Abstract
Ethylene plays essential roles in adaptive growth of rice (Oryza sativa). Understanding of the crosstalk between ethylene and auxin (Aux) is limited in rice. Here, from an analysis of the root-specific ethylene-insensitive rice mutant mao hu zi 10 (mhz10), we identified the tryptophan aminotransferase (TAR) MHZ10/OsTAR2, which catalyzes the key step in indole-3-pyruvic acid-dependent Aux biosynthesis. Genetically, OsTAR2 acts downstream of ethylene signaling in root ethylene responses. ETHYLENE INSENSITIVE3 like1 (OsEIL1) directly activated OsTAR2 expression. Surprisingly, ethylene induction of OsTAR2 expression still required the Aux pathway. We also show that Os indole-3-acetic acid (IAA)1/9 and OsIAA21/31 physically interact with OsEIL1 and show promotive and repressive effects on OsEIL1-activated OsTAR2 promoter activity, respectively. These effects likely depend on their EAR motif-mediated histone acetylation/deacetylation modification. The special promoting activity of OsIAA1/9 on OsEIL1 may require both the EAR motifs and the flanking sequences for recruitment of histone acetyltransferase. The repressors OsIAA21/31 exhibit earlier degradation upon ethylene treatment than the activators OsIAA1/9 in a TIR1/AFB-dependent manner, allowing OsEIL1 activation by activators OsIAA1/9 for OsTAR2 expression and signal amplification. This study reveals a positive feedback regulation of ethylene signaling by Aux biosynthesis and highlights the crosstalk between ethylene and Aux pathways at a previously underappreciated level for root growth regulation in rice.
Ethylene inhibition of root growth requires OsEIL1-OsAux/IAA interactions in a positive feedback loop to promote MHZ10/OsTAR2-mediated auxin biosynthesis in rice.
IN A NUTSHELL.
Background: The phytohormones ethylene and auxin (Aux) play important roles in developmental regulation and environmental adaption in plants. Ethylene promotes the accumulation of the master transcription regulators ETHYLENE INSENSITIVE3 (EIN3)/EIN3-like1 (EIL1). Aux triggers the degradation of Aux/indole-3-acetic acid (IAA) to release Aux RESPONSE FACTORs for downstream regulation. Although the biosynthesis and signaling pathways of these two phytohormones have been intensively studied, how, when, and where they crosstalk is still poorly understood, especially in rice.
Question: Are there more links between ethylene and Aux at the levels of biosynthesis and signaling?
Findings: Through an analysis of a root-insensitive ethylene response mutant, we identified the key tryptophan aminotransferase MHZ10/OsTAR2 participating in the Aux biosynthesis pathway. OsTAR2 is directly upregulated by OsEIL1. Interestingly, we detected no ethylene induction of OsTAR2 expression in the mutant roots, and discovered a positive feedback loop between Aux and ethylene for the regulation of OsEIL1-mediated OsTAR2 transcriptional activation. Further studies revealed that OsEIL1 interacts with OsIAA1/9/21/31. OsIAA21/31 repressed OsEIL1 activity through histone deacetylation at the OsTAR2 locus. The EAR motif and surrounding sequences of OsIAA1/9 recruit OsGCN5 for histone acetylation. This OsIAA1/9-mediated histone acetylation promotes OsEIL1-mediated OsTAR2 induction. Moreover, the Aux receptors OsTIR1/AFB2 mediate a faster degradation of OsIAA21/31 than of OsIAA1/9, thus allowing the release and promotion of OsEIL1 activity.
Next steps: More findings highlighting ethylene–Aux interactions are expected. In addition to the activity of the key transcription regulator OsEIL1 in this study, its protein turnover appears to be also modulated by the Aux pathway. Considering this point, is there a desensitization/balance mechanism after sufficient Aux biosynthesis has been attained for regulating long-term root growth in response to ethylene?
Introduction
The volatile plant hormone ethylene is involved in various biological processes in plants, including development, stress responses, and adaptive growth. Several key ethylene signaling components have been identified by isolating classical ethylene “triple response” mutants in the model plant Arabidopsis (Arabidopsis thaliana). Ethylene perception is initiated with ethylene binding to endoplasmic reticulum (ER)-located ethylene receptors (Bleecker et al., 1988; Chang et al., 1993; Hua et al., 1995, 1998; Hua and Meyerowitz, 1998; Sakai et al., 1998; Wang et al., 2006), which attenuates the kinase activity of the negative regulator CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) likely due to conformational changes of these receptors (Kieber et al., 1993; Clark et al., 1998; Wang et al., 2006). The CTR1 phosphorylation-dependent suppression of the activity of the multifunctional ETHYLENE INSENSITIVE2 (EIN2) is then alleviated (Qiao et al., 2012). The F-Box proteins EIN2 TARGETING PROTEIN1 (ETP1) and ETP2, which mediate the degradation of EIN2 through the ubiquitination-proteasome pathway, are also downregulated (Qiao et al., 2009). Thus, EIN2 is stabilized, and its C-terminal end is cleaved and translocated into the nucleus and/or processing body to activate the ETHYLENE INSENSITIVE3/EIN3-like1 (EIN3/EIL1) transcriptional cascade directly or through the translational repression of EIN3 BINDING F-BOX1/2 (EBF1/2), which mediate the proteasomal degradation of EIN3/EIL1 (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004; An et al., 2010; Li et al., 2015; Merchante et al., 2015). The chromatin-associated protein EIN2 NUCLEAR-ASSOCIATED PROTEIN1 and the two histone deacetylases (HDACs) SIRTUIN1/2 also function in EIN3-regulated gene expression (Zhang et al., 2016, 2017, 2018).
Of the tryptophan-dependent pathways of auxin (Aux) biosynthesis, the indole-3-pyruvic acid (IPyA)-dependent TAA/YUCCA pathway with two chemical steps appears to be the primary biosynthesis route (Zhao, 2018). In this pathway, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) first converts tryptophan into IPyA (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009), after which flavin monooxygenases from the YUCCA family catalyze the conversion of IPyA to indole-3-acetic acid (IAA) in the last rate-limiting step (Mashiguchi et al., 2011; Won et al., 2011; Dai et al., 2013). The Aux receptor F-BOX TRANSPORT INHIBITOR RESPONSE1/Aux SIGNALING F-BOX PROTEIN (TIR1/AFB) (Dharmasiri et al., 2005; Kepinski and Leyser, 2005), the repressor proteins Aux/IAAs (Rouse et al., 1998), and the transcription factors Aux RESPONSE FACTORS (ARFs) are the three main components of the Aux signaling pathway. Aux/IAAs generally contain three conserved domains. Domain I harbors an ETHYLENE RESPONSE FACTOR (ERF)-associated amphiphilic repression (EAR) motif involved in repressing the transcription regulatory activity of ARFs (Tiwari et al., 2004). Domain II determines the ubiquitin/26S proteasome-mediated degradation of Aux/IAAs via the Skip, Cullin, F-box complex SCFTIR/AFB (Worley et al., 2000; Ramos et al., 2001). The third domain, Phox and Bem1 (PB1), is involved in homodimerization or heterodimerization with ARFs (Kim et al., 1997; Morgan et al., 1999; Guilfoyle, 2015). In the presence of Aux, the ubiquitin-ligase SCFTIR/AFB complex interacts with and degrades Aux/IAAs through the Aux-dependent ubiquitination-mediated degradation pathway (Ulmasov et al., 1997; Gray et al., 2001; Tiwari et al., 2001). After Aux/IAA degradation, the released ARFs can regulate the expression of downstream genes and trigger Aux responses.
Rice (Oryza sativa L.) is a semi-aquatic monocotyledonous plant. Its etiolated seedlings show “double response” phenotypes in response to ethylene, namely inhibited root growth but promoted mesocotyl/coleoptile elongation (Zhou et al., 2020). Over the past several years, through an analysis of a set of ethylene-insensitive mao hu zi (mhz) mutants, we have identified conserved ethylene signaling components in rice, including MHZ12/OsERS2, MHZ7/OsEIN2, and MHZ6/OsEIL1 (Ma et al., 2013, 2018; Yang et al., 2015; Zhou et al., 2020). We also discovered new regulators (Zhao et al., 2021). For instance, the membrane protein MHZ3 stabilizes OsEIN2 through interacting with its N-terminal transmembrane natural resistance-associated macrophage proteins-like domain (Ma et al., 2018). The GDSL lipase MHZ11 modulates ethylene signaling by decreasing the interaction between ethylene receptors and OsCTR2 and by inhibiting OsCTR2 phosphorylation (Zhao et al., 2020a). The histidine kinase MHZ1, which physically interacts with and is inhibited by ethylene receptors, participates in ethylene regulation of root growth via phosphorelay in rice (Zhao et al., 2020b). Crosstalk between ethylene and other plant hormones has also been highlighted by the mhz mutants. Abscisic acid (ABA) represses ethylene biosynthesis and OsEIN2 expression to inhibit coleoptile elongation, while ethylene induces MHZ5 (OsCRTISO [CAROTENOID ISOMERASE])- and MHZ4 (OsABA4 [ABA-DEFICIENT4])-mediated ABA biosynthesis to inhibit root elongation (Ma et al., 2014; Yin et al., 2015). OsEIL2 suppresses GAOYAO1 transcription and reduces jasmonic acid accumulation to enhance mesocotyl/coleoptile elongation (Xiong et al., 2017). The E3 ligase MHZ2/SOIL-SURFACE ROOTING1 (SOR1) is involved in ubiquitination-mediated degradation of atypical OsIAA26 in root ethylene responses downstream of signaling from the OsTIR1/AFB2-Aux-OsIAA9 module (Chen et al., 2018).
In recent years, ethylene and Aux pathways have been extensively studied. However, the understanding of the relationship between these two phytohormones is still very limited especially in crops such as rice. In this study, we characterized the root-specific ethylene-insensitive rice mutant mhz10, which we previously identified (Zhou et al., 2020). MHZ10 encodes the tryptophan aminotransferase OsTAR2 (also named FISH BONE [FIB] and Tillering and Small Grain1 [TSG1]) and functions in ethylene-induced Aux biosynthesis in root ethylene response. The transcription factor MHZ6/OsEIL1 from the rice ethylene signaling pathway directly binds to the OsTAR2 promoter and activates its expression. The Aux/IAA proteins OsIAA1/9 and OsIAA21/31 showed promotion and repression regulatory activity, respectively, on OsEIL1 through interacting with the OsEIL1 C-terminal domain and recruiting histone acetylation or deacetylation-related components. These results expand our understanding of the interaction between ethylene and Aux in plants.
Results
Phenotypic analysis of mhz10 mutants and cloning of MHZ10
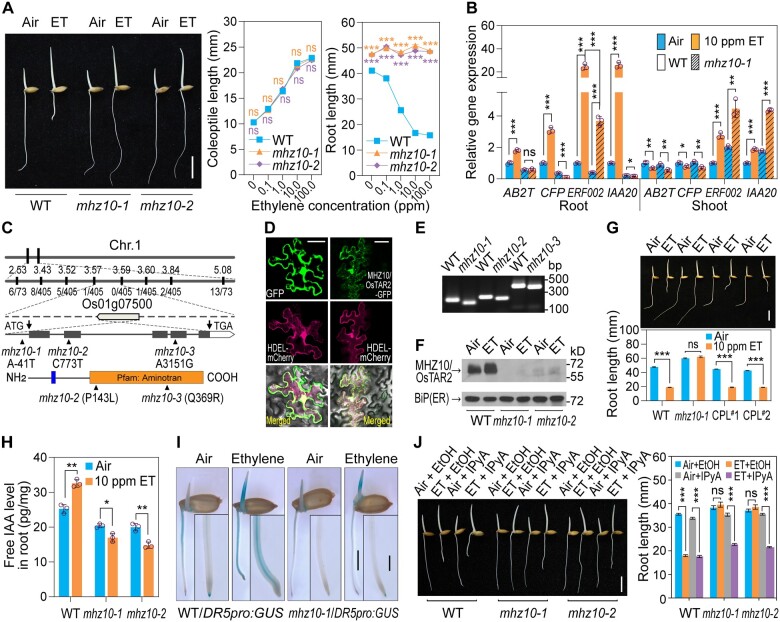
We analyzed the rice mutant mhz10 in this study; among three mhz10 mutant alleles, two (mhz10-1 and mhz10-2) were fertile. Upon ethylene treatment, coleoptile elongation of the mhz10-1 and mhz10-2 mutants was very similar to that of the wild-type (WT), whereas the root growth of the two mutants was insensitive to ethylene and these mutants had a slightly longer root than WT in air (Figure 1A). We also examined the expression of several ethylene-inducible genes. Ethylene treatment induced the expression of ABC-2 type transporter (AB2T), cyclase family protein (CFP), ERF002, and IAA20 in WT roots, while this induction was completely blocked or reduced in mutant roots (Figure 1B). In shoots, the expression levels of these genes responded to ethylene with a similar pattern in both WT and the mhz10-1 mutant (Figure 1B). The ethylene response and the responsive gene expression analysis indicated that mhz10 mutants are insensitive to ethylene in roots. We also investigated phenotypic changes of field-grown plants (Supplemental Figure S1). Compared to WT, the mhz10 mutants grew as shorter plants, with increased leaf angle, narrow and rolled leaves, with more tillers, and smaller panicles with fewer branches. The seed size, thousand-grain weight, and yield per plant of the mutants were also significantly reduced relative to WT. The mhz10-2 mutant showed more severe growth defects than mhz10-1, and mhz10-3 did not produce enough seeds for later propagation.
Figure 1.
Phenotype of mhz10 mutants and cloning of MHZ10/OsTAR2. A, Ethylene-response phenotype of mhz10. Representative seedlings grown in air or 10 ppm ethylene for 2.5 DAG are shown, scale bar, 10 mm. Data are means ± se (n ≥ 21). B, Expression of ethylene-responsive genes analyzed by RT-qPCR. One-DAG etiolated seedlings were treated with or without 10 ppm ethylene for 8 h. Three independent biological replicates were performed. Data are means ± sd. C, Map-based cloning of MHZ10/OsTAR2. Mutation sites and putative protein domains are shown in schematic diagrams. The first narrow box indicates a putative transmembrane domain and the second large box indicates a putative aminotransferase domain. Three allelic mutants are indicated, with two mutation sites in the coding region, and one upstream of the coding region. D, Subcellular localization of MHZ10/OsTAR2. ER-located protein HDEL-mCherry was used as a marker. More than 10 cells in different regions of infiltrated leaves were observed, and representative cells are shown. Scale bars, 20 μm. E, Identification of three allelic mutation sites in the genome by dCAPS. F, Immunoblot assay for MHZ10/OsTAR2 protein in roots. One-DAG etiolated seedlings were treated with or without 10 ppm ethylene for 12 h. MHZ10/OsTAR2 abundance in root tips (5 mm apical part of the roots) was analyzed with an antibody raised against MHZ10/OsTAR2. ER-located binding immunoglobulin protein served as a loading control. G, Phenotypic rescue by introducing a MHZ10/OsTAR2 genomic fragment into the mhz10-1 mutant. CPL, functional complemented line. Scale bar, 10 mm. Data are means ± se (n ≥ 18). H, Free IAA levels in roots in response to ethylene. Data are means ± sd (n = 3). I, GUS staining assay for ethylene-induced Aux in etiolated seedlings. DR5 promoter-driven GUS expression reflects Aux signaling output. Scale bar, 4 mm. Insets show enlargement of root tips, with scale bar, 1 mm. J, Effects of IPyA treatment on root length of mhz10 in ethylene response. Scale bar = 10 mm. Data are means ± se (n ≥ 21). *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control. Source data are provided in Supplemental Data Set 2.
The normal ethylene response of heterozygous progeny generated from a cross between mhz10-1 and WT suggested that a mhz10-1 is a recessive mutation (Supplemental Figure S2). Based on map-based cloning, we identified MHZ10 as Os01g07500, encoding an aminotransferase (Figure 1C and see below). Subcellular localization analysis showed that MHZ10 is an ER-located protein (Figure 1D). We confirmed the presence of mutations in the candidate gene in each of the three mutant alleles at the genome and protein levels (Figure 1, E and F). The mhz10-1 allele harbors an A-to-G base substitution 41-bp upstream from the start codon (Figure 1C). Both mhz10-2 and mhz10-3 have mutations resulting in amino acid residue substitutions at conserved sites in the aminotransferase domain, according to the amino acid sequence alignment (Figure 1C; Supplemental Figure S3). The mutants had no detectable or very low levels of MHZ10 protein and very little ethylene induction in roots, which may be due to the reduced transcriptional/translational efficiency of the mutated gene or instability of the mutant protein (Figure 1F). We introduced an ∼8.2-kb genomic fragment from WT containing the intact Os01g07500 sequence into mhz10-1, which fully complemented the abnormal root ethylene response, as well as the defects in field-grown plants (Figure 1G; Supplemental Figure S4). All these results indicate that MHZ10 corresponds to Os01g07500, which is essential for normal rice growth and root ethylene response. It should be mentioned that among the three mutant alleles, mhz10-1 was obtained from an EMS-mutagenized population, while mhz10-2 and 10-3 were identified from a screen for suppressors of OsEIN2-overexpressing plants.
MHZ10 encodes a putative tryptophan aminotransferase with a transmembrane domain and a catalytic domain (Figure 1C; Supplemental Figure S3). This gene has been named as OsTAR2/FIB/TSG1 in previous studies, and is homologous to Arabidopsis TAA1 and TAA1-RELATED1 (TAR1) and TAR2 involved in the IPyA-dependent Aux biosynthesis pathway in Arabidopsis (Stepanova et al., 2008; Abu-Zaitoon et al., 2012; Yoshikawa et al., 2014; Guo et al., 2020). MHZ10/OsTAR2 has three homologs named OsTAR1/FBL (Os05g07720), OsTARL1, and OsTARL2 in rice (Guo et al., 2020). While OsTAR1/FBL may be involved in grain development in rice, the OsTARL1 and OsTARL2 exhibited no aminotransferase activity (Yoshikawa et al., 2014; Guo et al., 2020). The severe defect in fertility in the mhz10-3 mutant is most likely related to the mutation in MHZ10 and is consistent with the abnormal development of flowers in fib (Yoshikawa et al., 2014).
Ethylene induces Aux biosynthesis in rice root via an OsTAR2-mediated IPyA-dependent pathway
The identification of the tryptophan aminotransferase MHZ10 (OsTAR2 thereafter) from the analysis of rice ethylene response mutants raised the possibility that OsTAR2 acts in ethylene-modulated Aux biosynthesis in the root ethylene response. We thus measured the levels of the free Aux IAA in roots (Figure 1H), and found that IAA levels are significantly increased in WT upon ethylene treatment. In contrast, we detected lower IAA levels in mhz10 roots in the presence or absence of ethylene. We crossed the Aux reporter line harboring a GUS gene, driven by a synthetic Aux-responsive promoter DR5 (Ulmasov et al., 1997; Yamamoto et al., 2007), into mhz10-1, and examined GUS staining in response to ethylene (Figure 1I). Ethylene induced GUS staining in both coleoptiles and roots of WT seedlings and in mhz10-1 coleoptiles, but the ethylene-induced staining was abolished in the mutant roots. These results indicate that OsTAR2 may play a major role in ethylene response of rice root.
To further elucidate the function of OsTAR2, we applied 0.1-mM IPyA to seedlings, a concentration that shows a different response in WT and mhz10 mutants (Supplemental Figure S5). Root growth in mhz10 mutants was significantly inhibited by ethylene treatment in the presence of IPyA (Figure 1J). These results demonstrate that OsTAR2 is specifically involved in rice root ethylene response by functioning at the conversion step from tryptophan to IPyA in the Aux biosynthesis pathway.
OsTAR2 acts downstream of ethylene signaling
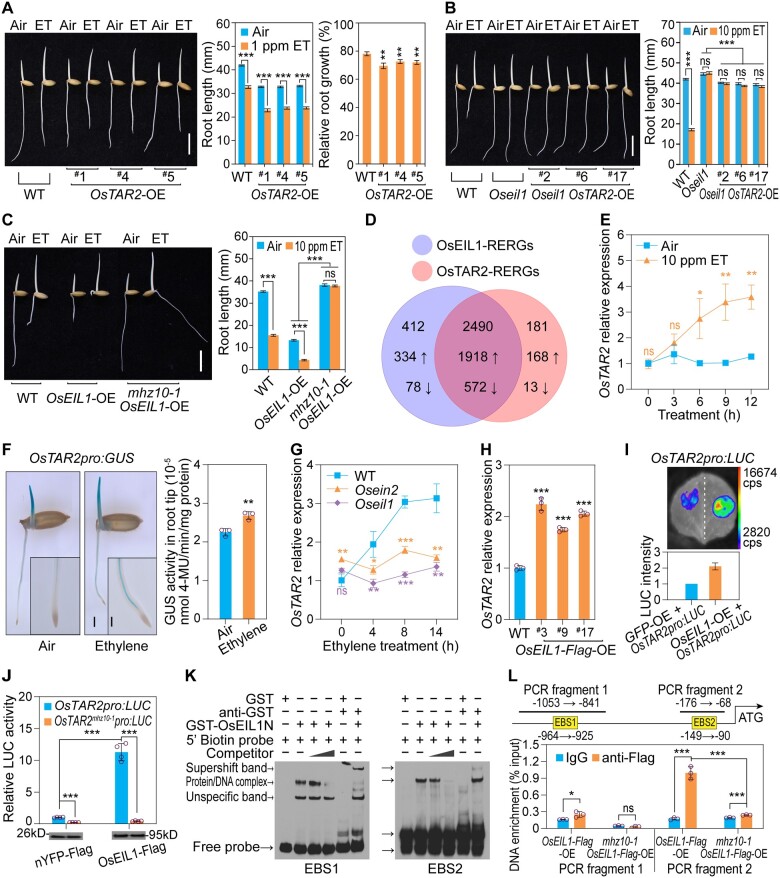
We examined the effects of OsTAR2 on ethylene responses in rice. Overexpression of OsTAR2 conferred a constitutive short root phenotype to etiolated seedlings compared to WT (Figure 2A; Supplemental Figure S6A). Ethylene treatment further reduced root growth in these transformants (Figure 2A). We investigated the genetic interaction between OsTAR2 and our previously identified ethylene signaling component MHZ6/OsEIL1 (Yang et al., 2015). Overexpression of OsTAR2 in the Oseil1 background reduced root length of the mutant, but did not exhibit significant response to ethylene in roots (Figure 2B; Supplemental Figure S6B). In mhz10-1 OsEIL1-OE plants, the mhz10-1 mutation almost fully suppressed the constitutive and hypersensitive root ethylene response of OsEIL1-OE transformants (Figure 2C), indicating that OsTAR2 is required for OsEIL1 function in regulating root growth. It should be noted that, upon ethylene treatment, although the root elongation was not significantly affected, the gravity response of mhz10-1 OsEIL1-OE roots was abnormal, as illustrated by the angle of the root growth direction, likely due to ethylene-induced alterations of the basal Aux distribution generated by residual OsTAR2 activity, further indicating that the ethylene/Aux crosstalk affects the root gravity response of rice. Together, these results suggest that OsTAR2 acts downstream of OsEIL1 in root ethylene response of rice.
Figure 2.
OsEIL1 activates OsTAR2 promoter activity through direct binding. A, Ethylene response of root length in OsTAR2-overexpression seedlings. Scale bar, 10 mm. Relative root growth is relative to length in air. Data are means ± se (n ≥ 17). B, Ethylene response of root length in Oseil1 OsTAR2-OE seedlings. Data are means ± se (n ≥ 18). C, Ethylene response of root length in mhz10-1 OsEIL1-OE seedlings. Please note that the root growth angle of mhz10-1 OsEIL1-OE was altered in ethylene treatment. Data are means ± se (n ≥ 16). D, OsTAR2 or OsEIL1-regulated gene expression in root ethylene response analyzed by RNA-seq. Total RNAs from root tips were used for the assay. Arrows pointing up and down indicate upregulated and downregulated genes, respectively. E, Response of OsTAR2 expression in root tips to ethylene. Data are means ± sd. F, Effects of ethylene on OsTAR2 promoter-driven GUS activity in etiolated seedlings. Representative seedlings are shown on the left. Scale bar, 2 mm. Insets show the enlargement of the root apical part, and scale bar, 0.5 mm. Root tips were harvested for quantification of GUS activity (shown on the right). Data are means ± sd (n = 3). G, Ethylene induction of OsTAR2 expression in Osein2 and Oseil1. Data are means ± sd (n = 3). H, OsTAR2 expression in root tips of OsEIL1-OE seedlings. Data are means ± sd (n = 3). I, Activation of OsEIL1 on OsTAR2pro:LUC activity in N. benthamiana leaves. OsTAR2 promoter-driven LUC was used as a reporter. OsEIL1-OE was used as an effector, and GFP-OE served as a control. Data are means ± sd (n = 8). J, Effects of OsEIL1 on OsTAR2pro:LUC or OsTAR2mhz10-1pro:LUC activity in rice protoplasts. OsEIL1-Flag was used as an effector, and nYFP-Flag was used as an effector control. Data are means ± sd (n = 4). Immunoblot shows Flag-tagged proteins. K, EMSA assay for OsEIL1 N-terminus binding to EBS elements in the OsTAR2 promoter. GST-tagged N-terminal domain (amino acids, aa 1–350) of OsEIL1 was used for testing the binding. Anti-GST antibody is also included in the assay, which leads to supershift band. L, ChIP-qPCR assay for OsEIL1 binding to the OsTAR2 promoter in vivo. OsEIL1-Flag-OE lines in WT or the mhz10-1 background were used for the ChIP analysis. PCR fragments 1 and 2 harboring ethylene binding sites EBS1 or EBS2 were examined. Data are means ± sd (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control.
Transcriptome deep sequencing (RNA-seq) analysis (NCBI database: PRJNA639684; Supplemental Data Set 1) was performed to investigate OsTAR2-regulated genes in root ethylene response (Figure 2D). Those genes that were regulated in WT root tips by ethylene but were not affected in mhz10 or Oseil1 mutants upon ethylene treatment were regarded as OsTAR2-regulated or OsEIL1-regulated ethylene-responsive genes, respectively. Most (93.2%) of the OsTAR2-regulated ethylene-responsive genes (designated OsTAR2-RERGs) were included in the OsEIL1-RERGs, and the common OsEIL1- and OsTAR2-RERGs genes represented 85.8% of all ethylene-responsive genes regulated by OsEIL1. These results not only support the essential role of OsTAR2-mediated Aux pathway in the root ethylene response of rice, but also suggest a close relationship between OsEIL1 and Aux signaling.
OsEIL1 binds to the OsTAR2 promoter and activates its transcription
We examined OsTAR2 expression in different tissues. OsTAR2 transcripts were abundant in vascular tissues of coleoptiles, roots, and shoots, as well as in florets (Supplemental Figure S7). Ethylene induced OsTAR2 expression, especially in root tips based on RT-qPCR analysis and OsTAR2 promoter GUS (OsTAR2pro:GUS) analysis, while we observed no significant change in transcript levels or GUS staining in shoots (Figure 2, E and F; Supplemental Figure S8), suggesting the specific role of OsTAR2 in root ethylene response at the seedling stage.
We also checked OsTAR2 expression in ethylene signaling mutants. Compared to the ethylene-mediated induction in WT, OsTAR2 expression was not induced by ethylene in Osein2 or Oseil1 (Figure 2G), indicating the essential role of ethylene signaling components in activating ethylene-induced OsTAR2 expression. In OsEIL1-Flag-overexpressing plants, OsTAR2 was more highly expressed in root tips compared to WT (Figure 2H; Supplemental Figure S6C). We further investigated if the transcription factor OsEIL1 directly regulated OsTAR2 promoter activity. In transient expression assays in both Nicotiana benthamiana leaves and rice protoplasts, OsEIL1 enhanced the reporter activity of OsTAR2 promoter-driven firefly luciferase (OsTAR2pro:LUC), indicating that OsEIL1 activates OsTAR2 transcription (Figure 2, I and J). However, when we used the mhz10-1 mutant promoter of OsTAR2, OsEIL1 failed to elevate the reporter activity, suggesting that the critical site for OsEIL1 activation on the promoter is disrupted by this mutation (Figure 2J). We also analyzed the OsTAR2 promoter region, and identified several putative OsEIL1-binding sites with the core sequence AYGWAYCT (Kosugi and Ohashi, 2000; Yamasaki et al., 2005). Electrophoretic mobility shift assay (EMSA) demonstrated that the N-terminal DNA-binding domain of OsEIL1 can bind to the sequences for their two predicted sites within the OsTAR2 promoter (Figure 2K; Supplemental Table S2). Chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) analysis provided additional evidence for OsEIL1 binding to the regions containing its cognate target sequences in vivo, and we detected a reduced affinity for OsEIL1 to the mhz10-1 promoter (Figure 2L). All these results demonstrate that the transcription factor OsEIL1 in the ethylene signaling pathway directly activates OsTAR2 expression to regulate root growth in response to ethylene.
Ethylene-induced OsTAR2 expression requires Aux pathway
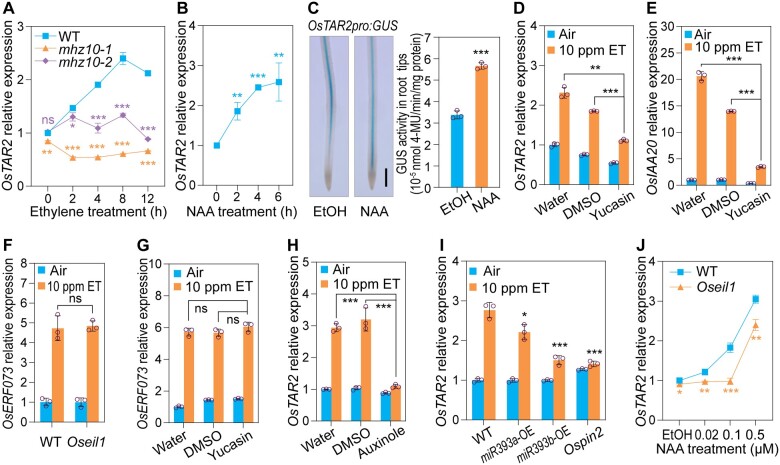
Ethylene induced the expression of OsTAR2 (Figure 2, E and F). We examined if mutations in OsTAR2 affected its own expression levels in response to ethylene. The mutation in mhz10-1 was located 41-bp upstream of the translation start site and abolished the ethylene induction (Figure 3A), most likely due to the inability of OsEIL1 to activate OsTAR2 transcription (Figure 2J). For mhz10-2, although the OsTAR2 upstream region is intact, its OsTAR2 expression levels also did not respond to ethylene (Figure 3A). These results indicate that ethylene-induced OsTAR2 expression requires OsTAR2 protein, a component of the Aux biosynthesis pathway. Considering that OsEIL1 can bind to the OsTAR2 promoter and activate its expression, the Aux pathway may integrate ethylene signaling at this level. We studied the effects of Aux on OsTAR2 expression further. Aux treatment also elevated OsTAR2 expression in root tips, as revealed from RT-qPCR and promoter-GUS analyses (Figure 3, B and C). Treatment with yucasin, an Aux biosynthesis inhibitor competing with the substrate IPyA for YUCCA (Nishimura et al., 2014), inhibited ethylene-induced OsTAR2 expression, as well as that of the ethylene-induced and Aux-responsive gene OsIAA20 (Chen et al., 2018), but had no obvious effect on the expression of the OsEIL1-independent ethylene-responsive gene OsERF073 (Figure 3, D–G). These analyses suggest that ethylene regulation of OsTAR2 is Aux dependent. We further investigated if the disruption of Aux signaling or polar transport would affect ethylene-induced OsTAR2 expression. Auxinole, an antagonist of IAA (Hayashi et al., 2012), drastically inhibited the ethylene induction of OsTAR2 expression in root tips (Figure 3H). In OsmiRNA393a/b-overexpression plants with reduced transcript levels for Aux receptor genes (Supplemental Figure S9) (Bian et al., 2012; Xia et al., 2012) and the Aux transporter mutant mhz8, a mutant in PIN-FORMED2 (OsPIN2) (Supplemental Figure S10) (Zhou et al., 2020), ethylene-induced OsTAR2 expression was partially or largely abolished (Figure 3I). These results indicate that the ethylene-mediated induction of OsTAR2 expression requires an active and functional Aux pathway.
Figure 3.
Aux pathway is required for ethylene-induced OsTAR2 expression. A, Response of OsTAR2 expression in root tips of mhz10 mutants to ethylene. Data are means ± sd (n = 3). B, Aux response of OsTAR2 expression in root tips. Two-DAG etiolated seedlings were treated with 0.1-μM NAA for the indicated duration. Data are means ± sd (n = 3). C, GUS activity assay for Aux-induced OsTAR2 expression in etiolated seedling roots. One-DAG etiolated seedlings were treated with or without 0.1-μM NAA for 4 h, and stained for 4 h. Scale bar, 1 mm. GUS activity was quantified (shown on the right). Data are means ± sd (n = 3). D, Ethylene-induced expression of OsTAR2 in the context of inhibiting Aux biosynthesis by yucasin. Etiolated seedlings were treated with or without 10 ppm ethylene and 50-μM yucasin for 8 h. Data are means ± sd (n = 3). E, Effects of yucasin treatment on ethylene-induced expression of OsIAA20. Data are means ± sd (n = 3). F, Ethylene response of OsERF073 expression in root tips of Oseil1. Data are means ± sd (n = 3). G, Effects of yucasin treatment on ethylene-induced OsERF073 expression. Data are means ± sd (n = 3). H, Ethylene induction of OsTAR2 expression in 50-µM auxinole treatment. Data are means ± sd (n = 3). I, Effects of genetic lesions in the Aux pathway on ethylene-induced OsTAR2 expression in root tips. OE lines had reduced expression of Aux receptor genes (Supplemental Figure S9). Ospin2 is a mutant identified in our study. Data are means ± sd (n = 3). J, Aux response of OsTAR2 expression in root tips of Oseil1. Data are means ± sd (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control.
To further elucidate the relationship between the Aux pathway and OsEIL1 in OsTAR2 regulation, we examined Aux-induced OsTAR2 expression in the Oseil1 mutant (Figure 3J). While all NAA concentrations tested induced OsTAR2 expression in WT roots, concentrations below 0.1 µM did not induce OsTAR2 expression in the Oseil1 mutant, suggesting that OsEIL1 is required for Aux-induced OsTAR2 activation at lower Aux concentrations. At higher Aux concentrations, the loss of OsEIL1 function also partially reduced the Aux induction of OsTAR2 expression. Considering that OsEIL1 directly binds to the OsTAR2 promoter and activates its expression, all the above results suggest that ethylene-induced OsEIL1 activation of OsTAR2 expression in root tips requires the participation of Aux signaling components.
Aux/IAAs interact with OsEIL1 to enhance or reduce OsTAR2 promoter activity
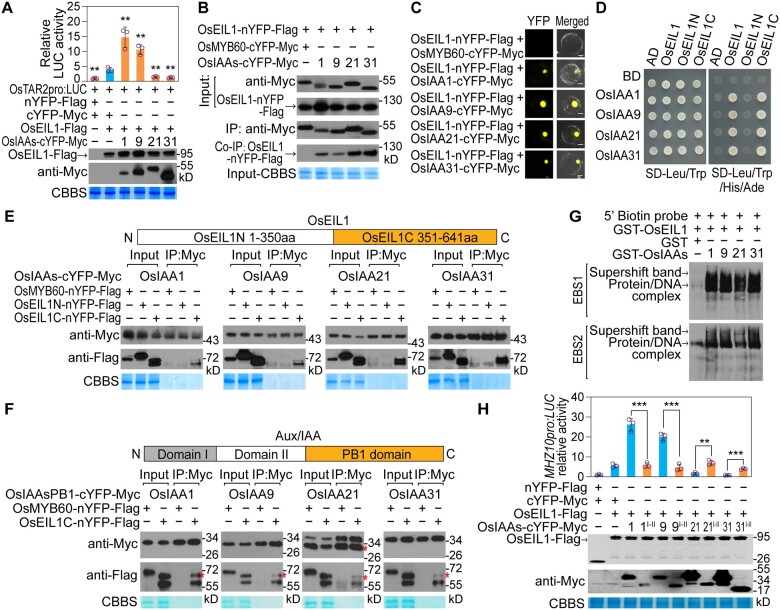
Since the transcription factor OsEIL1 is required for the induction of OsTAR2 expression by low concentrations of Aux (Figure 3J), and as OsEIL1-activated OsTAR2 expression requires the Aux pathway (Figure 3, A, D, H, and I), we hypothesized that OsEIL1 may cooperate with transcriptional regulators of the Aux pathway to control OsTAR2 expression. ARFs and Aux/IAAs are two such kinds of transcriptional regulators in Aux signaling. We first tested the effects of 14 OsARFs (Supplemental Table S1) expressed in root tips based on transcriptome data on OsTAR2 promoter activity using transfection of rice protoplasts. In this system, OsEIL1 activated OsTAR2 promoter activity, as revealed by the LUC activity from the OsTAR2pro:LUC reporter construct (Figure 2J). However, these 14 OsARFs did not significantly affect OsTAR2 promoter-driven LUC activity compared to OsEIL1, suggesting that these ARFs may not participate in the activation of OsTAR2 transcription (Supplemental Figure S11). We then examined the effects of 21 OsAux/IAAs expressed in root tips (Supplemental Table S1) on OsTAR2 promoter activity: again, we observed no significant effect (Supplemental Figure S12A). Considering that Aux/IAAs usually physically interact with ARFs to act as repressors to regulate gene expression, we investigated if these Aux/IAAs have any effects on OsEIL1-activated OsTAR2 promoter activity. While most of these Aux/IAAs did not have significant effects, we discovered that OsIAA1 and OsIAA9 did enhance OsEIL1-activated OsTAR2 promoter activity, while OsIAA21 and OsIAA31 showed inhibitory effects on OsEIL1-activated OsTAR2 promoter activity (Figure 4A; Supplemental Figure S12B).
Figure 4.
OsIAA1/9 and OsIAA21/31 regulate OsEIL1-activated OsTAR2 promoter activity through interacting with OsEIL1. A, Regulatory activity of OsIAA1/9/21/31 on OsEIL1-activated OsTAR2pro:LUC activity in rice protoplasts. Data are means ± sd (n = 3). Immunoblot assay shows the levels of the proteins encoded by the co-transfected constructs, and Coomassie Brilliant Blue Staining served as a loading control. **P < 0.01, as determined by a two-tailed Student’s t test compared to OsEIL1. B, Co-IP assay showing the interaction between OsIAA1/9/21/31 and OsEIL1. Nucleus-located protein OsMYB60 (Liu et al. 2018) served as a negative control. C, BiFC assay showing the interaction between OsIAA1/9/21/31 and OsEIL1 in rice protoplasts. More than five cells were observed in different microscope fields, and representative cells are shown. Scale bars, 10 μm. D, Yeast two-hybrid assay for the interaction between OsIAA1/9/21/31 and OsEIL1. AD-OsEIL1N and AD-OsEIL1C contain aa 1–350 and aa 251–641 of OsEIL1, respectively. E and F, Co-IP assay for the interaction between the C-terminus of OsEIL1 and the PB1 domain of OsIAA1/9/21/31. EIL1N-nYFP-Flag and EIL1C-nYFP-Flag contain aa 1–350 and aa 351–641 of OsEIL1, respectively. PB1 domains are aa 93–199, aa 91–182, aa 126–264, and aa 71–197 regions of OsIAA1, OsIAA9, OsIAA21, and OsIAA31, respectively. “*” indicates IAA21PB1-cYFP-Myc or EIL1C-nYFP-Flag band according to the predicted protein size. G, Enhanced effect of OsIAA1/9/21/31 on OsEIL1 binding to the EBS1 and EBS2 sites in the OsTAR2 promoter by EMSA-supershift assay. H, Activity of PB1 domain-deleted OsIAA1/9/21/31 in regulating OsEIL1-activated OsTAR2 promoter activity. Domains I and II are aa 1–92, aa 1–90, aa 1–125, and aa 1–70 regions of OsIAA1, OsIAA9, OsIAA21, and OsIAA31, respectively. Data are means ± sd (n = 3). **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control.
Since OsIAA1/9 and OsIAA21/31 showed regulatory effects on OsEIL1-activated OsTAR2 promoter activity, we asked if the four proteins regulate OsEIL1 activity through physical interaction. Co-immunoprecipitation (Co-IP) assay in rice protoplasts demonstrated that OsIAA1/9/21/31 all interact with OsEIL1 (Figure 4B). Bimolecular fluorescence complementation (BiFC) assay confirmed these interactions and localized them to the nucleus (Figure 4C). The transcription factor OsEIL1 is composed of an N-terminal DNA-binding domain and a C-terminal regulatory domain, and Aux/IAAs have a PB1 domain involved in protein–protein interaction, prompting us to map out which domain mediates the interaction between OsEIL1 and the OsIAAs. Yeast two-hybrid and Co-IP assays determined that the PB1 domain of OsIAA1/9/21/31 interacts with the C-terminal regulatory domain of OsEIL1 (Figure 4, D–F). EMSA supershift assay revealed that OsIAA1/9/21/31 enhances the affinity of OsEIL1 for the OsTAR2 promoter (Figure 4G). Furthermore, these Aux/IAA proteins lost their regulatory activity on OsEIL1 when the PB1 domain was deleted (Figure 4H), indicating that the regulation is dependent on protein–protein interaction. These results indicate that OsIAA1/9 and OsIAA21/31 exert their corresponding positive or negative effects on OsEIL1-activated OsTAR2 promoter activity through direct interaction with the OsEIL1 C-terminal regulatory domain.
OsIAA1/9 and OsIAA21/31 regulate OsEIL1-activated OsTAR2 expression in rice roots
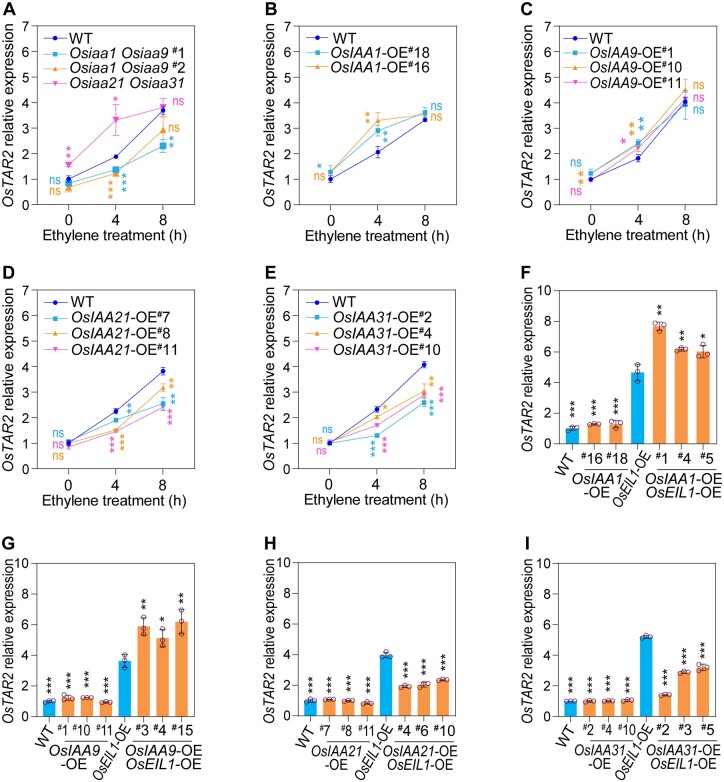
We further generated Osiaa1 Osiaa9 and Osiaa21 Osiaa31 double mutants using clustered regularly interspaced short palindromic repeats(CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing to assess the effects of these gene mutations on OsTAR2 expression (Supplemental Figure S13). OsTAR2 showed a lower transcriptional induction in the two Osiaa1 Osiaa9 double mutants but higher induction in the Osiaa21 Osiaa31 double mutant than that in WT, especially in the early ethylene response, consistent with a promotion role for OsIAA1/9 but an inhibitory role for OsIAA21/31 in OsTAR2 regulation (Figure 5A).
Figure 5.
Overexpression or mutation of OsIAA1/9/21/31 affects ethylene-induced OsTAR2 expression in root tips. A, Ethylene-induced OsTAR2 expression in root tips of Osiaa1 Osiaa9 and Osiaa21 Osiaa31 double mutants. One-DAG etiolated seedlings were treated with 10 ppm ethylene for 0, 4, or 8 h, and root tips were used for analyzing OsTAR2 expression. Three independent biological replicates were performed. Data are means ± sd. B–E, Ethylene-induced OsTAR2 expression in root tips of OsIAA1/9/21/31-OE seedlings. Data are means ± sd (n = 3). F–I, Effects of OsIAA1/9/21/31 overexpression on OsTAR2 expression in root tips of OsEIL1-OE seedlings. The data of OsTAR2 expression in WT and OsIAAs overexpression in WT background are from (B–E). Data are means ± sd (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to WT or OsEIL1-OE seedlings.
We also generated lines individually overexpressing OsIAA1/9 and OsIAA21/31 (OsIAA1/9-OE and OsIAA21/31-OE; Supplemental Figure S14, A–D), and OsTAR2 expression showed a moderately higher induction in OsIAA1/9-OE plants but exhibited a lower induction in OsIAA21/31-OE plants compared to WT in response to ethylene (Figure 5, B–E). We also overexpressed OsIAA1/9 or OsIAA21/31 into the OsEIL1-OE background (Supplemental Figure S14, E–H) (OsEIL1OE, Yang et al., 2015) by transgenic approach, and tested their effects on OsEIL1-activated OsTAR2 expression. OsIAA1/9 enhanced OsEIL1-activated OsTAR2 expression, whereas OsIAA21/31 has the opposite effect (Figure 5, F–I). These results support the notion that OsIAA1/9 promote and OsIAA21/31 reduce OsEIL1-activated OsTAR2 expression in rice roots.
We further examined the root ethylene response in the above Osiaa1 Osiaa9 and Osiaa21 Osiaa31 double mutants and in OsIAA1/9-OE and OsIAA21/31-OE plants in the WT or OsEIL1-OE background (Supplemental Figure S15). OsIAA21/31 was necessary and sufficient to confer reduced ethylene sensitivity in roots. The two genes also reduced the constitutive ethylene-response phenotype of short roots seen in the OsEIL1-OE line (Supplemental Figure S15, A, D, E, H, and I), consistent with their inhibitory role on OsEIL1 function. OsIAA1/9 showed similar functions in root ethylene response (Supplemental Figure S15, A, B, C, F, and G), probably reflecting their stronger roles in Aux signaling events in this process.
OsIAA1/9 and OsIAA21/31 regulate OsEIL1 activity by changing histone acetylation levels
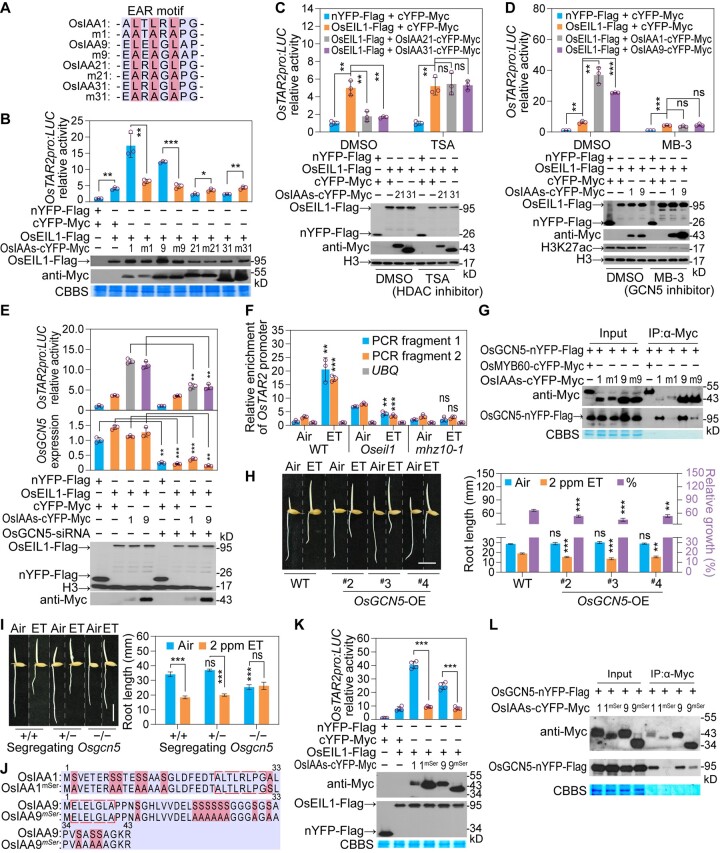
Since both OsIAA1/9 and OsIAA21/31 can interact with the C-terminal domain of OsEIL1 but show promotive and inhibitory effects, respectively, on OsEIL1-activated OsTAR2 promoter activity, we next investigated the mechanisms involved in these different effects. All four OsIAA proteins contain domains I, II, and PB1 (Supplemental Figure S16). Usually, the EAR motif within domain I determines the transcriptional regulatory activity of Aux/IAA proteins (Tiwari et al., 2001, 2004). Therefore, we generated mutations in the EAR motifs of these OsIAAs by changing three conserved leucine residues to alanine (Figure 6A; Szemenyei et al., 2008). These mutations in the EAR motifs eliminated both the promotive and inhibitory effects of OsIAA1/9/21/31 on OsEIL1-activated OsTAR2pro:LUC activity, even though the physical interaction between these EAR motif-mutated OsIAAs and OsEIL1 remained (Figure 6B; Supplemental Figure S17). This result indicates that the EAR motif is crucial for OsIAA1/9/21/31 to regulate OsEIL1 activity. According to previous studies, EAR motif-dependent transcriptional repression is related to chromatin remodeling through recruiting an HDAC complex (Kagale and Rozwadowski, 2011). Trichostatin A (TSA) (Vigushin et al., 2001), an HDAC inhibitor, was thus applied to investigate if the inhibitory activity of OsIAA21/31 on OsEIL1 is related to histone deacetylation: indeed, TSA treatment disrupted the inhibitory activity of OsIAA21/31 (Figure 6C). These results indicate that the inhibitory effect of OsIAA21/31 on OsEIL1-activated OsTAR2 expression depends on histone deacetylation.
Figure 6.
The EAR motif and its flanking sequences determine the regulatory activity of OsIAA1/9/21/31 on OsEIL1-activated OsTAR2 expression through affecting the histone acetylation/deacetylation. A, Generation of mutation in the EAR motifs of OsIAA1/9/21/31. Three conserved Leu (L) residues in the EAR motif of OsIAA1 (aa 24–31), OsIAA9 (aa 2–9), OsIAA21 (aa 23–30), and OsIAA31 (aa 8–15) were changed to Ala (A) residues, and mutant OsIAAs were designated m1, m9, m21, and m31, respectively. B, Effects of EAR motif mutations on OsIAA1/9/21/31-mediated regulation of OsEIL1 activity. Data are means ± sd (n = 3). C, Effects of HDAC inhibitor (TSA) on OsIAA21/31 inhibition of OsEIL1 activity. Data are means ± sd (n = 3). D, Effects of OsGCN5 inhibitor (MB-3) on OsIAA1/9-mediated promotion of OsEIL1 activity. Data are means ± sd (n = 3). E, Effects of repressing OsGCN5 expression by siRNA on OsIAA1/9-mediated promotion of OsEIL1 activity. Three biological replicates were performed in analyzing OsTAR2 promoter activity and OsGCN5 expression. Data are means ± sd. F, H3K27ac levels over the OsTAR2 promoter region in response to ethylene, as analyzed by ChIP. One-DAG etiolated seedlings were treated with 10 ppm ethylene for 8 h, and root tips were used for analysis. PCR fragments 1 and 2 are the same as in Figure 2F. Data are means ± sd (n = 3). G, Interaction between OsGCN5 and OsIAA1/9 or EAR motif-mutated OsIAA1/9, as analyzed by Co-IP. H, Ethylene response of root length in OsGCN5-OE seedlings. Etiolated seedlings were grown in air or 2 ppm ethylene for 1.5 DAG. Root length of kanamycin-resistant seedlings from segregating lines was measured. Dashed lines indicate the seams between images. Scale bar, 10 mm. Data are means ± se (n ≥ 9). I, Ethylene response of root length in Osgcn5 mutant seedlings. Etiolated seedlings were grown in air or 2 ppm ethylene for 2 DAG. Analysis was performed using the root length of seedlings from a segregating Osgcn5 line generated by CRISPR/Cas9. +/+, WT genotype of OsGCN5; +/–, heterozygous genotype; –/–, homozygous genotype. Scale bar, 10 mm. Data are means ± se (n ≥ 13). J, Artificial mutations in the EAR motif-flanking sequences in domain I of OsIAA1/9. Mutations (Ser to Ala) in OsIAA1/9 are indicated, and mutant OsIAAs were named OsIAA1mSer and OsIAA9mSer, respectively. K, Effects of the Ser-to-Ala substitution in domain I on OsIAA1/9 promotion of OsEIL1-activated OsTAR2 promoter activity. Data are means ± sd (n = 4). L, Reduced interaction between OsGCN5 and OsIAA1/9mSer as revealed by Co-IP. *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control.
To explore the promotive effect of OsIAA1/9 on OsEIL1, we also took the histone acetylation modification into consideration. Curcumin (Balasubramanyam et al., 2004) is a p300/CBP-specific histone acetyltransferase (HAT) inhibitor. We observed no obvious effect upon curcumin treatment (Supplemental Figure S18). However, treatment with MB-3 (Biel et al., 2004; Mai et al., 2006; Chen et al., 2017), a GCN5-specific HAT inhibitor, almost completely eliminated the promotive effect of OsIAA1/9 on OsEIL1-activated OsTAR2 promoter activity (Figure 6D). In addition, the promotive effect of OsIAA1/9 on OsEIL1-activated OsTAR2 promoter activity was largely alleviated when OsGCN5-specific short interfering RNAs (siRNAs) were co-transfected to knockdown OsGCN5 transcript levels (Figure 6E; Supplemental Table S2). Since ethylene cannot induce OsTAR2 expression in the Oseil1 and mhz10-1 mutants (Figures 2, G and 3, A), and as GCN5 positively regulates histone acetylation at H3K27, based on previous studies (Chen et al., 2017), we measured the levels of H3K27ac at the OsTAR2 promoter region using ChIP-qPCR. Ethylene treatment in WT increased H3K27ac levels in root tips, but not in Oseil1 or mhz10-1 (Figure 6F). A Co-IP assay demonstrated that OsIAA1/9 can interact with OsGCN5, but this interaction was reduced when the EAR motif was mutated (Figure 6G). In plants, overexpression of OsGCN5 conferred enhanced ethylene response in seedling roots (Figure 6H; Supplemental Figure S19), and Osgcn5 homozygous seedlings segregating from a population derived from a heterozygote generated by CRISPR/Cas9-mediated editing showed reduced or no ethylene sensitivity (Figure 6I; Supplemental Figure S20, A and B), indicating a positive role for OsGCN5-mediated histone acetylation in OsTAR2 expression and the rice root ethylene response. It should be noted that Osgcn5 homozygous seedlings showed short shoots and died after transfer to soil, likely due to essential roles of OsGCN5 in seedling establishment and/or environmental adaptation (Supplemental Figure S20C). These results indicate that OsIAA1/9 promote OsEIL1-activated OsTAR2 expression most likely through increasing OsGCN5-dependent histone acetylation.
Since the EAR motif is essential for both the promoting and repressive effects of these Aux/IAAs (Figure 6, A and B), we further investigated the reasons behind the opposing activities of OsIAA1/9 and OsIAA21/31. The EAR motifs in the four Aux/IAAs are not identical (Figure 6A). We thus exchanged eight core amino acid residues among the four OsIAAs to attempt to switch the activity type of OsIAA1/9 and IAA21/31. However, OsIAA1/9 with the EAR motif from OsIAA21/31, or OsIAA21/31 with the EAR motifs from OsIAA1/9, appeared to exert the same type of regulation on OsEIL1-activated OsTAR2pro:LUC activity as their intact counterparts (Supplemental Figure S21A). However, the OsIAA21/31 variants with domain I from OsIAA1/9 showed promotive effects (Supplemental Figure S21B). These results suggest that the promotive activity of OsIAA1/9 depends on both the EAR motif and its flanking sequences. Compared to the flanking sequences of OsIAA21/31, those of OsIAA1/9 are rich in serine residues (Supplemental Figure S16); we hence generated mutant OsIAA1/9 proteins with Ser-to-Ala substitutions in the flanking sequences for further studies (Figure 6J). Compared to intact OsIAA1/9, the proteins with the Ser-to-Ala substitution no longer promoted OsEIL1-activated OsTAR2pro:LUC activity (Figure 6K). To test whether the Ser residues are involved in recruiting OsGCN5, we assessed the interaction between the mutant OsIAA1/9 and OsGCN5 by Co-IP, which revealed that the interactions are largely abolished, very similar to the effects of the mutations in EAR motifs (Figure 6, G and L). These results indicate that the Ser residues in the flanking sequences near the EAR motif in domain I determine the specific promotive activity of OsIAA1/9 on OsEIL1. In addition, we noticed that the Ser-to-Ala OsIAA1/9 variants migrate faster than expected (Figure 6K), suggesting that posttranslational modifications occur on these Ser residues in the intact OsIAA1/9 proteins.
Differential slow degradation of OsIAA1/9 allows activation of OsEIL1-mediated OsTAR2 promoter activity
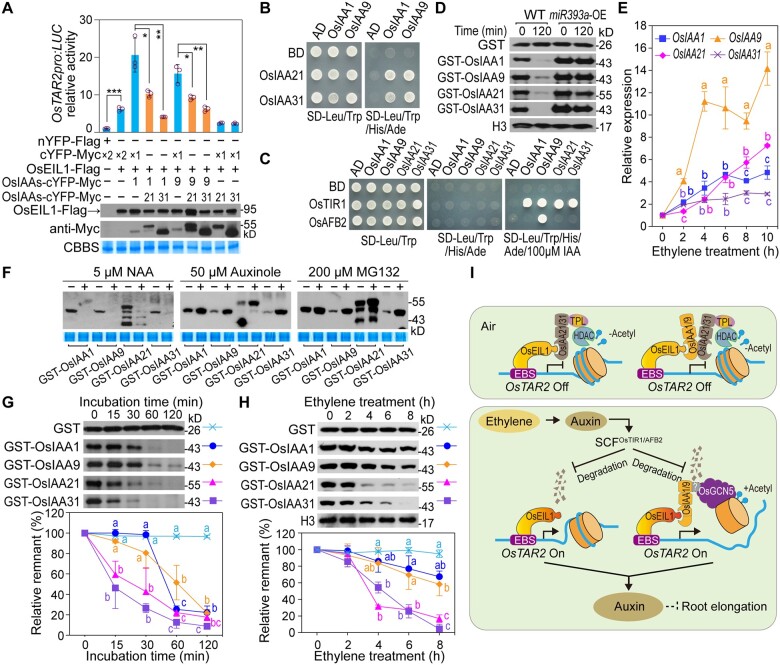
OsIAA1/9 and OsIAA21/31 play positive and negative roles, respectively, on OsEIL1-activated OsTAR2 promoter activity, so we next examined the effects of OsIAA21/31 on the promotive role of OsIAA1 or OsIAA9 in rice protoplasts. Co-transfection of OsIAA21 or OsIAA31 largely blocked or reduced the enhancement of OsIAA1 or OsIAA9 on OsEIL1 activity (Figure 7A), suggesting that OsIAA21/31 may “neutralize” the promotive role of OsIAA1/9. Aux/IAAs usually function as homodimers or heterodimers (Kim et al., 1997; Ouellet et al., 2001). We therefore tested the potential physical interaction between OsIAA1/9 and OsIAA21/31 by yeast two-hybrid: OsIAA1/9 did interact with OsIAA21/31 (Figure 7B). Such interactions may thus eliminate/reduce the promotive role of OsIAA1/9 on OsEIL1-activated OsTAR2 promoter activity.
Figure 7.
Interplay between OsIAA1/9/21/31 and OsEIL1 modulates OsEIL1-activated OsTAR2 expression in rice root. A, Roles of OsIAA1/9 and OsIAA21/31 in regulating OsEIL1-activated OsTAR2pro:LUC transcription. Data are means ± sd (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, as determined by a two-tailed Student’s t test compared to the corresponding control. B, Physical interaction between OsIAA1/9 and OsIAA21/31 in a yeast two-hybrid assay. C, Yeast two-hybrid assay for Aux-dependent interaction between OsIAA1/9/21/31 and the Aux receptors OsTIR1/AFB2. D, Degradation of OsIAA1/9/21/31 in root extracts of miR393a OE seedlings. Immunoblot evaluates the amount of the proteins, and histone H3 indicates the identical loading of root crude extracts in each reaction. E, Ethylene-induced expression of OsIAA1/9/21/31 in root tips. Data are means ± sd (n = 3). F, Effects of NAA, auxinole or MG132 treatments on OsIAA1/9/21/31 abundance in root extracts. For auxinole and MG132 treatments, seedlings used for root extracts preparation were pretreated with 10 ppm ethylene for 8 h. Each of the three chemicals or their corresponding solvents were preincubated with root extracts for 10 min, and then recombinant OsIAA was added and incubated for 30 min. G, Degradation of OsIAA1/9 and OsIAA21/31. Free GST protein served as a control. Data are means ± sd (n = 3). H, Comparison of the stability between OsIAA1/9 and OsIAA21/31 in response to ethylene. GST-tagged Aux/IAA proteins were incubated for 30 min with root crude extracts of etiolated seedlings treated with 10 ppm ethylene for different times. Data are means ± sd (n = 3). I, Working model for the OsEIL1-OsIAAs module in the regulation of OsTAR2 expression in root ethylene response. In air, OsEIL1 accumulates at a low level. OsIAA21/31 may interact with OsEIL1 to repress OsEIL1-activated OsTAR2 expression through recruiting the HDAC for potential histone deacetylation. OsIAA21/31 may also suppress the activity of the OsEIL1-OsIAA1/9 complex. OsTAR2 transcription stays at a normal level for basal Aux biosynthesis to sustain normal root growth. Upon ethylene treatment, the induced Aux levels lead to early degradation of OsIAA21/31, thus allowing the promotion of OsEIL1-activated OsTAR2 expression via OsIAA1/9 for signal amplification. The promotive roles of OsIAA1/9 involve the recruitment of the histone acetyltransferase OsGCN5 for histone modification and chromatin activation. Different lowercase letters indicate significance levels of P < 0.05, as determined by one-way ANOVA with Bonferroni correction (E, G, and H).
To investigate how these proteins interplay during signaling, we examined the possible fates of these proteins. Yeast two-hybrid assay showed that the four OsIAAs can interact with the Aux receptors OsTIR1/AFB2 in the presence of Aux (Figure 7C). We also assessed the protein degradation of OsIAA1/9/21/31 studied in root crude extracts. While most of the four OsIAAs were degraded in protein extracts from WT plants, their abundance was not significantly altered or only slightly decreased in root extracts from OsmiR393a-overexpressing plants in which the transcript levels of the Aux receptor genes OsTIR1/AFB2 are lower (Figure 7D; Supplemental Figure S9). These results demonstrate that the degradation of OsIAA1/9/21/31 is mediated by the Aux receptors OsTIR1/AFB2 in a typical Aux signaling pathway. We examined the expression of OsIAA1/9/21/31 in response to ethylene, and OsIAA9 exhibited the strongest inductive response, while OsIAA1/21/31 showed only mild or slight inductions (Figure 7E).
We also individually added NAA, auxinole, or MG132 to the root crude extracts to test their effects on the stability of the four OsIAAs. NAA promoted the degradation of all four OsIAAs, while auxinole or MG132 treatment stabilized the four OsIAA proteins (Figure 7F), indicating the effectiveness of the root extract assay system in studying the stability of these OsIAAs, consistent with our previous studies (Chen et al., 2018). We next compared the degradation of the OsIAAs using this system over a time course. Levels of all the four proteins decreased gradually. However, the degradation of OsIAA21/31 appeared to occur earlier than that of OsIAA1/9 (Figure 7G). Furthermore, OsIAA1/9 showed a higher stability in response to ethylene treatment than OsIAA21/31 (Figure 7H). Compared to OsIAA21/31, the expression of OsIAA1/9 had a stronger or similar ethylene induction but their protein turnover was slower (Figure 7, E, G, and H), hence allowing OsIAA1/9 to promote the activity of OsEIL1. It is likely that the earlier degradation of OsIAA21/31 relieves their inhibition on OsEIL1 and OsIAA1/9, and then OsIAA1/9 can promote OsEIL1-activated OsTAR2 promoter activity (Figure 7I).
Discussion
Through the analysis of the rice ethylene-insensitive mutants mhz10, we discovered that ethylene regulates root growth through the MHZ10/OsTAR2-mediated Aux biosynthesis pathway. OsTAR2 encodes a Trp aminotransferase converting Trp to IPyA that is further oxidized to IAA. The transcription factor MHZ6/OsEIL1 in the ethylene signaling pathway directly binds to the OsTAR2 promoter and activates its transcription. OsIAA1/9 and OsIAA21/31 interact with OsEIL1 to promote or inhibit OsEIL1-activated OsTAR2 promoter activity, respectively. Ethylene-induced Aux likely evokes Aux receptor-mediated degradation of the repressors OsIAA21/31, and allows the positive role of OsIAA1/9 on OsEIL1 for OsTAR2 activation through increasing chromatin histone acetylation (Figure 7I). Our study reveals a positive feedback regulatory mechanism facilitating Aux biosynthesis in the root ethylene response, and strengthens our understanding of the crosstalk between ethylene and Aux pathways.
Both the ethylene and Aux pathways are indispensable for ethylene-induced OsTAR2 expression. We propose that the two pathways may merge at this OsEIL1-Aux/IAAs module to synergistically control ethylene-inhibited root growth. This prediction is supported by the following evidence: (1) Ethylene specifically upregulates OsTAR2 expression in root tips, as judged from OsTAR2 promoter-GUS analysis (Figure 2, E and F); (2) Compared to WT, ethylene-induced OsTAR2 expression is abolished in Oseil1 and reduced in mutants of the Aux pathway (Figures 2, G, 3, A, D, H, and I); (3) Lower concentrations of Aux cannot induce OsTAR2 transcription in Oseil1, suggesting that OsEIL1 is required for OsTAR2 activation at low Aux levels (Figure 3J); (4) OsIAA1/9 and OsIAA21/31 can physically interact with OsEIL1 and affect OsEIL1-activated OsTAR2 expression in a positive and negative manner, respectively (Figure 4). We validated the effects of OsIAA1/9/21/31 on OsEIL1-activated OsTAR2 expression by using their corresponding overexpression and mutant lines (Figure 5); (5) We also observed physical interaction between OsIAA1/9 and OsIAA21/31, and they reciprocally influence their transcriptional regulatory activity, likely through these interactions (Figure 7, A and B); and (6) OsIAA21/31 may be subjected to earlier/faster degradation than OsIAA1/9 through the Aux receptor-dependent pathway (Figure 7, C–H).
It is worth wondering whether the expression of other genes in rice is also regulated by the OsEIL1-Aux/IAAs module as OsTAR2. OsYUC8 and OsHKT2;1 (High-affinity K+ transporter2;1) are ethylene-inducible genes, and both are directly regulated by OsEIL1 in rice roots according to previous studies (Yang et al., 2015; Qin et al., 2017). RT-qPCR analysis showed that these two genes cannot be induced in Oseil1 or mhz10-1 roots by ethylene (Supplemental Figure S22), suggesting that these two genes may be controlled by the same module. Considering that OsEIL1 and OsTAR2 co-regulate a large set of genes (Figure 2D), it is possible that at least a portion of the genes could be similarly regulated by the OsEIL1-Aux/IAAs module in rice roots.
He et al. (2011) identified the small molecule L-kynurenine (L-Kyn), a specific inhibitor of Arabidopsis TAA1/TARs involved in Aux biosynthesis. Their further studies revealed that Aux promotes the nuclear accumulation of EIN3 in an EBF1/EBF2-dependent manner in a positive feedback loop between Aux biosynthesis and ethylene signaling in Arabidopsis upon the application of L-Kyn (He et al., 2011). Whether Aux also promotes the nuclear accumulation of OsEIL1 in rice will be worth pursuing. We tested the effects of NAA, yucasin, L-Kyn, and auxinole treatment on Flag-tagged OsEIL1 protein levels in roots of OsEIL1-Flag-overexpressing seedlings. NAA drastically reduced OsEIL1-Flag abundance in root tips (Supplemental Figure S23A). The combined inhibition of Aux biosynthesis and Aux perception by treatment with chemicals such as yucasin, L-Kyn, or auxinole increased OsEIL1-Flag abundance in root tips (Supplemental Figure S23B). These analyses suggest that the Aux biosynthesis and signaling pathway facilitate OsEIL1 degradation. Such a potential regulatory mechanism may work at the desensitization stage of the ethylene response, when Aux biosynthesis and signaling have reached high levels. The regulation of OsEIL1 protein stability by Aux in rice may be different from that of EIN3 in Arabidopsis, and thus requires further investigation.
Several previous studies have demonstrated that ethylene-inhibited root elongation is largely dependent on the Aux pathway. TAA1 was identified from the isolation of the Arabidopsis mutant weak ethylene insensitive8 (wei8), which is involved in local Aux production and tissue-specific ethylene effects (Stepanova et al., 2008). OsYUC8 was identified as the mutant rice ethylene-insensitive (rein); OsYUC8 catalyzes the conversion of IPyA to IAA in response to ethylene by OsEIL1-activated expression (Qin et al., 2017). Our previous analysis also found that the E3 ubiquitin ligase MHZ2/SOR1 can target the nontypical Aux/IAA protein OsIAA26 for degradation, acting downstream of the OsTIR1/AFB-OsIAA9 module in root ethylene response (Chen et al., 2018). In this study, we characterized the rice root-specific ethylene-insensitive mutants mhz10, which harbor mutations in OsTAR2, encoding the tryptophan aminotransferase OsTAR2 that works at the first step of the IPyA Aux biosynthesis pathway, converting Trp to IPyA (Figure 1). Both MHZ10/OsTAR2 (this study) and OsYUC8 (Qin et al., 2017) were identified through the root ethylene response of rice seedlings, demonstrating that ethylene-inhibited root growth requires the IPyA Aux biosynthesis pathway in rice. Considering that the same mutant screen identified MHZ8/OsPIN2 (Aux transport) and MHZ2/SOR1 (Aux signaling) (Supplemental Figure S10) (Chen et al., 2018; Zhou et al., 2020), it is highly likely that the entire Aux pathway, from biosynthesis, transport, to signaling, is required for root ethylene responses in rice. It should be noted that while the three-member family TAA1, TAR1, and TAR2 had overlapping roles in Arabidopsis, the current OsTAR2 seems to have a major and decisive role, since the mhz10 mutants showed complete ethylene insensitivity in rice roots. Moreover, a systematic analysis of OsTAR2 sequences across 5,394 rice accessions only identified two polymorphisms converting amino acids in nonconserved sites (Supplemental Figure S3), suggesting that OsTAR2 may be highly conserved during rice evolution/domestication. In addition to the Aux pathway, the ABA biosynthesis pathway also participates in ethylene regulation of root growth in our previous studies (Ma et al., 2014; Yin et al., 2015).
Arabidopsis EIN3/EIL1 often acts as a crosstalk hub, connecting the ethylene pathway with many other biological processes. During plant development, the gibberellin repressor DELLA proteins interact with the DNA-binding domain of EIN3/EIL1, and repress EIN3/EIL1-regulated HOOKLESS1 (HLS1) expression (An et al., 2012). EIN3/EIL1-activated HLS1 expression is also repressed by the interaction between the salicylic acid receptor NONEXPRESSER OF PR GENES1 (NPR1) and EIN3/EIL1 (Huang et al., 2020). EIN3 and NPR1 also interact to affect leaf senescence (Wang et al., 2021; Yu et al., 2021). Additionally, EIN3 associates with MYC2, ROOT HAIR DEFECTIVE6, and PHYTOCHROME-INTERACTING FACTOR3 to regulate apical hook curvature (Song et al., 2014), root hair elongation (Feng et al., 2017), and chloroplast development (Liu et al., 2017). Moreover, JA-Zim domain proteins physically interact with EIN3 and repress EIN3/EIL1 through recruiting a corepressor reduced potassium dependency3-type histone deacetylase (HDA6) to regulate plant development and defense (Zhu et al., 2011). Presently, OsIAA1/9 and OsIAA21/31 can interact with OsEIL1, incorporating the primary Aux signal into the ethylene pathway.
Aux/IAAs usually play repressive roles on ARFs through EAR motif-mediated recruitment of TOPLESS (TPL)/TPL-RELATED (TPR) co-repressors and HDAC for chromatin inactivation and silencing of ARF target genes (Szemenyei et al., 2008). Inhibition of OsEIL1 activity by OsIAA21/31 is consistent with this mechanism, as revealed by treatment with the HDAC inhibitor TSA (Figure 6C). Different from this inhibitory feature, it is interesting to note that OsIAA1/9 play a promotive role on OsEIL1-activated OsTAR2 activity, likely through recruitment of the HAT OsGCN5 for chromatin activation (Figures 4–6). Such recruitment may require both the EAR motif and the surrounding Ser-rich sequence of OsIAA1/9. It is unknown whether this newly discovered promotive activity of OsIAA1/9 is specific to OsEIL1 or also has any effects on other factors. It cannot be ruled out that OsIAA1/9 may have a repressive activity toward some OsARFs through EAR motif-mediated recruitment of TPL/TPR co-repressors and HDAC, which would be consistent with the reduced sensitivity and moderate hypersensitivity to ethylene in roots of OsIAA1/9-OE and Osiaa1 Osiaa9 double mutant seedlings, respectively (Supplemental Figure S5, A, C, E, G, and I). Histone acetylation modification has been found to play roles in ethylene response in Arabidopsis (Zhang et al., 2017, 2018).
Taken together, we discovered that OsIAA1/9 and OsIAA21/31 interact with OsEIL1 and incorporate the primary Aux signal into the ethylene pathway to amplify signaling through Aux biosynthesis in the root ethylene responses of rice, revealing a crosstalk between ethylene and Aux pathway.
Materials and methods
Plant materials
Rice (O. sativa L.) plants were from the japonica variety Nipponbare. The mutants mhz7/Osein2 (Ma et al., 2013), mhz6/Oseil1 (Yang et al., 2015), and mhz8/Ospin2 (Zhou et al., 2020) were previously identified. A premature termination of translation in MHZ8/OsPIN2 was caused by a 568-bp DNA insertion from locus Os03g53480 at the 468-bp locus Os06g44970 (Supplemental Figure S10; Supplemental Table S2) (Olsen et al., 1984; Zhou et al., 2020). The miR393a OE line (Bian et al., 2012) and miR393b-OE line (Xia et al., 2012) were kindly provided by Prof. Mu-Yuan Zhu in ZHEJIANG University and Prof. Ming-Yong Zhang in South China Botanical Garden, CAS, respectively. The DR5pro:GUS line (Liu et al., 2015) was kindly provided by Prof. Jia-Yang Li in Institute of Genetics and Developmental Biology, CAS. Plants for epistasis analysis were prepared by crossing or Agrobacterium (Agrobacterium tumefaciens) (EHA105 strain)-mediated transformation. In transgenic plants overexpressing OsIAA1/9/21/31, the transgenes were driven by the OsACTIN promoter; in other overexpression lines, the transgenes were driven by the cauliflower mosaic virus (CaMV) 35S promoter. For ethylene responses in OsGCN5-overexpressing seedlings, kanamycin-resistant individuals from the T1 generation were phenotyped. The T2 generation of other overexpression plants was used for analysis. The Osiaa1 Osiaa9 and Osiaa21 Osiaa31 double mutants, and the Osgcn5 mutant were obtained by CRISPR/Cas9-mediated gene editing. For the analysis of Osgcn5, genotypes of the segregating individuals were identified by sequencing. ImageJ software was used to measure the root length of OsGCN5-OE and Osgcn5 seedlings. Nicotiana benthamiana was used for infiltration assays.
Plant growth conditions
For rice propagation and crossing, the plants were grown under the local natural light and temperature in the paddy field of the Experimental Stations of the Institute of Genetics and Developmental Biology in Beijing (116°23′E, 40°22′N) from May to October and in Hainan (110°04′E, 18°51′N) from November to next April. Tobacco seedlings were grown in a greenhouse at 22°C under a 15-h light/9-h dark photoperiod (light bulbs). Rice etiolated seedlings used for protoplasts isolation were grown on 1/2 MS medium (1/2 MS salt, 1% sucrose, and 0.3% phytagel) in dark at 28°C.
Statistical analysis
Calculations were performed using IBM SPSS Statistics version R24.0.0.2 software. For mean comparisons between two groups, a two-tailed Student’s t test was used for calculations. For multiple comparisons among groups, one-way analysis of variance (ANOVA) along with Bonferroni correction at a significance level of 0.05 was used for calculations. Details are provided in Supplemental Data Set 2.
Chemical treatments
Ethylene treatment was performed as previously described (Zhou et al., 2020). Briefly, in an incubator, rice seeds were sown on a stainless-steel mesh, and germinated in water at 37°C, then transferred into an air-tight box with the supplement of ethylene at 28°C for 2–3 days in the dark before phenotypic measurements. Air served as a control. Relative root growth was root length in ethylene treatment as a percentage of mean root length in air. One-day after germination (DAG) seedlings were used for short-term ethylene treatments. Roots of 1-DAG seedlings were immersed in NAA solutions for Aux treatment, and an equal volume of ethanol solution solvent served as a negative control. Co-treatments of ethylene with yucasin, IPyA, or auxinole solution are shown in Supplemental Figure S23. For histone acetylation/deacetylation inhibitor treatments, 80-μM curcumin (IC50 = 25 μM) (Balasubramanyam et al., 2004), 200-μM MB-3 (IC50 = 100 μM) (Aquea et al., 2017), or 3 μM TSA (IC50 = 2.4 nM) (Vigushin et al., 2001; Zhu et al., 2011) was applied to rice protoplasts cultures, and equal amounts of DMSO solvent served as negative controls.
Map-based cloning
Based on the recessive single mutation mhz10-1, individuals with the mhz10-1 phenotype in the F2 mapping population derived from a cross between mhz10-1 and the rice indica variety MH63 were selected. The mhz10 locus was first mapped to chromosome 1 within a 2.55-Mb region between markers Idl1-2.53 and Idl1-5.08 using 73 individuals, and further narrowed down to a 30-kb region between markers Idl1-3.57-AflII and Idl1-3.60 using 405 individuals (Figure 1C; Supplemental Table S2). Candidate genes in this region were sequenced, and mutations in mhz10 allelic mutants were confirmed by PCR-based dCAPS and immunoblot analyses (Figure 1, E and F; Supplemental Table S2).
Plasmid construction
To generate OsTAR2-complementation constructs, an 8.2-kb genomic DNA of OsTAR2 (2,969 bp of sequence upstream and 5,216 bp of sequence downstream from the start codon) was cloned into EcoRI/BamHI-digested pCAMBIA2300 vector. To construct OsTAR2pro:GUS, a 2,969-bp promoter fragment was cloned into PstI/XbaI-digested pCAMBIA2300-35S-GUS vector to replace the original CaMV 35S promoter. To construct OsTAR2pro:LUC, the promoter fragment was cloned into a premodified pEASY-LUC vector. The OsTAR2 coding sequence was cloned into KpnI/BamHI-digested pCAMBIA2300-35S vector to generate the OsTAR2-OE construct for OsTAR2 overexpression. For the construction of OsEIL1-Flag-OE, the OsEIL1 coding sequence fused to a Flag-tag sequence by overlapping PCR was integrated into BamHI/XbaI-linearized pCAMBIA2300-35S vector with a seamless assembly cloning kit (Clone Smarter, C5891). The coding sequences of OsIAA1/9/21/31 were cloned into pCAMBIA1300-OsACTINpro vector for overexpression. To construct OsEIL1-nYFP-FlagOE, OsARFs-nYFP-FlagOE, OsGCN5-nYFP-FlagOE and OsIAAs-cYFP-MycOE, the two vectors pCAMBIA2300-35S-nYFP-Flag and pCAMBIA2300-35S-cYFP-Myc were modified from pCAMBIA2300-35S, and then the coding sequences of the corresponding genes were introduced with a seamless assembly cloning kit. All primers used for cloning are listed in Supplemental Table S2.
IAA level measurements
About 2.5-cm apical part of roots from 2-DAG etiolated seedlings of WT and mhz10-1/2 with or without 8 h of ethylene treatment were collected for free IAA measurement. One biological replicate refers to an independent plant sampling. Three biological replicates were performed, and ∼200-mg powder (fresh weight) in each replicate was used for IAA extraction, purification, and measurements as previously described (Fu et al., 2012).
Gene expression assay
Total RNA was isolated using TRIzol reagent (Ambion, 15596018), and first-strand cDNAs were synthesized using a Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1642). Quantitative PCR (qPCR) assays were performed with three biological replicates using relative quantification (TransGen, AQ131). The expression of OsUBIQUITIN (OsUBQ, Os05g06770) served as a normalization control. qPCR primers are listed in Supplemental Table S2. For RNA-seq analysis, three biological replicates were performed. Total RNA was extracted from root tips, and mRNAs were isolated for preparing RNA sequencing libraries. An Illumina Nona 6000 platform was used for sequencing. After removing adaptors and low-quality reads, the software HISAT2 (Kim et al., 2019) was employed to build index and align paired-end reads to reference rice genome (O. sativa version 7.0 downloaded from Phytozome [https://phytozome-next.jgi.doe.gov/]). SAMtools (Li et al., 2009) was used to sort and convert the mapping results. Then, HTseq-count (Anders et al., 2015) was used to count the number of reads mapped to the reference genome. Differentially expressed genes were identified with the R package “edgeR” (version 3.12) (Robinson et al., 2010) with |Log2(fold-change)| ≥ 1 and q-value < 0.5. A biological replicate in RT-qPCR or RNA-seq refers to the RNA extracted from an independent plant sampling.
GUS staining assay and activity measurement
One-DAG seedlings were treated with 10 ppm ethylene for 8 h or with 0.1-μM NAA for 4 h, and then GUS staining and activity assay were performed as previously described (Ma et al., 2014; Huang et al., 2020). For GUS activity assays, root proteins were extracted with GUS extraction buffer (100-mM phosphate-buffered saline [PBS], pH 7.0, 10-mM Na2·EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] SDS, and 10-mM β-mercaptoethanol). Total proteins were quantified using a BCA Protein Assay Kit (Thermo, 23252). Each enzymatic reaction containing 2-mM 4-MUG substrate was incubated at 37°C for 30 min. The production of 4-MU was detected using a SpectraMax Paradigm Multi-Mode Detection Platform (Molecular Devices) with fluorescence measurement at 460 nm following excitation at 365 nm. The GUS activity was calculated as 4-MU produced per microgram total soluble protein per min.
Transcriptional regulatory activity assay in N. benthamiana leaves
Nicotiana benthamiana seedlings were grown in a greenhouse at 22°C under a 15-h light/9-h dark photoperiod for 5–6 weeks before Agrobacterium (EHA105 strain)-mediated infiltration of leaves. After infiltration, plants were kept under the same growth condition for 3 days before luciferase assay. The leaf surface was sprayed with a solution containing 100-mM beetle D-luciferin potassium salt (Promega, E Madison, WI, USA, 1605) and 0.01% (v/v) Triton X-100, and then luminescence was monitored and imaged with a plant imaging system (Berthold, NightSHADE LB 985). A biological replicate refers to two transformations consisting of the control group and the experimental group on either side of the main vein of a leaf.
Transcriptional regulatory activity assay in rice protoplasts
Plasmids for the effector (5-μg pCAMBIA2300-OsEIL1-Flag per transfection, 6-μg pCAMBIA2300-OsIAAs-cYFP-Myc per transfection), reporter (firefly luciferase, 4-μg pEASY-OsTAR2pro:LUC plasmid per transfection), and normalization control (Renilla luciferase, 1-μg PTRL plasmid per transfection) were co-transfected into rice protoplasts isolated from 7-day-old etiolated seedlings as previously described by polyethylene glycol (PEG)-mediated transfection (Bart et al., 2006). For repression of OsGCN5 expression, 30-μg OsGCN5-siRNA was applied per transfection reaction (Supplemental Table S2). After a 14-h culture period at 28°C in dark, luciferase activity assays were performed using a Dual-Luciferase Reporter Assay System (Promega, E1960), and measured on a GLOMAX 20/20 Luminometer (Promega). Relative LUC activity was defined as the ratio between firefly luciferase activity and Renilla luciferase activity, and then these values were further normalized relative to the control group. A biological replicate refers to one independent transfection event in one tube. Three or four biological replicates were performed in each experiment.
Preparation of recombinant proteins
Coding sequences of OsEIL1, OsIAA1/9/21/31 were individually cloned into pGEX-6P-1 vector, and then the plasmids were individually introduced into Escherichia coli BL21 (DE3). Escherichia coli cells were grown in LB liquid medium to an OD600 = 0.6. Recombinant protein production was induced by the addition of a 0.3-mM final concentration of isopropyl-β-d-thiogalactopyranoside for 8 h at 25°C under gentle shaking. Cells were collected and lysed by ultrasonic cell crusher (NINBO SCIENTZ BIOTECHNOLOGY CO.LTD) in 1 × PBS (pH 7.4). After centrifugation at 4°C for 30 min at 20,000 g, GST-fused proteins in the supernatant were purified using glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s protocol. The protein concentration was determined using a BCA Protein Assay Kit (Thermo, 23252).
EMSA/supershift-EMSA
Unlabeled and 5′-biotinylated complementary oligonucleotide pairs (Supplemental Table S2) were annealed to generate double-stranded unlabeled competitors and biotin-labeled probes by mixing equal amounts of sense and antisense oligonucleotides, and running the following temperature ramping program on a thermocycler: 95°C for 5 min; cooled down to 75°C by 0.1°C per second and kept it at 75°C for 30 min; and then cooled down to 10°C also by 0.1°C per second. According to the manufacturer’s protocol (Thermo Fisher Scientific, 20148), each EMSA contained 10-mM Tris–HCl (pH 7.5), 50-mM KCl, 1-mM DTT, 0.5-μg poly dI·dC, and 2-nM biotin-labeled probe in a 10-μL volume. One microgram recombinant GST, or GST-OsEIL1N protein, or anti-GST antibody was added into the reaction mixture. Ten-fold or 100-fold excess of unlabeled competitors relative to the biotin-labeled probe were preincubated with the protein for 30 min at 4°C, and then added the biotin-labeled probe and anti-GST antibody for the next 30 min of incubation. For supershift assays, 2-nM biotin-labeled probe was preincubated with 1-μg GST-OsEIL1 protein in PBS buffer for 30 min on ice and incubated for the next 30 min after the addition of 3 μg GST or GST-OsIAA protein. The protein-probe mixture was separated on a 6% native polyacrylamide gel and transferred to a nylon membrane (Thermo Fisher Scientific, LC2003). After UV light, the probes on the membrane were detected using a Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, 89880).
ChIP-qPCR assay
Root tissues from etiolated seedlings were collected, and the follow-up steps were executed according to the manufacturer’s protocol (Abcam, Cambridge, UK, ab117137). A biological replicate refers to an assay from an independent seedling sampling. Three biological replicates were performed, and 3 μg antibody (anti-Flag: CST, 14793S; anti-H3K27ac: Millipore, Burlington, MA, USA, 07-360) was used in each reaction. Mouse IgG served as a mock antibody control. DNA enrichment was measured relative to the Input (10% of chromatin relative to the ChIP sample) or relative to the OsUBIQUITIN genome fragments in the background. Primers are listed in Supplemental Table S2.
Protein isolation and immunoblot assay
RIPA buffer (Sigma, R0278) containing protease inhibitor cocktail was used for total protein isolation. Lysates were centrifuged at 20,000 g for 5 min at 4°C, and the supernatant was used for immunoblot assay. Proteins were heated with reducing loading buffer at 65°C for 10 min, and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Primary antibodies (Anti-BiP: Agrisera, AS09481; Anti-Myc: Abmart, M20019; Anti-Flag: MBL, M185; Anti-GST: CST, 2624S; Anti-H3: Agrisera, AS10710; Anti-H3K27ac: Millipore, 07-360-S) were diluted by 1:5,000 in PBS buffer containing 3% (w/v) skim milk and 0.05% (v/v) Tween-20. The OsTAR2 monoclonal antibody was prepared by immunizing mice from Abmart. Secondary antibodies (Abmart, M21001, M21002) were diluted by 1:10,000 in the same milk solution. Signals were detected by the chemiluminescence method (Thermo Scientific, 34580).
BiFC assay
For interactions between OsEIL1 and OsIAA1/9/21/31, the full-length coding sequences of OsEIL1 and each of the four OsIAAs were cloned in-frame with the sequence encoding a Flag-tagged N terminus (nYFP-Flag) and Myc-tagged C terminus (cYFP-Myc) of YFP, respectively. The nucleus-localized protein OsMYB60 (Liu et al., 2018) tagged with cYFP-Myc served as a negative control. Pairs of BiFC plasmids were co-transfected into rice protoplasts by the PEG-mediated method. The protoplasts were then incubated at 28°C for 14 h in the dark. YFP fluorescence was detected using a confocal microscopy (Carl Zeiss, TIRF3& Axioimager.Z2) with 514 nm/525–565 nm excitation/emission wavelengths.
Co-IP
Constructs encoding pairs of nYFP-Flag-tagged and cYFP-Myc-tagged proteins were co-transfected in Oseil1 shoot protoplasts, and the tagged OsMYB60 here also served as the negative control. After a 14-h incubation at 28°C in the dark, the protoplasts were lysed in RIPA buffer containing protease inhibitor cocktail. Five to 10 percent of the supernatant after 20,000 g centrifugation at 4°C for 5 min served as the input sample, and the rest was used for Co-IP with anti-Myc antibody, according to the manufacturer’s instructions (Sigma, E6654). Each Co-IP reaction was incubated at 4°C for 1 h, and then the beads were washed with RIPA buffer 5 times. Eluted immunoprecipitates and the previous input samples were immunoblotted using Myc or Flag antibodies.
Yeast two-hybrid assay
A GAL4-BD/AD system was used for yeast two-hybrid (Y2H) assay, and the coding sequences were cloned into the Y2H bait vector pGBKT7 or the prey vector pGADT7 (Clontech). Y2H Gold yeast strain (Saccharomyces cerevisiae) was transformed with bait and prey plasmids according to the protocol. Yeast colonies were grown on synthetic defined (SD) medium –Trp–Leu for 3 days at 30°C, and then were spotted onto the selective medium consisting of SD medium –Trp–Leu–His–Ade or 100 μM IAA. Plates were incubated for 3–5 days at 30°C. Yeast strains harboring the original pGADT7 or pGBKT7 plasmid were used as negative controls.
Protein degradation assay in vitro
Total proteins were extracted from 2-DAG etiolated rice seedling roots in a mild extraction buffer (50-mM Tris-MES, pH 8.0, 0.5-M sucrose, 1-mM MgCl2, 10-mM EDTA, 5-mM DTT, 0.1% [v/v] NP-40). Lysates were centrifuged at 12,000 g for 5 min at 4°C, and the supernatant was collected. Recombinant GST-OsIAA proteins purified from E. coli were co-incubated with the supernatant containing 10-mM ATP at 30°C, and reactions were terminated by adding the SDS–PAGE loading buffer. Samples were analyzed by immunoblot and the abundance of GST-OsIAA was measured using ImageJ software (V1.52a, National Institutes of Health) (Schneider et al., 2012). A biological replicate refers to an independent root crude protein extraction and a subsequent reaction. Relative quantification of the degradation rate from three biological repeats was performed.
Accession numbers
AB2T (Os06g36090); CFP (Os09g02270); OsERF002 (Os06g08340); OsIAA20 (Os06g07040); OsERF063: Os09g11480); OsERF073 (Os09g11460); FBL (Os05g07720); OsTARL1 (Os01g52010); OsTARL2 (Os01g51980); AtTAA1 (At1g70560); AtTAR1 (At1g23320); AtTAR2 (At4g24670); OsEIN2 (Os07g06130); OsEIL1 (Os03g20790); OsTIR1 (Os05g05800); OsAFB2 (Os04g32460); OsPIN2 (Os06g44970); OsIAA1 (Os01g08320); OsIAA9 (Os02g56120); OsIAA21 (Os06g22870); OsIAA31 (Os12g40900); OsGCN5 (Os10g28040); OsYUC8 (Os03g06654); and OsHKT2;1 (Os06g48810).
The raw RNA-seq reads were deposited at NCBI database under accession number PRJNA639684.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes and yield-related traits of field-grown WT and mhz10 mutant plants.
Supplemental Figure S2. Dominant/recessive analysis.
Supplemental Figure S3. Amino acid sequence alignment of OsTAR2 and its homologues in rice and Arabidopsis.
Supplemental Figure S4. Field-grown plants with the introduction of OsTAR2 genomic sequences.
Supplemental Figure S5. Tests for a proper concentration of IPyA treatment.
Supplemental Figure S6. Expression of OsTAR2 and OsEIL1 in transgenic lines.
Supplemental Figure S7. Profile of OsTAR2 expression in rice as revealed by GUS staining.
Supplemental Figure S8. Expression of OsTAR2 in response to ethylene in shoots.
Supplemental Figure S9. Expression level of OsTIR1 and OsAFB2 in roots of miR393a- and miR393b-overexpression seedlings.
Supplemental Figure S10. Phenotypic analysis of mhz8/Ospin2 and its gene identification.
Supplemental Figure S11. Regulatory activity of OsARFs on OsTAR2 promoter activity compared to OsEIL1.
Supplemental Figure S12. Examination of the transcriptional regulatory activity of OsIAAs on OsEIL1-activated OsTAR2 expression.
Supplemental Figure S13. Mutation sites in the Osiaa1 Osiaa9 and Osiaa21 Osiaa31 double mutants obtained by CRISPR/Cas9.
Supplemental Figure S14. Expression level of OsIAA1/9/21/31 in transgenic seedlings.
Supplemental Figure S15. Root length of OsIAA1/9/21/31-overexpression and mutants in response to ethylene.
Supplemental Figure S16. Amino acid sequence alignment of OsIAA1/9/21/31.
Supplemental Figure S17. Interaction between OsEIL1 and EAR motif-mutated OsIAA1/9/21/31 analyzed by Co-IP.
Supplemental Figure S18. Effects of curcumin on OsIAA1/9-promoting OsEIL1-activated OsTAR2 promoter activity.
Supplemental Figure S19. Overexpression level of OsGCN5 in plants.
Supplemental Figure S20. A Osgcn5 mutant prepared by CRISPR/Cas9.
Supplemental Figure S21. Domain/motif swapping tests for promotive effects of OsIAA1/9 on OsEIL1.
Supplemental Figure S22. Expression of OsYUC8 and OsHKT2;1 in Oseil1 and mhz10-1 root tips in response to ethylene.
Supplemental Figure S23. Regulation of Aux pathway on the stability of OsEIL1 in rice roots.
Supplemental Figure S24. Parts, assembly, and operation of the system for concomitant treatments of ethylene and soluble reagents.
Supplemental Table S1. OsARFs and OsIAAs expressed in root tips according to transcriptome analysis.
Supplemental Table S2. Primers/oligos used in this study.
Supplemental Data Set 1. RNA-seq analysis of mhz10 and WT.
Supplemental Data Set 2. Source files with statistical analysis.
Supplementary Material
Acknowledgments
We thank Prof. Mu-Yuan Zhu (Zhejiang University) and Prof. Ming-Yong Zhang (South China Botanical Garden, Chinese Academy of Sciences, CAS) for providing the seeds of OsmiR393a/b-OE lines, and thank Prof. Jia-Yang Li (Institute of Genetics and Developmental Biology, CAS) for providing the DR5pro:GUS line.
Funding
This work was supported by National Natural Science Foundation of China (32000220, 31530004, 31670274, and 31600980), and the State Key Lab of Plant Genomics.
Conflict of interest statement. None declared.
Contributor Information
Yang Zhou, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Biao Ma, College of Agriculture, South China Agricultural University, Guangzhou 510642, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou 510642, China.
Jian-Jun Tao, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Cui-Cui Yin, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Yang Hu, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yi-Hua Huang, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Wei Wei, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Pei-Yong Xin, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Jin-Fang Chu, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Wan-Ke Zhang, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Shou-Yi Chen, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Jin-Song Zhang, State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Y.Z., S.Y.C., and J.S.Z. designed the research and analyzed data. Y.Z., M.B., J.J.T., C.C.Y., Y.H.H., W.W., P.Y.X., J.F.C., and W.K.Z. performed research. B.M. and J.J.T. screened rice mutants. Y.H. and W.K.Z. performed bioinformatics analysis. Y.Z. and J.S.Z. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy in the Instructions for Authors (https://academic.oup.com/plcell) is: Jin-Song Zhang (jszhang@genetics.ac.cn).
References
- Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012) A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol Plant 146: 487–499 [DOI] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An FY, Zhao QO, Ji YS, Li WY, Jiang ZQ, Yu XC, Zhang C, Han Y, He WR, Liu YD, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquea F, Timmermann T, Herrera-Vasquez A (2017) Chemical inhibition of the histone acetyltransferase activity in Arabidopsis thaliana. Biochem Bioph Res Co 483: 664–668 [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK (2004) Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem 279: 51163–51171 [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian HW, Xie YK, Guo F, Han N, Ma SY, Zeng ZH, Wang JH, Yang YN, Zhu MY (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196: 149–161 [DOI] [PubMed] [Google Scholar]
- Biel M, Kretsovali A, Karatzali E, Papamatheakis J, Giannis A (2004) Design, synthesis, and biological evaluation of a small-molecule inhibitor of the histone acetyltransferase Gcn5. Angew Chem Int Edit 43: 3974–3976 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chen C, Li CL, Wang Y, Renaud J, Tian G, Kambhampati S, Saatian B, Nguyen V, Hannoufa A, Marsolais F, et al. (2017) Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3: 814–824 [DOI] [PubMed] [Google Scholar]
- Chen H, Ma B, Zhou Y, He SJ, Tang SY, Lu X, Xie Q, Chen SY, Zhang JS (2018) E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc Natl Acad Sci USA 115: 4513–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang XX, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XH, Mashiguchi K, Chen QG, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao YD (2013) The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288: 1448–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Feng Y, Xu P, Li BS, Li PP, Wen X, An FY, Gong Y, Xin Y, Zhu ZQ, Wang YC, et al. (2017) Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc Natl Acad Sci USA 114: 13834–13839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JH, Chu JF, Sun XH, Wang JD, Yan CY (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28: 1081–1087 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ (2015) The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Ye WW, Shan JX, Lin HX (2020) Tillering and small grain 1 dominates the tryptophan aminotransferase family required for local auxin biosynthesis in rice. J Integr Plant Biol 62: 581–600 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H (2012) Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem Biol 7: 590–598 [DOI] [PubMed] [Google Scholar]
- He WR, Brumos J, Li HJ, Ji YS, Ke M, Gong XQ, Zeng QL, Li WY, Zhang XY, An FY, et al. (2011) A small-molecule screen identifies L-Kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Dong Z, Guo P, Zhang X, Qiu Y, Li B, Wang Y, Guo H (2020) Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 32: 612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics-Us 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Ma MD, Feng Y, Li HJ, Wang YC, Ma YT, Li MZ, An FY, Guo HW (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683 [DOI] [PubMed] [Google Scholar]
- Liu LC, Tong HN, Xiao YH, Che RH, Xu F, Hu B, Liang CZ, Chu JF, Li JY, Chu CC (2015) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li DY, Zhang DL, Yin DD, Zhao Y, Ji CJ, Zhao XF, Li XB, He Q, Chen RS, et al. (2018) A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol 218: 774–788 [DOI] [PubMed] [Google Scholar]
- Liu XQ, Liu RL, Li Y, Shen X, Zhong SW, Shi H (2017) EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29: 3051–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Yin CC, He SJ, Lu X, Zhang WK, Lu TG, Chen SY, Zhang JS (2014) Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet 10: e1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Zhou Y, Chen H, He SJ, Huang YH, Zhao H, Lu X, Zhang WK, Pang JH, Chen SY, et al. (2018) Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proc Natl Acad Sci USA 115: 2520–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, He SJ, Duan KX, Yin CC, Chen H, Yang C, Xiong Q, Song QX, Lu X, Chen HW, et al. (2013) Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol Plant 6: 1830–1848 [DOI] [PubMed] [Google Scholar]
- Mai A, Rotili D, Tarantino D, Ornaghi P, Tosi F, Vicidomini C, Sbardella G, Nebbioso A, Miceli M, Altucci L, et al. (2006) Small-molecule inhibitors of histone acetyltransferase activity: Identification and biological properties. J Med Chem 49: 6897–6907 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu QW, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM (2015) Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697 [DOI] [PubMed] [Google Scholar]
- Morgan KE, Zarembinski TI, Theologis A, Abel S (1999) Biochemical characterization of recombinant polypeptides corresponding to the predicted βαα fold in Aux/IAA proteins. Febs Lett 454: 283–287 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366 [DOI] [PubMed] [Google Scholar]
- Olsen GM, Mirza JI, Maher EP, Iversen TH (1984) Ultrastructure and movements of cell organelles in the root cap of agravitropic mutants and normal seedlings of Arabidopsis thaliana. Physiol Plant 60: 523–531 [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen ZX, Huang SSC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Zhang ZJ, Wang J, Chen XB, Wei PC, Huang RF (2017) The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet 13: e1006955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QHG, Chang CR, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, DoleZal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, Coombes RC (2001) Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res 7: 971–976 [PubMed] [Google Scholar]
- Wang C, Dai S, Zhang ZL, Lao W, Wang R, Meng X, Zhou X (2021) Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J Integr Plant Biol 63: 828–833 [DOI] [PubMed] [Google Scholar]
- Wang WY, Esch JJ, Shiu SH, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB (2006) Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18: 3429–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen XL, Mashiguchi K, Zheng ZY, Dai XH, Cheng YF, Kasahara H, Kamiya Y, Chory J, Zhao YD (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]
- Xia KF, Wang R, Ou XJ, Fang ZM, Tian CG, Duan J, Wang YQ, Zhang MY (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7: 364–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ma B, Lu X, Huang YH, He SJ, Yang C, Yin CC, Zhao H, Zhou Y, Zhang WK, et al. (2017) Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell 29: 1053–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Terada T, et al. (2005) Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J Mol Biol 348: 253–264 [DOI] [PubMed] [Google Scholar]
- Yang C, Ma B, He SJ, Xiong Q, Duan KX, Yin CC, Chen H, Lu X, Chen SY, Zhang JS (2015) MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol 169: 148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin CC, Ma B, Collinge DP, Pogson BJ, He SJ, Xiong Q, Duan KX, Chen H, Yang C, Lu X, et al. (2015) Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell 27: 1061–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Ito M, Sumikura T, Nakayama A, Nishimura T, Kitano H, Yamaguchi I, Koshiba T, Hibara K, Nagato Y, et al. (2014) The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J 78: 927–936 [DOI] [PubMed] [Google Scholar]
- Yu X, Xu Y, Yan S (2021) Salicylic acid and ethylene coordinately promote leaf senescence. J Integr Plant Biol 63: 823–827 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LK, Ko EE, Shao K, Qiao H (2018) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qi B, Wang LK, Zhao B, Rode S, Riggan ND, Ecker JR, Qiao H (2016) EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat Commun 7: 13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LK, Qi B, Zhao B, Ko EE, Riggan ND, Chin KV, Qiao H (2017) EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc Natl Acad Sci USA 114: 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yin CC, Ma B, Chen SY, Zhang JS (2021) Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J Integr Plant Biol 63: 102–125 [DOI] [PubMed] [Google Scholar]
- Zhao H, Ma B, Duan KX, Li XK, Lu X, Yin CC, Tao JJ, Zhang WK, Xin PY, Man Lam S, et al. (2020a) The GDSL lipase MHZ11 modulates ethylene signaling in rice roots. Plant Cell 32: 1626–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Duan KX, Ma B, Yin CC, Hu Y, Tao JJ, Huang YH, Cao WQ, Chen H, Yang C, et al. (2020b) Histidine kinase MHZ1/OsHK1 interacts with ethylene receptors to regulate root growth in rice. Nat Commun 11: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YD (2018) Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Xiong Q, Yin CC, Ma B, Chen SY, Zhang JS (2020) Ethylene biosynthesis, signaling, and crosstalk with other hormones in rice. Small Methods 4: 1900278 [Google Scholar]
- Zhu ZQ, An FY, Feng Y, Li PP, Xue L, Mu A, Jiang ZQ, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.