In cotton, the basic helix–loop–helix transcription factor GhTCE1 forms part of a unique regulatory network involving reactive oxygen species homeostasis and cell wall metabolism that acts as a molecular switch to determine cell fate between unorganized callus growth versus organized development.
Abstract
Wounded plant cells can form callus to seal the wound site. Alternatively, wounding can cause adventitious organogenesis or somatic embryogenesis. These distinct developmental pathways require specific cell fate decisions. Here, we identify GhTCE1, a basic helix–loop–helix family transcription factor, and its interacting partners as a central regulatory module of early cell fate transition during in vitro dedifferentiation of cotton (Gossypium hirsutum). RNAi- or CRISPR/Cas9-mediated loss of GhTCE1 function resulted in excessive accumulation of reactive oxygen species (ROS), arrested callus cell elongation, and increased adventitious organogenesis. In contrast, GhTCE1-overexpressing tissues underwent callus cell growth, but organogenesis was repressed. Transcriptome analysis revealed that several pathways depend on proper regulation of GhTCE1 expression, including lipid transfer pathway components, ROS homeostasis, and cell expansion. GhTCE1 bound to the promoters of the target genes GhLTP2 and GhLTP3, activating their expression synergistically, and the heterodimer TCE1-TCEE1 enhances this activity. GhLTP2- and GhLTP3-deficient tissues accumulated ROS and had arrested callus cell elongation, which was restored by ROS scavengers. These results reveal a unique regulatory network involving ROS and lipid transfer proteins, which act as potential ROS scavengers. This network acts as a switch between unorganized callus growth and organized development during in vitro dedifferentiation of cotton cells.
Introduction
Developmental plasticity in response to environmental conditions is a key distinguishing feature of plants, and this is seen clearly in the response to mechanical wounding. Following wounding, differentiated somatic cells undergo redifferentiation either to form wound callus that seal the wound site to prevent infection or to undergo de novo organogenesis, such as adventitious rooting or, during tissue culture, somatic embryogenesis (SE) (e.g. Chen et al., 2014; Yu et al., 2017). A major question is how decisions are made at the molecular level to determine the choice of alternative pathways of either disorganized growth (callus formation) or organized growth, such as de novo meristem formation and SE.
External stimuli such as wounding are critical for callus initiation via cell division, and exogenous hormones are necessary for the propagation of callus in vitro and for the experimental induction of SE (Ikeuchi et al., 2017; Xu et al., 2018). Various environmental stresses such as drought, salt, and reactive oxygen species (ROS) can accelerate adventitious organogenesis or SE by repressing nonembryonic callus proliferation and promoting the transition from nonembryonic callus to callus with morphogenic potential. This can lead to the development of somatic embryos or root or shoot meristems (Jin et al., 2013; Fehér, 2015; Altpeter et al., 2016).
Adventitious organogenesis is under finely tuned regulation, whereby somatic cells re-enter the cell cycle and exhibit pluripotency via chromatin reprogramming (Lee and Seo, 2018). Callus induction is a prerequisite for this, and the callus induction rate, callus cell morphology, and the rate of callus proliferation all influence regeneration efficiency (Li et al., 2013; Zhang et al., 2019). Auxin and cytokinin signaling pathways are essential during organized development from callus (Yang et al., 2012; Liu et al., 2016). Gene expression related to stress responses, control of cell identity, and chromatin remodeling is also required (Ikeuchi et al., 2019). Several genes that function during SE and other types of organ regeneration have been identified via forward and reverse genetic approaches in Arabidopsis thaliana (Arabidopsis, Ikeuchi et al., 2017, 2019). More than half of these genes encode transcription factors (TFs), and given that TFs account for less than 6% of total protein-coding genes (Riechmann, 2002), it is likely that TF networks are of central importance as regulators during de novo organogenesis. To date, thousands of genes have been found to be expressed during SE or organogenesis (Yang et al., 2012; Du et al., 2019), but only a small number of them have been verified experimentally for function. Furthermore, callus has typically been studied as a single tissue even though it is heterogeneous in terms of cell morphology, metabolism, and developmental potential (Lindsey and Yeoman, 1983).
In a previous study in cotton (Gossypium hirsutum), we found that dedifferentiated callus cells are very different morphologically from highly organized somatic cells, and 466 TFs were identified as differentially expressed genes (DEGs) at different stages of SE (Yang et al., 2012). In the current study, we focused on the cell fate transition at an early stage of callus formation during in vitro dedifferentiation, and found that a basic helix–loop–helix (bHLH) family TF, GhTCE1, specifically promotes callus cell elongation by activating interacting lipid transfer and ROS homeostasis pathway genes and repressing adventitious organogenesis. Our results provide an important new role for lipid transfer proteins and ROS signaling in regulating early cell fate transitions following wound-induced cell division.
Results
GhTCE1 promotes callus cell elongation during dedifferentiation of cotton cells
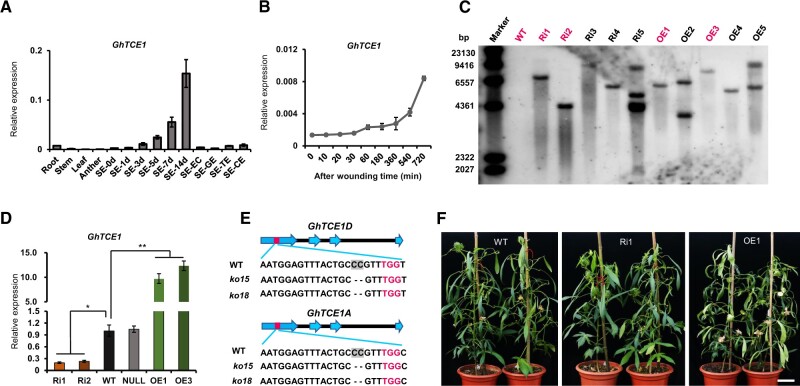
GhTCE1 (Gh_D06G2316) was identified during an RNA-seq profiling analysis during cotton SE (Yang et al., 2012), and was chosen for further study because, as a potential TF, it might play a role in early events during callus formation or SE. The 996-bp cDNA encodes a predicted bHLH family TF of 331 amino acid residues. The expression pattern of GhTCE1 was determined in cotton root, stem, leaf, anther, and at different time points during SE from cultured hypocotyl explants, and at earlier time points after explant wounding. GhTCE1 was expressed at low levels in vegetative and reproductive organs, but much more strongly during different stages of SE (Figure 1A). Moreover, it was induced at earlier time points after wounding, with the increased expression being detectable within 60 min, and very strong by 720 min, after wounding (Figure 1B), suggesting a role in the reprogramming process.
Figure 1.
Analysis of GhTCE1 expression and characterization of transgenic plants. A, Expression pattern of GhTCE1 in different tissues. SE-0d, -1d, -3d, -5d, -7d, or -14d means the explants of hypocotyls were induced for 1, 3, 5, 7, or 14 days during SE (embryogenic callus [SE-EC], globular stage embryo [SE-GE], torpedo stage embryo [SE-TE], and cotyledon stage embryo [SE-CE]). B, Expression pattern of GhTCE1 soon after wounding. The expression of GhUB7 was used as internal control. C, Immunoblot analysis of GhTCE1 transgenic lines. The first lane represents the λ DNA size marker. The DNA samples were digested with HindIII. The magenta front indicates the selected lines for study. D, Relative expression of GhTCE1 in transgenic lines 7 days post SE induction. WT, wild type; Ri1 and Ri2, Ri lines; OE1 and OE3, OE lines. Null, negative plants segregated from Ri1 in the T1 generation. The expression of GhUB7 was used as internal control. E, Sequence analysis of GhTCE1(A/D) double mutants ko15 and ko18 at the target sites. The PAM motif is marked in magenta and GhTCE1A and GhTCE1D can be distinguished by the SNP C/T (marked in green) downstream of the PAM motif. The black dotted line represents deletion compared with WT. F, Phenotypes of GhTCE1 transgenic plant lines grown on soil. Bars = 20 cm. Error bars in (A), (B), and (D) represent ± standard error of three biological replicates (∼20 explants were used as one replicate). Significance tests compared each transgenic line with WT plants. *P < 0.05, **P < 0.01, Supplemental Data Set S2.
Gh_D06G2316 and Gh_A06G0241 are paralogs on the D and A subgenomes of tetraploid cotton, sharing 98.8% sequence identity (Supplemental Figure S1). In order to explore the function of GhTCE1, we created RNAi (Ri) lines, overexpression (OE) lines, and CRISPR-mediated GhTCE1D/A (Gh_D06G2316/Gh_A06G0241) double mutants. Two Ri lines (Ri1 and Ri2), two OE lines (OE1 and OE3), a wild-type line (WT), as well as an Ri-negative line (Null) that segregated from Ri1 in the T1 generation, were selected for detailed analysis. Immunoblotting analyses revealed that each of these Ri and OE transgenic lines contain a single-copy T-DNA insertion (Figure 1C). The expression levels of GhTCE1 decreased to 24.0% of WT levels in Ri1 and to 21.6% in Ri2, and increased in the OE lines by 833.8% (OE1) and 1036.2% (OE3) compared with WT (Figure 1D). In addition, we created two double mutant lines ko15 and ko18 using CRISPR/Cas9-mediated gene editing. These were confirmed by high-throughput sequencing as frame-shift mutations and were selected for further study (Figure 1E). OE and Ri mature plants were morphologically similar to WT (Figure 1F).
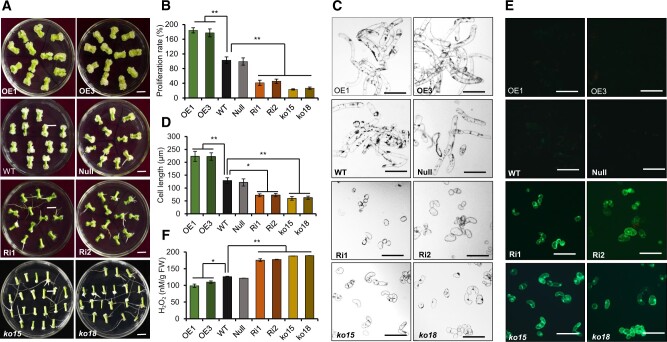
Wounded explants from different transgenic lines were cultured on MSB (MS medium plus B5 vitamins) medium according to our previous methods (Yang et al., 2012). At 20 days post wounding, significantly more adventitious roots were produced and a higher ratio of adventitious root regeneration was observed on Ri and mutant explants compared with WT plants. In contrast, the OE lines showed callus growth but no adventitious organogenesis (Figure 2A and Supplemental Figure S2). Both the callus proliferation rate (CPR), which is a measure of the fresh weight of callus tissue produced over time, and callus cell length were measured at 20 days post wounding. The CPR of lines Ri1 and Ri2 was reduced to 45.3% and 56.9%, respectively, compared with WT, and was even lower in ko15 (24.5%) and ko18 (30.7%) mutants (Figure 2B). However, the CPR increased 173.1% in OE1 and 182.9% in OE3, respectively, compared with WT (Figure 2B). Callus cell length was correspondingly reduced in loss-of-function lines compared with WT to 46.3% (Ri1), 37.9% (Ri2), 42.3% (ko15), and 51.8% (ko18), and it increased to 144% in OE1 and 135% in OE3 (Figure 2, C and D). These results show that GhTCE1 promotes callus cell growth and fresh weight (principally a reflection of increased cell length) but represses adventitious organogenesis.
Figure 2.
Phenotypic analysis of GhTCE1 transgenic tissues. A, The phenotypes of callus 20 days post wounding from different transgenic lines and mutants. ko15 and ko18, mutation lines. B, CPR of different lines 20 days post wounding. Error bars represent ± standard error of six biological replicates (∼20 explants each). C, Micrographs of callus cells of different lines 20 days post wounding. Bars = 200 μm. D, Statistical analysis of callus cells length of different lines 20 days post wounding. Error bars represent ± standard error of six biological replicates (∼50 cells each). E, DCFH-DA staining of ROS in GhTCE1 transgenic plants and mutants. F, H2O2 concentration in callus from GhTCE1 transgenic plants 20 days post wounding. Error bars represent ± standard errors of four biological replicates (∼20 explants each). Significance tests compared each transgenic line with WT plants. *P < 0.05, **P < 0.01, Supplemental Data Set S2.
GhTCE1 modulates ROS but not auxin homeostasis during cotton cell dedifferentiation
ROS and auxin play essential roles in cell fate determination and growth (Chapman et al., 2019). To examine the possible role for ROS, we monitored its accumulation by 2,7-dichloro-dihydrofluorescein diacetate (DCFH-DA) staining. DCFH-DA can permeate cell membranes and then be oxidized by ROS into impermeant DCF, which is fluorescent under UV light. The GhTCE1-Ri lines and mutant lines showed strong green fluorescence after DCFH-DA staining, but none was seen in WT or OE lines (Figure 2E). H2O2 concentrations were assayed and found to be significantly higher in Ri lines than in WT, and were significantly lower in OE lines than WT (Figure 2F). This demonstrates that GhTCE1 is a regulator of ROS accumulation, and that high levels of GhTCE1 expression are associated with reduced ROS accumulation, increased callus cell growth, and reduced adventitious organogenesis.
To determine the possible role of auxin in GhTCE1-mediated cell reprogramming, we measured IAA concentrations in the different lines at 20 days post wounding. Whereas one Ri line and one OE line had altered IAA levels, the IAA concentrations in all other lines were comparable, and on average similar to levels in WT (Supplemental Figure S3A). We also treated the explants with a synthetic auxin analog indole-3-butyric acid (IBA) to explore whether auxin treatment can mimic the phenotypes of wounded transgenic tissues. Whereas exogenous auxin promoted cell dedifferentiation and callus proliferation at 10 days post wounding, it promoted adventitious root formation in a concentration-dependent manner, and inhibited callus cell proliferation, at 20 days post wounding (Supplemental Figure S3B). Therefore, exogenous auxin treatments phenocopied the OE-like phenotype at 10 days and the Ri-like phenotype at 20 days post wounding (Supplemental Figure S3B). This suggests that auxin may function during organogenesis, but it is not likely be the cause of differential cell proliferation between GhTCE1 transgenic lines.
GhTCE1 targets and activates the expression of cell elongation-related genes during cotton cell dedifferentiation
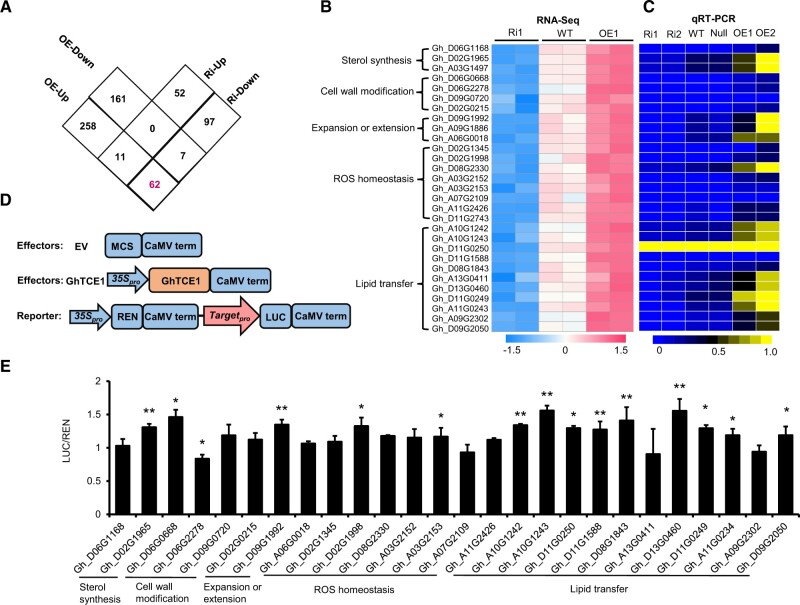
To explore the underlying mechanism by which GhTCE1 promotes disorganized cell growth, we conducted a transcriptomics analysis using the explants of the different transgenic lines at 7 days post wounding. Differences in cell dedifferentiation among WT, Ri, and OE were first apparent at this stage (Supplemental Figure S4). DEGs were defined based on a false discovery rate (FDR) of ≤0.05 and fold change of |log2| ≥ 1. We identified 331 up-regulated and 168 down-regulated DEGs in OE, and 63 up-regulated and 166 down-regulated DEGs in Ri compared with WT; 62 DEGs overlapped between OE-up and Ri-down DEGs, and included GhTCE1 (Figure 3A). GhTCE1 is expected to exhibit transcriptional activation activity, and so the remaining 61 DEGs identified were considered as potential GhTCE1 targets. Their expression levels as fragments per kilobase per million mapped (FPKM) are presented in Supplemental Data Set S1.
Figure 3.
Analysis of downstream targets of GhTCE1. A, Differential gene (DEG) analysis in GhTCE1 Ri and OE lines compared with WT. Screening for up or down regulated genes used a stringency of P ≤ 0.05 and a log2 fold change of ≥1 or ≤–1 compared with WT. Hypocotyls at 7 days post wounding were used for RNA-seq analyses, with two biological replicates (∼50 explants each) for each line. The magenta front indicates 62 DEGs overlapped between OE-up and Ri-down. B, Heat-map analysis of 29 DEGs from enriched pathways that were up-regulated or down-regulated in OE and Ri lines, respectively. C, Heat-map analysis of the 29 DEGs from (B) using qRT-PCR, with three biological replicates for each line. D, Schematic diagram of effector and reporter constructs. Orange rectangle, the effector; pink arrow, the reporter; blue rectangles, other vector components; blue arrows, 35S promoters. The empty vector (EV) was used as a negative control. E, Transactivation analysis between GhTCE1 and its candidate targets in cotton callus protoplasts using the LUC reporter system. Promoters from the candidate targets were fused to upstream of LUC in pGreenII 0800-LUC. 35Spro:REN in pGreenII 0800-LUC was used as internal control. GhTCE1 was fused downstream of the CaMV 35S promoter (35Spro) in pGreenII 62-SK. pGreenII 62-SK empty vector was used as a negative control and designated EV. Relative activation level represents the ratio of fluorescence intensity of LUC/REN with GhTCE1 to LUC/REN without GhTCE1. Error bars in (E) represent ± standard errors of three biological replicates (10 samples each). Significance tests compared the expression of Promoter:LUC+35Spro:REN+35Spro:GhTCE1 with Promoter:LUC+35Spro:REN+EV, P < 0.05 (*) or P < 0.01 (**), Supplemental Data Set S2.
Nearly half of the candidates (29 of 61) have annotated functions in sterol synthesis, cell wall modification, cell expansion, ROS homeostasis, and lipid transfer (Figure 3B and Supplemental Data Set S1). These functions are all potentially related to cell elongation, and were considered as candidate targets of GhTCE1. The expression levels of the 29 candidate targets were verified by qRT-PCR analysis, and all were upregulated in both OE1 and OE3, and 28 were downregulated in both Ri1 and Ri2 (Figure 3C). The expression levels from RNA-seq highly correlated with qRT-PCR results (Supplemental Figure S5, R2 = 0.9258). The remaining 32 DEGs are involved in various processes such as cellular metabolism, signal transduction, or protein modification (Supplemental Table S1). It is notable that no differentially expressed auxin metabolism- or signal transduction-related genes were found.
GhTCE1 is annotated as a bHLH TF and subcellular localization assays show that it is localized to the nucleus (Supplemental Figure S6A). bHLH family TFs can bind the E-box (5′-CANNTG-3′) or G-box (5′-CACGTG-3′) of target gene promoters, depending on sequences in the basic region (Toledo-Ortiz et al., 2003), and sequence analysis suggests that GhTCE1 belongs to the E-box binding sub-family. To confirm the transcriptional activity of GhTCE1, activation assays were carried out in yeast (Saccharomyces cerevisiae). The full length GhTCE1 coding sequence was fused to a GAL4 DNA binding domain (BD), designated Full:BD. The yeast strain Y2HGold harboring Full:BD can activate the GAL4 reporter gene (Supplemental Figure S6B), demonstrating that GhTCE1 is a transcriptional activator. To profile the region responsible for its transcriptional activation, we constructed a series of truncated GhTCE1:BD sequences in yeast Y2HGold and mated yeast strains containing them with yeast strain Y187 harboring the empty vector pGADT7 (Supplemental Figure S6C). In diploid yeast, the N-terminal region (1–160 aa) can activate the reporter gene while the C-terminal region (161–331 aa) cannot (Supplemental Figure S6D). Sequences comprising either 80–240 aa or 80–331 aa can also activate the reporter gene, suggesting that the region from 80 to 160 aa is critical for GhTCE1 transcriptional activation activity (Supplemental Figure S6D).
We downloaded 2-kb promoter region sequences (i.e. upstream of the translation start site) of 29 candidate target genes with potential roles in cell elongation from the cotton genome database (https://cottonfgd.org/) and then determined the presence and number of E-boxes. The number of E-boxes ranged from 2 to 13 (Supplemental Table S2). We cloned 26 promoters from the 29 candidates for testing in transactivation studies, which used a luciferase (LUC) reporter activation system in tobacco (Nicotiana benthamiana) leaves (Figure 3D). Of the 26 candidate promoters, 14 were activated by GhTCE1 to relatively low levels, less than two-fold above basal levels (Figure 3E). One possible explanation for this relatively low activation is that the full transcriptional activation of downstream targets may require protein partners which are absent in leaf.
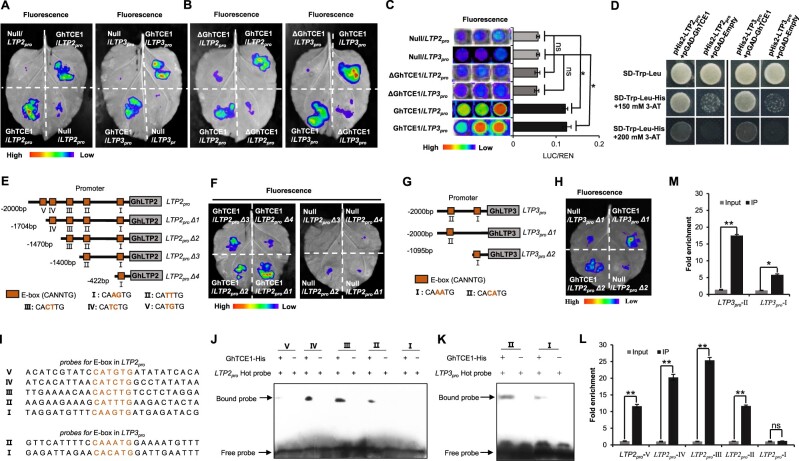
As GhLTPs are the most abundant members among the candidate GhTCE1 targets, and both GhLTP2 and GhLTP3 are highly expressed during SE, we selected the promoters of GhLTP2 and GhLTP3 for transcriptional activation assays with both full length GhTCE1 and the N-terminal truncated GhTCE1 (ΔGhTCE1). The results show that GhTCE1 can interact with the promoters of both GhLTP2 (LTP2pro) and GhLTP3 (LTP3pro), promoting LUC expression in tobacco leaves. In contrast, the N-terminal truncated version of GhTCE1 did not activate LTP2pro and LTP3pro (Figure 4, A and B). Meanwhile, we analyzed the activated expression of LTP2pro and LTP3pro by GhTCE1 and ΔGhTCE1 in cotton callus protoplasts using the LUC expression system, and our results show that GhTCE1 can activate LUC expression, but ΔGhTCE1 was unable to activate LUC expression (Figure 4C). To confirm that GhTCE1 can interact with the promoters of LTP2 and LTP3, we carried out yeast-one-hybrid (Y1H) experiments. There was no detectable interaction between GhTCE1 and the empty pGAD vector on SD/-Trp-Leu-His medium. However, in the presence of 3-amino-1,2,4-triazole (3-AT), GhTCE1 interacted with LTP2pro and LTP3pro (Figure 4D), confirming that GhTCE1 can bind to the promoters of LTP2 and LTP3.
Figure 4.
GhTCE1 binds to the promoters of GhLTP2 and GhLTP3 to activate their expression. A and B, LUC signals detected in tobacco leaves. Promoters of GhLTP2 and GhLTP3 (LTP2pro and LTP3pro) were separately fused upstream of LUC in the vector pGreenII 0800-LUC and designated LPT2pro:LUC and LPT3pro:LUC. As an internal control, 35Spro:REN in vector pGreenII 0800-LUC was used. GhTCE1 and ΔGhTCE1 were fused downstream of the 35S promoter in pGreenII 62-SK. The pGreenII 62-SK empty vector was used as negative control and named Null. For each tobacco leaf, half was used to express pGreenII 0800-LUC:LTPpro+pGreenII 62-SK:GhTCE1+35Spro:REN (named GhTCE1/LTPpro) and half was used to transiently express pGreenII 0800-LUC:LTPpro+pGreenII 62-SK empty vector:REN (named Null/LTPpro) following Agrobacterium-mediated transfection, ΔGhTCE1 in the same way. C, Activated expression of LTP2pro and LTP3pro by GhTCE1 in cotton callus protoplasts using the LUC expression system. Relative activation level represents the ratio of fluorescence intensity of LUC/REN with GhTCE1 to LUC/REN without GhTCE1. D, Y1H assays of GhTCE1 binding to LTP2pro and LTP3pro. The full-length coding sequence of GhTCE1 was fused to the pGADT7 vector, and LTP2pro and LTP3pro were cloned into the pHis2 vector. Interaction was determined on selective medium lacking Trp+Leu+His in the presence of 3-AT. E, A series of truncations (LTP2proΔ1 to LTP2proΔ4) with E-box deletions were used in the LUC activation system vector. F, GhTCE1 activates the expression of truncated LTP2pro by detected LUC signals on tobacco leaves. The truncated LTP2pro constructs were fused to the vector pGreenII 0800-LUC, and combined with pGreenII 62-SK:GhTCE1+35Spro:REN as a positive control. The pGreenII 62-SK empty vector was used as a negative control. They are labeled the same way as in (A) and (B). G, GhTCE1 activates the expression of truncated LTP3pro (LTP3proΔ1 andLTP3proΔ2) by LUC signals detected on tobacco leaves. H, GhTCE1 activates the expression of truncated LTP3pro (LTP3proΔ1 and LTP3proΔ2) by LUC signals detected on tobacco leaves. The process same to (F). I, The probe sequences of LTP2pro and LTP3pro are noted and the E-boxes are shown in red. J and K, EMSA showing the specificity of in vitro binding between GhTCE1 and the E-box elements of LTP2pro (J) and LTP3pro (K). The WT probes were subjected to biotin-labeling. No GhTCE1-His tag protein lines as control. The arrows indicate the up-shifted binds. L and M, Validation of the binding of GhTCE1 to the promoters E-box elements of LTP2pro (L) and LTP3pro (M) by ChIP-qPCR. Error bars in (C), (L), and (M) represent ± standard errors of three biological replicates (10 samples each in [C], three samples each in [L] and [M]). Statistical significance is indicated by *P < 0.05, **P < 0.01, Supplemental Data Set S2.
GhTCE1 belongs to the E-box (5′-CANNTG-3′) binding bHLH TF family, and in order to identify directly the transactivation region, we used a LUC reporter activation system in tobacco leaves and cotton callus protoplasts using the full-length and truncated GhLTP2 promoter (LTP2proΔ1 to LTP2proΔ4) regions and the E-boxes denoted as V–I, respectively (Figure 4E). The results showed that, in addition to the full-length promoter, LTP2proΔ1, LTP2proΔ2, and LTP2proΔ3 can each be activated for target LUC fusion expression in tobacco leaves (Figure 4F). Similarly, we examined LTP3pro and its two truncated forms (LTP3proΔ1 to LTP3proΔ2) and the E-boxes II and I (Figure 4G). The results show that both LTP3proΔ1 and LTP3proΔ2 can be activated for LUC expression (Figure 4H). To further verify the specificity of binding between GhTCE1 and the CANNTG motif, we generated a series of 26 bp probes for different E-boxes in the LTP2pro and LTP3pro to conduct electrophoretic mobility shift assays (EMSAs) (Figure 4I). Probes V–II, but not I, of LTP2pro, and both I and II of LTP3pro were bound by the GhTCE1 protein (Figure 4, J and K). In addition, chromatin-immunoprecipitation quantitative PCR (ChIP-qPCR) analysis shows that the promoter region of E-box II–IV of LTP2pro and E-box I–II of LTP3pro was enriched after immunoprecipitation of GhTCE1 (Figure 4, L and M).
The activation of other candidate targets of GhTCE1 in leaf or other tissues which undergo cell expansion or dedifferentiation following wounding was also investigated. The expression of GhTCE1 was barely detectable in young leaves of WT and Ri lines, whereas high expression levels (comparable to those seen in SE-7d tissues) were detected in the OE lines (Supplemental Figure S7A). Among the 29 candidate target genes, only 6 were expressed at higher levels in OE leaves than in WT (Supplemental Figure S7A). However, the expression levels of all the activated genes in young leaf were much lower than in callus at 7 days post wounding (SE-), and the average relative expression ratio of the activated candidates between young leaf and callus 7 days post wounding (Leaf/SE) ranged from 0.0008 to 0.0764 (Supplemental Figure S7A). This suggests that GhTCE1 cannot fully activate its candidate targets in young leaves, an idea consistent with the lack of morphological differences between GhTCE1 OE, Ri lines, and WT (Figure 1F). To investigate GhTCE1 expression during cell dedifferentiation, callus formation was induced by stem grafting (Supplemental Figure S7B). Expression of GhTCE1 changed a little during grafting-induced callus formation (Supplemental Figure S7C). Among the 29 candidate target genes of GhTCE1, 7 were induced during callus formation (Supplemental Figure S7C), suggesting possible roles in cell dedifferentiation and callus formation, but in a GhTCE1-independent manner. Given that OE of GhTCE1 inhibits organogenesis from wounded explants, we conclude it must be down-regulated after the early callus induction phase and prior to organogenesis.
Reduced expression of GhLTP2 and GhLTP3 results in increased ROS accumulation and reduced callus cell growth during dedifferentiation
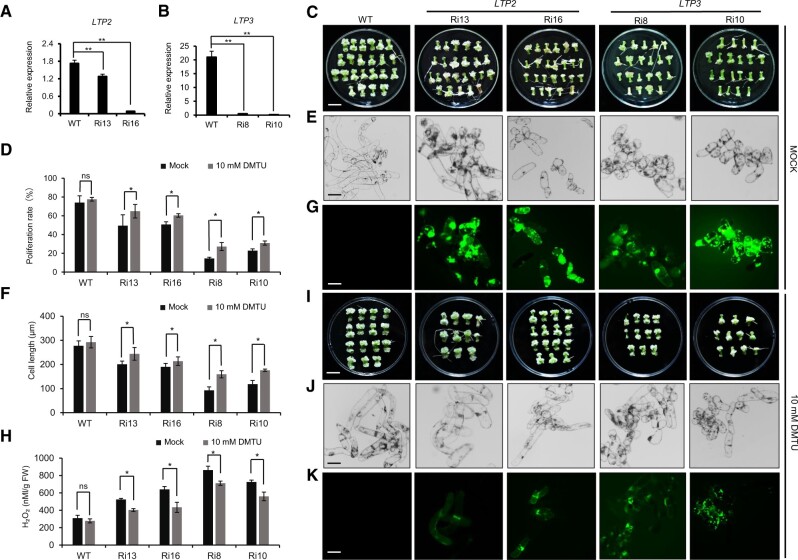
The above analysis shows that GhTCE1 activates the expression of both GhLTP2 and GhLTP3. To determine whether GhLTP2 and GhLTP3 (GhLTP2/3) also function in the control of callus formation or cell differentiation in cotton, GhLTP2-Ri13/Ri16 and GhLTP3-Ri8/Ri10 transgenic lines were obtained by Ri, and their expression was highly suppressed (Figure 5, A and B). The wounded explants from different transgenic lines showed significantly decreased CPR compared with WT, having smaller cell size during callus formation (Figure 5, C–F). DCFH-DA staining showed that callus from both GhLTP2 and GhLTP3 Ri lines accumulate more ROS than WT, and H2O2 concentrations increased significantly in the Ri lines (Figure 5, G and H).
Figure 5.
Phenotypic analysis of GhLTP2 and GhLTP3 Ri transgenic tissues. A and B, Relative expression of GhLTP2 and GhLTP3 in WT and respective Ri transgenic lines. Error bars represent ± standard error of three biological replicates as described in Figure 1A. C, Phenotype of callus 20 days post wounding for different transgenic lines following mock treatment. Bar = 1 cm. D, CPR of different lines 20 days post wounding following mock treatment or treatment with 10 mM DMTU. Error bars represent ± standard error of six biological replicates as described in Figure 2B. E, Micrographs of callus cells, under mock treatment. Bar = 100 μm. F, Statistic analysis of callus cell lengths of different lines 20 days post wounding under mock treatment and 10 mM DMTU. Error bars represent ± standard error of six biological replicates as described in Figure 2D. G, DCFH-DA staining of ROS in WT, GhLTP2, and GhLTP3 transgenic callus cells. Bar = 100 μm. H, H2O2 concentration in callus from transgenic plants 20 days post SE induction. Error bars represent ± standard errors of three biological replicates as described in Figure 2F. I–K, Phenotype, micrograph, and DCFH-DA staining of ROS in callus cells, 20 days post wounding from different transgenic lines under 10 mM DMTU treatment. Bar = 1 cm in I and 100 μm in J and K. Significance tests compared each transgenic line with WT plants. *P < 0.05, **P < 0.01. Results of statistical analyses are presented in Supplemental Data Set S2.
To further investigate the possible role for GhTCE1-GhLTP2/3 in the regulation of ROS homeostasis and a link with cell elongation, tissues were grown in the presence of the ROS scavenger 1,3-dimethylthiourea (DMTU) to determine any effects on callus formation or growth. All GhLTP2 and GhLTP3 (GhLTP2/3) Ri lines, but particularly GhLTP2-Ri13 and GhLTP2-Ri16, had more callus growth in the presence of 10 mM DMTU than did mock controls at 20 days post induction, but they had no additional lateral roots (Figure 5, D and I). The cell length of the Ri lines was also significantly greater in the presence of DMTU (Figure 5, F and J). GhLTP2/3 Ri lines both exhibited less ROS fluorescence in the presence of DMTU, with H2O2 concentrations also lower than those seen with mock control treatments (Figure 5, H and K). In addition, we also analyzed the ratio of adventitious root regeneration of GhLTP2/3 transgenic lines and WT plants, and the results were consistent with those for knockout or Ri GhTCE1 tissues. Compared with WT plants, LTP2/3 Ri lines showed a higher ratio of adventitious root regeneration, and this was significantly reduced in the presence of 10 mM DMTU (Supplemental Figure S8). These results further indicate that GhLTP2/3 expression is associated with low ROS accumulation and increased disorganized cell growth.
GhTCEE1 enhances the activation ability of GhTCE1 by forming heterodimers
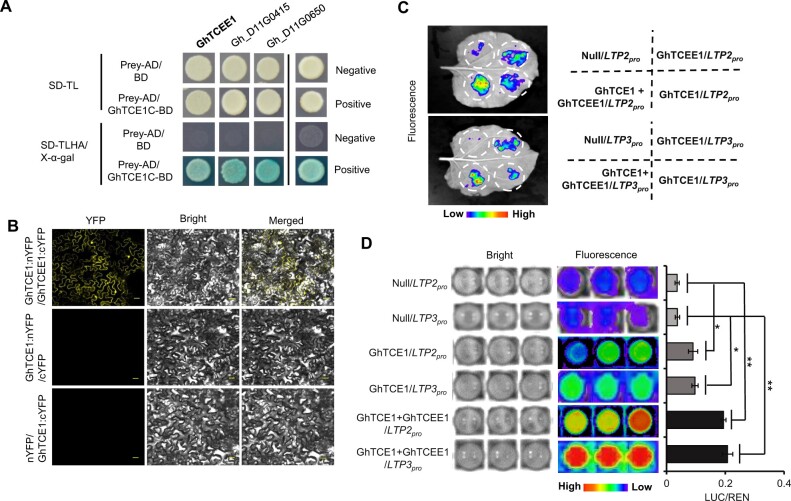
We have shown here that GhTCE1 activates cell elongation-related genes during somatic cell dedifferentiation and callus formation, whereas the activation of these genes by GhTCE1 is much weaker in cotton and tobacco leaves (Figure 3E). This suggests that GhTCE1 alone is not sufficient for full transcriptional activation and that other partners may be also involved during cell reprogramming. To identify potential partner proteins, the activation domain-truncated version of GhTCE1 (C-terminal from 161 to 331 aa), which contains the bHLH domain, was used as bait for yeast two-hybrid screening with the prey library constructed from callus cDNA isolated early after wounding. A total of 10 GhTCE1 candidate interacting proteins were identified (Supplemental Table S3). It has been reported that bHLHs usually regulate downstream targets by dimerization (Toledo-Ortiz et al., 2003), and so we focused on members of this family of TFs. We found that three bHLH proteins (Gh_D11G3523, Gh_D11G0415, and Gh_D11G0650) interacted with GhTCE1 (Figure 6A and Supplemental Figure S9). The interaction between GhTCE1 and GhTCEE1 (Gh_D11G3523) was further verified by bimolecular fluorescence complementation (BiFC) (Figure 6B). Consistent with interaction between the two proteins, the expression of GhTCEE1 was induced at earlier time-points after wounding, was detectable within 540 min after wounding, and increased strongly beyond this to 720 min after wounding, showing a co-expression pattern with GhTCE1 (Supplemental Figure S10).
Figure 6.
GhTCEE1 enhances the activation activity of GhTCE1 by forming heterodimers. A, Sequences encoding the GhTCE1 C-terminus from aa residues 160–331 without transactivation activity were fused to sequences encoding a GAL4 BD domain and designated GhTCE1-BD. The prey AD was linked to callus cDNAs. Negative represents no growth, positive represents growth. SD-TL/TLHA represents synthetically defined medium dropouts Trp, Leu or Trp, Leu, His, and Ade, respectively. B, BiFC assay between GhTCE1–nYFP and GhTCEE–cYFP in tobacco epidermal cells. Bars = 30 μm. C, LUC assays in tobacco leaves and cotton callus protoplasts. Promoters of GhLTP2 and GhLTP3 were, respectively, fused upstream of LUC. 35Spro:REN was used as an internal control. GhTCE1 and GhTCEE1 were fused downstream of 35Spro in pGreenII 62-SK. The pGreenII 62-SK empty vector was used as control named Null. For each tobacco leaf, four constructs were expressed: (i) pGreenII 0800-LUC:GhLTP2pro+35Spro:REN+pGreenII 62-SK empty (Null/LTP2pro): (ii) pGreenII 0800-LUC:GhLTP2pro+35Spro:REN+pGEreenII 62-SK:GhTCE1 (GhTCE1/LTP2pro); (iii) pGreenII 0800-LUC:GhLTP2pro+35Spro:REN+pGreenII 62-SK:GhTCEE1 (GhTCEE1/LTP2pro); and (iv) pGreenII 0800-LUC:GhLTP2pro+ 35Spro:REN+pGreenII 62-SK:GhTCE1+GhTCEE1 (GhTCE1+GhTCEE1/LTP2pro) following Agrobacterium-mediated transfection. D, Activation of LUC expression by GhTCE1/GhTCEE1 in cotton callus protoplasts. Relative activation level represents the ratio of fluorescence intensity of LUC/REN with GhTCE1 to LUC/REN without GhTCE1. Error bars represent ± standard errors of three biological replicates as described in Figure 3E. Statistical significance is indicated by *P < 0.05, **P < 0.01, Supplemental Data Set S2.
The LUC reporter activation assay was also used to verify the enhanced activation ability of GhTCE1 by GhTCEE1. The results showed that GhTCEE1 can activate GhLTP2pro and GhLTP3pro for LUC expression in a similar way as does GhTCE1 (Figure 6C). Furthermore, co-expression of GhTCE1 and GhTCEE1 led to significantly higher levels of GhLTP2pro and GhLTP3pro for LUC expression than either GhTCE1 or GhTCEE1 alone (Figure 6D), showing that GhTCE1 and its interacting protein GhTCEE1 function cooperatively to activate the expression of both GhLTP2 and GhLTP3.
Discussion
GhTCE1 regulates callus and adventitious root formation in an antagonistic manner
Callus and adventitious roots are typically induced following wounding or exogenous hormone application. The mutants alf4-1 and clf-50 swn-1 are defective in callus formation and also failed to produce adventitious roots, suggesting that callus and adventitious roots share some common genetic pathways (Liu et al., 2014). It has also been suggested that callus, formed from multiple organs, shares a gene expression pattern with some similarities to the root meristem (Sugimoto et al., 2010). LATERAL ORGAN BOUNDARIES DOMAIN (LBD) proteins are key transcriptional regulators for both lateral root development and callus induction, and their expression is directly regulated by WUSCHEL RELATED HOMEOBOX11 (WOX11) and WOX12 (Fan et al., 2012; Liu et al., 2014, 2018). In addition, WOX13 also plays a key role in callus formation and organ reconnection following grafting by regulating cellular reprogramming and cell wall modification (Ikeuchi et al., 2021). LBDs can interact with specific bHLH proteins to co-regulate downstream targets (Husbands et al., 2007). GhTCE1 acts as a bHLH protein and was found to interact with multiple TFs in yeast two-hybrid screens (Supplemental Figure S7). Future studies could determine whether GhTCE1 also interacts with LBDs during callus or adventitious organ development.
Although callus and adventitious roots are regulated by some common factors, there exists an antagonism between extensive callus formation, in which differentiated cells reprogramme and undergo elongation as they lose their original differentiated state, and adventitious root formation, in which cells form de novo meristems which, in contrast to callus cells, are small and highly organized. High concentrations of the auxin IBA promote callus production while lower concentrations promote adventitious roots (Supplemental Figure S2B), consistent with previous reports for indole acetic acid (IAA) treatment (Liu et al., 2014). Previously we found that GhL1L1 promotes adventitious root development rather than callus proliferation (Xu et al., 2019). In the current study, we show that explants expressing reduced levels of GhTCE1 showed reduced callus proliferation but enhanced adventitious root formation (Figure 2A), suggesting that GhTCE1 has an opposite or competing function to GhL1L1, further illustrating the antagonistic relationship between callus formation and adventitious root development.
GhTCE1 regulates callus cell proliferation and elongation by regulating multiple pathways
The transcriptomic analysis presented shows that multiple pathways linked to cell proliferation and elongation are regulated by GhTCE1. Among the 61 DEGs activated by GhTCE1, genes related to sterol synthesis, cell wall modification, cell extension/expansion, ROS homeostasis, and lipid transfer were highly enriched. Sterols such as brassinosteroids (BRs) are required for cell elongation and plant growth (Clouse, 2002). BR treatment of cultured cotton ovules results in a significant increase in fiber length, while the BR synthesis inhibitor BRZ strongly inhibits fiber elongation (Sun et al., 2005). BR-defective mutants show extremely dwarfed phenotypes with reduced cell length (Azpiroz, 1998). The BR-inducible gene EXORDIUM (EXO) was also found among the DEGs. EXO was originally identified in a promoter trap screen for early embryo genes in Arabidopsis (Farrar et al., 2003), and it encodes an extracellular protein required for cell expansion in Arabidopsis (Schroder et al., 2009).
Cell wall modification and cell expansion pathways regulate cell elongation. The identified DEGs contain genes predicted to be involved in cell wall metabolism, including pectin methylesterase, putative pectin acyl transferase, β-1,3-glucanase, and BBE-like enzyme for monolignol oxidation, expansion, and extension. Pectin de-methylesterification is important for hypocotyl and internode cell elongation in Arabidopsis (Pelletier et al., 2010; Peaucelle et al., 2011). Also, pectin acetylation is involved in hypocotyl elongation, and mutation of pectin acyltransferase results in short hypocotyls under dark growth conditions (Sinclair et al., 2017). Callose (β-1,3-glucan) can accumulate under stress conditions to impregnate the cell wall and restrict cell growth (Piršelová and Matušíková, 2012). Degradation of callose by β-1,3-glucanase can increase cell wall flexibility and promote pollen tube germination and elongation (Parre and Geitmann, 2005). BBE-like was reported to participate in the oxidation and mobilization of monolignols (Daniel et al., 2015), and a direct target of bZIP59-LBD16 in Arabidopsis is required for callus initiation during early SE (Xu et al., 2018), suggesting that cell wall metabolism is important for cell dedifferentiation and callus growth. Extensins and expansins are involved in cell wall assembly and are required for elongation of root hairs and hypocotyl cells (Cannon et al., 2008; Zdanio et al., 2020). We found that two homologous genes, encoding an extensin and an expansin, were activated by GhTCE1 during dedifferentiation of cotton somatic cells (Figure 3E). Ri of the extensin gene (Gh_D09G1992) resulted in decreased callus proliferation and cell elongation (Supplemental Figure S11). These data show that GhTCE1 likely promotes callus cell growth, at least in part, by regulating the cell wall modification machinery.
LTPs participate in cell fate switching via ROS homeostasis
A key feature of this work is the discovery that GhTCE1 regulates the transcription of the lipid transfer protein genes LTP2 and LTP3 through promoter binding (Figure 4), and data strongly suggest a link with ROS homeostasis in cell fate determination. ROS, including hydrogen peroxide (H2O2), singlet oxygen (1O2), the superoxide anion (), and the hydroxyl radical (OH•), can either promote or repress cell growth according to the particular species and distribution (Passardi et al., 2004). H2O2 promotes cell differentiation and strengthens the cell wall by promoting the crosslinking of cell wall components (Raggi et al., 2015). In contrast, OH• promotes cell elongation by cleaving polysaccharides or activating calcium channels (Foreman et al., 2003; Muller et al., 2009). In the current study, two NADPH oxidase and six peroxidase genes were identified as candidate targets of GhTCE1. NADPH oxidase participates in the production of , which can be converted into H2O2 spontaneously or by superoxide dismutase (SOD). Peroxidases are involved in H2O2 scavenging by substrate oxidation or in the production of OH• via the Haber–Weiss reaction (Shigeto and Tsutsumi, 2016). We provide evidence that GhTCE1 is also involved in ROS homeostasis, and potentially promotes cell elongation by activating the expression of peroxidases and NADPH oxidase.
LTPs represent other potential participants in cell elongation, but their functions have not been well characterized. A number of possible mechanisms by which they influence cell elongation should be considered. LTPs are small compact proteins with lipid binding and transport activity (Edqvist et al., 2018). They are usually localized to extracellular spaces, with targeting mediated by N-terminal signal peptides (Boutrot et al., 2008). LTPs are involved in the deposition of wax and lipid-based polymers in the defense response, as well as in cell wall loosening during cell elongation (Liu et al., 2015). LTPs can interact with, and promote the activity of the pectin-degrading enzyme polygalacturonase (PG), thereby loosening the cell wall (Tomassen et al., 2007). The lipid binding hydrophobic cavity of LTP may also interact with hydrophobic cell wall compounds, thereby disrupting their noncovalent bonding and promoting cell wall extension (Nieuwland et al., 2005). In the current study, reduced expression of LTP2 and LTP3 by Ri resulted in ROS accumulation and restricted callus cell growth, demonstrating a role for LTPs in mediating dedifferentiation control via ROS. Of particular significance here is the previously reported potential role for LTPs as ROS scavengers (e.g. Wang et al., 2014; Gangadhar et al., 2016; Xu et al., 2018). These studies describe how LTPs are upregulated under a range of abiotic stress conditions and can confer stress tolerance by scavenging ROS. Our data are consistent with a role for LTPs in scavenging ROS, regulating the transition from differentiated cell type (in the explant) to disorganized callus growth following wounding.
In vitro dedifferentiation of somatic cells post wounding is orchestrated by TFs
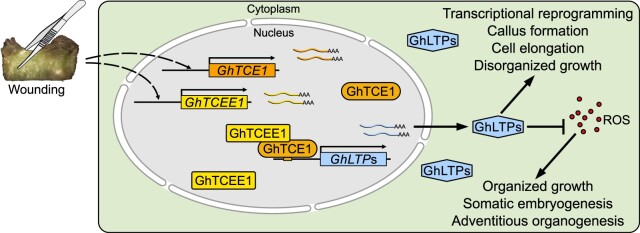
Callus induction is a clear example of plant developmental plasticity in which wounding leads to cell dedifferentiation, requiring a reprogramming of gene expression by induced TFs. Auxin response factor (ARF)- and cytokinin response factor (CRF)-mediated auxin and cytokinin responses are essential for callus formation under appropriate auxin/cytokinin ratios. The arf7 arf9 double mutant displays defective callus initiation, but ectopic expression of their downstream targets LBD16/17/18/29 is sufficient to induce autonomous callus production in the absence of exogenous phytohormones (Fan et al., 2012; Xu et al., 2018). Ectopic expression of CRF3 also results in autonomous callus formation (Xu et al., 2012). Furthermore, ARF10 represses the expression of ARABIDOPSIS RESPONSE REGULATOR 15 (ARR15) to promote callus initiation, highlighting the interplay between auxin and cytokinin (Liu et al., 2016). To date, more than 50 TFs have been reported to function during regeneration from callus in Arabidopsis, and nearly half of them participate in the regulation of callus growth itself (Ikeuchi et al., 2019). In the current study, we show that GhTCE1, with its partner protein GhTCEE1, is a part of a critical regulatory module involving LTPs and ROS. This module promotes cell dedifferentiation and callus growth in an auxin-independent manner, and it acts as a molecular switch to determine cell fate control by promoting callus growth and repressing more organized meristematic growth (Figure 7).
Figure 7.
Model outlining the proposed relationship between mechanical wounding, GhTCE1 and GhTCEE1 (bHLH dimers), LTPs, ROS, and cell fate determination. We propose that following wounding and callus initiation, GhTCE1 and GhTCEE1 are simultaneously at the transcriptional level, and the proteins interact to activate the transcription of LTP genes GhLTP2 and GhLTP3. The LTP proteins repress ROS activity, possibly by acting as ROS scavengers, as suggested by previous LTP studies in relation to roles in abiotic stress tolerance. Repression of ROS leads to the maintenance of callus cell identity and growth, whereas an increased level of ROS activity is associated with reduced cell growth and cell differentiation. We propose that the TCE1/TCEE1 transcriptional module regulates cell fate determination through these effects on ROS-mediated cell growth and differentiation control.
Materials and methods
Plant materials, vector construction, and genetic transformation
The cotton used in this study is G. hirsutum cv YZ1. Transgenic lines were planted in the greenhouse at Huazhong Agricultural University, Wuhan, China. The greenhouse was kept at 26°C–32°C, 14-h light/10-h dark photoperiod. The explants from transgenic lines and tobaccos that were used for LUC reporter assays were grown at 25°C in a light cultivation room (16-h light/8-h dark photoperiod, 3,000 lex). Ri vectors for GhTCE1 and GhLTP2/3 were constructed in the pHellsgate 4 vector (Wesley et al., 2001). The OE construct for GhTCE1 was in the pK2GW7 (Karimi et al., 2002) vector. The CRISPR-Cas9-mediated gene editing vector was constructed as described previously (Wang et al., 2018). All the primers used for vector construction are listed in Supplemental File S1. Transgenic plants were created by Agrobacterium-mediated transformation as described previously (Jin et al., 2006). The expression levels of GhTCE1 and GhLTP2/3 in transgenic plants were determined by qRT-PCR, using explants at 7 days post wounding, using GhUB7 as an internal control. T-DNA insertion copy numbers were determined by immunoblotting as described previously (Wang et al., 2018). The mutation analysis of GhTCE1 CRISPR-Cas9 transgenic plants was achieved by the Hi-TOM method as described previously (Liu et al., 2019). Mutants of GhTCE1(A/D) carrying frameshift mutations were chosen for phenotypic analysis.
Callus induction, CPR, cell length analysis, and adventitious root formation
Hypocotyls of etiolated seedlings were cut into 5–7 mm sections as explants for culture on MSB (MS medium plus B5 vitamins) medium and growth conditions of explants as described previously (Yang et al., 2012). The CPR represents the gained weight of per unit weight explants at 20 days post wounding induction. Callus cells were separated in 1× PBS (10 mM sodium phosphate, 137 mM NaCl, 2.7 mM KCl, and pH 7.5) and photographed under a light microscope (ZEISS, AXIO Scope A1). Cell length was measured from the micrograph using Image J software (Rha et al., 2015). Adventitious root formation was measured as a ratio of the frequency of roots formed per explant cultured.
DCFH-DA treatment, ROS staining, and H2O2 measurement
Hypocotyls of etiolated seeding were cut into 5–7 mm sections as explants, 10 mM DMTU was added to MSB medium as the treatment group, and MSB medium lacking DMTU was the negative control.
For ROS staining, fresh callus cells were collected 20 days post wounding and stained in the DCFH-DA solution described below for 5 min. Stained cells were rinsed three times in ddH2O and examined under a fluorescence microscope (Nikon, SMZ25) with 480 nm excitation, and 100 ms exposure time was used for each image. The DCFH-DA storage solution (pH 7.5) was 10 mmol/L prepared in dimethylsulfoxide, and it was diluted 1,000-fold with 1× PBS for ROS staining. The concentration of H2O2 from fresh callus was determined using a H2O2 quantification kit (#C500069, Sangon Biotech, Shanghai, China).
Endogenous IAA concentration assays and IBA treatment
Explants were collected 20 days post wounding and the IAA levels were determined by HPLC-MS as reported previously (Xu et al., 2019). In standard MSB medium, the IBA concentration (0.5 mg/L) was defined as 1×. The final concentrations of IBA used were 0, 0.5, 1, 2.5, 5, and 10 mg/L in MSB medium and defined as 0×, 1×, 2×, 5×, 10×, and 20×, respectively, for IBA treatments. Explants were photographed 10 and 20 days post wounding.
RNA-seq
Hypocotyls of WT and GhTCE1 Ri and OE lines were used for RNA extraction 7 days post wounding. RNA-seq was performed by the Beijing Genomics Institute. DEGs were determined using the threshold of P ≤ 0.05 and the absolute value of the log2 fold change ≥ 1. The annotations of DEGs were based on the functions of their homologues in Arabidopsis unless known in cotton.
Subcellular localization
Full length GhTCE1 was cloned into the vector pMDC43 with GFP fused to the N-terminus (Cai and Goodman, 2015). The 35Spro:GFP-GhTCE1 construct was transiently expressed in tobacco epidermal cells following Agrobacterium-mediated transfection. The pMDC43 empty vector harboring 35Spro:GFP was used as control. GFP fluorescence was detected under a confocal microscope (Olympus FV1200) at 2 days following Agrobacterium transfection.
BiFC assay
For the BiFC assay, the full-length sequences of GhTCE1 and GhTCEE1 were cloned into pDONR vector (Invitrogen), then into the destination vector pBiFCt-2in1-NC by Gateway Technology (Grefen and Blatt, 2012). The recombinant vectors were transformed into Agrobacterium tumefaciens strain GV3101, and transient expression was performed in tobacco epidermal cells. The YFP fluorescence in BiFC assays was examined by confocal microscopy (Olympus FV1200).
Autoactivation analysis of GhTCE1
The full-length sequence of GhTCE1 was inserted into the pGBKT7 vector to produce the Full:BD construct. Full:BD was transferred into the Y2HGold yeast strain and cultured on SD-T/X-α-gal plates for 3 days to detect its autoactivation (based on colony color). To further investigate the autoactivation in detail, four different truncated GhTCE1 versions were constructed in pGBKT7 and designated 1–160, 161–331, 80–240, and 80–331 according to the amino acid fragments contained in each. The different constructions of GhTCE1 were transferred into the Y2HGold yeast strain and mated with Y187 yeast harboring the pGADT7 empty vector. The positive and negative controls used were according to the manufacturer’s instructions (Clontech, PT4084-1). The diploid yeasts were cultured in SD-TL and SD-TLHA/X-α-gal plates for 3 days to detect autoactivation. SD means synthetically defined medium, SD-T/TL/TLHA represent SD dropouts Trp, Trp/Leu, and Trp/Leu/His/Ade, respectively.
Transactivation assays using the LUC reporter system
Transactivation assays were conducted as reported previously (Hu et al., 2018). In brief, the promoters of GhTCE1 candidate targets were cloned and inserted upstream of sequences encoding the LUC reporter in the pGreenII 0800-LUC vector. 35Spro:REN in the same vector was used as an internal control. GhTCE1 and GhTCEE1 were cloned downstream of the cauliflower mosaic virus 35S promoter (35Spro) in pGreenII 62-SK and the same vector without GhTCE1 was used as control. Activation of E-box motifs assays was conducted using the 2,000-bp full-length GhLTP2 promoter with five E-box motifs (I, II, III, IV, and V) and the GhLTP3 promoter with two E-box motifs (I and II). The truncation promoters were created by successively deleting one E-box in turn from the 5′-end and were named LTP2proΔ1 to LTP2proΔ4 for deletions of GhLTP2pro and named LTP3proΔ1 to LTP3proΔ2 for GhLTP3pro. The truncated promoters were cloned into the pGreenII 0800-LUC vector. All plasmids were transformed individually into A. tumefaciens strain GV3101 for transient expression in tobacco (N. benthamiana). The concentration of A. tumefaciens harboring the pGreenII 0800-LUC or pGreenII 62-SK vectors was adjusted to the same concentration (e.g. pGreenII 62-SK:GhTCE1, OD = 0.2, pGreenII 0800:LTP2pro, OD = 0.2) and mixed in volumes of 1:1 (pGreenII 0800-LUC: pGreenII 62-SK), and all of the pairs of the suspensions carrying the appropriate constructs were adjusted to the same OD600 = 0.6 prior to coinfiltration into tobacco leaves with same infiltrated areas to ensure comparability. The infiltrated plants were grown at 25°C in a light cultivation room (16-h light/8-h dark photoperiod, irradiance of 135 µmol m−1·s−1). After 60–72 h, the LUC luminescence of leaves was detected using a cryogenically cooled CCD camera (Lumazome PyLoN 2048B). For each tobacco leaf, one half was transfected with experimental vectors and the other half with control vectors by agroinfection. However, for four truncations, 62-SK:GhTCE1+ pGreenII 0800:LTP2proΔ constructs were evenly distributed on one leaf. Three biological replicates were carried out for each combination.
The LUC assays in cotton protoplasts were performed as described (Hellens et al., 2005). In brief, vector each of effector (1:1) and reporter constructs (total effectors:reporter = 1:1) were cotransformed into protoplasts using 40% polyethylene glycol 4000 (v/v) (Sigma), then cultured at 25°C in darkness for 16 h. LUC and REN activity were detected (the relative ratio of the LUC/REN ratio with GhTCE1 and GhTCEE1 versus the LUC/REN ratio without GhTCE1s and GhTCEE1) for transactivation assays using GhTCE1 and its candidate targets by using the dual-LUC assay reagents (Promega, Madison, WI) with the Multimode Plate Reader (Perkin-Elmer).
Yeast two-hybrid screening
The activation domain of truncated GhTCE1 (C-terminal from 160 to 331 aa) was cloned into pGBKT7 (designated pGBKT7-ΔGhTCE1) and transformed into yeast strain Y2HGold. The cDNA library from early time points after wounding were constructed in pGADT7 and transformed into yeast strain Y187. Yeast two-hybrid screening was performed according to the manufacturer’s instructions (Clontech, PT1172-1, PT4084-1). The vectors from positive clones were recovered and transferred into Escherichia coli for sequencing. After removing the unmatched or frameshift preys, the remaining protein coding preys (designated prey-AD) were transferred into the Y187 strain separately and mated with the Y2H strain containing either the pGBKT7 empty vector (BD) or pGBKT7-ΔGhTCE1 (ΔGhTCE1-BD). Both prey-AD/BD and prey-AD/ΔGhTCE1-BD combinations showing activated reporter genes were considered as false interactions. Only prey-AD + ΔGhTCE1-BD activated reporter genes while the prey-AD/BD did not were considered as true interactions.
Electrophoretic mobility shift assays
Biotin-labeled single-stranded DNA was synthesized and annealed to produce double strands. For EMSA, a sequence encoding full-length GhTCE1 was cloned into the pET28a (Novagen) vector, and His-tagged proteins were induced and expression in E. coli and proteins were purified by chromatography. Binding reactions were carried out using the LightShift Chemiluminescent EMSA Kit (Thermo scientific, 20148). Signals were captured by X-ray film and the film was developed according to the manufacturer’s instructions. The primers used for vector construction are listed in Supplemental File S1.
Chromatin-immunoprecipitation quantitative PCR
For ChIP experiments, GhTCE1-GFP transgenic lines were used for chromatin extraction and immunoprecipitation as described previously by Huang et al. (2018). Briefly, the samples were fixed with 1% (v/v) formaldehyde (Sigma, Cat. # F8775) for 10 min at room temperature, and neutralized with 0.125 M glycine for 5 min. The samples were collected by centrifuging (Beckman Coulter, Avanti JXN-26) at 1,000 g at 4°C for 5 min. A Bioruptor (KQ5200DE) was used at high power with 30-s on/30-s off cycles for 15 times until the average chromatin size was approximately 300 bp. Anti-GFP (ABclonal, AE012) antibodies (5 µg antibodies were added per 25 µg chromatin DNA) were used to perform immunoprecipitations. The immunoprecipitated chromatin fragments were isolated and preabsorbed with sheared salmon sperm DNA/protein A-agarose (Sigma-Aldrich) to remove nonspecific binding (IP). After DNA purification, the GhTEC1-binding sites on LTP2pro and LTP3pro were evaluated using qPCR and normalized by total chromatin (Input). The fold enrichment calculated as the ratio between the samples of input and IP samples. GhUBQ7pro was used as reference promoter and the primers are listed in Supplemental File S1.
Y1H assay
Y1H screening was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System User Manual (Clontech, PT4087-1). The full-length sequence of GhTCE1 was cloned into the pGADT7 vector and transformed into yeast strain Y187, the promoter sequences of LTP2pro and LTP3pro were cloned into the pHis2 vector and transformed into yeast strain AH109, and the pGAD-empty vector was used as negative control vector. Ten microliters of each for combined yeast strains Y187 and AH109 were cultured in 200 µL YPDA liquid medium (1 L YPDA contains 10 g yeast extract, 20 g tryptone, 20 g glucose, 100 mg adenine, fill ddH2O to 1 L, pH = 6.5) at 30°C for 20–24 h. Then, 6 µL yeast that mated successfully was spotted onto SD-Trp-Leu-His containing 200 mM 3-AT solid medium at 30°C for 3–5 days, then analyzed the interaction between GhTCE1 and LTP2pro and LTP3pro. The primers used for vector construction are listed in Supplemental File S1.
Accession numbers
Sequence information for the cotton genes in this study can be found in the Cotton Genome Database (https://cottonfgd.org/, G. hirsutum AD1, upland cotton, NAU assembly) according to the accession numbers as shown in Supplemental Data Set S1 and Supplemental Tables S1–S3.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein sequence alignment of Gh_D06G2316 (GhTCE1D) and GhA_06G0241 (GhTCE1A).
Supplemental Figure S2. GhTCE1 is involved in adventitious root regeneration during wound-induced callus formation.
Supplemental Figure S3. Auxin detection in GhTCE1 transgenic plants.
Supplemental Figure S4. Callus formation in GhTCE1 transgenic lines.
Supplemental Figure S5. Correlation analysis of 29 DEGs comparing RNA-seq versus qRT-PCR.
Supplemental Figure S6. Subcellular localization and autoactivation analysis of GhTCE1.
Supplemental Figure S7. Expression analysis of GhTCE1 and its candidate targets in different tissues.
Supplemental Figure S8. GhLTP2 and GhLTP3 are involved in adventitious root regeneration during wound-induced callus formation.
Supplemental Figure S9. Yeast two-hybrid screening for proteins interacting with GhTCE1.
Supplemental Figure S10. Expression pattern of GhTCEE1 at early time points following wounding.
Supplemental Figure S11. EXT1A and EXT1D are involved in cell elongation during SE.
Supplemental Data Set S1. FPKM values of 62 DEGs identified by RNA-seq analysis.
Supplemental Data Set S2. Statistical analysis tables.
Supplemental Table S1. Function annotation of 32 left candidate targets of GhTCE1.
Supplemental Table S2. Characterization of 29 GhTCE1 candidate targets.
Supplemental Table S3. Annotation of proteins interacting with GhTCE1 from yeast two-hybrid screening.
Supplemental File S1. List of primers used in this study.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Key Project of Research and the Development Plan of China (2018YFD1000907) and Fundamental Research Funds for the Central Universities (2662020ZKPY011).
Conflict of interest statement. The authors declare that there is no conflicts of interest.
Contributor Information
Jinwu Deng, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Weinan Sun, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Boyang Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Simin Sun, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Linjie Xia, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Yuhuan Miao, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Liangrong He, College of Plant Science, Tarim University, Alaer, Xinjiang 843300, China.
Keith Lindsey, Department of Biosciences, Durham University, South Road, Durham DH1 3LE, UK.
Xiyan Yang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Xianlong Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
X.Y., L.H., and X.Z. designed and guided the project. J.D. and W.S. performed the experiments and wrote the draft manuscript. B.Z., S.S., and L.X. helped perform the LUC assays. Y.M. helped in measuring ROS and auxin contents. K.L. organized the results and revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Xiyan Yang (yxy@mail.hzau.edu.cn).
References
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, et al. (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28: 1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Chantret N, Gautier MF (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Goodman JM (2015) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Qi H, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport D, Chen Y, Kieliszewski MJ (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105: 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JM, Muhlemann JK, Gayomba SR, Muday GK (2019) RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem Res Toxicol 32: 370–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qu Y, Sheng L, Liu J, Huang H, Xu L (2014) A simple method suitable to study de novo root organogenesis. Front Plant Sci 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD (2002) Brassinosteroids. Arabidopsis Book 1: e0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Pavkov-Keller T, Steiner B, Dordic A, Gutmann A, Nidetzky B, Sensen CW, van der Graaff E, Wallner S, Gruber K, et al. (2015) Oxidation of monolignols by members of the berberine bridge enzyme family suggests a role in plant cell wall metabolism. J Biol Chem 290: 18770–18781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Fang T, Liu Y, Huang L, Zang M, Wang G, Liu Y, Fu J (2019) Transcriptome profiling predicts new genes to promote maize callus formation and transformation. Front Plant Sci 10: 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist J, Blomqvist K, Nieuwland J, Salminen TA (2018) Plant lipid transfer proteins: are we finally closing in on the roles of these enigmatic proteins? J Lipid Res 59: 1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar K, Evans IM, Topping JF, Souter MA, Nielsen JE, Lindsey K (2003) EXORDIUM—a gene expressed in proliferating cells and with a role in meristem function, identified by promoter trapping in Arabidopsis. Plant J 33: 61–73 [DOI] [PubMed] [Google Scholar]
- Fehér A (2015) Somatic embryogenesis—stress-induced remodeling of plant cell fate. BBA (Gene Regul Mech) 1849: 385–402 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell J, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones J (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Gangadhar BH, Sajeesh K, Venkatesh J, Baskar V, Abhinandan K, Yu JW, Prasad R, Mishra RK (2016) Enhanced tolerance of transgenic potato plants over-expressing non-specific lipid transfer protein-1 (StnsLTP1) against multiple abiotic stresses. Front Plant Sci 7: 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Blatt MR (2012) A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Zhu LF, Zhang XN, Guan QQ, Xiao SH, Min L, Zhang XL (2018) GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol 178: 876–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Fang C, Yi K, Liu X, Qi H, Tan Y, Zhou J, Li Y, Liu M, Zhang Y, et al. (2018) The role of PTRF/Cavin1 as a biomarker in both glioma and serum exosomes. Theranostics 8: 1540–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HM, Springer PS (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Favero DS, Sakamoto Y, Iwase A, Coleman D, Rymen B, Sugimoto K (2019) Molecular mechanisms of plant regeneration. Annu Rev Plant Biol 70: 377–406 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Ito T, Tanaka H, Favero DS, Kawamura A, Sakamoto S, Wakazaki M, Tameshige T, Fujii H, et al. (2021) Wound-inducible WUSCHEL RELATED HOMEOBOX 13 is required for callus growth and organ reconnection. Plant Physiol 181: 425–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Lambolez A, Kojima M, Takebayashi Y, Heyman J, Watanabe S, Seo M, De Veylder L, et al (2017) Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol 175: 1158–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Hu L, Yuan D, Xu J, Gao W, He L, Yang X, Zhang X (2013) Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol J 12: 161–173 [DOI] [PubMed] [Google Scholar]
- Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H (2006) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant 50: 519–524 [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Lee K, Seo PJ (2018) Dynamic epigenetic changes during plant regeneration. Trends Plant Sci 23: 235–247 [DOI] [PubMed] [Google Scholar]
- Li S, Yan S, Wang AH, Zou G, Huang X, Han B, Qian Q, Tao Y (2013) Identification of QTLs associated with tissue culture response through sequencing-based genotyping of RILs derived from 93-11 × Nipponbare in rice (Oryza sativa). Plant Cell Rep 32: 103–116 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Yeoman MM (1983) The relationship between growth, differentiation and alkaloid accumulation in plant cell cultures. J Exp Bot 34: 1055–1065 [Google Scholar]
- Liu F, Zhang X, Lu C, Zeng X, Li Y, Fu D, Wu G (2015) Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J Exp Bot 66: 5663–5681 [DOI] [PubMed] [Google Scholar]
- Liu J, Hu X, Qin P, Prasad K, Hu Y, Xu L (2018) The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol 59: 734–743 [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang C, Jiao X, Zhang H, Song L, Li Y, Gao C, Wang K (2019) Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci China (Life Sci) 62: 1–7 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li J, Wang L, Li Q, Lu Q, Yu Y, Li S, Bai MY, Hu Y, Xiang F (2016) Repression of callus initiation by the miRNA-directed interaction of auxin–cytokinin in Arabidopsis thaliana. Plant J 87: 391–402 [DOI] [PubMed] [Google Scholar]
- Muller K, Linkies A, Vreeburg RA, Fry SC, Krieger-Liszkay A, Leubner-Metzger G (2009) In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol 150: 1855–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, Derksen J, Mariani C (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17: 2009–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parre E, Geitmann A (2005) More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol 137: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Hofte H, Laufs P, et al. (2011) The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 138: 4733–4741 [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, Vissenberg K, Delacourt J, Ndong YA, Pelloux J, Bischoff V, Urbain A, Mouille G, et al. (2010) A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol 188: 726–739 [DOI] [PubMed] [Google Scholar]
- Piršelová B, Matušíková I (2012) Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol Plant 35: 635–644 [Google Scholar]
- Raggi S, Ferrarini A, Delledonne M, Dunand C, Ranocha P, De Lorenzo G, Cervone F, Ferrari S (2015) The Arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage. Plant Physiol 169: 2513–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha EY, Kim JM, Yoo G (2015) Volume measurement of various tissues using the Image J Software. J Craniofac Surg 26: e505–06 [DOI] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. Arabidopsis Book 1: e0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder F, Lisso J, Lange P, Mussig C (2009) The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J, Tsutsumi Y (2016) Diverse functions and reactions of class III peroxidases. New Phytol 209: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Sinclair SA, Larue C, Bonk L, Khan A, Castillo-Michel H, Stein RJ, Grolimund D, Begerow D, Neumann U, Haydon MJ, et al. (2017) Etiolated seedling development requires repression of photomorphogenesis by a small cell-wall-derived dark signal. Curr Biol 27: 3403–3418.e3407 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD (2005) Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol 46: 1384–1391 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassen MM, Barrett DM, van der Valk HC, Woltering EJ (2007) Isolation and characterization of a tomato non-specific lipid transfer protein involved in polygalacturonase-mediated pectin degradation. J Exp Bot 58: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Wang F, Zang XS, Kabir MR, Liu KL, Liu ZS, Ni ZF, Yao YY, Hu ZR, Sun QX, Peng HR (2014) A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis. Gene 550: 18–26 [DOI] [PubMed] [Google Scholar]
- Wang P, Zhang J, Sun L, Ma Y, Xu J, Liang S, Deng J, Tan J, Zhang Q, Tu L, et al. (2018) High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J 16: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Xu C, Cao H, Xu E, Zhang S, Hu Y (2018) Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol 59: 744–755 [DOI] [PubMed] [Google Scholar]
- Xu C, Cao H, Zhang Q, Wang H, Xin W, Xu E, Zhang S, Yu R, Yu D, Hu Y (2018) Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat Plants 4: 108–115 [DOI] [PubMed] [Google Scholar]
- Xu J, Yang X, Li B, Chen L, Min L, Zhang X (2019) GhL1L1 affects cell fate specification by regulating GhPIN1-mediated auxin distribution. Plant Biotechnol J 17: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Liu J, Fan M, Xin W, Hu Y, Xu C (2012) A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 100: 116–124 [DOI] [PubMed] [Google Scholar]
- Xu Y, Zheng X, Song Y, Zhu L, Yu L, Gan L, Zhou S, Liu H, Wen F, Zhu C (2018) NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci Rep 8: 8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J (2012) Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol 12: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liu W, Liu J, Qin P, Xu L (2017) Auxin control of root organogenesis from callus in tissue culture. Front Plant Sci 10: 3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanio M, Boron AK, Balcerowicz D, Schoenaers S, Markakis MN, Mouille G, Pintelon I, Suslov D, Gonneau M, Hofte H, et al. (2020) The proline-rich family protein EXTENSIN33 is required for etiolated Arabidopsis thaliana hypocotyl growth. Plant Cell Physiol 61: 1191–1203 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhao H, Li W, Wu J, Zhou Z, Zhou F, Chen H, Lin Y (2019) Genome‐wide association study of callus induction variation to explore the callus formation mechanism of rice. J Integr Plant Biol 61: 1134–1150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.