Abstract
The granular dorsolateral prefrontal cortex (dlPFC) is an evolutionary specialization of primates that is centrally involved in cognition. Here, we assessed over 600,000 single-nucleus transcriptomes from adult human, chimpanzee, macaque, and marmoset dlPFC. While most transcriptomically-defined cell subtypes are conserved, we detected several only in some species and substantial species-specific molecular differences across homologous neuronal, glial and non-neural subtypes. The latter are exemplified by human-specific switching between expression of the neuropeptide somatostatin (SST) and tyrosine hydroxylase (TH), the rate-limiting enzyme in dopamine production, in certain interneurons, and also by expression of the neuropsychiatric risk gene FOXP2, which is human-specific in microglia and primate-specific in layer-4 granular neurons. We generated a comprehensive survey of dlPFC cellular repertoire and its shared and divergent features in anthropoid primates.
One-Sentence Summary:
Single-cell transcriptomics reveals prefrontal cell taxonomy and species differences in human, chimpanzee, macaque and marmoset.
The expansion and evolutionary specializations of the cerebral cortex, and especially the prefrontal cortex (PFC), are thought to underlie the rich and complex nature of cognition in humans and other primates (1, 2). The PFC is larger and anatomically more complex in anthropoid primates (i.e., monkeys and apes) compared to other analyzed mammals, covering the entire dorsolateral two-thirds of the frontal lobe and having a well-defined granular layer (L) 4 (1–6). The granular dorsolateral PFC (dlPFC) exhibits a broad connectivity with other brain regions (1, 2). Dysfunction of the dlPFC also has been implicated in the etiology of many neuropsychiatric disorders (7, 8). Accordingly, nonhuman primates have been used as model species because their complex cognition, expanded PFC, and genetic makeup best approximate that of humans (9). Single-cell genomics has emerged as a central tool in understanding cell taxonomy and evolution of the brain, but prior efforts to characterize human PFC were not extensive in the scope of cell type classification and nonhuman primates compared (10–15).
To better understand the cellular repertoire of the dlPFC and its conserved and divergent features, we performed single-nucleus RNA sequencing (snRNA-seq) of the dlPFC in adult humans (Homo sapiens), chimpanzees (Pan troglodytes), rhesus macaques (Macaca mulatta) and common marmosets (Callithrix jacchus), representing major anthropoid phylogenetic groups: Hominini (here represented by humans and chimpanzees), Catarrhini (humans, chimpanzees, and macaques) and Platyrrhini (marmosets). The inclusion of chimpanzee, one of the closest human living relatives, as well as two outgroup species enabled robust identification of human-specific features. The computational analysis of snRNA-seq data revealed diverse neuronal, glial and non-neural transcriptomically-defined cell types (hereafter referred to as “subtypes”) and uncovered multiple conserved and divergent features, including those unique to humans. To delineate regulatory mechanisms underlying human-specific changes, we conducted single-nucleus multiome (sn-multiome) characterizing chromatin accessibility (ATAC sequencing [ATAC-seq]) and gene expression (RNA-seq) simultaneously on adult human dlPFC. This resource is interactively accessible at http://resources.sestanlab.org/PFC/.
Transcriptomic classification of dlPFC cells in anthropoid primates
We performed snRNA-seq on histologically validated adult postmortem dlPFC samples from neurotypical male and female humans, chimpanzees, rhesus macaques and common marmosets (Fig. 1A, fig. S1A, and table S1). Each species-group included four donors, with four technical replicates per donor, which significantly increased analytic power (fig. S1B–D). Analysis with the consensus genome annotation (fig. S2; Materials and Methods) followed by stringent quality control yielded transcriptomic data from 610,719 high-quality nuclei comparably distributed across the four species (fig. S1E).
Fig. 1. Transcriptomically-defined cell taxonomy of anthropoid primate dlPFC.
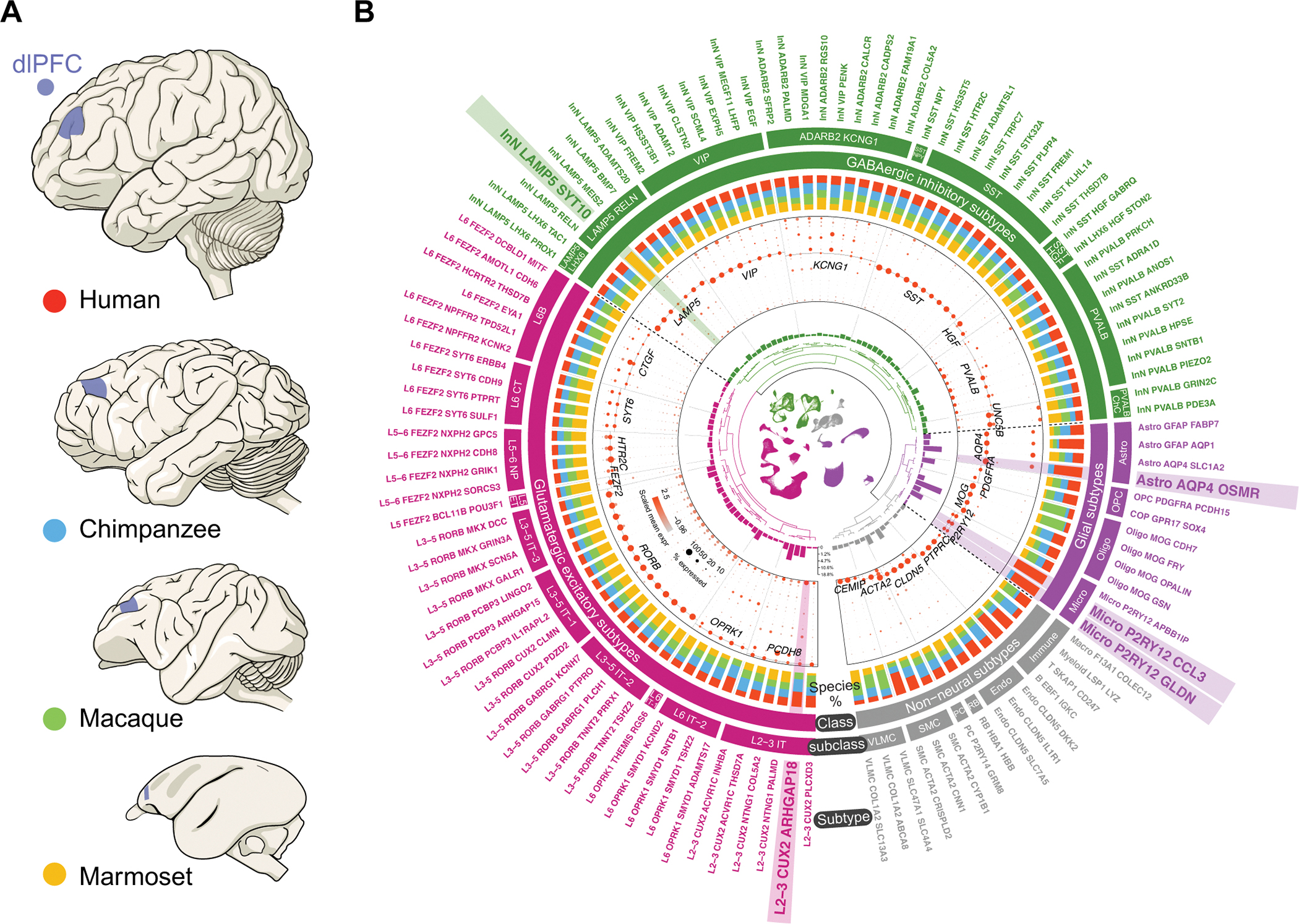
(A) Species (human – red; chimpanzee – blue; macaque – green; marmoset – yellow) and homologous dlPFC areas (blue) analyzed using snRNA-seq. (B) Overview of the snRNA-seq dataset, depicting information from innermost to outermost as: UMAP of all nuclei; dendrogram of hierarchically related subtypes; subtype proportions; marker gene expression; species representation in each subtype (color conforming to panel A); 4 classes; 29 subclasses; 114 refined subtypes. The subtypes detected only in some species are highlighted. See fig. S3A for a more detailed description. IT, intratelencephalic; ET, extratelencephalic; NP, near-projecting; CT, corticothalamic; ChC, chandelier cells; Astro, astrocyte; Micro, microglia; Oligo, oligodendrocytes; OPC: oligodendrocyte precursor cells; Endo, endothelial cells; RB, red blood lineage cells; PC, pericyte; SMC, smooth muscle cells; VLMC, vascular leptomeningeal cell.
Based on unsupervised clustering, expression profiling of marker genes, and transcriptomic integration with published human and mouse single-cell and spatial transcriptomic datasets (16–24) (Materials and Methods), we defined 3 levels of hierarchically-related taxonomy of neuronal, glial and non-neural subtypes (Fig. 1B and fig. S3A). Level 1 included 4 major classes, with 209,824 glutamatergic excitatory neurons, 101,845 GABAergic inhibitory neurons, 247,660 glial cells and 51,390 non-neural cells. Level 2 included 29 subclasses identified as 10 excitatory neuron subclasses characterized by their layer (L)- and axon projection properties (intratelencephalic, extratelencephalic, near projecting, corticothalamic and L6B); 9 inhibitory neuron subclasses characterized by established marker genes; 4 glial cell subclasses (astrocytes, oligodendrocyte precursor cells, oligodendrocytes and microglia); and 6 non-neural cell subclasses (immune cells, endothelial cells, red blood lineage cells, pericytes, smooth muscle cells, and vascular leptomeningeal cells). Level 3 included 114 further refined subtypes within each of the subclasses. These subtypes have a balanced contribution from donors in each species and their transcriptomic stability are generally not affected after randomly removing one donor in a given species (fig. S3A, B).
The 29 subclasses and most of the 114 subtypes were detected in all four primate species (Fig. 1B). Cell homologies were unaffected when integrating data using different methods (fig. S3C) or clustering nuclei within each species separately (figs. S11A, S13A and S16A). However, five subtypes were detected only in some species (hereafter referred to as “species-specific subtypes”), with no bias being introduced by donors of that given species (fig. S4A–D). The five subtypes included the excitatory neuron subtype L2–3 CUX2 ARHGAP18 absent in marmosets, the inhibitory neuron subtype InN LAMP5 SYT10 detected only in marmosets, the astroglial subtype Astro AQP4 OSMR detected only in humans and chimpanzees, the microglial subtype Micro P2RY12 CCL3 detected only in humans, and the microglial subtype Micro P2RY12 GLDN detected only in humans and chimpanzees.
Cellular and transcriptomic changes across anthropoid primate dlPFC subtypes
The multifaceted functions of the dlPFC require the orchestration of diverse cell types that have undergone substantial specialization, which could include the emergence of new cell subtypes, changes in the abundance of a shared or conserved cell type, variation in inter-cell heterogeneity, or reorganization of molecular features in conserved cell types (Fig. 2A).
Fig. 2. Shared and divergent features of dlPFC subtypes across anthropoid primates.
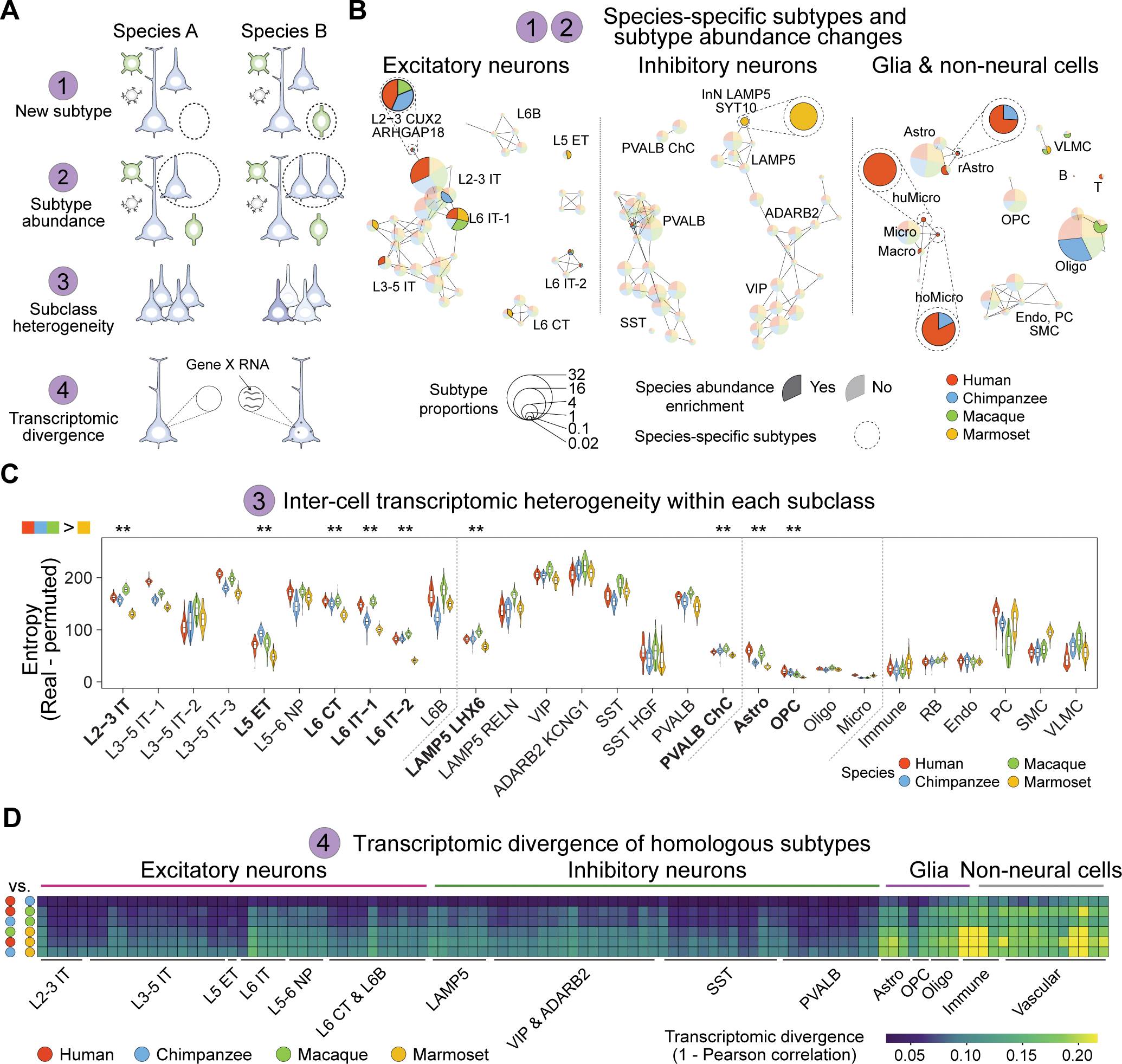
(A) Diagram illustrating possible models of species cellular and transcriptomic differences. (B) Subtype abundance comparisons across species. Each pie represents a subtype with size indicating the average subtype proportions across the four species (Materials and Methods). Links represent the expression correlation among subtypes. Subtypes showing species-enrichment in abundance are colored in opaque, otherwise in transparent. Species-specific subtypes are highlighted by dashed circles. (C) Transcriptomic heterogeneity among cells of the same subclasses. Significance was tested via pairwise Wilcoxon Rank Sum test with Bonferroni correction (**: adjusted p < 0.01). (D) Raw transcriptomic divergence across subtypes (columns) in all species pairs (rows). IT, intratelencephalic; ET, extratelencephalic; NP, near-projecting; CT, corticothalamic; ChC, chandelier cells; Astro, astrocyte; Micro, microglia; Oligo, oligodendrocytes; OPC: oligodendrocyte precursor cells; Endo, endothelial cells; RB, red blood lineage cells; PC, pericyte; SMC, smooth muscle cells; VLMC, vascular leptomeningeal cell. huMicro, human-specific microglia (Micro P2RY12 CCL3); hoMicro, Hominini-specific microglia (Micro P2RY12 GLDN). rAstro: reactive astrocyte (Astro AQP4 OSMR).
We first assessed whether the five species-specific subtypes (Fig. 2B) were transcriptomically unique for those species. For this, we calculated the markers for each species-specific subtype and visualized their expression enrichment across species on the Uniform Manifold Approximation and Projection (UMAP) layout (fig. S4A–D). The homologous cells of each species-specific subtype, if present in one species, were found to be marked by high enrichment scores and clustered together on the UMAP layout (fig. S4A–D). In contrast, no cells or only a scarce number of cells sparsely distributed on the UMAP were labeled by high enrichment scores in the species in which the species-specific subtypes were absent. By leveraging available cross-species snRNA-seq data (22, 25), we confirmed the species-specific presence of subtypes L2–3 CUX2 ARHGAP18, InN LAMP5 SYT10 and Micro P2RY12 CCL3, and demonstrated that the detection of species-specific subtypes is not limited to the donors analyzed in this study (figs. S4E and S17F). To further corroborate these results, we conducted single-molecule fluorescence in situ hybridization for representative marker genes and validated the species-specificity of the Catarrihini-specific upper layer intratelencephalic subtype L2–3 CUX2 ARHGAP18, the marmoset-specific inhibitory neuron subtype InN LAMP5 SYT10, and the Hominini-specific enrichment in astroglial subtype Astro AQP4 OSMR (figs. S5–7). Similar to the observation in the snRNA-seq data (fig. S4), we observed only a limited number of cells labeled by in situ hybridization in the species lacking the species-specific subtypes.
Although most subtypes are shared across the four analyzed primates, changes in their relative abundance can contribute to distinct cellular networks. An L2–3 intratelencephalic subtype (L2–3 CUX2 ACVR1C THSD7A), the most abundant subtype among all L2–3 intratelencephalic subtypes, was found to be enriched in humans compared to other species, and enriched in Catarrhini compared to marmosets (Fig. 2B and fig. S8). In contrast, we observed notable reduction of L5 extratelencephalic subtype and a L6 corticothalamic subtype in Catarrhini compared to marmosets (Fig. 2B).
Differences in cellular diversity across species can also arise when cell subpopulations within a subclass become transcriptomically more diversified or more homogenous within a given species. To delineate transcriptomic heterogeneity among cells within a given subclass, we adopted the concept of entropy, a measurement of uncertainty, in analyzing gene expression variability (fig. S9A; Materials and Method). Larger entropy values associate with higher gene expression variability, and vice versa. By subtracting the entropy in permutated data from that in observed data, we were able to reveal the overall transcriptomic heterogeneity among cells within each homologous subclass across species (Fig. 2C). We found a considerable heterogeneity increase in Catarrhini compared to marmosets in multiple neuronal and glial subclasses such as L2–3 intratelencephalic, PVALB chandelier cell, and astrocyte subclasses (Fig. 2C). Such heterogeneity arising in Catarrhini L2–3 intratelencephalic excitatory neurons confirms previous reports (16, 26) and extends our observations of expanded diversity and abundance in upper layer neurons in Catarrhini (Fig. 2B). Patterns of increased heterogeneity in Catarrhini were further supported by the augmented cell separation in principal component dimensions and the expansion of marker expression profiles distinguishing subtypes in Catarrhini (fig. S9B–C). Although many of these subtype markers shared across Catarrhini were also expressed broadly in marmosets, their expression was not distinguishable among the same subtypes (fig. S9D–E). This result suggests a possible mechanism underlying cell-type heterogeneity whereby genes originally shared among different subtypes become variable, resulting in greater inter-subtype transcriptomic differences.
Species differences can also arise from divergent gene expression in homologous types. We defined transcriptomic divergence based upon expression correlation of highly variable genes between homologous subtypes for each species-wise pair (Materials and Methods). Observed patterns of transcriptomic divergence mirrored evolutionary distances between the four species, with the human-chimpanzee pair showing the smallest transcriptomic divergence, whereas the chimpanzee-marmoset pair showing the largest (Fig. 2D). This analysis also showed that glia cell and non-neural cell subtypes were the most divergent in all species pairs (Fig. 2D), consistent with studies comparing primate species at the level of bulk tissue and cell-sorted populations (27, 28). We confirmed that these observations were not influenced by unequal cell numbers across subtypes or using a different highly variable gene set (fig. S10A–B) and were commensurate with species differences measured by the proportion of differentially expressed genes (fig. S10C). Together, this revealed that multiple sources of variation contributed to neuronal, glial and non-neural cell type differences across anthropoids.
Conserved and divergent features of excitatory neuron subtypes
We next investigated each major cell class to better delineate conserved and divergent features among subtypes. Excitatory neurons in each species were comprised by a multitude of subtypes predicted to have distinct properties (Fig. 3A and fig. S11A). Imputation of layer distribution and long-range projection types by integrating our data with published snRNA-seq data from human middle temporal gyrus (MTG) (16), spatial transcriptomic data from human PFC (18), and data from mouse primary visual cortex including projection identity (17) showed that excitatory neuron subtypes can be classified into 10 subclasses. These subclasses are characterized according to their predicted L2 to L6 layer localization and projection types (Fig. 1B and fig. S11B–D). The intratelencephalic subclass that accounted for more than 80% of the excitatory neurons was predicted to span layers L2 to L6 and was marked by layer-specific markers (L2–3: PCDH8; L3–5: RORB; L6: OPRK1; Fig. 1B). The extratelencephalic, corticothalamic and other subclasses were predicted to be localized in deep layers and were transcriptomically distinct (L5 extratelencephalic: FEZF2, BCL11B and POU3F1; L6 corticothalamic: SYT6; L5–6 near-projecting: HTR2C; L6B: CTGF; Fig. 1B).
Fig. 3. Taxonomy and divergent features of dlPFC excitatory neuron subtypes.
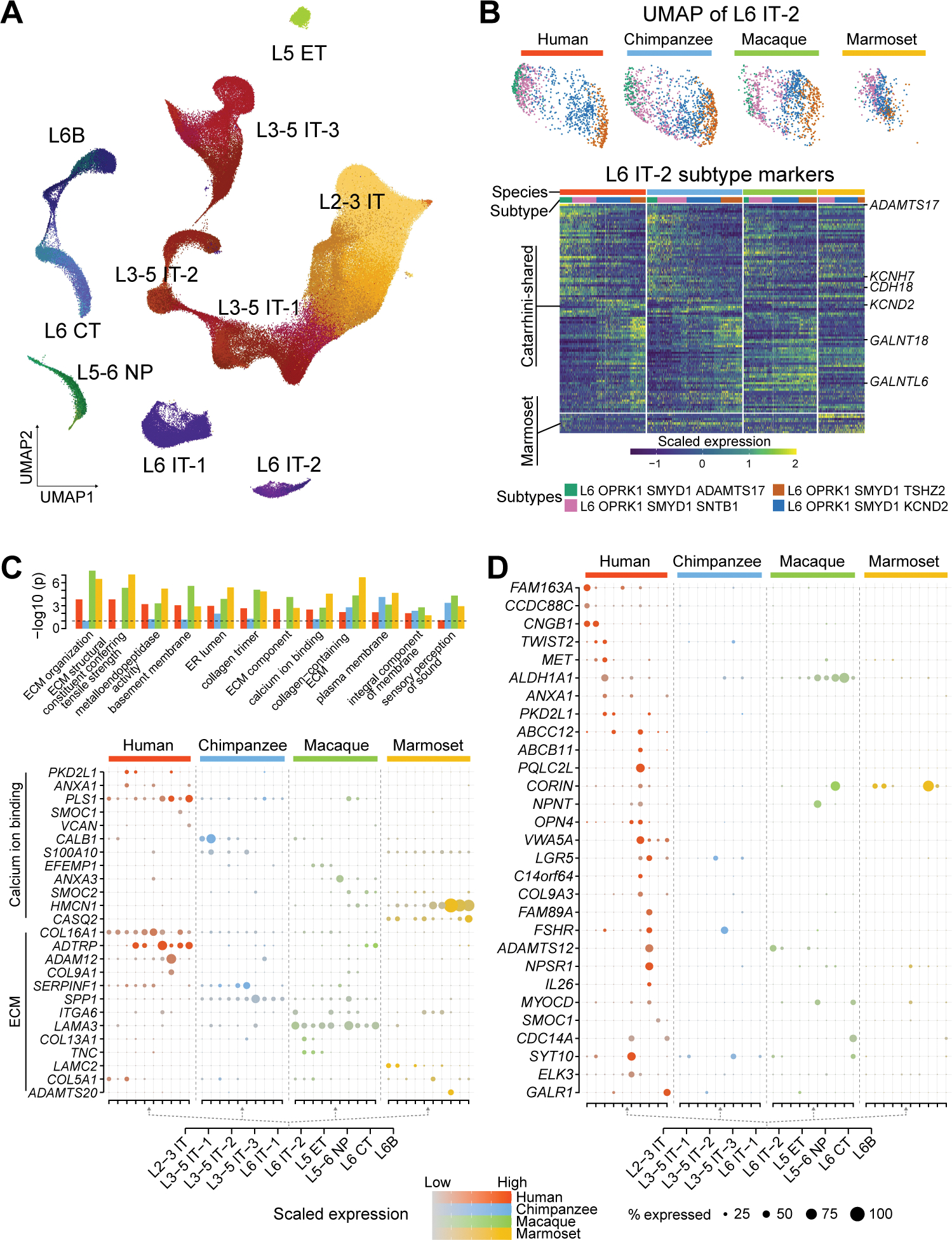
(A) UMAP visualization of all glutamatergic excitatory neuron subtypes with subclasses labeled. (B) Top: UMAP showing all L6 IT-2 subtypes. Bottom: Expression of subtype markers shared in Catarrhini (≥ 2 species) or specific to marmosets. (C) Top: Enrichment of Gene Ontology (GO) terms in the species-enriched genes from each species, with colors indicating the species. Only the GO terms enriched in at least three species are displayed. Bottom: Expression of the selected species-enriched genes involved in the calcium ion binding and extracellular matrix (ECM). ER, endoplasmic reticulum. (D) Expression of genes with high subclass and human specificity. IT, intratelencephalic; ET, extratelencephalic; NP, near-projecting; CT, corticothalamic.
Assessment of subclass heterogeneity across species revealed one L6 intratelencephalic subclass (L6 IT-2) having the greatest increase of inter-cell transcriptomic differences in Catarrhini compared to marmosets (Fig. 2C). In line with this observation, marmoset subtypes of this subclass were less separated on the UMAP and expressed fewer genes distinguishing them when compared to humans, chimpanzees, and macaques (Fig. 3B). Genes delineating the subtypes of this subclass in Catarrhini also mapped onto important biological pathways, including extracellular matrix and cell-cell interaction (ADAMTS17, CDH18), calcium ion binding proteins (KCNH7, KCND2) and protein glycosylation (GALNT18, GALNTL6), suggesting more diversified cellular functions.
We next identified transcriptomic changes restricted to each excitatory neuron subtype (fig. S12A). Within each species, we identified more species-enriched genes than species-depleted genes. As expected, marmosets had both more upregulated and downregulated genes compared to humans, chimpanzees and macaques. Gene Ontology enrichment of the exclusively enriched genes for each species converged upon processes related to extracellular matrix and calcium ion binding functions involved in synaptic transmission (Fig. 3C). For instance, genes within the annexin family, which play a critical role in calcium ion binding, were used differentially across primate brains: humans selectively expressed ANXA1 in upper-layer intratelencephalic subclasses, while macaques specifically expressed ANXA3 in deep-layer intratelencephalic subclasses. This analysis also highlighted human L3–5 intratelencephalic subclass enrichment of PKD2L1, which encodes a calcium-regulated cation channel. These results suggest that the components of the same biological pathways are differentially recruited across species, resulting in divergent molecular and cellular functions.
We further filtered genes with species- and subclass-restricted expression patterns using more stringent criteria (Materials and Methods), resulting in 20 to 51 genes in each species with potentially higher functional relevance (Fig. 3D and fig. S12B). The 29 genes detected in the human lineage included genes encoding transcription factors (TWIST2 and ELK3), genes associated with autism spectrum disorder (ASD) in SFARI gene list (https://gene.sfari.org/; CCDC88C, MET, ANXA1) as well as genes encoding critical retinoic acid signaling components (ALDH1A1; Fig. 3D and fig. S12C–D), which have recently been shown to regulate gene expression, spinogenesis and connectivity in PFC (29). We also found other retinoic acid signaling-associated genes exhibiting divergent expression across species: CYP26B1, enriched in Hominini L5–6 near-projecting and L6 corticothalamic subclasses, and CBLN2, globally upregulated in multiple Hominini excitatory neuron subclasses. These findings validate recently reported Hominini-specific changes in the retinoic acid-responsive enhancer predicted to upregulate CBLN2 expression in PFC excitatory neurons (30).
Conserved and divergent features of interneuron subtypes
Based on marker gene expression and transcriptomic integration with other human and mouse single-cell RNA-seq data (12, 16, 17, 19, 20) (Fig. 4A, S13B–D and S14A–C), we classified inhibitory neurons into 9 subclasses predicted as: long-range projecting neurons (SST NPY), Martinotti and non-Martinotti cells (SST), basket cells (PVALB), chandelier cells (PVALB ChC), Ivy cells (LAMP5 LHX6), LAMP5 neurogliaform cells (LAMP5 RELN), VIP inhibitory neurons (VIP), homologs of human TH-expressing inhibitory neurons (SST HGF), and homologs of mouse Sncg inhibitory neurons (ADARB2 KCNG1). We detected two PVALB chandelier subtypes marked by distinct expression profiles, showing specific enrichment of NMDA glutamate receptors regulating excitatory synaptic communication as well as non-clustered protocadherins mediating cell adhesion (fig. S14D–E). This pattern was observed in all the four primates analyzed (fig. S14D–E), suggesting conserved functional diversification between the two PVALB chandelier subtypes.
Fig. 4. Taxonomy and divergent features of dlPFC inhibitory neuron subtypes.
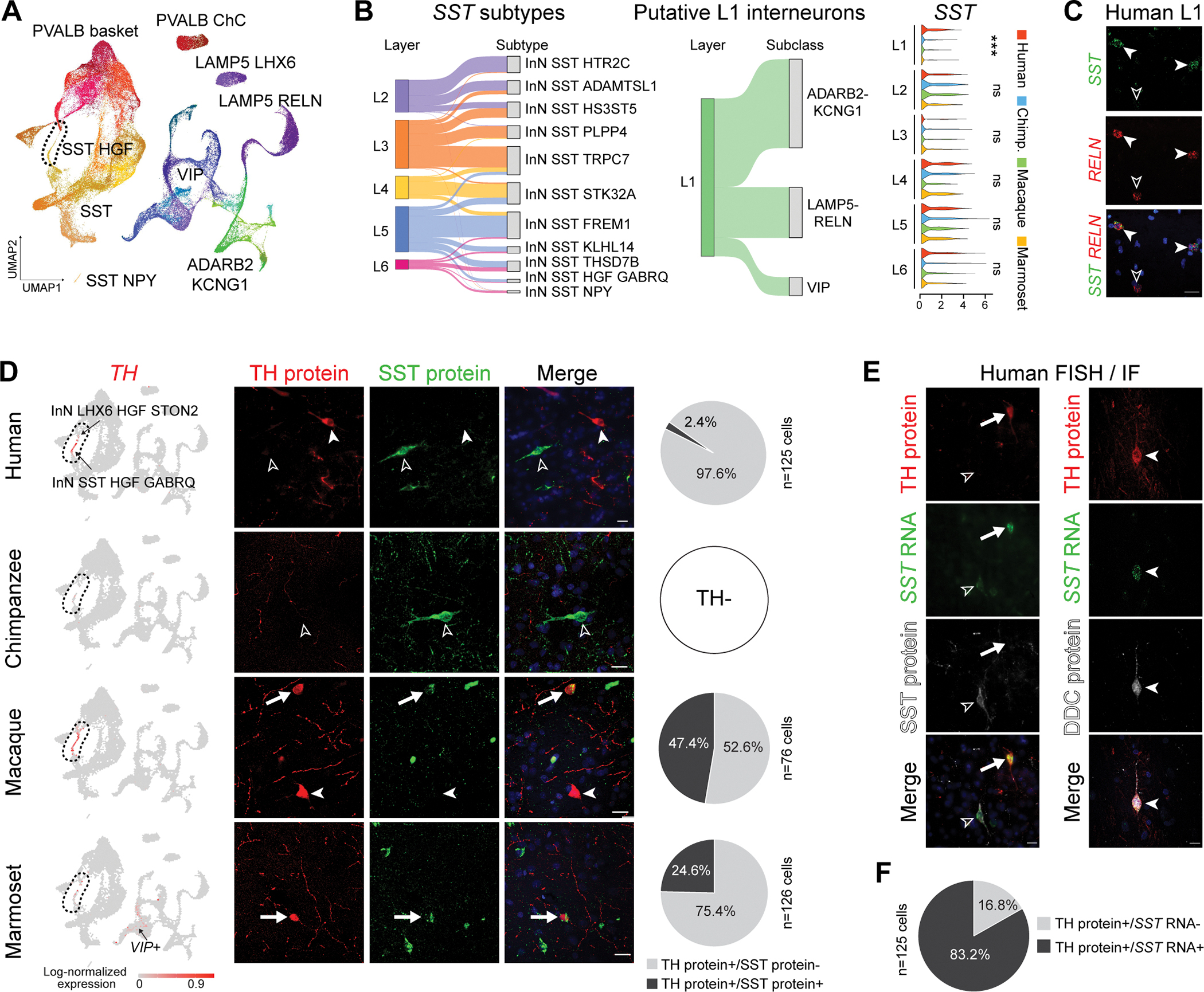
(A) UMAP layout of all GABAergic inhibitory neuron subtypes with subclasses labeled. ChC, chandelier cells. (B) Left: Sankey plots showing correspondence between predicted laminar organization and subtypes/subclasses. Right: SST expression across putative cortical layers. Significance was tested via one-sided Wilcoxon Rank Sum test (***: p < 0.001; ns: not significant). (C) colocalization of SST and RELN (arrowhead) in human dlPFC L1 revealed by double RNA in situ hybridization. Scale bar = 20 μm. (D) Left: log-normalized expression of TH. Middle: Double immunostaining for SST and TH. Right: Proportion of SST+ and SST− cells within TH+ neurons. Scale bars: 20μm in all species. (E) Immunofluorescence staining for TH and SST (left) or DDC (right), combined with SST RNA in situ hybridization. Scale bars: 20μm. (F) Proportion of SST+ and SST− cells in human TH+ neurons.
These inhibitory neuron subtypes were predicted to have different developmental origins based on lineage-related marker expression (medial ganglionic eminence [MGE]: LHX6; caudal ganglionic eminence [CGE]: ADARB2, NR2F2 and PROX1, figs. S13D and S14A–B). We found that the LAMP5 LHX6 subclass consisted of two subtypes marked by LHX6 expression, among which one subtype also expressed PROX1 (LHX6+/PROX1+; fig. S13D). The LAMP5 LHX6 subclass was previously reported to be enriched in primates (12, 16) and was considered to derive from MGE (31). Here, we found that the markers of the LHX6+/PROX1+ subtype showed expression enrichment in CGE-derived inhibitory neurons, whereas the markers of the LHX6+/PROX1- subtype showed enrichment in MGE-derived inhibitory neurons (fig. S14F). This pattern was conserved across the four primates (fig. S14F), suggesting a potential overlap of PROX1 and LHX6 molecular mechanisms commonly present in all the analyzed primates.
Imputation of layer distribution by transcriptomic integration with human MTG inhibitory neurons (fig. S14G) (16) showed that SST and PVALB inhibitory neuron subtypes were preferentially located from L2 to L6, while L1 inhibitory neurons included only ADARB2 KCNG1, VIP and LAMP5 subtypes (Fig. 4B). Some putative L1 inhibitory neuron subtypes in human also expressed SST (Fig. 4B and fig. S14G), but they lacked expression of other MGE-related genes including LHX6 and PVALB. However, few putative L1 inhibitory neurons from other species expressed SST (Fig. 4B). We confirmed the presence of these inhibitory neurons by performing in situ hybridization in human tissue against SST and RELN, a gene marking human L1 SST-expressing inhibitory neurons in the snRNA-seq data (Fig. 4C). These results suggest the presence SST-expressing inhibitory neurons in human L1.
Within homologous inhibitory neuron subtypes, we detected pronounced transcriptomic changes across species that followed patterns similar to excitatory neurons, with marmosets having the most distinct expression profiles, commensurate with phylogenetic relationships of the four primates (fig. S14H). MELTF, which encodes melanotransferrin and is involved in cellular iron uptake, was specifically upregulated in PVALB chandelier subtypes in humans (fig. S14H), suggesting novel roles of iron homeostasis in fast-spiking inhibitory neurons. Analysis of gene families (https://www.genenames.org/; Materials and Methods) overrepresented in species-enriched genes highlighted collagens in almost all species (fig. S14I), a pattern also observed in excitatory neurons (Fig. 3C). This analysis also revealed genes encoding neuropeptides (e.g., NMU and PENK) and carbonic anhydrases overrepresented in human-enriched genes; the former is a critical component orchestrating cell-cell communications, and the latter can regulate neuronal signaling through modulation of pH transients (fig. S14I). Cumulatively, these results suggested possible reorganization of synaptic communication across primate inhibitory neurons.
Human-specific switching between expression of SST and TH in interneurons
We have previously found that inhibitory neurons expressing tyrosine hydroxylase (TH), the rate-limiting enzyme in the biosynthesis of catecholamine neurotransmitters such as dopamine, are absent from the neocortex of nonhuman African apes (i.e., chimpanzees, bonobos and gorillas) (32, 33). Given the functional significance of dopamine in PFC function (7), we further profiled these inhibitory neurons using our dataset. We found TH expression in a small subset of dlPFC inhibitory neurons in humans and macaques. Expression was notably less in marmosets, and absent chimpanzees. Human and macaque TH-expressing inhibitory neurons co-expressed SST and were transcriptomically classified as the SST HGF subclass that was hierarchically split into two subtypes: InN LHX6 HGF STON2 subtype with low TH expression and InN SST HGF GABRQ subtype with high TH expression (Fig. 4D). Homologous subtypes were also found in chimpanzees (Fig. 4D), indicating that these inhibitory neurons are not absent in chimpanzees, but simply do not express TH. Expression of TH was also detected in the two homologous subtypes in marmosets, and some VIP-expressing inhibitory neurons, but generally showed much lower expression compared to humans and macaques (Fig. 4D), indicating a more complex pattern of species-specific regulation of TH expression in these inhibitory neurons.
Since these two subtypes are readily distinguishable by levels of SST expression (high in subtype InN SST HGF GABRQ and extremely low in subtype InN LHX6 HGF STON2; fig. S15A), we next characterized these two subtypes by immunofluorescence staining for TH and SST. TH-immunopositive inhibitory neurons were found in deep layers and the adjacent white matter of dlPFC in humans, macaques and marmosets, but not in chimpanzees (Fig. 4D). Many TH-immunopositive inhibitory neurons were also co-immunolabeled for SST in macaques (47.4%) and marmosets (24.6%; Fig. 4D). The two populations or states of TH-immunopositive inhibitory neurons distinguished by SST immunolabeling in macaques likely represent the two TH-expressing subtypes detected in our snRNA-seq data (fig. S15A). Furthermore, the lower proportion of TH-immunopositive inhibitory neurons co-immunolabelled for SST in marmosets may be due to the TH expression in marmoset VIP inhibitory neurons (Fig. 4D). Consistent with this, a subset of marmoset TH-immunopositive inhibitory neurons also expressed VIP in upper layers (28.1%, fig. S15C).
In contrast, only a few TH-immunopositive inhibitory neurons were co-immunolabeled for SST in humans (2.4%; Fig. 4D). Even with tyramide signal amplification, we could only detect trace SST-immunolabelling signals in human TH-immunopositive inhibitory neurons (fig. S15D–E). Such low SST-immunopositivity among human TH-immunopositive inhibitory neurons was inconsistent with our snRNA-seq data, as one human TH-expressing subtype (InN SST HGF GABRQ) exhibited prominent SST RNA expression comparable to the homolog subtype in macaques and marmosets (fig. S15A). Such inconsistency prompted us to investigate SST RNA expression in TH-immunopositive inhibitory neurons using additional tissue samples and methods. We complemented the immunofluorescence findings with in situ hybridization-based detection of SST RNA in human dlPFC tissue samples, and found that most human TH-immunopositive cells expressed SST RNA transcripts (83.2%, Fig. 4E–F), as predicted by our snRNA-seq data. This proportion is notably greater than the proportion of inhibitory neurons that are co-immunolabeled for both TH and SST (2.4%), suggesting that certain posttranscriptional mechanisms exist in humans to downregulate SST protein expression. Additionally, we profiled SST and TH immunohistochemistry in mouse frontal cortex, which revealed a proportion of inhibitory neurons co-immunolabeled for both SST and TH (57.9%; fig. S15F) similar to macaques (47.4%), but not humans (2.4%). Together, these results indicate that the switching between expression of SST and TH protein, a pattern reminiscent of populations of interneurons in the adult rat hypothalamus with switch between SST and TH and, subsequently, dopamine production, (34), is also present in certain dlPFC interneurons in a human-specific manner, at least in the context of the primates that we analyzed.
Since TH expression itself does not necessarily effectuate active dopamine production or signaling, this prompted us to profile other dopamine signaling pathway components in our dlPFC dataset (fig. S15A–B). We found that many genes orchestrating key dopamine signaling pathway processes, including dopamine synthesis (TH, GCH1), transport (SLC18A2), and binding to presynaptic receptors (DRD2) (7), were expressed in the human subtype InN SST HGF GABRQ. Yet, these genes exhibited either low or no expression in the other subtype InN LHX6 HGF STON2 or in other analyzed species (fig. S15A). DDC was not detected in the human InN SST HGF GABRQ subtype, likely due to its low expression, as we could detect its expression in the matched inhibitory neuron subtype in the human MTG data (Inh L5–6 SST TH; fig. S15A), which has higher sequencing depth using SMART-seq (16). In situ hybridization also confirmed the presence of RNA transcripts for both TH and DDC in the same human dlPFC cells that exhibited typical interneuronal morphology (fig. S15K). We further validated the protein expression of DDC in TH+/SST+ inhibitory neurons by performing double immunofluorescent staining for TH and DDC combined with in situ hybridization for SST in human dlPFC tissue samples (Fig. 4E). In contrast, in mouse, DDC immunolabeling was detected in putative dopaminergic neurons in the substantia nigra and ventral tegmental area, but not in neocortical TH-immunopositive inhibitory neurons (fig. S15L). Although expression of SLC6A3 was not detected in either our dlPFC or published MTG single cell dataset, SLC47A1, which encodes a non-canonical dopamine transporter (35), was uniquely expressed in both TH-expressing inhibitory neuron subtypes in humans (fig. S15A). These results suggest the emergence of key dopamine signaling pathway components in a subset of TH-expressing inhibitory neurons in human.
Conserved and divergent features of glial and non-neural subtypes
Within glia cells and non-neural cells, the observed taxonomy of oligodendroglial, astroglial, microglial, immune and vascular cells corresponded to 28 subtypes in humans, 27 subtypes in chimpanzees, 25 subtypes in macaques, and 25 subtypes in marmosets (Fig. 5A and fig. S16A). While these subtypes were labeled by conserved subclass markers (fig. S16B), prominent species-specific expression profiles were detected in each subtype, with a pattern consistent with the phylogenetic relationship among the four primates (Fig. 5B).
Fig. 5. Taxonomy and divergent features of dlPFC glia cell and non-neural cell subtypes.
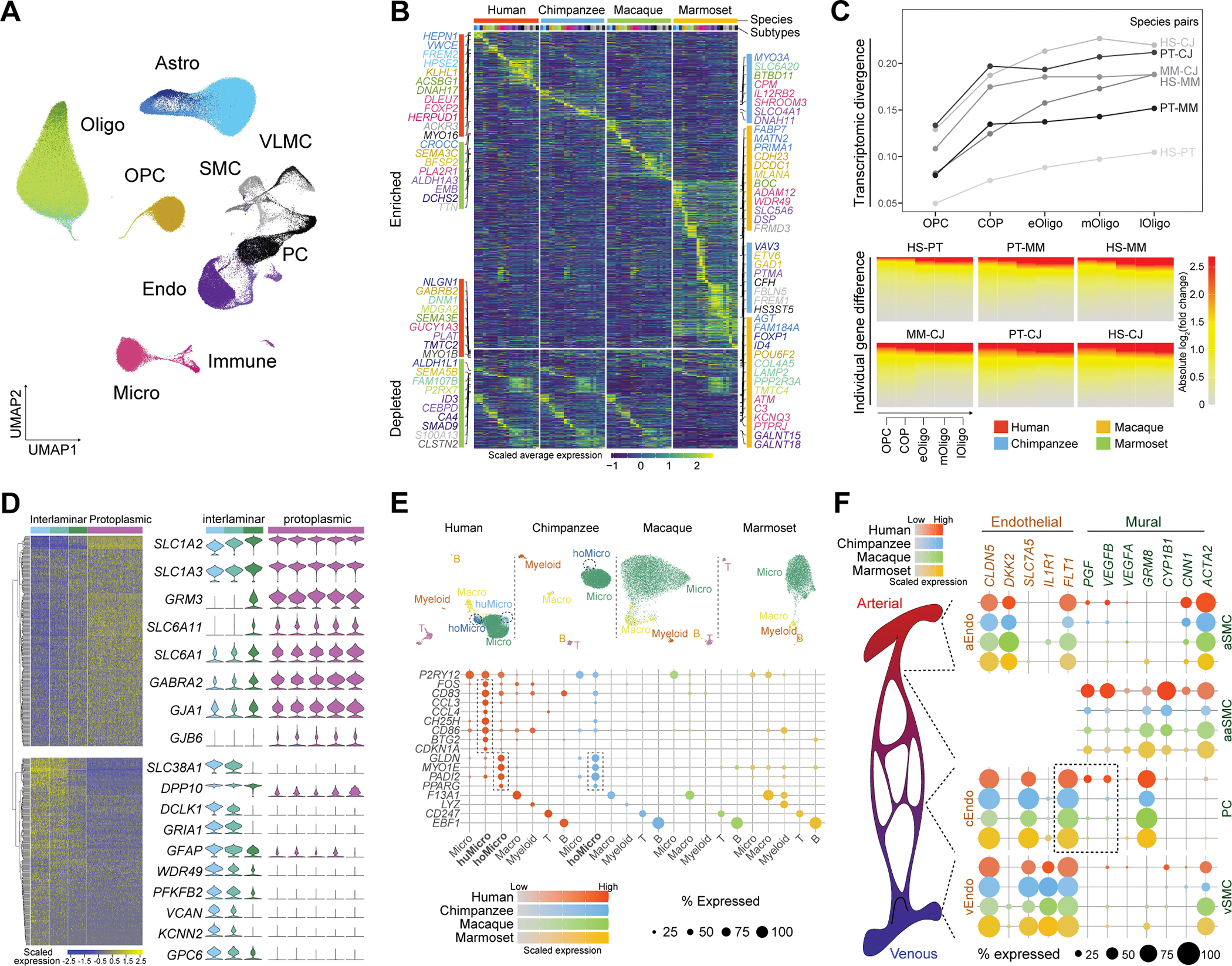
(A) UMAP showing all glia cell and non-neural cell subtypes in the four primates with subclasses labeled. (B) Standardized average expression of genes (rows) which are species-enriched (top) or -depleted (bottom) across subtypes (columns). Subtypes are color-barcoded on the top with color scheme conforming to panel A. Genes were colored according to the subtypes where they show enrichment or depletion. (C) Transcriptomic divergence (top) and gene expression differences (bottom) of subtypes in the oligodendrocyte lineage between each pair of species. HS: human; PT: chimpanzee; MM: macaque; CJ: marmoset. (D) Left: the two gene modules differentially expressed among the three interlaminar astrocyte subclusters. Right: Expression of the selected genes across the interlaminar and protoplasmic astrocyte subclusters. (E) Top: UMAP plots illustrating microglia and immune subtypes detected in the four primates. Bottom: expression of manually selected subtype markers genes. Marker expression labeling species-specific subtypes are highlighted by dashed rectangles. (F) Left: Illustration of the brain vascular architecture from arteries (red), arterioles, capillaries to veins (blue). Right: Vascular subtypes divided into endothelial cells (left) and mural cells (pericytes and smooth muscle cells, right) located along the arterial-venous axis. Dot plots show expression of marker genes and FLT1 signaling genes. Astro, astrocyte; Micro, microglia; Oligo, oligodendrocytes; OPC: oligodendrocyte precursor cells; Endo, endothelial cells; PC, pericyte; SMC, smooth muscle cells; VLMC, vascular leptomeningeal cell. huMicro, human-specific microglia subtype (Micro P2RY12 CCL3); hoMicro, Hominini-specific microglia subtype (Micro P2RY12 GLDN).
Subtypes related to the oligodendrocyte lineage represented a cellular differentiation and maturation trajectory spanning from oligodendrocyte precursor cells to mature oligodendrocytes (fig. S16C). This trajectory was shaped by conserved gene expression cascades across the four primates, and transcriptomically resembled that described in mice (21) (fig. S16C–D). However, prominent species differences were also observed, with transcriptomic divergence and individual gene expression differences among the four primates increasing along the trajectory (Fig. 5C). Evolutionarily divergent genes along the trajectory were particularly enriched for ligand and receptor binding features such as semaphorins and their neuropilin and plexin receptors (fig. S17A–B), which highlights the role of semaphorin signaling in regulating oligodendrocyte precursor cell recruitment and differentiation, as well as oligodendrocyte myelination.
Based on marker gene expression and imputation of layer distribution by integration of our data with human MTG and motor cortex data (16, 22), astrocytic subtypes were characteristic of known astrocyte subpopulations, including interlaminar astrocytes localizing in L1, protoplasmic astrocytes populating the grey matter, fibrous astrocytes residing in the white matter and reactive astrocytes (figs. S16E, H) (36). Since there is limited characterization of interlaminar astrocytes, we further sub-clustered these cells to probe their transcriptomic heterogeneity in greater detail. The resulting three subclusters displayed substantial expression differences in 612 genes that were organized into two co-expression modules (Fig. 5D). Module 1 included many genes encoding the major effectors for astrocyte-mediated clearance of glutamate and GABA from the extracellular space, such as glutamate transporters (SLC1A2 and SLC1A3) and GABA transporters (SLC6A11 and SLC6A1; Fig. 5D). Preferential expression of these genes in a subset of interlaminar astrocytes - juxtaposed to their broad expression in protoplasmic astrocytes - suggests a role in the clearance of neurotransmitters in cortical L1 by a subset of interlaminar astrocytes cells that adopted machineries similar to protoplasmic astrocytes.
Subtypes of Microglia, macrophages, myeloid cells, T cells and B cells formed the immune populations in these data (fig. S16A) (37). While we found broad conservation of immune subtypes across species, we highlighted one microglia subtype specific to humans (Micro P2RY12 CCL3) and one microglia subtype specific to Hominini (Micro P2RY12 GLDN, Fig. 5E). The human-specific microglial subtype expressed signatures of preactivation (FOS, CD83), chemokine secretion (CCL3, CCL4), anti-viral defense (CH25H), co-stimulation (CD86) and proliferation (BTG2, CDKN1A; Fig. 5E) and was very similar to a previously reported pre-active immune-sensing subpopulation, which emerges as early as the 10th gestational weeks and expands to mid-gestation in the absence of inflammation (38). Here we show that this cell type likely persists into adulthood in the human cerebral cortex (fig. S17E–F) (25). Its presence was also unrelated to sample age (table S1 and fig. S4I). The early developmental rise and persistence of this cell type, independent of different pathological states, may suggest it could have a specific and essential immune homeostatic and/or senescence-associated role, in contrast to specifically induced disease associated microglial subtypes (25, 37). The Hominini-specific microglial type showed high expression of GLDN, MYO1E, PADI2 and PPARG (Fig. 5E).
In addition to microglia, we identified 1,416 putative T cells, with the majority (885 or 62.5%) from human samples (fig. S16I) and their abundance is consistent across the human donors analyzed (8.24% ± 1.66% among all immune cells; fig. S4I). This number greatly exceeded those of myeloid cells (81 cells) and B cells (53 cells), surpassing proportions in peripheral circulation that would be expected for contaminants rather than a brain-resident T-cell population. Such cells likewise had expression of multiple genes not detected during the T-cell development in the human thymus (fig. S16K).
Through combinatorial marker gene expression and transcriptomic integration with mouse snRNA-seq data (23, 24), we found that vascular subtypes were predicted to locate along the arterial-arteriolar-capillary-venous axis including three endothelial subtypes (arterial, capillary, and venous endothelial subtypes), four mural subtypes (pericyte subtype, and arterial, arteriolar, and venous smooth muscle subtypes), three vascular leptomeningeal-like cell subtypes, and one putative red blood cell lineage subtype (fig. S16L; Materials and Methods). Some genes involved in key biological pathways displayed species-specific profiles along this vascular axis. This is exemplified by FLT1, encoding the vascular endothelial growth factor receptor, with conserved expression marking the capillary endothelial subtype. Among the three genes encoding its signaling molecules, VEGFA was sparsely expressed in pericytes of all species while VEGFB and PGF were enriched in human pericytes (Fig. 5F), suggesting the presence of human-specific signaling shaping vascular cell identity and communication.
Primate- and human-specific expression patterns of FOXP2
By assessing the intersection of genes showing human-enriched expression in microglia and those showing microglia-enriched expression compared to macrophages, we found 19 genes. FOXP2 was of particular interest among those genes, given that it encodes a transcription factor mutated in developmental verbal dyspraxia (39, 40), and is implicated in a number of neuropsychiatric disorders. While previous studies have shown that FOXP2 is expressed in neurons of the basal ganglia and neocortical layers 6 and 5 (40), its expression in microglia is unexpected. Some FOXP2-related disorders have been linked to alterations in microglia and neuroinflammation (37), highlighting the potential functional importance of human-specific FOXP2 expression in the context of microglia. We next performed in situ hybridization for FOXP2 RNA, combined with immunofluorescent staining against IBA1, and validated FOXP2 expression in human microglia (Fig. 6B). Similarly, we confirmed its protein expression in human microglia by performing triple-labeling immunofluorescence for RBFOX3 (NeuN), IBA1, and FOXP2 in human tissue, which revealed FOXP2 expression in both RBFOX3-positive neurons and IBA1-positive microglia (fig. S18A). Using multiple published datasets, including bulk tissue RNA-seq data of sorted microglia from the cerebral cortices of eight species (human, macaque, marmoset, sheep, mouse, rat, hamster and chicken, fig. S18B) (25), snRNA-seq data from mouse primary visual cortex (Fig. 6A) (17), cross-species snRNA-seq data of motor cortex (human, marmoset and mouse, fig. S18C) (22), and cross-species snRNA-seq data of hippocampus (human, macaque, pig and mouse, fig. S18D) (41), we independently validated human specificity of FOXP2 expression in microglia and found that it was a common feature shared by multiple brain regions (fig. S18B–E). In contrast, FOXP2 expression was not detected in macrophages in brain or other tissues (fig. S18F), nor in fetal microglia (fig. S18G–H).
Fig. 6. Species- and cell type-specific expression of FOXP2.
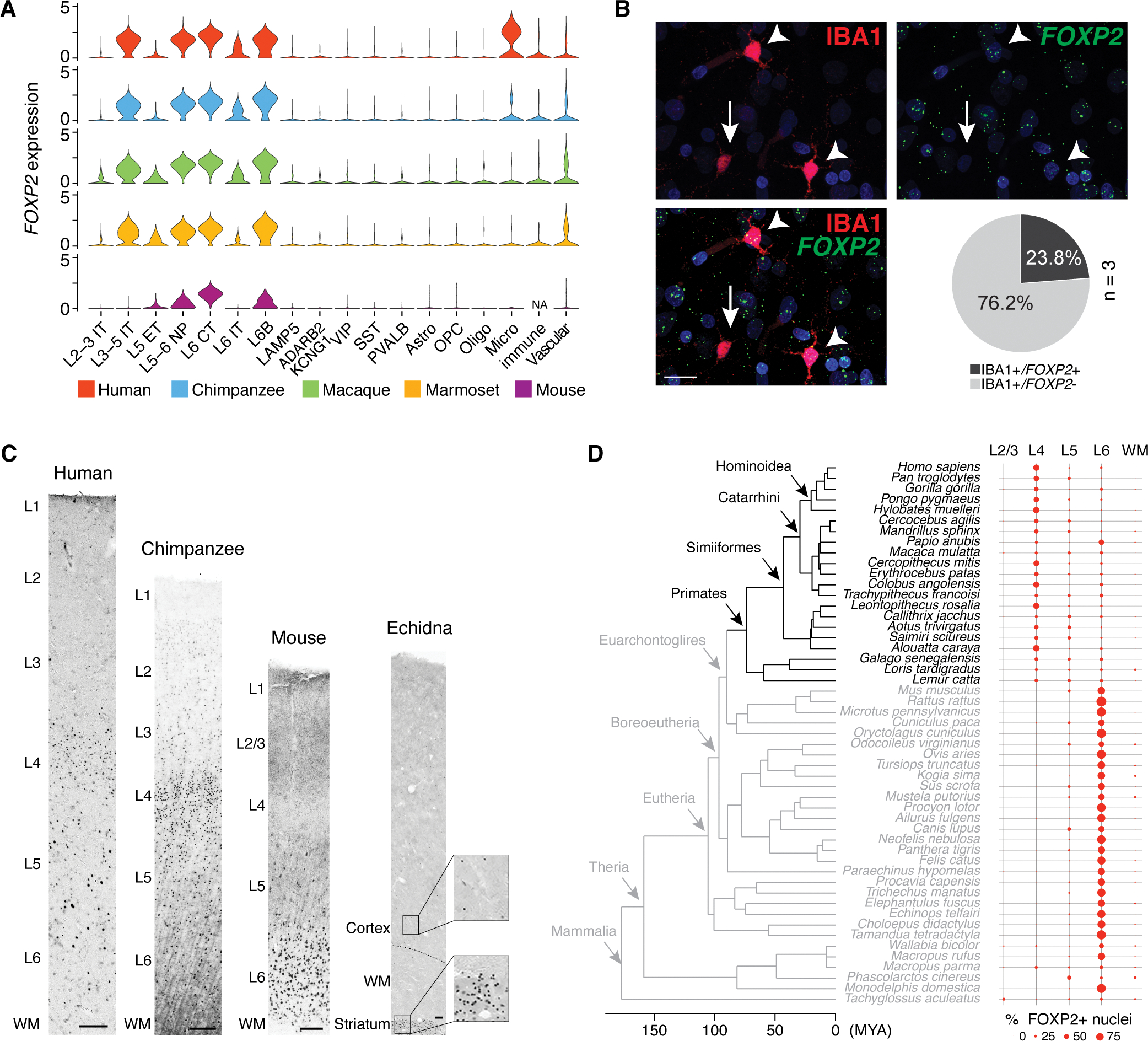
(A) Violin plots of FOXP2 expression across all subclasses in the four primates and mouse neocortex (17). (B) Immunofluorescent staining against IBA1 (red) combined with RNA in situ hybridization for FOXP2 RNA (green) in L6 of human dlPFC. Arrowheads indicate FOXP2+/IBA1+ microglia, whereas the arrow indicates FOXP2−/IBA1+ microglia. The pie chart summarizes the proportion of FOXP2-expressing cells among IBA1-immunopositive cells. Nuclei are stained with DAPI (blue). Scale bar: 20 μm. (C) Representative images of FOXP2 immunohistochemistry throughout cortical columns in human, chimpanzee, mouse, and echidna. The position of layers and white matter (WM) are indicated. Insets for echidna highlight numerous FOXP2-immunopositive nuclei in the striatum and scarce FOXP2-immunopositive nuclei in the deep neocortical layers. Scale bars: 100 μm for all species. (D) Phylogeny dendrogram of the 51 mammals and their corresponding laminar distribution of FOXP2-immunopositive nuclei.
We found that FOXP2 is highly expressed in several major L5 and L6 excitatory neuron subclasses across primates in our data (Fig. 6A), which matches previous studies in mice (40). Yet, we also observed a previously unknown enrichment of FOXP2 expression in L3–5 intratelencephalic excitatory neurons across primates, that was not detected in mice (Fig. 6A). The L3–5 intratelencephalic excitatory neurons likely encompass small neurons that give the primate dlPFC L4 its characteristically granular appearance (with a high density of small excitatory neurons). To test if FOXP2 is expressed in neurons of the granular L4, we conducted immunohistochemistry in the four primates analyzed in this study (Fig. 6C and table S3). We found that the majority of neuronal nuclei in granular L4 of the dlPFC were immunolabeled for FOXP2. To test if this expression pattern is specific to primates, which are the only mammals having the granular dlPFC, we extended our immunohistochemical study of FOXP2 expression to 21 primates and 30 non-primate mammals. This represented at total of 19 Orders, including Monotremes (represented by echidna), the lineage with the most ancient divergence from other mammals (table S3), and allowed us to identify the evolutionary conservation of this expression profile. We found that the pattern of prominent L4 FOXP2 expression was only observed in primates (Fig. 6D and figs. S19 and S20A) and was notable in granular PFC and other analyzed granular association areas involved in higher, integrative functions (table S3). Overall, this indicates that broad expression of FOXP2 in L4 excitatory neurons is an evolutionary specialization of primates.
To assess temporal expression patterns that might inform developmental regulation of primate FOXP2 specificity in L4, we performed immunohistochemistry on frontal cortices from fetal, perinatal, and adult humans, and observed FOXP2-immunopositive nuclei in L5 and L6 (fig. S20B) across all the analyzed developmental periods. However, human L4 FOXP2 immunoreactivity did not emerge until after 10 days postnatal age (fig. S20B), suggesting that FOXP2 is expressed well after L4 neurons have reached their laminar position, and myelination and synaptogenesis are at their peak (42). This suggests that FOXP2 is likely to be controlled by transcriptional programs that differ from those controlling FOXP2 expression in L5 and L6 excitatory neurons.
Shared and cell type-specific FOXP2 regulatory mechanisms
To identify possible regulatory mechanisms and the functional significance underlying the cell type- and species-specific FOXP2 expression patterns, we next performed sn-multiome (snATAC-seq ad snRNA-seq), profiling chromatin accessibility and gene expression simultaneously on the dlPFC of five additional neurotypical male and female adult human donors (table S1). Following stringent quality control, we obtained transcriptome data from 56,938 nuclei and chromatin accessibility data from 41,591 nuclei (fig. S21A–B). Using the human snRNA-seq data as the reference, we were able to define the same cell subclasses that were similarly distributed across donors (fig. S21B).
Through investigation of chromatin accessibility and physical contacts (43) in microglia from the sn-multiome data and multiple independent studies, we found that one cis-regulatory element topologically interacted with FOXP2 and was unique to microglia (fig. S21C–D). Similarly, using the sn-multiome data, we identified several cis-regulatory elements proximal to FOXP2 selectively enriched in L3–5 intratelencephalic excitatory neurons (fig. S21D; table S4).
By integrating the DNA co-accessibility and gene co-expression, we constructed gene regulatory networks connecting upstream regulators that positively regulate FOXP2 as well as downstream targets that are either positively or negatively regulated by FOXP2, in each of the FOXP2-expressing subclasses (fig. S22A; table S5). While the upstream regulators scarcely overlap between subclasses, the downstream targets are substantially shared among excitatory neuron subclasses (fig. S22B–C). This analysis highlighted NFIA, a previously reported FOXP2 cofactor (44), having both upstream and downstream regulatory roles in L5–6 non-intratelencephalic excitatory neurons (Fig. S22C). Furthermore, DSCAM implicated in Down syndrome, was predicted to be positively regulated by FOXP2 in microglia and exhibited human-specific expression in microglia as well (fig. S22C–D). Several genes (PHLDB2, PLCH1 and IL1RAPL2) positively regulated by FOXP2 in L3–5 intratelencephalic excitatory neurons also displayed primate-specific expression in these neurons compared to mice (fig. S22C–D). Among these targets, IL1RAPL2 has been reported to play a key role in dendritic spine formation (45). Using an independent algorithm identifying the FOXP2-related regulon (a module consisting of a transcription factor and its targets) across all cell subclasses rather than in a subclass-wise manner, we found that the FOXP2-regulon ranked as the top regulon of those specific to L3–5 intratelencephalic neurons (fig. S22E–F). This suggests a central role for FOXP2 in the transcriptional regulation in L3–5 intratelencephalic excitatory neurons (fig. S22G).
To validate the downstream targets in excitatory neurons independently, we electroporated either GFP or FOXP2 (human or mouse sequence) into the ventricular walls of embryonic day 14.5 mice, followed by RNA-seq analysis of neocortices at postnatal day 7. From this, we were able to validate several targets positively regulated by FOXP2, including PHLDB2, CALCRL, CNR1, CALD1, as well as SHISA6, which is negatively regulated by FOXP2 (fig S22C; table S7).
Taken together, our results highlight cellular, phylogenetic and developmental features in the cortical expression pattern of FOXP2 as well as potential regulatory mechanisms that might underlie its functional relevance to speech, language, and multiple neuropsychiatric disorders.
Certain brain disorder risk genes exhibit cell type- and species-specific expression
The species-divergent expression of the neuropsychiatric disease risk gene FOXP2 prompted us to characterize the expression of 906 genes previously associated with twelve major brain disorders systematically in this setting (Materials and Methods). By calculating the coefficients of variation of gene expression across species, we found the risk genes of many disorders, such as ASD, developmental delay disorder and epilepsy, are more conserved than background genes (fig. S23A). Based on gene expression conservation, these genes were parcellated into conserved (320 genes, 35%) or divergent (586 genes, 65%) categories (fig. S23B), followed by co-expression network analysis within each category to group these genes into different modules. In the divergent category, we identified multiple modules exhibiting phylogenetic group- or species-specific (including human-specific) patterns (fig. S23C, table S8). For example, two Parkinson’s disease risk genes (GBA and GOSR2, module 22) that regulate the degradation and secretion of alpha-synuclein displayed Hominini-enriched expression in a subset of excitatory neuron and inhibitory neuron subclasses (fig. S23D).
To gain a more detailed view into the cell type-specific expression of the 586 genes with species-divergent expressions, we performed differential expression analysis across subclasses. Of those genes, 157 (27%) exhibited subclass- and species-specific expression patterns (table S9). These are exemplified by the multiple sclerosis risk gene, MERTK, enriched in human L3–5 IT excitatory neurons and the ASD risk gene, CACNA1D, enriched in human microglia. MERTK showed conserved expression in microglia, consistent with its known function in myelination equilibrium regulation by modulating microglia activities (fig. S23F). However, its human-specific expression in L3–5 intratelencephalic neurons suggests an uncharacterized role in human excitatory neurons. Similarly, the ASD risk gene, NR4A2, which encodes a transcription factor, exhibited human-specific expression in smooth muscle cells (fig. S23E–F), suggesting possible reorganization of transcriptional regulation in anthropoids.
Conclusions
We generated a comprehensive transcriptomic survey of dlPFC neuronal, glial and non-neural cells in the most commonly studied anthropoid primates. We also revealed and characterized species differences in transcriptomic profiles, composition and diversity of cell types, including those associated with evolutionary specializations and functionality of dlPFC.
While the majority of cell subtypes are shared across the four species, we identified 5 species-specific subtypes. These subtypes showed balanced proportions in all donors of a given species and some were also detected in independent studies, corroborating the robustness in identifying cellular differences across species. Nevertheless, detection of a subtype in a subset of species does not necessarily implicates species-specificity, but alternatively could represent potential differences in cell type abundance or individual variability across species. To address this, we performed marker gene enrichment and in situ hybridization (figs. S4–7) and revealed that even in species where we did not detect a cell subtype, rare cells expressing markers of those subtypes may exist. While such results still substantiated species differences, species-specificity can be better resolved by profiling more cells and donors. Alternatively, species specificity might represent different cell states of homologous subtypes, and their species-specific presence could be associated with the differences in the living environment and lifestyles between these species. This might associate with baseline inflammation differences across species, which is particularly important for immune populations, such as microglia, and astrocytes.
Furthermore, we found that certain genes associated with key biological pathways exhibited cell type- and species-specific expression differences, featured by the switch of SST and TH expression in a subset of human inhibitory neurons. A possible mechanism underlying the posttranscriptional regulation of SST expression is that mRNA sequence changes lead to gain or loss of regulation from ribosome-binding proteins. Consistent with this, SST protein sequence changes are highly conserved in primates, and the mRNA sequences included one non-synonymous mutation specific to humans and multiple other synonymous mutation specific to apes (fig. S15G–J).
These putative dopaminergic interneurons have been previously described (32) but their molecular characterization and developmental origin remained unknown. Our study demonstrated that these interneurons express SST, as well as other key genes that are essential for dopaminergic function, and likely originate in the medial ganglionic eminence. Interestingly, it has been reported that humans, as well as other great apes, have an enrichment of dopaminergic afferents in the prefrontal cortex (33) and our results suggest that human have an extra population of dopaminergic interneurons in these cortical areas. This potential increase in dopamine levels might have important functional relevance for the evolution of human cognitive abilities and behavior (46).
In addition, notable molecular differences across species were unveiled in many neuropsychiatric diseases risk genes, exemplified by the human-specific FOXP2 expression in microglia and primate-specific FOXP2 expression in L4 excitatory neurons. While such expression patterns are associated with cell type-specific cis-regulatory elements, future studies are needed to determine their species-specificity. As a transcription factor, the species-specific expression of FOXP2 also leads to species-specific transcriptional changes in the L4 excitatory neurons or microglia (fig. S22), including upregulation of IL1RAPL2 involved in dendritic spine formation and DSCAM implicated in Down syndrome. Together, our results demonstrated this data as a resource for future studies in selecting primate models or developing human-specific platforms to model diseases.
Summary of methods
In the snRNA-seq experiment, the brain cell nuclei were isolated according to our previous protocol (10, 13, 41) and the following nuclei capture and library preparation were conducted using Chromium Single Cell 3′ Solution v3. The snRNA-seq reads were preprocessed with CellRanger using the consensus genome built in this study based on reciprocal exon liftOver. Doublet removal was carried out using scrublet (47) and custom AUCell-based (48) doublet detection scripts. Downstream batch correction, dimension reduction and cell clustering were performed via Seurat (49). Cell subtype separability was measured based on AUROC scores evaluating cell type identity prediction in down-sampled data where one donor was randomly removed. Detection of cell type markers and species-specific genes were realized using Wilcoxon Rank Sum test. Cell cluster identity annotation was based on marker gene expression and transcriptomic comparisons with multiple published data sets (16–24).
In assessing the global cellular and transcriptomic changes across species, AUCell-based (48) cell type marker enrichment was used to evaluate species-specificity of the detected species-specific subtypes. Cell subtype abundance comparisons were carried out using the scCODA algorithm (50), transcriptomic heterogeneity assessment was calculated via custom scripts based on Shannon entropy, and transcriptomic divergence was defined as the Pearson correlation coefficients subtracted from 1.
In the sn-multiome experiment, the brain cell nuclei isolation is a comparable modification of our previous protocol (10, 13, 41) followed by library construction using Chromium Single Cell Multiome ATAC + Gene Expression (10x Genomics PN 1000283). The sn-multiome data was preprocessed with CellRanger and MACS2 (51) followed by downstream filtering and dimension reduction using Seurat and Signac (49, 52). Cell type annotation was achieved via the label transfer algorithms in Seurat using the human snRNA-seq data as the reference. Detection of differentially accessible peaks was performed using the logistic regression implemented in Seurat. FOXP2 regulatory networks were constructed via scanpy (53), cicero (54), CellOracle (55) and pySCENIC (56) by integrating the DNA co-accessibility and gene co-expression information.
In the mouse in utero electroporation experiment, either FOXP2 or GFP was electroporated into embryonic day 14.5 mice ventricular walls and RNA-seq was performed on neocortices at postnatal day 7. Analysis of the RNA-seq data included reads alignment via STAR (57), reads counting via HTSeq (58) and differential gene expression via DESeq2 (59).
In analysis of FOXP2 cortical expression across species, immunohistochemistry for FOXP2 was conducted in 21 primate and 30 non-primate mammals with subsequent quantification of FOXP2 laminar distribution.
In the validation of species-specific subtypes, RNA in situ hybridization was performed with RNAscope on human, chimpanzee, and marmoset dorsolateral prefrontal cortices with probes targeting LAMP5 and CD8A, CUX2 and PRLR, or AQP4 and CHI3L1.
In characterization of SST expression in TH-expressing interneurons across species, immunohistochemistry of TH and SST was performed in human, chimpanzee, macaque, and marmoset dorsolateral prefrontal cortices and the percentage of SST+/TH+ interneurons was quantified. RNA in situ hybridization with SST probe was combined with immunohistochemistry of TH and SST protein in human dorsolateral prefrontal cortices utilizing RNAscope Co-detection.
Supplementary Material
Acknowledgments:
We thank Steven Wilson for assistance with tissue acquisition and processing; Rachel Chen and Mikihito Shibata for assistance with immunostaining; Adriana Cherskov and lab colleagues for comments. We thank the Traumatic Stress Brain Research Group for tissue acquisition.
Funding:
Medical Scientist Training Program grant: T32 GM140935 (RDR)
NINDS Ruth L. Kirschstein National Research Service Award: F32NS117780 (RK)
Agencia Estatal de Investigación (AEI) Spain grant PRE2020–093064 (XDM)
Institutional Ruth L. Kirschstein National Research Service Award: T32 GM141013 (Z.G.-S.)
National Institute on Aging R56 AG059284 (MD)
National Primate Research Centers P51 OD011133 (MD)
National Institutes of Health grants NS092988 and HG011641 (CCS)
National Science Foundation EF-2021785 (CCS)
Instituto de Salud Carlos III Spain and European Social Fund grant MS20/00064 (GS)
Agencia Estatal de Investigación (AEI) Spain grant PID2019–104700GA-I00 (GS)
National Institutes of Health grant R01HG010898–01 (GS and NS)
National Institutes of Health grant U01AG058608, R01AG066165 and P30 AG066508 (SMS)
NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation 28721 (AMMS)
National Institute of Child Health and Human Development core grant P50HD105353 (Waisman Center)
National Institutes of Health grant U01MH124619, U01MH116488 and U01 DA053628 (NS)
Footnotes
Data and materials availability:
The PFC snRNA-seq data were deposited in the NEMO Archive (RRID:SCR_002001) under identifier nemo:dat-zkgd0it accessible at https://assets.nemoarchive.org/dat-zkgd0it. The data can also be interactively visualized at http://resources.sestanlab.org/PFC. The human PFC sn-multiome data were deposited at NCBI GEO under the accession number: GSE207334. The mouse in utero electroporation RNA-seq data were deposited at NCBI GEO under the accession number: GSE206994. Scripts used in this study are available at Github repository: https://github.com/sestanlab/Cross-species-PFC-snRNA-seq and at Zenodo https://doi.org/10.5281/zenodo.6824096 (60). All other data are available in the main paper or supplement.
References and Notes
- 1.Barbas H, General cortical and special prefrontal connections: principles from structure to function. Annu Rev Neurosci 38, 269–289 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Preuss TM, Wise SP, Evolution of prefrontal cortex. Neuropsychopharmacology 47, 3–19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semendeferi K, Lu A, Schenker N, Damasio H, Humans and great apes share a large frontal cortex. Nat Neurosci 5, 272–276 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Schoenemann PT, Sheehan MJ, Glotzer LD, Prefrontal white matter volume is disproportionately larger in humans than in other primates. Nat Neurosci 8, 242–252 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Smaers JB, Gomez-Robles A, Parks AN, Sherwood CC, Exceptional Evolutionary Expansion of Prefrontal Cortex in Great Apes and Humans. Curr Biol 27, 714–720 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Donahue CJ, Glasser MF, Preuss TM, Rilling JK, Van Essen DC, Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc Natl Acad Sci U S A 115, E5183–E5192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cools R, Arnsten AFT, Neuromodulation of prefrontal cortex cognitive function in primates: the powerful roles of monoamines and acetylcholine. Neuropsychopharmacology 47, 309–328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis DA, Mirnics K, Transcriptome alterations in schizophrenia: disturbing the functional architecture of the dorsolateral prefrontal cortex. Prog Brain Res 158, 141–152 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Izpisua Belmonte JC et al. , Brains, genes, and primates. Neuron 86, 617–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M et al. , Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathys H et al. , Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krienen FM et al. , Innovations present in the primate interneuron repertoire. Nature 586, 262–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y et al. , Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanton S et al. , Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Lake BB et al. , Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodge RD et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasic B et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maynard KR et al. , Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci 24, 425–436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi D et al. , Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer C et al. , Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques S et al. , Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakken TE et al. , Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalucka J et al. , Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779 e720 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Vanlandewijck M et al. , A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Geirsdottir L et al. , Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 179, 1609–1622 e1616 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Hutsler JJ, Lee DG, Porter KK, Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res 1052, 71–81 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Pembroke WG, Hartl CL, Geschwind DH, Evolutionary conservation and divergence of the human brain transcriptome. Genome Biol 22, 52 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berto S et al. , Accelerated evolution of oligodendrocytes in the human brain. Proc Natl Acad Sci U S A 116, 24334–24342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata M et al. , Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 598, 483–488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata M et al. , Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 598, 489–494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y et al. , Mouse and human share conserved transcriptional programs for interneuron development. Science 374, eabj6641 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Sousa AMM et al. , Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghanti MA et al. , Species-specific distributions of tyrosine hydroxylase-immunoreactive neurons in the prefrontal cortex of anthropoid primates. Neuroscience 158, 1551–1559 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC, Neurotransmitter switching in the adult brain regulates behavior. Science 340, 449–453 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Kajiwara M, Ban T, Matsubara K, Nakanishi Y, Masuda S, Urinary Dopamine as a Potential Index of the Transport Activity of Multidrug and Toxin Extrusion in the Kidney. Int J Mol Sci 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben Haim L, Rowitch DH, Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18, 31–41 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Salter MW, Stevens B, Microglia emerge as central players in brain disease. Nat Med 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Kracht L et al. , Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science 369, 530–537 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP, A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA, Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol 460, 266–279 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Franjic D et al. , Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron 110, 452–469 e414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N, The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nott A et al. , Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hickey SL, Berto S, Konopka G, Chromatin Decondensation by FOXP2 Promotes Human Neuron Maturation and Expression of Neurodevelopmental Disease Genes. Cell Rep 27, 1699–1711 e1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valnegri P et al. , The X-linked intellectual disability protein IL1RAPL1 regulates excitatory synapse formation by binding PTPdelta and RhoGAP2. Hum Mol Genet 20, 4797–4809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raghanti MA et al. , A neurochemical hypothesis for the origin of hominids. Proc Natl Acad Sci U S A 115, E1108–E1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolock SL, Lopez R, Klein AM, Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291 e289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aibar S et al. , SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buttner M, Ostner J, Muller CL, Theis FJ, Schubert B, scCODA is a Bayesian model for compositional single-cell data analysis. Nat Commun 12, 6876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T, Srivastava A, Madad S, Lareau CA, Satija R, Single-cell chromatin state analysis with Signac. Nat Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pliner HA et al. , Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol Cell 71, 858–871 e858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamimoto K, Hoffmann CM, Morris SA, CellOracle: Dissecting cell identity via network inference and in silico gene perturbation. bioRxiv, 2020.2002.2017.947416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van de Sande B et al. , A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma S et al. , Molecular and cellular changes underlying evolutionary specializations of the primate dorsolateral prefrontal cortex. (2022). 10.5281/zenodo.6824096 [DOI]
- 61.Pockrandt C, Alzamel M, Iliopoulos CS, Reinert K, GenMap: ultra-fast computation of genome mappability. Bioinformatics 36, 3687–3692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haghverdi L, Lun ATL, Morgan MD, Marioni JC, Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim L, Mi D, Llorca A, Marin O, Development and Functional Diversification of Cortical Interneurons. Neuron 100, 294–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paul A et al. , Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539 e520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tricoire L et al. , Common origins of hippocampal Ivy and nitric oxide synthase expressing neurogliaform cells. J Neurosci 30, 2165–2176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharifi K et al. , FABP7 expression in normal and stab-injured brain cortex and its role in astrocyte proliferation. Histochem Cell Biol 136, 501–513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang X, Ransom BR, Ma JF, The role of AQP4 in neuromyelitis optica: More answers, more questions. J Neuroimmunol 298, 63–70 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Zamanian JL et al. , Genomic analysis of reactive astrogliosis. J Neurosci 32, 6391–6410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Artegiani B et al. , A Single-Cell RNA Sequencing Study Reveals Cellular and Molecular Dynamics of the Hippocampal Neurogenic Niche. Cell Rep 21, 3271–3284 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Hutchinson JN et al. , A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkler EA et al. , A single-cell atlas of the normal and malformed human brain vasculature. Science, eabi7377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeisel A et al. , Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014 e1022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Y et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arendsee Z et al. , phylostratr: a framework for phylostratigraphy. Bioinformatics 35, 3617–3627 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Tautz D, Domazet-Loso T, The evolutionary origin of orphan genes. Nat Rev Genet 12, 692–702 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Weber JA et al. , The whale shark genome reveals how genomic and physiological properties scale with body size. Proc Natl Acad Sci U S A 117, 20662–20671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Domazet-Loso T et al. , No Evidence for Phylostratigraphic Bias Impacting Inferences on Patterns of Gene Emergence and Evolution. Mol Biol Evol 34, 843–856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar S, Stecher G, Suleski M, Hedges SB, TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol Biol Evol 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Madeira F et al. , Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karczewski KJ et al. , The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iqbal S et al. , Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci U S A 117, 28201–28211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sayers EW et al. , Database resources of the national center for biotechnology information. Nucleic Acids Res 50, D20–D26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao J et al. , The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corces MR et al. , Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat Genet 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwan KY et al. , SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A 105, 16021–16026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shim S, Kwan KY, Li M, Lefebvre V, Sestan N, Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fertuzinhos S et al. , Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep 6, 938–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Werling DM et al. , Whole-Genome and RNA Sequencing Reveal Variation and Transcriptomic Coordination in the Developing Human Prefrontal Cortex. Cell Rep 31, 107489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malik R et al. , Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 50, 524–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boehme AK, Esenwa C, Elkind MS, Stroke Risk Factors, Genetics, and Prevention. Circ Res 120, 472–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guerreiro R, Bras J, Hardy J, SnapShot: Genetics of ALS and FTD. Cell 160, 798–798 e791 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Nguyen HP, Van Broeckhoven C, van der Zee J, ALS Genes in the Genomic Era and their Implications for FTD. Trends Genet 34, 404–423 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Abrahams BS et al. , SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4, 36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lambert SA et al. , The Human Transcription Factors. Cell 175, 598–599 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Korsunsky I et al. , Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer AL, Ousman SS, Astrocytes and Aging. Front Aging Neurosci 10, 337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tasic B et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruusuvuori E et al. , Neuronal carbonic anhydrase VII provides GABAergic excitatory drive to exacerbate febrile seizures. EMBO J 32, 2275–2286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nagai T et al. , The rewards of nicotine: regulation by tissue plasminogen activator-plasmin system through protease activated receptor-1. J Neurosci 26, 12374–12383 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van Nostrand EL et al. , A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gemechu JM, Bentivoglio M, T Cell Recruitment in the Brain during Normal Aging. Front Cell Neurosci 6, 38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park JE et al. , A cell atlas of human thymic development defines T cell repertoire formation. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y et al. , Decoding the temporal and regional specification of microglia in the developing human brain. Cell Stem Cell 29, 620–634 e626 (2022). [DOI] [PubMed] [Google Scholar]
- 106.Lavin Y et al. , Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dominguez Conde C et al. , Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhaduri A et al. , An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trevino AE et al. , Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069 e5023 (2021). [DOI] [PubMed] [Google Scholar]
- 110.Bian Z et al. , Deciphering human macrophage development at single-cell resolution. Nature 582, 571–576 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Zhang K et al. , A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001 e5919 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bush EC, Lahn BT, A genome-wide screen for noncoding elements important in primate evolution. BMC Evol Biol 8, 17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pollard KS et al. , Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2, e168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bird CP et al. , Fast-evolving noncoding sequences in the human genome. Genome Biol 8, R118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lindblad-Toh K et al. , A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atkinson EG et al. , No Evidence for Recent Selection at FOXP2 among Diverse Human Populations. Cell 174, 1424–1435 e1415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maricic T et al. , A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Mol Biol Evol 30, 844–852 (2013). [DOI] [PubMed] [Google Scholar]
- 118.Vermunt MW et al. , Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci 19, 494–503 (2016). [DOI] [PubMed] [Google Scholar]
- 119.Gittelman RM et al. , Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res 25, 1245–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kozlenkov A et al. , Evolution of regulatory signatures in primate cortical neurons at cell-type resolution. Proc Natl Acad Sci U S A 117, 28422–28432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prabhakar S, Noonan JP, Paabo S, Rubin EM, Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Yu F et al. , Detecting natural selection by empirical comparison to random regions of the genome. Hum Mol Genet 18, 4853–4867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morabito S et al. , Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat Genet 53, 1143–1155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen K et al. , Multiple sclerosis risk gene Mertk is required for microglial activation and subsequent remyelination. Cell Rep 34, 108835 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PFC snRNA-seq data were deposited in the NEMO Archive (RRID:SCR_002001) under identifier nemo:dat-zkgd0it accessible at https://assets.nemoarchive.org/dat-zkgd0it. The data can also be interactively visualized at http://resources.sestanlab.org/PFC. The human PFC sn-multiome data were deposited at NCBI GEO under the accession number: GSE207334. The mouse in utero electroporation RNA-seq data were deposited at NCBI GEO under the accession number: GSE206994. Scripts used in this study are available at Github repository: https://github.com/sestanlab/Cross-species-PFC-snRNA-seq and at Zenodo https://doi.org/10.5281/zenodo.6824096 (60). All other data are available in the main paper or supplement.


