Abstract
Pathogenic strains of bacteria are often highly adept at evading serum-induced cell death, which is an essential complement-mediated component of the innate immune response. This phenomenon, known as serum-resistance, is poorly understood, and as a result, no effective clinical tools are available to restore serum-sensitivity to pathogenic bacteria. Here, we provide evidence that exogenous glycine reverses defects in glycine, serine and threonine metabolism associated with serum resistance, restores susceptibility to serum-induced cell death, and alters redox balance and glutathione (GSH) metabolism. More specifically, in Vibrio alginolyticus and Escherichia coli, exogenous glycine promotes oxidation of GSH to GSH disulfide (GSSG), disrupts redox balance, increases oxidative stress and reduces membrane integrity, leading to increased binding of complement. Antioxidant or ROS scavenging agents abrogate this effect and agents that generate or potentiate oxidation stimulate serum-mediated cell death. Analysis of several clinical isolates of E. coli demonstrates that glutathione metabolism is repressed in serum-resistant bacteria. These data suggest a novel mechanism underlying serum-resistance in pathogenic bacteria, characterized by an induced shift in the GSH/GSSG ratio impacting redox balance. The results could potentially lead to novel approaches to manage infections caused by serum-resistant bacteria both in aquaculture and human health.
Keywords: Serum resistance, Glycine, Oxidative stress, Glutathione metabolism, GSH/GSSG, Complement
1. Introduction
Innate immunity plays an essential role in the response to bacterial pathogens and other infectious agents. The complement system is a complex component of the innate immune response that involves at least 30 proteins. Complement proteins recognize and coat the bacterial membrane, leading to phagocytosis, a proteolytic cascade, a potent inflammatory response and subsequent lysis and destruction of the bacterial pathogen. The complement system is evolutionarily-conserved from lower vertebrates through mammals [[1], [2], [3], [4], [5]].
Many pathogenic bacteria evade killing by complement system-mediated mechanisms, a phenomenon referred to as serum-resistance. Serum-resistant bacteria directly interact with and/or inhibit the activity of complement proteins, for example, by shielding with antibody to lipopolysaccharide (LPS) to prevent attack by other antibodies [[6], [7], [8], [9], [10], [11]]. LPS and capsular polysaccharide are critical for bacterial survival in serum [[12], [13], [14], [15], [16]]. Bacteria also remodel the structure of the bacterial outer membrane to reduce complement binding via the two component regulatory system Cpx and extra cytoplasmic function RNA polymerase sigma-E factor pathways [17]. Other mechanisms that maintain bacterial membrane integrity include the two component regulatory system Rcs, Tol-Pal system, phosphate transport, major outer membrane lipoprotein Lpp, outer membrane protein assembly complex (BamA-E), tail-specific protease, and membrane-bound inhibitor of C-type lysozyme [18,19]. Despite extensive research, the mechanisms underlying serum-resistance remain poorly understood. Because serum-resistant bacterial pathogens are also frequently resistant to antibiotics [20], novel and effective approaches to treat and prevent infections with serum-resistant pathogens are urgently needed.
Recently, we proposed that the bacterial metabolic state plays an important role in serum resistance [21,22]. More specifically, evidence suggests that glycine, serine and threonine metabolism is downregulated in serum-resistant bacteria, and that exposure to exogenous glycine, serine or threonine restores susceptibility to serum-induced cell death by a mechanism involving reduced synthesis of ATP, inhibition of the cAMP/CRP complex and increased expression of complement-binding proteins [22].
Here, we investigate the mechanism by which glycine reverses serum-resistance in Vibrio alginolyticus and clinical isolates of Escherichia coli. The results suggest that exogenous glycine may alter the bacterial redox balance, stimulates oxidation of glutathione and increases oxidative stress.
2. Results
2.1. Glycine potentiates serum-induced killing of Vibrio alginolyticus
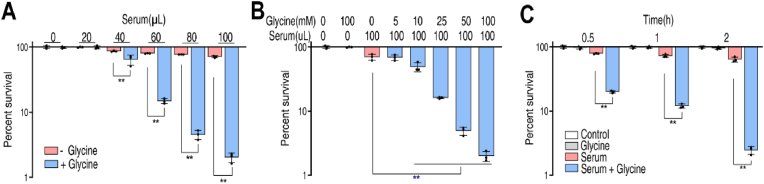
Exogenous glycine restores the ability of serum to kill pathogenic V. alginolyticus in a serum- and glycine-dose dependent manner. To demonstrate this, cultures of V. alginolyticus 12G01 were incubated with a variable amount of serum (0 μL, 20 μL, 40 μL, 60 μL, 80 μL, or 100 μL) in the presence or absence of 100 mM glycine, or in the presence of 100 μL serum and 5–100 mM glycine. The number of viable cells was quantified over time and the results are shown in Fig. 1A. In the presence of 100 μL serum, cell viability was 35.5-fold lower with 100 mM glycine than without glycine (Fig. 1A). Similarly, cell survival decreased with increasing concentrations of glycine. Control experiments show that 100 mM glycine alone does not inhibit bacterial growth (Fig. 1B). In the presence of serum and glycine, cell viability decreased with increasing time of incubation (Fig. 1C).
Fig. 1.
Glycine increased serum-induced killing of V. alginolyticus. A. Percent survival of V. alginolyticus 12G01 incubated with 100 mM glycine plus serum (0–100 μL) or without glycine B. Synergetic effects of 100 μL serum and glycine on viability of V. alginolyticus 12G01 were measured in a glycine dose-dependent manner (0–100 mM). C. Percent survival of V. alginolyticus 12G01 in the presence of 100 mM glycine or/and 100 μL serum for the indicated length of time. Results are displayed as mean ± standard errors of the means (SEM) (N = 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times.
2.2. Glycine reprograms the metabolome of V. alginolyticus
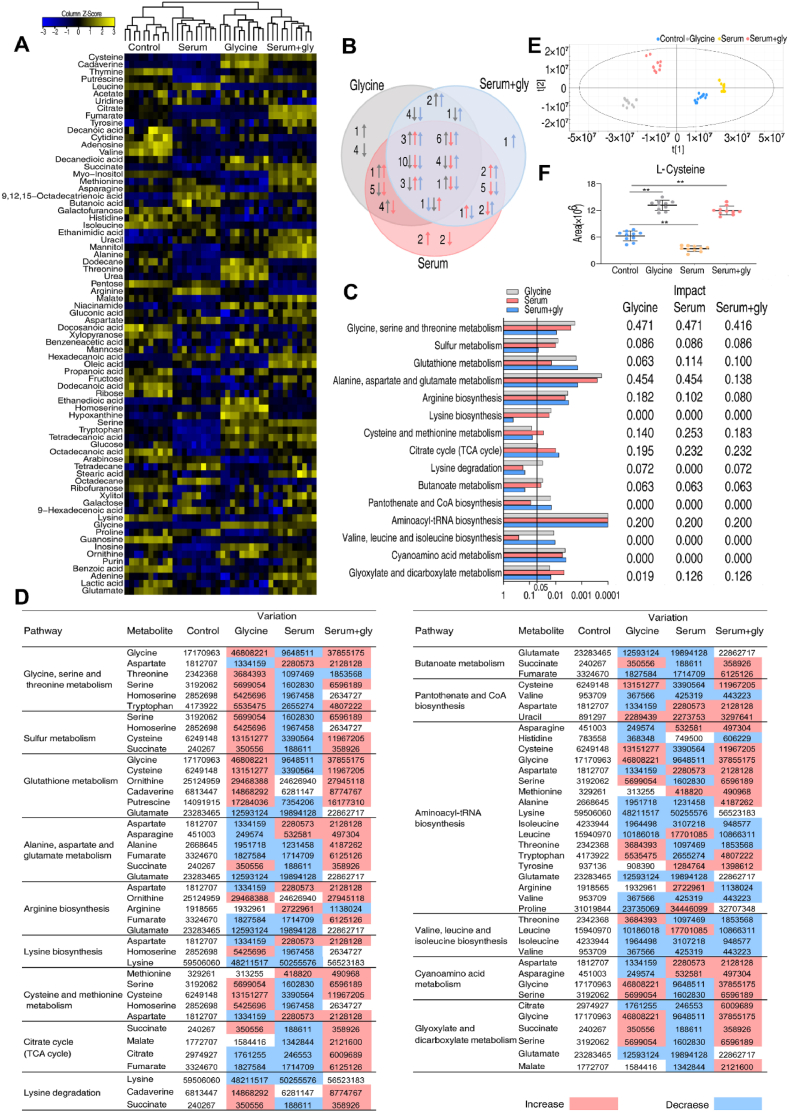
Our previous study indicated that glycine-induced depletion of ATP lowers bacterial viability in the presence of serum and plays a role in reverting the phenotype of serum-resistance [22]. Here, we explore other possible mechanisms, by performing reprogramming metabolomic analysis of V. alginolyticus in the presence of glycine, serum or glycine plus serum. After 2 h incubation, cell extracts were prepared and analyzed by GC-MS, with five independent biological replicates for each experimental group and two technical replicates for each biological replicate (N = 40 data points). Pearson correlation coefficient for this data set was 0.992, suggesting a high degree of reproducibility (Fig. S1A, Supporting Information). The data were corrected by removing the internal standard, known solvents, and duplicate signals from the same metabolite were merged. As shown in Fig. 2A, a total of 74 metabolites were identified. They were 22% carbohydrate, 36% amino acid, 22% lipid, 16% nucleotide and 4% uncategorized molecules (Fig. S1B, Supporting Information). Statistical differences were identified by Mann-Whitney U test with adjustment for a presumed false discovery rate between experimental groups and control [23]. Relative to the control, the abundance of 50, 52 and 46 metabolites were changed in the presence of glycine, serum or glycine plus serum, respectively (Fig. S2A, Supporting Information). Z-scores with variance were in the range −12.95 to 14.62, −10.03 to 39.79, −10.80 to 18.13 for the glycine, serum and glycine plus serum groups, respectively (Fig. S2B, Supporting Information). The abundance of 18 metabolites increased and 32 metabolites decreased in the presence of glycine, 12 metabolites increased and 40 metabolites decreased in the presence of serum, and 26 metabolites increased and 20 metabolites decreased in the presence of glycine and serum (Fig. S2B, Supporting Information). The abundance of 28 metabolites was changed in all three experimental groups (Fig. 2B).
Fig. 2.
Metabolomic profiling of V. alginolyticus in the presence of glycine, serum or serum plus glycine (serum + gly). A. Heat map of metabolites. Yellow and blue color indicate increase and decrease of metabolites relative to the median metabolite level, respectively (see color scale). B. Venn diagram comparing metabolites enriched in each experimental group: glycine (gray), serum (red) and serum + gly (blue). C. D. Enriched pathways and abundance of differential metabolites. E. PCA analysis of control, glycine, serum, and serum + gly group. F. Normalized abundance of l-cysteine. Results are displayed as mean ± SEM, and significant differences are identified. **p < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Metabolic pathways were identified using Metaboanalyst 5.0 [24]. This analysis identified 15 pathways (Fig. 2C), including glycine, serine and threonine metabolism, tricarboxylic acid cycle (TCA cycle) [22], glutathione (GSH) metabolism and sulfur metabolism (Fig. 2C). The abundance of metabolites involved in GSH metabolism and sulfur metabolism was higher in the presence of glycine or serum plus glycine than in the presence of serum (Fig. 2D). We note that GSH metabolism and sulfur metabolism are intertwined with glycine, serine and threonine metabolism, suggesting that exogenous glycine may fuel the two metabolic pathways and play roles in serum-resistance/serum-susceptibility.
Furthermore, an orthogonal partial least-squares discriminate analysis (OPLS-DA) was applied to identify biomarkers. The results show four groups clearly separate from each other (Fig. 2E). S-plot showed the covariance and correlation between the metabolites and the modeled class designation [25], allowing 26 biomarkers to be identified including l-cysteine (Figs. S3 and S4, Supporting Information). l-cysteine is a crucial metabolite in GSH metabolism and sulfur metabolism, whose abundance is higher in the presence of glycine or glycine plus serum and lower in the presence of serum than control (Fig. 2F). Therefore, we explored possible roles for l-cysteine, GSH and sulfur metabolism in glycine-stimulated serum-mediated killing of V. alginolyticus.
2.3. GSH biosynthetic pathway is critical for serum-induced cell death in the presence of glycine
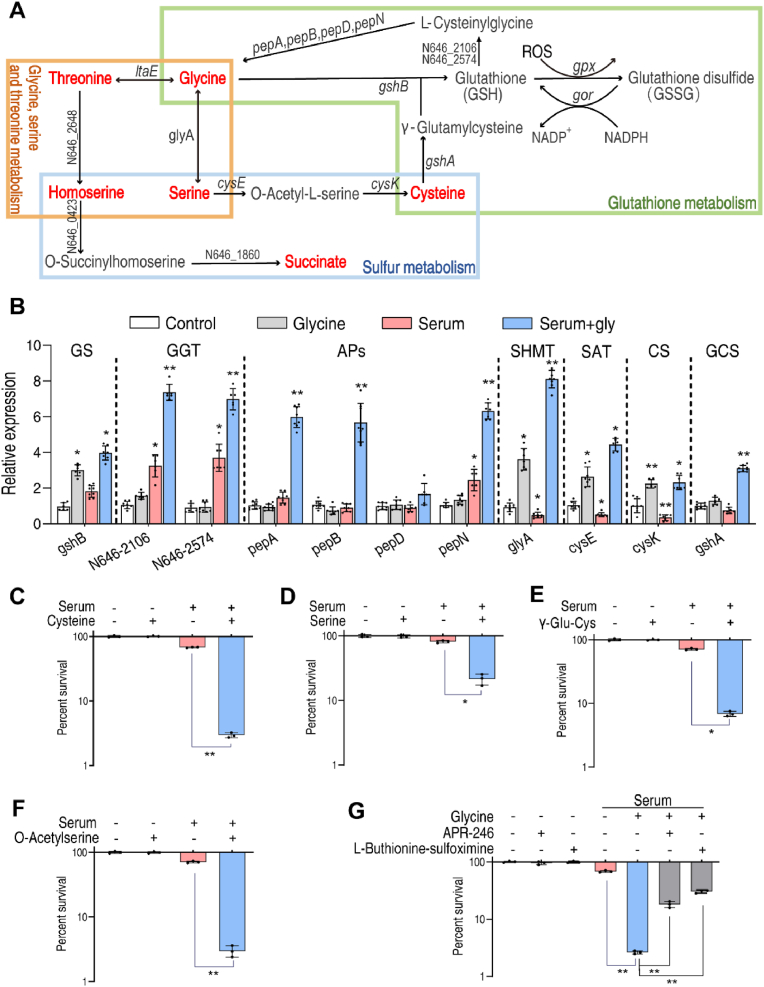
To confirm the hypothesis that exogenous glycine promotes GSH metabolism and sulfur metabolism, transcriptomic studies were performed to quantify expression of these pathways. As shown in Fig. 3A, glycine can be directly metabolized to GSH via GSH synthetase, encoded by gshB. Alternatively, it can be metabolized sequentially to serine, O-acetyl-l-serine, cysteine and GSH, catalyzed sequentially by glyA, cysE, cysK and gshA. Expression of gshB increased in the presence of glycine and glycine plus serum but did not increase in the presence of serum alone (Fig. 3B). More importantly, expression of glyA, cysE and cysK was specifically repressed by serum, and induced by glycine or glycine plus serum (Fig. 3B). These data indicate that glycine and glycine plus serum fuel GSH metabolism and enhance production of GSH, while serum has the opposite effect on expression of these genes. Glycine plus serum promoted higher expression of pepA/B/D/N in the metabolic pathway from GSH to glycine via l-cysteinylglycine than serum or glycine alone (Fig. 3B).
Fig. 3.
Glycine regulates GSH and sulfur metabolism. A. Metabolic pathways for glycine, serine, threonine, sulfur and GSH. Red indicates metabolites increased by glycine. B. Quantitative reverse transcription-PCR (qRT-PCR) of genes in GSH and sulfur metabolism. C–F. Percent survival of V. alginolyticus 12G01 in the presence of the indicated concentration of l-cysteine (12.5 mM) (C), serine (100 mM) (D), γ-Glu-Cys (250 μM) (E) and O-Acetylserine (50 mM) (F) plus100 μL serum. G. Percent survival of V. alginolyticus 12G01 in the presence of 100 mM glycine with 100 μL serum in the presence of l-buthionine-sulfoximine (50 mM) or APR-246 (1 mM). GS: GSH synthetase; GGT: gamma-glutamyltranspeptidase; Aps: aminopeptidase; SHMT: serine hydroxymethyl transferase; SAT: serine acetyltransferase; CS: cysteine synthase; GCL: glutamate cysteine ligase. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The above results suggest that glycine-stimulated GSH might play a role in serum-induced killing of bacteria. This hypothesis was tested by treating serum-resistant bacteria with l-cysteine. The results show 3.9- or 22.9-fold higher levels of serum-induced cell death when V. alginolyticus was incubated in the presence of serine or l-cysteine, respectively (Fig. 3C, D), consistent with previous observations in E. coli [22] (Fig. S5, Supporting Information). O-acetylserine and γ-glutamylcysteine, two
additional intermediates in GSH synthesis also increased serum-induced cell death (Fig. 3E, F). This effect was partially blocked by l-buthinine-sulfoximine (an inhibitor of GshB) or APR-246 (a small molecule of reducing GSH content) (Fig. 3G). In addition, expression of γ-glutamyltranspeptidase (GGT), encoded by N646_2106 and N646_2574, was increased by glycine plus serum (Fig. 3B). Furthermore, V. alginolyticus was treated with the GGT specific inhibitor, GGsTop, or deferoxamine, an iron chelating agent that sequesters iron required for GGT activity. Both compounds partially decreased glycine-potentiated serum killing but were less potent than l-buthinine-sulfoximine or APR-246. (Fig. S6, Supporting Information). Together, these results suggest that GSH metabolism plays major role in glycine-potentiated serum-induced killing.
2.4. Shift in GSH/GSSG ratio is critical for serum-mediated killing by glycine
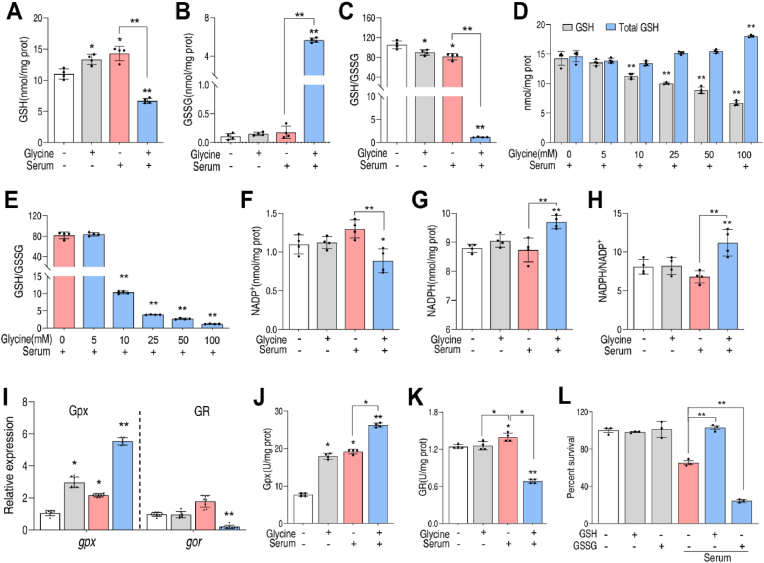
The impact of glycine on GSH metabolism was investigated by quantifying intracellular GSH in the presence of glycine, serum, or glycine plus serum. Total GSH and reduced GSH were quantified directly and the abundance of GSH disulfide (GSSG) was calculated. For example, in the control sample, the concentration of total GSH was 11.2193 nmol/mg protein (6.8122 μM GSH in 0.6071 mg/mL total protein), reduced GSH was 11.0100 nmol/mg protein, so GSSG was calculated to be 0.1046 nmol/mg (see details in Experimental Section). Data were normalized to a control sample, with no additions to culture media. The results indicate that both GSH and total GSH increased 1.2-fold in the presence of glycine and 1.3-fold in the presence of serum (Fig. 4A, Fig. S7, Supporting Information). Surprisingly, GSH was 1.6-fold lower but total GSH 1.6-fold higher in the presence of glycine plus serum (Fig. 4A, Fig. S7, Supporting Information). Because transcription of the genes involved in GSH metabolism remained high, and GSH functions as a major low molecular weight thiol, which maintains the cellular redox state [26], the data suggest increased oxidization of GSH to GSSG. Therefore, GSSG and the GSH/GSSG ratio were quantified in each sample. The content of GSSG was similar in the presence of glycine or serum but increased 54.4-fold in the presence of serum and glycine (Fig. 4B). The GSH/GSSG ratio was approximately 105.2:1 in the control (Fig. 4C), which is consistent with previous reports (e.g., 100:1 for most cells [27]). Addition of glycine or serum decreased the ratio 14.7% or 22.4%, respectively, while glycine plus serum reduced the ratio to 98.9% of control, indicating decreased redox potential accompanied by increased oxidative stress (Fig. 4C). This data was further demonstrated by glycine dose-dependent experiment. Addition of glycine slightly increased both GSH and total GSH to a similar level in the absence of serum, thereby slightly decreasing GSH/GSSG (Fig. S8, Supporting Information). However, when serum was added, total GSH increased and GSH decreased, thereby reducing the GSH/GSSG ratio in a glycine dose-dependent manner (Fig. 4D, E). These results suggest that glycine and serum synergistically promote oxidation of GSH to GSSG. In addition, the peroxiredoxins and thioredoxin/thioredoxin reductase systems are ubiquitous redox systems. Activities of peroxiredoxin and thioredoxin are stimulated by factors in serum, but not by glycine (Fig. S9, Supporting Information). Moreover, NADP+, NADPH, and their ratio were investigated, which are coupled to the GSH/GSSG system (Fig. 3A) [28]. NADP+ and NADPH content were barely changed by glycine, while total NADP(H) was increased in glycine, serum, and glycine plus serum groups. The increased fold was ranked by glycine < serum < glycine plus serum, (Fig. 4F, G; Fig. S10, Supporting Information). The NADPH/NADP+ ratio increased in glycine plus serum but did not in the presence of glycine or serum alone (Fig. 4H). Thus, GSSG and NADPH accumulate in cells treated with glycine plus serum. These results suggest that glycine plus serum drives oxidation of GSH primarily through the GSH metabolic pathway.
Fig. 4.
Impact of glycine and serum on redox balance in V. alginolyticus. A.B.C. Quantification of reduced GSH (A), GSSG (B) and the ratio of GSH to GSSG(C) in control, glycine, serum and serum plus glycine group. D.E. GSH, total GSH and GSH/GSSG (E) were measured with 100 μL serum in the presence of 0–100 mM glycine. F–H. Quantification of NADP+(F), NADPH(G) and NADPH/NADP+ (H) in control, glycine, serum and serum plus glycine group. I. Quantification of gene expression. J.K. The specific activity of Gpx (J) and GR (K) in control, glycine, serum and serum plus glycine groups. L. Percent survival of V. alginolyticus 12G01 in the presence of indicated concentration of GSH or GSSG. Serum volume was 100 μL and the concentration of glycine was 100 mM unless otherwise indicated. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times.
To analyze this further, the transcription of gpx encoding GSH peroxidase (Gpx) and gor encoding GSH reductase (GR) was quantified. Interestingly, the expression of the gene products, gpx, that catalyze oxidation of GSH were slightly increased in the presence of glycine or serum, and significantly increased in the presence of glycine plus serum (Fig. 4I). However, the expression of gor was unchanged in the presence of glycine, increased by serum but decreased in the presence of glycine plus serum (Fig. 4I). Consistent with this, the specific enzyme activity of Gpx was higher in cells treated with glycine or serum, and still higher in cells treated with serum plus glycine (Fig. 4J). Specific activity of GR was unchanged or slightly increased in cells treated with glycine or serum, respectively, but lower in cells treated with serum plus glycine (Fig. 4K).
Accumulation of GSSG can be toxic [29], because it tends to form mixed disulfides with and inactivate certain enzymes [30]. To confirm this interpretation of the data, bacteria were pretreated with GSH or GSSG for 10 min before being exposed to serum. When serum was added, GSH pre-treatment improved survival 1.6-fold, whereas GSSG pre-treatment decreased survival 2.7-fold (Fig. 4L). Therefore, the results indicate that glycine and serum act together to increase oxidation of GSH to GSSG, which is toxic to the treated bacteria.
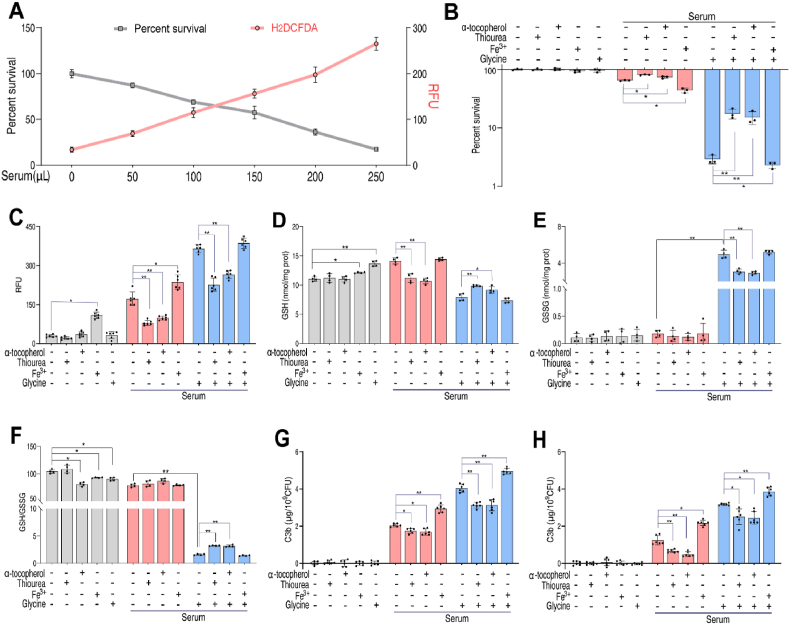
2.5. Glycine disrupts redox state to stimulate serum-induced killing
The involvement of redox system implied that oxidative stress may be required for the killing of bacteria by serum. Therefore, V. alginolyticus were incubated with different amounts (50 μL, 100 μL, 150 μL, 200 μL and 250 μL) of serum, which reduced the viability of bacteria in a dose-dependent manner (Fig. 5A). Intracellular redox status was monitored with dichlorodihydrofluorescein diacetate (H2DCFDA), a non-specific redox indicator, which does not precisely indicate the intracellular concentration of hydrogen peroxide (H2O2) or other oxidants [31]. In contrast to bacterial survival, fluorescence intensity increased in a serum dose-dependent manner (Fig. 5A), suggesting that cell viability and oxidative stress are negatively-correlated. To test the role of oxidative stress, cells were treated as above with addition of α-tocopherol, thiourea or ferric ion (Fe3+). Thiourea was used to scavenge superoxide anions and hydroxyl radicals to enhance oxidative stress tolerance [32,33] in response to antibiotic-induced ROS in bacteria [30,33]. Fe3+ is imported into cells by the transferrin receptor and subsequently converted to Fe2+. Higher levels of Fe2+ generate ROS through the Fenton reaction [31]. α-tocopherol is one of the most potent endogenous antioxidants in vivo [32,33]. As shown in Fig. 5B, neither glycine, α-tocopherol, thiourea nor Fe3+ changed cell viability in the absence of serum. In contrast, in the presence of serum, α-tocopherol increased the survival by 15.6%, thiourea increased cell survival by 24.8% and Fe3+ decreased survival by 32.4% (Fig. 5B). In the presence of glycine plus serum, α-tocopherol increased viability 5.2-fold, thiourea increased viability 6.1-fold and Fe3+ decreased viability 1.5-fold (Fig. 5B). Consistent with these results, oxidative stress increased in cells treated with serum or glycine plus serum but was reduced by α-tocopherol (42.7% in presence of serum and 27.4% in presence of glycine plus serum) or thiourea (54.0% in presence of serum and 38% in presence of glycine plus serum), but was increased by Fe3+ (37.3% in the presence of serum and 5.7% in the presence of glycine plus serum) (Fig. 5C). Accordingly, α-tocopherol or thiourea decreased GSH and total GSH in the presence of serum but increased GSH and decreased GSSG in the presence of glycine and serum, thereby increasing the GSH/GSSG ratio (Fig. 5D–F and Fig. S11, Supporting Information). However, Fe3+ had little effect. Serum complement kills bacteria through the activity of the membrane attack complex (MAC). Increased oxidative stress was associated with increased deposition of MAC components C3b and C5b-9 in cells treated with serum, glycine plus serum, and Fe3+ (Fig. 5G, H), but this effect decreased in cells co-treated with α-tocopherol or thiourea. Together, these data suggest that serum induces oxidative stress and that glycine amplifies this effect.
Fig. 5.
Glycine amplifies serum-induced oxidative stress. A. Percent survival (Gray line) and fluorescence intensity of H2DCFDA dye (Red line) of V. alginolyticus 12G01 incubated with fish serum (0–250 μL) in parallel samples. B. Percent survival of V. alginolyticus 12G01 in the indicated concentrations of glycine (100 mM), Fe3+(40 μM), α-tocopherol (100 μM) or thiourea (80 mM) plus fish serum (100 μL). C. Quantification of ROS in V. alginolyticus 12G01 being treated with indicated concentrations of glycine (100 mM), Fe3+(40 μM), α-tocopherol (100 μM) or thiourea (80 mM) plus fish serum (100 μL). D-F. GSH(D), GSSG(E) and GSH/GSSG(F) were measured in the indicated concentrations of glycine (100 mM), Fe3+(40 μM), α-tocopherol (100 μM) or thiourea (80 mM) plus fish serum (100 μL). G.H. Quantification of C3b(G) or C5b-9(H) on V. alginolyticus 12G01 incubated with thiourea (80 mM), Fe3+(40 μM), α-tocopherol (100 μM) and/or glycine (100 mM) in the presence or absence of 100 μL fish serum. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.6. Glycine-potentiated serum killing via oxidative stress in E. coli
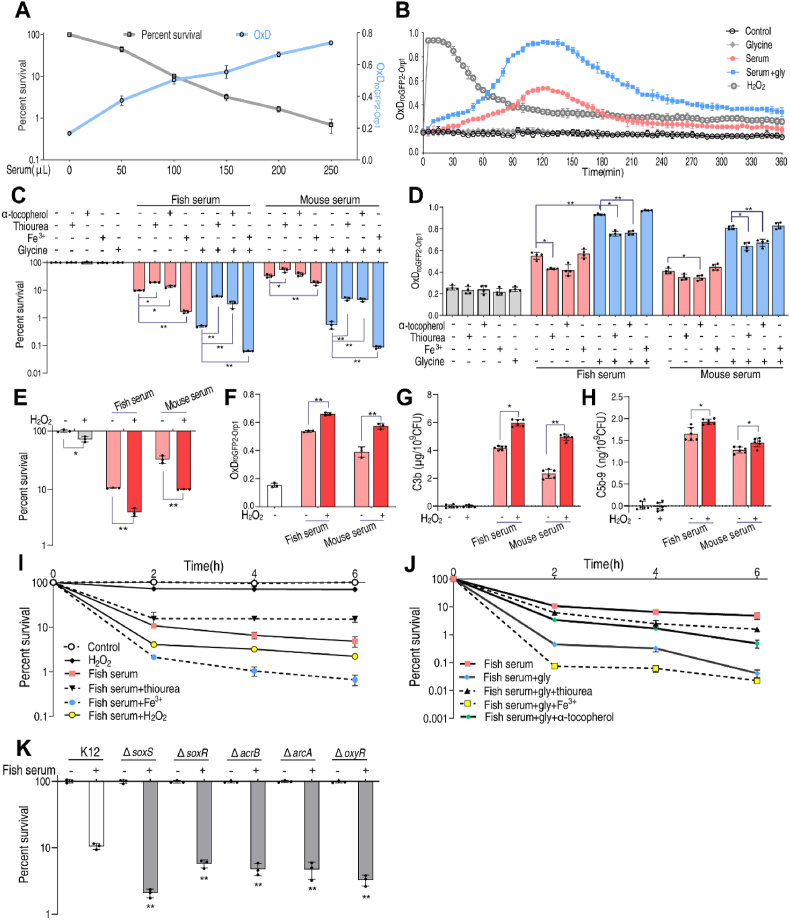
Since dye oxidation alone is insufficient evidence for detection of reactive species in biology, we explored the effects of glycine, serum on the survival and H2O2 generation in E. coli K12 using the genetically encoded roGFP2-Orp1 redox biosensor [35,36], and compared the results to those reported above in V. alginolyticus using the dye. The roGFP-Orp1 biosensor was previously developed to monitor the intracellular concentration of H2O2 in E. coli using ratiometric changes of the 405 and 488 nm excitation maxima of roGFP2 upon oxidation [37,38]. The 405/488 nm excitation ratio of the fully DTT-reduced and fully diamide oxidized probe was verified (Fig. S12A, Supporting Information). E. coli was killed by serum in a dose-dependent manner, and the serum-generated H2O2 resulted in an increased degree of oxidation (OxD) as demonstrated by the 405/488 nm excitation ratio of the biosensor, indicating an enhanced intracellular H2O2 level (Fig. 6A). The value of OxD varied in bacteria treated with saline, glycine, serum or glycine plus serum. The results suggest that H2O2 increased, reached a plateau from 90 to 150 min, and then decreased in cells exposed to serum or glycine plus serum. However, the maximal H2O2 concentration was higher in cells treated with glycine plus serum than in cells treated with serum (Fig. 6B). In contrast, in cells treated with serum and H2O2, the intracellular concentration of H2O2 increased immediately and then decreased back to baseline, indicating detoxification/scavenging of induced ROS (Fig. 6B). α-tocopherol reduced fish- or mouse serum-induced cell death 6.3-fold or 8.1-fold, and thiourea reduced fish- or mouse serum-induced cell death 11.8-fold or 8.8-fold and respectively. (Fig. 6C). Fe3+ increased fish or mouse serum-induced cell death 7.9-fold or 6.5-fold, respectively (Fig. 6C), and the OxD increased as bacterial survival decreased (Fig. 6D). H2O2 potentiated serum-induced cell death (Fig. 6E), which correlated with an increased OxD (Fig. 6F) and increased binding of complement proteins (Fig. 6G, H). E. coli survival decreased over time in the presence of serum, was increased by thiourea or α-tocopherol and decreased by addition of Fe3+ or H2O2 (Fig. 6I, J). These data are consistent with the hypothesis that redox imbalances play a role in glycine-potentiated serum-induced bacterial death.
Fig. 6.
The effect of reactive oxygen species on serum resistance is universal. A. Percent survival (gray line) and change in oxidation state of the roGFP2 probes (blue line) of E. coli incubated with fish serum (0–250 μL) in parallel samples. B. OxD kinetics of bacteria to H2O2(20 mM), glycine (100 mM), serum (100 μL) and serum (100 μL) plus glycine (100 mM) with biosensor, roGFP2-Orp1, expressed in E. coli were measured using the microplate reader. OxD was calculated using the 405/488 excitation ratio with emission at 510 nm. C. Percent survival of E. coli incubated with thiourea (150 mM), α-tocopherol (100 μM) or Fe3+(40 μM) in the presence or absence of 100 μL fish or mouse serum and glycine (100 mM). D. OxD of roGFP2-Orp1 expressed in E. coli were measured using the microplate reader after incubated with thiourea (150 mM), α-tocopherol (100 μM) or Fe3+(40 μM) in the presence or absence of 100 μL fish or mouse serum alone or in the presence of glycine (100 mM) for 2h. E. Percent survival of E. coli in the presence of H2O2 (1 mM), serum (100 μL) or both. F. OxD of roGFP2-Orp1 expressed in E. coli were measured using the microplate reader after incubated with H2O2 (1 mM), serum (100 μL), or both for 2h. G & H Quantification of C3b and C5b-9 on E. coli in the presence of H2O2(1 mM), serum (100 μL) or both. I & J Percent survival of E. coli incubated with thiourea (150 mM), α-tocopherol (100 μM), Fe3+(40 μM), H2O2 (1 mM) or glycine (100 mM) in the presence or absence of 100 μL serum for the indicated length of time. K. Percent survival of E. coli K12, ΔsoxS, ΔsoxR, ΔacrB, ΔacrA and ΔoxyR in the presence of 100 μL serum for 2h. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The role of oxidative stress was examined in mutant bacterial strains deleted for genes involved in the oxidative stress response. For this purpose, experiments were performed using strains carrying △soxS, △sosR, △acrA, △acrB, △marA, △marB, △arcA, △arcB, △cpxA, △cpxR and △oxyR. Viability was quantified in the presence and absence of serum. Interestingly, bacterial strains carrying △soxS, △sosR, △acrB, △arcA or △oxyR were more sensitive to serum than wildtype, while △cpxA was the most resistant strain (Fig. 6K; Fig. S13, Supporting Information). Because activated SoxS initiates the oxidative stress response pathway [34], it is not surprising that △soxS cells are more sensitive to serum than wildtype cells.
2.7. Glycine promotes GSSG accumulation to disrupt membrane integrity that facilitates complement binding
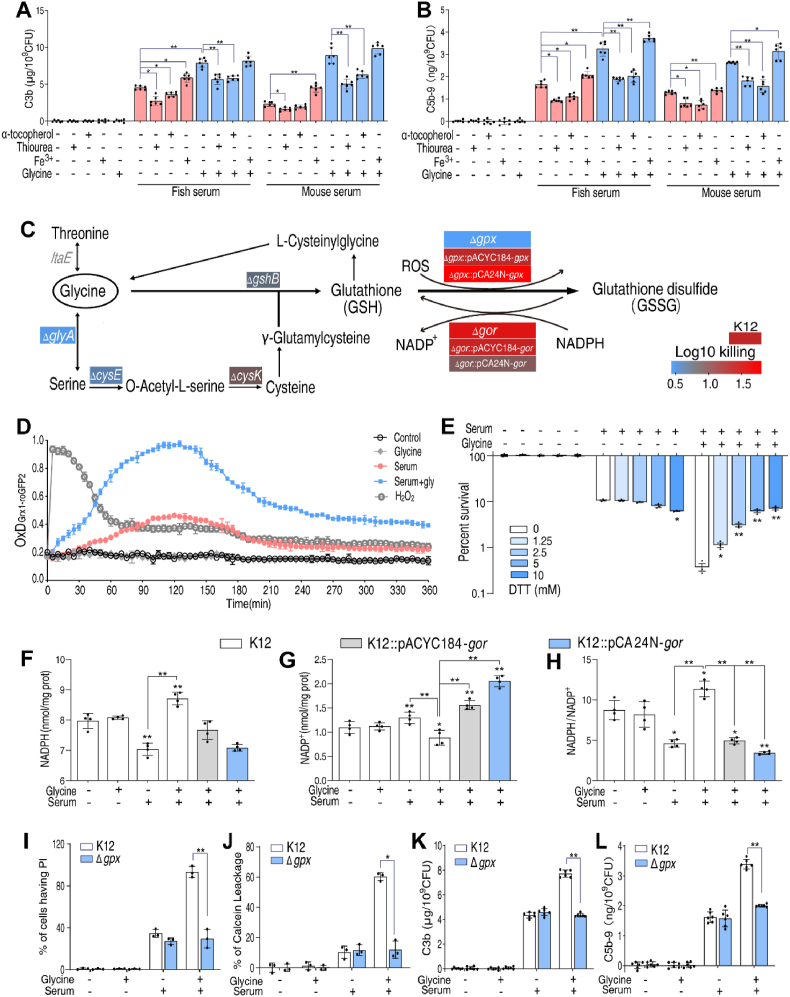
Deposition of complement proteins C3b and C5b-9 was similar in cells treated with fish serum or mouse serum (Fig. 7A, B). Complement binding decreased in cells treated with α-tocopherol or thiourea, while complement binding increased in cells treated with Fe3+.
Fig. 7.
GSSG accumulation, membrane integrity and complement deposition. A.B. Quantification of C3b(A) and C5b-9(B) on E. coli K12 incubated with thiourea (150 mM), α-tocopherol (100 μM) or Fe3+(40 μM) in the presence of 100 μL fish or mouse serum or in the presence of 100 μL fish or mouse serum with 100 mM glycine. C. Survival of the E. coli gene deletion mutants in the presence or absence of 100 μL serum plus 100 mM glycine. Heatmap scale (blue to red) represents the folds of killing. D. OxD kinetics of H2O2(20 mM), glycine (100 mM), serum (100 μL) and serum (100 μL) plus glycine (100 mM) of Grx1-roGFP2 expressed in E. coli were measured using the microplate reader. The degree of oxidation (OxD) was calculated using the 405/488 excitation ratio with emission at 510 nm. E. Percent survival of E. coli K12 incubated with 100 μL serum plus glycine in the presence of DTT (0–10 mM). F–H. Quantification of NADP+(F), NADPH(G) and NADPH/NADP+(H) of E. coli K12, K12:pACYC184-gor and K12:pCA42N-gor incubated with glycine (100 mM), serum (100 μL), or both. I.J. Percentage of PI uptake(I) or calcein leakage(J) in E. coli K12 and Δgpx in the presence of glycine (100 mM), serum (100 μL), or both for 1.5h quantified by flow cytometry. K.L. Quantification of C3b(K) and C5b-9(L) in E. coli K12 and Δgpx in the presence of glycine (100 mM), serum (100 μL), or both. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01. Each experiment was repeated independently at least three times. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Similar experiments were performed in mutant cells carrying deletions in genes involved in GSH metabolism, including gshB, gpx, glyA, cysE, and cysK. Deletions in these genes confer resistance to killing by glycine plus serum (Fig. 7C). However, the deletion of gor increased susceptibility to serum-induced killing (Fig. 7C). Deletions in gpx also decreased serum-mediating killing, implying that altered GSH redox balance (e.g. oxidation of GSH to GSSG) is critical.
To confirm that glycine promotes the oxidation of GSH to GSSG in the presence of serum, as observed in V. alginolyticus, we performed the following experiments. First, The E. coli Δgpx mutant cells were transformed with pACYC184:gpx, a low-copy plasmid to constitutively express Gpx, or pCA24N:gpx, a high-copy plasmid to overproduce Gpx induced by isopropyl β-D-thiogalactoside. The △gpx mutant cells were resistant to glycine-potentiated serum-induced killing, but sensitivity was restored by complementation or overexpression of Gpx (Fig. 7C). In contrast, the △gor deletion mutant was sensitive to glycine-potentiated serum-induced killing, but this sensitivity was abrogated by complementation or overexpression by transformation of the deletion strain with pACYC184:gor or pCA24N:gor, respectively (Fig. 7C). Second, in strains engineered to express Grx1-roGFP2, which allows intracellular GSH redox potential to be monitored [40], H2O2 induces a dramatic increase in GSSG, followed by a return the basal level within 45 min. In the presence of serum, the increase in GSSG was modest, similar to the level induced by H2O2. In contrast, in the presence of glycine plus serum, GSSG gradually increased and began to decrease after 120 min until reaching a “new” baseline level higher than the baseline in the presence of serum alone (Fig. 7D). This pattern is consistent with suggestion that glycine plus serum leads to an increase in intracellular H2O2 (Fig. 6B). Exposure of cells to glycine alone had no effect on the concentration of GSSG. Third, in bacteria treated with DTT to reduce GSSG to GSH, glycine-potentiated serum-induced killing was abolished (Fig. 7E). These data suggest that glycine plus serum promotes oxidation of GSH.
Our results also show that NAPDH accumulated in V. alginolyticus grown in the presence of glycine plus serum (Fig. 4G). NADPH is a cofactor for GR, and its higher abundance should promote reduction of GSSG to GSH. This possibility was tested and confirmed in E. coli (Fig. 7F). Thus, we hypothesize that reduced GR may be involved in the NAPDH accumulation. Furthermore, NADPH decreased, NADP+ increased and the NADPH/NADP+ ratio decreased in wildtype or △gpx mutant cells expressing GR from constitutive or inducible plasmid expression vectors (e.g., pACYC184-gor or pCA24N-gor, respectively), similar to the situation in serum-treated cells (Fig. 7F–H; Fig. S14, Supporting Information). These results suggest that glycine plus serum leads to lower GR activity, which in turn leads to higher NADPH.
The thioredoxin/thioredoxin reductase system is critical for defense against oxidative stress and involved in the reduction of oxidized protein disulfides [41]. E. coli K12 had two thioredoxins, namely thioredoxin A (TrxA) and thioredoxin C (TrxC). Oxidation of both of TrxA and TrxC was observed in serum and serum plus glycine but the oxidation level was not significant between each other. Glycine alone failed to cause oxidation of TrxA or TrxC (Fig. S15, Supporting Information), indicating the oxidative stress imposed by serum or serum plus glycine. In addition, the trxA, trxB, trxC, bcp and ahpC mutants did not affect serum- or glycine plus-serum-mediated killing. (Fig. S16, Supporting Information).
The role of GSSG in potentiating complement binding was investigated using propidium iodide staining and a calcium leakage assay to assess membrane integrity. Interestingly, in the presence of glycine plus serum, 93.2% of the control cells stained positive, but 29.7% were positive in △gpx mutant cells (Fig. 7I). Consistently, in the presence of glycine plus serum, there was massive cellular leakage of wildtype cells, but the fraction of leaking △gpx mutant cells was only 11.9% (Fig. 7J). Complement binding also decreased in △gpx mutant cells (Fig. 7K,L) .
Furthermore, pre-treatment of bacteria with GSH decreased killing while pre-treatment with GSSG increased killing (Fig. S17, Supporting Information). Treatment of bacteria with GSH increased intracellular GSH but not GSSG as indicated by GSH/GSSG ratio (Fig. S18, Supporting Information). Although how GSH is transported into bacteria is less explored, it has been used to counteract the antibiotics-induced ROS in several other studies [[35], [36], [37]]. Oligopeptide transporter family was suggested to be a potential GSH transporter in eukaryotes [38]. Thus, deletion mutants of oppB or oppC inhibited the influx of GSH to the bacteria and thereby abrogated the function of GSH in protecting cell death (Fig. S18, Supporting Information). OppB and OppC, two subunits of the ATP-binding cassette transporter oligopeptide permease (Opp), are responsible for the translocation of the substrate across the membrane. Thus, Opp may be a potential transporter of GSH in E. coli.
Glycine inhibits the reaction in which l-alanine is added to UDP-acetylmuramic acid by UDP-N-acetylmuramate-alanine ligase and the replacement of l-alanine residues which may weaken the structure of the cell wall [39,40]. To exclude this effect, we performed a cell leakage assay by including d-serine, which can be incorporated in the cell wall during bacterial growth [41]. d-serine or d-serine plus serum (with or without glycine) failed to cause cell leakage (Figs. S19A and B, Supporting Information). Meanwhile, d-serine had no synergistic effect with serum and did not alter cell survival (Fig. S19C, Supporting Information). These results demonstrate that neither glycine nor d-serine causes membrane damage under the experimental conditions tested. The results are consistent with previous reports showing that glycine does not sensitize bacteria to membrane targeting antibiotics, such as ampicillin, under the same conditions [22]. Meanwhile, 100 mM or higher glycine is required to induce expression of recombinant proteins in E. coli [42,43]. The tolerance of the bacteria to d-serine can be attributed to the presence of d-serine deaminase [44]. These data confirm that GSH metabolism and altered redox balance play roles in serum-mediated killing of bacteria in the presence of glycine.
2.8. Effect of GSH oxidation on serum-sensitivity of clinical isolates of E. coli
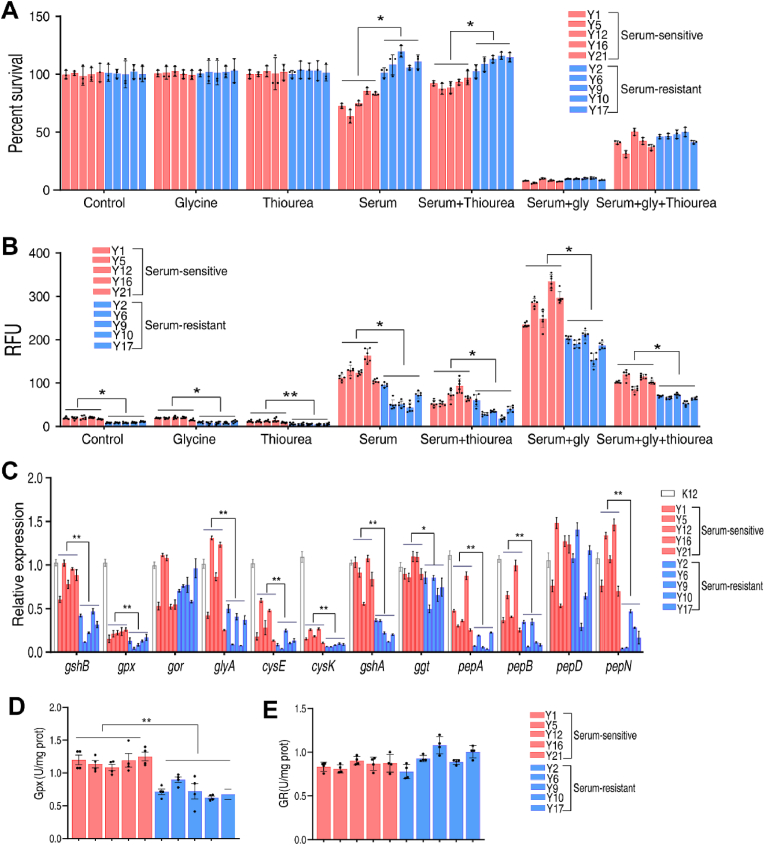
To explore the clinical relevance of the above results, we used ten multidrug-resistant clinical isolates of E. coli, including 5 serum-sensitive and 5 serum-resistant strains, as described previously [22]. Serum-sensitive and serum-resistant strains were equally susceptible to serum-induced killing in the presence of glycine (Fig. 8A), but cell death decreased 5.8-fold in the presence of thiourea (Fig. 8A). In addition, the clinical isolates of serum-resistant E. coli had lower level of oxidative stress than serum-sensitive strains (Fig. 8B). Glycine increased intracellular oxidative stress in serum-sensitive and serum-resistant strains (Fig. 8B), consistent with survival data. Expression of genes involved in GSH metabolism was lower in serum-resistant than in serum-sensitive strains (Fig. 8C). Consistent with this, the specific activity of Gpx was lower in serum-resistant strains, whereas specific activity of GR was similar (Fig. 8D, E). In the presence of glycine plus serum, Gpx activity increased and GR activity decreased (Fig. S20, Supporting Information).
Fig. 8.
GSH metabolism in clinical isolates. A. Percent survival of clinical serum-resistant and serum-sensitive strains in the presence of mouse serum (100 μL) or/and glycine (100 mM), thiourea (150 mM) or both. B. Quantification of ROS in clinical strains in the presence of mouse serum (100 μL) or/and Glycine (100 mM), thiourea (150 mM) or both. C. qRT-PCR for detection of expression of genes in GSH metabolism and sulfur metabolism of E. coli K12 and clinical strains. D.E. The specific activity of Gpx (D) and GR (E) in clinical serum-resistant and serum-sensitive strains. Results are displayed as mean ± standard errors of the means (SEM) (N ≥ 3 technical replicates per sample), and statistically significant differences are identified. *, p < 0.05, **, p < 0.01.
3. Discussion
Serum resistance is one of the most important traits of pathogenic bacteria, because it allows them to survive and proliferate in the host [[45], [46], [47]]. Although serum resistance has been studied for decades, it remains poorly understood, and effective strategies to manage infections with serum-resistant bacteria are urgently needed. While antibiotics are the drugs of choice to treat bacterial infections, there is some interest in developing alternatives to antibiotics, for example, by boosting the immune system [48,49]. The complement system, a critical component of the innate immune response to infection, is a potential target, in this regard.
One challenge is that the strategies used to evade the innate immune response differ from pathogen to pathogen [50]. Here, glycine-reprogrammed metabolome shows that glycine-induced metabolic flux promotes GSH metabolism and sulfur metabolism in V. alginolyticus, which was not detected in E. coli in our previous report [22]. Instead, activated TCA cycle was identified as the major metabolic pathway responsible for reverting serum resistance. Nonetheless, the gene, glyA, which mediates the metabolic flux of glycine, are shared by the two mechanism. In addition, it is important to recognize that serum resistance in E. coli may not be representative of serum resistance in other bacterial pathogens. The role of metabolites has also been underappreciated for many years [51]. We previously hypothesized that metabolic state plays a critical role in serum resistance and then demonstrated that exposure to exogenous metabolites can reverse serum resistance [22,52,53]. Furthermore, evidence support this hypothesis in several bacterial species.
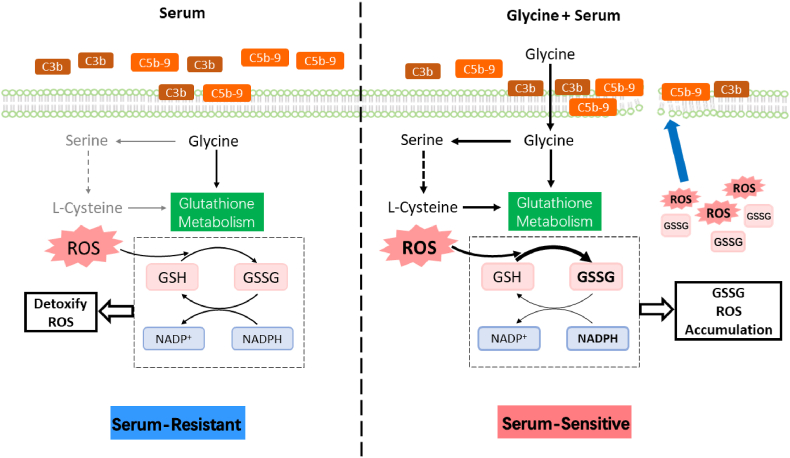
In this study, we explore the idea that serum resistance is influenced by glutathione metabolism-oxidative stress and redox balance, and compare results in V. alginolyticus and serum-resistant and serum-sensitive E. coli. Glycine, serine and threonine metabolism is altered in serum-resistant E. coli [22], glutathione metabolism and sulfur metabolism are repressed and GSH/GSSG redox balance is disrupted. Higher GSH/GSSG ratio correlates with ability to scavenge oxidants in serum-sensitive E. coli. Lower GSH/GSSG ratio and low oxidative stress correlates with serum resistance (see Fig. 9).
Fig. 9.
Proposed Model. When exposed to serum, bacteria experience oxidative stress, whose adverse effects can be mitigated by GSH metabolism, allowing bacterial survival. When exogenous glycine is added to serum, it can be metabolized directly or indirectly into serine and l-cysteine, and oxidation of GSH to GSSG increases while reduction of GSSG to GSH decrease. Thus, the accumulated GSSG and ROS might disrupt membrane integrity to facilitate complement binding.
The finding that oxidative stress is a determinant of serum sensitivity expands our understanding of oxidative stress function. Oxidative stress plays an important role in the immune response to bacterial infection [54,55], while their role in antibiotic efficacy is a matter of debate [56,57]. A possible role for oxidative in serum-induced bacterial cell death has not been previously recognized. However, Neisseria gonorrhoeae demonstrated 4-fold increase in oxygen consumption in the presence of serum [58], indicating a role for oxygen. Increased metabolism of oxygen generates ROS, as observed in this study and our previous study, as well as higher abundance of NADH [59,60]. Furthermore, deletion of the gene encoding antioxidant transcription factor SoxS increased bacterial sensitivity to serum. Other deletion mutants also showed altered sensitivity, but to a lesser extent than the soxS deletion mutant. This might be due to the complexity of the oxidative defense network. Thus, the simultaneous loss of several transcription factors might have a profound impact on sensitivity to serum. However, it is worth noting that addition of Fe3+ slightly increased the killing by both serum and glycine, but not as much as it increased killing in the presence of β-lactams, aminoglycosides and quinolones [57]. This suggests that the Fenton reaction might not contribute significantly to the effect, while toxicity of GSSG plays a greater role during glycine-potentiated serum-induced bacterial cell death. These data also suggest that oxidative stress plays a role in serum-induced killing.
Glycine reduces oxidative stress in the absence of serum but promotes oxidative stress in the presence of serum. In the absence of serum, glycine promotes biosynthesis of GSH, which scavenges oxidants [61,62]. Glycine inhibited ROS release in human neutrophils [63], and modulates ROS hemostasis in wheat seedlings stressed by high salt [64]. In the presence of serum, glycine promotes oxidation and suppresses reduction of GSH. Ultimately, this leads to toxic levels of GSSG [29], although the mechanism is not understood. However, it may involve impaired glutathionylation, which is pivotal for redox status [65,66]. Furthermore, glycine drives metabolic flux to the TCA cycle in the presence but not in the absence of serum [22]. These data suggest that the effects of a bacterial metabolite is influenced by the metabolic state of the bacteria.
This study also describes metabolic reprogramming, where exogenous compounds exert a metabolic shift, for example, a shift from an abnormal to a normal metabolome [67]. This approach has been used successfully to revert bacterial antibiotic-resistance and restore antibiotic sensitivity [67]. Metabolome-reprogramming molecules can be identified by comparing the two different metabolomes, one abnormal and one normal. Metabolome-reprogramming molecules may stimulate flux through a metabolic pathway that plays a critical role in restoring a normal phenotype, and the nature of the reprogrammed metabolic pathways may be a clue to understanding pathology and/or an “abnormal phenotype”. Indeed, the present study suggests that metabolome-reprogramming plays a role in glycine-stimulated serum-mediated bacterial cell death. In addition, a role for GSH metabolism-ROS and altered redox balance is proposed. Last, we propose that metabolomics and metabolic reprogramming could support development of novel therapeutic approaches for various phenotypes.
Another interesting observation is that the expression of GGT was induced in bacteria exposed to glycine plus serum. GGT is thought of as a prooxidant, because it cleaves the γ-glutamyl bond of GSH, degrading GSH, which is associated with atherosclerosis, cardiovascular disease, cancer, lung inflammation, neuroinflammation and bone disorders [68]. This is consistent with our observation that oxidative stress and expression of GGT increase in cells treated with glycine and serum. Although the role of GGT is less explored in bacteria than that in humans, GGT is thought to play a role in Helicobacter-associated changes in host cell metabolism leading to cancer [69,70]. We suggest that amino acids and peptides in serum may act as acceptors of the gamma-glutamyl residue of GGT, thereby accelerating GGT-dependent catabolism of extracellular GSH (i.e., see Fig. S18). This possibility was confirmed in experiments performed using the GGT inhibitor, GGsTop. More importantly, increased expression of GGT may cause S-thiolation of proteins associated with serum resistance, since it was described previously that GGT promoted S-thiolation of proteins including the receptors at the cell surface level [71,72]. Thus, it is highly possible that GGT-promoted protein S-thiolation occurs during glycine-potentiated serum-induced killing; however, the mechanism remains unknown and requires further investigation.
One technical limitation of this study is that the validity and normalization of qRT-PCR and metabolomic data might create artifacts, since bacteria might be alive but non-functional. However, in this study, we used deletion mutants to demonstrate that the relevant genes were functional and played a role during glycine-potentiated serum-induced killing at the population level.
In summary, this study demonstrates that glycine stimulates production of ROS in bacteria in the presence of serum, which in turn promotes oxidation of GSH and serum-induced bacterial cell death. Conversely, increased reduction of GSH causes lower abundance of ROS, which correlates with serum resistance. These results suggest a novel approach to reveal, identify and analyze serum-resistant pathogens, and provide mechanistic insight into the phenomenon of bacterial resistance to serum-induced cell death.
4. Experimental Section
Ethics statement: This study was conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health [73]. Animals were maintained according to the standard protocols. All experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (approval no. SYSU-IACUC-2020-B126716).
Bacterial strains and growth conditions: Bacterial strains used in this study were from the collection of our laboratory. V. alginolyticus 12G01 (GenBank accession Nos. AAPS01000007) from a single colony was cultured in 3% NaCl Luria-Bertani (LB) (tryptone [10 g/liter], yeast extract [5 g/liter], NaCl [30 g/liter]) at 30 °C and 200 rpm. Overnight cultures were diluted to 1:100 using fresh 3% NaCl LB medium and grew 30 °C and 200 rpm to OD600 approximately of 0.5. Bacterial cells were then collected by centrifugation at 8000×g for 5 min at 4 °C and washed three times with saline solution for the subsequent experiments. Unless otherwise indicated, 100 mM glycine was added at 2 h before the bacteria were collected for centrifugation in all experiments. E. coli strains were grown in LB medium at 37 °C. The other culture conditions were the same as those for V. alginolyticus. The parental strain E. coli K12 BW25113 and its knockout strains were obtained from the KEIO collection [74]. The multidrug-resistant E. coli used in this study was insensitive to at least three classes of antibiotics including β-lactamases, aminoglycoside, quinolones or/and sulfonamides.
Experimental animals and serum killing: Nile tilapia (Oreochromis niloticus) was obtained from Guangdong Tilapia Breeding Farm (Guangzhou, China) with equal number of male and female. The weight of each fish was around 500 ± 10 g. These animals were maintained in 25 L tanks under the following conditions as previously described [75]: water temperature at 28 °C; pH value of 7.0–7.5; carbon dioxide <10 mg/L; oxygen at 6–7 mg/L; nitrogen content at 1–2 mg/L. Tilapia was acclimated for four weeks. Six-week BALB/c mice were obtained from the experimental animal center of Sun Yat-sen University, Guangzhou, China, and were acclimatized for one week. Blood of Nile tilapia or BALB/c mice was collected via vein puncture and the serum was prepared by centrifugation. Sera from 50 fish and 20 mice respectively were packaged and kept at −80 °C for future use. Packaged sera were not allowed to undergo more than one round free-and-thaw cycle after they were taken from the refrigerator. Bacteria prepared as previously described were suspended with sterile saline and adjusted to OD600 of 1.0. An aliquot of 3 mL bacteria was collected in a 5 mL centrifugation tube and 100 μL serum or/and 100 mM glycine were added to the bacterial solution. An equal volume of sterile saline was added in the control group. The mixtures were cultured at 30 °C for 2 h at 200 rpm. Bacterial cells were collected by centrifugation at 8000 g for 10 min at 4 °C, and the bacterial pellet was then re-suspended by 3 mL sterile saline. To determine the colony-forming units (CFU) per mL, 100 μL of samples were 10-fold serially diluted and an aliquot of 5 μL of each dilution was spotted on the 3% NaCl LB agar plates and cultured at 30 °C for 10 h. Meanwhile, bacteria from the same culture were collected and washed twice with the same centrifugation protocol as described above for preparation of the GC-MS sample.
Metabolic profiling: Sample preparation was performed as described previously [22]. Briefly, bacterial cells were extracted with 1000 μL of cold methanol, which contained 10 μL of 0.1 mg/mL ribitol (Sigma) as internal analytical standard. The cells were lysed by sonication for 10 min at 30% intensity and were centrifuged for 10 min at 12,000 g at 4 °C. Then, 1000 μL of supernatant from the centrifugation was transferred into a new 1.5-ml tube and dried in a rotary vacuum centrifuge device (Labconco). The resulting samples were used for GC-MS analysis. Each sample had five biological replicates with two technical repeats.
GC-MS analysis was carried out with a variation on the two-stage technique as previously described [76]. First, protected carbonyl moieties of samples were exposed through a 90-min 37 °C reaction with 80 μL of 20 mg/ml methoxyamine hydrochloride in pyridine. This was followed by the derivatization of acid protons by a 30-min 37 °C reaction with the addition of 80 μL of N-methyl-N-trimethylsilytrifluoro-acetamide (MSTFA) (Sigma). Chemical analyses of samples were carried out by an Agilent G1701EAGC-MSD ChemStation (Agilent). The injection port was maintained at 270 °C. The derivatized sample of 1 μL was injected into a dodecyl benzene sulfonic acid (DBS) column (30-m length, 250-μm inner diameter [i.d.], 0.25-μm thickness) using the split less mode. The MS source temperature was maintained at 250 °C in the electron ionization (EI) (ionized directly) mode with 70 eV ionization energy and 8000 V acceleration voltage. The MS quad temperature was held constant at 150 °C. The initial temperature of the GC oven was programmed at 85 °C for 3 min, followed by an increase to 285 °C at a rate of 5 °C min−1. The temperature was then increased to 310 °C at a rate of 20 °C min−1 and held for 7 min. Helium was used as the carrier gas. The flow was kept constant at 1 ml min−1. MS was operated in a range of 50–600 m/z.
GC-MS data analysis: Date processing was conducted as previously described [77]. Mass fragmentation spectrum was analyzed by XCalibur software (Thermo fisher, version 2.1) to identify compounds by matching data with the National Institute of Standards and Technology (NIST) library and NIST MS search 2.0 program. The data were normalized according to total amount correction and standardized data containing metabolites, retention times, and peak areas, for further metabolomics analysis. The software IBM SPSS Statistics 19 was used to analyze the significant difference of the standardized data. The metabolites with differences were selected (P-value < 0.05). The R software (R × 64 3.6.1) was used for cluster analysis. Principal component analysis and S-plot analysis were performed by SIMCA-P + 12.0 software (Version 12; Umetrics, Umea, Sweden) and the metabolic pathway was done with MetaboAnalyst 5.0 enrichment [78].
qRT-PCR: Quantitative real-time PCR (qRT-PCR) was carried out as described previously [79]. Total RNA was isolated from V. alginolyticus using TRIzol reagent (Invitrogen Life Technologies, United States) according to the user manual. Electrophoresis in 1% (wt/vol) agarose gels was performed to check the quality of extracted RNA. By using an EvoM-MLV RT kit with gDNA clean for qPCR (AG11705; Accurate Biology), reverse transcription-PCR was carried out on 1 μg of total RNA according to the manufacturer's instructions. qRT-PCR was performed in 384-well plates with a total volume of 10 μL containing 5 μL 2 × SYBR green premix pro Taq HS qPCR kit (AG11701; Accurate Biology), 2.6 μL H2O, 2 μL cDNA template, and 0.2 μL each of forward and reverse primers (10 μM), and the reaction mixtures were run on a LightCycler 480 system (Roche, Germany). Data are shown as the relative mRNA expression compared with control with the endogenous reference 16S rRNA gene.
Measurement of redox status: Measurement of redox status was performed as previously described [80,81]. Bacteria were cultured as described previously and washed twice with pre-warmed phosphate-buffered saline (PBS). The bacterial suspensions were then incubated with 10 μM H2DCFDA (Sigma, United States) at 37 °C for 30 min in the dark. The samples were analyzed by a microplate reader (Varioskan LUX, Thermo fisher Scientific, United States) at an excitation and emission wavelength of 495 and 525 nm, respectively.
Measurement of total NADP(H), NADPH and NADP: The NADPH/NADP concentrations were measured with commercials kit (Abcam, No. 176724), using a 96-well plate reader [[82], [83], [84]]. Briefly, 1 × 109 CFU/mL bacterial cells were exposed to serum or/and glycine and incubated at 30 °C for 2 h. After incubation, bacteria were collected and re-suspended in PBS buffer to OD600 of 1.0. 1 mL of bacterial cells was collected at 10,000×g for 15 min at 4 °C, resuspend with 500 μL NADP/NADPH lysis buffer and incubated at room temperature for 15 min. Cell lysates were centrifuged at 2500 rpm for 5 min to remove cell debris, and supernatant were transferred to a new tube for assay. Incubate reaction at RT for 1 h protected from light, monitor fluorescence intensity at Ex/Em = 540/590 nm. The fluorescence in blank wells (with the PBS buffer only) is used as a control and is subtracted from the values for those wells with the NADPH reactions. Total NADPH and NAPDH was quantified according to manufacturer's protocol. Concentration of NADP in the test samples is calculated as: NADP = Total (NADP/NADPH) – NADPH
Measurement of specific activity of enzymes: The specific activity of GR, Gpx and thioredoxin reductase were measured with commercials kit (G0209F, Grace, Suzhou; A119992, Fusheng, Shanghai; BC1155, Solarbio, Beijing, respectively). Briefly, bacterial cells were exposed to serum or glycine or both for 2 h at 30 °C and were disrupted by sonic oscillation for 3 min for preparation of sample. GR activity was measured by DTNB reduction assays. GSSG is reduced to form GSH that reacts with DTNB to TNB. Units (U) of GR activity were calculated using an extinction coefficient of 1.36 × 104 L/mol/cm at 412 nm.
For Gpx activity, was based on the oxidation of GSH by Gpx and cumene hydroperoxide, coupled to the decrease of NADPH by GR. The decrease of NADPH (measured at OD = 340 nm) is proportional to Gpx activity. Deplete all GSSG in the sample prior to assay to ensure accurate assay results. The extinction coefficient of NADPH at 340 nm is 0.00622/μM/cm.
For thioredoxin reductase activity, TrxR catalyzes the reduction of DTNB by NADPH to generate TNB and NADP+, and 2-vinylpyridine is used to inhibit the original reduced GSH in the sample to ensure accurate assay results. The TrxR activity can be calculated by measuring the increase rate of TNB at 412 nm as described previously [85].
The specific activities of the above three enzymes were calculated by normalizing the units of GR, Gpx or thioredoxin reductase activity with the quantity of protein in each sample (BCA protein assay), which were expressed as U/mg protein.
Measurement of total GSH, reduced GSH (GSH) and GSH disulfide (GSSG): GSH concentration was assessed using a GSH/GSSG Ratio Detection Assay Kit (Abcam, No.205811), which uses a proprietary non-fluorescent water-soluble dye that becomes strongly fluorescent upon reacting with GSH [86,87]. Briefly, bacterial cells were exposed to serum or glycine or both for 2 h at 30 °C and were re-suspended in PBS buffer to OD600 = 1.0. An aliquot of treated cells (2 mL) was collected in 1 mL PBS/0.5% NP-40 and subject to sonic oscillation for 3 min to prepare bacterial cell lysates. An aliquot of the cellular extract was used for protein quantification by Bradford assay [88]. Ice cold 100% (w/v) trichloroacetic acid (TCA) was mixed with 5 vol of sample for deproteinization, followed by addition of NaHCO3 to pH 5. For GSH detection, add 50 μL of GSH Assay Mixture (GAM) into each GSH standard and sample well to make the total assay volume 100 μL/well, for total GSH (reduced and oxidized) detection, add 50 μL of Total Glutathione Assay Mixture (TGAM) into each GSSG standard and sample well to make total assay volume 100 μL/well. Incubate at room temperature for 30 min protected from light and read by a fluorescence microplate reader with excitation and emission at 490/520 nm. GSH and total GSH concentrations were calculated using a standard curve. GSSG was calculated as follows: GSSG = (Total GSH - GSH)/2.
Measurement of C3b/C5b-9 on the bacterial outer membrane: The commercial assay kits (FS-E63210, A119994, A119939, A119993, Fusheng, Shanghai) were used to quantify fish complement fragment 3B (C3b), fish complement protein 5b-9 (C5b-9), mouse complement fragment 3B (C3b) and mouse complement protein 5b-9 (C5b-9), respectively, on the bacterial outer membrane. These four kits use double antibodies sandwich method to determine the level of fish/mouse complement fragment 3B (C3b) or complement protein 5b-9 (C5b-9) in the sample. The absorbance (OD value) was measured with a microplate reader at the wavelength of 450 nm, and the concentration of complement fragment 3B or complement protein 5b-9 in the sample was calculated according to the standard curve. Briefly, the cultured bacterial cells were collected and washed three times with sterile saline. Then, the cells were harvested and adjusted to OD600 at 1.0. An aliquot of 50 μL cells were transferred into the encapsulated micropores precoated with captured antibodies, and the C3b/C5b9 was determined according to the user manuals.
Measurements of degree of oxidation (OxD): Plasmids pQE-60 roGFP2-Orp1-His and pQE-60 Grx1-roGFP2-His were gifts from Tobias Dick (RRID: Addgene_64976; RRID: Addgene_64799). E. coli strains were constructed using standard molecular and genetic techniques [89,90]. E. coli strains transformed with roGFP2 fused probes (Grx1-roGFP2 and roGFP2-Orp1) were cultivated in LB medium with 100 μM IPTG overnight to induce expression of the probes. Then, the cells were harvested and adjusted to OD600 at 1.0. Samples for fully reduced and oxidized controls were treated for 5 min with 8 mM diamide and 40 mM dithiothreitol (DTT), respectively [91]. The biosensor fluorescence emission at 510 nm was stimulated by excitation at 405 and 488 nm and measured using a microplate reader. OxD was calculated as previously described [[92], [93], [94]].
Detection of redox state of Trxs in E. coli bacteria:The redox state of Trxs was determined as described previously [95]. In brief, bacteria were exposed to serum or glycine or both for 2 h at 37 °C and were collected as described above for preparation of proteins sample. Proteins were precipitated with 10% trichloroacetic acid (TCA) and pellets were washed with ice-cold acetone 3 times. The pellet was dissolved in 50 mM Tris-HCl, pH 8.5, and 1% sodium dodecyl sulfate (SDS) containing 15 mM AMS by incubation at 37 °C for 1h. The protein concentration of the samples was determined by the BCA protein assay (Beyotime, Shanghai, China). Equal amounts of protein were separated on 15% SDS- PAGE gels. TrxA and TrxC were detected by Western blotting using the corresponding specific primary goat antibodies. The primary antibody dilution was 1:1000 for anti-Flag (Zen-bio, Chengdu, China); the secondary antibody dilution was 1:3000 (CST, MA, USA). Specific bands were detected by the Chemiluminescence Reagent Plus method (EpiZyme, Shanghai, China).
Propidium iodide (PI) uptake and calcein leakage assay: To measure membrane integrity of E. coli K12 or Δgpx upon exposure to glycine or serum or both, flow cytometry was adopted as described previously [96,97]. Propidium iodide (PI) intercalates with bases of deoxyribonucleic acid (DNA) and gives fluorescence. The produced fluorescence emission can be detected by flow cytometer. The harvested cells were added to glycine or serum or their combination, and the mixtures were incubated at 30 °C for 1.5h. After incubation, 106 CFU/mL diluted cells were mixed with 2 μg/ml PI at 30 °C for 20 min in dark. The excitation and emission at 544 nm and 620 nm, respectively, were used to detect PI fluoresces. A total of 10,000 cells were acquired for each flow cytometry analysis (CytoFLEX, Beckman Coulter Ltd., USA). Experiments were repeated with at least three independent biological replicates. Calcein AM (C3099, Thermo fisher Scientific, USA) is a membrane-permeating, non-fluorescent derivative of calcein with the excitation and emission wavelengths at 490 nm and 517 nm, respectively. After entering the cell, calcein AM is cleaved by cytoplasmic esterases, releasing the fluorescent calcein, which is membrane-impermeable. Calcein can leak out of the cells only if their membrane is damaged. In brief, 109 CFU/ml cells were incubated with 10 μg/ml of calcein AM at 30 °C for 2h. The cells were then treated with glycine or serum or both for 1.5h at 30 °C. For detection of calcein leakage, 106 CFU/mL cells were collected and then analyzed on flow cytometry (CytoFLEX, Beckman Coulter Ltd., USA) with a record count of 10,000 cells. Experiments were repeated with at least three independent biological replicates.
Statistical analysis: Data were analyzed and graphed using Prism 8.0 (GraphPad). After testing for normality, a two-tailed paired Student's t-test was used for all pairwise statistical comparisons unless otherwise indicated. Data shown are the means ± SEM. At least three biological replicates per experiment were carried out. Significance is described in the figure legends as: ∗ p < 0.05, ∗∗ p < 0.01.
Funding
This work was sponsored by grants from the National Key Research and Development Program of China [2018YFD0900501]; National Natural Science Foundation of China [32061133007, 32273177], Project supported by Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) [No. 311020006], and The Youth Talent Support Program of Guangdong Province [2017GC010617] (to B. P.).
Conflicts and interests
Authors declare they have no conflicts of interests
Acknowledgements
TSK, JHW and XWC conducted the experiments. TSK, JHW, XWC and ZGC performed data analysis. TSK, XWC, JZ and BP interpreted the data. BP wrote the manuscript. BP conceptualized and designed the project. All the authors reviewed the manuscript and acknowledged the contributions. We also thanks Dr. D. X. Yang for the assistance of collecting fish serum; Prof. X. M. Lin for the insightful discussion of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102512.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sarma J.V., Ward P.A. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y., Thangamani S., Ho B., Ding J.L. The ancient origin of the complement system. EMBO J. 2005;24:382–394. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner E., Frank M.M. Therapeutic potential of complement modulation. Nat. Rev. Drug Discov. 2010;9:43–56. doi: 10.1038/nrd3011. [DOI] [PubMed] [Google Scholar]
- 4.Kang Y.H., Tan L.A., Carroll M.V., Gentle M.E., Sim R.B. Target pattern recognition by complement proteins of the classical and alternative pathways. Adv. Exp. Med. Biol. 2009;653:117–128. doi: 10.1007/978-1-4419-0901-5_8. [DOI] [PubMed] [Google Scholar]
- 5.Ricklin D., Hajishengallis G., Yang K., Lambris J.D. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacLennan C.A., Gilchrist J.J., Gordon M.A., Cunningham A.F., Cobbold M., Goodall M., Kingsley R.A., van Oosterhout J.J., Msefula C.L., Mandala W.L., Leyton D.L., Marshall J.L., Gondwe E.N., Bobat S., Lopez-Macias C., Doffinger R., Henderson I.R., Zijlstra E.E., Dougan G., Drayson M.T., MacLennan I.C., Molyneux M.E. Dysregulated humoral immunity to nontyphoidal Salmonella in HIV-infected African adults. Science. 2010;328:508–512. doi: 10.1126/science.1180346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diao J., Bouwman C., Yan D., Kang J., Katakam A.K., Liu P., Pantua H., Abbas A.R., Nickerson N.N., Austin C., Reichelt M., Sandoval W., Xu M., Whitfield C., Kapadia S.B. Peptidoglycan association of murein lipoprotein is required for KpsD-dependent group 2 capsular polysaccharide expression and serum resistance in a uropathogenic Escherichia coli isolate. mBio. 2017;8 doi: 10.1128/mBio.00603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clay C.D., Soni S., Gunn J.S., Schlesinger L.S. Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 2008;181:5568–5578. doi: 10.4049/jimmunol.181.8.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan L.A., Yang A.C., Kishore U., Sim R.B. Interactions of complement proteins C1q and factor H with lipid A and Escherichia coli: further evidence that factor H regulates the classical complement pathway. Protein Cell. 2011;2:320–332. doi: 10.1007/s13238-011-1029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith A.E., Kim S.H., Liu F., Jia W., Vinogradov E., Gyles C.L., Bishop R.E. PagP activation in the outer membrane triggers R3 core oligosaccharide truncation in the cytoplasm of Escherichia coli O157:H7. J. Biol. Chem. 2008;283:4332–4343. doi: 10.1074/jbc.M708163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho D.K., Jarva H., Meri S. Human complement factor H binds to outer membrane protein Rck of Salmonella. J. Immunol. 2010;185:1763–1769. doi: 10.4049/jimmunol.1001244. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y.F., Yan J.J., Lei H.Y., Teng C.H., Wang M.C., Tseng C.C., Wu J.J. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect. Immun. 2012;80:1815–1822. doi: 10.1128/IAI.06395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan M.D., Peters K.M., Sarkar S., Lukowski S.W., Allsopp L.P., Gomes Moriel D., Achard M.E., Totsika M., Marshall V.M., Upton M., Beatson S.A., Schembri M.A. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cremet L., Broquet A., Jacqueline C., Chaillou C., Asehnoune K., Corvec S., Caroff N. Innate immune evasion of Escherichia coli clinical strains from orthopedic implant infections. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:993–999. doi: 10.1007/s10096-016-2628-6. [DOI] [PubMed] [Google Scholar]
- 15.Coggon C.F., Jiang A., Goh K.G.K., Henderson I.R., Schembri M.A., Wells T.J. A novel method of serum resistance by Escherichia coli that causes urosepsis. mBio. 2018;9 doi: 10.1128/mBio.00920-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells T.J., Whitters D., Sevastsyanovich Y.R., Heath J.N., Pravin J., Goodall M., Browning D.F., O'Shea M.K., Cranston A., De Soyza A., Cunningham A.F., MacLennan C.A., Henderson I.R., Stockley R.A. Increased severity of respiratory infections associated with elevated anti-LPS IgG2 which inhibits serum bactericidal killing. J. Exp. Med. 2014;211:1893–1904. doi: 10.1084/jem.20132444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bury-Mone S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., Jacq A., Bouloc P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King L.B., Pangburn M.K., McDaniel L.S. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J. Infect. Dis. 2013;207:1128–1134. doi: 10.1093/infdis/jis939. [DOI] [PubMed] [Google Scholar]
- 19.Bolourchi N., Shahcheraghi F., Shirazi A.S., Janani A., Bahrami F., Badmasti F. Immunogenic reactivity of recombinant PKF and AbOmpA proteins as serum resistance factors against sepsis of Acinetobacter baumannii. Microb. Pathog. 2019;131:9–14. doi: 10.1016/j.micpath.2019.03.031. [DOI] [PubMed] [Google Scholar]
- 20.Ragheb M.N., Thomason M.K., Hsu C., Nugent P., Gage J., Samadpour A.N., Kariisa A., Merrikh C.N., Miller S.I., Sherman D.R., Merrikh H. Inhibiting the evolution of antibiotic resistance. Mol. Cell. 2019;73:157–165 e5. doi: 10.1016/j.molcel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Z., Gong Q.Y., Wang Z., Chen Z.G., Ye J.Z., Li J., Wang J., Yang M.J., Ling X.P., Peng B. Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front. Immunol. 2017;8:1706. doi: 10.3389/fimmu.2017.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z.X., Guo C., Chen Z.G., Yang T.C., Zhang J.Y., Wang J., Zhu J.X., Li D., Zhang T.T., Li H., Peng B., Peng X.X. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 2019;10:3325. doi: 10.1038/s41467-019-11129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu D., Wang J., Guo C., Peng X.X., Li H. Elevated biosynthesis of palmitic acid is required for zebrafish against Edwardsiella tarda infection. Fish Shellfish Immunol. 2019;92:508–518. doi: 10.1016/j.fsi.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 24.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.E., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalesinskas L., Cudone E., Fofanov Y., Putonti C., S-plot2 Rapid visual and statistical analysis of genomic sequences. Evol Bioinform Online. 2018;14 doi: 10.1177/1176934318797354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 27.Dalle-Donne I., Milzani A., Gagliano N., Colombo R., Giustarini D., Rossi R. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxidants Redox Signal. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 28.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franco R., Cidlowski J.A. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 30.Halliwell B. Biochemical mechanisms accounting for the toxic action of oxygen on living organisms: the key role of superoxide dismutase. Cell Biol. Int. Rep. 1978;2:113–128. doi: 10.1016/0309-1651(78)90032-2. [DOI] [PubMed] [Google Scholar]
- 31.Su Y., Zhao B., Zhou L., Zhang Z., Shen Y., Lv H., AlQudsy L.H.H., Shang P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–136. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Forman H.J., Davies K.J., Ursini F. How do nutritional antioxidants really work: nucleophilic tone and para-hormesis versus free radical scavenging in vivo. Free Radic. Biol. Med. 2014;66:24–35. doi: 10.1016/j.freeradbiomed.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forman H.J., Augusto O., Brigelius-Flohe R., Dennery P.A., Kalyanaraman B., Ischiropoulos H., Mann G.E., Radi R., Roberts L.J., 2nd, Vina J., Davies K.J. Even free radicals should follow some rules: a guide to free radical research terminology and methodology. Free Radic. Biol. Med. 2015;78:233–235. doi: 10.1016/j.freeradbiomed.2014.10.504. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X., Drlica K. Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 2014;21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dwyer D.J., Belenky P.A., Yang J.H., MacDonald I.C., Martell J.D., Takahashi N., Chan C.T., Lobritz M.A., Braff D., Schwarz E.G., Ye J.D., Pati M., Vercruysse M., Ralifo P.S., Allison K.R., Khalil A.S., Ting A.Y., Walker G.C., Collins J.J. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong F., Stokes J.M., Cervantes B., Penkov S., Friedrichs J., Renner L.D., Collins J.J. Cytoplasmic condensation induced by membrane damage is associated with antibiotic lethality. Nat. Commun. 2021;12:2321. doi: 10.1038/s41467-021-22485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopatkin A.J., Stokes J.M., Zheng E.J., Yang J.H., Takahashi M.K., You L., Collins J.J. Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat Microbiol. 2019;4:2109–2117. doi: 10.1038/s41564-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachhawat A.K., Thakur A., Kaur J., Zulkifli M. Glutathione transporters. Biochim. Biophys. Acta. 2013;1830:3154–3164. doi: 10.1016/j.bbagen.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 39.Hishinuma F., Izaki K., Takahashi H. Effects of Glycine and D-amino acids on growth of various microorganisms. Agric. Biol. Chem. 1969;33:1577–1586. [Google Scholar]
- 40.Hammes W., Schleifer K.H., Kandler O. Mode of action of glycine on the biosynthesis of peptidoglycan. J. Bacteriol. 1973;116:1029–1053. doi: 10.1128/jb.116.2.1029-1053.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitney J.G., Grula E.A. Incorporation of D-serine into the cell wall mucopeptide of Micrococcus lysodeikticus. Biochem. Biophys. Res. Commun. 1964;14:375–381. doi: 10.1016/s0006-291x(64)80013-9. [DOI] [PubMed] [Google Scholar]
- 42.Li B., Wang L., Su L., Chen S., Li Z., Chen J., Wu J. Glycine and Triton X-100 enhanced secretion of recombinant α-CGTase mediated by OmpA signal peptide in Escherichia coli. Biotechnol. Bioproc. Eng. 2013;17:1128–1134. [Google Scholar]
- 43.P. Y. Aristos. Aristidou A., Ka-Yiu San Effects of glycine supplement on protein production and release in recombinant Escherichia coli. Biotechnol. Lett. 1993;15:331–336. [Google Scholar]
- 44.Cosloy S.D., McFall E. Metabolism of D-serine in Escherichia coli K-12: mechanism of growth inhibition. J. Bacteriol. 1973;114:685–694. doi: 10.1128/jb.114.2.685-694.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindler M.K., Schutz M.S., Muhlenkamp M.C., Rooijakkers S.H., Hallstrom T., Zipfel P.F., Autenrieth I.B. Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J. Immunol. 2012;189:4900–4908. doi: 10.4049/jimmunol.1201383. [DOI] [PubMed] [Google Scholar]
- 46.Koenigs A., Stahl J., Averhoff B., Gottig S., Wichelhaus T.A., Wallich R., Zipfel P.F., Kraiczy P. CipA of acinetobacter baumannii is a novel plasminogen binding and complement inhibitory protein. J. Infect. Dis. 2016;213:1388–1399. doi: 10.1093/infdis/jiv601. [DOI] [PubMed] [Google Scholar]
- 47.Maruvada R., Prasadarao N.V., Rubens C.E. Acquisition of factor H by a novel surface protein on group B Streptococcus promotes complement degradation. Faseb. J. 2009;23:3967–3977. doi: 10.1096/fj.09-138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang M., Yang L., Chen Z.G., Lai S.S., Zheng J., Peng B. Exogenous maltose enhances Zebrafish immunity to levofloxacin-resistant Vibrio alginolyticus. Microb. Biotechnol. 2020;13:1213–1227. doi: 10.1111/1751-7915.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang D.X., Yang H., Cao Y.C., Jiang M., Zheng J., Peng B. Succinate promotes phagocytosis of monocytes/macrophages in teleost fish. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.644957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee H.H., Collins J.J. Microbial environments confound antibiotic efficacy. Nat. Chem. Biol. 2011;8:6–9. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miajlovic H., Smith S.G. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol. Lett. 2014;354:1–9. doi: 10.1111/1574-6968.12419. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z., Li M.Y., Peng B., Cheng Z.X., Li H., Peng X.X. GC-MS-Based metabolome and metabolite regulation in serum-resistant Streptococcus agalactiae. J. Proteome Res. 2016;15:2246–2253. doi: 10.1021/acs.jproteome.6b00215. [DOI] [PubMed] [Google Scholar]
- 53.Cheng Z.X., Gong Q.Y., Wang Z., Chen Z.G., Ye J.Z., Li J., Wang J., Yang M.J., Ling X.P., Peng B. Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front. Immunol. 2017;8:1706. doi: 10.3389/fimmu.2017.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong Q.Y., Yang M.J., Yang L.F., Chen Z.G., Jiang M., Peng B. Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb. Biotechnol. 2020;13:796–812. doi: 10.1111/1751-7915.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang D.X., Yang M.J., Yin Y., Kou T.S., Peng L.T., Chen Z.G., Zheng J., Peng B. Serine metabolism tunes immune responses to promote Oreochromis niloticus survival upon edwardsiella tarda infection. mSystems. 2021;6 doi: 10.1128/mSystems.00426-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keren I., Wu Y., Inocencio J., Mulcahy L.R., Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 57.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 58.Cohen M.S., Cooney M.H. A bacterial respiratory burst: stimulation of the metabolism of Neisseria gonorrhoeae by human serum. J. Infect. Dis. 1984;150:49–56. doi: 10.1093/infdis/150.1.49. [DOI] [PubMed] [Google Scholar]
- 59.Zhang S., Wang J., Jiang M., Xu D., Peng B., Peng X.X., Li H. Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ. Microbiol. 2019;21:4724–4739. doi: 10.1111/1462-2920.14811. [DOI] [PubMed] [Google Scholar]
- 60.Peng B., Su Y.B., Li H., Han Y., Guo C., Tian Y.M., Peng X.X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metabol. 2015;21:249–262. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz-Ramirez A., Ortiz-Balderas E., Cardozo-Saldana G., Diaz-Diaz E., El-Hafidi M. Glycine restores glutathione and protects against oxidative stress in vascular tissue from sucrose-fed rats. Clin. Sci. (Lond.) 2014;126:19–29. doi: 10.1042/CS20130164. [DOI] [PubMed] [Google Scholar]
- 62.Forman H.J., Zhang H., Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Aspect. Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giambelluca M.S., Gende O.A. Effect of glycine on the release of reactive oxygen species in human neutrophils. Int. Immunopharm. 2009;9:32–37. doi: 10.1016/j.intimp.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Badran E.G., Abogadallah G.M., Nada R.M., Nemat Alla M.M. Role of glycine in improving the ionic and ROS homeostasis during NaCl stress in wheat. Protoplasma. 2015;252:835–844. doi: 10.1007/s00709-014-0720-2. [DOI] [PubMed] [Google Scholar]
- 65.Garcia J., Han D., Sancheti H., Yap L.P., Kaplowitz N., Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J. Biol. Chem. 2010;285:39646–39654. doi: 10.1074/jbc.M110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fratelli M., Demol H., Puype M., Casagrande S., Eberini I., Salmona M., Bonetto V., Mengozzi M., Duffieux F., Miclet E., Bachi A., Vandekerckhove J., Gianazza E., Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao X.L., Chen Z.G., Yang T.C., Jiang M., Wang J., Cheng Z.X., Yang M.J., Zhu J.X., Zhang T.T., Li H., Peng B., Peng X.X. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj0716. [DOI] [PubMed] [Google Scholar]
- 68.Corti A., Belcastro E., Dominici S., Maellaro E., Pompella A. The dark side of gamma-glutamyltransferase (GGT): pathogenic effects of an 'antioxidant' enzyme. Free Radic. Biol. Med. 2020;160:807–819. doi: 10.1016/j.freeradbiomed.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 69.Franzini M., Corti A., Fierabracci V., Pompella A. Helicobacter, gamma-glutamyltransferase and cancer: further intriguing connections. World J. Gastroenterol. 2014;20:18057–18058. doi: 10.3748/wjg.v20.i47.18057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ricci V., Giannouli M., Romano M., Zarrilli R. Helicobacter pylori gamma-glutamyl transpeptidase and its pathogenic role. World J. Gastroenterol. 2014;20:630–638. doi: 10.3748/wjg.v20.i3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corti A., Paolicchi A., Franzini M., Dominici S., Casini A.F., Pompella A. The S-thiolating activity of membrane gamma-glutamyltransferase: formation of cysteinyl-glycine mixed disulfides with cellular proteins and in the cell microenvironment. Antioxidants Redox Signal. 2005;7:911–918. doi: 10.1089/ars.2005.7.911. [DOI] [PubMed] [Google Scholar]
- 72.Dominici S., Pieri L., Paolicchi A., De Tata V., Zunino F., Pompella A. Endogenous oxidative stress induces distinct redox forms of tumor necrosis factor receptor-1 in melanoma cells. Ann. N. Y. Acad. Sci. 2004;1030:62–68. doi: 10.1196/annals.1329.008. [DOI] [PubMed] [Google Scholar]
- 73.Council N.R. eighth ed. The National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 74.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang M.J., Jiang M., Peng X.X., Li H. Myo-inositol restores Tilapia's ability against infection by aeromonas sobria in higher water temperature. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.682724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang M.J., Xu D., Yang D.X., Li L., Peng X.X., Chen Z.G., Li H. Malate enhances survival of zebrafish against Vibrio alginolyticus infection in the same manner as taurine. Virulence. 2020;11:349–364. doi: 10.1080/21505594.2020.1750123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang M., Kuang S.F., Lai S.S., Zhang S., Yang J., Peng B., Peng X.X., Chen Z.G., Li H. Na(+)-NQR confers aminoglycoside resistance via the regulation of l-alanine metabolism. mBio. 2020;11 doi: 10.1128/mBio.02086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bala C.G., Rusu A., Ciobanu D., Bucsa C., Roman G. Amino acid signature of oxidative stress in patients with type 2 diabetes: targeted exploratory metabolomic research. Antioxidants. 2021;10 doi: 10.3390/antiox10040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y., Wang X., Ali F., Li Z., Fu Y., Yang X., Lin W., Lin X. Comparative extracellular proteomics of aeromonas hydrophila reveals iron-regulated secreted proteins as potential vaccine candidates. Front. Immunol. 2019;10:256. doi: 10.3389/fimmu.2019.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ye J.Z., Su Y.B., Lin X.M., Lai S.S., Li W.X., Ali F., Zheng J., Peng B. Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 2018;9:29. doi: 10.3389/fmicb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boulghobra D., Grillet P.E., Laguerre M., Tenon M., Fauconnier J., Fanca-Berthon P., Reboul C., Cazorla O. Sinapine, but not sinapic acid, counteracts mitochondrial oxidative stress in cardiomyocytes. Redox Biol. 2020;34 doi: 10.1016/j.redox.2020.101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J., Zhang Y., Wang X., Walk S.T., Wang G. Integrated metabolomics and targeted gene transcription analysis reveal global bacterial antimonite resistance mechanisms. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.617050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang C., Chao Y., Xu W., Liu Z., Wang H., Huang K. Myeloid FBW7 deficiency disrupts redox homeostasis and aggravates dietary-induced insulin resistance. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Villa-Bellosta R. Dietary magnesium supplementation improves lifespan in a mouse model of progeria. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.202012423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Z., Xu J., Sun S., Lin W., Li Y., Lu Q., Li F., Yang Z., Lu Y., Liu W. Mecheliolide elicits ROS-mediated ERS driven immunogenic cell death in hepatocellular carcinoma. Redox Biol. 2022;54 doi: 10.1016/j.redox.2022.102351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang K., Zhang Z., Tsai H.I., Liu Y., Gao J., Wang M., Song L., Cao X., Xu Z., Chen H., Gong A., Wang D., Cheng F., Zhu H. Branched-chain amino acid aminotransferase 2 regulates ferroptotic cell death in cancer cells. Cell Death Differ. 2021;28:1222–1236. doi: 10.1038/s41418-020-00644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mishina N.M., Bogdanova Y.A., Ermakova Y.G., Panova A.S., Kotova D.A., Bilan D.S., Steinhorn B., Arner E.S.J., Michel T., Belousov V.V. Which antioxidant system shapes intracellular H2O2 gradients? Antioxidants Redox Signal. 2019;31:664–670. doi: 10.1089/ars.2018.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kielkopf C.L., Bauer W., Urbatsch I.L. Bradford assay for determining protein concentration. Cold Spring Harb. Protoc. 2020;2020 doi: 10.1101/pdb.prot102269. [DOI] [PubMed] [Google Scholar]
- 89.Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 91.Reuter W.H., Masuch T., Ke N., Lenon M., Radzinski M., Van Loi V., Ren G., Riggs P., Antelmann H., Reichmann D., Leichert L.I., Berkmen M. Utilizing redox-sensitive GFP fusions to detect in vivo redox changes in a genetically engineered prokaryote. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meyer A.J., Dick T.P. Fluorescent protein-based redox probes. Antioxidants Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 93.Morgan B., Sobotta M.C., Dick T.P. Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic. Biol. Med. 2011;51:1943–1951. doi: 10.1016/j.freeradbiomed.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 94.Loi V.V., Harms M., Muller M., Huyen N.T.T., Hamilton C.J., Hochgrafe F., Pane-Farre J., Antelmann H. Real-time imaging of the bacillithiol redox potential in the human pathogen Staphylococcus aureus using a genetically encoded bacilliredoxin-fused redox biosensor. Antioxidants Redox Signal. 2017;26:835–848. doi: 10.1089/ars.2016.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kadokura H., Tian H., Zander T., Bardwell J.C., Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 96.Singh M., Mukhopadhyay K. C-terminal amino acids of alpha-melanocyte-stimulating hormone are requisite for its antibacterial activity against Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55:1920–1929. doi: 10.1128/AAC.00957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tyagi P., Singh M., Kumari H., Kumari A., Mukhopadhyay K. Bactericidal activity of curcumin I is associated with damaging of bacterial membrane. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.