Summary
Thirst emerges from a range of cellular changes that ultimately motivate an animal to consume water. Although thirst-responsive neuronal signals have been reported, the full complement of brain responses is unclear. Here, we identify molecular and cellular adaptations in the brain using single-cell sequencing of water-deprived Drosophila. Water deficiency primarily altered the glial transcriptome. Screening the regulated genes revealed astrocytic expression of the astray-encoded phosphoserine phosphatase to bi-directionally regulate water consumption. Astray synthesizes the gliotransmitter D-serine, and vesicular release from astrocytes is required for drinking. Moreover, dietary D-serine rescues aay-dependent drinking deficits while facilitating water consumption and expression of water-seeking memory. D-serine action requires binding to neuronal NMDA-type glutamate receptors. Fly astrocytes contribute processes to tripartite synapses, and the proportion of astrocytes that are themselves activated by glutamate increases with water deprivation. We propose that thirst elevates astrocytic D-serine release, which awakens quiescent glutamatergic circuits to enhance water procurement.
Keywords: thirst, single-cell transcriptomics, glia, astrocytes, gliotransmission, D-serine, NMDA receptors, Drosophila, behavior, water procurement
Graphical abstract
Highlights
-
•
Water deprivation alters the levels of many glial-expressed genes in Drosophila
-
•
Levels of astray-encoded phosphoserine phosphatase bi-directionally alter drinking
-
•
Astrocytic D-serine regulates water procurement via NMDA-type glutamate receptors
-
•
Astrocytes frequently contribute processes to tripartite glutamatergic synapses
Park et al. show that water deprivation primarily alters the glial transcriptome in Drosophila. Screening the regulated genes identified astrocytic expression of the astray-encoded phosphoserine phosphatase and astrocyte-released D-serine to be key elements of the brain’s response toward facilitating thirst-directed behaviors.
Introduction
The sensation of thirst is predominantly a manifestation of our body’s response to being water deprived. Water deficit reduces the volume and increases the osmolarity of an animal’s blood. These changes induce multiple adaptations throughout the body, such as elevated activity of osmosensory neurons in the brain, alteration of blood pressure and heart rate, and increased retention of water.
Osmosensory neurons in the subfornical organ (SFO) and organum vasculosum (OV) of the lamina terminalis (LT) of the mammalian forebrain respond directly and indirectly to changes in plasma osmolality and blood volume/pressure, via hormones such as Angiotensin II.1, 2, 3 These neurons then directly or indirectly release compensatory neuropeptide/hormone signals like Vasopressin/anti-diuretic hormone which promotes bodily responses to reduce water loss and restore blood pressure, and behavioral responses to restore fluid balance.4, 5, 6, 7 Artificial engagement of LT neurons can induce water consummatory behaviors.8, 9, 10, 11, 12, 13, 14, 15, 16, 17 However, the full range of nervous system mechanisms that accompany the water-deprived state, and how they modulate behavioral regimes and actions to satisfy thirst, is currently unknown.
In Drosophila, ion transport peptide (ITP) is the likely anti-diuretic functional analog of the mammalian vasopressin and renin-angiotensin systems.18 ITP is produced by neurosecretory neurons in the brain and abdominal ganglion; its expression increases with water deprivation. ITP increases water consumption, reduces water excretion in the malpighian tubules, and increases hindgut reabsorption. ITP induction also represses feeding. Although circuits regulated by ITP remain to be identified, other thirst-regulated circuits required for water seeking and consumption/homeostasis have been reported. Interoceptive sensory neurons (ISNs) in the subesophageal zone (SEZ) directly sense high osmolality (via the cation channel Nanchung) and their inhibition promotes drinking.19 Interestingly, ISN activation (via adipokinetic hormine [AKH]) suppresses drinking and instead promotes feeding. Two other classes of SEZ neurons, the Janu neurons, regulate water seeking behavior up a humidity gradient, but not total water consumption.20 The Janu neurons are either GABAergic or Allatostatin (AstA)-releasing. Their joint activation is rewarding, and the AstA group simultaneously inhibits feeding while promoting water seeking. Finally, different types of mushroom body innervating dopaminergic (DA) neurons have been implicated in water seeking, water reward learning,21 and thirst-dependent control of the expression of water-seeking memories.22 Some of these DA neurons are regulated by the leucokinin neuropeptide which is released from neurons that are activated by elevated osmolality.22 More complex interaction between neuromodulators allows DA neurons to selectively promote state appropriate expression of thirst- or hunger-dependent memories.
Brain responses to water deprivation need not be exclusively neuronal. A blood-brain barrier (BBB) shields most neurons from the circulatory environment. Notably, the osmosensory neurons of the mammalian SFO and OV have processes outside the BBB where they can directly sample circulatory status.23 The mammalian BBB is formed by microvascular endothelial cells, pericytes, and astrocytes. Since astrocytes also innervate the neuropil, where their processes contribute to tripartite synapses (TPSs), they are well positioned to sense and/or transmit metabolites and signals representing nutrient status to neurons.24, 25, 26, 27, 28, 29, 30 In Drosophila, perineurial and subperineurial glia form the BBB, whereas astrocytes tile the entire brain and permeate the neuropil.26,31 Astrocytes in the fly therefore also have potential to influence neuronal activity in response to nutritional state.32 However, evidence for such a role of fly astrocytes is currently lacking.
The Drosophila brain provides an unparalleled opportunity to investigate neural mechanisms of thirst with cellular and molecular resolution. Recent advances in single-cell transcriptomics have generated transcriptional profiles from most of the abundant cell types in the fly brain.33, 34, 35, 36, 37 Here, we used single-cell transcriptomics to identify brain-wide and cell-type restricted changes in gene expression triggered by water deprivation. Most changes occurred within glia, rather than neurons. Functional analyses of thirst responsive genes identified astrocyte expression of the astray (aay)-encoded phosphoserine phosphatase and gliotransmission of its product D-serine to be required for regulated water consumption. We show that astrocytes contribute processes to tripartite glutamatergic synapses in the fly brain and that D-serine promotes water procurement via fly NMDA-type glutamate receptors. These findings provide a new molecular and cellular framework to understand how thirst alters brain physiology and behavior.
Results
Single-cell transcriptomics in thirsty Drosophila
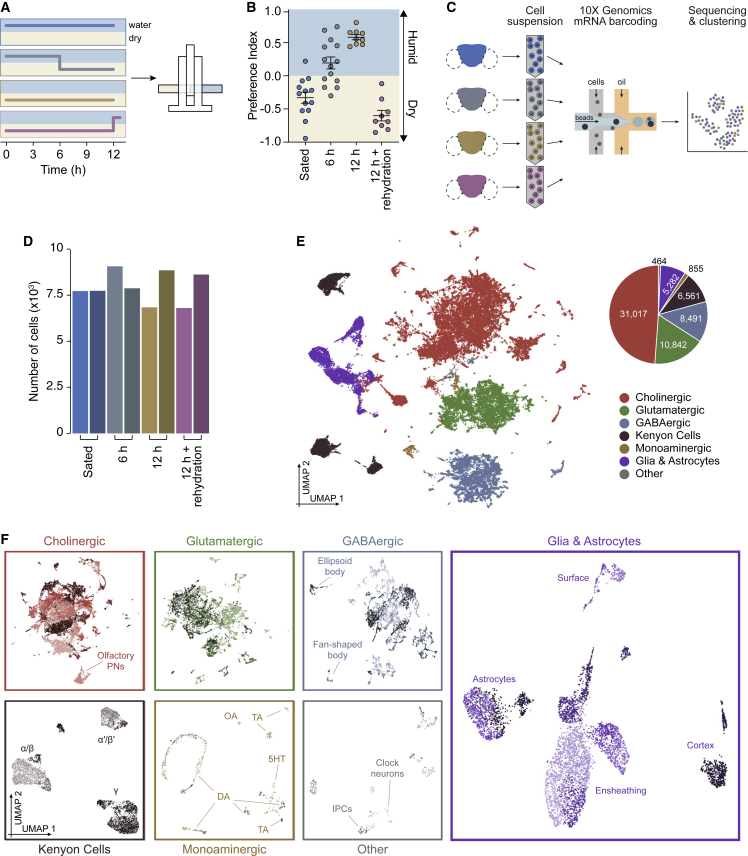
Drosophila prefer dry environments when water sated but seek humidity when water deprived, behaviors that serve fluid homeostasis.21,38,39 This behavioral switch is readily quantified by giving flies the choice in a T-maze between humid and dry chambers (Figure 1A). Although water-sated flies preferred the dry, those dehydrated for 6 or 12 h showed an increasing preference for humidity (Figure 1B). Flies water deprived for 12 h then permitted to drink reverted to preference for the dry chamber, demonstrating water attraction is rapidly reversed upon drinking (Figures 1A and 1B).
Figure 1.
Single-cell transcriptomics of thirsty Drosophila
(A) Schematic protocols for humidity preference assay. Flies were kept in vials with or without water for the indicated times, then given a T-maze choice between a humid and a dry chamber.
(B) Increasing dehydration converts humidity avoidance behavior into attraction. Attraction returns to avoidance with thirst quenching.
(C) Schematic of the process of single-cell transcriptomics analyses comparing flies from the four conditions in (B). Two independent samples were processed for each condition.
(D) Total number of cells obtained from each sample, after filtration of low-quality barcodes and doublets.
(E) Left: UMAP plot from first clustering step, and identification of seven main cell classes. Right: pie chart showing the number of cells obtained from each of these classes.
(F) UMAP plots showing sub-clustering of each of the seven cell classes shown in (E). Known cell types within each class are labeled. Kenyon cell labels represent known subtypes that innervate corresponding mushroom body lobes. Withing the monoaminergic cells labels are OA, octopaminergic; TA, tyraminergic; 5HT, serotonergic; DA, dopaminergic. Within other, IPCs, insulin-producing cells.
See also Figure S1.
To investigate cellular correlates of thirst, we used the 10X Genomics Chromium system to generate single-cell transcriptomic atlases of brains extracted from water-sated, 6 and 12 h dehydrated, and rehydrated flies (Figure 1C). After filtering out low-quality barcodes and putative cell doublets (Figure S1A), we retrieved a total of 63,512 cells. Importantly, each dehydration condition and experimental sample contributed a similar cell number to the total collection (Figure 1D). We used SCTransform and the canonical correlation anchor-based method in Seurat v340,41 to normalize and integrate data from each sample to help identify shared cell populations. An initial unsupervised clustering step was performed to partition cells into seven main classes, which we annotated based on the expression of known marker genes (Figure S1B). These classes include cholinergic, glutamatergic, GABAergic, mushroom body intrinsic Kenyon cells, monoaminergic neurons, glia, and “other” cells not in these previous categories (Figure 1E).33,40 Cells in each class were next independently subdivided into 184 clusters, each containing between 14 and 5,083 cells (Figure 1F). Monoaminergic clusters were smaller than others (median cells/cluster: 32), reflecting their discrete transmitters (DA, octopaminergic [OA], tyraminergic [TA], serotonergic [5HT]), functional specialization among neurons of each type, and relative scarcity in the brain. Conversely, Kenyon cell and other cholinergic neuron clusters were largest (median cells/cluster: 354.5 and 293.5, respectively; Figure S1C). Again, for each of these 184 clusters, we identified specific marker genes (Data S1; Figure S1F), with which we annotated known cell types (Figure 1F; see STAR Methods for detail). We noted that each experimental sample was evenly represented across cell clusters (Figures S1D and S1E), indicating that dehydration does not change the cellular composition of the adult fly brain, in contrast to in developing larvae where starvation reduced the number of undifferentiated headcase expressing neurons.41
The transcriptional signature of thirst in the fly brain
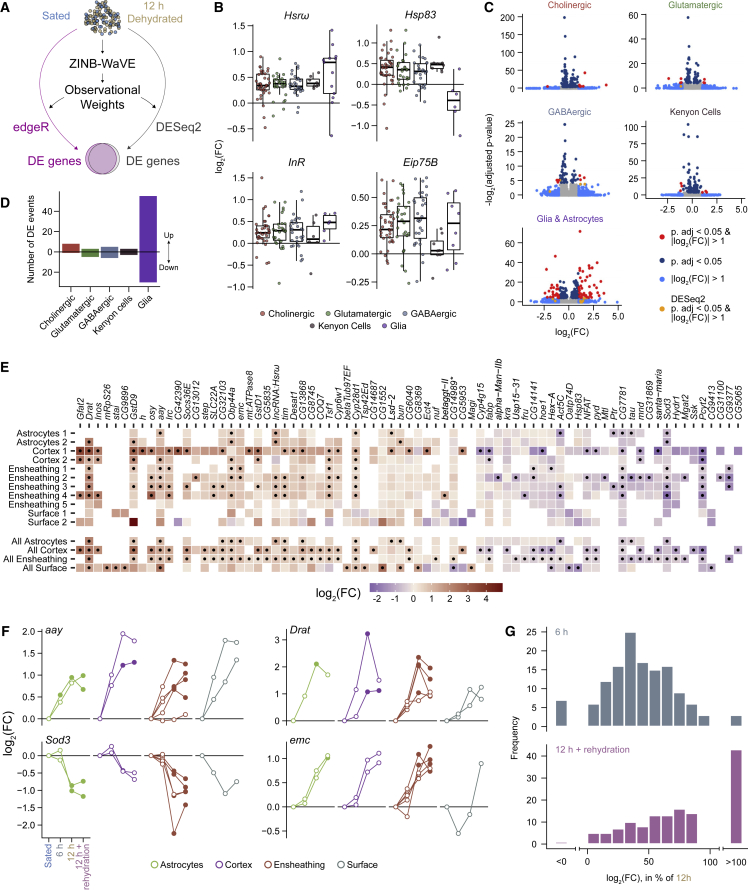
A characteristic of single-cell RNA sequencing (scRNA-seq) data is that expression of certain genes is stochastically undetectable in a fraction of the cells in which they are normally expressed. This artefact called “dropout” can result from low mRNA levels in single cells and inefficient mRNA capture during library preparation, and it produces a larger than expected number of zero read counts.42,43 Therefore, classic differential expression methods designed for negative binomial bulk transcriptome data often underperform when analyzing weakly expressed genes in scRNA-seq data.44,45 To correct for potential bias caused by zero read count inflation, we downweighed excess zeros using ZINB-WaVE,46,47 which enables a more accurate estimation of data dispersion and thereby improves detection of differentially expressed genes. Using ZINB-WaVE with both edgeR and DESeq2, two tools for quantifying differential expression,48,49 we calculated gene expression differences between water sated and 12 h dehydrated conditions across cell clusters (Figure 2A).
Figure 2.
The transcriptional signature of thirst in the fly brain
(A) Schematic of differential expression analysis. Observational weights calculated with ZINB-WaVE were used in edgeR and DESeq2 to correct for zero-inflation.
(B) Boxplots showing differential expression of the four genes most broadly regulated in each cluster after 12 h dehydration, grouped by main cell class.
(C) Volcano plots representing statistical significance against fold change for all genes tested in cholinergic neurons, glutamatergic neurons, GABAergic neurons, Kenyon cells, and glia, calculated with edgeR. Each plot represents pooled data from all clusters in each cell class. Genes with adjusted p value < 0.05 and |log2(FC)| > 1 in DESeq2 but not edgeR are labeled orange.
(D) Number of differential expression events identified in the five cell classes shown in (C). Values above the bar represent genes upregulated in thirsty flies and below the bar represents downregulated genes.
(E) Heatmap showing fold change for each of the most regulated genes in glia, for each cluster (top) and for clusters grouped by glia type (bottom), calculated with edgeR. Black dots: adjusted p value < 0.05. Empty tiles: transcript levels below detection threshold. ∗: CG14989 is the only gene significantly upregulated and downregulated in different clusters.
(F) Changes in expression for four differentially expressed genes across glia clusters, and through all four hydration conditions. In most cases, expression gradually increases or decreases during dehydration. After rehydration, mRNA levels tended to remain similar to levels measured in dehydrated flies. Full circles indicate adjusted p value < 0.05, and empty circles indicate adjusted p value ≥ 0.05.
(G) Comparisons of log2(FC) values (versus sated controls) after 6 h dehydration (top) or after rehydration (bottom), as a percentage of log2(FC) values after 12 h dehydration. Percentages were calculated for each of the significant gene-cluster pairs shown in (E) (black dots) and represented in histograms in 10% increments. Changes in expression are lower (<100%) after 6 h than after 12 h dehydration (top) for most genes. However, expression of many genes remains high (>100%) after thirsty flies are allowed to drink (bottom).
See also Figure S2.
We expected that general osmotic stress of dehydration might trigger a brain-wide transcriptional response. We identified four genes whose expression increased across most clusters, with an average log2(FC) across clusters above 0.25 (Figure 2B). Two of these, the chaperone encoding Hsp83 and the long non-coding RNA Hsrw, have been implicated in the cellular stress response and shown together to regulate transcription.50, 51, 52 Global upregulation of the insulin receptor (InR) may reflect the role of insulin signaling in balancing thirst and hunger,19,53 whereas the ecdysone-response gene Eip75B buffers circadian rhythms in stressful conditions.54
Cell-types exhibiting state-dependent changes in gene expression potentially play a role in the fly’s response to thirst, and the regulated genes may represent intra or intercellular mechanisms mediating physiological and behavioral responses. 84 genes showed strong differential expression (|log2(FC)| > 1, adjusted p value < 0.05) in at least one cell cluster, with either edgeR or DESeq2, which identified slightly different sets of genes (Figure S2A). Although we detected fewer genes in glia than in other cell clusters (Figure S2D), most differential expression events (85/119) occurred in glia, with the rest in cholinergic, glutamatergic, GABAergic, and Kenyon cell classes (Figures 2C and 2D). No individual cluster within monoaminergic and “other” classes contained enough cells to enable differential analysis with sufficient statistical power. No differences were apparent when clusters from these two classes were analyzed as a group. Within the glial clusters, 6 genes passed our differential expression criteria in astrocytes, 26 in ensheathing glia, and 33 in cortex glia. No differences were found in surface glia. However, pooling glial clusters by type provided enough statistical power to reveal 15 genes in surface glia and 2 additional genes in cortex glia (Figure 2E). With the exception of CG14989, which was upregulated in ensheathing glia and downregulated in surface glia, all other differentially expressed genes changed in a similar direction in multiple glial types. Gene Ontology analysis indicated that differentially expressed genes contribute to pathways related to metabolism, response to stress, or behavior (Figure S2E). In summary, the highest magnitude thirst-dependent changes in gene expression occur in glial cell-types, whereas neuronal transcriptomes remain comparatively stable.
We next tested whether expression of thirst-responsive genes changed with dehydration time. For several, including aay, Death resistor Adh domain containing target (Drat) and extra macrochaetae (emc), expression steadily increased as dehydration progressed (Figure 2F), tracking the gradual increase in behavioral preference for humidity (Figure 1B). Indeed, a majority of log2(FC) values for 6 h were between 30% and 70% of those at 12 h (Figure 2G). In contrast, superoxide dismutase 3 (Sod3) expression remained stable until 6 h but decreased by 12 h (Figure 2F, bottom left) consistent with its suppression, perhaps participating in the metabolic or stress response to severe dehydration. Quenching thirst tended to return some transcript levels toward their sated baseline, although with different magnitude for different genes and clusters. However, transcript levels of many genes continued to increase after rehydration (Figure 2F, bottom right, Figure 2G), suggesting prolonged action beyond the rehydration period. Together, these results illustrate a diversity of water deprivation-induced gene regulation patterns.
Astrocytic aay bi-directionally regulates water consumption
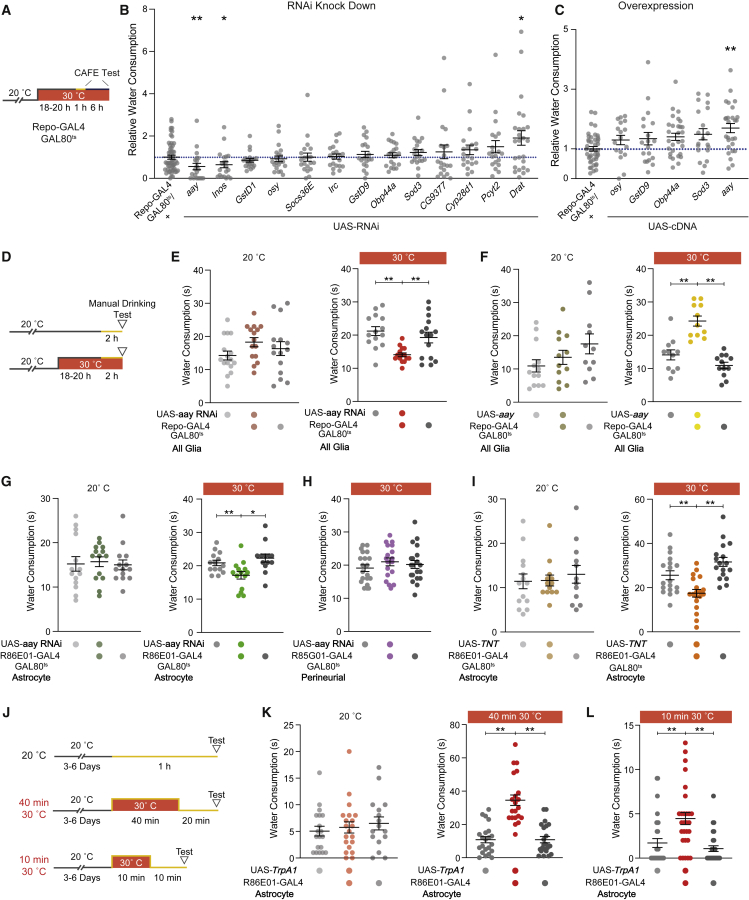
We tested whether glial genes identified by scRNA-seq regulated water consumption in the capillary feeding (CAFE) assay55 (Figures 3A and 3B). We targeted temporally restricted RNAi expression to adult glia using Repo-GAL4 combined with a ubiquitously expressed temperature-sensitive GAL8056 (Figure 3C). The strongest change in water consumption occured with aay. Adult-restricted RNAi knockdown of aay reduced water consumption, whereas overexpression increased it (Figures 3B, 3C, S3D, and S3E). Notably, aay RNAi appears to specifically regulate water consumption, as feeding was unaffected (Figure S3F).
Figure 3.
Astrocytic aay is a novel regulator of water consumption
(A) Schematic for temperature control of GAL80ts/GAL4 driven expression of UAS-RNAi or UAS-cDNA transgenes with CAFE test. Orange section of line indicates period of water restriction.
(B) Water consumption in the CAFE assay of flies with RNAi knock down of candidate genes. Dotted blue line indicates normalization to control Repo-GAL4 flies, equal to 1. ∗p < 0.05, ∗∗p < 0.01 two-tailed Mann-Whitney test nRNAi = 20–25, nRepo-GAL4 = 50.
(C) Water consumption in CAFE for UAS overexpression of targets. ∗∗p < 0.01 two-tailed Mann-Whitney test nRNAi = 17–29, nRepo-GAL4 = 43.
(D) Schematic for temperature control of GAL80ts/GAL4-driven UAS-RNAi or UAS-cDNA transgenes with manual water feeding assay. Orange section of line indicates period of water restriction.
(E) RNAi knockdown of aay reduces water consumption (n = 14–16).
(F) Overexpression of aay increases water consumption (n = 11–13).
(G) Astrocyte-specific RNAi knockdown of aay reduces water consumption (n = 13 and 14).
(H) RNAi knockdown of aay in perineurial glia does not alter water consumption (n = 18–20).
(I) Preventing vesicular transmission with tetanus-toxin (TNT) expression in astrocytes reduces water consumption (n = 12–19).
(J) Temperature regimen for TrpA1 activation of astrocytes in (K) and (L). Orange section of line indicates period of water restriction.
(K) Astrocyte activation for 40 min increases water consumption (n = 16–22). ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(L) Astrocyte activation for 10 min increases water consumption (n = 28–30). ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test. Normality was assessed using Shapiro-Wilk test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 Ordinary one-way ANOVA with Dunnett’s multiple comparisons test, unless otherwise stated. Individual data points are single flies. Data are mean ± standard error of the mean (SEM).
See also Figure S3.
We noticed significant death in the CAFE assay when manipulating aay (Figures S3G, S4D, and S4E), perhaps from sensitivity to stress of dehydration, or idiosyncrasies of the assay.57 To circumvent this issue, we employed a manual water consumption assay that is less challenging for the animals.19 These experiments revealed similar results to CAFE when knocking down or overexpressing aay in glia (Figures 3D–3F). We next tested whether aay-induced changes in water consumption could be assigned to a glial cell type.58 Expressing aay RNAi in astrocytes, but not in perineurial glia, reduced water consumption indicating the specific importance of astrocytic aay in controlling drinking (Figures 3G and 3H). Although baseline drinking of control flies varied between experiments, relative differences between genotypes and treatments remained consistent.
The aay gene encodes a phosphoserine phosphatase which converts phosphoserine into D- and L-serine.59, 60, 61 In mammals, the gliotransmitter D-serine is packaged into astrocytic vesicles and released in a SNARE and calcium-dependent manner.62, 63, 64, 65, 66 Drosophila astrocytes express vesicular machinery including synaptobrevin (Syb) and Syntaxin 1A (Syx1A) (Figures S3M and S3N). We tested whether vesicular release from astrocytes is necessary for water consumption by expressing a tetanus-toxin (TetX) transgene using an astrocyte-specific GAL4. Tetx abolishes evoked release by cleaving the vesicle-associated membrane protein synaptobrevin. Temporally restricted TetX expression in astrocytes reduced water consumption (Figure 3I).58
We also tested whether evoking Ca2+ entry into astrocytes promoted water consumption by expressing a transgene encoding the Ca2+-permeable temperature-sensitive TrpA1 channel in astrocytes (R86E01>TrpA1). We first tested flies with a 2 h “fictive dehydration” step at the TrpA1 activation temperature of 30°C; however, this manipulation reversibly paralyzed R86E01>TrpA1 flies, as previously described.67 We therefore shortened the 30°C stimulation to 40 min and allowed flies to recover at 20°C for 20 min (Figure 3J). This procedure substantially increased water consumption compared to controls carrying the R86E01-GAL4 or UAS-TrpA1 transgenes or those left at 20°C for the whole procedure (Figure 3K). Since vesicular release occurs promptly following astrocytic activation,62 we also further restricted the heat incubation to 10 min. This brief astrocyte activation was sufficient to increase water consumption (Figures 3L and S3L). Together, these data indicate that aay expression and vesicular release from activated astrocytes regulate water consumption.
D-serine facilitates water consumption via NMDA receptors
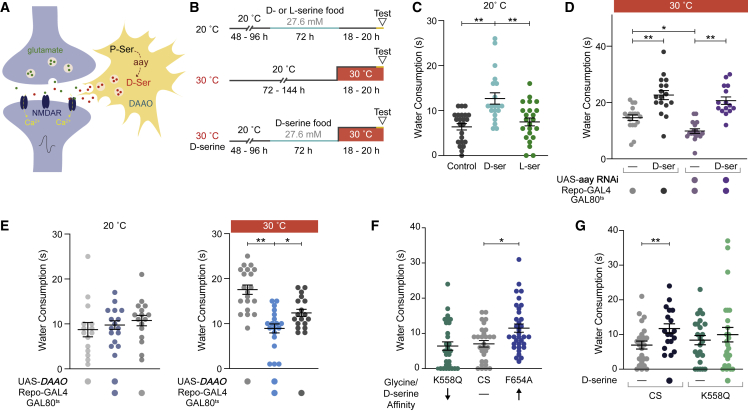
To further assess the role of D-serine as a gliotransmitter in water consumption, we supplemented the flies’ diet with D-serine and measured water intake (Figures 4A, 4B, and S4A). Feeding flies with D- but not L-serine increased water consumption (Figure 4C). D-serine is an enantiomer-specific co-agonist for NMDA-type glutamate receptors (NMDARs).68,69 We therefore tested if aay-dependent drinking defects could be rescued with dietary D-serine. Glial expression of aay RNAi was induced in flies previously fed with medium supplemented with D-serine (Figure 4B). D-serine feeding was sufficient to restore the aay-induced drinking deficit to a normal level (Figure 4D). Control experiments showed that flies did not prefer D-serine containing food and that D-serine did not alter feeding (Figures S4B and S4C).
Figure 4.
D-serine modulates water consumption via NMDARs
(A) Model of tripartite glutamatergic synapse showing aay-dependent synthesis of D-serine in astrocytes.
(B) Protocol for D-serine feeding and RNAi induction with water consumption experiments.
(C–G) Light blue section of lines indicate time on D- or L-serine food. Orange section of lines indicate period of water restriction.
(C) D- but not L-serine feeding increases water consumption (n = 20–23). ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(D) D-serine feeding rescues the water consumption defect in flies with aay knockdown (n = 15–18). ∗p < 0.05, ∗∗p < 0.01, ordinary one-way ANOVA with Dunnett’s multiple comparisons test.
(E) Glial overexpression of D-amino acid oxidase (DAAO) reduces water consumption (n = 16–20). ∗p < 0.05, ∗∗p < 0.01, ordinary one-way ANOVA with Dunnett’s multiple comparisons test.
(F) Flies harboring the F654A single site mutation in NMDAR1 exhibit increased water consumption. This mutation increases affinity for glycine/D-serine (n = 29–34). ∗p < 0.05, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(G) Flies harboring the K558Q amino acid substitution in NMDAR1 are insensitive to the D-serine induced increase in water consumption (n = 18–25). ∗∗p < 0.01, two-tailed Mann-Whitney test. Individual data points are single flies. Data are mean ± SEM.
See also Figure S4.
D-serine is degraded by D-amino acid oxidase (DAAO), which is encoded by the CG12338 and CG11236 genes in Drosophila.70,71 Since CG11236 expression is barely detectable in the brain, we focused on CG12338.70 Overexpressing a CG12338 DAAO transgene in glia suppressed water consumption (Figure 4E). Together, the aay manipulation, dietary supplementation, and DAAO data suggest that D-serine can bi-directionally regulate water consumption.
NMDARs are voltage and ligand-gated ion channels. Efficient channel activation requires binding of L-glutamate to the NR2 subunit and a co-agonist glycine or D-serine, which occupy the same site on the NR1 subunit.72 Prior neuronal depolarization is also required to remove a Mg2+ block, allowing ligand-gated channel conductance to occur. We therefore tested whether D-serine facilitated water consumption via NMDAR activation. We used flies harboring single-site mutations (K558Q and F654A) in the NMDAR1 gene which encodes the fly NR1 subunit. These amino acid substitutions are equivalent to K544Q and F639A in mammalian NR1.73, 74, 75 Glycine affinity is increased in receptors bearing F639A/F654A and decreased in those with K544Q/K558Q, whereas glutamate affinity is unaltered.76,77 Measuring water consumption revealed that flies harboring the F654A substitution in NR1 drank more than controls (Figure 4F). In addition, K558Q flies exhibited normal water consumption but were unable to acquire the D-serine induced increase in drinking (Figure 4G). Therefore, NMDAR co-agonist affinity directly impacts water consumption.
D-serine is a co-agonist of the Drosophila NMDA receptor
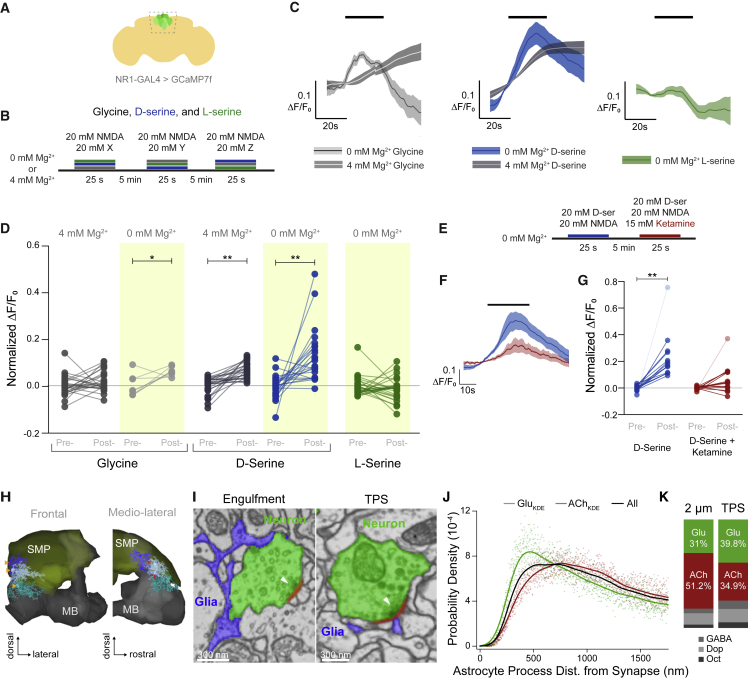
A prior study demonstrated that D- or L-Serine feeding regulated sleep in flies and that the effect depended on NMDARs.70 However, it is currently assumed that D-serine is a co-agonist of Drosophila NMDARs, as in mammals. Since NMDAR activation elicits increases in intracellular Ca2+, we expressed GCaMP7f broadly in NR1 positive (NR1+) neurons and performed in vivo calcium imaging in the brain of head fixed flies while bath-applying glycine, D-serine, or L-serine (Figures 5A and 5B). We recorded from NR1+ neurons (nmdar1-KIGAL4>GCaMP7f) in the pars intercerebralis (PI) because they are easily accessed and identified and therefore permitted reproducible measurements between flies (Figure 5A).70 In addition, responses were measured in Mg2+ free (0 mM Mg2+) and physiologically relevant concentrations of Mg2+ (4 mM Mg2+) (Figure 5B) to assess the importance of removing the Mg2+ occlusion from the channel pore.78,79 Bath application of NMDA with D-, but not L-, serine activated PI NR1+ neurons (Figures 5C and 5D). NMDA and glycine also produced excitation but only in Mg2+ free conditions (Figure 5D). D-serine and glycine in 4 mM Mg2+ promoted slower activation, likely due to the voltage-dependent Mg2+ block preventing fast activation. To validate that D-serine activation was occurring via NMDARs, we applied the non-competitive NMDAR antagonist ketamine with NMDA and D-serine (Figure 5E).80 Ketamine abrogated PI neuron responses to D-serine/NMDA (Figures 5F, 5G, and S5H) confirming that fly NMDARs are activated by D-serine.
Figure 5.
Astrocytes form tripartite synapses and D-serine is a co-agonist for NMDARs
(A) Illustration of imaging window for recording of Ca2+ responses in NMDAR1 expressing PI neurons.
(B) Protocol of drug application for (C) and (D). Order of application was randomized for each fly.
(C) Average traces for glycine, D-serine, and L-serine application in 0 or 4 mM Mg2+ (ngly = 8, 25; nD-ser = 25, 26; nL-ser = 28, respectively). Line is a moving average and shaded band is SEM.
(D) D-serine, but not L-serine, activates NR1+ PI neurons (from left to right, n = 25, 25, 8, 8, 25, 25, 23, 23, 28, and 28). ∗p < 0.05, ∗∗p < 0.001, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(E) Protocol for ketamine application with NMDA, TTX, and D-serine.
(F) Averaged traces for D-serine and NMDA (blue) and D-serine, NMDA, and ketamine (red) (n = 15).
(G) Ketamine inhibits D-serine induced activation of NR1+ PI neurons. ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test. Transparent dots indicate outliers that are >2 SD from the mean. When outliers are excluded from the analysis, the relationship still holds (see Figure S5F).
(H) Astrocytes tile the SMP. 3D representation of three astrocytes (shades of blue) reconstructed from EM data shown with the mushroom body neuropil (gray) and the SMP neuropil (yellow), shown in frontal and lateral views. Cell bodies are located outside the neuropil (yellow arrow heads), and processes in the neuropil have little overlap (∗).
(I) Astrocytes engulf synaptic boutons and contribute processes to tripartite synapses (TPSs). Grayscale EM data with labeled glutamatergic boutons (green) and glial processes (blue). Presynaptic densities (arrow) and synaptic cleft (red) are indicated. Left: example where glial processes contact a large proportion of a bouton’s membrane but not the synapse. Right: example of a glial process directly adjacent to the synaptic cleft and opposite the presynaptic density (white arrow head).
(J) Astrocytic processes are significantly closer to glutamatergic than cholinergic synapses in the SMP. Kernel density estimates (lines) of the probability distributions (points) of distances of Glu or Ach synapses or a random draw from both sets to 3 SMP based astrocytes. Only synapses in direct vicinity (2 μm radius around processes <600 nm thick) are considered. Statistical analyses shown in Figure S5.
(K) Glutamatergic synapses are overrepresented in tripartite synapses versus their representation in synapses in the general 2 μm vicinity of astrocytic processes. Ratios of synapses by predicted neurotransmitter usage are shown.
See also Figure S5 and Video S1 for anatomy of astrocyte processes engulfing a presynapse and contributing to TPS.
3D representations of an astrocyte (blue) in the SMP and glutamatergic neurons (green) with boutons containing annotated presynapses (red), which consist of synaptic vesicles (red spheres) and pre-synaptic T-bar densities (black). 00:00 to 00:55, When glial cells engulf axons, multiple processes of different sizes contact a substantial area of a presynaptic bouton’s membrane but do not extend into the synapse. 00:56-1:40, Fine astrocyte processes either pass through the synaptic cleft, or terminate within it, to form a tripartite synapse. In both cases the astrocyte processes contact the synaptic cleft alongside the postsynaptic neurons. Many closely spaced boutons of the same presynaptic neuron can be contacted by the same astrocyte, which therefore contributes processes to multiple tripartite synapses. Anatomical features are further highlighted in the video. Cellular meshes were retrieved from FlyWire and animations produced with blender.
Astrocytes contribute to tripartite synapses in the Drosophila brain
Processes from mammalian astrocytes infiltrate the synaptic cleft, forming TPSs.81 This allows glia to participate in neurotransmission through transmitter uptake and recycling and by release of gliotransmitters.82 Drosophila adult neuromuscular synapses have a tripartite structure,83 but the detailed anatomy of adult central nervous system (CNS) astrocytes is unclear. To address this, we analyzed astrocytes in the superior medial protocerebrum (SMP) neuropil within the FlyWire project (flywire.ai)84 which utilizes the full adult female brain (FAFB) transmission electron microscope dataset.85 We fully reconstructed 11 astrocytes and four were extensively reviewed to reconstruct their finest processes. These astrocytes tile the neuropil and closely associate with trachea, as previously reported in the larval brain and ventral nerve cord (VNC)31,86,87 (Figures 5H, S5J, and S5K). We identified astrocytic processes that engulfed synaptic boutons and that contacted synaptic clefts like processes from postsynaptic neurons—the latter providing evidence of canonical TPSs in the adult CNS (Figures 5I and S5L). Astrocytes also often contact the postsynaptic neuron within 500 nm of the synaptic cleft.
To determine whether astrocytes associate with a particular class of synapse, we used the prior computational predictions of neurotransmitter identity for neurons in the FAFB volume.88 We composed a vicinity profile by measuring distances of synapses from the fine processes of the 3 extensively reviewed SMP astrocytes and plotted their distance distributions categorized by presynaptic neurotransmitter. This analysis revealed astrocytic processes to be significantly closer to glutamatergic (962 ± 2 nm SD) and GABAergic (971 ± 3 nm) than cholinergic synapses (1,061 ± 1 nm) (Figures 5J, 5K, S5M, and S5N). Astrocytes are therefore ideally positioned to modulate glutamatergic synapses in the adult brain.
Astrocytes show heterogeneous responses to neurotransmitters
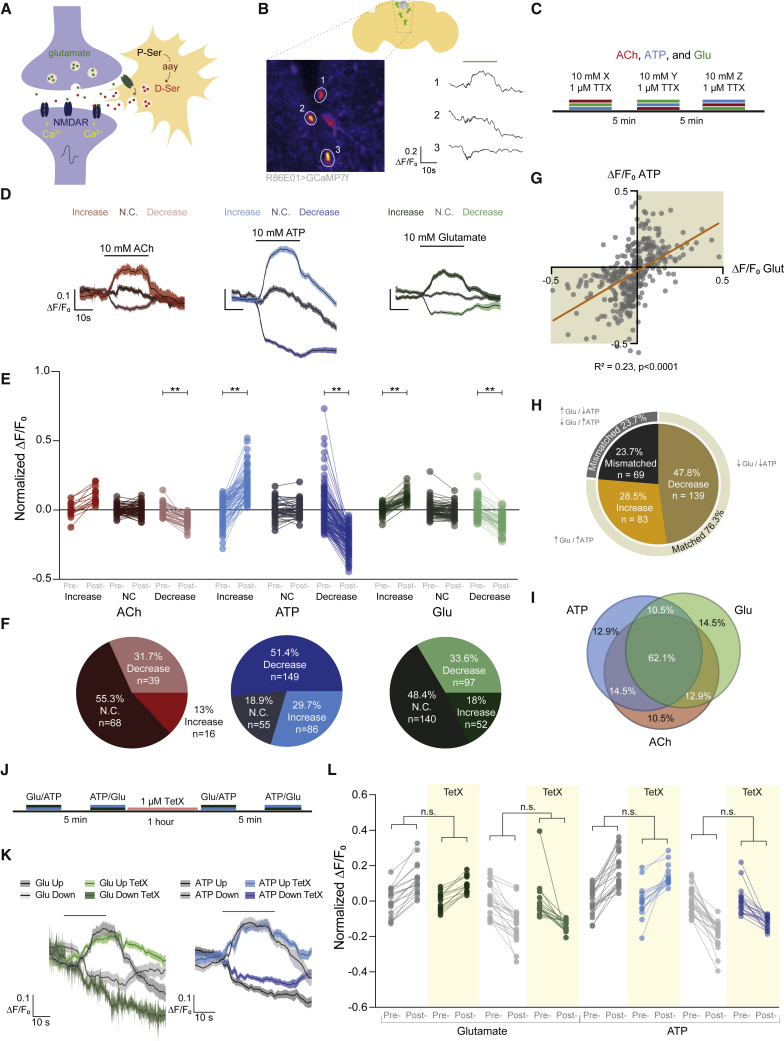
Astrocytes have been proposed to facilitate fast synaptic activity by releasing gliotransmitters such as glutamate, ATP, and D-serine.89 In mammals, glutamate binding to mGluR5 triggers astrocytic D-serine release90 and the combination of glutamate and D-serine produces maximal NMDAR activation. We therefore tested whether fly astrocytes were responsive to glutamate application by recording in vivo astrocytic Ca2+ responses (R86E01-GAL4>UAS-GCaMP7f) (Figures 6A and 6B). We included 1 μM tetrodotoxin (TTX) in the bath to block voltage-gated sodium channels and thereby inhibit polysynaptic neurotransmission. Surprisingly, we could classify astrocytes within individual flies depending on whether bath-applied glutamate evoked an increase, decrease, or no change in GCaMP fluorescence (Figure 6B). We also tested for astrocytic responses to acetylcholine (ACh), the predominant excitatory transmitter in the fly brain, and ATP which triggers Ca2+ waves in mammalian astrocytes.91,92 Astrocytes also exhibited differential responses to ACh and ATP (Figures 6C–6F). Moreover, the same astrocytes often exhibited similar responses to the different neurotransmitters. For instance, a substantial proportion of astrocytes were activated both by glutamate and ATP (Figures 6G, 6H, S6A, and S6B), and most of these had matched responses to all three transmitters (Figure 6I).
Figure 6.
Astrocytes are differentially responsive to neurotransmitters
(A) Illustration of possible glutamate-evoked D-serine release from astrocytes.
(B) Astrocytes adjacent to the pars intercerebralis show differential responses to glutamate application (green bar).
(C) Protocol for drug application with randomized order for acetylcholine (ACh), ATP, and glutamate (Glu).
(D) Average traces for ACh, ATP, and glutamate partitioned by the type of response (activated, no change, or inhibited). Responses were determined as follows: activated if μpost-drug > σpre-drug + μpre-drug, no change if μpost-drug fell within σpre-drug + μpre-drug and inhibited if μpost-drug < σpre-drug − μpre-drug. Line is smoothed average and shaded band is SEM.
(E) Neurotransmitters induce both excitatory and inhibitory responses in astrocytes. Paired datapoints represent responses from a single astrocyte, and group is assembled from multiple flies (nAch = 16, 68, 39; nATP = 86, 55, 149; nglut = 52, 140, 97). ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(F) Proportions of astrocytes sorted by response direction following acetylcholine, ATP, and glutamate application.
(G) Average ΔF/F0 during drug application for glutamate plotted against ΔF/F0 for ATP shows most astrocytes that are excited by glutamate are also excited by ATP and vice versa.
(H) Astrocytes are more likely to have matched responses between glutamate and ATP. Chi-square test between matched versus mismatched astrocytes. Fisher’s exact test matched versus mismatched p = 2.35e−11, OR = 3.24.
(I) Venn diagram showing considerable overlap of matched responses of astrocytes to all three neurotransmitters.
(J) Protocol for tetanus toxin (TetX) application.
(K) Average traces for glutamate- and ATP-evoked excitatory and inhibitory responses with and without TetX. Line is smoothed average and shaded band SEM.
(L) Blocking vesicular transmission does not suppress astrocyte responses to transmitter application (left to right, nPre-TetXATP = 21, 6; nPost-TetXATP = 16, 35; nPre-TetXGlut = 18, 11; nPost-TetXGlut = 14, 9). n.s. > 0.05 Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test. Individual data points are single astrocytes across multiple animals.
See also Figure S6.
We used TetX to eliminate all vesicular release and negate the possibility that evoking local transmission accounts for differential astrocytic responses (Figure S6C).93 Although there was no change in magnitude of astrocyte responses to either ATP or glutamate following 1 h TetX treatment, there was a slight reduction in percentage of ATP responsive astrocytes (Figures 6J–6L and S6F). We verified the effectiveness of TetX by demonstrating that a polysynaptic odor-evoked response in mushroom body output neuron (MBON)-γ5β′2a was eliminated (Figures S6D and S6E). Therefore, neurotransmitters evoke direct stereotyped activation or inhibition of astrocytes. Differential astrocytic responses to neuronal activation were reported in the larval VNC.94
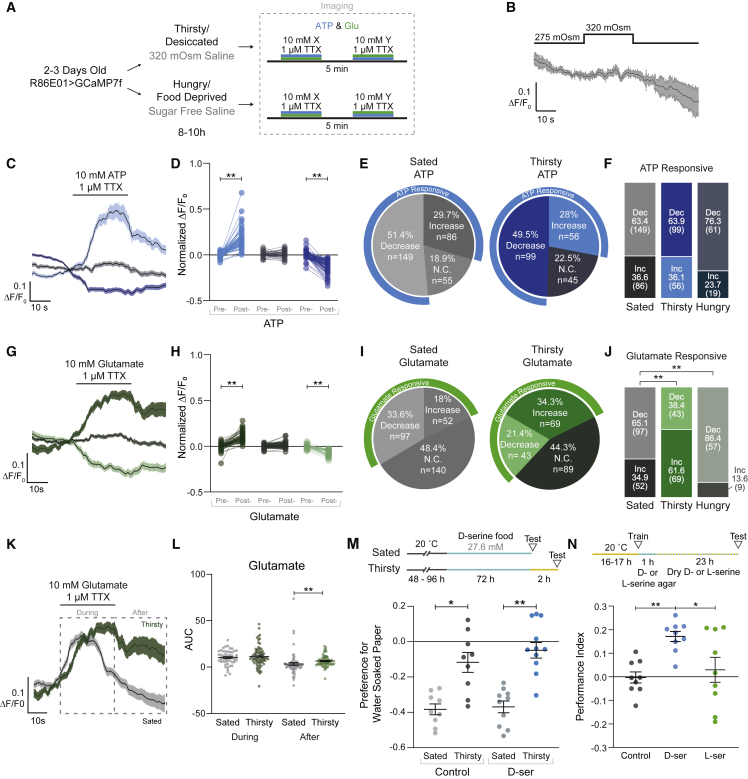
Water deprivation modulates functional properties of astrocytes
We next tested whether in vivo astrocytic responses to applied neurotransmitters changed in water- and food-deprived flies (Figure 7A). To prevent deprived flies from being “re-satiated” by application of normal extracellular saline, the saline composition was adjusted to reflect the desired physiological state of the animal.19,95 Astrocytes did not show a Ca2+ response to a brief increase in osmolarity (Figure 7B). Similar to sated flies, water- and food-deprived flies showed differential astrocytic responses to glutamate and ATP (Figures 7C–7J and S7A–S7C). However, only water-deprived flies showed an increase in the astrocyte proportion excited by glutamate, whereas ATP responsiveness remained the same (Figures 7F and 7J). Excitatory glutamate and ATP responses also appeared prolonged in thirsty flies compared with controls (Figures 7K, 7L, and S7D). Finally, thirsty flies had a higher proportion of mismatched astrocyte responses to ATP and glutamate, suggesting a loss of correlated responses (Figure S7G). In sum, water deprivation increases the proportion of glutamate-activated astrocytes and sustains glutamate-evoked Ca2+ responses.
Figure 7.
Water deprivation changes physiological properties of astrocytes
(A) Protocol for water and food deprivation with calcium imaging experiments.
(B) Acute exposure of high osmolarity saline does not induce a calcium response in astrocytes.
(C) Average traces for ATP partitioned by response type in thirsty flies. Line is smoothed average and shaded band is SEM.
(D) Responses pre- and post-treatment of ATP separated by categorical responses in water-deprived flies. ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(E) Proportion of astrocyte responses to ATP application in sated and thirsty flies.
(F) Astrocyte responses to ATP do not change with deprivation states. n.s. > 0.05 Fisher’s exact test with Bonferroni correction.
(G) Average traces for glutamate partitioned by response type in thirsty flies. Line is smoothed average and shaded band SEM.
(H) Responses pre- and post-treatment of glutamate separated by categorical responses in water-deprived flies. ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(I) Proportions of astrocyte responses type to glutamate application in sated and thirsty flies.
(J) Deprivation states differentially regulate astrocyte responsivity to glutamate application. The number of glutamate responsive astrocytes increases in thirsty flies and decreases in hungry flies. ∗∗p < 0.01, Fisher’s exact test with Bonferroni correction.
(K) Excitatory astrocyte responses to glutamate application are prolonged in thirsty animals.
(L) Area under the curve (AUC) of sections of traces marked in (K). ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(M) Flies pre-fed D-serine do not show differences in innate water seeking in the T-maze. ∗p < 0.05, ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test.
(N) Flies fed D-serine between training and testing show increased water memory performance. ∗p < 0.05, ∗∗p < 0.01, Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test. For (D), (H), and (L), individual data points are single astrocytes across multiple animals.
See also Figure S7.
D-serine promotes water-memory expression
Prior work implicated specific neuronal pathways in regulating water consumption and thirst-dependent control of innate and learned water seeking.19, 20, 21, 22 Since astrocytes broadly innervate the neuropil, we tested whether D-serine promoted water vapor seeking and memory expression in water-sated flies, like it did for drinking. Interestingly, the same 3-day feeding of D-serine that increased drinking did not promote attraction to water vapor (Figure 7M). We next tested thirst-dependent expression of water memory performance. Water-deprived flies were trained to associate an odor with water reward, and after training, they were housed in vials with agarose for 1 h then dry vials containing either nothing, D-serine, or L-serine powder. Only flies fed with D-serine showed 24 h water memory performance (Figure 7N). These data demonstrate that dietary D-serine promotes the expression of water-seeking memory, in addition to drinking behavior.
Discussion
Single-cell RNA-seq reveals a glial response to water deprivation
Our scRNA-seq studies of thirst revealed astrocytes and other glia to be the principal transcriptionally responsive cells in the fly brain. We did not identify significant transcriptional changes within peptidergic18, 19, 20 or monoaminergic neurons,21,22,96 perhaps due to the relative scarcity of these neurons in our datasets. Although glial expression of many genes changed in thirsty flies, our initial loss-of-function RNAi screen only revealed potential roles for aay, inos (myo-inositol-1-phosphate synthase), and Drat in the control of drinking behavior. Results for inos and Drat were not corroborated by additional testing. Nevertheless, induction of myo-inositol synthesis and uptake is a known cellular response to osmotic stress.97 Drat might also be involved in resistance to stress of dehydration.98 We similarly speculate that other thirst-regulated genes maintain glial function under stress, without impacting fluid intake in a measurable way.
Glial D-serine is required for motivated water-procuring behaviors
A key approach to verifying a role for D-serine in thirst was the ability to alter fly behavior by providing D-serine in the diet. Dietary D- but not L-serine restored the drinking defect of aay-deficient flies. Moreover, feeding D- but not L-serine to water-sated flies mimicked the behavioral effect of water-deprivation—promoting both drinking and water-seeking memory expression. We therefore conclude that gliotransmission of D-serine is an essential element of thirst-state behavioral modulation. Changes in hunger-directed feeding were not evident. At present we do not know if dietary D-serine floods the entire fly and bypasses the need for glial release. Interestingly, although our experiments demonstrated D-serine enantiomer specific aay rescue and gain-of-function effects on thirst-related behaviors, a previous study70 rescued a fly sleep defect with L- and D-serine feeding. However, rescue of sleep depended on intestinal serine racemase conversion of L- to D-serine. Since the authors also identified intestinal NMDAR1 expressing neurons,70 we speculate that L-serine conversion in the gut produces enough D-serine to alter sleep via that route, but it produces insufficient quantities to act in the brain.99
Does water-deprivation prime astrocytes to release D-serine?
We noted that increased aay expression resulted in detection of a larger proportion of astrocytes (74% versus 56%) expressing aay following water deprivation (Figure S7E). This potential broadening of expression might indicate that water deprivation induces aay to meet a forthcoming demand for D-serine release and/ or to replenish it following release from these astrocytes. Interestingly, a similar number of astrocytes (34% versus 18%) also gained sensitivity to exogenously applied glutamate in thirsty flies. Although we do not know if the same astrocytes are involved, it seems plausible that thirst increases astrocytic expression of aay and the number of glutamate responsive astrocytes, so that glutamate-releasing neurons can evoke their D-serine release. Although loss-of-function experiments revealed the importance of astrocytic aay, induction also occurred in cortex, ensheathing, and surface glia. The role of aay in these other glial types is currently unclear. Elevated expression of aay was attenuated when flies were permitted to quench their thirst, consistent with induction serving an enhanced need for D-serine synthesis.
Astrocytic D-serine facilitates the activity of glutamatergic circuits
We established that D-serine functions as an NMDAR co-agonist in Drosophila, like it does in mammals.69 Neuronal NMDA-evoked Ca2+ transients were potentiated by D-serine co-application and blocked by ketamine. Moreover, feeding D-serine did not promote drinking in flies harboring a K558Q point substitution in the NMDAR1 subunit that renders it insensitive to D-serine.74 Importantly, our ultrastructural analyses identified typical TPSs within the adult fly brain, as well as a new and more common anatomical motif where astrocytic processes engulf presynaptic boutons. Astrocytic processes are therefore well placed to potentially support synaptic transmission acting on either the pre- or post-synaptic neuron. Glial engulfment of presynaptic boutons may permit D-serine or other neuromodulators to regulate extra-synaptic NMDARs or other receptors to tune presynaptic transmitter release. Moreover, astrocytic processes in the SMP appear to be preferentially associated with glutamatergic synapses, despite cholinergic synapses being most abundant in this region. We therefore propose that thirst-regulated astrocyte-released D-serine facilitates NMDA-type currents so that the relevant glutamatergic synapses undergo short-term potentiation, allowing these water-procuring neural circuits to function more efficiently. If D-serine release depends on glutamate-evoked astrocytes at active TPSs, then its use will be need dependent and circuit specific. It is also conceivable that synaptic astrocyte processes can be remodeled by osmosensitive swelling and retraction.100 Further work is required to locate the critical D-serine modulated synaptic connections in the fly brain. We assume different circuits will underlie D-serine regulated learned water seeking and consumption.19, 20, 21, 22,96
Do glia directly sense water deprivation?
Although glia are ideally positioned for both integrating hemolymph metabolic and osmotic states and instructing neuronal activity, we did not observe obvious astrocytic Ca2+ responses when the osmolarity of the extracellular solution was briefly increased. It is possible that astrocyte activation requires a longer and more sustained exposure to hyperosmotic solution, that they are instead sensitive to hypovolaemic changes, or that water deprivation does not induce an astrocytic Ca2+ response (we also do not know whether thirst-driven glial transcriptional responses are Ca2+-dependent). In addition, although fly perineurial and subperieurial glia provide the barrier between the hemolymph and brain, cortex glia wrap neuronal cell bodies, and astrocytes permeate the neuropil and associate with synaptic compartments. It is therefore plausible that the glia layers permit transduction of osmosensing signals to astrocyes. For example, Ca2+ oscillations in cortex glia, which require the Na+/Ca2+, K+ exchanger zydeco,101 could influence astrocyte activity/Ca2+, as they are directly connected.102
Astrocytic Na(X) channels have been implicated in sodium sensing within the mouse SFO.103 Our scRNA-seq data reveal glial-specific expression of several potential osmo-sensing molecules. Solute carrier transporters such as Ncc69, NKCC, and inebriated (ine) localized to glial clusters (Data S2). Interestingly, the ine-encoded Na+/Cl− dependent neurotransmitter transporter is required for water homeostasis in the Malpighian tubules.104 Additionally, ine negatively regulates neuronal activity, which could be a mechanism by which astrocytes couple osmotic regulation with neurotransmission.105 Several TRP genes encode mechanosensitive channels responsive to changes in cell volume.106 The pain and wtrw TRP genes were exclusively detected in glia (Data S2). Finally, glia exhibit restricted expression of the inwardly rectifying potassium channel genes Irk2 and Irk3, which are necessary for proper fluid excretion and renal function in Drosophila.107 Therefore, glia may sense osmotic changes and regulate osmotic balance in the brain.
Thirst specificity
Although Drosophila are raised in the lab on food containing water, flies possess independent control mechanisms that permit selective food or water seeking.20, 21, 22,96,108 Thirst and hunger are however also coupled on a behavioral and neuronal level. For instance, prandial thirst is triggered by food ingestion to preemptively recalibrate blood osmolality.4,16,19,109 D-serine apparently distinguishes between water and food consumption. In addition, astrocytes showed opposing physiological changes with food compared with water deprivation (Figure 7J), and thirst-regulated genes barely changed when flies were hungry (Figure S2C). D-serine elevation, either by aay overexpression or D-serine feeding, promoted expression of water-seeking memory and water consummatory behaviors. Therefore, astrocytic aay-dependent D-serine signaling may provide a more general thirst modulation, as opposed to only regulating seeking20 or drinking.19 Further work is required to understand how thirst-driven D-serine interacts with other thirst-dependent modulatory signals. It will be interesting to determine whether D-serine influences sodium appetite and interacts with modulatory processes relevant to other motivational states.22,95,110, 111, 112, 113
Could D-serine be a conserved state-mechanism in mammals?
Thirst alters brain-wide population dynamics in the mouse.114 Since astrocytes permeate all regions of the brain, it is possible that D-serine contributes to the broad thirst-driven modulation of neural activity. In addition, we note that a lower D-serine serum concentration has been linked to patients suffering from Schizophrenia,115 6%–20% of whom are polydipsic.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Drierite, 8 mesh | ACROS Organics / Fisher Scientfic | Cat#10185130 |
| Schneider's culture medium | Gibco / Thermo Fisher Scientific | Cat#21720024 |
| d(–)-2-amino-5-phosphonovaleric acid | Sigma-Aldrich | Cat#A8054 |
| DNQX | Sigma-Aldrich | Cat#D0540 |
| Tetrodotoxin | Abcam | Cat#ab120054 |
| DPBS (calcium and magnesium free) | Gibco / Thermo Fisher Scientific | Cat#14190086 |
| Papain | Sigma-Aldrich | Cat#P4762 |
| Collagenase I | Sigma-Aldrich | Cat#C2674 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| BSA | New England Biolabs | Cat#B9000S |
| D-Serine | Tokyo Chemical Industry | Cat#S0033 |
| L-Serine | Tokyo Chemical Industry | Cat#S0035 |
| Glycine | Sigma-Aldrich | Cat#G8898 |
| N-methyl-D-aspartic acid | Sigma-Aldrich | Cat#M3262 |
| Acetylcholine | Sigma-Aldrich | Cat#A6625 |
| Adenosine Tri-Pphosphate | Sigma-Aldrich | Cat#A2383 |
| Glutamate | Sigma-Aldrich | Cat#G0355000 |
| Adenosine | Sigma-Aldrich | Cat#A9251 |
| Tetanus toxin | Sigma-Aldrich | Cat#T3194 |
| α,β-methylene adenosine 5’-diphosphate | Sigma-Aldrich | Cat#M3763 |
| 4-Methylcyclohexanol (98%) | Sigma-Aldrich | Cat#153095 |
| 3-octanol (99%) | Sigma-Aldrich | Cat#218405 |
| Critical commercial assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2, 4 rxns | 10X Genomics | Cat#PN-120267 |
| LightCycler 480 Probes Master | Roche | Cat#4887301001 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| SuperScript III First-Strand Synthesis SuperMix | Invitrogen | Cat#18080400 |
| Universal Probe Library | Roche | Cat#04683633001; Cat#04869877001 |
| LightCycler 480 Probes Master | Roche | Cat#4887301001 |
| Deposited data | ||
| Raw sequencing data and processed dataset | This study | GEO: GSE207799 |
| Drosophila melanogaster reference genome (dm6) and gene annotations (FlyBase, release 6.25, FB2018_06) | Thurmond et al.116 | https://flybase.org/ |
| Full Adult Female Brain (FAFB) transmission electron microscope (TEM) dataset | Zheng et al.85 | https://temca2data.org/ |
| Experimental models: Organisms/strains | ||
| Drosophila: 0273-GAL4 | Burke et al.117 and Gohl et al.118 | N/A |
| Drosophila: w; repo-GAL4, tub-GAL80ts; repo-GAL4 | S. Schirmeir & C. Klämbt | N/A |
| Drosophila: R86E01-GAL4 | Bloomington Drosophila Stock Center; Kremer et al.58 | RRID:BDSC_45914 |
| Drosophila: R85G01-GAL4 | Bloomington Drosophila Stock Center; Kremer et al.58 | RRID:BDSC_40436 |
| Drosophila: R66C08-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_49412 |
| Drosophila: UAS-mCherry | NA | Lab stock |
| Drosophila: UAS-aayRNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0454865 |
| Drosophila: InosRNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0472636 |
| Drosophila: UAS-GstD1RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0475105 |
| Drosophila: UAS-CG33970RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0462630 |
| Drosophila: UAS-Socs36ERNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0469597 |
| Drosophila: UAS-IrcRNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0472971 |
| Drosophila: UAS-GstD9RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0475656 |
| Drosophila: UAS-Obp44aRNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0464970 |
| Drosophila: UAS-Sod3RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0471232 |
| Drosophila: UAS-CG9377RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0464792 |
| Drosophila: UAS-Cyp28d1RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0470866 |
| Drosophila: UAS-Pcyt2RNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0477620 |
| Drosophila: UAS-DratRNAi | Vienna Drosophila RNAi Center | RRID:FlyBase_FBst0480136 |
| Drosophila: UAS-CG33970 (osy) | Bernard Moussian; Wang et al.119 | N/A |
| Drosophila: UAS-aay | FlyORF | RRID:FlyBase_FBst0501667 |
| Drosophila: UAS-GstD9 | FlyORF | F004171 |
| Drosophila: UAS-Obp44a | FlyORF | F003929 |
| Drosophila: UAS-Sod3 | FlyORF | F003855 |
| Drosophila: UAS-Daao1 | Dai et al.70 | N/A |
| Drosophila: UAS-Shits1 | Kitamoto120 | Lab stock |
| Drosophila: UAS-Kir2.1 | Paradis et al.121 | Lab stock |
| Drosophila: UAS-TNT E | Sweeney et al.122 | Lab stock |
| Drosophila: UAS-GCamP7f | Dana et al.123 | RRID:BDSC_79031; RRID:BDSC_80906 |
| Drosophila: NMDAR1K558Q | Nigel Atkinson; Troutwine et al.74 | N/A |
| Drosophila: NMDAR1F654A | Nigel Atkinson; Troutwine et al.74 | N/A |
| Oligonucleotides | ||
| TTCGCGAGGATGAATACGAT | This study | SdhA – FWD |
| CACGAGAGCGTGTGCTTG | This study | SdhA – REV |
| AAAAAGCTCCGGGAAAAGG | This study | GAPDH – FWD |
| AATTCCGATCTTCGACATGG | This study | GAPDH – REV |
| CACCATGAAGAACGCTGTTG | This study | Obp44 – FWD |
| GCTTGTAGTCGGAGGCAGAG | This study | Obp44 – REV |
| CCGTTTCCACGACATTGAGT | This study | Drat – FWD |
| GTTAATGCCTTGATGGGGAAC | This study | Drat – REV |
| TGGCTCCCTTTATCCCAAG | This study | GstD9 – FWD |
| AAAAACAGGCGCTGATTGAT | This study | GstD9 – REV |
| CGGTGACTCCCTTACCGTAG | This study | GstD1 – FWD |
| TTTGGCCACCTCGAATGT | This study | GstD1 – REV |
| AAAAAGCCAGCAAACCAAAA | This study | Socs36E – FWD |
| AGGTGATGACCCATTGGAAG | This study | Socs36E – REV |
| TGCAATGGGTGGAATTCAG | This study | Irc – FWD |
| ACCATTTCGAAGCAGGAATC | This study | Irc – REV |
| CCCCTGAAAAACGTCTATGC | This study | aay – FWD |
| AGCTATCGTATTCGCCCAAA | This study | aay – REV |
| AGTGCTGTAATATCCCCGATAAAC | This study | CG9377 – FWD |
| CAGAGATCATGGCGTCCTC | This study | CG9377 – REV |
| ACCGCTCTTTATGGACTTTGAG | This study | Pcyt2 – FWD |
| GCATGGCCTTGTCATCGTA | This study | Pcyt2 – REV |
| Software and algorithms | ||
| CellRanger | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome; RRID:SCR_017344 |
| R | R Development Core Team, 2008 | http://www.R-project.org/; RRID:SCR_001905 |
| Seurat v3 | Hafemeister and Satija124 and Stuart et al.125 | https://satijalab.org/seurat/; RRID:SCR_016341 |
| DoubletFinder | McGinnis et al.126 | https://github.com/chris-mcginnis-ucsf/DoubletFinder; RRID:SCR_018771 |
| ZINB-WaVE | Risso et al.46 and Van den Berge et al.47 | https://github.com/drisso/zinbwave |
| DESeq2 | Love et al.48 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID:SCR_015687 |
| edgeR | Robinson et al.49 and McCarthy et al.127 | https://bioconductor.org/packages/release/bioc/html/edgeR.html; RRID:SCR_012802 |
| ClusterProfiler | Yu et al.128 | https://github.com/YuLab-SMU/clusterProfiler |
| GOplot | Walter et al.129 | https://wencke.github.io/ |
| ScanImage 3.8 | Pologruto et al.130 | http://scanimage.vidriotechnologies.com |
| Fiji | NIH; Schindelin et al.131 | https://fiji.sc/ |
| FlyWire | Dorkenwald et al.84 | https://flywire.ai/ |
| Neuroglancer | Dorkenwald et al.84 | https://github.com/google/neuroglancer |
| CAVEclient | https://github.com/seung-lab/CAVEclient | |
| FAFBseg v.1.4 | https://github.com/navis-org/fafbseg-py | |
| Navis v.0.6 | https://github.com/navis-org/navis | |
| trimesh | https://github.com/mikedh/trimesh | |
| skeletor 1.1 | Au et al.132 | https://github.com/navis-org/skeletor |
| Blender 3.0 | Blender Community | https://www.blender.org/download/releases/3-0/ |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| Adobe Illustrator | Adobe Systems | https://www.adobe.com/uk/products/illustrator.html; RRID:SCR_010279 |
| Other | ||
| 10 mm CellTrix strainer | Sysmex | Cat#04-0042-2314 |
| Fuchs-Rosenthal haemocytometer | VWR | Cat#631–1096 |
| Fisherbrand chromatography paper 180g/m2 460x570 mm | Fisher Scientific | Cat#15649494 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Scott Waddell (scott.waddell@cncb.ox.ac.uk).
Correspondence regarding the single-cell transcriptomic data and its analysis should be addressed to Vincent Croset (vincent.croset@durham.ac.uk).
Material availability
This study did not generate new unique reagents.
Experimental model and subject details
Drosophila strains
GAL4 drivers used in this study are 0273-GAL4,117,118 R66C08-GAL4 (MBON-γ5β′2a),133 repo-GAL4, tub-GAL80ts; repo-GAL4,56,134,135 R86E01-GAL458 and R85G01-GAL4.58 UAS lines are w; +; UAS-mCherry, UAS-aayRNAi (VDRC, 23179), UAS-InosRNAi (VDRC, 100763), UAS-GstD1RNAi (VDRC, 103246), UAS-CG33970RNAi (VDRC, 38661), UAS-Socs36ERNAi (VDRC, 51821), UAS-IrcRNAi (VDRC, 101098), UAS-GstD9RNAi (VDRC, 103798), UAS-Obp44aRNAi (VDRC, 43203), UAS-Sod3RNAi (VDRC, 8760), UAS-CG9377RNAi (VDRC, 42835), UAS-Cyp28d1RNAi (VDRC, 7870), UAS-Pcyt2RNAi (VDRC, 105794), UAS-DratRNAi (VDRC, 108325), UAS-osy,119 UAS-GstD9 (FlyORF, F004171), UAS-Obp44a (FlyORF, F003929), UAS-Sod3 (FlyORF, F003855), UAS-aay (FlyORF, F002296), UAS-CG12338,70 UAS-Shits1,120 UAS-Kir2.1,121 UAS-TNT E,122 UAS-GCaMP-7f.123 Mutant strains are NMDAR1K558Q and NMDAR1F654A.74 Flies were raised on standard cornmeal food under a 12:12 light:dark cycle at 60% humidity and 25°C, unless otherwise stated. 3-day old mixed sex flies were used for single-cell transcriptomics, 3-6 day old mixed sex flies for water preference, RT-qPCR and imaging experiments, and 3-8 day old male flies for water consumption tests.
Method details
Water preference assays
Dehydration and water preference assays were performed as described.21 Briefly, groups of 50-100 flies were stored for a given time period in vials containing a ∼2 cm layer of Drierite topped with a piece of cotton and a dried sucrose-coated filter paper. Vials were kept in a sealed container with a layer of Drierite at the bottom. Flies were (re-)hydrated flies in vials containing 1% agarose and a wet piece of sucrose-coated filter paper. After the appropriate water-deprivation period, flies were transferred into a T-maze and given the choice between two chambers lined with either a dried or a wet piece of filter paper. Preference Index was calculated as the number of flies in the wet tube minus the number of flies in the dry tube, divided by the total number of flies in each experiment.
Brain dissociation and cell collection
Brains (0273-Gal4>mCherry) were dissected and dissociated as described.33 Briefly, 24 central brains from an equal number of male and female flies were individually dissected in ice-cold DPBS (Gibco, 14190–086) and immediately transferred into 1 mL toxin-supplemented Schneider’s medium (tSM: Gibco, 21720–001 + 50 mM d(-)-2- amino-5-phosphonovaleric acid, 20 mM 6,7-dinitroquinoxaline-2,3-dione and 0.1 mM tetrodotoxin) on ice. Brains were washed once with 1 mL tSM and incubated in tSM containing 1.11 mg/mL papain (Sigma, P4762) and 1.11 mg/mL collagenase I (Sigma, C2674). Brains were washed once more with tSM and subsequently triturated with flame-rounded 200 mL pipette tips. Dissociated brains were resuspended in 1 mL PBS + 0.01% BSA and filtered through a 10 mm CellTrix strainer (Sysmex, 04-0042-2314). Cell concentration was measured using a disposable Fuchs-Rosenthal haemocytometer (VWR, 631–1096) under a Leica DMIL LED Fluo microscope. A typical preparation from 24 brains yielded ∼600,000 cells.
Library preparation, sequencing, and processing
mRNA barcoding was performed using the Chromium Single Cell 3′ Reagent Kit v3 (10x Genomics), following the manufacturer’s instructions. For each sample, we targeted an 8,000 cell recovery. Two libraries were prepared for each condition (sated, 6h dehydrated, 12h dehydrated and 12h dehydrated + 45min rehydrated). Libraries were sequenced with NovaSeq 6000 (Illumina) at Oxford’s Wellcome Trust Centre for Human Genetics. We obtained 3.419 billion reads, and used CellRanger 3.1.0 to map these to the FB2018_06 Drosophila melanogaster genome assembly (v6.25),116 and create digital gene expression (DGE) matrices.
Filtering and doublet removal
Cell barcodes with <300 or >4,500 features, >20,000 UMIs, >15% mitochondrial RNA, >10% rRNA or >15% ribosomal proteins were discarded. DGE matrices were then merged by condition, normalized, and scaled using SCTransform in Seurat v3.124,125 This included regression for the effects replicate and sex – based on expression of the male-specific lncRNA roX1.136 DGE matrices were integrated using CCA anchor-based methods, and cells were clustered using the Louvain algorithm on the shared nearest neighbor graph with a resolution of 2 and an UMAP reduction performed for visualization, using the top 20 Principal Components (PCs). Doublets were removed using a hybrid method. First, DoubletFinder126 was run on data processed with the standard Seurat scaling method (without SCTransform), as we found that DoubletFinder failed to produce reliable BCmvn curves on SCtransformed data. This identified 3,493 potential doublets. Second, because the glial marker nrv2137 and markers for neurons releasing fast-acting neurotransmitters VAChT, VGlut or Gad1 are normally not co-expressed, we considered it parsimonious to classify “cells” expressing two or more of these genes as doublets. So as not to needlessly discard cells containing small contamination from these genes, we set thresholds of 3 (nrv2), 1.5 (VAChT), 2.1 (VGlut) and 2.3 (Gad1), above which we considered these genes to be highly expressed. These values were estimated from the normalized and scaled expression levels as the local minima in each gene’s bimodal distribution of non-zero values. We used the same principle to detect doublets in Kenyon Cells. We first identified Kenyon Cell clusters based on expression of Dop1R1, ey and mub.33 We then flagged all cells co-expressing markers for more than one Kenyon Cell subtype, namely Ca-alpha1T for αβ, ab for γ and CG8641 for α′β′,33 with thresholds of 1.5, 1.5 and 2.2, respectively (again estimated as the local minima in each gene’s bimodal distribution of non-zero values in their normalized and scaled expression levels). Overall, this co-expression strategy identified another 2,828 doublets. Doublets were evenly spread across clusters, with only six clusters comprised of >20% doublets. All doublets identified with either of these two methods (9.05% of all cells) were removed prior to subsequent analyses (Figure S1A).
Clustering
We developed a 4-step clustering pipeline with the aim of minimizing the number of PCs used for clustering and reducing the ‘curse of high dimensionality’ effects on nearest neighbor search.138,139 After doublet removal, data was clustered using the Louvain algorithm with the first 20 PCs and a resolution of 2 and an UMAP plot constructed using the same PC dimensions for visualization. Then each cluster was assigned to one of six major cell types present in the Drosophila brain, using expression of the following markers: VAChT (cholinergic), VGlut (glutamatergic), Gad1 (GABAergic), ey, Dop1R2, Pka-C1, mub (Kenyon Cells), Vmat (monoaminergic) and CG10433 (glia & astrocytes),33 with a seventh group containing non-assigned cells (“other”). Cells from each group were subsequently isolated. Data was normalized and scaled using SCTransform, and clustered and visualized on a UMAP plot, with 19, 17, 16, 12, 16, 14 and 12 PCs for each group, respectively. A resolution of 1 was used, except for the monoaminergic (4) and other (2) groups, with the aim of slightly over-clustering. Lastly, for each group a phylogenetic tree of all clusters was constructed in PCA space using Seurat’s BuildClusterTree function. Clusters on neighboring branches with less than 10 protein-coding genes differently expressed between them (Wilcoxon signed-rank test, adjusted p<0.05) were fused.
Annotation
Specific clusters were annotated based on the expression of the following known marker genes. Olfactory projections neurons (cholinergic): acj6, ct, Lim1,140, 141, 142 ellipsoid body (EB) large-field ring neurons (GABAergic): cv-c, Dh31, Octbeta2R, 5-HT7,143, 144, 145, 146 EB small-field ring neurons (GABAergic): cv-c, Dh31, Octbeta2R, not 5-HT7, ventral and dorsal fan-shaped body (GABAergic): cv-c, Dh31, sNPF, not Octbeta2R,147 medial fan-shaped body: cv-c, Dh31, not sNPF, not Octbeta2R, a/b Kenyon Cells: sNPF, Eip93F,33,148 g Kenyon Cells: sNPF, ab,33 a′/b′ Kenyon Cells: CG8641,33 dopaminergic neurons (monoaminergic): ple, DAT,149,150 serotonergic neurons (monoaminergic): SerT, Trh,151,152 octopaminergic neurons (monoaminergic): Tdc2, Tbh,153,154 tyraminergic neurons (monoaminergic): Tdc2, not Tbh, astrocytes: AANAT1, alrm, Gat, e,155, 156, 157 surface glia: Tret1-1, Mdr65,32,158 ensheathing glia: zyd, trol,35,101 cortex glia: zyd, wrapper.35 Thresholds for each gene were set manually. Top markers for each cluster were calculated (Data S1).
Differential expression (DE)
To account for zero-inflation triggered by dropout events and enable the use of DE tools initially created for analyzing bulk RNA-sequencing data, we generated cell-specific weights, using ZINB-WaVE.46,47 Both DESeq248 and edgeR49,127 were then used to calculate differential expression between pairs of conditions. Genes with |log2(FC)|>1 and adjusted p-value<0.05 calculated with either method were considered to be differentially expressed.
Pathway analysis
Gene Ontology Biological process over-representation test was performed for the differentially expressed genes shown in Figure 2E, using the Bioconductor package ClusterProfiler.128 Significantly enriched GO terms based on p-values were then visualized as a chord diagram with GOplot.129
RT-qPCR
Quantitative PCR experiments were performed as described.33 In brief, total RNA was extracted from groups of 40 fly heads using the RNeasy Mini kit (Qiagen 74104). mRNA was then reverse-transcribed using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen, 18080400) according to manufacturer’s instructions. qPCR was performed in a Light- Cycler 480 Instrument II (Roche, 05015243001) using the Universal Probe Library system (UPL; Roche, 04683633001 and 04869877001). Each 10 mL reaction contained 2.4 mL of pre-amplified cDNA, 0.4 mM of each primer (designed with Roche Assay Design Center), 0.2 mM of UPL probe, and 5 mL LightCycler 480 Probes Master (Roche, 4887301001). Cycles were as follows: 95°C, 10′; 45x [95°C, 10′; 60°C, 30′; Fluorescence acquisition; 72°C, 1′].
D-serine Feeding
For D-serine feeding 2.9 g/L or 27.6 mM of D-serine (S0033 Tokyo Chemical Industry) was mixed with 2% agarose and 5% sucrose. We used the same concentration of L-serine (S0035 Tokyo Chemical Industry). For control treated groups we used 2% agar and 5% sucrose.
CAFE Assay
CAFE assay was performed as described57 but adjusted for water consumption. Groups of 12 male flies were first dehydrated for 1 h in vials containing Drierite without indicator (ACROS Organics). Flies were then transferred to CAFE vials containing four glass capillaries loaded with 5 μL of deionized water and ∼0.1 μL of mineral oil. Vials were loaded into a tray covered with plastic wrap to reduce water loss due to evaporation. We also used 3 empty CAFE vials to assess the level of evaporation to subtract from the amount of water consumed by the flies. Flies were kept in either 20°C or 30°C incubators. Food consumption CAFE used 5% yeast extract and 5% sucrose solution and 10 male flies per vial measuring consumption across 24 h. Vials also contained 10 mL of 1% agarose. We kept track of the number of deaths for each vial and re-adjusted consumption values based on the number of flies alive by the conclusion of the assay. Water and food consumption in μL is recorded as total water consumption of 12 flies minus the average evaporation volume from the empty vials.
D-serine preference CAFE involved storing groups of 8 male flies per vial at 30°C for 18–20 h prior to the start of the assay. We then transferred the flies into CAFE preference vials containing two capillary tubes loaded with normal 5% yeast extract and 5% sucrose and two capillary tubes loaded with 2.9 g/L (27.6 mM) of D-serine (S0033 Tokyo Chemical Industry) with 5% yeast extract and 5% sucrose. Vials also contained 10 mL of 1% agarose. Flies were then given 24 h to feed then were removed. D-serine Preference Index was calculated as the volume of D-serine food minus volume of normal food divided by the total volume of food consumed.
Manual Feeding Assay
The water feeding assay was performed as described19 with minor adjustments. Flies were briefly anesthetized on ice and fixed to a glass slide using beeswax. The slide was then transferred to an empty pipette tip box containing Drierite without indicator (ACROS Organics), closed, then wrapped with parafilm at the seam. For experiments performed at 30°C we put the flies in an incubator for the 2 h dehydration step then performed the experiment in a temperature (30°C) and humidity-controlled (60-65%) booth. The flies were hand fed with a syringe containing deionized water. If the experiments were performed at 30°C the water was warmed to minimize temperature changes in the fly during feeding. We presented the water 10 times to each fly and measured cumulative time spent drinking the water. Presentations were made such that we briefly touched the fly’s legs with the water. Flies were excluded if they exhibited little to no movement when touched on their abdomen with the needle. For NMDAR single-site mutant flies we used a 2.5 h dehydration step instead of 2 h as the background Canton-S strain from the Atkinson lab (UT, Austin) showed greater dehydration resistance than Canton-S strain of the Waddell lab.
Hygrosensation/Water memory in the T-maze
We used 3-5 day old flies to assess hygrosensation in the water T-maze, as described.39 Briefly, 24 h prior to testing we group housed ∼100 flies per vial on standard cornmeal/agar food and a 20x60 mm piece of filter paper. If flies were water deprived, they were housed in vials containing desiccant for 2 h before testing. Testing was performed in a temperature (30°C) and humidity-controlled (60-65%) booth. Flies were loaded into a T-maze and given a 2 min choice between an arm containing wet filter paper or dry filter paper. Preference Index was calculated as the number of flies in the wet arm minus the number in the dry arm, divided by the total number of flies.
For water memory assays we used 3-5 day old flies. Prior to training, flies were water deprived for 16-17 h in vials containing dessicant. Water-deprived flies were trained to associate odor with water reward159 then were immediately transferred to vials containing 1% agarose (as a water source) supplemented with nothing, D-serine, or L-serine, 29 g/L (276mM). They were then housed for an additional 23 h in vials containing dry sucrose, dry sucrose and D-serine, or dry sucrose and L-serine, before being tested in the T-maze for water memory performance.
Two-Photon Calcium Imaging
Imaging experiments were performed using 3-7 day old flies as described previously.160,161 Briefly, flies were immobilized by cooling on ice and mounted in a custom-built chamber to allow free movement of their legs. For control conditions we used an external saline containing 103 mM NaCl, 3 mM KCl, 5 mM N-Tris, 10 mM trehalose, 10 mM glucose, 7 mM sucrose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2, 4 mM MgCl2, osmolarity 275 mOsm, pH 7.3. The head capsule was opened under room temperature carbogenated (95% O2, 5% CO2) external saline. The mounted fly was placed under the two-photon microscope (Scientifica). We used a Ti-Sapphire laser (Chameleon Ultra II, Coherent) to excite fluorescence using 140 fs pulses with 80 MHz repetition rate at 910 nm. We acquired 256 x 256 pixel images at 5.92 Hz controlled by ScanImage 3.8 software.130
For analysis, the two-photon images were manually segmented using Fiji.131 With a custom Fiji script we measured the average baseline fluorescence (F0) for 14 s prior to each drug treatment or stimulus delivery. F/F0 describes the fluorescence relative to the baseline. For drug delivery treatments we defined the “Pre-” treatment as the average F/F0 value for 14 s prior to the drug delivery and the “Post-” treatment as the average F/F0 in the 25 s from onset of drug delivery to the offset. To account for inter-cell baseline differences in F/F0 we normalized the “Pre-” treatment to equal 0 for each cell. For area under the curve (AUC) calculation we measured the approximated integral of F/F0 during the drug treatment (“During”) as well as after the treatment until the recording ended (“After”). To account for variance between individual cells, we normalized the beginning of the trace starting from the onset of drug delivery to equal 0 for each cell. For odor delivery AUC analysis we measured the F/F0 during the 5s odor presentation.
To image fly brains following desiccation or starvation, we starved and water deprived flies for 8-10 h prior to recordings. All recordings were performed between ZT 2-6 and deprived flies at ZT 18-22. Dehydration was performed as above.
Solutions
For acute drug application, we used a perfusion pump system (Fisher Scientific US 14-284-201) to continuously deliver saline at a rate of ∼0.043 mL/sec. All drugs were applied with in the presence of 1μM tetrodotoxin (TTX) to block voltage-gated sodium channels and propagation of action potentials.
NR1+ neuron and astrocyte recordings
Recordings of NR1+ neurons (nmdar1-KIGAL4>GCaMP7f) were performed using either 4 mM Mg2+ or 0 mM Mg2+ saline. To compensate for the change in osmolarity with Mg2+ free saline, we added 1.5 mM CaCl2 and 2.5 mM NaCl. We used 20 mM N-methyl-D-aspartic acid (Sigma-Aldrich M3262), D-serine (Tokyo Chemical Industry - S0033), L-serine (Tokyo Chemical Industry – S0035), and glycine (Sigma-Adlrich G8898). Drug mixtures were maintained at room temperature prior to application. Before recordings we pre-loaded the pump with 1 mL of saline then 1 mL of drug mixture and allowed the pump to run continuously throughout the recording so there were no discontinuities in flow during recording. We flushed the pump for 5 min with saline between each drug application. When multiple drug treatments were performed the application order was randomized for each animal to account for sensitization/desensitization effects. To block NMDARs, we applied 15 mM ketamine with NMDA, D-serine, and TTX.
Astrocyte recordings (R86E01-GAL4>GCaMP7f) were performed using normal external saline, hyperosmotic 320 mOsm saline,19 and sugar free saline.95 Hyperosmotic saline was made by adjusting the total amount of added solute in our original saline recipe, while maintaining the same ratios, so that the osmolarity was increased to 320 mOsm. We applied 10 mM acetylcholine (Sigma-Aldrich A6625), 10 mM ATP (Sigma-Aldrich A2383), 10 mM glutamate (Sigma-Aldrich G0355000), or 10 mM adenosine (Sigma-Aldrich A9251) with 1 μM TTX (Sigma-Aldrich T3194). When multiple drug treatments were performed the application order was randomized for each animal.
For MBON-γ5β′2a (M6) recordings (R66C08-GAL4>GCaMP7f), flies were exposed to a constant air stream containing mineral oil solvent (air). Flies were sequentially exposed to 3-octanol (OCT) and 4-methylcyclohexanol (MCH) for 5 s with 30 s inter-odor interval. To measure dendritic responses of MBON-γβ′2a signals were simultaneously acquired from both hemispheres and average responses were analyzed.
For tetanus toxin (TetX) application, we replaced the bath saline with saline containing 1 μM TetX and incubated the flies for 1 h at room temperature. Halfway through the incubation the bath solution was mixed 4X by pipetting.
For α,β-methylene adenosine 5’-diphosphate (AMPCP) (Sigma-Aldrich M3763) application we incubated the flies with 100 μM AMPCP in normal saline for at least 30 min. Flies were then imaged using saline containing AMPCP and ATP.
Identification of astrocytes in EM reconstructions
We used FlyWire (www.flywire.ai)84 to construct the morphology of astrocytes in the Full Adult Female Brain (FAFB) transmission electron microscope (TEM) dataset.85 Astrocytes were identified and distinguished from other glia in the raw EM data by first following cell processes that cross the ensheathing glial boundary, and following them to find their cell body. Astrocytic morphology is characteristically distinct from that of neuronal soma tracts, and ensheathing or cortex glia.
In FlyWire, 3D meshes from automatic A.I. based162 reconstructions of neurons and glia are readily available, but manual expert review is required to add missing, or remove extraneous, branches. Following revision neurons and glial cells had no false continuations, by standard criteria that are previously described.84,163,164 In brief, an expert reviewer (>1000 h experience) first inspected the astrocytes for false continuations, and obvious missing branches. Then a methodical visual inspection of all branches with neuroglancer84 allowed the user to identify and correct further smaller errors. A second expert reviewer then performed the same review to generate a consensus. In this study, 4 astrocytes were submitted for extensive review to two experienced reviewers following every process section by section from the tip inward toward. This review process only resulted in the correction of a few smaller branches with no more than 2 connections each.
Classification of astrocytic processes
Tripartite synapses (TPS) contain presynaptic, postsynaptic, and astrocytic process, with all compartments making direct contact with the synaptic cleft. We found that astrocytic processes sometimes also cover a large part of the perimeter of a presynaptic bouton without contacting the synaptic cleft – a motif we classified as ‘engulfing’. Lastly astrocytic processes were often also found to contact the post-synaptic neuron below the synaptic cleft (within 500 nm of a presynaptic terminal. These contacts to post-synaptic neurons were not included in the TPS analyses.
Analysis of astrocyte-neuronal type relationship
3D meshes of cells in FlyWire, information about synapses with pre and postsynaptic partners,165 and neurotransmitter predictions88 were downloaded with the CAVEclient (github.com/seung-lab/CAVEclient) and further analysed using functionalities from FAFBseg v.1.4 (https://github.com/navis-org/fafbseg-py) and Navis v.0.6 (https://github.com/navis-org/navis) and custom scripts (available upon request). Synapses with CleftScore ≥ 50 and ConnectionScore ≥ 33165 were considered for analyses, after thresholds were manually reviewed in a sample of 500. Synapses were classified as TPS, when an astrocyte was found as a postsynaptic partner to a neuronal presynapse.
To determine the vicinity profile of an astrocyte, it’s mesh was obtained and skeletonised with trimesh v.3.9.32 (https://github.com/mikedh/trimesh) and Skeletor 1.1.132
To exclude somata and main branches we only considered nodes with a radius of < 300 nm. To generate the vicinity profile, synapses within a 2 μm bounding box around the nodes were filtered for the above criteria and classified by neurotransmitter usage. Then the histogram of distances between the nodes and synapses of each neurotransmitter was generated and gaussian kernel density estimates and bootstrap samples of the means of the distance distributions computed with Python base functions for statistical analyses (Code available on request).
3D representations
3D representations and animations of astrocytes were created with Blender 3.0 (Blender Community, 2018) from meshes obtained from Flywire, via CAVEclient and neuroglancer within Flywire.
Quantification and statistical analysis
Single-cell transcriptomics
Statistical methods employed for single-cell analysis are described above. In brief, Wilcoxon signed-rank test was used for marker analysis and cluster annotation. For differential expression, the Wald test implemented in DESeq2 and the binomial generalized log-linear model implemented in edgeR’s glmWeightedF function were used. For these tests, N represents the number of cells in relevant clusters, and significance was defined using an adjusted p-value<0.05 after Benjamini-Hochberg correction. When relevant, details are found in the figure legends.
Behavioral Statistical Analysis
All behavioral data was analyzed using Prism GraphPad software. Data were analyzed for normality using the Shapiro-Wilk test. Depending on normality we used a one-way ANOVA with Dunnett’s multiple comparisons or Kruskal-Wallis ANOVA with Dunn’s multiple comparisons. Details on statistical analyses and the N for each experiment can be found in the figure legends. Significance was defined using an alpha < 0.05.
Two-Photon Calcium Imaging Analysis
For analysis, the two-photon images were manually segmented using Fiji and analyzed using a custom MATLAB script.131 With a custom Fiji script we measured the average baseline fluorescence (F0) for 14 s prior to each drug treatment or stimulus delivery. F/F0 describes the fluorescence relative to the baseline. For drug delivery treatments we defined the “Pre-” treatment as the average F/F0 value for 14 s prior to the drug delivery and the “Post-” treatment as the average F/F0 in the 25 s from onset of drug delivery to the offset. To account for inter-cell baseline differences in F/F0 we normalized the “Pre-” treatment to equal 0 for each cell. For area under the curve (AUC) calculation we measured the approximated integral of F/F0 during the drug treatment (“During”) as well as after the treatment until the recording ended (“After”). To account for variance between individual cells, we normalized the beginning of the trace starting from the onset of drug delivery to equal 0 for each cell. For odor delivery AUC analysis we measured the F/F0 during the 5s odor presentation.
Data were analyzed for normality using the Shapiro-Wilk test. Depending on normality we used a one-way ANOVA with Dunnett’s multiple comparisons or Kruskal-Wallis ANOVA with Dunn’s multiple comparisons. Details on statistical analyses and the N for each experiment can be found in the figure legends. Significance was defined using an alpha < 0.05. For categorical data statistical significance was determined using a chi-square test.
Analysis of astrocyte-neuronal type relationship
3D meshes of cells in FlyWire, information about synapses with pre and postsynaptic partners,165 and neurotransmitter predictions88 were downloaded with the CAVEclient (github.com/seung-lab/CAVEclient) and further analysed using functionalities from FAFBseg v.1.4 (https://github.com/navis-org/fafbseg-py) and Navis v.0.6 (https://github.com/navis-org/navis) and custom scripts (available upon request).
Synapses with CleftScore ≥ 50 and ConnectionScore ≥ 33165 were considered for analyses, after thresholds were manually reviewed in a sample of 500. Synapses were classified as TPS, when an astrocyte was found as a postsynaptic partner to a neuronal presynapse.
To determine the vicinity profile of an astrocyte, it’s mesh was obtained and skeletonised with trimesh v.3.9.32 (https://github.com/mikedh/trimesh) and Skeletor 1.1.132
To exclude somata and main branches we only considered nodes with a radius < 300 nm. To generate the vicinity profile, synapses within a 2 μm bounding box around the nodes were filtered for the above criteria and classified by neurotransmitter usage. Then histograms of distances between the nodes and synapses of each neurotransmitter were generated. For shortest distances between all astrocytes and synapses of specific neurotransmitters probability densities and their functions from gaussian kernel density estimates were created for data representation in Figure 5J (matplotlib.pyplot.hist(density = True); scipy.stats.gaussian_kde(), with bandwidth method = Scott’s rule (default) and equal weighting). For testing of statistical significance the distributions of means from 10000 samples of the shortest distances between all astrocytes and synapses using specific neurotransmitters obtained via bootstrapping (numpy.random.choice() for the all 3 astrocytes and all synapses) (shown in Figure S5N) were tested by a two sided Welch’s t-test assuming non equal variance (scipy.stats.ttest_ind(equal_var = False)) significance was determined when p < 0.05 (Code available on request).
Acknowledgments
We thank Bernard Moussian, Yi Rao, Stefanie Schirmeir, the Vienna Drosophila Resource Center, and Bloomington Drosophila Stock Center for flies. We are grateful to Hubert Slawinski at the Wellcome Trust Centre for Human Genetics for running single-cell transcriptomics and sequencing; Lucy Garner, Paul Brodersen, and the Bioconductor community for assistance and advice with single-cell transcriptomics analysis; and Ruth Brain for lab assistance. We thank the Princeton FlyWire team and Murthy and Seung labs for development and maintenance of FlyWire (supported by BRAIN Initiative grant MH117815), and the Princeton EM proofreading team for contributing neuronal edits. We acknowledge Philipp Schlegel for help implementing navis-related code and Joseph Hsu for proof reading astrocyte morphology. We thank members of Waddell, Sims, and Goodwin groups for discussion. A.P., V.C., C.D.T., and S.W. were supported by an ERC Advanced Grant to S.W. (789274). V.C. was additionally supported by starter funds from Durham University; N.O. was funded by Wellcome Collaborative awards to S.W. (203261/Z/16/Z and 221300/Z/20/Z). D.A. was supported by a Wellcome Collaborative Award to D.S. and S.W. (209235/Z/17/Z). E.M. was funded by an EMBO Long-Term Fellowship (ALTF 184-2019). S.W. is also supported by a Wellcome Principal Research Fellowship in Basic Biomedical Sciences (200846/Z/16/Z).
Author contributions
Designed research, A.P., V.C., N.O., D.A., C.D.T., E.M., and S.W.; performed research, A.P., V.C., N.O., D.A., C.D.T., and E.M.; analyzed data, A.P., V.C., N.O., D.A., C.D.T., D.S., and S.W.; resources, D.S., S.W., and V.C.; writing, S.W., A.P., V.C., and N.O.; supervision, D.S. and S.W.; funding acquisition, S.W. and D.S.
Declaration of interests
S.W. is a member of the advisory board of Current Biology.
Published: August 12, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.07.038.
Contributor Information
Vincent Croset, Email: vincent.croset@durham.ac.uk.
Scott Waddell, Email: scott.waddell@cncb.ox.ac.uk.
Supplemental Information
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Detailed code for scSeq analyses is available at https://github.com/sims-lab/FlyThirst. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All data reported in this paper will be shared by the lead contact upon request.
References
- 1.Bourque C.W. Central mechanisms of osmosensation and systemic osmoregulation. Nat. Rev. Neurosci. 2008;9:519–531. doi: 10.1038/nrn2400. [DOI] [PubMed] [Google Scholar]
- 2.Fitzsimons J.T. Angiotensin, thirst, and sodium appetite. Physiol. Rev. 1998;78:583–686. doi: 10.1152/physrev.1998.78.3.583. [DOI] [PubMed] [Google Scholar]
- 3.Pool A.-H., Wang T., Stafford D.A., Chance R.K., Lee S., Ngai J., Oka Y. The cellular basis of distinct thirst modalities. Nature. 2020;588:112–117. doi: 10.1038/s41586-020-2821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augustine V., Lee S., Oka Y. Neural control and modulation of thirst, sodium appetite, and hunger. Cell. 2020;180:25–32. doi: 10.1016/j.cell.2019.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson A.K., Thunhorst R.L. The neuroendocrinology of thirst and salt appetite: visceral sensory signals and mechanisms of central integration. Front. Neuroendocrinol. 1997;18:292–353. doi: 10.1006/frne.1997.0153. [DOI] [PubMed] [Google Scholar]
- 6.Rolls B.J. The effect of intravenous infusion of antidiuretic hormone on water intake in the rat. J. Physiol. 1971;219:331–339. doi: 10.1113/jphysiol.1971.sp009664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman C.A., Leib D.E., Knight Z.A. Neural circuits underlying thirst and fluid homeostasis. Nat. Rev. Neurosci. 2017;18:459–469. doi: 10.1038/nrn.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott S.B.G., Machado N.L.S., Geerling J.C., Saper C.B. Reciprocal control of drinking behavior by median preoptic neurons in mice. J. Neurosci. 2016;36:8228–8237. doi: 10.1523/JNEUROSCI.1244-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson B., McCann S.M. Drinking, antidiuresis and milk ejection from electrical stimulation within the hypothalamus of the goat. Acta Physiol. Scand. 1955;35:191–201. doi: 10.1111/j.1748-1716.1955.tb01277.x. [DOI] [PubMed] [Google Scholar]
- 10.Augustine V., Gokce S.K., Lee S., Wang B., Davidson T.J., Reimann F., Gribble F., Deisseroth K., Lois C., Oka Y. Hierarchical neural architecture underlying thirst regulation. Nature. 2018;555:204–209. doi: 10.1038/nature25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betley J.N., Xu S., Cao Z.F.H., Gong R., Magnus C.J., Yu Y., Sternson S.M. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature. 2015;521:180–185. doi: 10.1038/nature14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Lin Y.-C., Zimmerman C.A., Essner R.A., Knight Z.A. Hunger neurons drive feeding through a sustained, positive reinforcement signal. eLife. 2016;5 doi: 10.7554/eLife.18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gizowski C., Zaelzer C., Bourque C.W. Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature. 2016;537:685–688. doi: 10.1038/nature19756. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda T., Hiyama T.Y., Niimura F., Matsusaka T., Fukamizu A., Kobayashi K., Kobayashi K., Noda M. Distinct neural mechanisms for the control of thirst and salt appetite in the subfornical organ. Nat. Neurosci. 2017;20:230–241. doi: 10.1038/nn.4463. [DOI] [PubMed] [Google Scholar]
- 15.Oka Y., Ye M., Zuker C.S. Thirst driving and suppressing signals encoded by distinct neural populations in the brain. Nature. 2015;520:349–352. doi: 10.1038/nature14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmerman C.A., Lin Y.-C., Leib D.E., Guo L., Huey E.L., Daly G.E., Chen Y., Knight Z.A. Thirst neurons anticipate the homeostatic consequences of eating and drinking. Nature. 2016;537:680–684. doi: 10.1038/nature18950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerman C.A., Huey E.L., Ahn J.S., Beutler L.R., Tan C.L., Kosar S., Bai L., Chen Y., Corpuz T.V., Madisen L., et al. A gut-to-brain signal of fluid osmolarity controls thirst satiation. Nature. 2019;568:98–102. doi: 10.1038/s41586-019-1066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gáliková M., Dircksen H., Nässel D.R. The thirsty fly: ion transport peptide (ITP) is a novel endocrine regulator of water homeostasis in Drosophila. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jourjine N., Mullaney B.C., Mann K., Scott K. Coupled sensing of hunger and thirst signals balances sugar and water consumption. Cell. 2016;166:855–866. doi: 10.1016/j.cell.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landayan D., Wang B.P., Zhou J., Wolf F.W. Thirst interneurons that promote water seeking and limit feeding behavior in Drosophila. eLife. 2021;10 doi: 10.7554/eLife.66286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S., Owald D., Chandra V., Talbot C., Huetteroth W., Waddell S. Neural correlates of water reward in thirsty Drosophila. Nat. Neurosci. 2014;17:1536–1542. doi: 10.1038/nn.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senapati B., Tsao C.-H., Juan Y.-A., Chiu T.-H., Wu C.-L., Waddell S., Lin S. A neural mechanism for deprivation state-specific expression of relevant memories in Drosophila. Nat. Neurosci. 2019;22:2029–2039. doi: 10.1038/s41593-019-0515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persidsky Y., Ramirez S.H., Haorah J., Kanmogne G.D. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J. Neuroimmune Pharmacol. 2006;1:223–236. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 24.Fischer R., Schliess F., Häussinger D. Characterization of the hypo-osmolarity-induced Ca2+ response in cultured rat astrocytes. Glia. 1997;20:51–58. [PubMed] [Google Scholar]
- 25.Gourine A.V., Kasparov S. Astrocytes as brain interoceptors. Exp. Physiol. 2011;96:411–416. doi: 10.1113/expphysiol.2010.053165. [DOI] [PubMed] [Google Scholar]
- 26.Limmer S., Weiler A., Volkenhoff A., Babatz F., Klämbt C. The Drosophila blood-brain barrier: development and function of a glial endothelium. Front. Neurosci. 2014;8:365. doi: 10.3389/fnins.2014.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronaldson P.T., Davis T.P. Regulation of blood-brain barrier integrity by microglia in health and disease: A therapeutic opportunity. J. Cereb. Blood Flow Metab. 2020;40:S6–S24. doi: 10.1177/0271678X20951995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simard M., Nedergaard M. The neurobiology of glia in the context of water and ion homeostasis. Neuroscience. 2004;129:877–896. doi: 10.1016/j.neuroscience.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 29.Teschemacher A.G., Gourine A.V., Kasparov S. A role for astrocytes in sensing the brain microenvironment and neuro-metabolic integration. Neurochem. Res. 2015;40:2386–2393. doi: 10.1007/s11064-015-1562-9. [DOI] [PubMed] [Google Scholar]
- 30.de Tredern E., Rabah Y., Pasquer L., Minatchy J., Plaçais P.-Y., Preat T. Glial glucose fuels the neuronal pentose phosphate pathway for long-term memory. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stork T., Sheehan A., Tasdemir-Yilmaz O.E., Freeman M.R. Neuron-glia interactions through the heartless FGF receptor signaling pathway mediate morphogenesis of drosophila astrocytes. Neuron. 2014;83:388–403. doi: 10.1016/j.neuron.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkenhoff A., Weiler A., Letzel M., Stehling M., Klämbt C., Schirmeier S. Glial glycolysis is essential for neuronal survival in drosophila. Cell Metab. 2015;22:437–447. doi: 10.1016/j.cmet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Croset V., Treiber C.D., Waddell S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife. 2018;7 doi: 10.7554/eLife.34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft Ł., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C., et al. A single-cell transcriptome atlas of the aging drosophila brain. Cell. 2018;174:982–998.e20. doi: 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C. Phenotypic convergence: distinct transcription factors regulate common terminal features. Cell. 2018;174:622–635.e13. doi: 10.1016/j.cell.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Özel M.N., Simon F., Jafari S., Holguera I., Chen Y.-C., Benhra N., El-Danaf R.N., Kapuralin K., Malin J.A., Konstantinides N., et al. Neuronal diversity and convergence in a visual system developmental atlas. Nature. 2021;589:88–95. doi: 10.1038/s41586-020-2879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssens J., Aibar S., Taskiran I.I., Ismail J.N., Gomez A.E., Aughey G., Spanier K.I., De Rop F.V., González-Blas C.B., Dionne M., et al. Decoding gene regulation in the fly brain. Nature. 2022;601:630–636. doi: 10.1038/s41586-021-04262-z. [DOI] [PubMed] [Google Scholar]
- 38.Liu L., Li Y., Wang R., Yin C., Dong Q., Hing H., Kim C., Welsh M.J. Drosophila hygrosensation requires the TRP channels water witch and nanchung. Nature. 2007;450:294–298. doi: 10.1038/nature06223. [DOI] [PubMed] [Google Scholar]
- 39.Sayeed O., Benzer S. Behavioral genetics of thermosensation and hygrosensation in Drosophila. Proc. Natl. Acad. Sci. USA. 1996;93:6079–6084. doi: 10.1073/pnas.93.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnstedt O., Owald D., Felsenberg J., Brain R., Moszynski J.-P., Talbot C.B., Perrat P.N., Waddell S. Memory-relevant mushroom body output synapses are cholinergic. Neuron. 2016;89:1237–1247. doi: 10.1016/j.neuron.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunet Avalos C., Maier G.L., Bruggmann R., Sprecher S.G. Single cell transcriptome atlas of the Drosophila larval brain. eLife. 2019;8 doi: 10.7554/eLife.50354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mou T., Deng W., Gu F., Pawitan Y., Vu T.N. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front. Genet. 2019;10:1331. doi: 10.3389/fgene.2019.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 46.Risso D., Perraudeau F., Gribkova S., Dudoit S., Vert J.-P. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat. Commun. 2018;9:284. doi: 10.1038/s41467-017-02554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van den Berge K., Perraudeau F., Soneson C., Love M.I., Risso D., Vert J.P., Robinson M.D., Dudoit S., Clement L. Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol. 2018;19:24. doi: 10.1186/s13059-018-1406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakhotia S.C. Forty years of the 93D puff of Drosophila melanogaster. J. Biosci. 2011;36:399–423. doi: 10.1007/s12038-011-9078-1. [DOI] [PubMed] [Google Scholar]
- 51.Qin W., Neal S.J., Robertson R.M., Westwood J.T., Walker V.K. Cold hardening and transcriptional change in Drosophila melanogaster. Insect Mol. Biol. 2005;14:607–613. doi: 10.1111/j.1365-2583.2005.00589.x. [DOI] [PubMed] [Google Scholar]
- 52.Ray M., Acharya S., Shambhavi S., Lakhotia S.C. Over-expression of Hsp83 in grossly depleted hsrω lncRNA background causes synthetic lethality and l(2)gl phenocopy in Drosophila. J. Biosci. 2019;44:36. [PubMed] [Google Scholar]
- 53.Liu Y., Luo J., Carlsson M.A., Nässel D.R. Serotonin and insulin-like peptides modulate leucokinin-producing neurons that affect feeding and water homeostasis in Drosophila. J. Comp. Neurol. 2015;523:1840–1863. doi: 10.1002/cne.23768. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S., Chen D., Jang C., Nall A., Zheng X., Sehgal A. An ecdysone-responsive nuclear receptor regulates circadian rhythms in Drosophila. Nat. Commun. 2014;5:5697. doi: 10.1038/ncomms6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ja W.W., Carvalho G.B., Mak E.M., Rosa N.N. de la, Fang A.Y., Liong J.C., Brummel T., Benzer S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGuire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 57.Park A., Tran T., Atkinson N.S. Monitoring food preference in Drosophila by oligonucleotide tagging. Proc. Natl. Acad. Sci. U. S. A. 2018;115:9020–9025. doi: 10.1073/pnas.1716880115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kremer M.C., Jung C., Batelli S., Rubin G.M., Gaul U. The glia of the adult Drosophila nervous system. Glia. 2017;65:606–638. doi: 10.1002/glia.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prokopenko S.N., He Y., Lu Y., Bellen H.J. Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics. 2000;156:1691–1715. doi: 10.1093/genetics/156.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzberg A., D’Evelyn D., Schulze K.L., Lee J.K., Strumpf D., Tsai L., Bellen H.J. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 61.Wood P.L., Hawkinson J.E., Goodnough D.B. Formation of D-serine from L-phosphoserine in brain synaptosomes. J. Neurochem. 1996;67:1485–1490. doi: 10.1046/j.1471-4159.1996.67041485.x. [DOI] [PubMed] [Google Scholar]
- 62.Kang N., Peng H., Yu Y., Stanton P.K., Guilarte T.R., Kang J. Astrocytes release D-serine by a large vesicle. Neuroscience. 2013;240:243–257. doi: 10.1016/j.neuroscience.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martineau M., Galli T., Baux G., Mothet J.-P. Confocal imaging and tracking of the exocytotic routes for D-serine-mediated gliotransmission. Glia. 2008;56:1271–1284. doi: 10.1002/glia.20696. [DOI] [PubMed] [Google Scholar]
- 64.Mothet J.-P., Pollegioni L., Ouanounou G., Martineau M., Fossier P., Baux G. Glutamate receptor activation triggers a calcium-dependent and SNARE protein-dependent release of the gliotransmitter D-serine. Proc. Natl. Acad. Sci. USA. 2005;102:5606–5611. doi: 10.1073/pnas.0408483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mustafa A.K., Kim P.M., Snyder S.H. D-serine as a putative glial neurotransmitter. Neuron Glia Biol. 2004;1:275–281. doi: 10.1017/S1740925X05000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schell M.J., Brady R.O., Molliver M.E., Snyder S.H. D-serine as a neuromodulator: regional and developmental localizations in rat brain glia resemble NMDA receptors. J. Neurosci. 1997;17:1604–1615. doi: 10.1523/JNEUROSCI.17-05-01604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y.V., Ormerod K.G., Littleton J.T. Astrocyte Ca2+ influx negatively regulates neuronal activity. eNeuro. 2017;4 doi: 10.1523/ENEURO.0340-16.2017. 0340–16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleckner N.W., Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 1988;241:835–837. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- 69.Mothet J.-P., Parent A.T., Wolosker H., Brady R.O., Linden D.J., Ferris C.D., Rogawski M.A., Snyder S.H. d-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc. Natl. Acad. Sci. USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai X., Zhou E., Yang W., Zhang X., Zhang W., Rao Y. D-serine made by serine racemase in Drosophila intestine plays a physiological role in sleep. Nat. Commun. 2019;10:1986. doi: 10.1038/s41467-019-09544-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pollegioni L., Piubelli L., Sacchi S., Pilone M.S., Molla G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cell. Mol. Life Sci. 2007;64:1373–1394. doi: 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laube B., Hirai H., Sturgess M., Betz H., Kuhse J. Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron. 1997;18:493–503. doi: 10.1016/s0896-6273(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 73.Miller R.F. D-serine as a glial modulator of nerve cells. Glia. 2004;47:275–283. doi: 10.1002/glia.20073. [DOI] [PubMed] [Google Scholar]
- 74.Troutwine B., Park A., Velez-Hernandez M.E., Lew L., Mihic S.J., Atkinson N.S. F654A and K558Q mutations in NMDA receptor 1 affect ethanol-induced behaviors in drosophila. Alcohol. Clin. Exp. Res. 2019;43:2480–2493. doi: 10.1111/acer.14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoneda Y., Suzuki T., Ogita K., Han D. Support for radiolabeling of a glycine recognition domain on the N-methyl-D-aspartate receptor ionophore complex by 5,7-[3H]dichlorokynurenate in rat brain. J. Neurochem. 1993;60:634–645. doi: 10.1111/j.1471-4159.1993.tb03195.x. [DOI] [PubMed] [Google Scholar]
- 76.Dickinson R., Peterson B.K., Banks P., Simillis C., Martin J.C.S., Valenzuela C.A., Maze M., Franks N.P. Competitive inhibition at the glycine site of the N-methyl-D-aspartate receptor by the anesthetics xenon and isoflurane: evidence from molecular modeling and electrophysiology. Anesthesiology. 2007;107:756–767. doi: 10.1097/01.anes.0000287061.77674.71. [DOI] [PubMed] [Google Scholar]
- 77.Wafford K.A., Kathoria M., Bain C.J., Marshall G., Le Bourdellès B., Kemp J.A., Whiting P.J. Identification of amino acids in the N-methyl-D-aspartate receptor NR1 subunit that contribute to the glycine binding site. Mol. Pharmacol. 1995;47:374–380. [PubMed] [Google Scholar]
- 78.Ascher P., Nowak L. Electrophysiological studies of NMDA receptors. Trends Neurosci. 1987;10:284–288. [Google Scholar]
- 79.Mori H., Masaki H., Yamakura T., Mishina M. Identification by mutagenesis of a Mg(2+)-block site of the NMDA receptor channel. Nature. 1992;358:673–675. doi: 10.1038/358673a0. [DOI] [PubMed] [Google Scholar]
- 80.Martin D., Lodge D. Ketamine acts as a non-competitive N-methyl-D-aspartate antagonist on frog spinal cord in vitro. Neuropharmacology. 1985;24:999–1003. doi: 10.1016/0028-3908(85)90128-5. [DOI] [PubMed] [Google Scholar]
- 81.Ventura R., Harris K.M. Three-dimensional relationships between hippocampal synapses and astrocytes. J. Neurosci. 1999;19:6897–6906. doi: 10.1523/JNEUROSCI.19-16-06897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Santello M., Calì C., Bezzi P. Gliotransmission and the tripartite synapse. Adv. Exp. Med. Biol. 2012;970:307–331. doi: 10.1007/978-3-7091-0932-8_14. [DOI] [PubMed] [Google Scholar]
- 83.Danjo R., Kawasaki F., Ordway R.W. A tripartite synapse model in drosophila. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorkenwald S., McKellar C.E., Macrina T., Kemnitz N., Lee K., Lu R., Wu J., Popovych S., Mitchell E., Nehoran B., et al. FlyWire: online community for whole-brain connectomics. Nat. Methods. 2022;19:119–128. doi: 10.1038/s41592-021-01330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng Z., Lauritzen J.S., Perlman E., Robinson C.G., Nichols M., Milkie D., Torrens O., Price J., Fisher C.B., Sharifi N., et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018;174:730–743.e22. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Z., Freeman M.R. TrpML-mediated astrocyte microdomain Ca2+ transients regulate astrocyte–tracheal interactions. eLife. 2020;9 doi: 10.7554/eLife.58952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pereanu W., Spindler S., Cruz L., Hartenstein V. Tracheal development in the Drosophila brain is constrained by glial cells. Dev. Biol. 2007;302:169–180. doi: 10.1016/j.ydbio.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eckstein N., Bates A.S., Du M., Hartenstein V., Jefferis G.S.X.E., Funke J. Neurotransmitter classification from electron microscopy images at synaptic sites in Drosophila. Preprint at bioRxiv. 2020 doi: 10.1101/2020.06.12.148775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Savtchouk I., Volterra A. Gliotransmission: Beyond black-and-white. J. Neurosci. 2018;38:14–25. doi: 10.1523/JNEUROSCI.0017-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mustafa A.K., van Rossum D.B., Patterson R.L., Maag D., Ehmsen J.T., Gazi S.K., Chakraborty A., Barrow R.K., Amzel L.M., Snyder S.H. Glutamatergic regulation of serine racemase via reversal of PIP2 inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:2921–2926. doi: 10.1073/pnas.0813105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guthrie P.B., Knappenberger J., Segal M., Bennett M.V., Charles A.C., Kater S.B. ATP released from astrocytes mediates glial calcium waves. J. Neurosci. 1999;19:520–528. doi: 10.1523/JNEUROSCI.19-02-00520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee D., O’Dowd D.K. Fast excitatory synaptic transmission mediated by nicotinic acetylcholine receptors in Drosophila neurons. J. Neurosci. 1999;19:5311–5321. doi: 10.1523/JNEUROSCI.19-13-05311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamasaki S., Baumeister A., Binz T., Blasi J., Link E., Cornille F., Roques B., Fykse E.M., Südhof T.C., Jahn R. Cleavage of members of the synaptobrevin/VAMP family by types D and F botulinal neurotoxins and tetanus toxin. J. Biol. Chem. 1994;269:12764–12772. [PubMed] [Google Scholar]
- 94.MacNamee S.E., Liu K.E., Gerhard S., Tran C.T., Fetter R.D., Cardona A., Tolbert L.P., Oland L.A. Astrocytic glutamate transport regulates a Drosophila CNS synapse that lacks astrocyte ensheathment. J. Comp. Neurol. 2016;524:1979–1998. doi: 10.1002/cne.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheriyamkunnel S.J., Rose S., Jacob P.F., Blackburn L.A., Glasgow S., Moorse J., Winstanley M., Moynihan P.J., Waddell S., Rezaval C. A neuronal mechanism controlling the choice between feeding and sexual behaviors in Drosophila. Curr. Biol. 2021 doi: 10.1016/j.cub.2021.07.029. S0960–9822, 00984–00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu T.M., Bazzino P., Hurh S.J., Konanur V.R., Roitman J.D., Roitman M.F. Thirst recruits phasic dopamine signaling through subfornical organ neurons. Proc. Natl. Acad. Sci. USA. 2020;117:30744–30754. doi: 10.1073/pnas.2009233117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sacchi R., Li J., Villarreal F., Gardell A.M., Kültz D. Salinity-induced regulation of the myo-inositol biosynthesis pathway in tilapia gill epithelium. J. Exp. Biol. 2013;216:4626–4638. doi: 10.1242/jeb.093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen P., Tu X., Akdemir F., Chew S.K., Rothenfluh A., Abrams J.M. Effectors of alcohol-induced cell killing in Drosophila. Cell Death Differ. 2012;19:1655–1663. doi: 10.1038/cdd.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tomita J., Ueno T., Mitsuyoshi M., Kume S., Kume K. The NMDA receptor promotes sleep in the fruit fly, Drosophila melanogaster. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128101. e0128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y.-F., Parpura V. Astroglial modulation of hydromineral balance and cerebral edema. Front. Mol. Neurosci. 2018;11:204. doi: 10.3389/fnmol.2018.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Melom J.E., Littleton J.T. Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J. Neurosci. 2013;33:1169–1178. doi: 10.1523/JNEUROSCI.3920-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Farca Luna A.J., Perier M., Seugnet L. Amyloid precursor protein in Drosophila glia regulates sleep and genes involved in glutamate recycling. J. Neurosci. 2017;37:4289–4300. doi: 10.1523/JNEUROSCI.2826-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shimizu H., Watanabe E., Hiyama T.Y., Nagakura A., Fujikawa A., Okado H., Yanagawa Y., Obata K., Noda M. Glial Nax Channels control lactate signaling to neurons for brain [Na+] sensing. Neuron. 2007;54:59–72. doi: 10.1016/j.neuron.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 104.Luan Z., Quigley C., Li H.-S. The putative Na+/Cl--dependent neurotransmitter/osmolyte transporter inebriated in the Drosophila hindgut is essential for the maintenance of systemic water homeostasis. Sci. Rep. 2015;5:7993. doi: 10.1038/srep07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang Y., Stern M. In vivo properties of the drosophila inebriated-encoded neurotransmitter transporter. J. Neurosci. 2002;22:1698–1708. doi: 10.1523/JNEUROSCI.22-05-01698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pedersen S.F., Kapus A., Hoffmann E.K. Osmosensory mechanisms in cellular and systemic volume regulation. J. Am. Soc. Nephrol. 2011;22:1587–1597. doi: 10.1681/ASN.2010121284. [DOI] [PubMed] [Google Scholar]
- 107.Evans J.M., Allan A.K., Davies S.A., Dow J.A.T. Sulphonylurea sensitivity and enriched expression implicate inward rectifier K+ channels in Drosophila melanogaster renal function. J. Exp. Biol. 2005;208:3771–3783. doi: 10.1242/jeb.01829. [DOI] [PubMed] [Google Scholar]
- 108.Krashes M.J., DasGupta S., Vreede A., White B., Armstrong J.D., Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gutman Y., Krausz M. Regulation of food and water intake in rats as related to plasma osmolarity and volume. Physiol. Behav. 1969;4:311–313. [Google Scholar]
- 110.Jung Y., Kennedy A., Chiu H., Mohammad F., Claridge-Chang A., Anderson D.J. Neurons that function within an integrator to promote a persistent behavioral state in drosophila. Neuron. 2020;105:322–333.e5. doi: 10.1016/j.neuron.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McKinley M.J., Denton D.A., Ryan P.J., Yao S.T., Stefanidis A., Oldfield B.J. From sensory circumventricular organs to cerebral cortex: neural pathways controlling thirst and hunger. J. Neuroendocrinol. 2019;31 doi: 10.1111/jne.12689. [DOI] [PubMed] [Google Scholar]
- 112.Stricker E.M., Hoffmann M.L. Presystemic signals in the control of thirst, salt appetite, and vasopressin secretion. Physiol. Behav. 2007;91:404–412. doi: 10.1016/j.physbeh.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 113.Zhang S.X., Miner L.E., Boutros C.L., Rogulja D., Crickmore M.A. Motivation, perception, and chance converge to make a binary decision. Neuron. 2018;99:376–388.e6. doi: 10.1016/j.neuron.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Allen W.E., Chen M.Z., Pichamoorthy N., Tien R.H., Pachitariu M., Luo L., Deisseroth K. Thirst regulates motivated behavior through modulation of brainwide neural population dynamics. Science. 2019;364:253. doi: 10.1126/science.aav3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hons J., Zirko R., Vasatova M., Doubek P., Klimova B., Masopust J., Valis M., Kuca K. Impairment of executive functions associated with lower D-serine serum levels in patients with schizophrenia. Front. Psychiatry. 2021;12 doi: 10.3389/fpsyt.2021.514579. 514579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J., Matthews B.B., Millburn G., Antonazzo G., Trovisco V., et al. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Burke C.J., Huetteroth W., Owald D., Perisse E., Krashes M.J., Das G., Gohl D., Silies M., Certel S., Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gohl D.M., Silies M.A., Gao X.J., Bhalerao S., Luongo F.J., Lin C.-C., Potter C.J., Clandinin T.R. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y., Norum M., Oehl K., Yang Y., Zuber R., Yang J., Farine J.P., Gehring N., Flötenmeyer M., Ferveur J.F., et al. Dysfunction of Oskyddad causes Harlequin-type ichthyosis-like defects in Drosophila melanogaster. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008363. e1008363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 121.Paradis S., Sweeney S.T., Davis G.W. Homeostatic control of presynaptic release is triggered by postsynaptic membrane depolarization. Neuron. 2001;30:737–749. doi: 10.1016/s0896-6273(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 122.Sweeney S.T., Broadie K., Keane J., Niemann H., O’Kane C.J. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 123.Dana H., Sun Y., Mohar B., Hulse B.K., Kerlin A.M., Hasseman J.P., Tsegaye G., Tsang A., Wong A., Patel R., et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods. 2019;16:649–657. doi: 10.1038/s41592-019-0435-6. [DOI] [PubMed] [Google Scholar]
- 124.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. cels. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walter W., Sánchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 130.Pologruto T.A., Sabatini B.L., Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. OnLine. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Au O.K.-C., Tai C.-L., Chu H.-K., Cohen-Or D., Lee T.-Y. Skeleton extraction by mesh contraction. ACM Trans. Graph. 2008;27:1–10. [Google Scholar]
- 133.Aso Y., Sitaraman D., Ichinose T., Kaun K.R., Vogt K., Belliart-Guérin G., Plaçais P.Y., Robie A.A., Yamagata N., Schnaitmann C., et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3 doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee B.P., Jones B.W. Transcriptional regulation of the Drosophila glial gene repo. Mech. Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 135.Sepp K.J., Auld V.J. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kelley R.L., Kuroda M.I. The Drosophila roX1 RNA gene can overcome silent chromatin by recruiting the male-specific lethal dosage compensation complex. Genetics. 2003;164:565–574. doi: 10.1093/genetics/164.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun B., Salvaterra P.M. Two Drosophila nervous system antigens, Nervana 1 and 2, are homologous to the beta subunit of Na+,K(+)-ATPase. Proc. Natl. Acad. Sci. USA. 1995;92:5396–5400. doi: 10.1073/pnas.92.12.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aggarwal C.C., Hinneburg A., Keim D.A. In: Database Theory — ICDT 2001 Lecture Notes in Computer Science. Van den Bussche J., Vianu V., editors. Springer; 2001. On the surprising behavior of distance metrics in high dimensional space; pp. 420–434. [Google Scholar]
- 139.Bellman R. Princeton University Press; 1961. Adaptive Control Processes: A Guided Tour. [Google Scholar]
- 140.Komiyama T., Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr. Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- 141.Komiyama T., Johnson W.A., Luo L., Jefferis G.S.X.E. From lineage to wiring specificity. POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;112:157–167. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 142.Lai S.-L., Awasaki T., Ito K., Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- 143.Becnel J., Johnson O., Luo J., Nässel D.R., Nichols C.D. The serotonin 5-HT7Dro receptor is expressed in the brain of drosophila, and is essential for normal courtship and mating. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Donlea J.M., Pimentel D., Miesenböck G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kunst M., Hughes M.E., Raccuglia D., Felix M., Li M., Barnett G., Duah J., Nitabach M.N. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Z., Zhao X., He T., Wu X., Lv P., Zhu A.J., Du J. Epigenetic regulator Stuxnet modulates octopamine effect on sleep through a Stuxnet-Polycomb-Octβ2R cascade. EMBO Rep. 2021;22 doi: 10.15252/embr.201947910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nässel D.R., Enell L.E., Santos J.G., Wegener C., Johard H.A. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008;9:90. doi: 10.1186/1471-2202-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Johard H.A.D., Enell L.E., Gustafsson E., Trifilieff P., Veenstra J.A., Nässel D.R. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. J. Comp. Neurol. 2008;507:1479–1496. doi: 10.1002/cne.21636. [DOI] [PubMed] [Google Scholar]
- 149.Neckameyer W.S., Quinn W.G. Isolation and characterization of the gene for drosophila tyrosine hydroxylase. Neuron. 1989;2:1167–1175. doi: 10.1016/0896-6273(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 150.Pörzgen P., Park S.K., Hirsh J., Sonders M.S., Amara S.G. The antidepressant-sensitive dopamine transporter in Drosophila melanogaster: a primordial carrier for catecholamines. Mol. Pharmacol. 2001;59:83–95. doi: 10.1124/mol.59.1.83. [DOI] [PubMed] [Google Scholar]
- 151.Coleman C.M., Neckameyer W.S. Serotonin synthesis by two distinct enzymes in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 2005;59:12–31. doi: 10.1002/arch.20050. [DOI] [PubMed] [Google Scholar]
- 152.Demchyshyn L.L., Pristupa Z.B., Sugamori K.S., Barker E.L., Blakely R.D., Wolfgang W.J., Forte M.A., Niznik H.B. Cloning, expression, and localization of a chloride-facilitated, cocaine-sensitive serotonin transporter from Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1994;91:5158–5162. doi: 10.1073/pnas.91.11.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cole S.H., Carney G.E., McClung C.A., Willard S.S., Taylor B.J., Hirsh J. Two functional but noncomplementing drosophila tyrosine decarboxylase genes distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 2005;280:14948–14955. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- 154.Monastirioti M., Linn C.E., White K. Characterization of Drosophila tyramine beta-hydroxylase gene and isolation of mutant flies lacking octopamine. J. Neurosci. 1996;16:3900–3911. doi: 10.1523/JNEUROSCI.16-12-03900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Davla S., Artiushin G., Li Y., Chitsaz D., Li S., Sehgal A., van Meyel D.J. AANAT1 functions in astrocytes to regulate sleep homeostasis. eLife. 2020;9 doi: 10.7554/eLife.53994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Doherty J., Logan M.A., Taşdemir O.E., Freeman M.R. Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Muthukumar A.K., Stork T., Freeman M.R. Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat. Neurosci. 2014;17:1340–1350. doi: 10.1038/nn.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.DeSalvo M.K., Hindle S.J., Rusan Z.M., Orng S., Eddison M., Halliwill K., Bainton R.J. The Drosophila surface glia transcriptome: evolutionary conserved blood-brain barrier processes. Front. Neurosci. 2014;8:346. doi: 10.3389/fnins.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lin A.C., Bygrave A.M., de Calignon A., Lee T., Miesenböck G. Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat. Neurosci. 2014;17:559–568. doi: 10.1038/nn.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jacob P.F., Waddell S. Spaced training forms complementary long-term memories of opposite valence in drosophila. Neuron. 2020;106:977–991.e4. doi: 10.1016/j.neuron.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Owald D., Felsenberg J., Talbot C.B., Das G., Perisse E., Huetteroth W., Waddell S. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron. 2015;86:417–427. doi: 10.1016/j.neuron.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee K., Zung J., Li P., Jain V., Seung H.S. Superhuman accuracy on the SNEMI3D connectomics challenge. arXiv. 2017 Preprint at. arXiv:1706.00120. [Google Scholar]
- 163.Otto N., Pleijzier M.W., Morgan I.C., Edmondson-Stait A.J., Heinz K.J., Stark I., Dempsey G., Ito M., Kapoor I., Hsu J., et al. Input connectivity reveals additional heterogeneity of dopaminergic reinforcement in drosophila. Curr. Biol. 2020;30:3200–3211.e8. doi: 10.1016/j.cub.2020.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schneider-Mizell C.M., Gerhard S., Longair M., Kazimiers T., Li F., Zwart M.F., Champion A., Midgley F.M., Fetter R.D., Saalfeld S., et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife. 2016;5 doi: 10.7554/eLife.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Buhmann J., Sheridan A., Malin-Mayor C., Schlegel P., Gerhard S., Kazimiers T., Krause R., Nguyen T.M., Heinrich L., Lee W.-C.A., et al. Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nat. Methods. 2021;18:771–774. doi: 10.1038/s41592-021-01183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D representations of an astrocyte (blue) in the SMP and glutamatergic neurons (green) with boutons containing annotated presynapses (red), which consist of synaptic vesicles (red spheres) and pre-synaptic T-bar densities (black). 00:00 to 00:55, When glial cells engulf axons, multiple processes of different sizes contact a substantial area of a presynaptic bouton’s membrane but do not extend into the synapse. 00:56-1:40, Fine astrocyte processes either pass through the synaptic cleft, or terminate within it, to form a tripartite synapse. In both cases the astrocyte processes contact the synaptic cleft alongside the postsynaptic neurons. Many closely spaced boutons of the same presynaptic neuron can be contacted by the same astrocyte, which therefore contributes processes to multiple tripartite synapses. Anatomical features are further highlighted in the video. Cellular meshes were retrieved from FlyWire and animations produced with blender.
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Detailed code for scSeq analyses is available at https://github.com/sims-lab/FlyThirst. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All data reported in this paper will be shared by the lead contact upon request.