Abstract
The balanced homeostasis of the G protein–coupled receptor (GPCR), rhodopsin (Rho), is required for vision. Misfolding mutations in Rho cause photoreceptor death, leading to retinitis pigmentosa (RP) and consequently blindness. With no cure currently available, the development of efficient therapy for RP is an urgent need. Pharmacological supplementation with molecular chaperones, including flavonoids, improves stability, folding, and membrane targeting of the RP Rho mutants in vitro. Thus, we hypothesized that flavonoids by binding to P23H Rho and enhancing its conformational stability could mitigate detrimental effects of this mutation on retinal health. In this work, we evaluated the pharmacological potential of two model flavonoids, quercetin and myricetin, by using in silico, in vitro, and in vivo models of P23H Rho. Our computational analysis showed that quercetin could interact within the orthosteric binding pocket of P23H Rho and shift the conformation of its N-terminal loop toward the wild type (WT)-like state. Quercetin added to the NIH-3T3 cells stably expressing P23H Rho increased the stability of this receptor and improved its function. Systemic administration of quercetin to P23H Rho knock-in mice substantially improved retinal morphology and function, which was associated with an increase in levels of Rho and cone opsins. In addition, treatment with quercetin resulted in down regulation of the UPR signaling and oxidative stress-related markers. This study unravels the pharmacological potential of quercetin to slow down the progression of photoreceptor death in Rho-related RP and highlights its prospective as a lead compound to develop a novel therapeutic remedy to counter RP pathology.
Keywords: cone opsin, flavonoid, photoreceptor, retinal degeneration, retinitis pigmentosa, rhodopsin
1 ∣. INTRODUCTION
Retinitis pigmentosa (RP) is a heterogeneous inherited neurodegenerative disease, which presents with death of photoreceptors in millions of people worldwide. Most mutations in the rod opsin (RHO) gene are the predominant cause of blindness related to the autosomal dominant form of RP (adRP) (Kennan et al. 2005; Mendes et al. 2005). These mutations are classified into several classes depending on the protein impairment they cause (Rakoczy et al. 2011; Athanasiou et al. 2018). Specifically, Class 2 Rho mutations cause defective binding of 11-cis-retinal, leading to an imbalanced Rho homeostasis triggering photoreceptor degeneration. The substitution of Pro23 to His in Rho is the most prevalent Class 2 mutation in the human population, but its pathology remains not fully understood and treatment is lacking. P23H mutation disrupts the network of specific interactions between amino acid residues that are critical for Rho stability and function (Fanelli & Seeber 2010). Thus, targeting this mutant with stabilizing agents to prevent or slow down the progression of RP is a valid approach. Several recent studies showed that stabilization of the Rho mutants, including P23H Rho with retinoid-based and non-retinoid small molecules, improved the fitness of these Rho variants in vitro (Behnen et al. 2018; Chen et al. 2018; Mattle et al. 2018; Pasqualetto et al. 2020). In addition, we found that flavonoids positively modulate the stability of ligand-free rod opsin and improve membrane targeting of P23H Rho in vitro (Ortega et al. 2019). However, the effectiveness of these Rho modulators in RP has yet to be validated in vivo in animal models.
Another hallmark of RP linked to the P23H Rho mutation is the activation of cellular stress response due to over accumulation of misfolded Rho in the endoplasmic reticulum (ER) membranes followed by the activation of cell death mechanisms due to chronic stress caused by the mutant protein (Chiang et al. 2015; Walter & Ron 2011). Targeting the ER stress signaling to rescue photoreceptors in RP is one of the therapeutic strategies (Mostafa et al. 2019; Lin et al. 2007; Gupta et al. 2019). Interestingly, flavonoids in addition to their opsin stabilizing effect can modulate the ER stress pathways (Mostafa et al. 2019; Pandey et al. 2019) and exhibit neuroprotective effects on retinal cells in the RP mouse models (Liu et al. 2021; Perdices et al. 2020; Piano et al. 2019). Also, as we recently found, flavonoids prevent light-induced retinal degeneration, likely due to their ability to stabilize the emerged ligand-free opsin and inhibit the cellular events leading to photoreceptor death (Ortega et al. 2021). Thus, flavonoids could offer a dual beneficial effect of a structural chaperone and a modulator of cellular processes to mitigate the P23H Rho-linked pathology.
In this study, the primary objective was to assess if flavonoids improve the stability and function of P23H Rho in vitro and examine if these compounds decelerate photoreceptor loss in vivo in P23H Rho knock-in mice, a model of RP to provide a treatment option for this blinding disease. As we found, quercetin enhanced stability and signaling of P23H Rho in vitro and delayed retinal degeneration in these mice, thus suggesting that quercetin could be used as a preventive treatment or a lead compound to develop new therapeutics against the P23H Rho-associated RP.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Chemicals and reagents
4′6′-Diamidino-2-phenyl-indole (DAPI) for the nuclear staining was purchased from Life Technologies (Grand Island, NY). Dimethyl sulfoxide (DMSO) was obtained from Sigma (St. Louis, MO). Ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail tablets were purchased from Roche (Basel, Switzerland). 9-cis-retinal, quercetin, and myricetin were purchased from Sigma. NP40 lysis buffer was purchased from Invitrogen (Carlsbad, CA). Polyvinylidene difluoride (PVDF) membrane was obtained from Millipore (Burlington, MA). SYBR Green I Master mix for real-time quantitative polymerase chain reaction (RT-qPCR) was obtained from Thermo Fisher Scientific (Waltham, MA). Qiagen RNeasy Miniprep Kit and QuantiTect Reverse Transcription Kit were purchased from Qiagen (Venlo, Nederland's). RT2 Profiler™ PCR arrays were purchased from Qiagen (Germantown, MD).
2.2 ∣. Animals care and treatment
RhoP23H/P23H, RhoP23H/+ (Research Resource Identifier, RRID: IMSR_JAX:017628) (Sakami 2011), and C57BL/6J mice (RRID: IMSR_JAX:000664), Jackson Laboratory, Bar Harbor, ME) were used to test the protective effects of flavonoids against retinal degeneration at 14–21 days of age for RhoP23H/P23H and at 21–33 days of age for RhoP23H/+. Flavonoids were dissolved in DMSO/phosphate-buffered saline (PBS) (50/50 volume/volume) vehicle and administered to mice by intraperitoneal (i.p.) injection every other day at the same time of the day, three injections total at 20 mg/kg body weight (bw) according to the scheme shown below.
Both male and female mice were used in all experiments without any exclusion criteria. From each litter, one third of the animals were randomly assigned by simple randomization of flipping a coin to the control group and two thirds to the experimental group to test the effect of two different flavonoids—quercetin and myricetin. Mice were euthanized by cervical dislocation under deep anesthesia performed by i.p. injection of a cocktail containing ketamine (20 mg/ml) and xylazine (1.75 mg/ml) at a dose of 4 μl/g bw. All mice were housed in the Animal Resource Center at the School of Medicine, Case Western Reserve University (CWRU) and maintained in a 12-hr light/dark cycle at a constant temperature of 25°C. All animal procedures and experimental protocols were approved by the Institutional Animal Care and Use Committee at CWRU (Protocol number 2015-0124) and conformed to recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology, as well as the National Eye Institute Animal Care and Use Committee (NEI-ASP 682). Efforts were taken to minimize animal suffering.
This study was not preregistered. The total number of mice in this study was 148. Five or six mice per treatment group were used.
2.3 ∣. SD-OCT
The effect of flavonoids on the retina structure in RhoP23H/P23H and RhoP23H/+ mice we used ultrahigh-resolution spectral domain-optical coherence tomography (SD-OCT) (Bioptigen, Morrisville, NC) in vivo retina imaging. Mice pupils were dilated with 1% tropicamide and mice were anesthetized by i.p. injection of a cocktail containing ketamine (20 mg/ml) and xylazine (1.75 mg/ml) at a dose of 4 μl/g bw before imaging begins. The a-scan/b-scan ratio was set at 1200 lines. The OCT retinal images were obtained by scanning at 0 and 90° in the b-mode. Ten image frames were captured and averaged. The changes in the retinas of control nontreated mice and mice treated with flavonoid were determined by measuring the thickness of the retina 0.5–1.5 mm from the optic nerve head (ONH). Values of the retinal thickness were plotted using means and standard deviation (S.D.). Six mice were used in each experimental group.
2.4 ∣. Retinal histology
The effect of flavonoids on the retinal morphology in RhoP23H/P23H and RhoP23H/+ mice in comparison to vehicle-treated mice was determined by retinal histology analysis. Eyes were collected from euthanized RhoP23H/P23H and RhoP23H/+ mice at postnatal day 21 (P21) and P33, respectively, and fixed in 10% formalin in PBS for 48 h at room temperature (RT) followed by paraffin sectioning. Sections (5-μm thick) were stained with hematoxylin and eosin (H&E) and imaged with a ZEISS Axio Scan.Z1 slide scanner (Carl Zeiss Microscopy GmBH, Jena, Germany). The data were processed using Zeiss-Zen 3.2 software (blue edition). Six mice were used in each experimental group.
2.5 ∣. Immunohistochemistry
To detect the rod and cone photoreceptors, eyes were collected from flavonoid-treated or vehicle-treated euthanized RhoP23H/P23H mice 7 days after treatment at P21 and from RhoP23H/+ 12 days after treatment at P33. These eyes were fixed in 4% paraformaldehyde (PFA) for 24 hr, followed by their incubation in 1% PFA for 48 hr at RT, and then processed for cryo-sectioning. Sections (8-μm thick) were blocked with 10% normal goat serum (NGS) and 0.3% Triton X-100 in PBS for 1 hr at room temperature and then stained overnight at 4°C with a monoclonal 1D4 anti-Rho primary antibody to visualize rod photoreceptors and biotinylated peanut agglutinin (PNA) to visualize cones. The next day after washing with PBS, sections were incubated with Alexa Fluor 555-conjugated goat anti-mouse secondary antibody to detect rods and Fluor 488-conjugated streptavidin to detect cones for 2 hr at RT. Cell nuclei were detected by staining with DAPI. Slides were coverslipped with Fluoromount-G (SouthernBiotech).
2.6 ∣. Electroretinography
Electroretinography (ERG) recording was used to assess retinal function in both RhoP23H/P23H and RhoP23H/+ mice after treatment with flavonoids in comparison to vehicle-treated mice. Before ERG measurements, mice were anesthetized with a cocktail of 20 mg/ml ketamine and 1.75 mg/ml xylazine, and pupils were dilated with 1% tropicamide. Scotopic and photopic ERGs were recorded for both eyes of each mouse using a Celeris rodent ERG system and Espion Dyagnosys software Version 6 (Dyagnosys, LLC., Lowell, MA). The data were processed for each condition and ERG data were represented as means and S.D. for both a-wave and b-wave amplitudes. Each experimental group contained five mice.
2.7 ∣. RNA profiler array
The expression profile of genes associated with the mouse unfolded protein response (UPR) and its signal transduction pathways were assayed by using RT2 Profiler™ PCR Array (Qiagen, catalog number PAMM 089ZC). Total RNA was isolated from RhoP23H/P23H mouse eyes at P21 treated either with quercetin or vehicle, using Qiagen RNeasy kit following the manufacturer's protocol. Five mice per treatment group were used. Then, the RNA quality and quantification were carried out and a complementary DNA strand for each condition was amplified by using RT2 First-Strand Kit (Qiagen, catalog number 330401). The amplified cDNA was diluted with nuclease-free water and added to the RT2 qPCR SYBR Green Master Mix following the manufacture's protocol. The qPCR was performed on a StepOnePlus Real-Time PCR system (Applied Biosystems) according to the RT2 Profiler PCR Array instructions. The amplification was carried out under the following conditions: 95°C for 10 min, then 40 cycles at 95°C for 15 s, and 60°C for 1 min. Data analysis was conducted using raw Ct values for each gene and the RT2 Profiler PCR Array Data Analysis Spreadsheet. The built-in into an array plate housekeeping genes and amplification controls were used for normalization and data validation. The fold change of gene expression was shown as a heat map for each experimental condition.
2.8 ∣. Quantification of gene expression
The effect of flavonoids on the selected gene expression was validated in mouse eyes collected from RhoP23H/P23H mice at P21 and RhoP23H/+ mice at P33 treated either with flavonoid or with vehicle. Five mice per treatment group were used. Total RNA was isolated from the whole eye extracts by using the Qiagen RNeasy Miniprep Kit following the manufacturer's protocol. The obtained RNA samples were treated with DNase I to digest any residual chromosomal DNA. The RNA concentration was determined with a nanodrop spectrophotometer (Thermo Fisher Scientific). RNA was transcribed to cDNA by using the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer's protocol. Quantitative RT-PCR amplification was performed using LUNA NEB SYBR Green Master Mix (NEB) according to the manufacturer's protocol and using the StepOnePlus Real-Time PCR system (Applied Biosystems). The PCR conditions were as follows: 95°C for 3 min followed by 40 cycles of 95°C for 20 s and 60°C for 30 s. Fluorescence data were acquired at the step of 60°C. Gapdh was used as a control housekeeping gene. All data were normalized to Gapdh expression level, and the fold change was calculated for each gene. The amplified products were measured by amplification curve analysis using StepOne software version 2.3. The gene expression was measured using the comparative 2–(ΔΔCt) method. All primers used in this study are listed in Table 1.
TABLE 1.
The list of used primers
| Target | Species | Forward primer sequence 3′→5′ | Reverse primer sequence 3′→5′ |
|---|---|---|---|
| Rho | Mouse | CTTCCTGATCTGCTGGCTTC | ACAGTCTCTGGCCAGGCTTA |
| M Opsin | Mouse | GAGATTCAAGAAGCTGCGCC | TGTCCAGAACGAGTAGCC |
| S Opsin | Mouse | ACCATCTCCAGAATGCAAGC | ATGGTCAACAATCGGAACCA |
| Perk1 | Mouse | GTATTTCCCGGGTGAGCCAT | TCCATGTTTCTCACTGCCGC |
| Ire1 | Mouse | ACTGCCATTCACCATCCCAG | GTATTTCCCGGGTGAGCCAT |
| Chop1 | Mouse | CCTCTCAGTCAGCTGCGAAT | TGTGGGAAGCTTGGGCAAAA |
| Grp78 | Mouse | TGTGTGTGAGACCAGAACCG | TAGGTGGTCCCCAAGTCGAT |
| Atf6 | Mouse | AATTCTCAGCTGATGGCTGT | TTGTTGTGGGTGGTAGCTGG |
| Kit | Mouse | ATTCCCGGAGCCCACAATAG | GCCTGGATTTGCTCTTTGTTGT |
| Hmox-1 | Mouse | TCAAGGCCTCAGACAAATCC | ACAACCAGTGAGTGGAGCCT |
| Sod1 | Mouse | AAAATGAGGTCCTGCACTGG | ACCATCCACTTCGAGCAGAA |
| Catalase | Mouse | ACCACACATCCTGAACGAGGAG | GATGAAGCAGTGGAAGGAGC |
| Gapdh | Mouse | TTGAGGTCAATGAAGGGGTC | TCGTCCCGTAGACAAAATGG |
| Caspase-3 | Mouse | GGTGCCCTAAACCTGGCTA | CGTCCACATCGTACCAGAG |
| TNF-α | Mouse | GGTCTGGGCCATAGAACTGA | CAGCCTCTTCTCATTCCTGC |
| Mef2c | Mouse | TCCACCTCCCAGCTTTGAGAT | TGCCAGGTGGGATAAGAACG |
| Nr2e3 | Mouse | CCGGCTGAAGAAGTGCTTAC | TAAGGCTGGCCATAAAGTGG |
| Perk1 | Human | AGAGATTGAGACTGCGTGGC | GGTCTTGGTCCCACTGGAAG |
| Ire1 | Human | ACTGCCATTCACCATCCCAG | GTATTTCCCGGGTGAGCCAT |
| Chop1 | Human | TTGCCTTTCTCCTTCGGGAC | TCCTCCTCTTCCTCCTGAGC |
| Grp78 | Human | GAACGTCTGATTGGCGATGC | TCCCAAATAAGCCTCAGCGG |
| Kit | Human | CCTATCCTGGAATGCCGGTC | GTCTACCACGGGCTTCTGTC |
| Serp1 | Human | AAAGCTACCGTGGTGGAGTG | AACTTCGCAGTTGATGTGCC |
| Gapdh | Human | GAAAGCCTGCCGGTGACTAA | TGGAATTTGCCATGGGTGGA |
2.9 ∣. Immunoblotting
The proteins were extracted from the whole eyes collected from RhoP23H/P23H mice at P21 mice and RhoP23H/+ mice at P33 treated with either flavonoids or vehicle. Five mice per treatment group were used. The eyes were mechanically homogenized in an NP40 lysis buffer (Invitrogen) containing a 1% protease inhibitor cocktail (Roche). Then, the lysates were centrifuged at 12,000 g for 15 min at 4°C. The protein concentration was measured with a BCA Protein Assay Kit (Thermo Fisher Scientific) with bovine serum albumin as a standard. The protein extract (60 μg/lane) was mixed with a sample buffer, boiled for 5 min at 95°C, and loaded on an SDS-PAGE gel. Alternatively, protein extract was loaded on the SDS-PAGE gel immediately after mixing with a sample buffer without boiling. The protein samples were separated with 12% SDS-PAGE gel electrophoresis and then transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). The PVDF membrane was probed with the primary antibodies followed by the horse reddish peroxidase (HRP)-conjugated secondary antibody listed in Table 2. The immunoblots were developed with a ProSignal reagents kit and the Odyssey Imaging System (LI-COR, Biosciences). GAPDH was used as the loading control. The protein bands were quantified by densitometry analysis with ImageJ software. Band intensities were normalized to the intensity of GAPDH. The mean of data from three independent experiments is presented in the figures.
TABLE 2.
The list of used antibodies
| Antibody name | Species | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Anti-M Opsin | Rabbit polyclonal | Millipore | No: NP_064445 | 1:1000 |
| Anti-ATF6 | Rabbit polyclonal | Santa Cruz | No: sc-166659 | 1:500 |
| Anti-Grp78 (E-4) | Mouse monoclonal | Santa Cruz | No: sc-166490 | 1:500 |
| Anti-Perk1 | Rabbit polyclonal | Abcloal | No: A17940 | 1:250 |
| Anti-Catalase | Rabbit polyclonal | Abclonal | No: A11777 | 1:1000 |
| Anti-HMOX-1 | Rabbit polyclonal | Abclonal | No: A1346 | 1:1000 |
| Anti-GAPDH | Mouse monoclonal | Abclonal | NO: AC002 | 1:10,000 |
| Anti-mouse IgG, HRP conjugate | Goat | Promega | No: W4021 | 1:10,000 |
| Anti-rabbit IgG, HRP conjugated | Goat | Promega | No: W4011 | 1:10,000 |
| Anti-mouse IgG Alexa Fluor 555-conjugated | Goat | Thermofisher | No: A28180 | 1:400 |
| Anti-rabbit IgG Alexa Fluor 555-conjugated | Goat | Thermofisher | No: A27039 | 1:400 |
2.10 ∣. Retinoid analysis
Retinoids were extracted from RhoP23H/P23H and RhoP23H/+ mouse eyes collected under dim red light at P21 and P33, respectively, using previously published methods (Palczewski et al. 1999). Five mice per treatment group were used. Briefly, eyes were homogenized in methanol/PBS (50/50 volume/volume) solution, containing hydroxylamine (pH 7.5) added to 40-mM final concentration, followed by 20-min incubation at RT. The resulted retinal oximes were extracted with hexane and their isomeric content was determined by normal phase high-performance liquid chromatography (HPLC) with a Luna 10-μm PRE-Silica (3) 100 Å, 250 × 4.6-mm column (Beckman, San Ramon, CA). Retinoids were eluted isocratically with 10% ethyl acetate in hexane at a flow rate of 1.4 ml/min. Their signals were detected by absorption at 360 nm (Garwin & Saari 2000).
2.11 ∣. Cell culture
NIH-3T3 cells stably expressing WT or P23H rod opsin (Ortega et al. 2019) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and 1 unit/ml penicillin with 1 μg/ml streptomycin (Life Technologies) at 37°C under 5% CO2 according to the instructions from the ATCC Animal Cell Culture Guide.
2.12 ∣. Cell treatment with flavonoids, pigment reconstitution, and purification by 1D4 immunoaffinity chromatography
The NIH-3T3 cells expressing WT or P23H rod opsin were plated to 70 plates with a 10-cm diameter. At ~90% confluency, cells were treated with 10-μM quercetin for 4 hr before 5-μM 9-cis-retinal was added to the plates for 16 hr. Then, cells were harvested and pelleted by centrifugation at 800 g. The cell pellet was suspended in a buffer composed of 20-mM Bis-tris propane (BTP), 120-mM NaCl and protease inhibitor cocktail, pH 7.5, and 20-mM n-dodecyl-β-D-maltopyranoside (DDM). The cell suspension was incubated for 1 hr at 4°C on the rotating platform to solubilize membranes. The lysate was centrifuged at 100,000g for 1 hr at 4°C, and pigments were purified from the supernatant by immunoaffinity chromatography with an anti-Rho C-terminal 1D4 antibody immobilized on CNBr-activated agarose. Three hundred microliters of 6-mg 1D4/ml agarose beads was added to the supernatant and incubated for 1 hr at 4°C on the rotating platform. The resin was then transferred to a column and washed with 15 ml of 20-mM BTP, 120-mM NaCl, and 2-mM DDM, pH 7.5. Pigments were eluted with the same buffer, supplemented with 0.6 mg/ml of the TETSQVAPA peptide.
2.13 ∣. UV-visible spectroscopy of rho pigments
UV-visible spectra were measured in the dark from freshly purified samples using a UV-visible spectrophotometer (Cary 60, Varian, Palo Alto, CA), and their concentrations were quantified based on the extinction coefficient calculated for 9-cis-retinal regenerated pigment, ε 487 nm = 43,000 M−1 cm−1 (Spalink et al. 1983).
2.14 ∣. Thermal stability
Freshly purified WT Rho or P23H Rho samples from cells either treated with flavonoids or nontreated were incubated at 25°C or 37°C in the dark, and their spectra were recorded every 2 min for up to 1 hr. The absorbance at the maximum wavelength (485 nm for WT Rho and 479 nm for P23H Rho) was assumed 100% at the initial time point. The calculated percentages of the remaining pigments were plotted as a function of time. The half-times (t1/2) of the chromophore release were calculated from these plots. All samples were measured in triplicate.
2.15 ∣. cAMP detection
The NIH-3T3 cells stably expressing either WT Rho or P23H rod opsin were plated in two 96-well plates at a density of 50,000 cells per well in 85 μl of DMEM medium containing 10% FBS and antibiotics. The cells were treated with quercetin at different concentrations for 16 hr. Next, 9-cis-retinal was added for 2 hr to regenerate isoRho in cells treated or not with the flavonoid compound. Then, one plate was kept in the dark, while the second plate was exposed to bright (150 Watt) light for 15 min from a 10 cm-distance. The levels of accumulated cAMP were detected with the cAMP-Glo™ (Promega) following the manufacturer's protocol. The luminescence signal was recorded with a FlexStation 3 plate reader (Molecular Devices). A standard curve was developed using cAMP provided by the kit, and the percentage of cAMP was calculated for each condition. The values were expressed as a percentage, assuming the cAMP level detected in the nontreated cells as 100%. The experiment was performed in triplicate.
2.16 ∣. In silico analysis
The monomer of bovine rod opsin (PDB ID: 3CAP) was used, and the single point mutation P23H was introduced by using DUET software to produce the structural model suitable for in silico analysis. The structure of P23H rod opsin obtained was then submitted to the restrained molecular mechanics refinement with NAMD 2.12 software using the CHARMM22 force field. The structure quality was evaluated using ProCheck and ProSA programs. The molecular docking of quercetin to P23H rod opsin structural model was performed using VINA/VegaZZ 3.1.0.21 software as described in Ortega et al. (2019) running for 30 iterations. The resulting quercetin-opsin complex was visualized with the Biovia Discovery Studio Visualizer 17.2.0 software. The 2D diagrams were obtained by selecting the main atoms that interacted with the ligand. The WT rod opsin, P23H rod opsin, quercetin-bound WT rod opsin, and quercetin-bound P23H rod opsin structures were next used for the molecular dynamic (MD) simulations. The structures were submitted to the CABS-flex server individually and processed using the parameters set as default. The resolution of the structural models generated by the CABS-flex software allowed for the reconstruction of all-atom representation of physically realistic models. The output set of all-atom models was generated through trajectory clustering (by k-medoids method) and subsequent multistep reconstruction and optimization procedures available in the software. The data obtained from MD simulations were analyzed according to the root-mean-square fluctuations (RMSFs). In addition, MD simulations of the proteins and protein-quercetin complexes were carried (Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020). Protein and protein-quercetin complexes were inserted in a membrane of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosph ocholine) (Koynova & Caffrey 1998) at 300K, while the transmembrane region was obtained following the data of UniProt database (https://www.uniprot.org/). The complexes created were immersed in a box filled with water molecules using the simple point charge (SPC) scheme. The dimension of the box was set to 10 × 10 × 10 Å. Counter ions (6 Cl−) were added to neutralize charges, and additional Cl− and Na+ ions were added to obtain a final NaCl concentration of 0.15 M. Energy minimization was carried out by 2,000 steps using the steepest descent method with a threshold of 1.0 kcal/mol/Å. Periodic boundary conditions were used, and a cutoff of 9 Å was established for van der Waals interactions, and the Particle Mesh Ewald (PME) method with a tolerance of 10−9 was used in the electrostatic part. The NPT simulations were realized at 300 K with the Nose–Hoover algorithm (Nose 1984), and the pressure was maintained at 1 bar with the Martyna-Tobias-Klein barostat (Martyna et al., 1994). The same force field used to prepare the molecules was used in all runs (Roos et al. 2019). The simulation length was 100–1000 ns. The final states of the proteins were analyzed by the protein structure network (PSN) using the software of webPSN (Felline et al. 2020). This tool allows inferring the hubs, links, and clusters between the residues of the protein.
2.17 ∣. Statistical analyses
The values obtained for the ONL thickness measurements using the SD-OCT images, H&E-stained images, and for the ERG measurements were expressed as means ± standard deviations (S.D.). Based on previous reports from our group, six or five mice per treatment group were used in each experiment (Gao et al. 2018; Ortega et al. 2021). The specific experiments were designed to have groups of equal sizes, using randomization and masked analysis. Each experiment was repeated at least two times. Each in vitro experiment was performed in triplicate and was repeated at least two times. Quantification data of gene and protein expression are presented as a triplicate and a mean. For multiple comparisons, one or two-way ANOVA with Bonferroni post hoc tests were used. Degrees of freedom (F) are reported in the figure legends within parenthesis. All statistical calculations were performed using Prism GraphPad 7.02 software. Type1 error tolerance for the experiments was established at 5%. Values of P < 0.05 were considered statistically significant. The analysis was performed by a different person than the experimenter. No sample calculation was performed. No exclusion criteria were predetermined.
3 ∣. RESULTS
3.1 ∣. Flavonoids increase the conformational stability of P23H Rho
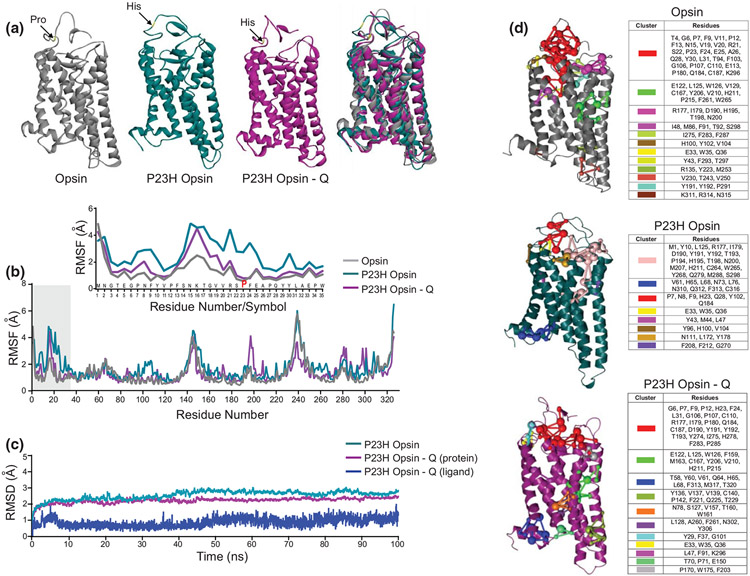
The P23H point mutation in the Rho N-terminal loop causes the conformational changes triggering decreased Rho structural stability. Previously, we found that flavonoids stabilize the ligand-free WT rod opsin (Ortega et al. 2019). Here we examined if flavonoids improve the conformational stability of P23H Rho. We developed the structural model of P23H rod opsin and performed molecular docking of quercetin to the orthosteric binding pocket of this mutant. Quercetin could bind to P23H rod opsin with a binding-free energy of −9.2 kcal/mol via π–π interactions and hydrogen bonds similar to WT rod opsin (Figure S2A and Ortega et al. 2019). The binding stability of quercetin and its effect on the structural conformation of P23H rod opsin were then examined by MD simulations (Figures 1a-c and S2b,c). We compared structures of the ligand-free and quercetin-bound P23H rod opsin to WT opsin. While the substitution of Pro23 to His increased the flexibility of the N-terminal loop, binding of quercetin resulted in the structural rearrangements shifting the conformation of this region toward WT-like (Figure 1a). The increased conformational Rho stability upon quercetin binding was also reflected in the RMSFs for the N-terminal residues (Figures 1b and S2c), indicating the potential of quercetin as a structural modulator. The root-mean-square deviation (RMSD) analysis showed that quercetin remained bound to the mutant or WT opsin (Figures 1c and S1). The PSN analysis indicated that the number of residue clusters that are important for the protein stability dramatically decreased in the P23H opsin as compared with WT opsin (Figure 1d, cyan and gray structures, respectively). However, binding of quercetin allowed to restore many of these interactions (Figure 1d, magenta structure). Altogether, these in silico results showed that flavonoids could bind to and modulate the structural stability of P23H rod opsin.
FIGURE 1.
The effect of flavonoids on the conformational stability of P23H Rho. (a) The ligand-free P23H rod opsin structure and the model of P23H rod opsin with quercetin bound were optimized and subjected to the molecular dynamic (MD) simulations carried out using Schrödinger software. A monomer of wild type (WT) rod opsin (PDB ID: 3CAP) was used as a control. The results of these MD simulations are shown. The extracellular N-terminal loop and the resident Pro23 residue are indicated with an arrow in the final pose obtained after MD simulations. The structural flexibility of the N-terminal loop increases upon the substitution of Pro23 to His residue. The binding of quercetin to P23H rod opsin increases its structural stability and facilitates the structural changes resembling the WT-like conformation. (b) The root-mean-square fluctuation (RMSF) for WT opsin (gray), P23H opsin (cyan), and P23H opsin with bound quercetin (magenta) plotted with the respect to the residue number of rod opsin. The N-terminal loop residues are highlighted with a gray bar. Inset, a close-up view of the N-terminal loop residues. (c) The root-mean-square deviation (RMSD) obtained for P23H opsin (cyan) and P23H opsin bound to quercetin (magenta) plotted with the respect to the initial pose. Also, the RMSD for the ligand bound to P23H opsin (dark blue) is shown. (d) The protein structure network (PSN) obtained for WT opsin, P23H opsin, and P23H opsin with quercetin bound to the orthosteric site. The specific residue interaction clusters are shown in the protein structures and specified in the tables
3.2 ∣. Flavonoids improve the stability and function of P23H Rho in vitro
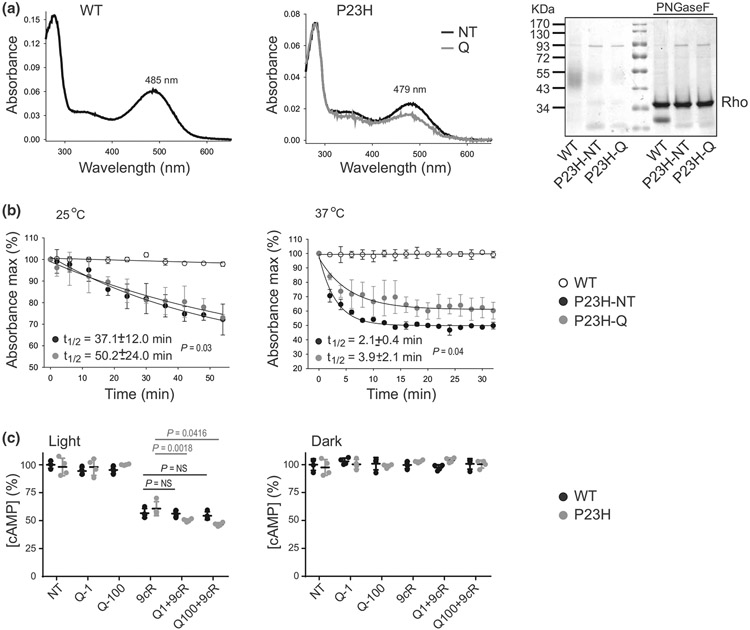
To validate our in silico findings, we examined if flavonoids increase the stability of P23H Rho in vitro. The NIH-3T3 cells expressing this mutant were incubated with quercetin followed by pigment regeneration with 9-cis-retinal and purification. 9-cis-Retinal was used because it exhibits greater stability than 11-cis-retinal and similar to the native retinal it enhances folding and membrane targeting of the P23H Rho mutant (Noorwez et al. 2004). The mutation caused a blue shift in the absorption maximum (λmax=479 nm) as compared with WT isorhodopsin (isoRho) (λmax=485 nm) (Figure 2a, middle and left panels, respectively). The stable binding of quercetin to P23H isoRho was evidenced by a small decrease in the λmax as compared with the protein purified from nontreated cells (Figure 2a, middle panel). Of note, treatment with quercetin did not change the mutant migration in the SDS-PAGE gel (Figure 2a, right panel). Quercetin bound to P23H isoRho provided a structural stabilization for the purified receptor and delayed its thermal decay at both 25°C and 37°C. The calculated half-times of quercetin-bound P23H isoRho were t1/2=50.2 ± 24.0 min at 25°C and 3.9 ± 2.1 min at 37°C as compared with nontreated P23H isoRho t1/2=37.1 ± 12.0 min at 25°C and 2.1 ± 0.4 min at 37°C (Figure 2b). Moreover, the binding of quercetin improved the functional signaling of this mutant. In the absence of its cognate G protein, transducin, Rho can couple to Gi/o signaling cascade. Using this ability, we examined the effect of quercetin on cAMP levels in the response to light stimulation in the NIH-3T3 cells stably expressing P23H isoRho. Cells were treated with quercetin at either 1- or 100-μM concentrations followed by isoRho regeneration with 9-cis-retinal (Figure 2c). The concentration of cAMP was substantially reduced in cells treated with either 9-cis-retinal or flavonoid followed by 9-cis-retinal and illuminated. Considerably, lower levels of cAMP were found in cells treated with both quercetin and 9-cis-retinal (Figure 2c, left panel, gray bars). Treatment with quercetin alone did not produce functional Rho. Consistent with previous observations, quercetin had no effect on cAMP levels in cells expressing WT isoRho (Figure 2c, left panel, black bars). Also, cAMP concentrations did not change in the dark (Figure 2c, right panel). Together, these results indicate the advantageous effect of flavonoids on the thermal stability and function of P23H isoRho in vitro.
FIGURE 2.
The effect of flavonoids on P23H Rho stability and function. Rho was purified from the NIH-3T3 cells stably expressing wild type (WT) Rho or the P23H Rho mutant. (a) UV-visible absorption spectra of WT Rho (left panel) and P23H Rho immunopurified from either nontreated (black line, middle panel) or quercetin-treated cells (gray line, middle panel) and regenerated with 9-cis-retinal. The representative coomassie blue-stained SDS-PAGE gel of the purified Rho samples either nontreated or treated with PNGaseF deglycosylase (right panel) indicates that treatment with quercetin did not change the migration pattern of P23H Rho. (b) Thermal stability of P23H Rho either nontreated or treated with quercetin in comparison to WT Rho upon incubation at 25°C (left panel) and 37°C (right panel) (n = 3, F3,8 = 66.10, p < 0.0001). The UV-visible absorption spectra were recorded every 2 min in the dark. The percentage of remaining pigments normalized to their initial concentrations was then plotted as a function of time. The half-time (t1/2) of chromophore release was calculated from these plots. Each measurement was performed in triplicate. The experiment was repeated twice. Error bars represent standard deviation (S.D.). (c) The effect of quercetin on the cAMP levels in the NIH-3T3 cells expressing P23H Rho either nontreated or treated with quercetin and exposed to light (n = 3, F5,36 = 235, p < 0.0001). Cells expressing WT Rho were used as a control. Cells were incubated with quercetin for 16 hr and 9-cis-retinal for 2 hr before the measurement. Forskolin was added to the cells to saturate their cAMP levels followed by light illumination. Control cells, nontreated, treated with quercetin, or treated with 9-cis-retinal underwent the same procedure. These measurements were also performed in the dark (n = 3, F5,36 = 0.52, p = 0.76). cAMP levels were detected as described in Section 2. Each condition was performed in triplicate, and the experiment was repeated twice. Statistical analysis was performed with the one-way ANOVA and Bonferroni post hoc tests. The P-values for statistically different changes are indicated in the figure. NT, nontreated; 9cR, treated with 5 μM 9-cis-retinal; Q-1, treated with 1-μM quercetin; Q-100, treated with 100-μM quercetin
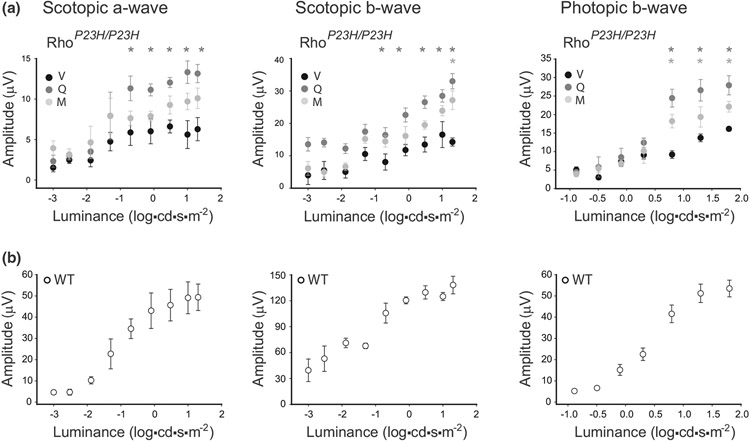
3.3 ∣. Flavonoids improve eye morphology, increase levels of visual receptors, and recover visual function in homozygous RhoP23H/P23H mice
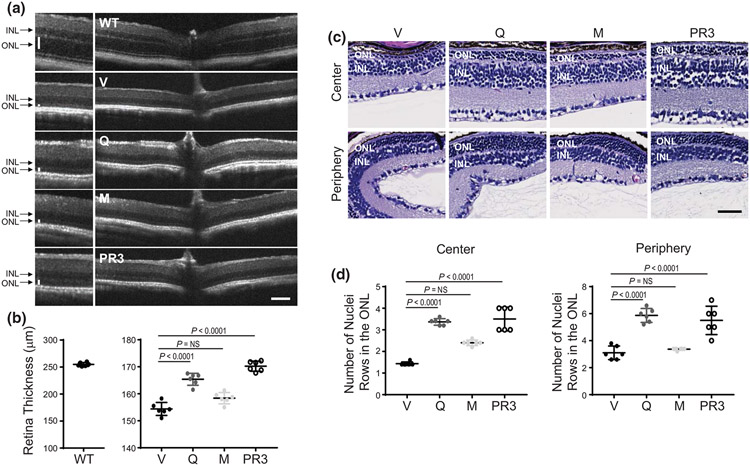
Our recent study revealed that quercetin and myricetin prevent retinal degeneration in mice injured with bright light (Ortega et al. 2021). To determine if flavonoids could prevent photoreceptor death in P23H Rho knock-in mice, we treated the homozygous RhoP23H/P23H mice with these two model flavonoids, quercetin, or myricetin, between P14 and P21 during the period of rapid photoreceptor degeneration. Control mice were treated with vehicle or photoregulin3 (PR3), which as shown by others slowed down retinal degeneration in RhoP23H/P23H mice (Nakamura et al. 2017). The OCT in vivo imaging indicated severe degeneration of the outer nuclear layer (ONL) in RhoP23H/P23H mice as compared with WT C57BL/6J mice (Figure 3a,b). Thus, to assess the effect of treatment with flavonoids, we measured the whole retina thickness and found considerable retinal thickening upon treatment with quercetin as compared with vehicle-treated mice (Figure 3a,b). In addition, this change was close to the improvement found for PR3. Treatment with myricetin was less effective, and the observed changes were not statistically different from vehicle-treated group. These results were confirmed by the histological assessment (Figure 3c). The vehicle-treated RhoP23H/P23H mice had only one to three remaining rows of photoreceptors within the ONL, 1.4 ± 1.4 in the center of the retina, and 3.0 ± 0.4 at the periphery. Of note, similar results were found in the nontreated mice. However, treatment with flavonoids resulted in an increased number of photoreceptor rows in the retina (center: 3.4 ± 0.1, periphery: 5.9 ± 0.5 for quercetin and center: 2.4 ± 0.1, periphery: 3.4 ± 0.1 for myricetin), indicating their therapeutic effect with higher effectivity of quercetin (Figure 3c,d).
FIGURE 3.
Protective effect of flavonoids against retinal degeneration in RhoP23H/P23H mice. Flavonoids (20 mg/kg bw) or dimethyl sulfoxide (DMSO) vehicle were administered intraperitoneally (i.p.) to homozygous RhoP23H/P23H mice at P14 every other day and mice were analyzed at P21. Wild type (WT) mice and RhoP23H/P23H mice treated with photoregulin3 (PR3) were used as controls. (a) Representative optical coherence tomography (OCT) images of the mouse eyes (n = 5, F4,25 = 1699, p < 0.0001). ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar, 100 μm. (b) The thickness of the retina measured at 500 μm from the optic nerve head (ONH). Error bars indicate S.D. Changes in the retina thickness observed between vehicle-treated and quercetin-treated but not myricetin-treated were statistically different. (c) Images of hematoxylin and eosin (H&E)-stained retinal sections visualizing the retina center and periphery. Scale bar, 50 μm. (d) The number of nuclei rows in the ONL counted in the retina center (n = 5, F3,20 = 64.32, p < 0.0001) and periphery (n = 5, F3,20 = 30.05, p < 0.0001). Error bars indicate standard deviation (S.D.). The changes found in mice treated with quercetin but not with myricetin were statistically different as compared with vehicle-treated control mice. Statistical analysis was performed with the one-way ANOVA and post hoc Bonferroni tests. The P-values for the statistically significant changes are indicated in the figure. The nonstatistically different changes are indicated as NS. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
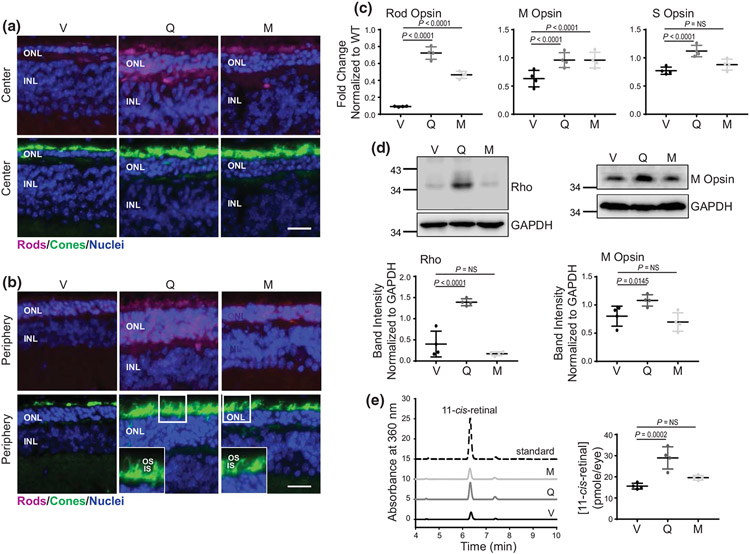
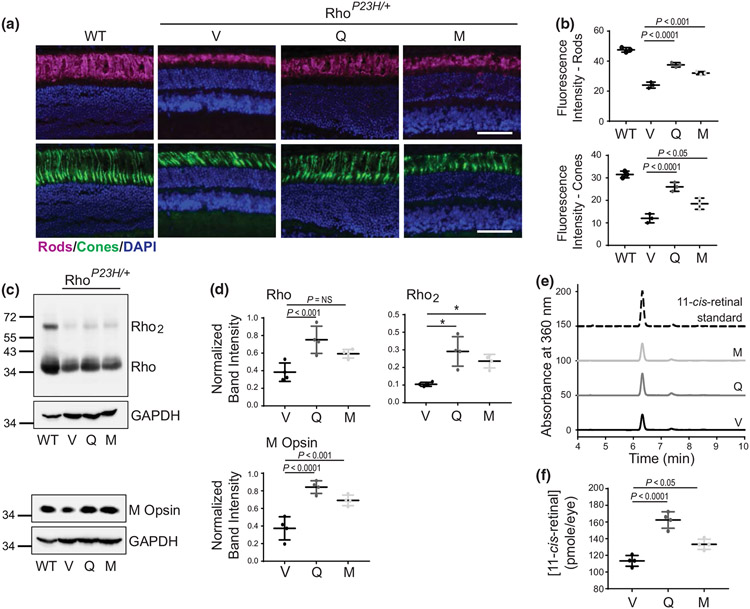
The levels of visual receptors, Rho, and cone opsins are directly related to photoreceptor homeostasis. By using immunostaining on the retinal cryosections from RhoP23H/P23H mice, we found that staining for Rho was hardly detectable and the labeling of cones was sporadic at both the center and periphery (Figure 4a,b), indicating severe photoreceptor degeneration. However, in mice treated with quercetin or myricetin, the levels of Rho and cones staining were considerably increased. Additionally, a complete cone layer with preserved anatomical structures, including the outer and inner photoreceptor segments (OS and IS), was present in the ONL, especially at the retina periphery (Figure 4b). The improvements were greater in response to treatment with quercetin. The gene and protein expression of Rho and cone opsins were also elevated upon treatment with flavonoids (Figures 4c,d and S3). The levels of Rho in mouse eyes can also be measured with high precision by retinoid analysis since most of the 11-cis-retinal in the eye is bound to opsin (Liang et al. 2004; McBee et al. 2001). In vehicle-treated mice, the concentration of 11-cis-retinal was 15.6 ± 1.2 pmoles/eye, while upon treatment with quercetin, it almost doubled reaching 27.0 ± 3.0 pmoles/eye, and in myricetin-treated eyes, it slightly increased to 19.6 ± 1.0 pmoles/eye, suggesting that treatment with flavonoids enriched the pool of functional receptors in the retina (Figure 4e). Mef2c (Myocyte enhancer factor 2 c) and Nr2e3 (Nuclear receptor subfamily 2 group e member 3) are key transcription factors involved in the regulation of the photoreceptor-specific gene expression under normal and pathological conditions (Hao et al. 2011; Nakamura et al. 2017). As we found previously, an increased expression of Mef2c correlates with the enhanced expression of Rho and cone opsins in response to flavonoid treatment (Ortega et al. 2021). Consistently, the levels of Mef2c were enhanced in RhoP23H/P23H mice treated with flavonoids (Figure S4). However, this treatment did not change the expression of Nr2e3.
FIGURE 4.
The effect of flavonoids on the expression of Rho and cone opsins in RhoP23H/P23H mice. (a, b) Immunohistochemistry in cryosections prepared from eyes collected from RhoP23H/P23H mice at P21 treated with either vehicle or flavonoids. Sections stained with an anti-Rho C terminus-specific antibody (magenta) indicate the expression level of Rho and the structural organization of rod photoreceptors. Peanut agglutinin (PNA) staining (green) shows the expression of cone opsins and the structural organization of cone photoreceptors. 4′6′-diamidino-2-phenyl-indole (DAPI) stained the nuclei (blue). The center of the retina is shown in (a) and the retina periphery in (b). Scale bar, 25 μm. (c) The expression levels of photoreceptor-specific genes encoding rod opsin, M cone opsin and S cone opsin were examined by RT-qPCR; three runs were performed. Rod opsin (n = 3, F2,9 = 168.20, p < 0.0001), M cone opsin (n = 3, F2,9 = 7.44, p = 0.0124), and S cone opsin (n = 3, F2,9 = 16.00, p = 0.0011). Total RNA was isolated from the eyes of RhoP23H/P23H mice treated with either vehicle or flavonoids. Relative fold change of these genes' expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The changes in the expression of the genes encoding rod opsin and M cone opsin were significantly different upon treatment with both flavonoids as compared with vehicle-treated control mice. The expression of S cone opsin was significantly upregulated only upon treatment with quercetin but not myricetin. The P-values for the statistically significant changes are indicated in the figure. The nonstatistically different changes are indicated as NS. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests. (d) Immunoblot analysis examining the changes in the protein expression of Rho (n = 3, F2,9 = 49.41, p < 0.0001) and M cone opsin (n = 3, F2,9 = 6.90, p = 0.0152), in the eyes of RhoP23H/P23H mice treated with either vehicle or flavonoids. Eyes from two mice from each treatment group were pooled to prepare protein extract. The representative immunoblots (upper panels) and quantification of Rho and M cone opsin protein expression levels (lower panel) are shown. Protein bands were quantified by densitometry analysis with ImageJ software. Band intensities were normalized to the intensity of GAPDH. Error bars indicate S.D. The mean of data from three independent experiments is shown. The P-values for the statistically significant changes are indicated in the figure. The nonstatistically different changes are indicated as NS. Statistical analysis was performed for each protein separately using one-way ANOVA and Bonferroni post hoc tests. (e) High-performance liquid chromatography (HPLC) elution profile of retinoid oximes extracted from eyes collected from dark-adapted RhoP23H/P23H mice at P21 treated either with quercetin (dark gray line), myricetin (light gray line), or vehicle (black line) (left panel); three HPLC runs were performed (n = 3, F2,9 = 25.16, p = 0.0002). 11-cis-retinal oxime was used as a standard control (dashed black line). Quantification of the 11-cis-retinal oxime concentration per eye in each treatment group (right panel). The P-values for the statistically significant changes are indicated in the figure. The nonstatistically different changes are indicated as NS. Statistical analysis was performed using one-way ANOVA analysis and Bonferroni post hoc tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
Flavonoids also improved the visual function of RhoP23H/P23H mice. The ERG a-wave and b-wave responses were minimal in vehicle-treated mice. The treatment with myricetin slightly improved the scotopic b-wave, photopic b-wave, and scotopic a-wave. However, in mice treated with quercetin, the amplitudes of both the scotopic and photopic ERG responses were substantially increased as compared with the responses recorded in vehicle-treated mice (Figure 5a). However, these improved ERG responses did not reach the levels of WT mice (Figure 5b).
FIGURE 5.
The effect of flavonoids on retinal function in RhoP23H/P23H mice. Retinal function was examined in RhoP23H/P23H mice at P21 treated with either flavonoids or vehicle (a) and compared with age-matched WT mice (b) by measuring the electroretinography (ERG) responses. Electroretinography (ERG) measurements were carried out in five mice per treatment group (scotopic a-wave, n = 5, F8,108 = 28.88, p < 0.0001; scotopic b-wave, n = 5, F8,108 = 34.81, p < 0.0001; photopic b-wave, n = 5, F6,84 = 78.38, p < 0.0001). The statistically different changes (P < 0.05) in the ERG responses compared between flavonoid-treated and vehicle-treated mice are indicated with asterisks (dark gray for quercetin treatment and light gray for myricetin treatment). Statistical analysis was performed with the two-way ANOVA and post hoc Bonferroni tests
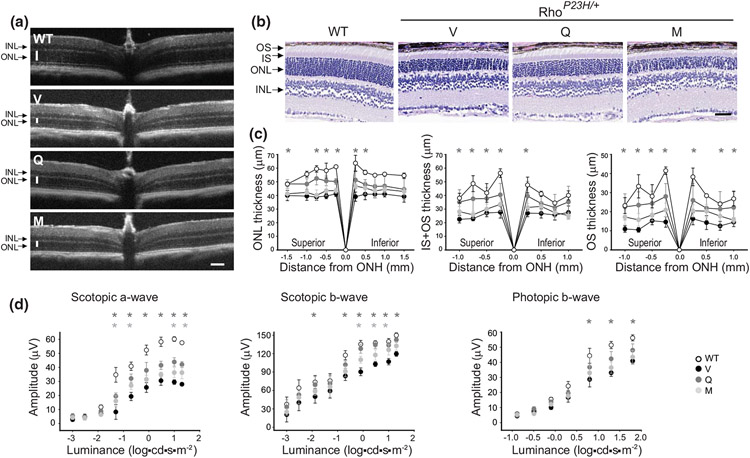
3.4 ∣. Flavonoids improve eye morphology, increase levels of visual receptors, and recover visual function in heterozygous RhoP23H/+ mice
The change of Pro23 to His in the single allele of the RHO gene results in adRP in humans. Thus, we also examined the effect of flavonoids in the heterozygous RhoP23H/+ mice in which the rate of photoreceptor death is slower than in RhoP23H/P23H mice. These mice were treated with flavonoids starting at P21 and analyzed at P33. The in vivo imaging and histological evaluation showed much thinner ONL and shorter outer segments (OS) in RhoP23H/+ mice than in WT mice. However, in mutant mice treated with flavonoids, retinal morphology was noticeably improved as compared with vehicle-treated mice (Figure 6a-c). The thickness of the ONL and the length of OS were increased. The visual responses were also improved in these mice treated with flavonoids. The ERG amplitudes of the scotopic a-wave, b-wave, and photopic b-wave were considerably higher in quercetin-treated RhoP23H/+ mice than in vehicle-treated mice but did not quite reach the response levels recorded in the WT mice (Figure 6d). Impressively, the amplitude of the scotopic b-wave in quercetin-treated mice closely resembled the ERG responses of the WT mice. The treatment with myricetin had a much lower effect on the improvement in the visual function in these mice. Although the scotopic a-wave and b-wave responses were slightly increased upon treatment with myricetin, this treatment only minimally improved the photopic b-wave responses. Collectively, these results strongly support the pharmacological potential of flavonoids, especially quercetin for the treatment of inherited RP related to Rho dysfunction.
FIGURE 6.
Protective effect of flavonoids against retinal degeneration in RhoP23H/+ mice. Flavonoids (20 mg/kg bw) or vehicle were administered intraperitoneally (i.p.) to heterozygous RhoP23H/+ mice at P21 every other day, and mice were analyzed at P33. Age-matched WT mice were used as a control. (a) Representative optical coherence tomography (OCT) images of the mouse eyes. ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar, 100 μm. (b) Images of the hematoxylin and eosin (H&E)-stained retinal sections. Scale bar, 50 μm. (c) The thickness of the retinal ONL, the inner and outer segments together (IS+OS), and the OS measured at 0.25, 0.5, 0.75, 1.0, and 1.5 mm from the ONH (ONL, n = 6, F10,175 = 303.50, p < 0.0001; IS+OS, n = 6, F8,140 = 53.72, p < 0.0001; OS, n = 6, F8,142 = 109.9, p < 0.0001). Error bars indicate standard deviation (S.D.). The changes found in mice treated with quercetin but not with myricetin were statistically different as compared with vehicle-treated control mice. The statistically different changes (P < 0.05) are indicated with an asterisk. Statistical analysis was performed with the two-way ANOVA and post hoc Bonferroni tests. (d), Retinal function was examined in RhoP23H/+ mice treated with vehicle or flavonoids at P33 and compared with the age-matched WT mice by measuring the electroretinography (ERG) responses. The ERG measurements were carried out in five mice per treatment group (scotopic a-wave, n = 5, F8,135 = 141.7, p < 0.0001; scotopic b-wave, n = 5, F8,135 = 94.14, p < 0.0001; photopic b-wave, n = 5, F6,105 = 177.4, p < 0.0001). Statistically different changes (P < 0.05) in the ERG responses after treatment with flavonoids compared with vehicle-treated mice were indicted with asterisks (dark gray for quercetin treatment and light gray for myricetin treatment). Statistical analysis was performed with the two-way ANOVA and post hoc Bonferroni tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
The improvement in retinal morphology and function is related to the increased survival of photoreceptors. Thus, we examined the effect of flavonoids on the levels of Rho and cone opsins. The staining with anti-Rho antibody visualizing rods and PNA staining showing cones performed on the retinal cryosections was substantially lower in RhoP23H/+ mice than in the WT mice, indicating degeneration of photoreceptors (Figure 7a,b). However, this progressive photoreceptor death was slowed down by treatment with flavonoids. Similarly, as found in RhoP23H/P23H mice, the greater effect on photoreceptor survival in RhoP23H/+ mice was produced by treatment with quercetin, while the effect of myricetin was more modest. The increased protein expression of Rho and cone opsins was confirmed by immunoblotting (Figures 7c,d and S5) and was associated with an enhanced gene expression of these receptors (Figure S6). Consistently, analysis of the retinoid content in RhoP23H/+ mouse eyes revealed the elevated concentration of 11-cis-retinal upon treatment with flavonoids (113.3 ± 5.6 pmoles/eye in vehicle-treated mice, 162.4 ± 8.5 pmoles/eye in quercetin-treated, and 133.3 ± 5.3 pmoles/eye in myricetin-treated mice). Together, these results indicate that flavonoids upsurge the levels of functional receptors in the retina of RhoP23H/+ mice (Figure 7e,f).
FIGURE 7.
Effect of flavonoids on photoreceptor survival in RhoP23H/+ mice. (a) Immunohistochemistry in cryosections prepared from eyes collected from RhoP23H/+ mice at P33 treated with either vehicle or flavonoids. Sections stained with an anti-Rho C terminus-specific antibody (magenta) show the expression level of Rho and the structural organization of rod photoreceptors. The peanut agglutinin (PNA) staining (green) shows the expression of cone opsins and the structural organization of cone photoreceptors. DAPI stained the nuclei (blue). Scale bar, 50 μm. (b) Quantification of magenta (upper panel) (n = 3, F3,8 = 123.1, p < 0.0001 or green (lower panel) fluorescence (n = 3, F3,8 = 52.97, p < 0.0001). The changes found in RhoP23H/+ mice treated with flavonoids were statistically different as compared with vehicle-treated control mice. The P-values for statistically different changes are indicated in the figure. The nonstatistically different changes are indicated as NS. Statistical analysis was performed with the one-way ANOVA and post hoc Bonferroni tests. (c) Immunoblot analysis examining the changes in the protein expression of Rho and M cone opsin in the eyes of RhoP23H/+ mice treated with either vehicle or flavonoids. Eyes from two mice from each treatment group were pooled to prepare protein extract. The representative immunoblots are shown. (d) Quantification of protein expression. The expression levels of Rho (Rho, n = 3, F2,9 = 12.80, p = 0.0023; Rho2, n = 3, F2,9 = 10.98, p = 0.0039) and M cone opsin (n = 3, F2,9 = 26.25, p = 0.0002) are shown. The protein bands were quantified by densitometry analysis with ImageJ software. Band intensities were normalized to the intensity of GAPDH. The mean of data from three independent experiments is shown. Error bars indicate standard deviation (S.D.). The P-values for statistically different changes between flavonoid-treated and vehicle-treated groups are indicated in the figure. The nonstatistically different changes are indicated as NS. Statistical analysis was performed for each gene separately using one-way ANOVA analysis and Bonferroni post hoc tests. (e) High-performance liquid chromatography (HPLC) elution profile of retinoid oximes extracted from mouse eyes collected from dark-adapted RhoP23H/+ mice at P33 treated either with quercetin (dark gray line), myricetin (light gray line), or vehicle (black line) (left panel). 11-cis-retinal oxime was used as a standard control (dashed black line). (f) Quantification of the 11-cis-retinal oxime concentration per eye in each treatment group (right panel); three HPLC runs were performed (n = 3, F3,8 = 41.50, p < 0.0001). The P-values for statistically different changes between flavonoid-treated and vehicle-treated groups are indicated in the figure. Statistical analysis was performed using one-way ANOVA analysis and Bonferroni post hoc tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
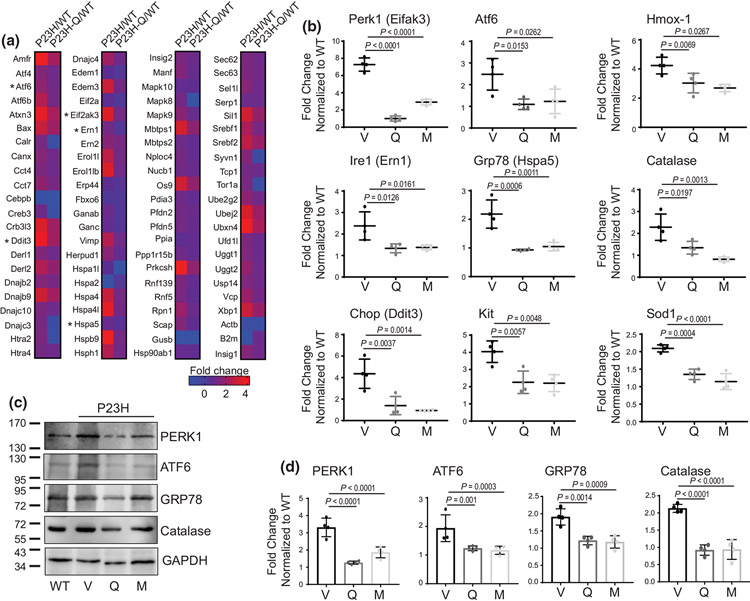
3.5 ∣. Flavonoids modulate cellular stress responses to decrease the apoptosis of photoreceptors in RhoP23H/P23H mice
The ER stress activation is a hallmark of the P23H Rho pathology. To gain a deeper understanding if stabilization of P23H Rho by flavonoids results in negative modulation of the cellular stress response, in particular, the UPR, we performed the RNA array and gene expression analysis using eyes collected from quercetin-treated or vehicle-treated RhoP23H/P23H mice and WT mice. Changes in the gene expression found in quercetin-treated or vehicle-treated RhoP23H/P23H mice were normalized to the expression of related markers in the WT mice and presented as a fold change in the form of a heat map (Figure 8a). Interestingly, treatment with quercetin resulted in the reduction of multiple UPR-related genes. The RNA array analysis was validated for several common markers linked to P23H Rho-induced UPR pathways, including the ER protein kinase 1 (Perk1), the activating transcription factor 6 (Atf6), the inositol requiring enzyme 1 (Ire1), the glucose-regulated protein 78 (Grp78), the DNA damage inducible transcription factor 3 (Ddit3/Chop), and the receptor tyrosine kinase (Kit) by RT-qPCR. The expression of these genes was substantially reduced in RhoP23H/P23H mice treated with quercetin as compared with vehicle-treated mice (Figure 8b). Similar changes were found in the protein expression levels for PERK1, ATF6, and GPR78 using immunoblotting (Figures 8c,d and S7). Additionally, the inhibitory effect of quercetin and myricetin on the expression levels of Perk1, Ire1, Grp78, Chop, Kit, and the stress-associated endoplasmic reticulum protein 1 (Serp1) genes was confirmed in vitro in the NIH-3T3 cells stably expressing P23H Rho (Figure S8). Similar results, indicating downregulation of the UPR markers by flavonoids, were obtained also for the heterozygous RhoP23H/+ mice (Figure S9a), thus confirming the ability of flavonoids to modulate the cellular stress response related to misfolding mutations.
FIGURE 8.
The effect of flavonoids on stress response in RhoP23H/P23H mice related to a P23H mutation in Rho. (a) RNA array of unfolded protein response (UPR) genes. The expression profile of the UPR-linked genes in the RhoP23H/P23H mice at P21 was compared between quercetin-treated and vehicle-treated mice in relation to the expression of these genes in age-matched WT mice. Five mice were used for each treatment group. Fold change is presented as a heat map. Asterisks indicate genes that were validated by RT-qPCR. (b) The expression levels of selected UPR-related and oxidative stress-related genes were examined by RT-qPCR; three runs were performed (Perk1, n = 3, F2,9 = 157.6, p < 0.0001; Atf6, n = 3, F2,9 = 7.69, p = 0.0113; Ire1, n = 3, F2,9 = 8.66, p < 0.008; Grp78, n = 3, F2,9 = 2.38, p = 0.1483; Chop, n = 3, F2,9 = 16.04, p = 0.0011; Kit, n = 3, F2,9 = 12.24, p = 0.0027; Hmox-1, n = 3, F2,9 = 17.03, p = 0.0009; Catalase, n = 3, F2,9 = 14.43, p = 0.0016; Sod1, n = 34, F2,9 = 35.31, p < 0.0001). Total RNA was isolated from the eyes of RhoP23H/P23H mice treated with either flavonoids or vehicle. Relative fold change of these genes' expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The change in the expression of the analyzed genes was significantly reduced upon treatment with both flavonoids. The P-values for statistically different changes are indicated in the figure. Statistical analysis was performed using one-way ANOVA and Bonferroni post hoc tests. (c) The changes in the protein expression of several UPR and oxidative stress-related markers in RhoP23H/P23H mice treated with either flavonoids or vehicle were examined by immunoblotting at P21. The representative immunoblots are shown. (d) Quantification of the protein bands by densitometric analysis using ImageJ software. The band intensities were normalized to the intensity of GAPDH. Error bars indicate S.D. The mean of data from three independent experiments is shown. (Perk1, n = 3, F2,9 = 252.8, p < 0.0001; Atf6, n = 3, F2,9 = 24.64, p < 0.0001; Grp78, n = 3, F2,9 = 19.40, p = 0.0005; Catalase, n = 3, F2,9 = 14.43, p < 0.0001). Statistical analysis was performed for each protein separately using one-way ANOVA and Bonferroni post hoc tests. The P-values for statistically different changes are indicated in the figure. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
The degeneration of rod photoreceptors in RP triggers the secondary death of cone photoreceptors, which is possibly associated with the toxic factors released by dying rods and oxidative stress. An increase in the reactive oxygen species (ROS) levels initiates the activation of well-known enzymes such as heme oxygenase (HMOX)-1 and catalase to neutralize these harmful reactive molecules. Flavonoids exhibit antioxidant properties; thus, we examined if treatment with flavonoids could modulate the oxidative stress response in RhoP23H/P23H mice. Indeed, the indirect effect of quercetin and myricetin administration was associated with a decrease in the gene and protein expression of HMOX-1 and catalase as compared with vehicle-treated mice (Figure 8b,c). In addition, the gene expression of detoxifying enzyme superoxide dismutase (Sod1) was also reduced (Figure 8b). The downregulation of these markers was also observed in the heterozygous RhoP23H/+ mice treated with flavonoids (Figure S9b).
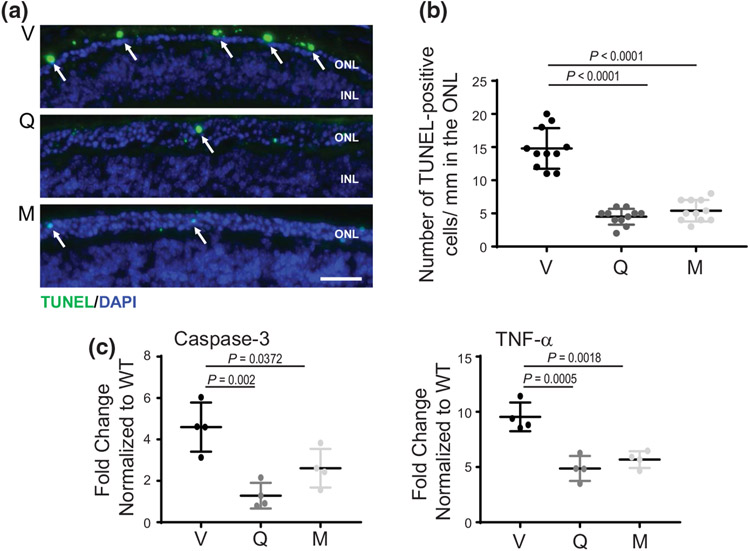
The prolonged activation of the UPR signaling caused by persistent ER stress related to the overload of misfolded P23H Rho induces apoptosis. Indeed, the TUNEL labeling that detects DNA fragmentation and measures apoptosis showed a high level of reactivity in the ONL in vehicle-treated RhoP23H/P23H mice (Figure 9a,b), which was associated with increased expression levels of apoptotic markers such as tumor necrosis factor (TNF)-α and caspase-3 as compared with WT mice (Figure 9c). Interestingly, treatment with flavonoids substantially decreased the number of apoptotic cells in the ONL and decreased the expression of TNF-α and caspase-3, indicating antiapoptotic properties of flavonoids important for prolonging the survival of photoreceptors (Figure 9a-c).
FIGURE 9.
The effect of flavonoids on the death of photoreceptors in RhoP23H/+ mice. (a) TUNEL staining performed in mouse eye cryosections. Dying photoreceptors are stained green. Nuclei stained with DAPI are blue. Scale bar, 50 μm. (b) Quantification of TUNEL-positive photoreceptor cells (n = 10, F2,30 = 79.85, p < 0.0001). The number of TUNEL-positive cells in the ONL was reduced in flavonoid-treated mice. Error bars indicate S.D. Statistical analysis was performed with the one-way ANOVA and Bonferroni post hoc tests. (c) The expression levels of cell death marker genes TNF-α and caspase-3 were examined by RT-qPCR; three runs were performed (TNF-α, n = 3, F2,9 = 21.13, p = 0.0004; caspase-3, n = 3, F2,9 = 12.51, p = 0.0025). Relative fold change of these genes' expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The P-values for statistically different changes in the expression of the analyzed genes upon treatment with both flavonoids as compared with vehicle-treated mice are indicated in the figure. Statistical analysis was performed for each gene separately, using one-way ANOVA and Bonferroni post hoc tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
4 ∣. DISCUSSION
The functional Rho is critical for proper vision (Jastrzebska 2013; Palczewski 2006). Mutations in the RHO gene, including the most frequently occurring P23H substitution, are the common cause of hereditary vision loss in humans (Athanasiou et al. 2018; Dryja et al. 1990). The substitution of Pro23 to His disrupts the network of local interactions, including the Cys110-Cys187 disulfide bond, which lessens the structural integrity of the N terminus and impairs the receptor stability and retinal binding (Behnen et al. 2018; McKibbin et al. 2007; Woods & Pfeffer 2020). Consequently, instability of the Schiff base linkage between the retinal chromophore and the receptor impacts its function. In addition, the regeneration of P23H Rho is slower as compared with WT Rho, which likely prolongs the receptor being in the ligand-free highly unstable and prone to aggregation state (Chen et al. 2014). Although P23H Rho can escape the ER quality control in vivo and traffic to the rod outer segments, insufficient regeneration of this mutant would diminish the supply of the functional receptor (Sakami et al. 2011). Thus, the supplementation with small molecule chaperones that would enhance the chromophore regeneration rates and increase the structural stability and folding appears to be a valid approach to improve the fitness of RP mutants (Chen et al. 2018; Behnen et al. 2018; Pasqualetto et al. 2020; Mattle et al. 2018).
Recently, we found that flavonoids enhance the structural stability of ligand-free WT rod opsin and increase the mobility of P23H Rho to the plasma membrane (Ortega et al. 2019; Ortega & Jastrzebska 2019). Importantly, flavonoids accelerated binding of 9-cis-retinal to regenerate the functional pigment. Although the allosteric modulation of opsin by flavonoids cannot be excluded, they exhibit higher affinity to the orthosteric site, which is large enough to accommodate both the retinal and a small molecule modulator (Ortega et al. 2019). Indeed, opsin co-crystallized with a nonretinal modulator revealed both a small molecule and octyl glucoside detergent occupying the orthosteric binding site (Mattle et al. 2018), confirming that flavonoids could modulate the orthosteric binding pocket allowing for faster entry and better accommodation of the natural ligand. Interestingly, a substantial increase in the regeneration rates with 9-cis-retinal was also observed previously for another RP-linked G90V Rho mutant upon treatment with quercetin (Herrera-Hernandez et al. 2017). In the current follow-up study, we found that binding of quercetin shifts the structural rearrangements in P23H Rho toward the WT-like conformation (Figures 1 and S2), restores multiple internal interactions (Figure 1d), and increases the receptor stability (Figure 2b) as compared with nontreated P23H Rho. Consequently, quercetin improves the signaling of this mutant receptor in vitro (Figure 2c). The enhanced signaling of P23H Rho by quercetin could potentially accelerate the receptor desensitization resulting in slightly decreased the overall protein levels of this receptor. Thus, the lower λmax of P23H pigment purified from cells treated with quercetin could be related to either structural changes affecting the binding of the retinal, increased receptor desensitization, or both. Thus, these results indicate that quercetin positively modulates the folding and structural stability of RP-related Rho mutants.
The effectiveness of two model flavonoids in RP was examined first in the homozygous RhoP23H/P23H mice to assure that the advantageous effects of flavonoids are directly related to the stabilization of the mutant receptor and confirmed in the heterozygous RhoP23H/− mice, which closely resemble human retinopathy. The treatment with flavonoids increased the survival of rod and cone photoreceptors in both mouse models, as evidenced by an increase in the levels of Rho and cone opsins in the retina, and improved the overall retinal morphology (Figures 3 and 4 and Figures 6 and 7, respectively), and function (Figures 5 and 6, respectively).
Interestingly, our data showed that quercetin was more effective than myricetin, suggesting that specific structural features of quercetin are likely important for restoring the native intramolecular interactions weekend by the mutation. In fact, the differences in the interaction pattern of quercetin and myricetin with Rho were found in our previous in silico and biochemical analyses (Ortega et al. 2019). Thus, our results strongly suggest that by binding to P23H Rho and modulating its structure, quercetin restores the native-like conformation of this Rho mutant, and consequently enhances its stability and transport to the targeted site to maintain the proper balance of the functional receptor. The improved fitness of the Rho mutant slows down the degeneration of rod photoreceptors circumventing the secondary death of cones with clear benefits for the structural and functional integrity of the retina. This also likely reduces the otherwise induced cellular stress response caused by the retention of misfolded mutant protein in the ER (Mendes et al. 2005; Walter & Ron 2011). Consistent with the previous report (Chiang et al. 2015), our RNA array analysis showed upregulation of many genes related to the UPR stress in the retinas of RhoP23H/P23H mice as compared with the WT mice. As emerged from our data, the expressions of the ER-resident main receptors of the UPR signaling, including PERK1, ATF6, and IRE1 were elevated in these mice. However, the treatment with quercetin resulted in a decrease in the levels of these markers (Figure 8). IRE1 and ATF6 are involved in the regulation of the ER chaperones, while activation of PERK1 leads to the induction of several cell death-related genes (Sizova et al. 2014). Consistently, we found many TUNEL-reactive photoreceptors in the retinas of RhoP23H/P23H mice accompanied by an increased expression of the apoptosis-related markers such as TNF-α and caspase-3, which were reduced in mice treated with either quercetin or myricetin (Figure 9). Modulation of the UPR signaling could be considered as a therapeutic target for inherited Rho-linked RP. However, inhibiting the UPR stress pathway might not be sufficient to avoid the disease progression, as the photoreceptor instability is the major factor causing the retinopathy. In fact, as shown previously, inhibition of PERK1 with its specific inhibitor exaggerated the RP pathology in the P23H Rho rats (Athanasiou et al. 2017). Additionally, the genetic ablation of CHOP, a transcription factor activated by PERK1, did not protect against RP associated with mutations in Rho (Chiang et al. 2015; Adekeye et al. 2014). Thus, the decrease in the activation of the UPR stress response markers by flavonoids likely reflects a secondary effect of the stabilizing effect of P23H Rho by these compounds. However, due to their antioxidant properties, an indirect effect of flavonoids on the ER stress pathways cannot be disregarded. Thus, flavonoids likely display a dual mechanism of protection against Rho-linked RP pathology. In fact, the elevated oxidative stress markers in the P23H Rho rats could be reduced by flavonoid epigallocatechin gallate (EGCG), which resulted in an improved visual function in these rats (Perdices et al. 2020). The involvement of oxidative stress in the progression of RP was also shown in rd10 mice, a model of RP-linked to the mutation in phosphodiesterase 6 (Piano et al. 2019). Interestingly, the treatment of these mice with nutraceuticals, including quercetin, slowed down photoreceptor degeneration. As reported, these improvements were related to a decrease in the levels of enzymes involved in redox homeostasis and cellular detoxification. Similarly, in our current study, systemic administration of quercetin to P23H Rho knock-in mice resulted in downregulation of antioxidant and detoxifying enzymes (Figure 8) that are activated in RP (Komeima et al. 2008; Léveillard et al. 2004).
5 ∣. CONCLUSIONS
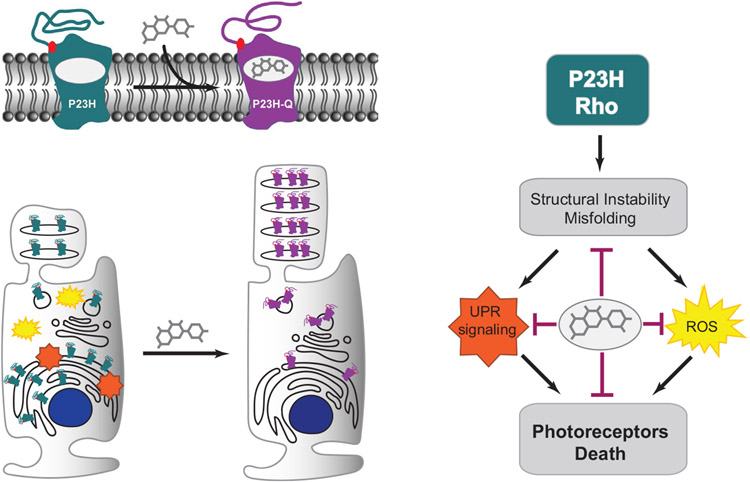
Collectively, our data revealed that flavonoids, especially quercetin, prevent or slow down photoreceptor degeneration in RP associated with misfolding mutations in Rho. The beneficial effect of quercetin is likely related to its dual action mechanism involving the conformational stabilization of the mutated Rho and consequent modulation of the cell stress processes (Figure 10). Hence, these findings open a new venue to explore the therapeutic potential of flavonoids as a preventive strategy to combat Rho-related RP.
FIGURE 10.
Schematic of dual protective effect flavonoids in RP associated with misfolding mutations in Rho. The substitution of Pro23, located in the Rho extracellular N-terminal loop, to His residue results in a structurally unstable receptor prone to aggregation in the endoplasmic reticulum (ER). An overload of misfolded protein induces the unfolded protein response (UPR) signaling and triggers reactive oxygen species (ROS) activation, which under continuous stress leads to the activation of cell death of photoreceptors. However, the binding of quercetin to the mutant Rho restores its WT-like conformation, which inhibits the UPR stress and prevents activation of photoreceptor cell death
Supplementary Material
FIGURE S1 The binding stability of quercetin within the orthosteric binding pocket of WT Rho. WT rod opsin (PDB ID: 3CAP) alone or in the complex with quercetin was subjected to the molecular dynamic (MD) simulations. A, The root-mean-square fluctuations (RMSF) were plotted with respect to the residue number of rod opsin. B, The root-mean-square deviations (RMSD) were plotted for WT opsin and WT opsin with bound quercetin (grey and green plots, respectively) with respect to the initial pose. Also, the RMSD for the ligand bound to WT opsin is shown (red plot)
FIGURE S2 The effect of flavonoids on the conformational stability of P23H Rho. A, The molecular docking of quercetin to P23H rod opsin model was conducted with VINA/VegaZZ using the orthosteric binding site parameters of rod opsin as described in (Ortega et al., 2019). The main interactions of quercetin with the residues in the orthosteric site of P23H rod opsin are shown. B, The ligand-free P23H rod opsin structure and the model of P23H rod opsin with quercetin bound were optimized and subjected to the molecular dynamic (MD) simulations carried out using CASB-Flex software. A monomer of WT rod opsin (PDB ID: 3CAP) was used as a control. The results of these MD simulations are shown. The extracellular N-terminal loop with the resident Pro23 residue is indicated with an arrow. The location of Pro23 or His23 in the final pose obtained after MD simulations is shown for each structure in the upper part of this panel. In the lower panel, ten superimposed final MD poses obtained for WT rod opsin, P23H rod opsin mutant and P23H rod opsin mutant with bound quercetin are shown. The structural flexibility of the N-terminal loop (indicated with arrow) increases upon the substitution of Pro23 to His residue. The binding of quercetin to P23H rod opsin increases its structural stability and facilitates the structural changes resembling WT-like conformation. C, The root-mean-square fluctuation (RMSF) plotted with respect to the residue number of rod opsin. The N-terminal loop residues are highlighted with a gray bar. Inset, a close-up view of the N-terminal loop residues
FIGURE S3 Immunoblot membranes. Whole eye extract from vehicle (V), quercetin (Q), or myricetin (M)-treated homozygous RhoP23H/P23H mice used for quantification of the protein expression of Rho and M cone opsin shown in Figure 4D. Quantified protein bands are indicated with red rectangular
FIGURE S4 The effect of flavonoids on the expression of transcription factors Mef2c and Ner2. The expression levels of Mef2c and Nr2e3 transcription factors that regulate the expression of Rho and cone opsins were examined with RT-qPCR; three runs were performed (Mef2c, n = 3, F2,9 = 16.74, P = 0.0009; Nr2e3, n = 3, F2,9 = 1.19, P = 0.3468). Total RNA was isolated from the eyes of RhoP23H/P23H mice treated with either vehicle or flavonoids collected at P21. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D). The expression of Mef2c was significantly different only upon treatment with quercetin but not myricetin. No change was found for the expression of Nr2e3. The P-value for statistically different changes between flavonoid-treated and vehicle-treated mice is indicated in the figure. The non-statistically different changes are indicated as NS. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
FIGURE S5 Immunoblot membranes. Whole eye extract from WT mice and vehicle (V), quercetin (Q), or myricetin (M)-treated heterozygous RhoP23H/+ mice used for quantification of the protein expression of Rho and M cone opsin shown in Figure 7D
FIGURE S6 The effect of flavonoids on the Rho and cone opsin gene expression in RhoP23H/+ mice. The expression levels of genes encoding rod opsin, M cone opsin, and S cone opsin were examined with RT-qPCR; three runs were performed (rod opsin, n = 3, F2,9 = 21.67, P = 0.0004; M opsin, n = 3, F2,9 = 15.30, P = 0.0013; S opsin, n = 3, F2,9 = 11.25, P = 0.0036). At P33 eyes were collected and total RNA was isolated from the eyes of RhoP23H/+ mice treated with either vehicle or flavonoids. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The change in the expression of the Rho and M cone opsin genes was significantly different upon treatment with quercetin, while the expression of the S cone opsin gene was significantly upregulated upon treatment with both flavonoids in comparison to vehicle-treated mice. The P-values for statistically different changes are indicated in the figure. The non-statistically different changes are indicated as NS. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
FIGURE S7 Immunoblot membranes. Whole eye extract from WT mice and vehicle (V), quercetin (Q), or myricetin (M)-treated heterozygous P23H Rho mice used for quantification of the UPR stress markers protein expression shown in Figure 8D. Quantified protein bands are indicated with red rectangular
FIGURE S8 The effect of flavonoids on stress response in P23H Rho-expressing cells. The expression levels of selected UPR stress-related markers were examined in the NIH-3T3 cells stably expressing P23H rod opsin treated with flavonoids by RT-qPCR; three runs were performed (Perk1, n = 3, F3,12 = 27.69, P < 0.0001; Ire1, n = 3, F3,12 = 4.59, P < 0.0232; Serp1, n = 3, F3,12 = 14.00, P = 0.0003, Grp78, n = 3, F3,12 = 5.04, P = 0.0137; Chop, n = 3, F3,12 = 22.24, P < 0.0001; Kit, n = 3, F3,12 = 60.79, P < 0.0001). Either total RNA was isolated from cells treated with flavonoids or the vehicle. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. The change in the expression of the analyzed genes was significantly reduced upon treatment with both flavonoids. Error bars indicate standard deviation (S.D.). The P-values for statistically different changes are indicated in the figure. NS, not statistically different. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
FIGURE S9 The effect of flavonoids on stress response in RhoP23H/+ mice related to a P23H mutation in Rho. The expression levels of selected UPR stress-related (A) and oxidative stress-related genes (B) were examined by RT-qPCR; three runs were performed (Perk1, n = 3, F2,9 = 19.07, P = 0.0006; Atf6, n = 3, F2,9 = 21.12, P = 0.0004; Ire1, n = 3, F2,9 = 21.24, P < 0.004; Grp78, n = 3, F2,9 = 2.37, P = 0.0004; Chop, n = 3, F2,9 = 16.33, P = 0.001; Kit, n = 3, F2,9 = 8.59, P = 0.0072; Hmox-1, n = 3, F2,9 = 17.03, P = 0.0066; Catalase, n = 3, F2,9 = 9.68, P = 0.0057). At P33 eyes were collected and total RNA was isolated from the eyes of heterozygous RhoP23H/+ mice treated with either flavonoids or vehicle. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The change in the expression of the analyzed genes was significantly reduced upon treatment with both flavonoids. The P-values for statistically different changes are indicated in the figure. NS, not statistically different. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
Significance.
Flavonoids bind to wild type (WT) rod opsin enhancing its stability and improve membrane targeting of retinitis pigmentosa (RP)-linked P23H rod opsin. Flavonoids exhibit antioxidant properties and modulate the activation of cellular stress pathways with beneficial effects in eye-related diseases. This study revealed that flavonoids could provide structural stabilization to P23H Rho and improve its function. Treatment with flavonoids slowed down retinal degeneration in vivo in the RP-related P23H Rho knock-in mice. RP is a blinding disease affecting millions of people worldwide with one out of ~3500 individuals. The results presented in this work open a new venue to explore nutraceuticals as a preventive treatment or lead compounds to develop new therapeutics against the P23H Rho-associated RP.
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (NIH) (EY025214 to B.J. The authors thank the Visual Science Research Center Core (supported by NIH grant P30 EY011373) with special gratitude directed to Catherine Doller for assistance with tissue sectioning and H&E staining. This work also was supported to H.P.-S. by the Fundación Seneca del Centro de Coordinación de la Investigación de la Región de Murcia (under Project 20988/PI/18) and by a grant from Ministerio de Economía y Competitividad de España (CTQ2017-87974-R). M.C.B. is a predoctoral employed to the training of research staff financed by the Plan Propio de Investigación de la UCAM. This research was partially supported by the supercomputing infrastructure of Poznan Supercomputing Center, and by the e-infrastructure program of the Research Council of Norway, and the supercomputer center of UiT - the Arctic University of Norway. The authors also thankfully acknowledge the computer resources and the technical support provided by the Plataforma Andaluza de Bioinformatica of the University of Malaga: Powered@NLHPC. This research also was partially supported by the supercomputing infrastructure of the NLHPC (ECM-02).
Funding information
Fundacion Seneca del Centro de Coordinacion de la Investigacion de la Region de Murcia; Ministerio de Economia y Competitividad de Espana; National Eye Institute; University of Malaga; the Research Council; Ministerio de Economía y Competitividad, Grant/Award Number: CTQ2017-87974-R; National Institute of Health
Abbreviations:
- adRP
autosomal dominant retinitis pigmentosa
- bw(s)
body weight(s)
- DAPI
4′6′-diamidino-2-phenyl-indole
- DDM
n-dodecyl-β-D-maltopyranoside
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- ER
endoplasmic reticulum
- ERG
electroretinography
- FBS
fetal bovine serum
- H&E
hematoxylin and eosin
- HPLC
high-performance liquid chromatography
- INL
inner nuclear layer
- i.p.
intraperitoneal
- ONH
optic nerve head
- ONL
outer nuclear layer
- OS
outer segments
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PNA
peanut agglutinin
- PVDF
polyvinylidene difluoride
- Rho
rhodopsin
- RMSD
root-mean-square deviation
- RMSF
root-mean-square fluctuation
- ROS
reactive oxygen species
- RP
retinitis pigmentosa
- RT
room temperature
- RT-qPCR
real-time quantitative polymerase chain reaction
- S.D.
standard deviation
- SD-OCT
spectral domain-optical coherence tomography
- UPR
unfolded protein response
- WT
wild type
Footnotes
DECLARATION OF TRANSPARENCY
The authors affirm that in accordance with the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest with the contents of this article.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
Supplementary Material Transparent Science Questionnaire for Authors
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Adekeye A, Haeri M, Solessio E, & Knox BE (2014). Ablation of the proapoptotic genes CHOP or Ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One, 9, e83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bellingham J, Kanuga N, Adamson P, & Cheetham ME (2017). The role of the ER stress-response protein PERK in rhodopsin retinitis pigmentosa. Human Molecular Genetics, 26, 4896–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bellingham J, Li W, McCulley C, Reeves PJ, & Cheetham ME (2018). The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Progress in Retinal and Eye Research, 62, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnen P, Felline A, Comitato A, Di Salvo MT, Raimondi F, Gulati S, & Fanelli F (2018). A small chaperone improves folding and routing of rhodopsin mutants linked to inherited blindness. iScience, 4, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen Y, Jastrzebska B, Golczak M, Gulati S, Tang H, & Palczewski K (2018). A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration. Nature Communications, 9, 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Jastrzebska B, Cao P, Zhang J, Wang B, Sun W, Yuan Y, Feng Z, & Palczewski K (2014). Inherent instability of the retinitis pigmentosa P23H mutant opsin. Journal of Biological Chemistry, 289, 9288–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT, & Lin JH (2015). Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Molecular Neurobiology, 52, 679–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, & Berson EL (1990). A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature, 343, 364–366. [DOI] [PubMed] [Google Scholar]
- Fanelli F, & Seeber M (2010). Structural insights into retinitis pigmentosa from unfolding simulations of rhodopsin mutants. The FASEB Journal, 24, 3196–3209. [DOI] [PubMed] [Google Scholar]
- Felline A, Seeber M, & Fanelli F (2020). webPSN v2.0: A webserver to infer fingerprints of structural communication in biomacromolecules. Nucleic Acids Research, 48, W94–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Parmar T, Palczewska G, Dong Z, Golczak M, Palczewski K, & Jastrzebska B (2018). Protective effect of a locked retinal chromophore analog against light-induced retinal degeneration. Molecular Pharmacology, 94, 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwin GG, & Saari JC (2000). High-performance liquid chromatography analysis of visual cycle retinoids. Methods in Enzymology, 316, 313–324. [DOI] [PubMed] [Google Scholar]
- Gupta S, Biswas J, Gupta P, Singh A, Tiwari S, Mishra A, & Singh S (2019). Salubrinal attenuates nitric oxide mediated PERK:IRE1alpha: ATF-6 signaling and DNA damage in neuronal cells. Neurochemistry International, 131, 104581. [DOI] [PubMed] [Google Scholar]
- Hao H, Tummala P, Guzman E, Mali RS, Gregorski J, Swaroop A, & Mitton KP (2011). The transcription factor neural retina leucine zipper (NRL) controls photoreceptor-specific expression of myocyte enhancer factor Mef2c from an alternative promoter. The Journal of Biological Chemistry, 286, 34893–34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Hernandez MG, Ramon E, Lupala CS, Tena-Campos M, Perez JJ, & Garriga P (2017). Flavonoid allosteric modulation of mutated visual rhodopsin associated with retinitis pigmentosa. Science Reporter, 7, 11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B (2013). GPCR: G protein complexes–the fundamental signaling assembly. Amino Acids, 45, 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan A, Aherne A, & Humphries P (2005). Light in retinitis pigmentosa. Trends in Genetics, 21, 103–110. [DOI] [PubMed] [Google Scholar]
- Komeima K, Usui S, Shen J, Rogers BS, & Campochiaro PA (2008). Blockade of neuronal nitric oxide synthase reduces cone cell death in a model of retinitis pigmentosa. Free Radical Biology & Medicine, 45, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova R, & Caffrey M (1998). Phases and phase transitions of the phosphatidylcholines. Biochimica et Biophysica Acta, 1376, 91–145. [DOI] [PubMed] [Google Scholar]
- Léveillard T, Mohand-Saïd S, Lorentz O, Hicks D, Fintz AC, Clérin E, & Sahel JA (2004). Identification and characterization of rod-derived cone viability factor. Nature Genetics, 36, 755–759. [DOI] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Maeda T, Maeda A, Modzelewska A, Filipek S, Saperstein DA, Engel A, & Palczewski K (2004). Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. The Journal of Biological Chemistry, 279, 48189–48196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, & Walter P (2007). IRE1 signaling affects cell fate during the unfolded protein response. Science, 318, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XB, Liu F, Liang YY, Yin G, Zhang HJ, Mi XS, & Xu Y (2021). Luteolin delays photoreceptor degeneration in a mouse model of retinitis pigmentosa. Neural Regeneration Research, 16, 2109–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattle D, Kuhn B, Aebi J, Bedoucha M, Kekilli D, Grozinger N, & Dawson RJP (2018). Ligand channel in pharmacologically stabilized rhodopsin. Proceedings of the National Academy of Sciences of the United States of America, 115, 3640–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, & Pepperberg DR (2001). Confronting complexity: The interlink of phototransduction and retinoid metabolism in the vertebrate retina. Progress in Retinal and Eye Research, 20, 469–529. [DOI] [PubMed] [Google Scholar]
- McKibbin C, Toye AM, Reeves PJ, Khorana HG, Edwards PC, Villa C, & Booth PJ (2007). Opsin stability and folding: The role of Cys185 and abnormal disulfide bond formation in the intradiscal domain. Journal of Molecular Biology, 374, 1309–1318. [DOI] [PubMed] [Google Scholar]
- Mendes HF, van der Spuy J, Chapple JP, & Cheetham ME (2005). Mechanisms of cell death in rhodopsin retinitis pigmentosa: Implications for therapy. Trends in Molecular Medicine, 11, 177–185. [DOI] [PubMed] [Google Scholar]
- Mostafa DG, Khaleel EF, Badi RM, Abdel-Aleem GA, & Abdeen HM (2019). Rutin hydrate inhibits apoptosis in the brains of cadmium chloride-treated rats via preserving the mitochondrial integrity and inhibiting endoplasmic reticulum stress. Neurological Research, 41, 594–608. [DOI] [PubMed] [Google Scholar]
- Nakamura PA, Shimchuk AA, Tang S, Wang Z, DeGolier K, Ding S, & Reh TA (2017). Small molecule Photoregulin3 prevents retinal degeneration in the Rho(P23H) mouse model of retinitis pigmentosa. eLife, 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorwez SM, Malhotra R, McDowell JH, Smith KA, Krebs MP, & Kaushal S (2004). Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. The Journal of Biological Chemistry, 279, 16278–16284. [DOI] [PubMed] [Google Scholar]
- Nose S (1984). A molecular-dynamics method for simulations in the canonical ensemble. Molecular Physics, 52, 255–268. [Google Scholar]
- Ortega JT, & Jastrzebska B (2019). The retinoid and non-retinoid ligands of the rod visual G protein-coupled receptor. International Journal of Molecular Sciences, 20, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JT, Parmar T, Golczak M, & Jastrzebska B (2021). Protective effects of flavonoids in acute models of light-induced retinal degeneration. Molecular Pharmacology, 99, 60–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JT, Parmar T, & Jastrzebska B (2019). Flavonoids enhance rod opsin stability, folding, and self-association by directly binding to ligand-free opsin and modulating its conformation. The Journal of Biological Chemistry, 294, 8101–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K (2006). G protein-coupled receptor rhodopsin. Annual Review of Biochemistry, 75, 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, & Saari JC (1999). Kinetics of visual pigment regeneration in excised mouse eyes and in mice with a targeted disruption of the gene encoding interphotoreceptor retinoid-binding protein or arrestin. Biochemistry, 38, 12012–12019. [DOI] [PubMed] [Google Scholar]
- Pandey VK, Mathur A, Khan MF, & Kakkar P (2019). Activation of PERK-eIF2alpha-ATF4 pathway contributes to diabetic hepatotoxicity: Attenuation of ER stress by Morin. Cell Signal, 59, 41–52. [DOI] [PubMed] [Google Scholar]
- Pasqualetto G, Schepelmann M, Varricchio C, Pileggi E, Khogali C, Morgan SR, & Bassetto M (2020). Computational studies towards the identification of novel rhodopsin-binding compounds as chemical chaperones for misfolded opsins. Molecules, 25, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdices L, Fuentes-Broto L, Segura F, Cuenca N, Orduna-Hospital E, & Pinilla I (2020). Epigallocatechin gallate slows retinal degeneration, reduces oxidative damage, and modifies circadian rhythms in P23H rats. Antioxidants (Basel), 9, 2–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano I, D'Antongiovanni V, Testai L, Calderone V, & Gargini C (2019). A nutraceutical strategy to slowing down the progression of cone death in an animal model of retinitis pigmentosa. Frontiers in Neuroscience, 13, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy EP, Kiel C, McKeone R, Stricher F, & Serrano L (2011). Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. Journal of Molecular Biology, 405, 584–606. [DOI] [PubMed] [Google Scholar]
- Roos K, Wu C, Damm W, Reboul M, Stevenson MJ, Lu C, & Harder ED (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15, 1863–1874. [DOI] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, & Palczewski K (2011). Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. The Journal of Biological Chemistry, 286, 10551–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova OS, Shinde VM, Lenox AR, & Gorbatyuk MS (2014). Modulation of cellular signaling pathways in P23H rhodopsin photoreceptors. Cell Signal, 26, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalink JD, Reynolds AH, Rentzepis PM, Sperling W, & Applebury ML (1983). Bathorhodopsin intermediates from 11-cis-rhodopsin and 9-cis-rhodopsin. Proceedings of the National Academy of Sciences of the United States of America, 80, 1887–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, & Ron D (2011). The unfolded protein response: From stress pathway to homeostatic regulation. Science, 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Woods KN, & Pfeffer J (2020). Conformational perturbation, allosteric modulation of cellular signaling pathways, and disease in P23H rhodopsin. Science Reporter, 10, 2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 The binding stability of quercetin within the orthosteric binding pocket of WT Rho. WT rod opsin (PDB ID: 3CAP) alone or in the complex with quercetin was subjected to the molecular dynamic (MD) simulations. A, The root-mean-square fluctuations (RMSF) were plotted with respect to the residue number of rod opsin. B, The root-mean-square deviations (RMSD) were plotted for WT opsin and WT opsin with bound quercetin (grey and green plots, respectively) with respect to the initial pose. Also, the RMSD for the ligand bound to WT opsin is shown (red plot)
FIGURE S2 The effect of flavonoids on the conformational stability of P23H Rho. A, The molecular docking of quercetin to P23H rod opsin model was conducted with VINA/VegaZZ using the orthosteric binding site parameters of rod opsin as described in (Ortega et al., 2019). The main interactions of quercetin with the residues in the orthosteric site of P23H rod opsin are shown. B, The ligand-free P23H rod opsin structure and the model of P23H rod opsin with quercetin bound were optimized and subjected to the molecular dynamic (MD) simulations carried out using CASB-Flex software. A monomer of WT rod opsin (PDB ID: 3CAP) was used as a control. The results of these MD simulations are shown. The extracellular N-terminal loop with the resident Pro23 residue is indicated with an arrow. The location of Pro23 or His23 in the final pose obtained after MD simulations is shown for each structure in the upper part of this panel. In the lower panel, ten superimposed final MD poses obtained for WT rod opsin, P23H rod opsin mutant and P23H rod opsin mutant with bound quercetin are shown. The structural flexibility of the N-terminal loop (indicated with arrow) increases upon the substitution of Pro23 to His residue. The binding of quercetin to P23H rod opsin increases its structural stability and facilitates the structural changes resembling WT-like conformation. C, The root-mean-square fluctuation (RMSF) plotted with respect to the residue number of rod opsin. The N-terminal loop residues are highlighted with a gray bar. Inset, a close-up view of the N-terminal loop residues
FIGURE S3 Immunoblot membranes. Whole eye extract from vehicle (V), quercetin (Q), or myricetin (M)-treated homozygous RhoP23H/P23H mice used for quantification of the protein expression of Rho and M cone opsin shown in Figure 4D. Quantified protein bands are indicated with red rectangular
FIGURE S4 The effect of flavonoids on the expression of transcription factors Mef2c and Ner2. The expression levels of Mef2c and Nr2e3 transcription factors that regulate the expression of Rho and cone opsins were examined with RT-qPCR; three runs were performed (Mef2c, n = 3, F2,9 = 16.74, P = 0.0009; Nr2e3, n = 3, F2,9 = 1.19, P = 0.3468). Total RNA was isolated from the eyes of RhoP23H/P23H mice treated with either vehicle or flavonoids collected at P21. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D). The expression of Mef2c was significantly different only upon treatment with quercetin but not myricetin. No change was found for the expression of Nr2e3. The P-value for statistically different changes between flavonoid-treated and vehicle-treated mice is indicated in the figure. The non-statistically different changes are indicated as NS. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
FIGURE S5 Immunoblot membranes. Whole eye extract from WT mice and vehicle (V), quercetin (Q), or myricetin (M)-treated heterozygous RhoP23H/+ mice used for quantification of the protein expression of Rho and M cone opsin shown in Figure 7D
FIGURE S6 The effect of flavonoids on the Rho and cone opsin gene expression in RhoP23H/+ mice. The expression levels of genes encoding rod opsin, M cone opsin, and S cone opsin were examined with RT-qPCR; three runs were performed (rod opsin, n = 3, F2,9 = 21.67, P = 0.0004; M opsin, n = 3, F2,9 = 15.30, P = 0.0013; S opsin, n = 3, F2,9 = 11.25, P = 0.0036). At P33 eyes were collected and total RNA was isolated from the eyes of RhoP23H/+ mice treated with either vehicle or flavonoids. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The change in the expression of the Rho and M cone opsin genes was significantly different upon treatment with quercetin, while the expression of the S cone opsin gene was significantly upregulated upon treatment with both flavonoids in comparison to vehicle-treated mice. The P-values for statistically different changes are indicated in the figure. The non-statistically different changes are indicated as NS. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
FIGURE S7 Immunoblot membranes. Whole eye extract from WT mice and vehicle (V), quercetin (Q), or myricetin (M)-treated heterozygous P23H Rho mice used for quantification of the UPR stress markers protein expression shown in Figure 8D. Quantified protein bands are indicated with red rectangular
FIGURE S8 The effect of flavonoids on stress response in P23H Rho-expressing cells. The expression levels of selected UPR stress-related markers were examined in the NIH-3T3 cells stably expressing P23H rod opsin treated with flavonoids by RT-qPCR; three runs were performed (Perk1, n = 3, F3,12 = 27.69, P < 0.0001; Ire1, n = 3, F3,12 = 4.59, P < 0.0232; Serp1, n = 3, F3,12 = 14.00, P = 0.0003, Grp78, n = 3, F3,12 = 5.04, P = 0.0137; Chop, n = 3, F3,12 = 22.24, P < 0.0001; Kit, n = 3, F3,12 = 60.79, P < 0.0001). Either total RNA was isolated from cells treated with flavonoids or the vehicle. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. The change in the expression of the analyzed genes was significantly reduced upon treatment with both flavonoids. Error bars indicate standard deviation (S.D.). The P-values for statistically different changes are indicated in the figure. NS, not statistically different. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests. V, treated with vehicle, Q, treated with quercetin, M, treated with myricetin
FIGURE S9 The effect of flavonoids on stress response in RhoP23H/+ mice related to a P23H mutation in Rho. The expression levels of selected UPR stress-related (A) and oxidative stress-related genes (B) were examined by RT-qPCR; three runs were performed (Perk1, n = 3, F2,9 = 19.07, P = 0.0006; Atf6, n = 3, F2,9 = 21.12, P = 0.0004; Ire1, n = 3, F2,9 = 21.24, P < 0.004; Grp78, n = 3, F2,9 = 2.37, P = 0.0004; Chop, n = 3, F2,9 = 16.33, P = 0.001; Kit, n = 3, F2,9 = 8.59, P = 0.0072; Hmox-1, n = 3, F2,9 = 17.03, P = 0.0066; Catalase, n = 3, F2,9 = 9.68, P = 0.0057). At P33 eyes were collected and total RNA was isolated from the eyes of heterozygous RhoP23H/+ mice treated with either flavonoids or vehicle. Relative fold change of these genes’ expression was normalized to the expression of Gapdh. Error bars indicate standard deviation (S.D.). The change in the expression of the analyzed genes was significantly reduced upon treatment with both flavonoids. The P-values for statistically different changes are indicated in the figure. NS, not statistically different. Statistical analysis was performed for each gene separately, using one-way ANOVA analysis and Bonferroni post hoc tests
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.