Abstract
Background
Genetics and biology may influence the age of onset of anorexia nervosa (AN). The aims of this study were to determine whether common genetic variation contributes to age of onset of AN and to investigate the genetic associations between age of onset of AN and age at menarche.
Methods
A secondary analysis of the Psychiatric Genomics Consortium genome-wide association study (GWAS) of AN was performed, which included 9335 cases and 31,981 screened controls, all from European ancestries. We conducted GWASs of age of onset, early-onset AN (<13 years), and typical-onset AN, and genetic correlation, genetic risk score, and Mendelian randomization analyses.
Results
Two loci were genome-wide significant in the typical-onset AN GWAS. Heritability estimates (single nucleotide polymorphism–h2) were 0.01–0.04 for age of onset, 0.16–0.25 for early-onset AN, and 0.17–0.25 for typical-onset AN. Early- and typical-onset AN showed distinct genetic correlation patterns with putative risk factors for AN. Specifically, early-onset AN was significantly genetically correlated with younger age at menarche, and typical-onset AN was significantly negatively genetically correlated with anthropometric traits. Genetic risk scores for age of onset and early-onset AN estimated from independent GWASs significantly predicted age of onset. Mendelian randomization analysis suggested a causal link between younger age at menarche and early-onset AN.
Conclusions
Our results provide evidence consistent with a common variant genetic basis for age of onset and implicate biological pathways regulating menarche and reproduction.
Keywords: Age of onset, Anorexia nervosa, Early-onset, Genetic risk score, Genetics, GWAS, Menarche, Mendelian randomization, Puberty
Anorexia nervosa (AN) is an eating disorder characterized by starvation, low body mass index (BMI), and a morbid fear of weight gain, affecting 0.9% to 1.4% of females and 0.1% to 0.3% of males (1,2). The etiology involves a complex interplay between genetics and the environment (3). Twin-based studies report a heritability of 50% to 60% (4). Common genetic polymorphisms account for a substantial portion of this heritability (single nucleotide polymorphism [SNP]-h2 = 11%–17%) (5). Peak age of onset is between 16 and 22 years in community-based epidemiological research (1) and 14 to 19 years in clinical populations (6), and onset after age 25 is atypical (7). To our knowledge, there are no genome-wide association study (GWAS) or heritability studies of age of onset of AN, and the factors that contribute to earlier rather than later onset are unknown (7).
The Psychiatric Genomics Consortium (PGC) GWAS of AN identified eight genomic regions associated with the risk of lifetime AN and implicated a psychiatric and metabolic etiology (5). Candidate gene studies have suggested that polymorphisms in serotonergic and appetite-regulating genes might be associated with age of onset (8, 9, 10, 11). However, candidate gene studies generally have been subject to important criticisms, including nonreplication. In illnesses such as schizophrenia and bipolar disorder, a higher genetic burden predicts earlier onset and age of onset for some psychiatric traits aggregates in families (12, 13, 14, 15). The Brainstorm Consortium combined molecular genetic data from 10 psychiatric disorders including AN and found a modest, significant correlation linking earlier age of onset to higher heritability (16). Meanwhile, twin studies of eating disorder symptoms suggest that genetic contributions change across development such that genetic effects explain negligible variance prepuberty and increase substantially in peripuberty (17). Because genetic factors influence AN risk, the first aim of this study is to investigate whether common genetic polymorphisms account for variation in age of onset (aim 1).
Although GWAS efforts have made tremendous contributions to our understanding of the genetic etiology of AN, phenotypic and genetic heterogeneity can hinder discovery of the genetic architecture of psychiatric traits (18). Insight can be improved by leveraging investigations of etiologically homogeneous illness subphenotypes, such as early-onset presentations. Differences in clinical presentation of AN by age are evident, although not well established because of limited research, with early-onset cases displaying predominantly non–binge/purge profiles, a faster rate of weight loss, less endorsement of psychological symptoms, more favorable long-term outcomes, and a higher male prevalence than typical-onset presentations (19, 20, 21, 22). The second aim of this study is to examine a subphenotype of AN, specifically early-onset, to aid in discovering the genetics and biology of AN and age of onset (aim 2).
Early pubertal timing has long been cited as a risk factor for AN, particularly early-onset AN, but the evidence base is weak and observational, and methodological issues complicate investigation (17). The relatively low prevalence of AN impedes prospective designs, and nutritional deficiencies in AN arrest pubertal development and complicate estimates of the causal influence of pubertal traits. Genetic designs, such as Mendelian randomization, can interrogate causality under specific assumptions while avoiding these measurement confounds. Population-based twin research has shown that shared genetic factors influence liability to earlier menarche and disordered eating (23). However, a significant genetic correlation between age at menarche (i.e., a commonly used measure of puberty timing) and AN was not evident in the largest AN GWAS to date (5). Large-scale genomic and phenotypic data collections and new analytic methods (i.e., genetic risk score [GRS] analysis, Mendelian randomization) have become available and present an excellent opportunity to examine whether puberty timing may be a causal risk factor for AN risk and age of onset (aim 3).
Methods and Materials
Design and Participants
This is a secondary analysis of individual-level data from a GWAS of AN (5), which we refer to as the parent study. Cohorts from the parent study were included here if they had cases with age of onset data. This resulted in 13 cohorts and 55% of cases (N = 9335) and 58% of controls (N = 31,981) included in this study from the parent study of 33 cohorts, 16,992 cases, and 55,525 controls (Table S1 in Supplement 2). For some secondary analyses using AN risk as a phenotype, all cases from the 13 cohorts above were included, resulting in 11,632 cases and 31,981 controls for those analyses. More details on recruitment, phenotyping, DNA collection, and genotyping are provided in Supplement 1 and other publications (5,24,25). Supplemental Methods in Supplement 1 and Table S2 in Supplement 2 provide phenotyping information for age of onset and early-onset AN, which was characterized as onset before age 13 years.
GWAS of Age of Onset of AN, Early-Onset AN, and Typical-Onset AN
Three GWASs were conducted: 1) a within-case GWAS on age of onset; 2) a case-control GWAS that stratified a subset of cases by the subphenotype of early-onset AN and compared these with ancestrally matched controls; and 3) a case-control GWAS of typical-onset AN for comparative purposes with respect to the genetic correlations and other secondary analyses, as detailed later (Table S3 in Supplement 2).
Quality control of genotype data is described in Supplement 1. GWASs were conducted using RICOPILI (26). Samples were of European ancestry, and genotypes were imputed to the 1000 Genomes reference (27). The first five principal components were included to capture ancestry-based population stratification. Linear regression for age of onset and logistic regression for early-onset AN and typical-onset AN were carried out on imputed variant dosages using additive models to test for associations between the markers and the phenotypes. Cohort-level GWAS analyses were combined with fixed-effects meta-analysis (including variants with imputation INFO scores > 0.7). The standard genome-wide cutoff (p < 5 × 10−8) was anticonservative, given three GWASs; therefore, results were interpreted at a Bonferroni-corrected threshold (p < 1.67 × 10−8). The GWAS of age of onset had >80% statistical power to detect genetic effects with 0.45% of the variance explained (R2), or βs between 1.06 and 1.59 (at minor allele frequency 0.05–0.5); the GWAS of early-onset AN had >80% power to detect an odds ratio (OR) between 1.32 and 1.70 (at minor allele frequency 0.05–0.5, assuming a lifetime prevalence of 0.1%) (28); and the GWAS of typical-onset AN had >80% power to detect an OR between 1.14 and 1.31 (at minor allele frequency 0.05–0.5, assuming a lifetime prevalence of 0.9%–4%) (29). Common variant heritability was estimated with linkage disequilibrium score regression (LDSC) (30) and the genomic-relatedness-based restricted maximum-likelihood (GREML) approach (31) implemented in GCTA (31). GREML analyses had 80% power to detect SNP-h2s ≥ 0.1 for age of onset, 0.07 for early-onset AN (liability scale), and 0.03 for typical-onset AN (liability scale) (32). Genetic correlation analyses were conducted on LD Hub (33), and genetic correlation analyses with two AN GWASs (5,34) not on LD Hub and between early- and typical-onset AN used LDSC (30,33). Gene mapping and tissue expression analyses were performed with FUMA (35).
GRSs as Predictors of Age of Onset of AN (GRSage of onset, GRSearly-onset AN, and GRSAN)
GRS analyses were conducted for aim 1 to investigate the evidence for a common variant–based genetic etiology. A GRS represents the combined effect of risk alleles carried by the individual and more powerfully predicts a complex trait than a single-SNP association analysis. Three sets of GRS at various p thresholds (pTs) were calculated for each individual with PRSice-2 (36) using the leave-one-cohort-out approach: 1) GRSage of onset using the age of onset GWAS from this study, 2) GRSAN using the GWAS results of the parent study (5), and 3) GRSearly-onset AN using the case-control GWAS for early-onset AN from this study (Supplemental Methods in Supplement 1). Linear regression quantified the association between GRSage of onset, GRSearly-onset AN, and GRSAN with age of onset with β and R2. Cohort-level analyses were combined with fixed-effects meta-analysis. p values were corrected using the false discovery rate (FDR) procedure (37).
GRSage at menarche as a Predictor of Age of Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
Large-scale genetic data have shown moderate to strong correlations between age at menarche and pubertal milestones across sexes, supporting the choice to capture puberty timing with age at menarche (38). GRSage at menarche was calculated using the summary statistics from Day et al. (38). GRS analyses were carried out using the procedure above to address aim 3 (Supplemental Methods in Supplement 1).
Causal Associations Between Puberty Timing and Age of Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
Mendelian randomization estimated the causal association between age at menarche and age of onset, early-onset AN, typical-onset AN, and AN risk, per aim 3. We used known genetic variants in GWAS summary statistics as instruments (5,38). Analyses were conducted with the TwoSampleMR package of MR-Base (39). We used the inverse-variance weighted estimator, which meta-analyzes the SNP-specific Wald estimates, and sensitivity approaches were applied (Supplement 1). Power calculations implied >80% power to detect a β of 1.01 for age at menarche on AN age of onset, OR of 0.90 for age at menarche on early-onset AN, OR of 1.16 for age at menarche on typical-onset AN, OR of 0.90 for age at menarche on AN risk, and β ≤ 0.99 for AN risk on age at menarche (40). Age at menarche was hypothesized to be inversely associated with early-onset AN and AN risk.
Results
Age of Onset Phenotype Summary
Table S1 in Supplement 2 describes the cohorts. The mean age of onset among the 9335 AN cases (99% female) in the 13 cohorts was 15.91 years (SD = 4.29, range 5–58), and approximately 15% had early-onset AN. A density plot of age of onset is shown in Figure S1 in Supplement 1.
GWASs of Age of Onset, Early-Onset AN, and Typical-Onset AN
The GWAS of age of onset (13 cohorts, 9335 cases) yielded no SNPs with p values < 1.67 × 10−8 (Figure S2 in Supplement 1). The SNP with the lowest p value was rs146976977 (p = 1.85 × 10−7). The phenotypic variance explained by GRSage of onset was R2 = 0.13%. SNP-h2 was 0.04 (SE = 0.05) with LDSC and 0.01 (SE = 0.02) with GREML.
No significant loci (p < 1.67 × 10−8) were observed for the GWAS of early-onset AN (5 cohorts, 1269 cases and 25,042 controls), although one false positive was observed at the standard genome-wide significance threshold (Figure S2 and Supplemental Results in Supplement 1). The false-positive designation was given because the SNP had no nearby linkage disequilibrium friends, an unrealistically large OR, was not genotyped (INFO score = 0.74), is low frequency, occurs in predominantly European populations, and based on the small sample size. GRSearly-onset AN explained 0.85% of the variance (liability scale R2), and observed scale SNP-h2 was 0.08 (SE = 0.02) with LDSC and 0.11 (SE = 0.01) with GREML. Liability scale SNP-h2 was 0.16–0.19 (SE = 0.04) with LDSC and 0.21–0.25 (SE = 0.02) with GREML, assuming a lifetime prevalence of 0.1% to 0.3% (28).
The GWAS of typical-onset AN (5 cohorts, 6998 cases and 25,042 controls) revealed two genome-wide significant loci (chromosome 3, rs3821875, OR = 1.21, 95% CI 1.14 to 1.29, p = 5.38 × 10−10; chromosome 9, rs4641158, OR = 1.23, 95% CI 1.15 to 1.31, p = 2.63 × 10−9) (Figure S2 in Supplement 1). The first was the top locus in the parent study and is complex and multigenic (i.e., >100 genes) with many chromatin and expression quantitative trait loci interactions. The second single-gene locus was not significant in the parent study and encodes CNTLN (centlein, centrosomal protein), which organizes microtubules (41) and is expressed in ovarian cells (42). Gene expression was most enriched in brain tissues, although no tissue expression p values (Figure S3 in Supplement 1), nor gene sets (Table S4 in Supplement 2), were Bonferroni significant. GRStypical-onset AN explained 0.25% of the variance (liability scale R2). Observed scale SNP-h2 was 0.21 (SE = 0.02) with LDSC and 0.19 (SE = 0.01) with GREML, and liability scale SNP-h2 assuming a lifetime prevalence of 0.9% to 4% (1,43,44) was 0.17–0.25 (SE = 0.02) with LDSC and 0.15–0.22 (SE = 0.02) with GREML.
The allelic effects at the eight genome-wide significant loci in the AN risk GWAS were investigated in the early- and typical-onset GWASs, and allelic effects were similar (Tables S5a–c in Supplement 2).
Genetic Correlations of Age of Onset, Early-Onset AN, and Typical-Onset AN
The genetic correlation between early- and typical-onset AN did not differ significantly from unity, rg = 0.81 (SE = 0.12). Owing to the low heritability of age of onset, the quantitative trait, we had insufficient power to explore genetic correlations with other traits, and none reached even nominal significance (p < .05).
We investigated genetic correlations between early-onset AN and 62 traits prioritized from six categories based on previous evidence from AN (5,17): psychiatric (i.e., schizophrenia, major depressive disorder), anthropometric (i.e., weight, height), glycemic (i.e., type 2 diabetes, insulin resistance), lipid related (i.e., high-density lipoprotein cholesterol, triglycerides, leptin), reproductive (i.e., age at menarche, age at menopause), and education and intelligence (i.e., IQ, college completion). Nominally significant (p < .05) genetic correlations were observed with eight traits (Table S6 in Supplement 2). Ranked in order of increasing p values, these were reproductive, education, glycemic, and lipid-related traits, but none were significant after FDR correction (37). We also considered the genetic correlations between early-onset AN, all 700+ traits on LD Hub, and previous GWAS of AN (Table S7 in Supplement 2). FDR-significant correlations were observed between early-onset AN and the three previous AN GWASs and a UK Biobank question on whether help was sought from a psychiatrist for nerves, anxiety, tension, or depression. The genetic correlations for typical-onset AN are in Table S8 in Supplement 2.
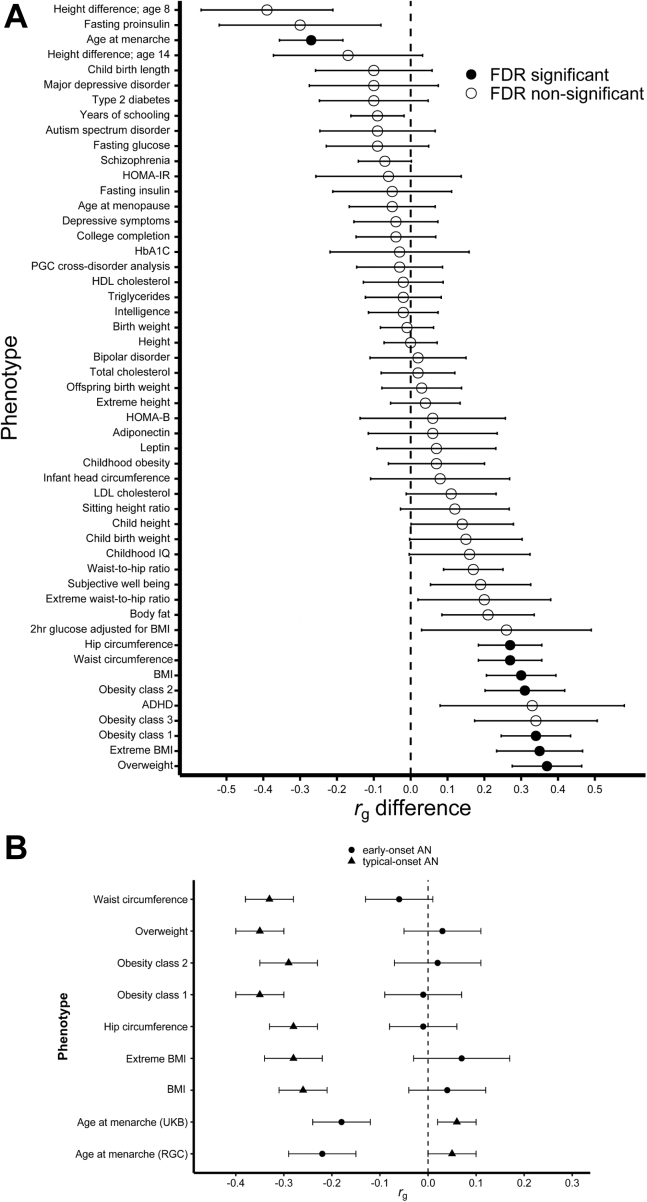
We compared the early-onset AN and typical-onset AN genetic correlations (Table S9 in Supplement 2). FDR-significant differences emerged for reproductive and anthropometric traits (Figure 1). Early-onset AN evidenced genetic overlap with younger age at menarche, whereas typical-onset AN did not. Early-onset AN did not genetically overlap with anthropometric traits, whereas typical-onset AN showed negative correlations.
Figure 1.
Early-onset and typical-onset AN show significantly different genetic correlation patterns with risk and comorbid traits. FDR-significant differences in genetic correlations were detected in two categories—anthropometric and reproductive—within six previously identified categories of risk or comorbid traits of interest for AN. (A) Sixty-two phenotypes were tested; duplicate phenotypes are not plotted (for duplicate phenotypes, we prioritized published summary statistics or the rg difference with the lowest SE). Full results are shown in Table S9 in Supplement 2. (B) Shows the rgs between the phenotypes and the age of onset subphenotypes. The error bars in both plots represent the SE. ADHD, attention-deficit/hyperactivity disorder; AN, anorexia nervosa; BMI, body mass index; FDR, false discovery rate; HbA1C, hemoglobin A1c; HDL, high-density lipoprotein; HOMA-B, homeostatic model assessment for beta cell function; HOMA-IR, HOMA for insulin resistance; LDL, low-density lipoprotein; PGC, Psychiatric Genomics Consortium; RGC, ReproGen Consortium; UKB, UK Biobank.
GRSage of onset, GRSearly-onset AN, and GRSAN as Predictors of Age of Onset of AN
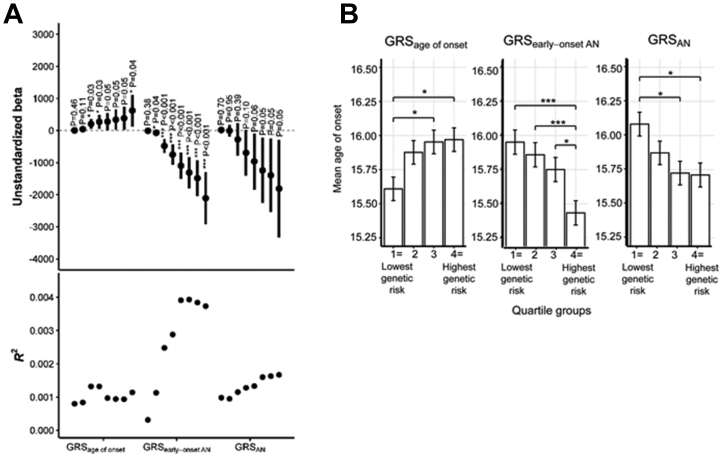
GRS analyses supported a common genetic basis of age of onset (Figure 2). Higher GRSage of onset significantly predicted higher age of onset at three pTs, and GRSearly-onset AN significantly predicted a younger age of onset at seven pTs. GRSAN did not significantly predict age of onset. The GRSs explained a small amount of phenotypic variance (R2) in age of onset (Figure 2). The highest R2s across pTs were 0.13% for GRSage of onset (at pT < .1), 0.39% for GRSearly-onset AN (pT < .4), and 0.17% for GRSAN (pT < 1). Cochrane Qs for the meta-analyses in Figure 2 (24 total) were nonsignificant (p < .05), indicating that despite methodological differences across cohorts (i.e., age of onset phenotyping, recruitment, and sampling), heterogeneity was not evident. The forest plots in Figure S4 in Supplement 1 depict β estimates across the cohorts; results are shown for the best-performing pTs for illustration. We also observed significant associations between GRS quartile groups and age of onset (Supplemental Results in Supplement 1; Figure 2). Descriptive information for the GRS leave-one-cohort-out analyses is provided in Table S10 in Supplement 2.
Figure 2.
GRSage of onset and GRSearly-onset AN significantly predict age of onset of AN. The p values are false discovery rate–corrected for multiple testing. ∗p < .05; ∗∗∗p < .001. (A) Unstandardized βs represent the average increase in age of onset (years) per 1-unit increase in GRS. The error bars are 95% confidence intervals. The eight data points in each field, from left to right, represent ascending p value selection thresholds (pT < .001, pT < .01, pT < .1, pT < .2, pT < .3, pT < .4, pT < .5, pT < 1) for inclusion of SNPs from the GWAS into the GRS (i.e., pT < 1 means that all SNPs were included in score calculation). Table S10 in Supplement 2 reports descriptive statistics for GRS. (B) Marginal means and standard errors are plotted. The tests of significant difference are from fixed-effects inverse-variance weighted meta-analyses of mean difference in age of onset by GRS quartile. The data include 13 cohorts (n = 9335) for GRSage of onset and GRSAN and 5 cohorts (n = 8267) for GRSearly-onset AN. AN, anorexia nervosa; GRS, genetic risk score; GRSAAO, GRS computed from the within-case age of onset GWAS; GRSAN, GRS computed from the case-control AN GWAS (2); GRSearly−onset AN, GRS computed from the case-control early-onset AN GWAS; GWAS, genome-wide association study; SNP, single nucleotide polymorphism.
GRSage at menarche as a Predictor of Age of Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
Per aim 3, we investigated the associations between GRSage at menarche and age of onset (13 cohorts, 9335 cases), early-onset AN (5 cohorts, 1269 cases and 25,042 controls), typical-onset AN (5 cohorts, 6998 cases and 25,042 controls), and AN risk (13 cohorts, 11,632 cases and 31,981 controls). GRSage at menarche significantly predicted age of onset of AN and early-onset AN at all pTs (Table 1). For instance, a 1 SD decrease in GRSage at menarche was associated with an age of onset decrease of 0.21 years (95% CI −0.31 to −0.12; at pT < .1) and a 20% higher odds of early-onset AN (95% CI 1.13 to 1.27; at pT < 1).
Table 1.
Association Between GRSage at menarche and Age at Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
| pT | Age of Onset of AN, 9335 Cases |
Early-Onset AN, 1269 Cases, 25,042 Controls |
Typical-Onset AN, 6998 Cases, 25,042 Controls |
AN Risk, 11,632 Cases, 31,981 Controls |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimates (95% CI) | p Value | Q | R2 | Estimates (95% CI) | p Value | Q | R2 | Estimates (95% CI) | p Value | Q | R2 | Estimates (95% CI) | p Value | Q | R2 | |
| 1 | −0.19 (−0.28 to −0.10) | 2.72 × 10−5a | 23.92a | 0.42% | 1.20 (1.13 to 1.27) | 4.13 × 10−9a | 2.35 | 0.30% | 0.99 (0.96 to 1.01) | .33 | 9.56 | 0.02% | 1.02 (0.99 to 1.04) | .21 | 19.96 | 0.03% |
| .5 | −0.19 (−0.28 to −0.10) | 2.48 × 10−5a | 22.63a | 0.42% | 1.20 (1.13 to 1.27) | 4.81 × 10−9a | 2.77 | 0.31% | 0.98 (0.96 to 1.01) | .30 | 9.58a | 0.02% | 1.01 (0.99 to 1.04) | .23 | 19.78 | 0.03% |
| .4 | −0.20 (−0.29 to −0.11) | 1.52 × 10−5a | 23.45a | 0.44% | 1.19 (1.12 to 1.27) | 8.94 × 10−9a | 2.96 | 0.30% | 0.98 (0.96 to 1.01) | .29 | 8.79 | 0.02% | 1.01 (0.99 to 1.04) | .24 | 19.64 | 0.03% |
| .3 | −0.21 (−0.30 to −0.12) | 8.00 × 10−6a | 23.53a | 0.46% | 1.20 (1.13 to 1.27) | 5.30 × 10−9a | 2.32 | 0.30% | 0.98 (0.95 to 1.01) | .16 | 8.01 | 0.02% | 1.01 (0.99 to 1.04) | .41 | 18.72 | 0.03% |
| .2 | −0.21 (−0.30 to −0.12) | 8.59 × 10−6a | 22.96a | 0.46% | 1.19 (1.12 to 1.26) | 2.24 × 10−8a | 2.47 | 0.27% | 0.98 (0.95 to 1.01) | .16 | 9.37 | 0.02% | 1.01 (0.99 to 1.04) | .39 | 20.34 | 0.03% |
| .1 | −0.21 (−0.31 to −0.12) | 6.13 × 10−6a | 28.06a | 0.51% | 1.19 (1.12 to 1.27) | 2.43 × 10−8a | 3.74 | 0.29% | 0.98 (0.95 to 1.00) | .10 | 10.50 | 0.02% | 1.01 (0.99 to 1.04) | .34 | 20.94 | 0.03% |
| .01 | −0.21 (−0.29 to −0.12) | 7.24 × 10−6a | 27.77a | 0.52% | 1.16 (1.09 to 1.23) | 1.22 × 10−6a | 2.03 | 0.23% | 0.98 (0.95 to 1.00) | .09 | 5.73 | 0.02% | 1.01 (0.99 to 1.04) | .33 | 9.95 | 0.02% |
| .001 | −0.18 (−0.27 to −0.09) | 4.96 × 10−5a | 16.30 | 0.35% | 1.13 (1.06 to 1.19) | 8.00 × 10−5a | 2.44 | 0.15% | 0.97 (0.94 to 1.00) | .04 | 4.68 | 0.02% | 1.01 (0.99 to 1.03) | .43 | 12.39 | 0.02% |
| 1 × 10−5 | −0.15 (−0.23 to −0.06) | 1.12 × 10−3a | 14.32 | 0.28% | 1.10 (1.03 to 1.16) | 2.57 × 10−3a | 8.93 | 0.18% | 0.96 (0.93 to 0.99) | .01 | 2.83 | 0.02% | 1.00 (0.98 to 1.03) | .91 | 15.25 | 0.02% |
| 5 × 10−8 | −0.12 (−0.20 to −0.03) | .01a | 10.69 | 0.19% | 1.11 (1.05 to 1.18) | 6.17 × 10−4a | 4.16 | 0.14% | 0.97 (0.94 to 1.00) | .05 | 1.40 | 0.01% | 1.00 (0.98 to 1.03) | .73 | 8.92 | 0.02% |
Estimates are β for age of onset of AN, and odds ratio for AN, early-onset AN, and typical-onset AN, and for interpretation purposes, reflect the effect of a standard deviation decrease in GRSage at menarche (from the mean). R2 estimates for early-onset AN, typical-onset AN, and AN risk are liability-scale estimates.
AN, anorexia nervosa; GRS, genetic risk score; pT, the p value threshold for single nucleotide polymorphism selection for each GRS score.
False discovery rate–adjusted p < .05.
Age at Menarche as a Causal Risk Factor for Age of Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
Mendelian randomization provided evidence consistent with a causal link between younger age at menarche and early-onset AN (β = −0.21, SE = 0.09, p = .02) (Table 2). For each 1-year decrease in age at menarche below the mean (in the observed range), the odds of early-onset AN increased by 23% (95% CI 3% to 48%). Genetically determined age at menarche did not significantly predict age of onset of AN—although the mostly positive, wide CI precludes definitive interpretation (β = 0.23, 95% CI −0.02 to 0.48, p = .08)—and did not predict typical-onset AN (β = 0.06, 95% CI −0.04 to 0.16, p = .26) or AN risk (β = −0.04, 95% CI −0.12 to 0.04, p = .27). In the opposite direction, AN risk was not a causal risk factor for age at menarche (β = 0.03, 95% CI −0.05 to 0.11, p = .50). Given the lack of SNP instruments from the age of onset, early-onset, and typical-onset AN GWASs, their causal effects on age at menarche could not be investigated. Sensitivity analyses complemented the findings of the inverse-variance weighted analyses (Supplement 1).
Table 2.
MR Analyses Testing the Causal Associations Between Age at Menarche and Age of Onset of AN, Early-Onset AN, Typical-Onset AN, and AN Risk
| Analysis | Age at Menarche→Age of Onset of AN | Age at Menarche→Early-Onset AN | Age at Menarche→Typical-Onset AN | Age at Menarche→AN Risk | AN Risk→Age at Menarche |
|---|---|---|---|---|---|
| IVW MR, β (SE) | 0.23 (0.13) | −0.21 (0.09)a | 0.06 (0.05) | −0.04 (0.04) | 0.03 (0.04) |
| Sensitivity Analyses | |||||
| Heterogeneity, Q | 238.38 | 212.28 | 295.16b | 395.54b | 3.39 |
| Egger regression, β (SE) | 0.29 (0.38) | −0.20 (0.25) | 0.17 (0.14) | 0.13 (0.11) | 0.41 (0.35) |
| Egger regression, intercept (SE) | −0.002 (0.02) | −0.0003 (0.01) | −0.005 (0.01) | −0.01 (0.004) | −0.03 (0.03) |
| Weighted median estimate, β (SE) | 0.21 (0.20) | −0.09 (0.15) | 0.07 (0.07) | 0.0002 (0.05) | 0.04 (0.04) |
| MR-PRESSO global | 240.42 | 214.14 | 297.76b | 399.03b | – |
| Steiger | Direction: truea | Direction: trueb | Direction: trueb | Direction: trueb | Direction: trueb |
| GSMR, β (SE) | 0.07 (0.10) | −0.11 (0.07) | 0.16 (0.03)b | 0.03 (0.02) | 0.03 (0.03) |
β values are differences in the outcome per year increase in age at menarche for the analyses with menarche timing as the exposure. Number of single nucleotide polymorphism instruments in IVW, Egger, weighted median, and GSMR analyses: menarche→age of onset of AN = 208, menarche→early-onset AN = 210, age at menarche→typical-onset AN = 210, age at menarche→AN risk = 206, AN risk→age at menarche = 3.
AN, anorexia nervosa; GSMR, generalized summary data-based Mendelian randomization; IVW, inverse-variance weighted; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier.
p < .05.
p < .001.
Discussion
This study provides evidence for a genetic basis for age of onset of AN, which is conferred at least partly through common genetic variants. GRSs capturing the effects of alleles associated with AN age of onset and early-onset AN significantly predicted age of onset. Furthermore, results suggested that the genetic architecture underlying earlier puberty, represented by age at menarche, may bring about an earlier onset of AN.
The SNP-h2 of the subphenotype of early-onset AN was similar to what has been reported for psychiatric diagnoses including AN (SNP-h2s 0.10–0.26). The SNP-h2 of age of onset, a quantitative trait, was low (SNP-h2s 0.01–0.04). Other psychological and behavioral quantitative traits have been found to have low heritabilities (i.e., depressive symptoms, subjective well-being, cigarette smoking, extraversion: SNP-h2s 0.05–0.06) (16). Twin-based upper bound estimates of heritability (twin-h2) of age of onset are lacking but would help to contextualize these results, as would SNP-h2 estimates from large, homogeneous datasets (>5000) (45). The heritabilities of early-onset and typical-onset AN were similar. This paints a different picture to the Brainstorm Consortium’s report combining several psychiatric disorders, which suggested earlier age of onset of psychiatric illness as an indicator for higher heritability (16). The Brainstorm Consortium analysis was a broad brushstroke view that relied on rough single estimates of average age of onset from experts and did not use phenotypic data. Our approach is data based, but the estimates are preliminary, given that the discovery GWASs have small samples for psychiatric GWAS.
No loci reached genome-wide significance in the age of onset and early-onset AN GWASs. Precise epidemiological estimates of early-onset AN are lacking, but prevalence is low and less than the lifetime prevalence of 0.3% in adolescents and 0.9% in adults (1,28). This limits GWAS statistical power at the current sample sizes. The genetic correlation between early-onset AN and AN risk is stronger than the genetic correlation of AN risk with other traits (i.e., psychiatric, anthropometric, metabolic: rgs −0.36 to 0.45) (5). The genetic correlation between early-onset and typical-onset AN was high, although distinct patterns of genetic correlations with other traits were observed. Early-onset AN correlated with lower age at menarche, and typical-onset AN correlated negatively with anthropometric traits. Studying homogeneous clinical subphenotypes such as early-onset AN may yield novel insight into the etiology of AN but is reliant on large phenotyping collections. Preliminary findings from this study suggest that a reproductive biology–based etiology for some patients may be worthy of further exploration.
Two loci were associated with typical-onset AN, one highly multigenic locus containing many brain-expressed genes and associated with AN risk (5), and a single-gene locus encoding a centrosomal protein and associated with phenotypes genetically correlated with AN (5), such as body fat distribution (adjusted-for-BMI) (46), high-density lipoprotein cholesterol level (47), and unipolar depression (48). The typical-onset AN GWAS was statistically powered for the initial implication of variants, given that ORs of 1.12 to 1.28 are in the range of the larger effects that we would expect to see in AN.
An important finding in this study was that GRSs for age of onset and early-onset AN significantly predicted age of onset. This provides evidence for a common variant genetic basis for age of onset, although the clinical significance is unclear. The age of onset difference was approximately 6 months between the lowest and highest GRSage of onset quartiles. Future studies characterizing the more extreme ends of the GRS distribution of age of onset (i.e., 5%) may see considerably greater differences in timing of AN onset. Irreversible growth stunting and long-term bone disease (49) are morbidities exacerbated by earlier onset and could be mitigated by risk prediction tools that predict not only illness risk but timing of onset.
The variance in age of onset accounted for by common genetic variants was low. The explanatory power of GRS is influenced by the underlying genetic architecture and heritability and sufficiently powered discovery GWASs. Even so, the ability to account for variance in psychiatric phenotypes has been limited. Schizophrenia, for example, has a large genetic component and a heritability estimate of 80% (50), but GRS has only been able to account for 7% of variation on the liability scale—the most of any psychiatric disorder—with this estimate increasing as GWAS power has increased (51). GRS broadly has proven useful in research applications—by predicting incidence, disease severity, and treatment response—and can yield benefit to personalized prevention and early intervention of disease even when accounting for low variance (52).
A prevailing difficulty in risk factor research for AN has been how to consider peripuberty as a puzzle piece. Early-onset AN was historically termed premenarchal AN (53), which failed to appreciate that the starvation emblematic of the disease arrests puberty (and omits males). The association between puberty timing and AN can be difficult to study because the low prevalence of AN renders prospective association studies impractical, and in treatment-seeking populations, patients typically have delays in help seeking. Genetic analyses circumvent some of these methodological difficulties. In this study, GRS for earlier age at menarche predicted early-onset AN and lower age of onset. Furthermore, there was evidence consistent with earlier age at menarche being a causal risk factor for early-onset AN. This could suggest a genetically distinct variant of AN, genetically linked to puberty that is predisposed to an earlier age of onset. Indeed, this aligns with previous hypotheses (54,55) that a subset of women with eating disorders may represent an ovarian hormone–sensitive phenotype. Furthermore, twin studies suggest that estrogen plays a role in genetic risk for disordered eating (17). Our results converge with clinical studies implicating early pubertal timing as a risk factor for eating disorders (17) and extend the literature by suggesting a shared or potentially causal genetic molecular basis.
Cases with early-onset AN did not show a genetic relationship with BMI or related anthropometric indices, in contrast to cases with typical-onset AN. Literature comparing premorbid BMI trajectory between these groups is lacking. Because metabolic factors in AN are a burgeoning area of study, this could be an interesting finding to follow up. Continued investigation of the subphenotype of early-onset AN may help to inform etiology and classification.
A limitation of this study is that the phenotyping of age of onset was not standardized across cohorts. The phenotype was age of onset of AN diagnosis in some cohorts and age of first symptoms in others. The reliance on retrospective recall of age of onset is also a limitation. The generalizability of the findings to non-Europeans and males is not known. Genetic effects may emerge earlier in males according to existing research (56). From the statistical power analyses, it seems reasonable to conclude that at this stage, the GWASs of age of onset and early-onset AN are underpowered to identify individual variants.
In conclusion, this study provides evidence that a genetic basis underlies AN age of onset and that reproductive biology may influence the early onset of AN. Larger, well-matched case-control samples with standardized age of onset data will help to further reveal the biological mechanisms that influence age of onset.
Acknowledgments and Disclosures
The PGC is supported by funding from the National Institute of Mental Health (Grant Nos. U01MH109528 and U01MH109514). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (Grant No. NWO 480-05-003, principal investigator: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam.
This content is the responsibility of the authors and does not represent the views of the funding bodies.
We are deeply thankful to the thousands of individuals who contributed their time and biological samples, and to the many collaborators who helped to collate these samples over the years. We are extremely grateful to members of the Eating Disorders Working Group of the Psychiatric Genomics Consortium for their contributions, efforts, and leadership (see Supplemental Appendix in Supplement 1). We thank the Anorexia Nervosa Genetics Initiative, which is an initiative of the Klarman Family Foundation. We thank the Children’s Hospital of Philadelphia, the Price Foundation Collaborative Group, Genetic Consortium for Anorexia Nervosa, and the Wellcome Trust Case Control Consortium-3. We thank the Stockholm Centre for Eating Disorders (SCÄ), the Swedish National Quality Register for Eating Disorders (Riksät), and Lifegene for their support of Anorexia Nervosa Genetics Initiative Sweden. We also thank the QSkin Sun and Health Study for control samples.
OAA is a consultant for HealthLytix. CMB is a grant recipient from Shire Pharmaceuticals and served on Shire Scientific Advisory Board; she receives royalties from Pearson. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.09.001.
Supplementary Material
References
- 1.Hudson J.I., Hiripi E., Pope H.G., Jr., Kessler R.C. The prevalence and correlates of eating disorders in the National Comorbidity Survey Replication. Biol Psychiatry. 2007;61:348–358. doi: 10.1016/j.biopsych.2006.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udo T., Grilo C.M. Prevalence and correlates of DSM-5-defined eating disorders in a nationally representative sample of U.S. adults. Biol Psychiatry. 2018;84:345–354. doi: 10.1016/j.biopsych.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zipfel S., Giel K.E., Bulik C.M., Hay P., Schmidt U. Anorexia nervosa: Aetiology, assessment, and treatment. Lancet Psychiatry. 2015;2:1099–1111. doi: 10.1016/S2215-0366(15)00356-9. [DOI] [PubMed] [Google Scholar]
- 4.Yilmaz Z., Hardaway J.A., Bulik C.M. Genetics and epigenetics of eating disorders. Adv Genomics Genet. 2015;5:131–150. doi: 10.2147/AGG.S55776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson H.J., Yilmaz Z., Thornton L.M., Hübel C., Coleman J.R.I., Gaspar H.A., et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51:1207–1214. doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Javaras K.N., Runfola C.D., Thornton L.M., Agerbo E., Birgegård A., Norring C., et al. Sex- and age-specific incidence of healthcare-register-recorded eating disorders in the complete Swedish 1979–2001 birth cohort. Int J Eat Disord. 2015;48:1070–1081. doi: 10.1002/eat.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favaro A., Busetto P., Collantoni E., Santonastaso P. In: Age of Onset of Mental Disorders: Etiopathogenetic and Treatment Implications. de Girolamo G., McGorry P.D., Sartorius N., editors. Springer International Publishing; Cham: 2019. The age of onset of eating disorders; pp. 203–216. [Google Scholar]
- 8.Kipman A., Bruins-Slot L., Boni C., Hanoun N., Adès J., Blot P., et al. 5-HT(2A) gene promoter polymorphism as a modifying rather than a vulnerability factor in anorexia nervosa. Eur Psychiatry. 2002;17:227–229. doi: 10.1016/s0924-9338(02)00657-0. [DOI] [PubMed] [Google Scholar]
- 9.Plana M.T., Torres T., Rodríguez N., Boloc D., Gassó P., Moreno E., et al. Genetic variability in the serotoninergic system and age of onset in anorexia nervosa and obsessive-compulsive disorder. Psychiatry Res. 2019;271:554–558. doi: 10.1016/j.psychres.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Dardennes R.M., Zizzari P., Tolle V., Foulon C., Kipman A., Romo L., et al. Family trios analysis of common polymorphisms in the obestatin/ghrelin, BDNF and AGRP genes in patients with anorexia nervosa: Association with subtype, body-mass index, severity and age of onset. Psychoneuroendocrinology. 2007;32:106–113. doi: 10.1016/j.psyneuen.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Ribasés M., Gratacòs M., Fernández-Aranda F., Bellodi L., Boni C., Anderluh M., et al. Association of BDNF with anorexia, bulimia and age of onset of weight loss in six European populations. Hum Mol Genet. 2004;13:1205–1212. doi: 10.1093/hmg/ddh137. [DOI] [PubMed] [Google Scholar]
- 12.Ferentinos P., Koukounari A., Power R., Rivera M., Uher R., Craddock N., et al. Familiality and SNP heritability of age at onset and episodicity in major depressive disorder. Psychol Med. 2015;45:2215–2225. doi: 10.1017/S0033291715000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilker R., Helenius D., Fagerlund B., Skytthe A., Christensen K., Werge T.M., et al. Is an early age at illness onset in schizophrenia associated with increased genetic susceptibility? Analysis of data from the Nationwide Danish Twin Register. EBiomedicine. 2017;18:320–326. doi: 10.1016/j.ebiom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin P.I., McInnis M.G., Potash J.B., Willour V., MacKinnon D.F., DePaulo J.R., Zandi P.P. Clinical correlates and familial aggregation of age at onset in bipolar disorder. Am J Psychiatry. 2006;163:240–246. doi: 10.1176/appi.ajp.163.2.240. [DOI] [PubMed] [Google Scholar]
- 15.Svensson A.C., Lichtenstein P., Sandin S., Öberg S., Sullivan P.F., Hultman C.M. Familial aggregation of schizophrenia: The moderating effect of age at onset, parental immigration, paternal age and season of birth. Scand J Public Health. 2012;40:43–50. doi: 10.1177/1403494811420485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brainstorm Consortium. Anttila V., Bulik-Sullivan B., Finucane H.K., Walters R.K., Bras J., et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360 doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klump K.L. Puberty as a critical risk period for eating disorders: A review of human and animal studies. Horm Behav. 2013;64:399–410. doi: 10.1016/j.yhbeh.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins N., Lewis C.M. Genetics of depression: Progress at last. Curr Psychiatry Rep. 2017;19:43. doi: 10.1007/s11920-017-0803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeb K.L., Brown M., Goldstein M.M. In: Eating Disorders in Children and Adolescents: A Clinical Handbook. Le Grange D., Lock J., editors. Guilford Press; New York: 2011. Assessment of eating disorders in children and adolescents; pp. 156–198. [Google Scholar]
- 20.Peebles R., Wilson J.L., Lock J.D. How do children with eating disorders differ from adolescents with eating disorders at initial evaluation? J Adolesc Health. 2006;39:800–805. doi: 10.1016/j.jadohealth.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Walker T., Watson H.J., Leach D.J., McCormack J., Tobias K., Hamilton M.J., Forbes D.A. Comparative study of children and adolescents referred for eating disorder treatment at a specialist tertiary setting. Int J Eat Disord. 2014;47:47–53. doi: 10.1002/eat.22201. [DOI] [PubMed] [Google Scholar]
- 22.Steinhausen H.C. Outcome of eating disorders. Child Adolesc Psychiatr Clin N Am. 2009;18:225–242. doi: 10.1016/j.chc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Baker J.H., Thornton L.M., Bulik C.M., Kendler K.S., Lichtenstein P. Shared genetic effects between age at menarche and disordered eating. J Adolesc Health. 2012;51:491–496. doi: 10.1016/j.jadohealth.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirk K.M., Martin F.C., Mao A., Parker R., Maguire S., Thornton L.M., et al. The Anorexia Nervosa Genetics Initiative: Study description and sample characteristics of the Australian and New Zealand arm. Aust N Z J Psychiatry. 2017;51:583–594. doi: 10.1177/0004867417700731. [DOI] [PubMed] [Google Scholar]
- 25.Thornton L.M., Munn-Chernoff M.A., Baker J.H., Juréus A., Parker R., Henders A.K., et al. The Anorexia Nervosa Genetics Initiative (ANGI): Overview and methods. Contemp Clin Trials. 2018;74:61–69. doi: 10.1016/j.cct.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson S.A., Crow S.J., Le Grange D., Swendsen J., Merikangas K.R. Prevalence and correlates of eating disorders in adolescents. Results from the National Comorbidity Survey Replication Adolescent Supplement. Arch Gen Psychiatry. 2011;68:714–723. doi: 10.1001/archgenpsychiatry.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauderman W.J., Morrison J.M. Quanto 1.2.4: A computer program for power and sample size calculations for genetic epidemiology studies. 2009. https://pphs.usc.edu/download-quanto/ Available at:
- 30.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visscher P.M., Hemani G., Vinkhuyzen A.A., Chen G.B., Lee S.H., Wray N.R., et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan L., Yilmaz Z., Gaspar H., Walters R., Goldstein J., Anttila V., et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry. 2017;174:850–858. doi: 10.1176/appi.ajp.2017.16121402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S.W., O’Reilly P.F. PRSice-2: Polygenic risk score software for biobank-scale data. GigaScience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 38.Day F.R., Thompson D.J., Helgason H., Chasman D.I., Finucane H., Sulem P., et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makino K., Umeda K., Uezu A., Hiragami Y., Sakamoto T., Ihn H., Nakanishi H. Identification and characterization of the novel centrosomal protein centlein. Biochem Biophys Res Commun. 2008;366:958–962. doi: 10.1016/j.bbrc.2007.12.050. [DOI] [PubMed] [Google Scholar]
- 42.Buckley M.A., Woods N.T., Tyrer J.P., Mendoza-Fandiño G., Lawrenson K., Hazelett D.J., et al. Functional analysis and fine mapping of the 9p22.2 ovarian cancer susceptibility locus. Cancer Res. 2019;79:467–481. doi: 10.1158/0008-5472.CAN-17-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keski-Rahkonen A., Mustelin L. Epidemiology of eating disorders in Europe: Prevalence, incidence, comorbidity, course, consequences, and risk factors. Curr Opin Psychiatry. 2016;29:340–345. doi: 10.1097/YCO.0000000000000278. [DOI] [PubMed] [Google Scholar]
- 44.Micali N., Martini M.G., Thomas J.J., Eddy K.T., Kothari R., Russell E., et al. Lifetime and 12-month prevalence of eating disorders amongst women in mid-life: A population-based study of diagnoses and risk factors. BMC Med. 2017;15:12. doi: 10.1186/s12916-016-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tropf F.C., Lee S.H., Verweij R.M., Stulp G., van der Most P.J., de Vlaming R., et al. Hidden heritability due to heterogeneity across seven populations. Nat Hum Behav. 2017;1:757–765. doi: 10.1038/s41562-017-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dede A.D., Lyritis G.P., Tournis S. Bone disease in anorexia nervosa. Hormones. 2014;13:38–56. doi: 10.1007/BF03401319. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 51.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 53.Cooper P.J., Watkins B., Bryant-Waugh R., Lask B. The nosological status of early onset anorexia nervosa. Psychol Med. 2002;32:873–880. doi: 10.1017/s0033291702005664. [DOI] [PubMed] [Google Scholar]
- 54.Hardin S.L., Thornton L.M., Munn-Chernoff M.A., Baker J.H. Premenstrual symptoms as a marker of ovarian hormone sensitivity in eating disorders. Int J Eat Disord. 2020;53:296–301. doi: 10.1002/eat.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker J.H., Girdler S.S., Bulik C.M. The role of reproductive hormones in the development and maintenance of eating disorders. Expert Rev Obstet Gynecol. 2012;7:573–583. doi: 10.1586/eog.12.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Culbert K.M., Burt S.A., Klump K.L. Expanding the developmental boundaries of etiologic effects: The role of adrenarche in genetic influences on disordered eating in males. J Abnorm Psychol. 2017;126:593–606. doi: 10.1037/abn0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.