Summary
Cells with latent stem ability can contribute to mammalian tissue regeneration after damage. Whether the central nervous system (CNS) harbors such cells remains controversial. Here, we report that DNGR-1 lineage tracing in mice identifies an ependymal cell subset, wherein resides latent regenerative potential. We demonstrate that DNGR-1-lineage-traced ependymal cells arise early in embryogenesis (E11.5) and subsequently spread across the lining of cerebrospinal fluid (CSF)-filled compartments to form a contiguous sheet from the brain to the end of the spinal cord. In the steady state, these DNGR-1-traced cells are quiescent, committed to their ependymal cell fate, and do not contribute to neuronal or glial lineages. However, trans-differentiation can be induced in adult mice by CNS injury or in vitro by culture with suitable factors. Our findings highlight previously unappreciated ependymal cell heterogeneity and identify across the entire CNS an ependymal cell subset wherein resides damage-responsive neural stem cell potential.
Keywords: ependymal cells, neural stem cells, tissue repair, CLEC9A, dendritic cells
Graphical abstract
Highlights
-
•
DNGR-1 is expressed early in mouse embryogenesis in a subset of ventricular progenitors
-
•
DNGR-1 tracing shows that those progenitors give rise to a subset of ependymal cells
-
•
DNGR-1-traced ependymal cells have latent regenerative potential
-
•
DNGR-1-traced ependymal cells can be mobilized by local injury
Frederico et al. show that embryonic DNGR-1 expression marks a population of ventricular progenitors committed to an ependymal cell subset yet endowed with damage-responsive neural stem cell potential in adulthood. Trans-differentiation can by induced in vitro with appropriate factors or in vivo after CNS injury.
Introduction
Tissues such as the intestine or skin undergo constitutive turnover sustained by professional stem cells that proliferate and continuously differentiate into the cell types that make up the organ (Barker et al., 2007; Blanpain and Fuchs, 2006). Other tissues such as the pancreas or lung display lower cell turnover; however, this can be greatly accelerated after injury (Tata et al., 2013; Zhou and Melton, 2018). Injury-triggered tissue renewal programs differ from those in the steady state in that they do not always require a dedicated stem cell pool. This is best exemplified by the liver, which lacks a professional stem cell population yet is capable of marked regeneration following injury, mediated by both hepatocyte replication and the emergence of facultative stem cells (Raven et al., 2017; Tarlow et al., 2014). Such facultative or latent stem cells frequently originate from quiescent or fully differentiated cells, and their contribution to tissue regeneration in response to injury can even overshadow that of professional stem cells in tissues that contain the latter (Clevers and Watt, 2018; Post and Clevers, 2019). Latent stem cells are operationally defined by the ability to replace cells lost upon injury, and, therefore, their identification requires retrospective analysis. In mouse models, this can be achieved using genetic tools such as lineage tracing (a.k.a. fate mapping), which allows one to assess whether a given cell population contributes to other lineages before and after application of tissue stress (Kretzschmar and Watt, 2012).
The adult mammalian central nervous system (CNS) constitutes one of the least regenerative organs, although it contains professional stem cells that sustain neurogenesis throughout life (Doetsch et al., 1999; Gage, 2000; Seri et al., 2001; Spalding et al., 2013). These adult neural stem cell (NSC) populations inhabit two discrete brain niches, the subventricular zone (SVZ) and the dentate gyrus (DG), and generate neurons that integrate neuronal circuits in the olfactory bulb (OB) and the hippocampus, respectively (Obernier and Alvarez-Buylla, 2019). Although these NSC compartments have been reported to reactively upregulate neurogenesis in response to brain insult (Dash et al., 2001; Liu et al., 2017; Parent and Lowenstein, 2002), the newly generated neurons show limited survival and circuit integration (Quadrato et al., 2014). Furthermore, no equivalent stem cell compartment has been identified in the spinal cord. Thus, whether CNS resident professional stem cells significantly contribute to CNS repair remains unclear and the prognosis for CNS damage, including for traumatic spinal cord injury, remains poor. This has provided impetus for assessing whether the CNS possesses facultative stem cells that can be mobilized for tissue repair.
Ependymal cells have been postulated to constitute one such latent stem cell compartment. Ependymal cells form a ciliated epithelial sheet lining cerebrospinal fluid (CSF)-filled compartments in the CNS. Coordinated beating of their cilia promotes CSF flow from the brain ventricles through the central canal of the spinal cord. The motile ciliogenesis program is governed by the transcription factor Forkhead Box protein J1 (FoxJ1) (Yu et al., 2008), and some mouse lineage-tracing studies based on FoxJ1 promoter activity have suggested that ependymal cells or their progeny can trans-differentiate into astrocytes upon spinal cord injury (Barnabé-Heider et al., 2010; Carlén et al., 2009; Llorens-Bobadilla et al., 2020; Meletis et al., 2008; Sabelström et al., 2013). However, this has not been seen in other FoxJ1-lineage-tracing models (Muthusamy et al., 2014, 2018; Ren et al., 2017). Furthermore, FoxJ1 can be expressed by non-ependymal cells, casting uncertainty on the fidelity of FoxJ1-based lineage tracing as a proxy for ependymal origin (Beckervordersandforth et al., 2010; Devaraju et al., 2013; Jacquet et al., 2009). Finally, a distinct lineage-tracing approach based on αSMA expression has failed to reveal facultative stem cell ability in ependymal cells (Shah et al., 2018). Thus, it remains unclear whether ependymal cells possess latent stem cell ability and whether this property is perhaps confined to a sub-population of these seemingly homogeneous cells.
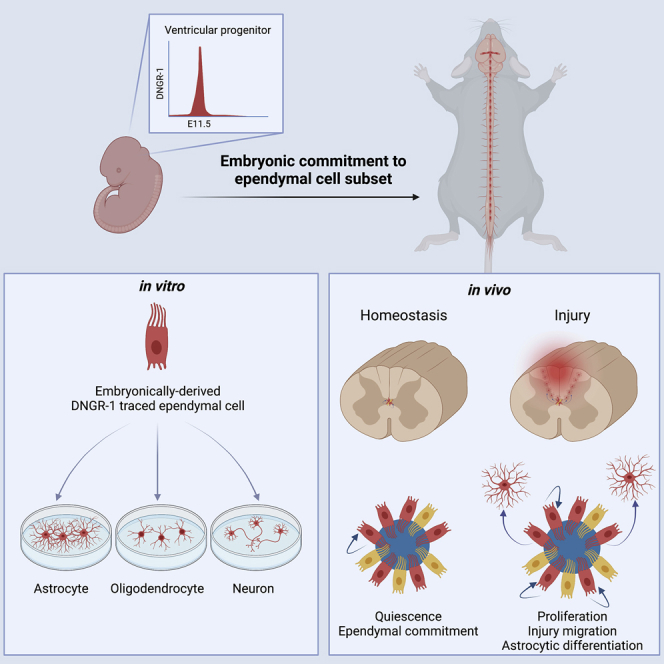
Here, we report that a subset of ependymal cells with latent stem cell potential can be defined in mice by its historical expression of DNGR-1, a receptor hitherto only found in cells of the immune system. DNGR-1-lineage-traced ependymal cells arise very early during embryogenesis to form a contiguous layer that extends from the brain ventricles through the entire spinal cord. Although these cells remain quiescent during homeostasis, they can undergo extensive proliferation and differentiation in response to spinal cord or brain damage. Therapeutic approaches aimed at harnessing their regenerative potential may hold promise in treating CNS injuries.
Results
DNGR-1 lineage tracing defines a non-dendritic cell compartment lining the brain ventricular system and spinal cord central canal
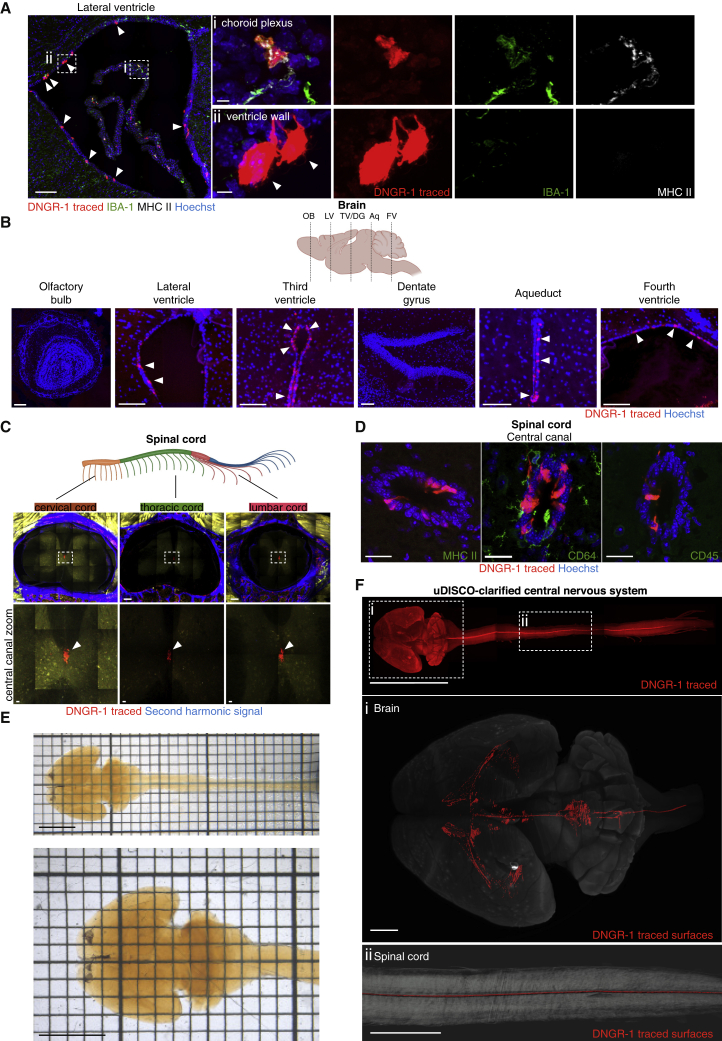
Dendritic cells are an important type of bone marrow-derived myeloid cell involved in immune regulation. DNGR-1 (also known as CLEC9A) is expressed by the conventional dendritic cell (cDC) subtype 1 (cDC1), as well as by cDC-committed progenitors in the bone marrow (Schraml et al., 2013). Taking advantage of the latter finding, we previously generated a DNGR-1 lineage tracer mouse (Clec9aCreRosaLSLtdTomato) in which the history of DNGR-1 expression defines the entire cDC lineage (Schraml et al., 2013). We initially set out to use Clec9aCreRosaLSLtdTomato lineage tracing to identify and study cDCs in the CNS. Immunohistological analysis of brains from adult mice readily revealed the presence of tdTomato+ cells displaying morphology and markers of cDCs (MHC class II [MHC II+] and IBA-1+) (Figure 1Ai). These DNGR-1-traced cDCs could be found in ample numbers in the meninges (data not shown) and choroid plexi (Figure 1Ai), as expected (Anandasabapathy et al., 2011; Quintana et al., 2015). Surprisingly, we also noticed the presence of DNGR-1-traced cells that lacked expression of canonical cDC markers lining the brain ventricles (tdTomato+, MHC II−, and IBA-1−; Figure 1Aii). These cells are henceforth termed non-cDC, DNGR-1-traced cells.
Figure 1.
DNGR-1 lineage tracing marks non-cDCs lining the brain ventricular system and the spinal cord central canal
(A) Brain cryosection from a Clec9aCreRosaLSLtdTomato animal stained with antibodies against IBA-1 (green), MHC II (white); zooms: (Ai) choroid plexus (Aii) ventricle wall. Arrowheads indicate DNGR-1-traced cells (red) in the ventricle wall lacking IBA-1 and MHC II staining.
(B) Brain coronal cuts showing DNGR-1-traced cells (red, arrowheads) in the ventricle walls of all CSF-filled compartments. No DNGR-1-traced cells (red) were observed in olfactory bulbs or dentate gyrus. OB, olfactory bulb; LV, lateral ventricle; TV, third ventricle; DG, dentate gyrus; Aq, aqueduct; FV, fourth ventricle.
(C) Multiphoton image of vibratome spinal cord sections showing DNGR-1-traced cells (red, arrowheads) around the central canal in different rostral-caudal levels. Second harmonic signal, blue.
(D) Spinal cord cryosections labeled with antibodies against MHC II, CD64, or CD45 (all green).
(E and F) Optically clarified (E) and light-sheet imaged (F) whole-CNS with DNGR-1-traced cells (red) lining the entire ventricular compartment. Bottom, zoomed view of indicated areas (Fi) brain (Fii) spinal cord.
Scale bars, 5 mm (E), 1 mm (F), 100 μm (A–D and zoom F), and 20 μm (zooms A and C). Sections from at least 3 animals were analyzed per experiment.
The brain ventricular system comprises four cisternae and extends caudally into the central canal of the spinal cord (Korzh, 2018). Serial brain sectioning demonstrated non-cDC, DNGR-1-traced cells embedded in the lining of all four ventricles (Figure 1B). Multiphoton imaging of different rostral-caudal regions of spinal cord demonstrated that non-cDC, DNGR-1-traced cells also lined the central canal (Figure 1C). As for the cells in brain ventricles, labeled cells in the central canal lacked expression of MHC II and CD64 (cDC and macrophage markers) or the pan-leucocyte marker, CD45 (Figure 1D). To assess if non-cDC, DNGR-1-traced cells formed a contiguous layer, we dissected the entire CNS from a DNGR-1-lineage-traced mouse and subjected it to optical clarification and light-sheet imaging (Figure 1E; Videos S1, S2, and S3). Non-cDC, DNGR-1-traced cells were evenly distributed from the brain ventricles to the sacral portion of the spinal cord in a pattern that corresponded to the boundaries of the CSF-filled network (Figure 1E; Video S3). The cells were also found in a DNGR-1 lineage tracer mouse strain made as a BAC transgenic (Schraml et al., 2013), excluding the possibility that labeling somehow results from disruption of the endogenous Clec9a locus (data not shown). Thus, in addition to marking bona fide cDCs in mice, DNGR-1 lineage tracing reveals a CNS cell population inhabiting the lining of the brain ventricular system and the spinal cord central canal.
Continuum of DNGR-1-traced cells (red) from the sacral part of the spinal cord to the brain ventricular compartment.
Dorsal perspective of the spinal cord counterstained with Hoechst (blue) demonstrating DNGR-1-traced cells (red) lining the central canal.
Brain DNGR-1-traced surfaces (red) lining the ventricular compartment. Surfaces were calculated with Imaris software. Brain contour was defined by tissue autofluorescence and DNGR-1-traced surfaces were defined by bright tdTomato fluorescence.
Non-cDC, DNGR-1-traced cells are unrelated to cDCs
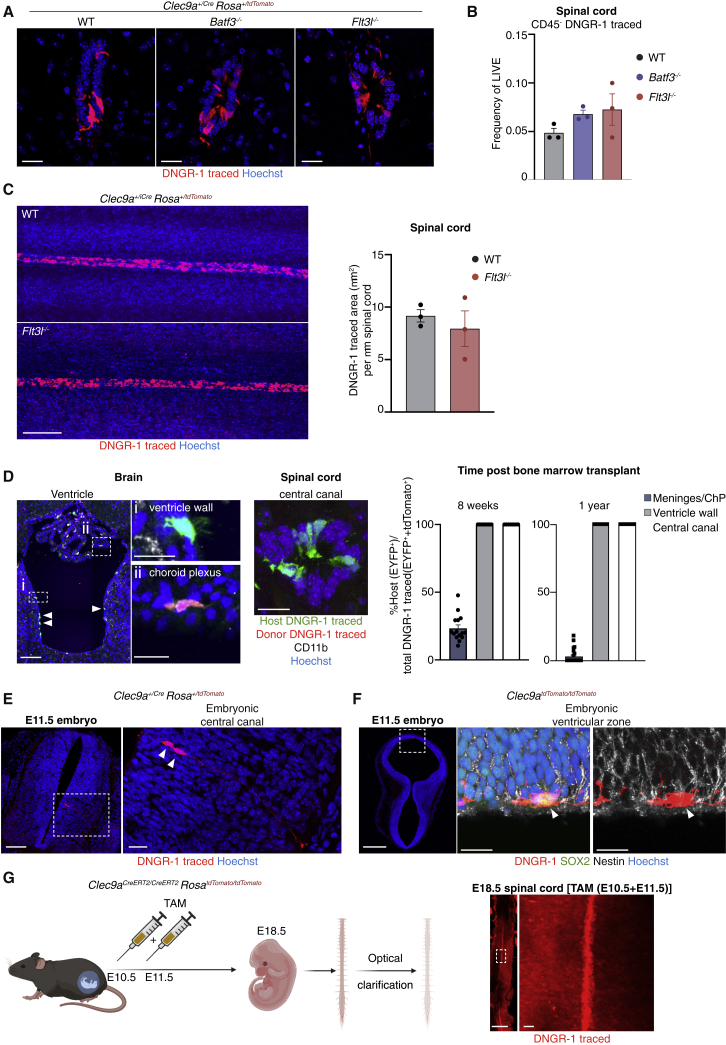
The absence of canonical markers argued that non-cDC, DNGR-1-traced cells were unrelated to their DNGR-1-traced cDC counterparts. To confirm this, we assessed the frequency of both DNGR-1-traced cellular populations in DNGR-1-traced animals crossed to mice deficient in BATF3 (a transcription factor required for the development of cDC1s) (Hildner et al., 2008) or FLT3L (a growth factor essential for development and survival of the entire cDC lineage) (Cabeza-Cabrerizo et al., 2021). As expected, numbers of DNGR-1-traced cDC1s and cDCs were severely reduced in mice lacking Batf3 and Flt3l, respectively (Figure S1A). In contrast, the frequency of non-cDC, DNGR-1-traced cells remained unaltered (Figures 2A and 2B). Light-sheet imaging of wholemount-clarified spinal cords confirmed that the absence of FLT3L did not affect the number, localization, or anatomical distribution of the non-cDC, DNGR-1-traced compartment (Figure 2C; Video S4).
Figure 2.
Non-cDC, DNGR-1-traced cells are embryonically derived
(A) Spinal cord central canal cryosections of Clec9aCreRosaLSLtdTomato animals lacking Batf3 or Flt3l.
(B) Flow cytometric quantification of CD45− DNGR-1-traced cells in spinal cords of Clec9aCreRosaLSLtdTomato animals lacking Batf3 or Flt3l.
(C) Maximum intensity projection of optically cleared spinal cords of Clec9aCreRosaLSLtdTomato animals lacking Flt3l or not (WT). Right, quantification of DNGR-1-traced area (μm2) per μm of spinal cord. Each dot represents one animal.
(D) Brain and spinal cord cryosections of irradiated Clec9aCreRosaLSLEYFP mouse 8 weeks after transplantation with Clec9aCreRosaLSLtdTomato bone marrow labeled with anti-CD11b (white). Host-derived radioresistant DNGR-1-traced cells (EYFP+, green) line the brain ventricle (white arrowheads) and the central canal of the spinal cord. (Di) Multi-ciliated host-derived DNGR-1-traced ependymal cell (green) in the ventricle wall; (Dii) CD11b+ (white) donor-derived hematopoietic DNGR-1-traced cell (red) in the choroid plexus. Right. Quantification of sections across 3–5 animals. ChP, choroid plexus.
(E) Spinal cord cryosection from an E11.5 Clec9aCreRosaLSLtdTomato embryo showing two DNGR-1-traced cells (red, arrowheads). DNGR-1-traced cells were not ciliated at this time point. Right, zoomed view.
(F) Brain cryosection from an E11.5 Clec9atdTomato embryo showing a DNGR-1-expressing cell embedded in the ventricular wall. Right, zoomed view and staining with SOX2 (green), nestin (white), and Hoechst (blue).
(G) In utero induction of lineage tracing. Right, wholemount optically clarified spinal cord from Clec9aCreERT2RosaLSLtdTomato E18.5 embryo after in utero exposure to tamoxifen at E10.5 and E11.5. Zoomed-in area shows DNGR-1 tracing (red) of the ependymal cell layer.
Scale bars, 500 μm (E, spinal cord, F, and G), 100 μm (A, C, D, and zooms E), and 20 μm (zoom D, F, and G). At least 3 animals or were analyzed per experiment. 1 embryo was analyzed per experiment.
Side-by-side comparison of spinal cords from Clec9aCreRosaLSLtdTomato and its cDC deficient counterpart (Clec9aCreRosaLSLtdTomatoFlt3l−/−) showing no reduction of DNGR-1- traced cells (red) lining the central canal of the spinal cord. Nuclei were counterstained with Hoechst (blue).
We also generated bone marrow chimeras in which irradiated DNGR-1 EYFP+ lineage-traced mice were reconstituted with DNGR-1 tdTomato+-lineage-traced bone marrow (Figure S1B). In this experimental system, tdTomato expression reports on bone marrow origin, whereas EYFP fluorescence reports host-derived radioresistant DNGR-1-traced cells. Consistent with hematopoietic origin, the vast majority of DNGR-1-traced cDCs in the meninges and choroid plexus (ChP) was tdTomato+ (Figure 2D). In contrast, 100% of nonDC, DNGR-1-traced cells lining the brain ventricles and the spinal cord central canal were EYFP+ (Figure 2D), reflecting host origin. Analysis more than 1 year post-transplantation (Figure S1B) revealed that meningeal and ChP DNGR-1-traced cDCs had been completely replenished by donor bone marrow-derived cells (tdTomato+), but even at this time point, their non-cDC, DNGR-1-traced counterpart remained 100% EYFP+ (Figures 2D and S1B). Thus, non-cDC, DNGR-1-traced cells do not originate from bone marrow-derived hematopoietic precursors.
Non-cDC, DNGR-1-traced cells emerge from DNGR-1-expressing ventricular progenitors at E11.5
Further underscoring their non-hematopoietic origin, the first non-cDC, DNGR-1-traced cells emerged at embryonic day 11.5 (E11.5), a developmental stage that precedes the advent of definitive hematopoiesis (Golub and Cumano, 2013). Analysis of optically clarified E11.5 DNGR-1-traced embryos (Figure S1C) complemented by immunohistological analysis (Figure 2E) demonstrated that some of these cells were already integrated in the walls of the brain ventricles and in the central canal of the spinal cord. As this developmental time point marked the emergence of DNGR-1-traced cells, it suggested that it also marked the inception of DNGR-1 lineage tracing by DNGR-1-expressing cells. To assess this, we generated a DNGR-1 reporter mouse by introducing tdTomato into the Clec9a locus (Clec9atdTomato). We first validated that Clec9a-driven tdTomato is expressed in DNGR-1-positive cells by examining the cDC lineage (cDC1s, which are DNGR-1+, were nearly 100% tdTomato+, whereas cDC2, which do not express DNGR-1, were <1% labeled) (Figure S1D). Analysis of E11.5 DNGR-1 reporter embryos showed rare DNGR-1+ cells embedded in the ventricular zone, which displayed concurrent expression of the progenitor marker SOX2 but not the neuroepithelial marker nestin (Figure 2F). In contrast, no DNGR-1+ cells were ever found in the ependymal layer of adult Clec9atdTomato reporter mice (Figure S1E), indicating that expression is confined to embryonic stages. To formally test whether the rare DNGR-1-expressing cell population at E11.5 could serve as the embryonic progenitors of non-cDC, DNGR-1-traced cells, we further developed a tamoxifen-inducible DNGR-1-lineage-tracing model (Clec9aCreERT2RosaLSLtdTomato). Validation of this transgenic strain was again carried out by monitoring labeling of the cDC lineage upon tamoxifen administration to adult animals, which led to the expected robust labeling of the cDC1 compartment and lower cDC2 marking (cDC2 lack DNGR-1 expression but arise from DNGR-1-positive progenitors [Schraml et al., 2013; Figure S1F]). In utero induction of DNGR-1 lineage tracing 1 day prior to and at the time when the first DNGR-1-expressing cells are detected (E10.5 + E11.5) and analysis of wholemount optically clarified spinal cords 1 week later (E18.5) recapitulated the spinal cord-labeling pattern observed in constitutive DNGR-1-lineage-tracing adult mice (Figure 2G). In contrast, such labeling was never seen in Clec9aCreERT2RosaLSLtdTomato mice treated with tamoxifen after birth (Figure S1G). Thus, transient DNGR-1 expression marks a population of ventricular embryonic progenitors fated to originate the non-cDC, DNGR-1-traced compartment found in the adult CNS.
The non-cDC, DNGR-1-traced compartment corresponds to ependymal cells
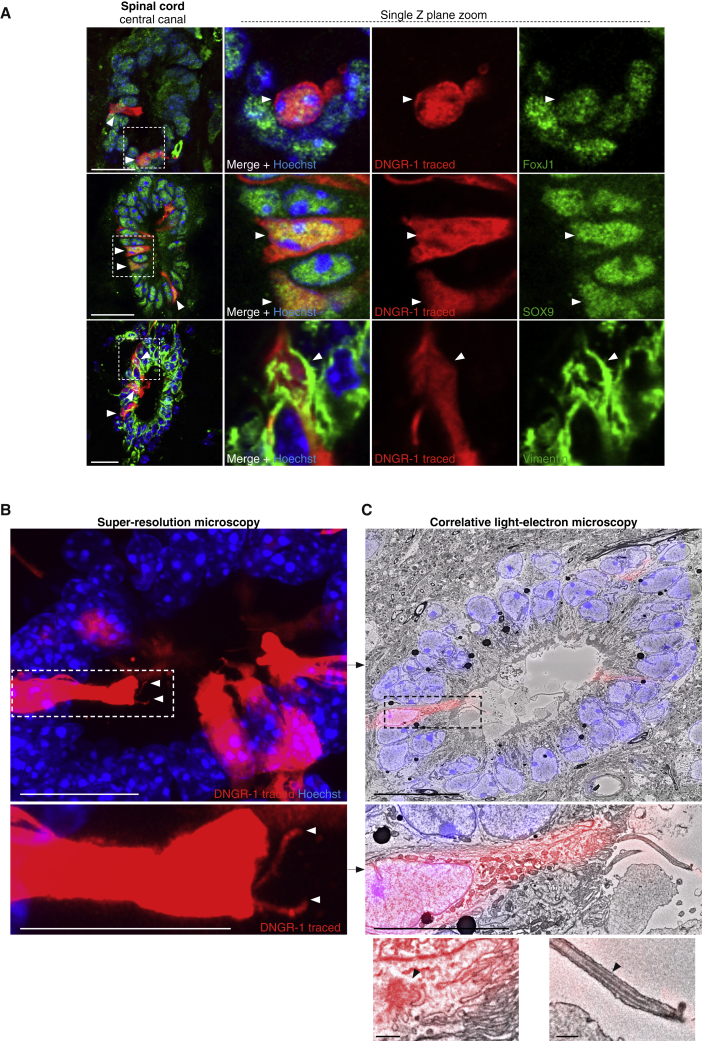
To assign an identity to non-cDC, DNGR-1-traced cells, we focused on the known cellular composition of the spinal cord. Non-cDC, DNGR-1-traced cells lacked expression of GFAP, NeuN, PKD2L1, NG2, or PDGFrβ, indicating that they were not astrocytes, neurons, CSF-contacting neurons, oligodendrocyte progenitors, or pericytes, respectively (Figure S2). However, non-cDC, DNGR-1-traced cells were uniformly positive for FoxJ1 and expressed SOX9 and vimentin (Figure 3A), consistent with being ependymal cells. Using correlative light (super-resolution) and electron (serial block-face) microscopy (Figures 3B and 3C), most non-cDC, DNGR-1-traced cells in the spinal cord central canal were found to have a simple/pseudostratified columnar epithelial cell shape (Figure S3A) and to possess two cilia (Figure 3B; Video S5), which displayed a 9+2 axoneme structure (Figures 3C and S3B), consistent with motile nature. Both cilia averaged a length of 11.86 ± 2.85 μm (Figure S3C) and could be further tracked through the image volume to their respective basal bodies (Figures 3C and S3D; Video S6). Thus, immunophenotypic and ultrastructural characterization identifies non-cDC, DNGR-1-traced cells in the CNS as ciliated ependymal cells.
Figure 3.
Non-cDC, DNGR-1-traced cells are ependymal cells
(A) Spinal cord central canal cryosections from Clec9aCreRosaLSLtdTomato mice labeled with antibodies against FoxJ1, SOX9, or vimentin (green). Zoomed-in panels show single optical Z slice of indicated areas. Arrowheads indicate DNGR-1-traced cells co-stained with the respective marker.
(B) Airyscan super-resolution of vibratome section from a DNGR-1-traced spinal cord central canal. Arrowheads in zoomed view indicate two cilia emerging from DNGR-1-traced ependymal cells.
(C) Composite of the super-resolution image shown in (B) on respective electron micrograph. Bottom, zoomed view of indicated area. Below left, basal body (arrowhead) and; below right, cilium displaying a 9 + 2 axoneme structure (arrowhead).
Scale bars, 20 μm (A–C), 10 μm (zooms B and C), and 0.5 μm (detailed views of C). Representative images of 2 animals analyzed.
Representative vibratome section imaged with the airyscan super-resolution module which reveals two cilia emerging from ependymal DNGR-1-traced cells (red). Nuclei were counterstained with Hoechst (blue).
Tracking of an individual cilium belonging to an DNGR-1-traced ependymal cell (red), from its central canal extension to its respective basal body.
DNGR-1-traced ependymal cells exist along a differentiation continuum
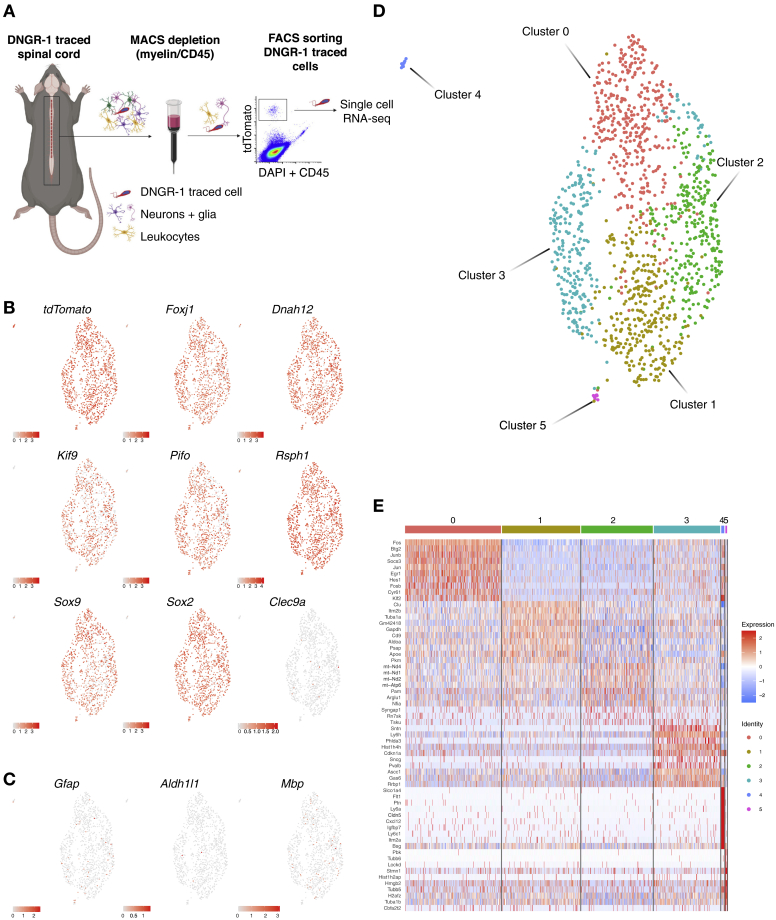
We carried out single-cell RNA sequencing (scRNA-seq) of CD45− DNGR-1-traced cells isolated from spinal cords (Figure 4A). After initial quality control, data from 1,241 cells were analyzed. Mapping of tdTomato reads onto the dataset confirmed that the cells originated from the DNGR-1-traced compartment (Figure 4B). In agreement with being ependymal cells, the vast majority expressed the genes encoding the master regulator of motile cilia (Foxj1), the structural components of cilia, such as dynein (Dnah12) and kinesin (Kif9) motors, the basal body that attaches cilia to the cell body (Pifo) and the radial spokes (Rsph1), as well as the transcription factors Sox9 and Sox2 (Figure 4B). Conversely, markers of astrocytes (Gfap, Aldh1l1) or oligodendrocytes (Mbp) were expressed at low levels by a negligible number of cells (Figure 4C). Other markers of oligodendrocytes (Olig1, Olig2, Mog, and Nkx2-2), oligodendrocyte progenitors (Sox10 and Pdgfra), or neuroblasts (Eomes and Tbr1) were completely absent from the dataset. Indeed, based on their global gene expression profile, 97% of the cells analyzed (1,204 cells of 1,241) matched the ependymal cell identity defined by the CNS single-cell reference atlas (Zeisel et al., 2018). Clec9a (which encodes DNGR-1) was not appreciably detected (Figure 4B; 3 cells of 1,241), consistent with the fact that it is only expressed in the embryonic precursors of the traced cells (see above).
Figure 4.
Single-cell RNA sequencing of non-cDC, DNGR-1-traced cells
(A) Purification of non-cDC, DNGR-1-traced cells. (n = 14 pooled spinal cords).
(B and C) (B) Feature plots showing projection of tdTomato, ependymal cell identity, or Clec9a and (C) glial cell transcripts on the UMAP space. Low expression (gray) and high expression (red).
(D) UMAP embedding of 1,241 DNGR-1-traced cells and unsupervised clustering showing high compactness of DNGR-1-traced ependymal cells.
(E) Heatmap representation of the top-10 differentially expressed genes defining each cluster. Low expression (blue) and high expression (red).
Analysis by UMAP dimensionality reduction and unsupervised clustering allowed sub-grouping of DNGR-1-traced ependymal cells into 6 clusters (Figures 4D and 4E). The latter did not correspond to groups of cells occupying distinct dorsal-ventral or rostral-caudal positions, as transcription factors governing such positioning (i.e., ZEB1, PAX6 and Hox-A5, Hox-A9, and Hox-A10, respectively) (Ghazale et al., 2019) were found across all clusters (data not shown). Cluster 4 (12 cells of 1,241) stood out by the expression of hallmark genes of vascular endothelial cells such as Cldn5, Flt1, and Pecam1 and concurrent absence/low expression of ependymal cell signature transcripts, including Foxj1, and was excluded from further analysis. Apart from cluster 4, the remaining clusters displayed high compactness within the UMAP space, suggesting that DNGR-1-traced ependymal cells are relatively homogeneous and that clusters likely represent cellular states along a lineage continuum rather than discrete subsets.
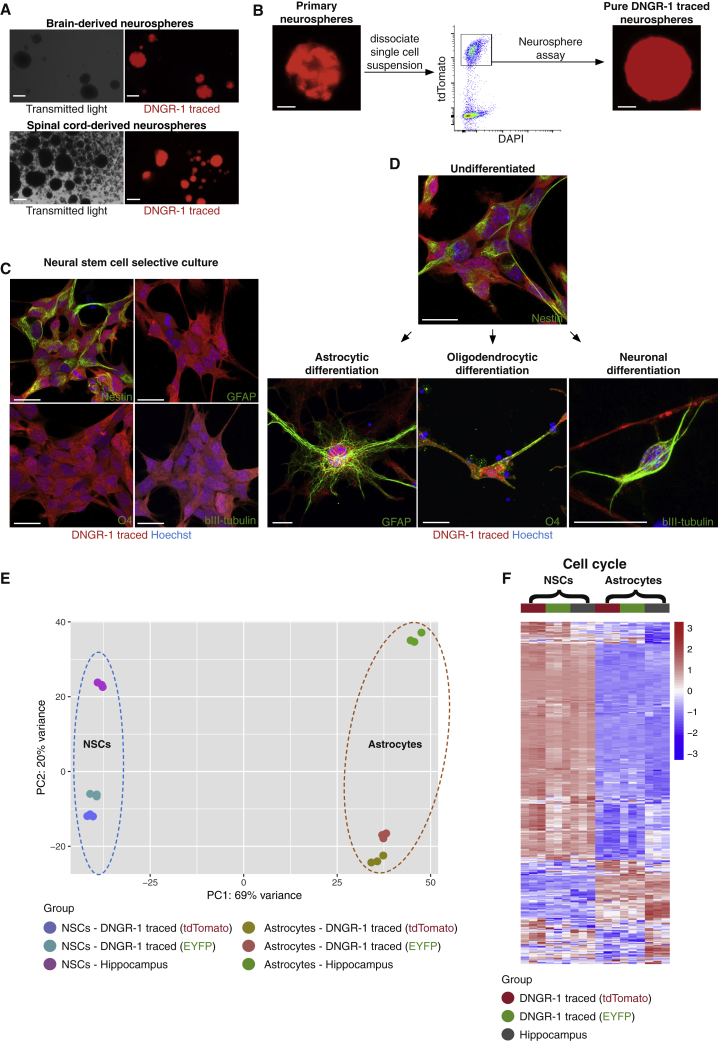
DNGR-1-traced ependymal cells display neural stem cell properties in vitro
Neurosphere assays allow for selective expansion of stem and progenitor cells in vitro (Reynolds and Weiss, 1992). Interestingly, preparations from brains and spinal cords of DNGR-1-lineage-traced mice gave rise to neurospheres containing DNGR-1-traced cells (Figure 5A). This was not due to upregulation of DNGR-1 ex vivo as no DNGR-1-traced neurospheres could be observed when grown from brains or spinal cords of Clec9aCreERT2RosaLSLtdTomato mice in the presence of 4-hydroxytamoxifen (Z-4OH-TAM) (Figure S4A). In contrast, Z-4OH-TAM-treated in vitro cDC cultures (Helft et al., 2015) grown from bone marrow of these animals yielded robust labeling of cDC1 (>98%), controlling for the sensitivity of the system (Figure S4B).
Figure 5.
DNGR-1-traced ependymal cells display neural stem cell properties in vitro
(A) Representative images from primary neurosphere cultures grown from brain or spinal cord of Clec9aCreRosaLSLtdTomato mice.
(B) Strategy for isolation of pure DNGR-1-traced neurospheres (tdTomato+).
(C and D) (C) Immunofluorescence of pure DNGR-1-traced neurosphere-derived cells (red) grown in adherence in the absence (C) or presence (D) of differentiation promoting factors and stained with antibodies against nestin, GFAP, O4, and βIII-tubulin (all green).
(E) Principal component analysis showing clustering of hippocampi-derived or DNGR-1-traced NSCs and their respective differentiated astrocytes. Each dot represents a biological replicate.
(F) Heatmap of differential expressed genes involved in cell cycle between NSC and astrocyte cell fates derived from the hippocampus or from DNGR-1-traced cells.
Scale bars, 200 μm (A), 100 μm (B), and 20 μm (C and D). Representative images of least 3 independent experiments.
On closer inspection, primary neurospheres contained both DNGR-1-traced and non-traced cells (Figure 5B). Primary neurospheres can be composed of stem and progenitor cells, the former being distinguished by their ability to self-renew over an extended period of time. Notably, when we FACS-sorted tdTomato+ cells from primary neurospheres and re-plated them, we were able to re-generate neurospheres that now contained 100% tdTomato+ cells and that could be grown and passaged repeatedly (Figure 5B). Thus, DNGR-1-traced cells in neurospheres are capable of prolonged self-renewal.
The proportion of actual NSCs within neurospheres is low (Reynolds and Rietze, 2005) but can be increased by adherent culture in NSC-favoring conditions (Conti et al., 2005; Pollard et al., 2006). In such cultures, neurosphere-derived DNGR-1-traced cells could be passaged for more than 6 months without cell loss and expressed the NSC marker nestin but not markers of differentiated CNS cells (Figure 5C). Remarkably, upon addition of suitable factors, the DNGR-1-traced cell cultures could be coaxed to differentiate into astrocytes, oligodendrocytes, or immature neurons (Figures 5D and S4C). We compared by bulk RNA-seq DNGR-1-traced cells and bona fide professional NSCs (isolated from the hippocampus rather than subventricular zone to avoid any possible contamination of the latter with ependymal layer), as well as their respective in vitro differentiated astrocyte progenies (Figures S4D and S4E). In principal component and cluster analysis, DNGR-1-traced NSCs (generated from two independent tracer mouse strains) clustered together with hippocampal NSCs both before and after differentiation, indicating that most of the variance was attributable to the differentiation process and not the cellular origin (Figure 5E). In their undifferentiated state, the similarity between the two NSC populations was also noticeable at the level of expression of genes involved in cell cycle, consistent with the ability of the cells to expand in vitro (Figure 5F). Thus, DNGR-1-traced ependymal cells display properties of stem cells when propagated and differentiated in vitro and are remarkably similar to bona fide NSCs under these culture conditions.
CNS injury induces differentiation of DNGR-1-traced ependymal cells
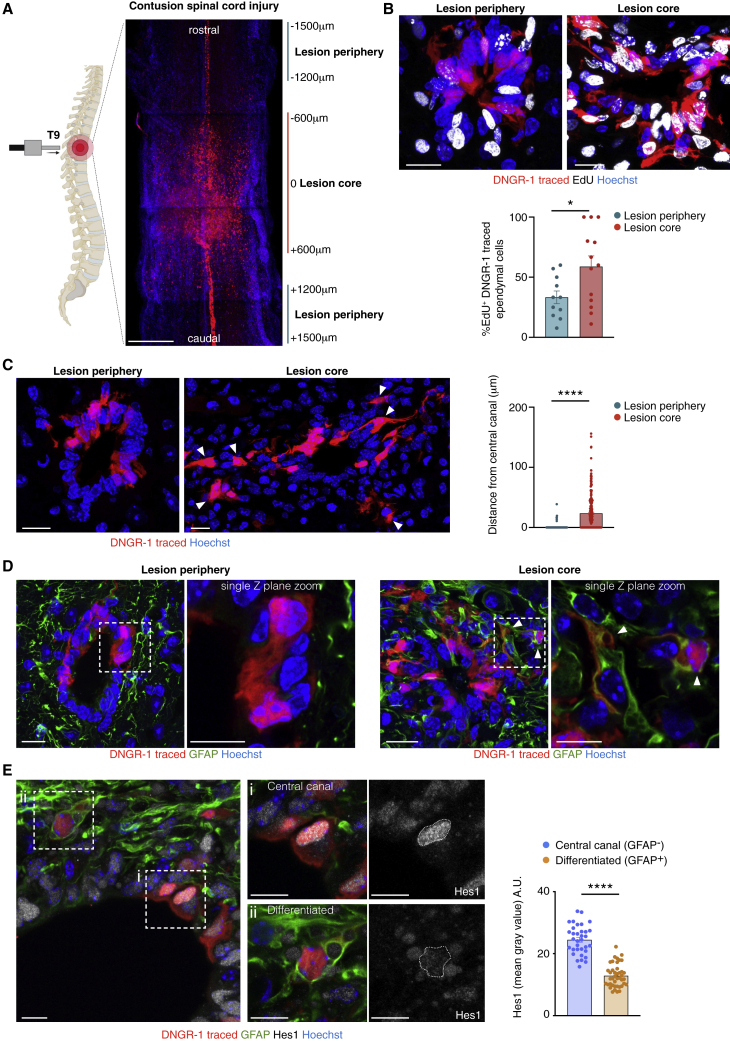
We assessed whether the differentiation and self-renewal potential of DNGR-1-traced ependymal cells could also be revealed in vivo. A fraction of DNGR-1-traced ependymal cells incorporated EdU over a 30-day period (Figure S5A), consistent with homeostatic proliferation. However, as mentioned, we did not find labeled neurons or glia in either spinal cord (Figure S2) or brain (Figure 1B) of DNGR-1-lineage-tracing mice, indicating lack of contribution to neuronal or glial lineages in the steady state. To determine whether this could be altered by injury, we subjected DNGR-1-lineage tracer mice to spinal cord contusion (Figure 6A). Histological examination identified an injury core comprising roughly 1,200 μm (Figure S5B) that extended symmetrically from the lesion epicentre (−600 μm rostral/+600 μm caudal). Rare DNGR-1-traced cDCs infiltrating the lesion could be distinguished from their traced ependymal cell counterparts by CD45 expression and dimmer tdTomato expression (Figure S5C). Light-sheet imaging revealed that the distribution pattern of DNGR-1-traced ependymal cells (tdTomatobright) remained unchanged in regions peripheral to injury but was significantly altered in the lesion core (Figure 6A; Video S7). There, DNGR-1-traced ependymal cells could be seen to have expanded and moved away from the central canal. Expansion appeared to be due to proliferation as EdU incorporation by DNGR-1-traced cells increased to approximately 60% in the lesion core (reaching 100% in some sections), compared with 30% in the lesion periphery (Figure 6B) 7 days after injury. Closer examination revealed that EdU incorporation correlated with displacement away from the central canal and acquisition of altered morphology (Figure 6C). Notably, a fraction of these displaced cells in the vicinity of the injury acquired GFAP immunoreactivity, consistent with astrocytic differentiation (Figure 6D). We did not observe differentiation into neurons or oligodendrocytes (Figure S5D). DNGR-1-traced cells away from the lesion epicenter were never found to express GFAP (Figure 6D), retained their usual morphology, and remained confined to the ependymal layer of the central canal (Figures 6A, 6C, and 6D), as suggested by the light-sheet images (Figure 6A; Video S7). We further tested a model of brain ventricular injury. Similar to spinal cord contusion, DNGR-1-lineage-traced ependymal cells lining the ventricles also reacted to local injury with morphological changes and migration toward the damage site, where some cells became GFAP+ (Figure S5E).
Figure 6.
Mobilization and differentiation of DNGR-1-traced ependymal cells after injury
(A) Maximum intensity projection of optically clarified spinal cord from an injured Clec9aCreRosaLSLtdTomato mouse. T9 refers to injured vertebra. Lesion coordinates showed result from white matter sparring determination (Figure S5B). Lesion periphery = −1,500–−1,200 μm rostral and +1,200–+1,500 μm caudal; lesion core = −600 μm rostral to +600 μm caudal.
(B–E) Spinal cord cryosections from Clec9aCreRosaLSLtdTomato mice, 7 days post injury, showing: (B) EdU (white) incorporation by DNGR-1-traced ependymal cells (red) in the the lesion periphery or lesion core. Below, quantification in both sites. Each dot represents one quantified image. (C) Proximity of DNGR-1-traced cells to the central canal in the lesion periphery or core. Arrowheads show DNGR-1-traced cells displaced from the central canal. Right, quantification of distance (μm) of DNGR-1-traced cells from the central canal. Each dot represents one cell. (D) Staining with the astrocytic marker GFAP (green) in the central canal of the lesion periphery (left) or core (right). Arrowheads, GFAP+DNGR-1-traced cells. (E) Spinal cord central canal cryosection from Clec9aCreRosaLSLtdTomato mice 21 days post injury stained labeled with anti-GFAP(green) and anti-Hes1 (white) antibodies. Right, zooms of (Ei) central canal or (Eii) peri-ependymal region. Right, quantification of Hes1 expression (a.u., arbitrary units). Error bars (SEM).
Scale bars, 400 μm (A), 20 μm (C and D), 15 μm (B), and 10 μm (E).
At least 3 animals were quantified per experiment. Groups were compared using unpaired t test. ∗p < 0.05, ∗∗∗∗p < 0.0001.
Location of DNGR-1-traced cells (bright red) in response to spinal cord injury around the lesion epicentre. DNGR-1-traced cells show displacement for their central canal location toward the injury site, which was applied dorsally. Nuclei were counterstained with Hoechst (blue).
To exclude the possibility of injury-driven upregulation of DNGR-1 expression in reactive astrocytes or other astrocytic progenitors, we repeated the ventricular injury experiments in Clec9aCreERT2RosaLSLtdTomato mice injected with tamoxifen immediately prior to injury (1–2 h) and during the initial recovery period (days 3 and 7 post injury). As a positive control for induction of DNGR-1 lineage tracing, DNGR-1-traced cDCs could be readily observed in the choroid plexi of injured animals (Figures 1A and S6A). In contrast, no DNGR-1-traced astrocytes were observed surrounding the injury scar or along the ependymal cell layer (Figure S6A). Clec9a single-molecule fluorescence in situ hybridization (smFISH) further confirmed that spinal cord injury does not induce DNGR-1 expression (Figure S6B). Additionally, in animals given EdU and subjected to spinal cord contusion, all DNGR-1-traced astrocytes were EdU+ (Figure S6C). Collectively, these data indicate that DNGR-1-traced astrocytes emerge from differentiation of the proliferative DNGR-1-traced ependymal compartment (Figure 6B). We conclude that DNGR-1-traced ependymal cells react to local CNS injury by increased proliferation and astrocytic differentiation.
To understand which signaling pathways may play a role in determining the homeostatic quiescence of DNGR-1-traced ependymal cells versus their mobilization in response to injury, we analyzed our single-cell dataset for expression of genes known to govern cell-fate decisions. Notch signaling has been shown to regulate quiescence of professional NSCs and the Notch2 transcript could be detected in multiples clusters (Figure S6D). Additionally, the Notch target gene Hes1, which inhibits differentiation (Ishibashi et al., 1995), was enriched in cluster 0 (Figures 4E and S6D). Consistent with the notion that the scRNA clusters reflect dynamic ependymal cell states rather than cellular subtypes, immunostaining of uninjured spinal cord revealed that all DNGR-1-traced ependymal cells expressed HES1 protein, albeit at different levels (Figure S6E.) In contrast, HES1 expression was much more heterogeneous in spinal cords from injured animals. DNGR-1-traced cells that moved away from the central canal and acquired GFAP expressed significantly lower HES1 levels than GFAP− DNGR-1-traced cells that remained in the ependymal canal (Figure 6E). Thus, DNGR-1-traced ependymal cell mobilization induced by tissue damage is associated with loss of HES1, consistent with release from a quiescent state and induction of differentiation.
DNGR-1-traced ependymal cells constitute a distinct ependymal subset wherein resides latent stem cell potential
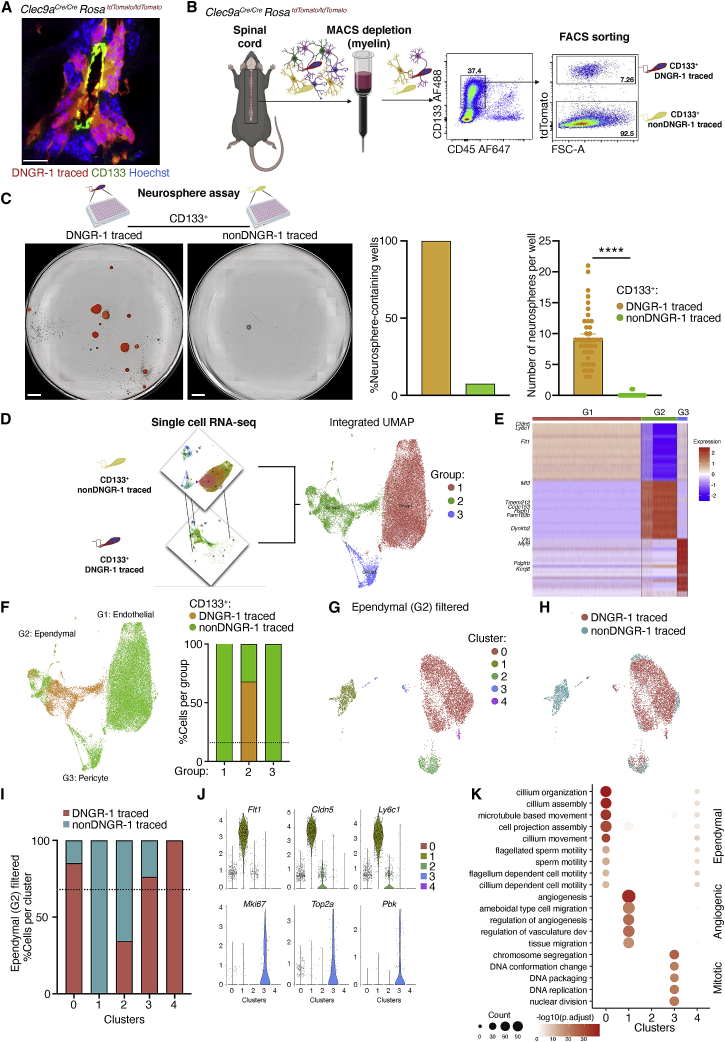
Although DNGR-1 lineage tracing faithfully marked ependymal cells, it did not label the entirety of the ependymal compartment. This could be due to incomplete penetrance of the Cre-mediated recombination event, resulting from a variable fraction of embryonic cells that transiently express DNGR-1 remaining unmarked. We bred DNGR-1-lineage-traced animals with the Cle9aCre and R26LSLtdTomato alleles in heterozygous or homozygous configuration and confirmed that the latter displayed increased ependymal cell labeling (Figure S7A). However, double-homozygous DNGR-1-lineage-tracing mice (Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato) with the maximum achievable penetrance still only displayed, on average, labeling of 60% of spinal cord ependymal cells as assessed by histology (Figure S7A). To test the possibility that traced and untraced cells constitute distinct ependymal cell subtypes, we used CD133 as a pan-ependymal cell marker (Alfaro-Cervello et al., 2012; Coskun et al., 2008; Meletis et al., 2008). We first confirmed that CD133 labels all DNGR-1-traced cells in the ependymal layer, as expected (Figures 7A and S7B). We then sorted CD133+ DNGR-1-traced cells and CD133+ non-DNGR-1-traced cells from spinal cords of Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato animals (Figure 7B) and tested them in the neurosphere assay (Figure 7C). Strikingly, 100% of the wells seeded with CD133+ DNGR-1-traced cells developed neurospheres, which expressed tdTomato and were present at an average of 10 neurospheres per well (Figure 7C). In contrast, only 7.5% of the wells seeded with CD133+ non-DNGR-1-traced cells developed neurospheres, which were tdTomato-negative and present on average as a single neurosphere per well (Figure 7C). Thus, the neurosphere-forming potential of CD133+ cells in the spinal cord is largely confined to the DNGR-1-traced compartment.
Figure 7.
DNGR-1-tracing marks a distinct class of ependymal cells, wherein resides latent stem cell ability
(A) Spinal cord central canal cryosection from a Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato mouse labeled with anti-CD133 (green). CD133 is expressed apically and marks all ependymal cells (traced and untraced).
(B) FACS-sorting of CD133+ DNGR-1-traced and CD133+ non-traced cells.
(C) Neurosphere assay of CD133+ DNGR-1-traced and CD133+ non-traced cells from 4 pooled spinal cords. Right, quantification of neurosphere-containing wells and number of neurospheres per well from each fraction. Data are from two independent experiments. Error bars (SEM). Groups were compared using unpaired t test. ∗∗∗∗p < 0.0001.
(D) Dataset integration and UMAP of CD133+ DNGR-1-traced cells and CD133+ non-traced cells into 3 groups.
(E) Heatmap of the top 20 differentially expressed genes in each group from (D).
(F) UMAP representation by cellular provenance and quantification of each cellular fraction per group. Orange, CD133+ DNGR-1-traced cells; Green, CD133+ non-traced cells. 97.89% of CD133+ DNGR-1-traced cells were unbiasedly assigned to ependymal cell group (group 2). The remaining CD133+ DNGR-1-traced cells were assigned to groups 1 (1.89%) and 3 (0.23%). Dashed line represents the proportion of CD133+ DNGR-1-traced cells within the UMAP space (15.95%—4,825 of 30,326 cells).
(G) UMAP of cells from group 2 (from F).
(H) UMAP shown in (G) indicating cellular provenance: CD133+ DNGR-1-traced ependymal cells in salmon and CD133+ non-traced cells in cyan.
(I) Percentage of traced and non-traced cells in each of the clusters 0–4 from (G). Dashed line represents the proportion of CD133+ DNGR-1-traced cells within UMAP space (67.9%—4,723 of 6,951 cells).
(J) Expression of a selected set of cluster markers from clusters 1 and 3 (from G).
(K) Distribution of the top 5 Gene Ontology biological processes defined by the differentially expressed genes in each cluster.
Scale bars, 500 μm (C) and 10 μm (A).
We then compared CD133+ DNGR-1-traced cells with CD133+ non-DNGR-1-traced cells by scRNA-seq (Figure 7D). Integration and embedding of 25,411 CD133+ non-DNGR-1-traced cells and 4,825 CD133+ DNGR-1-traced cells within the UMAP space resulted in unsupervised partitioning of the total 30,236 cells into 3 groups (Figure 7D) with distinct expression signatures (Figure 7E). Surprisingly, these did not all represent ependymal cells. Indeed, only cells in group 2 displayed a canonical ependymal cell signature defined by genes such as Ccdc153, Fam183b, Tmem212, or Mt3 (MacDonald et al., 2021; Shah et al., 2018; Zeisel et al., 2018; Figures 7E and S7C). CD133+ cells in groups 1 and 3 were defined by high expression of endothelial (Cldn5, Flt1, Pecam1, and Ly6c1) or pericyte (Pdgfrb, Kcnj8, Myl9, and Vtn) genes, respectively (Figures 7E and S7C). Notably, 97.9% (4,723 of 4,825) of DNGR-1-traced cells belonged to the ependymal cell group (group 2) (Figure 7F), representing 67.9% of the total number of cells in that group (4,723 of 6,951 cells), which matches the labeling ratio observed in histological sections (Figure S7A).
To determine whether DNGR-1-tracing marked a distinct ependymal cell population, we focused on the 98.2% of cells in group 2 that expressed ependymal cell signature genes and compared the DNGR-1-traced with the non-traced fractions. This revealed that 4,946 genes were significantly differentially expressed between the two populations (Figure S7D). Although both populations expressed canonical ependymal cell genes (Figure S7E), the non-DNGR-1-traced fraction displayed an additional enrichment for genes associated with vascular/angiogenic processes (Figures S7F and S7G). GSEA of all expressed genes in each population further highlighted the hybrid angiogenic/ependymal nature of the non-traced group 2 cells. Conversely, DNGR-1-traced ependymal cells were significantly enriched in genes involved in mitotic processes and bearing binding motifs for transcription factors involved in cell cycle, such as E2F, CHAMP1, or EVI1 (Figures S7G). Thus, DNGR-1-traced ependymal cells are distinct from their non-traced counterparts and display a mitotic signature.
To understand whether the above differences were reflective of ependymal cell heterogeneity, we reclustered cells in group 2 in a new UMAP (Figures 7G and S7H). This revealed 5 different clusters, with 3 closely apposed clusters (0, 3, and 4) sitting apart from clusters 1 and 2 (Figure 7G). Mapping cellular provenance onto the UMAP revealed that cluster 1 contained exclusively non-traced cells (Figure 7H). Conversely, the DNGR-1-traced cells were overrepresented in clusters 0, 3, and 4 and underrepresented in 2 (Figures 7H and 7I), further highlighting that DNGR-1-traced ependymal cells are not identical to non-traced ependymal cells (if DNGR-1-traced ependymal cells were identical to their non-traced counterparts, they would be expected to contribute equally to each cluster, in strict proportion [67.9%; dotted line in Figure 7I] to the number of cells that they contribute to the total of the cells in group 2 of the integrated UMAPs). Gene Ontology biological process categorization, revealed that cluster 1 (containing exclusively non-DNGR-1-traced cells) displayed an angiogenic/ependymal cell signature, consistent with its aforementioned hybrid cell phenotype (Figure 7K). Conversely, cluster 3, which contained an overrepresentation of DNGR-1-traced cells, displayed a cell division signature (Figures 7J, 7K, and S7D), marked by unique expression of the canonical cell cycle genes Mki67, Top2a, and Pbk. Taken together, these results suggest that the partial DNGR-1 tracing of CD133+ ependymal cells is not fully attributable to incomplete penetrance of the recombination event that initiates lineage tracing. Rather, they support the notion that the DNGR-1-traced cells represent a distinct ependymal cell population, wherein resides latent stem cell activity.
Discussion
Stem cell activity in ependymal cells has remained a contentious topic. Pioneering lineage-tracing studies using a human FoxJ1 promoter element first suggested the stem cell potential of ependymal cells (Barnabé-Heider et al., 2010; Carlén et al., 2009; Llorens-Bobadilla et al., 2020; Meletis et al., 2008; Sabelström et al., 2013). However, FoxJ1 expression is not confined to ependymal cells, (Beckervordersandforth et al., 2010; Benner et al., 2013; Devaraju et al., 2013; Jacquet et al., 2009) and a different FoxJ1 driver failed to reveal ependymal cell stemness (Muthusamy et al., 2018; Ren et al., 2017), perhaps because of FoxJ1 haploinsufficiency (Li et al., 2018). Here, we use a completely different driver, DNGR-1, previously studied in the context of cDC lineage tracing. DNGR-1 fate mapping showed high fidelity in ependymal cell marking, highlighted by our scRNA-seq of spinal cord DNGR-1-traced cells, which showed a 97% identity match to ependymal cells in the CNS reference atlas (Zeisel et al., 2018). Also, in the brain, DNGR-1 lineage tracing did not mark other progenitor populations, including homeostatic NSC pools in the SVZ or DG, as evidenced by the absence of traced OB interneurons or hippocampal granule neurons, respectively. Thus, the faithful ependymal cell labeling offered by the DNGR-1-lineage-tracing approach and the ability of the labeled cells to give rise to neurons, oligodendrocytes, and astrocytes in vitro confirms the presence of latent stem cell activity in the ependymal cell layer and corroborates the conclusions drawn by the studies using a human FoxJ1 promoter element.
Despite its accurate marking of the ependymal compartment, DNGR-1 lineage tracing did not label all ependymal cells. Fractional ependymal cell labeling could only be partly explained by incomplete penetrance of the recombination event that initiates lineage tracing. Indeed, comparison between DNGR-1-traced and non-traced cells revealed previously unrecognized ependymal cell heterogeneity. A major component of this heterogeneity was the expression of angiogenic factor-encoding genes by non-traced cells, which may correspond to the ependymal cells with a vascular signature identified previously (Luo et al., 2015). This vascular signature did not occur alongside stem cell or cell cycle genes, which were expressed predominantly in an ependymal cell cluster composed mostly of DNGR-1-traced cells and that likely corresponds to the mitotic fraction of the ependymal cell layer. It is tempting to speculate that this mitotic fraction is responsible for the self-renewal of the DNGR-1-labeled ependymal cell sub-population and is mobilized upon injury or ex vivo neurosphere culture. Thus, only a fraction of ependymal cells possesses latent stem cell potential and that fraction is contained within the DNGR-1-traced subset. Differential experimental capture of the DNGR-1-traced compartment may explain why some (Barnabé-Heider et al., 2010; Carlén et al., 2009; Llorens-Bobadilla et al., 2020; Meletis et al., 2008; Sabelström et al., 2013), but not other (Muthusamy et al., 2018; Ren et al., 2017; Shah et al., 2018), studies have found stem cell potential in ependymal cells. Interestingly, Troy (encoded by the Tnfrsf19 gene) has recently been shown to mark a sub-population of ependymal cells (Stenudd et al., 2022). However, in our dataset, Troy was expressed by all ependymal cells (Figures S7I and S7J).
Adult NSCs and ependymal cells share a common lineage, both emerging from radial glia (Aimone et al., 2014; Fuentealba et al., 2015; Ortiz-Álvarez et al., 2019). Radial glia to ependymal cell commitment has been reported to occur at E15.5, when the FoxJ1-dependent ciliated program is initiated (Barnabé-Heider et al., 2010; Li et al., 2018; Meletis et al., 2008; Ortiz-Álvarez et al., 2019). However, radial glia cells are highly heterogeneous and have been reported to display fate specification for glial lineages as early as E9.5 (Malatesta et al., 2000, 2003; McCarthy et al., 2001). We found DNGR-1 expression at E11.5 in a rare population of ventricular progenitor cells, which exclusively gave rise to the traced ependymal compartment seen in adulthood. Thus, DNGR-1 expression occurs in a subset of embryonic progenitors that has exited a neurogenesis and gliogenesis trajectory but retains latent stem cell potential throughout life. While this rare DNGR-1-expressing population displayed concomitant expression of SOX2, no staining by the neuroepithelial marker nestin was observed, and its lineage relationship to the heterogeneous radial glia pool remains to be fully dissected. Furthermore, the kinetics of expansion that must occur between the emergence of the embryonic DNGR-1-expressing subset and it populating the entire CSF-lining compartment remain to be determined. Finally, which elements of the Clec9a enhancer and promoter regulate DNGR-1 expression in ependymal cell-committed pool versus cDC progenitors and cDC1 is presently unknown and an area for future study.
The signals that dictate homeostatic quiescence of DNGR-1-traced ependymal cells versus injury-associated differentiation also remain to be determined. The oscillatory nature of Notch signaling regulates both active and quiescent states in somatic stem cells (Harris and Guillemot, 2019; Imayoshi et al., 2013; Sueda and Kageyama, 2020). Interestingly, ependymal cells have been reported as uniquely able to survive sustained Notch signaling (Baek et al., 2006; Ishibashi et al., 1994). We found global HES1 expression in DNGR-1-traced ependymal cells during homeostasis, in accordance with their quiescent state and lack of neurogenesis or gliogenesis. Notch signaling often occurs through lateral inhibition by cell-cell contact (Sueda and Kageyama, 2020), and we hypothesize that contact loss following injury could constitute an important determinant of HES1 downregulation associated with DNGR-1-traced ependymal cell differentiation. We did not observe differentiation of DNGR-1-traced ependymal cells in old animals (our unpublished observations), suggesting that the threshold of attrition required to activate ependymal cells is superior to that resulting from aging.
Current strategies to promote CNS repair focus on stem cell transplants or direct glial reprogramming in situ (Barker et al., 2018). Our work identifies an embryonically derived mammalian ependymal cell subset across the entire CNS endowed with latent stem cell potential. In contrast to mammals, anamniotes such as teleost fish species display exceptional spinal cord regeneration. This is orchestrated in part by cells inhabiting the ependymal cell niche termed ependymo-radial glial cells (ERGs), which, similarly to the DNGR-1-traced ependymal cell subset described here, proliferate in response to injury and migrate toward the lesion site. Despite these similarities, mammalian rehabilitation has a poor prognosis, with long-term chronic functional impairment. Although ERGs react to injury by increasing neurogenesis, differentiation of the DNGR-1-traced ependymal subset after injury in vivo was restricted to the astrocytic lineage, which in turn contributed to the glial scar. This is despite the fact that DNGR-1-traced ependymal cells show neuronal differentiation potential in vitro and is consistent with reports of a block in mammalian neurogenesis following injury (Barnabé-Heider et al., 2010; Becker et al., 2018; Meletis et al., 2008; Stenudd et al., 2015). Given that in anamniotes, ependymal to neuronal differentiation correlates with functional motor recovery after spinal cord injury (Becker and Becker, 2015; Becker et al., 2018; Cigliola et al., 2020), coaxing the ependymal damage-responsive stem cells described here to differentiate into neurons in vivo, perhaps through manipulation of signals within the injury microenvironment, could represent a therapeutic strategy for the treatment of CNS injury in mammals.
Limitations of the study
Our results argue for the presence of an embryonically fated sub-population of ependymal cells present throughout the CNS and with potential to differentiate into neurons, oligodendrocytes, or astrocytes. However, whether this means that those cells can be considered latent stem cells remains to be fully established. The tri-potency of DNGR-1-traced ependymal cells could only be revealed in vitro, and it will be important to assess why and how differentiation in vivo is restricted to astrocytes. Also, although they behave broadly similarly, we have not formally assessed possible differences in response to injury among the cells present in brain versus different parts of the spinal cord. Finally, our data are exclusively from mouse CNS. Whether analogous cells are present in other tissues and in humans remains to be investigated.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse/rat/human IBA-1 (polyclonal) | FUJIFILM Wako Shibayagi | Cat# 019-19741; RRID: AB_839504 |
| AF647 anti-mouse I-A/I-E (M5/114.15.2) | BioLegend | Cat# 107618; RRID: AB_493525 |
| AF488 anti-mouse I-A/I-E (M5/114.15.2) | BioLegend | Cat# 107616, RRID:AB_493523 |
| eF450 anti-mouse I-A/I-E (M5/114.15.2) | Thermo Fisher Scientific | Cat# 48-5321-82, RRID:AB_1272204 |
| Rabbit anti-mouse CD64 (#008) | Sino Biological | Cat# 50086-R008, RRID:AB_2860481 |
| FITC anti-mouse CD64 (X54-5/7.1) | BioLegend | Cat# 139316, RRID:AB_2566556 |
| AF488 anti-mouse CD45 (30-F11) | BioLegend | Cat# 103122, RRID:AB_493531 |
| AF647 anti-mouse CD45 (30-F11) | BioLegend | Cat# 103124, RRID:AB_493533 |
| Rat anti-mouse CD11b (5C6) | Bio-Rad | Cat# MCA711, RRID:AB_321292 |
| Rabbit anti-mouse/rat/human SOX-9 (polyclonal) | Merck Millipore | Cat# AB5535, RRID:AB_2239761 |
| Rabbit anti-mouse/rat/human Vimentin (EPR3776) | Abcam | Cat# ab92547, RRID:AB_10562134 |
| Mouse anti-mouse/human FOXJ1 (2A5) | eBioscience, Thermo Fisher Scientific | Cat# 14-9965-82, RRID:AB_1548835 |
| Chicken anti-GFAP (polyclonal) | Abcam | Cat# ab4674, RRID:AB_304558 |
| Mouse anti-NeuN (A60) | Merck Millipore | Cat# MAB377, RRID:AB_2298772 |
| Rabbit anti-mouse Polycystin-L (polyclonal) | Merck Millipore | Cat# AB9084, RRID:AB_571091 |
| Rabbit anti-NG2 (polyclonal) | Merck Millipore | Cat# AB5320, RRID:AB_1121367 |
| Goat anti-human Olig2 (polyclonal) | R and D Systems | Cat# AF2418, RRID:AB_2157554 |
| Rabbit anti-mouse/human/rat PDGFr beta (Y92) | Abcam | Cat# ab32570, RRID:AB_777165 |
| Rabbit anti-mouse HES1 (polyclonal) | Ryoichiro Kageyama, Institute for Frontier Life and Medical Sciences | N/A |
| AF488 anti-mouse SOX2 (Btjce) | Thermo Fisher Scientific | Cat# 53-9811-82, RRID:AB_2574479 |
| Mouse anti-mouse/human/rat Nestin (25/NESTIN) | BD Biosciences | Cat# 611659, RRID:AB_399177 |
| Mouse anti-oligodendrocyte marker O4 (O4) | R and D systems | Cat# MAB1326, RRID:AB_357617 |
| Mouse anti-mouse/human/rat tubulin beta 3 (TUJ1) | BioLegend | Cat# 801201, RRID:AB_2313773 |
| Rabbit anti-MAP2 (polyclonal) | Cell Signaling Technology | Cat# 4542, RRID:AB_10693782 |
| Rabbit anti-mouse/human/rat GABA(B)R2 (polyclonal) | Cell Signaling Technology | Cat# 3839, RRID:AB_2232133 |
| Mouse anti-GluN2B/NR2B (N59/36) | UC Davis/NIH | Cat# N59/36R, RRID:AB_275080 |
| Rabbit anti-mouse/human/rat Tyrosine Hydroxylase (E2L6M) | Cell Signaling Technology | Cat# 58844, RRID:AB_2744555 |
| Donkey anti-Rabbit IgG (H+L) AF488 conjugated | Thermo Fisher Scientific | Cat# A-21206, RRID:AB_2535792 |
| Donkey anti-Mouse IgG (H+L) AF488 conjugated | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 |
| Donkey anti-Goat IgG (H+L) AF647 conjugated | Thermo Fisher Scientific | Cat# A-21447, RRID:AB_141844 |
| Goat anti-Rat IgG (H+L) AF647 conjugated | Thermo Fisher Scientific | Cat# A-21247, RRID:AB_141778 |
| Goat anti-Rabbit IgG (H+L) AF647 conjugated | Thermo Fisher Scientific | Cat# A-21245, RRID:AB_2535813 |
| Goat anti-Chicken IgY (H+L) AF488 conjugated | Abcam | Cat# ab150173, RRID:AB_2827653 |
| Goat anti-Rabbit IgG (H+L) AF488 conjugated | Thermo Fisher Scientific | Cat# A-11034, RRID:AB_2576217 |
| Goat anti-Mouse IgG (H+L) AF488 conjugated | Thermo Fisher Scientific | Cat# A-11029, RRID:AB_2534088 |
| Donkey anti-rabbit IgG (H+L) AF555 conjugated | Thermo Fisher Scientific | Cat# A-31572, RRID:AB_162543 |
| AF488 anti-mouse CD133 (Prominin-1) (13A4) | Thermo Fisher Scientific | Cat# 53-1331-80, RRID:AB_529615 |
| Rat anti-mouse CD16/32 (2.4G2) | BD Biosciences | Cat# 553142, RRID:AB_394657 |
| PE/Cyanine7 anti-mouse CD45.2 antibody (104) | BioLegend | Cat# 109829, RRID:AB_1186103 |
| PerCP/Cy5.5 anti-mouse CD11c, (N418) | BD Biosciences | Cat# 560584, RRID:AB_1727422 |
| FITC anti-mouse CD103 (M290) | BD Biosciences | Cat# 557494, RRID:AB_396731 |
| APC-eF780 anti-mouse CD11b (M1/70) | eBioscience | Cat# 47-0112-82, RRID:AB_1603193 |
| AF647 anti-mouse XCR1 (ZET) | BioLegend | Cat# 148214, RRID:AB_2564369 |
| Rabbit anti-RFP (polyclonal) | Rockland | Cat# 600-401-379, RRID:AB_2209751 |
| Anti-mouse CD45 MicroBeads | Miltenyi Biotec | Cat# 130-052-301, RRID:AB_2877061 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| (Z)-4-Hydroxytamoxifen | Sigma-Aldrich | Cat# H7904 |
| Peanut oil | Sigma-Aldrich | Cat# P2144 |
| EdU (5-ethynyl-2’-deoxyuridine) | Thermo Fisher Scientific | Cat# E10187 |
| Tert-Butanol | Sigma-Aldrich | Cat# 360538 |
| Dichloromethane | Sigma-Aldrich | Cat# 270997 |
| Benzyl alcohol | Sigma-Aldrich | Cat# 305197 |
| Benzyl benzoate | Sigma-Aldrich | Cat# B6630 |
| Diphenyl ether | Alfa Aesar | Cat# A15791 |
| DL-alpha-Tocopherol | Alfa Aesar | Cat# A17039 |
| Laminin | Sigma-Aldrich | Cat# L2020 |
| Heparin | Sigma-Aldrich | Cat# H3393 |
| T3 hormone (3,3′,5-Triiodo-L-thyronine sodium salt) | Sigma-Aldrich | Cat# T2752 |
| Animal-Free Recombinant Murine FGF basic | Peprotech | Cat# AF-450-33 |
| Animal-Free Recombinant Murine EGF | Peprotech | Cat# AF-315-09 |
| Recombinant Murine IGF-I | Peprotech | Cat# 250-19 |
| Recombinant Murine BMP-4 | Peprotech | Cat# 315-27 |
| Critical commercial assays | ||
| Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 647 dye | Thermo Fisher Scientific | Cat# C10340 |
| RNAscope Multiplex FL V2 | Biotechne | Cat# 323110 |
| TSA plus Cyanine 5 | Perkin Elmer | Cat# NEL745001 |
| Neural Dissociation Kit (P) | Miltenyi biotec | Cat# 130-092-628 |
| Myelin removal beads II | Miltenyi biotec | Cat# 130-096-733, SCR_020279 |
| QIAShredder | QIAGEN | Cat# 79656 |
| RNeasy Mini kit | QIAGEN | Cat# 74004 |
| Deposited data | ||
| Mouse Brain Atlas | (Zeisel et al., 2018) | SRP135960 |
| Bulk RNA sequencing of DNGR-1 traced or hippocampal neural stem cells and astrocytic progenies | This paper | GSE145824 |
| Single Cell RNA sequencing of spinal cord DNGR-1 traced ependymal cells | This paper | GSE146226 |
| Single Cell RNA sequencing of spinal cord CD133+ DNGR-1 traced and CD133+ non-traced cells | This paper | GSE202959 |
| Experimental models: Organisms/strains | ||
| Clec9aCreRosaLSLtdTomato | (Schraml et al., 2013) | N/A |
| Clec9aCreRosaLSLEYFP | (Schraml et al., 2013) | N/A |
| Clec9aCreRosaLSLtdTomatoBatf3-/- | This paper | N/A |
| Clec9aCreRosaLSLtdTomatoFlt3l-/- | This paper | N/A |
| Clec9atdTomato | This paper | N/A |
| Clec9aCreERT2RosaLSLtdTomato | This paper | N/A |
| Oligonucleotides | ||
| RNAscope 2.5 LS Probe -Mm-Clec9a-01 | Biotechne | Cat# 537738 |
| Software and algorithms | ||
| Imaris (v.9.1.2) | Imaris software | https://imaris.oxinst.com/ |
| FlowJo (v10.8.1) | FlowJo | https://www.flowjo.com/ |
| ZEN black | ZEISS | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Prism (GraPad 9.2.0) | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| Seurat R package (v3) | Seurat | https://satijalab.org/seurat/ |
| Adobe InDesign (v17.1) | Adobe System | https://www.adobe.com/ |
| Biorender | Biorender | https://biorender.com/ |
| FIJI (v2.1.0/1.53c) | ImageJ | https://imagej.nih.gov/ij/ |
| R studio (3.5) | R software | https://www.rstudio.com/ |
| Other | ||
| Collagenase IV | Worthington | Cat# LS004188 |
| DNAse I | Roche | Cat# 11284932001 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L34962 |
| Fixation Medium A | Nordic MUbio | Cat# GAS-002A-1 |
| StemPro™ Accutase™ Cell Dissociation Reagent | Thermo Fisher Scientific | Cat# A1110501 |
| N-2 MAX Media Supplement | R and D Systems | Cat# AR009 |
| NeuroCult™ Proliferation Supplement | Stem Cell Technologies | Cat# 05701 |
| Matrigel | Corning | Cat# 354230 |
| B-27™ Supplement (50X), serum free | Thermo Fisher Scientific | Cat# 17504044 |
| Vybrant™ DiO Cell-Labeling Solution | Thermo Fisher Scientific | Cat# V22886 |
| FluoroMyelin™ Green Fluorescent Myelin Stain | Thermo Fisher Scientific | Cat# F34651 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Caetano Reis e Sousa (caetano@crick.ac.uk).
Material availability
All mouse lines generated in this study are available from the lead contact.
Experimental model and subject details
Mice
Clec9aCreERT2 (carrying allele Clec9atm4.1Crs) mice were generated by conventional gene targeting (knock-in) adding P2A CreERT2 to the Clec9a gene. Clec9atdTomato (carrying allele Clec9atm5.1Crs) mice were generated by knock-in of tdTomato with a BGH_pA at the ATG in exon 1 of the Clec9a gene. An FRT-flanked Neo cassette in the original constructs of both strains was removed by crossing to an FLPe expressing strain. Generation of Clec9aCreERT2 and Clec9atdTomato mice was contracted commercially (Ozgene, Perth, WA, Australia). Allele names refer to The Francis Crick Institute Nomenclature. Clec9aCre (Schraml et al., 2013), RosaLSLtdTomato, RosaLSLEYFP (Madisen et al., 2010), Batf3-/- (Hildner et al., 2008) and Flt3l-/- (McKenna et al., 2000) were described previously. All mice were bred at The Francis Crick Institute under specific pathogen-free conditions. All genetically modified mouse lines were backcrossed to C57BL/6j and six to fourteen-week-old mice were used in all experiments unless otherwise specified. Mouse genotypes from ear biopsies were determined using real time PCR with specific probes designed for each gene (Transnetyx, Cordova, TB). All animal experiments were performed in accordance with national and institutional guidelines for animal care and were approved by the Francis Crick Institute Biological Resources Facility Strategic Oversight Committee (incorporating the Animal Welfare and Ethical Review Body) and by the Home Office, UK.
Primary cell cultures
Neurosphere assay, neural stem cell adherent cultures and differentiation
Neurosphere assay. Clec9aCreRosaLSLtdTomato or Clec9aCreERT2RosaLSLtdTomato mice were sacrificed with i.p. injection of pentobarbital and perfused with 20ml of sterile PBS. Brains or spinal cords were digested into a single-cell suspension using Neural Tissue Dissociation kit (P) (Miltenyi) and 1.5x105 cells per cm2 were seeded per well (96 well plates) in DMEM:F12 (Thermo Fisher Scientific) supplemented with 0.015M KCl, 1% BSA, Neurocult (Stem Cell Technologies), bFGF (10ng/ml, Peprotech), EGF (20ng/ml, Peprotech), Heparin (2ug/ml, Sigma), antibiotics and glutamine (Neurosphere medium). 10-14 days after culture, plates were inspected using an EVOS XL Core microscope (Thermo Fished Scientific) and transmitted light and fluorescence images acquired. Wells containing tdTomato+ neurospheres were collected and neurospheres dissociated with StemPro Accutase (Thermo Fished Scientific) to generate a single-cell suspension. TdTomato+ cells were FACS sorted from primary neurospheres after StemPro Accutase dissociation and DAPI staining (to exclude dead cells) using an Aria Fusion (BD) with a 100μm nozzle. Purity checks were performed on sorted cells (<95% tdTomato+).
To assess potential DNGR-1 up-regulation triggered by in vitro culture, brains or spinal cords tissue from Clec9aCreERT2RosaLSLtdTomato mice were processed as above and further incubated with (Z)-4OH-TAM (500nM, Sigma) or DMSO at the onset of the assay and media was half-exchanged every 2/3 days.
For comparison of the neurosphere-forming potential between CD133+ DNGR-1-traced and CD133+ non-traced cells, spinal cords from Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato mice were digested and myelin-depleted as above and stained with anti-CD133 AF488 (1:100, clone 134A, BD) and anti-CD45 AF647 (1:100, clone 30F-11, Biolegend). CD133+CD45- DNGR-1-traced or non-traced cells were FACS-sorted on an Aria Fusion (BD) with a 100μm nozzle and 100 cells from each compartment seeded per well (96 well plate) in 100ul of neurosphere medium for 14 days.
Adherent cultures. Adherent cultures (Conti et al., 2005; Pollard et al., 2006) were established by plating (previously sorted) neurosphere-derived tdTomato+ single-cell suspensions in flasks pre-coated for at least 1 hour with DMEM:F12 (Thermo Fisher Scientific) supplemented with N-2 Max supplement (R&D systems), bFGF (20ng/ml, Peprotech), EGF (20ng/ml, Peprotech), Heparin (5ug/ml, Sigma), Laminin (2ug/ml, Sigma), antibiotics and glutamine. (NSC medium).
Astrocyte differentiation. To induce astrocyte differentiation (Bonaguidi et al., 2005), NSCs (5x104 cells) derived from adherent cultures were cultured in poly-l-lysine (20mg/ml) coated coverslips (13mm) in NSC medium overnight. On the following day, media was replaced with DMEM:F12 (Thermo Fisher Scientific) supplemented with BMP4 (20ng/ml, Peprotech) (astrocyte differentiation medium). After 3 days, media was replaced by freshly prepared astrocyte differentiation medium and cells incubated for another 3 days before cultures were terminated.
Oligodendrocyte differentiation. To induce oligodendrocyte differentiation (Hsieh et al., 2004; Panciera et al., 2016), NSCs (5x104 cells) derived from adherent cultures were cultured in poly-l-lysine (20mg/ml) coated coverslips (13mm) in NSC medium overnight. On the following day, media was replaced with Neurobasal medium (Thermo Fisher Scientific) supplemented with IGF-I (500ng/ml, Peprotech), 1xB27 supplement (Thermo Fisher Scientific), and T3 (30ng/ml, Sigma), antibiotics and glutamine (oligodendrocyte differentiation medium). After 3 days, media was replaced by freshly prepared oligodendrocyte differentiation medium and cells incubated for another 3 days before cultures were terminated.
Neuronal differentiation. To induce neuronal differentiation (Choi et al., 2014; Panciera et al., 2016), NSCs (5x104 cells) derived from adherent cultures were mixed with Matrigel (Corning) (diluted 1:15 in Neurobasal medium supplemented with 1x B27 + vitamin A (Thermo Fisher Scientific), antibiotics and glutamine (neuronal differentiation medium)) and added to a tissue culture well (24 wp) and allowed to form a thin-gel layer (1hr, 37C). After 3 days, media was replaced by freshly prepared neuronal differentiation medium and cells incubated for another 3 days before cultures were terminated.
In vitro cDC culture
In vitro cDC cultures were generated as described in Helft et al. (2015). Briefly, differentiated cDCs cells were prepared from day 9 bone marrow cultures grown from Clec9aCreERT2RosaLSLtdTomato mice in the presence of Flt3L (150 ng ml−1) exposed to (Z)-4OH-TAM (500nM, Sigma) or DMSO.
Method details
Radiation bone marrow chimeras
Clec9aCreRosaLSLEYFP mice were irradiated (two doses of 6.6 Gray separated by 4hours). On the following day, mice were i.v. injected with 2x106 total bone marrow cells isolated from Clec9aCreRosaLSLtdTomato mice. Animals were analyzed after 8 weeks or 1 year after reconstitution.
EdU administration in drinking water
10 week old Clec9aCreRosaLSLtdTomato mice were treated with EdU (Thermo Fisher Scientific) in drinking water (0.75mg/ml EdU + 1% sucrose) for a period of 30 days. This solution was protected from light and prepared fresh and changed every 3 days.
Immunofluorescence and confocal microscopy
Clec9aCreRosaLSLtdTomato mice were sacrificed with i.p. injection of pentobarbital and perfused with 20ml of PBS followed by 10 ml of 4% paraformaldehyde (PFA). Brains or spinal cords were dissected and further fixed in 4% PFA overnight (ON) at 4C. On the following day, tissues were washed in PBS for 1hr at 4C with agitation and transferred to 30% sucrose in PBS for 24hours at 4C. Tissues were finally embedded in O.C.T. (Tissue-Tek) and stored and -80C. Frozen tissue sections (10-20μm) were produced on a cryostat (Leica), mounted on SuperFrost Plus glass slides (Thermo Fisher Scientific) and stored at -80C. Before staining, sections were thawed at room temperature for 20 minutes, re-hydrated in PBS and blocked with 3% BSA, 0.3% Triton X-100 in PBS (blocking buffer) for 1 hour at room temperature in a humid chamber. Primary antibodies were diluted in blocking buffer and incubated ON at 4C in a humid chamber. TdTomato in direct reporter and inducible tracer was detected with anti-RFP.
For EdU incorporation detection, sections were permeabilized and incubated for 5 minutes with 3% BSA, 0.3% Triton X-100 and incubated with a Click-iT reaction cocktail solution (Click-iT Imaging kit, Thermo Fisher Scientific) containing 500μm AF647 PCA for 30 minutes at room temperature. Nuclei were counterstained with Hoechst. Images were quantified using ImageJ/FIJI for the proportion of DNGR-1-traced cells which had incorporated EdU per section in uninjured or injured spinal cords (lesion periphery and injury core).
For FoxJ1 and Hes1 staining, sections were subjected to epitope retrieval using citrate buffer (10mM NaCitrate, 0.05% Tween 20, pH 6, 3minutes at 80C) before blocking. Secondary antibodies were diluted in blocking buffer and incubated for 1 hour at room temperature in a humid chamber. Nuclei were counterstained with Hoechst and sections were mounted with ProLong Diamond Antifade (Thermo Fisher Scientific).
For the detection of DNGR-1 expressing cells in E11.5 Clec9atdTomato embryos or adult spinal cord tissue, sections were stained with a rabbit anti-RFP (rockland) polyclonal antibody followed by an AF555 donkey anti-rabbit secondary antibody.
Hes1 staining was quantified by drawing a nuclear mask defined by Hoechst staining and measuring the mean gray value intensity for Hes1 fluorescence channel on Fiji/ImageJ software (Schindelin et al., 2012).
To map the distribution of DNGR-1-traced cells across the brains of Clec9aCreRosaLSLtdTomato mice, brains were dissected and fixed in 10% neutral buffered formalin (NBF) ON at room temperature and moved to 70% ethanol on the following day. Tissues were embedded in paraffin and 4μm sections cut on a microtome. Paraffin was removed from sections by serial incubation through graded xylene and ethanol solutions.
Quantification of ependymal cell labeling in Clec9aCreRosaLSLtdTomato animals of different penetrance made by carrying Clec9aCre or RosaLSLtdTomato alleles in hetero- or homozygosity was performed in 20μm spinal cord sections by enumerating the fraction of DNRG-1-traced cells present in the ependymal cell layer.
For immunofluorescence of NSCs or astrocytes, oligodendrocytes or neuronal differentiated cultures, cells were cultured in poly-L-lysine coated (20mg/ml) coverslips, fixed (4%PFA, 30 minutes, room temperature), permeabilised (0.03% triton X-100, 30 minutes, room temperature) and blocked (3% BSA, 1 hour, room temperature) before being stained with primary antibodies (diluted in blocking buffer, 16 hours, 4C) and secondary antibodies (diluted in blocking buffer, 1 hour, room temperature). Coverslips were finally mounted in ProLong Diamond Antifade containing DAPI. Samples were imaged on a Zeiss LSM880 inverted confocal microscope and NSC cultures and their differentiated progeny samples were imaged using the Airyscan module.
Single molecule fluorescent in situ hybridisation (smFISH)
Clec9a mRNA expression was detected using the RNAscope Multiplex Fluorescent v2 assay combined with immunofluorescence (ACD) following manufacturer’s instructions. Lymph nodes or injured spinal cords from PBS and 4% paraformaldehyde (PFA) perfused Clec9a+/CreRosaLSLtdTomato mice were further fixed with 4% PFA (16h, 4C) and cryoprotected with 30% sucrose (16h, 4C) prior to making 10 μm thick frozen sections. Clec9a mRNA was detected using probe mm-Clec9a-01 (537731, ACD) in conjunction with TSA633 (NEL745001, Perkin Elmer). Sections were further stained with a rat anti-CD45 (1:100, clone 30-F11, Biolegend), detected with a donkey anti-rat AF488 (1:400, Thermo Fischer Scientific); and with a rabbit anti-RFP (1:600, polyclonal, Rockland), detected with a donkey anti-rabbit AF555 (1:400, polyclonal, Thermo Fischer Scientific). Nuclei were counterstained with Hoechst. Images were acquired on a LSM880 inverted confocal microscope (Zeiss). Number of Clec9a puncta per cell were quantified using ImageJ/FIJI. Background level of the assay was determined by counting the maximum number of Clec9a puncta in Clec9a-deficient animals (Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato).
Flow cytometry
Meninges, brains or spinal cords of PBS perfused Clec9aCreRosaLSLtdTomato mice were cut into small pieces and digested with Collagenase IV (200U/ml, Worthington) and DNase I (0.2mg/ml, Roche) in RPMI for 60 minutes at 37C. Digested tissues were strained though a 70μm cell strainer (BD Bioscience). Leukocytes were enriched in brain samples by Percoll gradient centrifugation (GE Healthcare). Nine parts Percoll were combined with one-part 10x PBS to obtain 100% Percoll. Cells were resuspended in 70% Percoll in PBS or HBSS/RPMI, overlaid with 37% and 30% Percoll and centrifuged at room temperature for 30min at 2000rpm without braking. Cells were collected at the 70/37% interface. DNGR-1 traced cDC1s or cDC2s were identified in meningeal and brain samples as singlets, LIVE, tdTomato+, CD45+ MHCII+, CD11c+ CD64-, CD103+ CD11b- (cDC1) or CD103- CD11b+ (cDC2). Spinal cord samples were not subjected to Percoll enrichment. All samples were Fc-blocked with anti-CD16/32 (clone 2.4G2, 1:100 BD Bioscience) prior to staining. Antibodies used: anti-CD45 PeCy7 (1:400, clone 104, Biolegend), anti-MHCII eF450 (1:200, clone M5.114.15.2, eBioscience), anti-CD11c PerCP Cy5.5 (1:100, clone N418, BD Bioscience), anti-CD64 AF647 (1:200, clone X54-5/7.1, Biolegend), anti-CD103 FITC (1:100, clone M290, BD Bioscience), anti-CD11b APCeF780 (1:200, clone M1/70, eBioscience). cDC1s were identified in in vitro cDC cultures by staining with anti-XCR1 AF647 (1:100, clone ZET, Biolegend). Dead cells were exclude by LIVE/DEAD Blue staining (Thermo Fisher).
Correlative light and electron microscopy
Fluorescence super-resolution microscopy
Clec9aCreRosaLSLtdTomato mice were sacrificed with pentobarbital and perfused with 20ml of pPBS followed by 10 ml of 4% paraformaldehyde (PFA). Spinal cords were dissected and further fixed in 4% PFA overnight (ON) at 4C. Spinal cords were embedded in 4% low melting point agarose (Life technologies) and 100μm vibratome sections were produced. Nuclei were counterstained with Hoechst and sections were imaged on a Zeiss LSM880 inverted confocal microscope using the Airyscan super-resolution module using Z-step correction (Z-step=100nm).
Once fluorescence microscopy was completed, the vibratome slices were further fixed in 2.5% glutaraldehyde and 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4) and processed according to the method of the National Centre for Microscopy and Imaging Research (Deerinck et al., 2010), before flat embedding in Durcupan resin between sheets of Aclar plastic.
SBF SEM data collection
Serial blockface scanning electron microscopy (SBF SEM) data was collected using a 3View2XP (Gatan, Pleasanton, CA) attached to a Sigma VP SEM (Zeiss, Cambridge). Flat embed vibratome slices were cut out and mounted on pins using conductive epoxy resin (Circuitworks CW2400). Each slice was trimmed using a glass knife to the smallest dimension in X and Y whilst retaining all the tissue, and the surface polished to reveal the tissue before sputter coating with a 2 nm layer of platinum, and loading in the 3View2XP. Two SBF SEM datasets were collected, both of which fully contained the fluorescence microscopy volume. Backscattered electron images were acquired using the 3VBSED detector at 8,192∗8,192 pixels with a dwell time of either 5 or 4 μs (10 nm reported pixel size for a horizontal frame width of 81.92 μm) and 50 nm slice thickness. The SEM was operated in variable pressure mode at a chamber pressure of either 10 or 5 pascals, with high current mode inactive. The 30 μm aperture was used, with an accelerating voltage of 2.5 kV. Dataset 1 comprised a total of 1,180 images, representing a depth of 59 μm, and volume of 395,942 μm3; dataset 2 comprised a total of 1,296 images, representing a depth of 64.8 μm, and volume of 434,865 μm3.
Image processing
Downstream image processing was carried out using Fiji/ImageJ (Schindelin et al., 2012). The images were batch converted to 8-bit tiff format, denoised using Gaussian blur (0.75 pixel radius), resharpened using two passes of unsharp mask (4 pixel radius 0.3 strength, 1 pixel radius 0.2 strength) and contrast enhanced (saturated pixels 0.2%, normalise); these parameters were tailored to suit the resolution and image characteristics of the datasets. Image registration was carried out using the ‘align virtual stack slices’ plugin, with a translation model used for feature extraction and registration. The aligned image stacks were calibrated for pixel dimensions, and cropped to regions of interest as required. To generate a composite of the volumes, Bigwarp (Bogovic et al., 2015; Russell et al., 2016) was used to map each fluorescence microscopy channel into the electron microscopy volume. After exporting the transformed fluorescence microscopy volumes from Bigwarp, the datasets were combined, and brightness/contrast adjusted for optimal on-screen presentation.
Multiphoton microscopy
300μm vibratome sections from spinal columns of Clec9aCreRosaLSLtdTomato mice were imaged using a Zeiss 710 NLO laser scanning multiphoton microscope equipped with a 20x 1.0 NA immersion lens. A pulsed Ti:sapphire laser (Spectra Physics MaiTai HP DeepSee) tuned to 900nm was used for excitation and emission wavelengths were detected through band-pass filters of 380-485nm (second harmonic signal) and 640-690nm (tdTomato). Images were analyzed and channels adjusted using Imaris (Bitplane).
Optical clearing of tissues
Whole central nervous system (CNS), spinal cords or E11.5 embryos from Clec9aCreRosaLSLtdTomatomice were optically cleared using uDISCO (Pan et al., 2016). Briefly, mice were sacrificed with pentobarbital and perfused with 20ml of PBS followed by 10 ml of 4% paraformaldehyde (PFA). Tissues were dissected and further fixed in 4% PFA overnight (ON) at 4C. On the following day, tissues were washed in PBS for 1 hour at 4C and nuclei were stained with Hoechst diluted in PBS and incubated ON at 4C. Tissues were dehydrated by serial incubation in tert-butanol solutions of increasing concentration (Sigma, 360538) 30% vol tert-butanol, 50% vol tert-butanol, 70% vol tert-butanol, 80% vol tert-butanol, 90% vol tert-butanol and 96% vol tert-butanol. Spinal cord and embryos were incubated in each solution for at least 4 hours at room temperature with agitation, protected from light. Whole CNS was further incubated with 100% vol tert-butanol and incubated in each solution for at least 16 hours at 35C with agitation, protected from light. Whole CNS was further incubated with a delipidation solution of pure dichloromethane (Sigma, 270997) for 70 minutes at room temperature with agitation. Finally, all tissues were incubated in BABB:D4 - prepared by mixing BABB (benzyl alcohol + benzyl benzoate 1:2, Sigma, 305197 and B6630) with diphenyl ether (DPE) (Alfa Aesar, A15791) at a ratio of 4:1 and adding 0.4% vol DL-alpha-tocopherol (Vitamin E) (Alfa Aesar, A17039) for at least 48hrs with agitation before imaging.
Light-sheet microscopy
uDISCO clarified CNS, spinal cord or embryo from Clec9acreRosaLSLtdtomato mice were imaged on a Miltenyi-LaVision BioTec Ultramicroscope II light-sheet microscope. Tissues were mounted on a sample holder and imaged in BABB-D4 solution with an Olympus MVPLAPO 2x /0.5 NA with a protective dipping cap (WD > 5.7 mm). Tissues were excited with a bi-directional 561nm wavelength distributed across three Gaussian light-sheets with a NA of 0.09 and exposed for 200ms. The step size between each image was 5 μm. Images from both light-sheets were acquired with an Andor Zyla 5.5 sCMOS camera and merged using the blend function in Imspector Pro software. A zoom of 1.25x-1.6x was used for imaging of E11.5 embryos. A zoom of 0.63x was used for imaging of whole CNS and volumes were stitched using BigStitcher (Hörl et al., 2019) in Fiji/ImageJ software (Schindelin et al., 2012). DNGR-1 traced surfaces were generated using Imaris software (Bitplane). Given the lower tdTomato fluorescence of DNGR-1 traced cDCs (Figures S1C and S5C) a low laser power was used to preferentially illuminate the bright tdTomato ependymal cell compartment in injured spinal cords (Figure 7A).
Embryonic tracking and in utero induction of DNGR-1 lineage tracing
Embryos were dated taking the day of the plugs as embryonic day (E) 0.5 and processed for histological analysis or wholemount clarification as detailed above.
For in utero induction of DNGR-1lineage tracing, Clec9aCreERT2RosaLSLtdTomato females at E10.5 and E11.5 stages of pregnancy were intraperitoneally injected with 5ul/g of body weight of a tamoxifen (T5648, Sigma) solution (20mg/ml) made in peanut oil (P2144, Sigma).
Spinal cord injury contusion model
Only female animals were used in spinal cord injury experiments. Two weeks prior to surgery mice went through a period of handling and acclimatization, during which body weight was assessed to ensure ideal surgical weight (18-20 g). Animals (10-11 weeks old) were anesthetized using a cocktail of ketamine (120 mg/kg) and xylazine (16 mg/kg) administered by intraperitoneal (ip) injection. All surgeries were performed under aseptic conditions. For spinal contusion injuries, a laminectomy of the ninth thoracic vertebra (T9), identified based on anatomical landmarks, was first performed (Harrison et al., 2013), followed by a moderate (75 kdynes; displacement: 600-700 μm) contusion using the Infinite Horizon Impactor (Precision Systems and Instrumentation, LLC.) (Scheff et al., 2003). After spinal cord injury, the muscle and skin were closed with 4.0 polyglycolic absorbable sutures (Safil, G1048213). Animals were injected with 0.5 ml of saline subcutaneously (s.c.) then placed onto warmed cages (35°C) until they recovered from anaesthesia and for the following recovery period (3 days). To prevent dehydration, mice were supplemented daily with saline (0.5 ml, s.c.) for the first 5 dpi. Bladders were manually voided twice daily for the duration of the experiment (Basso et al., 2006).
White matter sparing analysis to determine lesion epicentre
Segments from injured spinal cords containing the T9 region were processed for immunofluorescence as detailed above. 10μm-thick frozen serial sections spaced 100μm apart were stained with Fluoromyelin Green (Thermo Fisher Scientific) to identify white matter regions (myelinated) and counterstained with Hoechst. Sections were imaged on a Zeiss LSM880 inverted confocal microscope and images analysed with Fiji/ImageJ. Total cross-sectional area (TCA) and white matter area (WMA, fluoromyelin+) were calculated using Fiji/ImageJ and the proportional relationship WMA/TCA was calculated across the entire series of sections. The lowest WMA/TCA value (corresponding to the section with the smallest myelinated area) was defined as the lesion epicentre and lesion coordinates (rostrally and caudally) were extrapolated.
EdU administration to spinal cord injured animals
Mice were daily I.P. injected with 50mg/Kg EdU in 200ul of PBS for 7 consecutive days after contusion spinal cord injury.
Ventricular injury
Mice were anesthetized with 2% isofluorane, the head shaved and placed in a custom-built stereotaxic apparatus where anesthesia was maintained throughout the entire experiment with 0.5–1% isoflurane in O2. A craniotomy was performed bilaterally (0.5mm post-bregma, 1.5mm and -1.5mm mediolateral) and the dura was removed around the penetration site. A 30g needle (311.2μm outer diameter) was mounted on the stereotaxic apparatus, dipped into the green lipophilic dye DiOC18(3) (Thermo Fisher Scientific) and a 2.5mm dorsoventral incision was made bilaterally. Skin was sutured and mice allowed to recover in a heating chamber (35C) until fully recovered from anaesthesia. Clec9aCreERT2RosaLSLtdTomato mice were intraperitoneally injected with 5ul/g of body weight of a tamoxifen (T5648, Sigma) solution (20mg/ml) made in peanut oil (P2144, Sigma) 1-2 hours before injury and after 3- and 7-days post injury. 4 weeks after ventricular injury, mice were sacrificed as above and brains dissected. Serial 100μm vibratome sections were cut and stained as in described elsewhere (Cabeza-Cabrerizo et al., 2019).
Bulk RNA sequencing of NSC and astrocyte cultures
Sample preparation
Samples were prepared as illustrated in Figure S4D. Briefly, passage 10 NSCs adherent cultures generated from spinal cords of Clec9acreRosaLSLtdtomato, Clec9acreRosaLSLEYFP or from the hippocampi of GlastCreHuwe1flRosaLSLEYFPmice were either passaged, re-seeded as NSCs in adherent conditions or induced to differentiate into astrocytes (see above). NSCs seeded in adherent conditions were recovered with StemPro accutase (Thermo Fisher Scientific) after 4 days, whilst differentiated astrocytes were recovered after 6 days. Cells from either condition were lysed with RLT buffer (QIAGEN) supplemented with 2-mercapoethanol and cell lysates stored at -80C. Three replicates for each cell fate (3x106NSCs or astrocytes) generated from passages P10, P11 and P12 adherent NSCs were sequenced.
Library preparation and RNA-sequencing
Biological replicate libraries were prepared using the polyA KAPA mRNA HyperPrep Kit and sequenced on Illumina HiSeq 4000 platform, generating ∼26 million 75bp single-end reads per sample. The RSEM package (version 1.3.30) (Li and Dewey, 2011) in conjunction with the STAR alignment algorithm (version 2.5.2a) (Dobin et al., 2013) was used for the mapping and subsequent gene-level counting of the sequenced reads with respect to Ensembl mouse GRCm.38.89 version transcriptome. Normalisation of raw count data and differential expression analysis was performed with the DESeq2 package (version 1.18.1) (Love et al., 2014) within the R programming environment (version 3.4.3). Differentially expressed genes were defined as those showing statistically significant differences between the Stem Cell and Astrocytes Groups (FDR <0.05). Gene lists ranked by the Wald statistic were used to look for pathway and biological process enrichment using the Broad’s GSEA software (version 2.1.0) with genesets from MSigDB (version 6) (Subramanian et al., 2005).
Single-cell RNA sequencing
Single-cell sample preparation for spinal cord DNGR-1 traced cells:
Clec9aCreRosaLSLtdTomato mice were sacrificed with i.p. injection of pentobarbital, perfused with 20ml of sterile PBS, spinal cords dissected and digested into a single-cell suspension using Neural Tissue Dissociation kit (P) (Miltenyi Biotec) following manufacture instructions. Sample was enriched for ependymal cells by further processing with myelin removal magnetic beads II (Miltenyi Biotec) to remove myelinated cells by passage through a MACS column (Miltenyi Biotec). Cells were then stained with FITC-conjugated anti-CD45.2 (clone 104), incubated with anti-FITC magnetic MicroBeads (Miltenyi Biotec) and put through another MACS column to remove CD45+ cells. Cells were finally stained with APC-conjugated CD45 (clone 30-F11) and DAPI (to exclude dead cells) and single DNGR-1 traced (tdTomato+) CD45- DAPI- cells were FACS sorted on an Aria Fusion (BD) with a 100μm nozzle. QC confirmed viability <95% and cells were immediately loaded onto 10X Genomics Chromium according to manufacture instructions.
Single-cell sample preparation for comparison between CD133+ DNGR-1 traced and CD133+ non-traced cells from spinal cords
Spinal cords from Clec9aCre/CreRosaLSLtdTomato/LSLtdTomato mice were processed as above. Myelin-depleted single cell suspensions were stained with APC-conjugated CD45 (clone 30-F11) and AF488-conjugated CD133 (clone 13A4) and DAPI (to exclude dead cells). Single DAPI- CD45- CD133+ DNGR-1 traced or DAPI- CD45- CD133+ non- traced cells were FACS sorted on an Aria Fusion (BD) with a 100μm nozzle into two different Eppendorf tubes. QC confirmed viability <95% and cells were immediately loaded onto 10X Genomics Chromium according to manufacture instructions.
Single-cell library preparation and RNA-sequencing
Library generation for 10x Genomics analysis were performed following the Chromium Single Cell 3′ Reagents Kits (10x Genomics) and sequenced on an Hiseq4000 (Illumina), to achieve an average of ∼280,000 reads per cell. Raw reads were initially processed by the Cell Ranger v.3.0.2 pipeline, which deconvolved reads to their cell of origin using the UMI tags, aligned these to the mm10 transcriptome (to which we added the tdTomato-WPRE sequence to detect tdTomato-expressing cells) using STAR (v.2.5.1b) and reported cell-specific gene expression count estimates. All subsequent analyses were performed in R v.3.6.0 using the Seurat (v3) package (Butler et al., 2018). Genes were considered to be ‘expressed’ if the estimated (log10) count was at least 0.1. Primary filtering was then performed by removing from consideration: cells expressing fewer than 50 genes and cells for which mitochondrial genes made up greater than 10% of all expressed genes. PCA decomposition was performed and, after consideration of the eigenvalue ‘elbow-plots’, the first 20 components were used to construct the UMAP plot, showing 6 distinct clusters. Cluster specific gene markers were identified using a wilcoxon rank sum test and the top 10 or 20 genes ranked by logFC per cluster were used to generate a heatmap.
Samples CD133+ DNGR-1 traced and CD133+ non-traced were sequenced and analysed as detailed above. Samples were integrated using Seurat’s ‘IntegrateData’ function, after identifying 2000 anchor features (which did not include the tdTomato-WPRE sequence, excluding that its expression influenced UMAP construction and cellular clustering). Three clusters were designated using the expression of various markers. Cells belonging to cluster 2 were subsetted and reclustered, resulting in seven clusters. Clusters 4 and 6 were removed due to the absence of ependymal cell markers (135 cells in total, of which 7 were CD133+ DNGR-1 traced, corresponding to 1.8% of all cells in cluster 2). Cluster markers was identified using a Wilcoxon Rank Sum test and used the package ‘ClusterProfiler’ (Wu et al., 2021) used to identify enriched Gene Ontology biological processes per cluster.
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism software (GraphPad). Normal distribution within each group was first confirmed using the Kolmogorov-Smirnov test. Statistical significance between two groups was determined using an unpaired two-tailed Student’s t test. Significance was assumed with ∗p<0.05, ∗∗∗∗p<0.0001.
Acknowledgments
We thank the members of the Immunobiology Laboratory for helpful discussions and suggestions. We thank the BRF, Bioinformatics and Biostatistics, Advanced Light Microscopy, Experimental Histopathology, and Flow Cytometry STPs for their support throughout this project. We are also grateful to Ryoichiro Kageyama (Kyoto University) for kindly providing an anti-HES1 antibody. Cartoons and graphical abstract were created with BioRender.com. This work was supported by The Francis Crick Institute, which receives core funding from Cancer Research UK (FC001136), the UK Medical Research Council (FC001136), and the Wellcome Trust (FC001136), by an ERC Advanced Investigator grant (AdG 268670), by a Wellcome Investigator Award (WT106973/Z/15/Z), and by a prize from the Louis-Jeantet Foundation. This research was funded in whole, or in part, by the Wellcome Trust (grants FC001136 and 106973/Z/15/Z). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author contributions
B.F. conducted experiments with assistance from F. Gasparrini. I.M. and D.C. performed spinal cord injury model. T.A. performed ventricular injury model. B.A. and J.C. assisted with histology. C.P. and P.C. carried out bioinformatic analysis. C.J.P. performed electron microscopy. N.R. managed mouse stocks. J.B., F. Guillemot, A.T.S., and L.S. provided advice and contributed to experimental design. B.F. and C.R.S. designed the study, interpreted the data, and wrote the manuscript.
Declaration of interests
C.R.S. is a founder of Adendra Therapeutics and owns stock options and/or is a paid consultant for Adendra Therapeutics, Bicara Therapeutics, Montis Biosciences, Bicycle Therapeutics, Genor Biopharma, and Sosei Heptares, all unrelated to this work. C.R.S. also has an additional appointment as Visiting Professor in the Faculty of Medicine at Imperial College London and holds honorary professorships at University College London and King’s College London.
Published: August 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2022.07.012.
Contributor Information
Bruno Frederico, Email: bruno.frederico@crick.ac.uk.
Caetano Reis e Sousa, Email: caetano@crick.ac.uk.
Supplemental information
Data and code availability
-
•
This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
Bulk and single cell RNAseq datasets have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- Aimone J.B., Li Y., Lee S.W., Clemenson G.D., Deng W., Gage F.H. Regulation and function of adult neurogenesis: From genes to cognition. Physiol. Rev. 2014;94:991–1026. doi: 10.1152/physrev.00004.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Cervello C., Soriano-Navarro M., Mirzadeh Z., Alvarez-Buylla A., Garcia-Verdugo J.M. Biciliated ependymal cell proliferation contributes to spinal cord growth. J. Comp. Neurol. 2012;520:3528–3552. doi: 10.1002/cne.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandasabapathy N., Victora G.D., Meredith M., Feder R., Dong B., Kluger C., Yao K., Dustin M.L., Nussenzweig M.C., Steinman R.M., et al. Flt3L controls the development of radiosensitive dendritic cells in the meninges and choroid plexus of the steady-state mouse brain. J. Exp. Med. 2011;208:1695–1705. doi: 10.1084/jem.20102657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J.H., Hatakeyama J., Sakamoto S., Ohtsuka T., Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Barker N., Es J.H. van, Kuipers J., Kujala P., Born M. van den, Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker R.A., Götz M., Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. doi: 10.1038/s41586-018-0087-1. [DOI] [PubMed] [Google Scholar]
- Barnabé-Heider F., Göritz C., Sabelström H., Takebayashi H., Pfrieger F.W., Meletis K., Frisén J. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell. 2010;7:470–482. doi: 10.1016/j.stem.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Basso D.M., Fisher L.C., Anderson A.J., Jakeman L.B., McTigue D.M., Popovich P.G. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- Becker C.G., Becker T. Neuronal regeneration from ependymo-radial glial cells: cook, little pot, cook. Dev. Cell. 2015;32:516–527. doi: 10.1016/j.devcel.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Becker C.G., Becker T., Hugnot J.-P. The spinal ependymal zone as a source of endogenous repair cells across vertebrates. Prog. Neurobiol. 2018;170:67–80. doi: 10.1016/j.pneurobio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C., et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Benner E.J., Luciano D., Jo R., Abdi K., Paez-Gonzalez P., Sheng H., Warner D.S., Liu C., Eroglu C., Kuo C.T. Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature. 2013;497:369–373. doi: 10.1038/nature12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic J.A., Hanslovsky P., Wong A., Saalfeld S. Robust registration of calcium images by learned contrast synthesis. Preprint at arxiv. 2015 doi: 10.1109/isbi.2016.7493463. [DOI] [Google Scholar]
- Bonaguidi M.A., McGuire T., Hu M., Kan L., Samanta J., Kessler J.A. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development. 2005;132:5503–5514. doi: 10.1242/dev.02166. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza-Cabrerizo M., Blijswijk J. van, Wienert S., Heim D., Jenkins R.P., Chakravarty P., Rogers N., Frederico B., Acton S., Beerling E., et al. Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza-Cabrerizo M., Cardoso A., Minutti C.M., Pereira da Costa M., Reis e Sousa C. Dendritic cells revisited. Annu. Rev. Immunol. 2021;39:131–166. doi: 10.1146/annurev-immunol-061020-053707. [DOI] [PubMed] [Google Scholar]
- Carlén M., Meletis K., Göritz C., Darsalia V., Evergren E., Tanigaki K., Amendola M., Barnabé-Heider F., Yeung M.S.Y., Naldini L., et al. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci. 2009;12:259–267. doi: 10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D’Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigliola V., Becker C.J., Poss K.D. Building bridges, not walls: spinal cord regeneration in zebrafish. Dis. Model. Mech. 2020;13 doi: 10.1242/dmm.044131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Watt F.M. Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 2018;87:1015–1027. doi: 10.1146/annurev-biochem-062917-012341. [DOI] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun V., Wu H., Blanchi B., Tsao S., Kim K., Zhao J., Biancotti J.C., Hutnick L., Krueger R.C., Fan G., et al. CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc. Natl. Acad. Sci. USA. 2008;105:1026–1031. doi: 10.1073/pnas.0710000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P.K., Mach S.A., Moore A.N. Enhanced neurogenesis in the rodent hippocampus following traumatic brain injury. J. Neurosci. Res. 2001;63:313–319. doi: 10.1002/1097-4547(20010215)63:4<313::AID-JNR1025>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Deerinck T., Bushong E.A., Lev-Ram V., Shu X., Tsien R.Y., Ellisman M.H. Enhancing Serial Block-Face Scanning Electron Microscopy to Enable High Resolution 3-D Nanohistology of Cells and Tissues. Microsc. Microanal. 2010;16:1138–1139. [Google Scholar]
- Devaraju K., Barnabé-Heider F., Kokaia Z., Lindvall O. FoxJ1-expressing cells contribute to neurogenesis in forebrain of adult rats: evidence from in vivo electroporation combined with piggyBac transposon. Exp. Cell Res. 2013;319:2790–2800. doi: 10.1016/j.yexcr.2013.08.028. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics Oxf. Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Caillé I., Lim D.A., García-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F.H. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Ghazale H., Ripoll C., Leventoux N., Jacob L., Azar S., Mamaeva D., Glasson Y., Calvo C.-F., Thomas J.-L., Meneceur S., et al. RNA profiling of the human and mouse spinal cord stem cell niches reveals an embryonic-like regionalization with MSX1+ roof-plate-derived cells. Stem Cell Rep. 2019;12:1159–1177. doi: 10.1016/j.stemcr.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub R., Cumano A. Embryonic hematopoiesis. Blood Cells Mol. Dis. 2013;51:226–231. doi: 10.1016/j.bcmd.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Harris L., Guillemot F. HES1, two programs: promoting the quiescence and proliferation of adult neural stem cells. Genes Dev. 2019;33:479–481. doi: 10.1101/gad.325761.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M., O’Brien A., Adams L., Cowin G., Ruitenberg M.J., Sengul G., Watson C. Vertebral landmarks for the identification of spinal cord segments in the mouse. NeuroImage. 2013;68:22–29. doi: 10.1016/j.neuroimage.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., Reis e Sousa C. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörl D., Rojas Rusak F.R., Preusser F., Tillberg P., Randel N., Chhetri R.K., Cardona A., Keller P.J., Harz H., Leonhardt H., et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods. 2019;16:870–874. doi: 10.1038/s41592-019-0501-0. [DOI] [PubMed] [Google Scholar]
- Hsieh J., Aimone J.B., Kaspar B.K., Kuwabara T., Nakashima K., Gage F.H. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J. Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Ang S.L., Shiota K., Nakanishi S., Kageyama R., Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Moriyoshi K., Sasai Y., Shiota K., Nakanishi S., Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet B.V., Salinas-Mondragon R., Liang H., Therit B., Buie J.D., Dykstra M., Campbell K., Ostrowski L.E., Brody S.L., Ghashghaei H.T. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh V. Development of brain ventricular system. Cell. Mol. Life Sci. 2018;75:375–383. doi: 10.1007/s00018-017-2605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K., Watt F.M. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Floriddia E.M., Toskas K., Chalfouh C., Honore A., Aumont A., Vallières N., Lacroix S., Fernandes K.J.L., Guérout N., et al. FoxJ1 regulates spinal cord development and is required for the maintenance of spinal cord stem cell potential. Exp. Cell Res. 2018;368:84–100. doi: 10.1016/j.yexcr.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Liu Q., Jin W.-N., Liu Y., Shi K., Sun H., Zhang F., Zhang C., Gonzales R.J., Sheth K.N., La Cava A.L., et al. Brain ischemia suppresses immunity in the periphery and brain via different neurogenic innervations. Immunity. 2017;46:474–487. doi: 10.1016/j.immuni.2017.02.015. [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Chell J.M., Le Merre P.L., Wu Y., Zamboni M., Bergenstråhle J., Stenudd M., Sopova E., Lundeberg J., Shupliakov O., et al. A latent lineage potential in resident neural stem cells enables spinal cord repair. Science. 2020;370:137–138. doi: 10.1126/science.abb8795. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Coskun V., Liang A., Yu J., Cheng L., Ge W., Shi Z., Zhang K., Li C., Cui Y., et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161:1175–1186. doi: 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald A., Lu B., Caron M., Caporicci-Dinucci N., Hatrock D., Petrecca K., Bourque G., Stratton J.A. Single cell transcriptomics of ependymal cells Across age, region and species reveals cilia-related and metal ion regulatory roles as major conserved ependymal cell functions. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.703951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P., Hack M.A., Hartfuss E., Kettenmann H., Klinkert W., Kirchhoff F., Götz M. Neuronal or glial progeny regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Malatesta P., Hartfuss E., Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- McCarthy M., Turnbull D.H., Walsh C.A., Fishell G. Telencephalic neural progenitors appear to be restricted to regional and glial fates before the onset of neurogenesis. J. Neurosci. 2001;21:6772–6781. doi: 10.1523/jneurosci.21-17-06772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T.D., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- Meletis K., Barnabé-Heider F., Carlén M., Evergren E., Tomilin N., Shupliakov O., Frisén J. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 2008;6:e182. doi: 10.1371/journal.pbio.0060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N., Brumm A., Zhang X., Carmichael S.T., Ghashghaei H.T. Foxj1 expressing ependymal cells do not contribute new cells to sites of injury or stroke in the mouse forebrain. Sci. Rep. 2018;8:1766. doi: 10.1038/s41598-018-19913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N., Vijayakumar A., Cheng G., Ghashghaei H.T. A knock-in Foxj1(CreERT2::GFP) mouse for recombination in epithelial cells with motile cilia. Genesis. 2014;52:350–358. doi: 10.1002/dvg.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K., Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059. doi: 10.1242/dev.156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Álvarez G., Daclin M., Shihavuddin A., Lansade P., Fortoul A., Faucourt M., Clavreul S., Lalioti M.-E., Taraviras S., Hippenmeyer S., et al. Adult neural stem cells and multiciliated ependymal cells share a common lineage regulated by the geminin family members. Neuron. 2019;102:159–172.e7. doi: 10.1016/j.neuron.2019.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Cai R., Quacquarelli F.P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Ertürk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Fujimura A., Di Biagio D.D., Frasson C., Bresolin S., Soligo S., Basso G., Bicciato S., Rosato A., et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J.M., Lowenstein D.H. Seizure-induced neurogenesis: are more new neurons good for an adult brain? Prog. Brain Res. 2002;135:121–131. doi: 10.1016/S0079-6123(02)35012-X. [DOI] [PubMed] [Google Scholar]
- Pollard S.M., Conti L., Sun Y., Goffredo D., Smith A. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb. Cortex. 2006;16:i112–i120. doi: 10.1093/cercor/bhj167. [DOI] [PubMed] [Google Scholar]
- Post Y., Clevers H. Defining adult stem cell function at its simplest: the ability to replace lost cells through mitosis. Cell Stem Cell. 2019;25:174–183. doi: 10.1016/j.stem.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Quadrato G., Elnaggar M.Y., Giovanni S.D. Adult neurogenesis in brain repair: cellular plasticity vs. cellular replacement. Front Neurosci.-Switz. 2014;8:17. doi: 10.3389/fnins.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E., Fernández A., Velasco P., de Andrés B., Liste I., Sancho D., Gaspar M.L., Cano E. DNGR-1(+) dendritic cells are located in meningeal membrane and choroid plexus of the noninjured brain. Glia. 2015;63:2231–2248. doi: 10.1002/glia.22889. [DOI] [PubMed] [Google Scholar]
- Raven A., Lu W.-Y., Man T.Y., Ferreira-Gonzalez S., O’Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Ao Y., O’Shea T.M., Burda J.E., Bernstein A.M., Brumm A.J., Muthusamy N., Ghashghaei H.T., Carmichael S.T., Cheng L., et al. Ependymal cell contribution to scar formation after spinal cord injury is minimal, local and dependent on direct ependymal injury. Sci. Rep. 2017;7 doi: 10.1038/srep41122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds B.A., Rietze R.L. Neural stem cells and neurospheres--re-evaluating the relationship. Nat. Methods. 2005;2:333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Russell M.R.G., Lerner T.R., Burden J.J., Nkwe D.O., Pelchen-Matthews A., Domart M.-C., Durgan J., Weston A., Jones M.L., Peddie C.J., et al. 3D correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy. J. Cell Sci. 2017;130:278–291. doi: 10.1242/jcs.188433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelström H., Stenudd M., Réu P., Dias D.O., Elfineh M., Zdunek S., Damberg P., Göritz C., Frisén J. Resident neural stem cells restrict tissue damage and neuronal loss after spinal cord injury in mice. Science. 2013;342:637–640. doi: 10.1126/science.1242576. [DOI] [PubMed] [Google Scholar]
- Scheff S.W., Rabchevsky A.G., Fugaccia I., Main J.A., Lumpp J.E. Experimental modeling of spinal cord injury: characterization of a force-defined injury device. J. Neurotrauma. 2003;20:179–193. doi: 10.1089/08977150360547099. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml B.U., van Blijswijk J., Zelenay S., Whitney P.G., Filby A., Acton S.E., Rogers N.C., Moncaut N., Carvajal J.J., Reis e Sousa C. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Seri B., García-Verdugo J.M., McEwen B.S., Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J. Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.T., Stratton J.A., Stykel M.G., Abbasi S., Sharma S., Mayr K.A., Koblinger K., Whelan P.J., Biernaskie J. Single-cell transcriptomics and fate mapping of ependymal cells reveals an absence of neural stem cell function. Cell. 2018;173:1045–1057.e9. doi: 10.1016/j.cell.2018.03.063. [DOI] [PubMed] [Google Scholar]
- Spalding K.L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H.B., Boström E., Westerlund I., Vial C., Buchholz B.A., et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenudd M., Sabelström H., Frisén J. Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 2015;72:235–237. doi: 10.1001/jamaneurol.2014.2927. [DOI] [PubMed] [Google Scholar]
- Stenudd M., Sabelström H., Llorens-Bobadilla E., Zamboni M., Blom H., Brismar H., Zhang S., Basak O., Clevers H., Göritz C., et al. Identification of a discrete subpopulation of spinal cord ependymal cells with neural stem cell properties. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110440. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueda R., Kageyama R. Regulation of active and quiescent somatic stem cells by Notch signaling. Dev. Growth Differ. 2020;62:59–66. doi: 10.1111/dgd.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A., et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Ng C.P., Habacher H., Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., Zwan J. van der, Häring M., Braun E., Borm L.E., La Manno G.L., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Melton D.A. Pancreas regeneration. Nature. 2018;557:351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Continuum of DNGR-1-traced cells (red) from the sacral part of the spinal cord to the brain ventricular compartment.
Dorsal perspective of the spinal cord counterstained with Hoechst (blue) demonstrating DNGR-1-traced cells (red) lining the central canal.
Brain DNGR-1-traced surfaces (red) lining the ventricular compartment. Surfaces were calculated with Imaris software. Brain contour was defined by tissue autofluorescence and DNGR-1-traced surfaces were defined by bright tdTomato fluorescence.
Side-by-side comparison of spinal cords from Clec9aCreRosaLSLtdTomato and its cDC deficient counterpart (Clec9aCreRosaLSLtdTomatoFlt3l−/−) showing no reduction of DNGR-1- traced cells (red) lining the central canal of the spinal cord. Nuclei were counterstained with Hoechst (blue).
Representative vibratome section imaged with the airyscan super-resolution module which reveals two cilia emerging from ependymal DNGR-1-traced cells (red). Nuclei were counterstained with Hoechst (blue).
Tracking of an individual cilium belonging to an DNGR-1-traced ependymal cell (red), from its central canal extension to its respective basal body.
Location of DNGR-1-traced cells (bright red) in response to spinal cord injury around the lesion epicentre. DNGR-1-traced cells show displacement for their central canal location toward the injury site, which was applied dorsally. Nuclei were counterstained with Hoechst (blue).
Data Availability Statement
-
•
This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
Bulk and single cell RNAseq datasets have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.