Abstract
Genome-editing technologies, especially CRISPR (clustered regularly interspaced short palindrome repeats)/Cas9 (CRISPR-associated protein 9), endows researchers the ability to make efficient, simple , and precise genomic DNA changes in many eukaryotic cell types. CRISPR/Cas9-mediated efficient gene knockout holds huge potential to improve the efficacy and safety of chimeric antigen receptor (CAR) T cell-based immunotherapies. Here, we describe an optimized approach for a complete loss of endogenous T cell receptor (TCR) protein expression, by CRISPR/Cas9-mediated TCR α constant (TRAC) and TCR β constant (TRBC) gene knockout, followed by subsequent CD3 negative selection in engineered human ortho CAR19 T cells. We believe this method can be expanded beyond CAR T cell application, and target other cell surface receptors.
Graphical abstract:
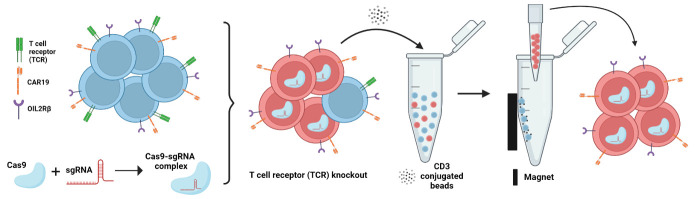
Schematic overview of the two-step process of endogenous TCR depletion in engineered human ortho CAR19 T cells using (1) CRISPR/Cas9-mediated gene knockout followed by (2) CD3 negative selection.
Keywords: CRISPR-Cas9 , TCR knockout , CD3 negative selection , ortho CAR T cells , TRAC/TRBC guide RNA , 4D-Nucleofector Lonza Electroporator
Background
CRISPR-Cas9 technology enables gene knockouts (KO) and complete loss of protein translation in human cells, especially in primary cells ( Seki and Rutz, 2018 ). In recently published research, we showed that combining an orthogonal human IL-2 and IL-2Rβ ( ortho -hIL-2/ ortho -hIL-2Rβ) system with CAR19 T cell therapy markedly enhanced the efficacy of the CAR T cell antitumor activity, in a well-established preclinical model of acute lymphoblastic leukemia (ALL) ( Zhang et al. , 2021 ). However, the unexpected deaths observed with high-dose ortho- hIL-2 led us to investigate mechanisms of toxicity. To explore whether T cell-mediated toxicity is related to acute xenogeneic graft-versus-host disease (GVHD) mediated through the T cell receptor (TCR), we knocked out the endogenous TCR of CAR T cells co-expressing ortho -hIL-2Rβ, using a CRISPR-Cas9 gene KO approach. We use guide RNAs (gRNA) targeting the TCR α constant (TRAC) and TCR β constant (TRBC) loci, followed by CD3 negative selection, resulting in TCR negative ortho CAR19 T cells.
Gene silencing is a powerful approach to study gene function and to discern molecular mechanisms underlying complex human diseases. Previous studies have utilized RNA interference (RNAi), small interfering RNA (siRNA), or short hairpin RNA (shRNA) technologies to effectively knock down genes of interest ( Berns et al. , 2004 ; O’ Keefe, 2013 ). However, these knockdown approaches do not result in complete loss of gene/protein expression or KO and can often result in off-target effects ( Jackson et al. , 2006 ).
The CRISPR-Cas9 genome editing system consists of two components: a “guide” RNA (gRNA) and a Cas9. The Cas9 protein is an endonuclease that uses gRNA to form base pairs with DNA target sequences, enabling Cas9 to introduce a site-specific double-stranded break in the DNA. Through RNA-directed Cas9 nucleases, the CRISPR-Cas9 system can modify DNA with greater precision than existing technologies, such as transcription activator-like endonuclease (TALEN), and zinc-finger nuclease (ZFN) ( Conant et al. , 2022 ). While both TALEN and ZFN are artificial structures formed by joining restriction endonucleases with DNA-binding protein domains, CRISPR-Cas9 technology takes advantage of the antiviral defense mechanism of prokaryotes to perform genome editing at the targeted DNA sequence ( Khan, 2019 ). Recent work done by Cui et al. revealed that both TALEN and ZFN inevitably generated massive off-target effects when targeting human papillomavirus 16 (HPV16), while the CRISPR-Cas9 system was shown to be more efficient and specific ( Cui et al. , 2021 ). Therefore, CRISPR-Cas9 technology shows better efficiency, feasibility, and practicality in multi-role clinical applications, as compared to TALEN and ZFN. Inference of CRISPR Edits (ICE) was used to analyze CRISPR editing data in this study. ICE is a free and easy-to-use software tool that offers fast and reliable analysis of CRISPR experiments, highly comparable to that of next-generation sequencing (NGS) data. CRISPR editing can be assessed and analyzed by uploading the Sanger sequencing data and specifying a guide sequence(s). A knockout score will be generated, which represents the proportion of cells that have either a frameshift or insertion-deletion mutation (indels). This score is a useful means to quantify the number of contributing indels that are likely to result in a functional KO of the targeted gene.
Our results showed that the CRISPR-Cas9 system is easy to operate, has a short cycle time, low cost, higher accuracy, and is more controllable in targeting TCR α and TCR β genes together than existing technologies such as TALEN and ZFN. The combination of CRISPR/Cas9 with CD3 negative selection leads to a complete TCR depletion in ortho CAR19 T cells.
Materials and Reagents
-
Tissue culture plates, flasks, and tubes
24 well cell tissue culture plates (Corning, catalog number: 3524)
T25 cell culture flasks (Thermo Fisher, catalog number: 156367)
T75 cell culture flasks (Thermo Fisher, catalog number: 156499)
15 mL centrifuge tubes (Corning, catalog number: 352095)
50 mL centrifuge tubes (Corning, catalog number: 430829)
Phosphate Buffered Saline solution (PBS) (Corning, catalog number: 21-031-CM)
RPMI 1640 medium (Gibco, catalog number: 11875085)
Bovine Calf Serum (Sigma, catalog number: 12306C-500ml)
Penicillin/Streptomycin (Thermo Fisher, catalog number: 10378016)
L-glutamine (Thermo Fisher, catalog number: A2916801)
Recombinant human Interleukin-2 (IL2) (R&D System, catalog number: 202-IL-050)
APC anti-human TCR α/β antibody (Biolegend, catalog number: 306718)
Dynabeads TM CD3 (Thermo Fisher, catalog number:11151D)
TrueCut TM Cas9 Protein v2 (Invitrogen, catalog number: A36497)
EasySet Magnet (Thermo Fisher, catalog number: NC9284020)
DNeasy Blood & Tissue Kit (Qiagen, catalog number: 69504)
Q5 Hot Start High-Fidelity DNA Polymerase Kit (BioLabs, catalog number: E0555S)
Trypan Blue Solution, 0.4% (Thermo Fisher, catalog number: 15250061)
QIAquick PCR Purification Kit (Qiagen, catalog number: 28106)
4D-Nucleofector TM X Kit (Lonza, catalog number: V4XC-2032)
Cell culture media (see Recipes)
Wash buffer for cells (see Recipes)
gRNA (see Recipes)
Equipment
4D-Nucleofector TM X Unit (Lonza, catalog number: AAF-1002X)
Flow Cytometer LSRFortessa (BD Biosciences, catalog number: 647800L6)
Eppendorf TM 5424 Microcentrifuges (Fisher, catalog number: 05-400-005)
Cell culture hood HeraSafe KS (Thermo Fisher, catalog number: 10110910)
CO2 incubator HeraCell VIOS 160i (Thermo Fisher, catalog number: 15381075)
VeritiPro TM 96-well Thermal Cycler (Applied Biosystems, catalog number: A47394)
Software
Genewiz (Azenta Life Sciences, https://www.genewiz.com/en )
ICE analysis (Synthego, https://ice.synthego.com/#/ )
CRISPOR (Tefor, http://crispor.tefor.net/ )
FlowJo (Tree Star Inc., version 10.1)
Prism 9 for macOS V.9.3.1(350) ( https://www.graphpad.com/scientific-software/prism/ )
BioRender ( https://biorender.com/ )
Procedure
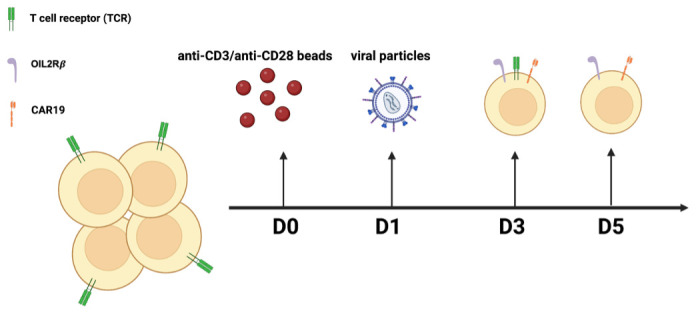
A general workflow for the overall experimental procedures can be found in Figure 1 .
Figure 1. General workflow of the overall experimental procedures.
CRISPR/Cas9-mediated gene knockout followed by CD3 negative selection results in a complete depletion of TCR in ortho CAR19 T cells.
-
Preparation of human ortho CAR19 T cells
Primary human total T cells were purchased from the Immunology Core of University of Pennsylvania.
Count cells and adjust to 1 × 10 6 cells/mL in RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, 100-U/mL penicillin, 100-g/mL streptomycin sulfate, and 10-mM HEPES. Assess cell viability using the Trypan Blue solution (0.4%, Thermo Fisher) as a cell stain for the dye exclusion test.
Wash the magnetic beads coated with anti-CD3/anti-CD28 three times with PBS, and once with cell culture media.
Mix T cells with the washed beads at a ratio of 1:3, for 16–24 h.
Transduce activated T cells with viral particles encoding ortho CAR19, as described by our previous work ( Zhang et al. , 2021 ). Briefly, both CD19BBz and ortho -hIL-2Rβ sequences were cloned in a p-TRPE lentiviral backbone, in frame with the restriction site XbaI and Sa1I for CD19BBz, ortho -hIL-2Rβ-T2A-mCherry, and CD19- ortho -hIL-2Rβ. Lentivirus was produced by transfecting HEK293T cells with a combination of the above transfer plasmid and three lentiviral package plasmids, pCI VSV-G, pRSV Rev, and pMDL gag/pol.RRE (Addgene).
Culture cells with 100 IU/mL human recombinant IL2, and feed them every day for three days, in preparation for CRISPR/Cas9-mediated TCR KO.
-
Preparation of the CRISPR-Cas9 complex
-
TCRα and TCRβ gRNA sequence (TRAC gRNA sequence: TGTGCTAGACATGAGGTCTA; TRBC gRNA sequence: GGAGAATGACGAGTGGACCC) were designed by using CRISPOR, an online program that helps design, evaluate, and clone guide sequences for the CRISPR/Cas9 system. The main considerations regarding potential gRNA candidates are for finding the gRNAs that (1) efficiently KO the gene close to ATG (translation start site) for maximum loss of function, or at the location of functional domain to disrupt the target function; (2) have no or less off-target genes.
The single human genomic sequence used in the TRAC and TRBC gRNA design was CTTCCTTTGTCCCCAATCTGGGCGCGCGCCGGCGCCCCCTGGCGGCCTAAGGACTCGGCGCGCCGGAAGTGGCCAGGGCGGGGGCGACCTCGGCTCACAGCGCGCCCGGCTATTCTCGCAGCTCACCATGGATGATGATATCGCCGCGCTCGTCGTCGACAACGGCTCCGGCATGTGCAAGGCCGGCTTCGCGGGCGACGATGCCCCCCGGGCCGTCTTCCCCTCCATCGTGGGGCGCC, with a protospacer adjacent motif (PAM) sequence of 20bp-NGG-Sp Cas9, SpCas9-HF1, eSpCas9 1.1.
Dissolve TCRα and TCRβ gRNAs (Agilent) in 1× TE buffer (IDT) at 10 μg/μL, and mix them together in equimolar concentrations.
Combine 1 μL of gRNA mixture, including 5 μg TCR α and 5 μg TCR β, with 2 μL (5 μg/μL) of Cas9 (Invitrogen).
Mix the gRNAs-Cas9 complex mixture by gently pipetting up and down twice; avoid creating air bubbles in the tube.
Incubate the complex mix at room temperature (RT: 15–25 °C) for 10 min.
-
-
Preparation of T cell suspension for electroporation
For each electroporation condition (including positive and negative controls), prepare 5 mL of cell expansion medium supplemented with 10% FBS, 2 mM L-glutamine, 100-U/mL penicillin, 100-g/mL streptomycin sulfate, 10-mM HEPES, and 100U IL2.
For each condition, add 2 mL of supplemented medium (prepared in Procedure A) to a 12-well plate (for 100-μL tip reactions), and place in a 37°C incubator.
Transfer 1.5 × 10 6 cells (for 100-μL cuvette reactions) from the ortho CAR19 T cells to a 15-mL tube. Centrifuge at 300 × g and RT for 5 min.
-
Electroporation of T cells with gRNA-Cas9 complex using the Lonza 4D system
A demonstration of how to perform electroporation using Lonza 4D system can be found in Figure 2 .
Aspirate the supernatant from the cell pellet (prepared in Procedure C). Resuspend cells in 100 μL of P3 buffer from the 4D-Nucleofector X Kit and pipette up and down to mix.
Using sterile 200-μL PCR tubes for each condition, combine the gRNA-Cas9 complex mixture (prepared in Procedure B) with cells in P3 buffer.
Mix gently by pipetting up and down carefully twice; avoid creating bubbles in the tube.
Transfer cells with the gRNA-Cas9 complex mixture into a cuvette from the 4D-Nucleofector X Kit, making sure that there are no air bubbles. Place the cuvette into the electroporation chamber.
Electroporate the cell mixture using the “human stim T-cell” preset program and a pulse code of EH-115.
-
Post-knockout culture
Immediately add culture media into the cuvette containing the electroporated cells, and incubate at 37 °C for 15 min.
Count cells, and adjust cell density to 0.8 × 10 6 cells/mL, by adding IL2-supplemented cell culture media.
Incubate at 37 °C for 72 h, for genome editing to occur.
-
Harvest cells for assessment of genome editing efficiency. Isolate and amplify the genomic DNA using primers that flank the target region. Sequence the PCR product.
PCR kit used: Q5 Hot Start High-Fidelity DNA Polymerase Kit
Harvest cells and check for TCR protein expression with flow cytometry, using APC human anti-TCRα/β.
-
Isolation of CD3 negative T cells
Negative selection of CD3 + cells is performed using Dynabeads CD3 (Thermo Fisher Scientific).
-
Wash the Beads
Resuspend the beads in the vial.
Transfer the desired volume of beads to a 5-mL tube.
Add the same volume of Isolation Buffer (PBS supplemented 0.1% BSA).
Place the tube in a magnet for 1 min, and discard the supernatant.
Remove the tube from the magnet and resuspend the washed beads in the same volume of Isolation Buffer as the initial volume of beads.
-
Prepare Cells
Prepare TCR KO T cells to 1 × 10 7 cells/mL in Isolation Buffer.
Transfer 1 mL of cells (1 × 10 7 ) to a tube, and add 25 µL of pre-washed and resuspended beads.
Incubate with gentle tilting and rotation at 2–8 °C for 30 min (depletion).
Place the tube in a Dynabeads TM magnetic rack for 2 min.
Transfer the supernatant to a new tube for further use, and discard the beads.
-
Flow cytometry
Wash the CD3 negative T cells twice with PBS, and stain with APC anti-human TCRα/β antibody (1:50, Biolegend) for 30 min, followed by two washes with PBS.
Quantify CD3 negative T cells using a BD LSRFortessa flow cytometer.
Perform analysis of T cell CD3 expression, using FlowJo software (Tree Star Inc., version 10.1).
Figure 2. A demonstration of how to perform electroporation using Lonza 4D system.

Data analysis
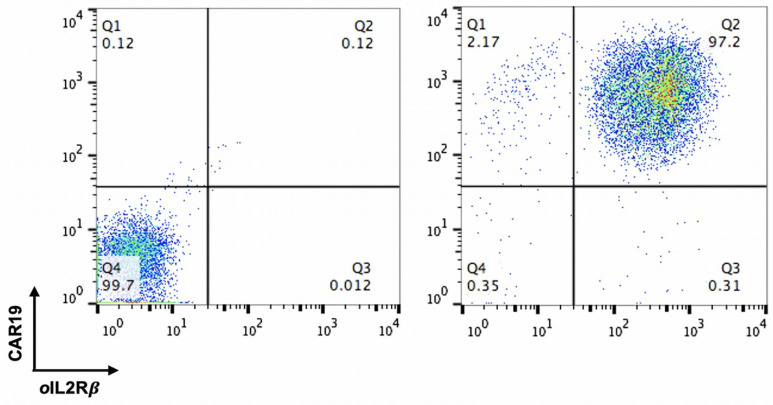
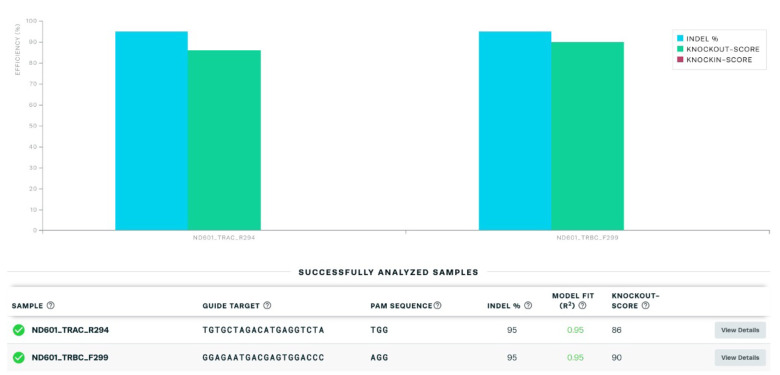
The expression of CAR19 and orthogonal IL-2Rβ was confirmed by flow cytometry before performing TCR KO ( Figure 3 ). Flow cytometry data were analyzed using the FlowJo software. For direct estimation of the efficiency and the type of mutations introduced by genome editing, genomic DNA was extracted from edited and non-edited T cells. PCR reactions were performed with primers surrounding the guide RNA, and the PCR products were visualized in 1.0% gel ( Figure 4 ), and purified for standard Sanger sequencing (Genewiz). Once Sanger sequencing of the DNA was complete, the sequence traces were analyzed using ICE ( Figures 5 and 6A -D). To completely deplete the unedited TCR positive cells, CD3 negative selection was performed using CD3 Dynabeads TM . The purity of the TCRα/β depletion was assessed by flow cytometry ( Figure 6E ); a demonstration of the flow cytometry gating strategy used in this study can be found in Video 1 .
Figure 3. FACS analysis of CAR19 and ortho -IL2Rβ expression in T cells.
Figure 4. Agarose gel electrophoresis results for the analysis of PCR products.
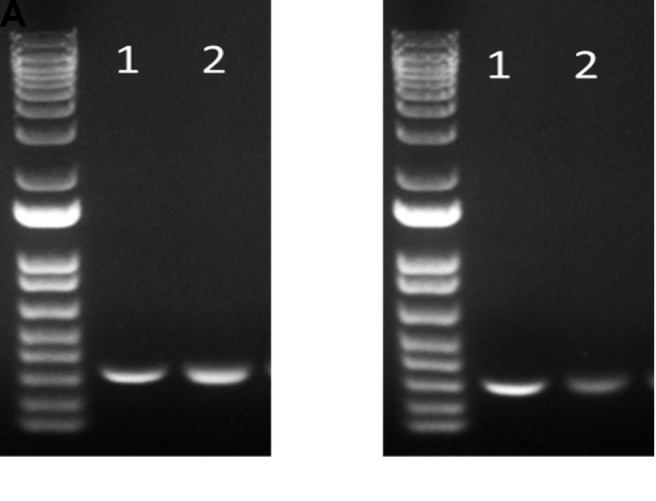
(A) TCRα. (B) TCRβ. Lane 1: Control (without TCR gRNA) Lane 2: TCR KO + CD3 negative selection.
Figure 5. Purification and sequencing of PCR products with a quantitative assessment of genome editing of TCRα/β knockout by using Synthego’s Inference of CRISPR Edits (ICE).
Figure 6. Sequencing results of TCRα/β KO visualized using ICE and flow cytometric assessment of TCRα/β depletion on day 4 after gene editing followed by CD3 negative selection.
Samples of TCRα and TCRβ KO were successfully analyzed with multiple parameters shown, including sample label, ICE score, R 2 score, knockout score, guide sequence, and PAM sequence. (A–B) The “contributions” tab shows the inferred sequences of TCRα (A) and TCRβ (B) that are present in an edited population, as well as their relative representation in the edited pool. The black vertical dotted line represents the cut site. (C–D) Sequencing chromatograms of TCRα KO vs. TCRα wild type (WT) (C) and TCRβ KO vs. TCRβ WT (D). (E) FACS analysis was used to confirm complete TCRα/β depletion after CD3 negative selection.
Video 1. Demonstration of the flow cytometry gating strategy for the confirmation of complete TCR depletion.
Recipes
-
Cell culture media
RPMI supplemented with 10% heat-inactivated FBS, 1% L-glutamine, 1% HEPES, 1% penicillin, and streptomycin.
-
Wash buffer for cells
PBS supplemented with 2% BSA
gRNA resuspended in TE buffer at 5 μg/μL
Acknowledgments
We thank the members of the following facilities at the University of Pennsylvania: Flow Cytometry Core for cell sorting, Human Immunology Core for providing T cells from normal donors.
This work was derived from the paper previously published by Zhang et al. (2021), called “A human orthogonal IL-2 and IL-2Rβ system enhances CAR T cell expansion and antitumor activity in a murine model of leukemia” in Sci Transl Med 13(625).
Competing interests
The authors have no financial and/or non-financial competing interests to disclose.
Ethics
T cells were collected under a University Institutional Review Board-approved protocol, and written informed consent was obtained from each healthy donor.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Berns K. , Hijmans E. M. , Mullenders J. , Brummelkamp T. R. , Velds A. , Heimerikx M. , Kerkhoven R. M. , Madiredjo M. , Nijkamp W. , Weigelt B. , et al. .( 2004 . ). A large-scale RNAi screen in human cells identifies new components of the p53 pathway . Nature 428 : 431 - 437 . [DOI] [PubMed] [Google Scholar]
- 2. Conant D. , Hsiau T. , Rossi N. , Oki J. , Maures T. , Waite K. , Yang J. , Joshi S. , Kelso R. , Holden K. , et al. .( 2022 . ). Inference of CRISPR Edits from Sanger Trace Data . CRISPR J 5 ( 1 ): 123 - 130 . [DOI] [PubMed] [Google Scholar]
- 3. Cui Z. , Liu H. , Zhang H. , Huang Z. , Tian R. , Li L. , Fan W. , Chen Y. , Chen L. , Zhang S. , et al. .( 2021 . ). The comparison of ZFNs, TALENs, and SpCas9 by GUIDE-seq in HPV-targeted gene therapy . Mol Ther Nucleic Acids , 26 : 1466 - 1478 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaj T. , Sirk S. J. , Shui S-I. and Liu J. ( 2016 . ). Genome-Editing Technologies: Principles and Applications . Cold Spring Harb Perspect Biol 8 ( 12 ): a023754 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson A. L. , Burchard J. , Schelter J. , Chau B. N. , Cleary M. , Lim L. and Linsley P. S. ( 2006 . ). Widespread siRNA"off-target" transcript silencing mediated by seed region sequence complementarity . RNA 12 ( 7 ): 1179 - 1187 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khan S. H. ( 2019 . ). Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application . Mol Ther Nucleic Acids , 16 : 326 - 334 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Keefe E. P. ( 2013 . ). siRNAs and shRNAs: Tools for protein knockdown by gene silencing . Mater Methods 3 : 197 . [Google Scholar]
- 8. Seki A. and Rutz S. ( 2018 . ). Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells . J Exp Med 215 ( 3 ): 985 - 997 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Q. , Hresko M. E. , Picton L. K. , Su L. , Hollander M. J. , Nunez-Cruz S. , Zhang Z. , Assenmacher C. A. , Sockolosky J. T. , Garcia K. C. , et al. .( 2021 . ). A human orthogonal IL-2 and IL-2Rβ system enhances CAR T cell expansion and antitumor activity in a murine model of leukemia . Sci Transl Med 13 ( 625 ): eabg6986 . [DOI] [PMC free article] [PubMed] [Google Scholar]