Abstract
Wetlands are one of the most threatened ecosystems in the world because more than 70% of the area worldwide has been lost since 1900. Wetland plant species rely greatly on water for seeds and propagules, which may lead to a downstream unidirectional dispersal and accumulation of genetic diversity downstream. However, several species show no support for unidirectional genetic diversity, revealing the complexity of population dynamics and gene flow in wetlands. Here, we used microsatellite loci to address how the past demographic dynamics shaped the contemporary spatial pattern in genetic diversity and population structure of Cyperus papyrus in wetlands of Southeast Africa. Using spatially explicit analysis and coalescent modelling, we found no support for unidirectional dispersal. Instead, we found higher genetic diversity in populations upstream than downstream in the river basin. We also found high admixture among populations, most likely due to connections between adjacent river basins during sporadic floods, and ongoing gene flow due to bird-mediated seed dispersal. Our results suggest stepping-stone migration due to strong isolation-by-distance, but not necessarily unidirectional. Moreover, the past demographic dynamics in the Holocene shaped the current pattern of genetic diversity and structure, leading to higher genetic diversity in populations upstream the Zambezi river basin. Our results also point to the very low genetic diversity of C. papyrus populations in Southeast Africa and the need for management and conservation strategies to guarantee the long-term persistence of the species in the region.
Subject terms: Genetics, Population genetics
Introduction
Wetlands are one of the most threatened ecosystems in the world. Originally covering ~10.3% of Earth's land surface (Kingsford et al. 2016), more than 70% of the wetlands worldwide have been lost since 1900 (Davidson 2014; Kingsford et al. 2016). Agriculture and urban area expansion leading to habitat loss, pollution and overharvesting are threatening the conservation of wetlands and their biodiversity and ecosystem services (Dudgeon et al. 2006; Vörösmarty et al. 2010). With increasing temperatures and climate unpredictability, water flow regimes in rivers and wetlands will change, imposing more challenges to wetland ecosystem conservation (Kingsford 2011).
In Africa, in the early twentieth century 5–7% of the continent was covered by wetlands, but 43% has already been lost (Junk et al. 2013; Kingsford et al. 2016). Most wetlands in Africa are situated in Central and East Africa and comprise four major river basins, the Nile, Niger, Congo and Zambezi, the Okavango Delta in Botswana, the Sudd in southern Sudan and Ethiopia, and several lakes, including the Lake Chad and the Rift Valley lakes, such as Victoria, Tanganyika, Malawi, Turkana, Mweru and Albert (Kabii 2021).
Wetland plant species rely greatly on the water for the dispersal of seeds and propagules, which may lead to a downstream unidirectional dispersal trend (Müller 1974; Boedeltje et al. 2004). The unidirectional dispersal trend may affect population demography and connectivity, leading to population extinction upstream (Anholt 1995; Honnay et al. 2010), and accumulation of genetic diversity downstream (the unidirectional diversity hypothesis, Ritland 1989). In contrast, populations located upstream may present low genetic diversity due to poor compensation from downstream (Honnay et al. 2010). In fact, some studies provide support for the unidirectional dispersal hypothesis. For instance, Myricaria laxiflora in the Three Gorges Valley of the Yangtze River (Liu et al. 2006), Heliconia metallica in Southeast Peru (Schleuning et al. 2011), Sparganium emersum in Niers River, Germany–The Netherlands (Pollux et al. 2009), and Impatiens glandulifera in Wales and Ireland (Love et al. 2013). On the other hand, several studies found no evidence of unidirectional dispersal in genetic diversity in plant species (e.g., Markwith and Scanlon 2007; Chen et al. 2009, 2017). For instance, Hymenocallis coronaria, a macrophyte of rocky rivers in Southeast USA, shows no consistent increase in genetic diversity and gene flow downstream (Markwith and Scanlon 2007), opposing the unidirectional dispersal hypothesis or Kimura’s one-directional stepping-stone model (Kimura and Weiss 1964). The “stepping-stone-model” advocates a decrease in allelic frequency correlation among populations with distance, similar to isolation-by-distance (Wright 1943), due to gene flow among adjacent populations (Kimura 1953; Kimura and Weiss 1964).
The lack of support for the unidirectional genetic diversity hypothesis may reveal the complexity of population dynamics and gene flow in wetlands. The migration dynamics, which may determine the evolutionary success of the species, results from intricate variables including life-history traits, geographical, environmental and human factors. Besides the pollination system that may promote long-distance dispersal, breaking the expected pattern in unidirectional genetic diversity due to water-mediated dispersal of seeds and vegetative propagules, the geological history and paleoclimatic changes may have affected demographic history and species distribution leading to more complex spatial patterns of genetic diversity distribution. The dynamics in effective population size through time and historical gene flow are essential to understand the current spatial patterns in genetic diversity, because past bottlenecks or population range retraction may decrease genetic diversity due to genetic drift (Holsinger and Weir 2009; Excoffier et al. 2009).
Most of the modern main rivers of Southeast Africa were derived from the Proto–Limpopo basin, which occupied an extensive area from Central African plateau towards the Indian Ocean (Goudie 2005; Tweddle 2010). In the Cretaceous/Tertiary transition, the uplift of Okavango, Kalahari and Zambezi axis changed the drainage leading to the formation of the Limpopo, Zambezi and Okavango river basins (Goudie 2005; Moore et al. 2008). The propagation of the East African rift system in the Paleogene c. 30–35 Ma caused the formation or affected the drainage system of the main modern river basins and lakes in East Africa (Grove 1986; Nyblade and Brazier 2002; Chorowicz 2005). Nevertheless, these river basins still have a strong biological connection considering the similarity of fish species (Tweddle 2010; Danley et al. 2012). Okavango and the upper Zambezi rivers, for instance, may be connected during extremely rainy years (Tweddle 2010).
During the Quaternary, several glaciations may have caused drier and colder climates in Africa (Shanahan and Zreda 2000; Ngomanda et al. 2009). The pollen fossil record indicates that Africa was cold and dry between ca. 35 and 15 kyr (Bonnefille et al. 1990), with lower lake levels. The driest period agrees with the LGM, ca. 21 kyr (Gasse et al. 1989). Temperature and precipitation started to increase ca. 15 kyr, also agreeing with the ending of the LGM (Bonnefille et al. 1990). Moreover, a period of strong aridity is recorded across East Africa ca. 13–11 cal. kyr (Bonnefille et al. 1995), matching the European Younger Dryas cold interval. A final glaciation occurred at 8.6 ± 0.5 kyr (Shanahan and Zreda 2000) corresponds to a widespread cold event in the early Holocene (Alley et al. 1997). The cycles of glaciation and interglaciation periods may have caused range contraction and expansion of wetland plant species, leading to an increase in the frequency of one or few alleles due to the dispersal of genotypes at the edge of expansion (Shine et al. 2011). This expansion may result in lower genetic diversity in newly colonized areas (Hewitt 1996), due to the spread of alleles that migrates on the forward motion (allele surfing, Excoffier and Ray 2008; Arenas et al. 2013). The fast colonization of new areas in addition to founder and density-dependent processes may cause low genetic diversity and gradients from the centre towards the periphery of the distribution range (Excoffier et al. 2009; Waters et al. 2013). In wetlands, this may cause higher genetic diversity in populations upstream than downstream, depending on which places were more suitable during glacial periods when populations retracted. Thus, understanding how the Quaternary climate changes affected the geographical distribution of lineages in wetlands may help understanding the dynamics and diversification of species to unravel biogeographical patterns in wetlands, as we address here for the paper reed sedge.
Cyperus papyrus L. (Cyperaceae) is a perennial rhizomatous species using C4 photosynthesis, which is dominant in wetlands across East and Southeast Africa, occurring in large and dense almost monospecific stands (Supplementary Fig. S1). The species evolved ca. 10.9 Ma (Besnard et al. 2009), and has been used by humans since ancient times and continues to be harvested by local riparian communities for fuel, handicraft and building. The species reproduces both asexually by creeping rhizomes, and sexually via pollination by wind. The dispersal unit is a small 1-seeded nutlet that is dispersed by water, birds and wind (Opio et al. 2014). The clearing of wetlands for agricultural and urban areas is decreasing and endangering papyrus populations and the ecosystem services provided by the species (Owino and Ryan 2007; Saunders et al. 2012). However, most studies about C. papyrus so far have been carried out in Kenya, shedding light on the ecology and conservation (e.g., Owino and Ryan 2007; Morrison and Harper 2009), use, management and impacts of anthropogenic disturbances (e.g., Jones et al. 2018), and populations genetics (e.g., Terer et al. 2012; Triest et al. 2014; Geremew et al. 2018a; Mwaniki et al. 2019). A comprehensive genetic study in other river basins is still lacking, hindering the understanding of species evolution and the processes driving the distribution of genetic diversity to support conservation strategies.
Here, we address how the past demographic dynamics shaped the contemporary spatial pattern in genetic diversity and population structure of C. papyrus in Southeast Africa river basins, (Zambezi, Okavango, Messalo, Inkomati, Maputo and Lugela). We specifically test the hypothesis of unidirectional dispersal, i.e., that populations downstream have higher genetic diversity than populations upstream. We tested the hypothesis at two levels, overall populations in different river basins and populations within a single river basin (Zambezi). If this hypothesis holds then we expect (i) that populations downstream have higher genetic diversity than upstream, (ii) higher admixture in populations downstream than upstream, and (iii) unidirectional migration from populations upstream towards populations downstream. On the other hand, because previous studies show strong isolation-by-distance in populations of C. papyrus in Kenya (Terer et al. 2012; Triest et al. 2014), if C. papyrus dispersal follows the stepping-stone model, we expect (iv) a stronger effect of isolation-by-distance than by-environment, (v) high admixture among populations due to stepping-stone gene flow, and (vi) higher migration between nearest neighbour populations. We also expect low genetic diversity within populations and high differentiation among populations due to the high frequency of vegetative reproduction (Opio et al. 2014).
Material and methods
Sampling sites and design
We sampled 1172 individuals of Cyperus papyrus, from 22 populations distributed across five countries in Southeast Africa, comprising six different major river basins (Fig. 1 and Supplementary Table S1). These populations were integrated into nine rivers (Zambezi, Shire, Kafui, São Filipe, Futi, Inkomati, Licungo, Messalo and Okavango), two lakes (Malawi and Gumbua) and four swamps (Nhamiconda, Nhamadiba, Muindi and Luabo).
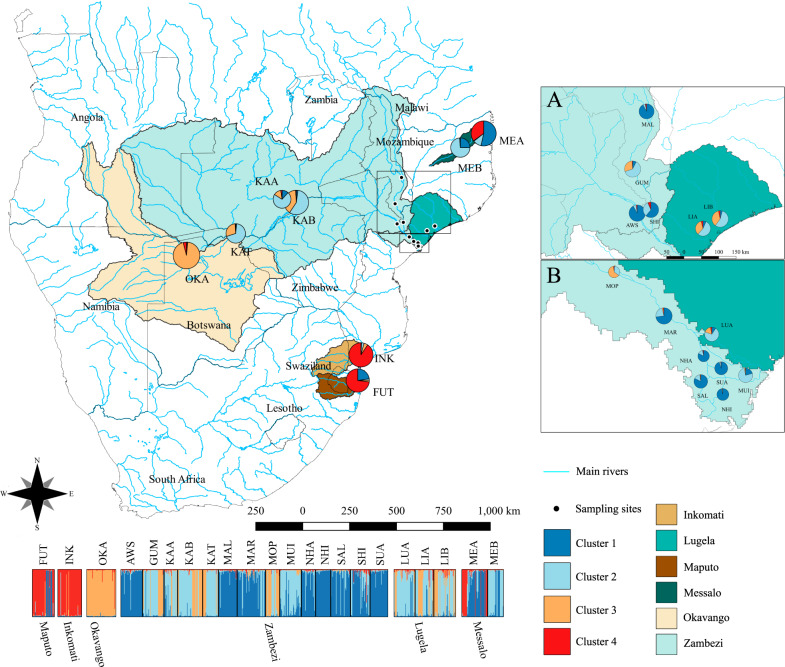
Fig. 1. Geographical distribution of the 22 sampling sites of Cyperus papyrus in Southeast Africa and the Bayesian genetic clustering of individuals based on 19 microsatellite loci (B).
A The geographical distribution of sampling sites onto the river basin shape file obtained from the Hydrologic Derivatives for Modelling and Analysis (HDMA, https://hidrosheds.cc.usgs.gov/hydro.php). Different river basins are highlighted by different colours following the legends. B Geographical distribution of the genetic clusters (K = 4). Each colour represents an inferred genetic cluster. The size of cluster chart section represents population co-ancestry for each cluster. The plot shows the individual ancestry for the most likely K = 4. Details of the populations are provided in Supplementary Table S1.
In each site, we collected leaves and shoots from at least 25 adults (up to 70 individuals in populations with more than 25), following a continuous and linear transect. To avoid sampling duplicates due to vegetative reproduction (Opio et al. 2014), the samples were collected at 3 m distance from each other, following Triest et al. (2014). We recorded the coordinates of the first and the last individuals of each transect using a Garmin GPS·, and mapped populations onto the river basin shape file obtained from the Hydrologic Derivatives for Modelling and Analysis (HDMA, https://hidrosheds.cc.usgs.gov/hydro.php), using the Geographic Information Systems QGis 3.16 (https://www.qgis.org/). Sampling was carried out from January to October 2017.
Genetic data
DNA was extracted from leaves dried in silica gel using the E.Z.N.A. SP Plant DNA Kit® (Omega BioTek, Norcross, GA). For genotyping, we used 19 microsatellites primers (C38, C4, C14, C52, C34, C10, C13F, C135, C5, C3, C28, C7, C56, C27, C12, C23, C15, C62 and C1) developed and optimized for C. papyrus (Triest et al. 2014). The polymerase chain reaction was performed following Triest et al. (2014), in thermal cyclers MJ PTC-200 (Marshall Scientific, Hampton, NH) and Bio-Rad MyCycler (Hercules, CA). The genotyping was performed using the Macrogen facilities (Seoul, Southern Korea). Microsatellite allele-size scoring was extracted using the software GeneMarker v3.0.0 (Holland and Parson 2011). Genotypes were first analysed to minimize genotyping errors due to stutter bands and drop using Micro-Checker v2.2.3 software (Van Oosterhout et al. 2004). The raw data showed no significant evidence of genotyping errors or null alleles.
Genetic diversity and population genetic structure
Because C. papyrus has vegetative reproduction, we first analysed multi-loci genotypes to discard potential clones using GenAlex software v.6.5 (Peakall and Smouse 2006). Our final dataset included 770 individuals (588 potential clones excluded).
For each population, we calculated the following genetic parameters with the software FSTAT 2.9.3.2 (Goudet 1995): A, mean number of alleles per locus; Ar, allelic richness (El Mousadik and Petit 1996); Ho, observed heterozygosity; He, expected heterozygosity under Hardy–Weinberg Equilibrium (Nei 1978); f inbreeding coefficient. We tested whether genetic diversity differs among river basins using permutation tests (10,000 permutations) implemented in FSTAT v2.9.3.2 software (Goudet 1995).
To analyse the genetic structure and population differentiation, we applied a hierarchal AMOVA implemented in Arlequin software v3.5 (Excoffier and Lischer 2010), partitioning the total variance in variance among river basins (FCT); among populations within river basins (FSC); among individuals in the total population (FIT); and among individuals within each population (FIS). A nonparametric test was performed to detect the statistical significance using 10,000 random permutations. We also estimated global FST (Wright 1965) and RST (Slatkin 1995), based on the variance in allele sizes to verify the contribution of stepwise-like mutations to genetic differentiation, and tested the hypothesis that FST = RST (Hardy et al. 2003) using the software SPAGeDi v1.4 (Hardy and Vekemans 2002).
We used Bayesian clustering simulation to assess the number of genetic clusters (K) and admixture among populations of C. papyrus from different river basins, using the software Structure v2.3.4 (Pritchard et al. 2000). We performed a burn-in period of 100,000 repetitions and then, 1,000,000 Markov Chain Monte Carlo (MCMC) repetitions of data collection, with admixture model of ancestry and correlated allele frequencies. To detect the consistency of results, ten independent runs were performed for each K value (K = 1–22). We assessed the most likely K supported by the data using the ΔK method (Evanno et al. 2005) implemented in Structure Harvester v0.6.94 (Earl and VonHoldt 2012).
Demographic dynamics across the river basins in Southeast Africa
We first estimated contemporary effective population size (Ne) using the linkage disequilibrium method implemented in NeEstimator v2.1 and selecting a threshold lowest allele frequency value of 0.05 (Do et al. 2014).
We then estimated demographic parameters using coalescent analysis implemented in the software Migrate-N v5.0.4 (Beerli 2009), to understand the demographic dynamics through time and migration. We estimated theta and effective population size (θ = 4μNe; coalescent or mutation parameter for a diploid genome, where Ne is effective population size), the number of migrants per generation from a scaled migration rate (M = 4Nem/θ, where m is migration rate), and population divergence time. To test the hypothesis of migration, we performed the coalescent analysis with eight groups of populations, based on the river basins and the Structure clusters (see results, Fig. 1): Okavango (OKA population, cluster 3), Inkomati and Maputo (INK and FUT, cluster 4), Lugela (LUA, LIA and LIB, mainly clusters 2 and 3), Messalo (MEA and MEB, clusters 1, 2 and 4), middle Zambezi river basin (KAA, KAB, KAT, mainly clusters 2 and 3), lower Zambezi, Shire river (AWS, SHI, NHA, NHI, cluster 1), lower Zambezi, Gumbua (GUM, MAL, MOP, clusters 1, 2 and 3), lower Zambezi, delta (SAL, SUA, MAR, MUI, clusters 1 and 2). Using Brownian motion model, we ran two long chains, sampling 1,000,000 steps and recorded 10,000 steps with 100,000 Burn-in per replicate. We included four independent chains with different temperatures (1.00, 1.50, 3.00 and 1,000,000.00) in a Metropolis-coupled MCMC procedure (Geyer and Thompson 1995). Bayesian estimates were computed individually for the 19 loci and summarized as weighted values over all loci, we selected the mean values. We ran two independent analyses to check results consistency using Tracer v1.7.1 (Rambaut et al. 2018). To estimate divergence time and demographic parameters we used the microsatellite mutation rate reported for a closely related lineage, Zea mays (Poaceae), of 7.7 × 10−4 (95% CI = 5.2 × 10−4 to 1.1 × 10−3) mutations per allele per generation (Vigouroux et al. 2002). For generation time we used 1 year, because C. papyrus’ steams takes ~6 months to reach maturity and 9–12 months to flowering and senescence (Terer et al. 2012). However, considering the individual lifetime, because it is a perennial sedge, we also used generation times of 2 and 5 years.
Spatial pattern in genetic diversity and structure
We applied multiple regression analyses to test whether populations are isolated-by-distance or by-environment. Thus, to infer the effect of geographical and environmental distance on genetic differentiation, we performed Multiple Matrix Regression with Randomization method, following Wang (2013). We calculated pairwise linearized FST (Slatkin 1995) using Arlequin software, and calculated the logarithm of the geographical distance between pairs of populations. For populations in the same river, the distance was calculated based on the watercourse distance. For environment variables, we obtained the 19 bioclimatic variables from WorldClim (https://www.worldclim.org/bioclim), and selected the bioclimatic variables from the highest loading on each of the three first factors, after performing a factorial analysis with Varimax rotation. We selected (Supplementary Table S2): BIO4 (temperature seasonality), BIO10 (mean temperature of the warmest quarter) and BIO14 (precipitation of driest month). In addition, we obtained the African data of digital elevation model (slope) with 3” resolution from the Digital Elevation Model database from the HDMA (https://www.sciencebase.gov/catalog/item/591f6d02e4b0ac16dbdde1c7).
To test whether populations downstream have higher genetic diversity due to unidirectional dispersal model, we also calculated the distance of each population to the most upstream population (KAA) only for the Zambezi River, due to the higher number of populations and to the distribution of populations up and downstream (Fig. 1). We regressed the genetic diversity (He) and allelic richness (Ar) against geographical distance using Minitab® software. We also regressed the genetic differentiation among populations up and downstream against geographical distance to test for bidirectional stepping-stone model.
To visualize the spatial pattern in population genetic structure and identify corridors and barriers to gene flow, we used the Estimated Effective Migration Surfaces (EEMS) method that explicitly represents genetic differentiation as a function of the migration rates based on an isolation-by-distance model. The effective migration surface represents migration rates that, would produce genetic dissimilarities similar to those observed in the data (Petkova1 et al. 2016). To run EEMS (https://github.com/dipetkov/eems), we used 22 demes and ran the analyses with a burn-in of 500,000 and MCMC length of 10,000,000. We optimized EEMS parameters by adjusting the qEffctProposalS2 (0.1), qSeedsProposalS2 (1.5), mEffctProposalS2 (4.0), mSeedsProposalS2 (4.0) and mrateMuProposalS2 (0.5) such that the acceptance proportions for all parameters, except degrees of freedom, were within 10–40%, as suggested by Petkova et al. (2016). Using rEEMSplots R package (Petkova et al. 2016), we plotted the surfaces of effective migration rates (m) and effective diversity (q), i.e., the expected genetic dissimilarity of two individuals sampled within the same deme, on Southeast Africa map.
Results
Genetic diversity and population genetic structure
Overall, we found low genetic diversity (He < 0.5) and allelic richness (Ar < 2.0) in all populations (Table 1). We also found a high and significant inbreeding coefficient in most populations (Table 1). He ranged from 0.280 to 0.497, but we found no significant difference among river basins (permutation test, p = 0.294). Although Maputo, Inkomati and Okavango had seemingly lower He, only one population was sampled in these river basins (Table 1). We also found no significant difference in allelic richness (p = 0.433) that was also apparently lower in those three river basins (Table 1). Inbreeding coefficient was also statistically similar among river basins (p = 0.345).
Table 1.
Genetic diversity of 22 populations of Cyperus papyrus in Southeast Africa, based on 19 microsatellite loci.
| River basin | Population | N | N-MLGs | He | Ne | 95% CI | A | Ar | f | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Maputo | FUT | 57 | 39 | 0.281 | 2.6 | 2.0–3.4 | 2.6 | 1.55 | 0.468 | <0.001 |
| Inkomati | INK | 51 | 42 | 0.289 | 10.7 | 6.2–17.4 | 2.7 | 1.57 | 0.295 | <0.001 |
| Okavango | OKA | 70 | 51 | 0.339 | 19.6 | 12.5–32.6 | 2.5 | 1.67 | 0.264 | <0.001 |
| Zambezi | AWS | 58 | 38 | 0.393 | 91.9 | 37.8–inf | 2.8 | 1.78 | 0.265 | <0.001 |
| GUM | 62 | 37 | 0.497 | 1.9 | 1.6–2.2 | 3.0 | 2.00 | 0.345 | <0.001 | |
| KAA | 50 | 23 | 0.487 | 7.5 | 4.4–11.7 | 2.9 | 1.98 | 0.253 | <0.001 | |
| KAB | 62 | 43 | 0.478 | 1.4 | 1.2–1.6 | 2.9 | 1.94 | 0.515 | <0.001 | |
| KAT | 34 | 27 | 0.449 | 1.2 | 1.0–1.4 | 2.7 | 1.88 | 0.389 | <0.001 | |
| MAL | 44 | 32 | 0.379 | 2.5 | 2.0–3.2 | 2.5 | 1.73 | 0.179 | <0.001 | |
| MAR | 60 | 49 | 0.452 | 7.8 | 5.1–10.8 | 2.8 | 1.91 | 0.236 | <0.001 | |
| MOP | 44 | 24 | 0.424 | 0.5 | 0.4–0.6 | 2.6 | 1.84 | 0.206 | <0.001 | |
| MUI | 55 | 37 | 0.377 | 1.6 | 1.3–1.9 | 2.4 | 1.70 | 0.180 | <0.001 | |
| NHA | 48 | 24 | 0.341 | 6.5 | 2.9–12.3 | 2.6 | 1.66 | 0.037 | 0.244 | |
| NHI | 44 | 26 | 0.350 | 2.7 | 1.9–5.8 | 2.2 | 1.68 | 0.022 | 0.329 | |
| SAL | 56 | 34 | 0.422 | 2.4 | 2.0–3.0 | 2.7 | 1.83 | 0.167 | <0.001 | |
| SHI | 61 | 34 | 0.337 | 5.1 | 3.2–9.2 | 2.5 | 1.67 | 0.209 | <0.001 | |
| SUA | 59 | 31 | 0.302 | 1.5 | 1.2–1.9 | 2.4 | 1.58 | −0.084 | 0.973 | |
| Lugela | LUA | 60 | 37 | 0.459 | 2.3 | 1.9–2.8 | 2.8 | 1.91 | 0.279 | <0.001 |
| LIA | 59 | 32 | 0.479 | 0.7 | 0.6–0.9 | 2.9 | 1.95 | 0.480 | <0.001 | |
| LIB | 39 | 38 | 0.441 | 1.7 | 1.4–2.0 | 2.9 | 1.89 | 0.467 | <0.001 | |
| Messalo | MEA | 51 | 45 | 0.384 | 2.6 | 2.2–3.2 | 2.7 | 1.76 | 0.391 | <0.001 |
| MEB | 48 | 27 | 0.415 | 2.6 | 2.1–3.2 | 2.7 | 1.81 | 0.312 | <0.001 | |
| Mean | 53.3 | 35.0 | 0.434 | 2.7 | 1.79 | 0.267 | ||||
| SD | 8.7 | 7.9 | 0.065 | 0.2 | 0.14 | 0.154 | ||||
| Total | 1172 | 770 | 0.499 | 0.276 | <0.001 |
N number of individuals sampled, N-MLGs sample size without repeated multilocus genotypes, He expected heterozygosity under Hardy–Weinberg equilibrium, A mean number of alleles per locus, Ar allelic richness, Ne effective population size based on linkage disequilibrium, 95% CI 95% confidence interval for Ne, f inbreeding coefficient, SD standard deviation.
We found high genetic differentiation among populations based on FST (0.204; p < 0.001) and RST (0.262; p < 0.001), that did not differ statistically (p = 0.112), suggesting that stepwise mutations did not cause a shift in mean allele sizes between populations. We found high and significant genetic differentiation among river basins (FCT = 0.070, p < 0.001) and among populations within river basins (FSC = 0.153, p < 0.001). However, variation was higher among individuals in total populations (FIT = 0.373, p < 0.001) and among individuals within each population (FIS = 0.204, p < 0.001). We also performed a hierarchical AMOVA based on Structure clusters (4 clusters) and found higher variation among clusters (FCT = 0.112, p < 0.001), and lower among populations within clusters (FSC = 0.099, p < 0.001), but inbreeding in total population (FIT = 0.363, p < 0.001) and within populations (FIS = 0.204, p < 0.001) were very similar to the former values obtained with the original populations.
Bayesian clustering showed that K = 4 was most likely to explain our data (Fig. 1 and Supplementary Fig. S2), but with high admixture among populations either from the same or different river basins. The unique populations from Maputo (FUT) and Inkomati (INK) river basins were grouped in cluster 4 (Fig. 1), although some individuals showed admixture with clusters 1 and 3. The Okavango (OKA) population was assigned to cluster 3, and Zambezi populations showed high admixture, with individuals assigned to the four clusters (Fig. 1). Populations from Lugela (LUA, LIA and LIB) river basin were assigned mainly to clusters 2 and 3, and Messalo populations (MEA and MEB) showed high admixture, with individuals assigned to clusters 1, 2 and 4.
Demographic dynamics across the river basins in Southeast Africa
Contemporary effective population size (Ne) based on the NE Estimator was low in most populations, ranging from 0.5 to 91.9 (Table 1). Population AWS, from Zambezi river basin (Shire River), had the highest Ne (91.9, 95% CI = 37.8–inf). However, this may be a result of sampling error, because the estimation of Ne converged to infinite; therefore, the highest Ne was observed in the Okavango population (Ne = 19.6; 95% CI = 12.5–32.6), followed by Inkomati (Ne = 10.7; 95% CI = 6.2–17.4).
Coalescent simulations showed relatively high effective population sizes (Table 2). Populations from the Middle Zambezi had the highest Ne (1666), followed by Lower Zambezi Delta (Ne = 1340), and Lower Zambezi Gumbua (Ne = 1184). Lineages from the Middle Zambezi diverged first, at ca. 6.6 kyr (considering 1 year generation time) to 33.3 kyr (considering 5 years generation time), followed by populations Lower Zambezi (ca. 5.3–26.8 kyr). Okavango, Lugela, Inkomat and Messalo lineages diverged only recently (Table 2).
Table 2.
Demographic parameters of Cyperus papyrus groups of populations in Southeast Africa, based on the coalescent analysis performed with Migrate-N.
| Population/group | θ | 95% HPD | Ne | TMRCA 1 | TMRCA 2 | TMRCA 5 |
|---|---|---|---|---|---|---|
| Inkomat | 1.813 | 0.000–3.733 | 589 | 2355 | 4709 | 11,773 |
| Lower Zambezi Shire | 3.084 | 0.467–5.667 | 1001 | 4006 | 8011 | 20,027 |
| Lower Zambezi Delta | 4.127 | 1.200–7.067 | 1340 | 5360 | 10,720 | 26,800 |
| Lower Zambezi Gumbua | 3.649 | 0.600–6.600 | 1185 | 4739 | 9478 | 23,696 |
| Middle Zambezi | 5.133 | 2.733–7.333 | 1667 | 6666 | 13,332 | 33,330 |
| Lugela | 1.624 | 0.000–3.600 | 527 | 2109 | 4218 | 10,545 |
| Messalo | 2.205 | 0.133–4.267 | 716 | 2864 | 5728 | 14,319 |
| Okavango | 1.467 | 0.000–3.400 | 28 | 1906 | 3811 | 9528 |
θ mutation parameter, 95% HPD highest probability density, Ne effective population size, TMRCA time of the most recent common ancestor in years for 1 year generation time (TMRCA 1), 2 years (TMRCA 2) and 5 years (TMRCA 5).
Overall, we found moderate to high migration between river basins or groups (Nem > 1.0 migrants per generation; Supplementary Table S3). Overall, Inkomat, Lugela and Okavango were sinks of migration, i.e., received immigrants from the other basins but sent no migrants (Supplementary Table S3). Messalo and the Lower and Middle Zambezi populations were important sources of migrants and also receive immigrants from each other.
Spatial pattern in genetic diversity and structure
We found high and significant genetic differentiation (FST) among all pairs of populations (Supplementary Table S4). Overall, Okavango population was the most divergent, with the highest values of pairwise FST. Differentiation between populations was significantly related to geographical distance (r2 = 0.192, p = 0.003, Supplementary Table S5). Although BIO14 (precipitation of the driest month, r2 = 0.192, p < 0.036) had a significant relationship with pairwise FST, the regression coefficient was negative (b = −0.187), implying no isolation-by-environment. We found no significant relationship between FST and BIO4 (temperature seasonality), mean BIO10 (temperature of a warmest quarter) and slope (all p > 0.05, Supplementary Table S5).
We found a significant relationship between genetic diversity and allelic richness and distance from the most upstream population in Zambezi river basin. However, genetic diversity (r2 = 0.369, p = 0.021, Supplementary Fig. S3a) and allelic richness (r2 = 0.394, p = 0.016, Supplementary Fig. S3b) decreased with geographical distance. Genetic differentiation was not significantly related to the geographical distance (r2 = 0.016, p = 0.227, Supplementary Fig. S3c) among populations from Zambezi river basin.
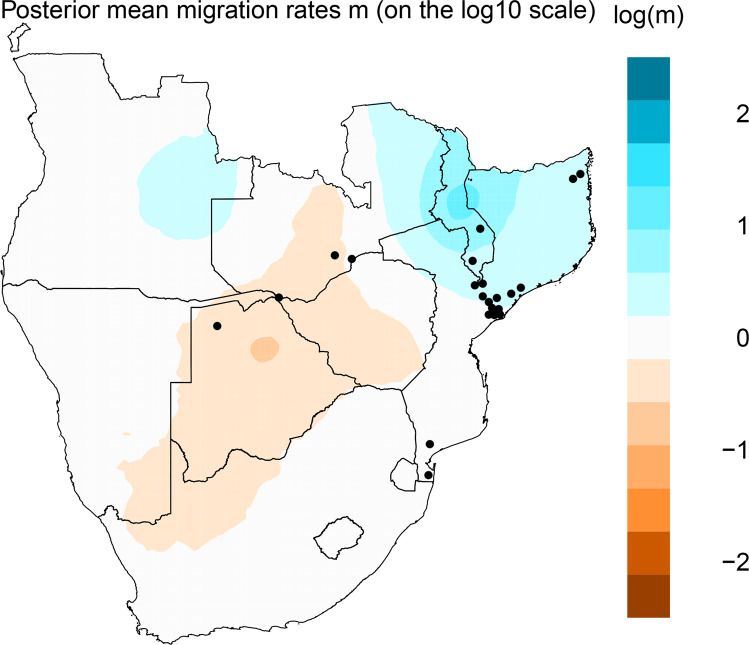
Our results showed that genetic similarities between demes tended to decay faster with distance, in the Middle Zambezi (populations KAA, KAB and KAT), Okavango (OKA), Maputo (FUT) and Inkomati (INK), and thus these demes have lower effective migration (Fig. 2). Populations of Lower Zambezi (see Fig. 1), Lugela and Maputo, showed slightly higher effective migration (Fig. 2). The diagnostic plot of the EEMS showed no deviations from the fitted model isolation-by-distance (Supplementary Fig. S4).
Fig. 2. Estimated effective migration for 22 populations of Cyperus papyrus in Southeast Africa.
The colour contour plot shows the posterior distribution of the migration rates on a log10 scale, relative to the overall migration rate across the geographical distribution. For instance, log(m) = 1 corresponds to an effective migration that is tenfold faster than the average.
Discussion
Seed dispersal and river basin connections partially explain the lack of unidirectional dispersal
Our findings show no support for a unidirectional dispersal. Populations upstream and downstream from different river basins showed similar values of genetic diversity (He) or allelic richness (Ar), and admixture was similar in populations upstream or downstream, contrary to predictions i and ii, respectively. Migration was similar in both directions upstream or downstream opposing the expected under unidirectional dispersal (prediction iii).
We found high genetic differentiation among populations from the same river basin, and among river basins. However, we found high admixture, but not only between neighbour populations (prediction v) nor higher admixture among populations downstream (ii). Notably, our results showed admixture among populations from Zambezi, Lugela and Messalo river basins, and Zambezi and Okavango. Zambezi, Lugela and Messalo river basins are connected during extremely rainy years, leading to gene flow among populations from these different river basins. Similarly, the Okavango river basin can be connected to the middle Zambezi, throughout the Chobe and Magwekwana rivers during exceptional flood years (Tweddle 2010), explaining the clustering in similar groups and common ancestry. Zambezi delta populations may also be connected due to flood episodes. The connection between river basins may also explain the high migration between river basins such as Zambezi, Messalo and Lugela, and the lack of unidirectional migration (prediction iii). This connection was also evidenced by coalescent analysis and effective migration surface (EEMS method). Population KAT is upstream and KAB is downstream Victoria falls in Zambezi, but they were assigned to the same Structure genetic clusters (2 and 3). Environmental conditions such as the high-water velocity at the confluence between Kafue and Zambezi rivers (Pinay 1988) may increase seed carry over long distances leading to high connectivity among populations and admixture.
The high admixture is evidence of ancient gene flow but also ongoing gene flow among some populations. It is important to note that C. papyrus reproduces mainly by vegetative rhizome growth, and is wind-pollinated. Wind pollination may potentially promote long-distance pollen dispersal, although in fragmented populations wind pollination may be limited to short distances (Koenig and Ashley 2003). This local pollen dispersal may lead to highly variable seed production among populations, but a masting seed production (sporadic production of large seed yields by a population) that are characteristics of wind-pollinated species (Satake and Iwasa 2002). The overharvesting of C. papyrus’ populations and loss of habitat (Nerima and Orikiriza 2013), may decrease the effectiveness of wind-pollen dispersal due to population fragmentation and isolation. We hypothesize that vegetative reproduction and limited pollen dispersal may contribute to the high differentiation among populations evidenced here. On the other hand, most long-distance gene flow may be due to seed dispersal. Hydrochory facilitates the connectivity among populations of C. papyrus along the river basins (Triest et al. 2014; Terer et al. 2015). Water may be the major disperser agent for C. papyrus, leading to long-distance dispersal, but mostly unidirectional. However, birds are also important seed dispersers and may carry seeds over a long distance, potentially assuaging the tendency of unidirectional water-mediated dispersal. The bird seed dispersal together with the sporadic connection between river basins during extremely rainy years may break the expected pattern of unidirectional dispersal and partially explain the opposite direction in the distribution of genetic diversity in relation to that expected by unidirectional dispersal. Our results showed that populations upstream the Zambezi river basin had higher genetic diversity than populations downstream the Zambezi delta, opposing the unidirectional dispersal hypothesis (Honnay et al. 2010). In addition, we found a tendency for higher migration between nearest groups or rivers, supporting a stepping-stone dispersal (prediction vi). Moreover, the prevailing of Tropical easterlies’ winds in Southeast Africa (Miller et al. 2019) may also account for pollen movement from populations downstream towards upstream rivers, leading to admixture and accumulation of genetic diversity upstream.
Following our expectation (iv), we found no evidence of isolation-by-environment and a strong effect of isolation-by-distance overall river basins, which is probably mediated by hydrochory leading to higher gene flow among neighbouring populations, expected by the stepping-stone model (prediction iv). In fact, C. papyrus show isolation-by-distance in other river basins, such as in lakes and wetlands in Kenya (Terer et al. 2015).
Demographic dynamics due to climate changes during the Holocene may also explain the lack of unidirectional dispersal footprint
Our findings show that demographic dynamics during the Holocene may have shaped the spatial patterns in genetic diversity and differentiation in populations of C. papyrus from Southeast Africa. The coalescent model indicated that lineages from the Middle Zambezi were the first to diverge from the MRCA, at ca. 6.6 kyr, suggesting that the recent Holocene climate changes are more related to the contemporary spatial pattern in genetic diversity and differentiation in C. papyrus than the geological history of the river basins. For instance, the capture of the middle Zambezi by the lower Zambezi during the transition of Pliocene and Pleistocene (Nugent 1990; Moore et al. 2008) is more ancient than the contemporary lineages of C. papyrus.
In fact, the MRCAs coincide with the last glaciation detected in Africa (Shanahan and Zreda 2000) that corresponds to a widespread cold event in the early Holocene (Alley et al. 1997), which may have caused a retraction in C. papyrus range and divergence of the main groups of Zambezi River. The spread of these ancestral lineages may have caused the lower genetic diversity in the new colonizing areas downstream and the isolation-by-distance. Moreover, the climate became drier and more unpredictable in Southeast Africa in the last ca. 5 cal kyr BP, probably due to the return of the westerlies from the equator, the high-pressure cell, and the South Indian Ocean Convergence Zone, with the most pronounced arid event at ca. 2.8 cal kyr BP (Miller et al. 2019). The drier climatic conditions and unpredictability may have caused cycles of populations retraction and expansion of C. papyrus, leading to loss of genetic diversity in some populations, and also lineage sorting and admixture, leading to higher genetic diversity in populations upstream than downstream, depending on which places were more suitable during glacial periods when populations retracted.
It is possible that populations upstream the Zambezi river basin were in more stable sites and were the source of propagules to new areas downstream, leading to an opposite pattern expected by the unidirectional dispersal model. In addition, several episodes of colder and drier climates occurred in Southeast Africa (Tyson 1999), ca. 4700 to 4200 BP and then from 2500 to 3200 BP, the Neoglacial period, a widespread event reported elsewhere in the world. The climate was very variable in the last 2000 BP. From 2000 to 900 BP the climate was characterized by cooling, warming from about 250 to 600 AD, and cooling again until the year 900 AD. From 900 to 1300 AD the climate was warm and variable, corresponding to the Medieval Warm Epoch (Tyson 1999). The five-century cooling from 1300 to 1850 AD, corresponds to the Little Ice Age in Southeast Africa. These cycles of warm and colder periods may explain the low genetic diversity and the variable migration among populations. While some river basins show more recent gene flow, in the last 570 years, most populations show negligible migration (<1.0 migrants per generation). The migration between far and apparently isolated river basins, such as Messalo and Maputo and Inkomati may be an effect of stepping-stone dispersal mediated by the connection between Messalo and Zambezi, but also raises the hypothesis of human-assisted migration.
The scenario of cycles of population retraction and expansion due to drier and colder periods in the African continent still holds if we consider a longer generation time such as 5 years (Table 2). The African continent, including East Africa, was cold and dry between ca. 35 and 15 kyr, with the driest period occurring at ca. 21 kyr, agreeing with the LGM (Bonnefille et al. 1990). Fossil pollen records also indicate large variability in precipitation during this time, which is consistent with the record of multiple glacial advances and retreats during the last glaciation (Shanahan and Zreda 2000). A period of strong aridity is also recorded across East Africa at 13–11 cal. kyr, coinciding with the European Younger Dryas cold interval (Bonnefille et al. 1990), followed by another glacial period at the Early Holocene ca. 8.6 kyr, the last glaciation detected in Africa (Shanahan and Zreda 2000) that corresponds to a widespread cold event in the early Holocene (Alley et al. 1997).
Life-history and environmental conditions may explain the spatial pattern in genetic diversity
Our results reveal that the geographical location and environmental conditions of the site play an important role in the patterns of genetic diversity and differentiation. In fact, populations of C. papyrus have low genetic diversity, similar to Kenya populations in East Africa (He = 0.412, SD = 0.149; Terer et al. 2015), and slightly lower than populations in the lake Naivasha, Kenya (He = 0.545, SD = 0.022, Triest et al. 2014). The species reproduces mainly by vegetative growth, which may lead to low genetic diversity, polymorphism and high differentiation among populations.
The highest level of genetic diversity in the Gumbua population (He = 0.497) can be explained mainly by the physical-geographical conditions of the site. The Gumbua lake (transitory lake) is crossed by the Shire River (Palamuleni et al. 2010). These conditions may allow genetic richness accumulation considering the contribution of individuals from upstream Shire River and small tributaries around the lake. In contrast, the low level of genetic diversity in the population at the Inkomati river basin may be a result of intense anthropogenic disturbances in southern Mozambique during the last 42 years (Smith et al. 2008). Anthropogenic disturbances may also affect the spatial pattern in the population genetic diversity of C. papyrus. In wetlands, changes in landscape features may disrupt gene flow and affect spatial patterns in genetic diversity. For instance, dams for crop irrigation and urban development may affect genetic differentiation and bidirectional gene flow in Miscanthus lutarioriparius in the Yangtze River in China (Yan et al. 2016).
Similarly, populations downstream the Zambezi river basin showed lower genetic diversity than populations upstream. Those populations downstream are located in the most disturbed area of Mozambique, close to urban and industrial areas and agriculture. Thus, the low contemporary effective population sizes and genetic diversity may be due to recent impacts on populations of C. papyrus. For instance, C. papyrus population INK showed very low genetic diversity (0.289) and is intensively harvested by the local community for building material, cattle grazing and other household objects (Jones et al. 2018). The construction of three dams along the middle Zambezi 70 years ago (Tumbare 2000) may have contributed to the genetic differentiation between populations, given the short generation time of C. papyrus, and may partially explain the low connectivity. Cyperus papyrus has a short life cycle (less than 2 years), seeds may germinate promptly after dispersal, and plants may flower within approximately 6 months (Rongoei and Outa 2016). Although the vegetative reproduction system of the species may affect generation time (Rongoei and Outa 2016), given the fast life cycle and time to first flowering and reproduction, anthropogenic impacts on genetic diversity are feasible. It is important to note that the analysis-based NeEstimator retrieved low values of Ne. This is most likely due to deviation from the models’ assumptions, which assumes isolated and random mating populations. In addition, we found a high inbreeding coefficient indicating selfing or biparental inbreeding. Nevertheless, C. papyrus may have strong fine-scale spatial genetic structure due to limited seed dispersal, with very low neighbourhood sizes ranging from 5 to 11 (Geremew et al. 2018b). Thus, it is likely that the Ne estimates based on linkage disequilibrium reflect more the effective size of local neighbourhoods than the effective size of a whole population. Also, the low genetic diversity may constrain a reliable estimate of Ne.
Concluding remarks
Our findings show that the spatial pattern in genetic diversity and population structure of C. papyrus in Southeast Africa is the outcome of intricate factors. We found no evidence of unidirectional dispersal, yet our data supports stepping-stone dispersal, but not necessarily unidimensional. On one hand, the past demographic dynamics in the Holocene, caused by several episodes of cool and dry climate interposed by wet and hot periods shaped the current pattern of genetic diversity and population structure among the adjacent river basins, leading to higher genetic diversity in populations upstream the Zambezi river basin. The high level of admixture may reveal past and ongoing gene flow among C. papyrus populations due to seed dispersal and connection among adjacent river basins due to floods during extremely wet years.
It is important to highlight the very low genetic diversity of C. papyrus populations in Southeast Africa, which may hamper the long-term viability of these populations. This may be due to the demographic dynamics, but also due to anthropogenic disturbances, such as overharvesting decreasing population sizes (Nerima and Orikiriza 2013), pollution and loss of habitat due to urban and agriculture expansion. It is also important to acknowledge that the number of populations sampled in the river basins was very different, which may affect the comparison of genetic diversity among river basins and the analysis of unidirectional dispersal in different river basins. Unfortunately, we could not sample more populations in Okavango due to the Botswana government's authorization to access sample sites and in other river basins in Mozambique due to civil war. Future studies may include more populations from these other river basins, and the upper Zambezi to better understand the spatial distribution of genetic diversity. It is also important to highlight that this is the first study about the spatial pattern of genetic diversity in the Southeast Africa River basins, and the first study about C. papyrus in the region. The Okavango Delta is unique in the world that flows towards the interior of the continent, to the Kalahari Desert in Botswana (Ramberg et al. 2006). Thus, a more in-depth study of this river basin is still needed. The Zambezi is an important river in the region, with a catchment area of ca. 1.32 million km2 in eight countries, providing ecosystem services to hundreds of thousands (Tweddle 2010). The low genetic diversity detected for C. papyrus may give a clue about the impacts of human disturbances along the catchment of this river basin and may provide important data to design sound conservation and management strategies.
Supplementary information
Acknowledgements
ELM had a fellowship from CAPES in the International Collaboration Program Science for Development (PGCD), which we gratefully acknowledge. LDV receive a CAPES fellowship. RGC is continuously supported by a productive grant from CNPq (project # 301300/2016-0) that partially supported this project.
Author contributions
ELM, LT and RGC conceived, supervised and funded the project. ELM collected samples and extracted genomic DNA. ELM and TS performed lab work. ELM, LDV and RGC performed genetic analyses. LDV and RGC performed the spatial analyses and designed figures. RGC wrote the manuscript draft. All authors read and approved the final manuscript.
Data availability
SSR genotypes have been deposited with Dryad 10.5061/dryad.t76hdr840 and additional data are provided as supporting information in the online version of this article.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor Olivier Hardy.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-022-00563-9.
References
- Alley RB, Mayewski PA, Sowers T, Stuiver M, Taylor KC, Clark PU. Holocene climatic instability: a prominent, widespread event 8200 yr ago. Geology. 1997;25:483–486. [Google Scholar]
- Anholt BR. Density dependence resolves the stream drift paradox. Ecology. 1995;76:2235–2239. [Google Scholar]
- Arenas M, François O, Currat M, Ray N, Excoffier L. Influence of admixture and paleolithic range contractions on current European diversity gradients. Mol Biol Evol. 2013;30:57–61. doi: 10.1093/molbev/mss203. [DOI] [PubMed] [Google Scholar]
- Beerli P (2009) How to use MIGRATE or why are Markov chain Monte Carlo programs difficult to use? In: Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C (eds). Population genetics for animal conservation, 1. Cambridge University Press, p 42–79
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol Biol Evol. 2009;26:1909–1919. doi: 10.1093/molbev/msp103. [DOI] [PubMed] [Google Scholar]
- Boedeltje G, Bakker JP, Brinke AT, Van Groenendael JM, Soesbergen M. Dispersal phenology of hydrochorous plants in relation to discharge, seed release time and buoyancy of seeds: the flood pulse concept supported. J Ecol. 2004;92:786–796. [Google Scholar]
- Bonnefille R, Riollet G, Buchet G, Icole M, Lafont R, Arnold M, et al. Glacialanterglacial record from intertropical Africa, high resolution pollen and carbon data at Rusaka, Burundi. Quat Sci Rev. 1995;14:917–936. [Google Scholar]
- Bonnefille R, Roeland JC, Guiot J. Temperature and rainfall estimates for the past 40,000 years in equatorial Africa. Nature. 1990;346:347–349. [Google Scholar]
- Chen Y, Li X, Yin L, Cheng Y, Li W. Genetic diversity and migration patterns of the aquatic macrophyte Potamogeton malaianus in a potamo-lacustrine system. Freshw Biol. 2009;54:1178–1188. [Google Scholar]
- Chen Y, Liu Y, Fan X, Li W, Liu Y. Landscape-scale genetic structure of wild rice Zizania latifolia: the roles of rivers, mountains and fragmentation. Front Ecol Evol. 2017;5:1–17. [Google Scholar]
- Chorowicz J. The East African rift system. J Afr Earth Sci. 2005;43:379–410. [Google Scholar]
- Danley PD, Husemann M, Ding B, DiPietro LM, Beverly EJ, Peppe DJ. The impact of the geologic history and paleoclimate on the diversification of East African Cichlids. Int J Evol Biol. 2012;2012:1–20. doi: 10.1155/2012/574851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NC. How much wetland has the world lost? Long-term and recent trends in global wetland area. Mar Freshw Res. 2014;65:934–941. [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- Dudgeon D, Arthington AH, Gessner MO, Kawabata ZI, Knowler DJ, Lévêque C, et al. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biol Rev Camb Philos Soc. 2006;81:163–182. doi: 10.1017/S1464793105006950. [DOI] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Foll M, Petit RJ. Genetic consequences of range expansions. Annu Rev Ecol Evol Syst. 2009;40:481–501. [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Gasse F, Lédée V, Massault M, Fontes JC. Water-level fluctuations of Lake Tanganyika in phase with oceanic changes during the last glaciation and deglaciation. Nature. 1989;342:57–59. [Google Scholar]
- Geremew A, Stiers I, Sierens T, Kefalew A, Triest L. Clonal growth strategy, diversity and structure: a spatiotemporal response to sedimentation in tropical Cyperus papyrus swamps. PLoS One. 2018;13:1–15.. doi: 10.1371/journal.pone.0190810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremew A, Woldemariam MG, Kefalew A, Stiers I, Triest L. Isotropic and anisotropic processes influence fine-scale spatial genetic structure of a keystone tropical plant. AoB Plants. 2018;10:plx076. doi: 10.1093/aobpla/plx076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CJ, Thompson EA. Annealing Markov chain Monte Carlo with applications to ancestral inference. J Am Stat Assoc. 1995;90:909–920. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Goudie AS. The drainage of Africa since the Cretaceous. Geomorphology. 2005;67:437–456. [Google Scholar]
- Grove AT (1986) Geomorphology of the African Rift System. Geological Society, London, Special Publications, 25, p 9–16
- Hardy OJ, Charbonnel N, Fréville H, Heuertz M. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics. 2003;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- Holland MM, Parson W. GeneMarker® HID: a reliable software tool for the analysis of forensic STR data. J Forensic Sci. 2011;56:29–35. doi: 10.1111/j.1556-4029.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnay O, Jacquemyn H, Nackaerts K, Breyne P, Van Looy K. Patterns of population genetic diversity in riparian and aquatic plant species along rivers. J Biogeogr. 2010;37:1730–1739. [Google Scholar]
- Jones MB, Kansiime F, Saunders MJ. The potential use of papyrus (Cyperus papyrus L.) wetlands as a source of biomass energy for sub-Saharan Africa. GCB Bioenergy. 2018;10:4–11. [Google Scholar]
- Junk WJ, An S, Finlayson CM, Gopal B, Květ J, Mitchell SA, et al. Current state of knowledge regarding the world’s wetlands and their future under global climate change: a synthesis. Aquat Sci. 2013;75:151–167. [Google Scholar]
- Kabii FM. Factors influencing tour guides job performance in selected tourist circuits in Kenya. Moi Univ. 2021;1:1–201. [Google Scholar]
- Kimura M. ‘Stepping stone’ model of population. Annu Rep Natl Inst Genet Jpn. 1953;3:62–63. [Google Scholar]
- Kimura M, Weiss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–76. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford RT. Conservation management of rivers and wetlands under climate change—a synthesis. Mar Freshw Res. 2011;62:217–222. [Google Scholar]
- Kingsford RT, Basset A, Jackson L. Wetlands: conservation’s poor cousins. Aquat Conserv Mar Freshw Ecosyst. 2016;26:892–916. [Google Scholar]
- Koenig WD, Ashley MV. Is pollen limited? The answer is blowin’ in the wind. Trends Ecol Evol. 2003;18:157–159. [Google Scholar]
- Liu Y, Wang Y, Huang H. High interpopulation genetic differentiation and unidirectional linear migration patterns in Myricaria laxiflora (Tamaricaceae), an endemic riparian plant in the Three Gorges Valley of the Yangtze River. Am J Bot. 2006;93:206–215. doi: 10.3732/ajb.93.2.206. [DOI] [PubMed] [Google Scholar]
- Love HM, Maggs CA, Murray TE, Provan J. Genetic evidence for predominantly hydrochoric gene flow in the invasive riparian plant Impatiens glandulifera (Himalayan balsam) Ann Bot. 2013;112:1743–1750. doi: 10.1093/aob/mct227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwith SH, Scanlon MJ. Multiscale analysis of Hymenocallis coronaria (Amaryllidaceae) genetic diversity, genetic structure, and gene movement under the influence of unidirectional stream flow. Am J Bot. 2007;94:151–160. doi: 10.3732/ajb.94.2.151. [DOI] [PubMed] [Google Scholar]
- Miller C, Finch J, Hill T, Peterse F, Humphries M, Zabel M, et al. Late Quaternary climate variability at Mfabeni peatland, eastern south Africa. Clim Past. 2019;15:1153–1170. [Google Scholar]
- Moore AE, Woody Cotterill FPD, Main MPL, Williams HB (2008) The Zambezi River. In: Large rivers: geomorphology and management, John Wiley & Sons, Ltd, Chichester, UK, p 311–332
- Morrison E, Harper D. Ecohydrological principles to underpin the restoration of Cyperus papyrus at Lake Naivasha, Kenya. Ecohydrol Hydrobiol. 2009;9:83–97. [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Müller K. Stream drift as a chronobiological phenomenon in running water ecosystems. Annu Rev Ecol Syst. 1974;5:309–323. [Google Scholar]
- Mwaniki P, Taita T, Sierens T, Triest L. Barriers to genetic connectivity of smooth flatsedge (Cyperus laevigatus) among alkaline-saline lakes of Eastern Rift Valley (Kenya) Aquat Bot. 2019;155:38–44. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerima G, Orikiriza LJB. Abundance and conservation of Cyperus papyrus in the Nakivubo wetland, Uganda. Int J Sci Basic Appl Res. 2013;7:75–82. [Google Scholar]
- Ngomanda A, Chepstow-Lusty A, Makaya M, Favier C, Schevin P, Maley J, et al. Western equatorial African forest-savanna mosaics: a legacy of late Holocene climatic change? Clim. 2009;5:647–659. [Google Scholar]
- Nugent C. The Zambezi River: tectonism, climatic change and drainage evolution. Palaeogeogr Palaeoclimatol Palaeoecol. 1990;78:55–69. [Google Scholar]
- Nyblade AA, Brazier RA. Precambrian lithospheric controls on the development of the East African rift system. Geology. 2002;30:755–758. [Google Scholar]
- Opio A, Jones MB, Kansiime F, Otiti T. Growth and development of Cyperus papyrus in a tropical wetland. Open J Ecol. 2014;4:113–123. [Google Scholar]
- Owino AO, Ryan PG. Recent papyrus swamp habitat loss and conservation implications in western Kenya. Wetl Ecol Manag. 2007;15:1–12. [Google Scholar]
- Palamuleni LG, Annegarn HJ, Landmann T. Land cover mapping in the upper shire river catchment in Malawi using Landsat satellite data. Geocarto Int. 2010;25:503–523. [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova D, Novembre J, Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 2016;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinay G. Hydrobiological assessment of the Zambezi River System: a review. Laxenburg: IIAASA Working paper. IIASA; 1988. [Google Scholar]
- Pollux BJA, Luteijn A, Van Groenendael JM, Ouborg NJ. Gene flow and genetic structure of the aquatic macrophyte Sparganium emersum in a linear unidirectional river. Freshw Biol. 2009;54:64–76. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramberg L, Hancock P, Lindholm M, Meyer T, Ringrose S, Sliva J, et al. (2006) Species diversity of the Okavango Delta, Botswana. In: Aquatic Sciences, Birkhäuser-Verlag, Vol 68, p 310–337
- Ritland K. Genetic differentiation, diversity, and inbreeding in the mountain monkeyflower (Mimulus caespitosus) of the Washington Cascades. Can J Bot. 1989;67:2017–2024. [Google Scholar]
- Rongoei PJK, Outa NO. Cyperus papyrus L. growth rate and mortality in relation to water quantity, quality and soil characteristics in Nyando Floodplain Wetland, Kenya. Open J Ecol. 2016;06:714–735. [Google Scholar]
- Satake A, Iwasa Y. Spatially limited pollen exchange and a long-range synchronization of trees. Ecology. 2002;83:993–1005. [Google Scholar]
- Saunders MJ, Kansiime F, Jones MB. Agricultural encroachment: Implications for carbon sequestration in tropical African wetlands. Glob Chang Biol. 2012;18:1312–1321. [Google Scholar]
- Schleuning M, Becker T, Vadillo GP, Hahn T, Matthies D, Durka W. River dynamics shape clonal diversity and genetic structure of an Amazonian understorey herb. J Ecol. 2011;99:373–382. [Google Scholar]
- Shanahan TM, Zreda M. Chronology of Quaternary glaciations in East Africa. Earth Planet Sci Lett. 2000;177:23–42. [Google Scholar]
- Shine R, Brown GP, Phillips BL. An evolutionary process that assembles phenotypes through space rather than through time. Proc Natl Acad Sci USA. 2011;108:5708–5711. doi: 10.1073/pnas.1018989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:1–15. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Easton J, Nhancale BA, Armstrong AJ, Culverwell J, Dlamini SD, et al. Designing a transfrontier conservation landscape for the Maputaland centre of endemism using biodiversity, economic and threat data. Biol Conserv. 2008;141:2127–2138. [Google Scholar]
- Terer T, Muasya AM, Triest L. Strong isolation by distance revealed among Cyperus papyrus populations in the Rift Valley lakes, Lake Victoria, and isolated wetlands of Kenya. Aquat Bot. 2015;121:57–66. [Google Scholar]
- Terer T, Triest L, Muthama Muasya A. Effects of harvesting Cyperus papyrus in undisturbed wetland, Lake Naivasha, Kenya. Hydrobiologia. 2012;680:135–148. [Google Scholar]
- Triest L, Sierens T, Terer T. Diversity and fine-scale spatial genetic structure of Cyperus papyrus populations in Lake Naivasha (Kenya) using microsatellite markers. Hydrobiologia. 2014;737:131–144. [Google Scholar]
- Tumbare MJ (2000) Management of river basins and dams: the Zambezi River Basin. CRC Press
- Tweddle D. Overview of the Zambezi River System: its history, fish fauna, fisheries, and conservation. Aquat Ecosyst Health Manag. 2010;13:224–240. [Google Scholar]
- Tyson PD. Late-Quaternary and Holocene palaeoclimates of southern Africa: a synthesis. S Afr J Geol. 1999;102:335–349. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular ecology notes 4:535–538
- Vigouroux Y, Jaqueth JS, Matsuoka Y, Smith OS, Beavis WD, Smith JSC, et al. Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol. 2002;19:1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186. [DOI] [PubMed] [Google Scholar]
- Vörösmarty CJ, McIntyre PB, Gessner MO, Dudgeon D, Prusevich A, Green P, et al. Global threats to human water security and river biodiversity. Nature. 2010;467:555–561. doi: 10.1038/nature09440. [DOI] [PubMed] [Google Scholar]
- Wang IJ. Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution (NY) 2013;67:3403–3411. doi: 10.1111/evo.12134. [DOI] [PubMed] [Google Scholar]
- Waters JM, Fraser CI, Hewitt GM. Founder takes all: density-dependent processes structure biodiversity. Trends Ecol Evol. 2013;28:78–85. doi: 10.1016/j.tree.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution (NY) 1965;19:395. [Google Scholar]
- Yan J, Zhu M, Liu W, Xu Q, Zhu C, Li J, et al. Genetic variation and bidirectional gene flow in the riparian plant Miscanthus lutarioriparius, across its endemic range: implications for adaptive potential. GCB Bioenergy. 2016;8:764–776. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SSR genotypes have been deposited with Dryad 10.5061/dryad.t76hdr840 and additional data are provided as supporting information in the online version of this article.