Summary
Aphids secrete diverse repertoires of salivary effectors into host plant cells to promote infestation by modulating plant defence. The greenbug Schizaphis graminum is an important cereal aphid worldwide. However, the secreted effectors of S. graminum are still uncharacterized. Here, 76 salivary proteins were identified from the watery saliva of S. graminum using transcriptome and proteome analyses. Among them, a putative salivary effector Sg2204 was significantly up‐regulated during aphid feeding stages, and transient overexpression of Sg2204 in Nicotiana benthamiana inhibited cell death induced by BAX or INF1. Delivering Sg2204 into wheat via the type III secretion system of Pseudomonas fluorescens EtAnH suppressed pattern‐triggered immunity (PTI)‐associated callose deposition. The transcript levels of jasmonic acid (JA)‐ and salicylic acid (SA)‐associated defence genes of wheat were significantly down‐regulated, and the contents of both JA and SA were also significantly decreased after delivery of Sg2204 into wheat leaves. Additionally, feeding on wheat expressing Sg2204 significantly increased the weight and fecundity of S. graminum and promoted aphid phloem feeding. Sg2204 was efficiently silenced via spray‐based application of the nanocarrier‐mediated transdermal dsRNA delivery system. Moreover, Sg2204‐silenced aphids induced a stronger wheat defence response and resulted in negative impacts on aphid feeding behaviour, survival and fecundity. Silencing of Sg2204 homologues from four aphid species using nanocarrier‐delivered dsRNA also significantly reduced aphid performance on host plants. Thus, our study characterized the salivary effector Sg2204 of S. graminum involved in promoting host susceptibility by suppressing wheat defence, which can also be regarded as a promising RNAi target for aphid control.
Keywords: Schizaphis graminum, salivary proteins, wheat, defence responses, nanocarrier‐mediated RNAi, aphid performance
Introduction
Greenbug Schizaphis graminum is one of the most important and devastating cereal aphids in the world because it draws phloem sap and serves as a vector for transmitting viruses (Blackman and Eastop, 2000). In contrast to most other aphid species, S. graminum is a phytotoxic aphid, and its feeding can rapidly induce obvious leaf chlorosis in susceptible plants, resulting in the deterioration of plant quality and even plant death (Al‐Mousawi et al., 1983; Blackman and Eastop, 2000; Nicholson and Puterka, 2014; Zhang et al., 2019). A previous study showed that induced hydrogen peroxide accumulation is predicted to be involved in the induction of chlorosis symptoms by S. graminum feeding (Zhang et al., 2020). As global warming continues, the potential risk of S. graminum infestation will increase, especially in the Northern Hemisphere, which could increase global food insecurity and poverty by invading economically important crops (Aljaryian and Kumar, 2016).
Plants have evolved complicated defence systems against pathogens and insect herbivores (Schuman and Baldwin, 2016). However, during infection, pathogens produce an arsenal of effectors that are thought to be major determinants of pathogen virulence in host plants (Dou and Zhou, 2012; Jones and Dangl, 2006). Effectors secreted by pathogens enter host plant cells to suppress plant defence responses and promote plant susceptibility (Giraldo and Valent, 2013). Recent evidence suggests that aphids, similar to plant pathogens, present conserved molecules in their saliva similar to pathogen‐associated molecular patterns (PAMPs), which are perceived by pattern recognition receptors (PRRs) on the plant cell membrane and trigger PAMP‐triggered immunity (PTI). Aphids then secrete effectors that suppress this and other types of plant defences and promote effector‐triggered susceptibility (ETS). Nevertheless, effectors that are deployed to suppress host defences are recognized by plant resistance (R) proteins, activating a strong immune response called effector‐triggered immunity (ETI), typically including a hypersensitive response (Hogenhout and Bos, 2011; Kaloshian and Walling, 2005; Rodriguez and Bos, 2013; van Bel and Will, 2016).
During probing and feeding, aphid secreted saliva containing virulence proteins (effectors) into plant cytoplasm and modulate plant defence pathways (Mugford et al., 2016). Silencing of C002, which is specifically expressed in the salivary gland of aphids, leads to lethality in the pea aphid Acyrthosiphon pisum (Mutti et al., 2006, 2008). Transit overexpression of salivary effectors MpC002, Mp1/PIntO1 and PIntO2 of the green peach aphid Myzus persicae in tobacco Nicotiana benthamiana or Arabidopsis promotes aphid fecundity (Bos et al., 2010; Pitino and Hogenhout, 2013). The salivary effector Mp55 of M. persicae has been reported to promote aphid virulence upon overexpression in transgenic Arabidopsis lines by reducing the accumulation of 4‐methoxyindol‐3‐ylmethylglucosinolate, callose and hydrogen peroxide (H2O2) in response to aphid feeding (Elzinga et al., 2014). Transient overexpression of candidate effectors Me10 and Me23 of the potato aphid Macrosiphum euphorbiae enhances aphid performance, indicating their roles in the suppression of plant defence (Atamian et al., 2013). Me10 was recently reported to interact with tomato 14‐3‐3 isoform 7 (TFT7) to regulate plant susceptibility to aphids (Chaudhary et al., 2019). Transgenic barley plants expressing the candidate effector Rp1 of the bird‐cherry aphid Rhopalosiphum padi showed suppressed expression levels of jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) defence‐related genes and significantly promoted aphid fecundity (Escudero‐Martinez et al., 2020).
The eliciting activity of aphid salivary proteins in plant defence responses was also detected. The infiltration of the watery saliva of M. persicae and the grain aphid Sitobion avenae induced plant defence responses and had adverse effects on aphid performance and feeding behaviour (De Vos and Jander, 2009; Zhang et al., 2017). Transient overexpression of Mp10 and Mp42 activated plant defence in N. benthamiana and reduced aphid fecundity (Rodriguez et al., 2014). In addition, Armet, a salivary protein of A. pisum, has been shown to induce an increase in SA accumulation by regulating the expression of SAMT and SABP2, which contribute to pathogen resistance in plants (Cui et al., 2019). The molecular chaperonin proteins derived from the aphid bacterial symbiont Buchnera aphidicola, GroEL of A. pisum and GroES of the grain aphid Sitobion miscanthi were found to trigger plant defence responses (Chaudhary et al., 2014; Li et al., 2022a).
RNA interference (RNAi) is widely used for gene function research in insects. It was developed as a promising approach for pathogen and pest control (Chung et al., 2021). Among various double‐stranded RNA (dsRNA) delivery methods, the spray application of dsRNA formulations may be one of the most suitable and promising dsRNA delivery methods for pathogen and pest control. A transdermal dsRNA delivery system with star polyamine nanocarriers was successfully established, improving the dsRNA penetration of the body wall of insects and the efficiency of gene silencing through spray‐based application (Li et al., 2019, 2022b). The involvement of vestigial and Ultrabithorax genes in the wing development of M. persicae and CYP6CY3 in the host adaptation of cotton aphid Aphis gossypii was demonstrated via nanocarrier‐mediated RNAi through spray application (Wei et al., 2021; Zhang et al., 2022). This indicates that this method could be applied to identify the roles of aphid salivary proteins in modulating aphid‐plant interactions. Moreover, the greenhouse application of nanocarrier‐delivered RNA pesticides targeting V‐type proton ATPase subunits d (ATP‐d) and g (ATP‐g) of M. persicae had a high control efficacy against aphids; this further shows the great potential of nanocarrier‐mediated RNAi in aphid control (Ma et al., 2022).
Although the roles of salivary effectors are an important topic in the study of aphid virulence, few studies have been performed to precisely identify the composition salivary proteins of the cereal aphid S. graminum and investigate the roles of potential effectors in wheat‐aphid interactions. In the present study, we combined RNA‐Seq transcriptome sequencing analysis and proteomic analysis of watery saliva to identify the secreted proteins in aphid saliva. Using a Pseudomonas fluorescens effector‐to‐host analyser (EtHAn)‐mediated delivery system, we found that the putative salivary effector Sg2204 suppresses basal immunity in wheat and promotes aphid performance. Sg2204‐silenced aphids induce stronger defence responses in wheat, and silencing of Sg2204 and its homologues by nanocarrier‐mediated RNAi has deterrent effects on aphid performance, suggesting its essential role in aphid feeding as an effector.
Results
Identification of salivary proteins and candidate effectors of S. graminum
In total, 62 276 916 bp raw reads were acquired from salivary glands of S. graminum (SAR accession number: SRR18308524). After removing adapters, ambiguous nucleotides and low‐quality sequences, 61 594 644 bp clean reads (9.24 G clean reads) remained. Subsequently, the transcriptome of S. graminum salivary glands was de novo assembled using the short reads assembly program Trinity; these reads were then clustered into 14 628 unigenes (Tables S1 and S2). These transcripts ranged from 301 to over 15 093 bp with an average size of 1631 bp (Figures S1–S4). All sequences of the unigenes in this study are provided in Data S1.
The raw MS/MS data of saliva were analysed using the MaxQuant software suite (version 1.5.2.8) and searched against the S. graminum salivary gland transcriptomic database. A total of 76 proteins were identified from the aphid watery saliva using LC–MS/MS (Table S3). Among them, 43 proteins were successfully annotated, with 33 proteins uncharacterized. Some digestive and detoxification enzymes were detected in aphid saliva, such as peroxidase and lipase. Several candidate effectors previously characterized in other aphid species were also identified in S. graminum saliva, including glucose dehydrogenase, C002 and Me10.
A specific set of criteria was used to prioritize the proteins and generate a list of candidate effectors (Figure 1). The criteria (molecular weight < 30 kDa, predicted to be secreted, number of unique peptides more than 2, functionally unknown) were based on the characteristics of the previously identified fungi and aphid candidate effectors. This process led us to identify nine candidate effectors from S. graminum saliva, designated Sg1350, Sg1625, Sg1655, Sg1670, Sg1695, Sg1762, Sg1920, Sg1993 and Sg2204, and all these proteins were functionally unknown (Table 1).
Figure 1.
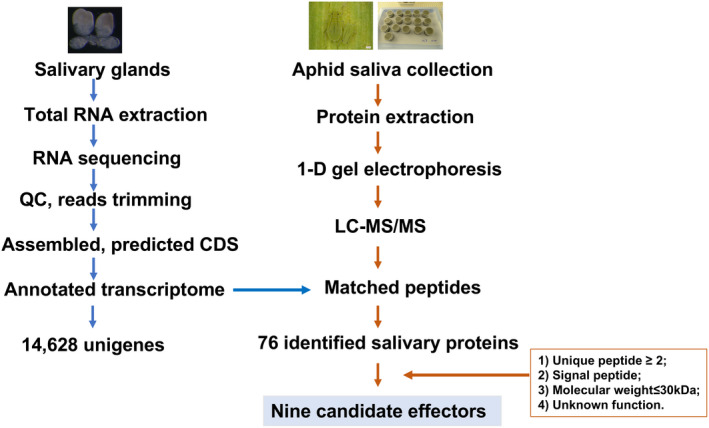
Schematic diagram of the bioinformatic analysis for the identification of salivary proteins and candidate effectors from the greenbug Schizaphis graminum.
Table 1.
List of nine candidate effectors identified from S. graminum saliva
| Protein name | Transcript ID | NCBI accession number | Unique peptides | Signal peptide (aa) | Molecular weight (kDa) | Annotation |
|---|---|---|---|---|---|---|
| Sg1350 | Cluster‐5206.1350 | XP_001950433.1 | 5 | 1–20 | 18.24 | Uncharacterized protein LOC100168922 |
| Sg1625 | Cluster‐5206.1625 | XP_001951405.1 | 5 | 1–22 | 22.68 | Uncharacterized protein LOC100166545 |
| Sg1655 | Cluster‐5206.1655 | XP_022165725.1 | 2 | 1–21 | 14.90 | Uncharacterized protein LOC111030505 |
| Sg1670 | Cluster‐5206.1670 | BAH70796.1 | 12 | 1–21 | 30.20 | ACYPI000490 |
| Sg1695 | Cluster‐5206.1695 | XP_022172257.1 | 7 | 1–20 | 23.87 | Uncharacterized protein LOC111035063 |
| Sg1762 | Cluster‐5206.1762 | XP_022160825.1 | 5 | 1–16 | 29.92 | Uncharacterized protein LOC111026940 |
| Sg1920 | Cluster‐5206.1920 | / | 8 | 1–22 | 20.32 | / |
| Sg1993 | Cluster‐5206.1993 | XP_015373648.1 | 2 | 1–25 | 27.16 | Uncharacterized protein LOC107168676 |
| Sg2204 | Cluster‐5206.2204 | XP_003246292.1 | 3 | 1–25 | 16.71 | Uncharacterized protein LOC100570017 |
‘/’ indicates that no homologues of Sg1920 were identified.
Identification and characterization of candidate effector Sg2204 in S. graminum
The expression of the nine candidate effectors in the salivary glands, heads, thorax, abdomen and midguts of apterous aphids was detected using RT‐qPCR. The results showed that all nine candidate effectors were specifically expressed in aphid salivary glands (Figure S5). For example, the relative expression of Sg1350 and Sg2204 in the salivary gland was 838.85 ± 92.76 and 1952.08 ± 0.74 times higher than that in the whole body, respectively.
To further characterize the specific transcript profiles of these nine candidate effectors at different aphid feeding time points, the transcript levels of candidate effectors were analysed using RT‐qPCR. As shown in Figure 2a, the expression levels of Sg1625, Sg1670, Sg1762, Sg1695 and Sg1993 showed no significant changes during aphid infestation. The expression levels of Sg1350, Sg1655 and Sg1920 were slightly increased, with a 1.86–3.51‐fold change at some time points. However, the abundance of Sg2204 was significantly highly induced during the infestation of wheat host plants and peaked at 12 hpi with an approximately 7.38‐fold increase. These results indicated that Sg2204 was highly induced during aphid feeding and may contribute significantly to aphid virulence.
Figure 2.
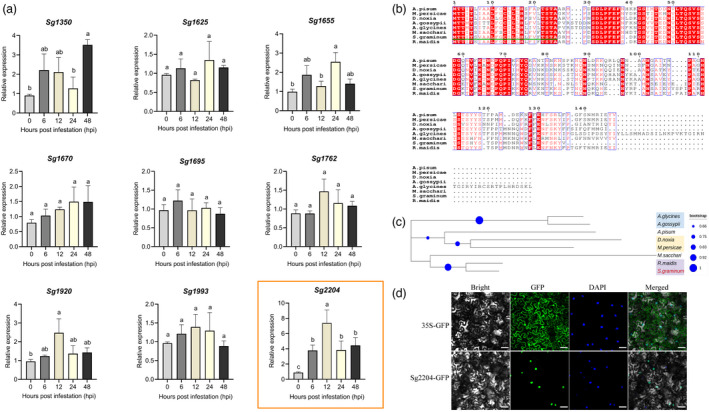
Characterization of Sg2204 in Schizaphis graminum. (a) RT‐qPCR results of the relative expression levels of nine candidate effectors of S. graminum after infestation on wheat plants at different time points. Data are presented as the mean ± SE (n = 3). Different lowercase letters above the bars indicate significant differences among groups (one‐way ANOVA followed by Duncan's multiple range tests, P < 0.05). (b) Multiple sequence alignment of the Sg2204 protein and orthologs from other aphid species. The deduced amino acid sequences from seven aphid species, including Acyrthosiphon pisum (NP_001156070.1), Myzus persicae (XP_022163343.1), Diuraphis noxia (XP_015374910.1), Aphis gossypii (XP_027851062.1), Aphis glycines (KAE9525059.1), Melanaphis sacchari (XP_025207447.1) and Rhopalosiphum maidis (XP_026819965.1). The sequence of Sg2204 was submitted to NCBI GenBank under the accession number OM676656. Red shades indicate identical amino acids. Alignment was created using Clustal Omega. (c) Phylogenetic tree constructed by comparing the amino acid sequences of Sg2204 and the orthologs from other aphid species. A phylogenetic tree was constructed by the neighbour‐joining method using MEGA7.0. Bootstrap values calculated as a percentage for 1000 replications are shown at the nodes. The amino acid sequence with green underline indicates the signal peptide of the Sg2204 protein. (d) Subcellular localization of Sg2204. GFP (empty vector, pCAMBIA1300) and effector Sg2204 were transiently overexpressed in N. benthamiana leaves by Agrobacterium infiltration. The images were taken 3 days after agroinfiltration using confocal microscopy. Bar = 20 μm.
We obtained a full‐length cDNA for the Sg2204 gene, which contains a 414‐bp open reading frame and encodes a polypeptide of 137 amino acid residues (Figure 2b). The first 25 amino acids constitute the signal peptide, with cleavage predicted between residues 25 and 26 (Figure 2b). BLAST analyses revealed that Sg2204 has no strong matches to any protein of known function or to any predicted protein outside of the family Aphididae. The amino acid sequence of Sg2204 was aligned with the amino acid sequences of orthologs in seven other aphid species. The results showed that Sg2204 has the highest identity (90%) with the R. madis ortholog but only 65% and 55% sequence identity to M. persicae and Diuraphis noxia orthologs, respectively. Phylogenetic analysis showed that Sg2204 and the R. madis ortholog were closely related, clustering into an independent clade (Figure 2c).
To identify the subcellular localization of Sg2204, transient expression analyses of the Sg2204‐GFP fusion protein in N. benthamiana leaves were performed. The control groups expressing only GFP with the cauliflower mosaic virus 35S promoter exhibited fluorescence throughout the cytoplasm and nucleus (Figure 2d). But the fluorescence of the Sg2204‐GFP fusion protein was only observed in the nucleus of N. benthamiana cells, which suggested that Sg2204 encodes a nucleus‐targeting protein.
Overexpression of Sg2204 in Nicotiana benthamiana suppresses cell death
The potential roles of Sg2004 in the suppression of BAX and PAMP INF1‐induced programmed cell death (PCD), which resembles the defence‐related HR in plant cells, were examined. As shown in Figure 3a–g, transient overexpression of GFP and Sg2204 did not induce PCD, but overexpression of both BAX and INF1 in tobacco activated obvious PCD and overexpression of Sg2204 could significantly suppress BAX and INF1‐induced PCD in N. benthamiana. As a negative control, overexpression of GFP did not affect the induced PCD symptoms.
Figure 3.
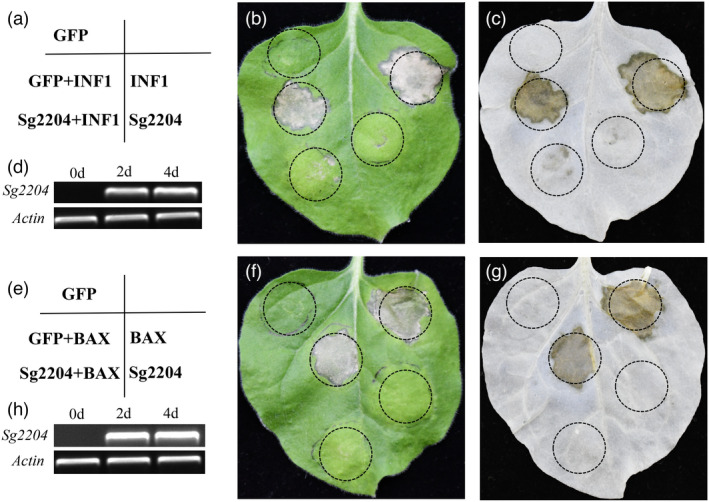
Transient overexpression of Sg2204 in Nicotiana benthamiana suppressed programmed cell death triggered by PAMP‐INF1 (a–d) and BAX (e–h). N. benthamiana leaves were infiltrated with recombinant strains of Agrobacterium tumefaciens cells carrying GFP (negative control) or Sg2204, and A. tumefaciens cells carrying INF1 (a, b) or BAX (e, f) were then infiltrated into the same region of leaves after 12 h, respectively. The leaves were decolorized with ethanol (c, g) and photographed 5 days after infiltration. Detection of the transcripts of Sg2204 was performed using RT‐PCR in transient‐overexpressing N. benthamiana leaves inoculated with A. tumefaciens carrying Sg2204 at 2 and 4 days, and the reference gene β‐actin of N. benthamiana served as a control (d, h). All experiments were performed with three biological replications.
Delivery of Sg2204 in wheat suppresses plant defence
To investigate the function of Sg2204 in inhibiting host plant immunity, Sg2204 was cloned into the expression vector pEDV6 and then delivered into wheat leaves using the P. fluorescens EtAnH‐mediated delivery system. Previous studies reported that infiltration of EtHAn expressing AvrRpt2, a well‐characterized TTSS effector from Pseudomonas syringae was sufficient to induce in planta cell death and noticeable chlorotic phenotype in wheat leaves (Mindrinos et al., 1994; Mudgett and Staskawicz, 1999; Yin and Hulbert, 2011). As presented in Figure 4a,b, the wheat leaves infiltrated with EtAnH delivering the AvrRpt2 (positive control) showed obvious chlorosis symptoms and H2O2 accumulation after 2 days, suggesting that the T3SS system was successfully established in this study. The infiltration of EtAnH expressing Sg2204 did not induce any obvious chlorosis symptoms or H2O2 accumulation on the leaves. The H2O2 content in wheat leaves treated with AvrRpt2 was significantly increased compared with that in the control groups, but the H2O2 content in pEDV6 (empty vector)‐ and pEDV6:Sg2204‐inoculated wheat leaves was not significantly different from that in the control groups (Figure 4d).
Figure 4.
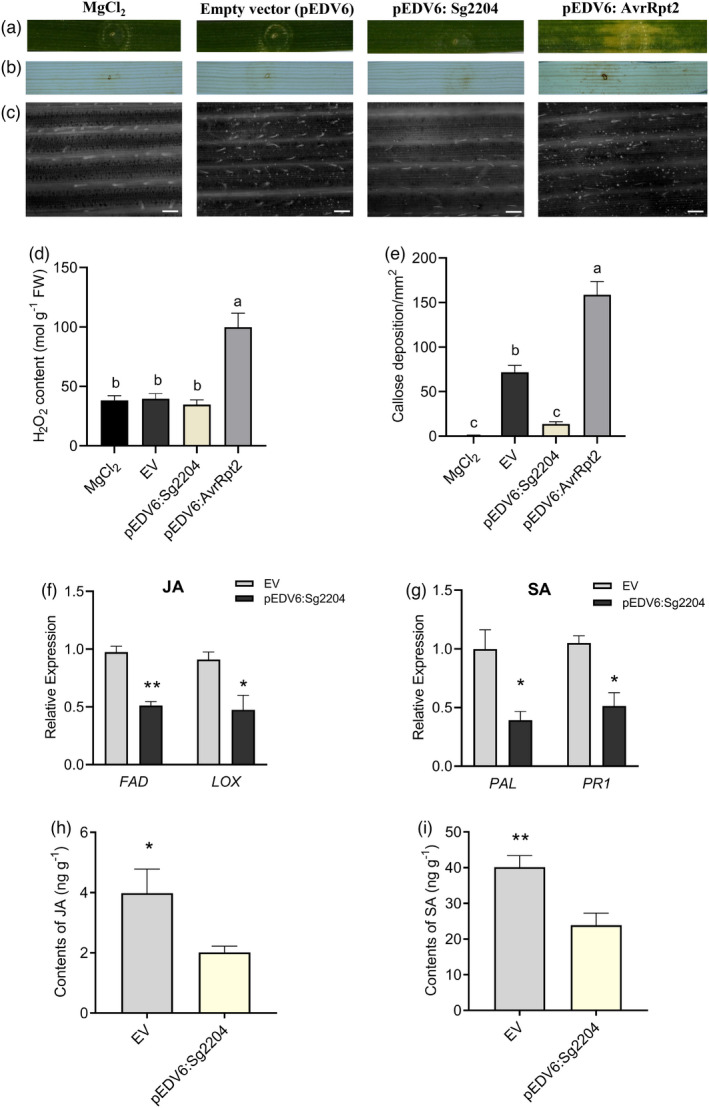
Delivery of Sg2204 into wheat suppressed plant defence. (a) Phenotypes of wheat leaves inoculated with MgCl2 buffer (mock), Pseudomonas fluorescens EtHAn carrying pEDV6 (empty vector, EV), pEDV6:Sg2204 and pEDV6:AvrRpt2 (positive control) at 72 h after infiltration. (b) DAB staining for H2O2 accumulation in wheat leaves infiltrated with MgCl2 buffer, P. fluorescens EtHAn carrying EV, pEDV6:Sg2204 and pEDV6:AvrRpt2 at 72 h after infiltration. (c) Callose deposition observed by fluorescence microscopy in wheat leaves infiltrated with MgCl2 buffer, P. fluorescens EtHAn carrying EV, pEDV6:Sg2204 and pEDV6:AvrRpt2 at 72 h after infiltration after aniline blue staining. Bars = 200 μm. (d) H2O2 content of wheat leaves inoculated with MgCl2 buffer, EV, pEDV6:Sg2204 or pEDV6:AvrRpt2. (e) Average number of callose deposits/mm2 in wheat leaves treated with MgCl2 buffer, EV, pEDV6:Sg2204 or pEDV6:AvrRpt2. Three biological replicates were performed for each treatment. Data are presented as the mean ± SE. Different lowercase letters above the bars indicate significant differences among groups (one‐way ANOVA followed by Duncan's multiple range tests, P < 0.05). (f) The expression levels of JA synthesis‐related genes FAD and LOX in wheat leaves after infiltration with P. fluorescens EtAnH carrying an empty vector (EV) or pEDV6:Sg2204 at 72 h. (g) The expression levels of the SA synthesis‐related gene PAL and the SA signalling pathway marker gene PR1 in wheat leaves after infiltration with an empty vector (EV) or pEDV6:Sg2204 at 72 h. (h) Change in endogenous JA contents in wheat leaves treated with P. fluorescens EtAnH carrying an empty vector (EV) or pEDV6:Sg2204. (i) Change in endogenous SA contents in wheat leaves inoculated with P. fluorescens EtAnH carrying an empty vector (EV) or pEDV6:Sg2204. Three biological replicates were performed for each treatment. Data are presented as the mean ± SE. Asterisks above the bars indicate significant differences between controls and treatments (*P < 0.05; **P < 0.01; Student's t test).
As a nonpathogen, P. fluorescens has been shown to induce PTI‐related defence genes and callose deposition following infiltration into the leaves of several plant species (Nguyen et al., 2010). Aniline blue staining showed that many callose deposits were observed in wheat leaves challenged by P. fluorescens EtHAn carrying the pEDV6 empty vector and positive control pEDV6:AvRpt2, indicating the activation of PTI in wheat. However, significantly less callose deposition was detected in wheat leaves delivering Sg2204 (Figure 4c–e). Taken together, these results suggested that Sg2204 suppressed PTI‐related callose deposition in wheat plants.
Considering that the SA‐ and JA‐ pathways are major signalling pathways involved in most plant defences against insect herbivores, the key genes associated with SA and JA and the levels of plant defence hormones were studied upon delivery of Sg2204 in wheat for 2 and 4 days. Our results showed that the transcript levels of the SA biosynthesis enzyme PAL and SA pathway marker gene PR1 were significantly down‐regulated after treatment with Sg2204 at 2 and 4 days, and the transcript levels of FAD and LOX, which are involved in the JA signalling pathway, were also significantly reduced at 4 days post‐infiltration of P. fluorescens EtAnH carrying Sg2204 (Figure 4f,g). Furthermore, LC–MS/MS results showed that delivery of Sg2204 in wheat significantly reduced the levels of JA and SA at 4 dpi compared with the control, suggesting that Sg2204 is involved in the suppression of both the SA and JA defence pathways in wheat plants (Figure 4h,i).
Delivering Sg2204 into wheat promotes aphid feeding behaviour and enhances aphid performance
As shown in Figure 5a, treating wheat with Sg2204 had a significant impact on the feeding behaviour of aphids. The duration of phloem ingestion (E2) was significantly higher in wheat treated Sg2204 compared with that infiltrated with the empty vector (control). In contrast, the durations of stylet probing activity (C) and nonprobing (Np) waveforms in Sg2204‐treated leaf areas were significantly higher than those in the control group. Additionally, aphids had a shorter latency to reach the first sustained E2 (>10 min) from the start of EPG recording when feeding on Sg2204‐infiltrated wheat leaves.
Figure 5.
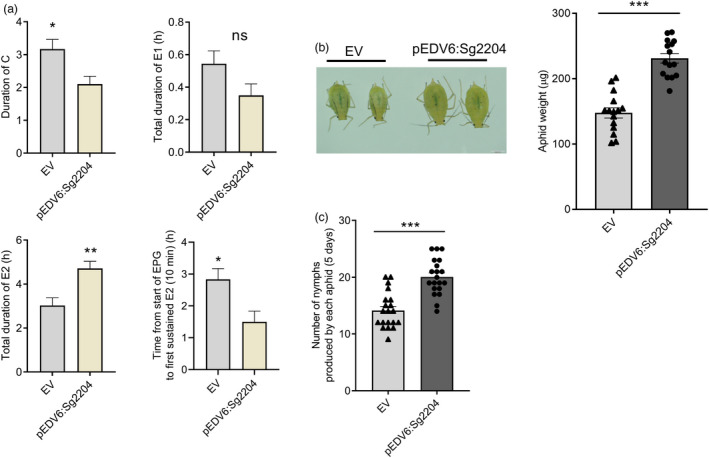
Overexpression of Sg2204 in wheat promoted Schizaphis graminum feeding and enhanced aphid body weight and fecundity. (a) Feeding behaviour parameters of S. graminum, including durations of stylet probing (C), salivation (E1), phloem ingestion (E2) and time from the start of EPG to the first sustained E2, when feeding on wheat leaves treated with empty vector (EV) and pEDV6:Sg2204. At least 10 biological replicates were performed for each group. (b) Body weight of S. graminum after feeding on wheat leaves inoculated with EV and pEDV6:Sg2204 for 7 days (n = 15). (c) Number of nymphs produced by S. graminum after feeding on wheat leaves infiltrated with EV and pEDV6:Sg2204 for 5 days (n = 20). Data are presented as the mean ± SE. Asterisks above the bars indicate significant differences between controls and treatments (*P < 0.05; **P < 0.01; ***P < 0.001; Student's t test).
We found that the weight of aphids feeding on wheat leaves treated with Sg2204 was significantly greater than that on the control leaves (Figure 5b). The number of nymphs produced by aphid adults was also significantly higher on Sg2204‐treated leaves than on the control leaves (Figure 5c). These results suggested that Sg2204 enhanced aphid performance and host susceptibility by promoting aphid feeding.
Sg2204 homologues from other aphid species have conserved functions in promoting wheat susceptibility to aphids
To investigate whether homologues of Sg2204 have conserved roles in suppressing PTI and promoting host susceptibility, Sg2204 and its homologues found in four other aphid species, S. avenae (Sa2204), R. midis (Rm2204), M. persicae (Mp2204) and A. pisum (Ap2204), were infiltrated into wheat using the P. fluorescens EtAnH‐mediated delivery system. As presented in Figure 6a,b, Sg2204 and all homologues inhibited callose deposition in treated wheat leaves compared with the control, and the number of callose deposits in wheat leaves infiltrated with the Sg2204 homologues was significantly less than that induced by nonpathogenic bacteria P. fluorescens carrying the empty vector. Furthermore, the results presented in Figure 6c show that the number of nymphs produced by S. graminum feeding on Sg2204 homologue‐treated wheat leaves was significantly higher than those feeding on the control, indicating that Sg2204 homologues have conserved functions in suppressing plant PTI and promoting aphid performance.
Figure 6.
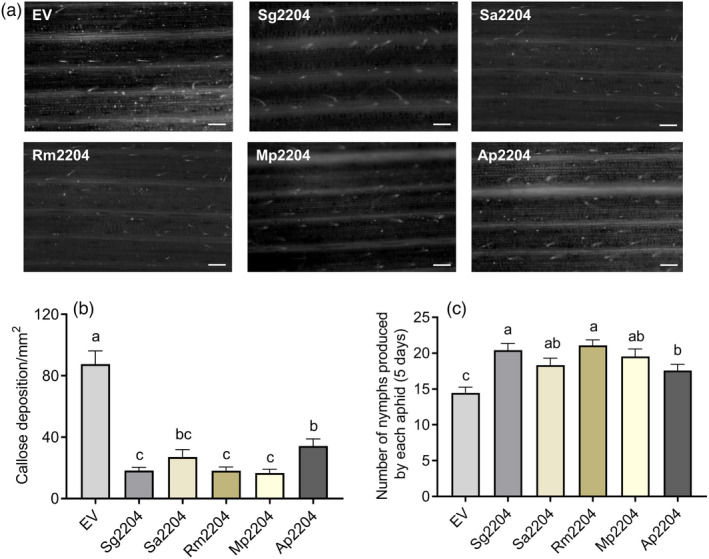
Sg2204 orthologs from Sitobion avenae (Sa2204) (NCBI accession no. OM831273), Rhopalosiphum maidis (Rm2204), Myzus persicae (Mp2204) and Acyrthosiphon pisum (Ap2204) have conserved functions in inhibiting wheat PTI defence and prompting aphid performance. (a) Callose deposition staining in wheat leaves after infiltration with Pseudomonas fluorescens EtHAn carrying an empty vector (EV), Sg2204, Sa2204, Rm2204, Mp2204 and Ap2204 at 2 days. Bars = 200 μm. (b) Average number of callose deposits/mm2 in wheat leaves inoculated with EV, Sg2204, Sa2204, Rm2204, Mp2204 and Ap2204 at 2 days (n = 3). (c) Number of nymphs produced by Schizaphis graminum after feeding on wheat leaves infiltrated with EV, Sg2204, Sa2204, Rm2204, Mp2204 and Ap2204 for 5 days (n = 15). Data are presented as the mean ± SE. Different lowercase letters above the bars indicate significant differences among groups (one‐way ANOVA followed by Duncan's multiple range tests, P < 0.05).
Silencing of Sg2204 by a nanocarrier‐mediated dsRNA delivery system reduces aphid performance and feeding
A schematic diagram of the transdermal dsRNA delivery system with nanocarriers is shown in Figure 7a, and the dsRNA formulation was sprayed onto aphids and wheat plants. After 12 h of treatment, obvious labelled‐dsRNA fluorescence was detected in the salivary glands and guts of aphids (Figure 7c). The RNAi efficiency of nanocarrier‐mediated RNAi on the Sg2204 gene was detected using RT‐qPCR. As shown in Figure 7b, the expression levels of Sg2204 in S. graminum were reduced significantly after treatment with 200 ng/μL dsSg2204 for 24 h. After 48 h of treatment, the transcript levels of Sg2204 were further decreased to 0.36 ± 0.065‐fold, which were significantly lower than those in the control.
Figure 7.
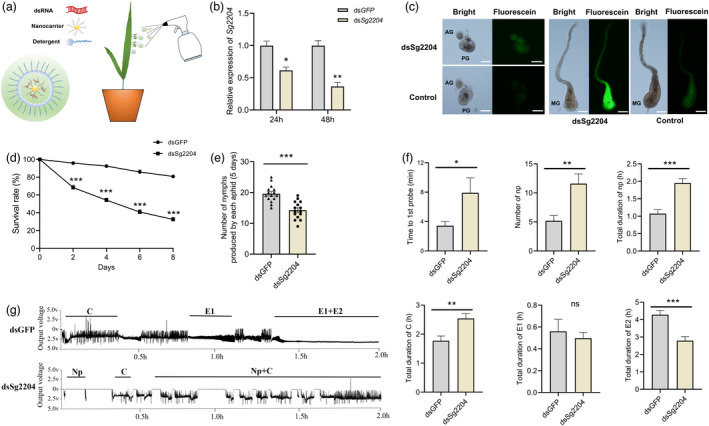
Silencing of Sg2204 by nanocarrier‐mediated RNAi negatively affected aphid performance and feeding behaviour. (a) Schematic diagram of the spray method of application of the nanocarrier‐mediated transdermal dsRNA delivery system. (b) Relative expression levels of Sg2204 at 24 and 48 h after dsSg2204/nanocarrier/detergent or dsGFP/nanocarrier/detergent treatment. Three biological replicates were conducted for each treatment (n = 3, Student's t test). (c) Examination of dsRNA uptake efficiencies in the salivary glands and guts of S. graminum using fluorescein‐labelled Sg2204‐dsRNA. Images were acquired at 12 h after spraying with fluorescein‐labelled Sg2204‐dsRNA/nanocarrier/detergent or H2O/nanocarrier/detergent (control) formulations using a fluorescence microscope. AG: accessory gland, HG: hindgut, MG: midgut and PG: principal gland. Scale bars = 100 μm. (d) Survival rate of dsGFP‐ or dsSg2204‐treated S. graminum after feeding on wheat plants at various time points (n = 10, Student's t test). (e) Number of nymphs produced by dsGFP‐ or dsSg2204‐treated S. graminum after feeding on wheat plants for 5 days (n = 15, Student's t test). (f) Effects of Sg2204 RNAi knockdown on the feeding behaviours of S. graminum based on EPG recordings (n ≥ 10, Mann–Whitney U test). (g) Representative EPG waveforms of Sg2204‐silenced aphids (dsSg2204) and control aphids (dsGFP) on wheat plants. Np, nonprobing; C, stylet probing; E1, salivation and E2, phloem ingestion. The data shown are the mean ± SE. Asterisks above the bars indicate significant differences between controls and treatments (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001).
The effects of silencing Sg2204 on aphid survival, fecundity and feeding behaviour were also investigated. The results in Figure 7d,e show that the survival rate of S. graminum treated with dsSg2204 was reduced to 54% after feeding on wheat plants for 4 days, which was significantly lower than that of the control group (dsGFP); this rate further decreased to 32% at 8 days. In addition, the number of nymphs produced by Sg2204‐silenced aphids was also significantly less than that produced by aphids in the control groups.
Aphid feeding behaviour was negatively affected after the expression of Sg2204 was inhibited. As shown in Figure 7f,g, the time to the first probe activity of aphids treated with dsSg2204 was significantly longer than that spent of the control. The number of nonprobing waveforms of Sg2204‐silenced aphids on wheat plants was greater than that on the control. In addition, the durations of the nonprobing and C phases of aphids treated with dsSg2204 were longer, but the duration of phloem ingestion (E2) was shorter, than those of aphids treated with dsGFP.
Sg2204‐silenced aphids activate stronger wheat defence responses
As shown in Figure 8a,b, we found that more callose was deposited in wheat leaves induced by Sg2204‐silenced aphid feeding compared with the control (without aphid infestation) and treatment groups induced by dsGFP‐fed aphids. The changes in the expression levels of the JA biosynthesis pathway‐associated genes FAD and LOX and the SA biosynthesis/signalling pathway‐related genes PAL and PR1 were also examined in wheat leaves infested with Sg2204‐silenced S. graminum using RT‐qPCR. As shown in Figure 8c, the expression levels of LOX, PAL and PR1 in wheat leaves infested with Sg2204‐silenced aphids were significantly higher than those in wheat leaves infested with aphids fed dsGFP.
Figure 8.
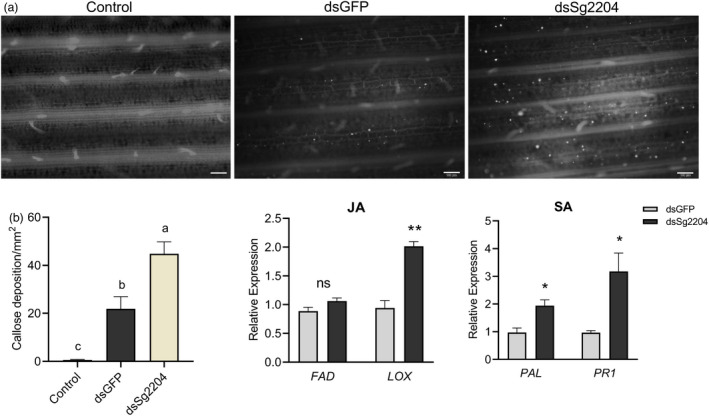
Infestation with Sg2204‐silenced Schizaphis graminum induced a stronger wheat defence response. (a) Callose deposition staining in wheat leaves after infestation with no aphids (control), dsGFP‐treated aphids or dsSg2204‐treated aphids. Bars = 200 μm. (b) Average number of callose deposits/mm2 in wheat leaves infested with no aphids (control), dsGFP‐treated aphids or dsSg2204‐treated aphids. (c) Relative expression levels of the JA‐related genes FAD and LOX and the SA marker genes PAL and PR1 in wheat leaves infested with Sg2204‐silenced aphids (dsSg2204) or control aphids (dsGFP). The data shown are the mean ± SE. Three biological replicates were performed for each group. Asterisks above the bars indicate significant differences between controls and treatments (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; Student's t test).
Silencing of Sg2204 homologues from other aphid species through a nanocarrier dsRNA delivery system reduces aphid survival and fecundity
Using the spray application method for nanocarrier‐based transdermal dsRNA delivery system, the expression levels of Sg2204 homologues from S. avenae, R. maidis, A. pisum and M. persicae were all significantly down‐regulated, which decreased to 28%–41% of that of control groups (P < 0.01) (Figure 9a). Additionally, RNAi‐mediated silencing of Sg2204 homologues in aphids significantly reduced the survival and fecundity of S. avenae, R. maidis and A. pisum (P < 0.001), and the percentages of aphid survival after gene silencing varied among different species. For example, the survival rates of A. pisum were reduced to 61% after gene silencing, but the survival rate of R. maidis was only 41%. However, gene silencing had no significant impacts on the survival of M. persicae (P = 0.052), but it significantly reduced the fecundity of four aphid species (P < 0.001) (Figure 9b,c).
Figure 9.
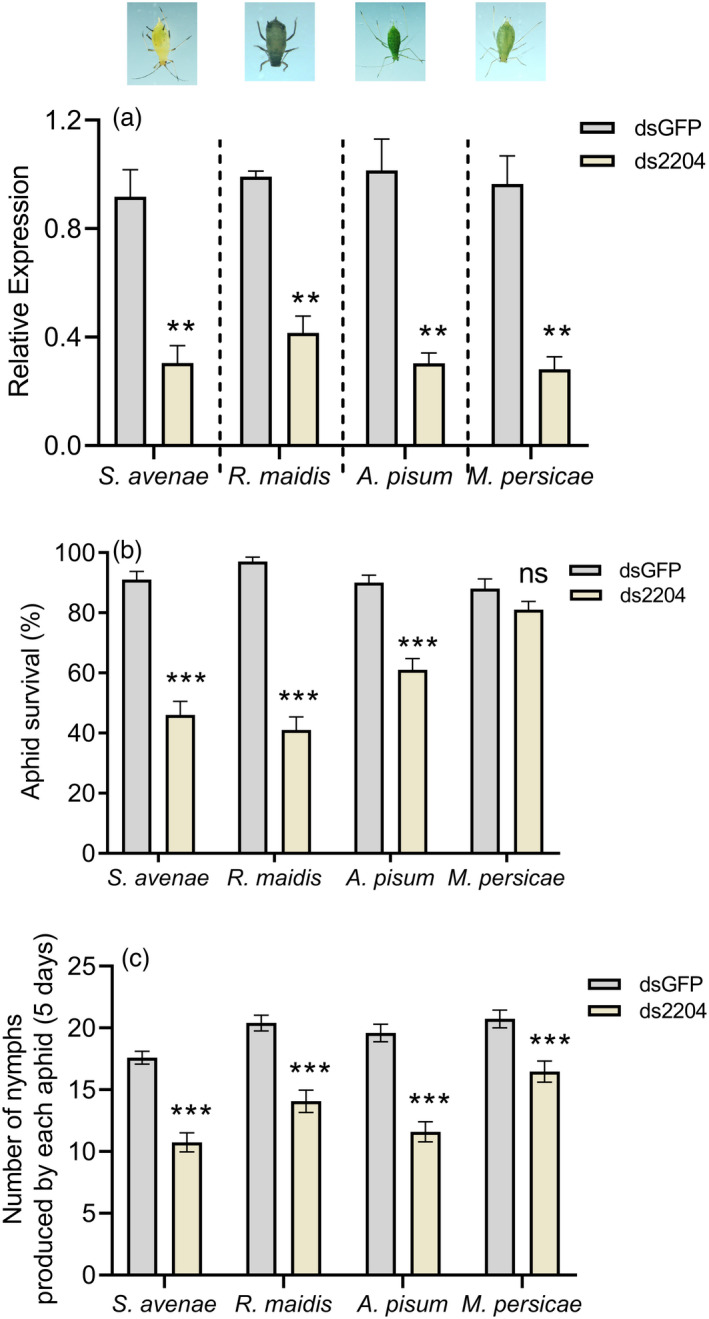
Silencing of Sg2204 homologues from Sitobion avenae, Rhopalosiphum maidis, Acyrthosiphon pisum and Myzus persicae via a transdermal dsRNA delivery system with nanocarriers decreased aphid survival and fecundity. (a) Relative expression levels of Sg2204 homologues after treatment with ds2204/nanocarriers/detergent or dsGFP/nanocarriers/detergent at 48 h (n = 3, Student's t test). (b) Survival rate of ds2204‐ or dsGFP‐treated S. avenae, R. maidis, A. pisum and M. persicae at 6 days after feeding on wheat, maize, broad bean and tobacco plants, respectively (n = 10, Student's t test). (c) Number of nymphs produced by ds2204‐ or dsGFP‐treated S. avenae, R. maidis, A. pisum and M. persicae when feeding on wheat, maize, soybean and tobacco plants for 5 days (n = 15, Student's t test). Data are presented as the mean ± SE. Asterisks above the bars indicate significant differences between controls and treatments (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; Student's t test).
Discussion
Due to the poor efficiency of Agrobacterium‐mediated transformation in cereal plants, few studies have been reported on the roles of salivary proteins from cereal aphids in modulating plant defence. The greenbug S. graminum is a major phytotoxic aphid pest of wheat, sorghum and barley (Blackman and Eastop, 2000); therefore, it is crucial to identify and functionally characterize salivary effectors from S. graminum to uncover the mechanisms employed by aphids when infesting cereals.
In our study, a total of 76 S. graminum watery salivary proteins were identified using integrated transcriptomic and proteomic analysis. Consistent with a previous study (Nicholson and Puterka, 2014), the presence of several proteins, such as glucose dehydrogenase, peroxidase, glutathione peroxidase and carbonic anhydrase, was also detected in S. graminum saliva in our study. We also identified several salivary proteins of S. graminum that had sequence similarity to several previously identified putative aphid effectors. For example, two salivary proteins with similarity to effector C002 from M. persicae and Me10 from M. euphorbiae were detected in S. graminum saliva. These proteins have previously been shown to suppress plant defence and promote aphid virulence (Atamian et al., 2013; Bos et al., 2010). In addition, we also identified cathepsin B protein in S. graminum saliva, which has been demonstrated to be involved in eliciting plant defence as an elicitor of M. persicae and to affect host adaptability (Guo et al., 2020). Whether these candidate effectors of S. graminum display conserved functions in modulating plant defence remains to be explored.
Delivering candidate effectors into cereal plants, such as wheat or barley, using the P. fluorescens EtHAn‐mediated delivery system has been successfully applied for the functional characterization of effectors from the wheat stripe rust fungus P. striiformis f. sp. tritici (Bai et al., 2021; Cheng et al., 2017; Upadhyaya et al., 2014; Xu et al., 2019a). In our study, delivery of Sg2204 into wheat using the P. fluorescens EtHAn system enhanced aphid fecundity and body weight and promoted aphid phloem feeding. Some researchers have proven that the salivary effectors of aphids and other hemipteran insects enhance host susceptibility by modulating plant defence pathways. For example, salivary effector Bt56 of Bemisia tabaci interacting with NTH202 promotes the susceptibility of tobacco to whitefly by affecting SA‐mediated plant defences (Xu et al., 2019b). Salivary proteins, such as BtFer1 from B. tabaci, glutathione peroxidase from the mirid bug Apolygus lucorum and vitellogenin from the small brown planthopper Laodelphax striatellus, attenuate H2O2‐mediated plant defence and improve insect feeding performance as effectors (Dong et al., 2020; Ji et al., 2021; Su et al., 2019).
A previous study demonstrated that callose deposition induced by aphids is involved in the sealing of sieve pores as a phloem defence mechanism that prevents the flow of nutrients to piercing‐sucking insects (Will and van Bel, 2006). In addition, the plant defence hormones JA and SA are important signalling molecules conferring plant resistance against aphids (Li et al., 2006; Yates and Michel, 2018; Züst and Agrawal, 2016). The expression levels of genes involved in both the JA‐ and SA‐dependent defence pathways of wheat, such as LOX, FAD, PAL and PR1, are significantly up‐regulated in response to S. graminum infestation (Reddy et al., 2013; Zhang et al., 2020; Zhu‐Salzman et al., 2004). Exogenous application of SA and JA on Arabidopsis, wheat, tomato and Brassica plants had negative effects on aphid population growth (Ali et al., 2021; Boughton et al., 2006; Bruce et al., 2003; Feng et al., 2021). Our results showed that delivery of Sg2204 into wheat suppressed callose deposition and JA‐ and SA‐dependent defence responses induced by nonpathogenic P. fluorescens. Therefore, the effects of Sg2204 on enhanced plant susceptibility to S. graminum are proposed to be associated with the suppression of both the JA‐ and SA‐dependent plant defence signalling pathways in wheat. Although an antagonistic relationship between SA and JA signalling pathways is evident (Thaler et al., 2012), some components of plants can activate or suppress the SA and JA signalling pathways simultaneously. For example, the expression of a mannose‐specific JRL‐like gene (TaJRLL1) of wheat in Arabidopsis thaliana increased the levels of both JA and SA and enhanced its resistance against pathogens (Xiang et al., 2011). WRKY70, a transcriptional factor, inhibited plant immunity by suppressing the expression of SA and JA signalling pathway genes, such as NPR1, VSP1 and VSP2 (Chakraborty et al., 2020; Despres et al., 1995; Li et al., 2004). It is speculated that Sg2204 interacts with the components of wheat plants that are involved in modulating both JA and SA defensive pathways. A previous study also showed that transgenic barley lines expressing the R. padi effector Rp1 reduced expression levels of JA‐, SA‐ and ET‐responsive genes and had enhanced plant susceptibility to R. padi (Escudero‐Martinez et al., 2020).
We also found that Sg2204 orthologs from five other aphid species inhibited wheat defence and promoted aphid fecundity, suggesting the functional conservation of the effector in Aphidinae. A previous study showed that M. persicae effector Mp10 orthologs from diverse plant‐feeding hemipteran insects have evolved conserved functions for suppressing ROS and Ca2+‐burst in N. benthamiana leaf discs upon elicitation with flg22 (Drurey et al., 2019). In addition, the M. euphorbiae effector Me10 and its homologue in A. gossypii, Ag10k, have been proven to interact with TFT7 in planta and interfere with the function of the TFT7 immune complex to promote aphid virulence (Chaudhary et al., 2019). However, the aphid protein effector C002 was previously reported to promote aphid colonization in a plant species‐specific manner, likely due to the presence or lack of NDNQGEE repeat motifs (Pitino and Hogenhout, 2013). Whether the Sg2204 orthologs from the other aphid species, which were not detected in this study, are involved in the suppression of host plant defence remains to be investigated.
Delivery of dsRNA into cells to silence the genes has been successfully applied to analyse the function of candidate effectors of hemipterans (Pitino et al., 2011; Xu et al., 2019b). Nanocarrier‐mediated RNAi is a practical delivery system for silencing the target genes of insects such as the soybean aphid Aphis glycines and M. persicae and is regarded as a promising strategy for gene function characterization and pest control (Li et al., 2022b; Niu et al., 2021; Yan et al., 2020). In this study, to further examine the roles of Sg2204 in suppressing plant defence, the Sg2204 gene was silenced through spray application of a nanocarrier‐mediated transdermal dsRNA delivery system. Our results showed that the expression levels of Sg2204 were significantly inhibited after the application of dsSg2204/nanocarriers/detergent, and its expression levels were reduced by more than 60% compared with those of the control groups. Previous study showed that the transcript levels of ATP‐d and ATP‐g in M. persicae decreased to 60%–70% after spray application of nanocarrier/dsRNA formulations (Ma et al., 2022), suggesting that the efficiency of nanocarrier‐mediated RNAi through spray method is stable in different aphid species. As expected, the expression levels of JA‐ and SA‐responsive genes were increased, and more callose deposits were observed in wheat after Sg2204‐silenced aphid infestation, which further suggested the involvement of Sg2204 in the suppression of plant basal defence. It was also found that the duration of nonprobing phase and stylet‐probing activity of aphids was significantly prolonged after silencing Sg2204. We cannot exclude the possibility that the mechanical damage caused by the frequent puncturing of the stylet lead to a stronger defence response of wheat. Interestingly, the silencing of Sg2204 homologues from four aphid species had deterrent effects on aphid survival and fecundity, indicating that this gene has conserved functions in aphid adaptability to host plants. Aphid salivary proteins which are essential for the infestation of plants appear to be a promising target for RNAi‐mediated pest control. For example, the silencing of salivary protein C002 in M. persicae and structural sheath protein in S. avenae via host‐induced gene silencing (HIGS) led to a significant decline in the reproduction and survival rates of aphids (Abdellatef et al., 2015; Pitino et al., 2011). Thus, Sg2204 and its homologues could be used as ideal RNAi target genes for aphid control through HIGS and spray‐induced gene silencing (Ma et al., 2022; Qiao et al., 2021; Sun et al., 2019).
Additionally, subcellular localization showed that Sg2204 protein localized to the nucleus in N. benthamiana, suggesting that this effector may interact with nuclear proteins of wheat to inhibit plant immunity. Some researchers have demonstrated that effectors of Fusarium head blight Fusarium graminearum and L. striatellus could interact with proteins localized in the plant cell nucleus, such as the transcription factor WRKY71 and protein kinase TaSnRK1α28, to suppress plant defence (Ji et al., 2021; Jiang et al., 2020). Therefore, it is worth identifying the proteins that interact with Sg2204 in wheat plants using a yeast two‐hybrid system in future studies.
Overall, our findings provide the first evidence that S. graminum employs the effector protein Sg2204 to enhance aphid performance on wheat by suppressing JA‐ and SA‐related signalling pathways to manipulate plant defence responses. Our findings also demonstrate that silencing of Sg2204 and its homologues by nanocarrier‐mediated RNAi was lethal to aphids. These results provide new insights into the molecular mechanisms underlying cereal aphid‐plant interactions and RNAi‐based strategies for aphid control.
Experimental procedures
Plants and insects
Clones of S. graminum, S. avenae, A. pisum and the corn aphid Rhopalosiphum maidis were initially established from a single aphid collected from a field in Langfang city, Hebei Province, northern China, and clones of M. persicae were established from a single aphid collected from a tobacco field in Kunming city, Yunan Province, southwestern China. All aphid clones were maintained under laboratory conditions (L : D = 16 h : 8 h; 20 ± 1 °C) on corresponding host plants, including wheat, broad bean, maize and tobacco plants for more than 4 years.
RNA isolation and library construction
Approximately 500 paired salivary glands of alate and apterous adult aphids were dissected in cold phosphate‐buffered saline (pH 7.2; HyClone, Thermo Scientific, MA). Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA concentration and integrity were measured using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA), respectively. Total RNA samples (3 μg) with standard quality were used to generate the sequencing libraries with the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (New England Biolabs, Beverly, MA) following the manufacturers' instructions. The library preparations were sequenced using an Illumina HiSeq 2500/MiSeq platform and paired‐end reads (the sequencing strategy was PE150). After removing adaptor sequences, ambiguous ‘N’ nucleotides and low‐quality sequences, the clean reads were assembled using Trinity r20140413p1 min_kmer_cov:2 and the other default parameters as described for de novo transcriptome assembly without a reference genome to generate transcripts and unigenes. The unigenes were searched against the nonredundant protein database (Nr), NCBI nucleotide database (Nt) and Swiss‐Prot protein database with an E value < 10−5 and against euKaryotic Ortholog Groups of proteins (KOG) with an E value < 10−3. The unigenes were annotated by Blast2GO in the Gene Ontology (GO) database with an E‐value threshold of 1.0−6 and annotated by the KEGG Automatic Annotation Server (KAAS) based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) with an E value threshold of 1.0−10.
Identification of aphid salivary proteins using liquid chromatography−mass spectrometry/mass spectrometry (LC–MS/MS)
Schizaphis graminum watery saliva was collected as described previously (Zhang et al., 2017). Proteins were electrophoresed on a 12.5% acrylamide separating gel with a 4% stacking gel for 1.5 h at 120 V. The gel was stained using the PlusOne Silver Staining Kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) following the manufacturer's instructions. Protein bands were excised from the prepared 1D gels and were then destained, reduced, alkylated and trypsin‐digested as described by Harmel et al. (2008). The shotgun analysis was conducted on an Easy nLC 1000 (Thermo Fisher Scientific, Waltham, MA) coupled with a Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The raw MS data were analysed using the MaxQuant software suite (version 1.5.2.8) and searched against the S. graminum salivary gland transcriptomic database. For the initial search, the precursor ion mass tolerance was 15 ppm. The search followed the enzymatic cleavage rule for trypsin; a maximum of two missed cleavage sites and 20 mmu mass tolerance for the fragment ions were allowed. For database searching, cysteine carbamidomethylation was considered a static modification, while N‐terminal acetylation and methionine oxidation were considered dynamic modifications. The cutoff for the global false discovery rate (FDR) in the peptide‐spectrum match (PSM) and protein identification was <0.01.
Subcellular localization
The coding sequence of Sg2204 (without a signal peptide) was constructed in pCAMBIA1300 with TagGFP fused at the C‐terminus. All constructs, including the empty vector pCAMBIA1300, were transformed into Agrobacterium tumefaciens GV3101 and cultured overnight in LB medium (50 μg/mL kanamycin, 25 μg/mL rifampicin) at 28 °C. The cells were harvested and resuspended in infiltration medium (10 mM MgCl2, 10 mM 2‐(N‐morpholino) ethanesulphonic acid, 200 μM acetosyringone, pH 5.6) to an OD600 of 0.5. Agrobacterium carrying the effector‐GFP constructs were then infiltrated into the leaves of 6‐week‐old N. benthamiana plants using a 1‐mL syringe without a syringe needle. The infiltrated leaves were collected at 3 days after infiltration and observed under a Zeiss LSM 880 laser confocal microscope (Zeiss, Jena, Germany). Cell nuclei were stained with 4,6‐diamidino‐2‐phenylindole (DAPI). GFP was excited at 488 nm, and DAPI signals were analysed at 405 nm.
Agrobacterium tumefaciens infiltration assays for the suppression of BAX/INF1‐induced programmed cell death
The coding sequence of Sg2204 excluding signal peptide was cloned into pGR107, and A. tumefaciens GV3101 carrying pGR107‐Sg2204 constructs was infiltrated into N. benthamiana leaves as described above. A. tumefaciens cells carrying BAX/INF1 were infiltrated into the same site 24 h later. The photos of leaf symptoms were taken 3 days after inoculation with BAX/INF1. The leaves were decolorized using ethanol. Three biological replicates were performed for each treatment. A. tumefaciens cells carrying GFP were used as negative controls.
Bacterial T3SS‐mediated overexpression in wheat plants
The coding sequence of Sg2204 and its orthologs with the predicted signal peptide excluded and the final stop codon included were amplified using Phusion High‐Fidelity DNA Polymerase (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. The PCR products were inserted into the entry vector pDONER221 by BP cloning and then introduced into the Gateway destination vector pEDV6 by an LR clonase reaction according to the manufacturer's instructions. The inserted PCR products were confirmed by sequencing. The primers used in this study are listed in Table S4.
The pEDV6:Sg2204 construct was transformed into the P. fluorescens strain EtHAn by electroporation. For the infiltration of the leaves, recombinant strains of EtHAn were grown in KB liquid medium (25 μg/mL rifampicin, 30 μg/mL gentamicin) for 24 h and resuspended in 10 mM MgCl2 solution. EtHAn suspensions (OD600 = 1.5) were infiltrated into the second leaves of wheat seedlings at the two‐leaf stage using a syringe without a needle. The infiltrated plants were grown and maintained in a climate chamber at 20 ± 1 °C for 2 days.
Detection of callose deposition and H2O2 accumulation in wheat leaves
Callose deposition in wheat leaves was visualized according to previously reported protocols (Hood and Shew, 1996). Callose deposition was observed and photographed with an Echo Revolve Hybrid Microscope (Echo Laboratories, San Diego, CA) using a DAPI filter. Thirty sites (1 mm2/site) were selected from infiltrated areas of each treated leaf, and the number of callose deposits was counted from each site. Three independent biological replicates were conducted. H2O2 accumulation was examined in wheat leaves by 3′‐diaminobenzidine (DAB) staining according to the histochemical methods described by Zhao et al. (2018). The stained leaves were imaged using an Olympus SZX‐16 (Olympus Corporation, Tokoyo, Japan). The endogenous H2O2 content in the wheat leaves was determined using the protocols reported by Ferguson et al. (1983).
RT‐qPCR analysis
The expression levels of candidate effector genes in aphid tissues and feeding periods on wheat plants were examined using RT‐qPCR analysis. RT‐qPCR was performed on an ABI 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA) as described previously (Zhang et al., 2017). Changes in the expression levels of genes involved in the JA and SA defence signalling pathways, including FAD, LOX, PAL and PR1, in wheat after infiltration with Sg2204 or infestation with Sg2204‐silenced aphids were detected. All primers for RT‐qPCR are presented in Table S4. All reactions were performed with three biological replicates, and the differential expression was calculated using the method (Livak and Schmittgen, 2002).
Measurement of phytohormones in wheat leaves
The phytohormones were analysed as described previously with some modifications (Xiang et al., 2011). In brief, 0.5 g of wheat leaves infiltrated with P. fluorescens EtHAn carrying Sg2204 or the empty vector at 2 days were harvested and homogenized in liquid nitrogen. Phytohormone extraction was performed by adding 10 mL of ethyl acetate, followed by ultrasound extraction for 20 min and centrifugation at 16000 g for 20 min at 4 °C. The supernatants were evaporated under a stream of nitrogen at 40 °C. The final extracts were redissolved in 1 mL of 70% methanol and centrifuged at 16000 g for 10 min. The supernatants were then collected and used as samples for further analysis with a liquid chromatography–tandem mass spectrometry system (LC–MS/MS). Three biological replicates were performed.
Aphid performance on wheat infiltrated with P. fluorescens EtHAn carrying Sg2204 and its homologues
After 2 days of P. fluorescens EtHAn infiltration, 10 apterous adult aphids were transferred to the infiltration sites of wheat leaves and restricted to a clip cage (2.5 × 2.5 × 2.5 cm). The number of nymphs produced by the aphids on each plant was recorded daily for 5 days, and 15 replicates were conducted in each group. Ten newborn nymphs were reared on wheat leaves infiltrated with P. fluorescens EtHAn, and each aphid was weighed on a microbalance (Sartotius, GPC, Germany) after 7 days. Fifteen replicates were performed for each group. The wheat plants were replaced by new seedlings after 4 days; these seedlings received the same treatment as above. Wheat plants treated with P. fluorescens EtHAn carrying an empty vector were used as controls.
RNAi application through the spray method of the transdermal dsRNA delivery system
The dsRNA specific for Sg2204 and the homologous genes was synthesized and purified using a TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, CA) according to the manufacturer's instructions with the primers listed in Table S4. All dsRNA sequences were labelled using the Fluorescein RNA Labeling Mix Kit (Sigma‐Aldrich, St Louis, MO). The star polycation (SPc) nanocarrier was gently mixed with dsRNA at a recommended mass ratio of 1 : 1, and 0.05% detergent (surfactant and softened water) was then added to reduce the surface tension of hydrophilic nanocomplexes (Li et al., 2019; Yan et al., 2020). The dsRNA/nanocarrier/detergent formulations with 200 ng/μL dsRNA were sprayed on the aphids and leaf surface. Aphids were then collected at 24 or 48 h after treatment with dsRNA to detect the transcript levels of target genes as described above. The primers used for RT‐qPCR are presented in Table S4.
To investigate the uptake of fluorescein labellling dsRNA in S. graminum, aphids were washed in distilled water for five times, and then the salivary glands and midguts of aphids were dissected in cold phosphate buffered saline (pH 7.2, HyClone, Thermo Scientific, MA) using a microscope (Olympus SZX16). The fluorescent signal in tissues was observed at 12 h after the spray of fluorescein‐labelled dsRNA/nanocarrier/detergent solution on aphids using a fluorescence microscope (Olympus BX63). Aphids sprayed with a H2O/nanocarrier/detergent formulation were used as a control.
To examine the effects of gene silencing on aphid performance, ten apterous adult aphids were transferred to each plant and sprayed with 200 ng/μL dsRNA. The number of surviving aphids on plants was counted after 6 days of dsRNA treatment, and ten biological replicates were performed. Five apterous adult aphids on each plant were also treated with dsRNA, and the number of nymphs produced by the aphids was recorded daily for 5 days. Fifteen replicates were conducted for each group. DsGFP was included as the control.
Aphid feeding behaviour monitoring by electronic penetration graph (EPG)
The feeding behaviour of the adult apterous aphids on wheat plants was recorded using EPG (DC‐EPG Giga‐8). EPG monitoring was recorded continuously for 8 h in a Faraday cage in the laboratory (20 ± 1 °C, L : D = 16 : 8 photoperiod, 70% RH). Each aphid and plant were used only once. The visualization and manual labeling of the various feeding waves were carried out using Stylet+a. The characteristics of the aphid feeding waveform patterns were identified as described previously (Prado and Tjallingii, 1994). At least 10 independent biological replicates were conducted for each treatment.
Statistical analysis
All data were analysed using SPSS Statistics 20.0 software (SPSS Inc., Chicago, IL). The differences among groups were examined using Student's t test and one‐way analysis of variance (ANOVA) test, followed by Duncan's multiple range test. EPG data were analysed by a Mann–Whitney U test. P values < 0.05 were considered to be statistically significant.
Conflicts of interests
The authors declare no conflict of interest.
Author contributions
J. L. C. and Y. Z. conceived and designed the research. Y. Z., X. B. L. and Y. S. carried out the experiment. Y. Z., F. J., Q. W. and H. L. analysed the data. Y. Z. and J. L. C. wrote the manuscript. F. F., J. L. C. and H. C. X. revised the manuscript. All authors read and approved the final manuscript.
Supporting information
Figure S1 Length distribution of transcripts and unigenes in transcriptome assembly for S. graminum salivary glands.
Figure S2 Results of similarity search of unigenes against Nr database.
Figure S3 Functional annotation of unigenes from S. graminum salivary glands using Gene Ontology (GO).
Figure S4 Metabolic pathway analysis of by unigenes from S. graminum salivary glands using Kyoto Encyclopedia of Genes and Genomes (KEGG).
Figure S5 RT‐qPCR results of the relative gene expression of nine candidate effectors in different tissues of S. graminum.
Table S1 The quality of S. graminum salivary glands unigene sequences and assembly.
Table S2 Number of unigenes annotated in seven public databases.
Table S3 Identification of watery salivary proteins of S. graminum using LC–MS/MS.
Table S4 All primers used in this study.
Data S1 All unigenes of salivary gland of Schizaphis graminum.
Acknowledgements
We thank Prof. Dr. Zhensheng Kang (State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A & F University) for providing the pEDV6 vector, pEDV6:AvrRpt2 constructs, and Pseudomonas fluorescens effector‐to‐host analyser (EtHAn) strain, and Prof. Dr. Jie Shen and Assoc. Prof. Dr. Shuo Yan (Department of Entomology and MOA Key Laboratory for Monitory and Green Control of Crop Pest, China Agricultural University) for providing star polycation nanocarrier. This study was financially supported by the National Natural Science Foundation of China (Nos. 31901881 and 31871979), China's Donation to the CABI Development Fund (IVM10051).
References
- Abdellatef, E. , Will, T. , Koch, A. , Imani, J. , Vilcinskas, A. and Kogel, K.H. (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae . Plant Biotechnol. J. 13, 849–857. [DOI] [PubMed] [Google Scholar]
- Ali, J. , Covaci, A.D. , Roberts, J.M. , Sobhy, I.S. , Kirk, W.D.J. and Bruce, T.J.A. (2021) Effects of cis‐jasmone treatment of Brassicas on interactions with Myzus persicae aphids and their parasitoid Diaeretiella rapae . Front. Plant Sci. 12, 711896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljaryian, R. and Kumar, L. (2016) Changing global risk of invading greenbug Schizaphis graminum under climate change. Crop Prot. 88, 137–148. [Google Scholar]
- Al‐Mousawi, A.H. , Richardson, P.E. and Burton, R.L. (1983) Ultrastructural studies of greenbug (Hemiptera: Aphididae) feeding damage to susceptible and resistant wheat cultivars. Ann. Entomol. Soc. Am. 76, 964–971. [Google Scholar]
- Atamian, H.S. , Chaudhary, R. , Cin, V.D. , Bao, E. , Girke, T. and Kaloshian, I. (2013) In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant Microbe Interact. 26, 67–74. [DOI] [PubMed] [Google Scholar]
- Bai, X.X. , Zhan, G.M. , Tian, S.X. , Peng, H. , Cui, X.Y. , Islam, M.A. , Goher, F. et al. (2021) Transcription factor BZR2 activates chitinase Cht20.2 transcription to confer resistance to wheat stripe rust. Plant Physiol. 187, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.L. and Eastop, V.F. (2000) Aphids on the World's Crops: An Identification and Information Guide. New York, NY: John Wiley & Sons Ltd. [Google Scholar]
- Bos, J.I.B. , Prince, D.C. , Pitino, M. , Maffei, M.E. , Win, J. and Hogenhout, S.A. (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (Green peach aphid). PLoS Genet. 6, e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton, A.J. , Hoover, K. and Felton, G.W. (2006) Impact of chemical elicitor applications on greenhouse tomato plants and population growth of the green peach aphid, Myzus persicae . Entomol. Exp. Appl. 120, 175–188. [Google Scholar]
- Bruce, T.J.A. , Martin, J.L. , Pickett, J.A. , Pye, B.J. , Smart, L.E. and Wadhams, L.J. (2003) cis‐Jasmone treatment induces resistance in wheat plants against the grain aphid, Sitobion avenae (Fabricius) (Homoptera: Aphididae). Pest Manag. Sci. 59, 1031–1036. [DOI] [PubMed] [Google Scholar]
- Chakraborty, J. , Sen, S. , Ghosh, P. , Jain, A. and Das, S. (2020) Inhibition of multiple defense responsive pathways by CaWRKY70 transcription factor promotes susceptibility in chickpea under Fusarium oxysporum stress condition. BMC Plant Biol. 20, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, R. , Atamian, H.S. , Shen, Z. , Briggs, S.P. and Kaloshian, I. (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. USA, 111, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, R. , Peng, H.C. , He, J. , MacWilliams, J. , Teixeira, M. , Tsuchiya, T. , Chesnais, Q. et al. (2019) Aphid effector Me10 interacts with tomato TFT7, a 14‐3‐3 isoform involved in aphid resistance. New Phytol. 221, 1518–1528. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.L. , Wu, K. , Yao, J.N. , Li, S.M. , Wang, X.J. , Huang, L.L. and Kang, Z.S. (2017) PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 19, 1717–1729. [DOI] [PubMed] [Google Scholar]
- Chung, S.H. , Feng, H. and Jander, G. (2021) Engineering pest tolerance through plant‐mediated RNA interference. Curr. Opin. Plant Biol. 60, 102029. [DOI] [PubMed] [Google Scholar]
- Cui, N. , Lu, H. , Wang, T. , Zhang, W. , Kang, L. and Cui, F. (2019) Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philos. Trans. R. Soc. Lond. B Biol. Sci. 4, 20180314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos, M. and Jander, G. (2009) Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana . Plant Cell Environ. 32, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Despres, C. , Subramaniam, R. , Matton, D.P. and Brisson, N. (1995) The activation of the potato PR‐l0a gene requires the phosphorylation of the nuclear factor PBF‐1. Plant Cell, 7, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y.M. , Jing, M.F. , Shen, D.Y. , Wang, C.Y. , Zhang, M.Q. , Liang, D. , Nyawira, K.T. et al. (2020) The mirid bug Apolygus lucorum deploys a glutathione peroxidase as a candidate effector to enhance plant susceptibility. J. Exp. Bot. 71, 2701–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, D. and Zhou, J.M. (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe, 12, 484–495. [DOI] [PubMed] [Google Scholar]
- Drurey, C. , Mathers, T.C. , Prince, D.C. , Wilson, C. , Caceres‐Moreno, C. , Mugford, S.T. and Hogenhout, S.A. (2019) Chemosensory proteins in the CSP4 clade evolved as plant immunity suppressors before two suborders of plant‐feeding hemipteran insects diverged. BioRxiv, 173278.
- Elzinga, D.A. , De Vos, M. and Jander, G. (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 27, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero‐Martinez, C. , Rodriguez, P.A. , Liu, S. , Santos, P.A. , Stephens, J. and Bos, J.I.B. (2020) An aphid effector promotes barley susceptibility through suppression of defence gene expression. J. Exp. Bot. 71, 2796–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J.L. , Zhang, J. , Yang, J. , Zou, L.P. , Fang, T.T. , Xu, H.L. and Cai, Q.N. (2021) Exogenous salicylic acid improves resistance of aphid‐susceptible wheat to the grain aphid, Sitobion avenae (F.) (Hemiptera: Aphididae). Bull. Entomol. Res. 111, 544–552. [DOI] [PubMed] [Google Scholar]
- Ferguson, I.B. , Watkins, C.B. and Harman, J.E. (1983) Inhibition by calcium of senescence of detached cucumber cotyledons effect on ethylene and hydroperoxide production. Plant Physiol. 71, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo, M.C. and Valent, B. (2013) Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Guo, H.J. , Zhang, Y.J. , Tong, J.H. , Ge, P.P. , Wang, Q.Y. , Zhao, Z.H. , Zhu‐Salzman, K.Y. et al. (2020) An aphid‐secreted salivary protease activates plant defense in phloem. Curr. Biol. 30, 4826–4836. [DOI] [PubMed] [Google Scholar]
- Harmel, N. , Létocart, E. , Cherqui, A. , Giordanengo, P. , Mazzucchelli, G. , Guillonneau, F. , De Pauw, E. et al. (2008) Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae . Insect Mol. Biol. 17, 165–174. [DOI] [PubMed] [Google Scholar]
- Hogenhout, S.A. and Bos, J.I.B. (2011) Effector proteins that modulate plant‐insect interactions. Curr. Opin. Plant Biol. 14, 422–428. [DOI] [PubMed] [Google Scholar]
- Hood, M.E. and Shew, H.D. (1996) Applications of KOH‐aniline blue fluorescence in the study of plant‐fungal interactions. Phytopathology, 86, 704–708. [Google Scholar]
- Ji, R. , Fu, J.M. , Shi, Y. , Li, J. , Jing, M.F. , Wang, L. , Yang, S.Y. et al. (2021) Vitellogenin from planthopper oral secretion acts as a novel effector to impair plant defenses. New Phytol. 232, 802–817. [DOI] [PubMed] [Google Scholar]
- Jiang, C. , Hei, R.N. , Yang, Y. , Zhang, S.J. , Wang, Q.H. , Wang, W. , Zhang, Q. et al. (2020) An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 11, 4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaloshian, I. and Walling, L.L. (2005) Hemipterans as plant pathogens. Annu. Rev. Phytopathol. 43, 491–521. [DOI] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate mediated signals in plant defense. Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Fu, Y. , Liu, X. , Sun, J. , Hou, M. , Zhang, Y. and Chen, J.L. (2022a) Activation of wheat defense response by Buchnera aphidicola‐derived small chaperone protein GroES in wheat aphid saliva. J. Agric. Food Chem. 70, 1058–1067. [DOI] [PubMed] [Google Scholar]
- Li, M. , Ma, Z. , Peng, M. , Li, L. , Yin, M. , Yan, S. and Shen, J. (2022b) A gene and drug co‐delivery application helps to solve the short life disadvantage of RNA drug. Nano Today, 43, 101452. [Google Scholar]
- Li, J. , Qian, J. , Xu, Y. , Yan, S. , Shen, J. and Yin, M. (2019) A facile‐synthesized star polycation constructed as a highly efficient gene vector in pest management. ACS Sustain. Chem. Eng. 7, 6316–6322. [Google Scholar]
- Li, Q. , Xie, Q. , Smith‐Becker, J. , Navarre, D.A. and Kaloshian, I. (2006) Mi‐1Mediated aphid resistance involves salicylic acid and mitogen‐activated protein kinase signaling cascades. Mol. Plant Microbe Interact. 19, 655–664. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2002) Analysis of relative gene expression data using real‐time quantitative PCR. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Zhang, Y.H. , Li, M.S. , Chao, Z.J. , Du, X.G. , Yan, S. and Shen, J. (2022) A first greenhouse application of bacteria‐expressed and nanocarrier‐delivered RNA pesticide for Myzus persicae control. J. Pest Sci. 10.1007/s10340-022-01485-5 [DOI] [Google Scholar]
- Mindrinos, M. , Katagiri, F. , Yu, G.L. and Ausubel, F.M. (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide‐binding site and leucine‐rich repeats. Cell, 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. and Staskawicz, B.J. (1999) Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32, 927–941. [DOI] [PubMed] [Google Scholar]
- Mugford, S.T. , Barclay, E. , Drurey, C. , Findlay, K.C. and Hogenhout, S.A. (2016) An immuno‐suppressive aphid saliva protein is delivered into the cytosol of plant mesophyll cells during feeding. Mol. Plant Microbe Interact. 29, 854–861. [DOI] [PubMed] [Google Scholar]
- Mutti, N.S. , Louis, J. , Pappan, L.K. , Begum, K. , Chen, M.S. , Park, Y. , Dittmer, N. et al. (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA, 105, 9965–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti, N.S. , Park, Y. , Reese, J.C. and Reeck, G.R. (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . J. Insect Sci. 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.P. , Chakravarthy, S. , Velásquez, A.C. , McLane, H.L. , Zeng, L. , Nakayashiki, H. , Park, D.H. et al. (2010) Methods to study PAMP‐triggered immunity using tomato and Nicotiana benthamiana . Mol. Plant Microbe Interact. 23, 991–999. [DOI] [PubMed] [Google Scholar]
- Nicholson, S.J. and Puterka, G.J. (2014) Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum Rondani) biotypes. J. Proteomics, 105, 186–203. [DOI] [PubMed] [Google Scholar]
- Niu, D. , Hamby, R. , Sanchez, J.N. , Cai, Q. , Yan, Q. and Jin, H. (2021) RNAs‐ a new frontier in crop protection. Curr. Opin. Biotech. 70, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino, M. , Coleman, A.D. , Maffei, M.E. , Ridout, C.J. and Hogenhout, S.A. (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS One, 6, e25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino, M. and Hogenhout, S.A. (2013) Aphid protein effectors promote aphid colonization in a plant species‐specific manner. Mol. Plant Microbe Interact. 26, 130–139. [DOI] [PubMed] [Google Scholar]
- Prado, E. and Tjallingii, W.F. (1994) Aphid activities during sieve element punctures. Entomol. Exp. Appl. 72, 157–165. [Google Scholar]
- Qiao, L. , Lan, C. , Capriotti, L. , Ah‐Fong, A. , Nino Sanchez, J. , Hamby, R. , Heller, J. et al. (2021) Spray‐induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, S.K. , Weng, Y. , Rudd, J.C. , Akhunova, A. and Liu, S.Y. (2013) Transcriptomics of induced defense responses to greenbug aphid feeding in near isogenic wheat lines. Plant Sci. 212, 26–36. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P.A. and Bos, J.I.B. (2013) Toward understanding the role of aphid effectors in plant infestation. Mol. Plant Microbe Interact. 26, 25–30. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P.A. , Stam, R. , Warbroek, T. and Bos, J.I.B. (2014) Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant Microbe Interact. 27, 30–39. [DOI] [PubMed] [Google Scholar]
- Schuman, M.C. and Baldwin, I.T. (2016) The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 61, 373–394. [DOI] [PubMed] [Google Scholar]
- Su, Q. , Peng, Z.K. , Tong, H. , Xie, W. , Wang, S.L. , Wu, Q. , Zhang, J.M. et al. (2019) A salivary ferritin in the whitefly suppresses plant defenses and facilitates host exploitation. J. Exp. Bot. 70, 3343–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Sparks, C. , Jones, H. , Riley, M. , Francis, F. , Du, W. and Xia, L. (2019) Silencing an essential gene involved in infestation and digestion in grain aphid through plant‐mediated RNA interference generates aphid‐resistant wheat plants. Plant Biotechnol. J. 17, 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. and Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Upadhyaya, N.M. , Mago, R. , Staskawicz, B.J. , Ayliffe, M.A. , Ellis, J.G. and Dodds, P.N. (2014) A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant Microbe Interact. 27, 255–264. [DOI] [PubMed] [Google Scholar]
- van Bel, A.J. and Will, T. (2016) Functional evaluation of proteins in watery and gel saliva of aphids. Front. Plant Sci. 7, e1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, L.Y. , Zhang, L.J. , Liu, N. , Gao, X.W. and Liu, X.M. (2021) Effect of RNAi targeting CYP6CY3 on the growth, development and insecticide susceptibility of Aphis gossypii by using nanocarrier‐based transdermal dsRNA delivery system. Pestic. Biochem. Physiol. 177, 104878. [DOI] [PubMed] [Google Scholar]
- Will, T. and van Bel, A.J.E. (2006) Physical and chemical interactions between aphids and plants. J. Exp. Bot. 57, 729–737. [DOI] [PubMed] [Google Scholar]
- Xiang, Y. , Song, M. , Wei, Z.Y. , Tong, J.H. , Zhang, L.X. , Xiao, L.T. , Ma, Z.Q. et al. (2011) A jacalin‐related lectin‐like gene in wheat is a component of the plant defence system. J. Exp. Bot. 62, 5471–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H.X. , Qian, L.X. , Wang, X.W. , Shao, R.X. , Hong, Y. , Liu, S.S. and Wang, X.W. (2019a) A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid‐signaling pathway. Proc. Natl. Acad. Sci. USA, 116, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Tang, C.L. , Wang, X.D. , Sun, S.T. , Zhao, J.R. , Kang, Z.S. and Wang, X.J. (2019b) An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10, 5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S. , Qian, J. , Cai, C. , Ma, Z.Z. , Li, J.H. , Yin, M.Z. , Ren, B.Y. et al. (2020) Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines . J. Pest Sci. 93, 449–459. [Google Scholar]
- Yates, A.D. and Michel, A. (2018) Mechanisms of aphid adaptation to host plant resistance. Curr. Opin. Insect Sci. 26, 41–49. [DOI] [PubMed] [Google Scholar]
- Yin, C. and Hulbert, S. (2011) Prospects for functional analysis of effectors from cereal rust fungi. Euphytica, 179, 57–67. [Google Scholar]
- Zhang, Y. , Fan, J. , Francis, F. and Chen, J.L. (2017) Watery saliva secreted by the grain aphid Sitobion avenae stimulates aphid resistance in wheat. J. Agric. Food Chem. 65, 8798–8805. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Fu, Y. , Fan, J. , Li, Q. , Francis, F. and Chen, J.L. (2019) Comparative transcriptome and histological analyses of wheat in response to phytotoxic aphid Schizaphis graminum and non‐phytotoxic aphid Sitobion avenae feeding. BMC Plant Biol. 19, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Fu, Y. , Wang, Q. , Liu, X.B. , Li, Q. and Chen, J.L. (2020) Transcriptome analysis reveals rapid defence responses in wheat induced by phytotoxic aphid Schizaphis graminum feeding. BMC Genomics, 21, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.H. , Ma, Z.Z. , Zhou, H. , Chao, Z.J. , Yan, S. and Shen, J. (2022) Nanocarrier‐delivered dsRNA suppresses wing development of green peach aphids. Insect Sci. 9, 669–682. [DOI] [PubMed] [Google Scholar]
- Zhao, M.X. , Wang, J. , Ji, S. , Chen, Z. , Xu, J. , Tang, C. , Chen, S. et al. (2018) Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici . Front. Plant Sci. 9, 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu‐Salzman, K.Y. , Salzman, R.A. , Ahn, J.E. and Koiwa, H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem‐feeding aphid. Plant Physiol. 134, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst, T. and Agrawal, A.A. (2016) Mechanisms and evolution of plant resistance to aphids. Nat. Plants, 2, 15206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Length distribution of transcripts and unigenes in transcriptome assembly for S. graminum salivary glands.
Figure S2 Results of similarity search of unigenes against Nr database.
Figure S3 Functional annotation of unigenes from S. graminum salivary glands using Gene Ontology (GO).
Figure S4 Metabolic pathway analysis of by unigenes from S. graminum salivary glands using Kyoto Encyclopedia of Genes and Genomes (KEGG).
Figure S5 RT‐qPCR results of the relative gene expression of nine candidate effectors in different tissues of S. graminum.
Table S1 The quality of S. graminum salivary glands unigene sequences and assembly.
Table S2 Number of unigenes annotated in seven public databases.
Table S3 Identification of watery salivary proteins of S. graminum using LC–MS/MS.
Table S4 All primers used in this study.
Data S1 All unigenes of salivary gland of Schizaphis graminum.


