Abstract
The purpose of this study was to evaluate, revise, and extend the Informed Consent Ontology (ICO) for expressing clinical permissions, including reuse of residual clinical biospecimens and health data. This study followed a formative evaluation design and used a bottom-up modeling approach. Data were collected from the literature on US federal regulations and a study of clinical consent forms. Eleven federal regulations and fifteen permission-sentences from clinical consent forms were iteratively modeled to identify entities and their relationships, followed by community reflection and negotiation based on a series of predetermined evaluation questions. ICO included fifty-two classes and twelve object properties necessary when modeling, demonstrating appropriateness of extending ICO for the clinical domain. Twenty-six additional classes were imported into ICO from other ontologies, and twelve new classes were recommended for development. This work addresses a critical gap in formally representing permissions clinical permissions, including reuse of residual clinical biospecimens and health data. It makes missing content available to the OBO Foundry, enabling use alongside other widely-adopted biomedical ontologies. ICO serves as a machine-interpretable and interoperable tool for responsible reuse of residual clinical biospecimens and health data at scale.
Keywords: Knowledge Bases, Evaluation Study, Informed Consent, Biological Specimen Banks, Informatics
1. BACKGROUND AND SIGNIFICANCE
Informed consent is a foundational requirement in both clinical care and research studies. Informed consent forms serve as the primary source of evidence that an informed consent process occurred, and that the consenter has received enough information to make an informed decision regarding the permissions they are being asked to either grant or deny (The Joint Commission, Division of Healthcare Improvement, 2016). While efforts are underway for tracing such permissions for biospecimens collected during research studies, significantly less attention has been given to clinical consent forms and permissions to reuse residual clinical biospecimens. Residual clinical biospecimens are the portions or derivatives of blood or tissue collected during clinical care process that remain after their clinical indications are fulfilled. These specimens are increasingly recognized as a valuable resource.
There is a need for information systems to facilitate discovery, access, and responsible reuse of stored biospecimens and data, and to facilitate data integration and knowledge discovery within a contemporary connected research environment. This vision requires development of technology which supports expectations around FAIR (Findable, Accessible, Interoperable, Reusable) Principles and includes metadata to support discovery of biospecimens and data according to their permitted and restricted uses (Wilkinson et al., 2016). Semantic web technologies such as ontologies hold great promise as infrastructure solution for scalable, interoperable approaches in health care and research (Kock-Schoppenhauer et al., 2017). Ontologies structurally enable integration of heterogeneous data sources by semantically representing core entities and relationships of a given domain and have been widely successful in biomedical data sciences (Smith et al., 2007).
The Basic Formal Ontology (BFO) is a realism-based, upper level ontology comprised of terms that represent objects (i.e., continuants) and processes (i.e., occurrents) (Arp et al., 2015). This common structure is the basis for enforcing logical rules across all ontologies which import or refer to BFO, including those of the Open Biomedical Ontologies (OBO) Foundry. All OBO Foundry ontologies share core principles, including (1) use of a shared logical structure (i.e., BFO) as the source for common classes and relationships, (2) open access and use, (3) community-based collaborative development, (4) and non-overlapping, strictly scoped content. The OBO Foundry enforces these design principles to achieve scientific accuracy and semantic interoperability through a formal logical basis (Smith et al., 2007).
The Informed Consent Ontology (ICO) is part of the OBO Foundry and serves as a reference ontology for representing informed consent (He, 2015/2020; Lin et al., 2014). ICO was designed to represent documents and processes specifically relevant to biomedical and health research. ICO contains representations of processes such as signing an informed consent form and Institutional Review Board (IRB) approval of the consent form, as well as for the investigator and participant roles. Although ICO was not developed to represent the nuanced information specific to informed consent in health care processes, ICO offers the ability to model individuals’ decisions during a process of informed consent. While some of the existing classes may be transferable to this new domain, ICO must be formally evaluated for reuse in clinical consent processes and extended or revised as necessary.
2. OBJECTIVES
The purpose of this study was to evaluate ICO for its completeness in expressing clinical permissions, including the reuse of residual clinical biospecimens and health data. ICO was subsequently revised and extended to broaden the reference terminology artifact and to make interoperable representations of clinical permissions available.
3. METHODS
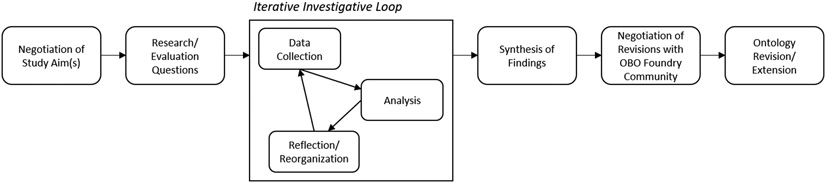
This study follows a formative evaluation design in which we examined an information resource (ICO) under development (Friedman & Wyatt, 2006). Figure 1 depicts our multistep evaluation process which includes: identification of study aim(s), development of research or evaluation questions, the iterative investigative loop – comprised of data collection, data analysis, and reflection and reorganization – synthesis of findings, community negotiation of revisions, and ontology revision and extension. Our analysis was also guided by evaluation methods and questions abstracted from the literature, including the National Institute of Standards and Technology’s (NIST) Ontology Summit’s guidance for evaluating ontologies across ontology life cycles (Neuhaus et al., 2014). Stakeholder involvement, including ICO developers and the OBO foundry community members, was solicited throughout the evaluation process.
Figure 1.
Multistep, Iterative Ontology Evaluation Process.
3.1. Identify Life Cycle Phase
Our evaluation focuses on adaptation of ICO to include a new domain. This aligns with Neuhaus et al.’s (2014) Ontology Development & Reuse Phase. Four evaluation tasks are recommended at this phase: informal modeling, formalization of competency questions, formal modeling, and operational adaptation (Neuhaus et al., 2014). Our analysis focuses on graphically modelling classes and relationships of content identified from relevant regulations and clinical consent forms through concept maps (i.e., informal modeling), followed by formal modeling within Web Ontology Language (OWL).
3.2. Determine Evaluation Aims and Questions
Evaluation largely depends on contribution and feedback from domain experts (Gelernter & Jha, 2016; Neuhaus et al., 2014). We developed evaluation aims and questions together with a community of ICO developers and OBO Foundry stakeholders. The questions guiding this evaluation were:
Does ICO contain the necessary classes and relationships to represent permissions from clinical consent forms?
Does ICO contain the necessary classes and relationships to represent permissions to reuse residual clinical biospecimens and health data, both from US federal regulations and clinical consent forms?
3.3. Iterative Investigative Loop
The Iterative Investigative Loop was comprised of three steps: data collection, analysis and modeling, and reflection and negotiation (Friedman & Wyatt, 2006). These steps were performed both iteratively and in tandem, enabling continuous revisiting of the data, revision of the models, and collaboration with stakeholders.
3.3.1. Data Collection
Data collected and analyzed for this evaluation study included permissions for clinical processes generally and reuse of residual clinical biospecimens and health data specifically. These permissions were abstracted from US federal regulations and clinical consent forms. First, permissions to reuse residual clinical biospecimens within US federal regulations were identified through a review of biomedical, legal, and health policy literature. The full methods and results of this review are described elsewhere (Umberfield, Kardia, et al., 2021). By identifying these permissions through the literature, we reduced introducing our own biases into ICO by instead extracting the interpretations of what is legally permissible by experts in their respective fields and incorporating a range of perspectives. The included regulations were:
The 21 st Century Cures Act
Clinical Laboratory Improvement Act, 1992
Health Insurance and Portability and Accountability Act, 1992
Protection of Human Subjects (i.e., Common Rule), 2019
Research and Investigations Generally (2011) (as cited in Public Health Services Act, 2019)
Second, an annotation study of clinical consent forms was conducted to identify permission-sentences. Permission-sentences are statements within the consent form that, when the form was signed by the patient or their legally authorized representative, permitted the health care facility or its clinicians to do some action or activity. Clinical consent forms were collected via direct contribution by health care facilities and systematic web searching, the methods of which are described elsewhere (Umberfield, Jiang, et al., 2021).
Permission-sentences were eligible for inclusion if they were positively identified by all three annotators of the consent forms. A sample of 15 permission-sentences were included in this evaluation. Four sentences were selected to for the heterogeneity of the activities they permitted (e.g. videotaping, surgery, anesthesia). Six permission-sentences were purposively selected because they explicitly permitted sharing or reuse of residual clinical biospecimens or health data. Five additional permission-sentences were randomly selected to reduce bias potentially introduced through purposive sampling. The sample was not further extended as the design pattern for permission-sentences (shown in Figure 2) remained stable when modeling the five randomly selected sentences.1
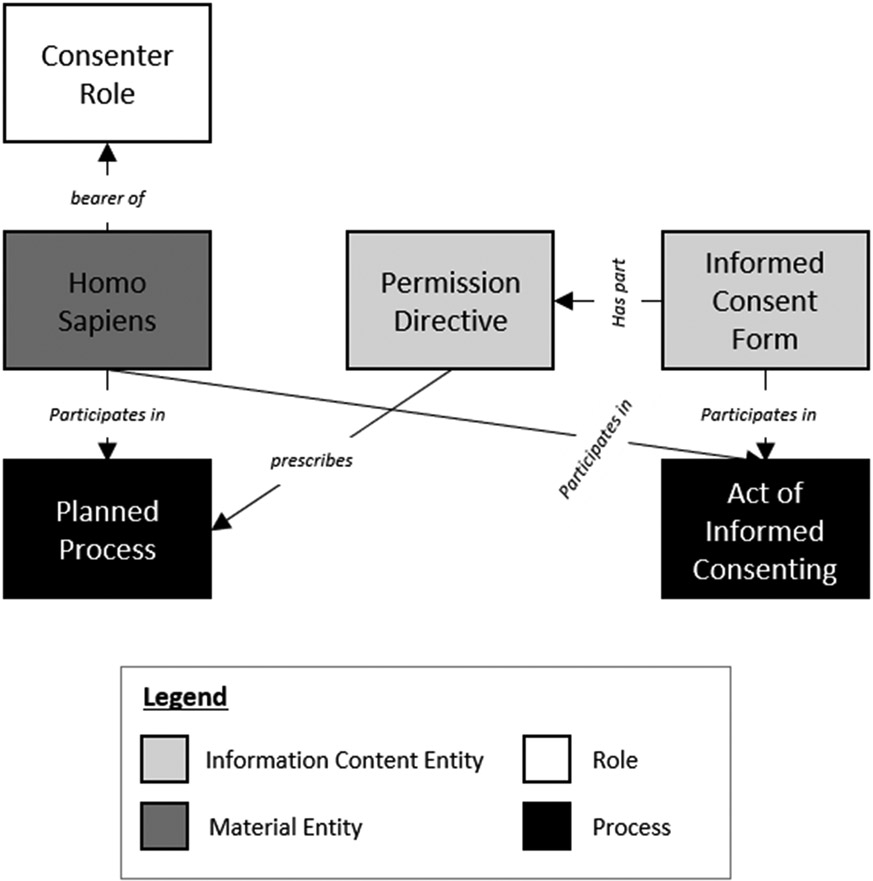
Figure 2.
Context for a Permission Directive (Permission-Sentence).
3.3.2. Analysis and Modeling
The goal of informal modeling is to identify all relevant ontological entities (i.e., classes and relationships), the entities’ important attributes, and appropriate terminology for the new domain. By informally modelling in concept maps, the models’ content is made understandable to both ontologists and domain experts (Neuhaus et al., 2014).
First, we identified all entities and their relationships within each permission from the source data. We then referenced key parent classes modeled in ICO – including information content entity, material entity, process, and role – and began sorting the identified entities into these categories without defining them or their hierarchies. At the same time, we graphically modeled the loosely defined classes from the consent forms using Mindjet MindManager and PowerPoint.2 We carefully examined ICO and OBO Foundry ontology classes in terms of their labels, textual definitions, and formal definitions for fidelity to their use in the context of US federal regulations and clinical consent forms. These steps were iteratively performed until a final model for each permission from the source data was developed and vetted by team members and collaborators.
3.3.3. Reflection and Negotiation
Ontology evaluation methods are not yet standardized. Therefore, we abstracted evaluation questions from the literature to guide verification (i.e., did we build it right?) and validation (i.e., did we build the right thing?) of ICO revisions and extensions. These reflection questions were used to guide negotiation during regular meetings with ICO team members and biweekly calls with members of the Ontology for Biobanking (OBIB) community. The list of evaluation questions includes:
Verification
Are the revisions adherent to OBO Foundry principles (OBO Foundry, 2020)?
Are classes consistent across the ontology’s hierarchy (Zhu et al., 2009)?
Are classes non-redundant (Zhu et al., 2009)?
Does the model capture only entities within the specified scope of the ontology (Neuhaus et al., 2014)?
Is the documentation sufficiently unambiguous to enable a consistent use of the terminology (Neuhaus et al., 2014)?
Validation
-
6.
Does the ontology contain the necessary and sufficient information (identified through the source data) to make it fit for our particular purpose? Are all entities within the scope of the ontology captured (Neuhaus et al., 2014; Zhu et al., 2009)?
-
7.
Do the domain experts agree with the ontological analysis (Neuhaus et al., 2014)?
3.3.4. Ontology Revision and Extension
ICO was revised and extended following all evaluation steps, synthesis of findings, and negotiations of proposed revisions with members of the OBO Foundry community. We categorize these revisions into three categories: (1) classes or relations within ICO that needed modification (2) classes or relations required for modeling clinical permissions, but whose content more appropriately belonged to the domains of other OBO Foundry ontologies, and (3) classes required for modeling clinical permissions that were not available in ICO or another OBO Foundry ontology. For the first category, we worked with ICO developers to revise the semantic labels, definitions, and the logical position of the entity within the ontology. Once completed, we performed automated artifact evaluation including lightweight reasoning using ROBOT, an open-source ontology tool (Jackson et al. 2019), to ensure structural consistency with OBO Foundry design principals. For the second category, we submitted requests to other OBO Foundry ontologies or worked directly with the developers to implement updates necessary to cover clinical permissions. Finally, for the third category, we added these classes to ICO’s import specification before running automated evaluation using ROBOT.
4. RESULTS
Prior to this evaluation, ICO contained 893 classes and 95 object properties (relations). Table 2 provides summary counts of all classes used in the modeling process, including those ICO classes that were transferable to this new domain and those that should either be imported or added to ICO or the OBO Foundry suite of ontologies for representing clinical permissions.
Table 2.
Summary Counts and Sources for Terms Used in Modelling
| Classes | Relationships | |
|---|---|---|
| Present in ICO | 52 | 12 |
| Recommend Import from | ||
| Other Ontologies | 26 | -- |
| New Classes | 12 | -- |
Modeling permissions abstracted from US federal regulations and clinical consent forms used 52 classes and 12 object properties already within ICO, demonstrating the appropriateness of extending ICO for the clinical domain rather than developing a new ontology. Five of these classes from ICO were flagged for revision. Revisions were regarding either the classes’ formal definitions (i.e., position within the hierarchy) or human-readable definitions, which were ambiguous or inaccurate.
Evaluation also revealed that extension of ICO is necessary to represent permissions from the source data. Twenty-six classes were recommended for import into ICO from other OBO Foundry ontologies. Additionally, we recommended twelve new classes to either be added to ICO or another OBO Foundry ontology in order to express all content within the included source data. Appendix A includes three tables, each summarizing:
A1) terms used that were already present in ICO,
A2) terms used that were not present in ICO but were identified in another OBO Foundry ontology (recommended for import), and
A3) new terms which were not identified in ICO or another OBO Foundry Ontology.
Following these changes, ICO contains 20 new classes or relations via import and 9 de novo ICO classes. Remaining classes not imported required further discussion and are documented as requests. In total ICO contains 1000 classes and 93 relations. Table 3 provides the ontology abbreviations used throughout this paper as well as the full PURL for that ontology. Appendix B contains supplementary tables reporting the respective classes and relationships used for each of the fifteen permission-sentences from clinical consent forms and mapping classes to sentence content.
Table 3.
Summary of source ontologies of classes in ICO, their abbreviations, PURLs, and citations. Abbreviations are Compact URIs (CURIE) that shorten the full resource identifier for terms in a given ontology. For example, ‘ICO:’ refers to the prefix ‘http://purl.obolibrary.org/obo/ICO_’ for classes and relations defined in the Informed Consent Ontology at http://purl.obolibrary.org/obo/ico.owl.
Early in the modeling process, a design pattern for the context of permission-sentences emerged. Figure 2 is the informal model of this base design pattern: The individual who is granting permission (‘homo sapiens’ (NCBITaxon:9606); ‘consenter role’ (ICO:0000086)) participates in a process of consenting (‘informed consent process’ (OBI:0000810)) by using a consent form (‘informed consent form’ (ICO:0000001)) which contains a permission-sentence (‘permission directive’ (ICO:0000244)) that prescribes some process (‘planned process’ (OBI:0000011)). It should be noted that this is the most simplified version of this design pattern, and there is significant heterogeneity and added complexity as each permission-sentence was modeled. As an example, the real-world person (i.e., instance) who is the consenter may also have a range of other important roles including being the patient (‘patient role’ (OBI:0000093)) or the patient’s legally authorized representative (‘legal guardian role’ (OMRSE:00000038)). Likewise, the processes that are prescribed by permission-sentences also varied widely, but most often included a ‘health care process’ (OGMS:0000096) like surgery or blood product administration, a ‘specimen collection process’ (OBI:0000659), or an ‘act of data sharing’ (ICO:0000228).
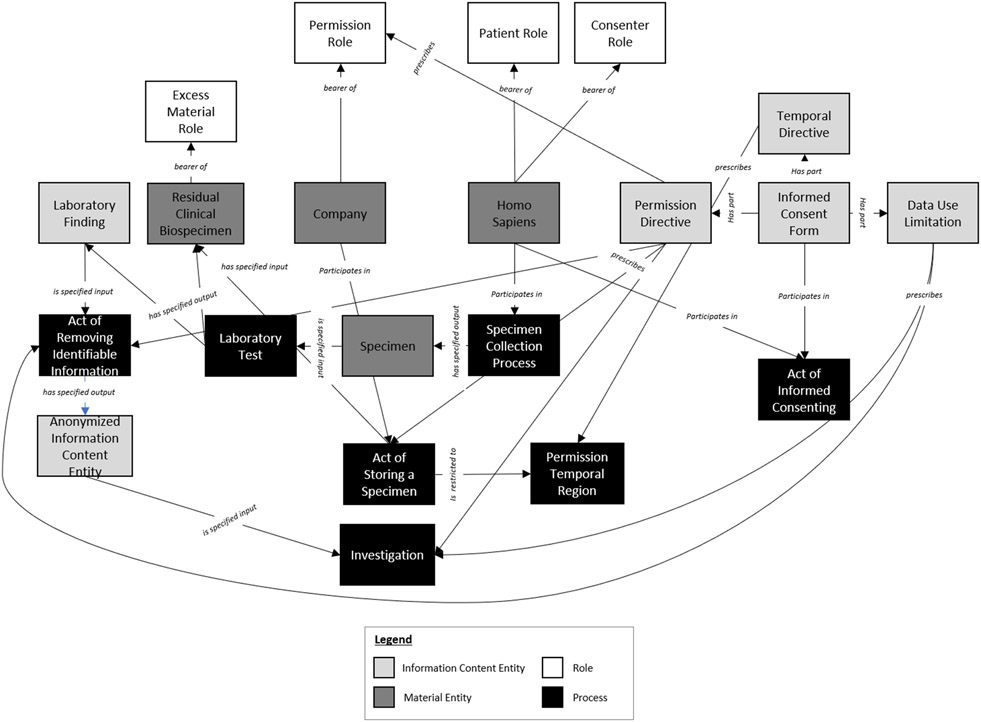
As the permission-sentences became more complex, so too did their graphic models. Figure 3 demonstrates this complexity. In this example, not only was some process prescribed, but also specifications on how the data that emerged from that process may be used (‘data use limitation’(DUO:0000001)) and the timeline by which these processes may occur (‘temporal restriction directive’ (new class, ICO:0000363)). Additionally, the flow of a given biospecimen becoming a residual clinical biospecimen which bears an ‘excess material role’(ICO:0000313) was fleshed out to ensure that all necessary classes were modeled.
Figure 3.
Graphic Model for the following permission sentence: “…I give permission for GeneDx to retain any remaining sample longer than 60 days after completion of testing and use my de-identified data for scientific and medical research purposes.”
As the source data were modeled and reviewed with team members, we asked the following evaluation questions to evaluate the revisions and extensions of ICO in terms of verification and validation:
4.1. Verification
1. Are the revisions adherent to OBO Foundry principles (OBO Foundry, 2020)?
OBO Foundry principles are centered on openness and reusability to a range of users and applications, and facilitating reuse through shared best practices such as common formats, clear definitions, community collaborations, and regular maintenance of the ontologies (OBO Foundry, 2020). ICO is released under a Creative Commons 3.0 license and written in OWL. The revisions from this evaluation do not affect ICO’s adherence to OBO Foundry principles, and any added classes will follow naming, defining, and documentation requirements.
2. Are classes consistent across the ontology’s hierarchy (Zhu et al., 2009)?
We aimed to appropriately map new classes at the same level of granularity as existing classes across the ontology. Three of the five revisions to existing ICO classes revolved around editing the location within the class hierarchy.
3. Are classes non-redundant (Zhu et al., 2009)?
During the informal modeling phase, we attempted to use existing ICO classes prior to suggesting extension of the ontology by adding classes. We also searched Ontobee (Ong et al., 2017) and Ontology Lookup Service (Côté et al., 2006) to identify classes and their definitions (formal and textual) that could be imported into ICO. The shared logical structure and hierarchy of classes in the OBO Foundry facilitates interoperability and ease of importing individual classes or entire branches of the ontologies’ trees.
4. Does the model capture only entities within the specified scope of the ontology (Neuhaus et al., 2014)?
Permissions from the source data and their relevant entities largely included terms specific to consent processes, their forms, and the individuals involved in the consent process; however, they also included entities relevant outside of consent processes, including health care processes such as surgeries, organizations such hospitals, and processes like the act of selling or owning some material. For this reason, negotiation with representatives from a range of OBO Foundry ontologies is ongoing to determine where this content fits best. As an example, efforts have already been completed to add and define the term ‘owner role’ in The Document Acts Ontology (D-Acts; Ceusters, 2012). Even though ownership was identified as a necessary concept, ownership extends beyond the context of informed consent (e.g., deeds and land ownership) which merits modeling in a broader reference ontology that provides content inherited by ‘downstream’ ontologies, including but not limited to ICO.
5. Is the documentation sufficiently unambiguous to enable a consistent use of the terminology (Neuhaus et al., 2014)?
Text definitions for all newly added terms were proposed based on existing and credible sources. Additionally, these text definitions have been double-checked to ensure that they match the terms’ formal definitions.
4.2. Validation
1. Does the ontology contain the necessary and sufficient info (identified through the source data) to make it fit for our particular purpose? Are all entities within the scope of the ontology captured (Neuhaus et al., 2014; Zhu et al., 2009)?
Our purpose was to represent permissions relevant to clinical contexts, including reuse of residual clinical biospecimens and health data for secondary purposes. By systematically extracting entities from all relevant US federal regulations and a sample of permission-sentences from clinical consent forms, we aimed to represent all necessary and sufficient classes for our purposes in ICO. After modeling a few permission-sentences, a distinct and consistent design pattern emerged for representing the context of permission-sentences (see Figure 2). By following our progression of modeling permission-sentences – from those purposively sampled for our most narrow use case, then purposively selected for heterogenous clinical permissions, and lastly a random sample of permission-sentences – we achieved saturation when identifying new classes, which demonstrates that a sufficient and representative sample of permission-sentences were modeled to capture most relevant classes.
2. Do the domain experts agree with the ontological analysis (Neuhaus et al., 2014)?
We presented our questions about modeling decisions and tentative models to the ICO development team and OBIB community during their weekly and biweekly meetings respectively. Graphic models were iteratively refined and presented back to these communities until members agreed that further changes were not needed. Likewise, we brought lists of new and revised classes to these meetings. We adopted their suggestions for class labels, formal definitions, textual definitions, and which OBO Foundry ontology each class should be placed in.
5. DISCUSSION
This evaluation revealed substantial overlap and therefore appropriateness of using ICO to represent permissions expressed in clinical consent forms. It further identified gaps and inconsistencies for representing such permissions and summarizes necessary extensions and revisions of ICO and other OBO Foundry ontologies. In their study to identify the minimum metadata necessary for biobank information systems to share their samples and data, Norlin et al. (2012) recognized the need for ontologies which represent the “ethical standards under which the samples are collected, any restrictions on research use, and access requirements to the samples” (p. 344). This work addresses a critical gap in formally representing how residual clinical biospecimens and health data can be responsibly reused, which is of increasing value in data-intensive health sciences (e.g., population health, translational science, precision health, etc.) which require access to such information. This work produces a machine-readable semantic resource, grounded in real-world sources included regulations and clinical consent forms. It further demonstrates the importance of continued development of reference terminologies to solve real-world problems.
One important feature of this work is demonstration of a bottom-up modeling approach to evaluate and extend an existing information resource using real-world source data. Such an approach has historically been used to map out the structure of a domain as a step towards developing information infrastructure (Harris et al., 2015; Schleyer et al., 2007). It enables discovery of the information needs of users (Shang et al., 2018) and – by fitting such modeling within an existing top-level structure – builds out content that is interoperable with existing, in-use classes and promotes infrastructure reuse (Freimuth et al., 2012). By using real world source data (regulations and consent forms), we were able to identify content coverage gaps in ICO and other OBO Foundry ontologies that may not have been otherwise identifiable. It also adds this missing content to the suite of ontologies, enabling use alongside widely adopted biomedical ontologies and acting as a valuable contribution for biobank information management.
It is our hope that ICO will serve as a reference terminology to facilitate responsible reuse of residual clinical biospecimens and health data, by both increasing their discoverability by entities that will advance scientific knowledge related to health and also protecting the agency and rights of patients, whose expressed choices regarding the disposition of their biospecimens and health data should be respected. Revising and extending ICO to include representation of this new domain enables future system interoperability of permissions and obligations for sharing and use which can be adopted by a range of systems and applications. As examples, ICO may also be used for assessing informed consent tools such as eConsent forms and as a resource for mapping and annotating text in future natural language processing tasks. It may also be used to build query tools or decision support systems to support covered entities, biorepositories, federated research networks, institutional review boards, and other individuals in identifying eligible biospecimen and data resources that meet their needs or deciding if and when certain biospecimen and data resources should be shared.
Ontology evaluation is a well-recognized challenge. Despite more than a decade of publication on the issue, there remains no standard methodology for ontology evaluation (Burton-Jones et al., 2005; Gelernter & Jha, 2016). Among the criteria by which ontologies are evaluated are “its coverage of a particular domain and the richness, complexity, and granularity of that coverage; the specific use cases, scenarios, requirements, applications, and data sources it was developed to address; and formal properties such as the consistency and completeness of the ontology and the representation language in which it is modeled” (Obrst et al., 2007). Ontology evaluation is underutilized, leading to the release of poor ontologies and ultimately hindering “the successful deployment of ontologies as a technology” (Neuhaus et al., 2014). Currently, systematic evaluation of ontologies requires that evaluators assemble methods from across various evaluation schemas. One strength of this study is its systematic evaluation of ICO, including both verification and validation.
Importantly, our evaluation and modeling process provides a roadmap for improving and expanding domain knowledge within ontologies, and addressing knowledge-representation gaps that hinder their successful uptake (Harris et al., 2015). This role is particularly well-suited to clinical research informaticists, an emerging and recognized specialty that leverages informatics to discover and manage new knowledge relating to health and disease and their use in research (American Medical Informatics Association, 2020). We also wish to amplify the need for collaboration and transparency in ontology development. There are hundreds to thousands of ontologies across repositories, but these ontologies are not necessarily interoperable. It is only with a shared semantic structure and collaborative negotiation of ontology structures that dynamic and changing knowledge can successfully be modeled in interoperable families of ontologies. This work must occur in open and collaborative communities to achieve transparency in knowledge representation.
Several limitations of this evaluation are recognized. First, deliberation with other OBO Foundry ontology communities is ongoing to determine the most appropriate home for some of the new terms which may be out of scope for ICO. Citations for correspondences regarding proposed and actual changes to ICO and other OBO Foundry ontologies are listed in Appendix C. Second, while we have worked with ontologists and clinical research informaticists, we have not yet presented our models to domain experts from the legal and compliance, health information management, or biobanking domains. However, the extensive literature review and systematic collection and analysis of clinical consent forms mitigates this limitation; the literature and approved consent forms are a form of the collective voice of domain experts. Future refinements will include further checking with experts to ensure we have correctly interpreted the literature and the permissions.
6. CONCLUSION
Representing permissions to reuse residual clinical biospecimens and health data in an information resource that is interoperable within a family of recognized biomedical ontologies is valuable for facilitating the responsible reuse of these resources at scale. By evaluating and extending ICO, we make a meaningful step in this direction. Our methods demonstrate the use of a bottom-up approach to modeling content from their respective domains’ perspectives. We propose such methods as a valuable way for clinical research informaticists and domain experts to engage in the development and revision of semantic information resources.
Supplementary Material
Table 1.
List of Permission-Sentences from Clinical Consent Forms which were included in this Evaluation. Note: By agreement with recruited facilities, we obscured facility names whose consent forms were not publicly available using “XXX”. For all facilities whose consent forms were publicly available, we did not remove identifiers.
| Purposively Sampled Permission-Sentences | |
|---|---|
| Reuse of Residual Clinical Biospecimens and/or Health Data | |
| 1. | I hereby authorize XXX to retain, preserve and use for scientific or teaching purposes, or to dispose at its discretion or convenience, any specimen or tissues taken from my body during my visit. |
| 2. | I am a New York state resident and I give permission for GeneDx to retain any remaining sample longer than 60 days after completion of testing and use my de-identified data for scientific and medical research purposes. |
| 3. | I DONATE and authorize XXX to own, use, retain, preserve, manipulate, analyze, or dispose of any excess tissues, specimens, or parts of organs that are removed from my body during the procedures described above and are not necessary for my diagnosis or treatment. |
| 4. | I agree that any excess tissue, fluids or specimens removed from my body during my outpatient visit or hospital stay ( my specimens ) that would otherwise be disposed of by the Hospital may be used for such educational purposes and research, including research on the genetic materials (DNA). |
| 5. | I authorize the pathologist, at his or her discretion, to retain, preserve, use, or dispose of any tissues, organs, bones, bodily fluid or medical devices that may be removed during the operation(s) or procedure(s). |
| 6. | I hereby consent to the use and disclosure of my protected health information as described in the Notice of Privacy Practices. |
| Other Clinical Procedures and Activities | |
| 7. | By signing this form, I am requesting and giving my consent for MHSM and the doctors and/or nurses to give me blood and/o [sic] blood products during this admission or series of treatments. |
| 8. | Your signature below indicates that you understand to your satisfaction the information about the genetic testing ordered by your health care provider and that you consent to having this testing performed. |
| 9. | I consent to the Facility videotaping, photographing, video monitoring, or taking other recordings of me or parts of my body for diagnosis, treatment, research, or patient safety purposes. |
| 10. | I also consent to diagnostic studies, tests, anesthesia, x-ray examinations and any other treatment or courses of treatment relating to the diagnosis or procedure described herein. |
| Randomly Sampled Permission-Sentences | |
| 11. | I, , request and consent to the start or induction of my labor by my provider: and other assistants as may be selected by him/her. |
| 12. | I voluntarily consent to receive medical and health care services that may include diagnostic procedures, examination, and treatment. |
| 13. | I consent to the use of closed-circuit television, taking of photographs (including videos), and the preparation of drawings and similar illustrative graphic material for scientific purposes providing my identity is not revealed. |
| 14. | I hereby consent to engaging in virtual health/telemedicine services, where available, as part of my treatment. |
| 15. | In the event a healthcare worker is exposed to my blood or body fluids in connection with my procedure, or during my hospital stay, I agree to the collection and testing of my blood for HIV. |
9. ACKNOWLEDGEMENTS
The authors would like to thank members of the Ontology for Biobanking (OBIB) community for their support and feedback throughout the evaluation process, including but not limited to Chris Stoeckert, Jihad Obeid, Frank Manion, and Mathias Brochhausen, and Sarah Bost.
11. FUNDING STATEMENT
EU was supported in part by the Robert Wood Johnson Foundation Future of Nursing Scholar’s Program predoctoral training program. The study was further supported by the National Human Genome Research Institute of the National Institutes of Health under award number U01HG009454, the Rackham Graduate Student Research Grant, and the University of Michigan Institute for Data Science. EU is presently funded as a Postdoctoral Research Fellow in Public & Population Health Informatics at Fairbanks School of Public Health and Regenstrief Institute, supported by the National Library of Medicine of the National Institutes of Health under award number T15LM012502. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the Robert Wood Johnson Foundation, the National Institutes of Health, the University of Michigan, Indiana University, or Regenstrief Institute.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests in the research.
While these sentences pointed to a range of processes (e.g., telemedicine, induction of labor), all processes fit under the umbrella class of ‘health care process’ (OGMS:0000096) or its parent, ‘planned process’ (OBI:0000011). Similarly, although a new role, ‘Pathologist role’ (OBI:0000145) further specified who might also bear the ‘permission role’ (ICO:0000199). The list of all permitted processes and all roles of those granted permission in all instances of consent forms is innumerable. Inclusion of these processes and roles is more apt in application ontology, rather than in ICO or other reference ontology. All permission-sentences included in this analysis are listed in Table 1.
For regulations, we focused just on identifying classes rather than graphically modeling their relationships, as there is not yet a legal ontology in the OBO Foundry. While important, a formal legal ontology is out of scope for the present work and is beyond the domain scope of ICO.
REFERENCES
- 21st Century Cures Act, Pub. L. No. 114-255, 130 Stat. 1049 (2016).
- American Medical Informatics Association. (2020). Clinical Research Informatics. AMIA. https://www.amia.org/applications-informatics/clinical-research-informatics [Google Scholar]
- Americans with Disabilities Act of 1990, 42 U.S.C. §12111 et seq.
- An Overview of the Common Core Ontologies. (2019). http://paper/An-Overview-of-the-Common-Core-Ontologies/ecac54098980b853f7558572247336084f6fdf3b
- Arp R, Smith B, & Spear AD (2015). Building Ontologies with Basic Formal Ontology. MIT Press. [Google Scholar]
- Ashburner M, & Schriml LM (2020). Gazetteer. http://purl.obolibrary.org/obo/gaz.owl [Google Scholar]
- Balhoff J (2017). Tailoring the NCI Thesaurus for Use in The OBO Library. ICBO. https://compasify.com/paper/Tailoring-the-NCI-Thesaurus-for-Use-in-The-OBO-Library.p-2b07a50776436851e72692a58c90c8e2727f356a [Google Scholar]
- Bandrowski A, Brinkman R, Brochhausen M, Brush MH, Bug B, Chibucos MC, Clancy K, Courtot M, Derom D, Dumontier M, Fan L, Fostel J, Fragoso G, Gibson F, Gonzalez-Beltran A, Haendel MA, He Y, Heiskanen M, Hernandez-Boussard T, … Zheng J (2016). The Ontology for Biomedical Investigations. PloS One, 11(4), e0154556. 10.1371/journal.pone.0154556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor C (2020). Chemical Methods Ontology. http://purl.obolibrary.org/obo/chmo.owl [Google Scholar]
- Brochhausen M, Zheng J, Birtwell D, Williams H, Masci AM, Ellis HJ, & Stoeckert CJ (2016). OBIB-a novel ontology for biobanking. Journal of Biomedical Semantics, 7(1), 23. 10.1186/s13326-016-0068-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton-Jones A, Storey VC, Sugumaran V, & Ahluwalia P (2005). A semiotic metrics suite for assessing the quality of ontologies. Data & Knowledge Engineering, 55(1), 84–102. 10.1016/j.datak.2004.11.010 [DOI] [Google Scholar]
- Buttigieg PL, Jensen M, Walls RL, & Mungall CJ (2016). Environmental semantics for sustainable development in an interconnected biosphere. International Conference on Biomedical Ontology and BioCreative (ICBO BioCreative 2016). http://ceur-ws.org/Vol-1747/IT201_ICBO2016.pdf [Google Scholar]
- Ceusters W (2012). An information artifact ontology perspective on data collections and associated representational artifacts. Studies in Health Technology and Informatics, 180, 68–72. [PubMed] [Google Scholar]
- Ceusters W, & Smith B (2015). Aboutness: Towards Foundations for the Information Artifact Ontology. In Proceedings of the Sixth International Conference on Biomedical Ontology (ICBO) (pp. 1–5). CEUR; vol. 1515. [Google Scholar]
- Clinical Laboratory Improvement Amendments, 42 U.S.C. §493 (1992).
- Côté RG, Jones P, Apweiler R, & Hermjakob H (2006). The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics, 7(1), 97. 10.1186/1471-2105-7-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, & Steinbeck C (2010). Chemical Entities of Biological Interest: An update. Nucleic Acids Research, 38(suppl_1), D249–D254. 10.1093/nar/gkp886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AD, Meehan TF, Bradford YM, Brush MH, Dahdul WM, Dougall DS, He Y, Osumi-Sutherland D, Ruttenberg A, Sarntivijai S, Van Slyke CE, Vasilevsky NA, Haendel MA, Blake JA, & Mungall CJ (2016). The Cell Ontology 2016: Enhanced content, modularization, and ontology interoperability. Journal of Biomedical Semantics, 7(1), 44. 10.1186/s13326-016-0088-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontier M, Baker CJ, Baran J, Callahan A, Chepelev L, Cruz-Toledo J, Del Rio NR, Duck G, Furlong LI, Keath N, Klassen D, McCusker JP, Queralt-Rosinach N, Samwald M, Villanueva-Rosales N, Wilkinson MD, & Hoehndorf R (2014). The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery. Journal of Biomedical Semantics, 5(1), 14. 10.1186/2041-1480-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke SOM, Philippakis AA, Rambla De Argila J, Paltoo DN, Luetkemeier ES, Knoppers BM, Brookes AJ, Spalding JD, Thompson M, Roos M, Boycott KM, Brudno M, Hurles M, Rehm HL, Matern A, Fiume M, & Sherry ST (2016). Consent Codes: Upholding Standard Data Use Conditions. PLOS Genetics, 12(1), e1005772. 10.1371/journal.pgen.1005772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund N, Andrianarisoa NH, van Enckevort E, Anton G, Debucquoy A, Müller H, Zaharenko L, Engels C, Ebert L, Neumann M, Geeraert J, T’Joen V, Demski H, Caboux É, Proynova R, Parodi B, Mate S, van Iperen E, Merino-Martinez R, … Silander K (2020). Extending the Minimum Information About BIobank Data Sharing Terminology to Describe Samples, Sample Donors, and Events. Biopreservation and Biobanking, 18(3), 155–164. 10.1089/bio.2019.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S (2012). The NCBI Taxonomy database. Nucleic Acids Research, 40(Database issue), D136–143. 10.1093/nar/gkr1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth RR, Freund ET, Schick L, Sharma MK, Stafford GA, Suzek BE, Hernandez J, Hipp J, Kelley JM, Rokicki K, Pan S, Buckler A, Stokes TH, Fernandez A, Fore I, Buetow KH, & Klemm JD (2012). Life sciences domain analysis model. Journal of the American Medical Informatics Association, 19(6), 1095–1102. 10.1136/amiajnl-2011-000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CP, & Wyatt JC (2006). Evaluation as a Field. In Evaluation Methods in Medical Informatics (2nd ed.). Springer-Verlag. 10.1007/978-1-4757-2685-5 [DOI] [Google Scholar]
- Food and Drug Administration Amendments Act of 2007, Pub. L. No. 110-85, 121 Stat. 823 (2007).
- Gelernter J, & Jha J (2016). Challenges in Ontology Evaluation. Journal of Data and Information Quality, 7(3), 1–4. 10.1145/2935751 [DOI] [Google Scholar]
- The Gene Ontology Consortium. (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research, 47(D1), D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Information Nondiscrimination Act of 2008, Pub. L. No. 110-223, 122 Stat. 881 (2008). [PubMed]
- Gkoutos GV, Schofield PN, & Hoehndorf R (2012). The Units Ontology: A tool for integrating units of measurement in science. Database: The Journal of Biological Databases and Curation, 2012, bas033. 10.1093/database/bas033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MR, Langford LH, Miller H, Hook M, Dykes PC, & Matney SA (2015). Harmonizing and extending standards from a domain-specific and bottom-up approach: An example from development through use in clinical applications. Journal of the American Medical Informatics Association: JAMIA, 22(3), 545–552. 10.1093/jamia/ocu020 [DOI] [PubMed] [Google Scholar]
- He Y (2020). Informed Consent Ontology [OWL]. ICO-ontology. https://github.com/ICO-ontology/ICO (Original work published 2015) [Google Scholar]
- He Y, Sarntivijai S, Lin Y, Xiang Z, Guo A, Zhang S, Jagannathan D, Toldo L, Tao C, & Smith B (2014). OAE: The Ontology of Adverse Events. Journal of Biomedical Semantics, 5, 29. 10.1186/2041-1480-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Insurance Portability and Accountability Act of 1996, Pub. L. No. 104-191, 110 Stat. 1936 (1996). [PubMed]
- Hicks A, Hanna J, Welch D, Brochhausen M, & Hogan WR (2016). The ontology of medically related social entities: Recent developments. Journal of Biomedical Semantics, 7(1), 47. 10.1186/s13326-016-0087-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan WR, Diller M, & Hicks A (2016). Graphical Entity Ontology. http://purl.obolibrary.org/obo/geo.owl [Google Scholar]
- Huntley RP, Harris MA, Alam-Faruque Y, Blake JA, Carbon S, Dietze H, Dimmer EC, Foulger RE, Hill DP, Khodiyar VK, Lock A, Lomax J, Lovering RC, Mutowo-Meullenet P, Sawford T, Van Auken K, Wood V, & Mungall CJ (2014). A method for increasing expressivity of Gene Ontology annotations using a compositional approach. BMC Bioinformatics, 15(1), 155. 10.1186/1471-2105-15-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, & Schriml LM (2015). Disease Ontology 2015 update: An expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Research, 43(Database issue), D1071–1078. 10.1093/nar/gku1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock-Schoppenhauer A-K, Kamann C, Ulrich H, Duhm-Harbeck P, & Ingenerf J (2017). Linked Data Applications Through Ontology Based Data Access in Clinical Research. Studies in Health Technology and Informatics, 131–135. 10.3233/978-1-61499-753-5-131 [DOI] [PubMed] [Google Scholar]
- Lin Y, Harris MR, Manion FJ, Eisenhauer E, Zhao B, Shi W, Karnovsky A, & He Y (2014). Development of a BFO-based Informed Consent Ontology (ICO). ICBO Conference Proceedings. International Conference on Biomedical Ontology. [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, & Haendel MA (2012). Uberon, an integrative multi-species anatomy ontology. Genome Biology, 13(1), R5. 10.1186/gb-2012-13-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Vasilevsky N, Matentzoglu N, Osumi-Sutherland D, Dahdul W, Balhoff J, Robb S, Haendel M, Buttigieg PL, Köhler S, Clare72, Harris M, Meier A, Hoehndorf Robert, euniceyi, Gkoutos G, Walls R, & Bellow S (2020). pato-ontology/pato: V2020-08-02. 10.5281/zenodo.3970080 [DOI] [Google Scholar]
- Neuhaus F, Ray S, & Sriram RD (2014). Toward ontology evaluation across the life cycle (NIST IR 8008). National Institute of Standards and Technology. [Google Scholar]
- Norlin L, Fransson MN, Eriksson M, Merino-Martinez R, Anderberg M, Kurtovic S, & Litton J-E (2012). A Minimum Data Set for Sharing Biobank Samples, Information, and Data: MIABIS. Biopreservation and Biobanking, 10(4), 343–348. 10.1089/bio.2012.0003 [DOI] [PubMed] [Google Scholar]
- Noy NF, & Rubin DL (2008). Translating the Foundational Model of Anatomy into OWL. Web Semantics (Online), 6(2), 133–136. 10.1016/j.websem.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OBO Foundry. (2020). Principles: Overview. The OBO Foundry. http://www.obofoundry.org/principles/fp-000-summary.html [Google Scholar]
- Obrst L, Ceusters W, Mani I, Ray S, & Smith B (2007). The Evaluation of Ontologies. In Baker CJO & Cheung K-H (Eds.), Semantic Web (pp. 139–158). Springer US. 10.1007/978-0-387-48438-9_8 [DOI] [Google Scholar]
- Ong E, Xiang Z, Zhao B, Liu Y, Lin Y, Zheng J, Mungall CJ, Courtot M, Ruttenberg A, & He Y (2017). Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Research, 45(D1), D347–D352. 10.1093/nar/gkw918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient Protection and Affordable Care Act, Pub. L. No. 111-148, 124 Stat. 119 (2010) (codified as amended in scattered sections of 21, 25, 26, 28, 29, 36, and 42 U.S.C.).
- Protection of Human Subjects, 45 C.F.R. § 46 (2019).
- Public Health Services Act § 301, 42 U.S.C. § 241 (2019).
- Rehabilitation Act of 1973, Pub. L. No. 93-112, 87 Stat. 355 (1973).
- Scheuermann RH, Ceusters W, & Smith B (2009). Toward an Ontological Treatment of Disease and Diagnosis. Summit on Translational Bioinformatics, 2009, 116–120. [PMC free article] [PubMed] [Google Scholar]
- Schleyer T, Spallek H, & Hernández P (2007). A Qualitative Investigation of the Content of Dental Paper-based and Computer-based Patient Record Formats. Journal of the American Medical Informatics Association, 14(4), 515–526. 10.1197/jamia.M2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang N, Weng C, & Hripcsak G (2018). A conceptual framework for evaluating data suitability for observational studies. Journal of the American Medical Informatics Association, 25(3), 248–258. 10.1093/jamia/ocx095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, The OBI Consortium, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, & Lewis S (2007). The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology, 25(11), 1251–1255. 10.1038/nbt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Joint Commission, Division of Healthcare Improvement. (2016). Informed Consent: More than getting a Signature (Issue 21; Quick Safety). The Joint Commission. https://www.jointcommission.org/-/media/deprecated-unorganized/imported-assets/tjc/system-folders/joint-commission-online/quick_safety_issue_twenty-one_february_2016pdf.pdf?db=web&hash=5944307ED39088503A008A70D2C768AA [Google Scholar]
- Torniai C, Brush M, Vasilevsky N, Segerdell E, Wilson M, Johnson T, Corday K, Shaffer C, & Haendel M (2011). Developing an application ontology for biomedical resource annotation and retrieval: Challenges and lessons learned. CEUR Workshop Proceedings, 833, 101–108. https://ohsu.pure.elsevier.com/en/publications/developing-an-application-ontology-for-biomedical-resource-annota-2 [Google Scholar]
- Umberfield EE, Jiang Y, Fenton SH, Stansbury C, Ford K, Crist K, Kardia SLR, Thomer AK, & Harris MR (2021). Lessons Learned for Identifying and Annotating Permissions in Clinical Consent Forms. Applied Clinical Informatics, 12(3), 429–435. 10.1055/s-0041-1730032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umberfield EE, Kardia SLR, Jiang Y, Thomer AK, & Harris MR (2021). Regulations and Norms for Reuse of Residual Clinical Biospecimens and Health Data. Western Journal of Nursing Research. 10.1177/01939459211029296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utecht J, Ball J, Bowman SM, Dodd J, Judkins J, Maxson RT, Nabaweesi R, Pradhan R, Sanddal ND, Winchell RJ, & Brochhausen M (2019). Development and Validation of a Controlled Vocabulary: An OWL Representation of Organizational Structures of Trauma Centers and Trauma Systems. Studies in Health Technology and Informatics, 264, 403–407. 10.3233/SHTI190252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg Ij. J., Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, … Mons B (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3, 160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Fan J-W, Baorto DM, Weng C, & Cimino JJ (2009). A review of auditing methods applied to the content of controlled biomedical terminologies. Journal of Biomedical Informatics, 42(3), 413–425. 10.1016/j.jbi.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.