PURPOSE
Profiling of circulating tumor DNA (ctDNA) is increasingly adopted in the management of solid tumors, concurrent with increased availability of more comprehensive ctDNA panels. However, variable ctDNA shed can result in variable assay sensitivity. We studied the relationship between ctDNA tumor fraction (TF) and detection of actionable alterations across cancer types.
METHODS
A total of 23,482 liquid biopsies (LBx) submitted between September 2020 and October 2021 were sequenced using a hybrid capture panel that reports genomic alterations (GAs) and genomic biomarkers across 324 cancer-related genes. The primary end points were the prevalence of targetable GAs by cancer type and detection in relationship to ctDNA TF. Sensitivity of detection in LBx was assessed in 1,289 patients with available tissue results.
RESULTS
94% (n = 22,130) of LBx had detectable ctDNA, with a median TF of 2.2%. LBx profiling detected GAs in National Comprehensive Cancer Network category 1 genes in 37% of lung, 30% of prostate, 36% of breast, and 51% of colon cancer cases. Potential germline GAs flagged on clinical reports were detected in genes including BRCA1/2, PALB2, CHEK2, and ATM. Polyclonal mutations in genes associated with resistance such as AR, ESR1, RB1, and NF1 were detected. The sensitivity of LBx to detect driver alterations identified in tissue biopsy from the same patient ranged from 58% to 86% but was consistently at or near 100% in cases with TF ≥ 10%.
CONCLUSION
Elevated ctDNA shed is associated with both high sensitivity and negative predictive value for detection of actionable GAs. The presence of elevated TF suggests adequate tumor profiling and may reduce the value of subsequent reflex to confirmatory tissue testing in patients with negative LBx results.
INTRODUCTION
Genomic profiling is an increasingly routine component in the care of patients with solid tumor malignancies. At the time of writing, the National Comprehensive Cancer Network (NCCN) guidelines recommend genomic testing as a component of the care of 17 different solid tumors, including nine cancers where it is recommended for early-stage disease.1 However, logistical challenges in obtaining routine tumor genomic testing for all patients have created a need for additional diagnostic approaches.
CONTEXT
Key Objective
This study examined a large clinical liquid biopsy cohort with algorithmically estimated levels of plasma tumor fraction (TF) to assess the relationship between circulating tumor DNA (ctDNA) shed and detection of actionable genomic alterations (GAs) in a range of cancer types and a comparison with matched tissue.
Knowledge Generated
GAs in National Comprehensive Cancer Network–listed actionable genes were detected commonly in lung, prostate, breast, and colon cancers (30%-51%). Potential germline variants in 24 actionable cancer susceptibility genes were detected in 7% of cases. In patients with tissue results available, sensitivity was associated with ctDNA shed and was 58%-85% overall and 94%-100% when elevated TF was present.
Relevance
Liquid biopsy represents a pragmatic alternative to tissue next-generation sequencing for detection of guideline-adherent GA in a range of cancer types. When TF is elevated, high negative predictive value for targetable alterations suggests high confidence in negative results, enabling prioritization of tissue reflex for low ctDNA samples.
Liquid biopsy (LBx) of circulating tumor DNA (ctDNA) has emerged as a compelling and pragmatic alternative to genomic analysis of tissue. With the US Food and Drug Administration (FDA) approval of several multigene assays offering tumor profiling of ctDNA,2,3 LBx is becoming a routine component of the care of a range of cancer types.4,5 However, the available FDA-approved LBx assays are quite different, with some covering a single gene and the most comprehensive spanning more than 300 genes. Yet, these assays are similar in that they share one limitation, which is that inadequate ctDNA shed into the blood can produce false-negative results. Because ctDNA shed is variable in patients with cancer, the FDA labeling dictates that a negative LBx should be confirmed with tumor testing to mitigate the risk of false-negative results.2,3,6,7
Here, we study this relationship between ctDNA shed and detection of actionable alterations on LBx testing. With over a year passed since the FDA approval of the largest FDA-approved LBx panel, FoundationOne Liquid CDx (F1LCDx), we reviewed the experience with this assay across > 23,000 specimens tested within our research data set. We hypothesized that ctDNA shed would be variable across cancer types and that this would affect the detection of guideline-associated actionable alterations on comprehensive genomic profiling (CGP) of ctDNA.
METHODS
Study Cohorts
We analyzed a consecutive series of LBx (F1LCDx) ordered within the United States between September 2020 and October 2021 during routine clinical care. Samples from patients with hematologic malignancies and with solid tumors having fewer than 50 LBx tested were excluded from the analysis, and patients with multiple LBx had one specimen chosen on the basis of quality metrics for a final cohort of LBx from 23,482 unique patients representing 25 solid tumor types (the CONSORT diagram is given in the Data Supplement).
For putative acquired resistance mutation analysis, LBx results were compared with tissue biopsies from patients with non–small-cell lung cancer (NSCLC), prostate cancer, breast cancer, and colorectal cancer (CRC) taken from local or metastatic sites (N = 92,932: local site, n = 55,944; metastatic site, n = 36,988). Many of the biopsies from primary sites are archival/resection specimens submitted for sequencing later than collection, in the advanced/metastatic setting, and thus may be biased toward more aggressive biology.
Concordance of tissue and liquid CGP results was performed on the subset of patients who had results for both, provided that blood collection for liquid CGP was within 0-5 years after tissue collection: NSCLC (n = 613), breast cancer (n = 292), CRC (n = 279), and pancreatic cancer (n = 105; the CONSORT diagram is given in the Data Supplement). A cohort from a previously published prospective trial (IMpower130, ClinicalTrials.gov identifier: NCT02367781)8 was used for validation of concordance of EGFR driver detection in NSCLC, where tissue results were available from local testing and/or central testing and LBx had been profiled with F1LCDx (n = 620).
Approval for this study, including a waiver of informed consent and Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (protocol 20152817). IMpower130 was performed in accordance with the International Conference on Harmonisation E6 and E2A and the Declaration of Helsinki.8
Comprehensive Genomic Profiling
CGP was performed in a Clinical Laboratory Improvement Amendment–certified, College of American Pathologists–accredited, New York State–approved laboratory (Foundation Medicine, Cambridge, MA).
LBx were profiled using a validated, FDA-approved next-generation sequencing panel assay F1LCDx.9 Circulating cell-free DNA was extracted from whole blood. CGP was performed using hybridization-captured, adaptor ligation–based libraries to a median unique coverage depth of 6,181× (range: 569-16,087×). F1LCDx reports single-nucleotide variants, insertions/deletions, genomic rearrangements, copy number amplifications, and losses in 324 cancer-related genes (see the Data Supplement for the gene list). F1LCDx also reports genomic signatures including blood tumor mutational burden (bTMB), high microsatellite instability (MSI-H), and tumor fraction (TF). bTMB was determined on the basis of 0.8 megabases, and microsatellite instability was measured using 1,800 loci.9
Tissue biopsies were analyzed using FoundationOne CDx as previously described.10,11 Briefly, the pathologic diagnosis of tissue biopsy was confirmed on routine hematoxylin and eosin–stained slides. Samples with a minimum of 20% tumor nuclei underwent DNA extraction, and 50-1,000 ng of DNA was used for hybrid capture of the same 324 cancer-related genes interrogated by F1LCDx.
Tumor Fraction Quantification
The levels of ctDNA shed for each LBx were quantified by calculating an investigational composite TF, which merges two methods for estimation of TF12. When TF is elevated (≥ 10%), an estimate is returned on the basis of a measure of tumor aneuploidy that incorporates observed deviations in coverage across the genome (Data Supplement). When lack of detectable tumor aneuploidy limits the ability to estimate TF, a variant-based calculation is made by identifying the highest allele fraction nongermline variant, excluding specific clonal hematopoiesis (CH)–associated alterations.12 This aneuploidy-based approach avoids erroneous inference of elevated TF because of the presence of germline variants detected at high variant allele frequency (VAF). See the Data Supplement for more details on the aneuploidy-based TF estimation method.
Refer to the Data Supplement for statistical methods, end points, and germline variant prediction.
RESULTS
Circulating Tumor DNA Shed
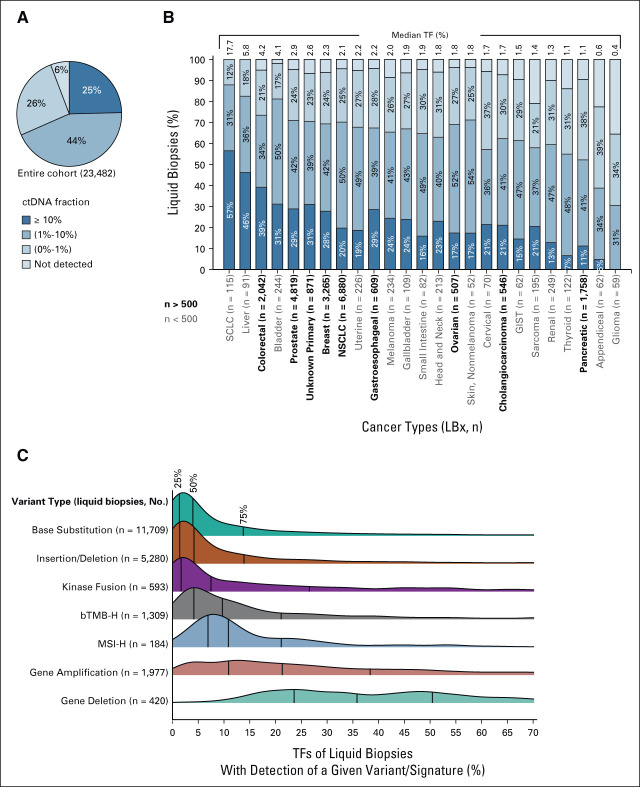
A total of 23,482 LBx were analyzed from 25 cancer types included in the analysis (Data Supplement). Focusing on 3,300 LBx reported in the most recent 2 months before the data lock, the median turnaround time (excluding cases with administrative holds) was 7.3 days from accessioning to report release (Data Supplement). Of the entire cohort of 23,482 LBx, 22,130 (94%) had detectable ctDNA (Fig 1A), and the median TF was 2.2%. ctDNA content of LBx varied somewhat by cancer type, with SCLC, liver, colon, and bladder cancers having a median ctDNA more than 4% on the high end, whereas cancers such as glioma and appendiceal cancer had median TFs below 1% (Fig 1B).
FIG 1.
ctDNA content of liquid biopsies from 23,482 patients. (A) 94% of samples had some level of detectable tumor content. (B) Variable ctDNA shed was seen across a range of cancer types, listed in order of median ctDNA fraction; the most common cancer types in the cohort (n > 500) are given in bold. (C) Variant/signature detection is somewhat dependent on ctDNA fraction, with short variants and kinase fusions commonly detected at low ctDNA fraction, but higher ctDNA levels seen in cases with detection of genomic signatures (bTMB-H [≥ 10 mutations/Mb], MSI-H) or copy number alterations. Quartiles are indicated on the density plots with vertical lines. Variant types are arranged in order of median TF of samples. bTMB-H, high blood tumor mutational burden; ctDNA, circulating tumor DNA; MSI-H, high microsatellite instability; LBx, liquid biopsy; TF, tumor fraction.
Detection of Actionable Alterations
It has been reported previously that higher levels of ctDNA content may be needed for sensitive detection of some genomic alterations such as copy number gain.13,14 We found that base substitutions, insertions/deletions (indels), and kinase fusions (Data Supplement) were detected in samples with median TFs of 4.0%, 4.2%, and 7.5%, respectively (Fig 1C), with detection of all variant types at VAFs as low as 0.1%, consistent with expected analytic performance.9 LBx in which bTMB ≥ 10 mutations/Mb (bTMB-High) or MSI-H was detected tended to have higher TFs (median 9.7% and 10.9%, respectively), consistent with higher tumor content enabling detection of more complex genomic signatures. Despite the higher median, bTMB-High and MSI-H were detected in biopsies with TFs as low as 0.93% and 0.44%, respectively. Copy number amplifications and gene deletions were detected in samples with even higher TF (amplifications: median 21.3%, interquartile ranger [IQR] [10.9%-38.4%]; deletions: median 35.8%, IQR [23.6%-50.5%]), consistent with the analytic expectations of the assay.9 Nevertheless, copy number amplifications were sometimes detected in samples with no other alterations or estimable aneuploidy (suggesting a TF < 10%). This phenomenon may reflect detection of high copy number amplifications in low TF samples.
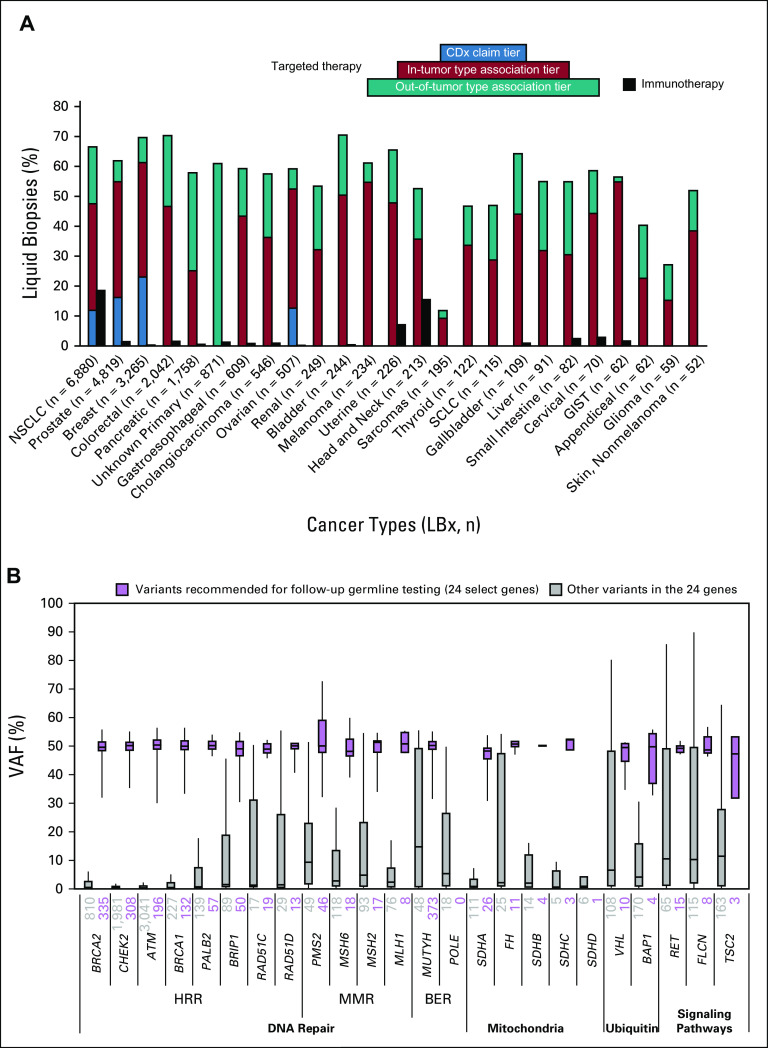
CGP of LBx detected variants with associated targeted therapies in most samples across many cancer types (Fig 2A). In the four most prevalent cancer types (NSCLC, prostate, breast, and CRC), genomic variants with in-tumor type–targeted therapy associations were identified in 47%-61% of samples. Of note, among samples where the primary cancer diagnosis was uncertain, 531 of 871 (61%) harbored variants with associated targeted therapies, highlighting the ability of LBx to help select targeted therapy in a tumor-agnostic fashion.
FIG 2.
Detection of targetable alterations and potential germline variants in liquid biopsies. (A) The percentage of liquid biopsies with clinical reports with in-tumor type therapy associations ranged from 61% of breast cancer specimens to 12% of sarcoma specimens, ordered by cancer type cohort size. The black immunotherapy bar indicates the percentage of samples with detected microsatellite instability (MSI-H) in any tumor type and/or detected bTMB-H in NSCLC and head and neck cancer samples. The percentage of samples with CDx claims on the report are shown in NSCLC, prostate, breast, and ovarian cancer. (B) Focusing on pathogenic variants detected 24 actionable cancer susceptibility genes, and qualifying ClinVar variants detected at > 30% VAF are reported with a banner recommending consideration of germline testing (purple). BER, base excision repair; bTMB-H, high blood tumor mutational burden; CDx, companion diagnostic; HRR, homologous recombination repair; LBx, liquid biopsy; MMR, mismatch repair; MSI-H, high microsatellite instability; NSCLC, non–small-cell lung cancer; VAF, variant allele frequency.
The percentage of samples with in-tumor type–associated therapies is dependent on several factors: the availability of pharmaceuticals approved for use in the cancer type, the prevalence of cancer drivers that are targetable, the ctDNA shed rate of the cancer type, and whether germline variants are associated with therapies in the cancer type. For example, although SCLC has a tendency to shed much more ctDNA than NSCLC (median TF 17.7% v 2.1%), this cancer type rarely harbors targetable drivers such as EGFR or ALK, resulting in a low (29%) fraction of samples with associated therapies. Ovarian cancer (52% of samples with in-tumor type–associated therapies) was found to have modest levels of ctDNA shed (median TF 1.8%), but there was nonetheless frequent detection of actionable BRCA1/2 alterations, many of which are of potential germline origin.
Focusing on 24 cancer susceptibility genes (Fig 2B), we found high rates of potential pathogenic germline variants in MUTYH (373), BRCA2 (335), CHEK2 (308), ATM (196), BRCA1 (132), PALB2 (57), and BRIP1 (50). Overall, 7% of samples had a potential pathogenic germline variant detected. Although potential germline variants tended to have a VAF that clustered around 50%, some truncating variants with similar VAFs were not found in ClinVar, highlighting the need for continuous accumulation of epidemiologic and functional evidence in public databases, especially from populations that are less well represented in the genomic literature.
Studying the four most represented cancer types in this study, detection of NCCN biomarkers was common. In NSCLC specimens, 37% carried a variant in NCCN category 1 biomarker genes and an additional 5% were positive for variants in emergent biomarkers defined as ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) level II-III15 biomarker genes (Fig 3A). In prostate cancer specimens, 30% were positive for variants in an NCCN category 1 biomarker gene (Fig 3B). The increased prevalence of alterations in ATM and CHEK2 is likely attributable to CH.16 ESCAT level II-III biomarker gene variants were detected in 12% of additional cases. 36% of breast cancer samples had variants in NCCN category 1 genes, and 18% additional cases had an ESCAT level II-III biomarker (Fig 3C). In CRC samples, 51% were positive for variants in an NCCN category 1 biomarker gene and 8% additional cases had an ESCAT level II-III biomarker (Fig 3D). For each of these cancers, a diversity of significant comutations and emerging targets were also detected (Data Supplement).17
FIG 3.
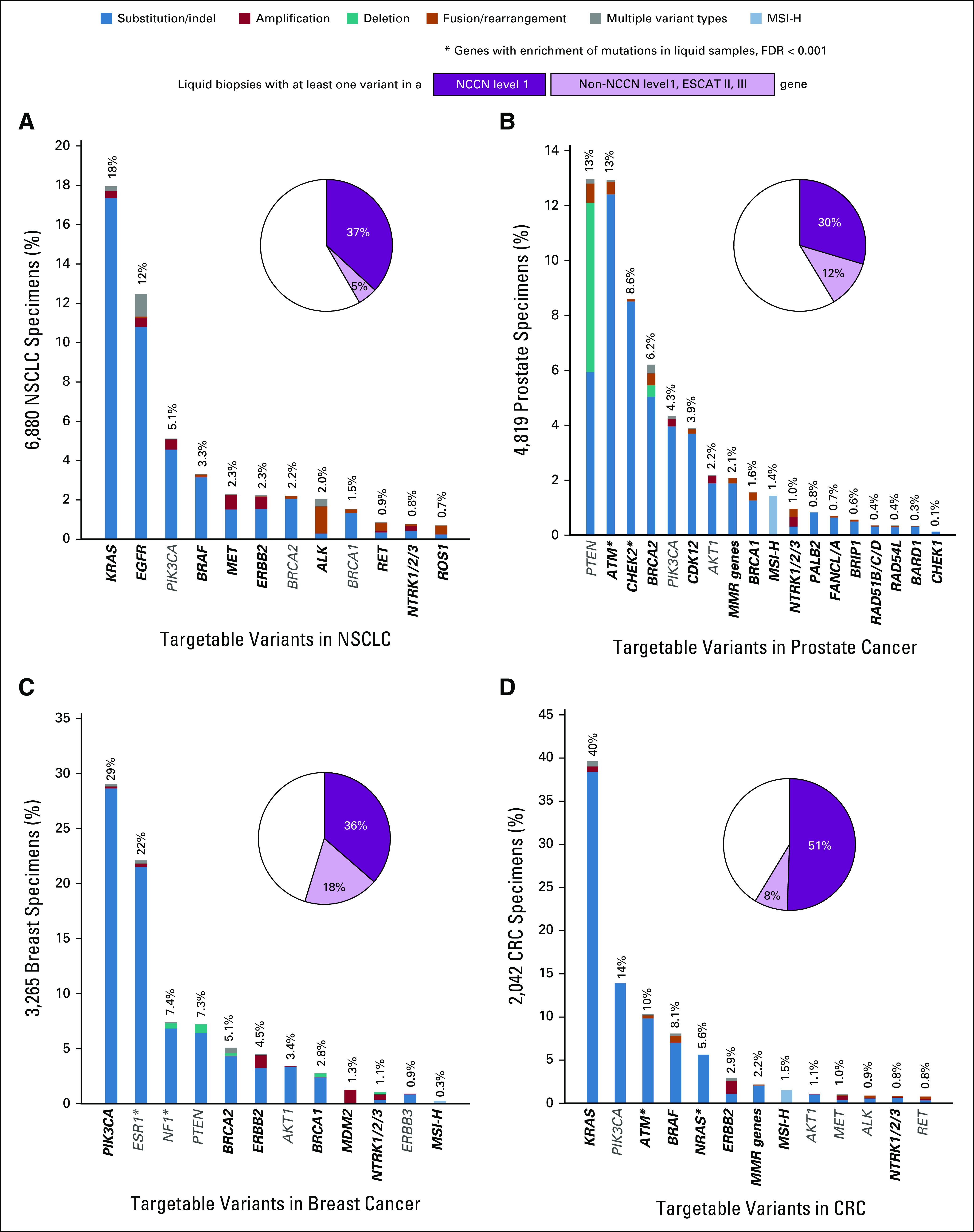
Pathogenic genomic alterations detected in actionable genes across four major cancer types. Plots include NCCN level 1 (dark purple, bold text) and other ESCAT level II-III genes (light purple, gray text) for (A) NSCLC, (B) prostate cancer, (C) breast cancer, and (D) colon cancer (NCCN/ESCAT genes). An asterisk indicates genes that have a significantly higher proportion of alterations detected in liquid biopsies than tissue biopsies (FDR < 0.001). MMR genes refer to MLH1, MSH2, MSH6, and PMS2. CRC, colorectal cancer; ESCAT, ESMO Scale for Clinical Actionability of Molecular Targets; FDR, false discovery rate; MMR, mismatch repair; MSI-H, high microsatellite instability; NCCN, National Comprehensive Cancer Network; NSCLC, non–small-cell lung cancer.
Enrichment in Liquid Biopsy of Polyclonal Alterations Suggestive of Acquired Resistance
We studied detection of polyclonality in ctDNA by examining genes where three or more pathogenic variants were detected in a given sample. We compared detection of multiple mutations in the same gene across tissue biopsies from primary sites, tissue biopsies from metastatic sites, and LBx. For some genes, enrichment of polyclonality in LBx was apparent across cancer types, presumably because of CH (DNMT3A, TET2, and ASXL1, Data Supplement).18 Other genes showed enrichment of polyclonal variants only in specific cancer types with established targeted therapy paradigms (Fig 4), suggesting that the multiple mutations could be caused by different treatment resistance mutations arising in separate tumor subclones. Indeed, such polyclonality was identified in genes with well-established roles in resistance: ALK in NSCLC, AR in prostate cancer, ESR1 in breast cancer, BRCA2 reversions in prostate and breast cancer, and EGFR and KRAS in CRC. Disease-specific analysis also identified polyclonality in more emergent resistance genes including RB1 and NF1 in breast cancer.19,20 Interestingly, for a different subset of actionable genes such as EGFR in NSCLC and PIK3CA in breast cancer, multiple mutations were observed across specimen types without an obvious enrichment for polyclonality in liquid over tissue specimens (Data Supplement).
FIG 4.
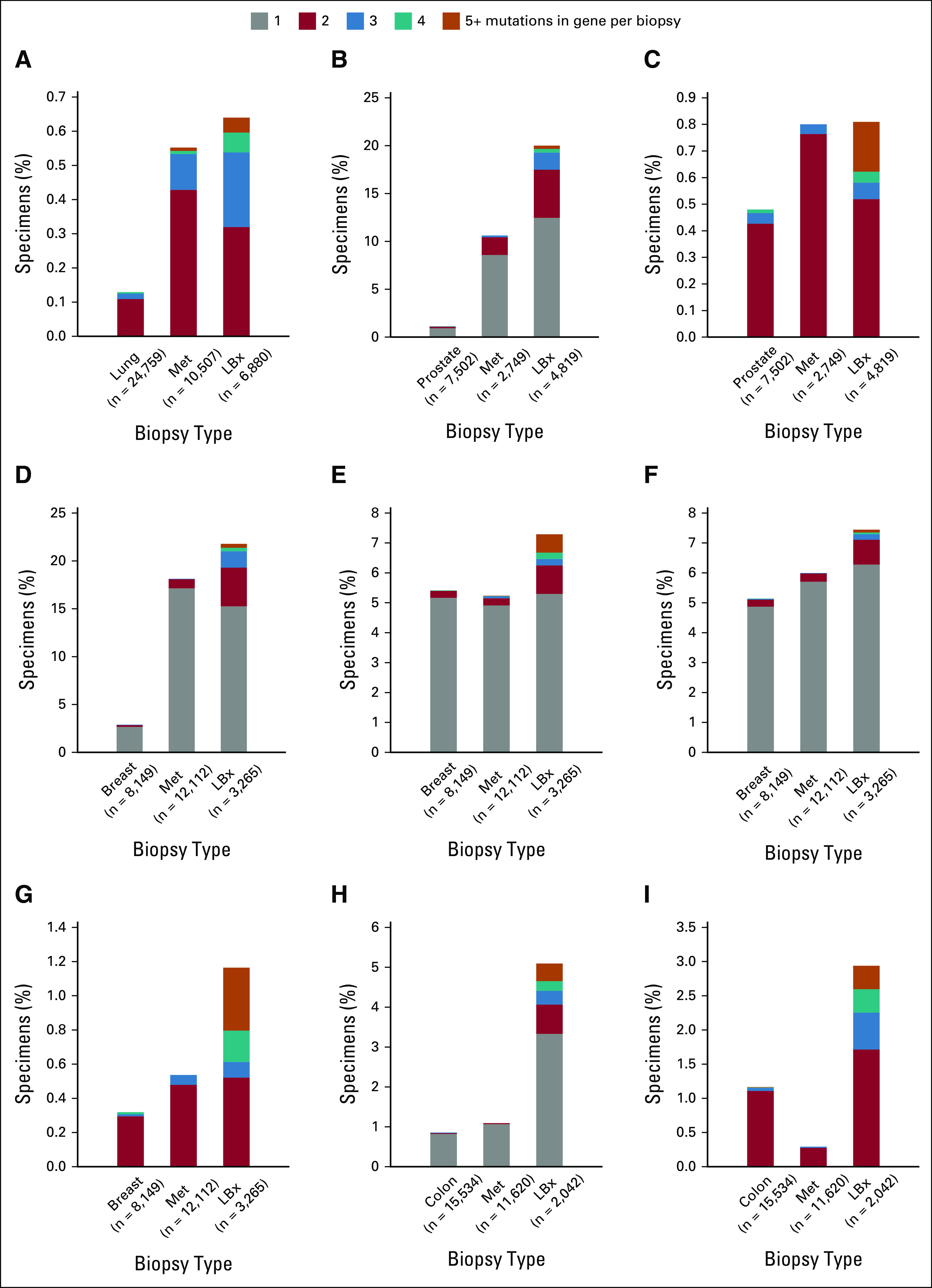
Polyclonality detected in genes associated with resistance. For each of these genes, the detection of multiple mutations in a single NSCLC biopsy is more common in LBx than in tissue biopsies collected at the primary/local site (first bar) and metastatic tissue biopsies (Met); (A) ALK in NSCLC, (B) AR in prostate, (C) BRCA2 in prostate, (D) ESR1 in breast, (E) RB1 in breast, (F) NF1 in breast, (G) BRCA2 in breast, (H) EGFR in CRC, and (I) KRAS in CRC. The number of each cohort is provided in parentheses. (A, C, G, and I) For driver alterations, samples with only one mutation were omitted from the plot. CRC, colorectal cancer; LBx, liquid biopsy; NSCLC, non–small-cell lung cancer.
Sensitivity for Detection of Actionable Alterations in Liquid
Sensitivity for detection of actionable alterations in LBx was assessed in a subset of 1,289 consecutive patients with NSCLC, breast cancer, CRC, or pancreatic cancer, who underwent tissue CGP followed by LBx testing as part of routine cancer care. Blood was collected at a median of 304 days after tissue (IQR: 27-670 days; Fig 5A). Positive percent agreement (PPA; sensitivity) was calculated for actionable biomarkers detected in > 20 tissue samples (Fig 5B). PPA values ranged from 58% to 86% (Fig 5C) but were consistently at or near 100% in the cases with elevated TF (≥ 10%), suggesting that ctDNA content is the primary determinant of tissue-liquid concordance. Indeed, the lowest PPA of 58% was observed for KRAS mutations in pancreatic cancer, the lowest shedding cancer type in this analysis (Fig 1B).
FIG 5.
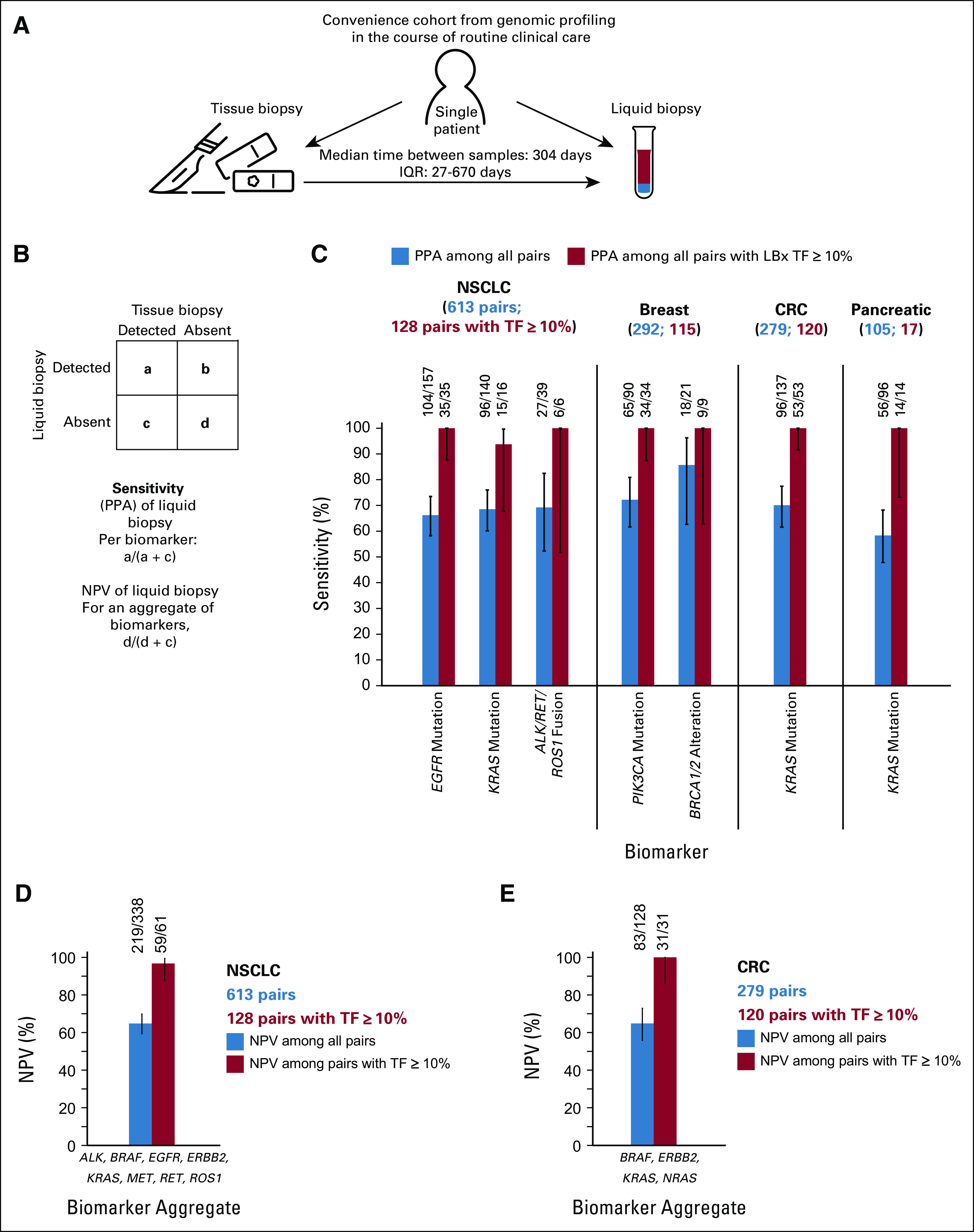
Sensitivity (PPA) and NPV of LBx profiling. Schematic of the convenience cohorts selected from the larger cohort. (A) A consecutive cohort of patients who had results from both tissue and liquid CGP was analyzed. Sensitivity of LBx (PPA) was defined for each individual biomarker with tissue as the reference standard. (B) For NPV of LBx, tissue negative for all tested biomarkers was used as the reference standard. (C) The sensitivity of LBx for detecting mutations and fusions ranged from 58% for KRAS in pancreatic cancer to 86% for BRCA1/2 alterations in breast cancer (blue bars); in the subsets of paired liquid and tissue where LBx ctDNA TF was ≥ 10%, the sensitivity of LBx was > 90% (red bars). (D) The NPV for mutations and fusions in actionable NSCLC biomarkers in eight NCCN genes (EGFR, ERBB2, KRAS, MET, ALK, BRAF, RET, and ROS1) was calculated for 613 NSCLC pairs. (E) Similarly, the NPV for mutations in actionable CRC biomarkers in four genes (BRAF, ERBB2, KRAS, and NRAS) was calculated for 279 CRC pairs. CGP, comprehensive genomic profiling; CRC, colorectal cancer; ctDNA, circulating tumor DNA; IQR, interquartile ranger; LBx, liquid biopsy; NCCN, National Comprehensive Cancer Network; NPV, negative predictive value; NSCLC, non–small-cell lung cancer; PPA, positive percent agreement; TF, tumor fraction.
To ascertain whether the PPA observed in the convenience cohort is representative of an unselected first-line population, we analyzed results from a cohort of 620 patients who had no prior treatment for stage IV nonsquamous NSCLC and had local and/or central tissue testing results from prospective phase III trial IMpower130, where the PPA was 100% (25 of 25; 95% CI, 83 to 100) in 25 patients with EGFR-positive tissue. Negative percent agreement was 98.3% (585 of 595; 95% CI, 96.8 to 99.1); some of the discordance with local testing may reflect a lack of coverage of exon 20 insertions by local tests (Data Supplement).
Given the excellent sensitivity in the setting of elevated TF, we studied whether LBx could be used to rule out the presence of an actionable alteration. We examined the negative predictive value (NPV) of LBx across all NCCN-recommended biomarkers in two cancer types: NSCLC where patients with wild-type status for certain key driver alterations can potentially benefit from immunotherapy and CRC where patients with wild-type status for KRAS and NRAS mutations may benefit from EGFR inhibition. Among 613 NSCLC pairs, the NPV was 65% (95% CI, 59 to 70) overall for actionable variants in eight NCCN genes (BRAF, EGFR, ERBB2, KRAS, MET, ALK, RET, and ROS1), yet the NPV rose to 97% (95% CI, 88 to 99) in the subset of 129 pairs with elevated TF (Fig 5D). Similarly, among 279 CRC pairs, the NPV was 65% (95% CI, 56 to 73) overall for actionable short variants in four NCCN genes (BRAF, ERBB2, KRAS, and NRAS), yet the NPV rose to 100% (95% CI, 86 to 100) in the subset of 120 pairs with elevated TF (Fig 5E).
DISCUSSION
These data suggest that CGP of ctDNA represents a pragmatic approach for detection of guideline-associated actionable alterations across a range of cancer types. Focusing on four common solid tumor types, we found that 30%-51% of LBx were positive for an alteration in an NCCN-listed actionable gene. In parallel, 7% of all LBx were positive for a possible germline variant in one of 24 cancer susceptibility genes and thus may be candidates for germline testing in the appropriate clinical context. Together, these findings highlight the actionability of LBx for patients with advanced solid tumors.
We identified a clear association between detection of actionable alterations and levels of ctDNA shed, which is variable across cancer types. Previous analytical validation and real-world studies of this assay have demonstrated that short variants and fusions can be detected at extremely low VAF (0.1%) with high sensitivity.9,21 In comparison, more complex alterations such as gene amplifications and deletions tended to be detected in patients with higher ctDNA levels. LBx is therefore likely to be most useful in patients with an increased likelihood of ctDNA shed, generally patients with advanced cancer who are untreated or progressing on therapy. For example, in patients with prostate cancer receiving LBx testing, it was recently shown that higher levels of ctDNA shed were associated with a higher prostate‐specific antigen level and collection of LBx within 60 days of new treatment initiation, whereas prostate‐specific antigen < 5 was associated with lower ctDNA shed.22 Optimal clinical use of LBx will require additional efforts to identify clinical features that identify patients who may be better served by tumor tissue profiling.
Importantly, when elevated TF was detected, sensitivity for short variants and fusions was nearly 100% compared with paired tissue from the same patient. In NSCLC and CRC, this resulted in a high NPV for ruling out the presence of certain driver alterations in patients with elevated TF and a negative LBx. Yet, the overall sensitivity for such targetable driver alterations was variable and ranged between 58% and 86%. These data point to the importance of the guidance on the FDA label, which directs that negative results from a LBx are confirmed using tissue testing. However, these data also suggest that the value of such a reflex to tissue testing may be less immediate in the setting of elevated TF. Leveraging ctDNA levels to inform the need for tissue reflex will require clinical grade reporting of TF estimation to guide such decisions. The aneuploidy-based method using germline single nucleotide polymorphisms reported here for estimating TF (Data Supplement) is already reported to clinicians; continuous development is focused on improving the dynamic range of this TF estimation.
We identified recurring evidence of polyclonal resistance mutations in LBx specimens, highlighting how LBx is uniquely suited for capturing the heterogeneity of resistance mechanisms in patients with acquired drug resistance. Across several cancer types and treatment pathways, we found that ctDNA enriches for multiple mutations in putative resistance genes, including in emerging targets such as AR and ESR1. The limited clinical annotation of this cohort makes discovery of novel resistance mechanisms challenging although in other reports, we have identified novel resistance mechanisms such as acquired fusions through analysis of ctDNA.16,21,23,24 We are hopeful that ctDNA-based CGP assays with genomic coverage across a range of cancer types may enable therapeutic targeting of such resistance mechanisms while reducing the burden of repeat biopsies on patients with cancer.
A limitation of the study is that it is a retrospective, real-world analysis of a consecutive cohort of eligible LBx patients and cannot speak directly to whether LBx profiling improves clinical outcomes as a part of routine clinical care. Yet, with a growing number of clinical trials with LBx-driven enrollment, there is a large evidence base demonstrating compelling clinical outcomes in patients with ctDNA-detected biomarkers, comparable with the outcomes in similar patients with tumor tissue–detected biomarkers.25-28 Combined with the regulatory approval of LBx assays, national guidelines including the NCCN are increasingly including LBx as a pragmatic option for tumor profiling. The validity of these assays for detecting such a broad range of actionable alterations offers an opportunity to steadily increase testing rates and increase access to precision therapy options and clinical trials.
ACKNOWLEDGMENT
We thank the patients, families, and physicians who ordered comprehensive genomic profiling from Foundation Medicine Inc. We thank Craig Cummings and Wei Zou for curation of the IMpower130 data and Conor O'Brien for facilitating data transfer. We thank Brandon Kocher and Angela Chen for feedback on analyses.
Hatim Husain
Consulting or Advisory Role: AstraZeneca, Foundation Medicine, PierianDx, Janssen, Blueprint Medicines, NeoGenomics Laboratories, Turning Point Therapeutics
Speakers' Bureau: AstraZeneca, Janssen
Research Funding: Pfizer (Inst), Bristol Myers Squibb (Inst), Regeneron (Inst), Lilly (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Janssen, Foundation Medicine
Dean C. Pavlick
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Bernard J. Fendler
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Patents, Royalties, Other Intellectual Property: The method to estimate the tumor fraction, used in the paper submission, currently has a patent pending. The original submission was on 26.11.2020. The submission number can be provided if needed.
Russell W. Madison
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Brennan Decker
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche, Vaccitech
Consulting or Advisory Role: Foundation Medicine, Avidea Technologies
Patents, Royalties, Other Intellectual Property: Foundation Medicine
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Christine A. Parachoniak
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Molly McLaughlin-Drubin
Employment: Foundation Medicine, Precision for Medicine (I)
Stock and Other Ownership Interests: Roche
Travel, Accommodations, Expenses: Foundation Medicine
Rachel L. Erlich
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Contributed to several patents and patents pending at Foundation Medicine
Travel, Accommodations, Expenses: Foundation Medicine
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Meghna Das Thakur
Employment: Roche/Genentech
Stock and Other Ownership Interests: Roche/Genentech
Research Funding: Roche/Genentech
Patents, Royalties, Other Intellectual Property: Method of treating a proliferative disease
Travel, Accommodations, Expenses: Roche/Genentech
Geoffrey R. Oxnard
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Hanna Tukachinsky
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
No other potential conflicts of interest were reported.
SUPPORT
Supported by the funding of Foundation Medicine Inc.
H.H. and D.C.P. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Hatim Husain, Dean C. Pavlick, Bernard J. Fendler, Garrett M. Frampton, Geoffrey R. Oxnard, Hanna Tukachinsky
Provision of study materials or patients: Hatim Husain, Meghna Das Thakur
Collection and assembly of data: Dean C. Pavlick, Bernard J. Fendler, Russell W. Madison, Garrett M. Frampton, Christine A. Parachoniak, Molly McLaughlin-Drubin, Rachel L. Erlich, Meghna Das Thakur, Hanna Tukachinsky
Data analysis and interpretation: Hatim Husain, Dean C. Pavlick, Brennan Decker, Ole Gjoerup, Christine A. Parachoniak, Molly McLaughlin-Drubin, Rachel L. Erlich, Alexa B. Schrock, Garrett M. Frampton, Geoffrey R. Oxnard, Hanna Tukachinsky
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hatim Husain
Consulting or Advisory Role: AstraZeneca, Foundation Medicine, PierianDx, Janssen, Blueprint Medicines, NeoGenomics Laboratories, Turning Point Therapeutics
Speakers' Bureau: AstraZeneca, Janssen
Research Funding: Pfizer (Inst), Bristol Myers Squibb (Inst), Regeneron (Inst), Lilly (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Janssen, Foundation Medicine
Dean C. Pavlick
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Bernard J. Fendler
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Patents, Royalties, Other Intellectual Property: The method to estimate the tumor fraction, used in the paper submission, currently has a patent pending. The original submission was on 26.11.2020. The submission number can be provided if needed.
Russell W. Madison
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Brennan Decker
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche, Vaccitech
Consulting or Advisory Role: Foundation Medicine, Avidea Technologies
Patents, Royalties, Other Intellectual Property: Foundation Medicine
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Christine A. Parachoniak
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Molly McLaughlin-Drubin
Employment: Foundation Medicine, Precision for Medicine (I)
Stock and Other Ownership Interests: Roche
Travel, Accommodations, Expenses: Foundation Medicine
Rachel L. Erlich
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Contributed to several patents and patents pending at Foundation Medicine
Travel, Accommodations, Expenses: Foundation Medicine
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Meghna Das Thakur
Employment: Roche/Genentech
Stock and Other Ownership Interests: Roche/Genentech
Research Funding: Roche/Genentech
Patents, Royalties, Other Intellectual Property: Method of treating a proliferative disease
Travel, Accommodations, Expenses: Roche/Genentech
Geoffrey R. Oxnard
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Hanna Tukachinsky
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Mateo J, Steuten L, Aftimos P, et al. : Delivering precision oncology to patients with cancer. Nat Med 28:658-665, 2022 [DOI] [PubMed] [Google Scholar]
- 2.US Food and Drug Administration P190032 : FoundationOne® Liquid CDx Technical Information. https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190032C.pdf [Google Scholar]
- 3.US Food and Drug Administration P200010/S001 : Guardant360® CDx Technical Information. https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010S001C.pdf [Google Scholar]
- 4.Rolfo C, Mack P, Scagliotti GV, et al. : Liquid biopsy for advanced NSCLC: A consensus statement from the International Association for the Study of Lung Cancer. J Thorac Oncol 16:1647-1662, 2021 [DOI] [PubMed] [Google Scholar]
- 5.Hasenleithner SO, Speicher MR: A clinician's handbook for using ctDNA throughout the patient journey. Mol Cancer 21:81, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.US Food and Drug Administration P150047C : cobas® EGFR Mutation Test v2. https://www.accessdata.fda.gov/cdrh_docs/pdf15/p150047c.pdf [Google Scholar]
- 7.US Food and Drug Administration P190001C : therascreen® PIK3CA RGQ PCR. https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190001C.pdf [Google Scholar]
- 8.West H, McCleod M, Hussein M, et al. : Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol 20:924-937, 2019 [DOI] [PubMed] [Google Scholar]
- 9.Woodhouse R, Li M, Hughes J, et al. : Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 15:e0237802, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frampton GM, Fichtenholtz A, Otto GA, et al. : Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023-1031, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milbury CA, Creeden J, Yip WK, et al. : Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 17:e0264138, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Pavlick D, Chung JH, et al. : Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy. J Gastrointest Oncol 10:831-840, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrock AB, Frampton GM, Suh J, et al. : Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol 11:1493-1502, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Chung JH, Pavlick D, Hartmaier R, et al. : Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol 28:2866-2873, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosele F, Remon J, Mateo J, et al. : Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann Oncol 31:1491-1505, 2020 [DOI] [PubMed] [Google Scholar]
- 16.Tukachinsky H, Madison RW, Chung JH, et al. : Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res 27:3094-3105, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey MH, Tokheim C, Porta-Pardo E, et al. : Comprehensive characterization of cancer driver genes and mutations. Cell 173:371-385.e18, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Ulrich BC, Supplee J, et al. : False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 24:4437-4443, 2018 [DOI] [PubMed] [Google Scholar]
- 19.Condorelli R, Spring L, O'Shaughnessy J, et al. : Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol 29:640-645, 2018 [DOI] [PubMed] [Google Scholar]
- 20.Sokol ES, Feng YX, Jin DX, et al. : Loss of function of NF1 is a mechanism of acquired resistance to endocrine therapy in lobular breast cancer. Ann Oncol 30:115-123, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JK, Hazar-Rethinam M, Decker B, et al. : The pan-tumor landscape of targetable kinase fusions in circulating tumor DNA. Clin Cancer Res 28:728-737, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonarakis ES, Tierno M, Fisher V, et al. : Clinical and pathological features associated with circulating tumor DNA content in real-world patients with metastatic prostate cancer. Prostate 82:867-875, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awad MM, Liu S, Rybkin II, et al. : Acquired resistance to KRAS(G12C) inhibition in cancer. N Engl J Med 384:2382-2393, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokol E, Sivakumar S, Jin D, et al. : Patient-matched tissue and liquid biopsies identify shared and acquired genomic alterations in breast cancer. J Clin Oncol 38:1050-1050, 2020. 32017652 [Google Scholar]
- 25.Heist RS, Garon EB, Tan DS, et al. : Abstract LB056: Accurate detection of MET exon 14 skipping using a liquid biopsy assay in NSCLC patients in the GEOMETRY mono-1 study. Cancer Res 81:LB056, 2021 [Google Scholar]
- 26.Loehr A, Patnaik A, Campbell D, et al. : Response to rucaparib in BRCA-mutant metastatic castration-resistant prostate cancer identified by genomic testing in the TRITON2 study. Clin Cancer Res 27:6677-6686, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxnard GR, Thress KS, Alden RS, et al. : Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 34:3375-3382, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juric D, Ciruelos E, Rubovszky G, et al. : Abstract GS3-08: Alpelisib + fulvestrant for advanced breast cancer: Subgroup analyses from the phase III SOLAR-1 trial. Cancer Res 79:GS3-08, 2019 [Google Scholar]