Abstract
Petal senescence is a crucial determinant for ornamental quality and economic value of floral crops. Salicylic acid (SA) and reactive oxygen species (ROS) are two prominent factors involved in plant senescence regulation. In this study, tulip TgNAP (NAC-like, activated by APETALA3/PISTILLATA) was characterized as positively regulating tulip petal senescence through dually regulating SA biosynthesis and ROS detoxification pathways. TgNAP was upregulated in senescing petals of tulip while exogenous SA and H2O2 treatments substantially promoted petal senescence in tulip. Silencing of TgNAP by VIGS assay delayed SA and H2O2-induced petal senescence in tulip, whereas overexpression of TgNAP promoted the senescence process in Arabidopsis (Arabidopsis thaliana) plants. Additionally, inhibition of SA biosynthesis prolonged the lifespan of TgNAP-silenced petal discs. Further evidence indicated that TgNAP activates the transcriptions of two key SA biosynthetic genes ISOCHORISMATE SYNTHASE 1 (TgICS1) and PHENYLALANINE AMMONIA-LYASE 1 (TgPAL1) through directly binding to their promoter regions. Meanwhile, TgNAP repressed ROS scavenging by directly inhibiting PEROXIDASE 12 (POD12) and POD17 expression. Taken together, these results indicate that TgNAP enhances SA biosynthesis and ROS accumulation to positively regulate petal senescence in tulip.
NAC transcription factor TgNAP promotes petal senescence through regulating salicylic acid and removing reactive oxygen species in Tulipa gesneriana.
Introduction
Plant senescence is the last stage during the whole developmental process, eventually resulting in death of cells, tissues, and organisms (Gan and Amasino, 1997; Zhang et al., 2021). Senescence generally occur in different organs, such as leaves, stems, roots, and petals (Schippers et al., 2015; Woo et al., 2019). Petal senescence is a complicated process and closely related to economic value of ornamental plants (Tripathi and Tuteja, 2007; Rogers, 2013; Ma et al., 2018). Plant senescence is regulated not only by internal factors such as natural aging, reproduction, reactive oxygen species (ROS), hormones, and sugar, but also by external factors including dehydration, temperature, light, ionic accumulation, nutrition limitation, and pathogen attack or wounding (Gan and Amasino, 1997; Woo et al., 2013; Zhao et al., 2016; Mhamdi and Van Breusegem, 2018). Among plant hormones, cytokinin (CTK) and gibberellins (GAs) are considered to retard senescence, while abscisic acid (ABA), ethylene (ETH), and salicylic acid (SA) are commonly regarded as aging-promoting hormones (Gan and Amasino, 1997; Morris et al., 2000; He et al., 2002; Zhao et al., 2016; Lee et al., 2021). As crucial plant hormones, ethylene and ABA have been well studied to participate in leaf senescence. Moreover, further studies have revealed that NAC TF family plays a vital role in ethylene- and ABA-induced leaf senescence. In maize (Zea mays), ZmNAC126 acts as a positive regulator in ethylene-induced leaf senescence functioning downstream of ZmEIN3 (ETHYLENE-INSENSITIVE3; Yang et al., 2020). ORESARA1 (ORE1), a NAC transcription factor in Arabidopsis (Arabidopsis thaliana), is also able to promote leaf senescence in ethylene signaling pathway by activating the expression of 1-AMINOCYCLOPROPANE-1-CARBOXYLIC SYNTHASE2 (ACS2; Qiu et al., 2015). Tomato (Solanum lycopersicum) SlNAP2 directly upregulates expression of SlSAG113 (SENESCENCE-ASSOCIATED GENE113) and ABA biosynthesis-related gene SlNCED1 (9-cis-EPOXYCAROTENOID DIOXYGENASE 1) to promote leaf senescence and establish ABA homeostasis (Ma et al., 2018). OsNAC2 increases ABA levels via regulating expression of ABA biosynthetic genes (Mao et al., 2017).
A growing body of evidence indicates that SA plays important roles in natural senescence process (Rivas-San Vicente and Plasencia, 2011; Zhang et al., 2013). SA level was 4-fold higher in senescing leaves when compared to young leaves (Morris et al., 2000), and SA accumulation further led to premature senescence in leaves (Vogelmann et al., 2012; Li et al., 2016). SA induced the expression of a couple of senescence-associated genes (SAGs; Wang et al., 2021a). Meanwhile, about 20% of senescence-enhanced genes exhibit substantially decreased expression in SA-deficient NahG transgenic Arabidopsis (Morris et al., 2000). Arabidopsis mutants including pad4 and npr1, which are defective in SA signaling pathway, showed delayed senescence phenotype and a great reduction of SAGs expression (Morris et al., 2000; Woo et al., 2019). Moreover, transcriptome analysis shows an intriguing observation that SA treatment has similar genomic change patterns with age-mediated leaf senescence (Pyung et al., 2007), further suggesting the positive role of SA in senescence progress.
ROS not only acts as toxic by-product, but also as signaling molecules to regulate development, growth, and defense response of plants (Miller et al., 2010; Huang et al., 2016). The excessive ROS production might damage macromolecules stabilization resulting in senescence initiation and cell death (Miller et al., 2010). H2O2 contents increase in different senescing plants, such as senescing peach (Prunus persica) fruit (Wu et al., 2016), ripening tomato (Jimenez et al., 2002), and Arabidopsis aged yellow leaf (Niu et al., 2020; Wang et al., 2020a). The cross-talk between oxidative burst and SA has been extensively reported. Recent studies have revealed that SA and ROS pathways are incorporated to modulate senescence process in plants (Guo et al., 2017). SA can induce the ROS accumulation (Rao et al., 1997; Wang et al., 2021b) and H2O2 also elicits the generation of SA (León et al., 1995). Altering SA biosynthesis resulted in a modulation of the catalase (CAT) expression, indicating that ROS function as upstream regulator of SA pathway (Han et al., 2013). Additionally, SA conversely regulates ROS accumulation through inhibiting antioxidant enzymes activities and reducing glutathione biosynthesis (Chen et al., 1993; Vlot et al., 2009). Therefore, the interactions between SA and ROS are complicated.
Transcriptional analysis has proved that a number of TFs are involved in senescence process in various plant species, including WRKYs, ERFs, MYBs, bHLH, homeobox, and NACs (NAM/ATAF/CUC), which further regulate expressions of downstream senescence associate genes (Lü et al., 2014; Song et al., 2014; Chen et al., 2017; Guo et al., 2017; Niu et al., 2020; Wang et al., 2020a; Yu et al., 2022). In Arabidopsis, 57.5% (65/113) of NAC genes exhibited expression changes during leaf senescence. Genetic study revealed that ANAC002/ATAF1, ANAC016, ANAC017/NTL7, ANAC019, ANAC029/AtNAP, ANAC059/ORS1, ANAC081/ATAF2, ANAC092/ORE1, and ANAC096 function as positive regulators of leaf senescence (Guo and Gan, 2006; Balazadeh et al. 2011; Wu et al., 2012; Kim et al., 2013; Nagahage et al., 2020). Balazadeh et al. (2010) identified 170 downstream genes induced by ANAC092/ORE1 and found that 46% of them are known to be involved in senescence pathway. ANAC059/ORS1 was induced by H2O2 treatment to trigger expression of senescence-associated gene regulatory networks which accelerated plant senescence (Balazadeh, et al. 2011). Other NACs, including ANAC042/JUB1, ANAC075/NAC075, and ANAC083/VNI2 in Arabidopsis, and LpNAL in perennial ryegrass (Lolium perenne L.) were reported to be negative regulators of leaf senescence (Yang et al., 2011; Wu et al., 2012; Kan et al., 2021; Yu et al., 2022). NAC transcription factor VNI2 was induced by ABA and delayed leaf aging through binding to the promoters of COR and RD genes (Yang et al., 2011). NAC genes NTL9, ATAF2, and CaNAC1 were substantially induced by SA and involved in biotic stress and leaf senescence (Delessert et al., 2005; Oh et al., 2005; Zheng et al., 2015; Nagahage et al., 2020). Recently, ANAC017, ANAC082, and ANAC090 were reported to negatively regulate leaf senescence by suppressing SA and ROS pathways (Kim et al., 2018). It is suggested that NAC TF may play a crucial role in SA signaling pathway. NAC075 activates CAT2 to reduce ROS accumulation and negatively regulates leaf senescence (Kan et al., 2021). H2O2 treatment was reported to induce the expression of several senescence-associated NACs, such as ANAC029/AtNAP, ANAC002/ATAF1, ANAC042/JUB1, and ANAC059/ORS1 (Guo and Gan, 2006; Balazadeh, 2011; Wu et al., 2012). However, the detailed regulatory networks among NAC, SA, and ROS remain largely unknown.
ANAC090 of Arabidopsis negatively regulates leaf senescence by suppressing SA and ROS responses (Kim et al., 2018). In this study, however, we found that TgNAP promotes petal senescence in tulip (Tulipa gesneriana) through activation of SA biosynthesis and ROS detoxification pathways. TgNAP was identified as one of the substantially upregulated NACs in senescing tulip petals. The leaf senescence was promoted in TgNAP overexpressed transgenic plants, while TgNAP silencing expression delayed petal disc aging. TgNAP modulated SA biosynthesis and H2O2 accumulation by directly activating TgICS1 and TgPAL1 transcription, and inhibiting expressions of ROS scavenging genes TgPOD12 and TgPOD17. We report here that TgNAP functions as a positive regulator during tulip petal senescence partially in SA- and H2O2-dependent manners.
Results
Transcriptomic analysis during petal senescence in tulip
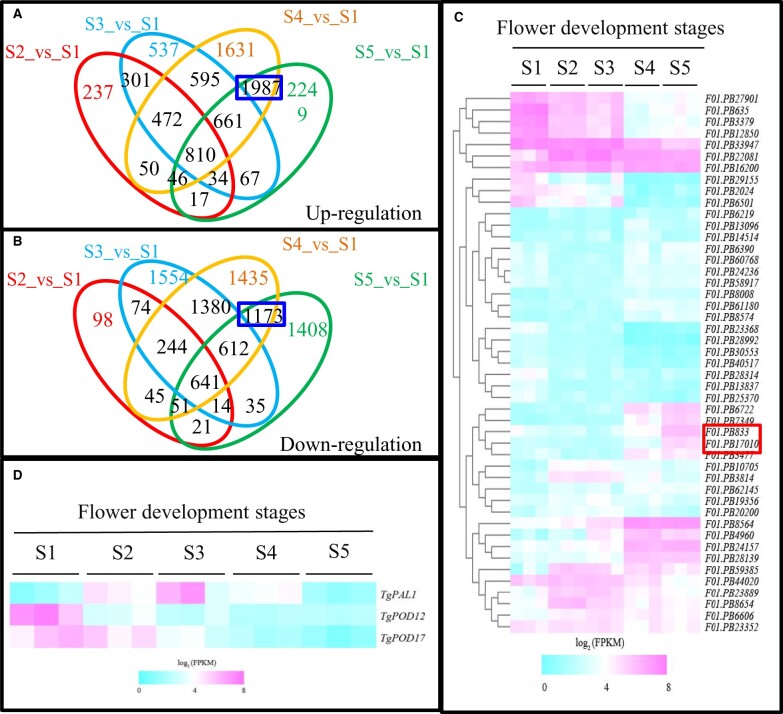
We previously investigated physiological and metabolite changes during tulip petal senescence (Wang et al., 2020b). RNA sequencing was performed to further dissect transcriptomic changes of tulip petal at five developmental stages. The results showed that 1967/1188, 3477/2644, 6252/5581 and 5871/3955 unigenes exhibited significant up-/downregulation from S2 to S5 stages when compared to S1 stage (Figure 1, A and B; Supplemental Table S1). Among them, 810 and 641 unigenes were commonly upregulated and downregulated at all developmental stages, respectively. Meanwhile, 1,987 and 1,173 unigenes were co-upregulated and co-downregulated only at petal senescence stages (S4_vs_S1 and S5_vs_S1; Figure 1, A and B; Supplemental Table S2). Protein interaction network analysis revealed that NAC transcription factor together with ROS, SA, ABA, and ET pathways-related genes functioned as the core nodes (Supplemental Figure S1a). We further obtained the expression patterns of NAC family genes in tulip. Totally 52 annotated NAC unigenes were identified. Unigenes F01.PB833 and F01.PB17010 were annotated as NAC029 and displayed relatively low expression during early blooming stages (S1–S3 stages) but had substantially increase in tulip petal at senescence stage (S4 and S5 stages; Figure 1C; Supplemental Table S3).
Figure 1.
RNA sequencing analysis during petal senescence of T. gesneriana. A and B, Overlapping analysis of differentially expressed unigenes. (A) Upregulation, (B) Downregulation. C, Expression profiles of 52 NAC genes in T. gesneriana during petal senescence. Two unigenes F01.PB17010 and F01.PB833 annotated as NAC029 were highlighted with red box. GFB (the first stage, S1), CFB (the second stage, S2), flowers in FB (the third stage, S3), flowers in ESP (the fourth stage, S4), and flowers in LSP (the fifth stage, S5). D, Expression profiles of TgPAL1, TgPOD12, and TgPOD17 in T. gesneriana during petal senescence.
As SA and ROS pathways are closely related with differential NACs during petal senescence in tulip, the expression of two SA biosynthesis-related genes and ROS scavenging genes were examined, respectively. TgPAL1 showed an obvious elevation from S2 to S4, peaking at S3 (Figure 1D; Supplemental Figure S1b). RT-qPCR results showed that TgICS1 exhibited steady upregulation during petal senescence (Supplemental Figure S1c). Heatmap and RT-qPCR results showed that TgPOD12 and TgPOD17 exhibited a marked decrease during senescence stage (Figure 1D; Supplemental Figure S1, d and e).
TgNAP is a petal senescence-related gene induced by SA
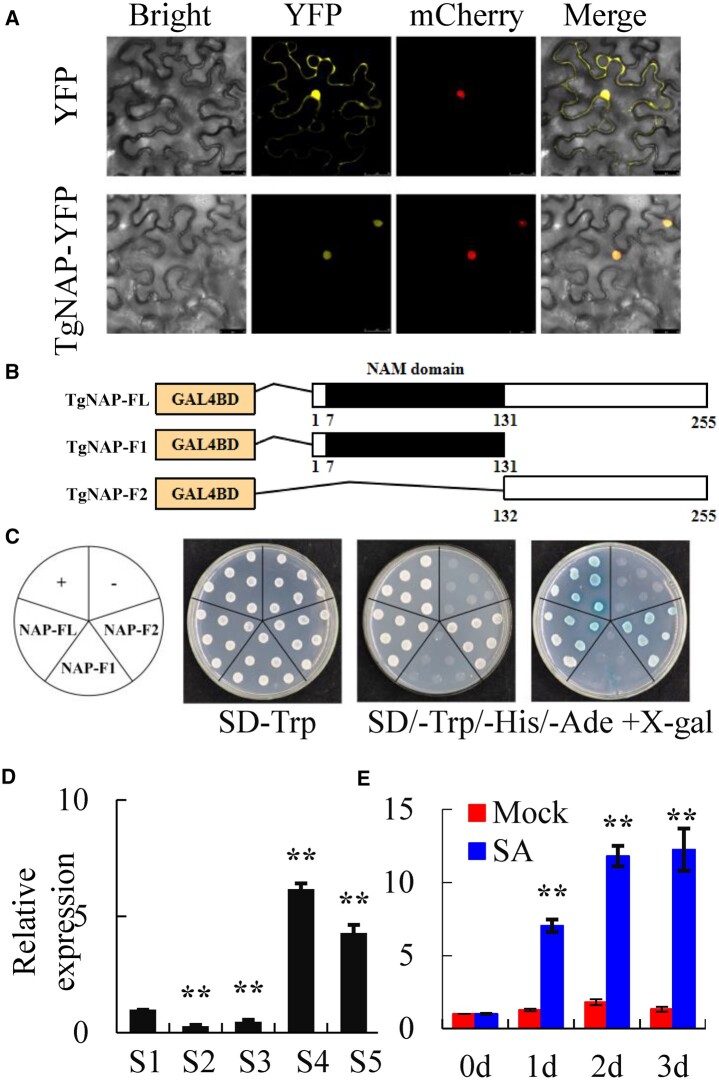
The open reading frame (ORF) of F01.PB833 is 765 bp, encoding a protein of 255 amino acids. Phylogenetic tree analysis indicated that F01.PB833 obtained the most similarity to AtNAC29 (AtNAP; Supplemental Figure S2a), which was thereby named as TgNAP. Protein domain analysis showed that TgNAP contains a conserved N-terminal region, including A–E five subdomains (Supplemental Figure S2b). TgNAP was localized to the nucleus in Nicotiana tabacum epidermal cells (Figure 2A). Truncated assay indicated that C terminus is essential for TgNAP transcriptional activation in yeast (Figure 2, B and C). Transcription level of TgNAP was confirmed by RT-qPCR. The results showed relatively lower expression at Stages 2 and 3 but significantly higher levels at Stages 4 and 5 (Figure 2D). Furthermore, TgNAP expression was induced greatly and significantly in tulip petals by exogenous SA treatment (Figure 2E). These results demonstrate that TgNAP is a tulip petal senescence-related gene induced by SA.
Figure 2.
Characterization of TgNAP during petal senescence in T. gesneriana. A, Subcellular location of TgNAP. The fusion construct TgNAP-YFP or YFP empty vector was co-transformed with VirD2NLS-mCherry (a nucleus marker) in N. benthamiana leaves. Confocal microscopic images showed bright field, yellow (for YFP), red (for mCherry) fluorescence signals in epidermal cells. Scale bars = 25 μm. B, Schematic diagrams of the full length (FL) and two truncated fragments (F1 and F2) of TgNAP used for constructing vectors. All constructed vectors were introduced in pGBKT7 vector with GAL4 DBD domain. The numbers below the bars indicate the positions of amino acid. C, Transcription activity assay. Growth of yeast cell (AH109 strain) transformed with various constructed vectors on SD/-Trp and SD/-Trp/-His/-Ade with or without X-α-gal selective medium. Transformation of pGBKT7-p53 and pGBKT7 vector was used as a positive and negative control, respectively. D and E, RT-qPCR analysis of TgNAP expression levels in T. gesneriana during petal senescence (D) or treated with 200 μM SA for 3 d (E). S1–S5 are abbreviations of first stage to fifth stage. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. Error bars represent ± SE (n = 3). Asterisks indicate significant difference between WT and two transgenic lines based on the LSD values (**P < 0.01).
Silencing of TgNAP delays petal senescence and inhibits SA synthesis in tulip
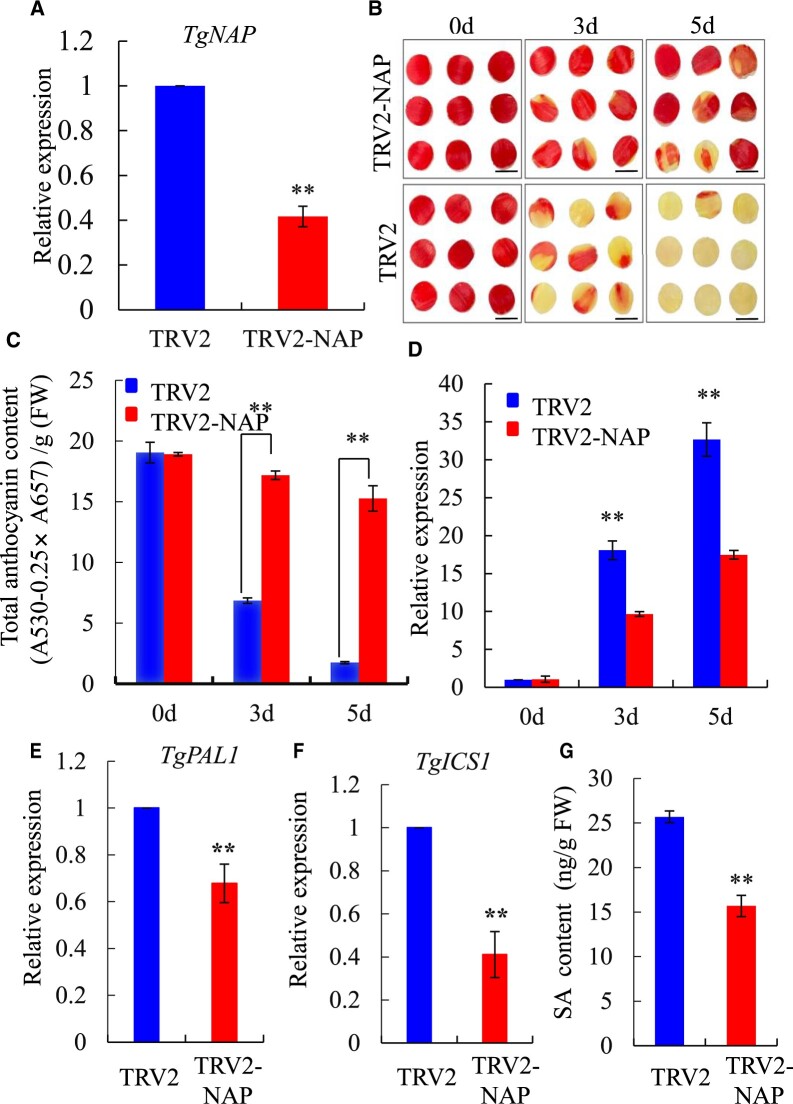
To further explore the potential role of TgNAP in petal senescence in tulip, TgNAP was silenced using virus-induced gene silencing (VIGS) method. The downregulated level of TgNAP was verified by genomic PCR (Supplemental Figure S3a) and RT-qPCR in TgNAP silenced petal discs and flowers (Figure 3A; Supplemental Figure S3b). The control petal discs exhibited color fading at Day 3 and completely turned yellow on Day 5 (Figure 3B). Conversely, silencing of TgNAP visibly delayed the petal senescence and only showed slight color fading on Day 5 (Figure 3B). In line with the phenotypic differences, significantly higher anthocyanin content and reduced transcript level of TgSAG6, which is the marker gene of senescence in tulip (Wang et al., 2020b), were observed in TgNAP silenced petal discs when compared to TRV2 control discs (Figure 3, C and D). In addition, SA biosynthesis genes TgICS1 and TgPAL1 were significantly repressed in TRV2-TgNAP (Figure 3, E and F) which was evidenced by reduced SA content in TRV2-TgNAP petal discs when compared to TRV2 control (Figure 3G). The flower longevity of TRV2 control flowers was significantly longer than that of TRV2-TgNAP (Supplemental Figure S3, c and d). The above results illustrated that TgNAP is involved in senescence process and SA biosynthesis in tulip petals.
Figure 3.
Silencing of TgNAP delayed senescence in tulip petal discs. A, Expression levels of TgNAP in the TRV2 control and TgNAP-silenced petals at Day 3 by RT-qPCR analysis. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. B, Phenotypes of tulip petal discs detached from TRV2 control and TRV2-TgNAP line under ambient conditions. Scale bar, 1 cm. The samples in each boxed area were photographed at the same time and are part of a single image. C, Total anthocyanin levels of TRV control and TgNAP-silenced petals. D, Expression levels of TgSAG6 of TRV2 control and TgNAP-silenced petals. E and F, Expression levels of TgPAL1 (E) and TgICS1 (F) of TRV2 control and TgNAP-silenced petals at Day 3. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. G, SA content in TRV2 control and TgNAP-silenced petals at Day 5. Error bars indicate SE (n = 3). Asterisks indicate significant difference between the TgNAP-silenced petals and the TRV control based on the LSD values (**P < 0.01).
Ectopic overexpression of TgNAP facilitates SA synthesis and leaf senescence
To confirm the potential role of TgNAP in plant senescence, transgenic Arabidopsis lines overexpressing TgNAP under the control of the cauliflower mosaic virus (CaMV) 35S promoter, were generated (Supplemental Figure S4, a–c). Under normal growth condition, no morphological differences were observed between the transgenic and wild-type plants during vegetative stage (Supplemental Figure S4a). However, TgNAP-OE transgenic lines exhibited a precocious rosette leaf senescence phenotype with more yellow mature leaves than WT plants on the 28th day after planting (Supplemental Figure S4b). Consistently, the chlorophyll content in TgNAP-OE rosette leaves was notably lower than that in WT (Supplemental Figure S4f). Additionally, the expressions of two senescence marker genes AtSAG12 and AtSAG13 were significantly induced in transgenic plants relative to WT (Supplemental Figure S4, d and e). TgNAP-OE transgenic plants also exhibited significantly higher H2O2 content when compared to WT (Supplemental Figure S4g).
Since SA level significantly increased in senescence tulip petal and TgNAP expression was markedly upregulated after SA treatment (Figure 2E), TgNAP could putatively promote petal senescence via SA-associated pathway. To verify the speculation, SA contents were measured in WT and TgNAP-OE transgenic Arabidopsis plants. The results revealed that SA level was significantly enhanced in TgNAP-OE lines when compared to WT (Supplemental Figure S4h). Moreover, the expressions of SA biosynthesis-related genes AtSID2, AtPAL1, AtEPS5, and AtEPS1 were significantly higher in TgNAP-OE than those in WT (Supplemental Figure S4, i–l). These results indicated that overexpression of TgNAP notably promoted leaf senescence in Arabidopsis via activating SA biosynthesis pathway.
TgNAP promotes senescence in an SA-dependent manner
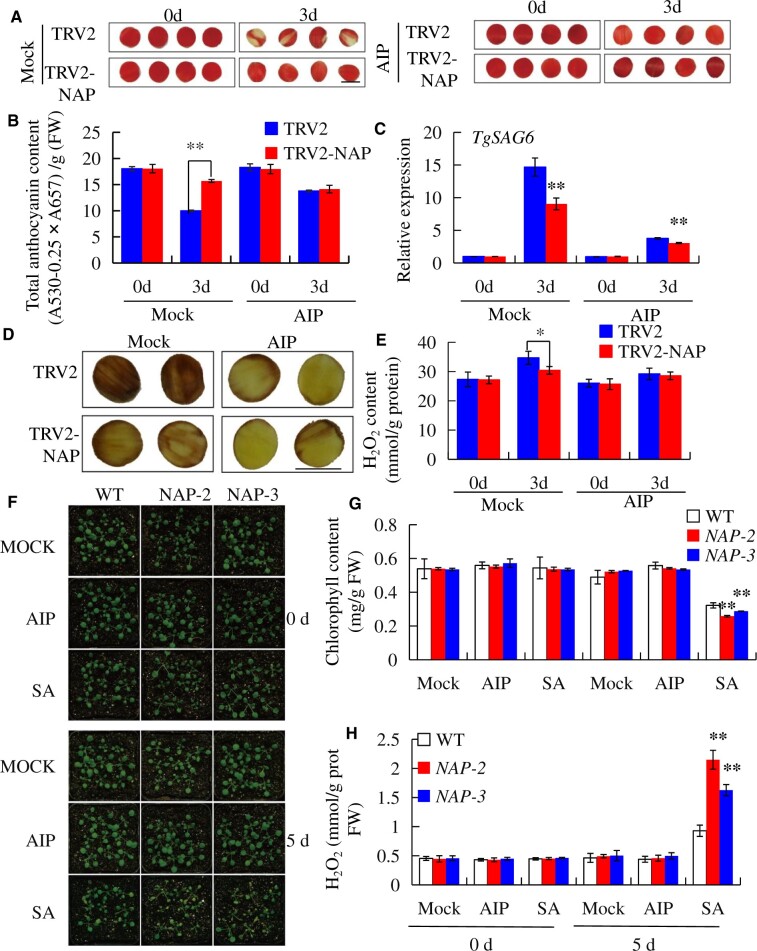
To explore if SA accumulation is necessary for TgNAP-promoted tulip petal senescence, the effect of SA and SA synthesis inhibitor (2-aminoindan-2-phosphonic acid, AIP; Solecka and Kacperska, 2013) on tulip petal and Arabidopsis leaf were further investigated. In the presence of AIP, petal senescence was obviously retarded as evidenced by similar color fading between TRV2-TgNAP and TRV2 (Figure 4A). Consistently, AIP treatment delayed anthocyanin degradation in TRV2 control petal discs as evidenced by similar anthocyanin contents between TRV2 and TRV2-TgNAP petal discs in the presence of AIP (Figure 4B). Additionally, TgSAG6 transcript was significantly reduced in both TRV2 and TRV2-TgNAP petal discs after AIP treatment (Figure 4C). TRV2 petal discs obtained significantly higher H2O2 accumulation in the absence of AIP than TRV2-TgNAP, however, AIP treatment resulted in similar H2O2 contents in TRV2-TgNAP and TRV2 petal discs (Figure 4, D and E). Furthermore, the 7–8 rosette leaves from 28-d-old WT and TgNAP-OE transgenic Arabidopsis were detached and treated with AIP and SA, respectively. After SA treatment for 3 d, the leaves of TgNAP-OE exhibited more yellowish than the leaves of WT (Supplemental Figure S5a). However, AIP treatment could obviously arrest the leaf senescence of TgNAP-OE (Supplemental Figure S5a). Consistently, the chlorophyll contents displayed decreased or increased levels after SA and AIP treatment, respectively, in either detected genotypes (Supplemental Figure S5b). Moreover, AIP treatment reduced H2O2 accumulation either in WT or in TgNAP-OE leaves (Supplemental Figure S5, c and d). Additionally, the whole plants of 2-week-old WT and TgNAP-OE transgenic Arabidopsis were also sprayed with AIP and SA. Upon 5 d of SA treatment, TgNAP-OE showed a larger portion of yellowing than WT (Figure 4F). Consistent with these phenotypes, the chlorophyll contents were lower and H2O2 levels were higher in TgNAP-OE transgenic Arabidopsis than WT (Figure 4, G and H). All these data indicated that TgNAP promotes plant senescence in an SA- and/or H2O2-dependent manner and SA biosynthesis potentially functions upstream of H2O2 accumulation in this process.
Figure 4.
Effects of SA biosynthesis on the senescence of tulip petals and 35S:TgNAP transgenic Arabidopsis. A, The phenotypes of TgNAP-silenced petals and the TRV control treated with 30 μM AIP. The samples in each boxed area were photographed at the same time. B and C, The total anthocyanin content (B) and expression level of TgSAG6 (C) of TRV control and TgNAP-silenced petals treated with AIP. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. Error bars indicate SE (n = 3). Asterisks indicate significant difference between WT and two transgenic lines based on the LSD values (**P < 0.01). Different letters denote significant differences at P < 0.05 analyzed by Tukey’s test. D, In situ detection of H2O2 in TRV control and TgNAP-silenced plants after AIP treatment, as revealed by histochemical staining with DAB. The samples in each boxed area were photographed at the same time and are part of a single image. E, H2O2 content in TRV control and TgNAP-silenced plants before and after AIP treatment. Error bars indicate SE (n = 3). Asterisks indicate significant difference between WT and two transgenic lines based on the LSD values (*P < 0.05). F, The phenotypes of 2-week-old WT and transgenic lines (#2 and #3) sprayed with or without 30 μM AIP and 200 μM SA for 5 d. G–I, Measurement of chlorophyll contents (G), DAB staining (H), and H2O2 contents (H) in the leaves shown in (F) on Day 5. Error bars indicate SE (n = 3). Asterisks indicate significant difference between WT and two transgenic lines based on the LSD values (*P < 0.05, **P < 0.01). Scale bar, 1.0 cm.
TgNAP directly activates TgPAL1 and TgICS1 expressions
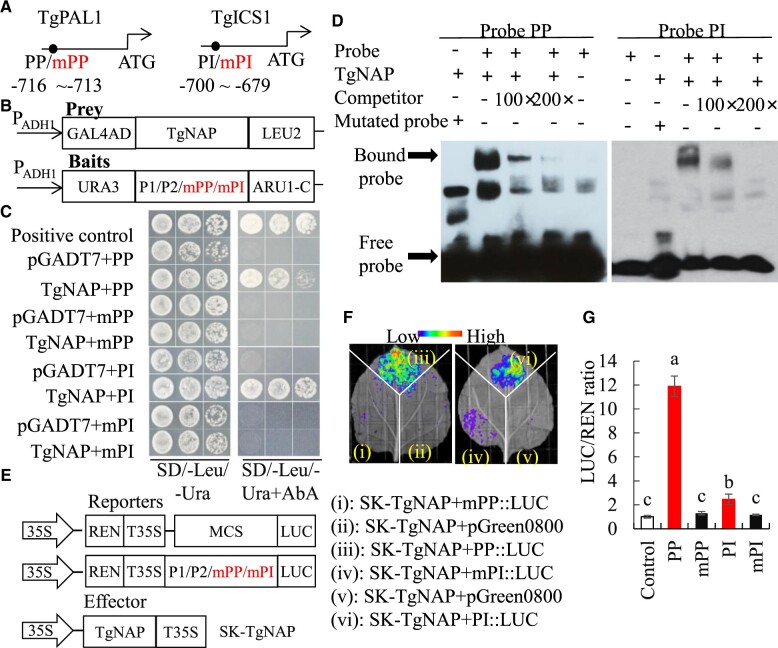
Isochorismate synthase (ICS) and phenylalanine ammonia-lyase (PAL) are two key enzymes in SA biosynthesis pathway, which are responsible for converting branched acid and phenylalanine into isochoric acid and trans-cinnamic acid, respectively (Chen et al., 2009; Dempsey et al., 2011). The promoter sequences of TgPAL1 and TgICS1 were cloned and both contain a core NAC-binding element located at −716 ∼−713 bp and −700 ∼−679 bp, respectively (Figure 5A). The binding sequence is very similar to the region of AtSAG113, AtSAG202, and AtCKX3 that AtNAP bound to (Zhang and Gan, 2012; Hu et al., 2021; Wang et al., 2022). To explore whether TgNAP directly promotes TgPAL1 and TgICS1 expression, yeast-one-hybrid (Y1H) assay was employed using TgNAP protein as the prey (Figure 5B). All transformed yeast grew well on SD/-Leu/-Ura media, whereas only positive control and yeast cells harboring TgNAP prey protein and wild-type TgNAP core binding sequence (PP and PI) survived on media supplemented with Aureobasidin A (AbA; Figure 5C). Electrophoretic mobility shift assay (EMSA) was carried out to document the interactions between TgNAP and core binding sequence. Gel-shift assay indicated His-TgNAP fusion protein is able to bind to the biotin-labeled probes PP and PI containing TgNAP core binding sequence. The protein-DNA complex binding was abolished when His-TgNAP fusion protein was incubated with mutated probes AAAG (mPP and mPI). After adding the unlabeled competitor probes, the binding capacity decreased gradually in a dosage-dependent manner (Figure 5D). These findings suggested that TgNAP directly and specifically binds to the promoter regions of TgPAL1 and TgICS1, respectively. A dual LUC reporter assay was further performed to examine the in vivo interactions of TgNAP protein with TgPAL1 and TgICS1 promoters in N. benthamiana leaves (Figure 5E). Substantial LUC fluorescence signals were observed in the presence of the TgNAP-SK effector and wild-type PP and PI reporters, whereas very low signals were found for mutated mPP and mPI reporters (Figure 5F). The LUC/REN ratio was significantly higher in co-transformation of TgNAP-SK and reporters containing TgNAP core binding sequence when compared to the control and mutated reporters (Figure 5G). Taken together, these results revealed that TgNAP could activate the TgPAL1 and TgICS1 promoter-driven transcription by binding to core-binding motif.
Figure 5.
Direct binding of TgNAP to promoters of TgPAL1 and TgICS1. A, Schematic diagram of the TgPAL1 and TgICS1 promoter. The black circles indicate the position of the partial promoter fragments within CACG motifs. PP: promoter of TgPAL1; PI: promoter of TgICS1. The numbers shown below indicated the position of specific binding of TgNAP in the promoters ahead of ATG. B, The prey and bait constructs used for Y1H assay. mPP and mPI is a mutated version of PP and PI, in which the CACG was replaced with AAAG. PADH1, promoter of alcohol dehydrogenase1. GAL4AD, galactose-specific transcription enhancing factor 4 activation domain. C, Growth of yeast cells co-transformed with various prey plus bait combinations on selective medium (SD/-Ura/-Leu) with (right) or without (left) AbA. AbA, Aureobasidin A. Positive control: p53-AbAi+pGAD-p53; Negative control: bait+pGADT7. D, EMSA analysis of specific binding of TgNAP to the promoter of TgPAL1 and TgICS1. The purified His-TgNAP protein and biotin-labeled probe of designed fragments containing CACG motif or mutated AAAG motif were used. Competitor was unlabeled probe at 100- and 200-fold. +: presence; −: absence. E, The schematic of the TgNAP effector and pTgICS1::LUC, pTgPAL1::LUC reporters. T35S, the terminator of CaMV 35S, respectively. MCS, multiple cloning sites. LUC, firefly luciferase. REN, Renilla luciferase. F, Live image of transcriptional activation of the TgICS1 and TgPAL1 promoters by TgNAP in N. benthamiana leaves using Dual-LUC system. G, Quantitative analysis of dual-LUC transient expression assays of the promoter activity in N. benthamiana protoplasts. LUC/REN ratio of SK-TgNAP + pGreen0800 empty vector was used as the negative control and was considered as 1 for data normalization. Error bars indicate SE (n = 3). Different letters denote significant differences at P < 0.05 analyzed by Tukey’s test.
Silencing of TgNAP delayed H2O2-induced tulip petal senescence
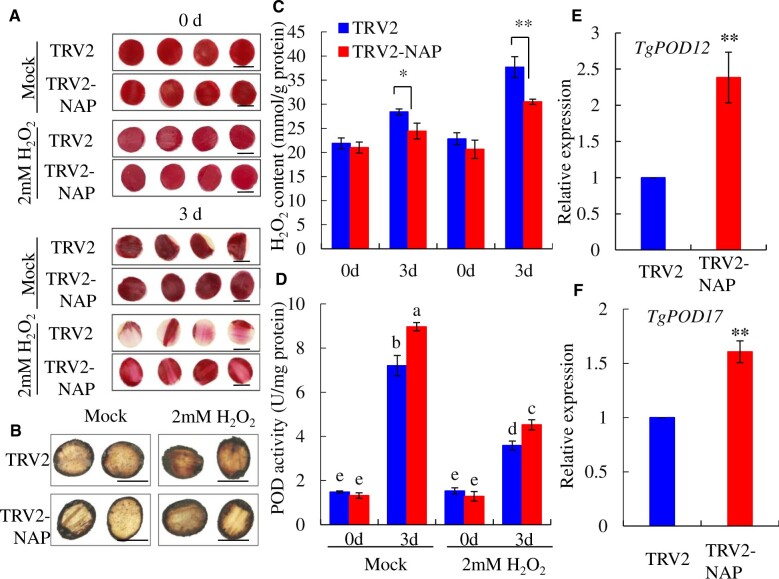
To obtain more insight about the ROS roles during tulip petal senescence, an exogenous treatment of H2O2 was performed using the tulip petal discs and Arabidopsis leaves. With 2.0 mM H2O2 treatment for 3 d in dark, the senescing phenotypes in TRV control samples including color fading and petal curly were substantially enhanced when compared with control treatment, while TRV2-TgNAP petals largely remained fresh and red color (Figure 6A). TRV2-TgNAP petals had less H2O2 accumulation than TRV2 control based on DAB staining results (Figure 6C). Quantification of H2O2 content showed that the levels of H2O2 in TRV2 control were significantly higher than that in TRV2-TgNAP silenced petal discs with or without H2O2 treatment (Figure 6C). The activity of H2O2 scavenging enzyme POD increased following petal senescence and was higher in TRV2-TgNAP silenced petal discs when compared to TRV2 control (Figure 6D). Consistently, the expression levels of TgPOD12 and TgPOD17 were upregulated after TgNAP silencing (Figure 6, E and F).
Figure 6.
Effect of H2O2 on petal senescence in TgNAP-silenced and the TRV2 control discs. A, The phenotypes of TgNAP-silenced petals and the TRV2 control treated with two mM H2O2. The samples in each boxed area were photographed at the same time. B, In situ detection of H2O2 in TRV2 control and TgNAP-silenced plants after ddH2O or H2O2 treatment, as revealed by histochemical staining with DAB. The samples in each boxed area were photographed at the same time and are part of a single image. C, The H2O2 contents of TRV2 control and TgNAP-silenced petals treated with or without two mM H2O2. D, The POD enzyme activity levels of TRV2 control and TgNAP-silenced petals treat with or without two mM H2O2. Blue and red columns indicate the TRV2 control and TgNAP-silenced petals, respectively. E and F, Expression levels of TgPOD12 (E) and TgPOD17 (F) in TRV2 control and TgNAP-silenced petals. Error bars indicate SE (n = 3). Asterisks indicate significant difference between the TgNAP-silenced petals and the TRV control based on the LSD values (*P < 0.05, **P < 0.01). Different letters denote significant differences at P < 0.05 analyzed by Tukey’s test. Scale bar, 2.0 cm.
TgNAP promotes H2O2-induced senescence
In transgenic Arabidopsis, both detached leaves and whole plants of TgNAP-OE lines and WT were used for H2O2-induced leaf senescence assays. Leaves of TgNAP-OE lines were prone to be senescing and turned yellow earlier than those of WT after H2O2 treatment (Supplemental Figure S6, a and e). Quantitative measurement and DAB staining indicated that TgNAP-OE lines accumulated higher H2O2 content than WT (Supplemental Figure S6, b, d, and g). POD activity decreased in TgNAP-OE lines which were consistent with the H2O2 levels and leaf phenotype (Supplemental Figure S6, c and f). Additionally, the expression levels of ROS-related genes in transgenic Arabidopsis and WT were investigated. The results showed that the expressions of ROS production-related genes (AtRBOHA, AtRBOHD, and AtRBOHF) showed significant upregulation (Supplemental Figure S7, a–c), while those of ROS scavenging-related genes (AtPRX4, AtPRX33, and AtPRX72) were significantly downregulated (Supplemental Figure S7, d–f). These data indicated that TgNAP is a positive regulator in H2O2-induced petal or leaf senescence.
TgNAP binds to promoters of TgPOD12 and TgPOD17 to suppress gene expression
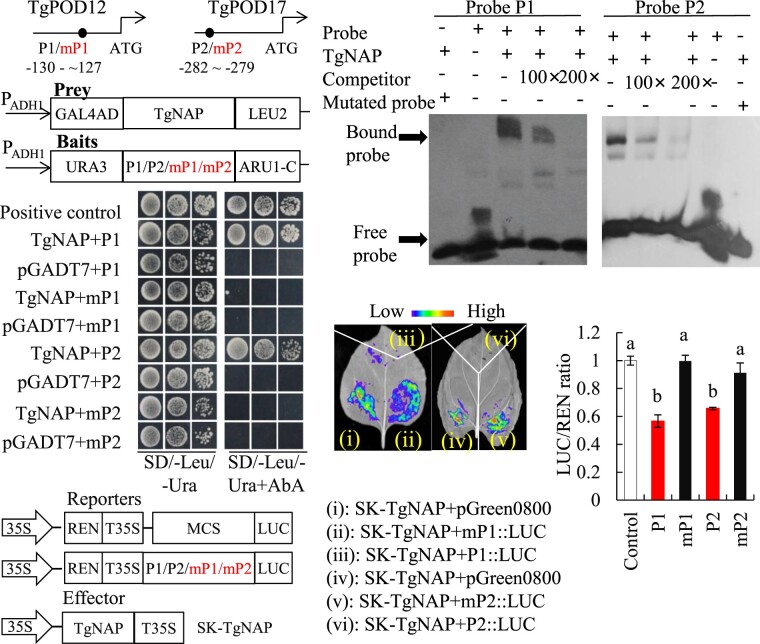
To further investigate the relationship between TgNAP and ROS homeostasis during petal senescence, the promoter sequences of TgPOD12 and TgPOD17 were amplified from tulip genomic DNA. There was one core-binding motif in the promoters of TgPOD12 and TgPOD17, respectively (Figure 7A). Y1H was conducted to investigate the interaction between TgNAP and promoters of TgPOD12/TgPOD17. The prey and bait vectors were constructed as indicated (Figure 7B). The results showed that only positive control and yeast cells co-transformed with TgNAP-AD prey and the wild-type P1/P2 bait grew normally on media supplemented with AbA. However, when the P1/P2 motif sequence was mutated from CACG to AAAG, the growth of yeast was completely inhibited (Figure 7C). Next, EMSA was performed to verify the binding of TgNAP to TgPOD12 and TgPOD17 promoters. Shifted bands were observed when His-TgNAP fusion protein was incubated with the biotin-labeled probes P1 and P2 containing core-binding motif, respectively (Figure 7D). Dual LUC reporter assays were conducted to further verify the transcriptional effect of TgNAP on TgPOD12 and TgPOD17 genes. Quantitative measurement of LUC/REN ratio and observation of LUC fluorescence indicated that TgNAP is able to suppress the promoters of TgPOD12 and TgPOD17 (Figure 7, E–G). These results were consistent with gene expression level changes of TgPOD12 and TgPOD17 in TRV2-TgNAP petals (Figure 6, E and F). Taken together, these results reveal that TgNAP represses TgPOD12 and TgPOD17 transcription through binding to the core-binding motif.
Figure 7.
Direct binding of TgNAP to promoters of TgPOD12 and TgPOD17. A, Schematic diagram of the TgPOD12 and TgPOD17 promoters. The black circles indicate the position of the partial promoter fragments within CACG motifs. P1: promoter of TgPOD12; P2: promoter of TgPOD17. B, The prey and bait constructs used for Y1H assay. mP1 and mP2 are mutated versions of P1 and P2, in which the CACG was replaced with AAAG. PADH1, promoter of alcohol dehydrogenase1. GAL4AD, galactose-specific transcription enhancing factor 4 activation domain. C, Growth of yeast cells co-transformed with various prey+bait combinations on selective medium (SD/-Ura/-Leu) with (right) or without (left) AbA. AbA, Aureobasidin A. Positive control: p53-AbAi+pGAD-p53; Negative control: bait+pGADT7. D, EMSA analysis of specific binding of TgNAP to the promoters of TgPOD12 and TgPOD17. The purified His-TgNAP protein and biotin-labeled probe of designed fragments containing CACG motif or mutated AAAG motif were used. Competitor was unlabeled probe at 100- and 200-fold. +: presence; −: absence. E, The schematic of the TgNAP effector and pTgPOD12::LUC, pTgPOD17::LUC reporters. T35S, the terminator of CaMV 35S, respectively. MCS, multiple cloning sites. LUC, firefly luciferase. REN, Renilla luciferase. F, Live image of transcriptional activation of the TgPOD12 and TgPOD17 promoters by TgNAP in N. benthamiana leaves using Dual-LUC system. G, Quantitative analysis of dual-LUC transient expression assays of the promoter activity in N. benthamiana protoplasts. LUC/REN ratio of SK-TgNAP + pGreen0800 empty vector was used as the negative control and was considered as 1 for data normalization. Error bars indicate SE (n = 3). Different letters denote significant differences at P < 0.05 analyzed by Tukey’s test.
Discussion
Flower senescence is the terminal step and irreversible process of flower development (van Doorn and Woltering, 2008; Ma et al., 2018). The petals, as the most important organ in flower, are able to attract pollinators and promote pollination (van Doorn and Woltering, 2008; Rogers, 2013; Ma et al., 2018). Petal senescence is accompanied by color fading, wilting, loss of scent, and abscission, that impacts economic and ornamental values of the flower (van Doorn and Woltering, 2008; Rogers, 2013; Ma et al., 2018; Sun et al., 2021). Different from leaf senescence, the molecular mechanisms of flower or petal senescence are much less studied and remain largely elusive (Woo et al., 2013; Bresson et al., 2018).
Previous reports have indicated that NAC TF family is prominent among plant leaf senescence-associated transcriptional regulators. Arabidopsis AtNAP, which is most closely related to TgNAP, has been demonstrated to cause precocious leaf senescence (Guo and Gan, 2006). Moreover, AtNAP is able to bind the promoter of its target genes AtSAG113 and AtCKX3 to mediate ABA and CK pathways to facilitate leaf senescence (Zhang and Gan, 2012; Hu et al., 2021). Rice OsNAP was shown to play a positive role in ABA-induced leaf senescence (Mao et al., 2017). Rose RhNAP has been reported to accelerate senescence in mature petals and enhance dehydration tolerance in young rose petals by activating the expression of RhCKX6 (Zou et al., 2021). Meanwhile, knocked down GnNAP in cotton also caused a greatly delay in leaf senescence (Fan et al., 2015). Interestingly, we found that TgNAP promotes tulip petal senescence through activating SA biosynthesis-related genes TgPAL1 and TgICS1, but suppresses expression of antioxidant-related genes TgPOD12 and TgPOD17 (Figures 5 and 7). These results are different from what observed by Kim et al. (2018) indicating NAC TFs play dual roles in regulating plant senescence through SA and ROS pathways. In this study, silencing TgNAP in tulip greatly delayed senescence process in petal discs (Figure 3), whereas overexpression of TgNAP remarkably accelerated leaf senescence in Arabidopsis (Figure S4). These data were consistent with the results of TgNAP putatively orthologous genes from other plant species. All these results revealed that TgNAP functioned as a positive regulator during tulip petal senescence.
To date, numerous studies have dissected the different signaling pathways mediated by TFs to regulate senescence process, including SA and H2O2 (Guo et al., 2017; Niu et al., 2020; Wang et al., 2020a). In line with this fact, we found that SA and H2O2 contents were significantly reduced in TRV2-TgNAP petals but increased in TgNAP-OE lines (Figure 3; Supplemental Figure S4). H2O2 and SA pathways were reported to form an elegant regulatory feedback loop to stimulate each other’s accumulation in multiple plant physiological processes including aging and senescence (León et al., 1995; Kauss and Jeblick, 1995; Rao et al., 1997; Guo et al., 2017). Our data showed that supplement with SA synthesis inhibitor AIP could greatly inhibit H2O2 accumulation in TRV2-TgNAP petals during petal senescence (Figure 4). Meanwhile, SA treatment enhanced H2O2 levels whereas AIP treatment reduced H2O2 accumulation in TgNAP overexpression and WT Arabidopsis (Figure 4). Therefore, SA promotes H2O2 production whereas H2O2 also elicits SA biosynthesis, confirming the roles of both molecules during tulip petal senescence.
Most TFs function as positive or negative regulators through binding to the cis-acting elements in the promoter of downstream target genes to modulate gene expression. Recently, several TFs were reported to regulate SA and ROS pathways following plant senescence. Arabidopsis AtWRKY75 plays a positive role in promoting leaf senescence through activation of AtSID2 and repression of AtCAT2 expression to enhance SA and H2O2 accumulation (Guo et al., 2017). AtWRKY42 and AtWRKY55 also participate in SA and ROS pathways to accelerate leaf senescence by activating AtSID2 and AtRBOH genes (Niu et al., 2020; Wang et al., 2020a). In Arabidopsis, ANAC019, ANAC055, and ANAC072 function as positive regulator of plant senescence partially through SA and JA pathways in terms of GO enrichment analysis (Hickman et al., 2013). Previous researches have defined that CACG is one of the core binding motif of NAC TFs (Olsen et al., 2005; Zhang and Gan, 2012; Hu et al., 2021; Wang et al., 2022). We further found that TgNAP could directly bind to the core-binding motif containing CACG in TgICS1 and TgPAL1 promoters, which is similar to AtNAP binding motif (Zhang and Gan, 2012; Hu et al., 2021; Wang et al., 2022), to activate their transcription in tulip (Figure 5). Meanwhile, the transcript levels of TgICS1 and TgPAL1 and the SA content decreased in the TgNAP silenced petal discs (Figure 3, E–G), but increased in TgNAP overexpression plants in relative to WT (Supplemental Figure S4, h–l). Our results are in line with a recent report about a positive feedback regulatory loop, SA-AtNAP-SAG202-ICS1-SA, that modulates SA-associated leaf senescence (Wang et al., 2022). However, other Arabidopsis NAC TFs, such as ANAC017, ANAC082, and ANAC090 troika, negatively regulate leaf senescence by suppressing SA and ROS responses (Kim et al., 2018), which was consistent with our results that exogenous supply of AIP (inhibitor of SA) could retard the senescence of tulip petals and Arabidopsis leaves (Figure 4; Supplemental Figure S5). Consequently, NAC transcription factors play divergent roles in plant senescence depending upon the specific regulation of downstream targets.
Although SA is synthesized from both ICS and PAL pathways, and TgICS1 and TgPAL1 are both activated by TgNAP, PAL pathway is responsible for synthesis of roughly 5% of SA in pathogen-infected or UV-treated Arabidopsis (Chen et al., 2009). PAL catalyzes phenylalanine to trans-cinnamate. It is the first step in the phenylpropanoid pathway and a crucial regulation point between primary and secondary metabolism, which eventually generate various phenylpropanoids, including SA and other secondary metabolite, such as flavonoids, lignin, stilbenes, anthocyanins, etc. (Huang et al., 2010; Vogt, 2010; Kong, 2015). Hence, PAL is involved in various signaling pathways to response to environmental stresses, plant growth, and development (Dixon and Paiva, 1995; MacDonald and D’Cunha, 2007; Chen et al., 2009). Therefore, TgNAP probably activates TgPAL1 to regulate other compounds synthesis, not only SA, which also participates in petal senescence regulation.
ROS plays crucial roles during natural course of senescence, and the accumulation of which is able to trigger senescence (Del Río and López-Huertas, 2016; Rogers and Munné-Bosch, 2016; Singh et al., 2016). As a relatively diffusible and stable ROS molecular, H2O2 increases during plant senescence, and the accumulated H2O2 resulted in a positive feedback regulation to senescence progress (Vanacker et al., 2006; Khanna-Chopra, 2012). Excessive levels of ROS, especially H2O2 content, can cause detrimental damage to cellular components, which lead to accelerated senescence process (Miller et al., 2010; Huang et al., 2016). Thus, antioxidant enzymes, such as CAT, POD, and SOD, are critical players for ROS scavenging to maintain the equilibrium of ROS accumulation (Gill and Tuteja, 2010). In addition to promote SA synthesis, TgNAP was also found to be involved in H2O2 accumulation to promote petal senescence in tulip. We discovered that two H2O2 scavenging-related genes, TgPOD12 and TgPOD17, were greatly upregulated in TRV-TgNAP petals and considerably downregulated in TgNAP overexpressed plants (Figure 6, C and D; Supplemental Figure S7). Further analysis revealed that TgNAP could directly recognize and bind to the core-binding motif in TgPOD12 and TgPOD17 promoters to repress their expression (Figure 7), and then lead to more ROS accumulation (Figure 6; Supplemental Figure S6). Recently, AtNAC075 was reported to repress CATALASE 2 (CAT2) expression through directly binding to its promoter region to negatively regulate plant senescence (Kan et al., 2021). Moreover, several other NAC TFs such as JUB1 and ORS1 are transcriptionally induced by H2O2 (Balazadeh et al., 2011; Wu et al., 2012), indicating a complicated regulation between NAC TFs and ROS pathway. These data collectively demonstrated that NAC TFs regulate plant senescence through integrating internal ROS signals.
Collectively, the expression of TgNAP was induced in senescing petals as well as by exogenous SA treatment. In turn, TgNAP further promotes the accumulation of SA and H2O2, which are closely related (Kauss and Jeblick, 1995; León et al., 1995; Rao et al., 1997), to trigger petal senescence. It is predictable that each of three factors (TgNAP, SA, and ROS) mutually activates the other two to initiate and accelerate the petal senescence process. Therefore, we propose that TgNAP-SA-ROS feedback loops promote tulip petal senescence in an irreversible manner. Nevertheless, the interactions between SA and H2O2 signaling pathways during plant senescence are still not well understood. Even though retarding SA synthesis inhibits intracellular H2O2 accumulation in tulip petals, the exact regulation effect of H2O2 treatment on SA pathway remains elusive. Further investigation into the core members harboring both SA and ROS homeostasis will be necessary to dissect the senescence mechanism and facilitate prolonging cut flower’s vase life.
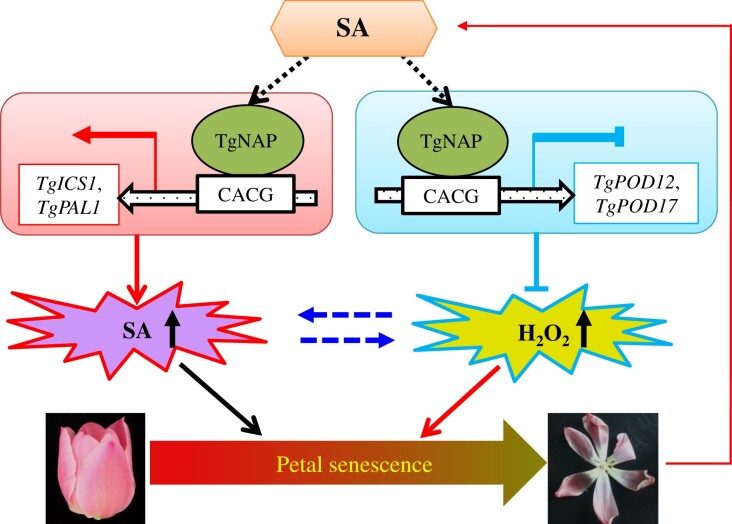
In conclusion, our study showed that a senescence-associated NAC TF TgNAP from T. gesneriana acts as a positive regulator of petal senescence. We propose a work model to illustrate how TgNAP functions in petal senescence in tulip (Figure 8). TgNAP obtains a dual regulation to SA biosynthesis and H2O2 accumulation during petal senescence in tulip. It is able to active SA synthesis gene (TgPAL1 and TgICS1) expressions and represses POD-encoding gene (TgPOD12 and TgPOD17) expressions, which resulted in more SA and H2O2 accumulation to promote petal and leaf senescence either in tulip or in transgenic Arabidopsis plants. Moreover, a mutual enhancement between SA and H2O2 could be responsible for irreversible progress during petal senescence in tulip flowers. The upstream regulator of TgNAP and other signaling pathways regulated by TgNAP remains to be investigated in the future.
Figure 8.
A proposed working model for the regulatory network of TgNAP during tulip petal senescence. TgNAP is induced by petal senescence and exogenous SA treatment, which subsequently activates TgICS1 and TgPAL1 transcription to enhance SA accumulation. In parallel, H2O2 scavenging genes TgPOD12 and TgPOD17 are repressed by TgNAP resulting in increasing ROS levels. Finally, SA and H2O2 work together to accelerate petal senescence in tulip.
Materials and methods
Plant materials and growth conditions
Tulipa gesneriana ‘World Favorite’ were planted in the greenhouse at Huazhong Agricultural University (Wuhan, China) under natural light at 18∼22°C. The flower tissues were sampled at different stages from bud coloring to petal senescence as previously described (Wang et al., 2020b). In previous study, five stages were selected for transcriptome analysis to obtain a comprehensive insight during flower opening and senescence process (RNA seq accession No. GSE136183), including green floral buds (GFB, Stage 1), colored floral buds (CFB, Stage 2), flowers in full bloom (FB, Stage 3), flowers in early senescent phase (ESP, Stage 4), and flowers in late senescent phase (LSP, Stage 5). SA and AIP (an inhibitor of PAL to decrease SA content) were used for petal discs treatments (Solecka and Kacperska, 2013). Intact tulip flower or petal discs (∼1.0 cm2) were placed in 200 µM SA or 30 μM AIP for indicated time courses. Mock samples were treated with distilled water. The A. thaliana ecotype Col-0 was used for transformation. Plants were grown in growth room under 16 h light/8 h dark photoperiod condition. To generate TgNAP overexpression lines, the ORF of TgNAP was clone into pCAMBIA1300 vector using the Xba I and Spe I restriction sites. The resulting pCAMBIA1300-TgNAP was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Arabidopsis by flower dip method (Clough and Bent, 1998). Nicotiana benthamiana was used for transient transformation for Subcellular location and Dual-LUC assays.
H2O2 measurements and DAB staining
H2O2 contents were measured using commercial detection kits (Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer’s instructions. In brief, 0.15 g of frozen flower samples were fully ground with 1.5 mL PBS buffer (0.1 mM, pH 7.8) and centrifuged at 4,000 rpm for 20 min at 4°C. And the supernatant was collected and used for H2O2 contents analysis. For histochemical staining, samples were incubated in 1 mg/mL DAB solution (in 0.01 mM PBS, pH 3.8) in dark for 10 h and then decolorized in 75% (v/v) ethanol until photographing.
Physiological measurements
Chlorophyll contents were measured as previously described. In brief, the plant leaves were cut and incubated in 95% ethanol for 1 d in dark. The absorbance was measured at 649 and 665 nm by using spectrophotometer. The chlorophyll contents were calculated following a formula as previously described (Tian et al., 2020). For anthocyanin contents measurement, the flower samples were dipped into acidic MeOH solution for 24 h at 4°C. The absorbance was measured at 530 and 657 nm as previously described (Luo et al., 2016; Shen et al., 2019). The total anthocyanin contents were determined according to a specific formula: QAnthocyanins = (A530–0.25×A657)/M. For SA measurement, 100 mg of samples were collected and frozen in liquid nitrogen, and SA contents were extracted and determined on UPLC-MS/MS system as previously described (Pan et al., 2010).
Senescence phenotype analysis
For natural leaf senescence, the third and fourth leaves of 28-d-old seedlings were used for chlorophyll content analysis and detection of gene expression. For H2O2-induced detached leaf senescence, seventh to eighth rosette leaves were detached and put in MES buffer (2 mM, pH 5.8) with or without 2 mM H2O2 for 3 d. For H2O2 induced seedlings senescence, 2-week-old Arabidopsis grown in soil separated with or without 200 mM H2O2 for 5 d under 16 h light/8 h dark photoperiod condition. For SA and AIP treatment, seventh to eighth rosette leaves were detached and incubated in MES buffer (2 mM, pH 5.8) with 200 µM SA or 30 µM AIP for 3 d, 2-week-old Arabidopsis grown in soil separated with or without 200 µM SA or 30 µM AIP for 5 d under 16 h light/8 h dark photoperiod condition. For VIGS samples, the phenotypes of petal discs were observed daily until senescence and color fading were occurred. For AIP treatment, petal discs were put in distilled water with or without 30 µM AIP. For H2O2 treatment, petal discs were incubated in distilled water with or without 2 mM H2O2 for 3 d in dark.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from tulip flower and Arabidopsis leaves using commercial RNA purification kit (Aidlab, RN33, China). One microgram of RNA was used for reverse transcription to generate cDNA by using the First Strand cDNA Synthesis Kit (Toyobo, Japan) based on instruction manual. RT-qPCR was performed on a QuantStudio 7 Flex PCR machine (Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) using ChamQ Universal SYBR qPCR Master Mix (Vazyme). The 20 µL reaction mixture contained 10 µL 2 × ChamQ Universal SYBR qPCR Master Mix, 200 ng cDNA, 0.2 µM forward and reverse primers. TgUBQ10-like was served as the internal reference gene (Wang et al., 2020b). All primer sequences were listed in Supplemental Table S1. The reaction program was 95°C for 3 min, 40 cycles of 10 s at 95°C and 30 s at 60°C, followed by 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. Relative gene expression levels were analyzed by 2–ΔΔCt method (Livak and Schmittgen, 2001).
RNA sequencing analysis
cDNA library was generated using the NEBNext Ultra II RNA Library Prep Kit (NEB, USA) and sequenced using an Illumina HiSeq 4000 sequencing system (Illumina, USA) with the paired-end sequencing method. The raw reads were filtered and clean data were mapped to the assembled tulip PacBio full-length transcriptome with the accession number PRJNA703083. Fragments per kilobase of transcript per million (FPKM) were used for the quantification of gene expression. Differential expression changes from three biological replicates were analyzed using the DESeq R package (1.10.1). Genes with FDR ≤ 0.05 and log2 |fold change| ≥1 were assigned as significantly differential expressed genes.
Cluster and protein interaction network analyses
Cluster analysis was conducted using the CLUSTER program (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) with uncentered matrix and complete linkage method (de Hoon et al., 2004). The resulting tree figure was visualized using Java Treeview (http://jtreeview.sourceforge.net/) as previously described (Chan, 2012).
Protein interaction networks were analyzed using the STRING version 10.5 (https://string-db.org/; Szklarczyk et al., 2015). The resulting data were imported into CytoScape software (https://cytoscape.org/) to generate protein interaction networks.
Cloning and sequence analysis of TgNAP
The full length of TgNAP was amplified by using specifically designed primers based on previous transcriptome data of T. gesneriana (GSE136183). Homologous proteins from other species were searched and obtained from NCBI database. A phylogenetic tree between TgNAP and all Arabidopsis NAC proteins was constructed using MEGA 7.0 software with neighbor-joining method. Multiple sequence alignment results were shown in GENEDOC software.
Subcellular location of TgNAP
The ORF of TgNAP without stop codon was amplified and fused to the 101LYFP vector, and the fused plasmid NAP-YFP was transformed into A. tumefaciens strain GV3101. For transient transformation, N. benthamiana leaves were infiltrated with GV3101 carrying NAP-YFP fused vector or empty vector (YFP) as previously described (Geng and Liu, 2018), and VirD2NLS inserted into mCherry was used as a nuclear marker. YFP signal (excitation at 514 nm, scanning at 520–540 nm) and RFP signal (excitation at 552 nm, scanning at 590–640 nm) were observed by a confocal laser scanning microscope (Leica TCSSP8, Germany).
Transcriptional activation activity assay
For transcriptional activation analysis, the full length of TgNAP and two truncated fragments, F1 (1–132 aa) and F2 (133–270 aa), were amplified and cloned into pGBKT7 vector to fuse with GAL4 DNA-binding domain. All resulting plasmids were introduced into yeast strain AH109 according to manufacturer’s protocol. The transformed yeast cells were plated onto SD/-Trp medium and SD/-Trp/-His/-Ade medium with or without 4 mg/mL X-α-gal at 30°C for 48–72 h. The transformed yeast with pGBKT7-p53 and empty vector pGBKT7 were used as positive control and negative control, respectively.
VIGS assay in tulip petals
To silence TgNAP in tulip petals, a non-conserved fragment of TgNAP with 330 bp length located at 425∼754 bp downstream of start codon (ATG) was amplified and inserted into pTRV2 vector. The specificity of target region in VIGS assay was verified by sequence alignment. The fused pTRV2-TgNAP vector, empty pTRV1 and pTRV2 were individually introduced into A. tumefaciens GV3101. The bacterial suspensions containing pTRV2-TgNAP and pTRV1, or pTRV2 and pTRV1 were prepared for transformation of tulip petals as previously described (Wu et al., 2017; Wang et al., 2020b). In brief, the pTRV1 bacterial suspension (OD600=1.5) was mixed with pTRV2 or pTRV2-TgNAP in 1:1 ratio (v/v) and suspended in MES buffer (200 mM acetosyringone, 10 mM MgCl2, and 10 mM MES, pH 5.8). The mixtures of bacterial solutions were incubated in dark at 28°C for 3∼4 h. 1.0 cm diameter discs of tulip petals were vacuum-infiltrated in bacterial solutions at 0.7 MPa. The infiltrated discs were washed with distilled water and incubated in darkness at 8°C for first 3 d, and then were transferred to growth chamber (16 h light/8 h dark) for phenotype observation (Wu et al., 2017). At least 20 tulip tepals were used for each treatment.
EMSA assay
The coding sequence of TgNAP was cloned into pHMGWA vector which contains a His tag, and the resulting plasmid was transformed into Escherichia coli strain Rosetta (DE3) to generate the His-TgNAP fusion protein. After induction by IPTG (0.5 mM) and expression at 37°C for 3 h, the HIS-TgNAP fusion protein was purified. The His-TgNAP fusion protein was induced by IPTG (0.5 mM) at 37°C for 3 h and purified using Qiagen Ni-NTA Agarose kit. The 40-bp promoter fragments of TgPAL1, TgICS1, TgPOD12, and TgPOD17 were synthesized as probes with biotin-label at the 3′-end. Competitors were unlabeled DNA with same sequence. The mutant probes were labeled, in which the core binding elements changed from CACG to AAAG. The EMSA assay was conducted using the LightShift Chemiluminescent EMSA Kit (Thermo) following the methods previously described (Geng and Liu, 2018).
Yeast one-hybrid (Y1H) assay
The promoter fragments of TgPAL1, TgICS1, TgPOD12, and TgPOD17, which contain CACG, were amplified and fused to pAbAi vector as baits vectors. In addition, mutant baits were created by replacing CACG with AAAG. All resulting bait and mutant bait plasmids were linearized and transformed into Y1H Gold yeast strain. The full length of TgNAP was amplified and inserted into pGADT7 to generate prey vector. The prey vector was transformed into bait and mutant bait yeast strains. The transformed yeast clones were spotted on SD/-Leu/-Ura medium with or without AbA. The yeast strains harboring baits and empty pGADT7 vector were used as negative control, whereas the yeast strains harboring pAbAi-p53 and pGADT7-p53 were used as positive control.
Dual-LUC assays
The CDS of TgNAP was amplified and cloned into pGreen II 62-SK vector to act as effector. The promoter original and mutated sequences of TgPAL1, TgICS1, TgPOD12, and TgPOD17 were amplified and cloned into pGreen II 0800-LUC vector to be used as reporters. All of the effector and reporters were introduced into A. tumefaciens strain GV3101 and transiently expressed in N. benthamiana leaves as previously described (Hu et al., 2018; Wang et al., 2019). The activities of LUC and REN were determined by using the Dual-Luciferase® Reporter Assay System (Promega, USA) following the instructions. For observation of LUC fluorescence, 1 mM D-Luciferin solution was used to spray on the infiltrated leaves that then were placed in dark for 5 min. After darkness, the luminescence was detected under a cooled CCD camera by plant living imaging system (Nightshade LB 985, Germany).
Statistical analysis
All experiments were performed at least twice with triplicate. All data were presented as means ± SE. Student’s t test and ANOVA were used for statistical difference analysis, and significant level was defined as P < 0.05 (*), P < 0.01 (**).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtSAG12 (AT3G20770), AtSAG13 (AT2G29350), AtSID2 (AT1G74710), AtPAL1 (AT2G37040), AtEPS1 (AT5G67160), AtEDS5 (AT4G39030), AtRbohA (AT5G07390), AtRbohD (AT5G47910), AtRbohF (AT1G64060), AtPrx33 (AT3G49110), AtPrx4 (AT1G14540), AtPrx72 (AT5G66390), TgNAP and RNA seq accession No. GSE136183.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. RNA sequencing analysis and gene expression profiles during petal senescence of T. gesneriana.
Supplemental Figure S2. Phylogenetic tree analysis and multiple alignments of NACs from diverse plants.
Supplemental Figure S3. Silencing of TgNAP delayed flower senescence.
Supplemental Figure S4. Overexpression of TgNAP promoted leaf senescence in Arabidopsis.
Supplemental Figure S5. Effect of exogenous SA on the senescence of TgNAP transgenic Arabidopsis and WT.
Supplemental Figure S6. Effect of exogenous H2O2 treatment on TgNAP transgenic Arabidopsis and WT.
Supplemental Figure S7. Expression of ROS-related genes in TgNAP transgenic Arabidopsis and WT analyzed by RT-qPCR.
Supplemental Table S1. Significantly changed unigenes during five senescence stages of tulip petal.
Supplemental Table S2. Significantly changed unigenes during S4 and S5 senescence stages in tulip petal.
Supplemental Table S3. FPKM values of TgNACs from transcriptome data.
Supplemental Table S4. List of all primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Jihong Liu for critical discussions and sharing lab equipment.
Funding
This work was supported by National Natural Science Foundation of China (32170372), National Key Research and Development Program (2020YFD1000400), and the Fundamental Research Funds for the Central Universities (Program No. 2662020YLPY010).
Conflict of interest statement. The authors declare that they have no conflict of interests.
Data availability statement
Tulip petal RNA-seq data at five different developmental stages were deposited into NCBI GEO database with the accession number GSE136183. The sequence of TgNAP was submitted to NCBI GenBank with the accession number MK239329.
Contributor Information
Lin Meng, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, PR China; National R&D Centre for Citrus Preservation, Huazhong Agricultural University, Wuhan 430070, PR China.
Haipo Yang, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, PR China; National R&D Centre for Citrus Preservation, Huazhong Agricultural University, Wuhan 430070, PR China.
Lin Xiang, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, PR China.
Yanping Wang, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, PR China; National R&D Centre for Citrus Preservation, Huazhong Agricultural University, Wuhan 430070, PR China.
Zhulong Chan, Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan 430070, PR China.
L.M. conducted the experiments. H.Y. collected the data. Y.W., Z.C., and L.M. designed the experiments and wrote the paper. Z.C., Y.W., and L.X. revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Zhulong Chan (zlchan@mail.hzau.edu.cn).
References
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bresson J, Bieker S, Riester L, Doll J, Zentgraf U (2018) A guideline for leaf senescence analyses: From quantification to physiological and molecular investigations. J Exp Bot 69: 769–786 [DOI] [PubMed] [Google Scholar]
- Chan Z (2012) Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100: 110–115 [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D (2017) Arabidopsis WRKY45 interacts with the DELLA Protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig DF (1993) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262: 1883–1886 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43: 745–757 [DOI] [PubMed] [Google Scholar]
- Del Río LA, López-Huertas E (2016) ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol 57: 1364–1376 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K, Bibi N, Gan S, Li F, Yuan S, Ni M, Wang M, Shen H, Wang X (2015) A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J Exp Bot 66: 4669–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1997) Making sense of senescence. Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Liu JH (2018) The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J Exp Bot 69: 2677–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G (2013) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Sign 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Hill C, Penfold CA, Breeze E, Bowden L, Moore JD, Zhang P, Jackson A, Cooke E, Bewicke-Copley F, et al. (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J 75: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Zhu L, Zhang X, Guan Q, Xiao S, Min L, Zhang X (2018) GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR31. Plant Physiol 178: 876–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Aken O, Van Schwarzländer M, Belt K, Millar AH (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol 171: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu B, Ren H, Chen L, Watkins CB, Gan S (2021). The leaf senescence-promoting transcription factor AtNAP activates its direct target gene CYTOKININ OXIDASE 3 to facilitate senescence processes by degrading cytokinins. Mol Hortic 1: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A, Creissen G, Kular B, Firmin J, Robinson S, Verhoeyen M, Mullineaux P (2002) Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214: 751–758 [DOI] [PubMed] [Google Scholar]
- Kan C, Zhang Y, Wang HL, Shen Y, Xia X, Guo H, Li Z (2021) Transcription factor NAC075 delays leaf senescence by deterring reactive oxygen species accumulation in Arabidopsis. Front Plant Sci 12: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Jeblick W (1995) Pretreatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiol 108: 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Chopra R (2012) Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249: 469–481 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Park JH, Kim JJ, Kim JJ, Hong S, Kim J, Kim JH, Woo HR, Hyeon C, Lim PO, et al. (2018) Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115: E4930–E4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC (2013) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 54: 1660–1672 [DOI] [PubMed] [Google Scholar]
- Kong JQ (2015) Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Adv 5: 62587–62603 [Google Scholar]
- Lee JH, Park YJ, Kim JY, Park CM (2021) Phytochrome B conveys low ambient temperature cues to the ethylene-mediated leaf senescence in Arabidopsis. Plant Cell Physiol 63: 326–339 [DOI] [PubMed] [Google Scholar]
- León J, Lawton MA, Raskin I (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol 108: 1673–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chang Y, Zhao C, Yang H, Ren D (2016) Expression of the inactive ZmMEK1 induces salicylic acid accumulation and salicylic acid-dependent leaf senescence. J Integr Plant Biol 58: 724–736 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J (2014) RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78: 578–590 [DOI] [PubMed] [Google Scholar]
- Luo P, Ning G, Wang Z, Shen Y, Jin H, Li P. et al. (2016) Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. Red color flower formation in plants. Front Plant Sci 6: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Ma C, Liu Y, Shahid MO, Wang C, Gao J (2018) Petal senescence: a hormone view. J Exp Bot 69: 719–732 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Y, Turečková V, Xue GP, Fernie AR, Mueller-Roeber B, Balazadeh S (2018) The NAC transcription factor SLNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol 177: 1286–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald MJ, D’Cunha GB (2007) A modern view of phenylalanine ammonia lyase. Biochem Cell Biol 85: 273–282 [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice nac transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Van Breusegem F (2018) Reactive oxygen species in plant development. Development 145: dev164376. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Morris K, Mackerness SAH, Page T, Fred John C, Murphy AM, Carr JP, et al. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Nagahage ISP, Sakamoto S, Nagano M, Ishikawa T, Mitsuda N, Kawai-Yamada M, Yamaguchi M (2020) An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Plant Physiol 170: 299–308 [DOI] [PubMed] [Google Scholar]
- Niu F, Cui X, Zhao P, Sun M, Yang B, Deyholos MK. et al. (2020) WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J 104: 171–184 [DOI] [PubMed] [Google Scholar]
- Oh SK, Lee S, Yu SH, Choi D (2005) Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 222: 876–887 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79-87 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protocols 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Pyung OL, Hyo JK, Hong GN (2007) Leaf senescence. Ann Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, et al. (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP, Murr DP, Watkins CB (1997) lnfluence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes (salicylic acid-mediated oxidative damage requires H2O2). Plant Physiol 115: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Rogers H, Munné-Bosch S (2016) Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: Similar but different. Plant Physiol 171: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ (2013) From models to ornamentals: how is flower senescence regulated? Plant Mol Biol 82: 563–574 [DOI] [PubMed] [Google Scholar]
- Schippers JHM, Schmidt R, Wagstaff C, Jing HC (2015) Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol 169: 914–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Sun T, Pan Q, Anupol N, Chen H, Shi J, Liu F, Deqiang D, Wang C, Zhao J, et al. (2019) RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol J 17: 2078–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Singh S, Parihar P, Mishra RK, Tripathi DK, Singh VP, Chauhan DK, Prasad SM (2016) Reactive oxygen species (ROS): beneficial companions of plants’ developmental processes. Front Plant Sci, 7: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecka D, Kacperska A (2013) Phenylpropanoid deficiency affects the course of plant acclimation to cold. Plant Physiol 119: 253–262 [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Qin M, Yu Q, Huang Z, Xiao Y, Li Y, Ma N, Gao JP (2021) Molecular understanding of postharvest flower opening and senescence. Mol Hortic 1: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015) STRING v10: protein-protein interaction networks integrated over the tree of life. Nucleic Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Ma L, Liu Y, Xu D, Chen Q, Li G (2020) Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SK, Tuteja N (2007) Integrated signaling in flower senescence: an overview. Plant Signal Behav 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Sandalio LM, Jiménez A, Palma JM, Corpas FJ, Meseguer V, Gómez M, Sevilla F, Leterrier M, Foyer CH, et al. (2006) Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. J Exp Bot 57: 1735–1745 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogelmann K, Drechsel G, Bergler J, et al. (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159: 1477–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wang C, Dai S, Zhang ZL, Lao W, Wang R, Meng X, Zhou X (2021a) Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J Integr Plant Biol 63: 828–833 [DOI] [PubMed] [Google Scholar]
- Wang M, Dai W, Du J, Ming R, Dahro B, Liu JH (2019) ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol J 17: 1316–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cui X, Yang B, Xu S, Wei X, Zhao P, Niu F, Sun M, Wang C, Cheng H, et al. (2020a) WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 147, dev189647 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu B, Hu Y, Gan S (2022) A positive feedback regulatory loop, SA- biosynthesis involved in leaf senescence but not defense response. Mol Hortic 2: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao H, Liu C, Cui G, Qu L, Bao M, Wang J, Chan Z, Wang Y (2020b) Integrating physiological and metabolites analysis to identify ethylene involvement in petal senescence in Tulipa gesneriana. Plant Physiol Biochem 149: 121–131 [DOI] [PubMed] [Google Scholar]
- Wang Z, Rong D, Chen D, Xiao Y, Liu R, Wu S, Yamamuro C (2021b) Salicylic acid promotes quiescent center cell division through ROS accumulation and down-regulation of PLT1, PLT2, and WOX5. J Integ Plant Biol 63: 583–596 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J Cell Sci 126: 4823–4833 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Lim PO, Nam HG (2019) Leaf senescence: systems and dynamics aspects. Annu Rev Plant Biol 70: 347–376 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ma N, Jia Y, Zhang Y, Feng M, Jiang CZ, Ma C, Gao J (2017) An ethylene-induced regulatory module delays flower senescence by regulating cytokinin content. Plant Physiol 173: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Jiang L, Yu M, An X, Ma R, Yu Z (2016) Proteomic analysis of changes in mitochondrial protein expression during peach fruit ripening and senescence. J Proteom 147: 197–211 [DOI] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang C, Qiu K, Chen H, Li Z, Li X, Song J, Wang X, Gao J, Kuai B, et al. (2020) The transcription factor ZmNAC126 accelerates leaf senescence downstream of the ethylene signalling pathway in maize. Plant Cell Environ 43: 2287–2300 [DOI] [PubMed] [Google Scholar]
- Yu G, Xie Z, Lei S, Li H, Xu B, Huang B (2022) The NAC factor LpNAL delays leaf senescence by repressing two chlorophyll catabolic genes in perennial ryegrass. Plant Physiol 10.1093/plphys/kiac070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Gan S (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Guo P, Xia X, Guo H, Li Z (2021) Multiple Layers of Regulation on Leaf Senescence: New Advances and Perspectives. Front Plant Sci 12: 788996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, Dong X (2015) Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci USA 112: 9166–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Lü P, Jiang L, Liu K, Zhang T, Chen J, Yao Y, Cui Y, Gao J, Zhang C (2021) Regulation of rose petal dehydration tolerance and senescence by RhNAP transcription factor via the modulation of cytokinin catabolism. Mol Hortic 1: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tulip petal RNA-seq data at five different developmental stages were deposited into NCBI GEO database with the accession number GSE136183. The sequence of TgNAP was submitted to NCBI GenBank with the accession number MK239329.