Abstract
Gain-of-function (GOF) mutations in CXCR4 cause WHIM syndrome characterized by infections, leukocyte retention in bone marrow (BM), and blood leukopenias. B-lymphopenia is evident at early progenitor stages, yet why CXCR4 GOF mutations cause B (and T) lymphopenia remains obscure. Using a CXCR4 R334X GOF mouse model of WHIM syndrome, we showed that lymphopoiesis is reduced due to dysregulated mesenchymal stem cell (MSC) transcriptome characterized by a switch from an adipogenic to an osteolineage-prone program with limited lymphopoietic activity. We identify Lymphotoxin beta receptor (LTβR) as a critical pathway promoting IL7 downregulation in MSCs. Blocking LTβR or CXCR4 signaling restored IL7 production and B cell development in WHIM mice. LTβR blocking also increased IL7 and BAFF production in secondary lymphoid organs (SLOs), increasing B and T cell numbers in the periphery. These studies revealed LTβR signaling in BM MSCs and SLO stromal cells limits the lymphocyte compartment size.
One sentence summary:
Increased LTβR signaling in mesenchymal stem cells reduces IL7 production and causes lymphopenia in WHIM syndrome.
Introduction.
Blood cells develop from hematopoietic stem cells (HSCs) through intermediate stages of increasingly proliferative lineage restricted progenitors. The diversity of hematopoietic lineages produced in the bone marrow (BM) is regulated by common (e.g. Kit ligand) and lineage-restricted cytokines (e.g. IL7, IL15, MCSF, GCSF) that are produced by perivascular Leptin-receptor expressing mesenchymal stem cells, periarteriolar pericytes, and endothelial cells (1–3). Recent studies employing single cell RNA sequencing revealed a significant overlap between mesenchymal and endothelial cell that produce myeloid and lymphoid lineage cytokines (4–6). Defects in local cytokine production by conditional gene targeting result in significant reductions in hematopoietic cell production (7–11). Likewise, hematopoiesis is severely reduced when hematopoietic progenitors lack the chemokine receptor CXCR4, or when niche cells are rendered deficient in CXCL12, the ligand for CXCR4 (8, 11–18). Collectively, these findings led us to propose that hematopoietic stem and progenitor cells depend on localization cues, namely CXCR4 and CXCL12, for cell-cell interactions and proximity to specialized niches that produce common and lineage-specific differentiation cytokines (19).
Of all hematopoietic cell lineages, B cells are among the most dependent on CXCR4 signaling for proper development in the BM (8, 13). Early B-lineage committed progenitors upregulate CXCR4 expression and remain in close contact with IL7-producing MSCs (20). Upon productive rearrangement of the preBCR, CXCR4 expression is further increased while α4β1 integrin-mediated adhesion is reduced, which confers increased random motility to preB cells and reduces exposure to IL7 (20, 21). Paradoxically, gain-of-function mutations in CXCR4, which causes WHIM (Warts, Hypogammaglobulinemia, Infections, Myelokathexis) syndrome in humans, also reduces BM B lymphopoiesis (22–26). This is particularly intriguing because hyperresponsive CXCR4 signaling to CXC12 would predict increased exposure to IL7 and consequently increased B cell progenitor proliferation. Instead, studies using a mouse model of a WHIM causing mutation (CXCR4 S338X) showed reduced proB and preB cell development in vivo, and also reduced naïve B and T cell numbers in the periphery (24). These unexpected observations led us to consider the possibility that the BM environment of WHIM patients and mice might be defective in supporting lymphoid lineage development, and to search for mechanisms controlling lymphopoietic niche activity in vivo.
Using single cell RNA sequencing, here we show altered Lepr+ MSC transcriptomic heterogeneity in WHIM mice that is characterized by a switch from a predominant adipogenic to an osteolineage-primed state. These changes reduced IL7 expression by Lepr+ MSCs, which in turn reduced lymphopoietic niche activity in vivo. We further show that Lepr+ MSCs express the Tumor Necrosis Factor (TNF) superfamily member lymphotoxin beta receptor (LTβR), and that excessive LTβR signaling in MSCs is responsible for reduced IL7 production and reduced lymphopoiesis in WHIM mice. In secondary lymphoid organs (SLOs) of WT and WHIM mice, LTβR signaling also reduced IL7 expression and BAFF production by lymph node fibroblastic reticular cells (FRCs). In vivo treatment with an LTβR antagonist or with a CXCR4 antagonist restored IL7 production and rescued B cell development in the BM. LTβR signaling blockade, but not CXCR4 blockade, also elevated IL7 and BAFF production by stromal cells in SLOs and restored naïve B and T cell numbers in the periphery. In summary, these studies revealed LTβR signaling as a negative regulator of B-lymphopoiesis by controlling IL7 expression in the BM, and of B and T cell homeostasis by adjusting IL7 and BAFF production in SLOs.
Results
Altered hematopoietic niche activity in WHIM mice.
To gain insights into mechanisms driving peripheral lymphopenia in WHIM syndrome we generated a mouse model of the most prevalent WHIM-causing R334X mutation in CXCR4 (Supplementary Figure 1A and 1B). Cxcr4WHIM/+ mice had significant reductions in the number of developing B cell subsets in the BM (Fig. 1A), a near 2-fold reduction in splenic transitional B cells (Fig. 1B), and a 2-3 fold reduction in naive B cells in blood and SLOs (Fig. 1C–E). CXCR4 protein was elevated on the surface of developing B cell subsets in the BM and spleen (Fig. 1F and 1G), presumably due to defective CXCL12-mediated CXCR4 desensitization (Fig. 1H). Increased CXCR4 signaling resulted in increased immature B cell retention in BM parenchyma and reduced localization in sinusoids (Fig. 1I and 1J), in agreement with prior studies (27–29). Neutrophils were equally represented in BM and blood, and a trend towards reduced numbers in the spleen of WHIM mice (Supplementary Figure 1C). Monocyte numbers in WHIM mice trended towards reduction in BM and in circulation (Supplementary Figure 1D). CXCR4 protein is also increased on the cell surface of immature neutrophils and monocytes (Supplementary Figure 1F). In WHIM patients, plasmacytoid dendritic cells (PDCs) numbers are reduced in peripheral blood (30), and in CXCR4 WHIM mice PDCs are significantly reduced in the spleen (Supplementary Figure 1G). Collectively, these results were consistent with prior studies using an independently generated WHIM mouse model (24, 25).
Figure 1: Increased lymphocyte retention in BM and reduced lymphopoiesis in WHIM mice.
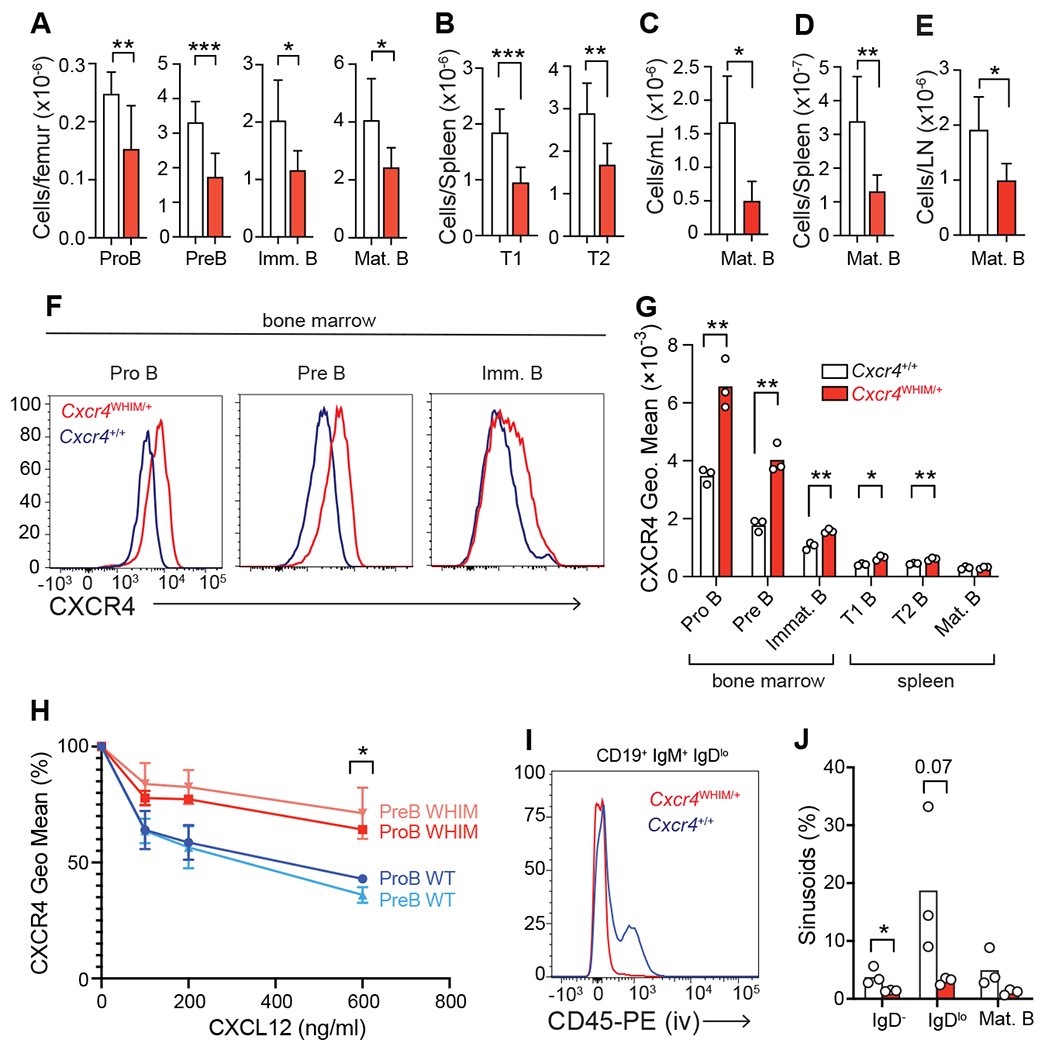
(A) BM B cell numbers of Cxcr4+/+ and Cxcr4WHIM/+ mice. (B) Numbers of transitional (T)1 and 2 B cells in the spleen. (C-E) Numbers of mature follicular (FO) B cells in blood (C), spleen (D), and peripheral lymph node (E). In panels A-E n=6-8 mice. (F and G) CXCR4 surface expression on developing B cells: (F) representative CXCR4 histogram; (G) quantification of CXCR4 geo. mean on developing B cells. (H) CXCL12-mediated CXCR4 desensitization measured on proB and preB cells from WT and WHIM mice. (I and J) BM sinusoidal versus parenchymal localization of B cell subsets in WT and WHIM mice: (I) representative histograms of CD45-PE labeling on Immature B cells 2 minutes after injection intravenously; (J) quantification of developing B cell subsets in sinusoids and parenchymal BM of WT and WHIM mice. Data in all panels are representative of at least 2 independent experiments. Bars indicate mean, error bars show standard deviation, circles depict individual mice. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 by unpaired two-sided Student’s t-test.
The fact that lymphopenia is significant at early IL-7-dependent stages suggested that lymphopoietic niches in the BM (8, 20) might be altered in WHIM mice. To test this possibility, we analyzed the non-hematopoietic cells that form lymphopoietic niches of Cxcr4WHIM/+ and wild-type (WT) littermate mice by droplet-based single cell RNA sequencing (scRNAseq). Non-hematopoietic cells were sorted based on absence of CD45, CD71, CD19 and Ter119 expression, and surface expression of leptin receptor (Lepr) and endothelial cell markers CD31 and CD144 (Supplementary Figure 1H), and mRNA libraries (10x Genomics) were prepared and sequenced (see Methods). A total of 20,837 cells, of which 10,819 cells were Cxcr4WHIM/+ and 10,018 cells were Cxcr4+/+, were sequenced at a depth of 44,395 and 41,108 mean reads/cell, respectively. The mean genes per cell were 1,912 and 1,450 for Cxcr4+/+ and Cxcr4WHIM/+ respectively. After quality control utilizing Seurat (31) and removal of hematopoietic contaminants (see Methods) 7,156 Cxcr4WHIM/+ and 5,442 Cxcr4+/+ cells were analyzed. Cells were visualized using Uniform Manifold Approximation and Projection (UMAP) (32). Unsupervised clustering analysis of the total of 12,598 cells revealed 12 clusters (Table 1). Of these, we identified 5 mesenchymal lineage cells (MSC) characterized by Lepr expression (clusters 0-3 and 5), 1 osteoblast cluster (cluster 4), 2 endothelial cell clusters (1 sinusoidal (cluster 6) and 1 arterial (cluster 7)), 1 fibroblast cluster (cluster 9), 1 chondrocyte cluster (cluster 10) (Fig. 2A, and Supplementary Figure 1I–L), and 2 hematopoietic progenitor cell clusters identified by the expression of genes Pf4, Ppbp (cluster 8) and Rgs1, Ifitm1, Adgrg1 (cluster 11). Since our sorting protocol enriched for Lepr+ MSCs, the number of identified MSC clusters was comparable to or even slightly exceeded that of previous studies (4–6).
Figure 2: Transcriptional heterogeneity of BM non-hematopoietic stromal and endothelial cells of WHIM mice.
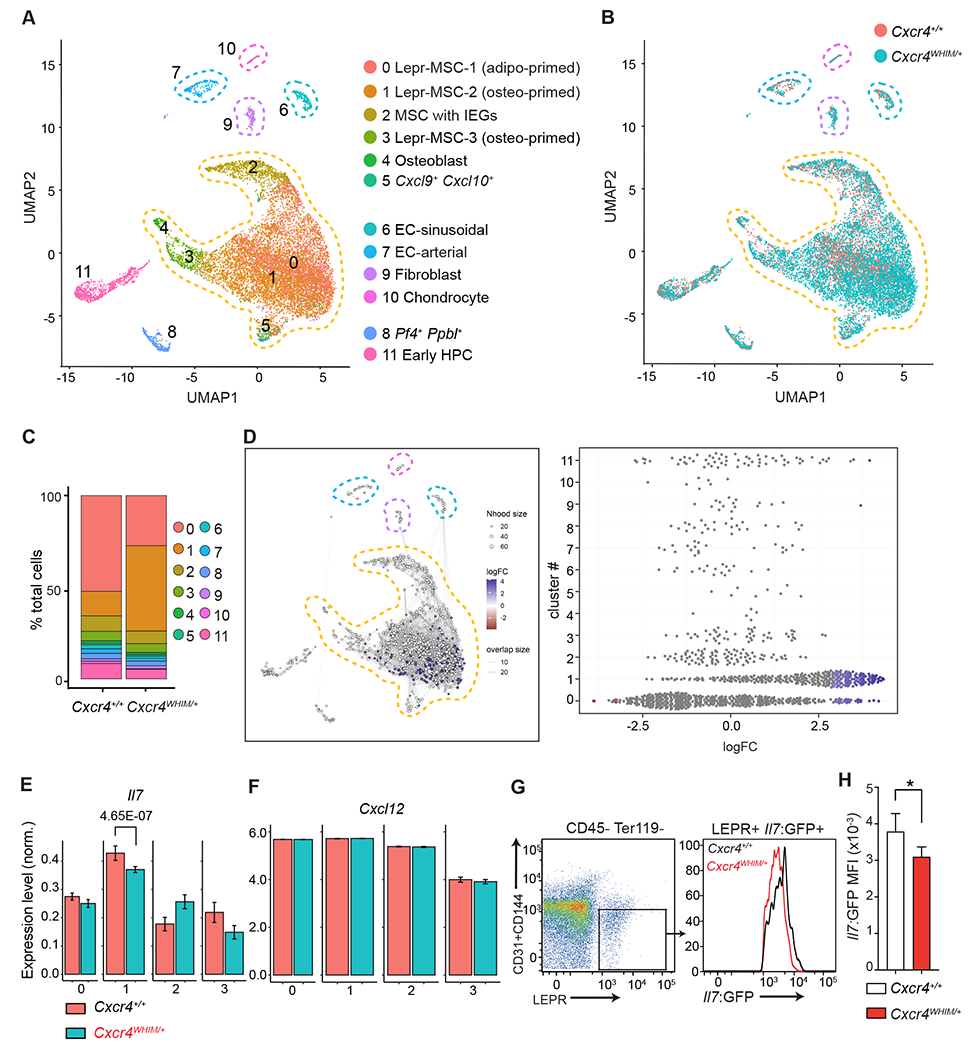
(A) UMAP visualization of scRNAseq data from BM endothelial and stromal cells.
Corresponding clusters are color coded, non-hematopoietic cell populations are marked with dashed lines. (B) Overlay of BM endothelial and stromal cell clusters as in (A) from Cxcr4+/+ and Cxcr4WHIM/+ mice. (C) Cluster representation in Cxcr4+/+ and Cxcr4WHIM/+ samples (D) Differential abundance test with Milo. Graphical representation (left). Beeswarm plot of logFC distribution in neighborhoods of cells from different Seurat clusters (right). Nodes represent neighborhoods colored by logFC between Cxcr4+/+ and Cxcr4WHIM/+. Node size corresponds to number of cells in each neighborhood. Graph edges show the number of cells shared between adjacent neighborhoods. (E) Normalized expression of Il7 in MSC clusters 0-3 of Cxcr4+/+ and Cxcr4WHIM/+ mice. Adjusted p-value is shown. (F) Normalized expression of Cxcl12 in MSC clusters 0-3 of Cxcr4+/+ and Cxcr4WHIM/+mice. (G) Cytometric analysis of Il7 expression in CD45− Ter119− CD31− CD144− LEPR+ MSC of Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ mice. (H) Il7-GFP MFI in Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ MSC. Data in panels G and H are representative of 3 independent experiments with 4-5 mice in each group/experiment. Bars indicate mean, error bars show standard deviation. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 by unpaired two-sided Student’s t-test.
Within MSC clusters, we identified a large cluster of Lepr+ cells with an adipogenic transcriptomic signature (cluster 0), consistent with prior studies (6), and two clusters of osteolineage-primed MSC (clusters 1 and 3). In addition, we identified an MSC cluster with increased expression of several immediate early genes (33) (Fosb, Fos, Zfp36, Jun, Junb, cluster 2), and one MSC cluster characterized by increased expression of the chemokines Cxcl9 and Cxcl10 (cluster 5). Comparison of Cxcr4WHIM/+ and Cxcr4+/+ BM cells confirmed the presence of all clusters in both samples (Fig. 2B and 2C). Differential abundance test between Cxcr4+/+ and Cxcr4WHIM/+ datasets using Milo (34) revealed that the abundance of cellular states is significantly different in clusters 0 and 1 (Fig. 2C and 2D; spatial False Discovery Rate (FDR) < 0.05). Specifically, we found a reduced representation of adipo-primed Lepr+ MSC (cluster 0) and an increase in osteo-primed Lepr+ MSC (cluster 1) in Cxcr4WHIM/+ mice (Fig. 2B and 2C). This alteration in MSC cluster composition was accompanied by statistically significant differential gene expression (DEG) in MSC clusters 0-3, with 210 DEGs in cluster 0, 187 DEGs in cluster 1, 197 DEGs in cluster 2 and 243 DEGs in cluster 3 (|log2FC| >0.2, adjusted p-value (padj) <0.05; Table 2). Among DEGs, Il7 expression was significantly downregulated in cluster 1 (Lepr-MSC-2; Fig. 2E and Supplementary Figure 1M), whereas Cxcl12 expression was similar in all MSC clusters (Fig. 2F and Supplementary Figure 1N). Using an Il7GFP/+ reporter allele, we could also measure significant Il7 downregulation in bulk Lepr+ MSCs isolated from Cxcr4WHIM/+ Il7GFP/+ mice by flow cytometry (Fig. 2G and 2H). Thus, these data suggested that the lymphopoietic activity of BM niches in Cxcr4WHIM/+ mice is reduced.
Lymphotoxin β receptor signaling in MSCs controls Il7 expression.
When examining the transcriptome of BM niche cells, we noted that MSCs, and in particular Lepr+ MSC-2 cells, express lymphotoxin β receptor (LTβR, Fig. 3A and 3B). Given the well-known role for LTβR in the organization of lymphoid compartments in SLOs (35, 36) we considered the possibility that LTβR signaling might also control lymphopoietic niche activity in the BM. To examine if LTβR signaling regulates lymphopoietic niches, we analyzed global transcriptomic changes by RNA sequencing of Lepr+ MSC isolated from C57BL6/J mice treated with a soluble LTβR-Ig decoy (a fusion between LTβR ectodomain and the Fc domain of a mouse IgG1 recognizing Hen Egg Lysozyme) or with control IgG1 (anti-Hen Egg Lysozyme, HEL-Ig) for 3 weeks (Fig. 3C), a period of time that is necessary for the disappearance of follicular dendritic cells from SLOs (37–39). This analysis revealed 199 DEGs between LTβR-Ig and HEL-Ig treated samples (Fig. 3D and 3E, padj < 0.05, and Table 3) of which 113 were downregulated and 86 were upregulated. Among the genes significantly downregulated were several chemokines (Cxcl16, Ccl19, Ccl7, and Ccl2) but not Cxcl12 (Table 3). Importantly, Il7 was significantly upregulated in LTβR-Ig treated mice (Fig. 3E and 3F). Treatment of Il7GFP/+ mice with LTβR-Ig for 3 weeks increased Il7 expression as measured by Il7-GFP expression on gated Lepr+ MSCs (Fig. 3G). The Il7 upregulation was associated with a statistically significant increase in the number of IL7-dependent proB and preB cells in the BM (Fig. 3H). To further examine if LTβR signaling was required in Lepr+ MSCs we generated Il7GFP/+ reporter mice with Ltbr conditionally deficient Lepr+ MSCs (Il7GFP/+ LeprCre/+ Ltbrfl/fl; Fig. 3I). Importantly, Il7-GFP expression was significantly increased in LTβR-deficient MSCs (Fig. 3J), which led to an increase in proB and preB cell numbers in the BM (Fig. 3K).
Figure 3: LTβR signaling in MSCs and control of Il7 expression.
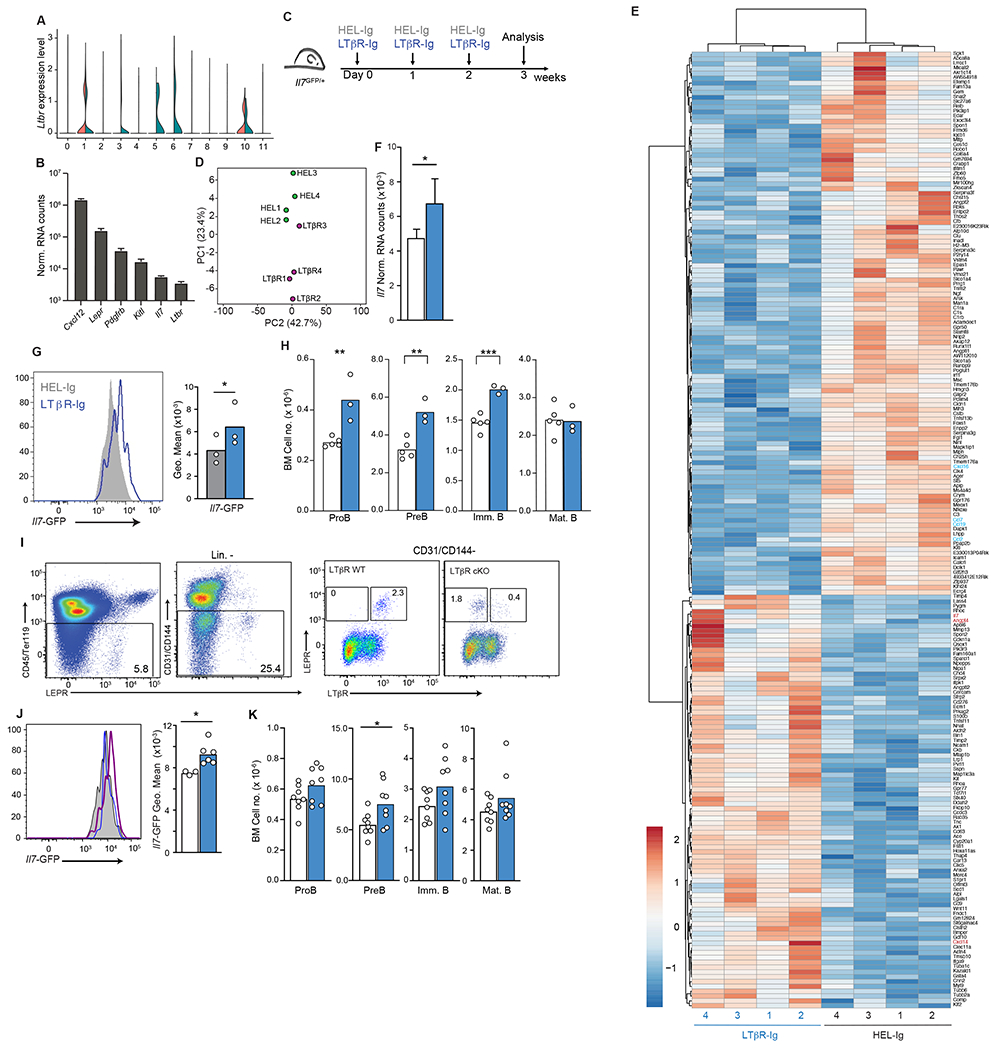
(A) Expression of Ltbr in BM endothelial and stromal cells of Cxcr4+/+ and Cxcr4WHIM/+ mice as violin plots. Data from clusters 0-11 of scRNAseq analysis as shown in Figure 1. (B) Normalized mRNA counts of Ltbr and key niche genes in sorted MSC measured by bulk RNA sequencing. (C) Experimental setup for LTβR signaling inhibition in Il7GFP/+ mice for 3 weeks. (D) PCA distribution plot (E) Differentially expressed genes (LogFC < 0.5 and adjust p< 0.05) between MSC from LTβR-Ig treated mice and HEL-Ig treated C57BL6/J mice. (F) Normalized Il7 expression in MSC from LTβR-Ig treated mice (blue, n=4) and HEL-Ig treated controls (white, n=4). (G) Histogram of Il7-GFP in MSCs of Il7GFP/+ mice treated with LTβR-Ig (blue) or HEL-Ig (grey). (H) Cell numbers of B cell developmental stages in the BM of LTβR-Ig treated (blue) and HEL-Ig treated (grey) WT mice. (I) Cytometric analysis of surface LTβR in MSCs of Lepr+/+ Ltbrfl/fl (LTβR WT) and LeprCre/+ Ltbrfl/fl mice (LTβR cKO). (J) Histogram of Il7-GFP in MSC of LTβR cKO and LTβR WT Il7GFP/+ mice. Histograms (left), grey: LTβR WT, blue: LTβR cKO, LTβR+ MSC, purple: LTβR cKO, LTβR− MSC. Bar graph (right), white: LTβR WT MSC, blue: LTβR cKO, LTβR− MSC. (K) Cell numbers of B cell developmental stages in the BM of LTβR WT (white) and LTβR cKO (blue) mice. Data in panels G-K are representative of 3 independent experiments. Bars indicate mean, circles depict individual mice. * p ≤ 0.05; ** p≤ 0.005; *** p≤ 0.0005 by unpaired two-sided Student’s t-test.
LTβR signaling blockade restores Il7 expression and lymphopoiesis in Cxcr4WHIM/+ mice
The lymphotoxin heterotrimer LTα1β2 and LIGHT are membrane-bound ligands of LTβR expressed on multiple hematopoietic cells (35, 36). In the BM, early hematopoietic progenitors showed measurable amounts of LTα1β2 expression, including developing B cells and myeloid cell subsets (Supplementary Figure 2A–D). In Ltb−/− mice, MSCs expressed higher Il7-GFP than Ltb+/+ mice (Supplementary Figure 2E), which correlated with a trend towards increased developing B cell subsets in the BM (Supplementary Figure 2F). Because IL7-producing MSCs express the highest amounts of CXCL12 in BM (Ref. 8, and Supplementary Figure 1N), we reasoned that CXCR4 GOF mutations not only increases lymphocyte retention in the parenchyma (Fig. 1I and 1J) but might also increase interactions between lymphocytes (and other leukocytes) and MSCs. In turn, increased leukocyte/MSC interactions could result in increased LTβR signaling and consequently reduced IL7 production in BM MSCs (Fig. 4A). Of note, naïve B cells are major sources of LTα1β2 required for the maintenance of stromal cell networks in SLOs (40–42). CXCR4 signaling promotes LTα1β2 expression in B cells (40), and in WHIM mice B cells also express significantly more membrane-bound ligands of LTβR (Fig. 4B and 4C). Expression of membrane-bound LTβR ligands trended towards increased in neutrophils and monocytes of WHIM mice, but this difference did not reach statistical significance (Supplementary Figure 2G). LTβR protein expression was equivalent in WHIM and WT MSCs (Supplementary Figure 2H). Consistent with the possibility that LTβR signaling is altered in WHIM MSCs, the LTβR-regulated genes Ccl19, Cxcl13, Ch25h, and Cyp7b1 (39, 43) were significantly increased in MSCs from WHIM mice (Fig. 4D). To test if increased LTβR signaling in MSCs impairs lymphopoiesis, we treated Cxcr4WHIM/+ Il7GFP/+ and Cxcr4+/+ Il7GFP/+ mice with LTβR-Ig or with control HEL-Ig for 1 week (Fig. 4E) and analyzed Il7-GFP expression in MSCs. Il7 expression was significantly increased in LTβR-Ig treated mice (Fig. 4F and 4G) whereas Cxcl12 expression remained unchanged (Supplementary Figure 3A and 3B). Importantly, proB, preB and immature B cell numbers in the BM of LTβR-Ig treated Cxcr4WHIM/+ mice were significantly increased to levels comparable to that of HEL-Ig-treated Cxcr4+/+ mice (Fig. 4H). Prolonging LTβR-Ig treatment to 3 weeks restored naive B cell numbers in the blood and spleen of Cxcr4WHIM/+ mice, but it also increased naive B cell numbers in Cxcr4+/+ mice (Fig. 4I and 4J). This change, however, was accompanied by a more than 2-fold reduction in B cell numbers in peripheral lymph nodes exclusively in Cxcr4+/+ mice (Fig. 4K), in agreement with a role for LTβR signaling in maintenance of high endothelial venules (44). The overall effect of LTβR-Ig treatment restored B cell numbers in Cxcr4WHIM/+ mice to similar numbers as in control treated Cxcr4+/+ mice (Fig. 4L). Similar findings were obtained in Cxcr4WHIM/+ mice carrying conditional Ltbr deletion in Lepr+ MSCs (Fig. 4M and 4N), even though Lepr-cre mediated Ltbr deletion also impacted the development of follicular dendritic cells (Supplementary Figure 3L). Taken together, these results showed that inhibition of LTβR signaling normalized BM Il7 expression, rescued B cell progenitor numbers in the BM of Cxcr4WHIM/+ mice, and elevated naïve B cell numbers in the periphery.
Figure 4: LTβR signaling blockade and effects on Il7 expression and lymphopoiesis in WHIM mice.
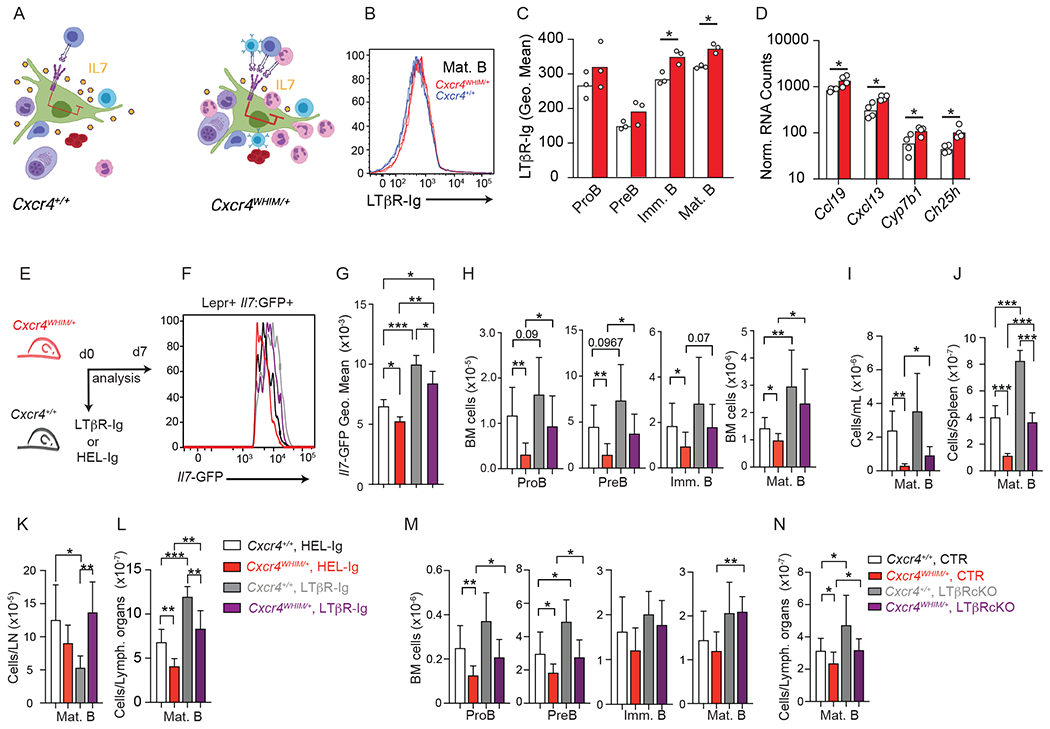
(A) Model of interactions between lymphotoxin-expressing leukocytes and LTβR-expressing MSCs in WT and WHIM mice generated with Biorender. (B and C) LTβR ligand expression on WT an WHIM B-lineage cells in BM: (B) Histogram of LTβR-Ig binding to mature B cells in BM of WT and WHIM mice; (C) Quantification of LTβR-Ig binding to developing B cells. (D) Normalized RNA counts of LTβR-regulated genes in WT and WHIM MSCs; data analyzed from bulk RNA sequencing. (E) Experimental setup for LTβR signaling inhibition in Cxcr4+/+ and Cxcr4WHIM/+ mice. (F) histogram of Il7-GFP in MSCs of Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ mice after 1 week of LTβR-Ig or HEL-Ig treatment. (G) Geo. Mean of Il7-GFP in MSCs of Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ mice. (H) Cell numbers of B cell developmental stages in the BM of Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ mice after 1 week of LTβR-Ig and HEL-Ig treatment. Mature B cell numbers in blood (I), spleen (J), lymph nodes (K) and combined SLOs (L) of Cxcr4+/+ and Cxcr4WHIM/+ mice after 3 weeks of LTβR-Ig or HEL-Ig treatment. (M and N) Cell numbers of B cell developmental stages in the BM (M), and peripheral mature B cell numbers (N) of Ltbrfl/fl LeprCre/+ (LTβR cKO) or control littermate mice (CTR) carrying Cxcr4+/+ or Cxcr4WHIM/+ alleles. Data in all panels are representative of at least 3 independent experiments with n= 4-6 mice. Bars indicate mean, error bars show standard deviation. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 by unpaired two-sided Student’s t-test.
Besides restoring B cell numbers, LTβR-Ig treatment significantly increased the number of CD4+ and CD8+ T cells in the spleen (Supplementary Figure 3C), while having less effects in T cell subsets in blood and lymph nodes (Supplementary Figure 3D and 3E). In Cxcr4WHIM/+ BM, CD4+ and CD8+ T cell numbers were increased by ~2- to 3-fold (Supplementary Figure 3F) presumably due to increased trafficking of Cxcr4WHIM/+ T cells to the BM (Supplementary Figure 3M–P). The numbers of developing T cell subsets in the thymus were unaffected by LTβR-Ig treatment (Supplementary Figure 3G–I). LTβR-Ig treatment restored the overall total number of CD4+ and CD8+ T cells present in combined SLOs to levels equivalent to that of control mice treated with HEL-Ig (Supplementary Figure 3J). T cell subsets also recovered in LTβR cKO Cxcr4WHIM/+ mice although with reduced magnitude compared to LTβR-Ig treatment (Supplementary Figure 3K), possibly due to Ltbr deletion in some Lepr-expressing stromal cells in SLOs (Supplementary Figure 3L).
CXCR4 antagonism normalizes Il7 expression and rescues B and T cell lymphopenia in Cxcr4WHIM/+ mice
To further establish a relationship between CXCR4 GOF mutations and increased LTβR signaling in BM MSCs, we treated Cxcr4WHIM/+ mice with an orally bioavailable CXCR4 antagonist by daily gavage (45). Remarkably, this treatment phenocopied the effects seen with LTβR-Ig treatment. Specifically, Il7-GFP expression in BM MSCs was significantly increased in Cxcr4WHIM/+ mice to levels seen in control treated Cxcr4+/+ mice, and rescued proB cell numbers to that of wild-type levels (Fig. 5A–C). The effects on preB and downstream developing B cell subsets were less evident due to the fact that these subsets are more dependent on active CXCR4 signaling for BM retention than proB cells (Supplementary Figure 4A) (27, 29, 46, 47). Prolonged CXCR4 antagonism also elevated naïve B cell numbers in circulation and in SLOs (Fig. 5D–F). Similarly, CXCR4 antagonist treatment increased CD4+ and CD8+ T cell numbers in spleen and blood of Cxcr4WHIM/+ mice (Supplementary Figure 4B and 4C), and CD8+ T cells in pLN (Supplementary Figure 4D), and reduced homing to the BM (Supplementary Figure 4E). Thymocyte development was not significantly affected by CXCR4 antagonism treatment (Supplementary Figure 4F–H). Furthermore, Il7 expression in cortical and medullary epithelial cells was equivalent in WT and WHIM mice and was unaffected by CXCR4 antagonism (Supplementary Figure 4I and 4J). Interestingly, LTβR-Ig treatment increased Il7-GFP expression in medullary thymic epithelial cells but not cortical thymic epithelial cells of both WT and WHIM mice (Supplementary Figure 4K). This finding agrees with prior studies showing a role for LTβR signaling in mTEC differentiation (48).
Figure 5: CXCR4 signaling blockade and effects on Il7 expression and lymphopoiesis in WHIM mice.
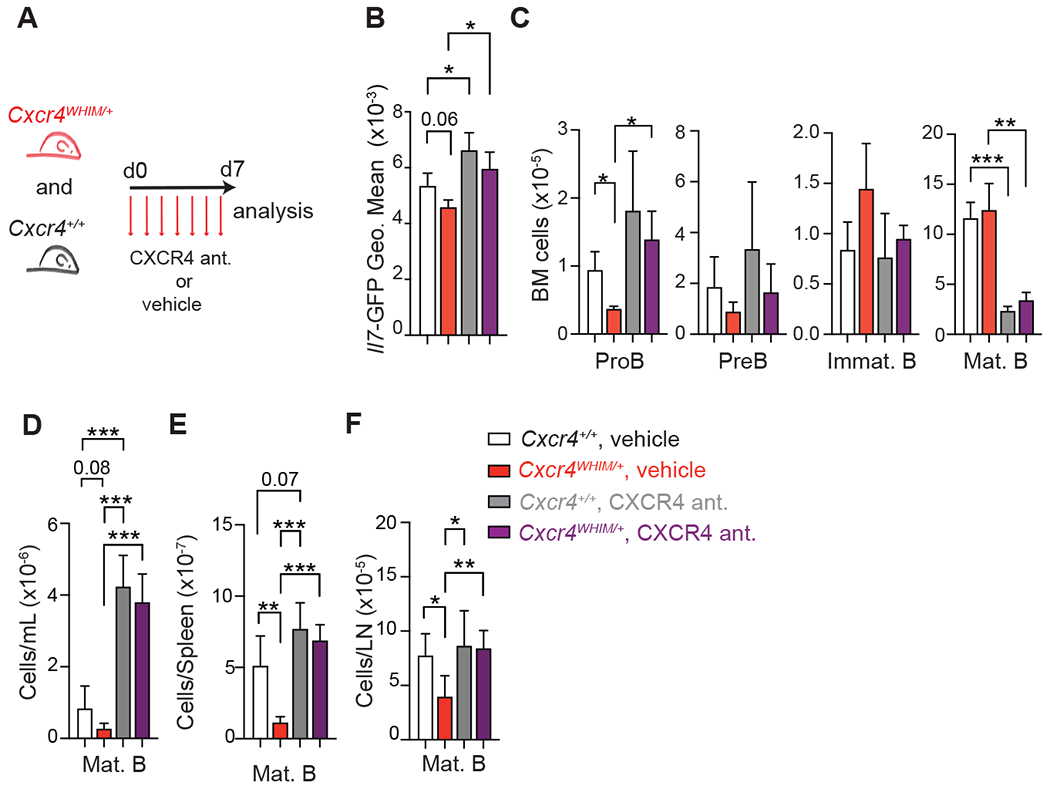
(A) Experimental setup for CXCR4 signaling inhibition in Cxcr4+/+ and Cxcr4WHIM/+ mice for 1 week. (B) Geo. Mean of Il7-GFP in MSC of Cxcr4+/+ and Cxcr4WHIM/+ Il7GFP/+ mice after 1 week of CXCR4 signaling inhibition. (C) Cell numbers of B cell developmental stages in the BM of Cxcr4+/+ and Cxcr4WHIM/+ mice after 1 week of CXCR4 signaling inhibition. Mature B cell numbers in blood (D), spleen (E), and in peripheral lymph nodes (F) of Cxcr4+/+ and Cxcr4WHIM/+ mice after 3 weeks of CXCR4 signaling inhibition. Data are representative of 2 independent experiments with n=4-6 mice. Bars indicate mean, error bars show standard deviation. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 by unpaired two-sided Student’s t-test.
Peripheral niches for lymphocyte homeostasis in Cxcr4WHIM/+ mice are intact.
Naïve B and T cell numbers are maintained by BAFF and IL7, respectively, and both cytokines are predominantly produced by fibroblastic reticular cells (FRCs) in SLOs. As FRCs also express CXCL12, we considered the possibility that dysregulated stromal cells in SLOs contribute to peripheral B and T cell lymphopenia in Cxcr4WHIM/+ mice. To test this possibility, we compared splenic and lymph node stromal cell transcriptomes of Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq. We FACS purified non-hematopoietic and non-endothelial cells based on absence of CD45, Ter119, CD71, CD31 and CD144 expression and generated mRNA libraries (10x Genomics) for high throughput deep sequencing. We analyzed 14,516 and 6,386 cells from Cxcr4WHIM/+ and Cxcr4+/+ spleens at a depth of 30,045 and 57,145 mean reads/cell, respectively. After quality control and removal of hematopoietic cells, 1283 Cxcr4WHIM/+ and 2201 Cxcr4+/+ cells were selected for further analyses. Unsupervised Leiden clustering analysis revealed 12 stromal cell clusters overlaid on UMAP plot (Fig. 6A, and Supplementary Figure 5A and Table 4). Specifically, cluster 0 is characterized by high expression of small leucin-rich proteoglycans Decorin and Lumican, and collagen 1 type 1a, that form the extracellular matrix (Fig. 6A and Supplementary Figure 5A and 5B). These cells are transcriptionally similar to red pulp and sub-capsular fibroblasts (49). Cells in cluster 1, characterized by high expression of aorta smooth muscle actin 2 (Acta2) and Transgelin (Tagln), also express abundant amounts of canonical mural cell markers (e.g. Pdgfrb, Cspg4). Cluster 2, marked by Upk3b, Igfbp6 and Krt19 expression, shares a transcriptional gene expression signature with Mesothelin-expressing mesothelial cells that form the outer layer of the splenic capsule (Fig. 6A and Supplementary Figure 5A, 5B, and 5F) (49). Cluster 3 represents a subset of red pulp fibroblasts that express the highest amount of Cxcl12 in the spleen (Fig. 6A and Supplementary Figure 5A–D), express Stem Cell Factor (Kitl, Supplementary Figure 5C), and resemble perivascular stromal cells of the red pulp that form a niche for hematopoietic stem cells (50). Cluster 4 cells express high amounts of genes associated with proliferative hematopoietic progenitors (e.g. Mik67, Kit, Lmo2, Ikzf1, Hmgb2, Stmn1, Fig. 6A and Supplementary Figure 5E). The increased representation of cluster 4 cells in WHIM samples (Fig. 6A and 6C) agrees with prior studies showing increased extramedullary hematopoiesis in the spleen of Cxcr4WHIM/+ mice (25). Cluster 5 also shares transcriptional similarities with cells in Cluster 1 and Mural cells with the notable exception that they lack Cspg4 expression (Fig 6A and Supplementary Figure 5A–C). Cluster 8 represents stromal cells associated with white pulp regions of the spleen as indicated by abundant expression of T-zone stromal cells (TRCs) defined by Ccl19 and Ccl21 expression; and genes defining B cell follicle stromal cell subsets (e.g. Cxcl13, Cr2, Ch25h, and Pthlh, Fig. 6A and Supplementary Figure 5A–B and 5G) (51). Finally, we also detected cells with a transcriptional signature of Schwann cells (cluster 6, Fig 6A and Supplementary Figure 5A–B), endothelial cells (cluster 7, Supplementary Figure 5A–B), and traces of other cell contaminants (clusters 9-11, Supplementary Figure 5A–B, Table 4).
Figure 6: Transcriptional heterogeneity of splenic stromal cells of WHIM mice.
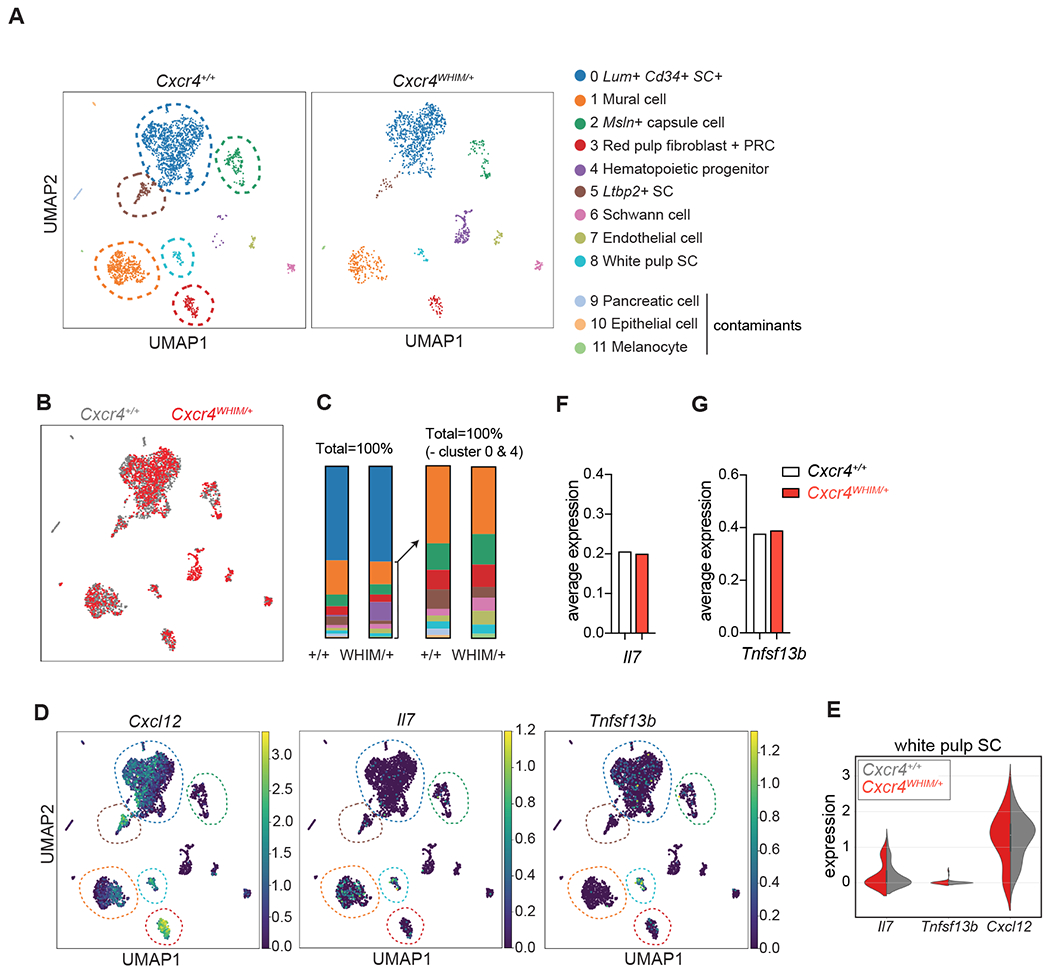
(A) UMAP visualization of scRNAseq data from CD45− Ter119− CD71− LIN− CD31− splenic stromal cells of Cxcr4+/+ and Cxcr4WHIM/+ mice. Corresponding clusters are color coded, stromal cell populations are marked with dashed lines. (B) Overlay of splenic stromal cell clusters of Cxcr4+/+ and Cxcr4WHIM/+ mice. (C) Cluster representation in Cxcr4+/+ and Cxcr4WHIM/+ samples; Left graph shows all clusters, right graph shows clusters 1-11 excluding cluster 4 (hematopoietic progenitors). (D) Cxcl12, Il7 and Tnfsf13b expression visualized on UMAP of data pooled from Cxcr4+/+ and Cxcr4WHIM/+ samples. Colors indicate average expression value. (E) Violin plots for Cxcl12, Il7 and Tnfsf13b in cluster 8 of Cxcr4+/+ and Cxcr4WHIM/+ mice. (F and G) Average expression of Il7 (F) and Tnfsf13b (G) in white pulp stromal cells (cluster 8) of Cxcr4+/+ and Cxcr4WHIM/+ mice.
In contrast to BM MSCs, the datasets obtained from Cxcr4WHIM/+ and Cxcr4+/+ splenic stromal cells showed minimal differences in cluster representation (Fig. 6B and 6C). Among non-hematopoietic clusters, only the Ltbp2high cell frequency was reduced in the Cxcr4WHIM/+ sample. Furthermore, expression profiles of Cxcr4WHIM/+ and Cxcr4+/+ clusters were largely comparable, with some DEGs detected (padj <0.05; cluster 0: 92 DEGs, cluster1: 10 DEGs, and cluster 2: 13 DEGs; Table 5), but none of these genes regulate B or T cell numbers in the periphery. Furthermore, white pulp stromal cells (cluster 8) expressed similar amounts of Il7 and Tnfsf13b (encodes BAFF; Fig. 6D–G) indicating that the defect seen in peripheral B and T cells could not be explained by altered production of homeostatic B and T cell cytokines. It should be noted that the total number of cells in cluster 8 (white pulp stromal cells) was too small for reaching a definitive conclusion.
To gain in depth resolution of differences in stromal cell composition (and gene expression) that regulate peripheral lymphocyte survival and homeostasis, we also examined stromal cells isolated from peripheral lymph nodes from Cxcr4WHIM/+ and Cxcr4+/+ mice by scRNAseq. Leiden cluster analysis revealed 7 clusters, including 1 large cluster of TRC/MRC/FDC (cluster 1) which was identified based on expression of classical markers such as Ccl19, Ccl21a, Enpp2, Tnfsf11 and Cr2 (Supplementary Figure 6A, 6D–E, 6H; and Table 6). We also detected previously described clusters of perivascular cells (PvC) and 3 clusters of Cd34+ SC (cluster 0, 2 and 3; Supplementary Figure 6A, and Table 6) (51), as well as 1 epithelial cell cluster (cluster 5) and 2 Schwann cell clusters (cluster 6 and 7). The cluster composition was largely comparable in Cxcr4WHIM/+ and Cxcr4+/+ lymph nodes (Supplementary Figure 6B–C), with relatively small numbers of DEGs detected between Cxcr4WHIM/+ and Cxcr4+/+ samples (Table 7) for each cluster. None of these genes are known to regulate lymphocyte homeostasis. Furthermore, neither Il7, Tnfsf13b, or Cxcl12 were differentially expressed between Cxcr4WHIM/+ and Cxcr4+/+ stromal cell clusters (Supplementary Figure 7A–C). Combined, these data suggested strongly that the peripheral B and T cell lymphopenia was independent of measurable changes in stromal cell composition or gene expression in Cxcr4WHIM/+ mice.
Altered B and T cell trafficking through SLOs in WHIM mice.
T cell recirculation between blood and SLOs is critical for T cell survival due to their dependency on IL-7 presumably produced by FRCs (51–53), and on S1P (54). Likewise, transitional and mature B cells require BAFFR signaling for differentiation and survival, and the cellular sources of BAFF are radiation resistant stromal cells (55, 56). When examining the relationship between cell clusters expressing CXCL12 and clusters capable of producing the B and T cell survival cytokines BAFF and IL7 respectively, we noted that CXCL12 high cells (cluster 3) did not express either cytokine (Fig. 6D). Even though Il7 and Tnfsf13b expression in stromal cell subsets of the spleen and lymph nodes were normal in Cxcr4WHIM/+ mice, we considered the possibility that B and T cell access to survival niches was affected by CXCR4 GOF mutations. To test this possibility, we transferred fluorescently labelled Cxcr4WHIM/+ (CFSE+) and Cxcr4+/+ B cells (CMTMR+) into C57BL6/J recipients and examined cell localization by flow cytometry and confocal microscopy 16 hours after adoptive transfer (Fig. 7A). We found Cxcr4WHIM/+ B cells preferentially home to the BM, while homing to spleen and pLN was significantly reduced (Fig. 7B). Of the fewer cells reaching the spleen, the majority localized in the red pulp (Fig. 7C and 7D). Within the white pulp, WHIM and WT B cells localized normally in the T-zone and follicular areas, with no significant differences found in their localization throughout distinct niches (Fig. 7E). Likewise, even though fewer WHIM B cells entered peripheral lymph nodes, WT and WHIM B cells distributed equivalently throughout follicular and T-zone niches (Supplementary Figure 7D and 7E). Transitional differentiation of splenic B cells expressing the WHIM allele was significantly reduced under competitive conditions (Fig. 7F). Consistent with reduced access to BAFF-producing stromal niches, we noted increased expression of apoptotic markers in WHIM-expressing transitional B cell subsets, which normalized after treatment with LTβR-Ig (Fig. 7G). LTβR-Ig treatment did not correct the abnormal trafficking of Cxcr4WHIM/+ B cells in vivo (Fig. 7H–K), which is expected given the fact that LTβR signaling controls CXCL13, CCL19 and CCL21 expression in SLO stromal cells (38). This observation raised the possibility that systemic LTβR signaling blockade corrected transitional B cell differentiation, and possibly mature B cell survival, through a mechanism independent of increased accessibility to lymphocyte survival niches in the periphery and increased IL-7 production in the BM.
Figure 7: B cell trafficking and transitional B cell differentiation in WHIM mice.
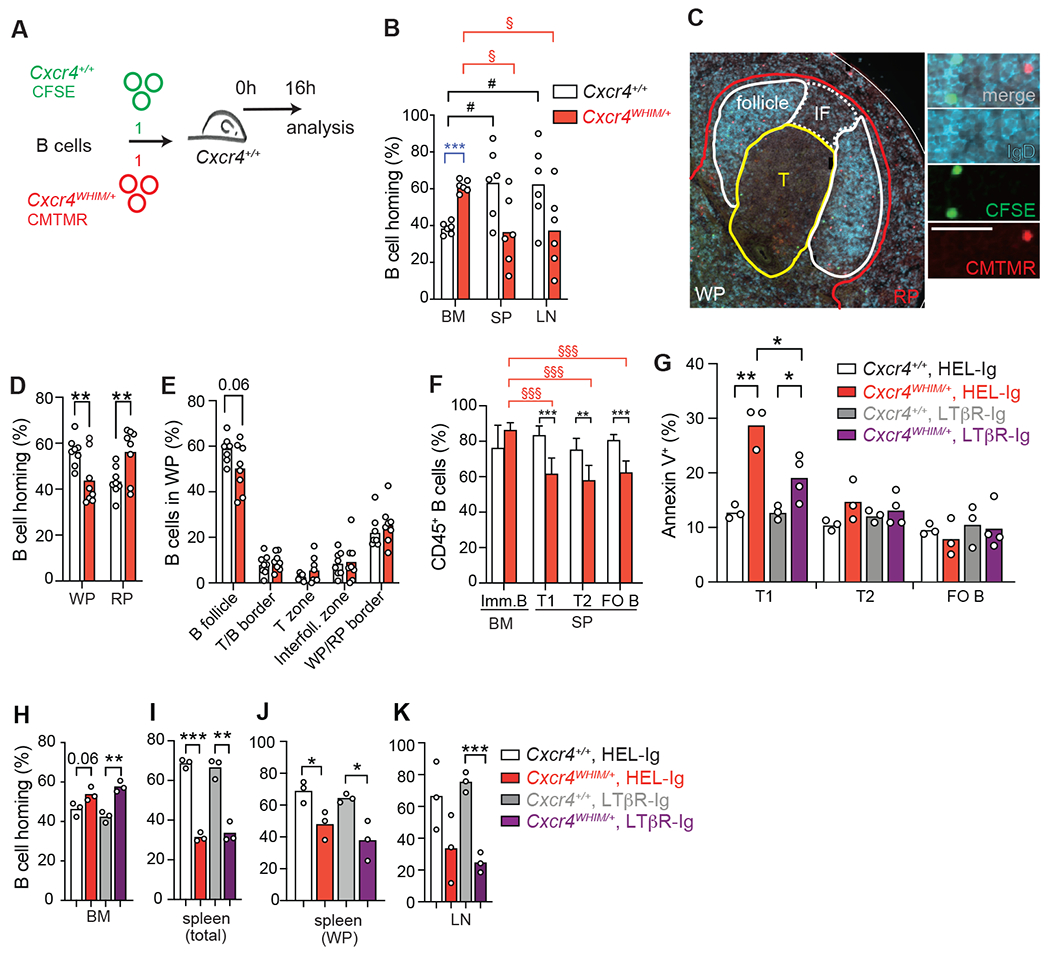
(A) Experimental setup for adoptive transfer of fluorescently labelled Cxcr4WHIM/+ (CFSE+) and Cxcr4+/+ (CMTMR+) B cells into wildtype recipients. (B) Distribution of Cxcr4WHIM/+ and Cxcr4+/+ B cells 16 hours after adoptive transfer to C57BL6/J recipient mice. BM (BM), spleen (SP) and peripheral lymph nodes (LN). Statistical comparisons were as follows: * = for Cxcr4+/+ vs. Cxcr4WHIM/+ B cells; § = between organs of Cxcr4WHIM/+ mice. (C) Histology of splenic Cxcr4+/+and Cxcr4WHIM/+ follicles. Sections were stained for IgD (blue) and nuclei (DAPI, grey); WP = white pulp, RP = red pulp, T = T zone, IF = interfollicular zone. Scale bar is 20μm. (D) Distribution of homed Cxcr4WHIM/+ (CFSE, green) and Cxcr4+/+ (CMTMR, red) B cells in splenic red (RP) and white pulp (WP). (E) Distribution of Cxcr4+/+ and Cxcr4WHIM/+ B cells within splenic lymphoid niches. Data pooled from four experiments. For panels B, D and E, bars indicate mean, circles represent individual mice. (F) B cell chimerism 8 weeks after mixed BM transplantation of CD45.2+ Cxcr4+/+ or Cxcr4WHIM/+ and CD45.1+ cells (8:2 ration), n=5 mice/group. (G) Frequency of Annexin V+ cells in transitional (T1, T2) and mature follicular B cell subsets (FO B) in Cxcr4+/+ and Cxcr4WHIM/+ spleens from mice pre-treated with LTβR-Ig or HEL-Ig (black) for 24h. (H-K) Distribution of Cxcr4WHIM/+ and Cxcr4 B cells 24h after transfer into C57BL6/J mice pre-treated with LTβR-Ig or HEL-Ig (black) for 24h. (H) BM; (I) total spleen; (J) splenic white pulp (WP); and lymph node (K). Bars indicate mean, error bars show standard deviation. Data in all panels are representative of 2-4 independent experiments. * = p ≤ 0.05, ** = p ≤ 0.01, *** = p ≤ 0.001 by unpaired two-sided Student’s t-test.
LTβR signaling regulates IL-7 and BAFF production in SLOs
While systemic LTβR and CXCR4 blockade acted equivalently in restoring lymphoid niche activity in BM MSCs, these treatments seemed to differ in mode of action in the periphery. Specifically, while CXCR4 blocking reduced B and T cell trafficking through the BM and facilitated trafficking through the periphery, LTβR-Ig treatment had no effect on CXCL12 expression by BM MSCs (Supplementary Figure 3A and 3B) and by splenic and lymph node stromal cells (Fig. 8A and 8B) consistent with prior observations (38). This observation led us to consider the possibility that LTβR signaling controls IL7 and BAFF production in SLOs such that it could compensate for reduced B and T cell trafficking to niches supporting lymphocyte survival. To test this possibility, we measured Il7 and Tnfsf13b transcripts in FACS sorted lymph node FRCs of C57BL6/J mice treated with HEL-Ig or LTβR-Ig. Indeed, both transcripts were upregulated by 2-3 fold when LTβR signaling was blocked for 1 week (Fig. 8E–G). Although we could not reliably detect IL7, we could measure increased BAFF protein by flow cytometry analyses of permeabilized lymph node FRCs of LTβR-Ig treated mice (Fig. 8H and 8I). Increased BAFF production could be monitored by staining BAFF bound to BAFF receptors on B cells in lymph nodes of WT and WHIM mice treated with LTβR-Ig (Fig. 8J and 8K), but not with CXCR4 antagonist (Fig. 8L). B cells from untreated or control treated WHIM mice also showed higher BAFF staining on the surface (Fig. 8J and 8K). This finding was consistent with a small but significant increase in BAFF concentration in the serum of WHIM patients when compared to healthy volunteers, but lower than the BAFF concentration measured in patients with a partial RAG deficiency (Fig. 8L). Prior studies demonstrated an inverse correlation between the magnitude of lymphopenia seen in primary immune deficient patients and serum BAFF concentration (57). Likewise, the severe lymphopenia seen in partial RAG deficient patients correlates with high BAFF concentration in serum of these patients (Fig. 8L) and suggests that WHIM patients are also afflicted by a mild, systemic, lymphopenia as seen in WHIM mice. In summary, these findings showed that LTβR signaling controls the production of homeostatic cytokines IL7 and BAFF by SLO stromal cells and thereby controls the size of the peripheral lymphocyte compartment.
Figure 8: LTβR signaling blockade and effects on Il7 and Tnfsf13b expression by fibroblastic reticular cells.
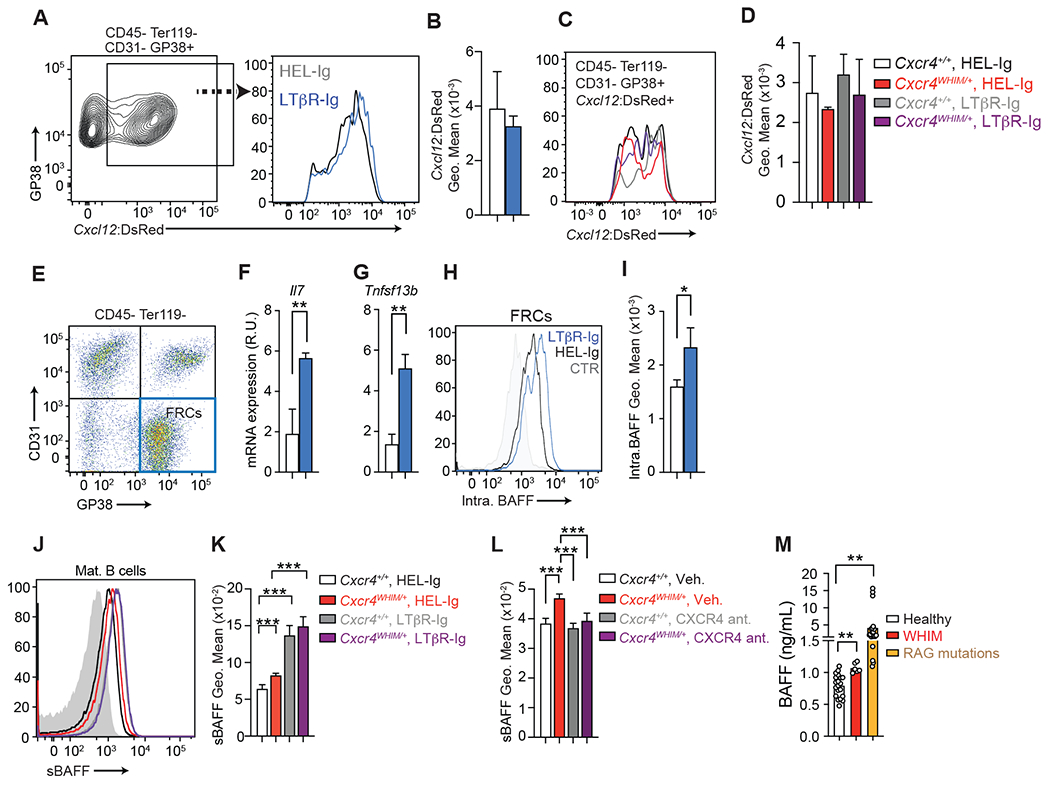
(A) Cxcl12-DsRed expression in GP38+ CD31− lymph node stromal cells after one week of LTβR-Ig (blue) and HEL-Ig (black) treatment. Left: gating strategy; Right: Cxcl12-DsRed histogram on gated FRCs (B). Cxcl12-DsRed geo. mean on FRCs described in panel A. (C) Cxcl12-DsRed histogram overlay of FRCs isolated from Cxcr4+/+ and Cxcr4WHIM/+ lymph nodes HEL-Ig or LTβR-Ig treated for 1 week. (D) Cxcl12-DsRed geo. mean on FRCs described in panel C. Data in panels B-D generated from n=3-4 mice/group. (E) Sorting gate of GP38+ CD31− lymph node stromal cells. (F and G) FRC gene expression: Il7 (F) and Tnfsf13b (G). mRNA expression is relative to Hprt. Data in panels F and G were generated with FRCs sorted from individual mice (n=3-4 mice/group). (H) Intracellular BAFF staining in lymph node FRCs after one week of LTβR-Ig (blue) and HEL-Ig (black) treatment. Control (grey) shows staining with anti-BAFF antibody pre-incubated with recombinant BAFF. (I) BAFF geo. mean on FRCs described in panel H. Data in panels H-I generated from n=3-4 mice/group. (J and K) Histogram of BAFF bound to mature B cell plasma membrane in lymph nodes of Cxcr4+/+ and Cxcr4WHIM/+ mice 3 weeks after treatment with LTβR-Ig and HEL-Ig (J) or with CXCR4 antagonist (K); data generated from n=3-4 mice/group. In panels B, D, F, G, L, J and K, bars indicate mean, error bars show standard deviation. (L) BAFF concentration (ng/mL) in human sera obtained from healthy donors, WHIM patients, and from patients with mutations in RAG gene causing partial RAG deficiency. Bars indicate mean, circles depict individual donors. For panels (B-G), bars indicate mean, error bars show standard deviation. For all panels, * = p ≤ 0.05, ** = p ≤ 0.01, *** = p ≤ 0.001 by unpaired two-sided Student’s t-test.
Discussion.
HSCs and hematopoietic progenitors rely predominantly on CXCR4/CXCL12 to encounter common and lineage-specific hematopoietic cytokines produced by niche cells (19). Consequently, hematopoietic cells engage niche cells for extended periods of time (20, 58, 59), which offers opportunities for cellular crosstalk and delivery of regulatory signals. In this study we showed that LTβR expressed in Lepr+ MSCs inhibits IL7 expression in vivo, but the magnitude of IL7 modulation, even though significant under homeostatic conditions, only modestly reduced B cell development. When the CXCR4/CXCL12 pathway is activated at supra-physiological levels, as seen in CXCR4 GOF that cause WHIM syndrome, the cross-talk between hematopoietic and niche cells is presumably intensified such that it causes a switch in the MSC pro-adipogenic gene expression program to an osteolineage program characterized by low IL7 production and reduced lymphopoietic activity. Blocking LTβR or CXCR4 signaling completely restored IL7 production and B-lymphopoiesis in the BM of WHIM mice.
Previous studies indicated that hematopoietic stem and progenitor cells expressing the CXCR4 S338X GOF mutation are partially defective in undergoing differentiation into lymphoid lineages in vivo (25). Our findings here reported are consistent with these observations and extend our understanding of how hematopoietic cell-intrinsic CXCR4 GOF impacts the BM niche and reduces its lymphopoietic potential. However, the reduction in IL7 expression in BM MSCs is only a contributing factor for the peripheral B lymphopenia seen in Cxcr4WHIM/+ mice. Other factors include the fact that CXCR4 GOF mutations increase B and T cell retention in BM and in splenic red pulp while reducing access to the white pulp and reducing trafficking through peripheral lymph nodes. Prior studies showed that FRCs in peripheral lymph nodes are a major source of BAFF for mature B cell survival (56), and recent studies indicate that T-zone FRCs express highest amounts of BAFF (51, 60). T cell zone FRCs also secrete IL7 and promote T cell survival in lymph nodes (61), although other sources of IL7 can also contribute to T cell survival in the periphery (62). In Cxcr4WHIM/+ mice, the altered migratory behavior reduces B cell access to BAFF sources in the white pulp and lymph nodes, which impacts transitional B cell differentiation in the spleen, and reduces B cell survival in spleen and lymph nodes. For T cells, the reduced trafficking into splenic white pulp and peripheral lymph nodes combined with increased trafficking through the BM not only reduces T cell exposure to IL7+ niches in the periphery but may also increase competition with lymphoid progenitors for limiting amounts of IL7 in BM.
Studies from a single WHIM patient that was spontaneously cured of pan-leukopenia revealed chromotryptic deletions in a single copy of chromosome 2 that contained the disease-causing CXCR4 R334X allele (63). This rare genetic event caused CXCR4 haploinsufficiency presumably in a single hematopoietic stem cell, which conferred competitive advantage over hematopoietic stem and progenitor cells expressing the WHIM allele. Evidence for this comes from the fact that myeloid cell development, including neutrophils, largely originated from mutated CXCR4 haploinsufficient hematopoietic progenitors (64). However, this patient continued to show paucity of naïve B and T cells in circulation, consistent with defective lymphopoiesis, but the few lymphocytes developed from WHIM-expressing lymphoid progenitors (63). It is possible that the genetic mosaicism detected in this patient’s BM is sufficient for reducing lymphoipoietic niche function. Having said this, why lymphoid lineage cells developed exclusively from CXCR4 haploinsufficient hematopoietic stem and progenitor cells is not clear.
Whereas the MSC transcriptome is significantly altered in WHIM mice, stromal cells in SLOs are much less affected as indicated by minor differences in transcriptional heterogeneity of stromal cell subsets in spleen and lymph nodes. MSCs express significantly higher amounts of CXCL12 than SLO stromal cells, which may render MSCs more susceptible to regulatory signals delivered by lymphocytes and other hematopoietic cells, such as LTβR ligands. B and T cells that escape CXCL12-mediated retention in BM and splenic red pulp navigate into the white pulp normally, and presumably interact with stromal cell subsets within a normal physiological range. Evidence supporting this interpretation is the fact that BAFF and IL7 are expressed by SLO stromal cells within normal range in WHIM mice despite both cytokines also being negatively regulated by LTβR signaling. It is interesting to note that FDCs, which are located at the center follicle and are in tight contact with densely packed naïve B cells, express lower amounts of BAFF when compared to other follicular and T-zone stromal cell subsets (51). This observation is consistent with studies showing that FDCs are highly dependent on LTβR signaling for development, and with differentiation trajectory analyses of splenic stromal cells by single cell RNA sequencing which indicate that FDCs represent the most differentiated stromal cell subset in SLOs (49).
Multiple studies suggested that WHIM patients are B cell lymphopenic since B cell counts are significantly reduced in peripheral blood. However, peripheral blood lymphopenia could simply reflect reduced access to blood or lymph circulation without necessarily impacting lymphocyte numbers in the periphery. Our findings here of an increased BAFF concentration in the serum of WHIM patients most likely reflects reduced BAFF consumption due to peripheral B lymphopenia (57, 65) . Furthermore, the fact that treatment with CXCR4 antagonists not only corrected peripheral B lymphopenia but also normalized BAFF levels in WHIM mice suggests that BAFF concentration in the serum of WHIM patients should be monitored before and after treatment as a readout for therapeutic efficiency.
WHIM patients suffer from recurrent respiratory tract infections and from skin and genital tract infections with human papillomavirus (HPV). As a limitation, it is not possible to infer from this study whether blocking LTβR or CXCR4 signaling in vivo would completely restore immune protection against microbial infections. Thus, additional studies designed to evaluate the multiple parameters involved in immune protection are needed.
In summary, our studies elucidated the mechanisms underlying peripheral B and T lymphopenia in WHIM and revealed LTβR signaling in primary and SLOs as a molecular “rheostat” controlling the lymphocyte compartment size.
Materials and Methods
Patient samples.
Plasma samples from 6 WHIM patients, 21 patients with partial RAG deficiency and 20 healthy controls were obtained from Dr. Jolan Walter’s biobank at USF. All subjects were recruited according to protocols approved by local Institutional Review Boards (IRBs) as follows: USF-Pro00035468 (PI: J.E.W.), USF-Pro00025693 (PI: J.E.W.), JHMI-IRB00175372 (PI: J.E.W.), JHMI-IRB00097062 (PI: J.E.W.) and JHMI-IRB00097938 (PI: J.E.W.).
Mice.
Cxcr4WHIM/+ mice were generated at the Yale Genome Editing Center using CRISPR/Cas technology. RNA guides were designed to introduce c.1021C>T and c.1023G>A mutations in exon 2 of murine Cxcr4 leading to the exchange of arginine 341 with a STOP codon (R341X, see Fig. S1A). Mice used for experiments were backcrossed to C57BL/6J mice 5-10 generations (JAX, strain code 000664). Adult C57BL/6NCR mice (strain code 556, CD45.2+) and B6-Ly5.1/Cr (stain code 564, CD45.1+) for experiments were purchased from Charles River Laboratories. LeprCre/+ mice were purchased from JAX. Ltbrfl/fl and Ltb−/− mice (66), Il7GFP/+ mice (67) and Cxcl12DsRed/+ mice (68) were from our internal colony. Male and female adult mice at 50% ratios (6-21 weeks) were used. All mice were maintained under specific pathogen-free conditions at the Yale Animal Resources Center and were used according to the protocol approved by the Yale University Institutional Animal Care and Use Committee.
LTβR signaling inhibition.
100-150μg of LTβR-Ig or HEL-Ig was injected intravenously once and analyzed 1 week later, or once a week for 3 weeks.
CXCR4 signaling inhibition.
Mice were gavaged daily with the CXCR4 antagonist X4P-X4-185-P1 (X4 Pharmaceuticals) in 50 mM citrate buffer, pH 4.0 for 1 or 3 weeks at 100 mg/kg, or with vehicle (for control groups). X4P-X4-185-P1 belongs to a class of CXCR4 antagonists that is orally bioavailable (45).
Lymphocyte homing assay.
B cells or T cells were isolated from Cxcr4WHIM/+ and Cxcr4+/+ spleens and peripheral lymph nodes by magnetic separation using the EasySep Mouse B cell Isolation Kit or EasySep Mouse T cell Isolation Kit (STEMCELL Technologies). T and B cells were fluorescently labeled with CFSE or CMTMR and a total of 1×107 mixed B cells or 1.5×107 mixed T cells at a ratio of 1:1 were adoptive transferred iv into recipient mice. Mice were analyzed 16-20h after adoptive transfers.
Flow cytometry.
Single cell suspensions of spleen, peripheral lymph node (axillary, brachial and inguinal) and thymi were prepared as previously described (8). BM cells were flushed from long bones using the same media. BM stromal cells were isolated as previously described (8). Stromal cells from lymph nodes were isolated by Collagenase IV and Dnase1 digestion. Lymph nodes were cleaned from surrounding fat tissue and cut into ~1mm thick slices using a razor blade, placed into HBSS supplemented with 2% heat-inactivated FBS, 1% Penicillin/Streptomycin, 1% L-glutamine, and 1% HEPES and incubated for 20 min at 37°C. Then, 1800 U/ml Collagenase IV (Worthington Biochemical Corporation) and 80 μg/ml Dnase1 (Sigma Aldrich) in the same media was added at a ratio of 1:1 to reach final concentrations of 900 U/ml Collagenase IV and 40 μg/ml Dnase1, and lymph node slices were digested for 20 min at 37°C. Next, the tissue was gently disrupted by pipetting and incubated for another 10 min at 37°C. Cells were carefully resuspended, suspensions centrifuged at 1,200 rpm for 10 min and resuspended in HBSS supplemented as above. Cell concentrations were determined with a Coulter counter (Beckman Coulter). Cells were stained with antibody cocktails diluted in FACS buffer on ice, at a concentration of 25 μl /1 × 106 cells.
The following antibodies were used (Biolegend unless indicated otherwise): anti-mouse CD3e (1452C11), CD4 (GK1.5), CD8 (53-5.8), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD23 (B3B4), CD25 (PC61), CD44 (IM7), CD45.1 (A20), CD45.2 (104), B220 (RA3-6B2), CD71 (RI7217), CD93 (AA4.1), CD115 (AFS98), CD117 (cKit, 2B8), IgM (II/41, Thermo Fisher Scientific), IgD (11-26c.2a), Gr1 (RB6-8C5), Ter119 (Ter119). Hematopoietic cell populations were identified as follows: Pro B: CD19+ CD93+ IgM− cKit+ or CD19+ CD93+ IgM− FSChigh; Pre B: CD19+ CD93+ IgM− c-Kit− or CD19+ CD93+ IgM− FSClow; Immature B: CD19+ CD93+ IgM+; Mature B: CD19+ (or B220+) CD93− IgM+ IgD+ or CD19+ (or B220+) CD93− CD23+; T1 B cells: CD19+ (or B220+) CD93+ IgM+ CD23− (or IgD−); T2 B cells: CD19+ (or B220+) CD93+ CD23+ (or IgD+); CD4+ T cells: CD3e+ CD4+; CD8+ T cells: CD3e+ CD8+; thymocytes: double negative (DN) 1: CD4− CD8− CD44+ CD25−, DN2: CD4− CD8− CD44+ CD25+, DN3: CD4− CD8− CD44− CD25+, DN4: CD4− CD8− CD44− CD25−, double positive (DP): CD4+ CD8+, CD4 single positive (SP4): CD4+, CD8 single positive (SP8): CD8+ ; Neutrophils: CD11b+ CD115− Gr1high; Monocytes: CD115+ Gr1int; The following antibodies were used to identify non-hematopoietic cell populations: anti-mouse GP38 (8.1.1), CD31 (390), CD144 (BV13), LEPR (goat polyclonal, R&D), LTβR (5G11). MSC: CD45− Ter119− CD31− CD144− LEPR+; lymph node GP38+ SC: CD45− Ter119− CD31− GP38+; For live/dead cell discrimination, cells were stained with DAPI. For detection of apoptotic cells, cells were first stained for surface proteins, then cells were washed twice with FACS buffer and stained for 15 min at RT with fluorescently labeled Annexin V in Annexin V Binding Buffer (Biolegend). Samples were recorded with LSRII cytometers (BD) and FACS Diva™ software (BD) and analyzed with FlowJo™ software (BD).
Flow cytometric detection of BAFF.
Cells were stained with anti-BAFF (clone 121808, R&D Systems, FITC-coupled). For intracellular staining cells were fixed with Cytofix/Cytoperm solution (BD) for 20 min on ice and washed twice with PermWash buffer (BD). Then cells were stained for 30 min at RT with anti-BAFF and then for 20 min at RT with a goat anti-rat IgG-BV421 antibody (Biolegend).
Immunofluorescence microscopy.
Tissue fixation and cryosectioning was performed as described (8). Sections were rehydrated in PBS for 20 mins, blocked with 5% FBS in PBS/0.1% TWEEN®20 for 20 mins, and stained with primary antibodies for 1 h. Sections were washed 5 times with PBS/0.1% TWEEN®20 and secondary antibodies applied for 1 h. Slides were mounted with Fluorescence Mounting Medium (DAKO). Images were acquired on a Zeiss Z1 Observer fluorescent microscope equipped with Colibri LED light sources, or with Leica SP8 confocal microscope. The following primary antibodies were used: anti-MAdCAM1 (MECA-367, Biolegend), rabbit anti-RFP (polyclonal, Rockland), CD3-biotin (1452C11), CD35-biotin (8C12, BD). The following secondary reagents were used: donkey anti-rat-IgG (H+L) Alexa Fluor 488 (Jackson Immuno Research), donkey anti-rabbit-IgG (H+L) Alexa Fluor 555 (Thermo Fisher Scientific), Streptavidin-Alexa Fluor 488 and Streptavidin-Alexa Fluor 555 (Thermo Fisher Scientific). Conjugated antibodies: IgD-Alexa Fluor 647 (11-26c.2a, Biolegend), CD21/35-FITC (7E9). Nuclei were labeled with DAPI. Images were analyzed with ImageJ (69) and Zen (Zeiss).
Preparation of MSCs for RNA sequencing.
Long bones were flushed with HBSS supplemented with 2% heat-inactivated FBS, 1% Penicillin/Streptomycin, 1% L-glutamine, 1% HEPES and 200 U/ml Collagenase IV. The flush out was digested for 30 min at 37°C, cell clumps were dissociated by gentle pipetting, and cells were filtered through a 100 μm nylon mesh and washed with HBSS 2% FBS, 1% Penicillin/Streptomycin, 1% L-glutamine, 1% HEPES. Hematopoietic cells were stained with biotin-conjugated CD45 and Ter119 antibodies and magnetically depleted using Dynabeads ® Biotin Binder (Thermo Fisher Scientific). The remaining cells were stained with antibodies against CD31, CD144 and LEPR, fluorochrome-conjugated Streptavidin and DAPI. Sorting was performed using a BD FACS Aria II and MSCs were identified as CD45− Ter119− CD31− CD144− LEPR+ cells. Cells were sorted into DMEM/10% FBS and re-sorted with the same strategy directly into 350 μL RLT plus buffer (Qiagen).
RNA sequencing of BM stromal cells.
RNA sequencing was performed using the Illumina HiSeq2500 system, with paired-end 2 × 76 bp read length. Using the TopHat v2.1.0 software, the sequencing reads were aligned onto the Mus musculus GRCm38/mm10 reference genome. Using HTSeq v0.8.0 software, the mapped reads were converted into the count matrix with default parameters, followed by the variance stabilizing transformation (VST) offered by DESeq2. followed by the variance stabilizing transformation (VST) offered by DESeq2. Differentially expressed genes (DEGs) were identified using the same software based on a negative binomial generalized linear models and visualized in hierarchically-clustered heatmaps using the pheatmap package in R.
Single-cell RNA sequencing of BM stromal cells.
ScRNAseq was performed by the Yale Center for Genome Analysis. The libraries were prepared using the Chromium Single Cell 3ʹ Reagent Kits v3.0 according to the protocol and run on an Illumina NovaSeq system with 100-bp paired-end reads to a coverage of ~ 40,000 - 44,000 mean reads per cell and ~ 80% saturation. The sequencing reads were aligned onto the Mus musculus GRCm38 (mm10) reference genome.
Preparation of secondary lymphoid organ stromal cells for single-cell RNA sequencing.
Peripheral lymph node stromal cell suspensions were prepared from 2 mice per genotype and pooled. Cells were stained with biotinylated antibodies against CD45 and Ter119 and hematopoietic cells were depleted using Dynabeads® Biotin Binder (Thermo Fisher Scientific). The leftover cells were stained with Streptavidin-BV711 (Biolegend), DAPI and antibodies against CD31, CD71 and GP38. Lymph node stromal cells were sorted as CD45− Ter119− CD71− CD31− cells into 350 μL DMEM with 20% FBS. GP38 was used to control for the presence of stromal cells in the suspension. Splenic stromal cells suspensions were prepared from 3 mice per genotype and pooled. The leftover cells were stained for CD45, Ter119, LIN (CD3e, B220, CD19), CD31, GP38 and PDGFRα. Splenic stromal cells were sorted as CD45− Ter119− LIN− CD71− CD31− into 350 μL DMEM with 20% FBS. GP38 and PDGFRα was used to control for the presence of stromal cells in the suspension. Single-cell RNA sequencing was performed as described for BM stromal cells, to a coverage of ~ 65,000 mean reads per cell and > 90% saturation for lymph node stromal cells and to a coverage of ~ 30,000 - 57,000 mean reads per cell and > 80% saturation for splenic stromal cells. The sequencing reads were aligned onto the Mus musculus GRCm38 (mm10) reference genome.
BM stromal cell single cell RNA sequencing
Pre-processing of scRNA-seq data – The Gene-Barcode matrix generated from 10x Genomic Cell Ranger was preprocessed using the Seurat (version 3.2.3) R package (70). Briefly, cells that had fewer than 200 or more than 6,000 detected features and more than 25% of mitochondrial gene content were excluded to remove low quality cells. Subsequently, contaminating hematopoietic cells were filtered out from the downstream analysis. After quality control, SCTransform (71) was used to normalize the count data and stabilize variance.
Dimension reduction and clustering - Highly variable genes were identified and utilized for dimensionality reduction of the entire 12,598 cell dataset with principal component analysis (PCA). IKAP (Identifying K mAjor cell Population groups in single-cell RNA-sequencing analysis,(72)) was used to estimate the number of optimal principal components (PCs) and clusters. Cell clustering was performed using FindClusters function of Seurat and visualized in a 2D space using the UMAP (Uniform Manifold Approximation and Projection,(73, 74)) visualization method. MSC clusters were further subjected to Markov Affinity-based Graph Imputation of Cells (MAGIC, (75)) to restore the structure of the data by imputing plausible gene expression in each cell.
Cell type annotation - Pairwise differential expression analysis was performed for each cluster using the MAST method (76) to identify marker genes. Cluster-specific marker genes were matched to the cell types based on the previously identified expression signature and the known marker expression in the publicly available databases, PanglaoDB (77) and the Human Protein Atlas (78). Top 10 significant positive marker genes were selected to generate feature plots and heatmap plots.
Differential abundance test - To conduct differential abundance analysis of neighborhoods, R implementation of Milo (https://github.com/MarioniLab/miloR, (79)) was applied. Cell neighborhoods were defined under parameters (k=12, d=20) and the proportion of randomly sampled graph vertices was set to prop = 0.1. Cell neighborhoods having statistically significant differential abundance between two groups were determined and plotDAbeeswarm was used to display abundance differences.
Secondary lymphoid organ stromal cell scRNA-seq data preprocessing and analysis.
scRNA sequencing data from lymph node and splenic stromal cells was analyzed using SCANPY (80). For this scatter plots were first manually inspected for total gene expression count (500 to 20,000), the number of expressed genes (> 100), and mitochondrial gene expression fraction (< 0.2 for the lymph node data and < 0.1 for the spleen data) to filter and preprocess the data. The lymph node data resulted in 12,521 cells (Cxcr4WHIM/+: 7,332; Cxcr4+/+: 5,189) and 17,944 genes. The spleen data resulted in 19,741 cells (Cxcr4WHIM/+: 13,660; Cxcr4+/+ 6,081) and 18,362 genes. Log-normalization and PCA with 50 PCs was performed and 10 neighbors were used for UMAP visualization. Next, all contaminating cells with non-zero expression of any of 44 hematopoietic marker genes were removed. Gene expression distributions were manually inspected in histograms, ordered dot plots, and UMAP plots. The filtering resulted in 3,113 cells for the lymph node data (Cxcr4WHIM/+: 2,131; Cxcr4+/+: 982) and 3,484 cells for the spleen data (Cxcr4WHIM/+: 1,283; Cxcr4+/+: 2,201). For unsupervised clustering, Leiden clustering with resolution = 0.2 and 0.1 for the lymph node and spleen data, respectively was performed (after manual inspection of several resolution choices in UMAP plots). 8 and 12 clusters were identified for the lymph node and spleen data, respectively. For differential gene expression analysis, Wilcoxon rank-sum tests were performed in two ways: (1) for each cluster against all the rest and (2) for the Cxcr4WHIM/+group against the Cxcr4+/+ group in each cluster.
Marker genes used for excluding hematopoietic contaminants: general hematopoietic markers (Ptprc, Ptpn22, Itga4, Gpr183), B-lineage (Igkc, Igha, Ighm, Ighd, Jchain, Iglv1, Iglv2, Iglv3, Iglc1, Iglc2, Iglc3, Igll1, Rag1, Rag2, Il7r, Vpreb1, Vpreb2, Vpreb3, Cd19, Cd79a, Cd79b, Dntt), T- and NK-lineage (Ccr7, Cd3e, Cd2, Gmza, Gmzb), myeloid lineage (Il1b, Ccr1, Cxcr2, Siglech, Ccr9, Lair1, Csf3r, Trem1, Cd300c), erythroid lineage (Ciita, Slc4a1, Rhd, Alas2).
Lymph node stromal cell preparation for Quantitative Real Time PCR.
Lymph node stromal cell suspension were prepared as described and sorted as CD45− Ter119− CD31− GP38+ cells directly into 350 μL RLT plus buffer with 3.5 μL β-mercaptoethanol (Qiagen) using a BD FACS Aria II. RNA was extracted from the lysates using the RNeasy® Plus Micro Kit (Qiagen).
RNA isolation and Quantitative Real Time PCR.
Total RNA was isolated from sorted stromal cells using the RNeasy® Plus Micro Kit (Qiagen). cDNA was synthetized from the isolated RNA using SuperScript™ III Reverse Transcriptase, Oligo(dT)12-18 primer, dNTPs and RNAseout™ RNase inhibitors (all Thermo Fisher Scientific). Q-PCR was performed with the SensiFAST™ SYBR Lo- ROX Kit (Bioline) and the CFX Touch™ Real-Time PCR detection system (BioRad). Hprt mRNA levels were used as control. PCR primer sequences: Hprt forward AGG TTG CAA GCT TGC TGG T; Hprt reverse TGA AGT ACT CAT TAT AGT CAA GGG CA; Il7 forward GCT GCC TGT CAC ATC ATC TG; Il7 reverse CAG CAC GAT TTA GAA AAG CAG C; Tnfsf13b forward: ACA CTG CCC AAC AAT TCC TG; Tnfsf13b reverse: TCG TCT CCG TTG CGT GAA ATC;
Human BAFF serum ELISA.
BAFF plasma concentrations were measured using commercial ELISA (R&D Systems, Minneapolis, MN).
Statistical analysis.
Statistical analyses were performed using Prism 8 (GraphPad). Unpaired two tailed Student’s t-test was used to determine statistical significance.
Supplementary Material
Table 3- LTβR-regulated genes in Lepr+ MSPCs. Differentially expressed genes analyzed by bulk RNA sequencing of Lepr+ MSPCs isolated from wild-type mice treated with control or LTβR-Ig (100μg/mouse/week) for 3 weeks. Related to Figure 2.
Table 2- Differentially expressed genes between BM non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 1.
Table 1- Average gene expression for BM non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice. Related to Figure 1.
Table 5- Differentially expressed genes between splenic non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 5.
Table 4- Average gene expression for splenic non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq. Related to Figure 5.
Table 6- Average gene expression for lymph node non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq. Related to Figure 6.
Table 7- Differentially expressed genes between lymph node non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 6.
Table 8- Age and sex distribution of healthy controls, WHIM, and partial RAG deficient patients.
Supplementary Figure 1: Characterization of Cxcr4WHIM/+ mice.
Supplementary Figure 2: Expression of lymphotoxin α1β2 in hematopoietic stem, progenitors, and differentiated cells in the BM.
Supplementary Figure 3: Effects of LTβR blocking in lymphocyte homeostasis.
Supplementary Figure 4: Effects of CXCR4 signaling blockade in lymphocyte development and homeostasis.
Supplementary Figure 5: Comparison of splenic stromal cells between Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Supplementary Figure 6: Comparison of lymph node stromal cells between Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Supplementary Figure 7: Continued characterization of lymph node stromal cells from Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Table 9- Raw data
Supplementary data file S1. Raw data excel file
Acknowledgements
We acknowledge the patients, clinicians who provided human patient samples and Boglarka Ujhazi and Maryssa Ellison for patient samples’ management and protocols.
Funding
These studies were funded by the NIH (RO1AI113040 and R21AI133060-01A1) and by X4 Pharmaceuticals (JPP). Jeffrey Modell Foundation, Robert A. Good Endowment at University of South Florida, and X4 Pharmaceuticals (JEW).
Footnotes
Competing interests. LD was an employee of X4 Pharmaceuticals. All other authors declare that they have no competing interests.
Data availability.
Bulk and single cell RNA sequencing data are publicly available on Gene Expression Omnibus under the accession code GSE190086.
References
- 1.Morrison SJ, Scadden DT, The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinho S, Frenette PS, Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol 20, 303–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugiyama T, Omatsu Y, Nagasawa T, Niches for hematopoietic stem cells and immune cell progenitors. Int Immunol 31, 5–11 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, Kokkaliaris KD, Mercier F, Tabaka M, Hofree M, Dionne D, Papazian A, Lee D, Ashenberg O, Subramanian A, Vaishnav ED, Rozenblatt-Rosen O, Regev A, Scadden DT, A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 177, 1915–1932 e1916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Dominguez A, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot M, Gutkin MC, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman DR, Butler JM, Economides A, Frenette PS, Adams RH, Satija R, Tsirigos A, Aifantis I, The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baccin C, Al-Sabah J, Velten L, Helbling PM, Grunschlager F, Hernandez-Malmierca P, Nombela-Arrieta C, Steinmetz LM, Trumpp A, Haas S, Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol 22, 38–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, Saunders TL, Enikolopov G, Morrison SJ, Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordeiro Gomes A, Hara T, Lim VY, Herndler-Brandstetter D, Nevius E, Sugiyama T, Tani-Ichi S, Schlenner S, Richie E, Rodewald HR, Flavell RA, Nagasawa T, Ikuta K, Pereira JP, Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 45, 1219–1231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asada N, Kunisaki Y, Pierce H, Wang Z, Fernandez NF, Birbrair A, Ma’ayan A, Frenette PS, Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol 19, 214–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comazzetto S, Murphy MM, Berto S, Jeffery E, Zhao Z, Morrison SJ, Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell 24, 477–486 e476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Wu Q, Johnson CB, Pham G, Kinder JM, Olsson A, Slaughter A, May M, Weinhaus B, D’Alessandro A, Engel JD, Jiang JX, Kofron JM, Huang LF, Prasath VBS, Way SS, Salomonis N, Grimes HL, Lucas D, In situ mapping identifies distinct vascular niches for myelopoiesis. Nature 590, 457–462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiyama T, Kohara H, Noda M, Nagasawa T, Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Nie Y, Han YC, Zou YR, CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med 205, 777–783 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T, The Essential Functions of Adipo-osteogenic Progenitors as the Hematopoietic Stem and Progenitor Cell Niche. Immunity 33, 387–399 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Noda M, Omatsu Y, Sugiyama T, Oishi S, Fujii N, Nagasawa T, CXCL12-CXCR4 chemokine signaling is essential for NK-cell development in adult mice. Blood 117, 451–458 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, Lai DM, Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 117, 429–439 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC, CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, Ledergor G, Jung Y, Milo I, Poulos MG, Kalinkovich A, Ludin A, Kollet O, Shakhar G, Butler JM, Rafii S, Adams RH, Scadden DT, Lin CP, Lapidot T, Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532, 323–328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao R, Lim VY, Kothapalli N, Ma Y, Fossati J, Zehentmeier S, Sun R, Pereira JP, Hematopoietic Stem Cell Niches and Signals Controlling Immune Cell Development and Maintenance of Immunological Memory. Front Immunol 11, 600127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fistonich C, Zehentmeier S, Bednarski JJ, Miao R, Schjerven H, Sleckman BP, Pereira JP, Cell circuits between B cell progenitors and IL-7(+) mesenchymal progenitor cells control B cell development. The Journal of experimental medicine 215, 2586–2599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehentmeier S, Pereira JP, Cell circuits and niches controlling B cell development. Immunol Rev 289, 142–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, Klotman ME, Diaz GA, Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet 34, 70–74 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Balabanian K, Lagane B, Pablos JL, Laurent L, Planchenault T, Verola O, Lebbe C, Kerob D, Dupuy A, Hermine O, Nicolas JF, Latger-Cannard V, Bensoussan D, Bordigoni P, Baleux F, Le Deist F, Virelizier JL, Arenzana-Seisdedos F, Bachelerie F, WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood 105, 2449–2457 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Balabanian K, Brotin E, Biajoux V, Bouchet-Delbos L, Lainey E, Fenneteau O, Bonnet D, Fiette L, Emilie D, Bachelerie F, Proper desensitization of CXCR4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood 119, 5722–5730 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Freitas C, Wittner M, Nguyen J, Rondeau V, Biajoux V, Aknin ML, Gaudin F, Beaussant-Cohen S, Bertrand Y, Bellanne-Chantelot C, Donadieu J, Bachelerie F, Espeli M, Dalloul A, Louache F, Balabanian K, Lymphoid differentiation of hematopoietic stem cells requires efficient Cxcr4 desensitization. The Journal of experimental medicine 214, 2023–2040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDermott DH, Murphy PM, WHIM syndrome: Immunopathogenesis, treatment and cure strategies. Immunol Rev 287, 91–102 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Pereira JP, An J, Xu Y, Huang Y, Cyster JG, Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat Immunol 10, 403–411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira JP, Cyster JG, Xu Y, A role for S1P and S1P1 in immature-B cell egress from mouse bone marrow. PLoS One 5, e9277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck TC, Gomes AC, Cyster JG, Pereira JP, CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J Exp Med 211, 2567–2581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tassone L, Moratto D, Vermi W, De Francesco M, Notarangelo LD, Porta F, Lougaris V, Facchetti F, Plebani A, Badolato R, Defect of plasmacytoid dendritic cells in warts, hypogammaglobulinemia, infections, myelokathexis (WHIM) syndrome patients. Blood 116, 4870–4873 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInnes, J LH.; Saul N; Großberger L, UMAP: Uniform Manifold Approximation and Projection. . Journal of Open Source Software 3, 861 (2018). [Google Scholar]
- 33.Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM, Immediate-early and delayed primary response genes are distinct in function and genomic architecture. The Journal of biological chemistry 282, 23981–23995 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dann E, Henderson NC, Teichmann SA, Morgan MD, Marioni JC, Milo: differential abundance testing on single-cell data using k-NN graphs. bioRxiv, 2020.2011.2023.393769 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Fu YX, Chaplin DD, Development and maturation of secondary lymphoid tissues. Annu Rev Immunol 17, 399–433 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Cyster JG, Lymphoid organ development and cell migration. Immunol Rev 195, 5–14 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Mackay F, Majeau GR, Lawton P, Hochman PS, Browning JL, Lymphotoxin but not tumor necrosis factor functions to maintain splenic architecture and humoral responsiveness in adult mice. Eur J Immunol 27, 2033–2042 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Ngo VN, Korner H, Gunn MD, Schmidt KN, Riminton DS, Cooper MD, Browning JL, Sedgwick JD, Cyster JG, Lymphotoxin alpha/beta and tumor necrosis factor are required for stromal cell expression of homing chemokines in B and T cell areas of the spleen. J Exp Med 189, 403–412 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira J, Kelly L, Xu Y, Cyster J, EBV induced molecule-2 mediates B cell segregation between outer and center follicle. Nature 460, 1122–1126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG, A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406, 309–314 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Cyster JG, Ansel KM, Reif K, Ekland EH, Hyman PL, Tang HL, Luther SA, Ngo VN, Follicular stromal cells and lymphocyte homing to follicles. Immunol Rev 176, 181–193 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Tumanov A, Kuprash D, Lagarkova M, Grivennikov S, Abe K, Shakhov A, Drutskaya L, Stewart C, Chervonsky A, Nedospasov S, Distinct role of surface lymphotoxin expressed by B cells in the organization of secondary lymphoid tissues. Immunity 17, 239–250 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR, The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 17, 525–535 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Browning JL, Allaire N, Ngam-Ek A, Notidis E, Hunt J, Perrin S, Fava RA, Lymphotoxin-β receptor signaling is required for the homeostatic control of HEV differentiation and function. Immunity 23, 539–550 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Dale DC, Firkin F, Bolyard AA, Kelley M, Makaryan V, Gorelick KJ, Ebrahim T, Garg V, Tang W, Jiang H, Skerlj R, Beaussant Cohen S, Results of a phase 2 trial of an oral CXCR4 antagonist, mavorixafor, for treatment of WHIM syndrome. Blood 136, 2994–3003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nie Y, Waite J, Brewer F, Sunshine MJ, Littman DR, Zou YR, The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J Exp Med 200, 1145–1156 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandal M, Okoreeh MK, Kennedy DE, Maienschein-Cline M, Ai J, McLean KC, Kaverina N, Veselits M, Aifantis I, Gounari F, Clark MR, CXCR4 signaling directs Igk recombination and the molecular mechanisms of late B lymphopoiesis. Nature immunology 20, 1393–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm T, Scheu S, Pfeffer K, Bleul CC, Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTbetaR. J Exp Med 198, 757–769 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng HW, Onder L, Novkovic M, Soneson C, Lutge M, Pikor N, Scandella E, Robinson MD, Miyazaki JI, Tersteegen A, Sorg U, Pfeffer K, Rulicke T, Hehlgans T, Ludewig B, Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat Commun 10, 1739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inra CN, Zhou BO, Acar M, Murphy MM, Richardson J, Zhao Z, Morrison SJ, A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature 527, 466–471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, Cyster JG, Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 48, 1014–1028 e1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, Cyster JG, Luther SA, Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nature immunology 8, 1255–1265 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Hara T, Shitara S, Imai K, Miyachi H, Kitano S, Yao H, Tani-ichi S, Ikuta K, Identification of IL-7–producing cells in primary and secondary lymphoid organs using IL-7–GFP knock-in mice. The Journal of Immunology 189, 1577–1584 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Mendoza A, Fang V, Chen C, Serasinghe M, Verma A, Muller J, Chaluvadi VS, Dustin ML, Hla T, Elemento O, Chipuk JE, Schwab SR, Lymphatic endothelial S1P promotes mitochondrial function and survival in naive T cells. Nature 546, 158–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackay F, Figgett WA, Saulep D, Lepage M, Hibbs ML, B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol Rev 237, 205–225 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Cremasco V, Woodruff MC, Onder L, Cupovic J, Nieves-Bonilla JM, Schildberg FA, Chang J, Cremasco F, Harvey CJ, Wucherpfennig K, Ludewig B, Carroll MC, Turley SJ, B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat Immunol 15, 973–981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesley R, Xu Y, Kalled SL, Hess DM, Schwab SR, Shu HB, Cyster JG, Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity 20, 441–453 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Christodoulou C, Spencer JA, Yeh SA, Turcotte R, Kokkaliaris KD, Panero R, Ramos A, Guo G, Seyedhassantehrani N, Esipova TV, Vinogradov SA, Rudzinskas S, Zhang Y, Perkins AS, Orkin SH, Calogero RA, Schroeder T, Lin CP, Camargo FD, Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upadhaya S, Krichevsky O, Akhmetzyanova I, Sawai CM, Fooksman DR, Reizis B, Intravital Imaging Reveals Motility of Adult Hematopoietic Stem Cells in the Bone Marrow Niche. Cell Stem Cell 27, 336–345 e334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pezoldt J, Wiechers C, Erhard F, Rand U, Bulat T, Beckstette M, Brendolan A, Huehn J, Kalinke U, Mueller M, Strobl B, Deplancke B, Cicin-Sain L, Sitnik KM, Single-cell transcriptional profiling of splenic fibroblasts reveals subset-specific innate immune signatures in homeostasis and during viral infection. Commun Biol 4, 1355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, Cyster JG, Luther SA, Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol 8, 1255–1265 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Knop L, Deiser K, Bank U, Witte A, Mohr J, Philipsen L, Fehling HJ, Muller AJ, Kalinke U, Schuler T, IL-7 derived from lymph node fibroblastic reticular cells is dispensable for naive T cell homeostasis but crucial for central memory T cell survival. Eur J Immunol 50, 846–857 (2020). [DOI] [PubMed] [Google Scholar]
- 63.McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, Velez D, Yim E, Bryke CR, Hsu N, Dai Z, Marquesen MM, Stregevsky E, Kwatemaa N, Theobald N, Long Priel DA, Pittaluga S, Raffeld MA, Calvo KR, Maric I, Desmond R, Holmes KL, Kuhns DB, Balabanian K, Bachelerie F, Porcella SF, Malech HL, Murphy PM, Chromothriptic cure of WHIM syndrome. Cell 160, 686–699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao JL, Yim E, Siwicki M, Yang A, Liu Q, Azani A, Owusu-Ansah A, McDermott DH, Murphy PM, Cxcr4-haploinsufficient bone marrow transplantation corrects leukopenia in an unconditioned WHIM syndrome model. J Clin Invest 128, 3312–3318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreuzaler M, Rauch M, Salzer U, Birmelin J, Rizzi M, Grimbacher B, Plebani A, Lougaris V, Quinti I, Thon V, Litzman J, Schlesier M, Warnatz K, Thiel J, Rolink AG, Eibel H, Soluble BAFF levels inversely correlate with peripheral B cell numbers and the expression of BAFF receptors. J Immunol 188, 497–503 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Koroleva EP, Kruglov AA, Kuprash DV, Nedospasov SA, Fu Y-X, Tumanov AV, Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity 32, 403–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller CN, Hartigan-O’Connor DJ, Lee MS, Laidlaw G, Cornelissen IP, Matloubian M, Coughlin SR, McDonald DM, McCune JM, IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. International immunology 25, 471–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding L, Morrison SJ, Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive integration of single-cell data. Cell 177, 1888–1902. e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen YC, Suresh A, Underbayev C, Sun C, Singh K, Seifuddin F, Wiestner A, Pirooznia M, IKAP-Identifying K mAjor cell Population groups in single-cell RNA-sequencing analysis. Gigascience 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McInnes L, Healy J, Melville J, Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426, (2018). [Google Scholar]
- 74.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW, Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol, (2018). [DOI] [PubMed] [Google Scholar]
- 75.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, Bierie B, Mazutis L, Wolf G, Krishnaswamy S, Pe’er D, Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729 e727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology 16, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franzen O, Gan LM, Bjorkegren JLM, PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Dann E, Henderson NC, Teichmann SA, Morgan MD, Marioni JC, Milo: differential abundance testing on single-cell data using k-NN graphs. bioRxiv, (2020). [DOI] [PubMed] [Google Scholar]
- 80.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome biology 19, 1–5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 3- LTβR-regulated genes in Lepr+ MSPCs. Differentially expressed genes analyzed by bulk RNA sequencing of Lepr+ MSPCs isolated from wild-type mice treated with control or LTβR-Ig (100μg/mouse/week) for 3 weeks. Related to Figure 2.
Table 2- Differentially expressed genes between BM non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 1.
Table 1- Average gene expression for BM non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice. Related to Figure 1.
Table 5- Differentially expressed genes between splenic non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 5.
Table 4- Average gene expression for splenic non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq. Related to Figure 5.
Table 6- Average gene expression for lymph node non-hematopoietic cell clusters identified in Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq. Related to Figure 6.
Table 7- Differentially expressed genes between lymph node non-hematopoietic cell clusters from Cxcr4+/+ and Cxcr4WHIM/+ identified by scRNAseq. Related to Figure 6.
Table 8- Age and sex distribution of healthy controls, WHIM, and partial RAG deficient patients.
Supplementary Figure 1: Characterization of Cxcr4WHIM/+ mice.
Supplementary Figure 2: Expression of lymphotoxin α1β2 in hematopoietic stem, progenitors, and differentiated cells in the BM.
Supplementary Figure 3: Effects of LTβR blocking in lymphocyte homeostasis.
Supplementary Figure 4: Effects of CXCR4 signaling blockade in lymphocyte development and homeostasis.
Supplementary Figure 5: Comparison of splenic stromal cells between Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Supplementary Figure 6: Comparison of lymph node stromal cells between Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Supplementary Figure 7: Continued characterization of lymph node stromal cells from Cxcr4+/+ and Cxcr4WHIM/+ mice by scRNAseq.
Table 9- Raw data
Supplementary data file S1. Raw data excel file
Data Availability Statement
Bulk and single cell RNA sequencing data are publicly available on Gene Expression Omnibus under the accession code GSE190086.


