Abstract
Premature abscission of flowers and fruits triggered by low light stress can severely reduce crop yields. However, the underlying molecular mechanism of this organ abscission is not fully understood. Here, we show that a gene (SlCLV3) encoding CLAVATA3 (CLV3), a peptide hormone that regulates stem cell fate in meristems, is highly expressed in the pedicel abscission zone (AZ) in response to low light in tomato (Solanum lycopersicum). SlCLV3 knockdown and knockout lines exhibit delayed low light-induced flower drop. The receptor kinases SlCLV1 and BARELY ANY MERISTEM1 function in the SlCLV3 peptide-induced low light response in the AZ to decrease expression of the transcription factor gene WUSCHEL (SlWUS). DNA affinity purification sequencing identified the transcription factor genes KNOX-LIKE HOMEDOMAIN PROTEIN1 (SlKD1) and FRUITFULL2 (SlFUL2) as SlWUS target genes. Our data reveal that low light reduces SlWUS expression, resulting in higher SlKD1 and SlFUL2 expression in the AZ, thereby perturbing the auxin response gradient and causing increased ethylene production, eventually leading to the initiation of abscission. These results demonstrate that the SlCLV3-SlWUS signaling pathway plays a central role in low light-induced abscission by affecting auxin and ethylene homeostasis.
The SlCLV3-SlWUS module regulates phytohormone homeostasis in low light-induced abscission.
IN A NUTSHELL.
Background: The abscission of plant reproductive organs occurs along a specific structure called the abscission zone (AZ), is triggered by developmental and environmental cues, and contributes to low crop yields. Environmental stimuli such as darkness and shade accelerate the abscission of flowers, flower buds, leaves, and immature fruits; this is associated with alterations in auxin and ethylene contents. Peptides might play a role in initiating abscission and regulating auxin and ethylene homeostasis during abscission.
Question: How do low-light conditions affect CLAVATA3 (SlCLV3) and WUSCHEL (SlWUS) to regulate phytohormone homeostasis and induce abscission in the important fruit crop tomato (Solanum lycopersicum)?
Findings: Low light stimulates the accumulation of SlCLV3 in the AZ and this SlCLV3 is perceived by the receptor kinases SlCLV1 and BARELY ANY MERISTEM1. After a series of signaling steps, SlWUS expression is repressed in the AZ. SlWUS negatively regulates abscission. Upon activation of the SlCLV3-SlWUS signaling pathway, the expression of the WUS target genes KNOX-LIKE HOMEDOMAIN PROTEIN1 and FRUITFULL2 is induced, the auxin response gradient in the AZ is disturbed, and ethylene production increases, leading to abscission. SlCLE2 can compensate for SlCLV3 function in regulating abscission.
Next steps: For the plant, impaired CLV3 signaling would lead to larger fruits by increasing locule number. Our results showed that CLV3 signaling mutants drop fewer flowers under low-light conditions. More flowers, along with larger and more fruit locules, and lower incidence of abscission during low-light conditions could contribute to greater yield.
Introduction
The abscission of plant reproductive organs occurs along a specific structure called the abscission zone (AZ), is triggered by developmental and environmental cues, and contributes to low crop yields (Taylor and Whitelaw, 2001). Environmental stimuli such as darkness and shade accelerate the abscission of flowers, flower buds, leaves, and immature fruits, which are associated with alterations in auxin and ethylene contents (Taylor and Whitelaw, 2001). For example, shading with a 92% polypropylene net for 5 days increases the ethylene production and abscission rate in apple (Malus domestica) fruits (Zhu et al., 2011). Similarly, 90% polypropylene net shading for 12 days increases the concentrations of indole-3-acetic acid (IAA) and ethylene biosynthesis intermediates on the proximal side of inflorescences in grape (Vitis vinifera), consequently inducing berry drop (Domingos et al., 2015). In tomato (Solanum lycopersicum), the transcription factor KNOTTED1-LIKE HOMEOBOX PROTEIN1 (SlKD1) regulates auxin transport and the response gradient of the AZ, thereby affecting the abscission of flower pedicels and petioles (Ma et al., 2015). FRUITFULL (SlFUL) regulates tomato fruit ripening by regulating the expression of ethylene biosynthesis genes (Fujisawa et al., 2014; Wang et al., 2014). However, the mechanisms involved in AZ responses to low-light stress and whether and how low light affects auxin and ethylene homeostasis remain largely unknown.
Peptide signals participate in organ abscission induced by environmental stimuli (Patharkar and Walker, 2016; Reichardt et al., 2020; Li et al., 2021). In Arabidopsis (Arabidopsis thaliana), one example is the INFLORESCENCE DEFICIENT IN ABSCISSION, HAESA, HAESA-LIKE2 (IDA-HAE/HSL2) module, which is associated with drought-induced leaf abscission (Patharkar and Walker, 2016). This module is also required for the shedding of cauline leaves triggered by pathogens (Patharkar et al., 2017). In tomato, the peptides phytosulfokine and IDA-LIKE6 (IDL6) are involved in drought and low-light stress-induced pedicel abscission (Reichardt et al., 2020; Li et al., 2021). However, these peptides are thought to accelerate abscission by inducing the expression of cell wall hydrolase genes.
In addition, nutritional stress induced by insufficient photoassimilate supply due to shading, girdling, or defoliation treatment causes the abscission of developing fruit (Yuan and Huang, 1988; Hieke et al., 2002). Nutritional stress is the main cause of fruit abscission in litchi (Litchi chinensis) (Zhao and Li, 2020). INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE, HAESA-LIKE (LcIDL1-LcHSL2) signaling module regulates the abscission of developing fruit in litchi (Ying et al., 2016; Wang et al., 2019), and LcKNAT1, the litchi homolog of Arabidopsis BREVIPEDICELLUS (BP, also named KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 [KNAT1]), suppresses ethylene production, ethylene signaling, and fruit abscission (Shi et al., 2011; Zhao et al., 2020; Ma et al., 2021). However, the regulation of LcKNAT1 by the LcIDL1-LcHSL2 signaling module has yet to be demonstrated. Nevertheless, the fact that the relationship between the IDA-HAE/HSL2 signaling module and BP has been well defined in Arabidopsis strongly suggests the potential regulation of both ethylene biosynthesis and cell wall degradation in the fruit AZ. Furthermore, these peptides might play a role in the initiation of abscission and the regulation of auxin and ethylene homeostasis during abscission (Patharkar and Walker, 2016; Reichardt et al., 2020; Li et al., 2021).
Another well characterized group of peptide hormones belong to the CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION-RELATED (CLE) family (Cock and McCormick, 2001; Zhang et al., 2014). A classic feedback system, comprising the homeodomain transcription factor WUSCHEL (WUS) and the peptide CLV3, provides the basis for the maintenance of the stem cell population at the shoot apex meristem (Mayer et al., 1998; Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000). The mature CLV3 peptide is a 12- or 13-residue glycopeptide modified by three arabinosyltransferases on a hydroxyproline (Hyp) residue at position 7 (Ohyama et al., 2009; Ogawa-Ohnishi et al., 2013; Shinohara and Matsubayashi, 2013; Xu et al., 2015). CLV3 is perceived by different types of receptor complexes, including CLAVATA1 (CLV1) homomers, BARELY ANY MERISTEMs (BAMs), CLV2/CORYNE (CRN) heteromers, and RECEPTOR-LIKE PROTEIN KINASE2 (RPK2) (DeYoung and Clark, 2008; Ogawa et al., 2008; Bleckmann et al., 2010; Kinoshita et al., 2010; Zhu et al., 2010; Wang et al., 2021a, 2021b), which limits the expression of WUS to prevent over-proliferation of meristems (Schoof et al., 2000). In addition, CLV3-INSENSITIVE KINASEs (CIKs) are co-receptors of CLV1, CLV2/CRN, and RPK2 to regulate stem cell homeostasis (Hu et al., 2018; Zhu et al., 2021). In turn, WUS promotes CLV3 expression to regulate its own activity (Schoof et al., 2000). Notably, cells in the AZ have a meristem-like nature, and WUS is also expressed in the AZ in tomato (Patterson, 2001; Wang et al., 2013). In Arabidopsis, WUSCHEL-LIKE HOMEOBOX13 negatively regulates fruit dehiscence and alters gene expression associated with cell separation at the valve margin (Romera-Branchat et al., 2013). However, whether the CLV3-WUS module plays a role in mediating abscission is still largely unknown.
CLE peptides act as key signals in response to environmental stimuli and mediate phytohormone homeostasis (Fletcher, 2020). Light regulates meristem activity by activating cytokinin signaling and inhibiting CLV1 and CLV3 expression in Arabidopsis (Yoshida et al., 2011). Moreover, CLE genes were reported to be expressed in the AZ of Arabidopsis floral organs (Jun et al., 2010). However, whether CLE signals play a role in abscission induced by abiotic stress remains to be elucidated. Here, we show that low-light conditions stimulate the SlCLV3-SlWUS signaling module and regulate phytohormone homeostasis to induce abscission.
Results
SlCLV3 positively regulates abscission in response to low-light stress
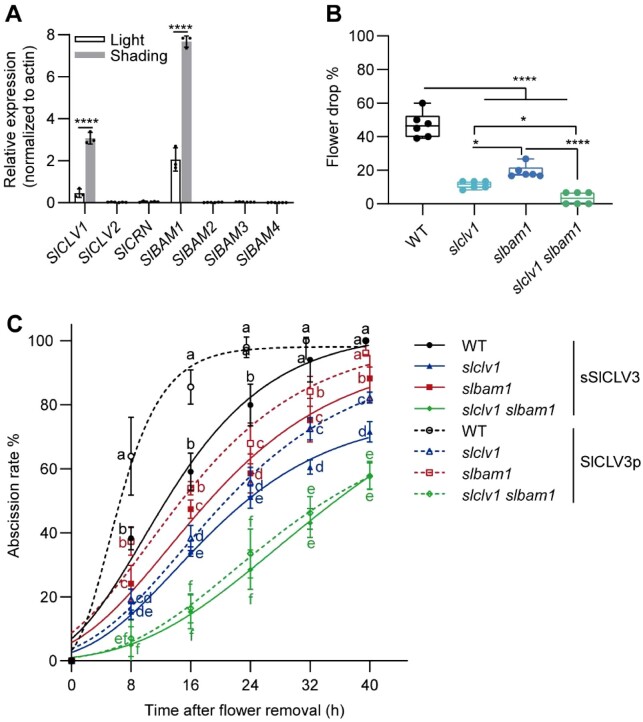
We reanalyzed a previously published transcriptome dataset generated from low light-treated tomato pedicels to look for SlCLE genes induced under these conditions (Li et al., 2021). After 70% shading for 7 days, only SlCLV3 was upregulated in the AZ (Supplemental Table S1). To confirm this result, we also investigated the expression levels of all 52 members of the SlCLE gene family in the AZ under shading treatment using reverse transcription quantitative PCR (RT-qPCR). We determined that the expression levels of SlCLV3, SlCLE5, SlCLE11, SlCLE12, SlCLE13, SlCLE14, and SlCLE32 increase following low-light stress (Figure 1A; Supplemental Figure S1) (Zhang et al., 2014; Carbonnel et al., 2022).
Figure 1.
Flower pedicel abscission is enhanced under low-light conditions in a SlCLV3-dependent manner. A, RT-qPCR analysis of relative SlCLV3 expression levels in the AZ after 0, 1, 3, 5, and 7 days under normal light or low-light conditions. Data are means ± standard deviation (SD) of three independent pools of AZ from different plants. Significant differences were determined by two-way ANOVA with Sidak’s test: *P < 0.05; ****P < 0.0001. B, Representative GUS staining pattern of CLV3pro:GUS inflorescences showing the expression of SlCLV3 in the AZ after growth under control and 70% shading conditions for 7 days. Three independent transgenic lines were stained for each condition. Scale bars, 1 cm. C, Representative inflorescence phenotype of the WT (tomato Ailsa Craig) and TAPG4pro:SlCLV3-RNAi lines 11, 17, and 33 under low-light conditions. Arrows indicate abscised flowers. Scale bar, 1 cm. D, Frequency of flower abscission in the WT and TAPG4pro:SlCLV3-RNAi lines 11, 17, and 33 under low-light conditions. Flower abscission was scored until fruit set. Five plants were scored for each genotype. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ****P < 0.0001. E, Frequency of flower abscission in the WT and the mutants Slclv3-8, Slclv3-13, and Slclv3-15 under low-light conditions. Flower abscission was scored until fruit set. Five plants were scored for each genotype. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ****P < 0.0001. F, Time course of pedicel abscission for WT, Slclv3, and TAPG4pro:SlCLV3-RNAi plants treated with H2O, sSlCLV3, or SlCLV3p peptide. Data are means ± SD of six independent treatments, with at least 10 pedicels per treatment. Different letters represent significant differences, as determined by two-way ANOVA with Tukey’s test, P < 0.05.
To explore their possible contribution to abscission, we generated a set of tomato lines individually silencing each of these CLE genes using virus-induced gene silencing (VIGS) (Supplemental Figure S2). We observed no significant difference in pedicel abscission rates between plants infiltrated with the pTRV2-empty vector control and the constructs pTRV2-SlCLE5, pTRV2-SlCLE11, pTRV2-SlCLE12, pTRV2-SlCLE13, pTRV2-SlCLE14, or pTRV2-SlCLE32 (Supplemental Figure S3, A–E and G). However, a reduction in SlCLV3 transcript levels delayed abscission significantly (Supplemental Figure S3F). One day after flower removal, 74% of the pedicel explants from the pTRV2-empty vector plants had abscised, whereas different pTRV2-SlCLV3 plants, in which SlCLV3 was silenced, only reached 21%–27% abscission at the same time (Supplemental Figure S3F). Under low-light conditions, 55% of the flowers of TRV2-empty vector plants had dropped, compared to 22%, 23%, and 35% in pTRV2-SlCLV3 #1, pTRV2-SlCLV3 #4, and pTRV2-SlCLV3 #7 plants, respectively (Supplemental Figure S3H).
To further characterize the role of SlCLV3 in the development of the AZ and abscission of the pedicel, we generated transgenic tomato plants (CLV3pro:GUS) harboring the SlCLV3 promoter driving the expression of the β-glucuronidase (GUS) reporter gene (Estornell et al., 2013). We observed that GUS staining derived from the CLV3pro:GUS reporter is induced in the pedicel AZ in response to low-light stress (Figure 1B). In situ hybridization analysis showed that the CLV3 mRNA accumulates in the pith near the inner side of the vascular bundle (Supplemental Figure S4).
We also generated SlCLV3 RNA interference (CLV3-RNAi) plants in which the RNAi cassette was expressed from the tomato abscission-specific promoter from POLYGALACTURONASE4 (TAPG4) (Ma et al., 2015) (Supplemental Figure S5A). Compared to the wild-type (WT), TAPG4pro:SlCLV3-RNAi plants exhibited unbranched inflorescences with normal flowers (Supplemental Figure S5B). Under shading conditions, 52% of WT flowers had dropped until fruit set, compared to 16% in TAPG4pro:SlCLV3-RNAi line 11, 24% in TAPG4pro:SlCLV3-RNAi line 17, and 20% in TAPG4pro:SlCLV3-RNAi line 33 (Figure 1, C and D).
In a complementary approach, we generated knockout mutants in SlCLV3 using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing. We obtained 10 primary transformants (T0s) with lesions in SlCLV3 (Supplemental Figure S6A). Seven of these plants exhibited severely fasciated flowers as well as abnormal AZs (Supplemental Figures S6, B–D). Importantly, we observed no abscission of flowers in these Slclv3 plants, even in the absence of pollination (Supplemental Figure S6B). Following morphological and histological analyses, we identified three independent fertile plants (Slclv3-8, -13, and -15) with a weak fasciated phenotype and normal AZ (Supplemental Figure S6, B–D). Upon exposure of WT and Slclv3 plants to shading conditions, 45% of WT flowers had dropped, compared to 14% for Slclv3-8, 10% for Slclv3-13, and 21% for Slclv3-15 mutant plants (Figure 1E). These results suggest that SlCLV3 is involved in low light-induced pedicel abscission.
Removing the auxin source by cutting off flowers induces pedicel explant abscission (Meir et al., 2010). We detected a 70% increase in SlCLV3 transcript levels in the AZ 0.5 h after flower removal, suggesting that SlCLV3 might also play a role in abscission induced by flower removal (Supplemental Figure S6E). To explore this possibility, we scored pedicel explant abscission in WT, weak Slclv3 mutants, and TAPG4pro:SlCLV3-RNAi plants 24 h after flower removal and treatment with water, which revealed that 77% of pedicel explants from the WT had abscised, but only 54% from Slclv3 and 52% from TAPG4pro:SlCLV3-RNAi plants had abscised (Figure 1F). These results further confirmed that SlCLV3 negatively regulates flower pedicel abscission.
To determine the contribution of the mature SlCLV3 peptide to abscission, we applied the peptide to pedicel explants. Specifically, we used a chemically synthesized peptide (SlCLV3p) that is hydroxylated at the Hyp residues Hyp4 and Hyp7 according to the mature Arabidopsis CLV3 peptide (Kondo et al., 2006). As a control, we created a peptide with the same but randomized amino acid sequence as SlCLV3p, which we designate sSlCLV3. At 24 h after flower removal, 75% of explants from WT plants treated with sSlCLV3 had abscised, which was the same abscission rate obtained with water treatment alone (Figure 1F). Notably, while 52% of explants from Slclv3 and TAPGpro:SlCLV3-RNAi plants treated with water or sSlCLV3 had abscised, 75% of explants from Slclv3 and TAPG4pro:SlCLV3-RNAi plants treated with SlCLV3p had abscised (Figure 1F). These results demonstrated that the application of SlCLV3p induces pedicel abscission and rescues the delayed abscission phenotype of Slclv3 and TAPG4pro:SlCLV3-RNAi plants, further confirming that SlCLV3 plays an essential role in mediating tomato pedicel abscission.
SlCLV3 function is dependent on the receptors SlCLV1 and SlBAM1 in the AZ
Multiple receptor complexes are required to perceive CLV3 signaling within the shoot apical meristem (Ogawa et al., 2008). To identify the specific SlCLV3 receptor(s) present in the AZ of tomato flower pedicels, we examined the expression of seven receptor-like genes by RT-qPCR (Kinoshita et al., 2010; Zhu et al., 2010; Xu et al., 2015; Hu et al., 2018). We detected only SlCLV1 and SlBAM1 transcripts in the AZ, and their expression levels were upregulated 9- and 4.4-fold under low-light conditions, respectively, relative to normal light conditions (Figure 2A). Furthermore, in situ hybridization indicated that CLV1 and BAM1 mRNAs accumulate in the vascular bundles (Supplemental Figure S7).
Figure 2.
SlCLV3 acts primarily through the receptor kinases SlCLV1 and SlBAM1. A, RT-qPCR analysis of SlCLV1, SlCLV2, SlCRN, SlBAM1, SlBAM2, SlBAM3, and SlBAM4 relative expression levels in the AZ. Data are means ± SD of three independent pools of AZ from different plants. Significant differences were determined by two-way ANOVA with Sidak’s test; ****P < 0.0001. B, Frequency of flower abscission in WT, Slclv1, Slbam1, and Slclv1 Slbam1 plants under low-light conditions. Flower abscission was scored until fruit set. Two plants were scored for each of the three independent plant lines. Significant differences were determined by one-way ANOVA with Tukey’s test; *P < 0.05; ****P < 0.0001. C, Time course of pedicel abscission for the WT, Slclv1, Slbam1, and Slclv1 Slbam1 plants treated with sSlCLV3 or SlCLV3p peptide. Data are means ± SD of six independent treatments, with at least 10 pedicels per treatment. Different lowercase letters represent significant differences, as determined by one-way ANOVA with Duncan’s test; P < 0.05.
We then isolated single (Slclv1 and Slbam1) and double mutants (Slclv1 Slbam1) using CRISPR/Cas9-mediated editing (Supplemental Figure S8, A–C). The Slbam1 single mutant had a normal AZ phenotype (Supplemental Figure S8F). The AZ of the first inflorescence in the Slclv1 single and Slclv1 Slbam1 double mutants exhibited an abnormal phenotype, but the AZ of subsequent inflorescences and inflorescences of lateral branches appeared normal (Supplemental Figure S8). We thus scored all single and double mutants for abscission in the 2nd and 3rd inflorescences. Compared to the WT, Slclv1, Slbam1, and Slclv1 Slbam1 plants showed delayed flower abscission under low-light conditions. While 46% of WT flowers had dropped, only 10% (Slclv1), 19% (Slbam1), and 3% (Slclv1 Slbam1) of flowers had dropped in the mutants (Figure 2B).
To test whether CLV1 and BAM1 function in CLV3 signaling in AZs, we treated pedicel explants from Slclv1 and Slclv1 Slbam1 plants with a solution of 30 nM sSlCLV3 or SlCLV3p. After 24 h of SlCLV3p application, 98% of pedicel explants from the WT had abscised, compared to 56% for Slclv1, 68% for Slbam1, and 34% for Slclv1 Slbam1 plants (Figure 2C), suggesting that SlCLV3p-induced abscission is impaired in the Slclv1 and Slbam1 single mutants and further repressed in Slclv1 Slbam1 double mutant plants. These results demonstrated that CLV3-mediated responses to low-light stress require CLV1 and BAM1 to accelerate abscission and that SlCLV3-associated SlCLV1 and SlBAM1 functions are not redundant during abscission of the tomato pedicel.
CLE2 can compensate for reduced CLV3 action
Multiple CLE genes have been reported to compensate for the loss of CLV3 function in Arabidopsis (Rodriguez-Leal et al., 2019). Similarly, in tomato Slclv3 mutants, SlCLE9 transcription rises as compensation to buffer stem cell homeostasis against reduced SlCLV3 levels (Rodriguez-Leal et al., 2019). Notably, we determined that only SlCLE2 transcript levels increase, by 2.1- and 3-fold in the AZ of CLV3-RNAi and Slclv3 mutants, respectively (Figure 3, A and B). As SlCLE2 is a close paralog to SlCLV3, we hypothesized that they might have similar functions (Xu et al., 2015). We observed that SlCLE2 expression does not change in response to low light or following abscission induced by flower removal (Figure 3B; Supplemental Figure S9C). However, SlCLE2 expression significantly increased in the Slclv3 mutants (Figure 3B).
Figure 3.
SlCLE2 compensates for the loss of SlCLV3 for abscission. A, RT-qPCR analysis of SlCLE2 relative expression levels in the AZ of WT, Slclv3-8, and Slclv3-15 plants. Data are means ± SD of three independent pools of the AZ. Significant differences were determined by two-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; ***P < 0.0005. B, RT-qPCR analysis of SlCLE2 relative expression levels in the AZ of WT, Slclv3, and TAPG4pro:SlCLV3-RNAi plants under normal and low-light conditions. Data are means ± SD of three independent pools of AZ from different plant lines. Significant differences were determined by two-way ANOVA with Sidak’s test compared to the WT grown in normal-light conditions (Light); *P < 0.05; **P < 0.01; ***P < 0.0005. C, Frequency of flower abscission in WT, Slclv3, Slcle2, and Slclv3 Slcle2 under low-light conditions. Flower abscission was scored until fruit set. Three biologically independent plant lines were treated. Two biologically independent treatments were performed for each plant line. Significant differences were determined by one-way ANOVA with Tukey’s test; ***P < 0.0005; ****P < 0.0001. D, Time course of WT flower pedicel explant abscission rates after treatment with different versions of SlCLV3 or SlCLE2 peptide at 8, 16, 24, 32, and 40 h after flower removal. SlCLV3p and SlCLE2p, triarabinosylated peptides. Control, water. Data are means ± SD of six independent treatments, with at least 10 pedicels per treatment. Different letters indicate significant differences, as determined by two-way ANOVA with Tukey’s test; P < 0.05. E, Time course of pedicel abscission for WT, Slclv1, Slbam1, and Slclv1 Slbam1 plants after treatment with sSlCLE2 or SlCLE2p peptide. Data are means ± SD of six independent treatments, with at least 10 pedicels per treatment. Different letters indicate significant differences, as determined by one-way ANOVA with Duncan’s test; P < 0.05.
To explore the role of SlCLE2 in low light-induced flower drop, we generated Slcle2 and Slclv3 Slcle2 mutant lines using CRISPR/Cas9-mediated editing (Supplemental Figure S9, A and B). The Slcle2 single mutants exhibited unbranched inflorescences with normal flowers. In contrast, the Slclv3 Slcle2 double mutants displayed greater branching of their inflorescences with fasciated flowers (Supplemental Figure S9, D and F). Under low-light conditions, 57% of WT flowers, 48% of Slcle2 flowers, and 21% of Slclv3 flowers had abscised, while Slclv3 Slcle2 plants showed only a 9% flower abscission rate (Figure 3C). These results indicated that loss of CLV3 stimulates the upregulation of CLE2 expression to compensate for the low light-induced abscission response in the AZ.
We also compared the effects of exogenous application of SlCLV3p or SlCLE2p peptide on the pedicel abscission of explants. At 24 h after flower removal, 72% of control pedicel explants treated with either random peptide (sSlCLV3 and sSlCLE2) had abscised, compared to 80% of pedicel explants treated with 30 nM SlCLE2p and 98% of those treated with 30 nM SlCLV3p (Figure 3D). Concomitant treatment with 15 nM SlCLV3p and 15 nM SlCLE2p resulted in 88% pedicel abscission 24 h after flower removal (Figure 3D). These results indicated that SlCLE2p treatment accelerates pedicel abscission, but to a lesser extent than seen with SlCLV3p.
To determine whether SlCLV1 and SlBAM1 are required for CLE2-regulated abscission, we analyzed the abscission rates of pedicel explants from WT and receptor mutants treated with SlCLE2p. At 24 h after SlCLE2p treatment, 81% of WT pedicel explants had abscised, compared to 52% for Slclv1, 69% for Slbam1, and 36% for Slclv1 Slbam1 plants (Figure 3E). These results demonstrated that SlCLE2 compensation acts through the SlCLV1 and SlBAM1 receptors.
CLV3 accelerates abscission through repression of WUS expression
The key function of CLV3 signaling in meristem maintenance is to limit the expression of WUS (Brand et al., 2000; Schoof et al., 2000). The finding that SlCLV3 functions in tomato pedicel abscission prompted us to determine if a similar SlCLV3-SlWUS regulatory mechanism exists in the AZ. Accordingly, we performed RT-qPCR with total RNA extracted from WT, Slclv3, Slclv1, Slbam1, Slclv3 Slcle2, and Slclv1 Slbam1 AZs. We observed that SlWUS expression is 2.7- to 4.7-fold higher in the mutant AZs compared to WT (Figure 4A).
Figure 4.
SlCLV3 regulates abscission by repressing SlWUS expression. A, Relative expression levels of SlWUS in the AZ of Slclv3, Slclv1, Slbam1, Slclv3 Slcle2, and Slclv1 Slbam1 plants. Data are means ± SD of three independent pools of AZ. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ****P < 0.0001. B and C, Time course of pedicel abscission for the WT, SlWUS-RNAi (lines 5, 7, and 8) (B), and SlWUS-OX lines (62, 65, and 68) (C). Each point is the mean ± SD of three biologically independent tests in each plant line, with at least 10 pedicels per test. Significant differences were determined by two-way ANOVA with Dunnett’s test; *P < 0.05; **P < 0.01; ****P < 0.0001. D, Frequency of flower drop in WT, SlWUS-OX (lines 62, 65, and 68), and SlWUS-RNAi (lines 5, 7, and 8) plants under low-light conditions. Flower abscission was scored until fruit set. Six plants were scored for each plant line. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ****P < 0.0001.
To determine whether WUS plays a role in tomato pedicel abscission, we attempted to construct Slwus mutants. However, knockout of the SlWUS gene causes premature termination of the seedling shoot apex meristem. We, therefore, resorted to generating RNAi plants to downregulate SlWUS transcript levels rather than fully abrogate SlWUS function (Supplemental Figure S10A). In WT plants, 70% of pedicels had abscised 1 day after flower removal, compared to 91% of pedicels in SlWUS-RNAi lines 5 and 8, and 89% in SlWUS-RNAi line 7 (Figure 4B). In addition, we constructed transgenic tomato plants in which SlWUS overexpression can be induced by dexamethasone application (Supplemental Figure S10B). Five days after flower removal, all WT pedicels had abscised, compared to 81% of SlWUS-OX-62 pedicels, 91% of SlWUS-OX-65 pedicels, and 73% SlWUS-OX-68 pedicels (Figure 4C). Under low-light conditions, 57% of WT flowers, 13% of SlWUS-OX-62 flowers, 17% of SlWUS-OX-65 flowers, 70% of SlWUS-RNAi line 5 flowers, and 67% of SlWUS-RNAi line 7 flowers had abscised (Figure 4D). These results demonstrated that overexpression of SlWUS strongly delays low light-induced abscission, while silencing SlWUS enhanced low light-induced abscission (Figure 4D). These results suggest that SlWUS negatively regulates pedicel abscission and that the SlCLV3-SlWUS regulatory mechanism also operates in the AZ.
SlWUS directly binds to the SlKD1 and SlFUL2 promoters and represses their expression in the AZ
To elucidate the mechanism by which the SlCLV3-SlWUS module regulates abscission, we performed a DNA affinity purification sequencing (DAP-seq) analysis to identify SlWUS targets. We obtained 24,032 highly reliable SlWUS-binding sites, of which ∼19% were located upstream from transcription start sites (Supplemental Figure S12A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis further showed that phytohormone signal transduction is significantly enriched among these putative target genes (Supplemental Figure S12B). We, therefore, tested the expression of genes encoding transcription factors belonging to the auxin and ethylene signaling pathways in the AZs of SlWUS-RNAi and SlWUS-OX plants by RT-qPCR. We established that SlKD1, a gene involved in the regulation of tomato pedicel auxin contents and response gradient (Ma et al., 2015), is bound by SlWUS and is more highly expressed in the AZ of SlWUS-RNAi plants and expressed to lower levels in SlWUS-OX plants relative to WT plants (Figure 5A).
Figure 5.
SlWUS directly represses SlKD1 and SlFUL2 expression in the tomato AZ. A, RT-qPCR analysis of the relative expression levels of auxin- and ethylene-related transcription factor genes in the AZ of WT, SlWUS-RNAi, and SlWUS-OX plants. Data are means ± SD of three independent pools of AZ from different plant lines. Significant differences were determined by two-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; **P < 0.01; ***P < 0.0005. B, RT-qPCR analysis of SlKD1 and SlFUL2 expression levels in the AZ of WT, Slclv3 Slcle2, and Slclv1 Slbam1 plants. Data are means ± SD of three independent pools of AZ from different plant lines. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ****P < 0.0001. C, Genome browser view of the distribution of DAP-seq reads (blue, input; red, WUS; pink, peak location) over the SlKD1 and SlFUL2 genomic regions. D, Yeast one-hybrid assay showing that SlWUS directly binds to the SlKD1 and SlFUL2 promoters. The combination of AD-Rec-P53 and P53-Promoter was used as the positive control, while AD-Empty and KD1-Promoter and FUL2-Promoter were the negative control. Aureobasidin A treatment (200 ng/L) is indicated at the bottom. E, Schematic diagram of the SlKD1 and SlFUL2 promoter fragments containing the binding sequence used for the Y1H assay and EMSA. The gel mobility shift assay revealed direct binding of SlWUS to the “GAATGATT” sequence in the SlKD1 promoter and “TTCATTCA” in the SlFUL2 promoter. The probe sequence from the SlKD1 and SlFUL2 promoters is shown, with red letters representing the binding motif. WT, probe with intact sequence. A mutated probe and a 200-fold excess amount of unlabeled probe were added as competitors to the binding reaction. The retarded bands and the free probes are indicated by arrowheads. F, LUC activity measured after transient infiltration of SlKD1pro:LUC or SlFUL2pro:LUC and 35S:SlWUS constructs in N. benthamiana leaves. Data are means ± SD of three biologically independent treatments. Significant differences were determined by one-way ANOVA with Dunnett’s test; ****P < 0.0001).
Interestingly, ethylene signaling-related genes were upregulated in the SlWUS-RNAi plants, while they were downregulated in the SlWUS-OX plants. In particular, the expression of the tomato fruit maturation regulatory gene SlFUL2 significantly increased in the AZ of SlWUS-RNAi plants and decreased in the SlWUS-OX plants (Figure 5A). These results suggested a role for SlWUS in inducing the auxin pathway and repressing the ethylene pathway in the AZ. We confirmed that the expression of SlKD1 and SlFUL2 decreases in the AZ of Slclv3 Slcle2 and Slclv1 Slbam1 mutant plants with lower SlCLV3 signaling (Figure 5B). Furthermore, we performed a yeast one-hybrid assay to test the binding of SlWUS to the SlKD1 and SlFUL2 promoters, which determined that SlWUS can indeed bind to both promoters (Figure 5D). Additionally, an electrophoretic mobility shift assay (EMSA) showed that a recombinant histidine-tagged SlWUS fusion protein (His-SlWUS) binds to the SlKD1 and SlFUL2 promoters, while the tag His alone did not (Figure 5E). Binding was substantially reduced by the addition of cold competitors with the same promoter sequences, while the addition of mutant competitors did not affect binding (Figure 5E).
We used a luciferase (LUC) transactivation assay in Nicotiana benthamiana leaves to confirm the direct regulation of SlKD1 and SlFUL2 transcription by SlWUS in planta. To this end, we placed the firefly LUC gene under the control of the SlKD1 or SlFUL2 promoters to be used as reporters and drove SlWUS expression by the cauliflower mosaic virus 35S promoter as an effector construct, with the empty vector being used as a negative control. When the effector and each reporter construct were co-infiltrated into N. benthamiana leaves, SlKD1 and SlFUL2 transcriptional activity, as determined by LUC activity, decreased compared to that in control leaves with the empty vector (Figure 5F). However, when the WUS binding sites in the FUL2 and KD1 promoter sequences were mutated, their expression was not significantly different from that in the control (Figure 5F). These results demonstrated that SlWUS suppresses the transcription of SlKD1 and SlFUL2 in tomato AZs.
SlFUL2 promotes tomato flower pedicel abscission
An RNAi construct targeting both FUL1 and FUL2 (FUL1/FUL2-RNAi) inhibits tomato fruit ripening and severely perturbs ethylene production (Bemer et al., 2012; Wang et al., 2014), but FUL1 and FUL2 function in pedicel abscission is not clear. Interestingly, we noticed that SlFUL1 is not expressed in the AZ (Figure 6A). To investigate the contribution of SlFUL2 to abscission, we measured the abscission rates of WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX transgenic plants. At 16 h after flower removal, 45% of WT pedicels, 18% of pedicels from SlFUL1/SlFUL2-RNAi line 11, and 32% of pedicels from SlFUL1/SlFUL2-RNAi line 36 had abscised (Figure 6B), while the corresponding values from the SlFUL2-OX line 2 and SlFUL2-OX line 13 were 95% and 77%, respectively (Figure 6B). These results indicated that SlFUL2 enhances flower pedicel abscission.
Figure 6.
SlFUL2 positively regulates flower pedicel abscission. A, RT-qPCR analysis of SlFUL1 and SlFUL2 relative expression levels at 0, 2, 4, 8, and 16 h after flower removal in the AZ. Data are means ± SD of three independent pools of AZ from different plants. B, Time course of pedicel abscission in the WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX plants at 8 and 16 h after flower removal. Data are means ± SD of three independent tests in each plant line, with at least 10 pedicels per treatment. Significant differences were determined by two-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; **P < 0.01; ***P < 0.0005; ****P < 0.0001). C, Ethylene production in the AZ of WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX plants, analyzed 8 h after flower removal. Data are means ± SD of three independent tests in each plant line. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; **P < 0.01.
We next measured ethylene production from the pedicels of WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX plants at 8 h after flower removal. Compared to that in the WT, ethylene production in SlFUL1/SlFUL2-RNAi line 11 and SlFUL1/SlFUL2-RNAi line 36 had dropped 3.9- and 2.1-fold, respectively (Figure 6C). In contrast, ethylene production in SlFUL2-OX line 2 and SlFUL2-OX line 13 was 2- and 1.6-fold higher than that in the WT (Figure 6C). Furthermore, we examined the expression of ethylene biosynthesis-related genes such as 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO) and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) in WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX AZs. Compared to that in WT plants, the expression of ACO5, ACO6, ACS1B, ACS2, ACS4, and ACS6 was lower in SlFUL1/SlFUL2-RNAi AZs and higher in SlFUL2-OX AZs (Supplemental Figure S13). These results were consistent with the notion that SlFUL2 accelerates pedicel abscission by promoting ethylene production in the AZ.
SlKD1 and SlFUL2 act downstream of the SlCLV3-SlWUS signaling module during low light-induced abscission
The above results prompted us to ask whether auxin response and ethylene production mediated by SlKD1 and SlFUL2 might be key to the SlCLV3-WUS regulatory module for low light-induced abscission. Accordingly, we compared GUS staining in the pedicels of plants harboring the auxin response reporter DR5:GUS and grown either under low-light or normal-light conditions (Mao et al., 1989; Wien et al., 1989; Taylor and Whitelaw, 2001; Dong et al., 2021). The presence of GUS staining indicates an active auxin response pathway (Ulmasov et al., 1997; Koenig et al., 2009). We observed a clear gradient in GUS staining between the distal and proximal sides of the pedicels under normal-light conditions, but no detectable staining in the AZs from plants under low-light conditions (Supplemental Figure S14B). To better understand the regulatory effect of the SlCLV3-SlWUS signaling module on auxin response in the AZ, we crossed the DR5:GUS reporter into the SlWUS-RNAi, Slclv3 Slcle2, and Slclv1 Slbam1 genotypes. We detected GUS staining in the distal side, but not the proximal side, of the AZ in SlWUS-RNAi DR5:GUS plants (Figure 7A), while GUS staining in the AZ of Slclv3 Slcle2 and Slclv1 Slbam1 plants was similar to that observed in WT plants (Figure 7A).
Figure 7.
SlKD1 and SlFUL2 act downstream of the low light-induced SlCLV3 signaling pathway. A, Expression pattern of DR5:GUS in the AZ of WT, Slclv3 Slcle2, Slclv1 Slbam1, and SlWUS-RNAi plants. Transverse sections were made from the distal region, the distal side of the AZ, the proximal side of the AZ, and the proximal region. Three independent plant lines were observed, with at least 10 pedicels per plant line. B and C, Ethylene production in the AZ of WT, Slclv3 Slcle2 (B), Slclv1 lbam1 (B), SlWUS-RNAi (C), and SlWUS-OX (C) plants, analyzed 8 h after flower removal. Data are means ± SD of three independent tests, with at least 10 pedicels from different plant lines per test. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; ***P < 0.0005. D and E, RT-qPCR analysis of SlKD1 (D) and SlFUL2 (E) expression levels in the AZ under normal light and shading conditions. Data are means ± SD of three independent pools of AZ from different plants. Significant differences were determined by Student’s t test; *P < 0.05; **P < 0.01. F, Frequency of flower abscission in WT, SlFUL1/SlFUL2-RNAi, SlKD1-RNAi, and SlFUL1/SlFUL2-RNAi SlKD1-RNAi plants under low-light conditions. Flower abscission was scored until fruit set. Six plants were scored for each genotype. Significant differences were determined by one-way ANOVA with Tukey’s test; *P < 0.05; ****P < 0.0001. G, Number of days to reach 50% abscised flowers in WT, Slclv3 Slcle2, Slclv3 Slcle2 SlFUL2-OX, Slclv3 Slcle2 SlKD1-OX, and Slclv3 Slcle2 SlFUL2-OX SlKD1-OX plants. Six independent tests were scored, with at least 10 pedicels per test. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; ****P < 0.0001. H, Number of days to reach 50% abscised flowers in WT, SlWUS-OX, SlWUS-OX SlFUL2-OX, SlWUS-OX SlKD1-OX, and SlWUS-OX SlFUL2-OX SlKD1-OX plants. Six independent tests were scored, with at least 10 pedicels per test. Significant differences were determined by one-way ANOVA with Dunnett’s test compared to the WT; *P < 0.05; ****P < 0.0001.
We noted that ethylene production in the AZs significantly increases under low-light conditions (Supplemental Figure S14C). We also investigated ethylene production in the AZs from SlWUS-RNAi, SlWUS-OX, Slclv3 Slcle2, and Slclv1 Slbam1 plants. Compared to that in the WT, ethylene production in SlWUS-OX, Slclv3 Slcle2, and Slclv1 Slbam1 plants decreased 3-, 6.3-, and 6.8-fold, respectively, at 8 h after flower removal (Figure 7, B and C). IN contrast, ethylene production increased two-fold in SlWUS-RNAi plants (Figure 7B). These results indicated that the SlCLV3-SlWUS signaling module is involved in low light-induced abscission by modulating the auxin response and ethylene production in the AZ.
We next measured the expression levels of SlKD1 and SlFUL2 under low-light conditions using RT-qPCR to obtain additional supporting evidence that they contribute to the SlCLV3-SlWUS signaling pathway. Under low-light conditions, the expression of SlKD1 and SlFUL2 in the AZ increased 17.7- and 4.5-fold, respectively, compared to that under normal-light conditions (Figure 7, D and E). Only 12% of flowers in the SlKD1-RNAi SlFUL1/SlFUL2-RNAi plants had dropped when grown under low-light conditions, which was significantly lower than that in KD1-RNAi and SlFUL1/SlFUL2-RNAi plants (Figure 7F). When we crossed SlKD1-OX or SlFUL2-OX to the Slclv3 Slcle2 double mutant (Supplemental Figure S15, B–E), we observed the partial rescue of the delayed abscission phenotype seen in Slclv3 Slcle2, while it was completely rescued in the Slclv3 Slcle2 SlKD1-OX SlFUL2-OX lines (Figure 7G). There was no significant difference between the WT and SlWUS-OX SlKD1-OX SlFUL2-OX plants (Figure 7H). These results thus indicated that SlKD1 and SlFUL2 function downstream from the SlCLV3-SlWUS signaling pathway.
Discussion
SlCLV3 is essential for low light-induced tomato pedicel abscission
CLV3 and CLE peptides play an essential role in stem cell maintenance (Somssich and Simon, 2012). AZ cells can remain in a quiescent, nondividing state and behave as meristematic cells (Butenko and Simon, 2015). Moreover, CLEs act as stress response signals and are involved in plant stress adaptation (Araya et al., 2014; Gutierrez-Alanis et al., 2017; Takahashi et al., 2018). Here, we present findings on the critical role of SlCLV3 in mediating low light-induced abscission. Our results showed that SlCLV3 is specifically expressed in the AZ, and its expression increases upon shading treatments and flower removal (Figures 1, A and B; Supplemental Figure S6E). Knockdown of SlCLV3 transcript levels in TAPG4pro:SlCLV3-RNAi transgenic plants, using the tissue-specific TAPG4 promoter, which is only expressed in the AZ during abscission (Meir et al., 2010), resulted in a significant delay in pedicel abscission and low light-induced flower drop. These results suggest that AZ-derived SlCLV3 plays a role in abscission (Figure 1, D and E; Supplemental Figure S5C). Moreover, the results obtained from the Slclv3 knockouts further validated a role for SlCLV3 in abscission. Indeed, we observed an aberrant inflorescence morphology (fasciation phenotype) (Supplemental Figure S6B) and defective pedicel AZ development in several Slclv3 mutants (Supplemental Figure S6D). The tomato AZ develops simultaneously with the emerging of lateral organs from the shoot apical meristem (SAM) (Szymkowiak et al., 1999). Thus, we observed alterations in lateral organ development in most Slclv3 knockout lines, as manifested by abnormally broadened (fasciated) inflorescences and irregular cellular architecture of the AZ. In these Slclv3 knockout lines, the CLE motif had disappeared from the SlCLV3 prepropeptide (Supplementary Figure S17) and there was no abscission because the AZ is highly disorganized. Those alterations in inflorescence and AZ development were not observed in TAPG4pro:SlCLV3-RNAi plants and flower abscission occurred (Figure 1D; Supplemental Figure S5C).
Moreover, we observed a much weaker fasciation phenotype and normal development of the AZ in the Slclv3-8, Slclv3-13, and Slclv3-15 mutants, which we thus used for further analysis in the study. Our data demonstrated that the abscission response under low-light conditions of three Slclv3 mutants is not blocked but greatly impaired (Figure 1E). Slclv3 mutants exhibit delayed pedicel abscission induced by flower removal (Figure 1F). We predicted that the three mutants (Slclv3-8, Slclv3-13, and Slclv3-15) with the weak fasciation phenotype and an apparently well-developed AZ may produce a truncated mature SlCLV3 peptide. The truncated mature SlCLV3 peptide might elicit weak signaling and thus account for the abscission response (Figure 1E). Interestingly, low light-induced abscission was more noticeable in opened flowers and less so in buds (Figure 1B). Fully opened flowers were suggested to be more sensitive to low light-induced abscission than flower buds (Dong et al., 2021). We speculate that the participation of SlCLV3 in mediating AZ abscission may vary as a function of developmental stage.
Eight Arabidopsis CLE genes are expressed in flower organ AZs (Jun et al., 2010). Furthermore, besides SlCLV3, ten soybean (Glycine max) CLE genes have been reported to be expressed in the petiole abscission layer and one litchi CLE gene in AZs in response to ethylene treatment (Li et al., 2015; Kim et al., 2016). A neighbor-joining dendrogram containing the mature tomato SlCLV3 peptide, five Arabidopsis CLEs, and five soybean CLEs showed more than 60% sequence similarity between the soybean CLEs and SlCLV3 (Supplemental Figure S18), suggesting that SlCLV3-mediated abscission might be conserved among different plant species.
SlCLV3 depends on SlCLV1 and SlBAM1 to regulate abscission, as well as transcriptional compensation by SlCLE2
In Arabidopsis, BAM1 and BAM2 function together with CLV1 as redundant CLV3 receptors (DeYoung and Clark, 2008). In WT Arabidopsis plants, CLV1 mRNA is expressed in the rib meristem, while BAM1 mRNA is expressed in the L1 cell layer of the shoot apical meristem, and BAM3 mRNA is expressed in developing vascular strands below emerging primordia. In the Arabidopsis clv1-8 mutant, ectopic expression of BAM1 and BAM3 at high levels in rib meristems can partially replace CLV1 function (Nimchuk et al., 2015). However, we established here that SlCLV1 and SlBAM1 are both expressed in vascular bundles in the pedicel AZ (Supplemental Figure S7) and both responded to low-light stress (Figure 2A). Knocking out only SlCLV1 or SlBAM1 delayed low light-induced abscission, and the effect was stronger when both genes were knocked out (Figure 2B). Compared to treatment with the random sSlCLV3 peptide, application of SlCLV3p induced abscission in the Slclv1 and Slbam1 mutants, but not in the Slclv1 Slbam1 double mutant (Figure 2C), suggesting that SlCLV1 and SlBAM1 are involved in SlCLV3-mediated abscission at least partially independently.
Compensation among ligand and receptor paralogs is critical for stem cell homeostasis (Diss et al., 2014; Nimchuk et al., 2015; El-Brolosy and Stainier, 2017). CLE compensation is an active mechanism in tomato shoot apical meristems, as SlCLE9 upregulation compensates for the loss of CLV3 to regulate stem cell homeostasis (Rodriguez-Leal et al., 2019). Similarly, we observed here that while SlCLE2 itself does not respond to low light, SlCLE2 expression did increase in the Slclv3 knockout plants (Figure 3, A and B). Knocking out CLE2 enhanced the delayed abscission phenotype of Slclv3 mutants under low-light conditions (Figure 3C). The SlCLE2 peptide function also acted through SlCLV1 and SlBAM1 to regulate abscission (Figure 3D). This compensation mechanism for the loss of CLV3 by transcriptional upregulation of one of the paralogs is essential for low light-induced abscission, supporting the theory that diverse genetic mechanisms buffer conserved developmental programs (Rodriguez-Leal et al., 2019).
SlWUS is a core regulatory element for abscission
The expression of WUS was upregulated in the AZ of mutants in constituents of the SlCLV3 signaling pathway. WUS is a key transcription factor and is required for maintaining stem cells in an undifferentiated state during stem cell maintenance and sexual organ development (Deyhle et al., 2007; Ma et al., 2019). In tomato, SlWUS is expressed in the AZ, and abscission initiated by auxin depletion is accompanied by a sharp downregulation of SlWUS expression (Meir et al., 2010; Wang et al., 2013). Our results reveal that SlWUS negatively regulates tomato pedicel abscission (Figure 4, B and C). These results demonstrate that the SlCLV3-SlWUS module operates in the AZ to regulate abscission in tomato.
Previous chromatin immunoprecipitation sequencing (ChIP-seq) analyses demonstrated that Arabidopsis WUS directly mediates the expression of phytohormone-, metabolism-, and development-related genes to maintain the stem cell niche (Busch et al., 2010). In our DAP-seq analysis, we determined that SlWUS binds to multiple DNA motifs (Supplemental Table S2), partially matching the Arabidopsis WUS consensus-binding site (Busch et al., 2010). We also identified the binding site TTCATTCA (Supplemental Table S2). Several transcription factors related to auxin and ethylene signaling were targeted by SlWUS. Notably, SlKD1, an important transcription factor that regulates auxin contents and the response gradient in tomato pedicel abscission, was such a target, and its expression was downregulated in SlWUS-OX AZs and upregulated in SlWUS-RNAi AZs. These findings were consistent with the observed disturbed auxin response gradient in the AZ of SlWUS-RNAi plants (Figures 5A and 7A). Our results suggest a direct link between SlWUS and the KNOX family gene SlKD1 in regulating tomato pedicel abscission.
We also identified SlFUL2 as a SlWUS target. A ChIP-chip study conducted by Fujisawa et al. (2014) showed that the expression of the ethylene biosynthesis pathway genes SlACS2, SlACS4, SlACO1, and SlACO6 are SlFUL1/SlFUL2-dependent during fruit ripening (Fujisawa et al., 2014). Synthesis of 1-aminocyclopropane-1-carboxylic acid is considered to be the rate-limiting step in ethylene production and FUL genes appear to modulate the expression of SlACS2 and SlACS4 during fruit ripening (Fujisawa et al., 2014). The expression of SlACS1A, SlACS2, SlACS6, SlACO3, and SlACO5 was upregulated in the AZ after flower removal (Meir et al., 2010). In this study, we also showed that SlFUL2 expression was upregulated and downregulated in the AZ of SlWUS-RNAi and SlWUS-OX plants, respectively (Figure 5A). Flower removal induced SlFUL2 but not SlFUL1 expression (Figure 6A), and pedicel abscission was delayed and accelerated in SlFUL1/SlFUL2-RNAi and SlFUL2-OX plants, respectively (Figure 6B). SlACS2, SlACS4, SlACS6, SlACO5, and SlACO6 were downregulated in the AZ of the SlFUL1/SlFUL2-RNAi plants and upregulated in the AZ of SlFUL2-OX plants (Supplemental Figure S13). Consistent with these observations, ethylene production decreased in the AZ of SlWUS-OX plants but increased in the SlWUS-RNAi lines (Figure 7C) and decreased in Slclv3 Slcle2 and Slclv1 Slbam1 plants (Figure 7B). The fact that neither knockout of SlCLV3 and SlCLE2 nor that of SlCLV1 and SlBAM1 completely blocked ethylene production (Figure 7B) suggests that pedicel abscission was not arrested, only delayed. This phenomenon was also observed when explants were pretreated with the ethylene action inhibitor 1-methylcyclopropene before flowers were removed (Meir et al., 2010). We also attempted to treat the pedicels of WT, Slclv3, and TAPG4pro:SlCLV3-RNAi plants directly with ethylene (Supplemental Figure S21). Ethylene treatment significantly accelerated pedicel abscission in all genotypes tested. However, ethylene treatment did not cause the Slclv3 mutant to return to the abscission rate seen for the WT. These data suggest that there are other components downstream of the CLV3-CLV1/BAM1 module that trigger cell separation in the AZ.
Overexpressing SlKD1 or SlFUL2 partially compensated for the delayed abscission of the SlWUS-OX transgenic plants (Figure 7G). SlWUS-OX SlKD1-OX SlFUL2-OX lines exhibited a complete rescue of the abscission defect caused by SlWUS-OX, suggesting that SlKD1 and SlFUL2 function downstream of SlWUS (Figure 7G). Arabidopsis WUS was reported to regulate auxin signaling and associated responses as well as cytokinin signaling to maintain SAM homeostasis (Ma et al., 2019). Our data indicate that SlWUS is a master regulator of auxin and ethylene homeostasis during abscission via its repression of SlKD1 and SlFUL2 expression. Moreover, the acquisition of ethylene sensitivity in the AZ was shown to be associated with altered expression of auxin-regulated genes during abscission induced by flower removal, which is consistent with the above notion (Meir et al., 2010). The acquisition of ethylene sensitivity in the AZ due to increased SlKD1 expression was associated with the upregulation of genes encoding auxin efflux carriers and auxin response factors and downregulation of IAA-amino acid hydrolases, Aux/IAA, and SMALL AUXIN UPREGULATED factors (Meir et al., 2015).
Low light triggers the SlCLV3-SlWUS module to disturb phytohormone homeostasis and induce tomato flower drop
Low light-induced plant organ abscission is associated with a change in auxin responses and ethylene production (Taylor and Whitelaw, 2001). However, the underlying mechanistic details remain unknown. Low light induces the expression of multiple genes encoding components of the CLV-WUS signaling pathway, including SlCLV3, SlCLV1, and SlBAM1. An analysis of the SlCLV3, SlCLE2, SlCLV1, and SlBAM1 promoter sequences using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) revealed the presence of a light response cis-acting regulatory G-box element in each of these promoters. This discovery suggests that a transcription factor related to light signaling may directly regulate the transcription of these genes, while blocking the SlCLV-SlWUS signaling module in tomato pedicels significantly delays low light-induced abscission (Figures 1 and 2; Supplemental Figure S22). Furthermore, compared to WT plants, the Slclv3 Slcle2 and Slclv1 Slbam1 mutants produced less ethylene and maintained the auxin response gradient in the AZ (Figure 7). Our results also showed that SlKD1 and SlFUL2 expression in the AZ is upregulated under low-light conditions but downregulated in the Slclv1 Slbam1 and Slclv3 Slcle2 mutants (Figures 5B and 7, D and E). Moreover, the Slclv3 Slcle2 abscission defect was rescued by overexpression of SlKD1 and SlFUL2 (Figure 7H). Collectively, the evidence presented here demonstrates that the SlCLV3-SlWUS signaling module is a key element connecting low-light stress and phytohormone signaling during abscission. Based on our results, we propose that the SlCLV3-SlWUS module controls the low light-induced abscission of the tomato flower pedicel (Figure 8).
Figure 8.
Model of low light-induced modulation of tomato flower pedicel abscission by SlCLV3- SlWUS. Low light stimulates the accumulation of SlCLV3 in the AZ, which is perceived by SlCLV1 and SlBAM1. After a series of signal transmissions, SlWUS expression is repressed in the AZ. In the absence of SlCLV3 (as in Slclv3 mutants), SlCLE2 can compensate for its function in regulating abscission. SlWUS acts as a negative regulator of abscission. Upon activation of the SlCLV3-SlWUS signaling pathway, the expression of SlKD1 and SlFUL2 is induced, the auxin response gradient in the AZ is disturbed, and ethylene production increases, leading to abscission.
Our previous study showed that SlIDL6 also regulates low light-induced tomato flower abscission (Li et al., 2021). SlIDL6 is expressed 16 h after flower removal, and SlCLV3 is thus induced prior to SlIDL6 in the AZ. Based on early work, tomato pedicel abscission was divided into an early and a later stage (Meir et al., 2010). SlIDL6 mainly functions in the later stage of abscission, while SlCLV3-SlWUS regulates the early stage of abscission. The expression of SlIDL6 decreased in the AZ of CLV signaling mutants 16 h after flower removal relative to the WT (Supplemental Figure S20A). We detected no significant difference in the expression of SlCLV3 between WT and Slidl6 knockout plants under low-light conditions (Supplemental Figure S20B). It is possible that SlIDL6 functions downstream of SlCLV3-SlWUS signaling. However, the Arabidopsis IDA-HAE/HSL2 signaling module blocks BP activity, which allows KNAT2 and KNAT6 expression to induce floral organ abscission (Shi et al., 2011). The litchi homolog of Arabidopsis KNAT1, LcKNAT1, inhibits the expression of ACSs, ACOs, and ETHYLENE-INSENSITIVE3-LIKEs (EILs), as well as that of genes encoding cell wall-remodeling enzymes that cause fruit abscission (Zhao et al., 2020; Ma et al., 2021). In tomato, a BP/KNAT1 homolog has yet to be reported to play a role in organ abscission, nor has it been shown that the IDA-HAE module regulates the expression of SlKD1 and ethylene production in the AZ. Therefore, there is still a possibility that the IDA-HAE and CLV3-WUS signaling modules regulate phytohormone signaling and cell separation in parallel pathways.
The different downstream components of SlCLV3-SlWUS signaling in the AZ and SAM are genetic engineering candidates for breeding larger tomato fruit
Shoot apical meristem development and abscission are two distinct developmental programs apparently regulated by the same SlCLV3-SlWUS signaling cascade; the underlying mechanism of this process deserves further discussion. First, the AZ is a meristem-like stem cell niche, and AZ cell separation may be conceived as a type of terminal differentiation. CLV3-WUS signaling is involved in this process by regulating the maintenance of the stem cell niche in the AZ. It is worth noting that three weak Slclv3 alleles showed mild fasciation phenotypes but considerable abscission defects. We cannot rule out the possibility that these three independent mutants still have a weak CLV3 signal, since they may still produce a truncated peptide. However, we also observed delayed abscission in the SlCLV3-RNAi lines, supporting the idea that even partial impaired CLV3 signaling will have a profound effect on abscission. The AZ might be more sensitive to the CLV3 signal than the SAM during inflorescence meristem development. Second, the genes acting downstream of CLV3-WUS in the AZ are distinct from those acting in the SAM. Although overexpression of CsFUL1A in cucumber (Cucumis sativus) increased flower organ number (petals and carpels), the encoded protein can directly bind to the CsWUS promoter to stimulate its transcription (Che et al., 2020). Although we detected the expression of SlFUL2 in the tomato SAM by RT-qPCR, SlFUL2-OX plants did not exhibit WUS-related phenotypes (Supplementary Figure S19). SlKD1 was also expressed in the SAM, as seen by RT-qPCR, but again its overexpression lines did not show a WUS-related phenotype (Supplemental Figure S19). It is perhaps not surprising that a transcription factor would display different functions in a tissue-dependent or developmentally regulated manner, with the possible contribution of different posttranscriptional and/or posttranslational regulation of SlWUS, SlFUL2, and SlKD1 function in the AZ and the shoot apical meristem. For the plant, impaired CLV3 signaling would lead to larger fruits by increasing locule number, which would require a concomitant strengthening of the AZ against fruit separation from the peduncle.
The use of the tomato jointless mutation or dipping flowers in an auxin solution prevents tomato flower and fruit abscission, thus increasing agricultural yield (Mao et al., 2000; Meir et al., 2010). The CLV-WUS pathway has been a target of selection during crop domestication to enhance agricultural yields (Chen et al., 2021; Fletcher, 2018; Li et al., 2018). Using CRISPR/Cas9 to edit maize (Zea mays) and tomato CLV3 promoters can enhance crop yield by increasing fruit size (Li et al., 2018; Liu et al., 2021). We showed that the Slclv3, Slclv1, Slclv1 Slbam1, and Slclv3 Slcle2 mutants have fewer flowers drop under low-light conditions. More flowers, larger and more fruit locules, and lower incidence of abscission during low-light conditions could contribute to greater yield.
Materials and methods
Plant material and growth conditions
Tomato (S. lycopersicum cultivar “Ailsa Craig”) and transgenic plants were grown in a greenhouse (16 h of light at 25°C, 8 h of dark at 15°C). Shading treatment was performed as previously described (Li et al., 2021). The photosynthetically active radiation levels under normal- and low-light conditions were 600 and 180 μmol m−2 s−1, respectively. For ethylene treatments, an ethylene concentration of 20 μL L−1 was used.
Nicotiana benthamiana plants were grown in a growth chamber (LPH-350S, NK system, Japan) under the following conditions: 25°C/18°C (day/night), a photoperiod of 16-h light/8-h dark, relative humidity of 70%, and a PPFD of 300 μmol m−2 s−1.
Vector construction and plant transformation
Two single guide RNAs (sgRNAs) were designed to target SlCLV3, SlCLE2, SlCLV1, and SlBAM1 to obtain genome-edited single mutants in the respective genes. To generate Slclv3 Slcle2 and Slclv1 Slbam1 double mutants, a single sgRNA per gene was designed (see Supplemental Table S3 for a list of the sgRNA sequences used in this study). The U6-26-SlCLE-gRNA cassettes were cloned into the CRISPR/Cas9 binary vector, pCBC-DT1T2-tomatoU6, to generate pCBC-DT1T2-tomatoU6-SlCLE. Each primary transformant (T0) was genotyped by sequencing PCR products amplified from the relevant target regions.
The WUS coding sequence was cloned into the pTA7002 vector digested with the XhoI and SpeI restriction enzymes to generate the pTA7002-WUS construct. pTA7002-WUS was then transformed into Agrobacterium (Agrobacterium tumefaciens) strain LBA4404 using electroporation. The resulting positive colonies were used to transform WT tomato Ailsa Craig by leaf disc cocultivation, as previously described (Wang et al., 2018a, 2018b). Genomic DNA was extracted using a Plant Genomic DNA Extraction Kit (TIANGEN, Beijing, China). Each plant was examined by PCR for the presence of the pTA7002-WUS construct with primers designed to recognize the hygromycin resistance gene. Positive transgenic plants were further examined for gene expression after spraying with a solution of 1 mg/L dexamethasone. The three T2 lines with the highest expression levels were selected for phenotypic investigation and follow-up experiments.
A 2,400-bp fragment of the TAPG4 promoter and a 282-bp fragment of the CLV3 coding sequence were amplified and subcloned into the binary vector GSA1285 (Ma et al., 2015). A 2,000-bp CLV3 promoter fragment was amplified from tomato genomic DNA and cloned into the pCAMBIA3301 vector (Biovector) digested with BamHI and NcoI. The TAPG4pro:CLV3-RNAi and CLV3pro:GUS vectors were transformed into Agrobacterium strain LBA4404 by electroporation and transformed into WT tomato as described above. Positive transformants were identified by PCR with primers designed to recognize the resistance marker gene. Positive TAPG4pro:SlCLV3-RNAi transformants were further identified for gene expression by RT-qPCR. Positive CLV3pro:GUS transformants were further identified by GUS staining. The three T2 lines were selected for follow-up experiments.
VIGS
The VIGS tool on the Sol Genomics Network (https://solgenomics.net/) homepage was used to design the SlCLE silencing fragments, which were amplified by PCR and ligated into the pTRV2 vector to generate the pTRV2-CLE constructs. The pTRV2-CLE, pTRV2 empty vector, and pTRV1 empty vectors were individually transformed into Agrobacterium strain GV3101. A mixture of cultures containing pTRV1 and pTRV2-CLE was used for infection as previously described (Liu et al., 2002). Silencing efficiency was determined when the flowers bloomed using RT-qPCR. The primers are listed in Supplemental Data Set S1.
Survey of abscission phenotypes
The flower drop percentage of the WT and mutants under normal-light and low-light conditions was calculated when each inflorescence set fruit. For pedicel explants, the abscission rate was determined as previously described (Wang et al., 2005). The SlWUS-OX pedicels were embedded into 1% (w/v) agar medium containing 1 mg/L dexamethasone and compared to pedicels embedded in 1% (w/v) agar medium containing the equivalent volume of ethanol (control). For peptide treatments, pedicel explants from WT and transgenic plants were placed into a 96-well microtiter plate with liquid containing different concentrations of CLV3p, CLE2p, or random peptides, making sure that the AZ was not exposed to the liquid. The peptides were synthesized by Saibaisheng (China).
RNA extraction and RT-qPCR analysis
Total RNA was extracted from the pedicel AZ using an RNA pure plant kit (CWBIO, Cambridge, MA, USA). One microgram of total RNA was used for first-strand cDNA synthesis with PrimeScriptRT Master Mix (Takara, Kusatsu, Japan) (20 μL reaction containing 4 μL 5 × PrimerScript RT Master Mix). qPCR was performed using gene-specific primers and the TB Green® Premix Ex Taq II (Takara) reaction system (a 20-μL reaction containing 2 μL cDNA (20 µL RT reaction system production was diluted four-fold in ddH2O), 200 nM of each primer, and 4 μL TB Green Premix EX Taq II) with a qTOWER3/G real-time system (Analytik Jena). The program consisted of 95°C for 30 s, 95°C for 5 s (40 cycles), and 60°C for 30 s (40 cycles). The tomato ACTIN gene (NCBI: NM_001330119.1) was used as an internal control. Primers are listed in Supplemental Data Set S1.
In situ hybridization
Unique sequences for SlCLV3, SlWUS, SlCLV1, and SlBAM1 transcripts were amplified from WT cDNA using TaKaRa Ex Taq polymerase and ligated into the pSPT18 and pSPT19 vectors (Roche, Basel, Switzerland). Constructs were linearized, and T7 or SP6 RNA polymerase was used for the RNA labeling reaction according to the standard protocol (Roche). Section preparation and in situ hybridization analysis were performed as previously described (Wang et al., 2018b).
DAP-seq and data analysis
The SlWUS coding sequence was fused to the Halo tag, and a DAP-seq genomic DNA library was prepared. Purification of the WUS protein and the DAP reaction were completed by Gene Denovo Biotechnology Co. (Guangzhou, China). The DNA libraries were sequenced on an Illumina HiSeq 4000 instrument by Gene Denovo Biotechnology Co. (Guangzhou, China). DAP-seq reads were aligned to the Tomato Genome version SL4.0 (https://solgenomics.net/organism/Solanum_lycopersicum/genome) using Bowtie2 (version: 2.2.5). MACS2 (version:2.1.2) software was used to identify read-enriched regions from the DAP-seq data. Peak-related genes were annotated using the CHIPseeker R package (Yu et al., 2015). The MEME suite (http://meme-suite.org/) was used to detect motifs. MEME (http://meme-suite.org/tools/meme) and DREME (http://meme-suite.org/tools/dreme) were used to detect long and short consensus sequences, respectively. Analysis of KEGG term enrichment was performed (Kanehisa et al., 2007).
Yeast one-hybrid assay
The SlWUS coding sequence was cloned into the pGADT7 vector (Clontech). The putative SlWUS target sites in the SlFUL2 and SlKD1 promoters were individually cloned into the pAbAi vector (Clontech). The assay was performed with the Matchmaker Gold Yeast One-Hybrid Library Screening System Kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions.
EMSA
The SlWUS coding sequence was cloned into the pET-30a vector (Biovector) to produce and purify recombinant His-WUS fusion protein. The resulting pET30a-WUS construct was transformed into Escherichia coli BL21 (DE3) cells (TIANGEN). Positive colonies were cultured in LB liquid medium containing 50 µg/mL kanamycin at 37°C until OD600 reached 0.6–0.8. Isopropyl β-D-1-thiogalactopyranoside was then added to the culture to a final concentration of 0.5 mM to induce recombinant protein production. Recombinant protein purification was performed using a His Tag Protein Purification Kit (CWBIO). EMSA was performed using a Chemiluminescent EMSA Kit (Beyotime). The 5′ biotin-labeled probe was prepared by the Saibaisheng Company (China). All oligonucleotide probes used in this study are listed in Supplemental Data Set S1.
LUC assay
Promoter fragments (2,600 bp) upstream of the SlFUL2 and SlKD1 coding sequences were individually cloned into the pGreenII-0800-LUC vector (Biovector) as the reporters, while the SlWUS coding sequence was cloned into pCAMBIA1300 (Biovector) as the effector. The reporter and effector constructs were individually transformed into Agrobacterium strain EHA105 (TIANGEN). Positive colonies were grown in YEP medium and resuspended in infiltration buffer (10 mM MES-KOH pH 5.2, 10 mM MgCl2, 100 μM acetosyringone) to an OD600 of 1.0 before being infiltrated into N. benthamiana leaves (Che et al., 2020). Firefly LUC and Renilla LUC activities were measured using the Dual-LUC Reporter Assay System (Promega, Madison, WI, USA). Renilla LUC activity was used as the internal control. The primers used are listed in Supplemental Data Set S1.
GUS staining
Tomato pedicels were placed into GUS staining solution (Real-Times Biotechnology Co. Beijing, China) and incubated at 37°C for 16 h in the dark. The treated pedicels were cleared from chlorophyll and stored in 70% (v/v) ethanol as previously described (Ma et al., 2015). Images shown are representative of >20 observed pedicels from three independent plants per genotype and construct.
Ethylene measurements
At least three biological replicates were analyzed. Fifteen flower pedicels were pooled for each replicate. The pedicels were cultured on 1% (w/v) agar medium for 8 h after their flowers were removed and then were weighed and placed in a sealed 10-mL glass jar at 23°C for 1 h. A syringe was then used to take out 1 mL of gas from the sealed jars. Ethylene production was measured as previously described (Wang et al., 2021a).
Phylogenetic analysis
Phylogenetic analysis of the Arabidopsis, soybean, and litchi CLEs and SlCLV3 amino acid sequences was performed using MEGA11 software with the neighbor-joining method.
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 8.0 software. In the case of comparisons between two sample groups, a Student’s t test was applied. For multiple comparisons, significance was determined by analysis of variance (ANOVA), followed by Dunnett’s, Sidak’s, or Tukey’s test (Supplemental Data Set S2).
Accession numbers
Sequence data from this article can be found in the Genome Database for Tomato (http://www.solgenomics.net/) or National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) under accession numbers SlCLV3 (Solyc11g071380); SlCLE1 (Solyc01g014100); SlCLE2 (Solyc01g098890); SlCLE3 (Solyc02g067550); SlCLE4 (Solyc02g087470); SlCLE5 (Solyc03g025960); SlCLE6 (Solyc05g006610); SlCLE6 (Solyc05g006610); SlCLE7 (Solyc05g007650); SlCLE8 (Solyc05g053630); SlCLE9 (Solyc06g074060); SlCLE10 (Solyc07g053370); SlCLE11 (Solyc07g062670); SlCLE12 (Solyc09g061410); SlCLE13 (Solyc09g091810); SlCLE14 (Solyc11g066120); SlCLV1 (Solyc04g081590); SlCLV2 (Solyc04g056640); SlCRN (Solyc05g023760); SlBAM1 (Solyc02g091840); SlBAM2 (Solyc03g043770); SlBAM3 (Solyc01g080770); SlBAM4 (Solyc01g103530); SlWUS (Solyc02g083950); SlKD1 (Solyc06g072480); SlFUL1 (Solyc06g069430); SlFUL2 (Solyc03g114830); SlARF1 (Solyc01g103050); SlARF3 (Solyc02g077560); SlARF10A (Solyc08g008380); SlARF11 (Solyc03g113410); SlEIL3-4 (Solyc01g006650); SlEIL3-2 (Solyc01g014480); SlEIL3 (Solyc01g096810); SlERF1 (Solyc05g051200); SlEIN4-like (Solyc05g055070); SlEIN4 (Solyc06g051610); SlEIL4 (Solyc06g073730); SlERF2-like (Solyc08g0072230); SlERF-C3 (Solyc09g066360); SlTSRF1 (Solyc09g089930); SlIDL6 (Solyc06g050140); SlERF-C7 (Solyc11g011740); SlACO1 (NM_001247095.2); SlACO2 (NM_001329913.1); SlACO3 (NM_001309213.1); SlACO4 (NM_001246938.2); SlACO5 (NM_001247108.1); SlACO6 (NM_001247709.2); SlACS1A (NM_001246993.2); SlACS1B (NM_001279342.3); SlACS2 (NM_001247249.3); SlACS3 (NM_001247097.2); SlACS4 (NM_001246946.1); SlACS5 (NM_001247227.2); SlACS6 (NM_001247235.2); SlACS7 (NM_001247417.1); and SlACS8 (NM_001247231.2). Raw data of DAP-seq have been deposited at NCBI GEO repository under the following accession number GSE210149.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of the SlCLE gene family under light and shading conditions.
Supplemental Figure S2. Determination of the silencing efficiency in SlCLE VIGS plants.
Supplemental Figure S3. Abscission phenotype of candidate SlCLE VIGS plants.
Supplemental Figure S4. In situ hybridization analysis of SlCLV3 in the AZ.
Supplemental Figure S5. Identification and inflorescence phenotype of TAPG4pro:SlCLV3-RNAi plants.
Supplemental Figure S6. Morphological, histological, and abscission analysis of pedicels from WT and Slclv3 plants.
Supplemental Figure S7. In situ hybridization analysis of SlCLV1 and SlBAM1 in the AZ.
Supplemental Figure S8. The 2rd and 3rd inflorescence phenotypes and AZ phenotype in receptor mutants.
Supplemental Figure S9. Inflorescence phenotypes and locule number in Slcle2 and Slclv3 Slcle2 plants.
Supplemental Figure S10. Expression analysis of SlWUS in the AZ of WT, SlWUS-RNAi, and SlWUS-OX plants.
Supplemental Figure S11. In situ hybridization analysis of SlWUS in the AZ.
Supplemental Figure S12. DAP-seq analysis of direct SlWUS target genes.
Supplemental Figure S13. Expression of ethylene biosynthesis genes in the AZ of WT, SlFUL1/SlFUL2-RNAi, and SlFUL2-OX plants.
Supplemental Figure S14. Low light-induced pedicel abscission and changes in ethylene production and auxin response.
Supplemental Figure S15. Identification of SlFUL2 and SlKD1 transgenic plants.
Supplemental Figure S16. Correlation analysis.
Supplemental Figure S17. Alignment of amino acid sequences from SlCLV3 for the 10 slclv3 mutants.
Supplemental Figure S18. Neighbor-joining phylogenetic tree of the tomato, Arabidopsis, and soybean CLE peptides.
Supplemental Figure S19. Phenotypic analysis of SlKD1 and SlFUL2.
Supplemental Figure S20 . Expression analysis of SlIDL6 in CLV3 signaling mutants and SlCLV3 in the Slidl6 mutant under low-light conditions.
Supplemental Figure S21. Abscission rate in the Slclv3 and TAPG4pro:SlCLV3-RNAi plants treated with ethylene.
Supplemental Figure S22. Schematic diagram of promoter fragments from members of the SlCLV3 signaling pathway.
Supplemental Table S1. Expression of SlCLE genes in a transcriptome database of low light-treated pedicels.
Supplemental Table S2. Motifs bound by WUS.
Supplemental Table S3. sgRNAs used for CRISPR/Cas9 mutagenesis and PCR primers used for genotyping.
Supplemental Data Set S1. Primers used in this study.
Supplemental Data Set S2. Summary of statistical analyses.
Supplemental Data Set S3. DAP-seq results for SlWUS.
Supplemental File S1. Multiple sequence alignment used for the phylogenetic tree shown in Supplemental Figure S18A.
Supplemental File S2. MEGA file of the alignment shown in Supplemental Figure S18A.
Supplementary Material
Acknowledgments
We thank Zhibiao Ye (Huazhong Agricultural University) for providing the FUL1/FUL2-RNAi and FUL2-OX transgenic tomato material. We thank Aide Wang (Shenyang Agricultural University) for providing the pTA7002 vector. We thank Ora Hazak (Fribourg University) for sharing the SlCLE16-52 coding sequences. We thank Plant Editors (planteditors.com) for editing this article. We thank the reviewers and editors for their valuable and insightful comments and suggestions.
Funding
This work was supported by the National Key Research and Development Program of China (grant numbers 2018YFD1000800), and the National Natural Science Foundation of China (grant numbers 31991184, 31861143045, U1708232, and 31672197).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Lina Cheng, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Ruizhen Li, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Xiaoyang Wang, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Siqi Ge, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Sai Wang, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Xianfeng Liu, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Jing He, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Cai-Zhong Jiang, Crops Pathology and Genetic Research Unit, United States Department of Agriculture Agricultural Research Service, Albany, California 95616, USA; Department of Plant Sciences, University of California, Los Angeles, California 95616, USA.
Mingfang Qi, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Tao Xu, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
Tianlai Li, College of Horticulture, Shenyang Agricultural University, Shenyang, 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang, China.
L.C., R.L., X.W., S.G., S.W., X.L., and J.H. performed the experiments. T.X. and T.L. designed the research. M.Q., C.J., T.X., and T. L. provided technical support, conceptual advice, and data analysis. L.C. and T.X. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the instructions for Authors (https://academic.oup.com/plcell) are: Tao Xu (syauxutao@syau.edu.cn) and Tianlai Li (tianlaili@126.com).
References
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, Von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PDB, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 841–853 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Simon R (2015) Beyond the meristems: similarities in the CLAVATA3 and INFLORESCENCE DEFICIENT IN ABSCISSION peptide mediated signalling pathways. J Exp Bot 66: 5195–5203 [DOI] [PubMed] [Google Scholar]
- Carbonnel S, Falquet L, Hazak O (2022) Conserved mechanism for perception of root-active CLE peptides. bioRxiv. preprint 10.1101/2022.01.21.477294. [DOI]
- Che G, Gu R, Zhao J, Liu X, Song X, Zi H, Cheng Z, Shen J, Wang Z, Liu R, et al. (2020) Gene regulatory network controlling carpel number variation in cucumber. Development 147: dev.184788. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li W, Gaines C, Buck A, Galli M, Gallavotti A (2021) Structural variation at the maize WUSCHEL1 locus alters stem cell organization in inflorescences. Nat Commun 12: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker EJ, Laux T (2007) WUSCHEL regulates cell differentiation during anther development. Dev Biol 302: 154–159 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Clark SE (2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diss G, Ascencio D, DeLuna A, Landry CR (2014) Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J Exp Zool B Mol Dev Evol 322: 488–499 [DOI] [PubMed] [Google Scholar]
- Dong X, Ma C, Xu T, Reid MS, Jiang CZ, Li T (2021) Auxin response and transport during induction of pedicel abscission in tomato. Hortic Res 8: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos S, Scafidi P, Cardoso V, Leitao AE, Di Lorenzo R, Oliveira CM, Goulao LF (2015) Flower abscission in Vitis vinifera L. triggered by gibberellic acid and shade discloses differences in the underlying metabolic pathways. Front Plant Sci 6: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Stainier DY (2017) Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet 13: e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell LH, Agusti J, Merelo P, Talon M, Tadeo FR (2013) Elucidating mechanisms underlying organ abscission. Plant Sci 199–200: 48–60 [DOI] [PubMed] [Google Scholar]
- Fletcher JC (2018) The CLV-WUS stem cell signaling pathway: a roadmap to crop yield optimization. Plants 7: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC (2020) Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci 25: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito YJTPC (2014) Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Alanis D, Yong-Villalobos L, Jimenez-Sandoval P, Alatorre-Cobos F, Oropeza-Aburto A, Mora-Macias J, Sanchez-Rodriguez F, Cruz-Ramirez A, Herrera-Estrella L (2017) Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Dev Cell 41: 555–570 e553 [DOI] [PubMed] [Google Scholar]
- Hieke S, Menzel CM, Doogan VJ, Lüdders P (2002) The relationship between yield and assimilate supply in lychee (Litchi chinensis Sonn.). J Hortic Sci Biotechnol 77: 326–332 [Google Scholar]
- Hu C, Zhu Y, Cui Y, Cheng K, Liang W, Wei Z, Zhu M, Yin H, Zeng L, Xiao Y, et al. (2018) A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat Plants 4: 205–211 [DOI] [PubMed] [Google Scholar]
- Jun J, Fiume E, Roeder AH, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. (2007) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yang J, Yang R, Sicher RC, Chang C, Tucker ML (2016) Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front Plant Sci 7: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N (2009) Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Li C, Wang Y, Ying P, Ma W, Li JJ (2015) Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front Plant Sci 6: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Shi CL, Wang X, Meng Y, Cheng L, Jiang CZ, Qi M, Xu T, Li T (2021) Inflorescence abscission protein SlIDL6 promotes low light intensity-induced tomato flower abscission. Plant Physiol 186: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu CJNB (2018) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36: 1160–1163 [DOI] [PubMed] [Google Scholar]
- Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu Q, Bartlett M, Jackson D (2021) Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants 7: 287–294 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SJTPJ (2002) Virus‐induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Ma C, Meir S, Xiao L, Tong J, Liu Q, Reid MS, Jiang CZ (2015) A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol 167: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ying P, He Z, Wu H, Li J, Zhao M (2021) The LcKNAT1-LcEIL2/3 regulatory module is involved in fruitlet abscission in litchi. Front Plant Sci 12: 802016–802016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K (2019) WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913 [DOI] [PubMed] [Google Scholar]
- Mao Z, Craker LE, Decoteau DR (1989) Abscission in Coleus: light and phytohormone control. J Exp Bot 40: 1273–1277 [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KV, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154: 1929–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Sundaresan S, Riov J, Agarwal I, Philosoph-Hadas S (2015) Role of auxin depletion in abscission control. Stewart Postharvest Rev 11: 1–15 [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM (2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294–294 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Osborne DJ, Morgan PW (1989) Abscission. Critic Rev Plant Sci 8: 103–129 [Google Scholar]
- Patharkar OR, Walker JC (2016) Core mechanisms regulating developmentally timed and environmentally triggered abscission. Plant Physiol 172: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Gassmann W, Walker JCJPG (2017) Leaf shedding as an anti-bacterial defense in Arabidopsis cauline leaves. PLoS Genet 13: e1007132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE (2001) Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol 126: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt S, Piepho HP, Stintzi A, Schaller A (2020) Peptide signaling for drought-induced tomato flower drop. Science 367: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Xu C, Kwon CT, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, Van Eck J (2019) Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat Genet 51: 786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480. e478 [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M, Ripoll JJ, Yanofsky MF, Pelaz S (2013) The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J 73: 37–49 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol 54: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Simon R (2012) Peptides regulating apical meristem development. Plant Signaling Peptides 16: 25–39 [Google Scholar]
- Szymkowiak, Eugene J, Irish EE (1999) Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 11: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, Shinozaki K (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556: 235–238 [DOI] [PubMed] [Google Scholar]
- Taylor JE, Whitelaw CA (2001) Signals in abscission. New Phytologist 151: 323–340 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker SJSPR (2009) Development of the abscission zone. Stewart Postharvest Rev 5: 5 [Google Scholar]
- Wang R, Li R, Cheng L, Wang X, Fu X, Dong X, Qi M, Jiang C, Xu T, Li T (2021a) SlERF52 regulates SlTIP1;1 expression to accelerate tomato pedicel abscission. Plant Physiol 185: 1829–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lu G, Hou Z, Luo Z, Wang T, Li H, Zhang J, Ye Z (2014) Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J Exp Bot 65: 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Aguirre L, Rodríguez-Leal D, Hendelman A, Benoit M, Lippman ZB (2021b) Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat Plants 7: 419–427 [DOI] [PubMed] [Google Scholar]
- Wang X, Liu D, Li A, Sun X, Zhang R, Wu L, Liang Y, Mao L (2013) Transcriptome analysis of tomato flower pedicel tissues reveals abscission zone-specific modulation of key meristem activity genes. PLoS One 8: e55238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yang H, Chen L, Yang S, Hua D, Wang J (2018a) Truncated BAM receptors interfere the apical meristematic activity in a dominant negative manner when ectopically expressed in Arabidopsis. Plant Sci 269: 20–31 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li T, Meng H, Sun X (2005) Optimal and spatial analysis of hormones, degrading enzymes and isozyme profiles in tomato pedicel explants during ethylene-induced abscission. Plant Growth Regul 46: 97–107 [Google Scholar]
- Wang F, Zheng Z, Yuan Y, Li J, Zhao M (2019) Identification and characterization of HAESA-like genes involved in the fruitlet abscission in litchi. Int J Mol Sci 20: 5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zou W, Xiao Y, Cheng L, Liu Y, Gao S, Shi Z, Jiang Y, Qi M, Xu T (2018b) MicroRNA1917 targets CTR4 splice variants to regulate ethylene responses in tomato. J Exp Bot 69: 1011–1025 [DOI] [PubMed] [Google Scholar]
- Wien H, Turner A, Yang S (1989) Hormonal basis for low light intensity-induced flower bud abscission of pepper. J Am Soc Hortic Sci 114: 981–985 [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu YH, Jiang K, Brooks C, Ogawa-Ohnishi M, Xiong G, Pauly M, et al. (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet 47: 784–792 [DOI] [PubMed] [Google Scholar]
- Xu P, Chen H, Cai WJE (2020) Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep 21: e48967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying P, Li C, Liu X, Xia R, Zhao M, Li J (2016) Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Sci Rep 6: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Mandel T, Kuhlemeier C (2011) Stem cell activation by light guides plant organogenesis. Genes Dev 25: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, He QY (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383 [DOI] [PubMed] [Google Scholar]
- Yuan R, Huang H (1988) Litchi fruit abscission: its patterns, effect of shading and relation to endogenous abscisic acid. Sci Hortic 36: 281–292 [Google Scholar]
- Zhang Y, Yang S, Song Y, Wang J (2014) Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato. BMC Genom 15: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li J (2020) Molecular events involved in fruitlet abscission in litchi. Plants 9: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li C, Ma X, Xia R, Chen J, Liu X, Ying P, Peng M, Wang J, Shi CL, et al. (2020) KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J Exp Bot 71: 4069–4082 [DOI] [PubMed] [Google Scholar]
- Zhu H, Dardick CD, Beers EP, Callanhan AM, Xia R, Yuan R (2011) Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol 11: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu C, Cui Y, Zeng L, Li S, Zhu M, Meng F, Huang S, Long L, Yi J, et al. (2021) Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis. Mol Plant 14: 1119–1134 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu CM, Lin J (2010) Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J 61: 223–233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.