Abstract
Monoclonal antibodies (mAbs) offer a treatment option for individuals with severe COVID-19 and are especially important in high-risk individuals where vaccination is not an option. Given the importance of understanding the evolution of resistance to mAbs by SARS-CoV-2, we reviewed the available in vitro neutralization data for mAbs against live variants and viral constructs containing spike mutations of interest. Unfortunately, evasion of mAb-induced protection is being reported with new SARS-CoV-2 variants. The magnitude of neutralization reduction varied greatly among mAb–variant pairs. For example, sotrovimab retained its neutralization capacity against Omicron BA.1 but showed reduced efficacy against BA.2, BA.4 and BA.5, and BA.2.12.1. At present, only bebtelovimab has been reported to retain its efficacy against all SARS-CoV-2 variants considered here. Resistance to mAb neutralization was dominated by the action of epitope single amino acid substitutions in the spike protein. Although not all observed epitope mutations result in increased mAb evasion, amino acid substitutions at non-epitope positions and combinations of mutations also contribute to evasion of neutralization. This Review highlights the implications for the rational design of viral genomic surveillance and factors to consider for the development of novel mAb therapies.
Subject terms: SARS-CoV-2, Viral evolution, Viral immune evasion, Antimicrobial responses, Infection
In this Review, Carabelli, Robertson and colleagues explore data on the neutralization of globally circulating variants of concern by monoclonal antibodies (mAbs) and discuss how knowledge of the dynamics of viral evasion of mAbs can contribute to viral surveillance and the development of novel mAb treatments, as well as inform predictions of resistance that may arise in the future.
Introduction
The SARS-CoV-2 pandemic has resulted in nearly 600 million confirmed cases of COVID-19 and more than 6 million deaths as of August 2022 (ref.1). Vaccines, antiviral drugs, monoclonal antibodies (mAbs) and non-pharmaceutical interventions such as lockdowns, contact-tracing, isolation, social distancing and intensive care and/or oxygen use have been deployed to control the spread of the virus and mitigate the harms of disease2–4. Global efforts to develop and deploy vaccinations have been unprecedented in their speed and scale and have contributed greatly to reducing transmission and severe disease in infected individuals. Antivirals such as remdesivir, molnupiravir and nirmatrelvir, and mAbs, administered either individually, such as sotrovimab or bebtelovimab, or as combination therapy ‘cocktails’, such as Evusheld (cilgavimab + tixagevimab), have provided much-needed additional treatment options for the clinically vulnerable and those who progress to severe disease3.
As of August 2022, a dozen mAbs that target the virus’s spike protein (Table 1) have been approved for clinical use in treating those infected by, or exposed to, SARS-CoV-2. mAbs that mediate the immune response by targeting host proteins also exist but are not considered further in this Review5. Virus-targeting mAbs are designed to reduce COVID-19 severity by binding to the SARS-CoV-2 spike glycoprotein leading to direct neutralization, antibody-dependent cellular phagocytosis, antibody-dependent cell-mediated cytotoxicity6 or complement activation. These prevent the virus from binding to the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of human cells, which is required for infection. The mAbs approved for clinical use and discussed here target the receptor binding domain (RBD) of the spike protein, but some mAbs in the early stages of development target other spike domains7,8.
Table 1.
Monoclonal antibodies approved for clinical use and SARS-CoV-2 variant evasion of neutralization
| Generic name | Other name | Manufacturer | mAb class | Spike epitopea | VOC resistanceb |
|---|---|---|---|---|---|
| Amubarvimab + romlusevimab85,86 | BRII-196 + BRII-198, P2C-1F11 + P2B-1G5 | Brii Biosciences |
Amubarvimab: class 1 Romlusevimab: no data |
Amubarivimab: 403, 415, 416, 417, 420, 421, 453, 455, 456, 457, 458, 459, 460, 473, 474, 475, 476, 477, 486, 487, 489, 493, 502, 505 Romlusevimab: no data |
Strong: BA.1.1, BA.4/5 Moderate: BA.1, BA.2, BA.2.12.1 |
| Bamlanivimab10 (withdrawn) | LYCoV555, LY3819253 | Eli Lilly | Class 2 | 351, 449, 450, 452, 455, 456, 470, 472, 481, 482, 483, 484, 485, 486, 487, 488, 489, 490, 492, 493, 494 | Strong: Beta, Gamma, Delta, BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4/5 |
| Bamlanivimab + etesevimab13,87,88 | Etesevimab: LY3832479, LY-CoV016 (see bamlanivimab) | Eli Lilly |
Etesevimab: class 1 Bamlanivimab: class 2 |
Etesevimab: 403, 405, 406, 408, 409, 415, 416, 417, 420, 421, 455, 456, 457, 458, 459, 460, 473, 474, 475, 476, 477, 486, 487, 489, 493, 494, 495, 500, 501, 502, 504, 505 (see bamlanivimab) | Strong: Beta, Gamma, BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4/5 |
| Bebtelovimab89 | LY-CoV1404, LY3853113 | Eli Lilly | Class 3 | 346, 439, 440, 441, 443, 444, 445, 446, 447, 448, 449, 450, 498, 499, 500, 501, 502, 506, 509 | |
| DXP-604 (ref.90) | NA | Singlomics + BeiGene | Class 1 | 403, 415, 416, 417, 420, 421, 453, 455, 456, 457, 458, 459, 460, 473, 474, 475, 476, 477, 486, 487, 489, 493, 496, 498, 500, 501, 502, 503, 505 |
Moderate: BA.1 Mild: Beta |
| Regdanvimab91 | Regkirona, CT-P59 | Celltrion Healthcare | Class 1 | 351, 403, 417, 446, 449, 450, 452, 453, 455, 456, 470, 483, 484, 485, 486, 489, 490, 492, 493, 494, 495, 496, 498, 505 |
Strong: BA.1, BA.2, BA.1.1 Moderate: Beta, Delta, Gamma |
| Ronapreve12,92 | REGEN-CoV2, casirivimab + imdevimab | Regeneron |
Casirivimab: class 1 Imdevimab: class 3 |
Casirivimab: 403, 417, 421, 453, 455, 456, 475, 476, 484, 485, 486, 487, 488, 489, 493 Imdevimab: 346, 439, 440, 441, 444, 445, 446, 447, 448, 449, 450, 498, 500 |
Strong: BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4/5 |
| Sotrovimab11,93,94 |
Xevudy, VIR-7831, GSK4182136 Parental antibody: S309 |
GlaxoSmith-Kline | Class 3 | 333, 334, 335, 337, 339, 340, 343, 344, 345, 346, 354, 356, 357, 359, 360, 361, 441, 509 |
Moderate: BA.2, BA.2.12.1, BA.4/5 Mild: BA.1 |
| Cilgavimab + tixagevimab95–97 |
Evusheld AZD7442, AZD1061/AZD8895 Parental antibodies: COV2-2130, COV2-2196 |
AstraZeneca |
Tixagevimab: class 1 Cilgavimab: class 2 |
Tixagevimab: 455, 456, 458, 475, 476, 477, 478, 479, 484, 485, 486, 487, 488, 489, 493 Cilgavimab: 345, 346, 439, 440, 441, 443, 444, 445, 446, 447, 448, 449, 450, 452, 484, 490, 492, 493, 494, 499 |
Strong: BA.1.1 Moderate: BA.1, BA.2.12.1, BA.4/5 Mild: BA.2 |
mAb, monoclonal antibody; mFRN, geometric mean fold reduction in neutralization; NA, not available; VOC, variant of concern. aEpitope positions are defined as those within 4.5 Å of the mAb when it is bound to the spike protein. bVOC strength of resistance is reported as strong (mFRN > 100), moderate (mFRN = 10–100) or mild (mFRN = 3–10), based on the data reported in Fig. 1. Although these designators are useful to the extent that they assign a qualitative judgement to the level of resistance, it must be emphasized that resistance of variants occurs across a continuum.
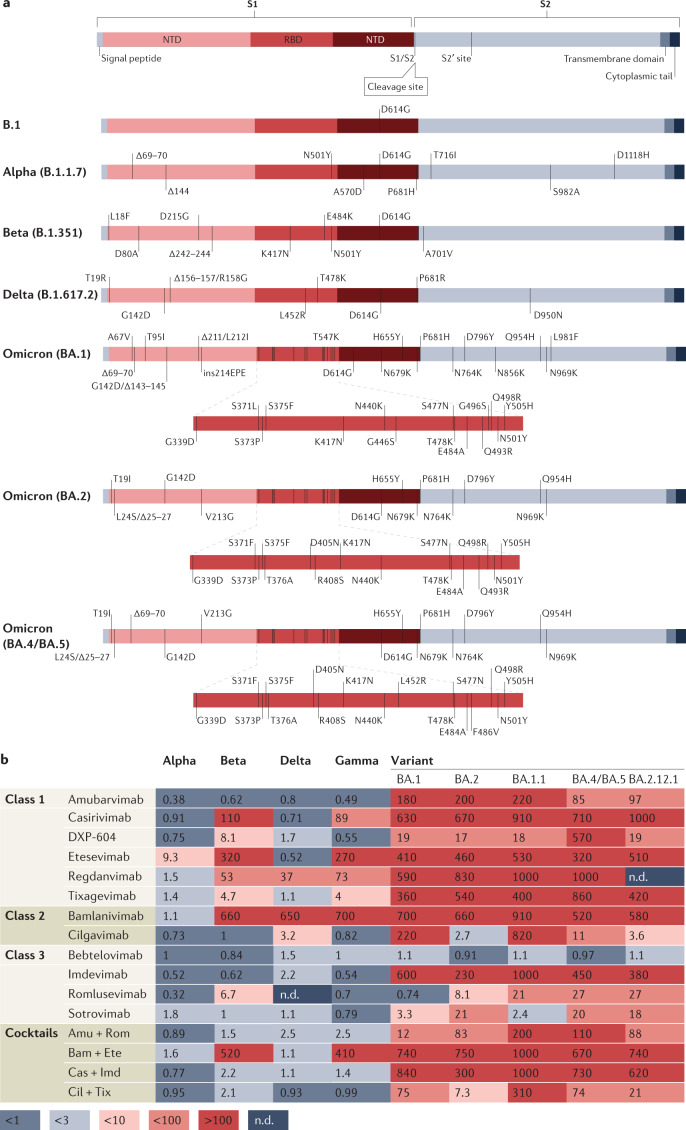
The efficacy of mAbs has been threatened by the emergence of SARS-CoV-2 variants resistant to existing treatments9. On 16 April 2021, the US Food and Drug Administration (FDA) ended the Emergency Use Authorization for bamlanivimab due to resistance shown by variants (Iota, Epsilon and others) carrying the L452R and E484K amino acid substitutions10. Since then, the mAb sotrovimab and the cocktails Ronapreve (casirivimab + imdevimab) and a combination of bamlanivimab + etesevimab have also had limitations placed on their use as resistant variants continue to emerge and spread11–13. Variants of concern (VOCs) Delta and Omicron, BA.1 and BA.2 lineages, have spread globally14–17, with Gamma and Beta VOCs previously associated mainly with regional spread18 (and sequencing bias) in Brazil and South Africa, respectively, and travel to these regions. At present, the Omicron lineages BA.4 and BA.5 (defined by the same spike mutations, in particular the RBD amino acid substitutions L452R and F486V, deletion 69–70, and the reversion R493Q, relative to BA.2), the BA.2.12.1 sub-lineage (defined by the additional RBD mutation L452Q, relative to BA.2) and others are emerging globally and causing new epidemic waves19–21 (Fig. 1a; see Supplementary Fig. 1). Unfortunately, all of these VOCs have shown resistance to available mAb treatments in vitro and so may be less susceptible to treatment in a clinical setting22–25.
Fig. 1. SARS-CoV-2 variant of concern evasion of neutralization by monoclonal antibodies.
a, Spike mutational profiles for each variant of concern (VOC) with the receptor binding domain (RBD) also shown for BA.1, BA.2, and BA.4 and BA.5. b, Geometric mean fold reduction in neutralization (mFRN) values of monoclonal antibodies (mAbs) against VOCs relative to reference or control variants. Means are calculated from published studies reporting neutralization data on clinically approved mAbs against VOCs. Data for the associated single studies are shown in Supplementary Fig. 2. Full data sets are available in Supplementary Data File 1, and confidence statistics in Supplementary Data File 2. Colours depict the strength of resistance: dark red, strong (mFRN > 100); red, moderate (mFRN = 10–100); light red, mild (mFRN = 3–10); light grey, no resistance (mFRN = 1–3); dark grey, increased sensitivity (mFRN < 1). All mFRN values are given to two significant figures. Amu + Rom, amubarvimab + romlusevimab; Bam + Ete, bamlanivimab + etesevimab; Cas + Imd, casirivimab + imdevimab; Cil + Tix, cilgavimab + tixagevimab; n.d., no neutralization data reported for the antibody–variant pair at the time of writing; NTD, amino-terminal domain.
The acquisition by SARS-CoV-2 of mutations conferring evasion properties to mAbs will almost certainly continue, leading to negative clinical outcomes and impacting the utility and longevity of mAbs in managing the ongoing pandemic. In this Review, we consider studies in the primary literature presenting data on the neutralization of globally circulating VOCs by mAbs, with the aim of identifying and characterizing the mutations that lead to this resistance. We discuss how knowledge of the dynamics of viral evasion of mAbs can contribute to viral surveillance and the development of novel mAb treatments, as well as inform predictions of resistance that may arise in the future.
SARS-CoV-2 variant evasion of mAbs
Neutralization assays are considered the ‘gold standard’ for high-throughput in vitro studies of antibody protection against viral infection26. The fold reduction in neutralization (FRN) is calculated by measuring the concentration of mAb required to prevent infection of cells by a virus or pseudovirus carrying a mutated SARS-CoV-2 spike protein and comparing the results with one or more wild type control sequences (usually the Wuhan-Hu-1 reference sequence sampled early in the pandemic), under the same experimental conditions. This allows comparisons among studies while mitigating the confounding effects of differences in experimental protocol. We reviewed the 118 published studies reporting neutralization data on clinically approved mAbs against VOCs and other mutants up to 1 August 2022 (see Supplementary Data File 1) and calculated the geometric mean FRN (mFRN) for each mAb–variant pair (Figs. 1b and 2; see Supplementary Figs. 2 and 3) to represent the extent of resistance of a variant relative to the ability of the mAb to neutralize wild type virus. The use of the geometric mean is considered the appropriate measure for averaging titres generated by serial dilution27. Geometric mean IC50 values were also calculated (see Supplementary Fig. 4) to show the absolute ability of each mAb to neutralize each variant in vitro. Below we discuss the main results for historical (that is, no longer circulating) and contemporary, summer 2022, SARS-CoV-2 VOCs.
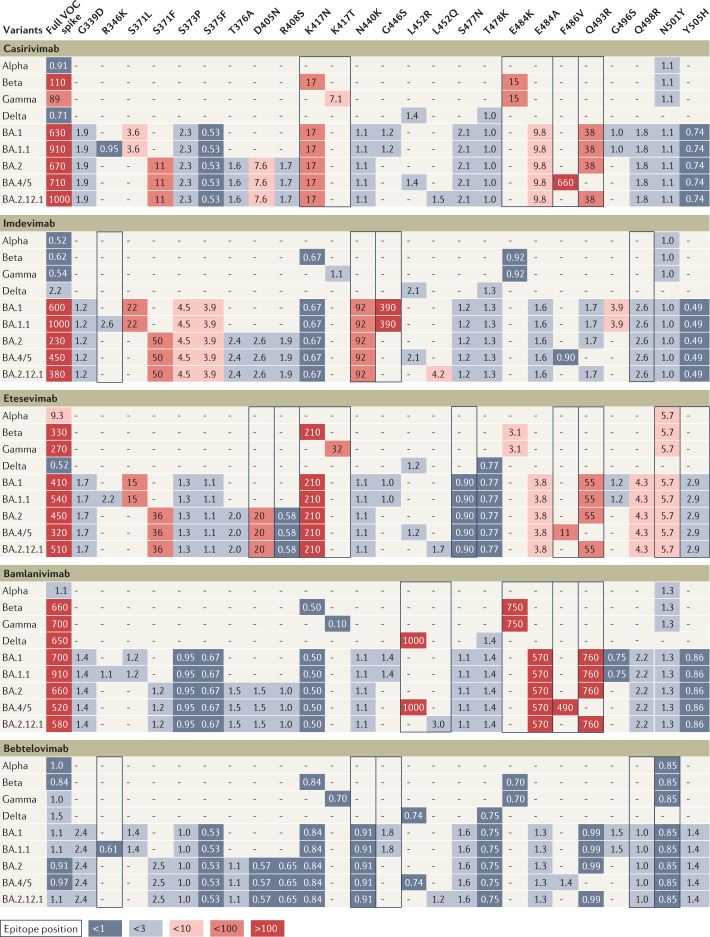
Fig. 2. Influence of individual spike mutations of interest on monoclonal antibody neutralization activity compared with antibody resistance of variants.
Heat map depicting fold change in monoclonal antibody (mAb) neutralization for variants of concern (VOCs) (left-most column) and individual mutations (other columns). Each row contains data for the full spike profile of a given VOC, as well as each definitive VOC mutation individually on a wild type background. Comparison of fold change values across a row indicates which mutations are responsible for any resistance shown by the full VOC. Values show geometric mean fold reduction in neutralization (mFRN). Boxes indicate epitope positions (see Supplementary Data File 3). Colours depict the strength of resistance: dark red, strong (mFRN > 100); red, moderate (mFRN = 10–100); light red, mild (mFRN = 3–10); light grey, no resistance (mFRN = 1–3); dark grey, increased sensitivity (mFRN < 1). ‘–’ indicates that the mutation is not present in the variant. All defining mutations at receptor binding domain (RBD) positions in VOCs are included. Supplementary Fig. 3 presents data for other mAbs for which less comprehensive data have been collected. The RBD is defined here as spike amino acid positions 319–541 (ref.65).
The Alpha VOC remains susceptible to most mAbs, with mild evasion of etesevimab neutralization relative to wild type (mFRN = 9.3; IC50 = 200 ng ml–1) (Fig. 1b; see Supplementary Fig. 4). The Delta VOC, despite being generally less resistant to mAbs overall (with mFRN < 4 for 13 out of 15 mAbs; IC50 < 50 ng ml–1 for 12 out of 14 for which IC50 data were available), demonstrated reduced neutralization by bamlanivimab (mFRN = 650; IC50 = 5,000 ng ml–1) and regdanvimab (mFRN = 37; IC50 = 67 ng ml–1).
Beta and Gamma VOCs exhibited reduced neutralization for 5 out of 16 mAbs with mFRN > 30 (Fig. 1b). This observation is likely due to both having mutations at the 417, 484 and 501 amino acid positions in the spike RBD (Fig. 1a; see Supplementary Fig. 1). However, considerable variability in FRN values from the different assays underpinning the mFRN values has been observed for these two variants (see Supplementary Fig. 2 and Supplementary Data File 2). For example, several studies reported only minimal or no resistance (FRN < 15, n = 5) of casirivimab by the Beta VOC, whereas others exhibited very high levels of evasion (FRN > 250, n = 13). This may be due to differences in experimental protocols among studies28,29 (for example, incubation time, viral system and target cells used). We compared the FRN results for different viral assay systems (virus isolate versus pseudovirus) and found the distribution of results to be similar in each group (see Supplementary Fig. 5), indicating that different viral systems do not have a confounding effect on our observations. This is in agreement with the high correlation among neutralization results using different viral systems observed elsewhere30. Although details on cell type and other experimental parameters were not readily available for all studies, making it possible that differences in these factors are confounding given that cell type and human ACE2 (hACE2) expression have been shown to affect the susceptibility of cells to the virus30,31, the overall trends in fold change are likely to be robust.
The Omicron BA.1, BA.2, BA.1.1, BA.2.12.1, and BA.4 and BA.5 sub-lineages were observed to have the strongest and broadest resistance to neutralization by mAbs, corresponding to high mFRN values against multiple mAbs. Despite this, bebtelovimab, romlusevimab, Evusheld (cilgavimab + tixagevimab) and sotrovimab retained their capacity for neutralization of BA.1 (mFRN = 1.1, 0.74, 75 and 3.3; IC50 = 2.6, 240, 270 and 290 ng ml–1, respectively) (Fig. 1b; see Supplementary Fig. 4), remaining viable options for treatment of those infected with BA.1. Sotrovimab also retained its neutralization capacity against BA.1.1 (mFRN = 2.4; IC50 = 180 ng ml–1), but showed decreased potency against BA.2, BA.4 and BA.5, and BA.2.12.1 (mFRN = 21, 20 and 18; IC50 = 1,400, 790 and 860 ng ml–1, respectively). Evusheld retained significant neutralization capacity against BA.2, BA.4 and BA.5, and BA.2.12.1 (mFRN = 7.3, 74 and 21; IC50 = 37, 180 and 59 ng ml–1, respectively), but was strongly evaded by BA.1.1 (mFRN = 310; IC50 = 810 ng ml–1). Cilgavimab retained its neutralization capacity against BA.2, BA.4 and BA.5, and BA.2.12.1 (mFRN = 2.7, 11 and 3.6; IC50 = 20, 67 and 25 ng ml–1, respectively), but was evaded by BA.1 and BA.1.1 (mFRN = 220 and 820; IC50 = 2,000 and 12,000 ng ml–1). Only the recently approved bebtelovimab retained its full neutralization capacity against all Omicron sub-lineages and the other VOCs, with Evusheld showing high levels of activity against all but BA.1.1. These data on the in vitro neutralization of Omicron variants by bebtelovimab and Evusheld, in combination with large clinical efficacy studies during waves dominated by other variants32 and smaller retrospective studies33–35 during Omicron waves, suggest that high doses of both treatments have an important role to play in treating and preventing infection with Omicron variants, although further clinical evidence is required to confirm this alongside the close monitoring of emergent resistance36.
The high fold change associated with Evusheld against BA.1 (mFRN = 75) (Fig. 1b) compared with bebtelovimab, romlusevimab and sotrovimab (mFRN = 1.1, 0.74 and 3.3), despite having an IC50 value (IC50 = 270 ng ml–1) (see Supplementary Fig. 4) within the range of the other antibodies (IC50 = 2.6, 240 and 290 ng ml–1), highlights an important limitation of the mFRN metric for comparing the neutralization capacities of different antibodies. Because fold change compares the mAb neutralization of the variant with neutralization of the wild type, mAbs that have very high levels of neutralization against wild types will have higher fold change values against a variant than another mAb that neutralizes the wild type less but the variant similarly. However, fold change is much better suited to our focus on the capacity of a given mAb to neutralize a specific variant relative to the neutralization of other variants.
We did not systematically analyse neutralization data on the recently emerged BA.2.75 variant, and early studies suggest that it exhibits some resistance to bebtelovimab but increased sensitivity to tixagevimab and casirivimab37–39.
Single mutation analysis to identify key resistance mutations
Neutralization assays using viruses containing the full complement of mutations that define a VOC cannot conclusively show the impact of specific mutations on mAb efficacy due to interactions among mutations in the spike protein. However, FRN data from viral constructs that contain single mutations of interest on a wild type background only assess the influence of individual mutations on mAb neutralization. For this reason, we compared published FRN values from assays involving viral constructs with single mutations with those using VOC spike proteins with the full complement of mutations (Fig. 2). Interestingly, single mutations had similar effects on resistance regardless of whether they were studied individually or in VOC representative sequences (Fig. 2). VOCs were found to be resistant to a mAb if they contained mutations that were resistant to that mAb in isolation, that is, on the reference or control background sequence.
Imdevimab showed a reduction in neutralizing activity in the presence of BA.1 (mFRN = 600), and in the presence of BA.1 RBD mutations G446S (mFRN = 390) and N440K (nFRN = 92) when studied individually (Fig. 2), and this trend was also seen for the other Omicron sub-lineages. Casirivimab was observed to be evaded by the Beta VOC (mFRN = 110) and by mutants containing RBD mutations found in the Beta VOC alone (E484K (mFRN = 15) and K417N (mFRN = 17)). The Gamma VOC (mFRN = 89) and single mutant constructs with K417T and E484K (mFRN = 7.1 and 15, respectively) — but not with N501Y (mFRN = 1.1) — also evaded casirivimab (Fig. 2). By contrast, it was observed that whenever variants lacked mutations that individually reduced neutralization, the mAbs generally retained their activity against the variants themselves. This was the case for the Delta VOC, which retained its susceptibility to casirivimab (mFRN = 0.71), and single mutants containing the Delta VOC amino acid substitutions L452R and T478K (mFRN = 1.4 and 1.0, respectively) (Fig. 2). Additionally, no resistance to bebtelovimab was displayed by any of the variants or any of the single mutant constructs (Fig. 2). For cocktails of mAbs consisting of non-competing antibodies that target different regions of SARS-CoV-2 spike protein, resistance was found to occur whenever the combination of mutations in a VOC results in resistance to each of the individual mAbs. For instance, BA.1 displayed strong resistance to Ronapreve (casirivimab + imdevimab) (mFRN = 840) (see Supplementary Fig. 3), with K417N and Q493R evading neutralization by casirivimab (mFRN = 17 and 38, respectively), and N440K, G446S and S371L resistant to imdevimab (mFRN = 92, 390 and 22, respectively).
The association between the mAb neutralization levels of single mutant constructs and full variants can also be found in the context of the bamlanivimab + etesevimab cocktail (see Supplementary Fig. 3). Beta and Gamma VOCs, and mutants containing E484K alone were observed to be resistant to the cocktail (mFRN = 510 and 410, and mFRN = 20, respectively) (see Supplementary Fig. 3). The greater reduction observed with Beta and Gamma VOCs in comparison with E484K in isolation could be due to the combination of E484K with K417N and K417T, found in Beta and Gamma VOCS, respectively. However, no loss of neutralization against the cocktail was seen for K417N and K417T in isolation (mFRN = 1.4 and 0.30). Similarly, the strong evasion of the cocktail by all of the Omicron sub-lineage variants is matched by that conferred by E484A in isolation (mFRN = 48), with BA.1, BA.1.1, BA.2 and BA.2.12.1 also carrying Q493R (mFRN = 100), and BA.4 and BA.5 carrying F486V (mFRN = 140). By contrast, the neutralization capacity of the cocktail was retained against the Delta VOC (mFRN = 1.1) as it was against the single mutants carrying the Delta VOC RBD mutations L452R (mFRN = 4.2) and T478K (mFRN = 1.5).
For other mAbs too, strong evasion of neutralization by all Omicron sub-lineage variants aligns with the resistance of viral constructs containing single Omicron RBD mutations. S371L, present in the BA.1 and BA.1.1 variants, conferred resistance to amubarvimab and etesevimab (mFRN = 17 and 15, respectively) (Fig. 2; see Supplementary Fig. 3). S371F, present in the BA.2, BA.4, BA.5, and BA.2.12.1 variants, conferred resistance to amubarvimab, casirivimab, etesevimab and regdanvimab (mFRN = 120, 11, 36 and 21, respectively). F486V, present in the BA.4 and BA.5 variants, conferred resistance to amubarvimab, casirivimab, etesevimab and bamlanivimab (mFRN = 12, 660, 11 and 490, respectively). Q493R, present in the BA.1, BA.1.1, BA.2, and BA.2.12.1 variants, conferred resistance to amubarvimab, casirivimab, etesevimab, regdanvimab, and bamlanivimab (mFRN = 11, 38, 55, 950 and 760, respectively). K417N, present in all of the Omicron variants, conferred resistance to casirivimab and etesevimab (mFRN = 17 and 210, respectively). E484A, present in all of the Omicron variants, conferred resistance to bamlanivimab (mFRN = 570), indicating that E484A confers resistance to the bamlanivimab + etesevimab cocktail in Omicron sub-lineages.
Although these observations support the hypothesis that, in general, the observed resistance of VOCs to specific mAbs is due to effects of individual resistance mutations acting in isolation40, in several cases the resistance observed for variants was greater than the sum of the evasive effects of the spike lineage-defining mutations alone. For instance, the neutralizing activity of amubarvimab against BA.1 and BA.1.1 was markedly reduced (mFRN = 180 and 220, respectively) although S371L and Q493R were individually found to confer only moderate to weak evasion of neutralization (mFRN = 17 and 11, respectively) (see Supplementary Fig. 3). Similarly, BA.1 and BA.1.1 strongly evaded both cilgavimab (mFRN = 220 and 820, respectively) and tixagevimab (mFRN = 360 and 400, respectively) administered alone, yet none of the constituent BA.1 RBD mutations conferred mFRN > 10 to either mAb. Thus, the strong evasion by BA.1 and BA.1.1 of these mAbs cannot be explained by the additive effects of their RBD mutations in isolation, indicating that other mechanisms may be at play. One possibility is that mutations outside the spike RBD contribute to resistance. Another is that synergistic effects among RBD mutations lead to enhanced resistance when they are present in combination.
Furthermore, there are examples where single mutations of interest show a higher reduction in neutralization than the full variant itself. For example, the mutant constructs bearing the BA.1 and BA.1.1 mutation S371L displayed moderate resistance to sotrovimab (mFRN = 20) (see Supplementary Fig. 3), whereas resistance observed with BA.1 and BA.1.1 was only mild (mFRN = 3.3 and 2.4). BA.1 mutations other than S371L, such as G496S41, might antagonistically dampen the effect of S371L leading to only mild resistance overall. This possibly explains why BA.2 — missing mutations at positions 446 and 496 (relative to BA.1) — was found to be less sensitive to sotrovimab (mFRN = 22 versus 3.3) than BA.1. Alternatively, amino-terminal domain (NTD) mutations may allosterically alter the BA.2 RBD leading to antigenic effects42. This mechanistic ambiguity of how mutations interact highlights the need for comparative studies where combinations of mutations are taken into consideration.
Epitope mutations drive mAb evasion
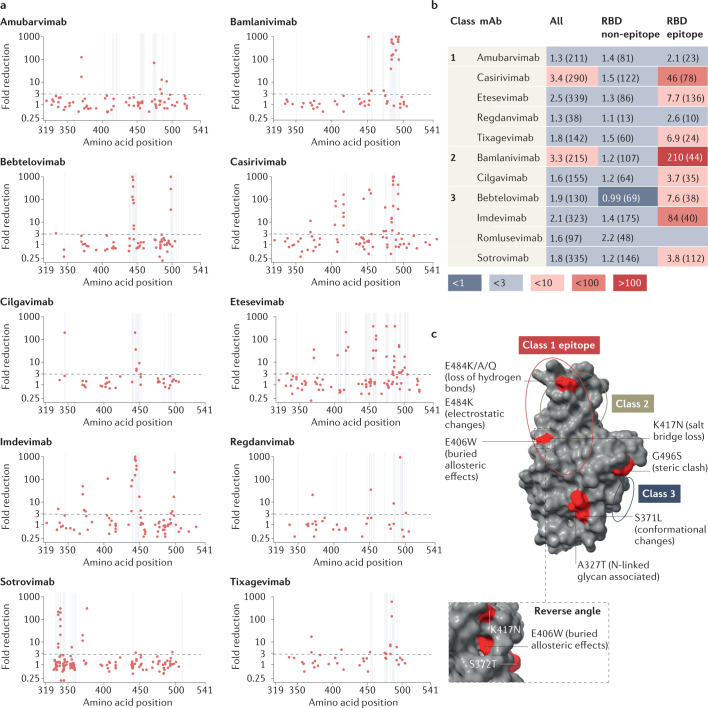
To better understand the effect of single mutations on mAb activity we investigated the role of mutations occurring at epitope and non-epitope positions (see Supplementary Data File 3) in the reduction of the neutralization by mAbs. As expected, mutations causing the strongest resistance were found within the binding footprint of the mAb at cognate epitope positions in the spike RBD (Fig. 3a,b). However, not all mutations at epitope positions were observed to cause mAb resistance, and a small number of strongly resistant mutations able to generate long-range allosteric perturbations were found at distal, non-epitope positions in the RBD (Fig. 3a,b) and in the NTD (see Supplementary Fig. 6a). Generally, mutations close to epitope positions, at the +/–1 proximal positions, but not inside the epitope, were observed to not cause reductions in neutralization (see Supplementary Fig. 6b).
Fig. 3. Assessment of monoclonal antibody resistance by mutations of interest occurring at epitope and non-epitope sites.
a, Geometric mean fold reduction in neutralization (mFRN) data for each monoclonal antibody (mAb) against viral constructs containing single mutations in the spike receptor binding domain (RBD) (positions 319–541). Epitope positions (see Supplementary Data File 3) indicated by vertical grey lines. Dashed line shows the mFRN = 3 threshold. Alternative substitutions at the same amino acid position are shown as separate points at the same x coordinate, for example E484K and E484Q are shown as two different points at the 484 position on the x axis. Most dots above the threshold coincide with the grey lines, indicating that mutations conferring resistance to neutralization tend to occur at epitope positions. However, some non-epitope mutations also confer resistance as shown by the dots above the threshold but not on grey lines. b, Pooled mFRN comparisons between epitope and non-epitope mutations. Values shown are mFRN for either all amino acid positions, all amino acid positions in the epitope of the mAb, or all amino acid positions in the RBD but not in the epitope of the mAb. The higher mFRN values of epitope positions indicate the increased contribution of mutations in the mAb epitope to the resistance of neutralization. Number in brackets indicates the number of assays contributing to each geometric mean value. Epitope unknown for romlusevimab. Colours depict the strength of resistance: dark red, strong (mFRN > 100); red, moderate (mFRN = 10–100); light red, mild (mFRN = 3–10); light grey, no resistance (mFRN = 1–3); dark grey, increased sensitivity (mFRN < 1). c, Isolated Omicron spike RBD structure [PDB:7TGW]98. Epitope regions for class 1, 2 and 3 mAbs are circled. Epitope and non-epitope mutations exemplifying mechanisms of mAb evasion are labelled: S371L, conformational changes and N-linked glycosylation43; A372T, N-linked glycosylation50; E406W, conformational changes in the epitope46; K417N, abolished salt bridges between mAb and RBD41; E484K/E484Q/E484A, loss of hydrogen bonds with mAb56; E484K, changes to electrostatic interactions54; G496S, steric clash99. Single mutant mFRN data across the full spike protein are shown in Supplementary Fig. 6a and pooled mFRN comparisons between epitope, non-epitope mutations, epitope proximal and RBD positions in Supplementary Fig. 6b.
At some sites, the level of evasion was found to depend on the specific amino acid substitution introduced by the mutation. For example, in the case of casirivimab and mutations at position 417, the substitution of a lysine (K) with either glutamic acid (E) or arginine (R) was associated with a marked reduction in neutralization activity (mFRN = 160 and 61, respectively) (see Supplementary Data File 1), whereas more moderate evasion was observed with asparagine (N) (mFRN = 17), with an even smaller effect observed with threonine (T) (mFRN = 7.1). Additionally, a single mutant carrying a valine (V) to threonine (T) substitution at position 445 showed strong resistance to casirivimab (mFRN = 110), whereas an alanine substitution (V445A) did not lead to evasion (mFRN = 1.7) (see Supplementary Data File 3). Similarly, sotrovimab was strongly evaded by mutants carrying P337L (mFRN = 160) and P337R (mFRN = 216) in isolation, but only mildly by those carrying P337T and P337H (mFRN = 8.5 and 5.8, respectively) and not at all by the P337S single mutant (mFRN = 1.3) (see Supplementary Data File 1).
There are some cases in which mutations outside the epitope contribute to resistance against the mAb, displaying a potential allosteric influence. The S371L single mutant conferred moderate evasion of amubarvimab (mFRN = 17), imdevimab (mFRN = 22), sotrovimab (mFRN = 20), and etesevimab (mFRN = 15) despite not being present in the epitopes bound by these mAbs (Fig. 3a). S371L also reduced neutralization by romlusevimab (mFRN = 17) but no epitope data are available. Similarly, mutants containing E406W alone markedly evaded both casirivimab (mFRN = 84) and imdevimab (mFRN = 110) (see Supplementary Data File 1); E406D was resistant to casirivimab (mFRN = 51); F377K evaded sotrovimab (mFRN = 300); and P499S evaded imdevimab (mFRN = 210). The ability to evade a broad range of mAbs conferred by S371L and mutations at 406 suggests that, given that each mAb targets different epitopes, those mutations might affect the global spike conformation rather than making local changes to specific epitopes only (Fig. 3c).
Several studies have been carried out to investigate the structural mechanisms through which non-epitope mutations can disrupt antibody binding. The S371L mutation was found to alter the conformation of both the 371–376 loop and the 365–370 helix, leading to disruptive interactions with mAbs at those sites43,44. At the same position, S371F leads to the repositioning of the N343 glycan, potentially causing resistance to sotroviamb45. E406W was observed to cause allosteric remodelling of sites contacted by cilgavimab and Ronapreve (casirivimab + imdevimab), reducing binding by these mAbs46, with allostery being central to spike dynamics more generally47,48. G496S may preclude mAb binding through steric clash, which may explain why a different mutation at the same site (G496R) was observed to cause evasion of etesivimab49. However, other studies showed that G496S stabilizes interactions with mAbs through the formation of hydrogen bonds, indicating that mutational effects are mAb-specific41. A study found that the D364N, A372S and A372T mutations may cause mAb evasion by supporting N-linked glycosylation at N370 (ref.50). Addition of a bulky glycan at this site obstructs mAb binding to the epitope directly and through stabilization of the RBD ‘down’ conformation that hides the epitope51 (Fig. 3c). Using an amino acid interaction approach, mutations at the non-epitope 373, 440, and 446 positions have been reported to disrupt sotrovimab binding through interactions with amino acids that are in the epitope52. Molecular dynamics-based approaches have also helped delineate how residues in the NTD and S2 domain may be able to allosterically affect antigenicity and infectivity42. Taken together, these observations show that comprehensive analysis of mAb evasion by SARS-CoV-2 must include non-epitope mutations in addition to the more prominent epitope mutation resistance effects.
Protein structure studies have reported on the mechanisms by which specific epitope substitutions affect the interactions between mAbs and the spike protein, shedding light on why some substitutions lead to resistance but others do not. The E484A mutation is likely to cause the loss of salt bridges between spike and class 2 mAbs (for example, bamlanivimab)41,53 and destabilize energy changes in bonds with class 1 casirivimab41. These different effects on bonding to antibodies of different classes may explain why E484A conferred strong evasion of bamlanivimab (class 2; mFRN = 570), but only mild resistance to casirivimab (class 1; mFRN = 9.8). Similarly, an explanation for the evasion of bamlanivimab (mFRN = 750), casirivimab (mFRN = 15), tixagevimab (mFRN = 6.8), and regdanvimab (mFRN = 8.7) by mutants containing E484K has been offered by computational modelling that reveals that the reversal of the side chain charge substantially alters the electrostatic complementarity of mAb binding54 (Fig. 3c). E484K, E484A and E484Q mutations all confer resistance to bamlanivimab and casirivimab to some extent, which, given the different biochemical properties of the amino acid substitutions, suggests that, in this case, it is the loss of glutamic acid (E) that is important, rather than the amino acid that replaces it. This idea is supported by the identification of E484 as a key binding site for some mAbs55, and by the loss of hydrogen bonds between E484 and an experimental mAb due to mutation56 (Fig. 3c). Structural analysis reveals L452R to disrupt a hydrophobic binding pocket, potentially explaining the loss of neutralization by antibodies targeting this site in Delta, BA.4 and BA.5 VOCs21. BA.4 and BA.5 also carry F486V, which involves the loss of phenylalanine from the binding site of multiple mAbs21. Linking the reduction in neutralization conferred by a mutation to its structural effects explains observed patterns of neutralization and may support the prediction of novel evasive mutations based on their putative structural effects.
The structural changes introduced by spike mutations have consequences not only for mAb evasion but also viral infectivity, replication, transmissibility and stability24,57–60. Some antigenic mutations decrease infectivity as they affect the ability of RBD to bind the hACE2 receptor required for entry into the human cell50,61–68 Other mutations can compensate for this detrimental effect by increasing hACE2 binding53, allowing the virus to retain its infectivity while reducing susceptibility to the antibody response. The BA.1 spike mutational profile demonstrates this as the antigenic evasion mutations G496S, Y505H, K417N, S371L, S373P and S375F alone decrease hACE2 binding affinity, but overall BA.1 infectivity is retained through the compensatory increase in affinity due to N501Y, S477N, G493K and Q498R23,53,61. It is possible that the affinity of BA.4 and BA.5 for hACE2 is further increased by the electrostatic complementarity provided by L452R, offering an explanation for the transmission advantage of both BA.4 and BA.5, and Delta VOCs21. Other mutations, such as S373P and S375F, do not have immune evasive effects in isolation but do so in combination52. These combinations are able to arise as the individual mutations have positive effects on other aspects of viral fitness and so experience positive selection before the full immune evasive combination is achieved52. Thus, the selection of other viral traits can have antigenic effects as some mutations impact multiple viral phenotypes.
Conclusions
Care must be taken when extrapolating between neutralization assays and the clinical efficacy of mAbs. Post-vaccination sera neutralization titres strongly correlate with protection from symptomatic infection and severe disease69,70, and mAb neutralization may be expected to follow a similar trend. A perfect correlation would not be expected due to mechanisms other than neutralization, such as antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis and complement activation, which are also important to the role of antibodies in combatting viral infection, and other aspects of the immune response such as T cells that are not captured in neutralization assays. Such effects likely contribute to the generally strong in vivo efficacy of sotrovimab71, despite its elevated IC50 values (see Supplementary Fig. 4). Some mAbs have their Fc domains altered, meaning that they are able to participate in effector functions to different degrees72,73. In addition, the variability in results from different studies must be taken into consideration. This is partly explained by the use of different neutralization assay protocols with alternative viral systems (for instance, authentic virus isolates, vesicular stomatitis virus or lentivirus-based pseudoviruses, or recombinant chimeric viruses), target cells (Vero E6, S-Fuse, 293T ACE2 and so on) and other experimental conditions that vary among studies (for example, incubation time, experimental output, cell type, ACE2 or TMPRSS2 expression and so on)30,74,75, and highlights the urgent need to build on the work of the World Health Organization (WHO) on the standardization of neutralization assays76,77. It will be important to improve understanding of the correlation between in vivo neutralization and clinical efficacy to prevent mAbs being withdrawn when they still have a benefit in patients31.
Although indications that Omicron infections are less clinically severe are encouraging78, the reduced susceptibility of Omicron lineages BA.1, BA.2, BA.4, and BA.5 to neutralization by many of the mAbs available for clinical use highlights the risk that novel variants pose to the efficacy of mAb treatments in the present and future. The more recently spreading BA.4 and BA.5 lineages, in particular, show the highest levels of mAb evasion of any VOC to date. Pandemic management strategies must account for the possibility that future variants will evade all currently available mAbs. There are several precautionary steps that can be taken (Fig. 4). Firstly, reliance must not be placed on any single control strategy so that the cost of any strategy failing is reduced. This is being achieved already with the combination of vaccination, mAbs, antivirals and non-pharmaceutical interventions such as self-isolation2–4. Secondly, mAb development strategies could minimize resistance to mAbs by targeting conserved epitopes79 and susceptible epitopes of future variants using knowledge of patterns of resistant mutations, such as those discussed in this Review. Maintaining a diverse range of viable drugs will ensure that effective treatments for COVID-19 are available, even if some variants are resistant to a subset of drugs. Thirdly, clinical mAb treatment needs to be conducted in a manner that mitigates the risk of directly causing the emergence and spread of resistant variants. Resistance mutations have been observed to emerge when individuals are treated with mAbs or convalescent plasma57,80–82. These resistant variants threaten to evade both mAb treatment and the polyclonal adaptive immune response as there is considerable overlap in the mutations that drive evasion in each case36,83.
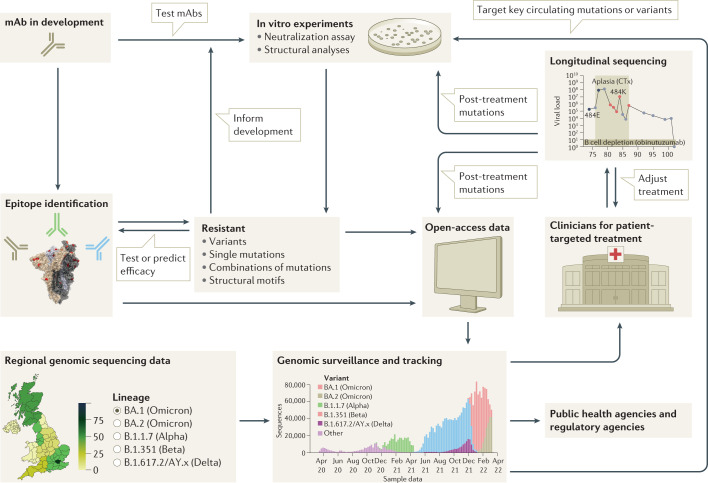
Fig. 4. Framework for the rational design of viral genomic surveillance for the development of efficient monoclonal antibody therapies.
A successful framework will integrate knowledge of the dynamics of monoclonal antibody (mAb) resistance by SARS-CoV-2 variants, including the central role played by epitope mutations, epistatic effects and evolutionary dynamics. mAb development must be supported by epitope identification, in vitro studies and the monitoring of resistance mutations. mAbs in early development may be ruled out if they target an epitope that contains mutations shown to cause resistance in vitro. Alternatively, low frequency of, or evolutionary barriers to, mutations in a given region may focus the development of mAbs that target that region. Resistance monitoring is a multifaceted process, spanning individual and combinatorial mutational effects in vitro, as well as genomic surveillance of circulating strains and longitudinal clinical studies that track genetic changes during treatment. Combining these different approaches will allow the development of mAbs highly effective against currently circulating strains, and robust to resistance by future variants.
Desirable clinical outcomes can be supported by virus genome sequencing to determine the variant causing an infection, allowing the most appropriate mAb treatment to be selected. Similarly, alignment of regional treatment guidelines with information about which variants are circulating and at what frequencies will support the effective logistical deployment of the available treatments. The FDA have already begun regulating on this basis with Emergency Use Authorizations for mAbs including a clause that prohibits a mAb being used in regions where the frequency of resistant variants is above 5%11–13. The UK Health Security Agency recommended longitudinal virus genome sequencing during therapeutic mAb usage to monitor for emerging resistance84. Additionally, continued exploitation of combination therapies, selection of appropriate dosages, and preventing the spread of virus from the patient to other individuals will make the development and onwards spread of resistant strains less likely.
Viral surveillance and mAb development efforts will benefit greatly from considering not only single epitope mutations but also non-epitope mutations alongside combinational and synergistic dynamics. These efforts must extend beyond antigenicity to the full range of viral traits, all of which play a role in determining the antigenic mutations that rise to prominence. The data discussed here depict mAb evasion to be dominated by the isolated effect of single mutations at epitope positions, and therefore that single mutational analysis can provide important insights into the mechanisms of mAb evasion for past and future variants. However, the evasive effects of non-epitope mutations and combinations of mutations mean that a complete understanding of mAb dynamics cannot be achieved by analysis of single epitope mutations alone. Rather, neutralization data should be combined with structural, combinatorial, molecular dynamic, and evolutionary studies to allow the pre-emption of novel future mutations that may have similar effects. With this in mind, public health agencies should continue to routinely survey sequence data generated locally and worldwide to detect viral mutations and variants that might impact adversely on the efficacy of therapeutics (Fig. 4).
Supplementary information
Acknowledgements
The COVID-19 Genomics UK (COG-UK) Consortium is supported by funding from the Medical Research Council (MRC) as part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) (grant code: MC_PC_19027) and Genome Research Limited, operating as the Wellcome Sanger Institute. The authors acknowledge use of data generated through the COVID-19 Genomics Programme funded by the Department of Health and Social Care. The views expressed are those of the author and not necessarily those of the Department of Health and Social Care or the UK Health Security Agency (UKHSA). The authors acknowledge support from the G2P-UK National Virology Consortium (MR/W005611/1) and MRC (MC_UU_12014/12).
Glossary
- Antibody-dependent cell-mediated cytotoxicity
A cytolytic process dependent on cooperation between cellular and humoral constituents of the immune system.
- Fc domains
The ‘fragment crystallizable region’ is a region of an antibody composed of constant domains of heavy chains and is involved in binding to Fc receptors and modulating immune activity.
- Geometric mean
An average, calculated as the nth root of the product of n numbers, that is most appropriate with a set of numbers that are exponential in nature, such as titres derived from a serial dilution.
- IC50 values
The minimal concentrations of monoclonal antibody (mAb) required for 50% neutralization of the virus in vitro.
Author contributions
M.C., T.P.P., W.T.H., D.W.W., B.J.W. and A.M.C. researched data for article. M.C., W.T.H., J.H., B.J.W., E.T., R.K.G., S.J.P., D.L.R. and A.M.C. substantially contributed to discussion of content. M.C., D.L.R. and A.M.C. wrote the article. T.P.P., W.T.H., J.H., B.J.W., E.T., R.K.G., S.J.P., D.L.R. and A.M.C. reviewed/edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Competing interests
R.K.G. has received honoraria for educational activities from Moderna, GSK and Janssen. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Contributor Information
David L. Robertson, Email: david.l.robertson@glasgow.ac.uk
Alessandro M. Carabelli, Email: alessandrocarabelli@gmail.com
Supplementary information
The online version contains supplementary material available at 10.1038/s41579-022-00809-7.
References
- 1.World Health Organization. WHO COVID-19 dashboard. World Health Organizationhttps://covid19.who.int/ (2022).
- 2.Uddin M, et al. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12:526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drożdżal S, et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist. Updat. 2021;59:100794. doi: 10.1016/j.drup.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L, Ayeh SK, Chidambaram V, Karakousis PC. Modes of transmission of SARS-CoV-2 and evidence for preventive behavioral interventions. BMC Infect. Dis. 2021;21:496. doi: 10.1186/s12879-021-06222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo L, Luo T, Du M, Mei H, Hu Y. Efficacy and safety of tocilizumab in hospitalized COVID-19 patients: a systematic review and meta-analysis. J. Infect. 2022;84:418–467. doi: 10.1016/j.jinf.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor PC, et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS ONE. 2012;7:e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suryadevara N, et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FDA. Bamlanivimab EUA. US Food and Drug Administrationhttps://www.fda.gov/media/147629/download (2021). This Letter explains that the Emergency Use Authorization for bamlanivimab was revoked by the FDA due to known resistance mutations circulating in high frequency.
- 11.FDA. Sotrovimab EUA. US Food and Drug Administrationhttps://www.fda.gov/media/149532/download (2022).
- 12.FDA. Casirivimab and imdevimab EUA. US Food and Drug Administrationhttps://www.fda.gov/media/145610/download (2022).
- 13.FDA. Bamlanivimab and etesevimab. US Food and Drug Administrationhttps://www.fda.gov/media/145801/download (2022).
- 14.Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of the SARS-CoV-2 delta variant, key spike mutations and immune escape. Front. Immunol. 2021;12:751778. doi: 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian D, Sun Y, Xu H, Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022;94:2376–2383. doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desingu PA, Nagarajan K. Omicron BA.2 lineage spreads in clusters and is concentrated in Denmark. J. Med. Virol. 2022;94:2360–2364. doi: 10.1002/jmv.27659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viana R, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 19.Tegally H, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1780. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuekprakhon A, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to Bamlanivimab. mAbs. 2021;13:1919285. doi: 10.1080/19420862.2021.1919285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng B, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlcochova P, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamasoba D, et al. Neutralisation sensitivity of SARS-CoV-2 Omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 2022;22:942–943. doi: 10.1016/S1473-3099(22)00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muruato AE, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brugh M. A simple method for recording and analyzing serological data. Avian Dis. 1978;22:362–365. doi: 10.2307/1589552. [DOI] [PubMed] [Google Scholar]
- 28.Planas D, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann M, et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36:109415. doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riepler L, et al. Comparison of four SARS-CoV-2 neutralization assays. Vaccines. 2021;9:13. doi: 10.3390/vaccines9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu MY, et al. WHO’s Therapeutics and COVID-19 Living Guideline on mAbs needs to be reassessed. Lancet. 2022 doi: 10.1016/S0140-6736(22)01938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dougan M, et al. Bebtelovimab, alone or together with bamlanivimab and etesevimab, as a broadly neutralizing monoclonal antibody treatment for mild to moderate, ambulatory COVID-19. medRxiv. 2022 doi: 10.1101/2022.03.10.22272100. [DOI] [Google Scholar]
- 33.Yetmar ZA, et al. Outcomes of bebtelovimab and sotrovimab treatment of solid organ transplant recipients with mild-to-moderate coronavirus disease 2019 during the Omicron epoch. Transpl. Infect. Dis. 2022;24:e13901. doi: 10.1111/tid.13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benotmane I, et al. Breakthrough COVID-19 cases despite prophylaxis with 150 mg of tixagevimab and 150 mg of cilgavimab in kidney transplant recipients. Am. J. Transplant. 2022 doi: 10.1111/ajt.17121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaminski H, et al. COVID-19 morbidity decreases with tixagevimab/cilgavimab preexposure prophylaxis in kidney transplant recipients non/low vaccine responders. Kidney Int. 2022;102:936–938. doi: 10.1016/j.kint.2022.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lusvarghi S, et al. SARS-CoV-2 BA.1 variant is neutralized by vaccine booster-elicited serum, but evades most convalescent serum and therapeutic antibodies. Sci. Transl Med. 2022;14:eabn8543. doi: 10.1126/scitranslmed.abn8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamasoba D, et al. Neutralization sensitivity of Omicron BA.2.75 to therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1101/2022.07.14.500041. [DOI] [Google Scholar]
- 38.Sheward DJ, et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, et al. Neutralizing antibody evasion and receptor binding features of SARS-CoV-2 Omicron BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.18.500332. [DOI] [Google Scholar]
- 40.Lazarevic I, Pravica V, Miljanovic D, Cupic M. Immune evasion of SARS‐CoV‐2 emerging variants: what have we learnt so far? Viruses. 2021;13:1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubin JH, et al. Structural models of SARS-CoV-2 Omicron variant in complex with ACE2 receptor or antibodies suggest altered binding interfaces. bioRxiv. 2021 doi: 10.1101/2021.12.12.472313. [DOI] [Google Scholar]
- 42.Ray D, Le L, Andricioaei I. Distant residues modulate conformational opening in SARS-CoV-2 spike protein. Proc. Natl Acad. Sci. USA. 2021;118:e2100943118. doi: 10.1073/pnas.2100943118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 44.Cerutti G, et al. Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep. 2022;38:110428. doi: 10.1016/j.celrep.2022.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park Y-J, et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. bioRxiv. 2022 doi: 10.1101/2022.05.08.491108. [DOI] [PubMed] [Google Scholar]
- 46.Addetia A, et al. Structural changes in the SARS-CoV-2 spike E406W mutant escaping a clinical monoclonal antibody cocktail. bioRxiv. 2022 doi: 10.1101/2022.01.21.477288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan ZW, et al. Allosteric perspective on the mutability and druggability of the SARS-CoV-2 Spike protein. Structure. 2022;30:590–607.e4. doi: 10.1016/j.str.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Díaz-Salinas MA, et al. Conformational dynamics and allosteric modulation of the SARS-CoV-2 spike. eLife. 2022;11:e75433. doi: 10.7554/eLife.75433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kannan SR, et al. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022;126:102779. doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nabel KG, et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2022;375:eabl6251. doi: 10.1126/science.abl6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson R, et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller NL, Clark T, Raman R, Sasisekharan R. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Rep. Med. 2022;3:100527. doi: 10.1016/j.xcrm.2022.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah M, Woo HG. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022;12:6031. doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreano E, et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl Acad. Sci. USA. 2021;118:e2103154118. doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding C, He J, Zhang X, Jiang C, Sun Y. Crucial mutations of spike protein on SARS-CoV-2 evolved to variant strains escaping neutralization of convalescent plasmas and RBD-specific monoclonal antibodies. Front. Immunol. 2021;12:693775. doi: 10.3389/fimmu.2021.693775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tortorici MA, et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kemp SA, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito A, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta D, et al. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell. Mol. Life Sci. 2021;78:7967–7989. doi: 10.1007/s00018-021-04008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar V, Singh J, Hasnain SE, Sundar D. Possible link between higher transmissibility of Alpha, Kappa and Delta variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. Int. J. Mol. Sci. 2021;22:9131. doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dejnirattisai W, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang L, et al. A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell. 2021;184:4392–4400. doi: 10.1016/j.cell.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray D, Quijano RN, Andricioaei I. Point mutations in SARS-CoV-2 variants induce long-range dynamical perturbations in neutralizing antibodies. Chem. Sci. 2022;13:7224–7239. doi: 10.1039/D2SC00534D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saville JW, et al. Structural and biochemical rationale for enhanced spike protein fitness in Delta and Kappa SARS-CoV-2 variants. Nat. Commun. 2022;13:742. doi: 10.1038/s41467-022-28324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valério M, Borges-Araújo L, Melo MN, Lousa D, Soares CM. SARS-CoV-2 variants impact RBD conformational dynamics and ACE2 accessibility. Front. Med. Tech. 2022;4:1009451. doi: 10.3389/fmedt.2022.1009451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wrobel AG, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, et al. SARS-CoV-2 variants increase kinetic stability of open spike conformations as an evolutionary strategy. mBio. 2022;13:e0322721. doi: 10.1128/mbio.03227-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Starr TN, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cromer D, et al. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microbe. 2022;3:e52–e61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khoury DS, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 71.Chen RE, et al. In vivo monoclonal antibody efficacy against SARS-COV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Case JB, et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. Nat. Commun. 2022;13:3824. doi: 10.1038/s41467-022-31615-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cathcart AL, et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2022 doi: 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- 74.Schmidt F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen RE, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Knezevic I, et al. WHO International Standard for evaluation of the antibody response to COVID-19 vaccines: call for urgent action by the scientific community. Lancet Microbe. 2022;3:e235–e240. doi: 10.1016/S2666-5247(21)00266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tao K, Tzou PL, Kosakovsky Pond SL, Ioannidis JPA, Shafer RW. Susceptibility of SARS-CoV-2 Omicron variants to therapeutic monoclonal antibodies: systematic review and meta-analysis. Microbiol. Spectr. 2022;10:e0092622. doi: 10.1128/spectrum.00926-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewnard JA, et al. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat. Med. 2022;28:1933–1943. doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang P, et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect. 2022;11:147–157. doi: 10.1080/22221751.2021.2011623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fenaux H, et al. Emergence of SARS-CoV-2 resistance mutations in a patient who received anti-SARS-COV2 spike protein monoclonal antibodies: a case report. BMC Infect. Dis. 2021;21:1223. doi: 10.1186/s12879-021-06902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jensen B, et al. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg. Health Eur. 2021;8:100164. doi: 10.1016/j.lanepe.2021.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rockett R, et al. Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use. N. Engl. J. Med. 2022;386:1477–1479. doi: 10.1056/NEJMc2120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lou F, et al. Understanding the secret of SARS-CoV-2 variants of concern/interest and immune escape. Front. Immunol. 2021;12:744242. doi: 10.3389/fimmu.2021.744242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.UK Health Security Agency. COVID-19: genomic surveillance of patients who are treated with neutralising monoclonal antibody or immunosuppressed. GOV.UKhttps://www.gov.uk/government/publications/covid-19-genomic-surveillance-of-patients-who-are-treated-with-neutralising-monoclonal-antibody-or-immunosuppressed (2021).
- 85.Brii Biosciences. Brii Bio announces amubarvimab/romlusevimab combination received approval from NMPA as first COVID-19 neutralizing antibody combination therapy in China. Brii Bioscienceshttps://www.briibio.com/news-detail.php?id=505 (2021).
- 86.Brii Biosciences. Brii Bio announces that National Health Commission of China adds amubarvimab/romlusevimab combination to its COVID-19 diagnosis and treatment guidelines (9th pilot edition). Brii Bioscienceshttps://www.briibio.com/news-detail.php?id=769#news (2022).
- 87.EMA. EMA issues advice on use of antibody combination (bamlanivimab/etesevimab). European Medicines Agencyhttps://www.ema.europa.eu/en/news/ema-issues-advice-use-antibody-combination-bamlanivimab-etesevimab (2021).
- 88.EMA. Bamlanivimab and etesevimab for COVID-19: withdrawal from the rolling review process. European Medicines Agencyhttps://www.ema.europa.eu/en/medicines/human/withdrawn-applications/bamlanivimab-etesevimab-covid-19 (2021).
- 89.FDA. Bebtelovimab EUA. US Food and Drug Administrationhttps://www.fda.gov/media/156151/download (2022).
- 90.The Science Times. COVID-19 antibody treatment now authorized for compassionate use in Beijing hospital; will this end the pandemic? The Science Timeshttps://www.sciencetimes.com/articles/34604/20211119/covid-19-antibody-treatment-now-authorized-compassionate-use-beijing-hospital.html (2021).
- 91.EMA. Regkirona. European Medicines Agencyhttps://www.ema.europa.eu/en/medicines/human/EPAR/regkirona (2021).
- 92.MHRA. Summary of the Public Assessment Report for Ronapreve. GOV.UKhttps://www.gov.uk/government/publications/regulatory-approval-of-ronapreve/summary-of-the-public-assessment-report-for-ronapreve (2021).
- 93.EMA. Xevudy. European Medicines Agencyhttps://www.ema.europa.eu/en/medicines/human/EPAR/xevudy#authorisation-details-section (2021).
- 94.MHRA. Summary of product characteristics for Xevudy. GOV.UKhttps://www.gov.uk/government/publications/regulatory-approval-of-xevudy-sotrovimab/summary-of-product-characteristics-for-xevudy#date-of-first-authorisationrenewal-of-the-authorisation (2021).
- 95.FDA. Evusheld EUA. US Food and Drug Administrationhttps://www.fda.gov/media/154704/download (2022).
- 96.EMA. Evusheld. European Medicines Agencyhttps://www.ema.europa.eu/en/medicines/human/EPAR/evusheld (2022).
- 97.MHRA. Regulatory approval of Evusheld (cilgavimab + tixagevimab). GOV.UKhttps://www.gov.uk/government/publications/regulatory-approval-of-evusheld-tixagevimabcilgavimab (2022).
- 98.RCSB PDB. 7TGW. RCSBhttps://www.rcsb.org/structure/7TGW (2022).
- 99.Islam SR, Prusty D, Manna SK. Structural basis of fitness of emerging SARS-COV-2 variants and considerations for screening, testing and surveillance strategy to contain their threat. medRxiv. 2021 doi: 10.1101/2021.01.28.21250666. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.