SUMMARY
Mycobacterium tuberculosis (Mtb) infects 25% of the world’s population and causes tuberculosis (TB), which is a leading cause of death globally. A clear understanding of the dynamics of immune response at the cellular level is crucial to design better strategies to control TB. We use the single-cell RNA sequencing approach on lung lymphocytes derived from healthy and Mtb-infected mice. Our results show the enrichment of the type I IFN signature among the lymphoid cell clusters, as well as heat shock responses in natural killer (NK) cells from Mtb-infected mice lungs. We identify Ly6A as a lymphoid cell activation marker and validate its upregulation in activated lymphoid cells following infection. The cross-analysis of the type I IFN signature in human TB-infected peripheral blood samples further validates our results. These findings contribute toward understanding and characterizing the transcriptional parameters at a single-cell depth in a highly relevant and reproducible mouse model of TB.
Graphical Abstract
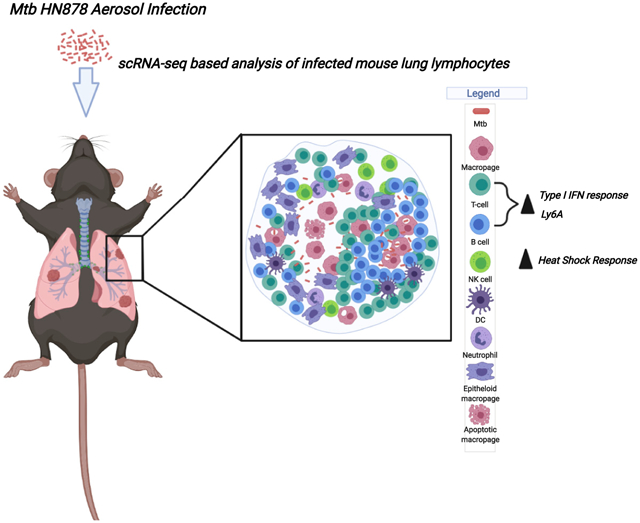
In brief
Akter et al. identify transcriptional signatures in Mtb-infected murine lungs using single-cell RNA sequencing. Lymphocytes show the enrichment of the type I IFN signature, while NK cells show the enrichment of heat shock responses in Mtb-infected lungs. Higher expression of Ly6A is observed on the surface of activated lymphocytes following Mtb infection.
INTRODUCTION
Mycobacterium tuberculosis (Mtb) infects almost 25% of the world’s population and causes tuberculosis (TB), which is a leading cause of death globally (World Health Organization, 2021). In most cases, infection leads to clinically asymptomatic latent TB infection (LTBI), but in 5%–10% of cases, it can cause active TB (ATB) disease. Despite the ongoing efforts to control TB, new challenges such as the increase in multi-drug resistance (MDR) TB, HIV-TB co-infection, and lack of effective vaccines impose roadblocks in controlling this disease (Sakamoto et al., 2019). To design better strategies to control TB, a clear understanding of the dynamics of immune response at cellular level is crucial.
Much of the work published has focused on specific cell types as central players in determining the outcome of TB infection. However, TB is a complex inflammatory disease with diverse cell types and interactions that lead to the generation of protective or pathological immune responses (Esaulova et al., 2021; de Martino et al., 2019). To understand the complex heterogeneity of this disease at unbiased single-cell depth, we have taken a single-cell RNA sequencing (scRNA-seq) approach on lung cells derived from healthy and Mtb-infected mice. The advantage of using high-throughput scRNA-seq techniques in understanding the immune interaction in complex inflammatory diseases is that we can study cell proportions, transcriptional heterogeneity, and activation status of cell subsets simultaneously (Hwang et al., 2018). Our analysis has identified the enrichment of type I interferon (IFN) signatures among the lymphoid cell clusters, as well as heat shock responses in natural killer (NK) cells from Mtb-infected mice lungs. We also identified lymphocyte antigen 6 complex, locus A (Ly6A) as a lymphoid cell activation marker and validated its upregulation in activated lymphoid cells following Mtb infection. To further identify cross-species correlates, we compared gene signatures from human TB whole-blood transcriptional profiles (Berry et al., 2010) with differential gene expression profiles generated from mouse immune cells by scRNA-seq. In concordance with the human transcriptional profiles, we found an enrichment of type I IFN signature in both TB-infected human peripheral blood samples and murine lung lymphoid cell immune clusters. Collectively, our study characterizes the transcriptional parameters at single-cell depth in the mouse model of TB, a highly relevant and reproducible model. These results will enable a better understanding of the immune cell landscape in rigorous animal models and can improve our fundamental understanding of the immunology of TB relevant for testing of TB vaccines and therapeutics.
RESULTS
Single-cell transcriptional profiling of lymphoid cells in the lung during Mtb infection
Lymphocytes are critical for Mtb control (Cooper and Khader, 2008). A recent study has examined the role of murine macrophage cell populations in the lung during Mtb infection at a single-cell level using an intranasal high dose of Erdman strain of Mtb (Pisu et al., 2021). However, thus far, the single-cell landscape of the lymphoid cells following more physiologically pertinent, low-dose aerosol Mtb (HN878) infection in the highly relevant and widely used mouse model has not been delineated. To characterize the lung lymphocyte landscape of mice infected with low-dose Mtb, we performed 10× scRNA-seq on single cells from the lungs of control mice (uninfected, n = 2), 50 days post-infection (dpi) (D50 Inf, n = 3), and 100 dpi (D100 Inf, n = 3) (Figure 1A). Following the single-cell alignment and quality control process, we found a total of 36,607 cells (3,215–5,211 cells per sample) that are high quality and comparable across the samples. The average number of cells per sample at 100 dpi (5,080) was slightly higher than the numbers at 50 dpi (4,933) and in uninfected (3,283) samples. We found that most of the cells were lymphoid cells due to the prior purification for live cells (Figures 1B and S1), which resulted in the enrichment of lymphoid populations to ~96% of the total cells; only 2.7% were myeloid and 0.82% were non-immune cells. The inclusion of live cell purification is especially efficient at the enrichment of lymphocytes in the lung single-cell suspensions (Figure S1A), shown by improved enrichment of CD3+ T cells, including CD4+ and CD8+ T cells (Figures S1B–S1D). Furthermore, the enrichment of the lymphoid population in the lung samples coincided with a decrease in several myeloid cell populations, including CD11c+, CD11b+, and CD11b+CD11c+ subsets (Figures S1E–S1H). Therefore, we focused on characterizing the single-cell transcriptomes of the detected lymphoid cell subsets in the lung following Mtb infection.
Figure 1. scRNA-seq transcriptional profiling of Mtb-infected murine lungs.
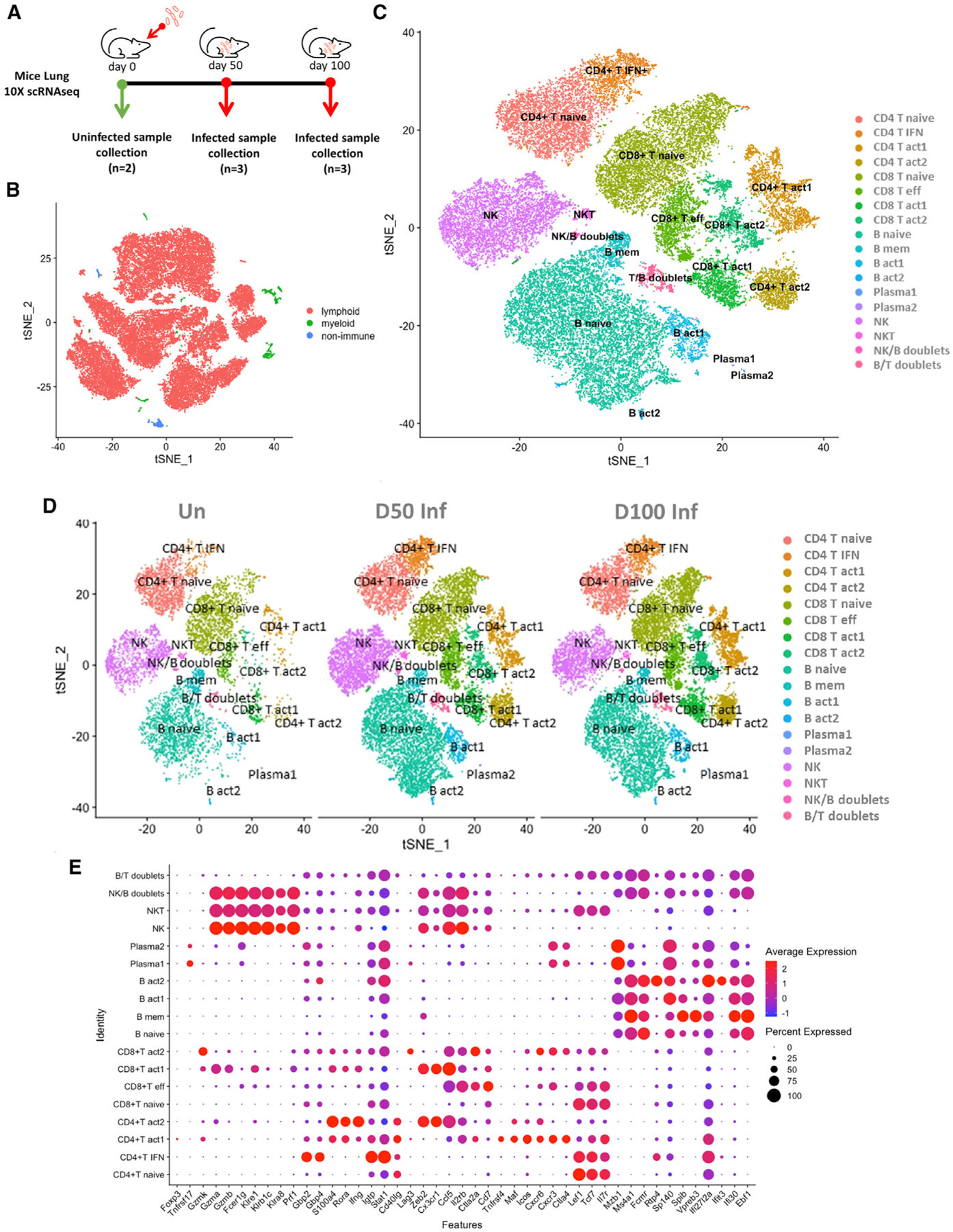
C57BL/6 mice were aerosol infected with Mtb HN878 and lungs were harvested at different dpi.
(A) Study design using 10x Genomics platform. For this study, single-cell suspensions from lungs were derived from mice at 0, 50, and 100 dpi and subjected to downstream analysis as described in the Method details.
(B) t-Distributed stochastic neighbor embedding (tSNE) visualization of the major cell types representing non-immune/lymphoid/myeloid cells in the dataset (all conditions together).
(C) tSNE plot of cell subtypes after re-clustering the lymphoid cells, colored according to cellular identity (all conditions together).
(D) tSNE plot of cell subtypes after re-clustering the lymphoid cells, colored according to cellular identity, split by condition.
(E) Dot plot indicating expression of key genes detected for each cell subtype. The dot color represents the expression level, and the dot size represents the percentage of cells in each cluster expressing a particular gene.
See also Figures S1 and S2.
Based on canonical gene expression, we identified three major types of lymphoid cells, namely, T cells (CD3d+), NK cells (Ncr1+Gzmb+), and B cells (CD79a+) (Figure S2). T cells were consistently the most abundant cell type (48%–55.9%) in all conditions, followed by B cells (27.8%–31.2%), NK cells (10.3%–16.7%), and NKT cells (CD3d+Ncr1+Gzmb+) (0.25%–0.54%) (Figure 2A). Re-clustering of the lymphoid cells yielded 18 sub-populations: 8 clusters of T cells, 4 B cell clusters, 2 clusters of plasma cells, 1 NK cell cluster, 1 NKT cluster, 1 T/B doublet, and 1 NK/B doublet cluster (Figures 1C and 1D). Identification of the cell subtypes of each cluster was based on the expression of known markers (Figure S2) and the canonical marker genes detected in each cluster (Figure 1E).
Activated T and B cell subsets accumulate in the lungs of Mtb-infected mice
Based on the enrichment of corresponding transcriptional signatures, T cell populations were defined as naive, memory, effector, and activated subpopulations (Figures 1C and 1D). Based on the expression and distribution of known and canonical T cell markers, we found 4 subtypes of CD4+ T cells (CD3e+ CD4+) and 4 subtypes of CD8+ T cells (CD3e+ CD8+). Among the 4 subtypes of CD4+ T cell clusters, 1 naive CD4+ (CD4+ T naive) T cell (Ccr7+ Sell+) cluster, 1 CD4+ IFN-responsive (CD4+ T IFN+) T cell (Igtp+ Stat1+) cluster, and 2 activated CD4+ (CD4+ T act1, CD4+ T act2) T cell (Ccr7− Sell− Ifng+ Rora+) clusters were present. The CD4+ T IFN+ cluster expressed classic IFN-inducible genes such as IFN-α-inducible protein 27-like 2A (Ifi27l2a), IFN-γ-induced GTPase (Igtp), signal transducer and activator of transcription 1-α/β (Stat1), and guanylate binding proteins (Gbp2, Gbp4) but low expression of genes associated with cytotoxicity (Figure 1E). Ifi27l2a is known to be expressed in lymphocytes in response to IFN during Mtb infection (Kang et al., 2011; María Irene et al., 2021), and Igtp and Stat1 are shown to be upregulated during Mtb infection through the IFN signaling pathway (Ehrt et al., 2001; Yi et al., 2020). GBPs are IFN-inducible proteins that execute distinct host defense activities against a wide variety of microbial pathogens, including Mtb (Tretina et al., 2019). The CD4+ act1 cluster harbored a relatively higher expression of genes associated with cytotoxic functions, including cytotoxic T lymphocyte-associated proteins (Ctla4 and Ctla2a), and chemokines, including chemokine (C-X-C motif) receptors (Cxcr3 and Cxcr6) (Figure 1E). The CD4+ T act2 cluster expressed high levels of IFN-γ (Ifng); zinc finger E-box binding homeobox 2 (Zeb2), which is associated with the terminal differentiation of T cells (Dominguez et al., 2015); S100a4 and interleukin-18 (IL-18) receptor accessory protein (Il18rap), components of the cytokine network that play a central role in protecting against Mtb infection (Robinson et al., 2012); integrin beta 1 (Itgb1); C-X3-C motif chemokine receptor 1 (Cx3cr1); and lectin-galactose binding soluble 1 (Lgals1)—genes that are involved in the migration of lymphocytes in response to infection and representing memory T cells. However, the expression of memory T cell-associated genes such as Cx3cr1 and S100a4 indicate representation of memory T cell-like features within this subset.
The four subtypes of CD8+ T cell clusters included a naive CD8+ T cell (CD8+ T naive expressing Ccr7+ Sell+), an effector CD8+ T cell (CD8+ T effector expressing CD44+ Cxcr3+ Ctla2a+), and two activated CD8+ T cells (CD8+ T act 1, CD8+ T act2, both expressing Ccr7− Sell− Ifng+ Rora+) clusters (Figures 1E and S2). The CD8+ T naive cluster is enriched with death-associated protein-like-1 (Dapl1), IL-7 receptor (Il7r), and T cell-specific transcription factor 7 (Tcf7). The CD8+ T effector cluster has a higher expression of chemokines (Cxcr3, Xcl1), Cd7, Ctla2a, Ctsw, IL-2 receptor-β chain (Il2rb), killer cell lectin-like receptors (Klra6, Klrd1), and lymphocyte antigen 6 complex-locus C2 (Ly6c2). The CD8+ T act1 cluster revealed a higher expression of Zeb2, along with Cx3cr1 and chemokine ligand 5 (Ccl5) genes, associated with the trafficking of immune cells during Mtb infection (Hall et al., 2009; Vesosky et al., 2010), whereas the CD8+ T act2 cluster exhibited a relatively higher expression of granzyme K (Gzmk), which is specifically induced in CD8+ T cells during the late stages of infection (Russell et al., 2019), as well as Ctla2a, chemokine receptors (Cxcr3 and Cxcr6), and lymphocyte activation gene 3 (Lag3).
To characterize changes in individual T cell subsets across three conditions, we further analyzed the eight T cell clusters. Naive CD4+/CD8+ T cells were the largest represented subtype of T cell population across all conditions (Figure 2B). The frequency of naive T cells was, however, the greatest in uninfected mice, in which, on average, approximately 36.5% and 39%, respectively, of the total CD4+ and CD8+ T cells were naive cells. In contrast, only fewer naive T cells—approximately 19.2% (CD4+) and 27.4% (CD8+)—were present in the infected lungs at 50 dpi. These numbers further decreased to 15.5% (CD4+) and 26.1% (CD8+) of the total T cells in the infected lungs at 100 dpi mice. Interestingly, all of the other types of T cells were lowest in the uninfected mice lungs compared to the infected ones, ranging from 1.4% to 5% in the CD4+ T clusters and 1.8%–7% in the CD8+ T clusters. In the CD4+ T IFN+ cluster, this subset increased from 3.9% of the total T cells in the uninfected mice to 10% in the Mtb-infected lungs at 50 dpi, then decreased to 8% in the Mtb-infected lungs at 100 dpi. In contrast, during the progression of the infection and disease, more activated cells accumulated within the Mtb-infected lungs when compared to uninfected lungs. In the CD4+ T act1 cluster, we identified the accumulation of cells during infection, resulting from 5% in the uninfected lungs to increases between 10.6% and 14.8% in the infected lungs at 50 and 100 dpi, respectively. There was also a large accumulation of cells in the CD4+ T act2 cluster, from 1.4% in the uninfected lungs to the 7.7% in the infected lungs at both 50 and 100 dpi.
Figure 2. Lymphoid clusters accumulate in the lungs during acute and chronic Mtb infection.
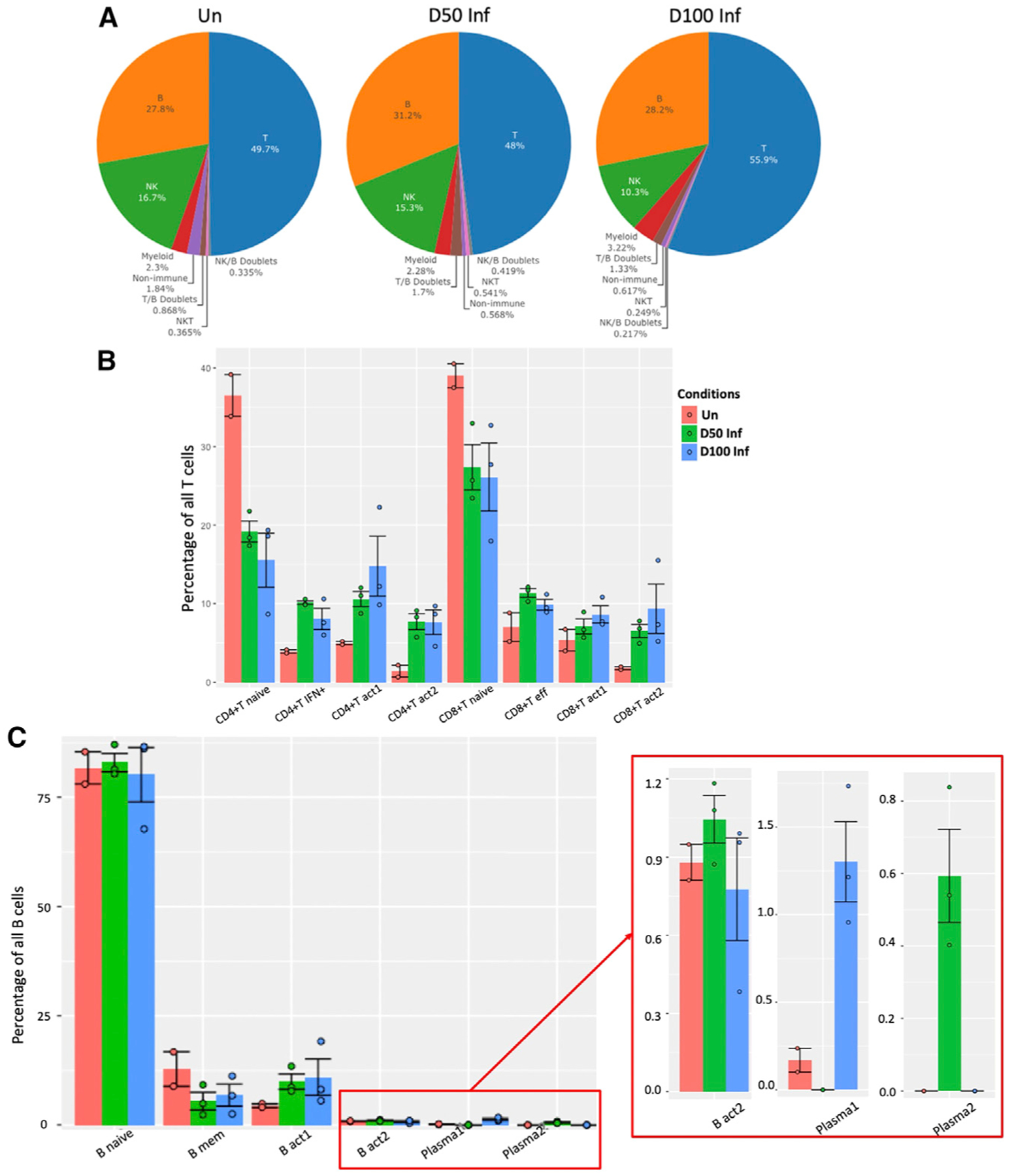
(A) Pie charts representing the distribution of identified lymphoid cell types across conditions (0, 50, and 100 dpi).
(B) Distribution of T cell subsets by cluster per condition per sample.
(C) Distribution of B cells by cluster per condition per sample. Error bars are based on standard error of the mean. Wilcoxon rank-sum exact test was used to test whether the differences are significant among conditions in each cluster.
Un, n = 2; D50 Inf, n = 3; D100 Inf, n = 3.
*p < 0.05, **p < 0.01, ***p < 0.001.
In the CD8+ T effector cluster, we observed an accumulation of cells from 7% of the total T cells in the uninfected lungs to 11.4% in the Mtb-infected lung at 50 dpi, which decreased to 9.9% at 100 dpi. In contrast, cell accumulations were observed in both CD8+ activated clusters during the progression of the infection and disease. In CD8+ T act1, there was an increased number of cells, from 5.4% in the uninfected lungs to 7.1%–8.6% in the Mtb-infected lungs at 50 and 100 dpi, respectively. However, a substantial increase was observed in the CD8+ T act2 cluster of Mtb-infected lungs, from 1.8% in the uninfected lung to 6.5%–9.4% in the infected mice lungs at 50 and 100 dpi, respectively. Our recent macaque study also found an accumulation of activated CD4+ and CD8+ T cells in the Mtb-infected lungs with ATB (Esaulova et al., 2021). These results together suggest that while the infected lungs still maintain naive T cell subsets, a key feature of the infection process and disease progression is the accumulation of activated T cells in the lung.
Based on the expression of canonical B cell markers, the B cell populations were defined into 4 B cell clusters (CD79a+ Ms4a1+) and 2 plasma cell clusters (CD79a+ Mzb1+) (Figures 1E and S2). The naive B cell (B naive) cluster was enriched with the expression of the Fc fragment of the immunoglobulin M (IgM) receptor (Fcmr), associated with antigen-driven immune responses (Choi et al., 2013), and major histocompatibility complex class II (MHC class II)-related transcripts such as H2-Aa, H2-Ab1, H2-Eb1, H2-Dmb2, and Cd74. The memory B cell cluster (B mem) exhibited a higher expression of Ms4a1, pre-B lymphocyte gene 3 (Vpreb3), and Spi-B transcription factor (Spib) genes, which are associated with B cell maturation (Figure 1E). Of the two activated B cell clusters, B act1 was enriched with the IFN-γ inducible gene IFN-γ-inducible protein 30 (Ifi30) (Gonzalez-Juarrero et al., 2009), B cell maturation marker cluster of differentiation 24a (Cd24a), and Sp140 nuclear body protein (Sp140), whereas B act2 was enriched with the higher expression of IFN-stimulated genes (ISGs), IFN-induced protein with tetratricopeptide repeats 3 (Ifit3), receptor transporter protein 4 (Rtp4), and Ifi27l2a. The major population of B cells (80%–83%), which was predominant across all conditions, was the naive subset (Figure 2C). In the B mem, we observed that cells decreased from 12.8% of the total B cells in uninfected lungs to 5.5% in Mtb-infected lungs at 50 dpi, followed by a slight increase to 6.8% in the Mtb-infected lung at 100 dpi. In contrast, activated B cell accumulation in the Mtb-infected lungs during infection and disease progression was comparable. In the B act1 cluster, we identified an increased percentage of cells from 4.5% in the uninfected lungs to 9.9%–10.9% in Mtb-infected lungs at 50 and 100 dpi, respectively. In the B act2 cluster, we observed an increased number of cells, from 0.88% in the uninfected lungs to 1.04% in the Mtb-infected lungs at 50 dpi, followed by a decrease to 0.77% in the Mtb-infected lungs at 100 dpi. These results suggest that like the T cells, naive B cells are well represented in the Mtb-infected lung. Furthermore, activated B cells also accumulate in the lungs during Mtb infection and disease progression.
Lymphoid cell clusters reflect a type I IFN signature following Mtb infection
To further investigate the differential transcriptomic changes in the lymphoid cells after Mtb infection, we compared the gene expression of lymphoid cell clusters in uninfected lungs with cells from Mtb-infected lungs at 50 and 100 dpi. Differential analysis identified several genes with consistently higher expression in different clusters of lymphoid cells in infected lungs, including Ly6A, Stat1, Igtp, Gbp2, and Ifng (Figures 3A–3C). Expression of Stat1, Igtp, Gbp2, and Ifng genes is regulated through the IFN pathway, and the Ly6A gene is expressed on the surface of hematopoietic stem cells as well as on activated T cells. Among these, the higher expression of Ly6A in lymphoid cells was not previously reported as related to Mtb infection. We also found that the differentially expressed genes (DEGs) that are upregulated in several lymphoid clusters in Mtb-infected lungs are involved in pathways targeting IFN and IL responses (Figures 4A–4C). Importantly, a higher expression of both the IFN and IL responses was observed in 3 out of 4 clusters of CD4+ T cells, 2 out of 4 clusters of CD8+ T cells, and 1 out of 4 clusters of B cells in the Mtb-infected lungs.
Figure 3. Transcriptional analysis of lymphoid cell clusters.
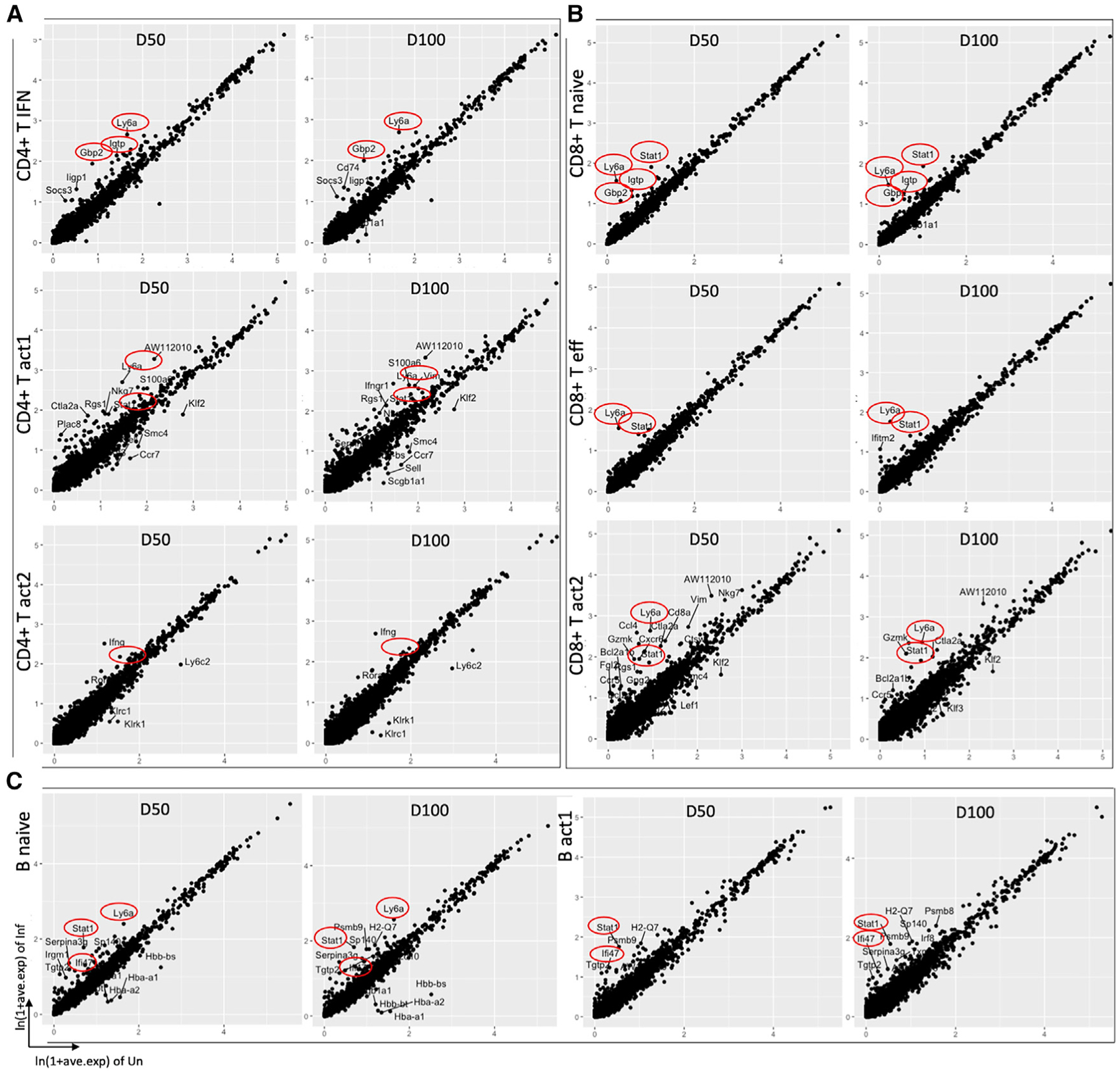
Scatterplots depicting the comparison of average expression of genes in infected (50/100 dpi) versus uninfected samples in (A) CD4+ T cell clusters (B) CD8+ T cell clusters, and (C) B cell clusters, highlighting the top DEGs (average logFC ≥ |±1|, adjusted p ≤ 0.05). IFN-related genes and Ly6A are highlighted in red circle. Each dot represents a gene.
Figure 4. IFN-related pathways are overrepresented in lymphoid cell clusters in Mtb-infected mice.
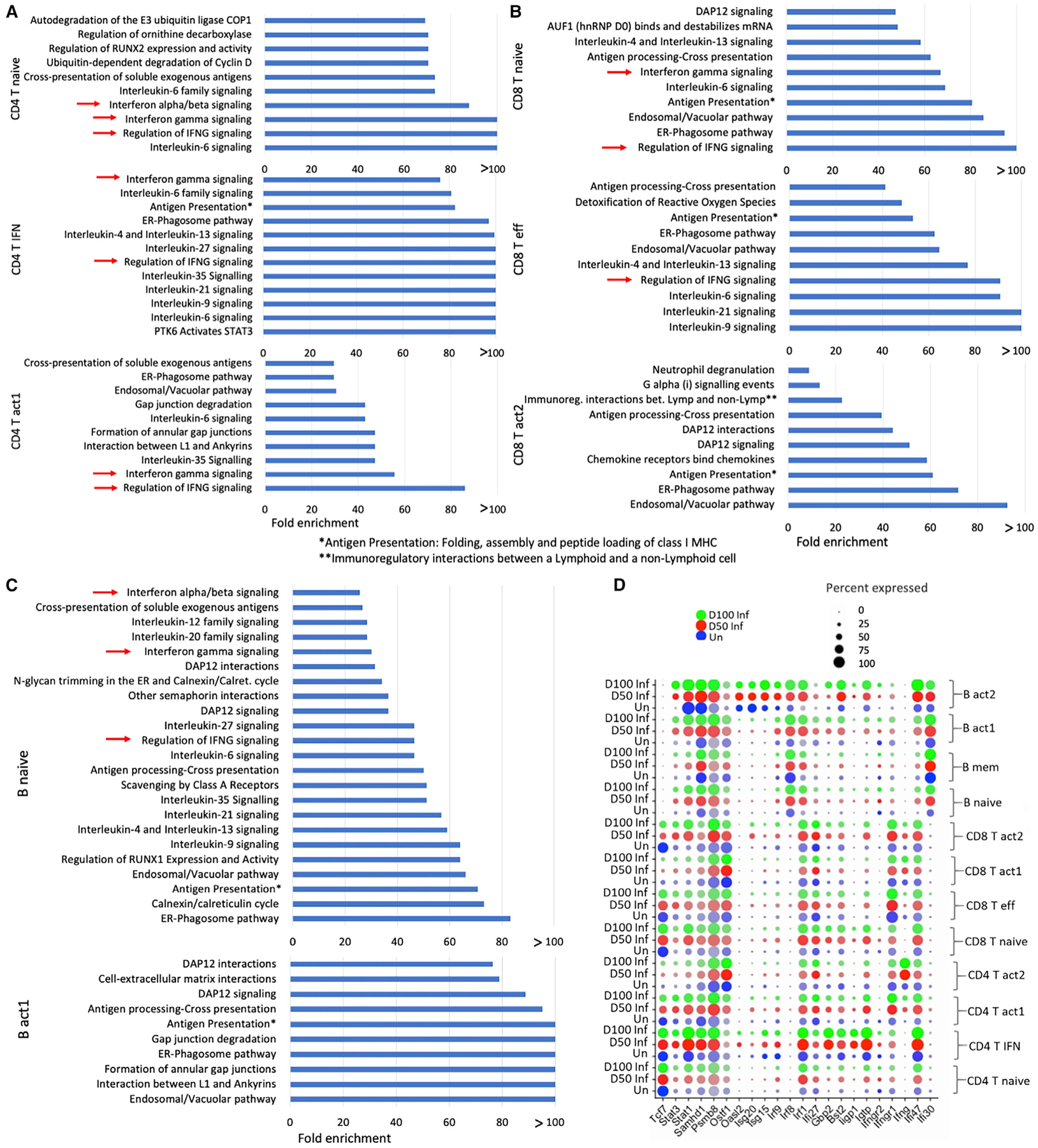
(A–C) Statistical overrepresentation test was performed with reactome pathways as annotation set. Top overrepresented pathways in Mtb-infected mice lungs at 50 dpi with respective fold enrichment in (A) CD4+ T cell clusters, (B) CD8+ T cell clusters, and (C) B cell clusters. False discovery rate (FDR) <0.05.
(D) Average expression of several IFN-related genes that were differentially expressed in respective lymphoid cell clusters. The dot size represents the percentage of cells in each cluster expressing a particular gene.
Among the CD4+ T cell clusters, we found higher numbers of DEGs in the CD4+ T act1 cluster following infection at both 50 and 100 dpi (158 and 207 DEGs, respectively) when compared to uninfected lungs. Approximately 23 of these genes are common with the DEGs of CD4+ T naive cells and 28 genes are common with DEGs of CD4+ T IFN+ cells while comparing infected lungs at 50 dpi versus uninfected mice. In comparison with Mtb-infected cells at 100 dpi versus uninfected cells, approximately 35 of these DEGs are common with CD4+ T naive cells and 28 are common with the CD4+ T IFN+ cluster (Table S1). Ly6A, Igtp, Gbp2, Socs3, Stat1, Ifi47, Iigp, and Irf1, genes that associate with the type I IFN signaling pathway, are among the top upregulated genes in the CD4+ T IFN+ cluster (Figures 3A and 4D). Ly6A; cathepsin W (Ctsw); IFN-associated genes Stat1, Igtp, Ifng, Ifngr1, and Ifi47; suppressor of cytokine signaling 3 (Socs3); Gbp2; cytotoxic genes Ctla2a and Gzmk; inducible T cell co-stimulator (Icos); Cxcr6; and regulator of G protein signaling 1 (Rgs1) are the top upregulated genes in the CD4+ T act1 cluster. Ifng and Rora are upregulated in the CD4+ T act2 cluster in Mtb-infected mice (Figures 3A and 4D). Pathway enrichment analysis using the upregulated DEGs in Mtb-infected lymphoid cells at 50 and 100 dpi revealed many important pathways related to TB disease, namely, IFN-γ signaling, IL-6 signaling, and regulation of IFN-γ signaling, as consistently overrepresented pathways in all 3 clusters: CD4+ T naive, CD4+ T IFN+, and CD4+ T act1 (Figure 4A). In addition, we identified several IL pathways signaling (IL-6, −35, −9, −21, −4, and −13), PTK6 activates STAT3, antigen presentation: folding, assembly, and peptide loading of MHC class I, and endoplasmic reticulum (ER)-phagosome pathway in the CD4+ T IFN+ cluster; IFN α/β signaling in the CD4+ T naive cluster; and IL-35 signaling and ER-phagosome pathway in the CD4+ T act1 cluster, which are among the top overrepresented pathways found in the Mtb-infected lungs compared to uninfected lungs.
While investigating the transcriptional changes after the infection in different CD8+ T cell clusters, we identified a higher number of genes that were differentially expressed in the CD8+ T naive cluster: 113 DEGs in infected lungs at 50 dpi and 117 DEGs in infected lungs at 100 dpi. Ly6A and several IFN-related genes, including Stat1, Igtp, Ifi47, Iigtp1, Socs3, and Gbp2 are among the top upregulated genes in infected mice (Figures 3B and 4D). Approximately 43 of these DEGs of the CD8+ T naive cluster are common with the DEGs of CD8+ T eff cluster and 20 DEGs of the naive cluster are common with the DEGs of CD8+ T act2 while comparing infected lungs at 50 dpi versus uninfected mice. In comparison with Mtb-infected cells at 100 dpi versus uninfected, 29 DEGs of the CD8+ T naive cluster are common with the DEGs of the CD8+ T eff cluster and 20 DEGs are common with CD8+ T act2 (Table S2). Genes such as Ly6A, Ctla2a, Cxcr6, Stat1, Gzmk, and Ccl4, which code for effector molecules, were the top highly expressed genes in Mtb-infected mice. In the CD8+ T act1 cluster, we found few DEGs, and their average log fold changes (FCs) were low (less than |±0.60|). In the CD8+ T act2 cluster, Ly6A, Stat1, Ctla2a, Gzmk, Nkg7, Cxcr6, Rgs1, Ccr5, and Ccl4 are among the top upregulated genes in Mtb-infected lungs compared to uninfected lungs. We identified IL-6 signaling, IL-4 and IL-13 signaling, and regulation of IFN-γ signaling among the top overrepresented pathways in CD8+ T naive and CD8+T eff clusters (Figure 4B). ER-phagosome pathway, endosomal/vacuolar pathway, antigen presentation: folding, assembly, and peptide loading of MHC class I, antigen processing-cross presentation, and DAP12 signaling are common top enriched pathways among these 3 CD8+ T clusters (CD8+ T naive, CD8+ eff, CD8+ T act2) using their respective upregulated DEGs in Mtb-infected lungs. Together, these results suggest type I IFN responses of both CD4+ T and CD8+T cells in the lungs of Mtb-infected mice.
IFN and IL responses are expressed in B cells in Mtb-infected mice lungs
In the B cell clusters, we identified more than 200 DEGs in the B naive clusters in both comparisons, namely, cells from infected lungs at 50 dpi versus uninfected and cells from infected lungs at 100 dpi versus uninfected. Irf1, AW112010, Pcbp2, Stat1, H2-K1, and H2-Q7 are the 6 DEGs (infected 50 dpi versus uninfected) that are common across the 3 clusters of B cells: B naive, B act1, and B mem. Ly6A, Stat1, Serpina3g, and Sp140 in B naive, Stat1, Ifi47, H2-Q7, Psmb8, and Psmb9 in B act1, and Stat1, Irf1, and H2-Q7 in B mem, are among the top upregulated DEGs in infected lungs (Figures 3C, 4D, and S3A). Statistical overrepresentation tests were carried out with the upregulated DEGs in infected lungs in different B cell clusters. ER-phagosome pathway, antigen presentation: folding, assembly, and peptide loading of MHC class I, endosomal/vacuolar pathway, and DAP12 signaling are among the top overrepresented pathways in B naive, B mem, and B act1 clusters (Figures 4C and S3B). In addition, in the B naive cluster, we observed several IFN responses such as regulation of IFN-γ signaling, IFN-γ signaling, and IFN-α/β signaling, and several IL signaling pathways (IL-9, −4, −13, −6, −12, −20, −21, −27, and −35 signaling) (Figure 4C). Therefore, in addition to T cells, robust type I IFN responses were observed in B cells in the lungs during Mtb infection and TB disease progression.
NK cell reduction occurs in the lungs of mice with chronic Mtb infection
We identified a cluster of NK cells that expressed classic NK cytotoxic genes such as perforin 1 (Prf1), natural cytotoxicity triggering receptor 1 (Ncr1), granzymes (Gzma and Gzmb), and Fc fragment of IgE receptor Ig (Fcer1g) (Figures 1E and S2) that induce downstream signaling, leading to the induction of T helper 2 cell (Th2) responses. In addition, several killer cell lectin-like receptors such as Klra8, Klrb1c, and Klre1 are also expressed this cluster (Figure 1E). We observed a steady decrease in NK cells from 16.7% of the total cells in uninfected mice lungs to 15.3% and 10.3% of the total cells in Mtb-infected lungs at 50 and 100 dpi, respectively (Figure 2A). Taken together, these results suggest a reduction of cytotoxic NK cells in the lung during chronic Mtb infection and TB disease progression. These results are consistent with recent studies from us and others that NK cell accumulation decreased during ATB disease (Esaulova et al., 2021, Roy Chowdhury et al., 2018).
Heat shock responses are expressed in NK cell clusters in Mtb-infected mice lungs
We performed differential analysis in the NK cell cluster to compare Mtb-infected mice lungs versus uninfected lungs. We identified 68 genes to be differentially expressed (13 were upregulated) in Mtb-infected lungs at 50 dpi and 92 genes to be differentially expressed (18 were upregulated) in Mtb-infected lungs at 100 dpi while comparing the expression pattern with uninfected lungs. Heat shock proteins (Hspa1a and Hspa1b), granzymes (Gzma and Gzmb), S100a6, Klrg1, and Cx3cr1 were observed to be upregulated in the Mtb-infected lungs at 50 dpi (Figure S3C). Although we could not identify any overrepresented pathways with the upregulated DEGs of Mtb-infected lungs at 50 dpi versus uninfected, we found HSF1-dependent transactivation, cellular response to heat stress, and regulation of HSF1-mediated heat shock response among the overrepresented pathways in Mtb-infected lungs at 100 dpi when compared with uninfected lungs (Figure S3D). Together, these results suggest that heat shock responses are expressed in lung NK cell clusters in the chronic phase of Mtb infection.
Upregulation of Stat1 and Ly6A expression in activated lymphoid cells in Mtb-infected lungs
We next validated our scRNA-seq results that identified Stat1 gene upregulation, a primary component of the IFN signaling pathway, in lymphoid cell clusters following Mtb infection (both at 50 and 100 dpi) (Figure 3). We used the Stat1 mRNA probe to detect mRNA expression within Mtb-infected lung granuloma. We found robust induction of Stat1 mRNA expression within lymphoid clusters in lung granulomas of infected mice (Figure 5A). Furthermore, our transcriptomic analysis also identified the induction of the Ly6A (lymphocyte antigen 6A) gene in lymphoid clusters following Mtb infection (Figure 3). Ly6A is a glycosyl phosphatidylinositol anchored molecule that works as a cell adhesion and cell signaling component and is expressed on the surface of activated lymphocytes (Stanford et al., 1997). Therefore, we quantitated the expression of Ly6A on different lymphoid cell populations in uninfected and Mtb-infected lungs (50 and 100 dpi). We found that Ly6A expression is increased on CD3+ T cells following Mtb infection, and was well reflected in both CD4+ and CD8+ T cell subsets, especially within CD44hiCD69+ activated T cells expressing IFN-γ. Thus, our results validated the expression of Ly6A as a marker of activation on both CD4+ and CD8+ T cells following Mtb infection (Figure 5B).
Figure 5. Induction of Stat1 and Ly6A expression in activated lymphoid cells in Mtb-infected lungs.
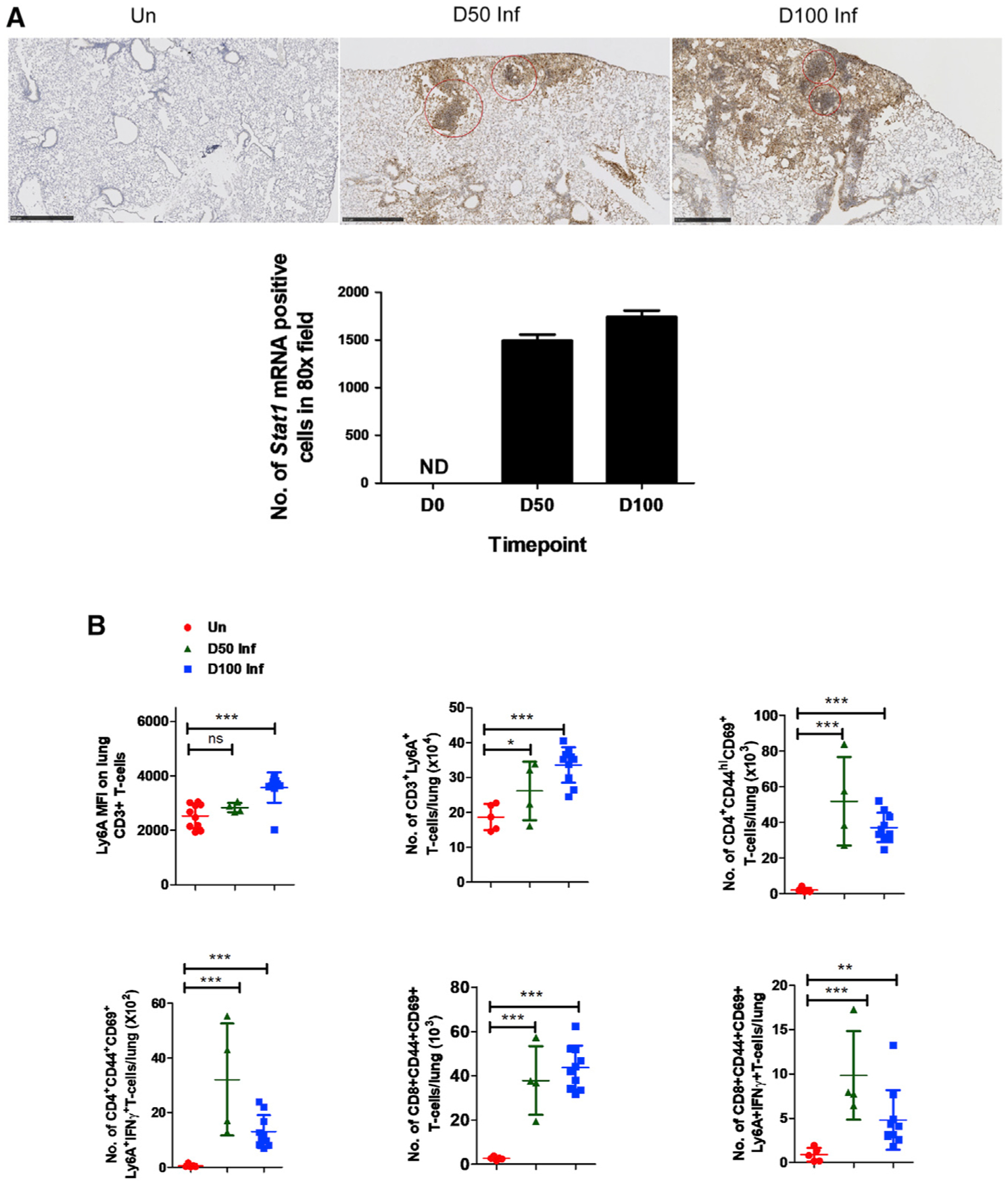
C57BL/6 mice were aerosol infected with Mtb HN878 and lungs were harvested at 0, 50, and 100 dpi.
(A) Histological analysis from paraffin-embedded sections from lungs of Mtb-infected mice showing induction of Stat1 mRNA using ISH (10×, upper panel) and its quantitations as number of Stat1 mRNA+ cells in 80× field (lower panel). ND, not detected. Red circles encompass regions with dense expression of Stat1 mRNA. Scale bar, 500 μM.
(B) Ly6A expression measured using flow cytometry among different effector lymphoid cell populations at 0, 50, and 100 dpi, as predicted by scRNA-seq analysis. Data points represent the mean ± SD values from 4 to 10 mice; significance assessed by Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
Cross-species expression of IFN signatures in murine and human TB
A previous study has found that whole-blood transcriptional signatures of TB are dominated by IFN-γ and type I IFN-α/β signaling in human whole-blood samples (Berry et al., 2010). Therefore, we compared the 393 signatures genes of ATB from that study (Berry et al., 2010) with our differential gene list in different lymphoid clusters to assess cross-species correlates of TB. We found that several genes are common across species—19 genes in CD4+ T clusters, 28 genes in CD8+ T clusters, and 18 genes in B cell clusters (Figure 6). The most common genes across all of the clusters are Stat1, Gbp2, Gbp4, Psme1, Psme2, Psmb9, Tap1, Tap2, and Zbp1. Moreover, in the whole-blood samples of ATB patients, the IFN-inducible transcripts were overexpressed in purified blood neutrophils only, not in CD4+ or CD8+ T cells while compared with healthy controls. However, in our study we found IFN signatures overexpressed in the CD4+ T and CD8+ T and B cells in the Mtb-infected mice lungs compared to the uninfected mice. These results suggest that while type I IFN responses in human peripheral blood may be well reflected within neutrophils, at the site of infection in the murine infected lungs, the type I IFN responses may also be induced within lymphoid clusters.
Figure 6. Type I IFN transcriptomic signatures are conserved across species post-Mtb infection in different clusters.
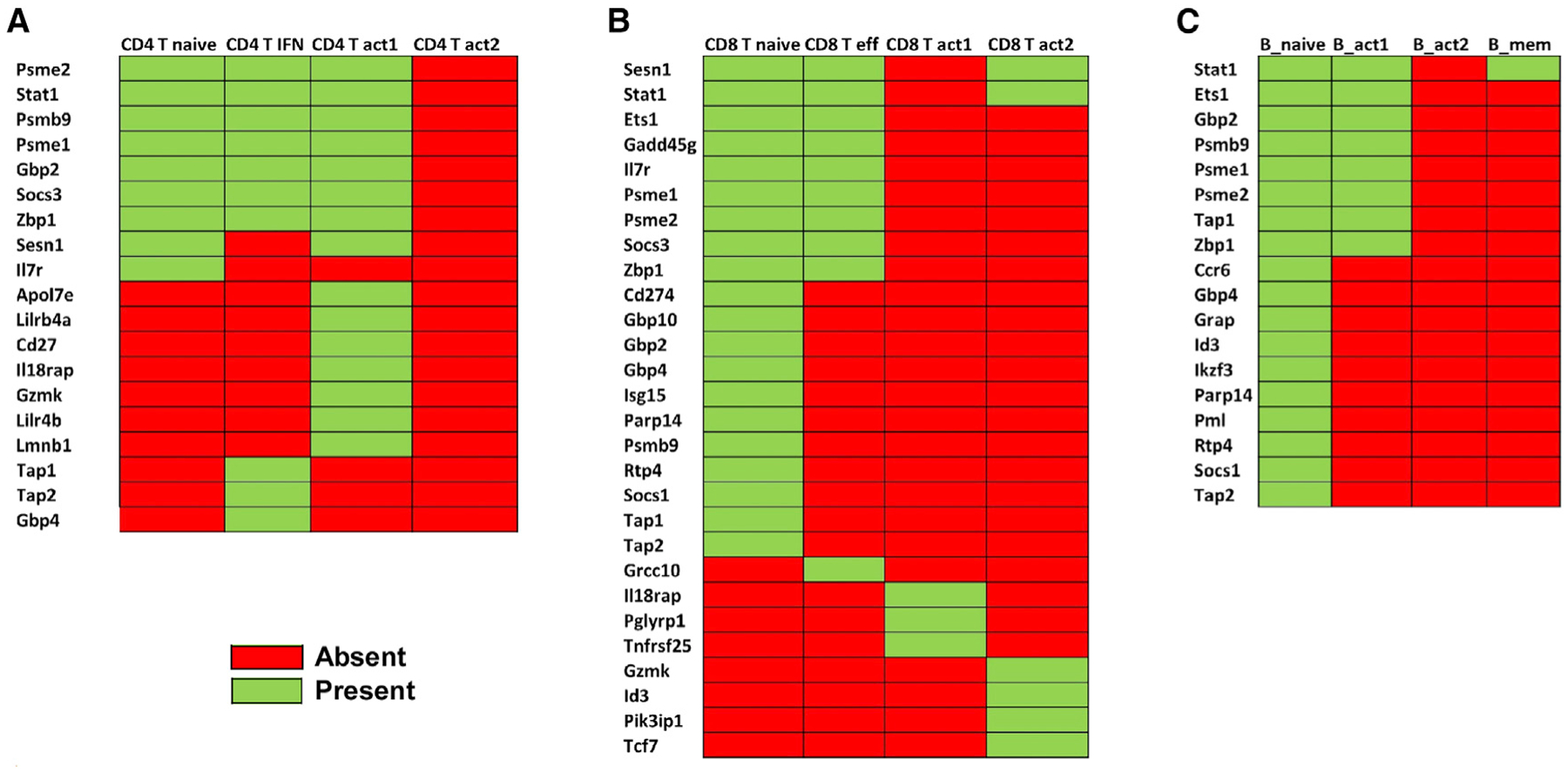
Our result from scRNA-seq data from Mtb-infected lung lymphoid clusters (this study) was compared with human whole-blood data from a published study (Berry et al., 2010) to identify DEGs shared among (A) CD4+ T cell clusters (B) CD8+ T cell clusters, and (C) B cell clusters. Green represents common in both datasets; red represents not found in respective cluster.
IFN-γ signaling in T cells is overrepresented in Bacille Calmette-Guérin (BCG)-vaccinated Mtb-infected lung lymphocytes
BCG vaccination induces accelerated lung lymphoid immune responses when compared to unvaccinated lungs (Gopal et al., 2012). To further delineate the early lymphoid cell changes in the Mtb-infected lungs of BCG vaccinated hosts, we carried out scRNA-seq of lung single cells from BCG-vaccinated Mtb-infected mice at the peak of the response (at 15 dpi, BCG, n = 2) and compared these responses to the peak of the lymphoid response in the lungs of Mtb-infected mice at 20 dpi (D20 Inf, n = 2). We chose D20 as this is the peak of the T cell responses in unvaccinated Mtb-infected lung (Gopal et al., 2012). The previous cluster analysis on this CD3+ scRNA-seq dataset found 8 clusters of T cells: 1 CD4+ T naive, 2 clusters of effector cells (CD4+ T eff1 CD4+ T eff2), 1 T regulatory (Treg), 1 CD8+ T naive, 1 cytotoxic CD8+ (CD8+ T cytotoxic), a mix of CD8+ and γδ T cells (CD8+ T and Tγδ), and an enriched γδ T cell cluster (Tγδ) (Das et al., 2021). We investigated the transcriptomic changes in different T cell clusters at the single-cell level. In both the CD4+ T naive and CD8+ T naive clusters, Ly6A, Stat1, Igtp, Gbp2, and Tgtp1 are among the top upregulated DEGs in the BCG-vaccinated Mtb-infected mice, when compared with the expression in early Mtb infection. In the CD4+ T naive cluster, more than 70% of the cells expressed Ly6a and Stat1 with an average FC of 1.8 in BCG-vaccinated Mtb-infected mice compared to 47% (Ly6a) and 52% (Stat1) cells in the unvaccinated mice (Figure 7A). In the CD8+ T naive cluster, 60% of the cells expressed Ly6a (average FC 2.2) in BCG-vaccinated mice compared to 18% cells in unvaccinated mice, while 80% cells expressed Stat1 (average FC 1.9) in BCG-vaccinated mice compared to 56% cells in the unvaccinated mice. Pathway enrichment analysis was carried out with the DEGs separately for each cluster. We identified enriched pathways only with the upregulated genes in BCG-vaccinated Mtb-infected mice of the clusters CD4+ T naive and CD8+ T naive. The important enriched pathways associated with both clusters include regulation of IFN-γ signaling, IFN-γ signaling, IL-4 and IL-13 signaling, Dap 12 signaling, and ER-phagosome pathway (Figure 7B). Many of these DEGs and pathways were found to be associated with Mtb-infected lungs in our study. These results together show that pathways associated with IFN-γ signaling are upregulated in lymphoid clusters in BCG-vaccinated Mtb-infected lungs when compared with early infection, suggesting that these could serve as correlates of protective immune responses following BCG vaccination.
Figure 7. IFN-γ transcriptional pathways are upregulated in lung lymphoid clusters in BCG-vaccinated mice.
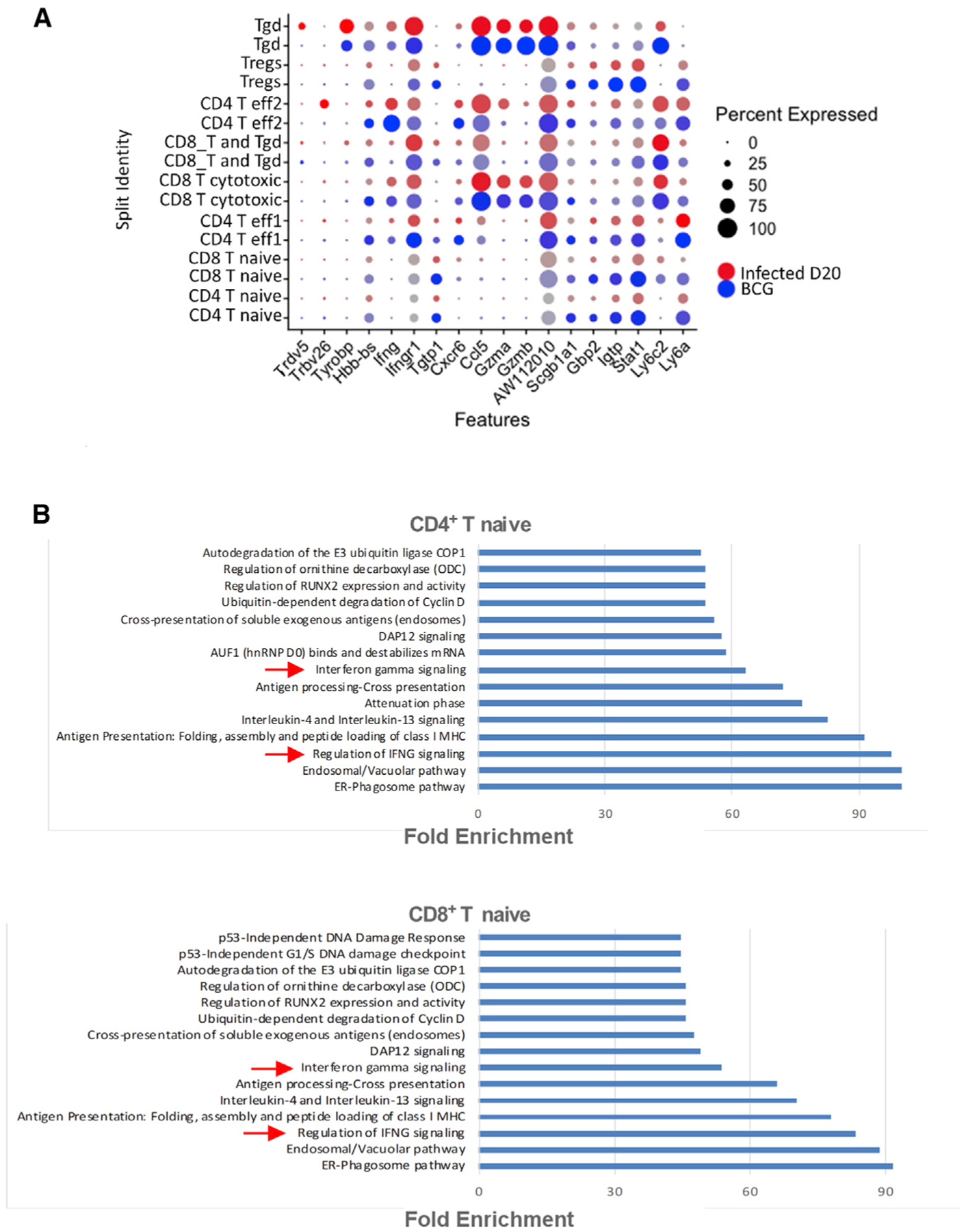
scRNA-seq was performed on lung single cells derived from BCG-vaccinated Mtb-infected mice at 15 dpi (n = 2) and compared to lungs of unvaccinated Mtb-infected mice at 20 dpi (n = 2).
(A) Dot plot represents the expression of top DEGs at different conditions in T cell clusters. The color of the dot represents the condition (red = unvaccinated infected, blue = BCG vaccinated infected).
(B) Top 15 significant overrepresented reactome pathways using the upregulated DEGs in BCG-vaccinated Mtb-infected lungs compared to unvaccinated Mtb-infected lungs in CD4+ T clusters. FDR <0.05.
DISCUSSION
Insights into the lung immune landscape following Mtb infection are limited. We recently described the single-cell landscape of Mtb-infected macaque lungs during TB and latency (Esaulova et al., 2021). Using scRNA-seq data, a recent study also identified and characterized the role of host macrophage cell populations in Mtb-infected murine lungs using high-dose Mtb infection (Pisu et al., 2021). Thus, with the advancement of scRNA-seq, our understanding of the molecular details of the immune response in healthy and Mtb-infected lungs can be significantly enhanced in both human samples and animal models such as mice and non-human primates (NHPs). In the present study, we have used scRNA-seq to understand in-depth the immune landscape of lymphocytes in the lungs at baseline and following Mtb infection in the murine model, which is widely used and is highly relevant for testing vaccines and therapeutics. We examined the transcriptional changes between individual lymphocyte clusters to identify commonalities and differences across populations during Mtb infection and following vaccination at high resolution. The method used in this study by enrichment of live cells provides a standard and reproducible technique for scRNA-seq generation of lymphoid cells in the lung. Our results show that activated T and B cells accumulate in the Mtb-infected lung, coinciding with a decrease in naive T cells during Mtb infection and TB disease progression. Furthermore, our results show overrepresentation of IFN and IL responses in both T and B cells and a reduction in NK cells during the Mtb infection and TB disease progression. scRNA-seq identified Ly6A as a marker for activation during TB; results were validated by the protein expression of Ly6A in different clusters of T cells in the Mtb-infected mice lung. These results enable a comprehensive understanding of the immune cell landscape in Mtb-infected murine lungs that can improve our fundamental understanding of the immunology of TB and characterize animal models relevant for testing TB vaccines and therapeutics.
The important role of T cells in mediating the control of Mtb infection is well documented (Cooper and Khader, 2008; Khader et al., 2007). Several murine studies have shown that the CD4+ and/or CD8+ T cell subset is required for the control of infection by antibody depletion of CD4+ T cells (Müller et al., 1987), by adoptive transfer (Feng and Britton, 2000; Orme and Collins, 1983, 1984), or in gene-deficient mice (Caruso et al., 1999; Mogues et al., 2001). Increased activated CD4+ and CD8+ T cells were found in the lung-draining lymph nodes of Mtb-infected mice (Feng et al., 1999; Serbina et al., 2000). Both CD4+ and CD8+ activated T cells migrate to the lungs and interact with APCs in Mtb-infected lungs between 14 and 28 dpi (Serbina et al., 2000). Consistent with a protective role of T cells in Mtb control, our study found that T cells are the most abundant lymphoid cells in the mice lung irrespective of the time point tested. Interestingly, we found that Mtb-infected mice lungs maintain a reasonable number of naive T and B cells during the course of experimental infection. This is an important finding of the present study, as the existence of naive T cells in the Mtb-infected lungs is often neglected. Consistent with our recent findings in macaque Mtb-infected lungs (Esaulova et al., 2021), our current findings also showed the progressive accumulation of activated T cells in the infected murine lungs.
NK cells play a central role in the innate immune response to microbial pathogens and may play a key role in the initial stages of the local immune response to Mtb infection (Vankayalapati et al., 2005). Recently, we reported that NK cells accumulate and localize in Mtb-infected lungs during TB latency in macaques, compared with their decreased accumulation during ATB disease (Esaulova et al., 2021). A recent multi-cohort study in human peripheral blood mononuclear cells (PBMCs) also showed an increase in the abundance of circulating cytotoxic NK cells in patients with latent TB compared to healthy individuals, which decreased during progression to active disease. This study also found that the NK cells returned to the baseline after TB treatment, suggesting that NK cells play a role in host resistance to Mtb infection (Roy Chowdhury et al., 2018). Furthermore, scRNA-seq revealed that cell death pathways were enriched in cytotoxic NK cells in patients with ATB compared to healthy controls or individuals with latent TB, suggesting that NK cell depletion in ATB may be due to their increased cytotoxicity (Cai et al., 2020). Our current findings, while showing the increased expression of pathways associated with cytotoxicity in NK cells in Mtb-infected lungs at 50 dpi, also showed that during chronic infection (100 dpi), NK cells are decreased compared to uninfected mice lungs. Furthermore, regulation of cell death pathways in NK cells was found highly enriched in Mtb-infected lungs at 100 dpi compared to uninfected lungs, suggesting that enhanced cytotoxicity may also result in NK cell reduction during chronic Mtb infection in mice. NK cells were found to play a protective role against Mtb infection in mice with T cell deficiencies (Feng et al., 2006). Our macaque and human studies also support the notion that NK cells are protective during TB. NK cells can contribute to the immune response against Mtb infection by directly killing the bacteria through releasing perforin and granulysin (Esaulova et al., 2021; Lu et al., 2014), upregulating CD8+T cell responses (Vankayalapati et al., 2005), and lysing Mtb-infected human monocytes and alveolar macrophages (AMs) (Vankayalapati et al., 2005; Venkatasubramanian et al., 2017). The present study also found perforin and granzyme-like cytotoxic molecule expression within the NK cell cluster. Thus, our results project a protective role for NK cells, likely through their cytotoxic effector mechanisms, but these same mechanisms result in NK cell reduction during chronic TB disease.
T cells, through the production of effector cytokines such as IFN-γ, IL-17, and IL-22, mediate the induction of protective immunity against TB in mice and humans (Cooper et al., 1993; Gopal et al., 2014; Khader et al., 2007; Treerat et al., 2017). Even though cytokines play pivotal roles in host protection against Mtb infection (Cooper et al., 1993; Gopal et al., 2014; Khader et al., 2007; Treerat et al., 2017), these cytokines can also be key mediators of inflammatory-mediated pathology. We identified a higher expression of chemokines, cytokines including IFN, and IL-associated genes in different lung T cell clusters in Mtb-infected mice when compared with expression within clusters in uninfected lungs. For example, our study found that both IFN-α/β (type I) and IFN-γ (type II IFN) transcripts are overexpressed in the lung lymphoid clusters in Mtb-infected mice compared to uninfected mice. Also, we identified that the IFN-γ transcript is overexpressed in the BCG-vaccinated mice compared to infected mice, especially in CD4+ T naive and CD8+ T naive cells, suggesting a protective role for IFN-γ during BCG vaccine-induced immunity.
Previous studies have indicated that BCG-induced IFN-γ production by T cells plays a protective role against Mtb (Cooper, 2009; Mathurin et al., 2009). The present study further establishes that following BCG vaccination, a type II IFN response is observed in lymphocytes. However, other studies, including those from our lab, have shown that deletion of IFN-γ-expressing CD4+ T cells has a minor role in providing protection against ATB, and may play a role in regulating protective responses at extrapulmonary sites (Connor et al., 2010; Gopal et al., 2012; Sakai et al., 2016). This is in contrast to the type I IFN response predominant within lymphocyte populations during primary Mtb infection. Both IFN-α/β and IFN-γ can have host-protective roles; however, IFN-α/β can also have detrimental effects, depending on the host immune environment that the pathogen encounters (Lalvani and Millington, 2008; Moreira-Teixeira et al., 2018, 2020). Therefore, our results suggest that type I and II IFN responses may play differential roles during primary infection and vaccine responses, which need to be evaluated further.
CD4+ T cells play a central role in regulating the immune response against Mtb through vast arrays of cytokines produced by different subsets, including Th1, Th2, and Th17. Th1 cells mainly induce protective responses by producing IFN-γ that acts on macrophages to induce anti-mycobacterial protection (Cooper and Khader, 2008). However, the Th2 subset, secreting IL-4, IL5, IL-6, and IL-10, participates in enhancing TB severity (Rook, 2007). Studies on pleotropic Th17 cells have suggested both protective and progressive roles (Cruz et al., 2010; Das and Khader, 2017; Gopal et al., 2014). Th17 cells induce the recruitment of inflammatory neutrophils and thus can induce TB pathology (Lyadova and Panteleev, 2015). In the present study, we observed the overrepresentation of the IFN-γ pathway in T cell clusters in BCG-vaccinated mice (compared to the Mtb-infected group), indicating that the generation of Th1 biased protective immune response post-vaccination may serve as an early correlate of protection. We also observed the type I IFN signature in all lymphoid cell clusters during early and late phases of Mtb infection. Type I IFN can enhance TB pathogenicity (Donovan et al., 2017; Dorhoi et al., 2014) but also play a role in regulating Th1 cell differentiation during the early phase of antigen recognition (Way et al., 2007). ILs are a group of cytokines involved in primary resistance to Mtb. IL-6 is involved in the host response to Mtb and can regulate inflammation, hematopoiesis and differentiation of T cells, and, most importantly, the suppression of T cell responses (VanHeyningen et al., 1997). In human PBMCs, the production of IL-4 was found to be significantly higher in TB patients compared to healthy controls (Van Crevel et al., 2000). Furthermore, an increase in IL-4 and IL-13 expression correlates with lung damage in humans with TB, indicating a role in countering protective immunity and contributing to Mtb replication and tissue pathology (Heitmann et al., 2014). Thus, our study has identified a higher expression of several IL and IFN pathways in different T and B cell clusters in Mtb-infected mice, suggesting that these pathways are induced in response to infection and can be either protective or immunomodulatory.
Glycosylphosphatidyl-inositol (GPI)-linked protein Ly6A is expressed on hematopoietic stem cells and activated lymphocytes (Li et al., 2014). Our study identified Ly6A expression as a marker induced in lymphocyte clusters post-Mtb challenge. Ly6A expression is regulated by type I IFN signaling (Khan et al., 1993), but its role during Mtb infection is still unclear. Higher expression of Ly6A was also found in interstitial macrophage populations in a recent TB study using scRNA-seq from mice lungs (Pisu et al., 2021). We further validated Mtb-induced expression of Ly6A on activated (CD44+CD69+) CD4+ and CD8+ T cells by flow cytometry. This leads us to believe that Ly6A is a marker for T cell activation during Mtb infection.
Increased IFN signature in Mtb-infected lung lymphoid clusters is consistent with the transcriptional signatures reported in human TB and in NHP lung following Mtb infection (Berry et al., 2010; Esaulova et al., 2021). The role for type I IFN in human TB was first revealed in a transcriptomic study of patients with ATB and Mtb-infected individuals (Berry et al., 2010). The reproducibility of type I IFN-inducible blood transcriptomic signature was later confirmed by several other studies on diverse human TB cohorts (Blankley et al., 2016; Sambarey et al., 2017; Singhania et al., 2018). Our recent study also identified the IFN-inducible transcriptional signature in Mtb-infected lungs with ATB in macaques (Ahmed et al., 2020). In addition, the overexpression of IFN response pathways, innate immune system, and several IL signaling pathways were detected across species in human TB progressors, macaques, and mice infected with Mtb (Ahmed et al., 2020; Berry et al., 2010). Comparing the transcriptomic signatures of Berry et al. with our transcriptomic signatures, we found several common IFN signature genes that are differentially expressed in different T and B cell clusters in our study. Stat1, Gbp2, Gbp4, Psme1, Psme2, Psmb9, Tap1, Tap2, and Zbp1 are the most common genes across different clusters in our present study. Among these, Gbp2, Gbp4, Stat1, and Tap1 were previously identified as prospective signatures of TB risk in the South African adolescent cohort study (ACS) (Zak et al., 2016) and were found highly expressed in progressors compared to naive in both mouse and macaque (Ahmed et al., 2020). Although the overexpression of IFN signatures was found in neutrophils from human peripheral blood only (Berry et al., 2010), our scRNA-seq study identified IFN signatures in CD4+ T cells, CD8+ T cells, and B cells in the infected mice lungs, suggesting that type I IFN responses may be well reflected within neutrophils in human peripheral blood, but in murine infected lungs, the response may also be reflected within lymphoid clusters.
In conclusion, we carried out an in-depth characterization of different subsets of lymphocytes in the mice lung during early and chronic phases of TB disease. Our results have demonstrated IFN signatures in lymphocyte cells and identified Ly6A as a marker that is induced upon Mtb infection in lymphocytes.
Limitations of the study
Our study has some limitations. For example, our work focuses mainly on the lymphoid compartment. Unfortunately, due to the loss of the myeloid compartment during live cell enrichment of the lung single-cell suspension, we were unable to define the transcriptional status of myeloid cells. However, we acknowledge that the myeloid cell compartment does play an important role in the generation of immune responses against TB, and the optimization of experimental protocols to safeguard these vital compartments needs to be addressed. Furthermore, although we find an increase in the type I IFN signature in lung lymphocytes, future studies should focus on elucidating the impact of this on the generation of protective immune response or pathogenesis during TB disease progression.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Shabaana A. Khader (sakhader@wustl.edu).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
The accession number of the raw and processed data for single cell RNA sequencing has been deposited in GEO. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Ly-6A, PerCP-Cyanine5.5 Monoclonal |
eBiosciences | Cat# 45-5981-82, RRID:AB_914372 |
| Serian Hamster anti-CD3e, AF700 Monoclonal, clone 500A2 |
BD Biosciences | Cat# 557984, RRID:AB_396972 |
| Rat anti-CD4, Pac Blue Monoclonal, clone RM4–5 |
BD Biosciences | Cat# 553050, RRID:AB_394586 |
| Rat anti-CD8a, APC-Cy7 Monoclonal, Clone 53–7.6 |
BD Biosciences | Cat# 557654, RRID:AB_396769 |
| Rat anti-CD44, PE-Cy7 Monoclonal, Clone IM7 |
Thermo Fisher Scientific | Cat# 25-0441-82, RRID:AB_469623 |
| Armenian Hamster anti-CD69, PE Monoclonal |
BD Biosciences | Cat# 553237, RRID:AB_394726 |
| anti-NK1.1, FITC Monoclonal, Clone PK136 |
Tonbo Biosciences | Cat# 35–5941, RRID:AB_2621722 |
| Rat Anti-CD11b, Allophycocyanin Conjugated Monoclonal Antibody, |
BD Biosciences | Cat# 553312, RRID:AB_398535 |
| Armenian Hamster Anti-CD11c Monoclonal Antibody, FITC Conjugated, Clone HL3 |
BD Biosciences | Cat# 553801, RRID:AB_395060 |
| Rat ant-Ly-6G and Ly-6C, BUV395 Monoclonal, Clone RB6–8C5 |
BD Biosciences | Cat# 563849, RRID:AB_2738450 |
| Rat ant-SiglecF, BV421 Monoclonal, clone E50–2440 |
BD Biosciences | Cat# 562681, RRID:AB_2722581 |
| Rat Anti-Mouse MHC Class II (I-A / I-E), eFluor650NC Monoclonal, Clone M5/114.15.2 |
Thermo Fisher Scientific | Cat# 95-5321-42, RRID:AB_1603293 |
| Armenian Hamster Anti-CD103 (Integrin alpha E), PE Monoclonal, clone 2E7 |
Thermo Fisher Scientific | Cat# 12-1031-82, RRID:AB_465799 |
| Rat anti-CD64, PE/Cyanine7 Monoclonal |
BioLegend | Cat# 139314, RRID:AB_2563904 |
| Rat Anti-IFN-gamma, Allophycocyanin Monoclonal, clone XMG1.2 |
BD Biosciences | Cat# 554413, RRID:AB_398551 |
| Bacterial and virus strains | ||
| Mycobacterium tuberculosis strain HN878 | BEI Resources, Manassas, VA | N/A |
| Deposited data | ||
| scRNA-seq, raw and analyzed data | This manuscript | GSE200639 |
| scRNA-seq, raw and analyzed data | PMID: 34253059 | GSE150657 |
| Experimental models: Organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| Software and algorithms | ||
| Cellranger 3.1 | 10x Genomics | https://support.10xgenomics.com/ |
| Seurat 3 | Stuart et al. (2019) | https://satijalab.org/seurat |
| ggplot2 | Wickham (2016) | https://cloud.r-project.org/package=ggplot2 |
| biomaRt | Durinck et al. (2009) | https://bioconductor.org/packages/biomaRt |
| PANTHER | Mi and Thomas (2009) | http://www.pantherdb.org/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 (B6) mice were obtained from Jackson Laboratory (Bar Harbor, ME) and bred at Washington University in St. Louis. Mice were age and sex-matched and used between 6–8 weeks of age. All mice were used and housed in accordance with the National Institute of Health guidelines for housing and care of laboratory animals. All the experiments in this study were granted by the Washington University in St. Louis Institutional Animal Care and Use Committee under protocol 20160129.
METHOD DETAILS
Bacterial infection
Mycobacterium tuberculosis W. Beijing strain, HN878 (BEI Resources) were grown to mid-log phase in Proskauer Beck medium containing 0.05% Tween 80 and frozen in at −80°C. Mice were infected with ~100 CFU Mtb HN878 via aerosol route using a Glas-Col airborne infection system.
Generation of single-cell suspensions from tissues
Lung single-cell suspensions from uninfected or Mtb-infected mice were isolated as previously described (Ardain et al., 2019). Briefly, mice were euthanized with CO2 and lungs were perfused with heparin in saline. Lungs were minced and incubated in Collagenase/DNAse for 30 minutes at 37°C. Lung tissue was pushed through a 70 μm nylon screen to obtain a single-cell suspension. Red blood cells were lysed and the cells were resuspended in suitable media or buffer for further use.
Single-cell RNA library generation and sequencing
Isolated total lung single-cell suspensions were enriched for live cells using dead cell depletion kit according to manufacturer’s instruction (Miltenyi Biotec) and subjected to droplet-based massively parallel single-cell RNA sequencing using Chromium Single Cell 5’ (v1) Reagent Kit as per manufacturer’s instructions (10x Genomics). Briefly, cell suspensions were loaded at 1,000 cells/μL with the aim to capture 10,000 cells/lane. The 10x Chromium Controller generated GEM droplets, where each cell was labeled with a specific barcode, and each transcript labeled with a unique molecular identifier (UMI) during reverse transcription. The barcoded cDNA underwent 16 cycles of amplification, and after SPRI bead purification, was removed from the BSL-3 space for library generation. Libraries were prepared from amplified cDNA and target enrichment products by fragmentation, end repair, A-tailing, adapter ligation, and sample index PCR as per the manufacturer’s instructions. Libraries were sequenced on a NovaSeq S4 (200 cycle) flow cell, targeting 50,000 read pairs/cell.
Single-cell RNA-seq data processing
The raw gene expression matrices were generated by the Cell Ranger software (10x Genomics, version 3) available on the 10x website. After demultiplexing, the resulting fastq files were aligned against the mouse reference genome mm10 with cellranger count. For each sample, the recovered-cell parameter was set to 10,000 cells that we expect to recover for each library. The output filtered gene count matrices were analyzed by R software (v.3.5.3) with the Seurat (Stuart et al., 2019) package (v.3.0.0). Cells that had less than 1,100 and more than 4,500 detected genes were filtered out. Different thresholds were chosen for mitochondrial genes because of their distinct distribution in each sample. Criteria for filtered cells were as follows: (1) had more than 5% (uninfected_2), (2) 6% (infected_d50_3, infected_d100_1), (3) 7% (infected_d100_2), (4) 8% (uninfected_1, infected_d50_1, infected_d100_3), and (5) 9% (infected_d50_2) of mitochondrial genes. Samples with similar condition were merged, data normalized with default parameters, and most variable genes were detected by the FindVariableFeatures function. All samples were combined using Seurat functions, FindIntegrationAnchors and IntegrateData. ScaleData was used to regresse out number of UMI’s and mitochondrial content and principal component analysis (PCA) was performed with RunPCA. The TSNE dimensionality reduction was performed on the scaled matrix using the first 17 PCA. For clustering, the FindNeighbors (17 PCA) and FindClusters (resolution 0.4) functions were used. FindAllMarkers was used to compare a cluster against all other clusters to identify the marker genes. For each cluster, the minimum required average log fold change in gene expression was set to 0.25 and the minimum percent of cells that must express gene in either cluster was set to 25%. To re-analyze lymphoid sub-populations, we pooled the clusters that we identified as lymphoid origin and re-run PCA, tSNE, and clustering to get a better resolution for further analysis. The function FindMarkers was used to compare the cells in a selected cluster across different conditions. The analysis was performed based on the Wilcoxon rank sum test with thresholds average logFC ≥0.25 and adj-pvalue ≤ 0.05 using Seurat. The over-representation test was performed on PANTHER website (Mi and Thomas, 2009) using Reactome pathway database with Fisher’s exact test and corrected for multiple tests using false discovery rate (threshold ≤0.05) as calculated by the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995). Several R packages were used to generate figures and intermediate data preprocessing, such as ggplot2 (Wickham, 2016) and biomaRt (Durinck et al., 2009).
Public scRNA-seq data re-analysis
We re-analyzed the publicly available GSE150657 dataset. The data alignment and clustering workflow are same as described in (Das et al., 2021). Differential analysis was carried out for each of the eight clusters: CD4+ naive, CD4+ eff1, CD4+ eff2, Tregs, CD8+ naive, CD8+ cytotoxic, ‘CD8+ T and Tgd’, and Tgd. The Seurat function FindMarkers was used to compare the gene expressions in a selected cluster of BCG vaccinated vs infected (20 dpi) mice. The analysis was performed using Wilcoxon rank sum test with default settings in Seurat. The over-representation test was performed using Reactome pathway database on PANTHER with default settings.
Histology
Mouse lung lobes were perfused and fixed with 10% neutral buffered formalin and embedded in paraffin. For determining the expression of Stat1 mRNA, formalin-fixed paraffin-embedded (FFPE) lung sections were subjected to in situ hybridization (ISH) with the mouse-Stat1 probe using the RNAscope 2.5HD Detection Kit (Brown staining) as per the manufacturer’s recommendations (Advanced Cell Diagnostics, Newark, CA). The representative pictures were captured with the Hamamatsu Nanozoomer 2.0 HT system with NDP scan image acquisition software.
Flow cytometry
Lung single-cell suspensions were prepared as described before (Khader et al., 2007), treated with Fc Block (CD16/CD32, 2.4G2, Tonbo Biosciences, San Diego, CA), and stained with appropriate fluorochrome-labeled specific or isotype control antibodies: CD11c (HL3, BD Biosciences), CD11b (M1/70, BD Biosciences and Tonbo Biosciences), Ly6A (D7, eBioscience), CD3 (145–2C11, Tonbo Biosciences), CD4 (GK1.5, BD Biosciences), CD44 (IM7, eBioscience), IFN-γ (XMG1.2, BD Biosciences), CD8 (53–6.7, BD Biosciences), CD69 (H1.2F3, BD Biosciences) and rat IgG1 (BD Biosciences). Cells were processed using a Becton Dickinson FACS LSR Fortessa X20 flow cytometer using FACSDiva software. Cells were gated based on their forward and side scatter characteristics and the frequency of specific cell types was calculated using FlowJo (FlowJo, LLC, Ashland, OR).
QUANTIFICATION AND STATISTICAL ANALYSIS
All of the statistical details of experiments can be found in the figure legends including the statistical tests used, exact value of n (number of animals or human samples used). Data represented as mean ± SD (standard deviation). Differences between the means of 2 groups were analyzed using the two-tailed Student’s t test in Prism 5 (GraphPad). *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant. No methods were applied to determine assumptions of the statistical approach.
Supplementary Material
Highlights.
scRNA-seq of lymphocytes derived from healthy and Mtb-infected murine lungs
Transcriptional analysis indicates enrichment of type I IFN signature in lymphocytes
Heat shock responses are expressed in NK cells in chronic phase of Mtb infection
The study identifies and validates Ly6A as a lymphocyte activation marker
ACKNOWLEDGMENTS
This work was supported by Washington University in St. Louis School of Medicine and NIH grants HL105427, AI111914, AI134236, AI155024, and AI123780 to S.A.K. and D.K. The authors wish to thank Amanda Swain, Washington University School of Medicine in St. Louis, for the biological data collection and Leah Mellett for helping in graphical abstract image creation with Biorender. com.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110983.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmed M, Thirunavukkarasu S, Rosa BA, Thomas KA, Das S, Rangel-Moreno J, Lu L, Mehra S, Mbandi SK, Thackray LB, et al. (2020). Immune correlates of tuberculosis disease and risk translate across species. Sci. Transl. Med 12, eaay0233. 10.1126/scitranslmed.aay0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardain A, Domingo-Gonzalez R, Das S, Kazer SW, Howard NC, Singh A, Ahmed M, Nhamoyebonde S, Rangel-Moreno J, Ogongo P, et al. (2019). Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature 570, 528–532. 10.1038/s41586-019-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. (2010). An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977. 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankley S, Graham CM, Levin J, Turner J, Berry MPR, Bloom CI, Xu Z, Pascual V, Banchereau J, Chaussabel D, et al. (2016). A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur. Respir. J 47, 1873–1876. 10.1183/13993003.02121-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Dai Y, Wang Y, Yang Q, Guo J, Wei C, Chen W, Huang H, Zhu J, Zhang C, et al. (2020). Single-cell transcriptomics of blood reveals a natural killer cell subset depletion in tuberculosis. EBioMedicine 53, 102686. 10.1016/j.ebiom.2020.102686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso AM, Serbina N, Klein E, Triebold K, Bloom BR, and Flynn JL (1999). Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. J. Immunol 162, 5407–5416. [PubMed] [Google Scholar]
- Choi S-C, Wang H, Tian L, Murakami Y, Shin D-M, Borrego F, Morse HC, and Coligan JE (2013). Mouse IgM Fc receptor, FCMR, promotes B cell development and modulates antigen-driven immune responses. J. Immunol 190, 987–996. 10.4049/jimmunol.1202227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor LM, Harvie MC, Rich FJ, Quinn KM, Brinkmann V, Gros GL, and Kirman JR (2010). A key role for lung-resident memory lymphocytes in protective immune responses after BCG vaccination. Eur. J. Immunol 40, 2482–2492. 10.1002/eji.200940279. [DOI] [PubMed] [Google Scholar]
- Cooper AM (2009). Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol 27, 393–422. 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AM, and Khader SA (2008). The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol. Rev 226, 191–204. 10.1111/j.1600-065x.2008.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, and Orme IM (1993). Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med 178, 2243–2247. 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A, Fraga AG, Fountain JJ, Rangel-Moreno J, Torrado E, Saraiva M, Pereira DR, Randall TD, Pedrosa J, Cooper AM, and Castro AG (2010). Pathological role of interleukin 17 in mice subjected to repeated BCG vaccination after infection with Mycobacterium tuberculosis. J. Exp. Med 207, 1609–1616. 10.1084/jem.20100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, and Khader S (2017). Yin and yang of interleukin-17 in host immunity to infection. F1000Research 6, 741. 10.12688/f1000research.10862.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Marin ND, Esaulova E, Ahmed M, Swain A, Rosa BA, Mitreva M, Rangel-Moreno J, Netea MG, Barreiro LB, et al. (2021). Lung epithelial signaling mediates early vaccine-induced cd4 + t cell activation and mycobacterium tuberculosis control. mBio 12, e0146821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino M, Lodi L, Galli L, and Chiappini E (2019). Immune response to Mycobacterium tuberculosis: a narrative Review. Front. Pediatr 7, 350. 10.3389/fped.2019.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, and Kaech SM (2015). The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med 211, 2113OIA258. 10.1083/jcb.2113oia258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan ML, Schultz TE, Duke TJ, and Blumenthal A (2017). Type I interferons in the pathogenesis of tuberculosis: molecular drivers and immunological consequences. Front. Immunol 8, 1633. 10.3389/fimmu.2017.01633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A, Yeremeev V, Nouailles G, Weiner J, Jörg S, Heinemann E, Oberbeck-Müller D, Knaul JK, Vogelzang A, Reece ST, et al. (2014). Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol 44, 2380–2393. 10.1002/eji.201344219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, and Nathan C (2001). Reprogramming of the macrophage transcriptome in response to interferon-γ and mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med 194, 1123–1140. 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaulova E, Das S, Singh DK, Choreño-Parra JA, Swain A, Arthur L, Rangel-Moreno J, Ahmed M, Singh B, Gupta A, et al. (2021). The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 29, 165–178.e8. 10.1016/j.chom.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CG, and Britton WJ (2000). CD4+and CD8+T cells mediate adoptive immunity to aerosol infection of Mycobacterium bovisBacillus Calmette-Guérin. J. Infect. Dis 181, 1846–1849. 10.1086/315466. [DOI] [PubMed] [Google Scholar]
- Feng CG, Bean AGD, Hooi H, Briscoe H, and Britton WJ (1999). Increase in gamma interferon-secreting CD8+, as well as CD4+, T cells in lungs following aerosol infection with Mycobacterium tuberculosis. Int. J. Lepr. Other Mycobact. Dis 67, 3242–3247. 10.1128/iai.67.7.3242-3247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CG, Kaviratne M, Rothfuchs AG, Cheever A, Hieny S, Young HA, Wynn TA, and Sher A (2006). NK cell-derived IFN-γ differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J. Immunol 177, 7086–7093. 10.4049/jimmunol.177.10.7086. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Juarrero M, Kingry LC, Ordway DJ, Henao-Tamayo M, Harton M, Basaraba RJ, Hanneman WH, Orme IM, and Slayden RA (2009). Immune response to mycobacterium tuberculosis and identification of molecular markers of disease. Am. J. Respir. Cell Mol. Biol 40, 398–409. 10.1165/rcmb.2008-0248oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal R, Lin Y, Obermajer N, Slight S, Nuthalapati N, Ahmed M, Kalinski P, and Khader SA (2012). IL-23-dependent IL-17 drives Th1-cell responses following Mycobacterium bovis BCG vaccination. Eur. J. Immunol 42, 364–373. 10.1002/eji.201141569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal R, Monin L, Slight S, Uche U, Blanchard E, Fallert Junecko A, Ramos-Payan R, Ramos-Payan R, Stallings CL, Reinhart TA, et al. (2014). Unexpected role for IL-17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathog. 10, e1004099. 10.1371/journal.ppat.1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Kurtz SL, Rigel NW, Gunn BM, Taft-Benz S, Morrison JP, Fong AM, Patel DD, Braunstein M, and Kawula TH (2009). The impact of chemokine receptor CX3CR1 deficiency during respiratory infections with Mycobacterium tuberculosis or Francisella tularensis. Clin. Exp. Immunol 156, 278–284. 10.1111/j.1365-2249.2009.03882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann L, Abad Dar M, Schreiber T, Erdmann H, Behrends J, Mckenzie ANJ, Brombacher F, Ehlers S, Hölscher C, and Hölscher C (2014). The IL-13/IL-4Rα axis is involved in tuberculosis-associated pathology. J. Pathol 234, 338–350. 10.1002/path.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B, Lee JH, and Bang D (2018). Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med 50, 1–14. 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DD, Lin Y, Moreno JR, Randall TD, and Khader SA (2011). Profiling early lung immune responses in the mouse model of tuberculosis. PLoS One 6, e16161. 10.1371/journal.pone.0016161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, et al. (2007). IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol 8, 369–377. 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- Khan KD, Shuai K, Lindwall G, Maher SE, Darnell JE, and Bothwell AL (1993). Induction of the Ly-6A/E gene by interferon alpha/beta and gamma requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc. Natl. Acad. Sci. U S A 90, 6806–6810. 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalvani A, and Millington KA (2008). T cells and tuberculosis: beyond interferon-γ. J. Infect. Dis 197, 941–943. 10.1086/529049. [DOI] [PubMed] [Google Scholar]
- Li Y, Esain V, Teng L, Xu J, Kwan W, Frost IM, Yzaguirre AD, Cai X, Cortes M, Maijenburg MW, et al. (2014). Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597–2612. 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-C, Wu T-S, Hsu Y-J, Chang C-J, Lin C-S, Chia J-H, Wu T-L, Huang T-T, Martel J, Ojcius DM, et al. (2014). NK cells kill mycobacteria directly by releasing perforin and granulysin. J. Leukoc. Biol 96, 1119–1129. 10.1189/jlb.4a0713-363rr. [DOI] [PubMed] [Google Scholar]
- Lyadova IV, and Panteleev AV (2015). Th1 and Th17 cells in tuberculosis: protection, pathology, and biomarkers. Mediators Inflamm. 2015, 1–13. 10.1155/2015/854507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- María Irene CC, Juan Germán RC, Gamaliel L-L, Dulce Adriana M-E, Estela Isabel B, Brenda Nohemí MC, Payan Jorge B, Zyanya Lucía ZB, Myriam B.d.V., Fernanda CG, et al. (2021). Profiling the immune response to Mycobacterium tuberculosis Beijing family infection: a perspective from the transcriptome. Virulence 12, 1689–1704. 10.1080/21505594.2021.1936432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathurin KS, Martens GW, Kornfeld H, and Welsh RM (2009). CD4 T-Cell-Mediated heterologous immunity between mycobacteria and Poxviruses. J. Virol 83, 3528–3539. 10.1128/jvi.02393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, and Thomas P (2009). PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol 563, 123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogues T, Goodrich ME, Ryan L, LaCourse R, and North RJ (2001). The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J. Exp. Med 193, 271–280. 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Teixeira L, Mayer-Barber K, Sher A, and O’Garra A (2018). Type I interferons in tuberculosis: Foe and occasionally friend. J. Exp. Med 215, 1273–1285. 10.1084/jem.20180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Teixeira L, Stimpson PJ, Stavropoulos E, Hadebe S, Chakravarty P, Ioannou M, Aramburu IV, Herbert E, Priestnall SL, Suarez-Bonnet A, et al. (2020). Type I IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and NETosis. Nat. Commun 11, 5566. 10.1038/s41467-020-19412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller I, Cobbold SP, Waldmann H, and Kaufmann SH (1987). Impaired resistance to Mycobacterium tuberculosis infection after selective in vivo depletion of L3T4+ and Lyt-2+ T cells. Infect. Immun 55, 2037–2041. 10.1128/iai.55.9.2037-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme IM, and Collins FM (1983). Protection against Mycobacterium tuberculosis infection by adoptive immunotherapy. Requirement for T cell-deficient recipients. J. Exp. Med 158, 74–83. 10.1084/jem.158.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme IM, and Collins FM (1984). Adoptive protection of the Mycobacterium tuberculosis-infected lung. Dissociation between cells that passively transfer protective immunity and those that transfer delayed-type hypersensitivity to tuberculin. Cell. Immunol 84, 113–120. 10.1016/0008-8749(84)90082-0. [DOI] [PubMed] [Google Scholar]
- Pisu D, Huang L, Narang V, Theriault M, Lê-Bury G, Lee B, Lakudzala AE, Mzinza DT, Mhango DV, Mitini-Nkhoma SC, et al. (2021). Single cell analysis of M. tuberculosis phenotype and macrophage lineages in the infected lung. J. Exp. Med 218, e20210615. 10.1084/jem.20210615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Jung JY, and Nau GJ (2012). Interferon-γ, tumor necrosis factor, and interleukin-18 cooperate to control growth of Mycobacterium tuberculosis in human macrophages. Cytokine 60, 233–241. 10.1016/j.cyto.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook GAW (2007). Th2 cytokines in susceptibility to tuberculosis. Curr. Mol. Med 7, 327–337. 10.2174/156652407780598557. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhury R, Vallania F, Yang Q, Lopez Angel CJ, Darboe F, Penn-Nicholson A, Rozot V, Nemes E, Malherbe ST, Ronacher K, et al. (2018). A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature 560, 644–648. 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Lamprecht DA, Mandizvo T, Jones TT, Naidoo V, Addicott KW, Moodley C, Ngcobo B, Crossman DK, Wells G, and Steyn AJ (2019). Compromised metabolic reprogramming is an early indicator of CD8+ T cell dysfunction during chronic Mycobacterium tuberculosis infection. Cell Rep. 29, 3564–3579.e5. 10.1016/j.celrep.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, Ganusov VV, and Barber DL (2016). CD4 T cell-derived IFN-γ plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must Be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog. 12, e1005667. 10.1371/journal.ppat.1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Lee S, Ishizuka A, Hinoshita E, Hori H, Ishibashi N, Komada K, Norizuki M, Katsuma Y, Akashi H, and Shibuya K (2019). Challenges and opportunities for eliminating tuberculosis – leveraging political momentum of the UN high-level meeting on tuberculosis. BMC Publ. Health 19, 76. 10.1186/s12889-019-6399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambarey A, Devaprasad A, Baloni P, Mishra M, Mohan A, Tyagi P, Singh A, Akshata J, Sultana R, Buggi S, and Chandra N (2017). Meta-analysis of host response networks identifies a common core in tuberculosis. Npj Syst. Biol. Appl 3, 4. 10.1038/s41540-017-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina NV, Liu C-C, Scanga CA, and Flynn JL (2000). CD8 + CTL from lungs of Mycobacterium tuberculosis -infected mice express perforin in vivo and lyse infected macrophages. J. Immunol 165, 353–363. 10.4049/jimmunol.165.1.353. [DOI] [PubMed] [Google Scholar]
- Singhania A, Verma R, Graham CM, Lee J, Tran T, Richardson M, Lecine P, Leissner P, Berry MPR, Wilkinson RJ, et al. (2018). A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun 9, 2308. 10.1038/s41467-018-04579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford WL, Haque S, Alexander R, Liu X, Latour AM, Snodgrass HR, Koller BH, and Flood PM (1997). Altered proliferative response by T lymphocytes of Ly-6A (Sca-1) null mice. J. Exp. Med 186, 705–717. 10.1084/jem.186.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treerat P, Prince O, Cruz-LAgunAs A, Muñoz-Torrico M, Salazar-Lezama MA, Selman M, Fallert-Junecko B, Reinhardt TA, Alcorn JF, Kaushal D, et al. (2017). Novel role for IL-22 in protection during chronic Mycobacterium tuberculosis HN878 infection. Mucosal Immunol. 10, 1069–1081. 10.1038/mi.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretina K, Park E-S, Maminska A, and MacMicking JD (2019). Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. J. Exp. Med 216, 482–500. 10.1084/jem.20182031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Crevel R, Karyadi E, Preyers F, Leenders M, Kullberg BJ, H Nelwan R, Nelwan RHH, van der Meer JM, and Van Der Meer JWM (2000). Increased production of interleukin 4 by CD4+ and CD8+ T cells from patients with tuberculosis is related to the presence of pulmonary cavities. J. Infect. Dis 181, 1194–1197. 10.1086/315325. [DOI] [PubMed] [Google Scholar]
- VanHeyningen TK, Collins HL, and Russell DG (1997). IL-6 produced by macrophages infected with Mycobacterium species suppresses T cell responses. J. Immunol 158, 330–337. [PubMed] [Google Scholar]
- Vankayalapati R, Garg A, Porgador A, Griffith DE, Klucar P, Safi H, Girard WM, Cosman D, Spies T, and Barnes PF (2005). Role of NK cell-activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J. Immunol 175, 4611–4617. 10.4049/jimmunol.175.7.4611. [DOI] [PubMed] [Google Scholar]
- Venkatasubramanian S, Cheekatla S, Paidipally P, Tripathi D, Welch E, Tvinnereim AR, Nurieva R, and Vankayalapati R (2017). IL-21-dependent expansion of memory-like NK cells enhances protective immune responses against Mycobacterium tuberculosis. Mucosal Immunol. 10, 1031–1042. 10.1038/mi.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesosky B, Rottinghaus EK, Stromberg P, Turner J, and Beamer G (2010). CCL5 participates in early protection against Mycobacterium tuberculosis. J. Leukoc. Biol 87, 1153–1165. 10.1189/jlb.1109742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way SS, Havenar-Daughton C, Kolumam GA, Orgun NN, and Murali-Krishna K (2007). IL-12 and type-I IFN synergize for IFN-γ production by CD4 T cells, whereas neither are required for IFN-γ production by CD8 T cells after Listeria monocytogenes infection. J. Immunol 178, 4498–4505. 10.4049/jimmunol.178.7.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2021). Global Tuberculosis Report 2021 (World Health Organization; ). [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; ). [Google Scholar]
- Yi XH, Zhang B, Fu YR, and Yi ZJ (2020). STAT1 and its related molecules as potential biomarkers in Mycobacterium tuberculosis infection. J. Cell Mol. Med 24, 2866–2878. 10.1111/jcmm.14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, et al. (2016). A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387, 2312–2322. 10.1016/s0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number of the raw and processed data for single cell RNA sequencing has been deposited in GEO. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Ly-6A, PerCP-Cyanine5.5 Monoclonal |
eBiosciences | Cat# 45-5981-82, RRID:AB_914372 |
| Serian Hamster anti-CD3e, AF700 Monoclonal, clone 500A2 |
BD Biosciences | Cat# 557984, RRID:AB_396972 |
| Rat anti-CD4, Pac Blue Monoclonal, clone RM4–5 |
BD Biosciences | Cat# 553050, RRID:AB_394586 |
| Rat anti-CD8a, APC-Cy7 Monoclonal, Clone 53–7.6 |
BD Biosciences | Cat# 557654, RRID:AB_396769 |
| Rat anti-CD44, PE-Cy7 Monoclonal, Clone IM7 |
Thermo Fisher Scientific | Cat# 25-0441-82, RRID:AB_469623 |
| Armenian Hamster anti-CD69, PE Monoclonal |
BD Biosciences | Cat# 553237, RRID:AB_394726 |
| anti-NK1.1, FITC Monoclonal, Clone PK136 |
Tonbo Biosciences | Cat# 35–5941, RRID:AB_2621722 |
| Rat Anti-CD11b, Allophycocyanin Conjugated Monoclonal Antibody, |
BD Biosciences | Cat# 553312, RRID:AB_398535 |
| Armenian Hamster Anti-CD11c Monoclonal Antibody, FITC Conjugated, Clone HL3 |
BD Biosciences | Cat# 553801, RRID:AB_395060 |
| Rat ant-Ly-6G and Ly-6C, BUV395 Monoclonal, Clone RB6–8C5 |
BD Biosciences | Cat# 563849, RRID:AB_2738450 |
| Rat ant-SiglecF, BV421 Monoclonal, clone E50–2440 |
BD Biosciences | Cat# 562681, RRID:AB_2722581 |
| Rat Anti-Mouse MHC Class II (I-A / I-E), eFluor650NC Monoclonal, Clone M5/114.15.2 |
Thermo Fisher Scientific | Cat# 95-5321-42, RRID:AB_1603293 |
| Armenian Hamster Anti-CD103 (Integrin alpha E), PE Monoclonal, clone 2E7 |
Thermo Fisher Scientific | Cat# 12-1031-82, RRID:AB_465799 |
| Rat anti-CD64, PE/Cyanine7 Monoclonal |
BioLegend | Cat# 139314, RRID:AB_2563904 |
| Rat Anti-IFN-gamma, Allophycocyanin Monoclonal, clone XMG1.2 |
BD Biosciences | Cat# 554413, RRID:AB_398551 |
| Bacterial and virus strains | ||
| Mycobacterium tuberculosis strain HN878 | BEI Resources, Manassas, VA | N/A |
| Deposited data | ||
| scRNA-seq, raw and analyzed data | This manuscript | GSE200639 |
| scRNA-seq, raw and analyzed data | PMID: 34253059 | GSE150657 |
| Experimental models: Organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| Software and algorithms | ||
| Cellranger 3.1 | 10x Genomics | https://support.10xgenomics.com/ |
| Seurat 3 | Stuart et al. (2019) | https://satijalab.org/seurat |
| ggplot2 | Wickham (2016) | https://cloud.r-project.org/package=ggplot2 |
| biomaRt | Durinck et al. (2009) | https://bioconductor.org/packages/biomaRt |
| PANTHER | Mi and Thomas (2009) | http://www.pantherdb.org/ |


