Summary
Stomata regulate plant water use and photosynthesis by controlling leaf gas exchange. They do this by reversibly opening the pore formed by two adjacent guard cells, with the limits of this movement ultimately set by the mechanical properties of the guard cell walls and surrounding epidermis.1,2 A body of evidence demonstrates that the methylation status and cellular patterning of pectin wall polymers play a core role in setting the guard cell mechanical properties, with disruption of the system leading to poorer stomatal performance.3, 4, 5, 6 Here we present genetic and biochemical data showing that wall arabinans modulate guard cell flexibility and can be used to engineer stomata with improved performance. Specifically, we show that a short-chain linear arabinan epitope associated with the presence of rhamnogalacturonan I in the guard cell wall is required for full opening of the stomatal pore. Manipulations leading to the novel accumulation of longer-chain arabinan epitopes in guard cell walls led to an increase in the maximal pore aperture. Using computational modeling combined with atomic force microscopy, we show that this phenotype reflected a decrease in wall matrix stiffness and, consequently, increased flexing of the guard cells under turgor pressure, generating larger, rounder stomatal pores. Our results provide theoretical and experimental support for the conclusion that arabinan side chains of pectin modulate guard cell wall stiffness, setting the limits for cell flexing and, consequently, pore aperture, gas exchange, and photosynthetic assimilation.
Keywords: stomata, guard cell, mechanics, computational modelling, cell wall, Arabidopsis
Graphical abstract
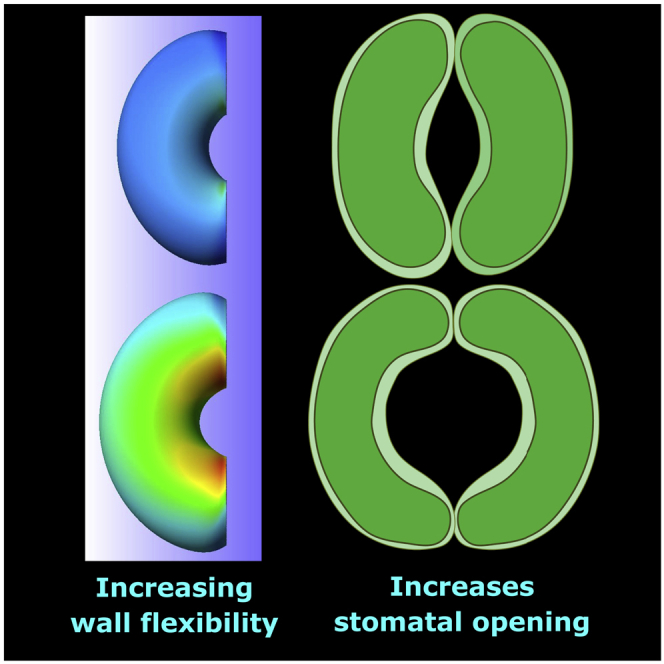
Highlights
-
•
Cell walls in stomata have a distinct composition of arabinans
-
•
Increasing the level of a specific arabinan makes the walls more flexible
-
•
Stomata with more flexible walls can open wider
-
•
Under high CO2, more flexible, wider stomata increase carbon assimilation rate
The degree of stomatal opening is set by the mechanical properties of the guard cell walls. Carroll et al. show that wall flexibility is set by the arabinan composition, and that by manipulating arabinan polymers it is possible to engineer stomata with increased opening under elevated CO2, leading to increased carbon assimilation.
Results and discussion
Guard cell walls are rich in short-chain linear arabinan epitopes
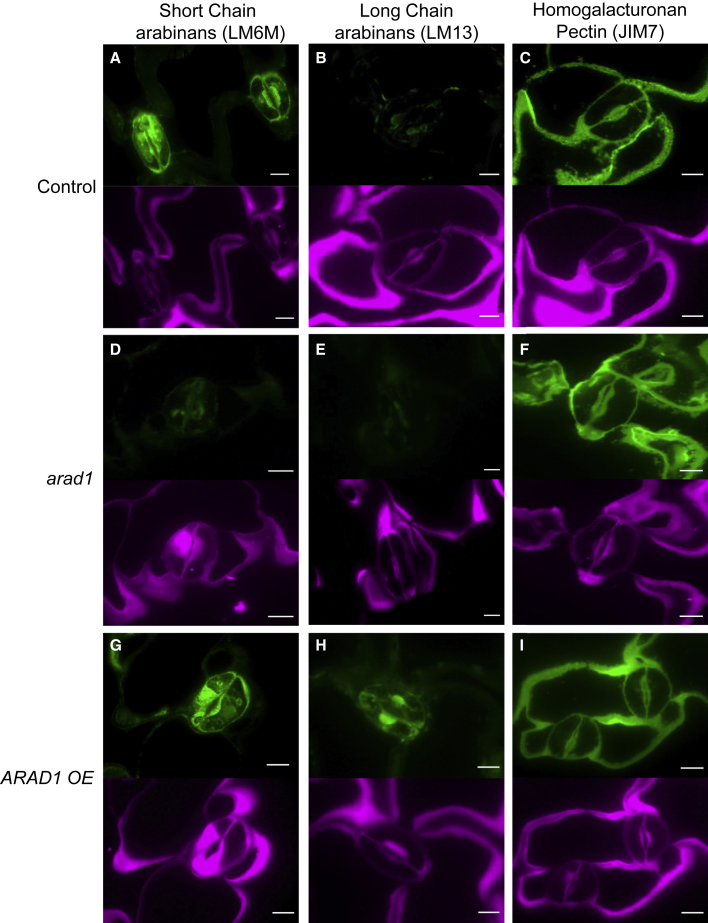
Arabidopsis leaf sections were screened with antibodies (mAb) against a range of cell wall epitopes. This revealed an elevated signal in guard cells for short-chain linear arabinan (SCL-arabinan) epitopes compared to neighboring epidermal cells (LM6M mAb) (Figure 1A, green signal), whereas the signal for longer-chain arabinan epitopes (LC-arabinans; revealed using LM13 mAb) was much lower and did not suggest enrichment in guard cells (Figure 1B). JIM7 mAb binding, which detects a range of homogalacturonan (HGA) polymers within the pectin network, was used as a positive cell wall binding control and was observed across all cell walls in the section (Figure 1C, green signal), supporting the idea that the SCL-arabinan epitope signal reflects a specific guard cell-related pattern of epitope distribution. This was further supported by the pattern of cell wall material shown by calcofluor staining of glucans (Figures 1A–1C, purple signal), which indicated that the patterns observed with LM6M and LM13 did not simply reflect the general distribution of cell wall material in the sections. These data are consistent with previous studies showing guard cell walls of Commelina communis, Vicia faba, and Zea mays are rich in arabinans.7,8 Our data show that Arabidopsis guard cells are specifically enriched in SCL-arabinan epitopes with very few LC-arabinan epitopes present.
Figure 1.
Antibody labeling reveals enrichment of arabinans in guard cells
(A–C) Paradermal sections of control tissue incubated with antibodies against SCL-arabinan epitopes (LM6M), LC-arabinan epitopes (LM13), or broad-spectrum pectin (JIM7).
(D–F) Paradermal sections of tissue from the arad1 mutant treated as in (A)–(C).
(G–I) Paradermal sections of tissue from ARAD1-OE leaves treated as in (A)–(C).
The upper part of each panel shows the antibody signal (green) indicating epitope distribution. The lower part of each panel (purple signal) shows the general distribution of cell wall material revealed by calcofluor staining. Scale bars, 10 μm.
The biosynthesis of cell wall arabinans depends upon the function of a set of arabinan synthase-encoding genes.9 Previous work has identified two sister genes, ARABINAN DEFICIENT 1 (ARAD1) and ARABINAN DEFICIENT 2 (ARAD2), which are proposed to play a core role in the synthesis of these carbohydrate polymers9,10 (Figure S2A). Analysis of expression databases indicated that while these two genes were not exclusively expressed in the guard cells, transcript levels were enriched in this cell type, notably ARAD1 (Figures S2B and S2C). We therefore identified and further characterized available mutants for these genes (arad1 and arad2) and generated a double-knockout mutant (arad1/arad2) (Figures S2D and S2E). When leaf sections of the arad1 mutant were screened with the same set of antibodies described in Figures 1A–1C, signal with LM6M (SCL-arabinan epitopes) was lost (Figure 1D), again with a lack of signal with LM13 (LC-arabinan epitopes) (Figure 1E), and a relatively uniform cell wall signal was observed with the JIM7 Ab (HGA) (Figure 1F). Similar experiments with arad2 did not reveal any loss of signal in guard cells incubated with LM6M, and the epitope patterns observed in the arad1/arad2 mutant were similar to those observed in the arad1 mutant alone (Figures S1A–S1F).
To confirm these data, we quantified the relative fluorescence signal of the various antibodies used (LM6-M, LM13, and JIM7) in sections for the range of genotypes analyzed (col-0, qrt1, arad1, arad2, and arad1/2). These data (Figures S1J–S1L) generally corroborated the image data shown in Figures 1 and S1. In particular, there was a loss of LM6-M signal in the arad1 and arad1/2 lines with a marginal change in the arad2 line (Figure S1J). The pattern for LM13 was similar, although there was a relatively high signal in the arad1/arad2 line (Figure S1K), with the JIM7 signal indicating no major difference in signal between any of the lines (Figure S1L). In addition, we performed an ELISA analysis on sequential extractions of wall material to further corroborate the change in arabinan epitopes patterns revealed by our immunolabeling experiments. As shown in Figures S1M–S1O, these data also indicated that there were increases in the level of shorter-chain arabinan epitopes (LM6-M) in the ARAD1-OE lines and a lower level in the arad1 and arad1/2 mutant lines. A similar pattern of decreased levels of longer-chain arabinan epitopes (LM13) was also observed in these backgrounds (Figures S1P–S1R). Both sets of data indicated that the arad1/2 double mutant line contained a similar pattern of depleted arabinan accumulation as the arad1 mutant that was not apparent in either the qrt1 or Col-0 controls. Finally, to investigate whether the altered patterns of arabinan were potentially linked to an altered pattern of hydroxycinnamic esters of pectic side chains (as suggested by previous research8), we visualized ester autofluorescence under UV illumination; however, no difference was apparent between control and arad1 mutant guard cells.
Taken together, our data show that the arad1 knockout mutants have very low levels of guard cell wall arabinan and suggest that the ARAD2 gene is not essential for the synthesis of guard cell wall arabinan. This is consistent with previous work showing that arad2 knockouts display an altered phenotype only in root tissues.9
Loss of SCL-arabinans impairs stomatal opening and decreases conductance
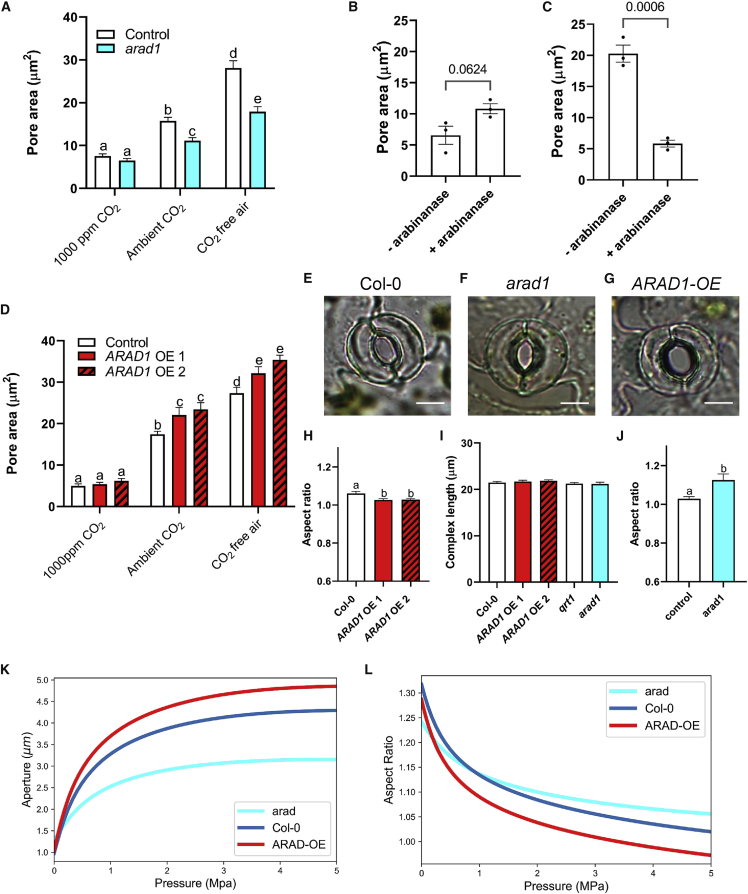
We then investigated the stomatal phenotype of these loss-of-function mutants. Direct measurement of stomatal apertures in epidermal strip bioassays showed that exposure to elevated CO2 (1,000 ppm) led to pore closure and CO2-free air led to pore opening in control samples (Figure 2A), as previously reported.3,11 In contrast, stomata in the arad1 mutant had an impaired opening response to low CO2 (p < 0.0001), whereas the closing response to elevated CO2 could not be distinguished from the control. It should be noted that the available arad1 mutant is a stock center SAIL mutant, which, as previously highlighted, are unexpectedly often in the qrt1 background,12 as was the case here. In all experiments reported with arad1, we used the appropriate qrt1 background as the control line. To further investigate the role of cell wall arabinans in stomatal opening, we pre-treated leaf samples with exogenous arabinanase for 2 h prior to testing the response of the stomata to altered CO2 levels. Consistent with the arad1 mutant data, the closing response of arad1 stomata to elevated CO2 did not show a major difference compared to the control following arabinanase treatment (p = 0.0624) (Figure 2B), but the opening response was significantly impaired (p = 0.0006) (Figure 2C).
Figure 2.
Guard cell arabinans set maximal stomatal aperture
(A–C) Bioassay data for (A) control (qrt1) (open bars) and arad1 (cyan) epidermal strips under different CO2 levels, as indicated. Columns indicate mean value and error bars = SEM (n = 6). A two-way ANOVA followed by Šidák test was performed. Samples that cannot be distinguished from each other at p < 0.05 are indicated by the same letter. (B) Col-0 epidermal strips pre-treated with arabinanase and observed under closing conditions (high CO2) or (C) under opening conditions (CO2 free air). Each point represents the mean aperture observed in a biological replicate (n = 3), with columns indicating mean value and error bars = SEM. Unpaired t tests were performed, and the calculated p value is shown in (B) and (C).
(D) Bioassay data for control (Col-0) (open bars) and two ARAD1-OE (red) epidermal strips under different CO2 levels, as indicated. Columns indicate mean value and error bars = SEM (n ≥ 8). A two-way ANOVA followed by Šidák test was performed. Samples that cannot be distinguished from each other at p < 0.05 are indicated by the same letter.
(E–G) Images of open stomata for (E) Col-0, (F) arad1, and (G) ARAD1-OE.
(H and J) Aspect ratio, as defined by stomatal complex length/complex width of (H) ARAD1-OE and (J) arad1 stomata.
(I) Complex length of ARAD1-OE, arad1, and controls (Col-0 and qrt1).
In (H)–(J), columns indicate mean value and error bars = SEM (n ≥ 6). For (H), data were analyzed by ANOVA followed by a Tukey test. For (I), ANOVA indicated that the samples could not be distinguished from each other at the 0.05 confidence limit. For (J), an unpaired t test mutant versus control was performed. For (H) and (J), samples that cannot be distinguished from each other at p < 0.05 are indicated by the same letter.
(K and L) Modeled change in pore aperture with pressure (K) or aspect ratio with pressure (L) for arad1 (cyan), ARAD1-OE (red), and control (Col-0) (dark blue) stomata.
See also Table S1.
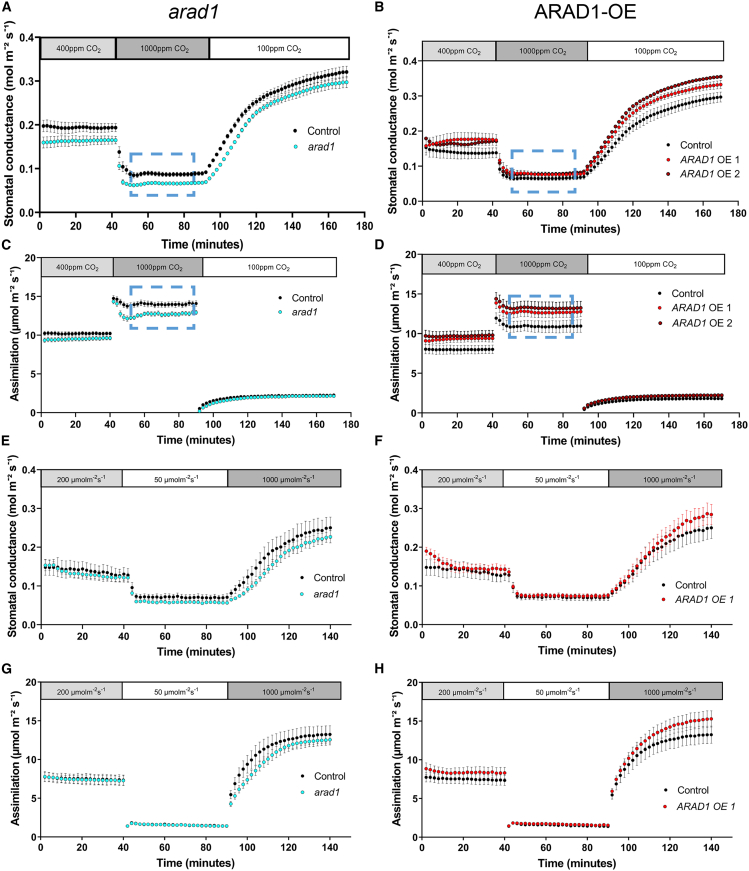
To test the functional outcome of the loss of arad1 at the whole-leaf level, infra-red gas exchange analysis was used. Under near-ambient CO2 level (400 ppm), arad1 leaves had a lower stomatal conductance, gs, than control leaves (Figure 3A). This lower gs was maintained under both elevated and decreased CO2 levels. The lower gs values at a range of CO2 levels were reflected in a lower assimilation rate, although at low CO2 (100 ppm) the difference was marginal (Figure 3C). In addition to CO2, stomatal aperture is known to respond to irradiance level.11 Under each of the three irradiances tested (50, 200, and 1,000 μmol m2 s−1), the arad1 leaves showed a lower gs relative to the control leaves (Figure 3E). With respect to assimilation rate, particularly under high irradiance, a condition expected to promote maximal stomatal aperture, there was a decrease in the arad1 leaves compared to the control (Figure 3G). These differences were not related to changes in stomatal density or index in the arad1 mutant (Figures S3A and S3C) and did not reflect any difference in theoretical anatomical gsmax (Figure S3G) or underlying photosynthetic performance, as analyzed by gas exchange analysis (Figure S3E). Thermal imaging of the arad1, arad2, and arad1/arad2 double mutant indicated that plants with the arad1 background had a higher rosette temperature than either the control or arad2 background (Figures S3H and S3I), consistent with loss of arad1 having a major outcome on guard cell function.
Figure 3.
Modulation of maximal stomatal aperture by altered guard cell arabinans leads to altered gas exchange
(A–D) Stomatal conductance (A and B) and assimilation rate (C and D) in leaves under a range of CO2 concentrations (as indicated) for arad1 (A and C) and ARAD1-OE (B and D) relative to controls.
(E–H) Stomatal conductance (E and F) and assimilation rate (G and H) in leaves under a range of irradiance levels (as indicated) for arad1 (E and G) and ARAD1-OE (F and H) relative to controls. n = 5 independent plants, with error bars = SEM.
In our analyses, no stomatal phenotype was observed with the qrt1 control line, suggesting that the arad1 allele had the major influence on the phenotype observed, although we cannot fully discount some contribution from the qrt1 background. Taken together, our analysis of leaves lacking ARAD1 expression or treated with arabinanase indicated that loss of arabinans in the guard cells led to stomata that had a smaller stomatal aperture and lower gas flux, especially under conditions known to promote maximal pore opening.
Overexpression of arabinan synthase leads to increased stomatal opening
To investigate the outcome of increasing arabinan synthesis on stomatal function, we created transgenic Arabidopsis plants in which the ARAD1 gene was ectopically overexpressed (ARAD1-OE; Figure S2F). Immunolabeling of ARAD1-OE leaves revealed an enrichment of SCL-arabinan epitopes (LM6M) in stomata relative to pavement cells (Figure 1G), as also observed in control samples (Figure 1A). However, LM13 labeling revealed that there was a marked increase in LC-arabinan epitopes in guard cells compared to control samples (Figure 1H). Labeling of HGA and glucans by JIM7 and calcofluor staining (Figures 1G–1I) suggested that the guard cell-enriched signals observed with LM6M and LM13 mAbs were not simply due to altered distribution of total cell wall material. We again corroborated these image data by quantifying the relative fluorescence signal in immunolabeled sections (Figures S1J–S1L). For LM6-M and LM13, there was an increase in signal relative to the Col-0 control in both ARAD1-OE lines analyzed, with no overt change in signal with the JIM7 control antibody. ELISA of cell wall extracts also supported an increased level of arabinan epitopes in the ARAD1-OE lines (Figures S1M–S1R).
The increased accumulation of LC-arabinan epitopes in the guard cells of ARAD1-OE leaves was associated with an altered performance in stomatal bioassays. Under conditions promoting stomatal opening (CO2-free air), the maximal pore area observed in both ARAD1-OE lines analyzed was greater than that observed in the control line (p < 0.0001) (Figure 2D). This difference was maintained under near-ambient levels of CO2 (p = 0.004), but under conditions of high CO2 (which promote stomatal closure) the ARAD1-OE stomata could not be distinguished from the control (Figure 2D). Closer examination of the stomata in ARAD1-OE lines under opening conditions revealed an apparent shape change, with the ARAD1-OE stomata appearing to be rounder than those in either control or arad1 leaves (Figures 2E–2G). Measurement of the length-to-width aspect ratio confirmed this, with ARAD1-OE stomata having a lower aspect ratio than control leaves (p = 0.0244) (Figure 2H). In contrast, open arad1 stomata had a greater aspect ratio than control leaves (p = 0.0154) (Figure 2J). This altered shape was not reflected in any difference in the length of the stomatal complexes (Figure 2I), suggesting that there was an altered lateral flexing of the guard cells in the mutant stomata.
Computational modeling and AFM analysis indicate changes in stiffness after modulation of guard cell wall arabinans
In previous work, we reported on the generation of a finite element (FE) model of guard cell mechanics that successfully recapitulated aspects of stomatal movement in response to turgor change.5,13 To investigate how a change in cell wall arabinan composition might mechanically influence guard cell performance to capture the phenotypes reported here, we explored parameter space to identify changes in the model that might plausibly simulate the observed changes in stomatal opening/closure in the arad1 and ARAD1-OE lines. Starting with the base model,13 we matched the model parameters to the mean dimensions of stomatal pore size and shape, and stomatal complex dimension measured in arad1, ARAD1-OE, and control plants (Table S1), then empirically explored whether parameters linked to either the modeled cellulose fiber network or the wall matrix led to outputs similar to the shape changes observed in these mutants. Altering fiber stiffness (with no other alteration to anisotropy or wall matrix properties) led to only minor changes in the pressure/aperture response curves. However, decreasing the stiffness of the cell wall matrix was sufficient to stretch the pressure/aperture response curve so that a greater final aperture was achieved (red line in Figure 2K; Table S1), similar to the phenotype observed in the ARAD1-OE transgenic plants. Conversely, increasing the Young’s Modulus, E, of the cell wall matrix led to a dampening of the aperture/pressure response curve, with a smaller final aperture at high pressure, recapitulating the phenotype of the arad1 mutant stomata (blue line in Figure 2K; Table S1). Reflecting the observed measurements used to parameterize the model, the stomatal apertures at low turgor pressure were similar in all cases. With respect to aspect ratio, at higher turgor pressure the model predicted a smaller aspect ratio (more circular stomata) in the ARAD-OE line and a higher aspect ratio (more elliptical stomata) in the arad1 mutant, successfully capturing this aspect of the phenotype (Figure 2L).
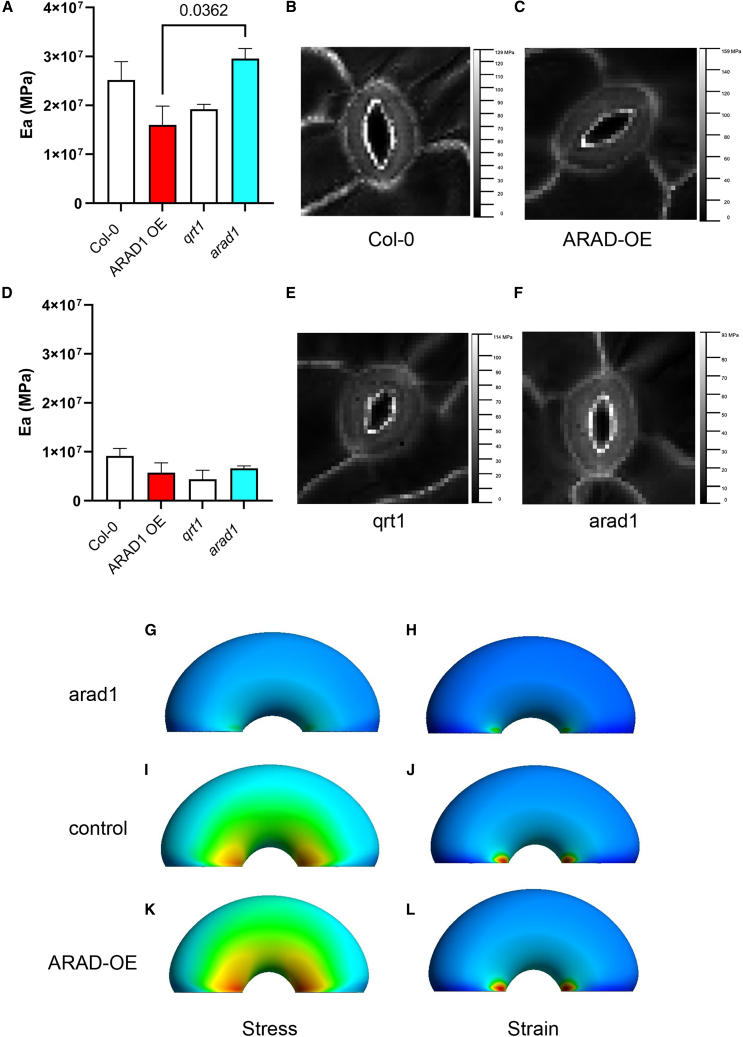
To test whether the mechanical properties of guard cells were altered in the arad1 and ARAD-OE lines, we performed a series of atomic force microscopy (AFM) measurements, using a similar approach to that previously described.5 The results indicated that the apparent modulus (EA) of the arad1 guard cells was significantly higher than that calculated for the ARAD1-OE guard cells (Figure 4A) (p = 0.0362), consistent with the shift required by the computational modeling summarized in Table S1. The relative increase in EA between arad1 and ARAD1-OE measured by AFM (2.4×) was of a similar magnitude to that predicted by the model (1.8×), again suggesting that the model was capturing a realistic element of guard cell mechanics. Analysis of EA values in the pavement cells of the same samples did not reveal any statistically significant difference between the different genotypes (Figure 4D), suggesting that the changes observed in the guard cells were having the major effect on the mechanical behavior of the system. Analysis of the AFM force maps did not reveal any overt differences in the spatial distribution of EA between the lines analyzed, consistent with a view of altered arabinan composition leading to bulk changes in guard cell wall stiffness to underpin the observed phenotype (Figures 4B, 4C, 4E, and 4F).
Figure 4.
Altered arabinans in the guard cell leads to altered wall stiffness
(A) Measured apparent Young’s modulus, Ea, in guard cells of col-0, ARAD-OE (red), qrt1, and arad1 (cyan) leaves. Columns indicate mean values for a pair of guard cells, with error bars = SEM and n ≥ 3. ANOVA followed by a Tukey test indicated that the mean Ea value for arad1 could be distinguished from that for ARAD1-OE (p = 0.0362).
(B and C) Force maps of stomata from (B) Col-0 and (C) ARAD-OE.
(D) Measured Ea in epidermal cells adjacent to guard cells of Col-0, ARAD-OE (red), qrt1, and arad1 (cyan) leaves. Columns indicate mean values. Error bars = SEM with n = 3. ANOVA indicated that none of the samples could be distinguished from each other at the 0.05 confidence limit.
(E and F) Force maps of stomata from (E) qrt1 and (F) arad1 plants.
(G–L) Modeled outputs of stress (G, I, and K) and strain (H, J, and L) for arad1 (G and H), control (I and J), and ARAD-OE (K and L) guard cells undergoing opening. The stress plots display the effective Cauchy stress (MPa = 106 N/m2) and the strain plots show the effective Lagrange strain (unitless), with warmer colors indicating higher values.
The FE modeling approach allows estimation of stress/strain patterns within the system at various points along the curves shown in Figure 2K. Considering stomata at high turgor pressure, the arad1 guard cells (Figures 4G and 4H) are predicted to experience relatively low stress and strain compared with the modeled Col-0 cells (Figures 4I and 4J), with ARAD1-OE cells showing slightly higher stress/strain values than Col-0 (Figures 4K and 4L). In all cells there is a radial gradient of stress/strain emanating from the inner point of the guard cell tip edge, but the model outputs suggest that the accumulation of long-chain arabinans leads to the inner walls of the ARAD-OE guard cells accommodating higher stress levels than the arad1 mutant while undergoing greater radial bending (compare Figures 4G and 4K).
Increased LC-arabinans in the guard cell wall leads to increased stomatal gas exchange
To investigate the outcome of the altered stomatal properties observed in the ARAD1-OE plants at the whole-organ level, we performed a series of gas exchange analyses (Figure 3B, 3D, 3F, and 3H). In contrast to arad1, leaves of ARAD1-OE showed a higher gs than control leaves under low and near-ambient CO2 conditions, although under elevated CO2 levels (expected to close stomata) this difference was not so marked (Figure 3B). The increased level of gs was reflected in a higher assimilation rate at all CO2 levels, though the difference under low CO2 was marginal (Figure 3D). Under differing irradiance levels, stomatal conductance in the ARAD1-OE leaves was slightly higher than that observed in control leaves, and at higher light levels (expected to maximally open stomata) this difference was more marked (Figure 3F). These differences in gs under different light regimes were reflected in the assimilation rates, with markedly higher rates being observed under high light conditions (Figure 3H). Our analysis indicated that the measured differences in gs did not reflect any significant difference in theoretical gsmax (Figure S3G) or underlying photosynthetic performance of the leaves, as analyzed by gas exchange analysis (Figure S3F).
One interesting observation from the gas exchange analyses shown in Figure 3 was that under conditions of elevated CO2, the assimilation rate of the ARAD1-OE line was higher than the control, whereas stomatal conductance under these conditions was similar for both transgenic and control (boxed regions in Figures 3B and 3D). To investigate this further, we calculated the intrinsic and instantaneous water use efficiency (iWUE and WUE) for leaves of arad1 and ARAD1-OE lines and compared them with the relevant controls under the range of CO2 conditions shown in Figure 3. As shown in Figure S3J, iWUE and WUE were slightly higher in arad1 leaves relative to controls at all CO2 levels (1.1×, 1.1×, and 1.2× control), whereas in ARAD1-OE leaves both were lower relative to control values at low and ambient CO2 levels (0.85× and 0.9× control, respectively). However, at elevated CO2 the iWUE and WUE values for ARAD1-OE leaves were higher relative to the control, with the increase being comparable to that calculated for arad1 leaves (1.1× versus 1.2× control). Under elevated CO2 the carbon assimilation rate was lower in the arad1 leaves compared to control (0.85× control), whereas for ARAD1-OE leaves under these conditions there was a relative increase in assimilation rate (1.2× control). Thus, under conditions of elevated CO2 the ARAD1-OE plants undergo a shift to improved water-use efficiency comparable to arad1, but this occurs against a background of a relative increase rather than decrease in carbon assimilation rate.
This study extends previous work suggesting that pectic arabinans are involved in setting guard cell wall properties7 and that, more generally, high levels of arabinan are associated with increased tissue flexibility.14,15 We provide functional genetic and biochemical evidence that demonstrate that arabinan composition dictates the parameters of stomatal opening/closing by modulating wall, and thus cell, flexibility. We show that shorter linear arabinan chains lead to a stiffer, less flexible guard cell wall, while longer chains lead to a less stiff wall with increased flexibility. Computational modeling, backed up by AFM measurements, suggests a plausible underpinning mechanism whereby a uniform influence on the guard cell wall Young’s modulus via altered arabinan composition leads to the specific cellular behavior observed. Exactly how arabinan chain length influences wall mechanical properties awaits elucidation. In addition, although our data strongly implicate ARAD1 as the key player in guard cells, we cannot exclude a role of ARAD2 in this process. Classical models of cell wall structure/function have come under scrutiny, with a body of data supporting models in which the degree of polymer entanglement plays an important role in setting matrix mechanical properties.16, 17, 18 In such models, decreasing the amount of short linear arabinan chains (as in the arad1 mutant) could promote a higher degree of polymer entanglement (leading to a stiffer matrix), while introducing longer arabinan chains (as in the ARAD1-OE lines) might act to physically distance load-bearing elements of the matrix, decreasing the degree of entanglement, thus decreasing wall stiffness. Other pectin modifications (e.g., demethylation patterns) might be super-imposed on the pattern/degree of polymer entanglement, influencing the ability of stretches of homogalacturan polymers to form, e.g., calcium bridges, thus further modulating wall mechanics and accounting for several reported outcomes of pectin modification on guard cell function.3,4,6 The development of more advanced methods of probing wall structure,19 coupled with continued advances in the application of methods to measure plant cell wall mechanical properties,20, 21, 22 provide the means to test such ideas.
Overall, the data presented here add to our understanding of how guard cell walls achieve the unique combination of strength and flexibility required as they undergo repeated extreme changes in shape while withstanding major changes in turgor pressure.23 In addition to the geometric properties provided by cellulose anisotropy and polar stiffening2 and the central role of pectins,3,4,6 modulation of wall modulus by arabinan side chains helps set the upper dynamic limit of the system. Our work further highlights the utility of the guard cell as a test bed for the elucidation of wall structure and function.
Finally, the mechanical properties of guard cells contribute to the dynamics and degree of stomatal opening5,24 and, therefore, play an important role in whole-plant water relations, crop stress resilience, and yield protection.25 Following success via manipulating stomatal density,26,27 improving crop stomatal function via targeted modification of guard cell walls is an attractive biotechnology target. Our data show that increasing guard cell LC-arabinan epitopes allows stomata to open to a greater extent but does not compromise the closing response. Gas exchange analysis indicates that although under present ambient CO2 levels plants with increased levels of LC-arabinan epitopes in their guard cells would have a lower WUE due to the altered dynamics of stomatal opening/closing, under elevated CO2 the change in WUE would be minimal and yet would come with the benefit of increased carbon assimilation, opening the door to potential increased yield for comparable water use. We suggest that the manipulation of guard cell flexibility via ARAD1 in crops could provide higher yields in a future scenario of further increases in atmospheric CO2 level.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| JIM7 | http://www.plantprobes.net/index.php | JIM7 |
| LM6M | http://www.plantprobes.net/index.php | LM6M |
| LM13 | http://www.plantprobes.net/index.php | LM13 |
| Anti-rat-IgG-FITC | Thermofisher | 31629; RRID: AB_228240 |
| Anti-rat-IgG-HRP | Thermofisher | A10549; RRID: AB_2534047 |
| Bacterial and virus strains | ||
| Agrobacterium GV3101 pMP90 | GoldBio | CC-207 |
| Chemicals, peptides, and recombinant proteins | ||
| Calcofluor white | Merck | 18909 |
| Endo-arabinase | Megazyme | E-EARAB |
| LR White Resin | London Resin Company | AGR1280 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana: col-0, qrt1, arad1 | NASC; https://www.arabidopsis.org/portals/mutants/stockcenters.jsp | Col-0, qrt1, arad1, arad2 |
| ARAD1-OE1 and ARAD1-OE2 | This Manuscript | ARAD1-OE |
| Arad1/arad2 | This Manuscript | Arad1/2 |
| Agrobacterium tumefaciens: GV3101 pMP90 | https://www.goldbio.com | CC-207 |
| Oligonucleotides | ||
| arad1 genotyping Fwd primer TATGTGTTCAGGGTGGAAAAGT | This Manuscript | N/A |
| arad1 genotyping Rev primer GGGAGACTTGACGCCAGATT | This Manuscript | N/A |
| arad2 genotyping Fwd primer TCGTTTATTTTGGTGGCAGTC | This Manuscript | N/A |
| arad2 genotyping Rev primer CGCCTCAGCCGGGTCAAAA | This Manuscript | N/A |
| Sail LB primer GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC | This Manuscript | N/A |
| Fwd primer for amplification of ARAD1 coding sequence caccATGGCGCGTAAATCTTCCCTCCTCAAAC | This Manuscript | N/A |
| Rev primer for amplification of ARAD1 coding sequence TTAAATGGAAGTGATAAGACCGGTTTGG | This Manuscript | N/A |
| RT PCR primer for arad1 FWD: GCTCCTCCACAGTCCAAAAG | This Manuscript | N/A |
| RT PCR primer for arad1 RED: ACGAGCTGCTACGAAAGGAA | This Manuscript | N/A |
| RT PCR primer for ARAD1-OE FWD: GAGTTGAGGATCGCAACACA | This Manuscript | N/A |
| RT PCR primer for ARAD1-OE REV: CGTAGCAGCTCGTCGATTCT | This Manuscript | N/A |
| Recombinant DNA | ||
| Plasmid: pENTR-D-TOPO | Thermofisher | K240020 |
| Plasmid:PMDC32 | Addgene | 32078 |
| Software and algorithms | ||
| FEBIO | FEBio Software Suite | https://febio.org |
| Finite Element Stomatal Model | GitHub | https://github.com/woolfeh/stomasimulator; http://doi.org/10.5281/zenodo.6546037 |
| ImageJ (Fiji) | NIH (Public Domain) | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew Fleming (a.fleming@sheffield.ac.uk).
Materials availability
All newly created Arabidopsis lines and the vectors used in their creation can be obtained by contacting the lead contact.
Experimental model and subject details
Arabidopsis seeds (Col-0, qrt1, arad1, arad2, arad1/arad2, ARAD1-OE1, ARAD1-OE2) were sown directly onto 60x60x80mm pots of pre-soaked M3 compost and perlite (3:1). The pots were stratified at 4°C for 1 week before being transferred to a controlled environment growth chamber (12 h light/12 h dark, 200 μmol m-2 s-1 PPFD, 22°C light/ 16°C dark, 60% humidity). The seedlings were thinned to 2 plants per pot at 10 days and further to 1 plant per pot at 14 days. Plants were harvested for experimental work after 5 weeks growth.
Method details
Molecular biology
The arad1 (SAIL_189_F10) and arad2 (SAIL_881_C10) T-DNA insertion lines were obtained from NASC (Nottingham, UK) and confirmed as homozygous for the insertion by PCR using the primers: arad1 5’-TATGTGTTCAGGGTGGAAAAGT- and 5’-GGGAGACTTGACGCCAGATT- arad2 5’-TCGTTTATTTTGGTGGCAGTC- and 5’-CGCCTCAGCCGGGTCAAAA- and the SAIL LB primer 5’-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-. The arad1/arad2 line was created by crossing the arad1 and arad2 T-DNA insertion lines, with homozygous T3 seed used for phenotypic characterization.
For the ARAD1-OE line the ARAD1 coding sequence was amplified from gDNA using primers 5’- caccATGGCGCGTAAATCTTCCCTCCTCAAAC – and 5’- TTAAATGGAAGTGATAAGACCGGTTTGG- and recombined into the pENTR-D-TOPO vector (Invitrogen). This was then cut by NsiI (NEB) and recombined by LR reaction into pMDC32. Plasmids were transformed into the GV3101 pMP90 agrobacterium strain by electroporation and transformed into the Col-0 background by floral dip.28 Transformants were selected on 0.5X MS (Murashige and Skoog) medium, 1.5% (w/v) sucrose containing 50mg/mL kanamycin and plants from the T3 generation were analyzed.
Gene expression levels were verified by RT-PCR following RNA extraction using the Spectrum RNA extraction kit as per manufacturer instructions. The following primer pairs were used: arad1 5’- GCTCCTCCACAGTCCAAAAG- and 5’- ACGAGCTGCTACGAAAGGAA-, ARAD1-OE 5’- GAGTTGAGGATCGCAACACA- and 5’- CGTAGCAGCTCGTCGATTCT.
Stomatal aperture measurements
Epidermal peels of mature leaves were removed at least 2 hours into the photoperiod and floated onto opening buffer (50 mM KCl, 10 mM MES, pH 6.2). Samples were maintained at 22°C with 200 mmol m-2 s-1 of light. Air was bubbled into the opening buffer containing either 0 ppm CO2 (CO2 free treatment), ambient CO2 or elevated CO2 (1000 ppm). Epidermal peels were imaged after 2 hours using an Olympus BX51 microscope and DP70 digital camera and stomatal apertures measured. For standard assays, 42 stomatal apertures were measured for each treatment in each of three independent experiments, with similar results being observed in each experiment. For each experiment epidermal peels were taken from at least 6 plants of each genotype. For the arabinanase pre-treatment epidermal peels were taken from plants and floated onto buffer (10 mM MES (pH6.2), 10 mM KCl, 0.1 mM CaCl2) containing 10 units/mL arabinanase (endo-/exo-Arabinanase from Cellvibrio japonicus, Megazyme) for 1 h before being introduced into the bioassay system.
Infrared gas exchange analysis
CO2 shifts and light shifts were carried out on 5-week-old plants. Measurements were taken with a LICOR-6800 infrared gas exchange analyzer with a 6 cm2 circular area for measurement. If the leaf did not fill the chamber the leaf area was measured and corrected for in the analysis. For both experimental set ups leaf temperature was maintained at 21°C, humidity at 60%, flow rate at 300 μmol s-1 and fan speed 10,000 rpm. For CO2 shifts photon flux density was maintained at 300 μmol m-2 s-1 with 10% blue light. For light shifts CO2 was maintained at 400 ppm.
For CO2 shifts a mature leaf was clamped into the chamber and allowed to acclimate at 400 ppm CO2 until stomatal conductance and assimilation rates had stabilized. The CO2 shift then began by supplying the leaf with 400 ppm CO2 for 40 min, followed by 1000 ppm CO2 for 50 min and finally 100 ppm CO2 for 80 min. Gas exchange measurements were recorded every 2 min throughout the experiment. Intrinsic water-use efficiency (iWUE) was calculated by dividing the assimilation rate by the stomatal conductance at a given time point and WUE (WUE) was calculated by dividing the assimilation rate by the transpiration rate at a given time point.
For light shifts a mature leaf was clamped into the chamber and allowed to acclimate at a photon flux density of 200 μmol m-2 s-1 with 10% blue light until the stomatal conductance and assimilation rate had stabilized. The light shift was then started with 200 μmol m-2 s-1 light for 40 min, followed by 50 μmol m-2 s-1 light for 50 min and finally 1000 μmol m-2 s-1 light for a further 50 min (all 10% blue light). Gas exchange measurements were taken every 2 min throughout the experiment.
Analysis of stomatal number
For stomatal density analysis fully expanded non-senescent leaves were harvested from 5 week old plants. Leaves were fixed in 4% (v/v) formaldehyde in PEM buffer (0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, adjusted to pH 7) for 8 hours. Leaves were then washed twice in 70% (v/v) ethanol for 30 minutes each wash. Tissue was then cleared by incubation in chloral hydrate (2.5 g mL-1) in 30% (v/v) glycerol twice for 8 h. Samples were then mounted in 30% (v/v) glycerol solution and imaged on an Olympus BX51 microscope under the 40x objective using Nomarski illumination, images were captured with an Olympus DP70 camera and the number of stomata counted. 4 viewpoints per leaf were analyzed.
Immunolabeling
For immunolabeling, leaf samples (3mm diameter leaf discs) were fixed in 4% (w/v) formaldehyde in PEM buffer (0.1MPIPES, 2mM EGTA, 1 mM MgSO4, adjusted to pH 7) by vacuum infiltration then dehydrated in an ethanol series (30 min each at 30%, 50%, 70%, 100% EtOH) and infiltrated with LR White Resin (London Resin Company) diluted in ethanol (45 min each at 10%, 20%, 30%, 50%, 70% & 90% resin then 3x8 h at 100%). Leaf discs were stacked vertically in gelatine capsules filled with resin and allowed to polymerize for 7 days at 37°C. Sections were cut to a thickness of 2 mm using a Reichert-Jung Ultracut E ultramicrotome using a glass knife. Further processing and incubation with the JIM7 and LM19 antibodies was as previously described.3 Briefly, sections were incubated with 3% (w/v) milk protein (Marvel, Premier Beverages, UK) in phosphate-buffered saline solution (PBS, pH 7.2) (hereafter known as PBS/MP). Sections were then incubated with a ten-fold dilution of primary monoclonal antibody in PBS/MP for 1 h at room temperature. Samples were washed 3 times with PBS and secondary antibody was added (anti-rat-IgG (whole molecule) coupled to fluorescein isothiocyanate FITC was used at 100-fold dilution in PBS/MP) for 1 h. Samples were kept in the dark from this step. Samples were counterstained with 0.25% (w/v) Calcofluor White solution diluted ten-fold in PBS for 5 min before mounting on slides with Citifluor AF1 anti-fade solution (Agar Scientific, UK). Images were captured using a DP51 camera. FITC was visualized using a filter set with 460-490 nm excitation filter, a 510-550 nm emission filter and a 505 nm dichroic mirror. Calcofluor White was visualized using a 395 nm excitation filter, a 460 nm emission filter and a 425 nm dichroic mirror.
Preparation of alcohol insoluble residues
Leaf tissue was flash frozen and freeze dried before being ground to a fine powder in a Qiagen TissueLyserII (Qiagen, Hilden, Germany) at 30Hz for 1 minute with two 3mm stainless steel ball bearings. To extract alcohol insoluble resides (AIR) 250 mg of ground tissue was sequentially incubated in 1 ml volumes of a solvent series consisting of ethanol (50%, 60%, 70%, 80%, 90% and 100% v/v) followed by acetone and a chloroform:methanol mixture (3:1) at each stage samples were incubated for 90 minutes on a rocking table at room temperature, sample pelleted by centrifugation and the solvent discarded. Following the final step, the sample was dried by evaporation leaving AIR which is enriched in cell walls.
ELISA of cell wall extracts
Cell walls were sequentially fractionated by the following procedure. 2 mg of AIR was incubated 1 ml of with 50 mM Cyclohexanediaminetetraacetic acid (CDTA), pH6 for 90 minutes and shook at 10 Hz in a Qiagen TissueLyserII. Undissolved sample was pelleted by centrifugation and the supernatant retained as the CDTA extract. This extraction process was repeated with 4 M Potassium Hydroxide and retained as the KOH extract. Any remaining residues were subjected to a cellulase digestion by incubating for 8 hours at 30°C with 1μg/ml of cellulase 5A (NZYTech, Lisbon, Portugal) in 20 mM Tric-HCl buffer pH 8.8 to give the cellulase extract. 20μl of each extract was diluted 5x using using 0.01 M Phosphate Buffer Saline (PBS), pH 7.4 in an immunosorp 96-well plate (Maxisorp, F96, Thermofisher) and incubated overnight at 4°C to coat the plates. After coating the plates were rinsed in tap water and blocked for 1 h at room temperature using 200 μL PBS containing 5% (w/v) nonfat bovine milk powder (Sigma) followed by extensive washing with tap water. 100 μL primary antibody (10x diluted in PBS containing 5% (w/v) milk powder) was added and incubated for 2 h at room temperature. Again the plates were washed, and 100 μL of anti-rat IgG coupled to horseradish peroxidase (Thermofisher), 1000-fold dilution in PBS containing 5% (w/v) milk powder, was added and incubated for 1 h followed by another washing step.
The plates were developed by adding 100 μL of substrate solution (to make 20 ml of substrate 2 ml of 1M sodium acetate pH 6; 200μl of 10 mg/ml of 3,3′,5,5′-tetramethylbenzidine in DMSO and 20 μl of 6% hydrogen peroxide were added to 17.78 ml of water) and incubate for 6 minutes. The reaction was stopped by adding 50 μL of 2.5 M H2SO4, resulting in the formation of a yellow color measured at 450 nm.
Atomic force microscopy
The method used was based on that described previously.5 Dissected and plasmolyzed (0.55 M mannitol; minimum 45 min) leaf blocks (approximately 5mm square) from 3-4 week old plants were indented using a Nano Wizard 3 AFM (JPK Instruments, DE) mounted with a 5 nm diameter pyramidal indenter (Windsor Scientific, UK) on a cantilever of nominal 45 N/m stiffness. Cantilever stiffness was determined by thermal tuning prior to experiment initiation. Tip sensitivity was calibrated by first performing indentations on a clean glass slide and varied between experiments. For each leaf, areas of 100x100 μm2 were indented with 128x128 points on the adaxial surface. Indentations were performed with 250 nN of force yielding an indentation depth range of 10-100 nm. Sample numbers for each experiment are given in the figure legends and text. Force indentation curves were analyzed using JPKSPM Data Processing software (JPK Instruments, DE; v. spm 5.0.69) using the following steps: voltage readings were converted to force using calibrated sensitivity and cantilever stiffness values, baseline subtraction and tilt correction, vertical displacement offset adjustment, indentation calculation by subtraction of cantilever bending from piezo position during indentation, and indentation modulus was calculated by fitting a Hertzian indentation model to the approach curve. The Hertz model assumes the indented surface is an infinite homogeneous half space, which is clearly not the case for the geometrically complex leaf surface. Hence the results of indentation experiments are quoted as an apparent modulus, Ea. Control experiments carried out at lower indentation rates and at lower indentation depths revealed similar results, and analysis did not reveal any surface topography which might easily account for the Ea patterns observed around or within the guard cells. Retraction curves were not analyzed due to numerous adhesion difficulties during tip removal from the surface. All AFM images shown are derived from force maps, with an indication of the calculated Ea values according to the heatmaps adjacent to the images.
Computational modelling
We used the finite element (FE) model of guard cell mechanics that was previously described5,13 and solved with FEBio.29 In brief, a guard cell pair is represented as two hollow, deformed tori that form ellipses for the pore and complex outlines. The two guard cells are connected by solid walls at the two poles. The initial geometry of the guard cell complex is described by the stoma length, pore length, pore width and guard cell width. These dimensions are matched to the mean data for arad1, col-0 control and ARAD1-OE cell types for the high CO2 conditions (Table S1). In all three cell types, the cell wall thickness and polar wall thickness was set to 0.1 and 0.3 μm, respectively, and the polar walls were fixed in space consistent with the findings previously reported.5 The guard cell wall was modelled using the transversely isotropic Veronda-Westmann material which is an uncoupled hyperelastic material that exhibits strain-stiffening.30 The Veronda-Westmann model for the isotropic cell wall matrix is described by two empirically determined coefficients C1 and C2. C1 (MPa) scales the magnitude of the stress-strain curve and C2 (dimensionless) defines the magnitude and nonlinearity of the stress-strain curve. These two parameters can be related to the Young’s Modulus, a measure of the stiffness of a material, by E=3(C1∗C2) which is valid only for small strains. For larger strains, the exponential term determined by C2 dominates and the strain-stiffening behavior is more pronounced with materials of a larger C2 becoming exponentially stiffer with increasing strain. The Bulk Modulus, a measure of a material’s resistance to compressibility, was set to 10 GPa, to make the material nearly incompressible. The Poisson ratio is a measure of the material’s deformation in a direction perpendicular to the load and is ≈0.5 for incompressible materials. The anisotropic direction, representing the circumferential cellulose microfibril (CMF) orientation, was calculated for the ∼18000 elements using an in-house meshing script. As previously reported,12 the fiber parameters were reduced to one value, C5 (MPa). In this study, C5 was found to have minimal effect on stomatal geometry and held constant for all cell types. The pressure/aperture graphs were achieved by increasing the pressure load (i.e., turgor pressure) from 0 to 5 MPa using the variable iterator which adjusts the incrementation value depending on the convergence data at the previous step. The stress/strain plots display the effective stress (MPa = 106 N/m2) and effective Lagrange strain (unitless) respectively. The effective Lagrange strain (unitless scalar), is calculated from the Lagrange strain tensor, E, and deviatoric strain, E’, where E’=E-(tr(E)/3)∗I, where tr(.) is the trace and I is the identity tensor. The effective Lagrange strain, e, is then given by where : indicates a double contraction of the tensor. A similar calculation is done for the stress tensor yielding the effective stress (von Mises stress).
Quantification and statistical analysis
Most of the experiments were comprised of 6 or more biological replicates with number of events measured indicated in the figure legends. Statistical differences were assessed by Student’s t test or ANOVA, using built-in functions of the statistical package GraphPad, with the exact test performed indicated in the figure legends. Statistical significance is indicated on the graphs and in the figure legends. Error bars represent standard error.
Acknowledgments
This work was supported by a BBSRC-White Rose DTP (BB/M011151) studentship to S.C. (A.J.F. and J.E.G.); S.A. was supported by a BBSRC Discovery Fellowship (BB/T009691/1); and A.J.F., J.E.G., R.J.M., R.S.S., and C.H.D. acknowledge support by BBSRC grant “Shape Shifting Stomata: The Role of Geometry in Plant Cell Function” (BB/T005041), with further support to R.J.M. via BBSRC’s Institute Strategic Programme “Plant Health” BB/P012574/1. Atomic force microscopy was performed at the Microscopy Facility of the Sainsbury Laboratory Cambridge University, which is supported by the Gatsby Charitable Foundation, with advice and support by Ross Carter and Sarah Robinson (SLCU).
Author contributions
Conceptualization, A.J.F. and J.E.G.; investigation, S.C., S.A., C.H.D., and A.J.F.; writing – original draft, A.J.F. and S.A.; writing – review & editing, all authors; supervision, A.J.F., J.E.G., S.A., R.J.M., and R.S.; funding acquisition, A.J.F., J.E.G., S.A., and R.J.M.
Declaration of interests
The authors declare no competing interests.
Published: June 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.05.042.
Supplemental information
Data and code availability
All of the scripts used to run these simulations, process the data and generate the graphs are available at Github: http://doi.org/10.5281/zenodo.6546037.
References
- 1.Franks P.J., Farquhar G.D. The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol. 2007;143:78–87. doi: 10.1104/pp.106.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolfenden H.C., Baillie A.L., Gray J.E., Hobbs J.K., Morris R.J., Fleming A.J. Models and mechanisms of stomatal mechanics. Trends Plant Sci. 2018;23:822–832. doi: 10.1016/j.tplants.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Amsbury S., Hunt L., Elhaddad N., Baillie A., Lundgren M., Verhertbruggen Y., Scheller H.V., Knox J.P., Fleming A.J., Gray J.E. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr. Biol. 2016;26:2899–2906. doi: 10.1016/j.cub.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Li W., Turner J.A., Anderson C.T. PECTATE LYASE LIKE12 patterns the guard cell wall to coordinate turgor pressure and wall mechanics for proper stomatal function in Arabidopsis. Plant Cell. 2021;33:3134–3150. doi: 10.1093/plcell/koab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter R., Woolfenden H., Baillie A., Amsbury S., Carroll S., Healicon E., Sovatzoglou S., Braybrook S., Gray J.E., Hobbs J., Morris R.J., Fleming A.J. Stomatal opening involves polar, not radial, stiffening of guard cells. Curr. Biol. 2017;27:2974–2983.e2. doi: 10.1016/j.cub.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rui Y., Xiao C., Yi H., Kandemir B., Wang J.Z., Puri V.M., Anderson C.T. POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell. 2017;29:2413–2432. doi: 10.1105/tpc.17.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones L., Milne J.L., Ashford D., McQueen-Mason S.J. Cell wall arabinan is essential for guard cell function. Proc. Natl. Acad. Sci. USA. 2003;100:11783–11788. doi: 10.1073/pnas.1832434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones L., Milne J.L., Ashford D., McCann M.C., McQueen-Mason S.J. A conserved functional role of pectic polymers in stomatal guard cells from a range of plant species. Planta. 2005;221:255–264. doi: 10.1007/s00425-004-1432-1. [DOI] [PubMed] [Google Scholar]
- 9.Harholt J., Jensen J.K., Verhertbruggen Y., Sogaard C., Bernard S., Nafisi M., Poulsen C.P., Geshi N., Sakuragi Y., Driouich A., et al. ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta. 2012;236:115–128. doi: 10.1007/s00425-012-1592-3. [DOI] [PubMed] [Google Scholar]
- 10.Harholt J., Jensen J.K., Sorensen S.O., Orfila C., Pauly M., Scheller H.V. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006;140:49–58. doi: 10.1104/pp.105.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assmann S.M., Jegla T. Guard cell sensory systems: recent insights on stomatal responses to light, abscisic acid, and CO2. Curr. Opin. Plant Biol. 2016;33:157–167. doi: 10.1016/j.pbi.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Nikonorova N., Yue K., Beeckman T., De Smet I. Arabidopsis research requires a critical re-evaluation of genetic tools. J. Exp. Bot. 2018;69:3541–3544. doi: 10.1093/jxb/ery161. [DOI] [PubMed] [Google Scholar]
- 13.Woolfenden H.C., Bourdais G., Kopischke M., Miedes E., Molina A., Robatzek S., Morris R.J. A computational approach for inferring the cell wall properties that govern guard cell dynamics. Plant J. 2017;92:5–18. doi: 10.1111/tpj.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhertbruggen Y., Marcus S.E., Chen J., Knox J.P. Cell wall pectic arabinans influence the mechanical properties of Arabidopsis thaliana inflorescence stems and their response to mechanical stress. Plant Cell Physiol. 2013;54:1278–1288. doi: 10.1093/pcp/pct074. [DOI] [PubMed] [Google Scholar]
- 15.Moore J.P., Farrant J.M., Driouich A. A role for pectin-associated arabinans in maintaining the flexibility of the plant cell wall during water deficit stress. Plant Signal. Behav. 2008;3:102–104. doi: 10.4161/psb.3.2.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Yu J., Wang X., Durachko D.M., Zhang S., Cosgrove D.J. Molecular insights into the complex mechanics of plant epidermal cell walls. Science. 2021;372:706–711. doi: 10.1126/science.abf2824. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T., Zheng Y., Cosgrove D.J. Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J. 2016;85:179–192. doi: 10.1111/tpj.13102. [DOI] [PubMed] [Google Scholar]
- 18.Wang T., Park Y.B., Cosgrove D.J., Hong M. Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol. 2015;168:871–884. doi: 10.1104/pp.15.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu J., Putnis C.V., Wang L. AFM imaging and single-molecule recognition of plant cell walls. Trends Plant Sci. 2022;27:412–413. doi: 10.1016/j.tplants.2021.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Milani P., Braybrook S.A., Boudaoud A. Shrinking the hammer: micromechanical approaches to morphogenesis. J. Exp. Bot. 2013;64:4651–4662. doi: 10.1093/jxb/ert169. [DOI] [PubMed] [Google Scholar]
- 21.Braybrook S.A., Jonsson H. Shifting foundations: the mechanical cell wall and development. Curr. Opin. Plant Biol. 2016;29:115–120. doi: 10.1016/j.pbi.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Robinson S., Huflejt M., Barbier de Reuille P., Braybrook S.A., Schorderet M., Reinhardt D., Kuhlemeier C. An automated confocal micro-extensometer enables in vivo quantification of mechanical properties with cellular resolution. Plant Cell. 2017;29:2959–2973. doi: 10.1105/tpc.17.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franks P.J., Buckley T.N., Shope J.C., Mott K.A. Guard cell volume and pressure measured concurrently by confocal microscopy and the cell pressure probe. Plant Physiol. 2001;125:1577–1584. doi: 10.1104/pp.125.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rui Y., Yi H., Kandemir B., Wang J.Z., Puri V.M., Anderson C.T. Integrating cell biology, image analysis, and computational mechanical modeling to analyze the contributions of cellulose and xyloglucan to stomatal function. Plant Signal. Behav. 2016;11:e1183086. doi: 10.1080/15592324.2016.1183086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAusland L., Vialet-Chabrand S., Davey P., Baker N.R., Brendel O., Lawson T. Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol. 2016;211:1209–1220. doi: 10.1111/nph.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caine R.S., Yin X., Sloan J., Harrison E.L., Mohammed U., Fulton T., Biswal A.K., Dionora J., Chater C.C., Coe R.A., et al. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2019;221:371–384. doi: 10.1111/nph.15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn J., Hunt L., Afsharinafar M., Meselmani M.A., Mitchell A., Howells R., Wallington E., Fleming A.J., Gray J.E. Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 2019;70:4737–4748. doi: 10.1093/jxb/erz248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Maas S.A., Ellis B.J., Ateshian G.A., Weiss J.A. FEBio: finite elements for biomechanics. J. Biomech. Eng. 2012;134:011005. doi: 10.1115/1.4005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veronda D.R., Westmann R.A. Mechanical characterization of skin-finite deformations. J. Biomech. 1970;3:111–124. doi: 10.1016/0021-9290(70)90055-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the scripts used to run these simulations, process the data and generate the graphs are available at Github: http://doi.org/10.5281/zenodo.6546037.