Abstract

Glycomics aims to identify the structure and function of the glycome, the complete set of oligosaccharides (glycans), produced in a given cell or organism, as well as to identify genes and other factors that govern glycosylation. This challenging endeavor requires highly robust, sensitive, and potentially automatable analytical technologies for the analysis of hundreds or thousands of glycomes in a timely manner (termed high-throughput glycomics). This review provides a historic overview as well as highlights recent developments and challenges of glycomic profiling by the most prominent high-throughput glycomic approaches, with N-glycosylation analysis as the focal point. It describes the current state-of-the-art regarding levels of characterization and most widely used technologies, selected applications of high-throughput glycomics in deciphering glycosylation process in healthy and disease states, as well as future perspectives.
1. Introduction
Glycosylation is the most common and complex post-translational protein modification. In addition to proteins, many lipids are glycosylated, and just recently it was shown that elaborated glycan structures can also be attached to RNA.1 In addition, the field of glycomics comprises polysaccharides such as glycosaminoglycans (GAGs). Regarding protein glycosylation, the attachment of different glycans to the same glycosylation site (alternative glycosylation or microheterogeneity) greatly contributes to the structural variability of these molecules and influences their function in a way that is analogous to the effects of changes in protein sequence caused by mutations in the corresponding gene.2
The most intensively studied example of the importance of alternative protein glycosylation for biological functions are immunoglobulins, which are among the main weapons in our arsenal for the multifacetted war against pathogens. They are an elaborate tool that can specifically recognize foreign structures. However, the binding to an antigen is only one aspect of their function. Namely, immunoglobulins have to activate proper molecular mechanisms to “deal with” this foreign, nonself object. The choice of how to react to a foreign antigen is one of the most complex decisions that has to be made, and these choices have to be made continuously throughout our lifetime. Alternation of immunoglobulin G (IgG) glycosylation appears to be a check point for initiation of specific effector functions directing immunoglobulins to different receptors, and in this way activating different branches of our immune system.3,4 Glycans attached to the fragment crystallizable (Fc) region are an integral part of the constant region domain (CH2) of antibodies, and as such represent an integral structural component that participates in the interaction with Fc receptors and other proteins. Attaching a different glycan to the polypeptide backbone changes the structure of the antibody and modifies its affinity for different receptors. The best currently known example is the role of core-fucose that acts as a “safety-switch” against antibody-dependent cellular cytotoxicity (ADCC) by attenuating binding of IgG to Fc-γ-receptor IIIA.5 Other effector functions modulated by IgG Fc glycosylation include antibody-dependent phagocytosis and complement activation with an often complex dependency of effector function on various glycosylation features.6,7 Glycosylation is an essential element in the development of different therapeutic monoclonal antibodies (mAbs), and glycoengineered drugs are already on the market.8 Interindividual differences in glycosylation are large and may be an important underlying element for the response or nonresponse to a given drug, ABO blood groups are a good example, but for the vast majority of drugs, data is still missing and this field needs further exploration.
Contrary to the polypeptide sequences of a protein that are predominantly changed by inducing changes in the sequence of the corresponding genes, glycans are encoded in a complex network of at least several dozen genes that are (beside allelic variants) also affected by epigenetics and the environment.9 This enables flexible and dynamic regulation of protein function and is extensively used to fine-tune functions of many proteins. More than 30 years ago, the initial discovery of changes in the IgG glycome composition in diseases was made,10 and until now, over 150 000 different glycomes have been analyzed in different diseases and physiological states (see section 6, Applications). Changes in glycosylation associate with numerous diseases, often even before any other symptoms of the disease are detectable, indicating that they might be a part of the molecular pathophysiology leading to the disease.11 With aging, the IgG N-glycome converts from a composition that is suppressing inflammation to an inflammation-promoting N-glycome that seems to be an underlying risk factor in many cardiometabolic and inflammatory diseases.4,12,13
The importance and critical role of carbohydrate post-translational modifications on many cell surfaces and secreted proteins’ structure as well as function have been studied for several decades. The majority of both high-throughput (HT) and functional glycomic studies of individual glycoproteins were performed on IgG, thus this is by far the most studied glycoprotein. Nevertheless, there’s still much more we don’t know about glycosylation, than what we do know. HT methods for the analysis of other proteins have been developed only recently, thus our level of understanding of the importance of interindividual difference in protein glycosylation is very low. Nevertheless, because glycosylation is an integral part of the proper functioning of every organism in nature the development of effective tools for detecting carbohydrate structures, and their changes under various physiological and pathological conditions is of utmost importance.14
This review provides a historical background of the field of glycomics with a focus on recent developments in methodology and applications of HT glycomics (roughly defined as the glycan analysis of thousand(s) of samples in large-scale studies). For the purpose of this review, we will define medium-throughput glycomics as analysis of hundred(s) of samples and low-throughput as methodology applicable to glycan analysis in small-scale studies (less than one hundred samples) taking into account the whole process from sample preparation to data analysis. Because of the complexity of glycan analysis workflows, analytical limitations of any of the steps ranging from study design via sample preparation, measurement, data processing, and data analysis may limit the advancement from low- to high-throughput applications. Therefore, significant methodological and technological efforts that are not necessarily at a HT level for the moment have also been highlighted with a promise of future implementation in HT glycomics workflows.
2. Levels of Characterization
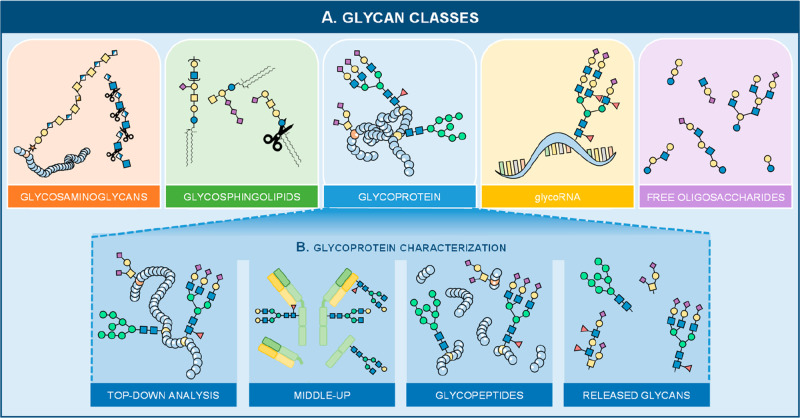
Glycosylation occurs on a diverse range of biomolecules, resulting in glycoproteins, proteoglycans, as well as glycosphingolipids (GSLs). Next to these glycoconjugates, other glycan classes exist such as free oligosaccharides, GAGs, and the just recently discovered glycosylated RNA1 (Figure 1A). Significant progress in analytical approaches for their analysis has been achieved in recent years (especially in the case of free oligosaccharides in, e.g., human milk.15−17 While all glycan classes are mutually important, the only glycan class that is currently analyzed in HT large-scale studies are the glycoproteins (Figure 1B) and will be the main focus of the next sections.
Figure 1.
(A) Various glycan classes exist such as glycosaminoglycans (GAGs), glycosphingolipids (GSLs), glycoRNA, free oligosaccharides, and glycoproteins. (B) A distinction can be made in various characterization categories of the latter glycan type. Namely, glycoproteins can be analyzed intact using a top-down method or a middle-up approach that can be used to study the subunits of (monoclonal) antibodies. Other methods include enzymatic digestions, which either cleave the protein into (glyco)peptides, also known as the bottom-up approach, or cleave the glycan portion from its conjugate (released glycan analysis). The latter two are currently the only characterization approaches that can be processed and analyzed in a HT manner. Several enzymes are available for GAGs which will result in disaccharides (indicated by the scissors), while enzymes available for GSL analysis will release the glycan headgroup from the lipid portion (indicated by the scissors).
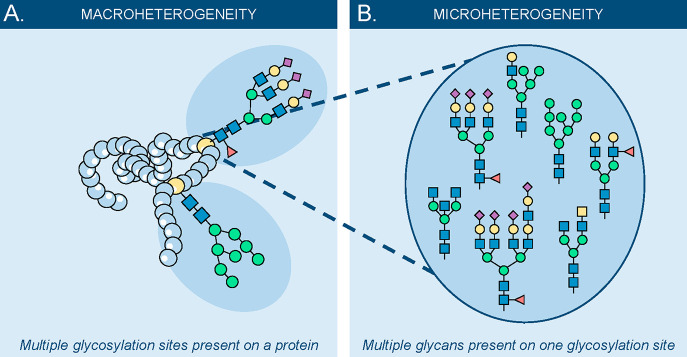
Glycoproteins can be further distinguished into those containing N-linked glycosylation (attachment through a nitrogen atom) and O-linked glycosylation (attachment through an oxygen atom). N-Linked glycosylation occurs only within a specific amino acid sequence (asparagine–X–serine or threonine (Asn–Xxx–Ser/Thr or N–X–S/T)). In this consensus sequence, X can be any of the amino acids except for proline (Pro or P). Unlike N-linked glycosylation, there is no clear amino acid sequence for O-linked glycosylation except that the glycan is attached to a Ser (S) or Thr (T). A glycoprotein can have several glycosylation sites (macroheterogeneity; Figure 2A) and various glycan species on a single site (microheterogeneity; Figure 2B). Whether or not the glycosylation site of a protein is fully occupied, partially occupied, or unoccupied depends on the conformation of the protein, spatial and temporal availability of glycosyltransferases, their activity, transcription factors, as well as the availability of sugar precursors.
Figure 2.
Macro- versus microheterogeneity of a glycoprotein. (A) Macroheterogeneity is the diversity of (multiple) glycosylation sites on a single glycoprotein. (B) Microheterogeneity is the variety on a single glycosylation site, where various glycan structures can be found.
The level of characterization can be selected dependent on the research question, varying from the intact analysis of a purified glycoprotein up to the analysis of the total released glycome of complex biological matrices (Figure 1B). The intact analysis allows, next to its glycosylation, to study the presence of other (post-translational) modifications, also known as proteoforms. While this provides insightful information about the protein it also requires pure substances, high sensitivity, high-resolution mass spectrometry (MS) analysis, and often additional separation techniques are applied. Moreover, the data analysis can be rather complicated as multiple modifications should be taken into account. In the case of biopharmaceuticals, the data analysis can be simplified by using a middle-up or middle-down approach. For this purpose, specific enzymes are available that perform a proteolytic cleavage in the hinge regions of the IgG resulting in specific Fc and antigen-binding fragment (Fab) domain. Dependent on the enzyme and/or additional reduction steps, individual subunits (Fc/2 or Fab/2) can be analyzed or the complete Fc2 and Fab2 domain can be investigated. This approach allows getting insights into the macroheterogeneity of the glycoprotein. However, it becomes complicated in the case of a large macroheterogeneity, and often it remains difficult to define glycoprotein microheterogeneity. HT glycomic studies on an intact level are still sparse on large sample sets (e.g., one of the largest intact HT glycomic studies was performed using 96 serum samples)18 and are more applied to glycopeptide and released glycan approaches.
The identification and characterization of glycopeptides can be established through a bottom-up approach. This method allows confirmation of the protein identity when a glycopeptide holds a unique peptide backbone as well as the occupancy of a specific glycosylation site. But also here, in the case of a large microheterogeneity, the complexity of the data can be a bottleneck, slowing down the throughput. By applying tandem MS, more information can be obtained about the structural composition of the glycan and the peptide. For HT analysis, the complexity of the sample needs to be condensed by efficient sample preparation steps such as affinity enrichment steps. Excellent examples for HT workflows are those for IgG, immunoglobulin A (IgA), and α-1-acid glycoprotein (AGP) enriched from serum or plasma19−21 or that of prostate-specific antigen (PSA) from seminal fluid,22 all allowing the complete sample preparation step in a 96-well plate format.
Released glycan analysis allows the analysis of the total glycomic profile of complex biomatrices by releasing all the glycans from the glycoproteins present in the sample. This is in great contrast to that of the intact, middle-down, and bottom-up approach where a rather pure sample is required prior to MS analysis. For the analysis of N-linked glycans, the enzyme peptide-N-glycosidase F (PNGaseF) is often utilized as it is able to release all N-linked glycans with a free reducing end from proteins (except when a core-fucose is present in α3-linkage) (Figure 3). More challenging is the release of O-linked glycans, as there is no universal enzyme available that can cleave all O-glycans, therefore, reductive β-elimination, a chemical approach, is often applied. Just recently, a workflow was published that allows the sequential release of N- and O-glycan in a 96-well format for approximately 500 000 cells using a PVDF membrane.23 However, this approach has not yet been performed on a larger sample set, which is mainly due to the time-consuming (manual) data interpretation and is therefore not yet to be considered a HT approach. Of note, this approach has been further optimized by de Haan et al.24 and employs additional 2-aminobenzamide (2-AB) labeling step, facilitating O-glycan isomer separation and enhancing sensitivity of detection.
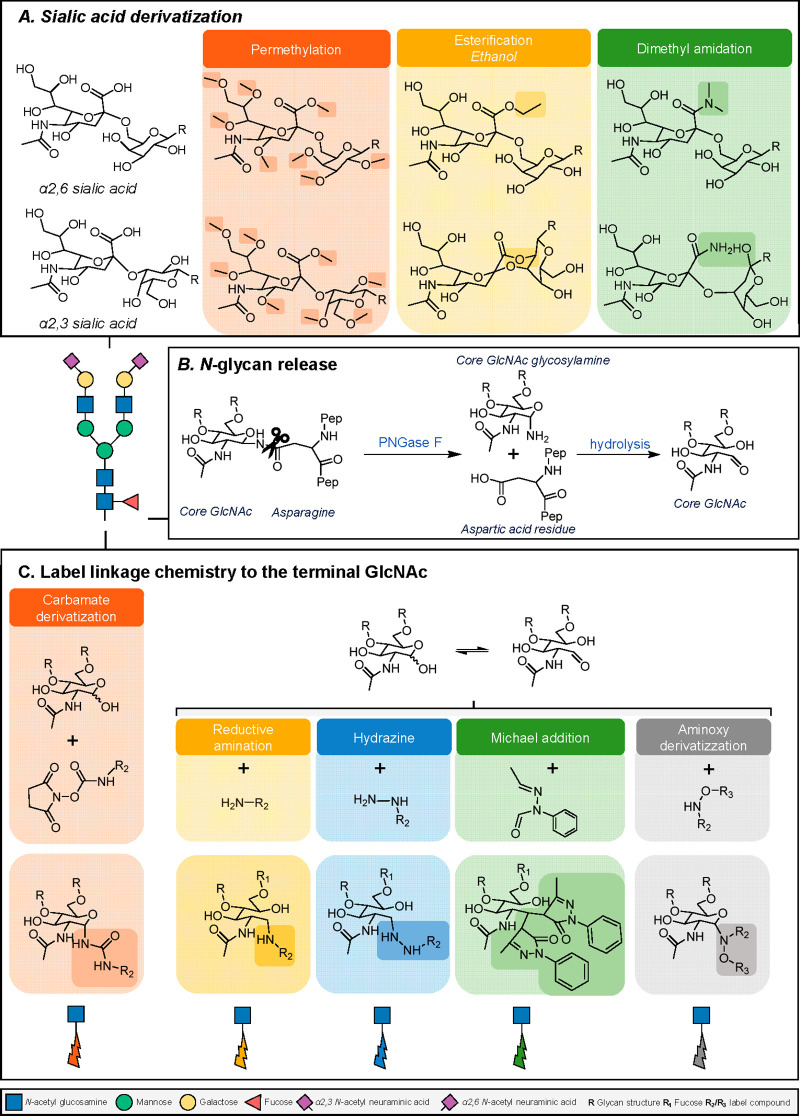
Figure 3.
A diversity of different chemistries is available to enable glycan analysis. (A) The most common derivatization strategies applied on terminal sialic acids, enabling stabilization and, in regard to esterification or dimethylamidation, also identification of the different isomers based upon mass difference by MS. In regard to the dimethylamidation procedure, the reaction consists of two parts. In the first step, α2,3-linked sialic acids react with the adjacent galactose to form a lactone, and the α2,6-linked sialic acids form a stable dimethylamide. The second step involves the addition of ammonia, with the lactone undergoing aminolysis, thereby transforming the carboxylic acid into a stable amide.26 (B) Illustration how an N-glycan attached to a protein (via an asparagine) is cleaved using the enzyme PNGase F. (C) Common procedures that are performed at the reducing end of the glycan: fluorescence detection can be enabled by introducing a label with a fluorophore, or MS ionization can be improved by adding a permanent positive charge (e.g., hydrazide labeling) or introducing a tertiary amine (e.g., carbamate chemistry), which could also allow the simultaneous analysis of glycans from different samples through the incorporation of stable isotopes (e.g., TMT-labeling).
In contrast, various workflows are developed to target solely the N-glycans, and most HT glycomic platforms result in two-dimensional (2D) data (intensity versus time or m/z), which can be used for the characterization and detection of the N-glycome. Next to the fact that the 2D format makes it easier to perform data analysis, established platforms with sizable glycan databases or repositories can be used (Table 1). Several extensive studies have been performed by using a separation platform coupled to a fluorescent detector (FLD) or by direct analysis using MS. However, it should be noted that all of these analytical platforms require extra sample preparation steps to enable their detection. For example, glycans themselves do not contain a fluorophore, which makes it impossible to detect them at high sensitivity by fluorescence. Therefore, the reducing end is often chemically modified by adding a label with fluorescent properties (Figure 3). For the detection by MS, it should be taken into account that sialic acids tend to be rather unstable in positive ionization mode when a time-of-flight (TOF) analyzer is being used. To avoid this, a derivatization approach is applied (Figure 3) and dependent on the chemistry a distinction can be made between the differently linked sialic acids as a mass difference is introduced (esterification or dimethylamidation).25
Table 1. Most Common Databases, Repositories and Software Tools for Glycomic Studiesa.
| database | description | URL |
|---|---|---|
| Carbohydrate Databases | ||
| Carbohydrate Structure Database (CSDB) | manually curated natural carbohydrate structures, taxonomy, bibliography, NMR, and other data from the literature (up to 2019). | http://csdb.glycoscience.ru/ |
| GAG-DB | a database that contains 3D structures of GAG binding proteins | https://gagdb.glycopedia.eu/ |
| GlycoBase (now GlycoStore) | over 650 N- and O-linked glycan structures available (exoglycosidase sequencing, U(H)PLC, and MS (MALDI-MS, LC-MS/MS) data | http://www.glycostore.org |
| GlycomeDB | contains glycan structures but has now been implemented in GlyTouCan | http://www.glycome-db.org/ |
| GlyConnect | contains glycan structures and their association with proteins, glycopeptide and glycosylation sties (curated); GlyConnect is integrated with GlyGen | https://glyconnect.expasy.org/ |
| Glycosciences.DB | a web portal that contains glycoinformatic databases and tools with a specific focus on 3D structures and 3D models; contains over 27 000 glycan entries, 13 900 3D structures, and almost 3500 NMR spectra | http://www.glycosciences.de/ |
| GlycoStore | a curated chromatographic, electrophoretic, and mass spectrometry composition database of N-, O-, and GSL glycans and free oligosaccharides associated with a range of glycoproteins, glycolipids, and biotherapeutics; approximately 850 unique glycan structure entries supported by over 10 000 retention positions determined (HILIC-FLD, U(H)PLC-FLD, PGC-LC-MS, and CGE-LIF). | https://www.glycostore.org/ |
| GlyGen | contains glycan structures as well as their association with proteins | https://www.glygen.org/ |
| GlyTouCan | a repository where more than 120 000 glycan structures are registered (uncurated); each glycan structure is assigned with a unique accession number | https://glytoucan.org/ |
| GlyXbase | database that contains the GU values of more than 400 glycan structures (glyXera) | Goldberg et al.34 |
| GUdatabase | library of APTS labeled N-glycans analyzed by C100 HT multicapillary electrophoresis system | https://lendulet.uni-pannon.hu/index.php/tools/2-uncategorised/46-c100htdatabase |
| KEGG | contains glycan structures present in KEGG pathways | http://www.genome.jp/ligand/kcam/ |
| Lipopolysaccharide | database for glycolipid structures (updated until 2007) | http://lipidbank.jp/cgi-bin/main.cgi?id=CLS |
| UniCarb-DB | a database and repository for glycomic MS data. Over 1100 structures and 1550 MS spectra are provided. | https://unicarb-db.expasy.org/ |
| Glycan Binding Proteins | ||
| Glycan Array Dashboard (GLAD) | automatically analyzes and visualizes glycan array data using glycan-binding intensities as input | https://glycotoolkit.com/GLAD/ |
| Glydin’ | visualizes and map the similarity of glycoepitopes in an interactive network | https://glycoproteome.expasy.org/epitopes/ |
| GlycoEpitopeDB | a database of antibodies that recognizes carbohydrates and glyco-epitopes (curated) | https://www.glycoepitope.jp/epitopes |
| MatrixDB | database that is focused on the interaction by ECM proteins, proteoglycan and GAGs (curated) | http://matrixdb.univ-lyon1.fr/ |
| SugarBindDB | database that provides information on carbohydrate sequences to which pathogenic organisms specifically adhere (curated) | https://sugarbind.expasy.org/ |
| Glycan Processing Pathways and Enzymes | ||
| Carbohydrate-Active enZYmes Database (CAZy) | a database that holds information about carbohydrate-active enzymes | http://www.cazy.org/ |
| GlycoGene DataBase (GGdb) | a database that provides information about which genes are associated with the biosynthetic pathway of glycans (e.g., glycosyltransferases, sugar nucleotide synthases, sugar-nucleotide transporters, sulfotransferases); over 180 human glycogenes are identified, cloned, and characterized | https://acgg.asia/ggdb2/ |
| Glycologue | prediction of glycosyltransferases and enzymes involved in the biosynthetic pathway of O- and N-glycans, human milk oligosaccharides (HMOs) as well as gangliosides | https://glycologue.org/ |
| SphinGOMAP | provides a pathway map for (glyco)sphingolipid biosynthesis | http://sphingolab.biology.gatech.edu/ |
| Software Tools | ||
| AutoGU | automated annotation and quantification of glycans using the GU index (HPLC-FLD data) | https://academic.oup.com/bioinformatics/article/24/9/1214/206953 |
| AutoGUI | automated annotation and quantification of glycans using the GU index (LC-MS data) | https://github.com/ruizhang84/GlycanGUIApp |
| Byonic | commercial tool for automated identification of glycopeptides using MS/MS data | https://www.proteinmetrics.com/products/byonic/ |
| Cartoonist | annotation of MS peaks with N-glycan cartoons | available at request to the authors35 |
| GlycanAnalysis | annotation of MS/MS spectra using glycan structures from GlycomeDB and KEGG glycan | https://www.shimadzu.co.jp/mass-research/soft.html |
| GlycanAnalyzer | automatically interprets exoglycosidase array by pattern matching of peak shifts of N-glycans after exoglycosidase digestion | https://glycananalyzer.neb.com |
| GlycoDigest | a software tool that simulates the exoglycosidase digestion (based upon GlycoBase) | https://glycoproteome.expasy.org/glycodigest/ |
| GlyConnect Compozitor | visualizes a set of glycan compositions and creates a network based upon shared monosaccharides | https://glyconnect.expasy.org/compozitor/ |
| GlycoForest | software tool that uses a partial de novo algorithm for sequencing glycan structures based on MS/MS spectra | https://glycoforest.expasy.org/ |
| GlycanMass | assists in the calculation of glycans masses (free reducing end, permethylated, or peracetylated) in Daltons | https://web.expasy.org/glycanmass/ |
| Glyco@Expasy | provides an overview of web-based glycoinformatic resources (portals, tools, and databases) | https://glycoproteome.expasy.org/glycomicsexpasy/ |
| GlycoMod | prediction of possible glycan compositions on proteins based upon mass (free, derivatized glycans and glycopeptides) | https://web.expasy.org/glycomod/ |
| Glycologue | a simulator of the enzymes involved in the biosynthesis of HMOs | https://glycologue.org/m/ |
| GlycoPAT | automated identification of glycopeptides of MS/MS data | https://virtualglycome.org/glycopat/ |
| GlycoPeptideGraphMS | automated identification of glycopeptides LC- and CE-MS data based upon known elution/migration criteria; at least one glycopeptide (node) should be assigned in the data using MS/MS. | https://bitbucket.org/glycoaddict/glycopeptidegraphms/src/master/ |
| GlycoReSoft | automated identification of glycopeptides of MS/MS data | https://github.com/mobiusklein/glycresoft |
| GlycoWorkbench | tool to assist in the interpretation of glycomic MS data | https://code.google.com/archive/p/glycoworkbench/ |
| Glynsight | visualizes and enables an interactive comparison of N- and O-glycan expression profiles | https://glycoproteome.expasy.org/glynsight/ |
| glyXtoolMS | automated identification of glycopeptides using MS/MS data | https://github.com/glyXera/glyXtoolMS |
| glyXtoolCE | commercial tool for the automatic annotation of CE-LIF data | https://www.separations.eu.tosohbioscience.com/OpenPDF.aspx?path=~/File%20Library/TBG/Products%20Download/Application%20Note/a21l02a.pdf |
| GRITS | assists in the interpretation of glycomics MS data | http://www.grits-toolbox.org/ |
| GUcal | integrated approach with GlycoStore for the analysis of N-glycans by CE-LIF | https://lendulet.uni-pannon.hu/index.php/tools |
| HappyTools | automatic annotation of LC- and CE fluorescence data | https://github.com/Tarskin/HappyTools |
| LaCyTools | automatic annotation of LC- and CE-MS data | https://github.com/Tarskin/LaCyTools |
| Mass Spectrometry-Based Automated Glycopeptide Identification Platform (MAGIC) | automatic annotation of glycopeptides using CID spectra | https://ms.iis.sinica.edu.tw/COmics/Software_MAGIC.html |
| MassyTools | automatic annotation of MS data | https://github.com/Tarskin/MassyTools |
| O-pair (MetaMorpheus) | automated identification of O-glycopeptides of MS/MS data | https://github.com/smith-chem-wisc/MetaMorpheus |
| Peptoonist | annotation of MS peaks with N-glycan cartoons including the peptide backbone | https://pubs.acs.org/doi/10.1021/pr070239f |
| PepSweetner | assists in the manual annotation of intact glycopeptide spectra, matching the molecular mass of the precursor mass to theoretical human N-glycopeptides | https://glycoproteome.expasy.org/pepsweetener/app/ |
| pGlyco | annotation of N-glycopeptides | https://github.com/pFindStudio/pGlyco3/releases |
| SimGlycan | commercial tool for the annotation of MS data using an internal database | http://www.premierbiosoft.com/glycan/ |
| Skyline | assists in the annotation and identification of glycan and glycopeptides using LC- or CE-MS data | https://skyline.ms/ |
| SwissMassAbacus | assists in the calculation of glycan and glycopeptide masses in Daltons | https://glycoproteome.expasy.org/swiss-mass-abacus/ |
| UNIFI | commercial tool for automatic assignment of glycans based upon GU values (LC) and m/z value (MS) | https://www.waters.com/nextgen/us/en.html |
NMR, nuclear magnetic resonance; 3D, three-dimensional; MALDI, matrix assisted laser desorption/ionization; LC, liquid chromatography; HILIC, hydrophilic-interaction liquid chromatography; PGC, porous graphitic carbon; LIF, laser-induced fluorescence detection; GU, glucose unit; APTS, 8-amino-1,3,6-pyrenetrisulfonic acid trisodium salt; ECM, extracellular matrix; HPLC, high-performance liquid chromatography.
3. Experimental Design of High-Throughput Glycomics Study
HT glycomics results in large amounts of data being generated during a course of usually several weeks or months and requires several steps of processing. Samples are analyzed in 96-sample (or 384-sample) batches which can be affected by experimental conditions, e.g., different batches of reagents, variable laboratory conditions, multiple analysts as well as different instruments. Moreover, glycosylation is confounded by different biological variables that have to be accounted for during the analysis. Proper experimental design is therefore of utmost importance to ensure the quality of generated data.
Human protein glycosylation is shown to be vastly variable between individuals of different ages, sex, body mass index (BMI), smoking habits, medication use, pregnancy, and inflammatory status, as well as a geographical origin.27−33 These variables are important confounders that should be taken into account in the design of a study, making sure they are equally distributed between studied biological groups (e.g., controls and patients, different therapy groups, etc). Moreover, although glycans are generally stable at common sample storage conditions, the differential sample preprocessing steps (especially for complex biological samples like plasma/serum, tissue samples, etc.) might have an impact on the obtained results due to loss/aggregation of specific glycoproteins if the analysis is done on the level of released glycans. Therefore, sample preprocessing, as well as storage information and location of the sample collection, should also be considered.
Proper experimental design of HT glycomics studies takes into account known biological confounders during blocked randomization and ensures that technical replicates of a representative standard sample are included in each batch of samples. Through blocked randomization, all known biological and experimental confounders should be equally distributed between each batch (block) of samples. In that case, any observed systematic shifts of individual batches most likely originate from experimental and not biological variation. Replication of a standard sample goes hand-in-hand with the blocked design of a study because it allows for the detection of potential batch effects (systematic shifts in measured data originating from the same 96- or 384-well plate) and any other technical variations caused by experimental conditions. When the abovementioned conditions in experimental design are met, it is possible to perform statistical batch correction which can somewhat aid in obtaining better quality data that would otherwise be skewed due to batch effects. Standard sample replicates also enable quality control of overall method variability during cohort analysis.
In addition to standard sample replicates added at the beginning of a study and processed together with the samples, it is common to add another set of standards (system suitability standards) during sample measurement. These depend on the technology being used for the glycan analysis, e.g., fluorescently labeled released N-glycans in ultrahigh-performance liquid chromatography (UHPLC) and capillary gel electrophoresis (CGE) analysis and are analyzed in parallel to cohort samples during every data acquisition sample set on each instrument used for the cohort analysis. This ensures that the instruments used in the study are working within the desired specifications and allow detection of any significant impact that deteriorating instrument components (e.g., UHPLC column, CGE polymer) might have on the analysis.
4. Historical Overview
Although a variety of analytical methods already exist for small-scale glycan studies, the demand for HT approaches that allow large-scale glycoprofiling continues to develop. These large-scale studies include from several hundred to several thousand samples of patients as well as healthy individuals. Methods used in such immense studies must possess the capacity to precisely analyze the glycome of many samples in a reasonable time frame and at a reasonable cost.
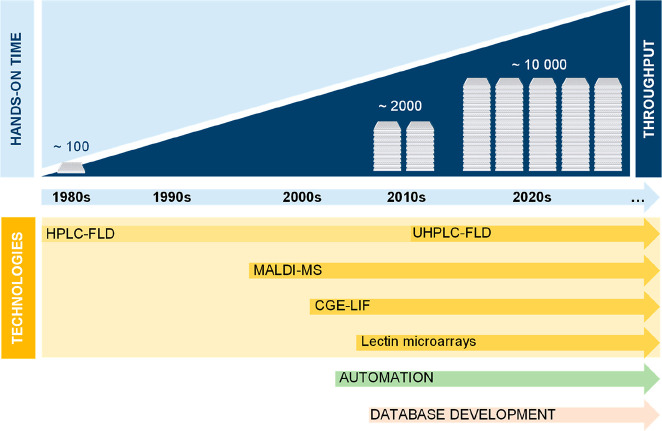
The development of HPLC, as one of the main techniques for glycan separation and detection, and a forerunner technique of UHPLC, began in the 1980s for small-scale studies, using laborious and time-consuming sample preparation protocols (Figure 4).36 These studies set the groundwork for the development of HT glycomics. Seminal work by Mullinax et al. in 1976,37 Parekh et al. in 198510 and 1988,38 and Ashford et al. in 198710,39, analyzing between 50 and 150 samples have set the stage for future HT applications. Glycans were released by hydrazinolysis developed by Takasaki et al. in 1982.40 For labeling of released glycans, the radioactive isotope NaB3H4 was used as described in 1974 by Takasaki and Kobata.41 Nowadays, a combination of these two sample preparation approaches is practically obsolete for HT analyses, as novel approaches are now available that avoid the use of radioactive compounds and come with less demanding sample preparation steps. Glycans were fractioned by Bio-Gel P-4 (400 mesh) gel filtration chromatography to separate neutral glycans based on their effective hydrodynamic volumes and coupled to HPLC radioactivity flow monitor to collect analogue signals.
Figure 4.
Historical overview of HT glycomic technologies applied for released N-glycan analysis.
Hase et al., first introduced fluorescent tagging of free oligosaccharides by reductive amination back in 1979,42 and glycans derivatized in such a way were analyzed by 2D paper electrophoresis. The sensitivity of fluorescent labeling was further improved by using HPLC with an FLD in 1981,43 and workflow for fluorescent labeling and reversed-phase (RP-)HPLC-FLD analysis of N-glycans was further optimized by Hase et al. in 1986,44 enabling glycan structure estimation from only several hundreds pmol of glycans. Glycans were released by hydrazinolysis and after subsequent N-acetylation, the reducing ends of glycans were coupled with 2-aminopyridine (2-PA) with sodium cyanoborohydride (NaBH3CN) as a reducing agent. The reaction of reductive amination was performed at 90 °C for 15 h, after which the obtained fluorescent derivatives were purified by Bio-Gel P-2 column chromatography and separated using RP-HPLC. In 1987, Takahashi et al., published a comparative structural study of the neutral N-linked glycans of human normal polyclonal IgG and pathological IgG1 isolated from the sera of patients with multiple myeloma.45 After desialylation using neuraminidase as well as pepsin digestion, IgG glycopeptides were digested with N-oligosaccharide glycopeptidase from almonds. The reducing ends of the obtained N-glycan chains were reductively aminated with the fluorescent reagent 2-PA and purified by gel filtration. The mixture of N-glycans was separated by RP-HPLC into 15 peaks. This study is one of the first comparative studies of IgG N-glycosylation employing today’s most widely used general workflow for LC analysis of released N-glycans consisting of enzymatic digestion, fluorescent labeling, cleanup, and analysis. In conjunction with hydrazinolysis, in most of the previously mentioned studies, labeled glycans were subjected to exoglycosidase (e.g., neuraminidase) treatment prior to oligosaccharide analysis, so that only particular glycosylation traits such as galactosylation were analyzed.
Glycan release by hydrazinolysis has several significant flaws, including the requirement of using anhydrous hydrazine, which is an extremely hazardous and explosive substance. In 1984 and 1991, Plummer et al., described the discovery46 and purification47 of a novel enzyme PNGase F, an amidase (amidohydrolase) that hydrolyzes the asparagine side chain amide bond of a wide variety of glycoprotein/glycopeptide substrates, generating an aspartic acid residue on a protein backbone and liberating the 1-amino oligosaccharide (glycosylamine). The latter slowly hydrolyzes nonenzymatically to ammonia and an oligosaccharide with a di-N-acetyl-chitobiose unit at the reducing end. The ability to cleave the major N-oligosaccharide classes of human glycoproteins, without the use of any other enzyme and under mild reaction conditions, makes the PNGase F a perfect candidate for obtaining released N-glycans in HT analysis.48
In the late 1980s, when Karas et al., were developing MALDI-MS, they already recognized its relevance for the MS-based analysis of saccharides. In the publication of 1987, which introduces MALDI for the analysis of nonvolatile compounds, it was demonstrated that ultraviolet laser desorption (UVLD)-MS analysis of stachyose (Gal(α1→6)Gal(α1→6)Glc(α1↔2β)Fru, where Gal, galactose; Glc, glucose; Fru, fructose) benefitted from the use of an ultraviolet (UV)-absorbing chemical matrix, nicotinic acid (NicAc), or tryptophan (Trp) to produce quasimolecular ions ([M + Na]+) of stachyose. The matrix-assisted analysis showed superior sensitivity compared to the UVLD-based analysis.49 A few years after, in 1993, Harvey showed the quantitative capabilities of MALDI-MS in oligosaccharide measurements.50 He confirmed a linear relation between oligosaccharide concentration and measured MS signal intensity over several orders of magnitude in analyte concentration. Additionally, it was shown that MALDI-MS is able to detect low femtomol amounts of various oligosaccharides using 2,5-dihydroxybenzoic acid (DHB) as a matrix, highlighting the usability of MALDI-MS as a comprehensive readout for complex oligosaccharide mixtures with large variations in dynamic range.
In 1997, Rudd et al., described the release of N-glycans directly from a band on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel using PNGase F.51 The reducing termini of the N-glycans were labeled with 2-AB to allow direct quantitation from the HPLC profiles. An amide-silica HPLC column enabled high-resolution separations of both charged and neutral N-glycans. The equivalent approach combined with exoglycosidase digestions was used by Küster et al. in 1997 to profile N-glycans by MALDI-MS and HPLC.52 In 2008, Royle et al. presented a robust, fully automatable technology platform, including software for the detailed analysis of N-linked glycans released from glycoproteins, which included sample immobilization in 96-well plates, glycan release with PNGase F, and fluorescent labeling.53 Relative quantitative HPLC analysis included the monosaccharide sequence, linkage, and arm-specific information for charged and neutral N-glycans, as well as an automatic structural assignment of peaks from HPLC profiles via web-based software that accessed the GlycoBase database of more than 350 N-glycan structures, including 117 present in human serum glycome. Another software (autoGU) was used to stepwise analyze data from exoglycosidase digests to generate a refined list of final structures. N-Glycans from a 96-sample plate could be released and purified in two or three days and profiled in two days. Although the SDS-PAGE and in-gel deglycosylation of IgG are unfavorable for HT analysis, these methods represented a milestone for the further development of HT approaches for released N-glycan analysis, especially the latter method in 96-well plate format. The first application of this method on a large cohort revealed variability, heritability, and key environmental factors that influence plasma N-glycome composition.27
The development of MALDI-MS instrumentation in the years following its introduction resulted in the recognition that the analytical tool is perfectly suited for HT analysis. Initially recognized by Hsieh et al. in the context of pharmaceutical compound screening, MALDI-MS was praised for its relative ease of the sample preparation and minute amounts of sample required, its high salt tolerance compared to electrospray ionization (ESI)-based platforms, the large mass range, and its measurement sensitivity.54 In the presented context and in the early days of HT MALDI-MS platforms, its use was able to increase throughput to approximately 10 s per sample. Hsieh et al. concluded the work with the mention that upon the introduction of MALDI-MS as a HT readout platform, the main throughput-limiting factor in the wet lab would become the sample preparation.54 However, the ease and repetitive nature of the MALDI-specific sample preparation steps and the similarities of the MALDI target plate to a well-based sample plate make the complete MALDI-MS workflow extremely compatible with automation strategies. As mentioned above, the conversion of sample preparation to the 96-well format was a major breakthrough for HT glycomics. One of the more common strategies for the MALDI-MS-based analysis of released N-glycans from complex protein mixtures, or purified proteins is through the capturing of proteins on a polyvinylidene fluoride (PVDF) membrane and subsequent enzymatic removal of the glycans from their carrier proteins. The use of PVDF membranes for protein sequencing55 and N-glycan analysis56,57 originates from the late 1980s to mid 1990s of the previous century. Papac et al. were the first to implement this workflow in a 96-well format for the effective deglycosylation of 50 μg of the glycoprotein recombinant tissue-type plasminogen activator (rt-PA).58 The basic workflow consisted of capturing the glycoproteins on the PVDF membrane, a reduction and alkylation of disulfide bonds disrupting the tertiary and quaternary of proteins as well as protein complexes, and blocking of all open PVDF protein binding sites using a polyvinylpyrrolidone polymer (PVP360). Prior to MALDI-MS analysis, a 3 h enzymatic deglycosylation step using PNGase F was performed and released N-glycans were desalted using cation exchange. This initial report showed the possibility of deglycosylating and analyzing 60 samples in 8 h, effectively being 8 min per glycoprotein sample.58
In parallel with developments of HT approaches for HPLC N-glycan analysis, innovative work by Callewaert et al. in the 2000s has set grounds for N-glycan analysis by CGE technology using widely available DNA sequencers.59−61 The workflow was equivalent to the one developed for HPLC N-glycan profiling consisting of enzymatic glycoprotein digestion by PNGaseF, desalting, fluorescent labeling with APTS, and cleanup procedure performed in 96-well plates. In 2004, Guttman et al. presented an automated 96-capillary array that allows the analysis and characterization of mono- and oligosaccharides.62,63 However, as this workflow did not involve the enzymatic release of N-glycans, an improved sample preparation method was introduced that could be applied to study the N-glycome of glycoproteins64 and to study for IgG specific glycosylation.65
Another breakthrough in HT N-glycan analysis was the deployment of the so-called in-solution N-glycan release and labeling instead of deglycosylation of immobilized proteins in SDS-PAGE gels as reported by Royle et al. in 2008.53 Not only was the in-gel method demanding in terms of invested labor, it also showed lower efficiency of recovering N-glycans from the gel, which affected the overall reproducibility of the method. In 2008, Ruhaak et al. published a protocol where plasma proteins were first denatured with SDS, followed by overnight incubation at 37 °C for the N-glycan release using PNGase F, all in the same 96-well plate.66 The samples were further processed in the same plate, released N-glycans were labeled with a mixture of a fluorescent dye 2-aminobenzoic acid (2-AA) and a reducing agent NaBH3CN in dimethyl sulfoxide (DMSO)/glacial acetic acid (10/3; v/v), a method developed a couple of years earlier by Bigge et al. in 1995.67 Labeled N-glycans were then purified using a HILIC-based solid-phase extraction (SPE) method with microcrystalline cellulose as the stationary phase. Eventually, the purified 2-AA labeled N-glycans were separated using normal phase HILIC-HPLC. This workflow was further optimized in 2010 to enable the separation and detection of released total plasma N-glycans by CGE-LIF.68N-Glycans were released enzymatically from denatured plasma glycoproteins in the same manner already described in 2008.66 However, labeling was performed by using a fluorescent dye 9-aminopyrene-1,4,6-trisulfonic acid in citric acid and 2-picoline borane (2-methylpyridine borane) complex as a nontoxic and efficient reducing agent in DMSO.
Large-scale studies on individual glycoprotein glycomes were partially hindered by the lack of methodologies that could enable fast and robust purification of a glycoprotein of interest from a large number of samples at an affordable price. Protein purification was therefore one of the major bottlenecks in large-scale proteomics and glycomics studies. IgG N-glycome analysis has largely been facilitated by the development of a Protein G monolithic 96-well plate. In 2011, Pučić et al. published the first large-scale population study of the IgG glycome applying a novel HT method for isolation of IgG from 2298 plasma samples and IgG N-glycan analysis using HILIC-UHPLC-FLD.69 The entire procedure for 96 samples, including the binding of plasma samples, washing, and elution of isolated IgG, was performed in less than 1.5 h and provided a significant milestone in HT analysis of IgG N-glycome, enabling subsequent large-scale population studies and genome-wide association studies (GWAS).70,71
In 2013, Burnina et al., developed a practical procedure to prepare fluorescently labeled N-glycans for both qualitative and quantitative analysis by MS and UHPLC.72 In a single 96-well filter plate with a hydrophobic membrane, antibodies (trastuzumab) samples were denatured, reduced, and deglycosylated by PNGase F digestion. The released N-glycans were then fluorescently labeled (InstantAB label for a rapid labeling procedure; 5 min at 37 °C) in a collection plate before being filtered using a hydrophilic membrane filter plate. The complete sample preparation method took less than 90 min and was done entirely in ready-to-use 96-well plates with simple buffer systems. These novel rapid labeling chemistries relied on the presence of glycosylamines, and therefore a fast deglycosylation step is required. On the basis of the research from Ruhaak et al.66 and Burnina et al. in 2015. Trbojević-Akmačić et al. further optimized a workflow for HT analysis of released N-glycans from plasma by HILIC-UHPLC-FLD73 by combining in solution deglycosylation, 2-AB labeling, and cleanup using only a hydrophilic membrane filter plate, which was the basis for some of the largest HT glycomic studies on total plasma/serum N-glycome and IgG N-glycome by the same technology.74−78
With the development of faster sample preparation protocols and analysis of large sample numbers, new challenges emerge regarding data processing and analysis. For separation-based techniques (e.g., UHPLC), manual peak integration is one of the most time-consuming tasks. Although (semi)automatic integration methods do exist in the proprietary software tools used for data acquisition, these are often not adapted for large cohorts due to small shifts in peak retention times that happen with the time of analysis. A significant shift in decreasing processing time occurred with the development of an automatic semisupervised method for peak integration.79
Although the detection and identification of N-glycans in HT glycomics, in general, predominantly relies on UHPLC, CGE, and MS methods,80 the drawbacks of these analytical platforms are the need for expensive equipment and complicated sample preparation steps, making lectin-based methods attractive for glycosylation research.81 Because of their high specificity and affinity toward glycans, lectins are another promising tool for glycan detection and the study of cells’ glycosylation. Using carbohydrate–lectin interactions, problems like separation and purifying carbohydrate-containing biomolecules are solved.
Lectins are a group of proteins that contain at least one noncatalytic site for specific and reversible binding to carbohydrates and the carbohydrate determinants of biopolymers without changing their structures.82,83 They are widely distributed in nature and have been isolated from viruses, fungi, bacteria, invertebrates, unicellular organisms, animals, and plants.84,85 Lectins play an important role in the immunological defense against pathogens,86 blocking of viral infections,87 regulation of microbial cell adhesion and migration,88 and control of protein levels in the blood.89
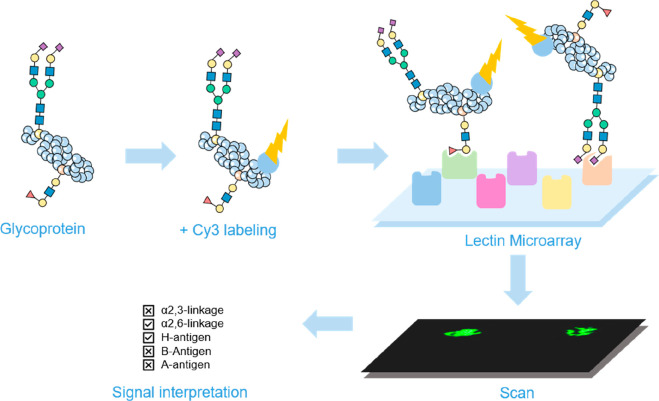
The carbohydrate-binding properties of lectins have historically been used for the separation and purification of glycoproteins, glycopeptides, and oligosaccharides by affinity chromatography (covalently bound to agarose, silica, or other polymer stationary phases),90−93 as histochemical and cytochemical reagents for detection of glycoconjugates in tissue sections, on cells and subcellular organelles,94,95 and in investigations of intracellular pathways of protein glycosylation and study of membrane transport,96 as well as for sorting cells using flow cytometry.97 Nowadays lectin microarrays are widely used as an analytical tool in various applications, e.g., to investigate the glycosylation in diverse cells during cell development and differentiation for analyzing cell processes, including cell differentiation and development, cell–cell communication, cell surface biomarker identification, and pathogen–host recognition.98
The first reports on lectin microarrays were published in 2005,99−102 and already in 2007 Rosenfeld et al. developed GlycoArray kits that enabled glycosylation analysis of intact glycoproteins within 4–6 h.103 This lectin microarray technology, utilizing nitrocellulose membrane-coated glass slides and labeled secondary antibodies with automated data analysis, showed high potential as a first-line tool for HT analysis directly from complex biological samples.
Development of sample preparation approaches for lectin microarray analysis allowed glycan profiling at the subnanogram level.104 However, lectin microarray application in HT glycomics has been lagging behind other, above-mentioned, techniques. Major reasons for that have been the limited repertoire of naturally occurring lectins (mostly from plants) that cannot detect all human glycosyl epitopes as well as insufficient sensitivity for some clinical samples and therefore rely on other techniques for structural elucidation.105 Even though they are currently not used in a HT glycomic analysis of hundreds to thousands of samples per cohort, lectin microarrays have recently been used for the analysis on the level of 50 to hundreds of samples. For example, this technology has been used in different applications from salivary glycoprotein analysis in type 2 diabetes mellitus,106 glycan analysis in hepatocellular carcinoma specimens107 and serum of prostate cancer (PCa) patients,108 IgG glycan analysis in primary biliary cholangitis,109 to seminal plasma glycome analysis in fertile and infertile subjects,110 demonstrating a perspective for future use on a larger scale. Lectins have also recently been employed in lectin-based biosensors.111,112
5. Technologies
HT glycomics relies on several technologies utilized for glycan profiling, mostly on the level of released glycans or glycopeptides. These technologies are often used in parallel because they offer different advantages and provide complementary information, e.g., UHPLC-FLD and LC-MS. Although general approaches in the sample preparation workflows are similar (Figure 5), each of these technologies also require some specific analytical conditions and data processing steps to obtain reliable and robust data (Figure 6).
Figure 5.
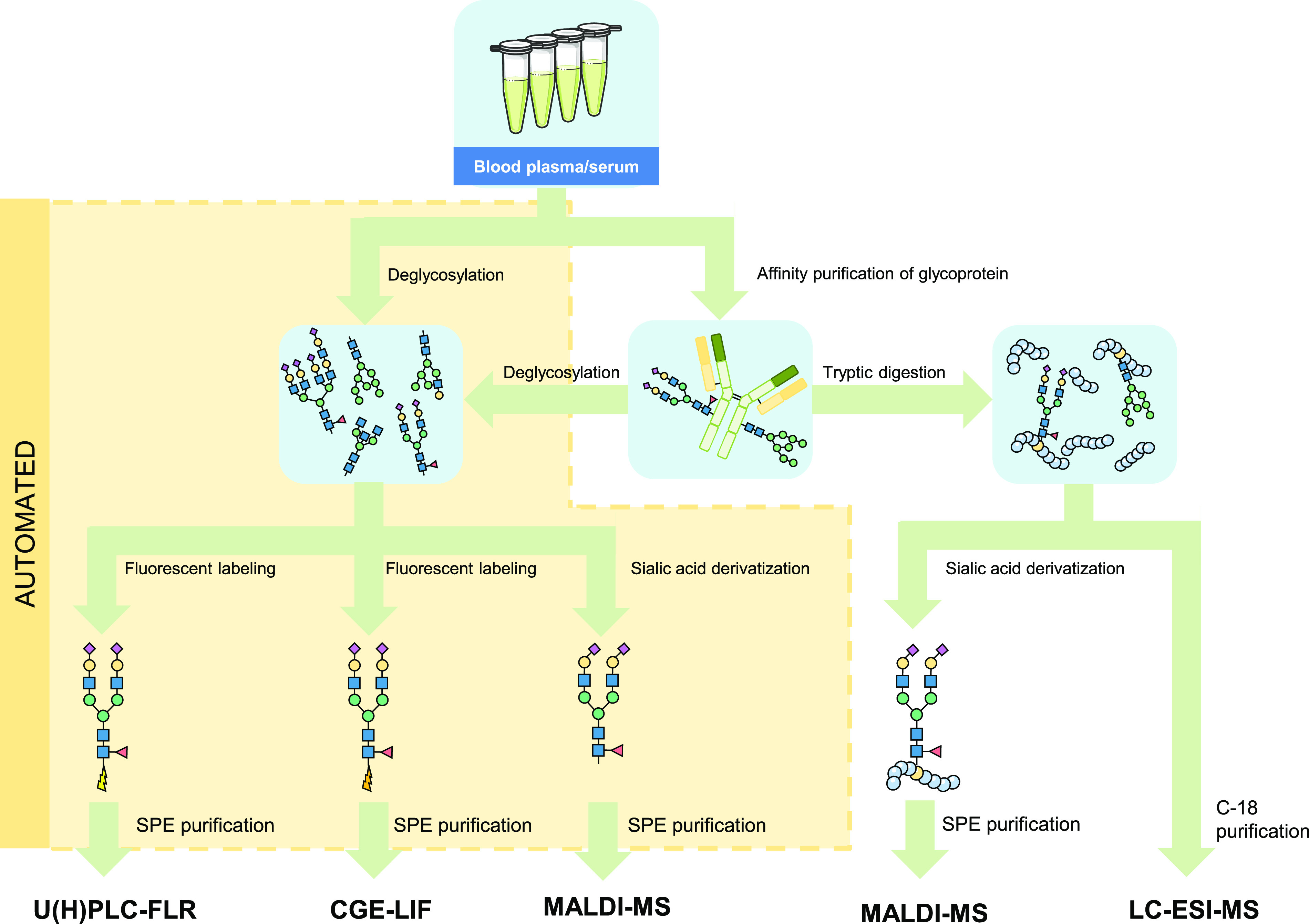
A diversity of workflows are available for HT glycomic analysis. For the different derivatization and labeling procedures as well as specific labels, see Figures 3 and 7, respectively. Please note that the sialic derivatization step of the glycopeptides will also modify the peptide backbone.
Figure 6.
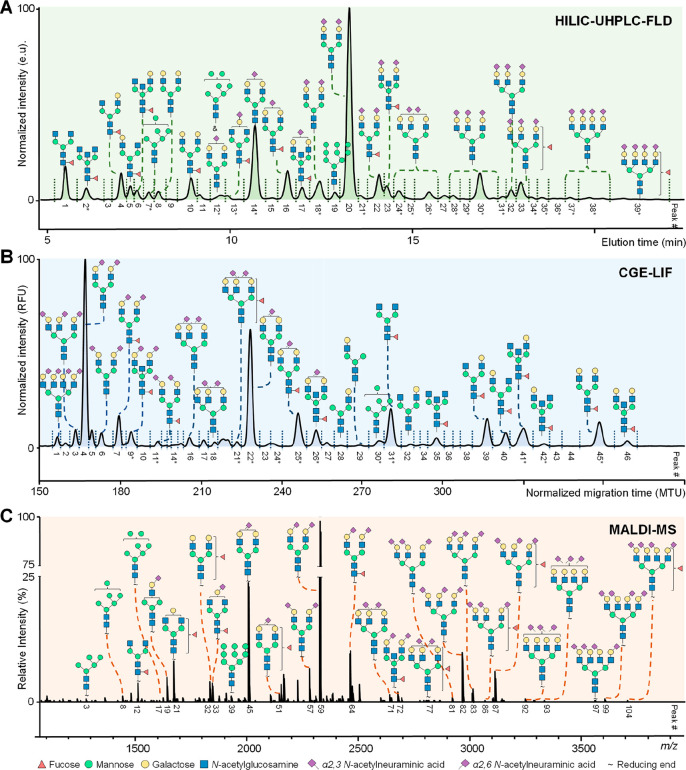
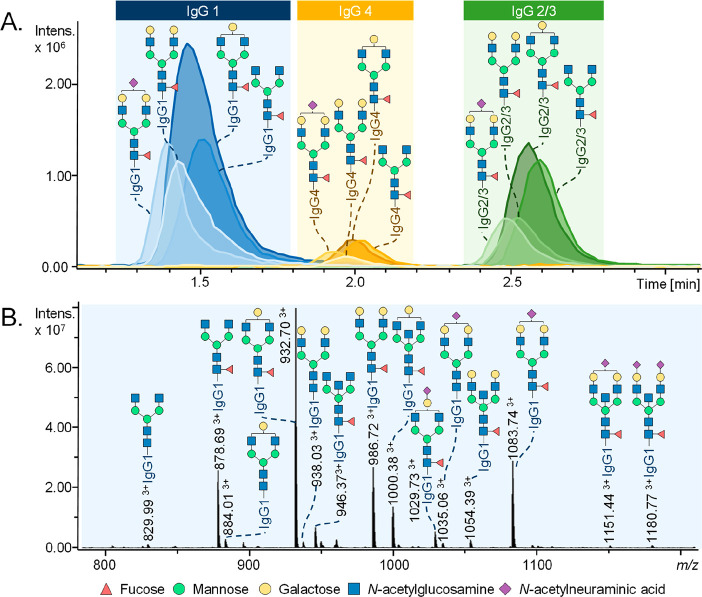
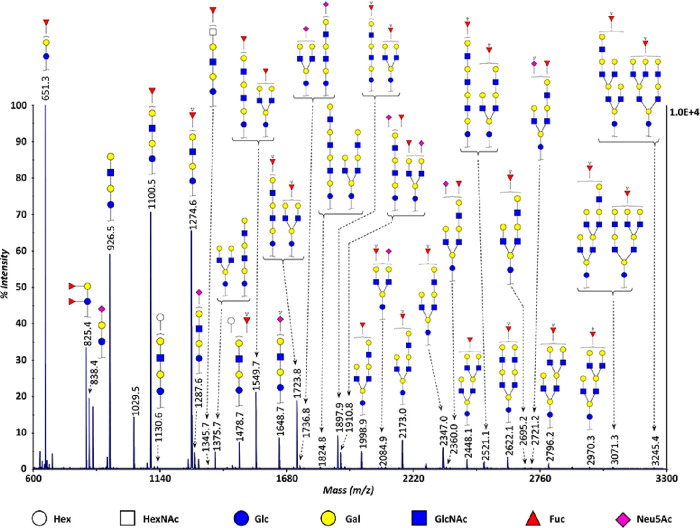
Exemplary profiles of the total serum N-glycome using (A) HILIC-UHPLC-FLD, (B) CGE-LIF, and (C) MALDI-MS. (A) Chromatogram after 2-AB labeling by HILIC-UHPLC-FLD. (B) Electropherogram after APTS labeling by CGE-LIF. (C) Mass spectrum after differential sialic acid esterification by MALDI-FT-ICR-MS. The assigned signals in the MS spectra correspond to [M + Na]+. Please note that HILIC-UHPLC-FLD and CGE-LIF provide (in some cases) isomer separation in regard to branching (galactose arm, bisection, and fucose position). Structures are assigned based on exoglycosidase treatment and/or tandem MS data as well as literature knowledge on biosynthetic pathway of N-glycans. *Some signals for the HILIC-UHPLC-FLD and CGE-LIF correspond to multiple N-glycan compositions for which the most abundant one is assigned in the figure.
5.1. Liquid Chromatography
Currently, several HT approaches for N-glycan analysis are in use, with HILIC-UHPLC-FLD being the most prevalent.113 While in the 1970s and 1980s glycan preparation workflows generally required sizable quantities of starting material, long hands-on time for sample preparation, and long analysis runs, the advancement of applicable chemistries, miniaturization of reaction volumes, and column particle technologies enabled UHPLC to become one of the most robust HT technologies used for glycan analysis. The UHPLC in combination with FLD allows complete characterization of complex glycan mixtures in a relatively short time and, although it requires high-end instrumentation, is much less expensive than MS. Thus, UHPLC is the method of choice for routine analysis of protein glycosylation with previously characterized glycan structures. For characterization of a novel glycan structure, UHPLC may be coupled with other methods that can provide further structural information, especially MS with LC-MS techniques.
5.1.1. Sample Preparation
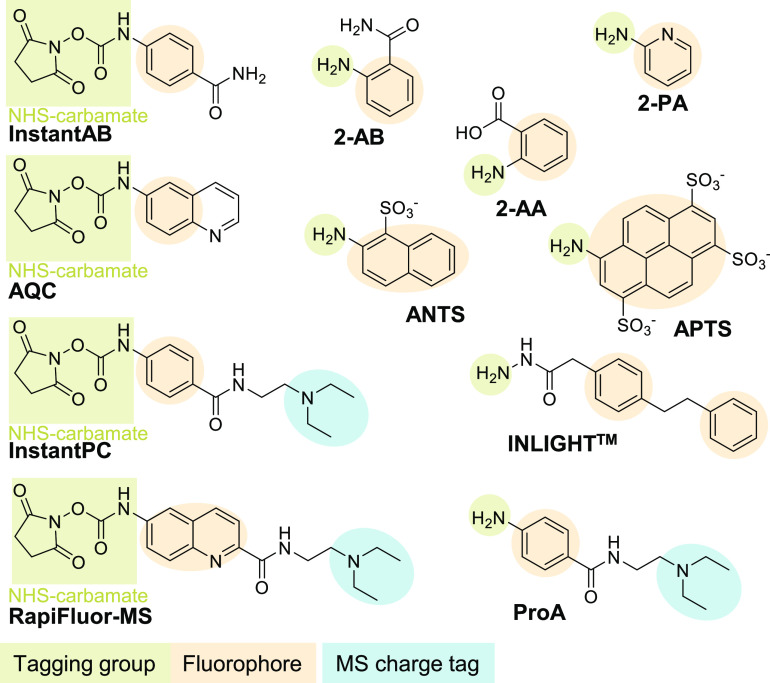
UHPLC is widely used for the analysis of released glycans, which means that prior to their separation and detection, glycans must be cleaved of their protein carrier. Samples are prepared usually by employing enzymatic deglycosylation by PNGase F, the most widely used enzyme in HT studies, fluorescent labeling of released glycans, cleanup procedure, and chromatographic analysis. Of note, while PNGase F is widely used in released N-glycan analysis, it appears to have poor efficiency for releasing highly truncated N-glycans with, e.g., only chitobiose or a single N-acetylglucosamine that appear to be rather common.114,115 Sometimes, an additional step of a cleanup procedure is being used after deglycosylation, facilitating the fluorescent labeling reaction efficiency.116,117 Historically, 2-AB has been the most widely used fluorescent label, starting from the first HT studies performed on the level of thousand samples.27,69,118,119 Although it results in high fluorescent signals, it is not easily ionizable, hindering MS characterization of labeled N-glycan species. Several other labels have recently been more and more applied, e.g., procainamide (ProA),120 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC),121 and RapiFluor-MS122 (Figure 7), showing equivalent or enhanced fluorescence compared to 2-AB, as well as ionization in the cases of ProA and RapiFluor-MS due to the introduction of a charged tertiary amine tag.117,120,123,124 Moreover, INLIGHT labeling strategy has been used to increase hydrophobicity and ionization of N-glycans for more efficient RP-LC-MS analysis.125,126 A typical fluorescent labeling reaction with 2-AB or ProA is performed at 65 °C for 2–3 h, creating a balance between labeling efficiency and the loss of sialic acids. With the development of rapid labeling chemistries (Figure 7), e.g., AQC,127 InstantAB,128 InstantPC,129 and RapiFluor-MS,130 the reaction time is significantly decreased to only 5 min, making this approach even more HT. Although rapid labeling chemistries significantly decrease the hands-on time, they suffer from a narrow range of starting sample quantity because all amine groups from the sample (N-glycosylamines, proteinaceous amines, other free amines) are being labeled, requiring a large excess of the labeling reagent. Therefore, rapid labels are currently more applicable to isolated glycoprotein N-glycan analysis (e.g., IgG) than for more complex samples like plasma/serum, where the level of amines can vary significantly between individual samples and might be more difficult to optimize before analysis. As the newly introduced labels also result in differential elution order of N-glycans, when it is FLD-based, it becomes complicated to compare the obtained results with previous studies as well as for data integration. Therefore, 2-AB is still widely used even though other labels have shown their advantages.
Figure 7.
A diversity of commonly used labels that can be attached to the reducing end of the glycan. Labeling is performed to enable fluorescence detection (all labels), to improve retention on RP-LC-FLD (2-AA, 2-AB, and 2-PA), to enable separation by introducing a negative charge for CGE-LIF (APTS, ANTS, 2-AA, or 2-PA), or to enhance MS ionization by introducing a tertiary amine through carbamate chemistry (InstantPC, RapiFluor-MS, ProA). 2-AB, 2-AA, and 2-PA can also be used as reactive MALDI matrices as these labels absorb UV light that is in the wavelength range of most common MALDI lasers (330–360 nm).
The cleanup procedure is generally performed by HILIC-SPE using different hydrophilic stationary phases, e.g., hydrophilic filters (in the form of 96-well plates)72,73 and hydrophilic bead-based cartridges or plates73,117 because these have proven to be effective in the removal of excess reagents and proteins from the previous sample preparation steps. Additionally, magnetic nanoparticles can also be used as a cleanup approach.120,131
Different variations of the standard glycan preparation workflows have recently become available in a kit format, e.g., GlycoWorks RapiFluor-MS N-Glycan Kit (Waters), AdvanceBio Gly-X N-Glycan Prep Kits (Agilent), and Glycoprofile Labeling Kits (Merck), expediting their application in basic HT glycomic research and biomarker discovery as well as automation.
5.1.2. Measurement and Data Processing
Fluorescently labeled glycans are generally analyzed in HT mode by HILIC-based LC due to its remarkable capabilities to separate polar and hydrophilic analytes in an aqueous–organic mobile phase. A linear gradient with an increasing percentage of 50 or 100 mM ammonium formate in acetonitrile enables efficient separation of N-glycan species depending on their charge, size, and linkages. Column chemistries are based on modified amide-based residues with 1.7–1.8 μm particles. Recent advancements in the column hardware surface preparation reduce the interactions between glycans and the metal surface of the column, resulting in improved peak recovery, shape, and reproducibility, which is especially important for sialylated species.79 After manual or automatic79 integration, total area normalization is usually used to extract glycan amounts as relative percentage areas (%area) used for further analysis, followed by batch correction and statistical analysis. Only recently, a systematic evaluations of other normalization methods for glycomics data, in addition to total area normalization, have been reported.132,133
5.1.3. Peak Assignment/Glycan Identification
GU values based on fluorescently labeled dextran ladder as an external standard are appointed to individual glycan structures separated by HILIC-UHPLC-FLD and used for the peak assignment. GU values are more stable than retention times of individual glycans, which can vary depending on the (U)HPLC system and chromatographic conditions used for the analysis and are usually used as reference values in the databases. Nowadays, extensive HPLC/UHPLC glycan databases containing GU values exist, although these are predominantly populated with 2-AB labeled glycans data, while migration information for alternatively used fluorescent labels is still largely missing and is one of the goals for future advancement of HT glycomic applications.
Exoglycosidase sequencing (glycan trimming with enzymes specific for different types of terminal sugar monomers) has been historically used as a complementary approach to indicate specific structural features found on glycans eluting in individual chromatographic peaks (Figure 8).51,134,135 Although it is worth noting that exoglycosidase sequencing has to be accompanied with MS characterization,69 either off-line after chromatographic peak collection or online by connecting UHPLC system to MS detector. Of note that, depending on the fluorescent label used for released glycan derivatization, exoglycosidase sequencing conditions might have to be optimized to achieve reliable results.136 In contrast to coupling CE to MS, the buffers used for HILIC separation are compatible with MS ionization techniques and recently used fluorescent labels have functional groups designed to facilitate ionization and UHPLC-MS analysis, making this approach highly desirable for structural characterization of glycans.
Figure 8.
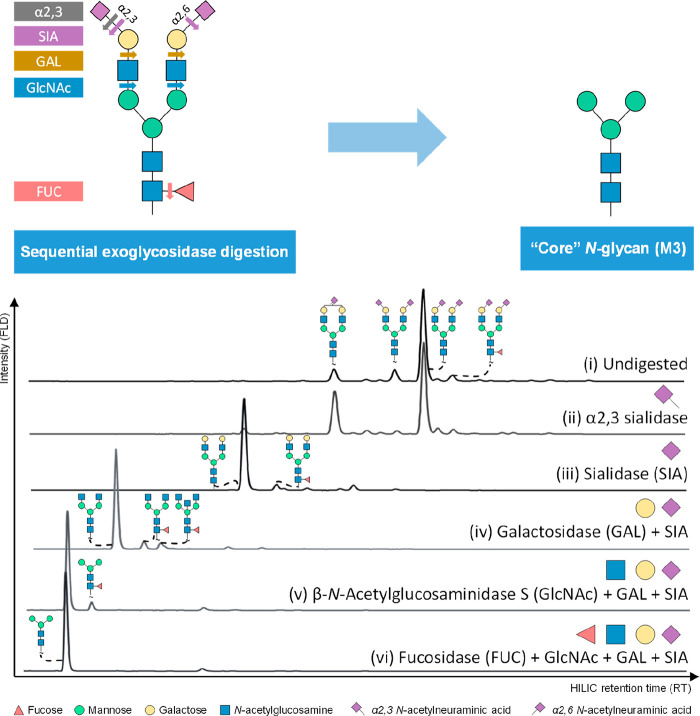
Exoglycosidase digestions of 2-AB labeled human plasma transferrin (Tf) N-glycans. (i) Undigested samples. (ii) Streptococcus pneumoniae α2–3 neuraminidase S. (iii) Arthrobacter ureafaciens α2-3,6,8,9 neuraminidase A (SIA). (iv) Streptococcus pneumoniae β1–4 galactosidase S (GAL) + SIA. (v) Streptococcus pneumoniae β-N-acetylglucosaminidase S (GlcNAc) + GAL + SIA. (vi) Omnitrophica bacterium α1-2,4,6 fucosidase O (FUC) + GlcNAc + Gal + SIA.
5.2. Capillary (Gel) Electrophoresis
Historically, CGE-LIF has been mostly employed for DNA sequencing.137 However, because it has been used for fetuin N-glycan analysis in 1996 by Guttman et al.138 and AGP N-glycan analysis in 2001 by Callewaert et al.,59 it has shown great potential for sensitive N-glycan analysis, and as 96 capillaries can be used in parallel, it theoretically allows for very HT. Practically, it has been mostly used in 4-, 8-, or 16-capillary setup due to capillary-to-capillary variations as well as the cost-effectiveness of analyzing smaller cohorts. Compared to UHPLC and MS, it offers advantages such as higher sensitivity and the separation of isobaric glycan structures, respectively, without sacrificing robustness and cost of analysis.61 However, its large-scale application in HT glycomics has been hindered by underdeveloped databases for glycan peak annotation and the lack of glycan standards. Moreover, coupling of CGE to MS is still challenging, making structural annotation of glycan peaks rather complex. On the other hand, capillary electrophoresis (CE) in combination with positive ion mode ESI-MS via a sheathless interface allows efficient droplet desolvation and analyte ionization. Although, because of longer analysis time and lower robustness, it has been mostly used for in-depth profiling and not in HT glycomics.139 Nevertheless, CGE-LIF has been routinely used for IgG N-glycan profiling both in biopharma studies140 and HT population and clinical studies141 and less routinely for other isolated glycoproteins and total plasma/serum protein N-glycome analysis.141,142
5.2.1. Sample Preparation
The sample preparation approach for CGE-LIF analysis is analogous to the approach used for UHPLC analysis of free N-glycans, and it consists of (usually enzymatic) deglycosylation, fluorescent labeling of free N-glycans, and cleanup procedure followed by CGE separation with LIF detection. In contrary to UHPLC fluorescent labels, the ones used for subsequent CGE-LIF analysis are charged, next to having a fluorophore, to allow electrophoretic separation of all glycans (not exclusively sialylated species).
The most commonly used dye is a triply negatively charged label, APTS, which is coupled to the N-glycans by reductive amination using NaBH3CN or nontoxic 2-picoline borane as a reducing reagent and acetic or citric acid. Acetic acid has been frequently used in the first studies and required a significant excess of APTS (more than 100-fold) to achieve a 95% glycan labeling efficiency.143 In addition to the high cost of analysis, excessive reagent amounts require extremely efficient cleanup procedures to obtain clean fluorescently labeled N-glycans and ensure reproducibility of analysis. Because temperature and duration of the derivatization procedure are a tradeoff between fast and efficient labeling on one hand and minimal desialylation (potentially skewing the results), on the other hand, this step requires optimization and fine-tuning. Váradi et al., have demonstrated that APTS glycan labeling in the presence of acetic acid has approximately 50% lower efficiency in a 2 h reaction compared to overnight labeling but decreases loss of sialic acids.144 Citric acid, which is stronger than acetic acid, enables a faster labeling reaction (50 min at 50 °C in contrast to an overnight incubation at 37 °C) with 10 times lower use of APTS and little or no loss of sialic acids.64 Therefore, labeling reactions in HT studies are performed in solution using citric acid. An alternative fluorescent label has been used for IgG N-glycan labeling and subsequent CGE-LIF analysis, e.g., singly negatively charged 2-amino-1-naphthalenesulfonic acid (2-ANTS), which resulted in drastically reduced glycan separation and more efficient ionization in CE-MS analysis compared to APTS-labeled IgG N-glycans.145
The introduction of a cleanup procedure using Sephadex G10 packed 96-well filter plates enabled N-glycan profiling at low picomolar amounts of the glycoprotein.59 Previously, N-glycans had been analyzed immediately after labeling and sensitivity of detection had been achieved by diluting the reaction mixture prior to analysis in order to lower the concentration of contaminants.138 The cleanup procedure is nowadays mostly done via HILIC-SPE68 as in the UHPLC-FLD N-glycan analysis. Alternative approaches like magnetic bead-based sample preparation have also been used.144 These are based on magnetic microparticles coated with carboxyl groups that reversibly bind to APTS labeled N-glycans with hydrophilic interactions in >80% acetonitrile environment (which acts as a crowding reagent), while deglycosylated proteins and excess reagents are washed away. Another mechanism of purification using carboxyl-coated magnetic beads that can readily be used is binding of released N-glycans (in positively charged glycosylamine form) by ionic interactions immediately after PNGase F release. In this later case, released N-glycans are eluted from the beads with aqueous APTS solution followed by the addition of the reducing agent to immediately initiate the labeling reaction without any interim concentration steps.144 This protocol enables faster sample preparation with the possibility for automation,146 although it has been mostly used on a lower scale for therapeutic antibodies N-glycan analysis due to higher cost per sample.
In recent years, HT application of CGE-LIF technology has been facilitated by the development of N-glycan preparation kits, e.g., Glycan Assure APTS Kit (Thermo Fisher), Fast Glycan Labeling and Analysis Kit (SCIEX), AdvanceBio Gly-X N-Glycan Prep Kit (Agilent), and glyXprep Sample Preparation Kit (glyXera), containing all reagents needed for a typical CGE-LIF workflow–glycoprotein deglycosylation, released N-glycan purification, APTS labeling, and removal of excess reagents. Although less cost-effective for large cohort analysis, sample preparation kits enable a more streamlined automatable solution in the case of N-glycan analysis in the standard types of samples, e.g., IgG N-glycans. This approach has been recently used for glycan analysis on IgG Fc fragment by CGE-LIF after IgG isolation and on-beads IgG digestion with IdeS to analyze IgG-Fc N-glycans in latent, active, and treated tuberculosis patients and healthy controls (n = 83).147 The same analytical approach has been used both for IgG N-glycan analysis and total plasma protein N-glycan analysis by CGE-LIF in a cohort of post-treatment controllers and post-treatment noncontrollers of human immunodeficiency virus (HIV) after antiretroviral therapy (ART) termination (n = 98),148 demonstrating a potential application of used technology for robust quantification of glycans as noninvasive plasma biomarkers.
5.2.2. Measurement and Data Processing
Fluorescently labeled negatively charged glycans are electrokinetically injected into capillaries by applying a low voltage for a short period of time. Injected glycans migrate in the applied electric field through capillaries and are being separated based on their hydrodynamic volumes and their mass-to-charge ratios or, as recently demonstrated for HMOs, based on the secondary equilibrium of the borate–vicinal diol complexation.149 Migration time alignment standards (coinjected bracketing standards) are used to minimize migration time shifts between samples and facilitate glycan identification and quantification, by enabling electropherogram alignment and GU unit assignation. After manual or automated peak integration, total area normalization is usually used to extract glycan amounts as relative %area used for further analysis, again followed by batch correction and statistical analysis. Alternatively, total height normalization can also be used to obtain relative peak height proportions (%rPHP).
5.2.3. Glycan Structure and Characterization
Analogous to UHPLC, structures of glycans separated by CGE-LIF are also elucidated by comparison of individual glycan peak glucose unit (GUCGE) with the GUCGE values of specific glycan structures in available databases and utilization of exoglycosidases sequencing.142,150
GUCGE values are assigned based on fluorescently (e.g., APTS) labeled standard oligosaccharide ladder, usually maltodextrin (homopolymer of α1,4-linked glucose), although dextran has also been used. The retention time of each unknown oligosaccharide correlates with the length of the sugar oligomer and is converted to a GUCGE scale used for a database search. It is of utmost importance that the same standard oligosaccharide ladder is used for analysis and the database buildup because CGE migration depends on hydrodynamic volumes affected by the molecular configuration and conformation.151 The development of databases containing CGE-LIF separated glycans has been lagging behind HPLC/UHPLC glycan databases due to more complex structural confirmation of individual glycans caused by difficulties of CGE coupling to MS. However, this is slowly changing, and nowadays several expanding databases, e.g., GUcal152 (recently broadened with the GlycoStore data)152,153 and glyXbase,154 exist (Table 1). Populating these databases with glycans labeled with alternative fluorescent labels and originating from glycoproteins other than human IgG will facilitate the use of CGE-LIF technology for low- and HT glycomic studies.
Exoglycosidase sequencing has been used as a complementary approach to assist the glycan structure characterization both for N-glycans analyzed by CGE-LIF and UHPLC. Although labor-intensive and less straightforward for more complex samples (e.g., total plasma/serum protein glycan pool),142 this approach has been often used for elucidation and confirmation of glycans originating from isolated glycoproteins (e.g., IgG)155 during HT method establishment. Exoglycosidase sequencing is shown to be automatable by using a temperature-controlled sample storage compartment of a capillary electrophoresis (CE) machine for enzymatic reactions and the separation capillary for delivery of the exoglycosidase enzymes, speeding up the process.155
While a fluorescent label used for glycan derivatization should enable efficient glycan separation in CGE and ionization in MS, the coupling to MS has been challenging due to the incompatibility of gel and buffers used for CGE separation with MS analysis as well as maintaining the closed electrical circuit which is needed for the CE analysis. Therefore, to enable subsequent MS analysis the CGE separation needs to be sacrificed, which complicates the one-on-one comparison between CGE-LIF and CE-MS for glycan structure confirmation. Several attempts to connect capillary zone electrophoresis (CZE) to MS in the past have shown some promise.156,157 Recently, CZE combined with positive ion mode ESI-MS has been successfully employed for N-glycan profiling after linkage-specific derivatization of sialic acids and labeling the reducing end of all N-glycans with Girard’s reagent P.139 This approach was shown to be highly sensitive and applicable to free N-glycan analysis of a complex biological sample, total human plasma glycoproteins. Although it allows in-depth profiling of low abundant glycans, this approach is currently more suitable for low- to semi-HT glycan analysis due to the long separation times as well as capillary to capillary variation.
5.3. Mass Spectrometry of Glycans
The use of MS is common in glycomics research and extends to all oligosaccharide classes (N- and O-linked glycosylation; Figure 9,24 glycolipids,158,159 GAGs,160,161 and free saccharides15) and to all levels of glycosylation characterization.162−166 MALDI-MS is very powerful when it comes to rapidly generating molecular fingerprints of complex samples, which makes it the preferred analytical strategy for HT profiling studies of released N-glycans from either purified glycoproteins or complex sample matrices like liquid biopsies (i.e., full blood, plasma, serum). As such, MALDI-MS is most commonly used as a stand-alone platform. ESI-MS is commonly hyphenated to molecular separation methods (i.e., HPLC or CE as described above), which is a limiting factor when it comes to throughput. Hence, ESI-MS-based platforms are more commonly used for the analysis of more complex analytes in which MALDI-MS does not provide sufficient resolving power, dynamic range, or sensitivity, like for example glycopeptides.167
Figure 9.
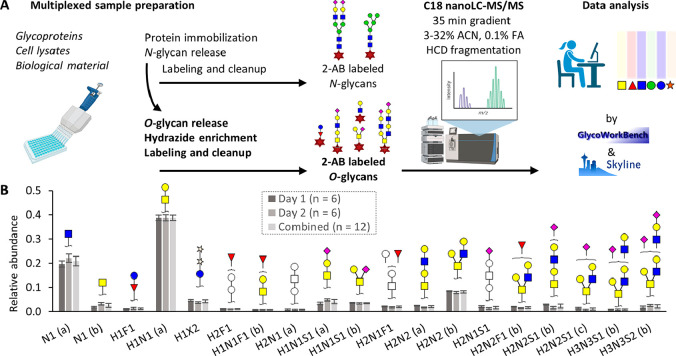
Multiplexed sample preparation workflow for N- and O-glycan profiling. Intra- and interday repeatability of the optimized method. (A) Proteins are immobilized on a polyvinylidene fluoride (PVDF) membrane by the addition of a (pure) glycoprotein, cell lysates, or derived from biological material (e.g., plasma). N-Glycans are released by the addition of PNGase F and eluted from the PVDF membrane. The O-glycans are released by adding a release agent and eluted from the PVDF membrane followed by a labeling procedure (2-AB). Eventually, the samples were analyzed using C18 nano-LC-MS/MS followed by data analysis. (B) Inter- and intraday repeatability of the O-glycan workflow. Average relative intensities for the O-glycans are displayed for those with a relative abundance above 1% per day. Error bars represent the standard deviations. Graphics in (A) were created using https://biorender.com/: H, hexose; N, N-acetylhexosamine; F, fucose; S, N-acetylneuraminic acid. Reproduced with permission from ref (24). Copyright 2022 de Haan et al.
Different LC-MS and CE-MS approaches have been successfully employed for the analysis of complex sugar mixtures, e.g., HMOs, and recent developments in this area have been reviewed by Auer et al.17 In addition to LC-MS and CE-MS that enable limited glycan isomer separation, hyphenation of MS to ion mobility (based on a gas-phase separation) has emerged as another technique enabling analysis of glycan and glycopeptide isomers. Ion mobility MS does not rely on derivatization or enzymatic reactions for isomer identification. Instead, isoforms are identified based on collision cross-sections differences of fragment ions facilitating elucidation of a complex and diverse glycan repertoire, not applicable exclusively to N-glycans but also to other glycan classes, e.g., O-glycans, HMOs, GAGs, etc.168,169 This technique has been applied for glycan analysis in smaller sample sets170−172 and has the potential for the analysis of glycan isomers in larger cohorts.
5.3.1. Sample Preparation
While the individual steps of the sample preparation workflow for hyphenated ESI-MS-platforms are largely defined by the molecular separation technology, the use of MALDI-MS brings some technology-specific aspects which include the spotting of the sample and MALDI matrix (or a mixture of both) on a target plate compatible with the MS platform.173 One additional step in the sample preparation that became very relevant upon the introduction of MALDI-MS analysis of released N-glycans, and the analysis of glycoconjugates by MS in general, was the stabilizing and linkage-specific derivatization of sialic acids through, for example, methyl174 or ethyl esterification175 and dimethylamidation.176,177 Beyond the scope of this review, a comprehensive review was published by De Haan et al., describing in great detail the different sialic acids, the different chemistries used for their derivatization, as well as their applications.178 However, the problems related to the MS-based analysis of sialylated N-glycans are multifaceted. First, N-glycans are most commonly analyzed as pseudomolecular cations, in the form of alkali-metal adducts. The negatively charged sialic acids have an adverse effect on the cation formation of sialylated species, which consequently results in quantitative biases of sialylated glycan species compared to neutral glycan species.179,180 Second, the sialylated glycan species are prone to forming a heterogeneous and unpredictable set of alkali-metal adducts. This results in a phenomenon called “peak splitting”, in which a single analyte population is represented by multiple ion species, different in their net mass through the incorporation of varying numbers or species of alkali-metal ions, and thus occupying different spaces in the m/z continuum. Typically, this results in ionization biases, a reduction of measurement sensitivity, and issues in peak annotation.181 Third, the sialic acid residue in sialylated glycan species is extremely fragile and prone to both in-source and post-source metastable fragmentation. Partial, as well as full loss of sialic acid residues, will result in loss of biologically relevant information and induce quantitative biases in complex glycan mixtures. Finally, sialylation introduces a large source of (biologically relevant) variation as the sialic acids can be bound to the rest of the glycan moiety through various linkages (i.e., α2,3, α2,6, α2,8, and α2,9). Sialylated glycans with multiple sialic acid residues often show linkage heterogeneity, resulting in a large number of potential isomeric glycan compositions. Without the exoglycosidase treatment (which cannot be considered HT), it is impossible to differentiate these in a typical MS1 analysis (which is common when using MALDI-MS), unless using chemical derivatization, which was shown to be feasible in HT fashion, for ethyl esterification by Reiding et al.175
To increase measurement sensitivity, substantial efforts were made to purify and concentrate glycans and/or glycopeptides prior to the spotting procedure. The most commonly used method in HT glycomics is the use of SPE applying the HILIC principle. The 96-well format adaptation of this method was pioneered by Selman et al.182 The presented method showed capable of analyzing 384 samples in less than 36 h using high-resolving power MALDI-Fourier transform ion cyclotron resonance (FTICR)-MS; less than 6 min per sample, of which approximately 18% (∼1 min) was consumed by the MALDI-FTICR-MS measurement.
Rather than using HILIC- or RP-SPE, glycoblotting has been proposed as an alternative for enrichment of released glycans or glycoconjugates from complex samples. Introduced by Nishimura et al. in 2005, the glycoblotting strategy is based on affinity purification of carbohydrates using beads coated with Fischer’s reagents.183 Following the mixing of the capture beads with a solubilized sample and the concomitant ligation of the carbohydrates to the Fischer’s polymers, the beads are separated from the liquid through spin filtration. Carbohydrates are then released from the beads and spotted with a MALDI matrix for MALDI-MS analysis. While the strategy has a high potential for HT application,184,185 it is limited by the high costs of the reagents.
An additional strategy to improve measurement sensitivity and decrease ionization bias, through reducing the metastable nature of the oligosaccharide ions, is the derivatization of the reducing end with a charge-containing or UV-reactive group. Mentioned previously as labels for fluorescence detection, 2-AA, 2-AB, and 2-PA absorb UV light in the wavelength range of the most common MALDI lasers (330–360 nm) and can thus be used as reactive MALDI matrices.186−189 Furthermore, reducing end derivatization using charge carriers Girard’s Reagent P and T have shown to have a beneficial effect on ionization in MALDI-MS applications.139,190−192
Major strides forward in increasing throughput in MALDI-MS-based studies were achieved through the automation of the sample preparation by robotization.193 Doherty et al. explored this for the analysis of released IgG N-glycans using HILIC-UHPLC-FLD.194 Here, the most time-consuming steps of the preparation, like disulfide bond reduction and alkylation, enzymatic N-glycan release, and analytical separation time, were optimized and reduced. The total length of the sample preparation protocol was reduced from 16 h when performed manually to 60 min when performed on the robotized platform. Bladergroen et al. were the first to integrate a complete robotized and HT sample preparation workflow for the MALDI-MS-based analysis of N-glycans released from total plasma (Figure 10).193 Minute amounts (6 μL) of total plasma were denatured using SDS, and N-glycans were released using PNGase F in a 96-well plate. Following enzymatic digestion, the sample preparation platform, based on the Hamilton MicrolabSTAR robot system, performed sialic acid derivatization through ethyl esterification, HILIC-SPE N-glycan purification, premixing of the purified and derivatized N-glycans with super-DHB matrix (9:1 (w/w) mixture of DHB and 2-hydroxy-5-methoxybenzoic acid) and sodium hydroxide, and subsequent spotting on a MALDI target plate with a 384-well format. The nature of the setup allowed for parallel processing of 384 samples in approximately seven hours (excluding N-glycan release), which comes close to an effective one minute per sample.
Figure 10.
Automation of released N-glycan analysis by MALDI-TOF-MS. A graphical representation of the setup is provided and the consecutive processing steps are as follows: (1) PNGase F release, (2) ethyl esterification, (3) glycan enrichment by hydrophilic polypropylene (GH Polypro, GHP) HILIC–SPE, (4) glycans are eluted from the GHP membrane, and (5) MALDI target spotting followed by (6) MALDI-TOF–MS analysis. Reprinted with permission from ref (193). Copyright 2015 American Chemical Society.
Alternative slide-based strategy to directly profile N-glycans from serum and plasma samples has been recently developed by Blaschke et al.195 The approach consists of 1 up to 2 μL of serum/plasma spotting on an amine-reactive slide, delipidation, and desalting steps followed by PNGase F deglycosylation by spraying the enzyme directly on the slide and analysis with MALDI-FTICR-MS. Equivalent approach based on MALDI-MS imaging (MSI) can also be used to profile N-glycome of more complex samples, e.g., urine, prostatic fluids, and expressed prostatic secretions.196 Although this novel rapid workflow has not yet been applied on a larger sample set, the approach does not require steps of glycan labeling, derivatization, and/or purification, making it attractive for future HT clinical applications.
5.3.2. Measurement and Data Processing
Although it was established early on that the introduction of MS in HT glycomics shifted the limits in throughput from analysis to sample workup and data processing, there have been substantial advances in (the use of) MS instrumentation that have pronounced effects on throughput. There are several ways of improving MS-related throughput: (i) directly through increasing the speed of the MS analysis/detection and (ii) indirectly through increasing the sensitivity of the MS detection and thus requiring less time for sampling.
The biggest impact for MALDI-MS-based HT glycomics is the recent release of high-speed MALDI-time-of-flight (TOF)-MS platforms.197−199 Both the linear MALDI-TOF/TOF-MS and the orthogonal MALDI-quadrupoleTOF-MS platforms operate using high-speed MALDI-stages and UV lasers with a 10k Hz repetition rate. This makes them capable of analyzing samples approximately 1 order of magnitude quicker than the previous generation of MALDI-MS equipment typically equipped with 2k Hz lasers and slower MALDI-stages.
For high-resolving power MALDI-FTICR-MS-based detection of released N-glycans, the major throughput affecting development is the introduction of 2Ω FTICR detection.200 Commercialized by Bruker Daltonics, 2Ω detection FTICR offers the ability to record double the number of datapoints within the same transient length of a conventional FTICR analysis and thus providing superior mass resolving power in the same analysis time. However, it can also be exploited to record data at the resolving power of a conventional FTICR analysis, albeit with half the transient length, increasing throughput by a factor of two.
Another fundamental development that has the potential to increase throughput for HT glycomics studies is the introduction of laser-induced postionization following MALDI, also referred to as MALDI-2.201,202 In short, MALDI-2 results in enhanced ionization for a number of analyte classes (including released N-glycans) and is based on the illumination of the developing MALDI-plume with a secondary UV laser following material ablation and desorption with the primary MALDI laser. Explorative studies into the effects of MALDI-2 of (oligo)saccharides have shown predominant enhancement of the formation of deprotonated ions in the negative polarity. A similar development has surfaced for ESI-MS-based approaches, where the enhancement of both desolvation and ionization is achieved through the use of dopant-enriched nitrogen (DEN) gas.139,203 The positive effect of DEN gas on measurement sensitivity of released N-glycans has been shown for both negative and positive ionization, and both CE-ESI-MS and (n)LC-ESI-MS approaches.
Optimal signal processing for HT approaches occurs on-the-fly, and generally data acquisition software has basic on-board signal processing capabilities like smoothing, baseline subtraction and recalibration. Nevertheless, reprocessing data or processing data offline can be valuable to have additional control over processing steps and parameters. Also, additional quality control measures are implanted which can aid in automatically removing outlier spectra or features. Although not specific for HT glycomics data, GeenaR is an open-source and publicly available R-package that performs spectral preprocessing, quality control, feature extraction, and spectral clustering for MALDI-TOF-MS spectra at substantial speed.204 The full GeenaR workflow takes approximately 6 s per spectrum, which is negligible when put in perspective of the time spent per sample on sample preparation, which is in the order of minutes. Important for HT clinical studies is the availability of a quality control module that identifies potential outliers and which takes into account the number of m/z values per spectrum in relation to the mass resolution, the intensity range of the m/z values, the presence of null or empty spectra, and resolution irregularities. Every processed spectrum is scored, and the user can decide in the postprocessing to discard (potential) outliers. Another open source solution to perform spectral outlier detection is the R-package MALDIrppa.205 The package assesses the multipeak structure of a MALDI-TOF-MS spectrum and provides it with an “atypical” (A-)score. The user can define or calculate A-score tolerances to identify outliers. Through removing outlier spectra prior to processing, precious time can be saved by only processing high-quality spectra.
5.3.3. Peak Assignment
The annotation of N-glycan compositions to m/z features is historically performed manually and based on accurate mass and isotopic pattern matching, following basic knowledge on N-glycan biosynthetic pathways. However, there has been a substantial effort in automating the peak annotation from MALDI-TOF-MS spectra to make it more reliable and faster. Cartoonist is a software tool that assigns m/z values to what the authors refer to as “cartoons” or glycan monosaccharide compositions.35,206 It includes the annotation of isomeric variants, although because the software works with MALDI-MS spectra, it cannot differentiate between the isomeric species.35,206 The mGIA method,207 and its improved version called GlycoMaid,208 published by Xu et al., are based on the matching of glycan isotope abundances to a previously constructed theoretical database. Uniquely, GlycoMaid is set up to deal with overlapping and isobaric glycan compositions and deconvolute the contribution of the individual N-glycan species to the measured isotopic distribution. Toolbox Accelerating Glycomics (TAG) is a tool for m/z annotation with N-glycan compositions and pathway analysis of MALDI-TOF-MS data.209
5.4. Mass Spectrometry of Glycopeptides
HT glycopeptide analysis has, hitherto, been largely focused on the analysis of IgG Fc glycopeptides, with recent expansions toward combined IgG and IgA glycosylation analysis19,210 as well as analysis of AGP (Figure 11).20 Fc glycopeptide analysis is achieved by LC-MS, with a few occasional MALDI-based analyses.211,212 RP-LC-MS is the main method used for HT IgG Fc glycopeptide profiling. LC-MS approaches for IgG glycosylation analysis at the glycopeptide level are already around for several decades, building on bottom-up proteomics workflows for the analysis of mAbs (peptide maps with MS detection) to monitor primary sequences of antibodies as well as a range of modifications including glycosylation.
Figure 11.
Results of the HT and site-specific N-glycosylation LC-MS analysis of human AGP. (A) Typical chromatogram with extracted ion traces of the most abundant glycopeptides from each glycosylation site. Trifluoroacetic acid was used in the mobile phase as an ion-pairing agent. (B) Summed mass spectrum for glycopeptide I1 with the most abundant glycan structures annotated. (C) MS/MS fragmentation spectrum for the peptide part of glycopeptide I1 with glycan composition N5H6S3. (D) MS/MS fragmentation spectrum for the glycan part of glycopeptide I1 N5H6S3. Reproduced with permission from ref (20). Copyright 2022 Keser et al. under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
5.4.1. Sample Preparation
In regard to sample preparation, the use of 96-well plate methods has been key for achieving HT.211,212 Workflows tend to start with complex matrices such as blood serum or plasma. The most commonly used initial step is an affinity enrichment of IgG using, e.g., immobilized protein G or protein A, with an acidic elution step.69,212,213 The choice of a volatile acid, such as formic acid, allows retrieving the eluted IgG using vacuum centrifugation. Sample preparation is continued by tryptic cleavage, generating for human IgGs a glycosylated nonapeptide covering the conserved N-glycosylation site of the CH2 domain of the IgG heavy chain.213
The acid elution step followed by vacuum centrifugation appears to have an important role in preparing the immunoglobulins for tryptic digestion. Importantly, this acid treatment appears sufficient to prime the antibody for the generation of tryptic Fc glycopeptides, without the need for reduction and alkylation steps.214 Tryptic cleavage can be achieved in low strength volatile buffer (e.g., 25 or 50 mM ammonium bicarbonate) by overnight incubation at 37 °C. The resulting IgG (glyco-)peptide mixtures can be directly used for LC-MS analysis but may also be stored for a longer period at −20 °C prior to MS analysis.215 Affinity purification with acidic elution has been successfully implemented not only for HT IgG glycoprofiling but also for other target glycoproteins such as IgA.19,210
In the past decade, HT IgG Fc glycosylation analysis has been expanded toward the analysis of antigen-specific IgG responses, such as autoantibodies, alloantibodies, antipathogen, and antivirus antibodies.216−220 For this, adaptations of enzyme-linked immunoassay (ELISA) procedures are most often applied: microtitration plates are coated with target antigens and used to capture the specific antibodies from serum or plasma, followed by extensive washes and elution of the adsorbed antibodies with an acidic solution. Further sample preparation is identical to that for affinity-purified, total IgG.
5.4.2. Measurement and Data Processing
Generally, no glycopeptide enrichment is being applied, and the complete tryptic IgG digests are analyzed by RP-LC-MS. Hereby, retention of polyclonal human IgG Fc glycopeptides is governed by sequence variations in the peptide portion and differs between subclasses, providing distinct IgG Fc glycopeptide clusters for the IgG1, IgG4, and IgG2/3 subclasses.221 Depending on the specific IgG3 allotype, IgG2 and IgG3 can result in an identical mass for their Fc glycopeptide portion. Notably, dependent on the ethnicity, other IgG allotypes may be dominant, with identical Fc glycopeptide signals for the IgG3 and IgG4 subclasses.221
Signal intensity in IgG Fc glycopeptide analysis can be boosted by applying a DEN gas, which, in combination with nanoscale separation (nanoESI), allows detection of low attomole amounts of IgG Fc glycopeptides on-column.213 Of note, this sensitivity is not needed for analysis of total serum or plasma IgG as the concentrations are approximately 10 mg/mL. However, the high sensitivity detection pays off with regard to method robustness, as high-quality measurements can be obtained by using only minute amounts of material, thereby minimizing contamination of the LC system and the ionization source. The high sensitivity is a prerequisite for the analyses of various specific IgG responses, e.g., some autoantibodies222 or alloantibodies219 of low abundance. Both high-resolution and low-resolution MS detection have been successfully applied,216,219 with high resolution having particular advantages in reducing or avoiding interference from coeluting, nonglycopeptide signals.
Next to LC-MS, MALDI-MS approaches have been developed for IgG Fc glycopeptide profiling, with various HT applications.211,212 Of note, by applying relatively cold MALDI matrices, these studies succeeded in limiting sialic acid loss and successfully detected sialylated IgG Fc glycopeptides, albeit with a still prominent loss of part of the sialic acids. Further developments have been made to largely or even completely prevent sialic acid loss. First, a particularly cold MALDI matrix was implemented, in combination with an intermediate-pressure MALDI source on an FTICR-MS, which allowed rapid collisional cooling of the protonated glycopeptide ions.165,223 Second, linkage-specific sialic acid derivatization was established for IgG Fc glycopeptides, allowing the facile, robust detection of both sialylated and nonsialylated IgG Fc glycopeptide.177 Both methodological innovations have, to our knowledge, not yet found their way into HT glycopeptide profiling, with HT LC-MS glycopeptide analysis approaches clearly prevailing.
Commonly data processing is performed in a targeted manner in batch mode using a glycopeptide list of the IgG subclasses of interest.224 Data curation focuses on confirming analyte identity on the basis of accurate mass, retention time, and isotopic pattern. The latter is also used for assessing and excluding potential interferences for high-precision quantification.
5.4.3. Peak Assignment
Analyte assignment is generally performed on the basis of accurate mass, retention time, and isotopic pattern using common glycobiological knowledge for a sanity check of the observed glycopeptide compositions and profiles. Analyte identities can be further confirmed for selected cases using tandem MS and released glycan analysis.225,226 With the increase of sensitivity and dynamic range of analytical methods, additional, minor glycopeptide variants such as hybrid-type glycans and monoantennary species are being included.227 The range of human IgG Fc glycans may be further increased with the detection of very low-abundant sulfated species, yet compelling evidence for their presence of the IgG Fc portion is still pending.228,229
5.5. Lectin Microarrays
Lectin microarrays was reported for the first time in 2005.99−102 The display of lectins in a microarray format enables multiple, distinct binding interactions to be observed simultaneously and therefore provides a complementary method for the HT screening of carbohydrates on glycoproteins or glycolipids.230 On the basis of the interaction and binding of the carbohydrate residues of the analyzed compounds with the corresponding lectins, lectin microarrays allow determining their specificity and the registration of interactions at very high accuracy, while the application of lectin microarrays in the analysis of hundreds or thousands of samples has been hurdled by the fact that they are mostly used for comparative analysis of glycan profiles (e.g., disease versus healthy) due to lack of quantitation, and rely on complementary techniques, most often MS,106 for the complete determination of glycan structures. However, in contrast to UHPLC and CE, which require the release of glycans from a glycoprotein, and MS, which mostly involves enzymatic digestion of a glycoprotein, lectin microarrays enable rapid and direct measurement of glycan profiles in complex biological samples, including O-glycans,108,231 on an intact glycoprotein232 and GSLs233,234 without the need for protein digestion and glycan release.
5.5.1. Sample Preparation and Measurement
Lectins with known and overlapping binding specificity are immobilized as microdots on a solid, usually glass, surface which has previously been activated by biochemical (e.g., streptavidin) or chemical (e.g., epoxy, N-hydroxysuccinimide (NHS), amino, gold) derivatization. After lectin immobilization, residual activated groups are blocked (e.g., using glycan-free serum albumin or amine). Interaction of carbohydrate residues with the corresponding lectins can be detected either directly through their prior labeling with fluorescent reagents (e.g., Cy3 monoreactive dye)106,235 or indirectly (e.g., by overlaying a fluorescently labeled antibody (if available))103 against the target glycoprotein (approach termed antibody-overlay lectin microarray) (Figure 12). Direct labeling is often preferred, however, it requires a relatively high amount of glycoproteins, it can disrupt glycoprotein–lectin interactions and it suffers from low sensitivity. On the other hand, indirect detection of glycan–lectin interactions using tyramide signal amplification for antibody-overlay lectin microarray (TSA-ALM), developed by Meany et al. in 2011 enabled detection of weak glycan–lectin interactions as a result of 100-fold increased sensitivity by using biotinylated tyramide.104 Indirect labeling, therefore, offers increased sensitivity due to signal amplification as well as high specificity due to low background labeling.105 Specific lectin–glycan interactions are detected with high accuracy after washing off the unbound probe98 to obtain the characteristic “glycan fingerprint”.
Figure 12.
Glycan microarray analysis. A graphical representation of a glycan microarray workflow, the consecutive processing steps are as follows: (1) glycoprotein is labeled with a fluorescent tag through Cy3 labeling, (2) the glycoprotein binds at the lectin microarray based upon the presented lectin on the microarray, and (3) the microarray is scanned, followed by (4) the interpretation of the signals.
The detection of interactions can be achieved using confocal and nonconfocal fluorescence98 and evanescent-field activated fluorescence.98,101,148 A bimolecular fluorescence quenching and recovery detection do not require the preliminary labeling of target carbohydrates.236
5.5.2. Data Processing/Peak Assignment/Identification
Fluorescent intensities of each lectin–glycoconjugate spot are usually extracted using appropriate software, e.g., Gene Pix,106 Mapix,231 GlycoStation ToolsPro,237 etc. Generally, the background is subtracted and signal values less than average background + two standard deviations are excluded from further analysis to eliminate the influence of nonspecific adsorption.106 Global normalization (to obtain normalized fluorescent intensities; NFIs) is further used to eliminate fluorescence bias, and processed data of the parallel data sets are compared with each other based on fold-changes. Normalized data is often analyzed using unsupervised average hierarchical cluster analysis (HCA) and principal component analysis (PCA).106
Automated data analysis for lectin microarray technology has been reported already back in 2007 by Rosenfeld et al., who developed an algorithm for glycan fingerprint interpretation based on the data obtained by analyzing hundreds of fingerprints.103 The fingerprint, a histogram representing the intensity of lectin–glycan binding, is finally represented as a table of glycan structures found in the sample and their relative abundances.
Identification of glycan epitopes largely depends on the repertoire of lectins used for microarray preparation. Historically, these have mostly been plant lectins whose specificity does not cover all human glycosyl epitopes and could lead to erroneous interpretation. Discovery and biochemical characterization of other natural lectins, as well as recombinant development of new lectins with more narrow specificity, can aid in obtaining more detailed information on structural glycan features facilitating their use in glycomic applications. Lectin microarrays have been successfully applied for glycan profiling of complex protein samples in highly heterogeneous matrices and could, in the future, find their application niche in cases where sample processing for detection with other techniques, e.g., MS, would be extremely laborious and complex. This approach could be useful for rapid assessment of the physiological status of cells or tissues by profiling cell surface carbohydrate expression patterns.
5.6. Data (Pre-)Processing and Analysis
In recent years, various computational tools have been developed to allow a faster and more user-friendly processing of HT glycomics data, e.g., for the identification of potential glycan biomarkers. Nonetheless, data processing still presents a major bottleneck in HT glycomics. Typically, HT glycomics data undergo various preprocessing steps as well as pretreatment steps before they can be used for data analysis as detailed in the following sections (Figure 13).
Figure 13.
Workflow for data (pre)processing and analysis. The various steps (1–8) involve preprocessing of raw data into clean data. Eventually, the data is normalized, and each analyte will be relatively quantified based upon the total summed area of all analytes observed in a measurement (%area), also known as direct glycosylation traits. To obtain insights in the biosynthetic pathway specific glycosylation features can be summed (e.g., galactosylation, sialylation, fucosylation). Either the direct or derived glycosylation traits can be used for statistical evaluation and the whole data set can be integrated with other data sets (e.g., proteomics or genomics).
5.6.1. Identification
Most of the current HT glycomics data processing methods are designed to target specific sets of analytes and prior to data processing, the glycomic analytes present in the sample should be explored and identified by employing strategies described earlier for the different HT glycomic technologies (see sections 5.1–5.5).167
Dependent on the applied technique, various libraries, tools, and databases are available to identify the glycan compositions based upon (normalized) retention time (HPLC-FLD; GlycoStore, previously GlycoBase),238 migration time (CGE-LIF; GlycoStore239 and glyXbase),154 accurate mass (MS; GlycoWorkbench, GlycanMass, UniCarb-DB (MS/MS), GlycoStore,239 and GlycoMod) or a combination of retention time and accurate mass data (UHPLC-FLR-MS, UNIFI).240 An extensive overview of available databases and repositories is provided in Table 1 and described more comprehensively in a recent review.241 Analyte identification can be supported by exoglycosidase treatments as well as the use of internal and external standards. For MS-based data, additional MS/MS experiments can be performed to further gain confidence in the glycan structure. For the analysis of released glycans, which are often analyzed by negative mode MS/MS, spectral libraries are available (Table 1),242 but often manual interpretation and identification is required which can, for example, be supported by GlycoWorkbench.243 Various software tools are available for glycoproteomics data (e.g., GlycoForest244 and Byonic; Table 1). However, also here manual interpretation and verification are recommended to contain the risk of false automatic assignments by the software tool. In cases where only accurate MS data is available, analytes can be assigned based on differences in retention time as well as mass increments relative to confidently identified glycans using tools such as GlycoMod,245 GlycoWorkbench,243 and GlycopeptideGraphMS246 (Table 1). It should be noted that current MS HT glycoproteomic approaches mainly provide monosaccharide compositional data and isomer differentiation is not achieved (except when sialic acid derivatization is employed).
5.6.2. Data Pre-Processing
Prior to data extraction, the raw data should be transformed into clean data and during this procedure often corrections are performed such as baseline correction or an alignment. This is to avoid any technical variation introduced by the analytical method that has been used and to match peaks across the sample set. Various software tools are available to perform these preprocessing steps in batches (Figure 13 and Table 1).
5.6.2.1. Alignment and Calibration
To ensure precision and minimize day-to-day and other potential technical variations introduced by the analytical platform, the first step for raw chromatographic and electrophoretic data involves the conversion of the observed retention/migration time to relative retention/migration times. For multiplexed CGE-LIF based data, an alignment algorithm is featured in glyXalign that identifies distinctive data points in the electropherogram.247 In addition, the alignment procedure can be followed live with an intuitive graphical user interface. Next to this tool, several tools are available that rely on the separate analysis of a glucose ladder across the sample set. Upon the basis of this ladder, the migration time can be standardized in GU. However, this calculation was, until a decade ago, a time-consuming task as no tools were available for the automatic assignment. This holdup disappeared with the release of the freely available tools such as the autoGU238 (recently replaced by GlycanAnalyzer)248 and the GUcal app.153,249
The introduction of a triple-internal standard (maltose, maltotriose, and maltopentadecaose; Agilent) avoids the need for additional measurements of a glucose ladder, as the internal standard can be coinjected with the samples.250 Here, the migration time will be transformed to a migration time unit (MTU), which will be assigned to each observed peak. However, caution is needed that the coinjected standards do not interfere with the analytes of interest (comigration), which could skew the MTU value. The commercially available software tool glyXtoolCE (glyXera)251 combines the coinjection of the triple-internal standard with the analysis of a GU ladder, resulting in a double migration time alignment (MTU′′).
Similar to glyXalign for CE-LIF data, a freely available computational tool called HappyTools, has been developed for LC-FLD data and allows automatic peak picking followed by the alignment of the retention times.252 While automated peak picking can be used, the user can also choose to provide a predefined list of analytes that will be used for peak picking and provides information about the peaks (e.g., retention time of the peak as well as the time window of the peak).
In the case of 2D MS-related data, MassyTools is capable of performing the calibration of the data when prior knowledge is available.253 Namely, specific masses need to be provided to perform the calibration of the spectra. Ideally, internal standards which cover the full m/z range can be used for this process. However, in most cases, predetermined analytes are selected that are known to be abundant in the samples as well as being previously identified by other techniques (e.g., tandem MS and exoglycosidase treatment) and a minimum of three m/z values should be provided. The alignment is also an important aspect for 3D MS data, where time is added as a third dimension (when compared to 2D MS data). For this purpose, other software tools are available such as LaCyTools, which is a targeted data processing package.224 Compared to MassyTools, there is no automated peak picking and prior knowledge is required. An alignment file should be provided that contains a list of masses with the desired/expected migration or retention times. For an appropriate alignment, at least three features with a unique retention time are required, preferably with a retention time covering the time window of interest. The alignment can then be followed up by a calibration step similar to that of MassyTools. However, this will not be performed on the total data set but only on those for specific clusters that will be predefined in the reference list. This list will contain all the analytes that should be integrated, and, depending on the separation technique, specific clusters can be defined. For example, RP-LC-MS analysis of IgG glycopeptides results in the (almost) coelution of the glycopeptides from the same IgG isoforms, as the separation is defined by the peptide portion (IgG subclass) and not so much by the glycan.213 This results in three distinct clusters of glycopeptides, which can be defined in the reference list with a specific retention time (± retention time range). It is important to note that, based upon each unique retention time and corresponding time window, LaCyTools will create 2D data (Figure 14B) from the 3D data by summing all spectra in the defined range per cluster (Figure 14A). The analytes that should be used for calibration can be selected in the last column of the reference list. However, please note that, if various clusters are defined, each cluster should contain at least three analytes that can be used for calibration. This calibration step could be considered a drawback of the software tool as, dependent on the analysis, not all clusters might have enough (confirmed) analytes that could be used for calibration.
Figure 14.
RP-LC-MS separation of tryptic Fc glycopeptides of a polyclonal serum IgG standard. (A) Extracted ion chromatograms (EIC) of the most abundant Fc glycopeptides per IgG subclass from polyclonal human serum IgG. The separation is mainly driven by the hydrophobicity of the peptide backbone and separate clusters are obtained where all glycoforms of a specific subclass elute. (B) Illustration of a summed mass spectrum of the cluster containing IgG Fc glycopeptides from subclass IgG1. Only the mass range of the triply charged species is displayed. IgG1 = EEQYNSTYR; IgG2/3 = EEQFNSTFR; IgG4 = EEQFNSTYR.
Exclusively for use with MALDI-MS data, rMSIproc is an open-source and publicly available R-package for spectral preprocessing and feature extraction.254 Although the package was developed to work with MALDI-MSI data (Bruker XMASS, imzML, and.tar formats), its open-source nature allows its application to XY-type files. The spectral preprocessing features Savitzky–Golay smoothing, label-free spectral alignment, recalibration of m/z values, peak picking, intensity normalization (to either total ion current/count (TIC), or using the root-means-squared (RMS) method) and signal-to-noise (S/N) ratio threshold peak picking. All processing can be applied to individual spectra, as well as batches of multiple spectra using the merged processing feature. The latter option provides a merged peak matrix, providing the intensities of all individual spectra projected on a common m/z axis.
To allow a faster identification process for HT glycomic data sets, new analytes can be identified by using a sample pool of analytes per investigated group (e.g., patients versus healthy controls) using exoglycosidase treatments or MS to assign unidentified peaks or m/z values. If new features are observed, it will be explored if these are indeed potential new analytes that can be added to the analytes/reference list by consulting the databases (Table 1).
5.6.2.2. Data Integration
A list of target analytes can be established, based upon the previous step, and used for the integration of the glycans or glycopeptides. While in some cases manual integration is necessary, to enable a HT glycomic workflow, several software tools can be considered for automated integration. Common tools are HappyTools for LC-FLD, glyXtoolCE for CGE-LIF, MassyTools for 2D MS data, and LaCyTools or Skyline for 3D MS data, and all make use of a target list and have the ability to subtract the background. For complex samples, more sophisticated approaches based on machine learning are also being developed.79
Whereas LaCyTools can be considered a bit more of a black box, Skyline allows visual interpretation of the automatically integrated data by using EICs/electropherograms.255 Moreover, it allows to assess the data quality, adjust the peak integration, and supports the raw data formats from almost every MS vendor (AB SCIEX, Agilent, Bruker, Thermo, and Waters), while LaCyTools requires a more generic form (mzXML). In the case of isomeric separation, caution is needed using LaCyTools. Namely, when the isomers are baseline resolved separate clusters can be created and can be integrated. If this is not the case and both isomers fall within the same time window of the cluster, the isomers will be considered the same analyte due to the same m/z value and integrated in such a way. Or if the time window is not set properly, only a part of an isomer might be taken into account for the integration.
5.6.2.3. Data Curation
To evaluate whether the individual analytes or measurements should be included for further processing, a (semi)automatic curation can be performed. Several parameters can be used for this; for example, HappyTools provides information about the observed retention time versus the expected retention time, observed S/N for each peak, and how well the observed peak fits with a Gaussian peak. Recently, an enhanced semiautomated computational approach based on electropherograms clustering, manual peak curation, and the HappyTools has also been reported.256 Both LaCyTools and Skyline provide valuable information about the integrated analytes such as S/N, ppm error, and the similarity between the observed isotopic pattern of each analyte (per charge state) versus the theoretical isotopic distribution. Specific values can be set for each parameter to decide whether the observed feature can be assigned as the expected analyte or that it might be an isobaric interference and that it should be removed from the data set. The same accounts for the measurement, when a large fraction of analytes is observed with an overall low S/N, it should be evaluated whether the specific measurement should be excluded from the data set.
It is recommended to upload the curated assigned chromatographic, electrophoretic, and mass spectrometric compositional data on GlycoStore (www.glycostore.org) to further support the resource that facilitates detailed glycan analysis for different workflows, or on their designated repositories (e.g., GlyTouCan; Table 1).
5.6.3. Normalization and Relative Quantification
Eventually, the final relative quantification of the detected glycoforms can be obtained, after the data preprocessing steps, by normalizing the data based upon the total area of all observed analytes (per protein and site, if applicable). This will result in %area, which will be further processed to assess whether a batch correction is needed and, eventually, to perform statistical analysis. Relative quantification of glycans remains the most widely used approach because it is more time- and cost-effective compared to absolute quantification, offering at the same time insights into aberrant glycan synthesis and regulation of the glycosylation process. On the other hand, absolute quantification may have an application niche in clinical diagnostics of free oligosaccharides, e.g., in milk or urine, as well as for glycosylation analysis of therapeutic glycoproteins.257 However, it should be noted that absolute quantification is not yet applied for HT glycomic workflows, even though advancements have been made in the CE field by the usage of an APTS labeled standard149 and in the MS field by the usage of isotopically labeled standard258 and labels.126,259
5.6.4. Glycosylation Trait Analysis
Directly measured glycosylation traits depend on the used technology and are generally individual fluorescently labeled glycan structures or individual glycopeptides that have been efficiently detected and quantified by UHPLC/CGE/MALDI analysis or LC-MS analysis, respectively. Even when the same quantification technology is used, differences in sample preparation protocols or analysis conditions affect separation (in the case of UHPLC and CGE) or ionization (in the case of MALDI and LC-MS) and can result in differentially quantified glycan structures. To enable integration of different studies and/or measurements, derived traits based on the shared glycan structural features (e.g., sialylation, galactosylation, fucosylation, branching, etc.) are being calculated from directly measured glycan species. Derived glycosylation traits are biologically more related to activities of specific enzymes in the glycosylation pathway and underlying genetic polymorphisms, making them beneficial for understanding the functional relevance of obtained results as well as for GWAS.
5.6.5. Data Analysis
Glycan measurements are not normally distributed (right-skewed distribution) and are usually log-transformed after normalization. Batch correction is then performed on log-transformed measurements by modeling the technical source of variation (which sample was analyzed on which plate) as a batch covariate.260 To correct the measurements for experimental noise, estimated batch effects are subtracted from log-transformed measurements and obtained results used for further statistical analysis. Because of the analysis of multiple (direct and/or derived) glycosylation traits correction of p-values for multiple testing should be applied.
5.6.6. Multi-Omic Data Integration
One of the challenges of vast glycomics data generation and integration with databases, is its complexity in terms of nonuniform glycan traits annotation that usually depends on the level of profiling, used analytical technology, originating glycoconjugates, etc. Although data generated by different technologies can be integrated, the same has been hindered by a lack of a digital standard to represent glycoproteins, consequently often leaving existing databases unpopulated with newly generated data.
Recent efforts of the global glycoinformatics community, through The GlySpace Alliance261 are directed toward providing relevant, trustable, and quality information regarding the glycan structures, their origin, biosynthesis, regulation, and functional roles. In line with that, the Minimum Information Required for A Glycomics Experiment (MIRAGE) commission aims to improve the quality of the glycomic data in the literature by implementing guidelines (e.g., sample preparation, glycan microarray analysis, LC, CE, and MS analysis)262−266 and templates that ensure that the most important details of a glycomic study are provided in their publications, making sure that these reported experiments can be repeated by others. Several journals now require these MIRAGE guidelines prior to submission of a manuscript. Whereas a standard semantic framework, GlycoConjugate Ontology (GlycoCoO), has been developed, solely on describing and representing glycoproteomics data.267
Efficient data integration is an essential prerequisite for some applications of HT glycomics, e.g., GWAS analysis, because they rely on the use of multiple cohorts data sometimes even generated with different technologies (e.g., UHPLC and LC-MS).268
6. Applications
The ability to reliably detect interindividual differences at the molecular level is the prerequisite of personalized medicine. Glycome composition integrates genetic, epigenetic, and environmental factors into chemical structures that can be reliably quantified, which makes it an ideal biomarker for personalized medicine.269 However, large studies need to be performed to properly evaluate and validate the biomarker potential of glycans. The total number of glycomic analyses is still very low if compared to the number of genetic, epigenetic, transcriptomic, metabolomic, or microbiome analyses, but thanks to the novel analytical approaches, more and more glycome data is being generated (Table 2).
Table 2. High-Throughput Glycomic Studies Involving More than 500 Samplesa.
| cohort description | sample no. | technology | manual or automated | data processing tool | ref |
|---|---|---|---|---|---|
| IgG | |||||
| RA | 1099 | MALDI-MS | manual | flexAnalysis | van de Geijn et al. (2009)212 |
| population study | 2298 | UHPLC-FLD, MALDI-MS, LC-MS | manual | GlycoWorkbench | Pucić et al. (2011)*69 |
| population study | 1709 | MALDI-MS | manual | flexAnalysis | Pučić Baković et al. (2013)211 |
| population study | 5117 | UHPLC-FLD | manual | NA | Krištić et al. (2013)70 |
| GWAS | 4095 | UHPLC-FLD, MALDI-MS | manual | NA | Lauc et al. (2013)*71 |
| population study, genetics and epigenetics | 1050 | UHPLC-FLD | manual | NA | Menni, Keser et al. (2013)270 |
| RA | 1800 | LC-MS | manual | Xtractor2D | Bondt et al. (2013)271 |
| population study, GWAS | 1201 | UHPLC-FLD, CGE-LIF, MALDI-MS, LC-MS | manual | flexAnalysis, DataAnalysis, Xtractor2D, glyXtool, glyXalign | Huffman et al. (2014)167 |
| population study | 3515 | UHPLC-FLD | manual | NA | Nikolac Perkovic et al. (2014)272 |
| SLE | 1020 | UHPLC-FLD | manual | NA | Vučković et al. (2015)*273 |
| IBD | 1114 | UHPLC-FLD | manual | NA | Trbojević-Akmačić et al. (2015)*274 |
| population study | 701 | UHPLC-FLD | manual | NA | Yu et al. (2016)275 |
| allergic diseases | 893 | LC-MS | manual | NA | Pezer et al. (2016)276 |
| CRC | 1298 | UHPLC-FLD | manual | NA | Vučković et al. (2016)*277 |
| kidney disfunction | 3274 | UHPLC-FLD | manual | NA | Barrios et al. (2016)278 |
| low back pain | 4511 | UHPLC-FLD | manual | NA | Freidin et al. (2016)279 |
| hypertension | 4757 | UHPLC-FLD | manual | NA | Wang et al. (2016)280 |
| population study | 1826 | LC-MS | manual | Xtractor 2D | Plomp et al. (2017)281 |
| population study, role of hormones | 1010 | HPLC-FLD, UPLC-FLD | manual | NA | Ercan et al. (2017)282 |
| glycosylation regulation | 5243 | LC-MS | manual | DataAnalysis, Xtractor2D | Benedetti et al. (2017)283 |
| GWAS | 8129 | UHPLC-FLD | manual | NA | Shen et al. (2017)284 |
| T2D | 5984 | UHPLC-FLD | manual | NA | Lemmers et al. (2017)285 |
| statin therapy | 4009 | UHPLC-FLD | manual | NA | Keser et al. (2017)286 |
| dyslipidaemia | 598 | UHPLC-FLD | manual | NA | Liu et al. (2018)287 |
| hypertension | 630 | LC-MS | manual | NA | Liu et al. (2018)288 |
| IBD | 3441 | LC-MS | manual | LaCyTools | Šimurina, de Haan, Vučković et al. (2018)*289 |
| population study | 637 | UHPLC-FLD | manual | NA | Russell et al. (2019)*74 |
| hyperuricemia | 635 | UHPLC-FLD | manual | NA | Hou et al. (2019)290 |
| cardiometabolic diseases | 701 | UHPLC-FLD | manual | NA | Wang et al. (2019)291 |
| T2D | 849 | UHPLC-FLD | manual | NA | Li et al. (2019)292 |
| GWAS | 12320 | UHPLC-FLD, LC-MS | manual | NA | Klarić et al. (2020)*268 |
| T2D | 1112 | UHPLC-FLD | manual | NA | Wu et al. (2020)293 |
| T2D | 1815 | UHPLC-FLD | manual | NA | Singh et al. (2020)31 |
| thyroid diseases | 4636 | UHPLC | manual | NA | Martin et al. (2020)77 |
| population study | 13061 | UHPLC-FLD, LC-MS | manual | NA | Štambuk et al. (2020)*29 |
| population study, weight intervention | 3841 | UHPLC-FLD | manual | NA | Greto et al. (2021)*294 |
| hypertension | 3452 | UHPLC-FLD | manual | NA | Kifer et al. (2021)*295 |
| hypertension and T2D | 883 | UHPLC-FLD | manual | NA | Meng et al. (2021)296 |
| perimenopause | 5080 | UHPLC-FLD | manual | NA | Deriš et al. (2022)297 |
| Total Serum Proteins | |||||
| population study | 594 | CE-LIF | manual | GeneMapper | Vanhooren et al. (2010)298 |
| hypertension | 972 | CE-LIF | manual | Genescan | Vilar-Bergua et al. (2015)299 |
| T2D | 1161 | CE-LIF | manual | Genescan | Testa et al. (2015)300 |
| breast cancer | 585 | UHPLC-FLD | automated | NA | Saldova et al. (2017)301 |
| RA | 1562 | MALDI-MS | manual and automated | flexAnalysis, MassyTools, GlycoWorkbench | Reiding et al. (2018)*302 |
| Total Plasma Proteins | |||||
| population study | 1008 | HPLC-FLD | manual | GlycoBase | Knežević et al. (2009)* 27 |
| population study | 1914 | HPLC-FLD | manual | GlycoBase | Knežević et al. (2010)28 |
| GWAS | 2705 | HPLC-FLD | manual | GlycoBase | Lauc et al. (2010)*118 |
| population study, lipidomics | 2035 | HPLC-FLD | manual | NA | Igl, Polašek et al. (2011)303 |
| GWAS | 3533 | HPLC-FLD | manual | GlycoBase | Huffman et al. (2011)*119 |
| metabolic syndrome | 732 | HPLC-FLD | manual | GlycoBase | Lu et al. (2011)304 |
| ADHD and autism | 525 | HPLC-FLD | manual | NA | Pivac et al. (2011)305 |
| medication | 2360 | HPLC-FLD | manual | NA | Saldova et al. (2012)306 |
| HNF1A-MODY | 783 | HPLC-FLD | manual | GlycoBase | Thanabalasingham et al. (2013)307 |
| population study | 2144 | MALDI-MS | manual | DataAnalysis, GlycoWorkbench | Reiding et al. (2017)302 |
| GWAS | 1338 | UHPLC-FLD | manual | NA | Suhre et al. (2017)308 |
| T2D | 505 | UHPLC-FLD | manual | NA | Adua et al. (2017)309 |
| T2D | 1614 | UHPLC-FLD | manual | NA | Keser et al. (2017)310 |
| T2D | 2281 | MALDI-MS | automated | flexAnalysis, MassyTools | Dotz et al. (2018)311 |
| IBD | 3631 | MALDI-MS | manual and automated | MassyTools | Clerc, Novokmet, Dotz et al. (2018)*312 |
| HNF1A-MODY | 989 | UHPLC-FLD | manual | NA | Juszczak et al. (2018)*78 |
| CRC | 1231 | UHPLC-FLD | manual and automated | GlycoBase | Doherty et al. (2018)*313 |
| GWAS | 3811 | UHPLC-FLD | manual | NA | Sharapov et al. (2019)*314 |
| population study | 2396 | HPLC-FLD | manual | Matlab | Ruhaak et al. (2020)315 |
| metformin and statin use in T2D | 3333 | MALDI-MS | automated | DataAnalysis | Singh et al. (2020)31 |
| cardiometabolic risk | 3140 | UHPLC-FLD | manual | NA | Wittenbecher et al. (2020)76 |
| GWAS | 4802 | UHPLC-FLD | manual | NA | Sharapov et al. (2021)*316 |
| T2D | 506 | UHPLC-FLD | manual | NA | Edua et al. (2021)317 |
| diabetes | 610 | UHPLC-FLD | manual | GlycoStore | Cvetko et al. (2021)318 |
| T2D complications | 3333 | MALDI-MS, FTICR-MS | manual and automated | MassyTools | Memarian et al. (2021)319 |
| IgG and Total Plasma Proteins | |||||
| kidney disease in T1D | 818 | UHPLC-FLD | manual | NA | Bermingham et al. (2017)75 |
| evaluation of glycomics normalization methods | 5139 | UHPLC-FLD, MALDI-MS, LC-MS | manual | DataAnalysis, GlycoWorkbench | Benedetti et al. (2020)133 |
| age-related macular degeneration | 2835 | HPLC-FLD, UHPLC-FLD | manual | NA | Bućan et al. (2022)320 |
| IgA | |||||
| RA | 1800 | MALDI-MS | manual | MassyTools | Bondt et al. (2017)321 |
| Antigen-Specific IgG | |||||
| RA | 703 | LC-MS | manual | LaCyTools | Bondt et al. (2018)222 |
| COVID-19 | 650 | LC-MS | manual | LaCyTools | Pongracz et al. (2021)*216 |
| viral infections | 2 × 400 | LC-MS | manual | Skyline | Larsen et al. (2021)217 |
| Apolipoprotein CIII | |||||
| population study | 771 | MALDI-MS | automated | MassyTools | Demus et al. (2021)322 |
| HMOs | |||||
| infant morbidity and inflammation | 659 | LC-MS | manual | MassHunter Qualitative Analysis | Jorgensen et al. (2021)*323 |
| AGP | |||||
| diabetes | 635 | LC-MS | manual | LaCyTools | Tijardović et al. (2022)324 |
| Tf and IgG | |||||
| GWAS | 1900 | UHPLC-FLD | manual | NA | Landini et al. (2022)*325 |
Studies denoted with an asterisk were highlighted in the main body of the manuscript. RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; IBD, inflammatory bowel disease; CRC, colorectal cancer; T2D, type 2 diabetes; ADHD, attention-deficit hyperactivity disorder; HNF1A-MODY, hepatocyte nuclear factor 1α maturity-onset diabetes of the young; T1D, type 1 diabetes; COVID-19, coronavirus disease 2019.
6.1. HT-Glycomics in Epidemiological Studies
In 1988, Parekh et al. were the first to report on the age-dependence of IgG glycosylation by analyzing the degree of galactosylation of total serum IgG N-glycans (both Fab and Fc) in a group of 151 healthy individuals of both sexes aged 1–70 years.38 Any heterogeneity in terms of attached sialic acids, N-acetylglucosamine, and fucose was eliminated by appropriate exoglycosidase treatments prior to oligosaccharide analysis so that galactosylation was the only trait analyzed. Glycans were released, isolated, and radioactively labeled, as described previously by Parekh et al., in 1985 and Ashford et al., in 1987.10,39 In these studies, it was discovered that the mean incidence of agalactosylated N-glycans on IgG, in which both outer arms terminate in N-acetylglucosamine, is over 30% at birth, reaches a minimum of about 20% in individuals at age 25, and then increases steadily until it reaches about 40% at age 70. The mean incidence of monogalactosylated N-glycans remains impressively consistent at about 40% across the age range, whereas digalactosylated N-glycans show a parabolic pattern inverse to the parabolic age-dependence curve of agalactosylated N-glycans. There were no sex-specific differences found. The first large-scale study (>500 samples) of N-glycans was carried out on 1008 individual plasma samples by Knežević et al. in 200927 and assessed whether there is any variability or heredity as well as key environmental factors that affect the human plasma N-glycome. The N-glycans from total plasma proteins were released and labeled with 2-AB following the procedure described by Royle et al.53 By combining HPLC analysis of fluorescently labeled glycans with sialidase digestion, glycans were separated and quantified into 33 chromatographic peaks. A high degree of variability was observed, with a median ratio of minimum to maximum values of 6.17 and significant age- and sex-specific differences. Heritability estimates varied widely for individual glycans, ranging from very low to very high. Glycan-wide environmental determinants with statistically significant effects of various variables such as diet, smoking, and cholesterol levels were also noted.
In 2011, Pučić et al., published the first large-scale population study of the IgG N-glycome applying a novel HT method for isolation of IgG from 2298 plasma samples.69 After IgG isolation, PNGase F was used to release the N-glycans attached to IgG followed by 2-AB labeling as described by Knežević et al.27 The results showed that nearly 96% of all neutral IgG glycans were core-fucosylated. Compared to other major plasma glycoproteins, IgG N-glycans are also less sialylated, with approximately 3% disialylated (A2G2S2) glycan structures. Agalactosylated and monogalactosylated structures were found to be 40% each, and digalactosylated structures represented 20% of the neutral IgG glycome. On average, 18% of neutral glycans contained a bisecting N-acetylglucosamine.
A study in Western Australia has shown that, in addition to the replicated association of BMI to agalactosylated IgG, measures of central adiposity explain the most variation in the IgG glycome and are associated with an increased abundance of pro-inflammatory IgG N-glycans (637 community-based individuals in the age range 45–69 years). This study suggests that the waist-to-height ratio or android/gynoid ratio should be considered instead of BMI when controlling for adiposity in IgG glycome biomarker studies.74
In the study by Greto et al., in 2021, an altered IgG N-glycome was observed after extensive weight loss following a low-calorie diet, bariatric surgery, or a decrease of BMI with time.294 The levels of bisecting N-acetylglucosamine substantially decreased after the low-calorie diet intervention following bariatric surgery, the core-fucosylated and agalactosylated glycans decreased, and an increase was observed for digalactosylated and monosialylated glycans, regardless of the type of surgery. The abundance of digalactosylated glycans also increased with a decrease in BMI, while the abundance of agalactosylated and high mannose glycans decreased with weight loss. These changes in relation to BMI drop for plasma-derived IgG N-glycan traits were observed by longitudinal monitoring of 1680 participants, and statistical analysis was performed on a subset of 3742 samples (measurements) that contained BMI information. These findings highlighted the effect of weight loss on IgG N-glycosylation. The largest population study, hitherto, holds the analysis of the IgG glycome over 10 000 individuals from 27 different populations.29 This study by Štambuk et al., confirmed strong associations between different IgG glycans (and Fc glycopeptides) and age but also the place of residence of an individual.29 Interestingly, several IgG glycans strongly correlated with the expected lifespan, suggesting that IgG glycome composition may correlate with all-cause mortality risk. Cardiovascular diseases (CVD) are the main cause of mortality, and Kifer et al., recently identified four IgG N-glycan traits that associate with an increased risk of incident hypertension.295 Three glycan traits, representing simple glycan structures containing a core fucose with only one or no galactose residues attached to the bisecting N-acetylglucosamine, significantly increase in individuals who developed hypertension during follow-up when compared with those defined as controls, whereas only one glycan structure decreases, a more complex digalactosylated structure with two sialic acid residues attached to the galactose molecules and without a bisecting N-acetylglucosamine. These results indicate that the IgG glycoprofile of individuals who would develop incidental hypertension during follow-up manifests a proinflammatory pattern and is associated with obesity years before diagnosis, which indeed supports a role of IgG glycosylation in the incident hypertension.
6.2. HT-Glycomics in Genetic Studies
The biosynthetic pathway of N-glycosylation is well defined, but very little is known about the genetic regulation of glycosylation. In 2010, Lauc et al. published the first GWAS of the N-glycome, which shed initial light on the associations between common genetic polymorphisms and protein N-linked glycosylation.118 The analysis of 2705 individuals from three population cohorts, from Croatia and Scotland, revealed that common variants in hepatocyte nuclear factor 1α (HNF1A) and fucosyltransferase genes FUT6 and FUT8 affect the N-glycan levels in human plasma. Subsequent functional studies confirmed that HNF1A regulates the expression of key fucosyltransferase and fucose biosynthesis genes and in this way acts as the main regulator of plasma protein fucosylation.118
As a sequel to the study of Lauc et al., the analysis was extended to 33 direct traits and 13 derived glycosylation traits in 3533 individuals by Huffman et al., using the same sample preparation protocol from 2011.119 In this European cohort, the previous findings were replicated and, additionally, three novel gene associations were identified (MGAT5, B3GAT1, and SLC9A9). While MGAT5 encodes a glycosyltransferase which is known to synthesize the associated glycans, neither B3GAT1 nor SLC9A9 had previously been functionally associated with glycosylation of plasma proteins. Because B3GAT1 has glucuronyltransferase activity, Huffman et al. have shown that glucuronic acid is present on the antennae of plasma glycoproteins and detected the corresponding HPLC peak. SLC9A9 encodes a proton pump that affects pH in the endosomal compartment. Recently, it has been reported that changes in the pH of the Golgi can affect protein sialylation,326 providing a possible mechanism for the observed association.
Sharapov et al., reported a GWAS of the composition of human plasma N-glycome in up to 3811 participants, which was measured by UHPLC.314 Starting with the 36 directly measured traits, an additional 77 derived traits were calculated, leading to a total of 113 glycan traits. From this, 12 different loci were discovered and replicated, the majority of loci contain genes encoding enzymes directly involved in glycosylation (FUT3/FUT6, FUT8, B3GAT1, ST6GAL1, B4GALT1, ST3GAL4, MGAT3, and MGAT5) and a known regulator of plasma protein fucosylation (HNF1A). Moreover, functional genomic annotation suggests a role for several other genes including DERL3, CHCHD10, TMEM121, IGH, and IKZF1. With an aim to verify the previous findings, a replication study was published using an independent set of 4802 individuals in 2021, where 36 directly measured traits and 81 derived glycan traits, resulting in a total of 117 glycan traits, were analyzed.316 Additionally, the association of three loci near the genes PRRC2A (HLA region), RUNX3/MAN1C1, and SLC9A9, which have not been identified before, was reported.
The first GWAS of an individual protein was performed on IgG, which is the most abundant plasma glycoprotein.71 This study implemented the IgG isolation procedure using monolithic protein G plates followed by the N-glycan release and labeling protocol as described by Pučić et al.69 HILIC-UHPLC was used as measurement and detection platform. Lauc et al. performed the first GWAS in 2013 to identify genetic loci associated with IgG N-glycosylation on 2247 individuals from the same European populations71 used for the GWAS study by Lauc et al. in 2010.118 Overall, nine genome-wide significant loci were identified of which four loci (ST6GAL1, B4GALT1, FUT8, and MGAT3) contained genes encoding glycosyltransferases. The remaining five loci (IKZF1, IL6ST-ANKRD55, ABCF2-SMARCD3, SUV420H1, and SMARCB1-DERL3) included genes that were previously not implicated in protein glycosylation, although most of them have strongly been associated with autoimmune and inflammatory conditions and/or hematological cancers. This study demonstrated that GWAS can be used to identify novel loci that control the glycosylation of a single plasma protein.
In 2020, the second and – up to now – largest GWAS study of IgG N-glycosylation was published by Klarić et al.268 Here, 8090 samples were analyzed from individuals of European ancestry and suggested how associated loci form a functional network. This new study doubled the number of associated loci compared to the first IgG N-glycome GWAS study in 2013.71 In total, 27 loci showed genome-wide significant associations, with another six loci showing suggestive significance. Thirteen of the genome-wide significant loci were found to validate prior findings, while 14 loci had never been linked to IgG glycosylation before. The same single nucleotide polymorphism (SNP) and glycan association was repeated in a meta-analysis of four different European cohorts (N = 2388) for 19 of 27 relevant loci (9 of 14 previously unassociated). There is no known role in glycosylation for any of the genes in the novelly associated loci.
Generally, GWAS studies indicated that regulatory networks that govern protein glycosylation include many other genes in addition to enzymes that are directly involved. These networks seem to be protein and/or cell-type specific, as indicated by the recent comparison of genes that regulate IgG and Tf glycosylation.325
6.3. HT-Glycomics in Clinical Studies
Glycome composition integrates genetic, epigenetic, and environmental factors, which makes glycans promising biomarkers for complex diseases. Furthermore, the glycocalyx, a thick layer of glycans on the surface of cells, engages in the contact of a cell and its environment and different cells with one another, including immune cells screening for danger.327 IgG glycans are involved in multiple humoral immune processes, such as ADCC, complement activation and complement-dependent cytotoxicity (CDC), antigen neutralization, target opsonization for phagocytosis, and hypersensitivity reactions. Significant alterations in the IgG glycome have also been reported in different diseases,273,274,328−332 making it a prospective biomarker for various diseases.
Rheumatoid arthritis (RA) is one of the most prevalent chronic inflammatory diseases which primarily involves the joints. Changes in IgG have been associated with RA, and most patients develop autoantibodies against IgG. Already in 1976, Mullinax et al. reported an apparent decrease in the galactose content of the Fc region of serum IgG from RA patients,37 which sparked the hypothesis that glycosylation of IgG might change in RA and could be part of the disease pathogenesis. On the basis of their work, Parekh et al. demonstrated in 1985 that the portion of IgG glycoforms that do not carry galactoses is particularly high for RA and primary osteoarthritis (OA) patients.10 This study was done on 42 IgG samples obtained from the serum of healthy controls and RA and OA patients.
Gindzienska-Sieskiewicz et al. analyzed IgG glycosylation from 50 patients with RA in comparison with 30 healthy controls and showed that galactosylation of IgG in patients with RA correlates with severity and duration of illness.328 Also, in patients with a long duration of RA, a significant decrease of galactose ratio in comparison with patients who have had arthritis for less than five years was observed. Patients with severe RA had a reduced IgG galactose content compared to the group of patients in remission. In the same study, it was demonstrated that a decrease of galactosylation is positively correlated with disease activity in patients with active RA.328 Gudelj et al., studied IgG glycosylation in RA in two prospective cohorts by measuring IgG N-glycan traits in 179 subjects who developed RA within 10 years using HILIC-UHPLC-FLD.329 In contrast to other studies, no correlation was observed in the decrease of galactosylation and sialylation in RA cases, with the time between recruitment to the study and RA diagnosis. These results indicate that decreased galactosylation might be a pre-existing risk factor involved in the disease development.
On the other hand, a HT glycomic analysis showed that serum protein N-glycosylation was strongly modified during pregnancy, both in RA patients and healthy controls.302 Using MALDI-TOF-MS, changes in the serum N-glycome were observed during 285 pregnancies (samples were collected before conception (only for RA patients), at three time points during pregnancy and at three time points after delivery). Specifically, an increase in galactosylation of diantennary N-glycans, an increase in tri- and tetraantennary species as well as an increase of α2,3-linked sialylation and a decrease in N-glycans with bisecting N-acetylglucosamine were found.302 The change in RA disease activity was shown to be negatively associated with the galactosylation of diantennary N-glycans and positively with the sialylation of triantennary fucosylated glycans.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that predominantly affects women and certain ethnic groups.333 SLE condition is influenced both by genes and by the environment, which results in the production of pathogenic autoantibodies, mainly of the IgG1 and IgG3 subclasses. Vučković et al., analyzed the composition of the IgG N-glycome in 261 SLE patients and 247 matched controls of Latin American Mestizo origin and in two independent replication cohorts of different ethnicity (108 SLE patients and 193 controls from Trinidad as well as 106 SLE patients and 105 controls from China) using the HILIC-UHPLC-FLD.273 A significant difference was observed in the IgG N-glycome composition between patients and controls. The most significant changes included a decrease in IgG galactosylation, sialylation, core-fucosylation, as well as an increase in bisecting N-acetylglucosamine. The magnitude of observed changes was associated with the intensity of the disease, indicating that aberrant IgG glycosylation may be important for the molecular mechanism in SLE.
Inflammatory bowel disease (IBD) is considered to be a result of environment, gut microbiota composition, and aberrant immune response in genetically susceptible individuals. Trbojević-Akmačić et al., showed significant differences in the IgG N-glycome composition between patients with ulcerative colitis (UC) or Crohn’s disease (CD) compared with controls.274 The IgG N-glycome was analyzed in 507 patients with UC, 287 patients with CD, and 320 controls by HILIC-UHPLC-FLD. IgG galactosylation was significantly decreased in both UC and CD patients, while IgG sialylation was significantly decreased in CD.274 Moreover, larger HT-studies on the level of IgG Fc glycopeptides289 and total plasma N-glycome312 showed that changes in N-glycosylation associated with the disease and clinical features of IBD, indicating that IgG and total plasma protein N-glycan profiles have translational potential as IBD biomarkers.289
As mentioned before, glycans play a major role in the immune system and represent one of the main defenses against various pathogens. The viruses use host-cell machinery to glycosylate their own proteins during replication. Viral envelope proteins from a variety of human pathogens including HIV-1, influenza virus, severe acute respiratory syndrome coronavirus (SARS-CoV)-1, and SARS-CoV-2, Zika virus, dengue virus, and Ebola virus have evolved to be extensively glycosylated. These viral host-cell-derived glycans facilitate various structural and functional roles, from immune evasion enabled by glycan shielding to the enhancement of immune cell infection. Giron et al., profiled circulating glycomes (plasma and bulk IgG) in 47 HIV-infected individuals.334 CGE-LIF and lectin microarray were used for glycomic analysis. They identified several total plasma protein N-glycome traits associated with a time-to-viral-rebound in two geographically distinct cohorts.334 IgG glycosylation was also recently studied in 167 patients with mild COVID-19 and 166 patients with severe COVID-19 using a HILIC-UHPLC-FLD.335 A significant decrease in IgG N-glycans containing bisecting N-acetylglucosamine was reported in severe, compared to mild, COVID-19 cases. Another study, conducted by Wang et al., showed a decreased level of total IgG fucosylation, sialylation, and galactosylation in severe COVID-19 cases compared to controls.336 Moreover, LC-MS analysis of IgG1 Fc glycopeptides in a longitudinal cohort of COVID-19 patients has demonstrated significant changes in anti-S IgG1 N-glycosylation near the disease onset compared to total plasma IgG1. These differences in N-glycosylation of anti-S IgG1 and total IgG1 were rapidly diminishing during the disease course.216
Recently, glycobiology has gained importance in cancer research given its role in understanding various cancer mechanisms and for diagnostic application.337−341 Also, advances in the field of proteomics and glycomics have helped glycobiologists decipher the link between glycan structures and disease progression.342 Glycosylation acts as a key regulatory mechanism, controlling several physiopathological processes and defects in glycosylation are associated with various diseases, including cancers.277,342−344 While glycoproteins are some of the most common clinically used biomarkers for diagnosis and monitoring of malignant progression, their glycosylation has not, yet, been taken into account.339,345−351 However, it has been widely shown that glycosylation changes can be associated with cancer, of which most are related to sialylation, fucosylation, mucin-type O-glycans, as well as N- and O-linked glycan branching.338,352,353
Sialylated carbohydrates have an important role in cellular recognition, cell adhesion, and cell signaling. An increase in α2,3- and α2,6-linked sialylation has been closely associated with cancer.354 A study by Cheeseman et al. revealed that elevated concentrations of sialic acids in plasma and serum correlated positively with the presence of CVD, diabetes, and the development of malignant tumors.344 Another study by Laider et al. found a significant correlation between increased α2,6-sialylation and adhesion abilities of metastatic melanoma cells.355 In line with these observations, Kremser et al., identified an altered N-glycome by MALDI-MS on integrins α3β1 and α5β3 and revealed that glycan modifications have been related with increased melanoma cell motility, impacting cell–ECM interactions.356 Fucosylation has also been associated with cancer.357
Overexpression of core-fucosylated glycans is an important feature of lung cancer and breast cancer,358,359 and it is also reflected in the serum activities of fucosyltransferases during the process of hepatocarcinogenesis.360 Noda et al., showed a highly significant increase in the levels of α1,6-fucosylated α-fetoprotein (AFP) in patients with hepatocellular carcinoma (HCC) in comparison to chronic liver diseases which was in part caused by up-regulation of the α1,6-fucosyltransferase.361 West et al., analyzed 138 HCC tissue samples by in situ N-glycan MALDI-MSI and showed that during malignant transformation levels of complex β1,6-branched N-linked glycans, as well as levels of fucosylation, are increased compared to adjacent untransformed tissue or tissue from patients with liver cirrhosis without cancer.362 Moreover, increased fucosylation was associated with reduced survival time. This analytical approach overrides the need for microdissection and tissue solubilization prior to analysis, making it an attractive approach for HT screening of tissue samples.
Another common feature of cancer is the overexpression of the GalNAc-type O-glycans, also called mucin-type O-glycans. Mucin-type O-glycans are mostly found in the transmembrane and on secreted glycoproteins. Also, during malignancy, the incomplete synthesis of O-glycans can occur and lead to abnormal expression of truncated O-glycans; disaccharide Thomsen–Friedenreich antigen (T antigen, Galβ1-3GalNAcα1-O-Ser/Thr), monosaccharide GalNAcα1-O-Ser/Thr (also known as Tn antigen), and their sialylated form (STn-antigen, Neu5Acα2-6GalNAcα1-O-Ser/Thr).363,364 The recent application of precise and stable glycan gene editing in mammalian cell lines combined with a HT MS approach has contributed to the characterization of the O-glycoproteome of cancer cells and identification of O-glycoproteins containing STn-antigen in sera of gastric cancer and healthy individuals.363,365 Two candidate biomarkers carrying the STn-antigen were identified, CD44 and GalNAc-T5, with CD44 validated as expressed in gastric cancer tissue. In addition, STn has been found on plasminogen in serum from patients with intestinal metaplasia and gastritis.366 Plasminogen O-glycans were analyzed by nano-HPLC-MALDI-TOF/TOF after lysine-sepharose affinity chromatography, in-gel de-O-glycosylation by reductive β-elimination and permethylation.366 A recent study that explored the serum protein O-glycosylation in 62 CRC patients and 20 healthy individuals by Gizaw et al.367 has revealed differences in O-glycan patterns between cancer samples and healthy controls that correlated with cancer progression. Moreover, O-glycan profiles differed between male and female patients during cancer progression.
Although all of the above-mentioned biomarkers have shown an aberrant glycosylation in relation to cancer,368−370 they have limited application due to their relatively low specificity and sensitivity. With the advent of new methods and technologies for glycan analysis, new biological information is generated, and potential disease biomarkers can be identified. Additionally, the newly developed HT platform technologies have enabled the analysis of large cohorts of samples in an efficient manner.371
For example, Miyoshi et al., observed an increased concentration of fucosylated haptoglobin in serum of patients with pancreatic cancer compared with healthy controls as well as with patients diagnosed with other types of cancer, e.g., HCC, gastric cancer, or CRC.372 Site-directed analysis of haptoglobin N-glycopeptides by RP-HPLC-ESI-MS showed a site-specific increase in fucosylation of diantennary glycans on two N-glycosylation sites and an increase in fucosylation of triantennary glycans on all four N-glycosylation sites in pancreatic cancer patients compared to chronic pancreatitis and healthy individuals. Further clinical investigation on 100 patients with CRC located near the liver showed higher levels of fucosylated haptoglobin,372 implicating cancer location as an important factor for changes in haptoglobin fucosylation. Moreover, haptoglobin glycosylation, along with Tf glycosylation, has shown to change in ovarian cancer,121 and along with IgG, Tf, and AGP glycosylation in patients bearing stomach adenocarcinoma.373
Glycosylation in CRC has also been studied on the level of total plasma N-glycome313 and IgG N-glycome.277 It was shown that CRC associated with a decrease in IgG galactosylation, IgG sialylation, and an increase in core-fucosylation of neutral N-glycans with a concurrent decrease of core-fucosylation of sialylated N-glycans. A model based on age, sex, and N-glycans showed a good discriminative power between CRC cases and controls (area under the curve = 0.755) implicating that variation in IgG N-glycosylation could be important for the prediction of disease course. Statistically significant differences were observed also on the level of the plasma N-glycome. A decrease in the core fucosylated diantennary glycans F(6)A2G2 and F(6)A2G2S(6)1 was highly significant at all stages of CRC. Additionally, stage 1 showed a unique biomarker signature compared to stages 2 up to 4.313
Altered total serum protein N-glycosylation (both fucosylation and sialylation) has been shown in a study on 13 sera with benign prostate hyperplasia (BPH) and 34 PCa samples, using HT normal phase (NP) and weak anion exchange (WAX)-HPLC N-glycan analysis in combination with exoglycosidase digestion.374 The levels of diantennary N-glycans containing core-fucose and N-glycans containing α2,3-linked sialic acids were significantly increased in PCa patients compared with patients with BPH. Triantennary trigalactosylated N-glycans and tetraantennary tetrasialylated N-glycans with antennary fucose were significantly decreased, and tetraantennary tetrasialylated N-glycans increased in Gleason score 7 compared with Gleason score 5 PCa patients. Identification of glycosylation patterns specific for BPH, Gleason 7, and Gleason 5 PCa patients with the used technology, implicated its potential use for noninvasive diagnosis. PCa, BPH patients, and healthy controls were also shown to have different total urine N-glycosylation profile (after desialylation) analyzed using a DNA sequencer-aided fluorophore-assisted carbohydrate electrophoresis (DSA-FACE) system. The urine N-glycome (after digital rectal examination) and prostate volume were combined into a urinary glycoprofile marker (UGM) that was able to discriminate between PCa and BPH.375,376 PSA has been recognized as a specific biomarker for PCa that can distinguish it from BPH. Measurement of a2,3-linked sialylation on PSA, which is characteristic for PCa patients, may improve the accuracy of early detection.377
HMOs are another clinically relevant glycan class with extensive nutritional and health benefits for infants.378,379 Because of their extreme complexity and diversity their comprehensive analysis, especially in HT mode, remains challenging. Significant progress in the quantification of this diverse glycan class has been made in the past decade16,17,380,381 (Figure 15), facilitating analysis in large-scale studies. Current knowledge on associations of HMO content with infant growth and health status is extensively reviewed by Sprenger et al.379 Most notably, a study done by HPLC-FLD analysis of 2-AB labeled oligosaccharides from 410 human milk samples showed that HMO concentrations and profiles differ between healthy women from different geographical regions.382 Analysis of HMOs by nano-LC-chip TOF-MS15 in two cohorts of 303 Malawian mother–infant sets showed a lower abundance of fucosylated and sialylated HMOs in breast milk of nonsecretor mothers (negative for the functional enzyme encoded by the fucosyltransferase 2 gene), whose infants were minimal compared to those showing normal growth.381 Another recent study employing the same analytical technology on 659 human milk samples collected at six months postpartum aimed to explore the association of HMOs and bioactive proteins with markers of inflammation and longitudinal prevalence of infant morbidity.323 Differences in the relative abundance of HMOs were observed between secretor and nonsecretor mothers, mainly a lower abundance of fucosylated HMOs and a higher abundance of sialylated HMOs in nonsecretors. Additionally, depending on the secretor status, significant associations of HMOs and bioactive proteins with inflammatory markers and infant morbidity were observed.323 Considering longitudinal changes in HMO composition during lactation383 and the heterogeneity of HMOs in women of, e.g., different geographical backgrounds or secretor status, more extensive longitudinal studies focused on different populations are needed.
Figure 15.
Representative MALDI-TOF-MS spectrum of permethylated HMOs. All masses correspond to fully permethylated, free reducing end, sodium adducts of HMOs. Possible structures for each mass are shown. Free lactose was excluded from the spectrum. HMOs were identified using GlycoMod. Reproduced with permission from ref (16). Copyright 2020 Oxford University Press.
Relevance of HMOs in the context of infant growth,381,384 allergies,385,386 and other diseases379 makes it an important glycan class also in the context of formula-fed infants’ health, as well as in the biopharmaceutical industry.387 Today, HMOs are synthesized artificially and added to infant milk formulations and baby food. As such, their analysis is subjected to stringent controls warranting highly robust and sensitive HT methods.
6.4. Glycoprofiling of Biopharmaceuticals
Two-thirds of therapeutic proteins currently on the market are glycoproteins. Glycans have been recognized as a critical quality attribute influencing function, efficacy, and safety of a drug, making their analysis essential throughout the screening of new drug candidates, product development, and manufacturing processes as well as in comparing biosimilars and biobetters to originator drug.388 Biotherapeutic glycoproteins are predominantly mAbs based on IgG1, Fc-fusion proteins, and cytokines.
The potency of therapeutic mAbs depends on effective antigen recognition combined with an apt effector function, which is largely dependent on the structure of Fc N-glycans. Core-fucose is known to decrease IgG binding to FcγRIIIA receptor reducing the ADCC,389,390 while terminal galactose increases the CDC, sialic acid affects inflammatory properties, and mannose affects pharmacokinetics.388 Glycosylation itself, if containing nonhuman glycans originating from, e.g., murine cell lines, can be immunogenic which compromises drug safety. Moreover, different host cellular production systems result in distinct glycosylation profiles while cell culture conditions, e.g., nutrients, oxygen level, and pH, also influence glycosylation,391 causing structural and functional changes and affecting therapeutic behavior. Upstream processing conditions and their effect on glycosylation must be monitored to ensure the final drug quality and safety as well as batch-to-batch consistency. Glycoengineering through culture conditions manipulations, gene knockouts, and expression of glycan-modifying enzymes is one of the approaches used in biotherapeutic glycoprotein development that also requires glycan characterization throughout the process.
Recent interlaboratory study on the glycosylation analysis of a NIST mAb reference material has demonstrated the applicability of a wide range of analytical methods for glycan analysis on the level of free glycans, glycopeptides, protein fragments, and intact mAb.392 However, not all of these methods have a high level of robustness necessary for HT applications and/or are not easily applicable in biopharmaceutical companies due to more stringent requirements resulting in the use of well-established and conservative methods, e.g., LC-FLD or LC-MS analysis of fluorophore-labeled glycans (Figure 16). Classical labeling approaches used in the biopharmaceutical industry (e.g., 2-AB and 2-AA labeling) are slowly being replaced by rapid deglycosylation and labeling of therapeutic glycoproteins using glycan analysis kits containing Rapid PNGase F and RapiFluor-MS240 or InstantPC labels.129,393 These novel labels provide increased sensitivity and higher ionization efficiency compared to traditional 2-AB and 2-AA, allowing for parallel analysis of the same sample with orthogonal techniques, e.g., LC-FLD and LC-MS, without the additional sample preparation steps. Moreover, increased sensitivity allows the detection of low abundant glycoforms. In line with the rapid labeling chemistries also goes robotization of sample preparation for less hands-on time, allowing the sample preparation of samples in a 96-well format in a few hours.240 Derived glycosylation traits are often used for the comparison of different biotherapeutics because they relate to differences in circulation half-time or mAbs effector functions.
Figure 16.
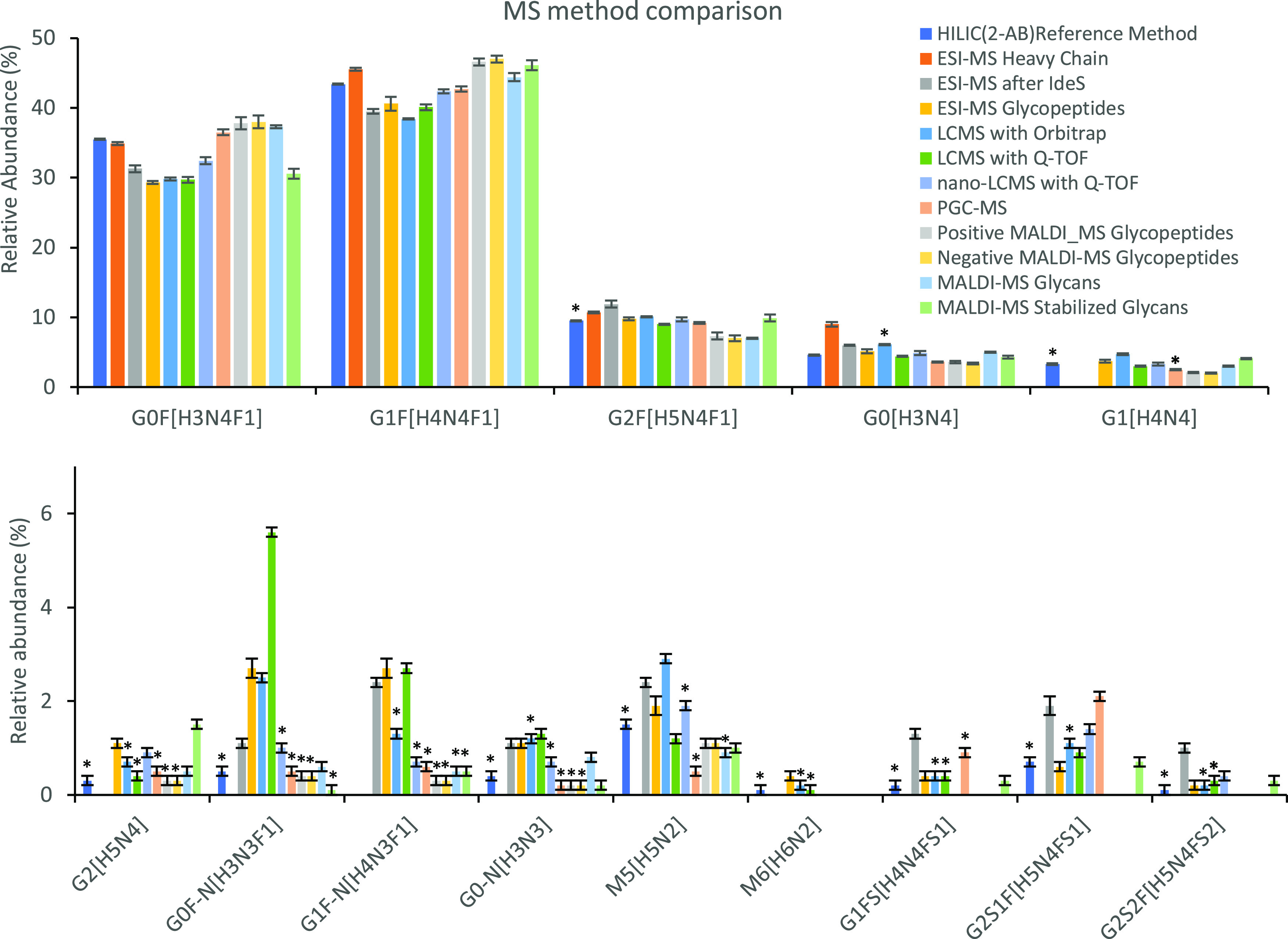
Relative quantitative evaluation of MS-based method performance in the analysis of therapeutic IgG Fc glycosylation. Each analytical method was applied in a batch of six replicates (the first set taken from the data set). Error bars represent the standard deviation. G1[H4N4] was not quantified for the ESI-MS after IdeS method. M6[H6N2], G1FS[H4N4FS1], and G2S1F [H5N4FF1S1] were not quantified for the analysis of glycopeptides in positive and negative ionization mode using MALDI-MS. All other missing bar graphs indicate those species were not detected with that specific method. Key: H, hexose; N, N-acetylhexosamine; F, deoxyhexose; S, N-acetylneuraminic acid (sialic acid); G0F-N, agalactosylated, core-fucosylated, monoantennary species. Data was obtained from ref (394) Copyright 2015 Reusch, et al.
Lectin microarrays have been shown to have the potential as a rapid orthogonal technique in glycosylation screening during therapeutic glycoprotein development.235,237 With relatively high throughput and sensitivity, lectin microarrays are especially advantageous during process development. However, because of the semiquantitative nature of results, for structural confirmation and more accurate quantitation, the use of standard methods such as U(H)PLC-FLD, CGE-LIF, and MS is warranted.
Lectin–glycan recognition has recently been coupled with the biolayer interferometry into a rapid HT screening platform based on biotinylated recombinant prokaryotic lectins and streptavidin-coated sensor.395
Some of the most widely used biotherapeutics, e.g., Fc-fusion protein etanercept, are also O-glycosylated, with glycosylation potentially affecting their biological function. Therefore, complete characterization of therapeutic glycoproteins’ glycosylation is of utmost importance. Reproducible and sensitive O-glycosylation profiling entails specific analytical challenges as already described above. Usually, sequential analysis of both N- and O-glycans is performed on the same sample, relying mostly on in-depth LC-MS profiling techniques. Comprehensive analysis of site-specific N- and O-glycosylation of etanercept by HILIC-UHPLC-FLR and LC-MS has been described by Houel et al.,396 while in-depth site-specific O-glycosylation profiling of glucagon-like peptide-1 (GLP1)-Fc fusion protein containing linker peptide has been recently described by Hashii et al.397O-Glycosylation analysis of therapeutic glycoproteins would benefit from future improvements of current strategies for O-glycan analysis and development of new ones.
6.5. Potential Diagnostic Applications
6.5.1. Liver Fibrosis
Liver fibrosis is a result of chronic liver damage accompanied by the accumulation of extracellular matrix proteins, which is a characteristic of most types of chronic liver diseases. The main causes of liver fibrosis are alcohol abuse, chronic hepatitis C virus (HCV) infection, and nonalcoholic steatohepatitis.398 If advanced, liver fibrosis leads to cirrhosis, liver failure, and portal hypertension, often requiring liver transplantation. Diagnosis of liver fibrosis in the early stages of the disease remains challenging due to the absence of symptoms until the disease already progressed, warranting the development of noninvasive early stage biomarkers.
In 2001, Callewaert et al., used a DSA-FACE to profile sialidase-treated N-glycans from the whole serum,59 which was the basis for the development of a cirrhosis biomarker (GlycoCirrhoTest)60 and a fibrosis biomarker (GlycoFibroTest)399 for patients with chronic HCV infection. Cao et al., adapted the DSA-FACE approach on a CE-based ABI 3500 system and simplified the procedure to four main steps: N-glycan release, N-glycan labeling, removal of sialic acids, and N-glycan profiling.400 The sera of 432 hepatitis B virus (HBV)-infected patients with liver fibrosis were analyzed to investigate the correlation between N-glycans and HBV-induced liver fibrosis and to verify a multiparameter diagnostic model related to changes in the serum N-glycome.400 Significantly changed N-glycan peaks in different fibrosis stages were selected in the modeling group, and multiparametric diagnostic models were established based on changed N-glycan levels by logistic regression analysis. N-Glycans models were compared with the aspartate aminotransferase to platelet ratio index (APRI), fibrosis index based on the four factors (FIB-4), glutamyltranspeptidase platelet albumin index (S index), GlycoCirrhoTest, and GlycoFibroTest. They found that the alterations of serum N-glycans are associated with HBV-related liver fibrosis and showed that multiparameter N-glycan models are powerful in diagnosing early stage fibrosis.
6.5.2. Diabetes
Diabetes mellitus is a high-prevalence heterogenic group of metabolic disorders characterized by the presence of hyperglycemia. This is a lifetime disease leading to reduced life expectancy, premature morbidity, and mortality. Because the disease burden is rising worldwide, many studies aim to identify predisposing factors, establish an early diagnosis, and correct classification of the disease as well as identify novel therapeutic targets.
In 2018, Juszczak et al. published research on 989 individuals diagnosed with diabetes when younger than 45 years of age (so-called “maturity-onset diabetes of the young” or MODY).78 MODY is caused due to variants in the HNF1A gene, and it is frequently misdiagnosed. Considering previous findings that plasma levels of antennary fucosylated N-glycans and high-sensitivity C-reactive protein (hs-CRP) are reduced in individuals with HNF1A-MODY, a nongenetic biomarker could improve subject selection for genetic testing and increase diagnostic rates. This study suggested the potential use of N-glycans and hs-CRP in discriminating individuals with damaging HNF1A alleles from those without HNF1A variants by identifying 29 individuals harboring 25 rare HNF1A alleles, of which 3 were novel and 12 were considered pathogenic. Antennary fucosylated N-glycans and hs-CRP were able to differentiate subjects with damaging HNF1A alleles from those without rare HNF1A alleles. N-Glycan release, labeling, and cleanup were performed as described previously by Trbojević-Akmačić et al. in 2015.73 Fluorescently labeled glycans were separated by HILIC-UHPLC-FLD into 42 chromatographic peaks, which enabled reliable quantification.
6.5.3. Breast Cancer
Breast cancer is currently the most prevalent cancer worldwide and the most dominant cause of cancer-related deaths in women.401 Early detection and diagnosis significantly improve the survival rate. However, currently used diagnostic markers are not fairly specific and sensitive, calling for more reliable biomarker identification. Glycosylation changes of serum proteins and tumor tissue have been associated with breast cancer progression having potential as new prominent diagnostic biomarkers.402,403
In 2016, Ju et al. reported five difucosylated N-glycans as potential biomarkers in initial breast cancer and recurrent breast cancer patients.404 They analyzed free permethylated N-glycans from serum glycoproteins in 134 samples, including 91 breast cancer sera and 43 healthy controls using linear ion-trap quadrupole (LTQ)-ESI-MS and observed significant changes in N-glycosylation between healthy controls, initial diagnostic patients, and recurrent patients. By comparing breast cancer patients and controls, a positive correlation was found between increased difucosylation and disease progression. Also, increased difucosylated N-glycans, containing both core and antennary fucose, could be a more reliable indicator of recurrent breast cancer because it was proved to outperform currently used breast cancer biomarkers, such as carcinoembryonic antigen (CEA), CA 15–3, and CA 125.
Recently, Terkelsen et al. have reported N-glycan signatures from tumor interstitial fluid (TIF, n = 85), paired normal interstitial fluids (NIF, n = 54), and serum of breast cancer patients (n = 28) as well as their association with clinical outcomes.405N-Glycans from the samples were released using a HT automated method by a liquid-handling robot (2-AB labeling) and analyzed by HILIC-UHPLC-FLR. An increase in the expression levels of nine N-glycans were reported, containing bisecting N-acetylglucosamine contributing to tumor suppression in NIF. Furthermore, levels of five N-glycans in TIF correlated with that in paired serum. To sum up, these results imply that profiling of N-glycans from breast tumor fluids is a promising biomarker for detection of tumor-derived glycan-signatures in the blood and may improve the diagnostic and prognostic stratification of patients with breast cancer.
Another recent age-matched case-control study explored the biomarker potential of total serum N-glycome for the detection of breast cancer. Analysis of total serum protein N-glycome by MALDI-FTICR-MS from 145 breast cancer patients and 171 healthy individuals showed that serum N-glycome differs between various cancer subtypes. Of note, some global total serum protein N-glycomic signatures that had previously been reported between breast cancer patients and controls were not replicated in the current cohort, implicating the effect of the heterogeneous character of the disease on N-glycome profiles.406
Heavily O-glycosylated mucin 1 is a recognized biomarker for the diagnosis of breast cancer.407,408 It has been known that the alterations of mucin O-glycans occur in the mammary gland during malignancy,408 and developing comprehensive O-glycan profiling workflows is an alternative strategy for the breast cancer biomarker discovery. Kirmiz et al., used the MALDI-FTICR-MS-based approach to analyze O-glycans extracted from breast cancer cell line supernatant, serum of a breast cancer mouse model, and human breast cancer patient samples.409 Observed differences in O-glycan profiles were sufficient to distinguish the patients with breast cancer from those without cancer by PCA, although the findings were tentative due to the small number of samples involved in the study. Also, similar glycan patterns were observed as in serum from ovarian cancer patients.410
6.5.4. Intact Analysis of Transferrin
Human Tf is a well-known biomarker of congenital disorders of glycosylation (CDGs),411−413 and the diagnosis is mainly based on the glycoform pattern observed for intact Tf by isoelectric focusing (IEF).414 However, using this method, it is difficult to discriminate between CDG-I and CDG-II and it often leads to false-negative results.415 Therefore, for fast and high sensitivity profiling and accurate characterization of heterogeneous glycan structures, high-performance separation techniques coupled to MS are used.416,417 Tegtmeyer et al., reported that using MS to analyze intact Tf in some patients results in a combination of Tf isoforms with a lack of glycans and abnormal glycan structures lacking galactose.418 Abu et al. analyzed plasma Tf from 19 CDG samples by high-resolution qTOF-MS, covering a broad range of biochemical and clinical severity.419 Also, to define normal Tf glycosylation, they analyzed 20 healthy volunteers by Tf-IEF and qTOF-MS. Total plasma glycoprofiling showed a decrease in galactosylation and sialylation in most phosphoglucomutase-1 (PGM1)-CDG patients, while fucosylation and high mannose glycans were increased. Also, using the qTOF-MS Tf glycoprofiling approach, Tf was revealed to be a highly sensitive and specific glycomarker for the diagnosis, as well as the primary test for PGM1-CDG.
More recently, Hipgrave et al., studied glycan profiles for Tf and IgG Fc from CDG patients and healthy controls using ESI-MS of intact proteins and (LC)-MS of tryptic glycopeptides.420 They reported lower levels of sialylated structures on plasma proteins as compared to healthy controls due to defects in proteins involved in Golgi trafficking and cytidine monophosphate (CMP)-sialic acid transport. Also, using MALDI-TOF-MS, they differentiated sialic acid linkage isomers via derivatization and reported unprecedented sialic linkage-specific effects, specifically, α2,3-sialylation might feature as a potential marker in future investigations.
7. Future Perspectives
Hitherto, HT glycomics is focused on N-glycan analysis and largely ignores other types of glycosylation. This is mostly due to a lack of universal enzymes that enable easy and HT workflows, as is the case for HT N-glycomics analyses, which are often accomplished at the PNGase F released N-glycan level. In the coming years, these relatively broadly disseminated HT N-glycomics analyses will expectedly benefit from the implementation of standards, both for identification and quantification purposes. Chemoenzymatic synthesis of N-glycans is rapidly evolving, including the development of stable isotope-labeled glycan standards.421−423 Implementation of glycan standards at various levels of the samples preparation and measurement workflows will not only allow highly accurate quantification but will also vastly improve the robustness of glycomic workflows, paving the way for the dissemination of glycoanalyses into nonglyco-specialist and routine laboratories. MS glycomic assays will particularly benefit from the broad use of standards, which will accelerate the implementation of glycomic assays in clinical diagnostics applications as well as the characterization of glycoprotein drugs in the biopharmaceutical industry.
Next to HT N-glycomics, the analysis of HMOs increasingly receives attention, and large numbers of samples are being analyzed at increasing depth, using a diverse range of methodologies, including HILIC-UHPLC-FLD, CE-LIF, and MS.383,424−427 HMO analysis contributes to deciphering the role and repertoire of bioactive carbohydrates and glycan-based neutraceuticals with potential health benefits, adding to our increasing insights into the link between metabolic disorders, glycome, and immune status.319,428
Regarding non-MS-based HT glycomics applications, relatively cheap, long-established labels such as 2-AB and APTS are still dominating. However, new labels with advantageous chromatographic and fluorescence, as well as MS detection, properties will increasingly be implemented. Also, the development of RP-UHPLC applications would allow HT-glycomics applications to be implemented on common nano-LC-high resolution-MS/MS platforms, which are broadly available in MS proteomics laboratories and will allow high-sensitivity detection of labeled glycans in HT mode. Expectedly, RP applications will gain ground relative to the currently dominating HILIC applications.
MS analysis of glycopeptides is rapidly advancing in HT glycomics applications, as it often allows protein- and site-specific glycosylation profiling. Similar to what was mentioned above, stable isotope-labeled glycopeptide standards will be very valuable for the smooth, further development and dissemination of these assays. Currently, HT glycopeptide analysis assays cover typically only a few proteins and not more than a dozen different peptide portions, with several hundred glycopeptide species quantified in an LC-MS run. With MS allowing multiplexed detection of complex glycopeptide mixtures featuring both profound peptide and glycan heterogeneity, the coverage of hundreds and potentially thousands of N-glycosylation sites should be feasible regarding analyte detection by LC-MS, and such analyses have repeatedly been performed in low-throughput mode. For HT glycopeptide-based analyses of complex mixtures, at the above-mentioned depth, robust HT sample preparation workflows will be needed, together with powerful tools for glycopeptide assignment and quantification from the complex LC-MS data, as well as suitable postprocessing data analysis strategies.
Intact glycoprotein analysis is another, very promising mode for HT protein glycosylation analysis, and while this approach has historically not been pursued a lot, it has vast potential. Importantly, in the biopharmaceutical industry, intact protein analysis is increasingly being implemented as a drug release assay, highlighting the power of these approaches. Similarly, intact glycoprotein analysis has already found its way into clinical diagnostics, showcasing its suitability for various types of applications. Strikingly, sample preparation workflows are often relatively simple, with a single affinity purification step sufficing, and increased implementation of such assays for HT glycoproteomics applications in clinical diagnostics and biopharma is foreseen.
Of note, both glycopeptide and intact glycoprotein-based HT glycomics workflows are amenable for the characterization of both N- and O-glycosylated proteins as demonstrated for human polyclonal IgG and IgA19 and for apoCIIIa analysis.322 While innovation on O-glycan release methods is continuing, the glycopeptide- and intact glycoprotein-based MS approaches will expectedly be the major drivers of HT O-glycomics in the coming years. Other fields, such as glycolipidomics and GAG analysis are still lagging behind concerning the development and implementation of HT glycomics applications.
MS-based HT glycosylation profiling methods can benefit from the implementation of ion mobility at the intact analyte level, i.e., right after analyte ionization.201 This extra dimension in glycan or glycopeptide analysis is beneficial in cases where isomer separation is achieved. Ion mobility is a promising addition for both MALDI and ESI ionization modes and is particularly paying off in combination with TOF analyzers.
Tissues and cell types differ wildly in their glycosylation repertoire, and only a very limited portion of this variety has hitherto been mapped. MSI has already for several years reached the level of spatial resolution that allows analysis of N-glycomic diversity in tissues at the cellular level.429−435 Approaches to assess O-glycans, glycolipids, and GAGs are still lagging behind, largely due to limitations in on-tissue sample preparation, with the lack of broad-specificity enzymes presenting a major bottleneck.436 Regarding on-tissue N-glycomics, recent improvements in ionization (MALDI-2) help to perform in-depth N-glycomic analysis with a decent dynamic range and spatial resolution.201 The implementation of specific derivatization techniques176,437 as well as ion mobility separation, provide big steps forward toward resolving glycan isomers, which is key for assessing the glycomic status of a tissue or cell type. Implementing these recent improvements in N-glycan MSI for the analysis of large tissue microarrays will bring HT N-glycomics to the next level and provide unprecedented insights into the glycobiology of tumor development as well as mucosal host–microbe and host–pathogen interactions.
For the years to come, glycomics and glycoproteomics analyses will come with making choices along the axes of throughput, analytical depth, and sensitivity. Future developments are sorely needed to further increase the speed, ease, and throughput of glycomics analyses for supporting dissemination into less specialized laboratories, including clinical diagnostics and various biopharma applications. At the same time, an increase in analytical depth is pursued, moving toward full structural definition of analytes with regard to glycan structure, glycosylation site, and contribution of glycosylation to the overall structural and functional proteoform repertoire. Additional depth is sought by covering thousands of glycosylation sites at the glycopeptide level,438 which is currently still at the cost of full structural elucidation of glycans as well as throughput. Highly promising are the high-sensitivity glycomics workflows, often relying on established proteomics and genomics hardware and platforms such as high-end nano-LC-MS/MS that can serve proteomics and glycomics purposes alike, as well as DNA analyzers for CE-LIF glycan analysis at utmost sensitivity and throughput. In fact, implementation of HT glycomics on HT platforms designed for other omics disciplines such as genomics and proteomics has proven a particularly successful approach, and further convergence and integration of HT omics approaches is envisioned.
Acknowledgments
This work has been supported by funding from the European Structural and Investments Funds for the projects Croatian National Centre of Research Excellence in Personalized Healthcare (contract no. KK.01.1.1.01.0010), Croatian National Centre of Competence in Molecular Diagnostics (contract no. KK.01.2.2.03.0006), and Development of personalized diagnostic tool for prevention and treatment of cardiometabolic disorders—CardioMetabolic (contract no. KK.01.2.1.02.0321).
Glossary
Abbreviations
- %area
relative percentage area
- %rPHP
relative peak height proportions
- 2-AA
2-aminobenzoic acid
- 2-AB
2-aminobenzamide
- 2-ANTS
2-amino-1-naphthalenesulfonic acid
- 2-PA
2-aminopyridine
- 2D
two-dimensional
- 3D
three-dimensional
- ADCC
antibody-dependent cellular cytotoxicity
- ADHD
attention-deficit hyperactivity disorder
- AFP
α-fetoprotein
- AGP
α-1-acid glycoprotein
- APRI
aspartate aminotransferase to platelet ratio index
- APTS
8-amino-1,3,6-pyrenetrisulfonic acid trisodium salt
- AQC
6-aminoquinolyl-N-hydroxysuccinimidyl carbamate
- ART
antiretroviral therapy
- BMI
body mass index
- BPH
benign prostate hyperplasia
- CD
Crohn’s disease
- CDC
complement-dependent cytotoxicity
- CDG
congenital disorder of glycosylation
- CE
capillary electrophoresis
- CEA
carcinoembryonic antigen
- CGE
capillary gel electrophoresis
- CMP
cytidine monophosphate
- COVID-19
coronavirus disease 2019
- CRC
colorectal cancer
- CVD
cardiovascular disease
- CZE
capillary zone electrophoresis
- DEN
dopant-enriched nitrogen
- DHB
2,5-dihydroxybenzoic acid
- DMSO
dimethyl sulfoxide
- DSA-FACE
DNA sequencer-aided fluorophore-assisted carbohydrate electrophoresis
- ECM
extracellular matrix
- EIC
extracted ion chromatogram
- ELISA
enzyme-linked immunoassay
- ESI
electrospray ionization
- Fab
antigen-binding fragment
- Fc
fragment crystallizable
- FIB-4
fibrosis index based on the four factors
- FLD
fluorescent detector
- Fru
fructose
- FTICR
Fourier transform ion cyclotron resonance
- GAGs
glycosaminoglycans
- Gal
galactose
- GHP
hydrophilic polypropylene, GH Polypro
- Glc
glucose
- GLP-1
glucagon-like peptide-1
- GSLs
glycosphingolipids
- GU
glucose unit
- GWAS
genome-wide association study
- HBV
hepatitis B virus
- HCA
hierarchical cluster analysis
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HILIC
hydrophilic-interaction liquid chromatography
- HIV
human immunodeficiency virus
- HMOs
human milk oligosaccharides
- HNF1A
hepatocyte nuclear factor 1α
- HPLC
high-performance liquid chromatography
- hs-CRP
high-sensitivity C-reactive protein
- HT
high-throughput
- IBD
inflammatory bowel disease
- IEF
isoelectric focusing
- IgA
immunoglobulin A
- IgG
immunoglobulin G
- LC
liquid chromatography
- LIF
laser-induced fluorescence
- LTQ
linear ion-trap mass spectrometer
- mAbs
monoclonal antibodies
- MALDI
matrix assisted laser desorption/ionization
- MIRAGE
Minimum Information Required for a Glycomics Experiment
- MODY
maturity-onset diabetes of the young
- MS
mass spectrometry
- MSI
mass spectrometry imaging
- MTU
migration time unit
- MTU′′
double migration time alignment
- NFIs
normalized fluorescent intensities
- NicAc
nicotinic acid
- NIF
normal interstitial fluids
- NHS
N-hydroxysuccinimide
- NMR
nuclear magnetic resonance
- NP
normal phase
- OA
osteoarthritis
- PCA
principal component analysis
- PCa
prostate cancer
- PGC
porous graphitic carbon
- PGM1
phosphoglucomutase-1
- PNGase F
peptide-N-glycosidase F
- ProA
procainamide
- PSA
prostate-specific antigen
- PVDF
polyvinylidene fluoride
- RA
rheumatoid arthritis
- RMS
root-means-squared
- RP
reversed-phase
- rt-PA
recombinant tissue-type plasminogen activator
- S/N
signal-to-noise
- S index
glutamyltranspeptidase platelet albumin index
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SLE
systemic lupus erythematosus
- SNP
single nucleotide polymorphism
- SPE
solid-phase extraction
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TAG
toolbox accelerating glycomics
- Tf
transferrin
- TIC
total ion current/count
- TIF
tumor interstitial fluid
- TOF
time-of-flight
- Trp
tryptophan
- TSA-ALM
tyramide signal amplification for antibody-overlay lectin microarray
- UC
ulcerative colitis
- UGM
urinary glycoprofile marker
- UHPLC
ultrahigh-performance liquid chromatography
- UV
ultraviolet
- UVLD
ultraviolet laser desorption
- WAX
weak anion exchange
Biographies
Irena Trbojević-Akmačić (1988) received her Ph.D. in Biochemistry in 2015 on high-throughput IgG glycosylation analysis by UHPLC in inflammatory bowel disease. She was appointed Deputy Head of Genos Glycoscience laboratory in 2015, Acting Head of Genos Glycoscience laboratory in 2016, and Head of the UPLC laboratory for glycan analysis in 2018. She currently works as Head of Laboratory for high-throughput glycomics and develops high-throughput methods for targeted glycoprotein purification and glycosylation analysis in large population, epidemiological and clinical studies.
Guinevere S. M. Lageveen-Kammeijer (1989) received her Ph.D. on exploring prostate-specific antigen, the well-known biomarker for prostate cancer, and its glycosylation by capillary electrophoresis and mass spectrometry. Currently, Guinevere performs her postdoctoral research at the Center for Proteomics and Metabolomics at the Leiden University Medical Center, in the group of Prof. Manfred Wuhrer. She currently works on further expanding a mass spectrometry-based prostate-specific antigen glycosylation assay, which she developed during her Ph.D. In addition, she explores the possibilities for the in-depth analysis of glycans and intact glycoproteins for biomarker discovery for other diseases as well as for the characterization of biopharmaceuticals.
Bram Heijs (1986) graduated in Life Science & Technology at the Faculty of Science of Leiden University (2013), focusing on biochemistry and analytical chemistry. During his Ph.D. (2018) at the Center for Proteomics and Metabolomics of the Leiden University Medical Center, he worked in close collaboration with the Pathology Department on developing, improving and applying MALDI-MS imaging methods for the proteomic and glycomic analysis of soft-tissue sarcomas. In 2019–2020, he worked as a postdoctoral researcher at the University Hospital in Münster (Germany) under the supervision of Prof. Klaus Dreisewerd and in close collaboration with Bruker Daltonics on the development of the timsTOF fleX MALDI-2 mass spectrometry platform. He now is principal investigator of MS imaging at the LUMC Center for Proteomics and Metabolomics, continuing the development and application of MS imaging methods for high-resolution multi-OMICs analysis in clinical tissues.
Tea Petrović (1992) graduated in Molecular Biology at the University of Zagreb, Faculty of Science, Department of Biology. She worked on her Master’s Thesis named “Effect of body mass index on in vitro fertilization outcomes in women” at the Clinic for Gynecology and Obstetrics, University Hospital Center, Zagreb. From April 2018, she is employed at Glycoscience Research Laboratory, Genos Ltd, Zagreb, Croatia, as a Laboratory analyst. Tea is also a Ph.D. student at the University of Zagreb, Faculty of Science, Department of Biology. Her current work is focused on protein glycosylation analysis using ultrahigh-performance liquid chromatography with a focus on IgG and plasma glycans in COVID-19.
Helena Deriš (1991) graduated Pharmacy—Research and Development of new drugs at the University of Zagreb, Faculty of Pharmacy and Biochemistry. During her studies, she worked on transferrin chemistry and glycosylation. She received University of Zagreb Rector’s award for her work on the spectroelectrochemical determination of the redox potential of holo-transferrin. Prior to graduating, she spent some time at Roche Diabetes Care’s Competitive Intelligence Department in Mannheim as an intern. After an internship in a public and clinical pharmacy, she started working at Genos in June 2018 as a Laboratory analyst. Her current work is focused on protein glycosylation analysis using UHPLC. Helena is also a Ph.D. student at the University of Zagreb, Faculty of Science, Department of Chemistry.
Manfred Wuhrer (1971) studied Biochemistry in Regensburg. He entered the field of glycosylation analysis during his Ph.D. (1999) and postdoc with Prof. Rudolf Geyer at Giessen University, Germany. In 2003, he joined the Leiden University Medical Center in The Netherlands. From 2013 to 2015, he worked in Amsterdam at the Vrije Universiteit as Professor of Analytics of Biomolecular Interactions. Since 2015, he is head of the Center for Proteomics and Metabolomics of the LUMC and Professor of Proteomics and Glycomics. He develops mass spectrometric methods for glycomics and glycoproteomics and applies them in biomedical research.
Gordan Lauc (1970) graduated in Molecular Biology in 1992 and got a Ph.D. in Biochemistry in 1995 at the University of Zagreb. Currently, he is a Professor of Biochemistry and Molecular Biology at the University of Zagreb, Director of the National Centre of Scientific Excellence in Personalised Healthcare, Honorary professor at the Kings College London, and member of the Johns Hopkins Society of Scholars. He is publishing in the field of glycobiology since 1991. In 2017, he initiated the launch of the Human Glycome Project and is one of its two codirectors. His research team is pioneering high-throughput glycomic analysis and the application of glycan biomarkers in the field of precision medicine. By combining glycomic data with extensive genetic, epigenetic, biochemical, and physiological data in a systems biology approach, they are trying to understand the role of glycans in normal physiology and disease. Professor Lauc coauthored over 200 research articles that are cited over 7000 times. In 2007, he founded Genos, a biotech company that is currently global leader in high-throughput glycomics. Research in Genos led to the development of the GlycanAge test of biological age.
The authors declare the following competing financial interest(s): GL is the founder and owner of Genos Ltd, a private research organization that specializes in high-throughput glycomic analysis and has several patents in this field. IT-A, TP, and HD are employees of Genos Ltd.
References
- Flynn R. A.; Pedram K.; Malaker S. A.; Batista P. J.; Smith B. A. H.; Johnson A. G.; George B. M.; Majzoub K.; Villalta P. W.; Carette J. E.; et al. Small RNAs Are Modified with N-Glycans and Displayed on the Surface of Living Cells. Cell 2021, 184, 3109–3124. 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristic J.; Zaytseva O. O.; Ram R.; Nguyen Q.; Novokmet M.; Vuckovic F.; Vilaj M.; Trbojevic-Akmacic I.; Pezer M.; Davern K. M.; et al. Profiling and Genetic Control of the Murine Immunoglobulin G Glycome. Nat. Chem. Biol. 2018, 14, 516–524. 10.1038/s41589-018-0034-3. [DOI] [PubMed] [Google Scholar]
- Seeling M.; Bruckner C.; Nimmerjahn F. Differential Antibody Glycosylation in Autoimmunity: Sweet Biomarker or Modulator of Disease Activity?. Nat. Rev. Rheumatol. 2017, 13, 621–630. 10.1038/nrrheum.2017.146. [DOI] [PubMed] [Google Scholar]
- Gudelj I.; Lauc G.; Pezer M. Immunoglobulin G Glycosylation in Aging and Diseases. Cell Immunol. 2018, 333, 65–79. 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Shields R. L.; Lai J.; Keck R.; O’Connell L. Y.; Hong K.; Meng Y. G.; Weikert S. H.; Presta L. G. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human Fcgamma RIII and Antibody-Dependent Cellular Toxicity. J. Biol. Chem. 2002, 277, 26733–26740. 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- Kuhns S.; Shu J.; Xiang C.; Guzman R.; Zhang Q.; Bretzlaff W.; Miscalichi N.; Kalenian K.; Joubert M. Differential Influence on Antibody Dependent Cellular Phagocytosis by Different Glycoforms on Therapeutic Monoclonal Antibodies. J. Biotechnol. 2020, 317, 5–15. 10.1016/j.jbiotec.2020.04.017. [DOI] [PubMed] [Google Scholar]
- van Osch T. L. J.; Nouta J.; Derksen N. I. L.; van Mierlo G.; van der Schoot C. E.; Wuhrer M.; Rispens T.; Vidarsson G. Fc Galactosylation Promotes Hexamerization of Human IgG1, Leading to Enhanced Classical Complement Activation. J. Immunol. 2021, 207, 1545–1554. 10.4049/jimmunol.2100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangeli R.; Palinsky W.; Bierau H. Glycoengineered Antibodies: Towards the Next-Generation of Immunotherapeutics. Glycobiology 2019, 29, 199–210. 10.1093/glycob/cwy092. [DOI] [PubMed] [Google Scholar]
- Stambuk T.; Klasic M.; Zoldos V.; Lauc G. N-Glycans as Functional Effectors of Genetic and Epigenetic Disease Risk. Mol. Aspects Med. 2021, 79, 100891. 10.1016/j.mam.2020.100891. [DOI] [PubMed] [Google Scholar]
- Parekh R. B.; Dwek R. A.; Sutton B. J.; Fernandes D. L.; Leung A.; Stanworth D.; Rademacher T. W.; Mizuochi T.; Taniguchi T.; Matsuta K.; et al. Association of Rheumatoid Arthritis and Primary Osteoarthritis with Changes in the Glycosylation Pattern of Total Serum IgG. Nature 1985, 316, 452–457. 10.1038/316452a0. [DOI] [PubMed] [Google Scholar]
- Rombouts Y.; Ewing E.; van de Stadt L. A.; Selman M. H.; Trouw L. A.; Deelder A. M.; Huizinga T. W.; Wuhrer M.; van Schaardenburg D.; Toes R. E.; et al. Anti-Citrullinated Protein Antibodies Acquire a Pro-Inflammatory Fc Glycosylation Phenotype Prior to the Onset of Rheumatoid Arthritis. Ann. Rheum. Dis. 2015, 74, 234–241. 10.1136/annrheumdis-2013-203565. [DOI] [PubMed] [Google Scholar]
- Dashti H.; Pabon Porras M. A.; Mora S. Glycosylation and Cardiovascular Diseases. Adv. Exp. Med. Biol. 2021, 1325, 307–319. 10.1007/978-3-030-70115-4_15. [DOI] [PubMed] [Google Scholar]
- Cindrić A.; Krištić J.; Martinić Kavur M.; Pezer M.. Glycosylation and Aging. In The Role of Glycosylation in Health and Disease; Lauc G., Trbojević-Akmačić I., Eds.; Springer International, 2021; pp 341–373. [DOI] [PubMed] [Google Scholar]
- Gabius H. J.; Andre S.; Kaltner H.; Siebert H. C. The Sugar Code: Functional Lectinomics. Biochim. Biophys. Acta 2002, 1572, 165–177. 10.1016/S0304-4165(02)00306-9. [DOI] [PubMed] [Google Scholar]
- Totten S. M.; Wu L. D.; Parker E. A.; Davis J. C. C.; Hua S.; Stroble C.; Ruhaak L. R.; Smilowitz J. T.; German J. B.; Lebrilla C. B. Rapid-Throughput Glycomics Applied to Human Milk Oligosaccharide Profiling for Large Human Studies. Anal. Bioanal. Chem. 2014, 406, 7925–7935. 10.1007/s00216-014-8261-2. [DOI] [PubMed] [Google Scholar]
- Porfirio S.; Archer-Hartmann S.; Moreau G. B.; Ramakrishnan G.; Haque R.; Kirkpatrick B. D.; Petri W. A. Jr.; Azadi P. New Strategies for Profiling and Characterization of Human Milk Oligosaccharides. Glycobiology 2020, 30, 774–786. 10.1093/glycob/cwaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer F.; Jarvas G.; Guttman A. Recent Advances in the Analysis of Human Milk Oligosaccharides by Liquid Phase Separation Methods. J. Chromatogr. B 2021, 1162, 122497. 10.1016/j.jchromb.2020.122497. [DOI] [PubMed] [Google Scholar]
- Nicolardi S.; van der Burgt Y. E. M.; Wuhrer M.; Deelder A. M. Mapping O-Glycosylation of Apolipoprotein C-III in MALDI-FT-ICR Protein Profiles. Proteomics 2013, 13, 992–1001. 10.1002/pmic.201200293. [DOI] [PubMed] [Google Scholar]
- Momčilović A.; de Haan N.; Hipgrave Ederveen A. L.; Bondt A.; Koeleman C. A. M.; Falck D.; de Neef L. A.; Mesker W. E.; Tollenaar R.; de Ru A.; et al. Simultaneous Immunoglobulin A and G Glycopeptide Profiling for High-Throughput Applications. Anal. Chem. 2020, 92, 4518–4526. 10.1021/acs.analchem.9b05722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keser T.; Tijardović M.; Gornik I.; Lukić E.; Lauc G.; Gornik O.; Novokmet M. High-Throughput and Site-Specific N-Glycosylation Analysis of Human Alpha-1-Acid Glycoprotein Offers a Great Potential for New Biomarker Discovery. Mol. Cell Proteomics 2021, 20, 100044. 10.1074/mcp.RA120.002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. F.; Shiao C. Y.; Wu M. Y.; Lin Y. C.; Chen H. H.; Chang W. C.; Wu M. S.; Kao C. C.; Tsai I. L. Quantitative Determination of Human IgA Subclasses and Their Fc-Glycosylation Patterns in Plasma by Using a Peptide Analogue Internal Standard and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8606 10.1002/rcm.8606. [DOI] [PubMed] [Google Scholar]
- Wang W.; Kałuża A.; Nouta J.; Nicolardi S.; Ferens-Sieczkowska M.; Wuhrer M.; Lageveen-Kammeijer G. S.; de Haan N. High-Throughput Glycopeptide Profiling of Prostate-Specific Antigen from Seminal Plasma by MALDI-MS. Talanta 2021, 222, 121495. 10.1016/j.talanta.2020.121495. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Madunić K.; Holst S.; Zhang J.; Jin C.; Ten Dijke P.; Karlsson N. G.; Stavenhagen K.; Wuhrer M. Development of a 96-Well Plate Sample Preparation Method for Integrated N- and O-Glycomics Using Porous Graphitized Carbon Liquid Chromatography-Mass Spectrometry. Mol. Omics 2020, 16, 355–363. 10.1039/C9MO00180H. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Narimatsu Y.; Koed Møller Aasted M.; Larsen I. S. B.; Marinova I. N.; Dabelsteen S.; Vakhrushev S. Y.; Wandall H. H. In-Depth Profiling of O-Glycan Isomers in Human Cells Using C18 Nanoliquid Chromatography-Mass Spectrometry and Glycogenomics. Anal. Chem. 2022, 94, 4343–4351. 10.1021/acs.analchem.1c05068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz T.; Wuhrer M.; de Haan N. Expanding the Reaction Space of Linkage-Specific Sialic Acid Derivatization. Molecules 2019, 24, 3617. 10.3390/molecules24193617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz T.; Verhoeven A.; Wuhrer M.; de Haan N. The Structure and Role of Lactone Intermediates in Linkage-Specific Sialic Acid Derivatization Reactions. Glycoconj. J. 2021, 38, 157–166. 10.1007/s10719-020-09971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic A.; Polasek O.; Gornik O.; Rudan I.; Campbell H.; Hayward C.; Wright A.; Kolcic I.; O’Donoghue N.; Bones J.; et al. Variability, Heritability and Environmental Determinants of Human Plasma N-Glycome. J. Proteome Res. 2009, 8, 694–701. 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- Knezevic A.; Gornik O.; Polasek O.; Pucic M.; Redzic I.; Novokmet M.; Rudd P. M.; Wright A. F.; Campbell H.; Rudan I.; et al. Effects of Aging, Body Mass Index, Plasma Lipid Profiles, and Smoking on Human Plasma N-Glycans. Glycobiology 2010, 20, 959–969. 10.1093/glycob/cwq051. [DOI] [PubMed] [Google Scholar]
- Štambuk J.; Nakić N.; Vučković F.; Pučić-Baković M.; Razdorov G.; Trbojević-Akmačić I.; Novokmet M.; Keser T.; Vilaj M.; Štambuk T.; et al. Global Variability of the Human IgG Glycome. Aging (Albany NY) 2020, 12, 15222–15259. 10.18632/aging.103884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur J. A.; Goetz J. A.; Alley W. R. Jr.; Novotny M. V. Smoking and Lung Cancer-Induced Changes in N-Glycosylation of Blood Serum Proteins. Glycobiology 2012, 22, 1684–1708. 10.1093/glycob/cws108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. S.; Naber A.; Dotz V.; Schoep E.; Memarian E.; Slieker R. C.; Elders P. J. M.; Vreeker G.; Nicolardi S.; Wuhrer M.; et al. Metformin and Statin Use Associate with Plasma Protein N-Glycosylation in People with Type 2 Diabetes. BMJ Open Diabetes Res. Care 2020, 8, e001230 10.1136/bmjdrc-2020-001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C.; Saldova R.; Wormald M. R.; Rudd P. M.; McElvaney N. G.; Reeves E. P. The Role and Importance of Glycosylation of Acute Phase Proteins with Focus on Alpha-1 Antitrypsin in Acute and Chronic Inflammatory Conditions. J. Proteome Res. 2014, 13, 3131–3143. 10.1021/pr500146y. [DOI] [PubMed] [Google Scholar]
- Jansen B. C.; Bondt A.; Reiding K. R.; Lonardi E.; de Jong C. J.; Falck D.; Kammeijer G. S. M.; Dolhain R. J. E. M.; Rombouts Y.; Wuhrer M. Pregnancy-Associated Serum N-Glycome Changes Studied by High-Throughput MALDI-TOF-MS. Sci. Rep. 2016, 6, 23296. 10.1038/srep23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D.; Bern M.; Parry S.; Sutton-Smith M.; Panico M.; Morris H. R.; Dell A. Automated N-Glycopeptide Identification Using a Combination of Single- and Tandem-MS. J. Proteome Res. 2007, 6, 3995–4005. 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- Brito A. E.; Kletter D.; Singhal M.; Bern M. Benchmark Study of Automatic Annotation of MALDI-TOF N-Glycan Profiles. J. Proteomics 2015, 129, 71–77. 10.1016/j.jprot.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam F. W.; Takahashi N. Structural Characterization of Glycoproteins. J. Chromatogr. 1988, 443, 267–284. 10.1016/S0021-9673(00)94799-X. [DOI] [PubMed] [Google Scholar]
- Mullinax F. Molecular Site and Enzymatic Origin of IgG Galactose Deficiency in Rheumatoid Arthritis and SLE. Arthritis Rheum. 1976, 19, 813. [Google Scholar]
- Parekh R.; Roitt I.; Isenberg D.; Dwek R.; Rademacher T. Age-Related Galactosylation of the N-Linked Oligosaccharides of Human Serum IgG. J. Exp. Med. 1988, 167, 1731–1736. 10.1084/jem.167.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford D.; Dwek R. A.; Welply J. K.; Amatayakul S.; Homans S. W.; Lis H.; Taylor G. N.; Sharon N.; Rademacher T. W. The Β1 → 2-D-Xylose and Α1 → 3-L-Fucose Substituted N-Linked Oligosaccharides from Erythrina Cristagalli Lectin. Eur. J. Biochem. 1987, 166, 311–320. 10.1111/j.1432-1033.1987.tb13516.x. [DOI] [PubMed] [Google Scholar]
- Takasaki S.; Mizuochi T.; Kobata A. Hydrazinolysis of Asparagine-Linked Sugar Chains to Produce Free Oligosaccharides. Methods Enzymol. 1982, 83, 263–268. 10.1016/0076-6879(82)83019-X. [DOI] [PubMed] [Google Scholar]
- Takasaki S.; Kobata A. Microdetermination of Individual Neutral and Amino Sugars and N-Acetylneuraminic Acid in Complex Saccharides. J. Biochem. 1974, 76, 783–789. 10.1093/oxfordjournals.jbchem.a130624. [DOI] [PubMed] [Google Scholar]
- Hase S.; Ikenaka T.; Matsushima Y. Analyses of Oligosaccharides by Tagging the Reducing End with a Fluorescent Compound. I. Application to Glycoproteins. J. Biochem. 1979, 85, 989–994. 10.1093/oxfordjournals.jbchem.a132431. [DOI] [PubMed] [Google Scholar]
- Hase S.; Ikenaka T.; Matsushima Y. A Highly Sensitive Method for Analyses of Sugar Moieties of Glycoproteins by Fluorescence Labeling. J. Biochem. 1981, 90, 407–414. 10.1093/oxfordjournals.jbchem.a133487. [DOI] [PubMed] [Google Scholar]
- Hase S.; Sugimoto T.; Takemoto H.; Ikenaka T.; Schmid K. The Structure of Sugar Chains of Japanese Quail Ovomucoid. The Occurrence of Oligosaccharides Not Expected from the Classical Biosynthetic Pathway for N-Glycans; a Method for the Assessment of the Structure of Glycans Present in Picomolar Amounts. J. Biochem. 1986, 99, 1725–1733. 10.1093/oxfordjournals.jbchem.a135649. [DOI] [PubMed] [Google Scholar]
- Takahashi N.; Ishii I.; Ishihara H.; Mori M.; Tejima S.; Jefferis R.; Endo S.; Arata Y. Comparative Structural Study of the N-Linked Oligosaccharides of Human Normal and Pathological Immunoglobulin G. Biochemistry 1987, 26, 1137–1144. 10.1021/bi00378a023. [DOI] [PubMed] [Google Scholar]
- Plummer T. H.; Elder J. H.; Alexander S.; Phelan A. W.; Tarentino A. L. Demonstration of Peptide:N-Glycosidase F Activity in Endo-β-N-Acetylglucosaminidase F Preparations. J. Biol. Chem. 1984, 259, 10700–10704. 10.1016/S0021-9258(18)90568-5. [DOI] [PubMed] [Google Scholar]
- Plummer T. H. Jr.; Tarentino A. L. Purification of the Oligosaccharide-Cleaving Enzymes of Flavobacterium Meningosepticum. Glycobiology 1991, 1, 257–263. 10.1093/glycob/1.3.257. [DOI] [PubMed] [Google Scholar]
- Tarentino A. L.; Plummer T. H. Enzymatic Deglycosylation of Asparagine-Linked Glycans: Purification, Properties, and Specificity of Oligosaccharide-Cleaving Enzymes from Flavobacterium Meningosepticum. Methods Enzymol. 1994, 230, 44–57. 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- Karas M.; Bachmann D.; Bahr U.; Hillenkamp F. Matrix-Assisted Ultraviolet Laser Desorption of Non-Volatile Compounds. Int. J. Mass Spectrom. Ion Processes 1987, 78, 53–68. 10.1016/0168-1176(87)87041-6. [DOI] [Google Scholar]
- Harvey D. J. Quantitative Aspects of the Matrix-Assisted Laser Desorption Mass Spectrometry of Complex Oligosaccharides. Rapid Commun. Mass Spectrom. 1993, 7, 614–619. 10.1002/rcm.1290070712. [DOI] [PubMed] [Google Scholar]
- Rudd P. M.; Gulle G. R.; Küster B.; Harvey D. J.; Opdenakker G.; Dwek R. A. Oligosaccharide Sequencing Technology. Nature 1997, 388, 205–207. 10.1038/40677. [DOI] [PubMed] [Google Scholar]
- Kuster B.; Wheeler S. F.; Hunter A. P.; Dwek R. A.; Harvey D. J. Sequencing of N-Linked Oligosaccharides Directly from Protein Gels: In-Gel Deglycosylation Followed by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry and Normal-Phase High-Performance Liquid Chromatography. Anal. Biochem. 1997, 250, 82–101. 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- Royle L.; Campbell M. P.; Radcliffe C. M.; White D. M.; Harvey D. J.; Abrahams J. L.; Kim Y. G.; Henry G. W.; Shadick N. A.; Weinblatt M. E.; et al. HPLC-Based Analysis of Serum N-Glycans on a 96-Well Plate Platform with Dedicated Database Software. Anal. Biochem. 2008, 376, 1–12. 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Hsieh F.; Keshishian H.; Muir C. Automated High Throughput Multiple Target Screening of Molecular Libraries by Microfluidic MALDI-TOF MS. J. Biomol. Screen 1998, 3, 189–198. 10.1177/108705719800300305. [DOI] [Google Scholar]
- Matsudaira P. Sequence from Picomole Quantities of Proteins Electroblotted onto Polyvinylidene Difluoride Membranes. J. Biol. Chem. 1987, 262, 10035–10038. 10.1016/S0021-9258(18)61070-1. [DOI] [PubMed] [Google Scholar]
- Weitzhandler M.; Hardy M.; Co M. S.; Avdalovic N. Analysis of Carbohydrates on IgG Preparations. J. Pharm. Sci. 1994, 83, 1670–1675. 10.1002/jps.2600831206. [DOI] [PubMed] [Google Scholar]
- Courchesne P. L.; Patterson S. D. Manual Microcolumn Chromatography for Sample Cleanup before Mass Spectrometry. BioTechniques 1997, 22, 244–250. 10.2144/97222bm10. [DOI] [PubMed] [Google Scholar]
- Papac D. I.; Briggs J. B.; Chin E. T.; Jones A. J. S. A High-Throughput Microscale Method to Release N-Linked Oligosaccharides from Glycoproteins for Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometric Analysis. Glycobiology 1998, 8, 445–454. 10.1093/glycob/8.5.445. [DOI] [PubMed] [Google Scholar]
- Callewaert N.; Geysens S.; Molemans P.; Contreras R. Ultrasensitive Profiling and Sequencing of N-Linked Oligosaccharides Using Standard DNA-Sequencing Equipment. Glycobiology 2001, 11, 275–281. 10.1093/glycob/11.4.275. [DOI] [PubMed] [Google Scholar]
- Callewaert N.; Van Vlierberghe H.; Van Hecke A.; Laroy W.; Delanghe J.; Contreras R. Noninvasive Diagnosis of Liver Cirrhosis Using DNA Sequencer-Based Total Serum Protein Glycomics. Nat. Med. 2004, 10, 429–434. 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- Laroy W.; Contreras R.; Callewaert N. Glycome Mapping on DNA Sequencing Equipment. Nat. Protoc. 2006, 1, 397–405. 10.1038/nprot.2006.60. [DOI] [PubMed] [Google Scholar]
- Khandurina J.; Anderson A. A.; Olson N. A.; Stege J. T.; Guttman A. Large-Scale Carbohydrate Analysis by Capillary Array Electrophoresis: Part 2. Data Normalization and Quantification. Electrophoresis 2004, 25, 3122–3127. 10.1002/elps.200406048. [DOI] [PubMed] [Google Scholar]
- Khandurina J.; Olson N. A.; Anderson A. A.; Gray K. A.; Guttman A. Large-Scale Carbohydrate Analysis by Capillary Array Electrophoresis: Part 1. Separation and Scale-Up. Electrophoresis 2004, 25, 3117–3121. 10.1002/elps.200406047. [DOI] [PubMed] [Google Scholar]
- Szabo Z.; Guttman A.; Rejtar T.; Karger B. L. Improved Sample Preparation Method for Glycan Analysis of Glycoproteins by CE-LIF and CE-MS. Electrophoresis 2010, 31, 1389–1395. 10.1002/elps.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Z.; Guttman A.; Bones J.; Karger B. L. Rapid High-Resolution Characterization of Functionally Important Monoclonal Antibody N-Glycans by Capillary Electrophoresis. Anal. Chem. 2011, 83, 5329–5336. 10.1021/ac2007587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Huhn C.; Waterreus W.-J.; de Boer A. R.; Neusüss C.; Hokke C. H.; Deelder A. M.; Wuhrer M. Hydrophilic Interaction Chromatography-Based High-Throughput Sample Preparation Method for N-Glycan Analysis from Total Human Plasma Glycoproteins. Anal. Chem. 2008, 80, 6119–6126. 10.1021/ac800630x. [DOI] [PubMed] [Google Scholar]
- Bigge J. C.; Patel T. P.; Bruce J. A.; Goulding P. N.; Charles S. M.; Parekh R. B. Nonselective and Efficient Fluorescent Labeling of Glycans Using 2-Amino Benzamide and Anthranilic Acid. Anal. Biochem. 1995, 230, 229–238. 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Hennig R.; Huhn C.; Borowiak M.; Dolhain R. J.; Deelder A. M.; Rapp E.; Wuhrer M. Optimized Workflow for Preparation of APTS-Labeled N-Glycans Allowing High-Throughput Analysis of Human Plasma Glycomes Using 48-Channel Multiplexed CGE-LIF. J. Proteome Res. 2010, 9, 6655–6664. 10.1021/pr100802f. [DOI] [PubMed] [Google Scholar]
- Pucic M.; Knezevic A.; Vidic J.; Adamczyk B.; Novokmet M.; Polasek O.; Gornik O.; Supraha-Goreta S.; Wormald M. R.; Redzic I.; et al. High Throughput Isolation and Glycosylation Analysis of IgG-Variability and Heritability of the IgG Glycome in Three Isolated Human Populations. Mol. Cell Proteomics 2011, 10, M111 010090. 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krištić J.; Vučković F.; Menni C.; Klarić L.; Keser T.; Beceheli I.; Pučić-Baković M.; Novokmet M.; Mangino M.; Thaqi K.; et al. Glycans Are a Novel Biomarker of Chronological and Biological Ages. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 779–789. 10.1093/gerona/glt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauc G.; Huffman J. E.; Pučić M.; Zgaga L.; Adamczyk B.; Mužinić A.; Novokmet M.; Polašek O.; Gornik O.; Krištić J.; et al. Loci Associated with N-Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers. PLoS Genet. 2013, 9, e1003225 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnina I.; Hoyt E.; Lynaugh H.; Li H.; Gong B. A Cost-Effective Plate-Based Sample Preparation for Antibody N-Glycan Analysis. J. Chromatogr. A 2013, 1307, 201–206. 10.1016/j.chroma.2013.07.104. [DOI] [PubMed] [Google Scholar]
- Trbojevic Akmačić I.; Ugrina I.; Štambuk J.; Gudelj I.; Vučković F.; Lauc G.; Pučić-Baković M. High-Throughput Glycomics: Optimization of Sample Preparation. Biochemistry (Moscow) 2015, 80, 934–942. 10.1134/S0006297915070123. [DOI] [PubMed] [Google Scholar]
- Russell A. C.; Kepka A.; Trbojević-Akmačić I.; Ugrina I.; Song M.; Hui J.; Hunter M.; Laws S. M.; Lauc G.; Wang W. Increased Central Adiposity Is Associated with Pro-Inflammatory Immunoglobulin G N-Glycans. Immunobiology 2019, 224, 110–115. 10.1016/j.imbio.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Bermingham M. L.; Colombo M.; McGurnaghan S. J.; Blackbourn L. A. K.; Vučković F.; Pučić Baković M.; Trbojević-Akmačić I.; Lauc G.; Agakov F.; Agakova A. S.; et al. N-Glycan Profile and Kidney Disease in Type 1 Diabetes. Diabetes Care 2018, 41, 79–87. 10.2337/dc17-1042. [DOI] [PubMed] [Google Scholar]
- Wittenbecher C.; Štambuk T.; Kuxhaus O.; Rudman N.; Vučković F.; Štambuk J.; Schiborn C.; Rahelić D.; Dietrich S.; Gornik O.; et al. Plasma N-Glycans as Emerging Biomarkers of Cardiometabolic Risk: A Prospective Investigation in the EPIC-Potsdam Cohort Study. Diabetes Care 2020, 43, 661–668. 10.2337/dc19-1507. [DOI] [PubMed] [Google Scholar]
- Martin T. C.; Šimurina M.; Za̧bczyń́ska M.; Martinic Kavur M.; Rydlewska M.; Pezer M.; Kozłowska K.; Burri A.; Vilaj M.; Turek-Jabrocka R.; et al. Decreased Immunoglobulin G Core Fucosylation, a Player in Antibody-Dependent Cell-Mediated Cytotoxicity, Is Associated with Autoimmune Thyroid Diseases. Mol. Cell Proteomics 2020, 19, 774–792. 10.1074/mcp.RA119.001860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczak A.; Pavić T.; Vučković F.; Bennett A. J.; Shah N.; Pape Medvidović E.; Groves C. J.; Šekerija M.; Chandler K.; Burrows C.; et al. Plasma Fucosylated Glycans and C-Reactive Protein as Biomarkers of HNF1A-MODY in Young Adult-Onset Nonautoimmune Diabetes. Diabetes Care 2019, 42, 17–26. 10.2337/dc18-0422. [DOI] [PubMed] [Google Scholar]
- Agakova A.; Vučković F.; Klarić L.; Lauc G.; Agakov F. Automated Integration of a UPLC Glycomic Profile. Methods Mol. Biol. 2017, 1503, 217–233. 10.1007/978-1-4939-6493-2_17. [DOI] [PubMed] [Google Scholar]
- Haab B. B. Using Lectins in Biomarker Research: Addressing the Limitations of Sensitivity and Availability. Proteomics Clin. Appl. 2012, 6, 346–350. 10.1002/prca.201200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius H. J.; Andre S.; Jimenez-Barbero J.; Romero A.; Solis D. From Lectin Structure to Functional Glycomics: Principles of the Sugar Code. Trends Biochem. Sci. 2011, 36, 298–313. 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Lakhtin V.; Lakhtin M.; Alyoshkin V. Lectins of Living Organisms. The Overview. Anaerobe 2011, 17, 452–455. 10.1016/j.anaerobe.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zeng X. Q.; Andrade C. A. S.; Oliveira M. D. L.; Sun X. L. Carbohydrate-Protein Interactions and Their Biosensing Applications. Anal. Bioanal. Chem. 2012, 402, 3161–3176. 10.1007/s00216-011-5594-y. [DOI] [PubMed] [Google Scholar]
- Peumans W. J.; Vandamme E. J. M. Lectins as Plant Defense Proteins. Plant Physiol 1995, 109, 347–352. 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E. J. M.; Peumans W. J.; Barre A.; Rouge P. Plant Lectins: A Composite of Several Distinct Families of Structurally and Evolutionary Related Proteins with Diverse Biological Roles. Crit. Rev. Plant Sci. 1998, 17, 575–692. 10.1016/S0735-2689(98)00365-7. [DOI] [Google Scholar]
- Mnich M. E.; van Dalen R.; van Sorge N. M. C-Type Lectin Receptors in Host Defense against Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 309. 10.3389/fcimb.2020.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Ratner D. M.; Ryan C. M.; Johnson P. J.; O’Keefe B. R.; Secor W. E.; Anderson D. J.; Robbins P. W.; Samuelson J. Anti-Retroviral Lectins Have Modest Effects on Adherence of Trichomonas Vaginalis to Epithelial Cells in Vitro and on Recovery of Tritrichomonas Foetus in a Mouse Vaginal Model. PLoS One 2015, 10, e0135340. 10.1371/journal.pone.0135340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordache F.; Ionita M.; Mitrea L. I.; Fafaneata C.; Pop A. Antimicrobial and Antiparasitic Activity of Lectins. Curr. Pharm. Biotechnol. 2015, 16, 152–161. 10.2174/138920101602150112151907. [DOI] [PubMed] [Google Scholar]
- Rydz N.; Swystun L. L.; Notley C.; Paterson A. D.; Riches J. J.; Sponagle K.; Boonyawat B.; Montgomery R. R.; James P. D.; Lillicrap D. The C-Type Lectin Receptor CLEC4M Binds, Internalizes, and Clears Von Willebrand Factor and Contributes to the Variation in Plasma Von Willebrand Factor Levels. Blood 2013, 121, 5228–5237. 10.1182/blood-2012-10-457507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly E. H.; Goldstein I. J. Glutaraldehyde-Insolubilized Concanavalin A: An Adsorbent for the Specific Isolation of Polysaccharides and Glycoproteins. Biochem. J. 1970, 118, 679–680. 10.1042/bj1180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham M.; Regnier F. E. Targeted Glycoproteomics: Serial Lectin Affinity Chromatography in the Selection of O-Glycosylation Sites on Proteins from the Human Blood Proteome. J. Chromatogr. A 2006, 1132, 165–173. 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- Sumi S.; Arai K.; Kitahara S.; Yoshida K. Serial Lectin Affinity Chromatography Demonstrates Altered Asparagine-Linked Sugar-Chain Structures of Prostate-Specific Antigen in Human Prostate Carcinoma. J. Chromatogr. B Biomed. Sci. Appl. 1999, 727, 9–14. 10.1016/S0378-4347(99)00069-9. [DOI] [PubMed] [Google Scholar]
- Pinto Lobo M. D.; Batista Moreno F. B. M.; Ferreira Souza G. H. M.; LIma Verde S. M. M.; de Azevedo Moreira R.; de Oliveira Monteiro-Moreira A. C. Label-Free Proteome Analysis of Plasma from Patients with Breast Cancer: Stage-Specific Protein Expression. Front. Oncol. 2017, 7, 14. 10.3389/fonc.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy J.; Goyal V.; Skutelsky E. Lectin Histochemistry of Mammalian Endothelium. Histochemistry 1987, 86, 603–607. 10.1007/BF00489554. [DOI] [PubMed] [Google Scholar]
- Manning J. C.; Romero A.; Habermann F. A.; Garcia Caballero G.; Kaltner H.; Gabius H. J. Lectins: A Primer for Histochemists and Cell Biologists. Histochem. Cell. Biol. 2017, 147, 199–222. 10.1007/s00418-016-1524-6. [DOI] [PubMed] [Google Scholar]
- Morgan G. W.; Kail M.; Hollinshead M.; Vaux D. J. Combined Biochemical and Cytological Analysis of Membrane Trafficking Using Lectins. Anal. Biochem. 2013, 441, 21–31. 10.1016/j.ab.2013.05.034. [DOI] [PubMed] [Google Scholar]
- Moriwaki K.; Miyoshi E. Basic Procedures for Lectin Flow Cytometry. Methods Mol. Biol. 2014, 1200, 147–152. 10.1007/978-1-4939-1292-6_13. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J.; Yamada M.; Kuno A.; Tateno H. Lectin Microarrays: Concept, Principle and Applications. Chem. Soc. Rev. 2013, 42, 4443–4458. 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]
- Angeloni S.; Ridet J. L.; Kusy N.; Gao H.; Crevoisier F.; Guinchard S.; Kochhar S.; Sigrist H.; Sprenger N. Glycoprofiling with Micro-Arrays of Glycoconjugates and Lectins. Glycobiology 2004, 15, 31–41. 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- Pilobello K. T.; Krishnamoorthy L.; Slawek D.; Mahal L. K. Development of a Lectin Microarray for the Rapid Analysis of Protein Glycopatterns. Chembiochem. 2005, 6, 985–989. 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- Kuno A.; Uchiyama N.; Koseki-Kuno S.; Ebe Y.; Takashima S.; Yamada M.; Hirabayashi J. Evanescent-Field Fluorescence-Assisted Lectin Microarray: A New Strategy for Glycan Profiling. Nat. Methods 2005, 2, 851–856. 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- Zheng T.; Peelen D.; Smith L. M. Lectin Arrays for Profiling Cell Surface Carbohydrate Expression. J. Am. Chem. Soc. 2005, 127, 9982–9983. 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- Rosenfeld R.; Bangio H.; Gerwig G. J.; Rosenberg R.; Aloni R.; Cohen Y.; Amor Y.; Plaschkes I.; Kamerling J. P.; Maya R. B. A Lectin Array-Based Methodology for the Analysis of Protein Glycosylation. J. Biochem. Biophys. Methods 2007, 70, 415–426. 10.1016/j.jbbm.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Meany D. L.; Hackler L.; Zhang H.; Chan D. W. Tyramide Signal Amplification for Antibody-Overlay Lectin Microarray: A Strategy to Improve the Sensitivity of Targeted Glycan Profiling. J. Proteome Res. 2011, 10, 1425–1431. 10.1021/pr1010873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang K.; Zhang W.; Jiang S.; Lin X.; Qian A. Application of Lectin Microarrays for Biomarker Discovery. ChemistryOpen 2020, 9, 285–300. 10.1002/open.201900326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Wang J.; Tang Z.; Li X.; Yin M.; Zhang F.; Shu J.; Chen W.; Yang S.; Li Z. Integrated Glycomics Strategy for the Evaluation of Glycosylation Alterations in Salivary Proteins Associated with Type 2 Diabetes Mellitus. RSC Advances 2020, 10, 39739–39752. 10.1039/D0RA05466F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama H.; Ohta M.; Iwashita Y.; Uchida H.; Shitomi Y.; Yada K.; Inomata M. Altered Glycosylation Associated with Dedifferentiation of Hepatocellular Carcinoma: A Lectin Microarray-Based Study. BMC Cancer 2020, 20, 192. 10.1186/s12885-020-6699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuno S.; Fujimura T.; Fujime M.; Miura Y.; Ueno T. O-Glycosylated Clusterin as a Sensitive Marker for Diagnosing Early Stages of Prostate Cancer. Prostate 2021, 81, 170–181. 10.1002/pros.24094. [DOI] [PubMed] [Google Scholar]
- Zeng X.; Li S.; Tang S.; Li X.; Zhang G.; Li M.; Zeng X.; Hu C. Changes of Serum IgG Glycosylation Patterns in Primary Biliary Cholangitis Patients. Front. Immunol. 2021, 12, 669137. 10.3389/fimmu.2021.669137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kołodziejczyk J.; Blixt O.; Olejnik B.; Zimmer M.; Ferens-Sieczkowska M. Application of Lectin Microarrays for the Analysis of Seminal Plasma Glycome. Andrologia 2018, 50, e13018 10.1111/and.13018. [DOI] [PubMed] [Google Scholar]
- Ganguly A.; Lin K.-C.; Muthukumar S.; Nagaraj V. J.; Prasad S. Label-Free Protein Glycosylation Analysis Using Nanomonitor—an Ultrasensitive Electrochemical Biosensor. Curr. Protoc. 2021, 1, e150 10.1002/cpz1.150. [DOI] [PubMed] [Google Scholar]
- Silva M. L. S. Lectin Biosensors in Cancer Glycan Biomarker Detection. Adv Clin Chem 2019, 93, 1–61. 10.1016/bs.acc.2019.07.001. [DOI] [PubMed] [Google Scholar]
- Trbojević-Akmačić I.; Vilaj M.; Lauc G. High-Throughput Analysis of Immunoglobulin G Glycosylation. Expert Rev. Proteomics 2016, 13, 523–534. 10.1080/14789450.2016.1174584. [DOI] [PubMed] [Google Scholar]
- Norris G. E.; Stillman T. J.; Anderson B. F.; Baker E. N. The Three-Dimensional Structure of PNGase F, a Glycosyl Asparaginase from Flavobacterium Meningosepticum. Structure 1994, 2, 1049–1059. 10.1016/S0969-2126(94)00108-1. [DOI] [PubMed] [Google Scholar]
- Chatterjee S.; Lee L. Y.; Kawahara R.; Abrahams J. L.; Adamczyk B.; Anugraham M.; Ashwood C.; Sumer-Bayraktar Z.; Briggs M. T.; Chik J. H. L.; et al. Protein Paucimannosylation is an Enriched N-Glycosylation Signature of Human Cancers. Proteomics 2019, 19, 1900010 10.1002/pmic.201900010. [DOI] [PubMed] [Google Scholar]
- Stöckmann H.; Adamczyk B.; Hayes J.; Rudd P. M. Automated, High-Throughput IgG-Antibody Glycoprofiling Platform. Anal. Chem. 2013, 85, 8841–8849. 10.1021/ac402068r. [DOI] [PubMed] [Google Scholar]
- Kozak R. P.; Tortosa C. B.; Fernandes D. L.; Spencer D. I. Comparison of Procainamide and 2-Aminobenzamide Labeling for Profiling and Identification of Glycans by Liquid Chromatography with Fluorescence Detection Coupled to Electrospray Ionization-Mass Spectrometry. Anal. Biochem. 2015, 486, 38–40. 10.1016/j.ab.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Lauc G.; Essafi A.; Huffman J. E.; Hayward C.; Knežević A.; Kattla J. J.; Polašek O.; Gornik O.; Vitart V.; Abrahams J. L.; et al. Genomics Meets Glycomics-the First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation. PLoS Genet. 2010, 6, e1001256 10.1371/journal.pgen.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman J. E.; Knezevic A.; Vitart V.; Kattla J.; Adamczyk B.; Novokmet M.; Igl W.; Pucic M.; Zgaga L.; Johannson Å.; et al. Polymorphisms in B3GAT1, SLC9A9 and MGAT5 Are Associated with Variation within the Human Plasma N-Glycome of 3533 European Adults. Hum. Mol. Genet. 2011, 20, 5000–5011. 10.1093/hmg/ddr414. [DOI] [PubMed] [Google Scholar]
- Váradi C.; Hajdu V.; Farkas F.; Gilányi I.; Oláh C.; Viskolcz B. The Analysis of Human Serum N-Glycosylation in Patients with Primary and Metastatic Brain Tumors. Life 2021, 11, 29. 10.3390/life11010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty R.; Muniyappa M.; Walsh I.; Stöckmann H.; Hilliard M.; Hutson R.; Saldova R.; Rudd P. M. A Robust and Versatile Automated Glycoanalytical Technology for Serum Antibodies and Acute Phase Proteins: Ovarian Cancer Case Study. Mol. Cell Proteomics 2019, 18, 2191–2206. 10.1074/mcp.RA119.001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriš H.; Cindrić A.; Lauber M.; Petrović T.; Bielik A.; Taron C. H.; van Wingerden M.; Lauc G.; Trbojević-Akmačić I. Robustness and Repeatability of Glycoworks RapiFluor-MS IgG N-Glycan Profiling in a Long-Term High-Throughput Glycomic Study. Glycobiology 2021, 31, 1062–1067. 10.1093/glycob/cwab050. [DOI] [PubMed] [Google Scholar]
- Keser T.; Pavić T.; Lauc G.; Gornik O. Comparison of 2-Aminobenzamide, Procainamide and RapiFluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-Glycan Analysis. Front. Chem. 2018, 6, 324. 10.3389/fchem.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S.; Veillon L.; Dong X.; Huang Y.; Mechref Y. Direct Comparison of Derivatization Strategies for LC-MS/MS Analysis of N-Glycans. Analyst 2017, 142, 4446–4455. 10.1039/C7AN01262D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. H.; Taylor A. D.; Muddiman D. C. Individuality Normalization When Labeling with Isotopic Glycan Hydrazide Tags (INLIGHT): A Novel Glycan-Relative Quantification Strategy. J. Am. Soc. Mass Spectrom. 2013, 24, 1376–1384. 10.1007/s13361-013-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar J. G.; Butler K. E.; Baker E. S.; Muddiman D. C. Enhanced Protocol for Quantitative N-Linked Glycomics Analysis Using Individuality Normalization When Labeling with Isotopic Glycan Hydrazide Tags (INLIGHT). Anal. Bioanal. Chem. 2020, 412, 7569–7579. 10.1007/s00216-020-02892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckmann H.; Duke R. M.; Millán Martín S.; Rudd P. M. Ultrahigh Throughput, Ultrafiltration-Based N-Glycomics Platform for Ultraperformance Liquid Chromatography (ULTRA3). Anal. Chem. 2015, 87, 8316–8322. 10.1021/acs.analchem.5b01463. [DOI] [PubMed] [Google Scholar]
- Cook K. S.; Bullock K.; Sullivan T. Development and Qualification of an Antibody Rapid Deglycosylation Method. Biologicals 2012, 40, 109–117. 10.1016/j.biologicals.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Segu Z.; Stone T.; Berdugo C.; Roberts A.; Doud E.; Li Y. A Rapid Method for Relative Quantification of N-Glycans from a Therapeutic Monoclonal Antibody During Trastuzumab Biosimilar Development. mAbs 2020, 12, 1750794. 10.1080/19420862.2020.1750794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber M. A.; Yu Y.-Q.; Brousmiche D. W.; Hua Z.; Koza S. M.; Magnelli P.; Guthrie E.; Taron C. H.; Fountain K. J. Rapid Preparation of Released N-Glycans for HILIC Analysis Using a Labeling Reagent That Facilitates Sensitive Fluorescence and ESI-MS Detection. Anal. Chem. 2015, 87, 5401–5409. 10.1021/acs.analchem.5b00758. [DOI] [PubMed] [Google Scholar]
- Váradi C.; Sikora E.; Vanyorek L.; Viskolcz B. Purification of Fluorescently Derivatized N-Glycans by Magnetic Iron Nanoparticles. Nanomaterials 2019, 9, 1480. 10.3390/nano9101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uh H.-W.; Klarić L.; Ugrina I.; Lauc G.; Smilde A. K.; Houwing-Duistermaat J. J. Choosing Proper Normalization Is Essential for Discovery of Sparse Glycan Biomarkers. Mol. Omics 2020, 16, 231–242. 10.1039/C9MO00174C. [DOI] [PubMed] [Google Scholar]
- Benedetti E.; Gerstner N.; Pučić-Baković M.; Keser T.; Reiding K. R.; Ruhaak L. R.; Štambuk T.; Selman M. H. J.; Rudan I.; Polašek O. Systematic Evaluation of Normalization Methods for Glycomics Data Based on Performance of Network Inference. Metabolites 2020, 10, 271. 10.3390/metabo10070271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk B.; Struwe W. B.; Ercan A.; Nigrovic P. A.; Rudd P. M. Characterization of Fibrinogen Glycosylation and Its Importance for Serum/Plasma N-Glycome Analysis. J. Proteome Res. 2013, 12, 444–454. 10.1021/pr300813h. [DOI] [PubMed] [Google Scholar]
- Saldova R.; Asadi Shehni A.; Haakensen V. D.; Steinfeld I.; Hilliard M.; Kifer I.; Helland A.; Yakhini Z.; Borresen-Dale A. L.; Rudd P. M. Association of N-Glycosylation with Breast Carcinoma and Systemic Features Using High-Resolution Quantitative UPLC. J. Proteome Res. 2014, 13, 2314–2327. 10.1021/pr401092y. [DOI] [PubMed] [Google Scholar]
- O’Flaherty R.; Harbison A. M.; Hanley P. J.; Taron C. H.; Fadda E.; Rudd P. M. Aminoquinoline Fluorescent Labels Obstruct Efficient Removal of N-Glycan Core α(1–6) Fucose by Bovine Kidney α-L-Fucosidase (BKF). J. Proteome Res. 2017, 16, 4237–4243. 10.1021/acs.jproteome.7b00580. [DOI] [PubMed] [Google Scholar]
- Saraswathy N.; Ramalingam P.. 5 - DNA Sequencing Methods. In Concepts and Techniques in Genomics and Proteomics, Saraswathy N., Ramalingam P. Eds.; Woodhead Publishing, 2011; pp 57-76. [Google Scholar]
- Guttman A.; Chen F.-T. A.; Evangelista R. A. Separation of 1-Aminopyrene-3,6,8-Trisulfonate-Labeled Asparagine-Linked Fetuin Glycans by Capillary Gel Electrophoresis. Electrophoresis 1996, 17, 412–417. 10.1002/elps.1150170221. [DOI] [PubMed] [Google Scholar]
- Lageveen-Kammeijer G. S. M.; de Haan N.; Mohaupt P.; Wagt S.; Filius M.; Nouta J.; Falck D.; Wuhrer M. Highly Sensitive CE-ESI-MS Analysis of N-Glycans from Complex Biological Samples. Nat. Commun. 2019, 10, 2137. 10.1038/s41467-019-09910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadouch M.; Ladner Y.; Perrin C. Analysis of Monoclonal Antibodies by Capillary Electrophoresis: Sample Preparation, Separation, and Detection. Separations 2021, 8, 4. 10.3390/separations8010004. [DOI] [Google Scholar]
- Reiding K. R.; Bondt A.; Hennig R.; Gardner R. A.; O’Flaherty R.; Trbojević-Akmačić I.; Shubhakar A.; Hazes J. M. W.; Reichl U.; Fernandes D. L.; et al. High-Throughput Serum N-Glycomics: Method Comparison and Application to Study Rheumatoid Arthritis and Pregnancy-Associated Changes. Mol. Cell Proteomics 2019, 18, 3–15. 10.1074/mcp.RA117.000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig R.; Cajic S.; Borowiak M.; Hoffmann M.; Kottler R.; Reichl U.; Rapp E. Towards Personalized Diagnostics Via Longitudinal Study of the Human Plasma N-Glycome. Biochim. Biophys. Acta 2016, 1860, 1728–1738. 10.1016/j.bbagen.2016.03.035. [DOI] [PubMed] [Google Scholar]
- Guttman A. High-Resolution Carbohydrate Profiling by Capillary Gel Electrophoresis. Nature 1996, 380, 461–462. 10.1038/380461a0. [DOI] [PubMed] [Google Scholar]
- Váradi C.; Lew C.; Guttman A. Rapid Magnetic Bead Based Sample Preparation for Automated and High Throughput N-Glycan Analysis of Therapeutic Antibodies. Anal. Chem. 2014, 86, 5682–5687. 10.1021/ac501573g. [DOI] [PubMed] [Google Scholar]
- Bunz S. C.; Cutillo F.; Neusüß C. Analysis of Native and APTS-Labeled N-Glycans by Capillary Electrophoresis/Time-of-Flight Mass Spectrometry. Anal. Bioanal. Chem. 2013, 405, 8277–8284. 10.1007/s00216-013-7231-4. [DOI] [PubMed] [Google Scholar]
- Szigeti M.; Lew C.; Roby K.; Guttman A. Fully Automated Sample Preparation for Ultrafast N-Glycosylation Analysis of Antibody Therapeutics. J. Lab. Autom. 2016, 21, 281–286. 10.1177/2211068215608767. [DOI] [PubMed] [Google Scholar]
- Grace P. S.; Dolatshahi S.; Lu L. L.; Cain A.; Palmieri F.; Petrone L.; Fortune S. M.; Ottenhoff T. H. M.; Lauffenburger D. A.; Goletti D.; et al. Antibody Subclass and Glycosylation Shift Following Effective Tb Treatment. Front. Immunol. 2021, 12, 679973. 10.3389/fimmu.2021.679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron L. B.; Palmer C. S.; Liu Q.; Yin X.; Papasavvas E.; Sharaf R.; Etemad B.; Damra M.; Goldman A. R.; Tang H. Y.; et al. Non-Invasive Plasma Glycomic and Metabolic Biomarkers of Post-Treatment Control of HIV. Nat. Commun. 2021, 12, 3922. 10.1038/s41467-021-24077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy D.; Borza B.; Domokos A.; Varadi E.; Szigeti M.; Meszaros-Matwiejuk A.; Molnar-Gabor D.; Guttman A. Ultrafast High-Resolution Analysis of Human Milk Oligosaccharides by Multicapillary Gel Electrophoresis. Food Chem. 2021, 341, 128200. 10.1016/j.foodchem.2020.128200. [DOI] [PubMed] [Google Scholar]
- Guttman A. Multistructure Sequencing of N-Linked Fetuin Glycans by Capillary Gel Electrophoresis and Enzyme Matrix Digestion. Electrophoresis 1997, 18, 1136–1141. 10.1002/elps.1150180719. [DOI] [PubMed] [Google Scholar]
- Mittermayr S.; Guttman A. Influence of Molecular Configuration and Conformation on the Electromigration of Oligosaccharides in Narrow Bore Capillaries. Electrophoresis 2012, 33, 1000–1007. 10.1002/elps.201100681. [DOI] [PubMed] [Google Scholar]
- Jarvas G.; Szigeti M.; Campbell M. P.; Guttman A. Expanding the Capillary Electrophoresis-Based Glucose Unit Database of the GUcal App. Glycobiology 2020, 30, 362–364. 10.1093/glycob/cwz102. [DOI] [PubMed] [Google Scholar]
- Jarvas G.; Szigeti M.; Guttman A. GUcal: An Integrated Application for Capillary Electrophoresis Based Glycan Analysis. Electrophoresis 2015, 36, 3094–3096. 10.1002/elps.201500397. [DOI] [PubMed] [Google Scholar]
- Cajic S.; Hennig R.; Burock R.; Rapp E.. Capillary (Gel) Electrophoresis-Based Methods for Immunoglobulin (G) Glycosylation Analysis. In Antibody Glycosylation, Pezer M. Ed.; Springer International Publishing, 2021; pp 137-172. [DOI] [PubMed] [Google Scholar]
- Szigeti M.; Guttman A. Automated N-Glycosylation Sequencing of Biopharmaceuticals by Capillary Electrophoresis. Sci. Rep. 2017, 7, 11663. 10.1038/s41598-017-11493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell E. J.; Ratnayake C.; Jayo R.; Zhong X.; Chen D. D. A Promising Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry Method for Carbohydrate Analysis. Electrophoresis 2011, 32, 2161–2166. 10.1002/elps.201100027. [DOI] [PubMed] [Google Scholar]
- Jayo R. G.; Thaysen-Andersen M.; Lindenburg P. W.; Haselberg R.; Hankemeier T.; Ramautar R.; Chen D. D. Y. Simple Capillary Electrophoresis-Mass Spectrometry Method for Complex Glycan Analysis Using a Flow-through Microvial Interface. Anal. Chem. 2014, 86, 6479–6486. 10.1021/ac5010212. [DOI] [PubMed] [Google Scholar]
- Teuber K.; Schiller J.; Jakop U.; Lüpold S.; Orledge J. M.; Blount J. D.; Royle N. J.; Hoodless A.; Müller K. MALDI-TOF Mass Spectrometry as a Simple Tool to Determine the Phospholipid/Glycolipid Composition of Sperm: Pheasant Spermatozoa as One Selected Example. Anim. Reprod. Sci. 2011, 123, 270–278. 10.1016/j.anireprosci.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Li J.; Martin A.; Cox A. D.; Moxon E. R.; Richards J. C.; Thibault P. Mapping Bacterial Glycolipid Complexity Using Capillary Electrophoresis and Electrospray Mass Spectrometry. Methods Enzymol. 2005, 405, 369–397. 10.1016/S0076-6879(05)05013-5. [DOI] [PubMed] [Google Scholar]
- Tissot B.; Gasiunas N.; Powell A. K.; Ahmed Y.; Zhi Z.-l.; Haslam S. M.; Morris H. R.; Turnbull J. E.; Gallagher J. T.; Dell A. Towards GAG Glycomics: Analysis of Highly Sulfated Heparins by MALDI-TOF Mass spectrometry. Glycobiology 2007, 17, 972–982. 10.1093/glycob/cwm072. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Young S. P.; Millington D. S. Quantification of Glycosaminoglycans in Urine by Isotope-Dilution Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry. Curr. Protoc. Hum. Genet. 2013, 76, 17.12.11–17.12.14. 10.1002/0471142905.hg1712s76. [DOI] [PubMed] [Google Scholar]
- Lippold S.; Thavarajah R.; Reusch D.; Wuhrer M.; Nicolardi S. Glycoform Analysis of Intact Erythropoietin by MALDI FT-ICR Mass Spectrometry. Anal. Chim. Acta 2021, 1185, 339084. 10.1016/j.aca.2021.339084. [DOI] [PubMed] [Google Scholar]
- Čaval T.; Buettner A.; Haberger M.; Reusch D.; Heck A. J. R. Discrepancies between High-Resolution Native and Glycopeptide-Centric Mass Spectrometric Approaches: A Case Study into the Glycosylation of Erythropoietin Variants. J. Am. Soc. Mass Spectrom. 2021, 32, 2099–2104. 10.1021/jasms.1c00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaysen-Andersen M.; Mysling S.; Højrup P. Site-Specific Glycoprofiling of N-Linked Glycopeptides Using MALDI-TOF MS: Strong Correlation between Signal Strength and Glycoform Quantities. Anal. Chem. 2009, 81, 3933–3943. 10.1021/ac900231w. [DOI] [PubMed] [Google Scholar]
- Selman M. H. J.; Hoffmann M.; Zauner G.; McDonnell L. A.; Balog C. I. A.; Rapp E.; Deelder A. M.; Wuhrer M. MALDI-TOF-MS Analysis of Sialylated Glycans and Glycopeptides Using 4-Chloro-Α-Cyanocinnamic Acid Matrix. Proteomics 2012, 12, 1337–1348. 10.1002/pmic.201100498. [DOI] [PubMed] [Google Scholar]
- Chen J.; Li X.; Edmondson A.; Meyers G. D.; Izumi K.; Ackermann A. M.; Morava E.; Ficicioglu C.; Bennett M. J.; He M. Increased Clinical Sensitivity and Specificity of Plasma Protein N-Glycan Profiling for Diagnosing Congenital Disorders of Glycosylation by Use of Flow Injection-Electrospray Ionization-Quadrupole Time-of-Flight Mass Spectrometry. Clin Chem 2019, 65, 653–663. 10.1373/clinchem.2018.296780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman J. E.; Pučić-Baković M.; Klarić L.; Hennig R.; Selman M. H. J.; Vučković F.; Novokmet M.; Krištić J.; Borowiak M.; Muth T.; et al. Comparative Performance of Four Methods for High-Throughput Glycosylation Analysis of Immunoglobulin G in Genetic and Epidemiological Research. Mol. Cell Proteomics 2014, 13, 1598–1610. 10.1074/mcp.M113.037465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; Gutierrez Reyes C. D.; Gautam S.; Yu A.; Cho B. G.; Goli M.; Donohoo K.; Mondello S.; Kobeissy F.; Mechref Y. MS-Based Glycomics and Glycoproteomics Methods Enabling Isomeric Characterization. Mass Spectrom. Rev. 2021, 21713. 10.1002/mas.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. J.; Thomas B.; Upton R.; Migas L. G.; Eyers C. E.; Barran P. E.; Flitsch S. L. Applications of Ion Mobility Mass Spectrometry for High Throughput, High Resolution Glycan Analysis. Biochim. Biophys. Acta 2016, 1860, 1688–1709. 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Isailovic D.; Kurulugama R. T.; Plasencia M. D.; Stokes S. T.; Kyselova Z.; Goldman R.; Mechref Y.; Novotny M. V.; Clemmer D. E. Profiling of Human Serum Glycans Associated with Liver Cancer and Cirrhosis by IMS-MS. J. Proteome Res. 2008, 7, 1109–1117. 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isailovic D.; Plasencia M. D.; Gaye M. M.; Stokes S. T.; Kurulugama R. T.; Pungpapong V.; Zhang M.; Kyselova Z.; Goldman R.; Mechref Y.; et al. Delineating Diseases by IMS-MS Profiling of Serum N-Linked Glycans. J. Proteome Res. 2012, 11, 576–585. 10.1021/pr200777u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaye M. M.; Valentine S. J.; Hu Y.; Mirjankar N.; Hammoud Z. T.; Mechref Y.; Lavine B. K.; Clemmer D. E. Ion Mobility-Mass Spectrometry Analysis of Serum N-Linked Glycans from Esophageal Adenocarcinoma Phenotypes. J. Proteome Res. 2012, 11, 6102–6110. 10.1021/pr300756e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ausdall D. A.; Marshall W. S. Automated High-Throughput Mass Spectrometric Analysis of Synthetic Oligonucleotides. Anal. Biochem. 1998, 256, 220–228. 10.1006/abio.1997.2498. [DOI] [PubMed] [Google Scholar]
- Wheeler S. F.; Domann P.; Harvey D. J. Derivatization of Sialic Acids for Stabilization in Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry and Concomitant Differentiation of Alpha(2 --> 3)- and Alpha(2 --> 6)-Isomers. Rapid Commun. Mass Spectrom. 2009, 23, 303–312. 10.1002/rcm.3867. [DOI] [PubMed] [Google Scholar]
- Reiding K. R.; Blank D.; Kuijper D. M.; Deelder A. M.; Wuhrer M. High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification. Anal. Chem. 2014, 86, 5784–5793. 10.1021/ac500335t. [DOI] [PubMed] [Google Scholar]
- Holst S.; Heijs B.; de Haan N.; van Zeijl R. J.; Briaire-de Bruijn I. H.; van Pelt G. W.; Mehta A. S.; Angel P. M.; Mesker W. E.; Tollenaar R. A.; et al. Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues. Anal. Chem. 2016, 88, 5904–5913. 10.1021/acs.analchem.6b00819. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Reiding K. R.; Haberger M.; Reusch D.; Falck D.; Wuhrer M. Linkage-Specific Sialic Acid Derivatization for MALDI-TOF-MS Profiling of IgG Glycopeptides. Anal. Chem. 2015, 87, 8284–8291. 10.1021/acs.analchem.5b02426. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Yang S.; Cipollo J.; Wuhrer M. Glycomics Studies Using Sialic Acid Derivatization and Mass Spectrometry. Nat. Rev. Chem. 2020, 4, 229–242. 10.1038/s41570-020-0174-3. [DOI] [PubMed] [Google Scholar]
- Huberty M. C.; Vath J. E.; Yu W.; Martin S. A. Site-Specific Carbohydrate Identification in Recombinant Proteins Using MALD-TOF MS. Anal. Chem. 1993, 65, 2791–2800. 10.1021/ac00068a015. [DOI] [PubMed] [Google Scholar]
- Pitt J. J.; Gorman J. J. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry of Sialylated Glycopeptides and Proteins Using 2,6-Dihydroxyacetophenone as a Matrix. Rapid Commun. Mass Spectrom. 1996, 10, 1786–1788. . [DOI] [Google Scholar]
- Grünwald-Gruber C.; Thader A.; Maresch D.; Dalik T.; Altmann F. Determination of True Ratios of Different N-Glycan Structures in Electrospray Ionization Mass Spectrometry. Anal. Bioanal. Chem. 2017, 409, 2519–2530. 10.1007/s00216-017-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M. H. J.; McDonnell L. A.; Palmblad M.; Ruhaak L. R.; Deelder A. M.; Wuhrer M. Immunoglobulin G Glycopeptide Profiling by Matrix-Assisted Laser Desorption Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2010, 82, 1073–1081. 10.1021/ac9024413. [DOI] [PubMed] [Google Scholar]
- Nishimura S.-I.; Niikura K.; Kurogochi M.; Matsushita T.; Fumoto M.; Hinou H.; Kamitani R.; Nakagawa H.; Deguchi K.; Miura N.; et al. High-Throughput Protein Glycomics: Combined Use of Chemoselective Glycoblotting and MALDI-TOF/TOF Mass Spectrometry. Angew. Chem., Int. Ed. 2005, 44, 91–96. 10.1002/anie.200461685. [DOI] [PubMed] [Google Scholar]
- Miura Y.; Hato M.; Shinohara Y.; Kuramoto H.; Furukawa J.-i.; Kurogochi M.; Shimaoka H.; Tada M.; Nakanishi K.; Ozaki M.; et al. BlotGlycoABC, an Integrated Glycoblotting Technique for Rapid and Large Scale Clinical Glycomics. Mol. Cell Proteomics 2008, 7, 370–377. 10.1074/mcp.M700377-MCP200. [DOI] [PubMed] [Google Scholar]
- Amano M.; Nishimura S.-I.; Fukuda M. Large-Scale Glycomics for Discovering Cancer-Associated N-Glycans by Integrating Glycoblotting and Mass Spectrometry. Methods Enzymol. 2010, 478, 109–125. 10.1016/S0076-6879(10)78004-6. [DOI] [PubMed] [Google Scholar]
- Smargiasso N.; Quinton L.; De Pauw E. 2-Aminobenzamide and 2-Aminobenzoic Acid as New MALDI Matrices Inducing Radical Mediated in-Source Decay of Peptides and Proteins. J. Am. Soc. Mass Spectrom. 2012, 23, 469–474. 10.1007/s13361-011-0307-5. [DOI] [PubMed] [Google Scholar]
- Anumula K. R.; Dhume S. T. High Resolution and High Sensitivity Methods for Oligosaccharide Mapping and Characterization by Normal Phase High Performance Liquid Chromatography Following Derivatization with Highly Fluorescent Anthranilic Acid. Glycobiology 1998, 8, 685–694. 10.1093/glycob/8.7.685. [DOI] [PubMed] [Google Scholar]
- Wuhrer M.; Deelder A. M. Negative-Mode MALDI-TOF/TOF-MS of Oligosaccharides Labeled with 2-Aminobenzamide. Anal. Chem. 2005, 77, 6954–6959. 10.1021/ac051117e. [DOI] [PubMed] [Google Scholar]
- Charlwood J.; Birrell H.; Gribble A.; Burdes V.; Tolson D.; Camilleri P. A Probe for the Versatile Analysis and Characterization of N-Linked Oligosaccharides. Anal. Chem. 2000, 72, 1453–1461. 10.1021/ac991268f. [DOI] [PubMed] [Google Scholar]
- Gao X.; Lu Y.; Wei M.; Yang M.; Zheng C.; Wang C.; Zhang Y.; Huang L.; Wang Z. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Analysis of Human Milk Neutral and Sialylated Free Oligosaccharides Using Girard’s Reagent P on-Target Derivatization. J. Agric. Food Chem. 2019, 67, 8958–8966. 10.1021/acs.jafc.9b02635. [DOI] [PubMed] [Google Scholar]
- Naven T. J. P.; Harvey D. J. Cationic Derivatization of Oligosaccharides with Girard’s T Reagent for Improved Performance in Matrix-Assisted Laser Desorption/Ionization and Electrospray Mass Spectrometry. Rapid Commun. Mass Spectrom. 1996, 10, 829–834. . [DOI] [Google Scholar]
- Zhang Y.; Wang B.; Jin W.; Wen Y.; Nan L.; Yang M.; Liu R.; Zhu Y.; Wang C.; Huang L.; et al. Sensitive and Robust MALDI-TOF-MS Glycomics Analysis Enabled by Girard’s Reagent T on-Target Derivatization (GTOD) of Reducing Glycans. Anal. Chim. Acta 2019, 1048, 105–114. 10.1016/j.aca.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladergroen M. R.; Reiding K. R.; Hipgrave Ederveen A. L.; Vreeker G. C.; Clerc F.; Holst S.; Bondt A.; Wuhrer M.; van der Burgt Y. E. Automation of High-Throughput Mass Spectrometry-Based Plasma N-Glycome Analysis with Linkage-Specific Sialic Acid Esterification. J. Proteome Res. 2015, 14, 4080–4086. 10.1021/acs.jproteome.5b00538. [DOI] [PubMed] [Google Scholar]
- Doherty M.; Bones J.; McLoughlin N.; Telford J. E.; Harmon B.; DeFelippis M. R.; Rudd P. M. An Automated Robotic Platform for Rapid Profiling Oligosaccharide Analysis of Monoclonal Antibodies Directly from Cell Culture. Anal. Biochem. 2013, 442, 10–18. 10.1016/j.ab.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Blaschke C. R. K.; Black A. P.; Mehta A. S.; Angel P. M.; Drake R. R. Rapid N-Glycan Profiling of Serum and Plasma by a Novel Slide-Based Imaging Mass Spectrometry Workflow. J. Am. Soc. Mass Spectrom. 2020, 31, 2511–2520. 10.1021/jasms.0c00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke C. R. K.; Hartig J. P.; Grimsley G.; Liu L.; Semmes O. J.; Wu J. D.; Ippolito J. E.; Hughes-Halbert C.; Nyalwidhe J. O.; Drake R. R. Direct N-Glycosylation Profiling of Urine and Prostatic Fluid Glycoproteins and Extracellular Vesicles. Front. Chem. 2021, 9, 734280. 10.3389/fchem.2021.734280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrinc Potočnik N.; Porta T.; Becker M.; Heeren R. M. A.; Ellis S. R. Use of Advantageous, Volatile Matrices Enabled by Next-Generation High-Speed Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Employing a Scanning Laser Beam. Rapid Commun. Mass Spectrom. 2015, 29, 2195–2203. 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- Boughton B. A.; Thomas O. R. B.; Demarais N. J.; Trede D.; Swearer S. E.; Grey A. C. Detection of Small Molecule Concentration Gradients in Ocular Tissues and Humours. J. Mass Spectrom. 2020, 55, e4460 10.1002/jms.4460. [DOI] [PubMed] [Google Scholar]
- Fu T.; Oetjen J.; Chapelle M.; Verdu A.; Szesny M.; Chaumot A.; Degli-Esposti D.; Geffard O.; Clément Y.; Salvador A.; et al. In Situ Isobaric Lipid Mapping by MALDI-Ion Mobility Separation-Mass Spectrometry Imaging. J. Mass Spectrom. 2020, 55, e4531 10.1002/jms.4531. [DOI] [PubMed] [Google Scholar]
- Thomas M. J.; Collinge E.; Witt M.; Palacio Lozano D. C.; Vane C. H.; Moss-Hayes V.; Barrow M. P. Petroleomic Depth Profiling of Staten Island Salt Marsh Soil: 2ω Detection FTICR MS Offers a New Solution for the Analysis of Environmental Contaminants. Sci. Total Environ. 2019, 662, 852–862. 10.1016/j.scitotenv.2019.01.228. [DOI] [PubMed] [Google Scholar]
- Heijs B.; Potthoff A.; Soltwisch J.; Dreisewerd K. MALDI-2 for the Enhanced Analysis of N-Linked Glycans by Mass Spectrometry Imaging. Anal. Chem. 2020, 92, 13904–13911. 10.1021/acs.analchem.0c02732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltwisch J.; Kettling H.; Vens-Cappell S.; Wiegelmann M.; Müthing J.; Dreisewerd K. Mass Spectrometry Imaging with Laser-Induced Postionization. Science 2015, 348, 211–215. 10.1126/science.aaa1051. [DOI] [PubMed] [Google Scholar]
- Madunic K.; Wagt S.; Zhang T.; Wuhrer M.; Lageveen-Kammeijer G. S. M. Dopant-Enriched Nitrogen Gas for Enhanced Electrospray Ionization of Released Glycans in Negative Ion Mode. Anal. Chem. 2021, 83, 6919. 10.1021/acs.analchem.1c00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete E.; Facchiano A.; Profumo A.; Angelini C.; Romano P. GeenaR: A Web Tool for Reproducible MALDI-TOF Analysis. Front. Genet. 2021, 12, 635814. 10.3389/fgene.2021.635814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palarea-Albaladejo J.; Mclean K.; Wright F.; Smith D. G. E. MALDIrppa: Quality Control and Robust Analysis for Mass Spectrometry Data. Bioinformatics 2018, 34, 522–523. 10.1093/bioinformatics/btx628. [DOI] [PubMed] [Google Scholar]
- Goldberg D.; Sutton-Smith M.; Paulson J.; Dell A. Automatic Annotation of Matrix-Assisted Laser Desorption/Ionization N-Glycan Spectra. Proteomics 2005, 5, 865–875. 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- Xu G.; Liu X.; Liu Q. Y.; Zhou Y.; Li J. Improve Accuracy and Sensibility in Glycan Structure Prediction by Matching Glycan Isotope Abundance. Anal. Chim. Acta 2012, 743, 80–89. 10.1016/j.aca.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Xu G.; Liu X.; Liu Q. Y.; Li J.; Zhou Y. Automatic Annotation and Visualization Tool for Mass Spectrometry Based Glycomics. Rapid Commun. Mass Spectrom. 2016, 30, 2471–2479. 10.1002/rcm.7735. [DOI] [PubMed] [Google Scholar]
- Miura N.; Hanamatsu H.; Yokota I.; Okada K.; Furukawa J.-I.; Shinohara Y. Toolbox Accelerating Glycomics (Tag): Glycan Annotation from MALDI-TOF MS Spectra and Mapping Expression Variation to Biosynthetic Pathways. Biomolecules 2020, 10, 1383. 10.3390/biom10101383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotz V.; Visconti A.; Lomax-Browne H. J.; Clerc F.; Hipgrave Ederveen A. L.; Medjeral-Thomas N. R.; Cook H. T.; Pickering M. C.; Wuhrer M.; Falchi M. O- and N-Glycosylation of Serum Immunoglobulin A is Associated with IgA Nephropathy and Glomerular Function. J. Am. Soc. Nephrol. 2021, 32, 2455–2465. 10.1681/ASN.2020081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baković M. P.; Selman M. H.; Hoffmann M.; Rudan I.; Campbell H.; Deelder A. M.; Lauc G.; Wuhrer M. High-Throughput IgG Fc N-Glycosylation Profiling by Mass Spectrometry of Glycopeptides. J. Proteome Res. 2013, 12, 821–831. 10.1021/pr300887z. [DOI] [PubMed] [Google Scholar]
- van de Geijn F. E.; Wuhrer M.; Selman M. H.; Willemsen S. P.; de Man Y. A.; Deelder A. M.; Hazes J. M.; Dolhain R. J. Immunoglobulin G Galactosylation and Sialylation Are Associated with Pregnancy-Induced Improvement of Rheumatoid Arthritis and the Postpartum Flare: Results from a Large Prospective Cohort Study. Arthritis Res. Ther. 2009, 11, R193. 10.1186/ar2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck D.; Jansen B. C.; de Haan N.; Wuhrer M. High-Throughput Analysis of IgG Fc Glycopeptides by LC-MS. Methods Mol. Biol. 2017, 1503, 31–47. 10.1007/978-1-4939-6493-2_4. [DOI] [PubMed] [Google Scholar]
- Amez-Martín M.; Wuhrer M.; Falck D. Immunoglobulin G Glycoprofiles Are Unaffected by Common Bottom-up Sample Processing. J. Proteome Res. 2020, 19, 4158–4162. 10.1021/acs.jproteome.0c00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amez Martín M.; Wuhrer M.; Falck D. Serum and Plasma Immunoglobulin G Fc N-Glycosylation Is Stable During Storage. J. Proteome Res. 2021, 20, 2935–2941. 10.1021/acs.jproteome.1c00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz T.; Nouta J.; Wang W.; van Meijgaarden K. E.; Linty F.; Vidarsson G.; Joosten S. A.; Ottenhoff T. H. M.; Hokke C. H.; de Vries J. J. C.; et al. Immunoglobulin G1 Fc Glycosylation as an Early Hallmark of Severe COVID-19. eBioMedicine 2022, 78, 103957. 10.1016/j.ebiom.2022.103957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. D.; de Graaf E. L.; Sonneveld M. E.; Plomp H. R.; Nouta J.; Hoepel W.; Chen H. J.; Linty F.; Visser R.; Brinkhaus M. Afucosylated IgG Characterizes Enveloped Viral Responses and Correlates with COVID-19 Severity. Science 2021, 371, eabc8378. 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. D.; Lopez-Perez M.; Dickson E. K.; Ampomah P.; Tuikue Ndam N.; Nouta J.; Koeleman C. A. M.; Ederveen A. L. H.; Mordmüller B.; Salanti A.; et al. Afucosylated Plasmodium Falciparum-Specific IgG Is Induced by Infection but Not by Subunit Vaccination. Nat. Commun. 2021, 12, 5838. 10.1038/s41467-021-26118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur R.; Kustiawan I.; Vestrheim A.; Koeleman C. A.; Visser R.; Einarsdottir H. K.; Porcelijn L.; Jackson D.; Kumpel B.; Deelder A. M.; et al. A Prominent Lack of IgG1-Fc Fucosylation of Platelet Alloantibodies in Pregnancy. Blood 2014, 123, 471–480. 10.1182/blood-2013-09-527978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer H. U.; Wang J.; Toes R. E.; van der Woude D.; Koeleman C. A.; de Boer A. R.; Huizinga T. W.; Deelder A. M.; Wuhrer M. Immunoglobulin 1 (IgG1) Fc-Glycosylation Profiling of Anti-Citrullinated Peptide Antibodies from Human Serum. Proteomics Clin. Appl. 2009, 3, 106–115. 10.1002/prca.200800098. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Falck D.; Wuhrer M. Monitoring of Immunoglobulin N- and O-Glycosylation in Health and Disease. Glycobiology 2020, 30, 226–240. 10.1093/glycob/cwz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A.; Hafkenscheid L.; Falck D.; Kuijper T. M.; Rombouts Y.; Hazes J. M. W.; Wuhrer M.; Dolhain R. ACPA IgG Galactosylation Associates with Disease Activity in Pregnant Patients with Rheumatoid Arthritis. Ann. Rheum. Dis. 2018, 77, 1130–1136. 10.1136/annrheumdis-2018-212946. [DOI] [PubMed] [Google Scholar]
- Selman M. H.; Hoffmann M.; Zauner G.; McDonnell L. A.; Balog C. I.; Rapp E.; Deelder A. M.; Wuhrer M. MALDI-TOF-MS Analysis of Sialylated Glycans and Glycopeptides Using 4-Chloro-Α-Cyanocinnamic Acid Matrix. Proteomics 2012, 12, 1337–1348. 10.1002/pmic.201100498. [DOI] [PubMed] [Google Scholar]
- Jansen B. C.; Falck D.; de Haan N.; Hipgrave Ederveen A. L. H.; Razdorov G.; Lauc G.; Wuhrer M. LaCyTools: A Targeted Liquid Chromatography-Mass Spectrometry Data Processing Package for Relative Quantitation of Glycopeptides. J. Proteome Res. 2016, 15, 2198–2210. 10.1021/acs.jproteome.6b00171. [DOI] [PubMed] [Google Scholar]
- Wuhrer M.; Stam J. C.; van de Geijn F. E.; Koeleman C. A.; Verrips C. T.; Dolhain R. J.; Hokke C. H.; Deelder A. M. Glycosylation Profiling of Immunoglobulin G (IgG) Subclasses from Human Serum. Proteomics 2007, 7, 4070–4081. 10.1002/pmic.200700289. [DOI] [PubMed] [Google Scholar]
- Stadlmann J.; Pabst M.; Kolarich D.; Kunert R.; Altmann F. Analysis of Immunoglobulin Glycosylation by LC-ESI-MS of Glycopeptides and Oligosaccharides. Proteomics 2008, 8, 2858–2871. 10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- de Haan N.; Boeddha N. P.; Ekinci E.; Reiding K. R.; Emonts M.; Hazelzet J. A.; Wuhrer M.; Driessen G. J. Differences in IgG Fc Glycosylation Are Associated with Outcome of Pediatric Meningococcal Sepsis. mBio 2018, 9, 545-18. 10.1128/mBio.00546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. R.; Gao W. N.; Grimm R.; Jiang S.; Liang Y.; Ye H.; Li Z. G.; Yau L. F.; Huang H.; Liu J.; et al. Reply to ’Trace N-Glycans Including Sulphated Species May Originate from Various Plasma Glycoproteins and Not Necessarily IgG’. Nat. Commun. 2018, 9, 2915. 10.1038/s41467-018-05082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauc G.; Vučković F.; Bondt A.; Pezer M.; Wuhrer M. Trace N-Glycans Including Sulphated Species May Originate from Various Plasma Glycoproteins and Not Necessarily IgG. Nat. Commun. 2018, 9, 2916. 10.1038/s41467-018-05173-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Shu J.; Li Z. Lectin Microarrays for Glycoproteomics: An Overview of Their Use and Potential. Expert Rev. Proteomics 2020, 17, 27–39. 10.1080/14789450.2020.1720512. [DOI] [PubMed] [Google Scholar]
- Bertok T.; Jane E.; Chrenekova N.; Hroncekova S.; Bertokova A.; Hires M.; Vikartovska A.; Kubanikova P.; Sokol R.; Fillo J.; et al. Analysis of Serum Glycome by Lectin Microarrays for Prostate Cancer Patients - a Search for Aberrant Glycoforms. Glycoconj. J. 2020, 37, 703–711. 10.1007/s10719-020-09958-4. [DOI] [PubMed] [Google Scholar]
- Chen S.; Qin R.; Mahal L. K. Sweet Systems: Technologies for Glycomic Analysis and Their Integration into Systems Biology. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 301–320. 10.1080/10409238.2021.1908953. [DOI] [PubMed] [Google Scholar]
- Du H.; Yu H.; Ma T.; Yang F.; Jia L.; Zhang C.; Zhang J.; Niu L.; Yang J.; Zhang Z.; et al. Analysis of Glycosphingolipid Glycans by Lectin Microarrays. Anal. Chem. 2019, 91, 10663–10671. 10.1021/acs.analchem.9b01945. [DOI] [PubMed] [Google Scholar]
- Du H.; Yu H.; Yang F.; Li Z. Comprehensive Analysis of Glycosphingolipid Glycans by Lectin Microarrays and MALDI-TOF Mass Spectrometry. Nat. Protoc. 2021, 16, 3470–3491. 10.1038/s41596-021-00544-y. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Luo S.; Zhang B. The Use of Lectin Microarray for Assessing Glycosylation of Therapeutic Proteins. mAbs 2016, 8, 524–535. 10.1080/19420862.2016.1149662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshi Y.; Nakata E.; Yamane H.; Hamachi I. A Fluorescent Lectin Array Using Supramolecular Hydrogel for Simple Detection and Pattern Profiling for Various Glycoconjugates. J. Am. Chem. Soc. 2006, 128, 10413–10422. 10.1021/ja0613963. [DOI] [PubMed] [Google Scholar]
- Roucka M.; Zimmermann K.; Fido M.; Nechansky A. Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC. Microarrays 2017, 6, 1. 10.3390/microarrays6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. P.; Royle L.; Radcliffe C. M.; Dwek R. A.; Rudd P. M. GlycoBase and AutoGU: Tools for HPLC-Based Glycan Analysis. Bioinformatics 2008, 24, 1214–1216. 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Walsh I.; Abrahams J. L.; Royle L.; Nguyen-Khuong T.; Spencer D.; Fernandes D. L.; Packer N. H.; Rudd P. M.; Campbell M. P. GlycoStore: A Database of Retention Properties for Glycan Analysis. Bioinformatics 2018, 34, 3231–3232. 10.1093/bioinformatics/bty319. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Reed C. E.; Birdsall R. E.; Yu Y. Q.; Chen W. High-Throughput Analysis of Fluorescently Labeled N-Glycans Derived from Biotherapeutics Using an Automated LC-MS-Based Solution. SLAS Technol.; Translating Life Sciences Innovation 2020, 25, 380–387. 10.1177/2472630320922803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams J. L.; Taherzadeh G.; Jarvas G.; Guttman A.; Zhou Y.; Campbell M. P. Recent Advances in Glycoinformatic Platforms for Glycomics and Glycoproteomics. Curr. Opin. Struct. Biol. 2020, 62, 56–69. 10.1016/j.sbi.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Campbell M. P.; Nguyen-Khuong T.; Hayes C. A.; Flowers S. A.; Alagesan K.; Kolarich D.; Packer N. H.; Karlsson N. G. Validation of the Curation Pipeline of UniCarb-DB: Building a Global Glycan Reference MS/MS Repository. Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844, 108–116. 10.1016/j.bbapap.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Damerell D.; Ceroni A.; Maass K.; Ranzinger R.; Dell A.; Haslam S. M. The GlycanBuilder and GlycoWorkbench Glycoinformatics Tools: Updates and New Developments. Biol. Chem. 2012, 393, 1357–1362. 10.1515/hsz-2012-0135. [DOI] [PubMed] [Google Scholar]
- Horlacher O.; Jin C.; Alocci D.; Mariethoz J.; Müller M.; Karlsson N. G.; Lisacek F. Glycoforest 1.0. Anal. Chem. 2017, 89, 10932–10940. 10.1021/acs.analchem.7b02754. [DOI] [PubMed] [Google Scholar]
- Cooper C. A.; Gasteiger E.; Packer N. H. GlycoMod - a Software Tool for Determining Glycosylation Compositions from Mass Spectrometric Data. Proteomics 2001, 1, 340–349. . [DOI] [PubMed] [Google Scholar]
- Choo M. S.; Wan C.; Rudd P. M.; Nguyen-Khuong T. GlycopeptideGraphMS: Improved Glycopeptide Detection and Identification by Exploiting Graph Theoretical Patterns in Mass and Retention Time. Anal. Chem. 2019, 91, 7236–7244. 10.1021/acs.analchem.9b00594. [DOI] [PubMed] [Google Scholar]
- Behne A.; Muth T.; Borowiak M.; Reichl U.; Rapp E. glyXalign: High-Throughput Migration Time Alignment Preprocessing of Electrophoretic Data Retrieved Via Multiplexed Capillary Gel Electrophoresis with Laser-Induced Fluorescence Detection-Based Glycoprofiling. Electrophoresis 2013, 34, 2311–2315. 10.1002/elps.201200696. [DOI] [PubMed] [Google Scholar]
- Walsh I.; Nguyen-Khuong T.; Wongtrakul-Kish K.; Tay S. J.; Chew D.; José T.; Taron C. H.; Rudd P. M. GlycanAnalyzer: Software for Automated Interpretation of N-Glycan Profiles after Exoglycosidase Digestions. Bioinformatics 2019, 35, 688–690. 10.1093/bioinformatics/bty681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvas G.; Szigeti M.; Guttman A. Structural Identification of N-Linked Carbohydrates Using the GUcal Application: A Tutorial. J. Proteomics 2018, 171, 107–115. 10.1016/j.jprot.2017.08.017. [DOI] [PubMed] [Google Scholar]
- Jarvas G.; Szigeti M.; Chapman J.; Guttman A. Triple-Internal Standard Based Glycan Structural Assignment Method for Capillary Electrophoresis Analysis of Carbohydrates. Anal. Chem. 2016, 88, 11364–11367. 10.1021/acs.analchem.6b03596. [DOI] [PubMed] [Google Scholar]
- Hennig R.; Reichl U.; Rapp E.. A Software Tool for Automated High-Throughput Processing of CGE-LIF Based Glycoanalysis Data, Generated by a Multiplexing Capillary DNA Sequencer. Poster presented at 5th Glycan Forum, Berlin, Germany, 2011.
- Jansen B. C.; Hafkenscheid L.; Bondt A.; Gardner R. A.; Hendel J. L.; Wuhrer M.; Spencer D. I. R. HappyTools: A Software for High-Throughput HPLC Data Processing and Quantitation. PLoS One 2018, 13, e0200280 10.1371/journal.pone.0200280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen B. C.; Reiding K. R.; Bondt A.; Hipgrave Ederveen A. L.; Palmblad M.; Falck D.; Wuhrer M. MassyTools: A High-Throughput Targeted Data Processing Tool for Relative Quantitation and Quality Control Developed for Glycomic and Glycoproteomic MALDI-MS. J. Proteome Res. 2015, 14, 5088–5098. 10.1021/acs.jproteome.5b00658. [DOI] [PubMed] [Google Scholar]
- Ràfols P.; Heijs B.; del Castillo E.; Yanes O.; McDonnell L. A.; Brezmes J.; Pérez-Taboada I.; Vallejo M.; García-Altares M.; Correig X. rMSIproc: An R Package for Mass Spectrometry Imaging Data Processing. Bioinformatics 2020, 36, 3618–3619. 10.1093/bioinformatics/btaa142. [DOI] [PubMed] [Google Scholar]
- Schilling B.; MacLean B. X.; D’Souza A.; Rardin M. J.; Shulman N. J.; MacCoss M. J.; Gibson B. W. MS1 Label-Free Quantification Using Ion Intensity Chromatograms in Skyline (Research and Clinical Applications). Quantitative Proteomics 2014, 154. 10.1039/9781782626985-00154.24565693 [DOI] [Google Scholar]
- Walsh I.; Choo M. S. F.; Chiin S. L.; Mak A.; Tay S. J.; Rudd P. M.; Yuansheng Y.; Choo A.; Swan H. Y.; Nguyen-Khuong T. Clustering and Curation of Electropherograms: An Efficient Method for Analyzing Large Cohorts of Capillary Electrophoresis Glycomic Profiles for Bioprocessing Operations. Beilstein J. Org. Chem. 2020, 16, 2087–2099. 10.3762/bjoc.16.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moh E. S. X.; Thaysen-Andersen M.; Packer N. H. Relative Versus Absolute Quantitation in Disease Glycomics. Proteomics Clin. Appl. 2015, 9, 368–382. 10.1002/prca.201400184. [DOI] [PubMed] [Google Scholar]
- Wang S.; Liu D.; Qu J.; Zhu H.; Chen C.; Gibbons C.; Greenway H.; Wang P.; Bollag R. J.; Liu K.; et al. Streamlined Subclass-Specific Absolute Quantification of Serum IgG Glycopeptides Using Synthetic Isotope-Labeled Standards. Anal. Chem. 2021, 93, 4449–4455. 10.1021/acs.analchem.0c04462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váradi C.; Mittermayr S.; Millán-Martín S.; Bones J. Quantitative Twoplex Glycan Analysis Using 12C6 and 13C6 Stable Isotope 2-Aminobenzoic Acid Labelling and Capillary Electrophoresis Mass Spectrometry. Anal. Bioanal. Chem. 2016, 408, 8691–8700. 10.1007/s00216-016-9935-8. [DOI] [PubMed] [Google Scholar]
- Leek J. T.; Johnson W. E.; Parker H. S.; Jaffe A. E.; Storey J. D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki-Kinoshita K. F.; Lisacek F.; Mazumder R.; York W. S.; Packer N. H. The GlySpace Alliance: Toward a Collaborative Global Glycoinformatics Community. Glycobiology 2020, 30, 70–71. 10.1093/glycob/cwz078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. P.; Abrahams J. L.; Rapp E.; Struwe W. B.; Costello C. E.; Novotny M.; Ranzinger R.; York W. S.; Kolarich D.; Rudd P. M.; et al. The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: LC Guidelines. Glycobiology 2019, 29, 349–354. 10.1093/glycob/cwz009. [DOI] [PubMed] [Google Scholar]
- Kolarich D.; Rapp E.; Struwe W. B.; Haslam S. M.; Zaia J.; McBride R.; Agravat S.; Campbell M. P.; Kato M.; Ranzinger R.; et al. The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Improving the Standards for Reporting Mass-Spectrometry-Based Glycoanalytic Data. Mol. Cell Proteomics 2013, 12, 991–995. 10.1074/mcp.O112.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; McBride R.; Stoll M.; Palma A. S.; Silva L.; Agravat S.; Aoki-Kinoshita K. F.; Campbell M. P.; Costello C. E.; Dell A. The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Improving the Standards for Reporting Glycan Microarray-Based Data. Glycobiology 2016, 27, 280. 10.1093/glycob/cww118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W. B.; Agravat S.; Aoki-Kinoshita K. F.; Campbell M. P.; Costello C. E.; Dell A.; Feizi T.; Haslam S. M.; Karlsson N. G.; Khoo K. H.; et al. The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Sample Preparation Guidelines for Reliable Reporting of Glycomics Datasets. Glycobiology 2016, 26, 907–910. 10.1093/glycob/cww082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageveen-Kammeijer G. S. M.; Rapp E.; Chang D.; Rudd P. M.; Kettner C.; Zaia J. The Minimum Information Required for a Glycomics Experiment (MIRAGE): Reporting Guidelines for Capillary Electrophoresis. Glycobiology 2022, 32, 580. 10.1093/glycob/cwac021. [DOI] [PubMed] [Google Scholar]
- Yamada I.; Campbell M. P.; Edwards N.; Castro L. J.; Lisacek F.; Mariethoz J.; Ono T.; Ranzinger R.; Shinmachi D.; Aoki-Kinoshita K. F. The Glycoconjugate Ontology (GlycoCoO) for Standardizing the Annotation of Glycoconjugate Data and Its Application. Glycobiology 2021, 31, 741–750. 10.1093/glycob/cwab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarić L.; Tsepilov Y. A.; Stanton C. M.; Mangino M.; Sikka T. T.; Esko T.; Pakhomov E.; Salo P.; Deelen J.; McGurnaghan S. J.; et al. Glycosylation of Immunoglobulin G Is Regulated by a Large Network of Genes Pleiotropic with Inflammatory Diseases. Sci. Adv. 2020, 6, eaax0301 10.1126/sciadv.aax0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štambuk T.; Klasić M.; Zoldoš V.; Lauc G. N-Glycans as Functional Effectors of Genetic and Epigenetic Disease Risk. Mol. Aspects Med. 2021, 79, 100891. 10.1016/j.mam.2020.100891. [DOI] [PubMed] [Google Scholar]
- Menni C.; Keser T.; Mangino M.; Bell J. T.; Erte I.; Akmacic I.; Vuckovic F.; Pucic Bakovic M. P.; Gornik O.; McCarthy M. I. Glycosylation of Immunoglobulin G: Role of Genetic and Epigenetic Influences. PloS One 2013, 8, e82558. 10.1371/journal.pone.0082558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A.; Selman M. H. J.; Deelder A. M.; Hazes J. M. W.; Willemsen S. P.; Wuhrer M.; Dolhain R. J. E. M. Association between Galactosylation of Immunoglobulin G and Improvement of Rheumatoid Arthritis During Pregnancy Is Independent of Sialylation. J. Proteome Res. 2013, 12, 4522–4531. 10.1021/pr400589m. [DOI] [PubMed] [Google Scholar]
- Nikolac Perkovic M.; Pucic Bakovic M.; Kristic J.; Novokmet M.; Huffman J. E.; Vitart V.; Hayward C.; Rudan I.; Wilson J. F.; Campbell H.; et al. The Association between Galactosylation of Immunoglobulin G and Body Mass Index. Prog. Neuropsychopharmacol Biol. Psychiatry 2014, 48, 20–25. 10.1016/j.pnpbp.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Vučković F.; Krištić J.; Gudelj I.; Teruel M.; Keser T.; Pezer M.; Pučić-Baković M.; Štambuk J.; Trbojević-Akmačić I.; Barrios C.; et al. Association of Systemic Lupus Erythematosus with Decreased Immunosuppressive Potential of the IgG Glycome. Arthritis Rheum. 2015, 67, 2978–2989. 10.1002/art.39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trbojević Akmačić I.; Ventham N. T.; Theodoratou E.; Vučković F.; Kennedy N. A.; Krištić J.; Nimmo E. R.; Kalla R.; Drummond H.; Štambuk J.; et al. Inflammatory Bowel Disease Associates with Proinflammatory Potential of the Immunoglobulin G Glycome. Inflamm. Bowel Dis. 2015, 21, 1237–1247. 10.1097/MIB.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Wang Y.; Kristic J.; Dong J.; Chu X.; Ge S.; Wang H.; Fang H.; Gao Q.; Liu D.; et al. Profiling IgG N-Glycans as Potential Biomarker of Chronological and Biological Ages: A Community-Based Study in a Han Chinese Population. Medicine (Baltimore) 2016, 95, e4112 10.1097/MD.0000000000004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer M.; Stambuk J.; Perica M.; Razdorov G.; Banic I.; Vuckovic F.; Gospic A. M.; Ugrina I.; Vecenaj A.; Bakovic M. P.; et al. Effects of Allergic Diseases and Age on the Composition of Serum IgG Glycome in Children. Sci. Rep. 2016, 6, 33198. 10.1038/srep33198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vučković F.; Theodoratou E.; Thaçi K.; Timofeeva M.; Vojta A.; Štambuk J.; Pučić-Baković M.; Rudd P. M.; Đerek L.; Servis D.; et al. IgG Glycome in Colorectal Cancer. Clin. Cancer Res. 2016, 22, 3078–3086. 10.1158/1078-0432.CCR-15-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios C.; Zierer J.; Gudelj I.; Štambuk J.; Ugrina I.; Rodríguez E.; Soler M. J.; Pavić T.; Šimurina M.; Keser T. Glycosylation Profile of IgG in Moderate Kidney Dysfunction. J. Am. Soc. Nephrol. 2016, 27, 933–941. 10.1681/ASN.2015010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidin M. B.; Keser T.; Gudelj I.; Štambuk J.; Vučenović D.; Allegri M.; Pavić T.; Šimurina M.; Fabiane S. M.; Lauc G.; Williams F. M. K. The Association between Low Back Pain and Composition of IgG Glycome. Sci. Rep. 2016, 6, 26815. 10.1038/srep26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Klarić L.; Yu X.; Thaqi K.; Dong J.; Novokmet M.; Wilson J.; Polasek O.; Liu Y.; Krištić J.; et al. The Association between Glycosylation of Immunoglobulin G and Hypertension: A Multiple Ethnic Cross-Sectional Study. Medicine (Baltimore) 2016, 95, e3379 10.1097/MD.0000000000003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomp R.; Ruhaak L. R.; Uh H.-W.; Reiding K. R.; Selman M.; Houwing-Duistermaat J. J.; Slagboom P. E.; Beekman M.; Wuhrer M. Subclass-Specific IgG Glycosylation Is Associated with Markers of Inflammation and Metabolic Health. Sci. Rep. 2017, 7, 12325. 10.1038/s41598-017-12495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan A.; Kohrt W. M.; Cui J.; Deane K. D.; Pezer M.; Yu E. W.; Hausmann J. S.; Campbell H.; Kaiser U. B.; Rudd P. M.; et al. Estrogens Regulate Glycosylation of IgG in Women and Men. JCI Insight 2017, 2, e89703 10.1172/jci.insight.89703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti E.; Pučić-Baković M.; Keser T.; Wahl A.; Hassinen A.; Yang J. Y.; Liu L.; Trbojević-Akmačić I.; Razdorov G.; Štambuk J.; et al. Network Inference from Glycoproteomics Data Reveals New Reactions in the IgG Glycosylation Pathway. Nat.Commun. 2017, 8, 1483. 10.1038/s41467-017-01525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.; Klarić L.; Sharapov S.; Mangino M.; Ning Z.; Wu D.; Trbojević-Akmačić I.; Pučić-Baković M.; Rudan I.; Polašek O.; et al. Multivariate Discovery and Replication of Five Novel Loci Associated with Immunoglobulin G N-Glycosylation. Nat. Commun. 2017, 8, 447. 10.1038/s41467-017-00453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R. F. H.; Vilaj M.; Urda D.; Agakov F.; Šimurina M.; Klaric L.; Rudan I.; Campbell H.; Hayward C.; Wilson J. F.; et al. IgG Glycan Patterns Are Associated with Type 2 Diabetes in Independent European Populations. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2240–2249. 10.1016/j.bbagen.2017.06.020. [DOI] [PubMed] [Google Scholar]
- Keser T.; Vučković F.; Barrios C.; Zierer J.; Wahl A.; Akinkuolie A. O.; Štambuk J.; Nakić N.; Pavić T.; Periša J.; et al. Effects of Statins on the Immunoglobulin G Glycome. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1152–1158. 10.1016/j.bbagen.2017.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.; Chu X.; Wang H.; Dong J.; Ge S.-Q.; Zhao Z.-Y.; Peng H.-L.; Sun M.; Wu L.-J.; Song M.-S.; et al. The Changes of Immunoglobulin G N-Glycosylation in Blood Lipids and Dyslipidaemia. J. Transl. Med. 2018, 16, 235. 10.1186/s12967-018-1616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. N.; Dolikun M.; Štambuk J.; Trbojević-Akmačić I.; Zhang J.; Wang H.; Zheng D. Q.; Zhang X. Y.; Peng H. L.; Zhao Z. Y.; et al. The Association between Subclass-Specific IgG Fc N-Glycosylation Profiles and Hypertension in the Uygur, Kazak, Kirgiz, and Tajik Populations. J. Hum. Hypertens. 2018, 32, 555–563. 10.1038/s41371-018-0071-0. [DOI] [PubMed] [Google Scholar]
- Šimurina M.; de Haan N.; Vučković F.; Kennedy N. A.; Štambuk J.; Falck D.; Trbojević-Akmačić I.; Clerc F.; Razdorov G.; Khon A.; et al. Glycosylation of Immunoglobulin G Associates with Clinical Features of Inflammatory Bowel Diseases. Gastroenterology 2018, 154, 1320–1333. 10.1053/j.gastro.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H.; Xu X.; Sun F.; Zhang X.; Dong H.; Wang L.; Ge S.; An K.; Sun Q.; Li Y.; et al. Hyperuricemia Is Associated with Immunoglobulin G N-Glycosylation: A Community-Based Study of Glycan Biomarkers. Omics 2019, 23, 660–667. 10.1089/omi.2019.0004. [DOI] [PubMed] [Google Scholar]
- Wang H.; Li X.; Wang X.; Liu D.; Zhang X.; Cao W.; Zheng Y.; Guo Z.; Li D.; Xing W.; et al. Next-Generation (Glycomic) Biomarkers for Cardiometabolic Health: A Community-Based Study of Immunoglobulin G N-Glycans in a Chinese Han Population. Omics 2019, 23, 649–659. 10.1089/omi.2019.0099. [DOI] [PubMed] [Google Scholar]
- Li X.; Wang H.; Russell A.; Cao W.; Wang X.; Ge S.; Zheng Y.; Guo Z.; Hou H.; Song M.; et al. Type 2 Diabetes Mellitus Is Associated with the Immunoglobulin G N-Glycome through Putative Proinflammatory Mechanisms in an Australian Population. Omics 2019, 23, 631–639. 10.1089/omi.2019.0075. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Li H.; Liu D.; Tao L.; Zhang J.; Liang B.; Liu X.; Wang X.; Li X.; Wang Y.; et al. IgG Glycosylation Profile and the Glycan Score Are Associated with Type 2 Diabetes in Independent Chinese Populations: A Case-Control Study. J. Diabetes Res. 2020, 2020, 5041346. 10.1155/2020/5041346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greto V. L.; Cvetko A.; Štambuk T.; Dempster N. J.; Kifer D.; Deriš H.; Cindrić A.; Vučković F.; Falchi M.; Gillies R. S.; et al. Extensive Weight Loss Reduces Glycan Age by Altering IgG N-Glycosylation. Int. J. Obes. 2021, 45, 1521–1531. 10.1038/s41366-021-00816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kifer D.; Louca P.; Cvetko A.; Deriš H.; Cindric A.; Grallert H.; Peters A.; Polašek O.; Gornik O.; Mangino M.; et al. N-Glycosylation of Immunoglobulin G Predicts Incident Hypertension. J. Hypertens. 2021, 39, 2527–2533. 10.1097/HJH.0000000000002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.; Song M.; Vilaj M.; Štambuk J.; Dolikun M.; Zhang J.; Liu D.; Wang H.; Zhang X.; Zhang J. Glycosylation of IgG Associates with Hypertension and Type 2 Diabetes Mellitus Comorbidity in the Chinese Muslim Ethnic Minorities and the Han Chinese. J. Pers. Med. 2021, 11, 614. 10.3390/jpm11070614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriš H.; Kifer D.; Cindrić A.; Petrović T.; Cvetko A.; Trbojević-Akmačić I.; Kolčić I.; Polašek O.; Newson L.; Spector T.; et al. Immunoglobulin G Glycome Composition in Transition from Premenopause to Postmenopause. iScience 2022, 25, 103897. 10.1016/j.isci.2022.103897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhooren V.; Dewaele S.; Libert C.; Engelborghs S.; De Deyn P. P.; Toussaint O.; Debacq-Chainiaux F.; Poulain M.; Glupczynski Y.; Franceschi C.; et al. Serum N-Glycan Profile Shift During Human Ageing. Exp. Gerontol. 2010, 45, 738–743. 10.1016/j.exger.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Vilar-Bergua A.; Riba-Llena I.; Vanhooren V.; Dewaele S.; Libert C.; Penalba A.; Montaner J.; Delgado P. N-Glycome Profile Levels Relate to Silent Brain Infarcts in a Cohort of Hypertensives. J. Am. Heart Assoc. 2015, 4, e002669 10.1161/JAHA.115.002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa R.; Vanhooren V.; Bonfigli A. R.; Boemi M.; Olivieri F.; Ceriello A.; Genovese S.; Spazzafumo L.; Borelli V.; Bacalini M. G.; et al. N-Glycomic Changes in Serum Proteins in Type 2 Diabetes Mellitus Correlate with Complications and with Metabolic Syndrome Parameters. PLoS One 2015, 10, e0119983 10.1371/journal.pone.0119983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R.; Haakensen V. D.; Rødland E.; Walsh I.; Stöckmann H.; Engebraaten O.; Børresen-Dale A.-L.; Rudd P. M. Serum N-Glycome Alterations in Breast Cancer During Multimodal Treatment and Follow-Up. Mol. Oncol. 2017, 11, 1361–1379. 10.1002/1878-0261.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiding K. R.; Vreeker G. C. M.; Bondt A.; Bladergroen M. R.; Hazes J. M. W.; van der Burgt Y. E. M.; Wuhrer M.; Dolhain R. Serum Protein N-Glycosylation Changes with Rheumatoid Arthritis Disease Activity During and after Pregnancy. Front. Med. 2018, 4, 241. 10.3389/fmed.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igl W.; Polašek O.; Gornik O.; Knežević A.; Pučić M.; Novokmet M.; Huffman J.; Gnewuch C.; Liebisch G.; Rudd P. M.; et al. Glycomics Meets Lipidomics—Associations of N-Glycans with Classical Lipids, Glycerophospholipids, and Sphingolipids in Three European Populations. Mol. Biosyst. 2011, 7, 1852–1862. 10.1039/c0mb00095g. [DOI] [PubMed] [Google Scholar]
- Lu J.-P.; Knežević A.; Wang Y.-X.; Rudan I.; Campbell H.; Zou Z.-K.; Lan J.; Lai Q.-X.; Wu J.-J.; He Y.; et al. Screening Novel Biomarkers for Metabolic Syndrome by Profiling Human Plasma N-Glycans in Chinese Han and Croatian Populations. J. Proteome Res. 2011, 10, 4959–4969. 10.1021/pr2004067. [DOI] [PubMed] [Google Scholar]
- Pivac N.; Knezević A.; Gornik O.; Pucić M.; Igl W.; Peeters H.; Crepel A.; Steyaert J.; Novokmet M.; Redzić I.; et al. Human Plasma Glycome in Attention-Deficit Hyperactivity Disorder and Autism Spectrum Disorders. Mol. Cell Proteomics 2011, 10, M110.004200. 10.1074/mcp.M110.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R.; Huffman J. E.; Adamczyk B.; Mužinić A.; Kattla J. J.; Pučić M.; Novokmet M.; Abrahams J. L.; Hayward C.; Rudan I.; et al. Association of Medication with the Human Plasma N-Glycome. J. Proteome Res. 2012, 11, 1821–1831. 10.1021/pr2010605. [DOI] [PubMed] [Google Scholar]
- Thanabalasingham G.; Huffman J. E.; Kattla J. J.; Novokmet M.; Rudan I.; Gloyn A. L.; Hayward C.; Adamczyk B.; Reynolds R. M.; Muzinic A.; et al. Mutations in HNF1A Result in Marked Alterations of Plasma Glycan Profile. Diabetes 2013, 62, 1329–1337. 10.2337/db12-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Arnold M.; Bhagwat A. M.; Cotton R. J.; Engelke R.; Raffler J.; Sarwath H.; Thareja G.; Wahl A.; DeLisle R. K.; et al. Connecting Genetic Risk to Disease End Points through the Human Blood Plasma Proteome. Nat.Commun. 2017, 8, 14357. 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adua E.; Roberts P.; Wang W. Incorporation of Suboptimal Health Status as a Potential Risk Assessment for Type II Diabetes Mellitus: A Case-Control Study in a Ghanaian Population. EPMA Journal 2017, 8, 345–355. 10.1007/s13167-017-0119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keser T.; Gornik I.; Vučković F.; Selak N.; Pavić T.; Lukić E.; Gudelj I.; Gašparović H.; Biočina B.; Tilin T.; et al. Increased Plasma N-Glycome Complexity Is Associated with Higher Risk of Type 2 Diabetes. Diabetologia 2017, 60, 2352–2360. 10.1007/s00125-017-4426-9. [DOI] [PubMed] [Google Scholar]
- Dotz V.; Lemmers R. F.; Reiding K. R.; Hipgrave Ederveen A. L. H.; Lieverse A. G.; Mulder M. T.; Sijbrands E. J.; Wuhrer M.; van Hoek M. Plasma Protein N-Glycan Signatures of Type 2 Diabetes. Biochim. Biophys. Acta, Gen. Subj. 2018, 1862, 2613–2622. 10.1016/j.bbagen.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Clerc F.; Novokmet M.; Dotz V.; Reiding K. R.; de Haan N.; Kammeijer G. S.; Dalebout H.; Bladergroen M. R.; Vukovic F.; Rapp E. Plasma N-Glycan Signatures Are Associated with Features of Inflammatory Bowel Diseases. Gastroenterology 2018, 155, 829–843. 10.1053/j.gastro.2018.05.030. [DOI] [PubMed] [Google Scholar]
- Doherty M.; Theodoratou E.; Walsh I.; Adamczyk B.; Stockmann H.; Agakov F.; Timofeeva M.; Trbojevic-Akmacic I.; Vuckovic F.; Duffy F.; et al. Plasma N-Glycans in Colorectal Cancer Risk. Sci. Rep. 2018, 8, 8655. 10.1038/s41598-018-26805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharapov S. Z.; Tsepilov Y. A.; Klaric L.; Mangino M.; Thareja G.; Shadrina A. S.; Simurina M.; Dagostino C.; Dmitrieva J.; Vilaj M.; et al. Defining the Genetic Control of Human Blood Plasma N-Glycome Using Genome-Wide Association Study. Hum. Mol. Genet. 2019, 28, 2062–2077. 10.1093/hmg/ddz054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Uh H.-W.; Beekman M.; Hokke C. H.; Westendorp R. G. J.; Houwing-Duistermaat J.; Wuhrer M.; Deelder A. M.; Slagboom P. E. Plasma Protein N-Glycan Profiles Are Associated with Calendar Age, Familial Longevity and Health. J. Proteome Res. 2011, 10, 1667–1674. 10.1021/pr1009959. [DOI] [PubMed] [Google Scholar]
- Sharapov S. Z.; Shadrina A. S.; Tsepilov Y. A.; Elgaeva E. E.; Tiys E. S.; Feoktistova S. G.; Zaytseva O. O.; Vuckovic F.; Cuadrat R.; Jäger S.; et al. Replication of 15 Loci Involved in Human Plasma Protein N-Glycosylation in 4802 Samples from Four Cohorts. Glycobiology 2021, 31, 82–88. 10.1093/glycob/cwaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adua E.; Memarian E.; Afrifa-Yamoah E.; Russell A.; Trbojević-Akmačić I.; Gudelj I.; Jurić J.; Roberts P.; Lauc G.; Wang W. N-Glycosylation Profiling of Type 2 diabetes Mellitus from Baseline to Follow-Up: An Observational Study in a Ghanaian Population. Biomarkn Med. 2021, 15, 467–480. 10.2217/bmm-2020-0615. [DOI] [PubMed] [Google Scholar]
- Cvetko A.; Mangino M.; Tijardović M.; Kifer D.; Falchi M.; Keser T.; Perola M.; Spector T. D.; Lauc G.; Menni C.; Gornik O. Plasma N-Glycome Shows Continuous Deterioration as the Diagnosis of Insulin Resistance Approaches. BMJ Open Diabetes Res Care 2021, 9, e002263. 10.1136/bmjdrc-2021-002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarian E.; ‘t Hart L. M.; Slieker R. C.; Lemmers R. F. L.; van der Heijden A. A.; Rutters F.; Nijpels G.; Schoep E.; Lieverse A. G.; Sijbrands E. J. G. Plasma Protein N-Glycosylation Is Associated with Cardiovascular Disease, Nephropathy, and Retinopathy in Type 2 Diabetes. BMJ Open Diabetes Res. Care 2021, 9, e002345. 10.1136/bmjdrc-2021-002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bućan I.; Škunca Herman J.; Jerončić Tomić I.; Gornik O.; Vatavuk Z.; Bućan K.; Lauc G.; Polašek O. N-Glycosylation Patterns across the Age-Related Macular Degeneration Spectrum. Molecules 2022, 27, 1774. 10.3390/molecules27061774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A.; Nicolardi S.; Jansen B. C.; Kuijper T. M.; Hazes J. M. W.; van der Burgt Y. E. M.; Wuhrer M.; Dolhain R. IgA N- and O-Glycosylation Profiling Reveals No Association with the Pregnancy-Related Improvement in Rheumatoid Arthritis. Arthritis Res. Ther. 2017, 19, 160. 10.1186/s13075-017-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demus D.; Naber A.; Dotz V.; Jansen B. C.; Bladergroen M. R.; Nouta J.; Sijbrands E. J. G.; Van Hoek M.; Nicolardi S.; Wuhrer M. Large-Scale Analysis of Apolipoprotein CIII Glycosylation by Ultrahigh Resolution Mass Spectrometry. Front. Chem. 2021, 9, 678883. 10.3389/fchem.2021.678883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen J. M.; Young R.; Ashorn P.; Ashorn U.; Chaima D.; Davis J. C. C.; Goonatilleke E.; Kumwenda C.; Lebrilla C. B.; Maleta K. Associations of Human Milk Oligosaccharides and Bioactive Proteins with Infant Morbidity and Inflammation in Malawian Mother-Infant Dyads. Curr. Dev. Nutr. 2021, 5, nzab072. 10.1093/cdn/nzab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijardović M.; Štambuk T.; Juszczak A.; Keser T.; Gasperikova D.; Novokmet M.; Tjora E.; Pape Medvidović E.; Stanik J.; Rasmus Njølstad P.; et al. Fucosylated AGP Glycopeptides as Biomarkers of HNF1A-Maturity Onset Diabetes of the Young. Diabetes Res. Clin. Pract. 2022, 185, 109226. 10.1016/j.diabres.2022.109226. [DOI] [PubMed] [Google Scholar]
- Landini A.; Trbojević-Akmačić I.; Navarro P.; Tsepilov Y. A.; Sharapov S. Z.; Vučković F.; Polašek O.; Hayward C.; Petrović T.; Vilaj M.; et al. Genetic Regulation of Post-Translational Modification of Two Distinct Proteins. Nat. Commun. 2022, 13, 1586. 10.1038/s41467-022-29189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellokumpu S. Golgi pH, Ion and Redox Homeostasis: How Much Do They Really Matter?. Front. Cell Dev. Biol. 2019, 7, 93. 10.3389/fcell.2019.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maverakis E.; Kim K.; Shimoda M.; Gershwin M. E.; Patel F.; Wilken R.; Raychaudhuri S.; Ruhaak L. R.; Lebrilla C. B. Glycans in the Immune System and the Altered Glycan Theory of Autoimmunity: A Critical Review. J. Autoimmun. 2015, 57, 1–13. 10.1016/j.jaut.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindzienska-Sieskiewicz E.; Klimiuk P. A.; Kisiel D. G.; Gindzienski A.; Sierakowski S. The Changes in Monosaccharide Composition of Immunoglobulin G in the Course of Rheumatoid Arthritis. Clin. Rheumatol. 2007, 26, 685–690. 10.1007/s10067-006-0370-7. [DOI] [PubMed] [Google Scholar]
- Gudelj I.; Salo P. P.; Trbojević-Akmačić I.; Albers M.; Primorac D.; Perola M.; Lauc G. Low Galactosylation of IgG Associates with Higher Risk for Future Diagnosis of Rheumatoid Arthritis During 10 Years of Follow-Up. Biochim. Biophys. Acta, Mol. Basis Dis. 2018, 1864, 2034–2039. 10.1016/j.bbadis.2018.03.018. [DOI] [PubMed] [Google Scholar]
- Shinzaki S.; Iijima H.; Nakagawa T.; Egawa S.; Nakajima S.; Ishii S.; Irie T.; Kakiuchi Y.; Nishida T.; Yasumaru M.; et al. IgG Oligosaccharide Alterations Are a Novel Diagnostic Marker for Disease Activity and the Clinical Course of Inflammatory Bowel Disease. Am. J. Gastroenterol. 2008, 103, 1173–1181. 10.1111/j.1572-0241.2007.01699.x. [DOI] [PubMed] [Google Scholar]
- Nakajima S.; Iijima H.; Shinzaki S.; Egawa S.; Inoue T.; Mukai A.; Hayashi Y.; Kondo J.; Akasaka T.; Nishida T.; et al. Functional Analysis of Agalactosyl IgG in Inflammatory Bowel Disease Patients. Inflamm. Bowel Dis. 2011, 17, 927–936. 10.1002/ibd.21459. [DOI] [PubMed] [Google Scholar]
- Fokkink W.-J. R.; Selman M. H. J.; Dortland J. R.; Durmuş B.; Kuitwaard K.; Huizinga R.; van Rijs W.; Tio-Gillen A. P.; van Doorn P. A.; Deelder A. M.; et al. IgG Fc N-Glycosylation in Guillain-Barré Syndrome Treated with Immunoglobulins. J. Proteome Res. 2014, 13, 1722–1730. 10.1021/pr401213z. [DOI] [PubMed] [Google Scholar]
- Barber M. R. W.; Drenkard C.; Falasinnu T.; Hoi A.; Mak A.; Kow N. Y.; Svenungsson E.; Peterson J.; Clarke A. E.; Ramsey-Goldman R. Global Epidemiology of Systemic Lupus Erythematosus. Nat. Rev. Rheumatol. 2021, 17, 515–532. 10.1038/s41584-021-00668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron L. B.; Papasavvas E.; Azzoni L.; Yin X.; Anzurez A.; Damra M.; Mounzer K.; Kostman J. R.; Sanne I.; Firnhaber C. S.; et al. Plasma and Antibody Glycomic Biomarkers of Time to HIV Rebound and Viral Setpoint. AIDS 2020, 34, 681–686. 10.1097/QAD.0000000000002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrović T.; Alves I.; Bugada D.; Pascual J.; Vučković F.; Skelin A.; Gaifem J.; Villar-Garcia J.; Vicente M. M.; Fernandes Â.; et al. Composition of the Immunoglobulin G Glycome Associates with the Severity of COVID-19. Glycobiology 2021, 31, 372–377. 10.1093/glycob/cwaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H.; Yang H.; Liu P.; Huang C.; Wang M.; Li Y.; Zhu M.; Wang J.; Xu Y.; Wang Y. Profile of Immunoglobulin G N-Glycome in COVID-19 Patients: A Case-Control Study. Front. Immunol. 2021, 12, 748566. 10.3389/fimmu.2021.748566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster M. M.; Esko J. D. The Sweet and Sour of Cancer: Glycans as Novel Therapeutic Targets. Nat. Rev. Cancer 2005, 5, 526–542. 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Glycosylation Defining Cancer Malignancy: New Wine in an Old Bottle. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 10231–10233. 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis C. A.; Osorio H.; Silva L.; Gomes C.; David L. Alterations in Glycosylation as Biomarkers for Cancer Detection. J. Clin. Pathol. 2010, 63, 322–329. 10.1136/jcp.2009.071035. [DOI] [PubMed] [Google Scholar]
- Taniguchi N.; Hancock W.; Lubman D. M.; Rudd P. M. The Second Golden Age of Glycomics: From Functional Glycomics to Clinical Applications. J. Proteome Res. 2009, 8, 425–426. 10.1021/pr801057j. [DOI] [PubMed] [Google Scholar]
- Freeze H. H. Understanding Human Glycosylation Disorders: Biochemistry Leads the Charge. J. Biol. Chem. 2013, 288, 6936–6945. 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D. H.; Bertozzi C. R. Glycans in Cancer and Inflammation — Potential for Therapeutics and Diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Pinho S. S.; Reis C. A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Cheeseman J.; Kuhnle G.; Stafford G.; Gardner R. A.; Spencer D. I.; Osborn H. M. Sialic Acid as a Potential Biomarker for Cardiovascular Disease, Diabetes and Cancer. Biomark. Med. 2021, 15, 911–928. 10.2217/bmm-2020-0776. [DOI] [PubMed] [Google Scholar]
- Gilgunn S.; Conroy P. J.; Saldova R.; Rudd P. M.; O’Kennedy R. J. Aberrant PSA Glycosylation-a Sweet Predictor of Prostate Cancer. Nat. Rev. Urol. 2013, 10, 99–107. 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- Locker G. Y.; Hamilton S.; Harris J.; Jessup J. M.; Kemeny N.; Macdonald J. S.; Somerfield M. R.; Hayes D. F.; Bast R. C. Jr. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. J. Clin. Oncol. 2006, 24, 5313–5327. 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- Zurawski V. R. Jr.; Orjaseter H.; Andersen A.; Jellum E. Elevated Serum CA 125 Levels Prior to Diagnosis of Ovarian Neoplasia: Relevance for Early Detection of Ovarian Cancer. Int. J. Cancer 1988, 42, 677–680. 10.1002/ijc.2910420507. [DOI] [PubMed] [Google Scholar]
- Goldstein M. J.; Mitchell E. P. Carcinoembryonic Antigen in the Staging and Follow-up of Patients with Colorectal Cancer. Cancer Invest. 2005, 23, 338–351. 10.1081/CNV-58878. [DOI] [PubMed] [Google Scholar]
- Ebeling F. G.; Stieber P.; Untch M.; Nagel D.; Konecny G. E.; Schmitt U. M.; Fateh-Moghadam A.; Seidel D. Serum CEA and CA 15–3 as Prognostic Factors in Primary Breast Cancer. Br. J. Cancer 2002, 86, 1217–1222. 10.1038/sj.bjc.6600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpulainen E. J.; Keskikuru R. J.; Johansson R. T. Serum Tumor Marker CA 15.3 and Stage Are the Two Most Powerful Predictors of Survival in Primary Breast Cancer. Breast Cancer Res. Treat. 2002, 76, 95–102. 10.1023/A:1020514925143. [DOI] [PubMed] [Google Scholar]
- Safi F.; Schlosser W.; Kolb G.; Beger H. G. Diagnostic Value of CA 19–9 in Patients with Pancreatic Cancer and Nonspecific Gastrointestinal Symptoms. J. Gastrointest. Surg. 1997, 1, 106–112. 10.1016/S1091-255X(97)80097-2. [DOI] [PubMed] [Google Scholar]
- Christiansen M. N.; Chik J.; Lee L.; Anugraham M.; Abrahams J. L.; Packer N. H. Cell Surface Protein Glycosylation in Cancer. Proteomics 2014, 14, 525–546. 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- Arnold J. N.; Saldova R.; Hamid U. M.; Rudd P. M. Evaluation of the Serum N-Linked Glycome for the Diagnosis of Cancer and Chronic Inflammation. Proteomics 2008, 8, 3284–3293. 10.1002/pmic.200800163. [DOI] [PubMed] [Google Scholar]
- Kim Y. J.; Varki A. Perspectives on the Significance of Altered Glycosylation of Glycoproteins in Cancer. Glycoconj. J. 1997, 14, 569–576. 10.1023/A:1018580324971. [DOI] [PubMed] [Google Scholar]
- Laidler P.; Lityńska A.; Hoja-Łukowicz D.; Łabedz M.; Przybyło M.; Ciołczyk-Wierzbicka D.; Pocheć E.; Trebacz E.; Kremser E. Characterization of Glycosylation and Adherent Properties of Melanoma Cell Lines. Cancer Immunol. Immunother. 2006, 55, 112–118. 10.1007/s00262-005-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremser M. E.; Przybyło M.; Hoja-Łukowicz D.; Pocheć E.; Amoresano A.; Carpentieri A.; Bubka M.; Lityńska A. Characterisation of Alpha3beta1 and Alpha(V)Beta3 Integrin N-Oligosaccharides in Metastatic Melanoma WM9 and WM239 Cell Lines. Biochim. Biophys. Acta 2008, 1780, 1421–1431. 10.1016/j.bbagen.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Carvalho A. S.; Harduin-Lepers A.; Magalhães A.; Machado E.; Mendes N.; Costa L. T.; Matthiesen R.; Almeida R.; Costa J.; Reis C. A. Differential Expression of Alpha-2,3-Sialyltransferases and Alpha-1,3/4-Fucosyltransferases Regulates the Levels of Sialyl Lewis a and Sialyl Lewis X in Gastrointestinal Carcinoma Cells. Int. J. Biochem. Cell Biol. 2010, 42, 80–89. 10.1016/j.biocel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Liu Y. C.; Yen H. Y.; Chen C. Y.; Chen C. H.; Cheng P. F.; Juan Y. H.; Chen C. H.; Khoo K. H.; Yu C. J.; Yang P. C.; et al. Sialylation and Fucosylation of Epidermal Growth Factor Receptor Suppress Its Dimerization and Activation in Lung Cancer Cells. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 11332–11337. 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapenko I. O.; Haakensen V. D.; Lüders T.; Helland A.; Bukholm I.; Sørlie T.; Kristensen V. N.; Lingjaerde O. C.; Børresen-Dale A. L. Glycan Gene Expression Signatures in Normal and Malignant Breast Tissue; Possible Role in Diagnosis and Progression. Mol. Oncol. 2010, 4, 98–118. 10.1016/j.molonc.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson W. L.; Du M. Q.; Johnson P. J.; Williams R. Fucosyltransferases: Differential Plasma and Tissue Alterations in Hepatocellular Carcinoma and Cirrhosis. Hepatology 1991, 13, 683–688. 10.1002/hep.1840130412. [DOI] [PubMed] [Google Scholar]
- Noda K.; Miyoshi E.; Uozumi N.; Yanagidani S.; Ikeda Y.; Gao C.; Suzuki K.; Yoshihara H.; Yoshikawa K.; Kawano K.; et al. Gene Expression of Alpha1–6 Fucosyltransferase in Human Hepatoma Tissues: A Possible Implication for Increased Fucosylation of Alpha-Fetoprotein. Hepatology 1998, 28, 944–952. 10.1002/hep.510280408. [DOI] [PubMed] [Google Scholar]
- West C. A.; Wang M.; Herrera H.; Liang H.; Black A.; Angel P. M.; Drake R. R.; Mehta A. S. N-Linked Glycan Branching and Fucosylation Are Increased Directly in HCC Tissue as Determined through in Situ Glycan Imaging. J. Proteome Res. 2018, 17, 3454–3462. 10.1021/acs.jproteome.8b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos D.; Freitas D.; Gomes J.; Magalhães A.; Steentoft C.; Gomes C.; Vester-Christensen M. B.; Ferreira J. A.; Afonso L. P.; Santos L. L.; et al. Probing the O-Glycoproteome of Gastric Cancer Cell Lines for Biomarker Discovery. Mol. Cell Proteomics 2015, 14, 1616–1629. 10.1074/mcp.M114.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka M. R.; Ju T.; Heimburg-Molinaro J.; Cummings R. D. Simple Sugars to Complex Disease-Mucin-Type O-Glycans in Cancer. Adv. Cancer Res. 2015, 126, 53–135. 10.1016/bs.acr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C.; Vakhrushev S. Y.; Vester-Christensen M. B.; Schjoldager K. T. B. G.; Kong Y.; Bennett E. P.; Mandel U.; Wandall H.; Levery S. B.; Clausen H. Mining the O-Glycoproteome Using Zinc-Finger Nuclease-Glycoengineered SimpleCell Lines. Nat. Methods 2011, 8, 977–982. 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Gomes C.; Almeida A.; Ferreira J. A.; Silva L.; Santos-Sousa H.; Pinto-de-Sousa J.; Santos L. L.; Amado F.; Schwientek T.; Levery S. B.; et al. Glycoproteomic Analysis of Serum from Patients with Gastric Precancerous Lesions. J. Proteome Res. 2013, 12, 1454–1466. 10.1021/pr301112x. [DOI] [PubMed] [Google Scholar]
- Gizaw S. T.; Gaunitz S.; Novotny M. V. Highly Sensitive O-Glycan Profiling for Human Serum Proteins Reveals Gender-Dependent Changes in Colorectal Cancer Patients. Anal. Chem. 2019, 91, 6180–6189. 10.1021/acs.analchem.9b00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K.; Satoh T.; Baba S.; Yamashita K. α1,2-Fucosylated and β-N-Acetylgalactosaminylated Prostate-Specific Antigen as an Efficient Marker of Prostatic Cancer. Glycobiology 2010, 20, 452–460. 10.1093/glycob/cwp197. [DOI] [PubMed] [Google Scholar]
- Jankovic M. M.; Milutinovic B. S. Glycoforms of CA125 Antigen as a Possible Cancer Marker. Cancer Biomark. 2008, 4, 35–42. 10.3233/CBM-2008-4104. [DOI] [PubMed] [Google Scholar]
- Saeland E.; Belo A. I.; Mongera S.; van Die I.; Meijer G. A.; van Kooyk Y. Differential Glycosylation of MUC1 and CEACAM5 between Normal Mucosa and Tumour Tissue of Colon Cancer Patients. Int. J. Cancer 2012, 131, 117–128. 10.1002/ijc.26354. [DOI] [PubMed] [Google Scholar]
- Adamczyk B.; Tharmalingam T.; Rudd P. M. Glycans as Cancer Biomarkers. Biochim. Biophys. Acta, Gen. Subj. 2012, 1820, 1347–1353. 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Miyoshi E.; Nakano M. Fucosylated Haptoglobin Is a Novel Marker for Pancreatic Cancer: Detailed Analyses of Oligosaccharide Structures. Proteomics 2008, 8, 3257–3262. 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- Bones J.; Mittermayr S.; O’Donoghue N.; Guttman A.; Rudd P. M. Ultra Performance Liquid Chromatographic Profiling of Serum N-Glycans for Fast and Efficient Identification of Cancer Associated Alterations in Glycosylation. Anal. Chem. 2010, 82, 10208–10215. 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- Saldova R.; Fan Y.; Fitzpatrick J. M.; Watson R. W.; Rudd P. M. Core Fucosylation and Alpha2–3 Sialylation in Serum N-Glycome Is Significantly Increased in Prostate Cancer Comparing to Benign Prostate Hyperplasia. Glycobiology 2011, 21, 195–205. 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- Vermassen T.; Van Praet C.; Vanderschaeghe D.; Maenhout T.; Lumen N.; Callewaert N.; Hoebeke P.; Van Belle S.; Rottey S.; Delanghe J. Capillary Electrophoresis of Urinary Prostate Glycoproteins Assists in the Diagnosis of Prostate Cancer. Electrophoresis 2014, 35, 1017–1024. 10.1002/elps.201300332. [DOI] [PubMed] [Google Scholar]
- Vermassen T.; Van Praet C.; Lumen N.; Decaestecker K.; Vanderschaeghe D.; Callewaert N.; Villeirs G.; Hoebeke P.; Van Belle S.; Rottey S.; et al. Urinary Prostate Protein Glycosylation Profiling as a Diagnostic Biomarker for Prostate Cancer. Prostate 2015, 75, 314–322. 10.1002/pros.22918. [DOI] [PubMed] [Google Scholar]
- Yoneyama T.; Ohyama C.; Hatakeyama S.; Narita S.; Habuchi T.; Koie T.; Mori K.; Hidari K. I.; Yamaguchi M.; Suzuki T.; et al. Measurement of Aberrant Glycosylation of Prostate Specific Antigen Can Improve Specificity in Early Detection of Prostate Cancer. Biochem. Biophys. Res. Commun. 2014, 448, 390–396. 10.1016/j.bbrc.2014.04.107. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Li T.; Xie J.; Zhang D.; Pi C.; Zhou L.; Yang W. Gold Standard for Nutrition: A Review of Human Milk Oligosaccharide and Its Effects on Infant Gut Microbiota. Microb. Cell Fact. 2021, 20, 108. 10.1186/s12934-021-01599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N.; Tytgat H. L. P.; Binia A.; Austin S.; Singhal A. Biology of Human Milk Oligosaccharides: From Basic Science to Clinical Evidence. J. Hum. Nutr. Diet. 2022, 35, 280–299. 10.1111/jhn.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albaladejo A. S.; Completo G. C.; Navarro B. R.; Nacario R.; Lebrilla C. Rapid-Throughput Analysis of Human Milk Oligosaccharides from Filipino Breastmilk. KIMIKA 2021, 32, 11–25. [Google Scholar]
- Charbonneau M. R.; O’Donnell D.; Blanton L. V.; Totten S. M.; Davis J. C.; Barratt M. J.; Cheng J.; Guruge J.; Talcott M.; Bain J. R.; et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell 2016, 164, 859–871. 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire M. K.; Meehan C. L.; McGuire M. A.; Williams J. E.; Foster J.; Sellen D. W.; Kamau-Mbuthia E. W.; Kamundia E. W.; Mbugua S.; Moore S. E.; et al. What’s Normal? Oligosaccharide Concentrations and Profiles in Milk Produced by Healthy Women Vary Geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plows J. F.; Berger P. K.; Jones R. B.; Alderete T. L.; Yonemitsu C.; Najera J. A.; Khwajazada S.; Bode L.; Goran M. I. Longitudinal Changes in Human Milk Oligosaccharides (HMOs) over the Course of 24 Months of Lactation. J. Nutr. 2021, 151, 876–882. 10.1093/jn/nxaa427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binia A.; Lavalle L.; Chen C.; Austin S.; Agosti M.; Al-Jashi I.; Pereira A. B.; Costeira M. J.; Silva M. G.; Marchini G.; et al. Human Milk Oligosaccharides, Infant Growth, and Adiposity over the First 4 Months of Lactation. Pediatr. Res. 2021, 90, 684–693. 10.1038/s41390-020-01328-y. [DOI] [PubMed] [Google Scholar]
- Miliku K.; Robertson B.; Sharma A. K.; Subbarao P.; Becker A. B.; Mandhane P. J.; Turvey S. E.; Lefebvre D. L.; Sears M. R.; Investigators t. C. S.; et al. Human Milk Oligosaccharide Profiles and Food Sensitization among Infants in the Child Study. Allergy 2018, 73, 2070–2073. 10.1111/all.13476. [DOI] [PubMed] [Google Scholar]
- Lodge C. J.; Lowe A. J.; Milanzi E.; Bowatte G.; Abramson M. J.; Tsimiklis H.; Axelrad C.; Robertson B.; Darling A. E.; Svanes C.; et al. Human Milk Oligosaccharide Profiles and Allergic Disease up to 18 Years. J. Allergy Clin. Immunol. 2021, 147, 1041–1048. 10.1016/j.jaci.2020.06.027. [DOI] [PubMed] [Google Scholar]
- Grabarics M.; Csernák O.; Balogh R.; Béni S. Analytical Characterization of Human Milk Oligosaccharides - Potential Applications in Pharmaceutical Analysis. J. Pharm. Biomed. Anal. 2017, 146, 168–178. 10.1016/j.jpba.2017.08.039. [DOI] [PubMed] [Google Scholar]
- Reusch D.; Tejada M. L. Fc Glycans of Therapeutic Antibodies as Critical Quality Attributes. Glycobiology 2015, 25, 1325–1334. 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; DiLillo D. J.; Bournazos S.; Giddens J. P.; Ravetch J. V.; Wang L.-X. Modulating IgG Effector Function by Fc Glycan Engineering. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 3485–3490. 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkawa T.; Nakamura K.; Yamane N.; Shoji-Hosaka E.; Kanda Y.; Sakurada M.; Uchida K.; Anazawa H.; Satoh M.; Yamasaki M.; et al. The Absence of Fucose but Not the Presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-Type Oligosaccharides Shows the Critical Role of Enhancing Antibody-Dependent Cellular Cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Hossler P.; Khattak S. F.; Li Z. J. Optimal and Consistent Protein Glycosylation in Mammalian Cell Culture. Glycobiology 2009, 19, 936–949. 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- De Leoz M. L. A.; Duewer D. L.; Fung A.; Liu L.; Yau H. K.; Potter O.; Staples G. O.; Furuki K.; Frenkel R.; Hu Y.; et al. NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods. Mol. Cell Proteomics 2020, 19, 11–30. 10.1074/mcp.RA119.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Mota L. M.; Bergin A.; O’Flaherty R.; Jones A.; Morgan B.; Butler M. High-Throughput and High-Sensitivity N-Glycan Profiling: A Platform for Biopharmaceutical Development and Disease Biomarker Discovery. Anal. Biochem. 2021, 623, 114205. 10.1016/j.ab.2021.114205. [DOI] [PubMed] [Google Scholar]
- Reusch D.; Haberger M.; Falck D.; Peter B.; Maier B.; Gassner J.; Hook M.; Wagner K.; Bonnington L.; Bulau P.; et al. Comparison of Methods for the Analysis of Therapeutic Immunoglobulin G Fc-Glycosylation Profiles-Part 2: Mass Spectrometric Methods. mAbs 2015, 7, 732–742. 10.1080/19420862.2015.1045173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Poza S.; Padros A.; Thompson R.; Butler L.; Islam M.; Mosely J. A.; Scrivens J. H.; F Rehman M.; Akram M. S. Tailor-Made Recombinant Prokaryotic Lectins for Characterisation of Glycoproteins. Anal. Chim. Acta 2021, 1155, 338352. 10.1016/j.aca.2021.338352. [DOI] [PubMed] [Google Scholar]
- Houel S.; Hilliard M.; Yu Y. Q.; McLoughlin N.; Martin S. M.; Rudd P. M.; Williams J. P.; Chen W. N- and O-Glycosylation Analysis of Etanercept Using Liquid Chromatography and Quadrupole Time-of-Flight Mass Spectrometry Equipped with Electron-Transfer Dissociation Functionality. Anal. Chem. 2014, 86, 576–584. 10.1021/ac402726h. [DOI] [PubMed] [Google Scholar]
- Hashii N.; Suzuki J.; Hanamatsu H.; Furukawa J.-i.; Ishii-Watabe A. In-Depth Site-Specific O-Glycosylation Analysis of Therapeutic Fc-Fusion Protein by Electron-Transfer/Higher-Energy Collisional Dissociation Mass Spectrometry. Biologicals 2019, 58, 35–43. 10.1016/j.biologicals.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Bataller R.; Brenner D. A. Liver Fibrosis. J. Clin. Investig. 2005, 115, 209–218. 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschaeghe D.; Laroy W.; Sablon E.; Halfon P.; Van Hecke A.; Delanghe J.; Callewaert N. GlycoFibroTest Is a Highly Performant Liver Fibrosis Biomarker Derived from DNA Sequencer-Based Serum Protein Glycomics. Mol. Cell Proteomics 2009, 8, 986–994. 10.1074/mcp.M800470-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.; Shang Q. H.; Chi X. L.; Zhang W.; Xiao H. M.; Sun M. M.; Chen G.; An Y.; Lv C. L.; Wang L.; et al. Serum N-Glycan Markers for Diagnosing Liver Fibrosis Induced by Hepatitis B Virus. World J. Gastroenterol. 2020, 26, 1067–1079. 10.3748/wjg.v26.i10.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S.; Zheng R.; Zhang S.; Wang S.; Chen R.; Sun K.; Zeng H.; Zhou J.; Wei W. Global Patterns of Breast Cancer Incidence and Mortality: A Population-Based Cancer Registry Data Analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. 10.1002/cac2.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd Hamid U. M.; Royle L.; Saldova R.; Radcliffe C. M.; Harvey D. J.; Storr S. J.; Pardo M.; Antrobus R.; Chapman C. J.; Zitzmann N. A Strategy to Reveal Potential Glycan Markers from Serum Glycoproteins Associated with Breast Cancer Progression. Glycobiology 2008, 18, 1105–1118. 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- Scott D. A.; Drake R. R. Glycosylation and Its Implications in Breast Cancer. Expert Rev. Proteomics 2019, 16, 665–680. 10.1080/14789450.2019.1645604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L.; Wang Y.; Xie Q.; Xu X.; Li Y.; Chen Z.; Li Y. Elevated Level of Serum Glycoprotein Bifucosylation and Prognostic Value in Chinese Breast Cancer. Glycobiology 2016, 26, 460–471. 10.1093/glycob/cwv117. [DOI] [PubMed] [Google Scholar]
- Terkelsen T.; Haakensen V. D.; Saldova R.; Gromov P.; Hansen M. K.; Stöckmann H.; Lingjaerde O. C.; Børresen-Dale A. L.; Papaleo E.; Helland Å.; et al. N-Glycan Signatures Identified in Tumor Interstitial Fluid and Serum of Breast Cancer Patients: Association with Tumor Biology and Clinical Outcome. Mol. Oncol. 2018, 12, 972–990. 10.1002/1878-0261.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeker G. C. M.; Vangangelt K. M. H.; Bladergroen M. R.; Nicolardi S.; Mesker W. E.; Wuhrer M.; van der Burgt Y. E. M.; Tollenaar R. A. E. M. Serum N-Glycan Profiles Differ for Various Breast Cancer Subtypes. Glycoconj. J. 2021, 38, 387–395. 10.1007/s10719-021-10001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S. R.; Ju T.; Cummings R. D. Protein Glycosylation in Cancer. Annu. Rev. Pathol. 2015, 10, 473–510. 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchell J. M.; Mungul A.; Taylor-Papadimitriou J. O-Linked Glycosylation in the Mammary Gland: Changes That Occur During Malignancy. J. Mammary Gland Biol. Neoplasia 2001, 6, 355–364. 10.1023/A:1011331809881. [DOI] [PubMed] [Google Scholar]
- Kirmiz C.; Li B.; An H. J.; Clowers B. H.; Chew H. K.; Lam K. S.; Ferrige A.; Alecio R.; Borowsky A. D.; Sulaimon S.; et al. A Serum Glycomics Approach to Breast Cancer Biomarkers. Mol. Cell Proteomics 2007, 6, 43–55. 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- An H. J.; Miyamoto S.; Lancaster K. S.; Kirmiz C.; Li B.; Lam K. S.; Leiserowitz G. S.; Lebrilla C. B. Profiling of Glycans in Serum for the Discovery of Potential Biomarkers for Ovarian Cancer. J. Proteome Res. 2006, 5, 1626–1635. 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- Guillard M.; Wada Y.; Hansikova H.; Yuasa I.; Vesela K.; Ondruskova N.; Kadoya M.; Janssen A.; Van den Heuvel L. P.; Morava E.; et al. Transferrin Mutations at the Glycosylation Site Complicate Diagnosis of Congenital Disorders of Glycosylation Type I. J. Inherit. Metab. Dis. 2011, 34, 901–906. 10.1007/s10545-011-9311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T. Diseases of Glycosylation Beyond Classical Congenital Disorders of Glycosylation. Biochim. Biophys. Acta 2012, 1820, 1306–1317. 10.1016/j.bbagen.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Mohamed M.; Guillard M.; Wortmann S. B.; Cirak S.; Marklova E.; Michelakakis H.; Korsch E.; Adamowicz M.; Koletzko B.; van Spronsen F. J.; et al. Clinical and Diagnostic Approach in Unsolved CDG Patients with a Type 2 Transferrin Pattern. Biochim. Biophys. Acta 2011, 1812, 691–698. 10.1016/j.bbadis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Jaeken J. Congenital Disorders of Glycosylation. Handb. Clin. Neurol. 2013, 113, 1737–1743. 10.1016/B978-0-444-59565-2.00044-7. [DOI] [PubMed] [Google Scholar]
- Cylwik B.; Naklicki M.; Chrostek L.; Gruszewska E. Congenital Disorders of Glycosylation. Part I. Defects of Protein N-Glycosylation. Acta Biochim. Pol. 2013, 60, 151–161. 10.18388/abp.2013_1965. [DOI] [PubMed] [Google Scholar]
- Hennrich M. L.; Gavin A. C. Quantitative Mass Spectrometry of Posttranslational Modifications: Keys to Confidence. Sci. Signal. 2015, 8, re5. 10.1126/scisignal.aaa6466. [DOI] [PubMed] [Google Scholar]
- Guillard M.; Morava E.; van Delft F. L.; Hague R.; Körner C.; Adamowicz M.; Wevers R. A.; Lefeber D. J. Plasma N-Glycan Profiling by Mass Spectrometry for Congenital Disorders of Glycosylation Type II. Clin Chem 2011, 57, 593–602. 10.1373/clinchem.2010.153635. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer L. C.; Rust S.; van Scherpenzeel M.; Ng B. G.; Losfeld M.-E.; Timal S.; Raymond K.; He P.; Ichikawa M.; Veltman J.; et al. Multiple Phenotypes in Phosphoglucomutase 1 Deficiency. N. Engl. J. Med. 2014, 370, 533–542. 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Bakar N.; Voermans N. C.; Marquardt T.; Thiel C.; Janssen M. C. H.; Hansikova H.; Crushell E.; Sykut-Cegielska J.; Bowling F.; MØrkrid L.; et al. Intact Transferrin and Total Plasma Glycoprofiling for Diagnosis and Therapy Monitoring in Phosphoglucomutase-I Deficiency. Transl. Res. 2018, 199, 62–76. 10.1016/j.trsl.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipgrave Ederveen A. L.; de Haan N.; Baerenfaenger M.; Lefeber D. J.; Wuhrer M. Dissecting Total Plasma and Protein-Specific Glycosylation Profiles in Congenital Disorders of Glycosylation. Int. J. Mol. Sci. 2020, 21, 7635. 10.3390/ijms21207635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L.; Edmunds G.; Gibbons C.; Zhang J.; Gadi M. R.; Zhu H.; Fang J.; Liu X.; Kong Y.; Wang P. G. Toward Automated Enzymatic Synthesis of Oligosaccharides. Chem. Rev. 2018, 118, 8151–8187. 10.1021/acs.chemrev.8b00066. [DOI] [PubMed] [Google Scholar]
- Malik A.; Seeberger P. H.; Varón Silva D.. Advances in the Chemical Synthesis of Carbohydrates and Glycoconjugates. In Advances in Glycobiotechnology, Rapp E., Reichl U. Eds.; Springer International, 2021; pp 201-230. [DOI] [PubMed] [Google Scholar]
- Delafield D. G.; Li L. Recent Advances in Analytical Approaches for Glycan and Glycopeptide Quantitation. Mol. Cell Proteomics 2021, 20, 100054. 10.1074/mcp.R120.002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. D.; Ruhaak L. R.; Lebrilla C. B.. Analysis of Milk Oligosaccharides by Mass Spectrometry. In High-Throughput Glycomics and Glycoproteomics: Methods and Protocols, Lauc G., Wuhrer M. Eds.; Springer; New York, 2017; pp 121-129. [DOI] [PubMed] [Google Scholar]
- Wrigglesworth D. J.; Goonatilleke E.; Haydock R.; Hughes K. R.; Lebrilla C. B.; Swanson K. S.; Jones P.; Watson P. High-Throughput Glycomic Analyses Reveal Unique Oligosaccharide Profiles of Canine and Feline Milk Samples. PLoS One 2020, 15, e0243323 10.1371/journal.pone.0243323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy D.; Borza B.; Domokos A.; Varadi E.; Szigeti M.; Meszaros-Matwiejuk A.; Molnar-Gabor D.; Guttman A. Ultrafast High-Resolution Analysis of Human Milk Oligosaccharides by Multicapillary Gel Electrophoresis. Food Chem. 2021, 341, 128200. 10.1016/j.foodchem.2020.128200. [DOI] [PubMed] [Google Scholar]
- Kottler R.; Mank M.; Hennig R.; Müller-Werner B.; Stahl B.; Reichl U.; Rapp E. Development of a High-Throughput Glycoanalysis Method for the Characterization of Oligosaccharides in Human Milk Utilizing Multiplexed Capillary Gel Electrophoresis with Laser-Induced Fluorescence Detection. Electrophoresis 2013, 34, 2323–2336. 10.1002/elps.201300016. [DOI] [PubMed] [Google Scholar]
- Masi A. C.; Embleton N. D.; Lamb C. A.; Young G.; Granger C. L.; Najera J.; Smith D. P.; Hoffman K. L.; Petrosino J. F.; Bode L.; et al. Human Milk Oligosaccharide DSLNT and Gut Microbiome in Preterm Infants Predicts Necrotising Enterocolitis. Gut 2021, 70, 2273–2282. 10.1136/gutjnl-2020-322771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. W.; Jones E. E.; Betesh L. R.; Romano P. R.; Gao P.; Copland J. A.; Mehta A. S.; Drake R. R. Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry Workflow for Spatial Profiling Analysis of N-Linked Glycan Expression in Tissues. Anal. Chem. 2013, 85, 9799–9806. 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. T.; Ho Y. Y.; Kaur G.; Oehler M. K.; Everest-Dass A. V.; Packer N. H.; Hoffmann P. N-Glycan Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging Protocol for Formalin-Fixed Paraffin-Embedded Tissues. Rapid Commun. Mass Spectrom. 2017, 31, 825–841. 10.1002/rcm.7845. [DOI] [PubMed] [Google Scholar]
- Powers T. W.; Holst S.; Wuhrer M.; Mehta A. S.; Drake R. R. Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules 2015, 5, 2554–2572. 10.3390/biom5042554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijs B.; Holst-Bernal S.; de Graaff M. A.; Briaire-de Bruijn I. H.; Rodriguez-Girondo M.; van de Sande M. A. J.; Wuhrer M.; McDonnell L. A.; Bovée J. Molecular Signatures of Tumor Progression in Myxoid Liposarcoma Identified by N-Glycan Mass Spectrometry Imaging. Lab. Invest. 2020, 100, 1252–1261. 10.1038/s41374-020-0435-2. [DOI] [PubMed] [Google Scholar]
- Gustafsson O. J. R.; Briggs M. T.; Condina M. R.; Winderbaum L. J.; Pelzing M.; McColl S. R.; Everest-Dass A. V.; Packer N. H.; Hoffmann P. MALDI Imaging Mass Spectrometry of N-Linked Glycans on Formalin-Fixed paraffin-Embedded Murine Kidney. Anal. Bioanal. Chem. 2015, 407, 2127–2139. 10.1007/s00216-014-8293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaval F.; Van Zeijl R.; Dalebout H.; Holst S.; van Pelt G. W.; Farina-Sarasqueta A.; Mesker W. E.; Tollenaar R.; Morreau H.; Wuhrer M. N-Glycomic Signature of Stage II Colorectal Cancer and Its Association with the Tumor Microenvironment. Mol. Cell Proteomics 2020, 20, 100057. 10.1074/mcp.RA120.002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black A. P.; Angel P. M.; Drake R. R.; Mehta A. S. Antibody Panel Based N-Glycan Imaging for N-Glycoprotein Biomarker Discovery. Curr. Protoc. Protein Sci. 2019, 98, e99 10.1002/cpps.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift C. L.; Drake R. R.; Mehta A.; Angel P. M. Multiplexed Imaging Mass Spectrometry of the Extracellular Matrix Using Serial Enzyme Digests from Formalin-Fixed Paraffin-Embedded Tissue Sections. Anal. Bioanal. Chem. 2021, 413, 2709–2719. 10.1007/s00216-020-03047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Shi X.; Vu N. Q.; Li G.; Li Z.; Shi Y.; Li M.; Wang B.; Welham N. V.; Patankar M. S.; et al. On-Tissue Derivatization with Girard’s Reagent P Enhances N-Glycan Signals for Formalin-Fixed Paraffin-Embedded Tissue Sections in MALDI Mass Spectrometry Imaging. Anal. Chem. 2020, 92, 13361–13368. 10.1021/acs.analchem.0c02704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J.; Taubenschmid J.; Wenzel D.; Gattinger A.; Dürnberger G.; Dusberger F.; Elling U.; Mach L.; Mechtler K.; Penninger J. M. Comparative Glycoproteomics of Stem Cells Identifies New Players in Ricin Toxicity. Nature 2017, 549, 538–542. 10.1038/nature24015. [DOI] [PMC free article] [PubMed] [Google Scholar]