Abstract
The gene encoding the pneumococcal surface adhesin A (PsaA) protein has been identified in three different viridans group streptococcal species. Comparative studies of the psaA gene identified in different pneumococcal isolates by sequencing PCR products showed a high degree of conservation among these strains. PsaA is encoded by an open reading frame of 930 bp. The analysis of this fragment in Streptococcus mitis, Streptococcus oralis, and Streptococcus anginosus strains revealed a sequence identity of 95, 94, and 90%, respectively, to the corresponding open reading frame of the previously reported Streptococcus pneumoniae serotype 6B strain. Our results confirm that psaA is present and detectable in heterologous bacterial species. The possible implications of these results for the suitability and potential use of PsaA in the identification and diagnosis of pneumococcal diseases are discussed.
Streptococcus pneumoniae infection is still a major cause of concern in human health. A recent report of the World Health Organization concluded that the impact of pneumococcal disease worldwide is similar to that of tuberculosis (25). It has been emphasized that the development of an improved pneumococcal vaccine is among the three vaccine priorities of industrialized countries (5). The 23-valent pneumococcal-polysaccharide vaccine provides only limited protection in young children, immunocompromised individuals, and elderly people (3, 6, 8, 14). Although the new polysaccharide-protein conjugate vaccine appears to be efficient in these poor responder groups, it will not protect against the capsular types of pneumococcal strains not included in the formulation. A promising approach in overcoming this problem is the use of third-generation vaccines composed of species-specific pneumococcal protein(s), which may elicit long-lasting, broadly protective T-cell-dependent immunity. One of these proteins currently considered as a vaccine candidate is the 37-kDa protein PsaA (pneumococcal surface adhesin A). This protein was first identified by Russell et al. (19) using monoclonal antibodies (MAbs) and has attracted a great deal of interest in recent years. Soon after the protein was identified, the psaA gene was cloned and sequenced (23). Although the two first psaA pneumococcal sequences reported from strains R36A and D39 showed high heterogeneity (1), PCR-restriction fragment length polymorphism analysis showed that psaA is highly conserved among the serotypes included in the 23-valent polysaccharide vaccine (22). In the same study (22), the authors sequenced a serotype 6B strain and concluded that the psaA sequences from D39 and the serotype 6B strain most likely represented the S. pneumoniae prototype sequences. More recently, Novak et al. reported that the psaA gene from a serotype 4 strain was 99.6% identical to the gene from strain D39 and 99.9% identical to the gene from the serotype 6B strain (17). Morrison and coworkers confirmed the presence of psaA in all of the 90 S. pneumoniae serotypes by PCR analysis (16). The specificity of the assay was proven by the lack of a similar signal when analyzing heterologous bacterial species (n = 30) and genera (n = 14), including the viridans group streptococci. This finding suggests that the psaA PCR assay might be successfully used for the detection of pneumococci and diagnosis of pneumococcal diseases (16).
The possible involvement of PsaA in the pathogenesis of pneumococcal disease was indicated by immunization studies performed with purified PsaA (24) and confirmed by insertion-duplication mutagenesis analysis of the psaA gene (1). Recently, Briles et al. (2) observed that immunization with PsaA reduces the carriage of pneumococci, suggesting that PsaA may be useful for the elicitation of herd immunity in humans.
During the search for protein antigens that could elicit protective immune responses against S. pneumoniae, we obtained an MAb (F1-1B) which bound specifically to a 37-kDa protein. Molecular studies have shown that MAb F1-1B binds to PsaA protein. We have also cloned and sequenced psaA from the unencapsulated pneumococcal strain R6 and from one serotype 3 clinical isolate. Moreover, the psaA gene has also been identified and sequenced in three viridans group streptococcal species: S. mitis, S. oralis, and S. anginosus. Fifty clinical isolates of S. mitis and S. oralis showed positive hybridization with a psaA probe. The demonstration of PsaA in heterologous organisms suggests that the effectiveness of this antigen as a useful diagnostic marker should be reconsidered.
MATERIALS AND METHODS
Bacterial strains.
The unencapsulated S. pneumoniae strain R6 was kindly provided by A. Tomasz (Rockefeller University, New York, N.Y.), and S. pyogenes strain 746/96 was provided by J. A. Sáez-Nieto (Centro Nacional de Microbiología, Madrid, Spain). Eleven strains of S. pneumoniae of serotypes 3, 4, 6, 9, 14, 15, 19, and 23 were taken from our laboratory collection. The other Streptococcus strains were S. mitis NCTC 12261, S. oralis NCTC 11427, S. anginosus NCTC 10713, S. sanguinis NCTC 7863, S. thermophilus NCDO 573, S. bovis NCDO 597, and S. mutans NCTC 10449. The Neisseria strains used were N. polysaccharea N 462, N. lactamica ATCC 10618, N. perflava ATCC 10555, N. cinerea ATCC 14685, and N. meningitidis C-11. We also used Escherichia coli ATCC 25922. In addition, we analyzed 50 viridans group streptococci isolated from pharynx exudates, sputum, and lower respiratory tract samples. These viridans group isolates were identified as S. mitis and S. oralis with the Rapid ID32 Strep system.
Protein analysis.
The MAb used in immunoblot analysis was obtained by immunization of female BALB/c Jico mice (Criffa, Lyon, France) with whole-cell suspensions of the S. pneumoniae strain R6. Mice were immunized by one intraperitoneal injection per week for 3 weeks, followed by an intravenous injection. The maximum number of cells injected was 107. Fusion was carried out on day 25 using standard procedures (4). Spleen cells from three mice were fused with P3X63-Ag8.653 myeloma cells (11). Supernatants of the growing hybridoma cultures were tested for antibodies against S. pneumoniae whole cells using an enzyme-linked immunosorbent assay. Polystyrene microtiter plates (Costar, Cambridge, Mass.) were coated with 50 μl of R6 cell suspension diluted 1:200 in 0.1 M carbonate buffer, pH 9.6, and incubated for 1 h at 37°C. After incubation, plates were blocked with 100 μl of 1% (wt/vol) bovine serum albumin (Sigma, St. Louis Mo.) in phosphate-buffered saline (PBS) for 1 h at 37°C and washed three times with PBS supplemented with 0.05% Tween 20 (PBST), and antibodies (50 μl/well) were added. After overnight incubation at 4°C, plates were washed three times with PBST, and then horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Bio-Rad, Richmond, Calif.) in PBST was added. Plates were incubated (37°C, 30 min) and then washed five times with PBST, and 50 μl of o-phenylenediamine (Sigma) (0.5 mg/ml in 0.2 M disodium phosphate–0.1 M citric acid, pH 5.0, with 0.02% hydrogen peroxide) was added. The reaction was terminated by the addition of 100 μl of 3 N H2SO4/well, and absorbance was determined at 492 nm.
The clone designated F1-1B was selected for future studies. The sensitivity of the F1-1B MAb was tested with pneumococcal strains of serotypes 3, 4, 6, 9, 14, 15, 19, and 23, using Western blot analysis. Specificity was also analyzed using Western blot analysis with MAb F1-1B on seven strains of viridans group streptococci, five strains of Neisseria species, and one strain of E. coli.
DNA manipulations and sequencing.
Standard techniques were used for the preparation and analysis of DNA (21). The pCR2.1 plasmid (Invitrogen, Carlsbad, Calif.) was used for cloning. The sequences of the primers used to amplify the psaA gene were generated from the nucleic acid sequence data of psaA from the serotype 6B strain. The sequences are 5′TCGCTCCCAAACAACGATAT3′ (B7) for the forward primer and 5′CGACGTGTTTGAGTTGAGCA3′ (B10) for the reverse primer. PCR amplification generated a 1,300-bp fragment that included the whole psaA gene and the first 217 nucleotides of psaD. The PCR fragments were sequenced on both strands in an Applied Biosystems 377 DNA sequencer using the dideoxynucleotide chain terminator method and appropriate oligonucleotide primers. Sequencing at least two independent templates eliminated possible errors that could have arisen from the PCR.
Hybridization with DNA probes.
The biotinylated PB710 probe was generated by PCR using the B7 and B10 primers (see above). Probe PP32 was obtained by PCR using S. pneumoniae R6 DNA as the template and the forward primer P3 (5′AGGATCTAATGAAAAAATTAG3′) and the reverse primer P2 (5′GCCTTCTTTACCTTGTTCTGC3′) used in the assay developed by Morrison and coworkers (16). This probe included only the psaA gene (930 bp). PCR was performed using biotinylated nucleotides, with annealing taking place at 52°C for 1 min and extension at 72°C for 2 min for 25 cycles. Before hybridization, the gels were transferred to nylon membranes using the capillary transfer method (21). The resulting membranes were processed with the Phototope-Star detection kit for nucleic acids (New England Biolabs) in accordance with the manufacturer's recommendations. The molecular weights of the hybridization signals were determined by comparison with a standard molecular weight ladder.
Analysis of sequence data.
Sequence analysis was performed with programs provided by the servers of the European Bioinformatics Institute and the Swiss Institute of Bioinformatics.
Nucleotide sequence accession numbers.
The sequences in this work have received the following EMBL accession numbers: S. pneumoniae strain R6, AF248230; S. pneumoniae type 3 strain 17912/93, AF248229; S. mitis NCTC 12261, AF248236; S. oralis NCTC 11427, AF248237; and S. anginosus strain NCTC 10713, AF248235.
RESULTS
Analysis with MAb F1-1B.
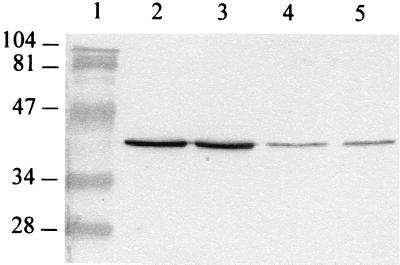
Among the MAbs obtained after the immunization of mice with strain R6, F1-1B bound to a 37-kDa protein antigen of this strain. This MAb also reacted with seven pneumococcal strains of different serotypes and, although with less intensity, with two viridans group streptococcal species, i.e., S. mitis and S. anginosus (Fig. 1). None of the other heterologous bacterial species tested reacted with the MAb (data not shown).
FIG. 1.
Western immunoblot analysis performed with the F1-1B MAb to whole-cell antigen preparations. Lanes: 1, protein standards; 2, S. pneumoniae strain R6; 3, S. pneumoniae serotype 3; 4, S. mitis; 5, S. anginosus. Numbers on the left are molecular masses, in kilodaltons.
DNA analysis and comparative studies.
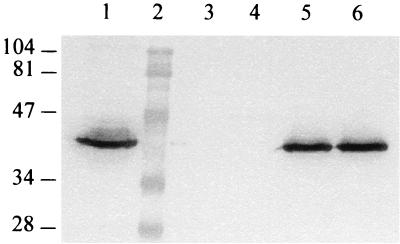
The size of the recognized protein indicated that PsaA might be the antigen recognized by the F1-1B MAb. In order to test this possibility, DNAs prepared from R6, a serotype 3 strain, and eight reference streptococcal species that belong to six different phylogenetic groups (9) were used as the template in PCRs, with the B7 and B10 oligonucleotides as primers. As expected, a 1.3-kb amplification product was recovered from the sample containing DNA from R6, all the pneumococci, and two streptococcal species (S. oralis and S. anginosus). A 1.4-kb DNA fragment from S. mitis was also amplified. Direct sequencing of the PCR fragments from R6 and from the serotype 3 strain revealed a 100% sequence homology with the psaA gene from strain D39 and its flanking sequences and a high degree of homology with other previously identified psaA genes: 90% for S. anginosus, 94% for S. oralis, and 95% for S. mitis. The PCR product from the reaction using R6 DNA was cloned in plasmid pCR2.1 (Invitrogen), and INVαF′ cells were transformed with the recombinant plasmid (pCRNB2). This process generated a recombinant plasmid containing a 1.3-kb insert. Lysates of positive and negative clones were used as antigens in Western blot analysis, and the F1-1B MAb bound only to the positive clones (Fig. 2), confirming that F1-1B recognizes specifically the PsaA protein.
FIG. 2.
Western immunoblot of PsaA protein performed with the F1-1B MAb. Lanes: 1, S. pneumoniae strain R6 whole-cell antigen preparations; 2, protein standards; 3 and 4, lysates from two clones of E. coli INVαF′ transformed with the vector plasmid pCR2.1; 5 and 6, lysates from two clones of E. coli INVαF′ harboring the recombinant plasmid pCRNB2. Numbers on the left are molecular masses, in kilodaltons.
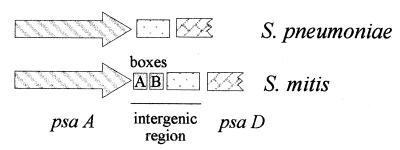
A comparison of the psaA-psaD intergenic regions is presented in Fig. 3. Analysis of S. mitis DNA showed two different regions. The first 105 bp of the 3′ end of psaA had significant similarity (92% homology) with pneumococcal boxes A and B and also with a sequence located downstream of the pneumococcal glpF gene (15). Upstream of box B, a 122-bp fragment sharing 94.3% homology with the psaA-psaD intergenic region from R6 could be detected. The sequence of this intergenic space in S. oralis and S. anginosus revealed a high degree of conservation compared to the corresponding fragment of strain R6 (120 and 122 bp with 79.7 and 76.4% homology, respectively).
FIG. 3.
Schematic representation of the psaA-psaD intergenic region in S. pneumoniae and in S. mitis. PCR direct sequencing of the psaA gene for S. mitis revealed 95% identity with the corresponding open reading frame of strain R6.
Comparison of the deduced amino acid sequences of PsaA among streptococci.
Figure 4 shows an alignment of the predicted type 3 PsaA protein sequence with those of strain R6 and the three type strains of viridans group streptococcal species. The sequence comparisons, using the SWISSPROT database and the FASTA 3 program (18), revealed significant similarities among them. Computed sequence homologies were 100% for PsaA from S. pneumoniae (strains R6 and ST3) and 98% for the pneumococcal PsaA and the homologous proteins from S. mitis, S. oralis, and S. anginosus.
FIG. 4.
Alignment of the deduced amino acid sequence of the PsaA protein from strain R6 with those from S. pneumoniae serotype 3, S. mitis, S. oralis, and S. anginosus. The putative signal peptidase II recognition site, the LXXC domain, is in boldface. Amino acid residues identical to those of R6 are indicated by hyphens.
Detection of the psaA gene in different viridans group streptococcal strains.
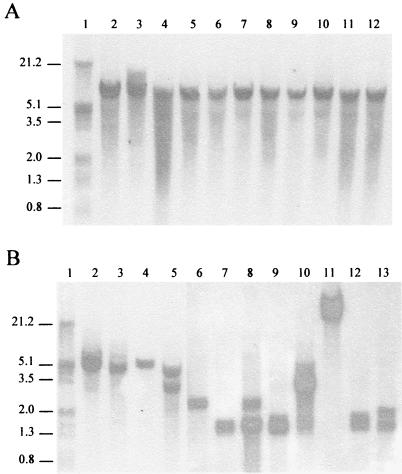
To check whether the psaA gene detected in the viridans group type strains was also present in clinical isolates of these species, 50 strains of S. mitis and S. oralis were analyzed. Genomic DNAs were digested with HindIII and electrophoresed, and a Southern hybridization was performed using the PB710 probe. All the pneumococci and viridans group strains showed hybridization with this probe, which includes the intergenic region between psaA-psaD and the first 217 nucleotides of psaD. To rule out a possible cross hybridization due to homology with these sequences, we constructed the PP32 probe. This probe recognized only the psaA gene. The results with the PP32 probe were identical, indicating that the bands corresponded to the psaA gene and not to the intergenic sequences. The results of the Southern hybridization with the PP32 probe for pneumococci are shown in Fig. 5A, and the results for a representative sample of viridans group streptococci are shown in Fig. 5B. While pneumococci produced a unique band of hybridization of the same size, heterogeneous patterns of hybridization were observed among viridans group streptococci.
FIG. 5.
Identification of the psaA gene by DNA-DNA hybridization. (A) Chromosomal DNAs from eight pneumococcal strains (types 3, 4, 6, 9, 14, 15, 19, and 23). (B) Chromosomal DNAs from 12 viridans group streptococcal strains. DNAs were digested with HindIII, electrophoresed, blotted, and hybridized with the PP32 probe. As size standards, HindIII-digested λ DNA was used (lane 1); molecular sizes (in kilobases) are indicated on the left.
DISCUSSION
A protein regarded as a pneumococcal vaccine candidate is required to be well conserved among all of the different isolates and should be species specific. Study results published until now seem to confirm that PsaA fulfills these requirements and could be regarded as a valid candidate for future vaccines. The results confirmed the presence of the protein in all of the 90 pneumococcal serotypes and its absence in all other streptococcal species. In this study, however, MAb F1-1B, which recognizes PsaA protein, reacted with S. mitis and S. anginosus strains as well and demonstrated that PsaA is also present in these viridans group streptococcal species, suggesting the ubiquity of this protein. Investigators have reported proteins in viridans group streptococci and Enterococcus faecalis that share common sequences with PsaA (12, 13). Some of these homologous proteins, such as ScaA from S. gordonii, SsaB from S. sanguinis, FimA from S. parasanguinis, and EfaA from E. faecalis, have been cloned and sequenced. Comparison of their nucleotide sequences with that of PsaA showed homology within the group that ranged from 57 to 82% (23). To rule out possible cross-reactions of these homologous proteins with the F1-1B MAb we analyzed the generation of amplification products in the viridans group streptococci. We found that whereas samples from some species failed to be amplified using the B7 and B10 psaA primers, S. mitis, S. oralis, and S. anginosus DNAs were successfully amplified. When the protein sequences of the amplification products of these species were analyzed, 98% homology was found for each of the three viridans group streptococcal species. Therefore, the presence of the psaA gene has been demonstrated in the type strains of three viridans group streptococcal species. Moreover, we have demonstrated by hybridization methods that clinical isolates of S. mitis and S. oralis also contain this gene. The variability of hybridization patterns observed among viridans group strains analyzed is in agreement with the extensive diversity demonstrated within these species in previous reports (9, 10).
In conclusion, we have demonstrated by Southern blotting and PCR analysis that, in contrast to current opinion, the psaA gene is in fact present in heterologous organisms.
ACKNOWLEDGMENTS
We thank J. Vázquez, R. López, and E. García for stimulating discussions and critical reading of the manuscript. We also thank J. A. Sáez-Nieto for kindly providing S. pyogenes strain 746/96 and F. Uruburu for providing the other Streptococcus species strains (Spanish Type Culture Collection).
This research was supported by grant 96/0460 from Fondo de Investigaciones Sanitarias (Ministerio de Sanidad y Consumo).
REFERENCES
- 1.Berry A M, Paton J C. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64:5255–5262. doi: 10.1128/iai.64.12.5255-5262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briles D E, Ades E, Paton J C, Sampson J S, Carlone G M, Huebner R C, Virolainen A, Swiatlo E, Hollingshead S K. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun. 2000;68:796–800. doi: 10.1128/iai.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler J C, Breiman R F, Campbell J P, Lipman H B, Broome C V, Facklam R R. Polysaccharide pneumococcal vaccine efficacy. JAMA. 1993;270:1826–1831. [PubMed] [Google Scholar]
- 4.Clafin L, Williams K. Mouse myeloma-spleen cell hybrids: enhanced hybridization frequencies and rapid screening procedures. Curr Top Microbiol Immunol. 1978;81:107–109. doi: 10.1007/978-3-642-67448-8_16. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J. Bumps on the vaccine road. Science. 1994;265:1371–1373. doi: 10.1126/science.8073271. [DOI] [PubMed] [Google Scholar]
- 6.Facklam R R, Breiman R F. Current trends in bacterial respiratory pathogens. Am J Med. 1991;91(Suppl. 6A):3S–11S. doi: 10.1016/0002-9343(91)90301-d. [DOI] [PubMed] [Google Scholar]
- 7.Fenoll A, Martínez-Suárez J V, Muñoz R, Casal J, García J L. Identification of atypical strains of Streptococcus pneumoniae by a specific DNA probe. Eur J Clin Microbiol Infect Dis. 1990;9:396–401. doi: 10.1007/BF01979468. [DOI] [PubMed] [Google Scholar]
- 8.Kajhty H, Eskola J. New vaccines for the prevention of pneumococcal infections. Emerg Infect Dis. 1996;2:289–298. doi: 10.3201/eid0204.960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 10.Kawamura Y, Whiley R A, Shu S-E, Ezaki T, Hardie J M. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 11.Kearney J F, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line which has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 12.Kolenbrander P E, Andersen R N, Ganeshkumar N. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect Immun. 1994;62:4469–4480. doi: 10.1128/iai.62.10.4469-4480.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mäkelä P H, Sibakov M, Herva E, Henrichsen J. Pneumococcal vaccine and otitis media. Lancet. 1980;ii:547–551. doi: 10.1016/s0140-6736(80)91989-3. [DOI] [PubMed] [Google Scholar]
- 15.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R, Morrison D A, Boulnois G J, Claverys J P. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison K E, Lake D, Crook J, Carlone G M, Ades E, Facklam R, Sampson J S. Confirmation of psaA in all 90 serotypes of Streptococcus pneumoniae by PCR and potential of this assay for identification and diagnosis. J Clin Microbiol. 2000;38:434–437. doi: 10.1128/jcm.38.1.434-437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 18.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell H, Tharpe J A, Wells D E, White E H, Johnson J E. Monoclonal antibody recognizing a species-specific protein from Streptococcus pneumoniae. J Clin Microbiol. 1990;28:2191–2195. doi: 10.1128/jcm.28.10.2191-2195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saluja S K, Weiser J N. The genetic basis of colony opacity in Streptococcus pneumoniae: evidence for the effect of box elements on the frequency of phenotypic variation. Mol Microbiol. 1995;16:215–227. doi: 10.1111/j.1365-2958.1995.tb02294.x. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sampson J S, Furlow Z, Whitney A M, Williams D, Facklam R, Carlone G M. Limited diversity of Streptococcus pneumoniae psaA among pneumococcal vaccine serotypes. Infect Immun. 1997;65:1967–1971. doi: 10.1128/iai.65.5.1967-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampson J S, O'Connor S P, Stinson A R, Tharpe J A, Russell H. Cloning and nucleotide sequence analysis of psaA, the Streptoccoccus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect Immun. 1994;62:319–324. doi: 10.1128/iai.62.1.319-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talkington D F, Brown B G, Tharpe J A, Koening A, Russell H. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA) Microb Pathog. 1996;21:17–22. doi: 10.1006/mpat.1996.0038. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. Investing in health research and development, a report of the Ad Hoc Committee on Health Research Relating to Future Intervention Options. TDR/Gen/96.1. Geneva, Switzerland: World Health Organization; 1996. [Google Scholar]