Abstract
Islands, and the particular organisms that populate them, have long fascinated biologists. Due to their isolation, islands offer unique opportunities to study the effect of neutral and adaptive mechanisms in determining genomic and phenotypical divergence. In the Canary Islands, an archipelago rich in endemics, the barn owl (Tyto alba), present in all the islands, is thought to have diverged into a subspecies (T. a. gracilirostris) on the eastern ones, Fuerteventura and Lanzarote. Taking advantage of 40 whole-genomes and modern population genomics tools, we provide the first look at the origin and genetic makeup of barn owls of this archipelago. We show that the Canaries hold diverse, long-standing and monophyletic populations with a neat distinction of gene pools from the different islands. Using a new method, less sensitive to structure than classical FST, to detect regions involved in local adaptation to insular environments, we identified a haplotype-like region likely under selection in all Canaries individuals and genes in this region suggest morphological adaptations to insularity. In the eastern islands, where the subspecies is present, genomic traces of selection pinpoint signs of adapted body proportions and blood pressure, consistent with the smaller size of this population living in a hot arid climate. In turn, genomic regions under selection in the western barn owls from Tenerife showed an enrichment in genes linked to hypoxia, a potential response to inhabiting a small island with a marked altitudinal gradient. Our results illustrate the interplay of neutral and adaptive forces in shaping divergence and early onset speciation.
Subject terms: Genetic variation, Genomics, Evolutionary ecology, Molecular ecology
Introduction
Due to the often-peculiar organisms that inhabit them, islands have always fascinated naturalists and scientists alike. Since Darwin’s first visit to the Galapagos in 1835, the study of insular populations has been crucial to the development of evolutionary theory (MacArthur and Wilson 1967; Grant 1998; Warren et al. 2015). Labelled nature’s test tubes, islands are home to a myriad of endemic (sub)species. Moreover, the combination of their relatively small size, discrete borders, geographical isolation and natural replication provides an excellent setting to study the evolutionary forces underlying population divergence and speciation (Losos and Ricklefs 2009). The divergence of insular populations from their founders and surrounding islands is the result of neutral and selective forces. Disentangling the respective impacts of these two forces is a challenging task, as they are interconnected and can both contribute to genetic and phenotypic differentiation.
Isolated and small populations, such as those frequently found on islands, are under a markedly strong influence of genetic drift. This process will alter the genetic makeup of the population by randomly removing rare alleles and fixating common ones, hence decreasing genetic diversity (Frankham 1997). This is a common occurrence on islands which, coupled with low gene flow, can lead to inbreeding (Keller and Waller 2002) and accelerate neutral divergence. Conversely, the absence of regular gene flow, and the often distinctive ecological conditions of the islands, may select beneficial alleles in the population (Lenormand 2002; Tigano and Friesen 2016). This process can lead to the emergence of ecomorphs via ecological divergence as populations adapt to unfilled insular niches (Losos and Ricklefs 2009), particularly so in remote islands that are colonised less often (MacArthur and Wilson 1963). Ecomorphs can occur in different islands or in the same one (Gillespie et al. 1997; Losos et al. 1998; Gillespie 2004), a concept mirrored in inland lakes (Malinsky et al. 2015), the aquatic homologous of islands. Eventually, ecomorphs can become new subspecies or even species and, in extreme cases, result in adaptive radiations as illustrated by Darwin’s finches (Grant 1999; Lamichhaney et al. 2015).
The Afro-European barn owl (Tyto alba) is a non-migratory, nocturnal raptor present from Scandinavia to Southern Africa. It is also found on numerous islands and archipelagos where subspecies have often been described (Uva et al. 2018). A recent study of the species’ genetic structure in the Western Palearctic (Cumer et al. 2021) described two main lineages occupying this region: the eastern lineage in the Levant and the western in Europe. In addition, Cumer et al. (2021) showed that barn owls from Tenerife (Canary Islands) were very distinct from both lineages. The Canaries are an active volcanic archipelago that started to form several million years ago (Anguita and Hernán 2000) about 100 km from the coast of north-western Africa and was never connected to the mainland. This long-term isolation (Norder et al. 2019), along with its subtropical climate and elevation gradients (Steinbauer et al. 2016), has resulted in a high level of endemicity, for example in plants (Carine et al. 2009), reptiles (Thorpe and Baez 1993; Nogales et al. 1998; Nogales et al. 2001; Molina-Borja 2003; Mateo et al. 2011), mammals (Hutterer et al. 1987; Pestano et al. 2003; Firmat et al. 2010; Masseti 2010) and birds (Illera et al. 2016; Lifjeld et al. 2016; Rodríguez et al. 2020; Senfeld et al. 2020). Among them, an endemic barn owl subspecies, T. a. gracilirostris (Hartert 1905), has been described based on its morphological traits, especially for its smaller size and darker coloration (Bannerman 1963; Clements et al. 2019). This subspecies is recorded in the eastern Canary Islands of Lanzarote and Fuerteventura, as well as in its surrounding islets (Lobos, Alegranza, Montaña Clara and La Graciosa), and is the only barn owl in this sector of the archipelago (Siverio 2007).
The presence of this subspecies on the eastern islands is surprising however, given that it is sandwiched between the western islands and the mainland which both harbour the nominal species T. alba. Lacking any evidence of different colonisation origins or timing, this could suggest that selection to local conditions acting on the eastern population has accelerated divergence in comparison to the western one. In both cases, neutral and adaptive microevolutionary processes promoting divergence of insular populations would leave traces on their genomic makeup. The advances in sequencing technology, and its decreasing costs, now allow to sequence the entire genomes of individuals. With the parallel development of sophisticated tools, it is possible to analyse changes in allelic frequencies at a high resolution to investigate the history of populations and inspect the genomic landscape for signals of local adaptation.
Here, we investigate the genomic bases of differentiation of barn owls from the Canary Islands. Making use of the whole-genome sequences from 40 individuals, we first describe the neutral genetic structure and diversity of the Canaries populations in contrast to the mainland, in order to retrace their history. Second, we employ a new relatedness-based method to probe the genomic landscape of these isolated populations for signals of local adaptation to the insular environment in regards to the mainland. Third, we characterise the climatic niches barn owls occupy on eastern and western islands, and explore how each population is diverging to adapt to their niches from a genomic perspective. Our results elucidate the genomic bases of the differentiation of insular populations, thus enlightening the classification of the Canaries barn owls.
Materials and methods
Whole-genome sequencing, SNP calling and identification of coding regions
For this study, 40 individual barn owls were sampled from four populations (Fig. 1; Sup. Table 1): Eastern Canary (EC, Fuerteventura and Lanzarote), Western Canary (WC, Tenerife), Morocco (MA), Portugal (PT) and Israel (IS). All but the EC and MA populations were published in previous work, including a North American owl (Tyto furcata) (Genbank accession JAEUGV000000000 (Cumer et al. 2021; Machado et al. 2021)). For EC and MA, we followed the same molecular and sequencing protocol as in the aforementioned publications. Succinctly, genomic DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) and individually tagged 100 bp TruSeq DNA PCR-free libraries (Illumina) were prepared according to manufacturer’s instructions. Whole-genome resequencing was performed on multiplexed libraries with Illumina HiSeq 2500 high-throughput paired-end sequencing technologies at the Lausanne Genomic Technologies Facility (GTF, University of Lausanne, Switzerland).
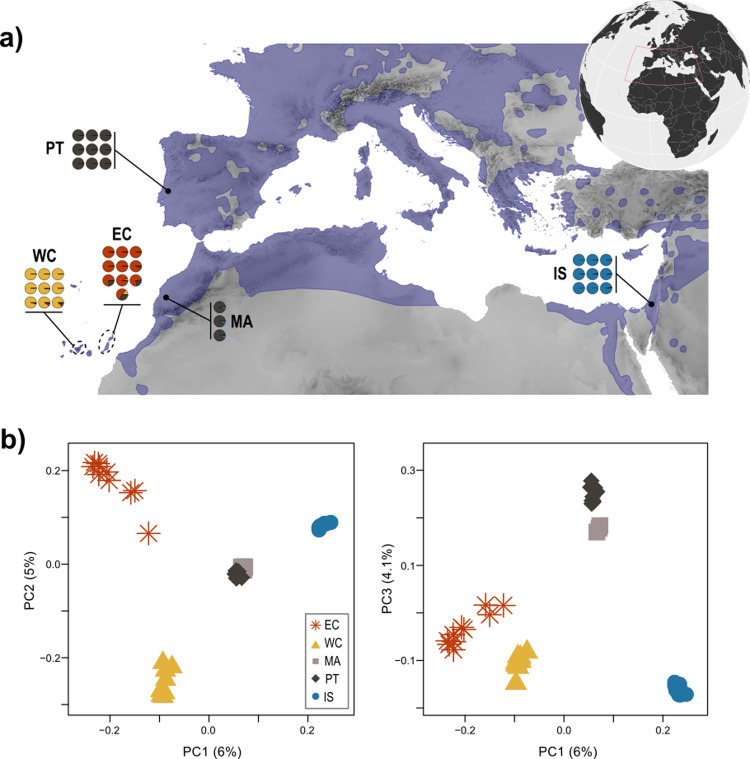
Fig. 1. Population structure of barn owls from the Mediterranean Basin and the Canary Islands.
a Individual admixture proportion of each of K = 4 lineages as determined by sNMF. Black dots are located at the approximate centroid of each sampled population. Dashed lines encircle the island(s) sampled for each Canary population. The current distribution of barn owls is plotted atop the map in blue (data from IUCN: BirdLife International 2019). b PCA of the 40 individuals. Principal component (axes) one and two (left panel) and one and three (right panel) are shown, values in parenthesis indicate the percentage of variance explained by each axis. Point shape and colour denote populations according to the legend.
The bioinformatics pipeline used to obtain analysis-ready SNPs from the 40 individuals plus the outgroup was the same as in Machado et al. (2021), adapted from the Genome Analysis Toolkit (GATK) Best Practices (van der Auwera et al. 2013) to a non-model organism following the developers’ recommendations. Briefly, reads were trimmed with Trimommatic v.0.36 (Bolger et al. 2014) and aligned with BWA-MEM v.0.7.15 (Li and Durbin 2009) to the barn owl reference genome (GenBank accession JAEUGV000000000; Machado et al. 2021). Base quality score recalibration (BQSR) was performed in GATK v.4.1.3 using high-confidence calls obtained from two independent callers: GATK’s HaplotypeCaller and GenotypeGVCF v.4.1.3 and ANGSD v.0.921 (Korneliussen et al. 2014). Following BQSR, variants were called with GATK’s HaplotypeCaller and GenotypeGVCFs v.4.1.3 from the recalibrated bam files. Genotype calls were filtered using GATK and VCFtools (Danecek et al. 2011) if they presented: low individual quality per depth (QD < 5), extreme coverage (600 > DP > 1000), mapping quality (MQ < 40 and MQ > 70), extreme hetero or homozygosity (ExcessHet > 20 and InbreedingCoeff > 0.9) and high read strand bias (FS > 60 and SOR > 3). We filtered further at the level of individual genotype for low quality (GQ < 20) and extreme coverage (GenDP < 10 and GenDP > 40). Lastly, we kept only bi-allelic sites with less than 5% of missing data across individuals and excluded SNPs on the sexual chromosome (Super scaffolds 13 and 42 (Machado et al. 2021), resulting in a dataset of 7′283′516 SNP. For analyses of neutral population structure and demography, an exact Hardy-Weinberg test was used to remove sites that significantly departed (p < 0.05) from the expected equilibrium using the R (R Development Core Team 2016) package HardyWeinberg (Graffelman and Morales-Camarena 2008; Graffelman 2015) yielding 6′827′220 SNP with a mean coverage of 19.9X (4.6 SD).
Population structure and genetic diversity
To investigate population structure among our samples, sNMF v.1.2 (Frichot et al. 2014) was run for K 2 to 5 in 25 replicates to infer individual clustering and admixture proportions. For this analysis, singletons were excluded and the remaining SNPs were pruned for linkage disequilibrium in PLINK v1.946 (Purcell et al. 2007; parameters -indep-pairwise 50 10 0.1) as recommended by the authors, retaining 288′775 SNP. The same dataset was used to perform a Principal Component Analysis (PCA) with the R package SNPRelate (Zheng et al. 2012). A second PCA was performed by merging the data in this study with that of (Cumer et al. 2021) that includes European populations (total of 2′036′320 SNP with no missing data) to assess where the Eastern Canary population (unsampled in Cumer et al. 2021) falls among the larger sampling.
Individual observed heterozygosity and population-specific private alleles were estimated using custom R scripts for each population. Individual-based relatedness (β; (Weir and Goudet 2017; Goudet et al. 2018), inbreeding coefficients for whole genome SNP data (FIS and FIT), overall, population-specific and pairwise FST (Weir and Goudet 2017) were calculated with hierfstat v.0.5-9 (Goudet 2005).
Demographic history
To investigate the demographic history of the insular populations and potential admixture events we used Treemix (Pickrell and Pritchard 2012). Using the LD-pruned dataset filtered further to include no missing data (228′980 SNP), Treemix was run for 10 replicates with 0 to 5 migration events, rooting the tree on the IS population, given its position on the PCA and what is known of the region from previous work (Cumer et al. 2021). The function get_f was used to estimate the variance explained by adding migration events.
We also run a Pairwise Sequentially Markovian Coalescent (PSMC) analysis using the 6′827′220 SNPs dataset, to infer changes in population sizes in the past. PSMC uses a hidden Markov model to identify recombination events across a single diploid genome. It infers the time to the most recent common ancestor (TMRCA) for each independent DNA segment and, based on the TMRCA distribution and the rate of coalescent events, infers ancestral Ne in a given time epoch (see Li and Durbin 2011 for details). The settings of the PSMC analysis were chosen according to a previous publication on the barn owl (Nadachowska-Brzyska et al. 2015). In brief, the upper limit of the TMRCA was set to 5 and the initial θ/ρ value to 5. Ne was inferred across 84 free atomic intervals (4 + 30*2 + 4 + 6 + 10), meaning that the first population-size parameter spans the first four atomic intervals, each of the next 30 parameters spans two intervals, while the last three parameters respectively span four, six and ten intervals. PSMC was ran independently for each individual. For plotting purposes, we used a generation time of 2 and a mutation rate of 9.5−10, according to Nadachowska-Brzyska et al. (2015).
Detection of genomic regions under selection
Insular vs mainland barn owls
In this study, we aimed to identify genomic regions potentially under selection at two different levels. First, to detect signatures of selection specific to barn owls of the Canary Islands, we grouped insular individuals (EC and WC) and compared them to the mainland ones (MA, PT and IS). A script by Simon Martin (https://github.com/simonhmartin/genomics_general/blob/master/popgenWindows.py) was used to estimate genome-wide patterns of relative (FSTILvsML) and absolute (dxy) divergence between the insular and mainland groups, and to calculate nucleotide diversity (π) per group, in windows of 100 kbp with 20 kbp steps.
SNPrelate was used to calculate a pairwise matrix of linkage disequilibrium (r) from SNPs with over 5% minor allelic frequency over all insular individuals (EC and WC) on one hand and all mainland individuals (PT, MA and IS) in the other, which was then squared to obtain r2. For plotting, we estimated the mean of non-overlapping 100 SNP windows.
The topology weighting method implemented in Twisst (Martin and Van Belleghem 2017), was used to quantify the relationships between the five populations in our dataset and visualise how they change along the genome. Twisst estimated the topology based on trees produced using Randomized Axelerated Maximum Likelihood (RAxML) v8.2.12 (Stamatakis 2014). RAxML inferred trees in sliding windows (100kbp of length, 20kbp of slide) using a generalised time-reversible (GTR) CAT model with Lewis ascertainment bias correction. Twisst then estimated the weighting of each taxon topology (defined as the fraction of all unique population sub-trees) per window.
Finally, we calculated population-specific FST from allele sharing matrices (β; (Weir and Goudet 2017; Goudet et al. 2018) in sliding windows of 100 kbp with 20 kbp steps, using hierfstat. For each matrix, we calculated i) the mean allele sharing for pairs of individuals from islands and ii) the mean allele sharing for pairs of individuals, one from an island and the other from the mainland, from which we obtain the islands specific FST. We shall refer to this estimate as FST island-specific (FSTCan; see Sup. Fig. 1 for a schematic representation). This method allowed us to identify genomic regions of high differentiation exclusively on the islands with no confounding effect from the mainland (see also Weir et al. 2005).
From the genome wide scans, we identified peaks of differentiation with at least two overlapping windows of FSTCan higher than 5 standard deviations (SD) from the mean (threshold: 0.377). This threshold of Five-sigma corresponds to a p value of 3 × 10−7 (or about 1chance in 3.5 million random observations) a value close to the 0.05 threshold after Bonferroni correction (0.05/555807 (number of windows) = 8.94e-07). Genes in these regions were extracted from the NCBI’s annotation of the reference genome (RefSeq accession: GCF_018691265.1). The gene list was then fed to ShinyGO v0.61 (Ge et al. 2020) to investigate potential enrichment of molecular pathways.
Eastern vs Western Canary Islands
On a second stage, to investigate potential genomic signals of differentiation, putatively linked with ecological adaptation to their distinct niches, we contrasted the two insular lineages (EC against WC). We estimated pairwise FST, dxy and π as for the island-mainland comparison described above.
Then, as for the island-mainland comparison above, we used hierfstat to calculate population specific FST in sliding windows of 100 kbp with 20 kbp steps based on a dataset including only the insular individuals. For each island, we consequently identified genomic regions with at least two overlapping windows of pairwise FST higher than 5 SD from the mean (regions highly differentiated between islands, threshold: 0.157) and above the 99th quantile of each population’s F (regions highly similar within each island, respective threshold for FSTEC = 0.404 and for FSTWC = 0.393) and extracted the genes in these regions as described above. This yielded two gene lists, one per island, which were input to ShinyGO as above.
Climatic niche analysis
To assess whether the barn owl populations of eastern and western Canary occupy different climatic niches, we used the Outlying Mean Index (OMI) approach, a multivariate method developed to investigate niche separation and niche breadth (Dolédec et al. 2000) as implemented in the R package ade4 v.1.7-16. This method measures the distance between the mean habitat conditions used by species (species centroid), and the mean habitat conditions of the sampling area (origin of the niche hyperspace). Thus, the position of the species will depend on their niche deviation from the reference, a theoretical ubiquitous species that tolerates the most general habitat conditions (i.e., a hypothetical species uniformly distributed among habitat conditions). Observation points for barn owls were compiled from three different sources: Global Biodiversity Information Facility (GBIF.org (2021)), samples sequenced in this study and in Burri et al. (2016). We kept records only from the islands sampled in this study, namely Tenerife (T. alba; N = 79) and Lanzarote and Fuerteventura (T. a. gracilirostris; N = 34 and N = 39, respectively).
Climatic variables for the Canary Islands were extracted from the WorldClim database (Hijmans et al. 2005) at 30 s resolution (~1 km2) using the R package rbioclim (Exposito-Alonso 2017). Redundant variables (correlation of 1) were trimmed and the final model was run with the following 13 variables: Mean Diurnal Range (BIO2), Isothermality (BIO3), Temperature Seasonality (BIO4), Max Temperature of Warmest Month (BIO5), Min Temperature of Coldest Month (BIO6), Temperature Annual Range (BIO7), Mean Temperature of Driest Quarter (BIO9), Annual Precipitation (BIO12), Precipitation of Driest Month (BIO14), Precipitation Seasonality (BIO15), Precipitation of Warmest Quarter (BIO18), Precipitation of Coldest Quarter (BIO19).
Results
Population structure and genetic diversity
The overall FST in our dataset was 0.0698. Individual ancestry analyses with sNMF distinguished four genetic clusters, separating each population into its own cluster, except Morocco (MA) and Portugal (PT) that clustered together (Fig. 1a). Similarly, PCA clustering clearly grouped individuals according to their population, except PT and MA (Fig. 1c). The first axis opposed the insular populations to the mainland, as did sNMF K = 2 (Sup. Fig. 2), with Eastern Canary (EC) and Israel (IS) at each extreme. The second axis contrasted the two insular populations EC and Western Canary (WC), as in K = 3 (Sup. Fig. 2), and finally the third axis segregated the mainland groups with PT and MA opposed to IS, as in K = 4 (Fig. 1a, c). Three individuals from EC (one from Fuerteventura and two from Lanzarote) showed small ancestry levels from both WC and PT in sNMF and were placed intermediately on axes 1 and 2 of the PCA. In the PCA including the European individuals, the first axis remained qualitatively the same with EC and WC together in opposition to IS (Sup. Fig. 3). Axes 2 and 3 switched positions, with MA, PT and the rest of Europe being first isolated from the rest and only then the two Canary split from each other (Sup. Fig. 3). In terms of differentiation, pairwise FST were highest between both islands and IS (EC-IS 0.092; WC-IS 0.088) as well as between islands (WC-EC 0.084). The mainland populations were overall less differentiated (all below 0.04), with PT and MA having the lowest FST with all other populations in accordance with their central position on the PCA (all below 0.06 for PT and 0.061 for MA), and the lowest between themselves (0.007) (Table 1).
Table 1.
Genetic diversity and population differentiation of barn owls from the Mediterranean Basin and the Canary Islands.
| Population | Abbrev. | N | #PA | #rPA | HO | FIS | FIT | FST | EC | WC | MA | PT | IS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eastern Canary | EC | 10 | 324458 | 265175 (3829) | 0.161 (0.013) | −0.015 (0.08) | 0.10 (0.07) | EC | 0.112 | 0.081 | 0.061 | 0.060 | 0.088 |
| Western Canary | WC | 9 | 288313 | 288313 | 0.167 (0.006) | −0.030 (0.04) | 0.07 (0.03) | WC | 0.081 | 0.105 | 0.055 | 0.054 | 0.083 |
| Morocco | MA | 3 | 234524 | 692508 (13287) | 0.190 (0.001) | −0.04 (0.01) | −0.06 (0.01) | MA | 0.061 | 0.055 | −0.013 | 0.007 | 0.034 |
| Portugal | PT | 9 | 669889 | 0.182 (0.008) | −0.013 (0.04) | −0.01 (0.04) | PT | 0.060 | 0.054 | 0.007 | 0.014 | 0.039 | |
| Israel | IS | 9 | 703392 | 703392 | 0.180 (0.002) | −0.036 (0.04) | −0.004 (0.01) | IS | 0.088 | 0.083 | 0.034 | 0.039 | 0.050 |
Right-hand-side of the table shows the matrix of population pairwise FST.
N number of sampled individuals, #PA private alleles in each population, #rPA private alleles (SD) rarefied to 9 individuals per population (note that PT and MA were merged), HO mean observed heterozygosity (SD), FIS population level inbreeding coefficient (SD), FIT mean individual inbreeding coefficient relative to the meta-population (SD), FST pairwise and population-specific FST.
Treemix yielded a population tree highly resembling the first PCA axis, with two main branches splitting from the IS root, one toward PT and MA and the second to WC and EC (Sup. Fig. 4). Most of the variance was explained by the tree with no migration (0.993). Nonetheless, the tree with two migration events displayed the highest likelihood (162.284), with one migration edge from the base of WC towards the tip of PT (edge weight 0.33) and the other from the base of PT towards the tip of EC (edge weight 0.13). Adding more than two migration events to the tree did not improve its fit to the data.
The Pairwise Sequentially Markovian Coalescent (PSMC) analysis showed similar fluctuations in population sizes in the past (from 106 to 105 years before the present), and more fluctuations in recent time (between 105 and 104, Sup. Fig. 5). In this recent time, PSMC did not provide consistent estimates of population sizes, with more variation between individuals from the same population than between populations.
Overall, mainland populations presented higher genetic diversity than the insular ones (Table 1). Nonetheless, both insular populations showed over 280′000 private polymorphic sites (Table 1). Accordingly, individual relatedness was higher within and between insular populations (Sup. Fig. 6), and PT had the lowest within population relatedness. All populations showed signs of random mating with FIS close to zero but slightly negative as expected of dioecious species (Balloux 2004), while the inbreeding levels of insular barn owls relative to the whole set of populations (FIT) were higher than those on the mainland (Table 1), a reflection of their isolation from the continent.
Island vs mainland genomic comparisons
In order to measure the differentiation between islands and mainland individuals, we computed two distinct FST along the genome (see methods for details). FSTCan allowed the detection of regions of high similarity in the islands as a whole, whereas FSTILvsML produced less clear results with population-specific allelic frequencies driving the overall signal instead of the defined groups. For example, FST ILvsML yielded a peak of differentiation between the islands and the mainland which, upon closer inspection of pairwise comparisons of the populations, turned out to be due to differences between EC and the mainland only, whereas WC did not produce the same signal (Sup. Fig. 7). The genomic comparisons of diversity and divergence between insular and mainland barn owls based on the FSTCan yielded 48 regions (478 100kbp overlapping windows) of high differentiation including 100 genes (Fig. 2a). ShinyGO analyses identified an enrichment of five functional categories related to morphogenesis in humans (Sup. Table 2). The largest enriched pathway—anatomical structure morphogenesis—included 25 of the genes in regions of high genomic differentiation. The remaining three categories—anatomical structure formation involved in morphogenesis, tube morphogenesis and blood vessel morphogenesis—were subsets of the largest pathway, including 15, 14, 12 and 10 of the 25 genes, respectively.
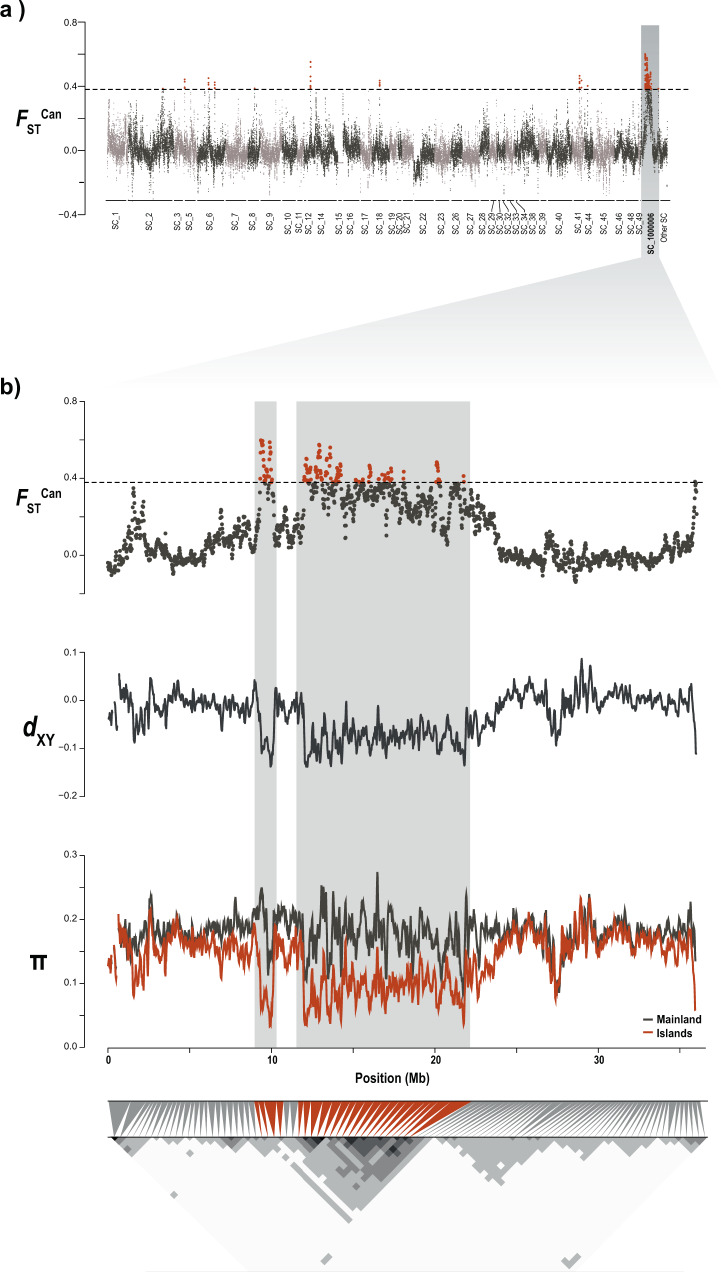
Fig. 2. Genomic landscape of differentiation between insular and mainland barn owls.
a Genome-wide beta comparison between individuals from the Canary Islands (EC & WC) and from the mainland (PT & IS). Each dot represents a 100 kbp window. Dashed line indicates the 5 SD threshold used to identify genomic regions of high differentiation, emphasised in red. Alternating grey colours denote different scaffolds. Shaded vertical bar highlights Scaffold 100006. b Zoom on Scaffold 100006 and, in particular, the ~15 Mb long highly differentiated genomic region (background shading) over windows of 100 kbp. From top to bottom, we see in this region a high FSTCan between insular and mainland barn owls, low absolute distance (dXY) between both islands (scaled with mean = 0) and reduction of nucleotide diversity (π) among insular individuals (red line) compared to the mainland (black line). The bottom triangular matrix shows pairwise LD (r2) between groups of 100 SNP along the chromosome in insular owls. Darker pixels show higher LD. Grey triangles match each pixel in the matrix diagonal to the region it spans on the chromosome above. Red triangles indicate pixels that overlap the region of high differentiation.
The largest of the peaks encompassed 15 Mb of Super-Scaffold_1000006. Of all the genes found in peaks of differentiation, 87 out of 100 were within this region. Here, insular owls showed a strong decrease in relative diversity compared to the mainland (FSTCan), as well as a drop of absolute divergence (dxy) between each other and nucleotide diversity (π). The region showed strong LD (r2) in all insular owls between neighbouring variants compared to the rest of the scaffold (Fig. 2b), which was not the case among continental ones (Sup. Fig. 8). In addition, FST was higher between island and mainland owls (Sup Fig. 7). Twisst showed roughly similar proportions of each tree along the genome, except for this region where there was a higher than average proportion of trees that joined EC and WC (Sup. Fig. 9). Of the 25 genes of the morphogenesis pathway, 18 were found in this closely-linked region of Super_Scaffold_1000006 (21% of the total 87 genes of the region).
Eastern vs Western Canary Islands
Climatic niche analysis
To investigate the genomic and ecological differentiation between island populations (EC and WC), we first depict different climate in the two set of islands based on barn owl observations in the Canary Islands. The climatic niche analyses (OMI) yielded two axes that explained the climatic variability in our study area (Fig. 3b), with the first axis (OMI1) explaining nearly all of it (99.3%). OMI1 was positively correlated with temperature and negatively correlated with precipitation (Sup. Fig. 11). The eastern population T. a. gracilirostris occupied a narrow niche of high temperature, low precipitation and low seasonal and daily variability. Tyto alba in Tenerife occupied a broader niche that covered most of OMI1, including some of the niche of T. a. gracilirostris. OMI2 explained little of the variability (0.7%), spreading slightly each population’s niche without segregating them.
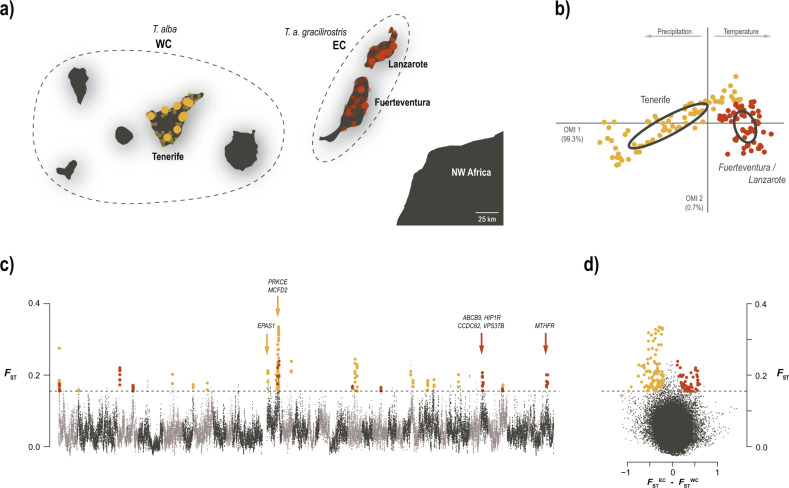
Fig. 3. Ecological divergence in barn owls from the Canary Islands.
a Sampling location of Western (WC; yellow) and Eastern (EC; red) Canary individuals, off the coast of north-western Africa. Sampled islands are named. Large dots indicate individuals sampled for WGS and population genomics analyses; small transparent dots are barn owl observations used in niche analysis. Dashed lines group islands according to barn owl taxonomy. b Climatic niche occupied by the two Canary populations. The first axis varies from cold and wet to hot arid environments. c Genome-wide FST comparison between individuals from each Canary population. Each dot represents a 100kbp window. Alternating grey colours denote different chromosomes. Dashed line indicates the 5 SD FST threshold used to identify genomic regions of high differentiation overlaid with FSTWC, highlighted in yellow and FSTEC in red. Arrows denote the location of genes linked with response to hypoxia in WC; and the cluster of four genes linked to morphological ratios and the main gene related to blood pressure in EC. d Same FST windows and threshold as in c plotted against the difference between FSTWC and FSTEC, clearly highlight the two sets of windows putatively under local adaptation.
Detection of differentially selected genomic regions
We then searched for regions under selection in each island, yielding to a total of 33 putatively adaptive regions (14 in EC and 19 in WC), with high differentiation (FST) between island and increased relatedness within each island (FSTEC and FSTWC; Fig. 3c, d, Sup. Fig. 12), from which we obtained two lists of genes. For EC, there were 30 such genes (Sup. Table 3) while for WC there were 25 genes (Sup. Table 4). Enrichment analyses found no link to a specific GO pathway for either island population. Nonetheless, on each island there were clusters of genes with similar identified functions, namely body proportions and blood pressure in EC, and cellular response to hypoxia in WC.
Discussion
Islands offer unique conditions for organisms to adapt and expand their niches. In the Canaries, an archipelago rich in endemics, the barn owl is one of the few raptors present and is thought to have diverged into a subspecies on the easternmost islands. Taking advantage of whole-genome sequences, we first investigate the origin of the population of barn owls in the Canary Islands, revealing that the latter are long-standing populations and allowing us to address their taxonomic classification. Using a new, more sensitive method based on excess of allele sharing within the islands, we detect 58 putatively locally adapted genomic segments, many of them grouped in a haplotype-like region seemingly under selection in the Canary Islands. A quarter of the tightly linked genes in this genomic region are enriched for genes contributing to a pathway of anatomical morphogenesis suggesting morphological adaptations to insularity. We also identify scattered genomic regions putatively locally adapted to either the eastern and western islands. For eastern Canaries, the identified genes in regions under selection belong to pathways linked to body proportions and blood pressure, consistent with the smaller owl size of this population living in a hot arid climate. In the western Canaries, barn owls from Tenerife display a potential signal of selection of genes related to hypoxia, a potential response to inhabiting an island with a steep elevation gradient.
Barn owls from the Canary Islands and Mediterranean Basin
Our work shows that, while each population in the study area has its own unique genetic composition (Fig. 1), barn owls from the Canary Islands are distinct from those on the mainland surrounding the Mediterranean Basin. Indeed, clustering methods constantly opposed insular individuals to mainland ones (Fig. 1c; Sup. Fig. 2) and were supported by the drift-based trees of Treemix. Thus, overall, our current results indicate that the two insular lineages—Eastern and Western Canaries—have a common origin.
In terms of genetic diversity, the islands were less diverse than the mainland, as expected, and showed slightly higher levels of inbreeding relative to the whole set (Frankham 1997). However, they presented nearly tenfold higher levels of private alleles than those reported for other islands in the Atlantic (British Isles; Machado et al. 2021) and in the Mediterranean Sea (Cyprus and Greek islands; Machado et al. 2021). Notably, the two Canaries insular populations, about 165 km apart, were more distant genetically from each other than either was from Portugal, over 1000 km away. Furthermore, they were more distant from each other even than WC was from any population in continental Europe in a recent genomic study (Cumer et al. 2021), as well as in an earlier one with microsatellites (Burri et al. 2016). The high private diversity could be partially explained by the small number of samples in north-western Africa, which may be masking any shared polymorphisms between the islands and the nearest mainland as private to the former. However, as we did sample PT and IS, the most diverse populations in the Western Palearctic assumed to meet in northern Africa (Cumer et al. 2021), much of this insular diversity is likely indeed private. Therefore, and despite the inconclusive PSMC analysis (Sup. Fig. 5), the divergence of the populations (Fig. 1, Table 1) suggest that the Canaries have been colonised much earlier than the other studied insular populations and have thus had the time for in situ mutations to accumulate in spite of genetic drift. These populations may also reflect a more ancient lineage from northern Africa that would have been replaced or simply evolved in a different direction. The addition of samples from African mainland (on top of the three MA samples), combined with more complex demographic modelling, would be needed to fully resolve the neutral history of these populations.
Genomic basis of insularity
We found multiple evidence of adaptation common to both barn owl lineages of the Canary Islands. To do so, we used an estimator of population specific FST (Weir and Goudet 2017; Goudet et al. 2018) to identify genomic regions with an excess of shared ancestry in all insular individuals relatively to the mean shared ancestry between islands and mainland individuals. Being a moment estimator, it can process efficiently a large amount of genomic loci, compared to maximum likelihood or Bayesian estimators (e.g. Foll and Gaggiotti 2008). More importantly, it does not rely on the F-model, which assumes independent populations. This island specific FST (FSTCan) identified regions of increased relatedness between insular individuals compared to the averaged relatedness along the genome (i.e. regions in which all insular individuals resemble each other more than expected). This population-specific, moment based and model-free estimator of FST provides a clearer result than classical pairwise FST scan, as it focuses on the diversity in the target population rather than taking an average over the set of populations, and should be a useful addition to the population genomic toolbox to detect nested signals of local adaptation, especially when there is substructure in the groups one wishes to compare. We thus considered regions highly similar in insular individuals as putatively under selection on the islands. The genomic landscape of FSTCan differentiation between insular and mainland owls yielded 58 windows putatively under selection (Fig. 2a). Among these windows, a particularly large and clear peak of differentiation stood out on Super-Scaffold 1000006. This region, ~15 Mb in length, was highly similar among insular individuals (Fig. 2, Sup. Fig. 10), and accompanied by a drop in dXY and π (Fig. 2b).
The joint analysis of these metrics allows to disentangle the evolutionary forces that drove this signal. Cruickshank and Hahn (2014) and Irwin et al. (2016, 2018) pointed that a pattern of high differentiation (Fst), combined with a decrease of diversity between (dXY) and within (π) populations, can be explained by either positive or background selection acting on the region. The increased linkage between alleles in this region suggests that it is transmitted in a haplotype-fashion. Combined with the fact that we do not see the slightest surge of LD in mainland individuals, it confirms that it is not a by-product of a region of low-recombination in this species (Sup. Fig. 8). Overall, these results provide evidence of selection in this genomic region in Canary Islands owls, possibly linked to an adaptation to insularity (Fig. 2).
A fifth of the genes in this haplotype (18 out of 87), in conjunction with 7 other genes in other potentially adaptive regions, significantly enriched the anatomical structure morphogenesis pathway (Sup. Table 2), a biological process related to the organisation and generation of anatomical structure during development. This suggests the selection on some morphological trait on insular individuals. Given that there is evidence of gene flow from the mainland into both islands (see admixed individuals in Fig. 1, Sup Fig, S2), we propose two hypotheses to explain how selection might act on this haplotype. First, it could confer a significant advantage to individuals carrying it on the island and prevent those that do not carry it from reproducing or surviving. In this scenario, immigrants from the mainland not carrying this haplotype would reproduce less in both islands. In the second scenario, selection would happen on the migrants themselves before reaching the island if, for example, the haplotype facilitates long flight over large spans of water. It is widely accepted that, given how dispersal capacity is highly variable across species, even among birds, some are more prone to colonising islands than others. Therefore, it is also conceivable that, within a species or population, some individuals are morphologically more predisposed or have better dispersal abilities than others. Since the barn owl generally avoids flying over open water, as demonstrated by its consistently higher differentiation on islands (Cumer et al. 2021; Machado et al. 2021, n.d.), this seems plausible. The absence of phenotypic measurements from the sequenced birds prevents us from establishing a link between phenotypes and genotypes on this data set, and we hence remain cautious on the speculation regarding the functional implications of this haplotype. Future work should verify the frequencies of this haplotype in a larger cohort as we only had 19 insular individuals in this study and, if possible, include detailed morphometric measurements to allow a GWAS-like approach.
Ecological divergence in the Canary Islands
In the Canary archipelago, both the eastern islands and Tenerife have many specific endemic species across multiple taxa. This is generally attributed to their intrinsic characteristics driving ecological speciation, namely the arid and windy conditions of Lanzarote and Fuerteventura, and the elevation gradient of the Teide volcano (up to 3718 m a.s.l.) in Tenerife. We quantified the climatic differences between the two environments with a niche analysis based on reported barn owl observations, and show that indeed, barn owls occupy significantly different niches on each group of islands (Fig. 3b). In the east, they are found on unvaryingly hot and dry locations, whereas Tenerife covers a wide range of temperature and precipitation (Fig. 3b).
We looked for evidence of differentiation and local adaptation in insular populations in the genomic data (Fig. 3c, d, Sup. Fig. 12). Within these populations (EC and WC), most of the genome shows levels of within-taxon diversity (πIL) that are only slightly smaller than between-taxon absolute nucleotide distance (dxy; Sup. Fig. 12a). indeed, the variation in both dxy and πIL across the genome shows the great majority of genomic windows cluster along or near the 1:1 line, the expectation in a single panmictic population. However, regions putatively under selection (with high differentiation (FST) and increased relatedness within each island [FSTEC and FSTWC], identified in Fig. 3) show a reduced nucleotide diversity (πIL) (Sup. Fig. 12a) with medium to high level of absolute divergence (dxy), since most of the windows have a dxy between mean ± 2sd or slightly above (Sup. Fig. 12b).
Different models have been proposed to explain regions with such higher differentiation, with regards to the within and between taxon diversity (see Irwin et al. 2018 for details). In case of divergence-with-gene-flow, the selection acting on a locus contributing to reproductive isolation between differentiating populations leads to a reduced gene flow between the two populations in the region closely linked to that locus, relatively to the rest of the genome. In this case, dxy is predicted to be higher in regions of high FST than in regions of low FST compared to the rest of the genome (Cruickshank and Hahn 2014). Selection in allopatry on distinct regions of the genome within each island population, would lead to lower within-population diversity (πEC and πWC) in those regions (as observed in Sup Fig. 12c), while dxy in these regions would be similar to the rest of the genome (as it’s the case in Sup. Fig. 12b). Thus, our scans support that selection in allopatry in each population, maybe combined with divergence with gene flow, drove the differentiation process of the owls from eastern and western Canaries.
From these scans, we observed that the eastern population had more genomic regions, and more genes, potentially under selection compared to the west (30 and 25 genes, respectively). Although no significant pathway enrichment was detected in either population, there were clear groups of genes with similar known functions in the putatively adapted regions. In the eastern population there were two such groups. The first, composed of seven genes, has significant links to body size and proportions in humans (see Sup. Table 3 and references therein). Among these, four genes—HIP1R, CCDC62, VPS37B and ABCB9—are tightly clustered in the barn owl genome (Fig. 3c), and have been linked to body height, body mass index and other body measurement ratios (Turcot et al. 2018; Kichaev et al. 2019; Vuckovic et al. 2020; Zhu et al. 2020) and may hint at a link with the smaller size of barn owls in the eastern population. The second group includes 11 genes related to numerous blood parameters (Sup. Table 3), a similar signal to that seen in chickens adapted to hot arid environments (Gu et al. 2020). In particular, the gene MTHFR has extensive connections to blood pressure (Newton-Cheh et al. 2009; Ehret et al. 2016; Liu et al. 2016; Surendran et al. 2016; Hoffmann et al. 2017; Wain et al. 2017; Kulminski et al. 2018; German et al. 2020), a trait known to vary with environmental temperature in mammals (Halonen et al. 2011) and birds (Darre and Harrison 1987), potentially suggesting barn owls circulatory systems could have adapted to the hot and dry conditions of the eastern Canaries.
In the western island population, regions putatively under selection contained an interesting group of four genes—EPAS1, PRKCE, MCFD2 and FZD8—with links to red blood cells, haemoglobin density and cellular response to hypoxia (i.e. low levels of oxygen; Sup. Table 4; Astle et al. 2016; Kichaev et al. 2019; Chen et al. 2020; Oskarsson et al. 2020; Vuckovic et al. 2020). Red blood cells, and the haemoglobin within, are responsible for transporting oxygen in the body and are direct targets of selection at high elevation (O’Brien et al. 2020). The gene EPAS1 in particular, is well known for being involved in adaptation to high altitude environments across vertebrates (Witt and Huerta-Sánchez 2019), overall hinting at a possible adaptation of barn owls to higher altitude in Tenerife. On this small island, barn owls are mostly concentrated at lower altitude (i.e. along the coast—0 to 300 m a.s.l.) and their repartition is reduced in the central part of the island due to the presence of the high mountains (about 2000 m a.s.l., that culminates up to the peak of Teide at 3718 m a.s.l.) (Siverio 1998). This is consistent with the known limited ability of this bird to live at high altitudes (Machado et al. 2018; Cumer et al. 2021). However, the observation of individuals and breeding couples at moderate altitudes (600–1200 m a.s.l., Siverio 1998), could support the hypothesis of an ongoing adaptation to higher altitude in this insular population. This hypothesis, which deserves further investigation, is also consistent with observations made in other populations in warm climates, where local barn owl populations expand their range by adapting to slightly higher altitudes (Romano et al. 2020).
Considering its wide distribution, even accounting for phenotypic plasticity, barn owls’ capacity to adapt to a variety of prey and environments is unquestionable. As such, detecting signals of local adaption in the Canary Islands is not surprising. Indeed, with islands generally being species-poor, the species that do inhabit them have to adapt to different or broader niches via ecological divergence (Losos and Ricklefs 2009). This is especially true of volcanic islands that arise isolated and uninhabited, in contrast to those intermittently connected to the mainland and more easily colonised. Moreover, the community of birds of prey in the Canary archipelago includes less than half the species found in the nearby mainland (eight diurnal and two nocturnal species; Martín and Lorenzo 2001; Rodríguez et al. 2018), likely due to the lack of suitable habitat, preys and/or the limited surface. A by-product of this is the reduction of inter-specific competition, which could have allowed the barn owl to maintain population sizes just large enough on the islands through time for selection to act and potentially expand its niche to better exploit the insular environment. Our results suggest this is happening in parallel on each island (Fig. 3c, 3d), consistent with their different niches (Fig. 3b) and relative genetic isolation, producing two distinct ecomorphs, adapted to distinct ecosystems.
Insular subspecies
The eastern islands of Lanzarote and Fuerteventura, and adjacent islets (Fig. 3a) are home to the barn owl subspecies T. a. gracilirostris. This classification is based on its smaller size and even on colouration pattern, although the latter is contested by ornithologists and inconsistent with reported phenotypical measurements (Burri et al. 2016). The reduction in size is actually a common pattern in insular barn owls (Romano et al. 2021), and could be an adaptation to nesting in very small cavities (i.e. cracks in lava walls) and/or to better navigate the strong winds in the eastern islands (Siverio 2007). The genomic data presented here is consistent with this population forming an endemic subspecies. It has diverged considerably from the mainland, with higher differentiation levels than barn owls from any other studied island in the Western Palearctic (Machado et al. 2021, n.d.). Moreover, we show it carries high levels of private genetic diversity and multiple genomic regions showing signs of local adaptation (Fig. 3c).
In contrast, barn owls from Tenerife and the remaining islands are considered to belong to the nominal T. a. alba found also on the mainland surrounding the Mediterranean Basin (Fig. 3a). However, it too has considerably diverged from the mainland and shows signs of potential ongoing adaptation in Tenerife and its elevation gradient (see previous section). Furthermore, it clusters with the other insular Canary population rather than the mainland (Fig. 1). While it is not the aim of this study to evaluate what constitutes a subspecies, we provide evidence that the Tenerife population is diverging significantly from its founding population, both neutrally and adaptively, albeit at a slower pace than the eastern population.
The reasons why the eastern population is more divergent than the western, a puzzling fact considering it is closer to the mainland, are not yet fully resolved. Neutral divergence in FST between these two insular populations suggest they are the result of two independent colonisation events rather than a strict east-to-west progression as described for other taxa (Juan et al. 2000). Although the islands themselves emerged from east to west, Tenerife is at least 11 million years old, twice the inferred time of formation of the T. alba species (Uva et al. 2018) and thus available for colonisation at the time. Nonetheless, an earlier settlement of the eastern islands would have given more time for both genetic drift and selection to promote divergence. Alternatively, a very small population size in the east, consistent with current census data (Palacios 2004; Siverio 2007), could account for the stronger drift in a scenario of simultaneous colonisation. However, it would strongly hinder local adaptation, making it a less likely hypothesis since we identified more regions putatively under selection in this population. Further work, including an extensive sampling of all island populations, as well as demographic modelling, would be needed to resolve this intriguing pattern and refine the history of the barn owl in the Canary archipelago.
Conclusion
Due to their intrinsic characteristics, islands house numerous endemics making them ideal systems to study the bases of ecological divergence. We provide empirical evidence that both neutral and adaptive evolutionary mechanisms shaped divergence from the mainland in barn owls from the Canary archipelago. Our results show clear signs of genome-wide differentiation (i.e. neutral), a combination of mutations (high private diversity; Table 1) and drift (high FST and FIT), consistent with theoretical expectations for populations established and isolated long ago despite some admixture. We also identify signals of local adaptation to common insular conditions (Fig. 2), as well as to each island’s niche, creating ecomorphs (Fig. 3). While the history and functional effect of the putatively adapted genomic regions identified here deserve further investigation, these observations highlight how selection can still act on small isolated populations. This study illustrates the capacity of a widespread bird to adapt to the local ecological conditions of small islands, an adaptative capacity which may prove essential in facing a changing global climate.
Data accessibility
Whole-genome resequencing data produced in this study were deposited at the NCBI Sequence Read Archive (SRA) under Bio project PRJNA774943. Sampling locations used for niche modelling are given in appendix 1.
Supplementary information
Acknowledgements
We are grateful to the following institutions and individuals that provided samples or aided in sampling to our study: Karim Rousselon, Gustavo Tejera, Sylvain Antoniazza, Reto Burri, Aurelio Martín, David P. Padilla, the “Association Marocaine pour la Protection des Rapaces AMPR”, the Natural History Museum of Tenerife and the Wildlife Rehabilitation Centre La Tahonilla (Cabildo Insular de Tenerife). We thank Céline Simon and Guillaume Dumont for their valuable assistance with molecular work. The Gobierno de Canarias provided all the corresponding permits for the collection and inter-island transport of samples. This study was funded by the Swiss National Science Foundation with grants 31003 A‐138180 & 31003A_179358 to JG and 31003A_173178 to AR.
Author contributions
APM, TC, AR, JG designed this study; FS, MC, IR, RL provided samples; APM produced whole-genome resequencing libraries; TC and APM conducted the analyses; APM and TC led the writing of the manuscript with input from all authors.
Funding
Open access funding provided by University of Lausanne.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Xiangjiang Zhan.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tristan Cumer, Ana Paula Machado.
These authors jointly supervised this work: Alexandre Roulin, Jérôme Goudet.
Contributor Information
Tristan Cumer, Email: tristan.cumer@unil.ch.
Jérôme Goudet, Email: Jerome.goudet@unil.ch.
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-022-00562-w.
References
- Cumer T, Machado AP, Dumont G, Bontzorlos VA, Ceccherelli R, Charter M, Dichmann K, Martens H-D, Kassinis N, Lourenço R, Manzia F, Ovari K, Prévost L, Rakovic M, Siverio F, Roulin A, and Goudet J (2021) Population genomics of barn owls in the Western Parlearctic; NCBI bio project PRJNA727977; 10.1093/molbev/msab343
- Machado AP, Cumer T, Iseli C, Beaudoing E, Dupasquier M, Guex N, Dichmann K, Lourenço R, Lusby J, Martens H-D, Prévost L, Ramsden D, Roulin A, and Goudet J (2021) Population genomics of barn owls in the British Isles; NCBI bio project PRJNA700797; 10.1111/mec.16250
- Anguita F and Hernán F (2000) The Canary Islands origin: A unifying model. J Volcanol Geotherm Res 103:1–26. Elsevier B.V
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen-Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JHA, Meacham S, Megy K, O’Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang SY, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo-de-Santa-Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido-Martín D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux F. Heterozygote excess in small populations and the heterozygote-excess effective population size. Evolution. 2004;58:1891–1900. doi: 10.1111/j.0014-3820.2004.tb00477.x. [DOI] [PubMed] [Google Scholar]
- Bannerman DA (1963) Birds of the Atlantic Islands. Vol. 1. A history of the birds of the Canary Islands and of the Salvages. Oliver & Boyd
- BirdLife International (2019) The IUCN Red List of Threatened Species. Version 6.2
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R, Antoniazza S, Gaigher A, Ducrest A-L, Simon C, Fumagalli L, Goudet J, Roulin A. The genetic basis of color-related local adaptation in a ring-like colonization around the Mediterranean. Evolution. 2016;70:140–153. doi: 10.1111/evo.12824. [DOI] [PubMed] [Google Scholar]
- Carine MA, Humphries CJ, Guma IR, Reyes-Betancort JA, Santos Guerra A. Areas and algorithms: evaluating numerical approaches for the delimitation of areas of endemism in the Canary Islands archipelago. J Biogeogr. 2009;36:593–611. [Google Scholar]
- Chen MH, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, Trivedi B, Jiang T, Akbari P, Vuckovic D, Bao EL, Zhong X, Manansala R, Laplante V, Chen M, Lo KS, Qian H, Lareau CA, Beaudoin M, Hunt KA, Akiyama M, Bartz TM, Ben-Shlomo Y, Beswick A, Bork-Jensen J, Bottinger EP, Brody JA, van Rooij FJ, Chitrala K, Cho K, Choquet H, Correa A, Danesh J, Di Angelantonio E, Dimou N, Ding J, Elliott P, Esko T, Evans MK, Floyd JS, Broer L, Grarup N, Guo MH, Greinacher A, Haessler J, Hansen T, Howson JM, Huang QQ, Huang W, Jorgenson E, Kacprowski T, Kähönen M, Kamatani Y, Kanai M, Karthikeyan S, Koskeridis F, Lange LA, Lehtimäki T, Lerch MM, Linneberg A, Liu Y, Lyytikäinen LP, Manichaikul A, Martin HC, Matsuda K, Mohlke KL, Mononen N, Murakami Y, Nadkarni GN, Nauck M, Nikus K, Ouwehand WH, Pankratz N, Pedersen O, Preuss M, Psaty BM, Raitakari OT, Roberts DJ, Rich SS, Rodriguez BAT, Rosen JD, Rotter JI, Schubert P, Spracklen CN, Surendran P, Tang H, Tardif JC, Trembath RC, Ghanbari M, Völker U, Völzke H, Watkins NA, Zonderman AB, Wilson PWF, Li Y, Butterworth AS, Gauchat JF, Chiang CWK, Li B, Loos RJF, Astle WJ, Evangelou E, van Heel DA, Sankaran VG, Okada Y, Soranzo N, Johnson AD, Reiner AP, Auer PL, Lettre G. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JF, Schulenberg TS, Iliff MJ, Billerman SM, Fredericks TA, Sullivan BL, and Wood CL (2019) The eBird/clements checklist of birds of the world: v2019
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Cumer T, Machado AP, Dumont G, Bontzorlos VA, Ceccherelli R, Charter M, Dichmann K, Martens H-D, Kassinis N, Lourenço R, Manzia F, Ovari K, Prévost L, Rakovic M, Siverio F, Roulin A, and Goudet J (2021) Landscape and climatic variations of the Quaternary shaped multiple secondary contacts among barn owls (Tyto alba) of the Western Palearctic. Mol Biol Evol, msab343, 10.1093/molbev/msab343.
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darre MJ, Harrison PC. Heart rate, blood pressure, cardiac output, and total peripheral resistance of single comb White Leghorn hens during an acute exposure to 35 C ambient temperature. Poult Sci. 1987;66:541–547. doi: 10.3382/ps.0660541. [DOI] [PubMed] [Google Scholar]
- Dolédec S, Chessel D, Gimaret-Carpentier C. Niche separation in community analysis: a new method. Ecology. 2000;81:2914–2927. [Google Scholar]
- Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA, Kanoni S, Petersen AK, Pihur V, Strawbridge RJ, Shungin D, Hughes MF, Meirelles O, Kaakinen M, Bouatia-Naji N, Kristiansson K, Shah S, Kleber ME, Guo X, Lyytikäinen LP, Fava C, Eriksson N, Nolte IM, Magnusson PK, Salfati EL, Rallidis LS, Theusch E, Smith AJP, Folkersen L, Witkowska K, Pers TH, Joehanes R, Kim SK, Lataniotis L, Jansen R, Johnson AD, Warren H, Kim YJ, Zhao W, Wu Y, Tayo BO, Bochud M, Absher D, Adair LS, Amin N, Arking DE, Axelsson T, Baldassarre D, Balkau B, Bandinelli S, Barnes MR, Barroso I, Bevan S, Bis JC, Bjornsdottir G, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Bornstein SR, Brown MJ, Burnier M, Cabrera CP, Chambers JC, Chang IS, Cheng CY, Chines PS, Chung RH, Collins FS, Connell JM, Döring A, Dallongeville J, Danesh J, De Faire U, Delgado G, Dominiczak AF, Doney ASF, Drenos F, Edkins S, Eicher JD, Elosua R, Enroth S, Erdmann J, Eriksson P, Esko T, Evangelou E, Evans A, Fall T, Farrall M, Felix JF, Ferrières J, Ferrucci L, Fornage M, Forrester T, Franceschini N, Franco OH, Franco-Cereceda A, Fraser RM, Ganesh SK, Gao H, Gertow K, Gianfagna F, Gigante B, Giulianini F, Goel A, Goodall AH, Goodarzi MO, Gorski M, Gräßler J, Groves CJ, Gudnason V, Gyllensten U, Hallmans G, Hartikainen AL, Hassinen M, Havulinna AS, Hayward C, Hercberg S, Herzig KH, Hicks AA, Hingorani AD, Hirschhorn JN, Hofman A, Holmen J, Holmen OL, Hottenga JJ, Howard P, Hsiung CA, Hunt SC, Ikram MA, Illig T, Iribarren C, Jensen RA, Kähönen M, Kang HM, Kathiresan S, Keating BJ, Khaw KT, Kim YK, Kim E, Kivimaki M, Klopp N, Kolovou G, Komulainen P, Kooner JS, Kosova G, Krauss RM, Kuh D, Kutalik Z, Kuusisto J, Kvaløy K, Lakka TA, Lee NR, Te Lee I, Lee WJ, Levy D, Li X, Liang KW, Lin H, Lin L, Lindström J, Lobbens S, Männistö S, Müller G, Müller-Nurasyid M, Mach F, Markus HS, Marouli E, McCarthy MI, McKenzie CA, Meneton P, Menni C, Metspalu A, Mijatovic V, Moilanen L, Montasser ME, Morris AD, Morrison AC, Mulas A, Nagaraja R, Narisu N, Nikus K, O’Donnell CJ, O’Reilly PF, Ong KK, Paccaud F, Palmer CD, Parsa A, Pedersen NL, Penninx BW, Perola M, Peters A, Poulter N, Pramstaller PP, Psaty BM, Quertermous T, Rao DC, Rasheed A, Rayner NW, Renström F, Rettig R, Rice KM, Roberts R, Rose LM, Rossouw J, Samani NJ, Sanna S, Saramies J, Schunkert H, Sebert S, Sheu WHH, Shin YA, Sim X, Smit JH, Smith AV, Sosa MX, Spector TD, Stančáková A, Stanton AV, Stirrups KE, Stringham HM, Sundstrom J, Swift AJ, Syvänen AC, Tai ES, Tanaka T, Tarasov KV, Teumer A, Thorsteinsdottir U, Tobin MD, Tremoli E, Uitterlinden AG, Uusitupa M, Vaez A, Vaidya D, Van Duijn CM, Van Iperen EPA, Vasan RS, Verwoert GC, Virtamo J, Vitart V, Voight BF, Vollenweider P, Wagner A, Wain LV, Wareham NJ, Watkins H, Weder AB, Westra HJ, Wilks R, Wilsgaard T, Wilson JF, Wong TY, Yang TP, Yao J, Yengo L, Zhang W, Zhao JH, Zhu X, Bovet P, Cooper RS, Mohlke KL, Saleheen D, Lee JY, Elliott P, Gierman HJ, Willer CJ, Franke L, Hovingh GK, Taylor KD, Dedoussis G, Sever P, Wong A, Lind L, Assimes TL, Njølstad I, Schwarz PEH, Langenberg C, Snieder H, Caulfield MJ, Melander O, Laakso M, Saltevo J, Rauramaa R, Tuomilehto J, Ingelsson E, Lehtimäki T, Hveem K, Palmas W, März W, Kumari M, Salomaa V, Chen YDI, Rotter JI, Froguel P, Jarvelin MR, Lakatta EG, Kuulasmaa K, Franks PW, Hamsten A, Wichmann HE, Palmer CNA, Stefansson K, Ridker PM, Loos RJF, Chakravarti A, Deloukas P, Morris AP, Newton-Cheh C, Munroe PB. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M (2017) rbioclim: Improved getData function from the raster R package to interact with past, present and future climate data from worldclim.org
- Firmat C, Gomes Rodrigues H, Renaud S, Claude J, Hutterer R, Garcia-Talavera F, Michaux J. Mandible morphology, dental microwear, and diet of the extinct giant rats Canariomys (Rodentia: Murinae) of the Canary Islands (Spain) Biol J Linn Soc. 2010;101:28–40. [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997;78:311–327. doi: 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. Fast and efficient estimation of individual ancestry coefficients. Genetics. 2014;196:973–983. doi: 10.1534/genetics.113.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBIF.org (2021) GBIF Occurrence Download 10.15468/dl.5pd26s
- Ge SX, Jung D, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German CA, Sinsheimer JS, Klimentidis YC, Zhou H, Zhou JJ. Ordered multinomial regression for genetic association analysis of ordinal phenotypes at Biobank scale. Genet Epidemiol. 2020;44:248–260. doi: 10.1002/gepi.22276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie R. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–359. doi: 10.1126/science.1091875. [DOI] [PubMed] [Google Scholar]
- Gillespie R, Croom H, Hasty G. Phylogenetic relationships and adaptive shifts among major clades of tetragnatha spiders (Araneae: Tetragnathidae) in Hawai’i. Pac Sci. 1997;51:380–394. [Google Scholar]
- Goudet J. HIERFSTAT, a package for R to compute and test hierarchical F -statistics. Mol Ecol Notes. 2005;5:184–186. [Google Scholar]
- Goudet J, Kay T, Weir BS. How to estimate kinship. Mol Ecol. 2018;27:4121–4135. doi: 10.1111/mec.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graffelman J. Exploring diallelic genetic markers: the {HardyWeinberg. } Package J Stat Softw. 2015;64:1–23. [Google Scholar]
- Graffelman J, Morales-Camarena J. Graphical tests for Hardy-Weinberg Equilibrium based on the ternary plot. Hum Hered. 2008;65:77–84. doi: 10.1159/000108939. [DOI] [PubMed] [Google Scholar]
- Grant PR (1999) Ecology and Evolution of Darwin’s Finches. Princeton University Press
- Grant PR. Evolution on Islands. Oxford, UK: Oxford University Press; 1998. [Google Scholar]
- Gu J, Liang Q, Liu C, Li S. Genomic analyses reveal adaptation to hot arid and harsh environments in native chickens of China. Front Genet. 2020;11:582355. doi: 10.3389/fgene.2020.582355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halonen JI, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Relationship between outdoor temperature and blood pressure. Occup Environ Med. 2011;68:296–301. doi: 10.1136/oem.2010.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer R, Lopez-Jurado LF, Vogel P. The shrews of the eastern Canary Islands: a new species (mammalia: Soricidae) J Nat Hist. 1987;21:1347–1357. [Google Scholar]
- Illera JC, Spurgin LG, Rodriguez-Exposito E, Nogales M, Rando JC. What are we learning about speciation and extinction from the Canary Islands? Ardeola. 2016;63:15–33. [Google Scholar]
- Irwin DE, Alcaide M, Delmore KE, Irwin JH, Owens GL. Recurrent selection explains parallel evolution of genomic regions of high relative but low absolute differentiation in a ring species. Mol Ecol. 2016;25:4488–4507. doi: 10.1111/mec.13792. [DOI] [PubMed] [Google Scholar]
- Irwin DE, Milá B, Toews DPL, Brelsford A, Kenyon HL, Porter AN, Grossen C, Delmore KE, Alcaide M, Irwin JH. A comparison of genomic islands of differentiation across three young avian species pairs. Mol Ecol. 2018;27:4839–4855. doi: 10.1111/mec.14858. [DOI] [PubMed] [Google Scholar]
- Juan C, Emerson BC, Oromí P, and Hewitt GM (2000) Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Elsevier Ltd. [DOI] [PubMed]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. [Google Scholar]
- Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinforma. 2014;15:1–13. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulminski AM, Huang J, Loika Y, Arbeev KG, Bagley O, Yashkin A, Duan M, Culminskaya I. Strong impact of natural-selection-free heterogeneity in genetics of age-related phenotypes. Aging. 2018;10:492–514. doi: 10.18632/aging.101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin CJ, Wang C, Zamani N, Grant BR, Grant PR, Webster MT, Andersson L. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends Ecol Evol. 2002;17:183–189. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifjeld JT, Anmarkrud JA, Calabuig P, Cooper JEJ, Johannessen LE, Johnsen A, Kearns AM, Lachlan RF, Laskemoen T, Marthinsen G, Stensrud E, García-Del-Rey E. Species-level divergences in multiple functional traits between the two endemic subspecies of Blue Chaffinches Fringilla teydea in Canary Islands. BMC Zool. 2016;1:1–19. [Google Scholar]
- Liu C, Kraja AT, Smith JA, Brody JA, Franceschini N, Bis JC, Rice K, Morrison AC, Lu Y, Weiss S, Guo X, Palmas W, Martin LW, Chen YDI, Surendran P, Drenos F, Cook JP, Auer PL, Chu AY, Giri A, Zhao W, Jakobsdottir J, Lin LA, Stafford JM, Amin N, Mei H, Yao J, Voorman A, Larson MG, Grove ML, Smith AV, Hwang SJ, Chen H, Huan T, Kosova G, Stitziel NO, Kathiresan S, Samani N, Schunkert H, Deloukas P, Li M, Fuchsberger C, Pattaro C, Gorski M, Kooperberg C, Papanicolaou GJ, Rossouw JE, Faul JD, Kardia SLR, Bouchard C, Raffel LJ, Uitterlinden AG, Franco OH, Vasan RS, O’Donnell CJ, Taylor KD, Liu K, Bottinger EP, Gottesman O, Daw EW, Giulianini F, Ganesh S, Salfati E, Harris TB, Launer LJ, Dörr M, Felix SB, Rettig R, Völzke H, Kim E, Lee WJ, Te Lee I, Sheu WHH, Tsosie KS, Edwards DRV, Liu Y, Correa A, Weir DR, Völker U, Ridker PM, Boerwinkle E, Gudnason V, Reiner AP, Van Duijn CM, Borecki IB, Edwards TL, Chakravarti A, Rotter JI, Psaty BM, Loos RJF, Fornage M, Ehret GB, Newton-Cheh C, Levy D, Chasman DI. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB, Jackman TR, Larson A, De Queiroz K, Rodríguez-Schettino L. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–6. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO. An equilibrium theory of insular zoogeography. Evolution. 1963;17:373–387. [Google Scholar]
- MacArthur RH and Wilson EO (1967) The theory of island biogeography. Princeton University Press
- Machado AP, Clément L, Uva V, Goudet J, Roulin A. The Rocky Mountains as a dispersal barrier between barn owl (Tyto alba) populations in North America. J Biogeogr. 2018;45:1288–1300. [Google Scholar]
- Machado AP, Cumer T, Iseli C, Beaudoing E, Dupasquier M, Guex N, Dichmann K, Lourenço R, Lusby J, Martens H-D, Prévost L, Ramsden D, Roulin A, Goudet J (2021) Unexpected post-glacial colonisation route explains the white colour of barn owls (Tyto alba) from the British Isles. Mol Ecol 1–16. 10.1111/mec.16250 [DOI] [PMC free article] [PubMed]
- Machado AP, Topaloudis A, Cumer T, Lavanchy E, Bontzorlos VA, Ceccherelli R, Charter M, Kassinis N, Lymberakis P, Manzia F, Ducrest AL, Dupasquier M, Guex N, Roulin A, Goudet J (2022) Genomic consequences of colonisation, migration and genetic drift in barn owl insular populations of the eastern Mediterranean. Mol Ecol 31:1375–1388 [DOI] [PMC free article] [PubMed]
- Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, Miska EA, Durbin R, Genner MJ, Turner GF. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350:1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín A, Lorenzo JA. Aves del archipiélago canario. Francisco Lemus: Editor; 2001. [Google Scholar]
- Martin SH, Van Belleghem SM. Exploring evolutionary relationships across the genome using topology weighting. Genetics. 2017;206:429–438. doi: 10.1534/genetics.116.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masseti M. Mammals of the Macaronesian islands (the Azores, Madeira, the Canary and Cape Verde islands): redefinition of the ecological equilibrium. Mammalia. 2010;74:3–34. [Google Scholar]
- Mateo JA, Crochet PA, Afonso OM. The species diversity of the genus Gallotia (Sauria: Lacertidae) during the Holocene on La Gomera (Canary Islands) and the Latin names of Gomeran giant lizards. Zootaxa. 2011;2755:66–68. [Google Scholar]
- Molina-Borja M. Sexual dimorphism of Gallotia atlantica atlantica and Gallotia atlantica mahoratae (Lacertidae) from the Eastern Canary Islands. J Herpetol. 2003;37:769–772. [Google Scholar]
- Nadachowska-Brzyska K, Li C, Smeds L, Zhang G, Ellegren H. Temporal dynamics of avian populations during pleistocene revealed by whole-genome sequences. Curr Biol. 2015;25:1375–1380. doi: 10.1016/j.cub.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, Van Der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliot KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Döring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, De Jong PE, Snieder H, Van Gilst WH, Clarke R, Goel A, Hamsten A, Altshuler D, Jarvelin MR, Elliott P, Lakatta EG, Forouhi N, Wareham NJ, Loos RJF, Deloukas P, Lathrop GM, Zelenika D, Strachan DP, Soranzo N, Williams FM, Zhai G, Spector TD, Peden JF, Watkins H, Ferrucci L, Caulfield M, Munroe PB, Berglund G, Melander O, Matullo G, Uiterwaal CS, van der Schouw YT, Numans ME, Ernst F, Homuth G, Völker U, Elosua R, Laakso M, Connell JM, Mooser V, Salomaa V, Tuomilehto J, Laan M, Navis G, Seedorf U, Syvänen AC, Tognoni G, Sanna S, Uda M, Scheet P, Schlessinger D, Scuteri A, Dörr M, Felix SB, Reffelmann T, Lorbeer R, Völzke H, Rettig R, Galan P, Hercberg S, Bingham SA, Kooner JS, Bandinelli S, Meneton P, Abecasis G, Thompson JR, Braga Marcano CA, Barke B, Dobson R, Gungadoo J, Lee KL, Onipinla A, Wallace I, Xue M, Clayton DG, Leung HT, Nutland S, Walker NM, Todd JA, Stevens HE, Dunger DB, Widmer B, Downes K, Cardon LR, Kwiatkowski DP, Barrett JC, Evans D, Morris AP, Lindgren CM, Rayner NW, Timpson NJ, Lyons E, Vannberg F, Hill AVS, Teo YY, Rockett KA, Craddock N, Attwood AP, Bryan C, Bumpstead SJ, Chaney A, Ghori J, William RG, Hunt SE, Inouye M, Keniry E, King E, McGinnis R, Potter S, Ravindrarajan R, Whittaker P, Withers D, Bentley D, Groves CJ, Duncanson A, Ouwehand WH, Boorman JP, Cant B, Jolley JD, Knight AS, Koch K, Taylor NC, Watkins NA, Winzer T, Braund PS, Dixon RJ, Mangino M, Stevens S, Donnely P, Davidson D, Marchini JL, Spencer ICA, Cardin NJ, Ferreira T, Pereira-Gale J, Hallgrimsdottir IB, Howie BN, Su Z, Vukcevic D, Easton D, Everson U, Hussey JM, Meech E, Prowse CV, Walters GR, Jones RW, Ring SM, Prembey M, Breen G, St. Clair D, Ceasar S, Gordon-Smith K, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskovina V, Nikolov I, O’Donovan MC, Owen MJ, Craddock N, Collier DA, Elkin A, Farmer A, Williamson R, McGruffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop DT, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Lewis CM, Onnie CM, Prescott NJ, Mathew CG, Forbes A, Sanderson J, Mathew C, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, Barton A, Bruce IN, Donovan H, Eyre S, Gilbert PD, Hider SL, Hinks AM, John SL, Potter C, Silman AJ, Symmons DPM, Thomson W, Worthington J, Frayling TM, Freathy RM, Lango H, Perry JRB, Weedon MN, Hattersley AT, Shields BM, Hitman GA, Walker M, Newport M, Sirugo G, Conway D, Jallow M, Bradbury LA, Pointon JL, Brown MA, Farrar C, Wordsworth P, Franklyn JA, Heward JM, Simmonds MJ, Cough SCL, Seal S, Stratton MR, Ban M, Goris A, Sawcer SJ, Compston A. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales M, De León L, Gómez R. On the presence of the endemic skink Chalcides simonyi Steind. 1891 in Lanzarote (Canary Islands) Amphib-Reptilia. 1998;19:427–430. [Google Scholar]
- Nogales M, Rando JC, Valido A, Martín A. Discovery of a living giant lizard, genus Gallotia (Reptilia: Lacertidae), from La Gomera, Canary Islands. Herpetologica. 2001;57:169–179. [Google Scholar]
- Norder SJ, Proios K, Whittaker RJ, Alonso MR, Borges PAV, Borregaard MK, Cowie RH, Florens FBV, de Frias Martins AM, Ibáñez M, Kissling WD, de Nascimento L, Otto R, Parent CE, Rigal F, Warren BH, Fernández-Palacios JM, van Loon EE, Triantis KA, Rijsdijk KF. Beyond the Last Glacial Maximum: Island endemism is best explained by long-lasting archipelago configurations. Glob Ecol Biogeogr. 2019;28:184–197. [Google Scholar]
- O’Brien KA, Simonson TS, and Murray AJ (2020) Metabolic adaptation to high altitude. Elsevier Ltd.
- Oskarsson GR, Oddsson A, Magnusson MK, Kristjansson RP, Halldorsson GH, Ferkingstad E, Zink F, Helgadottir A, Ivarsdottir EV, Arnadottir GA, Jensson BO, Katrinardottir H, Sveinbjornsson G, Kristinsdottir AM, Lee AL, Saemundsdottir J, Stefansdottir L, Sigurdsson JK, Davidsson OB, Benonisdottir S, Jonasdottir A, Jonasdottir A, Jonsson S, Gudmundsson RL, Asselbergs FW, Tragante V, Gunnarsson B, Masson G, Thorleifsson G, Rafnar T, Holm H, Olafsson I, Onundarson PT, Gudbjartsson DF, Norddahl GL, Thorsteinsdottir U, Sulem P, Stefansson K. Predicted loss and gain of function mutations in ACO1 are associated with erythropoiesis. Commun Biol. 2020;3:1–10. doi: 10.1038/s42003-020-0921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios CJ. Current status and distribution of birds of prey in the Canary Islands. Bird Conserv Int. 2004;14:203–213. [Google Scholar]
- Pestano J, Brown RP, Suárez NM, Benzal J, Fajardo S. Intraspecific evolution of Canary Island Plecotine bats, based on mtDNA sequences. Heredity. 2003;90:302–307. doi: 10.1038/sj.hdy.6800240. [DOI] [PubMed] [Google Scholar]
- Pickrell J and Pritchard J (2012) Inference of population splits and mixtures from genome-wide allele frequency data. Nat Preced, 10.1038/npre.2012.6956.1. Springer Science and Business Media LLC [DOI] [PMC free article] [PubMed]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- Rodríguez B, Rodríguez A, Siverio F, Siverio M. Factors affecting the spatial distribution and breeding habitat of an insular cliff-nesting raptor community. Curr Zool. 2018;64:173–181. doi: 10.1093/cz/zox005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A, Rodríguez B, Montelongo T, Garcia‐Porta J, Pipa T, Carty M, Danielsen J, Nunes J, Silva C, Geraldes P, Medina FM, and Illera JC (2020) Cryptic differentiation in the Manx Shearwater hinders the identification of a new endemic subspecies. J Avian Biol 10.1111/jav.02633
- Romano A, Séchaud R, Roulin A. Geographical variation in bill size provides evidence for Allen’s rule in a cosmopolitan raptor. Glob Ecol Biogeogr. 2020;29:65–75. [Google Scholar]
- Romano A, Séchaud R, Roulin A. Evolution of wing length and melanin-based coloration in insular populations of a cosmopolitan raptor. J Biogeogr. 2021;48:961–973. [Google Scholar]
- Senfeld T, Shannon TJ, van Grouw H, Paijmans DM, Tavares ES, Baker AJ, Lees AC, Collinson JM. Taxonomic status of the extinct Canary Islands Oystercatcher Haematopus meadewaldoi. Ibis. 2020;162:1068–1074. [Google Scholar]
- Siverio F. Distribución y estatus de Tyto alba (Scopoli, 1769) en Tenerife, islas Canarias (Aves, Tytonidae) Vieraea. 1998;26:121–131. [Google Scholar]
- Siverio F. Lechuza común, Tyto alba. In: Lorenzo JA, editor. Atlas de las aves nidificantes en el archipiélago canario (1997–2003) Madrid: Dirección General de Conservación de la Naturaleza-Sociedad Española de Ornitología; 2007. pp. 304–310. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauer MJ, Field R, Grytnes JA, Trigas P, Ah-Peng C, Attorre F, Birks HJB, Borges PAV, Cardoso P, Chou CH, De Sanctis M, de Sequeira MM, Duarte MC, Elias RB, Fernández-Palacios JM, Gabriel R, Gereau RE, Gillespie RG, Greimler J, Harter DEV, Huang TJ, Irl SDH, Jeanmonod D, Jentsch A, Jump AS, Kueffer C, Nogué S, Otto R, Price J, Romeiras MM, Strasberg D, Stuessy T, Svenning JC, Vetaas OR, Beierkuhnlein C. Topography-driven isolation, speciation and a global increase of endemism with elevation. Glob Ecol Biogeogr. 2016;25:1097–1107. [Google Scholar]
- Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, Staley JR, Tragante V, Tukiainen T, Yaghootkar H, Masca N, Freitag DF, Ferreira T, Giannakopoulou O, Tinker A, Harakalova M, Mihailov E, Liu C, Kraja AT, Nielsen SF, Rasheed A, Samuel M, Zhao W, Bonnycastle LL, Jackson AU, Narisu N, Swift AJ, Southam L, Marten J, Huyghe JR, Stančáková A, Fava C, Ohlsson T, Matchan A, Stirrups KE, Bork-Jensen J, Gjesing AP, Kontto J, Perola M, Shaw-Hawkins S, Havulinna AS, Zhang H, Donnelly LA, Groves CJ, Rayner NW, Neville MJ, Robertson NR, Yiorkas AM, Herzig KH, Kajantie E, Zhang W, Willems SM, Lannfelt L, Malerba G, Soranzo N, Trabetti E, Verweij N, Evangelou E, Moayyeri A, Vergnaud AC, Nelson CP, Poveda A, Varga TV, Caslake M, De Craen AJM, Trompet S, Luan J, Scott RA, Harris SE, Liewald DCM, Marioni R, Menni C, Farmaki AE, Hallmans G, Renström F, Huffman JE, Hassinen M, Burgess S, Vasan RS, Felix JF, Uria-Nickelsen M, Malarstig A, Reilly DF, Hoek M, Vogt TF, Lin H, Lieb W, Traylor M, Markus HS, Highland HM, Justice AE, Marouli E, Lindström J, Uusitupa M, Komulainen P, Lakka TA, Rauramaa R, Polasek O, Rudan I, Rolandsson O, Franks PW, Dedoussis G, Spector TD, Jousilahti P, Männistö S, Deary IJ, Starr JM, Langenberg C, Wareham NJ, Brown MJ, Dominiczak AF, Connell JM, Jukema JW, Sattar N, Ford I, Packard CJ, Esko T, Mägi R, Metspalu A, De Boer RA, Van Der Meer P, Van Der Harst P, Gambaro G, Ingelsson E, Lind L, De Bakker PIW, Numans ME, Brandslund I, Christensen C, Petersen ERB, Korpi-Hyövälti E, Oksa H, Chambers JC, Kooner JS, Blakemore AIF, Franks S, Jarvelin MR, Husemoen LL, Linneberg A, Skaaby T, Thuesen B, Karpe F, Tuomilehto J, Doney ASF, Morris AD, Palmer CNA, Holmen OL, Hveem K, Willer CJ, Tuomi T, Groop L, Käräjämäki A, Palotie A, Ripatti S, Salomaa V, Alam DS, Majumder AAS, Di Angelantonio E, Chowdhury R, McCarthy MI, Poulter N, Stanton AV, Sever P, Amouyel P, Arveiler D, Blankenberg S, Ferrières J, Kee F, Kuulasmaa K, Müller-Nurasyid M, Veronesi G, Virtamo J, Deloukas P, Elliott P, Zeggini E, Kathiresan S, Melander O, Kuusisto J, Laakso M, Padmanabhan S, Porteous DJ, Hayward C, Scotland G, Collins FS, Mohlke KL, Hansen T, Pedersen O, Boehnke M, Stringham HM, Frossard P, Newton-Cheh C, Tobin MD, Nordestgaard BG, Caulfield MJ, Mahajan A, Morris AP, Tomaszewski M, Samani NJ, Saleheen D, Asselbergs FW, Lindgren CM, Danesh J, Wain LV, Butterworth AS, Howson JMM, Munroe PB. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe RS, Baez M. Geographic variation in scalation of the lizard Gallotia stehlini within the island of Gran Canaria. Biol J Linn Soc. 1993;48:75–87. [Google Scholar]
- Tigano A, Friesen VL. Genomics of local adaptation with gene flow. Mol Ecol. 2016;25:2144–2164. doi: 10.1111/mec.13606. [DOI] [PubMed] [Google Scholar]
- Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M, Guo X, Hendricks AE, Karaderi T, Lempradl A, Locke AE, Mahajan A, Marouli E, Sivapalaratnam S, Young KL, Alfred T, Feitosa MF, Masca NGD, Manning AK, Medina-Gomez C, Mudgal P, Ng MCY, Reiner AP, Vedantam S, Willems SM, Winkler TW, Abecasis G, Aben KK, Alam DS, Alharthi SE, Allison M, Amouyel P, Asselbergs FW, Auer PL, Balkau B, Bang LE, Barroso I, Bastarache L, Benn M, Bergmann S, Bielak LF, Blüher M, Boehnke M, Boeing H, Boerwinkle E, Böger CA, Bork-Jensen J, Bots ML, Bottinger EP, Bowden DW, Brandslund I, Breen G, Brilliant MH, Broer L, Brumat M, Burt AA, Butterworth AS, Campbell PT, Cappellani S, Carey DJ, Catamo E, Caulfield MJ, Chambers JC, Chasman DI, Chen YDI, Chowdhury R, Christensen C, Chu AY, Cocca M, Cook JP, Corley J, Corominas Galbany J, Cox AJ, Crosslin DS, Cuellar-Partida G, D’Eustacchio A, Danesh J, Davies G, Bakker PIW, Groot MCH, Mutsert R, Deary IJ, Dedoussis G, Demerath EW, Heijer M, Hollander AI, Ruijter HM, Dennis JG, Denny JC, Angelantonio E, Drenos F, Du M, Dubé MP, Dunning AM, Easton DF, Edwards TL, Ellinghaus D, Ellinor PT, Elliott P, Evangelou E, Farmaki AE, Farooqi IS, Faul JD, Fauser S, Feng S, Ferrannini E, Ferrieres J, Florez JC, Ford I, Fornage M, Franco OH, Franke A, Franks PW, Friedrich N, Frikke-Schmidt R, Galesloot TE, Gan W, Gandin I, Gasparini P, Gibson J, Giedraitis V, Gjesing AP, Gordon-Larsen P, Gorski M, Grabe HJ, Grant SFA, Grarup N, Griffiths HL, Grove ML, Gudnason V, Gustafsson S, Haessler J, Hakonarson H, Hammerschlag AR, Hansen T, Harris KM, Harris TB, Hattersley AT, Have CT, Hayward C, He L, Heard-Costa NL, Heath AC, Heid IM, Helgeland Ø, Hernesniemi J, Hewitt AW, Holmen OL, Hovingh GK, Howson JMM, Hu Y, Huang PL, Huffman JE, Ikram MA, Ingelsson E, Jackson AU, Jansson JH, Jarvik GP, Jensen GB, Jia Y, Johansson S, Jørgensen ME, Jørgensen T, Jukema JW, Kahali B, Kahn RS, Kähönen M, Kamstrup PR, Kanoni S, Kaprio J, Karaleftheri M, Kardia SLR, Karpe F, Kathiresan S, Kee F, Kiemeney LA, Kim E, Kitajima H, Komulainen P, Kooner JS, Kooperberg C, Korhonen T, Kovacs P, Kuivaniemi H, Kutalik Z, Kuulasmaa K, Kuusisto J, Laakso M, Lakka TA, Lamparter D, Lange EM, Lange LA, Langenberg C, Larson EB, Lee NR, Lehtimäki T, Lewis CE, Li H, Li J, Li-Gao R, Lin H, Lin KH, Lin LA, Lin X, Lind L, Lindström J, Linneberg A, Liu CT, Liu DJ, Liu Y, Lo KS, Lophatananon A, Lotery AJ, Loukola A, Luan J, Lubitz SA, Lyytikäinen LP, Männistö S, Marenne G, Mazul AL, McCarthy MI, McKean-Cowdin R, Medland SE, Meidtner K, Milani L, Mistry V, Mitchell P, Mohlke KL, Moilanen L, Moitry M, Montgomery GW, Mook-Kanamori DO, Moore C, Mori TA, Morris AD, Morris AP, Müller-Nurasyid M, Munroe PB, Nalls MA, Narisu N, Nelson CP, Neville M, Nielsen SF, Nikus K, Njølstad PR, Nordestgaard BG, Nyholt DR, O’Connel JR, O’Donoghue ML, Olde Loohuis LM, Ophoff RA, Owen KR, Packard CJ, Padmanabhan S, Palmer CNA, Palmer ND, Pasterkamp G, Patel AP, Pattie A, Pedersen O, Peissig PL, Peloso GM, Pennell CE, Perola M, Perry JA, Perry JRB, Pers TH, Person TN, Peters A, Petersen ERB, Peyser PA, Pirie A, Polasek O, Polderman TJ, Puolijoki H, Raitakari OT, Rasheed A, Rauramaa R, Reilly DF, Renström F, Rheinberger M, Ridker PM, Rioux JD, Rivas MA, Roberts DJ, Robertson NR, Robino A, Rolandsson O, Rudan I, Ruth KS, Saleheen D, Salomaa V, Samani NJ, Sapkota Y, Sattar N, Schoen RE, Schreiner PJ, Schulze MB, Scott RA, Segura-Lepe MP, Shah SH, Sheu WHH, Sim X, Slater AJ, Small KS, Smith AV, Southam L, Spector TD, Speliotes EK, Starr JM, Stefansson K, Steinthorsdottir V, Stirrups KE, Strauch K, Stringham HM, Stumvoll M, Sun L, Surendran P, Swift AJ, Tada H, Tansey KE, Tardif JC, Taylor KD, Teumer A, Thompson DJ, Thorleifsson G, Thorsteinsdottir U, Thuesen BH, Tönjes A, Tromp G, Trompet S, Tsafantakis E, Tuomilehto J, Tybjaerg-Hansen A, Tyrer JP, Uher R, Uitterlinden AG, Uusitupa M, Laan SW, Duijn CM, Leeuwen N, Van Setten J, Vanhala M, Varbo A, Varga TV, Varma R, Velez Edwards DR, Vermeulen SH, Veronesi G, Vestergaard H, Vitart V, Vogt TF, Völker U, Vuckovic D, Wagenknecht LE, Walker M, Wallentin L, Wang F, Wang CA, Wang S, Wang Y, Ware EB, Wareham NJ, Warren HR, Waterworth DM, Wessel J, White HD, Willer CJ, Wilson JG, Witte DR, Wood AR, Wu Y, Yaghootkar H, Yao J, Yao P, Yerges-Armstrong LM, Young R, Zeggini E, Zhan X, Zhang W, Zhao JH, Zhao W, Zhou W, Zondervan KT, Rotter JI, Pospisilik JA, Rivadeneira F, Borecki IB, Deloukas P, Frayling TM, Lettre G, North KE, Lindgren CM, Hirschhorn JN, Loos RJF. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50:26–35. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uva V, Päckert M, Cibois A, Fumagalli L, Roulin A. Comprehensive molecular phylogeny of barn owls and relatives (Family: Tytonidae), and their six major Pleistocene radiations. Mol Phylogenet Evol. 2018;125:127–137. doi: 10.1016/j.ympev.2018.03.013. [DOI] [PubMed] [Google Scholar]
- van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, Chen MH, Raffield LM, Tardaguila M, Huffman JE, Ritchie SC, Megy K, Ponstingl H, Penkett CJ, Albers PK, Wigdor EM, Sakaue S, Moscati A, Manansala R, Lo KS, Qian H, Akiyama M, Bartz TM, Ben-Shlomo Y, Beswick A, Bork-Jensen J, Bottinger EP, Brody JA, van Rooij FJA, Chitrala KN, Wilson PWF, Choquet H, Danesh J, Di Angelantonio E, Dimou N, Ding J, Elliott P, Esko T, Evans MK, Felix SB, Floyd JS, Broer L, Grarup N, Guo MH, Guo Q, Greinacher A, Haessler J, Hansen T, Howson JMM, Huang W, Jorgenson E, Kacprowski T, Kähönen M, Kamatani Y, Kanai M, Karthikeyan S, Koskeridis F, Lange LA, Lehtimäki T, Linneberg A, Liu Y, Lyytikäinen LP, Manichaikul A, Matsuda K, Mohlke KL, Mononen N, Murakami Y, Nadkarni GN, Nikus K, Pankratz N, Pedersen O, Preuss M, Psaty BM, Raitakari OT, Rich SS, Rodriguez BAT, Rosen JD, Rotter JI, Schubert P, Spracklen CN, Surendran P, Tang H, Tardif JC, Ghanbari M, Völker U, Völzke H, Watkins NA, Weiss S, Cai N, Kundu K, Watt SB, Walter K, Zonderman AB, Cho K, Li Y, Loos RJF, Knight JC, Georges M, Stegle O, Evangelou E, Okada Y, Roberts DJ, Inouye M, Johnson AD, Auer PL, Astle WJ, Reiner AP, Butterworth AS, Ouwehand WH, Lettre G, Sankaran VG, Soranzo N. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182:1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain LV, Vaez A, Jansen R, Joehanes R, Van Der Most PJ, Erzurumluoglu AM, O’Reilly PF, Cabrera CP, Warren HR, Rose LM, Verwoert GC, Hottenga JJ, Strawbridge RJ, Esko T, Arking DE, Hwang SJ, Guo X, Kutalik Z, Trompet S, Shrine N, Teumer A, Ried JS, Bis JC, Smith AV, Amin N, Nolte IM, Lyytikäinen LP, Mahajan A, Wareham NJ, Hofer E, Joshi PK, Kristiansson K, Traglia M, Havulinna AS, Goel A, Nalls MA, Sõber S, Vuckovic D, Luan J, Del Greco FM, Ayers KL, Marrugat J, Ruggiero D, Lopez LM, Niiranen T, Enroth S, Jackson AU, Nelson CP, Huffman JE, Zhang W, Marten J, Gandin I, Harris SE, Zemunik T, Lu Y, Evangelou E, Shah N, De Borst MH, Mangino M, Prins BP, Campbell A, Li-Gao R, Chauhan G, Oldmeadow C, Abecasis G, Abedi M, Barbieri CM, Barnes MR, Batini C, Beilby J, Blake T, Boehnke M, Bottinger EP, Braund PS, Brown M, Brumat M, Campbell H, Chambers JC, Cocca M, Collins F, Connell J, Cordell HJ, Damman JJ, Davies G, De Geus EJ, De Mutsert R, Deelen J, Demirkale Y, Doney ASF, Dörr M, Farrall M, Ferreira T, Frånberg M, Gao H, Giedraitis V, Gieger C, Giulianini F, Gow AJ, Hamsten A, Harris TB, Hofman A, Holliday EG, Hui J, Jarvelin MR, Johansson Å, Johnson AD, Jousilahti P, Jula A, Kähönen M, Kathiresan S, Khaw KT, Kolcic I, Koskinen S, Langenberg C, Larson M, Launer LJ, Lehne B, Liewald DCM, Lin L, Lind L, Mach F, Mamasoula C, Menni C, Mifsud B, Milaneschi Y, Morgan A, Morris AD, Morrison AC, Munson PJ, Nandakumar P, Nguyen QT, Nutile T, Oldehinkel AJ, Oostra BA, Org E, Padmanabhan S, Palotie A, Paré G, Pattie A, Penninx BWJH, Poulter N, Pramstaller PP, Raitakari OT, Ren M, Rice K, Ridker PM, Riese H, Ripatti S, Robino A, Rotter JI, Rudan I, Saba Y, Saint Pierre A, Sala CF, Sarin AP, Schmidt R, Scott R, Seelen MA, Shields DC, Siscovick D, Sorice R, Stanton A, Stott DJ, Sundström J, Swertz M, Taylor KD, Thom S, Tzoulaki I, Tzourio C, Uitterlinden AG, Völker U, Vollenweider P, Wild S, Willemsen G, Wright AF, Yao J, Thériault S, Conen D, Attia J, Sever P, Debette S, Mook-Kanamori DO, Zeggini E, Spector TD, Van Der Harst P, Palmer CNA, Vergnaud AC, Loos RJF, Polasek O, Starr JM, Girotto G, Hayward C, Kooner JS, Lindgren CM, Vitart V, Samani NJ, Tuomilehto J, Gyllensten U, Knekt P, Deary IJ, Ciullo M, Elosua R, Keavney BD, Hicks AA, Scott RA, Gasparini P, Laan M, Liu Y, Watkins H, Hartman CA, Salomaa V, Toniolo D, Perola M, Wilson JF, Schmidt H, Zhao JH, Lehtimäki T, Van Duijn CM, Gudnason V, Psaty BM, Peters A, Rettig R, James A, Jukema JW, Strachan DP, Palmas W, Metspalu A, Ingelsson E, Boomsma DI, Franco OH, Bochud M, Newton-Cheh C, Munroe PB, Elliott P, Chasman DI, Chakravarti A, Knight J, Morris AP, Levy D, Tobin MD, Snieder H, Caulfield MJ, Ehret GB. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 2017;70:e4–e19. doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren BH, Simberloff D, Ricklefs RE, Aguilée R, Condamine FL, Gravel D, Morlon H, Mouquet N, Rosindell J, Casquet J, Conti E, Cornuault J, Fernández-Palacios JM, Hengl T, Norder SJ, Rijsdijk KF, Sanmartín I, Strasberg D, Triantis KA, Valente LM, Whittaker RJ, Gillespie RG, Emerson BC, and Thébaud C (2015) Islands as model systems in ecology and evolution: prospects fifty years after MacArthur-Wilson [DOI] [PubMed]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Goudet J. A unified characterization of population structure and relatedness. Genetics. 2017;206:2085–2103. doi: 10.1534/genetics.116.198424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt KE, Huerta-Sánchez E (2019) Convergent evolution in human and domesticate adaptation to high-altitude environments. Phil. Trans. R. Soc. B 374:20180235 [DOI] [PMC free article] [PubMed]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Guo Y, Shi H, Liu CL, Panganiban RA, Chung W, O’Connor LJ, Himes BE, Gazal S, Hasegawa K, Camargo CA, Qi L, Moffatt MF, Hu FB, Lu Q, Cookson WOC, Liang L. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145:537–549. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome resequencing data produced in this study were deposited at the NCBI Sequence Read Archive (SRA) under Bio project PRJNA774943. Sampling locations used for niche modelling are given in appendix 1.