Abstract
Lipid droplets (LDs) are evolutionarily conserved organelles that serve as hubs of cellular lipid and energy metabolism in virtually all organisms. Mobilization of LDs is important in light-induced stomatal opening. However, whether and how LDs are involved in stomatal development remains unknown. We show here that Arabidopsis thaliana LIPID DROPLETS AND STOMATA 1 (LDS1)/RABC1 (At1g43890) encodes a member of the Rab GTPase family that is involved in regulating LD dynamics and stomatal morphogenesis. The expression of RABC1 is coordinated with the different phases of stomatal development. RABC1 targets to the surface of LDs in response to oleic acid application in a RABC1GEF1-dependent manner. RABC1 physically interacts with SEIPIN2/3, two orthologues of mammalian seipin, which function in the formation of LDs. Disruption of RABC1, RABC1GEF1, or SEIPIN2/3 resulted in aberrantly large LDs, severe defects in guard cell vacuole morphology, and stomatal function. In conclusion, these findings reveal an aspect of LD function and uncover a role for lipid metabolism in stomatal development in plants.
Rab protein RABC1, together with its effectors, SEIPIN2 and SEIPIN3, regulate lipid droplet dynamics and stomatal development.
Introduction
Stomata are microscopic pores mainly located in the leaf epidermis of most terrestrial plants. Their role is to simultaneously regulate CO2 uptake, water loss, and transpirational cooling, thus directly influencing photosynthetic capacity and transpiration rates (Hetherington and Woodward, 2003; Duckett and Pressel, 2018). Stomatal formation is a result of a tightly regulated developmental sequence culminating in the symmetrical division of a guard mother cell (GMC) and the ensuing separation of the cell walls of the two resultant guard cells (GCs) to form a central pore (Zhao and Sack, 1999; Lee and Bergmann, 2019; Torii, 2021). Various transcriptional regulators and signaling components have been demonstrated to be involved in the control of stomatal development and function. These allow the plant to adapt its gas exchange to suit the prevailing conditions (Zoulias et al., 2018; Lawson and Matthews, 2020). Growing concern over the detrimental effects of climate change on food security has prompted great interest in understanding the control of stomatal development and function. Manipulation of stomatal traits has resulted in crop improvement. For example, a moderate reduction in stomatal density improves stress resilience and yield stability in barley (Hordeum vulgare), wheat (Triticum aestivum), and rice (Oryza sativa; Yu et al., 2008; Franks et al., 2015; Hughes et al., 2017; Caine et al., 2019; Dunn et al., 2019; Lu et al., 2019; Buckley et al., 2020).
All eukaryotic and some prokaryotic cells accumulate lipid droplets (LD) to survive periods of scarcity, protect cells against lipotoxicity and, in oil seeds, to provide a source of triacylglycerols (TAGs) that are broken down to provide energy after germination (Martin and Parton, 2006; Pyc et al., 2017; Walther et al., 2017; Huang, 2018). Cytoplasmic LDs are endoplasmic reticulum (ER)-derived multifunctional organelles consisting of a hydrophobic core of neutral lipids, mainly TAGs and sterol esters, coated with a protein-studded phospholipid monolayer (Guo et al., 2009; Wilfling et al., 2014; Rotsch et al., 2017; Zhang and Liu, 2019). Oilseeds accumulate large numbers of LDs that are mobilized during post-germinative growth, to fuel seedling growth, and intriguingly, up to 20% of LD degradation during germination takes place in the vacuoles (Poxleitner et al., 2006; Siloto et al., 2006; Graham, 2008; Chapman et al., 2012; Fan et al., 2019; Ischebeck et al., 2020). Despite the fact that LDs were observed by Hanstein in plant cells as early as 1880 (Chapman et al., 2012), most research on LDs has focused on oil seeds (Huang, 1992) and the important roles of LDs in other tissues such as pollen tubes and leaves have come to light only recently (Ischebeck et al., 2020).
In nonseed tissues, for example, in growing pollen tubes, LDs may be used as a way to transport lipids to the tip region for the synthesis of membrane lipids as well as for the provision of energy. In leaves, many genes involved in TAG metabolism are rapidly upregulated in the presence of abiotic stress and pathogen infection, suggesting they are related to cellular stress responses (Chapman et al., 2012; Ischebeck, 2016; Yang and Benning, 2018). A functional study on roots suggested that plants transfer lipids rather than sugars to sustain colonization by mutualistic mycorrhizal and parasitic fungi (Jiang et al., 2017). LDs are common in GCs and their abundance is diurnally regulated (Rutter and Willmer, 1979; Wanner et al., 1981; Sack, 1987; Gidda et al., 2016; McLachlan et al., 2016). LD mobilization followed by β-oxidation of fatty acids (FAs) is an important source of ATP during light-induced stomatal opening, a process which is evolutionarily conserved from basal plant lineages to angiosperms (McLachlan et al., 2016).
Proteomic studies of LDs in a variety of organisms have revealed a general role for LDs as transient or permanent depots for storing or sequestering proteins and identified a large number of regulatory proteins residing at the surface of LDs. These surface proteins control LD functions including TAG synthesis and breakdown, nutrient sensing, signal integration, and membrane trafficking (Cermelli et al., 2006; Yang et al., 2012; Kretzschmar et al., 2018; Zhang and Liu, 2019; Kretzschmar et al., 2020; Roberts and Olzmann, 2020; Pyc et al., 2021; Krawczyk et al., 2022). Rab proteins are small membrane-associated GTP-binding proteins involved in vesicular trafficking pathways that act as molecular switches, cycling between GDP-bound (inactive) and GTP-bound (active) states to transmit upstream signals to downstream effectors (Takai et al., 2001; Zerial and McBride, 2001). Accumulating evidence suggests that Rab proteins regulate LD formation, catabolism, and interaction with other subcellular organelles (Rasineni et al., 2014; Li and Yu, 2016; Xu et al., 2021). More than 30 Rab proteins have been suggested to be associated with LDs (Turro et al., 2006; Larsson et al., 2012; Krahmer et al., 2013; Rasineni et al., 2014; Li and Yu, 2016), however, to date the direct involvement of Rab proteins in plant LD biology, particularly with respect to stomatal development, has yet to be investigated.
During the process of stomatal development, there is considerable cell expansion and this is associated with an increased requirement for the new membrane to increase surface area (Zhao and Sack, 1999; Kaiser and Scheuring, 2020). Given LDs are hubs of cellular lipid metabolism (Martin and Parton, 2006; Walther and Farese, 2012), we reasoned that plants mobilize LDs and liberate lipids for membrane synthesis during stomatal formation. Here we report that LD abundance is highly dynamic during stomatal development. In addition, an LD genetic screen in Arabidopsis thaliana resulted in the isolation of the lds1 (lipid droplets and stomata 1) mutant, also known as Rab18 and RABC1 (Vernoud et al., 2003), that breaks the programmed LD dynamics in stomatal lineage cells (SLCs). LDS1/RABC1/Rab18, which we will henceforth refer to as RABC1, is distinctly expressed in SLCs and its expression level is correlated with the different phases of stomatal development. RABC1 deficiency resulted in compromised LD size, stomatal morphology, and functionality. We conclude that the RABC1 protein, together with its partner, a Guanine Exchange Factor (GEF), controls the dynamics and size of LDs in young stomata. We show that RABC1 does this by regulating its effector factors SEIPIN2 and SEIPIN3.
Results
The lds1 mutant displays aberrant LD size and LD dynamics during stomatal development
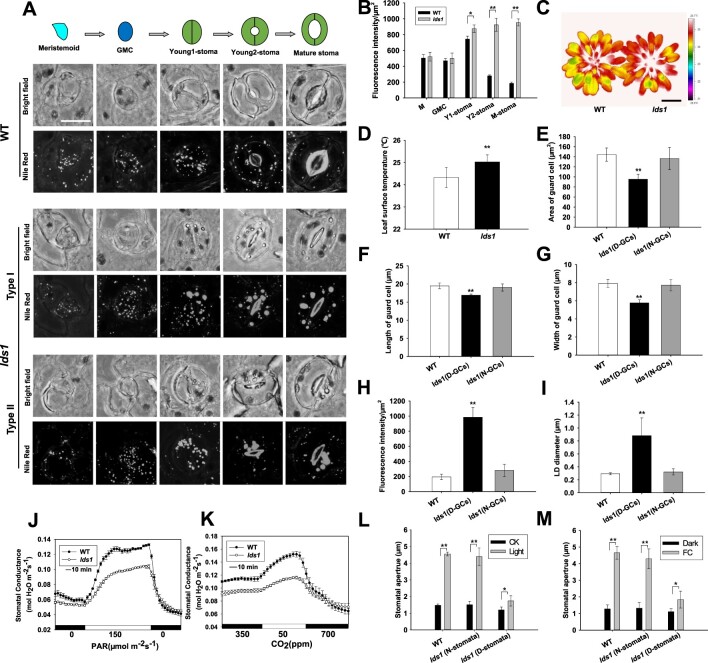
In Arabidopsis, stomata are produced by a tightly coordinated series of cell divisions that give rise to specialized SLCs, namely, M (meristemoid), GMC, and Young1-stoma (at this stage, symmetric division of GMC has been completed, stomatal length approximately equals stomatal width and the pore of the stoma does not appear or just starts to appear), Young2-stoma (at this stage, the stomatal complex has yet to achieve its full size but the stomatal pore is clearly visible), and M-stoma (Mature-stoma) according to the nomenclature used by Carter et al. (2017) and Torri (2021; Figure 1A). To test whether LDs are involved in stomatal development, we first used the neutral lipid stain, Nile red (NR) to monitor the dynamics of LDs in the SLCs (Fowler and Greenspan, 1985). As shown in Figure 1A and Supplemental Figure S1A, conspicuous staining of LDs was detected in the small triangular cell meristemoid (M), and the fluorescence intensity of the dye (as a proxy of the neutral lipid content) progressively increases during differentiation, reaching a peak in stage Young1 stomata, before decreasing rapidly at the Young2 stage. In mature stomata, the signal becomes much weaker (Figure 1, A and B). When the cells were stained with Bodipy 493/503 (Thumser and Storch, 2007), we observed similar LD dynamics in SLCs (Supplemental Figure S1B). A very recent finding by Liwen Jiang’s group lends strong support to our observation that LD abundance is higher in the young stomatal cells compared with other SLCs (Cao et al., 2022).
Figure 1.
The lds1 mutant displays aberrant LD size and LDs dynamics during stomatal development. A, LD dynamics in SLCs at different stages of stomatal development in WT and lds1. The representative stomatal images were Z-projections (maximal intensity) of confocal stacks from young leaves of 4-week-old plants. GMC, Scale bar, 10 μm (applies to all images in Figure 1A). B, Quantification of LD abundance at different stages of SLCs, including M (meristemoid), GMC, Y1 (young1)-stoma, Y2 (young2)-stoma, and M (mature) -stoma in lds1 and WT. Data are shown as means ± SD, n = 50. Asterisks represent Student’s t test significance (*P < 0.05; **P < 0.01). C and D, The leaf surface temperature of 5-week-old lds1 and WT under normal growth conditions in greenhouse. Scale bar, 2 cm. Data are shown as means ± SD, n = 40. Asterisks represent Student’s t test significance (**P < 0.01). E–G, Measurements of GCs area (E), length (F), and width (G) of normal shaped GCs (N-GCs) and deformed GCs (D-GCs) in lds1. Data are shown as means ± SD, n = 300. Asterisks represent Student’s t test significance (**P < 0.01). H and I, Quantifications of LDs abundance (H) and LDs diameter (I) in the normal shaped GCs (N-GCs) and deformed GCs (D-GCs) in lds1. Data are shown as means ± SD, n = 40. Asterisks represent Student’s t test significance (**P < 0.01). J and K, Light (J) and low CO2 (K)-induced increases in stomatal conductance were examined in 5-week-old lds1 and WT plants. Data are shown as means ± SD, n = 3. L and M, Stomatal opening responses induced by light (L) and fusicoccin (M) were investigated using leaf epidermal strips of 4-week-old lds1 and WT plants. N-stomata, normal-shaped stomata. D-stomata, deformed stomata. FC, fusicoccin. Data are shown as means ± SD, n≥60. Asterisks represent Student’s t test significance (*P < 0.05; **P < 0.01).
To identify the molecular regulator(s) involved in controlling the dynamics of LDs during stomatal development we performed a two-step, forward genetic screen, using infrared thermography and NR staining. Infrared thermography has been extensively used to noninvasively isolate mutant plants that are defective in stomatal function (Merlot et al., 2002; Wang et al., 2004; Xie et al., 2006; Hashimoto et al., 2006; He et al., 2018; Sun et al., 2022). We first screened 2,700 individual plants and isolated 32 candidates that displayed altered leaf surface temperature. Then we subjected these candidates to a second round of screening using NR to stain LDs in detached epidermis. This resulted in the isolation of one mutant plant designated lds1 (lipid droplets and stomata 1), which exhibited much enlarged GC LDs and increased leaf surface temperature compared with WT (Figure 1, A–D). Morphologically, more than half of the stomata examined (cotyledon 51.2 ± 5.5%, n = 500; rosette leaf 53.9 ± 3.5%, n = 500) were of irregular shape in lds1, with one or two deformed GCs (Figure 1A; Supplemental Figure S1C).
Since cotyledons and rosette leaves display a comparable phenotype, we focused on the stomata of rosette leaves. We divided the abnormal stomatal complexes into two categories based on the degree of deformity. When both GCs were deformed we classified these as “Type I” stomata (78.2% ± 5.2%, n = 500). The second category, “Type II” stomata (22.8% ± 3.4%, n = 500) consists of one seemingly normal and one impaired GC (Figure 1A). On closer inspection, in “Type II” stomata, we found that the irregular GC is smaller than the normal-shaped GC, and the symmetric layout of the two GCs was compromised (Figure 1, E–G; Supplemental Figure S1C). Type I stomatal complexes were characterized by a “thin shape” in contrast to the roundish shape of the normal stomata both in lds1 and in WT (Figure 1A). Importantly, only deformed GCs had enlarged LDs (Figure 1, A, H, and I). Not unexpectedly, the pattern of LD dynamics, during stomatal differentiation was also affected by the lds1 mutation. At M and GMC stages there was no detectable difference in either LD abundance or LD size between lds1 and WT (Figure 1A). However, on the basis of NR staining, at the Young1-stomata stage, we found that 27 of 50 stomata accumulated much more lipid and larger LDs in lds1 compared with wild type (Figure 1, A and B). In contrast to the progressive decrease of NR fluorescence in WT, the increase in NR fluorescence remained constant when the lds1 Young1-stomata differentiate into the Young2-stomata and subsequently into the M-stomata (Figure 1A). Consistently, we saw significant increases in the levels of 16:0, 18:0, 18:1, 18.2, and 18.3 FA molecular species in the lds1 mutant when compared with WT (Supplemental Figure S1D). These data indicate that LD defects and deformed GC morphology are two linked phenotypes in lds1. FDA staining (Krasnow et al., 2008) demonstrated that all deformed GCs were living, suggesting that the lds1 mutation does not result in significant damage to GC membrane integrity (Supplemental Figure S1E).
Given that SCAP1 is expressed from young stomata to mature stomata, and disruption of SCAP1 resulted in defective stomata, which are seemingly comparable to those in lds1 mutant (Negi et al., 2013), we therefore next investigated the relationship between the aberrant stomatal development and LD defects in the scap1 mutant. As shown in Supplemental Figure S1, O and P, there were no detectable LD defects in GCs of scap1 in contrast to lds1. Consistently, when we sequenced the SCAP1 gene in the lds1 mutant, we detected no mutation in SCAP1 gene. These results indicate that SCAP1 is not the causal gene for lds1 phenotypes, and by extension that stomatal deformations are not necessarily associated with LD defects.
We next investigated stomatal function and found that light and low [CO2]-induced changes in stomatal conductance were reduced substantially in the lds1 mutant compared with WT (Dark to light: 321.8% ± 41.2 in WT vs 231.3 ± 53.5% in lds1; high to low [CO2]: 236.4 ± 30.4% in WT vs. 163.6 ± 44.1% in lds1; Figure 1, J and K). The inhibitive effect of the lds1 mutation on stomatal opening was confirmed for both light and fusicoccin (Turner and Graniti, 1969; Yamauchi et al., 2016; Figure 1, L and M). In wild type, there is a rapid decrease in LD abundance during light-induced opening, this was not observed in the deformed GCs of lds1 (Supplemental Figure S1F; McLachlan et al., 2016). As we observed no appreciable difference in terms of stomatal density and stomatal index between lds1 and WT plants, we conclude that LDS1 is not involved in cell fate determination (Supplemental Figure S1, G and H). We also noted that biomass accumulation and stature of lds1 are reduced compared with WT (Supplemental Figure S1, I–K), but seed size and seed germination are not affected in the lds1 mutant (Supplemental Figure S1, L–N).
LDS1/RABC1 is highly expressed in young stomata and RABC1 targets to the surface of LDs
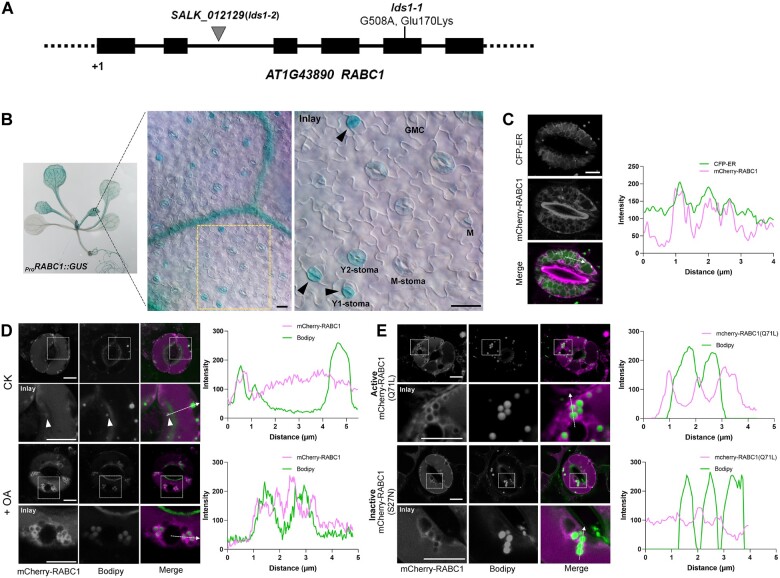
To identify the causal mutation of lds1, we undertook map-based gene cloning and next-generation DNA sequencing and identified a single point mutation (a G to A substitution) at position +508 in the At1g43890 (RABC1) open reading frame. This causes a substitution from a conserved acidic amino acid to a basic amino acid (E170K; Figure 2A; Supplemental Figure S2A). Next, the wild-type RABC1 cDNA was introduced into lds1 driven by RABC1 promoter and the transgene fully restored the phenotypic defects in LD size and stomatal morphology (Supplemental Figure S2, B–E). Furthermore, a T-DNA insertion mutant SALK_012129, which had no detectable RABC1 mRNA, exhibited similar defects to lds1, indicating that lds1 and SALK_012129 are allelic to each other (Supplemental Figure S2, C–E). These data demonstrate that a mutation in LDS1/RABC1 is responsible for the phenotypes described in this paper. For brevity, LDS1/RABC1 is hereafter referred to as RABC1. We renamed lds1 and SALK_012129 as lds1-1 and lds1-2, respectively, and subjected them to further investigation.
Figure 2.
LDS1/RABC1 is highly expressed in young stomata and RABC1 targets to the surface of LDs. A, Schematic view of mutation site determination of two independent lines of lds1 mutants. B, GUS staining of a 2-week-old seedling of ProRABC1:GUS transformant. RABC1 is highly expressed in leaf veins and SLCs. The inset image shows that RABC1 is highly expressed in young GCs. Arrowheads indicate Y1-stomata. M (meristemoid), GMC, Y1 (young1), Y2 (young2), M (mature) -stoma. Scale bars, 20 μm. C, mCherry-RABC1 transformants were used to determine whether RABC1 localized to the ER. mCherry-RABC1 co-localized with CFP-ER. The fluorescence intensity profile plot of mCherry-RABC1 and CFP-ER is quantified along the dotted arrow. Scale bar, 5 μm. D, mCherry-RABC1 transformants were used to determine the relationship between RABC1 localization and LDs. Oleic acid (OA) application induces LD production. Arrowheads indicate ring-like signals surrounding LDs. The fluorescence intensity profile plots of mCherry-RABC1 and Bodipy were quantified along the dotted arrows, respectively. Scale bar, 5 μm. E, Activity status of RABC1 dictates the subcellular localization of RABC1. The fluorescence intensity profile plots of mCherry-RABC1 and Bodipy were quantified along the dotted arrows, respectively. Scale bar, 5 μm.
Arabidopsis RABC1 has been suggested to be the counterpart of mammalian Rab18 (Vernoud et al., 2003). As shown in Supplemental Figure S2A, the putative amino acid sequence of RABC1 exhibited 63% identity and 61% identity to mouse MmRab18 and human HsRab18, respectively. Mutations in Rab18 impair lipid metabolism and result in enlarged LDs in several cell types, including HEK293T cells, human fibroblasts, and 3T3-L1 adipocytes. However, the causal basis for this is not clear (Li et al., 2017; Xu et al., 2018; Dejgaard and Presley, 2019). To investigate whether RABC1 and mammalian Rab18 have similar functions in vivo, we performed cross-species complementation tests. Ectopic expression of HsRab18 in Arabidopsis fully rescued the defects caused by lds1 mutation (Supplemental Figure S2, F–H). The Arabidopsis genome contains two homologs of RABC1. These are RABC2a (At5g03530) and RABC2b (At3g09910) which share 73% and 67% identity with RABC1, respectively (Supplemental Figure S2A). Neither single gene mutant caused defects in LD size or stomatal morphogenesis (Supplemental Figure S2, I–K). However, when the expression of either gene was driven by the RABC1 promotor in lds1, both LD size and stomatal morphology could essentially be restored to WT levels (Supplemental Figure S2, I–K). This result suggests that similarities in protein structure likely account for the interchangeable functions of RABC2a, RABC2b, and RABC1. Their different roles may be related to their tissue expression and, or abundance. Resolving whether this is the case must await further investigation.
Next, a transcriptional reporter, ProRABC1:GUS was constructed to evaluate RABC1 expression patterns. As shown in Figure 2B, RABC1 is widely expressed. In leaves, GUS production was most abundant in the veins and SLCs, with the highest signal intensity detected in Young1-stomata (Figure 2B), reminiscent of the dynamic changes of LDs in these cells (Figure 1A). RABC1 subcellular localization was evaluated using the ProRABC1:mCherry-RABC1 construct, which complemented the lds1 mutation (Supplemental Figure S2, B, D, and E). As shown in Figure 2C, mCherry-RABC1 localized to the ER. Next, we sought to establish whether RABC1 could also target to LDs. Under normal circumstances, only a small subset of LDs displayed mCherry fluorescence (Figure 2D). However, in response to treatment with oleic acid (18:1, unsaturated FA, OA), which is known to stimulate the formation of LDs (Xu et al., 2018), an increase in mCherry-RABC1 fluorescence was observed around most LDs (Figure 2D). We found that OA incubation not only significantly increased LD abundance, but also increased LD size in WT GCs. In the GCs of lds1, although the abundance and size of LDs also increased when incubated with OA, the increase was much less than seen in WT (Supplemental Figure S2, L and M).
Next, in order to study the effects of the state of the RABC1 protein on its intracellular localization, we used RABC1 (S27N; a dominant-negative form) and RABC1 (Q71L; a constitutively active form), two mutant forms of RABC1 that were generated based on the highly conserved molecular switch motif among Rab proteins (Ozeki et al., 2005; Lee et al., 2009). As shown in Figure 2, D and E, there was almost no OA-induced fluorescence of mCherry-RABC1 (S27N) surrounding LDs. This was in marked contrast to the obvious fluorescence in the constitutively active RABC1 (Q71L) or in OA-induced RABC1. This indicates that RABC1 trafficking to LDs (in response to the lipogenic stimulus, OA) depends on its activity state. This is consistent with the view that the activation status of Rab proteins impacts their subcellular localization (Bhuin and Roy, 2014; Cui et al., 2014).
RABC1GEF1 is a GEF for RABC1
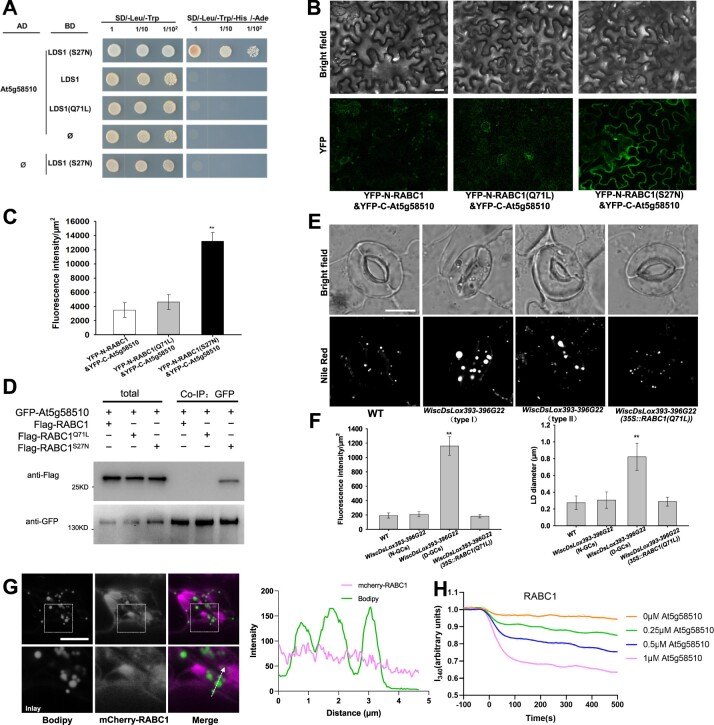
The conversion of Rab proteins from an inactive GDP-bound form to an active GTP-bound form relies on guanine nucleotide exchange factor proteins (GEFs; Zerial and McBride, 2001). To identify RABC1 protein regulators that interact with RABC1, we screened an Arabidopsis cDNA library in yeast using the dominant-negative form of RABC1 (S27N) as bait. Among the 30 candidate proteins that interact with the inactive RABC1, At5g58510 was repeatedly isolated, and a direct yeast two-hybrid (Y2H) assay confirmed that the protein encoded by At5g58510 could directly interact with RABC1 (S27N; Supplemental Data Set S1; Figure 3A; Supplemental Figure S3A). Based on its high sequence similarity to human Rab3GAP1 that works as a GEF for HsRab18 (Supplemental Figure S3B; Xu et al., 2018), it is likely that this protein has GEF activity for RABC1. Based on our characterization described below, we named this protein RABC1GEF1. The subsequent BiFC and Co-IP tests demonstrated that RABC1GEF1 and RABC1 interact in vivo in a RABC1 activity-dependent manner (Figure 3, A–D).
Figure 3.
At5g58510 is a GEF for RABC1. A, The interaction between At5g58510 and RABC1 was dependent on the activity status of RABC1 in the Y2H system. B, BiFC was used to verify the interactions between At5g58510 and RABC1. Scale bar, 30 μm. C, Fluorescence intensity of YFP in (B) was quantified. Data are shown as means ± SD, n = 3. Asterisks represent Student’s t test significance (**P < 0.01). D, Co-IP was used to verify the interaction between At5g58510 and RABC1. E and F, Both allelic mutants of At5g58510 showed defective LDs dynamics, LDs size, and stomatal morphology. Introduction of a dominant-active form of RABC1 (Q71L) into the mutants of At5g58510 rescued the defects. The representative stomatal images were single confocal planes from young leaves of 4-week-old plants (E). LD abundance and size of GCs were quantified (F). Data are shown as means ± SD, n = 20. Asterisks represent Student’s t test significance (**P < 0.01). Scale bar, 10 μm. G, Localization of RABC1 to LDs is blocked by RABC1GEF1 deficiency. The fluorescence intensity profile plots of mCherry-RABC1 and Bodipy are quantified along the dotted arrow. Scale bar, 10 μm. H, In vitro GEF assays of At5g58510. Nucleotide exchange of RABC1 was tested by monitoring tryptophan autofluorescence in the absence or presence of 0.25 mM, 0.5 mM, or 1 mM GST-At5g58510. The raw data are the jagged lines and the bold lines represent the trend lines.
Furthermore, two T-DNA insertion mutant lines (WiscDsLox393-396G22 and SALK_116404C) were identified that harbor no detectable mRNA of At5g58510 (Supplemental Figure S3C). WiscDsLox393-396G22 shows similar defects in LD size and stomatal morphology to lds1 (Figure 3, E and F). The ratio (55.6 ± 3.2%, n = 500) of deformed stomata caused by mutation in RABC1GEF1 closely resembles the ratio (53.9 ± 3.5%, n = 500) seen in lds1. RABC1GEF1 deficiency in SALK_116404C also causes similar defects to lds1 (Supplemental Figure S3, D and E).
In addition, the targeting of RABC1 to the LD surface is inhibited by mutation of RABC1GEF1 (Figure 3G). When a dominant-active form of RABC1 (Q71L) was introduced into either WiscDsLox393-396G22 or SALK_116404C, the defective LD size and stomatal morphology could be fully restored (Figure 3, E and F). Of note, although the protein encoded by At5g55060 in Arabidopsis resembles Rab3GAP1 (Supplemental Figure S3B), our data suggest that this protein exerts no GEF function for RABC1, as the corresponding loss-of-function mutant (SALK_058192C) displays no similar defects to those of lds1-1, lds1-2, WiscDsLox393-396G22 or SALK_116404C (Supplemental Figure S3, C–E).
Next, we measured the in vitro GEF activity of RABC1GEF1 towards RABC1 by monitoring the change in intrinsic tryptophan fluorescence upon nucleotide exchange as described (Pan et al., 1995; Antonny et al., 2001; Goh et al., 2007; Cui et al., 2014). RABC1GEF1 stimulates the guanine nucleotide exchange of RABC1 in a dose-dependent manner (Figure 3H), but shows almost no guanine nucleotide exchange activity for RHA1, a Rab GTPase used as the experimental control (Supplemental Figure S3F, Cui et al., 2014). Unlike RABC1GEF1, the protein encoded by At5g55060 shows no detectable GEF activity for RABC1 (Supplemental Figure S3F). These data demonstrate that RABC1GEF1 works as a GEF for RABC1.
SEINPIN2 and SEIPIN3 are two effectors for RABC1
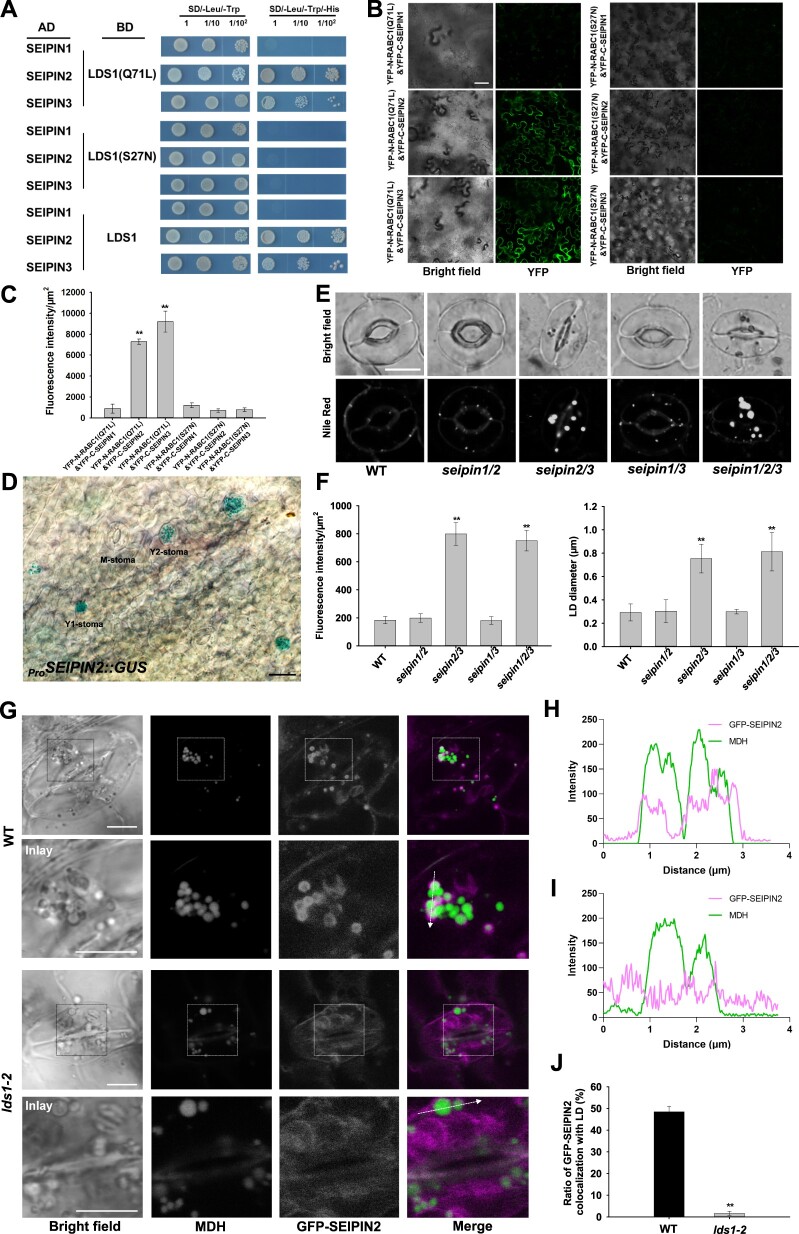
Rab GTPases generally work through their downstream effectors (Gillingham et al., 2014). To identify the effector(s) of RABC1, we conducted a second Y2H screen using a dominant-active RABC1 (Q71L) as bait and obtained SEIPIN2 as a possible effector for RABC1 among other 45 candidates (Figure 4A; Supplemental Data Set S2). SEIPIN2 is an orthologue of mammalian seipin (sei1 in yeast), which was originally identified as being deficient in a form of congenital lipodystrophy and has since been suggested to function in regulating the formation and size of LDs (Magre et al., 2001; Szymanski et al., 2007; Fei et al., 2008; Wang et al., 2016). Unlike yeast or mammals, Arabidopsis has three SEIPIN homologs, and their deficiency alters lipid contents and LD size, whereas overexpression of each homolog in either yeast cells or plant leaves influenced the number and size of LDs differently (Cai et al., 2015; Taurino et al., 2018; Huang, 2018; Greer et al., 2020; Pyc et al., 2021). As shown in Figure 4, A–C, SEIPIN2 physically interacts with RABC1 and the interaction depends on the RABC1 activity state.
Figure 4.
SEINPIN2 and SEIPIN3 are two effectors for RABC1. A, Interaction tests between SEIPINs and different status RABC1 in Y2H system. B, Interaction tests between SEIPINs and different status RABC1 in BiFC tests. Scale bar, 50 μm. C, Fluorescence intensity of YFP in (B) was quantified. Data are shown as means ± SD, n = 3. Asterisks represent Student’s t test significance (**P < 0.01). D, GUS staining of leaves from 2-week-old seedling of ProSEIPIN2:GUS transformant. SEIPIN2 is highly expressed in stomatal cells. Scale bar, 20 μm. E and F, Simultaneous disruption of SEIPIN2 and SEIPIN3 resulted in defective LD dynamics, aberrant LD size, and stomatal morphology. The representative stomatal images were single confocal planes from young leaves of 4-week-old plants (E). LD abundance and size of GCs were quantified (F). Data are shown as means ± SD, n = 20. Asterisks represent Student’s t test significance (**P < 0.01). Scale bar, 10 μm. G, Localization of GFP-SEIPIN2 to LDs is blocked by RABC1 deficiency. The LDs were stained with Monodansylpentane (MDH). Scale bar, 5 μm. H and I, Fluorescence intensity profile plots of GFP-SEIPIN2 and LDs in WT (H) and lds1-2 (I) are quantified along the dotted arrow in (G). J, Quantification of the proportion of GFP-SEIPIN2 localized on LDs in WT and lds1-2. Data are shown as means ± SD, n = 120. Asterisks represent Student’s t test significance (**P < 0.01).
Through constructing transgenic Arabidopsis that expresses a SEIPIN2 promoter-driven GUS reporter (ProSEIPIN2:GUS), we found that unlike SEIPIN1 and SEIPIN3, SEIPIN2 is preferentially expressed in GCs (Figure 4D; Supplemental Figure S4, A and B), consistent with a previous observation (Greer et al., 2020). Importantly, the highest expression of SEIPIN2 is in Young1 stomata resembling that of RABC1 (Figure 4D). RABC1 also interacts with SEIPIN3 but not SEIPIN1 (Figure 4, A–C). No LD defects were detected in seipin1, seipin2, or seipin3 individual gene mutant (Supplemental Figure S4, C–E). However, a comparable number of stomata in seipin2 seipin3 double mutant (13.37 ± 4.2%, n = 500) and seipin1 seipin2 seipin3 triple mutant (11.29 ± 2.5%, n = 500) displayed enlarged LDs and deformed GC morphology, whereas no such defects were observed in either seipin1 seipin2 or seipin1 seipin3 (Figure 4, E and F). This suggests that functional divergence has occurred in these paralogous SEIPIN proteins. It seems that SEIPIN2 acts as an important effector for RABC1 in GCs and SEIPIN3 can contribute to stomatal biology, whereas SEIPIN1 has distinct functions.
Considering that GTPases regulate the function of their effectors by affecting their localization (Pylypenko et al., 2018), we next examined whether mutations in RABC1 lead to abnormal localization of SEIPIN2/3. Consistent with the observation made in tobacco (Cai et al., 2015; Taurino et al., 2018; Greer et al., 2020; Pyc et al., 2021), we found that GFP-SEIPIN2/3 localizes to ER-like structures but hardly co-localize with LDs in the GCs of both WT and lds1 (Supplemental Figure S4G). When we used OA to promote LD production, we found that a large proportion of LDs had strong GFP-SEIPIN2/3 signals in WT, but the GFP signal was weak around the LDs in lds1 mutants (Figure 4, G–J; Supplemental Figure S4, H–K). This result suggests that the localization of SEIPIN2/3 to the LD surface is regulated by RABC1 activity. Since SEIPIN proteins are thought to be mainly responsible for the formation of LDs at the surface of ER, the regulation of their localization on LDs by RABC1 is likely to be related to the size of LDs, but the specific mechanism still needs further study. In addition, when overexpressing SEIPIN2 or SEIPIN3 in lds1, we found that defective LDs and stomatal morphology could not be restored in lds1 (Supplemental Figure S4, L and M). A possible explanation for this result is that RABC1 affects the function of SEIPIN2/3 by regulating their localization which might not be related to their expression levels.
DMP treatment mimics the phenotype of lds1 and overexpression of SDP1 partly restores the aberrant stomatal morphology of lds1
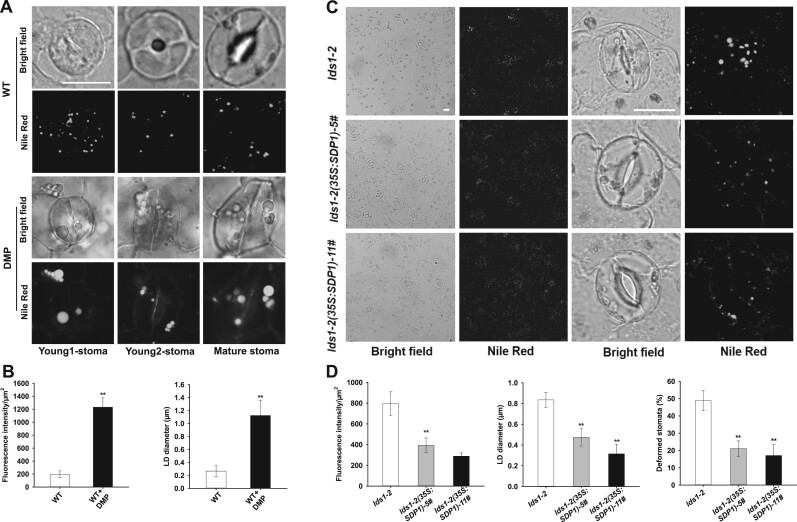
Diphenyl methylphosphonate (DMP) is a potent inhibitor of LD mobilization and in seedlings results in LD retention and the production of large LDs (Brown et al., 2013; Yu et al., 2018). In addition, DMP-treated isolated leaf epidermis exhibited delayed light-induced stomatal opening (McLachlan et al., 2016). When we grew Arabidopsis seedlings on half-strength Murashige and Skoog (1/2 MS) agar plates with 25 µM DMP, we found that many GCs contained aberrantly large LDs, and the corresponding stomata were irregular in shape (Figure 5, A and B). Similar to lds1, DMP treatment resulted in enlarged LDs in young GCs (Figure 5A), suggesting that blocking LD turnover during stomatal development causes defective stomatal morphology.
Figure 5.
DMP treatment mimics the phenotype of lds1 and overexpression of SDP1 partly restores the aberrant stomatal morphology of lds1. A and B, DMP treatment resulted in defective LD dynamics and stomatal morphology in WT. The representative stomatal images were single confocal planes from young leaves of 2-week-old plants (A). LD abundance and size of GCs were quantified (B). Data are shown as means ± SD, n = 30. Asterisks represent Student’s t test significance (**P < 0.01). Scale bar, 10 μm. C and D, Overexpression of SDP1 restored defects in LDs size and stomatal morphology in lds1-2. The representative stomatal images were single confocal planes from young leaves of 4-week-old plants (C). LD abundance and size (n = 30) of GCs, deformed stomata ratio (n = 400), were quantified (D). Data are shown as means ± SD. Asterisks represent Student’s t test significance (**P < 0.01). Scale bar, 20 μm.
To examine whether the enlarged LDs in lds1 could impair lipolysis, we developed transgenic plants that overexpress SUGAR DEPENDENT1 (SDP1), a major lipase for TAG degradation (Eastmond, 2006; Kelly et al., 2013). As shown in Figure 5, C and D and Supplemental Figure S5A, SDP1 overexpression largely restored LD size and stomatal morphology in lds1, suggesting that aberrant LDs caused by RABC1 deficiency likely slowed the access of lipases and, or other degradative enzymes to TAG substrates, reducing the production of free FAs with likely negative effects on ATP production.
Disruption of RABC1 function affects vacuole occupancy and OCL formation
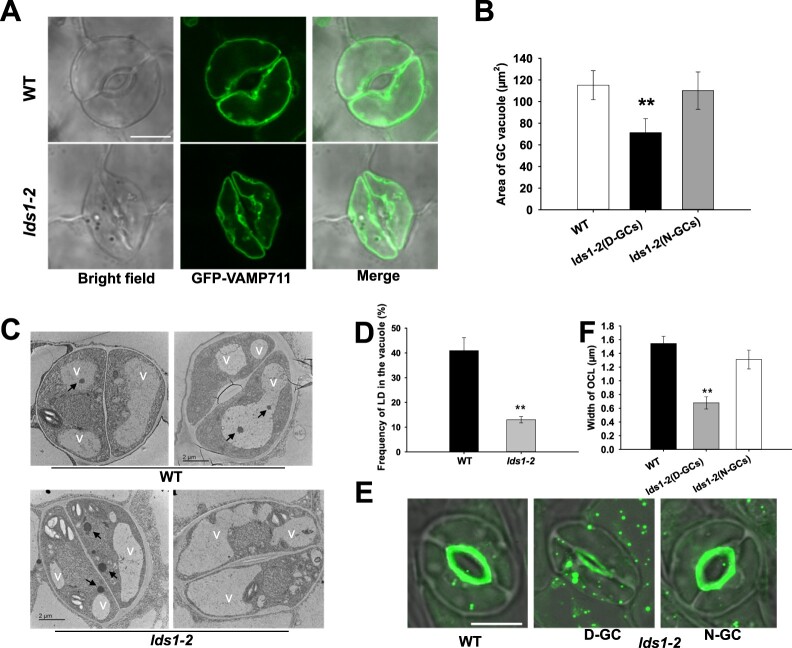
By taking advantage of a tonoplast marker, GFP-VAMP711 (Cui et al., 2014), we found that the area of vacuoles in deformed GCs is much smaller in lds1 than WT (Figure 6, A and B), suggesting an essential role of RABC1 in the intracellular expansion of the vacuoles. This observation was further substantiated using transmission electron microscopy (TEM) and FM4-64 staining (Supplemental Figure S5, B–E). Vacuole size defects were also clearly observed in the deformed GCs of lds1gef1 and seipin2 seipin3 (Supplemental Figure S5, D–G). Intriguingly, the frequency of LDs appearing in the vacuolar lumen of lds1 mutants was greatly reduced compared with WT (Figure 6, C and D), suggesting that RABC1 malfunction inhibits LD-vacuole interactions. Finally, the outer cuticular ledge (OCL), an extension of stomatal cell walls that forms an antechamber just above the stomatal pore and is involved in stomatal regulation (Merced and Renzaglia, 2014; Hunt et al., 2017; Hunt and Gray, 2020; Tang et al., 2020) was stained by NR (Figure 6, E and F). Whereas the OCL of deformed GCs in lds1 is much thinner, indicating that lds1 mutation impairs OCL formation. This observation reveals a previously unrecognized role of LDs related to OCL formation. In addition, when we examined the vacuole morphology and OCL structure in the GCs of scap1, we detected no significant difference between scap1 and WT (Supplemental Figure S5, D, E, H and I). This further supported our view that stomatal deformations are not necessarily associated with LD defects.
Figure 6.
Disruption of RABC1 function affects vacuole occupancy and OCL formation. A, Representative images of vacuoles in GCs of WT and lds1 plants. The representative stomatal images were single confocal planes from young leaves of 4-week-old plants. Scale bar, 10 μm. B, Quantification of the area of vacuoles in GCs of WT and lds1 plants. Data are shown as means ± SD, n = 50. Asterisks represent Student’s t test significance (**P < 0.01). C and D, The number of LDs in the vacuolar lumen of lds1 was reduced compared with WT. Representative TEM images of LDs in the vacuolar lumen (C), quantification of LDs in the vacuolar lumen (D). Arrows indicate LDs. V, vacuole. Data are shown as means ± SD, n = 30. Asterisks represent Student’s t test significance (**P < 0.01). E and F, The OCL of deformed GCs in lds1-2 is much thinner and narrower. The representative stomatal images were single confocal planes from young leaves of 4-week-old plants. Data are shown as means ± SD, n = 40. Asterisks represent Student’s t test significance (**P < 0.01).
Discussion
The acquisition of stomata was a critical evolutionary innovation that maximized plant survival and fitness through controlling the fundamental tradeoff between carbon dioxide uptake and water loss (Clark et al., 2022). To exploit stomata for crop improvement in the face of global climate change necessitates investigations that reveal essential regulators of stomatal development and function. In this study, we describe the cellular dynamics of LDs during stomatal development and identify RABC1 as a key regulator required for modulating LD dynamics and lipid metabolism during the establishment of functional stomata. Our data suggest a model in which RABC1 deficiency results in compromised LD size, deformed stomata, smaller GC vacuoles, and defective cuticle. We have also shown that RABC1GEF1 and SEIPIN2/3 exert GEF and effector function, respectively, for RABC1. RABC1GEF1 is required for the targeting of LDs by RABC1. Our findings expand the roles of LDs by revealing that LDs are likely required for GC morphogenesis, vacuole growth, and OCL establishment during stomatal development in plants (Figure 7).
Figure 7.
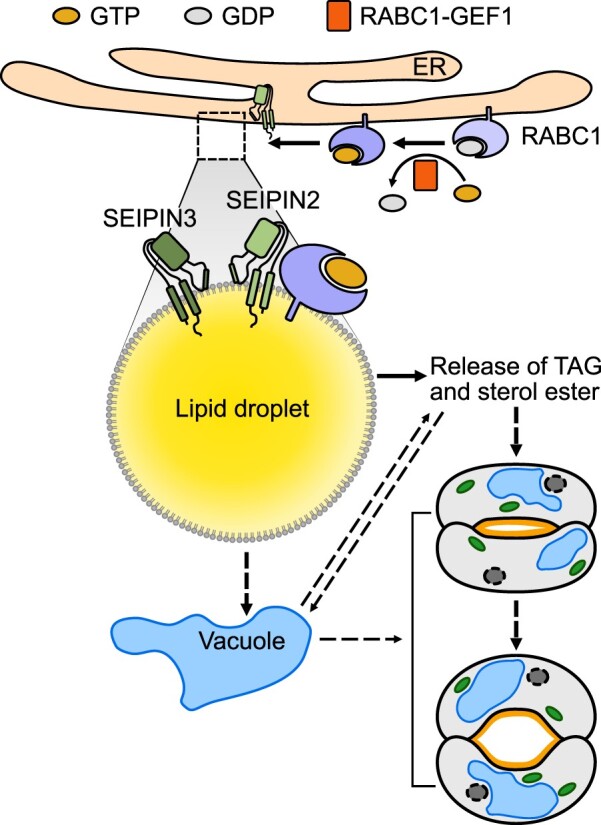
Proposed working model of RABC1 regulating LD dynamics and stomatal development. RABC1, a Rab protein that is preferentially expressed in young stomatal GCs, is activated by the binding of RABC1GEF1 (a specific guanine nucleotide exchange factor of RABC1) in response to a yet unknown developmental cue, and then targeted to the surface of LDs, where it interacts with SEIPIN2 and SEIPIN3, two ER-localized proteins that serve as the downstream effectors of RABC1 to regulate LD mobilization and lipid availability for the establishment of functional stomata. The dashed arrows indicate the possible involvement of the vacuole during RABC1-controlled LD homeostasis. ER, endoplasmic reticulum. For more details, please refer to the “Discussion” section.
Stomata are the end products of a specialized epidermal lineage, and each stoma possesses two seemingly identical GCs. The symmetric layout of the two GCs was disrupted in “Type II” stomata in lds1 (Figure 1A). Given that the ends of the two GCs of a stoma are connected, it is likely that this asymmetric layout results from the distortion imposed by the larger GC over its smaller neighbor. The distinct morphogenesis of the paired GCs demonstrated in “Type II” stomata confirms and extends previous evidence that while paired stomatal GCs are of same origin they behave autonomously (McAinsh et al., 1990; Pillitteri and Torii, 2012).
Our observation that the abundance of cytoplasmic LDs changes dramatically during stomatal development (Figure 1, A and B), and perturbing LD dynamics either by pharmacological or genetic means, compromises the formation of fully functional stomata (Figures 1A and 5A), suggests that maintaining LD homeostasis is indispensable for stomatal development. The lds1 mutant displayed enlarged LDs in the deformed GCs (Figure 1, A and I). This suggests that RABC1 is a negative regulator of LD size. This is likely to be of importance because LD size needs to be strictly controlled, as it is known that small LDs are suitable for fast lipid mobilization, whereas larger LDs are more suitable for long-term lipid storage (Suzuki et al., 2011). The size of LDs is the result of the combined action of LD growth and mobilization. Overexpression of SDP1 restored LD size and stomatal morphology (Figure 5, C and D), suggesting that the enlarged LDs, caused by the malfunctioning of RABC1, impact the access of the SDP1 lipase and the mobilization of LDs. It is likely that RABC1 also restricts LD growth as TAG content in the GC-enriched epidermis is significantly higher in lds1 than in WT (Supplemental Figure S1D). However, whether RABC1 functions in preventing LD coalescence or inhibiting localized synthesis of lipid (or in both processes) remains to be clarified. In the Rab18 knockout 3T3-L1 adipocytes of mammals, although the exact mechanism of LD enlargement remains unclear, the enlarged LDs were not a result of the fusion/coalescence of small LDs (Xu et al., 2018). In contrast to our finding, the TAG content of cultured Rab18 knockout 3T3-L1 adipocytes was decreased to the same extent as the reduction in the normal-sized LDs (Xu et al., 2018). Further dissecting the mechanisms by which RABC1 regulates LD homeostasis will not only help us better understand stomatal function and development but it will also help to provide essential insights into the production of vegetable oils in vegetative tissues which is a field of increasing interest to the biotechnology sector and biofuel industries (Xu and Shanklin, 2016).
The formation of the central vacuole allows metabolically inexpensive plant cell expansion, and interference with vacuolar function evokes severe developmental defects (Schumacher et al., 1999; Rojo et al., 2001; Li et al., 2005; Krüger and Schumacher, 2018). Studies of vacuoles in GCs have revealed a tight correlation between vacuole morphology and stomatal movement (Gao et al., 2005, 2009; Bak et al., 2013; Andrés et al., 2014; Cao et al., 2022). During stomatal development, there is a significant increase in cell size that will require an increase in plasma and vacuolar membrane surface area. Disruption of RABC1 causes defects in LD mobilization and vacuole size (Figures 1A and 6A), highlighting the relationship between these two organelles. What is less clear is the mechanism by which LDs are able to contribute toward the new membrane required for vacuolar growth. As it is the ER, rather than the vacuole that is the site of de novo phospholipid biosynthesis, it seems unlikely that LD TAGs could be used directly by the vacuole to synthesize the new phospholipids required for membrane expansion. Instead, perhaps the vacuole incorporates the phospholipid monolayer surrounding the LD so as to increase the surface area of the tonoplast. This is of course speculation but would be a topic to explore in the future. Another possibility is that RABC1 regulates LD homeostasis and vacuole growth separately, and, or the changes in vacuole occupancy and OCL formation are downstream effects of defective GC development. The final possibility is that LD vacuole interactions are related to lipophagy whereby lipids can be degraded and recycled (Ischebeck et al., 2020). We observed LDs within vacuoles in GCs and disruption of RABC1 led to a significant decrease of LDs entering the vacuole (Figure 6, C and D). These data indicate that there is an interaction of the LDs with the vacuole. If lipid recycling underlies the LD-vacuole interactions, it is possible that the lipids or their metabolites could be exported to sites of phospholipid (for membranes) or wax (for OCL) synthesis in the cell. Again, this possibility would be worth further investigation.
RABC1 localizes to the ER and weakly localizes to LDs under normal conditions, however, in OA-induced LD production, RABC1 targets to the surface of the LDs. This mirrors the targeting of Rab18 in mammalian cells. According to Kretzschmar et al. (2018), the tobacco (Nicotiana tabacum) counterpart of Rab18 was enriched in LD fractions of seedlings and pollen tubes but not as strongly enriched as canonical LD proteins. This is in line with the dual localization of RABC1 and suggests the localization of Rab18 proteins on LDs is transient and not very strong.
The finding that SEIPIN2 and SEIPIN3 serve as effectors of RABC1 is in accordance with the early work on Rab18 (which is a close homolog of RABC1 in mammalian cells; Li et al., 2019). In contrast to yeast or mammals, plants express three SEIPIN genes. The expansion of SEIPIN isoforms could have resulted in an expanded capacity to meet the distinct needs of land plants to adapt to an always-changing environment. This suggestion is supported by the fact that we only detected increased LD abundance and LD size in the GCs of the double mutant seipin2 seipin3 but not in seipin1 seipin2 and seipin1 seipin3. This suggests that SEIPIN1 is unlikely to be involved in the regulation of LDs by RABC1 in GCs. In support of this suggestion, all three SEIPIN proteins have been found to participate in the regulation of LD abundance in seeds/pollen grains (Taurino et al., 2018), whereas only the function of SEIPIN2 and SEIPIN3 depends on their interaction with the vesicle-associated membrane protein-associated protein family member AtVAP27-1 (Greer et al., 2020). One thing that might account for the distinct function of SEIPIN2, SEIPIN3, and SEIPIN1 is the sequence differences between SEIPIN1 and SEIPIN2/3, particularly between their N-terminal regions which have been proposed to be crucial for the functionality of SEIPIN proteins (Greer et al., 2020). It is important to note that the severity of deformed LDs and defective stomata in seipin2 seipin3 is much weaker than either lds1 or ldsgef1, and lds1 and seipin2 seipin3. This strongly suggests that there must be other effectors of RABC1.
In humans, Rab18 is correlated with Warburg Micro syndrome, a severe illness characterized by microcephaly, intellectual disability, optical atrophy, and hypogenitalism. However, whether there is a direct causal relationship between LD dysfunction and the disease is currently unknown (Bem et al., 2011; Dejgaard and Presley, 2019). Rab18 was detected at LD-ER contact sites, associated with the NRZ tethering complex and the Q-SNARES Use1, Syntaxin18, and BNIP1 to cooperatively support LD growth in adipocytes (Ozeki et al., 2005; Xu et al., 2018). In COS7 and HeLa cells, a complex of Rab18 and DFCP1 has been found to be important for efficient ER-LD tethering and LD expansion (Li et al., 2019). In addition to being a key factor regulating lipogenesis and lipolysis (Martin et al., 2005; Ozeki et al., 2005; Pulido et al., 2011), Rab18 is also involved in trafficking between the Golgi and ER (Dejgaard et al., 2008), regulating ER structure (Gerondopoulos et al., 2014), and autophagy (Feldmann et al., 2017; Bekbulat et al., 2020). Overall, the mechanistic details by which mutations in Rab18 cause enlarged LDs remain understudied. Arabidopsis Rab18 has not been previously ascribed an LD-related function. Our work reveals that RABC1 together with its GEF and effectors are involved in stomatal regulation by modulating LDs.
In summary, the results of the research described here reveal an unexpected role for LDs in the regulation of stomatal development. Underpinning this role is the Rab GTPase family member RABC1. We show that it interacts with SEIPIN2/3, which functions in LD formation. Future research will be directed to understanding whether, how LD-vacuole interactions result in the increase in vacuolar surface area seen during stomatal development.
Materials and methods
Plant materials, growth conditions, and plasmid construction
All Arabidopsis (A. thaliana L.) lines used in this study are in the Columbia (Col-0) ecotype background. The mutants lds1-2 (SALK_012129), rabc2a (SALK_061001C), rabc2b (WiscDsLoxHs095_05E), lds1gef1-1 (WiscDsLox393-396G22), lds1gef1-2 (SALK_116404C), SALK_058192C of At5g55060, seipin1-1, (SALK_Seq0958), seipin2-1 (GabiGK_183F09), seipin3-1 (SALK_019429C) and the marker line ER-ck (CS16250; Nelson et al., 2007) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University) and genotyped using specific primers, which are listed in the Supplemental Table S1. The scap1 mutant is as described by Negi et al. (2013) and was a kind gift from Professor Koh Iba (Kyushu University). The three single mutant lines of SEIPINs, and from which the corresponding double and triple mutants were derived through genetic crossings, are as described by Taurino et al. (2018).
Arabidopsis seeds were sown on half-strength Murashige and Skoog (1/2MS) media with 1% sucrose and 0.6% agar. After a 3-d stratification at 4°C in the dark, the seed plates were transferred to a greenhouse at 22°C with an average light intensity of 120 μmoles photons m−2 s−1 and relative humidity at 70%–80% with a 10-h-light/14-h-dark photoperiod. 7-d-old seedlings on the plates were transplanted into the potting soil/vermiculite mixture (Zhang et al., 2019). Nicotiana benthamiana seeds were directly sown at a depth of 0.5 cm below the soil/vermiculite mixture surface and the seedlings were grown in a greenhouse with 16-h-light/8-h-dark cycles at 28°C. The light source used in the greenhouses is wide-spectrum fluorescent bulb (YZ28-T5-28W, NVC).
The promoter sequence of RABC1 was amplified using the primers listed in Supplemental Table S1, while the promoter sequences of SEIPINs used in this study were as previously described (Taurino et al., 2018). The ProRABC1:GUS and ProSEIPINs:GUS transgenic plants were generated using pENTR/D-TOPO as an entry vector, pKGWFS7 plasmids as the destination vectors. Plasmid of ProRABC1:GUS was used to develop ProRABC1:mCherry-RABC1 (the natural or mutated RABC1), ProRABC1:RABC2a, and ProRABC1:RABC2bconstructs by replacing the GUS gene sequence with the coding sequences of mCherry-RABC1, RABC2a, and RABC2b, respectively. The nucleotide sequences of mCherry-RABC1 were generated by overlapping PCR using the CDS of mCherry and the natural or mutated RABC1. The coding sequences of RABC1(S27N) and RABC1(Q71L) were produced by site-directed mutagenesis PCR. pK2GW7 was used as the destination vector for Pro35S:MmRAB18, Pro35S:HsRAB18, and Pro35S:SDP1 constructs after introducing the corresponding CDS of MmRAB18, HsRAB18, and SDP1 to the entry vector, respectively. pGWB6 (Nakagawa et al., 2007) was the destination vectors for Pro35S:GFP-SEIPIN2/3 and Pro35S:GFP-VAMP711constructs, respectively. All primers used above are shown in Supplemental Table S1 and all generated constructs were verified by DNA sequencing.
Mutant screen
Ethyl methane sulfonate mutagenesis of Arabidopsis seeds was performed essentially as described (Kim et al., 2006). The M2 seeds were subjected to a two-step mutant screen based on infrared thermal imaging and NR staining to identify genes involved in LD dynamics and stomatal development/function. Thermal images were obtained using an SC660 (FLIR Systems, Wilsonville, USA). The camera was mounted vertically above the plants. Mean temperature of leaves no. 3, 4, 5, 6, and 7 below the apex were calculated using the customized region of interest (ROI) tool, according to the manufacturer’s instructions. Abaxial epidermal strips were detached from rosette leaves for LD staining. Briefly, strips were incubated in KCl/MES buffer (50 mM KCl, 10 mM MES/KOH, pH 6.15) containing 10 μg/mL NR (Sigma) or 10 μM Bodipy 493/503 (Invitrogen) for 10 min and then washed in KCl/MES buffer for 5 min before imaging. Confocal images were collected with a Leica SP8 confocal microscope. Following the identification of a mutant, it was backcrossed to WT 3 times.
Confocal microscopy
Images were collected using a Leica SP8 confocal microscope with a × 63/1.4 NA oil immersion objective lens. For LD staining, we used three dyes in this study, NR (10 μg/mL), Bodipy 493/503 (10 μM), and MDH (Monodansylpentane, Yang et al., 2012). The excitation wavelength was 405 nm and the emission wavelength was 440–485 nm for MDH; the excitation wavelength was 488 nm and the emission wavelength was 530–590 nm for NR while the emission wavelength was 500–530 nm for Bodipy 493/503. When detecting the LDs abundance and size in GCs, the Z-projections (maximal intensity) of confocal stacks from young leaves of 4-week-old plants was used. Z-series images were taken every 0.5 μm and the same settings of laser power and gain value for all samples examined. Z-projections were generated in Leica Advanced Fluorescence v3.1.0 (Leica) and Image J (NIH) was used for fluorescence intensity quantification. Briefly, images were first converted to 16-bit depth, and ROI (regions of interest) was defined by tracing the outline of cells in Image J. Raw integrated density (the sum of all pixel intensities within an ROI) and area were recorded. Fluorescence intensity was represented as a ratio of raw integrated density to area after minus the background fluorescence intensity. Given that the resolution of confocal microscopy is about 150 nm, it is difficult to observe and quantify smaller LDs, such as nascent LDs. However, this limitation does not have much influence on the conclusions of this paper.
Map-based mutant gene cloning
Candidate lds1 mutants were outcrossed to WT plants in the Landsberg erecta background (Ler) and the segregating F2 seedlings were screened using infrared thermography and microscopic observation following NR/Bodipy staining. Segregant analysis of totally 900 lds1 mutants was performed using simple sequence length polymorphism (SSLP) markers as previously described (Lukowitz et al., 2000; He et al., 2018). The mutation was narrowed down to a ca. 200 kb region of chromosome 1 between SSLP marker 125DW-G and 125DW-J2. T-DNA insertion lines representing all the annotated genes within this region were obtained from ARBC and screened. A T-DNA insertion line (SALK_012129) of At1g43890 which displayed similar LD accumulation defects to lds1 was identified. Allelism tests were performed using the F1 progeny of the lds1 x SALK_012129 which confirmed that lds1 and SALK_012129 are allelic to each other. The presence of a single point mutation in At1g43890 of the lds1 mutant and a T-DNA insertion in At1g43890 of the SALK_012129 line was validated using PCR-based genotyping and gene sequencing. We renamed the lds1 and SALK_012129 as lds1-1 and lds1-2, respectively.
Stomatal aperture measurements
Stomatal aperture was determined using the rapid imprinting technique described by Geisler et al. (2000). Light-bodied vinylpolysiloxane dental resin (eliteHD+, Zhermack Clinical, Badia Polesine, Italy) was attached to the abaxial side of the leaflet and then removed as soon as it dried. The resin epidermal imprints were covered with transparent nail polish, which was removed once it dried and served as a mirror image of the resin imprint. The nail-polish imprints were put on glass coverslips and photographed under a model ICC50 W bright-field inverted microscope (Leica Microsystem, Wetzlar, Germany). Stomatal images were later analyzed to determine aperture size using the ImageJ software fit-ellipse tool (http://rsb.info.nih.gov/ij/). A microscopic ruler (Olympus) was used for size calibration.
Stomatal conductance measurements
Stomatal conductance of intact leaves of 5-week-old plants to water vapor (gs) was measured with a Li-6400 gas exchange analyzer with a fluorometer chamber (Li-Cor, Lincoln, NE, USA) as described previously (Negi et al., 2013; He et al., 2018). The gs response to light was tested under 350 ppm CO2. Intact leaves were incubated in dark for 3 h first, then kept under light illumination (150 μmol m−2 s−1) for 2 h and another 1 h dark. The gs response to CO2 was measured at constant light intensity (150 μmol m−2 s−1). Temperature and relative humidity in the chamber were held at 22°C and 60%, respectively. All measurements were taken at a rate of one per minute.
Transgene expression analysis
GUS activity ofProRABC1:GUS and ProSEIPIN2:GUS transformants was performed as described previously (Negi et al, 2013; Taurino et al, 2018). Transformants were fixed in 90% ice-cold acetone for 20 min. After washing, transformants were incubated in 100 mM sodium phosphate (pH 7.4), 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronic acid at 37°C in the dark. Plants were bleached in 70% ethanol and mounted in chloral hydrate for imaging. Subcellular localization of mCherry-RABC1 was analyzed using a ProRABC1:mCherry-RABC1 transformant. After incubation with or without 200 μM OA (oleic acid, Xu et al., 2018) in buffer (50 mM KCl, 10 mM MES/KOH (pH 6.15)) for 6 h, leaf epidermal peels of the transformant were observed with a Leica SP8 confocal microscope. LDs were stained with Bodipy 493/503 (excitation laser 488 nm, emission 530–590 nm). mCherry was excited using a 552 nm laser and its emission is 590–650 nm.
Yeast two-hybrid analysis
The Y2H assay was based on the Matchmaker Gold Yeast Two-Hybrid System User Manual (Clontech). The cDNAs encoding different forms of RABC1 were cloned into pGBKT7 bait plasmid and introduced into Y2H Gold yeast strain. Possible autoactivation of RABC1 was determined according to the manufacturer’s protocol. We used the Mate and Plate Normalize Arabidopsis Universal Library (Clontech) for screening potential interacting proteins of RABC1. Briefly, we first prepared a concentrated overnight culture of the Bait Strain (cell density >1 × 108 cells/mL in SD/–Trp) before adding 1 mL of the Library Strain to the Bait Strain (4–5 mL) in a sterile 2 L flask. Next, we added 45 mL of 2×YPDA liquid medium, and incubated the reaction at 30°C for 20–24 h with gentle agitation (30–50 rpm). After centrifugation to pellet the cells, we resuspended all pelleted cells in 10 mL of 0.5×YPDA/Kan liquid medium before plating the culture on SD/-Leu/-Trp/-His/-Ade and incubated it at 30°C for 3–5 d. Yeast colonies that were alive on SD/-Leu/-Trp/-His/-Ade medium were streaked. Then plasmids of these colonies were isolated and sequenced. In addition, to further confirm the interactions in yeast, the plasmids pGADT7 and pGBKT7 with corresponding proteins were co-transformed into the yeast strain Y2H Gold. Empty vectors were used for negative controls.
Nucleotide exchange assays
The nucleotide exchange assays were performed to determine GEF for RABC1 as described (Pan et al., 1995; Antonny et al., 2001; Goh et al., 2007; Cui et al., 2014). Briefly, each purified GST-Rab protein was preloaded with a 25 M GDP at 25°C for 2 h. Then, each assay was incubated with or without GST-At5g58510 protein or GST-At5g55060 protein in reaction buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM MgCl2) for 100 s at 25°C. The nucleotide exchange reaction was initiated by the addition of 0.1 mM GMP-PNP (guanylyl imidodiphosphate). The Trp intrinsic fluorescence of Rab proteins was detected by fluorescence spectrophotometry at an excitation wavelength of 298 nm and an emission wavelength of 340 nm.
Expression and purification of GST fusion proteins
Vector pGEX-4T-1 was used for expressing GST fusion proteins. After inserting the respective CDS sequence into pGEX-4T-1 and confirming that the cloning was in-frame, the resultant plasmids of pGEX-GST-RABC1, pGEX-GST-RHA1, pGEX-GST-RABC1GEF1, and pGEX-GST-At5g55060 were transformed in BL21 Rosetta and recombinant proteins were purified according to published procedures (Goh et al., 2007; Fukuda et al., 2013; Cui et al., 2014). Briefly, the expression of recombinant proteins was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) then incubated overnight at 16°C. GST-tagged proteins were purified on a glutathione-Sepharose 4B column (GE Healthcare).
BiFC assays
The constructs for BiFC assays were generated by cloning the coding sequences of RABC1, RABC1GEF1, and SEIPINs into the series of pSAT expression vectors as described (Tzfira et al., 2005). The constructs were introduced into N. benthamiana leaves through Agrobacterium (Agrobacterium tumefaciens, strain GV3101)-mediated transformation. After incubated for 2–3 d, the leaves were examined using a Leica SP8 microscope (excitation 488 nm, emission 520–560 nm). Fluorescence intensity quantification was performed in a similar manner to that used for LDs as described above. Images were converted to 16-bit depth, and ROI was defined in Image J. The YFP intensity was represented as a ratio of raw integrated density to area after minus the background fluorescence intensity.
Co-IP reactions
The CDS of RABC1, RABC1(S27N), RABC1(Q71L), or RABC1GEF1 was each separately subcloned into pGWB12 or pGWB6 using LR reaction (Nakagawa et al., 2007). Then the plasmids were electroporated into the GV3101 strain of A. tumefaciens and transiently expressed in N. benthamiana. Co-IP reactions were performed according to published procedures (Roux et al., 2011; Fukuda et al., 2013). Briefly, leaves were ground in liquid nitrogen and suspended in the extraction buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 5 mM DTT, 0.5% NP40, 1 × protease inhibitor cocktail, pH 7.5) at 2 mL/g tissue powder. Samples were centrifuged at 16,000 g at 4°C for 10–15 min before the supernatant was transferred to a new tube and centrifuged at 16,000 g at 4°C for 5 min. The supernatant (1.5 mL) was incubated with 20 μL GFP Trap-A beads (Chromotek) for 2 h at 4°C. After washing 4 times with the immunoprecipitation buffer (the extraction buffer minus the protease inhibitor cocktail), the beads were then boiled in SDS sample buffer for 5 min before being subjected to western blot analysis using anti-GFP (ProteinTech, HRP-66002) and anti-Flag antibody (ProteinTech, HRP-66008) at a dilution of 1:5,000.
TAG analysis
GC-enriched epidermal strips were isolated essentially as described in McLachlan et al. (2016). Of the GCs, 97.3 ± 4.8% were intact and the epidermal pavement cell viability was 4.5% ± 0.8% using FDA staining. TAGs were extracted from 50 mg fresh-frozen epidermal strips, and analyzed by LC/MS/MS (Burgal et al., 2008).
TEM analysis
Leaf pieces from 3-week-old seedlings were fixed in 2% (v/v) glutaraldehyde in 100 mM phosphate buffer (pH7.4) at 4°C for overnight. The tissue was then rinsed, postfixed in 1% (w/v) osmium tetroxide for 2 h. After dehydration in an alcohol series (15%, 30%, 50%, 70%, 80%, 85%, 90%, and 95% alcohol, each treatment lasted for 30 min), the tissue was embedded in Spurr’s epoxy resin after a propylene oxide/Spurr’s epoxy resin incubation series (propylene oxide/Spurr’s epoxy resin = 2:1, 1:1, 1:2, every 12 h). After polymerizing at 60°C for 48 h, thin resin sections were prepared and stained with aqueous uranyl acetate and lead acetate. A JEM-1400Plus transmission electron microscope was used to visualize the sections.
Statistical analysis
All assays were conducted with three or more biological replicates and analyzed using SigmaPlot (version 12.0, Systat). Comparison of means was conducted using Student’s t tests.
Accession numbers
The GenBank accession numbers of the Arabidopsis genes used in this study are as follows: RABC1 (At1g43890), RABC1GEF1 (At5g58510), RABC2a (At5g03530), RABC2b (At3g09910), SEIPIN1 (At5g16460), SEIPIN2 (At1g29760), SEIPIN3 (At2g34380), SDP1 (At5g04040). The NCBI accession numbers for the proteins are as follows: RAB3GAP1 (NP_001165906), MmRab18 (NP_001265376), HsRab18 (NP_067075).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. lds1 mutation caused no alterations in stomatal density and stomatal index but did bring about growth arrest.
Supplemental Figure S2. RABC1-mediated LD regulation is conserved across species.
Supplemental Figure S3. Mutant identification, GEF activity analysis of At5g58510 and At5g55060.
Supplemental Figure S4. Localization analysis of SEIPIN2/3 and overexpression of SEIPIN2/3 in lds1-2.
Supplemental Figure S5. Vacuole area in GCs is affected by lds1, lds1gef1, and seipin2 seipin3 mutations.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set S1. Candidates identified during the Y2H screen using RABC1(S27N) as bait.
Supplemental Data Set S2 . Candidates identified during the Y2H screen using RABC1(Q71L) as bait.
Supplementary Material
Acknowledgments
The authors are grateful to Professor Maoteng Li (Huazhong University of Science and Technology) for the assistance with the neutral lipid analysis.
Funding
This work was supported by the National Natural Science Foundation of China (#31770282 and #31971811).
Conflict of interest statement. None declared.
Contributor Information
Shengchao Ge, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Ruo-Xi Zhang, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Yi-Fei Wang, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Pengyue Sun, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Jiaheng Chu, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Jiao Li, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Peng Sun, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Jianbo Wang, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Alistair M Hetherington, School of Biological Sciences, Life Sciences Building, University of Bristol, 24 Tyndall Avenue, Bristol, BS8 1TQ, UK.
Yun-Kuan Liang, State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Y.-K.L. conceived the project; S.G., Y.-F.W., P.Y.S., J.H.C., and J.L. conducted experiments; Y.-K.L., A.M.H., J.B.W., S.G., R.-X.Z., and P.S. analyzed data and wrote the manuscript. All authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yun-Kuan Liang (ykliang@whu.edu.cn).
References
- Andrés Z, Pérez-Hormaeche J, Leidi EO, Schlücking K, Steinhorst L, McLachlan DH, Schumacher K, Hetherington AM, Kudla J, Cubero B (2014) Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci USA 111: E1806–E1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol 3: 531–537 [DOI] [PubMed] [Google Scholar]
- Bak G, Lee EJ, Lee Y, Kato M, Segami S, Sze H, Maeshima M, Hwang JU, Lee Y (2013) Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell 25: 2202–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekbulat F, Schmitt D, Feldmann A, Huesmann H, Eimer S, Juretschke T, Beli P, Behl C, Kern A (2020) RAB18 loss interferes with lipid droplet catabolism and provokes autophagy network adaptations. J Mol Biol 432: 1216–1234 [DOI] [PubMed] [Google Scholar]
- Bem D, Yoshimura S, Nunes-Bastos R, Bond FC, Kurian MA, Rahman F, Handley MT, Hadzhiev Y, Masood I, Straatman-Iwanowska AA, et al. (2011) Loss-of-function mutations in RAB18 cause Warburg micro syndrome. Am J Hum Genet 88: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuin T, Roy JK (2014) Rab proteins: the key regulators of intracellular vesicle transport. Exp Cell Res 328: 1–19 [DOI] [PubMed] [Google Scholar]
- Brown LA, Larson TR, Graham IA, Hawes C, Paudyal R, Warriner SL, Baker A (2013) An inhibitor of oil body mobilization in Arabidopsis. New Phytol 200: 641–649 [DOI] [PubMed] [Google Scholar]
- Buckley CR, Caine RS, Gray JE (2020) Pores for thought: can genetic manipulation of stomatal density protect future rice yields? Front Plant Sci 10: 1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD (2015) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA (2019) Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Woolfenden H, Baillie A, Amsbury S, Carroll S, Healicon E, Sovatzoglou S, Braybrook S, Gray JE, Hobbs J, et al. (2017) Stomatal opening involves polar, not radial, stiffening of guard cells. Curr Biol 27: 2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Li Z, Huang S, Shi Y, Zhu Y, Lai MN, Lok PL, Wang X, Cui Y, Jiang L (2022) Correlation of vacuole morphology with stomatal lineage development by whole-cell electron tomography. Plant Physiol 188: 2085–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol 16: 1783–1795 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants: thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JW, Harris BJ, Hetherington AJ, Hurtado-Castano N, Brench RA, Casson S, Williams TA, Gray JE, Hetherington AM (2022) The origin and evolution of stomata. Curr Biol (in press; ). 10.1016/j.cub.2022.04.040 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L (2014) Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejgaard SY, Murshid A, Erman A, Kizilay O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R, Simpson JC, et al. (2008) Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci 121: 2768–81 [DOI] [PubMed] [Google Scholar]
- Dejgaard SY, Presley JF (2019) Rab18: new insights into the function of an essential protein. Cell Mol Life Sci 76: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett JG, Pressel S (2018) The evolution of the stomatal apparatus: intercellular spaces and sporophyte water relations in bryophytes—two ignored dimensions. Philos Trans R Soc B 373: 20160498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J, Hunt L, Afsharinafar M, Meselmani MA, Mitchell A, Howells R, Wallington E, Fleming AJ, Gray JE (2019) Reduced stomatal density in bread wheat leads to increased water-use efficiency. J Exp Bot 70: 4737–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yu L, Xu C (2019) Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 31: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann A, Bekbulat F, Huesmann H, Ulbrich S, Tatzelt J, Behl C, Kern A (2017) The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem Biophys Res Commun 486: 738–743 [DOI] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P (1985) Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem 33: 833–836 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Doheny‐Adams TW, Britton‐Harper ZJ, Gray JE (2015) Increasing water‐use efficiency directly through genetic manipulation of stomatal density. New Phytol 207: 188–195 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Wen L, Satoh-Cruz M, Kawagoe Y, Nagamura Y, Okita TW, Washida H, Sugino A, Ishino S, Ishino Y (2013) A guanine nucleotide exchange factor for Rab5 proteins is essential for intracellular transport of the proglutelin from the Golgi apparatus to the protein storage vacuole in rice endosperm. Plant Physiol 162: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Li CG, Wei PC, Zhang XY, Chen J, Wang XC (2005) The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol 139: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Wang XL, Ren F, Chen J, Wang XC (2009) Dynamics of vacuoles and actin filaments in guard cells and their roles in stomatal movement. Plant Cell Environm 32: 1108–1116 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A, Bastos RN, Yoshimura S, Anderson R, Carpanini S, Aligianis I, Handley MT, Barr FA (2014) Rab18 and a Rab18 GEF complex are required for normal ER structure. J Cell Biol 205: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Park S, Pyc M, Yurchenko O, Cai Y, Wu P, Andrews DW, Chapman KD, Dyer JM, Mullen RT (2016) Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol 170: 2052–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S (2014) Toward a comprehensive map of the effectors of Rab GTPases. Dev Cell 31: 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Uchida W, Arakawa S, Ito E, Dainobu T, Ebine K, Takeuchi M, Sato K, Ueda T, Nakano A (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Greer MS, Cai Y, Gidda SK, Esnay N, Kretzschmar FK, Seay D, McClinchie E, Ischebeck T, Mullen RT, Dyer JM (2020) SEIPIN isoforms interact with the membrane-tethering protein VAP27-1 for lipid droplet formation. Plant Cell 32: 2932–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV Jr, Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Negi J, Young J, Israelsson M, Schroeder JI, Iba K (2006) Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat Cell Biol 8: 391–397 [DOI] [PubMed] [Google Scholar]
- He J, Zhang RX, Peng K, Tagliavia C, Li S, Xue S, Liu A, Hu H, Zhang J, Hubbard K, et al. (2018) The BIG protein distinguishes the process of CO2-induced stomatal closure from the inhibition of stomatal opening by CO2. New Phytol 218: 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Huang AH (2018) Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol 176: 1894–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Biol 43: 177–200 [Google Scholar]
- Hughes J, Hepworth C, Dutton C, Dunn JA, Hunt L, Stephens J, Waugh R, Cameron DD, Gray JE (2017) Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol 174: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Amsbury S, Baillie A, Movahedi M, Mitchell A, Afsharinafar M, Swarup K, Denyer T, Hobbs JK, Swarup R, et al. (2017) Formation of the stomatal outer cuticular ledge requires a guard cell wall proline-rich protein. Plant Physiol 174: 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE (2020) How the stomate got his pore: very long chain fatty acids and a structural cell wall protein sculpt the guard cell outer cuticular ledge. New Phytol 228: 1698–1700 [DOI] [PubMed] [Google Scholar]
- Ischebeck T (2016) Lipids in pollen—they are different. Biochim Biophys Acta 1861: 1315–1328 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Krawczyk HE, Mullen RT, Dyer JM, Chapman KD (2020) Lipid droplets in plants and algae: Distribution, formation, turnover and function. Semin Cell Dev Biol 108: 82–93 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Kaiser S, Scheuring D (2020) To lead or to follow: contribution of the plant vacuole to cell growth. Front Plant Sci 11: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ (2013) The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Schumaker KS, Zhu JK (2006) EMS mutagenesis of Arabidopsis. Methods Mol Biol 323: 101–103 [DOI] [PubMed] [Google Scholar]
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV Jr, Walther TC (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 12: 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M, Matthews M, Shackel K (2008) Evidence for substantial maintenance of membrane integrity and cell viability in normally developing grape (Vitis vinifera L.) berries throughout development. J Exp Bot 59: 849–859 [DOI] [PubMed] [Google Scholar]
- Krawczyk HE, Sun S, Doner NM, Yan Q, Lim MSS, Scholz P, Niemeyer PW, Schmitt K, Valerius O, Pleskot R, et al. (2022) SEED LIPID DROPLET PROTEIN1, SEED LIPID DROPLET PROTEIN2, and LIPID DROPLET PLASMA MEMBRANE ADAPTOR mediate lipid droplet-plasma membrane tethering. Plant Cell 34: 2424–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar FK, Mengel LA, Muller AO, Schmitt K, Blersch KF, Valerius O, Braus GH, Ischebeck T (2018) PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30: 2137–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar FK, Doner NM, Krawczyk HE, Scholz P, Schmitt K, Valerius O, Braus GH, Mullen RT, Ischebeck T (2020) Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol 182:1326–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger F, Schumacher K (2018) Pumping up the volume-vacuole biogenesis in Arabidopsis thaliana. Semin Cell Dev Biol 80: 106–112 [DOI] [PubMed] [Google Scholar]
- Larsson S, Resjo S, Gomez MF, James P, Holm C (2012) Characterization of the lipid droplet proteome of a clonal insulin-producing beta-cell line (INS-1 832/13). J Proteome Res 11: 1264–1273 [DOI] [PubMed] [Google Scholar]
- Lawson T, Matthews J (2020) Guard cell metabolism and stomatal function. Annu Rev Plant Biol 71: 273–302 [DOI] [PubMed] [Google Scholar]
- Lee LR, Bergmann DC (2019) The plant stomatal lineage at a glance. J Cell Sci 132: jcs228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Mishra A, Lambright DG (2009) Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Luo X, Zhao S, Siu GK, Liang Y, Chan HC, Satoh A, Yu SS (2017) COPI-TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J 36: 441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yu SS (2016) Rab proteins as regulators of lipid droplet formation and lipolysis. Cell Biol Int 40: 1026–1032 [DOI] [PubMed] [Google Scholar]
- Li D, Zhao YG, Li D, Zhao H, Huang J, Miao G, Feng D, Liu P, Li D, Zhang H (2019) The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep 27: 343–358 [DOI] [PubMed] [Google Scholar]
- Li Z, Solomon JM, Isberg RR (2005) Dictyostelium discoideum strains lacking the RtoA protein are defective for maturation of the Legionella pneumophila replication vacuole. Cell Microbiol 7: 431–442 [DOI] [PubMed] [Google Scholar]
- Lu J, He J, Zhou X, Zhong J, Li J, Liang YK (2019) Homologous genes of epidermal patterning factor regulate stomatal development in rice. J Plant Physiol 234–235: 18–27 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre J, Delepine M, Khallouf E, Gedde-Dahl T, Van Maldergem L, Sobel E, Papp J, Meier M, Megarbane A, Bachy A (2001) Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet 28: 365–370 [DOI] [PubMed] [Google Scholar]
- Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG (2005) Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem 280: 42325–42335 [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7: 373–378 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM (1990) Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 343: 186–188 [Google Scholar]
- McLachlan DH, Lan J, Geilfus CM, Dodd AN, Larson T, Baker A, Hõrak H, Kollist H, He Z, Graham I, et al. (2016) The breakdown of stored triacylglycerols is required during light-induced stomatal opening. Curr Biol 26: 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merced A, Renzaglia K (2014) Developmental changes in guard cell wall structure and pectin composition in the moss Funaria: implications for function and evolution of stomata. Ann Bot 114: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto-Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S, et al. (2013) A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr Biol 23: 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T (2005) Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci 118: 2601–2611 [DOI] [PubMed] [Google Scholar]
- Pan JY, Sanford JC, Wessling-Resnick M (1995) Effect of guanine nucleotide binding on the intrinsic tryptophan fluorescence properties of Rab5. J Biol Chem 270: 24204–24208 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2012) Mechanisms of stomatal development. Annu Rev Plant Biol 63: 591–614 [DOI] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Samuels AL, Browse J, Rogers JC (2006) A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47: 917–933 [DOI] [PubMed] [Google Scholar]
- Pulido MR, Diaz-Ruiz A, Jimenez-Gomez Y, Garcia-Navarro S, Gracia-Navarro F, Tinahones F, Lopez-Miranda J, Fruhbeck G, Vazquez-Martinez R, Malagon MM (2011) Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One 6: e22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyc M, Cai Y, Greer MS, Yurchenko O, Chapman KD, Dyer JM, Mullen RT (2017) Turning over a new leaf in lipid droplet biology. Trends Plant Sci 22: 596–609 [DOI] [PubMed] [Google Scholar]
- Pyc M, Gidda SK, Seay D, Esnay N, Kretzschmar FK, Cai Y, Doner NM, Greer MS, Hull JJ, Coulon D (2021) LDIP cooperates with SEIPIN and LDAP to facilitate lipid droplet biogenesis in Arabidopsis. Plant Cell 33: 3076–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Hammich H, Yu IM, Houdusse A (2018) Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. Small GTPases 9: 22–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasineni K, McVicker BL, Tuma DJ, McNiven MA, Casey CA (2014) Rab GTPases associate with isolated lipid droplets (LDs) and show altered content after ethanol administration: potential role in alcohol-impaired LD metabolism. Alcohol Clin Exp Res 38: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MA, Olzmann JA (2020) Protein quality control and lipid droplet metabolism. Annu Rev Cell Dev Biol 36: 115–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Rotsch AH, Kopka J, Feussner I, Ischebeck T (2017) Central metabolite and sterol profiling divides tobacco male gametophyte development and pollen tube growth into eight metabolic phases. Plant J 92: 129–146 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter JC, Willmer CM (1979) A light and electron microscopy study of the epidermis of Paphiopedilum spp. with emphasis on stomatal ultrastructure. Plant Cell Environ 2: 211–219 [Google Scholar]
- Sack FD (1987) The development and structure of stomata. Stomatal Funct 59–89 10.1007/978-94-011-0579-8_3 [DOI] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H+–ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Isner JC, Ledru AC, Zhang Q, Pridgeon AJ, He Y, Menguer PK, Miller AJ, Sanders D, Mcgrath SP, et al. (2022) Countering elevated CO2 induced Fe and Zn reduction in Arabidopsis seeds. New Phytol (in press) 10.1111/nph.18290 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T (2011) Lipid droplets: size matters. J Electron Microsc 60: S101–S116 [DOI] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104: 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T (2001) Small GTP-binding proteins. Physiol Rev 81: 153–208 [DOI] [PubMed] [Google Scholar]
- Tang J, Yang X, Xiao C, Li J, Chen Y, Li R, Li S, Lü S, Hu H (2020) GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol 228: 1880–1896 [DOI] [PubMed] [Google Scholar]
- Taurino M, Costantini S, De Domenico S, Stefanelli F, Ruano G, Delgadillo MO, Sánchez-Serrano JJ, Sanmartín M, Santino A, Rojo E (2018) SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol 176: 1531–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumser AE, Storch J (2007) Characterization of a BODIPY-labeled fluorescent fatty acid analogue. Binding to fatty acid-binding proteins, intracellular localization, and metabolism. Mol Cell Biochem 299: 67–73 [DOI] [PubMed] [Google Scholar]
- Torii KU (2021) Stomatal development in the context of epidermal tissues. Ann Bot 128: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Graniti A (1969) Fusicoccin: a fungal toxin that opens stomata. Nature 223: 1070–1071 [Google Scholar]
- Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A (2006) Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic 7: 1254–1269 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131:1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV Jr(2017) Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33: 491–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV Jr (2012) Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81: 687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK (2016) Seipin is required for converting nascent to mature lipid droplets. eLife 5: e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Holroyd G, Hetherington AM, Ng CK (2004) Seeing ‘cool’and ‘hot’—infrared thermography as a tool for non‐invasive, high‐throughput screening of Arabidopsis guard cell signalling mutants. J Exp Bot 55: 1187–1193 [DOI] [PubMed] [Google Scholar]
- Wanner G, Formanek H, Theimer RR (1981) The ontogeny of lipid bodies (spherosomes) in plant cells: ultrastructural evidence. Planta 151: 109–123 [DOI] [PubMed] [Google Scholar]
- Wilfling F, Haas JT, Walther TC, Farese RV Jr(2014) Lipid droplet biogenesis. Curr Opin Cell Biol 29: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, Murchie E, Theobald J, Knight MR, William J, Davies WJ, et al. (2006) The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol 16: 882–887 [DOI] [PubMed] [Google Scholar]
- Xu C, Shanklin J (2016) Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu Rev Plant Biol 67: 179–206 [DOI] [PubMed] [Google Scholar]
- Xu D, Li P, Xu L (2021) Characterization of the role of Rab18 in mediating LD-ER contact and LD growth. Methods Mol Biol 2293: 229–241 [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Wu L, Li Y, Zhao D, Yu J, Huang T, Ferguson C, Parton RG, Yang H, et al. (2018) Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J Cell Biol 217: 975–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S, Takemiya A, Sakamoto T, Kurata T, Tsutsumi T, Kinoshita T, Shimazaki K (2016) The plasma membrane H+ -ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol 171: 2731–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Hsu CL, Yang JY, Yang WY (2012) Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One 7: e32693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H (2012) The proteomics of lipid droplets: structure, dynamics and functions of the organelle conserved from bacteria to humans. J Lipid Res 53: 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Benning C (2018) Functions of triacylglycerols during plant development and stress. Curr Opin Biotechnol 49: 191–198 [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang A, Li X, Kou M, Wang W, Chen X, Xu T, Zhu M, Ma D, Li Z, et al. (2018) Melatonin-stimulated triacylglycerol breakdown and energy turnover under salinity stress contributes to the maintenance of plasma membrane H+–ATPase activity and K+/Na+ homeostasis in sweet potato. Front Plant Sci 9: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu P (2019) The new face of the lipid droplet: lipid droplet proteins. Proteomics 19: 1700223. [DOI] [PubMed] [Google Scholar]
- Zhang RX, Ge S, He J, Li S, Hao Y, Du H, Liu Z, Cheng R, Feng YQ, Xiong L, et al. (2019) BIG regulates stomatal immunity and jasmonate production in Arabidopsis. New Phytol 222: 335–348 [DOI] [PubMed] [Google Scholar]
- Zhao L, Sack FD (1999) Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am J Bot 86: 929–939 [PubMed] [Google Scholar]
- Zoulias N, Harrison EL, Casson SA, Gray JE (2018) Molecular control of stomatal development. Biochem J 475: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.