Abstract
Amphibians and fish show considerable regeneration potential via dedifferentiation of somatic cells into blastemal cells. In terms of dedifferentiation, in vitro cellular reprogramming has been proposed to share common processes with in vivo tissue regeneration, although the details are elusive. Here, we identified the cytoskeletal linker protein desmoplakin (Dsp) as a common factor mediating both reprogramming and regeneration. Our analysis revealed that Dsp expression is elevated in distinct intermediate cells during in vitro reprogramming. Knockdown of Dsp impedes in vitro reprogramming into induced pluripotent stem cells and induced neural stem/progenitor cells as well as in vivo regeneration of zebrafish fins. Notably, reduced Dsp expression impairs formation of the intermediate cells during cellular reprogramming and tissue regeneration. These findings suggest that there is a Dsp-mediated evolutionary link between cellular reprogramming in mammals and tissue regeneration in lower vertebrates and that the intermediate cells may provide alternative approaches for mammalian regenerative therapy.
Desmoplakin may be an evolutionary link between cellular reprogramming in mammals and tissue regeneration in lower vertebrates.
INTRODUCTION
Ectopic expression of the reprogramming factors Oct3/4, Sox2, Klf4, and c-Myc (OSKM) promotes the dedifferentiation of somatic cells into induced pluripotent stem cells (iPSCs) (1). This so-called iPSC reprogramming (iPSCR) has great potential for modeling diseases, regenerative medicine, and studying the mechanisms of cell fate conversion. iPSCR involves the removal of somatic cell signatures and acquisition of pluripotency (2); however, some developmental genes, particularly epidermis-related genes, are temporally up-regulated (3–5), and distinct cell populations, different from iPSCs, occur transiently during reprogramming (6–8), indicating that iPSCR is not a simple reversion of normal developmental differentiation. In contrast to iPSCR, “temporary” up-regulation of OSKM can convert mammalian cells directly into lineage-specified cells (9, 10). This alternative process, known as pluripotency factor–mediated direct reprogramming (PDR), is thought to involve intermediate cells (ICs) that are distinct from iPSCs. However, this latter point is somewhat controversial and has been difficult to reconcile because the ICs are not yet fully characterized (9).
Amphibians and fish can regenerate entire missing body parts via dedifferentiation of wounded cells to form a blastema, which is a mass of de-differentiated multipotent cells that retain a memory of their tissue origin and redifferentiate into the lineage-specific cell types composing the damaged tissues or organs (11–13). Intriguingly, temporary up-regulation of the reprogramming factors or pluripotency factors is observed in the developing blastema, and these factors are required for regeneration (14–16), suggesting that there might be common mechanisms between reprogramming and regeneration. To elucidate the mechanism of OSKM-mediated reprogramming and potentially expand our understanding of in vivo regeneration, we sought to characterize the ICs.
RESULTS
Comparison of OSKM-mediated cellular reprogramming processes
To investigate cell status induced by temporary activation of OSKM in vitro, we conducted a comparative analysis of iPSCR and PDR using secondary mouse embryonic fibroblasts (2° MEFs) that express OSKM in response to doxycycline (dox). We reprogrammed the 2° MEFs to iPSCs, to induce neural stem/progenitor cells (iNPCs) and dopaminergic neuronal progenitors (iDPs) by PDR (Fig. 1A) (17, 18). These four reprogramming regimes generated appropriate target cells that expressed specific markers (Fig. 1B). The medium without Leukemia Inhibitory Factor (LIF) until day 5 as in PDR did not affect iPSCR (fig. S1, A to C). For comparative transcriptomic analysis of these four reprogramming processes, RNA samples were collected every 2 days (fig. S2A). The transcriptomic similarity determined by principal components analysis (PCA) revealed that all samples were similar until day 6 (cluster I) (Fig. 1C). Samples after day 6 fell into two distinct groups, iPSCR (cluster II) and PDR (cluster III) (Fig. 1C). Transcriptomic correlation analysis also identified these three clusters of samples (fig. S2B), suggesting that iPSCR and PDR are distinct but go through a common pathway until day 6.
Fig. 1. Substantial changes in gene expression occur immediately after day 6.
(A) Schematic of the four reprogramming regimes from 2° MEFs to iPSCs using two different protocols (top), or via PDR to iNPCs or iDPs (bottom). D, day; MEF, medium for mouse embryonic fibroblast (MEF); RepM-Ini, reprogramming initiation medium; RepM-PSC, reprogramming medium for pluripotent stem cell (PSC); RepM-NPC, reprogramming medium for neural stem/progenitor cell (NPC); RepM-DP, reprogramming medium for dopaminergic neuronal progenitor (DP). (B) Immunostaining images of cells from the four reprogramming regimes as in (A) on day 12 with markers for iPSC (Nanog and SSEA-1), iNPC (Pax6 and PLZF), or iDP (FoxA2). Scale bars, 200 μm. (C) Principal components analyses (PCA) of the microarray datasets. Each reprogramming sample is colored by the reprogramming regime. The numbers indicate the days after reprogramming when RNA was collected. F, 2° MEFs; C, common intermediates without LIF; P, iPSC reprogramming (LIF D0 to D12); P′, iPSCR (LIF D5 to D12); N, iNPC reprogramming; D, iDP reprogramming. (D) Heatmap showing gene expression of four groups in 2° MEFs (F), at day 6 samples (P6, P′6, N6, and D6), and day 12 samples (P12, P′12, N12, and D12). (E) The individual expression patterns of the four groups are superimposed. The number of genes in each group is presented. All values were normalized to the average expression of each gene and visualized by a line plot. Red line represents mean of gene expression.
Distinct signature of the intermediate phase
As somatic cell signatures were lost and the pluripotency-related genes were not yet activated (fig. S1B), we considered day 6 as the “intermediate phase” in our 2° MEF reprogramming system. To identify genes contributing to the establishment of the intermediate phase, we analyzed differentially expressed genes (DEGs) that changed by more than threefold between days 0 and 6 (fig. S2, C and D). Clustering DEGs into four groups by k-means clustering revealed that the expression of commonly down-regulated genes decreased rapidly until day 6 and remained low until the end of reprogramming. In contrast, the expression of commonly up-regulated genes until day 6 was differentially clustered as reprogramming proceeded (fig. S2, E and F). We classified the commonly up-regulated genes in early and intermediate phases (between days 0 and 6) into four groups based on their expression patterns after day 6 (Fig. 1, D and E, and fig. S2, G and H). The group IV genes that showed intermediate phase–specific expression covered more than 70% of the genes analyzed (Fig. 1, D and E). Similar results were also observed in another reprogrammable cell system (4F2A MEFs) (fig. S3, A to D). These results suggest that the ectopic expression of OSKM in the early phase of reprogramming might contribute mainly to establishing the intermediate phase.
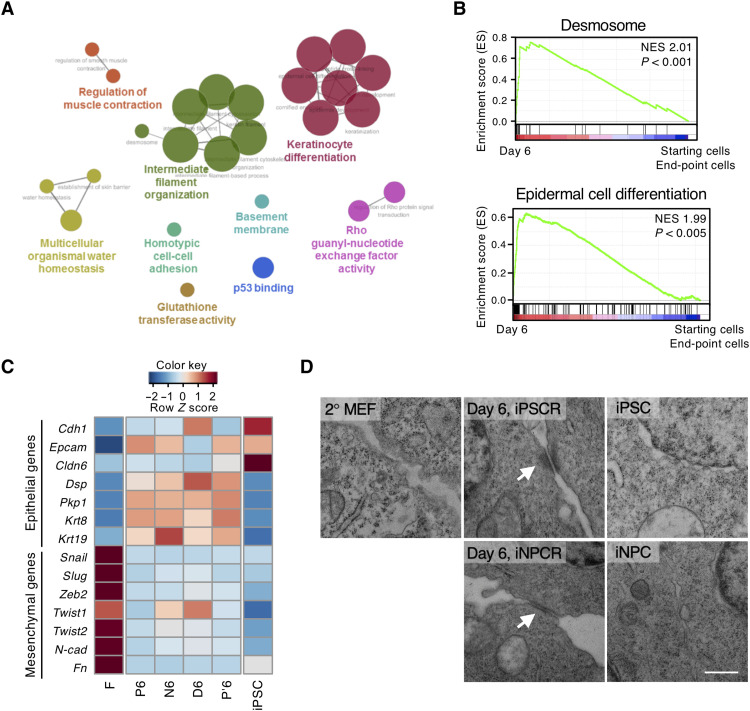
To characterize the group IV genes that showed an intermediate phase–specific expression pattern in both iPSCR and PDR, we performed network-based analyses of gene ontology (GO) terms using ClueGO on the common 244 genes of group IV in the 2° MEF and 4F2A MEF systems (Fig. 2A and fig. S4A). These 244 genes were rarely expressed in the starting fibroblasts and end-point cells, i.e., day 12 cells of iPSCR and PDR in the 2° MEF system and isolated iPSCs and iNPCs in the 4F2A MEF system, but were highly expressed on day 6 (fig. S4, B and C). The highest-ranking group included GO terms such as keratinocyte differentiation and epidermis development, and the next highest-ranking group included terms related to the desmosome, an intercellular junction in the epidermis (Fig. 2A and fig. S4D) (19). We used gene set enrichment analysis (GSEA) to further characterize the global expression patterns of intermediate phase–specific genes, comparing day 6 samples to starting fibroblasts and end-point cells in both the 2° MEF and 4F2A MEF systems. The ranking of the top 15 GO terms based on the normalized enrichment score (NES) revealed that they were related to the epidermis (fig. S4E), consistent with the ClueGO results (fig. S4D), with the highest-ranking gene set being “desmosome” and the next highest-ranking gene set being “epidermal cell differentiation” (Fig. 2B). Thus, epidermis-related signatures, particularly desmosome-related signatures, were common characteristics of intermediate phase–specific genes in both iPSCR and PDR.
Fig. 2. Desmosomes are formed only in the intermediate phase of reprogramming.
(A) Network-based analysis of GO terms under the classifications biological process, cellular component, immune system, and molecular function in the 244 common genes visualized by ClueGO. Major GO terms are represented at the node (P < 0.08). The linked lines between nodes were created on the basis of a predefined κ score level (>0.4). The node size represents the P value (P < 0.001; 0.001 < P < 0.05; 0.05 < P < 0.1). (B) Gene set enrichment analysis (GSEA) showing that “desmosome” and “epidermal cell differentiation” represent the highest normalized enrichment score (NES). GSEA was performed using transcriptomes of ICs (day 6 samples in 2° MEF and 4F2A MEF reprogramming) versus transcriptomes of starting cells (2° MEFs and 4F2A MEFs) and end-point cells (P12, P′12, N12, and D12 of 2° MEFs, iPSCs, and iNPCs of 4F2A MEFs). NESs and P values are shown. (C) Heatmap showing the expression of indicated genes, including epithelial and mesenchymal markers. (D) Representative TEM images of cellular junction regions in 2° MEFs, day 6 samples of iPSCR and iNPCR, iPSCs, and iNPCs. White arrows indicate mature desmosome. Scale bar, 0.5 μm. Data are representative of experimental duplicates.
Transient formation of desmosomes during reprogramming
Desmosomes are an important intercellular junction of the epidermis, which is a type of epithelial tissue. Since mesenchymal-to-epithelial transition (MET) is an essential process in the early phase of reprogramming (20), we compared the expression of desmosome-related genes with that of epithelial and mesenchymal genes to investigate whether the transient up-regulation of desmosome-related genes is part of this MET process (Fig. 2C). Among the epithelial genes, nondesmosomal component genes (Cdh1, Ep-cam, and Cldn6) showed increased expression in all samples on day 6 and in iPSCs, whereas mesenchymal genes (Snail, Slug, Zeb1, Twist1, Twist2, Cdh2, and Fn) showed decreased expression in all samples. However, desmosomal component genes (Dsp and Pkp1) and cytokeratins (Krt8 and Krt19) were increased in all samples on day 6 but reduced in iPSCs. This suggests that desmosome-related epithelial genes are regulated differently from the epithelial genes involved in MET, such as Cdh1.
Desmosomes are composed of desmosomal cadherins, i.e., desmocollins (Dsc) and desmogleins (Dsg), the armadillo proteins, i.e., plakoglobin and plakophilins, and the cytoskeletal linker protein, desmoplakin (Dsp). To resolve the relationship between desmosomes and reprogramming, we examined the expression of desmosomal components during reprogramming. The desmosomal genes were up-regulated mainly in early and intermediate phases of reprogramming, and some desmosomal proteins were specifically expressed on day 6 of iPSCR and iNPC reprogramming (iNPCR) by PDR, except desmoglein2 (Dsg2), which was mainly expressed in iPSCs (21, 22) (fig. S5, A to C). To determine whether this temporary expression of desmosomal components is also observed in other reprogramming systems, we analyzed reprogramming of 4F2A MEFs and 2° B cells into iPSCs (fig. S5, D and E). As expected, since reprogramming kinetics differ depending on the reprogramming system and the starting cell types, we did not observe identical expression patterns of desmosomal components to the 2° MEF system. Nevertheless, the expression of most desmosomal components was temporarily increased during reprogramming but decreased in iPSCs. In particular, the similar expressions in 2° B cells reprogramming obtained by analyzing public data (23) showed that temporary activation of desmosomal components might be a general event in OSKM-mediated reprogramming processes. We assessed the potential intermediate phase–specific formation of desmosomes by transmission electron microscopy (TEM) and found mature desmosomes in cells only on day 6 (Fig. 2D and fig. S5, F and G). We revisited the public data of different processes of reprogramming from human epithelial cells (24) and also found diminished DSP expression in the second half of the reprogramming (fig. S5H). These results suggest that a pivoting event in the desmosome occurs in the intermediate phase of the OSKM-mediated reprogramming process.
The role of Dsp in reprogramming
Dsp is essential for the formation of mature desmosomes (25), linking intermediate filament networks to the desmosomal plaque (26, 27). To investigate whether the formation of mature desmosomes is necessary for the progression of OSKM-mediated reprogramming, we suppressed Dsp expression using an isopropyl β-d-1-thiogalactopyranoside (IPTG)–inducible short hairpin RNA against Dsp (shDsp) during iPSCR and iNPCR (Fig. 3A). Dsp knockdown (KD) reduced the efficiency of iPSC generation, with the number of colonies positive for alkaline phosphatase (AP) and Nanog, markers for pluripotency, being significantly decreased (Fig. 3, B and C). Dsp KD also substantially reduced iNPC generation (Fig. 3D). To further investigate whether Dsp plays an essential role during reprogramming, we overexpressed DSP during iPSCR and iNPCR of human fibroblasts. DSP overexpression significantly increased the efficiency of iPSC and iNPC generation (fig. S6, A to E). Together, these results indicate that Dsp is a key factor affecting the progression of OSKM-mediated reprogramming, especially in the intermediate phase.
Fig. 3. Suppression of Dsp impedes OSKM-mediated reprogramming by controlling IC formation.
(A) Western blot for Dsp and Rpl7 during iPSCR and PDR of shDsp–green fluorescent protein (GFP)–positive sorted 2° MEFs. The bar graph indicates the quantitative expression of Dsp. Samples were harvested at day 6 in both iPSCR and iNPC. Four independent experiments were performed and Rpl7 was used as the loading control (n = 2 wells, P < 0.0001). D.W., distilled water; IPTG, isopropyl β-d-1-thiogalactopyranoside. (B and C) Whole-well imaging of alkaline phosphatase (AP) staining and quantitative analysis of the AP+ colonies (B) and Nanog+ colonies (C) at day 12 of iPSCR with D.W. or IPTG (B, n = 2 wells, P = 0.0112; C, n = 2 wells, P = 0.0056). (D) Quantitative analysis of Pax6+ colonies at day 12 of iNPCR with D.W. or IPTG (n = 2 wells; P = 0.0202). (E and F) Representative phase-contrast images and quantitative analysis of cell aggregates on day 6 of iPSCR (E) and iNPCR (F) with D.W. or IPTG. Scale bars, 200 μm [E, n = 2 wells; shDsp (−), P = 0.80; shDsp (+), P = 0.0047] [F, n = 2 wells; shDsp (−), P = 0.34; shDsp (+), P = 0.0015]. Error bars represent ± SEM (A) or ± SD (B to F). P values <0.05 were considered significant (*P < 0.05 and **P < 0.01), two-tailed, paired Student’s t test was used. Data are representative of two independent experiments.
Two types of IC populations
When we investigated the intermediate phase, we found cell aggregates distinct from iPSC or iNPC colonies around day 6 (fig. S1A), and the number of these cell aggregates decreased significantly upon Dsp KD (Fig. 3, E and F). Thus, we hypothesized that the cells expressing Dsp during the intermediate phase would be the critical ICs. To characterize the cell aggregates, we stained them with antibodies to the desmosomal components, Dsp and Dsc3 (Fig. 4, A and B). We found that there were two different cell populations: those that coexpressed Dsp and Dsc3, named “desmosomal component–expressing ICs (dICs)”, and those that did not express Dsp and Dsc3, which had a granular morphology, named “granular ICs (gICs)”. However, since gICs did not express Dsp but they disappeared after Dsp KD, we assumed that Dsp expression is a prerequisite condition to establish gICs. Furthermore, because these two types of ICs were mostly observed together in one cell aggregate and we found that gICs were not formed after Dsp KD even though they did not express Dsp (Figs. 3, E and F, and 4, A and B), we hypothesized that gICs might originate from dICs during reprogramming. Retrospective tracing from the established colonies by iPSCR and iNPCR supported the possibility of the same hierarchical relationship between dICs and gICs (fig. S7, A and B). These results suggest that there are two types of critical IC populations in reprogramming, and that they are related to Dsp expression.
Fig. 4. There are two types of ICs, depending on Dsp expression.
(A and B) Immunostaining and phase-contrast images with desmosomal components (Dsp and Dsc3) on day 6 of iPSCR (A) and iNPCR (B) under normal conditions. The black asterisk indicates positive cells and the white asterisk indicates negative cells of the indicated desmosomal components in one colony. Scale bars, 50 μm. (C) Schematic of two reprogramming regimes and sample collection for scRNA-seq. (D) The number of single-cell libraries at each reprogramming day. (E) UMAP plots of cells during iPSC or iNPC reprogramming colored by collection time points with bifurcating reprogramming trajectories inferred by Slingshot. The dotted line reveals the failed path of iNPC reprogramming.
To investigate the ultimate identity of the IC populations, we performed single-cell RNA sequencing (scRNA-seq) on 33,966 cells from iPSCR- and iNPCR-derived cells, as well as the starting fibroblasts, iPSCs, and iNPCs (Fig. 4, C and D, and fig. S8, A to G). As seen in the microarray analysis (Fig. 1C), reprogramming trajectories began to bifurcate from day 6 onward (Fig. 4E and fig. S9A). Since the two types of IC populations, dICs and gICs, were observed around day 6, we focused on cells that existed between days 5 and 7 (Fig. 5A and fig. S9B). We observed similar cellular distributions until day 6 in both iPSCR and iNPCR (Fig. 5A) and found that Shisa8, the early marker of the successful MET during iPSCR (28), was expressed in a subpopulation of cells between days 5 and 7 (Fig. 5B). These Shisa8-positive cells were divided into two subpopulations, distinguished by Dsp expression. Therefore, it is possible that the Shisa8+/Dsp+ cells on the uniform manifold approximation and projection (UMAP) are the dICs. In addition, since gICs were highly proliferative (movie S1), the elevated proliferation signature of Shisa8+/Dsp− cells (Fig. 5, B and C) suggests that the Shisa8+/Dsp− cells on the UMAP could be the gICs. In the isolated gICs, as expected, Dsp expression remained low, but Shisa8 and Cdk1 were highly expressed (Fig. 5D), confirming that the Shisa8+/Dsp− population on the UMAP were the gICs (Fig. 5B).
Fig. 5. gICs are derived from dICs.
(A) UMAP plots of scRNA-seq data showing cells between days 5 and 7 of iPSCR and iNPCR. (B and C) Signature score distribution for indicated genes (B) and identity gene sets (C) defining dICs and gICs on the UMAP. Scale bars represent the minimum (Min) and maximum (Max) signature scores during the whole reprogramming process. (D) Reverse-transcription quantitative PCR for indicated genes. 2° MEFs were used as a negative control (gray) and iPSCs were used as a positive control for the pluripotency marker (red). Day 6 samples included whole cells of day 6 and gICs were isolated on day 6 of iPSCR (black) and iNPCR (white). (E) Phase-contrast and immunostaining images of the same region of interest after re-attachment of gICs from day 6 of iPSCR or iNPCR (D0). Immunostaining was performed for Nanog on day 6 after reattachment (D+6). Scale bars, 100 μm. (F) Predicted reprogramming trajectories inferred by Slingshot showing the dICs branching to Shisa8-positive and Shisa8-negative paths. Cells are colored by clusters. Error bars represent ± SE. Data are representative of experimental triplicates. (G and H) UMAP plots of cells between days 5 and 7, highlighting the descendants of clusters 1 and 8 during iPSCR (E) and iNPCR (F). Color and intensity represent collection time point and descendant probability estimated by WOT.
gICs were easily detached from the culture dish and formed iPSC-like colonies after reattachment during iPSCR (movie S2). This feature is reminiscent of the “satellite iPSCs” that appear during reprogramming with the 2° MEF reprogramming system (29). When we isolated the floating gICs from the culture medium on day 6, reattached them, and continued reprogramming, the gICs were converted to iPSCs (Fig. 5E). Despite their potential to be iPSCs, the gICs rarely expressed pluripotency markers, i.e., Nanog, Rex1, and Esrrb (Fig. 5D and fig. S9C), suggesting that the gICs are a unique cell type distinct from iPSCs.
To investigate the lineage relationship between the dICs and gICs, we inferred their pseudo-temporal ordering along reprogramming trajectories using Slingshot (30). The analysis revealed that the dIC populations (clusters 1, 4, and 8; Shisa8+/Dsp+) were bifurcated into two trajectories by Shisa8 expression (Fig. 5F and fig. S9D). Cells along the Shisa8-negative path (clusters 2, 6, 7, and 11) expressed stromal signatures (e.g., MEF identity and senescence) (Figs. 5C and 4E and fig. S9A), implying that these cells are refractory to reprogramming. In the Shisa8-positive path, i.e., the successful reprogramming path (28), the dICs converted into the gIC populations (clusters 0, 3, and 12; Shisa8+/Dsp−). Waddington optimal transport (WOT) (28) analysis also showed similar results (Fig. 5, G and H), suggesting that gICs are derived from dICs. Thus, in the Shisa8-positive successful reprogramming path, Dsp expression is required to establish the first ICs, the dICs, and then Dsp expression is reduced to establish the second ICs, the gICs. Conclusively, we identified two types of sequentially linked ICs that are essential for the progress of cellular reprogramming.
The dsp-Akt signaling axis in reprogramming
Because Dsp is a junctional molecule, we wondered how Dsp could modulate reprogramming processes. Dsp expression is known to affect Akt (25, 31). We found that Dsp KD significantly reduced phosphorylated Akt in both iPSCR and iNPCR (fig. S10A). In addition, given that Dsp KD significantly reduced the number of IC aggregates (Fig. 3, E and F), we predicted that Dsp facilitates the formation of IC aggregates via Akt signaling. Consistent with this, inhibition of Akt signaling with the small-molecule MK2206 (32) during iPSCR and iNPCR substantially diminished the number of IC aggregates (fig. S10, B to D) and the abrogated subsequent reprogramming processes (fig. S10, E and F). However, MK2206 did not affect the self-renewal of iPSCs and iNPCs (fig. S10, G to J). Thus, our results suggest that Dsp-Akt signaling is specific and essential for the generation of the IC aggregates.
Absence of iPSC-like cells in the intermediate phase
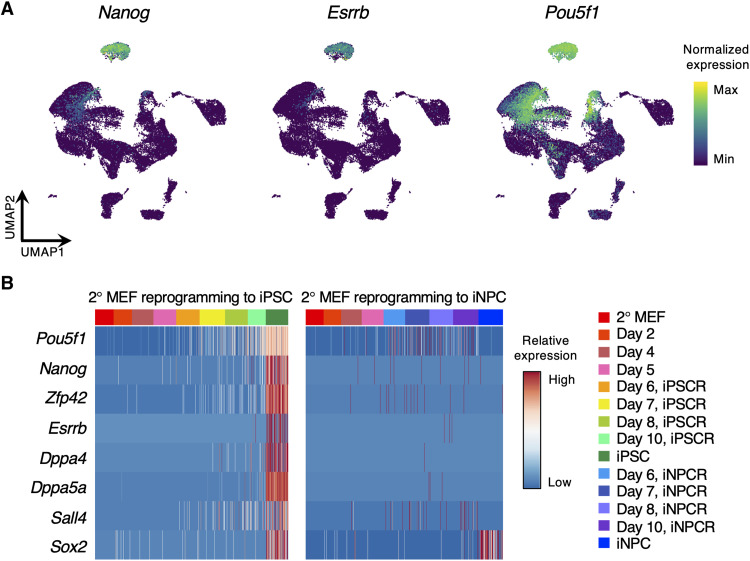
There have been concerns about the existence of iPSC-like intermediates in the PDR approach (33, 34). However, MK2206 abrogated the reprogramming processes (fig. S10, C to F), suggesting that there were no iPSC-like intermediates: If there were iPSCs in the intermediate phase, since MK2206 could not affect the iPSCs self-renewal (fig. S10, G and H), iPSC colonies would emerge eventually. In addition, we carefully examined our scRNA-seq data for the existence of iPSC-like cells during the intermediate phase of iPSCR and PDR. However, we could not detect any single cells showing iPSC-like expression of pluripotency genes (Fig. 6, A and B). Although we observed high expression of Pou5f1/Oct4 in some subset of cells, only a few cells expressed pluripotency genes including Oct4 from day 8, which is not considered the intermediate phase (Fig. 6, A and B, and table S1). Thus, we conclude that the IC aggregates that appear transiently in OSKM-mediated reprogramming are distinct cell populations that do not contain iPSCs.
Fig. 6. The expression of pluripotency markers during OSKM-mediated reprogramming.
(A) UMAP plots of cells during iPSC or iNPC reprogramming showing the expression of pluripotency marker genes. (B) Heatmaps showing the expression of the pluripotency marker gene signatures in cells from iPSCR and iNPCR. Reprogramming day is indicated by colors in the upper bar.
The role of Dsp in regeneration in vivo
Recently, short-term expression of OSKM has been reported to induce “rejuvenation” of tissues (35–37) or expression of youthful genes (38). In addition, it is already known that the OSKM reprogramming factors are transiently up-regulated during in vivo regeneration in amphibians and fish (14–16). Thereby, a link between cellular reprogramming and tissue regeneration has been proposed (15). Therefore, we sought to investigate whether Dsp is also required for in vivo regeneration, as in in vitro reprogramming. To assess whether dsp expression affects zebrafish larvae fin fold regeneration, we injected the dspa and dspb morpholino (dsp mix MO) into one-cell eggs. Dsp mix MO knocked down dspa and dspb expression at the transcriptional and protein levels (fig. S11, A and B) without inducing obvious nonspecific apoptosis at 1 day postfertilization (dpf) embryos (fig. S11C). dsp morphant embryos showed significant reduction of the fin fold regrowth (~26%) within 24 hours (Fig. 7A), while cell proliferation in the blastema region was also severely affected, as shown by 5-ethynyl-2′-deoxyuri-dine (EdU) labeling (fig. S11D) and phospho-histone H3 immunostaining (fig. S11E). The reduced fin fold regrowth after amputation and decreased cell proliferation in the blastema region were also consistently observed in mosaic knockout of dsp genes using dsp-specific single guide RNAs (sgRNAs; fig. S11, F to H). Whole-mount in situ hybridization (WISH) analysis revealed a substantial decrease in the blastema marker gene (junbb) in amputated dsp morphant embryos (Fig. 7B and fig. S12A). Moreover, other blastema marker genes (msx1b, fgf20a, her6, and hspd1) were also reduced in amputated dsp morphant embryos (fig. S12B). Consistent with larval fin fold regeneration phenotypes, an adult fin regeneration model also revealed that dsp KD in the amputated tips of dorsal fins impeded dorsal regrowth within 24 hours, to 68% of the regeneration seen in untreated ventral fin tips (Fig. 7C). This reduction was similar to that seen with injection of the pou5f3/oct4 morpholino (Fig. 7C), as previously reported (15). These results reveal that blastema-mediated regeneration also requires dsp expression.
Fig. 7. Loss of dsp impedes in vivo regeneration by controlling blastemal cell induction.
(A) Phase-contrast images of larvae fin folds after amputation. Fin fold amputation was done at 2 dpf and photographed 24 hours later. The graph shows the percentage of 1-dpa versus 0-dpa fin fold outgrowth (in gray) and average inhibition of 1-dpa versus 0-dpa fin fold (in red). Scale bar, 100 μm (n = 60, 56 embryos, P < 0.0001). dpa, days postamputation; ctrl, control; MO, morpholino; dsp mix MO, dspa MO + dspb MO. (B) Whole-mount in situ hybridization (WISH) analysis of larvae fin folds with control MO or dsp mix MO. Representative phase-contrast images and quantitative analysis show the expression of blastema marker gene (junbb) in regenerating fin folds. Uncut controls were equivalently stained. Dotted lines indicate the boundary of the fin folds. Scale bar, 100 μm (n = 40, 48 embryos, P < 0.0001). hdf, hours postfertilization; hpa, hours postamputation. (C) Phase-contrast and fluorescence merged images of adult fins after injection and electroporation with ctrl MO, pou5f3 MO (positive control), and dsp mix MO. Injection was done into the dorsal half of a 2-dpa blastema and photographed 24 hours later. The graph shows the percentage of dorsal versus ventral fin outgrowth (in gray) and average inhibition of dorsal versus ventral fin (in red). The line on each image denotes the fins at 2 dpa. Scale bar, 500 μm (n = 7 fishes, P < 0.0001). Error bars represent ± SD. P values <0.05 were considered significant (***P < 0.001), and two-tailed, paired Student’s t test was used. Data are representative of two independent experiments. See Materials and Methods for exact calculation.
Blastemal cells are mesenchymal cells producing bone, connective tissue, and dermis (12, 13). Dsp KD reduced both the number of IC aggregates (Fig. 3, E and F) and blastema marker expression (Fig. 7B and fig. S12B), suggesting that the IC populations and blastemal cells are similar in that they are affected by the expression of Dsp. Therefore, to evaluate the similarity between IC populations and blastemal cells, we further compared the cells in the intermediate phase of reprogramming with those in blastema-mediated regeneration (fig. S13, A to C) (39). We found that the expression of the blastema-specific markers was elevated in gICs (fig. S13A), and cluster 12 belonging to gICs showed the highest similarity to the blastemal cells (fig. S13C). To further investigate whether temporary activation of desmosome-related genes was also observed during in vivo tissue regeneration, we compared the publicly available zebrafish fin regeneration and axolotl limb regeneration data (12, 13) to our scRNA-seq data in terms of desmosome and pluripotency signatures in blastemal cells (fig. S13, D to M). To describe the emergence and disappearance of ICs, we analyzed our data from days 2 to 8 (fig. S13, D to F). During the reprogramming process, dICs emerged with the up-regulation of the desmosome signature and converted to gICs with the down-regulation of the desmosome signature (lineages 1 and 2 in fig. S13F). gICs arose with an increased pluripotency signature, but the expression level was far lower than iPSCs (fig. S13G). By comparing the dynamic expression patterns of our chosen gene signatures along the trajectories of cellular reprogramming with that along the trajectories of zebrafish and axolotl tissue regeneration, we found that the sequential expression patterns of desmosome and pluripotency signatures during cellular reprogramming are also observed in the regeneration process of zebrafish (fig. S13, H to J) and axolotl (fig. S13, K to M). These results imply that temporary up-regulation of desmosome-related genes may be a prerequisite for inducing de-differentiation in damaged tissue. Furthermore, our results suggest that the OSKM-mediated reprogramming process shares a Dsp-mediated de-differentiation mechanism with in vivo tissue regeneration.
DISCUSSION
It has long been questioned why mammals lost the regeneration potential observed in amphibians and fish. Since in vivo regeneration requires temporary induction of reprogramming factors, Christen et al. (15) suggested that in vivo regeneration may have a common mechanism with OSKM-mediate reprogramming in vitro. In this study, we found that the modulation of Dsp expression affected both in vitro cellular reprogramming and in vivo tissue regeneration. In particular, when dsp expression was suppressed by morpholino injection in zebrafish, blastema marker expression was substantially reduced (Fig. 7B and fig. S12B), and gICs were mapped to blastemal cells at the transcriptomic level (fig. S13, A to C). Thus, we propose that gICs may be the mammalian version of blastemal cells. Although we have not fully characterized the cellular identity of gICs, our findings on Dsp could provide a clue for solving the “key question” of the constraint of mammalian regeneration.
In addition, we confirmed that PDR does not pass through an iPSC state by analyzing the reprogramming process at the single-cell level. In PDR, we propose that short-term induction of the Yamanaka factors, i.e., OSKM, results in partial reprogramming and may establish a developmentally plastic state that then results in various lineage-specific cells (17). All three germ layer cells are generated by transient expression of the Yamanaka factors (10, 17, 40). However, there were reports that PDR passes through the iPSC state because the cells induced by PDR expressed pluripotency markers, i.e., Oct4 or Nanog (33, 34). In our study, we also observed some cells showing a high level of Oct4 expression (Fig. 6A), but only a few cells coexpressed other pluripotency marker genes with Oct4 (Fig. 6B and table S1). Thus, our findings clearly demonstrate that transient induction of OSKM establishes a unique state of cells that is distinct from iPSCs, which is a prerequisite for successfully progressing through both iPSCR and PDR.
Technically, PDR is short-term OSKM induction– and partial reprogramming–mediated cell fate conversion. Recently, the effect of partial reprogramming has been actively investigated. In vivo reprogramming often produces teratoma (41). However, short-term cyclic induction of OSKM enables partial reprogramming in vivo without teratoma formation (35). Recent studies have shown that transient induction of OSKM also ameliorates aging features in multiple tissues, improves tissue function, and promotes tissue regeneration (35, 37, 38, 42, 43), although the underlying mechanism is still elusive. Here, we suggest that there is a shared mechanism between in vitro cellular reprogramming and in vivo tissue regeneration. Therefore, our findings will be invaluable for understanding the mechanism of mammalian in vivo reprogramming, which might be evolutionary conserved.
In conclusion, our results suggest that Dsp is a common essential factor in IC-mediated in vitro reprogramming of mammalian cells and blastema-mediated in vivo regeneration of zebrafish. We believe that specifying the ICs of reprogramming could produce an unprecedented type of cells similar to blastemal cells, suggesting a novel paradigm for regenerative medicine.
MATERIALS AND METHODS
Reprogramming of MEF to iPSCs, iNPCs, and iDPs
Secondary MEFs (2° MEFs) and 4F2A MEFs were prepared as previously described (17, 44, 45). For iPSCR, 2° MEFs were thawed (p2) and plated (p3) on Geltrex (Thermo Fisher Scientific, Waltham, MA, USA)–coated culture dishes at 2 × 104 cells/cm2 in MEF derivation culture medium [MEF medium: Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), 1× GlutaMAX (Thermo Fisher Scientific), 1× MEM–nonessential amino acid (NEAA; Thermo Fisher Scientific), and 1% penicillin/streptomycin (Thermo Fisher Scientific)]. Dox (Sigma-Aldrich, St. Louis, MO, USA) induction was initiated the next day and continued the designated day. The cells were cultured in MEF medium for an additional day and the medium was changed to reprogramming initiation medium [RepM-Ini: KnockOut DMEM (Thermo Fisher Scientific) supplemented with 10% KnockOut serum replacement (Thermo Fisher Scientific), 5% fetal bovine serum, 1× GlutaMAX, 1× MEM-NEAA, 1% 1× penicillin/streptomycin, and 0.055 mM β-mercaptoethanol (Thermo Fisher Scientific)] or a pluripotent stem cell reprogramming medium [RepM-PSC: same as RepM-Ini containing mouse LIF (mLIF) (1000 U/ml); Millipore, Billerica, MA, USA]. For iNPC and iDP reprogramming, the medium was changed to neural stem cell reprogramming medium [RepM-NPC: advanced DMEM/F12 (Thermo Fisher Scientific) and neurobasal (Thermo Fisher Scientific) were mixed at 1:1 and supplemented with 0.05% bovine serum albumin (BSA), 1× N2 (Thermo Fisher Scientific), 1× B27 (Thermo Fisher Scientific), 1× GlutaMAX, 0.11 mM β-mercaptoethanol with fibroblast growth factor 2 (FGF2; 20 ng/ml; Peprotech, Rocky Hill, NJ, USA), FGF4 (2 ng/ml; Peprotech), and epidermal growth factor (EGF; 20 ng/ml; Peprotech)] and dopaminergic progenitor reprogramming medium [RepM-DP: advanced DMEM/F12 and neurobasal were mixed by 1:1 and supplemented with 0.05% BSA, 1× N2, 1× B27, 1× GlutaMAX, 0.11 mM β-mercaptoethanol with Sonic Hedgehog (SHH) (200 ng/ml; Peprotech), and FGF8b (100 ng/ml; Peprotech)] after day 5 (17, 18).
To assess the trilineage differentiation potential of iPSCs, we performed embryoid body (EB) formation assays. iPSCs were seeded at 1 × 104 cells on wells of ultralow attachment 96-well plates (Corning Incorporated, Kennebunk, ME, USA) and cultured in RepM-Ini medium with dox for 48 hours. After that, EBs were transferred to 35-mm petri dishes and cultured for 1 week in RepM-Ini medium without dox. The EBs were allowed to adhere to Geltrex-coated culture plates and cultured for an additional week.
For neuronal differentiation, iNPCs were seeded at 2 × 104 cells/cm2 onto poly-l-ornithine (Sigma-Aldrich) and Laminin (Sigma-Aldrich)–coated plastic coverslips (Thermo Fisher Scientific) with an appropriate maintenance medium. The next day, the culture medium was replaced with a neuronal differentiation medium comprising DMEM/F-12 (Thermo Fisher Scientific) containing 1× B27 without vitamin A (Thermo Fisher Scientific), brain-derived neurotrophic factor (20 ng/ml; Peprotech), glial cell-derived neurotrophic factor (20 ng/ml; Peprotech), 0.5 mM dibutyryl cyclic adenosine monophosphate (Enzo-Life Sciences, Basel, Switzerland), and 0.2 mM sodium butyrate (Sigma-Aldrich). Half of the culture medium was replaced every 3 days until analysis. For KD of Dsp, IPTG (1 mM; Sigma-Aldrich) was treated for 6 days from the day after plating.
gICs were isolated from the supernatant at day 6 of iPSCR and iNPCR. The harvested supernatant was centrifuged at 300g for 3 min, washed with Dulbecco’s phosphate-buffered saline (DPBS; WELGENE, Daegu, Korea), and reattached on Geltrex-coated culture plates at 2 × 104 cells/cm2 in RepM-PSC with dox. For MEF derivation, husbandry, animal care, and blastocyst injection were performed in accordance with guidelines from the Korea Research Institute of Bioscience and Biotechnology (KRIBB) and approved by KRIBB–Institutional Animal Care and Use Committee (IACUC; approval number: KRIBB-AEC-20044).
RNA preparation, cDNA synthesis, and quantitative PCR
Total RNA was extracted from the samples using the RNeasy Plus mini kit with QIAshredder (Qiagen, Hilden, Germany) and complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Quantitative polymerase chain reaction (PCR) was performed using a 1/50 concentration of the obtained cDNA with iQ SYBR Green Supermix (Bio-Rad) on an Applied Biosystems 7500 Fast Real-Time PCR instrument system (Thermo Fisher Scientific). The cycle threshold (Ct) value for each target gene was determined using software provided by the manufacturer. The expression data were normalized to the Ct value of Rpl7 (40). The primer sequences used in this study are listed in table S2.
AP staining
Samples were washed once with DPBS and fixed with 10% formalin solution (Sigma-Aldrich) for 30 s. AP staining was performed with the leukocyte alkaline phosphatase kit (Sigma-Aldrich) according to the manufacturer’s instruction. Briefly, fixed samples were washed once and then incubated with an AP substrate solution for 20 min in the dark.
Immunofluorescence staining
Cells were grown in 24- or 4-well tissue culture plates and washed once with DPBS. Samples were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) and 0.15% picric acid (Sigma-Aldrich) in DPBS for 15 min. Then, they were blocked and permeabilized with 3% BSA (Thermo Fisher Scientific) and 0.3% Triton X-100 (Sigma-Aldrich) in DPBS for 1 hour at room temperature. All primary antibodies were diluted in 1% BSA and incubated overnight at 4°C. After iterative washing with 0.1% BSA in DPBS, the samples were incubated with Alexa 594– or Alexa 488–conjugated secondary antibodies (Thermo Fisher Scientific) for 1 hour at room temperature. All fluorescent images were acquired using an Axio Vert.A1 microscope (Carl Zeiss, Oberkochen, Germany) and an Evos FL auto 2 imaging system (Thermo Fisher Scientific). The antibodies used in this study are listed in table S3.
Microarray analysis
Total RNA was extracted using the RNeasy Plus mini kit with QIAshredder (Qiagen), and global gene expression profiles were analyzed by an Agilent mouse whole-genome 4 × 44 K arrays (V2) chip (one-color platform; Agilent Technologies, Santa Clara, CA, USA). Briefly, the RNA quality of all samples was checked by the Agilent 2100 Bioanalyzer System, followed by amplification, labeling, and hybridization steps. All experiments were performed according to the manufacturer’s protocols.
The microarray data were processed using GeneSpring software (Agilent Technologies) and normalized using global scale normalization. To analyze functionally grouped GO terms, we used the Cytoscape software platform (v3.5.1; https://cytoscape.org/what_is_cytoscape.html). A functionally grouped GO term network was visualized by ClueGO plug-in (v2.3.4; http://apps.cytoscape.org/apps/cluego) and CluePedia (v1.3). The ClueGO analysis ranked the 244 genes into five annotation groups (group P value <0.002). The enrichment of genes in biological process, cellular component, and molecular function was analyzed by GSEA software (v3.0) (46). Parameters for GSEA were set as 1000 permutations of gene sets, a classic enrichment statistic and signal-to-noise separation metric. Starting cells contained 2° MEFs and 4F2A MEFs; day 6 samples contained P6, P′6, N6, and D6 in the 2° MEF system and P6 and N6 in 4F2A MEF system; end-point cells contained P12, P′12, N12, and D12 in the 2° MEF system and iPSC and iNPC in the 4F2A MEF system.
To analyze the iPSCR process from human bronchial epithelial cells and prostate epithelial cells (24), the normalized microarray matrices were obtained from National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE50206 using the GEO function of GEOquery (v.2.58.0) R package. Then, the matrices were scaled.
Western blot
Whole-cell extracts or deyolked zebrafish larvae at 3 dpf were prepared using RIPA buffer (Sigma-Aldrich) containing 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich) and a cocktail of protease inhibitors (Roche, Basel, Switzerland) and then centrifuged at 7000g for 5 min or 12,000g for 10 min at 4°C. The protein concentrations were determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific). An equal amount of total protein was separated on MP TGX precast gels (Bio-Rad) and transferred to polyvinylidene fluoride membranes (Bio-Rad). The membranes were blocked in tris-buffered saline (LPS Solution, Daejeon, Korea) containing 0.05% Tween 20 (TBST; Sigma-Aldrich) with 5% nonfat milk (BD Biosciences, Franklin Lakes, NJ, USA) or 1.5% BSA for 1 hour at room temperature and then incubated with specific primary antibodies overnight at 4°C. After washing with TBST six times for 30 min, the samples were incubated for 1 hour at room temperature with anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA; 1:5000). The blots were developed using ECL Select Western Blotting Detection Reagent (GE Healthcare, Little Chalfont, UK). The antibodies used in this study are listed in table S3.
Transmission electron microscopy imaging
To analyze desmosome formation, reprogramming was performed on Geltrex-coated coverslips (Electron Microscopy Sciences, Hatfield, PA, USA). Cells on the coverslip were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate solution (pH 7.0) for 1 hour, followed by 2% osmium tetroxide for 2 hours at 4°C. The cells were dehydrated with a graded acetone series and embedded into Spurr medium (Electron Microscopy Services). The samples were sectioned (60 nm) with an ultramicrotome (RMC MTXL; Boeckeler Instruments, Tuscon, AZ, USA), and double-stained with 2% uranyl acetate for 20 min and lead citrate for 10 min. The sections were then viewed under an H-7600 (HITACHI, Tokyo, Japan) transmission electron microscope at 80 kV (47).
Lentivirus production and transduction
The pLKO-GFP-IPTG-3xLacO construct containing shRNA against mouse Dsp (GCCTACAAGAAAGGTCTCATT) was designed by Sigma-Aldrich. The viruses were produced in human embryonic kidney 293T cells. Briefly, the cells at 60 to 80% confluence (16 hours after plating) were cotransfected with lentiviral plasmid along with vesicular stomatitis virus coat protein plasmid (pMD2.G; Addgene, Watertown, MA, USA) and packaging plasmid (psPAX2; Addgene) using TransIT-2020 transfection reagent (Mirus Bio, Madison, WI, USA). Transfected cells were washed with DPBS (16 hours after transfection) and grown for an additional 48 hours with MEF medium. Viral supernatants were collected and used to infect 2° MEFs with polybrene (6 μg/ml; Sigma-Aldrich).
Fluorescence-activated cell sorting
The infected 2° MEFs were isolated using fluorescence-activated cell sorting (FACS). Cells were dissociated in 0.05% trypsin-EDTA for 5 min, washed with DPBS, and resuspended with FACS buffer (0.5% BSA in DPBS) for isolation using a FACSaria cell sorter (BD Biosciences).
Human iPSC and iNPC reprogramming
Human iPSC and iNPC reprogramming were performed as previously described (48), with slight modifications. Briefly, human fibroblasts, CRL-2097 (American Type Culture Collection, Manassas, VA, USA), were plated into 24-well plates at a density of 30,000 cells/cm2 and then transduced with Sendai virus (SeV) mixtures (CytoTune-iPS 2.0 Sendai reprogramming kit; Thermo Fisher Scientific), according to the manufacturer’s instructions. The next day, the SeV mixtures were washed with DPBS. For human iPSCR, the cells were incubated in a fibroblast medium consisting of MEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, 1× sodium pyruvate (Thermo Fisher Scientific), and 1× MEM-NEAA for an additional 2 days. At 3 days posttransduction (dpt), the medium was replaced with iPSCR medium consisting of mTeSR-1 (STEMCELL Technologies, Vancouver, Canada) supplemented with 3.0 μM CHIR99021 (Tocris Bioscience, Bristol, UK), 0.5 μM A83-01 (Tocris), and 0.2 mM NaB (Sigma-Aldrich).
For human iNPC reprogramming, the SeV mixtures were washed with DPBS and replaced with a human neural reprogramming medium supplemented with 3.0 μM CHIR99021, 0.5 μM A83-01, and hLIF (10 ng/ml; Peprotech). The reprogramming medium was replaced every other day.
For DSP overexpression, human fibroblasts were transfected with 1 μg of DSP plasmid (Addgene, catalog no. 32227) and empty plasmid (pEGFP-N1; Takara Bio, Shiga, Japan) at 4 dpt. For reprogramming purposes, the use of human fibroblasts was exempted from institutional review board review by the Public Institutional Review Board Designated by Ministry of Health and Welfare (P01-201802-31-001).
Single-cell RNA sequencing
scRNA-seq libraries were generated using the Chromium Single Cell 3′ Reagent kit v2 (PN-120267, 10X Genomics, Seattle, WA, USA), Chromium Single Cell A Chip kit (PN-120236, 10X Genomics), and Chromium Single Cell i7 Multiplex kit (PN-120262, 10X Genomics). Cell viability was estimated to be more than 90% by trypan blue staining under the microscope. Cells were diluted to 2 × 105 to 2 × 106 cells/ml with 0.04% BSA in DPBS and loaded on the Chromium microfluidic platform, aiming to capture 3000 cells per channel. Subsequent library preparation was performed according to the manufacturer’s instructions. Libraries were sequenced on an Illumina HiSeq 4000 platform (2 × 100 bp).
scRNA-seq data analysis
Initial data processing and quality control
Raw FASTQ files were processed with the Cell Ranger software (v2.1.0) using default arguments. Reads were aligned to the mouse reference genome (GRCm38) with the Ensembl GRCm38.92 annotation. A gene-by-cell unique molecular identifier (UMI) count matrix for each condition was generated with “expect cells = 3000” and aggregated into a single count matrix. Empty droplets were identified and filtered out using the emptyDrops function of the DropletUtils (v0.99) R package (49) with false discovery rate (FDR) ≤ 0.05. By visually inspecting outliers in the PCA plot on the quality control metrics using the scater (v1.7.18) R package (50) as described previously (51), low-quality cells with less than 1000 UMIs, less than 102.5 detected genes (103 detected genes for iNPCs), and greater than 10% of UMIs mapped to mitochondrial genes were removed. The raw count matrix was normalized by cell-specific size factors estimated by the scran (v1.14.6) R package (52), and then log2-transformed with a pseudo-count of 1. Highly variable genes (HVGs) across all cells were identified using the same package with FDR ≤ 0.05.
Visualization and clustering
All cells were visualized in the UMAP plot using the RunUMAP function of the Seurat (v3.2.2) R package (53) with the first 50 PCs and n.neighbors = 10. Cells between days 5 and 7 were visualized in the UMAP plot from the first 25 PCs. Cells were clustered using the FindClusters function of the same package with the first 30 PCs and resolution = 0.6.
Signature scores
Gene set signature scores were computed using the AddModuleScore function of the Seurat with predefined lists of marker genes used in Schiebinger et al. (28). Blastema-specific markers (39) were used to calculate the blastema signature score. Desmosome (GO:CC) and EMT (Hallmark) gene lists were obtained from MSigDB.
scRNA-seq data analysis of killifish data
We obtained scRNA-seq data of killifish regeneration (39) from Sequence Read Archive (SRA) under BioProject PRJNA559885. Raw FASTQ files were processed using the Cell Ranger software (v.2.1.1) with the killifish reference genomes (Nfu_20140520) and Ensembl Nfu_20140520.102 annotation. We filtered out empty droplets using the emptyDrops function of the DropletUtils (v1.61) R package with FDR ≤ 0.01. We retained cells with at least 1000 UMIs and 400 detected genes. The raw count matrix normalization and HVGs selection (FDR ≤ 0.05) were done by the same method as above. For visualization and clustering, we used the same methods as above with 20 PCs and resolution = 0.3 but excluded the first PC showing the high correlation with the total UMI count. Cell types were manually annotated on the basis of the literature-based marker genes (39).
scRNA-seq data analysis of zebrafish data
Raw UMI count matrices were obtained from NCBI GEO under accession number GSE137971 (13). Cells with low total UMI counts (<102.5) and high UMIs mapped to mitochondrial genes (>10%) were removed. The normalization and HVG selection was done by the same method above. The clusters were identified and presented by UMAP using Seurat with batch-corrected 20 PCs by the harmony (v0.1.0) R package (54). Each cluster is annotated on the basis of the literature-based markers (13). Mesenchymal clusters were visualized by UMAP with the first 30 PCs and n.neighbors = 50 and clustered with 30PCs and resolution = 1.2 using Seurat R package.
scRNA-seq data analysis of axolotl data
Transcripts per million (TPM)–normalized matrix [early: 0 to 18 days postamputation (dpa)] and raw UMI count matrix (late: 18 to 38 dpa) were obtained from the supplementary material of Gerber et al. (12). TPM-normalized matrix (early) was transformed to UMI distribution by the quminorm function of quminorm (v.0.1.0) R package (55) with shape = 1. To match the cell number of the dataset (early, 802 cells; late, 21,819 cells), 800 cells from late time point data were used. The normalization and HVG selection were done by the same method above. The SelectIntegrationFeatures function of Seurat was used to select the genes to integrate datasets with nfeatures = 500. The FindIntegrationAnchors of Seurat was used to find a set of anchors between two datasets with the first 10 CCAs. Last, the dataset was integrated by IntegrateData with dim = 1:30. Then, an integrated dataset was visualized by UMAP and clustered with default arguments. The signature score was calculated by the batch-corrected gene expression matrix.
Trajectory analysis
The trajectories of each in the UMAP space were inferred using the slingshot function of the slingshot (v1.4.0) R package (30) with a predefined starting cluster (days 5 to 7, cluster 1; days 2 to 8, cluster 3; zebrafish, cluster 6; axolotl, cluster 7). The signature score along the trajectory was smoothed by locally estimated scatterplot smoothing regression. In WOT (28) analysis, transport maps were constructed on the HVG subset expression data using the optimal transport command-line interface with default arguments in the WOT (v1.0.8) Python package.
Cell type cluster matching analysis
Homologous genes between mouse and killifish were mapped using the biomaRt (v2.46.3) R package. We normalized homologous genes’ matched-expression matrix of two datasets using the SCTransform function from Seurat, setting the vars.to.regress parameter to the S phase and G2-M phase score due to the high cell cycle dependency. S phase and G2-M phase scores were calculated by the CellCycleScoring function of Seurat with literature-based cell cycle markers (56). We matched killifish data to mouse data using the SingleR function of the SingleR (v1.4.1) R package (57) with corrected count matrix by SCTransform and default arguments.
Maintaining zebrafish
Zebrafish (Danio rerio) AB (wild type) strain was maintained at 28.5°C under a standard condition. Fish were fed daily with a combination of dry food and brine shrimp and maintained under a 14-hour light/10-hour dark schedule. Zebrafish husbandry and animal care were performed in accordance with guidelines from the KRIBB and approved by KRIBB-IACUC (approval numbers: KRIBB-AEC-17073 and KRIBB-AEC-20056).
Adult caudal fin regeneration
Adult caudal fin regeneration experiments were performed with 6- to 9-month-old adults as previously described (58). Adult zebrafish that were adapted at 33°C for a day before experiments were anesthetized in tricaine before their caudal fin amputation experiments, and adult fin was amputated at the seven bony segments distal to the fin girdle. Following the surgery, the fish were returned to a 33°C tank. The following morpholinos (Gene Tools Inc., Philomath, OR, USA) were used in this study: pou5f3 MO, 5′-CGCTCTCTCCGTCATCTTTCCGCTA; dspa MO, 5′-AAACTAAAACCGAGGCTGACCTTCT; and dspb MO, 5′-CTGACTGTGTTTCAGACTGACCTGT.
Each morpholino contained a 3-fluorescein tag and was resuspended in water. Dspa and dspb morpholino were mixed by 1:1 ratio to knock down both desmoplakin genes simultaneously. At 48 hours postamputation, 1 mM morpholinos were injected into the regenerating tissue on the dorsal side of each zebrafish tail fin using a PV380 Pneumatic picopump (World Precision Instruments, Sarasota, FL, USA). Each morpholino injection was targeted to the regenerative tissue just distal to each bony ray and approximately 70 nl of morpholino solution was injected per bony ray.
Electroporation of both the dorsal and ventral (to control for a nonspecific electroporation effects) sides of the fin using NEPA21 Electroporator (Nepa Gene Co. Ltd., Chiba, Japan) was performed immediately following the injections. The electroporation parameter used three consecutive 50-ms pulses at 15 V with a 50-ms pause between pulses using a CUY647 15-mm-diameter platinum pate electrode (Protech International Inc., Cornelius, NC, USA). Electroporation was performed two times using the same parameters to increase electroporation efficiency. Fish were then returned to the same 33°C tank. At 72 hours postamputation, each fin was photographed using an Olympus SZX16 microscope equipped with a TUCSEN Dhyana 400 DC digital camera (Olympus, Tokyo, Japan). The area of both the dorsal (D) and ventral (V) fin regrowth was calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The percentage of regeneration was calculated by ((D3dpa − D2dpa) / (V3dpa − V2dpa)) × 100 and the statistical significance of the morpholino on regeneration was analyzed using Student’s t test.
Larvae fin fold regeneration
Zebrafish embryos of AB strain were maintained in E3 egg water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4) in a petri dish at 28.5°C. Control (0.4 mM) or dspa and dspb mix morpholinos at approximately 1 nl were injected into one-cell eggs. Zebrafish larvae at 2 dpf were anesthetized with tricaine in egg water and their fin fold was amputated using a surgical razor blade. The fin fold was carefully amputated at the same sites just posterior to notochords. For quantification of fin fold regeneration, the lengths from the notochord end to the posterior tip of fin fold at 0 hour postamputation and 24 hours postamputation were measured using ImageJ software and was analyzed using Student’s t test.
Zebrafish embryos at each stage were harvested with TRI Reagent Solution (Thermo Fisher Scientific), followed by purifying total RNA with the Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA, USA) and synthesizing cDNA with the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). The synthesized cDNA was amplified by semiquantitative PCR using the specific primers that are listed in table S2.
Whole-mount in situ hybridization
To make in situ probes, the DNA templates for zebrafish blastema marker gene, junbb, was amplified from cDNA of WT zebrafish using PCR (forward probe, 5′-TGGGTTACGGTCACAACGAC; reverse probe, 5′-CAGTGTCCGTTCTCTTCCGT). PCR products were agarose gel separated and purified and then cloned into pCRBlunt II-TOPO vector. Dig-labeled antisense probes were in vitro transcribed by SP6 or T7 RNA polymerase kit (Roche) and purified with NucAway spin columns (Thermo Fisher Scientific). Caudal fin of adult and larvae zebrafish for WISH were prepared by fixing with 4% paraformaldehyde in 1× PBS solution, dehydrating using methanol, stored at −20°C over 30 min, and serially rehydrated with 1× PBS containing 0.1% Tween 20 (PBST) solution. The rehydrated embryos were treated with proteinase K in 1× PBS and postfixed with 4% paraformaldehyde. The antisense probes were hybridized with the fixed embryos at each developmental stage in hybridizing solution [torula yeast RNA type VI (5 mg/ml), heparin (50 μg/ml), 50% formamide, 5× SSC (0.75 M NaCl, 0.075 M sodium citrate, 0.1% Tween 20, and adjusted to pH 6.0 with 1 M citric acid)] at 70°C overnight. The probes were washed serially using 2× SSCT-F (2× SSC, 0.1% Tween 20, and 50% formamide), 2× SSCT (2× SSC and 0.1% Tween 20), 0.2× SSCT (0.2× SSC and 0.1% Tween 20) at 70°C, and 1× PBST at room temperature. The embryos were blocked with the blocking solution (5% horse serum and 1× PBST) at room temperature, and the AP-conjugated anti-digoxigenin antibody (Roche) was added into the blocking solution at 4°C overnight. To detect the expression signal of the transcript, NBT/BCIP (nitro blue tetrazolium/5-Bromo-4-chloro-3-indolyl phosphate) solution (Roche) was used as an AP substrate. The expression pattern of transcripts was observed by using an Olympus SZX16 microscope and imaged with TUCSEN Dhyana 400 DC. The quantification of WISH was performed according to AP activity in the fin fold. We pixelated raw data using Photoshop (Adobe Systems Inc., San Jose, CA, USA) for the calculation of gene expression, and we counted the number of AP-positive (blue) pixels through Image J software. See table S7 for detailed calculation.
Whole-mount immunohistochemistry
To label phospho-histone H3–positive cells during regeneration, staining whole-mount immunohistochemistry was performed as follows. Zebrafish larvae at 3 dpf (1 dpa) were fixed with 4% paraformaldehyde overnight at 4°C. Larvae were washed thrice with PBST, dehydrated with methanol, and stored at −20°C. Larvae were rehydrated with PBST, treated with prechilled acetone at −20°C for 7 min. The larvae were rinsed with PBST, incubated with a blocking buffer (PBST with 10% bovine serum albumin) for 1 hour. Larvae were incubated overnight at 4°C with the primary antibodies diluted in blocking buffer. Larvae were washed thrice with PBST and then incubated for 1 hour at room temperature with the secondary antibody. The next day, they were washed with PBST, mounted, and imaged using an FV1000 confocal microscope (Olympus) with the identical fluorescence laser condition. Phospho-histone H3–positive cells were quantified from the acquired confocal images by counting the number of cells in the area of 200 μm from the amputation. Counting areas were indicated by the yellow dotted box in the representative imaging. The antibodies used in this study are listed in table S3.
EdU staining
To label proliferating cells during regeneration, EdU staining was performed by using the Click-iT Plus EdU Cell Proliferation Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Briefly, zebrafish larvae at 1 dpa were incubated with 500 μM EdU containing 10% dimethyl sulfoxide for 1 hour. The labeled larvae were fixed with 4% paraformaldehyde overnight at 4°C. Larvae were washed thrice with dH2O, treated with acetone at −20°C for 7 min, and permeabilized with PBS/1% dimethyl sulfoxide/1% Triton X-100 for 1 hour. For the fluorescence detection, the labeled larvae were incubated with Click-iT reaction cocktail containing Alexa Fluor 647 picolyl azide for 1 hour in the dark room temperature. They were washed with PBST, mounted, and imaged using an FV1000 confocal microscope (Olympus) with the identical fluorescence laser condition. EdU-positive cells were quantified from the acquired confocal images by counting the number of cells in the area of regeneration. Amputation parts were indicated by the yellow dotted line in the representative imaging.
sgRNA design and generation of dspa/dspb crispant
For the generation of dspa and dspb crispants, sgRNAs were designed by CRISPRscan (https://www.crisprscan.org/) with zebrafish genome version GRCz11/danRer11 and synthesized following the described protocol (59). dspa and dspb sgRNAs with an efficiency score >58 and no predicted off-targets were selected to target the dspa exon 1 and dspb exon 3, respectively. Three different sgRNAs were designed and used together to effectively knock out the target genes. Briefly, the sgRNA DNA templates were generated by fill-in PCR. A 52–nucleotide (nt) oligo (sgRNA primer) containing the T7 promoter, the 20-nt of the specific sgRNA DNA binding sequence, and a constant 15-nt tail for annealing was used in combination with an 80-nt reverse oligo to add the sgRNA-invariable 3′ end (tail primer). sgRNA DNA templates were generated by PCR according to the following protocol: two cycles of 2 min at 95°C, 10 min at 50°C, and 10 min at 72°C. PCR products were purified and used as a template for a T7 in vitro transcription reaction. In vitro transcribed sgRNAs were synthesized by using the MAXIscript T7 Transcription Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The sequences for sgRNAs used in this study are listed in table S2.
To generate dspa/dspb crispant, zebrafish embryos at the one-cell stage were injected with 1 to 2 nl of the total six different in vitro transcribed sgRNAs and EnGen Spy Cas9 NLS protein (New England Biolabs, EnGen Spy Cas9 NLS, M0646M) mixture containing sgRNAs (25 ng/μl) and Cas9 protein (300 ng/μl), respectively. To compare with dspa/dspb crispant larvae, a nonspecific scrambled sgRNA as previously described (60) was injected into zebrafish embryos following the same experimental procedure. Zebrafish injected with scrambled gRNA are referred to as sham-injected controls.
Statistical analysis
Statistical significance was calculated with the Student’s t test using Microsft Excel (Microsoft Office, WA, USA). Comparisons of two groups with equal variance were determined by the F test, and all statistical analyses were two-tailed. Statistically significant differences are indicated as *P < 0.05, **P < 0.01, or ***P < 0.001. No statistical methods were used to predetermine the sample size. Experimental triplicates were performed for all reverse transcription quantitative PCR or semi-PCR analyses.
Acknowledgments
Funding: This work was supported by grants from the National Research Foundation of Korea (2019R1A2C2087606 and 2018R1A5A1025511) to J.K. and J.K.K.; Samsung Research Funding Center of Samsung Electronics (SRFC-MA1601-06) to J.K.; National Research Council of Science and Technology (NST) of Ministry of Science and ICT of Korea (CRC-15-04-KIST) to J.-S.L.; KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (22-BR-01-03) to J.Y.M.; and the KRIBB research initiative program (KGM4722223 to J.K. and M.Y.S., KGM5362212 to J.K., KGM5192113 to M.K., and KGM9992211, KGM5352113, and KGM2112133 to J.-S.L.).
Author contributions: J.H. designed and performed the experiments, analyzed the data, and wrote the manuscript; B.S.K., B.M., and Y.H.C. performed bioinformatics analyses with scRNA-seq data; B.-H.Y. performed preliminary bioinformatics analyses with microarray data; I.I. prepared samples for 4F2A MEF microarray and scRNA-seq; J.K.K., S.-Y.K., and M.K. supervised the bioinformatics analyses; J.N. and J.-G.L. performed in vivo regeneration in zebrafish, with advice from J.-S.L.; M.L. performed human iPSC and iNPC reprogramming; H.C. performed TEM analysis, with advice from J.Y.M; J.S.P. and S.M.C. performed blastocyst injection for generation of 2° MEF with advice from Y.-K.K. and K.-H.N.; A.B. helped with genomic analyses; M.-O.L. and M.Y.S. provided helpful comments; J.K. supervised the overall project, analyzed the data, and wrote the manuscript.
Competing interests: A patent was filed by KRIBB and DGIST on 2 June 2021 (10-2021-0071679). The inventors are J.K., J.H., J.K.K., J.-S.L., M.Y.S., J.N., and A.B. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw and processed datasets generated in this study have been deposited in the NCBI GEO and SRA database under accession numbers GSE156888 (microarray) and PRJNA555736 (scRNA-seq), respectively.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
Data S1 to S8
REFERENCES AND NOTES
- 1.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Buganim Y., Faddah D. A., Jaenisch R., Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 14, 427–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polo J. M., Anderssen E., Walsh R. M., Schwarz B. A., Nefzger C. M., Lim S. M., Borkent M., Apostolou E., Alaei S., Cloutier J., Bar-Nur O., Cheloufi S., Stadtfeld M., Figueroa M. E., Robinton D., Natesan S., Melnick A., Zhu J., Ramaswamy S., Hochedlinger K., A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Malley J., Skylaki S., Iwabuchi K. A., Chantzoura E., Ruetz T., Johnsson A., Tomlinson S. R., Linnarsson S., Kaji K., High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 499, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuno A., Nishimura K., Takahashi S., Time-course transcriptome analysis of human cellular reprogramming from multiple cell types reveals the drastic change occurs between the mid phase and the late phase. BMC Genomics 19, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lujan E., Zunder E. R., Ng Y. H., Goronzy I. N., Nolan G. P., Wernig M., Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature 521, 352–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz B. A., Cetinbas M., Clement K., Walsh R. M., Cheloufi S., Gu H., Langkabel J., Kamiya A., Schorle H., Meissner A., Sadreyev R. I., Hochedlinger K., Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell 23, 289–305.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagawa H., Shimamoto R., Kim S. I., Oceguera-Yanez F., Yamamoto T., Schroeder T., Woltjen K., OVOL1 influences the determination and expansion of iPSC reprogramming intermediates. Stem Cell Rep. 12, 319–332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J., Ambasudhan R., Ding S., Direct lineage reprogramming to neural cells. Curr. Opin. Neurobiol. 22, 778–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu S., Wang H., Ding S., Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat. Protoc. 10, 959–973 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Jopling C., Boue S., Izpisua Belmonte J. C., Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 12, 79–89 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., Currie J. D., Camp J. G., Tanaka E. M., Treutlein B., Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou Y., Lee H. J., Chen Y., Ge J., Osman F. O. I., McAdow A. R., Mokalled M. H., Johnson S. L., Zhao G., Wan T., Cellular diversity of the regenerating caudal fin. Sci. Adv. 6, eaba2084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maki N., Suetsugu-Maki R., Tarui H., Agata K., del Rio-Tsonis K., Tsonis P. A., Expression of stem cell pluripotency factors during regeneration in newts. Dev. Dyn. 238, 1613–1616 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christen B., Robles V., Raya M., Paramonov I., Izpisua Belmonte J. C., Regeneration and reprogramming compared. BMC Biol. 8, 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M. T., Tsanov K. M., Nguyen L., Asara J. M., Cantley L. C., Daley G. Q., Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S. A., Zhang K., Ding S., Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H. S., Kim J., Jo Y., Jeon D., Cho Y. S., Direct lineage reprogramming of mouse fibroblasts to functional midbrain dopaminergic neuronal progenitors. Stem Cell Res. 12, 60–68 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Johnson J. L., Najor N. A., Green K. J., Desmosomes: Regulators of cellular signaling and adhesion in epidermal health and disease. Cold Spring Harb. Perspect. Med. 4, a015297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D., A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Eshkind L., Tian Q., Schmidt A., Franke W. W., Windoffer R., Leube R. E., Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur. J. Cell Biol. 81, 592–598 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Park J., Son Y., Lee N. G., Lee K., Lee D. G., Song J., Lee J., Kim S., Cho M. J., Jang J. H., Lee J., Park J. G., Kim Y. G., Kim J. S., Lee J., Cho Y. S., Park Y. J., Han B. S., Bae K. H., Han S., Kang B., Haam S., Lee S. H., Lee S. C., Min J. K., DSG2 is a functional cell surface marker for identification and isolation of human pluripotent stem cells. Stem Cell Rep. 11, 115–127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Stefano B., Sardina J. L., van Oevelen C., Collombet S., Kallin E. M., Vicent G. P., Lu J., Thieffry D., Beato M., Graf T., C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506, 235–239 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K., Yamanaka S., Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5, 3678 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Gallicano G. I., Kouklis P., Bauer C., Yin M., Vasioukhin V., Degenstein L., Fuchs E., Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143, 2009–2022 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Getsios S., Huen A. C., Green K. J., Working out the strength and flexibility of desmosomes. Nat. Rev. Mol. Cell Biol. 5, 271–281 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Vasioukhin V., Bowers E., Bauer C., Degenstein L., Fuchs E., Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3, 1076–1085 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Schiebinger G., Shu J., Tabaka M., Cleary B., Subramanian V., Solomon A., Gould J., Liu S., Lin S., Berube P., Lee L., Chen J., Brumbaugh J., Rigollet P., Hochedlinger K., Jaenisch R., Regev A., Lander E. S., Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith Z. D., Nachman I., Regev A., Meissner A., Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 28, 521–526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Street K., Risso D., Fletcher R. B., das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan H., South A. P., Hart I. R., Increased keratinocyte proliferation initiated through downregulation of desmoplakin by RNA interference. Exp. Cell Res. 313, 2336–2344 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Nishimura K., Aizawa S., Nugroho F. L., Shiomitsu E., Tran Y. T. H., Bui P. L., Borisova E., Sakuragi Y., Takada H., Kurisaki A., Hayashi Y., Fukuda A., Nakanishi M., Hisatake K., A role for KLF4 in promoting the metabolic shift via TCL1 during induced pluripotent stem cell generation. Stem Cell Rep. 8, 787–801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Nur O., Verheul C., Sommer A. G., Brumbaugh J., Schwarz B. A., Lipchina I., Huebner A. J., Mostoslavsky G., Hochedlinger K., Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 33, 761–768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maza I., Caspi I., Zviran A., Chomsky E., Rais Y., Viukov S., Geula S., Buenrostro J. D., Weinberger L., Krupalnik V., Hanna S., Zerbib M., Dutton J. R., Greenleaf W. J., Massarwa R., Novershtern N., Hanna J. H., Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat. Biotechnol. 33, 769–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ocampo A., Reddy P., Martinez-Redondo P., Platero-Luengo A., Hatanaka F., Hishida T., Li M., Lam D., Kurita M., Beyret E., Araoka T., Vazquez-Ferrer E., Donoso D., Roman J. L., Xu J., Esteban C. R., Nuñez G., Delicado E. N., Campistol J. M., Guillen I., Guillen P., Belmonte J. C. I., In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y. X., Blau H. M., Reversing aging for heart repair. Science 373, 1439–1440 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Lu Y., Brommer B., Tian X., Krishnan A., Meer M., Wang C., Vera D. L., Zeng Q., Yu D., Bonkowski M. S., Yang J. H., Zhou S., Hoffmann E. M., Karg M. M., Schultz M. B., Kane A. E., Davidsohn N., Korobkina E., Chwalek K., Rajman L. A., Church G. M., Hochedlinger K., Gladyshev V. N., Horvath S., Levine M. E., Gregory-Ksander M. S., Ksander B. R., He Z., Sinclair D. A., Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124–129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roux A., Zhang C., Paw J., Zavala-Solorio J., Vijay T., Kolumam G., Kenyon C., Kimmel J. C., Partial reprogramming restores youthful gene expression through transient suppression of cell identity. bioRxiv 2021.2005.2021.444556 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Wang W., Hu C. K., Zeng A., Alegre D., Hu D., Gotting K., Ortega Granillo A., Wang Y., Robb S., Schnittker R., Zhang S., Alegre D., Li H., Ross E., Zhang N., Brunet A., Sánchez Alvarado A., Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369, eaaz3090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S., Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Abad M., Mosteiro L., Pantoja C., Cañamero M., Rayon T., Ors I., Graña O., Megías D., Domínguez O., Martínez D., Manzanares M., Ortega S., Serrano M., Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Chen Y., Lüttmann F. F., Schoger E., Schöler H. R., Zelarayán L. C., Kim K. P., Haigh J. J., Kim J., Braun T., Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science 373, 1537–1540 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Wang C., Rabadan Ros R., Martinez-Redondo P., Ma Z., Shi L., Xue Y., Guillen-Guillen I., Huang L., Hishida T., Liao H. K., Nuñez Delicado E., Rodriguez Esteban C., Guillen-Garcia P., Reddy P., Izpisua Belmonte J. C., In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nat. Commun. 12, 3094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wernig M., Lengner C. J., Hanna J., Lodato M. A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R., A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26, 916–924 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carey B. W., Markoulaki S., Beard C., Hanna J., Jaenisch R., Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods 7, 56–59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamindago A., Lee N., Woo S., Choi H., Mun J. Y., Jang S. W., Yang S. I., Anton-Erxleben F., Bosch T. C. G., Yum S., Acute toxic effects of zinc oxide nanoparticles on Hydra magnipapillata. Aquat. Toxicol. 205, 130–139 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Lee M., Ha J., Son Y. S., Ahn H., Jung K. B., Son M.-Y., Kim J., Efficient exogenous DNA-free reprogramming with suicide gene vectors. Exp. Mol. Med. 51, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lun A. T. L., Riesenfeld S., Andrews T., Dao T. P., Gomes T.; participants in the 1st Human Cell Atlas Jamboree, Marioni J. C., EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarthy D. J., Campbell K. R., Lun A. T., Wills Q. F., Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ilicic T., Kim J. K., Kolodziejczyk A. A., Bagger F. O., McCarthy D. J., Marioni J. C., Teichmann S. A., Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 17, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lun A. T., Bach K., Marioni J. C., Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P. R., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Townes F. W., Irizarry R. A., Quantile normalization of single-cell RNA-seq read counts without unique molecular identifiers. Genome Biol. 21, 160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kowalczyk M. S., Tirosh I., Heckl D., Rao T. N., Dixit A., Haas B. J., Schneider R. K., Wagers A. J., Ebert B. L., Regev A., Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., Butte A. J., Bhattacharya M., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thummel R., Bai S., Sarras M. P., Song P., McDermott J., Brewer J., Perry M., Zhang X., Hyde D. R., Godwin A. R., Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev. Dyn. 235, 336–346 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Vejnar C. E., Moreno-Mateos M. A., Cifuentes D., Bazzini A. A., Giraldez A. J., Optimization strategies for the CRISPR-Cas9 genome-editing system. Cold Spring Harb. Protoc. 2016, pdb.top090894 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Bek J. W., Shochat C., de Clercq A., de Saffel H., Boel A., Metz J., Rodenburg F., Karasik D., Willaert A., Coucke P. J., Lrp5 mutant and crispant zebrafish faithfully model human osteoporosis, establishing the zebrafish as a platform for CRISPR-based functional screening of osteoporosis candidate genes. J. Bone Miner. Res. 36, 1749–1764 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S3
Movies S1 and S2
Data S1 to S8