Summary
Crocins are beneficial antioxidants and potential chemotherapeutics that give raise, together with picrocrocin, to the colour and taste of saffron, the most expensive spice, respectively. Crocins are formed from crocetin dialdehyde that is produced in Crocus sativus from zeaxanthin by the carotenoid cleavage dioxygenase 2L (CsCCD2L), while GjCCD4a from Gardenia jasminoides, another major source of crocins, converted different carotenoids, including zeaxanthin, into crocetin dialdehyde in bacterio. To establish a biotechnological platform for sustainable production of crocins, we investigated the enzymatic activity of GjCCD4a, in comparison with CsCCD2L, in citrus callus engineered by Agrobacterium‐mediated supertransformation of multi genes and in transiently transformed Nicotiana benthamiana leaves. We demonstrate that co‐expression of GjCCD4a with phytoene synthase and β‐carotene hydroxylase genes is an optimal combination for heterologous production of crocetin, crocins and picrocrocin in citrus callus. By profiling apocarotenoids and using in vitro assays, we show that GjCCD4a cleaved β‐carotene, in planta, and produced crocetin dialdehyde via C30 β‐apocarotenoid intermediate. GjCCD4a also cleaved C27 β‐apocarotenoids, providing a new route for C17‐dialdehyde biosynthesis. Callus lines overexpressing GjCCD4a contained higher number of plastoglobuli in chromoplast‐like plastids and increased contents in phytoene, C17:0 fatty acid (FA), and C18:1 cis‐9 and C22:0 FA esters. GjCCD4a showed a wider substrate specificity and higher efficiency in Nicotiana leaves, leading to the accumulation of up to 1.6 mg/g dry weight crocins. In summary, we established a system for investigating CCD enzymatic activity in planta and an efficient biotechnological platform for crocins production in green and non‐green crop tissues/organs.
Keywords: gardenia, crocus, carotenoid cleavage dioxygenase, metabolic engineering, crocins, apocarotenoid
GjCCD4a is an efficient tool for biotechnological production of crocins in green and non‐green plant tissues. It catalyses a new route for crocetin dialdehyde formation starting with β‐carotene and proceeding via C30 β‐apo‐8′‐carotenal intermediates.

Introduction
Carotenoids represent the largest class of natural isoprenoid pigments and are synthesized by all photosynthetic organisms and many non‐photosynthetic microorganisms (Moise et al., 2014; Rodriguez‐Concepcion et al., 2018). Humans cannot form carotenoids de novo, but need them as essential phytonutrients due to their provitamin A activity and antioxidant properties (Zheng et al., 2020a). Carotenoids are also responsible for the colour of fruits and flowers of many plant species, attracting animals for seed dispersal and pollination (Yuan et al., 2015). More importantly, carotenoids are essential for the photosynthetic apparatus, protecting it from photo‐oxidation and contributing to light harvesting (Hashimoto et al., 2016).
Carotenoids are susceptible to oxidation processes that cleave their conjugated double bond system at different sites, leading to diverse metabolites called apocarotenoids, which include pigments, volatiles, growth regulators, such as anchorene, β‐cyclocitral and zaxinone, and precursors of the phytohormones strigolactones (SLs) and abscisic acid (ABA) (Hou et al., 2016; Moreno et al., 2021; Schwartz et al., 1997; Wang et al., 2019). Apocarotenoid formation in plants is generally catalysed by carotenoid cleavage dioxygenases that are classified into two groups: the 9‐cis‐epoxycarotenoid dioxygenases (NCEDs) and carotenoid cleavage dioxygenases (CCD) (Auldridge et al., 2006; Giuliano et al., 2003). NCEDs catalyse the formation of the ABA precursors xanthoxin by cleaving 9′‐cis‐neoxanthin and/or 9‐cis‐violaxanthin (Schwartz et al., 1997). In Arabidopsis, CCDs are divided into four subfamilies: CCD1, CCD4, CCD7 and CCD8 (Jia et al., 2018; Walter and Strack, 2011). There are further CCD clades, such as CCD2L and zaxinone synthase (ZAS) that are present in the Crocus species, and mycorrhizal plants, respectively (Ahrazem et al., 2016; Frusciante et al., 2014; Wang et al., 2019). Plant CCD subfamilies are distinguished by their substrate specificity and cleavage sites, indicating different biological functions (Zheng et al., 2021b). CCD7 and CCD8 act sequentially to transform 9‐cis‐β‐carotene into carlactone, the precursor of the phytohormone SL (Al‐Babili and Bouwmeester, 2015; Alder et al., 2012). ZAS converts 3‐OH‐β‐apo‐10′‐carotenal into the growth regulator zaxinone (Wang et al., 2019). CCD1s have very relaxed substrate‐ and cleavage site specificity, mediating oxidative cleavage of many carotenoids and apocarotenoids at different sites, which leads to a plentitude of volatiles and dialdehyde products (Ilg et al., 2014; Simkin et al., 2004). The Crocus sativus CsCCD2L is closely related to the cytoplasm‐localized CCD1 subfamily, but it is localized in plastids. CsCCD2L cleaves both C7–C8/C7′–C8′ sites of zeaxanthin to produce crocetin dialdehyde, the precursor of crocetin and crocins (Ahrazem et al., 2016; Frusciante et al., 2014).
CCD4 enzymes are plastid localized. They are generally divided in two groups, based on their substrate and double bond specificity (Mi and Al‐Babili, 2019). For example, Arabidopsis and potato CCD4 enzymes cleave the C9′–C10′ and/or C9–C10 double bonds in bicyclic carotenoids, particularly β‐carotene, causing a decrease in carotenoid content and the production of C13 volatiles (Bruno et al., 2016; Gonzalez‐Jorge et al., 2013). The second group is represented by the Citrus CCD4b that targets the C7–C8 or C7′–C8′ site in different carotenoids, yielding C30 apocarotenoids responsible for red colour of citrus peel together with C10 volatiles (Ma et al., 2013; Rodrigo et al., 2013; Zheng et al., 2019, 2021a). The BdCCD4.1/3 from Buddleja davidii and GjCCD4a of Gardenia jasminoides are an example for a new CCD4 type that forms crocetin dialdehyde, the precursor of crocins. The activity of BdCCD4.1/3 is similar to that of the above‐mentioned CsCCD2L, producing crocetin dialdehyde by cleaving zeaxanthin as the only carotenoid substrate (Ahrazem et al., 2017). In contrast, GjCCD4a formed crocetin dialdehyde from lycopene, β‐carotene and zeaxanthin in carotenoid‐accumulating E. coli cells (Xu et al., 2020).
In vitro study of CsCCD2L activity indicated that crocetin dialdehyde biosynthesis occurs via two sequential reactions targeting the C7′–C8′/C7–C8 double bonds of zeaxanthin: the first cleavage forms 3‐OH‐β‐cyclocitral (C10) and 3‐OH‐β‐apo‐8′‐carotenal (C30); the C30 product is then cleaved into crocetin dialdehyde and a second 3‐OH‐β‐cyclocitral molecule (Frusciante et al., 2014). CsCCD2L cleaved also the shorter apocarotenoid 3‐OH‐β‐apo‐10′‐carotenal (C27) into C17‐dialdehyde in vitro (Frusciante et al., 2014). However, it is unclear whether CsCCD2L catalyses the latter reaction in planta. Similarly, it is still unknown whether GjCCD4a converts apocarotenoid substrates into crocetin dialdehyde, the precursor of crocins, and C17‐dialdehyde in vitro and in planta.
Crocins are the most valuable water‐soluble apocarotenoid pigments exhibiting strong colouring capacity and ranking first among natural food colourants (Gómez‐Gómez et al., 2010). Moreover, they are powerful free radical quenchers with broad range of human‐health benefits, including being neuroprotective, antitumoral, antidepressant and anti‐dementia, and having analgesic and sedative properties (Alavizadeh and Hosseinzadeh, 2014; Bukhari et al., 2018; Lopresti and Drummond, 2014; Mazidi et al., 2016). Other saffron apocarotenoids, such as picrocrocin (β‐D‐glucopyranoside of 3‐OH‐β‐cyclocitral) and crocetin (precursor of crocins), were shown to reduce the proliferation of human hepatocarcinoma adenocarcinoma and cells (Kyriakoudi et al., 2015), and to exert an anti‐inflammatory effect (Nam et al., 2010).
Due to the difficulty in obtaining crocins through chemical synthesis and the high labour costs in harvesting and processing hand‐picked stigmas of saffron and gardenia fruit, new biotechnological approaches to produce these compounds in a large amount are highly desirable (Ahrazem et al., 2016). Plant metabolic engineering holds considerable potential to create green factories for sustainable production of crocins (Ahrazem et al., 2022a,b; Martí et al., 2020). In this study, we perform functional comparison of GjCCD4a and CsCCD2L through multi‐gene supertransformation in citrus callus and transient expression in Nicotiana benthamiana leaves, aiming at identifying efficient enzyme and/or optimal multi‐gene combinations for biotechnological production of crocins and other valuable apocarotenoids in non‐green and green tissues/organs. Coupled with enzymatic assay and apocarotenoid profiling of engineered citrus callus, we provide direct evidence that the new pathway from β‐carotene to C20 crocetin dialdehyde likely proceeds via C30 β‐apo‐8′‐carotenal intermediates in planta and demonstrate a new activity of GjCCD4 in producing C17 dialdehyde. Furthermore, the engineered callus system, applied to identify an optimal combination for biotechnological production of crocetin, crocins and picrocrocin, also unravelled how perturbations of apocarotenoids/carotenoids pattern impact the carotenoid biosynthetic pathway, metabolome and plastid ultrastructure. More importantly, we found that GjCCD4a is a superior alternative to the CsCCD2L enzyme in engineering crocins and other apocarotenoids in different plant tissues, especially in N. benthamiana leaves in which transient expression of GjCCD4a triggered milligram‐scale production of crocins.
Results
Generation of metabolically engineered callus lines with distinct and stable phenotypes
First, we investigated the activity of GjCCD4a or CsCCD2L in Citrus calli. To increase the levels of carotenoid substrates and test different combinations of these pigments, we engineered multi‐transgenic callus lines expressing combinations of transgenes, including tpCrtB (a bacterial phytoene synthase fused with plastid transit peptide), OsBCH (rice β‐carotene hydroxylase), GjCCD4a or CsCCD2L. For this purpose, we used Agrobacterium‐mediated serial transformation, also referred to as supertransformation. We firstly transformed white‐coloured calli of Citrus paradisi Macf. (WT) containing a very low level of carotenoids with the plasmid pK2GW7‐TpCrtB conferring kanamycin resistance and enabling TpCrtB expression, which led to the strikingly orange‐red coloured transgenic line PSY (Figure 1a–d). Carotenoid analysis showed that the PSY line mainly accumulates carotenes (β‐carotene and α‐carotene), besides low amounts of downstream xanthophylls (Figure 1e). To increase the xanthophyll content, we supertransformed the PSY line with the plasmid pH2GW7‐OsBCH harbouring expression cassettes for hygromycin‐B resistance and OsBCH (Du et al., 2010), which resulted in the yellow‐coloured BCH/PSY line that accumulated much higher amounts of xanthophylls, but much less carotenes (Figure 1c–e). Colours and carotenoid pattern of the PSY and BCH/PSY lines remained unchanged, even after sub‐culturing for more than thirty‐six cycles, suggesting the stability required for functional studies of CCD genes.
Figure 1.
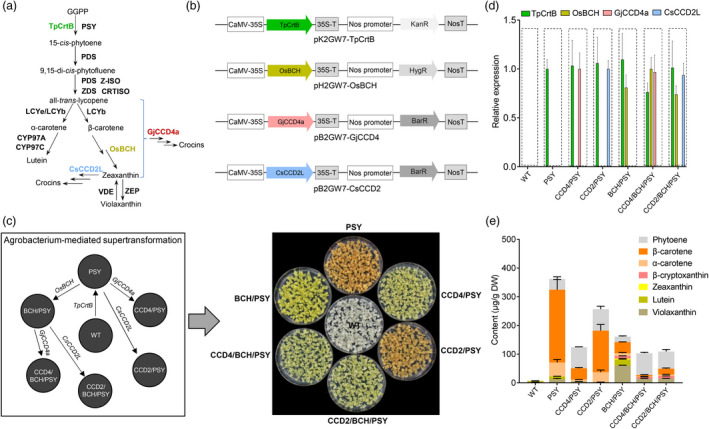
Agrobacterium‐mediated supertransformation generates various engineered callus lines with distinct colour phenotypes and carotenoid profiles. (a) Schematic carotenoid biosynthesis pathway. GGPP, geranylgeranyl diphosphate. The abbreviations of carotenoid pathway enzymes: CRTISO, carotenoid isomerase; CYP97, cytochrome P450‐type monooxygenase 97; LCYb, lycopene β‐cyclase; LCYe, lycopene ε‐cyclase; PDS, phytoene desaturase; PSY, phytoene synthase; VDE, violaxanthin de‐epoxidase; ZDS, ζ‐carotene desaturase; ZEP, zeaxanthin epoxidase; Z‐ISO, ζ‐carotene isomerase. Enzymes used in callus transformation: CsCCD2L, Crocus CCD2L; GjCCD4a, Gardenia CCD4a; OsBCH, rice β‐carotene hydroxylase; TpCrtB, bacterial phytoene synthase fused with plastid transit peptide. (b) Constructs generated and used to transform grapefruit calli. (c) Various engineered callus lines generated and their phenotypes. WT, wild‐type grapefruit callus; PSY, overexpression of tpCrtB in WT; CCD4/PSY, overexpression of GjCCD4a in PSY background; CCD2/PSY, overexpression of CsCCD2L in PSY background; BCH/PSY, overexpression of OsBCH in PSY background; CCD4/BCH/PSY, overexpression of GjCCD4a in BCH/PSY; CCD2/BCH/PSY, overexpression of CsCCD2L in BCH/PSY. (d) The relative expression levels of transgenes of engineered callus lines were quantified by Q‐RT‐PCR analysis. (e) The carotenoid composition and content of engineered callus lines were analysed by UHPLC‐DAD. The various coloured blocks represent different carotenoids as indicated in the key.
For the next supertransformation round, we relied on basta resistance for selection and used the plasmids pB2GW7‐CsCCD2L and pB2GW7‐GjCCD4 carrying expression cassettes for GjCCD4a and CsCCD2L, respectively, to transform the PSY and BCH/PSY callus, generating the supertransformed lines, CCD4/PSY (tpCrtB + GjCCD4a) and CCD4/BCH/PSY (tpCrtB + OsBCH + GjCCD4a), CCD2/PSY (tpCrtB + CsCCD2L) and CCD2/BCH/PSY (tpCrtB + OsBCH + CsCCD2L) (Figure 1c). Quantitative real‐time PCR confirmed the successful heterologous expression of the carotenoid biosynthetic (i.e. tpCrtB and OsBCH) and CCD genes in the engineered callus lines (Figures 1d and S1). As shown in Figure 1c, CCD4/PSY produced distinct pale‐yellow colour, whereas the corresponding CsCCD2L‐overexpressing line (CCD2/PSY) was similar to PSY line. Overexpression of either GjCCD4a or CsCCD2L in yellow‐coloured BCH/PSY background produced very pale‐yellow callus.
Overexpression of GjCCD4a strongly decreased the content of coloured carotenes and xanthophylls, but increased phytoene content
Carotenoid profiling showed a general reduction in total carotenoid content upon expression of either GjCCD4a or CsCCD2L in PSY background. However, the decrease in the levels of coloured carotenes (e.g. β‐carotene) in CCD2/PSY lines was much less pronounced than in CCD4/PSY lines (Figures 1e and S2), which is in agreement with the different intensity of pigmentation of the two types of lines (Figure 1c). The overexpression of either GjCCD4a or CsCCD2L in the xanthophyll‐rich line BCH/PSY dramatically decreased the levels of zeaxanthin and lutein (Figures 1e and S2), while the content of β‐cryptoxanthin was significantly reduced in three CCD4/BCH/PSY lines, but remained unchanged in the CCD2/BCH/PSY lines. These results demonstrate that GjCCD4a expression caused significantly more reduction in β‐carotene and β‐cryptoxanthin levels, compared with CsCCD2L, while the effect of both genes on di‐hydroxylated xanthophylls was similar. This observation is in line with the restricted substrate specificity of CsCCD2L in cleaving zeaxanthin and lutein (Frusciante et al., 2014) and indicates that GjCCD4a converts a wider range of carotenoids in planta. In contrast to other carotenoids, levels of the colourless phytoene were significantly increased in both GjCCD4a and CsCCD2L overexpressing lines in either PSY or BCH/PSY background (Figures 1e and S2), which indicates a positive effect of GjCCD4/CsCCD2L activity on phytoene accumulation and resembles the previously reported increase in phytoene content upon expression of CitCCD4b in citrus callus (Zheng et al., 2019).
Apocarotenoid profiling and in vitro assays uncovered the cleavage of apocarotenoids by GjCCD4a and the sequential steps in crocetin dialdehyde biosynthesis
To further investigate the cleavage activity of GjCCD4a and CsCCD2L in engineered callus system, we performed quantitative apocarotenoid profiling by using UHPLC‐Q‐Orbitrap‐MS (Mi et al., 2018). As shown in Figure 2a, all transgenic lines contained higher levels of apocarotenoids, compared with WT line, which is in line with their higher carotenoid contents (Figure 1e). The three independent CCD4/PSY lines showed significantly higher contents of β‐cyclocitral (C10) and β‐apo‐8′‐carotenal (C30), which arise through the C7′–C8′ cleavage double bond of β‐carotene, but lower levels of the C9′–C10′ cleavage product, β‐apo‐10′‐carotenal (C27), compared with PSY line (Figures 2a and S3). Moreover, crocetin dialdehyde (C20, m/z 329.1742 ([M + H]+)) was accumulated in CCD4/PSY lines, while it was almost nondetectable in the control line PSY (Figures 2a and S4). Considering that neither the PSY nor the CCD4/PSY lines contain significant amounts of zeaxanthin (Figure S2), this result suggests that GjCCD4a can also convert β‐carotene into crocetin dialdehyde in planta. To confirm this activity, we expressed GjCCD4a in β‐carotene‐ and zeaxanthin‐accumulating E. coli cells and observed the formation of crocetin dialdehyde (Figure S5), which is in line with previously reported in bacterio activity (Xu et al., 2020). As shown in Figures 1e and 2a, overexpression of CsCCD2L in PSY line also resulted in a slight decrease in β‐carotene and an increase in β‐apo‐8′‐carotenal, β‐cyclocitral and crocetin dialdehyde content, which were, however, much less pronounced compared with the overexpression of GjCCD4a. To further test whether the β‐apo‐8′‐carotenal formed by GjCCD4a in CCD4/PSY lines is an intermediate in crocetin dialdehyde biosynthesis, we incubated crude lysates of GjCCD4a producing E. coli cells with β‐apo‐8′‐carotenal in vitro. HPLC analysis confirmed the formation of crocetin dialdehyde from β‐apo‐8′‐carotenal through cleaving the C7–C8 double bond (Figure 2b). These results indicate that β‐apo‐8′‐carotenal is a substrate of GjCCD4a and that the new route of crocetin dialdehyde biosynthesis from β‐carotene by GjCCD4a occurs in two sequential steps.
Figure 2.
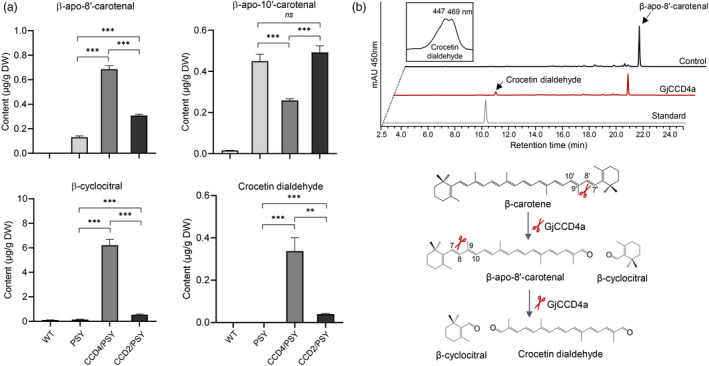
Analysis of β‐apocarotenoids and crocetin dialdehyde in different engineered callus lines and in vitro assay of GjCCD4a with C30 β‐apo‐8′‐carotenal. (a) Relative quantification of β‐apocarotenoids among various engineered callus lines. The identification and quantification of apocarotenoids were performed by UHPLC‐HR‐MS. Column names are defined in the legend of Figure 1. β‐apocarotenoid profile of other independent lines is shown in Figure S3. (b) UHPLC analysis of GjCCD4a activity and cleavage reactions for crocetin dialdehyde biosynthesis. Incubation of crude lysate of thioredoxin‐GjCCD4a expressing cells with β‐apo‐8′‐carotenal yielded C20 crocetin dialdehyde. Separation was performed in UHPLC system 2. Insets show UV/vis spectra of crocetin dialdehyde.
Analysis of the CCD4/BCH/PSY and CCD2/BCH/PSY lines, derived from the xanthophyll‐rich BCH/PSY line, unravelled a striking increase in the levels of the zeaxanthin cleavage products, 3‐OH‐β‐cyclocitral (C10) and 3‐OH‐β‐apo‐8′‐carotenal (C30) (Figures 3a and S6), accompanied by an almost complete absence of zeaxanthin (Figure S2). CCD4/BCH/PSY lines, but none of the three CCD2/BCH/PSY lines showed also a slight, but significant increase in 3‐OH‐β‐apo‐10′‐carotenal content compared with BCH/PSY line (Figures 3a and S6), consistent with a previous study showing the positive effect of CitCCD4 expression on 3‐OH‐β‐apo‐10′‐carotenal production (Zheng et al., 2021a). CCD4/BCH/PSY and CCD2/BCH/PSY lines accumulated crocetin dialdehyde, which was almost absent in the progenitor BCH/PSY (Figures 3a and S4), revealing the activities of both GjCCD4a and CsCCD2L in producing this crocin precursor in planta. We assumed that GjCCD4a cleaves the C30 intermediate 3‐OH‐β‐apo‐8′‐carotenal at the C7–C8 site to form crocetin dialdehyde, which we confirmed in in vitro assays (Figure 3b). These results indicate that GjCCD4a can convert zeaxanthin to crocetin dialdehyde through 3‐OH‐β‐apo‐8′‐carotenal, as it was previously shown in vitro for CsCCD2L (Frusciante et al., 2014).
Figure 3.
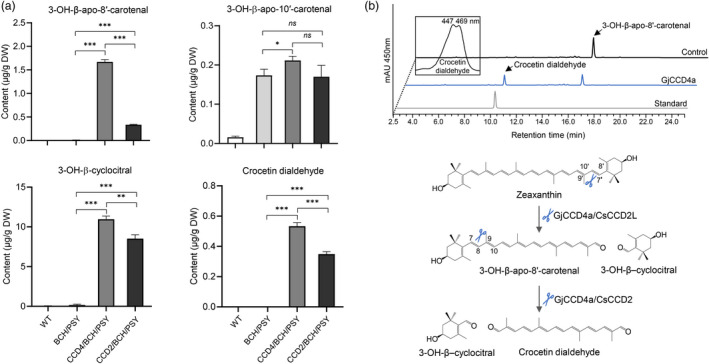
Analysis of hydroxylated apocarotenoids and crocetin dialdehyde in different engineered callus lines and in vitro assay of GjCCD4a with the C30 3‐OH‐β‐apo‐8′‐carotenal substrate. (a) Relative quantification of hydroxylated apocarotenoids among various engineered callus lines. Column names are defined in the legend of Figure 1. Analysis of hydroxylated apocarotenoids of other independent lines is shown in Figure S6. (b) UHPLC analysis of GjCCD4a activity and cleavage reactions for crocetin dialdehyde biosynthesis from 3‐OH‐β‐apo‐8′‐carotenal. Separation was performed in UHPLC system 2. The same HPLC‐DAD chromatogram of crocetin dialdehyde standard was also shown in Figure 2. The insets show UV/vis spectrum of GjCCD4a product crocetin dialdehyde.
Like CsCCD2L (Frusciante et al., 2014), our in vitro assay showed that GjCCD4a can also cleave 3‐OH‐β‐apo‐10′‐carotenal (C27), at C7–C8 site, yielding C17‐dialdehyde (m/z 257.1531 ([M + H]+)) (Figures 4a and S7), which explains the striking increase in C17‐dialdehyde observed upon overexpression of either GjCCD4a or CsCCD2L in BCH/PSY background (Figures 4b). Moreover, in vitro assay demonstrated that GjCCD4a also cleaves β‐apo‐10′‐carotenal (C27), producing a C17‐dialdehyde (Figure 4a), consistent with the decreased level of β‐apo‐10′‐carotenal and the increased content of C17‐dialdehyde in GjCCD4 overexpressing lines (Figures 2a, 4b and S3). The C17‐dialdehyde level in CCD4/PSY lines was much higher than that in CCD2/PSY lines (Figure 4b), further confirming that GjCCD4a cleaves the C7–C8 double bonds in non‐hydroxylated apocarotenoids, while CsCCD2L only targets hydroxylated apocarotenoids (Figure 4c; Frusciante et al., 2014).
Figure 4.
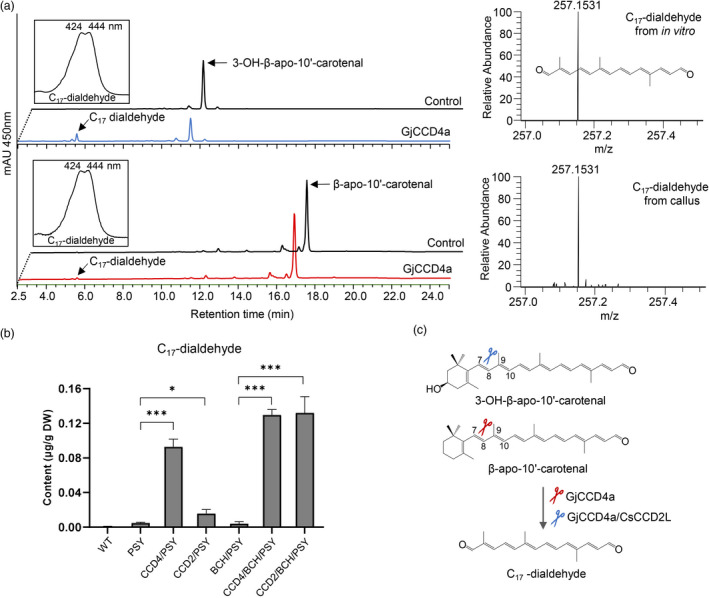
In vitro assay of GjCCD4a with C27 apocarotenoid substrates and relative quantification of C17‐dialdehyde among various engineered callus lines. (a) Incubation of crude lysate of thioredoxin‐GjCCD4a expressing cells with C27 β‐apo‐10′‐carotenal and 3‐OH‐β‐apo‐10′‐carotenal yielded a C17‐dialdehyde. Insets show UV/vis spectrum of C17‐dialdehyde product. The C17‐dialdehyde was identified by UHPLC‐HR‐MS analysis. (b) Level of C17‐dialdehyde in different engineered callus lines. Column names are defined in the legend of Figure 1. (c) Cleavage reactions catalysed by GjCCD4a for C17‐dialdehyde biosynthesis from C27 apocarotenoids.
Taken together, by integrating apocarotenoid profiling of engineered callus and in vitro assay, we uncovered the activity of GjCCD4a in cleaving apocarotenoids, which reinforces that β‐apo‐8′‐carotenal is the intermediate of a new route to crocetin dialdehyde from β‐carotene and points to the formation of C17 dialdehydes by GjCCD4.
Introduction of the CCD4 enzyme into citrus callus affected carotenoid pathway, plastid ultrastructure and levels of primary and secondary metabolites
Next, we investigated the effects of GjCCD4a expression on carotenoid pathway, cellular ultrastructure and metabolome in plant cells, using the CCD4/PSY callus lines that displayed the most pronounced phenotype in carotenoid/apocarotenoid content, compared with its control line PSY (Figure 1c–e). As shown in Figure S8, BCH transcript levels were significantly up‐regulated in both CCD4/PSY lines. This change may explain the striking increase in the content of hydroxyl apocarotenoids, such as β‐citraurinene and β‐citraurin (3‐OH‐β‐apo‐8′‐carotenal), in CCD4/PSY lines, compared with their PSY background, which accumulates only trace amounts of xanthophyll substrates (Figures S2 and S9). Transcripts of LCYB2 and CRTISO were significantly up‐regulated in only one of the two CCD4/PSY lines. We did not observe significant alterations in the expression of other carotenoid biosynthetic genes.
Next, we used transmission electron microscopy (TEM) to determine the effects of the transgenes at the cellular level. As shown in Figure 5a, the carotene‐rich PSY line showed chromoplast‐like structure containing visible plastoglobule, carotenoid crystals and characteristic internal membrane. The introduction of GjCCD4a in PSY background resulted in increased numbers of plastoglobuli within the chromoplast‐like plastids in CCD4/PSY line (Figure 5a), which might sequester the enhanced phytoene content (Figure S2), as described previously (Nogueira et al., 2013), and/or newly synthesized C30 apocarotenoids (Figure S9). Furthermore, GjCCD4a expression led to a clear reduction in carotenoid crystals, which is in agreement with the dramatic decrease in the content of β‐carotene assumed accumulate as carotenoid crystals in citrus callus (Cao et al., 2012).
Figure 5.
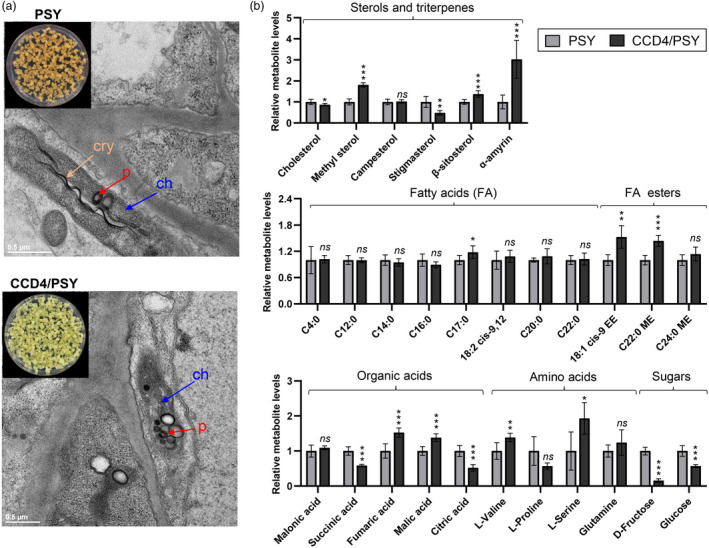
Transmission electron microscope images and GC–MS analysis of different engineered callus lines. (a) Changes in callus plastid ultrastructure resulting from the expression of GjCCD4a. Cry, carotenoid crystal and characteristic internal membrane; ch, chromoplast; p, plastoglobule. (b) Changes in metabolite pools of engineered callus resulting from the expression of GjCCD4a. EE, ethyl ester; ME, methyl ester. Different coloured blocks represent different engineered callus lines as indicated in the key.
The cellular ultrastructural differences between PSY and CCD4/PSY lines indicate that the overexpression of GjCCD4a may have caused metabolome perturbations beyond carotenoid/apocarotenoid pattern. To investigate this possibility, we performed a global GC–MS‐based metabolome analysis. As shown in Figure 5b, we observed in CCD4/PSY a significant increase in methyl sterol, β‐sitosterol and α‐amyrin, but a decrease in cholesterol and stigmasterol, compared with PSY line (Figure 5b). The GjCCD4a overexpressing CCD4/PSY line contained higher levels of heptadecanoic acid (C17:0), cis‐9‐octadecenoic acid (C18:1 cis‐9) ethyl ester (EE) and behenic acid (C22:0) methyl ester (ME) compared with PSY line. With respect to organic acids, CCD4/PSY line displayed significant higher levels of fumaric acid and malic acid, but contained significantly lower amounts of succinic acid and citric acid. The amounts of amino acids, L‐valine and L‐serine were significantly higher in CCD4/PSY, relative to PSY line. The sugars, D‐fructose and glucose showed a significant reduction upon the overexpression of GjCCD4a in PSY background. Overall, the overexpression of GjCCD4a had broad effects across the metabolome in citrus callus.
Determining the optimal multi‐gene metabolic engineering approach for production of crocetin, crocins and picrocrocin in citrus callus
Apocarotenoids are frequently subjected to enzymatic modifications in plant cells (Diretto et al., 2019; Koschmieder et al., 2021), and hence, we speculated that crocetin dialdehyde produced by the introduced CCDs may be converted into crocetin and crocins as final products. Using UHPLC‐DAD/HRMS analysis and an authentic standard, we identified crocetin in GjCCD4a or CsCCD2L‐overexpressing callus lines (Figures 6a and S10). The contents of crocetin were 9.88 ± 2.10, 2.96 ± 0.44, 2.67 ± 0.32 and 0.59 ± 0.02 μg/g DW in CCD4/BCH/PSY, CCD2/BCH/PSY, CCD4/PSY and CCD2/PSY, respectively (Figure 6a). We did not detect crocetin in WT, PSY and BCH/PSY lines. These differences in crocetin content are in agreement with variations in levels of its precursor crocetin dialdehyde in the different engineered callus lines (Figure S4).
Figure 6.
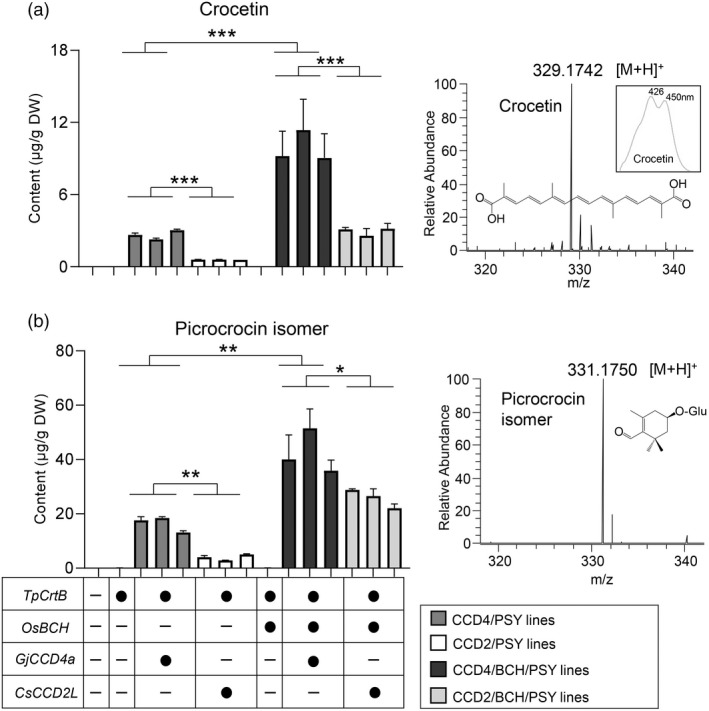
Identification and relative quantification of crocetin and picrocrocin in different engineered callus lines. Levels and high‐resolution MS spectra of crocetin (a) and picrocrocin isomer (b) in different engineered callus lines. The gene combinations are indicated below bars (black circle, was expressed; minus, was not included). Different coloured blocks represent different engineered callus lines as indicated in the key. Insets display structures and/or UV/vis spectrum of crocetin and picrocrocin.
UHPLC analysis of the polar extracts of callus samples showed three peaks with UV–vis absorption maximum from 432 to 439 nm and a shoulder from 454 to 462 nm, which resemble absorbance spectra of crocins (Figure S11a–c). The levels of these crocin‐like compounds were much higher in CCD4/PSY or CCD4/BCH/PSY lines compared with CCD2/PSY or CCD2/BCH/PSY, respectively (Figure S11d), suggesting that GjCCD4a was more active in producing crocins in citrus callus. In addition, CCD4/BCH/PSY and CCD2/BCH/PSY accumulated much higher levels of these three compounds than CCD4/PSY and CCD2/PSY, respectively (Figure S11d), revealing that β‐carotene hydroxylation is a limiting step for the production of crocins in citrus callus. As expected, these compounds were not detectable in polar extracts of WT, PSY and BCH/PSY lines.
Moreover, we identified a β‐D‐glucoside of 3‐OH‐β‐cyclocitral (picrocrocin isomer, m/z 331.17441 [M + H]+) in GjCCD4a or CsCCD2L‐overexpressing callus lines (Figure 6b). The relative contents of picrocrocin were 42.49 ± 8.72 and 25.82 ± 3.16 μg/g DW on average in the CCD4/BCH/PSY and CCD2/BCH/PSY lines, which is ~2.6 times higher than that in CCD4/PSY and ~7.1 times higher than that in CCD2/PSY, respectively. The picrocrocin isomer levels were very low in WT, PSY and BCH/PSY line (Figure 6b).
Taken together, our results revealed that GjCCD4a displayed a higher efficiency of crocetin, picrocrocin and crocin‐like compounds production in both PSY and BCH/PSY background, compared with CsCCD2L, and that simultaneous co‐expression of tpCrtB, OsBCH and GjCCD4a is an optimal combination for multi‐gene metabolic engineering of these compounds in citrus callus, a non‐green starch‐rich tissue.
GjCCD4a displayed a higher efficiency than CsCCD2L in apocarotenoid formation, and produced crocins in Nicotiana leaves at milligram‐scale
To further compare and explore the potential of GjCCD4a and CsCCD2L for biotechnological production of crocins and other apocarotenoids in green tissues, we transiently expressed these two genes in leaves of N. benthamiana. UHPLC‐HR‐MS analysis showed that the C7′–C8′ cleavage products, 3‐OH‐β‐apo‐8′‐carotenal and β‐apo‐8′‐carotenal, were strikingly increased in leaves infiltrated with pEAQ‐GjCCD4a, compared with those expressing empty vector (EV) (Figure S12). The content of 3‐OH‐β‐apo‐8′‐carotenal in PEAQ‐CsCCD2L leaves was also significantly increased compared with EV, but was ~4 times lower than that in pEAQ‐GjCCD4a leaves. β‐apo‐8′‐carotenal content showed only a slight increase in PEAQ‐CsCCD2L leaves, compared with EV, and was ~14 times lower than that in pEAQ‐GjCCD4a leaves (Figure S12). In contrast, there was no increase in the contents of 3‐OH‐β‐apo‐10′‐carotenal and β‐apo‐10′‐carotenal, neither in pEAQ‐GjCCD4a nor in PEAQ‐CsCCD2L leaves. Moreover, the contents of crocetin dialdehyde and crocetin were significantly higher in pEAQ‐GjCCD4a than in PEAQ‐CsCCD2L agroinfiltrated leaves. We also observed a striking increase in the content of a picrocrocin isomer in leaves infected with pEAQ‐GjCCD4a or PEAQ‐CsCCD2L (Figure S12).
In addition, we noticed distinctive yellow pigments visible to the naked eye in polar fractions of extracts of leaves agroinfiltrated with pEAQ‐GjCCD4a and PEAQ‐CsCCD2L, which is not observed in EV leaves (Figure 7a). UHPLC analysis of these yellow‐coloured polar fractions showed a series of peaks at 440 nm that were not detectable in those of EV leaves, including two peaks that eluted at 6.9 and 7.5 min and show UV absorption maxima at 439 and 462 nm, and 440 and 463 nm, which perfectly match crocin II (trans‐crocin 3) and crocin I (trans‐crocin 4) (Figures 7a,b and S13), the two most abundant crocins in saffron stigma and gardenia fruit (Carmona et al., 2006). Both PEAQ‐GjCCD4a‐ and PEAQ‐CsCCD2L‐agroinfiltrated leaves predominantly accumulated crocin II, followed by other crocins. Leaves expressing GjCCD4a displayed stronger yellow phenotype after immersion in 100% ethanol than those expressing CsCCD2L, while EV leaves were almost colourless (Figure 7c). Consistent with this observation, the contents of total crocins (1.61 ± 0.16 mg/g DW) and Crocin II in GjCCD4a‐expressed leaves were more than two times higher than those expressing CsCCD2L (Figure 7d).
Figure 7.
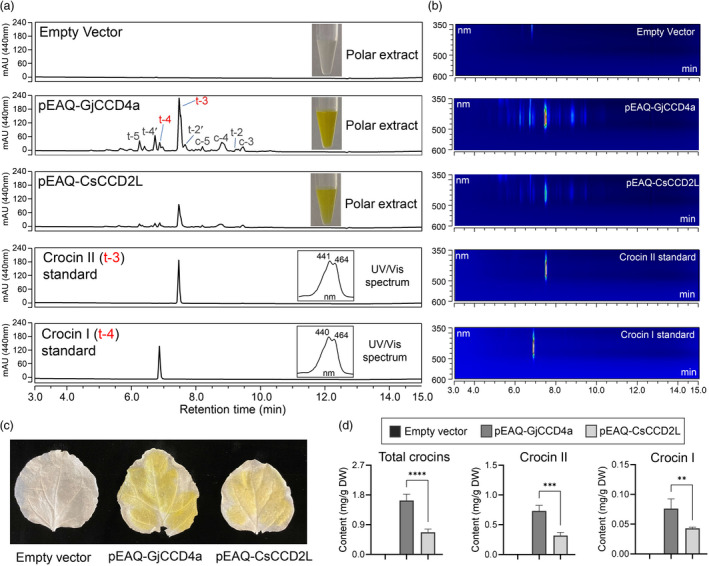
Crocin production in agroinfiltrated Nicotiana benthamiana leaves transiently expressing GjCCD4a or CsCCD2L. (a) Representative UHPLC chromatograms of polar extracts of agroinfiltrated N. benthamiana leaves. Peaks abbreviations correspond to: t‐5, trans‐crocin 5; t‐4′, trans‐crocin 4′; t‐4, trans‐crocin 4 (Crocin I); t‐3, trans‐crocin 3 (Crocin II); t‐2′, trans‐crocin 2′; c‐5, cis‐crocin 5; c‐4, cis‐crocin 4; t‐2, trans‐crocin 2; c‐3, cis‐crocin 3. Insets display the colour of polar extracts of agroinfiltrated leaves and the absorbance spectra of crocin I and crocin II standard. (b) UHPLC‐DAD/UV isoplot chromatogram of polar extracts. (c) The representative agroinfiltrated N. benthamiana leaf transiently expressing GjCCD4a or CsCCD2L display yellow colour after immersion in 100% ethanol for 2 days, while EV leaves are almost colourless. The yellow phenotype induced by GjCCD4a is stronger than that produced by CsCCD2L. (d) Quantification of crocins in agroinfiltrated N. benthamiana leaves was performed by UHPLC analysis. The column names are shown in the inset. Data are represented as mean ± SD of more than three independent pools of agroinfiltrated Nicotiana plants.
Carotenoid analysis showed that the levels of xanthophylls were significantly reduced in both pEAQ‐GjCCD4a‐ and PEAQ‐CsCCD2L‐agroinfiltrated leaves, at 6 days after agroinfiltration, while β‐carotene was significantly decreased only in pEAQ‐GjCCD4a leaves (Figure S14), which further reinforced the additional activity of GjCCD4a in targeting β‐carotene. We did not observe significant alterations in chlorophyll a and b in any of the leaf samples.
Overall, these results demonstrate that GjCCD4a has a higher activity than CsCCD2L in producing crocins and other valuable apocarotenoids, and triggers milligram‐scale production of crocins in N. benthamiana leaves.
Discussion
Most of our knowledge about CCDs enzymatic activity and substrate specificity originates from in vitro incubations and in bacterio assays utilizing E. coli engineered to produce different carotenoids. However, functional characterization in plants, for instance by using a transgenic approach, is important for deducing the role of CCDs in carotenoid metabolism and may provide insights about their real contribution to the carotenoid/apocarotenoid pattern. By integrating apocarotenoid profiling of the engineered callus lines and in vitro enzymatic assay, we provide direct evidence that GjCCD4a from G. jasminoides catalyses crocetin dialdehyde formation from β‐carotene and zeaxanthin through the β‐apo‐8′‐carotenal and 3‐OH‐β‐apo‐8′‐carotenal intermediates, respectively. It can be assumed that crocetin dialdehyde production from β‐carotene in G. jasminoides is a novel route for crocins production, which is not present in Crocus sativus, as CsCCD2L cleaves hydroxy‐apocarotenoids but not β‐apo‐8′‐carotenal (Frusciante et al., 2014). Indeed, overexpression of CsCCD2L in carotene‐rich PSY callus line produced much lower amounts of crocetin dialdehyde than in hydroxy‐carotenoid‐rich BCH/PSY background (Figures 2a and S4). The low amount of crocetin dialdehyde detected in CCD2/PSY line may be produced from trace amounts of zeaxanthin and 3‐OH‐β‐apo‐8′‐carotenal in PSY background. Furthermore, crocetin and crocins are produced by overexpression of GjCCD4a in engineered citrus callus and Nicotiana leaves (Figures 6 and 7), providing a first in planta evidence for the role of GjCCD4a in crocetin and crocins biosynthesis in G. jasminoides, for which the genetic transformation is not yet well established. Additionally, this result reinforced that oxidation of crocetin dialdehyde and glycosylation of crocetin can be efficiently complemented by non‐specific endogenous enzymes in various plant tissues (Frusciante et al., 2022; Martí et al., 2020).
CsCCD2L is a plastid enzyme; however, in the phylogenetic tree, it shows a close relationship to the cytosolic CCD1 subfamily characterized by relaxed cleavage site and substrate specificity (Figure 8; Frusciante et al., 2014; Ilg et al., 2014; Vogel et al., 2008). The evolution of crocin biosynthesis in Crocus species might be a result of gaining a plastid transient peptide, which provides CsCCD2L with access to zeaxanthin, and the evolutionary modification of the cleavage site specificity towards the C7–C8/C7′–C8′ double bonds. Phylogenetic analysis demonstrated that GjCCD4a constitutes a new independent sub‐class in CCD4 subfamily, which is closely related to B. davidii BdCCD4.1 and BdCCD4.3 (Figure 8). Indeed, all of these three CCD4s are able to catalyse the conversion of zeaxanthin to crocetin dialdehyde (Figure S5b; Ahrazem et al., 2017; Xu et al., 2020). However, GjCCD4a displayed an additional activity in converting β‐carotene and β‐apo‐8′‐carotenal into crocetin dialdehyde (Figures 2b and S5a), which is not present in B. davidii (Ahrazem et al., 2017; Xu et al., 2020), suggesting the independent evolution of an alternative route for crocins synthesis in G. jasminoides.
Figure 8.
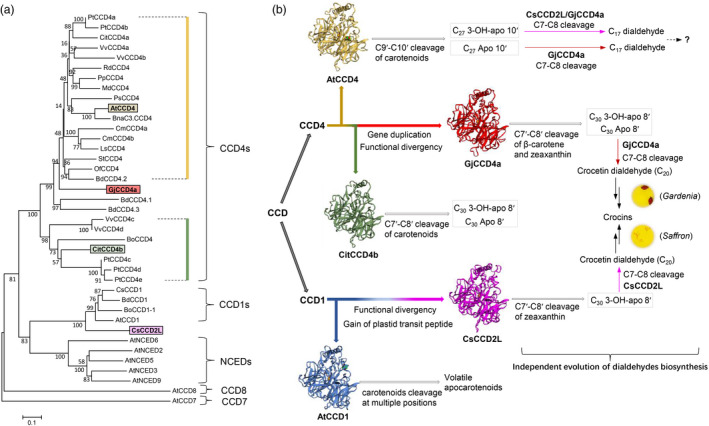
Model for independent evolution of dialdehydes and crocins biosynthesis pathways in different plants. (a) Phylogenetic analysis of carotenoid cleavage carotenoid (CCD) proteins from different species. CsCCD2L is a plastid enzyme; however, in the phylogenetic tree, it shows a close relationship to the cytosolic CCD1 subfamily. GjCCD4a constitutes a new independent sub‐class in CCD4 subfamily. The accession numbers were presented in previous studies (Ahrazem et al., 2016; Zheng et al., 2015) except for GjCCD4a (Gardenia jasminoides, KY631925.1); BnaC3.CCD4 (Brassica napus, KP658825); Buddleja davidii BdCCD4.1 (KX374547), BdCCD4.2 (KX374548), BdCCD4.3 (KX374549) and BdCCD1 (KX816559); Bixa orellana BoCCD1 (KT359018) and BoCCD4 (KT359024). (b) This figure displays the model of the convergent evolution of crocins, crocetin dialdehyde and C17‐dialdehyde biosynthesis in Crocus species (Saffron) and G. jasminoides (Gardenia). The evolution of crocin and dialdehyde biosynthesis in Saffron and Gardenia might be a result of the evolutionary modification of the cleavage site specificity of GjCCD4a and CsCCD2L towards the C7–C8/C7′–C8′ double bonds. GjCCD4a showed additional activities in converting other carotenoids (e.g. β‐carotene) into crocetin dialdehyde and forming C17‐dialdehyde from β‐apo‐10′‐carotenal, which is not present in Saffron. The 3D structure of CCDs protein was modelled using the SWISS MODEL online platform (https://swissmodel.expasy.org/) by using maize 9‐cis‐epoxycarotenoid dioxygenase (VP14) (PBD name: 3npe.1.A) as template.
Besides the formation of C20 crocetin dialdehyde, our data confirmed the in vitro activity of CsCCD2L in producing C17‐dialdehyde by cleaving 3‐OH‐β‐apo‐10′‐carotenal in planta (Frusciante et al., 2014), which again indicates the utility of engineered callus system for characterizing CCD enzymes. We also proved that GjCCD4a can cleave β‐apo‐10′‐carotenal, besides 3‐OH‐β‐apo‐10′‐carotenal, at the C7–C8 site, to yield C17‐dialdehyde (Figure 4). The C7–C8 cleavage of C27 apocarotenoids by GjCCD4a represents a new activity of plant CCD4s in producing C17‐dialdehyde that was not reported before.
Although supertransformation of callus used in this study is more time‐consuming, it can enable functional comparisons of CCD genes from different plant species in the same stable transgenic callus background containing the required carotenoid and apocarotenoid substrates. By using this system, we clearly demonstrated that GjCCD4a is more efficient than CsCCD2L in producing crocetin, crocins and picrocrocin in both PSY and BCH/PSY callus background (Figures 6 and S11), which is in agreement with the higher cleavage activity of GjCCD4a in producing their precursors, crocetin dialdehyde and 3‐OH‐β‐cyclocitral in these backgrounds (Figures 3 and S4).
The additional overexpression of OsBCH in CCD4/PSY or CCD2/PSY resulted in a significant increase in levels of crocetin and crocins (Figure 6), suggesting that hydroxylation of β‐carotene is one of the limiting steps that can enhance the yield of these compounds in non‐green citrus callus system. Consistent with this observation, previous studies revealed that crocins production in Nicotiana leaves was increased when CsCCD2L was co‐expressed with Crocus BCH2 (Martí et al., 2020). We found that the increase in the levels of crocetin and crocins in CCD2/BCH/PSY relative to CCD2/PSY is more pronounced than in CCD4/BCH/PSY relative to CCD4/PSY (Figure 6), revealing that BCH is more important for CsCCD2L‐based metabolic engineering of these compounds. This conclusion is in agreement with the restricted substrate specificity of CsCCD2L that utilizes zeaxanthin (Frusciante et al., 2014).
Among different combinations, the simultaneous co‐expression of tpCrtB, OsBCH and GjCCD4a was identified as an optimal one for heterologous production of crocetin, crocins and picrocrocin in non‐green citrus callus system (Figures 6 and S11). A previous study used maize callus transformation system to screen highly active PSYs from different plant sources, and the identified powerful enzyme was successfully applied to enhance β‐carotene content in Golden Rice (Paine et al., 2005). Our engineered callus system can likely provide a platform for ‘proof of concept’ testing of efficient enzymes and multi‐gene engineering approaches for efficient production of high‐value apocarotenoids in non‐green starch‐rich tissues/organs of crops, such as cereal endosperm and tubers, for which stable genetic transformation is still time‐consuming and work‐intensive. In addition, we show that these engineered citrus callus lines can also be grown as suspension cell cultures in big flasks on shakers (Figure S15), which allows cell culture growth in scale‐up and is applicable for optimization strategies developed for microbial cell systems, aiming at increasing metabolic output for efficient bio‐production of valuable apocarotenoids.
In addition to enzymatic activity, engineered callus system unravelled pleiotropic effects of perturbations to carotenoids/apocarotenoids on carotenoid biosynthesis pathway, metabolome and plastid ultrastructure. For example, we found that introducing GjCCD4a in carotene‐rich PSY background caused a significant increase in endogenous citrus BCH transcript levels (Figure S8), which is in agreement with the striking increase in levels of hydroxy‐apocarotenoids in PSY background that accumulates only trace amounts of zeaxanthin (Figures S2 and S9). Furthermore, overexpression of GjCCD4a in PSY background also increased the plastoglobuli number within chromoplast‐like plastids and led to a clear reduction in carotenoid crystals (Figure 5a), suggesting a possible subcellular adaptation to the enhancement of phytoene that was shown to be sequestered in plastoglobuli (Nogueira et al., 2013), and dramatic decrease in the content of β‐carotene (Figure S2) assumed to accumulate in form of carotenoid crystals in citrus callus (Cao et al., 2012). Moreover, we showed that the perturbations to carotenoids/apocarotenoids have broad effects across the metabolome, especially increasing the content of some fatty acids and their esters, several tricarboxylic acid (TCA) cycle intermediates and amino acids (Figure 5b).
In N. benthamiana leaves, we demonstrated that A. tumefaciens‐mediated transient expression of sole pEAQ‐GjCCD4a is sufficient to trigger remarkable accumulation of crocins (1.63 ± 0.16 mg/g DW) in only 6 days after agroinfiltration, which is more than two times higher than those obtained by transient expression of pEAQ‐CsCCD2L (0.67 ± 0.08 mg/g DW) (Figure 7). This amount is higher than that produced with recently reported transient expression of pBI121‐CsCCD2L and stable expression of CsCCD2L in Nicotiana plants (Ahrazem et al., 2022b; Frusciante et al., 2022). PEAQ vectors were shown to enable easy, rapid and extremely high‐level transient production of heterologous protein in plants with no reliance on viral replication (Sainsbury et al., 2009; Sainsbury and Lomonossoff, 2008) and had been used for milligram‐scale production of etoposide intermediates and triterpene compounds in leaves of N. benthamiana (Reed et al., 2017; Schultz et al., 2019). This study further highlights the opportunity to exploit pEAQ vectors for increasing the output of valuable metabolites in transient system. Moreover, the efficient production of crocins in N. benthamiana leaves by only expressing one single gene reveals an advantage of plant hosts that contain basic precursors of target metabolites and endogenous enzymes required for modification and stability of metabolites, compared with microbes.
The higher activity of GjCCD4a found in both green and non‐green plant tissues might be due to the broader substrate specificity of GjCCD4a that utilizes zeaxanthin and carotenes for crocin biosynthesis, compared with CsCCD2L that only cleaves zeaxanthin (Ahrazem et al., 2016, 2017; Frusciante et al., 2014). This broader specificity is particularly important because β‐carotene is present at much higher content than zeaxanthin in engineered callus and Nicotiana leaves (Figure S2; Frusciante et al., 2021). Recent studies showed that virus‐driven transient expression of CsCCD2L also resulted in efficient production of crocins (~2 mg/g DW) in Nicotiana leaves in 13 days (Martí et al., 2020), and stable expression of CsCCD2L, in combination with CsUGT2 and CsUGT709, in tomato fruits led to remarkable amounts of crocins and other valuable apocarotenoids (Ahrazem et al., 2022a). It can be assumed that crocins production in N. benthamiana leaves by using virus‐driven expression system and in tomato fruit by stable transformation could be further optimized by employing the GjCCD4a enzyme.
In conclusion, this study paves the way for an efficient biotechnological production of crocins and other high‐value apocarotenoid derivatives as pharmaceuticals in green tissues as well as non‐green starch‐rich organs of crops, but also highlights the contribution of functional diversification among CCD4 genes to the independent evolution of alternative apocarotenoids biosynthesis routes in different plants.
Experimental procedures
Plant materials
The wild‐type callus was propagated from abortive ovule embryogenic calli of C. paradise Macf and was subcultured at 20 days intervals and grown in the dark at room temperature. N. benthamiana were grown at growth room at 22–24 °C, under a 12‐h day–night photoperiod.
Plasmid construction and callus transformation
The CDS of tpCrtB (Zheng et al., 2021a) and OsBCH (GenBank: NM_001055350.1) was cloned into pK2GW7 and pH2GW7 gateway vector, respectively, by using ‘BP’ or ‘Topo’ and ‘LR’ reaction kit (Invitrogen, Carlsbad, CA, USA). The CDS of CsCCD2L (KP887110.1) or GjCCD4a (ARU08109.1) were cloned into pB2GW7 vector. The primers used for cloning are listed in Table S1. These constructs were electroporated into Agrobacterium tumefaciens EHA105 for following callus transformation.
The twenty‐day‐old wild‐type or transgenic callus were incubated with agrobacterium suspension (OD = 0.3) in liquid MS B5 medium for 10 min. After 3 days co‐cultivation in solid MS B5 medium, the transformed callus was selected with 60 mg/L kanamycin, 50 mg hygromycin or 50 mg/L glufosinate ammonium (Sigma‐Aldrich) for 4–5 weeks. To obtain stable transgenic callus, independent small piece of each recovered callus was subsequently transferred onto selective solid medium with antibiotics and subcultured for at least six cycles in the dark at 25 °C. The details of solid and liquid MS B5 medium used for subculture and transformation are shown in Method S1. Twenty‐day‐old transgenic lines gown in solid medium without antibiotic were harvested and stored at −80 °C for later metabolite and molecular analysis.
Transient expression in N. benthamiana leaves
The CDS of GjCCD4a or CsCCD2L was cloned into the PEAQ‐HT‐DEST1 gateway vector (Sainsbury et al., 2009). The primers used for gateway cloning are listed in Table S1. These two constructs and empty vector (EV) were transformed into Agrobacterium tumefaciens GV3101 using electroporation. The Agrobacterium‐mediated transient expression in 5‐week‐old N. benthamiana leaves was performed according to a previous description (Zheng et al., 2021a). pEAQ‐GjCCD4a, pEAQ‐CsCCD2L and EV agrobacterium cell pellets were resuspended in infiltration buffer to an OD600 of 1.0. The agroinfiltrated leaves were harvested after 6 days, then frozen in liquid nitrogen and stored at −80 °C until following metabolites analysis.
Quantitative real‐time PCR
Total RNA was extracted by using TRIzol and Direct‐zol™ RNA MiniPrep kit (Zymo, Irvine, CA, USA), and treated with DNaseI digestion. The first‐strand cDNA synthesis was performed by using iScript™ cDNA synthesis Kit (Bio‐Rad, Hercules, CA, USA). Quantitative real‐time PCR (qRT‐PCR) was performed on a CFX384 Touch Real‐Time PCR Detection System (Bio‐Rad) following the instruction of SsoAdvanced™ Universal SYBR® Green Supermix (Bio‐Rad, Hercules, CA, USA) Kit. The method was used to calculate gene relative expression. The qRT‐PCR primers are listed in Table S1.
UHPLC‐HR‐MS analysis of apocarotenoids
Extraction of apocarotenoids from lyophilized leaf and callus samples was performed according to a previous description (Mi et al., 2018; Zheng et al., 2021a). The extracts were re‐dissolved in acetonitrile/water (90/10, v/v), followed by filtration using a 0.22 μm filter for LC–MS analysis. The separation and detection of crocetin and apocarotenoids (except picrocrocin and 3‐OH‐β‐cyclocitral) were performed using UHPLC system coupled with a high‐resolution (HR) Q‐Orbitrap‐MS system, as previously described (Mi et al., 2018). The picrocrocin and 3‐OH‐β‐cyclocitral were detected by another UHPLC‐HR‐MS method used for profiling of glycosylated apocarotenoids (Mi et al., 2019). The identification and quantification of apocarotenoids were achieved by comparison with authentic standards and labelled internal apocarotenoid standards obtained from Buchem B.V. (Minden 60 7327 AW Apeldoorn, the Netherlands), as previously described (Mi et al., 2018).
UHPLC analysis of carotenoids, chlorophylls and crocins
A 10 or 20 mg aliquot of lyophilized nicotiana leaves or callus powder was extracted with chloroform and methanol (2/1, v/v), incubated on ice for 20 min, according to a previous description (Fraser et al., 2000). Then one volume of water was added to the crude extract to create a discrete partition by centrifugation. The lower chloroform phases were dried in a vacuum centrifuge and subjected to analysis of carotenoids and chlorophylls. The upper aqueous phases were collected for analysis of crocins.
The separation and detection of carotenoids and chlorophylls were conducted by using Dionex Ultimate 3000 UHPLC‐DAD with a C30 reversed‐phase column (250 × 4 mm, 5 μm; YMC Europa, Schermbeck, Deutschland). Three mobile phases used and the gradient condition was described in a previous study (Zheng et al., 2020b). Carotenoids and chlorophylls were identified by a comparison of spectral properties and typical retention time based on authentic standards and literatures (Fraser et al., 2007). Their contents were measured by comparison with calibration curves of standards.
For crocins, the separation and detection were conducted by using Dionex Ultimate 3000 UHPLC‐DAD with an Eclipse XDB‐C18 reversed‐phase column (4.6 × 150 mm, 3.5 μm; Agilent, Santa Clara, CA, USA). The mobile phase A (water/acetonitrile, 90/10, v/v/v) and mobile phase B (100% acetonitrile) were used to elute polar extracts with a flow rate of 0.8 mL/min. The gradient elution was from 100% A to 50% A/50% B in 10 min, stepped to 100% B over 4 min and held until 19 min, after which it returned to 100% A over 1 min, and held to the end of analysis (25 min). Crocins were identified by comparing retention times and UV‐visible spectra properties with authentic standards and literatures (Carmona et al., 2006; Martí et al., 2020). The quantification of crocins was performed by comparison with calibration curves of crocin II standards (Shanghai Huicheng Biotech Co., Ltd, Shanghai, China).
GC–MS analysis
Extraction and gas chromatography–mass spectrometry (GC–MS) analysis of metabolites were performed according to previous descriptions (Drapal et al., 2018; Zheng et al., 2020b). The ribitol and d27‐myristic acid were used as internal standard for polar and non‐polar metabolites, respectively. GC–MS was performed using Agilent HP6890 gas chromatograph with a 5973MSD and a 10 : 1 split injector. A mixture of alkanes, ranging from 10 to 30 carbons, was used for retention index external calibration. The GC–MS data were converted to analysis base file format (.abf) through the Reifycs Abf Converter software. The MS‐DIAL software (Tsugawa et al., 2015) was used for component peak identification, spectral deconvolution and integration of the peak area. The metabolites were identified by comparing the acquired Mass and MS/MS information with those in the publicly available retention‐indexed spectral library. The data were then exported into Microsoft Excel for calculating relative abundance.
Enzymatic activity assays
The CDS of GjCCD4a without chloroplast transit peptide (cTP) sequence was cloned in the pTHIO‐DAN1 vector in frame with N‐terminal thioredoxin. pThio‐GjCCD4a or empty vector was transformed into zeaxanthin or β‐carotene accumulating E. coli. The in bacterio activity assays were carried out according to a previous study (Bruno et al., 2015). The detailed reaction mixtures and metabolites extraction are described in Method S2. The extracts of in bacterio assays are subjected to UHPLC analysis with system 1.
For in vitro assay, the pThio‐GjCCD4a plasmid was transformed into E. coli BL21 competent cells harbouring the pGro7 plasmid. Bacterial induction, preparation of crude lysates and substrates, incubation and metabolites extraction were performed according to a previous study (Bruno et al., 2016). A detailed description of in vitro assay was provided in Method S3. The extracts of in vitro assay are subjected to UHPLC analysis with system 2.
Analysis of GjCCD4a in bacterio and in vitro products was performed on UHPLC‐DAD system with a reversed phase YMC Carotenoid C30 column (150 × 3 mm, 5 μm). Mobile phase A (MeOH: MTBE (1 : 1)) and mobile phase B (MeOH : MTBE : H2O (30 : 1 : 10)) were used to elute carotenoids and apocarotenoids with a flow rate of 0.6 mL/min. The gradient elution for system 1 was from 100% B to 0% B within 15 min, held until 24 min. The eluting gradient programme for system 2 was from 100% B to 45% B within 15 min, stepped to 0% B within 5 min and held until 24 min, after which it returned to 100% B within 1 min, and held until 33 min.
TEM analysis
The 10‐day‐old transgenic callus were transferred on MS B5 medium without sucrose for 1 week before transmission electron microscope (TEM) analysis. The callus samples were fixed with 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) containing 0.025 M L‐Ascorbic acid for a minimum of 48 h. Osmication was performed using reduced osmium (1 : 1 mixture of 2% osmium tetroxide and 3% potassium ferrocyanide). After pre‐embedding in 1% agar, samples were dehydrated in ethanol series and embedded in epoxy resin. Thin sections (80–110 nm thickness) were collected on copper grids and contrasted with lead citrate.
Imaging was performed using a TEM operating at 300 kV (Titan Cryo Twin, FEI Company, Hillsboro, OR). Images were recorded on a 4 k × 4 k CCD camera (Gatan Inc., Pleasanton, CA).
Phylogenetic analyses
The phylogenetic tree was built in MEGA5 using the Neighbour‐joining method as previously described (Tamura et al., 2011). The bootstrap test (1000 replicates) value is shown at the tree nodes. The NCBI accession numbers of CCD proteins used for phylogenetic analysis were displayed in Figure 8 legend.
Statistical analysis
All the data are displayed by mean ± Standard deviation (SD) with at least three biological replications unless stated otherwise. The statistical analysis of data was performed using GraphPad Prism v.8 and Excel 2010 (Microsoft, Redmond, WA, USA) software. Asterisk indicate significance of difference at *P value < 0.05, **P < 0.01 and ***P < 0.001, respectively.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
S.A‐B. conceived the project and supervised the experiments. X.Z. and S.A‐B. designed the experiments. X.Z performed most of the experiments and analysed the data with contributions from J.M., A.B., K.X.L. J.M. and K.X.L. helped to perform metabolite analysis. A.B. helped to perform in vivo and in vitro assays. R.S. performed TEM analysis. A.A. helped to perform qRT‐PCR experiments. X.Z and S.A‐B wrote the article;
Supporting information
Figure S1 Relative expression levels of transgenes of other two independent lines of each engineered callus line.
Figure S2 Carotenoid contents of three independent lines of each engineered callus line.
Figure S3 β‐apocarotenoid profiling of other two independent lines of each engineered callus lines.
Figure S4 The identification and relative quantification of crocetin dialdehyde among various engineered callus lines.
Figure S5 GjCCD4a cleaves β‐carotene and zeaxanthin to yield crocetin dialdehyde in E. coli.
Figure S6 Analysis of hydroxylated apocarotenoids in other two independent lines of each engineered callus lines.
Figure S7 The MS/MS spectra of C17 dialdehyde produced by in vitro assay.
Figure S8 Relative expression levels of carotenoid biosynthetic genes in PSY line and two lines of CCD4/PSY callus.
Figure S9 Relative quantification of hydroxy‐apocarotenoids in PSY line and two lines of CCD4/PSY callus.
Figure S10 The MS/MS spectra of crocetin standard and endogenous crocetin identified in engineered callus.
Figure S11 The identification and relative quantification of crocin‐like compounds among various engineered callus lines.
Figure S12 Apocarotenoid profiling of agroinfiltrated Nicotiana benthamiana leaves transiently expressing GjCCD4a or CsCCD2L.
Figure S13 Absorbance spectra of several crocins detected in the polar extracts of agroinfiltrated Nicotiana benthamiana leaves transiently expressing GjCCD4a or CsCCD2L.
Figure S14 Carotenoid analysis of agroinfiltrated Nicotiana benthamiana leaves by transiently expressing GjCCD4a or CsCCD2L.
Figure S15 Suspension culture of different engineered callus lines.
Table S1 Sequences of primers used in this study.
Methods S1 The MS B5 medium used for callus subculture and transformation.
Methods S2 In bacterio assay.
Methods S3 In vitro assays.
Acknowledgements
We thank Prof. Xiuxin Deng for providing the wild‐type calli of grapefruit (C. paradisi Macf.). We thank Dr. Elliott J. Price for technical support, and Dr. Juan C. Moreno and Dr. Yajun Wang for valuable discussions. This work was supported by baseline funding and Competitive Research Grants (CRG 2017 and CRG 2020) given to Salim Al‐Babili from King Abdullah University of Science and Technology (KAUST).
References
- Ahrazem, O. , Diretto, G. , Argandoña, J. , Rubio‐Moraga, Á. , Julve, J.M. , Orzáez, D. , Granell, A. et al. (2017) Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii . J. Exp. Biol. 68, 4663–4677. [DOI] [PubMed] [Google Scholar]
- Ahrazem, O. , Diretto, G. , Rambla, J.L. , Rubio‐Moraga, Á. , Lobato, M. , Frusciante, S. , Argandoña, J. et al. (2022a) Engineering high levels of saffron apocarotenoids in tomato. Hortic. Res. 9, uhac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrazem, O. , Rubio‐Moraga, A. , Berman, J. , Capell, T. , Christou, P. , Zhu, C. and Gómez‐Gómez, L. (2016) The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 209, 650–663. [DOI] [PubMed] [Google Scholar]
- Ahrazem, O. , Zhu, C. , Huang, X. , Rubio‐Moraga, A. , Capell, T. , Christou, P. and Gómez‐Gómez, L. (2022b) Metabolic engineering of crocin biosynthesis in Nicotiana species. Front. Plant Sci. 13, 861140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavizadeh, S.H. and Hosseinzadeh, H. (2014) Bioactivity assessment and toxicity of crocin: a comprehensive review. Food Chem. Toxicol. 64, 65–80. [DOI] [PubMed] [Google Scholar]
- Al‐Babili, S. and Bouwmeester, H.J. (2015) Strigolactones, a novel carotenoid‐derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Alder, A. , Jamil, M. , Marzorati, M. , Bruno, M. , Vermathen, M. , Bigler, P. , Ghisla, S. et al. (2012) The path from β‐carotene to carlactone, a strigolactone‐like plant hormone. Science, 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Auldridge, M.E. , McCarty, D.R. and Klee, H.J. (2006) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9, 315–321. [DOI] [PubMed] [Google Scholar]
- Bruno, M. , Beyer, P. and Al‐Babili, S. (2015) The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β‐ionone ring‐containing carotenes and non‐epoxidated xanthophylls. Arch. Biochem. Biophys. 572, 126–133. [DOI] [PubMed] [Google Scholar]
- Bruno, M. , Koschmieder, J. , Wuest, F. , Schaub, P. , Fehling‐Kaschek, M. , Timmer, J. , Beyer, P. et al. (2016) Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Biol. 67, 5993–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari, S.I. , Manzoor, M. and Dhar, M. (2018) A comprehensive review of the pharmacological potential of Crocus sativus and its bioactive apocarotenoids. Biomed. Pharmacother. 98, 733–745. [DOI] [PubMed] [Google Scholar]
- Cao, H. , Zhang, J. , Xu, J. , Ye, J. , Yun, Z. , Xu, Q. , Xu, J. et al. (2012) Comprehending crystalline β‐carotene accumulation by comparing engineered cell models and the natural carotenoid‐rich system of citrus. J. Exp. Bot. 63, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona, M. , Zalacain, A. , Sánchez, A.M. , Novella, J.L. and Alonso, G.L. (2006) Crocetin esters, picrocrocin and its related compounds present in Crocus sativus stigmas and Gardenia jasminoides fruits. Tentative identification of seven new compounds by LC‐ESI‐MS. J. Agric. Food Chem. 54, 973–979. [DOI] [PubMed] [Google Scholar]
- Diretto, G. , Ahrazem, O. , Rubio‐Moraga, Á. , Fiore, A. , Sevi, F. , Argandoña, J. and Gómez‐Gómez, L. (2019) UGT709G1: a novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus). New Phytol. 224, 725–740. [DOI] [PubMed] [Google Scholar]
- Drapal, M. , Barros de Carvalho, E. , Ovalle Rivera, T.M. , Becerra Lopez‐Lavalle, L.A. and Fraser, P.D. (2018) Capturing biochemical diversity in cassava (Manihot esculenta Crantz) through the application of metabolite profiling. J. Agric. Food Chem. 67, 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Wang, N. , Cui, F. , Li, X. , Xiao, J. and Xiong, L. (2010) Characterization of the β‐carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 154, 1304–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P.D. , Enfissi, E.M. , Halket, J.M. , Truesdale, M.R. , Yu, D. , Gerrish, C. and Bramley, P.M. (2007) Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell, 19, 3194–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P.D. , Pinto, M.E.S. , Holloway, D.E. and Bramley, P.M. (2000) Application of high‐performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 24, 551–558. [DOI] [PubMed] [Google Scholar]
- Frusciante, S. , Demurtas, O.C. , Sulli, M. , Mini, P. , Aprea, G. , Diretto, G. , Karcher, D. et al. (2022) Heterologous expression of Bixa orellana cleavage dioxygenase 4‐3 drives crocin but not bixin biosynthesis. Plant Physiol. 188, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frusciante, S. , Diretto, G. , Bruno, M. , Ferrante, P. , Pietrella, M. , Prado‐Cabrero, A. , Rubio‐Moraga, A. et al. (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA, 111, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, G. , Al‐Babili, S. and Von Lintig, J. (2003) Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 8, 145–149. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. , Rubio‐Moraga, A. and Ahrazem, O. (2010) Understanding carotenoid metabolism in saffron stigmas: unravelling aroma and colour formation. Funct Plant Sci. Biotechnol. 4, 56–63. [Google Scholar]
- Gonzalez‐Jorge, S. , Ha, S.H. , Magallanes‐Lundback, M. , Gilliland, L.U. , Zhou, A. , Lipka, A.E. , Nguyen, Y.N. et al. (2013) Carotenoid cleavage dioxygenase 4 is a negative regulator of β‐carotene content in Arabidopsis seeds. Plant Cell, 25, 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H. , Uragami, C. and Cogdell, R.J. (2016) Carotenoids and photosynthesis. Subcell. Biochem. 79, 111–139. [DOI] [PubMed] [Google Scholar]
- Hou, X. , Rivers, J. , León, P. , McQuinn, R.P. and Pogson, B.J. (2016) Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 21, 792–803. [DOI] [PubMed] [Google Scholar]
- Ilg, A. , Bruno, M. , Beyer, P. and Al‐Babili, S. (2014) Tomato carotenoid cleavage dioxygenases 1A and 1B: relaxed double bond specificity leads to a plenitude of dialdehydes, mono‐apocarotenoids and isoprenoid volatiles. FEBS Open Bio. 4, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, K.‐P. , Baz, L. and Al‐Babili, S. (2018) From carotenoids to strigolactones. J. Exp. Biol. 69, 2189–2204. [DOI] [PubMed] [Google Scholar]
- Koschmieder, J. , Wüst, F. , Schaub, P. , Álvarez, D. , Trautmann, D. , Krischke, M. , Rustenholz, C. et al. (2021) Plant apocarotenoid metabolism utilizes defense mechanisms against reactive carbonyl species and xenobiotics. Plant Physiol. 185, 331–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakoudi, A. , O'Callaghan, Y.C. , Galvin, K. , Tsimidou, M.Z. and O'Brien, N.M. (2015) Cellular transport and bioactivity of a major saffron apocarotenoid, picrocrocin (4‐(β‐D‐glucopyranosyloxy)‐2, 6, 6‐trimethyl‐1‐cyclohexene‐1‐carboxaldehyde). J. Agric. Food Chem. 63, 8662–8668. [DOI] [PubMed] [Google Scholar]
- Lopresti, A.L. and Drummond, P.D. (2014) Saffron (Crocus sativus) for depression: a systematic review of clinical studies and examination of underlying antidepressant mechanisms of action. Hum. Psychopharmacol. 29, 517–527. [DOI] [PubMed] [Google Scholar]
- Ma, G. , Zhang, L. , Matsuta, A. , Matsutani, K. , Yamawaki, K. , Yahata, M. , Wahyudi, A. et al. (2013) Enzymatic formation of β‐citraurin from β‐cryptoxanthin and Zeaxanthin by carotenoid cleavage dioxygenase 4 in the flavedo of citrus fruit. Plant Physiol. 163, 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí, M. , Diretto, G. , Aragonés, V. , Frusciante, S. , Ahrazem, O. , Gómez‐Gómez, L. and Daròs, J.‐A. (2020) Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus‐driven system. Metab. Eng. 61, 238–250. [DOI] [PubMed] [Google Scholar]
- Mazidi, M. , Shemshian, M. , Mousavi, S.H. , Norouzy, A. , Kermani, T. , Moghiman, T. , Sadeghi, A. et al. (2016) A double‐blind, randomized and placebo‐controlled trial of Saffron (Crocus sativus L.) in the treatment of anxiety and depression. J. Compl. Integr. Med. 13, 195–199. [DOI] [PubMed] [Google Scholar]
- Mi, J. and Al‐Babili, S. (2019) To color or to decolor: that is the question. Mol. Plant, 12, 1173–1175. [DOI] [PubMed] [Google Scholar]
- Mi, J. , Jia, K.‐P. , Balakrishna, A. , Wang, J.Y. and Al‐Babili, S. (2019) An LC‐MS profiling method reveals a route for apocarotene glycosylation and shows its induction by high light stress in Arabidopsis. Analyst, 144, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Mi, J. , Jia, K.‐P. , Wang, J.Y. and Al‐Babili, S. (2018) A rapid LC‐MS method for qualitative and quantitative profiling of plant apocarotenoids. Anal. Chim. Acta, 1035, 87–95. [DOI] [PubMed] [Google Scholar]
- Moise, A.R. , Al‐Babili, S. and Wurtzel, E.T. (2014) Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, J.C. , Mi, J. , Alagoz, Y. and Al‐Babili, S. (2021) Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J. 105, 351–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.N. , Park, Y.‐M. , Jung, H.‐J. , Lee, J.Y. , Min, B.D. , Park, S.‐U. , Jung, W.‐S. et al. (2010) Anti‐inflammatory effects of crocin and crocetin in rat brain microglial cells. Eur. J. Pharmacol. 648, 110–116. [DOI] [PubMed] [Google Scholar]
- Nogueira, M. , Mora, L. , Enfissi, E.M. , Bramley, P.M. and Fraser, P.D. (2013) Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell, 25, 4560–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine, J.A. , Shipton, C.A. , Chaggar, S. , Howells, R.M. , Kennedy, M.J. , Vernon, G. , Wright, S.Y. et al. (2005) Improving the nutritional value of golden rice through increased pro‐vitamin A content. Nat. Biotechnol. 23, 482–487. [DOI] [PubMed] [Google Scholar]
- Reed, J. , Stephenson, M.J. , Miettinen, K. , Brouwer, B. , Leveau, A. , Brett, P. , Goss, R.J. et al. (2017) A translational synthetic biology platform for rapid access to gram‐scale quantities of novel drug‐like molecules. Metab. Eng. 42, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo, M.J. , Alquezar, B. , Alos, E. , Medina, V. , Carmona, L. , Bruno, M. , Al‐Babili, S. et al. (2013) A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit‐specific apocarotenoid pigments. J. Exp. Bot. 64, 4461–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Concepcion, M. , Avalos, J. , Bonet, M.L. , Boronat, A. , Gomez‐Gomez, L. , Hornero‐Mendez, D. , Limon, M.C. et al. (2018) A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 70, 62–93. [DOI] [PubMed] [Google Scholar]
- Sainsbury, F. and Lomonossoff, G.P. (2008) Extremely high‐level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury, F. , Thuenemann, E.C. and Lomonossoff, G.P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. [DOI] [PubMed] [Google Scholar]
- Schultz, B.J. , Kim, S.Y. , Lau, W. and Sattely, E.S. (2019) Total biosynthesis for milligram‐scale production of etoposide intermediates in a plant chassis. J. Am. Chem. Soc. 141, 19231–19235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.H. , Tan, B.C. , Gage, D.A. , Zeevaart, J.A. and McCarty, D.R. (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science, 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Simkin, A.J. , Schwartz, S.H. , Auldridge, M. , Taylor, M.G. and Klee, H.J. (2004) The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β‐ionone, pseudoionone, and geranylacetone. Plant J. 40, 882–892. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa, H. , Cajka, T. , Kind, T. , Ma, Y. , Higgins, B. , Ikeda, K. , Kanazawa, M. et al. (2015) MS‐DIAL: data‐independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods, 12, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.T. , Tan, B.‐C. , McCarty, D.R. and Klee, H.J. (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 283, 11364–11373. [DOI] [PubMed] [Google Scholar]
- Walter, M.H. and Strack, D. (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 28, 663–692. [DOI] [PubMed] [Google Scholar]
- Wang, J.Y. , Haider, I. , Jamil, M. , Fiorilli, V. , Saito, Y. , Mi, J. , Baz, L. et al. (2019) The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 10, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Pu, X. , Gao, R. , Demurtas, O.C. , Fleck, S.J. , Richter, M. , He, C. et al. (2020) Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 18, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H. , Zhang, J. , Nageswaran, D. and Li, L. (2015) Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2, 15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Giuliano, G. and Al‐Babili, S. (2020a) Carotenoid biofortification in crop plants: citius, altius, fortius . Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 1865, 158664. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Mi, J. , Deng, X. and Al‐Babili, S. (2021a) LC–MS‐based profiling provides new insights into apocarotenoid biosynthesis and modifications in citrus fruits. J. Agric. Food Chem. 69, 1842–1851. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Xie, Z. , Zhu, K. , Xu, Q. , Deng, X. and Pan, Z. (2015) Isolation and characterization of carotenoid cleavage dioxygenase 4 genes from different citrus species. Mol. Genet. Genomics, 290, 1589–1603. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Yang, Y. and Al‐Babili, S. (2021b) Exploring the diversity and regulation of apocarotenoid metabolic pathways in plants. Front. Plant Sci. 12, 787049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Zhu, K. , Sun, Q. , Zhang, W. , Wang, X. , Cao, H. , Tan, M. et al. (2019) Natural variation in CCD4 promoter underpins species‐specific evolution of red coloration in citrus peel. Mol. Plant, 12, 1294–1307. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Zhu, K. , Ye, J. , Price, E.J. , Deng, X. and Fraser, P.D. (2020b) The effect of β‐cyclocitral treatment on the carotenoid content of transgenic Marsh grapefruit (Citrus paradisi Macf.) suspension‐cultured cells. Phytochemistry, 180, 112509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Relative expression levels of transgenes of other two independent lines of each engineered callus line.
Figure S2 Carotenoid contents of three independent lines of each engineered callus line.
Figure S3 β‐apocarotenoid profiling of other two independent lines of each engineered callus lines.
Figure S4 The identification and relative quantification of crocetin dialdehyde among various engineered callus lines.
Figure S5 GjCCD4a cleaves β‐carotene and zeaxanthin to yield crocetin dialdehyde in E. coli.
Figure S6 Analysis of hydroxylated apocarotenoids in other two independent lines of each engineered callus lines.
Figure S7 The MS/MS spectra of C17 dialdehyde produced by in vitro assay.
Figure S8 Relative expression levels of carotenoid biosynthetic genes in PSY line and two lines of CCD4/PSY callus.
Figure S9 Relative quantification of hydroxy‐apocarotenoids in PSY line and two lines of CCD4/PSY callus.
Figure S10 The MS/MS spectra of crocetin standard and endogenous crocetin identified in engineered callus.
Figure S11 The identification and relative quantification of crocin‐like compounds among various engineered callus lines.
Figure S12 Apocarotenoid profiling of agroinfiltrated Nicotiana benthamiana leaves transiently expressing GjCCD4a or CsCCD2L.
Figure S13 Absorbance spectra of several crocins detected in the polar extracts of agroinfiltrated Nicotiana benthamiana leaves transiently expressing GjCCD4a or CsCCD2L.
Figure S14 Carotenoid analysis of agroinfiltrated Nicotiana benthamiana leaves by transiently expressing GjCCD4a or CsCCD2L.
Figure S15 Suspension culture of different engineered callus lines.
Table S1 Sequences of primers used in this study.
Methods S1 The MS B5 medium used for callus subculture and transformation.
Methods S2 In bacterio assay.
Methods S3 In vitro assays.


