Abstract
Background:
Difficulties in the timely identification of patients in need of colorectal cancer (CRC) screening contribute to the low overall screening rates observed nationally.
Objective:
To use data within Electronic Health Record (EHR) to identify patients with prior CRC testing.
Design:
We modified a locally-developed clinical natural language processing (NLP) system to identify four CRC tests (colonoscopy, flexible sigmoidoscopy, fecal occult blood testing, and double contrast barium enema) within electronic clinical documentation. Text phrases in clinical notes and procedure reports which included references to CRC tests were interpreted by the system to determine whether testing was planned or completed, and to estimate the date of completed tests.
Setting:
Large academic medical center.
Patients:
200 patients ≥ 50 years old who had completed at least two non-acute primary care outpatient visits within a one-year period.
Measures:
We compared the recall (sensitivity) and precision (positive predictive value) of the NLP system, billing records, and manual review of electronic records, using a reference standard of human review of all available information sources.
Results:
For identification of all CRC tests, recall and precision were as follows: NLP system (recall 93%, precision 94%), manual chart review (74%, 98%), and billing records review (44%, 83%). Recall and precision for identification of patients in need of screening were: NLP system (recall 95%, precision 88%), manual chart review (99%, 82%), and billing records review (99%, 67%).
Limitations:
This study was performed in one medical center on a limited set of patients, and requires a robust EHR for implementation.
Conclusions:
Applying NLP to EHR records detected more CRC tests than either manual chart review or billing records review alone. NLP had better precision but marginally lower recall to identify patients who were due for CRC screening than billing record review.
Keywords: colorectal cancer screening, cancer screening, preventive health, natural language processing, electronic health records, electronic medical records
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and second leading cause of cancer death for both men and women in the United States. In 2009, an expected 146,970 new cases of colorectal cancer will lead to an estimated 49,920 deaths.1 Timely screening and removal of pre-cancerous adenomatous polyps can prevent many CRC cases.2 The United States Preventive Services Task Force recommends that all average-risk (asymptomatic, age 50 years and older, having no personal or family history of CRC or of adenomatous polyps, no history of inflammatory bowel disease, and no family history of a genetic syndrome of colorectal neoplasia) undergo scheduled screenings for CRC with an approved test.3 Despite widespread public health knowledge about the benefits of CRC screening, performance rates are low nationally. Only 40–60% of eligible patients have been found to have received appropriate screening.4;5
Accurate and timely identification of patients due for CRC screening constitutes the first critical step toward increasing screening rates. Traditional methods of identifying these individuals, including patient self-report, physician report, and use of billing data are frequently inaccurate, unreliable, or incomplete.6–10 Manual chart abstraction, often accepted to be the gold standard, is costly, time consuming, and limited by the thoroughness of individual abstractors. An automated approach based on available Electronic Health Record (EHR) data would potentially improve results while requiring no additional chart review or data entry. Increasingly, EHR records contain sufficient information to determine whether CRC testing is due, since EHR systems integrate laboratory results, procedure and radiology reports, and clinical narratives, such as primary care and gastroenterology clinic notes. Although EHR systems provide quick access to an individual patient’s documents, the volume of data recorded for thousands of patients can hinder rapid location of references to CRC testing. Furthermore, much relevant patient information exists as unstructured free text, which for computational purposes must be converted into structured content. The field of natural language processing (NLP) creates approaches and tools that can “recognize” concepts from free text narratives, including clinical documents.11–16 For this study, a locally developed NLP system17–20 (KnowledgeMap Concept Identifier – KMCI) was applied to a large set of clinical documents from EHR records to quickly identify references to CRC tests. The NLP system was modified to detect four common forms of CRC testing: colonoscopy, sigmoidoscopy (FSIG), double-contrast barium enema (DCBE), and fecal occult blood testing (FOBT). Our system also identified and recorded key contextual elements for each CRC test, such as the timing of the test and its status (e.g., completed vs. planned). We hypothesized that the NLP-based approach would outperform the traditional methods of CRC test status determination, including billing code queries and manual chart review, in precision and recall. We also examined NLP-identified references to uncompleted CRC tests to explore reasons that patients had not completed testing.
METHODS
Study Setting
The study was conducted at four Vanderbilt University Medical Center (VUMC) affiliated ambulatory health care clinics in Nashville, Tennessee. Collectively, professional staff at these sites includes over 30 attending physicians, 75 resident physicians, and 10 nurse practitioners. For more than a decade, clinical practices within VUMC have used a common, internally-developed EHR system which provides integrated access to inpatient and outpatient free-text clinical notes, reports of radiology and pathology studies, procedural (e.g., endoscopy) notes, and laboratory results. In addition, each EHR record includes a free-text, multidisciplinary “patient summary” in which providers enter brief descriptions of the patient’s past medical history (problems), procedural history, preventive health maintenance events (e.g., CRC testing or vaccinations), medications, allergies, family medical history, and social history.
Patient Eligibility
Entry criteria included age 50 years and older with a minimum of two non-acute primary care outpatient visits to one of the four ambulatory health clinics during a one-year period (October 1, 2006-September 30, 2007). From over 15,000 patients meeting inclusion criteria, we randomly selected 500 patients’ complete EHR records, which captures all health care documentation occurring within any VUMC inpatient or outpatient setting. Of note, we made no requirement that these patients received subspecialist care at VUMC (e.g., gastroenterology). We divided the 500 EHR records into 2 cohorts, one for development (300 in the training cohort) and one for evaluation (200 in the test cohort). The Vanderbilt Institutional Review Board approved this study.
Manual EHR Abstraction
Manual EHR abstraction involved review of clinical EHR patient records by at least one of two board-certified physicians in Internal Medicine trained on use of a standardized case abstraction form. Physician reviewers each examined 110 records randomly selected from the test cohort of 200 patient EHRs. Prior to the study start, both physicians completed training using 5 sample charts and the standardized case abstraction form. The study constrained physicians to review each patient’s record in the Vanderbilt clinical EHR interface (the same format available to clinicians during patient care). The manual physician review thus omitted review of billing records or access to any KMCI abstractions of EHR records. Reviewers recorded information regarding patient age, ethnicity, family history of CRC, and personal history of CRC. They recorded all references to completed CRC tests, associated dates, test results, and data source (e.g., clinic note, problem list). To calculate inter-rater reliability, physician reviewers abstracted 20 of the same patient records. The physician reviewers were not involved in development of the NLP algorithms described below.
Development of NLP System to Detect CRC Testing
The KMCI system, used in this study, is a general-purpose medical NLP system developed by several authors and colleagues at VUMC.17–20 The KMCI system identifies Unified Medical Language System (UMLS) concepts from biomedical text documents, and produces XML-tagged output containing lists of UMLS concepts found in each sentence with relevant context (e.g., is the concept negated?). The UMLS is composed of more than 100 individual vocabularies such as SNOMED-CT, the International Classifications of Diseases, and Medical Subject Headings (MeSH). Common identifiers link about eight million strings (synonyms) into more than two million “concepts”. For example, KMCI would encode the document phrase “no evidence of colon cancer” as “C0699790, Carcinoma of the colon, negated”, indicating that the concept was not present in the patient. The KMCI algorithm employs rigorous NLP techniques and document- and context-based disambiguation methods to accurately identify UMLS concepts. While KMCI was originally developed for medical curriculum documents,17 it has been used in research and production for a wide variety of clinical documents.19–22 KMCI is similar to several other systems that also identify UMLS concepts, such as the National Library of Medicine’s MetaMap23 or the Medical Language Extraction and Encoding system (MedLEE).13
To improve KMCI’s ability to recognize CRC-related concepts, we added 23 synonyms (e.g., “flex sig”, “guaiac card”) related to CRC testing to the existing UMLS Metathesaurus. Synonyms were added by physician review of sample training records augmented by queries of all words found in the training corpus of documents. The algorithmic modifications of the NLP system for use in this study have been previously described in detail.24 Briefly, we developed an algorithm that identifies and interprets time and date descriptors, and then associates them with identified CRC tests (e.g., “colonoscopy in 2005” or “flexible sigmoidoscopy 5 years ago”). Relative date references such as the latter (“5 years ago”) were calculated by subtracting the relative time period from the date of note as a reference point by the NLP system. We also created a status indicator algorithm that could identify negated phrases (i.e., “no” or “never”) as well as common verbs and other modifiers that change the status of CRC related testing (e.g., refused, declined, scheduled). A prior evaluation of these algorithms applied to colonoscopies found the date detection algorithm had a recall of 0.91 and a precision of 0.95, and the status algorithm had a recall of 0.82 and a precision of 0.95.24 The NLP methods for time/date interpretation and recognition of status modifiers were applied unchanged from prior studies.24
The output of the NLP system included CRC test concepts and their associated date and status information in each clinical note for each patient. To identify actually-completed CRC tests, we selected all CRC test concepts with identified dates of either “today” or dates occuring in the past, and ignored all CRC test concepts with status modifiers other than “completed”. Thus, discussions of CRC test scheduling, a patient’s need of CRC testing, or tests that were declined by the patient were not marked as “completed” CRC tests.
Many patient records contained multiple references to each unique CRC test. To aggregate these into a single set of unique CRC events for each patient, we developed algorithms that combined multiple date references for each procedure type (e.g., colonoscopy, FSIG, DCBE, FOBT). First, the algorithm collapsed exact date matches and overlapping date ranges for each procedure type to the most specific date retrieved by the system (e.g., “2005” and “2005–03-05” would be combined into two references to the same event). Second, the algorithm also combined any date reference (or range) overlapping another reference to the same procedure type if their dates occurred within 30 days of each other. This limit was chosen empirically through review of records in the training set. No EHR records from the test cohort were used in development of these algorithms.
Adjudicated Reference Standard
The study examined four CRC-related tests: colonoscopy, FSIG, DCBE, and serial FOBT. Multiple information resources contributed to our reference standard determination of the status of each test (completed or not completed; date of completion). The reference standard was created for each patient record in the test cohort by adjudicated review of all available primary data sources (results of previous physicians’ manual EHR record abstraction, institutional billing records, KMCI automated chart abstraction output, and access the primary EHR records to resolve questions). Two physicians reviewed all discrepancies among manual abstraction, KMCI, and billing records to score each as either a true positive (i.e., a completed CRC test validated by presence of an EHR document) or a false positive (i.e., a reported CRC test for which no supporting EHR records could be found). In each case, the EHR was taken as the gold standard, limiting the level of accuracy to that which was recorded in the patient’s record. The study used this adjudicated reference standard determination to classify whether KMCI correctly identified each unique reference to CRC-related testing for the patient during the study interval of interest. If the date determined by the adjudicated reference standard included the date of an individual CRC-related test, the instance was considered correct. We also used the adjudicated reference standard to determine if each patient was up to date for recommended CRC screening, according to current guidelines available at the time of the study25; a patient was considered up to date with a colonoscopy in the previous 10 years, a FSIG or DCBE in the previous 5 years, or three FOBTs in the previous year.
Statistical Analysis
We determined recall and precision to evaluate the performance of the following methods: manual EHR review, billing record review, and the NLP system. We calculated recall (or sensitivity) as the proportion of reference standard tests correctly identified by each method. McNemar’s chi-squared test enabled comparison of recall metrics among the different methods. We calculated precision (or positive predictive value) as the proportion of reference standard tests correctly identified by each method divided by the number of unique CRC-related test instances identified by each respective method. To compare precision among the methods, we applied a 2-sample test for equality of proportions. The F-measure was calculated as the harmonic mean between the recall and precision (2 × Recall × Precision / [Recall + Precision]). We calculated inter-rater reliability for the two physicians’ manual EHR reviews using Cohen’s Kappa.
RESULTS
Patient Characteristics
The study population for the 200 EHR test cohort was 62% female with a median age of 64 years (Table 1). Patients attended the primary care clinics for a median of five years; 77% of patients had attending physician caregivers, and 23% had care provided primarily by resident physicians. Forty-five patients had documented risk factors for CRC;26 of these, four patients had personal histories of previous CRC, one had inflammatory bowel disease, 22 had personal histories of adenomatous polyps, and 18 had documented family histories of colorectal cancer or polyps in first-degree relatives.
Table 1.
Characteristics of the Study Patients
| Patient Characteristics N=200 | |
|---|---|
|
| |
| Age in years (median [interquartile range]) | 64 [51–97] |
|
| |
| Gender, female (%) | 62 |
|
| |
| Race (%) | |
| White | 78 |
| Black | 16 |
| Asian | 3 |
|
| |
| Personal history of (%)* | |
| Adenomatous polyps | 11 |
| Inflammatory bowel disease | 0.5 |
| Colorectal cancer | 2 |
| Colectomy | 4 |
|
| |
| Family history of (%)* | |
| 1st degree relative with polyps | 3 |
| 1st degree relative with colorectal cancer | 6 |
| 2nd degree relative with colorectal cancer | 3 |
|
| |
| Number of colonoscopies performed per patient (%) | |
| 0 | 40 |
| 1 | 39 |
| 2 or more | 21 |
|
| |
| Number of flexible sigmoidoscopies performed per patient (%) | |
| 0 | 94 |
| 1 | 5 |
| 2 or more | 0 |
|
| |
| Number of 3-home fecal occult blood testing completed per patient (%) | |
| 0 | 86.5 |
| 1 | 8 |
| 2 or more | 5.5 |
|
| |
| Number of double contrast barium enemas performed per patient (%) | |
| 0 | 92.5 |
| 1 | 5.5 |
| 2 or more | 2 |
Percentages not mutually exclusive
Detection of CRC Test Results
The inter-rater agreement regarding patient need for CRC testing in study patients was high, with a kappa value of 0.80. Within these twenty patient charts, the adjudicated reference standard identified 29 CRC-related completed tests. Agreement between the two reviewers was 79% regarding identification of any completed CRC tests (kappa 0.54). Upon review of the six disagreements by a third reviewer, all were judged false negatives by one of the primary reviewers. On average, it took physician reviewers 11 minutes to complete one manual chart review (range 3–35 minutes).
Table 2 indicates the recall and precision for each of KMCI, manual chart review, and billing record review in identifying references to individually completed CRC-related tests, and for all four CRC-related tests combined. For both individual and combined CRC tests, KMCI had higher recall (93%) than chart review (74%) and billing record review (44%). Precision was higher for chart review (98%) and for billing record review (99%) than for KMCI (94%) for detecting references to colonoscopy. KMCI’s precision (94%) was higher than billing record review (83%) but lower than chart review (98%) for detecting references to any CRC testing.
Table 2.
Comparison of Methods to Detect Colorectal Cancer (CRC) Tests Compared to Adjudicated Reference Standard in 200 Patients
| All CRC Tests (n=265) | Colonoscopy (n=190) | FSIG (n=10) | DCBE (n=19) | FOBT (n=46) | |
|---|---|---|---|---|---|
|
| |||||
| NLP system | |||||
| Recall | 93% * | 92% | 100% | 100% | 96% |
| Precision | 94% †,‡ | 94% | 91% | 100% | 92% |
| F-measure | 94% | 93% | 95% | 100% | 94% |
|
| |||||
| Chart review | |||||
| Recall | 74% | 72% | 80% | 53% | 91% |
| Precision | 98% | 98% | 89% | 100% | 98% |
| F-measure | 84% | 83% | 84% | 69% | 94% |
|
| |||||
| Billing records | |||||
| Recall | 44% | 56% | 20% | 42% | 2% |
| Precision | 83% | 99% | 67% | 100% | 4% |
| F-measure | 58% | 71% | 31% | 59% | 3% |
p<0.001 when comparing recall of NLP to both chart review and billing records
p=0.1 when comparing precision of NLP to chart review
p=0.001 when comparing precision of NLP to billing records
NLP=natural language processing; CRC = colorectal cancer; FSIG = flexible sigmoidoscopy; DCBE = double contrast barium enema; FOBT = fecal occult blood testing
To highlight how each method performed to detect CRC test receipt for an individual patient, we compared the performance of each method using only the most recent CRC screening test for each patient. For this evaluation, each patient had a maximum of one CRC test, which minimized bias towards any method that detected multiple CRC tests better than other methods. Recall and precision for detecting the most recent CRC test of all CRC test types were 91% and 95% for NLP, 79% and 99% for manual review, and 50% and 85% for billing records. Performance to detect the most recent of each individual CRC test type was not significantly different from the test-level data presented in Table 2.
KMCI Performance by Note Type
Table 3 shows the recall and precision of KMCI for identifying references to CRC-related testing in different clinical note types available in the EHR. These note types include clinical narratives (inpatient and outpatient notes) as well as procedure reports (lower endoscopy reports, radiology reports, or laboratory reports). The precision of KMCI to correctly identify references to any CRC testing in semi-structured reports was consistently above 95%; recall varied from 55% to 90% depending on the CRC test reference. In all cases, recall and F-measure improved by combining both note types. Poorer recall from semi-structured reports resulted from an absence of the corresponding document type describing the given events (e.g., no operative report for a colonoscopy performed at another institution). Most precision errors resulted from physician errors in date estimation (e.g., “three years ago” when the test was actually four years prior) or from failure of the NLP algorithm to identify a status word in the context of the sentence (e.g., “Her last colonic evaluation was five years ago when Dr. [Name] attempted to perform a colonoscopy”). When we applied KMCI to the combination of clinical narratives and semi-structured reports, precision was highest for references to DCBE (100%), followed by references to colonoscopy (94%), FOBT (92%) and FSIG (91%). Recall was highest for references to DCBE and FSIG (both 100%) when KMCI was applied to the combination of clinical narratives and procedure reports.
Table 3.
Recall and Precision of Natural Language Processing (NLP) Algorithms
| NLP of Clinical Narratives only | NLP of Semi-Structured Reports only | NLP of All Notes | Adjudicated Reference Standard | |
|---|---|---|---|---|
|
| ||||
| Unique colonoscopy tests | ||||
| Discovered (n) | 157 | 105 | 185 | 190 |
| Correctly identified (n) | 146 | 105 | 174 | -- |
| Recall | 77% | 55% | 92% | -- |
| Precision | 93% | 100% | 94% | -- |
| F-measure | 84% | 71% | 93% | -- |
|
| ||||
| Unique FSIG tests | ||||
| Discovered (n) | 9 | 6 | 11 | 10 |
| Correctly identified (n) | 8 | 6 | 10 | -- |
| Recall | 80% | 60% | 100% | -- |
| Precision | 89% | 100% | 91% | -- |
| F-measure | 84% | 75% | 95% | -- |
|
| ||||
| Unique DCBE tests | ||||
| Discovered (n) | 6 | 17 | 19 | 19 |
| Correctly identified (n) | 6 | 17 | 19 | -- |
| Recall | 32% | 89% | 100% | -- |
| Precision | 100% | 100% | 100% | -- |
| F-measure | 48% | 94% | 100% | -- |
|
| ||||
| Unique FOBT tests | ||||
| Discovered (n) | 11 | 42 | 48 | 46 |
| Correctly identified (n) | 9 | 40 | 44 | -- |
| Recall | 20% | 87% | 96% | -- |
| Precision | 82% | 95% | 92% | -- |
| F-measure | 32% | 91% | 94% | -- |
CRC = colorectal cancer; FSIG = flexible sigmoidoscopy; DCBE = double contrast barium enema; FOBT = fecal occult blood testing
Status for Patients in need of Colonoscopy and Metrics
Among the 200 test cohort patients, 83 patients (42%) were not up to date for recommended CRC screening (assuming average risk) as determined by the adjudicated reference standard (Figure). Using the NLP methods, 90 patients would be recommended for screening, including 11 patients that were actually up to date and did not need screening (Table 4). Four patients would be missed for screening (falsely recorded as being up to date) by NLP alone. Using billing records alone, 122 patients would be recommended for CRC testing, 40 of whom did not need screening. One patient would be missed for screening. Using manual chart review, screening would be recommended for 100 patients, 18 of whom who did not need screening, and missing one patient who needed screening. In summary, for detecting patients in need of screening, NLP had a recall of 95% and precision of 88%; billing records had a recall of 99% and precision of 67%; and manual chart review had a recall of 99% and precision of 82%.
Figure.
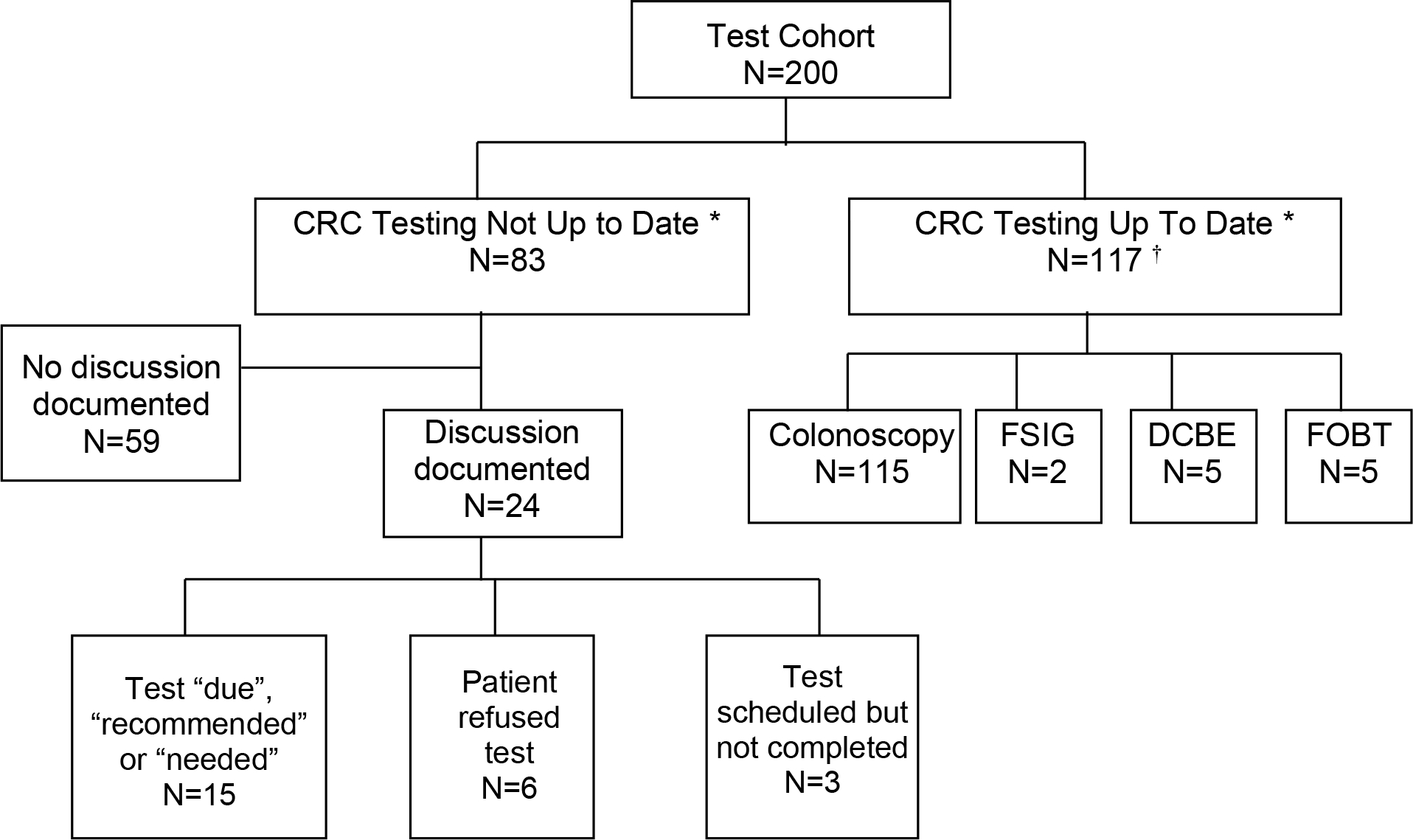
Detection of CRC Testing and Discussions by Natural Language Processing
* According to 2002 United States Preventive Task Force Guidelines for average risk individuals22
† Tests not mutually exclusive
Table 4.
Number of Patients Recommended for CRC Screening by Each Method.
| Number of Patients: | NLP | Billing Records | Chart Review |
|---|---|---|---|
|
| |||
| Recommended for screening (all positives) | 90 | 122 | 100 |
| Correctly labeled “not up to date” (True positives) | 79 | 82 | 82 |
| Correctly labeled “up to date” (True pegatives) | 106 | 77 | 99 |
| Incorrectly labeled “not up to date” (False positives) | 11 | 40 | 18 |
| Incorrectly labeled “up to date” (False negatives) | 4 | 1 | 1 |
| Recall | 95%† | 99% | 99% |
| Precision | 88%* | 67% | 82% |
| F-measure | 91% | 80% | 90% |
p=0.17 when comparing recall of NLP to either chart review or billing records
p<0.001 when comparing precision of NLP to billing records
For the patients not up to date, the majority (59 patients; 71%) contained no documentation regarding CRC-related testing (Figure). For the remaining 24 patients not up to date, EHR review indicated that CRC-related testing was “needed,” “recommended,” or “due” for 15. Six patients had refused CRC testing, while three patients had scheduled but not completed CRC testing.
DISCUSSION
In the current study, NLP of electronic health records outperformed the use of billing records to identify patients who received previous CRC-related testing. Many patients’ CRC-related tests performed at other institutions did not have corresponding billing records at our institution. This is a common scenario when cross-institutionally linked electronic health records, such as Health Information Exchanges (HIEs) do not exist. The NLP system also detected more CRC tests than physician chart review with only a modest number of false positives. The NLP system categorized the context of decisions to initiate CRC testing, such as whether the test was recommended, scheduled, or declined. Such details are typically not available in admininstrative data, yet are valuable to quality metrics which evaluate whether physicians are appropriately recommending CRC screening to their patents.
Although NLP detected prior CRC screening tests better than either physician chart review or billing records, the clinical utility of the NLP method is mildly diminished when identifying patients in need of screening. NLP methods have superior precision (fewer patients were incorrectly identified as due for screening) but slightly poorer recall (95% vs. 99%, p=0.17). In our cohort of 200 patients, a system based on NLP alone would have recommended screening for eleven people who did not need it, and failed to recommend screening for four people who were due. In contrast, billing records would have recommended screening on 40 people who did not need it but would have missed only one person who was due. Future efforts should work on improving NLP methods or combing them with billing records to improve recall while maintaining high precision.
Several previous studies have documented that NLP can identify a variety of important clinical events documented within EHR systems. These include, among others, adverse events in discharge summaries27 and clinical conditions from radiology reports.28–30 Our study demonstrates the potential of NLP in CRC testing. For a busy primary care provider, tracking CRC test status while also trying to address patients’ current medical problems can be challenging. Implementing a comprehensive EHR is no panacea, as demonstrated by the large gap between observed and ideal rates of CRC testing. Organized and highly accessible data is required to make informed CRC testing decisions. NLP technology can rapidly and efficiently extract information about previous CRC testing from each patient’s electronic record, sometimes with better recall than trained physician abstractors. Indeed, physician abstractors performed especially poorly in finding DCBE testing, likely because of the heterogenous ways in which these were recorded in the chart (e.g., non-standardized note titles and results reported via physician referral letters instead of radiology reports, etc.). The studied NLP system has many potential applications when linked to other types of electronic systems that address CRC testing. NLP could enhance procedure completion tracking systems, quality metric monitoring systems, and reminder systems that feed back to patients, providers, or institutions. For example, a clinical reminder system using NLP could provide real-time recommendations for CRC screening to providers as they access the patient’s electronic chart.
Our NLP system is one of the first to combine concept detection, temporal extraction and application, and status identification in the context of CRC testing. Required NLP features included detection of events, their timing, and their status across many clinical note types. Of note, KMCI achieved high precision despite the “multiple reference problem.” For example, in any given chart, there may exist 10 references to the same event, making it easy for one or more references to be misinterpreted. Many NLP system evaluation approaches would report this as 90% precision (assuming 1 of 10 references interpreted incorrectly); the current study would report this as 50% precision because the 9 similar references collapsed into one event occurrence. Despite this stringent constraint, KMCI system precision was high enough to allay concerns that false positive events might prevent future systems from categorizing patients incorrectly as having received CRC-related testing when they actually had not.
Our study has limitations. It was performed at a single institution with a comprehensive, locally-developed EHR system, and results may not be generalizable to other institutions. There was moderate inter-rater reliability (kappa 0.54) between physician chart reviewers regarding completed CRC tests despite physicians receiving training regarding standardized data collection. This finding reflects the difficulty of identifying test results scattered among electronic documentation and highlights the importance of automated approaches. In terms of limitations of our NLP system, KMCI can only be applied if medical text exists in an electronic format; it will not work for any portions of the chart that are handwritten or scanned. In addition, KMCI was tested on only a small number of test charts. Our reference standard incorporated the manually-verified correct output from KMCI; as we were testing the accuracy of KMCI, this could represent a form of incorporation bias. However, it would be unrealistic to expect an accurate manual review (the traditional gold standard) of every single line of text present in an EHR; thus, development of most NLP systems allow for incorporation of the system’s output into the reference standard. Finally, the ability of KMCI to detect CRC tests is limited by the information available in the EHR. If CRC testing is done at an outside site, it may not be documented in the EHR.
We must develop simple, fast, and cost-effective interventions to ameliorate currently low CRC screening rates in the United States. The literature describes a spectrum of potential interventions, including: provider education,31;32 assessing physicians’ health beliefs regarding screening,33 incorporating new screening technologies,34 and direct patient communication via print, email, and office encounters.35–37 The current study documents that an additional viable method for improving screening rates is the incorporation of automated data analysis systems into daily care practice. By improving identification of past receipt of CRC-related testing via automated data systems, it becomes readily transparent which patients still require screening. Our study results suggest that a robust system to identify CRC-related testing in EHR systems should incorporate NLP methods. Our NLP system allows for precise and timely identification of references to CRC testing in the EHR compared to traditional identification methods.
ACKNOWLEDGEMENTS
We would like to thank Dr. Jennifer K. Green at Vanderbilt University Medical Center for assisting in chart reviews. This work was supported by grants R21 CA116573 from the National Cancer Institute, and, in part, by R01 LM007995 from the National Library of Medicine and Vanderbilt CTSA grant 1 UL1 RR024975 from the National Center for Research Resources, National Institutes of Health.
Footnotes
CONFLICTS OF INTEREST
None of the authors have dual commitments or conflicts of interest.
Reference List
- (1).American Cancer Society. Cancer Facts and Figures 2009. 2009. Atlanta, GA, American Cancer Society. [Google Scholar]
- (2).Winawer SJ, Zauber AG, Ho MN et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. New England Journal of Medicine 1993; 329:1977–1981. [DOI] [PubMed] [Google Scholar]
- (3).U.S.Preventive Services Task Force. Screening for colorectal cancer: recommendation and rationale. Ann Intern Med 2002; 137:129–131. [DOI] [PubMed] [Google Scholar]
- (4).Use of colorectal cancer tests--United States, 2002, 2004, and 2006. MMWR Morb Mortal Wkly Rep 2008; 57:253–258. [PubMed] [Google Scholar]
- (5).Swan J, Breen N, Coates RJ et al. Progress in cancer screening practices in the United States: results from the 2000 National Health Interview Survey. Cancer 2003; 97:1528–1540. [DOI] [PubMed] [Google Scholar]
- (6).Freeman JL, Klabunde CN, Schussler N et al. Measuring breast, colorectal, and prostate cancer screening with medicare claims data. Med Care 2002; 40(8 Suppl):IV–42. [DOI] [PubMed] [Google Scholar]
- (7).Gordon NP, Hiatt RA, Lampert DI. Concordance of self-reported data and medical record audit for six cancer screening procedures. J Natl Cancer Inst 1993; 85:566–570. [DOI] [PubMed] [Google Scholar]
- (8).Montano DE, Phillips WR. Cancer screening by primary care physicians: a comparison of rates obtained from physician self-report, patient survey, and chart audit. Am J Public Health 1995; 85:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Payne TH, Murphy GR, Salazar AA. How well does ICD9 represent phrases used in the medical record problem list? Proc Annu Symp Comput Appl Med Care 1992;654–657. [PMC free article] [PubMed] [Google Scholar]
- (10).Zack DL, DiBaise JK, Quigley EM et al. Colorectal cancer screening compliance by medicine residents: perceived and actual. Am J Gastroenterol 2001; 96:3004–3008. [DOI] [PubMed] [Google Scholar]
- (11).Aronsky D, Fiszman M, Chapman WW et al. Combining decision support methodologies to diagnose pneumonia. Proc AMIA Symp 2001;12–16. [PMC free article] [PubMed] [Google Scholar]
- (12).Chapman WW, Fiszman M, Dowling JN et al. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap. Stud Health Technol Inform 2004; 107(Pt 1):487–491. [PubMed] [Google Scholar]
- (13).Friedman C Towards a comprehensive medical language processing system: methods and issues. Proc AMIA Annu Fall Symp 1997;595–599. [PMC free article] [PubMed] [Google Scholar]
- (14).Friedman C, Hripcsak G. Evaluating natural language processors in the clinical domain. Methods Inf Med 1998; 37:334–344. [PubMed] [Google Scholar]
- (15).Friedman C, Hripcsak G, Shablinsky I. An evaluation of natural language processing methodologies. Proc AMIA Symp 1998;855–859. [PMC free article] [PubMed] [Google Scholar]
- (16).Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Work in progress. Radiology 1990; 174:543–548. [DOI] [PubMed] [Google Scholar]
- (17).Denny JC, Smithers JD, Miller RA et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003; 10:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Denny JC, Peterson JF. Identifying QT prolongation from ECG impressions using natural language processing and negation detection. Stud Health Technol Inform 2007; 129(Pt 2):1283–1288. [PubMed] [Google Scholar]
- (19).Denny JC, Bastarache L, Sastre EA et al. Tracking medical students’ clinical experiences using natural language processing. J Biomed Inform 2009; 42: 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Denny JC, Miller RA, Waitman LR et al. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform 2009; 78 Suppl 1:S34–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Denny JC, Arndt FV, Dupont WD et al. Increased hospital mortality in patients with bedside hippus. Am J Med 2008; 121:239–245. [DOI] [PubMed] [Google Scholar]
- (22).Ritchie MD, Denny JC, Crawford DC et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010; 86:560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001;17–21. [PMC free article] [PubMed] [Google Scholar]
- (24).Denny JC, Peterson JF, Choma NN et al. Extracting timing and status descriptors for colonoscopy testing from electronic redical records. J Am Med Inform Assoc. 2010; 17:383–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Screening for colorectal cancer: recommendation and rationale. Ann Intern Med 2002; 137:129–131. [DOI] [PubMed] [Google Scholar]
- (26).Levin B, Lieberman DA, McFarland B et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 2008; 134:1570–1595. [DOI] [PubMed] [Google Scholar]
- (27).Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005; 12:448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fiszman M, Chapman WW, Aronsky D et al. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 2000; 7:593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hripcsak G, Friedman C, Alderson PO et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995; 122:681–688. [DOI] [PubMed] [Google Scholar]
- (30).Mendonca EA, Haas J, Shagina L et al. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform 2005; 38:314–321. [DOI] [PubMed] [Google Scholar]
- (31).Walsh JM, Salazar R, Terdiman JP et al. Promoting use of colorectal cancer screening tests. Can we change physician behavior? J Gen Intern Med 2005; 20:1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lane DS, Messina CR, Cavanagh MF et al. A provider intervention to improve colorectal cancer screening in county health centers. Med Care 2008; 46(9 Suppl 1):S109–S116. [DOI] [PubMed] [Google Scholar]
- (33).Shieh K, Gao F, Ristvedt S et al. The impact of physicians’ health beliefs on colorectal cancer screening practices. Dig Dis Sci 2005; 50:809–814. [DOI] [PubMed] [Google Scholar]
- (34).Zauber AG, Levin TR, Jaffe CC et al. Implications of new colorectal cancer screening technologies for primary care practice. Med Care 2008; 46(9 Suppl 1):S138–S146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Rawl SM, Champion VL, Scott LL et al. A randomized trial of two print interventions to increase colon cancer screening among first-degree relatives. Patient Educ Couns 2008; 71:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Carcaise-Edinboro P, Bradley CJ. Influence of patient-provider communication on colorectal cancer screening. Med Care 2008; 46:738–745. [DOI] [PubMed] [Google Scholar]
- (37).Chan EC, Vernon SW. Implementing an intervention to promote colon cancer screening through e-mail over the Internet: lessons learned from a pilot study. Med Care 2008; 46(9 Suppl 1):S117–S122. [DOI] [PubMed] [Google Scholar]


