Abstract
The ability of flow cytometry to resolve multiple parameters was used in a microsphere-based flow cytometric assay for the simultaneous determination of several cytokines in a sample. The flow cytometer microsphere-based assay (FMBA) for cytokines consists of reagents and dedicated software, specifically designed for the quantitative determination of cytokines. We have made several improvements in the multiplex assay: (i) dedicated software specific for the quantitative multiplex assay that processes data automatically, (ii) a stored master calibration curve with a two-point recalibration to adjust the stored curve periodically, and (iii) an internal standard to normalize the detection step in each sample. Overall analytical performance, including sensitivity, reproducibility, and dynamic range, was investigated for interleukin-4 (IL-4), IL-6, IL-10, IL-12, gamma interferon (IFN-γ), and tumor necrosis factor alpha. These assays were found to be reproducible and accurate, with a sensitivity in the picograms-per-milliliter range. Results obtained with FMBA correlate well with commercial enzyme-linked immunosorbent assay data (r > 0.98) for all cytokines assayed. This multiplex assay was applied to the determination of cytokine profiles in whole blood from atopic and nonatopic patients. Our results show that atopic subjects' blood produces more IL-4 (P = 0.003) and less IFN-γ (P = 0.04) than the blood of nonatopic subjects. However, atopic asthmatic subjects' blood produces significantly more IFN-γ than that of atopic nonasthmatic subjects (P = 0.03). The results obtained indicate that the FMBA technology constitutes a powerful system for the quantitative, simultaneous determination of secreted cytokines in immune diseases.
It has been known for years that fluorescent flow cytometric detection combined with the use of sized latex microspheres allows one to perform specific and quantitative immunoassays of soluble analytes (9). The ability of the flow cytometer to discriminate between individual microspheres on the basis of size, fluorescent intensity, and/or fluorescent wavelength makes possible multianalytical assays. The use of microspheres of different sizes for multiplex assays has been described for different analytes in numerous publications (1, 15, 16, 18, 23, 24). However, discrimination of microspheres by fluorescence has been documented only recently (8, 14).
The routine use of this attractive technology faces three distinct hurdles. First, the software commercialized with cytometers is complex and more appropriate for the qualitative cellular analysis of individual samples than for the batch mode of sampling required for the quantitative assay of several analytes. Second, reagent development faces unique analytical difficulties, such as the calibration of each individual assay in a multiplex assay and the quality of complex reagents with multiple components. Third, the concept of multiplex quantitative assays, albeit very attractive in principle, has yet to demonstrate its usefulness compared with well-accepted technologies such as the enzyme-linked immunosorbent assay (ELISA).
Two approaches to simultaneous cytokine assays have been reported recently (3, 4). The publications showed calibration curves but did not provide analytical details such as accuracy or reproducibility. Furthermore, to date no study has demonstrated the usefulness of flow cytometric multiplex analysis in a fully integrated system.
We designed both reagents and software for the flow cytometric multiplex analysis of soluble cytokines on a commercial flow cytometer. Our flow cytometer microsphere-based assay (FMBA) uses green fluorescence intensity measurement to discriminate between microspheres. Microspheres in each category are coated with a specific anticytokine monoclonal antibody. The red fluorescent intensity allows the sensitive quantitation of the immune complexes formed at the surface of each microsphere. We improved the calibration step by use of stored master curves, and we improved the reliability of the assay with an internal standard for the adjustment of the fluorescent signal from anticytokine microspheres in each sample. To evaluate the analytical performance of FMBA technology and investigate the cytokine profiles of in vitro-activated whole blood from atopic and nonatopic patients, we designed a six-cytokine multiplex assay for interleukin-4 (IL-4), IL-6, IL-10, IL-12, gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α).
T cells play a major role in inflammation via cytokine secretion. Atopic asthma is characterized by an impaired balance in the production of cytokines by T lymphocytes. Inflammation is associated with the T-helper-2 cytokine profile, with an increase in IL-4 and IL-5 secretion (13).
In T-cell cultures from atopic adults with asthma, an increase in IL-4 production and a decrease in IFN-γ production, compared to cultures from nonatopic adults with asthma, were observed. (26, 30). A contribution by several other cytokines, responsible for a proinflammatory response, such as IL-6 and TNF-α, or responsible for an anti-inflammatory response, such as IL-10, has been suspected (11, 12, 21). Most studies have been performed on peripheral blood mononuclear cells (PBMC) and macrophages; only a few have been performed on whole blood (6, 7). Using the FMBA we investigated the concentration of cytokines in whole blood of atopic and nonatopic asthmatics and in atopic and nonatopic controls. In parallel, cytokine expression at the single-cell level was also investigated by the intracellular staining of IFN-γ.
We illustrate here the analytical and informative potential of the FMBA technology as applied to the determination of the cytokine profile of the whole blood of atopic asthmatic patients.
MATERIALS AND METHODS
Microspheres.
Polystyrene microspheres, 5.5 μm (coefficient of variation [CV], 2.7%) in diameter, dyed with various amounts of green fluorochrome (excitation at 488 nm and emission at 525 nm), were obtained from Beckman Coulter (Miami, Fla.).
Covalent coupling of capture monoclonal antibodies.
Monoclonal antibodies were used for IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α (Immunotech, Marseille, France). Each capture antibody was applied to a given microsphere category via a covalent linkage based on a thiol-maleimide interaction as described below. Microspheres were washed twice in phosphate-buffered saline (PBS) by centrifugation (at (800 × g for 5 min) and then resuspended thoroughly prior to use by a combination of vortexing and sonication (Branson 1210; Branson Ultrasonics Corporation, Danbury, Conn.). Ten microliters of 10-mg/ml SMCC [4-(N-maleimidomethyl) cyclohexane 1-carboxylate] (Sigma, St Louis, Mo.) in anhydrous dimethyl sulfoxide (Aldrich, Milwaukee, Wis.) was added with mixing to 10 mg of microspheres. The microspheres were shaken for 2 h at room temperature. The activated microspheres were then washed twice with PBS as above. An anticytokine antibody (5 mg/ml) in 20 mM borate (pH 8.2) buffer was reduced with dithiothreitol (DTT; Merck, Darmstadt, Germany) prepared in distilled water just before use. The antibody solution was then brought to 10 mM DTT by adding 10 μl of fresh 1 M DTT. This solution was gently mixed and incubated for 30 min at room temperature. The reduced immunoglobulin G (IgG) was purified on a PD10 column in 50 mM HEPES–2 mM EDTA (pH 7) and immediately coupled to the activated microspheres; microspheres (107) were incubated overnight in PBS with 30 μg of reduced capture antibody with shaking at room temperature. The coated microspheres were then washed twice in PBS containing 5 g of bovine serum albumin (BSA)/liter, resuspended in PBS blocking buffer with 5 g of BSA/liter, and incubated overnight at 4°C. The microspheres were counted and adjusted to a concentration of 107/ml. Equal numbers of each category of coated microspheres were mixed to a final concentration of 106/ml of PBS containing 1 g of BSA/liter and 0.02% NaN3 and were stored at 4°C in the dark.
Detection antibody.
Detection antibodies for IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α (Immunotech) were biotinylated using NHS–long-chain biotin [N-hydroxy-sulfosuccinimidyl-6-(biotinamide) hexanoate] according to the specifications of the manufacturer (Pierce, Rockford, Ill.). The biotinylated antibodies were diluted to a final concentration of 1 μg/ml each in PBS containing 1 g of BSA/liter and 0.02% NaN3 and were stored at 4°C.
FMBA cytokine protocol.
Fifty microliters of whole-blood supernatants was incubated in membrane filter-bottomed microplates (Nunc, Roskilde, Denmark) with 50 μl of a biotinylated anticytokine antibody mixture and 10 μl of the microsphere mixture (anticytokine-coated and calibration microspheres) with shaking at room temperature for 2 h in the dark. Microspheres were then washed twice with 250 μl of wash solution (IM0425′, Immunotech) by aspiration through membrane filters; 100 μl of streptavidin-PC5 (1 μg/ml) (Immunotech) was added to each well, followed by incubation for 30 min in the dark with shaking at room temperature. Microspheres were then washed twice more, following the same protocol as above, and after resuspension in phosphate buffer, they were transferred to tubes for acquisition on the flow cytometer.
Flow cytometer standardization and acquisition.
Analysis was performed on a Beckman Coulter Epics (Hialeah, Fla.) XL-MCL flow cytometer; fluorescence excitation was at 488 nm with a 15-mW argon laser. The flow cytometer was controlled daily with DNA-Check beads (Beckman Coulter), according to the specifications of the manufacturer, to check the stability of the optical and fluidic systems. Briefly, 10,000 events were acquired with the DNA check protocol, with histograms for all parameters and regions that include the main population. Intensity peak and half-peak coefficient variation (HPCV) for all parameters gave a CV of <2% according to expected values. For a quantitative flow-cytometric assay, FMBA acquisition must be performed on a precisely standardized instrument, and forward scatter, side scatter, and FL1 (525 nm) and FL4 (675 nm) photomultiplier (PMT) voltages must be adjusted properly; to adjust the PMT voltages we used a set of standardization microspheres (Flow set; Beckman Coulter) and the automatic calibration feature of the SYSTEM-II software. Peak position target values were defined experimentally for the Flow set so that anticytokine microspheres were well discriminated on FL1 with the best dynamic range in FL4. Briefly, during Flow set microsphere acquisition (10,000 events), high voltage and gain were adjusted by SYSTEM-II software to ensure that the Flow set peak position gave a CV of <2.5% compared to the expected target values. After standardization, all the anticytokine microspheres within the set gate of the FMBA cytokine acquisition protocol were acquired. No compensation for FL1 and FL4 was applied. Acquisition was performed at high speed, and forward- and side-scatter gating was used to ensure that only single beads were analyzed. Data acquisition of about 500 events per cytokine microsphere set (4,000 gated events for our assay) was collected for each sample.
Reagent calibration.
For the calibration of the FMBA, computer-stored standard curves were used. For each cytokine immunoassay, these standard master curves were established at Immunotech by assaying dilutions (4,000 to 3.9 pg/ml) of international cytokine standards with the FMBA protocol. International standards used were as follows: IL-4, 88/656; IL-6, 89/548; IL-10, 92/516; IL-12, 86/504; TNF-α, 87/650 (National Institute for Biological Standards and Control, Hertfordshire, England); and IFN-γ, Gxg01-9025535 (Boston Biomedica, Inc., West Bridgewater, Md.). After acquisition on the cytometer, cytokine standard curves were collected and fitted, using a spline-fitting curve model. Curve fit parameters were then stored in a postanalysis database file. These master curves were used to analyze the immunoassay data. The stored master curve is valid for a given expiration date lot of reagents. The memorized curves may be revalidated at the time of the expiration date by a two-point recalibration performed with a standard solution containing a mixture of cytokines.
A separated set of unstained microspheres called “calibration microspheres” was biotinylated with two concentrations of NHS–long-chain biotin (Pierce). This category of microspheres, included in each test, captures the streptavidin-PC5 conjugate independently of cytokine-specific immunological reactions. The red signal emitted by this set of calibration microspheres is determined upon manufacturing and is stored with the standard curve fit parameters in the database file specific for a given reagent lot. The FL4 signal from these calibration microspheres is determined for each assay. This value is compared to the database, and the ratio is used to normalize the FL4 signal from the anticytokine microspheres (patent pending).
Data processing.
List mode files were processed automatically with dedicated post acquisition software. Data processing was achieved in discrete steps, as follows. (i) An automatic gating of each microsphere category separated populations according to the green fluorescence intensity. (ii) The mean red fluorescence intensity was determined for each microsphere set. This value is related directly to the amount of complex formed on the microsphere. (iii) FL4 signals from anticytokine microspheres were normalized according to the red fluorescent signal of the calibration microspheres run in parallel. (iv) Analyte concentration was determined using the stored master curves.
Study of fluorescent signal normalization with calibration microspheres.
We examined the effect of correction of the anticytokine microspheres' fluorescent signal by calibration microspheres on cytokine quantitation, either by using the optimal FL4 PMT voltage (1,110 V) or with the PMT adjusted to 1,120 to 1,150 V.
Follow-up of stored calibration.
We examined the stability of stored curves by performing assays of controls at 3, 6, 9, and 12 months. At each date, the data were processed either against the initial stored calibration or after recalibration of stored master curves by a two-point recalibration.
Analytical studies.
The analytical performance of the FMBA was determined as follows.
(i) Sensitivity.
The sensitivities of the individual assays were determined by performing 10 measurements of the 0- and 3.9-pg/ml cytokine standard solutions following the FMBA assay protocol. Sensitivity is defined as the lowest concentration significantly different from the zero standard with a probability of 98%.
(ii) Cross-reactivity.
Interference and cross-reactivity between cytokines were assessed by using a mixture of the different microspheres and detection antibodies with each cytokine standard, in turn, at 10 ng/ml in the FMBA.
(iii) Intra- and interassay precision.
Intra-assay precision was determined by assaying samples 10 times in the same assay. Interassay precision was determined by assaying the same sample in duplicate in 10 independent assays. The CV was determined.
(iv) FMBA-ELISA correlation.
A total of 40 whole-blood samples from healthy subjects were collected in sodium heparin tubes and were diluted 1:4 in RPMI culture medium (BioWhittaker, Verviers, France). For each sample 1 ml of diluted whole blood was cultured in a 6-well culture plate (Nunc) and stimulated with anti-CD3 and anti-CD28 monoclonal antibodies (clones UCHT1 and CD28.2; Immunotech) at 1 μg/ml. Supernatants were harvested after 24 h of culture and centrifuged at 300 × g for 10 min, and cytokine concentrations were determined in parallel with FMBA and ELISA kits (Immunotech) according to the manufacturer's instructions. Linear regression analysis was used to compare FMBA and ELISA results.
(v) Correlation of FMBA with external calibration and FMBA with stored calibration.
A total of 37 whole-blood samples from healthy subjects were collected in sodium heparin tubes and were diluted 1:4 in RPMI culture medium (BioWhittaker). For each sample 1 ml of diluted whole blood was cultured in a 6-well culture plate (Nunc) and stimulated with anti-CD3 and anti-CD28 monoclonal antibodies (clones UCHT1 and CD28.2; Immunotech) at 1 μg/ml. Supernatants were harvested after 24 h of culture and centrifuged at 300 × g for 10 min, and cytokine concentrations were determined in parallel by FMBA using cytokine standard solutions run in parallel with the sample and by FMBA using the stored master curve. Linear regression analysis was used to compare results.
Subjects.
A total of 61 subjects (20 males and 41 females; mean age, 39 ± 13 years) were included in this study. Thirty-six were patients referred to the allergy clinic for a history compatible with asthma. Among these, 30 were atopic asthmatics and 6 were nonatopic asthmatics. None of the asthmatics were treated with oral or parenteral steroids. Ten subjects were atopic nonasthmatics, and 15 were healthy controls. A blood sample was collected from each patient for culture. The protocol was approved by the ethics committee of Marseille-I, and each subject gave informed consent.
Diagnosis of atopy.
A diagnosis of atopy was based on the positivity of at least one skin prick test against a common environmental allergen from a standard battery of extracts (Laboratoire des Stallergenes, Paris, France). In all cases, a positive-control (codeine phosphate 9%) and a negative-control test was performed.
Diagnosis of asthma.
The diagnosis of asthma was confirmed on the basis of a history of dyspnea and wheezing, by either reversible airflow obstruction or a positive methacholine challenge test. Reversible airway obstruction was characterized by a 20% increase in forced expiratory volume in one second (FEV 1) after inhalation of 200 μg of albuterol. Methacholine challenge tests were performed when spirometric data were normal. Cumulative doses of methacholine were administered through a ME-FAR dosimeter (Electromedically, Brescia, Italy), and specific airway resistance (SRaw) measurements were made after each dose in an 830-liter constant body plethysmograph (model Master Lab; Jaeger, Würzburg, Germany). Bronchial hyperreactivity was defined as a 100% increase in SRaw at 200 μg of methacholine or less.
Whole-blood culture.
Each blood sample collected in sodium heparin tubes was divided into two parts, one for the immunoassays of cytokines in culture supernatants using the FMBA technique, and the other for the detection of intracytoplasmic cytokines. For microsphere-based immunoassays, 1 ml of whole blood diluted 1:1 with RPMI was cultured in 6-well plates either with or without 100 ng of phorbol 12-myristate acetate (PMA)/ml and 2 μg of ionomycin (Sigma, l'Isle d'Abeau Chesnes, France)/ml. After 6 h of culture, samples were harvested and centrifuged at 300 × g for 10 min, and the supernatants were frozen and kept at −20°C until assay. For intracytoplasmic staining, 50 μl of cells from whole blood was cultured in 96-well plates either with or without PMA and ionomycin at the concentrations given above, as well as in the presence of brefeldin (20 μg/ml) and monensin (2 μmol/liter) (Sigma, France). After 6 h of culture, cells were harvested, centrifuged at 300 × g for 10 min, washed, and resuspended in PBS.
Immunofluorescent staining of cells.
Cells were first incubated with 10 μl of phycoerythrin (PE)- and Cy5-conjugated anti-CD3 monoclonal antibody (clone UCHT1; Immunotech) or anti-CD8–PE–Cy5 (clone 13B8.2; Immunotech) for 15 min in the dark and then fixed and permeabilized using the IntraPrep reagents (Immunotech) as indicated by the manufacturer. Cells were then incubated for 15 min with 40 ng of fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ antibody (clone 4S B3; IgG1; Pharmingen, San Diego, Calif.) and with either 20 μl of PE-coupled anti-IL4 antibody (clone 4D9; Immunotech) or 20 μl of FITC- and PE-conjugated isotypic control antibodies (IgG1; clone 679.1 Mc7; Immunotech). After a wash in PBS by centrifugation at 300 × g for 10 min, cells were resuspended in 250 μl of PBS containing 0.5% formaldehyde. The specificity of the staining was assessed by blocking the reaction with increasing concentrations of recombinant cytokines (Pharmingen). Optimal concentrations of antibodies were determined in preliminary experiments.
Flow cytometry of cells.
Data were acquired directly on a Coulter EPICS XL cytometer, using XL SYSTEM II software. Dead cells, monocytes, and polymorphonuclear cells were excluded by forward- and side-scatter gating. Acquisition was then gated on the CD3+ cells or the CD8+ bright cells, and a minimum of 20,000 cells were acquired. Statistical markers were set using the negative control as a reference. Results are expressed as the percentage of IFN-γ- or IL-4-positive CD3+ or CD8+ cells.
Statistical analysis of patient samples.
Intracytoplasmic staining results are expressed as percentages ± standard deviations (SD). The average percentage of CD3+ cytokine-positive cells was compared between groups using the Student t test.
FMBA results are expressed as the mean of the concentration (in picograms per milliliter) ± the standard error of the mean (SEM). Statistical analysis was performed using Wilcoxon's test or the Mann-Whitney U test. A probability value of less than 0.05 was taken as statistically significant.
RESULTS
FMBA acquisition.
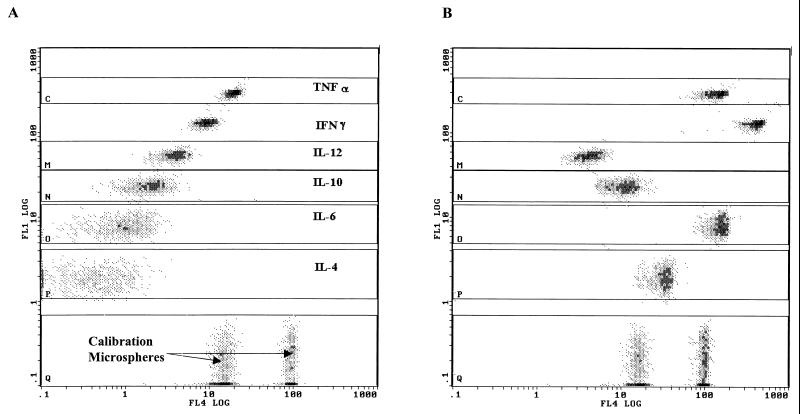
Figure 1 shows a typical example of the FL4/FL1 dot plots obtained with unstimulated (Fig. 1A) and stimulated (Fig. 1B) diluted whole-blood supernatants, assayed with the FMBA reagents. The individual anticytokine microsphere sets (IL-4 to TNF-α) were separated into discrete populations by FL1 (analyte discrimination) intensity versus FL4 (quantification). The red fluorescence intensity (FL4) emitted by anticytokine microspheres is a function of the cytokine concentration in the sample. At the bottom, the Q gate contains the calibration microsphere sets stained with streptavidin-PC5 conjugate independently of the other anticytokine-specific microsphere categories run simultaneously.
FIG. 1.
Dot plots of PC5 fluorescence (ordinate) versus FITC fluorescence (abscissa) for FMBA. Six cytokines were simultaneously measured in unstimulated whole blood (A) and in PMA- and ionomycin-stimulated whole blood (B). Dots in gate Q represent calibration microspheres.
In this example, no cytokine was detected in unstimulated whole-blood samples. In Fig. 1A, the FL4 signal observed is due to the leakage of FL1 intensity in the FL4 channel because no compensation was applied in the acquisition protocol. In stimulated whole-blood samples, all assayed cytokines, except IL-12 in this case, were observed.
FMBA performance for cytokines.
Multiplex assays have yet to demonstrate their usefulness for routine assays. We focused our efforts, therefore, on stored calibration curves, quality maintenance, and the stability of reagents so as to maximize the robustness of the system and simplify its use.
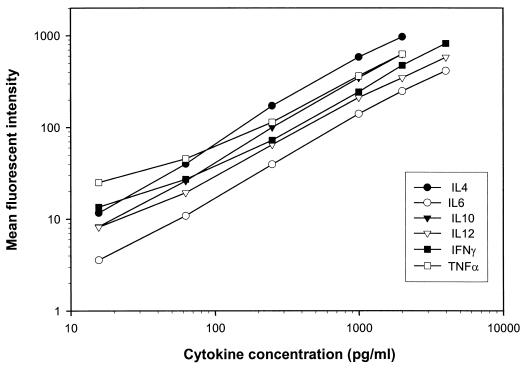
The standard curve for each cytokine FMBA was established by determining the mean fluorescence intensity in FL4 for each anticytokine microsphere set, incubated at a known concentration as determined by the cognate international standard. The ranges were 0 to 2,000 pg/ml for IL-4, IL-10, and TNF-α, and 0 to 4,000 pg/ml for IL-6, IL-12, and IFN-γ. Figure 2 shows, on a log-log graph, standard curves generated by the FMBA multiplex technology
FIG. 2.
Standard curves of IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α international standards obtained by FMBA. Assays were run in triplicate; results are means. Curves are presented as mean channel fluorescence versus concentration of standard.
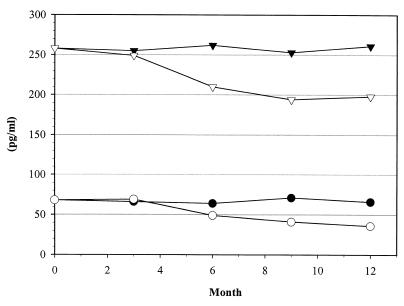
Figure 3 shows the results of a study of the stability of the stored calibration for IL-6. When data were processed on the basis of the initial stored calibration, correct results were obtained for as long as 3 months. Beyond that point, the concentration of the controls was underestimated. When the stored calibration was recalibrated with two-point recalibration, the concentration obtained was correct and the assay was valid for as long as 1 year. The aging of the reagents is a limiting factor in the validity of the stored standard that is corrected by the two-point recalibration.
FIG. 3.
Follow-up of two IL-6 control samples read on the stored master curve without (open symbols) or with (solid symbols) recalibration by a two-point recalibration procedure.
The results of the improvement of the reliability of the assay by the use of calibration micropheres are shown in Table 1. We have simulated slight variation in the setting of the cytometer by increasing the voltage of the PMT. With no signal correction, the sample concentration increased with the voltage and the concentration was overestimated. With the signal correction by calibration microspheres, fluorescent signal variation from anticytokine-coated microspheres was adjusted and the concentration was shown to remain the same whatever the PMT voltage. The calibration microspheres function as an internal standard normalizing signal variation.
TABLE 1.
Evaluation of effect of FL4 PMT voltage variation on concentrations of two controls with or without correction by calibration microspheres
| FL4 PMT voltage (V) | Concn (pg/ml) of:
|
|||
|---|---|---|---|---|
| Control 1
|
Control 2
|
|||
| No correction | Correction | No correction | Correction | |
| 1,110 | 194.9 | 194.9 | 529.3 | 529.3 |
| 1,120 | 205.3 | 195.0 | 539.4 | 521.4 |
| 1,130 | 214.3 | 189.1 | 552.7 | 526.4 |
| 1,140 | 226.3 | 194.7 | 563.7 | 522.6 |
| 1,150 | 232.3 | 190.7 | 574.2 | 525.3 |
| Mean | 214.62 | 192.88 | 551.86 | 525.00 |
| SD | 15.21 | 2.78 | 18.06 | 3.13 |
The sensitivities of the individual assays, defined as the lowest signal significantly different from the zero standard with 5 SD, are 0.3 pg/ml for IL-4, 0.6 pg/ml for IL-6, 0.3 pg/ml for IL-10, 1.6 pg/ml for IL-12, 4 pg/ml for IFN-γ, and 0.6 pg/ml for TNF-α. In all cases, intra- and interassay precision, evaluated on 10 culture supernatants, was excellent, with CVs below 5% (Tables 2 and 3).
TABLE 2.
Intra-assay precisiona
| Cytokine and sample | Mean concn (pg/ml) | CV (%) |
|---|---|---|
| IL-4 | ||
| S1 | 44.9 | 4.2 |
| S2 | 452.0 | 4.3 |
| IL-6 | ||
| S1 | 146.1 | 3.5 |
| S2 | 1,460 | 4.4 |
| IL-10 | ||
| S1 | 16.5 | 4.3 |
| S2 | 692.4 | 3.6 |
| IL-12 | ||
| S1 | 18.6 | 4.2 |
| S2 | 206.4 | 3.0 |
| IFN-γ | ||
| S1 | 39.0 | 4.3 |
| S2 | 466.4 | 2.6 |
| TNF-α | ||
| S1 | 41.9 | 4.1 |
| S2 | 506.7 | 2.2 |
Determined by assaying two samples 10 times.
TABLE 3.
Interassay precisiona
| Cytokine and sample | Mean concn (pg/ml) | CV (%) |
|---|---|---|
| IL-4 | ||
| S1 | 12.2 | 4.6 |
| S2 | 459.1 | 4.4 |
| IL-6 | ||
| S1 | 155.8 | 4.3 |
| S2 | 1,509.0 | 3.5 |
| IL-10 | ||
| S1 | 16.9 | 4.7 |
| S2 | 718.8 | 2.3 |
| IL-12 | ||
| S1 | 21.0 | 4.9 |
| S2 | 216.5 | 4.2 |
| IFN-γ | ||
| S1 | 43.9 | 4.5 |
| S2 | 495.5 | 3.8 |
| TNF-α | ||
| S1 | 49.0 | 4.8 |
| S2 | 542.3 | 4.9 |
Determined by assaying two samples in duplicate in 10 independent assays.
Table 4 shows the equations for correlation between sample concentrations read against either cytokine standard solutions run at the same time or the stored master curve. Results obtained with stored master curves are in very good agreement with those obtained with cytokine standard solutions.
TABLE 4.
Correlation between cytokine concentrationsa
| Cytokine | Equation | r | n |
|---|---|---|---|
| IL-4 | y = 1.01x − 4.37 | 0.99 | 37 |
| IL-6 | y = 1.00x − 4.65 | 0.99 | 37 |
| IL-10 | y = 1.00x + 0.49 | 0.98 | 37 |
| IL-12 | y = 1.02x + 5.31 | 0.99 | 37 |
| IFN-γ | y = 0.99x + 1.66 | 0.98 | 37 |
| TNF-α | y = 1,03x + 4.86 | 0.99 | 37 |
Determined by reading the sample against a fresh standard curve (dilution of cytokine standard solutions) or a stored master curve with FMBA for IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α. x corresponds to values determined on a stored master curve, and y corresponds to values determined on a fresh standard curve.
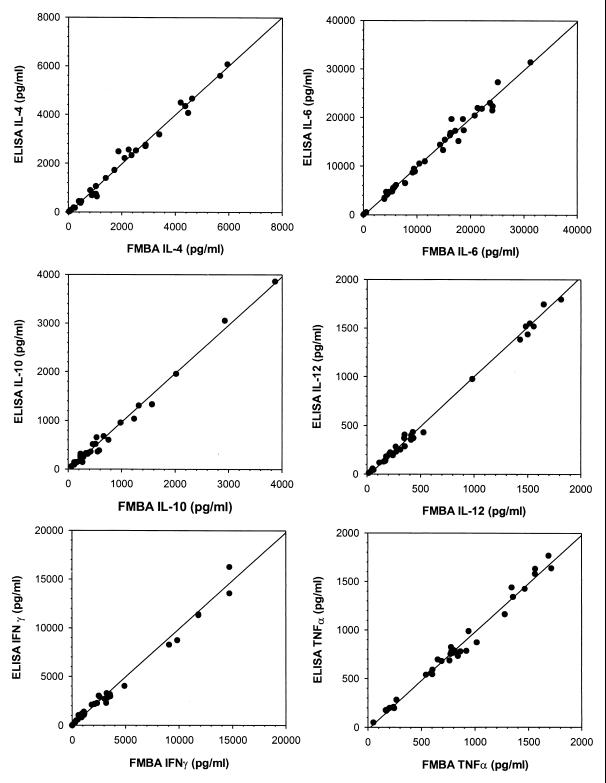
Figure 4 shows the correlation between the FMBA and the corresponding ELISAs. The results demonstrate excellent correlation between the FMBA and the ELISA (r > 0.98) for each of the six cytokines assayed.
FIG. 4.
Correlation between FMBA and Immunotech ELISA for IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α on 40 samples. Linear regressions were calculated as follows: for IL-4, y = 0.96x − 10.07 and r = 0.982; for IL-6, y = 0.96x − 4.33 and r = 0.987; for IL-10, y = 0.1.16x − 4.15 and r = 0.989; for IL-12, y = 0.92x − 11.43 and r = 0.994; for IFN-γ, y = 1.17x − 8.33 and r = 0.989; for TNF-α, y = 0.93x + 3.38 and r = 0.986.
Cross-reactivity.
Potential cross-reactivities were studied. Results, expressed as percentages, are shown in Table 5. Very low cross-reactivity (<0.3%) was observed only between IL-6 microspheres and IL-4 cytokine at concentrations much higher than those usually occurring in biological samples. These results demonstrate the high specificity of our FMBA reagents. Specificity in multiplex assays is guaranteed by careful selection of antibodies.
TABLE 5.
Cross-reactivitya
| Microsphere | % Cross-reactivity with incubated cytokine
|
|||||
|---|---|---|---|---|---|---|
| IL-4 | IL-6 | IL-10 | IL-12 | IFNγ | TNFα | |
| IL-4 | 100 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 |
| IL-6 | 0.28 | 100 | 0.01 | 0.03 | 0.02 | 0.05 |
| IL-10 | 0.01 | 0.00 | 100 | 0.00 | 0.01 | 0.02 |
| IL-12 | 0.00 | 0.00 | 0.00 | 100 | 0.00 | 0.00 |
| IFN-γ | 0.00 | 0.00 | 0.00 | 0.00 | 100 | 0.00 |
| TNF-α | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 100 |
Determined by assays in which each cytokine in turn was incubated, at a concentration of 10 ng/ml, with the microsphere mixture. Results, given in percentages, are calculated as the ratio of the concentration found to the concentration incubated.
Cytokine secretion.
Concentrations of IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α from patients and healthy subjects were measured in unstimulated and in PMA- and ionomycin-stimulated whole-blood samples.
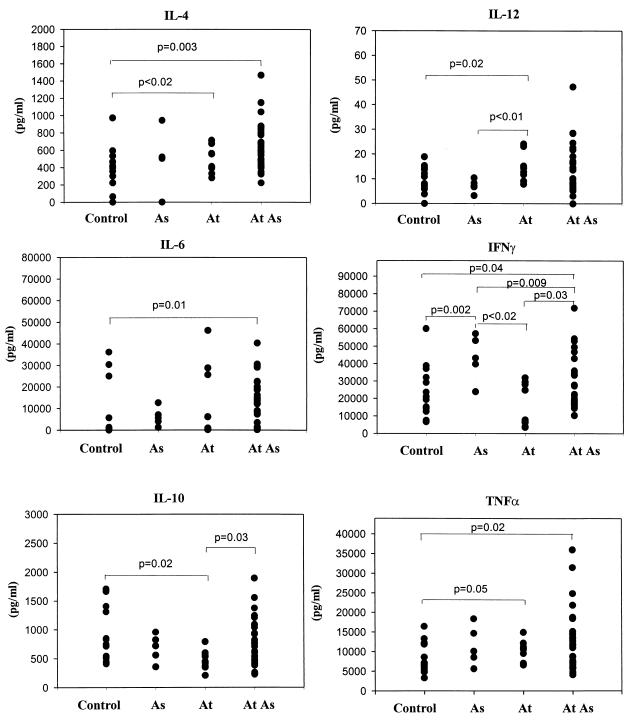
In unstimulated whole-blood samples IL-4 was not detected (data not shown). After stimulation, whole-blood samples from atopic nonasthmatic or asthmatic patients showed significantly more IL-4 (503.79 ± 51.00 pg/ml [P < 0.02] and 628.41 ± 48.41 pg/ml [P = 0.003], respectively) than samples from control patients (314.17 ± 67.77 pg/ml) (Fig. 5). IL-6 was detected in atopic patients' samples even without stimulation (data not shown). Following stimulation, IL-6 secretion was higher in samples from atopic asthmatics than in control samples (11,774 ± 2,058 versus 6,475 ± 3,045 pg/ml; P = 0.01). Spontaneous IL-10 secretion was significantly higher in atopic asthmatics' whole-blood samples than in control samples (P = 0.002) (data not shown). Conversely, after stimulation, samples from atopic nonasthmatics showed significantly less IL-10 than controls (476.59 ± 55.81 versus 949.02 ± 133.92 pg/ml; P = 0.02). IL-10 production was higher for asthmatic atopics than for nonasthmatic atopics (782.60 ± 74.88 versus 476.59 ± 55.81 pg/ml; P = 0.03). IL-12 secretion after stimulation was significantly higher in samples from atopic nonasthmatics than in samples from asthmatics (14.25 ± 1.76 versus 7.17 ± 1.17 pg/ml; P < 0.01) and controls (P = 0.02). Atopic asthmatics produced significantly less IFN-γ (29,519 ± 2,793 versus 43,509 ± 5,824 pg/ml; P = 0.009) than did asthmatic subjects without atopy. In contrast, samples from asthmatics with or without atopy had significantly more IFN-γ than control samples (29,519 ± 2,793 and 43,509 ± 5,824 versus 22,643 ± 3,803 pg/ml, respectively; P = 0.04 and P = 0.002). Furthermore, whole blood from asthmatic subjects produced significantly more IFN-γ after stimulation (29,519 ± 2,793 versus 16,807 ± 3,780 pg/ml; P = 0.03) than that from atopic patients who did not have asthma. Finally, whole blood from atopic asthmatic subjects secreted more TNF-α (14,213 ± 1,740 versus 8,520 ± 966 pg/ml; P = 0.02) than did blood from controls.
FIG. 5.
Secretion of IL-4, IL-6, IL-10, IL-12, IFN-γ, and TNF-α in PMA- and ionomycin-stimulated whole blood from six nonatopic asthmatic (As), 10 atopic nonasthmatic (At), 30 atopic asthmatic (At As), and 15 nonatopic, nonasthmatic (control) subjects.
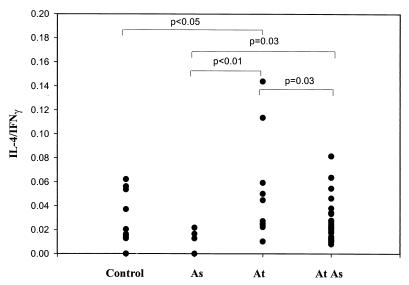
Furthermore, it was observed (Fig. 6) that samples from atopic asthmatics had IL-4/IFN-γ ratios higher than those of samples from asthmatic patients without atopy (P = 0.03).
FIG. 6.
Ratio of IL-4 to IFN-γ production in PMA- and ionomycin-stimulated whole blood from nonatopic asthmatic (As), atopic nonasthmatic (At), atopic asthmatic (At As), and nonatopic, nonasthmatic (control) subjects.
IL-4-producing T cells.
In unstimulated T cells, no IL-4 was detected by intracellular staining (number of positive cells, <0.05%). After stimulation the proportion of IL-4 producing T cells varied from 0.05 to 1% of T cells, (mean, 0.32 ± 0.03%). The proportion of IL-4-positive T cells did not differ significantly between samples from different patient groups. This proportion was 0.35 ± 0.06% for controls, 0.30 ± 0.07% for atopic nonasthmatics, 0.27 ± 0.08% for nonatopic asthmatics and 0.32 ± 0.04% for atopic asthmatics. CD8+ T cells did not produce IL-4 either spontaneously or after stimulation.
IFN-γ-producing T cells.
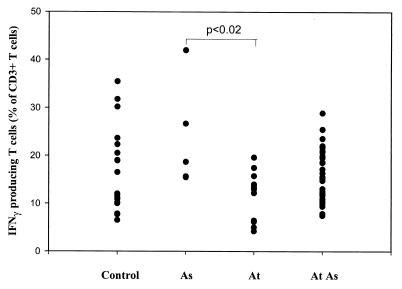
In unstimulated T cells, no IFN-γ was detected by intracellular staining. After stimulation, IFN-γ was detected in all samples. The proportion of IFN-γ-producing T cells varied from 4.2 to 41.9% of T cells (mean, 15.8 ± 7.3%).
The percentage of T cells producing IFN-γ was significantly lower in atopic nonasthmatic subjects (11.7 ± 5.1 versus 16.7 ± 8.9%; P < 0.02) than in asthmatics (Fig. 7). Among asthmatics, the proportion of IFN-γ-producing T cells tended to be higher for nonatopic asthmatics (25.7 ± 11.7%), but it was not significantly different from that for controls. The average proportion of IFN-γ-positive T cells in atopic asthma was 15.6 ± 5.4%.
FIG. 7.
Percentage of CD3+ cells producing IFN-γ in PMA- and ionomycin-stimulated whole blood from nonatopic, nonasthmatic (Control), nonatopic asthmatic (As), atopic nonasthmatic (At), and atopic asthmatic (At As) individuals.
Reassuringly, FMBA and intracellular staining gave similar IFN-γ secretion profiles for the different groups (Fig. 5 and 7).
DISCUSSION
An effective immune response requires the properly orchestrated activity of hundreds of molecules. In this complex system of immunological control, cytokines play a central role in the regulation of communication between cells. Specific tools are required to study the interplay of this multitude of regulatory species, and FMBA meets this requirement. We have shown here, for the six different cytokines assayed simultaneously, excellent correlation with results obtained in individual ELISAs.
The FMBA technology with its multianalytical capability presents several significant advantages. First, a small sample volume in a single tube is sufficient for analysis of a cytokine profile. With ELISA, the same volume would suffice only for a single assay, permitting the assay of only one cytokine at a time. This is particularly relevant to analyses of samples from infants or mice, where very small sample volumes are available. FMBA saves reagents, samples, and repetitive manipulation. Second, the FMBA is highly accurate and sensitive because each fluorescence signal represents the mean of several hundred measurements of single microspheres, and the measurement of each microsphere constitutes an assay by itself. A reduction to a mere 100 events would not affect the precision of the assay (3).
Certain limitations of flow cytometric multiplex quantitative analysis are overcome by original features of our FMBA design. First, the software responds to the quantitative requirements of multiplex immunoassays, unlike classical cytometry postanalysis software (SYSTEM-II, or EXPO; Beckman Coulter, Hialeah, Fla.) suitable for qualitative cell analysis. This postacquisition software automatically extracts and processes all data files generated by the cytometer to give the final results as concentrations (like ELISA microplate reader software). Second, the FL4 signal is normalized by the target signal provided by the calibration microspheres present in the same tube as the anticytokine-coated microspheres. This feature corrects for optical parameters such as filter efficiency, as well as slight variations in the protocol, aging of reagents, and detection by monitors. Such normalization significantly improves the precision and reproducibility of the assays. Another innovative feature concerns the calibration of the FMBA. The classical way of calibrating an assay involves the use, for every experiment, of standard solutions containing known amounts of the analyte run in parallel with the sample to be assayed so as to produce a standard curve. Here we used an internal database for FMBA calibration instead of standard solutions. The database contains the master curve parameters generated at the time of the manufacture of the reagents for each analyte using international standards and other information related to the reagents used. This initial calibration is valid for 1 month. It is adjusted by monthly assaying a two-point calibration mixture that provides respective target signals for nonspecific and specific FL4 values for each cytokine so as to extend validity. A 1-year study showed excellent reliability and accuracy and demonstrated that two-point recalibration corrects for variations that may occur as a result of the aging of the reagents. Furthermore, this system gave results comparable to those obtained with the use of standard solutions in the same run. The decrease in reagent costs combined with quality performance and increased throughput makes the internal calibration and two-point adjustment an attractive feature. The technology simplifies immunoassays, improves the practicability with stored calibration, and increases the reliability of multiplex assays by fluorescent signal correction, tube by tube.
We illustrated the usefulness of this highly informative technology by determining the cytokine profile in atopy. We demonstrated that whole-blood culture samples from atopic subjects with or without asthma showed increased IL-4 production compared with samples from nonatopic subjects who did not have asthma. Moreover, IFN-γ levels in atopic subjects were significantly lower than in nonatopic subjects. These results, obtained with stimulated whole blood, are similar to those described earlier for PBMC (25). The present study showed that the level of IFN-γ in whole cultured blood from atopic asthmatic patients is higher than that in atopic nonasthmatics. This indicates that the reduction in IFN-γ production in atopic patients may differ according to the type of asthmatic disease. IFN-γ profiles determined by the FMBA and by intracellular staining of CD3+ cells are similar for the different types of asthma (see Fig. 5 and 7).
Another aspect of interest is the balance between proinflammatory and anti-inflammatory cytokines. The proinflammatory mediators IL-6 and TNF-α occur in higher concentrations in atopic asthmatics than in normal controls. This is in accordance with the findings of previous studies performed on bronchoalveolar lavage fluids (2, 22), where secretion of proinflammatory cytokines is usually accompanied by an anti-inflammatory response. In our study, we found elevated levels of IL-10 in atopic asthmatics. Increased secretion of IL-10 by alveolar macrophages was previously reported by Magnan et al. (17). A recent study by Tillie-Leblond et al. showed an increase in anti-inflammatory mediators in bronchial lavage fluids from patients with asthma, but the net activity was proinflammatory (27).
The multiplex assay provides a unique tool for the investigation of the roles of different cytokines in various pathologic states. Refinements of the technique can also be adapted to other assays relevant to allergy or serology (10, 19, 28) or, for example, to nucleic acid-based tests (5, 20, 29).
We have described here an application to the study of asthma. The simultaneous analysis of samples for several cytokines allows the determination of cytokine profiles in many areas. This new assay technology provides high information output, suitable for the profiling of secreted compounds in pathological states, drug screening in the pharmaceutical industry, or basic research.
ACKNOWLEDGMENTS
This work was performed with the help of a grant from the Comité National de Lutte contre les Maladies Respiratoires et la Tuberculose.
We particularly thank H. Rickenberg and E. Rouvier for critical reading of the manuscript and R Hamelik for writing the data processing software.
REFERENCES
- 1.Bishop J E, Davis K A. A flow cytometric immunoassay for β2-microglobulin in whole blood. J Immunol Methods. 1997;210:79–87. doi: 10.1016/s0022-1759(97)00179-8. [DOI] [PubMed] [Google Scholar]
- 2.Broide D H, Lotz M, Cuomo A J, Coburn D A, Federman E C, Wasserman S I. Cytokines in sympomatic asthma airways. J Allergy Clin Immunol. 1992;5:958–967. doi: 10.1016/0091-6749(92)90218-q. [DOI] [PubMed] [Google Scholar]
- 3.Carson R T, Vignali D A A. Simultaneous quantitation of 15 cytokines using multiplexed flow cytometric assay. J Immunol Methods. 1999;227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 4.Collins D P, Luebering B J, Shaut D M. T-lymphocyte functionality assessed by analysis of cytokine receptor expression, intracellular cytokine expression, and femtomolar detection of cytokine secretion by quantitative flow cytometry. Cytometry. 1998;33:249–255. doi: 10.1002/(sici)1097-0320(19981001)33:2<249::aid-cyto21>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 5.Defoort J P, Martin M, Casano B, Prato S, Camilla C, Fert V. Simultaneous detection of multiplex-amplified human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA using a flow cytometer microsphere-based hybridization assay. J Clin Microbiobiol. 2000;38:1066–1077. doi: 10.1128/jcm.38.3.1066-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Groot D, Zangerle P F, Gevaert V, Fassotte M F, Beguin Y, Noizat-Pirenne F, Pirenne J, Gathy R, Lopez M, Dehart I, et al. Direct stimulation of cytokines (IL-1-β, TNF-α, IFN-γ, and GM-CSF) in whole blood in comparison with isolated PBMC stimulation. Cytokine. 1992;4:239–248. doi: 10.1016/1043-4666(92)90062-v. [DOI] [PubMed] [Google Scholar]
- 7.Ferry B, Antrobus P, Huzicka I, Fanel A, Lane A, Chapel H. Intracellular cytokine expression in whole blood preparation from normals and patients with atopic dermatitis. Clin Exp Immunol. 1997;110:410–417. doi: 10.1046/j.1365-2249.1997.4361452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulton R J, McDade R L, Smith P L, Kienker L J, Kettman J R. Advanced multiplexed analysis with the FlowMetrics system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 9.Fulwyler, M. J. July 1976. Method for detecting and separating antigen and antibodies in blood or other samples. British patent 1561042 (owned by Coulter).
- 10.Gordon R F, McDade R L. Multiplex quantitation of IgG, IgA and IgM with the Flow Metrix system. Clin Chem. 1997;43:1799–1801. [PubMed] [Google Scholar]
- 11.Gosse P, Tillie-Leblond I, Oundin S, Parmentier O, Wallaert B, Joseph M, Tonnel A B. Production of chemokines and proinflammatory and antiinflammatory cytokines by human alveolar macrophage activation by IgE receptors. J Allergy Clin Immunol. 1999;103:289–297. doi: 10.1016/s0091-6749(99)70504-x. [DOI] [PubMed] [Google Scholar]
- 12.Gosset P, Tsicopoulos A, Wallaert B, Joseph M, Capron A, Tonnel A B. Tumor necrosis factor alpha and interleukin-6 production by human mononuclear phagocytes from allergic asthmatics after IgE-dependent stimulation. Am Rev Respir Dis. 1992;146:768–774. doi: 10.1164/ajrccm/146.3.768. [DOI] [PubMed] [Google Scholar]
- 13.Hoekstra M O, Hoekstra Y, De Reus D, Rutgers B, Gerritsen J, Kauffman H F. Interleukin-4, interferon-gamma and interleukin-5 in peripheral blood of children with moderate atopic asthma. Clin Exp Allergy. 1997;27:1254–1260. [PubMed] [Google Scholar]
- 14.Kettman J R, Davies T, Chandler D, Oliver K G, Fulton R J. Classification and properties of 64 multiplexed microsphere sets. Cytometry. 1998;33:234–243. [PubMed] [Google Scholar]
- 15.Lindmo T, Bormer O, Ungelstad J, Nustad K. Immunometric assay by flow cytometry using mixtures of two particle types of different affinity. J Immunol Methods. 1990;126:183–189. doi: 10.1016/0022-1759(90)90149-p. [DOI] [PubMed] [Google Scholar]
- 16.Lisi P J, Huang C W, Hoffman R A, Teipel J W. A fluorescence immunoassay for soluble antigens employing flow cytometric detection. Clin Chem Acta. 1982;120:171–179. doi: 10.1016/0009-8981(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 17.Magnan A, Van Pee D, Bongrand P, Vervloet D. Alveolar macrophage interleukin IL-10 and IL-12 production in atopic asthma. Allergy. 1998;53:1092–1095. doi: 10.1111/j.1398-9995.1998.tb03821.x. [DOI] [PubMed] [Google Scholar]
- 18.McHugh T M, Miner R C, Logan L H, Stites D P. Simultaneous detection of antibodies to cytomegalovirus and herpes simplex virus by using flow cytometry and microsphere-based fluorescence immunoassay. J Clin Microbiol. 1988;26:1957–1961. doi: 10.1128/jcm.26.10.1957-1961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McHugh T M, Viele M K, Chase E S, Recktenwald D J. The sensitive detection and quantitation of antibody to HCV by using a microsphere-based immunoassay and flow cytometry. Cytometry. 1997;29:106–112. [PubMed] [Google Scholar]
- 20.Mehrpouyan M, Bishop J E, Osterova N, Van Cleve M, Lohman K L. A rapid and sensitive method for non-isotopic quantitation of HIV-1 RNA using thermophilic SDA and flow cytometry. Mol Cell Probes. 1997;11:337–347. doi: 10.1006/mcpr.1997.0123. [DOI] [PubMed] [Google Scholar]
- 21.Ohem J D, Hanfin J M, Nickoloff B J. Overexpression of IL-10 in atopic dermatitis. J Immunol. 1995;154:1956–1963. [PubMed] [Google Scholar]
- 22.Robinson D S, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bertley A M, Corrigan C, Durhan S, Kay A B. Predominant TH2-like bronchoalveolar T lymphocyte population in atopic asthma. N Engl J Med. 1994;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 23.Saunders G C, Jett J H, Martin J C. Amplified flow-cytometric separation-free fluorescence immunoassays. Clin Chem. 1985;31:2020–2033. [PubMed] [Google Scholar]
- 24.Stewart M W, Etches W S, Russell A S, Percy J S, Johnston C A, Chew C K. Detection of antiphospholipid antibodies by flow cytometry: rapid detection of antibody isotype and phospholipid specificity. Thromb Haemost. 1993;70:603–607. [PubMed] [Google Scholar]
- 25.Tang C, Rolland J M, Ward C, Bish R, Thien F, Walters E H. Seasonal comparison of cytokine profiles in atopic asthmatics and atopic non-asthmatics. Am J Respir Crit Care Med. 1996;154:1615–1622. doi: 10.1164/ajrccm.154.6.8970344. [DOI] [PubMed] [Google Scholar]
- 26.Tang M L, Colman J, Kemp A S. Interleukin-4 and interferon-gamma production in atopic and non-atopic children with asthma. Clin Exp Allergy. 1994;25:515–521. doi: 10.1111/j.1365-2222.1995.tb01088.x. [DOI] [PubMed] [Google Scholar]
- 27.Tillie-Leblond I, Pugin J, Marquette C H, Lamblin C, Saulnier F, Bricher A, Wallaert B, Tonnel A B, Gosset P. Balance between proinflammatory cytokines and their inhibitors in bronchial lavage from patients with status asthmaticus. Am J Respir Crit Care Med. 1999;159:487–494. doi: 10.1164/ajrccm.159.2.9805115. [DOI] [PubMed] [Google Scholar]
- 28.Utgaard J O, Frengen J, Stingbran T, Ullen A, Schmid R, Lidmo T. Analyte and label binding assay read by flow cytometry. Clin Chem. 1996;42:1702–1708. [PubMed] [Google Scholar]
- 29.Van Cleve M, Ostrerova N, Tietgen K, Cao W, Chang C, Collins M L, Kolberg J, Urdea M, Lohman K. Direct quantitation by flow cytometry using branched DNA signal amplification. Mol Cell Probes. 1998;12:243–247. doi: 10.1006/mcpr.1998.0179. [DOI] [PubMed] [Google Scholar]
- 30.Walker C, Bode E, Boer L, Hansel T T, Blaser K, Virchow J C., Jr Allergic and nonallergic asthmatics have distinct patterns of T-cell activation and cytokine production in peripheral blood and broncho-alveolar lavage. Am Rev Respir Dis. 1992;146:109–115. doi: 10.1164/ajrccm/146.1.109. [DOI] [PubMed] [Google Scholar]