on race for all patients included in the TARGET and the Italian cohort, and we were not able to include race as a covariate in our analysis for these two studies.
In conclusion, we provide evidence of rs4864950 (T>A) in KDR as a marker to predict common toxicities induced by sorafenib and regorafenib, namely hypertension, diarrhea, and dermatologic toxicities. Variant rs4864950 (T>A) in KDR can be genotyped to identify patients at risk of developing these toxicities when treated with sorafenib and regorafenib and might be used to provide a better risk assessment of these drugs. This genetic marker should be further evaluated in patients treated with other VEGFR TKIs to validate its application as a predictor of drug-induced toxicities of the entire class of VEGFR TKIs.
Funding:
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882 and U24CA196171 (to the Alliance for Clinical Trials in Oncology), UG1CA233373 and UG1CA233290. https://acknowledgments.alliancefound.org. Also supported in part by Bayer Healthcare/Berlex. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests: FI is an AbbVie employee and receives stocks from AbbVie; this work was conceived when FI was a faculty at the University of North Carolina at Chapel Hill, and this work does not represent a potential conflict of interest.
Ethics approval: The clinical studies were conducted in accordance with recognized ethical guidelines. The studies were performed in accordance with the Declaration of Helsinki and were approved by the local IRB. All participants provided written informed consent for sample collection and pharmacogenetic analysis.
ClinicalTrials.gov Identifier: NCT00073307 (TARGET), NCT01015833 (Alliance/CALGB 80802), and NCT01298570 (LCCC 1029).
Data Availability Statement:
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Qin S, Li A, Yi M, Yu S, Zhang M, Wu K. Recent advances on anti-angiogenesis receptor tyrosine kinase inhibitors in cancer therapy. J Hematol Oncol. 2019;12:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng H, Liu W, He T, Hong Z, Yi F, Wei Y, et al. Comparative Efficacy, Safety, and Costs of Sorafenib vs. Sunitinib as First-Line Therapy for Metastatic Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. Front Oncol. 2019; 9:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin X, Yin Y, Shen C, Chen H, Wang J, Cai Z, et al. Adverse events risk associated with regorafenib in the treatment of advanced solid tumors: Meta-analysis of randomized controlled trials. Onco Targets Ther. 2018; 11:6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidinger M Understanding and managing toxicities of vascular endothelial growth factor (VEGF) inhibitors. EJC. 2013; Suppl11:172–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLellan B, Ciardiello F, Lacouture ME. Regorafenib-associated hand-foot skin reaction: Practical advice on diagnosis, prevention, and management. Ann Oncol. 2015; 26:2017–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crona DJ, Skol AD, Leppanen VM, Glubb DM, Etheridge AS, Hilliard E, et al. Genetic variants of VEGFA and FLT4 are determinants of survival in renal cell carcinoma patients treated with sorafenib. Cancer Res. 2019; 79:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glubb DM, Paré-Brunet L, Jantus-Lewintre E, Jiang C, Crona D, Etheridge AS, et al. Functional FLT1 Genetic Variation is a Prognostic Factor for Recurrence in Stage I-III Non-Small-Cell Lung Cancer. J Thorac Oncol. 2015; 10:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y, Fu Y, Xie Q. Anti-angiogenic Agents in Combination With Immune Checkpoint Inhibitors: A Promising Strategy for Cancer Treatment Front Immunol. 2020; 11:1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidinger M, Danesi R. Management of Adverse Events Associated with Cabozantinib Therapy in Renal Cell Carcinoma. Oncologist. 2018; 33:306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin F, Yu H, Xu C, Chen HH, Bai JL. Safety of axitinib and sorafenib monotherapy for patients with renal cell carcinoma: A meta-analysis. J Biomed Res. 2018; 32:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo A, Nannini M, Novelli M, Dalia Ricci A, Scioscio VD, Pantaleo MA. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020; 12:1758835920936932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007; 356:125–134. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: Study designs and statistical tests. Am J Hum Genet. 2014; 95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abou-Alfa GK, Shi Q, Knox JJ, Kaubisch A, Niedzwiecki D, Posey J, et al. Assessment of Treatment With Sorafenib Plus Doxorubicin vs Sorafenib Alone in Patients With Advanced Hepatocellular Carcinoma: Phase 3 CALGB 80802 Randomized Clinical Trial. JAMA Oncol. 2019; 5:1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanoff HK, Goldberg RM, Ivanova A, O’Reilly S, Kasbari SS, Kim RD, et al. Multicenter, randomized, double-blind phase 2 trial of FOLFIRI with regorafenib or placebo as second-line therapy for metastatic colorectal cancer. Cancer. 2018; 124:3118–3126. [DOI] [PubMed] [Google Scholar]
- 16.Falcone A, Cremolini C, Loupakis F. Pharmacodynamic and analysis of angiogenesis-related factors during regorafenib therapy for metastatic colorectal cancer: the REGOLAND project. wwwfondazionearcoorg 2015.
- 17.Machiela MJ, Chanock SJ. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015; 31:3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent JW, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal gnomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012; 40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schütz G, et al. Regorafenib (BAY 73–4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer. 2011; 129:245–255. [DOI] [PubMed] [Google Scholar]
- 23.Abhinand CS, Raju R, Soumya SJ. VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal. 2016; 10:347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherry ST. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Mattia E, Cecchin E, Guardascione M, Foltran L, Di Raimo T, Angelini F, et al. Pharmacogenetics of the systemic treatment in advanced hepatocellular carcinoma. World J Gastroenterol. 2019; 25:3870–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C, Cao Q, Li P, Wang S, Wang J, Wang M, et al. The influence of genetic variants of sorafenib on clinical outcomes and toxic effects in patients with advanced renal cell carcinoma. Sci Rep. 2016; 6:200089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein A, Voigt W, Jordan K. Review: Chemotherapy-induced diarrhea: Pathophysiology, frequency and guideline-based management. Ther Adv Med Oncol. 2010; 2:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwakman JJM, Elshot YS, Punt CJA, Koopman M. Management of cytotoxic chemotherapy-induced hand-foot syndrome. Oncol Rev 2020; 14:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lou Y, Wang Q, Zheng J, Hu H, Liu L, Hong D, et al. Possible Pathways of Capecitabine-Induced Hand-Foot Syndrome. Chem Res Toxicol. 2016; 29:1591–1601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


