Abstract
Background
Researchers have long investigated a hypothesized interaction between genetic risk and stressful life events in the etiology of depression, but studies on the topic have yielded inconsistent results.
Methods
We conducted a genome-wide by environment interaction study (GWEIS) in 18,532 patients with depression from hospital-based settings and 20,184 population controls. All individuals were drawn from the iPSYCH2012 case-cohort study, a nationally representative sample identified from Danish national registers. Information on stressful life events including family disruption, serious medical illness, death of a first-degree relative, parental disability, and child maltreatment was identified from the registers and operationalized as a time-varying count variable. Hazard ratios for main and interaction effects were estimated using Cox regressions weighted to accommodate the case-cohort design. Our replication sample included 22,880 depression cases and 50,378 controls from the UK Biobank.
Results
The GWEIS in the iPSYCH2012 sample yielded three novel, genome-wide–significant (p < 5 × 10−8) loci located in the ABCC1 gene (rs56076205, p = 3.7 × 10−10), the AKAP6 gene (rs3784187, p = 1.2 × 10−8), and near the MFSD1 gene (rs340315, p = 4.5 × 10−8). No hits replicated in the UK Biobank (rs56076205: p = .87; rs3784187: p = .93; rs340315: p = .71).
Conclusions
In this large, population-based GWEIS, we did not find any replicable hits for interaction. Future gene-by-stress research in depression should focus on establishing even larger collaborative GWEISs to attain sufficient power.
Keywords: Case-cohort studies, Depression, Genetics, GXE, Register-based research, Stress
Major depression is a common, highly burdensome mental illness that effects as many as 21% of people at some point during their lifetimes (1,2). Studies suggest that major depression is around 30% to 40% heritable (3), meaning that a moderate amount of the population-level variability in major depression can be attributed to genetic factors. However, environment also plays an important role in determining who develops major depression and who does not. In particular, experiencing a stressful life event (SLE) in childhood or adulthood has been shown to increase depression risk (4,5). SLEs include childhood physical, sexual, or emotional abuse; death of a relative; severe illness; divorce or separation; economic deprivation; and forced exit from the workforce. Events can cause stress if they occur to the individual (e.g., child abuse, divorce, severe illness) or if they happen to a close relative, particularly during childhood (e.g., divorce or severe illness in a parent). Each of these events has been shown to be associated with increased risk for depression (6,7); however, the cumulative burden of stress is particularly relevant for determining depression risk. Studies have consistently shown that as the number of SLEs increases, risk for depression also increases, and individuals with over four SLEs experience depression risks 3 to 5 times those of individuals with no SLEs (6,8,9).
Historically, there has been great interest in the possibility of an interaction between SLEs and genetic liability as risk factors for depression. Such an interaction, if present, not only could lead to a better understanding of the underlying etiology of depression, but also could potentially be useful for identifying individuals at particularly high risk for developing depression. An early twin study (10) found that risk for depression after an SLE was only elevated among individuals with high genetic liability. Subsequently, researchers selected candidate genes that they believed were associated with depression risk and examined whether variants in these genes interacted with SLEs to predict depression (11, 12, 13, 14, 15, 16, 17, 18, 19). These studies yielded inconsistent results, with even meta-analyses reaching different conclusions regarding the validity of the associations (20, 21, 22, 23, 24, 25, 26, 27). Research examining the interaction between polygenic risk scores and SLEs has also yielded inconsistent results, with some finding evidence for interaction (28, 29, 30) and some failing to do so (29,31, 32, 33).
The hypothesis-driven (i.e., candidate gene) approach for identifying specific variants associated with a given outcome has not been successful in psychiatric research (34, 35, 36). This has led to the embrace of the genome-wide-association study (GWAS) as a method for identifying variants associated with psychiatric disorders in a theoretically agnostic fashion. In a GWAS, single nucleotide polymorphisms (SNPs) in sufficient linkage disequilibrium to tag the entire genome are tested for association with the outcome of interest. Significance is evaluated based on an adjusted alpha level to avoid false positive results. This method has been highly successful in psychiatric genetics and has led to the identification of over 100 variants associated with major depression at the genome-wide–significant alpha level (37). Thus far, GWASs have failed to replicate any findings from candidate gene-by-stress interaction studies (38).
To our knowledge, four prior studies have used this theoretically agnostic, genome-wide approach to evaluate whether individual genetic variants interact with SLEs as risk factors for depressive symptoms measured using symptoms scales including the Beck Depression Inventory, the General Health Questionnaire, and the Centers for Epidemiological Studies Depression Scale. Dunn et al. (39) conducted a genome-wide by environment interaction study (GWEIS) of depressive symptoms in a sample of 7179 African American and 3138 Hispanic/Latina women. They identified one genome-wide–significant SNP in the African American sample near the CEP350 gene (rs4652467, p = 4.10 × 10−10); however, this association did not replicate. Ikeda et al. (40) conducted a GWEIS of depressive symptoms and SLEs in 1088 individuals recruited from among employees of the Fujita Health University Hospital in Japan. The authors reported a significant interaction for a SNP near the BMP2 gene (rs10485715, p = 8.2 × 10−9); however, no attempts were made to replicate this result. Otowa et al. (41) conducted a GWEIS of depressive symptoms and SLEs in 320 Japanese individuals, with no genome-wide–significant results. Most recently, Arnau-Soler et al. (42) conducted GWEISs of depressive symptoms and SLEs in 4919 Europeans from the Generation Scotland cohort and 99,057 Europeans from the UK Biobank. The authors found two SNPs significant for interaction at the genome-wide level in the Generation Scotland sample: one near the PIWIL4 gene (p = 4.95 × 10−9) and one intronic to the ZCCHC2 gene (p = 1.46 × 10−8). They found no genome-wide–significant hits in the UK Biobank, and the significant hits from the Generation Scotland Sample did not replicate in the UK biobank.
Most of these GWEISs had sample sizes that most likely left them underpowered to detect significant interaction results. In addition, the outcome of all of these studies was depressive symptoms, rather than clinically defined major depression. Although depressive symptoms are highly genetically correlated with major depressive disorder (43), they nevertheless are a distinct outcome with, potentially, distinct associations with individual SNPs. Furthermore, all of these studies relied, out of necessity, on measures of SLEs that were retrospective and therefore potentially subject to recall bias (44,45). Finally, prior GWEISs were not able to account for the time-dependent nature of both SLEs and depression. SLEs can occur at multiple points during the lifespan, and analytic strategies that fail to account for this can potentially be subject to bias. GWAS has traditionally used logistic regressions to calculate odds ratios for the associations between individual SNPs and the odds of being a case. However, this approach does not measure risk for developing the disorder, which is arguably more useful from a clinical and public health standpoint (46). A different methodological approach is therefore needed to determine the associations between individual SNPs and risk for developing major depression, as well as potential interactions between SNPs and SLEs as risk factors for developing major depression.
Our aim in this study was to examine interactions between individual SNPs and a time-dependent, prospective measure of SLEs as risk factors for major depression in the general population. To accomplish this, we used data from the iPSYCH2012 case-cohort sample—a population-based cohort of individuals born in Denmark that includes information on psychiatric diagnoses from hospital-based settings. In addition, we also conducted a GWAS of major depression using survival analysis, rather than logistic regression, as the underlying statistical methodology to examine the associations between individual SNPs and risk for developing major depression in the general population.
Methods and Materials
Study Design and Sample
Data were drawn from the iPSYCH2012 study, which has a case-cohort design (47). In this design, the study sample is nested within a larger base population and includes all cases from the full cohort but only a subset of noncases (48). This reduces the cost and burden associated with collecting biological specimens (in the case of iPSYCH, DNA for genetic analysis). The subset used as the comparison group is typically a random sample of individuals drawn from the full cohort (i.e., the subcohort). Because it is random, some cases will by chance be selected as part of the subcohort. The great benefit of this design over a nested case-control design is that it enables the unbiased calculation of risk and hazard ratios, as in a cohort study (49). Because not all noncases from the full cohort are included, this design can be more efficient and cost-effective than a cohort study, particularly when the collection of biological samples is involved (48, 49, 50). For a detailed overview on case-cohort designs, see Barlow et al. (48), and for a brief tutorial, see Musliner et al. (51) (Supplement).
The iPSYCH2012 case-cohort sample includes a subcohort of 30,000 individuals (i.e., the subcohort) selected randomly from the base population of all individuals born in Denmark between 1981 and 2005 who survived to their first birthday and had known mothers (n = 1,472,762). To this random sample all additional cases from the base population (n = 56,189) were added, i.e., individuals who received a diagnosis of affective disorder, schizophrenia, autism, or attention-deficit/hyperactivity disorder between 1994 and 2012 in inpatient, outpatient, or emergency room settings in Danish psychiatric hospitals. Records of psychiatric diagnoses are stored in the Danish Psychiatric Central Research Register (52). Around 4% of individuals in the subcohort (n = 1188) also received one of the above psychiatric diagnoses, bringing the total number of individuals with a psychiatric diagnosis to 57,377. Biological material for DNA analysis was linked to information from national population-based registers using the unique, personal identification number assigned to all Danish citizens and legal residents since 1968 by the Danish Civil Registration System (53). The Danish Civil Registration System also includes parents’ personal identification numbers, allowing establishment of all known first-degree relatives (parents, siblings, half-siblings, and offspring).
For this study, we selected all individuals in the iPSYCH2012 subcohort and the remaining patients with depression (ICD-10 codes F32–F33) from the full cohort 1) who were of European ancestry based on principal component analysis, 2) who were successfully genotyped, and 3) for whom follow-up data starting at 10 years of age was available. We also removed at random 1 person from each pair of relatives (second degree or closer, > 0.2). The final study sample included 38,716 individuals: 20,563 individuals from the subcohort (of whom 379 had a depression diagnosis) and 18,153 additional individuals from the full cohort with a depression diagnosis (total number of patients with depression = 18,532).
Measures
Stressful Life Events
SLEs included death of a parent, sibling, or child; serious medical illness in the individual or one of their first-degree relatives; family disruption owing to divorce or separation; parental disability; and child maltreatment. SLE variables were obtained from Danish national population-based registers (52,54,55). A detailed description of how each SLE was measured is shown in Table S1. Dahl et al. (6) examined these events in the Danish registers and found that all were associated with depression risk individually, and that the number of SLEs was associated with depression in a dose-response fashion (6). Information on SLEs was combined into a time-varying count variable, such that individuals contributed person-time to the analyses within whichever category of SLE that they were in at that time, and switched to contribute person-time within a different SLE category when they experienced a subsequent SLE.
Genetic Data
DNA was obtained from blood spots collected at birth as part of routine clinical screening and stored in the Danish Newborn Screening Biobank (56). Bloodspots were located for 80,422 (93%) members of the iPSYCH2012 sample. Samples were genotyped at the Broad Institute of Harvard and MIT (Cambridge, MA) in 23 waves using the Infinium PsychChip v1.0 array (Illumina). Quality control and imputation were performed using the RICOPILI pipeline (57). The filtering process excluded variants with call frequency <0.98 or a Hardy-Weinberg equilibrium p value <1 × 10−6. Ninety percent (n = 77,639) of the sample passed quality control.
Analyses
Main and interaction effects for the associations between individual SNPs, SLEs, and depression were estimated using a series of Cox regressions. Owing to undersampling of noncases in a case-cohort design, weights must be applied to obtain accurate estimates (48). These weights ensure that only members of the random subcohort contribute person-time to the survival analyses, while cases outside the cohort enter the analyses a moment before their time of failure. For this study, we used the weighting method proposed by Prentice (50), in which members of the subcohort (including cases) receive a weight of 1, and depression cases outside the subcohort receive a weight of 0 before their failure date and 1 when they enter the risk set in which they themselves fail. This method has been shown to produce estimates that most closely resemble those obtained from the full cohort (58).
Persons in the study sample were followed from 10 years of age until first depression diagnosis, death, emigration, or December 31, 2012, whichever came first. The underlying time metric was age in days. The time-dependent SLE count variable was analyzed as a continuous variable. All analyses were adjusted for sex, birth year, and the first 5 ancestral principal components. Wald statistics were used to test for interaction. Analyses were conducted in R (version 3.1.2; R Foundation for Statistical Computing). Regional visualizations of results from GWEIS analyses were plotted with LocusZoom (59).
There are approximately 11 million directly genotyped and imputed SNPs available for members of the iPSYCH2012 sample. However, according to Danish law, some register-based data are available only at dedicated servers at Statistics Denmark. Because this study includes variables that can only be accessed through these servers, we were required to conduct the analyses in a Windows environment (Microsoft Corp.), which created some computational challenges that made it impossible to run our GWAS and GWEIS analysis in the full set of 11 million SNPs. To get around these challenges, we conducted our GWEIS of SLEs and depression in two stages: first, we selected a subset of SNPs in which minor allele frequency (MAF) was >0.01 and missing rate was <0.1. From there, we conducted linkage disequilibrium pruning with various r2 thresholds and found that an r2 value of 0.7 left us with 496,162 high-quality SNPs distributed across the genome. These SNPs were then uploaded onto the Statistics Denmark servers and merged with the register-based data for GWAS and GWEIS analysis. Based on the GWEIS analysis using these 496,162 SNPs, we identified all SNPs with interaction p values below p = 1 × 10−5. We then went back to the original sample of 11 million SNPs and identified all additional SNPs located 500 kb upstream or downstream of these SNPs and uploaded them onto the server at Statistics Denmark. This enabled a second stage of analysis in which there was dense coverage of the areas with suggestive evidence for interaction. For this second stage, statistical significance was evaluated at the genome-wide–significant α level of p < 5 × 10−8. Given the actual number of SNPs included in our GWEIS and the fact that the second stage of SNP selection specifically aimed to increase coverage of specific genomic areas, we posit p < 5 × 10−8 to be a conservative threshold.
Replication Attempt
We attempted to replicate our top findings in a case-control sample of depression drawn from the UK Biobank (60). The UK Biobank includes more than 500,000 persons 40–69 years of age at recruitment and holds a variety of biological measurements, lifestyle indicators, and biomarkers, including genome-wide genotype data on all participants. The current replication analyses were based on a sample of 73,258 genetically unrelated persons of European ancestry (22,880 depression cases and 50,378 controls) for whom SNP data as well as information on trauma exposure were available (61). Lifetime depression was assessed with questions from the Composite International Diagnostic Interview. Trauma exposure was operationalized as a dichotomous variable based on self-report of severe trauma experiences in childhood and adulthood. For detailed information on the replication sample, see Coleman et al. (61) (Supplement). We tested for interaction between the dichotomous trauma exposure and all available SNPs located within ±500 kb of the most significant SNP from each of the three genome-wide–significant loci identified in the iPSYCH2012 GWEIS. In total, 7745 SNPs were tested for interaction using PLINK2a (62). We assessed the number of independent loci tested for interaction at varying r2 (0.1–0.5) and differently sized windows (250–3000 kb) yielding 443 to 1252 independent loci (see Table S2).
Results
Sample Characteristics
Sample characteristics are shown in Table 1. Patients with depression inside and outside the population-based random subcohort showed similar characteristics. Sixty-nine percent of patients with depression and 49% of subcohort members were female. Mean age at first depression diagnosis was 19.6 years (19.7 years for patients outside the subcohort) (SD = 4.1 years inside the subcohort and 4.2 years outside the subcohort). SLEs were common—by 10 years of age, 48% of patients with depression (49% for patients outside the subcohort) and 39% of population-based control subjects had experienced at least one SLE.
Table 1.
Sample Characteristics
| Characteristic | MD Cases Outside the Subcohort (n = 18,153) | MD Cases Inside the Subcohort (n = 379) | Noncases From the Subcohort (n = 20,184) |
|---|---|---|---|
| Gender, n (%) | |||
| Female | 12,430 (68.5%) | 263 (69.4%) | 9848 (48.8%) |
| Male | 5723 (31.5%) | 116 (30.6%) | 10,336 (51.2%) |
| Birth Cohort, n (%) | |||
| 1981–1985 | 5953 (32.8%) | 126 (33.3%) | 3585 (17.8%) |
| 1986–1990 | 6670 (36.7%) | 150 (39.6%) | 4570 (22.6%) |
| 1991–2002 | 5530 (30.5%) | 103 (27.2%) | 12,029 (59.6%) |
| >1 SLE Before 10 Years of Age, n (%) | 8712 (48.0%) | 185 (48.8%) | 7857 (38.9%) |
| Age at First MD Diagnosis, Years, Mean (SD) | 19.7 (4.2) | 19.6 (4.1) | NA |
MD, major depression; NA, not applicable; SLE, stressful life event.
GWAS Results
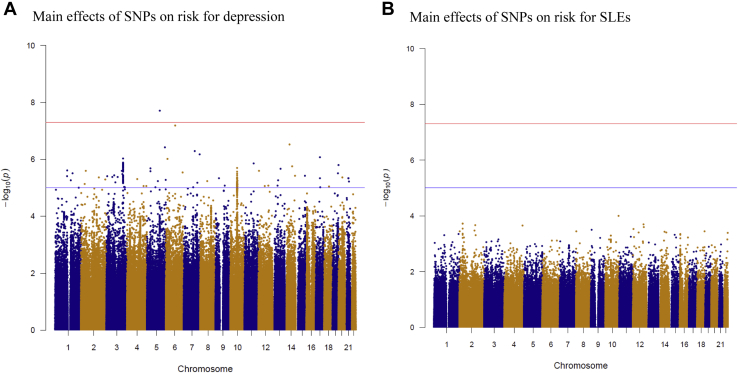
Figure 1 shows results from GWASs examining the main effects of 496,162 SNPs on the hazard of depression (Figure 1A) and the hazard of experiencing at least one SLE (Figure 1B). The GWAS of the risk for developing depression yielded 1 genome-wide–significant hit (rs7700661, p = 1.99 × 10−8) and 52 hits in which p < 1 × 10−5 (Figure 1A). No individual SNPs had p values <1 × 10−5 for the main effect of SNPs on the hazard of SLEs (Figure 1B).
Figure 1.
Manhattan plots for main effects of 496,162 SNPs on risk for depression and SLEs in 18,532 patients with major depression and 20,184 population-based control subjects. (A) Main effects of 496,162 individual SNPs on risk for major depression diagnosis in hospital-based settings in Denmark from 1995 to 2012. (B) Main effects of 496,162 individual SNPs on risk for experiencing at least one SLE. SLE, stressful life event; SNP, single nucleotide polymorphism.
GWEIS Results
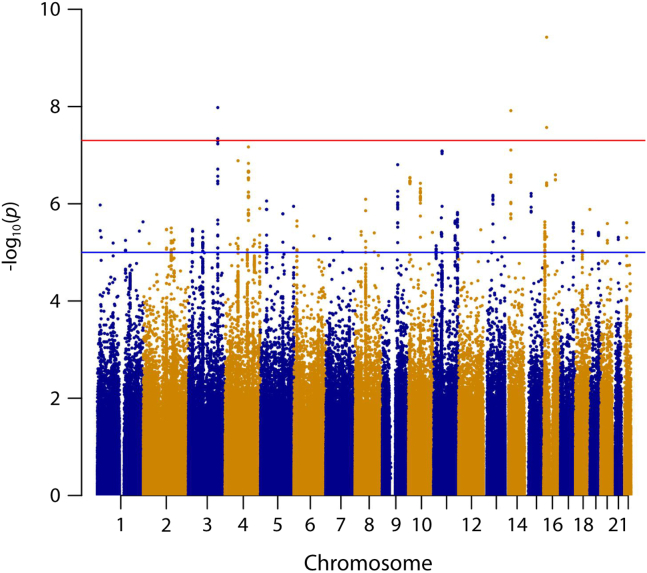
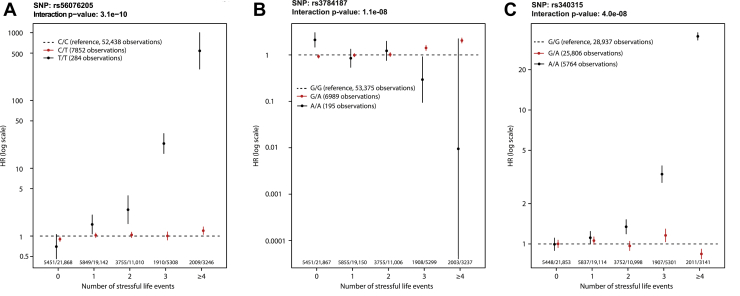
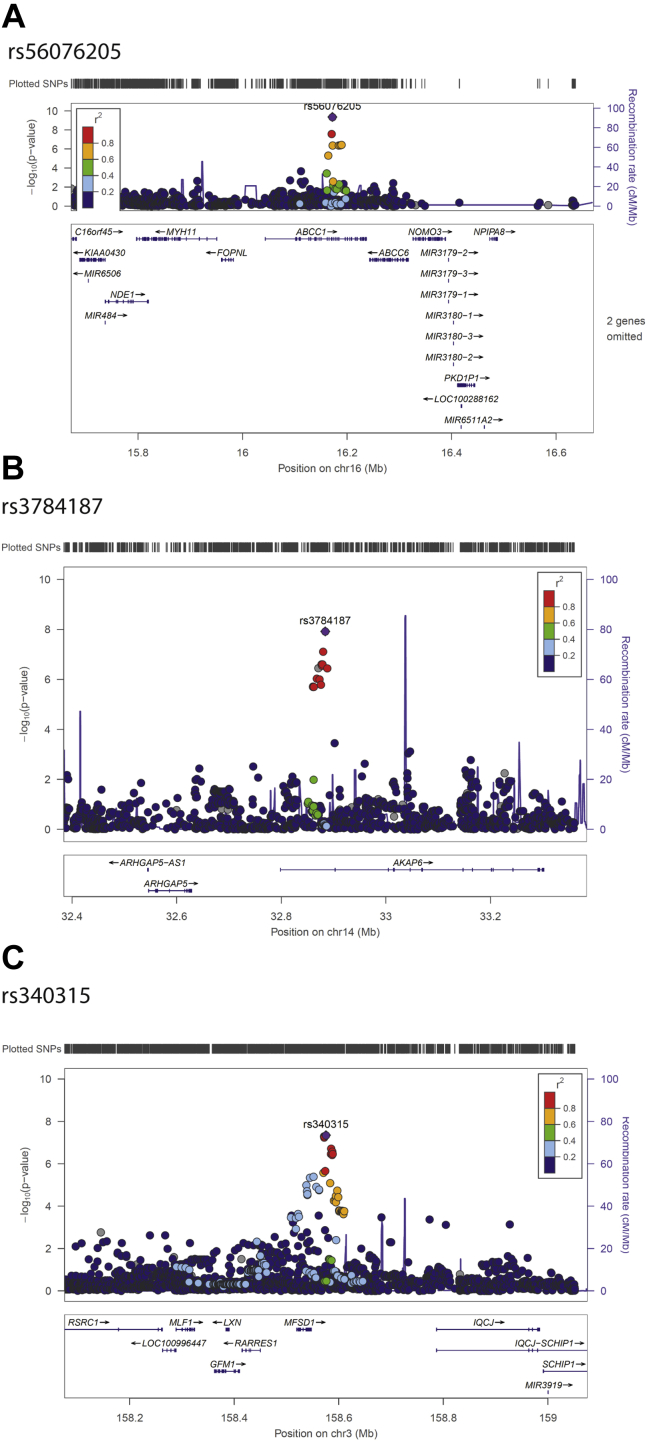
The GWEIS analysis of 496,162 SNPs yielded 60 SNPs in which p < 1 × 10−5 (Table 2). After rerunning the GWEIS including all SNPs located within 500 kb of these 60 SNPs, three independent loci reached genome-wide significance (Figure 2). Hazard ratios for the three top hits are shown in Figure 3, and region plots are shown in Figure 4. The top hit, rs56076205 (p = 3.7 × 10−10), was located in an intron of the ABCC1 gene. Compared with homozygotes for the major allele, homozygotes for the minor allele (MAF = 0.07) had a hazard for depression >20 times greater than homozygotes for the major allele at 3 SLEs, and >500 times greater at 4+ SLEs (see Figure 3A). ABCC1 is known as a multidrug resistance protein and has a range of commonly used drugs as substrate (63). Mice studies report a strong influence of ABCC1 on cerebral accumulation of amyloid-β (64). The second hit, rs3784187 (p = 1.2 × 10−8), was located in an intron of the AKAP6 gene. For this SNP, homozygotes for the minor allele (MAF = 0.06) showed a negative interaction such that as SLEs increased, risk for depression decreased (see Figure 3B). The protein transcribed from the AKAP6 gene is involved in intracellular signaling in the protein kinase A pathway (65). In 2015, a meta-analysis from the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium found a genome-wide–significant association between an SNP in the AKAP6 gene and general cognitive functioning (66). The final hit, rs340315 (p = 4.5 × 10−8), was located near the MFSD1 gene. MFSD1 is a membrane-bound solute carrier present in a wide range of human tissues (65). A recent mice study reported MFSD1 to be abundant in the plasma membrane of neurons (67). Further, the study found alterations in gene expression in response to environmental stress. Homozygotes for the minor allele (MAF = 0.31) showed a similar pattern to the first hit, such that the hazard for depression was >3 times higher at 3 SLEs and >30 times higher at 4+ SLEs compared with homozygotes for the major allele (see Figure 3C).
Table 2.
Sixty SNPs With p Values <1 × 10−5 for Interaction With SLEs on Risk for Major Depression Tested Among 496,162 SNPs in 18,532 Patients With Major Depression and 20,184 Population-Based Control Subjects
| Chromosome | Location (bp) | SNP | Interaction, p | Main Effect Depression, p | Main Effect SLEs, p | A1 | A2 | MAF | Gene Context |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 16650609 | rs149334507 | 1.05 × 10−6 | .0042 | .42 | A | C | 0.02 | ARHGEF19---[FBXO42]--SZRD1 |
| 1 | 21937317 | rs12083062 | 4.80 × 10−6 | .81 | .72 | T | C | 0.05 | ALPL--[RAP1GAP]--USP48 |
| 1 | 86628279 | rs150960662 | 6.38 × 10−6 | .92 | .59 | A | C | 0.02 | COL24A1-[]---ODF2L |
| 1 | 153310297 | rs821433 | 5.60 × 10−6 | .0014 | .56 | G | A | 0.10 | PGLYRP3--[PGLYRP4]--S100A9 |
| 1 | 228022150 | rs182670935 | 3.59 × 10−6 | .13 | .17 | G | A | 0.01 | SNAP47--[PRSS38]--WNT9A |
| 1 | 247711911 | chr1:247711911 | 2.34 × 10−6 | .14 | .93 | TGTT | CGTT | 0.17 | OR2C3--[GCSAML]--OR2G2 |
| 2 | 31549959 | rs207426 | 6.48 × 10−6 | .61 | .74 | C | A | 0.35 | FADS1--[FADS2]--FADS3 |
| 2 | 125009457 | rs79653267 | 3.30 × 10−6 | .77 | .91 | A | G | 0.02 | [CNTNAP5]---MTND5P22 |
| 2 | 150854878 | rs149282157 | 5.78 × 10−6 | .07 | .43 | A | G | 0.01 | MMADHC---[]---RND3 |
| 2 | 159027173 | rs10804390 | 6.21 × 10−6 | .10 | .47 | T | C | 0.32 | UPP2--[]CCDC148 |
| 2 | 166023849 | rs62174951 | 5.51 × 10−6 | .19 | .97 | G | A | 0.11 | SLC38A11---[SCN3A]--SCN2A |
| 3 | 20622558 | rs9846696 | 9.89 × 10−6 | .0001 | .69 | G | C | 0.05 | SGOL1---[] |
| 3 | 22055173 | rs61553318 | 3.29 × 10−6 | .06 | .41 | A | C | 0.04 | ZNF385D-AS2--[ZNF385D]---HMGB1P5 |
| 3 | 77675638 | rs876675 | 6.40 × 10−6 | .21 | .61 | C | T | 0.50 | VDAC1P7---[ROBO2]---RP11-354H21.1 |
| 3 | 158545195 | rs6792827 | 4.51 × 10−6 | .04 | .85 | T | G | 0.13 | RARRES1--[MFDS1]---IQCJ-SCHIP1 |
| 3 | 158551731 | rs61796809 | 4.34 × 10−6 | .11 | .98 | A | G | 0.22 | MFSD1-[]---IQCJ-SCHIP1 |
| 3 | 158583584 | rs340284 | 8.02 × 10−6 | .0001 | .86 | G | A | 0.36 | MFSD1--[]--IQCJ-SCHIP1 |
| 4 | 27574252 | rs75065309 | 6.72 × 10−6 | .08 | .23 | A | G | 0.01 | RP11-415C15.2---[]--IGBP1P5 |
| 4 | 59684939 | rs116510933 | 6.43 × 10−6 | .01 | .74 | A | G | 0.07 | RP11-577G20.2---[] |
| 4 | 69821738 | rs1841036 | 5.07 × 10−6 | .0031 | .21 | T | G | 0.16 | UGT2A3-[]--UGT2B11 |
| 4 | 126117627 | rs13110472 | 2.16 × 10−7 | .13 | .60 | T | C | 0.06 | ANKRD50---[]---FAT4 |
| 4 | 151061659 | rs72730361 | 7.41 × 10−6 | .09 | .99 | T | C | 0.02 | RP11-423J7.1---[DCLK2]---LRBA |
| 4 | 158141677 | rs28545562 | 5.47 × 10−6 | .03 | .69 | C | T | 0.02 | GLRB--[GRIA2]---RP11-364P22.1 |
| 4 | 186959486 | rs6818787 | 7.66 × 10−6 | .30 | .08 | C | A | 0.40 | SORBS2--[]--TLR3 |
| 5 | 33216242 | rs28566539 | 7.23 × 10−6 | .12 | .43 | T | C | 0.15 | NPR3---[CTD-2066L21.3]---TARS |
| 5 | 121067170 | rs7735996 | 6.14 × 10−6 | .16 | .75 | G | A | 0.06 | RP11-510I6.3---[]---FTMT |
| 5 | 178981060 | rs72822583 | 8.08 × 10−6 | .31 | .67 | T | C | 0.04 | ADAMTS2---[RUFY1]--HNRNPH1 |
| 6 | 15849887 | rs72823483 | 2.85 × 10−6 | .24 | .45 | A | G | 0.01 | DTNBP1---[]---MYLIP |
| 6 | 107310381 | rs9486484 | 4.53 × 10−6 | .30 | .53 | G | A | 0.15 | QRSL1---[]--C6orf203 |
| 7 | 21144220 | rs73277532 | 5.16 × 10−6 | .87 | .88 | G | T | 0.01 | ABC5B-SP8---[]---SP4 |
| 7 | 91011858 | rs73220765 | 9.71 × 10−6 | .51 | .57 | T | C | 0.01 | FZD1---[RP11-115N4.1][RP11-142A5.1]---MTERF1 |
| 8 | 32516140 | rs35955476 | 4.40 × 10−6 | .05 | .84 | C | CAG | 0.47 | NRG1-IT3---[NRG1]---RP11-11N9.4 |
| 8 | 56535514 | rs6474006 | 7.85 × 10−6 | .49 | .59 | C | T | 0.40 | XKR4--[]--TMEM68 |
| 8 | 103203727 | rs4102400 | 3.89 × 10−6 | .49 | .43 | T | C | 0.47 | NCALD--[]--RRM2B |
| 9 | 83000507 | rs7861030 | 1.56 × 10−7 | .0049 | .51 | T | C | 0.50 | NPAP1P4-[]---RP11-117O7.2 |
| 9 | 83023317 | rs10780394 | 6.22 × 10−6 | .0002 | .50 | G | A | 0.32 | NPAP1P4--[]---RP11-117O7.2 |
| 10 | 8286974 | rs1796867 | 2.85 × 10−7 | .0032 | .68 | A | G | 0.06 | PRPF38AP1--[]--LINC00708 |
| 10 | 64266748 | rs10995178 | 4.87 × 10−7 | .00001 | .89 | A | G | 0.45 | RTKN2---[ZNF365]---ADO |
| 10 | 129586689 | rs1926181 | 3.86 × 10−6 | .17 | .34 | A | C | 0.20 | FOXI2--[]--CLRN3 |
| 11 | 13920438 | rs61884777 | 8.35 × 10−6 | .12 | .54 | G | A | 0.09 | FAR1---[]--SPON1 |
| 11 | 41939498 | rs142799494 | 1.93 × 10−6 | .83 | .16 | T | A | 0.01 | LRRC4C---[]--RP11-148I19.1 |
| 11 | 44032917 | rs118008313 | 4.63 × 10−6 | .53 | .56 | T | C | 0.03 | C11orf96--[]--ACCSL |
| 11 | 44452139 | rs10769047 | 4.06 × 10−6 | .05 | .73 | A | T | 0.50 | ALX4---[]--CD82 |
| 11 | 45881397 | rs139670444 | 1.12 × 10−6 | .24 | .95 | A | AG | 0.05 | SLC35C1--[CRY2]--MAPK8IP1 |
| 11 | 116604070 | rs180353 | 5.53 × 10−6 | .45 | .88 | C | T | 0.20 | AP000770.1--[]--BUD13 |
| 11 | 128996355 | rs7944939 | 1.48 × 10−6 | .02 | .71 | C | T | 0.31 | TP53AIP1---[ARHGAP32]---BARX2 |
| 12 | 119758130 | rs140437928 | 3.41 × 10−6 | .02 | .23 | C | T | 0.02 | HSPB8--[]--CCDC60 |
| 13 | 51272084 | rs797498 | 6.55 × 10−7 | .06 | .58 | A | G | 0.08 | DLEU1-AS1---[DLEU1]--DLEU7 |
| 13 | 114591051 | rs9550266 | 4.89 × 10−6 | .01 | .28 | A | G | 0.16 | GAS6--[]LINC00452 |
| 14 | 32860927 | rs1951185 | 1.60 × 10−6 | .08 | .94 | T | C | 0.06 | ARHGAP5---[AKAP6][RP11-320M16.2]--RN7SL660P |
| 15 | 35002935 | rs16959528 | 6.12 × 10−7 | .94 | .59 | G | A | 0.11 | GOLGA8B---[]--GJD2 |
| 16 | 6338673 | rs1344474 | 9.41 × 10−6 | .86 | .39 | G | A | 0.12 | [RBFOX1][RB11-420N3.3] |
| 16 | 16172008 | rs56076205 | 3.74 × 10−10 | .05 | .55 | T | C | 0.07 | FOPNL---[ABCC1]--ABCC6 |
| 16 | 63680366 | rs12448930 | 3.17 × 10−7 | .09 | .86 | A | C | 0.25 | RP11-368L12.1--[]--RP11-370P15.1 |
| 17 | 70291156 | rs1967304 | 5.85 × 10−6 | .67 | .11 | C | A | 0.25 | SOX9---[]---SLC39A11 |
| 18 | 37425523 | rs2048647 | 3.52 × 10−6 | .03 | .48 | G | C | 0.21 | RP11-244M2.1--[RP11-636021.1]---LINC01477 |
| 18 | 77985650 | rs111447074 | 1.30 × 10−6 | .12 | .59 | T | C | 0.02 | ADNP2--[PARD6G] |
| 19 | 46785290 | rs112087991 | 4.06 × 10−6 | .0005 | .60 | C | T | 0.05 | IGFL1--[]--HIF3A |
| 20 | 35329303 | rs62206150 | 6.36 × 10−6 | .01 | .35 | G | A | 0.02 | SLA2--[NDRG3]--DSN1 |
| 21 | 31449079 | rs117181045 | 4.86 × 10−6 | .03 | .98 | G | T | 0.01 | GRIK1---[]--CLDN17 |
The 492,162 included SNPs were selected according to the following criteria: MAF >0.01 and missing rate <0.1; subsequently, linkage disequilibrium pruning with an r2 value of 0.7 was implemented. The gene context column lists the SNP location within brackets. Most closely located genetic variants 500 kb upstream or downstream for the index SNP are listed as well with any genes prioritized over long intergenic noncoding RNA, pseudogenes, etc. Distance from the index to other listed variants is denoted by dashes: no dash indicates <1 kb, one dash indicates <10 kb, two dashes indicates <100 kb, and three dashes <500 kb.
MAF, minor allele frequency; SLE, stressful life event; SNP, single nucleotide polymorphism.
Figure 2.
Manhattan plot of genome-wide by environment interaction analyses based on 18,532 patients with major depression and 20,184 population-based control subjects. The figure presents results of a GWEIS conducted in two stages. In stage 1, a GWEIS was conducted using 496,162 SNPs distributed across the genome. In stage 2, all SNPs located 500 kb up- or downstream from 60 SNPs with p values <10−5 in stage 1 were added to the analyses. The Manhattan plot shows results from both stages. GWEIS, genome-wide by environment interaction study; SNP, single nucleotide polymorphism.
Figure 3.
Interaction effects for stressful life events and top SNPs from 3 genome-wide–significant loci. Note. For each SNP, the HR for depression is plotted by number of stressful life events. Vertical bars represent 95% CI. Hazards were compared within each level of stressful life events with major allele homozygotes as reference. Wald statistics were used to test interactions, comparing linear trends for HR between genotypes. The small differences in the total number of observations are due to differences in the number of persons successfully genotyped for each SNP. Owing to the time-varying nature of the stressful life events variable, study participants could contribute person-time for different numbers of stressful life events. Therefore, the total number of observations exceeds the total number of participants in the study. HR, hazard ratio; SNP, single nucleotide polymorphism.
Figure 4.
Region plots for three top hits from a genome-wide by environment interaction study based on 18,532 patients with major depression and 20,184 population-based control subjects. The color of the dots indicates the linkage disequilibrium (r2) of SNPs with the top SNP of each loci. The r2 was based on the 1000 Genomes Project November 2014 European population. SNP, single nucleotide polymorphism.
Analysis of Top SNPs in UK Biobank
None of the three top SNPs were statistically significant in the replication attempt using UK Biobank data (rs56076205, p = .87; rs3784187, p = .93; rs340315, p = .71). The most significant interactions involved the following SNPs: rs190869692 (p = 3.2 × 10-5) in the ABCC1 gene 38,653 bp upstream from the iPSYCH2012 hit in the same gene (r2 = 0.002, p = .58); rs111284027 (p = 9.4 × 10−5) in the ARHGAP5 gene 259,273 bp downstream from our hit in the AKAP6 gene (r2 = 0.003, p = .44); rs146472082 (p = 5.1 × 10−5) in the RARRES1 gene 155,569 bp downstream from our hit near the MFSD1 gene (r2 = 0.053, p = .0011) (see Figure S1). Thus, all three SNPs identified in the replication analyses represented independent loci from the three genome-wide–significant loci identified in the iPSYCH2012 GWEIS.
Discussion
In this study, we report results from the first comprehensive, population-based GWEIS investigating the interaction between individual SNPs and a time-varying measure of SLEs as risk factors for a diagnosis of depression treated in inpatient, outpatient, or emergency room settings. The GWEIS yielded genome-wide–significant effects in three independent loci located in the ABCC1, AKAP6, and MSFD1 genes, as well as 50 hits in which p < 1 × 10−5. We attempted to replicate our top hits in a large sample of depression cases and controls from the UK Biobank; however, none of the hits were significant in the replication sample. This suggests that the original hits were false positives. However, there are notable differences between iPSYCH2012 and UK Biobank in terms of sampling, measurement, and design. The fact that different statistical methods were used (survival analysis vs. logistic regression) could also have contributed. However, it is not straightforward to isolate the impact of the statistical method alone, because conducting a logistic regression in our own sample would require us to make substantial changes to the design and sample composition. Thus, it would be difficult to tell if any difference in the results was due to the different statistical method or to the different design. Ultimately, it remains a possibility that one or more of these hits might replicate in a sample in which the measurement, design, and analysis are more comparable; however, unless such evidence becomes available, these hits should not be considered robust.
To our knowledge, this is the largest single-sample GWEIS conducted to date examining the interaction between individual variants and SLEs. Nevertheless, the presented analyses are still likely underpowered to detect most single-SNP gene-environment interactions (68). For years, GWASs were similarly underpowered to detect significant SNPs, until the development of large-scale international consortia allowed for the accumulation of enough samples to pass the inflection point for consistent findings (69). In comparison, the study of gene-environment interaction in psychiatric disorders has only begun to enter into its big data phase. The requirement for assessment of a complex, composite environment exposure in the large study populations necessary for studying interactions makes these studies challenging endeavors. Extrapolating from the history of GWASs in psychiatry, we believe that the inflection point for studies of gene-environment interaction will only be reached through international collaborations that combine studies with information on genetic variation and environment exposures.
Methodological Considerations
The following are additional methodological aspects of the study that should kept in mind when interpreting these results. First, the oldest depression cases in the iPSYCH2012 sample were diagnosed by 30 years of age. As such, they represent a cohort of early-onset depression cases, and therefore these results may not generalize to individuals who develop depression at older ages. Second, the depression cases in iPSYCH are all identified in hospital-based settings; therefore, these results may not generalize to individuals with untreated depression or individuals treated solely by their primary care doctors, who make up the majority of depression cases in Denmark (70). Third, although some of the SLEs included in this study are measured with high accuracy (e.g., death of a relative), others, particularly child maltreatment, are measured less accurately because they are based solely on register data. It is sadly very likely that some individuals in the sample experienced child maltreatment that was never recorded in the register, although the opposite (that individuals registered as having experienced child maltreatment did not experience it) is unlikely to be true. Fourth, we included a diverse range of stressful events in our study. Consequently, it is possible that some observed interactions relate to very specific types of SLEs. For example, it is plausible that risk for depression in relation to somatic disease is associated with the seriousness of the course of disease. Therefore, genetic variants associated with prognosis and/or treatment response could emerge as part of gene-environment interaction in the present study, e.g., ABCC1 has a range of anticancer and anti-HIV drugs as substrates, thus rendering somatic treatment less effective, thereby possibly increasing risk for depression.
Conclusions
In this population-based cohort of European ancestry, we identified three novel genetic loci that interacted with a time-varying measure of SLEs to predict hospital-treated depression at a genome-wide–significant level. However, none of these hits replicated in a large sample of depression cases and controls from the UK Biobank. Future gene-by-stress research in depression should focus on efforts to establish large collaborative GWEISs to generate sufficient statistical power to identify significant variants.
Acknowledgments and Disclosures
The iPSYCH project is funded by the Lundbeck Foundation (Grant Nos. R102-A9118, R155-2014-1724, and R248-2017-2003) and the universities and university hospitals of Aarhus and Copenhagen. KLM is funded by a postodoc fellowship from the Lundbeck Foundation (Grant No. R303-2018-3551). Genotyping of the iPSYCH2012 samples was supported by grants from the Lundbeck Foundation, the Stanley Foundation, the Simons Foundation (Grant No. SFARI 311789), and the National Institute of Mental Health (Grant No. 5U01MH094432-02). The Danish National Biobank resource is supported by the Novo Nordisk Foundation. The UK Biobank (Project ID 16577) represents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. High-performance computing facilities at the NIHR Biomedical Research Centre were funded with capital equipment grants from the Guy’s and St Thomas’ NHS Foundation Trust Charity (Grant No. TR130505) and Maudsley Charity (Grant No. 980).
The authors gratefully acknowledge the Broad Institute for genotyping. Initial genetic analyses were performed on the GenomeDK high-performance computing facility supported by the Centre for Genomics and Personalized Medicine and Center for Integrative Sequencing, Aarhus University.
A previous version of this article was published as a preprint on medRxiv: https://doi.org/10.1101/2021.09.03.21262452.
TW has served as scientific advisor to H. Lundbeck A/S. GB has received consultancy and speaker fees from Eli Lilly, Otsuka, and Illumina. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.11.003.
Supplementary Material
References
- 1.Kessler R.C., Bromet E.J. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119–138. doi: 10.1146/annurev-publhealth-031912-114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasin D.S., Sarvet A.L., Meyers J.L., Saha T.D., Ruan W.J., Stohl M., et al. Epidemiology of Adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiatry. 2018;75:336–346. doi: 10.1001/jamapsychiatry.2017.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan P.F., Neale M.C., Kendler K.S. Genetic epidemiology of major depression: Review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 4.Kessler R.C. The effects of stressful life events on depression. Annu Rev Psychol. 1997;48:191–214. doi: 10.1146/annurev.psych.48.1.191. [DOI] [PubMed] [Google Scholar]
- 5.Hammen C. Stress and depression. Annu Rev Clin Psychol. 2005;1:293–319. doi: 10.1146/annurev.clinpsy.1.102803.143938. [DOI] [PubMed] [Google Scholar]
- 6.Dahl S.K., Larsen J.T., Petersen L., Ubbesen M.B., Mortensen P.B., Munk-Olsen T., et al. Early adversity and risk for moderate to severe unipolar depressive disorder in adolescence and adulthood: A register-based study of 978,647 individuals. J Affect Disord. 2017;214:122–129. doi: 10.1016/j.jad.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Kessler R.C., Davis C.G., Kendler K.S. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol Med. 1997;27:1101–1119. doi: 10.1017/s0033291797005588. [DOI] [PubMed] [Google Scholar]
- 8.Anda R.F., Felitti V.J., Bremner J.D., Walker J.D., Whitfield C., Perry B.D., et al. The enduring effects of abuse and related adverse experiences in childhood. A convergence of evidence from neurobiology and epidemiology. Eur Arch Psychiatry Clin Neurosci. 2006;256:174–186. doi: 10.1007/s00406-005-0624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman D.P., Whitfield C.L., Felitti V.J., Dube S.R., Edwards V.J., Anda R.F. Adverse childhood experiences and the risk of depressive disorders in adulthood. J Affect Disord. 2004;82:217–225. doi: 10.1016/j.jad.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Kendler K.S., Kessler R.C., Walters E.E., MacLean C., Neale M.C., Heath A.C., et al. Stressful life events, genetic liability, and onset of an episode of major depression in women. Am J Psychiatry. 1995;152:833–842. doi: 10.1176/ajp.152.6.833. [DOI] [PubMed] [Google Scholar]
- 11.Caspi A., Sugden K., Moffitt T.E., Taylor A., Craig I.W., Harrington H., et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Li X., McGue M. Interacting effect of BDNF Val66Met polymorphism and stressful life events on adolescent depression. Genes Brain Behav. 2012;11:958–965. doi: 10.1111/j.1601-183X.2012.00843.x. [DOI] [PubMed] [Google Scholar]
- 13.Drachmann Bukh J., Bock C., Vinberg M., Werge T., Gether U., Vedel Kessing L. Interaction between genetic polymorphisms and stressful life events in first episode depression. J Affect Disord. 2009;119:107–115. doi: 10.1016/j.jad.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Elovainio M., Jokela M., Kivimäki M., Pulkki-Råback L., Lehtimäki T., Airla N., et al. Genetic variants in the DRD2 gene moderate the relationship between stressful life events and depressive symptoms in adults: Cardiovascular risk in young Finns study. Psychosom Med. 2007;69:391–395. doi: 10.1097/psy.0b013e31806bf365. [DOI] [PubMed] [Google Scholar]
- 15.Juhasz G., Hullam G., Eszlari N., Gonda X., Antal P., Anderson I.M., et al. Brain galanin system genes interact with life stresses in depression-related phenotypes. Proc Natl Acad Sci U S A. 2014;111:E1666–E1673. doi: 10.1073/pnas.1403649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J.M., Stewart R., Kim S.W., Yang S.J., Shin I.S., Kim Y.H., et al. Interactions between life stressors and susceptibility genes (5-HTTLPR and BDNF) on depression in Korean elders. Biol Psychiatry. 2007;62:423–428. doi: 10.1016/j.biopsych.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Liu W., Yao L., Yang C., Xiao L., Wan Q., et al. Negative life events and corticotropin-releasing-hormone receptor1 gene in recurrent major depressive disorder. Sci Rep. 2013;3:1548. doi: 10.1038/srep01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandelli L., Serretti A., Marino E., Pirovano A., Calati R., Colombo C. Interaction between serotonin transporter gene, catechol-O-methyltransferase gene and stressful life events in mood disorders. Int J Neuropsychopharmacol. 2007;10:437–447. doi: 10.1017/S1461145706006882. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann P., Bruckl T., Nocon A., Pfister H., Binder E.B., Uhr M., et al. Interaction of FKBP5 gene variants and adverse life events in predicting depression onset: Results from a 10-year prospective community study. Am J Psychiatry. 2011;168:1107–1116. doi: 10.1176/appi.ajp.2011.10111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bleys D., Luyten P., Soenens B., Claes S. Gene-environment interactions between stress and 5-HTTLPR in depression: A meta-analytic update. J Affect Disord. 2018;226:339–345. doi: 10.1016/j.jad.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 21.Culverhouse R.C., Saccone N.L., Horton A.C., Ma Y., Anstey K.J., Banaschewski T., et al. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. 2018;23:133–142. doi: 10.1038/mp.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosang G.M., Shiles C., Tansey K.E., McGuffin P., Uher R. Interaction between stress and the BDNF Val66Met polymorphism in depression: A systematic review and meta-analysis. BMC Med. 2014;12:7. doi: 10.1186/1741-7015-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karg K., Burmeister M., Shedden K., Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: Evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risch N., Herrell R., Lehner T., Liang K.Y., Eaves L., Hoh J., et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: A meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Ijzendoorn M.H., Belsky J., Bakermans-Kranenburg M.J. Serotonin transporter genotype 5HTTLPR as a marker of differential susceptibility? A meta-analysis of child and adolescent gene-by-environment studies. Transl Psychiatry. 2012;2:e147. doi: 10.1038/tp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., Shelton R.C., Dwivedi Y. Interaction between early-life stress and FKBP5 gene variants in major depressive disorder and post-traumatic stress disorder: A systematic review and meta-analysis. J Affect Disord. 2018;225:422–428. doi: 10.1016/j.jad.2017.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao M., Chen L., Yang J., Han D., Fang D., Qiu X., et al. BDNF Val66Met polymorphism, life stress and depression: A meta-analysis of gene-environment interaction. J Affect Disord. 2018;227:226–235. doi: 10.1016/j.jad.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Peyrot W.J., Milaneschi Y., Abdellaoui A., Sullivan P.F., Hottenga J.J., Boomsma D.I., et al. Effect of polygenic risk scores on depression in childhood trauma. Br J Psychiatry. 2014;205:113–119. doi: 10.1192/bjp.bp.113.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullins N., Power R.A., Fisher H.L., Hanscombe K.B., Euesden J., Iniesta R., et al. Polygenic interactions with environmental adversity in the aetiology of major depressive disorder. Psychol Med. 2016;46:759–770. doi: 10.1017/S0033291715002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colodro-Conde L., Couvy-Duchesne B., Zhu G., Coventry W.L., Byrne E.M., Gordon S., et al. A direct test of the diathesis-stress model for depression. Mol Psychiatry. 2018;23:1590–1596. doi: 10.1038/mp.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musliner K.L., Seifuddin F., Judy J.A., Pirooznia M., Goes F.S., Zandi P.P. Polygenic risk, stressful life events and depressive symptoms in older adults: A polygenic score analysis. Psychol Med. 2015;45:1709–1720. doi: 10.1017/S0033291714002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyrot W.J., Van der Auwera S., Milaneschi Y., Dolan C.V., Madden P.A.F., Sullivan P.F., et al. Does Childhood Trauma Moderate Polygenic Risk for Depression? A Meta-analysis of 5765 Subjects From the Psychiatric Genomics Consortium. Biol Psychiatry. 2018;84:138–147. doi: 10.1016/j.biopsych.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musliner K.L., Andersen K.K., Agerbo E., Albiñana C., Vilhjalmsson B.J., Rajagopal V.M., et al. Polygenic liability, stressful life events and risk for secondary-treated depression in early life: A nationwide register-based case-cohort study [published online ahead of print May 5] Psychol Med. 2021 doi: 10.1017/S0033291721001410. [DOI] [PubMed] [Google Scholar]
- 34.Bosker F.J., Hartman C.A., Nolte I.M., Prins B.P., Terpstra P., Posthuma D., et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry. 2011;16:516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- 35.Duncan L.E., Keller M.C. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson E.C., Border R., Melroy-Greif W.E., de Leeuw C.A., Ehringer M.A., Keller M.C. No evidence that schizophrenia candidate genes are more associated with schizophrenia than noncandidate genes. Biol Psychiatry. 2017;82:702–708. doi: 10.1016/j.biopsych.2017.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Border R., Johnson E.C., Evans L.M., Smolen A., Berley N., Sullivan P.F., et al. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am J Psychiatry. 2019;176:376–387. doi: 10.1176/appi.ajp.2018.18070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn E.C., Wiste A., Radmanesh F., Almli L.M., Gogarten S.M., Sofer T., et al. Genome-wide association study (GWAS) and genome-wide by environment interaction study (GWEIS) of depressive symptoms in African American and Hispanic/Latina Women. Depress Anxiety. 2016;33:265–280. doi: 10.1002/da.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikeda M., Shimasaki A., Takahashi A., Kondo K., Saito T., Kawase K., et al. Genome-wide environment interaction between depressive state and stressful life events. J Clin Psychiatry. 2016;77:e29–e30. doi: 10.4088/JCP.15l10127. [DOI] [PubMed] [Google Scholar]
- 41.Otowa T., Kawamura Y., Tsutsumi A., Kawakami N., Kan C., Shimada T., et al. The first pilot genome-wide gene-environment study of depression in the Japanese population. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnau-Soler A., Macdonald-Dunlop E., Adams M.J., Clarke T.-K., MacIntyre D.J., Milburn K., et al. Genome-wide by environment interaction studies of depressive symptoms and psychosocial stress in UK Biobank and Generation Scotland. Transl Psychiatry. 2019;9:14. doi: 10.1038/s41398-018-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin J.R., Reuben A., Newbury J.B., Danese A. Agreement between prospective and retrospective measures of childhood maltreatment: A systematic review and meta-analysis. JAMA Psychiatry. 2019;76:5845–5893. doi: 10.1001/jamapsychiatry.2019.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colman I., Kingsbury M., Garad Y., Zeng Y., Naicker K., Patten S., et al. Consistency in adult reporting of adverse childhood experiences. Psychol Med. 2016;46:543–549. doi: 10.1017/S0033291715002032. [DOI] [PubMed] [Google Scholar]
- 46.Murray G.K., Lin T., Austin J., McGrath J.J., Hickie I.B., Wray N.R. Could polygenic risk scores be useful in psychiatry?: A review. JAMA Psychiatry. 2021;78:210–219. doi: 10.1001/jamapsychiatry.2020.3042. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen C.B., Bybjerg-Grauholm J., Pedersen M.G., Grove J., Agerbo E., Baekvad-Hansen M., et al. The iPSYCH2012 case-cohort sample: New directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlow W.E., Ichikawa L., Rosner D., Izumi S. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52:1165–1172. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 49.Self S.G., Prentice R.L. Asymptotic distribution theory and efficiency results for case-cohort studies. Ann Stat. 1988;16:64–81. [Google Scholar]
- 50.Prentice R.L. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 51.Musliner K.L., Mortensen P.B., McGrath J.J., Suppli N.P., Hougaard D.M., Bybjerg-Grauholm J., et al. Association of polygenic liabilities for major depression, bipolar disorder, and schizophrenia with risk for depression in the Danish population. JAMA Psychiatry. 2019;76:516–525. doi: 10.1001/jamapsychiatry.2018.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mors O., Perto G.P., Mortensen P.B. The Danish Psychiatric Central Research Register. Scand J Public Health. 2011;39:54–57. doi: 10.1177/1403494810395825. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen C.B. The Danish Civil Registration System. Scand J Public Health. 2011;39:22–25. doi: 10.1177/1403494810387965. [DOI] [PubMed] [Google Scholar]
- 54.Lynge E., Sandegaard J.L., Rebolj M. The Danish National Patient Register. Scand J Public Health. 2011;39:30–33. doi: 10.1177/1403494811401482. [DOI] [PubMed] [Google Scholar]
- 55.Petersson F., Baadsgaard M., Thygesen L.C. Danish registers on personal labour market affiliation. Scand J Public Health. 2011;39:95–98. doi: 10.1177/1403494811408483. [DOI] [PubMed] [Google Scholar]
- 56.Norgaard-Pedersen B., Hougaard D.M. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Disord. 2007;30:530–536. doi: 10.1007/s10545-007-0631-x. [DOI] [PubMed] [Google Scholar]
- 57.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onland-Moret N.C., van der A.D.L., van der Schouw Y.T., Buschers W., Elias S.G., van Gils C.H., et al. Analysis of case-cohort data: A comparison of different methods. J Clin Epidemiol. 2007;60:350–355. doi: 10.1016/j.jclinepi.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 59.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coleman J.R.I., Peyrot W.J., Purves K.L., Davis K.A.S., Rayner C., Choi S.W., et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol Psychiatry. 2020;25:1430–1446. doi: 10.1038/s41380-019-0546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loscher W., Potschka H. Drug resistance in brain diseases and the role of drug efflux transporters. Nat Rev Neurosci. 2005;6:591–602. doi: 10.1038/nrn1728. [DOI] [PubMed] [Google Scholar]
- 64.Krohn M., Lange C., Hofrichter J., Scheffler K., Stenzel J., Steffen J., et al. Cerebral amyloid-beta proteostasis is regulated by the membrane transport protein ABCC1 in mice. J Clin Invest. 2011;121:3924–3931. doi: 10.1172/JCI57867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 66.Davies G., Armstrong N., Bis J.C., Bressler J., Chouraki V., Giddaluru S., et al. Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949) Mol Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perland E., Hellsten S.V., Lekholm E., Eriksson M.M., Arapi V., Fredriksson R. The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient intake. J Mol Neurosci. 2017;61:199–214. doi: 10.1007/s12031-016-0867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas D. Gene--environment-wide association studies: Emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan P.F., Agrawal A., Bulik C.M., Andreassen O.A., Borglum A.D., Breen G., et al. Psychiatric genomics: An update and an agenda. Am J Psychiatry. 2018;175:15–27. doi: 10.1176/appi.ajp.2017.17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Musliner K.L., Liu X., Gasse C., Christensen K.S., Wimberley T., Munk-Olsen T. Incidence of medically treated depression in Denmark among individuals 15-44 years old: A comprehensive overview based on population registers. Acta Psychiatr Scand. 2019;139:548–557. doi: 10.1111/acps.13028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.