Abstract
Monoclonal antibodies (MAbs) which react with heat-resistant proteins with molecular masses of 32 to 33 kDa of 14 different Bartonella species were produced. These antibodies did not react with antigens of 26 diverse bacterial strains by microimmunofluorescence assay except MAb B3D4, which reacted with Chlamydia psittaci and Chlamydia trachomatis at low titers. The identification of a common Bartonella antigenic protein will make it possible to later produce a diagnostic antigen by cloning and expressing it in Escherichia coli. Moreover, these MAbs allow all Bartonella species to be identified to the genus level.
The genus Bartonella currently comprises 14 species. Human infections due to Bartonella species are widely considered emerging diseases, although they also include long-recognized diseases such as Carrión's disease, trench fever, and cat scratch disease (15, 17, 23). Newer clinical manifestations, such as bacillary angiomatosis, peliosis hepatis, chronic lymphadenopathy, and endocarditis, which are sometimes due to uncommonly encountered species such as Bartonella elizabethae, Bartonella vinsonii subsp. berkhoffii, or Bartonella vinsonii subsp. arupensis, have been recently identified (1, 23, 25, 28, 32). Serologic diagnosis of Bartonella spp. is mostly based on microimmunofluorescence (MIF) serology that detects antibodies to B. quintana and B. henselae only (21, 23). A serologic test that detects antibodies against all species is not available. Such a test needs to detect an epitope common to, but also specific to, all Bartonella spp. A monoclonal antibody (MAb) that can recognize this epitope would be the first step towards detecting this antigen after cloning and expressing the Bartonella genome in Escherichia coli in order to produce it for use in an enzyme-linked immunosorbent assay. Bartonella spp. may be isolated from clinical samples by using cell culture systems with endothelial cells or blood- or hemin-containing axenic media (21, 29). When isolated, identification of Bartonella is mostly based on molecular methods. The availability of a MAb that could screen Bartonella at the genus level would avoid the use of expensive and time-consuming molecular procedures on non-Bartonella bacteria. We thus decided to produce and characterize Bartonella genus-specific MAbs.
The sources of Bartonella strains used to screen hybridomas and test the specificity of MAbs are presented in Table 1. Bartonella strains were harvested and suspended in deionized water for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or in phosphate-buffered saline (PBS) for the MIF assay after 5 to 7 days of culture on blood agar plates. The procedure for the production of MAbs has been detailed elsewhere (12, 22). Briefly, 6-week-old female BALB/c mice were inoculated with B. henselae Houston-1 suspended in 0.5 ml of PBS. The supernatants of the hybridomas were screened for antibodies to B. henselae by MIF. Representative hybridomas were subcloned twice by limiting dilution. Isotypes of MAbs were determined with an Immuno Type mouse monoclonal antibody isotyping kit with antisera to mouse immunoglobulin M (IgM), IgA, IgG1, IgG2a, IgG2b, and IgG3 (Sigma). Ascitic fluids were produced by injecting about 3 × 106 cells of hybridoma (B2D3 and B3D4) suspended in 0.5 ml of PBS into the mice 1 week after an intraperitoneal injection of 0.5 ml of pristane (2,6,10,14-tetramethylpentadecane; Sigma). The MIF assay (26) was used to screen hybridoma clones and to determine the specificity of the MAbs. Blind testing of 45 bacteria by MIF with MAbs B2D3 and B3D4 was carried out on 19 Bartonella strains, 3 Chlamydia strains, and 23 bacterial strains isolated in our laboratory from clinical samples (Table 1). Sera from immunized mice were used as positive controls, and sera from healthy mice were used as negative controls. SDS-PAGE and Western blotting were performed according to a modification of the method described by Laemmli (19, 22). Five human body lice from a laboratory colony were infected with a B. quintana strain by feeding on a bacteremic rabbit previously infected intravenously by 108 B. quintana cells. B. quintana bacteremia at the time the lice were fed was assessed by blood culture as previously described for cats (3). After being crushed and smeared onto microscope slides the lice were tested for Bartonella by MIF as described above with ascitic fluid of hybridoma B2D3 diluted 1:1,000.
TABLE 1.
Reactivity of MAbs with Bartonella antigens
| Species | Strain | Sourcea | Titer of MAb:
|
|
|---|---|---|---|---|
| B2D3 | B3D4 | |||
| B. henselae | Houston-1 (ATCC 4988) | Bacteremia (27) | 6,400 | 12,800 |
| B. henselae | San Ant 2 (SA2) | Cat scratch disease (6) | 25,600 | 25,600 |
| B. henselae | CAL-1 | Septicemia, United States | 6,400 | 3,200 |
| B. henselae | URBHLLY8 (CIP 104756) | Cat scratch disease (7) | 6,400 | 3,200 |
| B. henselae | URBHLIE9 | Endocarditis (7) | 6,400 | 6,400 |
| B. quintana | URBQMLY15 | Chronic lymphadenopathy (9, 21) | 6,400 | 12,800 |
| B. quintana | Fuller (ATCC VR-358) | Trench fever (30) | 6,400 | 12,800 |
| B. quintana | SH-PERM | Trench fever, Russia | 6,400 | 12,800 |
| B. clarridgeiae | URBCMNHC26 | Blood of cat, France | 6,400 | 12,800 |
| B. elizabethae | F9251 (ATCC 49927) | Endocarditis (5) | 6,400 | 3,200 |
| B. grahamii | V2 (NTCC 12860) | Blood of Clethrionomys glareolus (2) | 6,400 | 3,200 |
| B. taylorii | M6 (NTCC 12861) | Blood of Apodemus spp. (2) | 3,200 | 6,400 |
| B. doshiae | R18 (NTCC 12862) | Blood of Microtus agrestis (2) | 6,400 | 12,800 |
| B. vinsonii subsp. vinsonii | Baker (ATCC VR-152) | Spleen of Microtus pennsylvanicus (31) | 3,200 | 3,200 |
| B. vinsonii subsp. arupensis | OK 94-513 (ATCC 700727) | Bacteremia (32) | 6,400 | 12,800 |
| B. vinsonii subsp. berkhoffii | NCSV93-CO1 (ATCC 51672) | Blood of dog (18) | 1,600 | 3,200 |
| B. alsatica | IBS 383 (CIP 105477) | Blood of rabbit (13) | 6,400 | 12,800 |
| B. koehlerae | C-29 (ATCC 700693) | Blood of cat (10) | 6,400 | 12,800 |
| B. tribocorum | IBS 506 (CIP 105476) | Blood of rat (14) | 1,600 | 6,400 |
| B. bacilliformis | Monzon 812 | Blood of bartonellosis patient, Peru | 3,200 | 12,800 |
| C. psittaci | 50 | 800 | ||
| C. trachomatis | <25 | 400 | ||
| C. pneumoniae | <25 | <25 | ||
| 23 speciesb | <25 | <25 | ||
Geographic origin is given if the isolation of the strain is not detailed elsewhere.
Includes E. coli, Klebsiella pneumoniae, Enterobacter aerogenes, Yersinia enterocolitica, Shigella dysenteriae, Salmonella enterica, Campylobacter jejuni, Brucella melitensis, Ochrobactrum anthropi, Haemophilus influenzae, Kingella kingae, Neisseria meningitidis, Bacteroides fragilis, Desulfovibrio fairfieldensis, Fusobacterium necrophorum, Enterococcus faecalis, Afipia clevelandensis, Afipia felis, Pseudomonas aeruginosa, Pseudomonas putida, Burkholderia cepacia, Stenotrophomonas maltophilia, and Coxiella burnetii.
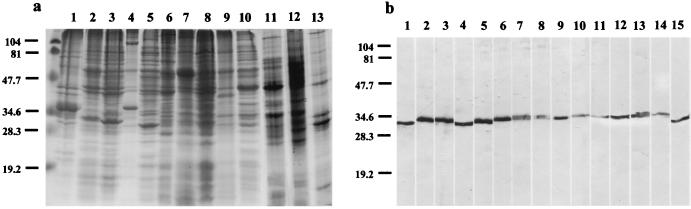
SDS-PAGE analysis of Bartonella antigens demonstrated distinct profiles of Bartonella species. Depending on species, 12 to 35 bands were observed. Proteins of 85, 71, 54, 44 to 47, 40, 36, 32 to 33, 30, and 18 to 19 kDa were common to all Bartonella strains studied (Fig. 1a). Both MAbs reacted with all tested Bartonella species. The immunofluorescence assay titers of MAbs with different Bartonella bacteria showed obvious differences. Titers from the homologous strain Houston-1 were the highest. The isotypes of B2D3 and B3D4 were identified as subclass IgG1. MAbs B2D3 and B3D4 showed reactivity with 32- or 33-kDa protein bands (Fig. 1b). The MAbs were directed against heat-resistant proteins because digestion with proteinase K completely destroyed the antigen's reactivities and heat treatment at 100°C for 10 min did not. The ascitic fluid from hybridomas B2D3 and B3D4 reacted with all of the Bartonella strains tested, but it did not react with the 23 other bacteria tested. Cross-reactivity was observed with Chlamydia psittaci and Chlamydia trachomatis. Nevertheless, the immunofluorescence assay titers of MAbs to Chlamydia spp. were much lower (Table 1). Bartonella spp. were demonstrated in four of the five infected lice by MIF with MAbs B2D3 and B3D4.
FIG. 1.
(a) SDS-PAGE analysis of different Bartonella species. Lanes: 1, B. bacilliformis; 2, B. henselae Houston-1; 3, B. henselae URBHLLY8; 4, B. clarridgeiae; 5, B. quintana; 6, B. elizabethae; 7, B. grahamii; 8, B. taylorii; 9, B. doshiae; 10, B. vinsonii subsp. vinsonii; 11, B. tribocorum; 12, B. koehlerae; 13, B. alsatica. (b) Western immunoblotting of MAb B2D3 with Bartonella antigens. Lanes: 1, B. bacilliformis; 2, B. henselae Houston-1; 3, B. henselae URBHLLY8; 4, B. clarridgeiae; 5, B. quintana; 6, B. elizabethae; 7, B. grahamii; 8, B. taylorii; 9, B. doshiae; 10, B. vinsonii subsp. vinsonii; 11, B. vinsonii subsp. berkhoffii; 12, B. vinsonii subsp. arupensis; 13, B. tribocorum; 14, B. koehlerae; 15, B. alsatica. Molecular masses (in kilodaltons) are shown at left.
The clinical manifestations of infections due to Bartonella, Coxiella, and Chlamydia can often be confused, especially in cases of infectious endocarditis. However, differential diagnosis of the diseases is important because their treatments are different. As Chlamydia spp. and Coxiella burnetii are strictly intracellular bacteria and Bartonella spp. are fastidious slowly growing organisms, they are difficult to isolate. Therefore, diagnosis of these infections continues to rely mainly on serology in spite of the serological cross-reactions among members of these genera that have been described (11, 20, 24). Moreover, because recently described species such as B. elizabethae, B. vinsonii subsp. berkhoffii, B. vinsonii subsp. arupensis, and B. clarridgeiae may be encountered in humans, a specific serologic test that recognizes all Bartonella infections is needed. In our work, we have obtained, as a first step, MAbs that recognize a protein antigen common to all Bartonella species. After cloning the Bartonella sp. genome in E. coli in order to obtain an expression bank, these MAbs could be used for screening products of clones in order to obtain a protein antigen common to all Bartonella spp. that could be used in an enzyme-linked immunosorbent assay for the detection of antibodies to all Bartonella spp. The anti-Bartonella genus-specific MAbs obtained in this study are highly specific, as they did not cross-react with 26 other bacterial species, except that MAb B3D4 cross-reacted at low titers with C. trachomatis and C. psittaci. Interestingly, none of the MAbs obtained reacted with Chlamydia pneumoniae or Coxiella burnetii, in spite of the cross-reactivity of these two genera and Bartonella spp. which has been previously described (11, 20, 24). Serological cross-reactivity between B. bacilliformis and C. psittaci antigens has been demonstrated previously (16). It was associated with a cross-reacting lipopolysaccharide antigen. Cross-reactivity between B. quintana, C. psittaci, and C. pneumoniae was later demonstrated in patients with B. quintana endocarditis (8). The serological cross-reactivity of Bartonella sp. and Chlamydia sp. antigens in the sera of patients infected by a member of these genera was also demonstrated to be due to cross-reacting protein antigens with molecular masses ranging from 30 to 90 kDa (24). Thus, our MAb B3D4 could also be used to investigate cross-reacting epitopes between Bartonella spp. and Chlamydia spp.
In conclusion, our Bartonella genus-specific MAbs recognized specifically all the tested species of Bartonella, and they successfully detected B. quintana in body lice. Thus, our MAbs may provide a tool to identify, at the genus level, isolated bacteria for which presumptive identification is compatible with Bartonella spp. or to detect such bacteria within arthropods, avoiding the use of molecular techniques for screening (4, 21).
Acknowledgments
We are grateful to R. Birtles for reviewing the manuscript.
REFERENCES
- 1.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Brenner S A, Rooney J A, Manzewitsch P, Regnery R L. Isolation of Bartonella (Rochalimaea) henselae: effects of methods of blood collection and handling. J Clin Microbiol. 1997;35:544–547. doi: 10.1128/jcm.35.3.544-547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouqui P, La Scola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. N Engl J Med. 1999;340:184–189. doi: 10.1056/NEJM199901213400303. [DOI] [PubMed] [Google Scholar]
- 5.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan M J, Wong M T, Regnery R L, Jorgensen J H, Garcia M, Peters J, Drehner D. Syndrome of Rochalimaea henselae adenitis suggesting cat scratch disease. Ann Intern Med. 1993;118:331–336. doi: 10.7326/0003-4819-118-5-199303010-00002. [DOI] [PubMed] [Google Scholar]
- 7.Drancourt M, Birtles R, Chaumentin G, Vandenesch F, Etienne J, Raoult D. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet. 1996;347:441–443. doi: 10.1016/s0140-6736(96)90012-4. [DOI] [PubMed] [Google Scholar]
- 8.Drancourt M, Mainardi J L, Brouqui P, Vandenesch F, Carta A, Lehnert F, Etienne J, Goldstein F, Acar J, Raoult D. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med. 1995;332:419–423. doi: 10.1056/NEJM199502163320702. [DOI] [PubMed] [Google Scholar]
- 9.Drancourt M, Moal V, Brunet P, Dussol B, Berland Y, Raoult D. Bartonella (Rochalimaea) quintana infection in a seronegative hemodialyzed patient. J Clin Microbiol. 1996;34:1158–1160. doi: 10.1128/jcm.34.5.1158-1160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Droz S, Chi B, Horn E, Steigerwalt A G, Whitney A M, Brenner D J. Bartonella koehlerae sp. nov., isolated from cats. J Clin Microbiol. 1999;37:1117–1122. doi: 10.1128/jcm.37.4.1117-1122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupon M, Savin de Larclause A-M, Brouqui P, Drancourt M, Raoult D, De Mascarel A, Lacut J Y. Evaluation of serological response to Bartonella henselae, Bartonella quintana and Afipia felis antigens in 64 patients with suspected cat-scratch disease. Scand J Infect Dis. 1996;28:361–366. doi: 10.3109/00365549609037920. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Monoclonal antibodies, growing hybridomas. In: Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 139–282. [Google Scholar]
- 13.Heller R, Kubina M, Mariet P, Riegel P, Delacour G, Dehio C, Lamarque F, Kasten R, Boulouis H J, Monteil H, Chomel B, Piemont Y. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol. 1999;49:283–288. doi: 10.1099/00207713-49-1-283. [DOI] [PubMed] [Google Scholar]
- 14.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piémont Y. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 15.Jerris R C, Regnery R L. Will the real agent of cat-scratch disease please stand up? Annu Rev Microbiol. 1996;50:707–725. doi: 10.1146/annurev.micro.50.1.707. [DOI] [PubMed] [Google Scholar]
- 16.Knobloch J, Bialek R, Müller G, Asmus P. Common surface epitope of Bartonella bacilliformis and Chlamydia psittaci. Am J Trop Med Hyg. 1988;39:427–433. doi: 10.4269/ajtmh.1988.39.427. [DOI] [PubMed] [Google Scholar]
- 17.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kordick D L, Swaminathan B, Greene C E, Wilson K H, Whitney A M, O'Connor S, Hollis D G, Matar G M, Steigerwalt A G, Malcolm G B, Hayes P S, Hadfield T L, Breitschwerdt E B, Brenner D J. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol. 1996;46:704–709. doi: 10.1099/00207713-46-3-704. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Scola B, Raoult D. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998) J Clin Microbiol. 1999;37:1899–1905. doi: 10.1128/jcm.37.6.1899-1905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Z, Raoult D. Species-specific monoclonal antibodies for rapid identification of Bartonella quintana. Clin Diagn Lab Immunol. 2000;7:21–24. doi: 10.1128/cdli.7.1.21-24.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurin M, Birtles R J, Raoult D. Current knowledge of Bartonella species. Eur J Clin Microbiol Infect Dis. 1997;16:487–506. doi: 10.1007/BF01708232. [DOI] [PubMed] [Google Scholar]
- 24.Maurin M, Eb F, Etienne J, Raoult D. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J Clin Microbiol. 1997;35:2283–2287. doi: 10.1128/jcm.35.9.2283-2287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurin M, Raoult D. Bartonella (Rochalimaea) quintana infections. Clin Microbiol Rev. 1996;9:273–292. doi: 10.1128/cmr.9.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philip R N, Casper E A, Burgdorfer W, Gerloff R K, Hughes L E, Bell E J. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J Immunol. 1978;121:1961–1968. [PubMed] [Google Scholar]
- 27.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux V, Eykyn S J, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–1700. doi: 10.1128/jcm.38.4.1698-1700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartzman W A, Nesbit C A, Baron E J. Development and evaluation of a blood-free medium for determining growth curves and optimizing growth of Rochalimaea henselae. J Clin Microbiol. 1993;31:1882–1885. doi: 10.1128/jcm.31.7.1882-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varela G, Vinson J W, Molina-Pasquel C. Trench fever. II. Propagation of Rickettsia quintana on cell-free medium from the blood of two patients. Am J Trop Med Hyg. 1969;18:708–712. [PubMed] [Google Scholar]
- 31.Weiss E, Dasch G A, Woodman D R, Williams J C. Vole agent identified as a strain of the trench fever rickettsia, Rochalimaea quintana. Infect Immun. 1978;19:1013–1020. doi: 10.1128/iai.19.3.1013-1020.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch D F, Carroll K C, Hofmeister E K, Persing D H, Robison D A, Steigerwalt A G, Brenner D J. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]