Abstract
Lipid droplets (LDs) stored during seed development are mobilized and provide essential energy and lipids to support seedling growth upon germination. Triacylglycerols (TAGs) are the main neutral lipids stored in LDs. The lipase SUGAR DEPENDENT 1 (SDP1), which hydrolyzes TAGs in Arabidopsis thaliana, is localized on peroxisomes and traffics to the LD surface through peroxisomal extension, but the underlying mechanism remains elusive. Here, we report a previously unknown function of a plant-unique endosomal sorting complex required for transport (ESCRT) component FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1) in regulating peroxisome/SDP1-mediated LD turnover in Arabidopsis. We showed that LD degradation was impaired in germinating free1 mutant; moreover, the tubulation of SDP1- or PEROXIN 11e (PEX11e)-marked peroxisomes and the migration of SDP1-positive peroxisomes to the LD surface were altered in the free1 mutant. Electron tomography analysis showed that peroxisomes failed to form tubules to engulf LDs in free1, unlike in the wild-type. FREE1 interacted directly with both PEX11e and SDP1, suggesting that these interactions may regulate peroxisomal extension and trafficking of the lipase SDP1 to LDs. Taken together, our results demonstrate a pivotal role for FREE1 in LD degradation in germinating seedlings via regulating peroxisomal tubulation and SDP1 targeting.
A plant-unique ESCRT component FREE1 mediates lipid droplet degradation in germinating seedlings by interacting with PEX11e and SDP1 to regulate peroxisomal tubulation and targeting of SDP1 to the lipid droplet.
IN A NUTSHELL.
Background: Lipid droplets (LDs) are major reservoirs providing energy and lipids for seedling establishment. In vascular plants, the peroxisome-localized lipase SUGAR DEPENDENT 1 (SDP1) has been shown to play a major role in modulating LD metabolism, while the retromer and endosomal sorting complex required for transport (ESCRT) III components play a role in LD metabolism via regulating peroxisomal extension and peroxisomal inner membrane formation, respectively, in Arabidopsis. However, how the peroxisome-localized SDP1 traffics to the LD surface for turnover remains elusive. We previously characterized a plant-unique ESCRT component FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1) and showed multiple functions in the endomembrane system and autophagy. Here we report a new role of FREE1 in LD turnover.
Question: Depletion of FREE1 led to accumulation of LDs in germinating seedlings with unknown mechanism. This study aims to illustrate the underlying mechanism of FREE1 function in regulating LD turnover.
Findings: We showed that both the formation of SDP1- or PEROXIN 11e (PEX11e)-positive tubules and the engulfment of LDs by the peroxisomes were impaired in free1 mutants. The degradation of LDs was also impaired in free1 mutants. We further showed that FREE1 interacts specifically with the amphipathic helix (AH) motif of PEX11e and SDP1 to regulate peroxisomal tubulation and SDP1 targeting to the LD surface.
Next steps: Future work should investigate the possible involvements and mechanisms of ESCRT-I and other ESCRT components in regulating peroxisome tubule formation and LD mobilization in plants.
Introduction
In seed plants, lipid droplets (LDs) are major reservoirs providing energy and essential lipid-derived molecules such as phospholipids, phytohormones, and oxylipins, that support seedling growth during germination and survival under environmental stresses (Shimada and Hara-Nishimura, 2015; Pyc et al., 2017a). In plants, triacylglycerols (TAGs) are the major neutral storage lipids in LDs, whereas oleosins (Siloto et al., 2006; Huang and Huang, 2015; Huang and Huang, 2017), caleosins (Naested et al., 2000; Poxleitner et al., 2006), LD-associated proteins (LDAPs; Gidda et al., 2016; Kim et al., 2016), and LDAP-interacting protein (Pyc et al., 2017b) are the major decorators on the LD surface. These surface decorators are highly dynamic during LD mobilization. Recently, it has been revealed that the ubiquitinated oleosins on the LD surface are disassociated from the surface by cooperation of PLANT UBX-DOMAIN CONTAINING FAMILY PROTEIN10 and the AAA ATPase CELL DIVISION CYCLE 48, followed by lipolysis to release energy for seedling growth (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). In land plants, SUGAR DEPENDENT 1 (SDP1), a patatin domain-containing protein lipase, plays a major role in the lipolytic pathway to modulate TAG metabolism and release free fatty acid (FA) for maintaining seedling growth during seed germination (Eastmond, 2006; Kelly et al., 2011, 2013). In mammals, the delivery of lipase to the LD surface is coordinated by coat protein complex I (COPI) and coat protein complex II (COPII) vesicles; however, in plants, SDP1 is localized on peroxisomes and moved to the LD surface likely through retromer-regulated peroxisomal extension (Soni et al., 2009; Thazar-Poulot et al., 2015; Huang et al., 2019b).
Peroxisomes are ubiquitous, membrane-bound organelles that contain enzymes essential for multiple metabolic pathways to support cell growth (Hu et al., 2012; Farre et al., 2019). In plants, β-oxidation of FAs, hormone biosynthesis, and nitrogen metabolism occur in peroxisomes (Hu et al., 2012). Peroxisomes are in close contact with LDs, and the peroxisome–LD contact sites are thought to facilitate FA trafficking to peroxisomes to prevent oxidative toxicity arising from the accumulation of cytosolic FAs (Binns et al., 2006; Barbosa et al., 2015; Pyc et al., 2017a). Peroxins, referred to as PEXs, have been implicated in peroxisome biogenesis through division of preexisting peroxisomes or de novo formation from the endoplasmic reticulum (Farre et al., 2019). The division of peroxisomes is initiated by a membrane remodeling process (Opalinski et al., 2011b, 2011a). Moreover, PEX11s contain a conserved N-terminus sequence that adopts an amphipathic helix (AH) structure essential for peroxisomal membrane remodeling in animals (Opalinski et al., 2011b; Yoshida et al., 2015). In Arabidopsis thaliana, PEX11s likely play roles in modulating peroxisomal tubule formation and fission (Lingard and Trelease, 2006; Orth et al., 2007). Nevertheless, the molecular mechanisms underlying peroxisomal extension and migration to the LD surface for LD homeostasis remain unknown in plants.
The endosomal sorting complex required for transport (ESCRT) machinery consisting of ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, with several accessory components, are evolutionarily conserved in eukaryotes and participate in many cellular processes (Gao et al., 2017). Recent evidence has shown that ESCRTIII components are required to release preperoxisomal vesicles from the ER to mediate peroxisome biogenesis to regulate LD metabolism in yeast (Mast et al., 2018). Membrane-shaping ESCRTIII components are recruited by M1 spatstin (a membrane-bound AAA ATPase) to reshape the LD membrane and facilitate FA trafficking from LD to peroxisomes in mammalian cells (Chang et al., 2019; Henne, 2019). Other ESCRT components, such as Vps27, play functional roles in regulating LD degradation through the incorporation of LDs into the vacuole via an interaction with clathrin to translocate into the vacuole in yeast (Oku et al., 2017). In plants, Arabidopsis ESCRTIII components are essential for peroxisomal inner membrane formation and LD metabolism (Wright and Bartel, 2020).
We have recently characterized a plant-unique ESCRT component termed FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1)/FYVE1, which is necessary for seedling development and plays important roles in regulating multivesicular body (MVB) biogenesis, MVB–vacuole trafficking, autophagosome–vacuole fusion, and vacuole formation in Arabidopsis (Gao et al., 2014, 2015; Reyes et al., 2014; Kolb et al., 2015). A detailed examination of transmission electron microscope (TEM) images indicated extensive accumulation of LDs in the root cells of germinating free1 mutant compared with those of the wild-type (WT; Gao et al., 2015), indicating a possible role for FREE1 in regulating LD degradation in germinating Arabidopsis seedlings.
In this study, we confirmed that LDs accumulated in germinating seedlings of the free1 mutants as compared with those of WT. We also generated transgenic Arabidopsis plants expressing GFP–SDP1 or mCherry-PEROXIN 11e (PEX11e) in WT or the free1 mutant background to follow the fate and dynamics of SDP1 and PEX11e in germinating seedlings. Distinct from WT, GFP–SDP1 or mCherry-PEX11e in germinating free1 mutants failed to form visible tubules, and the migration of SDP1 to the LD surface was also impaired. Electron tomography (ET) analysis with nanometer resolution showed that peroxisomes failed to form tubules to engulf LDs in free1 mutant, which is distinct from the WT in which LDs were encapsulated by peroxisomes. Further pull down and co-immunoprecipitation (co-IP) analysis showed that FREE1 interacted directly with both PEX11e and SDP1 to regulate peroxisomal tubulation and trafficking of lipase SDP1 to LD. We have thus revealed a pivotal role of FREE1 in mediating LD degradation in germinating seedlings via promoting peroxisomal tubulation and SDP1 targeting. This work also demonstrates that a plant-specific ESCRT component functions in a nonendosomal organelle.
Results
Peroxisome tubulation and migration to LD is defective in free1
We have previously shown that the plant-unique ESCRT component FREE1 regulates MVB and vacuole formation in plants, as the free1 mutant shows defects in ILV formation and exhibits fragmented vacuoles in root cells (Gao et al., 2014, 2015; Kolb et al., 2015). A subsequent closer examination of the TEM images indicated a possibility of LD accumulation in germinating free1 mutants as compared with the WT (Gao et al., 2015). Indeed, further detailed TEM analysis on germinating seedlings of WT and free1 mutants confirmed the accumulation of LDs in free1 mutants (Supplemental Figure S1). These results indicate a possible role of FREE1 in regulating LD degradation in Arabidopsis.
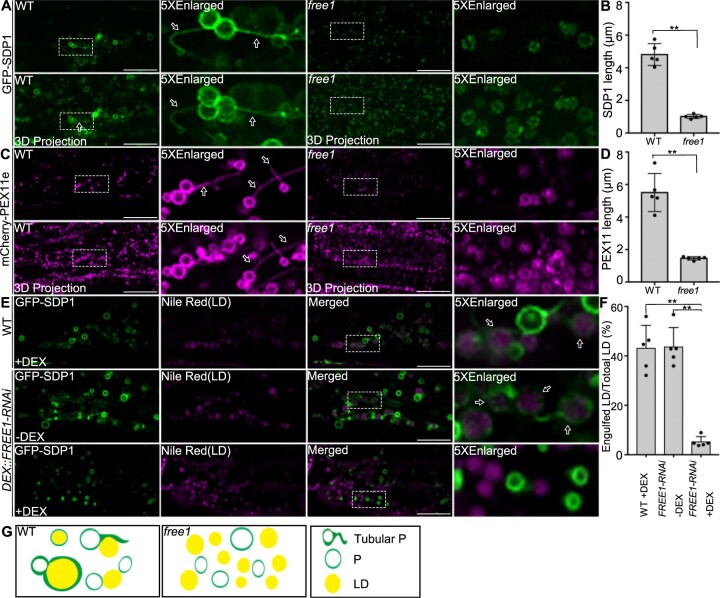
The lipolytic pathway, which involves the lipase SDP1 and peroxisomal tubulation for engulfing LDs, is mainly responsible for LD mobilization during seed germination in Arabidopsis (Eastmond, 2006; Kelly et al., 2011, 2013). In mammals and yeast, lipophagy is also an important pathway for LD turnover, in which LDs are sequestrated into lysosomes via macrolipohagy or microlipophagy (Singh et al., 2009; Velikkakath et al., 2012; Schepers and Behl, 2021). In plants, LDs can be delivered to vacuoles for degradation via the autophagy-dependent pathway in Arabidopsis leaves under dark conditions (Fan et al., 2019). To explore the possible role of lipophagy in LD turnover, we performed mobilization profile analysis of LDs in germinating seedlings of autophagy-deficient mutants atg5 and atg7 under normal conditions. Our results showed that LD degradation was not affected in both atg5 and atg7 mutants (Supplemental Figure S2), indicating that lipophagy plays a minor role in LD mobilization in comparison with the lipolytic pathway under normal growth conditions in Arabidopsis. Furthermore, to investigate if FREE1 depletion would affect the proper targeting of SDP1 and peroxisomal tubulation, we generated transgenic Arabidopsis plants expressing GFP–SDP1 in the WT or the free1 mutant background or the DEX::FREE1-RNAi line for DEX-induced knockdown (KD) of FREE1 (Gao et al., 2015) for investigation of the SDP1-positive peroxisomal tubulation. In the hypocotyl cells of 4-day-old transgenic Arabidopsis seedlings expressing GFP–SDP1, both ring-like structures and tubular structures, which likely represent peroxisomes and peroxisomal tubulation, respectively, were detected in WT cells (Figure 1A, left; Supplemental Figure S3A, upper and middle parts). These results are consistent with a previous study (Thazar-Poulot et al., 2015). In contrast, in hypocotyl cells of transgenic Arabidopsis free1 GFP–SDP1 or DEX-treated DEX::FREE1-RNAi GFP–SDP1 seedlings, albeit showing similar ring-like structures, GFP–SDP1 did not show obvious tubular structures in free1 mutant or DEX::FREE1-RNAi with DEX induction (Figure 1A, right; Supplemental Figure S3A, lower). Further quantification analysis confirmed that the SDP1-positive tubules were significantly longer in WT than those in free1 mutants or in DEX-treated DEX::FREE1-RNAi lines (Figure 1B; Supplemental Figure S3B). Taken together, these results suggested that FREE1 is required for the formation of SDP1-positive tubules.
Figure 1.
FREE1 regulates peroxisomal tubule formation and peroxisome encapsulated LDs. A, The formation of SDP1-positive tubules is defective in free1 mutants. GFP–SDP1 in WT and free1 mutants were germinated on MS medium for 4 days (D), followed by confocal imaging of the seedling hypocotyl region. Fifteen slices were collected in a total thickness of 15 μm for generating the 3D projection image. Arrows indicate examples of peroxisome tubulation. Dotted box indicates the area of enlargement. D, days after germination. Scale bars, 20 μm. B, Quantification of SDP1-positive tubule size in WT and free1 mutants in germinating seedlings shown in (A). The lengths of SDP1-positive tubules were analyzed using ImageJ software (NIH). The total number of 92 and 41 tubules were counted from the whole images of five individual hypocotyls of WT and free1 mutant, respectively. Error bars represent the S.D. from five individual seedlings, **P < 0.01 in Student’s t test. C, The formation of PEX11e-positive tubules is defective in free1 mutants. Transgenic Arabidopsis WT mCherry-PEX11e and free1 mCherry-PEX11e seeds were germinated in MS medium for 4 days (D), followed by confocal imaging of the hypocotyl region. Fifteen slices were collected in a total thickness of 15 μm for generating the 3D projection image. Dotted box indicated the area of enlargement. Scale bars, 20 μm. D, Quantification of PEX11e-positive tubule size in WT and free1 mutants in germinating seedlings shown in (C). The lengths of PEX11e-positive tubules were analyzed using ImageJ software (NIH). The total number of 112 and 72 tubules were counted from the whole images of five individual hypocotyls of WT and free1 mutant, respectively. Error bars represent the sd from five individual seedlings, **P < 0.01 in Student’s t test. E, The trafficking of SDP1 to LDs is defective in free1 mutants. Transgenic DEX::FREE1-RNAi GFP–SDP1 or WT GFP–SDP1 seeds were germinated in MS medium with or without DEX for 4 days (D), followed by LD staining using Nile Red dye and subsequent confocal imaging of hypocotyl regions showing GFP–SDP1 and LD. Dotted box indicates the area of enlargement. Arrows indicate examples of peroxisome-engulfed LD. Scale bars, 20 μm. F, The percentage of GFP–SDP1 engulfed LDs in DEX::FREE1-RNAi GFP–SDP1 shown in (E) are quantified. The total number of 267, 236, and 665 LDs were counted from the whole images of five individual hypocotyls of WT (+DEX), DEX::FREE1-RNAi (−DEX) and DEX::FREE1-RNAi (+DEX), respectively. The total number of 118, 101, and 33 SDP1-engulfed LDs were counted from the whole images of five individual hypocotyls of WT (+DEX), DEX::FREE1-RNAi (−DEX) and DEX::FREE1-RNAi (+DEX), respectively. Error bars represent the sd from five individual seedlings, **P < 0.01 in Student’s t test. G, Working model of FREE1 function in regulating peroxisome tubulation in germinating seedlings. FREE1 regulates the tubulation of peroxisome and the trafficking of SDP1 to LD surface for TAG hydrolysis. However, the peroxisome tubulation and the trafficking of SDP1 to LD surface were impeded in free1 mutant. P, Peroxisome; Tubular P, tubular peroxisome.
To further explore if the defective SDP1 tubules in the free1 mutants were caused by impaired peroxisomal tubulation, we also crossed the free1 mutant or DEX::FREE1-RNAi line with a plant expressing mCherry-PEX11e, which is a peroxisomal membrane protein belonging to the PEX11 family and plays key roles in peroxisomal proliferation and tubulation (Orth et al., 2007). Indeed, mCherry-PEX11e mainly labeled long tubules along with punctae and ring-like structures in WT background (Figure 1C, WT—left; Supplemental Figure S3C, upper and middle parts), as expected for peroxisomal tubulation and proliferation (Orth et al., 2007). However, mCherry-PEX11e in the free1 mutant background or in DEX-treated DEX::FREE1-RNAi line showed mainly punctae and ring-like structures but few tubules (Figure 1C, right; Supplemental Figure S3C, lower). Further quantification analysis showed that the length of mCherry-PEX11e tubules in WT was significantly longer than those in free1 mutant or in DEX-treated DEX::FREE1-RNAi line (Figure 1D; Supplemental Figure S3D). Taken together, these results (Figure 1, A–D; Supplemental Figure S3, A–D) indicated that FREE1 is required for peroxisomal tubulation in Arabidopsis.
To further study the effects of FREE1 depletion on the relationship between LDs and peroxisomes in germinating seedlings, we used Nile Red to stain LDs in germinating seedlings of DEX::FREE1-RNAi GFP–SDP1 in the presence or absence of DEX or free1 GFP–SDP1. As shown in Figure 1E, in DEX-treated WT GFP–SDP1 or non-DEX-treated DEX::FREE1-RNAi GFP–SDP1 or WT GFP–SDP1, GFP–SDP1 showed typical ring-like and tubular structures, and interestingly, many of the Nile Red-stained LDs were engulfed by the GFP–SDP1 (Figure 1E, upper and middle parts; Supplemental Figure S3E, upper), indicating the engulfment of LDs by the peroxisomes. In contrast, in DEX-induced KD of FREE1 or free1 mutant background, GFP–SDP1 exhibited mainly ring-like structures with little tubulation and the Nile Red-stained LDs were found largely outside and separated from the GFP–SDP1-positive ring-like structures (Figure 1E, lower; Supplemental Figure S3E, lower). Further quantification analysis confirmed these observations on the distinct relationship between SDP1 and LDs in WT versus KD (+DEX) or free1 mutant (Figure 1F; Supplemental Figure S3F). In summary, our confocal imaging data showed that both the formation of SDP1- or PEX11e-positive tubules and the engulfment of LDs by the peroxisomes were impaired in free1 mutants (Figure 1G).
ET analysis of relationship between peroxisomes and LDs in WT and free1 mutant
To further explore the above confocal results on the peroxisome–LD relationship at the ultracellular level, we first performed TEM analysis in hypocotyls of 4-day-old germinating seedlings of WT GFP–SDP1 and free1 GFP–SDP1; however, we failed to obtain complete sections containing sufficient numbers of LDs and peroxisomes for quantification, likely owing to the existence of large vacuoles in the hypocotyl cells. We next performed TEM analysis in root tip cells of 1-day-old germinating seedlings of WT GFP–SDP1 and free1 GFP–SDP1. We first performed TEM analysis of multiple cells of WT GFP–SDP1 and free1 GFP–SDP1, showing defective peroxisomal tubulation in free1 GFP–SDP1 (Supplemental Figure S4). We also performed TEM analysis on the DEX::FREE1-RNAi GFP–SDP1 line with or without DEX treatment. Our data showed that few peroxisome-engulfed LDs were observed in DEX::FREE1-RNAi GFP–SDP1 line upon DEX induction in comparison with the control (Supplemental Figure S5). To further explore the nature of LD–peroxisome association, we performed immunogold-TEM analysis with GFP antibodies on ultrathin sections of high-pressure frozen/freeze-substituted 1-day-old WT GFP–SDP1 root tip cells, showing specific gold-particle labeling on peroxisomes that engulf LDs (Supplemental Figure S6).
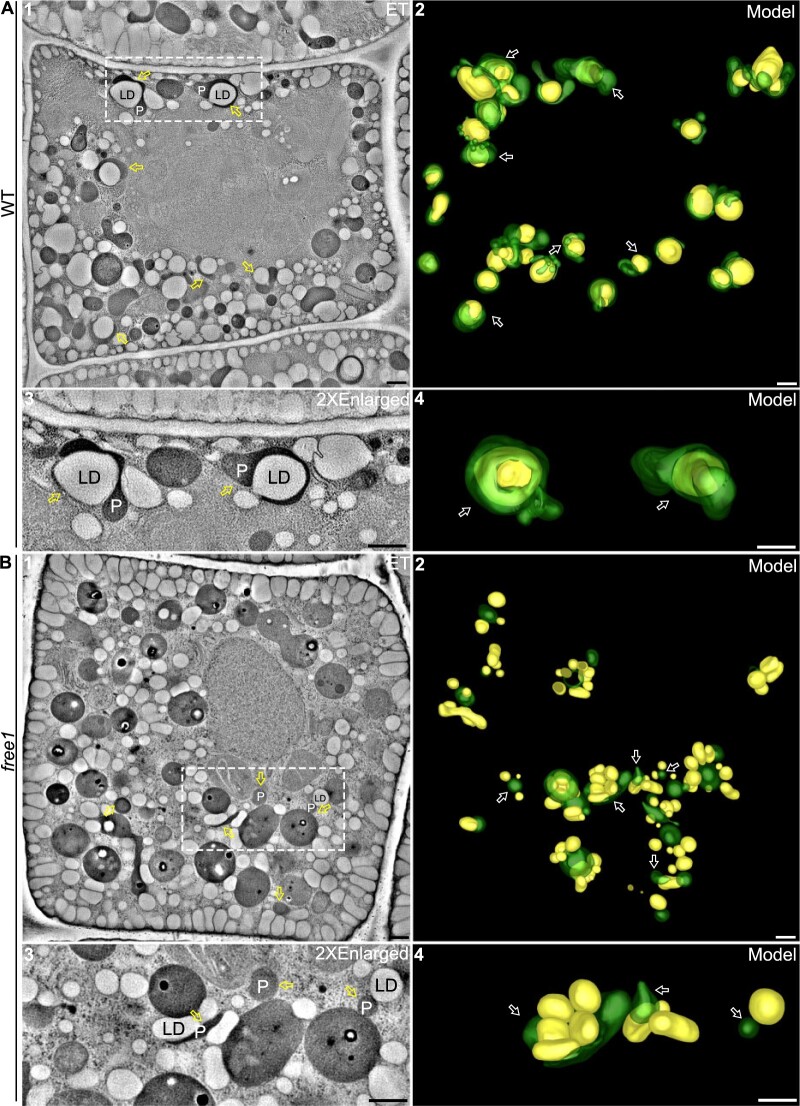
ET analysis with nanometer resolution has become a powerful tool to elucidate organelle relationships and organelle biogenesis (Zhuang et al., 2017; Cui et al., 2019; Cao et al., 2022). We performed ET analysis on two representative cells of 14 continuous 300-nm-thick sections from the TEM-analyzed WT GFP–SDP1 and free1 GFP–SDP1, respectively. As shown in Figure 2A, a representative tomographic slice of a WT cell showed the presence of numerous LDs and peroxisomes, with various profiles of tubulation as well as LD–peroxisome association (Figure 2A, parts 1 and 3), and the corresponding ET models further illustrated various stages or profiles of possible engulfment of LDs by the peroxisomal tubules and their tight association (Figure 2A, parts 1–4; Supplemental Movie S1). Moreover, we observed different stages of contact between peroxisomes and LDs, such as peroxisomes with short tubules enwrapping only a small surface of LDs, peroxisomes with long tubules wrapping around half of the LD surface, and peroxisomes nearly completely engulfing the whole surface of LDs, confirming that peroxisome–LD groupings are tightly associated with profiles of engulfment of LDs by peroxisomes in WT (Figure 2A). In addition, serial sections of representative samples showed the nearly complete engulfment of LDs by peroxisomes in WT (Supplemental Figure S7A).
Figure 2.
ET analysis of peroxisome–LD relationship in germinating seedlings of WT and free1 mutant. A, ET analysis of 14 continuous 300-nm-thick sections revealed relationships between peroxisomes and LDs in germinating seedlings of 1-day-old WT GFP–SDP1 root tip cells. The corresponding 3D model (part 2) shows the various profiles and stages of possible engulfment of LDs (yellow) by the tubular peroxisomes (green) in WT cells. The dashed box (part 1) and the corresponding 3D model are enlarged (parts 3 and 4). Arrows indicate examples of peroxisome-engulfed LDs. Peroxisomes adjacent to LDs are depicted in the 3D models. ET data were collected at 6.3099 nm/pixel. Scale bars, 500 nm. B, ET analysis of 14 continuous 300-nm-thick sections revealed relationships between peroxisomes and LDs in germinating seedlings of 1-day-old free1 GFP–SDP1 mutant root tip cells. The corresponding 3D model (part 2) shows the largely separated profiles of LDs (yellow) and nontubulated or less-tubulated peroxisomes (green) in free1 mutant cells. The dashed box (part 1) and the corresponding 3D model are enlarged (parts 3 and 4). Arrows indicate examples of no engulfment of LDs by the peroxisomes. Peroxisomes adjacent to LDs are depicted in the 3D models. ET data were collected at 6.3099 nm/pixel. Scale bars, 500 nm.
In contrast, when similar ET analysis was performed in root tip cells of 1-day-old germinating seedlings of the free1 mutant, different results were obtained (Figure 2B). Both the representative tomographic slice and the corresponding models of a free1 mutant cell showed few elongated peroxisomes and little surface tethering between peroxisomes and LDs (Figure 2B; parts 1–4; Supplemental Movie S2), albeit with rare observations of peroxisomes surrounding with multiple LDs with few contact areas (Figure 2B, part 2, indicated by asterisk). Additional enlarged serial sections of representative samples further confirmed that little tethering or engulfing between peroxisomes and LDs was seen in the in free1 mutant (Supplemental Figure S7B). These few surface attachments between peroxisomes and LDs in the free1 mutant may decrease the efficiency of LD mobilization, leading to the delay or impairment of LD degradation in free1 mutants. Taken together, the results of the ET analysis (Figure 2) are consistent with the confocal data (Figure 1; Supplemental Figure S3) and support the notion that FREE1 is required for the peroxisomal tubulation and the peroxisomal engulfment of LDs as well as proper SDP1 targeting to LDs.
FREE1 interacts directly with both PEX11e and SDP1 to mediate LD degradation
Our results so far showed that both SDP1-positive tubules and PEX11e-positive tubules were defective in free1 mutants (Figure 1; Supplemental Figure S3), and such defects of peroxisomal tubulation in the free1 mutant were further verified by ET analysis (Figure 2). To investigate the possible dynamic relationship between FREE1 and SDP1 or PEX11e in living cells, we also generated double transgenic Arabidopsis plants co-expressing GFP-FREE1 mCherry-SDP1 or GFP-FREE1 mCherry-PEX11e. Indeed, FREE1 showed a close association with both SDP1-positive and PEX11e-positive tubules in live-cell confocal imaging analysis (Supplemental Figure S8).
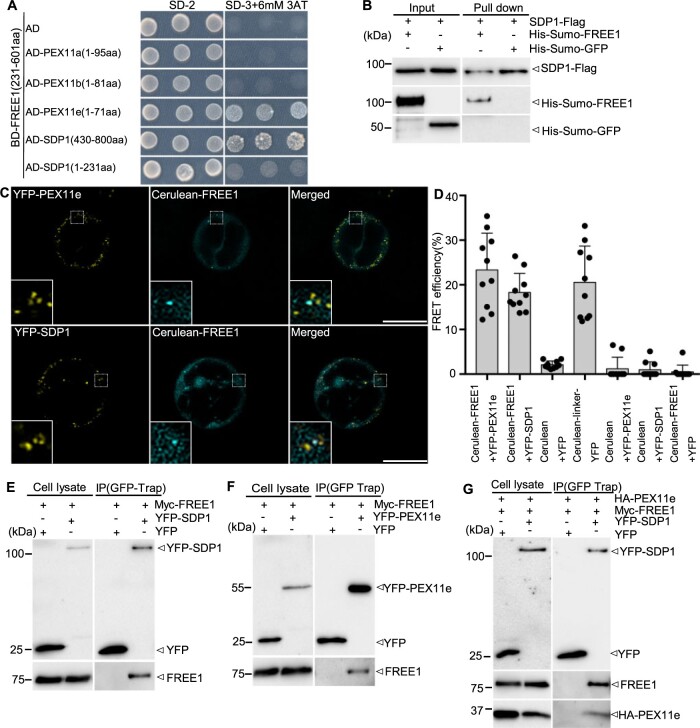
We next performed biochemical analysis to explore the possible molecular links among FREE1, PEX11e, and SDP1. A protease protection assay using microsomes from Arabidopsis cells expressing either SDP1–GFP or YFP–SDP1 confirmed the predicted topology of SDP1 that both the C-terminus and the N-terminus of SDP1 face the cytosol (Supplemental Figure S9, A and B). Further fractionation analysis of Arabidopsis cells expressing SDP1–GFP demonstrated that SDP1 is an integral membrane protein (Supplemental Figure S9C). We then expressed and purified recombinant proteins of His-Sumo-FREE1, His-Sumo-GFP, and Flag–SDP1 for further in vitro binding assays (Supplemental Figure S9D). PEX11s are peroxisomal membrane proteins that play important roles in peroxisomal proliferation and tubulation (Orth et al., 2007). Phylogenetic analysis showed that the N-termini of PEX11e, PEX11c, and PEX11d are highly conserved among themselves, but both PEX11a and PEX11b are less similar to PEX11e, PEX11c, and PEX11d (Lingard and Trelease, 2006; Supplemental Figure S10). Based on these results, and since SDP1 and PEX11 proteins are transmembrane proteins, we truncated the N-terminus or C-terminus of SDP1 and N-terminus of PEX11s which are facing in the cytosol for yeast two-hybrid (Y2H) analysis. Since the full-length FREE1 protein showed high self-activation activity, we used the truncated FREE1 (amino acids 231–601) as bait (Li et al., 2019). Y2H analysis showed that FREE1 interacted with both the C-terminus of SDP1 and the N-terminus of PEX11e, but not with the N-terminus of SDP1 nor the N-terminus of PEX11a and PEX11b (Figure 3A; Supplemental Figure S11A). We also generated several FREE1 domain truncations for Y2H analysis, showing that the truncated FYVE domain and the coiled-coil domain of FREE1 cannot interact with SDP1 and PEX11e (Supplemental Figure S11, B–D). Further in vitro binding assays showed that Flag–SDP1 pulls down with His-Sumo-FREE1 but not with His-Sumo-GFP, indicating a direct interaction of FREE1 with SDP1 (Figure 3B). The interaction between FREE1 and SDP1 or PEX11e was further confirmed by acceptor photobleaching fluorescence resonance energy transfer (FRET-AB) analysis, as FREE1 showed an interaction with PEX11e or SDP1 in the colocalized punctae, respectively (Figure 3, C and D). Moreover, a co-IP assay further confirmed the specific interaction of FREE1 with SDP1, FREE1 with PEX11e in planta, as FREE1 was immunoprecipitated by both YFP–SDP1 (Figure 3E) and YFP-PEX11e (Figure 3F), and PEX11e and FREE1 were also immunoprecipitated together by YFP–SDP1 (Figure 3G).
Figure 3.
FREE1 interacts with both PEX11e and SDP1. A, Y2H analysis of the binary interaction between FREE1 and PEX11e or SDP1. Transformed yeast cells were grown on synthetic complete medium lacking leucine and tryptophan (SD-2) as a control, or on SD-3 (without histidine, −His) +3-AT for interaction assays, and three colonies were picked for interaction assay at the same dilution. 3-AT is a chemical used to suppress self-activation of the BD genes. B, In vitro pull-down assay demonstrated the direct interaction between SDP1 and FREE1. His-Sumo-FREE1 and His-Sumo-GFP were incubated with SDP1–Flag and pulled down by Flag beads. Samples of input (left) and pull-down (right) were separated by SDS–PAGE, followed by immunoblot analysis with indicated antibodies. Arrowhead indicated His-Sumo-FREE1 protein pulled down by SDP1–Flag. C and D, FRET analysis showed direct interaction between FREE1 and PEX11e or SDP1 in vivo. Panels as well as 5 × enlarged views in the dashed box are images before FRET. FRET efficiency was quantified by using the (similar) punctae shown in the left bottom inlets of the merged panel. FRET efficiency was quantified as follows: FRETeff = (Dpost − Dpre)/Dpost where Dpost is the fluorescence intensity of the donor after photobleaching and Dpre is the fluorescence intensity of the donor before photobleaching. For each group, 10 individual protoplasts were used for FRET efficiency quantification and statistical analysis. Error bars are the sd of FRET efficiency. Scale bars, 25 μm. E–G, Co-IP assay showing interaction among SDP1, FREE1, and PEX11e. Transformed Arabidopsis protoplasts expressing the indicated constructs were subjected to protein extraction and IP with GFP-Trap. Samples of cell lysate (left) and GFP-Trap IP (right) were separated by SDS–PAGE, followed by immunoblot analysis with indicated antibodies.
To further illustrate whether PEX11e could interact directly with SDP1 in the absence of FREE1, we also performed additional Y2H and in vitro pull-down assays. The Y2H assay showed that neither the N-terminus nor the C-terminus of SDP1 could interact directly with the N-terminus of PEX11e (Supplemental Figure S12A). In addition, in vitro pull-down assay using PEX11e synthetic peptides (Supplemental Figure S12B) as well as recombinant FREE1 and SDP1 proteins showed that the PEX11e could not interact directly with SDP1 in the absence of FREE1 (Supplemental Figure S12C), but the PEX11e peptides P2 (amino acids 48–71) showed interaction with SDP1 in the presence of FREE1 (Supplemental Figure S12D). Taken together, these results demonstrated that FREE1 interacts directly with both PEX11e and SDP1, and that PEX11e could not interact directly with SDP1, suggesting that FREE1 may act as a scaffold bridging PEX11e and SDP1 during LD metabolism.
FREE1 interacts with the putative AH motif of PEX11e
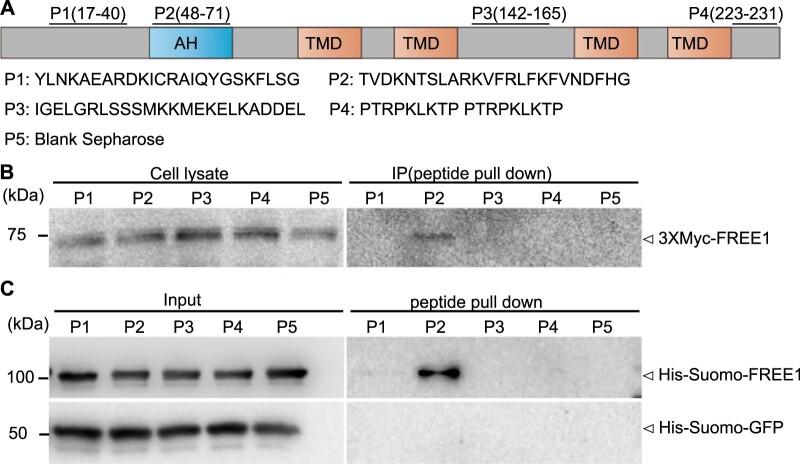
Sequence alignment analysis of PEX11s in plants and other eukaryotes by ClustalW showed that AtPEX11e contains a putative amphipathic helix (AH) motif at its N-terminus (amino acids 44–75) (Supplemental Figure S13A), whose counterpart was designated as PEX11-Amph in Penicillium chrysogenum PEX11 (Opalinski et al., 2011b). This AH motif with amphipathic properties was further confirmed by Heliquest analysis with its hydrophobic moments (µH) value >0.3 (Supplemental Figure S13, B–D). To gain further insight into the specific interaction between FREE1 and PEX11e, we performed pull-down assays using synthetic peptides corresponding to different regions of PEX11e (P1–P4, Figure 4A) and blank sepharose (P5) together with protein extracts isolated from transgenic Arabidopsis cells expressing Myc-FREE1 or recombinant His-Sumo-FREE1 proteins. FREE1 was pulled down specifically by Peptide 2 (amino acids 48–71), which is within the predicted AH motif of PEX11e (Figure 4, B and C). However, recombinant His-Sumo-FREE1 proteins could not be pulled down by the Peptides of putative AH motifs of PEX11a and PEX11b predicted by Heliquest, indicating that FREE1 interacts specifically and directly with the AH motif of PEX11e (Supplemental Figure S14).
Figure 4.
FREE1 interacts with the AH motif of PEX11e. A, Diagram of PEX11e with indicated positions and amino acids sequences of the four synthetic peptides (P1–P4) used in the peptide pull-down assays. TMD: Transmembrane domain. B and C, Peptide pull-down assays showed the specific interaction between FREE1 and the AH motif of PEX11e. B, Soluble proteins were extracted from Arabidopsis protoplasts expressing Myc-FREE1 and incubated with Sepharose beads conjugated with the four synthetic peptides corresponding to different regions of PEX11e (P1–P4 as indicated) separately and the blank control (P5) for pull-down assays, followed by immunoblot detection of bound proteins using anti-Myc antibodies. Arrowheads indicate the positions of the detected proteins. C, His-Sumo-FREE1 and His-Sumo-GFP were incubated with Sepharose beads conjugated with the four synthetic peptides corresponding to different regions of PEX11e (P1–P4 as indicated) separately and the blank control (P5) for pull-down assays, followed by immunoblot detection of bound proteins using anti-His antibodies. Arrowheads indicate the positions of the detected proteins.
Loss of FREE1 impairs the degradation of LDs in germinating seedlings
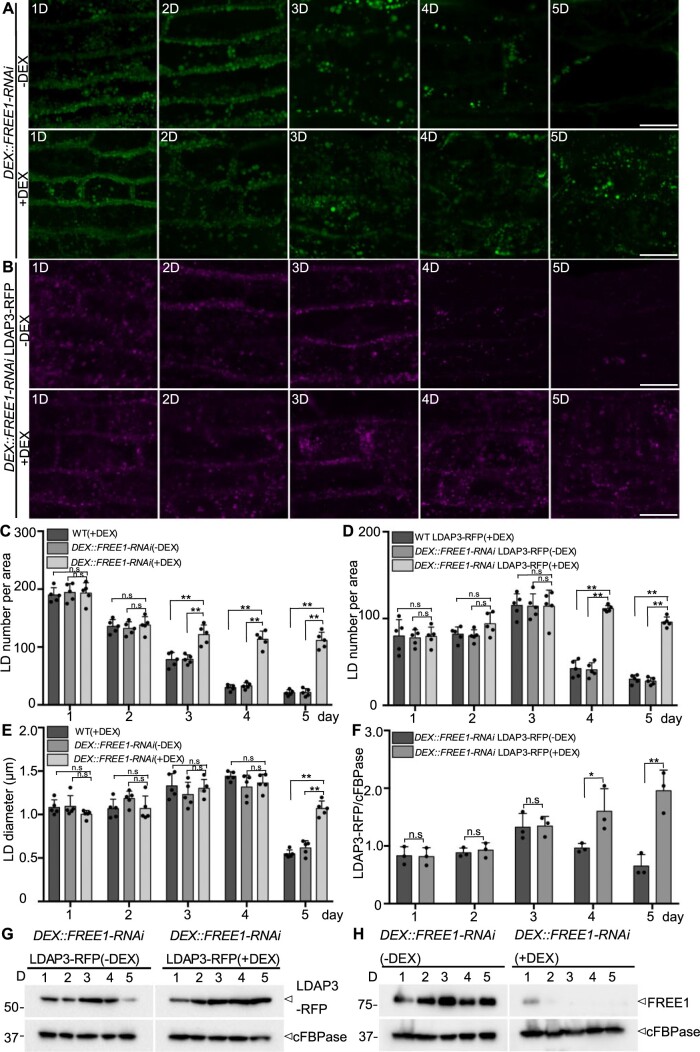
To further test the effects of FREE1 depletion on degradation of LDs, we examined the mobilization profiles of LDs in germinating seedlings of two different DEX::FREE1-RNAi lines in the presence or absence of DEX (Gao et al., 2015). Indeed, in germinating seedlings of two different DEX::FREE1-RNAi lines in the absence of DEX or WT with DEX treatment, LDs, visible as punctae by BODIPY staining, gradually disappeared, and most signals were lost at 5 days after germination (D) (Figure 5A, upper; Supplemental Figure S15A; Supplemental Figure S16A, upper and middle parts). However, in germinating seedlings of two different DEX::FREE1-RNAi lines in the presence of DEX, numerous LDs were still detected at 5 D (Figure 5A, lower; Supplemental Figure S16A, lower). We also examined the mobilization profiles of LDs in germinating seedlings of free1 T-DNA mutant with similar results, showing that numerous LDs were still detected at 5 D in free1 T-DNA mutant (Supplemental Figure S16B). To further explore these observations, we also generated a transgenic DEX::FREE1-RNAi LDAP3-RFP line by crossing the LD marker LDAP3-RFP into the DEX::FREE1-RNAi line, as LDAP3 belongs to LDAPs family targeting to the LD surface (Gidda et al., 2016). Indeed, in germinating seedlings of DEX::FREE1-RNAi LDAP3-RFP line in the absence of DEX or WT LDAP3-RFP with DEX, LDAP3-RFP-positive structures showed a similar decrease with much less signals at 5 D (Figure 5B, upper; Supplemental Figure S15B), whereas numerous LDAP3-RFP-positive structures were still detected at 5 D in germinating seedlings in the presence of DEX (Figure 5B, lower). Further quantification of LD number confirmed that LDs accumulated in germinating seedlings of both DEX::FREE1-RNAi lines with DEX treatment and free1 T-DNA mutant (Figure 5, C and D; Supplemental Figure S16C). Interestingly, the average size of LDs in free1 T-DNA line was significantly smaller than that of WT (Supplemental Figure S16D), but no such obvious differences between DEX-treated and non-DEX-treated DEX::FREE1-RNAi lines were observed at early germination stages (Figure 5E).
Figure 5.
LDs accumulated in germinating seedlings of free1 mutants. A, LDs accumulated in germinating Arabidopsis DEX::FREE1-RNAi lines upon DEX induction. Arabidopsis DEX::FREE1-RNAi seeds were germinated in MS medium with or without DEX for 1–5 D, followed by LD staining using BODIPY dye at indicated days and subsequent confocal imaging of the hypocotyl region. Fifteen slices were collected in a total thickness of 15 μm for generating the 3D projection image. D, days after germination. Scale bars, 20 μm. B, LDAP3-RFP accumulated in germinating Arabidopsis DEX::FREE1-RNAi lines upon DEX induction. Transgenic DEX::FREE1-RNAi LDAP3-RFP seeds were germinated in MS medium with or without DEX for 1–5 D, followed by confocal imaging of the hypocotyl region at indicated days. Fifteen slices were collected in a total thickness of 15 μm for generating the 3D projection image. D, days after germination. Scale bars, 20 μm. C, Quantification of BODIPY-stained LD numbers in germinating seedlings of Arabidopsis WT (as shown in Supplemental Figure S14A) or DEX::FREE1-RNAi as shown in A. LDs numbers of each 150 μm × 150 μm × 15 μm area from five individual seedlings were quantified. Error bars represent the sd from five individual seedlings, **P < 0.01 in Student’s t test, n.s., not significant. D, Quantification of LDAP3-RFP-positive LDs in germinating seedlings of Arabidopsis WT LDAP3-RFP (as shown in Supplemental Figure S14B) or DEX::FREE1-RNAi LDAP3-RFP line as described in (B). LDs numbers of each 150 μm × 150 μm × 15 μm area were quantified from five individual seedlings. Error bars represent the sd from five individual seedlings, **P < 0.01 in Student’s t test. E, Quantification of BODIPY-stained LD diameter in germinating seedlings of Arabidopsis WT (as shown in Supplemental Figure S14A) or DEX::FREE1-RNAi as shown in A. LD diameter in each 150 μm × 150 μm × 15 μm area from five individual seedlings were quantified. Error bars represent the sd from five individual seedlings, **P < 0.01 in Student’s t test. F, Quantification of LDAP3-RFP protein profiles. The intensity of the LDAP3-RFP was normalized by the cytosolic protein FBPase intensity. Error bars represent the sd. from three individual experiments, **P < 0.01 in Student’s t test. G, The LDAP3-RFP protein profiles in germinating Arabidopsis DEX::FREE1-RNAi LDAP3-RFP line. Transgenic Arabidopsis DEX::FREE1-RNAi LDAP3-RFP seeds were germinated in MS medium with or without DEX for 1–5 D, followed by protein extraction of seedlings at indicated days and immunoblot analysis with indicated antibodies. H, FREE1 protein profiles in germinating Arabidopsis DEX::FREE1-RNAi line. Arabidopsis DEX::FREE1-RNAi seeds were germinated in MS medium with or without DEX for 1–5 D, followed by protein extraction of seedlings at indicated days and immunoblot analysis with indicated antibodies.
These confocal observations were also supported by immunoblot analysis at the protein level. In addition, the protein level of LDAP3-RFP in seedlings of DEX::FREE1-RNAi LDAP3-RFP was significantly reduced at 5 D in the absence of DEX; however, upon DEX-induced KD of FREE1, the protein levels of LDAP3-RFP remained relatively constant from 2 D (days after germination) to 5 D (Figure 5, F and G). As shown in Figure 5H, DEX treatments knocked-down the FREE1 effectively during the 5-day germination of DEX::FREE1-RNAi lines as very little FREE1 protein was detected in seedlings of 2 D–5 D (Figure 5H). Taken together, these data further support that FREE1 is required for efficient LD metabolism in germinating seedlings.
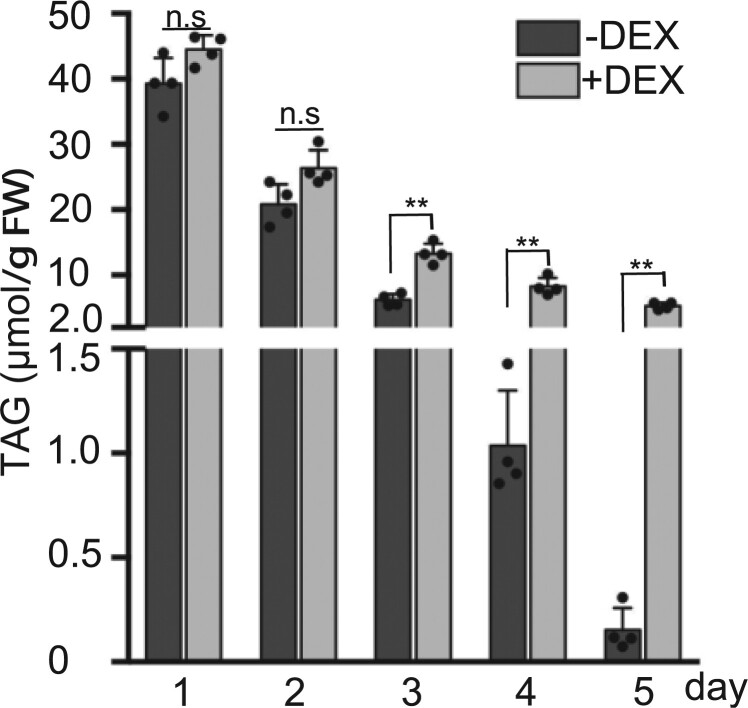
Given that TAGs are the major neutral lipids stored in LDs, we analyzed the amount and composition of TAGs in germinating seedlings. Total lipids were extracted from the germinating DEX::FREE1-RNAi lines in the presence or absence of DEX at 1 D–5 D for subsequent identification and quantification of TAGs using tandem mass spectrometry (Li et al., 2014; Xie et al., 2015; Liu et al., 2020b). As shown in Figure 6, the total TAG contents of 1 D–5 D germinating DEX::FREE1-RNAi seedlings without DEX induction were gradually reduced from 1 D to 4 D and barely detectable at 5 D. However, in the presence of DEX, the total TAG contents were only reduced slightly at 3 D–5 D, and relatively large amounts of the total TAG contents were still detected at 3 D–5 D, significantly higher than those without DEX treatments (Figure 6). Similar results were obtained when different TAG species contents were further analyzed in these experiments (Supplemental Figure S17). Taken together, these data reinforce that lipid mobilization is impaired in free1 mutants.
Figure 6.
Analysis of TAGs in germinating seedlings of free1 mutants. Arabidopsis DEX::FREE1-RNAi seeds were germinated in MS medium with or without DEX for 1–5 days (D), followed by sample collections at indicated days for lipid extraction and subsequent analysis of total TAGs (μmol g−1 FW). D, Days after germination. FW, fresh weight. Error bars represent the sd from four biological replicates, **P < 0.01 in Student’s t test.
Figure 7.
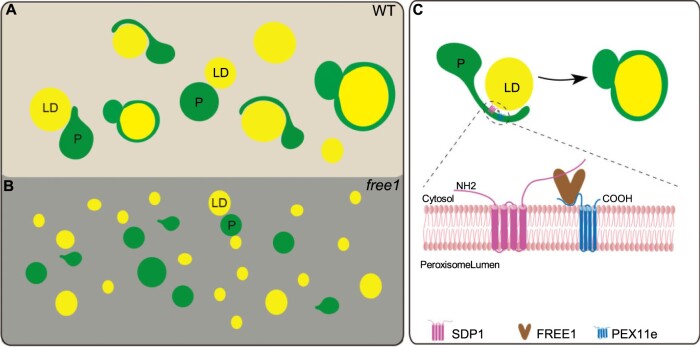
Working model of FREE1 function in mediating LD turnover in germinating seedlings of WT and free1 mutant. A, In germinating WT seedlings, LDs are engulfed by the tubular peroxisomes and degraded by the SDP1-mediated pathway. B, In germinating Arabidopsis free1 mutant, the formation of peroxisome tubulation is impeded and SDP1 is mis-targeted, resulting in accumulation of LDs. C, Possible mechanisms of PEX11e–FREE1–SDP1 interaction in mediating LDs degradation in germinating Arabidopsis seedlings. FREE1 interacts directly with both PEX11e and SDP1, which is required for the PEX11e-mediated peroxisome tubulation and proper targeting of SDP1 to the LD surface. P, peroxisome.
Discussion
FREE1 is required for peroxisomal tubulation
In eukaryotes, ESCRTs are known to be involved in many cellular processes, including MVB biogenesis, cytokinetic abscission, RNA virus replication, HIV budding, plasma membrane repair, exosome formation, defective nuclear pore complexes clearance, lysosome repair, phagophore closure, and microautophagy (Gao et al., 2017). Recent studies have also demonstrated the unconventional function of ESCRT components in mediating peroxisome biogenesis. For example, the yeast ESCRTIII regulates peroxisome biogenesis thus affecting peroxisome-mediated LD degradation (Mast et al., 2018), whereas the Arabidopsis ESCRTIII components are necessary for peroxisomal inner membrane formation to regulate LD metabolism (Wright and Bartel, 2020). Distinct from the well-known functions of the ESCRT components in peroxisome biogenesis, our studies have revealed an unexpected role of FREE1, a plant-specific ESCRT component, in regulating peroxisomal tubulation and thus LD mobilization in germinating seedlings. However, whether peroxisome tubule formation is regulated independently by the ESCRT component FREE1 or by the ESCRT-I complex in plants remains to be elucidated in future study.
ESCRT components function in membrane remodeling events by stabilizing negative membrane curvature, such as during endosomal intralumenal vesicle formation, enveloped virus budding, nuclear envelope closure, and cytokinetic abscission (Hurley, 2015; McCullough et al., 2015). In Arabidopsis, the PEX11 family proteins promote peroxisomal proliferation and tubulation (Orth et al., 2007) and the AH motif of PEX11s, conserved in eukaryotes, has membrane remodeling capacity (Opalinski et al., 2011b; Yoshida et al., 2015). In this study, we showed that both SDP1- and PEX11e-positive peroxisomal tubulation were impaired in free1 mutants (Figure 1, A–D; Supplemental Figure S3, A–D), and that FREE1 interacted directly with PEX11e, suggesting that the interaction may regulate peroxisomal membrane extension (Figure 3). Intriguingly, retromer was previously shown to play a role in regulating peroxisomal membrane extension in Arabidopsis (Thazar-Poulot et al., 2015); thus, it would be interesting to clarify the relationship between retromer and FREE1 in regulating peroxisomal tubulation in future study.
Recently, tumor susceptibility 101 (TSG101), an ESCRT-I component, was shown to cooperate with vacuolar protein sorting-associated protein 13D (VPS13D) to regulate the transfer of FAs from LDs to mitochondria to mediate the LD mobilization in mammalian cells (Wang et al., 2021). In Arabidopsis, FREE1 interacts with the ESCRT-I component Vps23 (Gao et al., 2014), and the FREE1 loss-of-function mutant shows a similar phenotype to those of other ESCRT mutants (Cai et al., 2014; Shen et al., 2016; Nagel et al., 2017; Liu et al., 2020a, 2021), in which the assembly or dissociation of the ESCRT machinery is disrupted and the formation of ILVs in MVBs is defective (Buono et al., 2017; Otegui, 2018). Interestingly, the Arabidopsis ESCRTIII component SNF7 is necessary for peroxisomal inner membrane formation, and LDs accumulate in plants expressing a SNF7L22W dominant-negative ESCRTIII variant (Wright and Bartel, 2020). In future study, it will be interesting to investigate the possible roles of ESCRT-I and other ESCRT components in regulating peroxisome tubule formation and LD mobilization in plants.
FREE1 is required for proper targeting of SDP1 to LDs
Lipolysis, an essential pathway for LD turnover in Arabidopsis, is a process whereby non-LD-localized lipases are transferred to the LD surface for LD metabolism (Eastmond, 2006; Kelly et al., 2011, 2013; Pyc et al., 2017a). A conserved patatin-like phospholipase domain-containing (PNPLA) is present in eukaryotic lipases, such as adipose triglyceride lipase (ATGL) in mammals, TAG lipases (TGLs) in yeast, and SDP1 in plants (Athenstaedt and Daum, 2005; Eastmond, 2006; Smirnova et al., 2006). Intriguingly, unlike the lipase ATGL delivery by COPI and COPII vesicles in the cytosol of animal cells (Soni et al., 2009), the major plant lipase SDP1 is localized on the peroxisome and transported to the LD surface via retromer-regulated peroxisomal tubular extension (Thazar-Poulot et al., 2015). However, how the lipase SDP1 is transferred to LDs remains elusive in plants. In this study, we showed that the transfer of SDP1 to LDs surface is impaired in free1 mutants (Figure 1, E and F; Supplemental Figure S3, E and F), and that FREE1 interacts directly with SDP1 and PEX11e (Figure 3), suggesting that FREE1 remodels peroxisome membrane via interacting with PEX11e to promote peroxisomal tubulation and regulate SDP1 targeting to the LD surface. In addition, the interaction of PEX11e and SDP1 depends on the presence of FREE1 (Figure 3; Supplemental Figure S12), suggesting that FREE1 may act as a scaffold bridging PEX11e and SDP1. Future study using genetic approaches can be carried out to test for the PEX11e–FREE1–SDP1 interaction complex in plants.
LD metabolism is impaired in the autophagy-deficient mutants of mammals and yeast (Singh et al., 2009; Velikkakath et al., 2012; Schepers and Behl, 2021), and our study showed that LD degradation was not defective in germinating seedlings of both atg5 and atg7 mutants under normal growth conditions (Supplemental Figure S2). However, under dark conditions, LDs can be delivered to vacuoles for degradation via autophagy-dependent pathway in Arabidopsis leaves (Fan et al., 2019). In future study, it will be interesting to explore the possible roles of lipophagy and lipolysis in lipid metabolism under stresses conditions in plants.
FREE1 is required for engulfment of LDs by tubulating peroxisomes
LD–peroxisome contact has been studied. In yeast, the high-curvature peroxisomal protrusions were found to penetrate into the lipid core of LDs at LD–peroxisome contact sites by TEM (Binns et al., 2006). Similarly, in mammals, elongated peroxisomes were visualized tightly associated to the LD surface by conventional TEM, correlative light electron microscopy and ET analysis, indicating that peroxisomes are in close contact with a large surface area of LDs at LD–peroxisome contact sites (Chang et al., 2019). Just how SDP1 becomes localized on the peroxisome to perform its TAG hydrolysis activity on the LD surface is also unclear. In this study, we have provided evidence obtained by ET analysis for a distinct peroxisome–LD association in WT compared to the free1 mutant. The ET data showed different stages of peroxisomal contact with LDs, including peroxisomes encapsulating entire LDs, wrapped around half of the LD surface or enwrapping a small part of LD surface in WT (Figure 2A; Supplemental Figure S4, upper; Supplemental Figure S5, upper; Supplemental Figure S7A). The large contact area between peroxisomes and LDs may improve the efficiency of TAG metabolism and also facilitate the trafficking of free FA to peroxisomes effectively for the subsequent metabolism (Yang et al., 2012). However, in free1 mutants, few elongated peroxisomes or large surface area tethering between peroxisomes and LDs were observed (Figure 2B; Supplemental Figure S4, lower; Supplemental Figure S5, lower; Supplemental Figure S7B). The little surface area attachment between peroxisomes and LDs may reduce the efficiency LD metabolism, leading to the delay of LD degradation in free1 mutants. Further confocal imaging and TAG analysis confirmed the presence of LDs and accumulation of TAGs in free1 mutants even at late stages of germination (Figures 5 and 6; Supplemental Figures S15–S17). Compared to the continuous reduction in TAG levels from 1D to 5 D (Figure 6), there were discrepancies in the LD diameter and LDAP3 levels (Figure 5, E and F). A slight increase in LD diameters at 3 D and 4 D during seed germination may be due to LD–LD fusion (Yang et al., 2012) albeit with a yet-unknown mechanism in plants (Figure 5E). The LDAP3 proteins were slightly increased at 3 D in both WT and FREE1-deficient seedlings, and then decreased at 4 D and 5 D in WT seedlings but remained at similar level in FREE1-deficient seedlings (Figure 5F). The abundance of TAGs continuously decreased from 1D to 5 D in WT, but the TAGs remained at similar level without further significant reduction from 3 D to 5 D in FREE1-deficient seedlings (Figure 6). The different metabolic processes of proteins and lipids in seedlings may contribute to the slight increase of LDAP3 proteins at 3 D. We conclude that the depletion of FREE1 affected the proper trafficking of SDP1 to LD surface for LD mobilization, thus resulting in a significantly higher LD numbers and LDAP3 levels in free1 mutant in comparison with WT (Figure 5).
A working model for FREE1 function in mediating LD turnover
In this study, we have shown that (1) PEX11e- or SDP1-marked peroxisome tubulation and proper targeting of SDP1 to LD are defective in free1 mutants; (2) The engulfment of LDs by peroxisomes is normal in WT, but very little surface area tethering exists between peroxisomes and LDs in the free1 mutant as revealed by ET analysis; (3) FREE1 interacts directly with PEX11e via the AH motif to promote peroxisome tubulation and with SDP1 for its proper targeting to the LD surface; and (4) LD degradation is impaired in free1 mutants. Figure 7 shows a working model of FREE1 function in regulating LD turnover in germinating Arabidopsis seedlings. In germinating WT seedlings, FREE1 directly interacts with both PEX11e and SDP1, suggesting that these interactions may regulate the peroxisomal tubules extension and the trafficking of SDP1 to the LD surface for TAG hydrolysis to release FAs to the peroxisome for further seedling development (Figure 7A). In free1 mutants, the formation of peroxisomal tubules is impeded and thus SDP1 fails to traffic to LD surface, leading to accumulation of LDs in free1 mutants (Figure 7B). Interestingly, the size of LDs in the free1 T-DNA mutant was smaller than that in the WT, although the mechanism of this difference is unclear (Supplemental Figure S16, B and D). Biochemically, FREE1 interacts directly with both PEX11e and SDP1, thus cooperating with PEX11e to mediate proper SDP1 trafficking (Figure 7C). Peroxisomal tubulation is a highly dynamic membrane remodeling process and PEX11 plays key roles in peroxisomal proliferation and tubulation via its putative AH motif (Opalinski et al., 2011b; Yoshida et al., 2015). We hypothesize that the interaction among FREE1, PEX11, and SDP1 may regulate the formation of SDP1-positive tubules that eventually enwrap the LDs, thereby facilitating SDP1 translocation to catabolize TAGs. In conclusion, our study sheds light on the function of the plant-specific ESCRT component FREE1 in nonendosomal organelles, namely peroxisomes, mediating peroxisomal tubulation and proper targeting of SDP1 to regulate LD metabolism in germinating seedlings.
Materials and methods
Plant materials and growth conditions
The A. thaliana transposon insertion line (15-1960-1) of free1 was obtained from the RIKEN. The DEX-induced FREE1-RNAi transgenic line was described previously (Gao et al., 2014). The Arabidopsis pK7WGF2-35Spro::GFP–SDP1 was kindly provided by Thierry Gaude (Thazar-Poulot et al., 2015). For the free1 GFP–SDP1 and DEX::FREE1-RNAi GFP–SDP1, the pK7WGF2-35Spro::GFP–SDP1 was crossed separately with free1 and DEX::FREE1-RNAi.
To generate the mCherry-PEX11e transgenic plants, PEX11e was amplified and cloned into pCAMBIA1300-UBQpro::mCherry-GW backbone for plant transformation. For the cross material free1 mCherry-PEX11e, the mCherry-PEX11e was crossed with free1. To generate the LDAP3-RFP transgenic plant, LDAP3 was amplified and cloned into the pCAMBIA1300-UBQpro::GW-RFP backbone under the UBQ10 promoter for plant transformation. To generate DEX::FREE1-RNAi LDAP3-RFP, the LDAP3-RFP line was crossed into the DEX::FREE1-RNAi line. To generate transgenic double lines of GFP-FREE1 mCherry-PEX11e and GFP-FREE1 mCherry-SDP1, the GFP-FREE1 line was crossed into the mCherry-PEX11e or mCherry-SDP1. The seeds were surface-sterilized and grown vertically on plates with Murashige and Skoog (MS) salts, pH 5.7, containing 1% sucrose and 0.8% agar at 22°C under a long day (16-h light (120 s−1 m−2 light intensity provided by white fluorescent lamps)/8-h dark cycle) condition. To induce the DEX::FREE1-RNAi plants, 10-μM DEX (10-mM stock dissolved in ethanol) was added in MS medium.
Plasmid construction
For the constructs used for the transgenic plants, the coding sequence was amplified from cDNA and cloned into the pDONR/Zeo vector using the Gateway cloning system (Invitrogen, Waltham, MA, USA, cat.no. 12535-035) and further cloned into pCAMBIA1300-UBQ10pro::mCherry-GW or pCAMBIA1300-UBQ10pro::GW-RFP vectors separately. For pCAMBIA1300-UBQ10pro::mCherry-PEX11e transgenic plants, PEX11e was cloned into the pDONR/Zeo and further cloned into pCAMBIA1300-UBQ10pro::mCherry-GW backbone to construct mCherry-PEX11e for generating transgenic plants. For pCAMBIA1300-UBQ10pro::LDAP3-RFP transgenic plants, LDAP3 was cloned into pDONR/Zeo and further cloned into the pCAMBIA1300-UBQ10pro::GW-RFP backbone to construct LDAP3-RFP for generating transgenic plants. For the constructs used for Y2H analysis, cDNAs were cloned into pGBKT7 and pGADT7 vectors separately. For the constructs used for recombinant protein expression, the SDP1–Flag was cloned into pEGFP-N1 vectors through KpnI and NotI double digestion. The p-His-SUMO–FREE1 and p-His-SUMO-GFP vectors have been previously described (Gao et al., 2014). All constructs were confirmed by Sanger sequencing. Primers used in this study are listed in Supplemental Table S1.
Generation of transgenic plants by Agrobacterium-mediated stable transformation
Arabidopsis WT plants were transformed by floral dip with the Agrobacterium tumefaciens strain GV3101 containing the pCAMBIA1300-UBQ10pro::LDAP3-RFP or pCambia1300-UBQpro::mCherry-PEX11e or pCambia1300-UBQpro::mCherry-SDP1 binary vector (Clough and Bent, 1998). The transgenic plants were screened on kanamycin or hygromycin, and kanamycin or hygromycin-resistant transgenic T1 plants were selected and planted to obtain T2 progenies. The T2 progenies were screened on kanamycin- or hygromycin-containing MS medium to obtain plants with a single insertion based on 3:1 segregation rules. Seeds from homozygous T3 plants were used for confocal observations and immunoblot analysis.
Recombinant protein purification
The pEGFP–N1–SDP1–Flag vector was expressed in suspension HEK293F mammalian cells as previously described (Lai et al., 2019). The suspension HEK293F cells were continuously incubated in an orbital shaker at 37°C, a humidified atmosphere of 8% CO2 in air, and rotated at 125 rpm for 3 days. Mammalian cells were stained by trypan blue dye to measure cell density and viability. HEK293F cells were diluted to 2.5 × 106 cells mL−1 with cell viability >95%. Upon transfection, pEGFP–N1–SDP1–Flag plasmids (after endotoxin removal) and polyethylenimine was diluted in Opti-MEM I Reduced Serum Medium (Thermo Fisher, Waltham, MA, USA) and mixed to add into HEK293F cells for two more days of culture. Then cells were harvested at 500g centrifugation for 10 min.
Cell pellets were resuspended in 15-mL lysis buffer (50-mM Tris–HCl pH 8.0, 300-mM NaCl, 10% glycerol, and 1 × complete protease inhibitor cocktail [Roche, Basel, Switzerland]), lysed using a homogenizer and ultracentrifuged at 100,000g for 1 h. The pellet was collected, homogenized, incubated in 10 mL lysis buffer with 1% n-dodecyl-β-D-maltopyranoside (DDM) for 2 h, and ultracentrifuged at 100,000g for 1 h. The supernatant was collected and incubated with anti-FLAG M2 magnetic beads for 2 h, after which the beads were washed 5 times using lysis buffer with 0.05% DDM, and bound proteins were eluted with lysis buffer with 0.05% DDM and 0.2-mg mL−1 3 × Flag. His-Sumo-FREE1, His-Sumo-GFP fusion proteins were expressed in Escherichia coli (BL21) upon treatment with 0.4-mM isopropyl β-D-1-thiogalactopyranoside at 20°C overnight and purified with Ni-NTA resin as previously described (Gao et al., 2014).
Both eluted proteins were dialyzed against pull-down buffer (50-mM Tris–HCl, 300-mM NaCl, and 10% glycerol, pH 7.5). Protein concentration was determined using Bio-Rad Protein Assay (cat. no. 5000006) and Coomassie Brilliant Blue staining of proteins SDS–PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) using the bovine serum albumin as a standard.
In vitro pull-down assay
For the in vitro pull-down assay, Flag magnetic beads were saturated with SDP1–Flag (Lai et al., 2019). Beads with SDP1–Flag were incubated with His-Sumo-FREE1 and His-Sumo-GFP in 1 mL cold pull-down buffer (50-mM Tris–HCl [pH 7.5], 150-mM NaCl, 10% glycerol) containing 1% GDN (glycogen diosgenin). After incubation at 4°C for 2 h in a top to end rotator, the beads were washed 7 times with pull-down buffer containing 0.02% GDN and then eluted by 0.1-M glycine buffer. The samples were boiled in SDS sample buffer, separated by SDS–PAGE gel and further analyzed by immunoblots using the appropriate antibodies.
Transient expression in Arabidopsis protoplasts
The Arabidopsis PSB-D suspension cultured cells were subcultured every 5 days at 25°C, with shaking at 130 rpm in the dark (Miao and Jiang, 2007). First, the 5-day-old Arabidopsis PSB-D suspension cells were digested by enzyme solution (1% cellulase “ONOZUKA” RS, 0.05% pectinase, and 0.2% driselase from Basidiomycetes sp in 25-mL TEX buffer) for 2 h at 25°C, with shaking at 130 rpm under dark. Second, electroporation buffer (0.4-M sucrose, 2.4-g L−1 HEPES, 6-g L−1 KCl, and 600-mg L−1 CaCl2·2H2O, pH 7.2) was used to wash the cells twice. Then, the protoplasts were transformed with plasmids via electroporation. The protoplasts were then incubated for 10–16 h prior to confocal imaging analysis or protein extraction.
co-IP analysis
pBI221–YFP–SDP1, pBI221-YFP-PEX11e, pBI221-YFP-FREE1, pBI221-YFP, pBI221-3Xmyc-FREE1, and pBI221-3XHA-PEX11e were separately transformed to Arabidopsis PSB-D protoplasts. After 16 h incubation, 2.5 mL transformed protoplasts were diluted three-fold with 250-mM NaCl in 15-mL Falcon tubes and then centrifuged at 700g for 5 min to harvest cells, followed by resuspension in ice-cold lysis buffer (25-mM HEPES [pH 7.2], 150-mM NaCl, 2-mM EDTA, 1-mM MgCl2, 0.5% Triton X-100, 0.5-mM dithiobis [succinimidyl propionate] and 1 × complete protease inhibitor cocktail [Roche]). The protoplasts were then lysed by passing through a 1-mL syringe with needle and incubated at 4°C for 30 min. Total lysates were centrifuged at 10,000g for 15 min at 4°C and supernatants were incubated with GFP-Trap magnetic beads for 4 h at 4°C in a top to end rotator. After incubation, the beads were washed with ice-cold washing buffer (25-mM HEPES [pH 7.2], 150-mM NaCl, 2-mM EDTA, 1-mM MgCl2, 0.05% Triton) four times and then eluted by boiling in SDS buffer. The samples of supernatant or IP were separated by SDS–PAGE gel and further analyzed by immunoblot using the appropriate antibodies.
High pressure freezing and transmission electron microscopy
The general procedures for transmission electron microscopy sample preparation and thin sectioning were performed as described previously (Cui et al., 2019). One-day-old seedlings of WT GFP–SDP1 and free1 GFP–SDP1 were frozen in a high-pressure freezer (Leica, Wetzlar, Germany), followed by freeze substitution performed in anhydrous acetone containing 2% OsO4/0.5% uranyl acetate at −85°C in an AFS freeze-substitution unit (Leica, Wetzlar, Germany) for 48 h and then gradually warmed to −45°C at a 1°C/1 h rate over a 40 h period. The samples were incubated at −45°C for over 12 h and incubated at −20°C for over 12 h and then warmed to 4°C gradually over a 12 h period. The roots were removed from the planchets after three acetone rinses at room temperature and slowly infiltrated stepwise with increasing concentrations of Epon resin (TedPella) over 96 h and then embedded in Epon at 65°C overnight. For immunogold TEM analysis, 1-day-old WT GFP–SDP1 seedlings were frozen in a high-pressure freezer, followed by freeze substitution in dry acetone containing 0.1% uranyl acetate at −85°C in an AFS freeze-substitution unit (Leica, Wetzlar, Germany) and infiltrated with HM20. Embedding and UV polymerization were performed stepwise at −10°C. Immunogold labeling was performed as previously described (Cui et al., 2019) with anti-GFP antibodies diluted at 40 μg/mL in 1% BSA, and gold-coupled rabbit secondary antibodies at 1:40 dilution in 1% BSA. Samples were observed under Hitachi H-7650 TEM with a charge-coupled device camera (Hitachi High-Technologies Corporation, Japan) operating at 80 kV.
ET analysis
The procedures for ET including sample sectioning, post-staining, and electron microscopy imaging were as previously described (Cui et al., 2019). In brief, 14 continuous 300-nm-thick sections of 1-day-old WT GFP–SDP1 and free1 GFP–SDP1 mutant root tips not occupied by large central large vacuoles were cut and poststained using uranyl acetate and lead citrate. ET images were collected with an FEI Tecnai F20 TEM (FEI Company) operating at 200 kV. For each grid, a tilt image stack (81 images) from +60° to −60° with 1.5° increments was collected, and the second tilt image stack was collected by rotating the grid by 90°. Dual-axis tomograms were calculated from pairs of image stacks with the etomo program of the IMOD software package (version 4.9.7, bio3d.colorado.edu). The 3D models were generated using the 3dmod program of the IMOD software package.
Confocal microscopy and FRET analysis
Confocal fluorescence images were collected with a Leica SP8 laser scanning confocal system with a 63× water lens. All fluorophore emissions were collected sequentially in single or double-labeling experiments. FRET analysis was performed on a Leica SP8 confocal system according to the manufacturer’s instructions using 405- and 514-nm laser (Gao et al., 2014; Shen et al., 2018).
LD staining
LDs (for Arabidopsis) were stained with 2-μg mL−1 BODIPY 493/503 (4-mg mL−1 stock in DMSO, excitation: 488-nm laser, emission: 500–540 nm) or 1-μg mL−1 Nile red (1-mg mL−1 stock in DMSO, excitation: 543-nm laser, emission: 590–640 nm) diluted in 1 × PBS buffer (pH 7.0) for 10 min followed by three washes with 1 × PBS buffer (Gidda et al., 2016). Thereafter, materials were observed under confocal microscopy SP8 (Leica) for imaging.
TAG amount and composition analysis
For lipid extraction, 70 mg of DEX-treated or non-DEX-treated DEX::FREE1-RNAi germinating seedlings at 1D–5D were freshly collected for analysis of TAGs by the Lipidall Technologies (China) (http://www.lipidall.com/). The samples were inactivated with 0.01% (w/v) butylated hydroxytoluene in isopropanol (prewarmed to 75°C to inactivate the solvent) (Liu et al., 2020b). The extraction buffer containing chloroform:methanol:300-mM ammonium acetate (30:41.5:3.5) (v/v/v) was added to the samples and shaken for 24 h at 150 rpm at room temperature. Samples were then centrifuged and supernatant were transferred to new tubes and dried in a SpeedVac. Lipid extracts were stored at −80°C until liquid chromatography–mass spectrometry (LC–MS) analyses.
For TAGs, lipidomic analyses were performed with an Exion UPLC-QTRAP 6500 Plus (Sciex) LC–MS (curtain gas = 20, ion spray voltage = 5,500 V, temperature = 400°C, ion source gas 1 = 35, ion source gas 2 = 35) (Lipidall Technologies, China). For reverse phase (RP) LC/MS, lipids were analyzed using a modified version of RP-high-performance liquid chromatography/electrospray ionization tandem mass spectrometry (HPLC/ESI/MS/MS). In brief, lipids were separated on a Phenomenex Kinetex 2.6 µm-C18 column (internal diameter 4.6 × 100 mm) using an isocratic mobile phase chloroform:methanol:0.1M ammonium acetate (100:100:4) (v/v/v) at a flow rate of 300-µL min−1 for 10 min. TAGs were quantitated using TAG(14:0)3-d5, TAG(15:0)3-d29, and TAG(18:0)3-d5 obtained from CDN isotopes (Quebec, Canada). For the relative quantitation of the TAGs in germinating seedlings, data are expressed in micromole per gram fresh weight as described previously (Huang et al., 2019a). Error bars represent the sd from four biological replicates. **P < 0.01 in Student’s t test (Supplemental File S1).
Y2H
For Y2H analysis, the corresponding genes were amplified from cDNA and cloned into the pGBKT7 or pGADT7 vectors (Clontech). Plasmids of each pair were co-transformed into the yeast AH109 strain and plated on synthetic drop-out medium lacking Trp and Leu (SD-2) for 3 days at 30°C (Gao et al., 2014). Positive colonies were further selected on SD medium lacking His, Trp, Leu (SD-3) containing 6 mM 3-amino-1,2,4-triazole for 2 days at 30°C. The experiments were performed twice independently with similar results.
Protein extraction and immunoblot analysis
The samples of 0.1 g Arabidopsis seedlings at different germination stages were grounded to powder in liquid nitrogen and homogenized in 100-μL protein extraction buffer containing 50-mM Tris–HCl (pH 7.5), 150-mM NaCl, 1-mM EDTA, 10% glycerol, 1% SDS (sodium dodecyl sulfate), and 1 × complete protease inhibitor cocktail (Roche). Total lysates were centrifuged at 700g for 15 min at 4°C, followed by SDS–PAGE and immunoblot analysis using the appropriate antibodies.
Topology analysis and protease protection assay
The isolation of microsomes and the analysis of protein topology were performed as previously described (Gao et al., 2012). YFP–SDP1 and SDP1–GFP were separately transformed to Arabidopsis PSB-D protoplasts. After 16-h incubation, 2.5-mL transformed protoplasts were diluted three-fold with 250-mM NaCl in 15-mL Falcon tubes and then centrifuged at 700g for 5 min to harvest cells, followed by resuspension in ice-cold extraction buffer (40-mM HEPES-KOH, pH 7.5, 1-mM EDTA, 10-mM KCl, and 0.4-M sucrose, and 1 × complete protease inhibitor cocktail [Roche]) and lysing by passing through a 1-mL syringe with needle several times. The samples were centrifuged at 600g for 3 min to remove large cellular debris, and the supernatants were further centrifuged at 100,000g for 30 min at 4°C. The pellet was assigned as the microsomal-enriched membranes and was resuspended in the extraction buffer to be divided into three parts with equal volume for protease protection assays with three distinct conditions: without trypsin, containing 100-μg mL−1 trypsin and containing 100-μg mL−1 trypsin with 1% Triton X-100. After incubation at 37°C for 30 min, the samples were boiled in SDS sample buffer, separated by SDS–PAGE gel and further analyzed by immunoblot using the appropriate antibodies.
For microsome isolation and membrane binding ability assays (Gao et al., 2014), the Arabidopsis PSB-D protoplasts transformed with SDP1–GFP were extracted in ice cold extraction buffer (40-mM HEPES-KOH at pH 7.5, 1-mM EDTA, 10-mM KCl, 0.4-M sucrose, 1 × complete protease inhibitor cocktail [Roche]). After centrifugation at 600g for 3 min to remove large cellular debris, the supernatant was further centrifuged at 100,000g for 30 min at 4°C. The supernatant (CS) and microsomal-enriched membranes fractions (CM) were separated from Arabidopsis cells. The membrane pellet was then resuspended in extraction buffer with 0.1-M Na2CO3 (pH 10), 1-M KCl, 1% (v/v) Triton X-100, or 1% (v/v) SDS, and incubated at 4°C for 1 h before separation as CS and CM fractions for immunoblot analysis with indicated antibodies.
In vitro peptide pull-down assay
The AH motifs of PEX11s were predicted by HELIQUEST software (http://heliquest.ipmc.cnrs.fr/) and PSIPRED software (http://bioinf.cs.ucl.ac.uk/psipred/). The peptides used in peptide pull-down assay were synthesized by GeneScript (https://www.genscript.com/). Synthetic peptides of 24–26 amino acid length were used because of synthesis limitations. We selected the region of amino acids 48–71 because of their highest hydrophobic moment (µH) values among the region of amino acids 44–75.
In vitro pull-down and peptide-binding assay were performed as previously described (Contreras et al., 2004; Suen et al., 2010). Synthetic peptides corresponding to the different regions of PEX11e, PEX11a, and PEX11b were coupled, via N-terminal NH2 group to cyanogen bromide-activated Sepharose 4B (3-mg peptides per milliliter of beads) according to standard procedures. The coupling reaction was quenched through incubation with 1-M glycine, pH 8.0, at room temperature for 2 h in a top to end rotator. Peptides coupling efficiencies were monitored by measuring 280-nm absorbance.
For the peptide-binding assay, the Arabidopsis PSB-D protoplasts transformed with 3 × myc-FREE1 were extracted in ice-cold lysis buffer (50-mM Tris–HCl [pH 7.5], 150-mM NaCl, 1-mM EDTA, 10% glycerol, 0.5% Triton X-100, and 1 × complete protease inhibitor cocktail [Roche]) and then lysed by passing through a 1-mL syringe with needle and incubated at 4°C for 30 min. The supernatants were then incubated with Sepharose beads coupled with the peptides for 4 h at 4°C in a top to end rotator. After incubation, the beads were washed six times with 1 × lysis buffer and then eluted by boiling in SDS sample buffer. The samples were boiled in SDS sample buffer, separated by SDS–PAGE gel and further analyzed by immunoblotting with the appropriate antibodies.
For the peptide pull-down binding assay, sepharose beads coupled with the peptides were incubated with His-Sumo-FREE1, His-Sumo-GFP, or Flag–SDP1 in 1-mL cold pull-down buffer (50-mM Tris–HCl [pH 7.5], 150-mM NaCl, 10% glycerol) containing 1% Triton. After incubation at 4°C for 1 h in a top to end rotator, the beads were washed seven times with pull-down buffer. The samples were boiled in SDS sample buffer, separated by SDS–PAGE, followed by immunoblot detection using the appropriate antibodies.
Antibodies
Primary antibodies used in this study were anti-FREE1 (homemade) (Gao et al., 2014), anti-GFP (homemade) (Cui et al., 2019), anti-GFP (1:1,000 dilution) (Chromotek, 029762), anti-RFP (1:1,000 dilution) (Abcam, Cambridge, UK ab125244), anti-HA (1:1,000 dilution) (Abcam, ab18181), anti-MYC (1:500 dilution) (Santa Cruz, SC789), anti-Flag (1:8,000 dilution) (Sigma, F3165), anti-His (1:1,000 dilution) (GE, ZT-4710.01), and anti-FBPase (1:1,000 dilution) (Agrisera, AS04043).
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are FREE1 (At1G20110), SDP1 (At5G04040), LDAP3 (At3G05500), PEX11e (At3G61070), PEX11a (At1G47750), and PEX11b (At3G47430).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. LDs accumulated in germinating free1 mutants (supports Figure 1).
Supplemental Figure S2. LDs did not accumulate in germinating seedlings of atg5 or atg7 mutants (supports Figure 1).
Supplemental Figure S3. FREE1 regulates peroxisomal tubule formation and peroxisome-encapsulated LDs (supports Figure 1).
Supplemental Figure S4. Electron micrographs of relationships between peroxisomes and LDs in WT GFP–SDP1 and free1 GFP–SDP1 mutant (supports Figure 2).
Supplemental Figure S5. Electron micrographs of relationships between peroxisomes and LDs (supports Figure 2).
Supplemental Figure S6. Immunogold-TEM analysis of relationships between peroxisomes and LDs (supports Figure 2).
Supplemental Figure S7. ET analysis of relationships between peroxisomes and LDs in WT and free1 mutant (supports Figure 2).
Supplemental Figure S8. FREE1 is closely associated with the peroxisome tubule (supports Figure 3).
Supplemental Figure S9. Analysis of SDP1 topology (supports Figure 3).
Supplemental Figure S10. Sequence similarity analysis of AtPEX11s (supports Figure 3).
Supplemental Figure S11. FREE1 interacts with both PEX11e and SDP1 (supports Figure 3).
Supplemental Figure S12. PEX11e could not interact directly with SDP1 in the absence of FREE1 (supports Figure 3).
Supplemental Figure S13. AtPEX11e contains a conserved N-terminal AH motif (supports Figures 3 and 4).
Supplemental Figure S14. FREE1 interacts with the AH motif of PEX11e (supports Figure 4).
Supplemental Figure S15. LD degradation was not affected by DEX treatment in the WT and WT LDAP3-RFP seedlings (supports Figure 5).
Supplemental Figure S16. LDs accumulated in germinating seedlings of free1 mutants (Supports Figure 5).
Supplemental Figure S17. Analysis of TAGs in germinating seedlings of free1 mutants (supports Figure 6).
Supplemental Table S1. Primers used in this study.
Supplemental Movie S1. ET analysis of relationships between peroxisomes and LDs in WT. (supports Figure 2A).
Supplemental Movie S2. ET analysis of relationships between peroxisomes and LDs in free1 mutant. (supports Figure 2B).
Supplemental File S1. Details of statistical analyses.
Supplementary Material
Acknowledgments
We thank Prof. Thierry Gaudea (Université de Lyon) for providing the GFP–SDP1 transgenic plants.
Funding
This work was supported by grants from the National Natural Science Foundation of China (91854201), the Research Grants Council of Hong Kong (AoE/M-05/12, CUHK 14100818, 14101219, C4033-19E, C4002-17G, C4002-20W, C4002-21EF, C2009-19G and R4005-18), and The Chinese University of Hong Kong (CUHK) Research Committee and CAS-Croucher Funding Scheme for Joint Laboratories to L.J., the National Natural Science Foundation of China (32061160467, 31870171) and Fok Ying-Tong Education Foundation for Young Teachers in the Higher Education Institutions of China (171014) to C.G.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Shuxian Huang, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Zhiqi Liu, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Wenhan Cao, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Hongbo Li, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University (SCNU), Guangzhou, 510631, China.
Wenxin Zhang, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Yong Cui, School of Life Sciences, State Key Laboratory of Cellular Stress Biology, Xiamen University, Xiamen, 361102, China.
Shuai Hu, State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China.
Mengqian Luo, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Ying Zhu, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China.
Qiong Zhao, School of Life Sciences, East China Normal University, Shanghai, 200062, China.
Lijuan Xie, College of Plant Protection, State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Caiji Gao, Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University (SCNU), Guangzhou, 510631, China.
Shi Xiao, School of Life Sciences, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, Sun Yat-sen University, Guangzhou, 510275, China.
Liwen Jiang, School of Life Sciences, Centre for Cell & Developmental Biology and State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, China; CUHK Shenzhen Research Institute, Shenzhen, 518057, China; Institute of Plant Molecular Biology and Agricultural Biotechnology, The Chinese University of Hong Kong, Shatin, China.
S.H. and L.J. conceived and designed research. S.H., Z.L., W.C., H.L., W.Z., S.H., Y.Z., L.X., and S.X. performed experiments. S.H., Z.L., W.C., and Y.C. generated the electron tomography models. S.H. and H.L. extracted lipid and analyzed data. S.H., W.Z., and M.L. generated constructs. S.H., C.G., and Q.Z. generated resources. S.H., Z.L., W.C., and L.J. analyzed data. S.H. and L.J. wrote the article. L.J. supervised the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Liwen Jiang (ljiang@cuhk.edu.hk).
References
- Athenstaedt K, Daum G (2005) Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem 280: 37301–37309 [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Savage DB, Siniossoglou S (2015) Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Curr Opin Cell Biol 35: 91–97 [DOI] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM (2006) An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buono RA, Leier A, Paez-Valencia J, Pennington J, Goodman K, Miller N, Ahlquist P, Marquez-Lago TT, Otegui MS (2017) ESCRT-mediated vesicle concatenation in plant endosomes. J Cell Biol 216: 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhuang XH, Gao CJ, Wang XF, Jiang LW (2014) The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol 165: 1328–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao WH, Li ZP, Huang SX, Shi YW, Zhu Y, Lai MN, Lok PL, Wang XF, Cui Y, Jiang LW (2022) Correlation of vacuole morphology with stomatal lineage development by whole-cell electron tomography. Plant Physiol 188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Weigel AV, Ioannou MS, Pasolli HA, Xu CS, Peale DR, Shtengel G, Freeman M, Hess HF, Blackstone C, et al. (2019) Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. J Cell Biol 218: 2583–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contreras I, Yang YD, Robinson DG, Aniento F (2004) Sorting signals in the cytosolic tail of plant p24 proteins involved in the interaction with the COPII coat. Plant Cell Physiol 45: 1779–1786 [DOI] [PubMed] [Google Scholar]
- Cui Y, Cao WH, He YL, Zhao Q, Wakazaki M, Zhuang XH, Gao JY, Zeng YL, Gao CJ, Ding Y, et al. (2019) A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat Plants 5: 95–105 [DOI] [PubMed] [Google Scholar]
- Deruyffelaere C, Purkrtova Z, Bouchez I, Collet B, Cacas JL, Chardot T, Gallois JL, D'Andrea S (2018) PUX10 is a CDC48A adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in Arabidopsis. Plant Cell 30: 2116–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JL, Yu LH, Xu CC (2019) Dual role for autophagy in lipid metabolismin Arabidopsis. Plant Cell 31: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Mahalingam SS, Proietto M, Subramani S (2019) Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep 20: e46864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L (2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24: 2556–2563 [DOI] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Cui Y, Fu X, He Y, Zhao Q, Zeng Y, Shen J, Luo M, Jiang L (2015) Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci USA 112: 1886–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L (2017) Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci 22: 986–998 [DOI] [PubMed] [Google Scholar]
- Gao CJ, Yu CKY, Qu S, San MWY, Li KY, Lo SW, Jiang LW (2012) The golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and golgi retention signals at its C terminus. Plant Cell 24: 2086–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Park S, Pyc M, Yurchenko O, Cai YQ, Wu P, Andrews DW, Chapman KD, Dyer JM, Mullen RT (2016) Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol 170: 2052–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM (2019) Spastin joins LDs and peroxisomes in the interorganelle contact ballet. J Cell Biol 218: 2439–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JP, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-Y, Huang AHC (2017) Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol 174: 2248–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DQ, Sun YB, Ma ZM, Ke MY, Cui Y, Chen ZC, Chen CF, Ji CY, Tran TM, Yang L, et al. (2019a) Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc Natl Acad Sci USA 116: 21274–21284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Huang AH (2015) Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol 169: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Jiang LW, Zhuang XH (2019b) Possible roles of membrane trafficking components for lipid droplet dynamics in higher plants and green algae. Front Plant Sci 10: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH (2015) ESCRTs are everywhere. EMBO J 34: 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ (2013) The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Park KY, Seo YS, Kim WT (2016) Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol 170: 2494–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb C, Nagel MK, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E (2015) FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol 167: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar FK, Mengel LA, Muller AO, Schmitt K, Blersch KF, Valerius O, Braus GH, Ischebeck T (2018) PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30: 2137–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LTF, Yu C, Wong JSK, Lo HS, Benlekbir S, Jiang L, Lau WCY (2019) Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy 16: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Li YZ, Zhao Q, Li TT, Wei J, Li BY, Shen WJ, Yang C, Zeng YL, Rodriguez PL, et al. (2019) The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat Plants 5: 512–524 [DOI] [PubMed] [Google Scholar]
- Li MY, Baughman E, Roth MR, Han XL, Welti R, Wang XM (2014) Quantitative profiling and pattern analysis of triacylglycerol species in Arabidopsis seeds by electrospray ionization mass spectrometry. Plant J 77: 160–172 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Trelease RN (2006) Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci 119: 1961–1972 [DOI] [PubMed] [Google Scholar]
- Liu CL, Zeng YL, Li HB, Yang C, Shen WJ, Xu M, Xiao ZD, Chen TS, Li BY, Cao WH, et al. (2021) A plant-unique ESCRT component, FYVE4, regulates multivesicular endosome biogenesis and plant growth. New Phytol 231: 193–209 [DOI] [PubMed] [Google Scholar]
- Liu JY, Wang YN, Cheng YF (2020a) The ESCRT-I components VPS28A and VPS28B are essential for auxin-mediated plant development. Plant J 104: 1617–1634 [DOI] [PubMed] [Google Scholar]
- Liu NJ, Wang N, Bao JJ, Zhu HX, Wang LJ, Chen XY (2020b) Lipidomic analysis reveals the importance of GIPCs in Arabidopsis leaf extracellular vesicles. Mol Plant 13: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Mast FD, Herricks T, Strehler KM, Miller LR, Saleem RA, Rachubinski RA, Aitchison JD (2018) ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J Cell Biol 217: 2087–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. (2015) Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350: 1548–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Jiang L (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Nagel MK, Kalinowska K, Vogel K, Reynolds GD, Wu ZX, Anzenberger F, Ichikawa M, Tsutsumi C, Sato MH, Kuster B, et al. (2017) Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci USA 114: E7197–E7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, Maeda Y, Kagohashi Y, Kondo T, Yamada M, Fujimoto T, Sakai Y (2017) Evidence for ESCRT- and clathrin-dependent microautophagy. J Cell Biol 216: 3263–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Kiel JAKW, Williams C, Veenhuis M, van der Klei IJ (2011b) Membrane curvature during peroxisome fission requires Pex11. EMBO J 30: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Veenhuis M, van der Klei IJ (2011a) Peroxisomes: membrane events accompanying peroxisome proliferation. Int J Biochem Cell B 43: 847–851 [DOI] [PubMed] [Google Scholar]
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS (2018) ESCRT-mediated sorting and intralumenal vesicle concatenation in plants. Biochem Soc T 46: 537–545 [DOI] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC (2006) A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47: 917–933 [DOI] [PubMed] [Google Scholar]
- Pyc M, Cai YQ, Gidda SK, Yurchenko O, Park S, Kretzschmar FK, Ischebeck T, Valerius O, Braus GH, Chapman KD, et al. (2017b) Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J 92: 1182–1201 [DOI] [PubMed] [Google Scholar]
- Pyc M, Cai YQ, Greer MS, Yurchenko O, Chapman KD, Dyer JM, Mullen RT (2017a) Turning over a new leaf in lipid droplet biology. Trends Plant Sci 22: 596–609 [DOI] [PubMed] [Google Scholar]
- Reyes FC, Buono RA, Roschzttardtz H, Di Rubbo S, Yeun LH, Russinova E, Otegui MS (2014) A novel endosomal sorting complex required for transport (ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J Biol Chem 289: 4980–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers J, Behl C (2021) Lipid droplets and autophagy-links and regulations from yeast to humans. J Cellular Biochem 122: 602–611 [DOI] [PubMed] [Google Scholar]
- Shen JB, Gao CJ, Zhao Q, Lin YS, Wang XF, Zhuang XH, Jiang LW (2016) AtBRO1 functions in ESCRT-I complex to regulate multivesicular body protein sorting. Mol Plant 9: 760–763 [DOI] [PubMed] [Google Scholar]
- Shen JB, Zhao Q, Wang X, Gao CJ, Zhu Y, Zeng Y, Jiang L (2018) A plant Bro1 domain protein BRAF regulates multivesicular body biogenesis and membrane protein homeostasis. Nat Commun 9: 3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Hara-Nishimura I (2015) Leaf oil bodies are subcellular factories producing antifungal oxylipins. Curr Opin Plant Biol 25: 145–150 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL (2006) ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep 7: 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS (2009) Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 122: 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen PK, Shen JB, Sun SSM, Jiang LW (2010) Expression and characterization of two functional vacuolar sorting receptor (VSR) proteins, BP-80 and AtVSR4 from culture media of transgenic tobacco BY-2 cells. Plant Sci 179: 68–76 [Google Scholar]
- Thazar-Poulot N, Miquel M, Fobis-Loisy I, Gaude T (2015) Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc Natl Acad Sci USA 112: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N (2012) Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Fang N, Xiong J, Du YJ, Cao Y, Ji WK (2021) An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. Nat Commun 12: 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ZJ, Bartel B (2020) Peroxisomes form intralumenal vesicles with roles in fatty acid catabolism and protein compartmentalization in Arabidopsis. Nat Commun 11: 6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie LJ, Yu LJ, Chen QF, Wang FZ, Huang L, Xia FN, Zhu TR, Wu JX, Yin J, Liao B, et al. (2015) Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J 81: 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, Galea A, Sytnyk V, Crossley M (2012) Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol 24: 509–516 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Niwa H, Honsho M, Itoyama A, Fujiki Y (2015) Pex11mediates peroxisomal proliferation by promoting deformation of the lipid membrane. Biol Open 4: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang XH, Chung KP, Cui Y, Lin WL, Gao CJ, Kang BH, Jiang LW (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA 114: E426–E435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.