Abstract
Staphylococcus aureus is an important human and livestock pathogen that is well-protected against environmental insults by a thick cell wall. Accordingly, the wall is a major target of present-day antimicrobial therapy. Unfortunately, S. aureus has mastered the art of antimicrobial resistance, as underscored by the global spread of methicillin-resistant S. aureus (MRSA). The major cell wall component is peptidoglycan. Importantly, the peptidoglycan network is not only vital for cell wall function, but it also represents a bacterial Achilles’ heel. In particular, this network is continuously opened by no less than 18 different peptidoglycan hydrolases (PGHs) encoded by the S. aureus core genome, which facilitate bacterial growth and division. This focuses attention on the specific functions executed by these enzymes, their subcellular localization, their control at the transcriptional and post-transcriptional levels, their contributions to staphylococcal virulence and their overall importance in bacterial homeostasis. As highlighted in the present review, our understanding of the different aspects of PGH function in S. aureus has been substantially increased over recent years. This is important because it opens up new possibilities to exploit PGHs as innovative targets for next-generation antimicrobials, passive or active immunization strategies, or even to engineer them into effective antimicrobial agents.
Keywords: Staphylococcus aureus, peptidoglycan hydrolase, cell wall, subcellular protein localization, pathogenesis, antimicrobial susceptibility
Introduction
Staphylococcus aureus is a small and spherical Gram-positive bacterium (Fig. 1), belonging to the phylum of the Firmicutes. As a commensal, S. aureus frequently colonizes asymptomatically the nares, skin and gut of humans (Mainous et al. 2006, Raineri, Altulea and van Dijl 2022). Nonetheless, S. aureus is also one of the most renowned opportunistic pathogens, causing many different diseases that range from relatively mild skin and soft tissue infections to life-threatening diseases, such as bacteremia, pneumonia and endocarditis. The invasive behavior of S. aureus is often triggered through injury or medical interventions, where the barrier function of the host cells or tissues is compromised. However, S. aureus also produces a variety of virulence factors that allow this bacterium to invade the human body. Beyond the epithelial or endothelial barriers, S. aureus is able to bind to the extracellular matrix of host cells and tissues, to invade host cells, or to form biofilms on tissues and medical implants (Raineri, Altulea and van Dijl 2022). At the same time, it is able to effectively evade innate and adaptive immune defenses of the host, either through the secretion of particular virulence factors or the formation of biofilms (Thammavongsa et al. 2015, Goldmann and Medina 2018). Ultimately, this may result in persistent or chronic infections (Josse et al. 2017). In recent years, the treatment of S. aureus infections has become increasingly difficult due to the fact that this pathogen has acquired many different antibiotic resistances, as critically exemplified by the emergence of methicillin-resistant S. aureus (MRSA) lineages in hospitals and the community (Turner et al. 2019).
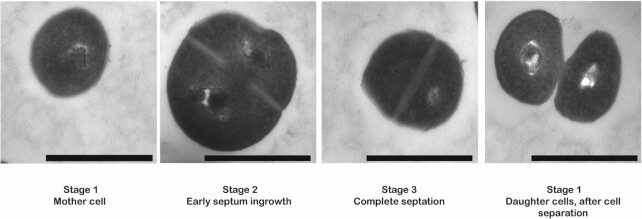
Figure 1.
Different stages in the cell cycle of Staphylococcus aureus as visualized by transmission electron microscopy. During the initial phase of the cell cycle (stage 1), a spherical S. aureus mother cell will become slightly enlarged and forms a septum at the mid-cell position (stage 2). Once the synthesis of the septum is complete and the cell is further enlarged (stage3), bacterial autolytic enzymes will promote cell division, resulting in two daughter cells (stage 1). After cell separation, the septum of the daughter cells is reshaped from a flat surface into a hemisphere. Images were recorded by transmission electron microscopy. The magnification is indicated by scale bars (1 μm).
Gram-positive bacteria, like S. aureus, are surrounded by a thick cell wall, which plays vital roles in maintaining cell shape, cell integrity, cell viability and the protection against osmotic stress under different environmental conditions (Vollmer et al. 2008, Silhavy et al. 2010). The structure and composition of the cell wall varies between different Gram-positive bacterial species (Do et al. 2020). However, peptidoglycan is invariably the major cell wall component that surrounds the bacterial cytoplasmic membrane (Turner et al. 2014). S. aureus peptidoglycan contains relatively short glycan strands that are highly cross-linked through species-specific peptide bridges; 80%–90% of the so-called stem peptides are cross-linked via a pentaglycine bridge (Snowden and Perkins 1990, Monteiro et al. 2019, Sobral and Tomasz 2019). These cross-links are important for the integrity and physical strength of the peptidoglycan. Importantly, the peptidoglycan network surrounding the cell represents a highly dynamic structure that is maintained not only through the synthesis and modification of novel strands but also through the cleavage and degradation of existing strands. These different activities are indispensable for bacterial growth and cell division, and they involve the concerted action of peptidoglycan synthases and hydrolases (Monteiro et al. 2015). Notably, an appropriate balance between bacterial peptidoglycan synthesis and hydrolysis is crucial for the bacterial cell viability. In particular, when peptidoglycan synthesis is slowed down while peptidoglycan hydrolysis proceeds, autolysis of the staphylococcal cell will occur (Ghuysen et al. 1966).
The mechanism of peptidoglycan synthesis has been investigated for a long time. The process starts with the synthesis of Lipid II precursors carrying disaccharide-peptide units inside the cell (Ruiz 2008, Sham et al. 2014), and their subsequent ‘flipping’ across the cytoplasmic membrane by specific transporters known as ‘flippases’ (Fig. 2) (Ruiz 2008). Afterwards, glycosyltransferases, such as the penicillin-binding protein 2 (PBP2), MGT and SgtA of S. aureus will polymerize the translocated Lipid II precursors into nascent glycan strands (Lovering et al. 2007, Reed et al. 2011, Lovering et al. 2012). These nascent strands will be cross-linked to stem peptides in the existing peptidoglycan matrix through the action of transpeptidases, such as PBP1, PBP3 and PBP4 (Sauvage et al. 2008). Because of its vital role in bacterial growth and survival, the process of peptidoglycan synthesis has been a preferred target for antibiotic development. This is underscored by important antibiotics like β-lactams (e.g. penicillin or next-generation cephalosporins) and vancomycin which, respectively, target penicillin-binding proteins and acyl-D-alanyl-D-alanine in the S. aureus cell wall (Nagarajan 1991, El-Shaboury et al. 2007, Rajagopal and Walker 2017). On the other hand, the equally important process of cell wall hydrolysis has, so far, received relatively little attention as a possible therapeutic target for antibiotics (Do et al. 2020).
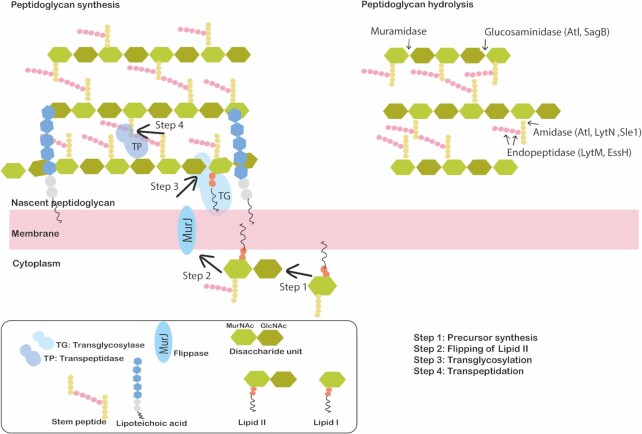
Figure 2.
Schematic representation of the S. aureus cell envelope structure. The S. aureus cell envelope is composed of a cytoplasmic membrane that is surrounded by a thick layer of peptidoglycan. For peptidoglycan synthesis (left), Lipid II units carrying disaccharides are synthesized in the cytoplasm (Step 1) and exported across the cytoplasmic membrane by a flippase (MurJ) (Step 2). Subsequently, transglycosylases (TG) insert the disaccharides into a new glycan strand (Step 3). The stem peptides of the glycan strands are then cross-linked by transpeptidases (TP) to the existing peptidoglycan matrix (Step 4). Peptidoglycan hydrolysis (right) occurs due to the action of peptidoglycan hydrolases which are classified as glycosidases (glucosaminidase and muramidase), amidases or endopeptidases. The specificity of the cleavage sites of these PGH's of S. aureus are indicated by arrows.
The maturation of peptidoglycan requires not only the action of peptidoglycan synthases but also that of peptidoglycan hydrolases (PGHs) (Shockman and Höltje 1994, Shockman et al. 1996, Vollmer et al. 2008, Do et al. 2020). In general, Gram-positive bacteria contain multiple PGHs, which cleave specific sites in the peptidoglycan and have specific roles in cell wall biogenesis (Smith et al. 2000). Depending on the specific cleavage sites, three main types of PGHs can be distinguished, namely glycosidases, amidases and endopeptidases (Fig. 3) (Rigden et al. 2003, Fenton et al. 2010, Szweda et al. 2012). The physiological roles of PGHs are highly diverse, since they include the regulation of glycan strand length, control over the level of peptidoglycan cross-linking, the remodelling of peptidoglycan after cell separation, and the recycling of peptidoglycan fragments (Vollmer et al. 2008, Rajagopal and Walker 2017). The precise numbers and essential features of PGHs differ widely between bacteria. In particular, S. aureus genomes encode at least 18 PGHs with known or putative hydrolytic activities to support fundamental cellular processes during the cell cycle. Interestingly, none of these enzymes is essential for S. aureus survival with the exception of SsaA (SA2093), which is essential for strain NTCT8325, but not for strain N315 (Chaudhuri et al. 2009). The latter strain dependency with respect to the possible essentiality of PGHs implies that a precise distinction of the individual and concerted (inter)actions of PGHs in S. aureus is rather challenging. Accordingly, the fundamental functions of individual PGHs and their coordination with peptidoglycan synthesis are still poorly understood. However, disentangling the different roles of PGHs will be important, since they are potentially druggable targets for novel antibiotics, or passive or active anti-staphylococcal immunization approaches (Wang et al. 2021). PGHs are particularly attractive targets for drugs or immunization not only because they are exposed on the bacterial cell surface and have shown high immunogenicity (Pastrana et al. 2018, Dreisbach et al. 2020, Wang et al. 2021), but also because they play vital roles in cell growth and division. In view of this potential opportunity to develop novel preventive or therapeutic approaches to fight staphylococcal infections, the present review is aimed at providing a detailed overview of what is currently known about the physiological roles and specificity of the different PGHs of S. aureus and how they are expressed and function during different stages in the bacterial cell cycle and during infection. Of note, an important requirement for novel drug targets is that they are expressed by all the different lineages of a particular pathogen. Therefore, this review will be focused on the 18 PGHs encoded by the S. aureus core genome (Table 1). In contrast, PGHs encoded by plasmids and bacteriophages, such as LytP2 and LytP4 from the S. aureus Newman prophages ϕNM2 and ϕNM4 (Bae et al. 2006), respectively, will not be addressed as this topic has recently been reviewed (Fabijan et al. 2020, Rai and Khairnar 2021, Walsh et al. 2021).
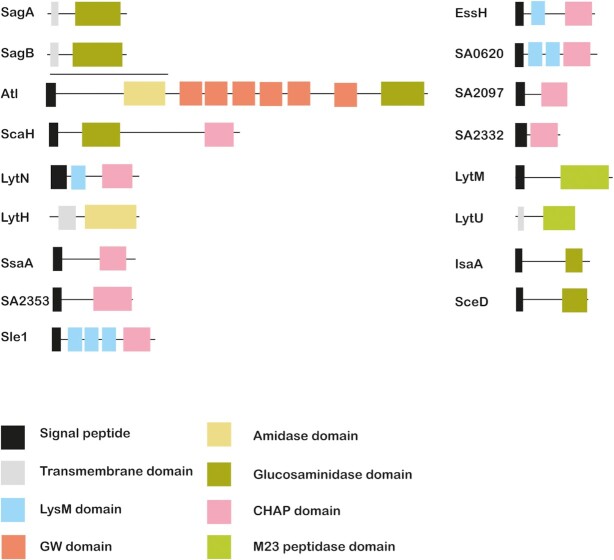
Figure 3.
Modular structure of S. aureus peptidoglycan hydrolases. S. aureus peptidoglycan hydrolases have a modular structure. The different domains that make up these enzymes include catalytic domains, such as the amidase, glucosaminidase, CHAP, M23 peptidase and/or lytic transglycosylase domains. In addition, peptidoglycan hydrolases contain signal peptides, transmembrane domains and/or cell wall binding domains (e.g. LysM, GW) for subcellular localization.
Table 1.
Overview of S. aureus peptidoglycan hydrolases.
| Protein | Mw (kDa) | AA | N315 | USA300 | NCTC8325 |
|---|---|---|---|---|---|
| SagA | 29.6 | 258 | SA2100 | SAUSA300_2256 | SAOUHSC_02580 |
| SagB | 32.5 | 284 | SA1593 | SAUSA300_1720 | SAOUHSC_01895 |
| Atl | 136.7 | 1248 | SA0905 | SAUSA300_0955 | SAOUHSC_00994 |
| Aly/ScaH | 69.2 | 619 | SA2437 | SAUSA300_2579 | SAOUHSC_02979 |
| LytN | 43.1 | 383 | SA1090 | SAUSA300_1140 | SAOUHSC_01219 |
| LytH | 32.7 | 291 | SA1458 | SAUSA300_1588 | SAOUHSC_01739 |
| SsaA | 29.3 | 267 | SA2093 | SAUSA300_2249 | SAOUHSC_02571 |
| EssH | 33.1 | 297 | SA0270 | SAUSA300_0277 | SAOUHSC_00256 |
| SA2353* | 27.6 | 255 | SA2353 | SAUSA300_2503 | SAOUHSC_02883 |
| Sle1/Aaa | 35.8 | 334 | SA0423 | SAUSA300_0438 | SAOUHSC_00427 |
| SA0710* | 30.2 | 279 | SA0710 | SAUSA300_0739 | SAOUHSC_00773 |
| SA0620* | 28.1 | 265 | SA0620 | SAUSA300_0651 | SAOUHSC_00671 |
| SA2097* | 17.4 | 166 | SA2097 | SAUSA300_2253 | SAOUHSC_02576 |
| SA2332* | 16.8 | 143 | SA2332 | SAUSA300_2482 | SAOUHSC_02855 |
| LytM | 34.3 | 316 | SA0265 | SAUSA300_0270 | SAOUHSC_00248 |
| LytU | 22 | 192 | SA0205 | SAUSA300_0207 | SAOUHSC_00174 |
| IsaA | 24.2 | 233 | SA2356 | SAUSA300_2506 | SAOUHSC_02887 |
| SceD | 24.1 | 231 | SA1898 | SAUSA300_2051 | SAOUHSC_02333 |
Protein names and the respective accession codes for the S. aureus strains N315, USA300 and NCTC8325 are presented. *Functionally uncharacterized PGHs are named by their S. aureus N315 accession codes. Mw, molecular weight; AA, total numbers of amino acids including the signal peptide according to the annotation for S. aureus N315.
Staphylococcal cell wall composition
The cell wall of Gram-positive bacteria contains not only multiple layers of peptidoglycan, but also other polymers like wall teichoic acids (WTAs), lipoteichoic acids (LTAs) and attached proteins (Strominger et al. 1967, Kojima et al. 1985, Schneewind et al. 1995). The wall may be coated with capsular polysaccharides (Keinhoerster et al. 2019). Typically, the thickness of the cell wall is 20 to 40 nm (Giesbrecht et al. 1998, Matias and Beveridge 2006). The peptidoglycan is composed of multiple long and stiff cross-linked glycan strands, with each strand consisting of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units and attached stem peptides that are relatively short and flexible (de Pedro and Cava 2015). Different Gram-positive bacteria vary in the composition of the stem peptides and the respective cross-linked peptide bridges. The stem peptides of S. aureus peptidoglycan consist of L-alanine, D-isoglutamine, L-lysine, D-alanine and a terminal D-alanine (Vollmer et al. 2008, Silhavy et al. 2010). Further, the stem peptides are covalently cross-linked by cleavage of the amide bond of D-Ala-D-Ala and the generation of a new amide bond between the fourth D-Ala of a stem peptide and the first glycine of a pentaglycine bridge on a neighbouring glycan strand (de Pedro and Cava 2015). Notably, the pentaglycine interpeptide bridge is a unique and essential component of the S. aureus peptidoglycan, which permits the generation of a three-dimensional mesh that allows for cell plasticity. However, it also provides the cell wall with sufficient strength and rigidity needed under different environmental and infectious conditions (Delaune et al. 2011). The glycan strands of S. aureus are relatively short with, on average, 6 to 10 disaccharides (Boneca et al. 2000, Wheeler et al. 2015). This is apparently important for cell wall flexibility, as longer glycan strands resulting from inactivation of the S. aureus glucosaminidase SagB were shown to cause an increased cell wall stiffness (Wheeler et al. 2015). The level of glycan strand cross-linking also affects the cell wall stiffness. This view is underscored by experiments where S. aureus was treated with lysostaphin, an enzyme from Staphylococcus simulans that targets the pentaglycine bridge of the S. aureus cell wall, which resulted in decreased cell wall stiffness due to a reduced level of peptidoglycan cross-linking (Chan et al. 2016). Importantly, prolonged treatment with lysostaphin was shown to result in the formation of osmotically fragile cells (Schindler and Schuhardt 1964, Francius et al. 2008). Apart from variations in the peptidoglycan strand length, the peptidoglycan can also be chemically modified. Such peptidoglycan modifications play crucial roles in protecting S. aureus against innate host defenses. For instance, O-acetylation of the MurNAc C6 hydroxyl and N-deacetylation of the GlcNAc C2 acetyl will protect the bacteria against lysozyme produced by the host (Bera et al. 2006, Moynihan et al. 2014, Rajagopal and Walker 2017).
As indicated above, S. aureus incorporates complex teichoic acids (TAs) into its cell wall (Weidenmaier and Peschel 2008). These TAs serve several important roles related to cell wall maintenance and bacterial fitness under different environmental challenges. The TAs are either covalently bound to glycan strands in the case of WTAs, or anchored to the cytoplasmic membrane in the case of LTAs. The WTAs are glycopolymers consisting of repeating ribitol-5-phosphate units that are covalently linked to peptidoglycan via phosphodiester bonds between the GlcNAc-1-phosphate and the C6 position of the MurNAc of the peptidoglycan, which may be tailored by different amino acids and sugars (Brown et al. 2013). In the context of cell wall biogenesis, WTAs play important roles in peptidoglycan metabolism by regulating the localization of PBPs which, in turn, facilitates the cross-linking of peptidoglycan (Atilano et al. 2010, Sewell and Brown 2014, Rajagopal and Walker 2017). The WTA polymers have a negatively charged backbone, which provides proton-binding sites, thereby contributing to the regulation of autolytic activity and the localization of wall-associated PGHs (Jolliffe et al. 1981, Penyige et al. 2002). However, WTAs are involved in many more processes. For instance, WTAs impact directly or indirectly on protein secretion, and they provide a reservoir of metal ions that are needed in the bacterial physiology and the post-translocational folding of extracellular proteins (Sarvas et al. 2004). WTAs also serve roles in bacterial pathogenesis, including adhesion to host cells and tissues, and biofilm formation (Gross et al. 2001, Sieradzki and Tomasz 2003, Frankel and Schneewind 2012, Brown et al. 2013). In addition, WTAs regulate the localization of the transpeptidase PBP4 to control the crosslinking level of peptidoglycan (Atilano et al. 2010), and WTAs protect the bacteria against heat, low osmolarity, cationic antimicrobial peptides and antimicrobial fatty acids (Brown et al. 2013). Lastly, a connection between WTAs, autolysis, and vancomycin susceptibility of S. aureus was recently reported (Hort et al.2021). An important conclusion from this study was that β-glycosylated WTAs result in decreased affinity of the major autolysin AtlA for binding to the cell wall, thereby reducing bacterial cell lysis.
On the other hand, LTAs are zwitterionic polymers composed of repeating glycerol-phosphate units that are anchored to the cytoplasmic membrane through fatty acids (Xia et al. 2010). Like the WTAs, the LTAs have been implicated in the control of cell morphology, growth, division and pathogenesis (Oku et al. 2009, Campbell et al. 2011).
The Gram-positive bacterial cell wall contains many proteins with roles in host adhesion, colonization and infection. These surface proteins are translocated across the cytoplasmic membrane via specific secretion pathways and they are subsequently either covalently or non-covalently attached to the cell wall (Schneewind et al. 1995, Navarre and Schneewind 1999). In particular, peptidoglycan provides the attachment sites for the covalent anchoring of surface proteins, which is mediated by transpeptidases that are better known as sortases (Navarre and Schneewind 1999, Tsompanidou et al. 2012). These sortases recognize a C-terminal LPXTG motif in nascent wall-associated proteins, cleave between the threonine and glycine residues of this motif, and couple the resulting terminal threonine residue to the terminal glycine of a pentapeptide bridge (Navarre and Schneewind 1999). Non-covalently cell wall-bound proteins interact with cell wall polymers, especially peptidoglycan, with the help of specific cell wall-binding domains. Alternatively, such proteins associate with the cell wall through specific physico-chemical features, including electrostatic and hydrophobic interactions (Chhatwal 2002, Dreisbach et al. 2011). Many of the covalently and non-covalently cell wall-bound proteins have roles in peptidoglycan metabolism and pathogenesis (Schneewind et al. 1995, Wardenburg et al. 2007).
Peptidoglycan hydrolases
PGHs are enzymes capable of cleaving the covalent bonds of glycan strands or stem peptides. These enzymes are widely conserved in bacteria where they are required for accurate cell growth, division and separation. However, genes encoding PGHs are highly redundant in most bacterial genomes (Vollmer et al. 2008). Accordingly, they are by themselves not essential for S. aureus growth and survival, at least under laboratory conditions, with exception of the aforementioned SsaA (SA2093) of S. aureus NTCT8325 (Antignac et al. 2007).
Depending on their specific cleavage sites in peptidoglycan, the PGHs are grouped into three classes. First, the glycosidases cleave the glycan strand, and here two different types of enzymes can be distinguished; the glucosaminidases cleave the β-1,4 glycosidic bond between GlcNAc and MurNAc whereas the lytic transglycosylases (muramidases) cleave the β-1,4 glycosidic bond between MurNAc and GlcNAc resulting in the formation of MurNAc with 1,6-anhydromuramic acid residues (Höltje et al. 1975). Second, the amidases cleave the amide bond between MurNAc and the first L-Ala residues of the stem peptide. Thirdly, the endopeptidases cleave the bond between two amino acid residues within a stem peptide or the pentaglycine bridge.
Notably, S. aureus uses N- acetylglucosaminidases instead of N-acetylmuramidases to cleave its own glycan strands. This may relate to the aforementioned O-acetylation of the MurNac residues, which S. aureus employs to protect itself from peptidoglycan cleavage by the host-derived N-acetylmuramidase lysozyme, which cleaves the MurNAc-β-1,4-GlcNAc bond of the glycan strand (Bera et al. 2005). Previous studies characterized various PGHs of S. aureus, including four glucosaminidases (Atl, SagA, SagB, ScaH), two lytic transglycosylases (IsaA, SceD), four putative amidases (Atl, Sle1, LytN, EssH) and five putative endopeptidases (LytN, LytH, LytM, LytU, EssH). Here it should be noted that this listing refers twice to some particular proteins because these proteins are associated with two distinct PGH activities. For instance, Atl has both a glucosaminidase domain and an amidase domain, which become physically separated by proteases produced by S. aureus during growth (Komatsuzawa et al. 1997). The amidase domain of Atl specifically targets the muramyl-tetrapeptide MurNAc-L-Ala-D-iGln-L-Lys-NHA-D-Ala-NH2 (MtetP) of the peptidoglycan of S. aureus (Büttner et al. 2014). Interestingly, another study showed that binding of the stem peptides of peptidoglycan is important for the specific association of the Atl homolog of S. epidermidis AmiE with its substrates (Lützner et al. 2009). The features of the different PGHs from S. aureus are summarized in Supplementary Table 1. The specific peptidoglycan cleavage sites of some of these PGHs, like those that contain cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domains have not yet been characterized. CHAP domain-containing PGHs are commonly present in many different bacteria, where they can act as amidases and/or endopeptidases (Bateman and Rawlings 2003). For example, the CHAP domain of the S. aureus PGH Sle1 only has amidase activity, while the CHAP domains of the S. aureus PGHs LytN and EssH have both amidase and endopeptidase activities (Bobrovskyy et al. 2018).
Frequently, PGHs show a modular architecture that combines different catalytic domains and/or cell wall-binding domains, and these can be located in the N-terminus or the C-terminus of a PGH (Fig. 3). The wall-binding domains allow the binding of PGHs to the peptidoglycan at an adequate concentration. They also properly position the respective PGH active sites towards the PG substrate cleavage site for efficient formation of enzyme-substrate complexes (Vermassen et al. 2019). The type and number of cell wall-binding domains differ among PGHs (Steen et al. 2005). This is exemplified by the cell wall-binding Lysin Motif (LysM), which is widely distributed in both prokaryotes and eukaryotes (Buist et al. 2008). Bacterial LysM domains recognize and non-covalently bind to the N-acetylglucosamine moiety of peptidoglycan (Frankel and Schneewind 2012). LysM domains also direct the murein hydrolases Sle1 and LytN to the cross wall of S. aureus, where they assume different positions (Frankel et al. 2011, Frankel and Schneewind 2012, Pastrana et al. 2017). LytN contains a YSIRK/GS signal which also directs this protein to the septum of dividing cells (Frankel et al. 2011). SH3b, another cell wall-binding domain, targets proteins to the pentaglycine bridge with high affinity and specificity, and SH3b-mediated peptidoglycan binding is actually a key step in peptidoglycan hydrolysis (Visweswaran et al. 2014, Gonzalez-Delgado et al. 2020). The GW domain of Atl is a repeating cell wall-binding domain that binds to LTA (Sugai et al. 1995, Baba and Schneewind 1998, Zoll et al. 2012), which is responsible for the correct protein docking (Komatsuzawa et al. 1997, Frankel and Schneewind 2012, Zoll et al. 2012). Some PGHs are capable of cleaving cross-links of soluble muropeptides and intact peptidoglycan sacculi. For example, mutanolysin from Streptomyces globisporus cleaves the MurNAc-GlcNAc that carries a stem peptide (Kawata et al. 1983). In contrast, the glucosaminidase domain of Atl specifically cleaves the glycan strand without attached stem peptides (Nega et al. 2020). Compared to Atl, the glucosaminidase SagB prefers longer glycan strands as substrates which, in fact, implies an important role for SagB in controlling the glycan strand length (Wheeler et al. 2015, Chan et al. 2016). The amidase LytH exclusively cleaves peptides from non-cross-linked nascent peptidoglycan (Do et al. 2020).
Altogether, studies on the PGHs of S. aureus have shown that these enzymes are exported proteins that localize either to the bacterial septum (Sle1 [Frankel and Schneewind 2012], Atl [Schlag et al. 2010], LytN [Frankel et al. 2011], IsaA [Sakata et al. 2005]) or the peripheral wall region (LytM [Ramadurai et al. 1999] and LytU [Raulinaitis et al. 2017]) during cell growth and division. Some PGHs, like Atl and Sle1, are also secreted into the extracellular milieu and, subsequently, these PGHs cleave the septum at the cross wall (Komatsuzawa et al. 1997, Frankel and Schneewind 2012). Moreover, PGHs may even contain both a membrane anchor and an extracytoplasmic catalytic domain, as exemplified by LytU, LytH, SagA, and SagB. Their positioning close to the extra-cytoplasmic side of the membrane suggests that the latter four enzymes preferentially target nascent peptidoglycan.
Physiological roles of PGHs
PGHs have diverse functions, including the regulation of cell growth, cell enlargement, remodelling of peptidoglycan, peptidoglycan turnover and recycling, assembly of secretion systems, widening of pores in the wall to allow secretory protein passage and release of surface proteins, cell separation during cell division, and autolysis (Fig. 4) (Scheurwater et al. 2008, Vollmer et al. 2008). Some of the S. aureus PGHs play multiple functions in these processes (Vollmer et al. 2008, Frirdich and Gaynor 2013), and this probably explains why the loss of individual PGHs in Gram-positive bacteria is in most cases neither lethal, nor associated with major morphological phenotypes (Vollmer et al. 2008, Frirdich and Gaynor 2013).
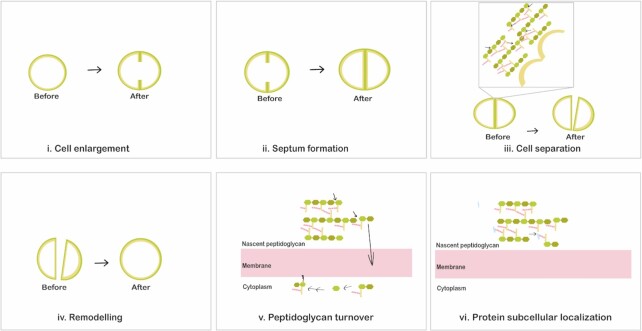
Figure 4.
Physiological roles of S. aureus peptidoglycan hydrolases. Peptidoglycan hydrolases play different essential roles during bacterial growth and division. PGHs are involved in the entire cell cycle including cell enlargement (i), cell enlargement during septum formation (ii), daughter cell separation upon cell division (iii), and remodelling of the peptidoglycan network after daughter cell separation (iv). In addition, peptidoglycan hydrolases serve in peptidoglycan turnover (v) and they modulate cell wall passage of cell surface-located proteins (vi).
PGH contributions to cell separation
One of the main functions of PGHs is to separate the daughter cells at the final stage in the cell division process. In the division phase, cells form a dynamic Z-ring through polymerization of the tubulin homologue FtsZ, which is an essential cytoplasmic cell division protein that recruits other proteins of the cell division machinery to synthesize the division septum (Ortiz et al. 2016, den Blaauwen et al. 2017). The two resulting hemispherical daughter cells are interlinked through a narrow peripheral ring, have their own cytoplasmic membranes, but share the septum, which is a layer of peptidoglycan that needs to be separated after cell division through the hydrolysis of crosslinked peptidoglycan by PGHs (Matias and Beveridge 2007, Vollmer et al. 2008). To this end, PGHs first create perforation holes in the peripheral ring that links the daughter cells. The resolution of the peripheral ring (‘popping’) occurs within milliseconds, which most likely depends on alterations in the cell surface stress (Zhou et al. 2015). Nonetheless, upon separation of the daughter cells there are no apparent changes in the volume of each daughter cell. Prior to cell separation, a large belt of peptidoglycan is present at the division sites, which may serve to protect the peripheral wall from hydrolysis by PGHs that participate in the ‘pre-splitting’ of the septal cell wall (Matias and Beveridge 2007, Turner et al. 2010). The concerted and precise action of PGHs in cell separation is thus very important because incorrect cell separation could affect proper division or delay the separation of daughter cells.
Amidases such as Atl, Sle1 and LytN have been implicated more prominently in cell separation by contributing to peptidoglycan cleavage at the septum. In particular, Atl localizes at the peripheral ring region of the surface of the septum prior cell separation (Yamada et al. 1996). In atl deletion mutants and mutants in which the amidase or glucosaminidase domains were inactivated, the cell wall of the ‘cracked’ daughter cell was shown to be smooth and flat, while the outer wall was rough (Nega et al. 2020). The rough surface was mostly detectable in the atl deletion mutant and the mutant with an inactivated glucosaminidase domain. Cluster formation was observed for all three mutants. Nonetheless, cell division was not principally affected in atl mutants with inactivated amidase or glucosaminidase domains (Nega et al. 2020). This may relate to the fact that in an atl mutant, the expression of Sle1 and other PGHs (i.e. SagA, SsaA-SA2093, ScaH, LytH, IsaA, LytM, SceD, EssH, SA2097, SA0620, SA2353, SA2332) was increased, likely compensating for a possible defect in cell separation due to the absence of Atl (Heilmann et al. 2005, Pasztor et al. 2010, Hirschhausen et al. 2012). Nevertheless, loss of Atl or Sle1 increased the duration of the cell cycle from 66 ± 9 min for the parental wild-type strain, to 82 ± 8 min or 86 ± 15 min, respectively (Monteiro et al. 2015). Although most upregulated genes in the atl mutant were shown to be controlled by the two-component system WalKR, no difference in expression of this regulatory system was detected in the atl mutant compared to the wild-type strain (Pasztor et al. 2010).
The amidase Sle1 plays important roles in cell separation by degrading the outer edge of the septum during cell division. Sle1 is not required for the resolution of the outer septum, but inactivation of Sle1 delays the onset of daughter cell separation, while elevated levels of Sle1 accelerate the onset of daughter cell separation and lead to a reduced cell size (Thalsø-Madsen et al. 2019). Of note, the aforementioned perforations in the peripheral ring at mid-cell, which are introduced prior to cell separation, are closely correlated to the Sle1 levels (Thalsø-Madsen et al. 2019). Furthermore, the lack of some amidases results in a partial dysregulation of peptidoglycan synthesis, and the respective mutant bacterial cells increase in size and show cell division defects. The loss of glucosaminidases, such as Atl and SagB, impact cell separation (Wheeler et al. 2015, Chan et al. 2016, Sutton et al. 2021). Bacteria lacking SceD and IsaA displayed impaired cell separation and clumping (Sakata et al. 2005, Stapleton et al. 2007).
PGH contributions to cell enlargement
During the cell cycle, the expansion of cells is important for maintaining the bacterial shape. This expansion probably starts at the cell periphery and also occurs at the division sites (Pinho et al. 2013, Lund et al. 2018). The proper subcellular location of FtsZ determines the final cell size (Jorge et al. 2011, Veiga et al. 2011). After cell division, S. aureus undergoes slight elongation at mid-cell along the lateral wall. This is mediated by PBP3 and RodA, which promote the sidewall insertion of peptidoglycan (Reichmann et al. 2019). S. aureus reshapes the flat septum into the curved hemispheres of two daughter cells once the cell separation has been completed (Pinho et al. 2013). PGHs drive the peptidoglycan remodelling after splitting by breaking existing bonds in the peptidoglycan, thereby increasing the peptidoglycan surface area and allowing the peptidoglycan to stretch. In turn, this allows the incorporation of new peptidoglycan units into the nascent glycan strands. A group of PGHs with glucosaminidase activity (i.e. Atl, SagA, ScaH and SagB) is required for cell enlargement after cell separation, and for reaching the mature cell shape (Wheeler et al. 2015). As mentioned above, physical properties, such as the cell wall stiffness and glycan strand length, may affect the cell wall elasticity, thereby impacting cell enlargement. Peptidoglycan strand length and cross-linking levels also affect the cell wall stiffness (Loskill et al. 2014, Wheeler et al. 2015). Accordingly, cell enlargement may depend on the reduction of cell wall stiffness. One model proposes that initially a dense and stiff peptidoglycan is formed. However, hydrolysis by PGHs makes it less dense, less cross-linked and less stiff, thereby allowing the insertion of new peptidoglycan and enabling the enlargement of cells into their mature shape (Loskill et al. 2014, Wheeler et al. 2015). SagB is dominantly responsible for determining the length of glycan strands, producing a GlcNAc at the end of one resulting strand and a MurNAc at the start of the other strand (Schaefer et al 2021, Willing et al. 2021). Accordingly, the inactivation of SagB results in long unprocessed glycan strands, which cause increased cell wall stiffness and decreased cell wall elasticity. Other glucosaminidases, like Atl, SagA and ScaH have an additional, though modest, impact on glycan strand length and cell enlargement, which may be due to reduced peptidoglycan cross-linking, especially by SagA (Wheeler et al. 2015, Chan et al. 2016, Pasquina-Lemonche et al. 2020).
PGH contributions to cell wall turnover
The turnover or recycling of bacterial peptidoglycan is constantly progressing during cell growth. Different bacteria have distinct strategies to breakdown the peptidoglycan and reutilize the liberated cell wall fragments for peptidoglycan recycling (Mayer et al. 2019). In fact, it has been reported that around 45% of the peptidoglycan is turned over in one generation in bacteria (Goodell 1985). In S. aureus, Atl plays an important role in cell wall turnover. In particular, Atl hydrolyzes peptidoglycan, thereby producing peptides and the disaccharide MurNAc-GlcNAc (Kluj et al. 2018). This unique disaccharide unit is transported into the bacterial cell and phosphorylated by the phosphotransferase system transporter MurP. The intracellular 6-phospho-N-acetylmuramidase MupG cleaves the MurNAc 6-phosphate-GlcNAc and produces MurNAc 6-phosphate and GlcNAc (Kluj et al. 2018). The MurQ esterase then converts MurNAc 6-phosphate into GlcNAc 6-phosphate (Borisova et al. 2016). Such cell wall turnover products may subsequently be perceived as signalling molecules for bacterial recognition by other (micro)organisms or mammalian hosts. In some bacteria this will lead to the induction of β-lactamase production (Jacobs et al. 1997). Endopeptidases could also mediate the turnover of peptidoglycan. Importantly, the peptidoglycan turnover caused by PGHs regulates the release of some surface proteins that may be involved in host cell adhesion and invasion (Boneca 2005). The immunodominant staphylococcal antigen A (IsaA) and its paralogue SceD with putative soluble lytic transglycosylase domains may participate in peptidoglycan turnover and cell wall hydrolysis (Stapleton et al. 2007).
Role of PGHs in extracellular protein localization
The Sec pathway is the primary secretion pathway of S. aureus, which directs signal peptide-dependent translocation of proteins across the cytoplasmic membrane into the cell wall and extracellular milieu (Sibbald et al. 2006, Dreisbach et al. 2011, Schneewind and Missiakas 2014). However, among the surface-associated and extracellular proteins of S. aureus, many typically cytoplasmic proteins can be encountered, which are nowadays referred to as extracellular cytoplasmic proteins (ECPs) (Sibbald et al. 2006). PGHs have been implicated in the extracellular localization of these ECPs by modulating the peptidoglycan complexity and cross-linking (Ebner et al. 2015, 2017, Ebner et al. 2016, Ebner and Götz 2019, Zhao et al. 2020). For instance, this applies to SagB, which is evidenced by the fact that sagB mutant bacteria display defects in cell wall anchoring and protein secretion, whereas the liberation of ECPs is enhanced (Chan et al. 2016). Nonetheless, the release of ECPs by sagB mutant bacteria does not seem to be due to autolysis or membrane leakage (Chan et al. 2016). On the other hand, atl mutant bacteria display a significant decrease in the release of ECPs to the bacterial cell surface and into the extracellular milieu. This suggests a role of Atl-mediated cell wall weakening in the release of ECPs (Pasztor et al. 2010). In this context it is noteworthy that various ECPs have known roles in staphylococcal virulence, such as the adhesion to host cells. Importantly, there are also PGHs known to operate in close concert with dedicated protein secretion systems. This is exemplified by the EssH protein, which is a CHAP domain-containing PGH encoded by the ess locus. The activity of EssH is associated with the ESAT-6-like secretion system, also referred to as the type 7 secretion system (T7SS). In particular, EssH is required for assembly of the T7SS secretion system and secretion of the EsxC and EssD proteins by the T7SS-dependent pathway (Bobrovskyy et al. 2018). Through its amidase and endopeptidase activities EssH cleaves the stem peptides and pentaglycine bridges of the peptidoglycan to create space for protein passage across the bacterial cell wall. Likewise, PGHs involved in the assembly of secretion systems have been reported in other bacteria, such as Escherichia coli (Koraimann 2003).
Possible cross-talk of PGHs and peptidoglycan synthases
The peptidoglycan synthases and PGHs of Gram-positive bacteria are located at distinct subcellular positions, which sets limits to direct interactions between these enzymes (Vollmer et al. 2008). In particular, peptidoglycan synthases are positioned in the inner peptidoglycan layer, while PGHs are mostly positioned in the outer layers. However, there are examples of indirect cross-talk between peptidoglycan synthases and PGHs. For instance, the amidase LytH is a determinant for the subcellular localization of peptidoglycan synthases by controlling the density of peptidoglycan assembly sites. Consequently, the inactivation of LytH results in a displacement of division sites (Do et al. 2020). This can be compensated by a reduced activity of the PBP2 polymerase, which implies that LytH controls cell growth by regulating the PBP2-mediated synthesis of new peptidoglycan (Do et al. 2020). FtsZ is frequently mislocalized in lytH mutant cells, which implies that LytH is also important for FtsZ assembly (Do et al. 2020). Similarly, deletion of the glucosaminidase domain of Atl was also shown to result in a defective localization of the division septum, since in 25%–35% of glucosaminidase-mutated bacteria an asymmetric cell division was detected (Nega et al. 2020).
Regulation of PGHs
Control of PGH expression by gene regulatory systems
To maintain the rigid shape and structure of the peptidoglycan network, the expression and activity of PGHs needs to be spatially and temporally controlled (Egan et al. 2017). Several direct and indirect mechanisms involved in the regulation of PGHs are known to exist (Typas et al. 2012, Egan et al. 2017). These involve transcriptional control by gene regulatory systems of S. aureus (e.g. sigma factors, RNA binding proteins and small regulatory RNAs), altered biological activity of PGHs and their substrate binding by the presence of LTA and WTA or peptidoglycan modification, proteolytic processing by proteases, protein-protein interactions that activate or block the activity of PGHs, as well as environmental factors.
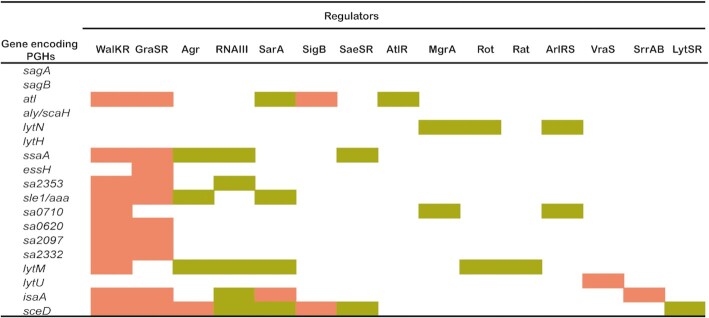
At the transcriptional level, two-component signal transduction systems and global regulators are involved in regulating the product of PGHs (Table 2 and Fig. 5). S. aureus is known to possess 16 so-called two-component regulatory systems (TCSs; e.g. WalKR, AgrAC, and ArlRS), which are composed of a sensor module with histidine kinase activity and a cognate DNA-binding response regulator that modulates transcription. Such TCSs monitor particular stresses or extracellular stimuli (Szurmant et al. 2007) and regulate the appropriate physiological responses through autophosphorylation of the sensor module and subsequent transfer of the phosphoryl group to the cognate response regulator, which leads to activation of the regulator (Kleerebezem et al. 1997). Particular TCSs may control the expression of multiple PGH-encoding genes. For instance, the WalR response regulator of the essential WalKR (YycGF) TCS specifically binds to the promoter regions of genes encoding the S. aureus PGHs Atl, LytM, SsaA (SA2093), IsaA, SceD, EssH, the three CHAP domain-containing proteins SA0620, SA2097, and SA2353) (Table 2) (Dubrac and Msadek 2004, Dubrac et al. 2007), Sle1 and SA2332 (Delauné et al. 2012), thereby positively controlling their expression. In addition, WalKR also positively regulates peptidoglycan biosynthesis and turnover (Dubrac et al. 2007). Importantly, WalKR can both regulate and be regulated by other regulatory components. For example, the YycH and YycI proteins stimulate WalK activity through WalKR phosphorylation (Gajdiss et al. 2020). Also, the WalR regulator stimulates the SaeSR TCS (Delauné et al. 2012), whereas SaeSR negatively regulates the ssaA gene (SA2093) (Rogasch et al. 2006). The expression of ssaA is positively regulated by GraSR (Falord et al. 2011), a TCS that has an important function in the regulation of the bacterial metabolism under oxidative stress conditions, at low pH and at higher temperature. Nine genes encoding PGHs are regulated by both GraSR and WalKR (Dubrac et al. 2007, Falord et al. 2011). Thus, the TCSs play multiple roles in the regulation of S. aureus cell wall metabolism (Dubrac et al. 2007, Delaune et al. 2011).
Table 2.
Peptidoglycan hydrolase regulation at the transcriptional level.
| Gene encoding PGHs | Main transcriptional regulators |
|---|---|
| sagA | - |
| sagB | - |
| atl | WalKR (↑) (Dubrac et al. 2007), SigB (↑) (Houston et al. 2011), AtlR (↓) (Houston et al. 2011), SarA (↓) (Trotonda et al. 2009), GraSR (↑) (Herbert et al. 2007, Falord et al. 2011) |
| aly/scaH | - |
| lytN | ArlRS (↓) (Memmi et al. 2012), MgrA (↓) (Luong et al. 2006), Rot (↓) (Chu et al. 2013), Rat (↓) (Ingavale et al. 2003) |
| lytH | - |
| ssaA | WalKR (↑) (Dubrac et al. 2007), GraSR (↑) (Falord et al. 2011), SaeSR (↓) (Rogasch et al. 2006), Agr (↓) (Dunman et al. 2001), putative RNAIII (↓) (Lioliou et al. 2016) |
| essH | GraSR (↑) (Falord et al. 2011) |
| sa2353 | WalKR (↑) (Dubrac et al. 2007), RNAIII (↓) (Boisset et al. 2007), GraSR (↑) (Falord et al. 2011) |
| sle1/aaa | WalKR (↑) (Delauné et al. 2012), Agr (↓) (Hirschhausen et al. 2012), SarA (↓) (Hirschhausen et al. 2012), GraSR (↑) (Herbert et al. 2007, Falord et al. 2011) |
| sa0710 | WalKR (↑) (Dubrac et al. 2007), ArlRS (↓) (Crosby et al. 2020), MgrA (↓) (Luong et al. 2006) |
| sa0620 | WalKR (↑) (Dubrac et al. 2007), GraSR (↑) (Falord et al. 2011) |
| sa2097 | WalKR (↑) (Dubrac et al. 2007), GraSR (↑) (Falord et al. 2011) |
| sa2332 | WalKR (↑) (Delauné et al. 2012), GraSR (↑) (Falord et al. 2011) |
| lytM | WalKR (↑) (Dubrac et al. 2007), Rot (↓) (Chu et al. 2013), Agr (↓) (Ramadurai et al. 1999), SarA (↓) (Ramadurai et al. 1999), RNA III (↓) (Chunhua et al. 2012), Rat (↓) (Ingavale et al. 2003) |
| lytU | VraSR (↑) (Pietiäinen et al. 2009) |
| isaA | WalKR (↑) (Dubrac et al. 2007, Stapleton et al. 2007), SarA (↑) (Stapleton et al. 2007), SrrAB (↑) (Stapleton et al. 2007), putative RNAIII (↓) (Lioliou et al. 2016), GraSR (↑) (Falord et al. 2011) |
| sceD | WalKR (↑) (Dubrac et al. 2007, Stapleton et al. 2007), SigB (↑) (Ziebandt et al. 2001, Stapleton et al. 2007), Agr (↑) (Stapleton et al. 2007), putative RNAIII (↓) (Lioliou et al. 2016), LytSR (↓) (Stapleton et al. 2007), SaeSR (↓) (Stapleton et al. 2007), GraSR (↑) (Falord et al. 2011), SarA (↓) (Stapleton et al. 2007) |
Genes for functionally uncharacterized PGHs are named by their S. aureus N315 accession codes; (↑) and (↓), up- or down-regulation of the respective PGH-encoding gene; -, unknown regulation mechanism.
Figure 5.
Peptidoglycan hydrolase gene regulation by different transcriptional regulators. The Figure provides an overview of PGH-encoding genes of S. aureus and translational regulators that are known to modulate the PGH gene expression. Up-regulation or down-regulation by particular gene regulators are indicated in pink or olive green, respectively.
One of the key TCSs of S. aureus, the accessory gene regulation (agr) quorum-sensing system that modulates the expression of over 100 genes, is also involved in the regulation of cell wall metabolism (Dunman et al. 2001). Various genes for cell wall-associated proteins are repressed in the late exponential growth phase by the agr system (Benito et al. 1998), or indirectly regulated via the small RNA III which is a key effector of the agr system (Abdelnour et al. 1993, Benito et al. 1998, Huntzinger et al. 2005). RNA III also targets other regulatory proteins and indirectly regulates the expression of several genes, including PGH-encoding genes. Thus, it was reported that lytM is negatively regulated by agr (Ramadurai et al. 1999). In particular, it was shown that the expression of lytM is negatively regulated by RNA III through interaction with the 5’UTR of the lytM mRNA and by blocking the ribosome-binding site of lytM at the post-transcriptional stage (Chunhua et al. 2012, Lioliou et al. 2016). Furthermore, RNA III was shown to negatively regulate SA2353 (Boisset et al. 2007), and this small RNA was predicted to bind to the promoter regions of sceD, isaA and ssaA (SA2093) (Lioliou et al. 2016). Lastly, the agr system regulates the expression of protease genes, which negatively affect the activity of Atl and other PGHs at the post-transcriptional level.
The expression of various PGH-encoding genes of S. aureus is repressed by the TCSs LytSR (Brunskill and Bayles 1996a) and ArlRS (Fournier and Hooper 2000, Crosby et al. 2020), and by the transcriptional regulators MgrA (Rat) (Ingavale et al. 2003, Ingavale et al. 2005), Rot (Chu et al. 2013), SarA (Cheung et al. 2004, Trotonda et al. 2009) and AtlR (Houston et al. 2011). LytSR negatively regulates PGH activity by regulating the expression of the lytSR operon and its downstream lrgAB operon (Brunskill and Bayles 1996b). In turn, the lrgAB operon, and also the paralogous cidABC operon apparently control the transport of PGHs across the membrane, thereby facilitating PGH regulation at the post-transcriptional level (Groicher et al. 2000, Rice et al. 2003, Rice and Bayles 2008, Bayles 2014). Here it should be noted that cidA encodes a putative holin that may disrupt the membrane integrity thereby promoting bacterial cell death and lysis, while lrgA encodes an anti-holin that counteracts cell death and lysis (Groicher et al. 2000, Rice et al. 2003, Rice and Bayles 2008). ArlRS and MgrA constitute a regulatory cascade that controls expression of a large number of genes (Crosby et al. 2016). ArlRS regulates autolysis by activating the expression of mgrA (Crosby et al. 2016, Crosby et al. 2020), where ArlRS and MgrA jointly mediate the repression of lytN (Luong and Lee 2006, Memmi et al. 2012). Furthermore, ArlRS, is a negative regulator of the aforementioned essH gene, but this does not involve MgrA (Crosby et al. 2020).
The regulator Rot, a DNA-binding protein from the SarA family of S. aureus regulators, can bind to the promoter regions of lytM and lytN to directly regulate the expression of these PGH-encoding genes (Chu et al. 2013). Rot also regulates the transcription of lryAB, which leads to an indirect regulation of PGH production (Chu et al. 2013). The negative regulation of autolysis by MgrA and SarA may be mediated indirectly by downregulation of sarV, which encodes a positive regulator of several PGHs (Manna et al. 2004, Trotonda et al. 2009). MgrA represses the expression of at least two transcriptional regulators, SarV and AtlR (Crosby et al. 2016). The repressor AtlR downregulates the transcription of atl (Houston et al. 2011), whereas the sigma factor SigB negatively regulates the agr locus, thereby positively influencing atl transcription (Houston et al. 2011). Lastly, the TCS VraSR is able to sense perturbations in cell wall synthesis and negatively regulates autolytic activity (Kuroda et al. 2003, Antignac et al. 2007).
Several factors have been reported to positively regulate the expression of PGH-encoding genes. For instance, the TCS SrrAB regulates gene expression in S. aureus in response to oxygen availability (Yarwood et al. 2001) and it positively regulates the transcription of isaA (Stapleton et al. 2007). Like the agr system, the regulators SarA, Rot, MgrA and SaeR also regulate the expression of extracytoplasmic proteases that can modulate PGH activity through proteolysis of, for instance, Atl (Gimza et al. 2019).
Lastly, some regulators may exert both positive and negative regulation of the expression of PGH-encoding genes. This is exemplified by the small RNA SprX of S. aureus, which exerts its regulatory function by base-pairing with cis- or trans-encoded target mRNAs, thereby influencing the mRNA stability and translation (Caldelari et al. 2013). Thus, SprX can directly interact with the WalR mRNA to positively regulate the expression of lytM and atl. On the other hand, SprX also binds to the isaA mRNA, thereby negatively influencing its stability (Buchad and Nair 2021). Another target of SprX is the spoVG gene, which encodes a site-specific DNA-binding protein responsible for the inhibition of cell wall degradation (Eyraud et al. 2014). In particular, it was shown that SpoVG can directly bind to the putative promoter region of lytN, femA and lytSR to repress their transcription (Liu et al. 2016).
Regulation of PGH activity by cell wall components
WTA and LTA are known to influence the activity of PGHs and their substrate binding capability. In particular, WTA was shown to act as a temporal and spatial regulator of peptidoglycan metabolism (Atilano et al. 2010, Schlag et al. 2010), being a key determinant for the localization and activity of Atl (Weidenmaier et al. 2004). WTA also prevents the association of the GW cell wall-binding domain of Atl with the side wall (also referred to as the lateral wall), but not with the septum where WTA is less abundant or altered in structure, which results in a more acidic milieu that reduces the activity of Atl (Schlag et al. 2010, Biswas et al. 2012, Tiwari et al. 2018). TagO is an N-acetylglucosamine-phosphate transferase, which catalyzes the first step of WTA biosynthesis. Accordingly, a tagO mutant was shown to be completely devoid of WTA. The absence of WTA resulted in an even distribution of Atl on the surface of a tagO mutant leading to increased autolytic activity (Schlag et al. 2010). WTA also mediates the specific binding of Sle1 and LytN to the cross-wall of the septum (Schlag et al. 2010) and, accordingly, a lack of WTA results in a disordered localization of Sle1 and LytN (Chan et al. 2013). Consistent with this finding, it was shown that Sle1 lacking the LysM domain is unable to fulfill its function. In contrast, the function of LytN was not affected by absence of the LysM domain (Frankel and Schneewind 2012). The latter finding can be explained by the presence of the YSIRK/GS motif in the signal peptide of LytN, which contributes to the localization of LytN to the cross-wall (DeDent et al. 2008). Of note, the GW cell wall-binding domain of Atl can also bind to LTA, which is highly abundant at the septum, resulting in Atl targeting to the septum (Biswas et al. 2012, Zoll et al. 2012). Similar to WTA, LTA has been implicated in controlling the activity of various PGHs (Fedtke et al. 2007, Tiwari et al. 2018).
Regulation of PGH activity by protein–protein interactions
Protein-protein interactions can affect the activity and localization of PGHs. For instance, PGHs can form complexes with other proteins, such as peptidoglycan synthases, to execute their biological functions (Höltje 1998, Typas et al. 2012). Thus, SagB and the cytoplasmic membrane protein SpdC (LyrA) were shown to form a complex and cleave nascent glycan strands to release newly synthesized glycan strands from the cell membrane and to produce lipid II-attached oligomers for further peptidoglycan elongation (Schaefer et al. 2021). The SagB partner protein SpdC orients the active site of SagB for cleaving glycan strands (Schaefer et al. 2021). SpdC also antagonizes the WalR-dependent expression of SceD, LytM, Atl, and SA2097 (Poupel et al. 2018). Another example of protein-protein interactions relevant for PGH activity concerns the membrane-localized amidase LytH, which requires catalytic activation by the polytopic membrane protein ActH (Schaefer et al. 2021). The LytH-ActH amidase complex can also regulate the activity of peptidoglycan synthases. In particular, this complex removes the stem peptides from a nascent glycan strand at the cell periphery and promotes the relocation of peptidoglycan synthases to the septum during cell division (Do et al. 2020). Intramolecular protein-protein interactions also appear to determine PGH activity. This is exemplified by the LytM protein that shows weak PGH activity (Osipovitch et al. 2015). In contrast, the PGH activity is strongly increased upon truncation of LytM to the so-called M23 endopeptidase domain (Fig. 3). This suggests that the activity of LytM's M23 domain is suppressed by the so-called occluding domain of LytM (Odintsov et al. 2004). Until now, it is not known how the intact LytM becomes activated, but this most likely occurs through the interaction with partner proteins rather than proteolytic processing since the full-size LytM is readily detectable in the growth medium of S. aureus (Wang et al. 2021).
Regulation of PGHs by proteolytic activity
Staphylococcus aureus possesses several cytoplasmic and secreted proteolytic enzymes that play important functions in regulating the expression and activities of PGHs (Shaw et al. 2004, Kolar et al. 2013). For example, Sle1 is a substrate of the highly conserved protease ClpXP, which is composed of the peptidase subunit ClpP and the chaperone subunit ClpX. Inactivation of ClpXP was shown to result in increased Sle1 activity (Feng et al. 2013, Thalsø-Madsen et al. 2019), which seems to be due to ClpXP mediated processing of the signal peptide-bearing Sle1 precursor protein (Feng et al. 2013, Liu et al. 2017). Likewise, enhanced levels of IsaA, SceD, LytM, LytN, SsaA (SA2093) were detected in both clpP and clpX mutant strains (Kirsch et al. 2020). Cytoplasmic proteases, such as ClpXP, have been implicated in the activation of some regulatory factors, such as Rot, thereby indirectly regulating the expression of PGHs (Frees et al. 2005). Importantly, also extracellular proteases seem to modulate the PGH levels by proteolytic processing, in order to dispose of damaged or no longer needed proteins, or in response to external or internal signals. These extracellular proteases include the metalloprotease Aur, the serine proteases SspA, SplA, and SplC, the serine protease-like proteins SplB, SplD, SplE, and SplF, as well as the cysteine proteases ScpA and SspB (Dubin 2002, Kolar et al. 2013). In particular, the SspA protease was shown to be involved in the proteolytic processing of Atl, thereby regulating Atl activity and autolysis (Chen et al. 2013, Sahukhal et al. 2015).
Regulation of PGHs by environmental factors
Lastly, the activity of PGHs can be modulated by environmental factors, such as the salt and metal concentrations, or the pH (Gilpin et al. 1972, Foster 1995, Calamita and Doyle 2002). The impact of metals is exemplified by the M23 domain of LytM, LytU and the amidase domain of Atl, which all display zinc-dependent enzymatic activity (Büttner et al. 2014, Raulinaitis et al. 2017). The PGH activity of LytM is strongly inhibited in the presence of Fe2+, Cu2+, and Mg2+ ions and low concentrations of NaCl or KCL (Ramadurai et al. 1999). In contrast, the presence of Cu2+ or Co2+ was shown to increase the activity of LytU (Raulinaitis et al. 2017). A clear pH dependency was reported for SagB, which is more active at pH 5 than at pH 7.5 (Wheeler et al. 2015). LytM shows optimal activity at pH values between 5 and 8 (Ramadurai et al. 1999), whereas Atl has optimal activity above pH 6.5. Of note, Atl activity seems to be enhanced at low temperature and high NaCl concentrations (Foster 1995). Accordingly, it has been proposed that changes in the ionic composition of the cell wall affect PGH activity (Cheung and Freese 1985). In particular, it was shown that WTA affects the concentration of proton-binding sites in the cell wall, thereby modulating the activity of Atl (Biswas et al. 2012). The optimal pH for activity may relate to the specific localization of a PGH within the cell wall, where a high concentration of protons is encountered at the membrane-cell wall interface due to the proton-motive force, while the proton concentration can be lower towards the cell surface (Calamita et al. 2001).
Impact of antibiotics on PGH activity
Inhibition of peptidoglycan synthesis by antibiotics can lead to bacterial autolysis. This was exemplified by perturbations of the peptidoglycan synthesis in S. aureus, either by growing the bacteria in the presence of sub-inhibitory concentrations of β-lactam antibiotics, or by downregulating the expression of the peptidoglycan synthase PBP2 (Antignac et al. 2007). However, in response to the respective cell wall stress, the synthesis of PGHs is repressed at the transcriptional level, leading to a decreased susceptibility to autolysis (Antignac et al. 2007). In turn, this may result in a reduced susceptibility for β-lactams, as evidenced by the observation that PGH inactivation typically enhances the resistance to β-lactam antibiotics (Tomasz et al. 1970). In different staphylococcal lineages, the antibiotic resistance profiles can be very different, which will impact on the correlation of antibiotic activity and PGHs to different extents. Accordingly, some PGH mutants show actually an increased susceptibility to antibiotics. For example, a sagB mutant of the MRSA USA300 LAC strain was shown to be more susceptible to the β-lactam antibiotic oxacillin (Chan et al. 2016). This may be due to the increased length of glycan strands as a consequence of the SagB-deficiency. The increased glycan strand length may, in fact, cause a perturbed recognition and transpeptidation by the PBP2a that is responsible for the methicillin resistance of the USA300 LAC strain (Chan et al. 2016). Furthermore, β-lactam resistance can be correlated to the level of Sle1 in MRSA isolates of the USA300 lineage (Thalsø-Madsen et al. 2019). In particular, in oxacillin-treated cells the expression of sle1 is decreased, which leads to a cell separation defect. Conversely, high Sle1 levels accelerate the splitting of daughter cells and decrease the cell size under oxacillin stress conditions. This implies that the Sle1 level may be positively coupled to the transpeptidase activity of PBPs, and that an increased Sle1 level contributes to oxacillin resistance by promoting cell separation. In contrast, sle1 mutant cells are more susceptible to oxacillin, which may be linked to a synthetically lethal effect on septum synthesis (Thalsø-Madsen et al. 2019). Another example where an antibiotic impacts on PGH activity concerns targocil, which targets the TarG subunit of the WTA translocase. Targocil treatment of methicillin-susceptible S. aureus was shown to decrease the cell lysis by sequestering Atl at the membrane instead of the septal surface, which was a consequence of the accumulation of untranslocated WTA molecules (Tiwari et al. 2018).
Role of PGHs in Pathogenesis
S. aureus can thrive and survive inside and outside a wide variety of eukaryotic cells. Accordingly, the peptidoglycan structure and dynamics can be altered in different niches provided by the host, which seems to be very important for bacterial survival and pathogenesis (Chamaillard et al. 2003, Boneca 2005). For instance, during biofilm formation, which frequently occurs on tissues and implanted biomaterials, the cross-linking of the peptidoglycan was found to be reduced (Kim et al. 2018). Also here the non-crosslinked pentaglycine bridges were shown to provide a support for surface proteins that promote the biofilm formation (Kim et al. 2018). During a S. aureus kidney infection, smaller bacterial cells with thicker cell walls and less crosslinking of the peptidoglycan were observed compared to in vitro cultured bacteria. It was suggested that the non-crosslinked pentaglycine bridges could be used as attachment sites for surface proteins that are needed in abscess formation (Sutton et al. 2021). In addition, during infection, the structure of peptidoglycan showed less complexity, which may relate to an adjustment of the peptidoglycan metabolism. This implies that the energy gained by the adjusted peptidoglycan metabolism could be used for other processes in order to allow the bacterial proliferation in a limited environment (Sutton et al. 2021). In extreme cases, S. aureus may even partially or completely lose its cell wall, thereby converting to the L-form as has been reported for recurrent and persistent infections. However, the L-form bacteria are unstable under normal growth conditions, so when the environment changes or the inducer (e.g. penicillin or oxacillin) is removed, these bacteria will convert again to the normal state with a protective cell wall (Xu et al. 2020).
Importantly, peptidoglycan fragments can be perceived by the host as bacterial virulence factors. For instance, peptidoglycan turnover mediated by PGHs was shown to lead to the shedding of peptidoglycan fragments into the host environment, thereby eliciting a proinflammatory response in the host (Girardin et al. 2003). In particular, it is known that the products of peptidoglycan turnover, muropeptides, can target the human intracellular immune receptors Nod1 and Nod2 which, in turn, leads to NF-κB activation and the release of proinflammatory factors, such as cytokines, chemokines and various antimicrobial compounds (Davis and Weiser 2011).
The pathogenicity of S. aureus largely relies on secreted and cell wall-localized proteinaceous virulence factors that promote the interaction between bacteria and the host. Accordingly, various PGHs exposed on the bacterial cell surface also play important roles in bacterial pathogenesis, including host cell adhesion, host cell internalization, inflammatory responses, immune activation and the aforementioned biofilm formation (McCarthy et al. 2016, Schlesier et al. 2020, Sutton et al. 2021). PGH activities may alter the overall charge of the bacterial surface, or they may modify the exposure of adhesins which, in turn, leads to altered bacterial aggregation, adhesive properties or altered biofilm formation. For example, the major autolysin Atl also acts as a host adhesin and interacts with the extracellular matrix of host cells during attachment, colonization and infection. Atl has been shown to bind to fibronectin and vitronectin mainly via its GW-peptidoglycan binding domain (Kohler et al. 2014, Porayath et al. 2018), and it also binds to the human heat shock cognate protein (Hsc70) with high affinity (Schlesier et al. 2020), thereby promoting the efficient internalization into host cells, including non-professional phagocytes (Hirschhausen et al. 2010), such as endothelial cells (Schlesier et al. 2020) and human epithelial cells (Dziewanowska et al. 2000). Atl also binds to the extracellular matrix components heparin and gelatine, thereby promoting bacterial binding to multiple different host cells during an invasive infection (Porayath et al. 2018). An atl mutant was shown to be attenuated in nanopore propagation in implant materials in vitro (which mimics infection of bone canaliculi) and in abscess formation in vivo, which may be attributed to an altered promotion of cell clustering and rough cell surface formation (Masters et al. 2021). Intriguingly, atl mutant bacteria excreted almost no ECPs, such as aldolase (FbaA) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). This could also explain the attenuation of atl mutants at least in part, because FbaA and GAPDH are known to promote the adherence of S. aureus to host cells (Pasztor et al. 2010, Ebner et al. 2016). The PGH Sle1 can act as an adhesin by binding to human fibrinogen, fibronectin and vitronectin (Heilmann et al. 2005). Both the LysM domain and CHAP domain of Sle1 have been shown to play roles in host adhesion (Scheurwater et al. 2008, Kim et al. 2018). Furthermore, both the sle1 and the atl mutant bacteria showed a significant decrease in pathogenesis in acute murine infection models (Kajimura et al. 2005). In addition, loss of the glucosaminidase domain of Atl leads to cell clustering and attenuation in a murine model of osteomyelitis (Varrone et al. 2014). Nonetheless, another study obtained no evidence for a role of Atl in murine infection (McCarthy et al. 2016), and the lack of Atl by itself was not sufficient to cause attenuation in a Zebrafish infection model (Sutton et al. 2021). On the other hand, inactivation of SagA and/or ScaH did lead to attenuation in a Zebrafish infection model (Sutton et al. 2021). A sagB mutant also displayed significant attenuation in a murein sepsis model, which implies that the absence of SagB significantly reduces the fitness of S. aureus (Sutton et al. 2021). Altogether, the reason(s) why PGH mutations cause attenuation of S. aureus appear to be rather complex. For instance, inactivation of SagB results initially in thinner cell walls that take longer transition time to reach maturity which, in turn, may lead to altered, less effective interactions with the host (Balraadjsing et al. 2019, Pasquina-Lemonche et al. 2020, Sutton et al. 2021). However, sagB mutants also display aberrant excretion of cytoplasmic proteins and reduced release of membrane and secreted proteins (Chan et al. 2016), which may also be a cause of attenuation. Bacteria with isaA or sceD mutations did not show significant attenuation in a murine infection model, whereas an isaA sceD double mutant displayed clear attenuation (Stapleton et al. 2007). On the other hand, isaA single mutant bacteria were attenuated in a Galleria mellonella larval infection model (Zhao et al. 2019), whereas SceD was shown to be important for nasal colonization and dissemination of S. aureus in host tissues (Stapleton et al. 2007).
PGHs are frequently involved in the aforementioned formation of biofilms that, in general, consist of proteins, polysaccharides and extracellular DNA (i.e. eDNA) (Schilcher and Horswill 2020) and that also contribute to the bacterial pathogenesis (Archer et al. 2011). Importantly, PGHs can mediate the release of eDNA which is a key factor in biofilm formation. Both through its amidase and glucosaminidase domains, Atl is involved in the primary attachment of the bacteria to surfaces and, consequently, this PGH serves an essential function in biofilm formation (Biswas et al. 2006, Bose et al. 2012). Specifically, unprocessed Atl promotes the primary attachment of S. aureus to surfaces, while the proteolytically activated Atl promotes cell lysis, the release of eDNA and cell accumulation for early biofilm development (Biswas et al. 2006). Atl plays an essential role in the development of so-called ica-independent biofilms. The formation of such biolfilms is brought about by a diverse array of surface proteins, such as Aap and Bap. These proteins serve as adhesins that bind to the host's extracellular matrix, instead of extracellular polysaccharide adhesins, such as the polysaccharide intracellular adhesin or polymeric N-acetyl-glucosamine that are synthesized by enzymes encoded in the ica operon (O'Gara 2007, Houston et al. 2011). Next to Atl, previous research has also shown that Sle1 may have a role in the formation of bacterial biofilms (Liu et al. 2017). Lastly, an elevated expression of isaA as induced by the anti-biofilm reagent tannic acid or by protein overexpression may also lead to biofilm formation (Payne et al. 2013).
PGHs as potential targets for active or passive immunization
PGHs are commonly considered as promising targets for active and passive immunization against S. aureus infections, because most of these proteins are exposed on the bacterial cell surface, where they play important roles in cell wall metabolism. Proteomic analyses of the cell surface of S. aureus have shown that ScaH (Aly), LytM, IsaA, SA0620, SA2097 are nearly always present at this subcellular location of clinical S. aureus isolates (Ziebandt et al. 2010). For active immunization, PGHs could thus potentially be used to elicit protective host immune responses either through the generation of specific antibodies that opsonize S. aureus and lead to its elimination by complement and/or professional phagocytes, or by inducing protective cellular immune responses (Bredius et al. 1993). Importantly, there is an increasing need for vaccines against S. aureus, due to the wide-spread antibiotic resistance displayed by this pathogen. The idea that PGHs are suitable targets for vaccination is corroborated by the observation that the amidase domain of the major PGH Atl can elicit protective immunity against S. aureus in a murine infection model (Nair et al. 2015). It is, however, presently unknown whether this protection takes place through the staphylococcal opsonization by anti-Atl antibodies and subsequent immune clearance, or through the inactivation of Atl by such antibodies. Another study showed that the amidase domain of Atl in combination with an Alum adjuvant efficiently protects mice against an S. aureus infection (Suresh et al. 2021). An octa-valent S. aureus antigen mixture was shown to induce significant IgG responses against LytM, IsaA and the propeptide of Atl in immunized mice, although this did not result in a protection against death upon S. aureus infection (van den Berg et al. 2015b). However, patients with the genetic blistering disease epidermolysis bullosa, whose wounds are highly colonized with multiple S. aureus lineages and who rarely develop serious invasive staphylococcal infections, do show high IgG levels against these antigens (van der Kooi-Pol et al. 2012,van der Kooi-Pol et al. 2013, van der Kooi-pol et al. 2013, van der Kooi-pol et al.2014, van den Berg et al. 2015b, Pastrana et al. 2018). This seems to suggest that IgGs against LytM, IsaA and Atl may also confer some degree of protection against S. aureus infections in humans. Likewise, in a screen for immunogenic non-covalently cell wall-bound proteins, it was observed that Atl, IsaA, EssH, Sle1 and SsaA (SA2093) elicit high IgG levels in sera of patients with EB (Pastrana et al. 2018). These findings imply that surface-exposed PGHs could be potential targets for anti-staphylococcal vaccines. This idea is also supported by passive immunization experiments. For instance, the human monoclonal antibody 1D9, which binds to the N-terminal region of IsaA, can protect mice at least partially against S. aureus bacteraemia and skin infection (van den Berg et al. 2015a). Likewise the humanized monoclonal antibody hUK-66 that specifically binds IsaA was shown to protect against killing by S. aureus in murine infection models, and this monoclonal antibody also effectively enhanced S. aureus killing in whole blood (Lorenz et al. 2011, Oesterreich et al. 2014).
Interestingly, so-called lysibodies that were engineered by fusing the cell wall-binding domain of Atl and the Fc region of human IgG were reported to efficiently bind the S. aureus cell wall. In turn, this was shown to lead to the fixation of complement on the bacterial surface, the promotion of phagocytosis by neutrophils and macrophages, and the protection of mice from an MRSA infection (Raz et al. 2017). Another lysibody composed of the cell wall-binding domain of lysostaphin and the Fc region of human IgG was shown to promote phagocytosis by neutrophils and macrophages even more efficiently, as well as the protection of mice against MRSA infection (Raz et al. 2018). These observations indicate that PGHs can be effectively applied as targets for active or passive immunization and also for the development of completely new generations of engineered antimicrobials.
Conclusion
The Gram-positive bacterial cell wall is essentially one molecule of cross-linked peptidoglycan that shelters a protoplast within its boundaries, thereby providing solid support against high turgor pressures. On the other hand, the cell wall is highly dynamic, allowing the cell to grow and divide and, at the same time, it serves as a semi-porous filter that allows the uptake of essential nutrients, the exclusion of toxic compounds from the bacterial environment, and the excretion of metabolic wastes. Yet another function of the cell wall is to serve as a matrix that binds and retains proteins with key functions in nutrient acquisition, cell adhesion to surfaces and bacterial virulence (van Dijl and du Teil Espina 2021). To make all these vital cell wall functions happen, PGHs play central roles as highlighted in our present review. The PGHs are key in peptidoglycan modulation and, therefore, essential for the bacterial survival in widely differing environments where staphylococci can thrive, ranging from fomites to contaminated food products and different niches in the bodies of livestock and humans. To avoid the destruction of their essential cell wall structures and functions, bacterial cells need to carefully regulate the expression of PGHs both spatially and temporally. This is not a trivial challenge, as underscored by S. aureus, which needs to carefully control the activities of its 18 core PGHs. Our present review summarizes the current understanding of how S. aureus manages to control PGH activity, how the bacterium takes advantage of the different properties and localizations of its PGHs, and how it suppresses their potentially cytotoxic effects. Importantly, PGHs are much more than enzymes that degrade cell walls, since they serve multiple biological functions in bacterial growth and survival in widely varying environments. Moreover, they participate in the release of ECPs needed for the bacterial adhesion to surfaces of medical implants or host tissues, or in the regulated bacterial cell death to generate eDNA needed for biofilm formation. PGHs are also critical for the cell cycle and allow S. aureus to execute various functions in human pathogenesis. Altogether, these vital functions of PGHs imply that they can potentially be used as targets for the development of anti-staphylococcal therapies. This is exemplified by the possible application of PGHs to lyse bacteria, to target PGHs through passive or active anti-staphylococcal immunization approaches, or even to use parts of the staphylococcal PGHs in the engineering of novel antimicrobial drugs. Yet, in spite of the fact that our understanding of PGH functions has substantially increased over recent years, there are still many open questions that need to be answered before we can fully appreciate their possible application for novel antimicrobial therapies. For instance, when and where exactly in the bacterial cell do the different PGHs play their physiological roles, or how exactly do they facilitate the sorting and secretion of virulence factors (van Dijl and du Teil Espina 2021)? Also, we need to know more about the plethora of either cooperative or antagonistic interactions between these enzymes that are responsible for cell wall homeostasis, and about how PGH function is regulated by the levels of peptidoglycan crosslinking and other major cell wall components like WTA and LTA. A better understanding of this complex interplay between cell wall synthesis, the synthesized polymers and PGHs may ultimately allow us to define new targets for new-generation antibiotics, vaccines and therapeutic antibodies that can help us to disrupt the essential processes of cell growth and division in important multi-drug resistant pathogens like S. aureus.
Availability of data and Materials
Not applicable
Authors’ contributions
M.W. drafted the manuscript. MW, GB, and JMvD collected literature and contributed manuscript sections. MW prepared the Figures. GB and JMvD supervised the project. All authors, critically revised the manuscript, gave final approval and agree to be accountable for all aspects of the review.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dennis GAM Koedijk, F Romero Pastrana and Rianne C Prins for collecting the relevant literature and helpful discussions.
Contributor Information
Min Wang, Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Hanzeplein 1, PO Box 30001, 9700 RB Groningen, the Netherlands.
Girbe Buist, Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Hanzeplein 1, PO Box 30001, 9700 RB Groningen, the Netherlands.
Jan Maarten van Dijl, Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Hanzeplein 1, PO Box 30001, 9700 RB Groningen, the Netherlands.
Conflicts of interest
The authors have no conflicts of interest to declare.
Funding
This research was supported by the China Scholarship Council grant 201708110184 and the Graduate School of Medical Sciences of the University of Groningen (to MW).
References
- Abdelnour A, Arvidson S, Bremell Tet al. The accessory gene regulator (agr) controls staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignac A, Sieradzki K, Tomasz A.. Perturbation of cell wall synthesis suppresses autolysis in staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. J Bacteriol. 2007;189:7573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer NK, Mazaitis MJ, Costerton JW. et al. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilano ML, Pereira PM, Yates Jet al. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in staphylococcus aureus. Proc Natl Acad Sci. 2010;107:18991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Schneewind O.. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of staphylococcus aureus. EMBO J. 1998;17:4639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Baba T, Hiramatsu Ket al. Prophages of staphylococcus aureus newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–47. [DOI] [PubMed] [Google Scholar]
- Balraadjsing PP, Lund LD, Souwer Yet al. The nature of antibacterial adaptive immune responses against staphylococcus aureus is dependent on the growth phase and extracellular peptidoglycan. Infect Immun. 2019;88:e00733–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Rawlings ND.. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28:234–7. [DOI] [PubMed] [Google Scholar]
- Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12:63–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito Y, Lina G, Greenland Tet al. Trans-complementation of a staphylococcus aureus agr mutant by staphylococcus lugdunensis agr RNAIII. J Bacteriol. 1998;180:5780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A, Biswas R, Herbert Set al. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006;74:4598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A, Herbert S, Jakob Aet al. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of staphylococcus aureus. Mol Microbiol. 2005;55:778–87. [DOI] [PubMed] [Google Scholar]
- van den Berg S, Bonarius HPJ, van Kessel KPMet al. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against staphylococcus aureus bacteremia. Int J Med Microbiol. 2015a;305:55–64. [DOI] [PubMed] [Google Scholar]
- van den Berg S, Koedijk DGAM, Back JWet al. Active immunization with an octa-valent staphylococcus aureus antigen mixture in models of s . aureus bacteremia and skin infection in mice. PLoS One. 2015b;10:e0116847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R, Martinez RE, Göhring Net al. Proton-binding capacity of staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. PLoS One. 2012;7:e41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R, Voggu L, Simon UKet al. Activity of the major staphylococcal autolysin atl. FEMS Microbiol Lett. 2006;259:260–8. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T, Hamoen LW, Levin PA.. The divisome at 25: the road ahead. Curr Opin Microbiol. 2017;36:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovskyy M, Willing SE, Schneewind Oet al. EssH peptidoglycan hydrolase enables staphylococcus aureus type VII secretion across the bacterial cell wall envelope. J Bacteriol. 2018;200:e00268–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E. et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator rot by an antisense mechanism. Genes Dev. 2007;21:1353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol. 2005;8:46–53. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Huang Z-H, Gage DAet al. Characterization of staphylococcus aureus cell wall glycan strands, evidence for a new β-N-acetylglucosaminidase activity. J Biol Chem. 2000;275:9910–8. [DOI] [PubMed] [Google Scholar]
- Borisova M, Gaupp R, Duckworth Aet al. Peptidoglycan recycling in Gram-positive bacteria is crucial for survival in stationary phase. MBio. 2016;7:e00923–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Lehman MK, Fey PDet al. Contribution of the staphylococcus aureus atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One. 2012;7:e42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredius RG, De Vries CE, Troelstra Aet al. Phagocytosis of staphylococcus aureus and haemophilus influenzae type b opsonized with polyclonal human igg1 and igg2 antibodies. Functional hFc gamma RIIa polymorphism to IgG2. J Immunol. 1993;151:1463–72. [PubMed] [Google Scholar]
- Brown S, Santa Maria JP Jr, Walker S. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW.. Identification and molecular characterization of a putative regulatory locus that affects autolysis in staphylococcus aureus. J Bacteriol. 1996a;178:611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW.. Identification of lytsr-regulated genes from staphylococcus aureus. J Bacteriol. 1996b;178:5810–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchad H, Nair M.. The small RNA SprX regulates the autolysin regulator WalR in staphylococcus aureus. Microbiol Res. 2021;250:126785. [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok Jet al. LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol Microbiol. 2008;68:838–47. [DOI] [PubMed] [Google Scholar]
- Büttner FM, Zoll S, Nega Met al. Structure-function analysis of staphylococcus aureus amidase reveals the determinants of peptidoglycan recognition and cleavage. J Biol Chem. 2014;289:11083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamita HG, Doyle RJ.. Regulation of autolysins in teichuronic acid-containing bacillus subtilis cells. Mol Microbiol. 2002;44:601–6. [DOI] [PubMed] [Google Scholar]
- Calamita HG, Ehringer WD, Koch ALet al. Evidence that the cell wall of bacillus subtilis is protonated during respiration. Proc Natl Acad Sci. 2001;98:15260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari I, Chao Y, Romby Pet al. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med. 2013;3:a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J, Singh AK, Santa Maria JP Jret al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in staphylococcus aureus. ACS Chem Biol. 2011;6:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Yet al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–7. [DOI] [PubMed] [Google Scholar]
- Chan YGY, Frankel MB, Dengler V. et al. Staphylococcus aureus mutants lacking the lytr-cpsa-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol. 2013;195:4650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YGY, Frankel MB, Missiakas Det al. SagB glucosaminidase is a determinant of staphylococcus aureus glycan chain length, antibiotic susceptibility, and protein secretion. J Bacteriol. 2016;198:1123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Allen AG, Owen PJet al. Comprehensive identification of essential staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics. 2009;10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Krishnan V, Macon Ket al. Secreted proteases control autolysin-mediated biofilm growth of staphylococcus aureus. J Biol Chem. 2013;288:29440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Bayer AS, Zhang Get al. Regulation of virulence determinants in vitro and in vivo in staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. [DOI] [PubMed] [Google Scholar]
- Cheung HY, Freese E.. Monovalent cations enable cell wall turnover of the turnover-deficient lyt-15 mutant of bacillus subtilis. J Bacteriol. 1985;161:1222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatwal GS. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 2002;10:205–8. [DOI] [PubMed] [Google Scholar]
- Chu X, Xia R, He Net al. Role of rot in bacterial autolysis regulation of staphylococcus aureus NCTC8325. Res Microbiol. 2013;164:695–700. [DOI] [PubMed] [Google Scholar]
- Chunhua M, Yu L, Yaping Get al. The expression of LytM is down-regulated by RNAIII in staphylococcus aureus. J Basic Microbiol. 2012;52:636–41. [DOI] [PubMed] [Google Scholar]
- Crosby HA, Schlievert PM, Merriman JAet al. The staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 2016;12:e1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby HA, Tiwari N, Kwiecinski JMet al. The staphylococcus aureus ArlRS two-component system regulates virulence factor expression through mgrA. Mol Microbiol. 2020;113:103–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Weiser JN.. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect Immun. 2011;79:562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDent A, Bae T, Missiakas DMet al. Signal peptides direct surface proteins to two distinct envelope locations of staphylococcus aureus. EMBO J. 2008;27:2656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauné A, Dubrac S, Blanchet Cet al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun. 2012;80:3438–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune A, Poupel O, Mallet Aet al. Peptidoglycan crosslinking relaxation plays an important role in staphylococcus aureus walkr-dependent cell viability. PLoS One. 2011;6:e17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijl JM, du Teil Espina M.. Proteins sorted by ‘chaos and disorder.’. Nature Microbiology. 2021;6:973–4. [DOI] [PubMed] [Google Scholar]
- Do T, Page JE, Walker S.. Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. J Biol Chem. 2020;295:3347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T, Schaefer K, Santiago AG. et al. Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nat Microbiol. 2020;5:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisbach A, van Dijl JM, Buist G.. The cell surface proteome of staphylococcus aureus. Proteomics. 2011;11:3154–68. [DOI] [PubMed] [Google Scholar]
- Dreisbach A, Wang M, van der Kooi-Pol MMet al. Tryptic shaving of staphylococcus aureus unveils immunodominant epitopes on the bacterial cell surface. J Proteome Res. 2020;19:2997–3010. [DOI] [PubMed] [Google Scholar]
- Dubin G. Extracellular proteases of staphylococcus spp. Biol Chem. 2002;383:1057–86. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Boneca IG, Poupel Oet al. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in staphylococcus aureus. J Bacteriol. 2007;189:8257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Msadek T.. Identification of genes controlled by the essential YycG/YycF two-component system of staphylococcus aureus. J Bacteriol. 2004;186:1175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunman P áM, Murphy E, Haney Set al. Transcription profiling-based identification of staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:17341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewanowska K, Carson AR, Patti JMet al. Staphylococcal fibronectin binding protein interacts with heat shock protein 60 and integrins: role in internalization by epithelial cells. Infect Immun. 2000;68:6321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner P, Götz F.. Bacterial excretion of cytoplasmic proteins (ECP): occurrence, mechanism, and function. Trends Microbiol. 2019;27:176–87. [DOI] [PubMed] [Google Scholar]
- Ebner P, Luqman A, Reichert Set al. Non-classical protein excretion is boosted by PSMα-induced cell leakage. Cell Rep. 2017;20:1278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner P, Prax M, Nega Met al. Excretion of cytoplasmic proteins (ECP) in staphylococcus aureus. Mol Microbiol. 2015;97:775–89. [DOI] [PubMed] [Google Scholar]
- Ebner P, Rinker J, Götz F.. Excretion of cytoplasmic proteins in staphylococcus is most likely not due to cell lysis. Curr Genet. 2016;62:19–23. [DOI] [PubMed] [Google Scholar]
- Ebner P, Rinker J, Nguyen MTet al. Excreted cytoplasmic proteins contribute to pathogenicity in staphylococcus aureus. Infect Immun. 2016;84:1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Cleverley RM, Peters Ket al. Regulation of bacterial cell wall growth. FEBS J. 2017;284:851–67. [DOI] [PubMed] [Google Scholar]
- El-Shaboury SR, Saleh GA, Mohamed FAet al. Analysis of cephalosporin antibiotics. J Pharm Biomed Anal. 2007;45:1–19. [DOI] [PubMed] [Google Scholar]
- Eyraud A, Tattevin P, Chabelskaya Set al. A small RNA controls a protein regulator involved in antibiotic resistance in staphylococcus aureus. Nucleic Acids Res. 2014;42:4892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabijan AP, Lin RCY, Ho Jet al. Safety of bacteriophage therapy in severe staphylococcus aureus infection. Nat Microbiol. 2020;5:465–72. [DOI] [PubMed] [Google Scholar]
- Falord M, Mäder U, Hiron Aet al. Investigation of the staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One. 2011;6:e21323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedtke I, Mader D, Kohler Tet al. A staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol. 2007;65:1078–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Michalik S, Varming ANet al. Trapping and proteomic identification of cellular substrates of the ClpP protease in staphylococcus aureus. J Proteome Res. 2013;12:547–58. [DOI] [PubMed] [Google Scholar]
- Fenton M, McAuliffe O, O'Mahony Jet al. Recombinant bacteriophage lysins as antibacterials. Bioengineered Bugs. 2010;1:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SJ. Molecular characterization and functional analysis of the major autolysin of staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, Hooper DC.. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of staphylococcus aureus. J Bacteriol. 2000;182:3955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius G, Domenech O, Mingeot-Leclercq MPet al. Direct observation of staphylococcus aureus cell wall digestion by lysostaphin. J Bacteriol. 2008;190:7904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Hendrickx APA, Missiakas DMet al. LytN, a murein hydrolase in the cross-wall compartment of staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem. 2011;286:32593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel MB, Schneewind O.. Determinants of murein hydrolase targeting to cross-wall of staphylococcus aureus peptidoglycan. J Biol Chem. 2012;287:10460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frees D, Sørensen K, Ingmer H.. Global virulence regulation in staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect Immun. 2005;73:8100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frirdich E, Gaynor EC.. Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr Opin Microbiol. 2013;16:767–78. [DOI] [PubMed] [Google Scholar]
- Gajdiss M, Monk IR, Bertsche Uet al. YycH and YycI regulate expression of staphylococcus aureus autolysins by activation of WalRK phosphorylation. Microorganisms. 2020;8:870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J-M, Tipper DJ, Strominger JL.et al. (eds.) Methods in Enzymology. New York & London: Academic Press, 1966,685–99. [Google Scholar]
- Giesbrecht P, Kersten T, Maidhof Het al. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin RW, Chatterjee AN, Young FE.. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of staphylococcus aureus. J Bacteriol. 1972;111:272–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimza BD, Larias MI, Budny BGet al. Mapping the global network of extracellular protease regulation in staphylococcus aureus. Msphere. 2019;4:e00676–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala Jet al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–72. [DOI] [PubMed] [Google Scholar]
- Goldmann O, Medina E.. Staphylococcus aureus strategies to evade the host acquired immune response. Int J Med Microbiol. 2018;308:625–30. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Delgado LS, Walters-Morgan H, Salamaga Bet al. Two-site recognition of staphylococcus aureus peptidoglycan by lysostaphin SH3b. Nat Chem Biol. 2020;16:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell E. Recycling of murein by escherichia coli. J Bacteriol. 1985;163:305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groicher KH, Firek BA, Fujimoto DFet al. The staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Götz Fet al. Key role of teichoic acid net charge in staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Hartleib J, Hussain MSet al. The multifunctional staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect Immun. 2005;73:4793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S, Bera A, Nerz Cet al. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007;3:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhausen N, Schlesier T, Peters Get al. Characterization of the modular design of the autolysin/adhesin aaa from staphylococcus aureus. PLoS One. 2012;7:e40353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhausen N, Schlesier T, Schmidt MAet al. A novel staphylococcal internalization mechanism involves the major autolysin atl and heat shock cognate protein hsc70 as host cell receptor. Cell Microbiol. 2010;12:1746–64. [DOI] [PubMed] [Google Scholar]
- Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J V, Mirelman D, Sharon Net al. Novel type of murein transglycosylase in escherichia coli. J Bacteriol. 1975;124:1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hort M, Bertsche U, Nozinovic Set al. The role of β-glycosylated wall teichoic acids in the reduction of vancomycin susceptibility in vancomycin-intermediate staphylococcus aureus. Microbiol Spect. 2021;9:e0052821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston P, Rowe SE, Pozzi Cet al. Essential role for the major autolysin in the fibronectin-binding protein-mediated staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Boisset S, Saveanu C. et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale S, van Wamel W, Luong TTet al. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in staphylococcus aureus. Infect Immun. 2005;73:1423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale SS, Van Wamel W, Cheung AL.. Characterization of RAT, an autolysis regulator in staphylococcus aureus. Mol Microbiol. 2003;48:1451–66. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Frère J-M, Normark S.. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell. 1997;88:823–32. [DOI] [PubMed] [Google Scholar]
- Jolliffe LK, Doyle RJ, Streips UN.. The energized membrane and cellular autolysis in bacillus subtilis. Cell. 1981;25:753–63. [DOI] [PubMed] [Google Scholar]
- Jorge AM, Hoiczyk E, Gomes JPet al. EzrA contributes to the regulation of cell size in staphylococcus aureus. PLoS One. 2011;6:e27542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse J, Laurent F, Diot A.. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Frontiers in Microbiology. 2017;8:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura J, Fujiwara T, Yamada Set al. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase sle1 involved in cell separation of staphylococcus aureus. Mol Microbiol. 2005;58:1087–101. [DOI] [PubMed] [Google Scholar]
- Kawata S, Takemura T, Yokogawa K.. Characterization of two N-acetylmuramidases from streptomyces globisporus 1829. Agric Biol Chem. 1983;47:1501–8. [Google Scholar]
- Keinhoerster D, George SE, Weidenmaier Cet al. Function and regulation of staphylococcus aureus wall teichoic acids and capsular polysaccharides. Int J Med Microbiol. 2019;309:151333. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Chang J, Rimal Bet al. Surface proteins and the formation of biofilms by staphylococcus aureus. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2018;1860:749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch VC, Fetzer C, Sieber SA.. Global inventory of clpp-and clpx-Regulated proteins in staphylococcus aureus. J Proteome Res. 2020;20:867–79. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Quadri LEN, Kuipers OPet al. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. [DOI] [PubMed] [Google Scholar]
- Kluj RM, Ebner P, Adamek Met al. Recovery of the peptidoglycan turnover product released by the autolysin atl in staphylococcus aureus involves the phosphotransferase system transporter MurP and the novel 6-phospho-N-acetylmuramidase mupG. Frontiers in Microbiology. 2018;9:2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler TP, Gisch N, Binsker Uet al. Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J Biol Chem. 2014;289:4070–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima N, Araki Y, Ito E.. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J Bacteriol. 1985;161:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar SL, Antonio Ibarra J, Rivera FEet al. Extracellular proteases are key mediators of s taphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen. 2013;2:18–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuzawa H, Sugai M, Nakashima Set al. Subcellular localization of the major autolysin, ATL and its processed proteins in staphylococcus aureus. Microbiol Immunol. 1997;41:469–79. [DOI] [PubMed] [Google Scholar]
- van der Kooi-Pol MM, Duipmans JC, Jonkman MFet al. Host–pathogen interactions in epidermolysis bullosa patients colonized with staphylococcus aureus. Int J Med Microbiol. 2014;304:195–203. [DOI] [PubMed] [Google Scholar]
- van der Kooi-Pol MM, Sadaghian Sadabad M, Duipmans JCet al. Topography of distinct staphylococcus aureus types in chronic wounds of patients with epidermolysis bullosa. PLoS One. 2013;8:e67272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooi-Pol MM, De Vogel CP, Westerhout-Pluister GNet al. High anti-staphylococcal antibody titers in patients with epidermolysis bullosa relate to long-term colonization with alternating types of staphylococcus aureus. J Invest Dermatol. 2013;133:847–50. [DOI] [PubMed] [Google Scholar]
- van der Kooi-Pol MM, Veenstra-Kyuchukova YK, Duipmans JCet al. High genetic diversity of staphylococcus aureus strains colonizing patients with epidermolysis bullosa. Exp Dermatol. 2012;21:463–6. [DOI] [PubMed] [Google Scholar]
- Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci (CMLS). 2003;60:2371–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Kuroda H, Oshima Tet al. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in staphylococcus aureus. Mol Microbiol. 2003;49:807–21. [DOI] [PubMed] [Google Scholar]
- Lioliou E, Fechter P, Caldelari Iet al. Various checkpoints prevent the synthesis of staphylococcus aureus peptidoglycan hydrolase LytM in the stationary growth phase. RNA Biology. 2016;13:427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang X, Qin Jet al. The ATP-dependent protease ClpP inhibits biofilm formation by regulating agr and cell wall hydrolase sle1 in staphylococcus aureus. Front Cell Infect Microbiol. 2017;7:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang S, Sun B.. SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant staphylococcus aureus strain N315. Antimicrob Agents Chemother. 2016;60:3455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U, Lorenz B, Schmitter Tet al. Functional antibodies targeting IsaA of staphylococcus aureus augment host immune response and open new perspectives for antibacterial therapy. Antimicrob Agents Chemother. 2011;55:165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loskill P, Pereira PM, Jung Pet al. Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the staphylococcus aureus cell envelope. Biophys J. 2014;107:1082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, De Castro LH, Lim Det al. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–5. [DOI] [PubMed] [Google Scholar]
- Lovering AL, Safadi SS, Strynadka NCJ.. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–78. [DOI] [PubMed] [Google Scholar]
- Lund VA, Wacnik K, Turner RDet al. Molecular coordination of staphylococcus aureus cell division. Elife. 2018;7:e32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong TT, Lee CY.. The arl locus positively regulates staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology. 2006;152:3123–31. [DOI] [PubMed] [Google Scholar]
- Luong TT, Dunman PM, Murphy Eet al. Transcription profiling of the MgrA regulon in staphylococcus aureus. J Bacteriol. 2006;188:1899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lützner N, Pätzold B, Zoll Set al. Development of a novel fluorescent substrate for autolysin e, a bacterial type II amidase. Biochem Biophys Res Commun. 2009;380:554–8. [DOI] [PubMed] [Google Scholar]
- Mainous AG, Hueston WJ, Everett CJet al. Nasal carriage of staphylococcus aureus and methicillin-resistant s aureus in the united states, 2001–2002. Ann Family Med. 2006;4:132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna AC, Ingavale SS, Maloney Met al. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in staphylococcus aureus. J Bacteriol. 2004;186:5267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters EA, Muthukrishnan G, Ho L. et al. Staphylococcus aureus cell wall biosynthesis modulates bone invasion and osteomyelitis pathogenesis. Front Microbiol. 2021;12:723498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VRF, Beveridge TJ.. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in staphylococcus aureus. J Bacteriol. 2006;188:1011–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VRF, Beveridge TJ.. Cryo-electron microscopy of cell division in staphylococcus aureus reveals a mid-zone between nascent cross walls. Mol Microbiol. 2007;64:195–206. [DOI] [PubMed] [Google Scholar]
- Mayer C, Kluj RM, Muehleck Met al. Bacteria's different ways to recycle their own cell wall. Int J Med Microbiol. 2019;309:151326. [DOI] [PubMed] [Google Scholar]
- McCarthy H, Waters EM, Bose JLet al. The major autolysin is redundant for staphylococcus aureus USA300 LAC JE2 virulence in a murine device-related infection model. FEMS Microbiol Lett. 2016;363:fnw087. [DOI] [PubMed] [Google Scholar]
- Memmi G, Nair DR, Cheung A.. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant staphylo coccus aureus strains. J Bacteriol. 2012;194:759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JM, Covas G, Rausch Det al. The pentaglycine bridges of staphylococcus aureus peptidoglycan are essential for cell integrity. Sci Rep. 2019;9:5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro JM, Fernandes PB, Vaz Fet al. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun. 2015;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Sychantha D, Clarke AJ.. Chemical biology of peptidoglycan acetylation and deacetylation. Bioorg Chem. 2014;54:44–50. [DOI] [PubMed] [Google Scholar]
- Nagarajan R. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob Agents Chemother. 1991;35:605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N, Vinod V, Suresh MKet al. Amidase, a cell wall hydrolase, elicits protective immunity against staphylococcus aureus and s . epidermidis. Int J Biol Macromol. 2015;77:314–21. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O.. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nega M, Tribelli PM, Hipp Ket al. New insights in the coordinated amidase and glucosaminidase activity of the major autolysin (Atl) in staphylococcus aureus. Communications Biology. 2020;3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gara JP. ica and beyond: biofilm mechanisms and regulation in staphylococcus epidermidis and staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–88. [DOI] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak Met al. Latent LytM at 1.3 Å resolution. J Mol Biol. 2004;335:775–85. [DOI] [PubMed] [Google Scholar]
- Oesterreich B, Lorenz B, Schmitter Tet al. Characterization of the biological anti-staphylococcal functionality of hUK-66 igg1, a humanized monoclonal antibody as substantial component for an immunotherapeutic approach. Hum Vaccin Immunother. 2014;10:926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku Y, Kurokawa K, Matsuo Met al. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of staphylococcus aureus cells. J Bacteriol. 2009;191:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C, Natale P, Cueto Let al. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev. 2016;40:57–67. [DOI] [PubMed] [Google Scholar]
- Osipovitch DC, Therrien S, Griswold KE.. Discovery of novel s . aureus autolysins and molecular engineering to enhance bacteriolytic activity. Appl Microbiol Biotechnol. 2015;99:6315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquina-Lemonche L, Burns J, Turner RDet al. The architecture of the Gram-positive bacterial cell wall. Nature. 2020;582:294–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana FR, Neef J, van Dijl JMet al. A lactococcus lactis expression vector set with multiple affinity tags to facilitate isolation and direct labeling of heterologous secreted proteins. Appl Microbiol Biotechnol. 2017;101:8139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana FR, Neef J, Koedijk DGAMet al. Human antibody responses against non-covalently cell wall-bound staphylococcus aureus proteins. Sci Rep. 2018;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasztor L, Ziebandt A-K, Nega Met al. Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J Biol Chem. 2010;285:36794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D E, Martin NR, Parzych KRet al. Tannic acid inhibits staphylococcus aureus surface colonization in an isaa-dependent manner. Infect Immun. 2013;81:496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Cava F.. Structural constraints and dynamics of bacterial cell wall architecture. Front Microbiol. 2015;6:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penyige A, Matkó J, Deák Eet al. Depolarization of the membrane potential by β-lactams as a signal to induce autolysis. Biochem Biophys Res Commun. 2002;290:1169–75. [DOI] [PubMed] [Google Scholar]
- Pinho MG, Kjos M, Veening J-W.. How to get (a) round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–14. [DOI] [PubMed] [Google Scholar]
- Porayath C, Suresh MK, Biswas Ret al. Autolysin mediated adherence of staphylococcus aureus with fibronectin, gelatin and heparin. Int J Biol Macromol. 2018;110:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupel O, Proux C, Jagla Bet al. SpdC, a novel virulence factor, controls histidine kinase activity in staphylococcus aureus. PLoS Pathog. 2018;14:e1006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Khairnar K.. Overview of the risks of staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Brazilian J Microbiol. 2021;52:2031–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri EJM, Altulea D, van Dijl JM.. Staphylococcal trafficking and infection—from ‘nose to gut’ and back. FEMS Microbiol Rev. 2022;46:fuab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal M, Walker S.. Envelope structures of Gram-positive bacteria. Curr Top Microbiol Immunol. 2017;404:1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadurai L, Lockwood KJ, Lockwood Jet al. Characterization of a chromosomally encoded glycylglycine endopeptidase of staphylococcus aureus. Microbiology. 1999;145:801–8. [DOI] [PubMed] [Google Scholar]
- Raulinaitis V, Tossavainen H, Aitio Oet al. Identification and structural characterization of LytU, a unique peptidoglycan endopeptidase from the lysostaphin family. Sci Rep. 2017;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Serrano A, Lawson Cet al. Lysibodies are IgG fc fusions with lysin binding domains targeting staphylococcus aureus wall carbohydrates for effective phagocytosis. Proc Natl Acad Sci. 2017;114:4781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Serrano A, Thaker Met al. Lysostaphin lysibody leads to effective opsonization and killing of methicillin-resistant staphylococcus aureus in a murine model. Antimicrob Agents Chemother. 2018;62:e01056–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed P, Veiga H, Jorge AMet al. Monofunctional transglycosylases are not essential for staphylococcus aureus cell wall synthesis. J Bacteriol. 2011;193:2549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann NT, Tavares AC, Saraiva BMet al. SEDS–bPBP pairs direct lateral and septal peptidoglycan synthesis in staphylococcus aureus. Nature Microbiology. 2019;4:1368–77. [DOI] [PubMed] [Google Scholar]
- Rice KC, Bayles KW.. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Firek BA, Nelson JBet al. The staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden DJ, Jedrzejas MJ, Galperin MY.. Amidase domains from bacterial and phage autolysins define a family of γ-d, l-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28:230–4. [DOI] [PubMed] [Google Scholar]
- Rogasch K, Rühmling V, Pané-Farré Jet al. Influence of the two-component system SaeRS on global gene expression in two different staphylococcus aureus strains. J Bacteriol. 2006;188:7742–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in escherichia coli. Proc Natl Acad Sci. 2008;105:15553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahukhal GS, Batte JL, Elasri MO.. msaABCR operon positively regulates biofilm development by repressing proteases and autolysis in staphylococcus aureus. FEMS Microbiol Lett. 2015;362:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata N, Terakubo S, Mukai T.. Subcellular location of the soluble lytic transglycosylase homologue in staphylococcus aureus. Curr Microbiol. 2005;50:47–51. [DOI] [PubMed] [Google Scholar]
- Sarvas M, Harwood CR, Bron Set al. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:311–27. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak Met al. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–58. [DOI] [PubMed] [Google Scholar]
- Schaefer K, Owens TW, Page JEet al. Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization. Nat Microbiol. 2021;6:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater E, Reid CW, Clarke AJ.. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–91. [DOI] [PubMed] [Google Scholar]
- Schilcher K, Horswill AR.. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol Rev. 2020;84:e00026–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler CA, Schuhardt VT.. Lysostaphin: a new bacteriolytic agent for the staphylococcu s. Proc Natl Acad Sci. 1964;51:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag M, Biswas R, Krismer Bet al. Role of staphylococcal wall teichoic acid in targeting the major autolysin atl. Mol Microbiol. 2010;75:864–73. [DOI] [PubMed] [Google Scholar]
- Schlesier T, Siegmund A, Rescher Uet al. Characterization of the Atl-mediated staphylococcal internalization mechanism. Int J Med Microbiol. 2020;310:151463. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Fowler A, Faull KF.. Structure of the cell wall anchor of surface proteins in staphylococcus aureus. Science. 1995;268:103–6. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D.. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochimica Et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843:1687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell EWC, Brown ED.. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot (Tokyo). 2014;67:43–51. [DOI] [PubMed] [Google Scholar]
- Sham L-T, Butler EK, Lebar MDet al. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L, Golonka E, Potempa Jet al. The role and regulation of the extracellular proteases of staphylococcus aureus. Microbiology. 2004;150:217–28. [DOI] [PubMed] [Google Scholar]
- Shockman GD, Daneo-Moore L, Kariyama Ret al. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microb Drug Resist. 1996;2:95–8. [DOI] [PubMed] [Google Scholar]
- Shockman GD, Höltje J-V.. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J.-M., et al. (eds) New Comprehensive Biochemistry. Amsterdam: Elsevier Science B.V, 1994,131–66. [Google Scholar]
- Sibbald M, Ziebandt AK, Engelmann Set al. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol Rev. 2006;70:755–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieradzki K, Tomasz A.. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of staphylococcus aureus. J Bacteriol. 2003;185:7103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S.. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Blackman SA, Foster SJ.. Autolysins of bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146:249–62. [DOI] [PubMed] [Google Scholar]
- Snowden MA, Perkins HR.. Peptidoglycan cross-linking in staphylococcus aureus: an apparent random polymerisation process. Eur J Biochem. 1990;191:373–7. [DOI] [PubMed] [Google Scholar]
- Sobral R, Tomasz A.. The staphylococcal cell wall. Microbiology Spectrum. 2019;7:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton MR, Horsburgh MJ, Hayhurst EJet al. Characterization of IsaA and SceD, two putative lytic transglycosylases of staphylococcus aureus. J Bacteriol. 2007;189:7316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Horsburgh GJet al. AcmA of lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 2005;272:2854–68. [DOI] [PubMed] [Google Scholar]
- Strominger JL, Izaki K, Matsuhashi Met al. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed Proc. 1967;26:9–22. [PubMed] [Google Scholar]
- Sugai M, Komatsuzawa H, Akiyama Tet al. Identification of endo-beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase as cluster-dispersing enzymes in staphylococcus aureus. J Bacteriol. 1995;177:1491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh MK, Kumar VA, Biswas Let al. Protective efficacy of alum adjuvanted amidase protein vaccine against staphylococcus aureus infection in multiple mouse models. J Appl Microbiol. 2021. [DOI] [PubMed] [Google Scholar]
- Sutton JAF, Carnell OT, Lafage L. et al. Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction. PLoS Pathog. 2021;17:e1009468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, White RA, Hoch JA.. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17:706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweda P, Schielmann M, Kotlowski Ret al. Peptidoglycan hydrolases-potential weapons against staphylococcus aureus. Appl Microbiol Biotechnol. 2012;96:1157–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J, Komatsuzawa H, Yamada Set al. Molecular characterization of an atl null mutant of staphylococcus aureus. Microbiol Immunol. 2002;46:601–12. [DOI] [PubMed] [Google Scholar]
- Thalsø-Madsen I, Torrubia FR, Xu Let al. The sle1 cell wall amidase is essential for β-lactam resistance in community-acquired methicillin-resistant staphylococcus aureus USA300. Antimicrob Agents Chemother. 2019;64:e01931–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammavongsa V, Kim HK, Missiakas Det al. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13:529–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari KB, Gatto C, Walker Set al. Exposure of staphylococcus aureus to targocil blocks translocation of the major autolysin atl across the membrane, resulting in a significant decrease in autolysis. Antimicrob Agents Chemother. 2018;62:e00323–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A, Albino A, Zanati EVE.. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–40. [DOI] [PubMed] [Google Scholar]
- Trotonda MP, Xiong YQ, Memmi Get al. Role of mgrA and sarA in methicillin-resistant staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis. 2009;199:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompanidou E, Denham EL, Sibbald MJJBet al. The sortase a substrates FnbpA, FnbpB, ClfA and ClfB antagonize colony spreading of staphylococcus aureus. PLoS One. 2012;7:e44646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NA, Sharma-Kuinkel BK, Maskarinec SAet al. Methicillin-resistant staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17:203–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RD, Ratcliffe EC, Wheeler Ret al. Peptidoglycan architecture can specify division planes in staphylococcus aureus. Nat Commun. 2010;1:1–9. [DOI] [PubMed] [Google Scholar]
- Turner RD, Vollmer W, Foster SJ.. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CAet al. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrone JJ, de Mesy Bentley KL, Bello-Irizarry SNet al. Passive immunization with anti-glucosaminidase monoclonal antibodies protects mice from implant-associated osteomyelitis by mediating opsonophagocytosis of staphylococcus aureus megaclusters. J Orthop Res. 2014;32:1389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga H, Jorge AM, Pinho MG.. Absence of nucleoid occlusion effector noc impairs formation of orthogonal FtsZ rings during staphylococcus aureus cell division. Mol Microbiol. 2011;80:1366–80. [DOI] [PubMed] [Google Scholar]
- Vermassen A, Leroy S, Talon Ret al. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol. 2019;10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GRR, Leenhouts K, van Roosmalen Met al. Exploiting the peptidoglycan-binding motif, LysM, for medical and industrial applications. Appl Microbiol Biotechnol. 2014;98:4331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, De Pedro MA.. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–67. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier Pet al. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–86. [DOI] [PubMed] [Google Scholar]
- Walsh L, Johnson CN, Hill Cet al. Efficacy of phage-and bacteriocin-based therapies in combatting nosocomial MRSA infections. Front Mol Biosci. 2021;8:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, van den Berg S, Hernández YMet al. Differential binding of human and murine iggs to catalytic and cell wall binding domains of staphylococcus aureus peptidoglycan hydrolases. Sci Rep. 2021;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardenburg JB, Patel RJ, Schneewind O.. Surface proteins and exotoxins are required for the pathogenesis of staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier C, Kokai-Kun JF, Kristian SAet al. Role of teichoic acids in staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–5. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A.. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–87. [DOI] [PubMed] [Google Scholar]
- Wheeler R, Turner RD, Bailey RGet al. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. MBio. 2015;6:e00660–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing S, Schneewind O, Missiakas D.. Regulated cleavage of glycan strands by the murein hydrolase SagB in staph ylococcus aureus involves a direct interaction with LyrA (SpdC). J Bacteriol. 2021;203:e00014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Kohler T, Peschel A.. The wall teichoic acid and lipoteichoic acid polymers of staphylococcus aureus. Int J Med Microbiol. 2010;300:148–54. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang B, Wang Let al. Unusual features and molecular pathways of staphylococcus aureus L-form bacteria. Microb Pathog. 2020;140:103970. [DOI] [PubMed] [Google Scholar]
- Yamada S, Sugai M, Komatsuzawa Het al. An autolysin ring associated with cell separation of staphylococcus aureus. J Bacteriol. 1996;178:1565–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, McCormick JK, Schlievert PM.. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of staphylococcus aureus. J Bacteriol. 2001;183:1113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chlebowicz-Flissikowska MA, Wang Met al. Exoproteomic profiling uncovers critical determinants for virulence of livestock-associated and human-originated staphylococcus aureus ST398 strains. Virulence. 2020;11:947–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Palma Medina LM, Stobernack Tet al. Exoproteome heterogeneity among closely related staphylococcus aureus t437 isolates and possible implications for virulence. J Proteome Res. 2019;18:2859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Halladin DK, Rojas ERet al. Mechanical crack propagation drives millisecond daughter cell separation in staphylococcus aureus. Science. 2015;348:574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebandt A, Weber H, Rudolph Jet al. Extracellular proteins of staphylococcus aureus and the role of SarA and σB. Protomecis. 2001;1:480–93. [DOI] [PubMed] [Google Scholar]
- Ziebandt A, Kusch H, Degner Met al. Proteomics uncovers extreme heterogeneity in the staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics. 2010;10:1634–44. [DOI] [PubMed] [Google Scholar]
- Zoll S, Schlag M, Shkumatov AVet al. Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J Bacteriol. 2012;194:3789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable