Abstract
Wild rice (Oryza rufipogon) has a lower panicle seed setting rate (PSSR) and gamete fertility than domesticated rice (Oryza sativa), but the genetic mechanisms of this phenomenon remain unknown. Here, we cloned a null allele of OsMLH1, an ortholog of MutL-homolog 1 to yeast and mammals, from wild rice O. rufipogon W1943 and revealed a 5.4-kb retrotransposon insertion in OsMLH1 is responsible for the low PSSR in wild rice. In contrast to the wild-type, a near isogenic line NIL-mlh1 exhibits defective crossover (CO) formation during meiosis, resulting in reduced pollen viability, partial embryo lethality, and low PSSR. Except for the mutant of mismatch repair gene postmeiotic segregation 1 (Ospms1), all other MutL mutants from O. sativa indica subspecies displayed male and female semi-sterility similar to NIL-mlh1, but less severe than those from O. sativa japonica subspecies. MLH1 and MLH3 did not contribute in an additive fashion to fertility. Two types of MutL heterodimers, MLH1-PMS1 and MLH1-MLH3, were identified in rice, but only the latter functions in promoting meiotic CO formation. Compared to japonica varieties, indica cultivars had greater numbers of CO events per meiosis. Our results suggest that low fertility in wild rice may be caused by different gene defects, and indica and japonica subspecies have substantially different CO rates responsible for the discrepancy between the fertility of mlh1 and mlh3 mutants.
A retrotransposon insertion at MUTL-HOMOLOG 1 is responsible for low seed set in wild rice and reveals the discrepancy of crossover rates of indica and japonica subspecies.
Introduction
Rice (Oryza sativa L.) is one of the most important staple foods for more than half of the world’s population. Panicle seed setting rate (PSSR) is one of the main determinants of rice grain yield (Li et al., 2013). Rice domestication is mainly associated with an increase in fertility and PSSR. It is believed that cultivated rice has higher fertility and PSSR than its close progenitor wild rice (Oryza rufipogon). It is revealed that the average pollen fertility was 25.6%, and PSSR under natural conditions was 3.0%–53.1% through investigation on 40 wild populations of O. rufipogon collected from the north part of Hainan Province (Yan et al., 2014). Whole-mount stain-clearing confocal laser scanning microscopy (WCLSM) analysis revealed that the mean percentage of abnormal embryo sacs of 141 O. rufipogon accessions collected from Guangdong Province was 11.1% and the highest was 67.9% (Yang et al., 2006). Because of the high genetic diversity and variation, the mechanisms underlying the semi-sterility in O. rufipogon remain poorly understood.
The low PSSR in rice could result from spikelet sterility due to abnormal floret structures, defective pollen grain or embryo sac development, impaired anther dehiscence, gametophytic incompatibility, or inappropriate temperature at the reproductive stage (Xu et al., 2017). Meiosis is an essential and conserved process for eukaryotic sexual reproduction (Mercier et al., 2015; Serra et al., 2018). Several key events including homologous pairing, synapsis, and recombination during prophase of the first meiotic division are essential for efficient reciprocal exchange of homologs, termed crossover (CO) (Mercier et al., 2015). Loss of many meiotic genes will result in low PSSR, even complete sterility (Zhou et al., 2011, 2017; Wang et al., 2012; Ren et al., 2019; Zhang et al., 2020a, 2020b). Meiotic CO initiating from DNA double-strand breaks induced by sporulation 11 transesterases (Mercier et al., 2015), occurs via at least two distinct pathways in most species (Higgins et al., 2004; Lhuissier et al., 2007; Li et al., 2018). In plants, the majority of COs are formed via the Class I pathway, which depends on the ZMM proteins (zipper1-4, meiotic recombination 3 [MER3], MutS-homolog 4 [MSH4], MSH5), and MLH1–3 (MutLγ) nuclease, and are subject to interference (Copenhaver et al., 2002; Lhuissier et al., 2007; Falque et al., 2009; Wang et al., 2012; Wang and Copenhaver, 2018). The remaining COs are formed via a minor noninterfering Class II pathway, orchestrated by the methyl methanesulfonate and UV-sensitive protein 81-essential meiotic endonuclease1 structure-specific endonuclease (Boddy et al., 2001; Mercier et al., 2005; Wang and Copenhaver, 2018).
In eukaryotes, including yeast and mammals, four MutL homolog (MutL) proteins (postmeiotic segregation 1 [PMS1] and MLH1–3) are identified and involved in multiple pathways of DNA repair and recombination (Kolodner and Marsischky, 1999; Wang et al., 1999; Kadyrova et al., 2020). In yeast, mutating each of these genes results in reduced spore viability by 3.4%–21.2% depending on the mutant, compared with wild-type (WT) (Wang et al., 1999). In contrast, Mus musculus mutants of MutL genes show severe infertility (Lipkin et al., 2002; Toledo et al., 2019). Three types of complexes are revealed among yeast and mammal MutL homologs: those between MLH1 and each of the other three family members (Wang et al., 1999; Kadyrova et al., 2020). Among these, only the MLH1–3 heterodimer (MutLγ) is implicated specifically in meiotic CO and promotes the CO formation by interacting with MutSβ (MSH2–3) (Kadyrova et al., 2020) and/or MutSγ (Msh4–Msh5) (Cannavo et al., 2020). In plants, only three MutL homologs are known to be present (Dion et al., 2007). The phenotypes of single-gene mutants via T-DNA insertion (Jackson et al., 2006; Dion et al., 2007; Li et al., 2009), over-expressing mutated alleles (Alou et al., 2004; Xu et al., 2012a), or gene editing (Mao et al., 2021; Xin et al., 2021), reveal distinct functions for the three MutL homologs in plants, but their biological functions remain to be characterized. Moreover, a thorough investigation on protein–protein interactions among three MutL genes in plants has not been carried out, although recently, an interaction between the C-termini of OsMLH1 and OsMLH3 was reported (Mao et al., 2021; Xin et al., 2021).
In this work, we used a classical forward-genetics approach to clone a quantitative trait locus (QTL), PSSR1, from a wild rice W1943 and showed that the semi-sterile phenotype of pssr1 was the consequence of mutation of MutL-homolog 1 (OsMLH1). Although the functions of MutL genes have been revealed in yeast and mammals using single-gene mutants (Wang et al., 1999; Lipkin et al., 2002; Kadyrova et al., 2020), their functions in plants, especially in rice, need to be characterized. So, we utilized yeast two-hybrid, bimolecular fluorescence complementation assay (BiFC), and genetic assays via clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein9 (Cas9) (CRISPR–Cas9) technology to address: (1) if the mutation of each MutL gene can cause fertility defects in rice as reported in other taxa? (2) do MutL genes have divergent functions and have an additive effect on fertility? (3) which protein(s) of MutL homologs can assemble and involve in MER in rice? We unraveled that mutating mlh1/3 in japonica leads to more severe reduced fertility and lower percentage of meiocytes harboring 12 bivalents than in indica, indicating that these mutants from indica may have more residual COs than those from japonica under destruction of partial class I COs by the mutation of MLH1/3. This discovery prompted us to address whether indica has more CO events per meiosis, that is a higher CO rate than japonica.
Results
Morphological characterization of the mlh1 mutant and map-based cloning of MLH1
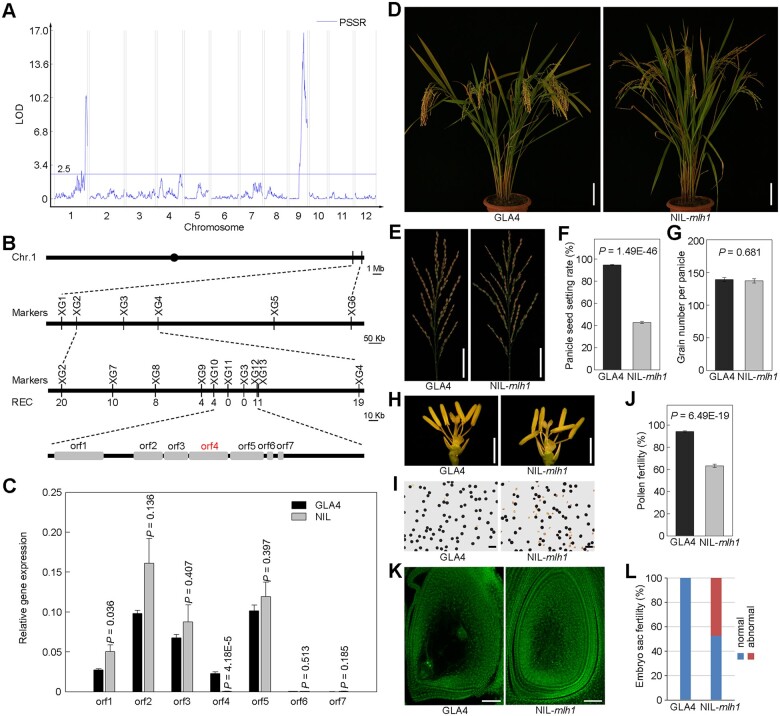
By examining the PSSR of each line and subsequent QTL analysis, two major QTLs for low PSSR were identified with a threshold of logarithm of the odds (LOD) ≥ 2.5 in the population from a cross between an indica variety GLA4 and a wild rice accession W1943 (Figure 1A), with the PSSR1 locus located at the distal region of the long arm of chromosome 1. To fine map this locus, one backcross inbred line BIL86 with low PSSR, harboring the PSSR1 locus and carrying several other segments from W1943, was selected for further backcrossing to GLA4. The F2 progeny showed a 1:3 (121:404, x2 = 0.8 < 3.84) segregation ratio between semi-sterile and fertile lines, suggesting that the semi-sterile phenotype was controlled by a single recessive gene. We then carried out high-resolution mapping using 1,152 BC1F3 individuals, and the locus was finally delimited to a 49-kb region between the two markers XG10 and XG12, containing seven predicted genes in total (Figure 1B). We measured the expression levels of these genes in the panicle (Figure 1C) and flag leaf (Supplemental Figure S1) by reverse transcription quantitative PCR (RT–qPCR), and found that only orf4/LOC_Os01g72880 markedly differentially expressed in the panicle and leaf of GLA4 and NIL plants (P < 0.0001), with nearly no expression in the NIL plants. Therefore, we deduced that the gene LOC_Os01g72880 encoding a DNA mismatch repair protein MLH1 (hereafter referred to as OsMLH1) is PSSR1.
Figure 1.
Mapping and characterization of OsMLH1. A, QTL mapping for PSSR. B, The positional cloning of MLH1. The candidate region was mapped to a ∼49-kb genomic DNA region lying between markers XG10 and XG12 and co-segregated with XG11 and XG3 in a large population (n = 1,536). In total, seven genes were predicted within the 49-kb region. Rec, number of recombinants. C, The relative expression levels of the seven candidate genes in panicles of GLA4 and NIL-mlh1 like plants. Values are means ± sem, n = 4. Significance was assessed by Student’s t test. D, Comparison of a GLA4 plant and a NIL-mlh1 plant. Bar = 10 cm. E–G, Comparison of panicles of GLA4 and NIL-mlh1 (E). Bar = 5 cm. Histograms of PSSR (F) and grain number per panicle (G) are shown beside. Values are means ± sem, n = 30. Significance was assessed by Student’s t test. H, Comparison of a GLA4 and a NIL-mlh1 spikelet after removing the lemma and palea. Bar = 2 mm. I, KI-I2 staining of GLA4 pollen and NIL-mlh1 pollen. Bar = 100 μm. J, Quantifications of the pollen fertility in GLA4 and NIL-mlh1. Values are means ± sem, n = 20. Significance was assessed by Student’s t test. K, Comparison of GLA4 mature embryo sac and NIL-mlh1 abnormal mature embryo sac spikelet. Bar = 50 μm. L, Quantifications of the fertility of embryo sac in GLA4 and NIL-mlh1.
The NIL-mlh1 exhibited normal plant architecture and could not be distinguished from the WT (GLA4) during the vegetative stage, but with low PSSR at the mature stage (Figure 1, D and E). Under normal field conditions, the PSSR of GLA4 was >90%, while it was 42.7% in NIL-mlh1 (Figure 1F). The PSSR in the heterozygotes is not reduced, similar to that of GLA4 (Supplemental Figure S2). In addition, the grain number per panicle showed no difference between GLA4 and NIL-mlh1 (Figure 1G). Other agricultural traits also exhibited no obvious differences between them (Supplemental Table S1). NIL-mlh1 had normal floral organs (Figure 1H) but produced more abortive pollens (Figure 1I). The pollen fertility was 94.2% in GLA4. In contrast, the value was 63.2% in NIL-mlh1 (Figure 1J). Some spikelets of NIL-mlh1 did not form the normal eight-nuclear embryo sac (Figure 1K), and the embryo sac fertility of NIL-mlh1 was 52.4% (Figure 1L). These results suggested that both male and female gametophytes of NIL-mlh1 might develop abnormally.
To clarify the causation of male semi-sterility, we first examined anther transverse section to characterize developmental defects in NIL-mlh1 (Supplemental Figure S3). The early stages of anther meiosis were indistinguishable between GLA4 and NIL-mlh. The microsporocytes of both are located at the center of each anther locule, surrounded by four layers of the anther wall (Supplemental Figure S3, A and F). However, eventually, only about half of the microspores showed normal development in the NIL-mlh1.
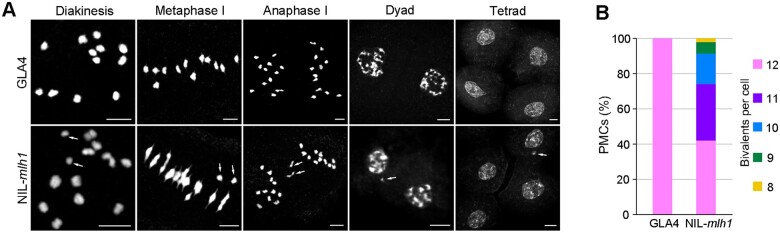
The aberrant pollen grains might suggest meiotic defects. To further investigate the cellular cause of the male semi-sterility in NIL-mlh1, we compared the meiotic chromosome behavior of meiocytes in GLA4 and NIL-mlh1 (Figure 2A). We found that several chromosomal defects were apparent from diakinesis to the following meiotic stages in NIL-mlh1. We observed no univalents in GLA4 meiocytes containing 12 bivalents in each cell (n = 142) (Figure 2B). However, in NIL-mlh1, the number of bivalents varied from 8 to 12, and ∼60% of pollen mother cells (PMCs) had less than or equal to 11 bivalents (n = 172). This result suggested that CO formation was affected in NIL-mlh1, and MLH1 is required for CO formation.
Figure 2.
Cytological Analysis of GLA4 and NIL-mlh1. A, Meiotic chromosome behaviors in GLA4 and NIL-mlh1. Some unpaired chromosomes in NIL-mlh1 are indicated with white arrows. Bar = 5 μm. B, Quantification of the number of bivalents in GLA4 and NIL-mlh1.
Complementary experiment and association analysis
To confirm that the putative gene is responsible for the mutant phenotype, a complementary construct containing an 8,768-bp genomic DNA fragment covering the entire LOC_Os01g72880 gene region was transformed into the NIL-mlh1. The transgene could rescue the semi-sterility phenotype of the NIL-mlh1 (Supplemental Figure S4, A–C). Compared to NIL-mlh1, transgene-positive plants showed a significant increase in the PSSR and pollen fertility with both being more than 90% (Supplemental Figure S4, D and E). These results demonstrated that LOC_Os01g72880 (OsMLH1) was the gene locus of PSSR1.
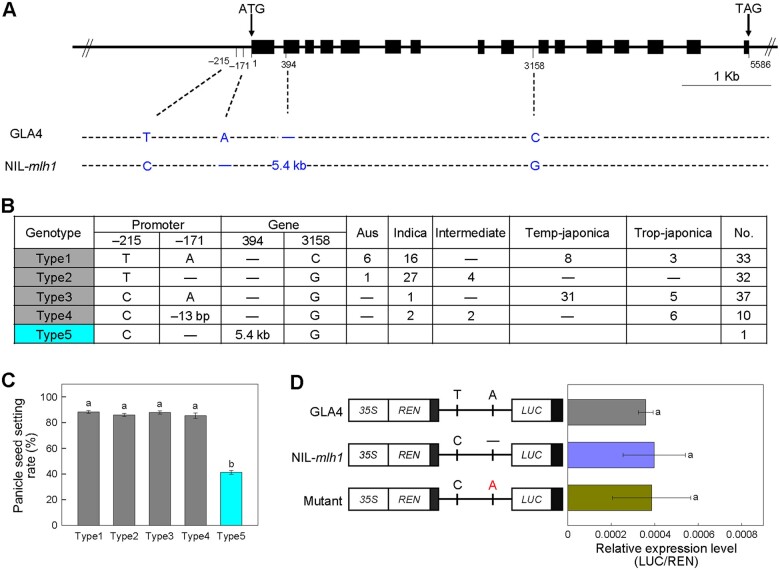
Four nucleotide polymorphisms between the genomic regions of OsMLH1 of GLA4 and NIL-mlh1, composed of two single-nucleotide polymorphisms (SNPs) in the promoter, one 5.4-kb insertion in the second exon and one SNP in the ninth intron were detected (Figure 3A). To reveal the association between these mutations and PSSR, we sequenced the OsMLH1 gene fragments of 112 diverse accessions, grouped them into four types based on haplotype analysis, and examined their seed setting rates (Figure 3B; Supplemental Data Set 1). Compared to NIL-mlh1 (type5), all four types have higher PSSR (Figure 3C).
Figure 3.
Association Mapping. A, Schematic of the gene structure and allelic variation on OsMLH1 between GLA4 and NIL-mlh1 indicated by vertical lines at bottom. B, Haplotype analysis of the OsMLH1 gene region from 112 rice cultivars and NIL-mlh1. C, The PSSR of the five haplotypes. Values are means ± sem. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way analysis of variance (ANOVA). D, LUC reporter gene transient assay of promoter activity in rice protoplasts. Left, three constructs, one of which is a site-directed mutation at one SNP in the promoter region. Right, the relative expression was quantified according to LUC/REN values. Values are means ± sem, n = 4. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way ANOVA.
Furthermore, we conducted transient expression assays of the site-directed mutated promoter fragments of OsMLH1 to test the effects of the two SNPs in the promoter region. Both the promoter fragment with one mutation (−171 position) and the NIL-mlh1 promoter have almost equal relative activities, same as GLA4 (Figure 3D). Moreover, compared with the WT, both the transgenic plants with large fragment deletion (−170 to −349; mlh1CR-pro-1) and small fragment deletion (−146 to −173; mlh1CR-pro-2) in the promoter of Nipponbare by CRISPR–Cas9 showed no difference in the PSSR and pollen viability (Supplemental Figure S5). Taken together, these results suggested that among the four variations, the 5.4-kb insertion resulted in the loss-of-function of OsMLH1 and subsequent low PSSR in NIL-mlh1.
The 5.4-kb insertion was subsequently amplified from the NIL-mlh1 genome (Supplemental Figure S6, A–C) and sequenced. Eight other backcross inbred lines having the mlh1 locus and low PSSR were also verified to harbor the 5.4-kb insertion (Supplemental Figure S6D). We obtained a 5,429-bp insertion sequence (Supplemental Data Set 2), which is a typical LTR-retrotransposon (TE) as a member of the Gypsy family. We searched for the TE in the genomic sequences of 3,000 rice varieties (Wang et al., 2018), the 32 high-quality genomes of O. sativa (Qin et al., 2021) and the 16 diverse genomes of O. rufipogon from our lab (Zhao et al., 2018). There is only a single copy of this TE in domesticated rice (Supplemental Table S2), but single or two copies of that in wild rice, strongly implying that lineage-specific transpositional bursts have not occurred for it (Supplemental Table S3). The proportion of cultivated rice harboring this TE was ∼20%, which might be an underestimate due to incompletely sequenced elements. We detected this TE in 13 accessions accounting for ∼40.6% of the 32 accessions with high-quality genomes (Qin et al., 2021). Surprisingly, the position of the TE was almost specific to each cultivated varietal group (Supplemental Table S2). In contrast, the position of the TE in nine wild rice accessions differed and presented tremendous diversity. Furthermore, we found that no PCR products were amplified from the 250 other diverse O. rufipogon accessions using the primers NIL-F/5′-R and 3′-F/NIL-R.
MLH1 interacts with two other MutL genes and the chromatin remodeling complex subunit SWIB
Three MutL paralogs (OsMLH1, OsPMS1, and OsMLH3) were detected in rice via comparison of homologous sequences. Phylogenetic analysis across some plant species also revealed three subgroups of MutL proteins corresponding to MLH1, PMS1, and MLH3 clades, respectively (Supplemental Figure S7).
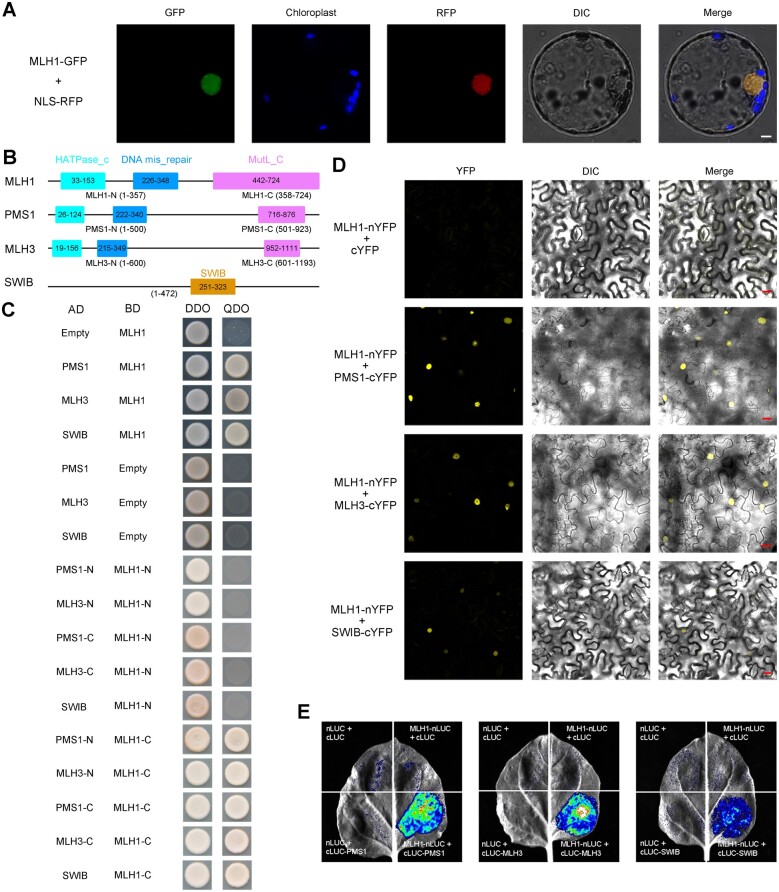
To gain deep insight into the molecular mechanism underlying MLH1 during meiosis, we screened for MLH1-interacting proteins via yeast library screening. Many positive clones were first obtained, but only three full-length coding sequence (CDS) clones were further verified to have actual interactions with OsMLH1, two of which are its homologs, OsPMS1 and OsMLH3. Another is OsSWIB/OsBAF60/LOC_Os04g31320, a subunit of the SWI/SNF (SWITCH/SUCROSE NONFERMENTING) chromatin remodeling complex and an putative ortholog of mammalian BRAHMA-associated factors (BAF60s), called CHC1, SWP73B, or BAF60 in Arabidopsis (Sacharowski et al., 2015), belonging to the SWIB/MDM2 domain superfamily protein in rice.
All four GFP fusion proteins (OsMLH1-GFP, OsPMS1-GFP, OsMLH3-GFP, and OsSWIB-GFP) were detected in the nucleus (Figure 4A; Supplemental Figure S8). RT–qPCR analysis revealed that OsMLH1 transcripts were highly accumulated in seedlings, flag leaves, and young panicles during meiosis stage (Supplemental Figure S9). To further examine the possible interaction domains among these proteins, we performed two-hybrid assay with their different truncated fragments. The assay indicated that the N-terminal region of OsMLH1 did not interact with any truncated OsPMS1 and OsMLH3, whereas the C-termini of OsMLH1 could interact with each of truncated OsPMS1 and OsMLH3 (Figure 4, B and C). It was also revealed that the interaction between OsMLH1 and OsSWIB was mediated by the MutL domain of OsMLH1 (Figure 4, B and C). We further conducted BiFC assays to confirm the interactions in Nicotiana benthamiana epidermal cells. Yellow fluorescent protein (YFP) signals were detected in cells co-expressing each vector pair, except for the negative control (Figure 4D). Then, luciferase (LUC) assay also verified these interactions (Figure 4E). Due to no interaction between OsPMS1 and OsMLH3 revealed by our BiFC test, it was suggested that none of the three MutL proteins in rice interactions with members of the family other than OsMLH1, and two types of heterodimers OsMLH1–OsPMS1 and OsMLH1–3 exist in rice, supporting the previous report from yeast that MLH1 playing a pivotal, coordinating role, functions in combination with each of the other MutL proteins (Wang et al., 1999).
Figure 4.
Protein sub-cellular localization and protein–protein interactions. A, Sub-cellular localization of OsMLH1-GFP fusion protein in rice protoplasts. Bar = 5 μm. B, The illustration of the conserved domains of three MutL proteins and OsSWIB in rice. C, Yeast two-hybrid assay to examine interactions among MutL proteins and chromatin remodeling complex subunit SWIB. Transformed yeast cells were grown on a synthetic medium lacking Trp and Leu (DDO) or Trp, Leu, His, and Ade (QDO). AD, GAL4 activation domain; BD, GAL4 DNA-binding domain. D, BiFC analysis of the interaction between MLH1 and each of PMS1, MLH3, and SWIB. Bar = 20 μm. E, LUC complementation assay between MLH1 and each of PMS1, MLH3, and SWIB.
Targeted disruption of MutL genes reveals lower male and female fertility in mutants of japonica than indica
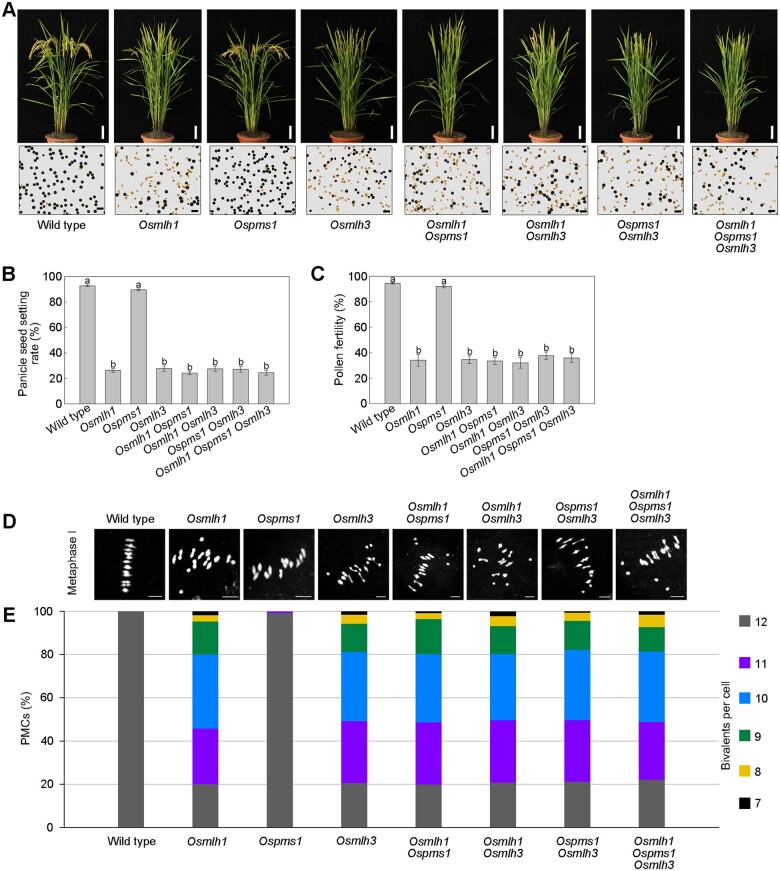
To further clarify the function of MutL genes in rice, firstly, we generated single, double, and triple MutL gene mutants in the japonica variety Dongjing, due to the relative simplicity of genetic transformation of japonica compared with indica varieties. Our analysis showed that among the three single mutants of MutL genes, Osmlh1 and Osmlh3 displayed significantly low PSSR, while that was almost unaffected in Ospms1. All double and triple mutant plants had almost equal and low PSSR, similar to single Osmlh1 and Osmlh3 mutants (Figure 5, A and B). In WT and Ospms1, the pollen fertility was no less than 90%, while that in all other mutants was ∼40% (Figure 5C).
Figure 5.
Targeted mutation of MutL genes in japonica (Dongjing). A, Gross morphology of mature plants (upper parts) and pollen grains with KI-I2 solution (lower parts) of WT and mutants. Bar = 10 cm (upper). Bar = 100 μm (lower). B, Histograms of PSSR. Values are means ± sem, n = 40. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way ANOVA. C, Histograms of pollen fertility. Values are means ± sem, n = 20. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way ANOVA. D, Representative images of metaphase I cells are shown. Bar = 5 μm. E, Quantification of the number of bivalents in WT and mutants. Bivalents, as a readout for CO formation, were assessed in chromosome spreads of the indicated mutant lines.
Using 4’,6-diamidino-2-phenylindole (DAPI) staining on chromosomes of PMC spreads, we observed no univalents in WT meiocytes (n = 109). Except for the Ospms1, the number of bivalents varied from 7 to 12 in the other six mutants. We found univalents in ∼80% of all analyzed cells of Osmlh1 (n = 105), Osmlh3 (n = 122), Osmlh1 Ospms1 (n = 111), Osmlh1 Osmlh3 (n = 131), Ospms1 Osmlh3 (n = 157), and Osmlh1 Ospms1 Osmlh3 (n = 123), with on average 10.39–10.48 bivalents per meiocyte in these mutants (Figure 5, D and E). However, it is perplexing that compared to NIL-mlh1 with indica background, loss of MLH1 in japonica variety Dongjing leads to more severely reduced viable pollens and lower PSSR. In comparison with only ∼20% of the meiocytes with 12 bivalents in Osmlh1 of japonica, this percentage was two-fold higher in indica (NIL-mlh1).
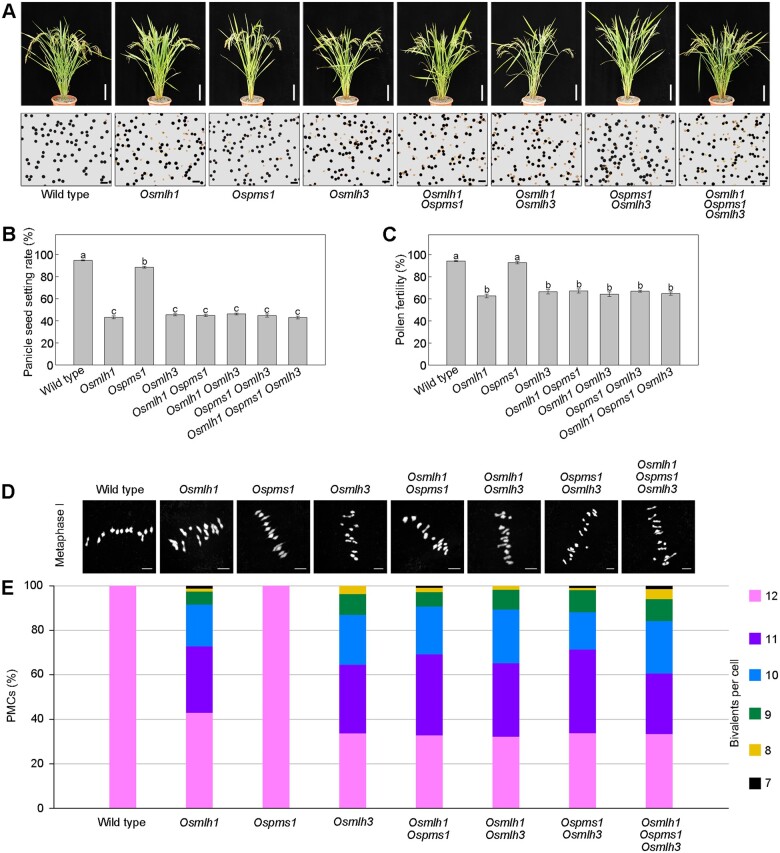
To confirm that these discrepancies between Osmlh1 (japonica background) and NIL-mlh1 (indica background) were indeed the results with no statistical bias, we further generated all corresponding mutants in indica GLA4 and examined their phenotypes. The data showed that Ospms1 exhibited high fertility identical to WT, and all other mutants from indica had similar phenotypes as NIL-mlh1, with the PSSR and pollen viability reduced to a little >40% and 60%, respectively (Figure 6, A–C). Except for Ospms1, analysis of DAPI-stained meiotic chromosomes of all other mutants revealed 10.70–11.03 bivalents per cell (Figure 6, D and E), consistent with the above observations in NIL-mlh1.
Figure 6.
Targeted mutation of MutL genes in indica (GLA4). A, Gross morphology of mature plants (upper) and pollen grains with KI-I2 solution (lower) of WT and mutants. Bar = 10 cm (upper). Bar = 100 μm (lower). B, Histograms of PSSR. Values are means ± sem, n = 40. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way ANOVA. C, Histograms of pollen fertility. Values are means ± sem, n = 20. The presence of the same lower letter denotes a nonsignificant difference among them (P > 0.01). P-values were calculated by one-way ANOVA. D, Representative images of metaphase I cells are shown in WT and mutants of indica. Bar = 5 μm. E, Quantification of the number of bivalents in WT and mutants of indica. Bivalents, as a readout for CO formation, were assessed in chromosome spreads of the indicated mutant lines.
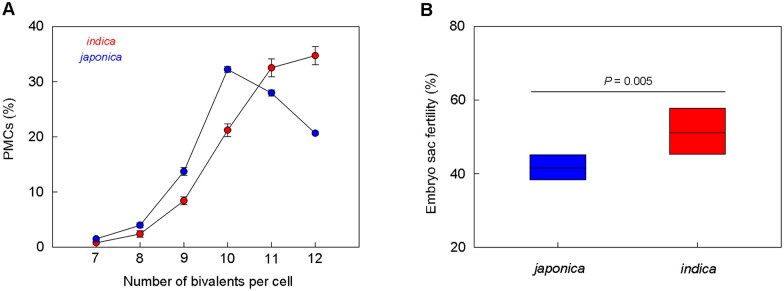
We calculated averages of the percentage of PMCs with different numbers of bivalents under loss of mlh1/3 (including Osmlh1, Osmlh3, Osmlh1 Ospms1, Osmlh1 Osmlh3, Ospms1 Osmlh3, and Osmlh1 Ospms1 Osmlh3, the same below) in both indica and japonica. We found that the majority (60%–70%) of PMCs had 12 or 11 bivalents in indica. In comparison, ∼60% of PMCs had ten or 11 bivalents in japonica (Figure 7A), suggestive of a more severe defect of male meiosis in japonica than indica under mlh1/3 mutation.
Figure 7.
Comparison of fertility between indica and japonica with lack of MLH1/3. A, Averages of the number of bivalents per meiocyte in all other mutants of indica and japonica, exclusive of Ospms1. Values are means ± sem, n = 5. B, Averages of the embryo sac fertility in all other mutants of indica and japonica, exclusive of Ospms1. Middle horizontal lines in the box represent the means. Significance was assessed by Student’s t test.
Because the meiosis process of megaspore mother cells (MMCs) is challenging to observe, instead, we examined the fertility of mature embryo sacs in WT and all mutants. We observed only a few abnormal embryo sacs in WT and Ospms1 of both subspecies. In contrast, we detected many abnormal embryo sacs in other mutants (Supplemental Table S4; Supplemental Figure S10), with most of them (>90%) being abortive with no cell differentiation (Supplemental Figure S10, A and B). No function of MLH1/3 resulted in lower embryo sac fertility in japonica than indica (41.5% versus 51.4%; Figure 7B), indicating that mutants of japonica also had more severe defects in female meiosis.
Taken together, we could conclude that three MutL genes in rice have divergent functions, only MLH1 and MLH3 were involved in MER; meiocytes from mlh1/3 mutants of japonica displayed fewer bivalents than those of indica, strongly indicating that mutants from indica had more residual COs than those from japonica under destroying of partial class I COs due to lack of MLH1/3. Because both MLH1 and MLH3 participate in the formation of class I COs (Lhuissier et al., 2007; Mercier et al., 2015), the phenomenon motivated us to speculate that the number of COs per meiosis is higher in indica than japonica.
CO rates in indica and japonica
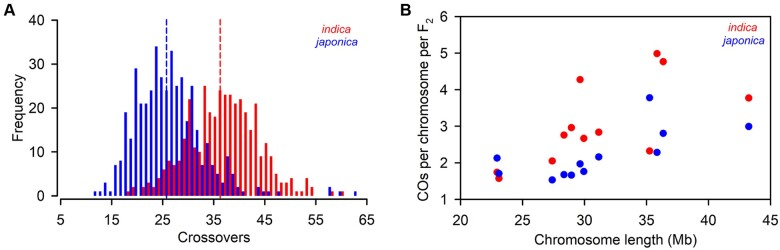
To confirm the above hypothesis, we analyzed the CO number per meiosis of both cultivated rice subspecies. As a first step in analyzing CO events, the newly generated 435 japonica F2 plants were genotyped, and an indica F2 population with 384 individuals from our previous report (Huang et al., 2016) was also re-genotyped. Then, we constructed two bin maps based on the recombination CO breakpoints detected in these two F2 populations, which generated 2,812 and 2,559 bins in the indica (Supplemental Data Set 3) and japonica (Supplemental Data Set 4) population, respectively. The physical length of each recombination bin ranged from 100 kb to 3 Mb in the indica map with an average of 132.13 kb, and from 100 kb to 2.95 Mb in the japonica map with an average of 144.96 kb. From these two crosses, a total of 25,600 COs were detected in 819 F2 plants, 14,093 and 11,507 COs for indica and japonica populations, respectively. The average number of CO events (36.7, ranging from 18 to 60) per F2 individual of indica was markedly higher than that (26.5, ranging from 12 to 63) in japonica F2 individuals (t test, P < 0.001; Figure 8A). The mean COs for each chromosome pair varied from 1.6 to 5.0 in the indica population, and from 1.5 to 3.8 in the japonica population (Figure 8B). As expected, the average CO number per chromosome pair was positively correlated with the physical length of the chromosome (r = 0.68, P < 0.05 for indica; r = 0.70, P < 0.05 for japonica), indicating that longer chromosomes have more CO events, consistent with previous studies (Salome et al., 2012; Si et al., 2015). The total amount of observed COs per F2 corresponded to a total genetic distance of 1,834.9 cM in indica population, higher than that in japonica population (1,322.6 cM). By assuming constant genome size in indica and japonica, we could estimate their average CO rates were 4.92 and 3.55 cM Mb−1, respectively.
Figure 8.
CO frequencies in indica and japonica. A, Histogram of CO number per F2 individual in indica and japonica populations. Vertical dash lines indicate mean values. B, CO s per chromosome per F2 compared with chromosome length in indica and japonica.
Discussion
pssr1 is a rare allele
In this study, we report the positional cloning of pssr1 causing low fertility from the wild rice and show that the semi-sterile phenotype of pssr1 is the consequence of a 5.4-kb LTR-retrotransposon insertion in the exon 2 of OsMLH1 which leads to a failed cDNA transcript. Because the pssr1 allele was not detected in other collected wild rice accessions, we speculated that it was a rare allele, suggesting that the low PSSR and fertility in different wild rice populations or accessions might be caused by reasons, which remain to be further elucidated.
Functional divergence of MutL genes
Previous studies on the function of MutL genes in plants and nonplant organisms did not focus on double and multiple MutL gene mutants. In this study, we analyzed the effects of single, double, and triple MutL gene mutants in two rice subspecies, indica and japonica. Loss of function analysis demonstrates functional divergence of MutL genes in rice. In contrast to Osmlh1 and Osmlh3, Ospms1 confers no marked defects in either chromosome segregation or male and female fertility. The spore viability in pms1 mutant of yeast showed only ∼10% reduction (Wang et al., 1999). However, Arabidopsis plants lacking PMS1 displayed a severely reduced seed set and a high proportion of collapsed pollen (Li et al., 2009). In rice, the heterozygotes of Osmlh1 mutant exhibited unreduced PSSR (Supplemental Figure S2). However, the seed set rate in heterozygotes of Atmlh1 mutant was reduced by ∼50% (Dion et al., 2007). These results also suggest that some functional specificity of MutL genes exists among organisms.
In comparison with Osmlh1 or Osmlh3, the Osmlh1 Osmlh3 double mutant shows indistinguishable reductions in fertility and the number of bivalents per meiocyte, indicating that OsMLH1 and OsMLH3 do not have an additive effect on fertility. Immunolocalization studies have revealed that AtMLH3 and HvMLH3 localize as discrete foci at zygotene of meiosis, and the loading of AtMLH1 is dependent on AtMLH3, as AtMLH1 foci are not observed in the Atmlh3 mutants, indicating that MLH3 is recruited earlier than MLH1 to the newly formed axes potentially during synapsis (Jackson et al., 2006; Colas et al., 2016). Irrespective of indica or japonica, the fertility and the mean numbers of bivalents in PMCs of Osmlh1/3 are much higher than those in ZMM protein mutants like Osmsh4, Osmsh5, and Oshei10 (Wang et al., 2012, 2016; Luo et al., 2013), supporting previous studies that both MLH1 and MLH3 function downstream of MSH4/5 and HEI10. Taken together with the interaction studies described above, it was strongly suggested that OsMLH1 and OsMLH3 act in the same pathway, and form an OsMLH1–3 heterodimer implicated in MER and CO formation.
Osmlh1/3 mutants have similar male and female semi-sterility
More recently, two independent labs reported Osmlh1 and Osmlh3 single mutants’ phenotypes(Mao et al., 2021; Xin et al., 2021). A spontaneous mutant of Osmlh3, also called female sterile variation 1, in Mao et al.’ study exhibited different male and female gamete fertility, that is, severe female gamete sterility (86.5% of embryo sacs are nonfunctional) and slightly reduced pollen fertility (no statistical data) (Mao et al., 2021). In contrast, the Osmlh1 and Osmlh3 single mutants in Xin et al. displayed substantially reduced male fertility (∼40%) and partially destroyed female gametes (no statistical data), so the authors thought that the markedly reduced PSSR in these mutants was largely attributed to the abnormal development of pollens, that is, male defects (Xin et al., 2021). However, in our study, both male and female gametes in Osmlh1/3 single and double mutants have similar semi-sterile phenotypes, which were less severe in indica mutants than in japonica mutants. About 40%–60% pollen fertility in NIL-mlh1 and Osmlh1/3 mutants should be sufficient for seed set due to excessive pollens, as revealed in Mao et al.’ (2021) study, suggesting the low PSSR in NIL-mlh1 and Osmlh1/3 mutants may be mainly attributed to the substantial reduction of embryo sac fertility. Few PMCs having 12 bivalents were found in the mutants of their study. In our study, however, we revealed that ∼20%–40% of PMCs had 12 bivalents in Osmlh1/3 mutants of indica and japonica with on average 10.39–11.03 bivalents per meiocyte, corresponding to 35%–65% viable pollens (Figures 5 and 6), indicating that the semi-sterility of male gametes should be maintained by a considerable percentage of meiocytes with 12 bivalents.
Inter-subspecific difference of COs per meiosis may be driven by a near-uniform increase in recombination across the genome
Meiotic CO is not only a major force in evolution and creates genetic diversity and is also an important facilitator during plant breeding by combining favorable alleles into elite varieties (Zelkowski et al., 2019; Kivikoski et al., 2021). To our knowledge, no detailed comparative analysis on CO rate of both rice O. sativa indica and japonica subspecies has been carried out until now. Only the historical recombination maps of indica and japonica using 150 re-sequenced genome data from the 3000 Rice Genome Project was constructed (Marand et al., 2019), providing a historical measure of meiotic COs regarding multiple generations of both subspecies. In this study, we revealed on average 36.7 and 26.5 CO events per F2 individual per meiosis in indica and japonica hybrids, respectively. These values are very similar to previous reports or the estimates calculated from other rice maps. An average of 33 COs were detected in 24 F2 unstressed individuals from F1 seeds of a super hybrid rice LYP9 obtained from the cross between indica cultivars PA64s and 9311 (Si et al., 2015). Based on 44,049 high-quality SNPs between two indica varieties ZS97 and MH63, a map with 1,850 recombination bins was generated in the ZS97 × MH63 F2 population with 163 individuals, corresponding to a total genetic map length of 2,081 cM (Li et al., 2017), which is equivalent to ∼ 41.6 COs per nucleus. This CO number is slightly higher than the CO frequency of indica F1 hybrid in our study. From the genotyped data of an intra-subspecific F2 population with 150 plants from the cross between japonica Nipponbare and Dongjing (Mieulet et al., 2018), we quantified on average 31.8 COs per F2 plant per meiosis in the japonica population (Supplemental Data Set 5), which was also higher than that in our japonica population, indicating the variations of CO rate among intra-subspecific individuals. These data also showed that indica has a higher CO rate than japonica, supporting our hypothesis. Previous studies revealed that the nucleotide diversity and linkage disequilibrium (LD) decay rates were higher in indica than japonica (Huang et al., 2010; Xu et al., 2012b; Wang et al., 2018). Because recombination can break down LD and is mutagenic (Nachman, 2001), it is expected that indica population may have a higher CO rate than japonica population, contributing to higher nucleotide diversity and LD decay rate in the former. This is verified by this study.
Our genetic analysis demonstrates that indica has, on average, ∼39% higher CO rates than japonica in chromosome level. Some previous works have reported that the variation of inter-populations and inter-individuals in CO rate may be the result of allelic variation in meiosis-related gene(s) (Kong et al., 2008; Ziolkowski et al., 2017; Lawrence et al., 2019). Further research is needed to discover the modifier(s), resulting in the difference in CO rate, and then utilize it/them to elevate the genetic CO rate and increase genetic variation during rice breeding, especially japonica breeding.
Materials and methods
Plant materials and QTL analysis
To understand the genetic mechanisms of low PSSR in wild rice, we developed a set of backcross recombinant inbred lines derived from a cross between a cultivated rice variety O. sativa ssp. indica Guangluai4 (GLA4) and a wild rice accession O. rufipogon (W1943) with low fertility (Supplemental Figure S11). We investigated the PSSR of all 250 lines, with three panicles per plant and six plants per line, that is 18 panicles per line. Subsequent QTL analysis revealed the presence of two QTLs of PSSR1 and PSSR9, both of which are responsible for low PSSR.
The 112 cultivated rice varieties used in the association mapping analysis are listed in Supplemental Data Set 1. Sequences of all primers used in genetic mapping are listed in Supplemental Data Set 6. All plant materials were grown in paddy fields in Shanghai or Hainan Province, China.
Plasmid construction
To generate the complementation construct, an entire 8768-bp PSSR1/MLH1/LOC_Os01g72880 genomic region was amplified from WT (GLA4) and then inserted into the binary vector pCAMBIA1300. The resulting plasmid was then transformed into NIL-mlh1.
For the CRISPR–Cas9 targeting of three MutL genes, the intermediate vector SK-gRNA and the CRISPR–Cas9 binary vector pC1300-cas9 were used. We first generated a single mutant (Osmlh1), two double mutants (Osmlh1 Ospms1 and Osmlh1 Osmlh3), and a triple mutant (Osmlh1 Ospms1 Osmlh3) in japonica variety (Dongjing), and then three single mutants (Osmlh1, Ospms1, Osmlh3) and a triple mutant (Osmlh1 Ospms1 Osmlh3) in indica variety (GLA4). The other corresponding single or double mutants were segregated from the triple heterozygous Osmlh1 Ospms1 Osmlh3. Finally, for each subspecies, seven types of mutants were generated.
For mutation in promoter sequences of MLH1, a construct carrying two gRNAs targeting the promoter of a japonica variety Nipponbare was generated.
The target sequences used in CRISPR/Cas9 are listed in Supplemental Data Set 6. The resulting plant lines and F1, F2, or F3 plants from crosses were genotyped by PCR using primers described in Supplemental Data Set 6.
RNA extraction and RT–qPCR analysis
For gene expression analysis, samples were ground in liquid nitrogen, and total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). In vitro reverse transcription was performed using ReverTra Ace qPCR RT Master Mix (TOYOBO, Osaka, Japan). RT–qPCR analysis was performed on an ABI QuantStudio 5 instrument using the THUNDERBIRD SYBR qPCR Master Mix (TOYOBO). Rice Actin was used as the internal control. Primers are listed in Supplemental Data Set 6.
Semithin section
Observation of anther development was performed as previously described by Zhou et al. (2011). In brief, spikelets of different developmental stages were fixed in formaldehyde: acetic acid (FAA: 50% [v/v] ethanol = 5: 5: 90) for >24 h and then dehydrated through an ethanol series. Samples were embedded in Eponate 12 (Ted Pella) and sectioned. Images were captured using Zeiss Axio imager.M2.
Fertility examination
Six plants of the WT and mutants were used to estimate the pollen and embryo-sac fertility. At the heading stage, the spikelets from three panicles per plant were fixed in FAA for >24 h at room temperature, after which they were stored in 70% (v/v) ethanol at 4°C. Pollen fertility was examined with the I2-KI staining method, using mixed pollens from all six anthers of each spikelet. Three independent microscope fields were observed by light microscopy, and the average of percentage of stained pollens from three fields was expressed as the fertility of each spikelet. To evaluate the embryo-sac fertility, 100–200 ovaries were inspected using WCLSM method (Zhou et al., 2011). Three or four panicles per plant at maturity were harvested to investigate PSSR.
Meiotic chromosome preparation
Young panicles of all the WT and mutant lines were fixed in Carnoy’s solution (ethanol:glacial acetic acid = 3:1). Meiotic chromosome was prepared as described previously (Cheng, 2013). After counterstaining with DAPI (Vector Laboratories; H-1200), cytological analysis was performed under confocal laser scanning microscopy (Leica SP8).
Dual-LUC assay
To generate LUC expression constructs, the promoters of MLH1 amplified from NIL-mlh1 and GLA4, together with a directed-mutant promoter (“A” insertion) from NIL-mlh1 were cloned into the pGreenII 0800-Luc expression vector. The plasmids were introduced into rice stem protoplasts, and then the transformed protoplasts were incubated overnight. The dual-LUC assay is performed by sequentially quantifying the firefly LUC and Renilla LUC activities of the same sample using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Subcellular localization
To determine the subcellular localization of the MLH1, PMS1, MLH3, and SWIB proteins, we cloned their full-length CDSs from GLA4 into the p1300-GFP vector. The NLS sequence was fused with RFP into PA7-35S-RFP as a nuclear marker. We co-transformed the p1300-MLH1-GFP and PA7-35S-NLS-RFP into rice protoplasts. We also transformed the four generated plasmids with GFP fusion protein into N. benthamiana leaves via infiltration. The subcellular localization and colocalization of these proteins were examined using confocal microscopy (Leica SP8). GFP signals were excited at 488 nm and emission was detected at 515–545 nm. RFP signals were excited at 552 nm and emission was detected at 580–620 nm. The relevant primers are listed in Supplemental Data Set 6.
Library screening and Y2H assay
The yeast prey library was constructed using the purified total RNA from the young rice panicles during the meiotic stage. The full-length CDS of MLH1 was cloned into the pGBKT7 vector to construct the bait plasmid pGBKT7-MLH1, then which was transformed into yeast strain Y2H. Transformed cells were further transformed with plasmids from the prey library. Co-transformants were selected on SD/Trp–Leu–His–Ade culture medium. Surviving clones were further screened on SD/Trp–Leu–His–Ade/X-a-gal medium to examine the LacZ activity. Then, positive clones were used for colony PCR, and the products were sequenced and analyzed. For the interaction verification, the full-length or truncated CDSs of related genes were inserted into the vector pGADT7 or pGBKT7, and then the plasmids were transformed into Y2H Gold. These verification assays were repeated 3 times. The relevant primer sequences are shown in Supplemental Data Set 6.
BiFC assay
To conduct BiFC assays, the full-length CDS of MLH1 was cloned into pUC-nYFP, and the full-length CDSs of PMS1, MLH3, and SWIB were cloned into pUC-cYFP. Constructs were transformed into Agrobacterium tumefaciens strain GV3101 (pSoup-p19), and then infiltrated into young but fully expanded N. benthamiana leaves. After 48-h dark incubation, leaves were excised and fluorescence was examined using a confocal microscope (Leica SP8). YFP signals were excited at 514 nm and emission was detected at 520–545 nm. The relevant primer sequences used in this assay are listed in Supplemental Data Set 6.
Split-LUC complementation assay
The cDNA of MLH1 was cloned to the N-terminal Luc fusion vector JW771-nLUC, and the coding region sequences of PMS1, MLH3, and SWIB were cloned to the C-terminal Luc fusion vector JW772-cLUC. LUC assays were performed as described previously (Chen et al., 2008). LUC activity was measured using a cooled CCD-image system (Tanon 5200). These assays were independently repeated >3 times. Primers used in this assay are listed in Supplemental Data Set 6.
Genotyping by whole-genome resequencing and CO number counting
Two sets of F2 population from japonica–japonica cross (Wuyungeng7A × R254) and indica–indica cross (TianfengA × Guanghui998) were applied to estimate the CO rate and CO numbers in japonica and indica, respectively. The indica–indica cross included 384 plants and sequencing data was reported previously (Huang et al., 2016). The japonica–japonica cross was newly generated and contained 435 individuals. Genomic DNA was extracted from the fresh leaf of each F2 plant. A sequence library was constructed and sequencing was conducted at Illumina Hiseq2500 platform as described previously (Huang et al., 2016). Each F2 plant was resequenced with ∼0.2× genome coverage. Genotyping was conducted following a previous report (Huang et al., 2009). The sequencing data of the parental lines were aligned against the reference genome sequence (IRGSP release build 1.0) to identify high-quality SNPs by BWA (version 0.7.17) and GATK (version 4.1.8.1). The custom-made parental pseudomolecules were generated by replacing the bases of the reference genome sequence (IRGSP release build 1.0) with identified SNPs. Raw reads of each F2 individual were aligned back to corresponding parental pseudomolecules using BWA (version 0.7.17) and recombination map was constructed by a customized Perl script. CO rate and CO numbers were counted based on the recombination map using customized Perl script.
Statistical analyses
Statistical significance was determined by Student’s t test. Statistical comparisons among samples were performed using one-way analysis of variance. The multiple sequence alignment was conducted using MAFFT version 7 online service (https://mafft.cbrc.jp/alignment/server/index.html). The neighbor-joining method was used to construct phylogenetic tree with 1,000 bootstrap replicates in MEGA version 6.0. The protein domains were predicted by SMART (http://smart.embl-heidelberg.de/).
Accession numbers
Sequence data of the major genes from this article can be found in the GenBank data libraries under the following accession numbers: OsMLH1, Os01g0958900; OsPMS1, Os02g0592300; OsMLH3, Os09g0551900; OsSWIB, Os04g0382100. Accession numbers of the homologs of MLH1, PMS1, and MLH3 used for phylogenetic analysis are as follows: Aegilops tauschii subsp. tauschii, XP_020166257.1, XP_020182843.1, XP_020180293.1; A. thaliana, NP_567345.2, NP_567236.1, NP_001328253.1; Brachypodium distachyon, XP_003565112.1, XP_003575262.1, XP_010238476.1; Glycine soja, XP_028230240.1, XP_028207543.1, XP_028240586.1; Hordeum vulgare subsp. vulgare, BAJ99147.1, KAE8773953.1, AFP73613.1; Oryza brachyantha, XP_015691074.1, XP_006647441.1, XP_015696863.1; O. sativa subsp. japonica, XP_015649867.1, XP_015627646.1, XP_015612513.1; Populus trichocarpa, XP_024446715.1, XP_002321013.1, XP_024456729.1; Prunus persica, XP_020410162.1, ONH93891.1, XP_020423821.1; Setaria viridis, TKW18956.1, TKW40036.1, XP_034578389.1; Sorghum bicolor, XP_002456964.1, XP_021316187.1, XP_021310121.1; Triticum turgidum subsp. durum, VAH85715.1, VAI57965.1, VAI20910.1; Vitis vinifera, XP_003633884.2, XP_010652175.1, XP_010649441.1; Zea mays, XP_008656853.1, XP_008678747.1, XP_020404133.1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The relative expression levels of the seven candidate genes in flag leaves of GLA4 and NIL-mlh1 like plants.
Supplemental Figure S2. Comparison of WT (GLA4) and heterozygote plant.
Supplemental Figure S3. Comparison of male gametogenesis in the WT and NIL-mlh1.
Supplemental Figure S4. Complementary test of MLH1.
Supplemental Figure S5. The mutation in the promoter of OsMLH1 by CRISPR-Cas9.
Supplemental Figure S6. Identification of the 5.4-kb retrotransposon insertion in mlh1 allele of NIL-mlh1 line.
Supplemental Figure S7. Phylogenetic analysis of MutL family proteins.
Supplemental Figure S8. Subcellular localization of MutL family proteins and OsSWIB.
Supplemental Figure S9. Expression pattern of OsMLH1.
Supplemental Figure S10. Representative abnormal embryo sacs.
Supplemental Figure S11. KI-I2 staining of GLA4 and W1943 pollens.
Supplemental Table S1. Comparison of morphological traits between WT (GLA4) and NIL-mlh1.
Supplemental Table S2. Tracking results for the retrotransposon in 3000 rice genomes.
Supplemental Table S3. Tracking results for the retrotransposon in wild rice genomes.
Supplemental Table S4. Examination of mature embryo sacs in WT and MutL genes knockout lines.
Supplemental Data Set 1. PSSR of rice accessions.
Supplemental Data Set 2. The 5,429-bp retrotransposon sequence inserted in mlh1 gene.
Supplemental Data Set 3. The bin map of indica population.
Supplemental Data Set 4. The bin map of japonica population.
Supplemental Data Set 5. The CO number per chromosome pair of 150 F2 plants from cross between Nipponbare and Dongjing.
Supplemental Data Set 6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank professors Zhukuan Cheng (Institute of Genetics and Developmental Biology, CAS) and Wanqi Liang (Shanghai Jiao Tong University) for helpful suggestions and discussions. We thank Jiqin Li, Shuining Yin and Wenjuan Cai (CAS Center for Excellence in Molecular Plant Sciences) for technical support.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31788103) and the Chinese Academy of Sciences (XDB27010301).
Data availability statement
All data are available from the corresponding author upon reasonable request.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Kun Liu, College of Life Sciences, Anhui Normal University, Wuhu, 241000, China; National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Erwang Chen, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Zhoulin Gu, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Bingxin Dai, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China; School of Life Science and Technology, Shanghai Tech University, Shanghai, 201210, China.
Ahong Wang, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Zhou Zhu, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Qi Feng, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Congcong Zhou, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Jingjie Zhu, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Yingying Shangguan, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Yongchun Wang, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Zhen Li, College of Life Sciences, Anhui Normal University, Wuhu, 241000, China; National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Qingqing Hou, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Danfeng Lv, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Changsheng Wang, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Tao Huang, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Zixuan Wang, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
Xuehui Huang, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
Bin Han, National Center for Gene Research, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, 200233, China.
B.H. conceived the research. K.L., E.C., B.D., J.Z., Y.S., Q.H., D.L., and C.W. performed the wet experiments. K.L. analyzed the majority of data. Z.G., Z.Z., and X.H. did the informatics analysis. K.L., E.C., A.W., Y.W., Z.L., and Z.W. were responsible for field management and phenotype investigation. T.H. managed server station. Q.F. and C.Z. performed library construction and whole genome sequencing. K.L. and B.H. wrote the article. K.L., E.C., and B.H. supervised and completed the writing.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Bin Han (bhan@ncgr.ac.cn).
References
- Alou AH, Jean M, Domingue O, Belzile FJ (2004) Structure and expression of AtPMS1, the Arabidopsis ortholog of the yeast DNA repair gene PMS1. Plant Sci 167: 447–456 [Google Scholar]
- Boddy MN, Gaillard PHL, McDonald WH, Shanahan P, Yates JR, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran-Guiu X, Charbonnier JB, et al. (2020) Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586: 618–622 [DOI] [PubMed] [Google Scholar]
- Chen HM, Zou Y, Shang YL, Lin HQ, Wang YJ, Cai R, Tang XY, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZK (2013) Analyzing meiotic chromosomes in rice. Methods Mol Biol 990: 125–134 [DOI] [PubMed] [Google Scholar]
- Colas I, Macaulay M, Higgins JD, Phillips D, Barakate A, Posch M, Armstrong SJ, Franklin FCH, Halpin C, Waugh R, et al. (2016) A spontaneous mutation in MutL-Homolog 3 (HvMLH3) affects synapsis and crossover resolution in the barley desynaptic mutant des10. New Phytologist 212: 693–707 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Housworth EA, Stahl FW (2002) Crossover interference in Arabidopsis. Genetics 160: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion E, Li LL, Jean M, Beizile F (2007) An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant J 51: 431–440 [DOI] [PubMed] [Google Scholar]
- Falque M, Anderson LK, Stack SM, Gauthier F, Martin OC (2009) Two types of meiotic crossovers coexist in maize. Plant Cell 21: 3915–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XH, Feng Q, Qian Q, Zhao Q, Wang L, Wang AH, Guan JP, Fan DL, Weng QJ, Huang T, et al. (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19: 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XH, Wei XH, Sang T, Zhao QA, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genetics 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Huang XH, Yang SH, Gong JY, Zhao Q, Feng Q, Zhan QL, Zhao Y, Li WJ, Cheng BY, Xia JH, et al. (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537: 629–633 [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH (2006) Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Gujar V, Burdett V, Modrich PL, Kadyrov FA (2020) Human MutL gamma, the MLH1-MLH3 heterodimer, is an endonuclease that promotes DNA expansion. Proc Natl Acad Sci USA 117: 3535–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivikoski M, Rastas P, Löytynoja A, Merilä J (2021) Effects of crossover interference on genomic recombination landscape. bioRxiv, doi.org/10.1101/2020.1112.1114.422614 [Google Scholar]
- Kolodner RD, Marsischky GT (1999) Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9: 89–96 [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM, Gudjonsson SA, Sverrisson S, Thorlacius T, et al. (2008) Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319: 1398–1401 [DOI] [PubMed] [Google Scholar]
- Lawrence EJ, Gao HB, Tock AJ, Lambing C, Blackwell AR, Feng XQ, Henderson IR (2019) Natural variation in TBP-ASSOCIATED FACTOR 4b controls meiotic crossover and germline transcription in Arabidopsis. Curr Biol 29: 2676–2686 [DOI] [PubMed] [Google Scholar]
- Lhuissier FGP, Offenberg HH, Wittich PE, Vischer NOE, Heyting C (2007) The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 19: 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Li XT, Wang Y, Mi JM, Xing F, Zhang DH, Dong QY, Li XH, Xiao JH, Zhang QF, et al. (2017) Three representative inter and intra-subspecific crosses reveal the genetic architecture of reproductive isolation in rice. Plant J 92: 349–362 [DOI] [PubMed] [Google Scholar]
- Li LL, Dion E, Richard G, Domingue O, Jean M, Belzile FJ (2009) The Arabidopsis DNA mismatch repair gene PMS1 restricts somatic recombination between homeologous sequences. Plant Mol Biol 69: 675–684 [DOI] [PubMed] [Google Scholar]
- Li SC, Li WB, Huang B, Cao XM, Zhou XY, Ye SM, Li CB, Gao FY, Zou T, Xie KL, et al. (2013) Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat Commun 4: 2793. [DOI] [PubMed] [Google Scholar]
- Li YF, Qin BX, Shen Y, Zhang FF, Liu CZ, You HL, Du GJ, Tang D, Cheng ZK (2018) HEIP1 regulates crossover formation during meiosis in rice. Proc Natl Acad Sci USA 115: 10810–10815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. (2002) Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31: 385–390 [DOI] [PubMed] [Google Scholar]
- Luo Q, Tang D, Wang M, Luo WX, Zhang L, Qin BX, Shen Y, Wang KJ, Li YF, Cheng ZK (2013) The role of OsMSH5 in crossover formation during rice meiosis. Mol Plant 6: 729–742 [DOI] [PubMed] [Google Scholar]
- Mao BG, Zheng WJ, Huang Z, Peng Y, Shao Y, Liu CT, Tang L, Hu YY, Li YK, Hu LM, et al. (2021) Rice MutL gamma, the MLH1-MLH3 heterodimer, participates in the formation of type I crossovers and regulation of embryo sac fertility. Plant Biotechnol J 19: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Zhao HN, Zhang WL, Zeng ZX, Fang C, Jiang JM (2019) Historical meiotic crossover hotspots fueled patterns of evolutionary divergence in rice. Plant Cell 31: 645–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogue F, Doutriaux MP, Horlow C, Grelon M, et al. (2005) Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr Biol 15: 692–701 [DOI] [PubMed] [Google Scholar]
- Mercier R, Mezard C, Jenczewski E, Macaisne N, Grelon M (2015) The molecular biology of meiosis in plants. Ann Rev Plant Biol 66: 297–327 [DOI] [PubMed] [Google Scholar]
- Mieulet D, Aubert G, Bres C, Klein A, Droc G, Vieille E, Rond-Coissieux C, Sanchez M, Dalmais M, Mauxion JP, et al. (2018) Unleashing meiotic crossovers in crops. Nat Plants 4: 1010–1016 [DOI] [PubMed] [Google Scholar]
- Nachman MW (2001) Single nucleotide polymorphisms and recombination rate in humans. Trends Genet 17: 481–485 [DOI] [PubMed] [Google Scholar]
- Qin P, Lu HW, Du HL, Wang H, Chen WL, Chen Z, He Q, Ou SJ, Zhang HY, Li XZ, et al. (2021) Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184: 3542–3558 [DOI] [PubMed] [Google Scholar]
- Ren Y, Chen D, Li WJ, Zhou D, Luo T, Yuan GQ, Zeng J, Cao Y, He ZS, Zou T, et al. (2019) OsSHOC1 and OsPTD1 are essential for crossover formation during rice meiosis. Plant J 98: 315–328 [DOI] [PubMed] [Google Scholar]
- Sacharowski SP, Gratkowska DM, Sarnowska EA, Kondrak P, Jancewicz I, Porri A, Bucior E, Rolicka AT, Franzen R, Kowalczyk J, et al. (2015) SWP73 subunits of Arabidopsis SWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development. Plant Cell 27: 1889–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Bomblies K, Fitz J, Laitinen RAE, Warthmann N, Yant L, Weigel D (2012) The recombination landscape in Arabidopsis thaliana F-2 populations. Heredity 108: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H, Lambing C, Griffin CH, Topp SD, Nageswaran DC, Underwood CJ, Ziolkowski PA, Seguela-Arnaud M, Fernandes JB, Mercier R, et al. (2018) Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc Natl Acad Sci USA 115: 2437–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si WN, Yuan Y, Huang J, Zhang XH, Zhang YC, Zhang YD, Tian DC, Wang CL, Yang YH, Yang SH (2015) Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F-2 plants. New Phytologist 206: 1491–1502 [DOI] [PubMed] [Google Scholar]
- Toledo M, Sun X, Brieno-Enriquez MA, Raghavan V, Gray S, Pea J, Milano CR, Venkatesh A, Patel L, Borst PL, et al. (2019) A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLgamma during crossover formation in meiotic prophase I. PLoS Genet 15: e1008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Wang Y, Cheng ZJ, Zhao ZG, Chen J, Sheng PK, Yu Y, Ma WW, Duan EC, Wu FQ, et al. (2016) The role of OsMSH4 in male and female gamete development in rice meiosis. J Exp Bot 67: 1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KJ, Wang M, Tang D, Shen Y, Miao CB, Hu Q, Lu TG, Cheng ZK (2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N (1999) Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Mauleon R, Hu ZQ, Chebotarov D, Tai SS, Wu ZC, Li M, Zheng TQ, Fuentes RR, Zhang F, et al. (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Copenhaver GP (2018) Meiotic recombination: mixing it up in plants. Ann Rev Plant Biol 69: 577–609 [DOI] [PubMed] [Google Scholar]
- Xin XD, Li XW, Zhu JK, Liu XB, Chu ZH, Shen JL, Wu CY (2021) OsMLH1 interacts with OsMLH3 to regulate synapsis and interference-sensitive crossover formation during meiosis in rice. J Genet Genom 48: 485–496 [DOI] [PubMed] [Google Scholar]
- Xu J, Li MR, Chen L, Wu GJ, Li HQ (2012a) Rapid generation of rice mutants via the dominant negative suppression of the mismatch repair protein OsPMS1. Theor Appl Genet 125: 975–986 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu FY, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, et al. (2012b) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30: 105–111 [DOI] [PubMed] [Google Scholar]
- Xu Y, Yang J, Wang YH, Wang JC, Yu Y, Long Y, Wang YL, Zhang H, Ren YL, Chen J, et al. (2017) OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genetics 13: e1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XW, Tang QJ, Wang HJ, Yun Y, Yan F, Xing FN (2014) Investigation and analysis of fertility characteristics of Oryza rufipogon Griff. in North part of hainan province. J Plant Genetic Resource 15: 882–887 [Google Scholar]
- Yang PZ, Guo HB, Zhao XJ, Li JQ, Liu XD, Lu YG (2006) Reproductive characteristics of Oryza rufipogon Griff. in Gaozhou, Guangdong Province II.Embryo Sac Fertility, Embryo Sac development, embryogenesis and endosperm development. J Plant Genet Resource 7: 136–143 [Google Scholar]
- Zelkowski M, Olson MA, Wang MH, Pawlowski W (2019) Diversity and determinants of meiotic recombination landscapes. Trends Genet 35: 359–370 [DOI] [PubMed] [Google Scholar]
- Zhang FF, Ma LJ, Zhang C, Du GJ, Shen Y, Tang D, Li YF, Yu HX, Ma BJ, Cheng ZK (2020a) The SUN domain proteins OsSUN1 and OsSUN2 play critical but partially redundant roles in meiosis. Plant Physiol 183: 1517–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Shen Y, Miao CB, Cao YW, Shi WQ, Du GJ, Tang D, Li YF, Luo Q, Cheng ZK (2020b) OsRAD51D promotes homologous pairing and recombination by preventing nonhomologous interactions in rice meiosis. New Phytologist 227: 824–839 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu HY, Li Y, Wang A, Tian QL, Zhan QL, Lu YQ, Huang T, Wang YC, et al. (2018) Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50: 278–284 [DOI] [PubMed] [Google Scholar]
- Zhou L, Han JL, Chen YL, Wang YX, Liu YG (2017) Bivalent formation 1, a plant-conserved gene, encodes an OmpH/coiled-coil motif-containing protein required for meiotic recombination in rice. J Exp Bot 68: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SR, Wang Y, Li WC, Zhao ZG, Ren YL, Wang Y, Gu SH, Lin QB, Wang D, Jiang L, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23: 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Underwood CJ, Lambing C, Martinez-Garcia M, Lawrence EJ, Ziolkowska L, Griffin C, Choi K, Franklin FCH, Martienssen RA, et al. (2017) Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev 31: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.
Conflict of interest statement. The authors declare no conflict of interest.