Abstract
Current screening methods for ovarian cancer have failed to demonstrate a significant reduction in mortality. Uterine lavage combined with TP53 ultradeep sequencing for the detection of disseminated ovarian cancer cells has emerged as a promising tool, but this approach has not been tested for early-stage disease or non-serous histologies. In addition, lavages carry multiple background mutations, the significance of which is poorly understood. Uterine lavage was collected preoperatively in 34 patients undergoing surgery for suspected ovarian malignancy including 14 patients with benign disease and 20 patients with ovarian cancer [6 non-serous and 14 high-grade serous-like (serous)]. Ultradeep duplex sequencing (∼3,000×) with a panel of common ovarian cancer genes identified the tumor mutation in 33% of non-serous (all early stage) and 79% of serous cancers (including four early stage). In addition, all lavages carried multiple somatic mutations (average of 25 mutations per lavage), more than half of which corresponded to common cancer driver mutations. Driver mutations in KRAS, PIK3CA, PTEN, PPP2R1A, and ARID1A presented as larger clones than non-driver mutations and with similar frequency in lavages from patients with and without ovarian cancer, indicating prevalent somatic evolution in all patients. Driver TP53 mutations, however, presented as significantly larger clones and with higher frequency in lavages from individuals with ovarian cancer, suggesting that TP53-specific clonal expansions are linked to ovarian cancer development. Our results demonstrate that lavages capture cancer cells, even from early-stage cancers, as well as other clonal expansions and support further exploration of TP53 mutation burden as a potential ovarian cancer risk factor.
Significance:
Cancer driver mutations are found in uterine lavage DNA in all individuals, but driver TP53 mutations presented as significantly larger clones and with higher frequency in lavages from individuals with ovarian cancer. This suggests that TP53-specific clonal expansion plays a role in tumorigenesis and presents opportunities for early detection.
Introduction
Ovarian cancer is a prominent cause of cancer-related mortality, with more than 200,000 annual deaths worldwide (1, 2). Because of the indistinct symptoms experienced during disease progression, ovarian cancer is most often diagnosed at an advanced stage, unfortunately leading to high mortality rates. Five-year survival for advanced stage disease is still only 30%, while survival for early stage is >90% (3). This has prompted efforts to develop screening and early detection methods, including transvaginal ultrasound and cancer antigen 125 (CA-125) serum testing (4, 5), but none has thus far demonstrated a significant reduction in mortality (6). Explorations into liquid biopsies using pap tests have shown limited sensitivity for ovarian cancer detection (7–9).
Lavage of the uterine cavity with 10 mL of saline using a three-way catheter can detect exfoliated cancer cells from the ovary or fallopian tube (10, 11) and be safely performed in an outpatient setting (12), suggesting a novel noninvasive screening tool. Using next-generation sequencing (NGS) and droplet digital PCR to identify the known tumor mutation, Maritschnegg and colleagues demonstrated that 80% of uterine lavages from individuals with ovarian cancer carried tumor DNA (10). Our group then performed blinded uterine lavage analysis using duplex sequencing (DS), an ultra-accurate ultrasensitive NGS approach (13, 14), and demonstrated detection of cancer-specific TP53 mutations in 80% of samples from high-grade serous cancer (HGSC) cases (11). An advantage of DS is that it allows for the detection of not only tumor DNA but also low-frequency mutant clones (9, 11, 15, 16), which are now recognized as the result of prevalent somatic clonal evolution (17, 18) and might be linked to cancer development (19, 20). Supporting this notion, we have shown that Pap test DNA carries more non-tumor TP53 pathogenic mutant clones in individuals with ovarian cancer (9), but this has not been properly tested in lavage DNA. In addition, the combined approach of uterine lavage plus DS has not been tested for non-serous ovarian cancer or early-stage disease, which are important aspects for widespread implementation of a clinical test for early ovarian cancer detection.
While 75% of epithelial ovarian cancer cases have high-grade serous histology, the remaining 25% include low-grade serous, clear cell, endometrioid, and mucinous histologies. Non-serous carcinomas are typically driven by a variety of genetic alterations including activation of PIK3CA and the Wnt-β-catenin pathway, and inactivation of ARID1A and PTEN (21). Furthermore, while the majority of HGSC harbor a TP53 mutation, up to 20% of cases may not (22–25). Thus, any ovarian cancer screening method utilizing detection of cancer-driving mutations must expand beyond TP53 to improve sensitivity. In this study, we aimed to pilot the combination of uterine lavage with ultradeep sequencing using an expanded gene panel to improve detection of both early-stage and non-serous ovarian cancer. In addition, we aimed to leverage the extreme sensitivity of ultradeep DS to characterize background somatic mutations in lavages and determine potential associations with ovarian cancer.
Materials and Methods
Patients and Samples
The study included 34 patients who underwent surgery with preoperative concern for an ovarian malignancy at the University of Washington (Seattle, WA) between November 2019 and October 2020. Inclusion criteria included the presence of a uterus, and at least one ovary and fallopian tube. Patients undergoing neoadjuvant chemotherapy before surgery were excluded because of the potential impact on sequencing findings. The study was designed in accordance with recognized ethical guidelines for patient participation. Patients were enrolled prior to surgery under an Institutional Review Board–approved protocol at the University of Washington (Seattle, WA) and provided informed written consent for tissue collection, including tumor and a preoperative uterine lavage. Uterine lavages were collected after induction of anesthesia and vaginal antiseptic preparation using a transcervical catheter (Ovartec) as described previously (10, 12). The clinicopathological characteristics of the patients are included in Supplementary Table S1 and further described in Supplementary Materials and Methods. In most cases, carcinoma was detected on intraoperative pathology and a staging procedure was performed per the surgeon's usual practice. One case of stage IB HGSC was not staged as intraoperative pathology was benign. Patient sample numbering for the article was assigned on the basis of histology and age. During this time period, which included the first year of the SARs-Co-V2 pandemic, many patients with advanced ovarian cancer received neoadjuvant chemotherapy to reduce perioperative morbidity and mortality, and were thus excluded from this study. This led to a higher proportion of early-stage cancers in this sampled population.
Samples were stored at the University of Washington Gynecologic Oncology Tissue Bank. Lavages were mixed with an ethanol-based stabilization medium and filtered with a gravity flow 100 μm cell strainer to remove potential clusters of endometrial cells. Filtered samples were then centrifuged at 300 × g for 10 minutes and cell pellets were frozen at −80°C. A subset of samples underwent additional centrifugation to increase size of cell pellet. Genomic DNA was extracted from cell pellets using the Dneasy Blood & Tissue Kit (Qiagen) with proteinase K digestion at 37°C for 2 hours and including RNAse treatment. DNA was quantified by Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific) and stored at −80°C until library preparation. Sartorius Vivacon 500 DNA concentrators were used if needed prior to library preparation.
Lavage DNA Sequencing
A total of 200 ng of lavage DNA were processed for DS using commercially available kits (TwinStrand Biosciences). Library preparation consisted of sonication, end-repair, A-tailing, ligation to duplex adapters, fragment amplification, hybridization capture with 120 bp biotinylated probes (TP53 human panel v1.0 from TwinStrand Biosciences, and xGen Hyb probes from Integrated DNA Technology, for the rest of genes), and library amplification. The capture panel was designed to target candidate ovarian cancer driver genes previously identified as the most common drivers in endometrial, clear-cell and high-grade serous carcinomas according to the Catalogue of Somatic Mutations in Cancer (COSMIC; v95, accessed on January 24, 2022; ref. 26). The panel included TP53, ARID1A, PTEN, PPP2R1A, CDKN2A, KRAS (whole genes; Supplementary Table S2); and CTNNB1, PIK3CA, and BRAF (hotspots only; Supplementary Table S3). The total size of the coding region captured was 12.1 Kb. Given the small size of the panel, two rounds of hybridization capture were performed to increase efficiency (27). Proper library fragment size was confirmed by Agilent 4200 TapeStation. Libraries were quantified using the Qubit dsDNA HS Assay kit, diluted, and pooled for sequencing. Libraries were sequenced using 150 PE reads on a HiSeq at Genewiz, allocating approximately 13 million clusters per sample.
Data Analysis
Sequencing reads were analyzed as described previously (13) using pipeline v2.1.2 from https://github.com/Kennedy-Lab-UW/Duplex-Seq-Pipeline. Variant Call Format (VCF) files were converted to MAF files, which were then postprocessed with R version 4.1.2 (ref. 28; Supplementary Materials and Methods). For each sample, the overall duplex depth per gene was calculated as the average depth of all on-target coding nucleotides sequenced (Supplementary Table S4). For each mutation, variant allele frequency (VAF) was calculated as the number of mutant duplex reads divided by the total duplex depth at the given position. To correct for the variability in sequencing depth across samples, sample comparisons were made based on mutation frequency (MF) and mutation burden (MB), calculated for each gene and overall for all the genes in the study. MF was calculated as the number of mutant positions divided by the total number of duplex nucleotides sequenced, and MB was calculated as the number of total mutant duplex reads (each mutant duplex read corresponds to a single DNA mutant molecule) divided by the total number of duplex nucleotides sequenced (Supplementary Table S4).
COSMIC data (26) were used to determine the codon location of substitutions reported for ovarian carcinomas for the genes and transcripts of interest. The histograms of mutation location for each gene were then compared with the histograms obtained with mutations identified in uterine lavage. In addition, COSMIC data were used to determine ovarian cancer hotspot codons, which were defined as codons with two or more substitutions and accounting for at least 1% of the reported substitutions for ovarian carcinoma in each gene (Supplementary Table S5). For oncogenes in the study (BRAF, CTNNB1, KRAS, and PIK3CA), cancer driver mutations were defined as missense mutations in ovarian cancer hotspot codons. For tumor suppressor genes (CDKN2A, PTEN, PPP2R1A, ARID1A, and TP53), cancer driver mutations were defined as missense mutations in ovarian cancer hotspot codons plus any nonsense, insertion/deletion (indel) or splice mutations. The list of annotated coding mutations is shown in Supplementary Table S6.
Tumor Sequencing
Tumor DNA was sequenced to identify driver mutations and compare with mutations found in the lavages. For five of the 13 high-grade serous cases in the study, the TP53 tumor driver mutation had been previously identified using the targeted BROCA panel as part of a larger institutional study (Table 1; refs. 29, 30). For the remaining eight high-grade serous tumors and all the non-serous tumors, DNA was extracted from formalin-fixed, paraffin-embedded tissue sections and duplex sequenced using a MiSeq Illumina platform on site as detailed in Supplementary Materials and Methods.
TABLE 1.
Tumor and uterine lavage mutation testing
| Tumor testing | Uterine lavage testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Ovarian cancer type | Histology | Age at surgery | Stage | Tumor testing platform | Mutation | Protein variant | Coding variant | Tumor mutation found | VAF | Mutant duplex reads | Duplex depth at position |
| P15 | Non-serous | Clear-cell carcinoma | 53 | IC3 | DS | ARID1A | p.W1023Vfs*10 | c.3066dup | yes | 0.0134 | 14 | 1046 |
| P16 | Non-serous | Clear-cell carcinoma | 64 | IIB | DS | TP53 | P.E221* | c.661G>T | no | |||
| P17 | Non-serous | Clear-cell carcinoma | 69 | IA | DS | ARID1A | p.K1808Nfs*4 | c.5423 5424insTTAC | no | |||
| P18 | Non-serous | Endometrioid carcinoma | 57 | IC1 | DS | CTNNB1 | P.S33C | c.98C>G | no | |||
| P19a | Non-serous | Endometrioid carcinoma | 58 | IC3 | DS | ARID1A CTNNB1 PIK3CA |
p.R1335* p.D32Y P.H1047R |
C.40030T c.94G>T c.3140A>G |
no | |||
| P20 | Non-serous | Endometrioid carcinoma | 83 | IA | DS | PIK3CA | P.N1044Y | c.3130A>T | yes | 0.0053 | 16 | 2997 |
| P21 | Serous | Carcinosarcoma | 51 | IB | DS | TP53 | p.X307 splice | c.919+1G>A | yes | 0.0075 | 42 | 5593 |
| P22 | Serous | HGSC | 52 | IIIC | BROCA | TP53 | P.G244V | c.731G>T | yes | 0.0050 | 21 | 4166 |
| P23 | Serous | HGSC | 56 | IIB | BROCA | TP53 | p.R342Tfs*4 | c.1023 1024insAC | yes | 0.0005 | 2 | 3835 |
| P24 | Serous | HGSC | 57 | IB | DS | TP53 | P.C275F | c.824G>T | yes | 0.0169 | 97 | 5723 |
| P25 | Serous | HGSC | 61 | IIA | DS | TP53 | P.V157F | C.469G>T | no | |||
| P26 | Serous | HGSC | 62 | IIIB | DS | TP53 | P.R273C | c.817C>T | yes | 0.0037 | 22 | 5905 |
| P27 | Serous | HGSC | 67 | IIIA1 | BROCA | TP53 | p.V274A | c.821T>C | yes | 0.0005 | 2 | 3938 |
| P28 | Serous | HGSC | 68 | IVB | DS | TP53 | P.V143M | c.427G>A | no | |||
| P29 | Serous | HGSC | 68 | IVB | DS | TP53 | p.S315Rfs*26 | c.942 954del | no | |||
| P30 | Serous | HGSC | 71 | IIIC | DS | TP53 | P.H193Y | c.577C>T | yes | 0.0091 | 56 | 6130 |
| P31 | Serous | HGSC | 72 | IIIC | BROCA | TP53 | P.S241F | C.722OT | yes | 0.0045 | 9 | 1999 |
| P32 | Serous | HGSC | 73 | IIIC | DS | TP53 | p.X126 splice | c.376-2A>G | yes | 0.0002 | 1 | 4291 |
| P33 | Serous | HGSC | 76 | IIA | BROCA | TP53 | P.P151H | c.452C>A | yes | 0.0962 | 372 | 3866 |
| P34 | Serous | HGSC | 83 | IIB | DS | TP53 | p.S261Vfs*84 | c.780del | yes | 0.1832 | 926 | 5055 |
Abbreviation: DS, duplex sequencing; HGSC, high-grade serous carcinoma.
aNote that patient P19 had three tumor driver mutations.
Statistical Analysis
Comparison of MF, MB, and VAF across groups of individuals was performed by Mann–Whitney U test. Correlations were tested with Spearman rank test. Associations between categorical variables were tested with Fisher exact test. Two logistic regression models were constructed, one including the standard ovarian cancer risk variables (age and CA-125) and an exploratory model including age, CA-125, and TP53 MB. The models equated the relationship between variables with the occurrence of ovarian cancer to estimate beta coefficients with 95% confidence intervals. Because of a heavy right-tailed distribution, CA-125 was log transformed. Age and TP53 MB were presented as a per SD increase. All tests were two sided at an alpha level (type 1 error rate) of 0.05. Statistical analyses were performed with SPSS version 26 (31), R version 4.1.1 (28), and Stata 16 (32).
Software Availability
The Duplex Sequencing Pipeline is available at https://github.com/Kennedy-Lab-UW/Duplex-Seq-Pipeline.
Data Availability Statement
Sequencing data from this study are available at the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under BioProject ID PRJNA879769.
Results
Uterine lavage was collected preoperatively from 34 patients that underwent gynecologic surgery for suspected ovarian cancer using commercial catheters (Supplementary Fig. S1A; Supplementary Table S1). Cell pellet DNA was analyzed with ultradeep DS using a panel of genes frequently mutated in the most common ovarian cancer histologies (Supplementary Fig. S1B; Supplementary Tables S2 and S3). Average depth of sequencing in target coding regions was 3,222x (minimum 1,203x, maximum 4,164x). Routine pathologic review of surgical specimens revealed that 14 patients had benign masses and 20 patients had ovarian cancer, including clear-cell carcinoma (3), endometrioid carcinoma (3), carcinosarcoma (1), and HGSC (13). The carcinosarcoma case had a significant serous component and was combined with the HGSC cases in a “serous” histology group. A total of 12 of the 20 patients with ovarian cancer (60%) had early-stage disease (FIGO stage I or II), providing a unique opportunity to test the sensitivity of this approach for early ovarian cancer detection (Supplementary Fig. S1C). One patient had prior exposure to chemotherapy for breast cancer and two patients with HGSC were later identified to carry germline BRCA2 mutations (Supplementary Table S1).
Uterine Lavage Detected the Tumor Mutation in More Than Two-thirds of Ovarian Cancer Cases
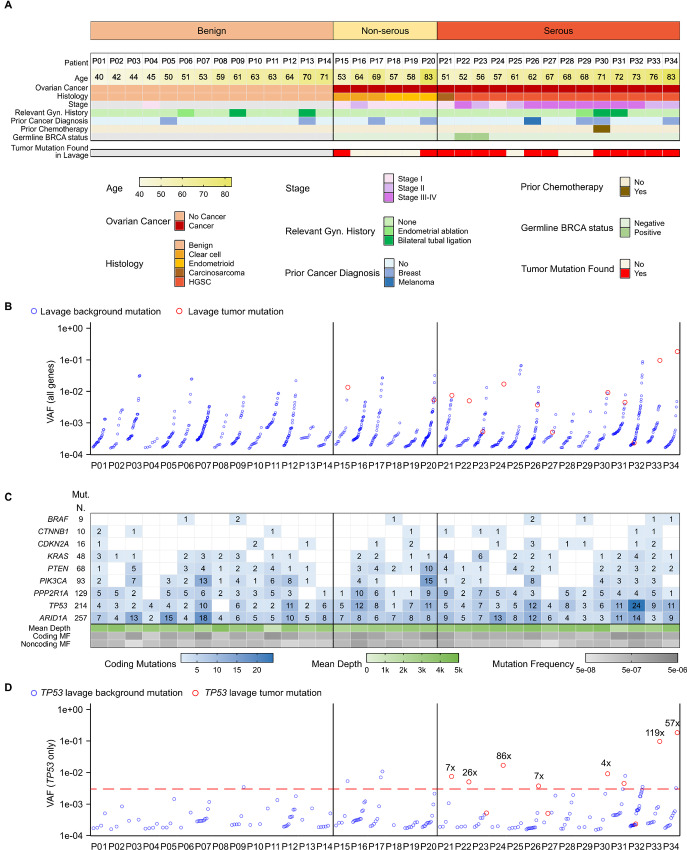
For the 20 patients with ovarian cancer, neoplastic DNA was sequenced to determine whether the ovarian cancer mutation was present in the lavage. Non-serous cancers had mutations in a variety of genes including ARID1A, CTNNB1, PIK3CA, and TP53, whereas the serous cancers were driven exclusively by TP53 mutations (Table 1). In total, 13 of 20 (65%) of the tumor mutations were identified in the corresponding lavage, but the rate of detection was higher in serous ovarian cancer (11/14, 79%) than in non-serous cancers (2/6, 33%). Because duplex reads correspond to unique DNA molecules, the VAF of mutations is a direct readout of clone size. The tumor clones identified in lavage ranged from 0.02% to 18% of the sequenced DNA. The VAF of the tumor mutation in lavage was not associated with tumor stage or blood levels of antigen CA-125 (Supplementary Fig. S2).
We next explored whether the identification of the tumor mutation in uterine lavage was related to clinicopathological characteristics (Fig. 1A). Of seven lavages in which the tumor mutation was not identified, four were in early-stage non-serous cancers, and three were in serous cancers. One of the serous cancers was early stage and another corresponded to an individual with a prior endometrial ablation. Remarkably, the tumor mutation was identified in five of six (83%) lavages from stage I–II serous ovarian cancer, indicating that early stage was not a factor preventing detection and suggesting early transit of cancerous clones. In addition, the tumor clone was also detected in the lavages of two patients that were in their 70s and had undergone a prior bilateral tubal ligation, which was unexpected. One patient had fallopian tube involvement, which may have locally spread to the proximal tubal fragment, though this could not be confirmed. The other patient had uterine myometrial involvement of tumor. Alternatively, cells carrying these mutations might have traveled not through the tubal lumen but via lymphatic or hematologic channels, or these clones may represent nonmalignant parallel somatic evolution.
FIGURE 1.
Summary of mutations identified in lavages from patients with and without ovarian cancer. A, Clinicopathological characteristics of the patients are color coded according to the legend. Patients are sorted by ascending age within each histologic group. For patients with ovarian cancer, it is indicated whether the tumor mutation was found in the uterine lavage. B, The VAF for all mutations detected in uterine lavage samples is displayed. Red circles correspond to tumor mutations and blue circles correspond to other background mutations, sorted by ascending VAF within each patient. C, The total number of mutations identified for each gene in lavage samples is indicated (Mut. N) as well as the number of mutations in each lavage (blue gradient boxes). The mean depth of sequencing for coding exons is indicated with a gradient scale in green. Coding and noncoding MF, corresponding to the count of unique mutations adjusted by nucleotides sequenced, are indicated with a gradient scale in gray. D, The VAF for all TP53 mutations detected in each uterine lavage sample is displayed, with the red circles indicating tumor mutations. The ratio of the VAF of the tumor mutation to the VAF of the largest background mutation is indicated if >4x. Red line indicates a potential VAF cutoff of 0.003.
In addition to tumor mutations, lavages carried other background mutations (Fig. 1B; Supplementary Table S6). These mutations were identified in all the genes sequenced and in patients with and without ovarian cancer (Fig. 1C). In a subset of patients with ovarian cancer, the tumor mutation had a VAF higher than background mutations (Fig. 1B), indicating that the tumor clone was the largest in the lavage. In lavages from other patients with ovarian cancer, however, background mutations obscured the tumor mutation. We observed that many of the large background clones corresponded to genes other than TP53, which are relevant for non-serous histologies but generally not for serous ovarian cancer (Supplementary Fig. S3). Thus, we restricted the analysis to TP53 to determine whether focusing on this main driver gene could help distinguish tumor mutations from background (Fig. 1D). In seven of 11 lavages from serous ovarian cancer cases in which the tumor mutation was identified (63%), the mutation was present with VAF greater than 0.003 and more than 4-fold higher VAF than the largest background mutation. Comparatively, only 1 of 14 patients without cancer had a TP53 mutation VAF greater than 0.003. These results demonstrate clear identification of TP53-mutant tumor DNA in a large fraction of lavages despite background TP53 mutations.
Lavages Had an Excess of Coding versus Noncoding Background Mutations and Showed Higher Frequency of TP53 Mutations in Patients with Ovarian Cancer
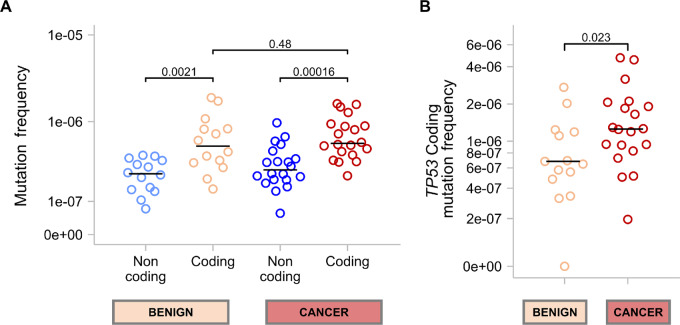
We next explored whether background mutations harbored relevant biological information that could help distinguish patients with and without ovarian cancer. All lavages carried multiple mutations in at least two or more genes (Fig. 1C) with an average of 25 mutations per lavage (min = 6, max = 54). The genes with the least mutations were BRAF and CTNNB1 and the ones with the most mutations were TP53 and ARID1A, although these differences were partially due to variation in the size and depth of the regions sequenced for each gene. In general, lavages that showed large numbers of mutations in one gene also showed large numbers of mutations in other genes. Gene-specific MF were calculated to adjust for depth of sequencing (Supplementary Table S4). The MF of all the genes were highly correlated (Supplementary Fig. S4) confirming that samples with high levels of background mutations carried them across multiple genes.
We then determined the overall coding MF (all genes) and noncoding MF because the target panel also captured intronic regions that contained mutations in all samples. Coding MF was significantly higher than noncoding MF in lavages from patients with and without ovarian cancer (Fig. 2A). When ovarian cancer cases were separated into non-serous and serous, the difference remained significant for serous ovarian cancer (Supplementary Fig. S5A). While noncoding mutations reflect mutagenic processes, the excess of coding mutations in lavage suggests clonal expansions of cells with functional mutations, as described previously (11, 19). The overall coding MF was not significantly different between patients with and without ovarian cancer, indicating that clonal expansions in the selected genes, as a whole, occur similarly in both groups. However, when comparing coding MF by gene, we observed that background mutations in TP53 were more abundant in lavages of patients with ovarian cancer than those without cancer (Fig. 2B). When separating cancer cases into serous and non-serous, there were not significant differences in lavage TP53 MF between the two groups (Supplementary Fig. S5B). Both cancer groups, however, had increased TP53 MF compared with the benign group although the difference was significant only for serous cases (Supplementary Fig. S5B). For the rest of the genes in the study, there were not significant differences between the MF in lavages from cancer and benign cases (Supplementary Fig. S6). TP53 was also the only gene whose coding MF was positively correlated with age (Supplementary Fig. S7A), consistent with prior studies (9, 11, 15). This association, however, was influenced by the fact that the oldest patients in the study had ovarian cancer and high TP53 MF (Supplementary Fig. S7B).
FIGURE 2.
Comparison of MF in patients with and without ovarian cancer. MF is calculated by dividing the number of mutations detected by the number of nucleotides sequenced. Each circle represents the MF for an individual sample. P values for Mann–Whitney U tests are shown for each comparison. The median MF for each group is indicated with a horizontal black bar. A, Comparison of coding and noncoding MF for all sequenced genes combined in uterine lavages from benign and cancer cases. B, Comparison of TP53-specific coding MF between benign and cancer cases.
More Than Half of the Mutations in Uterine Lavage are Common Ovarian Cancer Driver Mutations, Many of Which are Expanded in Larger Clones Compared to Non-driver Mutations
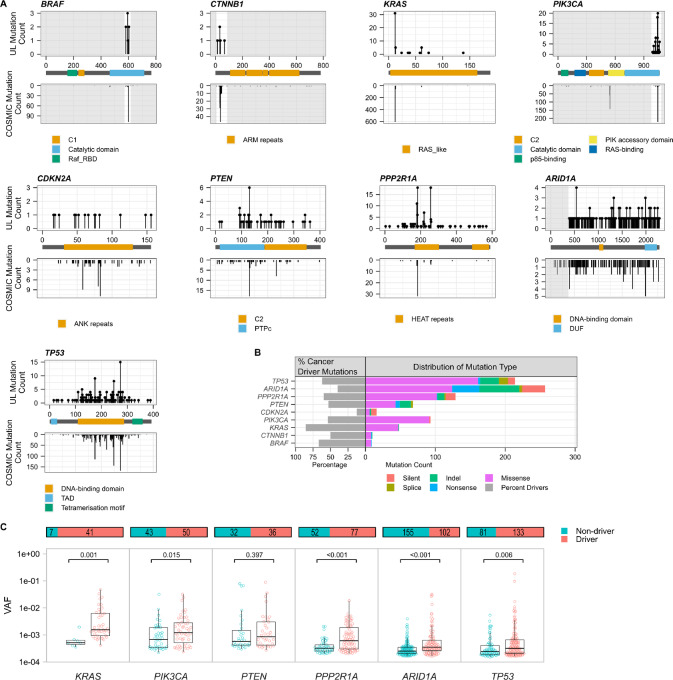
To gain further insight into the nature of lavage mutations, we plotted them along the coding region of each gene and compared their distribution with the distributions obtained for ovarian cancer mutations reported in COSMIC (Fig. 3A). Overall, the distribution of mutations was very similar in lavages and in COSMIC, indicating that lavage mutations are not random, but mimic mutations found in ovarian cancer, even in the absence of ovarian cancer. One remarkable exception was BRAF p.V600E, which accounts for 78.8% of BRAF mutations in ovarian cancer but was not observed in lavages. Lavages, however, carried mutations in other BRAF hotspot codons common in ovarian cancer albeit at much lower frequency (codons 594, 581, and 597, representing 2.9%, 2.2%, and 2.2% of BRAF mutations, respectively).
FIGURE 3.
Characterization of uterine lavage mutations by gene. A, The location and distribution of mutations in uterine lavage samples mirror those identified in ovarian carcinomas. Top panels show the location by codon position of somatic mutations identified in uterine lavage, with mutation counts plotted on the Y axis. Lower panels show mutations identified in ovarian carcinomas in COSMIC. Indels are excluded as they might span multiple codon locations. Areas of the gene not captured in the DS panel are grayed out. Gene domains are highlighted in the legends. B, Percentage of cancer driver mutations identified for each gene in the entire cohort (left) and distribution of mutation type per gene (right). Cancer driver mutations are defined, in oncogenes, as substitutions occurring in common hotspots codons and, in tumor suppressor genes, as substitutions occurring in common hotspots codons plus insertion/deletions, nonsense, and splice mutations. C, The VAF of driver versus non-driver mutations identified in the entire cohort is displayed by gene. Only genes that exhibited mutations in >50% of the uterine lavage samples are shown. Each circle represents a unique mutation. Overlying box plots display the quartiles with whiskers extending up to 1.5× the interquartile range. Bar plots above display the total number of mutations in each group and the distribution between driver and non-driver. P values correspond to Mann–Whitney U tests.
On the basis of ovarian cancer data from COSMIC, we then determined what proportion of the mutations identified in lavages could be considered cancer driver mutations. Of 844 coding mutations identified in lavages, more than half (452, 54%) qualified as cancer driver mutations (Supplementary Table S6). With the exception of CDKN2A, which carried few mutations overall, all genes carried high levels of cancer driver mutations ranging from 40% in ARID1A to 85% in KRAS (Fig. 3B). While these proportions showed some variation across samples, especially for tumor suppressor genes (Supplementary Fig. S8A), they were not significantly different between lavages from patients with and without ovarian cancer (Supplementary Fig. S8B) indicating that clonal expansions of driver mutations in lavage are prevalent irrespective of ovarian cancer progression.
The types of mutations observed for each gene corresponded to expectations based on their roles as oncogenes or tumor suppressor genes. Oncogenes (PIK3CA, KRAS, CTNNB1, and BRAF) carried mostly missense mutations whereas tumor suppressor genes (TP53, ARID1A, PPP2R1A, PTEN, and CDKN2A) were enriched for indels, nonsense and splice mutations (Fig. 3B). The analysis of the overall mutational spectrum also demonstrated a high resemblance between lavage mutations and mutations observed in ovarian cancers (Supplementary Fig. S9). The spectrum was characterized by an enrichment of C>T mutations in lavages from patients with non-serous and serous ovarian cancer as well as lavages from patients older than 50 years of age, consistent with the pattern observed in ovarian cancer and the age-related origin of C>T mutations (33).
We hypothesized that cells carrying cancer driver mutations might be more likely to clonally expand than cells without driver mutations, resulting in overall higher VAF for driver mutations. To test this hypothesis, for the six genes that exhibited mutations in >50% of the uterine lavage samples, we compared the VAF of non-driver versus driver mutations (Fig. 3C). Despite most mutations being present at very low VAF (<0.01), we observed that for all the genes except PTEN, the VAF of driver mutations was significantly higher than the VAF of non-driver mutations. These results demonstrate that lavage DNA carries large clones driven by common cancer driver mutations and that ultradeep sequencing can quantify their size, and thus demonstrate their expansion, compared with non-driver mutations.
TP53 MB in Uterine Lavage is Higher in Patients with Ovarian Cancer and has Significant Predictive Value Over Age and CA-125
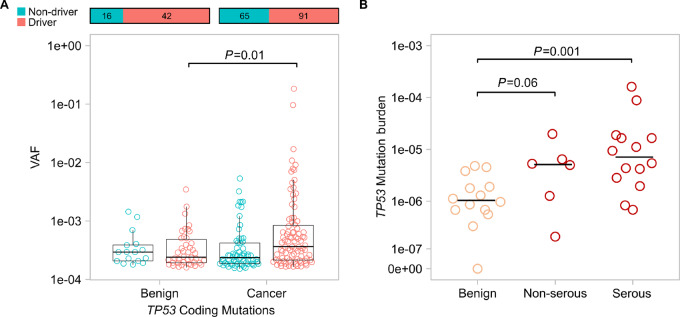
We then wondered whether the size of clones driven by cancer driver mutations was larger in patients with ovarian cancer than in those without. For all the genes with mutations in more than 50% of lavages, we plotted the VAF of cancer driver mutations in patients with and without ovarian cancer. Interestingly, we did not observe significant differences in any of the genes (Supplementary Fig. S10), with the exception of TP53 (Fig. 4A). Patients with and without ovarian cancer had similar proportions of cancer driver mutations, but the VAF of TP53 driver mutations was significantly higher in those with ovarian cancer, indicating larger clonal expansions.
FIGURE 4.
Uterine lavages from patients with ovarian cancer carry more large clones with TP53 driver mutations and show higher TP53 mutation burden than lavages from patients without cancer. A, Comparison of VAF of TP53 driver mutations in lavage DNA from patients with and without ovarian cancer. Cancer driver mutations in TP53 include substitutions occurring in common hotspots codons plus insertion/deletions, nonsense, and splice mutations. Each circle corresponds to a unique mutation. Overlying box plot displays the quartiles with whiskers extending up to 1.5× the interquartile range. Bar plots above display the total number of mutations in each group and the distribution between driver and non-driver. P values correspond to Mann–Whitney U test comparing the distribution of VAF of cancer driver mutations between patients with benign and cancer. B,TP53 mutation burden, calculated as the total number of TP53-mutant molecules identified in a lavage divided by the total number of nucleotides sequenced, is compared between patients with benign disease, non-serous ovarian cancer, and serous ovarian cancer (high-grade serous and carcinosarcoma). Each circle corresponds to an individual uterine lavage sample. Horizontal bars indicate the median for each group and P values correspond to Mann–Whitney U tests.
Given that ovarian cancer was associated with the number of TP53 mutations as well as the size of mutant clones, we reasoned that TP53 MB should be the best metric to discriminate patients with and without ovarian cancer because it counts not only the number of mutated positions, but the number of mutant molecules in each position, reflecting both number and size of clones. Thus, for all the lavages in the study, we calculated the MB of each gene (Supplementary Fig. S11). As expected, we did not observe significant differences in the MB of cancer and benign lavages except for TP53. Patients with ovarian cancer had significantly higher TP53 MB than those without ovarian cancer (P = 0.001). Notably, when separating by histology, we observed that TP53 mutation burden tended to be higher in serous as well as non-serous ovarian cancer (Fig. 4B). These results suggest that TP53 clonal evolution might be related not only to the development of HGSC, but also non-serous cancer despite not being the most common driver of those cancer types.
As with TP53 MF, TP53 MB was also significantly associated with age (Spearman correlation, P = 0.003). Because samples were consecutively collected and not age matched, we performed a sensitivity study restricting the comparison of TP53 MB with patients with and without cancer in the same age range (>50 and <72). TP53 MB was higher in patients with ovarian cancer with borderline significance (P = 0.055).
Finally, to test whether TP53 MB could provide clinical value for the detection of ovarian cancer in patients with pelvic masses, we built two logistic regression models to compare the predictive value of CA-125 and age versus CA-125, age, and TP53 MB (Table 2). We found that TP53 MB was significant (P = 0.036) even when accounting for CA-125 and age, suggesting potential value of TP53 MB to improve the predictive value of the current markers. This significant association was retained even when restricting to serous ovarian cancer, despite the smaller sample size. Given the strong associations observed for TP53 MB and ovarian cancer despite low numbers, future larger studies are warranted to confirm these findings.
TABLE 2.
Logistic regression for ovarian cancer prediction
| All patients (n = 34) | Non-serous cancer excluded (n = 28) | |||
|---|---|---|---|---|
| β (95% Cl) | P | β (95% Cl) | P | |
| Current predictive variables | ||||
| Age (per SD increase)a | 1.08 (0.01–2.15) | 0.048 | 1.16 (−0.05 to 2.37) | 0.061 |
| CA-125 (log tranformed) | 0.66 (0.09–1.23) | 0.024 | 0.73 (0.10–1.35) | 0.023 |
| Exploratory model | ||||
| TP53 mutation burden (per SD increase)b | 19.14 (1.29–36.98) | 0.036 | 24.82 (0.39–49.26) | 0.046 |
| Age (per SD increase)a | 1.15 (−0.63 to 2.92) | 0.205 | 2.05 (−0.98 to 5.07) | 0.185 |
| CA-125 (log tranformed) | 0.99 (0.15–1.83) | 0.021 | 1.09 (0.10–2.09) | 0.031 |
aAge per SD increase = 10 years.
b TP53 mutation burden per SD increase = 3.05E-5.
Discussion
We have demonstrated that the combination of uterine lavage with ultradeep sequencing enables detection of ovarian cancer at two levels. First, tumor DNA was identified in more than two-thirds of lavages, even when present at very low frequency (<0.001), and from patients with early-stage cancers. Second, in addition to tumor cells, lavages also carried abundant TP53 clonal expansions, which were more frequent and larger in patients with ovarian cancer. TP53 MB, which captures both the number and size of clonal expansions, was associated with ovarian cancer independently of age and CA-125, suggesting potential as an ovarian cancer biomarker. Interestingly, while other ovarian cancer genes also showed extensive clonal expansions in lavage DNA, these expansions were observed at similar levels in patients with and without ovarian cancer, pointing to the unique role of TP53 clonal expansions in association with ovarian cancer development.
The detection rate of TP53 tumor driver mutations in lavages from patients with serous ovarian cancer was 79%, similar to the 80% detection rate that we reported in a prior study (11). For non-serous ovarian cancer, however, we detected the tumor mutation only in 33% of cases: 1/3 endometrioid carcinomas and 1/3 clear-cell carcinomas. While these numbers are small, they may reflect the different origins of disease. High-grade serous ovarian cancer often arises in fallopian tube epithelium, as can carcinosarcoma. However, clear-cell and endometrioid ovarian cancer generally arise in endometriosis. Therefore, uterine lavages might not capture cancer cells as frequently in cancers arising beyond the tubal lumen. Remarkably, the TP53 driver mutation could be detected in five of six early-stage serous ovarian cancer cases indicating potential for the detection of early lesions. There are multiple potential explanations for the finding of tumor-specific TP53 mutations in the uterine lavage, especially considering some cases involved cancer confined to an ovarian cyst. In some cases, tubal or myometrial involvement of the tumor can lead to dissemination of the TP53-mutant clones. Alternatively, cancer cells may travel through lymphatic or hematologic channels, or the captured clones may represent nonmalignant parallel somatic evolution. Furthermore, TP53 foci (also known as TP53 signatures) are common in fallopian tubes of women with HGSC or at high risk of HGSC (34) indicating that multiple TP53-mutant clones coexist and are potential precursors. TP53-mutant cells might exfoliate from these foci, in agreement with the precursor escape theory of HGSC carcinogenesis (35), and be collected in uterine lavage.
To clinically apply the detection of tumor-specific mutations for cancer diagnosis, tumor mutations must be distinguishable from background mutations without a priori knowledge of the tumor genetics. In our serous cohort, only 7 of 11 TP53 driver mutations were present at VAF higher than 0.003 (all but one non-cancer case were below that threshold) and with more than 4× the VAF of the largest background mutation. While this sensitivity is limited for the identification of cancer cases without prior knowledge of the tumor mutation, we have discovered that TP53 background mutations carry valuable information that could also be leveraged for ovarian cancer screening.
We found that uterine lavages detect multiple background mutations in cancer driver genes. More than half of these mutations were common cancer driver mutations, including canonical mutations in KRAS and PIK3CA. For two of the oncogenes (KRAS and PIK3CA) and three of the tumor suppressor genes (PPP2R1A, ARID1A, and TP53), cancer driver mutations were not only very abundant, but also had significantly higher VAF than non-driver mutations, indicating clonal expansion of mutant clones. On the basis of prior studies (36–39) and the nature of uterine lavage, it is likely that most of the mutations observed in lavage DNA originate in endometrium. Whole genome and target sequencing of endometrial glands have identified extensive clonal expansions in individuals without endometrial cancer, with more frequent somatic mutations seen in KRAS and PIK3CA specifically (36–38). In addition, a prior study using NGS of uterine lavage samples also identified somatic mutations in PIK3CA, KRAS, and PTEN (among other genes), representing clonal expansion in the normal endometrium (39). Overall, our results extend these prior findings by providing a high-resolution (VAF < 0.001) characterization of cancer driver mutations in uterine lavage of patients with and without ovarian cancer and provide strong evidence supporting the new paradigm of clonal evolution in normal tissue (19).
Interestingly, TP53 was the only tested gene with a different frequency and burden of mutations in uterine lavages from patients with and without ovarian cancer. TP53-driven clonal expansions appeared to be linked to the development of ovarian cancer, with increased driver mutation VAF, mutational frequency, and mutational burden in ovarian cancer cases compared with benign cases. Notably, the high burden of TP53 mutations in lavage was not always due to the specific tumor mutation alone but to other large TP53-mutant clones present in lavage DNA from serous as well as non-serous ovarian cancer. While we cannot determine the origin of the TP53-mutant clones in lavage, our data are consistent with the hypothesis that an increased burden of TP53-mutant clones is associated with the development of ovarian carcinoma. TP53 clonal evolution is key to the current understanding of the pathogenesis of high-grade serous ovarian cancer as an evolutionary process initiated from TP53-mutant cells in the fallopian tube (40). Aside from the possibility of early precursor escape of TP53-mutant cells from TP53 foci commonly found in fallopian tubes (34–35), excess of TP53 clones observed in lavages from patients with serous ovarian cancer may correspond to endometrial TP53 field effects extending to the fallopian tube epithelium. For non-serous cancers, the connection between TP53 clonal expansions and cancer development is not clear because these cancers are driven by mutations in multiple genes other than TP53 (21). The role of clonal expansions in carcinogenesis might not only be related to the direct growth of mutant cells but also to the generation of microenvironments that are permissive to the expansion of other mutant clones (19). Further studies with larger number of cases are warranted to explore this hypothesis. Nevertheless, from a clinical perspective, the measurement of TP53 MB in uterine lavage appears as a promising tumor agnostic, minimally invasive molecular test for screening or risk stratification of ovarian cancer. This is an urgent need especially in the subset of patients at high-risk of ovarian cancer due to inherited BRCA1 and BRCA2 mutations (41). While cost considerations must be addressed when applying this tool to a larger population, feasibility in the outpatient setting has already been demonstrated (12).
Our study has two important limitations. First, it was based on convenience collection of gynecologic cases given the pilot study design, and therefore was not age matched. To address this limitation, we performed logistic regression including TP53 MB, age and CA-125 as covariates and demonstrated that TP53 MB was significantly associated with ovarian cancer in the model. Second, because the number of cases is small, we have no power to test associations and interactions with other ovarian cancer risk factors such as prior chemotherapy, endometriosis, or germline BRCA mutations. Future studies with larger sample sizes matched for age that allow for adjustment of ovarian cancer factors are currently under way.
In conclusion, by performing deep sequencing of uterine lavage with a panel of common ovarian cancer genes, we have demonstrated a prevalent process of clonal expansion in patients with and without ovarian cancer in agreement with multiple findings of clonal evolution in normal endometrium. While TP53 tumor mutations can be found in lavages, the most relevant finding of our work is the discovery of increased TP53 mutation burden in lavage DNA of patients with ovarian cancer. This exploratory work expands upon prior utilization of uterine lavage DNA for ovarian cancer detection (10, 11) by focusing on the associations between mutational burden and risk and using higher-fidelity methods to improve the accuracy and detection of low-frequency mutations. Although the sample size is small, these findings support the emerging notion that clonal expansions of certain cancer susceptibility genes, in this case TP53, might be linked to the development of cancer and may harbor clinical value as a biomarker for cancer risk thus informing future study design.
Supplementary Material
Supplementary Methods
Table S1. Clinical patient information
Table S2. Duplex Sequencing gene targets
Table S3. Custom probes for hotspots
Table S4. Mutation Frequency and Mutation Burden by patient and gene
Table S5. Codons most frequently mutated in ovarian cancers
Table S6. List of coding mutations by patient and gene
S1. Experimental design
S2. Association between the variant allele frequency (VAF) of tumor mutations identified in uterine lavage with stage of the ovarian cancer (A) and preoperative CA-125 level (B)
S3. Gene distribution of the largest mutant clones identified in uterine lavage
S4. Correlations between Mutation Frequencies (MF) of genes commonly mutated in lavage DNA
S5. Comparison of Mutation Frequencies (MF) by patient group.
S6. Comparison of Mutation Frequency (MF) for individual genes in uterine lavages from patients with and without cancer
S7. Correlations between TP53 coding mutation frequency (MF) and age
S8. Distribution of driver and non-driver mutations identified in lavage DNA by gene and patient
S9. Mutation spectrum distribution of coding mutations detected in lavage DNA compared to ovarian cancer mutations reported in COSMIC
S10. Comparison of Variant Allele Frequency (VAF) of driver mutations in lavage DNA from patients with and without ovarian cancer
S11. Comparison of mutation burden (MB) of common ovarian cancer genes in lavage DNA from patients with and without ovarian cancer
Acknowledgments
We deeply thank all the patients who donated samples, without whom this research would not have been possible. Our team would also like to acknowledge the University of Washington gynecologic operating room staff members Rodrigo Hyas, Ronnel Jugarap, and Holly Olivieri for their assistance with this project and David L. Wells, M.D. for providing pathology review. This work was supported in part by T32 CA009515 (TSG), Rivkin Bridge Fund 649149 (RAR), R01 CA259384 (RAR), R21 CA240885 (RAR), the Minnesota Ovarian Cancer Alliance (BMN), the Tom and Nancy Jensen Gynecologic Cancer Research Fund (BMN), and the Northwest Biotrust Shared Resource of the Fred Hutch/University of Washington Cancer Consortium P30 CA015704.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
H.J. Gray reports other from UpToDate outside the submitted work. J.B. Liao reports grants from Merck, Precigen, AstraZeneca, Forty-Seven, Sumitomo Dainippon Pharma Oncology, Laekna Therapeutics, Instil Bio, Harpoon Therapeutics, and Genentech outside the submitted work. P. Speiser reports a patent to Three Lumen Balloon Catheter Apparatus US2015305725 issued and a patent to Non-Invasive Cancer Diagnosis EP51899 20130131 issued; and is shareholder of OVARTEC GmbH. E.M. Swisher reports grants from NIH during the conduct of the study. R.A. Risques reports grants from NCI, Rivkin Center, Minnesota Ovarian Cancer Alliance; and personal fees and other from TwinStrand Biosciences during the conduct of the study; grants from NCI and Rivkin Center; other from NanoString Technologies; and personal fees, non-financial support, and other from TwinStrand Biosciences outside the submitted work; in addition, R.A. Risques has a patent to WO/2018/175997 pending, licensed, and with royalties paid. No disclosures were reported by the other authors.
Authors’ Contributions
T.S. Ghezelayagh: Conceptualization, resources, formal analysis, investigation, visualization, methodology, writing-original draft, writing-review and editing. B. Kohrn: Data curation, formal analysis, visualization, methodology. J. Fredrickson: Investigation, methodology. E. Manhardt: Resources. M.R. Radke: Resources, data curation. R. Katz: Formal analysis, investigation. H.J. Gray: Resources. R.R. Urban: Resources. K.P. Pennington: Resources. J.B. Liao: Resources. K.M. Doll: Resources. E.J. Simons: Resources. J.K. Burzawa: Resources. B.A. Goff: Resources. P. Speiser: Resources. E.M. Swisher: Resources. B.M. Norquist: Conceptualization, resources, supervision, funding acquisition, investigation, project administration, writing-review and editing. R.A. Risques: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing-original draft, project administration, writing-review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 3. Ovary SEER 5-Year Relative Survival Rates, 2012–2018. Surveillance, Epidemiology, and End Results (SEER) Program Populations (1969–2020) (www.seer.cancer.gov/popdata), National Cancer Institute, DCCPS, Surveillance Research Program; 2022.
- 4. Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011;305:2295–303. [DOI] [PubMed] [Google Scholar]
- 5. Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 2016;387:945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson JT, Webber EM, Sawaya GF. Screening for ovarian cancer: updated evidence report and systematic review for the US preventive services task force. JAMA 2018;319:595–606. [DOI] [PubMed] [Google Scholar]
- 7. Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih I-M, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5:167ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Li L, Douville C, Cohen JD, Yen T-T, Kinde I, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med 2018;10:eaap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krimmel-Morrison JD, Ghezelayagh TS, Lian S, Zhang Y, Fredrickson J, Nachmanson D, et al. Characterization of TP53 mutations in Pap test DNA of women with and without serous ovarian carcinoma. Gynecol Oncol 2020;156:407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, Vergote I, et al. Lavage of the uterine cavity for molecular detection of mullerian duct carcinomas: a proof-of-concept study. J Clin Oncol 2015;33:4293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salk JJ, Loubet-Senear K, Maritschnegg E, Valentine CC, Williams LN, Higgins JE, et al. Ultra-sensitive TP53 sequencing for cancer detection reveals progressive clonal selection in normal tissue over a century of human lifespan. Cell Rep 2019;28:132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maritschnegg E, Heitz F, Pecha N, Bouda J, Trillsch F, Grimm C, et al. Uterine and tubal lavage for earlier cancer detection using an innovative catheter: a feasibility and safety study. Int J Gynecol Cancer 2018;28:1692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat Protoc 2014;9:2586–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012;109:14508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krimmel JD, Schmitt MW, Harrell MI, Agnew KJ, Kennedy SR, Emond MJ, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A 2016;113:6005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matas J, Kohrn B, Fredrickson J, Carter K, Yu M, Wang T, et al. Colorectal cancer is associated with the presence of cancer driver mutations in normal colon. Cancer Res 2022;82:1492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martincorena I. Somatic mutation and clonal expansions in human tissues. Genome Med 2019;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Risques RA, Kennedy SR. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet 2018;14:e1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kakiuchi N, Ogawa S. Clonal expansion in non-cancer tissues. Nat Rev Cancer 2021;21:239–56. [DOI] [PubMed] [Google Scholar]
- 20. Kennedy SR, Zhang Y, Risques RA. Cancer-associated mutations but no cancer: insights into the early steps of carcinogenesis and implications for early cancer detection. Trends Cancer 2019;5:531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurman RJ, Shih Ie-M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol 2016;186:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010;221:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghezelayagh TS, Pennington KP, Norquist BM, Khasnavis N, Radke MR, Kilgore MR, et al. Characterizing TP53 mutations in ovarian carcinomas with and without concurrent BRCA1 or BRCA2 mutations. Gynecol Oncol 2021;160:786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salani R, Kurman RJ, Giuntoli R 2nd, Gardner G, Bristow R, Wang TL, et al. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer 2008;18:487–91. [DOI] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmitt MW, Fox EJ, Prindle MJ, Reid-Bayliss KS, True LD, Radich JP, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods 2015;12:423–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R Core Team. R: A language and environment for statistical computing. 4.1.1. Vienna, Austria: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 29. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:18032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. IBM Corp. IBM SPSS Statistics for Windows. 26.0. Armonk, NY: IBM Corp.; 2019. [Google Scholar]
- 32. StataCorp. Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC; 2019. [Google Scholar]
- 33. Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Norquist BM, Garcia RL, Allison KH, Jokinen CH, Kernochan LE, Pizzi CC, et al. The molecular pathogenesis of hereditary ovarian carcinoma: alterations in the tubal epithelium of women with BRCA1 and BRCA2 mutations. Cancer 2010;116:5261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soong TR, Howitt BE, Horowitz N, Nucci MR, Crum CP. The fallopian tube, "precursor escape" and narrowing the knowledge gap to the origins of high-grade serous carcinoma. Gynecol Oncol 2019;152;426–33. [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi M, Nakaoka H, Suda K, Yoshihara K, Ishiguro T, Yachida N, et al. Spatiotemporal dynamics of clonal selection and diversification in normal endometrial epithelium. Nat Commun 2022;13:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore L, Leongamornlert D, Coorens THH, Sanders MA, Ellis P, Dentro SC, et al. The mutational landscape of normal human endometrial epithelium. Nature 2020;580:640–6. [DOI] [PubMed] [Google Scholar]
- 38. Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep 2018;24:1777–89. [DOI] [PubMed] [Google Scholar]
- 39. Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, Schumacher CA, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med 2016;13:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shih IM, Wang Y, Wang TL. The origin of ovarian cancer species and precancerous landscape. Am J Pathol 2021;191:26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017;317:2402–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Table S1. Clinical patient information
Table S2. Duplex Sequencing gene targets
Table S3. Custom probes for hotspots
Table S4. Mutation Frequency and Mutation Burden by patient and gene
Table S5. Codons most frequently mutated in ovarian cancers
Table S6. List of coding mutations by patient and gene
S1. Experimental design
S2. Association between the variant allele frequency (VAF) of tumor mutations identified in uterine lavage with stage of the ovarian cancer (A) and preoperative CA-125 level (B)
S3. Gene distribution of the largest mutant clones identified in uterine lavage
S4. Correlations between Mutation Frequencies (MF) of genes commonly mutated in lavage DNA
S5. Comparison of Mutation Frequencies (MF) by patient group.
S6. Comparison of Mutation Frequency (MF) for individual genes in uterine lavages from patients with and without cancer
S7. Correlations between TP53 coding mutation frequency (MF) and age
S8. Distribution of driver and non-driver mutations identified in lavage DNA by gene and patient
S9. Mutation spectrum distribution of coding mutations detected in lavage DNA compared to ovarian cancer mutations reported in COSMIC
S10. Comparison of Variant Allele Frequency (VAF) of driver mutations in lavage DNA from patients with and without ovarian cancer
S11. Comparison of mutation burden (MB) of common ovarian cancer genes in lavage DNA from patients with and without ovarian cancer
Data Availability Statement
Sequencing data from this study are available at the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under BioProject ID PRJNA879769.