Abstract
Genome editing tools such as CRISPR/Cas9 enable the rapid and precise manipulation of genomes. CRISPR-based genome editing has greatly simplified the study of gene function in cell lines, but its widespread use has also highlighted challenges of reproducibility. Phenotypic variability among different knockout clones of the same gene is a common problem confounding the establishment of robust genotype–phenotype correlations. Optimized genome editing protocols to enhance reproducibility include measures to reduce off-target effects. However, even if current state-of-the-art protocols are applied phenotypic variability is frequently observed. Here we identify heterogeneity of wild-type cells as an important and often neglected confounding factor in genome-editing experiments. We demonstrate that isolation of individual wild-type clones from an apparently homogenous stable cell line uncovers significant phenotypic differences between clones. Strikingly, we observe hundreds of differentially regulated transcripts (477 up- and 306 downregulated) when comparing two populations of wild-type cells. Furthermore, we show a variety of cellular and biochemical alterations in different wild-type clones in a range that is commonly interpreted as biologically relevant in genome-edited cells. Heterogeneity of wild-type cells thus contributes to variability in genome-edited cells when these are generated through isolation of clones. We show that the generation of monoclonal isogenic wild-type cells prior to genomic manipulation reduces phenotypic variability. We therefore propose to generate matched isogenic control cells prior to genome editing to increase reproducibility.
Subject terms: Cell biology, Nephrology
Introduction
In the last decade, genome editing tools such as CRISPR/Cas9 have transformed the ease and precision of site-specific genome modifications1–3. Among many other applications, these molecular tools enable efficient genome editing of somatic cell lines to study effects of loss- or gain-of-function alleles4. Cultured cell lines have since been widely used to study gene function and to develop cell-based models of human disease5,6. The increasing number of such studies has drawn attention to the difficulty of establishing robust genotype–phenotype correlations and reproducible results using genome-edited cell lines.
The general challenges of achieving reproducible results with cell culture experiments have been well documented7. A wide range of factors, among them growth conditions, passage number, genetic drift, undetected contaminations or differing cell line origins, can lead to phenotypic variability and influence reproducibility of results8–11. In genome-edited cells, off-target effects have been described as a cause of phenotypic variability among different knockout (KO) clones of the same gene12. Therefore, extensive efforts have been devoted to reducing off-target effects in genome-edited cells13–16. However, even if current state-of-the-art protocols for genome editing are employed, considerable phenotypic variability can be observed.
We have encountered this problem studying the genes causing Autosomal Dominant Polycystic Kidney Disease (ADPKD), the most common monogenic disorder resulting in kidney failure. Mutations in the genes Polycystic kidney disease 1 (PKD1) and Polycystic kidney disease 2 (PKD2) cause ADPKD. There is an unmet need for robust cell-based models for ADPKD to study pathogenic pathways and to identify drug targets through high-throughput screening. We therefore applied CRISPR/Cas9 to generate KO cells of Pkd1 and Pkd2 in different well-established kidney epithelial cell lines17. To reduce variability at the gene level, we generated null alleles of both genes by deleting the entire coding region. This eliminates uncertainty about putative residual protein function, which may confound the analysis of alleles harboring small indels18.
However, despite strictly adhering to current protocols for cell culture and genome editing, we still observed considerable variability of phenotypes when analyzing multiple KO clones of the same genes. We therefore decided to investigate additional factors contributing to clonal variability. After transfection of CRISPR constructs, standard protocols for the generation of stable genome-edited cell lines usually require clonal isolation of a successfully genome-edited monoclonal cell out of a pool of wild-type (WT) cells19. These parental WT cells are typically used as experimental controls, assuming genetically and phenotypically homogeneous WT cell populations. We hypothesized that heterogeneity of WT cells might contribute to the observed phenotypic variability in genome-edited cells.
Here we test this hypothesis by studying the knockout of the Pkd1 gene in stable kidney epithelial cells as a proof of concept. We demonstrate that WT cells display considerable clonal variability that contributes to phenotypic variability of genome-edited cells. Moreover, we show that the generation of isogenic WT subclones prior to genome editing reduces clonal heterogeneity of genome-edited cells.
Results
Genome-edited Pkd1-deficient cells show clonal variability
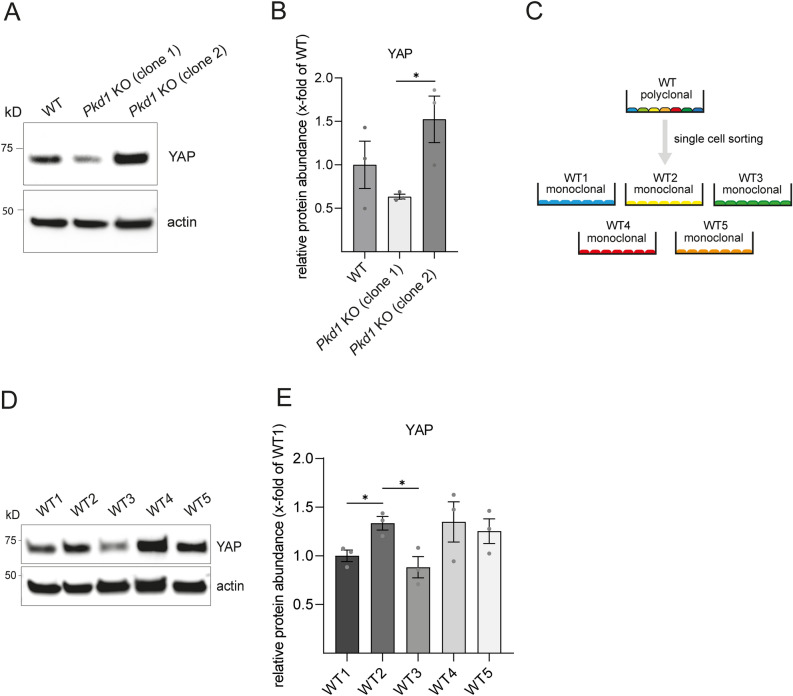
PKD1 encodes Polycystin-1 (PC-1), a transmembrane protein that has been implicated in the regulation of multiple signaling pathways. Recently, the Hippo pathway effector protein YAP has been identified as a downstream target of PC-1 and modulator of polycystic phenotype severity in murine ADPKD models20. Protein levels of YAP were shown to be upregulated in human and murine Pkd1-deficient kidneys. Since kidney cysts in ADPKD arise from tubular epithelial cells21, we analyzed YAP levels in a murine Pkd1-deficient kidney epithelial cell line (mIMCD-3) we had previously established17. Surprisingly, YAP levels were decreased in Pkd1-deficient cells (clone 1) compared to WT control in contrast to the published data (Fig. 1A). To verify the observed YAP reduction, we analyzed a second Pkd1-deficient clone (clone 2) generated in the same genome editing experiment. Yet, YAP levels of the second Pkd1 KO clone were increased compared to WT control and significantly higher compared to Pkd1 KO clone 1 (Fig. 1A,B).
Figure 1.
mIMCD-3 Pkd1 knockout clones and monoclonal mIMCD-3 wild-type clones vary in protein abundance of YAP (A) Protein abundance of YAP was analyzed in mIMCD-3 WT cells compared to two different Pkd1 KO clones after western blotting. (B) Densitometry shows significant differences in protein abundance for YAP in both Pkd1 KO clones. (C) WT cells were single cell sorted by FACS generating monoclonal WT cells. Five different monoclonal cell populations were arbitrarily labeled WT1–WT5. (D) Protein abundance of YAP was analyzed in monoclonal mIMCD-3 WT cells. (E) Densitometry shows varying levels of YAP with significant differences between WT1 and WT2, and WT2 and WT3, respectively. Three independent experiments were included in analysis. Error bars represent SEM. Statistical significance was evaluated using unpaired t-test. *Indicates p < 0.05. Unprocessed blots for (A and D) are presented in Supplementary Figs. S6 and S7.
One possible and often cited explanation for the observed clonal variability could be the occurrence of CRISPR/Cas9-related off-target events12. However, we had taken measures to reduce off-target events including the use of gRNAs with low off-target prediction as well as a Cas9 nickase. In addition, we also observed clonal heterogeneity in several TALEN-generated KO cell lines (data not shown) which are thought to have a lower probability of off-target effects22. Furthermore, all Pkd1 KO clones used in this study are confirmed null alleles. Thus allelic variants with different functional effects (gain- versus loss-of-function) and severity (hypomorphic versus complete loss-of-function) can be ruled out. Together this suggested that other factors may contribute to variable results in clonal genome-edited cells. We therefore hypothesized that phenotypic variability in genome-edited cells can be caused by intrinsic heterogeneity of the WT cells from which they arise.
Monoclonal wild-type mIMCD-3 cells display significant phenotypic variability
To address the potential influence of WT heterogeneity, we decided to perform single cell sorting of mIMCD-3 WT cells in order to generate monoclonal WT cell lines (Fig. 1C). We first screened monoclonal WT cells for the presence of primary cilia since ADPKD is a so-called ciliopathy—a group of disorders characterized by structural or functional defects of primary cilia23. Consequently, a cell-based model for ADPKD should incorporate cilia. Surprisingly, immunofluorescence staining for acetylated tubulin as a ciliary marker did not show any signal in some clones (data not shown). For further characterization, we only selected cilia-positive mIMCD-3 cell clones which we labeled WT 1–5. These clones were analyzed for protein abundance of YAP (Fig. 1D). Quantitative analysis showed significant alterations in YAP levels among these clones (Fig. 1E). Notably, the magnitude of differences was comparable to those in genome-edited cells (Fig. 1A). Moreover, alterations of protein levels in different wild-type clones were not restricted to YAP. We also conducted experiments with a set of kinases (pPERK, CaMKII delta and pAMPK) which have been shown to be linked to Pkd signaling24–26. Quantitative analysis of protein expression demonstrated significant alterations in protein levels for these kinases among monoclonal WT clones (Supplementary Fig. S1). In summary, substantial differences in protein levels were found between WT clones in a range which is often interpreted as biologically relevant.
To test whether clonal variability also manifests in other assays, we investigated cellular survival in the presence of cytotoxic compounds. Drug susceptibility was interrogated by generating dose–response curves for survival with three monoclonal and the parental polyclonal WT cell lines. Dose–response curves were generated for two commonly used nephrotoxic drugs, cisplatin and methotrexate. Drug susceptibility for cisplatin was not significantly altered between WT clones (Supplementary Fig. S2A,B). In contrast, treatment with the dihydrofolate reductase inhibitor methotrexate led to reproducible and significant changes in viability of multiple WT clones (Supplementary Fig. S2C,D).
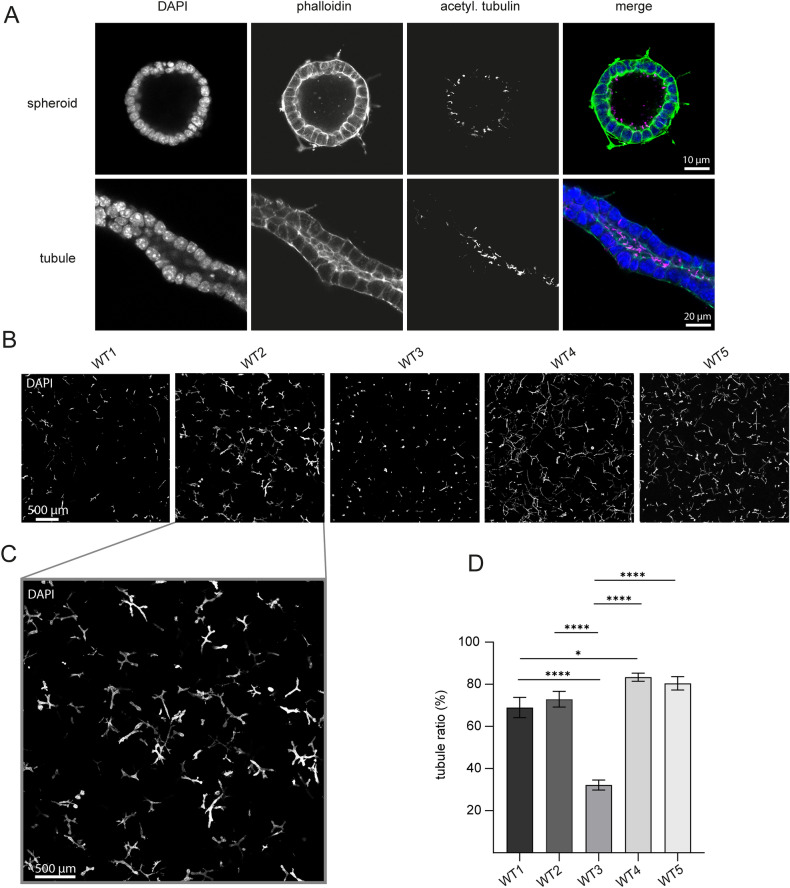
To study phenotypic variability in more complex biological programs, we analyzed tubulogenesis in 3D cell cultures. 3D cell cultures are increasingly used for disease modelling and dissection of mechanistic biological processes27. We and others use kidney tubule organoids to investigate Polycystin signal transduction and the mechanisms of tubulogenesis in vitro. Predominant structures of our 3D cell culture model can be divided into cyst-like epithelial structures (spheroids) and tubular epithelial structures (tubules) (Fig. 2A). Screening of monoclonal mIMCD-3 WT clones for their tubulogenic potential and proliferative features revealed significant phenotypic differences between distinct mIMCD-3 WT clones (Fig. 2B–D). Consequently, dissecting gene function in 3D cell cultures with polyclonal cell lines can be biased by heterogeneity of WT cells resulting in misinterpretation of gene deletion effects after isolation of subclones.
Figure 2.
Monoclonal mIMCD-3 wild-type cells show different morphology in 3D cell culture (A) Predominant structures of a kidney tubule organoid assay derived from mIMCD-3 WT cells. Representative spheroid (upper row) and representative kidney tubule (lower row) in a 3D-Matrigel-collagen scaffold. Images were acquired with confocal microscopy. (B) Representative images of five different monoclonal mIMCD-3 WT cells in a 3D-Matrigel-collagen scaffold. WT clones show different growth patterns, e.g. induction of kidney tubules is highly impaired in WT3 compared to other clones. (C) Magnification of WT2-image from panel B illustrates high number of differentiated kidney tubules in this clone. (D) Quantification of tubule ratio (tubules/ all structures) of five monoclonal mIMCD-3 WT clones. Tubule ratio varies significantly between clones. Bars represent mean tubule ratio of three independent experiments (four technical replicates each). Error bars represent SEM. Statistical significance was evaluated using unpaired t-test.*Indicates p < 0.05. ****Indicates p < 0.0001.
In summary, different WT subclones derived from the same well-characterized mIMCD-3 cell line show remarkably different characteristics regarding abundance of proteins, drug susceptibility and morphological patterns in 3D cell cultures. We thus concluded that WT heterogeneity might be a driver for phenotypic variability of genome-edited cell lines. This led us to modify our genome editing protocol and introduce an initial single cell sorting step to generate monoclonal WT cell lines prior to transfection of gRNAs and Cas9 (Supplementary Fig. S3).
KO clones derived from monoclonal isogenic wild-type cells show less variability and differences in protein levels compared to polyclonal KO clones
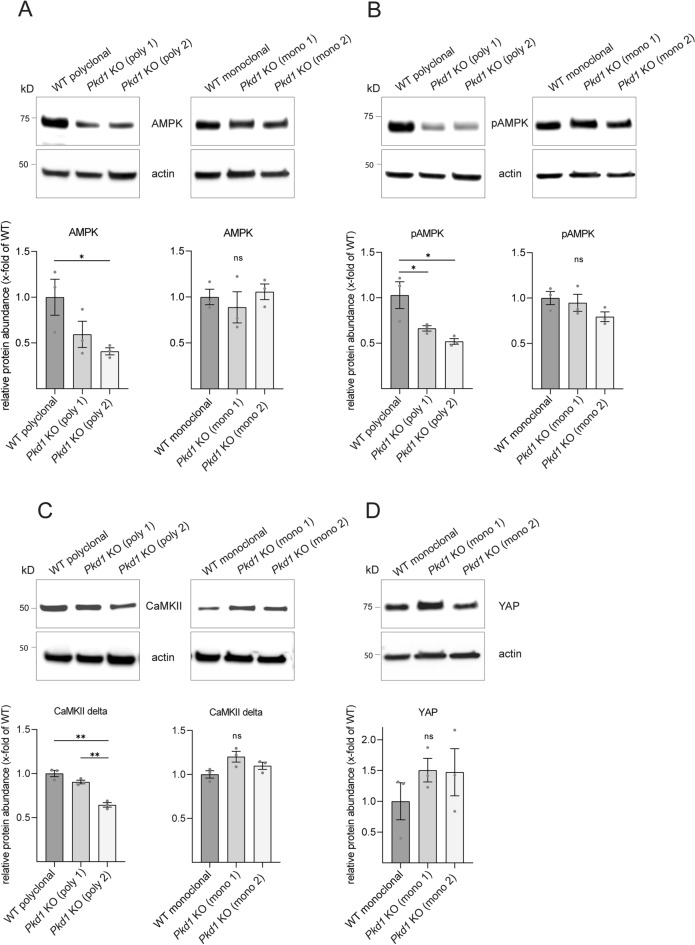
Using this modified genome editing workflow, we generated monoclonal Pkd1 KO clones from a matched isogenic WT control cell line. In the following, we refer to these cells as monoclonal Pkd1 KO cells. Cell lines generated from polyclonal parental cells are termed polyclonal KO cells. Again, we analyzed expression levels of polycystic kidney disease signaling related kinases in these new monoclonal Pkd1 KO cell lines and compared this to results obtained in polyclonal KO cell lines24,25. This comparison showed significant differences: while polyclonal Pkd1 KO clones showed a significant reduction of 5' adenosine monophosphate-activated protein kinase (AMPK) compared to polyclonal WT control, monoclonal Pkd1 KO clones showed no significant change in AMPK levels compared to an isogenic WT control (Fig. 3A). Similarly, levels of phosphorylated monophosphate-activated protein kinase (pAMPK) were significantly changed in polyclonal Pkd1 KO clones, but not in the monoclonal Pkd1 KO clones (Fig. 3B). Notably, investigation of genotype-dependent changes of protein levels of CaMKII delta in polyclonal Pkd1 KO clones showed variable results. Polyclonal Pkd1 KO clone 1 showed identical CaMKII delta levels compared to the WT control (Fig. 3C). In contrast, Pkd1 KO clone 2 showed a significant reduction compared to both WT and Pkd1 KO clone 1. In the monoclonal setting, levels of CaMKII were not significantly different. Similarly, YAP levels of both monoclonal Pkd1 KO clones were almost identical with a non-significant increase compared to WT control (Fig. 3D). In contrast, we observed a high degree of clonal variability with opposite results in polyclonal Pkd1 KO clones (Fig. 1). In summary, experiments using monoclonal Pkd1 KO cells with matched isogenic control cells led to reduced variability.
Figure 3.
Variability of protein levels for AMPK, pAMPK, CaMKII delta and YAP is reduced in monoclonal compared to polyclonal Pkd1 knockout clones (A) Protein abundance of AMPK is significantly reduced in Pkd1 KO clone 2 compared to polyclonal WT control. Monoclonal Pkd1 KO clones show no significant differences in protein abundance of AMPK compared to monoclonal WT control. (B) Protein abundance of pAMPK is significantly reduced in both polyclonal Pkd1 KO clones compared to polyclonal WT control. Monoclonal Pkd1 KO clones show no significant differences compared to isogenic WT control. (C) Protein abundance of CaMKII delta is significantly reduced in Pkd1 KO clone 2 compared to polyclonal WT control and Pkd1 KO clone 1. Monoclonal Pkd1 KO clones show no significant differences compared to isogenic WT control. (D) Monoclonal Pkd1 KO clones show no significant differences in protein abundance of YAP compared to isogenic WT control. Polyclonal setting is illustrated in Fig. 1. Three independent experiments were included in each analysis of protein levels. Error bars represent SEM. Statistical significance was evaluated using unpaired t-test. *Indicates p < 0.05. **Indicates p < 0.01. ***Indicates p < 0.001. ns means non-significant. Unprocessed blots for (A–D) are presented in Supplementary Figs. S8, S9, S10, and S11.
Monoclonal and polyclonal wild-type cells show profound transcriptomic differences
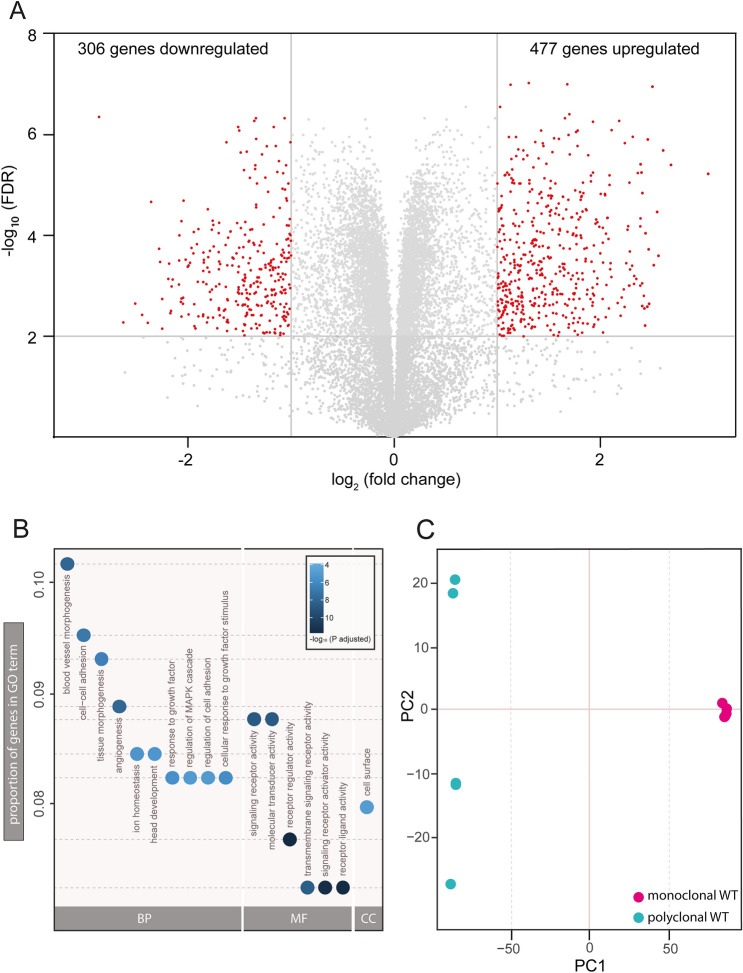
To obtain a more global view on differences between monoclonal and polyclonal WT cells as a possible confounding factor in genome editing experiments, we performed an unbiased transcriptome-wide analysis using RNA sequencing. Differential gene expression analyses are frequently conducted to compare genome-edited with WT cell lines. They allow for the identification of genes or pathways revealing information about physiological or pathophysiological processes28–30. We compared the transcriptomic profiles of mono- and polyclonal WT cells to uncover the potential contribution of clonal isolation to differential gene expression. Strikingly, transcriptomic analysis of mono- vs polyclonal WT cells revealed a large number of differentially regulated genes despite stringent cut-off criteria (Fig. 4A; Supplementary Data 1). In monoclonal WT cells 477 genes were significantly upregulated while 306 genes were downregulated compared to polyclonal WT cells (adjusted p < 0.01). Likewise, Gene Ontology (GO) term analysis yielded a variety of significantly enriched GO terms when comparing mono- and polyclonal WT cells (Fig. 4B). Of note, the most significant altered GO terms between mono- and polyclonal cells are linked to receptor and signaling activity. Such a result within the context of an experiment comparing Pkd1 KO and WT cells would easily be interpreted as meaningful, given the role of PC-1 as a transmembrane receptor. In fact, however, this significant enrichment was purely derived from comparing mono- and polyclonal WT cells. Finally, principal component analysis (PCA) confirmed that variability of biological replicates is drastically reduced when using monoclonal cell lines (Fig. 4C), a finding which is in line with our prior experiments. Taken together, these transcriptomic analyses identify WT heterogeneity as a significant confounding factor in experiments involving clonal isolation.
Figure 4.
RNA sequencing analysis reveals altered expression levels between mono- and polyclonal wild-type cell lines and confirms heterogeneity of polyclonal cells. RNA was extracted from poly- and monoclonal (WT5) mIMCD-3 WT cell lines. RNA sequencing was performed for four technical replicates each. (A) Differential gene expression between poly- and monoclonal (WT5) cell lines are illustrated in a volcano plot. Threshold for false discovery rate (FDR) was set to log10 ≥ 2. Threshold for fold change was set to log2 ± 1. Grey dots represent genes which do not meet threshold criteria (FDR log10 < 2 and/or log2 (fold change) between ± 1). Red dots indicate significant changes in expression. 306 genes are significantly downregulated, 477 genes are significantly upregulated in monoclonal compared to polyclonal mIMCD-3 WT cell lines (adjusted p-value < 0.01). (B) Gene ontology (GO) term analysis shows most significant changes in expression levels of genes in dependence of gene ontology classification. Y-axis depicts proportion of genes in GO term analysis. Each dot represents distinct ontologies. Colour of dots represents significance of changes in expression levels between mono- and polyclonal WT-cells. Colour scheme depicts significance levels (− log10 p-adjusted). BP biological process, MF molecular function, CC cellular components. (C) Principal component (PC) analysis shows variability between sequencing replicates. Monoclonal sequencing replicates cluster together in contrast to polyclonal sequencing replicates showing a heterogeneous pattern.
Monoclonal cell line and its subclones show similar transcriptomic profiles
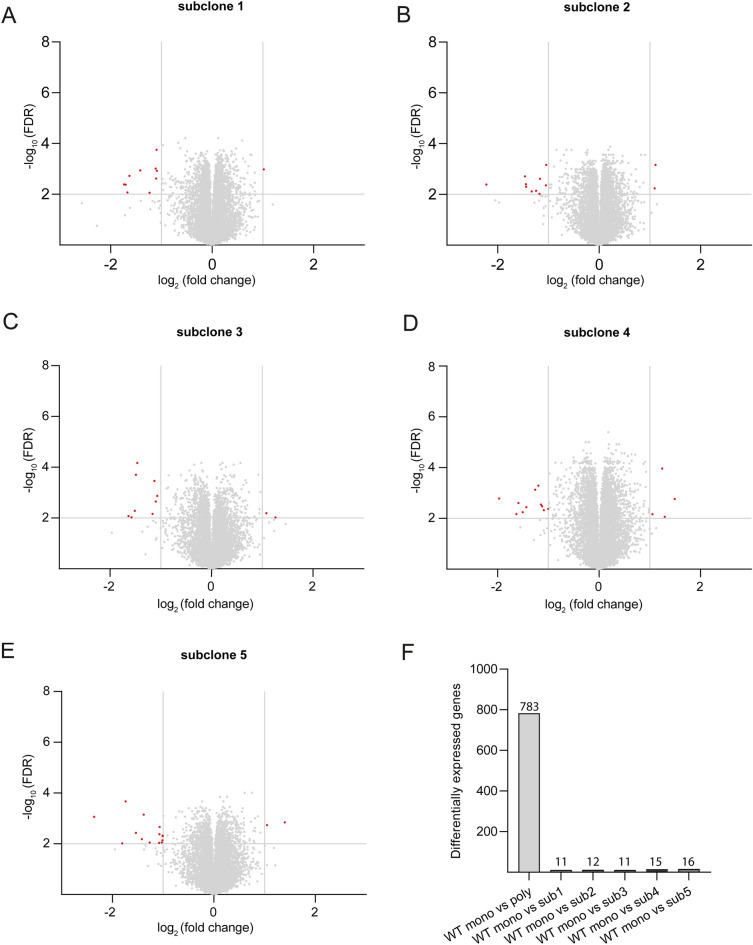
In a transcriptomic comparison of mono- and polyclonal wild-type cell lines we observed hundreds of differentially regulated genes (Fig. 4A). In western blot experiments we showed less variability in experiments involving monoclonal genome-edited cells and its corresponding control cell line compared to polyclonal cell lines. Based on these observations, we hypothesized that subclones derived from a monoclonal isogenic cell line should be more similar to their parental cell line than those derived from polyclonal cells. To test this hypothesis, we performed single cell sorting of WT clone 5 and expanded five resulting subclones over a period of approximately 6 weeks. After this timespan we isolated RNA of these subclones and performed RNA sequencing. Notably, all investigated subclones and their parental cell line had highly similar transcriptomic profiles (Fig. 5A–E; Supplementary Data 2) with a low amount of differentially expressed genes (mean differentially expressed genes: 13). In contrast, the transcriptomic comparison between monoclonal WT5 and the polyclonal WT cell line resulted in 783 differentially expressed genes (Fig. 5F). Therefore, the amount of differentially expressed genes is drastically reduced by using subclones generated from a monoclonal cell line.
Figure 5.
Monoclonal wild-type cell line and corresponding subclones show similar transcriptomic profiles in contrast to the polyclonal wild-type. Volcano plots with differentially expressed genes in five different subclone cell lines of the parental monoclonal cell line in comparison with the parental monoclonal cell line (A–E). A bar chart (F) summarizes total amount of differentially expressed genes comparing parental cell line with polyclonal wild-type cell line as well as subclones with parental cell line. Threshold for false discovery rate (FDR) was set to log10 ≥ 2. Threshold for fold change was set to log2 ± 1. Grey dots represent genes which do not meet threshold criteria (FDR log10 < 2 and/or log2 (fold change) between ± 1).
Phenotypic variability of monoclonal cell lines is not explained by chromosomal aberrations
Our results show that mIMCD-3 cells are polyclonal in nature and show distinct phenotypic characteristics. However, the underlying mechanism is unclear. Immortalized cell lines have been shown to be polyploid31,32. Polyploidy can result in genomic instability leading to structural aberrations33–35. Chromosomal instabilities could potentially explain the variation in phenotypes between monoclonal cell lines. We therefore asked if chromosomal aberrations are found in the monoclonal wild-type cell lines as well as in the Pkd1 KO cell lines. To address this question, we conducted multicolor fluorescence in situ hybridizations for these cell lines. A comparison of karyograms between monoclonal wild-type cells and between polyclonal and monoclonal Pkd1 KO cell lines showed similar karyograms without evidence for structural aberrations or instabilities (Supplementary Fig. S4). Of note, our analysis did not reveal an increase in polyploidy in Pkd1 KO clones as demonstrated in a previous study36. In conclusion, karyotype differences or chromosomal aberrations are probably not the cause for the observed phenotypic variability.
Wild-type heterogeneity is a cause for clonal variability in genome editing experiments
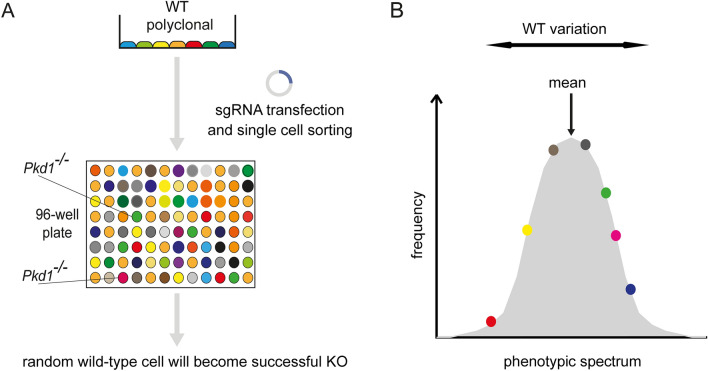
In conclusion, our data show that mIMCD-3 WT cells are polyclonal and heterogeneous despite their monoclonal origin37, possibly through genetic drift. Hence, the isolation of subclones from polyclonal cells after genomic manipulation leads to propagation of cells with distinct features (Fig. 6A). In the typical setting of such experiments, a random WT cell with distinct features is converted into a successful KO clone. This has two problematic implications: (i) KO clones will display phenotypic variability due to their polyclonal origin and (ii) KO clones lack proper controls. Since polyclonal WT control cells represent the mean for an investigated phenotype, phenotypic alterations observed in subsequent experiments with KO clones can originate from deviations of a specific WT clone from the mean instead of being a consequence of genetic loss-of-function (Fig. 6B). Our findings identify heterogeneity of WT cells as a major confounding factor in genome editing experiments involving clonal isolation and show that the establishment of matched isogenic control cells increases reproducibility and reduces erroneous results.
Figure 6.
Model explaining heterogeneity of polyclonal knockout cell lines. (A) Our data shows that mIMCD-3 cells are polyclonal in nature. Genome editing results in KO cells derived from a random WT cell. Subsequent experiments are conducted with a heterogenic WT control, therefore confounding the analysis of gene deletion effects. (B) Polyclonal mIMCD-3 cells represent the mean of a phenotypic spectrum. Our data shows that monoclonal WT cells are heterogeneous concerning distinct phenotypes. In experiments, KO cells derived from random WT cells are compared with polyclonal, heterogenic WT cells. This can lead to misinterpretation of findings, since the observed phenotype may result from heterogeneity of WT cells instead of gene deletion.
Discussion
Genome editing tools such as CRISPR/Cas9 have given cell biologists the opportunity to study genetic alterations such as loss- or gain-of-function mutations in cell culture models. Validity and reproducibility of results in cell culture models depend on strict adherence to an optimized experimental setting. Effect sizes are often small and hard to detect. Hence, to precisely dissect genotype–phenotype relationships, it is critical to minimize confounding factors.
Here, we study a frequently neglected confounding factor when using genome-edited cells. We demonstrate that the polyclonal nature of WT cells contributes to phenotypic variability in genome-edited cells. Cultured immortalized cell lines such as HeLa, CHO, HEK293, MDCK, or mIMCD-3 cells are extensively used for genome editing experiments38–42. They are well characterized, easy to propagate, and are considered to be rather stable phenotypically compared to primary cells. Therefore, these cell lines are suitable for many research applications including disease modeling and high-throughput screening for drug discovery. Some of these cell lines exhibit physiological characteristics of the tissue of origin. For example, mIMCD-3 cells used in this study retain many differentiated characteristics of the kidney collecting duct including apico-basal polarity, high transepithelial resistance and electrolyte transport37. Furthermore, mIMCD-3 cells are ciliated and therefore extensively used to study ciliopathies such as ADPKD43–45. Although mIMCD-3 cells were established as a monoclonal cell line, we now show that they are inhomogeneous and subclones feature a broad phenotypic spectrum regarding multiple traits. For instance, differential gene expression analysis of monoclonal vs. polyclonal WT cell lines leads to the detection of a large number of differentially expressed genes. After genome editing and subsequent single cell sorting, this phenotypic spectrum inherent to polyclonal WT cells can be misinterpreted as a consequence of CRISPR-induced genetic alterations. Of note, the issue of WT heterogeneity is likely further aggravated in cell types with higher inherent genetic and phenotypic variability such as reprogrammed human induced pluripotent stem cells, which are frequently used for genome editing experiments.
We further show that the confounding potential of heterogeneous WT cell lines can be reduced by establishing monoclonal cell lines by single cell sorting before genome editing. Our study demonstrates that this optimized workflow reduces (i) phenotypic variability of genome-edited cell lines as well as (ii) false-positive detection of genotype-dependent phenotypes. In principle, false-negative results are also possible, e.g. when the genetic manipulation results in a phenotype that is masked by an opposite phenotype from the WT spectrum of the selected genome-edited clone.
In addition to studying multiple genome-edited clones, rescue experiments or reversal of genome-editing can help to validate positive findings and eliminate false-positive results. This includes erroneous phenotypes due to variability of WT cell lines, but also those due to off-target effects of the genome editing process. Using monoclonal cell lines for genome editing has the potential to significantly minimize the time spent trying to verify results that ultimately prove not reproducible. Besides that, this approach might be useful in cases when rescue experiments are not feasible, e.g. when a narrow control of expression levels of a protein is necessary and technically not achievable or when investigating dominant negative mutations. We therefore propose to generate matched isogenic control cells prior to genome editing experiments involving isolation of clones. While this requires additional time, it will likely improve validity and reproducibility of such experiments.
When using monoclonal cell lines it should be considered that a single clone might not be representative for the totality of the cell line. Subsequently, generation and editing of several distinct clones should be considered. Furthermore, a specific clone might not be suited for the planned investigations, e.g. we encountered mIMCD-3 clones that did not develop primary cilia. Obviously, such a clone is not suited for further investigation of ciliopathies. Hence, phenotypic assessment of WT clones used for genome editing regarding desired features for further investigation is advised.
Although we demonstrated considerable phenotypic differences between monoclonal wild-type cell lines, the underlying mechanism for heterogeneity remains elusive. Karyogram analyses showed polyploidy for all investigated cell lines. However, structural aberrations were not observed. Genetic drift in cell lines due to spontaneous mutations has been frequently reported and does not necessarily require chromosomal aberrations46. It is therefore prudent to expand and freeze a high number of cells after clonal isolation as suggested in our protocol (Supplementary Fig. S3). This allows for future experiments at low passage numbers without significant genetic drift. Our transcriptomic analyses of a second generation of subclones from an isogenic monoclonal cell line (Fig. 5) demonstrates that clonal isolation from a monoclonal parental cell line results in very low variability of transcript levels even after multiple passages.
There are several limitations to this study. Firstly, we conducted experiments focusing on a single cell line. However, since heterogeneity has been frequently observed in different cell lines9–11,47, our findings are likely to apply to other cell lines as well. Secondly, investigated KO effects are restricted to a single gene (Pkd1). Yet, the underlying problem of WT heterogeneity is independent of the gene targeted via genome editing. Thus, similar findings are to be expected when targeting other genes.
In summary, this study identifies heterogeneity of WT cell lines as a significant cause for phenotypic variability of genome-edited cells. In addition, we demonstrate and validate an improved protocol for genome editing of stable cell lines.
Methods
Cell culture
mIMCD-3 wild-type cells were obtained from ATCC (Manassas, VA, USA, CRL-2123; Lot 5064816). Cells were cultured with DMEM:F12 (LONZA, cat.no. BE12-719F), fetal bovine serum was added to a final concentration of 10% and penicillin/streptomycin (Sigma, P0781) with a final concentration of 1%. Subculturing of cells was performed twice a week in a 1:10 ratio. Cells were maintained in a humidified incubator at 37°C and 10% CO2. Experiments were strictly performed with comparable and lowest possible passage numbers. In this manuscript, the term “wild-type” is used for mIMCD-3 cells that were not edited by CRISPR-Cas9. Of note, the original cell line obtained from ATCC was generated from an SV-40 transgenic mouse37.
Protein isolation, SDS-PAGE, Western blot and ECL detection
Cells were harvested five days after epithelial confluency. Proteins were isolated and processed as described previously48,49. Chemiluminescence was detected by a 16-bit ChemoCam system (Intas). Images were acquired within the dynamic range of the charge-coupled device sensor. None of the analyzed bands was saturated. Antibodies used for protein detection: anti-YAP (Cell Signaling, 4912), anti-CaMKII delta (Abcam, ab181052), anti-pPERK (Cell Signaling, 3179), anti-pAMPK (Cell Signaling, 2535), anti-AMPK (Cell Signaling, 2532) and anti-ß-actin (Sigma A1978). Primary antibodies were diluted 1:1000 (anti-ß-actin 1:5000). Secondary antibodies (anti-rabbit, GE Healthcare NA934; anti-mouse, Dako P0447) were used 1:10,000.
Cell sorting
Cells were single cell sorted with a BD FACS Aria III into 96-wells (Greiner Bio one, flat bottom, transparent). Cells were maintained in a humidified incubator at 37°C and 10% CO2, media was changed weekly. After 2.5 weeks, growth of clones was confirmed by visualization under an inverted microscope. Cells were expanded into 6-wells and finally 100 mm dishes.
Cytotoxicity assay
Cisplatin (Merck, 232120) and methotrexate (Merck, 454126) were dissolved in PBS and DMSO, respectively. 4 × 104 mIMCD-3 cells were seeded into 384-wells (Greiner bio-one, 781098) in 20 µl media. 5 µl of compounds were added at day 1 at indicated concentrations and incubated for 72 h. Negative control for cisplatin was conducted by incubating cells with media and PBS used for solving cisplatin at highest concentration (2%). Negative control for methotrexate was conducted by incubating cells with media and DMSO used for solving methotrexate at highest concentration (0.1% DMSO). Positive control was conducted by incubating cells for 72 h with digitonin (1000 µg/ml). At day 4, 25 µl of Promega’s CellTiter-Glo 2.0 solution were added per well to assess cell viability. Further steps were performed according to the manufacturer´s protocol. Luminescence was detected by a Tecan Spark reader.
Generation of Pkd1 knockout cells
Generation of polyclonal mIMCD-3 Pkd1 KO cells was described earlier17. For generation of monoclonal KO the same protocol using the same Nickase-expressing plasmid pX335-U6-Chimeric-BB-CBh-hSpCas9n (Addgene, cat. no. 42335) and the same gRNAs (Supplementary Fig S5) was applied. However, a single cell sorting step was introduced prior to CRISPR/Cas9 application. Pkd1 null alleles were confirmed by genomic sequencing, as well as complete absence of the respective mRNA and protein.
Kidney tubule organoid assay
Growth-factor reduced Matrigel (Corning, 356231) and collagen type I rat tail (Ibidi, 50201) were mixed on ice in a 1: 1.25 ratio. 350 µl of this mixture were diluted with 630 µl DMEM:F12 media without fetal bovine serum. 20 µl HEPES (Sigma, H 0887) and 10 µl HGF (4 ng/µl, Merck, GF116) were added. mIMCD-3 cells were harvested via trypsinization (0.25% trypsin), counted and transferred to the gel mixture (150 cells/µl gel mixture). 80 µl/well of cell-gel-mixture were seeded into 96-wells (Greiner bio-one, flat bottom, transparent), plates were transferred to an incubator (37 °C). After 30 min, 220 µl of DMEM:F12 with fetal bovine serum and HGF (40 ng/ml) were carefully added on top of the cell-gel-mixture. After incubation for 72 h (37°C), media was carefully removed and replaced by 220 µl DMEM:F12 with fetal bovine serum without HGF. After an additional 96 h, cells were fixed for immunofluorescence.
Immunofluorescence
mIMCD-3 cells grown in a Matrigel-collagen scaffold for 7 days were fixed with 4% PFA (200 µl/well, 30 min). After two washing steps (PBS 200 µl/well, 10 min) and blocking (5% horse serum, 0.1% Triton-X in PBS, 1 h), primary antibody or fluorescent dyes were added for 1 h. After three further washing steps (PBS 200 µl/well, 3 min), samples were incubated with the secondary antibody for 30 min. After washing an additional three times, samples were stored in PBS until imaging. Antibodies and fluorescent dyes used were anti-acetylated tubulin antibody (Sigma, T6793) Hoechst 33342 (Thermo Fisher Scientific, H1399, 1:1000), Alexa Fluor 488 phalloidin (Invitrogen, A12379), and anti-mouse Alexa Fluor 488 (Molecular Probes, A-11029).
Organoid microscopy and image analysis
Kidney tubule organoids were visualized using a confocal microscope (Zeiss LSM-880). 300 µm z-stacks were imaged per well and fused into 2D images (Maximum intensity projection, Zeiss Zen blue). Structures per 2D image were categorized by morphology: Round structures were classified as spheroids, elongated structures > 30 µm were classified as tubules.
RNA sequencing and data analysis
Total RNA of cells was extracted and sequenced at Novogene Co., Ltd. Sequencing of four biological replicates for each polyclonal and monoclonal cell lines was performed on an Illumina sequencer. Polyclonal cells were sequenced as 50 bp single-end reads and monoclonal cells as paired-end 150 bp reads. The raw sequencing data were quality controlled by Novogene and reads containing adapters and low quality base which is over 50% of the total bases were removed. The final fastq files were then aligned to mouse reference genome sequence GRCm38 using STAR 2.7.3a with default parameters50. The genome sequence and gene annotation files were downloaded from the Ensembl release 81. RSEM v1.3.1 was used to calculate gene expression values, which were represented as TPM (Transcripts Per Kilobase Million)51. Principal components analysis of the samples was performed using the R function prcomp on the log2(TPM) values of TPM. In order to perform differential gene expression analysis between polyclonal and monoclonal samples, which were sequenced at different time points, log2(TPM) values were z-score transformed. Welch t-test was used to compare gene expression between polyclonal and monoclonal samples as well as the monoclonal parental cell line with second-generation clones (two-sided comparison). Genes with Benjamini–Hochberg adjusted p-value < 0.01 and a z-score difference of < 1 or > 1 were defined as differentially expressed genes. Differentially expressed genes were tested for enrichment of Gene Ontology terms by the R package clusterProfiler v3.18.1 with default settings52.
Multicolor FISH
Metaphases were prepared from mICMD-3 cells according to standard procedure53,54. The analysis was carried out using a 24XCyte Multicolor FISH probe (mFISH) (Metasystems, Altlussheim, Germany) and the analysis was performed according to the manufacturer´s instructions. The ISIS software (V. 5.10.129) was used for analysis (Metasystems, Altlussheim, Germany; https://metasystems-international.com/). Whenever possible, 5 metaphases were analyzed per sample.
Statistics
All results are expressed as mean ± SEM. A p-value < 0.05 was considered significant unless otherwise specified. Statistical analysis was performed as described in the figure legends. Software used for statistics: Graph Pad Prism 8 for Windows and R v4.0.5 for Linux. Densitometry was performed with ImageJ 1.46R. For quantitative analysis, the intensity of the protein of interest band was normalized to the intensity of the corresponding actin band.
Supplementary Information
Acknowledgements
The authors acknowledge Simone Diederichsen and Céline Costa for expert technical assistance. We would like to thank the Life Imaging Center (LIC) of the University Freiburg and the Lighthouse Core Facility for their assistance with microscopy and cell sorting. CRISPR/Cas9 plasmids were a gift from Feng Zhang. M.K. is supported by the German Research Foundation (DFG): TRR 152 (Project-ID 239283807); SFB1453 (Project-ID 431984000) and Germany's Excellence Strategy (CIBSS—EXC-2189—Project ID 390939984). L.W. received support by Else Kröner-Fresenius-Stiftung (2016_Kolleg.03). A.K. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project-ID 431984000—SFB 1453. Y.L. is supported by DFG KO 2598/4-2.
Author contributions
M.K., T.B. and L.W. conceived the study. L.W., T.B., A.K. and M.K. wrote and edited the manuscript. L.W., T.B., A.H., I.C., M.N., K.R., B.N., N.M., Y.B. and F.S. planned and performed experiments. L.W., J.J., B.G., G.G. and Y.L. analyzed data. M.K. and T.B. supervised the study.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
RNA-sequencing data generated and analyzed during the current study will be available in the gene expression omnibus database under the Accession Number: GSE196762. All other datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-22885-8.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 6.Cruz NM, et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017;16:1112–1119. doi: 10.1038/nmat4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman LP, et al. Reproducibility: Changing the policies and culture of cell line authentication. Nat. Methods. 2015;12:493–497. doi: 10.1038/nmeth.3403. [DOI] [PubMed] [Google Scholar]
- 8.Masters JR, Stacey GN. Changing medium and passaging cell lines. Nat. Protoc. 2007;2:2276–2284. doi: 10.1038/nprot.2007.319. [DOI] [PubMed] [Google Scholar]
- 9.Kleensang A, et al. Genetic variability in a frozen batch of MCF-7 cells invisible in routine authentication affecting cell function. Sci. Rep. 2016;6:28994. doi: 10.1038/srep28994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torsvik A, et al. U-251 revisited: Genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med. 2014;3:812–824. doi: 10.1002/cam4.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, et al. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 2019;37:314–322. doi: 10.1038/s41587-019-0037-y. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, et al. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol. 2018;14:311–316. doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose JC, et al. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat. Commun. 2020;11:2697. doi: 10.1038/s41467-020-16542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JK, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofherr A, et al. Efficient genome editing of differentiated renal epithelial cells. Pflugers Arch. 2017;469:303–311. doi: 10.1007/s00424-016-1924-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuladhar R, et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019;10:4056. doi: 10.1038/s41467-019-12028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giuliano CJ, Lin A, Girish V, Sheltzer JM. Generating single cell-derived knockout clones in mammalian cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 2019;128:e100. doi: 10.1002/cpmb.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J, et al. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev. 2018;32:781–793. doi: 10.1101/gad.315127.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S, et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc. Natl. Acad. Sci. USA. 2007;104:18688–18693. doi: 10.1073/pnas.0708217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song X, Tsakiridis E, Steinberg GR, Pei Y. Targeting AMP-activated protein kinase (AMPK) for treatment of autosomal dominant polycystic kidney disease. Cell Signal. 2020;73:109704. doi: 10.1016/j.cellsig.2020.109704. [DOI] [PubMed] [Google Scholar]
- 25.Bracken C, et al. CaMKII as a pathological mediator of ER stress, oxidative stress, and mitochondrial dysfunction in a murine model of nephronophthisis. Am. J. Physiol. Renal Physiol. 2016;310:F1414–1422. doi: 10.1152/ajprenal.00426.2015. [DOI] [PubMed] [Google Scholar]
- 26.Liang G, et al. Polycystin-2 down-regulates cell proliferation via promoting PERK-dependent phosphorylation of eIF2alpha. Hum. Mol. Genet. 2008;17:3254–3262. doi: 10.1093/hmg/ddn221. [DOI] [PubMed] [Google Scholar]
- 27.Artegiani B, Clevers H. Use and application of 3D-organoid technology. Hum. Mol. Genet. 2018;27:R99–R107. doi: 10.1093/hmg/ddy187. [DOI] [PubMed] [Google Scholar]
- 28.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 29.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J. Transl. Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Toouli CD, et al. Comparison of human mammary epithelial cells immortalized by simian virus 40 T-Antigen or by the telomerase catalytic subunit. Oncogene. 2002;21:128–139. doi: 10.1038/sj.onc.1205014. [DOI] [PubMed] [Google Scholar]
- 32.Velicescu M, et al. Aneuploidy and telomere attrition are independent determinants of crisis in SV40-transformed epithelial cells. Cancer Res. 2003;63:5813–5820. [PubMed] [Google Scholar]
- 33.Frattini A, et al. High variability of genomic instability and gene expression profiling in different HeLa clones. Sci. Rep. 2015;5:15377. doi: 10.1038/srep15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Battini L, et al. Loss of polycystin-1 causes centrosome amplification and genomic instability. Hum. Mol. Genet. 2008;17:2819–2833. doi: 10.1093/hmg/ddn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauchman MI, Nigam SK, Delpire E, Gullans SR. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am. J. Physiol. 1993;265:F416–424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 38.Sergeeva D, Camacho-Zaragoza JM, Lee JS, Kildegaard HF. CRISPR/Cas9 as a genome editing tool for targeted gene integration in CHO cells. Methods Mol. Biol. 2019;1961:213–232. doi: 10.1007/978-1-4939-9170-9_13. [DOI] [PubMed] [Google Scholar]
- 39.Reuven N, Adler J, Myers N, Shaul Y. CRISPR co-editing strategy for scarless homology-directed genome editing. Int. J. Mol. Sci. 2021;22:3741. doi: 10.3390/ijms22073741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, et al. Establishment of a HEK293 cell line by CRISPR/Cas9-mediated luciferase knock-in to study transcriptional regulation of the human SREBP1 gene. Biotechnol. Lett. 2018;40:1495–1506. doi: 10.1007/s10529-018-2608-2. [DOI] [PubMed] [Google Scholar]
- 41.Karlgren M, et al. A CRISPR-Cas9 generated MDCK cell line expressing human MDR1 without endogenous canine MDR1 (cABCB1): An improved tool for drug efflux studies. J. Pharm. Sci. 2017;106:2909–2913. doi: 10.1016/j.xphs.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 42.Lakshmipathi J, et al. Identification of NFAT5 as a transcriptional regulator of the EDN1 gene in collecting duct. Am. J. Physiol. Renal Physiol. 2019;316:F481–F487. doi: 10.1152/ajprenal.00509.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh AK, Hurd T, Hildebrandt F. 3D spheroid defects in NPHP knockdown cells are rescued by the somatostatin receptor agonist octreotide. Am. J. Physiol. Renal Physiol. 2012;303:F1225–1229. doi: 10.1152/ajprenal.00135.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherpa RT, et al. Sensory primary cilium is a responsive cAMP microdomain in renal epithelia. Sci. Rep. 2019;9:6523. doi: 10.1038/s41598-019-43002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, et al. Ciliary membrane proteins traffic through the Golgi via a Rabep1/GGA1/Arl3-dependent mechanism. Nat. Commun. 2014;5:5482. doi: 10.1038/ncomms6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quevedo R, et al. Assessment of genetic drift in large pharmacogenomic studies. Cell Syst. 2020;11:393–401. doi: 10.1016/j.cels.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Yu MJ, et al. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc. Natl. Acad. Sci. USA. 2009;106:2441–2446. doi: 10.1073/pnas.0813002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofherr A, Wagner C, Fedeles S, Somlo S, Kottgen M. N-glycosylation determines the abundance of the transient receptor potential channel TRPP2. J. Biol. Chem. 2014;289:14854–14867. doi: 10.1074/jbc.M114.562264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofherr A, Wagner CJ, Watnick T, Kottgen M. Targeted rescue of a polycystic kidney disease mutation by lysosomal inhibition. Kidney Int. 2016;89:949–955. doi: 10.1016/j.kint.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlegelberger B, et al. Clinicopathogenetic significance of chromosomal abnormalities in patients with blastic peripheral B-cell lymphoma. Kiel-Wien-Lymphoma Study Group. Blood. 1999;94:3114–3120. [PubMed] [Google Scholar]
- 54.Schlegelberger B, et al. Diagnostic Cytogenetics. Springer; 1999. pp. 51–185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data generated and analyzed during the current study will be available in the gene expression omnibus database under the Accession Number: GSE196762. All other datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.