Abstract
Resistance to chemotherapeutic drugs is a major cause of treatment failure in acute myeloid leukemias (AML). To better characterize the mechanisms of chemoresistance, we first identified genes whose expression is dysregulated in AML cells resistant to daunorubicin or cytarabine, the main drugs used for induction therapy. The genes found to be activated are mostly linked to immune signaling and inflammation. Among them, we identified a strong upregulation of the NOX2 NAPDH oxidase subunit genes (CYBB, CYBA, NCF1, NCF2, NCF4 and RAC2). The ensuing increase in NADPH oxidase expression and production of reactive oxygen species, which is particularly strong in daunorubicin-resistant cells, participates in the acquisition and/or maintenance of resistance to daunorubicin. Gp91phox (CYBB-encoded Nox2 catalytic subunit), was found to be more expressed and active in leukemic cells from patients with the French-American-British (FAB) M4/M5 subtypes of AML than in those from patients with the FAB M0-M2 ones. Moreover, its expression was increased at the surface of patients’ chemotherapy-resistant AML cells. Finally, using a gene expression based score we demonstrated that high expression of NOX2 subunit genes is a marker of adverse prognosis in AML patients. The prognostic NOX score we defined is independent of the cytogenetic-based risk classification, FAB subtype, FLT3/NPM1 mutational status and age.
Introduction
Acute myeloid leukemias (AML) are a heterogeneous group of hematologic malignancies resulting from the transformation of hematopoietic stem- or progenitor cells. Prognosis is poor, in particular for old patients (>60 years).1 For fit patients, the standard treatment generally relies on intensive chemotherapy combining 3 days of an anthracycline (daunorubicin [DNR] or idarubicin) and 7 days of cytarabine (Ara-C) (the “3+7” regimen)2.
Resistance to chemotherapy remains the main cause of relapse and death. However, the mechanisms responsible for the acquisition and maintenance of chemoresistance are not fully elucidated. Those described include changes in drug uptake/efflux,3 modulation of the pro-drug activation process,4,5 greater resistance to apoptosis,6 enhanced DNA repair abilities7 and modulation of energy and redox metabolism.8,9 A better characterization of the pathways dysregulated in chemoresistant AML would be instrumental to find new therapeutic targets to overcome chemo-resistance. This could also provide new biomarkers allowing improvement of AML risk stratification, which is currently mostly based on the number and nature of the cytogenetic abnormalities.2 Such prognostic biomarkers might help clinicians in their therapeutic decisions. This is all the more important considering that new therapies, such as those targeting mutated FLT3 and IDH1/2 or BCL2, are emerging as promising alternatives to standard chemotherapies.10
Here, we have addressed the mechanisms underlying AML resistance through the identification of genes whose expression is dysregulated in AML cells resistant to DNR or Ara-C. We found that chemoresistant AML cells mostly activate genes related to the inflammatory response. In particular, we identified strong upregulation of the different subunits of the NADPH oxidase NOX2. NADPH oxidases constitute a seven-member family of multi-subunit oxidases, whose sole function is the production of reactive oxygen species (ROS).11 NOX2 is the main NOX isoform expressed in myeloid cells and was found to be over-expressed in AML.12 In line with their overexpression, we found a large increase in NOX-derived ROS production in chemoresistant AML cells, particularly in those resistant to DNR. This activation of NOX2 and the subsequent increase in ROS production participate in the acquisition/maintenance of the resistant phenotype. In addition, we found that NOX2 expression increases at the surface of chemotherapy-resistant AML blasts. Finally, we provide evidence that NOX2 is a marker of both chemoresistance and adverse prognosis in AML patients.
Methods
Additional details of the methods can be found in the Online Supplementary Material.
Patients’ samples
Bone marrow aspirates were collected at diagnosis or after induction chemotherapy (15 to 45 days after the beginning of the chemotherapeutic regimen comprising a combination of DNR and Ara-C). Written informed consent to participation in the study was obtained from patients after approval of the protocol by the institutional “Sud Méditerranée 1” Ethical Committee (ref-2013-A00260-45; HemoDiag collection). Immediately after collection, leukocytes were purified by density-based centrifugation using Histopaque 1077 (Sigma Aldrich) and submitted to flow cytometry analysis. The clinical characteristics of the patients are provided in Online Supplementary Table S3. When indicated, cells were sorted after CD45/SSC gating13 with a CD45-Pacific-Blue antibody (Beckman Coulter) and CD34-PerCP-Vio700 (Miltenyi Biotec) using an Aria IIU cell sorter (Becton Dickinson).
Flow cytometry
Cells were washed in phosphate-buffered saline containing 2% fetal bovine serum and incubated at 4°C for 30 min in the presence of FITC-conjugated gp91phox antibodies (D162-4; MBL), then washed and analyzed using an LSR Fortessa flow cytometer (Becton Dickinson). For the patients’ samples, leukemic cells were identified using CD45/SSC gating13 (see Online Supplementary Figure S6 for an example of gating). The median fluorescence intensities for gp91phox on the red blood cells present in the samples (negative control) were subtracted from the mean fluorescence intensities for gp91phox on the leukemic cells.
Preparation and sequencing of the RNA-sequencing libraries
Total RNA was purified using the GenElute Mammalian Total RNA kit (Sigma-Aldrich), treated with DNase I (4 U, New England Biolabs) and RNasin (2.5 U, Promega) and re-purified. RNA quality was assessed using a BioAnalyzer Nano 6000 chip (Agilent). Three independent experiments were performed for each cell line. Libraries were prepared using a TruSeq® Stranded mRNA Sample Preparation kit (Illumina). Libraries were sequenced using an Illumina Hiseq 2500 sequencer as single-end 125 base reads. Image analysis and base calling were performed using HiSeq Control Software, real-time analysis and bcl2fastq. The RNA-sequencing data are available on the Gene Expression Omnibus repository under accession number GSE193094
Generation of a prognostic score based on the expression of NOX2 genes
Gene expression data from three independent cohorts of adult patients diagnosed with AML were used.14-16 Patients without available survival data, with a diagnosis of myelodysplastic syndrome, or not treated with conventional chemotherapy were excluded. The first cohort (the Verhaak cohort)15 consisted of 504 patients. The second (the Metzeler cohort)16 comprised 242 patients with a normal karyotype (CN-AML). The third cohort (from The Cancer Gene Atlas [TCGA])14 included 148 patients. Gene Expression Omnibus accession numbers are GSE6891, GSE12417 and GSE68833. FLT3/NPM1 statuses for the Metzeler cohort of patients were kindly provided by Metzeler et al.16 Because data normalization was different, expression levels were normalized using the standard normal cumulative distribution (X’ = (X-μ)σ) for each gene. In the first cohort, cutpoints were determined for the six genes of interest (CYBA, CYBB, NCF1, NCF2, NCF4, and RAC2) using MaxStat analysis, therefore defining |3 coefficients. The first cohort was then randomly divided into two sub-cohorts of 252 patients each (training and test sets). As previously described,17-20 we created a risk score based on the levels of expression of all NOX2 constituent genes in the training set. It was defined for each patient as the sum of the |3 coefficients for all these genes weighted by +1 or -1 according to whether the patient’s sample signal was above or below/equal to the probeset MaxStat value. Thus, overexpression of genes associated with a poor prognosis increased the NOX score while overexpression of those associated with a good prognosis decreased it. Patients from the training cohort were ranked according to increased NOX score and, for a given score value Y, the difference in survival of patients with a NOX score <Y or >Y was computed using MaxStat. The NOX score was then individually computed for patients in the test set, using the cutoff value determined in the training cohort. Finally, we transposed our model in the validation cohorts (Metzeler and TCGA cohorts), using both cutpoints defined for each probeset and the cutoff score determined in the training set. Survival analyses were assessed using the Kaplan-Meier method and survival curves were compared using the log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazard model.
Results
Chemoresistant acute myeloid leukemia cells activate inflammation-related transcriptional programs
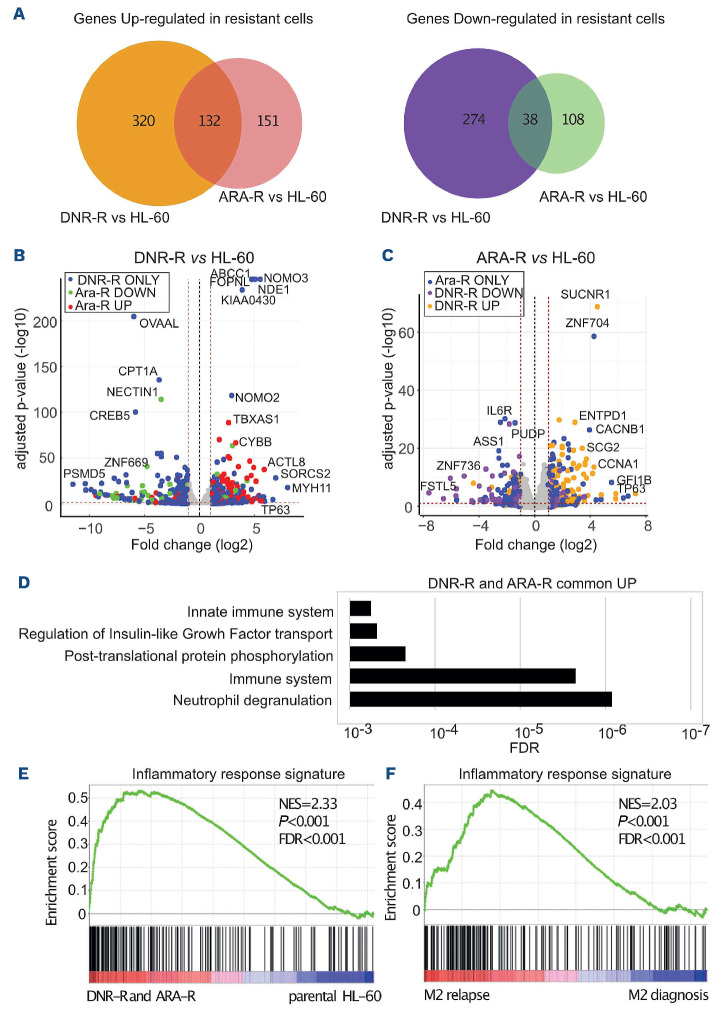
To study AML chemoresistance, we performed RNA-sequencing in the AML model cell line HL-60, using parental cells and DNR-resistant (DNR-R) or Ara-C-resistant (ARAR) cell populations.21 We identified 989 differentially expressed genes between drug-resistant and parental HL-60 cells (Figure 1A and Online Supplementary Table S1). Some were upregulated in resistant cells (452 for DNR-R and 283 for ARA-R) and others were downregulated (312 for DNR-R and 146 for ARA-R). One hundred thirty-two genes were commonly upregulated and 38 downregulated in both types of resistant cells compared to parental cells (Figure 1A). In general, the most deregulated genes showed the same trend of up- or down-regulated expression in both DNR-R and ARA-R cells (Figure 1B, C). However, many genes were preferentially modulated upon acquisition of resistance to one of the two drugs (594/764 for DNR-R and 259/429 for ARA-R) (Figure 1A and Online Supplementary Figure S1A).
Ontological analysis of the genes upregulated in both ARA-R and DNR-R cells showed a strong enrichment for immune signaling and inflammation (Figure 1D). Enrichment of a few pathways involved in the regulation of transcription was found for the genes downregulated in DNR-R cells, and no enrichment for specific pathways was found for the genes downregulated in ARA-R cells (Online Supplementary Table S2). Gene set enrichment analysis (GSEA) further showed enrichment of an inflammatory signature for the genes upregulated in resistant versus parental HL-60 cells (Figure 1E). This signature included various cytokines and chemokines, which are upregulated in both types of resistant cells, with, however, stronger induction in DNR-R cells (Online Supplementary Figure S1B).
We then wondered if the inflammatory signature could also be enriched in patients relapsing after chemotherapy. We thus used publicly available transcriptomic data obtained from nine patients at diagnosis and relapse.22 GSEA revealed that the inflammatory signature was enriched at relapse in the three patients with the French-American-British (FAB) M2 subtype of leukemia (Figure 1F), but not in those from the M1 and M4 subtypes (Online Supplementary Figure S2A, B). Accordingly, a signature containing all genes upregulated in both ARA-R and DNR-R HL-60 cells was found to be enriched at relapse in M2 patients, but not in M1 and M4 patients (Online Supplementary Figure S2C), suggesting that the mechanisms underlying chemoresistance differ depending on the degree of maturation of the leukemic clone. Finally, the genes upregulated specifically in DNR-R cells were also enriched in genes involved in inflammation while those enriched only in ARA-R cells were not enriched for any specific pathway (Online Supplementary Table S2). Thus, our data suggest that AML resistance to chemotherapies, in particular DNR, can be associated with transcriptional reprogramming involving the induction of an inflammatory response.
Figure 1.
Transcriptional reprogramming upon acquisition of chemoresistance by HL-60 acute myeloid leukemia cells. RNA-sequencing was performed on mRNA purified in three independent experiments conducted with HL-60 cells and their derived cytarabine-resistant (ARA-R) and daunorubicin-resistant (DNR-R) populations. (A) Venn diagram for upregulated (>2 fold, false decision rate [FDR] <0.05) and downregulated (>2 fold, FDR <0.05) genes in ARA-R and DNR-R cells compared to parental HL-60 cells. (B) Volcano plot for differentially expressed genes (DEG) between DNR-R and HL-60 cells. Blue dots correspond to the DEG in DNR-R but not in ARA-R cells, red dots to DEG in DNR-R cells that are upregulated in ARA-R cells and green dots to DEG that are downregulated in ARA-R cells. (C) Volcano plot for DEG between ARA-R and HL-60 cells. Blue dots correspond to the DEG in ARA-R but not in DNR-R cells, yellow dots to DEG in ARA-R cells that are upregulated in DNR-R cells and purple dots to DEG in ARA-R cells that are downregulated in DNR-R cells. (D) Gene ontology analysis of genes upregulated in both DNR-R and ARA-R cells compared to parental HL-60 cells. (E) Gene set enrichment analysis (GSEA) was performed using RNA sequencing data from ARA-R, DNR-R and parental HL-60 cells. The enrichment for the “inflammatory response” signature (175 genes) is shown. The normalized enrichment score (NES), nominal P-value and FDR are presented. (F) The inflammatory signature (175 genes) identified in (E) was used in GSEA with RNA-sequencing data from three patients from the FAB M2 subtype obtained from a publicly available cohort.22
Resistance to daunorubicin is associated with strong overexpression and activation of the NAPDH oxidase NOX2
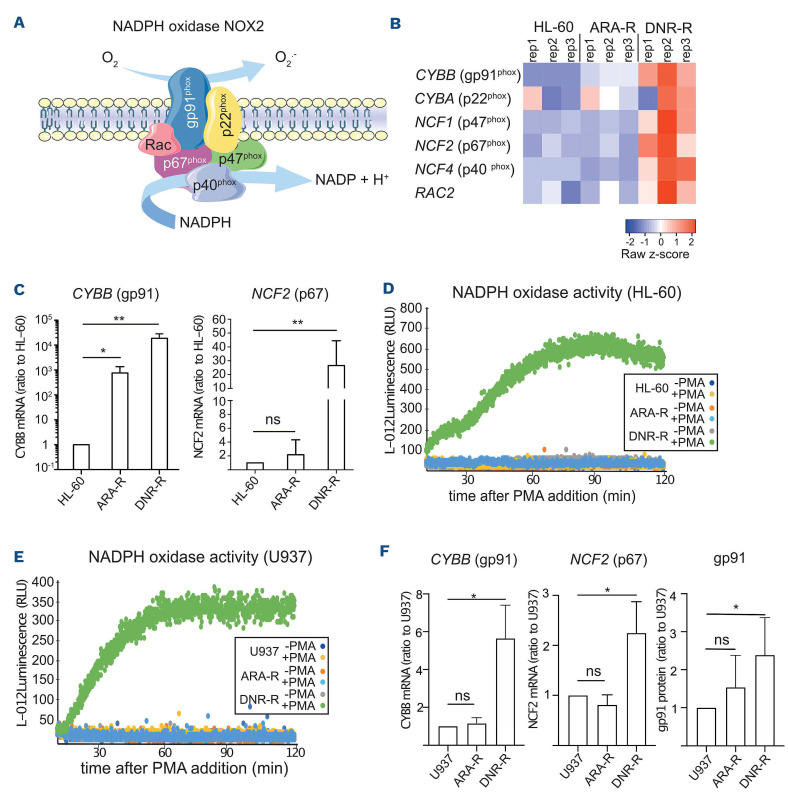
At the top of the list of genes upregulated in the inflammatory response signature of resistant cells, in particular those resistant to DNR, we identified CYBB (Online Supplementary Figure S1B). This gene encodes the gp91phox protein, the catalytic subunit of NOX2, the NADPH oxidase (NOX) family member principally expressed in the hematopoietic system and in AML.23 Two of the NOX2 subunits, gp91phox and p22phox, are associated with the plasma membrane (Figure 2A). Upon activation, the cytosolic subunits (p67phox, p47phox and p40phox) and the small GTPase Rac2 are recruited to the membrane and activate the oxidase.24 This leads to the production of extracellular superoxides, which can freely diffuse across the plasma membrane.
Further pointing to a link between NOX2 and DNR resistance, a NADPH oxidase signature was found to be enriched in DNR-R cells compared to parental cells (Online Supplementary Figure S2D). Indeed, although CYBB was upregulated in both ARA-R and DNR-R cells, the other NOX2 subunits were specifically upregulated in DNR-R as compared to parental HL-60 cells (Figure 2B). Quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) analysis confirmed that CYBB is strongly upregulated in DNR-R cells and, to a lesser extent, in ARA-R cells, as compared to parental HL-60 cells (Figure 2C). The NCF2 gene (encoding p67phox) was also overexpressed in DNR-R but not in ARA-R cells (Figure 2C). To assess whether higher gene expression translates into higher
NADPH oxidase activity, we measured extracellularly produced superoxides (.O2-). Basal NADPH oxidase activity is low under standard growth conditions because the cytosolic subunits are not bound to the membrane. We there-
Figure 2.
NOX2 expression and activity are increased in daunorubicin-resistant HL-60 cells. (A) The NAPDH oxidase NOX2 is composed of two membrane-associated proteins, gp91phox and p22phox. Upon activation, the cytosolic subunits p67phox, p47phox and p40phox translocate to the membrane together with the small GTPase Rac2 to form the full oxidase complex. (B) Heatmap of the RNA-sequencing data in parental, daunorubicin-resistant (DNR-R) and cytarabine-resistant (ARA-R) cells showing the expression of the genes encoding the NOX2 subunits (CYBB, CYBA, NCF1, NCF2, NCF4, and RAC2). (C) The expression of CYBB and NCF2 was analyzed by quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) on mRNA purified from parental, ARA-R and DNR-R cells and normalized to S26 mRNA levels. Data are presented as percentages of the HL-60 condition (n=5, mean ± standard deviation [SD]). (D, E) Parental, ARA-R and DNR-R HL-60 (D) or U937 (E) cells were treated or not with phorbol myristate acetate (PMA) and the production of reactive oxygen species was measured by following L-012 luminescence over 2 h (n=4 for HL-60, n=3 for U937, a representative experiment is shown). (F) The expression of CYBB and NCF2 was analyzed by RT-qPCR on mRNA purified from parental, ARA-R and DNR-R cells and normalized to S26 mRNA levels. Data are presented as percentages of the U937 condition (n=4, mean ± SD). gp91phox surface expression was measured by flow cytometry in parental, DNR-R and ARA-R U937 cells. The median fluorescence intensity (MFI) obtained with isotypic controls was subtracted from the gp91 MFI and normalized to the parental cells (n=6, mean ± SD). RLU: relative light units fore treated cells with phorbol myristate acetate (PMA), a well-known activator of NADPH oxidases. No superoxide production was detected in HL-60 cells, which are known to have low or no NADPH oxidase activity when not differentiated.25,26 Strong ROS production was measured in PMA-treated DNR-R but not in ARA-R cells (Figure 2D). To confirm these results, we used another cell line, U937, rendered resistant to Ara-C or DNR.21 DNR-R but not ARAR U937 cells had increased NOX activity (Figure 2E), over-expressed CYBB and NCF2 and expressed higher levels of gp91phox at their surface compared to parental cells (Figure 2F).
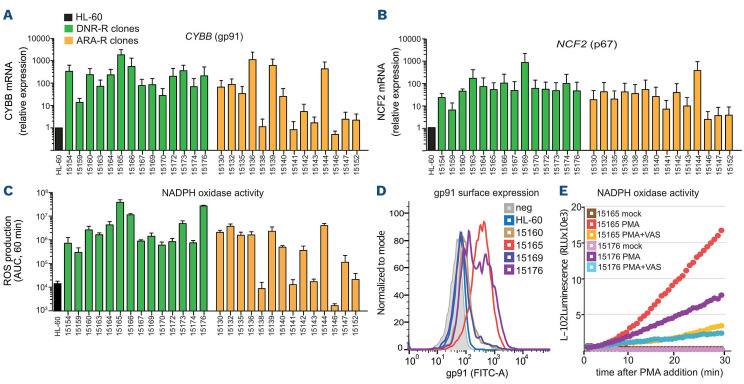
We then cloned the chemoresistant HL-60 cell populations. All DNR-R clones showed a strong increase in both CYBB (Figure 3A) and NCF2 (Figure 3B) mRNA levels (above 10 to more than 1000-fold compared to HL-60 cells). Accordingly, PMA-induced extracellular ROS production was increased in all DNR-R clones compared to parental HL-60 (Figure 3C). Although the ARA-R population showed no significant increase in ROS production (Figure 2E), a fraction of the ARA-R clones showed an increase in CYBB and NCF2 expression and high NOX activity in comparison to parental HL-60 cells (Figure 3A-C). The mean increase in NOX activity was however around 7-fold lower in ARA-R-compared to DNR-R clones, which might explain why it was not detected within the ARA-R population. In addition, ARA-R clones with high NOX activity might have a survival advantage during the cloning process. To exclude the possibility that increased NOX2 expression and activity were due to the cloning procedure, we cloned the parental HL-60 cells. None of the HL-60 clones showed an increase in NADPH oxidase-derived ROS production (Online Supplementary Figure S3). To determine if high NOX activity was sufficient to confer DNR resistance, we measured the IC50 for DNR of two ARA-R clones, one with low NOX activity (15138) and one with high NOX activity (15144). None of the ARA-R clone was resistant to DNR, indicating that high NOX expression in ARA-R cells is not sufficient to confer cross-resistance to DNR (Online Supplementary Figure S4A). In addition, higher NOX activity did not confer resistance to venetoclax, a BCL2 inhibitor now used in the treatment of AML patients (Online Supplementary Figure S4B). Finally, for both DNR-R and ARAR clones, increased NOX activity was not correlated with a significant increase in the differentiation of the chemo-resistant clones (Online Supplementary Figure S4C).
We then measured gp91phox membrane expression using flow cytometry of two high ROS-producing clones (15165 and 15176) and two low ROS-producing clones (15160 and 15169) from DNR-R cells. Overexpression of gp91phox at the cell surface was found to be high only in the clones producing high levels of ROS (15165 and 15176) (Figure 3D). VAS2870, a pan-NADPH oxidase inhibitor with preferential activity against NOX2,27 strongly reduced PMA-induced ROS production in DNR-R clones (Figure 3E), further demonstrating that increased ROS production in chemoresistant AML cells is due to NADPH oxidase activation.
Figure 3.
Overexpression of NADPH oxidase components is responsible for increased production of reactive oxygen species in chemoresistant acute myeloid leukemia cells. (A-C) Daunorubicin-resistant (DNR-R) and cytarabine-resistant (ARA-R) HL-60 cell populations were cloned and 14 clones for each were used to measure mRNA expression for CYBB (A) and NCF2 (B) using quantitative reverse transcriptase polymerase chain reaction analysis. Data were normalized to S26 mRNA levels and are presented as ratios to parental HL-60 cells (n=3, mean ± standard deviation [SD]). (C) Reactive oxygen species (ROS) production was measured after addition of phorbol myristate acetate (PMA) using L-012 luminescence. The area under the curve (AUC) was calculated after plotting L-012 luminescence over 1 h (n=3, mean ± SD). (D) gp91phox surface expression was measured by flow cytometry in parental HL-60 cells as well as in two DNR-R clones showing high NADPH oxidase activity (15165 and 15176) and two clones with low NADPH oxidase activity (15160 and 15169). (E) DNR-R clones 15165 and 15176 were stimulated or not with PMA with or without VAS2870 and ROS production was measured by luminometry (n=3, a representative experiment is shown).
Collectively, our data suggest that acquisition of chemoresistance, in particular to DNR, is associated with increased expression of the subunits of the NOX2 NADPH oxidase, its cell surface expression and activity in AML cells.
gp91phox overexpression does not affect the proliferation of chemoresistant cells but participates in their resistance to daunorubicin
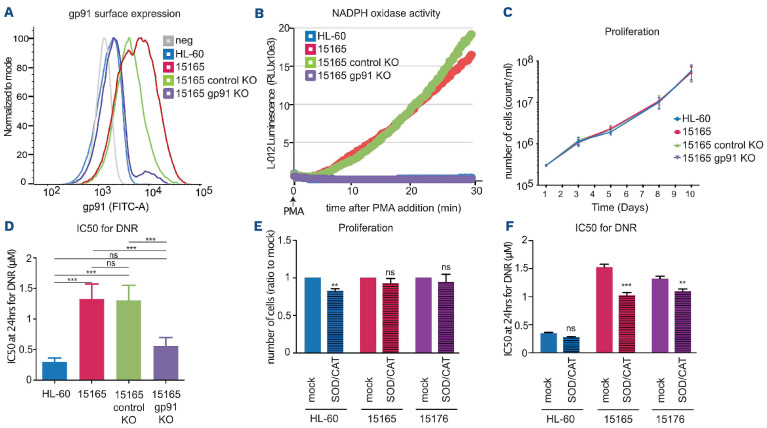
To address the role of NOX2 activation in chemoresistant cells, we measured the proliferation of four DNR-R and two ARA-R clones (one high and one low NOX2-expressing clone). No differences were observed in their proliferation compared to that of the parental HL-60 cells (Online Supplementary Figure S4D), suggesting that high NOX2 is not conferring a proliferative advantage to the chemoresistant cells. Then, using CRISPR/Cas9 technology, we knocked out the CYBB gene in the DNR-R clone showing the highest level of gp91phox expression (clone 15165). This led to a strong decrease in gp91phox cell surface expression (Figure 4A) and abolished NADPH oxidase activity (Figure 4B). No difference was observed between the proliferation rates of parental HL-60 and DNR-R cells expressing or not gp91phox (Figure 4C). DNR-R cells CRISPRed for gp91phox regained sensitivity to DNR compared to control CRISPRed cells (Figure 4D and Online Supplementary Figure S5C).
To further demonstrate the involvement of NOX2-derived ROS production in the maintenance of DNR resistance, we cultured the parental HL-60 cells and two DNR-R clones (15165 and 15176) for 3 weeks in medium supplemented with superoxide dismutase (SOD), which transforms superoxide anions to H2O2 and catalase, which degrades H2O2 to H2O.28,29 This efficiently removed NOX2-produced ROS from the medium (Online Supplementary Figure S5A, B). Although this did not affect the proliferation of the DNR-R clones (Figure 4E), it did decrease their resistance to DNR (Figure 4F). Similarly, long-term treatment of the DNR-R clones with the NOX inhibitor VAS2870 re-sensitized the clones to DNR (Online Supplementary Figure S5D). This suggests that overexpression of gp91phox and the ensuing increase in ROS production contribute to the acquisition and/or the maintenance of resistance to DNR in our model.
Figure 4.
NOX2 overexpression participates in the acquisition and/or maintenance of resistance to daunorubicin. (A) gp91phox surface expression was measured by flow cytometry in parental HL-60 cells, in daunorubicin-resistant (DNR-R) clones (15165) as well as 15165 cells subjected to CRISPR/Cas9 KO for gp91Phox and 15165 cells subjected to mock CRISPR/Cas9 mutagenesis. (B) NADPH oxidase activity was detected by measurement of reactive oxygen species (ROS) in the cells presented in (A) after addition of phorbol myristate acetate (PMA) using L 012 luminescence.(n=3, a representative experiment is shown). (C) Cell proliferation was measured using MTS (n=3, mean ± standard deviation [SD]). (D) IC50 for daunorubicin (DNR) was measured after 24 h of treatment using MTS (n=4, mean ± SD). (E, F) HL60 and two DNR-R clones (15165 and 15176) were cultured in the presence of SOD (30 U/mL) and catalase (25 µg/mL) for 3 weeks. (E) The number of cells was then measured using MTS and represented as a ratio to the mock-treated cells (n=5, mean ± SD). (F) IC50 for DNR was measured after 24 h of DNR treatment using MTS (n=4, mean ± SD). All P-values were calculated with one-way analysis of variance and the Tukey multiple comparison test.
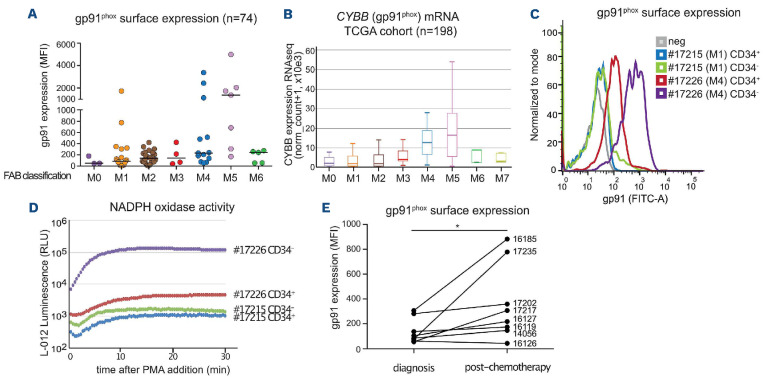
gp91phox expression and NOX2 activity are higher in FAB M4/M5 acute myeloid leukemia subtypes and increase on chemotherapy-resistant patients’ cells
To further characterize the importance of NOX2 in AML patients, we used flow cytometry to assay the expression of gp91phox on the surface of primary AML cells from 74 patients at diagnosis. Differences in gp91phox levels were neither associated with a specific group from the European LeukemiaNet 2017 classification2 (favorable, intermediate or adverse), nor with NPM1 or FLT3 mutational status (Online Supplementary Figure S7 and Online Supplementary Table S3). However, gp91phox levels were significantly higher in the M5 AML subtype than in normal CD34+ cells and leukemic blasts from the M0, M1 and M2 subtypes of the FAB classification (Figure 5A and Online Supplementary Table S3). Using transcriptomic data from the TCGA cohort,14 we confirmed that CYBB mRNA expression is higher in the M4/M5 subtypes (Figure 5B). To assess whether increased gp91phox cell surface expression is linked to higher NAPDH oxidase activity, we measured PMA-stimulated ROS production using sorted AML cells (bulk CD34- leukemic cells and leukemic stem cells [LSC]-containing CD34+ cells) from one patient with low gp91phox (M1 subtype, patient #17215) and another with high gp91phox cell surface expression (M4 subtype, patient #17226) (Figure 5C). Weak PMA-induced ROS production was detected in the low gp91phox-expressing patient’s cells, which contrasted with high production in the high gp91phox-expressing patient’s cells (Figure 5D). For the M4 patient’s cells, gp91phox level (Figure 5C) and ROS production (Figure 5D) were much higher in the bulk of leukemic cells (CD34-) than in the LSC-containing compartment (CD34+).
Figure 5.
NOX2 expression and activity are higher in patients with FAB M4/M5 acute myeloid leukemia. (A) gp91phox expression was measured by flow cytometry on the leukemic cells (CD45/SSC gating) present in bone marrow aspirates taken at diagnosis from 74 patients with acute myeloid leukemia (AML). Patients are classified according to the French-American-British classification. The median is represented as a horizontal line for each group. P=0.0057 in one-way analysis of variance (Kruskal-Wallis test) (B) mRNA expression for the CYBB gene was measured by RNA-sequencing in The Cancer Genome Atlas cohort comprising 198 AML patients. (C) gp91phox surface expression was measured by flow cytometry on AML cells from two patients sorted using CD45/SSC gating and separated according to their level of expression of the CD34 marker. (D) Sorted AML cells (CD34+ and CD34-) for each patient were used to measure production of reactive oxygen species after stimulation with phorbol myristate acetate (PMA) (10 nM). (E) gp91phox expression (MFI) was measured by flow cytometry on leukemic cells (CD45/SSC gating) present in bone marrow aspirates taken at diagnosis and after the induction chemotherapy (between 15 and 45 days after the beginning of the treatment) from eight AML patients (Online Supplementary Table S3, ID numbers of the patients indicated on the figure). Four of the patients reached complete remission (CR) after induction chemotherapy (16119, 16127, 16185, and 17235), two reached CR after allografting (14056 and 16126) and two never reached CR (17202 and 17217). All patients but one (17235, who was allografted) relapsed. MFI: mean fluorescence intensity, FAB: French-American-British; TCGA: The Cancer Genome Atlas.
Table 1.
List of the six probe sets included in the NOX score.
Finally, for eight patients of our cohort, we compared the mean gp91phox expression on leukemic blasts taken at diagnosis with that on blasts that resisted induction therapy in the same patients. An increase in gp91phox labeling after chemotherapy was found in seven of the eight patients (Figure 5E and Online Supplementary Table S3). This supported the idea that, similarly to the chemoresistant cell lines, chemotherapy-resistant AML cells generally express higher levels of gp91phox at their surface.
NOX2 overexpression is a marker of adverse prognosis in acute myeloid leukemia
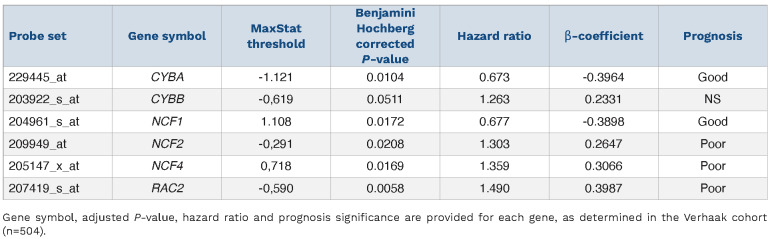
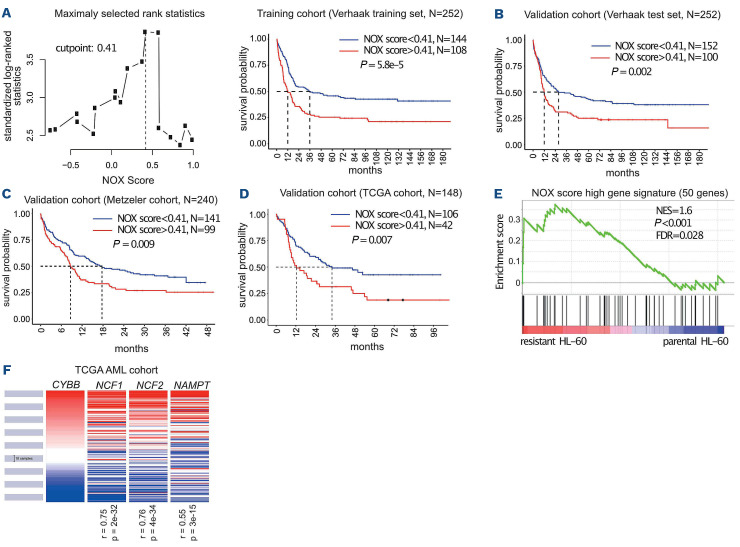
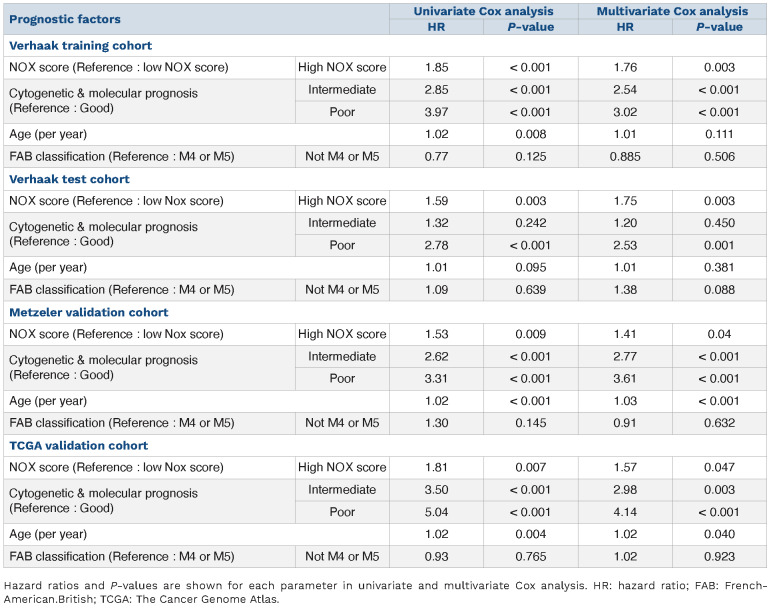
To study the link between NOX2 and AML response to treatments, we determined the individual prognostic value of the genes coding for the six NOX2 subunits (CYBB, CYBA, NCF1, NCF2, NCF4, and RAC2) using publicly available data from the Verhaak cohort.15 Increased expression was associated with a poor prognosis for four of them (CYBB, NCF2, NCF4, and RAC2) and with a good prognosis for the other two (CYBA and NCF1) (Table 1). To take into account the expression and prognostic value of all NOX2 subunits instead of individual ones, we developed a NOX2-subunit gene expression-based score. The contribution of individual NOX2 genes and the cutpoint were determined using a training cohort comprising 252 patients (Verhaak training set).15 Patients with a high “NOX score” had a worse prognosis with a hazard ratio of 1.85 in univariate Cox analysis (Figure 6A and Table 2). The prognostic significance of the NOX score was then confirmed in a test set (n=252) from the Verhaak cohort (Figure 6B and Table 2) as well as in two independent validation cohort: the Metzeler16 (Figure 6C, Table 2 and Online Supplementary Table S4) and TCGA cohorts14 (Figure 6D, Table 2 and Online Supplementary Table S6). The poor prognostic-based NOX score remained statistically significant in a multivariate Cox analysis including the cytogenetic-based classification, NPM1/FLT3 mutational status (for the normal karyotype cohort), and age (Table 2). The NOX score strongly correlated with the expression of subunits associated with poor prognosis (CYBB, NCF2, NCF4 and RAC2) while the correlation with CYBA and NCF1 was less significant (Online Supplementary Figure S8). Finally, in line with the higher expression of CYBB in M4/M5 FAB subtypes, the NOX score was generally higher in the M4/M5 compared to the other FAB subtypes (Online Supplementary Figure S9). Nevertheless, the prognostic value of the NOX score was independent of the FAB subtype, which did not have prognostic value on its own in any of the cohorts (Table 2).
To determine if the worse prognosis of high NOX score patients could be associated with higher chemoresistance, we selected the 10% of patients with the highest NOX score and compared their transcriptome to those of the 50% of patients with the lowest NOX scores. We then used the 50 most upregulated genes in the high NOX score patients (Online Supplementary Table S4) as a gene-set in a GSEA analysis of the upreguleted RNA-sequencing data from parental and chemoresistant HL-60 cells. The high NOX score gene set was significantly enriched in the chemoresistant compared to parental HL-60 cells (Figure 6E). Interestingly, the most upregulated gene in the high NOX score patients is NAMPT, encoding a critical enzyme in the recycling of NAD, an essential co-factor for NADPH oxidase activity. Accordingly, we found that NAMPT expression is highly correlated to that of the CYBB, NCF1 and NCF2 genes in AML patients (Figure 6F).
Together, these data suggest that the overexpression of the NOX2 NADPH oxidase is a marker of adverse prognosis in AML associated with enhanced chemoresistance of the leukemic cells.
Figure 6.
Identification of a NOX score with prognostic value in acute myeloid leukemia. (A) Patients (n=252) of the Verhaak training cohort were ranked according to increasing NOX score. The cutpoint of 0.41 was selected using the MaxStat R function to obtain a maximum difference in overall survival between the two groups. Kaplan-Meier survival curves were established using the cutpoint value of 0.41 in the training cohort. (B-D) The NOX-score was validated in three independent cohorts using the 0.41 cutpoint. (E) Gene set enrichment analysis was performed using RNA sequencing data from chemoresistant cells compared to parental HL-60 cells. The gene list comprises the 50 most upregulated genes in the 10% of patients with the highest NOX score, compared to the 50% patients with the lowest NOX score in the Verhaak cohort. Normalized enrichment score and nominal P-value are presented. (F) Expression of CYBB, NCF1, NCF2 and NAMPT in 200 patients’ samples from The Cancer Genome Atlas cohort. R represents the Pearson rho correlation coefficient between the expression of NCF1, NCF2 or NAMPT and CYBB. TCGA: The Cancer Genome Atlas; NES: normalized enrichment score; FDR: false discovery rate.
Discussion
Our work unveiled that NOX2 expression increases in chemoresistant AML and could participate in the acquisition and/or maintenance of chemoresistance. Moreover, NOX2 was found to be a marker of adverse prognosis in AML patients. NOX2 is the main NADPH oxidase complex expressed in the hematopoietic system. It is responsible for the respiratory burst, a release of superoxides (.O2-) in the phagosome, which is used to eliminate engulfed pathogens.30 In addition to their essential role in host protection, ROS produced by NADPH oxidases also function as second messengers in the regulation of numerous signaling pathways.31 Dysregulation of the expression and activity of NADPH oxidases has been linked to various cancers, including AML.32 NOX2 expression was found to be generally higher in leukemic blasts from AML patients than in normal CD34+ hematopoietic progenitors and involved in leukemic cell proliferation.12 NOX2 was shown to be mostly expressed in M4 and M5 subtypes of leukemia according to the FAB classification and rarely on leukemic blasts from M1 or M2 subtypes.26,33 By analyzing gp91phox, the catalytic subunit of NOX2, at the surface of blasts from 74 AML patients, we confirmed that NOX2 expression is higher in the M4/M5 subtypes than in the M0, M1 and M2 subtypes. In addition, within a given patient’s sample, gp91phox expression and NOX2 activity were higher in the bulk of leukemic cells (CD34-) than in the LSC-containing CD34+ compartment, which nevertheless showed basal activity. This is congruent with the fact that NOX2 is required for LSC self-renewal and, in turn, leukemogenesis.23 Moreover, our results indicate that the increase in NOX2 activity in chemoresistant cell lines is linked with the transcriptional activation of most, if not all, of its six subunits. Interestingly, their expression, in particular those of CYBB, NCF1 and NCF2, is highly correlated. Increased NOX2 expression could confer a selective advantage to the chemoresistant clones. Supporting this hypothesis, we could demonstrate, in patients, that therapy-resistant cells remaining after induction chemotherapy express higher levels of gp91phox at their surface. Moreover, preventing NOX2 activation in DNR-R cells through the deletion of CYBB, ROS scavenging or NADPH oxidase inhibition re-sensitized them to DNR. This indicates that NOX2 overexpression could be involved in the acquisition and/or maintenance of resistance. The underlying molecular mechanisms remain to be characterized. They might involve the regulation of specific signaling pathways through the reversible oxidation of cysteines present in the catalytic site of enzymes such as kinases and phosphatases34 or, as we showed previously, of SUMOylation enzymes.35,36 Their modulation would participate in the transcriptional reprogramming observed in chemoresistant AML cells and in the acquisition and/or maintenance of chemoresistance. NOX-derived ROS were also shown to promote AML cell proliferation,12,37 possibly by activating genes involved in glycolysis38 or through metabolic reprograming.39 In addition, oncogenes such as mutated RAS40,41 or FLT3-ITD42 activate NADPH-oxidase derived ROS production, which increases hematopoietic cell proliferation and leukemic transformation. Finally, NOX2-derived ROS stimulate the transfer of mitochondria from bone marrow stromal cells to the AML blasts, increasing their metabolic activity and proliferation.43 In our study, high NOX-expressing chemoresistant clones did not proliferate faster than chemosensitive parental cells and deletion of CYBB or ROS scavenging did not affect their doubling time. Thus, although the pro-proliferative effect of NOX-derived ROS might be involved in leukemogenesis, it does not seem to confer a proliferative advantage to chemoresistant cells. Finally, NADPH-oxidase-derived ROS are also involved in the interaction between leukemic cells and the immune system. They can induce apoptosis of natural killer cells and T cells33 and prevent the maturation of dendritic cells.44 Whether or not such an inactivation of antitumor immune cells could also confer a selective advantage to the chemoresistant AML cells that overexpress NOX2 remains an open question.
Table 2.
Cox analysis of overall survival in the Verhaak training set (n=252), the Verhaak test set (n=252), the Metzeler validation set (n=240) and The Cancer Genome Atlas validation set (n=148) according to NOX score, cytogenetic prognosis, age and French-American-British classification.
When analyzing the prognostic value of the individual NOX2 subunits, we found that four were associated with a poor prognosis (gp91phox, p67phox, p40phox and Rac2) and, more surprisingly, two with a good prognosis (p22phox and p47phox). The association of CYBA (p22phox) and NCF1 (p47phox) with a good prognosis might be due to NOX2-independent functions of these proteins. In particular, p22phox is also involved in the assembly of NOX1, NOX3 and NOX411 and p47phox can activate both NOX145 and NOX3.46 We thus developed the NOX score to take into account the expression and the prognostic value of all subunits. Patients with a high NOX score showed overexpression of CYBB, NCF2, NCF4 or RAC2. A high NOX score was associated with a poor prognosis in three independent AML cohorts. The NOX score was found to be independent of other prognostic factors such as age, cytogenetic risk or NPM1/FLT3 mutational status in normal karyotype AML. Consistent with a higher expression of CYBB in FAB M4/M5 subtypes, we found that the NOX score was higher in these subtypes. However, the NOX score was found to be independent of the FAB classification in multivariate analysis, indicating that its prognostic value is not linked to the FAB subtype. Although patients with a high NOX score showed enrichment in the chemoresistance signature, the prognostic value of the NOX score might not only be linked to the role of NOX2 in chemoresistance. The NOX score should now be validated in a prospective cohort to confirm its prognostic value. NAPDH-oxidase inhibitors have been proposed as anticancer drugs. Histamine, which inhibits NADPH-oxidase-derived ROS production, is already approved, in combination with interleukin-2, to prevent relapse in AML.47 This treatment was found to be more effective in M4/M5 patients.48 The NOX score might constitute another biomarker to select patients eligible for such therapy. Many other molecules targeting NADPH oxidases have been developed in the past few years.49
However, most of them suffer from a lack of specificity.50,51 Interestingly, we found that NAMPT, the gene coding for a rate-limiting enzyme in the recycling of NAD, is the most upregulated gene in patients with a high NOX score and its expression is highly correlated to NOX2 subunit levels in AML patients. In addition, NAMPT was more expressed in chemoresistant HL-60 cells than in parental cells, in line with recent findings showing its overexpression in chemoresistant versus chemosensitive LSC.52 Inhibitors of NAMPT have shown antileukemic activity in preclinical models52–54 and some of them are being tested in clinical trials (NCT02702492, NCT04281420). Even though the NADP/NADPH cycle relies on the activity of various enzymes, targeting NAMPT in patients with a high NOX score might represent another option to inhibit NOX2 activity in AML cells, by decreasing NADPH supply. In conclusion, our work points to a link between NOX2 and AML chemoresistance. Targeting NOX2 might constitute a new therapeutic strategy to overcome AML chemoresistance and improve the prognosis of patients.
Supplementary Material
Acknowledgments
We are grateful to the IGMM "Ubiquitin Family in Hematological Malignancies" group members for fruitful discussions and critical reading of the manuscript. We thank the Montpellier Ressources Imagerie (MRI) platforms for technical assistance.
Funding Statement
Funding: Funding was provided by the CNRS, the Fondation ARC pour la Recherche sur le Cancer, Ligue Nationale contre le Cancer (Programme Equipe Labellisée), the INCA (ROSAML), the ANR (“Investissements d’avenir” program; ANR-16-IDEX-0006), the Fondation pour la Recherche Médicale (FRM) to LG and the Ligue Nationale contre le Cancer to MB. The HEMODIAG_2020 collection of clinical data and patients’ samples was funded by the Montpellier University Hospital, the Montpellier SIRIC and the Languedoc Roussillon Region. MGX acknowledges financial support from the France Génomique National infrastructure, funded as part of “Investissements d’Avenir” program managed by the Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136-1152. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin JJG, Briz O, Rodríguez-Macias G, Díez-Martín JL, Macias RIR. Role of drug transport and metabolism in the chemoresistance of acute myeloid leukemia. Blood Rev. 2016;30(1):55-64. [DOI] [PubMed] [Google Scholar]
- 4.Galmarini CM, Thomas X, Graham K, et al. Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br J Haematol. 2003;122(1):53-60. [DOI] [PubMed] [Google Scholar]
- 5.Ax W, Soldan M, Koch L, Maser E. Development of daunorubicin resistance in tumour cells by induction of carbonyl reduction. Biochem Pharmacol. 2000;59(3):293-300. [DOI] [PubMed] [Google Scholar]
- 6.Sillar JR, Enjeti AK. Targeting apoptotic pathways in acute myeloid leukaemia. Cancers (Basel). 2019;11(11):1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearsall EA, Lincz LF, Skelding KA. The role of DNA repair pathways in AML chemosensitivity. Curr Drug Targets. 2018;19(10):1205-1219. [DOI] [PubMed] [Google Scholar]
- 8.Hosseini M, Rezvani H, Aroua N, et al. Targeting myeloperoxidase disrupts mitochondrial redox balance and overcomes cytarabine resistance in human acute myeloid leukemia. Cancer Res. 2019;79(20):5191-5203. [DOI] [PubMed] [Google Scholar]
- 9.Farge T, Saland E, de Toni F, et al. Chemotherapy resistant human acute myeloid leukemia cells are not enriched for leukemic Stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiorentini A, Capelli D, Saraceni F, Menotti D, Poloni A, Olivieri A. The time has come for targeted therapies for AML: lights and shadows. Oncol Ther. 2020;8(1):13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moghadam ZM, Henneke P, Kolter J. From flies to men: ROS and the NADPH oxidase in phagocytes. Front Cell Dev Biol. 2021;9:628991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hole PS, Zabkiewicz J, Munje C, et al. Overproduction of NOX-derived ROS in AML promotes proliferation and is associated with defective oxidative stress signaling. Blood. 2013;122(19):3322-3330. [DOI] [PubMed] [Google Scholar]
- 13.Brahimi M, Saidi D, Touhami H, Bekadja MA. The use of CD45/SSC dot plots in the classification of acute leukemias. J Hematol Thromb Dis. 2014;2:e107. [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhaak RGW, Wouters BJ, Erpelinck CAJ, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112(10):4193-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Combès E, Andrade AF, Tosi D, et al. Inhibition of ataxia-telangiectasia mutated and RAD3-related (ATR) overcomes oxaliplatin resistance and promotes antitumor immunity in colorectal cancer. Cancer Res. 2019;79(11):2933-2946. [DOI] [PubMed] [Google Scholar]
- 18.Gabellier L, Bret C, Bossis G, Cartron G, Moreaux J. DNA repair expression profiling to identify high-risk cytogenetically normal acute myeloid leukemia and define new therapeutic targets. Cancers. 2020;12(10):2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herviou L, Kassambara A, Boireau S, et al. PRC2 targeting is a therapeutic strategy for EZ score defined high-risk multiple myeloma patients and overcome resistance to IMiDs. Clin Epigenetics. 2018;10(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassambara A, Hose D, Moreaux J, et al. Genes with a spike expression are clustered in chromosome (sub)bands and spike (sub)bands have a powerful prognostic value in patients with multiple myeloma. Haematologica. 2012;97(4):622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gâtel P, Brockly F, Reynes C, et al. Ubiquitin and SUMO conjugation as biomarkers of acute myeloid leukemias response to chemotherapies. Life Sci Alliance. 2020;3(6):e201900577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopher MJ, Petti AA, Rettig MP, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med. 2018;379(24):2330-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adane B, Ye H, Khan N, et al. The hematopoietic oxidase NOX2 regulates self-renewal of leukemic stem cells. Cell Rep. 2019;27(1):238-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schröder K. NADPH oxidases: current aspects and tools. Redox Biol. 2020;34:101512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy R, Rotrosen D, Nagauker O, Leto TL, Malech HL. Induction of the respiratory burst in HL-60 cells. Correlation of function and protein expression. J Immunol. 1990;145(8):2595-2601. [PubMed] [Google Scholar]
- 26.Dakik H, El Dor M, Leclerc J, et al. Characterization of NADPH oxidase expression and activity in acute myeloid leukemia cell lines: a correlation with the differentiation status. Antioxidants. 2021;10(3):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dao VT, Elbatreek MH, Altenhöfer S, et al. Isoform-selective NADPH oxidase inhibitor panel for pharmacological target validation. Free Radic Biol Med. 2020;148:60-69. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto T, Sakaguchi N, Hachiya M, Nakayama F, Yamakawa M, Akashi M. Role of catalase in monocytic differentiation of U937 cells by TPA: hydrogen peroxide as a second messenger. Leukemia. 2009;23(4):761-769. [DOI] [PubMed] [Google Scholar]
- 29.Chamulitrat W, Schmidt R, Tomakidi P, et al. Association of gp91phox homolog Nox1 with anchorage-independent growth and MAP kinase-activation of transformed human keratinocytes. Oncogene. 2003;22(38):6045-6053. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DC. The phagocyte respiratory burst: historical perspectives and recent advances. Immunol Lett. 2017;192:88-96. [DOI] [PubMed] [Google Scholar]
- 31.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15(6):411-421. [DOI] [PubMed] [Google Scholar]
- 32.Sillar JR, Germon ZP, De Iuliis GN, Dun MD. The role of reactive oxygen species in acute myeloid leukaemia. Int J Mol Sci. 2019;20(23):6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aurelius J, Thorén FB, Akhiani AA, et al. Monocytic AML cells inactivate antileukemic lymphocytes: role of NADPH oxidase/gp91phox expression and the PARP-1/PAR pathway of apoptosis. Blood. 2012;119(24):5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50-64. [DOI] [PubMed] [Google Scholar]
- 35.Bossis G, Sarry J-E, Kifagi C, et al. The ROS/SUMO axis contributes to the response of acute myeloid leukemia cells to chemotherapeutic drugs. Cell Rep. 2014;7(6):1815-1823. [DOI] [PubMed] [Google Scholar]
- 36.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21(3):349-357. [DOI] [PubMed] [Google Scholar]
- 37.Reddy MM, Fernandes MS, Salgia R, Levine RL, Griffin JD, Sattler M. NADPH oxidases regulate cell growth and migration in myeloid cells transformed by oncogenic tyrosine kinases. Leukemia. 2011;25(2):281-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson AJ, Hopkins GL, Rastogi N, et al. Reactive oxygen species drive proliferation in acute myeloid leukemia via the glycolytic regulator PFKFB3. Cancer Res. 2020;80(5):937-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson AJ, Davies S, Darley RL, Tonks A. Reactive oxygen species rewires metabolic activity in acute myeloid leukemia. Front Oncol. 2021;11:632623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hole PS, Pearn L, Tonks AJ, et al. Ras-induced reactive oxygen species promote growth factor–independent proliferation in human CD34+ hematopoietic progenitor cells. Blood. 2010;115(6):1238-1246. [DOI] [PubMed] [Google Scholar]
- 41.Aydin E, Hallner A, Grauers Wiktorin H, Staffas A, Hellstrand K, Martner A. NOX2 inhibition reduces oxidative stress and prolongs survival in murine KRAS - induced myeloproliferative disease. Oncogene. 2019;38(9):1534-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayavelu AK, Müller JP, Bauer R, et al. NOX4-driven ROS formation mediates PTP inactivation and cell transformation in FLT3ITD-positive AML cells. Leukemia. 2016;30(2):473-483. [DOI] [PubMed] [Google Scholar]
- 43.Marlein CR, Zaitseva L, Piddock RE, et al. NADPH oxidase-2 derived superoxide drives mitochondrial transfer from bone marrow stromal cells to leukemic blasts. Blood. 2017;130(14):1649-1660. [DOI] [PubMed] [Google Scholar]
- 44.Martner A, Wiktorin HG, Lenox B, et al. Histamine promotes the development of monocyte-derived dendritic cells and reduces tumor growth by targeting the myeloid NADPH oxidase. J Immunol. 2015;194(10):5014-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Youn JY, Gao L, Cai H. The p47phox- and NADPH oxidase organiser 1 (NOXO1)-dependent activation of NADPH oxidase 1 (NOX1) mediates endothelial nitric oxide synthase (eNOS) uncoupling and endothelial dysfunction in a streptozotocin-induced murine model of diabetes. Diabetologia. 2012;55(7):2069-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng G, Ritsick D, Lambeth JD. Nox3 regulation by NOXO1, p47phox, and p67phox.. J Biol Chem. 2004;279(33):34250-34255. [DOI] [PubMed] [Google Scholar]
- 47.Brune M, Castaigne S, Catalano J, et al. Improved leukemia-free survival after postconsolidation immunotherapy with histamine dihydrochloride and interleukin-2 in acute myeloid leukemia: results of a randomized phase 3 trial. Blood. 2006;108(1):88-96. [DOI] [PubMed] [Google Scholar]
- 48.Aurelius J, Martner A, Brune M, et al. Remission maintenance in acute myeloid leukemia: impact of functional histamine H2 receptors expressed by leukemic cells. Haematologica. 2012;97(12):1904-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altenhöfer S, Radermacher KA, Kleikers PWM, Wingler K, Schmidt HHHW. Evolution of NADPH oxidase inhibitors: selectivity and mechanisms for target engagement. Antioxid Redox Signal. 2015;23(5):406-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Dor M, Dakik H, Polomski M, et al. VAS3947 induces UPR-mediated apoptosis through cysteine thiol alkylation in AML cell lines. Int J Mol Sci. 2020;21(15):5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Q-A, Hess DT, Wang B, Miyagi M, Stamler JS. Off-target thiol alkylation by the NADPH oxidase inhibitor 3-benzyl-7-(2-benzoxazolyl)thio-1,2,3-triazolo[4,5-d]pyrimidine (VAS2870). Free Radic Biol Med. 2012;52(9):1897-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones CL, Stevens BM, Pollyea DA, et al. Nicotinamide metabolism mediates resistance to venetoclax in relapsed acute myeloid leukemia stem cells. Cell Stem Cell. 2020;27(5):748-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerner RR, Macheiner S, Reider S, et al. Targeting NAD immunometabolism limits severe graft-versus-host disease and has potent antileukemic activity. Leukemia. 2020;34(7):1885-1897. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell SR, Larkin K, Grieselhuber NR, et al. Selective targeting of NAMPT by KPT-9274 in acute myeloid leukemia. Blood Adv. 2019;3(3):242-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.