Abstract
Cold tolerance at the booting (CTB) stage is a major factor limiting rice (Oryza sativa L.) productivity and geographical distribution. A few cold-tolerance genes have been identified, but they either need to be overexpressed to result in CTB or cause yield penalties, limiting their utility for breeding. Here, we characterize the function of the cold-induced transcription factor WRKY53 in rice. The wrky53 mutant displays increased CTB, as determined by higher seed setting. Low temperature is associated with lower gibberellin (GA) contents in anthers in the wild type but not in the wrky53 mutant, which accumulates slightly more GA in its anthers. WRKY53 directly binds to the promoters of GA biosynthesis genes and transcriptionally represses them in anthers. In addition, we uncover a possible mechanism by which GA regulates male fertility: SLENDER RICE1 (SLR1) interacts with and sequesters two critical transcription factors for tapetum development, UNDEVELOPED TAPETUM1 (UDT1), and TAPETUM DEGENERATION RETARDATION (TDR), and GA alleviates the sequestration by SLR1, thus allowing UDT1 and TDR to activate transcription. Finally, knocking out WRKY53 in diverse varieties increases cold tolerance without a yield penalty, leading to a higher yield in rice subjected to cold stress. Together, these findings provide a target for improving CTB in rice.
WRKY53 reduces gibberellin levels and seed setting under cold stress at the booting stage; whereas wrky53 results in cold tolerance without a yield penalty, making it a good target for rice breeding.
IN A NUTSHELL.
Background: Rice (Oryza sativa L.) is a staple food for over half of the global population. Cold stress at the booting stage leads to serious yield losses and has become a major limiting factor for rice production in regions of higher altitude and latitude. Therefore, isolating cold tolerance genes to facilitate breeding cultivars with higher cold tolerance at the booting (CTB) stage remains an urgent concern. Rice cold tolerance is a complex trait that is controlled by multiple loci. Due to the difficulties in identifying CTB phenotypes, only a few genes regulating CTB have been functionally characterized thus far.
Question: We aimed to characterize a CTB-related gene, WRKY53, and investigate its underlying mechanism, then use it to breed rice varieties with greater CTB.
Findings: We identified the cold-induced WRKY transcription factor WRKY53 and found that WRKY53 negatively regulates rice CTB. WRKY53 fine-tuning gibberellic acid (GA) levels in anthers via directly transcriptionally repressing GA biosynthesis genes in anthers. Moreover, we demonstrate that the possible mechanism of GA regulating male fertility in which a DELLA protein interacts with two critical transcription factors for tapetum development, and GA promotes the degradation of this DELLA protein, allowing the transcription factors to activate downstream genes. Importantly, knocking out WRKY53 in diverse varieties increases cold tolerance without a yield penalty, leading to a higher yield in rice subjected to cold stress. Therefore, WRKY53 is a valuable target for cold-tolerance breeding.
Next steps: WRKY53 negatively regulates resistance to Xanthomonas oryzae pv. oryzae sheath blight, striped stem borer, and CTB in rice. We are interested in evaluating the practical value of wrky53 mutants by assessing their yield under multiple conditions, including pathogen infection, insect pests, and low temperature or other stresses in the field.
Introduction
Rice (Oryza sativa L.) is an important food crop; cultivated rice in Asia consists of two major subspecies, indica (O. sativa ssp. indica) and japonica (O. sativa ssp. japonica) (Kovach et al., 2007; Sang and Ge, 2007). Typical japonica cultivars belong to the temperate japonica group and are grown at lower ambient temperatures typically found at higher latitudes or altitudes. By contrast, indica and tropical japonica cultivars can only be planted at higher ambient temperatures (Huang et al., 2012). Accordingly, after long-term selection, temperate japonica cultivars are more tolerant to cold exposure than indica and tropical japonica cultivars (Ma et al., 2015b; Liu et al., 2018). Indeed, in the northern part of the japonica growing range, cold stress at the booting stage has become a major limiting factor for rice production in regions at higher altitudes and latitudes (Zhang et al., 2017; Guo et al., 2020). It is estimated that low temperature (LT) in the fall is associated with a loss in rice production of around 3–5 million tons annually in China (Zhu et al., 2015). Therefore, isolating cold-tolerance genes to facilitate the breeding of cultivars with higher cold tolerance remains an urgent concern.
Rice cold tolerance is a complex trait that is controlled by multiple loci. Cold tolerance at different growth stages is regulated by distinct sets of genes and diverse mechanisms. Although many quantitative trait loci (QTL) have been identified that affect cold tolerance at diverse stages, only a few genes have been functionally characterized to date. For instance, the QTL for LT germinability on chromosome 3 (qLTG3-1) confers cold tolerance during seed germination (Fujino et al., 2008). Similarly, CHILLING-TOLERANCE DIVERGENCE1 (COLD1), HAN1 (which means “chilling” in Chinese), and the basic leucine zipper transcription factor bZIP73 positively regulates cold tolerance during the vegetative stage (Ma et al., 2015b; Liu et al., 2018; Mao et al., 2019). Evaluating cold tolerance at the booting (CTB) stage is difficult, as scoring requires the dissection of the main stem to observe the developing panicle. Therefore, although multiple QTL have been identified for this growth stage, only a few genes have been cloned (Saito et al., 2001; Andaya and Mackill, 2003; Saito et al., 2004; Kuroki et al., 2007; Zhou et al., 2010). For example, CTB1 is likely an F-box protein that controls CTB stage, but the underlying mechanism remains elusive (Saito et al., 2004, 2010). A naturally occurring allele in the CTB4a promoter in japonica rice enhanced CTB4a expression, leading to higher ATP contents, and improved cold tolerance of japonica rice (Zhang et al., 2017). bZIP73 is a highly diverged gene between japonica and indica that controls cold tolerance at both the seedling and booting stages by regulating abscisic acid (ABA) and reactive oxygen species (ROS) levels in plant tissues (Liu et al., 2018, 2019). CTB4a and bZIP73 have been the target of both natural selection for local adaptation and artificial selection for domestication, as the more cold-tolerant haplotypes contribute to adaptation to the colder climates north of the temperate japonica growth region. However, some varieties that carry the functional single-nucleotide polymorphisms (SNPs) in CTB4a and bZIP73 are still sensitive to cold at the booting stage (Zhang et al., 2017; Liu et al., 2019; Guo et al., 2020). Additionally, CTB4a and bZIP73 positively regulate CTB but need to be overexpressed to enhance CTB, which limits their usefulness in rice breeding (Zhang et al., 2017; Liu et al., 2019). LT TOLERANCE1 (LTT1) negatively regulates CTB stage by maintaining ROS homeostasis, but the ltt1 mutation is accompanied by a lesion mimic phenotype that affects its potential application in rice breeding programs (Xu et al., 2020). The identification of new CTB-related genes with better practical value would therefore be advantageous to the breeding rice varieties with greater CTB stage.
The phytohormone gibberellin (GA) is an essential and natural regulator of growth and development in rice (Olszewski et al., 2002; Sun and Gubler, 2004; Chen et al., 2019). The GA biosynthetic pathway consists of at least 13 genes in rice, encoding the enzymes that catalyze 4 enzymatic steps (Chen et al., 2019). Loss of function in 11 of these 13 genes results in sterility or a lower seed setting rate, indicating that reduced GA contents severely impair fertility and seed setting in rice (Chen et al., 2019). In addition, overexpression of GA 20-OXIDASE1 (GA20ox1) also results in lower seed setting rates (Oikawa et al., 2004), suggesting that normal GA levels are essential for normal seed setting, while higher or lower GA levels both affect grain yield. Exposure to LTs at the booting stage leads to a drastic reduction in mature pollen and slows down the growth of anthers, resulting in a severe decline in seed setting (Saito et al., 2001, 2004). Notably, mutants defective in GA signaling or biosynthesis display aborted pollen development with an abnormal enlargement of tapetal cells, which is very similar to the phenotypes observed when plants are subjected to LT at the booting stage (Aya et al., 2009; Oda et al., 2010). These observations were confirmed by the fact that GA contents decrease in cold-treated anthers and that the exogenous application of GA can effectively rescue cold-induced male sterility (Sakata et al., 2014). These results indicate that GA might play a positive role in rice CTB (Sakata et al., 2014). However, little is known about the underlying mechanism or how to enhance CTB by manipulating rice GA levels during breeding. In addition, many genes have been shown to regulate rice male sterility, among them, TAPETUM DEGENERATION RETARDATION (TDR) and UNDEVELOPED TAPETUM1 (UDT1) encode two critical basic helix and helix transcriptional factors controlling tapetal cell development and degradation (Jung et al., 2005; Zhang et al., 2008). As described above, both GA and cold stress at the booting stage regulate male fertility via affecting tapetal cell development (Aya et al., 2009; Oda et al., 2010; Sakata et al., 2014). Whether cold stress at the booting stage and GA signaling regulate male fertility through modulating UDT1 or TDR activity is an open question worth exploring.
In this study, we characterized the cold-induced WRKY transcription factor gene WRKY53 and determined that its loss of function ameliorated CTB. WRKY53 negatively regulates the GA content of anthers by repressing the expression of GA biosynthesis genes. The DELLA protein SLENDER RICE1 (SLR1) interacts with UDT1 and TDR, and GA releases the sequestration of SLR1, which allows UDT1 and TDR to carry out their functions as transcription factors. Importantly, knocking out WRKY53 in multiple genetic backgrounds by genome editing showed that the cold tolerance of several elite japonica cultivars can be significantly improved without compromising yield, demonstrating that WRKY53 is a valuable target for cold-tolerance breeding.
Results
The wrky53 mutant displays higher CTB stage
CTB stage decreases the seed setting rate, which severely threatens rice production, especially at high latitude regions (Zhu et al., 2015; Liu et al., 2019; Guo et al., 2020; Xu et al., 2020). To identify cold tolerance-related genes at the booting stage, we performed a transcriptome deep sequencing (RNA-seq) analysis of young rice panicles exposed to the LT of 15°C at the booting stage. We thus identified around 200 transcription factor genes as being either upregulated or downregulated by LT treatment. We systematically overexpressed 87 LT upregulated transcription factor genes in the japonica cultivar “Longjing11” (LJ11) and used the resulting transgenic plants to assess their LT stress response. Plants overexpressing WRKY53 (WRKY53-OE) and grown under normal conditions (NCs) exhibited a lower seed setting rate relative to the wild type. Importantly, this lower seed setting rate was comparable to that seen in wild-type plants subjected to cold stress (Supplemental Figure S1, A and B), suggesting that WRKY53 might be involved in CTB. We therefore selected WRKY53 for further characterization in this study.
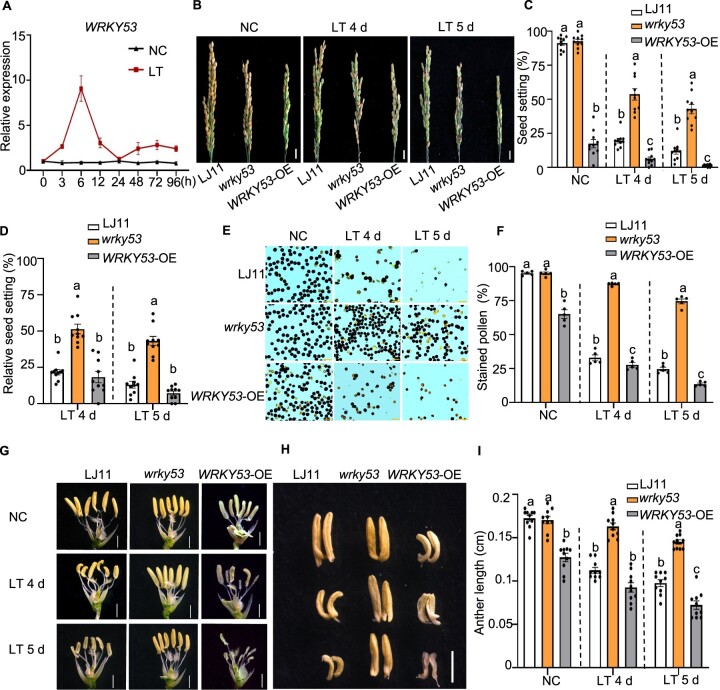
We measured the expression levels of WRKY53 during the booting stage and in response to LT. WRKY53 expression was highly induced within hours of LT treatment and then declined. Notably, WRKY53 displayed higher expression levels throughout the LT treatment compared with NC (Figure 1A). We used the WRKY53-OE lines and wrky53 mutants generated by genome editing via clustered regularly interspaced short palindromic repeats (CRISPRs)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing from our previous study (Tian et al., 2017; Supplemental Figure S1, C and D). Under NC and LT, the seed setting rate of WRKY53-OE was significantly lower than that of the wild-type LJ11 or the wrky53-2 mutant (Figure 1, B and C). However, after 4 or 5 days of LT at 15°C, the wrky53 mutant (wrky53-2 unless specified otherwise thereafter) exhibited much higher relative seed setting rates compared with LJ11 and WRKY53-OE plants (Figure 1, B–D). Indeed, the relative seed setting rate of the wrky53 mutant was 51.2% and 42.6% after 4 and 5 days of exposure to 15°C, respectively, while that of LJ11 only reached 21.7% (after 4 days) and 12.9% (after 5 days) (Figure 1D). To validate the higher seed setting rate of wrky53 mutants under LTs, we performed seven independent replications using treatments of varying lengths and at diverse temperatures over five consecutive years from 2017 to 2021. We established that, under LT, the seed setting rate of the wrky53 mutant is reproducibly higher than that of LJ11 (Supplemental Figure S2, A–G). In addition, the wrky53-1 mutant (another wrky53 mutant allele) (Tian et al., 2017) also displayed higher seed setting rates than LJ11 under LT (Supplemental Figure S3, A–C). Collectively, these results indicate that WRKY53 negatively regulates rice CTB.
Figure 1.
WRKY53 negatively regulates CTB stage in rice. A, Time course of WRKY53 relative expression levels in panicles of the cultivar LJ11 exposed to a 15°C cold treatment during the booting stage. The expression level on day 0 was set to 1. Data are shown as means ± se (n = 3). B–D, Representative images of panicles (B), seed setting rate (C), and relative seed setting rate (D) of LJ11, wrky53, and WRKY53-OE plants grown under NCs and after 4 or 5 days of LT. Scale bar, 1 cm. Data are shown as means ± se (n = 10). E and F, Pollen fertility (E) and stained pollen ratio (F) of LJ11, wrky53, and WRKY53-OE pollen from plants grown under NC or after 4 and 5 days of LT. Scale bar, 20 μm. Data are shown as means ± se (n = 5). G, Gross morphology of anthers from LJ11, wrky53, and WRKY53-OE plants grown under NC or after 4 or 5 days of LT. Scale bar, 1 mm. H and I, Anthers (H) and anther length (I) in LJ11, wrky53, and WRKY53-OE plants grown under NC or after 4 or 5 days of LT. Scale bar, 1 mm. Data are shown as means ± se (n = 10). Dotted lines separate NC and LT treatments, and the comparisons are among different genotypes in the same treatment. Each dot represents the result from one biological replicate, error bars indicate means ± se. Significant differences are indicated by different lowercase letters (P < 0.05, one-way ANOVA with Tukey’s significant difference test).
WRKY53 regulates pollen fertility and tapetum development
LT at the booting stage results in anther and pollen injuries that lead to anther growth arrest and pollen sterility (Saito et al., 2004; Sakata et al., 2014). In addition, anther length was proposed to be significantly and positively correlated with CTB, thus providing a good proxy for measuring CTB across rice varieties (Saito et al., 2001). To investigate how WRKY53 negatively regulates rice CTB, we quantified pollen fertility and examined anther morphology of WRKY53-OE, wrky53, and LJ11 plants. Under NC, the pollen fertility of WRKY53-OE plants was significantly lower than that of wrky53 or LJ11 plants. Under LT, the pollen fertility of the wrky53 mutant was also much higher than that of either LJ11 or WRKY53-OE plants (Figure 1, E and F), which was consistent with the higher seed setting rate seen in wrky53 plants (Figure 1, B and C). In addition, under NC, the WRKY53-OE anthers were shorter, shriveled, and pale, compared with the normal anthers of LJ11 and wrky53 plants. After exposure to LT for 4–5 days, LJ11 plants had short and shrunken anthers that turned white, which was consistent with their lower seed setting rate under these conditions (Figure 1, G–I). By contrast, the anthers of wrky53 plants remained bright yellow long after 5 days of treatment at 15°C (Figure 1, G–I).
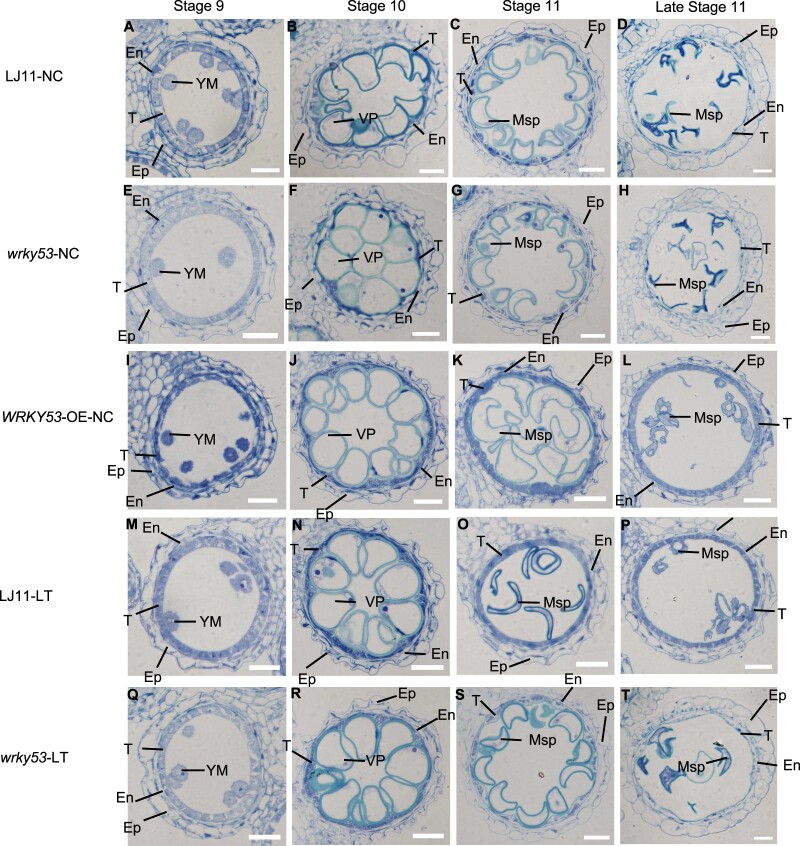
Tapetum cells can provide the necessary nutrients for pollen development, and premature or delayed degradation of the tapetum is often associated with male sterility (Jung et al., 2005; Li et al., 2006; Zhang et al., 2008). LT at the booting stage results in delayed tapetum degeneration, with cold tolerance correlating with the degree of tapetum degeneration (Oda et al., 2010). Given that WRKY53-OE plants displayed decreased pollen fertility, a phenotype similar to LJ11 plants subjected to LT, and that normal tapetum development is required for pollen fertility, we next examined tapetum development and tapetum cell layers of WRKY53-OE, wrky53, and LJ11 plants under NC and LT. Anther development is divided into 14 stages, and tapetum degradation takes place during stages 9–11, which are the young microspore (YM) stage (stage 9), vacuolated pollen (VP) stage (stage 10), and pollen mitosis stage (stage 11 and late stage 11) (Zhang and Wilson, 2009). Therefore, we observed anther development from stages 9 to 11. Under NC, at stage 9, the microspores in the LJ11, wrky53 mutant, and WRKY53-OE plants were released into the locules and the tapetal layer concentrated faster and showed no difference between genotypes (Figure 2, A, E, and I). At stage 10, microspores of LJ11 and wrky53 mutant plants expanded and vacuolized, and the tapetal layer degenerated and became thinner (Figure 2, B and F). By contrast, the tapetal layer of WRKY53-OE plants was slightly thicker than that of LJ11 and wrky53 mutant plants (Figure 2, B, F, and J). At stage 11 and late stage 11, in LJ11 and wrky53 mutant plants, vacuoles contracted; the originally round microspores changed into crescent-shaped microspores; tapetal cells continued to degenerate and almost disappeared (Figure 2, C, D, G, and H). By contrast, the tapetal cells in WRKY53-OE plants degraded very slowly and remained clearly visible; they were even thicker than those of LJ11 and wrky53 mutant plants (Figure 2, C, D, G, H, K, and L).
Figure 2.
Histological analysis of anther development in LJ11, wrky53 mutant, and WRKY53-OE plants under NCs or LT treatment. Anther development in wild-type (LJ11), the wrky53 mutant, and WRKY53-OE plants at stage 9, stage 10, stage 11, and late stage 11. Ep, epidermal cell layer; En, endothelial cell layer; VP, vacuole pollen; Msp, microspore. Bars = 20 μm. A, E, I, M, and Q, Transverse sections of anthers at stage 9 for wild-type (A, M), wrky53 (E, Q), and WRKY53-OE (I) plants under NC or LT treatment. B, F, J, N, and R, Transverse sections of anthers at stage 10 for wild-type (B, N), wrky53 (F, R), and WRKY53-OE (J) plants under NC or LT treatment. C, D, G, H, K, L, N, O, P, S, and T, Transverse sections of anthers at stage 11 and late stage 11 for wild-type (C, D, O, P), wrky53 (G, H, S, T), and WRKY53-OE (K, L) plants under NC or LT treatment.
Under LT, at stage 9, the microspores and the tapetal layer in LJ11 and wrky53 mutant plants showed little difference (Figure 2, M and Q). However, at stage 10, the tapetum of LJ11 plants became thicker compared with that of the wrky53 mutant (Figure 2, N and R). At stage 11 and late stage 11, the tapetum of LJ11 plants did not degenerate timely (Figure 2, O and P). However, the tapetum of wrky53 mutant plants degenerated normally, suggesting that the tapetum of wrky53 mutant plants is not damaged by LT, which was consistent with their higher CTB (Figures 1, B and C, and 2, S and T; Supplemental Figures S2 and S3). Collectively, these results showed that WRKY53-OE tapetum does not degrade normally under NC, whereas wrky53 mutant tapetum degenerates normally under LT, suggesting that WRKY53 negatively regulates CTB via regulating tapetum development.
Together, these results indicate that the healthy anthers and higher fertile pollen ratio in the wrky53 mutant under cold stress might contribute to the stronger CTB observed in this genotype.
WRKY53 negatively regulates GA content in anthers
GA biosynthesis and GA signaling mutants were reported to be hypersensitive to LT at the booting stage, and exogenous application of GA can improve rice CTB (Sakata et al., 2014). We conducted this assay on the LJ11 variety and determined that irrigation of plants with 5 µM GA3 slightly decreased the seed setting rate of plants grown under NC compared with plants irrigated with water (Supplemental Figure S4, A–E). However, the same GA3 application significantly and consistently increased the seed setting rate of LJ11 and WRKY53-OE plants exposed to LT (Supplemental Figure S4, A–G), thus validating the previous finding that GA positively regulates CTB in rice (Sakata et al., 2014).
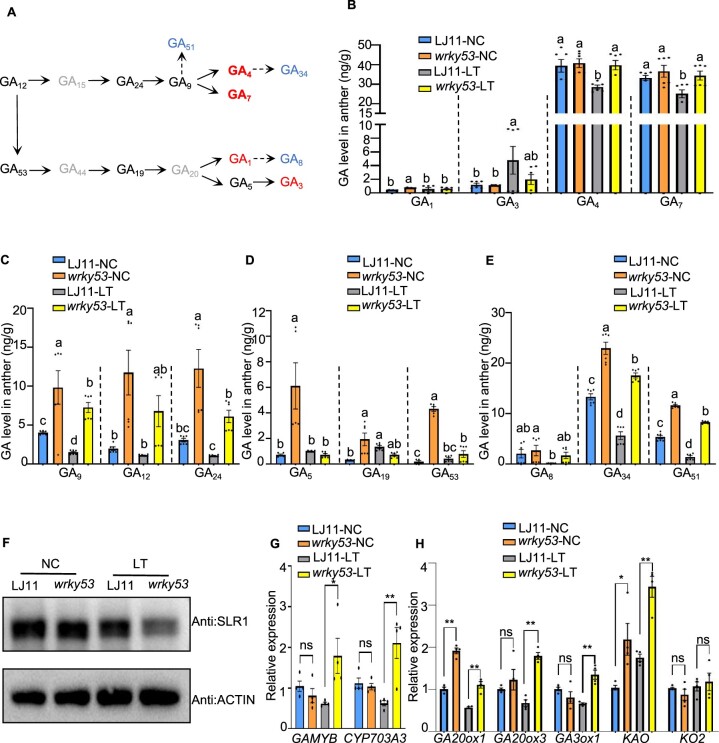
To investigate the physiological mechanism underlying the increased cold tolerance phenotype of the wrky53 mutant, we measured the contents of diverse GA components in anthers from LJ11 and wrky53 plants grown under NC or LT (Figure 3A). GA1, GA3, GA4, and GA7 are bioactive GAs, with GA4 and GA7 being the predominant GAs in reproductive organs, while GA1 and GA3 are the main GAs in vegetative organs (Masatoshi et al., 1991; Hirano et al., 2008; Yamaguchi, 2008; Hedden, 2012; Sakata et al., 2014; Hedden, 2020). In agreement with previous studies, we also observed that the GA4 and GA7 contents in anthers are around 20–100 times higher than the GA1 and GA3 contents regardless of the temperature (Figure 3B). Importantly, under NC, the wrky53 mutant and LJ11 plants accumulated similar levels of GA4 and GA7 (Figure 3B). Following a LT treatment consisting of 15 days at 18°C, the levels of GA4 and GA7 significantly decreased in LJ11 plants relative to those under NC; however, GA4 and GA7 levels did not decline in the wrky53 mutant (Figure 3B). Consistent with these results, the immediate precursors of GA4 and GA7, including GA12, GA24, and GA9, accumulated to higher levels in the wrky53 mutant than in LJ11 plants under both NC and LT (Figure 3, A and C). By contrast, LJ11 mutant plants had similar GA1 and GA3 contents to wrky53 mutants, and the GA1 and GA3 contents of LJ11 and wrky53 mutant plants did not significantly decrease under LT compared with NC (Figure 3B). Levels of the immediate GA1 and GA3 precursors, including GA53, GA19, and GA5, were higher in the wrky53 mutant than in LJ11 plants under NC but were comparable between LJ11 and wrky53 mutant plants under LT (Figure 3, A and D). In addition, the inactivate GAs, GA34, and GA51 showed higher levels in the wrky53 mutant than in LJ11 plants (Figure 3, A and E). Considering that GA4 and GA7 are the dominant bioactive GAs, and that they accumulated to higher levels in the wrky53 mutant than in LJ11 plants under LT compared with NC (Figure 3, A and B), we speculated that LT-treated wrky53 mutant plants might accumulate more bioactive GAs than LT-treated LJ11 plants. To test this hypothesis, we examined the abundance of the rice DELLA protein SLR1 in panicles, as SLR1 degradation is promoted by GA (Ueguchi-Tanaka et al., 2008). We determined that SLR1 protein levels are comparable between LJ11 and wrky53 mutant plants under NC; however, under LT, the wrky53 mutant had markedly lower SLR1 levels than LJ11 plants (Figure 3F and Supplemental Figure S5, A–C). We also examined the expression levels of the GA-regulated MYB domain transcription factor gene GAMYB and its target gene CYTOCHROME P450 703A3 (CYP703A3), which is induced by GA and specifically expressed in anthers (Aya et al., 2009). We observed that GAMYB and CYP703A3 transcript level plants are similar in LJ11 and the wrky53 mutant under NC; however, under LT, the expression levels of GAMYB and CYP703A3 in the wrky53 mutant were significantly higher than in LJ11 plants (Figure 3G). Collectively, these results suggest that the wrky53 mutant accumulates more bioactive GAs under LT than LJ11 plants, which also indicates that WRKY53 might negatively regulate rice CTB by modulating GA levels in anthers.
Figure 3.
WRKY53 negatively regulates GA content and GA biosynthesis gene expression in anthers. A, Schematic diagram of the GA-biosynthetic pathway. Undetected GAs are shown in gray, bioactive GAs are in red, and inactivated GAs are in blue. B, C, D, and E, GA contents in developing anthers of LJ11 and wrky53 plants grown under NC or LT. The concentrations and identity of endogenous GAs were determined using a Thermo Scientific Ultimate 3000 UHPLC. Data are shown as means ± se (n = 6). Dotted lines separate treatments and the comparisons are among different genotypes in the same treatment. Each dot represents the result from one biological replicate. Significant differences are indicated by different lowercase letters (P < 0.05, one-way ANOVA with Tukey’s significant difference test). F, SLR1 protein abundance in panicles of LJ11 and wrky53 plants grown under NC or after 4 days of LT treatment, as detected by immunoblot with an anti-OsSLR1 antibody. ACTIN, detected with anti-ACTIN antibody, was used as the loading control. G, Relative expression levels of GAMYB and CYP703A3 in developing panicles of LJ11 and wrky53 plants grown under NC or after 4 days of LT treatment. Data are shown as means ± se (n = 4). H, Relative expression levels of GA biosynthesis genes in developing panicles of LJ11 and wrky53 plants grown under NC or after 4 days of LT treatment. Data are shown as means ± se (n = 4). Each dot represents the result from one biological replicate. P-values were calculated by Student’s t test: **P < 0.01, *P < 0.05, and ns indicates no significant difference.
In rice, GA20ox1 is a major GA biosynthesis gene that is expressed across multiple tissues and is highly expressed in reproductive organs (Suuplemental File S1; Monna et al., 2002; Sasaki et al., 2002). In addition to GA20ox1, GA20ox3 and GA3ox1 are two other GA biosynthesis genes that are expressed specifically in anthers (Supplemental Data Set S2; Sakamoto et al., 2004; Chen et al., 2019). To investigate whether the lower GA contents induced by LT were caused by a reduced expression of GA biosynthesis genes, we examined their expression levels in panicles under LT and NC. We established that the transcript levels of GA20ox1, GA20ox3, and GA3ox1 increase gradually with panicle development; however, LT resulted in a marked drop in their expression levels compared with NC (Supplemental Figure S6, A–C). We further compared GA biosynthesis gene expression between LJ11 and the wrky53 mutant plant: under LT, the transcript levels of GA20ox1, GA20ox3, and GA3ox1 were significantly higher in the wrky53 mutant than in LJ11 plants (Figure 3H). Given that GA12, the first C20-GA in the GA biosynthetic pathway, had higher accumulation in the wrky53 mutant relative to LJ11 plants under both NC and LT (Figure 3C), we also examined the expression of ent-KAURENE OXIDASE2 (KO2) and ent-KAURENOIC ACID OXIDASE (KAO), which play key roles in the early steps of GA biosynthesis (Chen et al., 2019). We determined that KAO, but not KO2, shows higher expression in the wrky53 mutant relative to in LJ11 plants under both NC and LT, which partially explains why there was more GA12 in wrky53 mutants than in LJ11 plants (Figure 3H). In addition, because the wrky53 mutant accumulated more inactivate GAs (GA34 and GA51) than LJ11 plants (Figure 3E), we also measured the expression of GA2-oxidases, which deactivate bioactive GAs (Sakamoto et al., 2004). We determined that GA2ox1, GA2ox2, and GA2ox3 display higher expression levels in LT-treated wrky53 mutant plants than LT-treated LJ11 plants (Supplemental Figure S7). Collectively, WRKY53 might negatively regulate both GA biosynthesis genes and catabolism genes directly or indirectly, and then fine-tune GA contents during cold stress at the booting stage, consequently regulating rice CTB.
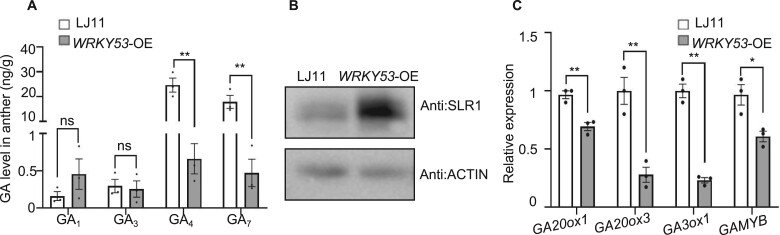
To confirm that WRKY53 negatively regulates GA content, we further examined the GA content of WRKY53-OE anthers under NC. The contents of GA4 and GA7 in WRKY53-OE anthers were dramatically lower than those of LJ11 anthers, while the contents of GA1 and GA3 were comparable between LJ11 and WRKY53-OE anthers (Figure 4A). Consistent with this result, SLR1 protein abundance in WRKY53-OE plants was significantly higher than in LJ11 plants, and the expression of GAMYB in WRKY53-OE plants was significantly lower than that in LJ11 plants (Figure 4, B and C and Supplemental Figure S8). We also investigated the expression levels of GA biosynthesis genes in LJ11 and WRKY53-OE plants and found that the transcript levels of GA20ox1, GA20ox3, and GA3ox1 are significantly higher in LJ11 plants than in WRKY53-OE plants (Figure 4C). Collectively, these results suggest that WRKY53 negatively regulates GA biosynthesis.
Figure 4.
WRKY53-OE anthers have lower GA contents and GA biosynthesis gene expression. A, GA contents in developing anthers of LJ11 and WRKY53-OE plants grown under NC. The concentrations and the identity of endogenous GAs were determined using a Thermo Scientific Ultimate 3000 UHPLC. Data are shown as means ± se (n = 3). B, SLR1 protein abundance in panicles of LJ11 and WRKY53-OE plants grown under NC, as detected by immunoblot with an anti-OsSLR1 antibody. ACTIN, detected with an anti-ACTIN antibody, were used as the loading control. C, Relative expression levels of GA biosynthesis genes in developing panicles of LJ11 and WRKY53-OE plants grown under NC. Data are shown as means ± se (n = 4). Each dot represents the result from one biological replicate, error bars indicate means ± se. P-values were calculated by Student’s t test: **P < 0.01, *P < 0.05, and ns indicates no significant difference.
WRKY53 directly represses the expression of GA biosynthesis genes
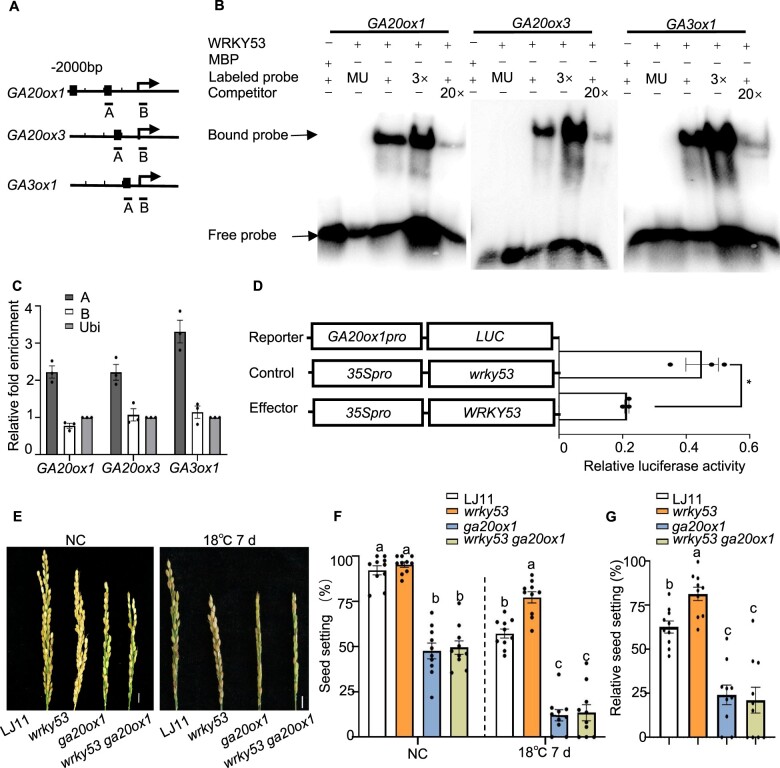
In previous studies, WRKY53 has been shown to function as a transcriptional repressor during brassinosteroid (BR) signaling and in response to herbivory in rice (Hu et al., 2016; Tian et al., 2017, 2021). We therefore tested whether WRKY53 can directly repress the expression of GA biosynthesis genes. WRKY-type transcription factors bind to the W-box (C/TTGACC/T) in their target promoters, and sequence analysis indicated the presence of the W-box element in the promoter regions of GA20ox1, GA20ox3, and GA3ox1 (Figure 5A). We then employed electrophoretic mobility shift assays (EMSAs) to test the binding of WRKY53 to these W-box-containing regions in vitro. A recombinant fusion protein consisting of maltose-binding protein (MBP) and WRKY53 efficiently bound to the promoter regions of GA20ox1, GA20ox3, and GA3ox1 containing the W-box (Figure 5B). In addition, chromatin immunoprecipitation (ChIP) assays confirmed that WRKY53 can bind to the W-box region of the GA20ox1, GA20ox3, and GA3ox1 promoters in vivo (Figure 5C), and LT treatment did not affect the binding capacity of WRKY53 to these targets (Supplemental Figure S9).
Figure 5.
WRKY53 binds to the promoters of GA biosynthesis genes and represses their transcription. A, Schematic diagrams of the GA20ox1, GA20ox3, and GA3ox1 promoters. The arrows indicate the position of the translation start codon. Scale, 500 bp. DNA fragments (A and B) were used for ChIP and the black squares indicate the W-box regions (T)TGACC. B, EMSA showing that WRKY53 binds to the conserved W-box of the GA20ox1, GA20ox3, and GA3ox1 promoters. A 52-bp W-box sequence containing DNA fragments in the A region in (A) was used as a probe. Versions of the probe with the W-box mutated to AAAAAA served as mutant probes; unlabeled probes served as competitive probes. MBP was used as a negative control. C, ChIP assays showing that WRKY53 binds to the promoters of GA20ox1, GA20ox3, and GA3ox1 in vivo. Immunoprecipitation was performed with anti-OsWRKY53 antibody. Immunoprecipitated chromatin was analyzed by qPCR using primers indicated in (A). qPCR enrichment was calculated by normalizing to ACTIN and to the total input of each sample. Data are shown as means ± se (n = 3). D, Transient assay in rice protoplasts showing that WRKY53 suppresses the transcription of GA20ox1. Schematic diagrams of the effector and reporter plasmids are indicated. The mutated WRKY53 coding sequence in wrky53, causing a frameshift in the coding sequence, was used as the control. Reporter together with control (35Spro:wrky53) or 35Spro:WRKY53 effector constructs were transfected into rice protoplasts. Relative LUC activity was calculated as the ratio between firefly LUC and REN LUC. Data are shown as means ± se (n = 3). Each dot represents the result from one biological replicate and error bars indicate means ± se. P-values were calculated by Student’s t test, *P < 0.05. E, F, and G, Representative images of panicles (E), seed setting rate (F), and relative seed setting rate (G) of LJ11, wrky53, ga20ox1, and wrky53 ga20ox1 mutants grown under NC and after LT stress. Scale bar is 1 cm. Data are shown as means ± se (n = 10). Dotted lines were drawn to separate treatments, and comparisons are among different genotypes in the same treatment. Each dot represents the result from one biological replicate. Significant differences are indicated by different lowercase letters (P < 0.05, one-way ANOVA with Tukey’s significant difference test).
We also performed transient expression assays in rice protoplasts using a construct WRKY53-OE (35Spro:WRKY53) as an effector and the firefly luciferase (LUC) gene driven by the promoters of GA biosynthesis genes as reporters. The GA20ox3pro:LUC and GA3ox1pro:LUC reporter constructs resulted in very low LUC activity in protoplasts, possibly due to their anther-specific expression characteristics; we therefore focused on the GA20ox1pro:LUC reporter construct for rice protoplast transient assays. The transient expression of WRKY53 lowered LUC activity derived from the GA20ox1pro:LUC reporter to approximately 50% of the control, which was an effector construct overexpressing the WRKY53 coding region from the wrky53 mutant allele (Figure 5D). Together, these results indicated that WRKY53 can directly repress the transcription of GA biosynthesis genes, and thus negatively regulate GA content in anthers.
Next, we generated ga20ox1, ga20ox3, and ga3ox1 single mutants via CRISPR/Cas9-mediated genome editing (Supplemental Figure S10). The single mutants ga20ox1, ga20ox3, and ga3ox1 exhibited a dwarf stature to various levels depending on the inactivated gene, likely resulting from GA deficiency (Supplemental Figure S11, A–C). Additionally, the ga20ox1 single mutant was characterized by a much lower seed setting rate (around 48%) relative to the wild-type LJ11 (91.8%), while neither ga20ox3 nor ga3ox1 set seed (Supplemental Figure S11, D and E). Detailed analysis showed that the fertile pollen rate in the ga20ox1 mutant is around 66.7%, while the ga20ox3 and ga3ox1 mutants had no fertile pollen (Supplemental Figure S11, F and G). Moreover, we observed that the tapetum degeneration at stage 10, stage 11, and late stage 11 in the ga20ox1 mutant is largely delayed, which may have contributed to the decreased pollen fertility in this mutant (Supplemental Figure S12). These results indicated that GA biosynthesis is essential for pollen fertility and seed setting, which is consistent with previous studies (Chen et al., 2019). We also generated wrky53 ga20ox1, wrky53 ga20ox3, and wrky53 ga3ox1 double mutants via CRISPR/Cas9-assisted gene editing by co-targeting the GA biosynthesis genes and WRKY53 in the LJ11 background (Supplemental Figure S10). All double mutants showed phenotypes similar to the ga20ox1, ga20ox3, and ga3ox1 single mutants under NC (Supplemental Figure S11, A–G). In addition, we assessed the CTB of ga20ox1 and wrky53 ga20ox1 mutants: the mutants were hypersensitive to LT relative to the wild type, which was in contrast to the wrky53 mutant (Figure 5, E–G and Supplemental Figure S11H). These results suggest that WRKY53 may act genetically upstream of GA biosynthesis, as might be expected for a transcription factor that regulates the expression of GA biosynthesis genes.
Rice DELLA protein SLR1 interacts with UDT1 and TDR
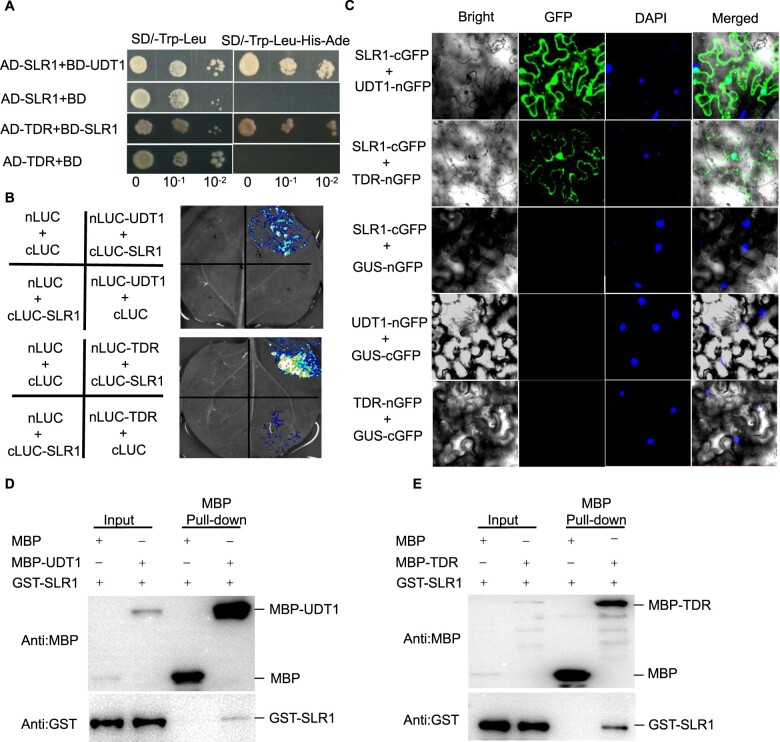
GA biosynthesis is essential for male fertility (Chen et al., 2019; Supplemental Figure S11, D–G). Therefore, we next explored how GA regulates this biological process. Given that DELLAs are key components of GA signaling and mediate diverse GA-regulated responses through interacting with and sequestrating partner proteins to block their activity (Daviere and Achard, 2013; Huang et al., 2015), we were interested in whether SLR1 (a rice DELLA) can directly interact with the transcription factors that regulate male fertility. The sequestration of the bHLH PHYTOCHROME INTERACTING FACTORS by DELLAs is an important molecular link between light and GA signaling (Feng et al., 2008). We selected the two bHLH proteins UDT1 and TDR, because they play key roles in regulating tapetum development and degeneration, and udt1 and tdr mutants are male sterile (Jung et al., 2005; Li et al., 2006; Zhang et al., 2008). We first performed yeast two-hybrid assays and showed that SLR1 can interact with UDT1 and TDR directly (Figure 6A). Split-LUC complementation assays also revealed an interaction of SLR1 with UDT1 and TDR in Nicotiana benthamiana leaves (Figure 6B). Bimolecular fluorescence complementation (BiFC) analysis in N. benthamiana confirmed that SLR1 interacts with UDT1 and TDR in the nuclei of living plant cells (Figure 6C). Moreover, pull-down assays also showed that the glutathione S-transferase (GST)-SLR1 fusion protein can interact with the MBP-UDT1 or MBP-TDR fusion protein in vitro (Figure 6, D and E and Supplemental Figure S13). These results indicate that SLR1-UDT1 and SLR1-TDR may mediate GA-regulated male sterility.
Figure 6.
SLR1 interacts with UDT1 and TDR. A, Interaction between SLR1 and UDT1/TDR in a yeast two-hybrid assay. BD-UDT1, AD-SLR1, BD-SLR1, and AD-TDR were used as indicated. Clones that grew on SD–Trp–Leu–His–Ade medium indicate protein interaction in yeast cells. The empty AD-SLR1/BD and AD-TDR/BD were used as negative controls. B, LUC complementation imaging assay showing that SLR1 interacts with UDT1/TDR in N. benthamiana leaves. Co-infiltration of cLUC-SLR1 and nLUC-UDT1 or nLUC-TDR constructs leads to the reconstitution of the LUC signal, whereas no signal was detected when cLUC-SLR1 and nLUC, cLUC and nLUC-UDT1 or nLUC-TDR, or cLUC and nLUC were co-infiltrated. In each experiment, at least five independent N. benthamiana leaves were infiltrated and analyzed. C, BiFC assay showing that SLR1 interacts with UDT1/TDR in N. benthamiana leaves. Co-infiltration of cGFP-SLR1 and nGFP-UDT1 or nGFP-TDR leads to the reconstitution of the GFP signal, whereas no signal was detected when cGFP-SLR1 and nGFP, or cGFP and nGFP-UDT1 or nGFP-TDR were co-infiltrated. DAPI was used to visualize the nuclei. In each experiment, at least five independent N. benthamiana leaves were infiltrated and analyzed. D and E, Pull-down assay showing the interaction between UDT1/TDR and SLR1. GST-SLR1 was pulled down by MBP-UDT1 and MBP-TDR immobilized on amylose resin beads and was detected with anti-MBP and anti-GST antibody, respectively.
GA regulates male fertility through SLR1-UDT1 and SLR1-TDR cascades
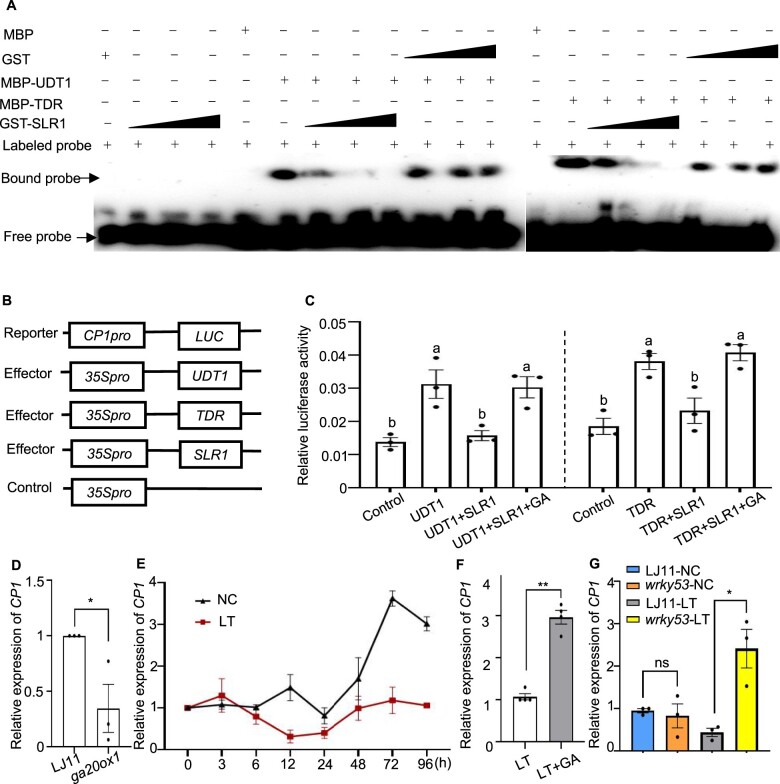
UDT1 and TDR modulate tapetum development and degradation via directly regulating the expression of tapetal marker genes (Osc4, Osc6, and CYSTEINE PROTEASE1 [OsCP1]) (Jung et al., 2005; Li et al., 2006). These three genes are dramatically downregulated in the udt1 and tdr mutants; TDR can bind to the OsCP1 promoter in vivo; Oscp1 mutants show male sterility (Jung et al., 2005; Li et al., 2006). To address how the SLR1-UDT1 and SLR1-TDR cascades might affect male sterility, we asked whether SLR1 affects the DNA binding capacity of UDT1 and TDR. We determined that UDT1 and TDR efficiently bind to the E-box region in the OsCP1 promoter, whereas this binding was significantly impaired in the presence of SLR1 (Figure 7A and Supplemental Figure S14). We further examined the effect of the SLR1–UDT1/TDR interaction on the expression of OsCP1 in a rice protoplast transient expression system (Figure 7B). We observed that TDR and UDT1 can activate the transcription of OsCP1 (Figure 7C). However, the upregulated OsCP1 levels in cells expressing TDR and UDT1 were markedly attenuated by the co-expression of SLR1 (Figure 7, B and C). Notably, the application of GA activated OsCP1 transcription (Figure 7, B and C). Furthermore, we found that OsCP1 levels are significantly lower in the ga20ox1 mutant than in LJ11 plants (Figure 7D). Collectively, the in vitro and in vivo evidence indicated that SLR1 directly interacts with UDT1 and TDR to sequester them and block their function, which represents a previously unreported mechanism by which GA regulates male sterility besides GAMYB.
Figure 7.
SLR1 represses the transcriptional activity of UDT1 and TDR via physical sequestration, and GA alleviates this repression. A, EMSA showing that direct binding of UDT1 and TDR to the CP1 promoter is inhibited by the addition of increasing amounts of recombinant GST-SLR1 protein. Increasing amounts of GST protein were used as a negative control. B, Schematic diagrams of the effector and reporter plasmids used in the transient assay in rice protoplasts. C, Relative LUC activity in rice protoplasts co-transfected with the reporter and different combinations of effectors. Relative LUC activity was calculated as the ratio between firefly LUC and REN LUC. Data are shown as means ± se (n = 3). Each dot represents the result from one biological replicate. Significant differences are indicated by different lowercase letters (P < 0.05, one-way ANOVA with Tukey’s significant difference test). D, Relative CP1 expression levels in developing panicles of LJ11 and ga20ox1 plants grown under NC. Data are shown as means ± se (n = 3). E, Time course of relative CP1 expression levels in panicles of the cultivar LJ11 exposed to a 15°C cold treatment during the booting stage. The expression level on day 0 was set to 1. Data are shown as means ± se (n = 3). F, Relative CP1 expression levels in developing panicles of LJ11 plants grown under LT treatment with or without GA3 application. Data are shown as means ± se (n = 4). G, Relative CP1 expression levels in developing panicles of LJ11 and wrky53 plants grown under NC or after 4 days of LT treatment. Data are shown as means ± se (n = 3). Each dot represents the result from one biological replicate. P-values were calculated by Student’s t test: **P < 0.01 and *P < 0.05.
To investigate whether the SLR1-UDT1 and SLR1-TDR cascades were involved in the CTB response, we examined the expression levels of OsCP1 in panicles under LT and NC. We determined that OsCP1 expression gradually increases with the panicle development, and OsCP1 expression was lower under LT than under NC (Figure 7E). This result indicated that decreased expression of OsCP1 under LT might be one means by which LT regulates male sterility. In addition, GA application can raise OsCP1 expression under LT (Figure 7F). Additionally, to investigate whether the higher expression of OsCP1 can be responsible for better CTB in the wrky53 mutant, we measured OsCP1 transcript levels under LT: OsCP1 expression was significantly higher in the wrky53 mutant relative to LJ11 plants (Figure 7G). Consistent with this result, OsCP1 showed markedly decreased expression levels in WRKY53-OE plants than in LJ11 plants (Supplemental Figure S15). Collectively, these results suggest that the SLR1-UDT1 and SLR1-TDR cascades are indeed involved in rice CTB responses, and support the notion that WRKY53 regulates CTB via regulating GA levels in anthers.
Loss of WRKY53 function shows great potential for cold tolerance breeding
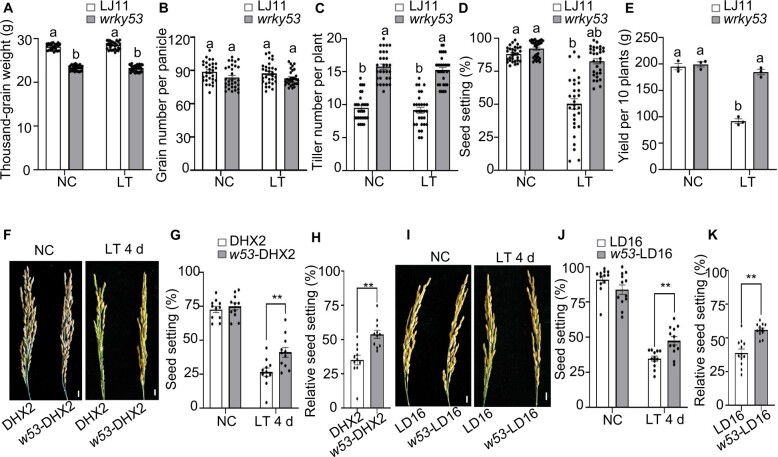
‘Since wrky53 significantly improved CTB in the LJ11 background (Figure 1, B–D and Supplemental Figures S2 and S3), we evaluated agronomic traits and application potential in rice breeding by fully phenotyping the wrky53 mutant in the LJ11 background. The thousand-grain weight of the wrky53 mutant was significantly lower than that of LJ11 plants, likely resulting from the smaller seeds of wrky53 mutant plants (Supplemental Figure S16, A–D). The grain number per panicle was comparable between the wrky53 mutant and LJ11 plants (Supplemental Figure S16, A–D). Notably, the wrky53 mutant produced more tillers per plant than LJ11 (Supplemental Figure S16, A–D), which may compensate for the lower thousand-grain weight of the mutant, thus resulting in a comparable final yield per plant between the two genotypes (Supplemental Figure S16, A–D). These results indicate that the wrky53 mutation does not grossly negatively affect yield of rice plants grown under NCs (Supplemental Figure S16).
To further evaluate the potential of wrky53 for application in cold tolerance breeding, we cultivated LJ11 and wrky53 mutant plants in a paddy field under NC or with LT treatment consisting of 17°C deep water irrigation during the booting stage. For both LJ11 and wrky53 mutant plants, values for the thousand-grain weight, grain number per panicle, and tiller number per plant were comparable between NC and LT (Figure 8, A–C). In the wrky53 mutant, the seed setting rate was slightly lower under LT than under NC; however, it was significantly (∼37.5%) lower under LT than under NC in LJ11 (Figure 8D). In effect, the seed setting rate of wrky53 mutant plants was 32% higher than that of LJ11 plants under LT (Figure 8D) and the yield per 10 plants of the wrky53 mutant was about 43.8% higher than that of LJ11 under LT (Figure 8E). To validate this result, we repeated the field experiment with 17°C deep water irrigation in 2021 (Supplemental Figure S17). We found that the wrky53 mutant reproducibly display a significantly higher seed setting rate than LJ11 (Supplemental Figure S17E), and the yield per plot of the wrky53 mutant was 30.8% higher than that of LJ11 (Supplemental Figure S17F).
Figure 8.
Loss-of-function in WRKY53 can improve CTB stage without yield penalty. A–E, Agronomic traits scored in LJ11 and wrky53 plants grown in a paddy field under LT or NCs at the booting stage. Data are shown as means ± se (n = 30 in A–D and n = 3 in E). Each dot represents the result from one biological replicate. Significant differences are indicated by different lowercase letters (P < 0.05, one-way ANOVA with Tukey’s significant difference test). F, G, and H, Panicles (F), seed setting (G), and relative seed setting rate (H) of DHX2 and w53-DHX2 plants grown under NC or after 4 days of LT treatment. Data are shown as means ± se (n = 11). I, J, and K, Panicles (I), seed setting (J), and relative seed setting rate (K) of Longdao16 (LD16) and w53-LD16 plants grown under NC or after 4 days of LT treatment. Data are shown as means ± se (n = 12). Scale bar, 1 cm (F, I). Each dot represents the result from one biological replicate. P-values were calculated by Student’s t test: **P < 0.01.
Daohuaxiang 2 (DHX2) and Longdao 16 (LD16) are two elite cultivars with good grain quality that are grown in the Heilongjiang province, the northernmost rice cultivation region of China, where they are frequently exposed to LTs (Li et al., 2015, 2017; Zhang et al., 2017). Unfortunately, DHX2 and LD16 are very sensitive to LT stress at the booting stage, which greatly restricts their growth region and can threaten their yields during years with LTs (Guo et al., 2020). To globally validate the usefulness of nonfunctional WRKY53 in enhancing CTB of elite cultivars, we generated loss-of-function alleles for WRKY53 in the DHX2 and LD16 backgrounds by genome editing, which we named wrky53-DHX2 and wrky53-LD16 (or w53-DHX2 and w53-LD16 for simplicity) (Supplemental Figures S18, S19, A and B, and S20, A and B). Grains from w53-DHX2 and w53-LD16 plants were slightly shorter than grains from the respective wild types, DHX2 and LD16 (Supplemental Figures S19 and S20, C–G), which was in agreement with the negative role of WRKY53 in regulating seed size already observed in the LJ11 background (Tian et al., 2017). In addition, when compared with DHX2 and LD16, the thousand-grain weight in w53-DHX2 and w53-LD16 decreased by 8.9%–18.9%, while the tiller number of w53-DHX2 and w53-LD16 increased by 21.9%–19.6% (Supplemental Figures S19 and S20, H–J). Therefore, the yield per plant for w53-DHX2 and w53-LD16 was comparable to that of DHX2 and LD16 under NC (Supplemental Figures S19 and S20, K). However, after 4 days of an LT treatment consisting of 15°C at the booting stage, the seed setting and relative seed setting rates for both w53-DHX2 (Figure 8, F–H) and w53-LD16 (Figure 8, I–K) mutants were markedly higher than those of their respective wild types. The relative seed setting rates of w53-DHX2 and w53-LD16 were around 18.86% and 17.1% higher than those of DHX2 and LD16, respectively (Figure 8, H and K). Three independent experiments consistently showed similar results (Supplemental Figure S21, A–L). In parallel, we also examined the expression levels of GA20ox1, GA20ox3, GA3ox1, and GAMYB in these materials: all four genes were more highly expressed in the w53-DHX2 and w53-LD16 mutants than in their respective wild-type backgrounds under LT (Supplemental Figure S21, M and N). These results support the notion that WRKY53 negatively regulates GA biosynthesis (Figure 3). Together, these results indicate that editing WRKY53 will be a valuable approach to improve rice CTB without incurring a yield penalty.
Discussion
WRKY53 regulates rice CTB stage
Cold stress at the booting stage is a major factor that limits rice production in two distinct ways: first, cold stress causes direct loss of yield by negatively affecting pollen fertility and seed setting and second, cold stress determines the geographical limits of rice growth into higher altitudes and latitudes (Zhu et al., 2015; Liu et al., 2019; Guo et al., 2020; Xu et al., 2020). Because of the difficulty in evaluating CTB and the genetic complexity of the cold tolerance trait, only a few genes have been characterized in rice to date (Kuroki et al., 2007; Saito et al., 2010; Zhou et al., 2010; Zhang et al., 2017; Liu et al., 2019; Guo et al., 2020). In this study, we demonstrated that WRKY53 negatively regulates CTB by fine-tuning GA levels in anthers. First, WRKY53 expression was rapidly induced by cold treatment at the booting stage (Figure 1A). Second, WRKY53 overexpression led to lower seed setting rates, suggesting that WRKY53 might negatively regulate seed setting (Figure 1, B and C and Supplemental Figure S1, A and B). Third, wrky53 mutants generated by genome editing in diverse cultivar backgrounds consistently showed higher seed setting rates than their respective parents when exposed to LT, indicating that WRKY53 is a key negative regulator of CTB in rice (Figures 1, B–I, 2, and 8, F–K and Supplemental Figures S2, S3, and S21, A–L). Therefore, WRKY53 may provide a suitable target for improving rice CTB. Several studies have previously shown that WRKY53 functions in diverse developmental processes (seed size control and BR signaling) and biotic stress responses (pathogen infections and herbivore attacks) (Chujo et al., 2007; Hu et al., 2016; Tian et al., 2017, 2021; Yuan et al., 2019; Xie et al., 2021). This study demonstrates that WRKY53 plays a key role in cold tolerance responses, likely by modulating GA biosynthesis in anthers. We previously showed that WRKY53 positively regulates rice BR signaling, which is also involved in pollen fertility (Ye et al., 2010; Tian et al., 2017, 2021; Yan et al., 2020). Therefore, to rule out the possibility that WRKY53-regulated CTB is dependent on BR signaling, we applied BRs during LT, but observed no effect on rice cold tolerance. In addition, we also examined the CTB of rice lines with defects in two major BR signaling components, bzr1-D (a dominant allele of the transcription factor gene BRASSINAZOLE-RESISTANT1) and GSK2-RNA interference (RNAi knockdown of GLYCOGEN SYNTHASE KINASE2), and found that these two genotypes displayed similar cold tolerance to the wild type (Supplemental Figure S22). Given that WRKY53 is a functional transcriptional repressor, it will be informative to identify and characterize its various direct targets to dissect the multifaced functions of WRKY53.
WRKY53 regulates CTB via modulating the GA-SLR1-UDT1/TDR module
GA biosynthesis and signaling mutants display aborted pollen and result in impaired fertility and even sterility in rice (Oikawa et al., 2004; Sakamoto et al., 2004; Chen et al., 2019). Mutants of the GA-inducible anther-regulator gene GAMYB, the GA20ox3 regulator SWOLLEN ANTHER WALL1 (SAW1), and the GA-biosynthesis gene GA20ox1 showed male sterility with defective tapetal cell development (Supplemental Figure S12; Aya et al., 2009; Wang et al., 2020). Interestingly, LT at the booting stage also causes male sterility due to abnormal enlargement of tapetal cells (Figure 2, B–D and M–O; Oda et al., 2010). In addition, GA contents decreased in cold-treated anthers and exogenous application of GA effectively rescued cold-induced male sterility (Figure 3B and Supplemental Figure S4; Sakata et al., 2014). Moreover, GA biosynthesis and signaling mutants are hypersensitive to LT at the booting stage (Sakata et al., 2014). In this study, we propose that GA regulates male fertility by alleviating the sequestration imposed by SLR1 on the transcriptional activity of UDT1 and TDR (two critical transcription factors for tapetum development) (Figures 6 and 7). Together with results of previous studies, our current results indicate that cold stress leads to a decrease in GA content in anthers, which slows the degradation of the anther tapetum layer, decreasing pollen fertility and the seed setting rate (Figures 1–3). These results suggest that GA plays a positive role in rice CTB through maintaining timely tapetal cell degradation to ensure normal pollen fertility and increasing the seed setting rate under cold stress.
In this study, we established that WRKY53 can bind to the promoter regions of GA biosynthesis genes to suppress their transcription in anthers, leading to the higher GA content seen in wrky53 relative to the wild type under LT (Figures 3 and 5). This model is supported by higher GAMYB transcript levels and lower SLR1 protein abundance in the wrky53 mutant than in the wild type under LT (Figure 3, F and G). In addition, we found that SLR1 can interact with UDT1 and TDR, and repress their transcriptional activity toward OsCP1, whereas GA application alleviated this repressive sequestration (Figures 6 and 7). Accordingly, expression of OsCP1 was downregulated in ga20ox1 mutants and by LT treatment, and can be upregulated by GA application (Figure 7). These results suggest that GA-alleviated repression of UDT1/TDR imposed by SLR1 may represent a possible mechanism by which GA regulates male fertility besides GAMYB (Figures 6 and 7). Moreover, under LT, wrky53 mutant anthers contained more bioactive GA than wild-type anthers, and OsCP1 was more highly expressed in wrky53 anthers compared with wild-type anthers (Figures 3, A–E and 7, G). These results support the idea that WRKY53 negatively regulates CTB by modulating the GA biosynthetic pathway (Figure 9). By contrast, GA application reduced the seed setting rate and overexpression of GA20ox1 similarly decreased the seed setting rate (Supplemental Figure S4; Oikawa et al., 2004), suggesting that both a higher and a lower GA content in anthers will affect seed setting, and that GA contents must be precisely controlled to within a specific range to maintain normal seed setting. In this scenario, the fact that WRKY53 expression is induced by LT treatment and that WRKY53 suppresses GA biosynthesis in anthers indicates that WRKY53 is a suitable target that can be employed to mitigate the LT-mediated repression of GA biosynthesis in anthers (Figures 1, 3, and 5). We therefore propose a model in which WRKY53 is induced in wild-type plants under cold stress, leading to the repression of GA biosynthesis and a decline in GA content. Lower GA leads to higher SLR1 protein abundance and lower GAMYB transcript levels. SLR1 then interacts with and suppresses TDR/UDT1 transcriptional activity, and finally results in impaired pollen development and a decreased seed setting rate (Figure 9). By contrast, in wrky53 mutant plants subjected to cold stress, GA biosynthesis is no longer repressed by WRKY53, such that GA content does not significantly decline and is instead maintained at a suitable physiological level. Moreover, SLR1-mediated repression of TDR/UDT1 is relieved to facilitate the normal development of anthers and pollen, and GAMYB transcript levels do not significantly decrease, thus contributing to normal seed setting (Figure 9).
Figure 9.
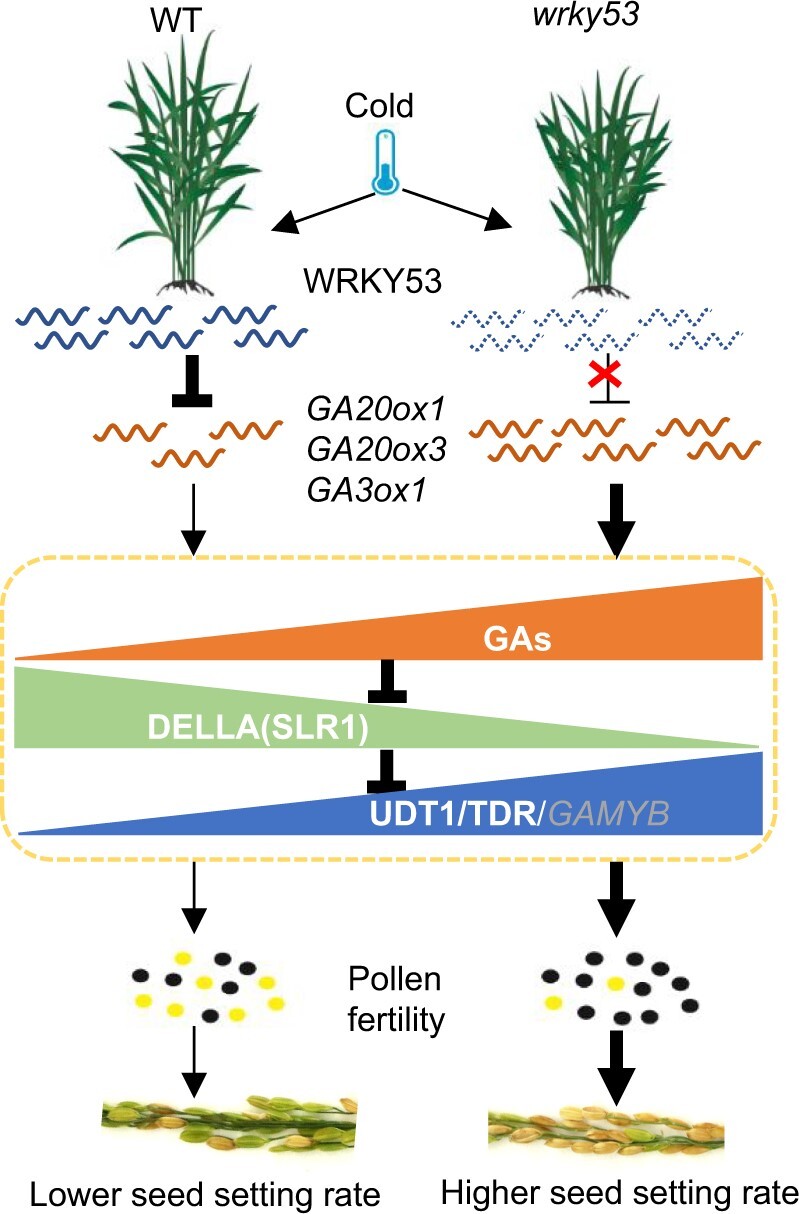
Working model explaining how WRKY53 regulates rice CTB stage. In wild-type plants under cold stress, WRKY53 expression is induced, leading to the repression of GA biosynthesis and a decline in GA contents. Lower GA leads to higher SLR1 protein abundance and lower GAMYB transcript levels. SLR1 then interacts with and suppresses TDR/UDT1 transcriptional activity, finally resulting in impaired pollen development and a decreased seed setting rate. In the wrky53 mutant subjected to cold stress, GA biosynthesis is no longer repressed by WRKY53; GA contents do not significantly decline; SLR1-mediated suppression of TDR/UDT1 is alleviated to facilitate normal development of anthers and pollen; and GAMYB transcript levels are not significantly decreased. Therefore, normal seed setting occurs.
Knocking out WRKY53 in rice has promising breeding potential
To date, four genes have been identified with functions in CTB stage in rice: CTB1, CTB4a, bZIP73, and LTT1. CTB1 encodes an F-box protein, but its target for degradation is unknown; bZIP73 and CTB4a have been shown to positively regulate CTB stage, and have undergone strong selection during domestication and northward expansion of rice growth (Saito et al., 2010; Zhang et al., 2017; Liu et al., 2019; Xu et al., 2020). Although both bZIP73 and CTB4a contain functional SNPs that can be used in pyramid breeding, the fact that they must be overexpressed to significantly increase cold tolerance has greatly limited their application value (Zhang et al., 2017; Liu et al., 2019). In addition, although the ltt1 mutant is more tolerant to cold than the wild type, its associated lesion mimic phenotype negatively affects yield (Xu et al., 2020). Moreover, bZIP73 has been reported to regulate cold tolerance both at the vegetative stage and booting stage via modulating ABA biosynthesis. We also examined the cold tolerance of the wrky53 mutant at the seedling stage and found that the wrky53 mutant and LJ11 seedlings display comparable cold tolerance at the vegetative stage and similar expression levels of NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) and NCED5, suggesting that WRKY53 does not play roles in cold tolerance at the vegetative stage and that ABA might not mediate the WRKY53-regulated CTB response (Supplemental Figure S23, A–D). Here and in previous work, we have demonstrated that although the seed size and thousand-grain weight of the wrky53 mutant is lower than that of the wild type, this defect was fully compensated by an increased tiller number per plant in the mutant, and therefore the final yield for the wrky53 mutant is comparable to that of the wild type when grown under NCs (Figure 8, A–E and Supplemental Figures S16 and S17). It will be worth investigating the mechanism by which WRKY53 regulates tiller number. Interestingly, IDEAL PLANT ARCHITECTURE1 (IPA1), a critical regulator of rice plant architecture and grain yield, is also a transcriptional activator (Jiao et al., 2010; Lu et al., 2013). WRKY78, which is highly similar to WRKY53, is a putative IPA1 target, which suggests that WRKY53 might function downstream of IPA1 to regulate tiller development (Lu et al., 2013).
In this study, we found that WRKY53-OE plants under NCs exhibited lower seed setting rates than wild-type plants, indicating that over expression of WRKY53 might have no application value (Figure 1 and Supplemental Figures S1 and S2). By contrast, wrky53 mutants show better cold tolerance than wild-type plants, as described in this study, and also an increased resistance to Xanthomonas oryzae pv. oryzae (Xoo) sheath blight and the striped stem borer (Hu et al., 2016; Yuan et al., 2019; Xie et al., 2021). Although the 1,000-grain weight of wrky53 mutants was lower than that of wild-type plants, they had more tillers and therefore their yield per plant was not lower under NCs (Supplemental Figure S16). These results suggest that wrky53 has great putative application value for improving rice yield in natural rice production subjected to diverse biotic and abiotic stress. For example, under cold stress conditions, the seed setting rate and yield of wrky53 mutants was significantly higher than that of wild-type plants, suggesting that inactivating WRKY53 will be a valuable strategy to improve tolerance to cold stress in rice (Figure 8, D and E and Supplemental Figure S17, E and F). It will also be interesting to evaluate the comprehensive practical application value of wrky53 mutants by assessing their yield under multiple pathogen infections insect pest attacks, LTs, and other stresses in the field. Knocking out WRKY53 will be facilitated by CRISPR/Cas9 technology, which bypasses the generation of genetically modified organisms, thus alleviating consumer concerns and minimizing the associated regulatory burden.
Materials and methods
Plant materials and growth conditions
The rice (O. sativa L.) genotypes used in this study were the cultivars LJ11, DHX2, and Longdao16 (LD16). They were grown in a field (under natural long-day conditions) or in a greenhouse at 30°C for 14 h (day) and 24°C for 10 h (night).
Generation of transgenic rice plants
The WRKY53 overexpression line (WRKY53-OE) and the wrky53 mutants, both in the LJ11 background, were described previously (Tian et al., 2017). In this study, the mutants wrky53-1 and wrky53-2 were used; wrky53-2 was used for most experiments and labeled wrky53 if not indicated otherwise. Previously published gene editing vectors targeting WRKY53 for transformation were used to generate a loss of function wrky53 mutant in the DHX2 and Longdao16 cultivars (Tian et al., 2017). To generate the ga20ox1, ga20ox3, and ga3ox1 single mutants and the wrky53 ga20ox1, wrky53 ga20ox3, and wrky53 ga3ox1 double mutants in the LJ11 background, the target sequences for GA20ox1, GA20ox3, GA3ox1, and WRKY53 (Supplemental Data Set S1) were synthesized and ligated into respective single-guide RNA (sgRNA constructs), and then were sequentially ligated into the CRISPR/Cas9 binary vector pYLCRISPR/Cas9Pubi-H as described (Ma et al., 2015a).
CTB stage assays
For CTB assays in the greenhouse, each plant was grown in a cylindrical plastic pot 15 cm in height and 15 cm in diameter containing dry soil and watered with fertilizer. Approximately 10 days before booting, when the distance between the auricles of the flag leaf and the penultimate leaf of the tiller was –5 cm to 0 cm (between the meiotic stage and the uninucleate pollen stage), tillers were tagged and a subset of the pots were moved to a LT greenhouse (15°C or 18°C constant temperature and 14-h light and 10-h dark photoperiod). After the indicated number of days of cold treatment, the treated pots were returned to the greenhouse for normal growth (NC; 30°C for 14-h light and 24°C for 10-h dark) until maturity, at which point plants were harvested and traits of tagged tillers were measured. The seed setting rate was determined as the percentage of full grains per panicle relative to the total grain number per panicle. The relative seed setting rate was determined as the percentage of the seed setting rate under LT relative to that under NC. More than 10 plants per line were used for treatment in each biological replicate. For CTB assays in paddy fields, plants were grown in a normal field with a cold-water irrigation system (Acheng Farm, Harbin, China). Plants were treated with cool water (25 cm depth) kept at 17°C from the primordium stage until the completion of heading.
For reverse transcription quantitative PCR (RT-qPCR) analysis of cold-regulated gene expression, ChIP assay and detection of SLR1 protein abundance, approximately 10 days before booting, when the distance between the auricles of the flag leaf and the penultimate leaf of the tiller was within –5 cm to 0 cm (between the meiotic stage and the uninucleate pollen stage), tillers were tagged and pots were moved to an LT greenhouse (15°C constant temperature, 14-h light, and 10-h dark photoperiod). NC samples were taken on the first day of LT treatment and LT samples were taken 4 days into LT treatment.
The LT treatment was as follows for measuring GA contents in cold-treated anthers. Plants were transferred to LT when the distance between the auricles of the flag leaf and the penultimate leaf of the main tiller was –8 to –6 cm (meiotic stage). Plants remained in the LT greenhouse (18°C, 14-h light, and 10-h dark photoperiod) until 1 day before flowering (around 15 days). For the NC treatment, plants were grown under NC until 1 day before flowering (around 11 days).
For semi-thin sections, plants were transferred to LT when the distance between the auricles of the flag leaf and the penultimate leaf of the main tiller was –8 cm to –6 cm (meiotic stage). Plants were grown in the LT greenhouse (15°C, 14-h light, and 10-h dark photoperiod) for 4 days. Anther of NC and LT samples was taken from panicle with the –5 cm pulvinus distance to 1 day before flowering.
Cold tolerance assays at seedling stage
Four-week-old LJ11 and wrky53 plants were cold treated at 4°C for 48 h or 72 h in a growth chamber (RXZ-0405) and then recovered at 28°C for 7–10 days. The survival rates were determined as the percentage of surviving seedlings.
GA and BR application experiment
At the booting stage, when the distance between the auricles of the flag leaf and the penultimate leaf of the tiller was –10 cm to –5 cm, plants were irrigated with water containing 5 μM GA3 four times in 3-day intervals or 4.16 μM brassinolide two times in 3-day intervals.
Microscopy
Starch staining of pollen grains and observations of anther morphology were performed 1 day before flowering. For pollen viability assays, anthers were fixed in a formalin–acetic acid–alcohol (FAA) solution (5% formaldehyde, 5% glacial acetic acid, 63% ethanol, and 27% water, 5/5/63/27) Fixed anthers were ground to release pollen grains, stained with 1% (w/v) I2-KI Lugol’s iodine solution, then observed under a light microscope (Olympus BX53). Pollen grains that were stained blue were counted to score pollen fertility. Anther morphology was observed directly under a microscope (Olympus SZX16) without staining.
For semi-thin sections, tissues were fixed in FAA solution. After dehydration through a graded ethanol series, samples were placed in resin and then cut into 1-µm-thick sections with a microtome (Leica UC7). The sections were stained with 0.5% (w/v) toluidine blue, rinsed with distilled water, dried overnight, and examined with an optical microscope (OLYMPUS BX43).
Quantification of GAs
The contents of endogenous GAs in anthers were measured by the Wuhan Greensword Creation Technology Company (http://www.greenswordcreation.com) by UHPLC-MS/MS. In brief, anther samples (100 mg) were ground to a fine powder and extracted with 1 mL 80% (v/v) methanol at 4°C for 12 h. Then, 1.0 ng/g diverse [2H2]-labeled GA components ([2H2] GA1 [1 ng/g], [2H2] GA3 [1 ng/g], [2H2] GA4 [1 ng/g], [2H2] GA5 [1 ng/g], [2H2] GA7 [1 ng/g), [2H2] GA8 [1 ng/g), [2H2] GA9 [1 ng/g], [2H2] GA12 [1 ng/g], [2H2] GA19 [1 ng/g], [2H2] GA24 [1 ng/g], [2H2] GA34 [1 ng/g], [2H2] GA51 [1 ng/g], and [2H2] GA53 [1 ng/g]) were added as internal standards for quantification. The analysis of GA contents was performed on a Thermo Scientific Ultimate 3000 UHPLC coupled with a TSQ Quantiva. For this assay, due to the limited amounts of each sample, the anthers from around 30 plants were collected and mixed, and then the mixed samples were split into three technical repeats with two biological replicates. A representative GA chromatogram is shown in Supplemental File S2.
Total RNA isolation and RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with Dnase I. First-strand cDNA was synthesized from 2 µg of total RNA using Superscript II Reverse Transcriptase (Invitrogen). Quantitative PCR was performed with SYBR Green PCR master mix (Takara). Data were collected using a Bio-Rad chromo 4 real-time PCR system. All expression values were normalized against UBIQUITIN (Os01g0328400). The primers used are listed in Supplemental Data Set S1. Four biological repeats with three technical repeats per biological repeat were performed for each analysis. Values are means ± se of three or four biological repeats.
ChIP assay
Wild-type and WRKY53-OE plants were used for ChIP assays, as previously described (Tian et al., 2017). Briefly, 2 g of rice plant leaves collected at the booting stage for each treatment (LT and NC) was crosslinked with crosslinking solution containing 1% (w/v) formaldehyde under a vacuum; crosslinking was then stopped by the addition of 5 mM glycine for 5 min. The samples were ground to a fine powder in liquid nitrogen and used to isolate nuclei. The protein–DNA complexes were immunoprecipitated with anti‐WRKY53 antibody (Beijing Protein Innovation, Abp80095) and the precipitated DNA was recovered and analyzed by qPCR. Chromatin precipitated without antibody was used as a negative control. The primers used for qPCR are listed in Supplemental Data Set S1. Data are presented as means ± se of three biological replicates, each consisting of three technical replicates.
EMSAs
The full-length coding sequences of WRKY53, UDT1, TDR, and SLR1 were amplified from Nipponbare cDNA and cloned into the pVP13 vector or pDEST15 vector via LR recombination to generate the MBP-WRKY53, MBP-UDT1, MBP-TDR, and GST-SLR1 fusion protein constructs, respectively. These constructs were transformed into Escherichia coli strain BL21 (DE3) for producing the fusion proteins. The recombinant proteins were affinity-purified using Dextrin Beads 6FF (SMART LIFESCIENCE, Cat No. SA026010) and Glutathione Beads (SMART LIFESCIENCE, Cat No. SA008050). Oligonucleotide probes of ∼52 bp containing the wild-type W-box (TGACC) or a mutated W-box (AAAAA) motif and the wild-type E-box (CANNTG) were synthesized and labeled with biotin using the EMSA Probe Biotin Labeling Kit (Beyotime, Cat No. GS008). For unlabeled probe competition, unlabeled probe was added to the reactions. EMSA was performed using a Chemiluminescent EMSA kit (Beyotime, Cat No. GS009). Probe sequences are shown in Supplemental Data Set S1.
Transient transcription dual-LUC assays
The 35Spro:WRKY53, 35Spro:UDT1, 35Spro:TDR, and 35Spro:SLR1 constructs were generated and used as effectors. The 35Spro:wrky53 construct overexpressing the WRKY53 coding region from the wrky53-2 mutant allele was used as control. The promoter regions (upstream of the ATG) of GA20ox1 and CP1 were cloned into the pGreenII 0800-LUC vector and used as reporters (Supplemental Data Set S1). The resulting effector and reporter constructs were co-transfected into protoplasts (Tian et al., 2017). To investigate the effects of the SLR1–UDT1/TDR interaction, protoplast cells co-transfected with the indicated reporter and effector constructs were treated with 10 μmM GA3 for 6 h. Then, the protoplasts were collected and subjected to LUC activity assays. Renilla (REN) LUC driven by the 35S promoter in the pGreenII 0800-LUC vector was used as an internal control. Firefly LUC and REN activities were measured with a dual-LUC reporter assay kit using a GloMax 20/20 luminometer (Promega). LUC activity was normalized to REN activity. For each plasmid combination, three independent transformations were performed with three technical replicates per independent transformation. Values are means ± se of three independent transformations.
Protein gel blot analysis
For immunoblots, anti-OsSLR1 antibody (Huang et al., 2015) and anti-ACTIN antibody (Abmart, M20009M, 1:2,500) were used as primary antibodies, and peroxidase-labeled goat anti-rabbit antibody (Abcam, ab6721) or goat anti-mouse antibody (Abcam, ab6789) were utilized as secondary antibodies. Membranes were developed with a Super signal west pico chemiluminescent substrate kit (Pierce Biotechnology) and the signal was detected by chemiluminescence imaging (Tanon 5200).
Yeast two-hybrid assay
The coding sequences of UDT1 and SLR1 were cloned into the EcoRI and PstI sites of the pGBKT7 vector to generate the BD-UDT1 and BD-SLR1 constructs. The coding sequences of SLR1 and TDR were cloned into the EcoRI and XhoI sites of the pGADT7 vector to generate the AD-SLR1 and AD-TDR constructs. The resulting constructs were transformed into yeast strain Y2H Gold. The presence of the plasmids was confirmed by growth on synthetic defined (SD) medium lacking Trp and Leu (SD–Trp–Leu). To assess protein interactions, the positive yeast clones were suspended in liquid SD–Trp–Leu medium to an OD600 = 1.0. The suspended cells and dilution series were spotted onto plates containing SD–Trp–Leu–His–Ade- medium. Interactions were observed after 3 days of incubation at 30°C.
LUC complementation imaging assays
The cLUC-SLR1, nLUC-UDT1, and nLUC-TDR constructs were generated using the primers listed in Supplemental Data Set S1. Agrobacterium (Agrobacterium tumefaciens) colonies harboring different combinations of constructs were co-infiltrated into N. benthamiana leaves, and the infiltrated leaves were analyzed for LUC activity using chemiluminescence imaging (Tanon 5200) 48 h after infiltration. The leaf was sprayed with 1 mmol d-luciferin and then imaged 10 min later.
BiFC assay
For BiFC assays, the OsSLR1, OsUDT1, and OsTDR coding sequences were cloned in-frame with the sequence encoding the N-terminal or C-terminal half of green fluorescent protein (GFP), and generated cGFP-OsSLR1, nGFP-OsUDT1, and nGFP-OsTDR constructs using the primers listed in Supplemental Data Set S1. The Agrobacterium strain GV3101 carrying corresponding vector combinations was infiltrated into young N. benthamiana leaves. The fluorescence was observed by confocal microscopy (Leica) after 3 days of growth.
Pull-down assay
The full-length coding sequence of SLR1 in pENTR/D-TOPO was subcloned into the expression vector pDEST15 to generate the GST-SLR1 fusion vector. The coding sequences of UDT1 and TDR were cloned into the pVP13 vector to generate the MBP-UDT1 and MBP-TDR constructs using the primers listed in Supplemental Data Set S1. The resulting vectors were transformed into E. coli (strain BL21) to produce the protein, and the fusion proteins were purified using the appropriate affinity chromatography. Recombinant MBP, MBP-UDT1, or MBP-TDR was incubated with beads for 2 h at 4°C and subsequently incubated with GST-SLR1 for 2 h. Beads were then washed thoroughly in PBS buffer, boiled in 1× SDS-PAGE sample buffer, and analyzed by immunoblot using anti-GST antibody (Abmart, M20007) and anti-MBP antibody (CWBIO, CW0288M), respectively.
Yield experiment
To investigate the potential use of WRKY53 for rice breeding, seeds for LJ11, w53-LJ11, Daohuaxiang2 (DHX2), w53-DHX2, Longdao16 (LD16), and w53-LD16 were sown in a paddy field under natural conditions in Heilongjiang and Hainan provinces. The following agronomic traits were investigated: thousand-grain weight, grain number per panicle, tiller number, seed setting rate, yield per plant, and yield per plot. Values are means ± se of the indicated number of plants.
Statistical analyses
Student’s two-tailed t test was used for significant difference analysis between two samples. One-way ANOVA analyses followed with Tukey’s test (P < 0.05) were used for pairwise multiple comparisons. All the analyses were performed using SPSS software. Data for all statistical analyses are shown in Supplemental Data Set S2.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: WRKY53 (Os05g0343400); GA20ox1 (Os03g0856700); GA20ox3 (Os07g0169700); GA3ox1 (Os05g0178100); GAMYB (Os03g0578900); CYP703A3 (Os08g0131100); UBIQUITIN (Os01g0328400); ACTIN1 (Os03g50885); GA2ox1 (Os05g0158600); GA2ox2 (Os01g0332200); GA2ox3 (Os01g0757200); KAO (Os06g0110000); KO2 (Os06g0570100); SLR1 (Os03g0707600); UDT1 (Os07g0549600); TDR (Os02g0120500); CP1(Os04g0670500); NCED3 (Os03g0645900); and NCED5 (Os12g0617400).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Panicles and seed setting rate of LJ11, wrky53, and WRKY53-OE plants.
Supplemental Figure S2. The wrky53 mutant shows enhanced CTB in multiple independent replications.
Supplemental Figure S3. The wrky53-1 mutant shows enhanced CTB in multiple independent replications.
Supplemental Figure S4. Exogenous application of GA improves rice CTB.
Supplemental Figure S5. SLR1 protein abundance in LJ11 and wrky53 plants under NCs and LT treatment.
Supplemental Figure S6. LT repressed the expression of GA20ox1, GA20ox3, and GA3ox1.
Supplemental Figure S7. Expression of GA catabolism genes in LJ11 and wrky53 plants.
Supplemental Figure S8. SLR1 protein abundance in LJ11 and WRKY53-OE plants grown under NCs.
Supplemental Figure S9. LT treatment does not affect the binding ability of WRKY53 to the promoters of GA biosynthesis genes.
Supplemental Figure S10. Generation and identification of ga20ox1, ga20ox3, ga3ox1, wrky53 ga20ox1, wrky53 ga20ox3, and wrky53 ga3ox1 mutants.
Supplemental Figure S11. Phenotypic analysis of ga20ox1, ga20ox3, ga3ox1, wrky53 ga20ox1, wrky53 ga20ox3, and wrky53 ga3ox1 mutants.
Supplemental Figure S12. Histological analysis of anther development in LJ11 and ga20ox1 mutant plants under NCs.
Supplemental Figure S13. SLR1 interacts with UDT1 and TDR in pull-down assays (supports Figure 6).
Supplemental Figure S14. UDT1 can bind to the CP1 promoter directly in vitro.
Supplemental Figure S15. Expression of CP1 in LJ11 and WRKY53-OE plants.
Supplemental Figure S16. wrky53 shows no accompanying yield penalty.
Supplemental Figure S17. wrky53 has higher yield under cold stress at the booting stage.
Supplemental Figure S18. Generation and identification of wrky53 mutants in DHX2 and LD16.
Supplemental Figure S19. Phenotypic analysis of wrky53 mutants in DHX2.
Supplemental Figure S20. Phenotypic analysis of wrky53 mutants in LD16.
Supplemental Figure S21. wrky53 mutants in both DHX2 and LD16 show increased CTB in multiple independent replications.
Supplemental Figure S22. BRs are not involved in rice CTB.
Supplemental Figure S23. WRKY53 does not play a role in cold tolerance at the seedling stage.
Supplemental Data Set S1. Primers used in this study.
Supplemental Data Set S2. ANOVA data for this study.
Supplemental File S1. Tissue-specific expression of diverse GA biosynthesis genes.
Supplemental File S2. Representative chromatograms for diverse GA components.
Supplementary Material
Acknowledgments
We thank Dr Xiangbing Meng for rice transformation and Prof. Yihua Zhou for providing antibodies.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31871591), Natural Science Foundation of Heilongjiang (Grant No. JQ2020C003), National Natural Science Foundation of China-Heilongjiang Joint Fund (Grant No. U20A2025), Youth Innovation Promotion Association CAS (Grant No. 2021229), and Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA24040102).
Conflict of interest statement. The authors declare no conflicts of interest.
Contributor Information
Jiaqi Tang, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Xiaojie Tian, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China.
Enyang Mei, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Mingliang He, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Junwen Gao, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Jun Yu, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Min Xu, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jiali Liu, Key Laboratory of Saline-Alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
Lu Song, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Xiufeng Li, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China.
Zhenyu Wang, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China.
Qingjie Guan, Key Laboratory of Saline-Alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
Zhigang Zhao, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Chunming Wang, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing 210095, China.
Qingyun Bu, Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Harbin 150081, China; The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Q.B. and X.T. conceived and designed the experiments. X.T. and J.T. performed most of the experiments with help from E.M., M.H., J.G., J.Y., M.X., J.L., L.S., X.L., Z.W., Q.G., and Z.Z. C.W., Q.B., and X.T. analyzed the data and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Qingyun Bu (buqingyun@iga.ac.cn).
References
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tian X, Xue L, Zhang X, Yang S, Traw MB, Huang J (2019) CRISPR-based assessment of gene specialization in the gibberellin metabolic pathway in rice. Plant Physiol 180: 2091–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, et al. (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Daviere JM, Achard P (2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA 105: 12623–12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zeng Y, Li J, Ma X, Zhang Z, Lou Q, Li J, Gu Y, Zhang H, Li J, et al. (2020) Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol J 18: 2491–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P (2012) Gibberellin Biosynthesis. eLS.
- Hedden P (2020) The current status of research on gibberellin biosynthesis. Plant Cell Physiol 61: 1832–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, Hasegawa Y, Ueguchi-Tanaka M, Matsuoka M (2008) Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol 49: 1429–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Ye M, Li R, Lou Y (2016) OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant Signal Behav 11: e1169357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, et al. (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice undeveloped tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23: 578–587 [DOI] [PubMed] [Google Scholar]
- Kuroki M, Saito K, Matsuba S, Yokogami N, Shimizu H, Ando I, Sato Y (2007) A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8. Theor Appl Genet 115: 593–600 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou W, Ren Y, Tian X, Lv T, Wang Z, Fang J, Chu C, Yang J, Bu Q (2017) High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J Genet Genomics 44: 175–178 [DOI] [PubMed] [Google Scholar]
- Li X, Liu H, Wang M, Liu H, Tian X, Zhou W, Lu T, Wang Z, Chu C, Fang J, et al. (2015) Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J Integr Plant Biol 57: 698–707 [DOI] [PubMed] [Google Scholar]
- Liu C, Schlappi MR, Mao B, Wang W, Wang A, Chu C (2019) The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol J 17: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Cao S, Schlappi MR, Zhao B, Xiao G, et al. (2018) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun 9: 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, et al. (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015a) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. (2015b) COLD1 confers chilling tolerance in rice. Cell 160: 1209–1221 [DOI] [PubMed] [Google Scholar]
- Mao D, Xin Y, Tan Y, Hu X, Bai J, Liu ZY, Yu Y, Li L, Peng C, Fan T, et al. (2019) Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc Natl Acad Sci USA 116: 3494–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masatoshi N, Isomaro Y, Satoru K, Noboru M, Nobutaka TJP, Physiology C (1991) Semi-quantification of GAX and GA4 in male-sterile anthers of rice by radioimmunoassay. Plant Cell Physiol 32: 511–513
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9: 11–17 [DOI] [PubMed] [Google Scholar]
- Oda S, Kaneko F, Yano K, Fujioka T, Masuko H, Park JI, Kikuchi S, Hamada K, Endo M, Nagano K, et al. (2010) Morphological and gene expression analysis under cool temperature conditions in rice anther development. Genes Genet Syst 85: 107–120 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M (2004) A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol 55: 687–700 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hayano-Saito Y, Kuroki M, Sato YJPE (2010) Map-based cloning of the rice cold tolerance gene Ctb1 179: 97–102 [Google Scholar]
- Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109: 515–522 [DOI] [PubMed] [Google Scholar]
- Saito K, Miura K, Nagano K, Hayano-Saito Y, Araki H, Kato A (2001) Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor Appl Genet 103: 862–868 [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T, Oda S, Tsunaga Y, Shomura H, Kawagishi-Kobayashi M, Aya K, Saeki K, Endo T, Nagano K, Kojima M, et al. (2014) Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol 164: 2011–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Ge S (2007) Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev 17: 533–538 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Tian X, Li X, Zhou W, Ren Y, Wang Z, Liu Z, Tang J, Tong H, Fang J, Bu Q (2017) Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol 175: 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, He M, Mei E, Zhang B, Tang J, Xu M, Liu J, Li X, Wang Z, Tang W, et al. (2021) WRKY53 integrates classic brassinosteroid signaling and the mitogen-activated protein kinase pathway to regulate rice architecture and seed size. Plant Cell 33: 2753–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaya V, Mackill D (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theor Appl Genet 106: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Wang B, Fang R, Chen F, Han J, Liu YG, Chen L, Zhu Q (2020) A novel CCCH-type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice. J Integr Plant Biol 62: 1594–1606 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chinese Sci Bull 54: 2342–2353 [Google Scholar]
- Xie W, Ke Y, Cao J, Wang S, Yuan M (2021) Knock out of transcription factor WRKY53 thickens sclerenchyma cell walls, confers bacterial blight resistance. Plant Physiol 187: 1746–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang R, Wang Y, Zhang L, Yao S (2020) A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ 43: 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yan MY, Xie DL, Cao JJ, Xia XJ, Shi K, Zhou YH, Zhou J, Foyer CH, Yu JQ, et al. (2020) Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J 102: 931–947 [DOI] [PubMed] [Google Scholar]
- Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X (2010) Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan DP, Zhang C, Wang ST, Liu Y, Xuan Y (2019) Transcriptome analysis of rice leaves in response to Rhizoctonia solani infection.
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Pan Y, Li J, Zhou L, Shi H, Zeng Y, Guo H, Yang S, Zheng W, et al. (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun 8: 14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zeng Y, Zheng W, Tang B, Yang S, Zhang H, Li J, Li Z (2010) Fine mapping a QTL qCTB7 for cold tolerance at the booting stage on rice chromosome 7 using a near-isogenic line. Theor Appl Genet 121: 895–905 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Chen K, Mi X, Chen T, Ali J, Ye G, Xu J, Li Z (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS ONE 10: e0145704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.