Abstract
Maternal-to-filial nutrition transfer is central to grain development and yield. nitrate transporter 1/peptide transporter (NRT1-PTR)-type transporters typically transport nitrate, peptides, and ions. Here, we report the identification of a maize (Zea mays) NRT1-PTR-type transporter that transports sucrose and glucose. The activity of this sugar transporter, named Sucrose and Glucose Carrier 1 (SUGCAR1), was systematically verified by tracer-labeled sugar uptake and serial electrophysiological studies including two-electrode voltage-clamp, non-invasive microelectrode ion flux estimation assays in Xenopus laevis oocytes and patch clamping in HEK293T cells. ZmSUGCAR1 is specifically expressed in the basal endosperm transfer layer and loss-of-function mutation of ZmSUGCAR1 caused significantly decreased sucrose and glucose contents and subsequent shrinkage of maize kernels. Notably, the ZmSUGCAR1 orthologs SbSUGCAR1 (from Sorghum bicolor) and TaSUGCAR1 (from Triticum aestivum) displayed similar sugar transport activities in oocytes, supporting the functional conservation of SUGCAR1 in closely related cereal species. Thus, the discovery of ZmSUGCAR1 uncovers a type of sugar transporter essential for grain development and opens potential avenues for genetic improvement of seed-filling and yield in maize and other grain crops.
Zea mays Sucrose and Glucose Carrier1 (ZmSUGCAR1) encodes a sugar transporter that functions in grain filling.
IN A NUTSHELL.
Background: Grain development depends on the efficient transport of nutrients from maternal to filial seed tissues. Sucrose is the most abundant form of photo-assimilate in long-distance phloem transport. Early work with maize indicated that a considerable portion of sucrose is hydrolyzed to hexose prior to entering newly forming seeds. However, much of this sucrose could also move directly into the endosperm without being hydrolyzed. To date, evidence supporting such a direct sucrose transporter has been lacking.
Question: Are there any transporters responsible for the direct uptake of sucrose in maize endosperms during grain filling? If so, how does this work?
Findings: We report here an unexpected transporter mediating direct sucrose transfer in maize. The causal gene for a defective kernel phenotype was identified as an NRT1/PTR-type transporter, which typically transports nitrate, peptides, and ions. This protein demonstrated an additional capacity to transport both sucrose and glucose, thus representing a sugar transporter, which we named sucrose and glucose carrier (SUGCAR). Its activity was systematically verified by tracer-labeled sugar uptake and serial electrophysiological studies. Specifically expressed in the basal endosperm transfer layer, a loss-of-function mutation of ZmSUGCAR1 caused a marked decrease in sucrose and glucose and subsequent kernel shrinkage in maize. Notably, orthologs of ZmSUGCAR1 from wheat and sorghum displayed similar sugar transport activities, thus support the functional conservation of this transporter in closely related cereal species.
Next steps: Analyze the structure of ZmSUGCAR1 to identify the potential binding sites of different substrates. If possible, generate mutants affecting the transport of specific substrates to better dissect this multi-functional transporter.
Introduction
Grain development depends on the efficient transport of nutrients from maternal to filial seed tissues (Lalonde et al., 2004; Bihmidine et al., 2013). Key steps occur at the base of the newly forming seed, where soluble sugars predominate in the feeding process essential to grain filling and yield. To date, several distinct families of sugar transporters are known: the monosaccharide transporter (MST)-like gene family including seven subclades, such as sugar transporter proteins (STPs), polyol transporters (PLTs), and tonoplast monosaccharide transporters (TMTs) (Klepek et al., 2005; Buttner, 2007; Schulz et al., 2011; Lalonde and Frommer, 2012), the sucrose transporters (SUT)-type transporters (Carpaneto et al., 2005; Braun and Slewinski, 2009; Kühn and Grof, 2010), and the sugar facilitators of the sugars will eventually be exported transporters (SWEETs) family (Chen et al., 2010; Kühn and Grof, 2010; Baker et al., 2012; Chen et al., 2012; Eom et al., 2015).
Sugar import into developing grains of major crops, however, remains poorly understood (Braun et al., 2014). Seed development can be temporally divided into the pre-storage and storage phases (Zhang et al., 2007). During the pre-storage phase, the basic structure of a crop kernel progressively gains shape through rapid cellular differentiation, a developmental step programmed by hierarchical molecular cascades (Weber et al., 2005; Zhang et al., 2007; Milne et al., 2018). In grains such as in maize (Zea mays), a major phase change accompanies the establishment of the basal endosperm transfer layer (BETL). The BETL mediates nutrient uptake from the extracellular space between the maternal phloem and the newly forming seed (Leroux et al., 2014).
In the developing maize grain, sugar uptake can be best described by the well-supported Shannon hypothesis: (i) sucrose is hydrolyzed by cell wall invertase Z. mays Miniature1 (ZmMN1) in the phloem-unloading zone, (ii) glucose and fructose are released and in turn taken up by the endosperm (Shannon, 1972), and (iii) sucrose is re-synthesized in the endosperm from the two monosaccharides (Shannon et al., 1986; Chourey et al., 1995). The entry of hexoses into endosperm is facilitated at least in part by ZmSWEET4c (Sosso et al., 2015) and associated with the induction of Z. mays Myb-related protein 1 (ZmMRP1), a putative master regulator of BETL cell fate (Zheng and Wang, 2010; Doll et al., 2017). The observation that maize mutants lacking either ZmMN1 or ZmSWEET4c were unable to fully differentiate BETL regions points to these genes constituting a glucose-mediated feed-forward network likely orchestrating BETL development (Barrero et al., 2009; Monjardino et al., 2013; Sosso et al., 2015).
The BETL, once formed, facilitates rapid progression of the storage phase in kernel development and accumulation of sugar-based kernel dry matter. During this phase, the activity of cell-wall invertase in the extracellular space drops. The endosperm then must import sucrose via proton-driven sugar symporters (Cheng et al., 1996; Patrick and Offler, 2001; Chourey et al., 2006; Zhang et al., 2007; Wang et al., 2008; Milne et al., 2018).
The nitrate transporter 1/peptide transporter (NRT1/PTR) family belongs to the major facilitator superfamily (MFS) (Yan, 2015), which is ubiquitously present in all kingdoms of life. In plants, this family is referred to as the NRT1/PTR family (NPF) (Léran et al., 2014). Compared with their counterparts in animals and bacteria, plant NPF members have been shown to transport a wider variety of substrates (Dietrich et al., 2004; Jeong et al., 2004; Rentsch et al., 2007; Tsay et al., 2007; Krouk et al., 2010; Nour-Eldin et al., 2012; Saito et al., 2015; Tal et al., 2016); however, so far sugars were not among these substances. Notably, ZmNPF7.9/Mn2/Dek54 (Zm00001d019294) encodes a low-affinity nitrate transporter expressed in the BETL (Wei et al., 2021), and its knockout mutant bears small kernels with abnormal starch deposition (Guan et al., 2020; Zhou et al., 2021; Wei et al., 2021); metabolite profiling suggested that ZmNPF7.9 may regulate seed development by affecting lipid and amino acid homeostasis (Wei et al., 2021). However, the innate linkage of kernel shrinkage and hindered nitrate transport remains elusive.
Here, using a forward genetic approach, we identified a distinct mutation in the maize sugar transporter gene Sucrose and Glucose Carrier 1 (ZmSUGCAR1) as a variant of ZmNPF7.9/NRT1.5 that was responsible for a poorly filled defective-kernel phenotype. The ZmSUGCAR1 gene is a paralog of AtNPF7.3 and is specifically expressed in the BETL of maize kernels. Our discovery that ZmSUGCAR1 is capable of transporting sucrose and glucose adds sugars as substrate to the spectrum of NPF-class proteins. Given that ZmSUGCAR1 is essential for proper grain development, it is therefore a potential target in future breeding programs for superior seed sugar-filling.
Results
Cloning and characterization of the gene responsible for poorly filled kernels
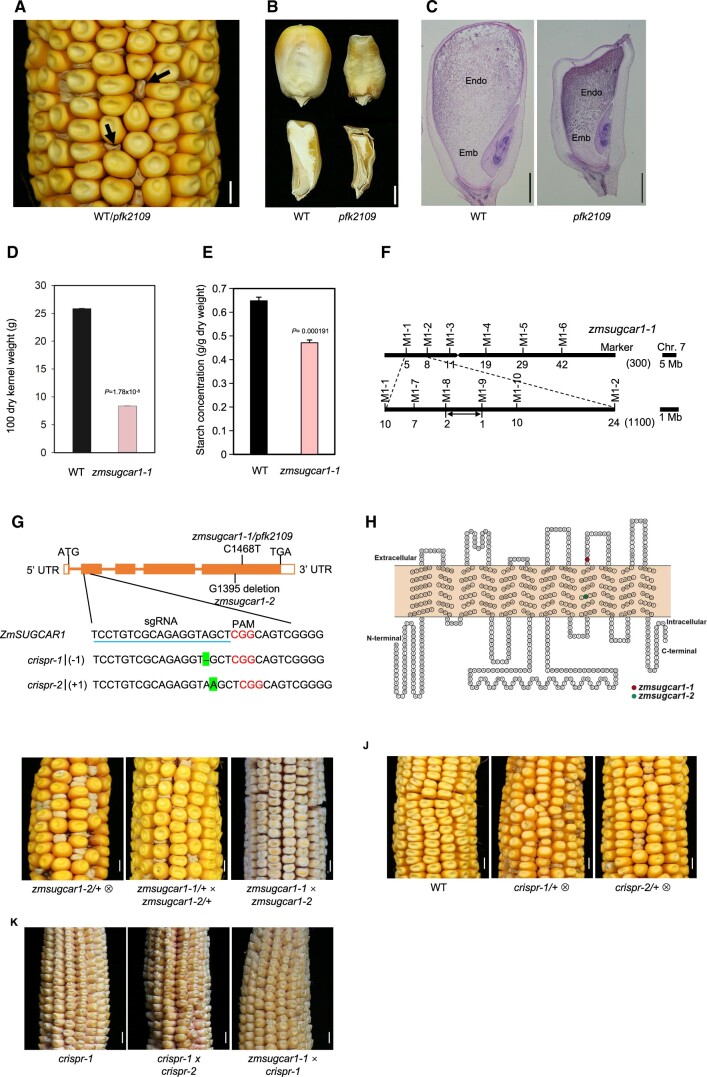
To dissect the molecular mechanisms regulating seed development in maize, we screened an ethyl methanesulfonate (EMS)-mutagenized library for homozygous mutants with altered kernel size. One mutant called poorly filled kernel2109 (pfk2109) showed severe defects in kernel development (Figure 1, A–C and Supplemental Figure S1A). Mutant embryos were able to germinate and develop into morphologically normal and fertile plants (Supplemental Figure S2, A–E). At later developmental stages, however, grain filling and endosperm enlargement were impaired resulting in poorly filled kernels with empty pericarps (Figure 1, A–C and Supplemental Figure S1A). The 100-kernel weight of the mutant line was reduced by approximately 64% compared with that of the wild-type (WT), and the endosperm at maturity contained 27% less starch (Figure 1, D and E).
Figure 1.
Mutant-kernel phenotypes and map-based cloning of ZmSUGCAR1. A, Segregation of the pfk2109 phenotype in a maize ear at 45 DAP. The magnification highlighted poorly filled kernels with empty pericarps (arrows). B, Mature kernels (upper panel) with sagittal sections (lower panel) of WT (left) or pfk2109 (right). C, Histological analysis of WT or pfk2109 kernels at 20 DAP, stained with eosin. Emb, embryo; Endo, endosperm. D, 100 grain weight of WT and zmsugcar1-1 lines at 45 DAP. E, Starch measurement of WT and zmsugcar1-1 kernels at 45 DAP. F, Map-based cloning of the ZmSUGCAR1 variant. The ZmSUGCAR1 was located between the molecular markers M1-8 and M1-9 on chromosome 7. Molecular markers and number of recombinants were indicated, respectively. G, Gene structure of ZmSUGCAR1 and mutation sites of different zmsugcar1 mutant lines. The filled boxes represented exons and the lines represented introns. The point mutation (C1468T) in zmsugcar1-1 and single base-pair deletion (G1395 deletion) in zmsugcar1-2 were shown on the gene-structure diagram. The sgRNA target sequence was underlined and the PAM motif was colored. The single base pair (A) deletion of the crispr-1 allele and a single base pair (A) insertion of the crispr-2 allele were highlighted. H, A 2-D schematic of ZmSUGCAR1 based on OCTOPUS (http://octopus.cbr.su.se/) prediction transmembrane (TM) secondary structure. The red and green colored amino acid was prematurely stopped, respectively, in the zmsugcar1-1 and zmsugcar1-2 mutant. I, A representative self-pollinated ear of zmsugcar1-2 heterozygotes (left) and allelic tests using heterozygotes (middle) and homozygotes (right) of zmsugcar1-1 and zmsugcar1-2. J, The phenotype and genetic test of two independent crispr mutant lines. Representative self-pollinated ears of WT (left) and crispr-1 (middle) and crispr-2 (right) heterozygotes. K, The allelic test of zmsugcar1-1 and crispr-1 (left) mutants. The offspring annotated as crispr1 × crispr-2 (middle) and zmsugcar1-1 × crispr-1 (right). Scale bars, 5 mm for A, I, J, and K and 1 mm for B and C. P < 0.05 indicated significant differences between WT and mutant lines.
The pfk2109 mutation is a recessive mutation since the F2 ears segregated at a 3:1 ratio of normal and defective kernels (Figure 1A and Table 1). To test whether the poorly filled kernel phenotype in zmsugcar1-1 is maternally controlled, we performed reciprocal crosses between the WT and zmsugcar1-1. There was no defective kernel phenotype observed whether the zmsugcar1-1 was used as the maternal parent or not (Supplemental Figure S1B).
Table 1.
Allelic test of ZmSUGCAR1 mutants
| Number of normal kernels | Number of small kernels | χ2 | P | |
|---|---|---|---|---|
| The segregation ratio of zmsugcar1-1/+⊗ | ||||
| 1 | 283 | 94 | 0.000884 | 0.976278 |
| 2 | 300 | 119 | 2.584726 | 0.107899 |
| 3 | 281 | 94 | 0.000889 | 0.976215 |
| 4 | 293 | 101 | 0.084602 | 0.771155 |
| The segregation ratio of zmsugcar1-2/+ × zmsugcar1-1/+ | ||||
| 1 | 310 | 106 | 0.051282 | 0.820847 |
| 2 | 289 | 98 | 0.021533 | 0.883336 |
| 3 | 296 | 101 | 0.041142 | 0.839264 |
| The segregation ratio of crispr-1/+⊗ | ||||
| 1 | 306 | 96 | 0.268657 | 0.604234 |
| 2 | 299 | 97 | 0.053872 | 0.816458 |
| 3 | 315 | 101 | 0.115385 | 0.734095 |
| The segregation ratio of crispr-2/+⊗ | ||||
| 1 | 320 | 106 | 0.00313 | 0.955385 |
| 2 | 293 | 98 | 0.000853 | 0.976707 |
| 3 | 309 | 99 | 0.117647 | 0.731601 |
Map-based cloning narrowed down the locus responsible for the mutant phenotype to the short arm of Chromosome 7, between the markers M1-8 and M1-9 (Figure 1F). By sequencing the candidate genes, the mutant gene was identified to be Zm00001d019294, with a premature stop codon in the fifth exon caused by the mutation C1468T (Figure 1, G and H). Zm00001d019294 was previously annotated as ZmNPF7.9/NRT1.5 of the NPF transporter family with a characteristic PTR2 domain (Léran et al., 2014; Guan et al., 2020; Zhou et al., 2021; Wei et al., 2021). We later renamed this gene ZmSUGCAR1 and the pfk2109 mutant zmsugcar1-1 based on our identification of this variant and its critical function in sugar loading during grain development.
In addition, we identified a spontaneous mutant with a poorly filled defective-kernel phenotype that highly resembled zmsugcar1-1 kernels (Figure 1I). Fine-mapping-based sequencing of the ZmSUGCAR1 locus revealed a G1395 deletion, resulting in another premature stop codon in the fifth exon (Figure 1, G–I and Supplemental Figure S1C). The mutant was subsequently named zmsugcar1-2 (Figure 1I). A genetic complementation test of zmsugcar1-1 and zmsugcar1-2 confirmed that the severely defective kernel phenotype was due to loss-of-function mutations of the same gene (Figure 1I and Table 1).
CRISPR-Cas9-mediated knockouts of ZmSUGCAR1 produced the identical kernel phenotype and genetic analyses revealed the mutation to be recessive (Figure 1, G, J, and K and Table 1). The independent crispr line, crispr-1, was used to reconfirm the results in the abovementioned physiological analyses performed with zmsugcar1-1. Both the 100-kernel weight and starch content in crispr-1 were as low as in zmsugcar1-1 when compared with the WT kernels (Supplemental Figure S3, A and B).
ZmSUGCAR1, encoding a plasma-membrane protein, was specifically expressed in the BETL of maize kernels
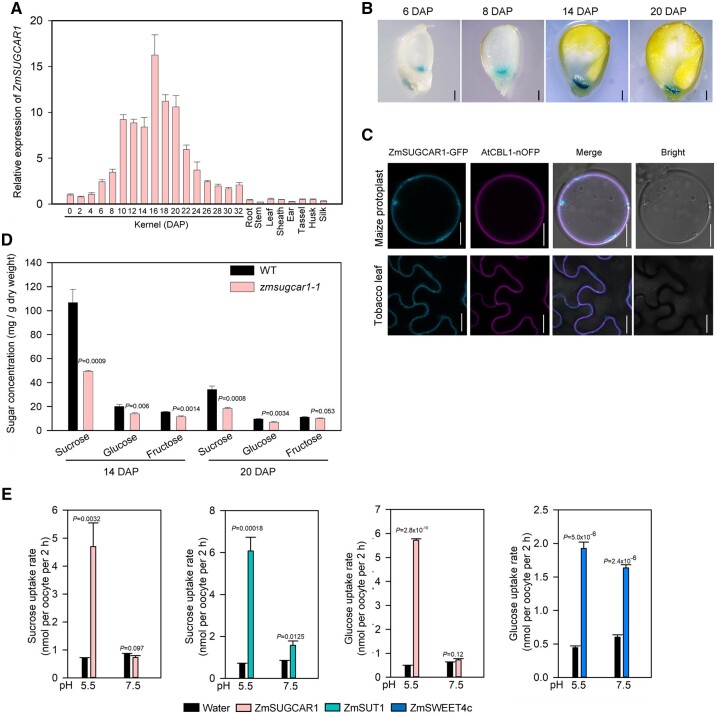
Reverse transcription quantitative PCR (RT-qPCR) analysis indicated that ZmSUGCAR1 was predominantly expressed in kernels, with only very low expression detected in other tissues (Figure 2A). ZmSUGCAR1 showed the highest expression 16 days after pollination (DAP), coinciding with the most prominent stage of active grain filling (Figure 2A). GUS staining of transgenic maize lines, using kernels from 6 DAP to 20 DAP expressing the GUS gene under control of the ZmSUGCAR1 promoter, confirmed that ZmSUGCAR1 was specifically expressed in the BETL of maize kernels (Figure 2B). To further validate the tissue specificity of ZmSUGCAR1 in maize kernels, we cryo-dissected WT kernel tissues, quantified transcript abundance by mRNA sequencing, and established that ZmSUGCAR1 expression at the mRNA level was around 10-fold greater in the BETL than in the neighboring maternal transfer zones (MATZs) (Supplemental Figure S4A).
Figure 2.
Expression and localization of ZmSUGCAR1 and its sugar transport activities in oocytes. A, Relative expression of ZmSUGCAR1 in different maize organs/tissues. Kernels were collected at different development stages. Root, stem, leaf, sheath, tassel, ear, husk, and silk were sampled at 65 days after sowing. B, GUS staining of ProZmSUGCAR1:GUS transgenic kernels at four representative stages during seed development. C, Subcellular localization of ZmSUGCAR1 in maize protoplasts and tobacco leaves. AtCBL1-OFP was used as a marker to indicate the plasma membrane. The GFP and OFP signals were visualized using confocal microscopy. Scale bars, 1 mm in (B) and 20 μm in (C). D, Sugar concentrations in the endosperm of WT and zmsugcar1-1 mutants at 14 and 20 DAP. E, Sugar uptake assays of ZmSUGCAR1 in oocytes. Oocytes expressing ZmSUT1 or ZmSWEET4c were used as positive controls for sucrose and glucose uptake, respectively. Oocytes injected with water were used as the negative control. P < 0.05 indicated significant differences between WT and mutant lines (D) or significant uptake (E).
Subcellular localization analyses in maize mesophyll protoplasts and Nicotiana benthamiana leaves indicated that ZmSUGCAR1-GFP co-localized with the plasma membrane marker protein AtCBL1-OFP (Figure 2C). When expressed in Xenopus laevis oocytes, a ZmSUGCAR1-GFP fusion protein localized to the plasma membrane of oocytes (Supplemental Figure S4B).
ZmSUGCAR1 acted as a sugar transporter importing glucose and sucrose into the endosperm
Neofunctionalization is well-known among paralogous proteins, where gain of previously unrecognized regulatory or functional features can aid adaptation to biological or environmental challenges (Krouk et al., 2010; Nour-Eldin et al., 2012; Léran et al., 2014; Saito et al., 2015; Tal et al., 2016). Given that sugar delivery is closely associated with grain filling and endosperm development (Lalonde et al., 2004; Bihmidine et al., 2013), we had a closer look the NPF mutant. Indeed, sugar accumulation in the endosperms of zmsugcar1-1 and crispr-1 was substantially lower than that in the WT (Figure 2D and Supplemental Figure S3C). The sucrose concentration during grain filling was reduced by ∼50% and the glucose level dropped by ∼30% in the zmsugcar1-1 endosperm comparing with that in WT, resulting in an abrupt decrease in the sucrose-to-hexose ratio (Supplemental Figure S4C). We therefore hypothesized that ZmSUGCAR1 may directly transport sugars into the endosperm, an unexpected function for the NPF family proteins otherwise known for their transport of various charged substrates (Léran et al., 2014; Watanabe et al., 2020; Kazachkova et al., 2021).
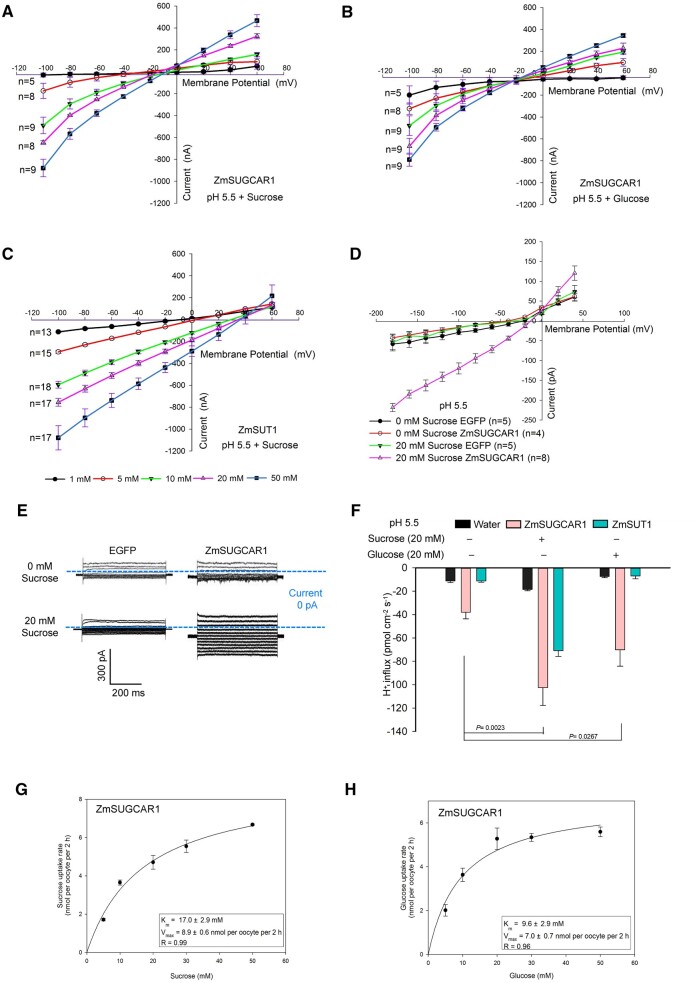
When expressed in X. laevis oocytes, ZmSUGCAR1 was able to take up both (14C-labeled) sucrose and glucose, depending on the extracellular pH (Figure 2E). Low extracellular pH promoted sugar uptake, while at neutral pH values sugar uptake disappeared (Figure 2E), suggesting that ZmSUGCAR1-mediated sugar uptake was coupled with H+. ZmSUGCAR1 did not mediate sugar efflux in oocytes in contrast to the sugar facilitators AtSWEET1 and AtSWEET12 that mediated glucose and sucrose effluxes, respectively (Supplemental Figure S5). Our finding of ZmSUGCAR1 likely operating as an H+/sugar co-transporter was further tested by electrophysiological recordings. Using the two-electrode voltage-clamp (TEVC) technique, oocytes expressing ZmSUGCAR1 were challenged by membrane potential changes. In the presence of sugars and response to long voltage pulses to negative membrane potentials, large ionic currents were elicited (Figure 3, A and B and Supplemental Figure S4, D and E). Both sucrose and glucose could trigger ZmSUGCAR1-mediated and sugar concentration-dependent ionic currents of similar magnitudes (Figure 3, A and B and Supplemental Figure S4, D and E). As an internal control for the oocyte system, we also studied substrate dependence of the well-characterized SUT1/SUC2 SUT ZmSUT1 (Carpaneto et al., 2005). In contrast to ZmSUGCAR1 and in line with the literature, we found ZmSUT1 transported sucrose but not glucose (Figure 3C and Supplemental Figure S4F). Thus, our voltage–clamp experiments with the oocyte system confirmed the results of the sugar uptake studies.
Figure 3.
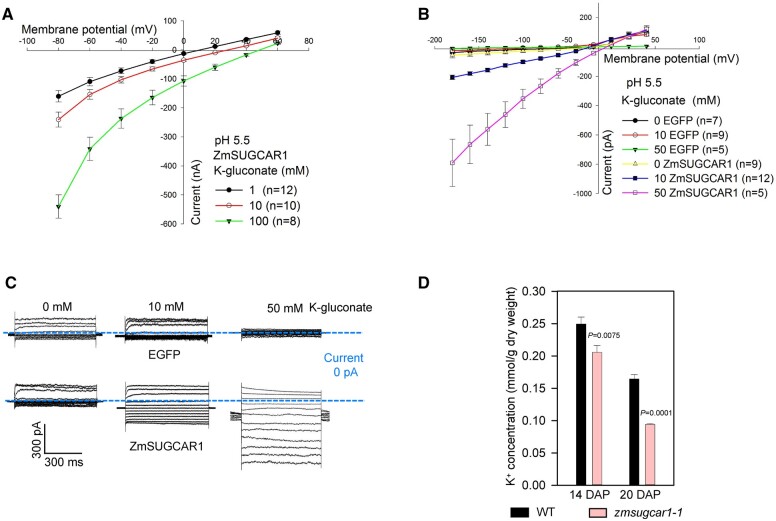
ZmSUGCAR1 mediated both sucrose and glucose transport by electrophysiology assays. A–C, Current/voltage (I/V) relationships of steady-state currents for ZmSUGCAR1 (A, B) and ZmSUT1 (C) triggered by sucrose and glucose in oocytes. The ionic currents were recorded using the TEVC technique. The oocyte membrane potential was held at −60 mV and test potentials ranging from +60 to −100 mV were applied for 800 ms using a step recording mode. The I/V relationships were obtained by subtracting the I/V curves defined in the absence of sugar from those obtained in the presence of sugar. D, Current/voltage (I/V) relationships of the steady-state currents derived from the whole-cell patch-clamp recordings in HEK293T cells. The cells transfected with empty vector (EGFP) were used as the negative control. Data were presented as mean ± se, and n indicated biologically independent cells. E, Whole-cell patch-clamp recordings in HEK293T cells expressing ZmSUGCAR1. F, H+ flux rate in oocytes using NMT/MIFE assays. Oocytes expressing ZmSUT1 were used as positive control for the sucrose-mediated H+ influx, and oocytes injected with water were used as negative control. G and H, Kinetics of ZmSUGCAR1 for sucrose and glucose uptake in Xenopus oocytes. The Km and Vmax values were determined from the Michaelis–Menten fit. P < 0.05 indicated significant differences.
To better control the cytoplasmic composition as sugar-free, we applied the patch clamp technique to ZmSUGCAR1 expressing HEK293T cells. In this heterologous mammalian expression system, sucrose-induced ionic currents were also evoked at negative membrane potentials. ZmSUGCAR1-mediated currents were observed at pH 5.5 but not pH 7.5 (Figure 3, D and E and Supplemental Figures S4G and S6, A–C). This fact again points to the ability of ZmSUGCAR1 to operate as an H+-coupled sugar transporter.
In the next step, we used non-invasive micro-test technology (NMT)/microelectrode ion flux estimation (MIFE) technique to test in oocytes expressing ZmSUGCAR1 for sugar associated H+ flux (Figure 3F and Supplemental Figure S6D). At pH 5.5, oocytes expressing ZmSUGCAR1 had a background H+ influx activity of −38.1 pmol·cm−2·s−1 in the absence of sugar. Upon sugar stimulation, the H+ influx was enhanced up to 2.7-fold for sucrose and 1.8-fold for glucose, respectively (Figure 3F). In contrast to ZmSUGCAR1, in the oocytes expressing ZmSUT1 only sucrose but not glucose triggered the H+ influx (Figure 3F).
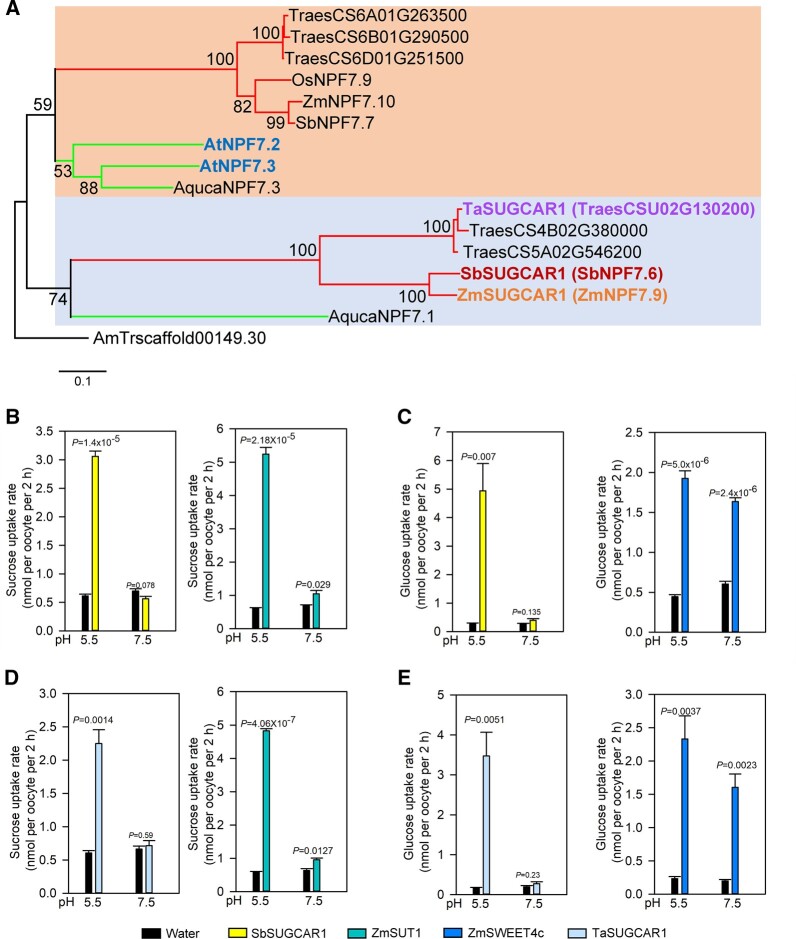
Studies on the uptake kinetics of ZmSUGCAR1 via 14C-labeled sucrose and glucose revealed Michaelis–Menten constants (Km) of 17 mM for sucrose and 9.6 mM for glucose, identifying ZmSUGCAR1 as a low-affinity sugar transporter (Figure 3, G and H). The SbSUGCAR1 encoded by NPF7.6 in Sorghum bicolor and TaSUGCAR1 in wheat (Triticum aestivum) are the closest orthologs of ZmSUGCAR1 in those species (Figure 4A; Supplemental Figure S7 and Supplemental Files S1–S4). SbSUGCAR1 and TaSUGCAR1 have similar gene structure as ZmSUGCAR1 (Supplemental Figure S8). The expression of SbSUGCAR1 and TaSUGCAR1 closely resembled the expression pattern of ZmSUGCAR1 (Supplemental Figure S8). When tested in oocytes via 14C-labeled sugar uptake, SbSUGCAR1 and TaSUGCAR1 transported sucrose and glucose in an H+-dependent manner just like ZmSUGCAR1 (Figure 4, B–E). In contrast, the paralogous proteins ZmNPF7.10 in maize and AtNPF7.3 in Arabidopsis thaliana were unable to transport these sugars (Supplemental Figure S9).
Figure 4.
Phylogenetic analysis of SUGCAR1 homologous proteins and sugar uptake by SbSUGCAR1 and TaSUGCAR1 in Xenopus oocytes. A, A simplified phylogenetic tree of SUGCAR1 homologous proteins from different plant species. The protein sequences of A. trichopod was used as outgroup. Two large, color-shaded blocks indicated major clades in this superfamily. The upper and lower branches represented monocots and dicots in each color-shaded clade, respectively. ZmSUGCAR1 (ZmNPF7.9), SbSUGCAR1 (SbNPF7.6), TaSUGCAR1, AtNPF7.3 and AtNPF7.2 were highligted in bold fonts, respectively. All bootstraps (1,000 replicates) above 50 were listed on the branches. The scale bar represented branch length. Species abbreviations were as follows: Ta or Traes, T. aestivum; Os, O. sativa; Zm, Z. mays; Sb, S. bicolor; At, A. thaliana; and Aquca, A. coerulea. Accession numbers of NPF proteins were the same as in Léran et al. (2014). B–E, Sugar uptake by SbSUGCAR1 and TaSUGCAR1 in oocytes. Oocytes injected with water were used as the negative control, and oocytes injected with ZmSUT1 or ZmSWEET4c were used as positive controls. P < 0.05 indicated significant differences.
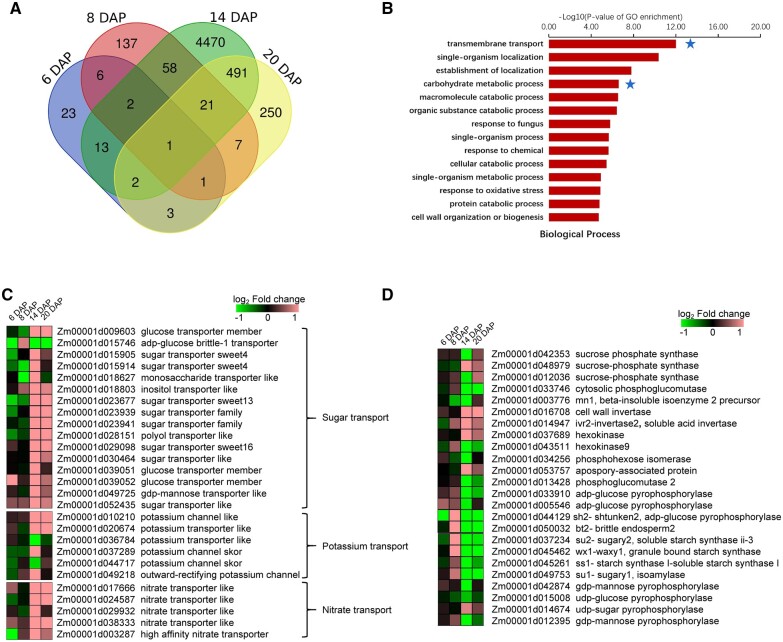
Given the sucrose concentration in basal regions of maize kernels is many-fold higher (100–150 mM) (Doehlert and Kuo, 1990) than the ZmSUGCAR1 Km, the properties of ZmSUGCAR1 make this NFP an ideal candidate transporter for driving sucrose uptake and accumulation in maize kernels. Loss-of-function mutation of ZmSUGCAR1 and subsequent reduction of sugar accumulation in zmsugcar1-1 kernels may induce compensatory expression of other sugar-provision or -conversion genes. Transcriptome profiling revealed more differentially expressed genes (DEGs) on 14 and 20 DAP than two earlier time points, with significant GO term enrichment involved in transmembrane transport and carbohydrate metabolism (Figure 5, A and B and Supplemental Data Set S1). In particular, 12 sugar transporters and 6 related metabolism genes in the mutant kernels were mostly up-regulated at the critical grain filling stage (from 14 to 20 DAP) (Figure 5, C and D and Supplemental Data Set S2), such as ZmSWEET13a (Zm00001d023677; Zhan et al., 2015; Bezrutczyk et al., 2018) and ZmSWEET4a,b (Zm00001d015905 and Zm00001d015914) expressed in the placenta-chalazal region or the pericarp of maize kernels (Zhan et al., 2015), glucose transporter (Zm00001d039052), and sucrose phosphate synthase (Zm00001d048979). Such enhanced expression of an array of sugar transporters indicated that ZmSUGCAR1 knockout caused a substantial impact on sugar transport at the transcription level in response to sugar shortage during grain filling. On the other hand, expression of three starch synthesis genes was down-regulated (Figure 5D), probably preconditioning the severely unnourished endosperm in the mutant line.
Figure 5.
Transcriptome profiling showing gene differential expression and GO term enrichment during early grain development in WT and zmsugcar1-1. A, Venn diagram of DEGs in WT and zmsugcar1-1 kernels at 6, 8, 14, and 20 DAP. B, GO term enrichment analysis of up-regulated genes in the biological process in WT and zmsugcar1-1 kernels at 20 DAP. The stars indicated enrichment of genes involved in TM transport and carbohydrate metabolism. C and D, Heat mapping of differential expression of sugar, potassium, and nitrate transporters (C) and sugar and starch metabolism-related genes (D) in WT and zmsugcar1-1 kernels at 6, 8, 14, and 20 DAP.
ZmSUGCAR1 contributed to K+ loading of kernels
Although Arabidopsis lacks an ortholog of ZmSUGCAR1, its paralog AtNPF7.3 (Figure 4A and Supplemental Figure S7) was found to reside on the plasma membrane and operate as a dual transporter for and K+ loading into the xylem (Lin et al., 2008; Li et al., 2017). Differential expression of six K+ and five transporter genes in the zmsugcar1-1 kernels also pointed to ZmSUGCAR1’s involvement in potassium and nitrate transport (Figure 5C). As expected, ZmSUGCAR1 did show a K+ transport side activity at pH 5.5 and 7.5 (Figure 6, A–C and Supplemental Figures S6E and S10, A and B). To examine the physiological relevance of K+ transport activities of ZmSUGCAR1 in maize, the K+ concentration in the zmsugcar1-1 and crispr-1 endosperm was examined. The K+ concentration in the mutant endosperm, indicating real accumulation levels of K+, significantly decreased by 18%–43% (Figure 6D and Supplemental Figure S3D), suggesting that ZmSUGCAR1 contributes to K+ loading into the maize endosperm and subsequent accumulation.
Figure 6.
ZmSUGCAR1 mediated K+ transport. A, Current/voltage (I/V) relationships of steady-state currents for ZmSUGCAR1 triggered by K+ in oocytes. The ionic currents were recorded using the TEVC technique. The oocyte membrane potential was held at −60 mV and test potentials ranging from +60 to −80 mV were applied for 800 ms using a step recording mode. B, I/V relationships of steady-state currents derived from the whole-cell patch-clamp recordings in HEK293T cells. The cells transfected with empty vector (EGFP) were used as the negative control. Data were presented as mean ± se, and n indicated biologically independent cells. C, Whole-cell patch-clamp recordings in HEK293T cells expressing ZmSUGCAR1. D, K+ concentration in the endosperm of WT and zmsugcar1-1 at 14 and 20 DAP during grain filling. P < 0.05 indicated significant differences.
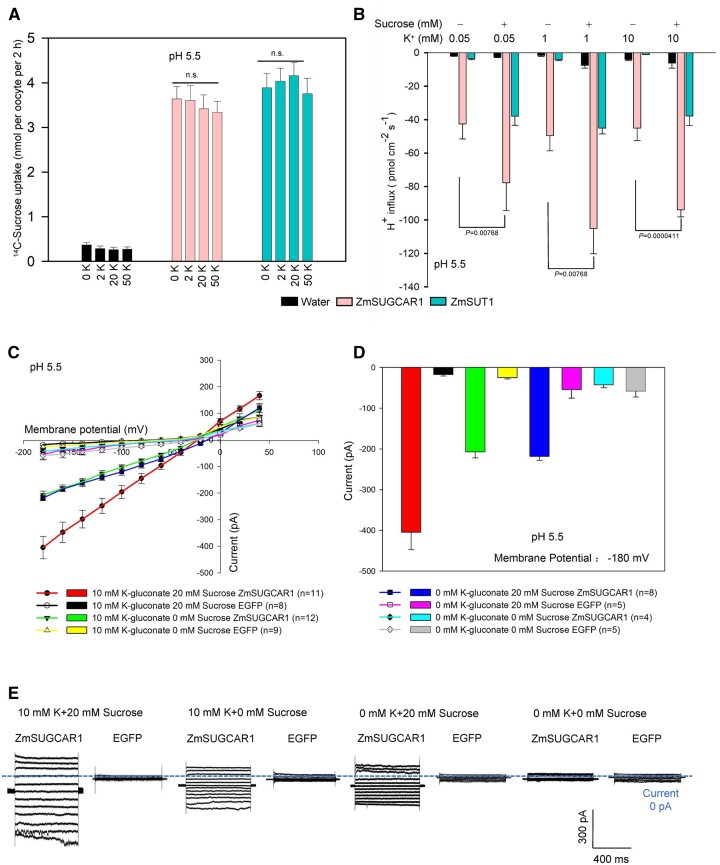
Considering that sugars and K+ are all substrates of ZmSUGCAR1, we performed competition experiments in oocytes. External K+ concentrations did not significantly affect ZmSUGCAR1-mediated sugar uptake and H+ influx in oocytes (Figure 7, A and B). Such independent transport for sugar and K+ was also confirmed in HEK293T cells (Figure 7, C–E). The poorly filled phenotype of zmsugcar1-1 kernels could be due to the lack of both, the sugar and K+ transport function through ZmSUGCAR1. However, ZmSUGCAR1’s independence in sugar transport favors another plausible argument that interference of sugar loading during a critical sugar mobilization period results in severe endosperm-filling defects.
Figure 7.
Sucrose uptake by ZmSUGCAR1 independent of the extracellular K+. A, ZmSUGCAR1 mediates sucrose uptake independent of extracellular K+ using 14C-labeled sugar uptake assay in oocytes. Oocytes expressing ZmSUT1 were used as positive controls for sucrose uptake. Oocytes injected with water were used as the negative controls. B, ZmSUGCAR1 mediates proton influx induced by sucrose independent of the extracellular K+ using NMT in the oocytes at pH 5.5. Oocytes expressing ZmSUT1 were used as the positive control for the sucrose-mediated H+ influx, and oocytes injected with water were used as the negative control. C, I/V relationships of steady-state currents for ZmSUGCAR1 triggered by sucrose, independent of extracellular K+, according to the whole-cell patch-clamp recordings in HEK293T cells. D, ZmSUGCAR1-mediated currents in HEK293T cells at −180 mV. The cells transfected with empty vector (EGFP) were used as negative control; data are presented as mean ± se, and n indicated biologically independent cells in (C) and (D). E, Whole-cell patch-clamp recordings in HEK293T cells expressing ZmSUGCAR1. P < 0.05 indicated significant differences.
ZmSUGCAR1 did not affect nitrate accumulation in the endosperm
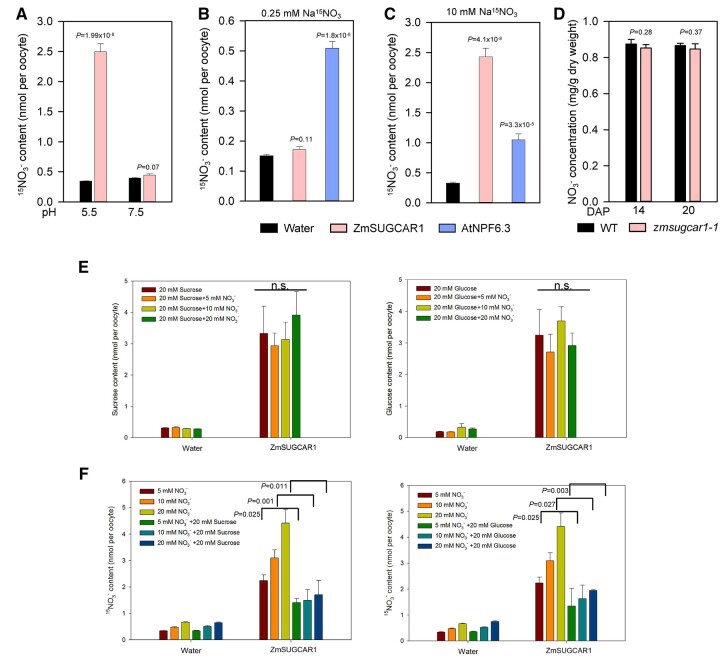
In agreement with previous work (Wei et al., 2021), uptake was only detected at high levels, suggesting that ZmSUGCAR1 has low-affinity transporter side activity (Figure 8, A–C). However, in the presence of with either sucrose or glucose in the extracellular buffer, nitrate did not affect sucrose and glucose uptake, while both sugars significantly reduced nitrate uptake (Figure 8, E and F and Supplemental Figure S10, C and D), suggesting that ZmSUGCAR1 preferentially transported sucrose or glucose over . More importantly, the dry weight-based nitrate concentration in the zmsugcar1-1 and crispr-1 endosperm was not significantly different from that in WT kernels during grain filling (Figure 8D and Supplemental Figure S3E) although smaller kernels led to a less amount of nitrate in the entire zmnpf7.9 seed in an earlier report (Wei et al., 2021). Therefore, ZmSUGCAR1 may function as a genetically redundant transporter in vivo or at most a minor player in relation to nitrate storage in the endosperm. Alternatively, nitrate transport of ZmSUGCAR1 or nitrate itself does not seem to have significant biological relevance to grain filling.
Figure 8.
Nitrate () transport by ZmSUGCAR1 in oocytes and accumulation in the endosperm. A, The pH dependence of ZmSUGCAR1-mediated nitrate uptake in oocytes. Oocytes injected with water were used as a negative control. B and C, Nitrate uptake activity of ZmSUGCAR1 in oocytes under low (B) or high (C) nitrate conditions. Oocytes injected with AtNPF6.3 and water were used as positive and negative controls, respectively. D, Nitrate concentration in the endosperm of WT and zmsugcar1-1 at 14 and 20 DAP. E and F, Competition assays between sugar and nitrate () uptake by ZmSUGCAR1. Oocytes injected with water were used as a negative control. P < 0.05 indicated significant differences.
To characterize the function of ZmSUGCAR1 further, we tested a number of other reported NPF substrates including chloride, oligopeptides, and hormones. In TEVC studies with oocytes, no ionic currents were detectable in the presence of chloride and dipeptides (Supplemental Figure S10, E and F). Using uptake or efflux studies, we did not spot plant hormones such as auxin (indole-3-acetic acid [IAA]), gibberellin, or jasmonic acid as ZmSUGCAR1 substrates either (Supplemental Figure S11).
ZmSUGCAR1 functions after BETL establishment
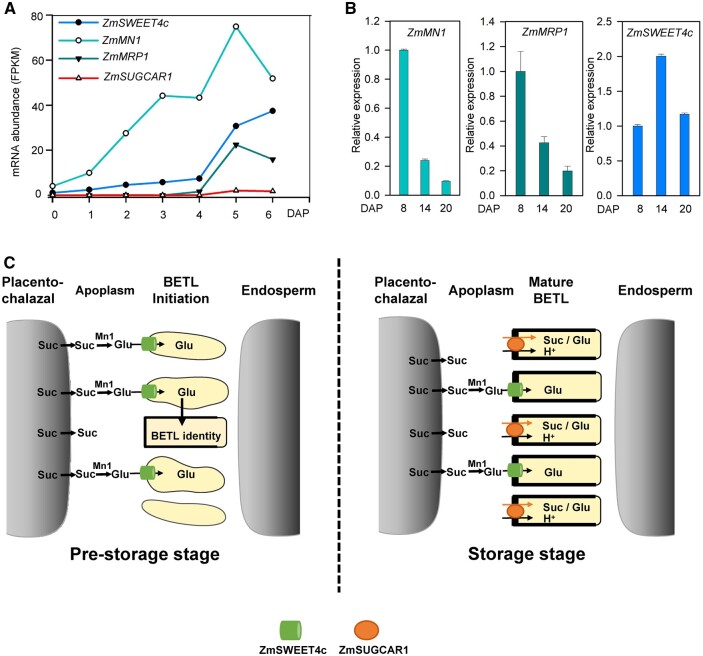
To find out whether disruption of ZmSUGCAR1 feeds back on the reported genes in regulating BETL development, the expression of ZmMN1, ZmSWEET4c, and ZmMRP1 (Chourey et al., 2006; Barrero et al., 2009; Zheng and Wang, 2010; Sosso et al., 2015; Doll et al., 2017) was monitored. All three critical BETL genes showed early expression induction from 0 to 6 DAP favoring BETL establishment; meanwhile, ZmSUGCAR1 had undetectable or extremely low levels of expression (Figure 9A). Transcript abundance of these three BETL foundation genes gradually declined over grain filling (Figure 9B) and was not affected in zmsugcar1-1 during kernel development at 6 and 8 DAP (Supplemental Data Set S1). Further, zmsugcar1-1 did not display apparent structural alterations of the BETL (Supplemental Figure S2, F and G). Thus, ZmSUGCAR1 most likely does not affect BETL cell-type differentiation, but functions after BETL establishment.
Figure 9.
Expression of ZmSUGCAR1, ZmSWEET4c, ZmMN1, and ZmMRP1 during seed development and a schematic model of ZmSUGCAR1 functioning in grain filling. A, mRNA abundance (FPKM) of ZmSUGCAR1, ZmSWEET4c, ZmMN1, and ZmMRP1 based on RNA-seq. FPKM, fragments per kilobase of transcript per million mapped reads. B, Relative expression of ZmMN1, ZmMRP1, and ZmSWEET4c in WT kernels by quantitative PCR. C, A working model of ZmSUGCAR1 in regulating grain filling in maize. At the pre-storage stage of seed development, the sucrose in the apoplasm unloaded from the maternal placenta–chalazal region is hydrolyzed to hexose by the cell-wall invertase Mn1; then the ZmSWEET4c (cylinder) mediates hexose import and initiates BETL formation. During the storage stage, ZmSUGCAR1 (oval) acts as an H+ and sucrose (Suc)/glucose (Glu) symporter to transfer sugars from the maternal placenta–chalazal region into the endosperm across the BETL, circumventing sucrose hydrolysis. Meanwhile, SWEET4c also operates for glucose transport after BETL establishment.
Taken together, these results suggest that ZmSUGCAR1 functions as a unique proton-coupled sucrose and glucose transporter. Loss-of-function of ZmSUGCAR1 hampered the efficient translocation of sugars across the BETL, resulting in drastically reduced sucrose concentrations, starch accumulation, and subsequent kernel shrinkage (Figures 1 and 2D). We conclude that the BETL-localized ZmSUGCAR1 mediates sugar transport into the endosperm and directly impacts grain filling in maize.
Discussion
Sucrose, the most abundant form of photo-assimilate in long-distance phloem transport, is unloaded in the maternal tissue adjacent to developing grains (Patrick, 1997; Zhang et al., 2007; Braun and Slewinski, 2009; Kühn and Grof, 2010; Braun et al., 2014; Milne et al., 2018). Early work with maize indicated that a considerable portion of this sucrose was hydrolyzed to hexose prior to uptake by newly forming seeds (Shannon, 1972). However, much of the sucrose could also move directly into the endosperm without alteration (Schmalstig and Hitz, 1987; Shannon, et al., 1993). The existence of an active sucrose/proton-coupled co-transport system for this direct sucrose import was first suggested against previous models for transport of un-hydrolyzed sucrose into the endosperm (Porter et al., 1985, 1987; Patrick, 1990). For many years, experimental evidence supporting such a direct sucrose transfer model has been lacking. Here, the ZmSUGCAR1-mediated direct sucrose uptake into endosperm cells provides a means for concurrent entry of both sucrose and hexoses. Our data suggest that two modes of sugar supply to the endosperm exist depending on the varying phases of kernel development.
During early kernel development, the ZmMN1, ZmSWEET4c, and ZmMRP1 representing a glucose-responsive feed-forward system are involved in the hexose transport and BETL formation (Sosso et al., 2015), which precedes ZmSUGCAR1 expression (Figure 9A; Yi et al., 2019). The scenario reflects the significance of hexoses resulted from cell-wall invertase activity to the structural development of this key tissue layer during nutrient import (Barrero et al., 2009; Monjardino et al., 2013; Sosso et al., 2015). ZmSUGCAR1 shows high specificity of expression in the BETL but does not have a marked effect on BETL cell fate. The difference is likely due to the delayed onset of ZmSUGCAR1 expression until 5 DAP, when much of the BETL structure is already established (Figure 9A). The seemingly intact BETL of ZmSUGCAR1-knockout mutants, despite their empty pericarp phenotype, could potentially result from the undisturbed function of ZmSWEET4c and other players of the BETL.
At later stages of kernel development, the content of sucrose was much higher than that of glucose and fructose in the WT endosperms when measured at 14 DAP (Figure 2D), possibly indicating that the disaccharide sucrose may be directly transported into the endosperm without the need of pre-hydrolysis into monosaccharides. This is supported by the progressively declining expression of ZmMN1 and ZmMRP1 at 8, 14, and 20 DAP (Sosso et al., 2015; Figure 9B) and a corresponding decline in cell-wall invertase activity (Cheng et al., 1996), suggesting that kernel development shifts to the storage phase. Parallel observations from rice (Oryza sativa) and faba bean (Vicia faba) also indicate that cell-wall invertase activity declines at the onset of the storage phase in kernel development, while active sucrose uptake by transporters in filial tissues becomes predominant (Weber et al., 1997; Hirose et al., 2002). We observed a marked reduction in sucrose versus hexose levels in the zmsugcar1-1 endosperm by 14–20 DAP (Figure 2D and Supplemental Figures S3C and S4C), which demonstrates that ZmSUGCAR1 primarily functions in transporting the major photoassimilate sucrose into the endosperm during the storage phase of grain filling (Figure 9C).
ZmSUGCAR1, SbSUGCAR1, and TaSUGCAR1 show functional conservation in sucrose transport capability (Figures 2–4). SbSUGCAR1 and TaSUGCAR1 all have high levels of expression in endosperm, suggesting that their encoded proteins have possible roles in grain filling similar to ZmSUGCAR1 (Supplemental Figure S8).
To date, the SUT family transporters in crops have been the only viable candidates for sucrose transport (Bai et al., 2016; Radchuk et al., 2017; Milne et al., 2018). In maize, ZmSUT1 is the best-characterized member and serves to load sucrose into the sieve element–companion cell complex (SE-CCC) of leaves (Slewinski et al., 2009; Baker et al., 2016). In kernels, however, the relatively low expression of ZmSUT1 is consistent with a minor role in that structure (Davidson et al., 2011; Sekhon et al., 2011; Li et al., 2014), although other unidentified SUT proteins may still be important to grain filling.
Our data demonstrate that ZmSUGCAR1 under physiological conditions represents a proton-coupled sugar transporter that also mediates sugar-independent K+ currents, which is well in line with dramatically reduced sucrose and significantly less K+ accumulation in the zmsugcar1 endosperm (Figures 2D and 6D and Supplemental Figure S3, C and D). These results are compatible with the widely recognized conception that sugar translocation from source to sink organs within plants is facilitated by K+ at the physiological level (Pettigrew, 2008; Gajdanowicz et al., 2011; Zörb et al., 2014). ZmSUT1 is a strictly H+-coupled sucrose symporter, thus the reversal potential of ZmSUT1-expressing oocytes shifts to more positive membrane potentials (close to the predicted Nernst potential for H+) in the presence of sucrose (Figure 3C; Carpaneto et al., 2005). Therefore, ZmSUT1 requires the co-expression of K+ channel ZMK2 for efficient sucrose loading in the phloem tissue (Deeken et al., 2002; Philippar et al., 2003). Due to the weak voltage dependence of ZMK2, the K+ background currents prevent sucrose-induced membrane depolarization mediated by ZmSUT1 in the presence of sucrose. Our electrophysiological data suggest that ZmSUGCAR1-mediated K+ currents are independent of sugar and H+ transport (Figure 7). Thus, there is no H+/K+ symport activity for ZmSUGCAR1. One would expect an intermediate reversal potential between the K+ conductance and the H+-coupled sucrose-induced conductance, dissimilar to that of the aforementioned H+/sucrose symporter SUT1. Especially, the background K+ conductance would stabilize/clamp the membrane potential to the reversal potential of K+, so that a large sugar-induced depolarization of the membrane potential is prevented. Here, ZmSUGCAR1 seems to combine the functions of the SUT (e.g. SUCs/SUTs) and the K+ channel (e.g. AKT2/ZMK2) in one protein, which makes the sucrose transport more efficient in BETL cells. However, inherent linkage of sugar loading to grain filling and dramatic reduction in sucrose accumulation in the mutant endosperm indicate that disruption of sugar transport activities convincingly interprets the phenotype of kernel shrinkage in mutant lines, regardless of with or without K+ facilitation, which may be further solidified by genetic complementation of the Zmsugcar1 mutant with a well-characterized sugar specific transporter driven by the ZmSUGCAR1 promotor. New loss-of-function alleles affecting K+ or nitrate transport will help fully decipher the functions of this multi-functional transporter in the future.
In summary, the collective evidence presented here demonstrates that the maize BETL-specific ZmSUGCAR1 gene (1) represents another type of active sugar transporters, (2) provides a mechanism for the direct uptake of sucrose and glucose into endosperm cells, and (3) is critical to grain filling. The unexpected capacity for sugar transport by ZmSUGCAR1 distinguishes an additional functional group among the many NPF genes in plants and indicates potentially diverse roles in flux of sugars and nutrient resources. The identification and analyses of ZmSUGCAR1 as a direct sucrose and glucose transporter well complement actions of SWEETs, SUTs, and MSTs. Thus, our findings provide important insights into sugar intake by grains and may lead to new strategies for improving grain filling and agricultural output.
Materials and methods
Plant materials and growth conditions
The zmsugcar1-1 mutant was obtained from the EMS-mutagenized WT Z. mays B73 inbred miniature seed germplasm bank generated in our laboratory. The zmsugcar1-2 allele was a natural mutant obtained from Professor Jin (China Agricultural University). Allelism of zmsugcar1-1 and zmsugcar1-2 was confirmed by genetic complementation tests with reciprocal crosses between heterozygous and homozygous plants. Homozygous seeds were germinated on sterile filter paper with sterile water 3 days in the dark at 30°C. WT or homozygous plants were grown side by side for each experiment, either in summer field conditions (Shangzhuang Experiment Field of China Agricultural University). Sagittal sections of seeds at four developmental stages were cut with a utility knife for phenotype observation. Nicotiana benthamiana plants were grown at 22°C in an environmentally controlled growth room under long photoperiod conditions (16-h light/8-h dark). The relative humidity during the day and night was 50%. Light intensity was 100–130 mmol photons m−2 s −1 PPFD.
Map-based cloning of ZmSUGCAR1
The F2 mapping population was derived from a cross between pfk2109 and the Mo17 inbred. The F2 kernels with the pfk2109 phenotype were used for gene mapping. Map-based cloning was performed using more than 1,300 mutant individuals from the F2 mapping population. We developed molecular markers according to sequence polymorphisms between B73 and Mo17 including InDels. The zmsugcar1-1 locus was mapped to an interval between the markers M7-1 and M7-2 on the short arm of Chromosome 7. Fine mapping anchored the mutation site to a 2-Mb genomic region between the markers M7-8 and M7-9.
The zmsugcar1-2 locus was mapped to an interval between the markers M2-1 and M2-2 on the short arm of Chromosome 7. Fine mapping anchored the mutation site to a 2-Mb genomic region between the markers M2-8 and M2-9. The primers used for marker generation and candidate gene (Zm00001d019294) amplification are listed in Supplemental Data Set S3.
Constructs for CRISPR-Cas9 mutagenesis plants
To create ZmSUGCAR1 CRISPR lines, a 19-bp sgRNA (TCCTGTCGCAGAGGTAGCT) targeting the second exon of ZmSUGCAR1 was cloned into the CRISPR-Cas9 expression vector (pCAMBIA3301) (Frame et al., 2002; Zhu et al., 2016). The resulting construct was transformed into the strain EHA105 and introduced into embryos of B73-329 (12 days after pollination) by Agrobacterium-mediated transformation as described (Zhu et al., 2016). After 3-day culture at 22°C, embryos were transferred to selection media for 2-week growth in darkness at 28°C. The calli were next moved to regeneration media for shooting and rooting, and positive transgenic events were identified via herbicide resistance and verified by DNA sequencing at the seedling stage. Knockout lines were transferred to a standard greenhouse, backcrossed twice to B73-329, and then self-pollinated to obtain homozygous knockout lines without the CRISPR/Cas9 cassette by bialaphos (bar) resistance and PCR sequencing. The F2 zmsugcar1 homozygotes were used for genotyping and phenotyping. The related primers are listed in Supplemental Data Set S3.
GUS staining assays
For promoter activity analysis, a 2,100-bp genomic fragment upstream of the ATG start codon was PCR-amplified from WT genomic DNA with the primer pair promoter-F and promoter-R (Supplemental Data Set S4) and fused to the GUS reporter gene in the binary vector pCAMBIA1391. The resulting construct pZmSUGCAR1 promoter-GUS was introduced into the WT by the Agrobacterium-mediated transformation method. Histochemical staining of GUS activity in the transgenic plants was performed as previously described (Jefferson, 1987; Barrero et al., 2009). Images were captured using Leica Application Suite 3.3.0 software.
Light microscopy of cytological sections
Immature WT and zmsugcar1-1 mutant kernels were collected from the homozygous plants at four development stages, respectively. The kernel was cut along the longitudinal axis and the central slice containing the embryo was fixed in 4% paraformaldehyde at 4°C overnight. The fixed material was dehydrated in a gradient ethanol series (50%, 70%, 80%, 95%, and 100% ethanol). After clearing with HistoChoice Clearing Agent (Sigma, H2779) and paraffin wax infiltration, the sample was embedded and sectioned at 8 μm with a Leica RM2235 slicer. The sections were stained with eosin and observed with Olympus SZ51 microscopy. Approximately 20 sections from each line were observed with representative results presented in Figure 1 and Supplemental Figure S2.
Subcellular localization of ZmSUGCAR1 in maize protoplasts and N. benthamiana leaves
The coding sequence of ZmSUGCAR1 fused with GFP at the C-terminal was cloned into the pSuper1300 vector and then transfected into Agrobacterium tumefaciens (GV3101) via electroporation. The AtCBL1-OFP construct was used as a plasma membrane marker (Batistic et al., 2010). Bacterial cultures were grown, pelleted, and dissolved in infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone) to a cell density as measured by OD600 of approximately 0.5–0.7 for 2 h. A syringe was used to infiltrate the mixture into the lower surface of N. benthamiana leaves. Plants were incubated for 3 days after infiltration before imaging.
Transient expression of ZmSUGCAR1-GFP and AtCBL1-nOFP in maize mesophyll protoplasts was performed according to the protocol described in (Wen et al., 2017). Briefly, mesophyll protoplasts were isolated from pooled etiolated leaves of 12-day-old maize plants and diluted to 6 × 106 protoplasts/mL. Protoplasts were then transformed with ZmSUGCAR1-GFP and AtCBL1-OFP constructs using the PEG method and resuspended to 2 × 105 protoplasts/mL. After 24 h dark incubation, protoplasts were visualized using a Zeiss 880 confocal microscope.
GFP and nOFP were detected using a Zeiss 880 confocal microscope with simultaneous acquisition at 522–572 nm (GFP) and 667–773 nm (OFP). Laser excitation was at 488 nm for GFP and 561 nm for OFP and RFP, respectively. Image analysis was performed using the Zeiss 880 suit imaging software.
RNA extraction and RT-qPCR analyses
Six individual plants per line were selected for RNA isolation using Trizol (Thermo Fisher) according to the manufacturer’s instructions. After DNaseI treatment, the first-strand cDNA was synthesized from 2 μg of total RNA with oligo (dT) as the primer, using a reverse transcription kit (TaKaRa). RT-qPCR was carried out using the ABI7500 real-time PCR system with the SYBR Premix Ex Taq (TaKaRa; RR041A), following the manufacturer’s instructions. The maize 18s rRNA gene was used as an internal control. Gene expression levels were determined from at least three biological replicates and three technical replications per reaction. The 2−ΔΔCt method was used for relative quantification. The qPCR experiments repeated independently twice with comparable results. Gene-specific primers are shown in Supplemental Data Set S4.
Hundred-kernel weight and starch quantification
Representative ears were harvested from field-grown WT and mutant (zmsugcar1-1 and cripr-1) lines and dried to constant weight. One hundred kernels from each ear were weighed on a laboratory scale. Five ears from each line were used as individual biological replicates. WT, homozygous zmsugcar1-1, and cripr-1 seeds were harvested at 14, 20, or 45 DAP, and were dried at 80°C to a constant weight, then ground to a fine powder with a pestle and mortar. Fifty milligrams of tissue was incubated with 1 mL of 70% ethanol for 1 h on ice, with frequent vigorous vortexing. Subsequently, the samples were spun for 15 min at 4°C at 13,000 g, and the supernatant was removed. This extraction was repeated again. The pellet was dried in a vacuum concentrator and resuspended in water. Starch was quantified with the Total Starch Assay kit using 510 nm spectrophotometer method (Megazyme, K-TSTA). The experiments were repeated twice independently with similar results. Each independent experiment contained three individual ears as biological replicates.
Potassium, nitrate, and soluble sugar measurement
Maize kernels from WT or homozygous zmsugcar1-1 and cripr-1 ears were collected at 14, 20, or 45 DAP, respectively, then the pericarp and embryo were removed with a razor blade, and endosperms were dried at 80°C to a constant weight, using 50 endosperms at each stage pooled as one biological replicate.
For potassium measurements, pooled endosperm samples were treated in a muffle furnace at 575°C for 6 h. Approximately 0.1 g endosperm samples were dissolved in 0.1 N HCl. K+ contents were measured using the 4100-MP AES system (Microwave Plasma Atomic Emission Spectroscopy, Agilent). The measured values were normalized on the basis of dry weight.
For nitrate and soluble sugar measurements, endosperms were dried at 80°C for 24 h and ground to a fine powder with a mortar and pestle. Add 0.1 g dry endosperm samples into a 15-mL polypropylene tube and suspended in 10 mL deionized water. After treatment in an ultrasonic bath for 30 min, the mixture was centrifuged for 10 min at 12,000 g. The resulting suspension was 100× times diluted and filtered through a 0.22-μm filter. Twenty-five microliters of extracted sample solution was analyzed by a high-performance ion chromatography system (HPIC, Dionex ICS3000, Thermo, USA). Nitrate and sugars were, respectively, eluted with 12 and 25 mM NaOH at a flow rate of 1.0 mL min−1. AS11 analytical column (250 × 4 mm) and an AG11 guard column (50 × 4 mm) were used for nitrate determination. The CarboPac PA10 pellicular anion-exchange resin column (250 × 4 mm) connected to a CarboPac PA10 guard column (50 × 4 mm) was used for sucrose, glucose, and fructose separation. The measured values were normalized based on dry weight or the entire endosperm. The experiments were repeated twice with similar results independently. Each independent experiment contained three individual ears as biological replicates.
Sequence BLAST and phylogenetic tree construction
ZmSUGCAR1 (Zm00001d019294) protein was used as the query for BLAST searches against Z. mays, S. bicolor, Setaria italica, Setaria viridis, O. sativa, Brachypodium stacei, Brachypodium distachyon, Musa acuminata, Arabidopsis thaliana, Solanum lycopersicum, Aquilegia coerulea, Amborella trichopod genomes on phytozome (https://phytozome.jgi.doe.gov), Aegilops tauschii, T. aestivum, and Hordeum vulgare genomes on EnsemblPlants (http://plants.ensembl.org/index.html), Elaeis guineensis and Pinus pinaster on NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and Gnetum montanum on ONEKP (https://db.cngb.org/onekp/).
Protein sequences were aligned by using MUSCLE 3.6 (http://www.drive5.com/muscle/), manually adjusted in GeneDoc 3.2 (https://www.soft-pedia.com/get/Science-CAD/GeneDoc.shtml), sequentially scored using CLUSTALX programs, and then protein matrix scoring 12 was used to construct the phylogenetic tree. The maximum-likelihood phylogenetic trees were generated by the MEGA 6.0 (https://www.megasoftware.net/history.php). Bootstrapping was performed with 1,000 replications.
Library construction and RNA-seq analyses
Two biological replicates were used for RNA isolation, library construction, and later sequencing. Total RNA was extracted from WT ovules every day from 0 to 6 DAP and 6, 8, 14, and 20 DAP kernels of WT and zmsugcar1-1. The RNA-Seq libraries were constructed and then subjected to Illumina sequencing in Hiseq2000 (Chen et al., 2014). After sequencing, raw reads were aligned to the B73 reference genome (v4) using Tophat 2.0.6 (Trapnell and Schatz, 2009) with default settings for all parameters. The DEGs were defined by the thresholds of a P-value < 0.05 (Student’s t test) and a log2FC (fold change) > 1.0. The relative expression levels of DEGs in the RNA-Seq analysis were denoted in fragments per kilobase of exon per million fragments mapped. Two biological replicate samples were performed for the zmsugcar1-1 mutant and the WT, and three technical validations were performed for each biological replication. VennDiagram (https://bioinformatics.psb.ugent.be/webtools/Venn/) was employed to analyze differential gene expression over four sampling time points. Gene Ontology (GO) enrichment analysis was performed with the maize sequence v4 ontologies by agriGO v2.0 (http://bioinfo.cau.edu.cn/agriGO/). Significantly enriched GO terms were identified as those with a P-value <0.05. Heat map configuration was then generated accordingly (http://cran.r-project.org/package=pheatmap).
For the transcriptome analysis of kernel sub-regions, kernel tissues were cryo-dissected at −22°C in a Leica CM1950 cryostat using different ears for each biological replicate. All samples were harvested from W22 inbred plants grown under field conditions in Florida (UF-PSRU, Citra). The FPKM values (mean ± sd, n = 2) were calculated for two biological replicates from RNA-seq analysis.
Xenopus oocyte isolation and RNA injection
Coding sequences of ZmSUGCAR1, SbSUGCAR1, TaSUGCAR1, AtNPF7.3, ZmNPF7.10, and AtNPF6.3 were codon optimized and synthesized. These cDNAs were then cloned into the expression vector pT7TS. After linearization of pT7TS plasmids with BamHI, RNA was transcribed in vitro using an mRNA synthesis kit (mMACHINE T7 kit; Ambion). Xenopus laevis oocytes were isolated in 25 mL Ca2+-free ND96 solution (in mM: 96 NaCl, 2 MgCl2,1 KCl, and 5 HEPES, pH 7.5 adjusted with NaOH with 0.1 mg mL−1 gentamycin and 0.1 mg mL−1 streptomycin) containing 43 mg collagenase and 12.5 mg trypsin inhibitor at 23°C for 1.5 h and were then recovered in ND96 solution (in mM: 96 NaCl, 2 MgCl2,1 KCl, 1.8 CaCl2, and 5 HEPES, pH 7.5 adjusted with NaOH with 0.1 mg mL−1 gentamycin and 0.1 mg mL−1 streptomycin) for 24 h. Oocytes were injected with cRNA (25 ng in 25 nL) after recovery and were incubated at 18°C in ND96 solution. Incubation buffer was changed every 12 h. Oocytes injected with water were used as the negative control.
14C-labeled sugar uptake and efflux in Xenopus oocytes
The assay was performed with modification as described previously (Chen et al., 2010, 2012). For sugar uptake assay, 40 h after cRNA injection, groups of 9–12 oocytes were transferred into 500 μL Na-Ringer (in mM: 115 NaCl, 2 KCl, 1 MgCl2, 1.8 CaC12, 10 MES-Tris, pH 5.5 or 10 HEPES-NaOH, pH 7.5) with 0.1 mg mL−1 gentamycin and d-glucose (4 μCi mL−1 D-[1-14C]-glucose; PerkinElmer) or sucrose (4 μCi mL−1 [14C(U)]-sucrose; PerkinElmer). After incubation at 18°C for 2 h, oocytes were transferred to ice-cold Na-Ringer, washed four times, solubilized with 100 μL 1% (w/v) SDS, and three oocytes were measured as one sample.
For tracer sugar efflux assay, 36 h after cRNA injection, oocytes were injected with 50 nL solution containing 50 mM glucose (0.18 μ Ci μL−1 D-[1-14C]-glucose) or 50 mM sucrose (0.18 μ Ci μL−1 [14C(U)]-sucrose). Oocytes were immediately washed once in Na-Ringer and then the oocytes were transferred into 1 mL Na-Ringer reaction buffer. At defined time points, reaction buffer (950 μL) was removed for scintillation counting. Oocytes were solubilized with 1% SDS and analyzed for retained radioactivity. The experiments were repeated using three different batches of oocytes independently with similar results.
15 uptake assay in Xenopus oocytes
The assay was performed with modification as described previously (Lin et al., 2008), 40 h after cRNA injection oocytes were incubated for 2 h in a solution containing 230 mM mannitol, 0.3 mM CaCl2, 10 mM MES-Tris, pH 5.5 or 10 mM HEPES-NaOH, pH 7.5, and the required concentration of K15NO3 (0.25 mM for high-affinity uptake assays and 10 mM for low-affinity uptake experiments), washed five times with ND96 buffer, and individually dried at 80°C for 24 h, and then the retained 15N was measured. The experiments were repeated twice independently with similar results.
3H-labeled IAA transport assay in Xenopus oocytes
The ZmSUGCAR1 CDS was cloned into the vector pBF1 and linearized 3′ prime of the poly(A) sequence. cRNA was in vitro transcribed using the mMACHINE SP6 kit (Ambion). Oocytes were isolated in Modified Barth’s Medium (MBM, in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.66 NaNO3, 0.75 CaCl2, and 10 HEPES) containing 2 mg/mL collagenase and 0.1 mg/mL trypsin inhibitor at 34°C for 20 min. Oocytes were manually sorted for stages V and VI. After recovery in MBM for 24 h at 16°C oocytes were injected with 50 nL cRNA (1 μg/μL) or water (negative control) and incubated at 16°C in -free Barth’s solution (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 1.57 MgSO4, 0.75 CaCl2, 10 HEPES, pH 7.4 adjusted with NaOH) with 50 μg/mL gentamicin, 100 μg/mL streptomycin, and 100 units/mL penicillin for 30 h. The incubation buffer was changed once. IAA uptake and efflux assays were performed 40 h after injection (3H-IAA, specific activity 25 Ci/mmol, concentration 1 μCi/μL, company ARC).
For IAA uptake assays, oocytes were transferred into 500 μL Na-Ringer (in mM: 115 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 MES [pH 6.0], or 5 HEPES (pH 7.5), respectively; pH adjusted with NaOH or HCl) containing 100 nM IAA (labeled with 40 nM 3H-IAA). After incubation at 20°C for 3 h in pH 6 firstly, a batch of oocytes were transferred to IAA-free Na-Ringer in pH 6 or pH 7.5, respectively, for 0.5 or 1 h. After the particular incubation times, oocytes were transferred to ice-cold Na-Ringer with the respective pH, washed four times, and sampled individually. Oocytes were solubilized with 100 μL 10% (w/v) SDS and the amount of radioactivity was determined by liquid scintillation counting. The experiments were repeated twice independently with similar results.
IAA efflux assays were performed with modifications as described previously (Fastner et al., 2017). Oocytes were injected with 50 nL containing 1 µM 3H-IAA or 300 µM IAA (containing 1 µM 3H-IAA). Oocytes injected with water were used as negative controls. Injected oocytes were transferred into -free Barth’s pH 7.5 (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 1.57 MgSO4, 0.75 CaCl2, 10 HEPES, pH 7.5 adjusted with NaOH). The retained 3H-IAA was determined by liquid scintillation counting.
The competition uptake assay between sugar and in Xenopus oocytes
For 15 uptake assay, 40 h after injection, oocytes expressing ZmSUGCAR1 were incubated in a solution containing 210 mM mannitol, 0.3 mM CaCl2, 10 mM MES-Tris, pH 5.5, 20 mM sucrose or glucose, and the required concentration of K15NO3 (mannitol was used to regulate the osmotic potential) for 2 h, and then the retained 15N was measured. For the [14C]-labeled sugar uptake, the oocytes were incubated in the Na-Ringer-based solutions with 0.1 mg mL−1 gentamycin and 20 mM d-glucose (4 μCi mL−1 d-[1-14C]-glucose) or sucrose (4 μCi mL−1 [14C(U)]-sucrose) and 5 mM, 10 mM, or 20 mM CsNO3 (NaCl was used to regulate the osmotic potential) for 2 h and then the 14C in oocyte was measured. Oocytes expressing AtNPF6.3 were used as positive controls for nitrate uptake. Oocytes injected with water were used as the negative control. The experiments were repeated using three different batches of oocytes independently with similar results.
The competition uptake assay between sugar and K+ in Xenopus oocytes
Forty hours after injection, oocytes expressing ZmSUGCAR1 were incubated in the Na-Ringer-based bath solutions with 0.1 mg mL−1 gentamycin containing 20 mM sucrose (4 μCi mL−1 [14C] sucrose) under different extracellular KCl concentrations (NaCl was used to regulate the osmotic potential), respectively, for 2 h. The 14C content in oocytes was subsequently determined. Oocytes expressing ZmSUT1 were used as positive controls for sucrose uptake. Oocytes injected with water were used as the negative control. The experiments were repeated using three different batches of oocytes independently with similar results.
TEVC recording from Xenopus oocytes
For whole-cell recordings, the TEVC technique was applied using a Gene-Clamp 500B amplifier (Molecular Devices) at room temperature (20°C). The microelectrodes were filled with 3 M KCl. Whole-cell currents digitized through a Digidata 1322A AC/DC converter using Clampex 9.0 software (Axon Instruments). For sugar assays, the bath solution contained 90 mM Na-gluconate, 2 mM K-gluconate, 1 mM Mg-gluconate2, 1.8 mM Ca-gluconate2, 1 mM LaCl3, 10 mM MES (pH 5.5 adjusted with Tris), and different concentrations of sugar as indicated. The osmolality of bath solutions was adjusted to 260 mmol/kg with sorbitol. The ionic currents were recorded using the TEVC technique. The oocyte membrane potential was held at −60 mV and test potentials ranging from +60 to −100 mV were applied for 800 ms using a step recording mode. The I/V relationships were obtained by subtracting the I/V curves defined in the absence of sugar from those obtained in the presence of sugar.
For potassium, the bath solution contained 15 mM Na-gluconate, 1 mM Mg-gluconate2, 1.8 mM Ca-gluconate2, 1 mM LaCl3, 10 mM MES (pH 5.5 adjusted with Tris), and different concentrations of K-gluconate. The osmolality of bath solutions was adjusted to 260 mmol/kg with sorbitol. The ionic currents were recorded using the TEVC technique. The oocyte membrane potential was held at −60 mV and test potentials ranging from +60 to −80 mV were applied for 800 ms using a step recording mode.
For nitrate, chloride, and dipeptides, the bath solution was based in 210 mM mannitol, 10 mM MES (pH 5.5 adjusted with Tris), containing the following candidate substrates 10 mM Gly–Gly, 10 mM HCl, and 10 mM Ala–Phe or CsNO3, respectively. The oocytes expressing ZmSUGCAR1 were incubated in bath solutions as indicated and the ionic currents were recorded using the TEVC technique. The oocyte membrane potential was held at −60 mV and test potentials ranging from −20 to −120 mV were applied for 800 ms using a step recording mode.
The experiments were repeated using three different batches of oocytes independently with similar results. More than five oocytes were used for uptake and electrophysiological analysis and only three were examined when there was no positive result at a given working condition.
HEK293T cell culture and whole cell patch-clamping experiments
HEK293T cells were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin as described by Gao et al. (2016). For patch-clamp experiments in HEK293T cells, the CDS of ZmSUGCAR1 was fused upstream of enhanched green fluorescent protein (EGFP) sequences with an IRES linker and constructed into PIRES2-EGFP vector. Plasmids for HEK293T transfection were extracted from Escherichia coli (DH5α) using a QIAGEN Plasmid kit. HEK293T cells were plated at a density at 60% confluence in 35-mm-diameter dishes and transfected using a Lipofectamine 2000 Transfection Reagent kit (Invitrogen, USA) in OPTI-MEM (Gibco, ThermoFisher). Cells were transfected with 0.8 μg of ZmSUGCAR1-pIRES2-EGFP or pIRES2-EGFP. Cells were analyzed 24 h after transfection. HEK293T cells of a similar size showing bright EGFP fluorescence were used for patch-clamp experiments and the HEK293T cells expressing EGFP were used as mock control. Standard whole-cell recording techniques were applied (Hamill et al., 1981). Pipettes were pulled with a glass capillary puller (Model PC-10; Narishige, Japan) and polished using a micro-forge (Model MF-830; Narishige). Their resistance was 4–6 MΩ. Whole-cell patch-clamp experiments were conducted using an Axopatch-200B patch-clamp setup (Axon Instruments, California, USA) with a Digitata1440A digitizer combined with an inverted microscope (Model A1; Carl Zeiss, Germany). pClamp 10.2 software (Axon Instruments) was used for data acquisition and analysis. Voltage protocol: 1 s pulses from −180 mV to + 40 mV (20 mV steps), holding potential −60 mV. The standard pipette solution contained 100 mM K-gluconate, 2 mM Mg-gluconate2, 2.5 mM Mg-ATP, and 10 mM HEPES (pH 7.4 adjusted with Tris).
For Figure 6, B and C, the bath solution contained 50 mM Na-gluconate, 10 mM MES (pH 5.5 adjusted with Tris), and K-gluconate was added to the bath solution by substituting 50 mM Na-gluconate to increase external potassium concentration to 10 and 50 mM successively. For Figure 3, D and E and Supplemental Figures S4, G and S6, A–C, the bath solution contained 50 mM Na-gluconate, 0/20 mM sucrose, and 10 mM MES (pH 5.5 or 7.5 adjusted with Tris). For Figure 7, C–E and Supplemental Figure S10B, the bath solution contained 50 mM Na-gluconate, 0/20 mM sucrose, 10 mM MES (pH 5.5 adjusted with Tris), and K-gluconate was added to the bath solution by substituting 50 mM Na-gluconate to increase external potassium concentration to 10 mM. The osmolality of bath and pipette solutions was adjusted to 350 mmol/kg with d-sorbitol.
NMT/MIFE assays
Oocytes were injected with water, ZmSUGCAR1, or ZmSUT1 cRNA and incubated in MBS solution for 30 h. Prior to H+ flux rate measurement, oocytes were equilibrated in the measuring solution for 1 min. The measuring solution for H+ contained (in mM) 88.95 NaCl, 0.05 KCl, 0.71 CaCl2, 0.82 MgSO4, and 0.03 MES/HEPES, pH 5.5/7.5 (Li et al., 2017). For Figure 3F and Supplemental Figure S6D, the H+ flux rate induced by sucrose/glucose, when 20 mM sucrose/glucose was added into the measuring solution, the concentration of NaCl was adjusted to 78.95 mM to maintain the osmolarity. For Figure 7B, KCl was added to the bath solution by substituting NaCl to increase external potassium concentration to 1 and 10 mM. After equilibrium 1 min, the sample oocyte was transferred to the container containing measuring solution. The H+ flux rates across the oocyte membrane were measured continuously for 2 min when the flux rate reached to the steady state. The mean net H+ flux was calculated based on all the transient H+ flux data recorded during the period of measuring time. The value obtained from NMT indicates net ion flux and the negative values in the figures represent H+ influx.
Hormone uptake assays in oocytes
Hormone uptake assays were performed with modifications as described previously (Tal et al., 2015). GA3 and JA were obtained from Sigma-Aldrich. Oocytes injected with cRNA were incubated for 48 h at 17°C in Kulori-based buffer (in mol: 90 NaCl, 1 KCl, 2 MgCl2, 10 HEPES-NaOH, pH 7.5) before assaying. Xenopus oocytes uptake assays were carried out as follows: oocytes were preincubated in Kulori-based buffer (in mol: 90 NaCl, 1 KCl, 2 MgCl2, 10 MES-Tris, pH 5.5) for 5 min, then transferred to Kulori-based buffer in pH 5.5 or 7.5 with 100 μM JA or 100 μM GA3 at 18°C for 60 min incubation, followed by four washes and transferred to Eppendorf tubes (3 oocytes per tube). Excess washing buffer was removed and oocytes were burst in 50 μL of 50% MeOH and the homogenate was left in the freezer for 2 h. This was followed by centrifugation at 20,000 g for 15 min to pellet remaining protein. The supernatant was transferred to new tubes and diluted with 60 μL H2O. The diluted samples were then filtered through a 0.45-μm PVDF-based filter plate (MSHVN4550, Merck Millipore) and subsequently analyzed by analytical LC-MS.
Analyses of hormones and hormone analogs by LC-MS
Compounds in extracts of oocytes were directly analyzed by LC-MS/MS. GA3 and JA analysis was performed on a quadrupole linear ion trap hybrid mass spectrometer (QTRAP 5500, AB SCIEX, Foster City, California, USA) equipped with an electrospray ionization source coupled with a UPLC (Waters, Milford, Massachusetts, USA). Five microliters of each sample were injected onto a BEH C18 column (100 mm * 2.1 mm, 1.7 μm). The UPLC inlet method and ESI source parameters were set as reported previously (Xin et al., 2020). GA3 and JA were detected in negative multiple reaction monitoring (MRM) mode. The MRM transitions for GA3 and JA are 345.2 > 143.1.1 and 209.1 > 59.0. Extracted data were analyzed using Microsoft Excel, statistical analysis and data plotting were done using SigmaPlot version 13.0 (Systat Software, USA).
Statistical analysis
Data processing with simple calculations (e.g., mean, standard deviation/error, maximum, and minimum) was performed by Microsoft Excel (version 16.55). One-way analysis of variance (ANOVA) was performed by the R program (version 3.6.1) and P < 0.05 was considered statistically significant across treatments, which was alternatively subject to the Mann–Whitney U test by the function “wilcox.test” in the R program (version 3.6.1) at the same significance level. All statistical results were listed in Supplemental Data Set S5.
Accession numbers
Sequence data of ZmSUGCAR1 can be found in the Maize Genetics and Genomics Database (https://maizegdb.org/) under the following accession number: Zm00001d019294. Sequence data of A. thaliana AtNPF7.3/NRT1 and AtNPF7.25 from this article can be found in TAIR under accession numbers AT1G32450.1 and AT4G21680.1, respectively; S. bicolor SbSUGCAR1 following accession number Sb02g007605.1 is from Gramene database; T. aestivum TaSUGCAR1 under accession number TraesCSU02G130200 is from http://www.wheat-expression.com. The other accession numbers used for phylogenetic analysis can be found in Figure 4 and Supplemental Figure S7. General sequence data in this manuscript can be found in the GenBank/EMBL database and RNA-Seq data generated in this study have been deposited in the NCBI SRA database with an accession number PRJNA824795.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genetic analysis of the zmsugcar1-1 mutant and map-based cloning of zmsugcar1-2.
Supplemental Figure S2. Morphological and histological comparison of WT and zmsugcar1-1 plants, kernels, or the BETL at different growth stages.
Supplemental Figure S3. Physiological measurements of WT and crispr plants.
Supplemental Figure S4. Expression and localization of ZmSUGCAR1, sugar concentration, and sugar transport activities of ZmSUGCAR1.
Supplemental Figure S5. ZmSUGCAR1 did not mediate sugar efflux in Xenopus oocytes.
Supplemental Figure S6. K+ rather than sugar uptake by ZmSUGCAR1 at pH 7.5.
Supplemental Figure S7. Phylogenetic analysis of SUGCAR1 homologous proteins from different plant species.
Supplemental Figure S8. Gene structure and expression profile of SbSUGCAR1 (A) and TaSUGCAR1 (B).
Supplemental Figure S9. Homologous proteins AtNPF7.3 and ZmNPF7.10 of ZmSUGCAR1 did not transport sugar.
Supplemental Figure S10. Current/voltage (I/V) relationships of currents for ZmSUGCAR1 triggered by different substrates in oocytes and related competition assay.
Supplemental Figure S11. ZmSUGCAR1 did not mediate auxin (IAA), JA, and GA transport in Xenopus oocytes.
Supplemental File S1. Full-length sequence alignment used for phylogenetic analysis shown in Figure 4A.
Supplemental File S2. The Newick format of the phylogenetic tree shown in Figure 4A.
Supplemental File S3. Full-length alignment used for phylogenetic analysis shown in Supplemental Figure S7.
Supplemental File S4. The Newick format of the phylogenetic tree shown in Supplemental Figure S7.
Supplemental Data Set S1. Significant gene differential expression in zmsugcar1-1 and WT kernels at four different stages.
Supplemental Data Set S2. A list of genes differentially expressed in zmsugcar1-1 and WT kernels on 14 and 20 DAP for heat map illustration.
Supplemental Data Set S3. Molecular marker primers used for map-based cloning.
Supplemental Data Set S4. Primers used for plasmid construction and functional analysis.
Supplemental Data Set S5. Statistical analyses.
Supplementary Material
Acknowledgments
We thank Dr. Jinfang Chu at Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for technical assistance in hormone transport assays.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2021YFF1000500, 2021YFD1200700, 2018YFA0901000), National Natural Science Foundation of China (91935303, 31972491) and Beijing Outstanding University Discipline Program.
Conflict of interest statement. The authors declare no conflicts of interest.
Contributor Information
Bo Yang, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Jing Wang, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Miao Yu, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB), College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Meiling Zhang, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Yanting Zhong, The Key Laboratory of Plant–Soil Interactions (MOE), Department of Plant Nutrition, College of Resources and Environmental Sciences, China Agricultural University, Beijing 100193, China.
Tianyi Wang, National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Peng Liu, Plant Molecular and Cellular Biology Program, Horticultural Sciences Department, Genetics Institute, University of Florida, Gainesville, Florida, USA.
Weibin Song, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Haiming Zhao, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
Astrid Fastner, Institute of Plant Sciences, University of Bern, Bern 3013, Switzerland.
Marianne Suter, Institute of Plant Sciences, University of Bern, Bern 3013, Switzerland.
Doris Rentsch, Institute of Plant Sciences, University of Bern, Bern 3013, Switzerland.
Uwe Ludewig, Institute of Crop Science, Nutritional Crop Physiology (340h), University of Hohenheim, Stuttgart 70593, Germany.
Weiwei Jin, National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China; Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University, Beijing 100193, China.
Dietmar Geiger, Department of Molecular Plant Physiology and Biophysics, Julius-von-Sachs-Institute for Biosciences, University of Würzburg, Würzburg 97082, Germany.
Rainer Hedrich, Department of Molecular Plant Physiology and Biophysics, Julius-von-Sachs-Institute for Biosciences, University of Würzburg, Würzburg 97082, Germany.
David M Braun, Division of Biological Sciences, Interdisciplinary Plant Group, and Missouri Maize Center, University of Missouri, 116 Tucker Hall, Columbia, Missouri 65211, USA.
Karen E Koch, Plant Molecular and Cellular Biology Program, Horticultural Sciences Department, Genetics Institute, University of Florida, Gainesville, Florida, USA.
Donald R McCarty, Plant Molecular and Cellular Biology Program, Horticultural Sciences Department, Genetics Institute, University of Florida, Gainesville, Florida, USA.
Wei-Hua Wu, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB), College of Biological Sciences, China Agricultural University, Beijing 100193, China; Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University, Beijing 100193, China.
Xuexian Li, The Key Laboratory of Plant–Soil Interactions (MOE), Department of Plant Nutrition, College of Resources and Environmental Sciences, China Agricultural University, Beijing 100193, China; Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University, Beijing 100193, China.
Yi Wang, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB), College of Biological Sciences, China Agricultural University, Beijing 100193, China; Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University, Beijing 100193, China.
Jinsheng Lai, State Key Laboratory of Plant Physiology and Biochemistry (SKLPPB) and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China; Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University, Beijing 100193, China.
J.L., B.Y., Y.W., and J.W. designed the research. B.Y., J.W., M.Y., M.Z., Y.Z., T.W., P.L., W.S., H.Z., A.F., and M.S. conducted the experiments. B.Y., J.L., Y.W., and X.L. wrote the article. R.H., D.G., W.-H.W., D.R., U.L., W.J., D.M.B., K.E.K., and D.R.M. revised the article.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Jinsheng Lai (jlai@cau.edu.cn), Yi Wang (yiwang@cau.edu.cn), Xuexian Li (steve@cau.edu.cn)
References
- Bai AN, Lu XD, Li DQ, Liu JX, Liu CM (2016) NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res 26: 384–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RF, Leach KA, Braun DM (2012) SWEET as sugar: new sucrose effluxers in plants. Mol Plant 5: 766–768 [DOI] [PubMed] [Google Scholar]
- Baker RF, Leach KA, Boyer NR, Swyers MJ, Benitez-Alfonso Y, Skopelitis T, Luo A, Sylvester A, Jackson D, Braun DM (2016) Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiol 172: 1876–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero C, Royo J, Grijota-Martinez C, Faye C, Paul W, Sanz S, Steinbiss HH, Hueros G (2009) The promoter of ZmMRP-1, a maize transfer cell-specific transcriptional activator, is induced at solute exchange surfaces and responds to transport demands. Planta 229: 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Bezrutczyk M, , HartwigT, , HorschmanM, , CharSN, , YangJ, , YangB, , FrommerWB, , Sosso D (2018) Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol 218: 594–603 [DOI] [PubMed] [Google Scholar]
- Bihmidine S, Hunter CT 3rd, Johns CE, Koch KE, Braun DM (2013) Regulation of assimilate import into sink organs: update on molecular drivers of sink strength. Front Plant Sci 4: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL (2009) Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol 149: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Wang L, Ruan YL (2014) Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signaling to enhance crop yield and food security. J Exp Bot 65: 1713–1735 [DOI] [PubMed] [Google Scholar]
- Buttner M (2007) The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett 581: 2318–2324 [DOI] [PubMed] [Google Scholar]
- Carpaneto A, Geiger D, Bamberg E, Sauer N, Fromm J, Hedrich R (2005) Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. J Biol Chem 280: 21437–21443 [DOI] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J (2014) Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol 166: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1996) The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8: 971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Cheng WH, Taliercio EW, Im KH (1995) Genetic aspects of sucrose metabolizing enzymes in developing maize seed. In Madore MA and Luca WJ, eds, Carbon Partitioning and Sucrose–Sink interactions in Plants. American Society of Plant Physiologists, Rockville, Maryland, pp 239–245 [Google Scholar]
- Chourey PS, Jain M, Li QB, Carlson SJ (2006) Genetic control of cell wall invertases in developing endosperm of maize. Planta 223: 159–167 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M (2011) Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May ST, Hedrich R (2002) Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334–344 [DOI] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter Grotemeyer M, Flückiger R, Slusarenko AJ, Ward JM, Rentsch D (2004) AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. Plant J 40: 488–499 [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Kuo TM (1990) Sugar metabolism in developing kernels of starch-deficient endosperm mutants of maize. Plant Physiol 92: 990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll NM, Depege-Fargeix N, Rogowsky PM, Widiez T (2017) Signaling in early maize kernel development. Mol Plant 10: 375–388 [DOI] [PubMed] [Google Scholar]
- Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB (2015) SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol 25: 53–62 [DOI] [PubMed] [Google Scholar]
- Fastner A, Absmanner B, Hammes UZ (2017) Use of Xenopus laevis oocytes to study auxin transport. In Kleine-Vehn J, Sauer M, eds, Plant Hormones. Methods in Molecular Biology, Vol 1497. Humana Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdanowicz P, Michard E, Sandmann M, Rocha M, Corrêa LG, Ramírez-Aguilar SJ, Gomez-Porras JL, González W, Thibaud JB, van Dongen JT, et al. (2011) Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc Natl Acad Sci USA 108: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QF, Gu LL, Wang HQ, Fei CF, Fang X, Hussain J, Sun SJ, Dong JY, Liu H, Wang YF (2016) Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc Natl Acad Sci USA 113: 3096–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Dong Y, Lu S, Liu T, He C, Liu C, Liu Q, Dong R, Wang J, Li Y, . et al. (2020) Characterization and map-based cloning of miniature2-m1, a gene controlling kernel size in maize. J Integr Agric 19: 1961–1973 [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ (1981) Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch 391: 85–100 [DOI] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T (2002) Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol 43: 452–459 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jeong J, Suh S, Guan C, Tsay YF, Moran N, Oh CJ, An CS, Demchenko KN, Pawlowski K, Lee Y (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol 134: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazachkova Y, Zemach I, Panda S, Bocobza S, Vainer A, Rogachev I, Dong Y, Ben-Dor S, Veres D, Kanstrup C et al. (2021) The GORKY glycoalkaloid transporter is indispensable for preventing tomato bitterness. Nat Plants 7: 468–480 [DOI] [PubMed] [Google Scholar]
- Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N (2005) Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-Symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell 17: 204–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Kühn C, Grof CP (2010) Sucrose transporters of higher plants. Curr Opin Plant Biol 13: 288–298 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Frommer WB (2012) SUT sucrose and MST monosaccharide transporter inventory of the Selaginella genome. Front Plant Sci 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Leroux BM, Goodyke AJ, Schumacher KI, Abbott CP, Clore AM, Yadegari R, Larkins BA, Dannenhoffer JM (2014) Maize early endosperm growth and development: from fertilization through cell type differentiation. Am J Bot 101: 1259–1274 [DOI] [PubMed] [Google Scholar]
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y (2017) NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29: 2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang D, Yang R, Logan K, Chen H, Zhang S, Skaggs MI, Lloyd A, Burnett WJ, Laurie JD, et al. (2014) Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc Natl Acad Sci USA 111: 7582–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RJ, Grof CP, Patrick JW (2018) Mechanisms of phloem unloading: shaped by cellular pathways, their conductance and sink function. Curr Opin Plant Biol 43: 8–15 [DOI] [PubMed] [Google Scholar]
- Monjardino P, Rocha S, Tavares AC, Fernandes R, Sampaio P, Salema R, da Câmara Machado A (2013) Development of flange and reticulate wall ingrowths in maize (Zea mays L.) endosperm transfer cells. Protoplasma 250: 495–503 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534 [DOI] [PubMed] [Google Scholar]
- Patrick JW, Offler CE (2001) Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52: 551–564 [PubMed] [Google Scholar]
- Patrick JW (1997) Phloem unloading: sieve element unloading and post-sieve element transport. Annu Rev Plant Physiol Plant Mol Biol 48: 191–222 [DOI] [PubMed] [Google Scholar]
- Patrick JW (1990) Sieve element unloading: cellular pathway, mechanisms and control. Physiol Plant 78: 298–308 [Google Scholar]
- Pettigrew WT (2008) Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol Plant 133: 670–681 [DOI] [PubMed] [Google Scholar]
- Philippar K, Büchsenschutz K, Abshagen M, Fuchs I, Geiger D, Lacombe B, Hedrich R (2003) The K+ channel KZM1 mediates potassium uptake into the phloem and guard cells of the C4 grass Zea mays. J Biol Chem 278. [DOI] [PubMed] [Google Scholar]
- Porter GA, Knievel DP, Shannon JC (1985) Sugar efflux from maize (Zea mays L.) pedicel tissue. Plant Physiol 77: 524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter GA, Knievel DP, Shannon JC (1987) Assimilate unloading from maize (Zea mays L.) pedicel tissues. Plant Physiol 83: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V, Riewe D, Peukert M, Matros A, Strickert M, Radchuk R, Weier D, Steinbiß HH, Sreenivasulu N, Weschke W, et al. (2017) Down-regulation of the sucrose transporters HvSUT1 and HvSUT2 affects sucrose homeostasis along its delivery path in barley grains. J Exp Bot 68: 4595–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 12: 2281–2289 [DOI] [PubMed] [Google Scholar]
- Saito H, Oikawa T, Hamamoto S, Ishimaru Y, Kanamori-Sato M, Sasaki-Sekimoto Y, Utsumi T, Chen J, Kanno Y, Masuda S, et al. (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6: 6095–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, et al. (2011) Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J 68: 129–136 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM (2009) Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot 6: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalstig JG, Hitz WD (1987) Transport and metabolism of a sucrose analog (1′-fluorosucrose) into Zea mays L. endosperm without invertase hydrolysis. Plant Physiol 85: 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC (1972) Movement of C-Labeled assimilates into kernels of Zea mays L: I. Pattern and rate of sugar movement. Plant Physiol 49: 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Knlevel DP, Chourey PS, Liu SY, Liu KC (1993) Carbohydrate metabolism in the pedicel and endosperm of miniature maize kernels. Plant Physiol 102: 42 [Google Scholar]
- Shannon JC, Porter GA, Knlevel DP (1986) Phloem unloading and transfer of sugars into developing corn endosperm. In Cronshaw J, Lucas WJ, Giaquinta RT, eds, Phloem Transport, Alan Liss, Inc., New York, pp 265–277 [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, et al. (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47: 1489–1493 [DOI] [PubMed] [Google Scholar]
- Tal I, Zhang Y, Jørgensen ME, Pisanty O, Barbosa IC, Zourelidou M, Regnault T, Crocoll C, Olsen CE, Weinstain R, et al. (2016) The Arabidopsis NPF3 protein is a GA transporter. Nat Commun 7: 11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Schatz MC (2009) Optimizing data intensive GPGPU computations for DNA sequence alignment. Parallel Comput 35: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Takahashi N, Kanno Y, Suzuki H, Aoi Y, Takeda-Kamiya N, Toyooka K, Kasahara H, Hayashi KI, Umeda M, et al. (2020) The Arabidopsis NRT1/PTR FAMILY protein NPF7.3/NRT1.5 is an indole-3-butyric acid transporter involved in root gravitropism. Proc Natl Acad Sci USA 117: 31500–31509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U (1997) A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell 9: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YM, Ren ZJ, Wang BH, Zhang L, Zhao YJ, Wu JW, Li LG, Zhang XS, Zhao XY (2021) A nitrate transporter encoded by ZmNPF7.9 is essential for maize seed development. Plant Sci 308: 110901. [DOI] [PubMed] [Google Scholar]
- Wen Z, , TyermanSD, , DechorgnatJ, , OvchinnikovaE, , DhuggaKS, , KaiserBN (2017) . Maize NPF6 Proteins Are Homologs of Arabidopsis CHL1 That Are Selective for Both Nitrate and Chloride. Plant Cell 29: 2581–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin P, Guo Q, Li B, Cheng S, Yan J, Chu J (2020) A tailored high-efficiency sample pretreatment method for simultaneous quantification of 10 classes of known endogenous phytohormones. Plant Commun 1: 100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N (2015) Structural biology of the major facilitator superfamily transporters. Annu Rev Biophys 44: 257–283 [DOI] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, et al. (2019) High-temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31: 974–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, , ThakareD, , MaC, , LloydA, , NixonNM, , ArakakiAM, , BurnettWJ, , LoganKO, , WangD, , Wang X et al. (2015) RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27: 513–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Zhou Y, Dibley K, Tyerman S, Furbank R, Patrick J (2007) Nutrient loading of developing seeds. Funct Plant Biol 34: 314–331 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang Z (2010) Current opinions on endosperm transfer cells in maize. Plant Cell Rep 29: 935–942 [DOI] [PubMed] [Google Scholar]
- Zhou L, Liu CX, Chen QL, Wang WQ, Yao S, Zhao ZK, Zhu SY, Hong XD, Xiong YH, Cai YL (2021) Fine mapping and candidate gene analysis of maize defective kernel mutant dek54. Acta Agron Sin 47: 1903–1912 [Google Scholar]
- Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J (2016) Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J Genet Genomics 43: 25–36 [DOI] [PubMed] [Google Scholar]
- Zörb C, Senbayram M, Peiter E (2014) Potassium in agriculture-status and perspectives. J Plant Physiol 171: 656–669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.